Abstract
Metal ions provide considerable functionality across biological systems, and their utilization within biomolecules has adapted through changes in the chemical environment to maintain the activity they facilitate. While ancient earth's atmosphere was rich in iron and manganese and low in oxygen, periods of atmospheric oxygenation significantly altered the availability of certain metal ions, resulting in ion replacement within biomolecules. This adaptation mechanism has given rise to the phenomenon of metal cofactor interchangeability, whereby contemporary proteins and nucleic acids interact with multiple metal ions interchangeably, with different coordinated metals influencing biological activity, stability, and toxic potential. The ability of extant organisms to adapt to fluctuating metal availability remains relevant in a number of crucial biomolecules, including the superoxide dismutases of the antioxidant defense systems and ribonucleotide reductases. These well-studied and ancient enzymes illustrate the potential for metal interchangeability and adaptive utilization. More recently, the ribosome has also been demonstrated to exhibit interchangeable interactions with metal ions with impacts on function, stability, and stress adaptation. Using these and other examples, here we review the biological significance of interchangeable metal ions from a new angle that combines both biochemical and evolutionary viewpoints. The geochemical pressures and chemical properties that underlie biological metal utilization are discussed in the context of their impact on modern disease states and treatments.
Keywords: metals, iron, manganese, magnesium, ribosome, superoxide dismutase, reactive oxygen species, redox regulation, metalloprotein, interchangeability
Abbreviations: cam-SOD, cambialistic SOD; Co, cobalt; COVID-19, coronavirus disease 2019; Cu, copper; Fe, iron; Fe–S, iron–sulfur; Fe-SOD, Fe-containing SOD; H2O2, hydrogen peroxide; ISCU, Iron–Sulfur Cluster; MCO, metal-catalyzed oxidation; MCT, metal chelation therapy; Mg, magnesium; Mn, manganese; Mn-SOD, manganese-containing SOD; Ni, nickel; O2•−, superoxide; •OH, hydroxyl radical; RdRp, RNA-dependent RNA polymerase; RNR, ribonucleotide reductase; ROS, reactive oxygen species; Rpe, ribulose-5-phosphate 3-epimerase; SARS-CoV2, severe acute respiratory syndrome coronavirus 2; SOD, superoxide dismutase; ZF, zinc finger; Zn, zinc
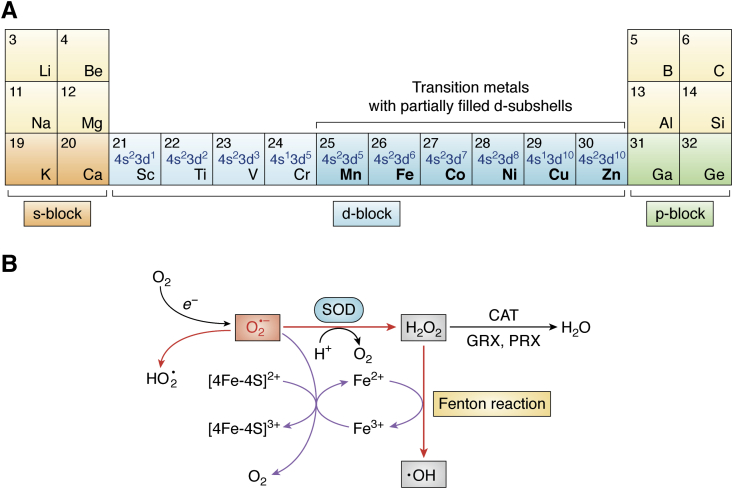
The unique chemical properties of numerous metal ions facilitate extensive interactions with biomolecules, with impacts across all areas of cellular activity, including fundamental processes, such as respiration, metabolism, nitrogen fixation, photosynthesis, DNA replication, transcription, and protein synthesis (1, 2, 3, 4, 5, 6, 7, 8). At least ten metal elements are considered essential for most forms of life (9), including six of the d-block elements of the periodic table: manganese (Mn), iron (Fe), cobalt (Co), nickel (Ni), copper (Cu), and zinc (Zn) ((10) and Fig. 1A). These metals are characterized by the ability to form ions with partially filled d-subshells (shown in Fig. 1A, blue). This electron configuration facilitates multiple oxidation states, defining many of their chemical properties. These metals are called transition metals, with the most biologically relevant examples appearing in the first row of the d-block in the periodic table (Fig. 1A, dark cyan squares). Although biologically important, Zn is excluded from the transition metals by some definitions because of possession of a complete d-subshell (Fig. 1A).
Figure 1.
Transition metals and oxidative stress.A, transition metals. A section of the periodic table showing s-block (orange), d-block (cyan), and p-block (green) chemical elements. Transition metals of the first row, characterized by partially filled d-subshells, are shown in dark cyan squares in bolded black lettering. Zn (which possesses a complete d-subshell) is also shown in this group. The electronic structures of the d-block elements are shown in blue lettering. B, reactions of the superoxide anion; iron and redox cycling. Negatively charged free radical superoxide (O2•−, shown in red lettering) is the product of one-electron (e−) reduction of dioxygen (O2). Upon protonation, O2•− can form the hydroperoxyl radical (HO2•). Superoxide dismutase (SOD, cyan oval) catalyzes the dismutation (disproportionation) of O2•−, thereby generating O2 and hydrogen peroxide (H2O2). H2O2 is converted to H2O by various antioxidant enzymes, such as catalases (CAT), glutathione peroxidases (GPX), and peroxiredoxins (PRX). Redox-active Fe2+ ions are oxidized by H2O2, generating highly reactive hydroxyl radicals (OH•) and Fe3+ through the Fenton reaction. Fe3+ can be reduced to Fe2+ by O2•−, resulting in redox cycling (purple arrows). By itself, O2•− can reduce Fe3+ to Fe2+ within iron–sulfur cluster proteins, resulting in enzyme inactivation and accumulation of Fe2+, which further powers Fenton chemistry. Modified from Ref. (271).
The human body contains amounts of magnesium (Mg), Fe, and Zn in the gram range, whereas milligram amounts of Mn, Cu, Co, and molybdenum are present (9, 11). These metals perform both catalytic and structure-stabilizing roles and are predominantly available as divalent cations (possessing two fewer electrons than the neutral state). In fact, approximately 40% of enzymes with known structures depend upon at least one metal cofactor for catalytic activity (12). Analysis of metal-binding domains in the proteome suggests that metal-mediated folds are proportional to proteome size across the kingdoms of life, whereas the specific metals predicted to be utilized reveal distinct changes through evolutionary history (13). The utilization of metal ions for life on earth may predate protein-oriented extant biology, as nucleic acids in a metal-rich prebiotic environment are hypothesized to generate the earliest enzymatic mechanisms. Much as in proteins, metal ions are employed as catalytic cofactors in RNA species (14) and coordinated by both the anionic sugar phosphate backbone and the nucleotide bases (4, 14). Metal ions similarly interact with the DNA backbone and bases (15). The high charge density of the metal ions allows large RNAs to form complexes and closely packed folds and tertiary interactions, facilitating elaborate and dynamic structures, such as the ribosome—the essential protein-synthesizing machine that operates in every living cell. Transition metal ions bind throughout rRNA both loosely and at specific sites, with Mg being the major metal ion contributing to present structures of large and small ribosomal subunits (4, 14, 16).
Despite an effort, drawing definitive conclusions about the physiological utilization of metal ions by biomolecules within cells is a technically challenging task, hindered by several experimental issues. These include the dissociation of ions during biomolecule purification, physical properties that limit detection in structural models, and broader impacts of overexpressing metal-binding species on metal availability (17). Further complications in the elucidation of metal ions usage for biomolecules functionality arise from the phenomenon of metal ion interchangeability, wherein one of several different ions are able to occupy a specific biomolecular binding site. Metal ions, in many cases, self-assemble into complexes with biomolecules (18) and, as such, can be greatly influenced by intracellular metal availability dictated by a particular physiological or environmental condition. Therefore, the flexibility in metal ion preference may be more prevalent than currently understood. Thus, the view of a single native metal for a given binding site may, at least in some cases, be an unfavorably strict categorization. In a healthy cell, cytosolic and organellar metal cation levels are tightly regulated as a means of protection against the undesired activity of certain metal elements, while metal imbalances manifest in numerous human diseased states. This identified link between the ability of certain biomolecules to interact with a non-native metal ion(s), which may abolish their correct functionality leading to mitigation of disease, supports the need for further knowledge of the molecular mechanisms governing metal ion interchangeability. Such investigation will advance our understanding of disease etiology and progression, with a potential for new therapeutical intervention strategies.
Particular progress in revealing transition metal interchangeability in biomolecular structure and function has been made recently because of new technological developments. An emerging example of particular interest is the utilization of metals by the ribosome. While the high Mg content of contemporary ribosomes supports a preference of the metal-binding sites for Mg2+, a recently published study that replicated a prebiotic environment of anoxic earth rich in Fe and Mn and low in oxygen revealed that Mg2+ on ribosomes can be replaced by Fe2+ or Mn2+ without affecting protein-synthesizing activity (19, 20). Besides an important impact on new biochemical features of a ribosome and medicine-related translational science, this technically advanced approach provided the scientific community with an elaborate biological model crucial for investigating the origin of life and evolution of biological molecules. In fact, the recapitulated conditions mimicking the anoxic earth environment supported a hypothesis that a ribosome represents an extraordinarily well-conserved RNA–protein structure that existed in a complex with Fe ions when earth's atmosphere was depleted of oxygen (3, 20). These studies were corroborated with biochemical assays conducted with ribosomes from Saccharomyces cerevisiae, wherein it was demonstrated that eukaryotic ribosomes maintained an ability to interact with Fe2+ at the selected sites under normal physiological conditions. This suggests a possibility that this biological atavism might play an essential role in the regulation of protein synthesis, the faultless processivity of which contributes to protein homeostasis and protection against neurodegenerative diseases (21). These recent developments in understanding ribosome biology in the context of transition metal interchangeability open up many questions requiring further investigation.
Flexibility in metal ion interactions with biomolecules is not only limited to ribosomes but also has been documented to be prevalent in metalloproteins. This is commonly observed in in vitro assays, wherein enzyme activity is assessed in the presence of different metals to identify which confers the greatest catalytic activity. In addition, certain metal ions, including Mg, are not directly detectable in spectroscopic studies and difficult to identify in crystallographic studies (22, 23) and can be readily substituted with alternative ions for the purposes of structural assessment (24, 25), further indicating the relative ease with which certain ions can replace one another.
Large-scale environmental changes accompanied with the accumulation of molecular oxygen in ancient earth's atmosphere, the groundbreaking event that occurred in the course of evolution dated billions of years ago, drove adaptation of biomolecules by selecting organisms that had the capability to defend against highly reactive chemical products derived from the incomplete reduction of oxygen (known as reactive oxygen species [ROS]) and utilize alternative metals to catalyze crucial biochemical reactions. One of the prominent examples of how these pressures are mirrored in extant biology is the host–pathogen interface. The innate immune system orchestrates challenging chemical assaults upon invading pathogens that in many cases display flexibility in metal utilization in their responses. For example, the connection between metal biochemistry and oxidative stress is central to the phagocytic immune response, which employs both oxidant assault and nutrient metal limitation in the defense against pathogens (26). Many pathogens are sensitive to metal levels on either side of a relatively narrow window, and host systems exploit this (27). During infection of a host organism, bacterial pathogens will commonly experience challenging conditions, including both overload and limitation of trace metal elements, as well as severe oxidative stress (28, 29, 30).
Transition metals are also known to play an important role in several viruses' survival and pathogenesis. Relevant to the present time, much of the research has been conducted investigating the role of Fe and other transition metals in the severe acute respiratory syndrome coronavirus 2 (SARS-CoV2)–related pathologies. Although many questions remain unanswered, it is clear that metals play an important role during SARS-CoV2 infection and propagation, whereas multiple manifestations of coronavirus disease 2019 (COVID-19), such as immune dysfunction, inflammation, hypercoagulation, hyperferritinemia, have been linked to Fe overload (31).
The possible ambiguity in assignments of metal ion associations with biomolecules, along with emerging examples of physiologically relevant metal interchangeability, indicates an existing gap in knowledge with an impact on diverse biological topics briefly mentioned previously. To gain an up-to-date picture of the transition metal interchangeability phenomenon, here, we discuss the current progress that has been made.
As such, we outline the deeply embedded nature of divalent metal cations in biological macromolecules and place this in the context of the risk of oxidative damage, which is present in an oxygen-rich environment. The geochemical changes that occurred on earth since the establishment of life are then discussed as they relate to metal availability and utilization. As some of the best-studied examples that have had recent developments in understanding metal interchangeability, we describe the role of metal ions in the superoxide dismutases (SODs), the R2 subunit of the ribonucleotide reductases (RNRs), and the ribosome. We choose these biomolecule examples as they remain in the frontline of scientific research, providing new information on how metals can replace each other to meet various physiological cues. Finally, we describe how flexible metal utilization impacts both bacterial and viral infections and immunity. While we focus on Fe and Mn as the most prominent examples of physiological metal cofactors, other biologically important metals are also discussed.
Metal ions provide powerful biochemical functionality to biomolecules
As stated previously, ∼40% of biomolecules utilize metals as cofactors, suggesting that metal ions are essential for cellular physiology, with the first-row transition metals being of particular importance (Fig. 1A). For their interactions with biomolecules, metal ions can be considered in terms of several properties, including charge density, radii, and reactivity. Redox activity is also of particular relevance to biology because it prescribes some catalytic capabilities. Redox inactive metal cations, of which Mg and Zn are the most common in biomolecules, tend to be utilized in structures to stabilize negative charges, as well as functioning as Lewis acids to activate substrates by accepting lone pair electrons with no net change in oxidation state (12, 32, 33). Many of the transition metals are redox active, such as Fe, Mn, Cu, and Ni, as electrons of their incomplete d-subshells (Fig. 1A) can be lost, allowing for several oxidation states. While such cations can act as Lewis acids, they are commonly used in the catalysis of redox reactions, in which electron transfer results in a change of oxidation state.
Both the intracellular availability and the stability of formed complexes are important factors in metal cofactor binding. The predicted complex stability of divalent metal cations is described by the Irving–Williams series (Mg2+ < Mn2+ < Fe2+ < Ni2+ < Co2+ < Cu2+ > Zn2+) (34), which illustrates that stability of complexes increases with atomic number across the divalent metal cations until reaching Zn, which does not possess unpaired d-shell electrons and thus forms less stable interactions than Cu2+ (35). However, many biomolecules associate with less competitive cations, such as Mg2+, Fe2+, and Mn2+ (18), as well as monovalent cations of potassium or sodium, which form even less stable interactions.
Despite tight regulation of cellular concentrations of Mg2+, Fe2+, and Mn2+ accomplished by a coordinated effort of metal transporters and buffering chaperones in regulating free ion levels (36), the high affinity of these metals toward biomolecules ensures their activity even at low concentrations. For example, Fe is central to the heme and Fe–sulfur (Fe–S) cluster complexes, which are crucial cofactors in electron transport chain reactions, oxygen transport, translation termination, and antioxidant pathways. The background and current understanding of Fe–S cluster assembly and function have been detailed in several informative recent reviews (37, 38). The synthesis of these ancient cofactors can be catalyzed by UV light from the reduced Fe–S species, which were prevalent in the prebiotic earth, supporting an early role for Fe–S clusters in the evolution of life (39).
The propensity for biomolecules to be flexible in their metal binding partner may reflect two features of the interactions. First, while biomolecular structures may evolve to prefer a metal cofactor by excluding similar ions, which are suboptimal, nonfunctional, or deleterious, there are overlapping characteristics of cations, which in many cases impede absolute specificity. Second, experimental evidence of selective advantages gained by retaining or acquiring the ability to exchange cofactors suggests that tolerance of alternative metal ions may be beneficial in certain circumstances. In other examples, divergent or convergent evolution has produced multiple distinct biomolecules within organisms, allowing consistent activity in environments of varying metal availability and limitations.
Change of earth's atmosphere as a driving force of the evolution of biomolecules
Great oxidation events, molecular oxygen, and ROS
Early in the history of earth, volcanic processes were the major contributors to the composition of the atmosphere and oceans. This ancient earth's atmosphere was anoxic and reducing, with oceans rich in soluble divalent transition metal ions (40, 41, 42). While the time line of changes in earth's atmosphere remains under discussion (43, 44), it is thought that 2 to 3 billion years ago, the accumulation of molecular oxygen in the atmosphere occurred in what is known as the great oxidation event. A further oxidation event is likely to have transpired less than 1 billion years ago (neoproterozoic oxidation event), which had a more significant impact on oxygen levels in the ocean (45). These shifts in atmospheric composition occurred subsequent to biogenesis (44, 45), involving complex fluctuations of oxygen stores with biological processes likely being the primary source of the molecular oxygen (43, 46, 47). It has been proposed that prevalent methanogenic archaea, which depended on Ni for metabolic catalysis, declined following a decrease in volcanic sources of Ni (48). This led to a decrease in methane production and allowed proliferation of species requiring less Ni, such as photosynthetic marine cyanobacteria (40, 49).
Abundant oxygen led to the expansion of organisms utilizing oxidative metabolism, with oxygen acting as an electron acceptor, as it contains two unpaired electrons with parallel spins and is metabolized by a univalent (single electron) mechanism, generating several ROS as intermediates. Among others (50), ROS include the superoxide (O2•−), hydroxyl (•OH) radicals, and hydrogen peroxide (H2O2) (51). H2O2, while itself not a radical, is prone to univalent reduction by Fe and Cu ions, making it a significant contributor to oxidative damage to various biomolecules, along with the O2•− and •OH radicals. O2•− radicals are generated from the electron transport chain reactions and are converted to the less reactive H2O2 by SODs (43). Other antioxidant enzymes, such as catalase, glutathione peroxidase, and/or peroxiredoxins then convert H2O2 to water (52). Alternatively, the •OH radical can be generated from H2O2 by high-energy radiation, or by metal ion catalysis reactions, such as the Fenton reaction that is discussed later (Fig. 1B).
Oxidation of biomolecules and role of metal ions
ROS, especially highly reactive •OH radical, can oxidize most biological targets, resulting in short lifetimes and very limited diffusion distances (53). ROS broadly damage proteins and amino acids via a range of modifications to amino acid side chains that lead to protein inactivation and degradation, the induction of polypeptide cleavages, and promotion of cross-linking and aggregation (54). In many cases, damage to the proteins can occur following exposure to xenobiotic metals, such as heavy metals (55). Another metal-induced harmful protein modification mechanism is related to site-specific metal-catalyzed oxidation (MCO) of amino acids at metal-binding sites. Specifically, MCO systems were found to target a wide variety of essential cellular enzymes and structural proteins, including glutamine synthetase, chymotrypsin, myosin, α-synuclein, catalases, and SODs. It was found that the activity of most of the MCO systems is dependent on ions of Fe or Cu, both of which are also involved in Fenton chemistry, whereby Fe2+, and in some instances, Cu2+, react with H2O2 to produce a •OH radical and a hydroxide ion ((56) and Fig. 1B). Fenton-generated •OH radical damages amino acids within proteins (57, 58).
The oxidative damage of proteins is implicated in a plethora of human pathologies, including neurodegenerative diseases, such as Alzheimer's (59) and Parkinson's disease (60), amyotrophic lateral sclerosis (61); muscular dystrophy (62), pulmonary emphysema (63), atherosclerosis (64), and age-related clinical pathologies, such as age-related macular degeneration (65), and cataractogenesis (66). For example, inactivation of SODs via oxidation (67, 68) or mutations (61, 69) results in enzymatic incompetency, degradation, or aberrant cellular localization. This leads to increased levels of ROS, causing protein damage and aggregation (which manifest in the progression of amyotrophic lateral sclerosis, Alzheimer's disease, and Parkinson's disease (70)) and DNA lesions (which can promote tumorigenesis (71)). Oxidation and cleavage of protein chaperones, such as Hsp90, has also been demonstrated to occur by an Fe-mediated mechanism, compounding the direct damage of ROS to proteins by disrupting the folding and stability of the targets of this chaperone (72, 73), leading to neurodegeneration and prionopathies (74). The scope of protein oxidation and the various mechanisms are reviewed in detail in Ref. (53).
