Abstract
The effective management of biliary tract cancers (BTCs) has been hampered by limited options for systemic therapy. In recent years, the focus on precision medicine has made technologies such as next-generation sequencing (NGS) accessible to clinicians to identify targetable mutations in BTCs in tumor tissue (primarily) as well as blood, and to treat them with targeted therapies when possible. It has also expanded our understanding of functional pathways associated with genetic alterations and opened doors for identifying novel targets for treatment. Recent advances in the precision medicine approach allowed us to identify new molecular markers in BTCs, such as epigenetic changes (methylation and histone modification) and non-DNA markers such as messenger RNA, microRNA, and long non-coding RNA. It also made detecting these markers from non-traditional sources such as blood, urine, bile, and cytology (from fine-needle aspiration and biliary brushings) possible. As these tests become more accessible, we can see the integration of different molecular markers from all available sources to aid physicians in diagnosing, assessing prognosis, predicting tumor response, and screening BTCs. Currently, there are a handful of approved targeted therapies and only one class of immunotherapy agents (immune checkpoint inhibitors or ICIs) to treat BTCs. Early success with new targets, vascular endothelial growth factor receptor (VEGFR), HER2, protein kinase receptor, and Dickkopf-1 (DKK1); new drugs for known targets, fibroblast growth factor receptors (FGFRs) such as futabatinib, derazantinib, and erdafitinib; and ICIs such as durvalumab and tremelimumab is encouraging. Novel immunotherapy agents such as bispecific antibodies (bintrafusp alfa), arginase inhibitors, vaccines, and cellular therapy (chimeric antigen receptor—T cell or CAR-T, natural killer cells, tumor-infiltrating lymphocytes) have the potential to improve outcomes of BTCs in the coming years.
Keywords: biliary tract cancer, cholangiocarcinoma, mutation, methylation, targeted therapy, immunotherapy, biomarker, circulating DNA
Introduction
Biliary tract cancers (BTCs) are a heterogeneous group of aggressive malignancies that arise from the epithelium of the biliary tract, which includes the bile ducts and gallbladder (1). Cholangiocarcinoma (CCA) includes tumors arising from the bile ducts and are classified anatomically as either intrahepatic and extrahepatic depending on the part of the biliary tract they originated from (2). BTCs are rare and aggressive tumors with a 5-year survival rate for metastatic disease being only 2% (3). In 2017, around 200,000 BTC cases were reported worldwide (4). The incidence and mortality increased by 76% and 65%, respectively, in the last quarter-century (1997–2017). It is difficult to estimate the incidence and mortality of BTC in the United States as epidemiologic data on intrahepatic cholangiocarcinoma (IHC) and hepatocellular cancers (HCC) are reported together, while extrahepatic cholangiocarcinoma (EHC) and gallbladder cancers (GBC) are reported as one group (5). In 2021, it is estimated that there will be approximately 42,000 new cases and 30,000 deaths from HCC & IHC. For EHC & GBC this estimate approximately 12,000 and 4,000, respectively (5). Peri-hilar CCAs are usually classified under EHC.
While surgery is the only curative treatment, unfortunately, the majority of patients with BTC (60%–70%) present with advanced or metastatic disease, and therefore, palliative locoregional and systemic therapy are the only options for treatment (6). Based on the results of the ABC-02 trial, the combination of gemcitabine and cisplatin has become the standard first-line treatment for advanced biliary tract tumors demonstrating a median overall survival of 11.7 months (7). For the modest proportion of patients who go on to receive second-line chemotherapy, the guidelines are less clear about the appropriate therapy for patients who have progressed on Gem-cis and still maintain adequate functional and lab status to tolerate more therapy. Recently, the phase-3 ABC-06 trial showed a survival advantage of FOLFOX over active symptom control (ASC) for patients who had received Gem-cis in the first line (8). A systematic review of second-line therapies in biliary cancers demonstrated a mean PFS of 3.2 months and a mean overall survival (OS) of 7.2 months (9). However, for patients whose tumors harbor targetable mutations, targeted therapy is preferred over chemotherapy in the second line. These include pemigatinib and infigratinib for fusions or mutations in fibroblast growth factor receptor-2 (FGFR2), larotrectinib and entrectinib for neurotrophic tropomyosin receptor kinase (NTRK) fusions, and ivosidenib for isocitrate dehydrogenase 1 (IDH1) (10–14). Pembrolizumab, an immune checkpoint inhibitor (ICI), is recommended for patients with microsatellite instability-high (MSI-H) (15). Unfortunately, only a small proportion of patients have tumors harboring these specific mutations. There is, therefore, an urgent need to expand the arsenal of therapeutic options to treat BTCs and identify biomarkers with reliable prognostic and predictive value. It is imperative that precision medicine strategies should go beyond only somatic mutations to help realize this goal.
Precision Medicine in Biliary Tract Cancer: Current State and Future Directions
Precision medicine refers to tailoring an approach specific to an individual at the molecular level (16). It gained a greater role in oncology over the last two decades, partly driven by easier access to next-generation sequencing (NGS)-based comprehensive genomic profiling (CGP) that enables detection of alterations in the genome, including base-pair substitutions or single nucleotide polymorphisms (SNP), copy number variations (CNV), insertions/deletions, and rearrangements (17). Precision medicine enables us to understand the genomic landscape of BTCs that in turn shed light on the pathways responsible for the malignant transformation and drug resistance and ultimately in effective drug discovery. Moreover, growing evidence shows that IHC, EHC, and GBC have noteworthy differences in their respective genomic landscape (as discussed below).
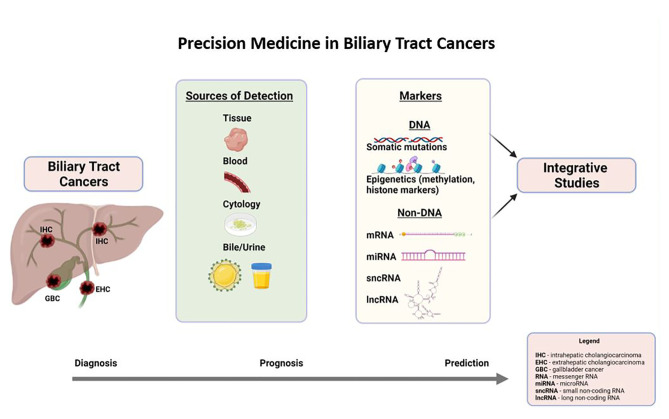
Precision medicine is synonymous with mutational profiling in tumor tissues, precisely for targetable mutations in current practice. In the last 5 years, there has been remarkable progress in identifying other DNA molecular markers such as epigenetic markers and non-DNA molecular markers such as RNA markers (coding and non-coding), metabolites, and protein markers in the tumor tissues. Moreover, the fruits of precision medicine were extended to other sources of tumor genetic material (DNA and RNA) such as blood, bile, urine, and cytology (biliary brushings), which is exciting. This part of the discussion will focus on DNA markers, including somatic mutations, associated signaling pathways, and epigenetic markers (DNA-methylation markers) in tumor tissue, blood, bile, and cytology (as illustrated in Figure 1 ). Non-DNA markers, such as messenger RNA and non-coding RNA, will be discussed briefly. Prognostic and predictive markers are presented in each section if the evidence is available.
Figure 1.
Precision medicine in biliary tract cancers.
Somatic Mutations and Functional Pathways in Biliary Tract Cancer in Tumor Tissue
The most frequently mutated genes in the tumor tissue of BTCs are TP53, KRAS, CDKN2A/B, and SMAD4 (18–24). The same studies reported low prevalence (usually <5%) of targetable mutations such as IDH1/2, FGFR2, BRAF, PIK3CA, and NTRK. It is difficult to estimate the frequency of each genetic alteration for the entire group (EHC, IHC, and GBC) as the study populations were different in these studies. Among BTCs, there are some noticeable differences in the detected mutations, which are summarized in Table 1 (22, 25–34).
Table 1.
Prominent differences in mutated genes among the BTCs.
| Specific BTC | Most likely exclusive | Most frequent | Least frequent |
|---|---|---|---|
| Intrahepatic cholangiocarcinoma | Present—IDH1/2 | BAP1, CDK2NA, ARD1A, FGFR1–3, MET | TP53, PIKCA, HER2 |
| Extrahepatic cholangiocarcinoma | Absent—FGFR1–3, MET EGFR | KRAS b | CDK2NA/B, ARD1A a |
| Gall bladder carcinoma | No exclusive mutations | TP53, PIK3CA, HER2, BRAF, EGFR | BAP1 |
Mutations less frequent in IHC compared with EHC along with KRAS, HER2, and SMAD4.
Mutations more frequent in IHC compared with EHC along with PBRM1.
Targetable mutations with approved therapies (FGFR2 and IDH1/2) are more common in IHC, and IDH1/2 mutations rarely occur in EHC or GBCs (32, 33). Eight genes identified as potential drivers for IHC are TP53, KRAS, IDH1, PTEN, ARID1A, EPPK1, ECE2, and FYN (30). In multifocal metastatic IHC, the SNPs and CNVs in the primary are often concordant with the intrahepatic metastasis (same segment and <2 cm from the primary) and satellite lesions (different segment and >2 cm from the primary) (35). Clinically, this may indicate that multifocal IHCs originate from common progenitor cells and can be considered for surgical resection.
EHCs rarely have targetable mutations and are more likely than other BTCs to harbor a KRAS mutation. Precursor lesions, intraductal tubular papillary neoplasm (ITPN), predominantly originate in intrahepatic ducts and have very few mutations (36). Intraductal papillary neoplasm of the bile duct (IPNB) is usually localized in extrahepatic ducts and has mutational profiles similar to EHC. Overall, the precursor lesions and invasive CCA have overlapping mutations with few exceptions: ROBO2 mutations exist only in invasive CCA, and CTNNB1 are identified in ITPN and IPNB (36). TP53 is the most common mutation in GBC, while PIK3CA is the least prevalent (32, 33). HER2 alterations are common in GBC compared with CCAs. CNVs in CDKN2A, TP53, MDM2 proto-oncogene, and CCD1 genes and HER2 amplifications increased with the development GBC from its precursor lesions (gallstones, low-grade/high-grade dysplasia) (37). These distinct mutational profiles among BTCs can help localize the origin of the tumor and tailor therapy for individual patients.
Mutation detection in tumor tissue or blood helps identify the cell signaling pathways that play a pivotal role in carcinogenesis, drug resistance, and prognosis. Tools such as Ingenuity Pathway Analysis (IPA) were used in previous studies to correlate the genomic variations with specific signaling pathways (27). The major pathways and associated gene alterations in BTCs are as follows: i) FGF pathway with FGFR mutations; ii) mTOR with mutations such as FBXW7, PIK3CA, PTEN, NF1, NF2, PIK3R1, STK11, TSC1, and TSC2; iii) MAP/ERK pathway with KRAS, MYC, BRAF, EGFR, MAP2K1, MAP3K1, and NRAS; iv) DNA damage repair (DDR) pathway with MSH6, BRCA1, BRCA2, BAP1, ATM, MLH1, and MSH2; and v) chromatin remodeling (CR) modification pathway with BAP1, ARID1A, and PBRM1 (27).
About 19% of BTCs have DDR gene alteration mutations and usually co-exist with CR alterations such as ARID1A and PBRM1 mutations (38). These tumors tend to have a high tumor mutational burden (TMB) and a worse prognosis (38, 39). cAMP-dependent signaling activation is another pathway common for all three types of BTCs (25). Nepal et al. classified IHCs based on the three common mutations, IDH, KRAS, and TP53, or undetermined, and showed their potential predictive value in cell lines. IDH-mutated IHCs are rich in metabolic pathways such as glutathione metabolism and the citrate cycle and respond to metabolic modulators such as IDH1 inhibitors. KRAS-mutated IHCs are rich in immune-related pathways and actin cytoskeleton rearrangement and may benefit from microtubule modulators or immunotherapy. TP53-mutated tumors are rich in cell cycle dysregulation (MAPK, WNT, and p53 signaling) and may benefit from topoisomerase inhibitors. In IHC without IDH, TP53, and KRAS mutations, the mTOR pathway is predominant and may benefit from mTOR inhibitors. The relation between IDH-mutated tumors and metabolic enzymes was observed in other studies too, but rigorous preclinical and clinical studies are needed before this classification can be used in clinical practice (32).
