ABSTRACT
Most swimming bacteria are capable of following gradients of nutrients, signaling molecules and other environmental factors that affect bacterial physiology. This tactic behavior became one of the most-studied model systems for signal transduction and quantitative biology, and underlying molecular mechanisms are well characterized in Escherichia coli and several other model bacteria. In this review, we focus primarily on less understood aspect of bacterial chemotaxis, namely its physiological relevance for individual bacterial cells and for bacterial populations. As evident from multiple recent studies, even for the same bacterial species flagellar motility and chemotaxis might serve multiple roles, depending on the physiological and environmental conditions. Among these, finding sources of nutrients and more generally locating niches that are optimal for growth appear to be one of the major functions of bacterial chemotaxis, which could explain many chemoeffector preferences as well as flagellar gene regulation. Chemotaxis might also generally enhance efficiency of environmental colonization by motile bacteria, which involves intricate interplay between individual and collective behaviors and trade-offs between growth and motility. Finally, motility and chemotaxis play multiple roles in collective behaviors of bacteria including swarming, biofilm formation and autoaggregation, as well as in their interactions with animal and plant hosts.
Keywords: chemotaxis, motility, Escherichia coli, environmental adaptation, physiology
This review summarizes the recent advances in understanding the impact of flagellar motility and chemotaxis on various behaviors of bacteria, from nutrient acquisition and population range expansion to interactions among bacteria and with their animal and plant hosts.
INTRODUCTION
Swimming bacteria are able to monitor changes in environmental conditions as they move and to adapt their swimming pattern accordingly, in order to swim towards their preferred environment. Such biased movement in chemical gradients, called chemotaxis, is one of the longest and most thoroughly studied bacterial behavioral responses. Understanding of the molecular mechanisms controlling the chemotactic behavior has become highly refined over the years, especially in the model organism Escherichia coli (Wadhams and Armitage 2004; Colin and Sourjik 2017; Bi and Sourjik 2018). As a consequence, the typical behavior of a bacterial cell in a simple gradient and the underlying biochemistry and biophysics are well understood, and they could be mathematically modelled down to minute quantitative details (Tu 2013; Micali and Endres 2016; Colin and Sourjik 2017; Waite, Frankel and Emonet 2018; Wong-Ng, Celani and Vergassola 2018).
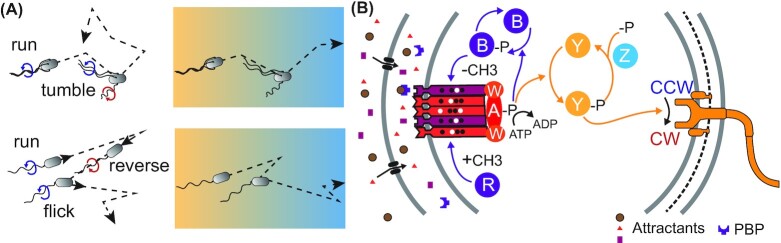
Flagellated bacteria typically swim in a series of more or less straight runs interrupted by short reorientations (Fig. 1A). In peritrichous bacteria like E. coli, runs occur when all flagella rotate unidirectionally (counterclockwise in the case of E. coli) and form a bundle that propels the cell body forward (Berg 1975; Macnab 1977). Tumbles, which result from transient reversal of the rotary direction of flagellar motors, cause the flagellar bundle to fall apart and lead to reorientation of the cell body. The strategies of reorientation in polarly flagellated bacteria are more complex and diverse, with several distinct cell motility states, which might have been evolutionary selected to match the respective bacterial habitat (Altindal, Xie and Wu 2011; Constantino et al. 2018; Grognot and Taute 2021; Stocker 2011; Taktikos, Stark and Zaburdaev 2013; Xie et al. 2011). Regardless of the specific mechanism of reorientation, such run-reorientation behavior results over long times in an active diffusion that enables bacteria to efficiently spread in their environment.
Figure 1.
Chemotactic behavior and signaling pathway. (A), Two prominent types of bacterial flagellar motility patterns, run-tumble and run-reverse-flick swimming. Both types of swimming lead to effective diffusion in homogeneous environments and get biased by the chemotaxis pathway to climb up physicochemical gradients. (B), Schematic representation of the chemotaxis pathway of E. coli, featuring clustered chemosensory complexes formed by receptors bound to histidine kinase CheA and adaptor protein CheW. Chemoreceptors detect chemical ligands, either directly via their ligand binding domains or indirectly upon interactions with periplasmic binding proteins (PBPs), and modulate activity of CheA. The signal is transmitted to flagellar motor by phosphorylation of the diffusible response regulator CheY, which modulates the direction of motor rotation. The signal is terminated by the phosphatase CheZ. Receptor methylation enzymes, the methyltransferase CheR and the methylesterase CheB carry out adaptation to steady stimulation and provide short-term memory for temporal concentration comparisons.
The chemotaxis system modulates the duration of the runs according to perceived changes in environmental conditions, making them longer or shorter, if conditions get better or worse, respectively, to bias the average cell motion towards favorable environments (Berg and Brown 1972; Larsen et al. 1974; Macnab and Koshland 1972). The signaling pathway controlling this behavior is highly conserved among bacteria and even archaea (Fig. 1B). Nevertheless, several distinct classes of chemotaxis pathways could be distinguished based on their detailed molecular composition and evolutionary relatedness, some of which control behaviors other than flagellar motility (Gumerov, Andrianova and Zhulin 2021; Wuichet and Zhulin 2010). In contrast to many other bacteria, the genome of E. coli encodes only a single motility and chemotaxis system, which moreover functions with a nearly minimal set of chemotaxis proteins. Such comparative simplicity turned E. coli into a preferred model for studying signal transduction in bacterial chemotaxis (Bi and Sourjik 2018; Parkinson, Hazelbauer and Falke 2015).
In general, bacterial chemotaxis pathways consist of two modules – one for rapid signal transduction and another for slower adaptation (Shimizu, Tu and Berg 2010). The signal transduction module is composed of transmembrane chemoreceptors that change conformation upon ligand binding or other environmental perturbations and together with the adaptor protein CheW modulate the activity of a histidine kinase CheA (Parkinson, Hazelbauer and Falke 2015). Together with CheA and CheW, chemoreceptors form stable supramolecular sensory complexes that primarily cluster at cell poles in E. coli and other bacteria (Yang and Briegel 2020). The kinase CheA phosphorylates the diffusible response regulator CheY, which, when phosphorylated, binds to the flagellar motor to induce its clockwise rotation and thus cell tumbling. This signaling core is highly conserved among all chemotaxis pathways (Wuichet and Zhulin 2010). Many bacterial systems, including that of E. coli, also possess a specific phosphatase CheZ that rapidly dephosphorylates CheY, thereby ensuring that the phosphorylation level of CheY reflects the kinase activity with only short delay. In other chemotaxis pathways, CheY dephosphorylation is carried out by alternative phosphatases, CheC or CheX (Silversmith 2010). Some of the chemotaxis pathways, including the closely related pathway in Salmonella enterica, include an additional component of the sensory complexes CheV, which has the CheW-like scaffolding domain and the CheY-like regulatory domain (Alexander et al. 2010).
The signal transduction module of the chemotaxis pathway belongs to a larger family of two-component systems (TCSs) that enable environmental sensing in prokaryotes and are also present in fungi and plants (Stock, Robinson and Goudreau 2000). One important difference between the canonical TCSs and the chemotaxis pathways is that in the former the sensory, kinase and phosphatase activities are typically executed by a single sensory kinase protein, whereas in chemotaxis these functions are carried out by different proteins within one stable complex (Gumerov, Andrianova and Zhulin 2021; Sourjik and Armitage 2010). Such segregation of sensory and signaling activities likely facilitates evolutionary adaptation of the chemotaxis pathway to new environmental niches with different chemoeffector requirements, where specific chemoreceptors could be rapidly acquired or lost without affecting the function of the signaling core. Indeed, both specificities and the number of chemoreceptors apparently correlate with the respective lifestyles of bacterial species (Ortega, Zhulin and Krell 2017).
The chemotaxis pathway further includes an adaptation module that is composed of two enzymes CheR and CheB, which respectively methylate and demethylate specific residues on the receptor and thus counterbalance the effect of ligand binding on receptor conformation (Goy, Springer and Adler 1977; Kehry and Dahlquist 1982; Terwilliger, Wang and Koshland 1986). The (de)methylation rates depend primarily on the current activity of the receptor-kinase complex and they are slow compared to the other reactions within the pathway (Block, Segall and Berg 1983; Sourjik 2004; Sourjik and Berg 2002). Consequently, CheR and CheB provide a delayed integral negative feedback, which allows the cell to respond to temporal changes in experienced conditions over a wide range of backgrounds (Barkai and Leibler 1997; Berg and Purcell 1977; Kalinin et al. 2009; Lazova et al. 2011; Mesibov, Ordal and Adler 1973; Segall, Block and Berg 1986; Yi et al. 2000). This methylation-dependent adaptation module is unique in comparison to other TCSs, and present in the vast majority of bacterial chemotaxis pathways. In addition to CheR and CheB, adaptation in other chemotaxis pathways such as that of Bacillus subtilis involves CheV and a receptor deamidase CheD that are absent in E. coli, but the interplay between these different levels of adaptation remains poorly understood (Walukiewicz et al. 2014).
Clustering of chemoreceptors and associated chemotaxis proteins appears to be a universal feature of all studied prokaryotic chemotaxis systems (Sourjik 2004; Yang and Briegel 2020). Clustering allows receptor-kinase complexes to respond cooperatively and thus highly sensitively to changes in environmental conditions (Sourjik 2004; Tu 2013). Since receptors with different ligand specificities are mixed within clusters, clustering further facilitates signal integration (Parkinson, Ames and Studdert 2005). Finally, many bacteria express multiple chemotaxis systems, and hence spatial segregation provided by clustering might help to separate proteins belonging to different systems and thus prevent their undesired interference (Sourjik and Armitage 2010).
In contrast to this highly detailed molecular understanding of signal processing and motility control in E. coli and several other bacteria, the physiological importance of chemotaxis and flagellar motility are not well established even for model bacterial systems. In this review, we thus aim to summarize the current state of knowledge about different physiological aspects of chemotactic behavior. These range from importance of chemotaxis for enhanced nutrient acquisition by individual bacteria and population range expansion to the role that chemotaxis plays in bacteria-bacteria and bacteria-host interactions. We further illustrate how better understanding of bacterial chemotactic behavior in its physiological context(s) might help to rationalize many of its observed properties, from ligand specificity of chemoreceptors to growth-dependent regulation of motility gene expression.
CHEMOTAXIS TOWARDS NUTRIENTS
Correlation between chemotactic and nutritional preferences of bacteria
Early studies of chemotaxis showed that bacteria are attracted to common nutrients, such as amino acids or sugars (Adler, Hazelbauer and Dahl 1973; Mesibov and Adler 1972; Pfeffer 1884), while being repelled from harmful conditions such as toxic levels of inorganic ions or extreme pH (Tso and Adler 1974), which led to the assumption that bacteria use chemotaxis to accumulate in environmental niches that provide optimal conditions for growth. Confirming this correlation between chemotactic and metabolic preferences, several studies have shown that, even within the same chemical class, the most potent chemoattractants are those compounds that are also preferentially consumed (Neumann, Grosse and Sourjik 2012; Somavanshi, Ghosh and Sourjik 2016; Yang et al. 2015) or give the shortest lag time in growth when used as a carbon source (Fernandez et al. 2017). Thus, bacteria might generally utilize chemotaxis to enhance acquisition of high-value nutrients in their environment, and many ligand preferences of bacterial chemoreceptors could be explained by such nutrient taxis. Consistently, bacteria that acquired capabilities to metabolize environmental pollutants also apparently evolved chemotaxis towards these chemicals (Krell et al. 2013; Parales and Harwood 2002). More generally, evolutionary selection for locating optimal physiological conditions could explain responses to unconventional chemoeffectors, which signal by affecting metabolism or perturbing cellular physiology or energy state (Alexandre, Greer-Phillips and Zhulin 2004; Bi and Sourjik 2018; Schweinitzer and Josenhans 2010).
However, correlation between chemotactic and metabolic or physiological preferences of bacteria is not always the case. For instance, B. subtilis appears to use gradients of amino acids (Yang et al. 2015) or ethanol (Tohidifar et al. 2020) as environmental cues in order to locate sources of nutrients, such as plant roots or decaying organic matter, rather than because of their immediate nutritional value. As discussed below, such tactic responses to gradients of small molecules excreted by animals, plants or other microbes, irrespective of the nutritional value, might be common in host–bacteria interactions and enable bacteria to orient themselves relative to their hosts or cooperation partners and also to locate specific niches, such as wound areas.
Adaptation of nutrient search strategies to respective environments
In the environment, bacteria are likely to encounter a variety of complex chemoeffector landscapes which result from different geometries of their release and diffusion, and from advection by flows and degradation (Raina et al. 2019). Typical examples are patches where an initial localized spike of attractant spreads by diffusion, e.g. after a burst of a phytoplankton cell. In turbulent marine environments, these patches can turn into filaments of nutrients (Taylor and Stocker 2012; Watteaux, Stocker and Taylor 2015). The simplest biologically relevant case is release of a freely diffusing attractant at a constant rate by a fixed point source – e.g. by a pore in a plant or animal epithelium or by a living cell aggregate – which results in a gradient where both concentration and the relative gradient of concentration (1/c dc/dx) decrease as the inverse of the distance to the source (Berg and Purcell 1977). In another relevant case, chemical concentration only varies in one direction, e.g. when an attractant is uniformly released from the flat surface of a large object or diffuses from an air-liquid interface. Uniform degradation of the attractant will result here in an exponential decay of attractant concentration away from the source, the steepness of which is determined by the degradation rate. In contrast, if the released chemical is absorbed by a distant sink, e.g. sessile (micro)organisms, a linear gradient of the chemical forms between source and sink. Importantly, these different gradient shapes will affect tactic behaviors. For instance, bacteria such as E. coli that respond to relative gradients (log-sensors) (Kalinin et al. 2009; Lazova et al. 2011; Menolascina et al. 2017; Mesibov, Ordal and Adler 1973; Sourjik and Berg 2002), will maintain constant velocity of chemotactic drift in an exponential gradient, while increasing drift velocity as they climb a point-source gradient or slowing down in a linear gradient. In the laboratory, most quantitative experiments probing chemotactic behaviors of swimming cells are carried out in steady linear gradients of chemoeffectors, which stems from the ease with which such gradients can be created within microfluidic devices (Ahmed, Shimizu and Stocker 2010; Colin, Zhang and Wilson 2014; Kalinin et al. 2009), as well as from the relative simplicity of the theoretical analysis of the cell behavior in that case. Nevertheless, more complex and time varying gradients can be mimicked under laboratory conditions by controlling flow in microfluidic devices (Ahmed, Shimizu and Stocker 2010; Englert, Manson and Jayaraman 2010; Stocker et al. 2008) or by releasing caged compounds to form transient patches (Brumley et al. 2019; Jikeli et al. 2015; Mccray and Trentham 1989).