Nucleic acids are also highly susceptible to damage by ROS, and oxidant damage to both RNA and DNA is implicated in many diseases, including neurodegenerative conditions and cancer (75, 76). The Fe- and/or Cu-driven Fenton reaction (Fig. 1B) has been identified as a source of oxidants leading to nucleic acid damage (77, 78, 79), and this is discussed in the following section. In addition, Fe and Mg can cleave RNA by a nonoxidative mechanism (termed in-line cleavage), as was recently demonstrated (3).
Oxidation of free RNA species can cause strand cleavages and oxidative base modifications. While oxidized mRNAs are recognized by ribosomes, the lesions are associated with decreased translational efficiency and an increase in truncated or misfolded protein products (80, 81). Besides damages to the transcripts, the translational machinery, which is constructed of proteins and RNAs, is also susceptible to oxidant-induced impairments. Ribosomes are large ribonucleotide–protein complexes that are at the center of the protein synthesis machinery. Ribosomes are composed of the small subunit (30S for prokaryotes and 40S for eukaryotes) and large subunit (50S for prokaryotes and 60S for eukaryotes). The subunits are assembled as intricately folded rRNAs for which Mg2+ ions play a critical role by coordinating rRNA folding and interaction with ribosomal proteins. Eukaryotes also have distinct ribosomes in the plastids and mitochondria, where they are likely to be particularly exposed to oxidative species derived from electron transport chain reactions (82). RNA oxidation leads to guanine modification (8-oxo-7,8-dihydroguanine), which is associated with wide range of pathologies, such as neurodegeneration, neuropsychiatric disorders, and atherosclerosis (82). Thus, oxidative stress negatively impacts translational processes (83), while maintenance of translational function can promote adaptation and survival responses (84, 85).
Fenton chemistry and role of Fe and Cu
Fe can act as an electron donor and acceptor owing to its reduced Fe2+ and oxidized Fe3+ states, which are important for its biological utility. The employment of Fe in biomolecules became established in biology in an aqueous earth environment, which was Fe rich and contained little oxygen.
The mononuclear or dinuclear ions of Fe are employed in the catalytically active centers of many enzymes, including the SODs and RNRs, both of which are susceptible to metal interchange and are the focus of this review. Much of the toxicity associated with Fe is a result of the Fenton reaction (Fig. 1B). In the reducing environment of the cytosol, O2•− can oxidize and destabilize Fe–S cluster complexes with crucial activity in the citric acid cycle, thereby blocking aerobic metabolism and generating free Fe, which can further participate in Fenton chemistry (Fig. 1B). The term “redox cycling” is used to refer to this propagation of ROS by a combination of the Fenton reaction and the activity of the O2•− anion in the regeneration of free Fe2+ (51, 86).
Similar to Fe, ions of Cu that exist in oxidized (Cu2+) and reduced (Cu+) states play roles in electron transport and many redox enzyme–driven mechanisms (87). Importantly, Cu+ also participates in Fenton-like reactions, and there is a partial overlap of biological activities and interactions between homeostatic mechanisms between Cu and Fe. Both are highly redox active, facilitating roles in many enzymatic reactions (88). Because of the oxidative damage associated with loss of homeostasis of these metals, regulation of free Fe and Cu in cells is important for protection from metal-dependent oxidative damage. Cu is especially toxic because of subsequent reactions involving the Cu2+ ion, including binding to thiol groups, generating large amounts of •OH radicals, and promoting further Fe2+-mediated Fenton reactions (89). For these reasons, free Cu is extremely limited within cells, being almost entirely bound by highly conserved Cu-binding proteins (90). Toxicity of Cu is mitigated in cells by metallothioneins that sequester Cu as well as Zn and non-nutrient heavy metals cadmium and mercury (91). Cu was likely not employed in primitive biomolecules because of limited availability but became required later, for example, in the catalytic center of certain SOD enzymes (Table 1), as an alternative to Fe or Mn (87).
Table 1.
Metal ions interchangeability within SODs
| Kingdom/order | Species | SOD | Native Me cofactor | Active site's Me replacement | Dismutation enzymatic activity | Cambialism | Reference |
|---|---|---|---|---|---|---|---|
| Bacteria/Bacillales | Staphylococcus aureus | SodA | Mn2+ | — | Active | — | (141) |
| SodMa | Mn2+/Fe2+a | Mn2+ or Fe2+a | Activea | Yesa | (141)a | ||
| smSoda | Mn2+a | Mn2+ or Fe2+a | Activea | Yesa | (257, 258)a | ||
| stSoda | Mn2+a | Mn2+ or Fe2+a | Activea | Yesa | (258, 259)a | ||
| Bacteria/Enterobacterales | Escherichia coli | SodA | Mn2+ | — | Active | — | (125, 260) |
| Fe2+ | Gain of function: peroxidasec | No | (141) | ||||
| Mutant SodAG165Ta | —a | Fe2+a | Activea | Yesa | (139)a | ||
| SodA | Mn2+ | Mn2+, Fe2+ hybridb | Partially activeb | Nob | (261)b | ||
| SodB | Fe2+ | — | Active | — | (125) | ||
| Mn2+ | Inactive | No | (125) | ||||
| Mutant SodBT165Ga | —a | Mn2+a | Activea | Yesa | (137)a | ||
| SodC | Cu2+Zn2+ | — | Active | — | (262) | ||
| Bacteria/Streptomycetales | Streptomyces coelicolor | SodN | Ni2+ | Ni2+ | Active | — | (196, 197) |
| Fe2+ | Inactive | No | (197) | ||||
| Zn2+ | Inactive | No | (197) | ||||
| Fe2+, Zn2+ hybridb | Activeb | Nob | (196)b | ||||
| Bacteria/Bacteroidales | Porphyromonas gingivalis | pgSoda | Fe2+a | Mn2+ or Fe2+a | Activea | Yesa | (258)a |
| Bacteria/Actinobacteria | Propionibacterium shermanii | psSoda | Fe2+a | Mn2+ or Fe2+a | Activea | Yesa | (258)a |
| Archaea/Sulfolobales | Acidianus ambivalens | FeSod | Fe2+ | — | Active | — | (263) |
| Co2+ | Inactive | No | (263) | ||||
| Ni2+ | Inactive | No | (263) | ||||
| Mn2+ | Inactive | No | (263) | ||||
| Fungi/Saccharomycetales | Saccharomyces cerevisiae | Sod1 | Cu2+Zn2+ | — | Active | — | (264) |
| Sod2 | Mn2+ | — | Active | — | (265) | ||
| Fe2+ | Inactive | No | (136, 266) | ||||
| Animalia/Mammalia | Mammals | SOD1 | Cu2+Zn2+ | — | Active | — | (190) |
| Cu2+Cu2+ | Inactive | No | (189) | ||||
| SOD2 | Mn2+ | — | Active | — | (120, 267) | ||
| Fe2+ | Gain of function: peroxidasec | No | (133) | ||||
| SOD3 | Cu2+Zn2+ | — | Active | — | (118, 267, 268, 269) | ||
| Plantae/laurales | Cinnamomum camphora | FeSODa | Fe2+a | Mn2+ or Fe2+a | Activea | Yesa | (270)a |
Highlight SODs with cambialistic properties.
Highlight hybrid SODs, whereby two different metal ions (indicated in the table) occupy two distinct enzyme dimer's subunits.
Highlight SODs with peroxidase activity gained upon metal ion replacement.
Chemical properties of Fe, Mn, and Mg dictate their interchangeability within biomolecules
Conserved and ancient biomolecules are associated with Mn and Fe as a result of the availability of these cations in the early earth environment, as well as the catalytic capabilities supplied by their redox activities, which provide essential functions. The utilization of either Fe or Mn within closely related SODs (Table 1) from the most ancient group of these enzymes reflects the chemical similarities of these two divalent ions for use in redox mechanisms. These transition metal elements are adjacent in the periodic table and have similar radii, ligand affinities, and coordination preferences, presenting a challenge for biomolecules to choose between (36, 92).
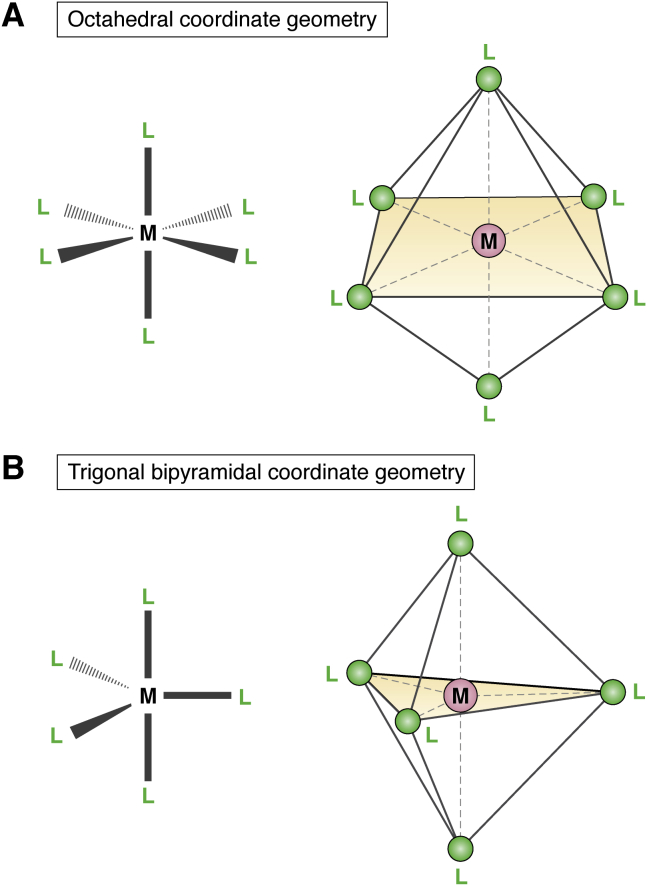
Unlike Fe, Mn does not participate in Fenton chemistry because of possessing a higher reduction potential (93), and as such, does not present the same toxicity risks in an oxidative environment. Mn also functions in enzymes as a Lewis acid in mechanisms that are more comparable to those catalyzed by Mg or Zn (94). Coordinated Mg ions can often be replaced with Mn (2, 95, 96), as they have similar binding site requirements (97). While Mg is an alkaline metal possessing no d-electrons, the Mg2+ ion shares a relatively similar ionic radius to Mn2+ and Fe2+. When forming complexes, all three cations prefer to coordinate six ligands (24) in an octahedral liganding geometry (Fig. 2A), and, in several examples, can occupy the same binding sites (19, 22, 25, 98). Mg forms very few covalent interactions with its ligands, making it able to be rapidly exchanged. The d-electrons of Mn2+ contribute to electrophilic interaction with ligands, allowing it to tolerate greater distortions of the bonding geometry than Mg2+, thereby lending itself better to catalytic mechanisms (99). While the Irving–Williams series predicts greater stability of Mn or Fe complexes, the substitution of Mg with either transition metal may be limited by the vastly greater intracellular availability of Mg. In support of this, it has been reported that disruption of Mg2+-dependent processes occurs with increased Mn2+ import under conditions of low environmental Mg (96).
Figure 2.
The six- and five-coordinate geometries. Diagrams (2D on the left; 3D on the right) of the six-coordinate octahedral geometry (A) and the atypical five-coordinate trigonal bipyramidal geometry (B) of various metal ions within biomolecules discussed in the article. M represents the ion of iron or manganese; L represents a ligand. Modified from Ref. (128).
Mg is fundamental in stabilizing protein, lipid, and nucleic acid structures (100) and is involved in many catalytic mechanisms (25). It is the most common metal found in enzymes according to systematic analyses of reported protein structures, appearing in 16% of all enzymes (12, 100). For comparison, Mn is identified as a cofactor in approximately 6% of all enzymes with known structure, whereas Fe is a cofactor in 8% (12, 101). As Mg is known to form less stable interactions than other metal ions and is readily replaced, it is possible that structural reports underestimate the physiological occupation of binding sites by this element (101). Zn is also utilized extensively in cells and appears in more enzyme structures than Mn or Fe (101). Divalent ions of Zn and Mn have very similar radii (0.74 and 0.75 Å, respectively) but otherwise have dissimilar binding profiles and biochemical behavior and do not typically replace one another (24).
Geochemical shifts drove extant metal biology: SOD as a prominent example
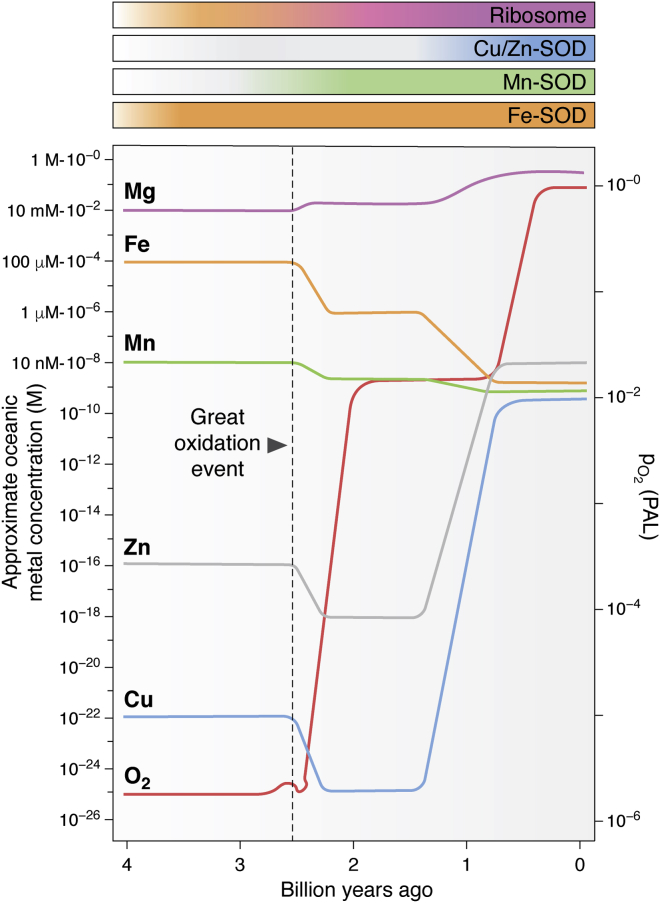
With the proliferation of oxygen and the potential for oxidative damage to cellular components by ROS, organisms that had evolved antioxidant defenses would have had a major advantage (43). Phylogenetic analysis suggests that ROS scavenging enzymes, such as SODs, peroxiredoxins, and catalases, had already emerged prior to the great oxidation event in response to the presence of low-level or localized oxygen (102). Antioxidant systems, thus, became increasingly valuable as oxygenation increased (43), whereas broad fluctuations in metal availability occurred concomitantly. The transition metals Mn, Fe, Co, and Ni were in solutions at relatively high levels in the early ocean environments because of the high sulfur content and low levels of oxygen, whereas precipitation of Cu and Zn would have rendered them unavailable for biochemistry (103). The increase in environmental oxygen led to the precipitation of Mn, Fe, Co, and Ni, and a major increase in the amount of available Zn, effectively reversing the availability of these metals (13, 42, 103, 104). These shifts in metal solubility were reflected by changes in their biological utilization. As such, comparison of structural motifs in the proteomes of organisms across the three domains of life supports the hypothesis that bioavailability of metals presented an evolutionary pressure resulting in the differences in their utilization (105).
One prominent example of geochemical shift–driven evolution of biomolecules is the appearance of alternatives to the primitive Fe-containing SOD (Fe-SOD), which utilize other metal cofactors (Fig. 3 and Table 1). SODs are the major enzymes responsible for the removal of O2•− anions that are unavoidably generated during aerobic metabolic reactions (51). British biochemist and writer Dr Lane characterized discovery of SODs as “the most important discovery of modern biology never to win a Nobel prize.” In fact, being discovered in 1969, SODs from various organisms remain of high interest for over last 5 decades.
Figure 3.
Schematic representation of concentrations of oxygen and selected metal cations in earth oceans. Estimates of the appearance of metal-utilizing biomolecules are shown above the graph in gradient-colored horizontal bars, indicating the early appearance of the ribosome. The most ancient superoxide dismutases (SODs), likely Fe-SODs, gave rise to Mn-SODs as oxygenation increased, with Cu/Zn-SODs appearing subsequently as copper and zinc became available. Oxygen levels (red line, right axis) are given as pO2 (partial pressure of oxygen) relative to PAL (present atmospheric level). The dotted line indicates the time of the great oxidation event (GOE). Modified from Refs. (103, 173, 272).
The SODs catalyze a dismutation (disproportionation) reaction to produce O2 and H2O2 from two O2•− via a cyclic oxidation–reduction electron transfer (106):
| (1) |
| (2) |
In this two-step reaction, the oxidized form of the metal (M) center ions (Mox-SOD) are first converted to the reduced form (Mred-SOD) with the formation of O2 (reaction 1), followed by oxidation of the reduced form of the metal ions into their oxidized form by O2•− with the release of H2O2 (reaction 2). Thus, one O2•− reduces the SOD, whereas another O2•− oxidizes the SOD in a so-called “ping–pong” mechanism (107). Several metal ion cofactors (such as Fe, Mn, Cu, Zn, and Ni) can be employed in the SOD active sites, which perform this mechanism, as summarized in Table 1. In this catalytic mechanism, the metal ion is utilized by the SODs as a source of protons by employing structural aspects of the metal binding site to adjust the redox potential, which also acts to regulate the access of anions (108). The activity of SODs requires them to exert tight control over the reactivity of the bound metals. The reduction midpoint potential (Em, a measure of the propensity of a chemical species to gain electrons in a redox reaction) of the metal cofactors is manipulated to around 300 mV in all known examples, despite the variety of metals employed across the enzyme families. The calculated reduction midpoint potential in aqueous solution compared with normal hydrogen electrode (a standard for zero redox potential as defined by the potential of a platinum electrode in 1 M acid solution) for the M3+/M2+ transition of Fe, Mn, and Ni are considerably different (0.77, 1.5, and 2.4 V, respectively) (109), requiring structural adaptation to bring the value into effective catalytic range (108, 110). The SODs demonstrate both the power of redox-active inorganic cofactors and the need to control their redox activity for function.
SODs are found in the archaea, prokarya, and eukarya ((43) and Table 1). Originally derived independently as three distinct families, abundance of SODs has undergone shift during course of evolution because of changes in earth's environment (Fig. 3). The first Mn/Fe-SOD arose in the low oxygen/high Fe environment (Fig. 3, orange and green curves). Thus, SODs represent a powerful and informative model to investigate metal ions interchangeability within biomolecules.
The Mn/Fe-SOD family utilizes Fe or Mn at their catalytic center, reflecting the high levels of these metals in the preoxidation earth in which they originated. Fe-SODs are found in some primitive eukaryotes, plants, and bacteria, whereas Mn-containing SODs (Mn-SODs) are widespread. Within bacteria, aerobes tend to contain Mn-SODs or both Mn-SOD and Fe-SOD, whereas strict anaerobes may have one Fe-SOD, or none at all (111). Mn-SOD (SodA) from Escherichia coli is flexible in binding Mn2+ or Fe2+ depending on growth conditions. As such, Mn-SOD prefers cognate Mn ion when cultivated in the presence of oxygen; whereas under anaerobic conditions, bacterial Mn-SOD accommodates ions of Fe resulting in dismutation-inactive enzyme. Excess Fe has also been linked to the formation of partially active Fe-Mn-SOD (hybrid SOD), wherein a dimer's subunits contain Mn2+ and Fe2+ in their active site resulting in partial activity (summarized in Table 1). Thus, it was proposed that the selectivity of a metal ion cofactor in the bacterial Fe/Mn-SODs is defined by the bioavailability of Mn or Fe (112).