Mutational profiling can also aid in identifying the etiology of BTCs. Fluke-positive (Opistharchis viverrine and Clonorchis sinensis related) tumors predominantly have KRAS, TP53, KMT2C (MLL3), ROBO2, RNF43, PEG3, GNAS, SMAD4, BRCA1/2, and HER2 compared with fluke-negative tumors (40–42). BAP1, IDH1/2, and FGFR mutations are frequent in the latter group (41, 42). In IHCs, HBsAg-seropositive patients most likely have TP53 mutations, and KRAS mutations are common in HBsAg-seronegative patients (30). IDH alteration-positive IHCs usually do not have any underlying risk factors such as infections, bile duct cysts, alcohol/tobacco, or Thorotrast exposure (33).
Prognostic and Predictive Value of Somatic Mutations in Biliary Tract Cancer
The somatic mutations with potential prognostic value are summarized in Table 2 (20, 22, 26–28, 32, 42–45). Detection of FGFR and/or IDH1 in IHC and PBRM1 and/or BAP1 in EHC is associated with a good outcome. Alternatively, PIK3CA in EHC and specific mutations in IHC, such as TP53, KRAS, CDK2NA/B, EGFR, and PBRM1, are bad prognostic markers. Mutations with prognostic value are not well established for GBCs. CCAs with mismatch repair (MMR) deficiency and low mesothelin levels have poorer outcomes compared with those with MMR proficient and high mesothelin expression (median OS: 14.5 vs. 30.0 months, p = 0.05) (46). In the same study, CCA with MMR deficiency alone had a trend toward the worst prognosis (median overall survival or OS: 19.2 vs. 28.1 months, p = 0.07). In IHC, Zhu et al. have demonstrated an interesting relation between mutations and pathological features: a) IDH1 alone or IDH/IDH2 is associated with bilobar invasion of the tumor; b) KRAS alone with positive margins (R1) and direct invasion of surrounding organs and KRAS, NRAS, or BRAF with R1; and c) NRAS with intrahepatic metastasis (47).
Table 2.
Somatic mutations and their prognostic value in biliary tract cancers.
| Tumor group | Worse | Better | No effect |
|---|---|---|---|
| Biliary tract cancers |
ARID1A
KRAS a Del at 7q22.1 High TMB |
FGFR2
PRB1A |
CDKN2A, CDKN2B, IDH, PIK3CA, MYC alteration |
| Intrahepatic cholangiocarcinoma |
TP53
KRAS CDK2NA/B EGFR BAP1 a ARID1A a PBRM1 BRCA1/2 |
FGFR
(point mutations and translocations) IDH1 b |
CDK2NA
BRAF PTEN HER2 |
| Extrahepatic cholangiocarcinoma |
PBRM1
BAP1 c |
PIK3CA |
TP53
KRAS CDK2NB A1RD1 PBRM1 SMAD4 |
| Gallbladder cancers | None identified | None identified |
TP53
KRAS CDK2NA/B ARID1A IDH1 PIK3CA SMAD4 MYC |
TMB, tumor mutational burden.
No effect in one study and bad prognosis in others.
No effect in some studies and good prognosis in others.
PFS is different but not OS.
Studies have shown that patients with targetable mutations have better response and survival advantages if treated with the appropriate targeted therapy (20, 21). Therefore, it is crucial to attempt to identify targetable mutations when possible. For patients with no targetable mutations, there are no established mutations with predictive value. EHCs with overexpression of programmed cell death protein 1 (PD-1)/programmed death-ligand 1 (PD-L1) and higher lymphocyte infiltration respond better to ICIs (48). There are no mutations that can predict response in patients receiving chemotherapy either. With limited treatment options and poor outcomes in BTCs, this area needs further study. One area where this would be used is in the neoadjuvant setting where the lack of treatment response to chemotherapy could be identified upfront, and alternate therapy such as targeted agents or immunotherapy could be used instead (49).
DNA-Methylation Markers and Integrative Approach in Biliary Tract Cancer
Epigenetic changes are modifications in the genome not involving the nucleotide sequence (50). They can be a) DNA hypo- or hypermethylation; b) histone modification including methylation, phosphorylation, acetylation, and SUMOylation; c) CR; and d) RNA-associated silencing. In this section, the focus will be on DNA-methylation markers. Comparing the methylation profiles of malignant vs. normal or precancerous tissues can provide insight into pathways of malignant transformation and cells of origin in BTCs (36). Integrating methylation profiles with clinical and/or mutational profiles has prognostic and predictive value (28, 29, 42, 51).
In CCAs, frequently methylated genes are APC, DAPK, E-cadherin/CDH1, GSTP, RASSF1A, hMHL1, MGMT, p15INK4b , p16INK4a , p14ARF , p73, 14-3-3 sigma, SOCS-3, EGFR, and RAR-β (52, 53). RASSF1A, HOXA1, HPP1, CDH1, and NEUROG1 are predominantly methylated genes in EHC, while CHFR, GSTP1, IGF2, MGMT, MINT31, p14, and RBP1 are predominant in IHC (52, 54). MGMT promoter methylation level is high, and protein expression is low in IHC compared with normal tissues (55). MGMT inhibition promotes cell proliferation via p21, P27, and Cyclin E. Low-expression MGMT in tumor tissues is associated with worse clinicopathological features and outcomes. Similarly, promoter methylation of APC, p16, and TIMP3 have a good prognostic value in IHC (56). EHCs with lymph node metastasis have higher CpG island loci and hypermethylation of T1G1 gene (54). A data mining study in an extensive database identified genes associated with methylation pathways in GBC (FGA, F2, HAO1, CFH, PIPOX, ITIH4, GNMT, MAT1A, MTHFD1, HPX, CTH, EPHX2, HSD17B6, AKR1C4, CFHR3, ENNP1, and NAT2) (57). Among them, the methylated genes were FGA, CFH, F2, HPX, and PIPOX. When validated in the clinical samples, FGA, CFH, F2, HPX, and PIPOX were high compared with controlled tissues but not significantly different. Multiple studies identified numerous genes methylated in GBC, and a set of genes consistently represented in these studies is as follows: APC, SHP1, 3-OST-2, FHIT, p16, SEMA3B, and CDH13 (58).
Integration of the genomic (somatic mutations, CNVs, and gene expression) and epigenomic data (methylation) has been attempted to better understand the disease processes. In such studies, the tumors are classified into groups, or clusters, to identify high-risk populations. In one such study, IHCs were divided into four clusters using the iClusterPlus platform (42). Cluster 1 mainly had fluke-positive IHCs rich in TP53, ARID1A, BRCA1/2, and H3K27me3 promoter mutations; had high expression of HER2, AKT1, and EZH2; and low expression of TET2 and CpG island hypermethylation. Clusters 2 and 3 had a mix of fluke-positive and fluke-negative tumors. Cluster 2 tumors were rich in TP53 mutations and had high expression of CTNNB1, WNT5B, and AKT1. Cluster 3 was rich in immune-related pathways and had the highest CNVs. Cluster 4 had tumors rich in BAP1, IDH1/2, and FGFR mutations; high FGFR1–4 gene expression; and CpG shore island hypermethylation. Cluster 4 had a better prognosis over clusters 1–3. Qui et al. divided BTCs into six clusters based on the degree of methylation (51). The cluster with a high methylation rate had the highest CNV and a worse prognosis. Lower methylation rate tumors had higher BCR/TCR diversity, immune cell infiltration, and PD-L1 and cytotoxic T-lymphocyte-associated protein 4 (CTLA-4) mRNA expression and, hence, potentially respond to ICI better. Interestingly, hypermethylated genes participate in DNA-binding transcription activity, and hypomethylated genes are involved in transmembrane receptor and ion binding.
In another study, the authors integrated gene expression data, signaling pathways, chromosomal abnormalities, mutations, and poor prognosis signatures derived from previous studies in HCC and tyrosine kinase inhibitors in CCA (28, 59–61). They divided the IHCs into two classes: “proliferative” and “inflammatory”. The proliferative class accounted for 62% of the study population. These tumors had KRAS and BRAF mutations; more oncogenic pathways such as RAS/MAPK and MET; poor prognostic signatures; specific CNVs, amplifications at 11q13.2, deletions at 14q22.1; and moderate to poor differentiation with intraneural invasion on histology and poor survival. In the inflammatory class, tumors had activated inflammatory signaling pathways, overexpression of cytokines (interleukin or IL-3, IL-4, IL-6, IL-10, IL-17A, and CCL19), and STAT 3 activation; no poor prognostic signatures, KRAS or BRAF mutations; and well-differentiated tumors with favorable features and good survival.
IDH-mutant CCAs have a distinct molecular profile, according to a study that integrated genomic (somatic mutations and CNV) and epigenomic data (DNA-methylation) with mRNA expression (29). They have higher mitochondrial DNA copy numbers, low expression of chromatin modifiers, and elevated expression of mitochondrial genes compared with IDH-WT tumors. Comparing 103 IHCs with matched controls, Zhou et al. identified three pathways (transforming growth factor-b/Smad signaling pathway along with known Ras/phosphatidylinositol-4,5-bisphosphate 3-kinase signaling and p53/cell cycle signaling) along with genes involved in epigenetic regulation and oxidative phosphorylation more frequent in IHCs (30). Using samples from patients in Chile (which has the highest incidence of GBC), Brägelmann et al. were able to identify methylation changes during the progression from gallstone disease to dysplasia and then to GBC (37). They identified stages of progression through a sequence of early (gallstone disease and low-grade dysplasia), intermediate (high-grade dysplasia), and late (GBC) stages. In particular, methylation of genes involved in WNT signaling, Hedgehog signaling, and tumor suppression increased with tumor grade. CNVs also increased along with tumor grade (as mentioned above).
Somatic Mutations in Blood, Bile, and Cytology Specimens in Biliary Tract Cancer
Circulating cell-free DNA (cfDNA) usually refers to nucleic acids (fragments of DNA) detected in the peripheral blood released secondary to apoptosis or necrosis (62, 63). In cancer patients, a significant proportion of cfDNA comes from normal cells of the body, a small part related to tumors, coming from primary tumors, metastatic sites, or circulating tumor cells (CTC), and is called circulating tumor-DNA (ctDNA) (64). In the last 5 years, liquid biopsies (detecting mutations in cfDNA) have gained popularity in oncology practice as they may be the only source for tumor DNA in certain cancers such as BTCs, where getting adequate tissue for sequencing is difficult. Among BTCs, acquiring tissue for EHCs is more difficult than for IHC and GBCs. In BTC, limited studies in this area indicate a high mutation detection rate (74%–85%) regardless of whether patients are treatment naive or on treatment at the time of sample collection and have reliable concordance with mutations in the tumor tissue (50%–74%) (21, 65–67). Concordance in the matched samples (blood and tissue from the same patient) depends on the source of the tissue (higher with metastatic vs. primary site) and also the tumor type [higher in IHC (92%) compared with EHC or GBC] (67).
The cfDNA has a dependable sensitivity and specificity in diagnosing BTC (68, 69). The frequency of the detected mutations in cfDNA in BTC patients is the same as those detected in the tumor tissue (21, 67). Due to limited studies, it is hard to conclusively identify cfDNA mutational profile differences between IHC, EHC, and GBCs. In one study with 69% IHCs, 18% EHC, and 13% GBC, FGFR and ARID1A were the most prevalent alteration in IHC and EHC, respectively. CDK6, APC, and SMAD4 alterations were common in GBCs. TP53 and KRAS were the most prevalent in all three groups (70). BTCs in younger patients (<50 years) have more FGFR2, PIK3CA, MET, and BRAF mutations compared with older patients (≥50 years) (70). TP53 (67% vs. 35%, p = 0.6) was predominant in the older population.
