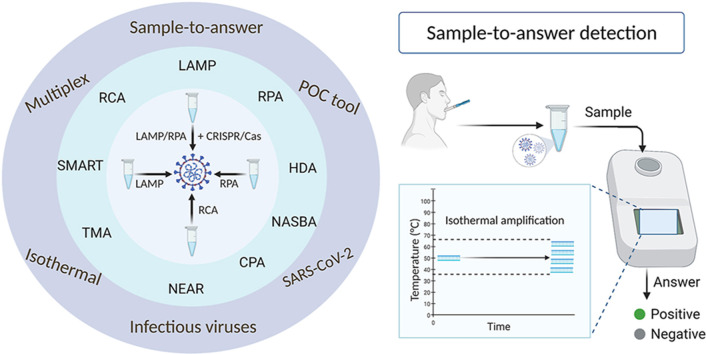
Abstract
As the COVID-19 pandemic continues to affect human health across the globe rapid, simple, point-of-care (POC) diagnosis of infectious viruses such as SARS-CoV-2 remains challenging. Polymerase chain reaction (PCR)-based diagnosis has risen to meet these demands and despite its high-throughput and accuracy, it has failed to gain traction in the rapid, low-cost, point-of-test settings. In contrast, different emerging isothermal amplification-based detection methods show promise in the rapid point-of-test market. In this comprehensive study of the literature, several promising isothermal amplification methods for the detection of SARS-CoV-2 are critically reviewed that can also be applied to other infectious viruses detection. Starting with a brief discussion on the SARS-CoV-2 structure, its genomic features, and the epidemiology of the current pandemic, this review focuses on different emerging isothermal methods and their advancement. The potential of isothermal amplification combined with the revolutionary CRISPR/Cas system for a more powerful detection tool is also critically reviewed. Additionally, the commercial success of several isothermal methods in the pandemic are highlighted. Different variants of SARS-CoV-2 and their implication on isothermal amplifications are also discussed. Furthermore, three most crucial aspects in achieving a simple, fast, and multiplexable platform are addressed.
Keywords: SARS-CoV-2 detection, Infectious viruses detection, Isothermal amplification, CRISPR/Cas, Variants, Pandemic
Graphical abstract
1. Introduction
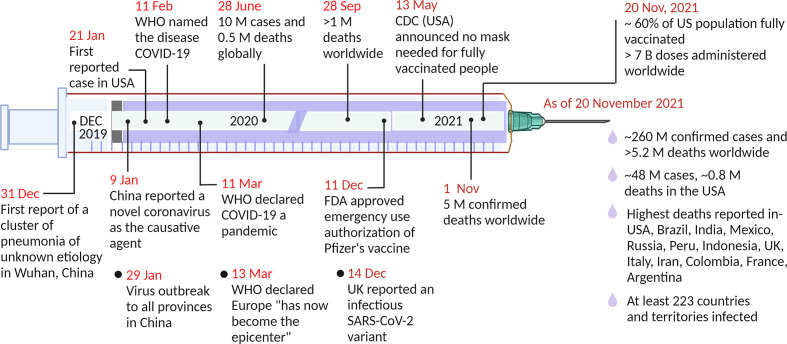
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has continued to affect many countries since the first case was reported in Wuhan, China, at the end of 2019 [1,2], Subsequently, local human-to-human transmission of the virus occurred rapidly. Within months, it had spread across the globe [3,4]. Within the next nine months, the pandemic caused more than 1 million deaths worldwide. Development of different vaccines and increased vaccination across the world have been going on since the beginning of 2021. Many countries, including the United States, United Kingdom, France, Italy, Australia, Spain, and Germany, have been able to control the pandemic to at least some extent, largely due to widespread testing and vaccination. However, the number of cases reported has fluctuated widely in highly populated countries, including India, Mexico, Bangladesh, Indonesia, Brazil, and Russia. As of November 20, 2021, more than 5 million people have died due to the current pandemic and approximately 260 million cases of infection have been reported across a total of at least 223 countries and territories [5]. Fig. 1 illustrates a timeline of some of the major events of the pandemic. This numbers are increasing as the virus is evolving with different infectious variants emerging in different countries. Vaccines can provide an avenue for mitigating the spread of the pandemic, however, breakthrough infections can still happen due to the emergence of variants and lessened vaccine effectiveness over time [6,7]. Thus, a system for rapid, simple, and high throughput diagnostic testing for the detection of SARS-CoV-2 has a crucial role to control the pandemic. Moreover, the development of such system can also contribute to the unrelenting demand of other infectious viruses detection.
Fig. 1.
Timeline of the major events of the COVID-19 pandemic. WHO, World Health Organization; FDA, Food and Drug Administration; CDC, Centers for Disease Control and Prevention. Created with BioRender.com.
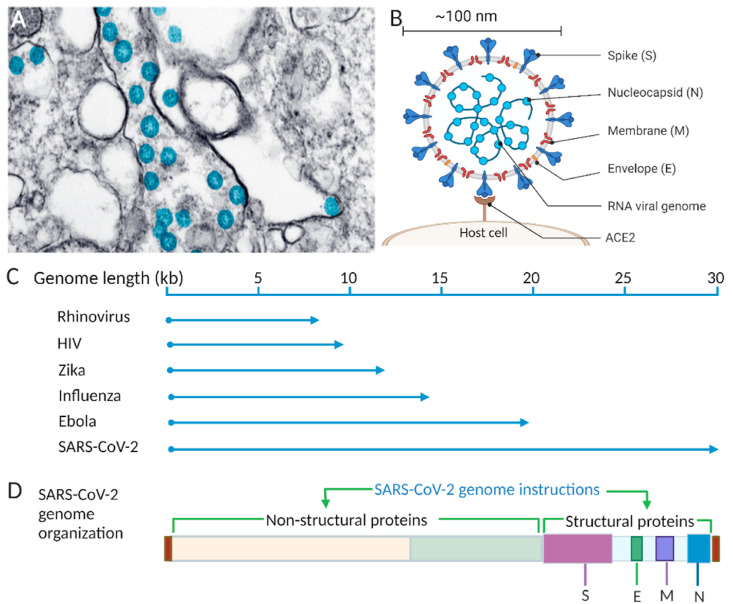
Different aspects of SARS-CoV-2, such as the structural properties, genomic information, mode of transmission, mechanism of binding to host cells and replication, have been well studied [[8], [9], [10], [11], [12]]. The virion is mostly spherical, with a diameter in the low nanometer range (∼100 nm) [13,14]. A transmission electron microscope image (Fig. 2 A) shows spherical SARS-CoV-2 particles that were obtained from an isolate of the first case in the USA provided by the CDC [15,16]. The main feature of SARS-CoV-2 is four structural proteins (spike (S), membrane (M), envelope (E), and nucleocapsid (N)) (Fig. 2B).
Fig. 2.
SARS-CoV-2 morphology and genomic illustrations. A) Transmission electron microscope image of SARS-CoV-2 viral particles (blue) [15], B) Schematic of SARS-CoV-2 with structural proteins, and its genome, binding interaction between host cell and virus spike protein, C) Comparison of the SARS-CoV-2 viral genome with genomes of other common RNA viruses. D) SARS-CoV-2 genome organization showing the relative positions of nonstructural and structural proteins. Created with BioRender.com. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
The virus genome is a positive-sense, single-stranded RNA 29,903 bases long [9], which makes it one of the largest genomes of the well-known RNA viruses, such as influenza (13.5 kb) [17], rhinovirus (7.2 kb) [18], Zika (10.8 kb) [19], HIV (9 kb) [20], and Ebola (∼19 kb) [21] (Fig. 2C). This relatively long genome of SARS-CoV-2 carries necessary genetic information to encode diverse structural and nonstructural proteins (Fig. 2D). The upstream region, near the 5′ end of the genome, contains two large open reading frames (∼21 kb), that encode a polyprotein that is cleaved by proteases (protease originated from the polyprotein) into nonstructural proteins of the virus, including an RNA-dependent RNA polymerase (RdRP) and an exonuclease [13]. Near the 3′ end, the genome encodes four structural proteins [22]: S, E, M, and N proteins. These proteins define the structure of the virus particle and thereby its ability to infect the human body. There are also interspaced regions between the four structural proteins for accessory proteins in the same 3’ end of the genome. The S protein of the coronavirus is the key to mediating the host-pathogen interaction. The virus infects people preferentially through the S protein by the combined action of its two functional subunits. One of the subunits, S1, binds between the virus particle and the host cell receptor angiotensin-converting enzyme 2 (ACE2) (Fig. 2B). The other subunit, S2, facilitates the fusion of the host cell membrane and the virus particle [23]. The overall role of the S protein in the efficient entry and infectivity of the virus has been reported elsewhere [24,25].
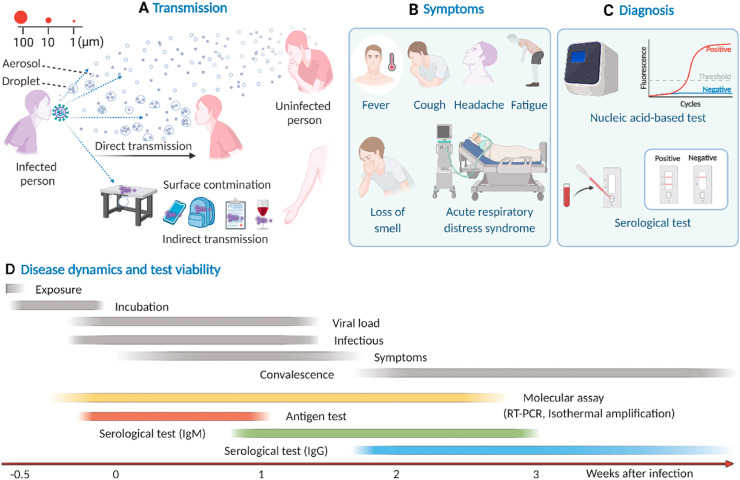
The virus transmits primarily by two main routes: directly via respiratory droplets containing the virus from an infected to an uninfected person in proximity and indirectly from various contaminated surfaces [26] (Fig. 3 A). Airborne transmission may occur through suspended fine droplets and aerosols generated from respiratory fluids exhaled by an infected person [27]. While human-to-human direct transmission occurs quickly and effectively, indirect transmission through contaminated surfaces depends on the stability and infectiousness of the virus particles. Stability depends on the nature of the surface on which the virus particles are deposited and even on the ambient temperature and humidity. Smooth surfaces provide the greatest stability over a wide range of temperature (4–37 °C) [28].
Fig. 3.
Viral epidemiology of SARS-CoV-2. A) Transmission, B) Clinical symptoms, C) Diagnosis, and D) Disease dynamics and test viability. Figure D is reprinted by permission from [Springer Nature] [Nature Reviews Genetics], Testing at scale during the COVID-19 pandemic, Tim R. Mercer et al., [31]. Copyright 2021. Created with BioRender.com.
SARS-CoV-2-infected people develop common respiratory illness symptoms including fever, shortness of breath, loss of smell and taste, headache, and fatigue (Fig. 3B). Older infected people (>60 years) are more likely to experience greater disease severity [29]. In particular, older people with pre-existing disease are prone to severe illness manifested by acute respiratory distress syndrome, which most often leads to death. However, many of the infected people show very mild or no clinical symptoms [30] but are still contagious, making it difficult to control the transmission.
Ideally, a routine testing practice prior to the development of symptoms and isolation of the suspected individual is the crucial way to control virus transmission. However, the lack of adequate testing in many countries limits their ability to correctly assess the extent of the pandemic, which catalyzes the fast spread of the virus in many parts of the world. In many resource-limited countries, tests are preferentially performed on people who have clinical symptoms or have been exposed to SARS-CoV-2, leading to difficulty assessing the true spread of the disease. After more than a year, promising vaccines have been developed and are now being administered [32]. However, routine testing for SARS-CoV-2 will still be necessary as we enter the post pandemic era for monitoring long-term vaccine efficacy.
Nucleic acid-based testing and rapid antigen tests are the two classes of viral testing methods widely employed as diagnostic tests. As the name suggests, rapid antigen tests detect the presence of viral proteins in respiratory specimens typically by a lateral flow immunoassay approach. Most of these type of detections only take minutes to complete, are inexpensive, and can be used in POC settings [33]. However, they are inherently less sensitive than nucleic acid-based test. Unlike rapid antigen tests, nucleic acid-based test involves target amplification, making it a highly sensitive detection method (Fig. 3C).
Antibody tests are also used in some cases. Antibody-based tests commonly detect anti-SARS-CoV-2 IgM and IgG antibodies in blood using lateral flow assays or enzyme-linked immunosorbent assay (ELISA) [34]. A detailed discussion of this type of test can be found elsewhere [[35], [36], [37]]. Viral infection triggers the generation of antibodies. For serological tests, a detectable level of the IgM antibody is generated after at least one week of infection and gradually deceases after three weeks of infection, while the IgG antibody is generated after two weeks of infection and may remain detectable for weeks (Fig. 3D).
Because antibody-based tests detect host immune response as opposed to viral components of SARS-CoV-2 and the less sensitive nature of rapid antigen tests; nucleic acid-based molecular test is considered the standard method of SARS-CoV-2 detection. Nucleic acid-based test sensitively detects genetic footprints in one or more specific target regions of the SARS-CoV-2 RNA. The ability of molecular test to sensitively detect viral RNA depends on several factors, such as the durations of the incubation period, viral shedding, and onset of symptoms, each of which has a time window with relative intensity [38,39] (Fig. 3D). Infection occurs with exposure to the virus and a subsequent incubation period of a few days [40] before clinical symptoms begin to surface in the body [41]. Viral shedding starts a few days before the onset of symptoms, and the viral load reaches its maximum level within the first week of symptoms onset; therefore, infectiousness corresponds to the same time window. In addition, the clinical sensitivity of molecular tests often depends on the time relative to the symptoms onset and on the viral load in an individual [42,43]. Typically, viral loads start to reach at peak just before symptoms and persist a little more than a week, although can be at peak for prolonged time [44]. It is when molecular tests can most accurately detect viral infection in symptomatic patients [31]. However, high viral load does not necessarily associate with disease symptoms. A study shows that asymptomatic patients can also have higher viral loads [45]. Moreover, it can be highly varied in different types of specimens [43] and those collected from asymptomatic, pre symptomatic and symptomatic patients [46].
The nucleic acid-based test of SARS-CoV-2 starts with the collection of specimens from suspected patients, which are then treated to extract viral RNA. If necessary, collected specimen can be stored for up to 2–3 days at 2–8 °C before subsequent extraction of viral RNA. Viral RNA is extracted and purified from deactivated virus particles after cell lysis using a standard protocol [47].
The current widely used nucleic acid-based detection of SARS-CoV-2 proceeds via reverse transcription-polymerase chain reaction (RT-PCR) mediated by reverse transcriptase and DNA polymerase enzymes respectively. Both enzymatic processes of the RT-PCR are performed in a single step using extracted and purified viral RNA. RNA is converted to complementary DNA (cDNA) that is subsequently amplified and detected in the presence of appropriate primers and probes. The thermal profile of the RT-PCR is initiated at ∼50 °C, to allow for reverse transcription by the often heat intolerant reverse transcriptase enzyme, followed by a standard polymerase chain reaction that relies on a series of temperature cycles where each cycle consists of switching between higher temperature (denaturation of the duplex structure at ∼ 95 °C) to lower temperature steps (primer hybridization and elongation at ∼ 55–60 °C). There are wide ranges of RT-PCR thermal profiles based on the duration of each step that might vary slightly depending mainly on specific test's polymerase, primers, and probes and instrument used. The most common readout mode of RT-PCR-based testing uses fluorescence-based detection system. A recent review on RT-PCR-based testing is published elsewhere [48]. It may be noted that RT-PCR does not directly detect the presence of SARS-CoV-2. RT-PCR positive results can confirm the presence of viral RNA from clinical samples but do not necessarily mean that the patient is infected with the disease at the time of testing [49]. Interpreting the detection of viral RNA presence as an active infection can thus be inaccurate. The test can detect residual viral RNA, which does not necessarily come from active virus in the body.
Nevertheless, RT-PCR remains the first choice for infectious virus diagnosis and has been widely employed for SARS-CoV-2 detection. However, several aspects of RT-PCR-based detection are not quite convenient for simple and fast diagnosis. The most obvious disadvantage is the requirement of an expensive thermocycler, which is also a deterrent for typical point-of-care (POC) facilities. RT-PCR can take 4–6 h turnaround time [50] and needs highly trained personnel to obtain the test results, creating severe limitations on disease management in low-resource countries, as testing centers are expensive to build and hard to staff [51]. Moreover, RT-PCR requires specific workplace standards to establish a high-efficiency assay. In particular, viral RNA detection via RT-PCR is very sensitive to contaminants present from raw clinical samples during RNA extraction [52]. Inhibitors affect DNA polymerase activity in the amplification steps and can significantly reduce the amplification efficiency.
One distinctive advantage of an isothermal-based amplification, as opposed to PCR, method is that it amplifies the target nucleic acid at a constant temperature, thus avoiding the requirement for an expensive thermal cycler. Ideally, any isothermal amplification can be performed with a very simple heating source such as a water bath or Peltier heater requiring minimal energy input. This feature of isothermal amplifications enables them to function as POC-based tests. Moreover, different amplification mechanisms of several emerging isothermal methods allow detection assays to be run at a wide range of temperatures. The fluorescence probes used in RT-PCR can be expensive, while colorimetric indicators used in isothermal amplification are readily available at low cost.
2. Isothermal amplification methods for the detection of SARS-CoV-2 and other infectious viruses
Promising isothermal amplification methods started to emerge in the early 1990s, i.e., within a few years after the invention of PCR [53]. New isothermal techniques have evolved over the years by versatile probe and primer design, exploring, and engineering new enzymatic activities, and investigating specific reaction conditions. Because of continuing research to improve analytical performance, each existing isothermal technique can suggest or transform into a new technique. However, all these methods are unified because they operate isothermally. Because of their versatility as both enzymatic [54,55] and nonenzymatic [56] systems, isothermal-based amplifications have been at the forefront of nucleic acid research for many years. Several isothermal amplification methods have gained widespread application in biosensing [[57], [58], [59], [60]]. Owing to their simplicity in instrumentation, and diverse detection methods, it is no surprise that a significant number of reports over the last year have been published on isothermal amplification-based SARS-CoV-2 detection. Some of the methods have found commercial applications as well as approval from the FDA to detect SARS-CoV-2, while some of the methods show great potential toward diagnostic application but have yet to find commercial success. Isothermal methods can potentially be applied to detect any virus infection based on nucleic acid-based molecular assays. In the following sections, several emerging isothermal-based amplifications and their potential as simple and rapid detection methods are emphasized. As the pandemic continues, different commercial platforms based on isothermal amplification have been developed and received emergency use authorization. Some of these platforms that are currently available in the market are highlighted. CRISPR/Cas coupled isothermal amplifications are also focused. Critical functions of different Cas enzymes and their further development toward efficient combined detection with emerging isothermal amplifications are discussed. Different infectious variants of SARS-CoV-2 and their implications on the isothermal-based detection are also discussed.
2.1. Reverse transcription loop-mediated amplification (RT-LAMP)
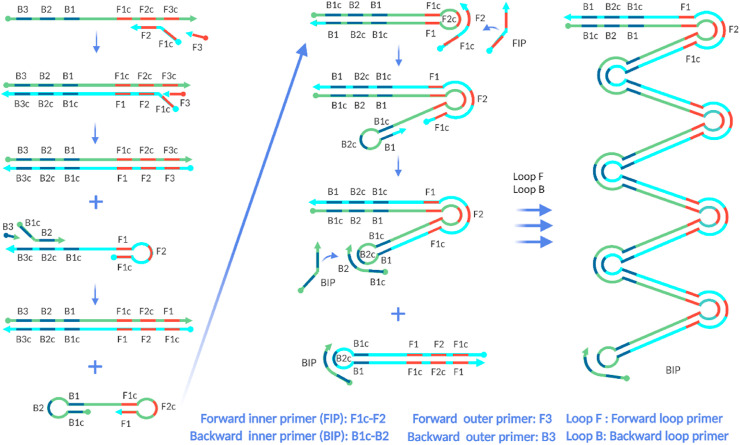
Loop-mediated amplification (LAMP) was first reported in 2000 by Notomi et al., [61]. LAMP is typically performed between 60 and 65 °C. Similar to RT-PCR, RT-LAMP also detects RNA-based viruses with a reverse transcription process prior to amplification in single tube assembly. However, unlike RT-PCR, both RT and LAMP reactions of RT-LAMP can be carried out at ∼65 °C. LAMP requires higher number of primers than PCR. Two pairs of primers, generally known as inner primers (forward and backward) and outer primers (forward and backward), are required. The two inner primers recognize four of six specific regions of the target nucleic acid, and the two outer primers recognize the other two regions. However, most current LAMP-based studies use two additional primers (loop primers) in order to accelerate amplification by annealing to the loop region in the stem-loop structure [62,63]. Fig. 4 shows a simple representation of LAMP mechanism.
Fig. 4.
Simple representation of the LAMP mechanism. In the first step, both inner and outer primer pairs and polymerase enzyme generate dumbbell-like structures from the target. Then, in cycling amplification step, the two inner primers and polymerase further amplify these structures. The elongation steps generate various sizes of the products (three arrows are used to represent various reaction pathways. The two Loop primers are also used in the elongation step. (Reproduced with permission from Ref. [64]). Created with BioRender.com.
LAMP consists of three main steps: formation of the stem-loop structure, amplification, and elongation of the product. It starts with generating a stem-loop structure with the target gene by the combined action of inner and outer primers and a strand displacing DNA polymerase upon the target. The stem loop structure serves as the “seed” for the next step, the loop-mediated cycling amplification step, where only the two inner primers are used. The products from cycling amplification step are used as template for the last step: the elongation and recycling step. Loop primers accelerate the product formation in this step. For clarity, Fig. 4 does not show the binding of loop primers. For elaborate discussion on the design and mechanism of loop primers, readers are directed to the original study by Notomi et al., [62].
As final products, LAMP rapidly generates various lengths of stem-loop DNA with cauliflower-like structures containing many “alternately inverted repeats of the target sequence” [61]. Careful, target-specific design of each primer is essential for highly specific amplification. Basic primer design considerations are the same for PCR and LAMP; however, primer design for LAMP is more challenging. Six LAMP primers are designed to recognize eight specific sites of the intended target gene region. Most often, the intended target region introduces constraints such as distances between binding sites. Two inner primers needed for LAMP are usually long sequences of approximately 45–50 nucleotides. Longer primers add complications, as they are prone to form undesired secondary structures resulting in nonspecific amplification. However, computational approaches [65] and several web-based tools are useful for LAMP primer design [[66], [67], [68]].
2.1.1. Detection of the LAMP product
The LAMP product can be detected by various real-time and endpoint readout modes. Among them, turbidity is one of the oldest readout modes, and it is based on the creation of a byproduct, magnesium pyrophosphate. Amplification progresses as the Bst polymerase (a moderately thermotolerant enzyme derived from Bacillus stearothermophilus with strand displacing activity—a crucial requirement for isothermal amplification) utilizes a high concentration of dNTPs. dNTPs are substrates of Bst polymerase enzymes, and their turnover leaves pyrophosphate anions in the system, which rapidly react with magnesium cations in the buffer [69,70]. This endpoint detection offers simple naked-eye monitoring of the LAMP products; however, commercial turbidimeters now allow quantitative detection in real-time [71,72].
Colorimetry, which permits simple visual monitoring, is another widely used readout mode of LAMP. Colorimetric indicators generate different colors as the LAMP reaction progresses due to the presence of auxiliary components of the reaction system (i.e., MnCl2, Mg2P2O7) and produce qualitative endpoint results. A few examples of such indicators are hydroxy naphthol blue [73] and calcein [74,75], and pH-dependent indicators are phenol red and neutral red [76]. Other indicators, such as malachite green [77] and leuco crystal violet [78], are also employed to detect LAMP products. One potential downside of traditional endpoint colorimetry is that the generated color can be hard to see in complex biological samples; moreover, a pair of “experienced eyes” are needed to interpret the color change, particularly where the LAMP target concentration is very low. Additionally, optimizing the reaction time is perhaps more important for endpoint colorimetric LAMP. Long reaction times can lead to false positives in negative sample controls probably due to the high activity of the polymerase in the LAMP mix. The WarmStart LAMP mix developed by New England Biolabs has been widely employed for LAMP-based detection. It inhibits enzyme activity at a lower temperature to ensure LAMP reaction only occurs at operational temperature >60 °C. This reagent has been used in kits developed by Color Genomics and others for SARS-CoV-2 detection [79]. Real-time and quantitative LAMP with a colorimetric readout mode are also possible. One such study used smartphones and open-source apps to monitor the dye Eriochrome Black T color change in real time in a miniaturized LAMP reaction [80]. A similar real-time colorimetric LAMP reaction was performed in a miniaturized platform using smartphone-based color detection of the phenol red indicator [81]. The study reportedly achieves a limit of detection (LOD) similar to fluorescence-based real-time PCR. Another recent study reported LAMP with a real-time colorimetry readout based on the UV light absorbance of the dye Eriochrome Black T [82].