The most recently evolved family, the Cu/Zn-SODs, utilize Cu along with Zn. The Cu/Zn-SODs are found only in certain bacterial species (113, 114) but are ubiquitous among higher eukaryotes (115, 116, 117). Humans express three Cu/Zn-SOD isoforms; the cytosolic SOD1 (117), the extracellular SOD3 (118), and the mitochondrial Mn-SOD (SOD2) (119, 120). They are absent in archaeal species and are considered to have appeared long after the evolution of the Mn/Fe-SOD family (107), correlating with the increase in bioavailable Cu and Zn (43, 103). The Ni-SODs are found in algae and predominantly marine species of bacteria and likely appeared after Mn/Fe-SODs, presenting a selective advantage in marine environments, as levels of Fe and Mn have diminished and concentrations of Ni remained relatively consistent (103, 121).
These examples illustrate that the appearance and accumulation of molecular oxygen throughout earth's evolution accompanied by a shift in the availability of transition metals induced a switch from Fe-containing SODs to new variants of this enzyme that adapted alternative metal ions as cofactors, as illustrated in Figure 3 (44, 122). Thus, the utilization of metal ions throughout extant biological systems is intrinsically linked to the employment of molecular oxygen for fundamental processes in aerobic biology and the management of the associated toxicity of ROS. In addition, extant systems can be informative of the chemical environments in which they arose, even when these are drastically different from those in which they now function.
Cation replacement examples: Focus on Fe, Mn, and Mg
The exchange of Fe and Mn in functional enzymes is illustrated by a few prominent examples. The mononuclear SODs demonstrate the flexible utilization of either Mn or Fe within an isoform, whereas the essential RNR enzyme family, which employs a dinuclear cation pair, includes a further interesting example of an ancient and conserved enzyme, which utilizes different metal ions, and the expression of distinct forms with differing specificities. Finally, recent studies have identified an ability of the ribosome to undergo interchange of bound metal ions (20, 123). These examples, discussed in detail later, suggest that the transition metal interchangeability occurs across different molecular structures and can be informative about the conditions under which they developed.
Mn-SOD, Fe-SOD, and cambialistic Mn/Fe-SOD
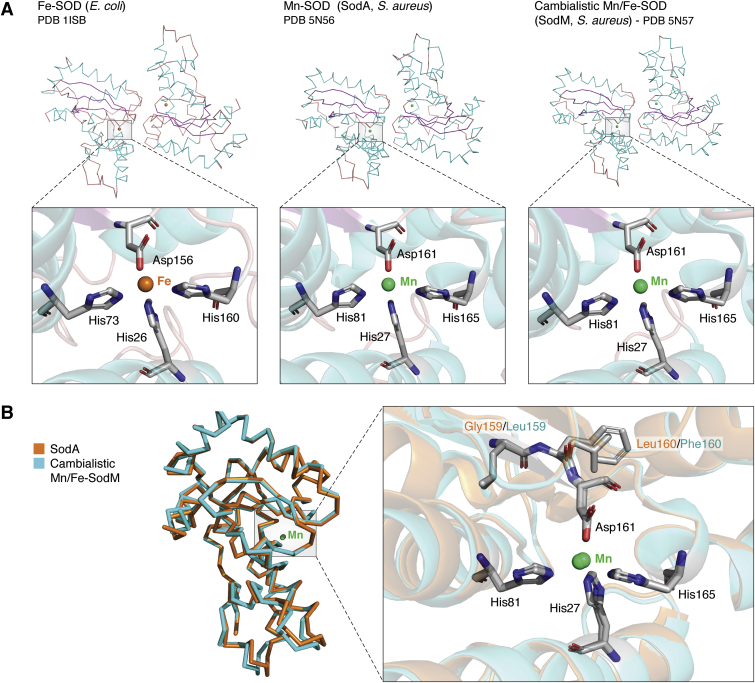
The Fe-SODs and Mn-SODs are highly homologous in sequence and three-dimensional structure (124, 125), whereby the amino acid residues and the funnel that allows a substrate (O2•−) access to the metal ion are identical. Catalytically active sites of both SODs contain three histidines and one aspartic acid that bind the metal ion cofactor (Fig. 4A, left and middle panels). In addition, the Fe and Mn ions also bind to a solvent molecule (water or hydroxide) that is engaged in the formation of hydrogen bonds and, together with histidines and aspartic acid, participates in the coordination of either cation in an unusual distorted trigonal bipyramidal geometry around the metal center, as depicted in Figure 2B (126, 127, 128).
Figure 4.
Structural properties of Mn-specific, Fe-specific, and cambialistic Mn/Fe-SODs.A, comparison of the crystal structures (top) and the metal-bound active sites (bottom) of Fe-SOD from Escherichia coli, Mn-SOD (SodA) and cambialistic Mn/Fe-SOD (cam-SOD, SodM) from Staphylococcus aureus. Metal ions (shown as colored spheres; orange for iron ion and green for manganese ion) are coordinated by three histidine (His) residues and one aspartic acid (Asp) residue and a solvent molecule (not shown). The figure is generated using PyMol; Protein Data Bank IDs are indicated in the figure. B, superimposed ribbon representation of Mn-specific SodA (orange) and cambialistic Mn/Fe-SodM (cyan) monomer structures from Staphylococcus aureus is shown on the left. Superimposed structures of active centers of SodA and SodM are shown on the right. Four residues from the primary coordination sphere (His27, His81, Asp161, and His165, black lettering) coordinate a metal ion (Mn is shown as a green sphere). Two residues from the secondary coordination sphere (SodA: Gly159, Leu160, orange lettering; SodM: Leu159, Phe160, cyan lettering) provide cambialistic properties to SodM. SOD, superoxide dismutase.
Despite this remarkable similarity, most Fe-SOD and Mn-SOD enzymes are only functional when bound to their cognate metal ion (Table 1), illustrating their high specificity to metal cofactor (129, 130, 131). Moreover, it has been revealed that the replacement of Mn with Fe in Mn-SODs from mammals (SOD2) and E. coli (SodA) results in the generation of an alternative isoform (Fe-SOD2s) with peroxidase prooxidant activity, thereby promoting oxidative stress, presumably via utilization of H2O2 ((132, 133) and Table 1). These findings demonstrate that, in some cases, incorporation of a noncognate metal ion does not disable an enzyme but switches its function, highlighting the biological significance of the metal selectivity process.
What is the mechanism behind the metal ions selectivity of SODs? Being structural components of SODs, Fe and Mn ions cycle between +2 and +3 oxidation states during O2•− turnover; however, these oxidation states correspond to different d-electron configurations for Mn and Fe, resulting in distinct +3 to +2 reduction characteristics. It is believed that these differences are compensated by protein components, such as specific amino acids of a secondary coordination sphere of Fe-SOD or Mn-SOD. Of note, the first (inner) coordination sphere refers to the array of direct interactions of a ligand with a metal ion, whereas the secondary (outer) coordination sphere consists of ions that interact with the first coordination sphere, without direct binding to a metal ion. Although Mn-SODs and Fe-SODs share identical metal ion–containing catalytically active sites (Fig. 4A, left and middle panels), different amino acid residues lie adjacent to it and are responsible for metal ion specificity and activity that occur without major structural reorganization of the active center. For example, glutamine from the second coordination sphere of SODs was found to be involved in determining the redox potential of the active site, thus, impacting the metal cofactor selection process. As such, Gln69/Fe-SOD and Gln146/Mn-SOD from E. coli promote metal ion specificity (130, 131, 134, 135), and a similar observation was made of Sod2 (Mn-Sod) from S. cerevisiae (136). Furthermore, the crystal structures of native Mn-Sod2 and artificial Fe-Sod2 from S. cerevisiae at 2.05 and 1.79 Å resolution, respectively, demonstrated no significant alteration in the active site or overall structure upon binding the non-native metal Fe, and identified Asp163 and Lys80 as those responsible for the metal specificity of Mn-SOD (136). Another residue important for the SOD specificity is Thr165 present in Fe-SODs from E. coli (whereas a Gly residue occurs at this position in the majority of Mn-SODs (137, 138)), whereby swapping Thr165 and Gly165 in Fe-SOD (SodB) and Mn-SOD (SodA) changes the metal cofactor preference ((137, 139) and Table 1).
Despite the high metal ion specificity of Mn-SODs and Fe-SODs, some organisms have acquired an alternative form of an enzyme that can accommodate Mn or Fe ions in the catalytic center and retain enzymatic activity. These metal cofactor flexible SODs have been termed cambialistic or cam-SOD (140). Cam-SODs were found in microorganisms adapted to different growth conditions, including microaerophiles, aerobes, obligate anaerobes, and thermophiles. This interesting phenomenon raises questions of how and why some of these enzymes developed cambialistic properties.
In this regard, SodM (cam-SOD) from the Gram-positive opportunistic pathogen Staphylococcus aureus has become an informative experimental model, as it allowed direct comparison with the strictly Mn-dependent SodA (Mn-SOD) isoform (141, 142, 143). Thus, it has been shown that Mn- and Fe-bound cam-SOD exhibits comparable enzymatic activities (141). Moreover, the X-ray diffraction analysis performed with Mn-loaded or Fe-loaded SodA and SodM demonstrated only minor deviations in the catalytic center architecture and metal binding physicochemical properties, whereby metal ions (Mn or Fe) are coordinated by His27, His81, Asp161, and His165 ((144) and Figure 4A, middle and right panels). These structural similarities suggest that cambialism is not provided by the inner sphere coordination geometry but relies on differences in the secondary coordination sphere. Mutational analysis has identified two amino acids present in positions 159 and 160 (Fig. 4B) that vary between SodA (possesses Gly159 and Leu160) and SodM (Leu159 and Phe160) and make no direct contact but are in close proximity (<10 Å) to a metal cofactor (144). Significantly, swapping these amino acids between SodA and SodM did not affect active center structures but enabled cambialistic properties to SodA (144). It was proposed that amino acid side chains in positions 159 and 160 are responsible for changes in the reduction potential of the metal ions, likely underlying the mechanism of catalysis governed by Mn and Fe ions (144). This example indicates that subtle sequence alterations near the active site impose metal specificity on one isoform or allow flexibility in the other. One possible explanation for the impact of the amino acid residues equivalent to 159 and 160 of S. aureus is their role in assembling the appropriate hydrogen-bonding network that includes a metal-coordinated solvent, as described in Ref. (139). However, other studies demonstrated that solvent proton positions are similar in the structure of Mn-SOD and cam-SOD (145). These discrepancies call for further evaluation of the mechanisms by which the secondary sphere amino acids control redox tuning in cooperation with Mn and Fe ions.
The key Leu159 and Phe160 residues of cam-SODs are highly conserved within the lineages of the S. aureus tree, including Staphylococcus argenteus and Staphylococcus schweitzeri (146). Thus, it has been proposed that cam-SOD arose from a redundant gene encoding a second Mn-SOD via evolutionary-enforced mutagenesis (144, 147). Such an Fe-to-Mn switch likely occurred in response to diminished Fe bioavailability during oxygenation of the atmosphere (13), accompanied by Fe engagement in Fenton chemistry with O2•−. Furthermore, the appearance of modern cam-SODs that are able to utilize both Mn and Fe allows adaptation to a plethora of stresses, including oxidative stress and nutrient starvation, such as Mn scarcity (141, 148), representing elegant stress-resistance strategies (149) that are discussed later.
The R2 subunit of class I RNRs
The RNRs are an ancient enzyme group uniquely responsible for the production of deoxyribonucleotides (dNTPs) from ribonucleotide precursors (150, 151). Different cation pair requirements have evolved in isoforms of the RNR R2 subunits. Fe or Mn cations are coordinated by this subunit, with the different metal ions facilitating activity in differing oxidative conditions. A high level of functional interchangeability between Fe or Mn cations has complicated the elucidation of the precise mechanism of the R2 subunit function. The chemically demanding redox reaction catalyzed by RNRs is essential in most organisms as a central controller of DNA replication, positioning these enzymes as potent targets for anticancer and antiviral drugs (152, 153). The implications of metal selection in these enzymes in pathogens are discussed later.
Three classes of RNR enzymes exist, all of which involve transition metal cofactors for radical generation and differ in their activity in aerobic or anaerobic environments. Class I RNRs, which are discussed here, are oxygen dependent and generate the catalytic radical via their dimetal binding R2 subunit.
The R2 subunits of class I RNRs are ferritin-like proteins (154, 155). These subunits contain a dinuclear metal binding site where the radical species are generated (17, 156). Metals are incorporated as divalent cations and oxidized to higher oxidation states as part of the activation mechanism. In the prototypic E. coli enzyme of the RNR family, the R2 subunit coordinates two Fe ions, defining class Ia subgroup, which also includes the human and yeast RNRs. The mechanism utilizes the electrons of the bound di-Fe in the reduction of molecular oxygen, resulting in an active state with a tyrosine radical (Tyr122) and an oxidized Fe3+ ion pair (17). While Mn2+ can bind in place of Fe2+, it does not support catalytic activity (157).
R2 proteins from other organisms have since been found to employ a pair of Mn ions (class Ib) or a “heterobimetallic” mixed Mn/Fe cofactor (class Ic) (158, 159, 160). While the di-Fe R2 proteins are damaged by H2O2, the Mn/Fe-binding R2 protein from the human pathogen Chlamydia trachomatis is resistant, and, in fact, becomes oxidized and activated in the presence of H2O2 (161). Interestingly, the C. trachomatis R2 subunit also has significant adaptations to the residues involved in the transfer of the radical from the metal site to the active site, lacking the highly conserved tyrosine, which usually harbors the radical following oxidation. It was proposed that because of the differing redox properties of the Mn in the metal ion pair, Mn can exist in an Mn2+ state upon oxidation and fulfill the oxidant function of the tyrosine radical. At the same time, Fe is unable to participate (162).
An interesting structural feature of the R2 proteins from the Mn/Fe or Mn/Mn binding RNRs allows selective self-assembly with the appropriate cations, despite the generally higher concentration of Fe in cells and the higher predicted stability of Fe2+ complexes over Mn2+ complexes (160, 163). Minor changes to coordinating residues can switch the binding preference between the two transition metals. A single mutation of a residue in the second coordination sphere alters the specificity of class Ib R2 subunit of Bacillus anthracis, such that under aerobic conditions, the protein is populated with an Mn/Fe ion pair (163).
The investigation of R2 proteins, which natively coordinate a di-Mn cofactor, proved problematic. Despite in vivo evidence that Mn was required, attempts to reconstitute the enzyme with Mn in vitro produced no activity, whereas introducing Fe was able to restore some enzymatic capacity. This is due to a requirement of the di-Mn mechanism for an additional cofactor, a flavin-mononucleotide coenzyme NrdI, which is present in all organisms expressing class Ib RNR. The oxidation of bound Mn2+ or Fe2+ is required for the activation of the R2 protein, and whereas Fe2+ reacts directly with O2 to become oxidized, Mn2+ requires NrdI to produce the oxidizing radical (164).
Ribosomal Mg is a subject to replacement
As discussed earlier, the geological data indicate that the changes in earth's atmosphere and metal availability correlate with the biological utilization of metal ions by various biomolecules, including ribosomes (Fig. 3). Similarly to ancient SOD enzymes that appeared early during earth's evolution and underwent a significant divergence over time, the ribosomal core serves remarkably well as a tool to investigate early molecular biology and biochemistry, as it remained largely invariant since the last universal common ancestor (165), yet appeared to go through significant changes in respect of the utilization of metal ions (20, 166).
The contemporary ribosome coordinates Mg2+ extensively for structural stabilization, with X-ray analysis indicating ∼200 Mg2+ ions are associated with the large subunit alone (4), and further studies indicating as many as 1000 Mg2+ sites on the entire ribosome (167). At least six distinct Mg2+ binding structures were evident (20, 167), aiding in folding and assembly of the rRNA (168), mediating interactions with tRNA, mRNA, and stabilizing the intersubunit interface (169). Mg ions maintain a kink between the P-site and the A-site of the ribosome (170), and microclusters of Mg2+ pairs within the large subunit stabilize the peptidyl transfer center (171).
The high level of conservation of the ribosome since its evolution 3 to 4 billion years ago (172) leads to the idea that Mg2+ may not have been the original cation utilized in ribosomal structures, which first appeared prior to oxidation of the environment with less abundant Mg2+ and more prevalent ions of Mn2+ or Fe2+ (20, 173). Direct proof for this hypothesis came from a recent study by Bray et al. (20), wherein an ancient earth's atmosphere was replicated in an anoxic chamber with a 98% Ar and 2% H2, and lyophilized ribosomes were reassembled in the presence of Mn2+ or Fe2+ instead of Mg2+ ions. This elegant approach demonstrated that ribosomes retained their translational competency when their structure was rebuilt in vitro in the presence of alternative rRNA-stabilizing cations, suggesting metals' interchangeability within a prokaryotic ribosome (20). The conservation of the ribosome may mean that Mn2+ or Fe2+ represent ancient binding partners, which remain functional in extant organisms. However, the physiological relevance is dependent on metal availability within the cell. In addition, the presence of oxidant species in the present environment, which would not have been a concern in an anoxic archaean earth, produces the risk of damage to RNA structures closely associating with Fe2+ by Fenton-induced ROS.
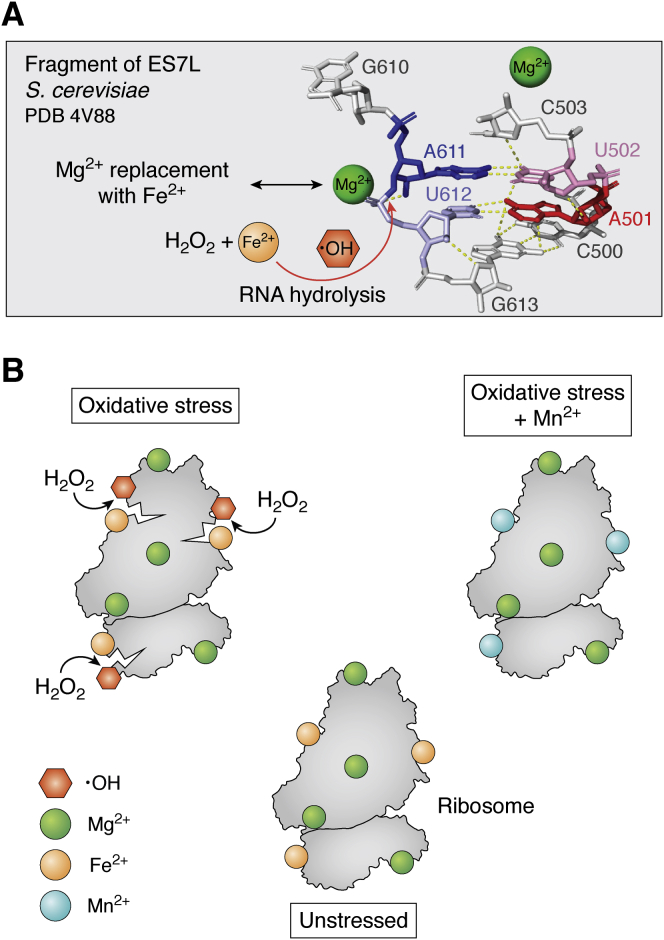
Indeed, a genetic screen of the S. cerevisiae deletion strains conducted in our laboratory has identified grx5Δ and yfh1Δ strains as highly susceptible to oxidant-induced rRNA scissions (21). Both these deletion strains contain a high level of labile Fe (21, 123), which prompted investigation of the possibility that rRNA hydrolysis is accomplished via the site-specific Fenton reaction. By devising an in vitro assay, the rRNA cleavage pattern observed in oxidant-treated grx5Δ cells was recapitulated in vitro with ribosomes purified from wildtype cells grown under normal nutrient-rich conditions, Fe(NH4)2(SO4)2 and ascorbic acid used as a prooxidant. The intensity and number of rRNA cleavage events were dependent on a concentration of Fe present in the in vitro reaction, as well as in strains carrying various levels of labile Fe, and correlated with cell viability. Interestingly, treatment of ribosomes with ascorbic acid alone still resulted in low-level cleavage within the expansion segment 7 of the large ribosomal subunit 60S (ES7L) of 25S rRNA (Fig. 5A), suggesting that even under normal growth conditions, ribosomes retained an ability to replace Mg2+ with Fe2+ at the selected sites. Similar low-intensity ES7L 25S rRNA cleavage has been detected upon treatment of cells with low doses of H2O2 (a condition, wherein H2O2 functions as a signaling molecule (174)), thus promoting resistance to subsequent acute oxidative stress (175). The Fe-dependent ES7L cleavage did not affect the translational activity of ribosomes, further suggesting a role of Mg2+-to-Fe2+ replacement within ES7L in the adaptive response to stress (175). Structural data (21) demonstrated that 2 Mg2+ ions exposed to the solvent side are located ∼6 to 8 Å away from the ES7L cleavage site (A611↓U612), implying that •OH radical generated during the Fenton reaction is in close proximity to the sugar phosphate backbone (Fig. 5A). Furthermore, we identified that Fe2+-mediated oxidant-dependent rRNA hydrolysis (175) was mitigated by increased Mn2+ availability, which seemingly can compete with Fe2+ for binding sites, resists induction of the Fenton reaction and, thus, accomplishes a protective role (123). The impact of Mn and Fe binding to ribosomes is outlined in Figure 5B. These results were consistent with the protective effect of replacing Fe2+ with Mn2+ described in E. coli under oxidative stress (176, 177).