Detection of clinically relevant and targetable mutations for approved therapies is better in ctDNA than tissue, and it ranges between 30% and 40% in advanced CCAs (65, 71). In a study with 71 BTCs with detectable ctDNA, 75% of the patients had targetable mutations (both on- and off-label for BTC), indicating that this will be a valuable tool in the future (21). Variant allelic frequency (VAF) represents the percentage of mutant reads divided by the total number of reads coverage at a specific genomic position (72). VAF in ctDNA at the baseline correlates well with the tumor burden in CCA but does not have any predictive or prognostic value (65, 67). cfDNA integrity (ratio of ALU247 and ALU115) had a reliable diagnostic and prognostic value (unfavorable clinicopathological features) in GBC (68). Short segments of DNA originally characterized by Arthrobacter luteus (Alu) restriction endonucleases are ALU units (69).
CTC detection is low in CCA, especially in early-stage (stages I and II) tumors, and detection rates vary with the thresholds for positive tests (CTC ≥ 2 vs. CTC ≥ 5). In a study reported in 2016, detection rates for the threshold of ≥2 and ≥5 were 17% and 9%, respectively, and most of them were advanced-stage cancers (73). Both the thresholds had significant prognostic value in the entire group. In subgroup analyses, both maintained prognostic value in metastatic CCA and EHC. A threshold of ≥5, in addition, had prognostic value in the IHC group and trended toward significance in non-metastatic CCAs. Li et al. proposed the concept of ctDNA fingerprinting in eight tumor types, including CCA (74). The authors began with whole exosome sequencing (WES) in patients before the treatment (surgery or systemic therapy or locoregional therapy) appropriate to the disease. Then, a patient-specific panel with high-frequency clonal population clusters was designed and followed in the patients after treatment. Two entities followed in subsequent tests were ctDNA content fraction (CCF) and the fold change in CCF. The CCF is higher, and the fold change increased in patients who progress compared with patients with stable disease or response. In patients receiving selective internal radiation, a reduction in the CNVs was observed in patients with IHC (75). Platinum-based therapy showed a potential benefit in BTCs with DDR gene mutations in a small study (65). The benefit was seen in partial response rate, radiological response, and increased overall survival.
In one of the earlier studies that proved the feasibility of detecting DNA-methylation markers in CCA, a four-gene panel with HOXA1, PKRCB, CYP26C1, and PTGDR was proposed to differentiate CCA from healthy controls with sensitivity/specificity of 83%/93% (76). A couple of years later, higher cfDNA methylation levels of OPCML and HOXA9 genes, individually or together, also proved to have a good diagnostic value in distinguishing CCA from benign biliary diseases (77). The sensitivity/specificity of a combined marker is 62%/100%. Non-specific methylation levels in cfDNA do not have good diagnostic utility in GBC, with sensitivity/specificity around 55%/50% (68). We need robust studies to take the methylation markers in cfDNA to clinical practice.
Examining cytology specimens derived from endoscopic retrograde cholangiopancreatography (ERCP) to identify malignant cells is the standard test to diagnose malignancy. The addition of fluorescence in situ hybridization (FISH) and mutational profile testing to cytology helps to diagnose BTCs in patients who present with biliary strictures (78–81). Mutational profiling with KRAS and other prominent tumor-suppressor genes alone can increase the sensitivity by 56% and diagnostic yield to 100% when done in conjunction with cytology (82).
DNA-methylation markers in the biliary brushings can also be a tool for diagnosing CCA, and many studies reported using four to six gene panels for this purpose. Andersen et al. suggested that a four-gene panel, namely, CDO1, CNRIP1, SEPT9, and VIM, detected in 45%–77% of the samples had a sensitivity and specificity of 85% and 98%, respectively (83). Prachayakul et al. reported using the methylation index (MI) of two genes, HOXA1 and NEUROG1 (84). Each has better sensitivity and accuracy than cytology and CA 19-9 in differentiating CCA from benign diseases. The methylation markers were positive even in patients with negative cytology and normal CA 19-9 levels. The combined sensitivity/accuracy of MI, cytology, and CA 19-9 was 97%/91%. Yang et al. reported good sensitivity and specificity of methylated EMX1 for differentiating EHC from primary sclerosing cholangitis (PSC) and non-PSC controls (85). In the same study, HOXA1 had a sensitivity of 100% differentiating CCA from non-PSC controls, but not in PSC controls. Parsi et al. showed that 80% (N = 10) of NTCs have positive methylation for one of three genes, CCND2, NPTX2, and TFPI2 (86).
The ctDNA isolated from the bile using the techniques employed for plasma cfDNA (NGS) has shown some encouraging results (87, 88). In bile, mutations can be detected in 60%–75% of BTC patients, with specificity close to 100%. Concordance rates between bile ctDNA and tissue DNA are higher than plasma cfDNA (88%). p16INK4a is a tumor-suppressor gene frequently associated with many cancers, including CCA (89). In bile samples, promoter methylation of p16INK4a is positive in over 50% of CCA compared with just 6% in benign diseases (cholelithiasis) and normal bile ducts (90). Concordance with the tissue samples for p16INK4a promoter methylation was as high as 86%. Just as with mutation profiling of biliary brushings, few panels for methylation markers in bile have been reported in the literature. In EHC, methylation of the five-gene panel with CCND2, CDH13, GRIN2B, RUNX3, and TWIST1 has a better sensitivity than cytology (83% with methylation markers vs. 46% in cytology) (91). Similarly, Zhang et al. proposed a six-gene panel (DKK3, p16, SFRP2, DKK2, NPTX2, and ppENK) to diagnose pancreatobilary cancers with sensitivity/specificity/accuracy of 77.27%/77.78%/77.50% (92). In summary, expanding precision medicine to bile and cytology brushings can improve diagnostic testing and help identify better prognostic and predictive biomarkers.
Non-DNA Molecular Markers in BTC Tissues
Non-DNA molecular markers include messenger RNA (mRNA), non-coding RNA (ncRNA), proteins, and metabolites. mRNA and ncRNA are within the scope of precision medicine. The ncRNA refers to RNA that is not translated into protein and can be detected in the blood and is being extensively studied in tumors for diagnostic and prognostic purposes (93, 94). They can be classified bases on their size into a) long non-coding RNAs or lcRNAs that are more than 200 bases; and b) small non-coding RNAs or sncRNAs that have up to 200 bases which are further classified into small nucleolar RNAs (snoRNAs), small nuclear RNAs (snRNAs), Piwi-RNAs (piRNAs), and microRNA or miR. Extracellular vesicles are lipid membrane-bound spheres released from cells into body fluids (blood, saliva, bile, and urine) comprising materials shed from the cells, including proteins, nucleic acids, and metabolites (95, 96). They are a good source for molecular markers in the blood. They are of three kinds: microvesicles, exosomes, and apoptotic bodies (97, 98).
Among the RNA molecular markers, miR is the preferred biomarker among all the ncRNAs in oncology as it has a proven role in pathways implicated in malignant transformation and can be detected/measured easily in blood, bile, and other body fluids; it is a more stable nucleic acid making it more reliable (94, 99–101). Even though it is still in the experimental stage, the current landscape of prominent non-DNA markers in blood and urine is summarized in Table 3 (44, 102–128).
Table 3.
Non-DNA markers in biliary tract cancers.
| Source | Non-DNA marker |
|---|---|
| Tissue | miR-22, -125a/b, -127, -199a, -376a/c, -142-3p, -25, -15a/b, -193, -17-5p, -374, -106a/b, -224, -130b, -19a, -331, -324-5p, -20, 17-3p, -223, -15b, -103, -98, -204, -338, -198, -302d, -328, -337, -302b, -184, -320, -371, -185, -222, -214, -373, -145, -200c, let-7a, let-7b, -21, -135b, -122, -27a, -29a, -429, -24, -203, -29b, -20a/-20b, -93, -30e, -30b, -151-3p, -10a, -181a, -96, -663b, -103, -221, -107, -424, -340, -451, -145, -99a, -630, let-7c, -144, -100, -139-5p, -337-3p, -1, -126, -376c, -517c+-519a, -520e, -30c, -96, -30b, -100, -145 lncRNA PANDAR 141, AFAP1‐AS1 140, CCAT-1, NEAT-1, MALT-1, CPS1-IT1 Prognostic value: miR-192, -675-5p, -652-3p, -338-3p, -126, -21, 192, -21, -214, -151-3p, -373 |
| Blood | In circulating-free RNA: mRNAH/PSC/UC: CMIP, GAD1, NME1, CSD1, NME1, CDS1, CK1B, CKS1B; miR-21HI/b, -221H/b, -194 with miR483-5pH/b, -222 with miR483-5pPSC, -122H, -192hI/Liverfluke, 26aH, -150H/I, -106AHL, -26aPSC, -122PSC, -1281PSC, -126PSC, -30bPSC
In EV: miR-604H, -1224H, -200c-3p, -96-5p, -151a-5p, -191-5p, -4732-3p, -551BH/PSC/UC, -200a/c-3p: lcRNA—LOC100134868, LOC643955. PTTg3P Prognostic value: seven mRNA signatures, namely, CD36, GGCX, UBASH3B, DBN1, PTTG1, CCNA2, and SPATS2. In resected tumors, postop decline in total miR level (by 20), miR-106a, -192, -26a, -150. miR-200a/c-3p (in EV) |
| Bile | Circulating-free miR9, -145, -105, -147b,let-7f-2,let-7i,-302c,199a-3p, -222
a
, -942, -640
PSC, -412PSC, -1537PSC, -3189PSC, -30d-5pBenign, -92a-3pBenign; higher methylation rates of miRNA-1247 and -200a In EV: miR-191, -486-3p, -1274b, -16, -484 |
| Urine | Circulating-free mRNA: UBE2C, SERPINB1; miRNA 21 and 192H
In EV: miR-483; snRNA—RNU11: miscRNA—LOC257358: vtRNA—RNA1-1 |
H, compared to healthy; PSC, compared to primary sclerosing cholangitis; UC, ulcerative colitis; I, exclusive for IHC; B, include all BTCs; Benign, compared to benign; mRNA, messenger RNA; miR, micro RNA; lncRNA, long non-coding RNA; EV, extra vesicles; miscRNA, miscellaneous RNA; vtRNA, vault RNAs; snRNA, small nuclear RNA.
miR122 is lower compared with PSC and higher in healthy.
In summary, NGS of the tumor tissue (when available) is widely used to identify the targetable mutations in the current clinical practice for managing BTCs. There is a strong clinical need to develop novel biomarkers. On the clinical side, we may see the expansion of precision medicine in three different directions soon: firstly, identifying more reliable DNA-molecular markers such as DNA-methylation markers and non-DNA markers such as mRNA, miRNA, and lncRNA; secondly, making detection of the molecular markers in non-invasive sources of genetic material such as blood (ctDNA and EV), bile, and urine feasible with acceptable reliability and accessible in day-to-day clinical practice; and finally, integrating different molecular makers from different sources and stratifying in high prognostic and predictive value. Non-invasive sources will also help in monitoring the treatment response. Novel techniques to detect ctDNA such as surface-enhanced Raman scattering (SERS) and biosensors are being studied, which may compete and replace traditional techniques (polymerase chain reaction and NGS) (129–133).
Treatment Updates
Current Management of Biliary Tract Cancers
Gemcitabine/cisplatin (GC) combination is the systemic therapy of choice in advanced BTC, and capecitabine is the recommended adjuvant treatment (in case of negative margins) (134, 135). The NCCN guidelines recommend FOLFOX based on the ABC-06 trial, and the difference in OS was minimal even though statistically significant (6.2 months in the FOLFOX group vs. 5.3 months in the supportive therapy group) (136). GC and the nab-paclitaxel combination seem promising, especially in the first line (49, 137). In a recently presented trial, nanoliposomal irinotecan (nal-IRI) plus 5-FU/LV was compared with 5-FU/LV in a phase IIb randomized trial (138). The PFS (7.1 months for the combination vs. 1.4 months, p = 0.019), OS (8.6 vs. 5.5 months, p > 0.001), and ORR (14% vs. 5.8%) were better for the combination therapy which is encouraging. The serious adverse events (≥ grade 3) were very high in the combination group (77% vs. 31%).