Fluorescence-based detection is also a widely used readout mode of LAMP. Intercalating dyes have been used to detect LAMP products. One of the advantages of using fluorescent dyes is that they enable both qualitative and quantitative real-time detection. In real-time fluorescence-based assays, the amplified LAMP product is characterized by the threshold time, Tt [83], which is similar to the cycle threshold Ct in PCR. The selection of dyes is crucial for fluorescence-based LAMP readouts. SYBR Green I has been commonly used [84,85] as an intercalating dye that emits green fluorescent light upon irradiation with UV light when bound to dsDNA; PicoGreen and ethidium bromide dyes are also used for the same purpose [[86], [87], [88], [89]]. However, some of these dyes inhibit the LAMP reaction. A detailed comparison of a large number of dyes and their inhibitory effects on the LAMP reaction shows that SYBR Green I significantly inhibits the amplification reaction, while most of the SYTO dyes show no or very little inhibition based on three metrics: the LOD, the signal-to-noise ratio and the Tt [90]. A similar study suggests that SYBR Green dyes are more inhibitory than SYTO dyes [91]. Although widely used in nucleic acid-based amplification reactions, SYBR Green I also inhibits PCR [92,93].
LAMP products can also be detected electrochemically. Different redox-active substances, such as methylene blue [94,95] and ruthenium hexamine [96,97], are employed for quantitative and qualitative detection. Other notable detection modes include gold nanoparticle (AuNP)-based detection [98], antibody-based lateral flow assay [99], and traditional postamplification analysis by gel electrophoresis. For a comprehensive review of different LAMP readout modes, the interested readers are directed to a study by Zhang et al., [100].
The mode of detection of LAMP or any other isothermal amplifications, endpoint or real time, is selected and implemented mainly based on the specific requirement of the assay and available resources. In principle, endpoint modes do not require expensive, sophisticated instrumentation; however, this benefit is offset by them providing only qualitative or less-quantitative information. On the other hand, real-time modes report amplification progress and provide quantitative information, thus generally requiring high-cost, complex instrumentation.
2.1.2. RT-LAMP-based detection of infectious human viruses
LAMP-based assays have shown promising results for detecting viral RNA of several highly infectious viruses. In the early 2000s, a Chikungunya fever outbreak in many parts of the world was caused by a positive-sense single-stranded RNA virus [[101], [102], [103], [104]]. A new isothermal technique based on RT-LAMP effectively diagnosed the virus based on simple visual detection [105].
There are several reports on applying RT-LAMP to mosquito-borne infectious and deadly viruses detection. Teoh et al. used this method to detect Dengue virus, a mosquito-borne RNA-based virus, with high sensitivity and specificity in suspected patient samples [106]. The study was validated with a large number of clinical samples. The LOD was 100 copies of viral RNA. In a recent LAMP study, Jong-Gil Kim and his coworkers were able to detect 3.5 copies/μl viral RNA of Dengue virus [107]. Another mosquito-borne viral pathogen is Zika virus [108]. Wang et al. reported highly sensitive RT-LAMP-based detection of Zika virus. They reported that the detection was more sensitive than RT-PCR by at least one order of magnitude [109].
The Ebola virus has emerged as a highly infectious and deadly virus in recent years in some parts of the world [110]. Benzine et al. reported an excellent RT-LAMP-based assay with a sensitivity of 10 RNA copies/μl to detect Ebola virus in whole blood samples [111]. This assay was equally sensitive and specific as conventional RT-PCR-based assays, and the researchers further developed a POC tool to perform rapid and simple detection on a small and portable testing device.
Following the pandemic in 2009 caused by H1N1 influenza A virus [112], a group of researchers from Japan [113] used a simple pocket warmer device as a heating source to make a potential diagnostic tool based on their modified RT-LAMP method. Their assay performance was evaluated by using nasal swab samples from a large group of suspected patients, and their RT-LAMP assay detected 26 positive samples compared to 27 positive samples detected by regular RT-PCR-based methods, indicated that the LAMP assay is almost equal in sensitivity (96.3%) to RT-PCR.
2.1.3. RT-LAMP-based detection of SARS-CoV-2
RT-LAMP has shown great promise in detecting different coronaviruses. The first successful RT-LAMP-based detection of coronavirus SARS-CoV was reported in 2004 by a group of scientists including Tsugunori Notomi [114], who initially developed the technique in 2000. Using clinical sample, they found that this technique has great potential to detect SARS-CoV, with an LOD of 0.01 PFU. Their novel study reported enhanced sensitivity and specificity compared to those of RT-PCR. Another coronavirus, Middle East respiratory syndrome coronavirus (MERS-CoV), was also detected with high sensitivity and specificity by the LAMP method [[115], [116], [117], [118]].
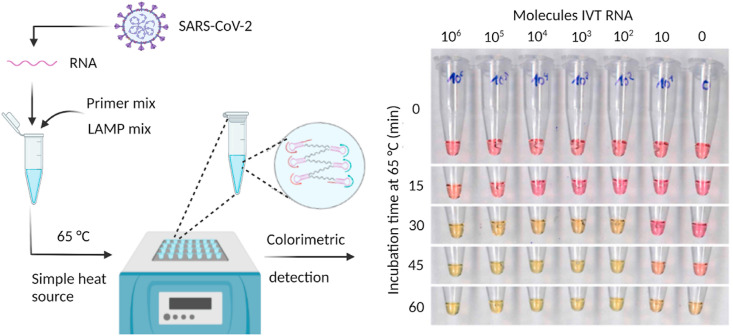
The great promise of LAMP in detecting infectious viruses, including coronaviruses, has inspired researchers worldwide to develop RT-LAMP-based assays for SARS-CoV-2 detection. RT-LAMP-based assay of the virus has the same preamplification steps as RT-PCR, i.e., the extraction of viral RNA to be amplified with a target gene-specific primer mix and necessary enzymatic reagents often commercially called the “LAMP mix” (Fig. 5 ). The fundamental aspects of the RT-LAMP and RT-PCR-based detection of SARS-CoV-2 and any RNA viruses are the same (reverse transcription and subsequent amplification in single step reaction). Moreover, the nature of the detection (endpoint or real-time, qualitative, or quantitative), analytical sensitivity and specificity, reaction time, simplicity of the assay (prior viral RNA extraction from the sample needed or not) are all essential.
Fig. 5.
RT-LAMP assay to detect SARS-CoV-2. The LAMP primer mix contain three pairs of target-specific primers: two forward primers, two backward primers, and two loop primers. The LAMP reaction mix contained DNA polymerase, reverse transcriptase, isothermal buffer, and signal reporter. A simple heating block can be used to amplify the target region of the viral RNA using colorimetric detection. The image on the right shows the time-dependent color change, adapted from Dao Thi et al., [125]. Reprinted with permission from AAAS. Created with BioRender.com. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
RT-LAMP assay starts with selecting suitable target gene(s) and designing specific primers and probes. An ideal target gene would be highly specific to and highly conserved in SARS-CoV-2. There are several target regions in the viral genome for nucleic acid-based tests, among which the ORF1ab, E, M, and N genes are commonly employed. Frequent mutations occur in the S gene [119], making it a less common target [120]. Compared to other structural genes, the N gene is highly conserved [121,122], thus less likely to be mutated. This gene is one of the best molecular targets for in vitro diagnostics [119]. Different authorities, such as the WHO, US CDC, and CDC China, developed different primer sets targeting different genomic regions for the RT-PCR test. Multiple targets are commonly used in the recommended RT-PCR tests. For example, the US CDC recommended that RT-PCR tests use two regions in the N gene, while in Germany, initially, regions from the E gene and RdRP were selected. Selecting multiple targets in nucleic acid amplification tests is a better analytical approach to reduce false negatives, as mutations occur in the virus over time. Especially, RNA-based virus mutate at higher rate than DNA-based virus [123]. However, interpreting the test results with multiple targets might be challenging when all targets do not amplify, in which case retesting [124] might be necessary.
Most of the literature reports LAMP primer sets specific to various regions of the N gene [126]. An N gene-targeted RT-LAMP assay was developed by several groups [122,125,127] by using a colorimetric readout. Interestingly, all three studies reported a similar LOD of approximately 100 copies of viral RNA. Among them, Dao Thi et al. used a large sample size of 768 suspected patients for clinical sample validation for their assay [125]. Using the phenol red indicator causes a gradual transition in visible color from red to yellow as the H+ concentration increases in the reaction over time. They reported excellent clinical sensitivity of the RT-LAMP method, with more than 98% sensitivity and specificity. However, the study suggested that RT-LAMP failed to effectively amplify the target from the sample, which exhibited Ct values above 30 by RT-PCR. Baek et al. reported N gene-targeted amplification [122] that could detect viruses within 30 min. The specificity was confirmed by no amplification of other coronavirus samples. They reported a very low number of false-positive results: just two from 154 clinical samples. Genomic region of open reading frame is also used as target. Laura E. Lamb and her coworkers reported an assay that targets the ORF1ab gene of the virus [128]. Their assay relied on simple naked-eye detection based on a fluorescent dye, SYBR Green I. The applicability of the assay on clinical samples was evaluated on 40 samples from COVID-19 patients. However, 5% false-negative and 10% false-positive results were reported. Another study reported RT-LAMP-based detection of the ORF1ab gene [129]. They obtained an LOD as low as 10 copies of the gene per reaction, but validation with clinical samples resulted in 25 false negatives out of 248 COVID-19 patient samples.
Targeting more than one gene region might result in better clinical sensitivity in the RT-LAMP assay [130,131], especially with low viral load samples targeting a single gene can result in false negatives. Several reports, including one by Yan et al. employed more than one gene region of SARS-CoV-2; in their study, the ORF1ab and S genes were targeted in a large number of clinical samples [132]. Notably, they evaluated the specificity of the method using 60 related respiratory pathogens and found excellent selectivity in favor of SARS-CoV-2 with no cross-reactivity for similar pathogens. Intriguingly, they found that the clinical sensitivity of the assay was ten times higher than that of RT-PCR for both targets, and detection for all 130 clinical samples they used for RT-LAMP and RT-PCR agreed 100%. Jiang et al. also reported an assay by targeting two genes, the ORF1ab and N genes, using the fluorescent dye SYTO-9 [133]. Huang et al. targeted three regions of the ORF1ab, S, and N genes [134], which resulted in rapid and good sensitivity and detected two copies of target RNA per reaction volume. By using four sets of primers (with six required primers in each set) for each genomic region (ORF1ab, S and two regions of the N gene), they showed the assay's potential to detect the virus without RNA isolation from the sample matrix. However, they tested the assay on only 16 clinical samples.
Another interesting assay was based on two distinct primer sets that selectively targeted both the ORF1ab and N genes [135]; it employed a nanoparticle-based colorimetric readout system for the simultaneous detection of both genes. Fluorescein (FITC)- and digoxigenin-modified primers were used to amplify the ORF1ab and N genes, respectively. The authors described the multiplexity of the test by amplifying both the target genes in a single reaction platform. They tested pre-confirmed samples of both positive and negative cases. The assay detected all 33 positive clinical samples when targeting both genes in parallel. When targeting only one gene, they found approximately 15% false-negative results, which showed that targeting more than one region in an RT-LAMP assay can provide better performance.
Using a single, constant operating temperature in the near 65 °C certainly has distinctive advantages over PCR-based viral detection. However, undesired cross-reactivity in LAMP reaction can be an issue because of a higher number of primers. With the emergence of different variants of SARS-CoV-2, the existing LAMP assay should be reconfigured with adding more variant specific primers. Adding one or more common mutations among the widely circulated infectious variants as additional target regions, LAMP's clinical sensitivity should be reevaluated. Designing and optimizing primers against multiple target regions of the viral RNA can increases the complexity. The primer sets against all the targets might be fully specific but not equally sensitive in LAMP reactions; thus, the interpretation of clinical tests from a multitarget LAMP test might be challenging.
2.2. Nicking enzyme amplification reaction (NEAR)
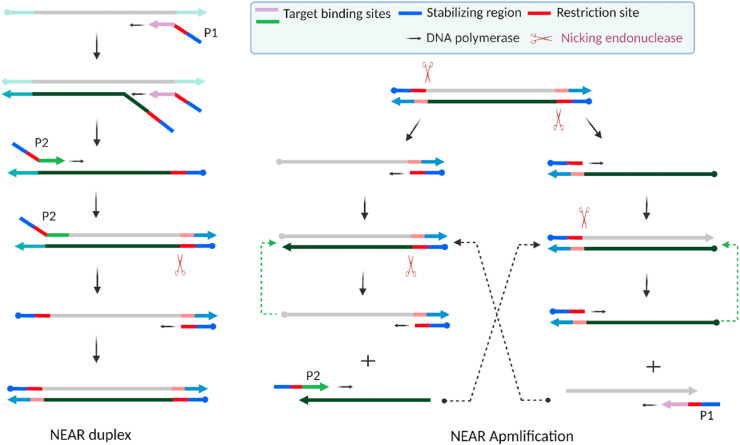
The nicking endonuclease amplification reaction (NEAR), also reported as nicking enzyme-assisted amplification (NEAA) or nicking enzyme-mediated amplification (NEMA), is an enzymatic isothermal amplification reaction [136]. Commercially, Abbott Diagnostics has been developing NEAR-based influenza virus as well as SARS-CoV-2 detection systems [137]. The key components of NEAR are a pair of target-specific primers, forward and reverse, each designed with three distinctive sites, and two highly efficient enzymes: strand-displacing DNA polymerase and nicking endonuclease. There are a wide range of commercially available nicking endonuclease enzymes active over a wide range of temperatures, each with a unique nicking sequence that they recognize [138,139]. Fig. 6 illustrates NEAR with two primers, P1 and P2, each containing a stabilizing region at the 5′ end, a target binding region at the 3’ end and a nick recognition site in the middle.
Fig. 6.
Schematic of the nicking endonuclease amplification reaction (NEAR). Two main steps are involved in the NEAR: duplex formation (left) and amplification (right). Key components of NEAR are a pair of target-specific primers (P1, P2) and two enzymes: strand-displacing polymerase and nicking endonuclease enzymes. Figure adapted with permission from Ref. [140]. Created with BioRender.com.
Therefore, the choice of nicking endonuclease largely relies on primer design or vice versa and ultimately on the intended target. NEAR starts with hybridization of the primer P1 to the target. DNA polymerase extends P1 from the 3’ end. Another P1 primer strand readily binds to the target and is also extended by highly processive and promiscuous DNA polymerase while efficiently releasing the prior extended strand. The primer P2 readily binds to this released strand. As extension occurs by the polymerase, a nicking recognition site is generated on the duplex. The nicking endonuclease readily recognizes the site and creates a nick, while the polymerase enzyme displaces the nicked strand to start another extension process. Therefore, from the primer-target interaction at the beginning and the subsequent combined action of the two enzymes, a NEAR duplex is generated. This duplex acts as the seed for the subsequent exponential amplification. The duplex undergoes nicking at both ends. Polymerization of the resulting templates generates shorter duplexes, which further undergo nicking to generate single-stranded products. The primers hybridize with these products, and the processes repeat to generate shorter duplexes in a positive feedback loop, resulting in exponential amplification.
However, the downside of this reaction is frequent nonspecific product formation, which limits the sensitivity of NEAR. Nonspecific product formation can be suppressed by carefully designing the primers, selecting appropriate enzymes, and optimizing the reaction conditions. Key experimental parameters, such as the experimental temperature, the relative concentrations of both polymerase and nicking endonuclease, the Mg2+ concentration enabling the activity of both enzymes, dNTPs, and multiple sets of primer candidates and their concentration, need to be optimized.
2.3. Nucleic acid sequence-based amplification (NASBA)
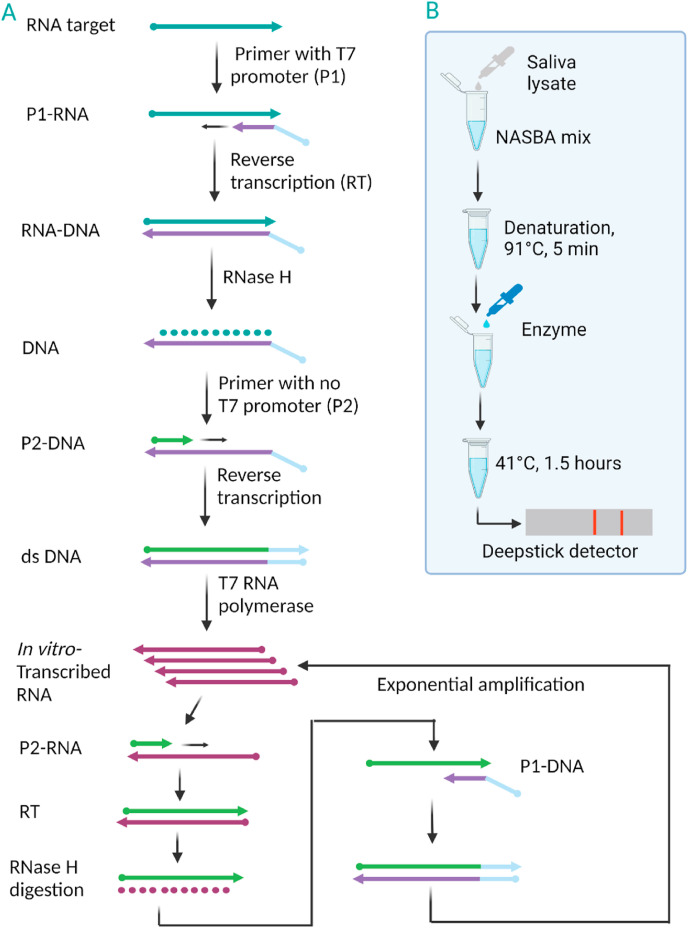
The Nucleic acid sequence-based amplification (NASBA) was first reported in 1991, making it one of the oldest isothermal amplification methods [141]. NASBA has been used to detect different RNA-based infectious viruses, such as hepatitis A virus (HAV) [142], human immunodeficiency virus type 1 (HIV-1) [143], and hepatitis C virus (HCV) [144]. To amplify target RNA [145], NASBA method requires three enzymes (reverse transcriptase, RNase H, and T7 RNA polymerase) and two target specific primers. One of the primers are designed to contain the T7 polymerase promoter sequence. Fig. 7 illustrates the amplification principle of NASBA. Detection of viral RNA via NASBA usually starts with a brief initial denaturation step at 65 °C to disrupt secondary RNA structure prior to isothermal amplification at 41 °C. The reaction starts with hybridization of the target RNA to the primer that contains the T7 polymerase promoter sequence (Fig. 7A). Extension of the primer by reverse transcriptase generates an RNA-DNA hybrid, which then creates single strand DNA by RNase H enzyme. The second primer binds to newly generated single strand DNA to create dsDNA. Due to the T7 promoter sequence in dsDNA, T7 RNA polymerase binds to it, making in vitro transcribed (IVT) RNA.
Fig. 7.
A) Simple mechanism of NASBA. B) Schematic of the single-tube experimental setup to detect SARS-CoV-2 RNA from saliva samples via NASBA-based INSIGHT. Figure adapted with permission from Ref. Wu, Q. et al., [146]. Created with BioRender.com.
IVT RNA undergoes cyclic steps of primer binding, RT, and dsDNA formation, resulting in exponential amplification of the ssRNA products. NASBA efficiently achieves more than 107-fold amplification in approximately 2 h [147]. The ssRNA product readily binds to complementary oligonucleotides and thus can be conveniently monitored by using fluorescent probes such as molecular beacons [148,149]. The amplification temperature (41 °C) is mild and favorable for developing a microfluidic device [150]-based POC NASBA approach. Such devices have been developed to integrate NASBA-based sensitive detection of pathogens [[151], [152], [153]]. NASBA-based detection of SARS-CoV-2 with substantial POC features, INSIGHT (isothermal NASBA sequencing-based high-throughput test), has been published recently [146]. An S gene-based amplified product was sensitively detected in both real-time and endpoint fashions by a fluorescent molecular beacon probe and a lateral flow readout, respectively. The authors employed saliva as a sample and achieved an approximately 50 viral RNA copy LOD. A simple workflow of INSIGHT is illustrated in Fig. 7B. However, in this study, patient samples were not used for clinical validation.
NASBA-based commercial diagnostic system is available. The fully automated NASBA-based diagnostic system NucliSENS EasyQ was developed by BioMerieux [154] and has been widely employed for virus detection [155,156]. The NASBA-based assay generates RNA products; therefore, it is crucial to maintain RNase-free conditions to ensure RNA stability. Another concern is that when amplification occurs at mild temperature, low-stringency reaction conditions may cause nonspecific amplification. Using an ice bath to assemble enzymatic reaction mixture or keeping enzymes separate and adding them to the remaining amplification mixture immediately before amplification can avoid nonspecific amplification.
2.4. Transcription-mediated amplification (TMA)
The principles of transcription-mediated amplification (TMA) and NASBA are very similar, and they have similar readout modes. Similar to NASBA, TMA also involves synthesis of cDNA from target RNA and reverse transcriptase interaction, formation of dsDNA and generation of IVT RNA amplicons by T7 RNA polymerase. However, TMA uses two enzymes, reverse transcriptase and RNA polymerase [145]. The unique reverse transcriptase enzyme also possesses RNase H activity; thus, TMA functions without an additional RNase H enzyme [157]. A TMA-based isothermal system detected the presence of RNA viruses in clinical samples [158] and was more sensitive than RT-PCR in hepatitis C virus detection in liver tissue [159]. Similar to NASBA, TMA also needs optimized reaction conditions and careful primer design to limit nonspecific amplification.
2.5. Recombinase polymerase amplification (RPA)
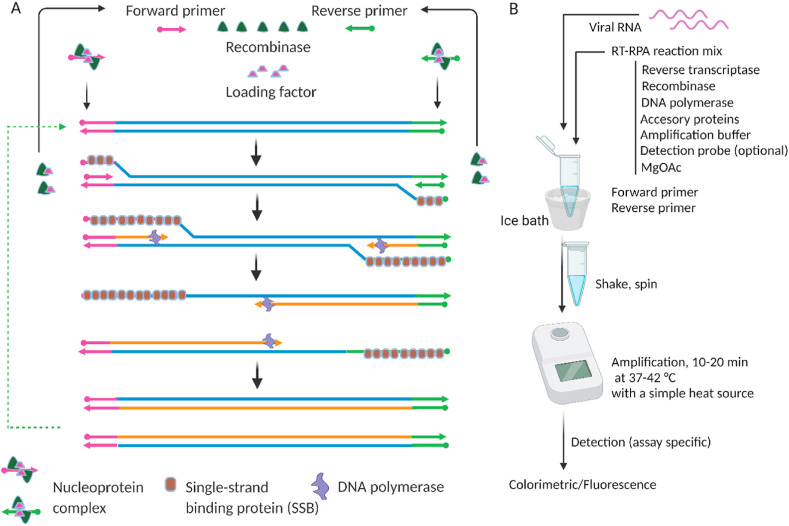
A newer isothermal technique, recombinase polymerase amplification (RPA), was first reported in 2006 [160]. RPA operates between 37 and 42 °C. Three key proteins, recombinase (UvsX), strand displacing polymerase (Bsu), and single-stranded binding (SSB) protein (gp32), and other accessory proteins are needed for RPA. RPA starts when the recombinase forms a nucleoprotein complex with two specific primers, i.e., forward and reverse primers designed based on a specific target. This binding of the recombinase protein to primers is assisted by the loading factor, UvsY. Creatine kinase is used to generate ATP and provides energy for the recombinase protein.
The nucleoprotein complex interacts with the target dsDNA to unwind the target at both primer sites to make a D-loop (Fig. 8 A). This initial D-loop opening is stabilized by an SSB protein. Therefore, the respective primer can bind to the complementary primer binding site on the already opened dsDNA target. Once the primer binds to the target, strand displacement polymerase starts to generate a new strand and new dsDNA. The newly generated dsDNA acts as the starting template for a new round of amplification to generate more dsDNA, and the amplification continues in an exponential manner. The operating temperature of RPA (37–42 °C) can be easily provided by a simple heating source. Fascinatingly, a study reported that RPA can be performed even with no equipment by using only body heat [162]. In that study, RPA was performed to detect HIV-1 DNA by incubating the reaction mixture near different parts of the human body.
