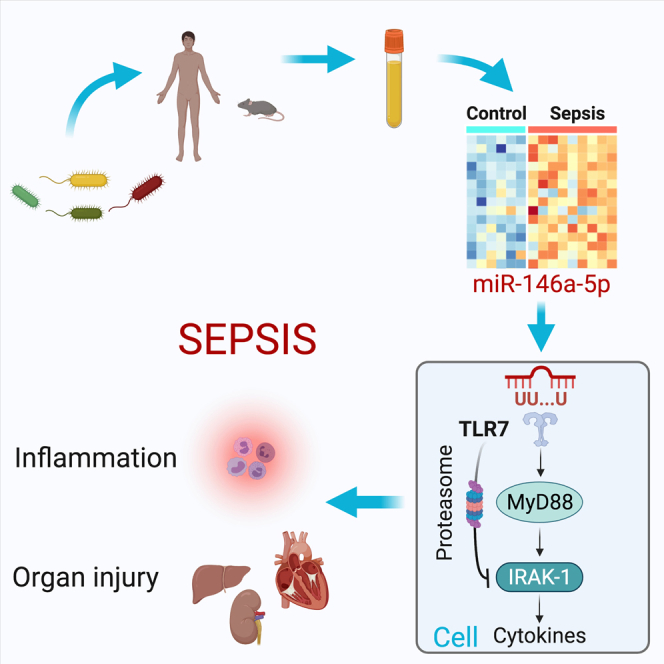
Summary
Extracellular miRNAs (ex-miRNAs) mediate intercellular communication and play a role in diverse physiological and pathological processes. Using small RNA sequencing, we identify that miRNAs are the most abundant RNA species in the plasma and differentially expressed in murine and human sepsis, such as miR-146a-5p. Exogenous miR-146a-5p, but not its duplex precursor, induces a strong immunostimulatory response through a newly identified UU-containing motif and TLR7 activation, and an immunotolerance by rapid IRAK-1 protein degradation via TLR7→MyD88 signaling and proteasome activation, whereas its duplex precursor acts by targeting 3′ UTR of Irak-1 gene via Ago2 binding. miR-146a knockout in mice offers protection against sepsis with attenuated interleukin-6 (IL-6) storm and organ injury, improved cardiac function, and better survival. In septic patients, the plasma miR-146a-5p concentrations are closely associated with the two sepsis outcome predictors, blood lactate and coagulopathy. These data demonstrate the importance of extracellular miR-146a-5p in innate immune regulation and sepsis pathogenesis.
Subject areas: Molecular biology, Immunology, Cell biology
Graphical abstract
Highlights
-
•
miRNAs are the predominant RNA biotype in the plasma and markedly altered in sepsis
-
•
Ex-miR-146a-5p stimulates innate immune response via a UU-motif and TLR7 activation
-
•
Ex-miR-146a-5p downregulates IRAK-1 protein through TLR7 and proteasome activation
-
•
Plasma miR-146a-5p is associated with sepsis predictors and plays a role in sepsis
Molecular biology; Immunology; Cell biology
Introduction
Sepsis is a critical condition induced by a dysregulated host response to infection (Singer et al., 2016). It is characterized by profound systemic inflammation, coagulopathy, cardiovascular collapse, and multiorgan failure. Despite decades of intensive research, sepsis remains a leading cause of the in-hospital mortality with few specific treatments (Angus and Poll, 2013; Rhee et al., 2017). Preclinical animal studies have established that dysregulated host innate immune responses play a pivotal role in sepsis pathogenesis (Angus and Poll, 2013). The pattern recognition receptors (PRRs) such as Toll-like receptors (TLRs) (Kawai and Akira, 2010) are an essential part of the innate immunity and recognize pathogen-associated molecular patterns (PAMPs) (Akira and Takeda, 2004), such as bacterial lipopolysaccharide, lipoproteins, and viral nucleic acids. Although these PAMPs are well known for their roles in triggering host immune response via various TLRs after infection (Chen et al., 2021), the contribution of endogenous danger-associated molecular patterns (DAMPs) released from host cells, in particular nucleic acids, in body's innate immune response and in sepsis pathogenesis remains poorly understood.
MicroRNAs (miRNAs) are evolutionarily conserved small noncoding RNAs that function within the cell to repress gene expression by binding to the 3′ UTR of target mRNAs (Ambros, 2001). In miRNA biogenesis, a primary miRNA (pri-miRNA) is transcribed from the gene locus and processed into a precursor miRNA (pre-miRNA) hairpin by the ribonuclease III (RNase III) enzyme Drosha in the nucleus (Denli et al., 2004). Pre-miRNA is then exported to cytoplasm and further cleaved by another RNase III, Dicer, to generate a double-stranded (ds) miRNA duplex (Denli et al., 2004; Zhang et al., 2004). Once formed, the ds miRNA duplex is loaded onto an Argonaute protein (Ago). Subsequently, the passenger strand of the duplex is expulsed and degraded, whereas the dominant guide strand forms the RNA-induced silencing complex (RISC) with Ago2, the effector protein of the RISC (Yoda et al., 2010), which subsequently binds to 3′ UTR of targeted genes.
miRNAs also widely exist in extracellular (ex) space including blood circulation and other body fluids (Turchinovich et al., 2016). These cell-free miRNAs are packaged into and carried by various plasma macromolecular complexes, including EVs (Ying et al., 2017), high-density lipoprotein (HDL) (Vickers et al., 2011), and Ago2 (Arroyo et al., 2011). These ex-miRNAs can act on the recipient cells in a paracrine/endocrine fashion and play a role in various pathological conditions (Turchinovich et al., 2016), such as cancer metastasis (Fabbri et al., 2012), neurodegeneration (Lehmann et al., 2012), and pain (Park et al., 2014). We and others have shown that host cellular RNAs are released into the blood during certain critical illness such as sepsis and ischemic injury in animals (Chen et al., 2014; Feng et al., 2015, 2017; Zou et al., 2016) and humans (Caserta et al., 2016). In cell-based in vitro systems, exogenous miRNAs can stimulate production of complement factor B (Zou et al., 2016), cytokines (Feng et al., 2017), and procoagulant tissue factor (Williams et al., 2019). However, how extracellular miRNAs regulate innate immunity and their role in sepsis pathogenesis remain unclear.
In this study, using small RNA sequencing, we profiled plasma miRNAs in mice and humans and discovered distinct miRNA expression patterns in septic animals and patients as compared with their healthy controls. We identified miR-146a-5p as one of the most abundant plasma miRNAs upregulated in both septic mice and humans. We tested the role of ex-miR-146a in inflammation in vitro and in vivo and delineated the underlying molecular mechanism by which miR-146a-5p activates TLR7 and regulates cellular IRAK-1, an enzyme critical for TLR signaling. Finally, we tested the contributory role of miR-146a-5p in the pathogenesis of sepsis and its association with clinical manifestations of septic patients.
Results
Extracellular miRNAs are the predominant RNA biotype in the plasma and markedly altered in septic animals and humans
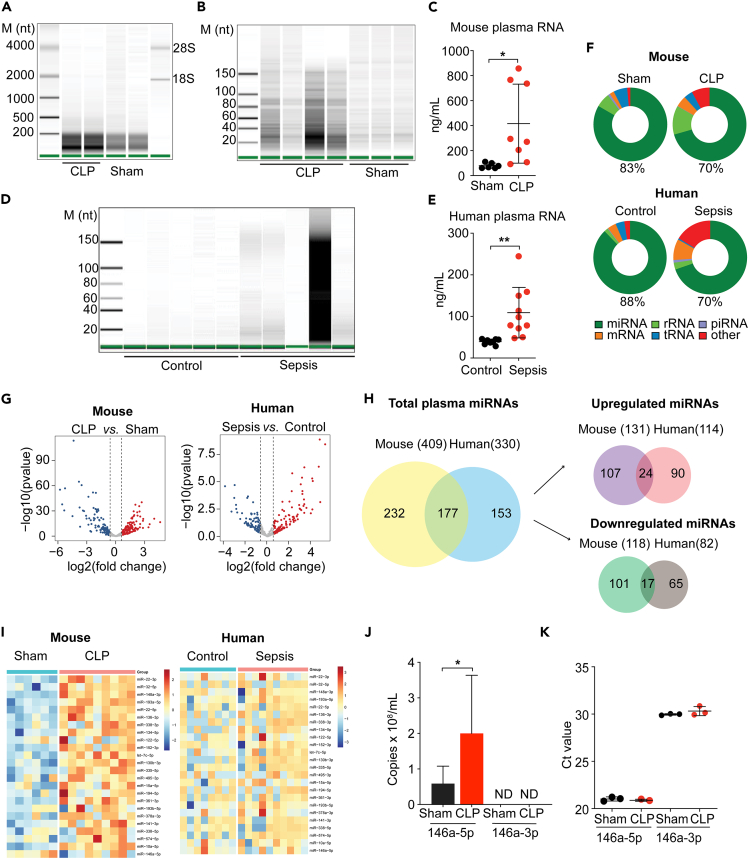
To characterize plasma RNAs in health and sepsis, we first performed a chip-based capillary electrophoresis analysis on the plasma RNAs isolated from healthy and septic mice and humans. We found that the majority of plasma RNA was between 10 and 200 nt in size (Figures 1A, 1B, 1C, 1D, and 1E). There was a more than 5-fold increase in total plasma RNA concentration in septic mice (created with cecal ligation and puncture, CLP) compared with sham mice (Figure 1C). Similarly, septic patients with abdominal infection and the Sequential Organ Failure Assessment (SOFA) scores between 7 and 12 exhibited coagulopathy, cytokine storm, and 2.7-fold increase in total plasma RNA concentration as compared with age- and gender-matched healthy volunteers (Table S1; Figure 1E). To profile the circulating RNA species, we performed small RNAseq on plasma RNAs isolated from septic mice and patients as well as age-matched healthy controls. Results indicated that miRNAs were the most abundant RNA species in the plasma, representing 83% and 88% of total reads in sham mice and healthy human controls, respectively, and 70% in both septic mice and humans (Figure 1F). Of the 1,978 known mouse miRNAs (miRbase release 22), 409 miRNAs with ≥5 raw reads were detected in the mouse plasma, among which 131 were upregulated and 118 downregulated by ≥1.5-fold in the CLP group compared with the sham group as illustrated in the volcano plot (Figure 1G) and detailed in the heatmaps (Figure S1) and Venn diagrams (Figure 1H). In humans, a total of 330 plasma miRNAs were detected, 177 (53.6%) of which shared with mouse miRNAs, 114 upregulated, and 82 downregulated by ≥1.5-fold as compared with the healthy human controls (Figure 1H). Of note, a total of 24 miRNAs were upregulated in and shared by both septic mice and humans as illustrated in Figures 1H and 1I. Among the top ten most abundant plasma miRNAs, the top four differentially expressed miRNAs, namely miR-146a-5p, miR-10a-5p, miR-22-3p, and miR-122-5p, were shared by septic mice and humans. Based on the reported functional annotation in Table S2, miR-146a-5p plays a role in regulation of hematopoiesis and immune function by targeting Irak-1 and Traf-6 genes (Boldin et al., 2011; Park et al., 2015; Taganov et al., 2006); miR-22-3p and miR-122-5p regulate multiple genes involved in cancer metastasis, cell development, and metabolism among other functions; miR-10a-5p is involved in cell differentiation, apoptosis, and wnt signaling. Given its potential role in immune regulation, we further quantified plasma miR-146a-5p, the dominant strand of miR-146a, and found it was significantly upregulated in the plasma of septic mice (Figure 1J). In contrast, miR-146a-3p, the passenger strand of miR-146a, was undetectable in the plasma and only detected at much lower levels in the liver (Figure 1K; high Ct value = low expression).
Figure 1.
Plasma RNA profiling in septic mice and humans
(A and B) Capillary electrophoresis analysis of mouse plasma RNA.
(C) Plasma RNA concentrations in sham and septic mice.
(D) Capillary electrophoresis analysis of human plasma RNA.
(E) Plasma RNA concentrations in healthy and septic humans.
(F) Pie charts illustrating various plasma RNA biotypes identified by RNAseq in mice and humans.
(G) Volcano plots of differentially expressed miRNAs between CLP versus Sham (Mouse) and Sepsis versus Control (Human).
(H) Venn diagrams of the number of total plasma miRNAs, miRNAs upregulated and downregulated in sepsis, and miRNAs shared by mice and humans.
(I) Heatmaps of the 24 upregulated plasma miRNA shared by septic mice and humans.
(J) qRT-PCR analysis of plasma miR-146a-5p (Sham: n = 6, CLP: n = 8) and miR-146a-3p (Sham: n = 5, CLP: n = 5) in Sham and CLP mice. ND = not detectable.
(K) Expression of miR-146a-5p and miR-146a-3p in the liver.
∗p < 0.05, ∗∗p < 0.01, Welch t test.
Data are represented as mean ± SD.
See also Table S1 for the demographic and laboratory data of septic patients and healthy controls and Figure S1 for the heatmap showing differential expression of the entire panels of plasma miRNAs between control and sepsis in mice and humans.