Targeted Therapy
In FGFR2 fusions or rearrangements, pemigatinib and infigratinib are recommended (139). IDH1 or IDH1 mutant BTCs can be treated with ivosidenib (14). Neurotrophic tyrosine receptor kinase (NTRK) gene rearrangement-positive tumors respond to NTRK inhibitors such as larotrectinib and entrectinib (10, 11). Dabrafenib and trametinib combination was recently approved for BRAF V600E mutation (140, 141). The list of targeted therapies is summarized in Table 4 . The published trials are cited, and the identifier is added to unpublished trials.
Table 4.
Targeted therapy in biliary tract cancer.
| Target | Drug | Approved | Early success | Ongoing |
|---|---|---|---|---|
| FGFR2 | Pemigatinib | Pemigatinib** | Pemigatinib + GC (NCT03656536)III * | |
| Infigratinib | Infigratinib** | Infigratinib vs. gemcitabine cisplatin (NCT03773302)III * | ||
| Futibatinib | Futibatinib** II (142) | Futibatinib vs. GC (NCT04093362)* III | ||
| Derazantinib | Derazantinib** I/II (143) | Ponatinib (NCT02265341)** II | ||
| Erdafitinib | Erdafitinib** II (144) | Deibo 137 (NCT03834220)** II | ||
| IDH1 | Ivosidenib | Ivosidenib** | ||
| LY3410738 (for IDH1 R132) | LY3410738 (NCT04521686)** I | |||
| Olutasidenib (FT-2102) | FT-2102 vs. FT-2102 and nivolumab (NCT03684811)** I/II | |||
| IDH1/2 | Dasatinib | Dasatinib (NCT02428855)** II | ||
| AG-881 | AG-881 (NCT02481154)** I | |||
| IDH2 | Enasidenib (NCT02273739)** I/II | |||
| NTRK | Larotectinib | Larotectinib** | ||
| Entrectinib | Entrectinib** | |||
| Protein kinase CK2 inhibitor | Silmitasertib | Silmitasertib + GC* I/II (145) | ||
| VEGF inhibitor | Regorafenib | Regorafenib** I (146) | Regorafenib (NCT02053376)** II | |
| Surufatinib | Surufatinib** II (147) | |||
| HER2 inhibitor | Zanidatamab | Zanidatamab** I/II (148, 149) | Zanidatamab (NCT04466891)** II | |
| Neratinib | Neratinib** II (150) | |||
| Trastuzumab | Trastuzumab + pertuzumab** II (151) | |||
| Trastuzumab + mFOLFOX** II (152) | ||||
| Dickkopf-1 (DKK1) | DKN-01 | DKN-01 + GC* I (128) | DKN-01 + N (NCT04057365)** II | |
| ATR inhibitor | Ceralasertib (Ce) (AZD6738) | Ce + D (NCT03780608)** II | ||
| Ce + olaparib (NCT03878095)** II | ||||
| BRCA1/2 inhibitor | Olaparib | Olaparib (NCT04042831)* II | ||
| Niraparib | Niraparib (NCT03207347)* II | |||
| CDK4/6 | Abemaciclib | Abemaciclib (NCT04003896)** II | ||
| HDAC | Entinostat | Entinostat + nivolumab (NCT03250273)** II |
I, phase 1 trials; II, phase 2 trials; III, phase 3 trials; *first line; **second line or more; NTRK, neurotrophic tyrosine receptor kinase; ATR, ataxia telangiectasia and Rad3-related; HDAC, histone deacetylase; BRCA, breast cancer gene; VEGF, vascular endothelial growth factor; HER2, human epidermal growth factor receptor 2; IDH, isocitrate dehydrogenase; FGFR, fibroblast growth factor receptor.
Immunotherapy
Treating disease by modulating (suppressing or activating) the immune system refers to immunotherapy (153). ICI is the most commonly used for immunotherapy in oncology and is used in almost all solid and hematological malignancies. In clinical practice, after progression in GC, targeted therapy or immunotherapy is considered if feasible. In patients with microsatellite instability (MSI-H), pembrolizumab is recommended based on the KEYNOTE-158 study with 22 advanced BTCs. The objective response rate was observed in 41% of BTCs with two patients with complete response (154). Correlation between TMB and response to ICI is difficult to assess as none of the BTCs enrolled in KEYNOTE-158 had higher TMB (≥10 mutations/Mb). PDL-1 overexpression does not seem to have any effect on responses either (155, 156). Nivolumab and ipilimumab combination did not have encouraging results either, with just 23% ORR (157). The current NCCN recommendation is to use nivolumab for refractory advance BTCs that are not MSI-H irrespective of TMB, and PDL-1 expression is weak (156).
Currently, many trials are combining ICI with either chemotherapy or targeted therapy or locoregional therapies such as transarterial chemoembolization (TACE), cryotherapy, radiofrequency ablation (RFA), and radiotherapy (as reported in Table 5 ). Traditional ICIs target either PD-1 or PD-L1 or CTLA-4. Bintrafusp alfa, a bifunctional antibody targeting TGF-β (transforming growth factor-β) and PD-L1, had success in the early phase trials (162). Vaccines with individual peptides such as MUC1, Wilms tumor 1 (WT1), or multiple in treating advanced BTC peptides had limited success previously but not enough to pursue it forward (170–175). There is some evidence that chimeric antigen receptor-modified T cells (CART) against epidermal growth factor receptor (EGFR) and CD133 and tumor-infiltrating lymphocytes are effective in managing refractory CCA (176–178).
Table 5.
Immunotherapy in biliary tract cancers.
| Drug class | Drug | Approved | Early success | Ongoing trials |
|---|---|---|---|---|
| Immune checkpoint inhibitor | Pembrolizumab (P) | P** in MSI-H | P + lenvatinib** II (158) | P + GC vs. placebo + GC* III (159) |
| P + sargramostim (NCT02703714)** II | ||||
| P + Olaparib (NCT04306367)** II | ||||
| XmAb®22841 monotherapy vs. XmAb®22841 + P (NCT03849469)** I | ||||
| P + XL888 (Hsp90 inhibitor)** I (NCT03095781) | ||||
| P + Peginterferon alpha-2b** II (NCT02982720) | ||||
| Nivolumab (N) | N** | N + GC vs. N + ipilimumab* II (160) | N + S-1+ G (NCT04172402)* II | |
| N + GC** I (161) | N + rucaparib (NCT03639935)* II | |||
| Nal-irinotecan/5-fluorouracil/leucovorin + N (NCT03785873)** I/II | ||||
| N + high-dose XRT vs. N + ipilimumab and high-dose XRT (NCT02866383)** II | ||||
| N + TPST-1120 (PPARα antagonist)* I (NCT03829436) | ||||
| Bintrafusp alfa | Bintrafusp alfa** II (162) | Bintrafusp alfa + GC* II/III (163) | ||
| Durvalumab (D) ± tremelimumab (T) | D ± T + GC (164) | D + GC (NCT03875235)* III | ||
| D and D + T** I (165) | D + olaparib (NCT03991832)** II | |||
| D + AZD6738 (ATP inhibitor) + olaparib (NCT04298021)** II | ||||
| D vs. D + T (NCT04238637)** II | ||||
| D + guadecitabine (NCT03257761)** I | ||||
| T + TACE or RFA** II (166) | D + T vs. D + T + TACE vs. D + T + RFA vs. D + T + cryotherapy (NCT02821754)** II |
|||
| D + SNDX-6352 (NCT04301778)** II | ||||
| D + T + XRT (NCT03482102)** II | ||||
| Toripalimab (To) | To + GC* (167) | |||
| To + lenvatinib + GEMOX (168) | To + GEMOX (NCT04191343)* II | |||
| To (IV) + HAIC infusion of oxaliplatin, 5-FU, and Bev (NCT04217954)* II | ||||
| Camrelizumab (C) | C + cryoablation (NCT04299581)** II | |||
| C + radiotherapy (NCT03898895)* II | ||||
| Sintlimab | Sintlimab (PD-1) + anlotinib (AL3818) (169) | |||
| STI-3031 | STI-3031 (NCT03999658)** II | |||
| SHR-1210 | SHR-1210 + capecitabine (NCT04295317)** II | |||
| Avelumab | Avelumab + regorafenib (NCT03475953)** I/II | |||
| Arginase inhibition | NCB001158 | NCB001158 + GC* I/II (13) | ||
| Natural killer (NK) cells | Allogenic NK cell | Allogenic NK cell (SMTNK) + P (NCT03937895)** I/I | ||
| Autologous cells | T cells | Tumor-infiltrating lymphocytes with high-dose aldesleukin** II (NCT03801083) | ||
| Central memory T cells + standard therapy after resection* II (NCT03820310) | ||||
| CAR-T cell therapy | CAR-T | MUC1 CAR-T cell therapy + fludarabine + cyclophosphamide* I/II (NCT03633773) | ||
| Anti-HER2 CAR-T cell* I (NCT04660929) | ||||
| Oncolytic virus | Adenovirus | Virus encoding immunostimulatory TMZ-CD40L and 4-1BBL with GC* I/II (NCT03225989) |
I, phase 1 trials; II, phase 2 trials; III, phase 3 trials; *, first line; **, second line or more; GC, gemcitabine/cisplatin; CAR-T, chimeric antigen receptor—T cell; MSI-H, microsatellite instability—high.
Conclusion
BTCs are rare cancers with a high mortality rate and limited systemic options. It is important to recognize the significant differences in the genomic landscape of IHC, EHC, and GBC. As noted above, we need to start investing in DNA markers other than somatic mutations such as methylation markers and non-DNA markers (miRNA, mRNA, and lncRNA) to help diagnose, screen, and predict the treatment response in BTCs. There is also a critical need to explore and refine biomarkers (DNA and non-DNA) in blood, bile, and cytology specimens, as they are more accessible than tissue in BTCs. The low prevalence of MSI-H and targetable mutations in BTC restricts the use of available/approved therapies. The success of drugs targeting new targets (such as PKCK2, HER2, and DKK1) and newer drugs for older targets (FGFR inhibitors such as futibatinib, derazantinib, and erdafitinib) is encouraging, and we may have a host of new drugs in the next 3–4 years. Traditional PD-1, PD-L1, and CTLA-4 inhibitors are being tested in various ways, as monotherapy, in combination with targeted therapy or chemotherapy, or locoregional therapies for treating BTCs, which might open up a whole new arsenal of drugs to choose. Newer immunotherapies such as bintrafusp alfa, arginase inhibitors, and T-cell-mediated treatments can further expand the horizon in the management of BTCs.