Figure 5.
rRNA is a substrate for the site-specific Fenton reaction.A, the highly conserved part of the expansion segment 7 (ES7L) of the large ribosomal subunit 60S from Saccharomyces cerevisiae. The depicted fragment corresponds to nucleotide pairs A501:U612 and U502:A611 of the ES7L and is shown in colors, surrounding bases are shown in gray. Dashed lines indicate predicted polar contacts between the four conserved bases (A501:U612 and U502:A611). Mg2+ ions are shown as green spheres. The Protein Data Bank file 4V88 was used. Replacement of Mg2+ with Fe2+ (shown as an orange sphere) powers the rRNA-localized Fenton reaction under oxidative conditions, whereupon the hydroxyl radical (•OH) is formed. •OH hydrolyses the sugar phosphate backbone at the specific site between A611 and U612. Modified from Ref. (21). B, Mg2+, Fe2+, and Mn2+ interchangeability on the ribosome. Unstressed cells contain active ribosomes, which bind divalent metal cations throughout their structure (bottom). These are predominantly Mg2+ but include a number of Fe2+ ions. In oxidative stress conditions, Fe2+ participates in Fenton reactions generating •OH radicals, which cleave the rRNA and fragment the ribosome (top left). However, if sufficient Mn2+ is available, the Fe2+ is displaced and Mn2+ occupies these sites. Under oxidative stress, a ribosome that coordinates Mn2+ in place of Fe2+ is not subjected to the generation of hydrolytic radical species and, thereby, is resistant to the stress-induced damages (top right). Modified from Ref. (123).
Taken together, it seems reasonable to propose that the translation machinery maintained an ability to associate with the transition metal cations that stabilized its structure. Given that eukaryotic ribosomes retained the capability to replace Fenton-resistant Mg2+ with Fenton-active Fe2+ through the course of evolution, it is possible that this newly identified quality of the protein-translating machinery plays a regulatory role during gene expression as a means of adjustment to environmental changes.
The work described in this section was conducted and published recently, and, therefore, many questions have remained unanswered. It will be important to map other cleavage sites, dissect the molecular mechanics of stress adaptation accomplished by Fenton-cleaved ribosomes, investigate site-specific Fenton cleavages within human ribosomes, and elucidate human disease relevance. Furthermore, we would like to highlight that the discovery of metal interchangeability within the ancient molecular structure of the ribosome identifies an overlooked yet promising model molecule to study the evolution of biomolecules. Such observations recontextualize the ribosome, which is often considered as a stable entity resistant to changes or environment-induced perturbations, as being subject to metal ion replacement depending on genetically or environmentally induced changes in metal homeostasis. Thus, researchers should take special considerations while studying fundamental processes, such as translation and translational control.
The largely invariant core of the ribosome and the persistence of the translation machinery since the last universal common ancestor are central to many hypotheses regarding cellular evolution (172, 178). The oldest and most conserved part of the ribosome is free of protein and supports an early phase of biology, which was dominated by RNA. It has been demonstrated that RNA species are capable of carrying out Fe and Mn-mediated redox mechanisms (179, 180). Under anoxic conditions, a single-electron transfer reaction, much like those fundamental to metabolism, can be performed by RNA coordinating Fe2+ instead of Mg2+ (180). It is possible to substitute Mn2+ for Mg2+ in the hammerhead ribozyme, which is a small self-cleaving RNA that catalyzes reversible cleavage and ligation reactions at a specific site. Using 2′-mercaptonucleosides as biochemical probe and the “metal specificity switch” approach, it was found that Mn2+-to-Mg2+ substitution enhances enzymatic activity of the hammerhead ribozyme (179). These findings indicate the capability of RNA molecules to associate with a greater range of metal ions and possess more diverse catalytic abilities than are frequently observed in extant biology.
Interchangeability with other metal ions
While much of the research on this topic focuses on Mn and Mg replacement by Fe at metal binding sites, flexible binding certainly extends to other biologically relevant metal ions. Zn is a widely utilized metal ion in proteins and may have replaced Fe as a cofactor in many enzymes where it presents less of a risk of toxicity (37).
The Zn finger (ZF) domain represents one of the most widespread and diverse structural motifs in biology, interacting with nucleic acid, protein, and lipid targets (181), and the bound Zn cation can be subject to replacement, such as in the estrogen receptor (182, 183). In this enzyme, Cu or Ni substitution results in loss of function, whereas DNA binding capacity is retained with Co or cadmium (183). In the case of Cu, this example illustrates the potential for damage caused by uncontrolled Cu availability as it can deactivate enzymes by displacement of Zn, in addition to its contribution to the generation of •OH radicals. In addition, replacement of Zn2+ with Fe2+ in the ZF domain of the estrogen receptor does not abolish DNA binding but can induce generation of oxidative radicals and cause DNA damage (184). The impact of metal ion replacement in ZF domains was recently reviewed in detail in Ref. (181).
Another interesting example of Zn2+-to-Fe2+ replacement was recently shown for the well-conserved scaffold protein ISCU (Iron–Sulfur Cluster assembly enzyme) that is central in 2Fe–2S and 4Fe–4S cluster synthesis and maturation, and, thus, has been the subject of extensive investigation (reviewed in Ref. (185)). Bound to Zn2+, Zn-ISCU catalyzes Fe–S clusters assembly inefficiently, whereas replacing Zn2+ with Fe2+, which can participate in an alternative redox-dependent reaction, generates “Fe-loaded” ISCU with a robust enzymatic activity (186). Gervason et al. (186) proposed that Fe-ISCU is the physiologically relevant form of the enzyme; however, further studies are required to dissect two mechanisms of Fe–S cluster assembly governed by Zn and Fe-loaded ISCUs. Nevertheless, this work revealed that, unlike for the ZF domain of the estrogen receptor (discussed previously (184)), replacement of Zn2+ with Fe2+ within ISCU might play a beneficial role in cellular physiology (186, 187).
In the Cu/Zn-SODs, the Cu ion plays the catalytic role, whereas Zn has a structural role, although it can be also required for catalytic activity that is maintained over a wide pH range (188, 189, 190). Cu ions have been shown to occupy empty Zn-binding sites within an SOD dimer, potentially inhibiting activity of the enzyme ((191) and Table 1). More recently, new members of this SOD class adapted to limited Zn availability conditions have been identified. These Cu-only SODs, found in both prokaryotes and eukaryotes, have an enhanced dimer interface, which provides stability and leaves them unable to bind Zn (192, 193), whereas other amino acid adaptations (Glu110) fulfill the electrostatic role of Zn (194).
Other metals with unpaired d-electrons, which have roles in biological systems, such as Co and Ni, may also replace Mg or Mn. Ni is not widely used as a cofactor in extant biochemistry, although it may have been an important catalyst in early biology. Within the small number of known Ni metalloenzymes are several interesting examples (reviewed in Ref. (195)), including Ni-SODs that are inactive upon Ni2+ replacements ((196, 197) and Table 1) and a conserved acireductone dioxygenase of the methionine salvage pathway, which binds either Ni2+ or Fe2+, and has distinct catalytic activities depending on the cofactor (198). Other examples are isoforms of mandelate racemase that prefer Mg but can also function with several other cation alternatives, including Mn, Ni, or Co (95, 199).
Adapting to changes in metal availability
Pathogenic bacteria depend on their antioxidant systems as they aim to maintain a foothold in the chemically challenging host environment. The distinct families of SOD can be employed in combination to facilitate an effective antioxidant defense through changing metal availabilities, or as in some examples described earlier, a single isoform can be functional with multiple metal ions, conferring a similar adaptability. The Streptomyces genus contains Ni-SODs, sometimes along with an Fe-SOD isoform, which is expressed only when Ni is unavailable (121, 200). In many clinically relevant strains, the Fe-containing SOD has been lost and only the Ni-SOD remains, possibly reflecting the advantage of lowering requirements for Fe in mammalian pathogens to avoid host-imposed Fe restrictions (121, 201).
Protective roles for Mn
The effect of metal limitation can be minimized by increased uptake. This is seen in S. aureus with two types of Mn uptake proteins, the ABC transporter MntABC and the Nramp-related MntH, which compete for Mn and enhance pathogen survival (202). Interestingly, these transporters are physiologically highly selective of Mn over Fe and other divalent cations (99), whereas Nramp family transporters are often associated with broad metal ion transport function. The exception to the Mn2+ specificity of MntH is related to the transport of Cd2+, which is also imported and contributes to toxicity (203), and which also inhibits the ABC-type Mn transporters (204), illustrating the challenges in selecting for strict metal targets. Studies in E. coli showed that MntH expression is regulated by OxyR as part of a battery of oxidative stress response genes that enhance Mn availability and limit Fe-mediated damage. These include catalase and peroxidase enzymes, the ferritin-like protein Dps that sequesters Fe2+, the Fe-uptake repressor Fur (205). It was shown that the activity of Mn-SOD is dependent on Mn import by MntH, and in unstressed cells, the low level of the transporter is a limiting factor of the activity of the SOD enzyme (205).
Mn-SOD from E. coli and human can also bind Fe2+ when it is present in excess relative to Mn2+. This metal cofactor replacement results, however, in formation of inactive Fe-bound enzyme ((206, 207) and Table 1). It was suggested that the cell's ability to appropriate Mn confers resistance through protection of numerous metalloproteins, which normally bind Fe2+ (177, 205). Another example for the selection of a particular metal ion by biosystems is an increased sensitivity of the pentose phosphate pathway to H2O2 that occurs because of the inactivation of an Fe-coordinating enzyme, ribulose-5-phosphate 3-epimerase (Rpe). It was shown that the enzyme-bound Fe2+ ion dissociates upon oxidation. Although restoring Fe2+ back to the binding site returns activity to most Rpe enzymes, a subset is damaged by the oxidation. This ultimately leads to total loss of Rpe activity through successive cycles of Fe binding and oxidation. The stress-induced increase in Mn import mitigates this mechanism, as Mn2+ can bind to and activate Rpe in place of Fe2+ without being sensitive to the presence of H2O2 (177). E. coli increase intracellular Mn2+ levels by over 10-fold in response to H2O2 (205), indicating that metal cofactor selection may be mediated by cellular control of relative concentrations. Interestingly, in vitro experiments showed that Zn2+ binds Rpe with greater affinity than Mn2+ or Fe2+ and is also present in the cell at higher concentrations. Thus, it is hypothesized that the wealth of other Zn2+-competing ligands in the cell, including glutathione, effectively reduces the availability of Zn to Rpe and other enzymes (177). Subsequent work established that this is not a unique case, as further nonredox metalloenzymes, which are likely to coordinate Fe2+ in vivo, are damaged by H2O2 and protected by increased Mn2+ import conducted by MntH, as well as by sequestration of Fe2+ by Dps (176).
The Mn-utilizing class Ib RNRs are well represented in pathogenic bacteria, including B. anthracis, S. aureus, and E. coli (158, 208), and likely arose through an evolutionary process of adaptation to oxidative stress and Fe restriction inflicted by hosts. Indeed, many human pathogen genomes encode RNRs from at least two classes (209, 210, 211). For example, the opportunistic pathogen Streptococcus sanguinis requires its class Ib RNR and Mn for aerobic growth and virulence. In contrast, class III RNR, which depends on an Fe–S cluster, is necessary for growth under anaerobic conditions (211).
Metal-free alternatives
An alternative response to circumvent metal limitation involves utilizing surrogate proteins, which can substitute for the usual metal-dependent species and maintain activity while forgoing the metal entirely. The ribosome has been linked to the response to Zn limitation in bacteria (212). In response to Zn depletion, a number of Zn-binding ribosomal proteins are replaced with Zn-free homologs, making a significant contribution to the available Zn content of the cell (212, 213), although, perhaps, with a loss of translational activity (214). Other similar examples of metal-free homologs include an Mn-independent variant of phosphoglycerate mutase expressed in S. aureus, which allows glycolysis to continue during host-imposed Mn limitation (215).
Cu-only SODs are present in the pathogens Mycobacterium tuberculosis and Candida albicans, where they contribute to virulence by detoxification of host-derived oxidant species (216, 217). The Cu binding site is also more open than in the Cu/Zn-SODs, and in C. albicans, the enzyme is secreted prior to binding the metal ion, whereas eukaryotic extracellular Cu/Zn-SODs are usually charged with Cu and Zn within the cell. The combination of independence from Zn availability and the capability to bind extracellular Cu ions may be advantageous in maintaining catalytic activity (217).
Bacterial infection and host metal sequestration mechanisms
Transition metals are essential for microorganisms like bacteria; thus, during nutrient limitation, mechanisms for metal acquisition become critical for bacterial cell survival. To overcome metal scarcity imposed by bacterial hosts, bacteria have evolved an elegant mechanism of synthetizing metal ion scavengers, known as metallophores, that possess high affinity to metal ions. Metallophores belong to a family of small molecules that bind various metal ions in the extracellular environment, following by active import of chelated metal complexes inside the bacterial cells. Normally, metallophores are divided into different groups based on their affinity toward a specific metal, such as siderophores for Fe, chalcophore for Cu, manganesophore for Mn, nickelophore for Ni, and zincophore for Zn ((218), references therein). Staphylopine produced by the pathogenic bacteria S. aureus stands out as a broad-spectrum affinity metallophore, as it is able of chelating various transition metals (219), thus efficiently overpowering host immunity.
Another nutrient limitation–induced strategy employed by the innate immune system involves calprotectin, which is released at infection sites from epithelial cells and neutrophils (212, 220). Calprotectin has previously been characterized as a chelator of Mn and Zn, while recent work establishes that it also actively sequesters Fe, and calprotectin-treated media were found to have reduced availability of all three metals (221). Neutrophils can contain very high calprotectin levels, sometimes accounting for as much as half of all protein content of the cytoplasm (222), which indicates the utility of the broad antimicrobial effect of metal sequestration. Proteins related to calprotectin of the S100 family of calcium-binding proteins are also important in host defense (27).
The metal sequestering action of calprotectin is exploited for competitive advantage in the gut pathogen Salmonella typhimurium, which expresses a high-affinity Zn transporter during infection. This allows it to survive the Zn restriction imposed by the host, whereas other commensal bacteria of the microbiota are more sensitive, resulting in reduced competition at the intestinal mucosa (223). The ability to adapt is particularly important for the transition between the different host environments exploited by opportunistic pathogens, such as group A Streptococcus. Recent work has highlighted the importance of metal homeostasis for survival of these bacteria in host organisms. The role of metals in the antioxidant defenses is important even while in a nonpathogenic state but becomes critical when the bacteria become invasive and induces an immune response (26, 30). The activity of calprotectin during S. aureus infection leads to a reduction in pathogen SOD activity, increased bacterial O2•− levels, and improved clearance by the host immune system (148). The metal binding site of calprotectin is required for the antimicrobial effect, whereas supplying excess Mn2+ protects the pathogen against oxidative stress (148).
Metals in viral infections
Viral genome replication and protein synthesis require trace metals, and their availability promotes the expansion of viral populations. Fe, in particular, has been studied for its association with viral infections, including those of HIV-1, human cytomegalovirus, hepatitis B and C viruses, and herpes simplex virus 1. Some viruses (arenaviruses and mouse mammary tumor virus) have been shown to target Fe-rich cells by utilizing Fe-import machinery for cell entry or manipulate host Fe homeostasis to their benefit. The availability of essential metals, including Fe, Mn, and Zn, can also influence the progression or resolution of many viral infections. This can occur as a result of both the immune system effects on metals and their direct impact on viral pathogenicity (224, 225, 226). Much is still unknown about the relevance of trace metal ions to virus–host interactions. Computational analysis has indicated that Zn-binding and Mn-binding domains are prevalent in viral proteomes and that many viral metal-binding proteins target host metal homeostasis regulators (227). The relevance of metal ions to the immune response to the SARS-CoV2—a cause of an unprecedented COVID-19 pandemic—has been discussed recently (228). As such, serum Fe levels have been linked to mortality risk in SARS-CoV2 patients (229, 230). Based on sequence analysis, it has been suggested that the cytoplasmic tail of the SARS-CoV2 spike protein may interact with the Fe exporter ferroportin (231). Whether Fe dysregulation in these patients is a result of inflammation or a contributing factor to pathogenesis remains currently unclear (230). However, most viruses require Fe as part of their replication cycle, and beneficial effects have been observed by treating other RNA virus infections with Fe chelators or inducing ferroportin expression to increase Fe efflux. The possibility of targeting SARS-CoV2 by limiting Fe availability is under investigation (232). At least two viral proteins encoded by the severe acute respiratory syndrome coronaviruses genomes (that are conserved in SARS-CoV2) bind Mn and function with other divalent metal cofactors. These include an endonuclease Nsp15, which promotes immune evasion and has shown activity with coordinated Mg2+ (233). Furthermore, an essential RNA-dependent RNA polymerase (RdRp) Nsp12, which shares binding site homology with an RdRp from poliovirus, was active with a broad range of divalent ions, such as Mg2+, Mn2+, and Fe2+ (234). Recent work has found that the catalytic subunit Nsp12 of the Nsp12–Nsp7–Nsp8 complex is able to accommodate two Fe–S clusters in the Zn-binding sites. Moreover, it was found that anoxically purified Fe–S–Nsp12 RdRp complex is even more prominent in the RNA template binding capacity and the polymerase activity than that assembled aerobically with two Zn ions (235). Thus, SARS-CoV2 RdRp is a bona fide subject of metal interchangeability relevant to the COVID-19 pandemic. Interestingly, nitroxide-enforced oxidation of Fe–S clusters caused their disassembly and blocked SARS-CoV2 replication in cell culture, raising a possibility that these clusters may serve as preferable cofactors for the SARS-CoV2 RdRp (235). Further investigation into roles for metal replacement strategies in viral infection may be beneficial for developing both specific and broadly applicable antiviral treatments.
Metal imbalances as a causative factor of human disease states and metal chelation therapy
As they are key elements of numerous cellular biosystems (12), metal ion imbalances contribute to human disease states of inherited or acquired origin, such as chronic or aging enforced. For example, Fe deficiencies manifest in different types of anemias, whereby insufficient amounts of consumed/available Fe result in low levels of hemoglobin leading to insufficient oxygenation of body organs (reviewed in Ref. (236)). Metal imbalances are linked to pathophysiology of cardiovascular diseases, the number one killer worldwide (237, 238). For example, Fe deficiency is widespread in heart failure patients (239), whereas excessive Fe is linked to atherosclerosis and coronary heart disease (240, 241). Zn deficiency manifests in some malignancies, multiple sclerosis, and sepsis, wherein Zn scarcity was found to shift metal ions homeostasis leading to Fe and Cu overload (242, 243). In inherited blood disorders, such as β-thalassemia and sickle cell disease, mutations in hemoglobin subunit beta prohibit accommodation of Fe causing Fe overload (244). Mutation in the FXN gene encoding frataxin, the protein involved in Fe–S cluster assembly (245), cause Friedreich's ataxia (spinocerebellar degeneration) that is also associated with Fe overload (246). Another autosomal recessive disease, the Wilson disease (also known as hepatolenticular degeneration), affects primarily the liver and basal ganglia of the brain, and is caused by mutations in the ATP7B gene and generation of defective Cu transport protein leading to Cu build up (247, 248). A growing body of evidence has revealed that metal imbalances, such as Fe and Cu overload (249), Zn deficiency (250), manifest in numerous neurodegenerative disorders, such as Parkinson's disease, Alzheimer's disease, and Huntington's disease and are linked to enhanced oxidative stress that causes protein aggregation.