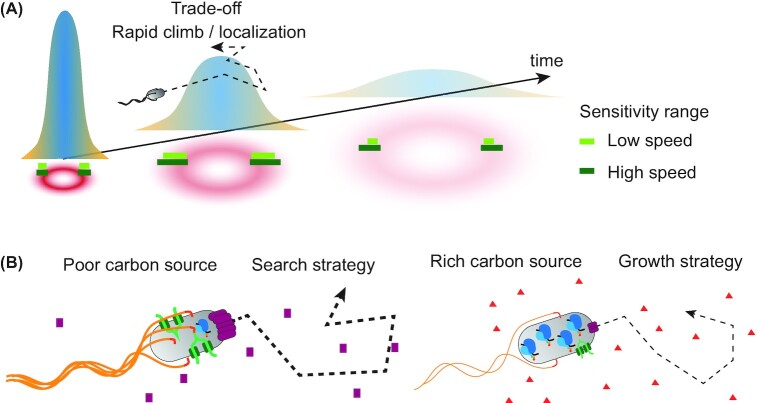
Responding to such dynamic gradients poses additional challenges to chemotactic bacteria (Fig. 2A), since they must not only climb the gradients rapidly but also localize near the maxima of attractant concentrations, and also react to the time evolution of the concentration profile, in order to maximize the efficiency of their chemotactic behavior (Blackburn and Fenchel 1999; Brumley et al. 2019; Stocker et al. 2008). Indeed, the pathway response of E. coli was suggested to meet a theoretical trade-off between efficient gradient climbing, ensured by the rapid tumble suppression upon stimulation, and localization at maximal concentrations, allowed by perfect adaptation (Clark and Grant 2005). This response was then shown to represent a generalist strategy to maximize the minimum nutrient uptake for any concentration profile, thus well-suited for unpredictable environments (Celani and Vergassola 2010). Consistent with this, the inferred distribution of gradient shapes that are most likely to be encountered by E. coli was found to be very wide (Clausznitzer et al. 2014). Another, complementary strategy to deal with spatiotemporally variable environments might be conferred by the large phenotypic variability of the chemotactic response observed even in clonal E. coli populations (Frankel et al. 2014; Karin and Alon 2021; Vladimirov et al. 2008).
Figure 2.
Trade-offs in chemotactic behavior and regulation of chemotaxis. (A), Chemotactic response to time varying concentration profiles that could result from diffusive spreading of attractant patch needs to balance rapid gradient climbing and localization at the peak. Higher swimming velocity expands sensitivity range of bacterial chemotaxis, particularly in shallow gradients (right), but incurs additional energetic costs. (B), Motility and nutrient uptake are regulated antagonistically with biosynthetic machinery dependent on the nutritional quality of the carbon source. During growth in poor carbon sources (left), motility is upregulated in proportion to potentially higher advantage provided by chemotaxis towards sources of additional nutrients (search strategy). In rich carbon sources (right), motility is downregulated to enable higher investment into biosynthetic machinery (growth strategy).
Other chemotactic bacteria, e.g. those in marine habitats, might have further improved on this generalist strategy (Brumley et al. 2020). It was argued that marine bacteria have specifically adopted the run-reverse-flick motility pattern (Xie et al. 2011), which differs from E. coli run-tumble behavior, to improve their localization at nutrient maxima without compromising gradient climbing (Stocker et al. 2008; Xie and Wu 2014), although it might reduce their ability to explore new nutrient patches. A generally higher swimming speed and chemokinetic ability of marine bacteria might enable more efficient exploitation of transient nutrient patches, both increasing the localization at maxima (Son, Menolascina and Stocker 2016) and resulting in higher sensitivity to shallow gradients (Brumley et al. 2019; Hein et al. 2016). Finally, bacterial chemotaxis systems seem to be able to detect about as small a change in concentrations as possible, given physical limitations imposed by the cell size and by diffusion of ligands and of bacteria themselves (Aquino et al. 2011; Berg and Purcell 1977; Bialek and Setayeshgar 2005; Brumley et al. 2019; Colin, Zhang and Wilson 2014; Micali and Endres 2016; Mora and Wingreen 2010).
The physical properties of the environment can also affect cell swimming and chemotaxis beyond the aforementioned effects of flow on gradient shapes. Aquatic bacteria are particularly exposed to fluid flows that exert mechanical shear, which can stir the swimming direction (Jing et al. 2020; Marcos et al. 2012) and drive swimming bacteria to regions of high flow shear (Bearon and Hazel 2015; Rusconi, Guasto and Stocker 2014). Because of this stirring, shear flows were predicted (Bearon and Pedley 2000; Locsei and Pedley 2009; Luchsinger, Bergersen and Mitchell 1999) and observed (Rusconi, Guasto and Stocker 2014) to reduce the efficiency of chemotaxis, even when the gradient is unaffected. Here as well, the run-reverse swimming pattern might bring the adaptive advantage of improving the chemotactic response in flow (Luchsinger, Bergersen and Mitchell 1999). Bacteria can also exploit physical properties of the environment to improve chemotactic navigation. A prominent example is provided by magnetotactic bacteria, which use needle-shaped magnetosomes to align with the earth magnetic field (Blakemore 1982; Faivre and Schuler 2008) and to follow it downwards (Blakemore, Frankel and Kalmijn 1980; Simmons, Bazylinski and Edwards 2006; Zhang et al. 2010). Combined with aerotaxis, such magnetotaxis enables these bacteria to position themselves in the growth-favorable microaerobic layer of their aqueous sediment habitats (Faivre and Schuler 2008; Lefevre and Bazylinski 2013; Mao et al. 2014; Spormann and Wolfe 1984; Yazi et al. 2018; Zhang et al. 2010).
Trade-offs between motility and growth
Although accumulation towards sources of nutrients may lead to increased nutrient uptake by bacteria and therefore to enhanced growth, swimming motility also requires high investment of cellular resources. Biogenesis of motility system and powering of flagellar motor rotation consume respectively up to several percent of total cellular protein and energy budget in E. coli (Berg 2003; Colin and Sourjik 2017; Milo et al. 2010) and these costs are likely similar or even higher in other bacteria. Consequently, expression of motility genes can significantly reduce bacterial growth under conditions where it provides no advantage (e.g. in a well-stirred environment), implying the existence of an environment-dependent fitness trade-off between the benefits and costs of resource investment in motility (Fraebel et al. 2017; Ni et al. 2020; Ni et al. 2017; Taylor and Stocker 2012; Yi and Dean 2016). Such adaptive trade-offs are common in bacteria, as well as in other organisms, which typically need to optimize different conflicting functions during evolution (Ferenci 2016). Indeed, besides trade-offs associated with gene expression, other trade-offs related to the costs of precise operation of the chemotaxis machinery have been recognized (Brumley et al. 2019; Govern and Ten Wolde 2014; Lan et al. 2012).
As a consequence, bacteria evolved multiple regulatory strategies to optimize cellular resource allocation dependent on their growth conditions (Molenaar et al. 2009; Schuetz et al. 2012; Scott et al. 2010).This is reflected in the multilevel regulation of expression of bacterial flagellar and chemotaxis genes by a variety of environmental and cellular cues (Amsler, Cho and Matsumura 1993; Chevance and Hughes 2008; Guttenplan and Kearns 2013; Pruss 2017). One of the most prominent mechanisms of this regulation in E. coli is by the carbon catabolite repression mediated by cyclic adenosine monophosphate (cAMP), which reflects growth rate and carbon uptake into the cell and is elevated during growth on poor carbon sources (Adler and Templeton 1967; Hui et al. 2015; Liu et al. 2005). High levels of cAMP under carbon-limited growth activate multiple pathways for uptake and catabolism of alternative carbon sources, as well as genes involved in the TCA cycle and amino acid synthesis (Hui et al. 2015; Liu et al. 2005). This regulation enhances nutrient uptake and catabolism at a cost of reduced allocation of resources in protein biosynthesis (You et al. 2013). The activation of flagellar and chemotaxis genes by cAMP might follow similar regulatory logic, enhancing carbon acquisition by active accumulation towards sources of nutrients in carbon-poor environments (Amsler, Cho and Matsumura 1993; Hui et al. 2015; Liu et al. 2005) (Fig. 2B). Indeed, active acquisition of nutrients by motile bacteria becomes increasingly important in carbon-poor environments, as demonstrated by co-culturing chemotactic and non-chemotactic E. coli in presence of nutrient gradients (Ni et al. 2020). Notably, the relative fitness benefit provided by chemotaxis exhibits the same dependence on the growth rate as expression of flagellar genes, indicating that E. coli invests resources in motile behavior in proportion to its anticipated benefit. A fitness benefit of chemotaxis in an unstirred co-culture was also observed in absence of artificially introduced gradients of nutrients, apparently due to the self-generation of gradients by bacteria through excretion and subsequent chemotaxis-mediated consumption of metabolites (Ni et al. 2020). Similar cross-feeding might contribute to positive selection for motility in natural microbial communities, and it might also explain the rapid accumulation of motility-activating mutations in a resting culture of E. coli (Parker, Demetci and Li 2019).
In bacteria, trade-offs associated with resource allocation are typically adaptive and can be tuned by mutations dependent on the environment (Ferenci 2016). Consistently, under experimental selection for enhanced chemotaxis, the balance between bacterial motility and growth could be easily and gradually shifted by a variety of mutations along a well-defined growth-motility trade-off line (Fraebel et al. 2017; Ni et al. 2017; Yi and Dean 2016). The main phenotypic change observed in these different studies was an enhancement of flagellar gene expression and thus of cell swimming velocity, likely because of the steep dependence of the chemotactic drift of individual bacteria on their velocity (Schauer et al. 2018). Moreover, a similar trade-off was observed between enhancement of motility and growth reduction in all studies. In contrast, genetic mutations underlying the evolved phenotypic changes differed largely between individual studies, presumably due to the epistatic effects of strain background and/or differences in selection protocols. This confirms the high plasticity of bacterial motility that enables it to evolutionarily adapt to novel environments, which might be a common property of bacterial networks (Hindre et al. 2012). Notably, it was proposed that such evolvability might be favored by the hierarchical design of bacterial flagellar gene regulatory networks (Ni et al. 2017).
IMPORTANCE OF VARIABILITY OF CHEMOTACTIC PERFORMANCE IN BACTERIAL POPULATIONS
Trade-offs between different conflicting functions might also explain the co-existence of individual cells with different physiological states within bacterial populations. The importance of such phenotypic heterogeneity that is observed for behaviors of individual cells even in genetically homogeneous microbial populations has been well recognized in recent years (Bettenworth et al. 2019; Jung et al. 2019; Veening, Smits and Kuipers 2008; Weigel and Dersch 2018). Bacterial swimming behavior provided one of the first examples of such behavioral individuality (Spudich and Koshland 1976). In E. coli, both the run-and-tumble bias and the chemotactic sensitivity are subject to cell-to-cell variability as well as temporal variability within each cell. Variations in the expression level of chemotactic proteins, particularly CheR and CheB (Dufour et al. 2016) appear to produce a cell-to-cell variability in the adaptation dynamics (Keegstra et al. 2017) and in the duration of runs (Min et al. 2009). Flagellar number, which varies among individual cells with the expression levels of motility genes, is another modulator of the tumbling rate (Mears et al. 2014; Vladimirov, Lebiedz and Sourjik 2010). The pathway gain, arising from cooperative responses of the chemoreceptor clusters and flagellar motors, also appears to show strong cell-to-cell variability (Salek et al. 2019).
This variability in the settings of the chemotaxis pathway strongly modulates the chemotactic performance of individual cells (Salek et al. 2019; Waite et al. 2016; Wong-Ng et al. 2016). This is thought to allow bet-hedging strategies in response to chemical gradients, with the population separating between adventurous strong responders and more sedentary weak responders. Such separation of the population has indeed been observed in self-generated chemical gradients (Fu et al. 2018; Salek et al. 2019). Since the set of pathway parameters eliciting the strongest response varies strongly with gradient shape and steepness (Dufour et al. 2014; Long, Zucker and Emonet 2017), it was argued that phenotypic heterogeneity in chemotactic behavior might have been evolutionarily selected to optimize chemotaxis in variable environments (Frankel et al. 2014; Karin and Alon 2021; Vladimirov et al. 2008). The variability in pathway activity was, however, found to be reduced at high levels of chemoattractant, allowing cell populations to combine the exploratory strategy in nutrient-poor media with the faithful and fairly homogeneous response once gradients are encountered (Kamino et al.2020).
Additionally, even within unstimulated single cells, slow but large temporal fluctuations of CheY phosphorylation and therefore of the tumbling rate could be observed (Colin et al. 2017; Keegstra et al. 2017; Min et al. 2012; Min et al. 2009). These temporal fluctuations apparently originate from the amplification of the noisy and slow receptor methylation dynamics as well as of thermal noise by strongly coupled receptor clusters (Colin et al. 2017), and they can largely explain the broad, power-law distributions of run durations observed in cell populations (Korobkova et al. 2004; Min et al. 2009; Park et al. 2010). Such long-term fluctuations are thought to enhance the effective diffusion of single cells and therefore their ability to explore the environment (Benichou et al. 2011; Matthaus, Jagodic and Dobnikar 2009; Matthaus et al. 2011), and they might also enhance the chemotactic drift (Flores et al. 2012). Additionally, the activity fluctuations were predicted to increase the coordination of the flagellar motors, thus improving chemotactic performance (Sneddon, Pontius and Emonet 2012).
Even more pronounced heterogeneity is observed in other cases, where only a fraction of cells in a population becomes motile. Such bimodality of flagellar gene expression has been described in B. subtilis (Kearns and Losick 2005), S. enterica serovar Typhimurium (Koirala et al. 2014) as well as in pathogenic E. coli strains (Laganenka et al. 2020), and it is likely to be a common phenomenon. Differentiation of the population into distinct subpopulations of motile and sessile non-flagellated cells might reflect previously discussed trade-offs/physiological conflicts between colonization and exploration of the environment (Koirala et al. 2014; Mukherjee and Kearns 2014) and between resource investment in growth and motility (Syvertsson et al. 2021). Yet another trade-off exists in bacterial pathogens, where flagellar motility and chemotaxis provide benefit at the early stage of infection (see below) but flagellation later becomes a burden since the flagellum is a major antigen recognized by the immune system (Sporing et al. 2018). Consistently, relative fractions of motile cells in bacterial populations are regulated by a variety of factors, from nutrient levels and stress response to physical properties of the environment, but the complex underlying mechanisms remain only partly understood (Koirala et al. 2014; Laganenka et al. 2020; Mukherjee and Kearns 2014; Sporing et al. 2018; Wang et al. 2020).
MOTILITY AND CHEMOTAXIS IN POPULATION AND COLLECTIVE BEHAVIORS
Motility-driven expansion of the population range
Colonization of a porous growth medium from a single inoculation site represents the simplest example of chemotactic behaviour at the level of a bacterial population. Such motility-dependent expansion of the bacterial population range is typically experimentally studied using a soft agar assay, where bacteria grow and propagate from a central inoculum into a low percentage agar gel supplied with nutrients (Adler 1966) (Fig. 3A). The agar mesh is loose enough for the cells to swim and navigate gradients, such that colony propagation results from a combination of growth, motility and chemotaxis. In contrast to liquid media, swimmers get stuck in the agar mesh, which they can only escape by tumbling, therefore restricting efficient cell propagation to cells with intermediate tumbling rates (Wolfe and Berg 1989). Although chemotaxis is not strictly required for spreading in soft agar, it can greatly accelerate the rate of bacterial colony expansion. By consuming nutrients and other chemicals inside the colony, metabolically active bacteria create gradients in the medium, which they can subsequently use to migrate outwards as expanding chemotactic rings of a constant high density (Adler 1966; Koster et al. 2012). This behavior can be mathematically captured by the classical Keller–Segel model of chemotaxis and its numerous variants (Keller and Segel 1971; Tindall et al. 2008). Similar behavior can also be observed in liquid media, for example in narrow straight channels (e.g. glass capillaries) where the population response to a self-generated gradient takes the form of travelling chemotactic bands (Adler 1966; Saragosti et al. 2011).
Figure 3.