Author Contributions
Conceptualization: AsM. Writing—original draft preparation: AsM. Writing—review and editing: ArM, EW, and AT. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
- 1. Valle JW, Kelley RK, Nervi B, Oh DY, Zhu AX. Biliary Tract Cancer. Lancet (2021) 397(10272):428–44. doi: 10.1016/s0140-6736(21)00153-7 [DOI] [PubMed] [Google Scholar]
- 2. Hennedige TP, Neo WT, Venkatesh SK. Imaging of Malignancies of the Biliary Tract- an Update. Cancer Imaging (2014) 14(1):14. doi: 10.1186/1470-7330-14-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Howlader N, Noone AM, Krapcho M, Miller D, Bishop K, Kosary CL, Yu M, Ruhl J, Tatalovich Z, Mariotto A, Lewis DR, Chen HS, Feuer EJ, Cronin KA. eds. SEER Cancer Statistics Review, 1975-2014. Bethesda, MD: National Cancer Institute; (2017). Available at: https://seer.cancer.gov/csr/1975_2014/. based on November 2016 SEER data submission, posted to the SEER web site. [Google Scholar]
- 4. Ouyang G, Liu Q, Wu Y, Liu Z, Lu W, Li S, et al. The Global, Regional, and National Burden of Gallbladder and Biliary Tract Cancer and Its Attributable Risk Factors in 195 Countries and Territories, 1990 to 2017: A Systematic Analysis for the Global Burden of Disease Study 2017. Cancer (2021) 127(13):2238–50. doi: 10.1002/cncr.33476 [DOI] [PubMed] [Google Scholar]
- 5. Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer Statistics, 2021. CA Cancer J Clin (2021) 71(1):7–33. doi: 10.3322/caac.21654 [DOI] [PubMed] [Google Scholar]
- 6. Tella SH, Kommalapati A, Borad MJ, Mahipal A. Second-Line Therapies in Advanced Biliary Tract Cancers. Lancet Oncol (2020) 21(1):E29–41. doi: 10.1016/s1470-2045(19)30733-8 [DOI] [PubMed] [Google Scholar]
- 7. Valle J, Wasan H, Palmer DH, Cunningham D, Anthoney A, Maraveyas A, et al. Cisplatin Plus Gemcitabine Versus Gemcitabine for Biliary Tract Cancer. N Engl J Med (2010) 362(14):1273–81. doi: 10.1056/NEJMoa0908721 [DOI] [PubMed] [Google Scholar]
- 8. Lamarca A, Palmer DH, Wasan HS, Ross PJ, Ma YT, Arora A, et al. Second-Line FOLFOX Chemotherapy Versus Active Symptom Control for Advanced Biliary Tract Cancer (ABC-06): A Phase 3, Open-Label, Randomised, Controlled Trial. Lancet Oncol (2021) 22(5):690–701. doi: 10.1016/s1470-2045(21)00027-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lamarca A, Hubner RA, David Ryder W, Valle JW. Second-Line Chemotherapy in Advanced Biliary Cancer: A Systematic Review. Ann Oncol (2014) 25(12):2328–38. doi: 10.1093/annonc/mdu162 [DOI] [PubMed] [Google Scholar]
- 10. Drilon A, Laetsch TW, Kummar S, Dubois SG, Lassen UN, Demetri GD, et al. Efficacy of Larotrectinib Intrkfusion–Positive Cancers in Adults and Children. N Engl J Med (2018) 378(8):731–9. doi: 10.1056/nejmoa1714448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Doebele RC, Drilon A, Paz-Ares L, Siena S, Shaw AT, Farago AF, et al. Entrectinib in Patients With Advanced or Metastatic NTRK Fusion-Positive Solid Tumours: Integrated Analysis of Three Phase 1-2 Trials. Lancet Oncol (2020) 21(2):271–82. doi: 10.1016/s1470-2045(19)30691-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Abou-Alfa GK, Bibeau K, Schultz N, Yaqubie A, Millang BM, Ren H, et al. Effect of FGFR2 Alterations on Survival in Patients Receiving Systemic Chemotherapy for Intrahepatic Cholangiocarcinoma. J Clin Oncol (2021) 39(3_suppl):303. doi: 10.1200/JCO.2021.39.3_suppl.303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Javle MM, Bridgewater JA, Gbolahan OB, Jungels C, Cho MT, Papadopoulos KP, et al. A Phase I/II Study of Safety and Efficacy of the Arginase Inhibitor INCB001158 Plus Chemotherapy in Patients (Pts) With Advanced Biliary Tract Cancers. J Clin Oncol (2021) 39(3_suppl):311. doi: 10.1200/JCO.2021.39.3_suppl.311 [DOI] [Google Scholar]
- 14. Abou-Alfa GK, Macarulla T, Javle MM, Kelley RK, Lubner SJ, Adeva J, et al. Ivosidenib in IDH1-Mutant, Chemotherapy-Refractory Cholangiocarcinoma (ClarIDHy): A Multicentre, Randomised, Double-Blind, Placebo-Controlled, Phase 3 Study. Lancet Oncol (2020) 21(6):796–807. doi: 10.1016/s1470-2045(20)30157-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Le DT, Durham JN, Smith KN, Wang H, Bartlett BR, Aulakh LK, et al. Mismatch Repair Deficiency Predicts Response of Solid Tumors to PD-1 Blockade. Science (2017) 357(6349):409–13. doi: 10.1126/science.aan6733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. National Research Council Committee on AFfDaNToD . The National Academies Collection: Reports Funded by National Institutes of Health. In: Toward Precision Medicine: Building a Knowledge Network for Biomedical Research and a New Taxonomy of Disease. Washington (DC: National Academies Press (US) Copyright © 2011, National Academy of Sciences; (2011). [PubMed] [Google Scholar]
- 17. Signorovitch J, Janku F, Wheler JJ, Miller VA, Ryan J, Zhou Z, et al. Estimated Cost of Anticancer Therapy Directed by Comprehensive Genomic Profiling (CGP) in a Single-Center Study. J Clin Oncol (2017) 35(15_suppl):6605. doi: 10.1200/JCO.2017.35.15_suppl.6605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Yi B, Liu L, Gu D, Chen R, Xia X, Jiang X. Comprehensive Molecular Characterization of Chinese Patients With Intrahepatic and Extrahepatic Cholangiocarcinoma. J Clin Oncol (2021) 39(15_suppl):e16128–e. doi: 10.1200/JCO.2021.39.15_suppl.e16128 [DOI] [Google Scholar]
- 19. Umemoto K, Yamamoto H, Oikawa R, Takeda H, Doi A, Horie Y, et al. Genomic Alterations for Novel Targeted Therapies in Pancreatobiliary Cancers From Real-World Data. J Clin Oncol (2021) 39(15_suppl):4124. doi: 10.1200/JCO.2021.39.15_suppl.4124 [DOI] [Google Scholar]
- 20. Hollebecque A, Tselikas L, Ducreux MP, Boileve A, Gelli M, Boige V, et al. 70p Genomic Landscape and Efficacy of Precision Medicine in Biliary Tract Cancers. Ann Oncol (2020) 31:S267–S8. doi: 10.1016/j.annonc.2020.08.048 [DOI] [Google Scholar]
- 21. Okamura R, Kurzrock R, Mallory RJ, Fanta PT, Burgoyne AM, Clary BM, et al. Comprehensive Genomic Landscape and Precision Therapeutic Approach in Biliary Tract Cancers. Int J Cancer (2021) 148(3):702–12. doi: 10.1002/ijc.33230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Javle M, Bekaii-Saab T, Jain A, Wang Y, Kelley RK, Wang K, et al. Biliary Cancer: Utility of Next-Generation Sequencing for Clinical Management. Cancer (2016) 122(24):3838–47. doi: 10.1002/cncr.30254 [DOI] [PubMed] [Google Scholar]
- 23. Demols A, Perez-Casanova L, Rocq L, Charry M, De Nève N, Verrellen A, et al. 71p NTRK Gene Fusions in Bilio-Pancreatic Cancers. Ann Oncol (2020) 31:S268. doi: 10.1016/j.annonc.2020.08.049 [DOI] [Google Scholar]
- 24. Ahn KS, Kang KJ. Molecular Heterogeneity in Intrahepatic Cholangiocarcinoma. World J Hepatol (2020) 12(12):1148–57. doi: 10.4254/wjh.v12.i12.1148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Nakamura H, Arai Y, Totoki Y, Shirota T, Elzawahry A, Kato M, et al. Genomic Spectra of Biliary Tract Cancer. Nat Genet (2015) 47(9):1003–10. doi: 10.1038/ng.3375 [DOI] [PubMed] [Google Scholar]
- 26. Wardell CP, Fujita M, Yamada T, Simbolo M, Fassan M, Karlic R, et al. Genomic Characterization of Biliary Tract Cancers Identifies Driver Genes and Predisposing Mutations. J Hepatol (2018) 68(5):959–69. doi: 10.1016/j.jhep.2018.01.009 [DOI] [PubMed] [Google Scholar]
- 27. Churi CR, Shroff R, Wang Y, Rashid A, Kang HC, Weatherly J, et al. Mutation Profiling in Cholangiocarcinoma: Prognostic and Therapeutic Implications. PloS One (2014) 9(12):e115383. doi: 10.1371/journal.pone.0115383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Sia D, Hoshida Y, Villanueva A, Roayaie S, Ferrer J, Tabak B, et al. Integrative Molecular Analysis of Intrahepatic Cholangiocarcinoma Reveals 2 Classes That Have Different Outcomes. Gastroenterology (2013) 144(4):829–40. doi: 10.1053/j.gastro.2013.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Farshidfar F, Zheng S, Gingras M-C, Newton Y, Shih J, Robertson AG, et al. Integrative Genomic Analysis of Cholangiocarcinoma Identifies Distinct IDH-Mutant Molecular Profiles. Cell Rep (2017) 18(11):2780–94. doi: 10.1016/j.celrep.2017.02.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zou S, Li J, Zhou H, Frech C, Jiang X, Chu JSC, et al. Mutational Landscape of Intrahepatic Cholangiocarcinoma. Nat Commun (2014) 5(1):5696. doi: 10.1038/ncomms6696 [DOI] [PubMed] [Google Scholar]
- 31. Ross JS, Wang K, Gay L, Al-Rohil R, Rand JV, Jones DM, et al. New Routes to Targeted Therapy of Intrahepatic Cholangiocarcinomas Revealed by Next-Generation Sequencing. Oncologist (2014) 19(3):235–42. doi: 10.1634/theoncologist.2013-0352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Jiao Y, Pawlik TM, Anders RA, Selaru FM, Streppel MM, Lucas DJ, et al. Exome Sequencing Identifies Frequent Inactivating Mutations in BAP1, ARID1A and PBRM1 in Intrahepatic Cholangiocarcinomas. Nat Genet (2013) 45(12):1470–3. doi: 10.1038/ng.2813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Borger DR, Tanabe KK, Fan KC, Lopez HU, Fantin VR, Straley KS, et al. Frequent Mutation of Isocitrate Dehydrogenase (IDH)1 and IDH2 in Cholangiocarcinoma Identified Through Broad-Based Tumor Genotyping. Oncologist (2012) 17(1):72–9. doi: 10.1634/theoncologist.2011-0386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Valle JW, Lamarca A, Goyal L, Barriuso J, Zhu AX. New Horizons for Precision Medicine in Biliary Tract Cancers. Cancer Discov (2017) 7(9):943–62. doi: 10.1158/2159-8290.cd-17-0245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lee SH, Simoneau EB, Karpinets T, Futreal PA, Zhang J, Javle M, et al. Genomic Profiling of Multifocal Intrahepatic Cholangiocarcinoma Reveals Intraindividual Concordance of Genetic Alterations. Carcinogenesis (2021) 42(3):436–41. doi: 10.1093/carcin/bgaa124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Goeppert B, Stichel D, Toth R, Fritzsche S, Loeffler MA, Schlitter AM, et al. Integrative Analysis Reveals Early and Distinct Genetic and Epigenetic Changes in Intraductal Papillary and Tubulopapillary Cholangiocarcinogenesis. Gut (2021) gutjnl–2020-322. doi: 10.1136/gutjnl-2020-322983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Brägelmann J, Barahona Ponce C, Marcelain K, Roessler S, Goeppert B, Gallegos I, et al. Epigenome-Wide Analysis of Methylation Changes in the Sequence of Gallstone Disease, Dysplasia, and Gallbladder Cancer. Hepatology (2021) 73(6):2293–310. doi: 10.1002/hep.31585 [DOI] [PubMed] [Google Scholar]