Having recognized that metal ions overload is a causative factor of disease etiology and progression, molecules that possess high affinity for metal ions are used in clinic to trap and neutralize excessive or toxic metals. Thus, the metal chelation therapy (MCT) has been administered as one of the effective ways to fight transition metals' overload, along with poisoning of heavy metals (251). MCT has been developed and used in clinical practice since the 1970s. By forming a complex with metal ion through ionic and coordination bonding (detailed in Ref. (218)), metal chelators alter metals' chemical properties, making metal ions unavailable for biological activities within metabolic pathways. As such, deferiprone, deferoxamine, and deferasirox are used for the removal of Fe during treatment of thalassemia, myelodysplasia, and sickle cell anemia, and penicillamine is used to deplete excessive Cu during Wilson's disease therapy (252). Animal studies have revealed a power of EDTA as an MCT agent in treatment of diabetic cataract, a condition that is associated with oxidative damage of lens cells (253). In addition, recently developed panel of small molecules, known as neuropeptides, which are produced and released by neurons, has shown to be effective during treatments of neurodegenerative diseases, providing neuroprotection. Neuropeptides are able to chelate excessive ions of Fe and Cu and, therefore, reduce formation of metal-mediated amyloid aggregates (254). Several excellent recent reviews (251, 255) discuss metal ion chelators in great details in a context of disease treatments, including newly developed drugs that are currently in clinical trials (255).
Conclusions and future directions
The idea of a preferred cation associated with metal binding sites on biomolecules is strictly context dependent, as many of the examples discussed previously show. In fact, the chemical environment exudes a significant influence on metal ion chemistry. Technical challenges are commonly encountered when investigating physiologically relevant associations of biomolecules with metals (198, 202, 256). In a setting where enzyme activity can be measured in vitro, the strategy of chelating metal ions and then reintroducing them back into the biomolecule's structure, followed by testing for activity, is highly informative.
Here, we provide a new perspective relevant to the investigation of metal-bound biomolecules in the context of the metal interchangeability phenomenon. As such, we advise researchers to consider the particular cation that provides the highest activity to a biomolecule and those that may be functionally interchangeable and allow any activity, as they may be physiologically relevant under circumstances where ratios of available metals are disrupted or during stress. In more complex examples, such as in vitro experimentation to probe metal requirements, it is not possible, and both purification techniques and structural analyses are limited in their capacity to elucidate physiological metal ion interactions. In addition, several examples indicate that expressing alternative isoforms of biomolecules can circumvent metal limitation or oxidant damage, either by coordinating alternative metals that reduce the risk of damage or by going without the metal component altogether. The recent work on binding multiple metal cations to the ribosome expands on the established adaptability of protein enzymes in utilizing metals. A mounting body of evidence demonstrates that numerous metal-binding proteins maintain an amazing ability to adapt to metal flux and oxidative stress. The ubiquitous and conservative nature of these proteins and the ribosome sheds further light on the evolutionary pressures and geochemical shifts, thus, defining the relationship between biological systems, oxygen, and metals.
Further work is needed to understand the role of metal substitution on ribosomal activity and stability, as disrupted protein synthesis has broad impacts on human health. Changes in metal homeostasis are associated with many human diseases, including anemias and neurodegenerative disorders, in addition to the impacts on many pathogenic species and immune responses described in this review. Of current relevance, there is promising potential in exploring whether metal ion replacements can combat the propagation of viral infections, such as SARS-CoV2. Future work should also consider that flexibility of metal coordination in biomolecules may be underestimated and could have a broad reach across biological systems. Investigation into possible roles for metal ion interchangeability in fine-tuning cell behaviors and responses may reveal novel adaptive mechanisms.
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
We are thankful to Dimitri Pestov, Brandon Trainor, and Russell Sapio for the critical evaluation of this work and constructive comments on the article. We express our gratitude to Arnab Ghosh for providing original structure for Figure 5A.
Author contributions
D. G. J. S. and N. S. conceptualization; N. S. resources; D. G. J. S. and N. S. visualization; D. G. J. S. writing–original draft; D. G. J. S. and N. S. writing–review and editing.
Funding and additional information
This work was supported by the National Institutes of Health grant R01GM114308 (to N. S.), the New Jersey Health Foundation grant PC13-21CV (to N. S.), and the Osteopathic Heritage Foundation grant 67294 (to N. S.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Edited by F. Peter Guengerich
Contributor Information
Daniel G.J. Smethurst, Email: dansmethurst@protonmail.com.
Natalia Shcherbik, Email: shcherna@rowan.edu.
References
- 1.Bellenger J.P., Wichard T., Xu Y., Kraepiel A.M.L. Essential metals for nitrogen fixation in a free-living N2-fixing bacterium: Chelation, homeostasis and high use efficiency. Environ. Microbiol. 2011;13:1395–1411. doi: 10.1111/j.1462-2920.2011.02440.x. [DOI] [PubMed] [Google Scholar]
- 2.Garcia-Diaz M., Bebenek K., Krahn J.M., Pedersen L.C., Kunkel T.A. Role of the catalytic metal during polymerization by DNA polymerase lambda. DNA Repair. 2007;6:1333–1340. doi: 10.1016/j.dnarep.2007.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Guth-Metzler R., Bray M.S., Frenkel-Pinter M., Suttapitugsakul S., Montllor-Albalate C., Bowman J.C., Wu R., Reddi A.R., Okafor C.D., Glass J.B., Williams L.D. Cutting in-line with iron: Ribosomal function and non-oxidative RNA cleavage. Nucleic Acids Res. 2020;48:8663–8674. doi: 10.1093/nar/gkaa586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Klein D.J. The contribution of metal ions to the structural stability of the large ribosomal subunit. RNA. 2004;10:1366–1379. doi: 10.1261/rna.7390804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li L., Yang X. The essential element manganese, oxidative stress, and metabolic diseases: Links and interactions. Oxid. Med. Cell Longev. 2018 doi: 10.1155/2018/7580707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Steffens G.C.M., Soulimane T., Wolff G., Buse G. Stoichiometry and redox behaviour of metals in cytochrome-c oxidase. Eur. J. Biochem. 1993;213:1149–1157. doi: 10.1111/j.1432-1033.1993.tb17865.x. [DOI] [PubMed] [Google Scholar]
- 7.Thiele D.J. Metal-regulated transcription in eukaryotes. Nucleic Acids Res. 1992;20:1183–1191. doi: 10.1093/nar/20.6.1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yruela I. Transition metals in plant photosynthesis. Metallomics. 2013;5:1090–1109. doi: 10.1039/c3mt00086a. [DOI] [PubMed] [Google Scholar]
- 9.Maret W. The metals in the biological periodic system of the elements: Concepts and conjectures. Int. J. Mol. Sci. 2016;17:1–8. doi: 10.3390/ijms17010066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Crans D.C., Kostenkova K. Open questions on the biological roles of first-row transition metals. Commun. Chem. 2020;3:1–4. doi: 10.1038/s42004-020-00341-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zoroddu M.A., Aaseth J., Crisponi G., Medici S., Peana M., Nurchi V.M. The essential metals for humans: A brief overview. J. Inorg. Biochem. 2019;195:120–129. doi: 10.1016/j.jinorgbio.2019.03.013. [DOI] [PubMed] [Google Scholar]
- 12.Andreini C., Bertini I., Cavallaro G., Holliday G.L., Thornton J.M. Metal ions in biological catalysis: From enzyme databases to general principles. J. Biol. Inorg. Chem. 2008;13:1205–1218. doi: 10.1007/s00775-008-0404-5. [DOI] [PubMed] [Google Scholar]
- 13.Dupont C.L., Butcher A., Valas R.E., Bourne P.E., Caetano-Anollés G. History of biological metal utilization inferred through phylogenomic analysis of protein structures. Proc. Natl. Acad. Sci. U. S. A. 2010;107:10567–10572. doi: 10.1073/pnas.0912491107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Freisinger E., Sigel R.K.O. From nucleotides to ribozymes-a comparison of their metal ion binding properties. Coord. Chem. Rev. 2007;251:1834–1851. [Google Scholar]
- 15.Morris D.L. DNA-bound metal ions: Recent developments. Biomol. Concepts. 2014;5:397–407. doi: 10.1515/bmc-2014-0021. [DOI] [PubMed] [Google Scholar]
- 16.Sigel R.K.O., Vaidya A., Pyle A.M. Metal ion binding sites in a group II intron core. Nat. Struct. Biol. 2000;7:1111–1116. doi: 10.1038/81958. [DOI] [PubMed] [Google Scholar]
- 17.Högbom M. Metal use in ribonucleotide reductase R2, di-iron, di-manganese and heterodinuclear - an intricate bioinorganic workaround to use different metals for the same reaction. Metallomics. 2011;3:110–120. doi: 10.1039/c0mt00095g. [DOI] [PubMed] [Google Scholar]
- 18.Griese J.J., Roos K., Cox N., Shafaat H.S., Branca R.M.M., Lehtiö J., Gräslund A., Lubitz W., Siegbahn P.E.M., Högbom M. Direct observation of structurally encoded metal discrimination and ether bond formation in a heterodinuclear metalloprotein. Proc. Natl. Acad. Sci. U. S. A. 2013;110:17189–17194. doi: 10.1073/pnas.1304368110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Athavale S.S., Petrov A.S., Hsiao C., Watkins D., Prickett C.D., Gossett J.J., Lie L., Bowman J.C., O'Neill E., Bernier C.R., Hud N.V., Wartell R.M., Harvey S.C., Williams L.D. RNA folding and catalysis mediated by iron (II) PLoS One. 2012;7 doi: 10.1371/journal.pone.0038024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bray M.S., Lenz T.K., Haynes J.W., Bowman J.C., Petrov A.S., Reddi A.R., Hud N.V., Williams L.D., Glass J.B. Multiple prebiotic metals mediate translation. Proc. Natl. Acad. Sci. U. S. A. 2018;115:12164–12169. doi: 10.1073/pnas.1803636115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zinskie J.A., Ghosh A., Trainor B.M., Shedlovskiy D., Pestov D.G., Shcherbik N. Iron-dependent cleavage of ribosomal RNA during oxidative stress in the yeast Saccharomyces cerevisiae. J. Biol. Chem. 2018;293:14237–14248. doi: 10.1074/jbc.RA118.004174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maguire M.E., Cowan J.A. Magnesium chemistry and biochemistry. Biometals. 2002;15:203–210. doi: 10.1023/a:1016058229972. [DOI] [PubMed] [Google Scholar]
- 23.Weston J. PATAI’S Chemistry of Functional Groups. John Wiley & Sons, Ltd; 2009. Biochemistry of magnesium; pp. 1–45. [Google Scholar]
- 24.Bock C.W., Katz A.K., Markham G.D., Glusker J.P. Manganese as a replacement for magnesium and zinc: Functional comparison of the divalent ions. J. Am. Chem. Soc. 1999;121:7360–7372. [Google Scholar]
- 25.Cowan J.A. Metal activation of enzymes in nucleic acid biochemistry. Chem. Rev. 1998;98:1067–1087. doi: 10.1021/cr960436q. [DOI] [PubMed] [Google Scholar]
- 26.Jakubovics N.S. An ion for an iron: Streptococcal metal homeostasis under oxidative stress. Biochem. J. 2019;476:699–703. doi: 10.1042/BCJ20190017. [DOI] [PubMed] [Google Scholar]
- 27.Hood M.I., Skaar E.P. Nutritional immunity: Transition metals at the pathogen-host interface. Nat. Rev. Microbiol. 2012;10:525–537. doi: 10.1038/nrmicro2836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Imlay J.A. Where in the world do bacteria experience oxidative stress? Environ. Microbiol. 2019;21:521–530. doi: 10.1111/1462-2920.14445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sultana S., Foti A., Dahl J.U. Bacterial defense systems against the neutrophilic oxidant hypochlorous acid. Infect. Immun. 2020;88:1–17. doi: 10.1128/IAI.00964-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Turner A.G., Djoko K.Y., Ong C., lynn Y., Barnett T.C., Walker M.J., McEwan A.G. Group A Streptococcus co-ordinates manganese import and iron efflux in response to hydrogen peroxide stress. Biochem. J. 2019;476:595–611. doi: 10.1042/BCJ20180902. [DOI] [PubMed] [Google Scholar]
- 31.Habib H.M., Ibrahim S., Zaim A., Ibrahim W.H. The role of iron in the pathogenesis of COVID-19 and possible treatment with lactoferrin and other iron chelators. Biomed. Pharmacother. 2021;136:111228. doi: 10.1016/j.biopha.2021.111228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Valasatava Y., Rosato A., Furnham N., Thornton J.M., Andreini C. To what extent do structural changes in catalytic metal sites affect enzyme function? J. Inorg. Biochem. 2018;179:40–53. doi: 10.1016/j.jinorgbio.2017.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Valdez C.E., Smith Q.A., Nechay M.R., Alexandrova A.N. Mysteries of metals in metalloenzymes. Acc. Chem. Res. 2014;47:3110–3117. doi: 10.1021/ar500227u. [DOI] [PubMed] [Google Scholar]
- 34.Miličević A., Branica G., Raos N. Irving-Williams order in the framework of connectivity index 3χv enables simultaneous prediction of stability constants of bivalent transition metal complexes. Molecules. 2011;16:1103–1112. doi: 10.3390/molecules16021103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Irving H., Williams R.J.P. Order of stability of metal complexes. Nature. 1948;162:746–747. [Google Scholar]
- 36.Foster A.W., Osman D., Robinson N.J. Metal preferences and metallation. J. Biol. Chem. 2014;289:28095–28103. doi: 10.1074/jbc.R114.588145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cardenas-Rodriguez M., Chatzi A., Tokatlidis K. Iron–sulfur clusters: From metals through mitochondria biogenesis to disease. J. Biol. Inorg. Chem. 2018;23:509–520. doi: 10.1007/s00775-018-1548-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Srour B., Gervason S., Monfort B., D'Autréaux B. Mechanism of iron–sulfur cluster assembly: In the intimacy of iron and sulfur encounter. Inorganics. 2020;8:55. [Google Scholar]
- 39.Bonfio C., Valer L., Scintilla S., Shah S., Evans D.J., Jin L., Szostak J.W., Sasselov D.D., Sutherland J.D., Mansy S.S. UV-light-driven prebiotic synthesis of iron-sulfur clusters. Nat. Chem. 2017;9:1229–1234. doi: 10.1038/nchem.2817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Konhauser K.O., Pecoits E., Lalonde S.V., Papineau D., Nisbet E.G., Barley M.E., Arndt N.T., Zahnle K., Kamber B.S. Oceanic nickel depletion and a methanogen famine before the Great Oxidation Event. Nature. 2009;458:750–753. doi: 10.1038/nature07858. [DOI] [PubMed] [Google Scholar]
- 41.Robbins L.J., Lalonde S.V., Planavsky N.J., Partin C.A., Reinhard C.T., Kendall B., Scott C., Hardisty D.S., Gill B.C., Alessi D.S., Dupont C.L., Saito M.A., Crowe S.A., Poulton S.W., Bekker A., et al. Trace elements at the intersection of marine biological and geochemical evolution. Earth-Sci. Rev. 2016;163:323–348. [Google Scholar]
- 42.Williams R.J.P., Fraústo Da Silva J.J.R. Evolution was chemically constrained. J. Theor. Biol. 2003;220:323–343. doi: 10.1006/jtbi.2003.3152. [DOI] [PubMed] [Google Scholar]
- 43.Case A.J. On the origin of superoxide dismutase: An evolutionary perspective of superoxide-mediated redox signaling. Antioxidants. 2017 doi: 10.3390/antiox6040082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nisbet E.G., Grassineau N.V., Howe C.J., Abell P.I., Regelous M., Nisbet R.E.R. The age of Rubisco: The evolution of oxygenic photosynthesis. Geobiology. 2007;5:311–335. [Google Scholar]
- 45.Canfield D.E. The early history of atmospheric oxygen: Homage to Robert M. Garrels. Annu. Rev. Earth Planet. Sci. 2005;33:1–36. [Google Scholar]
- 46.Fischer W.W., Hemp J., Johnson J.E. Evolution of oxygenic photosynthesis. Annu. Rev. Earth Planet. Sci. 2016;44:647–683. [Google Scholar]
- 47.Sessions A.L., Doughty D.M., Welander P.V., Summons R.E., Newman D.K. The continuing puzzle of the great oxidation event. Curr. Biol. 2009;19:R567–R574. doi: 10.1016/j.cub.2009.05.054. [DOI] [PubMed] [Google Scholar]
- 48.Thauer R.K. My lifelong passion for biochemistry and anaerobic microorganisms. Annu. Rev. Microbiol. 2015;69:1–30. doi: 10.1146/annurev-micro-091014-104344. [DOI] [PubMed] [Google Scholar]
- 49.Raymond J., Segre D. The effect of oxygen on biochemical networks and the evolution of complex life. Science. 2006;311:1764–1767. doi: 10.1126/science.1118439. [DOI] [PubMed] [Google Scholar]
- 50.Ghosh A., Shcherbik N. Effects of oxidative stress on protein translation: Implications for cardiovascular diseases. Int. J. Mol. Sci. 2020;21:2661. doi: 10.3390/ijms21082661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fridovich I. Oxygen: How do we stand it? Med. Princ. Pract. 2013;22:131–137. doi: 10.1159/000339212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen Y., Azad M.B., Gibson S.B. Superoxide is the major reactive oxygen species regulating autophagy. Cell Death Differ. 2009;16:1040–1052. doi: 10.1038/cdd.2009.49. [DOI] [PubMed] [Google Scholar]
- 53.Davies M.J. Protein oxidation and peroxidation. Biochem. J. 2016;473:805–825. doi: 10.1042/BJ20151227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Stadtman E.R., Levine R.L. Free radical-mediated oxidation of free amino acids and amino acid residues in proteins. Amino Acids. 2003;25:207–218. doi: 10.1007/s00726-003-0011-2. [DOI] [PubMed] [Google Scholar]
- 55.Solenkova N.V., Newman J.D., Berger J.S., Thurston G., Hochman J.S., Lamas G.A. Metal pollutants and cardiovascular disease: Mechanisms and consequences of exposure. Am. Heart J. 2014;168:812–822. doi: 10.1016/j.ahj.2014.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Winterbourn C.C. Toxicity of iron and hydrogen peroxide: The Fenton reaction. Toxicol. Lett. 1995;82–83:969–974. doi: 10.1016/0378-4274(95)03532-x. [DOI] [PubMed] [Google Scholar]
- 57.Cole N.B., Murphy D.D., Lebowitz J., Di Noto L., Levine R.L., Nussbaum R.L. Metal-catalyzed oxidation of α-synuclein. J. Biol. Chem. 2005;280:9678–9690. doi: 10.1074/jbc.M409946200. [DOI] [PubMed] [Google Scholar]