Ex miR-146a-5p, but not its ds duplex precursor, induces innate immune inflammation in vivo
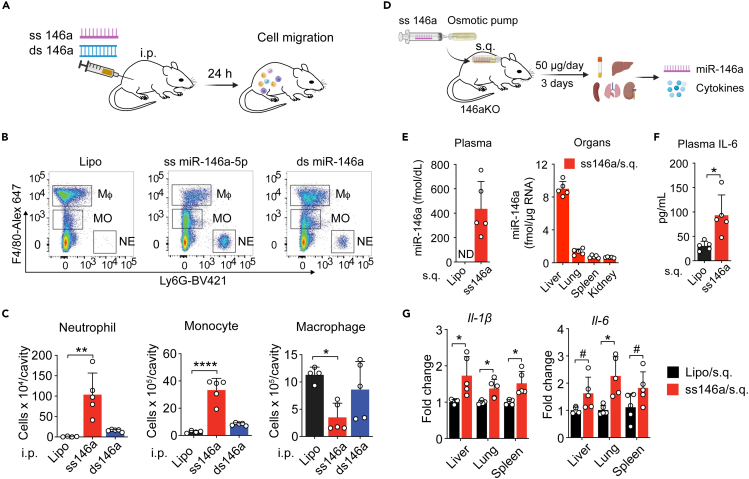
Twenty-four hours after i.p. injection (Figure 2A), animals treated with single-stranded (ss) miR146a-5p exhibited a marked increase in Ly6G+ neutrophils, and F4/80lo monocytes in the peritoneal cavity, as well as a decrease in peritoneal F4/80hi resident macrophages. In contrast, mice treated with the double-stranded (ds) miR-146a duplex precursor had minimal peritonitis, with the number of peritoneal neutrophils, monocytes, and macrophages similar to that of vehicle-injected mice (gating strategy—Figure S2A; Figures 2B and 2C). Systemically, when delivered via subcutaneously implanted osmotic pumps (50 μg/day/mouse) to miR146a-KO mice (Figure 2D), miR-146a-5p administration resulted in elevated miR-146a-5p expression in the plasma and various organs (Figure 2E) and induced a significant increase in plasma interleukin-6 (IL-6) (Figure 2F) and IL-1β and IL-6 gene expression in the liver, lung, and spleen (Figure 2G).
Figure 2.
Single-stranded (ss) miR-146a-5p induces local and systemic inflammation in vivo
(A) Experimental diagram of ss-miR-146a-5p (20 μg/mouse) and ds miR-146a duplex (20 μg/mouse) i.p. injection.
(B) Representative flow cytometry plots of residential macrophages (Mφ, CD45+F4/80highLy6G−), monocytes (MO, CD45+F4/80lowLy6G−), and neutrophils (NE, CD45+F4/80-Ly6G+) 18 h after peritoneal injection (i.p.) of lipofectamine, ss-miR-146a-5p, or ds miR-146a duplex. See also Figure S2A for the gating strategy.
(C) Numbers of total leukocytes, neutrophils, monocytes, and macrophages in the abdominal cavity 24 h after miR-146a or vehicle administration.
(D) Experimental diagram of osmatic pump delivery of miR-146a-5p (50 μg/mouse/day x 3 days).
(E) Plasma and various organ miR-146a-5p levels in miR-146a−/− mice following the osmatic pump delivery of miR-146a-5p or vehicle alone as quantified by qRT-PCR.
(F) Plasma IL-6 levels as determined by ELISA from the same mice as in E.
(G) IL-1β and IL-6 gene levels in the various organs as determined by qRT-PCR.
∗p < 0.05, ∗∗p < 0.01, ∗∗∗∗p < 0.0001, #p = 0.07, Welch t test.
Data are represented as mean ± SD.
Ex miR-146a-5p activates innate immune response via its U12U13-containing motif and direct association with TLR7
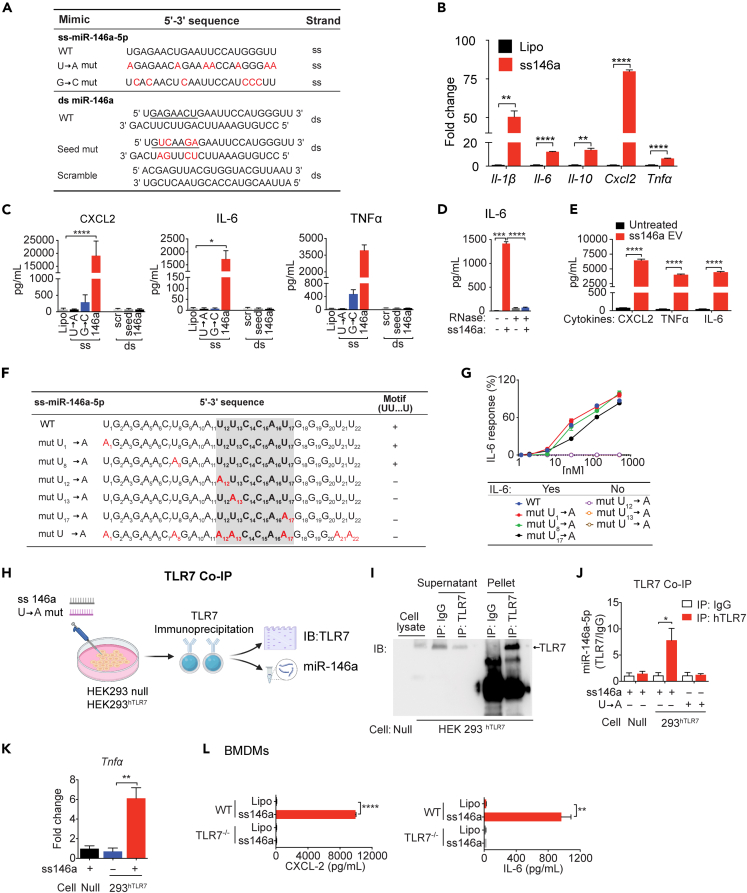
We performed a series of experiments to determine the molecular mechanism by which ss miR-146a-5p induces innate immune activation in a cell-based system. We treated mouse bone-marrow-derived macrophages (BMDMs) with ss miR-146a-5p, ds miR-146a duplex precursor, or various mutants as listed in Figure 3A. ss-miR-146a-5p led to a marked expression of several cytokine genes, i.e., Il-1β, Il-6, Il-10, Cxcl2, and Tnfα (Figure 3B), and proteins, i.e., CXCL2, IL-6, and tumor necrosis factor alpha (TNFα) (Figure 3C). On the other hand, the U→A or G→C mutant of ss miR-146a-5p failed to stimulate cytokine production (Figure 3C), demonstrating the importance of uridine and guanosine. In contrast, the duplex precursor, its seed region mutant, or a scrambled control failed to exhibit any stimulatory effect on cytokine production (Figure 3C). RNase A treatment completely abolished the effect of miR-146a-5p (Figure 3D). Extracellular vesicles (EVs) are natural carriers for plasma miRNAs (Thery et al., 2002). Our recent work has revealed that miR-146a-5p expression is upregulated in septic plasma EVs and may contribute to the proinflammatory effect of septic EVs (Kronstadt et al., 2021; Xu et al., 2018). Indeed, HEK293T EVs loaded ss miR-146a-5p mimics induced a robust CXCL2, TNFα, and IL-6 production in BMDMs (Figure 3E). Together, these data suggest that ss miR-146a-5p, not its ds miR-146a precursor, is a strong innate immune activator inducing multiple cytokine production in a U/G-dependent manner.
Figure 3.
Single-stranded (ss) miR-146a-5p induces proinflammatory cytokine production via its specific motif and TLR7 binding
(A) Nucleotide sequences of ss-miR-146a-5p, its duplex precursor (ds miR-146a), and their corresponding mutants. The seed region is underlined and mutations marked in red.
(B) ss-miR-146a-5p (50 nM) induces cytokine gene expression in BMDMs (n = 3).
(C) Production of cytokine proteins in BMDMs treated with miR-146a-5p (ss), miR-146a duplex (ds), their corresponding mutants at 50 nM, or lipofectamine only (Lipo) for 16 h.
(D) Effect of RNase A on ss-miR-146a-5p-induced IL-6 production. n = 3.
(E) EVs loaded with miR-146a-5p (0.3 pmol/16 μg EVs/100 μL) induces multiple cytokine productions in BMDMs.
(F) Nucleotide sequences of ss miR-146a-5p and its various U→A mutants. The U12U13…U17 motif sequence is highlighted in shade. U→A mutants are in red.
(G) Dose response of IL-6 production in BMDMs treated with miR-146a-5p or its various single U→A mutants (50 nM); n = 3.
(H) Schematic diagram of co-immunoprecipitation (Co-IP) experiments.
(I) Representative immunoprecipitation (IP) using IgG or anti-TLR7 Ab in HEK293-hTLR7 (293hTLR7) or null cells followed by immunoblot (IB) for TLR7.
(J) qRT-PCR analysis of ss-miR-146a-5p associated with hTLR7 IP (Null cell: n = 4, HEK293-hTLR7 cell: n = 6).
(K) ss-miR-146a-5p induces Tnfα gene expression in HEK293-hTLR7 cells but not in null cells. n = 3.
(L) WT, not TLR7−/−, BMDM’s response to ss-miR-146a-5p treatment with cytokine productions. n = 3.
∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001, Welch t test.
Data are represented as mean ± SD.
To identify any specific sequence preference or motif in ss miR-146a-5p structure that is essential for its innate immune activity, we first synthesized 33 plasma miRNAs that were identified in the septic patients' plasma and tested their abilities to stimulate IL-6 production in BMDMs (data not shown), a strong predictor of sepsis severity (Merx and Weber, 2007). We then analyzed their nucleotide sequences using a computer exhaustive search algorithm and searched for nucleotide sequences between 3- and 7-nucleotide long that were only present in those miRNAs that induced IL-6 production in our cell-based system. The computer algorithm identified UU … U (U = uridine) as the possible functional motif of ss miR-146a-5p (shown within the shaded region in Figure 3F). We then experimentally tested the relative importance of each U in miR-146a-5p structure and the necessity of the U12U13 … U17 motif for the miR-146a-5p-induced IL-6 production by individually mutating each U. We discovered that the mutations outside of the motif, i.e., U1→A or U8→A mutation, had no impact on IL-6 production (Figure 3G). However, either U12→A or U13→A, but not U17→A, all within the motif, abrogates IL-6 production (Figure 3G). As expected, miR-146a-mut U→A with all seven “U” replaced with “A” abolished the effect of miR-146a-5p. These data suggest that the consecutive U12 and U13 within the UU … U motif is absolutely essential for the ability of miR-146a-5p to activate IL-6 production and are consistent with a recent report that the consecutive UU is important for synthetic small ss RNA binding to and activation of crystal structure of TLR7 (Zhang et al., 2018).
To demonstrate whether ss miR-146a-5p acts by physical association to TLR7, HEK293 cells expressing human TLR7 or HEK293 null cells were incubated with miR-146a-5p or the miR-146a-5p U→A mutant overnight, and hTLR7 was immunoprecipitated using a specific anti-TLR7 antibody (Figure 3H). Western blotting of hTLR7 in Figure 3I showed effective pulldown of hTLR7 by anti-TLR7 but not by the control IgG. Subsequent qRT-PCR analysis of the TLR7 immunoprecipitates showed that ss miR-146a-5p was enriched in the immunoprecipitates of HEK293 hTLR7 cells but not that of null cells, whereas miR-146a-5p U→A mutant was undetectable in 293 hTLR7 immunoprecipitates (Figure 3J). Functionally, miR-146a-5p treatment also induced Tnfα gene induction in HEK293-hTLR7 cells but not in null cells (Figure 3K). This is consistent with the observation that miR-146a-5p induced CXCL2 and IL-6 production in WT but not TLR7−/− BMDMs (Figure 3L). Together, these data suggest that miR-146a-5p stimulates cytokine production by directly binding and activating TLR7.