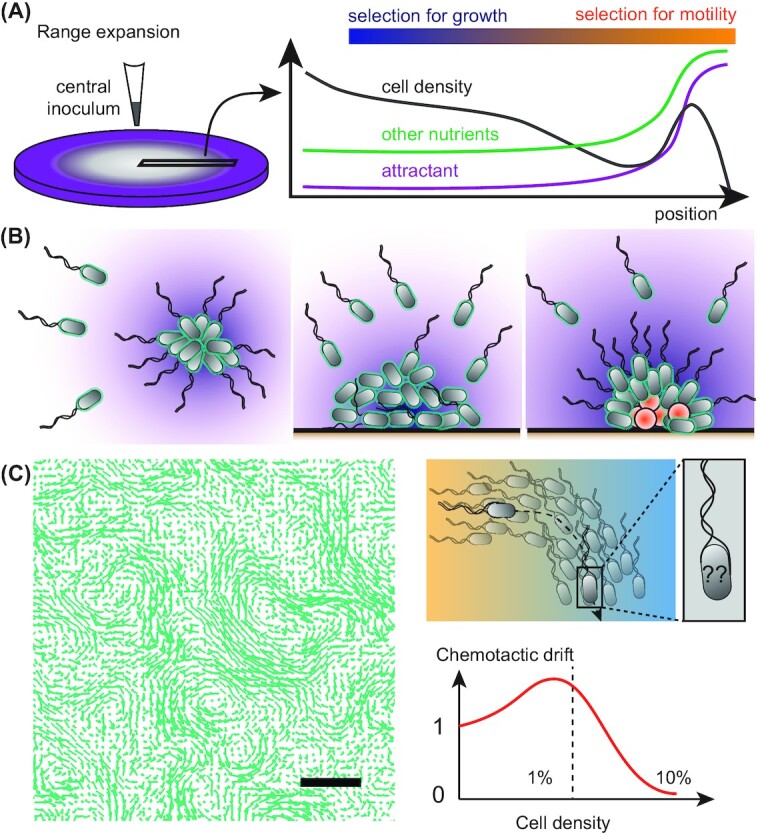
Collective chemotactic behaviors. (A), Population expansion driven by chemotaxis towards self-generated gradients produced by metabolite consumption in porous medium (left) results in a spatial organization of the cell population, with selection for motility at the front and for growth at the rear of the spreading colony (right). (B), Chemoattraction to quorum-sensing signals can enhance autoaggregation and biofilm formation in single or multi-species communities of bacteria that secrete an attractant. (C), Swirling collective motion emerges at high bacterial cell densities, as observed on maps of the local cell velocities (left). It impairs the chemotactic perception of gradients by inducing random reorientations on the time scale of gradient sensing (top right), thus reducing chemotactic drift above the cell density at which collective motion begin to emerge (bottom right, dashed line).
Although such traveling bands formed by bacterial monoculture are unlikely to be common in nature, they might nevertheless be relevant for population spreading in natural porous environments. Moreover, these assays have been used as a general model to investigate the interplay between growth and directed movement in collective range expansion of a population. In this model, the chemoattractants can serve as aroma-like cues that allow a population to migrate outward well before nutrients run out, even if these attractants make up only a small portion of the nutrients available in the medium (Cremer et al. 2019). Moreover, collectively spreading bacterial populations are able to cross and colonize maze-like or even fractal structures that mimic environments with complex geometries such as the soil or the lungs (Park et al. 2003; Phan et al. 2020). The importance of heterogeneity in individual chemotactic responses within spreading populations was also investigated. It was shown that the apparent conflict between this cell-to-cell variability and the coherent motion as a band of the population can be resolved by the stratification of the individuals in the self-generated chemical gradient according to their individual characteristics (Fu et al. 2018).
Motility and chemotaxis in a porous medium further play an important role when several populations compete for the same habitat. Trade-off between growth and expansion rate can in this case lead to phenotypic segregation, where fast swimming but slow growing cells occupy the outer rings, while fast growing but slow swimming cells keep the center and are at risk of being globally outcompeted over the whole plate (Gude et al. 2020; Liu et al. 2019) (Fig. 3A). Consistently, in an initially clonal population there is selection for mutations that favor swimming speed at the expense of growth in the outer edge of the spreading colony, whereas mutations favoring growth over speed are selected close to the center (Liu et al. 2019; Ni et al. 2017). Thus, the observed trade-offs between growth and motility likely play an important role in bacterial niche formation and evolution in structured environments. Strikingly, in E. coli these trade-offs lead to a negative frequency selection on strains with different levels of motility in a spreading population, resulting in stable coexistence of such strains (Gude et al. 2020). Spatiotemporal structuring of multispecies bacterial colonies can be further greatly enhanced by either antagonistic or cooperative regulatory interactions between species (Curatolo et al. 2020).
Autoaggregation of chemotactic bacteria
Besides these dynamic traveling bands that rely on nutrient gradient formation and reshaping by a spreading bacterial population, chemotactic bands can also emerge through several other mechanisms. At the levels of individual bacteria, accumulation towards a specific location can be observed when cells respond to opposite gradients of chemicals, as well as in response to oxygen (Alexandre, Greer and Zhulin 2000; Rebbapragada et al. 1997; Shioi et al. 1988), pH (Yang and Sourjik 2012) or temperature (Oleksiuk et al. 2011; Paulick et al. 2017; Salman and Libchaber 2007; Yoney and Salman 2015), for which the chemotactic response changes from repellent to attractant as a function of the level of stimulation. In this scenario, two opposing chemotactic ‘forces’ drive bacteria to accumulate in an intermediate region, where the net chemotactic velocity is null (Hu and Tu 2013; Zhang et al. 2019). Bidirectional sensing of many physical and chemical stimuli (e.g. pH or temperature) by chemotactic bacteria is thought to generally play an important role in locating physiologically optimal niches in chemically complex environments.
Emergence of higher complexity patterns, which can occur even in initially uniform environments, typically relies on tactic responses elicited by chemical interactions between bacteria, although other mechanisms such as swimming speed regulation might contribute, too (Cates 2012; Curatolo et al. 2020). Instead of (or in addition to) generating and sensing gradients of chemicals which were already present in the medium, bacteria release chemicals that elicit chemotactic responses by their peers, which can lead to chemotactic self-attraction and generate aggregative processes on various length scales. For example, E. coli forms regular lattices of millimeter-large high-density spots on soft agar plates, which could be explained by the chemotactic response to self-produced gradient of aspartate (Budrene and Berg 1991; Budrene and Berg 1995). Chemotaxis to self-produced attractants can further lead to accumulation of bacteria in small sub-millimeter cavities in microfluidic devices (Park et al. 2003).
Although in the aforementioned examples cluster formation does not require any physical interactions among bacteria, self-attraction may also enhance bacterial aggregation (clumping) that is mediated by various surface adhesins (Defoirdt 2011) (Fig. 3B). Indeed, chemotactic response to the quorum-sensing molecule autoinducer 2 (AI-2) secreted by cell aggregates can indeed largely enhance the autoaggregation of E. coli, mediated either by adhesin antigen 43 or by curli filaments (Hegde et al. 2011; Jani et al. 2017; Laganenka, Colin and Sourjik 2016; Song and Wood 2021). AI-2 is an interspecies communication signal produced by a wide variety of bacteria (Pereira, Thompson and Xavier 2013; Waters and Bassler 2005). Further supporting its potential role in establishing interspecies interactions within complex microbial communities, chemoattraction to AI-2 can mediate co-aggregation of different species (Laganenka and Sourjik 2017), and it is not restricted to AI-2-producing bacteria (Zhang et al. 2020). Besides AI-2, other quorum-sensing signals can also mediate bacterial self-attraction responses, for instance the S signal in Vibrio parahaemolyticus (Lamb, Trimble and McCarter 2019). Chemotaxis can also mediate self-repulsion. For example, the chemotaxis pathway of Azospirillum brasilense exerts a negative effect on cell clumping (Bible et al. 2008). The complex chemotactic response of E. coli to self-produced indole might serve both functions, mediating self-repulsion at low levels of secreted indole but leading to self-attraction when the levels are high (Yang et al. 2020). Given the large diversity of autoinducer signals produced by bacteria, all these different scenarios of intra- and interspecies attraction and repulsion are likely to be found in natural microbial communities where they might produce complex collective behaviors (Grauer et al. 2020).
Physical interactions between motile cells
In addition to chemical and adhesive interactions, at higher densities cell swimming itself leads to physical interactions between bacteria, both direct when cells collide (steric interactions) and through the fluid that they displace (hydrodynamic interactions). When the cell density increases, hydrodynamic interactions are the main contributor to the emergence of swirling collective motion made of intermittent jets and eddies of hundreds of cells (Cisneros et al. 2007; Dunkel et al. 2013; Koch and Subramanian 2011; Liu et al. 2000; Luchsinger, Bergersen and Mitchell 1999; Sokolov et al. 2007; Wensink et al. 2012; Wolgemuth 2008). These are observed in many bacterial species at the front of swarms of swimming bacteria propagating at the surface of semi-solid agar gels (Be'er and Ariel 2019; Kearns 2010; Partridge and Harshey 2013) or near air–water interfaces (Holscher et al. 2015). Interestingly, these physical interactions strongly modify the chemotactic response, both in two- and three-dimensional geometries: Although the response is slightly enhanced at moderate cell densities, the emergence of the collective motion annihilates the ability of E. coli to follow chemical gradients (Colin, Drescher and Sourjik 2019), due to rapid randomization of the direction of motion of swimming bacteria caused by collective motion that prevents gradient sensing through temporal comparisons (Fig. 3C). Therefore, there is a physical upper limit on the density at which bacteria can chemotactically accumulate near a source of attractant or in a travelling chemotactic band. Such reduction of chemosensing at high density must also affect bacterial swarms, where collective motion arises in the dense quasi-monolayer of swimming cells behind the colony edge (Darnton et al. 2010; Harshey 1994; Jeckel et al. 2019; Kearns and Losick 2003). Accordingly, chemotaxis is not necessary for swarming (Ariel et al. 2018; Be'er and Ariel 2019; Burkart, Toguchi and Harshey 1998; Sidortsov, Morgenstern and Be'er 2017). Nevertheless, a recent report suggested that an E. coli swarm may be able to bias its motion towards higher concentrations of an attractant (Tian et al. 2021), which would require specific mechanisms to counteract the physics-driven loss of chemotaxis. These could include a strongly reduced tumbling rate, which is observed not only for E. coli but also for other swarming bacteria (Ford et al. 2018; Mariconda, Wang and Harshey 2006; Partridge et al. 2019; Partridge et al. 2020), as well as cell elongation (Ilkanaiv et al. 2017; Kearns 2010) and modified fluid flows within the swarm compared to suspensions (Chen et al. 2017; Jeckel et al. 2019; Li et al. 2017). Importantly, these results hold for the flagella-propelled bacteria. In contrast, the chemotactic ability of bacteria that move at high density on semisolid surfaces using twitching or gliding motility remains poorly understood, although their collective migration up chemical gradients has been reported (Guzzo et al. 2018; Islam and Mignot 2015; Kearns, Robinson and Shimkets 2001; Oliveira, Fostera and Durham 2016; Sampedro et al. 2015).
Roles of flagella, motility and chemotaxis in biofilm formation
With the formation of a surface attached biofilm, bacteria adopt a sessile lifestyle which offers protection against harsh environments and enables division of labor in bacterial communities. The formation of submerged biofilms on liquid–solid interfaces typically proceeds through several stages, including surface attachment, growth and maturation of matrix-embedded communities, and finally biofilm dispersion (Rumbaugh and Sauer 2020; Stoodley et al. 2002). These sessile biofilm communities are commonly viewed in opposition to the explorative motile planktonic lifestyle. Indeed, genes required for production of biofilm matrix and those for motility are antagonistically regulated in E. coli and other bacteria, including their mutually exclusive expression (Besharova et al. 2016; Guttenplan and Kearns 2013; Pesavento et al. 2008; Pruss 2017; Serra et al. 2013). A major signal controlling this transition between gene expression profiles characteristic for motile and sessile states is the second messenger cyclic diguanosine monophosphate (c-di-GMP), which generally promotes biofilm formation and reduces motility (Guttenplan and Kearns 2013; Hengge 2009; Jenal and Malone 2006). Besides leading to repression of flagellar gene expression, c-di-GMP can also reduce motility at the post-translational level (Guttenplan and Kearns 2013), e.g. by activating the flagellar motor break protein YcgR in E. coli (Boehm et al. 2010; Paul et al. 2010; Ryjenkov et al. 2006).
Despite this generally antagonistic regulation, increasing evidence suggests that flagella, motility and chemotaxis play important roles at all stages of biofilm formation. Strains of E. coli defective in motility form smaller and sparser submerged biofilms (Pratt and Kolter 1998; Wood et al. 2006). Similar phenotypes were observed for non-motile mutants of diverse bacteria, including Pseudomonas, Shewanella, Agrobacterium and Bacillus species (Holscher et al. 2015; Merritt, Danhorn and Fuqua 2007; O'Toole and Kolter 1998; Thormann et al. 2004). Flagellar motility indeed strongly promotes the initial attachment to the surface (Berke et al. 2008; Elgeti and Gompper 2013; Li et al. 2011). Attachment might be further enhanced by a strong chemoattractant response that suppresses tumbles, thus physically favoring accumulation of swimming bacteria at the surface and increasing their chances of attachment (Berke et al. 2008; Li and Tang 2009; Suchanek et al. 2020). Motility and chemotaxis can further drive bacteria towards a favorable niche for attachment and/or biofilm formation if the latter is chemoattractive, e.g. an air–water interface (Ardre et al. 2015; Holscher et al. 2015), gut epithelial surface (Misselwitz et al. 2012) or plant root (Scharf, Hynes and Alexandre 2016). Besides enhancing accumulation and attachment to surfaces, chemotaxis to the self-produced attractant AI-2 (see above) promotes the formation of larger and more structured submerged E. coli biofilms (Jani et al. 2017; Laganenka, Colin and Sourjik 2016). Since AI-2 is produced by many bacteria and mediates cross-species chemical interactions (Pereira, Thompson and Xavier 2013), chemoattraction to AI-2 may also favor co-aggregation and mixed biofilm formation, as indeed observed in co-cultures of E. coli and E. faecalis (Laganenka and Sourjik 2017). In addition to the requirement of motility for such chemotaxis-dependent enhancement of biofilm formation, flagella can directly promote surface attachment, serving as adhesins (Friedlander, Vogel and Aizenberg 2015).
In the mature biofilm, motility is repressed but the flagellum could be repurposed as an important structural element of the biofilm matrix (Besharova et al. 2016; Serra et al. 2013; Wood et al. 2006). Finally, reactivation of motility may be important at the stage of biofilm dispersal (Rumbaugh and Sauer 2020), which might be further enhanced by chemotactic self-repulsion. Interestingly, in Helicobacter pylori this self-repulsion is mediated by AI-2 (Anderson et al. 2015; Rader et al. 2011; Sweeney et al. 2019), in contrast to its biofilm-promoting role as an attractant in E. coli.