- 58.Stadtman E.R. Metal ion-catalyzed oxidation of proteins: Biochemical mechanism and biological consequences. Free Radic. Biol. Med. 1990;9:315–325. doi: 10.1016/0891-5849(90)90006-5. [DOI] [PubMed] [Google Scholar]
- 59.Sultana R., Perluigi M., Butterfield D.A. Protein oxidation and lipid peroxidation in brain of subjects with Alzheimer's disease: Insights into mechanism of neurodegeneration from redox proteomics. Antioxid. Redox Signal. 2006;8:2021–2037. doi: 10.1089/ars.2006.8.2021. [DOI] [PubMed] [Google Scholar]
- 60.Rodgers K.J., Hume P.M., Morris J.G.L., Dean R.T. Evidence for L-dopa incorporation into cell proteins in patients treated with levodopa. J. Neurochem. 2006;98:1061–1067. doi: 10.1111/j.1471-4159.2006.03941.x. [DOI] [PubMed] [Google Scholar]
- 61.Hilton J.B., White A.R., Crouch P.J. Metal-deficient SOD1 in amyotrophic lateral sclerosis. J. Mol. Med. (Berl.) 2015;93:481–487. doi: 10.1007/s00109-015-1273-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Terrill J.R., Radley-Crabb H.G., Iwasaki T., Lemckert F.A., Arthur P.G., Grounds M.D. Oxidative stress and pathology in muscular dystrophies: Focus on protein thiol oxidation and dysferlinopathies. FEBS J. 2013;280:4149–4164. doi: 10.1111/febs.12142. [DOI] [PubMed] [Google Scholar]
- 63.McGuinness A.J.A., Sapey E. Oxidative stress in COPD: Sources, markers, and potential mechanisms. J. Clin. Med. 2017;6 doi: 10.3390/jcm6020021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Stocker R., Keaney J.F. Role of oxidative modifications in atherosclerosis. Physiol. Rev. 2004;84:1381–1478. doi: 10.1152/physrev.00047.2003. [DOI] [PubMed] [Google Scholar]
- 65.Ethen C.M., Reilly C., Feng X., Olsen T.W., Ferrington D.A. Age-related macular degeneration and retinal protein modification by 4-hydroxy-2-nonenal. Invest. Ophthalmol. Vis. Sci. 2007;48:3469–3479. doi: 10.1167/iovs.06-1058. [DOI] [PubMed] [Google Scholar]
- 66.Dean R.T., Dunlop R., Hume P., Rodgers K.J. Proteolytic “defences” and the accumulation of oxidized polypeptides in cataractogenesis and atherogenesis. Biochem. Soc. Symp. 2003 doi: 10.1042/bss0700135. [DOI] [PubMed] [Google Scholar]
- 67.Hodgson E.K., Fridovich I. The interaction of bovine erythrocyte superoxide dismutase with hydrogen peroxide: Inactivation of the enzyme. Biochemistry. 1975;14:5294–5299. doi: 10.1021/bi00695a010. [DOI] [PubMed] [Google Scholar]
- 68.Salo D.C., Lin S.W., Pacifici R.E., Davies K.J. Superoxide dismutase is preferentially degraded by a proteolytic system from red blood cells following oxidative modification by hydrogen peroxide. Free Radic. Biol. Med. 1988;5:335–339. doi: 10.1016/0891-5849(88)90105-0. [DOI] [PubMed] [Google Scholar]
- 69.Kang S.W. Superoxide dismutase 2 gene and cancer risk: Evidence from an updated meta-analysis. Int. J. Clin. Exp. Med. 2015;8:14647–14655. [PMC free article] [PubMed] [Google Scholar]
- 70.Flynn J.M., Melov S. SOD2 in mitochondrial dysfunction and neurodegeneration. Free Radic. Biol. Med. 2013;62:4–12. doi: 10.1016/j.freeradbiomed.2013.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hempel N., Carrico P.M., Melendez J.A. Manganese superoxide dismutase (Sod2) and redox-control of signaling events that drive metastasis. Anticancer Agents Med. Chem. 2011;11:191–201. doi: 10.2174/187152011795255911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Beck R., Dejeans N., Glorieux C., Creton M., Delaive E., Dieu M., Raes M., Levêque P., Gallez B., Depuydt M., Collet J.F., Calderon P.B., Verrax J. Hsp90 is cleaved by reactive oxygen species at a highly conserved N-terminal amino acid motif. PLoS One. 2012;7:1–9. doi: 10.1371/journal.pone.0040795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Castro J.P., Fernando R., Reeg S., Meinl W., Almeida H., Grune T. Non-enzymatic cleavage of Hsp90 by oxidative stress leads to actin aggregate formation: A novel gain-of-function mechanism. Redox Biol. 2019;21:101108. doi: 10.1016/j.redox.2019.101108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bohush A., Bieganowski P., Filipek A. Hsp90 and its co-chaperones in neurodegenerative diseases. Int. J. Mol. Sci. 2019;20 doi: 10.3390/ijms20204976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Cooke M.S., Evans M.D., Dizdaroglu M., Lunec J. Oxidative DNA damage: Mechanisms, mutation, and disease. FASEB J. 2003;17:1195–1214. doi: 10.1096/fj.02-0752rev. [DOI] [PubMed] [Google Scholar]
- 76.Kong Q., Lin C.-L.G. Oxidative damage to RNA: Mechanisms, consequences, and diseases. Cell. Mol. Life Sci. 2010;67:1817–1829. doi: 10.1007/s00018-010-0277-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Imlay J.A., Chin S.M., Linn S. Toxic DNA damage by hydrogen peroxide through the Fenton reaction in vivo and in vitro. Science. 1988;240:640–642. doi: 10.1126/science.2834821. [DOI] [PubMed] [Google Scholar]
- 78.Li Z., Chen X., Liu Z., Ye W., Li L., Qian L., Ding H., Li P., Aung L.H.H. Recent advances: Molecular mechanism of RNA oxidation and its role in various diseases. Front. Mol. Biosci. 2020;7:1–7. doi: 10.3389/fmolb.2020.00184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Poulsen H.E., Specht E., Broedbaek K., Henriksen T., Ellervik C., Mandrup-Poulsen T., Tonnesen M., Nielsen P.E., Andersen H.U., Weimann A. RNA modifications by oxidation: A novel disease mechanism? Free Radic. Biol. Med. 2012;52:1353–1361. doi: 10.1016/j.freeradbiomed.2012.01.009. [DOI] [PubMed] [Google Scholar]
- 80.Simms C.L., Hudson B.H., Mosior J.W., Rangwala A.S., Zaher H.S. An active role for the ribosome in determining the fate of oxidized mRNA. Cell Rep. 2014;9:1256–1264. doi: 10.1016/j.celrep.2014.10.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Tanaka M., Chock P.B., Stadtman E.R. Oxidized messenger RNA induces translation errors. Proc. Natl. Acad. Sci. U. S. A. 2007;104:66–71. doi: 10.1073/pnas.0609737104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Shcherbik N., Pestov D.G. The impact of oxidative stress on ribosomes: From injury to regulation. Cells. 2019;8:1379. doi: 10.3390/cells8111379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Shenton D., Smirnova J.B., Selley J.N., Carroll K., Hubbard S.J., Pavitt G.D., Ashe M.P., Grant C.M. Global translational responses to oxidative stress impact upon multiple levels of protein synthesis. J. Biol. Chem. 2006;281:29011–29021. doi: 10.1074/jbc.M601545200. [DOI] [PubMed] [Google Scholar]
- 84.Blevins W.R., Tavella T., Moro S.G., Blasco-Moreno B., Closa-Mosquera A., Díez J., Carey L.B., Albà M.M. Extensive post-transcriptional buffering of gene expression in the response to severe oxidative stress in baker's yeast. Sci. Rep. 2019;9:11005. doi: 10.1038/s41598-019-47424-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zhu M., Dai X. Maintenance of translational elongation rate underlies the survival of Escherichia coli during oxidative stress. Nucleic Acids Res. 2019;47:7592–7604. doi: 10.1093/nar/gkz467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Murray K.F., Messner D.J., Kowdley K.V. Physiology of the Gastrointestinal Tract: Sixth Edition. 4th Ed. Elsevier B.V.; Amsterdam, The Netherlands: 2006. Mechanisms of hepatocyte detoxification; pp. 1483–1504. [Google Scholar]
- 87.Festa R.A., Thiele D.J. Copper: An essential metal in biology. Curr. Biol. 2011;21:R877–R883. doi: 10.1016/j.cub.2011.09.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Gulec S., Collins J.F. Molecular mediators governing iron-copper interactions. Annu. Rev. Nutr. 2014;34:95–116. doi: 10.1146/annurev-nutr-071812-161215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Hong Enriquez R.P., Do T.N. Bioavailability of metal ions and evolutionary adaptation. Life. 2012;2:274–285. doi: 10.3390/life2040274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Rae A.T.D., Schmidt P.J., Pufahl R.A., Culotta V.C., Halloran T.V.O. Undetectable intracellular free copper: The requirement of a copper chaperone for superoxide dismutase. Science. 1999;284:805–808. doi: 10.1126/science.284.5415.805. [DOI] [PubMed] [Google Scholar]
- 91.Calvo J., Jung H., Meloni G. Copper metallothioneins. IUBMB Life. 2017;69:236–245. doi: 10.1002/iub.1618. [DOI] [PubMed] [Google Scholar]
- 92.Dudev T., Lim C. Competition among metal ions for protein binding sites: Determinants of metal ion selectivity in proteins. Chem. Rev. 2014;114:538–556. doi: 10.1021/cr4004665. [DOI] [PubMed] [Google Scholar]
- 93.Aguirre J.D., Culotta V.C. Battles with iron: Manganese in oxidative stress protection. J. Biol. Chem. 2012;287:13541–13548. doi: 10.1074/jbc.R111.312181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Schmidt S.B., Husted S. The biochemical properties of manganese in plants. Plants (Basel) 2019;8:381. doi: 10.3390/plants8100381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Harty M.L., Sharma A.N., Bearne S.L. Catalytic properties of the metal ion variants of mandelate racemase reveal alterations in the apparent electrophilicity of the metal cofactor. Metallomics. 2019;11:707–723. doi: 10.1039/c8mt00330k. [DOI] [PubMed] [Google Scholar]
- 96.Hohle T.H., O'Brian M.R. Magnesium-dependent processes are targets of bacterial manganese toxicity. Mol. Microbiol. 2014;93:736–747. doi: 10.1111/mmi.12687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Khrustalev V.V., Barkovsky E.V., Khrustaleva T.A. Magnesium and manganese binding sites on proteins have the same predominant motif of secondary structure. J. Theor. Biol. 2016;395:174–185. doi: 10.1016/j.jtbi.2016.02.006. [DOI] [PubMed] [Google Scholar]
- 98.Moon W.J., Liu J. Replacing Mg2+ by Fe2+ for RNA-cleaving DNAzymes. ChemBioChem. 2020;21:401–407. doi: 10.1002/cbic.201900344. [DOI] [PubMed] [Google Scholar]
- 99.Kehres D.G., Maguire M.E. Emerging themes in manganese transport, biochemistry and pathogenesis in bacteria. FEMS Microbiol. Rev. 2003;27:263–290. doi: 10.1016/S0168-6445(03)00052-4. [DOI] [PubMed] [Google Scholar]
- 100.Piovesan D., Profiti G., Martelli P.L., Casadio R. The human “magnesome”: Detecting magnesium binding sites on human proteins. BMC Bioinformatics. 2012 doi: 10.1186/1471-2105-13-S14-S10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Waldron K.J., Rutherford J.C., Ford D., Robinson N.J. Metalloproteins and metal sensing. Nature. 2009;460:823–830. doi: 10.1038/nature08300. [DOI] [PubMed] [Google Scholar]
- 102.Inupakutika M.A., Sengupta S., Devireddy A.R., Azad R.K., Mittler R. The evolution of reactive oxygen species metabolism. J. Exp. Bot. 2016;67:5933–5943. doi: 10.1093/jxb/erw382. [DOI] [PubMed] [Google Scholar]
- 103.Saito M.A., Sigman D.M., Morel F.M.M. The bioinorganic chemistry of the ancient ocean: The co-evolution of cyanobacterial metal requirements and biogeochemical cycles at the Archean-Proterozoic boundary? Inorg. Chim. Acta. 2003;356:308–318. [Google Scholar]
- 104.Feig A.L., Uhlenbeck O.C. Vol 37. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1999. 12 the role of metal ions in RNA biochemistry; pp. 287–319. (Cold Spring Harbor Monograph Archive). [Google Scholar]
- 105.Dupont C.L., Yang S., Palenik B., Bourne P.E. Modern proteomes contain putative imprints of ancient shifts in trace metal geochemistry. Proc. Natl. Acad. Sci. U. S. A. 2006;103:17822–17827. doi: 10.1073/pnas.0605798103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Miller A.-F. Superoxide dismutases: Active sites that save, but a protein that kills. Curr. Opin. Chem. Biol. 2004;8:162–168. doi: 10.1016/j.cbpa.2004.02.011. [DOI] [PubMed] [Google Scholar]
- 107.Miller A.F. Superoxide dismutases: Ancient enzymes and new insights. FEBS Lett. 2012;586:585–595. doi: 10.1016/j.febslet.2011.10.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Herbst R.W., Guce A., Bryngelson P.A., Higgins K.A., Ryan K.C., Cabelli D.E., Garman S.C., Maroney M.J. Role of conserved tyrosine residues in NiSOD catalysis: A case of convergent evolution. Biochemistry. 2009;48:3354–3369. doi: 10.1021/bi802029t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Uudsemaa M., Tamm T. Density-functional theory calculations of aqueous redox potentials of fourth-period transition metals. J. Phys. Chem. A. 2003;107:9997–10003. [Google Scholar]
- 110.Miller A.F. Redox tuning over almost 1 V in a structurally conserved active site: Lessons from Fe-containing superoxide dismutase. Acc. Chem. Res. 2008;41:501–510. doi: 10.1021/ar700237u. [DOI] [PubMed] [Google Scholar]
- 111.Takao M., Yasui A., Oikawa A. Unique characteristics of superoxide dismutase of a strictly anaerobic archaebacterium Methanobacterium thermoautotrophicum. J. Biol. Chem. 1991;266:14151–14154. [PubMed] [Google Scholar]
- 112.Mizuno K., Whittaker M.M., Bächinger H.P., Whittaker J.W. Calorimetric studies on the tight binding metal interactions of Escherichia coli manganese superoxide dismutase. J. Biol. Chem. 2004;279:27339–27344. doi: 10.1074/jbc.M400813200. [DOI] [PubMed] [Google Scholar]
- 113.Benov L.T., Fridovich I. Escherichia coli expresses a copper- and zinc-containing superoxide dismutase. J. Biol. Chem. 1994;269:25310–25314. [PubMed] [Google Scholar]
- 114.Schnell S., Steinman H.M. Function and stationary-phase induction of periplasmic copper-zinc superoxide dismutase and catalase/peroxidase in Caulobacter crescentus. J. Bacteriol. 1995;177:5924–5929. doi: 10.1128/jb.177.20.5924-5929.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Bermingham-McDonogh O., Gralla E.B., Valentine J.S. The copper, zinc-superoxide dismutase gene of Saccharomyces cerevisiae: Cloning, sequencing, and biological activity. Proc. Natl. Acad. Sci. U. S. A. 1988;85:4789–4793. doi: 10.1073/pnas.85.13.4789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Crapo J.D., Oury T., Rabouille C., Slot J.W., Chang L.Y. Copper,zinc superoxide dismutase is primarily a cytosolic protein in human cells. Proc. Natl. Acad. Sci. U. S. A. 1992;89:10405–10409. doi: 10.1073/pnas.89.21.10405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.McCord J.M., Fridovich I. Superoxide dismutase. An enzymic function for erythrocuprein (hemocuprein) J. Biol. Chem. 1969;244:6049–6055. [PubMed] [Google Scholar]
- 118.Marklund S.L. Human copper-containing superoxide dismutase of high molecular weight. Proc. Natl. Acad. Sci. U. S. A. 1982;79:7634–7638. doi: 10.1073/pnas.79.24.7634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Mondola P., Damiano S., Sasso A., Santillo M. The Cu, Zn superoxide dismutase: Not only a dismutase enzyme. Front. Physiol. 2016;7:1–8. doi: 10.3389/fphys.2016.00594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Weisiger R.A., Fridovich I. Superoxide dismutase. Organelle specificity. J. Biol. Chem. 1973;248:3582–3592. [PubMed] [Google Scholar]
- 121.Dupont C.L., Neupane K., Shearer J., Palenik B. Diversity, function and evolution of genes coding for putative Ni-containing superoxide dismutases. Environ. Microbiol. 2008;10:1831–1843. doi: 10.1111/j.1462-2920.2008.01604.x. [DOI] [PubMed] [Google Scholar]
- 122.Tripp B.C., Bell C.B., Cruz F., Krebs C., Ferry J.G. A role for iron in an ancient carbonic anhydrase. J. Biol. Chem. 2004;279:6683–6687. doi: 10.1074/jbc.M311648200. [DOI] [PubMed] [Google Scholar]
- 123.Smethurst D.G.J., Kovalev N., McKenzie E.R., Pestov D.G., Shcherbik N. Iron-mediated degradation of ribosomes under oxidative stress is attenuated by manganese. J. Biol. Chem. 2020;295:17200–17214. doi: 10.1074/jbc.RA120.015025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Stallings W.C., Pattridge K.A., Strong R.K., Ludwig M.L. Manganese and iron superoxide dismutases are structural homologs. J. Biol. Chem. 1984;259:10695–10699. [PubMed] [Google Scholar]
- 125.Vance C.K., Miller A.F. Novel insights into the basis for Escherichia coli superoxide dismutase's metal ion specificity from mn-substituted FeSOD and its very high Em. Biochemistry. 2001;40:13079–13087. doi: 10.1021/bi0113317. [DOI] [PubMed] [Google Scholar]
- 126.Miller A.F., Wang T. A single outer-sphere mutation stabilizes apo-Mn superoxide dismutase by 35 °C and disfavors Mn binding. Biochemistry. 2017;56:3787–3799. doi: 10.1021/acs.biochem.7b00175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Tabares L.C., Gätjens J., Un S. Understanding the influence of the protein environment on the Mn(II) centers in superoxide dismutases using high-field electron paramagnetic resonance. Biochim. Biophys. Acta. 2010;1804:308–317. doi: 10.1016/j.bbapap.2009.09.027. [DOI] [PubMed] [Google Scholar]
- 128.Borgstahl G.E., Pokross M., Chehab R., Sekher A., Snell E.H. Cryo-trapping the six-coordinate, distorted-octahedral active site of manganese superoxide dismutase. J. Mol. Biol. 2000;296:951–959. doi: 10.1006/jmbi.1999.3506. [DOI] [PubMed] [Google Scholar]
- 129.Kwasigroch J.M., Wintjens R., Gilis D., Rooman M. SODa: An Mn/Fe superoxide dismutase prediction and design server. BMC Bioinformatics. 2008;9:257. doi: 10.1186/1471-2105-9-257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Wintjens R., Noël C., May A.C.W., Gerbod D., Dufernez F., Capron M., Viscogliosi E., Rooman M. Specificity and phenetic relationships of iron- and manganese-containing superoxide dismutases on the basis of structure and sequence comparisons. J. Biol. Chem. 2004;279:9248–9254. doi: 10.1074/jbc.M312329200. [DOI] [PubMed] [Google Scholar]
- 131.Whittaker J.W. The irony of manganese superoxide dismutase. Biochem. Soc. Trans. 2003;31:1318–1321. doi: 10.1042/bst0311318. [DOI] [PubMed] [Google Scholar]
- 132.Ganini D., Petrovich R.M., Edwards L.L., Mason R.P. Iron incorporation into MnSOD A (bacterial Mn-dependent superoxide dismutase) leads to the formation of a peroxidase/catalase implicated in oxidative damage to bacteria. Biochim. Biophys. Acta. 2015;1850:1795–1805. doi: 10.1016/j.bbagen.2015.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Ganini D., Santos J.H., Bonini M.G., Mason R.P. Switch of mitochondrial superoxide dismutase into a prooxidant peroxidase in manganese-deficient cells and mice. Cell Chem. Biol. 2018;25:413–425.e6. doi: 10.1016/j.chembiol.2018.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Parker M.W., Blake C.C. Iron- and manganese-containing superoxide dismutases can be distinguished by analysis of their primary structures. FEBS Lett. 1988;229:377–382. doi: 10.1016/0014-5793(88)81160-8. [DOI] [PubMed] [Google Scholar]