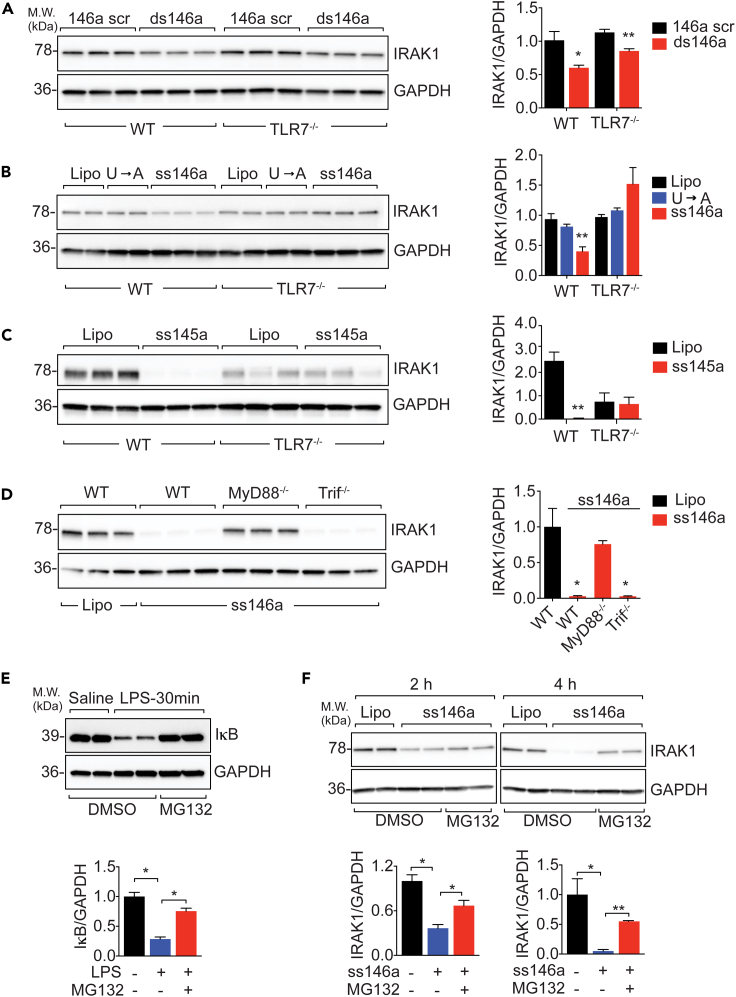
miR-146a-5p negatively regulates IRAK-1 protein expression through TLR7 signaling and cellular proteasome activation
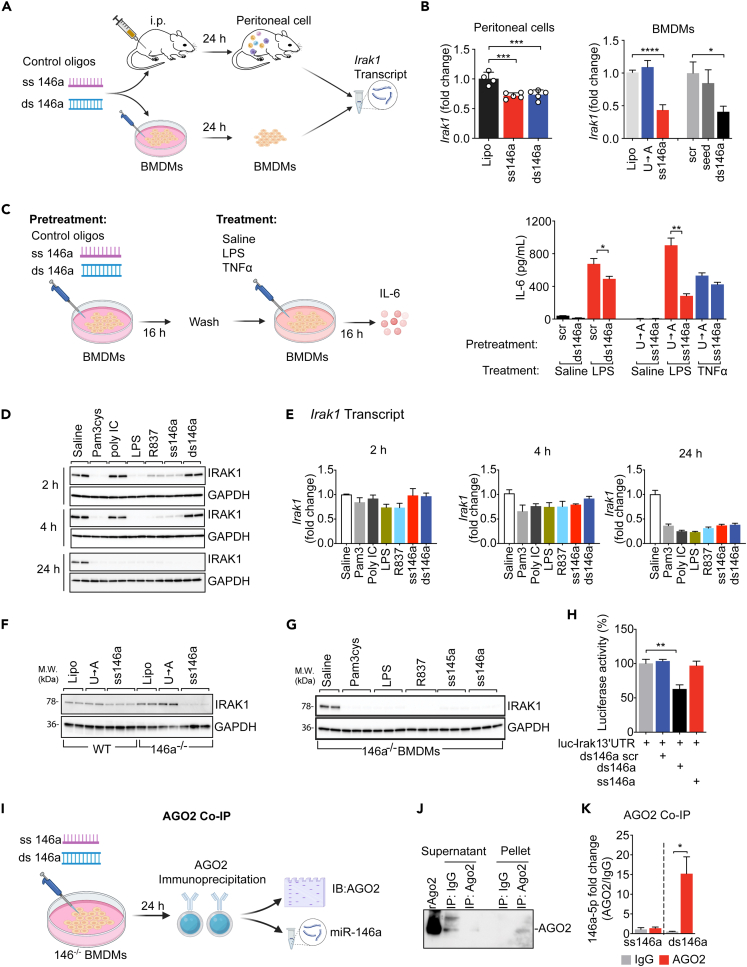
Cellular IRAK-1 is a key kinase involved in the proximal signaling of multiple pattern recognition receptors such as TLRs (Akira and Takeda, 2004). Its mRNA is reportedly targeted by intracellular miR-146a (Boldin et al., 2011; Taganov et al., 2006). Here we discovered a different mechanism that negatively regulates IRAK-1 protein expression via miRNA-TLR7 signaling and proteasome-mediated degradation. When administered in vivo (20 μg, i.p.) to mice or in vitro (50 nM, BMDMs), both ss miR-146a-5p and its ds precursor downregulated Irak-1 gene in the infiltrated peritoneal cells and in BMDM cultures after 24 h of treatment (Figures 4A and 4B). Pretreatment of BMDMs with ss miR-146a-5p or the duplex precursor (but not their mutants), followed by medium removal and cell washing, also attenuated functional response to subsequent LPS treatment, whereas it had no effect on that of TNFα (which acts via IRAK-1-independent signaling) (Figure 4C). Of interest, although they both downregulated Irak-1 gene after 24 h, their effects on cellular IRAK-1 protein expression followed quite different kinetics and mechanisms. As shown in Figure 4D, ss miR-146a-5p and various other TLR ligands, including Pam3Cys (TLR2), LPS (TLR4), and R837 (TLR7), but not ds miR-146a duplex or poly IC (TLR3), all led to a rapid and dramatic reduction of IRAK-1 protein within 2 h of treatment, hours before Irak-1 transcript degradation, which occurred between 4 and 24 h, following the same treatments (Figure 4E). These data suggest a posttranslational mechanism responsible for the rapid degradation of IRAK-1 protein following TLR2, TLR4, and TLR7 activation.
Figure 4.
ss-miR-146a-5p downregulates IRAK-1 protein expression through TLR7 signaling and cellular proteasome activation
(A) Schematic diagram of in vivo (20 μg, i.p.) and in vitro (50 nM, BMDMs) treatment with ss miR-146a-5p and ds miR-146a duplex.
(B) Downregulation of Irak-1 transcript in the peritoneal cells after i.p. injection of or BMDMs treated with ss-miR-146a-5p, dd miR-146a duplex, mutants, or vehicle alone (Lipo) for 24 h; n = 4. ∗p < 0.05, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001, one-way ANOVA.
(C) Pretreatment of BMDMs with ss-miR-146a-5p or ds miR-146a duplex desensitizes BMDMs to subsequent LPS, but not TNFα, stimulation. BMDMs were pretreated with ds miR-146a, ss miR-146a-5p, or the indicated mutants, all at 50 nM, for 16 h, washed, and then challenged with 10 ng/mL LPS or 10 ng/mL TNFα for 16 h n = 3. ∗p < 0.05, ∗∗p < 0.01, Welch t test.
(D and E) Time course of IRAK-1 protein (D) and transcript (E) downregulation after treatment with miR-146a mimics and TLR ligands. BMDMs were treated with Pam3cys (1 μg/mL), poly IC (0.5 μg/mL), LPS (0.1 μg/mL), R837 (0.5 μg/mL), ss-miR-146a-5p (50 nM), or ds miR-146a (50 nM) for 2, 4, or 24 h. At each time point, cellular IRAK-1 protein and mRNA (n = 3) were analyzed using western blot and qRT-PCR, respectively.
(F) Western blot analysis of IRAK-1 protein in WT and miR-146a−/− BMDMs treated with ss-miR-146a-5p, U→A mutant, or vehicles (Lipo) alone.
(G) Western blot analysis of IRAK-1 protein in miR-146a−/− BMDMs treated with Pam3cys (1 μg/mL), LPS (0.1 μg/mL), R837 (0.5 μg/mL), miR-145a-5p (50 nM), or miR-146a-5p (50 nM) for 24 h
(H) ds miR-146a duplex, but not ss miR-146a-5p, represses luciferase output of an IRAK-1-3′ UTR reporter assay. Irak-1-3′UTR luciferase reporter assay in the presence of scrambled ds miR-146a (ds146a scr), ds miR-146a (ds146a), or ss-miR-146a-5p (ss146a), all at 50 nM. The experiments were performed in triplicates and repeated twice. ∗∗p < 0.01, Student t test.
(I and J) Immunoprecipitation of Ago2 in miR-146a−/− BMDMs treated with ss miR-146a-5p and ds miR-146a duplex.
(K) miR-146a-5p associated with Ago2 protein. miR-146a−/− BMDMs were incubated with ss-miR146a-5p or ds miR-146a duplex (100 nM) for 24 h. The levels of miR-146a-5p associated with Ago2 were quantified using qRT-PCR following Ago2 IP (ss146a-IgG, n = 3; n = 4 for the rest groups). ∗p < 0.05, Student t test.
Data are represented as mean ± SD.
TLR activation is known to induce cellular miR-146a expression (Nahid et al., 2016). To determine the necessity of endogenous miR-146a in TLR ligand-induced IRAK-1 downregulation, we tested IRAK-1 expression in miR-146a−/− BMDMs and found that miR-146a-5p, miR-145a-5p (Zou et al., 2016), Pam3cys, LPS, and R837, all induced marked IRAK-1 protein downregulation even in the absence of endogenous miR-146a (Figures 4F and 4G). To test whether ss miR-146a-5p regulates Irak-1 gene expression through its binding to the 3′ UTR, we transiently transfected HEK293 cells with Irak-1-3′ UTR luciferase reporter vectors together with either miR-146a duplex or miR-146a-5p. We observed a significant decrease in luciferase activity in cells treated with miR-146a duplex, but not with ss miR-146a-5p (Figure 4H), suggesting that treatment with ds miR-146a duplex, but not ss miR-146a-5p, targets and represses Irak-1 mRNA via its 3′ UTR. Further, we examined the binding of miR-146a duplex or miR-146a-5p to components of RISC, which is critical for miRNA-mediated gene targeting (Yoda et al., 2010). In this experiment, ss miR-146a-5p or ds miR-146a duplex was transfected into miR-146a−/− BMDMs to eliminate the potential interference from endogenous miR-146a. Ago2, the effector protein of the RISC complex, was immunoprecipitated as demonstrated by Ago2 immunoblotting (Figures 4I and 4J). We found that only Ago2 immunoprecipitated from cells transfected with miR-146a duplex, but not with ss miR-146a-5p, contained ss miR-146a-5p as detected by qRT-PCR (Figure 4K). Together, these surprising data suggest that exogenous miR-146a duplex is capable of binding to Ago2 protein and downregulates Irak-1 gene via an RISC-dependent mechanism, whereas miR-146-5p downregulates IRAK-1 protein through a mechanism independent of Ago2/RISC and Irak-1 3′ UTR binding.
Because TLR7 is essential for ex-miR-146a-5p sensing, we next tested the necessity of TLR7 in miR-146a-5p-induced IRAK-1 protein degradation. ds miR-146a duplex downregulated IRAK-1 protein expression in both WT and TLR7−/− BMDMs (Figure 5A). In contrast, ss miR-146a-5p downregulated IRAK-1 in WT cells but failed to do so in TLR7−/− cells (Figure 5B). To determine whether the degradation of IRAK-1 protein was the result of TLR7 activation and not only limited to miR-146a-5p, we treated WT or TLR7−/− cells with miR-145a-5p, a miRNA identified previously in septic plasma with strong immune-stimulating activity but with no predicted Irak-1 targeting (Alexander et al., 2015; Feng et al., 2017; Zou et al., 2016). Similar to miR-146a-5p, miR-145a-5p led to marked downregulation of IRAK-1 protein expression in WT cells, and this effect was abolished in TLR7−/− cells (Figure 5C). Deletion of MyD88, an adaptor in TLR7 signaling and upstream of IRAK-1, completely prevented IRAK-1 downregulation induced by miR-146a-5p, whereas deletion of Trif, an adaptor in dsRNA-TLR3 and endotoxin-TLR4 signaling (O'Neill and Bowie, 2007), had no effect on miR-146a-5p-mediated IRAK-1 protein downregulation (Figure 5D). Finally, cellular IRAK-1 protein is reportedly ubiquitinated in response to TLR stimulation (Kong et al., 2017) and degraded by proteasomes following its phosphorylation (Li et al., 2000; Yamin and Miller, 1997). In consistent with this report, we found that the proteasomal inhibitor MG-132 prevented LPS-induced IκB protein degradation (Figure 5E) as well as ss miR-146a-5p-induced IRAK-1 protein degradation at 2 and 4 h (Figure 5F). All together, these data strongly suggest that TLR7-MyD88 signaling and cellular proteasome activation critically mediates extracellular miR-146a-5p-initiated IRAK-1 protein downregulation.
Figure 5.
Extracellular miR-146a-5p downregulates IRAK-1 protein via TLR7→MyD88 signaling and proteasome activation
(A) miR-146a duplex-mediated IRAK-1 downregulation is preserved in TLR7−/− BMDMs. Immunoblot analysis of IRAK-1 in WT and TLR7−/− BMDMs treated with miR-146a duplex (ds146a, 50 nM) or scrambled control (146a scr, 50 nM) for 48 h. Experiment was performed in triplicates and repeated twice. ∗p < 0.05, ∗∗p < 0.01, versus scrambled miR-146a duplex (146 scr). Welch t test.
(B) ss-miR-146a-5p-mediated downregulation of IRAK-1 protein is abolished in TLR7−/− BMDMs. Immunoblot analysis of IRAK-1 in WT and TLR7−/− BMDMs treated with miR-146a-5p or the U→A mutant (50 nM). ∗∗p < 0.01 versus U→A mutant in WT BMDMs. Welch t test. Experiments were done in duplicates or triplicates and repeated twice.
(C) Immunoblot analysis of IRAK-1 in WT and TLR7−/− BMDMs treated with ss miR-145a-5p (50 nM) or vehicle (Lipo). ∗∗p < 0.01 versus WT Lipo. Welch t test. Experiments were done in triplicates and repeated twice.
(D) IRAK-1 downregulation by ss-miR-146-5p requires MyD88 but not Trif. Immunoblot analysis of IRAK-1 in WT, MyD88−/−, and Trif−/− BMDMs treated with miR-146a-5p (50 nM). ∗p < 0.05 versus WT Lipo, Welch t test. Experiments were repeated twice.
(E) Proteosome-mediated IκB degradation after LPS stimulation is inhibited by MG132. IκB-α immunoblot in BMDMs stimulated with LPS (100 ng/mL) in the absence or presence of proteasome inhibitor MG-132 (1 μM). ∗p < 0.05, Welch t test.