Fig. 8.
Schematic of recombinase polymerase amplification (RPA) [161]. A) RPA mechanism (reproduced from Ref. [161] with permission from the Royal Society of Chemistry). B) A simple workflow of a typical RT-RPA. The amplification mix contains a pair of primers, recombinase, loading factors, reverse transcriptase, DNA polymerase, accessory proteins, and single-strand binding proteins that are available in commercial kits. Created with BioRender.com.
RPA has also been used to detect SARS-CoV-2 [163,164]. Xia et al. developed a very sensitive yet simple detection method based on a modified RPA with an LOD as low as one copy of RNA per reaction [164]. In this study, the N and S genes were detected via a fluorescence-based readout, and the researchers tested potential multiplex assays for detecting both targets in the same reaction tubes. The study demonstrated the assay's simplicity by using a cup of hot water as a heating source. This assay showed high specificity by distinguishing between SARS-CoV-2 and other coronaviruses such as MERS-CoV and SARS-CoV. However, validation using clinical samples, was not reported. Behrmann et al. reported another RPA-based sensitive assay of SARS-CoV-2 [163] that detected the N gene by fluorescence readout and validated the assay using clinical samples; comparison with RT-PCR results showed full accordance. The study reported results within 15–20 min. RT-RPA can detect target genes rapidly due to the exponential nature of the amplification. A typical RT-RPA workflow is illustrated in Fig. 8B. The reaction assembly primarily includes mixing of target viral RNA and the RT-RPA reaction mix. Commercial RPA kit that contains the components of the RPA reaction (TwistDx, Cambridge, UK) is available [161] to detect pathogens, including SARS-CoV-2 [165]. RPA reagents can be purchased from TwistDx in a liquid format as well as in a lyophilized pellet format, a great advantage for on-site application.
The downside is that RPA requires more proteins than LAMP. In principle, RPA has readout modes similar to those of LAMP, such as fluorescence-based, colorimetric, and lateral flow strips. Similar to LAMP, in fluorescence-based real-time RPA, the threshold time, Tt [163] can be used to monitor RPA reaction. The two primers needed for RPA can be designed with the computation tool PrimedRPA [166]. The smaller number of primers in RPA might make it simpler than LAMP; however, RPA often suffers from lower specificity than LAMP [167]. Since RPA is active at lower temperatures, using an ice bath to assemble the reaction mixture prior to amplification might restrict nonspecific amplification to a minimum.
2.6. Other potential isothermal amplification methods
There are some other promising isothermal methods that are well known in nucleic acid-based detection of infectious viruses, including but not limited to RAMP, RCA, CPA, HDA, and SMART. SARS-CoV-2 detection studies with these methods are rare; however, all these methods have great potential for use in viral and pathogenic detection, and it is worth discussing these methods briefly in this section.
2.6.1. RAMP, a combination of RPA and LAMP
RAMP is a two-stage isothermal amplification based on a combination of RPA and LAMP. RAMP reportedly showed at least tenfold higher sensitivity than either RPA or LAMP alone [168]. In this hybrid amplification process reported by Song et al. the target was amplified by RPA at 37 °C in the first stage, followed by second-stage amplification by LAMP at 63 °C. The assay generated multiplex detection of 16 different targets of different deadly viruses, including HIV, Zika, and HPV viruses, in less than 40 min. RAMP products were detected with either fluorescent or colorimetric dye.
Although not peer-reviewed yet, a recent report on a RAMP-based SARS-CoV-2 detection method called Penn-RAMP [169] describes RPA carried out at approximately 40 °C followed by LAMP at approximately 60 °C. Targeting the ORF1ab gene, the authors compared the performances of RAMP, PCR and LAMP and found that RAMP has at least ten times higher sensitivity. However, this promising result of the RAMP method was not validated with clinical samples. The downside of RAMP is that it is carried out in two stages at two different operating temperatures. While this combined isothermal process is an interesting technique, it increases complexity in assay design due to several different types of primers and enzymes involved in the two different isothermal methods.
2.6.2. Rolling circle amplification (RCA)
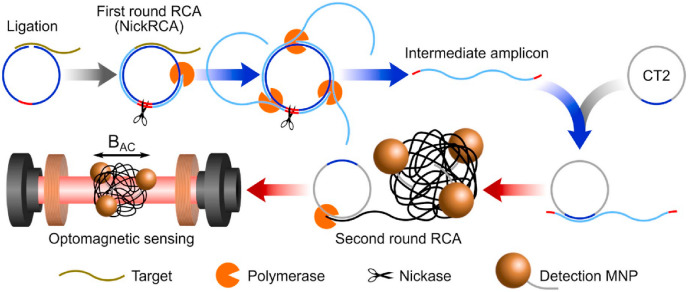
Rolling circle amplification (RCA) is a widely researched isothermal method [170]. As the name suggests, this amplification method requires a circular template. If the starting template is linear, then a circular template is made first with ligase. The target is hybridized on the template's complementary sequence at both ends to make a “padlock probe” prior to the formation of a ligase-mediated circular template. This padlock probe-initiated circular template might result in high specificity for the subsequent amplification. The resulting RCA product is a very long ssDNA or RNA concatemer containing many complementary repeating units of the circular template. Since its discovery, it has found diverse applications in bioanalysis [171], including detecting human viruses [172,173]. Wang et al. reported an RCA-based sensitive detection of SARS-CoV [174] in 2005, but RCA has not garnered as much attention as LAMP in detecting SARS-CoV-2. One study of SARS-CoV-2 detection was published based on modified RCA [175] using two rounds of RCA at 37 °C (Fig. 9 ). In this proof of principle study, the target-initiated RCA generated the first set of amplicons, which acted as targets for the next round of RCA. The amplicon coils from this second round RCA conjugated with magnetic nanoparticles for optomagnetic detection. However, this report used synthetic complementary DNA of the target viral gene, not actual viral RNA from clinical specimens.
Fig. 9.
Schematic of RCA-based detection based on synthetic complementary DNA of SARS-CoV-2. After two rounds of RCA, the amplified product was detected with an optomagnetic sensing platform. MNP-magnetic nano particles. Figure adapted with permission from Ref. [175].
Recently, another RCA-based study reported SARS-CoV-2 detection using two target gene regions of N and S [176]. The RCA products were hybridized with detection probes modified with redox dyes to be detected electrochemically (differential pulse voltammetry). While promising, the study had some limitations for the method to be further applied as a diagnostic tool. Although no thermal cycling is required, multiple operating temperatures were reported for different steps throughout the process. It requires post amplification washing steps, which might be a deterrent for a POC application.
2.6.3. Signal-mediated amplification of RNA technology (SMART)
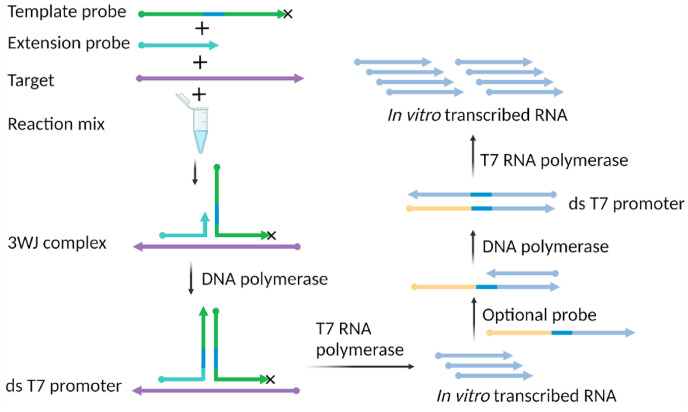
The signal-mediated amplification of RNA technology (SMART) is a simple isothermal amplification method [177]. SMART needs only two target-specific single-stranded probes and two enzymes to function. Both RNA and DNA can be detected [178]. SMART assays start with the formation of a complex of three strands comprising the target and the two probes. The specific design of the probes and their interaction with the target form “a three-way junction (3WJ)” or T-junction complex. The two probes, termed the extension probe and template probe, are designed to have complementary regions of the target that allow them to hybridize with the target at adjacent sites. The two probes also include a region that is much shorter and allows them to hybridize with one another. This specific design criterion allows the probes to form a 3WJ complex in the presence of the target of interest, which serves as the “seed” of the subsequent amplification. Fig. 10 illustrates the principle of SMART. The DNA polymerase, in the reaction mix, extends the shorter probe (or extension probe) using the template probe.
Fig. 10.
Simple illustration of signal-mediated amplification of RNA technology (SMART). Target-initiated 3WJ complex formation and subsequent extension and in vitro transcribed RNA product generation are accomplished by synergy between two enzymes-DNA polymerase and T7 RNA polymerase. The 3′ end of the template probe is modified (x) by phosphorylation to prevent extension by DNA polymerase. Created with BioRender.com.
The template probe contains the T7 RNA polymerase promoter. Therefore, upon polymerization, a double-stranded T7 functional promoter is created that enables the T7 RNA polymerase to transcribe RNA in vitro. To further increase the sensitivity of SMART, a third probe can be used [179]. The third probe does not include the complementary region of the target sequence and thus does not bind to the target. It includes the complementary region of the upstream IVT RNA product and the T7 promoter region. Although SMART was developed at the same time as LAMP, SMART has not received much attention in pathogen detection. However, given its simple probe design and minimal requirement of dual enzymes that are active at the same temperature, it has the potential to detect infectious viruses.
2.6.4. Helicase-dependent amplification (HDA)
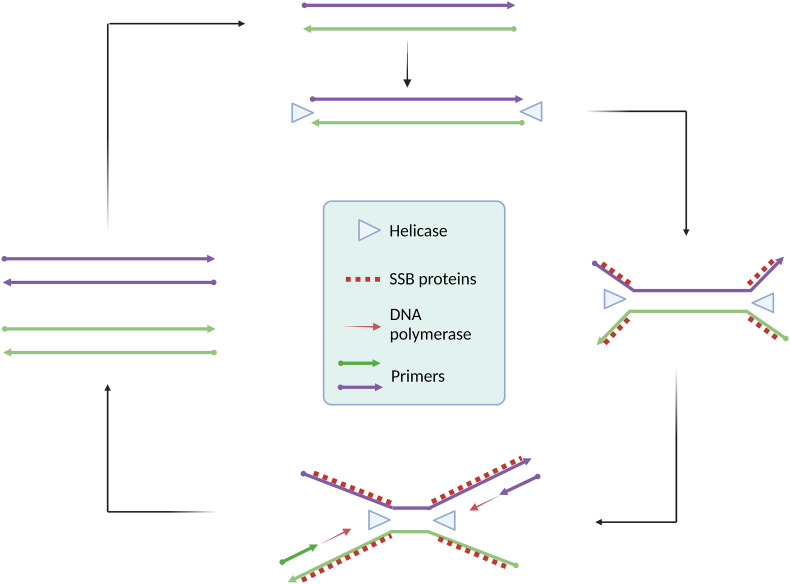
Helicase-dependent amplification (HDA) is another promising isothermal amplification for nucleic acid analysis of pathogens [180,181]. As the name suggests, the helicase enzyme plays a crucial role in the amplification. In vivo DNA replication uses helicase enzymes to unwind dsDNA. Based on the helicase-mediated denaturation of dsDNA, HDA was first reported [182] in 2004 with an operating temperature of 37 °C. The HDA-based commercial diagnostic tools AmpliVue and Solana were developed by Quidel [183]. HDA is a highly robust isothermal method that can analyze nucleic acids from clinical samples, including plasma [184], urine [185], vaginal swabs [186], blood [187], and stool [188,189]; thus, HDA can potentially be applied to detect SARS-CoV-2 RNA from various clinical samples. Fig. 11 illustrates a simple HDA principle.
Fig. 11.
Schematic of helicase-dependent amplification (HDA). HDA relies on helicase enzyme to unwind the target. A pair of primers, DNA polymerase, and single strand binding proteins ensure rapid accumulation of HDA products. Created with BioRender.com.
HDA starts with enzymatic denaturation of dsDNA as opposed to the thermal denaturation in PCR. The helicase enzyme, using free energy from ATP hydrolysis, generates ssDNA from dsDNA. SSB proteins are used to stabilize readily generated ssDNA. Two specific primers (forward and reverse) annealed with ssDNA and DNA polymerase enzyme extend the primer strands to form dsDNA products, which undergo further rounds of amplification by helicase, SSB, and polymerase enzymes, resulting in exponential amplification and sensitive detection within 1 h [190]. With the availability of thermostable helicase enzymes, HDA protocols have been developed to operate at higher temperatures (60–65 °C) for rapid amplification with improved sensitivity [191]. An excellent review was recently published on using HDA to detect pathogens [191]. HDA assays are some of the simplest rapid and sensitive isothermal techniques. The success of HDA largely depends on target-specific primer design. The primary concern of HDA-based assays is undesired primer-dimer formation and off-target primer binding leading to misleading or nonspecific amplification [192].
2.6.5. Cross-priming amplification (CPA)
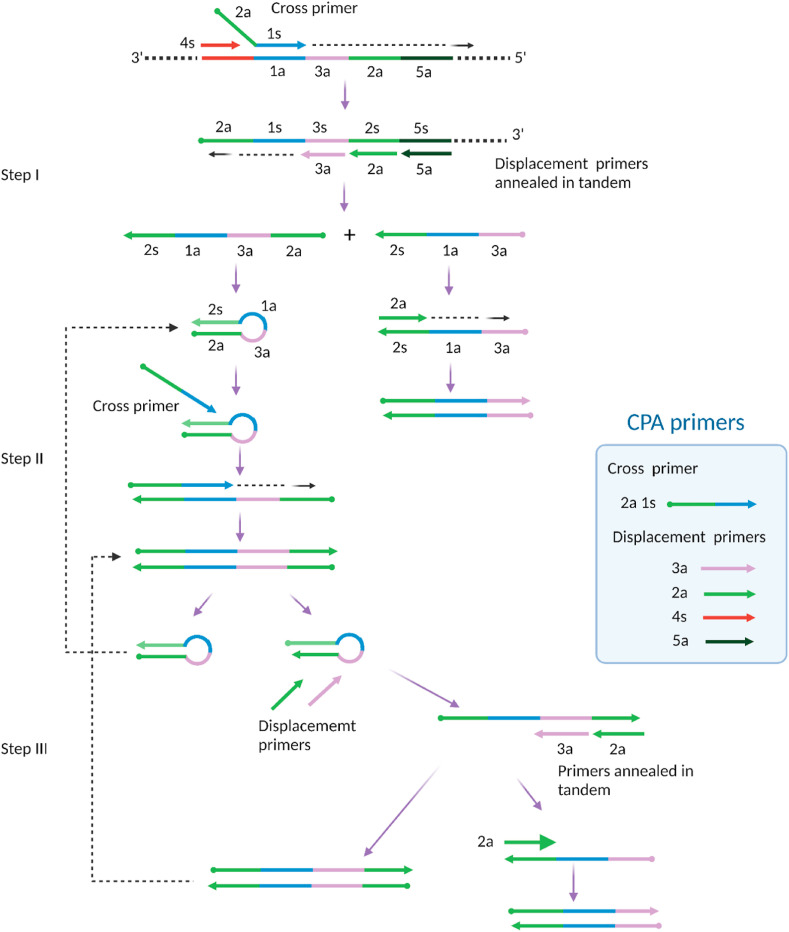
Cross-priming amplification (CPA) is another emerging isothermal amplification method for pathogen detection [[193], [194], [195]]. It relies on a higher number of primers and a smaller number of enzymes to amplify a target of interest. First developed and commercialized by Ustar Biotechnologies (Hangzhou, China) [196], CPA requires one enzyme-DNA polymerase with strand-displacing activity, which might be advantageous for low-cost assays. However, several primers are required for CPA, three to five with at least one specially designed longer primer is known as a cross primer [194]. There are two distinct domains in the cross primer: one at the 3′ end has a site complementary to the target and thus can bind to the target through this domain, and the other, noncomplementary domain is at the 5’ end. Fig. 12 illustrates that the CPA reaction is initiated by forming a complex of multiple primers and target strands. At an operational temperature of 60–65 °C, the cross primer (2a 1s) and an upstream displacement primer (4s) hybridize to the target, forming a complex of three strands. Bst polymerase extends the cross primer strand. Simultaneously, the upstream primer strand is also extended, displacing the extended cross primer strand from the complex.
Fig. 12.
Schematic of cross priming amplification (CPA). CPA relies on Cross primer and several Displacement primers. CPA consist initial products (step I), stem-loop products (step II), and subsequent amplification of the final products (step III). Adapted with permission from Ref. Xu, G. et al., [194].
Three additional primers (3a, 2a, 5a) in tandem hybridize with the released strand, forming a complex of four strands. Extensions and displacements from this complex result in two different ssDNA products, one of which undergoes direct primer extension to create dsDNA. The other ssDNA product from this step forms a hairpin-like structure followed by cross priming extension to create a dsDNA structure, which further generates two hairpin structures. High concentrations of primers and highly active Bst polymerase facilitate a series of annealing, extension, and displacement processes, leading to exponential amplification of dsDNA. The CPA product is dsDNA and thus can be read out by fluorescence mode. However, several recent studies used simple lateral flow strip-based detection, showing the diverse detection modes of CPA [[197], [198], [199]]. CPA is a relatively new isothermal approach and is being further developed [194].
2.7. Isothermal amplification methods combined with the CRISPR/Cas system
Often known as “genetic scissors,” the CRISPR/Cas system is a revolutionary discovery in molecular biology [200]. It was discovered in bacteria as a naturally occurring phenomenon in their genome. Some bacteria survive viral attacks and develop resistance against any such future attack by copying part of the genetic sequence of the invading virus and storing it in the immune system [201,202]. This defense mechanism of the bacteria is expressed as clustered regularly interspaced short palindromic repeats, termed CRISPR. It is the specific “spacer” sequence in the genome of bacteria located between palindromic sequence repeats that relate to viral genomes [203]. As an immune response against the attacking virus, this signature spacer sequence is transcribed into small RNA molecules [204,205]. RNA, along with certain enzymes in bacteria, forms the CRISPR/Cas-based adaptive immune system [204,206], which protects bacteria by destroying the DNA or RNA of the invading virus [207]. Interestingly, it was later found that this natural immune system of many bacteria (also archaea) can be engineered for use as a genetic tool in mammalian cells [208] and human cells [209,210]. Within a few years of its landmark discovery in 2012 by a group of researchers [211], this revolutionary tool has already proven to be extremely useful for diverse applications ranging from gene editing [208,212] to bioanalytical applications [213,214] to therapeutics [215,216] and has shown remarkable potential in nucleic acid-based detection of pathogens [[217], [218], [219], [220]].
A CRISPR/Cas system coupled with various isothermal amplification for diagnosis of pathogens is in place [[221], [222], [223]]. While isothermal amplification provides high sensitivity by exponential enrichment of the target gene, the incorporated CRISPR/Cas system provides high precision in detecting amplified target products. The CRISPR/Cas detection is based on the formation of an “effector complex” comprising two defining elements: Cas enzymes and guide RNA (gRNA). The effector complex leverages the cooperative relation between these two elements. While the recognition of the target nucleic acid is provided by gRNA, cleavage of the nucleic acid is executed by the Cas enzyme [217,224,225]. The programmable gRNA, for certain Cas enzymes, is basically a complex of two parts RNA: a CRISPR RNA (crRNA) and a trans-activating crRNA (TracrRNA) (Fig. 13 A, upper). While the TracrRNA part facilitates the pairing of the entire gRNA and the Cas enzyme and helps to obtain the active conformation of the effector complex, the crRNA part contains a short region named the spacer, which binds to the target. However, to simplify the experiment, both crRNA and TracrRNA of the gRNA can be combined in a single and larger RNA motif often known as single guide RNA (sgRNA) [226] (Fig. 13A, lower). Similar to the LAMP primer design tool, web-based gRNA design tools for CRISPR/Cas systems are available online with varying usability [[227], [228], [229], [230]]. However, manual adjustment still might be necessary. Most often, in vitro optimization of the sgRNA from a set of designed candidate sgRNAs is needed for specific assays. The spacer sequence is designed, modified, and changed as needed to contain the complementary base of the protospacer, a region of the intended target in the assay obtained as upstream isothermally amplified products. Hybridization between the spacer and protospacer ensures target specificity and activates the effector complex for that specific target. However, for some Cas enzymes, the effector complex also requires the presence of a very short (2–5 bases) [231] region in the target in close proximity to the protospacer named the protospacer adjacent motif (PAM) [232,233]. The required PAM sequence is different for different Cas enzymes [234] and is typically considered a constraint in CRISPR-based biosensing [235].
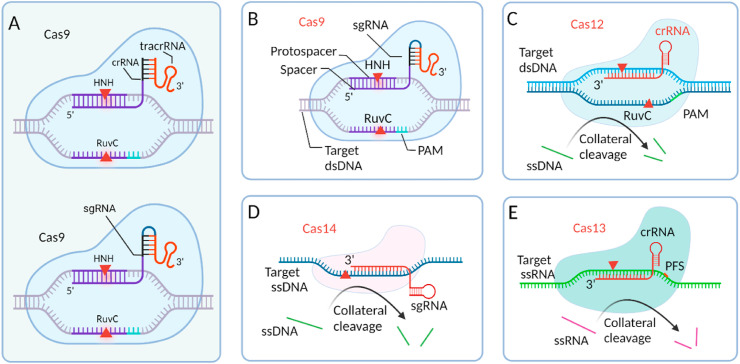
Fig. 13.
Class 2 Cas enzymes used in the CRISPR/Cas system. A) The CRISPR/Cas9 system uses a two-part gRNA (tracrRNA, crRNA (top)) or an sgRNA (bottom) with a linker between tracrRNA and crRNA. B) sgRNA-directed endonuclease activity of Cas9 on target dsDNA. C) Collateral cleavage and target cleavage activity of Cas12. D) Collateral cleavage and target (ssDNA) cleavage activity of Cas14. D) Collateral cleavage and target (ssRNA) cleavage activity of Cas13. All Cas enzymes are drawn by showing their relative size (created with BioRender.com). Figures B–E adapted with permission from Ref. [239].
CRISPR/Cas-based pathogen detection systems typically employ different Cas enzymes with their preferred target nucleic acids. Some Cas enzymes preferentially cleave dsDNA, ssDNA, or both, while other Cas enzymes exclusively cleave RNA. In fact, discovering various Cas enzymes and evolving the system as strong tools for biosensing as well as pathogen detection are inseparable. Based on the specific functional structure of Cas enzymes, CRISPR/Cas systems can be divided into class 1 and class 2 systems [236]. Class 1 systems include functional complexes of multiple effector Cas enzymes, while class 2 systems are a single and large effector Cas enzyme. To date, most of the discovered Cas enzymes belong to class 1 [237]. However, due to the simple and convenient assay design with only one effector protein in class 2 systems, this system has been the most used in detecting pathogens, including SARS-CoV-2 [238]. The most common class 2 Cas enzymes that are used in nucleic acid detection are Cas9 (type II), Cas12 (type V), and Cas13 (type VI).
Cas9 is a large enzyme varying from ∼950 to over 1600 amino acids based on the species from which it is isolated [236]. Upon binding sgRNA, in the presence of the signature PAM sequence NGG (N is any nucleotide), Cas9 unwinds and cleaves the dsDNA target at the specific site by using two nuclease domains: His-Asn-His (HNH) and RuvC [226]. The functional domain HNH cleaves the protospacer-containing strand (complementary to the spacer), while the RuvC domain cleaves the other strand of the dsDNA target [[240], [241], [242]]. Both nuclease domains are necessary for dsDNA cleavage by Cas9 (Fig. 13B). Likewise, the endonuclease activity of Cas9 can be controlled and even deactivated by mutating the amino acids of the functional domains. Mutation in one domain, either RuvC or HNH, transforms Cas9 into a nicking endonuclease (nCas9), meaning that it cleaves or nicks only one strand of the target dsDNA [241,243]. Amino acid mutation in both nuclease domains deactivates Cas9 (dCas9) to form a catalytically inactive version that no longer cleaves any of the strands even though it binds the target dsDNA [239,244]. All these structural variations of Cas9 make it useful for gene manipulation [245,246]. There are some published reports that use Cas9-, dCas9-, and nCas9-based CRISPR/Cas detection of nucleic acids [221,[247], [248], [249]]. However, Cas9 is a well-known enzyme for CRISPR/Cas-based genome editing [218,250].