MOTILITY AND CHEMOTAXIS IN HOST-MICROBE INTERACTIONS
Colonization and infection of animal hosts
Although only a fraction of bacteria associated with animal hosts are motile, flagellar motility and chemotaxis are common among bacterial pathogens, and typically important for successful host colonization and infection (Chaban, Hughes and Beeby 2015; Erhardt 2016; Matilla and Krell 2018). Motility might have several functions in the animal-microbe interactions (Chaban, Hughes and Beeby 2015; Erhardt 2016), and it might be particularly important in the gastrointestinal (GI) tract, where most of the animal microbiota resides (Fan and Pedersen 2021) (Fig. 4A). Successful colonization of the GI tract by enteric bacteria mostly depends on their ability to penetrate (or disrupt) the viscous mucus layer to reach a favorable niche. The importance of the mucus barrier in maintaining gut homeostasis is underscored by studies showing that MUC2-deficient mice are prone to spontaneous inflammation (Van der Sluis et al. 2006) and less resistant to bacterial infection (Bergstrom et al. 2010; Zarepour et al. 2013). Flagellar motility enables bacteria to increase their rate of encounter with the mucus surface (Misselwitz et al. 2012) and attach to and penetrate the mucous layer (Arora et al. 1998; Baban et al. 2013; Lane et al. 2005; Pichon et al. 2009; Tamar, Koler and Vaknin 2016; Wright, Seed and Hultgren 2005). Whereas some bacteria degrade or modify the mucus to facilitate penetration (Celli et al. 2009; Szabady et al. 2011), others (like S. Typhimurium) preferentially invade epithelial cells in the gut regions devoid of a continuous mucus layer (Furter et al. 2019). The attachment rate can be further enhanced by epithelium-produced molecules like mucins and their degradation products (Hugdahl, Beery and Doyle 1988; Nelson et al. 1990) or, potentially, AI-2 mimics (Ismail, Valastyan and Bassler 2016), which is consistent with a growing appreciation for the role of compounds released into the lumen by endocrine and immune systems of animal hosts in modulating host-microbe interactions during gut colonization (Neuman et al. 2015; Pacheco and Sperandio 2009; Rhee, Pothoulakis and Mayer 2009). It is likely that some of these molecules also mediate chemotactic responses. Specifically, E. coli has been shown to sense a number of human hormones, including norepinephrine (NE), 3,4-dihydroxymandelic acid (DHMA), dopamine, melatonin, as well as other chemicals that might be secreted into the gut lumen by animal hosts and/or microbiota, such as spermidine and indole (Bansal et al. 2007; Lopes and Sourjik 2018; Pasupuleti et al. 2014; Pasupuleti et al. 2018; Sule et al. 2017). Interestingly, responses to several of these compounds mediated by two major E. coli chemoreceptors, Tar and Tsr, are opposite (Lopes and Sourjik 2018; Yang et al. 2020). As discussed above, such opposing responses could lead to bacterial accumulation at an intermediate point within a gradient, which could be at a certain distance from gut epithelial surface, possibly enabling E. coli bacteria to escape antimicrobial activities of the mucous layer while remaining in proximity to the epithelium (Lopes and Sourjik 2018). Alternatively, a bimodal response could result in avoidance of intermediate concentrations, as appears to be the case for indole response of E. coli and was proposed to split bacteria into two subpopulations, one attracted towards the source of chemoeffector and the other repelled from it (Yang et al. 2020). Indeed, chemotaxis towards hormones or repulsion from indole was proposed to enhance attachment of E. coli to HeLa cells (Bansal et al. 2007; Bansal et al. 2008).
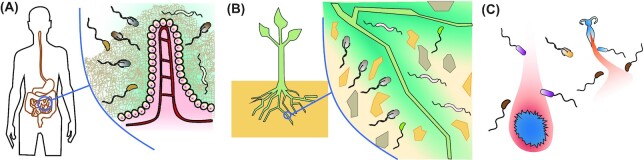
Figure 4.
Relevance of chemotactic motility in natural bacterial habitats. (A), In the gut, bacteria navigate in the lumen according to chemical gradients emanating from the epithelium. Motility is further used to penetrate the mucus layer that surrounds the epithelium. (B), In the rhizosphere, bacteria navigate through the complex structure of the soil and follow chemical gradients released by plant roots. (C), In the marine environment, bacteria follow chemical gradients released by planktonic and larger organisms and face turbulent flows which stir both the gradients and the cells as they swim.
The involvement of motility and chemotaxis in the establishment and maintenance of infection is well documented for a number of human pathogens, see (Matilla and Krell 2018) for a recent comprehensive review. The best studied examples of chemotaxis in motile pathogenic bacteria include Helicobacter pylori (Aihara et al. 2014; Collins et al. 2018; Hanyu et al. 2019; Huang et al. 2015; Johnson and Ottemann 2018; Perkins et al. 2019; Schweinitzer et al. 2008; Williams et al. 2007), Campylobacter jejuni (Elgamoudi et al. 2021; Khan et al. 2020; Korolik 2019; Li et al. 2014), S. Typhimurium (Olsen et al. 2013; Rivera-Chavez et al. 2016; Stecher et al. 2008; Stecher et al. 2004), Vibrio cholerae (Echazarreta and Klose 2019; Freter, O'Brien and Macsai 1981), and Pseudomonas aeruginosa (Corral-Lugo et al. 2018; Garvis et al. 2009; Martin-Mora et al. 2018; Matilla et al. 2021; Reyes-Darias et al. 2015; Rico-Jimenez et al. 2016; Schwarzer, Fischer and Machen 2016). All of these pathogens exhibit chemotactic responses to at least some of the common metabolites, such as amino acids, sugars and organic acids. This indicates that, similar to E. coli, their chemoeffector preferences could be at least partly explained by the role of chemotaxis in enhanced nutrient acquisition, although some of these metabolites might play double roles as nutrients and as cues released by the host. Among these pathogenic bacteria, the most specialized spectrum of chemoeffector metabolites is observed for H. pylori that infects gastric epithelium and is therefore adapted to a highly specific ecological niche (Johnson and Ottemann 2018), whereas the broadest spectrum of metabolites is recognized by the opportunistic and highly versatile pathogen P. aeruginosa (Matilla and Krell 2018; Ortega, Zhulin and Krell 2017). Nevertheless, many tactic responses of pathogens appear to primarily or exclusively serve as orientation cues within their animal hosts, enabling bacteria to locate sites of infection. These include taxis to urea, pH and bicarbonate by H. pylori (Cerda, Rivas and Toledo 2003; Huang et al. 2017; Huang et al. 2015), to deoxycholate by C. jejuni (Li et al. 2014), and to histamine, gamma-aminobutyrate (GABA) and inorganic phosphate by P. aeruginosa (Corral-Lugo et al. 2018; Reyes-Darias et al. 2015; Rico-Jimenez et al. 2016), as well as aero- and energy taxes exhibited by most pathogens (Behrens et al. 2013; Horne, Mattson and Pruss 2009; Rivera-Chavez et al. 2013; Vegge et al. 2009). It is worth noting that the ability of at least some bacterial pathogens to benefit from chemotaxis during infection depends on the environmental context. The role of chemotaxis in S. Typhimurium infection only becomes apparent with the advent of intestinal inflammation (Stecher et al. 2008) and V. cholerae strains lacking particular chemotaxis gene clusters show higher fitness compared to the wild type cells in the proximal small intestine (Butler and Camilli 2004; Millet et al. 2014). Given that much of the complexity of metabolic interactions and chemical communication between animal hosts and their microbiota remains to be uncovered, the aforementioned host-specific tactic responses likely represent only a fraction of cues and signals perceived by pathogenic as well as by non-pathogenic motile bacteria inhabiting animal guts.
Interactions in the rhizosphere and in aquatic environments
With its porous structure and variable water and nutrient content, soil represents a much more heterogeneous and complex environment. Since it typically sustains less fluid flow and has a lower density of microorganisms compared to the animal GI tract, soil can support stable long-range gradients of nutrients and signaling molecules. Together with the fact that soil is typically also more nutrient-poor, it is therefore not surprising that swimming motility and chemotaxis are common among soil bacteria (Fig. 4B). Motility seems to play a particularly important role in the rhizosphere, the region of the soil immediately surrounding plant roots that is enriched in root exudates and active microbial communities. Accordingly, plant pathogens and symbionts are nearly all motile and possess on average nearly twice as many chemoreceptors as motile animal/human pathogens (Lacal et al. 2010; Matilla and Krell 2018; Scharf, Hynes and Alexandre 2016). Chemotaxis in the rhizosphere can indeed enhance both symbiotic and pathogenic interactions of bacteria with their plant hosts through chemotaxis towards a variety of root- or leaf-secreted chemicals, including organic acids, carbohydrates, sugar alcohols, amino acids and plant hormones, for recent reviews see (Matilla and Krell 2018; Scharf, Hynes and Alexandre 2016). Flavonoids, phenolic compounds secreted by plant roots, have also been proposed to serve as specific chemoattractants (Dharmatilake and Bauer 1992), although these findings were recently questioned (Compton et al. 2020).
The importance of chemotaxis towards root exudates and specific compounds for colonization is well documented in several host–symbiont systems, most prominently Sinorhizobium meliloti (Caetano-Anolles, Crist-Estes and Bauer 1988; Caetano-Anolles, Wrobel-Boerner and Bauer 1992; Gulash et al. 1984), Rhizobium leguminosarum (Miller et al. 2007; Yost, Rochepeau and Hynes 1998) and A. brasilense (Greer-Phillips, Stephens and Alexandre 2004; O'Neal, Vo and Alexandre 2020). Chemotaxis has also been reported to enhance infection by several plant pathogens (Matilla and Krell 2018). Several studied examples of virulence-related chemotactic responses include Pseudomonas syringae pathovars (Cerna-Vargas et al. 2019; Melotto et al. 2006), Ralstonia solanacearum (Corral et al. 2020; Tans-Kersten, Huang and Allen 2001; Yao and Allen 2007) and Dickeya dadantii (Antunez-Lamas et al. 2009; Rio-Alvarez et al. 2015). Similar to animal pathogens, chemotaxis not only enables general attraction of these plant pathogens towards their plant hosts, but it also helps them to localize their preferred sites of infection, such as wounds or open stomata. More generally, other potentially beneficial or opportunistically pathogenic bacterial species in the rhizosphere, such as various Pseudomonas and Bacillus species use chemotaxis to accumulate towards the nutrient-rich environment around plant roots (Feng et al. 2018; Lopez-Farfan et al. 2019; Massalha et al. 2017). Moreover, it is assumed that chemotaxis towards a specific set of chemicals might enable host plant selection, but how exactly such specific recognition could be achieved remains to be determined.
Similar to these host-microbe interactions in the rhizosphere, motility and chemotaxis might be generally beneficial for the establishment of symbiotic communities and nutrient acquisition by bacteria living in aquatic environments (Fig. 4C). Indeed, active migration and colonization might aid transmission of microbial symbionts between hosts, and the importance of motility has been demonstrated for colonization of several marine animals by their bacterial symbionts (Bright and Bulgheresi 2010; Raina et al. 2019). For instance, in the squid-vibrio symbiosis system, Vibrio fischeri follows a chitin gradient through ducts and antechambers and actively migrates toward the pores of the light organ in a corkscrew-like motion (Aschtgen et al. 2019; Mandel et al. 2012). Other squid and cuttlefish species have similar symbiotic consortia composed of bacterial genera like Roseobacter, Pseudoalteromonas, Vibrio and Shewanella, which are known for their motility and chemotaxis (Barbieri et al. 2001), and chemotaxis might help these bacteria to colonize their hosts and to counter intestinal flow (Wiles et al. 2020). Chemotaxis towards planktonic chitin is also thought to be an important component of the ecology of chitin-degrading Vibrio species (Erken, Lutz and McDougald 2015). Marine phytoplankton were also shown to secret a range of compounds including dimethylsulfoniopropionate (DMSP), amino acids, sugars and organic acids that attract chemotactic bacterial symbionts (Miller and Belas 2006; Sonnenschein et al. 2012; Tout et al. 2017). Similarly, the marine macroalga Ulva mutabilis (Chlorophyta) releases DMSP to attract chemotactic marine bacteria (Kessler et al. 2018).
CONCLUDING REMARKS
Although motility is among the most studied bacterial behaviors under defined laboratory conditions, its multifaceted importance for the physiology of individual bacteria and microbial communities only recently became appreciated. In this review, we provided an overview of multiple functions of motility, with a primary focus on chemotaxis. As evident from studies of the E. coli model, even for the same species chemotaxis might make multiple contributions to physiology, including nutrient acquisition, expansion of the population range, biofilm formation and host colonization. Importantly, these different functions of motility and chemotaxis are not mutually exclusive but context-dependent. Even a single E. coli chemoreceptor Tsr can mediate chemotaxis to the preferentially consumed amino acid serine, to bacterial signaling molecules AI-2 and indole, and to animal hormones (Hegde et al. 2011; Lopes and Sourjik 2018; Mesibov and Adler 1972; Orr et al. 2020; Yang et al. 2020). Consequently, knockouts of individual receptors frequently show pleiotropic defects, from reduced growth fitness under conditions where chemotaxis is important to reduced biofilm formation and virulence. Moreover, the deletion of general chemotaxis genes not only impairs chemotaxis but also changes the swimming pattern of bacteria, either making them smooth swimming or tumbly, which can affect surface attachment, collective behaviors or spreading even in absence of specific chemotactic responses. These intertwined effects complicate the mechanistic understanding of the observed impacts of chemotaxis and motility in such complex environments as the rhizosphere or the GI tract, where grains, surfaces or the mucus affect swimming patterns (de Anna et al. 2020; Figueroa-Morales et al. 2019; Frangipane et al. 2019; Galajda et al. 2007; Makarchuk et al. 2019; Sipos et al. 2015; Spagnolie et al. 2015), and need to be kept in mind while interpreting such data.
Another outstanding challenge in understanding the physiological and environmental importance of bacterial chemotaxis lies in the characterization of ligand specificity for the many chemotaxis receptors present in different bacterial species. Whereas signaling domains of receptors are highly conserved and can be easily identified bioinformatically, their ligand binding domains and corresponding sensing mechanisms are highly diverse (Ortega, Zhulin and Krell 2017). Although several different approaches to systematically identify ligands for various chemoreceptors have been recently established (Bi et al. 2016; Boyeldieu et al. 2021; Lehning et al. 2017; Luu et al. 2019; Matilla, Martin-Mora and Krell 2020), only a tiny fraction of chemoreceptor ligands are currently known and even fewer have an established mode of binding (Ortega, Zhulin and Krell 2017). With the increasing number of characterized ligand-receptor interactions and better understanding of ligand binding by the major structural classes of ligand-binding domains of receptors, computational prediction of ligand specificity should ultimately become possible.
Contributor Information
Remy Colin, Max Planck Institute for Terrestrial Microbiology & Center for Synthetic Microbiology (SYNMIKRO), Karl-von-Frisch Strasse 16, Marburg D-35043, Germany.
Bin Ni, Max Planck Institute for Terrestrial Microbiology & Center for Synthetic Microbiology (SYNMIKRO), Karl-von-Frisch Strasse 16, Marburg D-35043, Germany; College of Resources and Environmental Science, National Academy of Agriculture Green Development, China Agricultural University, Yuanmingyuan Xilu No. 2, Beijing 100193, China.
Leanid Laganenka, Institute of Microbiology, D-BIOL, ETH Zürich, Vladimir-Prelog-Weg 4, Zürich 8093, Switzerland.
Victor Sourjik, Max Planck Institute for Terrestrial Microbiology & Center for Synthetic Microbiology (SYNMIKRO), Karl-von-Frisch Strasse 16, Marburg D-35043, Germany.
FUNDING
This work was supported by the Max Planck Gesellschaft and the Deutsche Forschungsgemeinschaft Grants CO1813/2-1 to RC and LA 4572/1-1 to LL.
Conflicts of interest
The authors declare no conflict of interest.