(F) Rapid miR-146a-5p-mediated IRAK-1 downregulation is inhibited by MG132. IRAK-1 immunoblot in BMDMs stimulated with 50 nM miR-146a-5p in the absence or presence of proteasome inhibitor MG-132 (1 μM). ∗p < 0.05, ∗∗p < 0.01, Student t test.
Data are represented as mean ± SD.
miR-146a KO mice are partially protected against sepsis with reduced inflammation and improved organ function
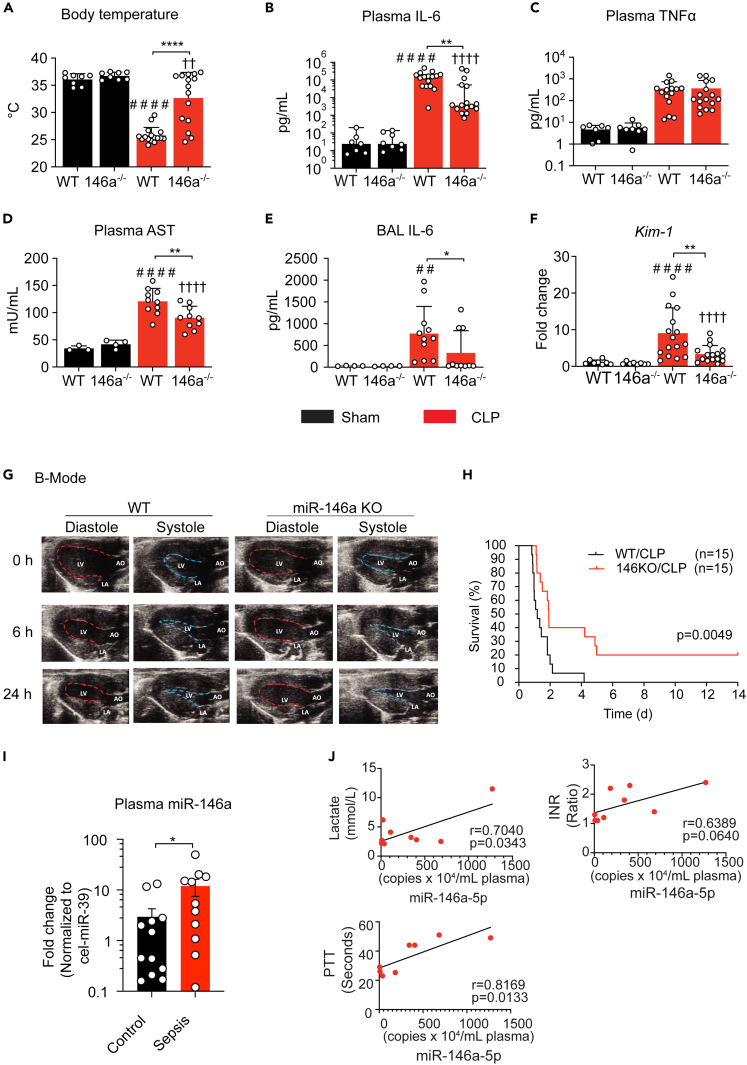
We next investigated the impact of systemic miR-146a deficiency on the pathophysiological changes in murine sepsis. Twenty-four hours after CLP, WT mice developed severe hypothermia with the rectal temperatures at 25.8 ± 1.5°C (versus Sham: 35.3 ± 0.9°C), a sign of circulatory collapse (Figure 6A). These septic mice also experienced cytokine storm with markedly elevated plasma levels of IL-6 (154,486 pg/mL) and TNFα (364 pg/mL) (Figures 6B and 6C), as well as elevated cytokine gene expressions in the kidney, lung, and liver (data not shown). In addition, sepsis caused acute liver injury with elevated plasma aspartate transaminase (AST), increased lung inflammation with marked elevation of bronchoalveolar lavage (BAL) IL-6, and acute kidney injury (AKI) with increased tissue biomarker kim-1 (Zhang et al., 2007) (Figures 6D, 6E, and 6F). In comparison, age-/gender-matched miR-146a−/− septic mice had warmer core temperature (33.7 ± 4.0°C versus 25.8 ± 1.5°C) (Figure 6A), markedly reduced plasma IL-6 (3,796 pg/mL versus 154,486 pg/mL) (Figure 6B), AST, BAL IL-6, and kidney kim-1 expression (Figures 6D, 6E, and 6F).
Figure 6.
Role of miR-146a in polymicrobial sepsis
(A–H) miR-146a−/− mice are protected against polymicrobial sepsis
(A) Core body temperature of WT and miR-146a−/− mice 24 h after sham or CLP procedures. ∗∗∗∗p < 0.0001 versus WT-Sham, ††p < 0.01 versus KO-Sham, ∗∗∗∗p < 0.0001; two-way ANOVA with Bonferroni's post-hoc test.
(B and C) Plasma IL-6 (B) and TNFα (C) level in WT and miR-146a−/− mice at 24 h after Sham or CLP procedures. ####p < 0.0001 versus WT-Sham, ††††p < 0.0001 versus KO-Sham, ∗∗p < 0.01; Mann Whitney test.
(D) Plasma AST activity in WT and miR-146a−/− mice. ####p < 0.0001 versus WT-Sham, ††††p < 0.0001 versus KO-Sham, ∗∗p < 0.01. two-way ANOVA with Bonferroni's post-hoc test.
(E) BAL IL-6 in WT and miR-146a−/− mice. ##p < 0.01 versus WT-Sham, ∗p < 0.05 versus WT-CLP; Mann Whitney test.
(F) Kim-1 was measured by qRT-PCR. ####p < 0.0001 versus WT-Sham, ††††p < 0.0001 versus KO-Sham, ∗∗p < 0.01; Mann Whitney test.
(G) Representative B-mode echocardiographic measurements of WT and miR-146a−/− mice at baseline, 6 h, and 24 h after CLP procedure. LV, left ventricle; LA, left atrium; AO, aorta. See also Figure S2B for representative M-mode echocardiography.
(H) Survival rate of WT and miR-146a−/− septic mice.
(I) Plasma miR-146a-5p levels in control and septic humans (Control: n = 8, Sepsis: n = 10). ∗p < 0.05, Welch t test.
(J) Pearson's correlation of the plasma miRNA copy numbers with the plasma lactate, international normalized ratio (INR), and partial thromboplastin time (PTT) in septic humans.
Data are represented as mean ± SD.
See also Figure S3 for Pearson's correlation between the plasma miRNA (miR-10a-5p, miR-122a-5p, miR-22-3p) concentrations, and the clinical parameters—serum lactate, blood international normalized ratio (INR), and partial thromboplastin time (PTT) in septic humans.
Acute cardiac dysfunction is a hallmark of cardiovascular collapse during septic shock and contributes to its high mortality (Merx and Weber, 2007; Parrillo, 2008). As illustrated by the B-Mode echocardiographic images in Figure 6G, M-mode in Figure S2B, and the quantitative data in Table 1, WT septic mice developed severe left ventricle (LV) dysfunction as early as 6 h after CLP as evidenced by much smaller LV dimensions at the end of diastole and systole (LVIDd and LVIDs), a 29% reduction in fraction shortening (FS), a 56% reduction in stroke volume (SV), and a 61% reduction in cardiac output (CO) when compared with their sham controls. Such cardiac dysfunction persisted up to 24 h after CLP surgery. In comparison, miR-146a−/− CLP mice had significantly improved LV performance when compared with WT CLP mice with significantly higher FS, SV, and CO at both 6 h and 24 h postsurgery (Figures 6G and Table 1). Finally, miR-146a KO mice had significantly improved survival between 2 and 14 days after CLP procedure (Figure 6H).
Table 1.
Echocardiographic measurements before and after CLP in WT and miR-146a−/− mice
| Group (n) | WT-Sham (4) | KO-Sham (4) | WT-CLP (11) | KO-CLP (10) | p1 | p2 | p3 |
|---|---|---|---|---|---|---|---|
| Baseline | |||||||
| LVIDd, mm | 2.8 ± 0.15 | 3.0 ± 0.05 | 3.0 ± 0.14 | 3.0 ± 0.15 | ns | ns | ns |
| LVIDs, mm | 1.6 ± 0.05 | 1.5 ± 0.13 | 1.6 ± 0.18 | 1.6 ± 0.13 | ns | ns | ns |
| HR, bpm | 679 ± 21 | 681 ± 17 | 694 ± 30 | 678 ± 22 | ns | ns | ns |
| FS, % | 44.7 ± 2.3 | 49.5 ± 3.2 | 46.5 ± 4.1 | 45.8 ± 3.4 | ns | ns | ns |
| EF, % | 77.6 ± 2.3 | 82.1 ± 3.0 | 79.1 ± 4.2 | 78.5 ± 3.5 | ns | ns | ns |
| CO, mL/min | 15.9 ± 2.4 | 19.7 ± 0.5 | 19.1 ± 1.9 | 18.1 ± 1.9 | ns | ns | ns |
| SV, μL | 23.4 ± 3.4 | 28.9 ± 0.6 | 27.5 ± 3.2 | 26.7 ± 3.3 | ns | ns | ns |
| 6h post-surgery | |||||||
| LVIDd, mm | 3.0 ± 0.19 | 3.1 ± 0.17 | 2.4 ± 0.30 | 2.6 ± 0.19 | <0.001 | 0.0027 | 0.0748 |
| LVIDs, mm | 1.6 ± 0.17 | 1.5 ± 0.13 | 1.5 ± 0.21 | 1.4 ± 0.14 | ns | ns | ns |
| HR, bpm | 570 ± 7 | 616 ± 15 | 489 ± 55 | 554 ± 34 | ns | ns | ns |
| FS, % | 48.2 ± 1.6 | 49.8 ± 2.4 | 34.3 ± 6.0 | 45.2 ± 3.8 | <0.0001 | ns | <0.0001 |
| EF, % | 80.9 ± 1.6 | 82.4 ± 2.2 | 65.3 ± 8.3 | 78.3 ± 3.9 | <0.0001 | ns | <0.0001 |
| CO, mL/min | 16.4 ± 2.1 | 19.3 ± 3.2 | 6.4 ± 2.4 | 10.8 ± 2.7 | <0.001 | <0.0001 | 0.0046 |
| SV, μL | 28.7 ± 3.5 | 31.2 ± 4.8 | 12.8 ± 3.9 | 19.3 ± 3.9 | <0.0001 | <0.0001 | 0.0044 |
| 24h post-surgery | |||||||
| LVIDd, mm | 2.9 ± 0.05 | 2.9 ± 0.15 | 2.4 ± 0.33 | 2.6 ± 0.30 | 0.0009 | ns | 0.0347 |
| LVIDs, mm | 1.5 ± 0.08 | 1.4 ± 0.05 | 1.5 ± 0.24 | 1.4 ± 0.12 | ns | ns | ns |
| HR, bpm | 677 ± 15 | 681 ± 25 | 338 ± 42 | 499 ± 154 | <0.0001 | <0.0001 | <0.0001 |
| FS, % | 48 ± 3.2 | 49.9 ± 1.2 | 37.1 ± 6.5 | 45.5 ± 5.8 | 0.0007 | ns | 0.0005 |
| EF, % | 80.9 ± 3.0 | 82.7 ± 1.0 | 68.9 ± 8.1 | 78.4 ± 6.0 | 0.0018 | ns | 0.0009 |
| CO, mL/min | 17.6 ± 1.4 | 17.6 ± 1.8 | 4.7 ± 1.9 | 11.0 ± 5.9 | <0.0001 | 0.0013 | <0.0001 |
| SV, μL | 25.9 ± 1.7 | 26 ± 3.7 | 13.8 ± 5.0 | 20.6 ± 6.7 | <0.0001 | ns | 0.0022 |
The echocardiographic data are presented as mean ± SD.
Two-way ANOVA with Tukey's post-hoc test for repeated measurements.
p1: WT-Sham versus WT-CLP.
p2: KO-Sham versus KO-CLP.
p3: WT-CLP versus KO-CLP.
ns, not significant.
Plasma miRNA-146a-5p concentrations correlate with lactate levels and coagulopathy in septic patients
We examined the correlation of plasma miR-146a-5p concentrations with plasma lactate and coagulopathy in septic patients, two standard clinical tests that are highly associated with sepsis in-hospital mortality (Casserly et al., 2015; Dhainaut et al., 2005; Rhodes et al., 2017). Figure 6I shows qRT-PCR validation of the plasma miR-146a-5p in septic humans compared with age-/gender-matched controls. Within the group of patients, we observed a strong positive correlation between the copy numbers of plasma miR-146a-5p and the plasma lactate levels as well as increased international normalized ratio (INR) and partial thromboplastin time (PTT) (Figure 6J). Similar high clinical correlation was observed with plasma miR-10a-5p levels but not with plasma miR-122-5p and miR-22-3p (Figure S3).