- 38. Xiao Z, Zhang J, Zhou Q, He C, Mao K, Chen T, et al. 68p Analysis of DNA Damage Repair (DDR) Pathway Genes in Biliary Tract Cancer and Correlation With Immunogenic Biomarker. Ann Oncol (2020) 31:S267. doi: 10.1016/j.annonc.2020.08.046 [DOI] [Google Scholar]
- 39. Zimmer K, Kocher F, Untergasser G, Puccini A, Xiu J, Wolf D, et al. Identification and Prognostic Impact of PBRM1 Mutations in Biliary Tract Cancers: Results of a Comprehensive Molecular Profiling Study. J Clin Oncol (2021) 39(15_suppl):4022. doi: 10.1200/JCO.2021.39.15_suppl.4022 [DOI] [Google Scholar]
- 40. Ong CK, Subimerb C, Pairojkul C, Wongkham S, Cutcutache I, Yu W, et al. Exome Sequencing of Liver Fluke-Associated Cholangiocarcinoma. Nat Genet (2012) 44(6):690–3. doi: 10.1038/ng.2273 [DOI] [PubMed] [Google Scholar]
- 41. Chan-On W, Nairismägi M-L, Ong CK, Lim WK, Dima S, Pairojkul C, et al. Exome Sequencing Identifies Distinct Mutational Patterns in Liver Fluke–Related and Non-Infection-Related Bile Duct Cancers. Nat Genet (2013) 45(12):1474–8. doi: 10.1038/ng.2806 [DOI] [PubMed] [Google Scholar]
- 42. Jusakul A, Cutcutache I, Yong CH, Lim JQ, Huang MN, Padmanabhan N, et al. Whole-Genome and Epigenomic Landscapes of Etiologically Distinct Subtypes of Cholangiocarcinoma. Cancer Discov (2017) 7(10):1116–35. doi: 10.1158/2159-8290.cd-17-0368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Graham RP, Barr Fritcher EG, Pestova E, Schulz J, Sitailo LA, Vasmatzis G, et al. Fibroblast Growth Factor Receptor 2 Translocations in Intrahepatic Cholangiocarcinoma. Hum Pathol (2014) 45(8):1630–8. doi: 10.1016/j.humpath.2014.03.014 [DOI] [PubMed] [Google Scholar]
- 44. Wang L-J, He C-C, Sui X, Cai M-J, Zhou C-Y, Ma J-L, et al. MiR-21 Promotes Intrahepatic Cholangiocarcinoma Proliferation and Growth In Vitro and In Vivo by Targeting PTPN14 and PTEN. Oncotarget (2015) 6(8):5932–46. doi: 10.18632/oncotarget.3465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Wan P, Chi X, Du Q, Luo J, Cui X, Dong K, et al. miR-383 Promotes Cholangiocarcinoma Cell Proliferation, Migration, and Invasion Through Targeting IRF1. J Cell Biochem (2018) 119(12):9720–9. doi: 10.1002/jcb.27286 [DOI] [PubMed] [Google Scholar]
- 46. Kunk PR, Obeid JM, Winters K, Pramoonjago P, Brockstedt DG, Giobbie-Hurder A, et al. Mismatch Repair Deficiency in Cholangiocarcinoma. J Clin Oncol (2018) 36(4_suppl):269. doi: 10.1200/JCO.2018.36.4_suppl.269 [DOI] [Google Scholar]
- 47. Zhu AX, Borger DR, Kim Y, Cosgrove D, Ejaz A, Alexandrescu S, et al. Genomic Profiling of Intrahepatic Cholangiocarcinoma: Refining Prognosis and Identifying Therapeutic Targets. Ann Surg Oncol (2014) 21(12):3827–34. doi: 10.1245/s10434-014-3828-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Montal R, Sia D, Montironi C, Leow WQ, Esteban-Fabró R, Pinyol R, et al. Molecular Classification and Therapeutic Targets in Extrahepatic Cholangiocarcinoma. J Hepatol (2020) 73(2):315–27. doi: 10.1016/j.jhep.2020.03.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Shroff RT, Javle MM, Xiao L, Kaseb AO, Varadhachary GR, Wolff RA, et al. Gemcitabine, Cisplatin, and Nab-Paclitaxel for the Treatment of Advanced Biliary Tract Cancers. JAMA Oncol (2019) 5(6):824. doi: 10.1001/jamaoncol.2019.0270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Bara AW, Braszewska A, Kwasniewska J. DNA Methylation—An Epigenetic Mark in Mutagen-Treated Brachypodium Distachyon Cells. Plants (2021) 10(7):1408. doi: 10.3390/plants10071408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Qiu Z, Ji J, Xu Y, Zhu Y, Gao C-F, Wang G, et al. Epigenomic Analysis of Biliary Tract Cancer Identifies Subtypes With Different Immune Characteristics and Clinical Outcomes. J Clin Oncol (2021) 39(15_suppl):e16153–e. doi: 10.1200/JCO.2021.39.15_suppl.e16153 [DOI] [Google Scholar]
- 52. Yang B, House MG, Guo M, Herman JG, Clark DP. Promoter Methylation Profiles of Tumor Suppressor Genes in Intrahepatic and Extrahepatic Cholangiocarcinoma. Mod Pathol (2005) 18(3):412–20. doi: 10.1038/modpathol.3800287 [DOI] [PubMed] [Google Scholar]
- 53. Stutes M, Tran S, Demorrow S. Genetic and Epigenetic Changes Associated With Cholangiocarcinoma: From DNA Methylation to microRNAs. World J Gastroenterol (2007) 13(48):6465. doi: 10.3748/wjg.v13.i48.6465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Kim BH, Cho NY, Choi M, Lee S, Ja JJ, Gyeong HK. Methylation Profiles of Multiple CpG Island Loci in Extrahepatic Cholangiocarcinoma Versus Those of Intrahepatic Cholangiocarcinomas. Arch Pathol Lab Med (2007) 131(6):923–30. doi: 10.5858/2007-131-923-MPOMCI [DOI] [PubMed] [Google Scholar]
- 55. Chen J, Li Z, Chen J, Du Y, Song W, Xuan Z, et al. Downregulation of MGMT Promotes Proliferation of Intrahepatic Cholangiocarcinoma by Regulating P21. Clin Trans Oncol (2020) 22(3):392–400. doi: 10.1007/s12094-019-02140-9 [DOI] [PubMed] [Google Scholar]
- 56. Lee S, Kim WH, Jung H-Y, Yang MH, Kang GH. Aberrant CpG Island Methylation of Multiple Genes in Intrahepatic Cholangiocarcinoma. Am J Pathol (2002) 161(3):1015–22. doi: 10.1016/s0002-9440(10)64262-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Yang C, Chen J, Yu Z, Luo J, Li X, Zhou B, et al. Mining of RNA Methylation-Related Genes and Elucidation of Their Molecular Biology in Gallbladder Carcinoma. Front Oncol (2021) 11:621806. doi: 10.3389/fonc.2021.621806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Tekcham DS, Tiwari PK. Epigenetic Regulation in Gallbladder Cancer: Promoter Methylation Profiling as Emergent Novel Biomarkers. Asia Pac J Clin Oncol (2016) 12(4):332–48. doi: 10.1111/ajco.12507 [DOI] [PubMed] [Google Scholar]
- 59. Hoshida Y, Nijman SMB, Kobayashi M, Chan JA, Brunet J-P, Chiang DY, et al. Integrative Transcriptome Analysis Reveals Common Molecular Subclasses of Human Hepatocellular Carcinoma. Cancer Res (2009) 69(18):7385–92. doi: 10.1158/0008-5472.can-09-1089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Boyault S, Rickman DS, De Reyniès A, Balabaud C, Rebouissou S, Jeannot E, et al. Transcriptome Classification of HCC is Related to Gene Alterations and to New Therapeutic Targets. Hepatology (2007) 45(1):42–52. doi: 10.1002/hep.21467 [DOI] [PubMed] [Google Scholar]
- 61. Andersen JB, Spee B, Blechacz BR, Avital I, Komuta M, Barbour A, et al. Genomic and Genetic Characterization of Cholangiocarcinoma Identifies Therapeutic Targets for Tyrosine Kinase Inhibitors. Gastroenterology (2012) 142(4):1021–31.e15. doi: 10.1053/j.gastro.2011.12.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Mouliere F, Thierry AR. The Importance of Examining the Proportion of Circulating DNA Originating From Tumor, Microenvironment and Normal Cells in Colorectal Cancer Patients. Expert Opin Biol Ther (2012) 12(sup1):S209–S15. doi: 10.1517/14712598.2012.688023 [DOI] [PubMed] [Google Scholar]
- 63. Jahr S, Hentze H, Englisch S, Hardt D, Fackelmayer FO, Hesch RD, et al. DNA Fragments in the Blood Plasma of Cancer Patients: Quantitations and Evidence for Their Origin From Apoptotic and Necrotic Cells. Cancer Res (2001) 61(4):1659–65. [PubMed] [Google Scholar]
- 64. Chen Q, Zhang Z-H, Wang S, Lang J-H. Circulating Cell-Free DNA or Circulating Tumor DNA in the Management of Ovarian and Endometrial Cancer. Onco Targets Ther (2019) 12:11517–30. doi: 10.2147/ott.s227156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Lamarca A, Kapacee Z, Breeze M, Bell C, Belcher D, Staiger H, et al. Molecular Profiling in Daily Clinical Practice: Practicalities in Advanced Cholangiocarcinoma and Other Biliary Tract Cancers. J Clin Med (2020) 9(9):2854. doi: 10.3390/jcm9092854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Zill OA, Greene C, Sebisanovic D, Siew LM, Leng J, Vu M, et al. Cell-Free DNA Next-Generation Sequencing in Pancreatobiliary Carcinomas. Cancer Discov (2015) 5(10):1040–8. doi: 10.1158/2159-8290.cd-15-0274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Ettrich TJ, Schwerdel D, Dolnik A, Beuter F, Blätte TJ, Schmidt SA, et al. Genotyping of Circulating Tumor DNA in Cholangiocarcinoma Reveals Diagnostic and Prognostic Information. Sci Rep (2019) 9(1). doi: 10.1038/s41598-019-49860-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Kumari S, Husain N, Agarwal A, Neyaz A, Gupta S, Chaturvedi A, et al. Diagnostic Value of Circulating Free DNA Integrity and Global Methylation Status in Gall Bladder Carcinoma. Pathol Oncol Res (2019) 25(3):925–36. doi: 10.1007/s12253-017-0380-6 [DOI] [PubMed] [Google Scholar]
- 69. Umetani N, Kim J, Hiramatsu S, Reber HA, Hines OJ, Bilchik AJ, et al. Increased Integrity of Free Circulating DNA in Sera of Patients With Colorectal or Periampullary Cancer: Direct Quantitative PCR for ALU Repeats. Clin Chem (2006) 52(6):1062–9. doi: 10.1373/clinchem.2006.068577 [DOI] [PubMed] [Google Scholar]
- 70. Mody K, Kasi PM, Yang J, Surapaneni PK, Bekaii-Saab T, Ahn DH, et al. Circulating Tumor DNA Profiling of Advanced Biliary Tract Cancers. JCO Precis Oncol (2019) 3):1–9. doi: 10.1200/po.18.00324 [DOI] [PubMed] [Google Scholar]
- 71. Kasi PM, Le AD, Barrett A. Comparative Landscape of Actionable Somatic Alterations in Advanced Cholangiocarcinoma From Circulating Tumor and Tissue-Based DNA Profiling. J Clin Oncol (2021) 39(3_suppl):342. doi: 10.1200/JCO.2021.39.3_suppl.342 [DOI] [Google Scholar]