REFERENCES
- Adler J, Hazelbauer GL, Dahl MM. Chemotaxis toward sugars in Escherichia coli. J Bacteriol. 1973;115:824–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adler J, Templeton B. The effect of environmental conditions on the motility of Escherichia coli. J Gen Microbiol. 1967;46:175–84. [DOI] [PubMed] [Google Scholar]
- Adler J. Chemotaxis in bacteria. Science. 1966;153:708–16. [DOI] [PubMed] [Google Scholar]
- Ahmed T, Shimizu TS, Stocker R. Bacterial chemotaxis in linear and nonlinear steady microfluidic gradients. Nano Lett. 2010;10:3379–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aihara E, Closson C, Matthis ALet al. Motility and chemotaxis mediate the preferential colonization of gastric injury sites by Helicobacter pylori. PLoS Pathog. 2014;10:e1004275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander RP, Lowenthal AC, Harshey RMet al. CheV: CheW-like coupling proteins at the core of the chemotaxis signaling network. Trends Microbiol. 2010;18:494–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexandre G, Greer SE, Zhulin IB. Energy taxis is the dominant behavior in Azospirillum brasilense. J Bacteriol. 2000;182:6042–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexandre G, Greer-Phillips S, Zhulin IB. Ecological role of energy taxis in microorganisms. FEMS Microbiol Rev. 2004;28:113–26. [DOI] [PubMed] [Google Scholar]
- Altindal T, Xie L, Wu XL. Implications of three-step swimming patterns in bacterial chemotaxis. Biophys J. 2011;100:32–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amsler CD, Cho M, Matsumura P. Multiple factors underlying the maximum motility of Escherichia coli as cultures enter post-exponential growth. J Bacteriol. 1993;175:6238–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson JK, Huang JY, Wreden Cet al. Chemorepulsion from the quorum signal autoinducer-2 promotes Helicobacter pylori biofilm dispersal. mBio. 2015;6:e00379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antunez-Lamas M, Cabrera E, Lopez-Solanilla Eet al. Bacterial chemoattraction towards jasmonate plays a role in the entry of Dickeya dadantii through wounded tissues. Mol Microbiol. 2009;74:662–71. [DOI] [PubMed] [Google Scholar]
- Aquino G, Clausznitzer D, Tollis Set al. Optimal receptor-cluster size determined by intrinsic and extrinsic noise. Phys Rev E. 2011;83:021914. [DOI] [PubMed] [Google Scholar]
- Ardre M, Henry H, Douarche Cet al. An individual-based model for biofilm formation at liquid surfaces. Phys Biol. 2015;12:066015. [DOI] [PubMed] [Google Scholar]
- Ariel G, Sidortsov M, Ryan SDet al. Collective dynamics of two-dimensional swimming bacteria: experiments and models. Phys Rev E. 2018;98:032415. [Google Scholar]
- Arora SK, Ritchings BW, Almira ECet al. The Pseudomonas aeruginosa flagellar cap protein, FliD, is responsible for mucin adhesion. Infect Immun. 1998;66:1000–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aschtgen MS, Brennan CA, Nikolakakis Ket al. Insights into flagellar function and mechanism from the squid-Vibrio symbiosis. Npj Biofilms Microbiomes. 2019;5:32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baban ST, Kuehne SA, Barketi-Klai Aet al. The role of flagella in Clostridium difficile pathogenesis: comparison between a non-epidemic and an epidemic strain. PLoS One. 2013;8:e73026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bansal T, Englert D, Lee Jet al. Differential effects of epinephrine, norepinephrine, and indole on Escherichia coli O157:H7 chemotaxis, colonization, and gene expression. Infect Immun. 2007;75:4597–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bansal T, Jesudhasan P, Pillai Set al. Temporal regulation of enterohemorrhagic Escherichia coli virulence mediated by autoinducer-2. Appl Microbiol Biotechnol. 2008;78:811–9. [DOI] [PubMed] [Google Scholar]
- Barbieri E, Paster BJ, Hughes Det al. Phylogenetic characterization of epibiotic bacteria in the accessory nidamental gland and egg capsules of the squid Loligo pealei (Cephalopoda:loliginidae). Environ Microbiol. 2001;3:151–67. [DOI] [PubMed] [Google Scholar]
- Barkai N, Leibler S. Robustness in simple biochemical networks. Nature. 1997;387:913–7. [DOI] [PubMed] [Google Scholar]
- Be'er A, Ariel G. A statistical physics view of swarming bacteria. Mov Ecol. 2019;7:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bearon RN, Hazel AL. The trapping in high-shear regions of slender bacteria undergoing chemotaxis in a channel. J Fluid Mech. 2015;771:R3. [Google Scholar]
- Bearon RN, Pedley TJ. Modelling run-and-tumble chemotaxis in a shear flow. B Math Biol. 2000;62:775–91. [DOI] [PubMed] [Google Scholar]
- Behrens W, Schweinitzer T, Bal Jet al. Role of energy sensor TlpD of Helicobacter pylori in gerbil colonization and genome analyses after adaptation in the gerbil. Infect Immun. 2013;81:3534–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benichou O, Loverdo C, Moreau Met al. Intermittent search strategies. Rev Mod Phys. 2011;83:81. [DOI] [PubMed] [Google Scholar]
- Berg HC, Brown DA. Chemotaxis in Escherichia coli analyzed by 3-dimensional tracking. Nature. 1972;239:500. [DOI] [PubMed] [Google Scholar]
- Berg HC, Purcell EM. Physics of chemoreception. Biophys J. 1977;20:193–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg HC. Bacterial behaviour. Nature. 1975;254:389–92. [DOI] [PubMed] [Google Scholar]
- Berg HC. The rotary motor of bacterial flagella. Annu Rev Biochem. 2003;72:19–54. [DOI] [PubMed] [Google Scholar]
- Bergstrom KS, Kissoon-Singh V, Gibson DLet al. Muc2 protects against lethal infectious colitis by disassociating pathogenic and commensal bacteria from the colonic mucosa. PLoS Pathog. 2010;6:e1000902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berke AP, Turner L, Berg HCet al. Hydrodynamic attraction of swimming microorganisms by surfaces. Phys Rev Lett. 2008;101:038102. [DOI] [PubMed] [Google Scholar]
- Besharova O, Suchanek VM, Hartmann Ret al. Diversification of gene expression during formation of static submerged biofilms by Escherichia coli. Front Microbiol. 2016;7:1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bettenworth V, Steinfeld B, Duin Het al. Phenotypic heterogeneity in bacterial quorum sensing systems. J Mol Biol. 2019;431:4530–46. [DOI] [PubMed] [Google Scholar]
- Bi S, Pollard AM, Yang Yet al. Engineering hybrid chemotaxis receptors in bacteria. ACS Synth Biol. 2016;5:989–1001. [DOI] [PubMed] [Google Scholar]
- Bi S, Sourjik V. Stimulus sensing and signal processing in bacterial chemotaxis. Curr Opin Microbiol. 2018;45:22–9. [DOI] [PubMed] [Google Scholar]
- Bialek W, Setayeshgar S. Physical limits to biochemical signaling. Proc Natl Acad Sci USA. 2005;102:10040–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bible AN, Stephens BB, Ortega DRet al. Function of a chemotaxis-like signal transduction pathway in modulating motility, cell clumping, and cell length in the alphaproteobacterium Azospirillum brasilense. J Bacteriol. 2008;190:6365–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackburn N, Fenchel T. Influence of bacteria, diffusion and sheer on micro-scale nutrient patches, and implications for bacterial chemotaxis. Mar Ecol Prog Ser. 1999;189:1–7. [Google Scholar]
- Blakemore RP, Frankel RB, Kalmijn AJ. South-seeking magnetotactic bacteria in the southern-hemisphere. Nature. 1980;286:384–5. [Google Scholar]
- Blakemore RP. Magnetotactic bacteria. Annu Rev Microbiol. 1982;36:217–38. [DOI] [PubMed] [Google Scholar]
- Block SM, Segall JE, Berg HC. Adaptation kinetics in bacterial chemotaxis. J Bacteriol. 1983;154:312–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehm A, Kaiser M, Li Het al. Second messenger-mediated adjustment of bacterial swimming velocity. Cell. 2010;141:107–16. [DOI] [PubMed] [Google Scholar]
- Boyeldieu A, Ali Chaouche A, Mejean Vet al. Combining two optimized and affordable methods to assign chemoreceptors to a specific signal. Anal Biochem. 2021;620:114139. [DOI] [PubMed] [Google Scholar]
- Bright M, Bulgheresi S. A complex journey: transmission of microbial symbionts. Nat Rev Microbiol. 2010;8:218–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brumley DR, Carrara F, Hein AMet al. Bacteria push the limits of chemotactic precision to navigate dynamic chemical gradients. Proc Natl Acad Sci USA. 2019;116:10792–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brumley DR, Carrara F, Hein AMet al. Cutting through the noise: bacterial chemotaxis in marine microenvironments. Front Mar Sci. 2020;7:527. [Google Scholar]
- Budrene EO, Berg HC. Complex patterns formed by motile cells of Escherichia coli. Nature. 1991;349:630–3. [DOI] [PubMed] [Google Scholar]
- Budrene EO, Berg HC. Dynamics of formation of symmetrical patterns by chemotactic bacteria. Nature. 1995;376:49–53. [DOI] [PubMed] [Google Scholar]
- Burkart M, Toguchi A, Harshey RM. The chemotaxis system, but not chemotaxis, is essential for swarming motility in Escherichia coli. Proc Natl Acad Sci USA. 1998;95:2568–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler SM, Camilli A. Both chemotaxis and net motility greatly influence the infectivity of Vibrio cholerae. Proc Natl Acad Sci USA. 2004;101:5018–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caetano-Anolles G, Crist-Estes DK, Bauer WD. Chemotaxis of Rhizobium meliloti to the plant flavone luteolin requires functional nodulation genes. J Bacteriol. 1988;170:3164–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caetano-Anolles G, Wrobel-Boerner E, Bauer WD. Growth and movement of spot inoculated Rhizobium meliloti on the root surface of Alfalfa. Plant Physiol. 1992;98:1181–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cates ME. Diffusive transport without detailed balance in motile bacteria: does microbiology need statistical physics?. Rep Prog Phys. 2012;75:042601. [DOI] [PubMed] [Google Scholar]
- Celani A, Vergassola M. Bacterial strategies for chemotaxis response. Proc Natl Acad Sci USA. 2010;107:1391–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celli JP, Turner BS, Afdhal NHet al. Helicobacter pylori moves through mucus by reducing mucin viscoelasticity. Proc Natl Acad Sci USA. 2009;106:14321–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerda O, Rivas A, Toledo H. Helicobacter pylori strain ATCC700392 encodes a methyl-accepting chemotaxis receptor protein (MCP) for arginine and sodium bicarbonate. Fems Microbiol Lett. 2003;224:175–81. [DOI] [PubMed] [Google Scholar]
- Cerna-Vargas JP, Santamaria-Hernando S, Matilla MAet al. Chemoperception of specific amino acids controls phytopathogenicity in Pseudomonas syringae pv. tomato. mBio. 2019;10:e01868–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaban B, Hughes HV, Beeby M. The flagellum in bacterial pathogens: for motility and a whole lot more. Semin Cell Dev Biol. 2015;46:91–103. [DOI] [PubMed] [Google Scholar]
- Chen C, Liu S, Shi XQet al. Weak synchronization and large-scale collective oscillation in dense bacterial suspensions. Nature. 2017;542:210–4. [DOI] [PubMed] [Google Scholar]
- Chevance FF, Hughes KT. Coordinating assembly of a bacterial macromolecular machine. Nat Rev Microbiol. 2008;6:455–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cisneros LH, Cortez R, Dombrowski Cet al. Fluid dynamics of self-propelled microorganisms, from individuals to concentrated populations. Exp Fluids. 2007;43:737–53. [Google Scholar]
- Clark DA, Grant LC. The bacterial chemotactic response reflects a compromise between transient and steady-state behavior. Proc Natl Acad Sci USA. 2005;102:9150–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clausznitzer D, Micali G, Neumann Set al. Predicting chemical environments of bacteria from receptor signaling. Plos Comput Biol. 2014;10:e1003870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colin R, Drescher K, Sourjik V. Chemotactic behavior of Escherichia coli at high cell density. Nat Commun. 2019;10:5329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colin R, Rosazza C, Vaknin Aet al. Multiple sources of slow activity fluctuations in a bacterial chemosensory network. Elife. 2017;6:e26796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colin R, Sourjik V. Emergent properties of bacterial chemotaxis pathway. Curr Opin Microbiol. 2017;39:24–33. [DOI] [PubMed] [Google Scholar]
- Colin R, Zhang R, Wilson LG. Fast, high-throughput measurement of collective behaviour in a bacterial population. J R Soc Interface. 2014;11:0486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins KD, Hu S, Grasberger Het al. Chemotaxis allows bacteria to overcome host-generated reactive oxygen species that constrain gland colonization. Infect Immun. 2018;86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Compton KK, Hildreth SB, Helm RFet al. An updated perspective on Sinorhizobium meliloti chemotaxis to alfalfa flavonoids. Front Microbiol. 2020;11:581482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Constantino MA, Jabbarzadeh M, Fu HCet al. Bipolar lophotrichous Helicobacter suis combine extended and wrapped flagella bundles to exhibit multiple modes of motility. Sci Rep. 2018;8:14415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corral J, Sebastia P, Coll NSet al. Twitching and swimming motility play a role in Ralstonia solanacearum pathogenicity. mSphere. 2020;5:e00740–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corral-Lugo A, Matilla MA, Martin-Mora Det al. High-affinity chemotaxis to histamine mediated by the TlpQ chemoreceptor of the human pathogen Pseudomonas aeruginosa. mBio. 2018;9:e01894–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cremer J, Honda T, Tang Yet al. Chemotaxis as a navigation strategy to boost range expansion. Nature. 2019;575:658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curatolo AI, Zhou N, Zhao Yet al. Cooperative pattern formation in multi-component bacterial systems through reciprocal motility regulation. Nat Phys. 2020;16:1152. [Google Scholar]
- Darnton NC, Turner L, Rojevsky Set al. Dynamics of bacterial swarming. Biophys J. 2010;98:2082–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Anna P, Pahlavan AA, Yawata Yet al. Chemotaxis under flow disorder shapes microbial dispersion in porous media. Nat Phys. 2020;17:68–73. [Google Scholar]
- Defoirdt T. Can bacteria actively search to join groups?. ISME J. 2011;5:569–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dharmatilake AJ, Bauer WD. Chemotaxis of Rhizobium meliloti towards nodulation gene-inducing compounds from alfalfa roots. Appl Environ Microbiol. 1992;58:1153–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dufour YS, Fu X, Hernandez-Nunez Let al. Limits of feedback control in bacterial chemotaxis. Plos Comput Biol. 2014;10:e1003694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dufour YS, Gillet S, Frankel NWet al. Direct correlation between motile behavior and protein abundance in single cells. Plos Comput Biol. 2016;12:e1005041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunkel J, Heidenreich S, Drescher Ket al. Fluid dynamics of bacterial turbulence. Phys Rev Lett. 2013;110:228102. [DOI] [PubMed] [Google Scholar]
- Echazarreta MA, Klose KE. Vibrio flagellar synthesis. Front Cell Infect Microbiol. 2019;9:131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elgamoudi BA, Andrianova EP, Shewell LKet al. The Campylobacter jejuni chemoreceptor Tlp10 has a bimodal ligand-binding domain and specificity for multiple classes of chemoeffectors. Sci Signal. 2021;14:eabc8521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elgeti J, Gompper G. Wall accumulation of self-propelled spheres. Europhys Lett. 2013;101:48003. [Google Scholar]
- Englert DL, Manson MD, Jayaraman A. Investigation of bacterial chemotaxis in flow-based microfluidic devices. Nat Protoc. 2010;5:864–72. [DOI] [PubMed] [Google Scholar]
- Erhardt M. Strategies to block bacterial pathogenesis by interference with motility and chemotaxis. Curr Top Microbiol Immunol. 2016;398:185–205. [DOI] [PubMed] [Google Scholar]
- Erken M, Lutz C, McDougald D. Interactions of Vibrio spp. with zooplankton. Microbiol Spectr. 2015;3:VE–0003-2014. [DOI] [PubMed] [Google Scholar]
- Faivre D, Schuler D. Magnetotactic bacteria and magnetosomes. Chem Rev. 2008;108:4875–98. [DOI] [PubMed] [Google Scholar]
- Fan Y, Pedersen O. Gut microbiota in human metabolic health and disease. Nat Rev Microbiol. 2021;19:55–71. [DOI] [PubMed] [Google Scholar]
- Feng HC, Zhang N, Du WBet al. Identification of chemotaxis compounds in root exudates and their sensing chemoreceptors in plant-growth-promoting rhizobacteria Bacillus amyloliquefaciens SQR9. Mol Plant Microbe In. 2018;31:995–1005. [DOI] [PubMed] [Google Scholar]