Discussion
We investigated a pivotal role of extracellular (ex) miR-145a-5p in host innate immune response and bacterial sepsis. We found that both CLP mice and ICU septic patients with abdominal source of infection had a significant increase in their plasma RNA. A vast majority of the plasma RNA in both healthy and septic subjects turned out to be extracellular miRNAs with substantial overlapping between mice and humans and differential expression patterns between health and sepsis. One of the most abundant plasma miRNAs shared by septic mice and humans was miR-146a-5p, which appeared to be the predominant strand of miR-146a in the plasma and tissues, as no or much lower level of miR-146-3p was detected in the plasma or liver, respectively. We discovered that extracellular miR-146a-5p is highly proinflammatory and plays a pivotal role in sepsis. When administered in vivo, ss miR-146a-5p, but not its precursor ds miR-146a duplex, induced acute peritonitis with robust monocyte/neutrophil infiltration after only a single dose of i.p. injection. Systemic administration of exogenous miR-146a-5p via continuous s.q. pump delivery in miR-146a−/− mice reached the plasma level at fmol/dL and led to significant plasma IL-6 and various organ expression of IL-1 and IL-6 genes. These data suggest that extracellular miR-146a-5p, but not its ds duplex precursor, is a potent innate immune activator capable of driving local and systemic proinflammatory response.
Our data also provide important insights into how ss miR-146a-5p works to activate proinflammatory responses. Using a computer predictive algorithm and a series of single U→A mutations in the miR-146a-5p molecule, we identified the essential role of the successive U12U13 in the U12U13xxxU17 motif for miR-146a-5p-mediated cellular cytokine response. These data are consistent with the report that consecutive UU is important for synthetic small ssRNA binding to and activation of crystal structure of TLR7 (Zhang et al., 2018). Moreover, using co-immunoprecipitation and TLR7-KO mice, we clearly demonstrated the physical association between ss miR-146-5p and TLR7 in cells expressing TLR7 and the necessity of TLR7 in miR-146a-5p-induced proinflammatory cytokine production in macrophages. In contrast, miR-146a duplex, a short dsRNA with a two-nucleotide overhang at 3′ end (Zhang et al., 2004), fails to exhibit any immune-stimulatory effect in macrophage cultures or after i.p. injection. This may be because it is not recognized by other intracellular ds RNA sensors, as RIG-I (retinoic-acid-inducible gene 1) preferentially recognizes blunt-end short ds RNAs, whereas MDA5 (melanoma-differentiation-associated gene 5) and TLR3 are only activated by long ds RNAs (Roers et al., 2016).
We carefully delineated the underlying molecular mechanism by which ss miR146a-5p regulates IRAK-1, a serine/threonine kinase important for all TLR signaling except that of TLR3. Previous studies have reported that endogenous intracellular miR-146a or transfection with exogenous ds miR-146a targets Traf6 and Irak-1 genes and attenuates host immune response to endotoxin (Boldin et al., 2011; Nahid et al., 2009, 2016; Taganov et al., 2006). As such, cellular miR-146a is generally considered a negative regulator of innate immunity (Mehta and Baltimore, 2016; O'Connell et al., 2012). Here, we identified two distinct mechanisms leading to cellular IRAK-1 downregulation in response to ss miR-146a-5p or ds miR-146a duplex. When incubated with BMDMs, exogenous miR-146a duplex, not ss miR-146a-5p, was co-immunoprecipitated with intracellular Ago2 in cells deficient of endogenous miR-146a and repressed Irak-1 3′UTR luciferase reporter activity. These data suggest that ds miR-146a duplex is associated with Ago2, a key component of the RISC complex, and likely downregulates Irak-1 gene via binding to its 3′ UTR. In stark contrast, the ability of ss miR-146a-5p to downregulate IRAK-1 protein appears to be Ago2/RISC-independent but completely dependent on TLR7→MyD88 signaling, as this effect was abolished in cells lacking TLR7 or MyD88 but remained unchanged in cells deficient of Trif. Moreover, the finding that the TLR7 ligands, R837 and miR-145a-5p, neither of which targets Irak-1 3′ UTR, also markedly attenuated IRAK-1 protein expression, further suggesting the pivotal role of TLR7 activation in IRAK-1 protein downregulation.
Importantly, our data suggest that TLR-mediated IRAK-1 protein downregulation involves a posttranslational event and is independent of RISC complex and its gene degradation. The finding that IRAK-1 protein level was rapidly decreased within 2 h, many hours before Irak-1 mRNA was downregulated, after treatment with ss miR-146a-5p or other TLR ligands, such as Pam3cys, LPS, and R837, supports this notion. This conclusion is further supported by two important observations: (1) IRAK-1 protein level was markedly reduced by ss miR-146a-5p and the TLR ligands even in BMDMs deficient of miR-146a gene, suggesting that the effect was independent of any endogenous miR-146a induction, and (2) IRAK-1 protein downregulation induced by miR-146a-5p was reversed by the proteasomal inhibitor MG-132, which is known to block LPS-induced and proteasome-mediated IκB degradation (Chen et al., 1995; DiDonato et al., 1996). IRAK-1 is critically involved in various MyD88-dependent TLR signaling (Akira and Takeda, 2004). Thus, our finding suggests that rapid IRAK-1 protein degradation, not only promoted by extracellular nucleic acids, such as miR-146a-5p and miR-145a-5p, but also other pattern molecules that signal through IRAK-1 pathway, such as LPS (TLR4) and lipoprotein (TLR2), may represent an important molecular mechanism of cellular tolerance after initial innate immune stimulation. Cellular tolerance phenomena have been widely reported in various receptor systems, immune or nonimmune, such as endotoxin tolerance (Seeley and Ghosh, 2017) and β-adrenergic receptor desensitization (Hausdorff et al., 1990; Lohse et al., 1990). Such innate immune tolerance may be important in self-limiting the scope of host proinflammatory response to danger signaling and attenuate potentially damaging effects although its clinical significance in the setting of infection is still unclear (Seeley and Ghosh, 2017). Recent studies suggest that a number of cellular miRNAs are differentially expressed during the course of endotoxin response and may play a role in LPS tolerance by regulating LPS-induced gene activity at posttranscriptional levels (Seeley and Ghosh, 2017). Our data suggest that extracellular miRNAs, such as miR-146a-5p, may induce a cross-tolerance to LPS at the posttranslational level via proteosomal IRAK-1 degradation. Altogether, extracellular miR-146a-5p seems to play a dual role—proinflammatory cytokine production and innate immune cell activation and rapid posttranslational IRAK-1 degradation—both via TLR7-MyD88 signaling pathway.
We found that miR-146a−/− mice were partially protected from severe polymicrobial sepsis with improved cardiac function and attenuated organ injury when compared with their WT counterpart. miR-146a−/− mice maintained higher core temperature and improved LV filling (higher LVIDd) and cardiac output, all indicating better cardiovascular performance during sepsis. Consistent with the improved cardiovascular function, miR-146a−/− septic mice had significantly lower plasma IL-6 storm, a known cardiac depressor and predictor in sepsis (Merx and Weber, 2007), than WT septic mice, attenuated liver, lung, and kidney injury, and significantly improved survival. Although the definite contribution of the proinflammatory function of miR-146a-5p to sepsis is yet to be determined, these findings clearly suggest that host endogenous miR-146a-5p functions as an innate immune activator and plays a contributory role in driving systemic inflammation and organ injury in sepsis. Supporting this are the recent reports that mice deficient of TLR7, the nucleic acid sensor for single-stranded RNA, are protected from sepsis-induced systemic cytokine storm, coagulopathy, and organ injury (Jian et al., 2019; Williams et al., 2019).
Circulating miRNAs are packaged into and carried by various plasma carriers, such as EVs (Ying et al., 2017), high-density lipoprotein (HDL) (Vickers et al., 2011), and Ago2 (Arroyo et al., 2011; Geekiyanage et al., 2020). These macromolecular complexes protect miRNAs from plasma RNase digestion and exhibit various biological activities by delivering their cargo miRNAs to target cells. One of such examples is EV-associated miR-146a-5p. EVs from mesenchymal stromal cells prevent lymphoid-cells-dominant allergic airway inflammation partially through their cargo miR-146a-5p (Fang et al., 2020). miR-146a-5p in EVs are released from activated macrophages and transported into endothelial cells, resulting in reduction of endothelial permeability (Hu et al., 2020). We report that plasma EVs isolated from septic mice or miRNA-loaded HEK293T EVs induce strong innate immune response in macrophages, cardiomyocytes, and in intact animals in part via their miRNA cargos such as miR-146a-5p (Jeyaram et al., 2020; Shimada et al., 2020; Xu et al., 2018). HDL is another carrier that can bind and transport plasma miRNAs (Vickers et al., 2011). However, the amount of HDL-bound plasma miRNAs is reportedly very small, e.g., less than 1% of plasma miR-146a-5p is HDL-bound in humans (Wagner et al., 2013). On the other hand, circulating Ago2 complexes reportedly serve as a major carrier of miRNAs in plasma (Arroyo et al., 2011). A recent report demonstrates that several Ago2-associated plasma miRNAs (miR-16 and let-7a) can base-pair sequence specifically to a target mimic oligonucleotide, suggesting their potential abilities to posttranscriptionally regulate genes if they enter the cells in sufficient quantities (Geekiyanage et al., 2020). However, the role and the significance of the Ago2-miRNA delivering route under normal and pathological conditions remain unclear. Finally, it is worth noting that different methods for plasma RNA isolation can strongly affect the type and amount of RNA associated with these carriers and data need to be carefully interpreted (Srinivasan et al., 2019).
In our limited number of septic patients, we observed a close correlation between their plasma miR-146a-5p copy numbers and blood lactate concentrations and severity of coagulopathy such as INR and PTT. Increase in the serum lactate levels may represent tissue hypoperfusion and hypoxia among other causes and is highly associated with organ injury and worse clinical outcome (Casserly et al., 2015; Rhodes et al., 2017). Lactate-guided resuscitation of patients with septic shock improves survival (Jansen et al., 2010; Jones et al., 2010). Similarly, coagulopathy, as evidenced by increased INR and PTT, is a sign of worse outcome in septic patients (Dhainaut et al., 2005). These data in the septic patients are consistent with our animal findings and support the hypothesis that plasma extracellular miR-146a-5p plays an important role in sepsis pathogenesis.
Given the pivotal role of miRNAs in innate immune regulation under both normal and pathological conditions (Lee et al., 2020; Mann et al., 2017; Mehta and Baltimore, 2016; O'Connell et al., 2012), various approaches have been investigated to modulate extracellular and intracellular miRNAs as potential strategies of novel treatments (Lee et al., 2020). These include various delivering vehicles of miRNAs or anti-miRNAs, such as virus, polymers, lipid, EVs, and RNA nanoparticles (Lee et al., 2020). However, how to control cell- and tissue-specific delivery of miRNAs or anti-miR after systemic administration to minimize nonspecific and unwanted side effect is a major challenge. To target miRNAs in sepsis, neutralizing proinflammatory plasma miRNAs such as miR-146a-5p using anti-miR inhibitors or TLR7 antagonists to block their downstream receptors may represent a reasonable next step to validate the finding of the deletion study and test the therapeutic efficacy of the pharmacological intervention targeting miRNA→TLR7 signaling.
In summary, RNA sequencing analysis demonstrates an upregulation of numerous plasma miRNAs, such as miR-146a-5p, in septic mice and humans. In septic patients, plasma miR-146a-5p concentrations are strongly associated with the two clinical outcome predictors, blood lactate and coagulopathy. Mechanistically, extracellular miR-146a-5p acts as an innate immune effector driving acute inflammatory response by binding to the nucleic acid sensor TLR7 via a UU-containing motif-dependent mechanism. Extracellular miR-146a-5p, via TLR7-MyD88 signaling, also negatively regulates cellular IRAK-1 protein, a key innate immune molecule, by proteasome-mediated protein degradation. In vivo, miR-146-5p proves to be sufficient to elicit both local and systemic innate immune response. Mice lacking miR-146a were protected against polymicrobial sepsis with reduced plasma IL-6 storm, organ injury, and markedly improved cardiovascular performance. These data support a critical role for extracellular miR-146a-5p in innate immune inflammation and sepsis pathogenesis and offer a rationale for selective targeting extracellular miRNAs as a novel strategy for sepsis intervention.