The Cas12 family of proteins are type V effector proteins. With 1100–1300 amino acids [251], Cas12 proteins are slightly smaller than Cas9 proteins. Cas12 is well known in CRISPR/Cas-based detection of infectious viruses, including SARS-CoV-2. Similar to Cas9, Cas12 is also directed by a gRNA, albeit a shorter one known as crRNA [252,253], to bind and cleave dsDNA targets in the presence of thymine-rich PAM sequences. With no HNH domain, Cas12a performs dsDNA cleavage using the endonuclease ability of the RuvC domain [254]. In addition to target cleavage, Cas12, unlike Cas9, also provides collateral cleavage activity (also referred to as trans-cleavage) on any ssDNA in the sample, a useful property in nucleic acid-based detection [255]. It enables a ssDNA reporter probe modified with fluorophore and quencher groups to be used to monitor an assay (Fig. 13C).
Cas14, another type V effector protein, has been reported recently [225,256]. Similar to Cas12, it can exhibit indiscriminate cleavage of ssDNA [239]. However, unlike Cas12, Cas14 does not require a PAM sequence in the target [225], and it is much smaller in size, with ∼400–700 amino acids [225]. The Cas14 family preferentially targets ssDNA (Fig. 13D). The PAM-independent target cleavage of this newer class 2 enzyme has great potential to enable detection of a wide variety of ssDNA-based pathogens. Although ssDNA is preferentially targeted by Cas14, dsDNA can also be targeted by Cas14 by providing a TTTA PAM sequence [257].
Cas13 family effector proteins, belonging to type VI, are unique in their target and collateral cleavage. Unlike the other class 2 Cas enzymes, Cas13 specifically targets and cleaves ssRNA [258] and does not require a PAM sequence in the target (Fig. 13E). However, for the target recognition it prefers a non-guanine base or protospacer flanking site (PFS) to be present next to the protospacer, which can be easily found. With a similar size (∼900–1300 amino acids) [251] to Cas12, the well-known subtype from the Cas13 family for pathogen detection is Cas13a [259]. Due to its ssRNA targets, Cas13 is especially suitable for RNA-based virus detection, including detecting SARS-CoV-2 [260,261].
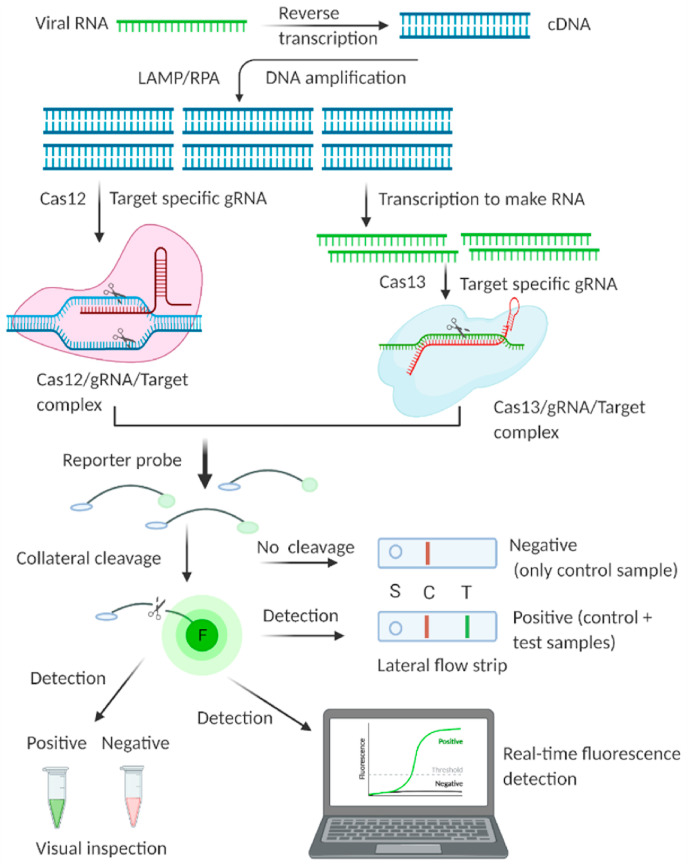
Isothermal amplifications such as LAMP and RPA are combined with different Cas enzymes involved with the CRISPR system for viral RNA (SARS-CoV-2) detection (Fig. 14 ). Isothermal-based CRISPR/Cas detection systems can be performed with simple equipment, even a water bath [262,263]. Ideally, CRISPR/Cas-based detection can be performed with fluorophore and quencher-modified nucleic acid reporters. This method generates a specific signal based on the cleavage of the reporter fluorescence probe, which allows simple visual detection [264]. Another simple way that is more amenable to POC detection is to interpret the result by employing a lateral flow assay, similar to a pregnancy test strip [[265], [266], [267], [268], [269]]. An assay developed by Broughton et al. is named DNA Endonuclease-Targeted CRISPR Trans Reporter (DETECTR) [270]; it combines RT-LAMP with CRISPR/Cas12 to give sensitive detection of the target genes E and N of SARS-CoV-2. RT-LAMP-based amplified products are recognized by the Cas12/gRNA complex, which activates Cas12 for collateral cleavage of the fluorophore-quencher-labeled reporter probe. A lateral flow strip-based readout confirmed the cleavage of the reporter ssDNA probe. Together, the isothermal amplification and CRISPR/Cas steps of the assay took approximately 45 min to detect SARS-CoV-2.
Fig. 14.
Simpler representation of a typical isothermal amplification based CRISPR/Cas detection system for viral nucleic acids. Typically, LAMP or RPA based amplification is involved to amplify target for subsequent class 2 Cas enzyme activity by Cas12 and Cas13. The system involves “Collateral cleavage” based qualitative and quantitative detection. Created with BioRender.com.
The LOD was as low as 10 copies per μl of the reaction, suggesting high sensitivity of the assay, and the selectivity of the assay was confirmed using other coronaviruses. Clinical samples were preidentified with RT-PCR, and DETECTR could detect 100% of the negative samples correctly and 95% of the positive samples. The researchers attempted detection with a crude sample to avoid the RNA extraction step but found that this resulted in a significantly lower sensitivity. The Cas12 used in DETECTR is constrained by the PAM site in the target, which typically deters a CRISPR/Cas system from wide applicability. Another concern with DETECTR is its post amplification tube opening. It is ideal for nucleic acid-based detection in a single pot with a sample. Another assay was developed by the Zhang group. The Zhang group published a detailed protocol of the assay named SHERLOCK [267] (Specific High Sensitivity Enzymatic Reporter Unlocking). SHERLOCK relies on several enzymatic processes: RT, RPA (RT-RPA), and T7 RNA transcription [267]. RT-RPA was used to amplify the target gene sequence, which resulted in dsDNA products. T7 RNA transcription makes RNA from the dsDNA products. SHERLOCK relies upon the cleavage of the Cas13 enzyme, which explicitly possesses RNA cleavage activity [271] and, thus, is convenient for the detection of RNA viruses such as SARS-CoV-2. The gRNA/Cas13/target ternary complex activates the system to execute collateral cleavage of reporter RNA. The readout is performed by the lateral flow strip. SHERLOCK was developed by extensive optimization of the reaction conditions to detect regions of the S and ORF1ab genes of SARS-CoV-2. SHERLOCK has also been used for the sensitive detection of other RNA viruses, such as Zika virus and Dengue virus [217,261]. The advantage of the Cas13-based SHERLOCK method over the Cas12-based DETECTR method is that Cas13 has much more target sequence flexibility; however, SHERLOCK requires an additional enzyme, T7 RNA polymerase, for Cas13 RNase activity.
CRISPR/Cas brings a high degree of specificity in isothermal amplification. CRISPR/Cas systems are designed considering target-specific gRNA, which directs them; thus, these systems act upon the amplified products from the target sequence, thus avoiding any carry over primer-dimer products. However, we must acknowledge the limitations and challenges of this promising detection system. CRISPR/Cas-based system coupled with isothermal amplification is essentially an enzyme-intensive process, which might be expensive. To incorporate the CRISPR/Cas system with isothermal amplification for pathogen detection, the choice of Cas enzymes must consider the operating temperature of the isothermal amplification. Table 1 shows different potential isothermal amplification methods combined with CRISPR/Cas system reported in the literature. In the literature, most CRISPR/Cas-based systems employ LAMP and RPA isothermal amplification. However, we also outlined other potential isothermal amplification methods that can be combined with CRISPR/Cas systems based on the Cas effector proteins preferred targets and the amplified products of the isothermal amplification methods.
Table 1.
Combinations of isothermal amplification and the CRISPR/Cas system for pathogen detection.
| Isothermal amplification | Amplification temp. (°C)c | Target | Amplicon | Class 2 Cas enzymed | Ref |
|---|---|---|---|---|---|
| LAMP | 60–65 | DNA, RNAa | DNA | Cas12a, Cas12b, Cas9 | [75,[275], [276], [277], [278], [279]] |
| RPA | 37–42 | DNA, RNAa | DNA | Cas12a, Cas13ab Cas13bb | [160,258,269,[280], [281], [282], [283]] |
| RCA | 30–60 | DNA, RNAa | DNA | Cas14, Cas9, Cas12a | [280,284,285] |
| NASBA | 41 | RNA (DNA) | RNA, DNA | Cas13a, Cas13b, Cas9 | [146,154,286] |
| NEAR | 55–60 | DNA, RNAa | DNA | Cas12a, Cas12b, Cas9 | [136,140] |
| TMA | ∼40–60 | RNA (DNA) | RNA, DNA | Cas13a, Cas13b, Cas9 | [159,287] |
| HDA | 37–65 | DNA, RNAa | DNA | Cas12a, Cas13ab Cas13bb | [191,192,280] |
| SMART | 41 | RNA, DNA | RNA | Cas13a, Cas13b | [178] |
| CPA | 60–65 | DNA, RNAa | DNA | Cas12a, Cas13ab Cas13bb | [193,194] |
RNA virus can be targeted using an additional enzyme, reverse transcriptase, to make cDNA from target RNA.
RNA target for Cas13 can be obtained by in vitro transcription of an RPA-generated DNA amplicon to use as a target for Cas13.
Typical amplification temperature: variability might exist.
Most commonly used Cas enzymes with suitable isothermal amplification for SARS-CoV-2 and other pathogen detection based on target types; however, variability might exist depending on the organism from which Cas enzymes are derived.
Typically, isothermal amplifications operate at temperatures between 40 and 65 °C, largely based on polymerase-and amplification-specific auxiliary enzymes. Finding an optimum operating temperature that satisfies both upstream isothermal amplification and downstream Cas activities is crucial and might warrant extensive empirical investigation. Conveniently, Class 2 Cas enzymes and their subtypes, work at a wide range of temperatures. For instance, type V Cas12 enzymes have commonly used subtypes Cas12a and Cas12b. Cas12a enzymes are most functional at approximately 28 °C, while Cas12b enzymes perform best at approximately 50 °C [268]. Therefore, the Cas12a enzyme appears to be a better enzyme for amplification methods with lower operating temperatures, such as RPA [263], while Cas12b is suitable for LAMP [223]-based CRISPR/Cas systems. However, these subtypes and other enzymes and their thermal activity and performance largely depend on the microorganisms from which they are derived; thus, their operating temperatures and suitability for different isothermal amplifications can vary [255,269,[272], [273], [274]].
Choice of Cas enzymes also depends on the target nucleic acid. The amplified product from an isothermal amplification reaction is employed as the target of downstream Cas enzymes (Table 1). The amplicons of LAMP and RPA is DNA. Therefore, Cas12 or Cas9 is preferred with LAMP and RPA combined CRISPR system. However, enzymes can convert RNA to DNA or vice versa during isothermal amplification, making these combined systems flexible for both RNA and DNA based viruses detection. Exploring novel enzymes with varying working temperatures is important.
In addition to the gRNA, most existing CRISPR/Cas systems derive their target specificity from PAM sequences. However, the PAM requirement for Cas9 and Cas12 adds an extra layer of constraint, especially when isothermal amplification is combined with CRISPR/Cas systems. For example, LAMP-based amplification already has somewhat inconvenient target selection-primer design issues. To use CRISPR/Cas12 with LAMP, the target needs to have its PAM sequence 5′(T)TTN-3’ close to the protospacer. A highly conserved region with suitable binding sites for primers and PAM sequence for downstream Cas12 may be hard to find in the pathogen genome. Li and coworkers demonstrated that if a PAM sequence is not present, designing PAM-containing LAMP primers to incorporate the PAM into amplicon dsDNA enables CRISPR/Cas detection [288]. Using Cas13 demands less constraint, as it does not need a PAM sequence to execute collateral cleavage. However, the protospacer region needs to be selected so that the first base immediately after the protospacer region avoids a guanine base, which can be frequently satisfied by careful selection of the protospacer. Similar to Cas13, Cas14 also executes PAM-independent cleavage. However, Cas13 and Cas14 are currently not commercially available [289]; thus, they are obtained by in-house protein expression and isolation in working labs.
2.8. Isothermal amplification method-based commercial devices
Since the pandemic began, emerging isothermal-based detection systems have garnered greater attention than ever previously from academia, industry, and government agencies. Many isothermal-based SARS-CoV-2 detection systems are commercially available and have obtained emergency use authorization (EUA) by the US FDA or are CE marked in Europe. Among them, commercial LAMP-based systems have achieved greater success because of their flexibility, robustness, and simplicity. EIKEN CHEMICAL (Tokyo, Japan) is one of the pioneers of commercial LAMP kits and turbidimetry-based real-time LAMP devices (LA-500 and LA-320C), which have been used for detecting pathogens [290]. For SARS-CoV-2 detection, they developed Loopamp™ kits, which need only a suitable heat source for end use [291]. Color Health Inc. was one of the first companies to obtain EUA [79]. This RT-LAMP-based kit reportedly achieved an LOD of approximately 0.75 copies/μl viral RNA via real time colorimetry. A bead-based RNA extraction step, however, is needed prior to amplification, which is time consuming.
Other RT-LAMP kits from SeaSun Biomaterials have obtained EUA [292]. Both RT and LAMP reactions occur at 60 °C, a large advantage in simplifying the workflow. However, the kit was validated with only certain PCR instruments (Applied Biosystems® 7500 Real-Time PCR instrument), limiting its large-scale application and somewhat undermining the desired feature of requiring simple instrumentation for isothermal detection. Detectachem [293], Mammoth Biosciences [294], and Sherlock Biosciences [295] are among companies that obtained EUA for their testing kits. All these kits were developed by adapting slightly different isothermal approaches and come with different advantages in one or more steps, from sample processing to amplification to detection, with the unifying goal of detecting viruses in a rapid, accurate, and simple manner. The Detectachem kit has the advantage of using direct samples for the assay without the RNA extraction step, while the kits of the other two companies involve isothermal detection systems based on the CRISPR/Cas system. In the United Kingdom, the Department of Health and Social Care authorized the “LamPORE COVID-19” assay developed by Oxford Nanopore Technologies [296]. This technology is promising for high-throughput detection. This CE marked RT-LAMP-based assay is designed to perform with a small palm-sized portable device and is reportedly capable of running 2000 tests in a day. Amazingly, this assay stands out by leveraging the sequencing of the RT-LAMP amplified target and thus can directly detect the presence of the target viral RNA sequence. Moreover, the sequencing of the target can accurately report false positives that might occur in the reaction.
Isothermal techniques other than LAMP have also enjoyed commercial success and have obtained EUA in the pandemic. The Hologic Panther system uses a TMA-based SARS-CoV-2 assay [297], and it is a large-scale clinical testing system capable of running over 1000 tests in a day. This fully automated platform has been used to sensitively detect different human coronaviruses (hCoVs) [298], and one study even found that its analytical performance was higher than that of RT-PCR [299].
NEAR-based isothermal amplification was employed by Abbott for a diagnostic system for the influenza virus [300]. Formerly known as “Alere i”, the isothermal system has been available on the market for a few years for detecting the flu virus [301,302]. An improved version, “Abbott ID Now”, was developed for SARS-CoV-2 detection and also received EUA from the FDA [303]. From direct swab, the “ID NOW COVID-19” testing platform can detect viral RNA in less than 15 min. ID NOW COVID-19 is an integrated diagnostic platform of a sample processing unit and detection system with a small device footprint for POC facilities (Fig. 15 A).
Fig. 15.
Flowchart of different commercial isothermal-based detection systems of SARS-CoV-2. A) ID NOW COVID-19 detection by Abbott. Sample processing units (left) contain a sample receiver, transfer cartridge, and test base. The sample receiver containing lysis buffer is used treat patient sample. Following quick mixing, the transfer cartridge is used to transfer the treated sample manually to the test base containing a lyophilized NEAR mix. Amplification and fluorescence-based detection are performed by the ID NOW instrument (right). Adapted with permission from Ref. [140]. B) “All-In-One Test Kit”—a commercial POC testing system developed by Lucira Health. The system involves a user-friendly, simple workflow of “self-test” virus detection on a small and portable device. (adapted with permission) [304].
Lucira Health developed an impressive isothermal-based detection system [304]. The kit includes all supplies to run the test in a small portable device powered by two AA batteries. This CLIA-waived kit enables at-home testing with very simple and nontechnical steps (Fig. 15B). The test result is interpreted with visually read positive or negative green light on the device. The kit uses an RT-LAMP-based test targeting the N gene of the virus and can give results within 30 min. This sample-to-answer kit obtained EUA to perform self-testing of SARS-CoV-2 outside of healthcare setting. Another test kit, “Cue COVID-19 Test Kit” was developed for similar use [305]. The user can obtain results within 20 min in a “sample-to-answer” manner by the automated testing platform containing three basic elements; Cue sample wand (for sample collection), Cue test cartridge (contain reagents), and Cue cartridge reader (for reporting the result). With self-collected sample, the test automatically starts when the sample wand with a nasal swab is inserted into the test cartridge that is connected to the Cue cartridge reader [305]. A smart phone app is used to interpret the test result.
While there are obvious benefits of different at-home testing kits, a concern might be the often delayed or missing test reporting by the consumer to the public health authority, which mainly depends on the willingness of the at-home user. For these commercial kits and other diagnostics systems, EUA is issued for a certain time in unusual situations such as a pandemic that demands widespread testing. Therefore, large-scale comparative studies of these kits using various sample types collected from symptomatic and asymptomatic patients are necessary before permanent approval and thus regular testing in the post-pandemic era.
For detecting SARS-CoV-2, along with these commercially successful isothermal amplification systems, the analytical performances of some isothermal-based studies in the literature are summarized in Table 2 .
Table 2.
Performance of recent isothermal amplification-based assays to detect SARS-CoV-2.
| Target Gene |
Amplification | Timea (min) | LODb (Copy) | Detection modec | Clinical specimens (sample size) | Comment |
|---|---|---|---|---|---|---|
| ORF1ab | RT-LAMP | 40 | 10 | C | Yes (248) | Some false negatives from clinical samples [129] |
| ORF1ab | RT-LAMP | 45 | 300 | F | Yes (40) | Small number of clinical samples were employed [128] |
| N, S | RT-LAMP | 40 | 100 | C, F | No | For optimization, 16 primer sets were employed [306] |
| N | RT-LAMP | 30 | 100 | C | Yes (154) | High specificity in clinical samples (98%) [122] |
| ORF1ab, N |
RT-LAMP | 30 | 12.5 | F | Yes (260) | Sensitivity in both simulated and actual clinical samples is high [133] |
| ORF1ab, S |
RT-LAMP | 30 | 20 | C, T | Yes (130) | No cross-reactivity against many related viruses [132] |
| N | RT-LAMP | 40 | 118 | C | Yes (56) | 93% consistency with RT-PCR in analyzing clinical samples [127] |
| ORF1ab, S, N | RT-LAMP | 30 | 2 | C | Yes (16) | RT-LAMP with no RNA isolation was attempted [134] |
| N | RT-LAMP | 30 | 100 | C | Yes (768) | Developed a rapid LAMP that avoids RNA isolation [125] |
| ORF1ab, N | RT-LAMP | 40 | 12 | L | Yes (129) | Multiplexing-LAMP targeting two genes in a single tube [135] |
| ORF3a, E | RT-LAMP | 60 | 42 | C | Yes (366) | Different clinical samples tested [307] |
| N | RT-RPA | 20 | 8 | F | Yes (19) | Similar performance to RT-PCR [163] |
| N, S | RT-RPA | 30 | 1 | F | No | Not validated with clinical samples [164] |
| E, N | RT-RPA | 15 | 15 | F | Yes (36) | Similar performance to RT-PCR [165] |
| N, S | RCA | 120 | 10 | E | Yes (106) | Similar performance to RT-PCR [176] |
| S | NASBA | 120 | 50 | F | No | Not validated with clinical samples [146] |
| E, N | RT-LAMP, CRISPR | 60 | 10 | F, L | Yes (24) | Similar performance to RT-PCR [276] |
| ORF1a | RT-RPA, CRISPR | 50 | 200 | F, C | Yes (50) | Good sensitivity (90%) and specificity (100%) compared to RT-PCR [308] |
| N, E | RT-LAMP | 70 | 0.75d | C | – | EUA from FDA (Color Genomics) [79] |
| N, E | RT-LAMP | 30 | 75d | C | – | EUA from FDA (Detectachem) [293] |
| N | RT-LAMP, CRISPR | 45 | 20d | F | – | EUA from FDA (Mammoth Biosciences) [294] |
| N | RT-LAMP | 35 | 0.9d | C | – | EUA from FDA, self-collected sample, at-home use (Lucira) [304] |
Approximate assay time.
LOD-limit of detection is expressed in target RNA copies/reaction from the literature.
F-Fluorescence, C-colorimetric, L-lateral flow assay, E-electrochemical.
LOD-limit of detection is expressed in target RNA copies/μl.
3. Isothermal amplification methods-based detection: Scope and challenges
Isothermal amplification-based assays can be improved to become better alternatives to PCR as a detection system for highly infectious viruses. The main argument in favor of isothermal detection methods is that unlike PCR-based detection, they are not based on repeated temperature switching. While this is true for the amplification part, when we consider the entire assay from sample treatment to detection, it often requires more than one temperature, which is not convenient for an ideal sample-to-answer approach. In some assays in the literature, analytical performance is compared to that of PCR by using mock samples with viral RNA spiked into a biological fluid [128,146,164]. Those analytical sensitivity tests with simulated samples may not yield the same results or even can give false negatives using clinical specimens. Clinical studies using different types of specimens are required to know the robustness and true sensitivity of the assay. It is particularly important as potential false-negative results can have a significant effect on disease containment. Getting accurate results from even a well-designed and sensitive isothermal assay is not always guaranteed. For example, an optimized and “highly specific” assay can generate false-positive results in different settings because of human error such as improper sample handling, reagent mixing, or even mislabeling. A simple and reduced workflow, sample-to-answer assay can prevent these potential sources of false positives and ensure diagnostic specificity.
3.1. Three crucial diagnostic challenges
An infectious virus such as SARS-CoV-2 demands moving the testing environment from the benchtop to the field, from the clinical laboratory to POC settings—making a next-generation diagnostic tool available for routine testing rather than being limited by cost, time, or laboratory facilities. Such a tool would be inexpensive, readily available to end users, operable with minimal technical knowledge, portable with a minimal instrument footprint, and able to detect quickly with fewer reagents and at a single temperature. Isothermal methods are faster and simpler than non isothermal PCR-based methods, and the amplified product can be detected visually with an unaided eye, making them good candidates for next-generation diagnostic tools. As the scientific community and commercial developer continue to make progress on this front, three questions must be asked: How easy is it to process clinical samples? Is concurrent detection of more than one virus possible? How simple are the detection system and the platform? These specific issues concerning SARS-CoV-2 and other infectious viruses are highlighted in Fig. 16 .
Fig. 16.
Characteristics of a next-generation diagnostic tool with the three most important criteria: easy sample treatment, POC tool, and multiplexable system.
3.1.1. Rapid sample treatment toward faster and easier detection
Detection of SARS-CoV-2 requires extraction of its RNA from clinical samples, which involves several time-consuming steps [309], is labor-demanding, and is often impossible to perform with shortages of extraction kits. An isolation-free RNA method would significantly reduce the turnaround time and cost of the test. Some reports have shown that RT-PCR can be performed using patient samples directly [[310], [311], [312], [313]]. Using raw clinical samples should be more convenient with LAMP than PCR, as LAMP is more tolerant to inhibitors [314]. Several studies have shown that LAMP assays can sensitively detect SARS-CoV-2 without RNA extraction from clinical samples [125,128,134,315]. Dao Thi et al. reported directly using nasopharyngeal swab samples to detect viral RNA [125]. However, the direct use of swab samples resulted in less sensitivity due to a strong matrix effect. The swab samples were exposed to brief heat treatment at 95 °C for 5 min. The brief heat treatment of the samples inactivates the endogenous RNases and, likely, disrupts the viral capsid to free the RNA [316]. Companies like Lucira Health, and Abbott have developed isothermal-based diagnostic systems which operate with minimal sample processing.