- 135.Whittaker J.W. Prokaryotic manganese superoxide dismutases. Methods Enzymol. 2002;349:80–90. doi: 10.1016/s0076-6879(02)49323-8. [DOI] [PubMed] [Google Scholar]
- 136.Kang Y., He Y.X., Zhao M.X., Li W.F. Structures of native and Fe-substituted SOD2 from Saccharomyces cerevisiae. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2011;67:1173–1178. doi: 10.1107/S1744309111029186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Yamakura F., Sugio S., Hiraoka B.Y., Ohmori D., Yokota T. Pronounced conversion of the metal-specific activity of superoxide dismutase from Porphyromonas gingivalis by the mutation of a single amino acid (Gly155Thr) located apart from the active site. Biochemistry. 2003;42:10790–10799. doi: 10.1021/bi0349625. [DOI] [PubMed] [Google Scholar]
- 138.Bachega J.F.R., Navarro M.V.A.S., Bleicher L., Bortoleto-Bugs R.K., Dive D., Hoffmann P., Viscogliosi E., Garratt R.C. Systematic structural studies of iron superoxide dismutases from human parasites and a statistical coupling analysis of metal binding specificity. Proteins. 2009;77:26–37. doi: 10.1002/prot.22412. [DOI] [PubMed] [Google Scholar]
- 139.Osawa M., Yamakura F., Mihara M., Okubo Y., Yamada K., Hiraoka B.Y. Conversion of the metal-specific activity of Escherichia coli Mn-SOD by site-directed mutagenesis of Gly165Thr. Biochim. Biophys. Acta. 2010;1804:1775–1779. doi: 10.1016/j.bbapap.2010.04.011. [DOI] [PubMed] [Google Scholar]
- 140.Meier B., Barra D., Bossa F., Calabrese L., Rotilio G. Synthesis of either Fe- or Mn-superoxide dismutase with an apparently identical protein moiety by an anaerobic bacterium dependent on the metal supplied. J. Biol. Chem. 1982;257:13977–13980. [PubMed] [Google Scholar]
- 141.Garcia Y.M., Barwinska-Sendra A., Tarrant E., Skaar E.P., Waldron K.J., Kehl-Fie T.E. A superoxide dismutase capable of functioning with iron or manganese promotes the resistance of Staphylococcus aureus to calprotectin and nutritional immunity. PLoS Pathog. 2017;13:1–19. doi: 10.1371/journal.ppat.1006125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Valderas M.W., Gatson J.W., Wreyford N., Hart M.E. The superoxide dismutase gene sodM is unique to Staphylococcus aureus: Absence of sodM in coagulase-negative staphylococci. J. Bacteriol. 2002;184:2465–2472. doi: 10.1128/JB.184.9.2465-2472.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Karavolos M.H., Horsburgh M.J., Ingham E., Foster S.J. Role and regulation of the superoxide dismutases of Staphylococcus aureus. Microbiology (Reading) 2003;149:2749–2758. doi: 10.1099/mic.0.26353-0. [DOI] [PubMed] [Google Scholar]
- 144.Barwinska-Sendra A., Garcia Y.M., Sendra K.M., Baslé A., Mackenzie E.S., Tarrant E., Card P., Tabares L.C., Bicep C., Un S., Kehl-Fie T.E., Waldron K.J. An evolutionary path to altered cofactor specificity in a metalloenzyme. Nat. Commun. 2020;11:2738. doi: 10.1038/s41467-020-16478-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Barwinska-Sendra A., Baslé A., Waldron K.J., Un S. A charge polarization model for the metal-specific activity of superoxide dismutases. Phys. Chem. Chem. Phys. 2018;20:2363–2372. doi: 10.1039/c7cp06829h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Tong S.Y.C., Schaumburg F., Ellington M.J., Corander J., Pichon B., Leendertz F., Bentley S.D., Parkhill J., Holt D.C., Peters G., Giffard P.M. Novel staphylococcal species that form part of a Staphylococcus aureus-related complex: The non-pigmented Staphylococcus argenteus sp. nov. and the non-human primate-associated Staphylococcus schweitzeri sp. nov. Int. J. Syst. Evol. Microbiol. 2015;65:15–22. doi: 10.1099/ijs.0.062752-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Suzuki H., Lefébure T., Bitar P.P., Stanhope M.J. Comparative genomic analysis of the genus Staphylococcus including Staphylococcus aureus and its newly described sister species Staphylococcus simiae. BMC Genomics. 2012;13:38. doi: 10.1186/1471-2164-13-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Kehl-Fie T.E., Chitayat S., Hood M.I., Damo S., Restrepo N., Garcia C., Munro K.A., Chazin W.J., Skaar E.P. Nutrient metal sequestration by calprotectin inhibits bacterial superoxide defense, enhancing neutrophil killing of Staphylococcus aureus. Cell Host Microbe. 2011;10:158–164. doi: 10.1016/j.chom.2011.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Wessling-Resnick M. Nramp1 and other transporters involved in metal withholding during infection. J. Biol. Chem. 2015;290:18984–18990. doi: 10.1074/jbc.R115.643973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Lundin D., Berggren G., Logan D.T., Sjöberg B.M. The origin and evolution of ribonucleotide reduction. Life. 2015;5:604–636. doi: 10.3390/life5010604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Torrents E., Aloy P., Gibert I., Rodríguez-Trelles F. Ribonucleotide reductases: Divergent evolution of an ancient enzyme. J. Mol. Evol. 2002;55:138–152. doi: 10.1007/s00239-002-2311-7. [DOI] [PubMed] [Google Scholar]
- 152.Aye Y., Li M., Long M.J.C., Weiss R.S. Ribonucleotide reductase and cancer: Biological mechanisms and targeted therapies. Oncogene. 2014;34:2011–2021. doi: 10.1038/onc.2014.155. [DOI] [PubMed] [Google Scholar]
- 153.Zimanyi C.M., Chen P.Y.T., Kang G., Funk M.A., Drennan C.L. Molecular basis for allosteric specificity regulation in class ia ribonucleotide reductase from Escherichia coli. Elife. 2016;5 doi: 10.7554/eLife.07141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Andrews S.C. The ferritin-like superfamily: Evolution of the biological iron storeman from a rubrerythrin-like ancestor. Biochim. Biophys. Acta. 2010;1800:691–705. doi: 10.1016/j.bbagen.2010.05.010. [DOI] [PubMed] [Google Scholar]
- 155.Hintze K.J., Theil E.C. Cellular regulation and molecular interactions of the ferritins. Cell. Mol. Life Sci. 2006;63:591–600. doi: 10.1007/s00018-005-5285-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Kolberg M., Strand K.R., Graff P., Kristoffer Andersson K. Structure, function, and mechanism of ribonucleotide reductases. Biochim. Biophys. Acta. 2004;1699:1–34. doi: 10.1016/j.bbapap.2004.02.007. [DOI] [PubMed] [Google Scholar]
- 157.Pierce B.S., Hendrich M.P. Local and global effects of metal binding within the small subunit of ribonucleotide reductase. J. Am. Chem. Soc. 2005;127:3613–3623. doi: 10.1021/ja0491937. [DOI] [PubMed] [Google Scholar]
- 158.Cotruvo J.A., Stubbe J. Escherichia coli class Ib ribonucleotide reductase contains a dimanganese(III)-tyrosyl radical cofactor in vivo. Biochemistry. 2011;50:1672–1681. doi: 10.1021/bi101881d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Cox N., Ogata H., Stolle P., Reijerse E., Auling G., Lubitz W. A tyrosyl-dimanganese coupled spin system is the native metalloradical cofactor of the R2F subunit of the ribonucleotide reductase of Corynebacterium ammoniagenes. J. Am. Chem. Soc. 2010;132:11197–11213. doi: 10.1021/ja1036995. [DOI] [PubMed] [Google Scholar]
- 160.Dassama L.M.K., Krebs C., Bollinger J.M., Rosenzweig A.C., Boal A.K. Structural basis for assembly of the MnIV/FeIII cofactor in the class Ic ribonucleotide reductase from Chlamydia trachomatis. Biochemistry. 2013;52:6424–6436. doi: 10.1021/bi400819x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Jiang W., Xie J., Nørgaard H., Bollinger J.M., Krebs C. Rapid and quantitative activation of Chlamydia trachomatis ribonucleotide reductase by hydrogen peroxide. Biochemistry. 2008;47:4477–4483. doi: 10.1021/bi702085z. [DOI] [PubMed] [Google Scholar]
- 162.Roos K., Siegbahn P.E.M. Density functional theory study of the manganese-containing ribonucleotide reductase from Chlamydia trachomatis: Why manganese is needed in the active complex. Biochemistry. 2009;48:1878–1887. doi: 10.1021/bi801695d. [DOI] [PubMed] [Google Scholar]
- 163.Grāve K., Griese J.J., Berggren G., Bennett M.D., Högbom M. The Bacillus anthracis class Ib ribonucleotide reductase subunit NrdF intrinsically selects manganese over iron. J. Biol. Inorg. Chem. 2020;25:571–582. doi: 10.1007/s00775-020-01782-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Cotruvo J.A., Stich T.A., Britt R.D., Stubbe J. Mechanism of assembly of the dimanganese-tyrosyl radical cofactor of class Ib ribonucleotide reductase: Enzymatic generation of superoxide is required for tyrosine oxidation via a Mn(III)Mn(IV) intermediate. J. Am. Chem. Soc. 2013;135:4027–4039. doi: 10.1021/ja312457t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Bernier C.R., Petrov A.S., Kovacs N.A., Penev P.I., Williams L.D. Translation: The universal structural core of life. Mol. Biol. Evol. 2018;35:2065–2076. doi: 10.1093/molbev/msy101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Bray M.S., Lenz T.K., Bowman J.C., Petrov A.S., Reddi A.R., Hud N.V., Williams L.D., Glass J.B. Ferrous iron folds rRNA and mediates translation. bioRxiv. 2018 doi: 10.1101/256958. [preprint] [DOI] [Google Scholar]
- 167.Zheng H., Shabalin I.G., Handing K.B., Bujnicki J.M., Minor W. Magnesium-binding architectures in RNA crystal structures: Validation, binding preferences, classification and motif detection. Nucleic Acids Res. 2015;43:3789–3801. doi: 10.1093/nar/gkv225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Petrov A.S., Bowman J.C., Harvey S.C., Williams L.D. Bidentate RNA-magnesium clamps: On the origin of the special role of magnesium in RNA folding. RNA. 2011;17:291–297. doi: 10.1261/rna.2390311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Selmer M. Structure of the 70S ribosome complexed with mRNA and tRNA. Science. 2006;313:1935–1942. doi: 10.1126/science.1131127. [DOI] [PubMed] [Google Scholar]
- 170.Demeshkina N., Jenner L., Westhof E., Yusupov M., Yusupova G. A new understanding of the decoding principle on the ribosome. Nature. 2012;484:256–259. doi: 10.1038/nature10913. [DOI] [PubMed] [Google Scholar]
- 171.Hsiao C., Williams L.D. A recurrent magnesium-binding motif provides a framework for the ribosomal peptidyl transferase center. Nucleic Acids Res. 2009;37:3134–3142. doi: 10.1093/nar/gkp119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Fox G.E. Origin and evolution of the ribosome. Cold Spring Harb. Perspect. Biol. 2010;2 doi: 10.1101/cshperspect.a003483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Jones C., Nomosatryo S., Crowe S.A., Bjerrum C.J., Canfield D.E. Iron oxides, divalent cations, silica, and the early earth phosphorus crisis. Geology. 2015;43:135–138. [Google Scholar]
- 174.Veal E.A., Day A.M., Morgan B.A. Hydrogen peroxide sensing and signaling. Mol. Cell. 2007;26:1–14. doi: 10.1016/j.molcel.2007.03.016. [DOI] [PubMed] [Google Scholar]
- 175.Shedlovskiy D., Zinskie J.A., Gardner E., Pestov D.G., Shcherbik N. Endonucleolytic cleavage in the expansion segment 7 of 25S rRNA is an early marker of low-level oxidative stress in yeast. J. Biol. Chem. 2017;292:18469–18485. doi: 10.1074/jbc.M117.800003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Anjem A., Imlay J.A. Mononuclear iron enzymes are primary targets of hydrogen peroxide stress. J. Biol. Chem. 2012;287:15544–15556. doi: 10.1074/jbc.M111.330365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Sobota J.M., Imlay J.A. Iron enzyme ribulose-5-phosphate 3-epimerase in Escherichia coli is rapidly damaged by hydrogen peroxide but can be protected by manganese. Proc. Natl. Acad. Sci. U. S. A. 2011;108:5402–5407. doi: 10.1073/pnas.1100410108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Root-Bernstein R., Root-Bernstein M. The ribosome as a missing link in prebiotic evolution III: Over-representation of tRNA-and rRNA-like sequences and plieofunctionality of ribosome-related molecules argues for the evolution of primitive genomes from ribosomal RNA modules. Int. J. Mol. Sci. 2019;20:140. doi: 10.3390/ijms20010140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Hamm M.L., Nikolic D., Van Breemen R.B., Piccirilli J.A. Unconventional origin of metal ion rescue in the hammerhead ribozyme reaction: Mn2+-assisted redox conversion of 2′-mercaptocytidine to cytidine. J. Am. Chem. Soc. 2000;122:12069–12078. [Google Scholar]
- 180.Hsiao C., Chou I.-C., Okafor C.D., Bowman J.C., O'Neill E.B., Athavale S.S., Petrov A.S., Hud N.V., Wartell R.M., Harvey S.C., Williams L.D. RNA with iron(II) as a cofactor catalyses electron transfer. Nat. Chem. 2013;5:525–528. doi: 10.1038/nchem.1649. [DOI] [PubMed] [Google Scholar]
- 181.Kluska K., Adamczyk J., Krężel A. Metal binding properties, stability and reactivity of zinc fingers. Coord. Chem. Rev. 2018;367:18–64. [Google Scholar]
- 182.Koch K.A., Peña M.M.O., Thiele D.J. Copper-binding motifs in catalysis, transport, detoxification and signaling. Chem. Biol. 1997;4:549–560. doi: 10.1016/s1074-5521(97)90241-6. [DOI] [PubMed] [Google Scholar]
- 183.Predki P.F., Sarkar B. Effect of replacement of “zinc finger” zinc on estrogen receptor DNA interactions. J. Biol. Chem. 1992;267:5842–5846. [PubMed] [Google Scholar]
- 184.Conte D., Narindrasorasak S., Sarkar B. In vivo and in vitro iron-replaced zinc finger generates free radicals and causes DNA damage. J. Biol. Chem. 1996;271:5125–5130. doi: 10.1074/jbc.271.9.5125. [DOI] [PubMed] [Google Scholar]
- 185.Maio N., Rouault T.A. Iron-sulfur cluster biogenesis in mammalian cells: New insights into the molecular mechanisms of cluster delivery. Biochim. Biophys. Acta. 2015;1853:1493–1512. doi: 10.1016/j.bbamcr.2014.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Gervason S., Larkem D., Mansour A.B., Botzanowski T., Müller C.S., Pecqueur L., Le Pavec G., Delaunay-Moisan A., Brun O., Agramunt J., Grandas A., Fontecave M., Schünemann V., Cianférani S., Sizun C., et al. Physiologically relevant reconstitution of iron-sulfur cluster biosynthesis uncovers persulfide-processing functions of ferredoxin-2 and frataxin. Nat. Commun. 2019;10:3566. doi: 10.1038/s41467-019-11470-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Iannuzzi C., Adrover M., Puglisi R., Yan R., Temussi P.A., Pastore A. The role of zinc in the stability of the marginally stable IscU scaffold protein: The stability determinants of IscU. Protein Sci. 2014;23:1208–1219. doi: 10.1002/pro.2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Ellerby L.M., Cabelli D.E., Graden J.A., Valentine J.S. Copper-zinc superoxide dismutase: Why not pH-dependent? J. Am. Chem. Soc. 1996;118:6556–6561. [Google Scholar]
- 189.Nedd S., Redler R.L., Proctor E.A., Dokholyan N.V., Alexandrova A.N. Cu,Zn-superoxide dismutase without Zn is folded but catalytically inactive. J. Mol. Biol. 2014;426:4112–4124. doi: 10.1016/j.jmb.2014.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Robinett N.G., Peterson R.L., Culotta V.C. Eukaryotic copper-only superoxide dismutases (SODs): A new class of SOD enzymes and SOD-like protein domains. J. Biol. Chem. 2018;293:4636–4643. doi: 10.1074/jbc.TM117.000182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Valentine J.S., Pantoliano M.W., McDonnell P.J., Burger A.R., Lippard S.J. pH-dependent migration of copper(II) to the vacant zinc-binding site of zinc-free bovine erythrocyte superoxide dismutase. Proc. Natl. Acad. Sci. U. S. A. 1979;76:4245–4249. doi: 10.1073/pnas.76.9.4245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Gleason J.E., Galaleldeen A., Peterson R.L., Taylor A.B., Holloway S.P., Waninger-Saroni J., Cormack B.P., Cabelli D.E., John Hart P., Culotta V.C. Candida albicans SOD5 represents the prototype of an unprecedented class of Cu-only superoxide dismutases required for pathogen defense. Proc. Natl. Acad. Sci. U. S. A. 2014;111:5866–5871. doi: 10.1073/pnas.1400137111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Spagnolo L., Töro I., D'Orazio M., O'Neil P., Pedersen J.Z., Carugo O., Rotilio G., Battistoni A., Djinović-Carugo K. Unique features of the sodC-encoded superoxide dismutase from Mycobacterium tuberculosis, a fully functional copper-containing enzyme lacking zinc in the active site. J. Biol. Chem. 2004;279:33447–33455. doi: 10.1074/jbc.M404699200. [DOI] [PubMed] [Google Scholar]
- 194.Peterson R.L., Galaleldeen A., Villarreal J., Taylor A.B., Cabelli D.E., Hart P.J., Culotta V.C. The phylogeny and active site design of eukaryotic copper-only superoxide dismutases. J. Biol. Chem. 2016;291:20911–20923. doi: 10.1074/jbc.M116.748251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Alfano M., Cavazza C. Structure, function, and biosynthesis of nickel-dependent enzymes. Protein Sci. 2020;29:1071–1089. doi: 10.1002/pro.3836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Kim F.J., Kim H.P., Hah Y.C., Roe J.H. Differential expression of superoxide dismutases containing Ni and Fe/Zn in Streptomyces coelicolor. Eur. J. Biochem. 1996;241:178–185. doi: 10.1111/j.1432-1033.1996.0178t.x. [DOI] [PubMed] [Google Scholar]
- 197.Youn H.D., Kim E.J., Roe J.H., Hah Y.C., Kang S.O. A novel nickel-containing superoxide dismutase from Streptomyces spp. Biochem. J. 1996;318:889–896. doi: 10.1042/bj3180889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Deshpande A.R., Wagenpfeil K., Pochapsky T.C., Petsko G.A., Ringe D. Metal-dependent function of a mammalian acireductone dioxygenase. Biochemistry. 2016;55:1398–1407. doi: 10.1021/acs.biochem.5b01319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Taylor Ringia E.A., Garrett J.B., Thoden J.B., Holden H.M., Rayment I., Gerlt J.A. Evolution of enzymatic activity in the enolase superfamily: Structural studies of the promiscuous o-succinylbenzoate synthase from amycolatopsis. Biochemistry. 2004;43:5716–5727. doi: 10.1021/bi0497897. [DOI] [PubMed] [Google Scholar]
- 200.Kim E.-J., Chung H.-J., Suh B., Hah Y.C., Roe J.-H. Transcriptional and post-transcriptional regulation by nickel of sodN gene encoding nickel-containing superoxide dismutase from Streptomyces coelicolor Müller. Mol. Microbiol. 1998;27:187–195. doi: 10.1046/j.1365-2958.1998.00674.x. [DOI] [PubMed] [Google Scholar]
- 201.Leclere V., Boiron P., Blondeau R. Diversity of superoxide-dismutases among clinical and soil isolates of streptomyces species. Curr. Microbiol. 1999;39:365–368. doi: 10.1007/s002849900473. [DOI] [PubMed] [Google Scholar]