Limitations of the study
A limitation to this study is that although we have identified the functional role of the UU-containing motif in miR-146a-5p-induced proinflammatory response and the association between miR-146a-5p and TLR7 protein, we do not have evidence that miR-146a-5p binds directly to TLR7. Purified or crystal TLR7 protein will be needed to establish the direct binding and the affinity between miR-146a-5p and the TLR7 structure (Zhang et al., 2018). Also, although exogenous miR-146a-5p is sufficient to induce innate immune response and miR-146a KO mice have attenuated inflammation and improved organ function in sepsis, the definite role of miR-146a-5p-mediated innate inflammation in sepsis-induced organ injury and mortality is yet to be determined. Based on our data, we conclude that host endogenous miR-146a-5p may function as an innate immune activator and play a contributory role in systemic inflammation and organ injury. Finally, the study only examined one of many upregulated plasma miRNAs, from low to high abundance, that contain UU … U motif and may be proinflammatory. How these circulating extracellular miRNAs regulate host innate immunity via TLR7 or other mechanisms and play a role in sepsis warrants further investigation. In this context, targeting these proinflammatory plasma miRNAs and their converging TLR7 signaling pathway may represent an appealing strategy for future sepsis intervention.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Toll-like Receptor 7 (D7) Rabbit mAb | Cell Signaling Technology | Cat#5632; RRID:AB_10692895 |
| IRAK1 (D51G7) Rabbit mAb | Cell Signaling Technology | Cat#4504; RRID:AB_1904032 |
| GAPDH (14C10) Rabbit mAb | Cell Signaling Technology | Cat#2118; RRID:AB_561053 |
| IκBα (44D4) Rabbit mAb | Cell Signaling Technology | Cat#4812; RRID:AB_10694416 |
| Normal rabbit IgG | Cell Signaling Technology | Cat#2729; RRID:AB_1031062 |
| Anti-rabbit IgG, HRP | Cell Signaling Technology | Cat#7074; RRID:AB_2099233 |
| Anti-mouse IgG, HRP | Cell Signaling Technology | Cat#7076; RRID:AB_330924 |
| Ago2/eIF2C2 mouse mAb (2E12-1C9) | Novus | Cat#H00027161-M01; RRID:AB_2277660 |
| CD45-PE, clone 30-F11 | BD Pharmingen | Cat#553081; RRID:AB_394611 |
| F4/80-Alex 647, clone T45-2342 | BD Pharmingen | Cat#565853; RRID:AB_2744474 |
| Ly6G-BV421, clone 1A8 | BD Biosciences | Cat#562737; RRID:AB_2737756 |
| Biological samples | ||
| Human plasma | University of Maryland Shock Trauma Center | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| Recombinant M-CSF | R&D System | 416-ML |
| Recombinant Ribonuclease Inhibitor | Sigma | R1158 |
| MG-132, ≥95% | Sigma | 474787 |
| Lipopolysaccharides | Sigma | L4391 |
| Imiquimod (R837) | InvivoGen | tlrl-imqs |
| Pam3Cys | Enzo Life Sciences | ALX-165-066-M002 |
| Poly(I:C) | Enzo Life Sciences | ALX-746-021-M002 |
| Critical commercial assays | ||
| Quant-it RNA Assay kit | Thermo Fisher | Q33140 |
| BCA Protein Assay kit | Thermo Fisher | 23225 |
| Agilent Small RNA kit | Agilent | 5067-1548 |
| Mouse IL-6 Duoset ELISA kit | R&D System | DY406 |
| Mouse TNFa Duoset ELISA kit | R&D System | DY410 |
| Mouse CXCL2 Duoset ELISA kit | R&D System | DY452 |
| GoTaq qPCR Master Mix | Promega | A6001 |
| Dual Luciferase Reporter Assay kit | Promega | E1910 |
| miRCURY LNA SYBR Green PCR kit | Qiagen | 339345 |
| Immobilon Forte Western HRP substrate | Millipore | WBLUF0100 |
| Deposited data | ||
| Small RNAseq mouse plasma samples | This paper | GEO: GSE133733 |
| Small RNAseq human plasma samples | This paper | GEO: GSE149764 |
| Experimental models: Cell lines | ||
| Human: HEK 293XL/null | InvivoGen | Cat# 293xl-null |
| Human: HEK 293XL/hTLR7 | InvivoGen | Cat# 293xl-htlr7 |
| Experimental models: Organisms/strains | ||
| Mouse: C57bl/6J | The Jackson Laboratory | Stock#000664 |
| Mouse: B6.Cg-Mir146tm1.1Bal/J | The Jackson Laboratory | Stock#016239 |
| Mouse: B6.Tlr7tm1Flv/J | The Jackson Laboratory | Stock#008380 |
| Oligonucleotides | ||
| ss-miR-146a-5p sequence: UGAGAACUG AAUUCCAUGGGUU |
IDT Technology | N/A |
| ss-miR-146a-5p U-A mut. sequence: AGAGAACAGAAAACCAAGGGAA | IDT Technology | N/A |
| ss-miR-146a-5p G-C mut. sequence: UCACAACUCAAUUCCAUCCCUU | IDT Technology | N/A |
| ss-miR-145a-5p sequence: GUCCAGUUUUCCCAGGAAUCCCU |
IDT Technology | N/A |
| ds-miR-146a mimic | Qiagen | Cat# 339173 |
| ds-miR-146a seed region mutation | Qiagen | Cat# 339173 |
| ds-miR-146a scrambled mutation | Qiagen | Cat# 339173 |
| Recombinant DNA | ||
| Plasmid: CMV-luciferase-Irak1-3’UTR | Addgene | Cat #15095 |
| Plasmid: pRL-SV40 | Addgene | Cat #27163 |
| Software and algorithms | ||
| Prism 9 | GraphPad | https://www.graphpad.com/scientific-software/prism/ |
| exceRpt small RNA-seq pipeline | Genboree Bioinformatics | http://genboree.org/theCommons/projects/exrna-tools-may2014/wiki/Small_RNA-seq_Pipeline |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Dr. Wei Chao (wchao@som.umaryland.edu), Professor and Co-Director, Center for Shock, Trauma and Anesthesiology Research, University of Maryland School of Medicine, Baltimore, MD 21201, USA.
Materials availability
Plasma or miRNAs generated in this study are available from the lead contact.
Experimental model and subject details
Human subjects
Healthy volunteers and ICU septic patients were enrolled under the protocol (HP-0081592) approved by the Institutional Review Board of University of Maryland School of Medicine. We have complied with all relevant ethical regulations. Septic patients at 18 years or older with radiographic or surgical evidence of abdominal sources of infection and age/gender-matched healthy controls were enrolled at University of Maryland Medical Center (Baltimore, MD, USA) after written informed consent was obtained from all study subjects. Plasma was prepared from blood samples by centrifugation (1,000x g for 10 min twice followed by 10,000x g for 10 min).
Animal subjects
C57BL/6J (stock no. 000664) and miR-146a KO (B6/miR-146atm1.1Bal/J, stock no. 016239) mice were purchased from Jackson Laboratory (Bar Harbor, ME). Adult male mice were between 9-12 weeks of age and weighed between 20-30 g. The animal experimental protocols were approved by the Institutional Animal Care and Use Committee, University of Maryland School of Medicine (Approval Number: 0918001, Baltimore, MD). All animal experiments were performed in compliance with the relevant guidelines and regulations of the National Institutes of Health (Bethesda, MD) and the ARRIVE guidelines of the National Centre for the Replacement, Refinement, and Reduction of Animals in Research (London, EN).
A murine model of polymicrobial sepsis was created by cecal ligation and puncture (CLP) (Zou et al., 2016). Briefly, the cecum of anesthetized mice (100 mg/kg ketamine and 4 mg/kg xylazine) was exposed, ligated at 1.5 cm from the tip and punctured twice with a 18G needle. Two mm of feces was gently squeezed out from the ligated cecum. Sham mice only received laparotomy. Pre-warmed sterile normal saline (0.03 ml/g body weight), bupivacaine (3 mg/kg body weight) and buprenorphine (0.1 mg/kg body weight) was administrated s.c. before and after surgery. Twenty-four hours after the Sham or CLP surgery, rectal temperature was recorded. Blood (via cardiac puncture), bronchoalveolar lavage (BAL), and organs were collected. For all in vivo experiments, the information of reagents and mouse strains was blinded to experimenters involved and was revealed only after all data analyses were completed.
Method details
Study design
The goal of the study was to examine the role of extracellular miRNAs as an innate immune regulator and in sepsis pathogenesis. To survey plasma miRNA landscape in health and sepsis, RNA sequencing was employed in mice with polymicrobial sepsis, ICU septic patients with confirmed abdominal sources of infection, and their age/gender-matched healthy cohorts. The Pearson correlation was tested between plasma miRNA copy numbers and serological outcomes of septic patients. To test the role of miRNAs in inflammation and in sepsis, miR-146a-5p mimics and KO mice were employed. Various in vitro and in vivo endpoints were tested based on their clinical relevance and pathological significance in human sepsis, such as core temperature, cytokines, innate immune cell activation, organ injury biomarkers, and echocardiography. To define the molecular mechanisms by which single-stranded miR-146a-5p regulates innate immune activities, various miR-146a-5p derivatives with single nucleotide mutations were tested and various in vitro assays were employed including immunoprecipitation, irak-1 reporter assay, and proteasome inhibitor. For animal studies, the minimal number of subjects used in any experiments was 5 per groups to attain a statistical significance of p < 0.05 with a power of at least 80%. All mice were assigned to a treatment groups using a randomized block experimental design. All in vitro assays were replicated in triplicate with fully independent runs on separate days. For all in vivo experiments, treatments and mouse strains were blinded to the operators and coded by an experimenter who was not otherwise involved in the project. Unblinding occurred only after all data collection and data-entry had been completed. Sample sizes, biological replicates, and statistical methods are provided in the corresponding figure legends.
Plasma small RNAseq
RNA was isolated using Trizol LS from 100 μl of cell-free plasma and was analyzed by an Agilent 2100 Bioanalyzer with the Small RNA kit (Agilent Technologies, Santa Clara, CA). Six microliters of each RNA solution were used for small RNA sequencing library preparation. Libraries were sequenced on an Illumina HiSeq2500 (Illumina, San Diego, CA) and analyzed by Norgen Bioteck Corp. The raw sequence data were analyzed using the exceRpt small RNAseq pipeline (Genboree Bioinformatics). Differential expression (DE) analysis was performed using R package edgeR v3.24.0, which incorporates empirical Bayes estimation into a negative binomial distribution(Robinson et al., 2010). The log2 fold change and log p values from DE analysis were used to draw heatmaps using R package pheatmap v1.0.12 (Kolde and Vilo, 2015). Target genes for selected miRNAs were predicted using the R package multiMiR (Ru et al., 2014) and functional analysis on these predicted targets was performed using the R package clusterProfiler (Yu et al., 2012). The small RNA sequencing data of both mouse and human plasma samples have been deposited in Gene Expression Omnibus (GEO: GSE133733 and GEO: GSE149764).
In vivo miRNA administration
Systemic administration of miR-146a-5p mimics was performed via osmotic pumps (ALZET Osmotic Pumps, Cupertino, CA). Briefly, pumps filled with lipofectamine (lipo) or lipo-packed miR-146a-5p mimics (50 μg/mouse/day x 3 days) were implanted into a subcutaneous pocket. In a separate group of mice, miR-146a (both ss and ds) mimics (20 μg) were delivered by direct intraperitoneal (i.p.) injection. Peritoneal cells were harvested 24 hours after i.p. injection and analyzed with a BD LSR II flow cytometer as detailed below.
Flow cytometry
Twenty-four hours after the i.p. injection of ss miR-146-5p or ds miR-146a, peritoneal cells were collected and counted using hematocytometer. The cell suspension was stained with CD16/CD32 Fc blocker (0.5 μg/sample) prior to the staining of the surface antibodies (CD45-PE, 1:1500; F4/80-Alexa 647, 1:100; Ly6G-BV421, 1:100) at 4°C for 30 minutes. Data was acquired in BD LSRII flow cytometer (BD Biosciences, Frankin Lakes, NJ), and analyzed by SpectroFlo software (Cytek Bioscience, Fremont, CA) and FlowJo (Ashland, OR). The gating strategy is shown in Figure S2A.
Liver function test
Plasma AST activity was detected using an AST kit (Sigma) following manufactory's instructions.
qRT-PCR
qRT-PCR was performed as previously described (Zou et al., 2016). The primer sequences are presented in Table S3. Gene expression in tissue and cell was normalized to GAPDH using the 2−ΔΔCt method and presented as relative fold change between treatment and control groups. A 20 μl RT reaction containing 3.3 pmol of miR-146a-5p mimic was performed to prepare a 1011 copies/μL stock. To determine plasma miRNA copy number, qPCR standard curves were generated with serially diluted cDNA solutions (from 102 to 109 copies/μl). For relative comparison, plasma miRNA expression was normalized to Cel-miR-39-3p (Qiagen) spike-in using the 2−ΔΔCt method.
Cell cultures
Bone marrow cells were isolated from the tibias and femurs of male mice, cultured, and differentiated into bone marrow-derived macrophages (BMDMs) in the presence of 10ng/mL recombinant mouse M-CSF (R&D Systems). Human embryonic kidney cells 293 (HEK293-null and HEK293-hTLR7) were maintained in DMEM high glucose, 10% fetal bovine serum and 1% penicillin/streptomycin at 37°C and 5% CO2.
Preparation of extracellular vesicles and cargo loading
EVs were isolated from HEK293T conditioned media and loaded as previous described (Jeyaram et al., 2020; Lamichhane et al., 2015). Pelleted EVs were resuspended in 1X PBS and filtered using a 0.2 μm syringe filter. Total EV protein concentration was measured. EVs (300 μg/mL) were first dehydrated in 70% ethanol for 12 hrs. EVs were rehydrated in 1mL of citrate buffer (150 mM, pH 2.5) for 1 hour and dialyzed in 1X HEPES buffered saline (1X HBS, pH 7.0) overnight. EV size distribution and concentration were evaluated by NanoSight LM10. 3x109 EVs were incubated with 1000 pmol of miRNA cargo for 2 hours before washing with 1X HBS.
ELISA
CXCL-2, IL-6 and TNFα levels in culture media, BAL, and plasma were measured using ELISA kits following the manufacturer's manuals.
Western blot
Cellular proteins were separated by SDS-PAGE and transferred to a PVDF membrane. Membrane was blocked with 5% non-fat milk and incubated with primary and secondary antibodies. Images were captured using Bio-rad ImageDoc system and quantified using ImageJ software (NIH, Bethesda, MD). Target protein expression level was normalized to GAPDH.