- Ferenci T. Trade-off mechanisms shaping the diversity of bacteria. Trends Microbiol. 2016;24:209–23. [DOI] [PubMed] [Google Scholar]
- Fernandez M, Matilla MA, Ortega Aet al. Metabolic value chemoattractants are preferentially recognized at broad ligand range chemoreceptor of Pseudomonas putida KT2440. Front Microbiol. 2017;8:990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueroa-Morales N, Dominguez-Rubio L, Ott TLet al. Mechanical shear controls bacterial penetration in mucus. Sci Rep. 2019;9:9713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores M, Shimizu TS, ten Wolde PRet al. Signaling noise enhances chemotactic drift of E. coli. Phys Rev Lett. 2012;109:148101. [DOI] [PubMed] [Google Scholar]
- Ford KM, Antani JD, Nagarajan Aet al. Switching and torque generation in swarming E. coli. Front Microbiol. 2018;9:2197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraebel DT, Mickalide H, Schnitkey Det al. Environment determines evolutionary trajectory in a constrained phenotypic space. Elife. 2017;6:e24669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frangipane G, Vizsnyiczai G, Maggi Cet al. Invariance properties of bacterial random walks in complex structures. Nat Commun. 2019;10:2442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frankel NW, Pontius W, Dufour YSet al. Adaptability of non-genetic diversity in bacterial chemotaxis. Elife. 2014;3:e03526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freter R, O'Brien PC, Macsai MS. Role of chemotaxis in the association of motile bacteria with intestinal mucosa: in vivo studies. Infect Immun. 1981;34:234–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedlander RS, Vogel N, Aizenberg J. Role of flagella in adhesion of Escherichia coli to abiotic surfaces. Langmuir. 2015;31:6137–44. [DOI] [PubMed] [Google Scholar]
- Fu X, Kato S, Long Jet al. Spatial self-organization resolves conflicts between individuality and collective migration. Nat Commun. 2018;9:2177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furter M, Sellin ME, Hansson GCet al. Mucus architecture and near-surface swimming affect distinct Salmonella Typhimurium infection patterns along the murine intestinal tract. Cell Rep. 2019;27:2665–78 e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galajda P, Keymer J, Chaikin Pet al. A wall of funnels concentrates swimming bacteria. J Bacteriol. 2007;189:8704–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garvis S, Munder A, Ball Get al. Caenorhabditis elegans semi-automated liquid screen reveals a specialized role for the chemotaxis gene cheB2 in Pseudomonas aeruginosa virulence. PLoS Pathog. 2009;5:e1000540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govern CC, Ten Wolde PR. Optimal resource allocation in cellular sensing systems. Proc Natl Acad Sci USA. 2014;111:17486–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goy MF, Springer MS, Adler J. Sensory transduction in Escherichia coli - Role of a protein methylation reaction in sensory adaptation. Proc Natl Acad Sci USA. 1977;74:4964–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grauer J, Lowen H, Be'er Aet al. Swarm hunting and cluster ejections in chemically communicating active mixtures. Sci Rep. 2020;10:5594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greer-Phillips SE, Stephens BB, Alexandre G. An energy taxis transducer promotes root colonization by Azospirillum brasilense. J Bacteriol. 2004;186:6595–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grognot M, Taute KM. More than propellers: how flagella shape bacterial motility behaviors. Curr Opin Microbiol. 2021;61:73–81. [DOI] [PubMed] [Google Scholar]
- Gude S, Pince E, Taute KMet al. Bacterial coexistence driven by motility and spatial competition. Nature. 2020;578:588–92. [DOI] [PubMed] [Google Scholar]
- Gulash M, Ames P, Larosiliere RCet al. Rhizobia are attracted to localized sites on legume roots. Appl Environ Microbiol. 1984;48:149–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gumerov VM, Andrianova EP, Zhulin IB. Diversity of bacterial chemosensory systems. Curr Opin Microbiol. 2021;61:42–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttenplan SB, Kearns DB. Regulation of flagellar motility during biofilm formation. FEMS Microbiol Rev. 2013;37:849–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzzo M, Murray SM, Martineau Eet al. A gated relaxation oscillator mediated by FrzX controls morphogenetic movements in Myxococcus xanthus. Nat Microbiol. 2018;3:948–59. [DOI] [PubMed] [Google Scholar]
- Hanyu H, Engevik KA, Matthis ALet al. Helicobacter pylori uses the TlpB receptor to sense sites of gastric injury. Infect Immun. 2019;87:e00202–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harshey RM. Bees arent the only ones - Swarming in gram-negative bacteria. Mol Microbiol. 1994;13:389–94. [DOI] [PubMed] [Google Scholar]
- Hegde M, Englert DL, Schrock Set al. Chemotaxis to the quorum-sensing signal AI-2 requires the Tsr chemoreceptor and the periplasmic LsrB AI-2-binding protein. J Bacteriol. 2011;193:768–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hein AM, Brumley DR, Carrara Fet al. Physical limits on bacterial navigation in dynamic environments. J R Soc Interface. 2016;13:20150844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hengge R. Principles of c-di-GMP signalling in bacteria. Nat Rev Microbiol. 2009;7:263–73. [DOI] [PubMed] [Google Scholar]
- Hindre T, Knibbe C, Beslon Get al. New insights into bacterial adaptation through in vivo and in silico experimental evolution. Nat Rev Microbiol. 2012;10:352–65. [DOI] [PubMed] [Google Scholar]
- Holscher T, Bartels B, Lin YCet al. Motility, chemotaxis and aerotaxis contribute to competitiveness during bacterial pellicle biofilm development. J Mol Biol. 2015;427:3695–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horne SM, Mattson KR, Pruss BM. An Escherichia coli aer mutant exhibits a reduced ability to colonize the streptomycin-treated mouse large intestine. Antonie Van Leeuwenhoek. 2009;95:149–58. [DOI] [PubMed] [Google Scholar]
- Hu B, Tu Y. Precision sensing by two opposing gradient sensors: how does Escherichia coli find its preferred pH level?. Biophys J. 2013;105:276–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang JY, Goers Sweeney E, Guillemin Ket al. Multiple acid sensors control Helicobacter pylori colonization of the stomach. PLoS Pathog. 2017;13:e1006118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang JY, Sweeney EG, Sigal Met al. Chemodetection and destruction of host urea allows Helicobacter pylori to locate the epithelium. Cell Host Microbe. 2015;18:147–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hugdahl MB, Beery JT, Doyle MP. Chemotactic behavior of Campylobacter jejuni. Infect Immun. 1988;56:1560–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hui S, Silverman JM, Chen SSet al. Quantitative proteomic analysis reveals a simple strategy of global resource allocation in bacteria. Molecular systems biology. 2015;11:784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilkanaiv B, Kearns DB, Ariel Get al. Effect of cell aspect ratio on swarming bacteria. Phys Rev Lett. 2017;118:158002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islam ST, Mignot T. The mysterious nature of bacterial surface (gliding) motility: a focal adhesion-based mechanism in Myxococcus xanthus. Semin Cell Dev Biol. 2015;46:143–54. [DOI] [PubMed] [Google Scholar]
- Ismail AS, Valastyan JS, Bassler BL. A host-produced autoinducer-2 mimic activates bacterial quorum sensing. Cell Host Microbe. 2016;19:470–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jani S, Seely AL, Peabody VGet al. Chemotaxis to self-generated AI-2 promotes biofilm formation in Escherichia coli. Microbiol. 2017;163:1778–90. [DOI] [PubMed] [Google Scholar]
- Jeckel H, Jelli E, Hartmann Ret al. Learning the space-time phase diagram of bacterial swarm expansion. Proc Natl Acad Sci USA. 2019;116:1489–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenal U, Malone J. Mechanisms of cyclic-di-GMP signaling in bacteria. Annu Rev Genet. 2006;40:385–407. [DOI] [PubMed] [Google Scholar]
- Jikeli JF, Alvarez L, Friedrich BMet al. Sperm navigation along helical paths in 3D chemoattractant landscapes. Nat Commun. 2015;6:7985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jing GY, Zottl A, Clement Eet al. Chirality-induced bacterial rheotaxis in bulk shear flows. Sci Adv. 2020;6:eabb2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson KS, Ottemann KM. Colonization, localization, and inflammation: the roles of H. pylori chemotaxis in vivo. Curr Opin Microbiol. 2018;41:51–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung K, Brameyer S, Fabiani Fet al. Phenotypic heterogeneity generated by histidine kinase-based signaling networks. J Mol Biol. 2019;431:4547–58. [DOI] [PubMed] [Google Scholar]
- Kalinin YV, Jiang L, Tu Yet al. Logarithmic sensing in Escherichia coli bacterial chemotaxis. Biophys J. 2009;96:2439–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamino K, Keegstra JM, Long Jet al. Adaptive tuning of cell sensory diversity without changes in gene expression. Sci Adv. 2020;6:eabc1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karin O, Alon U. Cell-Cell Variation In Chemotaxis Gain Implements A Simulated Tempering Strategy For Efficient Navigation In Complex Environments, available at SSNR: 10.2139/ssrn.3766494. 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearns DB, Losick R. Cell population heterogeneity during growth of Bacillus subtilis. Genes Dev. 2005;19:3083–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearns DB, Losick R. Swarming motility in undomesticated Bacillus subtilis. Mol Microbiol. 2003;49:581–90. [DOI] [PubMed] [Google Scholar]
- Kearns DB, Robinson J, Shimkets LJ. Pseudomonas aeruginosa exhibits directed twitching motility up phosphatidylethanolamine gradients. J Bacteriol. 2001;183:763–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearns DB. A field guide to bacterial swarming motility. Nature Reviews Microbiology. 2010;8:634–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keegstra JM, Kamino K, Anquez Fet al. Phenotypic diversity and temporal variability in a bacterial signaling network revealed by single-cell FRET. Elife. 2017;6:e27455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kehry MR, Dahlquist FW. The methyl-accepting chemotaxis proteins of Escherichia coli - identification of the multiple methylation sites on methyl-accepting chemotaxis protein-I. J Biol Chem. 1982;257:378–86. [PubMed] [Google Scholar]
- Keller EF, Segel LA. Model for chemotaxis. J Theor Biol. 1971;30:225–34. [DOI] [PubMed] [Google Scholar]
- Kessler RW, Weiss A, Kuegler Set al. Macroalgal-bacterial interactions: role of dimethylsulfoniopropionate in microbial gardening by Ulva (Chlorophyta). Mol Ecol. 2018;27:1808–19. [DOI] [PubMed] [Google Scholar]
- Khan MF, Machuca MA, Rahman MMet al. Structure-activity relationship study reveals the molecular basis for specific sensing of hydrophobic amino acids by the Campylobacter jejuni chemoreceptor Tlp3. Biomolecules. 2020;10:744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch DL, Subramanian G. Collective hydrodynamics of swimming microorganisms: living fluids. Annu Rev Fluid Mech. 2011;43:637–59. [Google Scholar]
- Koirala S, Mears P, Sim Met al. A nutrient-tunable bistable switch controls motility in Salmonella enterica serovar Typhimurium. mBio. 2014;5:e01611–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korobkova E, Emonet T, Vilar JMGet al. From molecular noise to behavioural variability in a single bacterium. Nature. 2004;428:574–8. [DOI] [PubMed] [Google Scholar]
- Korolik V. The role of chemotaxis during Campylobacter jejuni colonisation and pathogenesis. Curr Opin Microbiol. 2019;47:32–7. [DOI] [PubMed] [Google Scholar]
- Koster DA, Mayo A, Bren Aet al. Surface growth of a motile bacterial population resembles growth in a chemostat. J Mol Biol. 2012;424:180–91. [DOI] [PubMed] [Google Scholar]
- Krell T, Lacal J, Reyes-Darias JAet al. Bioavailability of pollutants and chemotaxis. Curr Opin Biotechnol. 2013;24:451–6. [DOI] [PubMed] [Google Scholar]
- Lacal J, Garcia-Fontana C, Munoz-Martinez Fet al. Sensing of environmental signals: classification of chemoreceptors according to the size of their ligand binding regions. Environ Microbiol. 2010;12:2873–84. [DOI] [PubMed] [Google Scholar]
- Laganenka L, Colin R, Sourjik V.. Chemotaxis towards autoinducer 2 mediates autoaggregation in Escherichia coli. Nat Commun. 2016;7:12984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laganenka L, Lopez ME, Colin Ret al. Flagellum-mediated mechanosensing and RflP control motility state of pathogenic Escherichia coli. mBio. 2020;11:e02269–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laganenka L, Sourjik V.. Autoinducer 2-dependent Escherichia coli biofilm formation is enhanced in a dual-species co-culture. Appl Environ Microbiol. 2017;84:e02638–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb E, Trimble MJ, McCarter LL. Cell-cell communication, chemotaxis and recruitment in Vibrio parahaemolyticus. Mol Microbiol. 2019;112:99–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan G, Sartori P, Neumann Set al. The energy-speed-accuracy tradeoff in sensory adaptation. Nat Phys. 2012;8:422–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane MC, Lockatell V, Monterosso Get al. Role of motility in the colonization of uropathogenic Escherichia coli in the urinary tract. Infect Immun. 2005;73:7644–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen SH, Reader RW, Kort ENet al. Change in direction of flagellar rotation is basis of chemotactic response in Escherichia coli. Nature. 1974;249:74–7. [DOI] [PubMed] [Google Scholar]
- Lazova MD, Ahmed T, Bellomo Det al. Response rescaling in bacterial chemotaxis. Proc Natl Acad Sci USA. 2011;108:13870–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefevre CT, Bazylinski DA. Ecology, diversity, and evolution of magnetotactic bacteria. Microbiol Mol Biol Rev. 2013;77:497–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehning CE, Heidelberger JB, Reinhard Jet al. A modular high-throughput in vivo screening platform based on chimeric bacterial receptors. ACS Synth Biol. 2017;6:1315–26. [DOI] [PubMed] [Google Scholar]
- Li GL, Bensson J, Nisimova Let al. Accumulation of swimming bacteria near a solid surface. Phys Rev E. 2011;84:041932. [DOI] [PubMed] [Google Scholar]
- Li GL, Tang JX. Accumulation of microswimmers near a surface mediated by collision and rotational Brownian motion. Phys Rev Lett. 2009;103:078101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Zhai H, Sanchez Set al. Noncontact cohesive swimming of bacteria in two-dimensional liquid films. Phys Rev Lett. 2017;119:018101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Lou H, Ojcius DMet al. Methyl-accepting chemotaxis proteins 3 and 4 are responsible for Campylobacter jejuni chemotaxis and jejuna colonization in mice in response to sodium deoxycholate. J Med Microbiol. 2014;63:343–54. [DOI] [PubMed] [Google Scholar]
- Liu JK, Deng GH, Yuan ZTet al. Turbulence under the microscope. J Biol Phys. 2000;26:77–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M, Durfee T, Cabrera JEet al. Global transcriptional programs reveal a carbon source foraging strategy by Escherichia coli. J Biol Chem. 2005;280:15921–7. [DOI] [PubMed] [Google Scholar]
- Liu WR, Cremer J, Li DJet al. An evolutionarily stable strategy to colonize spatially extended habitats. Nature. 2019;575:664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locsei JT, Pedley TJ. Run and tumble chemotaxis in a shear flow: the effect of temporal comparisons, persistence, rotational diffusion, and cell shape. B Math Biol. 2009;71:1089–116. [DOI] [PubMed] [Google Scholar]