Apart from minimal sample treatment, the type of sample, ideally noninvasive and self-collected, also has a great impact on the test in a pandemic. Chow et al. demonstrated that samples from nasopharyngeal swabs are more sensitive than throat swab samples for detecting the virus [307]; other similar reports on nasal and throat swab samples were also published recently [317]. However, both types of samples bring certain discomfort to patients and require special protocols, swabs, and collection by healthcare persons. The use of saliva as a self-collected sample overcomes these issues [318,319]. Recent isothermal amplification studies have demonstrated that using saliva samples can potentially make detection easier and faster [146,315]. However, using saliva with a pH-dependent colorimetric LAMP method might generate misleading results, as human saliva reportedly changes the pH from 7.4 to 6.8 [320]. Even these small changes in pH can induce phenol red, a common colorimetric indicator of LAMP, to change from red to yellow, even without the presence of a viral RNA target that initiates LAMP. To overcome potential false positives from saliva samples, Yang et al. reported RT-LAMP [321] that used a “saliva stabilization solution” that contained 14.5 mM NaOH to reduce the salivary acidity. The study reported that the saliva-based LAMP assay provided faster detection with fewer false positives than a RT-PCR test with EUA.
3.1.2. Point-of-care (POC) tool for potential self-detection
Along with minimal sample processing, the next important criterion for a next-generation diagnostic tool is that it would be scalable and user-friendly to allow routine testing at the POC. It must not require sophisticated technical knowledge to use, thereby permitting on-site detection. With these features, the tool would enable an individual to detect the virus with a self-collected sample. Ideally, a POC tool would be portable and require simple instrumentation. The miniaturized platform would perform the analysis with a microvolume of reagents, ensuring that it would be cost-effective.
Several reports have been published on microfluidic-based isothermal detection of infectious viruses [78,[322], [323], [324]], and most of these reports involve prototypes that are not fully developed for large-scale diagnosis. However, recently, a CRISPR/Cas-based preprint report named STOP (SHERLOCK Testing in One Pot) with potential POC advantages was published [275]. This advanced version of SHERLOCK detects SARS-CoV-2 from saliva samples in approximately 1 h using a small heating device. Unlike the previous version of RPA-based SHERLOCK [267], this version offers simpler operation with a single-step reaction based on LAMP amplification. The single-step assay reduces potential sample cross-contamination and achieves highly specific detection of SARS-CoV-2 against other coronaviruses, such as MERS-CoV and SARS-CoV. Recently, another report was published on POC-based SARS-CoV-2 detection [83]. The assay employed LAMP-based amplification with extraction-free RNA with a very brief heat treatment (95 °C, 1 min) of the clinical sample. Notably, the assay used smartphone-based portable detection platform. Both the LAMP mix and the heat-treated sample were loaded in two syringes (Fig. 17 A).
Fig. 17.
Smartphone-based handheld POC instrument. A) Simple workflow of SARS-CoV-2 sample preparation by brief heat treatment and transfer of the thermally lysed sample to a syringe. Another syringe was loaded with LAMP reaction mix. B) Photograph of the microfluidic chip integrated POC tool. C) Disposable microfluidic cartridge. Figure adapted with permission from Ganguli et al., [83].
The POC platform integrated the necessary heating elements, LEDs, dye-specific spectrum filters, and a smartphone to capture fluorescence images with real-time monitoring (Fig. 17B). A small microfluidic device made of polyurethane was used as the reaction chamber. The channel network for fluidic transmission in the device was designed to contain three distinctive regions: inlet, mixing and amplification regions (Fig. 17C). The two fluids enter the mixing zone before heading into a circular amplification region, where the reaction occurs. The microfluidic chip is then inserted into the platform where an integrated heater provides the operating temperature (65 °C). This system detects a positive sample in 30 min and has been verified by using a small number of clinical samples, and full agreement with RT-PCR results was obtained.
POC diagnostic tool occasionally faces a challenge with the need for cold storage of reaction mixes in low-resource settings. In particular, for a POC system that includes reagents in liquid format, operating instructions should include proper handling procedures of isothermal amplification mixes. For instance, colorimetric LAMP mixes in a liquid format tend to be unstable at room temperature [125]. Prolonged air exposure of the mixes should be avoided, as atmospheric CO2 can acidify the LAMP mixes. LAMP mixes need to be handled carefully by not opening the tube until the contents are added into the reaction chamber of the POC tool shortly before running the test. After multiple uses and repeated exposure to air, however, the color of the mix can change and become unusable for further use. The advantage of the POC tool can be realized by using a lyophilized LAMP mix. Lyophilization of reaction mixes improves their stability at room temperature [325], and it simplifies transport and storage, thus potentially eliminating the need for cold chain management. Many recent LAMP-based assays of SARS-CoV-2, including the commercially available Lucira Health's POC at-home test and Abbott's ID NOW COVID-19, use lyophilized reagents.
3.1.3. Multiplexable, high-throughput detection
Performing multiple genotyping tests on one virus or testing for several viruses in a single reaction is used in a multiplexable detection approach. Multiplexing is highly desired in bioanalysis and more so for pathogen detection since many viruses have similar genetic material and their infections have similar clinical symptoms; therefore, accurately detecting as many of these related pathogens as possible in a single reaction is desired. Respiratory pathogens such as SARS-CoV-2 have similar genetic material to other members of its Coronaviridae family. Early in the pandemic, a report was published by a group of researchers in China [326] who conducted a study on a large number of people in a hospital in Wuhan, China. They found that out of 307 patients, approximately 57% were infected by SARS-CoV-2 and at least one other influenza virus. Although non isothermal, PCR revolutionized the molecular biology field and biosensing applications because it is amenable to multiplexing, where more than one target can be amplified in a single reaction [[327], [328], [329], [330], [331]]. An RT-PCR-based multiplexed system was developed by US CDC named as “Flu SC2 Multiplex Assay” to codetect SARS-CoV-2 and influenza viruses [332]. In an isothermal-based assay, two basic approaches can be used to realize multiplex detection. One is to design an assay that is compatible with multiple primers and probes for multiple targets in a single reaction chamber. The other one is to utilize physical barriers among multiple isothermal reactions in microfluidic reaction platforms with multiple reaction chambers – the latter simplifies detection drastically by allowing the detection of only one target per reaction chamber.
In both cases, the greatest challenge is developing a convenient signal readout system for all intended targets (the probes and their chemistry, i.e., optical, electrochemical, and potentially label-free). Evaluating whether a single readout mode can be used for multiple targets or a combination of modes needs to be used [333] is central for multiplex assays. For simplicity, a convenient signal readout that can monitor multiple probes is preferred. In fluorescence readouts, multiple target-specific fluorescence probes with distinctive emission wavelengths can be used. Several double-modified probes labeled with multiple reporter dyes with unique spectral properties need to be designed carefully to specifically bind to their intended target sites. Such an approach was adapted in a promising LAMP-based multiplex method named detection of amplification by release of quenching (DARQ) [334]. In this method, in addition to other primers used in a typical single target LAMP, a modified forward inner primer and a shorter reporter oligonucleotide are used. A 5′ quencher-modified FIP primer is pre-annealed to a 3′ fluorophore labeled complementary shorter oligonucleotide to make a duplex probe. The 5′ end of the quencher-modified FIP is hybridized with a 3′ fluorophore-labeled reporter oligonucleotide, while the 3’ end of the FIP remains available to bind to the intended target site to initiate LAMP reaction. As the reaction progresses, subsequent strand generation displaces the fluorophore-labeled reporter oligonucleotide from the duplex probe. This displacement of the reporter generates a fluorescent signal. Using different fluorophore-labeled reporter oligonucleotides enables the DARQ method to detect different targets in single reaction which was first reported by Tanner et al., [334]. The same group from the New England Biolab developed a multiplex LAMP assay for detecting SARS-CoV-2 and influenza viruses [335]. Using synthetic RNA targets, the DARQ method detected all four targets within 40 min in a single reaction, suggesting its potential to detect viral RNA from clinical specimens.
Similar to DARQ, another multiplex LAMP approach has been reported by Ball et al. named quenching of unincorporated amplification signal reporters (QUASR) [336]. The QUASR reaction mixture uses 5′ fluorophore modified primers (loop or inner primers) and 3’ quencher-modified short oligonucleotide. Unlike the DARQ LAMP, QUASR allows only endpoint detection.
Another probe-based multiplex LAMP is based on a molecular beacon probe. Each end of the probe contains a short complementary region to make a hairpin-like structure [337]. The target-specific molecular beacon is a dual-modified probe with a quencher on one end and a fluorophore on the other. In its native state, the probe is non-fluorescent because of the proximity of the fluorophore and quencher groups. The opening of the molecular beacon probe and the disruption of its hairpin structure activates the probe to report fluorescence signal. This conformational change only occurs upon binding with a target specific strand. Using different molecular beacons with different fluorophores permits multiplex detection. A recent multiplex LAMP assay “Multiplex LAMP-BEAC” uses molecular beacon probes to detect multiple target genes of SARS-CoV-2 in saliva specimen [338].
Potential cross-binding of the probes and primers in the reaction can generate false-positive results. The microfluidic approach of nucleic acid detection might be a potential solution that can physically isolate multiplex isothermal reactions on chips, thereby minimizing cross-reactivity and nonspecific amplification. Multiplex isothermal amplification can potentially be performed on microfluidic chips by designing and assembling multiple channels. Recently, Shao et al. demonstrated a microfluidic approach for a multiplex CRISPR/Cas system for detecting four cancer related single nucleotide polymorphisms [339] where multiple microfluidic channels serve as reaction chambers. Nevertheless, multiplexed bioanalysis is inherently challenging in regard to detecting coinfection since testing accuracy also depends on the viral load. The viral load of SARS-CoV-2 may significantly differ from that of other coinfected flu viruses.
High-throughput detection is also desired in a pandemic like COVID-19. In this context, pooled testing is adapted for initial screening purposes. A pooled testing approach can improve existing isothermal-based tests with reduced consumption of testing reagents. In this approach, laboratories can combine the same type of specimen collected from several persons to perform only one test on the combined pool of specimens [340]. A negative result essentially confirms each of the specimens as negative. However, a positive result from a pooled test may warrants each of the specimens to be tested separately to determine which person is infected [341].
4. Emerging variants and their implication on the isothermal-based detection of the of SARS-CoV-2
Viruses naturally mutate over time, as characterized by changes in the genetic sequence. After nearly two years since the first case reported in Wuhan China, SARS-CoV-2 has also mutated, giving rise to different variants [342]. The WHO [343] has declared at least four Variant of Concern (VOC) of SARS-CoV-2; Alpha (or B.1.1.7), Beta (or B.1.351), Gamma (or P.1), and Delta (or B.1.617.2). These variants have unique mutations at different positions in the genome as well as mutations in common. For example, all of the four variants have a common mutation D614G in the spike protein and contain diverse mutations in both S1 (residues 1–686) and S2 subunits (residues 687–1273) of the spike protein (Fig. 18 ). Notably, the receptor-binding domain (RBD) in the S1 subunit contains several mutations, such as the K417 N mutation in Beta and Delta, E484K mutation in Beta and Gamma, N501Y (common mutation in Alpha, Beta, and Gamma), and L452R and T478K mutation in Delta variant [344]. These mutations in the RBD likely allow variants more effective in binding and infecting host cells and contribute toward high transmission and rapid spread in many countries [345,346]. For example, the recent surge in Delta variant cases in different countries including the United States indicates its high transmissibility and reportedly it is 60% more transmissible than the Alpha variant [347]. It is suggested that the presence of furin cleavage site in the spike protein might give SARS-CoV-2 and its variants an advantage to easily enter the host cells [348]. Higher infectivity of the Delta variant can be associated with the notable mutation P681R near the furin cleavage site.
Fig. 18.
Schematic showing amino acid changes to the spike (S) protein in the four variants of concern (VOCs) of SARS-CoV-2-Alpha, Beta, Gamma, and Delta [349]. S1 and S2 are the two functional domains of the S protein. RBD-receptor-binding domain, FCS-furin cleavage site.
For the specific detection of variants, genome sequencing is the primary method. It can provide detailed information about mutations; however, it is expensive and incompatible with large-scale testing at the point of need. The emergence of Delta and other variants can have profound effects on the nucleic acid-based SARS-CoV-2 detection as the accuracy of such assays can be compromised by mutations in the primer and probe binding sites. A combination of genomic surveillance of different VOCs and revisiting the design of an existing nucleic acid-based test is necessary.
In most isothermal-based systems, including commercially available kits, the detection of the virus includes multiple target genes. Multi-target isothermal assays offer higher accuracy. Assays that target single gene are the most vulnerable against VOCs. Some assays include S gene as the target, which might suffer from reduced sensitivity or even false-negative results because of the diverse mutations in the spike protein. Recently, failure of the S gene-targeted assay is reported for a EUA assay named “TaqPath COVID-19 Combo Kit” developed by Thermo Fisher Scientific [350]. While, in another report [351], it is suggested that the detection of other targets such as the ORF1ab and nucleocapsid (N) genes are not affected, which indicates that an isothermal-based detection of SARS-CoV-2 designed with multiple target genes are less likely to be affected by the presence of VOCs.
In a region of high prevalence of VOCs, the potential vulnerability of S gene targeted assay can be used as an indirect screening method for the presence of VOCs. A single run of a multi-target isothermal-based assay would be sufficient for the screening method. Targeting the S-gene along with N, E, and ORF1ab genes in the assay and getting negative or reduced sensitivity for the S gene while positive results for other genes can be used in this approach. However, such screening method for VOCs should be interpreted cautiously and, at times, must be validated using genome sequencing. A more direct approach for VOCs screening would be obtained by a multiplex isothermal-based assay that includes genes such as N, ORF1ab, E and one or more common mutations in the S protein of different VOCs as targets. By including common mutations such as D616G and or N501Y in the primer and probe design [352], S gene-targeted assay can be reconfigured. A DARQ-LAMP-based approach or similar multiple probes-based approach can be used to detect a large number of variants in a single test.
Infectious VOCs might continue to emerge with the course of pandemic and post-pandemic. Thus, it is crucial to identify new variants and monitor their mutations by genome sequencing and accordingly reconfigure existing primer and probes of any isothermal-based assay. “LamPORE COVID-19” assay, that combined an isothermal method LAMP, and sequencing, is a good example. With the emergence of SARS-CoV-2 variants, this sequencing-based isothermal technology has immense potential to detect the presence of variants in the sample.
5. Concluding remarks
RT-PCR, the current widely employed method for detecting SARS-CoV-2, is inconvenient for routine testing. It suffers from a long sample-to-result time and is performed by expensive equipment, thus being inconvenient for POC diagnosis. As a potential solution, different isothermal methods have been reported and developed. Isothermal-based assays of viral detection show robust performance with high potential at the POC. A significant number of isothermal-based commercial diagnostic kits have been EUA approved displaying their workability with minimal sample processing, and sample-to-answer diagnosis. Isothermal assays involving diverse CRISPR/Cas systems show great potential. However, isothermal-based sensitive multiplex assay is still nascent. A great advantage is that different types of isothermal assays with a wide range of temperatures can be performed using an inexpensive and simple heating device. We hope that these isothermal approaches in the post-COVID-19 era will expand into more advanced avenues toward efficient, simple, rapid sample-to-answer pathogen diagnostic tools.
Author contributions
As the contributing author, MMI planned and conceptualized the project, designed all figures, and searched literatures. MMI wrote the manuscript and addressed reviewers comments. MMI revised and edited the manuscript. Dipak Koirala helped with proofreading of the manuscript.
Declaration of competing interest
There is no competing interests.
Acknowledgments
All figures were created with BioRender.com. Fig. 1 was adapted with the design idea from YaleMedicine.
I would like to thank Dr. Peter B. Allen, my Ph.D. supervisor for his teaching and sincere effort to make me curious about nucleic acid-based technology. For their help and support, I would like to thank Dr. Jesse Clarke of the University of California, Santa Cruz, CA, USA, Dr. Roseann Berg of Green River College, Seattle, WA, Nabittun Nahar of Washington State University, WA, USA, and Wolfgang Brolley, Moscow, ID, USA.
Biographies

Md Mamunul Islam: Dr. Md Mamunul Islam obtained his Ph.D. from the Department of Chemistry, University of Idaho, Moscow, ID, USA. Currently he is working in a leading molecular diagnostic company in the USA, where he is working to develope novel isothermal-based detection systems. He is interested in bioanalysis with a focus on cancer biomarker miRNAs, diverse nucleic acid-based analytes, simple detection of infectious virus. His Ph.D. research involves design and application of enzymatic, semi-enzymatic and non-enzymatic isothermal DNA circuits, label-free fluorescent based biosensors, selecting novel aptamers, developing low-cost, simple microfluidic devices, and their application in bio-nanotechnology.

Dipak Koirala: Dipak Koirala is a graduate student working toward his Ph.D. at the Department of Chemistry, University of Idaho. His current research is focused on developing DNA-based biosensors, low-cost electrochemical biosensors, and energy storage.
References
- 1.Andersen K.G., Rambaut A., Lipkin W.I., Holmes E.C., Garry R.F. The proximal origin of SARS-CoV-2. Nat. Med. 2020;26:450–452. doi: 10.1038/s41591-020-0820-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Virus Origin/Origins of the SARS-CoV-2 Virus. https://www.who.int/health-topics/coronavirus/origins-of-the-virus
- 3.Chen N., et al. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet Lond. Engl. 2020;395:507–513. doi: 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li Q., et al. Early transmission dynamics in Wuhan, China, of novel coronavirus–infected pneumonia. N. Engl. J. Med. 2020 doi: 10.1056/NEJMoa2001316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Coronavirus Disease (COVID-19) – World Health Organization. https://www.who.int/emergencies/diseases/novel-coronavirus-2019
- 6.Goldberg Y., et al. Waning immunity after the BNT162b2 vaccine in Israel. N. Engl. J. Med. 2021 doi: 10.1056/NEJMoa2114228. null. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Naaber P., et al. Dynamics of antibody response to BNT162b2 vaccine after six months: a longitudinal prospective study. Lancet Reg. Health - Eur. 2021;10 doi: 10.1016/j.lanepe.2021.100208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhou P., et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu F., et al. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hoffmann M., et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280. doi: 10.1016/j.cell.2020.02.052. e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Letko M., Marzi A., Munster V. Functional assessment of cell entry and receptor usage for SARS-CoV-2 and other lineage B betacoronaviruses. Nat. Microbiol. 2020;5:562–569. doi: 10.1038/s41564-020-0688-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim D., et al. The architecture of SARS-CoV-2 transcriptome. Cell. 2020;181:914–921. doi: 10.1016/j.cell.2020.04.011. e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bar-On, Y. M., Flamholz, A., Phillips, R. & Milo, R. SARS-CoV-2 (COVID-19) by the numbers. eLife 9,. [DOI] [PMC free article] [PubMed]
- 14.Varga Z., et al. Electron microscopy of SARS-CoV-2: a challenging task – authors' reply. Lancet. 2020;395:e100. doi: 10.1016/S0140-6736(20)31185-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Udugama B., et al. Diagnosing COVID-19: the disease and tools for detection. ACS Nano. 2020;14:3822–3835. doi: 10.1021/acsnano.0c02624. [DOI] [PubMed] [Google Scholar]
- 16.Details - Public Health Image Library(PHIL) https://phil.cdc.gov/Details.aspx?pid=23354
- 17.Lee N., et al. Genome-wide analysis of influenza viral RNA and nucleoprotein association. Nucleic Acids Res. 2017;45:8968–8977. doi: 10.1093/nar/gkx584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stobart C.C., Nosek J.M., Moore M.L. Rhinovirus biology, antigenic diversity, and advancements in the design of a human rhinovirus vaccine. Front. Microbiol. 2017;8 doi: 10.3389/fmicb.2017.02412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang A., Thurmond S., Islas L., Hui K., Hai R. Zika virus genome biology and molecular pathogenesis. Emerg. Microb. Infect. 2017;6:e13. doi: 10.1038/emi.2016.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Watts J.M., et al. Architecture and secondary structure of an entire HIV-1 RNA genome. Nature. 2009;460:711–716. doi: 10.1038/nature08237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wawina-Bokalanga T., et al. Complete genome sequence of a new Ebola virus strain isolated during the 2017 likati outbreak in the democratic republic of the Congo. Microbiol. Resour. Announc. 2019;8 doi: 10.1128/MRA.00360-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu A., et al. Genome composition and divergence of the novel coronavirus (2019-nCoV) originating in China. Cell Host Microbe. 2020;27:325–328. doi: 10.1016/j.chom.2020.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang R., Hozumi Y., Yin C., Wei G.-W. Decoding SARS-CoV-2 transmission and evolution and ramifications for COVID-19 diagnosis, vaccine, and medicine. J. Chem. Inf. Model. 2020;60:5853–5865. doi: 10.1021/acs.jcim.0c00501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wrapp D., et al. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Belouzard S., Chu V.C., Whittaker G.R. Activation of the SARS coronavirus spike protein via sequential proteolytic cleavage at two distinct sites. Proc. Natl. Acad. Sci. U.S.A. 2009;106:5871–5876. doi: 10.1073/pnas.0809524106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Modes of Transmission of Virus Causing COVID-19: Implications for IPC Precaution Recommendations. https://www.who.int/news-room/commentaries/detail/modes-of-transmission-of-virus-causing-covid-19-implications-for-ipc-precaution-recommendations