Immunoprecipitation (IP)
HEK293-null and HEK293-hTLR7 cells transfected with ss miR-146a-5p (100 nM), or ss miR-146a-5p U→A (100 nM) mimics were harvested 16 hours post-transfection and lysed in IP lysis buffer (10mM Tris pH 7.4, 150 mM NaCl, 1% NP-40, supplemented with protease/RNase inhibitor) for 20 minutes on ice and centrifuged at 12000 x g, 4°C for 15 minutes. Cell lysate supernatant was incubated with anti-TLR7 or control IgG-conjugated protein A/G agarose beads at 4°C overnight with rotation and then washed four times with IP wash buffer (10mM Tris pH 7.4, 200 mM NaCl, 1% NP-40, supplemented with protease/RNase inhibitor). miR-146a-/- BMDMs transfected with ss miR-146a-5p (100nM) or ds miR-146a (100nM) mimics were harvested 24h post-transfection. IP was performed with mouse anti-Ago2 (clone 2E12-1C9) or control IgG-conjugated protein A/G agarose beads as described above. Immunoprecipitated Ago-miR-146a-5p complexes were eluted from bead and the eluant used as a source of template for detection of associated miR-146a-5p by qRT-PCR.
Echocardiography
Non-invasive transthoracic echocardiography was performed in non-anesthetized mice before surgery (0 hour) and then 6 hours and 24 hours after surgery. Images were acquired and analyzed by an operator (Y.Y.) who was blinded to mouse strain and group information. In brief, B-mode images were acquired from parasternal long axis view. M-mode images were collected from a parasternal short axis view at the mid-ventricular level with a clear view of papillary muscles using a Vevo 2100 Imaging System (VisualSonics, Inc, Toronto, ON). The following measurements were obtained: left ventricular (LV) internal dimension at end-systole (LVIDs), LV internal dimension at end-diastole (LVIDd), and heart rate (HR), end diastolic volume (EDV) and end systolic volume (ESV) were calculated as follows: EDV = [7/(2.4 + LVIDd)] x LVIDd3 and ESV = [7/(2.4 + LVIDs)] x LVIDs3. Stroke volume (SV) was defined as EDV–EVS. Cardiac output (CO) was calculated as HR x SV. Fractional shortening (FS) was determined as [(LVIDd–LVIDs)/LVIDd] x %. Ejection fraction (EF) was defined as (EDV–ESV)/EDVx100%. The values of three consecutive cardiac cycles were averaged.
IRAK-1 3′UTR luciferase reporter assay
HEK293 cells in 96-well plates were co-transfected with 100 ng of CMV-luciferase-Irak-1 3′UTR vector (Addgene, Watertown, MA), 5 pmol of ds or ss miR-146a mimics and 10 ng of pRL-SV40 (Addgene, Watertown, MA) for normalization of transfection efficiency. Luciferase activities were measured 48 hours later.
Quantification and statistical analysis
Operators of in vivo procedures were blinded to the strain, group, and treatment information. In vivo data were coded for analysis before group information was revealed. Statistical analysis was performed using GraphPad Prism 9 software (San Diego, CA). Data were expressed as mean ± SD unless stated otherwise. Normality of all numerical data was assessed using Shapiro-Wilk test. Student t-test was performed for two-group comparison if the hypothesis of normality was met. For multi-group comparison and when the data met the two assumptions – normal population distribution and equal variance, one-way ANOVA with Tukey's post-hoc test or two-way ANOVA with Bonferroni post-hoc test was performed. If the data did not meet the assumptions, we performed Kruskal-Wallis non-parametric ANOVA or two-way ANOVA after the data were transformed to the log or rank format. Difference was considered to be statistically significant if p < 0.05 with two tails. The survival distributions were presented as ratio variables and compared using log-rank (Mantel–Cox) test. Pearson's correlation coefficient was used to detect the associations between plasma miRNA concentrations and the lactate and coagulation function. Detailed statistical methods were described in the figure legends and tables.
Acknowledgments
This work was supported in part by the National Institutes of Health (NIH), United States, grants R01GM122908, R35GM140822 (WC), R35GM124775 (LZ), R01NS110567 (WC, LZ), K08HL153784 (BW), R01HL141611, R01NS110637, and R01HL141922 (SMJ) and by the Frontiers in Anesthesia Research Award from the International Anesthesia Research Society (WC). We thank Xiaoxuan Fan, PhD. and the Flow Cytometry Core of the Center for Innovative Biomedical Resources, University of Maryland School of Medicine, for their technical support. We also thank Roumen Vesselinov, PhD, Department of Epidemiology and Center for Shock, Trauma, and Anesthesiology Research, University of Maryland School of Medicine, for providing statistical consultation on human studies. Finally, we would like to thank Dr. Stefani Vogel for critically reading the manuscript. The graphic abstract was created with BioRender.com.
Author contributions
Conceptualization, L.Z. and W.C.; Methodology, S.W., Y.Y., A.S., J.Z., J.H., F.C., and A.J.; Investigation, S.W., B.W., R.K., S.S., S.M.J., L.Z., and W.C.; Writing—Original Draft, S.W. and W.C.; Writing—Review & Editing, L.Z. and W.C.; Funding Acquisition, L.Z. and W.C.; Resources, S.W., J.Z., and Z.L.; Supervision, L.Z. and W.C.
Declaration of interests
The authors have declared that no conflict of interest exists.
Published: December 17, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2021.103441.
Contributor Information
Lin Zou, Email: lzou@som.umaryland.edu.
Wei Chao, Email: wchao@som.umaryland.edu.
Supplemental information
Document S1. Figures S1–S3 and Table S1–S3
Data and code availability
-
•
Data - All data reported in the study will be shared by the lead contact upon request - the RNA sequence datasets of mouse and human subjects generated during this study are available at the Gene Expression Omnibus (GEO) site (https://www.ncbi.nlm.nih.gov/geo/) (GEO: GSE133733 and GEO: GSE149764, respectively).
-
•
Code - This study did not generate/analyze any code.
-
•
Any additional information related to the reagents, methods, and data reported in this paper is available from the lead contact upon request.
References
- Akira S., Takeda K. Toll-like receptor signalling. Nat. Rev. Immunol. 2004;4:499–511. doi: 10.1038/nri1391. [DOI] [PubMed] [Google Scholar]
- Alexander M., Hu R., Runtsch M.C., Kagele D.A., Mosbruger T.L., Tolmachova T., Seabra M.C., Round J.L., Ward D.M., O'Connell R.M. Exosome-delivered microRNAs modulate the inflammatory response to endotoxin. Nat. Commun. 2015;6:7321. doi: 10.1038/ncomms8321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambros V. microRNAs: tiny regulators with great potential. Cell. 2001;107:823–826. doi: 10.1016/s0092-8674(01)00616-x. [DOI] [PubMed] [Google Scholar]
- Angus D.C., Poll T.v.d. Severe sepsis and septic shock. New Engl. J. Med. 2013;369:840–851. doi: 10.1056/nejmra1208623. [DOI] [PubMed] [Google Scholar]
- Arroyo J.D., Chevillet J.R., Kroh E.M., Ruf I.K., Pritchard C.C., Gibson D.F., Mitchell P.S., Bennett C.F., Pogosova-Agadjanyan E.L., Stirewalt D.L., et al. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc. Natl. Acad. Sci. USA. 2011;108:5003–5008. doi: 10.1073/pnas.1019055108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boldin M.P., Taganov K.D., Rao D.S., Yang L., Zhao J.L., Kalwani M., Garcia-Flores Y., Luong M., Devrekanli A., Xu J., et al. miR-146a is a significant brake on autoimmunity, myeloproliferation, and cancer in mice. J. Exp. Med. 2011;208:1189–1201. doi: 10.1084/jem.20101823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caserta S., Kern F., Cohen J., Drage S., Newbury S.F., Llewelyn M.J. Circulating plasma microRNAs can differentiate human sepsis and systemic inflammatory response syndrome (SIRS) Sci. Rep. 2016;6:28006. doi: 10.1038/srep28006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casserly B., Phillips G.S., Schorr C., Dellinger R.P., Townsend S.R., Osborn T.M., Reinhart K., Selvakumar N., Levy M.M. Lactate measurements in sepsis-induced tissue hypoperfusion: results from the surviving sepsis campaign database. Crit. Care Med. 2015;43:567–573. doi: 10.1097/CCM.0000000000000742. [DOI] [PubMed] [Google Scholar]
- Chen C., Feng Y., Zou L., Wang L., Chen H.H., Cai J.Y., Xu J.M., Sosnovik D.E., Chao W. Role of extracellular RNA and TLR3-Trif signaling in myocardial ischemia-reperfusion injury. J. Am. Heart Assoc. 2014;3:e000683. doi: 10.1161/jaha.113.000683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z., Hagler J., Palombella V.J., Melandri F., Scherer D., Ballard D., Maniatis T. Signal-induced site-specific phosphorylation targets I kappa B alpha to the ubiquitin-proteasome pathway. Gene Dev. 1995;9:1586–1597. doi: 10.1101/gad.9.13.1586. [DOI] [PubMed] [Google Scholar]
- Chen F., Zou L., Williams B., Chao W. Targeting toll-like receptors in sepsis: from bench to clinical trials. Antioxid. Redox. Signal. 2021;35:1324–1339. doi: 10.1089/ars.2021.0005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denli A.M., Tops B.B., Plasterk R.H., Ketting R.F., Hannon G.J. Processing of primary microRNAs by the Microprocessor complex. Nature. 2004;432:231–235. doi: 10.1038/nature03049. [DOI] [PubMed] [Google Scholar]
- Dhainaut J.F., Shorr A.F., Macias W.L., Kollef M.J., Levi M., Reinhart K., Nelson D.R. Dynamic evolution of coagulopathy in the first day of severe sepsis: relationship with mortality and organ failure. Crit. Care Med. 2005;33:341–348. doi: 10.1097/01.ccm.0000153520.31562.48. [DOI] [PubMed] [Google Scholar]
- DiDonato J., Mercurio F., Rosette C., Wu-Li J., Suyang H., Ghosh S., Karin M. Mapping of the inducible IkappaB phosphorylation sites that signal its ubiquitination and degradation. Mol. Cell Biol. 1996;16:1295–1304. doi: 10.1128/mcb.16.4.1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabbri M., Paone A., Calore F., Galli R., Gaudio E., Santhanam R., Lovat F., Fadda P., Mao C., Nuovo G.J., et al. MicroRNAs bind to Toll-like receptors to induce prometastatic inflammatory response. Proc. Natl. Acad. Sci. 2012;109:E2110–E2116. doi: 10.1073/pnas.1209414109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang S.B., Zhang H.Y., Wang C., He B.X., Liu X.Q., Meng X.C., Peng Y.Q., Xu Z.B., Fan X.L., Wu Z.J., et al. Small extracellular vesicles derived from human mesenchymal stromal cells prevent group 2 innate lymphoid cell-dominant allergic airway inflammation through delivery of miR-146a-5p. J. Extracell Vesicles. 2020;9:1723260. doi: 10.1080/20013078.2020.1723260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y., Chen H., Cai J., Zou L., Yan D., Xu G., Li D., Chao W. Cardiac RNA induces inflammatory responses in cardiomyocytes and immune cells via Toll-like receptor 7 signaling. J. Biol. Chem. 2015;290:26688–26698. doi: 10.1074/jbc.M115.661835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y., Zou L., Yan D., Chen H., Xu G., Jian W., Cui P., Chao W. Extracellular MicroRNAs iInduce pPotent iInnate iImmune rResponses via TLR7/MyD88-dDependent mMechanisms. J. Immunol. 2017;199:2106–2117. doi: 10.4049/jimmunol.1700730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geekiyanage H., Rayatpisheh S., Wohlschlegel J.A., Brown R., Jr., Ambros V. Extracellular microRNAs in human circulation are associated with miRISC complexes that are accessible to anti-AGO2 antibody and can bind target mimic oligonucleotides. Proc. Natl. Acad. Sci. USA. 2020;117:24213–24223. doi: 10.1073/pnas.2008323117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hausdorff W.P., Lohse M.J., Bouvier M., Liggett S.B., Caron M.G., Lefkowitz R.J. Two kinases mediate agonist-dependent phosphorylation and desensitization of the beta 2-adrenergic receptor. Symp. Soc. Exp. Biol. 1990;44:225–240. [PubMed] [Google Scholar]
- Hu W., Xu B., Zhang J., Kou C., Liu J., Wang Q., Zhang R. Exosomal miR-146a-5p from Treponema pallidum-stimulated macrophages reduces endothelial cells permeability and monocyte transendothelial migration by targeting JAM-C. Exp. Cell Res. 2020;388:111823. doi: 10.1016/j.yexcr.2020.111823. [DOI] [PubMed] [Google Scholar]
- Jansen T.C., van Bommel J., Schoonderbeek F.J., Sleeswijk Visser S.J., van der Klooster J.M., Lima A.P., Willemsen S.P., Bakker J., group L.s. Early lactate-guided therapy in intensive care unit patients: a multicenter, open-label, randomized controlled trial. Am. J. Respir. Crit. Care Med. 2010;182:752–761. doi: 10.1164/rccm.200912-1918OC. [DOI] [PubMed] [Google Scholar]