- 72. Strom SP. Current Practices and Guidelines for Clinical Next-Generation Sequencing Oncology Testing. Cancer Biol Med (2016) 13(1):3–11. doi: 10.28092/j.issn.2095-3941.2016.0004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Yang JD, Campion MB, Liu MC, Chaiteerakij R, Giama NH, Ahmed Mohammed H, et al. Circulating Tumor Cells Are Associated With Poor Overall Survival in Patients With Cholangiocarcinoma. Hepatology (2016) 63(1):148–58. doi: 10.1002/hep.27944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Li J, Jiang W, Wei J, Zhang J, Cai L, Luo M, et al. Patient Specific Circulating Tumor DNA Fingerprints to Monitor Treatment Response Across Multiple Tumors. J Trans Med (2020) 18(1). doi: 10.1186/s12967-020-02449-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Winter H, Kaisaki PJ, Harvey J, Giacopuzzi E, Ferla MP, Pentony MM, et al. Identification of Circulating Genomic and Metabolic Biomarkers in Intrahepatic Cholangiocarcinoma. Cancers (2019) 11(12):1895. doi: 10.3390/cancers11121895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Yang JD, Yab TC, Taylor WR, Foote PH, Ali HA, Lavu S, et al. Detection of Cholangiocarcinoma by Assay of Methylated DNA Markers in Plasma. Gastroenterology (2017) 152(5):S1041–S2. doi: 10.1016/s0016-5085(17)33526-6 [DOI] [Google Scholar]
- 77. Wasenang W, Chaiyarit P, Proungvitaya S, Limpaiboon T. Serum Cell-Free DNA Methylation of OPCML and HOXD9 as a Biomarker That may Aid in Differential Diagnosis Between Cholangiocarcinoma and Other Biliary Diseases. Clin Epigenet (2019) 11(1). doi: 10.1186/s13148-019-0634-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Gonda TA, Viterbo D, Gausman V, Kipp C, Sethi A, Poneros JM, et al. Mutation Profile and Fluorescence In Situ Hybridization Analyses Increase Detection of Malignancies in Biliary Strictures. Clin Gastroenterol Hepatol (2017) 15(6):913–9.e1. doi: 10.1016/j.cgh.2016.12.013 [DOI] [PubMed] [Google Scholar]
- 79. Kipp BR, Barr Fritcher EG, Clayton AC, Gores GJ, Roberts LR, Zhang J, et al. Comparison of KRAS Mutation Analysis and FISH for Detecting Pancreatobiliary Tract Cancer in Cytology Specimens Collected During Endoscopic Retrograde Cholangiopancreatography. J Mol Diagn (2010) 12(6):780–6. doi: 10.2353/jmoldx.2010.100016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Sturm PD, Rauws EA, Hruban RH, Caspers E, Ramsoekh TB, Huibregtse K, et al. Clinical Value of K-Ras Codon 12 Analysis and Endobiliary Brush Cytology for the Diagnosis of Malignant Extrahepatic Bile Duct Stenosis. Clin Cancer Res (1999) 5(3):629–35. [PubMed] [Google Scholar]
- 81. Singhi AD, Nikiforova MN, Chennat J, Papachristou GI, Khalid A, Rabinovitz M, et al. Integrating Next-Generation Sequencing to Endoscopic Retrograde Cholangiopancreatography (ERCP)-Obtained Biliary Specimens Improves the Detection and Management of Patients With Malignant Bile Duct Strictures. Gut (2020) 69(1):52–61. doi: 10.1136/gutjnl-2018-317817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Kushnir VM, Mullady DK, Das K, Lang G, Hollander TG, Murad FM, et al. The Diagnostic Yield of Malignancy Comparing Cytology, FISH, and Molecular Analysis of Cell Free Cytology Brush Supernatant in Patients With Biliary Strictures Undergoing Endoscopic Retrograde Cholangiography (ERC): A Prospective Study. J Clin Gastroenterol (2019) 53(9):686–92. doi: 10.1097/mcg.0000000000001118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Andresen K, Boberg KM, Vedeld HM, Honne H, Jebsen P, Hektoen M, et al. Four DNA Methylation Biomarkers in Biliary Brush Samples Accurately Identify the Presence of Cholangiocarcinoma. Hepatology (2015) 61(5):1651–9. doi: 10.1002/hep.27707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Prachayakul V, Rugivarodom M, Nopjaroonsri P, Cheirsilpa K, Chang A, Kamolhan T, et al. Diagnostic Power of DNA Methylation Markers Suggestive of Cholangiocarcinoma in ERCP-Based Brush Cytology. Gastrointest Endosc (2021). doi: 10.1016/j.gie.2021.07.005 [DOI] [PubMed] [Google Scholar]
- 85. Yang JD, Ghoz H, Aboelsoud MM, Taylor WR, Yab TC, Berger CK, et al. DNA Methylation Markers for Detection of Cholangiocarcinoma: Discovery, Validation, and Clinical Testing in Biliary Brushings and Plasma. Hepatol Commun (2021) 5(8):1448–59. doi: 10.1002/hep4.1730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Parsi MA, Li A, Li CP, Goggins M. DNA Methylation Alterations in Endoscopic Retrograde Cholangiopancreatography Brush Samples of Patients With Suspected Pancreaticobiliary Disease. Clin Gastroenterol Hepatol (2008) 6(11):1270–8. doi: 10.1016/j.cgh.2008.07.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Kinugasa H, Nouso K, Ako S, Dohi C, Matsushita H, Matsumoto K, et al. Liquid Biopsy of Bile for the Molecular Diagnosis of Gallbladder Cancer. Cancer Biol Ther (2018) 19(10):934–8. doi: 10.1080/15384047.2018.1456604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Shen N, Zhang D, Yin L, Qiu Y, Liu J, Yu W, et al. Bile Cell−Free DNA as a Novel and Powerful Liquid Biopsy for Detecting Somatic Variants in Biliary Tract Cancer. Oncol Rep (2019) 42(2):549–60. doi: 10.3892/or.2019.7177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Liggett WH, Sidransky D. Role of the P16 Tumor Suppressor Gene in Cancer. J Clin Oncol (1998) 16(3):1197–206. doi: 10.1200/jco.1998.16.3.1197 [DOI] [PubMed] [Google Scholar]
- 90. Klump B, Hsieh CJ, Dette S, Holzmann K, Kiebetalich R, Jung M, et al. Promoter Methylation of INK4a/ARF as Detected in Bile-Significance for the Differential Diagnosis in Biliary Disease. Clin Cancer Res (2003) 9(5):1773–8. [PubMed] [Google Scholar]
- 91. Shin S-H, Lee K, Kim B-H, Cho N-Y, Jang J-Y, Kim Y-T, et al. Bile-Based Detection of Extrahepatic Cholangiocarcinoma With Quantitative DNA Methylation Markers and Its High Sensitivity. J Mol Diagn (2012) 14(3):256–63. doi: 10.1016/j.jmoldx.2012.01.014 [DOI] [PubMed] [Google Scholar]
- 92. Zhang Y, Yang B, Du Z, Gao Y-T, Wang Y-J, Jing X, et al. Identification and Validation of Specific Methylation Profile in Bile for Differential Diagnosis of Malignant Biliary Stricture. Clin Biochem (2010) 43(16-17):1340–4. doi: 10.1016/j.clinbiochem.2010.08.013 [DOI] [PubMed] [Google Scholar]
- 93. Kishikawa T. Circulating RNAs as New Biomarkers for Detecting Pancreatic Cancer. World J Gastroenterol (2015) 21(28):8527. doi: 10.3748/wjg.v21.i28.8527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Afonso M, Rodrigues P, Simão A, Castro R. Circulating microRNAs as Potential Biomarkers in Non-Alcoholic Fatty Liver Disease and Hepatocellular Carcinoma. J Clin Med (2016) 5(3):30. doi: 10.3390/jcm5030030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Zhang Y, Liu Y, Liu H, Tang WH. Exosomes: Biogenesis, Biologic Function and Clinical Potential. Cell Biosci (2019) 9(1). doi: 10.1186/s13578-019-0282-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Doyle L, Wang M. Overview of Extracellular Vesicles, Their Origin, Composition, Purpose, and Methods for Exosome Isolation and Analysis. Cells (2019) 8(7):727. doi: 10.3390/cells8070727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Borges FT, Reis LA, Schor N. Extracellular Vesicles: Structure, Function, and Potential Clinical Uses in Renal Diseases. Braz J Med Biol Res (2013) 46(10):824–30. doi: 10.1590/1414-431x20132964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Salem PES, Ghazala RA, El Gendi AM, Emara DM, Ahmed NM. The Association Between Circulating MicroRNA-150 Level and Cholangiocarcinoma. J Clin Lab Anal (2020) 34(11). doi: 10.1002/jcla.23397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Bartel DP. MicroRNAs. Cell (2004) 116(2):281–97. doi: 10.1016/s0092-8674(04)00045-5 [DOI] [PubMed] [Google Scholar]
- 100. Hayes J, Peruzzi PP, Lawler S. MicroRNAs in Cancer: Biomarkers, Functions and Therapy. Trends Mol Med (2014) 20(8):460–9. doi: 10.1016/j.molmed.2014.06.005 [DOI] [PubMed] [Google Scholar]
- 101. Xu L, Qi X, Duan S, Xie Y, Ren X, Chen G, et al. MicroRNAs: Potential Biomarkers for Disease Diagnosis. BioMed Mater Eng (2014) 24(6):3917–25. doi: 10.3233/bme-141223 [DOI] [PubMed] [Google Scholar]
- 102. Silakit R, Loilome W, Yongvanit P, Thongchot S, Sithithaworn P, Boonmars T, et al. Urinary microRNA-192 and microRNA-21 as Potential Indicators for Liver Fluke-Associated Cholangiocarcinoma Risk Group. Parasitol Int (2017) 66(4):479–85. doi: 10.1016/j.parint.2015.10.001 [DOI] [PubMed] [Google Scholar]
- 103. Han HS, Kim MJ, Han J-H, Yun J, Kim HK, Yang Y, et al. Bile-Derived Circulating Extracellular miR-30d-5p and miR-92a-3p as Potential Biomarkers for Cholangiocarcinoma. Hepatobiliary Pancreat Dis Int (2020) 19(1):41–50. doi: 10.1016/j.hbpd.2019.10.009 [DOI] [PubMed] [Google Scholar]
- 104. Shigehara K, Yokomuro S, Ishibashi O, Mizuguchi Y, Arima Y, Kawahigashi Y, et al. Real-Time PCR-Based Analysis of the Human Bile MicroRNAome Identifies miR-9 as a Potential Diagnostic Biomarker for Biliary Tract Cancer. PloS One (2011) 6(8):e23584. doi: 10.1371/journal.pone.0023584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Wang L-J, Zhang K-L, Zhang N, Ma X-W, Yan S-W, Cao D-H, et al. Serum miR-26a as a Diagnostic and Prognostic Biomarker in Cholangiocarcinoma. Oncotarget (2015) 6(21):18631–40. doi: 10.18632/oncotarget.4072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Silakit R, Loilome W, Yongvanit P, Chusorn P, Techasen A, Boonmars T, et al. Circulating miR-192 in Liver Fluke-Associated Cholangiocarcinoma Patients: A Prospective Prognostic Indicator. J Hepatobiliary Pancreat Sci (2014) 21(12):864–72. doi: 10.1002/jhbp.145 [DOI] [PubMed] [Google Scholar]