- 27.Dhand R., Li J. Coughs and sneezes: their role in transmission of respiratory viral infections, including SARS-CoV-2. Am. J. Respir. Crit. Care Med. 2020;202:651–659. doi: 10.1164/rccm.202004-1263PP. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chin A.W.H., et al. Stability of SARS-CoV-2 in different environmental conditions. Lancet Microbe. 2020;1:e10. doi: 10.1016/S2666-5247(20)30003-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mueller A.L., McNamara M.S., Sinclair D.A. Why does COVID-19 disproportionately affect older people? Aging. 2020;12:9959–9981. doi: 10.18632/aging.103344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shental N., et al. Efficient high-throughput SARS-CoV-2 testing to detect asymptomatic carriers. Sci. Adv. 2020;6 doi: 10.1126/sciadv.abc5961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mercer T.R., Salit M. Testing at scale during the COVID-19 pandemic. Nat. Rev. Genet. 2021;1:12. doi: 10.1038/s41576-021-00360-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Creech C.B., Walker S.C., Samuels R.J. SARS-CoV-2 vaccines. J. Am. Med. Assoc. 2021;325:1318. doi: 10.1001/jama.2021.3199. [DOI] [PubMed] [Google Scholar]
- 33.Kevadiya B.D., et al. Diagnostics for SARS-CoV-2 infections. Nat. Mater. 2021;20:593–605. doi: 10.1038/s41563-020-00906-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sethuraman N., Jeremiah S.S., Ryo A. Interpreting diagnostic tests for SARS-CoV-2. J. Am. Med. Assoc. 2020;323:2249–2251. doi: 10.1001/jama.2020.8259. [DOI] [PubMed] [Google Scholar]
- 35.Bastos M.L., et al. Diagnostic accuracy of serological tests for covid-19: systematic review and meta-analysis. BMJ. 2020;370:m2516. doi: 10.1136/bmj.m2516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Peeling R.W., et al. Serology testing in the COVID-19 pandemic response. Lancet Infect. Dis. 2020;20:e245–e249. doi: 10.1016/S1473-3099(20)30517-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Espejo A.P., et al. Review of current advances in serologic testing for COVID-19. Am. J. Clin. Pathol. 2020;154:293–304. doi: 10.1093/ajcp/aqaa112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Factors associated with prolonged viral RNA shedding in patients with coronavirus disease 2019 (COVID-19) | clinical infectious diseases. Oxford Academic. 2019 doi: 10.1093/cid/ciaa351. https://academic.oup.com/cid/article/71/15/799/5818308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.He X., et al. Temporal dynamics in viral shedding and transmissibility of COVID-19. Nat. Med. 2020;26:672–675. doi: 10.1038/s41591-020-0869-5. [DOI] [PubMed] [Google Scholar]
- 40.Lauer S.A., et al. The incubation period of coronavirus disease 2019 (COVID-19) from publicly reported confirmed cases: estimation and application. Ann. Intern. Med. 2020;172:577–582. doi: 10.7326/M20-0504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McAloon C., et al. Incubation period of COVID-19: a rapid systematic review and meta-analysis of observational research. BMJ Open. 2020;10 doi: 10.1136/bmjopen-2020-039652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pan Y., Zhang D., Yang P., Poon L.L.M., Wang Q. Viral load of SARS-CoV-2 in clinical samples. Lancet Infect. Dis. 2020;20:411–412. doi: 10.1016/S1473-3099(20)30113-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wölfel R., et al. Virological assessment of hospitalized patients with COVID-2019. Nature. 2020;581:465–469. doi: 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
- 44.Cevik M., et al. SARS-CoV-2, SARS-CoV, and MERS-CoV viral load dynamics, duration of viral shedding, and infectiousness: a systematic review and meta-analysis. Lancet Microbe. 2021;2:e13–e22. doi: 10.1016/S2666-5247(20)30172-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hasanoglu, I. et al. Higher viral loads in asymptomatic COVID-19 patients might be the invisible part of the iceberg. Infection 1 doi:10.1007/s15010-020-01548-8. [DOI] [PMC free article] [PubMed]
- 46.Walsh K.A., et al. SARS-CoV-2 detection, viral load and infectivity over the course of an infection. J. Infect. 2020;81:357. doi: 10.1016/j.jinf.2020.06.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tan S.C., Yiap B.C.D.N.A. RNA, and protein extraction: the past and the present. J. Biomed. Biotechnol. 2009;2009 doi: 10.1155/2009/574398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bustin S.A., Nolan T. RT-qPCR testing of SARS-CoV-2: a primer. Int. J. Mol. Sci. 2020;21 doi: 10.3390/ijms21083004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rasmussen A.L., Popescu S.V. SARS-CoV-2 transmission without symptoms. Science. 2021;371:1206–1207. doi: 10.1126/science.abf9569. [DOI] [PubMed] [Google Scholar]
- 50.Weissleder R., Lee H., Ko J., Pittet M.J. COVID-19 diagnostics in context. Sci. Transl. Med. 2020;12 doi: 10.1126/scitranslmed.abc1931. [DOI] [PubMed] [Google Scholar]
- 51.Homolka, S. et al. Two pandemics, one challenge—leveraging molecular test capacity of tuberculosis laboratories for rapid COVID-19 case-finding - volume 26, number 11—november 2020 - emerging infectious diseases journal - CDC. doi:10.3201/eid2611.202602. [DOI] [PMC free article] [PubMed]
- 52.Wiley Online Library; 2012. PCR Inhibitors – Occurrence, Properties and Removal - Schrader - 2012 - Journal of Applied Microbiology.https://sfamjournals.onlinelibrary.wiley.com/doi/full/10.1111/j.1365-2672.2012.05384.x [DOI] [PubMed] [Google Scholar]
- 53.Isothermal Amplification of Nucleic Acids | Chemical Reviews. https://pubs.acs.org/doi/10.1021/acs.chemrev.5b00428 [DOI] [PubMed]
- 54.Joneja A., Huang X. Linear nicking endonuclease-mediated strand displacement DNA amplification. Anal. Biochem. 2011;414:58–69. doi: 10.1016/j.ab.2011.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Islam M.M., Ghielmetti V.M., Allen P.B. Graphene oxide assisted light-up aptamer selection against Thioflavin T for label-free detection of microRNA. Sci. Rep. 2021;11:4291. doi: 10.1038/s41598-021-83640-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Damase T.R., Islam M.M., Shipley M., Allen P.B. Thioflavin T as a noncovalent reporter for a label-free, non-enzymatic, catalytic DNA amplifier. Methods Appl. Fluoresc. 2020 doi: 10.1088/2050-6120/aba357. [DOI] [PubMed] [Google Scholar]
- 57.Liu H., et al. Sensitive and rapid detection of microRNAs using hairpin probes-mediated exponential isothermal amplification. Biosens. Bioelectron. 2017;89:710–714. doi: 10.1016/j.bios.2016.10.099. [DOI] [PubMed] [Google Scholar]
- 58.Ness J.V., Ness L.K.V., Galas D.J. Isothermal reactions for the amplification of oligonucleotides. Proc. Natl. Acad. Sci. Unit. States Am. 2003;100:4504–4509. doi: 10.1073/pnas.0730811100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Du Y.-C., Zhu L.-N., Kong D.-M. Label-free thioflavin T/G-quadruplex-based real-time strand displacement amplification for biosensing applications. Biosens. Bioelectron. 2016;86:811–817. doi: 10.1016/j.bios.2016.07.083. [DOI] [PubMed] [Google Scholar]
- 60.Jia H., Li Z., Liu C., Cheng Y. Ultrasensitive detection of microRNAs by exponential isothermal amplification. Angew. Chem. Int. Ed. 2010;49:5498–5501. doi: 10.1002/anie.201001375. [DOI] [PubMed] [Google Scholar]
- 61.Notomi T., et al. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 2000;28:e63. doi: 10.1093/nar/28.12.e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nagamine K., Hase T., Notomi T. Accelerated reaction by loop-mediated isothermal amplification using loop primers. Mol. Cell. Probes. 2002;16:223–229. doi: 10.1006/mcpr.2002.0415. [DOI] [PubMed] [Google Scholar]
- 63.Wang J., et al. Colorimetric detection of horse meat based on loop-mediated isothermal amplification (LAMP) Food Anal. Methods. 2019;12:2535–2541. [Google Scholar]
- 64.Deng H., Gao Z. Bioanalytical applications of isothermal nucleic acid amplification techniques. Anal. Chim. Acta. 2015;853:30–45. doi: 10.1016/j.aca.2014.09.037. [DOI] [PubMed] [Google Scholar]
- 65.Huang X., Tang G., Ismail N., Wang X. Developing RT-LAMP assays for detection of SARS-CoV-2 in saliva. medRxiv. 2021;4(25) doi: 10.1101/2021.04.25.21256085. 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.LAMP Primer Designing Software PrimerExplorer. https://primerexplorer.jp/e/
- 67.Shirshikov F.V., Pekov Y.A., Miroshnikov K.A. MorphoCatcher: a multiple-alignment based web tool for target selection and designing taxon-specific primers in the loop-mediated isothermal amplification method. PeerJ. 2019;7 doi: 10.7717/peerj.6801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jia B., et al. GLAPD: whole genome based LAMP primer design for a set of target genomes. Front. Microbiol. 2019;10 doi: 10.3389/fmicb.2019.02860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mori Y., Notomi T. Loop-mediated isothermal amplification (LAMP): a rapid, accurate, and cost-effective diagnostic method for infectious diseases. J. Infect. Chemother. 2009;15:62–69. doi: 10.1007/s10156-009-0669-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mori Y., Nagamine K., Tomita N., Notomi T. Detection of loop-mediated isothermal amplification reaction by turbidity derived from magnesium pyrophosphate formation. Biochem. Biophys. Res. Commun. 2001;289:150–154. doi: 10.1006/bbrc.2001.5921. [DOI] [PubMed] [Google Scholar]
- 71.Mori Y., Kitao M., Tomita N., Notomi T. Real-time turbidimetry of LAMP reaction for quantifying template DNA. J. Biochem. Biophys. Methods. 2004;59:145–157. doi: 10.1016/j.jbbm.2003.12.005. [DOI] [PubMed] [Google Scholar]
- 72.Yuan X.-Y., et al. LAMP real-time turbidity detection for fowl adenovirus. BMC Vet. Res. 2019;15:256. doi: 10.1186/s12917-019-2015-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Goto M., Honda E., Ogura A., Nomoto A., Hanaki K.-I. Colorimetric detection of loop-mediated isothermal amplification reaction by using hydroxy naphthol blue. Biotechniques. 2009;46:167–172. doi: 10.2144/000113072. [DOI] [PubMed] [Google Scholar]
- 74.Hong M., et al. A modified visual loop-mediated isothermal amplification method for diagnosis and differentiation of main pathogens from Mycobacterium tuberculosis complex. World J. Microbiol. Biotechnol. 2012;28:523–531. doi: 10.1007/s11274-011-0843-y. [DOI] [PubMed] [Google Scholar]
- 75.Tomita N., Mori Y., Kanda H., Notomi T. Loop-mediated isothermal amplification (LAMP) of gene sequences and simple visual detection of products. Nat. Protoc. 2008;3:877–882. doi: 10.1038/nprot.2008.57. [DOI] [PubMed] [Google Scholar]
- 76.Tanner N.A., Zhang Y., Evans T.C. Visual detection of isothermal nucleic acid amplification using pH-sensitive dyes. Biotechniques. 2015;58:59–68. doi: 10.2144/000114253. [DOI] [PubMed] [Google Scholar]
- 77.Lucchi N.W., Ljolje D., Silva-Flannery L., Udhayakumar V. Use of malachite green-loop mediated isothermal amplification for detection of plasmodium spp. parasites. PLoS One. 2016;11 doi: 10.1371/journal.pone.0151437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Song J., et al. Instrument-free point-of-care molecular detection of Zika virus. Anal. Chem. 2016;88:7289–7294. doi: 10.1021/acs.analchem.6b01632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Color SARS-CoV-2 RT-LAMP Diagnostic Assay - EUA Summary. https://www.fda.gov/media/138249/download
- 80.Nguyen H.Q., Nguyen V.D., Van Nguyen H., Seo T.S. Quantification of colorimetric isothermal amplification on the smartphone and its open-source app for point-of-care pathogen detection. Sci. Rep. 2020;10:15123. doi: 10.1038/s41598-020-72095-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Papadakis G., et al. Real-time colorimetric LAMP methodology for quantitative nucleic acids detection at the point-of-care. bioRxiv. 2020;7(22) doi: 10.1101/2020.07.22.215251. 2020. [DOI] [Google Scholar]
- 82.Nguyen D.V., Nguyen V.H., Seo T.S. Quantification of colorimetric loop-mediated isothermal amplification process. BioChip J. 2019;13:158–164. [Google Scholar]
- 83.Ganguli A., et al. Rapid isothermal amplification and portable detection system for SARS-CoV-2. Proc. Natl. Acad. Sci. Unit. States Am. 2020;117:22727–22735. doi: 10.1073/pnas.2014739117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zhang X., et al. Development of a loop-mediated isothermal amplification assay for rapid detection of subgroup J avian leukosis virus. J. Clin. Microbiol. 2010;48:2116–2121. doi: 10.1128/JCM.02530-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Oliveira K. G. de, et al. Rapid molecular diagnostics of COVID-19 by RT-LAMP in a centrifugal polystyrene-toner based microdevice with end-point visual detection. Analyst. 2021;146:1178–1187. doi: 10.1039/d0an02066d. [DOI] [PubMed] [Google Scholar]
- 86.Safavieh M., et al. Emerging loop-mediated isothermal amplification-based microchip and microdevice technologies for nucleic acid detection. ACS Biomater. Sci. Eng. 2016;2:278–294. doi: 10.1021/acsbiomaterials.5b00449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kong X., et al. Development and application of loop-mediated isothermal amplification (LAMP) for detection of Plasmopara viticola. Sci. Rep. 2016;6:28935. doi: 10.1038/srep28935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Parida M., Posadas G., Inoue S., Hasebe F., Morita K. Real-time reverse transcription loop-mediated isothermal amplification for rapid detection of west nile virus. J. Clin. Microbiol. 2004;42:257–263. doi: 10.1128/JCM.42.1.257-263.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Tsai S.-M., et al. Development of a loop-mediated isothermal amplification for rapid detection of orf virus. J. Virol. Methods. 2009;157:200–204. doi: 10.1016/j.jviromet.2009.01.003. [DOI] [PubMed] [Google Scholar]
- 90.Quyen T.L., Ngo T.A., Bang D.D., Madsen M., Wolff A. Classification of multiple DNA dyes based on inhibition effects on real-time loop-mediated isothermal amplification (LAMP): prospect for point of care setting. Front. Microbiol. 2019;10 doi: 10.3389/fmicb.2019.02234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Oscorbin I.P., Belousova E.A., Zakabunin A.I., Boyarskikh U.A., Filipenko M.L. Comparison of fluorescent intercalating dyes for quantitative loop-mediated isothermal amplification (qLAMP) Biotechniques. 2016;61 doi: 10.2144/000114432. [DOI] [PubMed] [Google Scholar]
- 92.Gudnason H., Dufva M., Bang D.D., Wolff A. Comparison of multiple DNA dyes for real-time PCR: effects of dye concentration and sequence composition on DNA amplification and melting temperature. Nucleic Acids Res. 2007;35:e127. doi: 10.1093/nar/gkm671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Eischeid A.C. SYTO dyes and EvaGreen outperform SYBR Green in real-time PCR. BMC Res. Notes. 2011;4:263. doi: 10.1186/1756-0500-4-263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Luo J., et al. A real-time microfluidic multiplex electrochemical loop-mediated isothermal amplification chip for differentiating bacteria. Biosens. Bioelectron. 2014;60:84–91. doi: 10.1016/j.bios.2014.03.073. [DOI] [PubMed] [Google Scholar]
- 95.Nagatani N., et al. Semi-real time electrochemical monitoring for influenza virus RNA by reverse transcription loop-mediated isothermal amplification using a USB powered portable potentiostat. Analyst. 2011;136:5143–5150. doi: 10.1039/c1an15638a. [DOI] [PubMed] [Google Scholar]
- 96.Hashimoto K., Inada M., Ito K. Multiplex real-time loop-mediated isothermal amplification using an electrochemical DNA chip consisting of a single liquid-flow channel. Anal. Chem. 2019;91:3227–3232. doi: 10.1021/acs.analchem.8b05284. [DOI] [PubMed] [Google Scholar]
- 97.Hashimoto K., Inada M., Ito K. A novel voltammetric approach for real-time electrochemical detection of targeted nucleic acid sequences using LAMP. Anal. Biochem. 2017;539:113–117. doi: 10.1016/j.ab.2017.10.019. [DOI] [PubMed] [Google Scholar]
- 98.Ye X., Fang X., Li X., Kong J. Gold nanoparticle-mediated nucleic acid isothermal amplification with enhanced specificity. Anal. Chim. Acta. 2018;1043:150–157. doi: 10.1016/j.aca.2018.09.016. [DOI] [PubMed] [Google Scholar]
- 99.Njiru Z.K. Rapid and sensitive detection of human African trypanosomiasis by loop-mediated isothermal amplification combined with a lateral-flow dipstick. Diagn. Microbiol. Infect. Dis. 2011;69:205–209. doi: 10.1016/j.diagmicrobio.2010.08.026. [DOI] [PubMed] [Google Scholar]
- 100.Zhang X., Lowe S.B., Gooding J.J. Brief review of monitoring methods for loop-mediated isothermal amplification (LAMP) Biosens. Bioelectron. 2014;61:491–499. doi: 10.1016/j.bios.2014.05.039. [DOI] [PubMed] [Google Scholar]
- 101.Laras K., et al. Tracking the re-emergence of epidemic chikungunya virus in Indonesia. Trans. R. Soc. Trop. Med. Hyg. 2005;99:128–141. doi: 10.1016/j.trstmh.2004.03.013. [DOI] [PubMed] [Google Scholar]
- 102.Pastorino B., et al. Epidemic resurgence of Chikungunya virus in democratic Republic of the Congo: identification of a new central African strain. J. Med. Virol. 2004;74:277–282. doi: 10.1002/jmv.20168. [DOI] [PubMed] [Google Scholar]
- 103.Schuffenecker I., et al. Genome microevolution of chikungunya viruses causing the Indian Ocean outbreak. PLoS Med. 2006;3:e263. doi: 10.1371/journal.pmed.0030263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Powers A.M., Brault A.C., Tesh R.B., Weaver S.C. Re-emergence of Chikungunya and O’nyong-nyong viruses: evidence for distinct geographical lineages and distant evolutionary relationships. J. Gen. Virol. 2000;81:471–479. doi: 10.1099/0022-1317-81-2-471. [DOI] [PubMed] [Google Scholar]
- 105.Parida M.M., et al. Rapid and real-time detection of chikungunya virus by reverse transcription loop-mediated isothermal amplification assay. J. Clin. Microbiol. 2007;45:351–357. doi: 10.1128/JCM.01734-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Teoh B.-T., et al. Detection of dengue viruses using reverse transcription-loop-mediated isothermal amplification. BMC Infect. Dis. 2013;13:387. doi: 10.1186/1471-2334-13-387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Kim J.-G., et al. Rapid discriminative detection of dengue viruses via loop mediated isothermal amplification. Talanta. 2018;190:391–396. doi: 10.1016/j.talanta.2018.08.019. [DOI] [PubMed] [Google Scholar]
- 108.Zhao J., Feng R. Sensitive and rapid detection of Zika virus by loop-mediated isothermal amplification. Virus Gene. 2019;55:43–50. doi: 10.1007/s11262-018-1612-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Wang X., et al. Rapid and sensitive detection of Zika virus by reverse transcription loop-mediated isothermal amplification. J. Virol. Methods. 2016;238:86–93. doi: 10.1016/j.jviromet.2016.10.010. [DOI] [PubMed] [Google Scholar]
- 110.Oleribe O.O., et al. Ebola virus disease epidemic in West Africa: lessons learned and issues arising from West African countries. Clin. Med. 2015;15:54–57. doi: 10.7861/clinmedicine.15-1-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Benzine J.W., et al. Molecular diagnostic field test for point-of-care detection of Ebola virus directly from blood. J. Infect. Dis. 2016;214:S234–S242. doi: 10.1093/infdis/jiw330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Christman M., Kedwaii A., Xu J., Donis R., Lu G. Pandemic (H1N1) 2009 virus revisited: an evolutionary retrospective. Infect. Genet. Evol. J. Mol. Epidemiol. Evol. Genet. Infect. Dis. 2011;11:803–811. doi: 10.1016/j.meegid.2011.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Hatano B., et al. Mobile and accurate detection system for infection by the 2009 pandemic influenza A (H1N1) virus with a pocket-warmer reverse-transcriptase loop-mediated isothermal amplification. J. Med. Virol. 2011;83:568–573. doi: 10.1002/jmv.22031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Thai H.T.C., et al. Development and evaluation of a novel loop-mediated isothermal amplification method for rapid detection of severe acute respiratory syndrome coronavirus. J. Clin. Microbiol. 2004;42:1956–1961. doi: 10.1128/JCM.42.5.1956-1961.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Shirato K., et al. Development of fluorescent reverse transcription loop-mediated isothermal amplification (RT-LAMP) using quenching probes for the detection of the Middle East respiratory syndrome coronavirus. J. Virol. Methods. 2018;258:41–48. doi: 10.1016/j.jviromet.2018.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Shirato K., et al. Detection of Middle East respiratory syndrome coronavirus using reverse transcription loop-mediated isothermal amplification (RT-LAMP) Virol. J. 2014;11 doi: 10.1186/1743-422X-11-139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Bhadra S., et al. Real-time sequence-validated loop-mediated isothermal amplification assays for detection of Middle East respiratory syndrome coronavirus (MERS-CoV) PLoS One. 2015;10 doi: 10.1371/journal.pone.0123126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Lee S.H., et al. One-pot reverse transcriptional loop-mediated isothermal amplification (RT-LAMP) for detecting MERS-CoV. Front. Microbiol. 2017;7 doi: 10.3389/fmicb.2016.02166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Ascoli C.A. Could mutations of SARS-CoV-2 suppress diagnostic detection? Nat. Biotechnol. 2021;39:274–275. doi: 10.1038/s41587-021-00845-3. [DOI] [PubMed] [Google Scholar]
- 120.Li Q., et al. The impact of mutations in SARS-CoV-2 spike on viral infectivity and antigenicity. Cell. 2020;182:1284–1294. doi: 10.1016/j.cell.2020.07.012. e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Le T.K., et al. Nucleic acid-based technologies targeting coronaviruses. Trends Biochem. Sci. 2021;46:351–365. doi: 10.1016/j.tibs.2020.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Baek Y.H., et al. Development of a reverse transcription-loop-mediated isothermal amplification as a rapid early-detection method for novel SARS-CoV-2. Emerg. Microb. Infect. 2020;9:998–1007. doi: 10.1080/22221751.2020.1756698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Duffy S. Why are RNA virus mutation rates so damn high? PLoS Biol. 2018;16 doi: 10.1371/journal.pbio.3000003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Tang Y.-W., Schmitz J.E., Persing D.H., Stratton C.W. Laboratory diagnosis of COVID-19: current issues and challenges. J. Clin. Microbiol. 2020;58 doi: 10.1128/JCM.00512-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Dao Thi V.L., et al. A colorimetric RT-LAMP assay and LAMP-sequencing for detecting SARS-CoV-2 RNA in clinical samples. Sci. Transl. Med. 2020 doi: 10.1126/scitranslmed.abc7075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Dong Y., et al. Comparative evaluation of 19 reverse transcription loop-mediated isothermal amplification assays for detection of SARS-CoV-2. Sci. Rep. 2021;11:2936. doi: 10.1038/s41598-020-80314-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Lu R., et al. A novel reverse transcription loop-mediated isothermal amplification method for rapid detection of SARS-CoV-2. Int. J. Mol. Sci. 2020;21 doi: 10.3390/ijms21082826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Lamb L.E., Bartolone S.N., Ward E., Chancellor M.B. Rapid detection of novel coronavirus/Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) by reverse transcription-loop-mediated isothermal amplification. PLoS One. 2020;15 doi: 10.1371/journal.pone.0234682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Yu L., et al. Rapid detection of COVID-19 coronavirus using a reverse transcriptional loop-mediated isothermal amplification (RT-LAMP) diagnostic platform. Clin. Chem. 2020;66:975–977. doi: 10.1093/clinchem/hvaa102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Sharma K., Sharma M., Batra N., Sharma A., Dhillon M.S. Diagnostic potential of multi-targeted LAMP (loop-mediated isothermal amplification) for osteoarticular tuberculosis. J. Orthop. Res. 2017;35:361–365. doi: 10.1002/jor.23293. [DOI] [PubMed] [Google Scholar]