- Jeyaram A., Lamichhane T.N., Wang S., Zou L., Dahal E., Kronstadt S.M., Levy D., Parajuli B., Knudsen D.R., Chao W., Jay S.M. Enhanced loading of functional miRNA cargo via pH gradient modification of extracellular vesicles. Mol. Ther. 2020;28:975–985. doi: 10.1016/j.ymthe.2019.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jian W., Gu L., Williams B., Feng Y., Chao W., Zou L. Toll-like receptor 7 contributes to inflammation, organ injury, and mortality in murine sepsis. Anesthesiology. 2019;131:105–118. doi: 10.1097/ALN.0000000000002706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones A.E., Shapiro N.I., Trzeciak S., Arnold R.C., Claremont H.A., Kline J.A., Emergency Medicine Shock Research Network I. Lactate clearance vs central venous oxygen saturation as goals of early sepsis therapy: a randomized clinical trial. JAMA. 2010;303:739–746. doi: 10.1001/jama.2010.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawai T., Akira S. The role of pattern-recognition receptors in innate immunity: update on Toll-like receptors. Nat. Immunol. 2010;11:373–384. doi: 10.1038/ni.1863. [DOI] [PubMed] [Google Scholar]
- Kolde R., Vilo J. GOsummaries: an R package for visual functional annotation of experimental data. F1000Res. 2015;4:574. doi: 10.12688/f1000research.6925.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong F., Liu Z., Jain V.G., Shima K., Suzuki T., Muglia L.J., Starczynowski D.T., Pasare C., Bhattacharyya S. Inhibition of IRAK1 ubiquitination determines glucocorticoid sensitivity for TLR9-induced inflammation in macrophages. J. Immunol. 2017;199:3654–3667. doi: 10.4049/jimmunol.1700443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kronstadt S.M., Pottash A.E., Levy D., Wang S., Chao W., Jay S.M. Therapeutic potential of extracellular vesicles for sepsis treatment. Adv. Ther. (Weinh) 2021;4:2000259. doi: 10.1002/adtp.202000259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamichhane T.N., Raiker R.S., Jay S.M. Exogenous DNA loading into extracellular vesicles via electroporation is size-dependent and enables limited gene delivery. Mol. Pharm. 2015;12:3650–3657. doi: 10.1021/acs.molpharmaceut.5b00364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee T.J., Yuan X., Kerr K., Yoo J.Y., Kim D.H., Kaur B., Eltzschig H.K. Strategies to modulate MicroRNA functions for the treatment of cancer or organ injury. Pharmacol. Rev. 2020;72:639–667. doi: 10.1124/pr.119.019026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann S.M., Kruger C., Park B., Derkow K., Rosenberger K., Baumgart J., Trimbuch T., Eom G., Hinz M., Kaul D., et al. An unconventional role for miRNA: let-7 activates Toll-like receptor 7 and causes neurodegeneration. Nat. Neurosci. 2012;15:827–835. doi: 10.1038/nn.3113. [DOI] [PubMed] [Google Scholar]
- Li L., Cousart S., Hu J., McCall C.E. Characterization of interleukin-1 receptor-associated kinase in normal and endotoxin-tolerant cells. J. Biol. Chem. 2000;275:23340–23345. doi: 10.1074/jbc.M001950200. [DOI] [PubMed] [Google Scholar]
- Lohse M.J., Benovic J.L., Caron M.G., Lefkowitz R.J. Multiple pathways of rapid beta 2-adrenergic receptor desensitization. Delineation with specific inhibitors. J. Biol. Chem. 1990;265:3202–3211. [PubMed] [Google Scholar]
- Mann M., Mehta A., Zhao J.L., Lee K., Marinov G.K., Garcia-Flores Y., Lu L.F., Rudensky A.Y., Baltimore D. An NF-kappaB-microRNA regulatory network tunes macrophage inflammatory responses. Nat. Commun. 2017;8:851. doi: 10.1038/s41467-017-00972-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta A., Baltimore D. MicroRNAs as regulatory elements in immune system logic. Nat. Rev. Immunol. 2016;16:279–294. doi: 10.1038/nri.2016.40. [DOI] [PubMed] [Google Scholar]
- Merx M.W., Weber C. Sepsis and the heart. Circulation. 2007;116:793–802. doi: 10.1161/CIRCULATIONAHA.106.678359. [DOI] [PubMed] [Google Scholar]
- Nahid M.A., Benso L.M., Shin J.D., Mehmet H., Hicks A., Ramadas R.A. TLR4, TLR7/8 agonist-induced miR-146a promotes macrophage tolerance to MyD88-dependent TLR agonists. J. Leukoc.yte Biol. 2016;100:339–349. doi: 10.1189/jlb.2a0515-197r. [DOI] [PubMed] [Google Scholar]
- Nahid M.A., Pauley K.M., Satoh M., Chan E.K. miR-146a is critical for endotoxin-induced tolerance: implication in innate immunity. J. Biol. Chem. 2009;284:34590–34599. doi: 10.1074/jbc.m109.056317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connell R.M., Rao D.S., Baltimore D. microRNA regulation of inflammatory responses. Annu. Rev. Immunol. 2012;30:295–312. doi: 10.1146/annurev-immunol-020711-075013. [DOI] [PubMed] [Google Scholar]
- O'Neill L.A., Bowie A.G. The family of five: TIR-domain-containing adaptors in Toll-like receptor signalling. Nat. Rev. Immunol. 2007;7:353–364. doi: 10.1038/nri2079. [DOI] [PubMed] [Google Scholar]
- Park C.K., Xu Z.Z., Berta T., Han Q., Chen G., Liu X.J., Ji R.R. Extracellular microRNAs activate nociceptor neurons to elicit pain via TLR7 and TRPA1. Neuron. 2014;82:47–54. doi: 10.1016/j.neuron.2014.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park H., Huang X., Lu C., Cairo M.S., Zhou X. MicroRNA-146a and microRNA-146b regulate human dendritic cell apoptosis and cytokine production by targeting TRAF6 and IRAK1 proteins. J. Biol. Chem. 2015;290:2831–2841. doi: 10.1074/jbc.m114.591420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parrillo J.E. Septic shock--vasopressin, norepinephrine, and urgency. New Engl. J. Med. 2008;358:954–956. doi: 10.1056/NEJMe0800245. [DOI] [PubMed] [Google Scholar]
- Rhee C., Dantes R., Epstein L., Murphy D.J., Seymour C.W., Iwashyna T.J., Kadri S.S., Angus D.C., Danner R.L., Fiore A.E., et al. Incidence and Trends of sSepsis in US hHospitals uUsing cClinical vs Claims dData, 2009-2014. JAMA. 2017;318:1241–1249. doi: 10.1001/jama.2017.13836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhodes A., Evans L.E., Alhazzani W., Levy M.M., Antonelli M., Ferrer R., Kumar A., Sevransky J.E., Sprung C.L., Nunnally M.E., et al. Surviving sepsis campaign: international guidelines for management of sepsis and septic shock: 2016. Crit. Care Med. 2017;45:486–552. doi: 10.1097/CCM.0000000000002255. [DOI] [PubMed] [Google Scholar]
- Robinson M.D., McCarthy D.J., Smyth G.K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roers A., Hiller B., Hornung V. Recognition of endogenous nucleic acids by the innate immune system. Immunity. 2016;44:739–754. doi: 10.1016/j.immuni.2016.04.002. [DOI] [PubMed] [Google Scholar]
- Ru Y., Kechris K.J., Tabakoff B., Hoffman P., Radcliffe R.A., Bowler R., Mahaffey S., Rossi S., Calin G.A., Bemis L., Theodorescu D. The multiMiR R package and database: integration of microRNA-target interactions along with their disease and drug associations. Nucl. Acids Res. 2014;42:e133. doi: 10.1093/nar/gku631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeley J.J., Ghosh S. Molecular mechanisms of innate memory and tolerance to LPS. J. Leukoc. Biol. 2017;101:107–119. doi: 10.1189/jlb.3MR0316-118RR. [DOI] [PubMed] [Google Scholar]
- Shimada B.K., Yang Y., Zhu J., Wang S., Suen A., Kronstadt S.M., Jeyaram A., Jay S.M., Zou L., Chao W. Extracellular miR-146a-5p induces cardiac innate immune response and cardiomyocyte dysfunction. Immunohorizons. 2020;4:561–572. doi: 10.4049/immunohorizons.2000075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer M., Deutschman C.S., Seymour C.W., Shankar-Hari M., Annane D., Bauer M., Bellomo R., Bernard G.R., Chiche J.D., Coopersmith C.M., et al. The third international consensus definitions for sepsis and septic shock (sepsis-3) JAMA. 2016;315:801–810. doi: 10.1001/jama.2016.0287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan S., Yeri A., Cheah P.S., Chung A., Danielson K., De Hoff P., Filant J., Laurent C.D., Laurent L.D., Magee R., et al. Small RNA sequencing across diverse biofluids identifies optimal methods for exRNA isolation. Cell. 2019;177:446–462. doi: 10.1016/j.cell.2019.03.024. e416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taganov K.D., Boldin M.P., Chang K.J., Baltimore D. NF-kappaB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc. Natl. Acad. Sci. 2006;103:12481–12486. doi: 10.1073/pnas.0605298103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thery C., Zitvogel L., Amigorena S. Exosomes: composition, biogenesis and function. Nat. Rev. Immunol. 2002;2:569–579. doi: 10.1038/nri855. [DOI] [PubMed] [Google Scholar]
- Turchinovich A., Tonevitsky A.G., Burwinkel B. Extracellular miRNA: a collision of two paradigms. Trends Biochem. Sci. 2016;41:883–892. doi: 10.1016/j.tibs.2016.08.004. [DOI] [PubMed] [Google Scholar]
- Vickers K.C., Palmisano B.T., Shoucri B.M., Shamburek R.D., Remaley A.T. MicroRNAs are transported in plasma and delivered to recipient cells by high-density lipoproteins. Nat. Cell Biol. 2011;13:423–433. doi: 10.1038/ncb2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner J., Riwanto M., Besler C., Knau A., Fichtlscherer S., Roxe T., Zeiher A.M., Landmesser U., Dimmeler S. Characterization of levels and cellular transfer of circulating lipoprotein-bound microRNAs. Arterioscler Thromb. Vasc. Biol. 2013;33:1392–1400. doi: 10.1161/ATVBAHA.112.300741. [DOI] [PubMed] [Google Scholar]
- Williams B., Neder J., Cui P., Suen A., Tanaka K., Zou L., Chao W. Toll-like receptors 2 and 7 mediate coagulation activation and coagulopathy in murine sepsis. J. Thromb. Haemost. 2019;17:1683–1693. doi: 10.1111/jth.14543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J., Feng Y., Jeyaram A., Jay S.M., Zou L., Chao W. Circulating plasma extracellular vesicles from septic mice induce inflammation via microRNA- and TLR7-dependent mechanisms. J. Immunol. 2018;201:3392–3400. doi: 10.4049/jimmunol.1801008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamin T.T., Miller D.K. The interleukin-1 receptor-associated kinase is degraded by proteasomes following its phosphorylation. J. Biol. Chem. 1997;272:21540–21547. doi: 10.1074/jbc.272.34.21540. [DOI] [PubMed] [Google Scholar]
- Ying W., Riopel M., Bandyopadhyay G., Dong Y., Birmingham A., Seo J.B., Ofrecio J.M., Wollam J., Hernandez-Carretero A., Fu W., et al. Adipose tissue macrophage-derived exosomal mirnas can modulate in vivo and in vitro insulin sensitivity. Cell. 2017;171:372–384. doi: 10.1016/j.cell.2017.08.035. e312. [DOI] [PubMed] [Google Scholar]
- Yoda M., Kawamata T., Paroo Z., Ye X., Iwasaki S., Liu Q., Tomari Y. ATP-dependent human RISC assembly pathways. Nat. Struct. Mol. Biol. 2010;17:17–23. doi: 10.1038/nsmb.1733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu G., Wang L.G., Han Y., He Q.Y. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16:284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Kolb F.A., Jaskiewicz L., Westhof E., Filipowicz W. Single processing center models for human Dicer and bacterial RNase III. Cell. 2004;118:57–68. doi: 10.1016/j.cell.2004.06.017. [DOI] [PubMed] [Google Scholar]
- Zhang Z., Humphreys B.D., Bonventre J.V. Shedding of the urinary biomarker kidney injury molecule-1 (KIM-1) is regulated by MAP kinases and juxtamembrane region. J. Am. Soc. Nephrol. 2007;18:2704–2714. doi: 10.1681/ASN.2007030325. [DOI] [PubMed] [Google Scholar]
- Zhang Z., Ohto U., Shibata T., Taoka M., Yamauchi Y., Sato R., Shukla N.M., David S.A., Isobe T., Miyake K., Shimizu T. Structural analyses of toll-like receptor 7 reveal detailed rna sequence specificity and recognition mechanism of agonistic ligands. Cell Rep. 2018;25:3371–3381. doi: 10.1016/j.celrep.2018.11.081. e3375. [DOI] [PubMed] [Google Scholar]
- Zou L., Feng Y., Xu G., Jian W., Chao W. Splenic RNA and MicroRNA mimics promote complement factor B production and alternative pathway activation via innate immune signaling. J. Immunol. 2016;196:2788–2798. doi: 10.4049/jimmunol.1502106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Document S1. Figures S1–S3 and Table S1–S3
Data Availability Statement
-
•
Data - All data reported in the study will be shared by the lead contact upon request - the RNA sequence datasets of mouse and human subjects generated during this study are available at the Gene Expression Omnibus (GEO) site (https://www.ncbi.nlm.nih.gov/geo/) (GEO: GSE133733 and GEO: GSE149764, respectively).
-
•
Code - This study did not generate/analyze any code.
-
•
Any additional information related to the reagents, methods, and data reported in this paper is available from the lead contact upon request.