- 202.Kehl-Fie T.E., Zhang Y., Moore J.L., Farrand A.J., Hood M.I., Rathi S., Chazin W.J., Caprioli R.M., Skaar E.P. MntABC and MntH contribute to systemic Staphylococcus aureus infection by competing with calprotectin for nutrient manganese. Infect. Immun. 2013;81:3395–3405. doi: 10.1128/IAI.00420-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Kehres D.G., Zaharik M.L., Finlay B.B., Maguire M.E. The NRAMP proteins of Salmonella typhimurium and Escherichia coli are selective manganese transporters involved in the response to reactive oxygen. Mol. Microbiol. 2000;36:1085–1100. doi: 10.1046/j.1365-2958.2000.01922.x. [DOI] [PubMed] [Google Scholar]
- 204.Claverys J.-P. A new family of high-affinity ABC manganese and zinc permeases. Res. Microbiol. 2001;152:231–243. doi: 10.1016/s0923-2508(01)01195-0. [DOI] [PubMed] [Google Scholar]
- 205.Anjem A., Varghese S., Imlay J.A. Manganese import is a key element of the OxyR response to hydrogen peroxide in Escherichia coli. Mol. Microbiol. 2009;72:844–858. doi: 10.1111/j.1365-2958.2009.06699.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Beyer W.F., Fridovich I. In vivo competition between iron and manganese for occupancy of the active site region of the manganese-superoxide dismutase of Escherichia coli. J. Biol. Chem. 1991;266:303–308. [PubMed] [Google Scholar]
- 207.Whittaker M.M., Whittaker J.W. Metallation state of human manganese superoxide dismutase expressed in Saccharomyces cerevisiae. Arch. Biochem. Biophys. 2012;523:191–197. doi: 10.1016/j.abb.2012.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Johansson R., Torrents E., Lundin D., Sprenger J., Sahlin M., Sjöberg B.M., Logan D.T. High-resolution crystal structures of the flavoprotein NrdI in oxidized and reduced states - an unusual flavodoxin: Structural biology. FEBS J. 2010;277:4265–4277. doi: 10.1111/j.1742-4658.2010.07815.x. [DOI] [PubMed] [Google Scholar]
- 209.Cotruvo J.A., Stubbe J. NrdI, a flavodoxin involved in maintenance of the diferric-tyrosyl radical cofactor in Escherichia coli class Ib ribonucleotide reductase. Proc. Natl. Acad. Sci. U. S. A. 2008;105:14383–14388. doi: 10.1073/pnas.0807348105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Lundin D., Torrents E., Poole A.M., Sjöberg B.M. RNRdb, a curated database of the universal enzyme family ribonucleotide reductase, reveals a high level of misannotation in sequences deposited to Genbank. BMC Genomics. 2009;10:589. doi: 10.1186/1471-2164-10-589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Rhodes D.L.V., Crump K.E., Makhlynets O., Snyder M., Ge X., Xu P., Stubbe J.A., Kitten T. Genetic characterization and role in virulence of the ribonucleotide reductases of Streptococcus sanguinis. J. Biol. Chem. 2014;289:6273–6287. doi: 10.1074/jbc.M113.533620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Jordan M.R., Wang J., Capdevila D.A., Giedroc D.P. Multi-metal nutrient restriction and crosstalk in metallostasis systems in microbial pathogens. Curr. Opin. Microbiol. 2020;55:17–25. doi: 10.1016/j.mib.2020.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Chandrangsu P., Huang X., Gaballa A., Helmann J.D. Bacillus subtilis FolE is sustained by the ZagA zinc metallochaperone and the alarmone ZTP under conditions of zinc deficiency. Mol. Microbiol. 2019;112:751–765. doi: 10.1111/mmi.14314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Li Y., Sharma M.R., Koripella R.K., Yang Y., Kaushal P.S., Lin Q., Wade J.T., Gray T.A., Derbyshire K.M., Agrawal R.K., Ojha A.K. Zinc depletion induces ribosome hibernation in mycobacteria. Proc. Natl. Acad. Sci. U. S. A. 2018;115:8191–8196. doi: 10.1073/pnas.1804555115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Radin J.N., Kelliher J.L., Solórzano P.K.P., Grim K.P., Ramezanifard R., Slauch J.M., Kehl-Fie T.E. Metal-independent variants of phosphoglycerate mutase promote resistance to nutritional immunity and retention of glycolysis during infection. PLoS Pathog. 2019;15:1–21. doi: 10.1371/journal.ppat.1007971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Frohner I.E., Bourgeois C., Yatsyk K., Majer O., Kuchler K. Candida albicans cell surface superoxide dismutases degrade host-derived reactive oxygen species to escape innate immune surveillance. Mol. Microbiol. 2009;71:240–252. doi: 10.1111/j.1365-2958.2008.06528.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Robinett N.G., Culbertson E.M., Peterson R.L., Sanchez H., Andes D.R., Nett J.E., Culotta V.C. Exploiting the vulnerable active site of a copper-only superoxide dismutase to disrupt fungal pathogenesis. J. Biol. Chem. 2019;294:2700–2713. doi: 10.1074/jbc.RA118.007095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Ghssein G., Matar S. Chelating mechanisms of transition metals by bacterial metallophores “pseudopaline and staphylopine”: A quantum chemical assessment. Computation. 2018;6:56. [Google Scholar]
- 219.Ghssein G., Brutesco C., Ouerdane L., Fojcik C., Izaute A., Wang S., Hajjar C., Lobinski R., Lemaire D., Richaud P., Voulhoux R., Espaillat A., Cava F., Pignol D., Borezee-Durant E., et al. Biosynthesis of a broad-spectrum nicotianamine-like metallophore in Staphylococcus aureus. Science. 2016;352:1105–1109. doi: 10.1126/science.aaf1018. [DOI] [PubMed] [Google Scholar]
- 220.Zackular J.P., Chazin W.J., Skaar E.P. Nutritional immunity: S100 proteins at the host-pathogen interface. J. Biol. Chem. 2015;290:18991–18998. doi: 10.1074/jbc.R115.645085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Nakashige T.G., Zhang B., Krebs C., Nolan E.M. Human calprotectin is an iron-sequestering host-defense protein. Nat. Chem. Biol. 2015;11:765–771. doi: 10.1038/nchembio.1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Urban C.F., Ermert D., Schmid M., Abu-Abed U., Goosmann C., Nacken W., Brinkmann V., Jungblut P.R., Zychlinsky A. Neutrophil extracellular traps contain calprotectin, a cytosolic protein complex involved in host defense against Candida albicans. PLoS Pathog. 2009;5 doi: 10.1371/journal.ppat.1000639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Liu J.Z., Jellbauer S., Poe A.J., Ton V., Pesciaroli M., Kehl-Fie T.E., Restrepo N.A., Hosking M.P., Edwards R.A., Battistoni A., Pasquali P., Lane T.E., Chazin W.J., Vogl T., Roth J., et al. Zinc sequestration by the neutrophil protein calprotectin enhances salmonella growth in the inflamed gut. Cell Host Microbe. 2012;11:227–239. doi: 10.1016/j.chom.2012.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Drakesmith H., Prentice A. Viral infection and iron metabolism. Nat. Rev. Microbiol. 2008;6:541–552. doi: 10.1038/nrmicro1930. [DOI] [PubMed] [Google Scholar]
- 225.Chaturvedi U.C., Shrivastava R. Interaction of viral proteins with metal ions: Role in maintaining the structure and functions of viruses. FEMS Immunol. Med. Microbiol. 2005;43:105–114. doi: 10.1016/j.femsim.2004.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Chaturvedi U.C., Shrivastava R., Upreti R.K. Viral infections and trace elements: A complex interaction. Curr. Sci. 2004;87:1536–1554. [Google Scholar]
- 227.Chasapis C.T. Interactions between metal binding viral proteins and human targets as revealed by network-based bioinformatics. J. Inorg. Biochem. 2018;186:157–161. doi: 10.1016/j.jinorgbio.2018.06.012. [DOI] [PubMed] [Google Scholar]
- 228.De Jesus J.R., De Araújo Andrade T. Understanding the relationship between viral infections and trace elements from a metallomics perspective: Implications for COVID-19. Metallomics. 2020;12:1912–1930. doi: 10.1039/d0mt00220h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Mehta P., McAuley D.F., Brown M., Sanchez E., Tattersall R.S., Manson J.J. COVID-19: Consider cytokine storm syndromes and immunosuppression. Lancet. 2020;395:1033–1034. doi: 10.1016/S0140-6736(20)30628-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Edeas M., Saleh J., Peyssonnaux C. Iron: Innocent bystander or vicious culprit in COVID-19 pathogenesis? Int. J. Infect. Dis. 2020;97:303–305. doi: 10.1016/j.ijid.2020.05.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Ehsani S. COVID-19 and iron dysregulation: Distant sequence similarity between hepcidin and the novel coronavirus spike glycoprotein. Biol. Direct. 2020;15:19. doi: 10.1186/s13062-020-00275-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Liu W., Zhang S., Nekhai S., Liu S. Depriving iron supply to the virus represents a promising adjuvant therapeutic against viral survival. Curr. Clin. Microbiol. Rep. 2020;7:13–19. doi: 10.1007/s40588-020-00140-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Bhardwaj K., Guarino L., Kao C.C. The severe acute respiratory syndrome coronavirus Nsp15 protein is an endoribonuclease that prefers manganese as a cofactor. J. Virol. 2004;78:12218–12224. doi: 10.1128/JVI.78.22.12218-12224.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Jablonski S.A., Morrow C.D. Mutation of the aspartic acid residues of the GDD sequence motif of poliovirus RNA-dependent RNA polymerase results in enzymes with altered metal ion requirements for activity. J. Virol. 1995;69:1532–1539. doi: 10.1128/jvi.69.3.1532-1539.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Maio N., Lafont B.A.P., Sil D., Li Y., Bollinger J.M., Krebs C., Pierson T.C., Linehan W.M., Rouault T.A. Fe-S cofactors in the SARS-CoV-2 RNA-dependent RNA polymerase are potential antiviral targets. Science. 2021;373:236–241. doi: 10.1126/science.abi5224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Sankaran V.G., Weiss M.J. Anemia: Progress in molecular mechanisms and therapies. Nat. Med. 2015;21:221–230. doi: 10.1038/nm.3814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Bi Y., Ajoolabady A., Demillard L.J., Yu W., Hilaire M.L., Zhang Y., Ren J. Dysregulation of iron metabolism in cardiovascular diseases: From iron deficiency to iron overload. Biochem. Pharmacol. 2021;190:114661. doi: 10.1016/j.bcp.2021.114661. [DOI] [PubMed] [Google Scholar]
- 238.Rines A.K., Ardehali H. Transition metals and mitochondrial metabolism in the heart. J. Mol. Cell. Cardiol. 2013;55:50–57. doi: 10.1016/j.yjmcc.2012.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Rocha B.M.L., Cunha G.J.L., Menezes Falcão L.F. The burden of iron deficiency in heart failure: Therapeutic approach. J. Am. Coll. Cardiol. 2018;71:782–793. doi: 10.1016/j.jacc.2017.12.027. [DOI] [PubMed] [Google Scholar]
- 240.Cornelissen A., Guo L., Sakamoto A., Virmani R., Finn A.V. New insights into the role of iron in inflammation and atherosclerosis. EBioMedicine. 2019;47:598–606. doi: 10.1016/j.ebiom.2019.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Kraml P. The role of iron in the pathogenesis of atherosclerosis. Physiol. Res. 2017;66:S55–S67. doi: 10.33549/physiolres.933589. [DOI] [PubMed] [Google Scholar]
- 242.Evans G.W. Zinc and its deficiency diseases. Clin. Physiol. Biochem. 1986;4:94–98. [PubMed] [Google Scholar]
- 243.Bleackley M.R., Macgillivray R.T.A. Transition metal homeostasis: From yeast to human disease. Biometals. 2011;24:785–809. doi: 10.1007/s10534-011-9451-4. [DOI] [PubMed] [Google Scholar]
- 244.Taher A. Iron overload in thalassemia and sickle cell disease. Semin. Hematol. 2005;42:S5–S9. doi: 10.1053/j.seminhematol.2005.01.005. [DOI] [PubMed] [Google Scholar]
- 245.Klockgether T. Update on degenerative ataxias. Curr. Opin. Neurol. 2011;24:339–345. doi: 10.1097/WCO.0b013e32834875ba. [DOI] [PubMed] [Google Scholar]
- 246.La Rosa P., Petrillo S., Fiorenza M.T., Bertini E.S., Piemonte F. Ferroptosis in Friedreich's ataxia: A metal-induced neurodegenerative disease. Biomolecules. 2020;10 doi: 10.3390/biom10111551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Dusek P., Roos P.M., Litwin T., Schneider S.A., Flaten T.P., Aaseth J. The neurotoxicity of iron, copper and manganese in Parkinson's and Wilson's diseases. J. Trace Elem. Med. Biol. 2015;31:193–203. doi: 10.1016/j.jtemb.2014.05.007. [DOI] [PubMed] [Google Scholar]
- 248.Chaudhry H.S., Anilkumar A.C. StatPearls. StatPearls Publishing; Treasure Island, FL: 2021. Wilson disease. [Google Scholar]
- 249.Ward R.J., Zucca F.A., Duyn J.H., Crichton R.R., Zecca L. The role of iron in brain ageing and neurodegenerative disorders. Lancet Neurol. 2014;13:1045–1060. doi: 10.1016/S1474-4422(14)70117-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Uwitonze A.M., Ojeh N., Murererehe J., Atfi A., Razzaque M.S. Zinc adequacy is essential for the maintenance of optimal oral health. Nutrients. 2020;12 doi: 10.3390/nu12040949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Kontoghiorghes G.J., Efstathiou A., Ioannou-Loucaides S., Kolnagou A. Chelators controlling metal metabolism and toxicity pathways: Applications in cancer prevention, diagnosis and treatment. Hemoglobin. 2008;32:217–227. doi: 10.1080/03630260701727119. [DOI] [PubMed] [Google Scholar]
- 252.Członkowska A., Litwin T. Wilson disease - currently used anticopper therapy. Handb. Clin. Neurol. 2017;142:181–191. doi: 10.1016/B978-0-444-63625-6.00015-X. [DOI] [PubMed] [Google Scholar]
- 253.Zhang M., Shoeb M., Liu P., Xiao T., Hogan D., Wong I.G., Campbell G.A., Ansari N.H. Topical metal chelation therapy ameliorates oxidation-induced toxicity in diabetic cataract. J. Toxicol. Environ. Health A. 2011;74:380–391. doi: 10.1080/15287394.2011.538835. [DOI] [PubMed] [Google Scholar]
- 254.Ben-Shushan S., Miller Y. Neuropeptides: Roles and activities as metal chelators in neurodegenerative diseases. J. Phys. Chem. B. 2021;125:2796–2811. doi: 10.1021/acs.jpcb.0c11151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Roemhild K., von Maltzahn F., Weiskirchen R., Knüchel R., von Stillfried S., Lammers T. Iron metabolism: Pathophysiology and pharmacology. Trends Pharmacol. Sci. 2021;42:640–656. doi: 10.1016/j.tips.2021.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Papp-Wallace K.M., Maguire M.E. Manganese transport and the role of manganese in virulence. Annu. Rev. Microbiol. 2006;60:187–209. doi: 10.1146/annurev.micro.60.080805.142149. [DOI] [PubMed] [Google Scholar]
- 257.De Vendittis A., Amato M., Mickniewicz A., Parlato G., De Angelis A., Castellano I., Rullo R., Riccitiello F., Rengo S., Masullo M., De Vendittis E. Regulation of the properties of superoxide dismutase from the dental pathogenic microorganism Streptococcus mutans by iron- and manganese-bound co-factor. Mol. Biosyst. 2010;6:1973–1982. doi: 10.1039/c003557b. [DOI] [PubMed] [Google Scholar]
- 258.Russo Krauss I., Merlino A., Pica A., Rullo R., Bertoni A., Capasso A., Amato M., Riccitiello F., De Vendittis E., Sica F. Fine tuning of metal-specific activity in the Mn-like group of cambialistic superoxide dismutases. RSC Adv. 2015;5:87876–87887. [Google Scholar]
- 259.Andrus J.M., Bowen S.W., Klaenhammer T.R., Hassan H.M. Molecular characterization and functional analysis of the manganese-containing superoxide dismutase gene (sodA) from Streptococcus thermophilus AO54. Arch. Biochem. Biophys. 2003;420:103–113. doi: 10.1016/j.abb.2003.09.007. [DOI] [PubMed] [Google Scholar]
- 260.Privalle C.T., Fridovich I. Transcriptional and maturational effects of manganese and iron on the biosynthesis of manganese-superoxide dismutase in Escherichia coli. J. Biol. Chem. 1992;267:9140–9145. [PubMed] [Google Scholar]
- 261.Privalle C.T., Fridovich I. Inductions of superoxide dismutases in Escherichia coli under anaerobic conditions. Accumulation of an inactive form of the manganese enzyme. J. Biol. Chem. 1988;263:4274–4279. [PubMed] [Google Scholar]
- 262.Gabbianelli R., D'Orazio M., Pacello F., O'Neill P., Nicolini L., Rotilio G., Battistoni A. Distinctive functional features in prokaryotic and eukaryotic Cu,Zn superoxide dismutases. Biol. Chem. 2004;385:749–754. doi: 10.1515/BC.2004.091. [DOI] [PubMed] [Google Scholar]
- 263.Schäfer G., Kardinahl S. Iron superoxide dismutases: Structure and function of an archaic enzyme. Biochem. Soc. Trans. 2003;31:1330–1334. doi: 10.1042/bst0311330. [DOI] [PubMed] [Google Scholar]
- 264.Chang E.C., Crawford B.F., Hong Z., Bilinski T., Kosman D.J. Genetic and biochemical characterization of Cu,Zn superoxide dismutase mutants in Saccharomyces cerevisiae. J. Biol. Chem. 1991;266:4417–4424. [PubMed] [Google Scholar]
- 265.Luk E., Yang M., Jensen L.T., Bourbonnais Y., Culotta V.C. Manganese activation of superoxide dismutase 2 in the mitochondria of Saccharomyces cerevisiae. J. Biol. Chem. 2005;280:22715–22720. doi: 10.1074/jbc.M504257200. [DOI] [PubMed] [Google Scholar]
- 266.Yang M., Cobine P.A., Molik S., Naranuntarat A., Lill R., Winge D.R., Culotta V.C. The effects of mitochondrial iron homeostasis on cofactor specificity of superoxide dismutase 2. EMBO J. 2006;25:1775–1783. doi: 10.1038/sj.emboj.7601064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Zelko I.N., Mariani T.J., Folz R.J. Superoxide dismutase multigene family: A comparison of the CuZn-SOD (SOD1), Mn-SOD (SOD2), and EC-SOD (SOD3) gene structures, evolution, and expression. Free Radic. Biol. Med. 2002;33:337–349. doi: 10.1016/s0891-5849(02)00905-x. [DOI] [PubMed] [Google Scholar]
- 268.Antonyuk S.V., Strange R.W., Marklund S.L., Hasnain S.S. The structure of human extracellular copper-zinc superoxide dismutase at 1.7 A resolution: Insights into heparin and collagen binding. J. Mol. Biol. 2009;388:310–326. doi: 10.1016/j.jmb.2009.03.026. [DOI] [PubMed] [Google Scholar]
- 269.Shrestha P., Yun J.-H., Kim W.T., Kim T.-Y., Lee W. Cloning, purification, and characterization of recombinant human extracellular superoxide dismutase in SF9 insect cells. Mol. Cells. 2016;39:242–249. doi: 10.14348/molcells.2016.2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Chen H.-Y., Hu R.-G., Wang B.-Z., Chen W.-F., Liu W.-Y., Schröder W., Frank P., Ulbrich N. Structural studies of an eukaryotic cambialistic superoxide dismutase purified from the mature seeds of camphor tree. Arch. Biochem. Biophys. 2002;404:218–226. doi: 10.1016/s0003-9861(02)00299-0. [DOI] [PubMed] [Google Scholar]
- 271.Wang Y., Branicky R., Noë A., Hekimi S. Superoxide dismutases: Dual roles in controlling ROS damage and regulating ROS signaling. J. Cell Biol. 2018;217:1915–1928. doi: 10.1083/jcb.201708007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Moore E.K., Jelen B.I., Giovannelli D., Raanan H., Falkowski P.G. Metal availability and the expanding network of microbial metabolisms in the Archaean eon. Nat. Geosci. 2017;10:629–636. [Google Scholar]