- 131.Perchetti G.A., Nalla A.K., Huang M.-L., Jerome K.R., Greninger A.L. Multiplexing primer/probe sets for detection of SARS-CoV-2 by qRT-PCR. J. Clin. Virol. 2020;129 doi: 10.1016/j.jcv.2020.104499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Yan C., et al. Rapid and visual detection of 2019 novel coronavirus (SARS-CoV-2) by a reverse transcription loop-mediated isothermal amplification assay. Clin. Microbiol. Infect. Off. Publ. Eur. Soc. Clin. Microbiol. Infect. Dis. 2020;26:773–779. doi: 10.1016/j.cmi.2020.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Jiang M., et al. Development and validation of a rapid, single-step reverse transcriptase loop-mediated isothermal amplification (RT-LAMP) system potentially to Be used for reliable and high-throughput screening of COVID-19. Front. Cell. Infect. Microbiol. 2020;10 doi: 10.3389/fcimb.2020.00331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Huang W.E., et al. RT-LAMP for rapid diagnosis of coronavirus SARS-CoV-2. Microb. Biotechnol. 2020;13:950–961. doi: 10.1111/1751-7915.13586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Zhu X., et al. Multiplex reverse transcription loop-mediated isothermal amplification combined with nanoparticle-based lateral flow biosensor for the diagnosis of COVID-19. Biosens. Bioelectron. 2020;166 doi: 10.1016/j.bios.2020.112437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Wang L., et al. Technical aspects of nicking enzyme assisted amplification. Analyst. 2018;143:1444–1453. doi: 10.1039/c7an02037f. [DOI] [PubMed] [Google Scholar]
- 137.Valentin T., et al. Prospective evaluation of three rapid molecular tests for seasonal influenza in patients presenting at an emergency unit. J. Clin. Virol. 2019;111:29–32. doi: 10.1016/j.jcv.2019.01.003. [DOI] [PubMed] [Google Scholar]
- 138.Chan S.-H., Stoddard B.L., Xu S. Natural and engineered nicking endonucleases—from cleavage mechanism to engineering of strand-specificity. Nucleic Acids Res. 2011;39:1–18. doi: 10.1093/nar/gkq742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Zheleznaya L.A., et al. Nicking endonucleases. Biochem. Biokhimiia. 2009;74:1457–1466. doi: 10.1134/s0006297909130033. [DOI] [PubMed] [Google Scholar]
- 140.Kilic T., Weissleder R., Lee H. Molecular and immunological diagnostic tests of COVID-19: current status and challenges. iScience. 2020;23 doi: 10.1016/j.isci.2020.101406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Compton J. Nucleic acid sequence-based amplification. Nature. 1991;350:91–92. doi: 10.1038/350091a0. [DOI] [PubMed] [Google Scholar]
- 142.Abd El Galil K.H., et al. Real-time nucleic acid sequence-based amplification assay for detection of hepatitis A virus. Appl. Environ. Microbiol. 2005;71:7113–7116. doi: 10.1128/AEM.71.11.7113-7116.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.de Baar M.P., et al. Design and evaluation of a human immunodeficiency virus type 1 RNA assay using nucleic acid sequence-based amplification technology able to quantify both group M and O viruses by using the long terminal repeat as target. J. Clin. Microbiol. 1999;37:1813–1818. doi: 10.1128/jcm.37.6.1813-1818.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Damen M., Sillekens P., Cuypers H.T., Frantzen I., Melsert R. Characterization of the quantitative HCV NASBA assay. J. Virol. Methods. 1999;82:45–54. doi: 10.1016/s0166-0934(99)00079-8. [DOI] [PubMed] [Google Scholar]
- 145.Zhao Y., Chen F., Li Q., Wang L., Fan C. Isothermal amplification of nucleic acids. Chem. Rev. 2015;55 doi: 10.1021/acs.chemrev.5b00428. [DOI] [PubMed] [Google Scholar]
- 146.Wu Q., et al. INSIGHT: a population-scale COVID-19 testing strategy combining point-of-care diagnosis with centralized high-throughput sequencing. Sci. Adv. 2021;7 doi: 10.1126/sciadv.abe5054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Yan L., et al. Isothermal amplified detection of DNA and RNA. Mol. Biosyst. 2014;10:970–1003. doi: 10.1039/c3mb70304e. [DOI] [PubMed] [Google Scholar]
- 148.Zhao Y., Perlin D.S. In: Fungal Diagnostics: Methods and Protocols. O'Connor L., Glynn B., editors. 2013. Quantitative detection of Aspergillus spp. by real-time nucleic acid sequence-based amplification; pp. 83–92. Humana Press. [DOI] [PubMed] [Google Scholar]
- 149.Lamhoujeb S.L., Charest H.C., Fliss I.F., Ngazoa S.N., Jean J.J. Real-time molecular beacon NASBA for rapid and sensitive detection of norovirus GII in clinical samples. Can. J. Microbiol. 2009 doi: 10.1139/W09-105. [DOI] [PubMed] [Google Scholar]
- 150.Islam M.M., Loewen A., Allen P.B. Simple, low-cost fabrication of acrylic based droplet microfluidics and its use to generate DNA-coated particles. Sci. Rep. 2018;8:8763. doi: 10.1038/s41598-018-27037-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Dimov I.K., et al. Integrated microfluidic tmRNA purification and real-time NASBA device for molecular diagnostics. Lab Chip. 2008;8:2071–2078. doi: 10.1039/b812515e. [DOI] [PubMed] [Google Scholar]
- 152.Gulliksen A., et al. Real-time nucleic acid sequence-based amplification in nanoliter volumes. Anal. Chem. 2004;76:9–14. doi: 10.1021/ac034779h. [DOI] [PubMed] [Google Scholar]
- 153.Gulliksen A., et al. Parallel nanoliter detection of cancer markers using polymer microchips. Lab Chip. 2005;5:416–420. doi: 10.1039/b415525d. [DOI] [PubMed] [Google Scholar]
- 154.Gracias K.S., Mckillip J.L. Nucleic acid sequence-based amplification (nasba) in molecular bacteriology: a procedural guide. J. Rapid Methods Autom. Microbiol. 2007;15:295–309. [Google Scholar]
- 155.Boulet G.A.V., et al. Nucleic acid sequence-based amplification assay for human papillomavirus mRNA detection and typing: evidence for DNA amplification. J. Clin. Microbiol. 2010;48:2524–2529. doi: 10.1128/JCM.00173-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Tillmann R.L., Simon A., Müller A., Schildgen O. Sensitive commercial NASBA assay for the detection of respiratory syncytial virus in clinical specimen. PLoS One. 2007;2:e1357. doi: 10.1371/journal.pone.0001357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Langabeer S.E., et al. Transcription-mediated amplification and hybridisation protection assay to determine BCR-ABL transcript levels in patients with chronic myeloid leukaemia. Leukemia. 2002;16:393–399. doi: 10.1038/sj.leu.2402392. [DOI] [PubMed] [Google Scholar]
- 158.Sarrazin C., Teuber G., Kokka R., Rabenau H., Zeuzem S. Detection of residual hepatitis C virus RNA by transcription-mediated amplification in patients with complete virologic response according to polymerase chain reaction–based assays. Hepatology. 2000;32:818–823. doi: 10.1053/jhep.2000.17709. [DOI] [PubMed] [Google Scholar]
- 159.Hofmann W.P., et al. Comparison of transcription mediated amplification (TMA) and reverse transcription polymerase chain reaction (RT-PCR) for detection of hepatitis C virus RNA in liver tissue. J. Clin. Virol. 2005;32:289–293. doi: 10.1016/j.jcv.2004.08.011. [DOI] [PubMed] [Google Scholar]
- 160.Piepenburg O., Williams C.H., Stemple D.L., Armes N.A. DNA detection using recombination proteins. PLoS Biol. 2006;4 doi: 10.1371/journal.pbio.0040204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Li J., Macdonald J., Stetten F. von. Review: a comprehensive summary of a decade development of the recombinase polymerase amplification. Analyst. 2018;144:31–67. doi: 10.1039/c8an01621f. [DOI] [PubMed] [Google Scholar]
- 162.Crannell Z.A., Rohrman B., Richards-Kortum R. Equipment-free incubation of recombinase polymerase amplification reactions using body heat. PLoS One. 2014;9 doi: 10.1371/journal.pone.0112146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Behrmann O., et al. Rapid detection of SARS-CoV-2 by low volume real-time single tube reverse transcription recombinase polymerase amplification using an exo probe with an internally linked quencher (Exo-IQ) Clin. Chem. 2020;66:1047–1054. doi: 10.1093/clinchem/hvaa116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Xia S., Chen X. Single-copy sensitive, field-deployable, and simultaneous dual-gene detection of SARS-CoV-2 RNA via modified RT–RPA. Cell Discov. 2020;6:1–4. doi: 10.1038/s41421-020-0175-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.El Wahed A.A., et al. Suitcase lab for rapid detection of SARS-CoV-2 based on recombinase polymerase amplification assay. Anal. Chem. 2021;93:2627–2634. doi: 10.1021/acs.analchem.0c04779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Higgins M., et al. PrimedRPA: primer design for recombinase polymerase amplification assays. Bioinformatics. 2019;35:682–684. doi: 10.1093/bioinformatics/bty701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Kim Y., et al. Single-strand RPA for rapid and sensitive detection of SARS-CoV-2 RNA. medRxiv. 2020 doi: 10.1101/2020.08.17.20177006. [DOI] [Google Scholar]
- 168.Song J., et al. Two-stage isothermal enzymatic amplification for concurrent multiplex molecular detection. Clin. Chem. 2017;63:714–722. doi: 10.1373/clinchem.2016.263665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.El-Tholoth M., Bau H.H., Song J. A single and two-stage, closed-tube, molecular test for the 2019 novel coronavirus (COVID-19) at home, clinic, and points of entry. ChemRxiv. 2020 doi: 10.26434/chemrxiv.11860137.v1. [DOI] [Google Scholar]
- 170.Zhao W., Ali M.M., Brook M.A., Li Y. Rolling circle amplification: applications in nanotechnology and biodetection with functional nucleic acids. Angew. Chem. Int. Ed. 2008;47:6330–6337. doi: 10.1002/anie.200705982. [DOI] [PubMed] [Google Scholar]
- 171.Ali M.M., et al. Rolling circle amplification: a versatile tool for chemical biology, materials science and medicine. Chem. Soc. Rev. 2014;43:3324–3341. doi: 10.1039/c3cs60439j. [DOI] [PubMed] [Google Scholar]
- 172.Ciftci S., et al. Digital rolling circle amplification–based detection of Ebola and other tropical viruses. J. Mol. Diagn. 2020;22:272–283. doi: 10.1016/j.jmoldx.2019.10.014. [DOI] [PubMed] [Google Scholar]
- 173.Hamidi S.V., Ghourchian H., Tavoosidana G. Real-time detection of H5N1 influenza virus through hyperbranched rolling circle amplification. Analyst. 2015;140:1502–1509. doi: 10.1039/c4an01954g. [DOI] [PubMed] [Google Scholar]
- 174.Wang B., et al. Rapid and sensitive detection of severe acute respiratory syndrome coronavirus by rolling circle amplification. J. Clin. Microbiol. 2005;43:2339–2344. doi: 10.1128/JCM.43.5.2339-2344.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Tian B., Gao F., Fock J., Dufva M., Hansen M.F. Homogeneous circle-to-circle amplification for real-time optomagnetic detection of SARS-CoV-2 RdRp coding sequence. Biosens. Bioelectron. 2020;165 doi: 10.1016/j.bios.2020.112356. [DOI] [PubMed] [Google Scholar]
- 176.Chaibun T., et al. Rapid electrochemical detection of coronavirus SARS-CoV-2. Nat. Commun. 2021;12:802. doi: 10.1038/s41467-021-21121-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Wharam S.D., et al. Specific detection of DNA and RNA targets using a novel isothermal nucleic acid amplification assay based on the formation of a three-way junction structure. Nucleic Acids Res. 2001;29 doi: 10.1093/nar/29.11.e54. e54–e54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Wharam S.D., Hall M.J., Wilson W.H. Detection of virus mRNA within infected host cells using an isothermal nucleic acid amplification assay: marine cyanophage gene expression within Synechococcus sp. Virol. J. 2007;4:52. doi: 10.1186/1743-422X-4-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Khan P., Aufdembrink L.M., Engelhart A.E. Isothermal SARS-CoV-2 diagnostics: tools for enabling distributed pandemic testing as a means of supporting safe reopenings. ACS Synth. Biol. 2020 doi: 10.1021/acssynbio.0c00359. [DOI] [PubMed] [Google Scholar]
- 180.Andresen D., von Nickisch-Rosenegk M., Bier F.F. Helicase dependent OnChip-amplification and its use in multiplex pathogen detection. Clin. Chim. Acta Int. J. Clin. Chem. 2009;403:244–248. doi: 10.1016/j.cca.2009.03.021. [DOI] [PubMed] [Google Scholar]
- 181.Barreda-García S., et al. Attomolar quantitation of Mycobacterium tuberculosis by asymmetric helicase-dependent isothermal DNA-amplification and electrochemical detection. Biosens. Bioelectron. 2015;68:122–128. doi: 10.1016/j.bios.2014.12.029. [DOI] [PubMed] [Google Scholar]
- 182.Vincent M., Xu Y., Kong H. Helicase-dependent isothermal DNA amplification. EMBO Rep. 2004;5:795–800. doi: 10.1038/sj.embor.7400200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Helicase Dependent Amplification | Point of Care Testing from Quidel. https://www.quidel.com/molecular-diagnostics/helicase-dependent-amplification-tests
- 184.Tang W., et al. Nucleic acid assay system for tier II laboratories and moderately complex clinics to detect HIV in low-resource settings. J. Infect. Dis. 2010;201(Suppl 1):S46–S51. doi: 10.1086/650388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Tong Y., Lemieux B., Kong H. Multiple strategies to improve sensitivity, speed and robustness of isothermal nucleic acid amplification for rapid pathogen detection. BMC Biotechnol. 2011;11:50. doi: 10.1186/1472-6750-11-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.O'Neil D., Doseeva V., Rothmann T., Wolff J., Nazarenko I. Evaluation of Chlamydia trachomatis and Neisseria gonorrhoeae detection in urine, endocervical, and vaginal specimens by a multiplexed isothermal thermophilic helicase-dependent amplification (tHDA) assay. J. Clin. Microbiol. 2011;49:4121–4125. doi: 10.1128/JCM.00952-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Li Y., et al. Detection and species identification of malaria parasites by isothermal tHDA amplification directly from human blood without sample preparation. J. Mol. Diagn. JMD. 2013;15:634–641. doi: 10.1016/j.jmoldx.2013.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Chow W.H.A., et al. Application of isothermal helicase-dependent amplification with a disposable detection device in a simple sensitive stool test for toxigenic Clostridium difficile. J. Mol. Diagn. JMD. 2008;10:452–458. doi: 10.2353/jmoldx.2008.080008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Huang S., et al. Low cost extraction and isothermal amplification of DNA for infectious diarrhea diagnosis. PLoS One. 2013;8 doi: 10.1371/journal.pone.0060059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Ma F., Liu M., Tang B., Zhang C. Sensitive quantification of MicroRNAs by isothermal helicase-dependent amplification. Anal. Chem. 2017;89:6182–6187. doi: 10.1021/acs.analchem.7b01113. [DOI] [PubMed] [Google Scholar]
- 191.Barreda-García S., Miranda-Castro R., de-los-Santos-Álvarez N., Miranda-Ordieres A.J., Lobo-Castañón M.J. Helicase-dependent isothermal amplification: a novel tool in the development of molecular-based analytical systems for rapid pathogen detection. Anal. Bioanal. Chem. 2018;410:679–693. doi: 10.1007/s00216-017-0620-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Chen F., et al. Zero-background helicase-dependent amplification and its application to reliable assay of telomerase activity in cancer cell by eliminating primer–dimer artifacts. Chembiochem. 2016;17:1171–1176. doi: 10.1002/cbic.201500605. [DOI] [PubMed] [Google Scholar]
- 193.Fang R., et al. Cross-priming amplification for rapid detection of Mycobacterium tuberculosis in sputum specimens. J. Clin. Microbiol. 2009;47:845–847. doi: 10.1128/JCM.01528-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Xu G., et al. Cross priming amplification: mechanism and optimization for isothermal DNA amplification. Sci. Rep. 2012;2:246. doi: 10.1038/srep00246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Meng X.-Y., et al. Cross-priming amplification-based lateral flow strip as a novel tool for rapid on-site detection of wild-type pseudorabies virus. Sensor. Actuator. B Chem. 2018;259:573–579. [Google Scholar]
- 196.Cui L., et al. Detection of severe fever with thrombocytopenia syndrome virus by reverse transcription–cross-priming amplification coupled with vertical flow visualization. J. Clin. Microbiol. 2012;50:3881–3885. doi: 10.1128/JCM.01931-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Su Z.D., et al. Establishment and application of cross-priming isothermal amplification coupled with lateral flow dipstick (CPA-LFD) for rapid and specific detection of red-spotted grouper nervous necrosis virus. Virol. J. 2015;12:149. doi: 10.1186/s12985-015-0374-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Wang F.-X., et al. Reverse transcription cross-priming amplification–nucleic acid test strip for rapid detection of porcine epidemic diarrhea virus. Sci. Rep. 2016;6:24702. doi: 10.1038/srep24702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Huo Y.-Y., Li G.-F., Qiu Y.-H., Li W.-M., Zhang Y.-J. Rapid detection of prunus necrotic ringspot virus by reverse transcription-cross-priming amplification coupled with nucleic acid test strip cassette. Sci. Rep. 2017;7:16175. doi: 10.1038/s41598-017-16536-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Ledford H., Callaway E. Pioneers of revolutionary CRISPR gene editing win chemistry Nobel. Nature. 2020;586:346–347. doi: 10.1038/d41586-020-02765-9. [DOI] [PubMed] [Google Scholar]
- 201.Barrangou R., Marraffini L.A. CRISPR-Cas systems: prokaryotes upgrade to adaptive immunity. Mol. Cell. 2014;54:234–244. doi: 10.1016/j.molcel.2014.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Bradde S., Nourmohammad A., Goyal S., Balasubramanian V. The size of the immune repertoire of bacteria. Proc. Natl. Acad. Sci. Unit. States Am. 2020;117:5144–5151. doi: 10.1073/pnas.1903666117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Karvelis T., et al. crRNA and tracrRNA guide Cas9-mediated DNA interference in Streptococcus thermophilus. RNA Biol. 2013;10:841–851. doi: 10.4161/rna.24203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Loureiro A., da Silva G.J. CRISPR-Cas: converting A bacterial defence mechanism into A state-of-the-art genetic manipulation tool. Antibiotics. 2019;8 doi: 10.3390/antibiotics8010018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Amitai G., Sorek R. CRISPR–Cas adaptation: insights into the mechanism of action. Nat. Rev. Microbiol. 2016;14:67–76. doi: 10.1038/nrmicro.2015.14. [DOI] [PubMed] [Google Scholar]
- 206.Ma E., Harrington L.B., O'Connell M.R., Zhou K., Doudna J.A. Single-stranded DNA cleavage by divergent CRISPR-Cas9 enzymes. Mol. Cell. 2015;60:398–407. doi: 10.1016/j.molcel.2015.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Barrangou R., et al. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315:1709–1712. doi: 10.1126/science.1138140. [DOI] [PubMed] [Google Scholar]
- 208.Cong L., et al. Multiplex genome engineering using CRISPR/cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Cho S.W., Kim S., Kim J.M., Kim J.-S. Targeted genome engineering in human cells with the Cas9 RNA-guided endonuclease. Nat. Biotechnol. 2013;31:230–232. doi: 10.1038/nbt.2507. [DOI] [PubMed] [Google Scholar]
- 210.Mali P., et al. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Jinek M., et al. A programmable dual-RNA–guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Wang F., Qi L.S. Applications of CRISPR genome engineering in cell biology. Trends Cell Biol. 2016;26:875–888. doi: 10.1016/j.tcb.2016.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Xiong Y., et al. Functional DNA regulated CRISPR-cas12a sensors for point-of-care diagnostics of non-nucleic-acid targets. J. Am. Chem. Soc. 2020;142:207–213. doi: 10.1021/jacs.9b09211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Zhao X., et al. Rapid and sensitive exosome detection with CRISPR/Cas12a. Anal. Bioanal. Chem. 2020;412:601–609. doi: 10.1007/s00216-019-02211-4. [DOI] [PubMed] [Google Scholar]
- 215.Li H., et al. Applications of genome editing technology in the targeted therapy of human diseases: mechanisms, advances and prospects. Signal Transduct. Target. Ther. 2020;5:1–23. doi: 10.1038/s41392-019-0089-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Li B., Niu Y., Ji W., Dong Y. Strategies for the CRISPR-based therapeutics. Trends Pharmacol. Sci. 2020;41:55–65. doi: 10.1016/j.tips.2019.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Gootenberg J.S., et al. Nucleic acid detection with CRISPR-Cas13a/C2c2. Science. 2017;356:438–442. doi: 10.1126/science.aam9321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Aman R., Mahas A., Mahfouz M. Nucleic acid detection using CRISPR/cas biosensing technologies. ACS Synth. Biol. 2020;9:1226–1233. doi: 10.1021/acssynbio.9b00507. [DOI] [PubMed] [Google Scholar]
- 219.Wang M., Zhang R., Li J. CRISPR/cas systems redefine nucleic acid detection: principles and methods. Biosens. Bioelectron. 2020;165 doi: 10.1016/j.bios.2020.112430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Peng S., Tan Z., Chen S., Lei C., Nie Z. Integrating CRISPR-Cas12a with a DNA circuit as a generic sensing platform for amplified detection of microRNA. Chem. Sci. 2020;11:7362–7368. doi: 10.1039/d0sc03084h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Zhou W., et al. A CRISPR–Cas9-triggered strand displacement amplification method for ultrasensitive DNA detection. Nat. Commun. 2018;9:5012. doi: 10.1038/s41467-018-07324-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Huang M., Zhou X., Wang H., Xing D. Clustered regularly interspaced short palindromic repeats/cas9 triggered isothermal amplification for site-specific nucleic acid detection. Anal. Chem. 2018;90:2193–2200. doi: 10.1021/acs.analchem.7b04542. [DOI] [PubMed] [Google Scholar]
- 223.Li L., et al. HOLMESv2: a CRISPR-cas12b-assisted platform for nucleic acid detection and DNA methylation quantitation. ACS Synth. Biol. 2019;8:2228–2237. doi: 10.1021/acssynbio.9b00209. [DOI] [PubMed] [Google Scholar]
- 224.Makarova K.S., Koonin E.V. Annotation and classification of CRISPR-cas systems. Methods Mol. Biol. Clifton NJ. 2015;1311:47–75. doi: 10.1007/978-1-4939-2687-9_4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Harrington L.B., et al. Programmed DNA destruction by miniature CRISPR-Cas14 enzymes. Science. 2018;362:839–842. doi: 10.1126/science.aav4294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Jiang F., Doudna J.A. CRISPR–Cas9 structures and mechanisms. Annu. Rev. Biophys. 2017;46:505–529. doi: 10.1146/annurev-biophys-062215-010822. [DOI] [PubMed] [Google Scholar]
- 227.Cui Y., Xu J., Cheng M., Liao X., Peng S. Review of CRISPR/Cas9 sgRNA design tools. Interdiscipl. Sci. Comput. Life Sci. 2018;10:455–465. doi: 10.1007/s12539-018-0298-z. [DOI] [PubMed] [Google Scholar]
- 228.Montague T.G., Cruz J.M., Gagnon J.A., Church G.M., Valen E. CHOPCHOP: a CRISPR/Cas9 and TALEN web tool for genome editing. Nucleic Acids Res. 2014;42:W401–W407. doi: 10.1093/nar/gku410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Brazelton V.A., et al. A quick guide to CRISPR sgRNA design tools. GM Crops Food. 2016;6:266–276. doi: 10.1080/21645698.2015.1137690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Sun J., et al. CRISPR-Local: a local single-guide RNA (sgRNA) design tool for non-reference plant genomes. Bioinformatics. 2019;35:2501–2503. doi: 10.1093/bioinformatics/bty970. [DOI] [PubMed] [Google Scholar]
- 231.Palermo G., et al. Protospacer adjacent motif-induced allostery activates CRISPR-cas9. J. Am. Chem. Soc. 2017;139:16028–16031. doi: 10.1021/jacs.7b05313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Mojica F.J.M., Díez-Villaseñor C., García-Martínez J., Almendros C. Short motif sequences determine the targets of the prokaryotic CRISPR defence system. Microbiol. Read. Engl. 2009;155:733–740. doi: 10.1099/mic.0.023960-0. [DOI] [PubMed] [Google Scholar]
- 233.Wang J., et al. Structural and mechanistic basis of PAM-dependent spacer acquisition in CRISPR-cas systems. Cell. 2015;163:840–853. doi: 10.1016/j.cell.2015.10.008. [DOI] [PubMed] [Google Scholar]
- 234.Gleditzsch D., et al. PAM identification by CRISPR-Cas effector complexes: diversified mechanisms and structures. RNA Biol. 2018;16:504–517. doi: 10.1080/15476286.2018.1504546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Collias D., Beisel C.L. CRISPR technologies and the search for the PAM-free nuclease. Nat. Commun. 2021;12:555. doi: 10.1038/s41467-020-20633-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Shmakov S., et al. Discovery and functional characterization of diverse class 2 CRISPR-cas systems. Mol. Cell. 2015;60:385–397. doi: 10.1016/j.molcel.2015.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Liu T.Y., Doudna J.A. Chemistry of Class 1 CRISPR-Cas effectors: binding, editing, and regulation. J. Biol. Chem. 2020;295:14473–14487. doi: 10.1074/jbc.REV120.007034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Nk P., S C. Diverse class 2 CRISPR-cas effector proteins for genome engineering applications. ACS Chem. Biol. 2017;13:347–356. doi: 10.1021/acschembio.7b00800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Feng W., et al. CRISPR technology incorporating amplification strategies: molecular assays for nucleic acids, proteins, and small molecules. Chem. Sci. 2021 doi: 10.1039/D0SC06973F. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Nishimasu H., et al. Crystal structure of Cas9 in complex with guide RNA and target DNA. Cell. 2014;156:935–949. doi: 10.1016/j.cell.2014.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Gasiunas G., Barrangou R., Horvath P., Siksnys V. Cas9–crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc. Natl. Acad. Sci. Unit. States Am. 2012;109:E2579–E2586. doi: 10.1073/pnas.1208507109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Wang H., La Russa M., Qi L.S. CRISPR/Cas9 in genome editing and beyond. Annu. Rev. Biochem. 2016;85:227–264. doi: 10.1146/annurev-biochem-060815-014607. [DOI] [PubMed] [Google Scholar]