- Long J, Zucker SW, Emonet T. Feedback between motion and sensation provides nonlinear boost in run-and-tumble navigation. Plos Comput Biol. 2017;13:e1005429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopes JG, Sourjik V.. Chemotaxis of Escherichia coli to major hormones and polyamines present in human gut. ISME J. 2018;12:2736–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Farfan D, Reyes-Darias JA, Matilla MAet al. Concentration dependent effect of plant root exudates on the chemosensory systems of Pseudomonas putida KT2440. Front Microbiol. 2019;10:78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luchsinger RH, Bergersen B, Mitchell JG. Bacterial swimming strategies and turbulence. Biophys J. 1999;77:2377–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luu RA, Schomer RA, Brunton CNet al. Hybrid two-component sensors for identification of bacterial chemoreceptor function. Appl Environ Microbiol. 2019;85:e01626–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macnab RM, Koshland DE. Gradient-sensing mechanism in bacterial chemotaxis. Proc Natl Acad Sci USA. 1972;69:2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macnab RM. Bacterial flagella rotating in bundles: a study in helical geometry. Proc Natl Acad Sci USA. 1977;74:221–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarchuk S, Braz VC, Araujo NAMet al. Enhanced propagation of motile bacteria on surfaces due to forward scattering. Nat Commun. 2019;10:4110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandel MJ, Schaefer AL, Brennan CAet al. Squid-derived chitin oligosaccharides are a chemotactic signal during colonization by Vibrio fischeri. Appl Environ Microbiol. 2012;78:4620–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao XG, Egli R, Petersen Net al. Magneto-chemotaxis in sediment: first insights. Plos One. 2014;9:e102810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcos Fu HC, Powers TRet al. Bacterial rheotaxis. Proc Natl Acad Sci USA. 2012;109:4780–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariconda S, Wang QF, Harshey RM. A mechanical role for the chemotaxis system in swarming motility. Mol Microbiol. 2006;60:1590–602. [DOI] [PubMed] [Google Scholar]
- Martin-Mora D, Ortega A, Perez-Maldonado FJet al. The activity of the C4-dicarboxylic acid chemoreceptor of Pseudomonas aeruginosa is controlled by chemoattractants and antagonists. Sci Rep. 2018;8:2102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massalha H, Korenblum E, Malitsky Set al. Live imaging of root-bacteria interactions in a microfluidics setup. Proc Natl Acad Sci USA. 2017;114:4549–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matilla MA, Krell T. The effect of bacterial chemotaxis on host infection and pathogenicity. FEMS Microbiol Rev. 2018;42:fux052. [DOI] [PubMed] [Google Scholar]
- Matilla MA, Martin-Mora D, Gavira JAet al. Pseudomonas aeruginosa as a model to study chemosensory pathway signaling. Microbiol Mol Biol Rev. 2021;85:e00151–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matilla MA, Martin-Mora D, Krell T. The use of isothermal titration calorimetry to unravel chemotactic signalling mechanisms. Environ Microbiol. 2020;22:3005–19. [DOI] [PubMed] [Google Scholar]
- Matthaus F, Jagodic M, Dobnikar J. E. coli superdiffusion and chemotaxis-search strategy, precision, and motility. Biophys J. 2009;97:946–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthaus F, Mommer MS, Curk Tet al. On the origin and characteristics of noise-induced Levy walks of E. coli. Plos One. 2011;6:e18623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mccray JA, Trentham DR. Properties and uses of photoreactive caged compounds. Annu Rev Biophys Biophys Chem. 1989;18:239–70. [DOI] [PubMed] [Google Scholar]
- Mears PJ, Koirala S, Rao CVet al. Escherichia coli swimming is robust against variations in flagellar number. Elife. 2014;3:e01916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melotto M, Underwood W, Koczan Jet al. Plant stomata function in innate immunity against bacterial invasion. Cell. 2006;126:969–80. [DOI] [PubMed] [Google Scholar]
- Menolascina F, Rusconi R, Fernandez VIet al. Logarithmic sensing in Bacillus subtilis aerotaxis. npj Syst Biol Appl. 2017;3:16036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merritt PM, Danhorn T, Fuqua C. Motility and chemotaxis in Agrobacterium tumefaciens surface attachment and biofilm formation. J Bacteriol. 2007;189:8005–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesibov R, Adler J. Chemotaxis toward amino acids in Escherichia coli. J Bacteriol. 1972;112:315–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesibov R, Ordal GW, Adler J. Range of attractant concentrations for bacterial chemotaxis and threshold and size of response over this range - Weber law and related phenomena. J Gen Physiol. 1973;62:203–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Micali G, Endres RG. Bacterial chemotaxis: information processing, thermodynamics, and behavior. Curr Opin Microbiol. 2016;30:8–15. [DOI] [PubMed] [Google Scholar]
- Miller LD, Yost CK, Hynes MFet al. The major chemotaxis gene cluster of Rhizobium leguminosarum bv. viciae is essential for competitive nodulation. Mol Microbiol. 2007;63:348–62. [DOI] [PubMed] [Google Scholar]
- Miller TR, Belas R. Motility is involved in Silicibacter sp TM1040 interaction with dinoflagellates. Environ Microbiol. 2006;8:1648–59. [DOI] [PubMed] [Google Scholar]
- Millet YA, Alvarez D, Ringgaard Set al. Insights into Vibrio cholerae intestinal colonization from monitoring fluorescently labeled bacteria. PLoS Pathog. 2014;10:e1004405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milo R, Jorgensen P, Moran Uet al. BioNumbers-the database of key numbers in molecular and cell biology. Nucleic Acids Res. 2010;38:D750–D3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min TJL, Mears PJ, Golding Iet al. Chemotactic adaptation kinetics of individual Escherichia coli cells. Proc Natl Acad Sci USA. 2012;109:9869–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min TL, Mears PJ, Chubiz LMet al. High-resolution, long-term characterization of bacterial motility using optical tweezers. Nat Methods. 2009;6:831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misselwitz B, Barrett N, Kreibich Set al. Near surface swimming of Salmonella Typhimurium explains target-site selection and cooperative invasion. PLoS Pathog. 2012;8:e1002810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molenaar D, van Berlo R, de Ridder Det al. Shifts in growth strategies reflect tradeoffs in cellular economics. Molecular systems biology. 2009;5:323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mora T, Wingreen NS. Limits of sensing temporal concentration changes by single cells. Phys Rev Lett. 2010;104:248101. [DOI] [PubMed] [Google Scholar]
- Mukherjee S, Kearns DB. The structure and regulation of flagella in Bacillus subtilis. Annu Rev Genet. 2014;48:319–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson JW, Tredgett MW, Sheehan JKet al. Mucinophilic and chemotactic properties of Pseudomonas aeruginosa in relation to pulmonary colonization in cystic fibrosis. Infect Immun. 1990;58:1489–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuman H, Debelius JW, Knight Ret al. Microbial endocrinology: the interplay between the microbiota and the endocrine system. FEMS Microbiol Rev. 2015;39:509–21. [DOI] [PubMed] [Google Scholar]
- Neumann S, Grosse K, Sourjik V. Chemotactic signaling via carbohydrate phosphotransferase systems in Escherichia coli. Proc Natl Acad Sci USA. 2012;109:12159–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni B, Colin R, Link Het al. Growth-rate dependent resource investment in bacterial motile behavior quantitatively follows potential benefit of chemotaxis. Proc Natl Acad Sci U S A. 2020;117:595–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni B, Ghosh B, Paldy FSet al. Evolutionary remodeling of bacterial motility checkpoint control. Cell Rep. 2017;18:866–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Neal L, Vo L, Alexandre G. Specific root exudate compounds sensed by dedicated chemoreceptors shape Azospirillum brasilense chemotaxis in the rhizosphere. Appl Environ Microbiol. 2020;86:e01026–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Toole GA, Kolter R. Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Mol Microbiol. 1998;30:295–304. [DOI] [PubMed] [Google Scholar]
- Oleksiuk O, Jakovljevic V, Vladimirov Net al. Thermal robustness of signaling in bacterial chemotaxis. Cell. 2011;145:312–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira NM, Fostera KR, Durham WM. Single-cell twitching chemotaxis in developing biofilms. Proc Natl Acad Sci USA. 2016;113:6532–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen JE, Hoegh-Andersen KH, Casadesus Jet al. The role of flagella and chemotaxis genes in host pathogen interaction of the host adapted Salmonella enterica serovar Dublin compared to the broad host range serovar S. Typhimurium. BMC Microbiol. 2013;13:67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr AA, Yang J, Sule Net al. Molecular mechanism for attractant signaling to DHMA by E. coli Tsr. Biophys J. 2020;118:492–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortega A, Zhulin IB, Krell T. Sensory repertoire of bacterial chemoreceptors. Microbiol Mol Biol Rev. 2017;81:e00033–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacheco AR, Sperandio V. Inter-kingdom signaling: chemical language between bacteria and host. Curr Opin Microbiol. 2009;12:192–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parales RE, Harwood CS. Bacterial chemotaxis to pollutants and plant-derived aromatic molecules. Curr Opin Microbiol. 2002;5:266–73. [DOI] [PubMed] [Google Scholar]
- Park H, Pontius W, Guet CCet al. Interdependence of behavioural variability and response to small stimuli in bacteria. Nature. 2010;468:819–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S, Wolanin PM, Yuzbashyan EAet al. Influence of topology on bacterial social interaction. Proc Natl Acad Sci USA. 2003;100:13910–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker DJ, Demetci P, Li GW. Rapid accumulation of motility-activating mutations in resting liquid culture of Escherichia coli. J Bacteriol. 2019;201:e00259–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkinson JS, Ames P, Studdert CA. Collaborative signaling by bacterial chemoreceptors. Curr Opin Microbiol. 2005;8:116–21. [DOI] [PubMed] [Google Scholar]
- Parkinson JS, Hazelbauer GL, Falke JJ. Signaling and sensory adaptation in Escherichia coli chemoreceptors: 2015 update. Trends Microbiol. 2015;23:257–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Partridge JD, Harshey RM. Swarming: flexible roaming plans. J Bacteriol. 2013;195:909–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Partridge JD, Nhu NTQ, Dufour YSet al. Escherichia coli remodels the chemotaxis pathway for swarming. mBio. 2019;10:e00316–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Partridge JD, Nhu NTQ, Dufour YSet al. Tumble suppression is a conserved feature of swarming motility. mBio. 2020;11:e01189–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasupuleti S, Sule N, Cohn WBet al. Chemotaxis of Escherichia coli to norepinephrine (NE) requires conversion of NE to 3,4-dihydroxymandelic acid. J Bacteriol. 2014;196:3992–4000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasupuleti S, Sule N, Manson MDet al. Conversion of Norepinephrine to 3,4-dihydroxymandelic acid in Escherichia coli requires the QseBC quorum-sensing system and the FeaR transcription factor. J Bacteriol. 2018;200:e00564–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul K, Nieto V, Carlquist WCet al. The c-di-GMP binding protein YcgR controls flagellar motor direction and speed to affect chemotaxis by a “backstop brake” mechanism. Mol Cell. 2010;38:128–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulick A, Jakovljevic V, Zhang Set al. Mechanism of bidirectional thermotaxis in Escherichia coli. Elife. 2017;6:e26607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira CS, Thompson JA, Xavier KB.. AI-2-mediated signalling in bacteria. FEMS Microbiol Rev. 2013;37:156–81. [DOI] [PubMed] [Google Scholar]
- Perkins A, Tudorica DA, Amieva MRet al. Helicobacter pylori senses bleach (HOCl) as a chemoattractant using a cytosolic chemoreceptor. PLoS Biol. 2019;17:e3000395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pesavento C, Becker G, Sommerfeldt Net al. Inverse regulatory coordination of motility and curli-mediated adhesion in Escherichia coli. Genes Dev. 2008;22:2434–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeffer W. Locomotorische Richtungsbewegungen durch chemische Reize. Untersuch Bot Inst Tübingen. 1884;1:363–482. [Google Scholar]
- Phan TV, Morris R, Black MEet al. Bacterial route finding and collective escape in mazes and fractals. Phys Rev X. 2020;10:031017. [Google Scholar]
- Pichon C, Hechard C, du Merle Let al. Uropathogenic Escherichia coli AL511 requires flagellum to enter renal collecting duct cells. Cell Microbiol. 2009;11:616–28. [DOI] [PubMed] [Google Scholar]
- Pratt LA, Kolter R.. Genetic analysis of Escherichia coli biofilm formation: roles of flagella, motility, chemotaxis and type I pili. Mol Microbiol. 1998;30:285–93. [DOI] [PubMed] [Google Scholar]
- Pruss BM. Involvement of two-component signaling on bacterial motility and biofilm development. J Bacteriol. 2017;199:e00259–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rader BA, Wreden C, Hicks KGet al. Helicobacter pylori perceives the quorum-sensing molecule AI-2 as a chemorepellent via the chemoreceptor TlpB. Microbiol. 2011;157:2445–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raina JB, Fernandez V, Lambert Bet al. The role of microbial motility and chemotaxis in symbiosis. Nat Rev Microbiol. 2019;17:284–94. [DOI] [PubMed] [Google Scholar]
- Rebbapragada A, Johnson MS, Harding GPet al. The Aer protein and the serine chemoreceptor Tsr independently sense intracellular energy levels and transduce oxygen, redox, and energy signals for Escherichia coli behavior. Proc Natl Acad Sci USA. 1997;94:10541–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes-Darias JA, Garcia V, Rico-Jimenez Met al. Specific gamma-aminobutyrate chemotaxis in pseudomonads with different lifestyle. Mol Microbiol. 2015;97:488–501. [DOI] [PubMed] [Google Scholar]
- Rhee SH, Pothoulakis C, Mayer EA.. Principles and clinical implications of the brain-gut-enteric microbiota axis. Nat Rev Gastro Hepat. 2009;6:306–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rico-Jimenez M, Reyes-Darias JA, Ortega Aet al. Two different mechanisms mediate chemotaxis to inorganic phosphate in Pseudomonas aeruginosa. Sci Rep. 2016;6:28967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rio-Alvarez I, Munoz-Gomez C, Navas-Vasquez Met al. Role of Dickeya dadantii 3937 chemoreceptors in the entry to Arabidopsis leaves through wounds. Mol Plant Pathol. 2015;16:685–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivera-Chavez F, Lopez CA, Zhang LFet al. Energy taxis toward host-derived nitrate supports a Salmonella pathogenicity island 1-independent mechanism of invasion. mBio. 2016;7:e00960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivera-Chavez F, Winter SE, Lopez CAet al. Salmonella uses energy taxis to benefit from intestinal inflammation. PLoS Pathog. 2013;9:e1003267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rumbaugh KP, Sauer K. Biofilm dispersion. Nat Rev Microbiol. 2020;18:571–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusconi R, Guasto JS, Stocker R. Bacterial transport suppressed by fluid shear. Nat Phys. 2014;10:212–7. [Google Scholar]
- Ryjenkov DA, Simm R, Romling Uet al. The PilZ domain is a receptor for the second messenger c-di-GMP: the PilZ domain protein YcgR controls motility in enterobacteria. J Biol Chem. 2006;281:30310–4. [DOI] [PubMed] [Google Scholar]
- Salek MM, Carrara F, Fernandez Vet al. Bacterial chemotaxis in a microfluidic T-maze reveals strong phenotypic heterogeneity in chemotactic sensitivity. Nat Commun. 2019;10:1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salman H, Libchaber A. A concentration-dependent switch in the bacterial response to temperature. Nat Cell Biol. 2007;9:1098. [DOI] [PubMed] [Google Scholar]
- Sampedro I, Parales RE, Krell Tet al. Pseudomonas chemotaxis. FEMS Microbiol Rev. 2015;39:17–46. [DOI] [PubMed] [Google Scholar]
- Saragosti J, Calvez V, Bournaveas Net al. Directional persistence of chemotactic bacteria in a traveling concentration wave. Proc Natl Acad Sci USA. 2011;108:16235–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scharf BE, Hynes MF, Alexandre GM. Chemotaxis signaling systems in model beneficial plant-bacteria associations. Plant Mol Biol. 2016;90:549–59. [DOI] [PubMed] [Google Scholar]
- Schauer O, Mostaghaci B, Colin Ret al. Motility and chemotaxis of bacteria-driven microswimmers fabricated using antigen 43-mediated biotin display. Sci Rep. 2018;8:9801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuetz R, Zamboni N, Zampieri Met al. Multidimensional optimality of microbial metabolism. Science. 2012;336:601–4. [DOI] [PubMed] [Google Scholar]
- Schwarzer C, Fischer H, Machen TE. Chemotaxis and binding of Pseudomonas aeruginosa to scratch-wounded human cystic fibrosis airway epithelial cells. PLoS One. 2016;11:e0150109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweinitzer T, Josenhans C. Bacterial energy taxis: a global strategy?. Arch Microbiol. 2010;192:507–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweinitzer T, Mizote T, Ishikawa Net al. Functional characterization and mutagenesis of the proposed behavioral sensor TlpD of Helicobacter pylori. J Bacteriol. 2008;190:3244–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott M, Gunderson CW, Mateescu EMet al. Interdependence of cell growth and gene expression: origins and consequences. Science. 2010;330:1099–102. [DOI] [PubMed] [Google Scholar]
- Segall JE, Block SM, Berg HC. Temporal comparisons in bacterial chemotaxis. Proc Natl Acad Sci USA. 1986;83:8987–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serra DO, Richter AM, Klauck Get al. Microanatomy at cellular resolution and spatial order of physiological differentiation in a bacterial biofilm. mBio. 2013;4:e00103–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu TS, Tu Y, Berg HC. A modular gradient-sensing network for chemotaxis in Escherichia coli revealed by responses to time-varying stimuli. Molecular systems biology. 2010;6:382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shioi J, Tribhuwan RC, Berg STet al. Signal transduction in chemotaxis to oxygen in Escherichia coli and Salmonella typhimurium. J Bacteriol. 1988;170:5507–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidortsov M, Morgenstern Y, Be'er A. Role of tumbling in bacterial swarming. Phys Rev E. 2017;96:022407. [DOI] [PubMed] [Google Scholar]
- Silversmith RE. Auxiliary phosphatases in two-component signal transduction. Curr Opin Microbiol. 2010;13:177–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons SL, Bazylinski DA, Edwards KJ. South-seeking magnetotactic bacteria in the Northern Hemisphere. Science. 2006;311:371–4. [DOI] [PubMed] [Google Scholar]
- Sipos O, Nagy K, Di Leonardo Ret al. Hydrodynamic trapping of swimming bacteria by convex walls. Phys Rev Lett. 2015;114:258104. [DOI] [PubMed] [Google Scholar]
- Sneddon MW, Pontius W, Emonet T. Stochastic coordination of multiple actuators reduces latency and improves chemotactic response in bacteria. Proc Natl Acad Sci USA. 2012;109:805–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokolov A, Aranson IS, Kessler JOet al. Concentration dependence of the collective dynamics of swimming bacteria. Phys Rev Lett. 2007;98:158102. [DOI] [PubMed] [Google Scholar]
- Somavanshi R, Ghosh B, Sourjik V. Sugar influx sensing by the phosphotransferase system of Escherichia coli. PLoS Biol. 2016;14:e2000074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Son K, Menolascina F, Stocker R. Speed-dependent chemotactic precision in marine bacteria. Proc Natl Acad Sci USA. 2016;113:8624–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song S, Wood TK. The primary physiological roles of autoinducer 2 in Escherichia coli are chemotaxis and biofilm formation. Microorganisms. 2021;9:386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnenschein EC, Syit DA, Grossart HPet al. Chemotaxis of Marinobacter adhaerens and its impact on attachment to the diatom Thalassiosira weissflogii. Appl Environ Microbiol. 2012;78:6900–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sourjik V, Armitage JP. Spatial organization in bacterial chemotaxis. EMBO J. 2010;29:2724–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sourjik V, Berg HC.. Receptor sensitivity in bacterial chemotaxis. Proc Natl Acad Sci USA. 2002;99:123–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sourjik V. Receptor clustering and signal processing in E coli chemotaxis. Trends Microbiol. 2004;12:569–76. [DOI] [PubMed] [Google Scholar]
- Spagnolie SE, Moreno-Flores GR, Bartolo Det al. Geometric capture and escape of a microswimmer colliding with an obstacle. Soft Matter. 2015;11:3396–411. [DOI] [PubMed] [Google Scholar]
- Sporing I, Felgner S, Preusse Met al. Regulation of flagellum biosynthesis in response to cell envelope stress in Salmonella enterica serovar Typhimurium. mBio. 2018;9:e00736–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spormann AM, Wolfe RS. Chemotactic, magnetotactic and tactile behavior in a magnetic Spirillum. Fems Microbiol Lett. 1984;22:171–7. [Google Scholar]
- Spudich JL, Koshland DE Jr. Non-genetic individuality: chance in the single cell. Nature. 1976;262:467–71. [DOI] [PubMed] [Google Scholar]
- Stecher B, Barthel M, Schlumberger MCet al. Motility allows S. Typhimurium to benefit from the mucosal defence. Cell Microbiol. 2008;10:1166–80. [DOI] [PubMed] [Google Scholar]
- Stecher B, Hapfelmeier S, Muller Cet al. Flagella and chemotaxis are required for efficient induction of Salmonella enterica serovar Typhimurium colitis in streptomycin-pretreated mice. Infect Immun. 2004;72:4138–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stock AM, Robinson VL, Goudreau PN. Two-component signal transduction. Annu Rev Biochem. 2000;69:183–215. [DOI] [PubMed] [Google Scholar]
- Stocker R, Seymour JR, Samadani Aet al. Rapid chemotactic response enables marine bacteria to exploit ephemeral microscale nutrient patches. Proc Natl Acad Sci U S A. 2008;105:4209–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stocker R. Reverse and flick: hybrid locomotion in bacteria. Proc Natl Acad Sci USA. 2011;108:2635–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoodley P, Sauer K, Davies DGet al. Biofilms as complex differentiated communities. Annu Rev Microbiol. 2002;56:187–209. [DOI] [PubMed] [Google Scholar]
- Suchanek VM, Esteban-Lopez M, Colin Ret al. Chemotaxis and cyclic-di-GMP signalling control surface attachment of Escherichia coli. Mol Microbiol. 2020;113:728–39. [DOI] [PubMed] [Google Scholar]
- Sule N, Pasupuleti S, Kohli Net al. The Norepinephrine metabolite 3,4-dihydroxymandelic acid Is produced by the commensal microbiota and promotes chemotaxis and virulence gene expression in enterohemorrhagic Escherichia coli. Infect Immun. 2017;85:e00431–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweeney EG, Nishida A, Weston Aet al. Agent-based modeling demonstrates how local chemotactic behavior can shape biofilm architecture. mSphere. 2019;4:e00285–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Syvertsson S, Wang B, Staal Jet al. Different resource allocation in a Bacillus subtilis population displaying bimodal motility. J Bacteriol. 2021. DOI 10.1128/JB.00037-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabady RL, Yanta JH, Halladin DKet al. TagA is a secreted protease of Vibrio cholerae that specifically cleaves mucin glycoproteins. Microbiol. 2011;157:516–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taktikos J, Stark H, Zaburdaev V. How the motility pattern of bacteria affects their dispersal and chemotaxis. PLoS One. 2013;8:e81936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamar E, Koler M, Vaknin A. The role of motility and chemotaxis in the bacterial colonization of protected surfaces. Sci Rep. 2016;6:19616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tans-Kersten J, Huang HY, Allen C. Ralstonia solanacearum needs motility for invasive virulence on tomato. J Bacteriol. 2001;183:3597–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor JR, Stocker R. Trade-offs of chemotactic foraging in turbulent water. Science. 2012;338:675–9. [DOI] [PubMed] [Google Scholar]
- Terwilliger TC, Wang JY, Koshland DE. Kinetics of receptor modification. The multiply methylated aspartate receptors involved in bacterial chemotaxis. J Biol Chem. 1986;261:10814–20. [PubMed] [Google Scholar]
- Thormann KM, Saville RM, Shukla Set al. Initial phases of biofilm formation in Shewanella oneidensis MR-1. J Bacteriol. 2004;186:8096–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian M, Zhang C, Zhang Ret al. Collective motion enhances chemotaxis in a two-dimensional bacterial swarm. Biophys J. 2021. DOI 10.1016/j.bpj.2021.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tindall MJ, Maini PK, Porter SLet al. Overview of mathematical approaches used to model bacterial chemotaxis II: bacterial populations. B Math Biol. 2008;70:1570–607. [DOI] [PubMed] [Google Scholar]
- Tohidifar P, Bodhankar GA, Pei Set al. The unconventional cytoplasmic sensing mechanism for ethanol chemotaxis in Bacillus subtilis. mBio. 2020;11:e02177–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tout J, Astudillo-Garcia C, Taylor MWet al. Redefining the sponge-symbiont acquisition paradigm: sponge microbes exhibit chemotaxis towards host-derived compounds. Environ Microbiol Rep. 2017;9:750–5. [DOI] [PubMed] [Google Scholar]
- Tso WW, Adler J. Negative chemotaxis in Escherichia coli. J Bacteriol. 1974;118:560–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu Y. Quantitative modeling of bacterial chemotaxis: signal amplification and accurate adaptation. Annu Rev Biophys. 2013;42:337–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Sluis M, De Koning BA, De Bruijn ACet al. Muc2-deficient mice spontaneously develop colitis, indicating that MUC2 is critical for colonic protection. Gastroenterol. 2006;131:117–29. [DOI] [PubMed] [Google Scholar]
- Veening JW, Smits WK, Kuipers OP. Bistability, epigenetics, and bet-hedging in bacteria. Annu Rev Microbiol. 2008;62:193–210. [DOI] [PubMed] [Google Scholar]
- Vegge CS, Brondsted L, Li YPet al. Energy taxis drives Campylobacter jejuni toward the most favorable conditions for growth. Appl Environ Microbiol. 2009;75:5308–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vladimirov N, Lebiedz D, Sourjik V. Predicted auxiliary navigation mechanism of peritrichously flagellated chemotactic bacteria. Plos Comput Biol. 2010;6:e1000717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vladimirov N, Lovdok L, Lebiedz Det al. Dependence of bacterial chemotaxis on gradient shape and adaptation rate. Plos Comput Biol. 2008;4:e1000242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wadhams GH, Armitage JP. Making sense of it all: bacterial chemotaxis. Nature reviews Molecular cell biology. 2004;5:1024–37. [DOI] [PubMed] [Google Scholar]
- Waite AJ, Frankel NW, Dufour YSet al. Non-genetic diversity modulates population performance. Molecular systems biology. 2016;12:895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waite AJ, Frankel NW, Emonet T. Behavioral variability and phenotypic diversity in bacterial chemotaxis. Annu Rev Biophys. 2018;47:595–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walukiewicz HE, Tohidifar P, Ordal GWet al. Interactions among the three adaptation systems of Bacillus subtilis chemotaxis as revealed by an in vitro receptor-kinase assay. Mol Microbiol. 2014;93:1104–18. [DOI] [PubMed] [Google Scholar]
- Wang X, Koirala S, Aldridge PDet al. Two tandem mechanisms control bimodal expression of the flagellar genes in Salmonella enterica. J Bacteriol. 2020;202:e00787–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters CM, Bassler BL. Quorum sensing: cell-to-cell communication in bacteria. Annu Rev Cell Dev Biol. 2005;21:319–46. [DOI] [PubMed] [Google Scholar]
- Watteaux R, Stocker R, Taylor JR. Sensitivity of the rate of nutrient uptake by chemotactic bacteria to physical and biological parameters in a turbulent environment. J Theor Biol. 2015;387:120–35. [DOI] [PubMed] [Google Scholar]
- Weigel WA, Dersch P. Phenotypic heterogeneity: a bacterial virulence strategy. Microbes Infect. 2018;20:570–7. [DOI] [PubMed] [Google Scholar]
- Wensink HH, Dunkel J, Heidenreich Set al. Meso-scale turbulence in living fluids. Proc Natl Acad Sci USA. 2012;109:14308–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiles TJ, Schlomann BH, Wall ESet al. Swimming motility of a gut bacterial symbiont promotes resistance to intestinal expulsion and enhances inflammation. PLoS Biol. 2020;18:e3000661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams SM, Chen YT, Andermann TMet al. Helicobacter pylori chemotaxis modulates inflammation and bacterium-gastric epithelium interactions in infected mice. Infect Immun. 2007;75:3747–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe AJ, Berg HC. Migration of bacteria in semisolid agar. Proc Natl Acad Sci USA. 1989;86:6973–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolgemuth CW. Collective swimming and the dynamics of bacterial turbulence. Biophys J. 2008;95:1564–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong-Ng J, Celani A, Vergassola M. Exploring the function of bacterial chemotaxis. Curr Opin Microbiol. 2018;45:16–21. [DOI] [PubMed] [Google Scholar]
- Wong-Ng J, Melbinger A, Celani Aet al. The role of adaptation in bacterial speed races. Plos Comput Biol. 2016;12:e1004974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood TK, Barrios AFG, Herzberg Met al. Motility influences biofilm architecture in Escherichia coli. Appl Microbiol Biotechnol. 2006;72:361–7. [DOI] [PubMed] [Google Scholar]
- Wright KJ, Seed PC, Hultgren SJ. Uropathogenic Escherichia coli flagella aid in efficient urinary tract colonization. Infect Immun. 2005;73:7657–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wuichet K, Zhulin IB. Origins and diversification of a complex signal transduction system in prokaryotes. Sci Signal. 2010;3:ra50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie L, Altindal T, Chattopadhyay Set al. From the cover: bacterial flagellum as a propeller and as a rudder for efficient chemotaxis. Proc Natl Acad Sci USA. 2011;108:2246–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie L, Wu XL. Bacterial motility patterns reveal importance of exploitation over exploration in marine microhabitats. Part I: theory. Biophys J. 2014;107:1712–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Chawla R, Rhee KYet al. Biphasic chemotaxis of Escherichia coli to the microbiota metabolite indole. Proc Natl Acad Sci USA. 2020;117:6114–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W, Briegel A. Diversity of bacterial chemosensory arrays. Trends Microbiol. 2020;28:68–80. [DOI] [PubMed] [Google Scholar]
- Yang Y, A MP, Hofler Cet al. Relation between chemotaxis and consumption of amino acids in bacteria. Mol Microbiol. 2015;96:1272–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang YL, Sourjik V. Opposite responses by different chemoreceptors set a tunable preference point in Escherichia coli pH taxis. Mol Microbiol. 2012;86:1482–89. [DOI] [PubMed] [Google Scholar]
- Yao J, Allen C. The plant pathogen Ralstonia solanacearum needs aerotaxis for normal biofilm formation and interactions with its tomato host. J Bacteriol. 2007;189:6415–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yazi SR, Nosrati R, Stevens CAet al. Migration of magnetotactic bacteria in porous media. Biomicrofluidics. 2018;12:011101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi TM, Huang Y, Simon MIet al. Robust perfect adaptation in bacterial chemotaxis through integral feedback control. Proc Natl Acad Sci USA. 2000;97:4649–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi X, Dean AM. Phenotypic plasticity as an adaptation to a functional trade-off. Elife. 2016;5:e19307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoney A, Salman H. Precision and variability in bacterial temperature sensing. Biophys J. 2015;108:2427–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yost CK, Rochepeau P, Hynes MF. Rhizobium leguminosarum contains a group of genes that appear to code for methyl-accepting chemotaxis proteins. Microbiology. 1998;144:1945–56. [DOI] [PubMed] [Google Scholar]
- You C, Okano H, Hui Set al. Coordination of bacterial proteome with metabolism by cyclic AMP signalling. Nature. 2013;500:301–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarepour M, Bhullar K, Montero Met al. The mucin Muc2 limits pathogen burdens and epithelial barrier dysfunction during Salmonella enterica serovar Typhimurium colitis. Infect Immun. 2013;81:3672–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Li S, Liu Xet al. Sensing of autoinducer-2 by functionally distinct receptors in prokaryotes. Nat Commun. 2020;11:5371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang WJ, Chen CF, Li Yet al. Configuration of redox gradient determines magnetotactic polarity of the marine bacteria MO-1. Env Microbiol Rep. 2010;2:646–50. [DOI] [PubMed] [Google Scholar]
- Zhang X, Si G, Dong Yet al. Escape band in Escherichia coli chemotaxis in opposing attractant and nutrient gradients. Proc Natl Acad Sci USA. 2019;116:2253–58. [DOI] [PMC free article] [PubMed] [Google Scholar]