- 243.Ran F.A., et al. Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell. 2013;154:1380–1389. doi: 10.1016/j.cell.2013.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Qi L.S., et al. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell. 2013;152:1173–1183. doi: 10.1016/j.cell.2013.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Doi G., et al. Catalytically inactive Cas9 impairs DNA replication fork progression to induce focal genomic instability. Nucleic Acids Res. 2021;49:954–968. doi: 10.1093/nar/gkaa1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Chen X., et al. In trans paired nicking triggers seamless genome editing without double-stranded DNA cutting. Nat. Commun. 2017;8:657. doi: 10.1038/s41467-017-00687-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Eki R., et al. A robust CRISPR–Cas9-based fluorescent reporter assay for the detection and quantification of DNA double-strand break repair. Nucleic Acids Res. 2020;48 doi: 10.1093/nar/gkaa897. e126–e126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Moon J., et al. Colorimetric detection of SARS-CoV-2 and drug-resistant pH1N1 using CRISPR/dCas9. ACS Sens. 2020;5:4017–4026. doi: 10.1021/acssensors.0c01929. [DOI] [PubMed] [Google Scholar]
- 249.Wang T., Liu Y., Sun H.-H., Yin B.-C., Ye B.-C. An RNA-guided Cas9 nickase-based method for universal isothermal DNA amplification. Angew. Chem. Int. Ed. 2019;58:5382–5386. doi: 10.1002/anie.201901292. [DOI] [PubMed] [Google Scholar]
- 250.Pickar-Oliver A., Gersbach C.A. The next generation of CRISPR-Cas technologies and applications. Nat. Rev. Mol. Cell Biol. 2019;20:490–507. doi: 10.1038/s41580-019-0131-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Rahimi H., et al. CRISPR systems for COVID-19 diagnosis. ACS Sens. 2021;6:1430–1445. doi: 10.1021/acssensors.0c02312. [DOI] [PubMed] [Google Scholar]
- 252.McCarty N.S., Graham A.E., Studená L., Ledesma-Amaro R. Multiplexed CRISPR technologies for gene editing and transcriptional regulation. Nat. Commun. 2020;11:1281. doi: 10.1038/s41467-020-15053-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Ghorbani A., et al. A short overview of CRISPR-Cas technology and its application in viral disease control. Transgenic Res. 2021;30:221–238. doi: 10.1007/s11248-021-00247-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Chen J.S., et al. CRISPR-Cas12a target binding unleashes indiscriminate single-stranded DNase activity. Science. 2018;360:436–439. doi: 10.1126/science.aar6245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Freije C.A., Sabeti P.C. Detect and destroy: CRISPR-based technologies for the response against viruses. Cell Host Microbe. 2021;29:689–703. doi: 10.1016/j.chom.2021.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Aquino-Jarquin G. CRISPR-Cas14 is now part of the artillery for gene editing and molecular diagnostic. Nanomed. Nanotechnol. Biol. Med. 2019;18:428–431. doi: 10.1016/j.nano.2019.03.006. [DOI] [PubMed] [Google Scholar]
- 257.Karvelis T., et al. PAM recognition by miniature CRISPR–Cas12f nucleases triggers programmable double-stranded DNA target cleavage. Nucleic Acids Res. 2020;48:5016–5023. doi: 10.1093/nar/gkaa208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Freije C.A., et al. Programmable inhibition and detection of RNA viruses using Cas13. Mol. Cell. 2019;76:826–837. doi: 10.1016/j.molcel.2019.09.013. e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Liu L., et al. The molecular architecture for RNA-guided RNA cleavage by Cas13a. Cell. 2017;170:714–726. doi: 10.1016/j.cell.2017.06.050. e10. [DOI] [PubMed] [Google Scholar]
- 260.Fozouni P., et al. Amplification-free detection of SARS-CoV-2 with CRISPR-Cas13a and mobile phone microscopy. Cell. 2021;184:323–333. doi: 10.1016/j.cell.2020.12.001. e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Myhrvold C., et al. Field-deployable viral diagnostics using CRISPR-Cas13. Science. 2018;360:444–448. doi: 10.1126/science.aas8836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Ding X., et al. Ultrasensitive and visual detection of SARS-CoV-2 using all-in-one dual CRISPR-Cas12a assay. Nat. Commun. 2020;11:4711. doi: 10.1038/s41467-020-18575-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Huang Z., et al. Ultra-sensitive and high-throughput CRISPR-p owered COVID-19 diagnosis. Biosens. Bioelectron. 2020;164 doi: 10.1016/j.bios.2020.112316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Wang B., et al. Cas12aVDet: a CRISPR/cas12a-based platform for rapid and visual nucleic acid detection. Anal. Chem. 2019;91:12156–12161. doi: 10.1021/acs.analchem.9b01526. [DOI] [PubMed] [Google Scholar]
- 265.Lee R.A., et al. Ultrasensitive CRISPR-based diagnostic for field-applicable detection of Plasmodium species in symptomatic and asymptomatic malaria. Proc. Natl. Acad. Sci. Unit. States Am. 2020;117:25722–25731. doi: 10.1073/pnas.2010196117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Yuan T., et al. A rapid and sensitive CRISPR/Cas12a based lateral flow biosensor for the detection of Epstein–Barr virus. Analyst. 2020;145:6388–6394. doi: 10.1039/d0an00663g. [DOI] [PubMed] [Google Scholar]
- 267.Kellner M.J., Koob J.G., Gootenberg J.S., Abudayyeh O.O., Zhang F. SHERLOCK: nucleic acid detection with CRISPR nucleases. Nat. Protoc. 2019;14:2986–3012. doi: 10.1038/s41596-019-0210-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.van Dongen J.E., et al. Point-of-care CRISPR/Cas nucleic acid detection: recent advances, challenges and opportunities. Biosens. Bioelectron. 2020;166 doi: 10.1016/j.bios.2020.112445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Gootenberg J.S., et al. Multiplexed and portable nucleic acid detection platform with Cas13, Cas12a, and Csm6. Science. 2018;360:439–444. doi: 10.1126/science.aaq0179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Broughton J.P., et al. CRISPR–Cas12-based detection of SARS-CoV-2. Nat. Biotechnol. 2020;38:870–874. doi: 10.1038/s41587-020-0513-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Li Y., Li S., Wang J., Liu G. CRISPR/Cas systems towards next-generation biosensing. Trends Biotechnol. 2019;37:730–743. doi: 10.1016/j.tibtech.2018.12.005. [DOI] [PubMed] [Google Scholar]
- 272.Nguyen L.T., Smith B.M., Jain P.K. Enhancement of trans-cleavage activity of Cas12a with engineered crRNA enables amplified nucleic acid detection. Nat. Commun. 2020;11:4906. doi: 10.1038/s41467-020-18615-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Tran M.H., et al. A more efficient CRISPR-Cas12a variant derived from Lachnospiraceae bacterium MA2020. Mol. Ther. Nucleic Acids. 2021;24:40–53. doi: 10.1016/j.omtn.2021.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Gao D.S., Zhu X., Lu B. Development and application of sensitive, specific, and rapid CRISPR-Cas13-based diagnosis. J. Med. Virol. 2021;93:4198–4204. doi: 10.1002/jmv.26889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Joung J., et al. Point-of-care testing for COVID-19 using SHERLOCK diagnostics. medRxiv. 2020 doi: 10.1101/2020.05.04.20091231. [DOI] [Google Scholar]
- 276.Ali Z., et al. iSCAN: an RT-LAMP-coupled CRISPR-Cas12 module for rapid, sensitive detection of SARS-CoV-2. Virus Res. 2020;288 doi: 10.1016/j.virusres.2020.198129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Curtis K.A., Rudolph D.L., Owen S.M. Sequence-specific detection method for reverse transcription, loop-mediated isothermal amplification of HIV-1. J. Med. Virol. 2009;81:966–972. doi: 10.1002/jmv.21490. [DOI] [PubMed] [Google Scholar]
- 278.Bao Y., et al. CUT-LAMP: contamination-free loop-mediated isothermal amplification based on the CRISPR/Cas9 cleavage. ACS Sens. 2020;5:1082–1091. doi: 10.1021/acssensors.0c00034. [DOI] [PubMed] [Google Scholar]
- 279.Wang R., et al. opvCRISPR: one-pot visual RT-LAMP-CRISPR platform for SARS-cov-2 detection. Biosens. Bioelectron. 2021;172 doi: 10.1016/j.bios.2020.112766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Zhao Y., Chen F., Li Q., Wang L., Fan C. Isothermal amplification of nucleic acids. Chem. Rev. 2015;115:12491–12545. doi: 10.1021/acs.chemrev.5b00428. [DOI] [PubMed] [Google Scholar]
- 281.Lutz S., et al. Microfluidic lab-on-a-foil for nucleic acid analysis based on isothermal recombinase polymerase amplification (RPA) Lab Chip. 2010;10:887–893. doi: 10.1039/b921140c. [DOI] [PubMed] [Google Scholar]
- 282.Hou T., et al. Development and evaluation of a rapid CRISPR-based diagnostic for COVID-19. PLoS Pathog. 2020;16 doi: 10.1371/journal.ppat.1008705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Patchsung M., et al. Clinical validation of a Cas13-based assay for the detection of SARS-CoV-2 RNA. Nat. Biomed. Eng. 2020;4:1140–1149. doi: 10.1038/s41551-020-00603-x. [DOI] [PubMed] [Google Scholar]
- 284.Wang R., et al. Rolling circular amplification (RCA)-Assisted CRISPR/Cas9 cleavage (RACE) for highly specific detection of multiple extracellular vesicle MicroRNAs. Anal. Chem. 2020;92:2176–2185. doi: 10.1021/acs.analchem.9b04814. [DOI] [PubMed] [Google Scholar]
- 285.Tian B., Minero G.A.S., Fock J., Dufva M., Hansen M.F. CRISPR-Cas12a based internal negative control for nonspecific products of exponential rolling circle amplification. Nucleic Acids Res. 2020;48 doi: 10.1093/nar/gkaa017. e30–e30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Pardee K., et al. Rapid, low-cost detection of Zika virus using programmable biomolecular components. Cell. 2016;165:1255–1266. doi: 10.1016/j.cell.2016.04.059. [DOI] [PubMed] [Google Scholar]
- 287.Kirkconnell B., et al. Design and validation of transcription-mediated-amplification nucleic acid amplification tests for mycoplasma genitalium. J. Clin. Microbiol. 2019;57 doi: 10.1128/JCM.00264-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288.Li S.-Y., et al. CRISPR-Cas12a-assisted nucleic acid detection. Cell Discov. 2018;4 doi: 10.1038/s41421-018-0028-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289.Ganbaatar U., Liu C. CRISPR-based COVID-19 testing: toward next-generation point-of-care diagnostics. Front. Cell. Infect. Microbiol. 2021;11 doi: 10.3389/fcimb.2021.663949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Song Q., et al. Identification of human metapneumovirus genotypes A and B from clinical specimens by reverse transcription loop-mediated isothermal amplification. J. Virol. Methods. 2014;196:133–138. doi: 10.1016/j.jviromet.2013.10.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291.EIKEN CHEMICAL CO.,LTD; 2021. LoopampTM SARS-CoV-2 Detection Kit | Molecular Test “LAMP” | Products.https://www.eiken.co.jp/en/products/lamp/covid19test/ [Google Scholar]
- 292.AQ-TOP COVID-19 Rapid Detection Kit. https://www.fda.gov/media/138307/download
- 293.Detectachem MobileDetect-BIO BCC19 Test Kit for SARS-CoV-2 Detection. https://www.fda.gov/media/141791/download
- 294.FDA. Emergency Use Authorization (EUA) Summary for Mammoth Biosciences SARS-CoV-2 RNA DETECTR Assay. https://www.fda.gov/media/139937/download
- 295.FDA. Emergnecy Use Authorization (EUA) Summary for Sherlock Biosciences CRISPR SARS-CoV-2 Kit. https://www.fda.gov/media/137746/download
- 296.Lampore COVID-19. http://oxfordnanoporedx.com/products/lampore-covid-19 Oxford Nanopore Technologies.
- 297.Aptima® SARS-CoV-2 Assay (Panther® System) https://www.fda.gov/media/138096/download
- 298.Cordes A.K., et al. Fully automated detection and differentiation of pandemic and endemic coronaviruses (NL63, 229E, HKU1, OC43 and SARS-CoV-2) on the Hologic Panther Fusion. medRxiv. 2020;8(31) doi: 10.1101/2020.08.31.20185074. 2020. [DOI] [PubMed] [Google Scholar]
- 299.Gorzalski A.J., et al. High-Throughput Transcription-mediated amplification on the Hologic Panther is a highly sensitive method of detection for SARS-CoV-2. J. Clin. Virol. 2020;129 doi: 10.1016/j.jcv.2020.104501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 300.Kanwar N., Michael J., Doran K., Montgomery E., Selvarangan R. Comparison of the id now influenza A & B 2, cobas influenza A/B, and xpert xpress flu point-of-care nucleic acid amplification tests for influenza A/B virus detection in children. J. Clin. Microbiol. 2020;58 doi: 10.1128/JCM.01611-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 301.Nie S., et al. Evaluation of Alere i Influenza A&B for rapid detection of influenza viruses A and B. J. Clin. Microbiol. 2014;52:3339–3344. doi: 10.1128/JCM.01132-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 302.Wang H., Deng J., Tang Y.-W. Profile of the Alere i Influenza A & B assay: a pioneering molecular point-of-care test. Expert Rev. Mol. Diagn. 2018;18:403–409. doi: 10.1080/14737159.2018.1466703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 303.Abbott I.D. NOW COVID-19 Test. https://www.fda.gov/media/136525/download
- 304.FDA. Emergnecy Use Authorization (EUA) Lucira COVID-19 All-In-One Test Kit. https://www.fda.gov/media/143808/download
- 305.Real-time COVID-19 Testing | Cue. https://cuehealth.com/products/how-cue-detects-covid-19/
- 306.Park G.-S., et al. Development of reverse transcription loop-mediated isothermal amplification assays targeting severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) J. Mol. Diagn. JMD. 2020;22:729–735. doi: 10.1016/j.jmoldx.2020.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 307.Chow F.W.-N., et al. A rapid, simple, inexpensive, and mobile colorimetric assay COVID-19-LAMP for mass on-site screening of COVID-19. Int. J. Mol. Sci. 2020;21 doi: 10.3390/ijms21155380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 308.Arizti-Sanz J., et al. Integrated sample inactivation, amplification, and Cas13-based detection of SARS-CoV-2. bioRxiv. 2020 doi: 10.1101/2020.05.28.119131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 309.Kemal K.S., Reinis M., Weiser B., Burger H. Methods for viral RNA isolation and PCR amplification for sequencing of near full-length HIV-1 genomes. Methods Mol. Biol. Clifton NJ. 2009;485:3–14. doi: 10.1007/978-1-59745-170-3_1. [DOI] [PubMed] [Google Scholar]
- 310.Lübke N., et al. Extraction-free SARS-CoV-2 detection by rapid RT-qPCR universal for all primary respiratory materials. J. Clin. Virol. 2020;130 doi: 10.1016/j.jcv.2020.104579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 311.Wee S.K., Sivalingam S.P., Yap E.P.H. Rapid direct nucleic acid amplification test without RNA extraction for SARS-CoV-2 using a portable PCR thermocycler. Genes. 2020;11 doi: 10.3390/genes11060664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 312.Bruce E.A., et al. DIRECT RT-qPCR DETECTION OF SARS-CoV-2 RNA FROM PATIENT NASOPHARYNGEAL SWABS WITHOUT AN RNA EXTRACTION STEP. bioRxiv. 2020;3(20) doi: 10.1101/2020.03.20.001008. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 313.Smyrlaki I., et al. Massive and rapid COVID-19 testing is feasible by extraction-free SARS-CoV-2 RT-PCR. medRxiv. 2020;4(17) doi: 10.1101/2020.04.17.20067348. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 314.Lee D., et al. Simple and highly sensitive molecular diagnosis of Zika virus by lateral flow assays. Anal. Chem. 2016;88:12272–12278. doi: 10.1021/acs.analchem.6b03460. [DOI] [PubMed] [Google Scholar]
- 315.Rabe B.A., Cepko C. SARS-CoV-2 detection using isothermal amplification and a rapid, inexpensive protocol for sample inactivation and purification. Proc. Natl. Acad. Sci. Unit. States Am. 2020;117:24450–24458. doi: 10.1073/pnas.2011221117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 316.Anahtar M.N., et al. Clinical assessment and validation of a rapid and sensitive SARS-CoV-2 test using reverse transcription loop-mediated isothermal amplification without the need for RNA extraction. Open Forum Infect. Dis. 2021;8 doi: 10.1093/ofid/ofaa631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 317.Ben-Assa N., et al. Direct on-the-spot detection of SARS-CoV-2 in patients. Exp. Biol. Med. 2020 doi: 10.1177/1535370220941819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 318.Wyllie A.L., et al. Saliva or nasopharyngeal swab specimens for detection of SARS-CoV-2. N. Engl. J. Med. 2020;383:1283–1286. doi: 10.1056/NEJMc2016359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 319.Vogels C.B.F., et al. SalivaDirect: a simplified and flexible platform to enhance SARS-CoV-2 testing capacity. Med. 2021;2:263–280. doi: 10.1016/j.medj.2020.12.010. e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 320.Cameron S.J.S., Huws S.A., Hegarty M.J., Smith D.P.M., Mur L.A.J. The human salivary microbiome exhibits temporal stability in bacterial diversity. FEMS Microbiol. Ecol. 2015;91 doi: 10.1093/femsec/fiv091. [DOI] [PubMed] [Google Scholar]
- 321.Yang Q., et al. Saliva TwoStep for rapid detection of asymptomatic SARS-CoV-2 carriers. medRxiv. 2020;7(16) doi: 10.1101/2020.07.16.20150250. 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 322.Kaarj K., Akarapipad P., Yoon J.-Y. Simpler, faster, and sensitive Zika virus assay using smartphone detection of loop-mediated isothermal amplification on paper microfluidic chips. Sci. Rep. 2018;8:12438. doi: 10.1038/s41598-018-30797-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 323.Yao Y., et al. Rapid detection of influenza virus subtypes based on an integrated centrifugal disc. ACS Sens. 2020;5:1354–1362. doi: 10.1021/acssensors.9b02595. [DOI] [PubMed] [Google Scholar]
- 324.de Puig H., Bosch I., Collins J.J., Gehrke L. Point-of-Care devices to detect Zika and other emerging viruses. Annu. Rev. Biomed. Eng. 2020;22:371–386. doi: 10.1146/annurev-bioeng-060418-052240. [DOI] [PubMed] [Google Scholar]
- 325.García-Bernalt Diego J., et al. Progress in loop-mediated isothermal amplification assay for detection of Schistosoma mansoni DNA: towards a ready-to-use test. Sci. Rep. 2019;9:14744. doi: 10.1038/s41598-019-51342-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 326.Yue H., et al. The epidemiology and clinical characteristics of co-infection of SARS-CoV-2 and influenza viruses in patients during COVID-19 outbreak. J. Med. Virol. 2020 doi: 10.1002/jmv.26163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 327.Khamrin P., et al. A single-tube multiplex PCR for rapid detection in feces of 10 viruses causing diarrhea. J. Virol. Methods. 2011;173:390–393. doi: 10.1016/j.jviromet.2011.02.012. [DOI] [PubMed] [Google Scholar]
- 328.Elnifro E.M., Ashshi A.M., Cooper R.J., Klapper P.E. Multiplex PCR: optimization and application in diagnostic virology. Clin. Microbiol. Rev. 2000;13:559–570. doi: 10.1128/cmr.13.4.559-570.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 329.Pham N.T.K., et al. A novel RT-multiplex PCR for detection of Aichi virus, human parechovirus, enteroviruses, and human bocavirus among infants and children with acute gastroenteritis. J. Virol. Methods. 2010;169:193–197. doi: 10.1016/j.jviromet.2010.07.038. [DOI] [PubMed] [Google Scholar]
- 330.Nguyen T.T., et al. Multiplex nested RT-PCR for detecting avian influenza virus, infectious bronchitis virus and Newcastle disease virus. J. Virol. Methods. 2013;188:41–46. doi: 10.1016/j.jviromet.2012.12.004. [DOI] [PubMed] [Google Scholar]
- 331.Liu P., et al. A novel multiplex PCR for virus detection by melting curve analysis. J. Virol. Methods. 2018;262:56–60. doi: 10.1016/j.jviromet.2018.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 332.CDC. Information for laboratories about coronavirus (COVID-19) Centers for Disease Control and Prevention. 2020 https://www.cdc.gov/coronavirus/2019-ncov/lab/multiplex.html [Google Scholar]
- 333.Bruch R., Urban G.A., Dincer C. CRISPR/Cas powered multiplexed biosensing. Trends Biotechnol. 2019;37:791–792. doi: 10.1016/j.tibtech.2019.04.005. [DOI] [PubMed] [Google Scholar]
- 334.Tanner N.A., Zhang Y., Evans T.C. Simultaneous multiple target detection in real-time loop-mediated isothermal amplification. Biotechniques. 2012;53:81–89. doi: 10.2144/0000113902. [DOI] [PubMed] [Google Scholar]
- 335.Zhang Y., Tanner N.A. Development of multiplexed reverse-transcription loop-mediated isothermal amplification for detection of SARS-CoV-2 and influenza viral RNA. Biotechniques. 2021 doi: 10.2144/btn-2020-0157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 336.Ball C.S., et al. Quenching of unincorporated amplification signal reporters in reverse-transcription loop-mediated isothermal amplification enabling bright, single-step, closed-tube, and multiplexed detection of RNA viruses. Anal. Chem. 2016;88:3562–3568. doi: 10.1021/acs.analchem.5b04054. [DOI] [PubMed] [Google Scholar]
- 337.Tyagi S., Kramer F.R. Molecular beacons: probes that fluoresce upon hybridization. Nat. Biotechnol. 1996;14:303–308. doi: 10.1038/nbt0396-303. [DOI] [PubMed] [Google Scholar]
- 338.Sherrill-Mix S., et al. Detection of SARS-CoV-2 RNA using RT-LAMP and molecular beacons. Genome Biol. 2021;22:169. doi: 10.1186/s13059-021-02387-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 339.Shao N., Han X., Song Y., Zhang P., Qin L. CRISPR-Cas12a coupled with platinum nanoreporter for visual quantification of SNVs on a volumetric bar-chart chip. Anal. Chem. 2019;91:12384–12391. doi: 10.1021/acs.analchem.9b02925. [DOI] [PubMed] [Google Scholar]
- 340.Hogan C.A., Sahoo M.K., Pinsky B.A. Sample pooling as a strategy to detect community transmission of SARS-CoV-2. J. Am. Med. Assoc. 2020;323:1967–1969. doi: 10.1001/jama.2020.5445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 341.Barak N., et al. Lessons from applied large-scale pooling of 133,816 SARS-CoV-2 RT-PCR tests. Sci. Transl. Med. 2021;13 doi: 10.1126/scitranslmed.abf2823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 342.Hu J., et al. Emerging SARS-CoV-2 variants reduce neutralization sensitivity to convalescent sera and monoclonal antibodies. Cell. Mol. Immunol. 2021;18:1061–1063. doi: 10.1038/s41423-021-00648-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 343.Tracking SARS-CoV-2 Variants. https://www.who.int/activities/tracking-SARS-CoV-2-variants
- 344.McCallum M., et al. Molecular basis of immune evasion by the delta and kappa SARS-CoV-2 variants. 2021. https://www.biorxiv.org/content/10.1101/2021.08.11.455956v2 (2021) 8,11. [DOI] [PMC free article] [PubMed]
- 345.Li F. Structure, function, and evolution of coronavirus spike proteins. Annu. Rev. Virol. 2016;3:237–261. doi: 10.1146/annurev-virology-110615-042301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 346.Harvey W.T., et al. SARS-CoV-2 variants, spike mutations and immune escape. Nat. Rev. Microbiol. 2021;19:409–424. doi: 10.1038/s41579-021-00573-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 347.Planas D., et al. Reduced sensitivity of SARS-CoV-2 variant Delta to antibody neutralization. Nature. 2021;596:276–280. doi: 10.1038/s41586-021-03777-9. [DOI] [PubMed] [Google Scholar]
- 348.Xia S., et al. The role of furin cleavage site in SARS-CoV-2 spike protein-mediated membrane fusion in the presence or absence of trypsin. Signal Transduct. Target. Ther. 2020;5:1–3. doi: 10.1038/s41392-020-0184-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 349.How Dangerous Is the Delta Variant (B.1.617.2)? https://asm.org/Articles/2021/July/How-Dangerous-is-the-Delta-Variant-B-1-617-2 ASM.org.
- 350.Health C., for D. 2021. R. SARS-CoV-2 Viral Mutations: Impact on COVID-19 Tests.https://www.fda.gov/medical-devices/coronavirus-covid-19-and-medical-devices/sars-cov-2-viral-mutations-impact-covid-19-tests FDA. [Google Scholar]
- 351.Guerra-Assunção J.A., et al. 2021. Reliability of Spike Gene Target Failure for Ascertaining SARS-CoV-2 Lineage B.1.1.7 Prevalence in a Hospital Setting.https://www.medrxiv.org/content/10.1101/2021.04.12.21255084v1 (2021) 04.12.21255084. [DOI] [Google Scholar]
- 352.Shen S., et al. 2021. A Rapid SARS-CoV-2 Variant Detection by Molecular-Clamping Based RT-qPCR.https://www.medrxiv.org/content/10.1101/2021.04.01.21254484v1 (2021) 04.01. [DOI] [Google Scholar]