- 107. Voigtländer T, Gupta SK, Thum S, Fendrich J, Manns MP, Lankisch TO, et al. MicroRNAs in Serum and Bile of Patients With Primary Sclerosing Cholangitis and/or Cholangiocarcinoma. PloS One (2015) 10(10):e0139305. doi: 10.1371/journal.pone.0139305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Bernuzzi F, Marabita F, Lleo A, Carbone M, Mirolo M, Marzioni M, et al. Serum microRNAs as Novel Biomarkers for Primary Sclerosing Cholangitis and Cholangiocarcinoma. Clin Exp Immunol (2016) 185(1):61–71. doi: 10.1111/cei.12776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Cheng Q, Feng F, Zhu L, Zheng Y, Luo X, Liu C, et al. Circulating miR-106a is a Novel Prognostic and Lymph Node Metastasis Indicator for Cholangiocarcinoma. Sci Rep (2015) 5(1):16103. doi: 10.1038/srep16103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Jiang W, Deng X, Zhu T, Wei Y, Lei Z, Guo M, et al. Identification of Cholangiocarcinoma Associated With Hepatolithiasis via the Combination of miRNA and Ultrasound. Cancer Manage Res (2020) 12:1845–53. doi: 10.2147/cmar.s241870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Correa-Gallego C, Maddalo D, Doussot A, Kemeny N, Kingham TP, Allen PJ, et al. Circulating Plasma Levels of MicroRNA-21 and MicroRNA-221 Are Potential Diagnostic Markers for Primary Intrahepatic Cholangiocarcinoma. PloS One (2016) 11(9):e0163699. doi: 10.1371/journal.pone.0163699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Loosen SH, Lurje G, Wiltberger G, Vucur M, Koch A, Kather JN, et al. Serum Levels of miR-29, miR-122, miR-155 and miR-192 Are Elevated in Patients With Cholangiocarcinoma. PloS One (2019) 14(1):e0210944. doi: 10.1371/journal.pone.0210944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Roderburg C, Loosen SH, Bednarsch J, Alizai PH, Roeth AA, Schmitz SM, et al. Levels of Circulating PD-L1 Are Decreased in Patients With Resectable Cholangiocarcinoma. Int J Mol Sci (2021) 22(12):6569. doi: 10.3390/ijms22126569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Wang S, Yin J, Li T, Yuan L, Wang D, He J, et al. Upregulated Circulating miR-150 is Associated With the Risk of Intrahepatic Cholangiocarcinoma. Oncol Rep (2015) 33(2):819–25. doi: 10.3892/or.2014.3641 [DOI] [PubMed] [Google Scholar]
- 115. Lapitz A, Arbelaiz A, O’Rourke CJ, Lavin JL, Casta AL, Ibarra C, et al. Patients With Cholangiocarcinoma Present Specific RNA Profiles in Serum and Urine Extracellular Vesicles Mirroring the Tumor Expression: Novel Liquid Biopsy Biomarkers for Disease Diagnosis. Cells (2020) 9(3):721. doi: 10.3390/cells9030721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Shen L, Chen G, Xia Q, Shao S, Fang H. Exosomal miR-200 Family as Serum Biomarkers for Early Detection and Prognostic Prediction of Cholangiocarcinoma. Int J Clin Exp Pathol (2019) 12(10):3870–6. [PMC free article] [PubMed] [Google Scholar]
- 117. Li L, Masica D, Ishida M, Tomuleasa C, Umegaki S, Kalloo AN, et al. Human Bile Contains MicroRNA-Laden Extracellular Vesicles That can be Used for Cholangiocarcinoma Diagnosis. Hepatology (2014) 60(3):896–907. doi: 10.1002/hep.27050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Xue X-Y, Liu Y-X, Wang C, Gu X-J, Xue Z-Q, Zang X-L, et al. Identification of Exosomal miRNAs as Diagnostic Biomarkers for Cholangiocarcinoma and Gallbladder Carcinoma. Signal Transduct Target Ther (2020) 5(1). doi: 10.1038/s41392-020-0162-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Collins AL, Wojcik S, Liu J, Frankel WL, Alder H, Yu L, et al. A Differential MicroRNA Profile Distinguishes Cholangiocarcinoma From Pancreatic Adenocarcinoma. Ann Surg Oncol (2014) 21(1):133–8. doi: 10.1245/s10434-013-3240-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Chen L, Yan HX, Yang W, Hu L, Yu LX, Liu Q, et al. The Role of microRNA Expression Pattern in Human Intrahepatic Cholangiocarcinoma. J Hepatol (2009) 50(2):358–69. doi: 10.1016/j.jhep.2008.09.015 [DOI] [PubMed] [Google Scholar]
- 121. Lu X, Zhou C, Li R, Deng Y, Zhao L, Zhai W. Long Noncoding RNA AFAP1-AS1 Promoted Tumor Growth and Invasion in Cholangiocarcinoma. Cell Physiol Biochem (2017) 42(1):222–30. doi: 10.1159/000477319 [DOI] [PubMed] [Google Scholar]
- 122. Xu Y, Jiang X, Cui Y. Upregulated Long Noncoding RNA PANDAR Predicts an Unfavorable Prognosis and Promotes Tumorigenesis in Cholangiocarcinoma. Onco Targets Ther (2017) 10:2873–83. doi: 10.2147/ott.s137044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Ma S-L, Li A-J, Hu Z-Y, Shang F-S, Wu M-C. Co-Expression of the Carbamoyl-Phosphate Synthase 1 Gene and Its Long Non-Coding RNA Correlates With Poor Prognosis of Patients With Intrahepatic Cholangiocarcinoma. Mol Med Rep (2015) 12(6):7915–26. doi: 10.3892/mmr.2015.4435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Tan X, Huang Z, Li X. Long Non-Coding RNA MALAT1 Interacts With miR-204 to Modulate Human Hilar Cholangiocarcinoma Proliferation, Migration, and Invasion by Targeting CXCR4. J Cell Biochem (2017) 118(11):3643–53. doi: 10.1002/jcb.25862 [DOI] [PubMed] [Google Scholar]
- 125. Parasramka M, Yan IK, Wang X, Nguyen P, Matsuda A, Maji S, et al. BAP1 Dependent Expression of Long Non-Coding RNA NEAT-1 Contributes to Sensitivity to Gemcitabine in Cholangiocarcinoma. Mol Cancer (2017) 16(1). doi: 10.1186/s12943-017-0587-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Jiang XM, Li ZL, Li JL, Zheng WY, Li XH, Cui YF, et al. LncRNA CCAT1 as the Unfavorable Prognostic Biomarker for Cholangiocarcinoma. Eur Rev Med Pharmacol Sci (2017) 21(6):1242–7. [PubMed] [Google Scholar]
- 127. Ohtsubo K, Miyake K, Arai S, Fukuda K, Yanagimura N, Suzuki C, et al. Aberrant Methylation of Tumor Suppressive miRNAs in Bile From Patients With Pancreaticobiliary Diseases. Anticancer Res (2019) 39(10):5449–59. doi: 10.21873/anticanres.13738 [DOI] [PubMed] [Google Scholar]
- 128. Goyal L, Sirard C, Schrag M, Kagey MH, Eads JR, Stein S, et al. Phase I and Biomarker Study of the Wnt Pathway Modulator DKN-01 in Combination With Gemcitabine/Cisplatin in Advanced Biliary Tract Cancer. Clin Cancer Res (2020) 26(23):6158–67. doi: 10.1158/1078-0432.ccr-20-1310 [DOI] [PubMed] [Google Scholar]
- 129. Lu X, Wang L, Chen Q, Wang Y, Cao Z, Zhou G, et al. A Biosensor Based on Multifunctional Allostery for Dynamic Analysis of Circulating Tumor DNA. Chem Commun (2020) 56(30):4184–7. doi: 10.1039/d0cc01560a [DOI] [PubMed] [Google Scholar]
- 130. Li M, Cushing SK, Zhou G, Wu N. Molecular Hot Spots in Surface-Enhanced Raman Scattering. Nanoscale (2020) 12(43):22036–41. doi: 10.1039/d0nr06579j [DOI] [PubMed] [Google Scholar]
- 131. Wang H-F, Ma R-N, Sun F, Jia L-P, Zhang W, Shang L, et al. A Versatile Label-Free Electrochemical Biosensor for Circulating Tumor DNA Based on Dual Enzyme Assisted Multiple Amplification Strategy. Biosens Bioelectron (2018) 122:224–30. doi: 10.1016/j.bios.2018.09.028 [DOI] [PubMed] [Google Scholar]
- 132. Liang Z, Zhou J, Petti L, Shao L, Jiang T, Qing Y, et al. SERS-Based Cascade Amplification Bioassay Protocol of miRNA-21 by Using Sandwich Structure With Biotin–Streptavidin System. Analyst (2019) 144(5):1741–50. doi: 10.1039/c8an02259c [DOI] [PubMed] [Google Scholar]
- 133. Darrigues E, Nima ZA, Majeed W, Vang-Dings KB, Dantuluri V, Biris AR, et al. Raman Spectroscopy Using Plasmonic and Carbon-Based Nanoparticles for Cancer Detection, Diagnosis, and Treatment Guidance.Part 1: Diagnosis. Drug Metab Rev (2017) 49(2):212–52. doi: 10.1080/03602532.2017.1302465 [DOI] [PubMed] [Google Scholar]
- 134. Valle J, Wasan H, Palmer DH, Cunningham D, Anthoney A, Maraveyas A, et al. Cisplatin Plus Gemcitabine Versus Gemcitabine for Biliary Tract Cancer. N Engl J Med (2010) 362(14):1273–81. doi: 10.1056/nejmoa0908721 [DOI] [PubMed] [Google Scholar]
- 135. Primrose JN, Fox R, Palmer DH, Prasad R, Mirza D, Anthoney DA, et al. Adjuvant Capecitabine for Biliary Tract Cancer: The BILCAP Randomized Study. J Clin Oncol (2017) 35(15_suppl):4006. doi: 10.1200/JCO.2017.35.15_suppl.4006 [DOI] [Google Scholar]
- 136. Lamarca A, Palmer DH, Wasan HS, Ross PJ, Ma YT, Arora A, et al. Second-Line FOLFOX Chemotherapy Versus Active Symptom Control for Advanced Biliary Tract Cancer (ABC-06): A Phase 3, Open-Label, Randomised, Controlled Trial. Lancet Oncol (2021) 22(5):690–701. doi: 10.1016/s1470-2045(21)00027-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Rizzo A, Salati M, Frega G, Merz V, Caputo F, Ricci AD, et al. Second-Line Chemotherapy (2L) in Elderly Patients With Advanced Biliary Tract Cancer (ABC): A Multicenter Real-World Study. J Clin Oncol (2021) 39(3_suppl):322. doi: 10.1200/JCO.2021.39.3_suppl.322 [DOI] [Google Scholar]
- 138. Yoo C, Kim K-P, Kim I, Kang MJ, Cheon J, Kang BW, et al. Liposomal Irinotecan (Nal-IRI) in Combination With Fluorouracil (5-FU) and Leucovorin (LV) for Patients With Metastatic Biliary Tract Cancer (BTC) After Progression on Gemcitabine Plus Cisplatin (GemCis): Multicenter Comparative Randomized Phase 2b Study (NIFTY). J Clin Oncol (2021) 39(15_suppl):4006. doi: 10.1200/JCO.2021.39.15_suppl.4006 [DOI] [Google Scholar]
- 139. Makawita S, Abou-Alfa GK, Roychowdhury S, Sadeghi S, Borbath I, Goyal L, et al. Infigratinib in Patients With Advanced Cholangiocarcinoma With FGFR2 Gene Fusions/Translocations: The PROOF 301 Trial. Future Oncol (2020) 16(30):2375–84. doi: 10.2217/fon-2020-0299 [DOI] [PubMed] [Google Scholar]
- 140. Salama AKS, Li S, Macrae ER, Park JI, Mitchell EP, Zwiebel JA, et al. Dabrafenib and Trametinib in Patients With Tumors With BRAF(V600E) Mutations: Results of the NCI-MATCH Trial Subprotocol H. J Clin Oncol (2020) 38(33):3895–904. doi: 10.1200/jco.20.00762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Subbiah V, Lassen U, Élez E, Italiano A, Curigliano G, Javle M, et al. Dabrafenib Plus Trametinib in Patients With BRAF(V600E)-Mutated Biliary Tract Cancer (ROAR): A Phase 2, Open-Label, Single-Arm, Multicentre Basket Trial. Lancet Oncol (2020) 21(9):1234–43. doi: 10.1016/s1470-2045(20)30321-1 [DOI] [PubMed] [Google Scholar]
- 142. Bridgewater J, Meric-Bernstam F, Hollebecque A, Valle JW, Morizane C, Karasic T, et al. 54p Efficacy and Safety of Futibatinib in Intrahepatic Cholangiocarcinoma (iCCA) Harboring FGFR2 Fusions/Other Rearrangements: Subgroup Analyses of a Phase II Study (FOENIX-CCA2). Ann Oncol (2020) 31:S261–S2. doi: 10.1016/j.annonc.2020.08.032 [DOI] [Google Scholar]
- 143. Mazzaferro V, El-Rayes BF, Droz Dit Busset M, Cotsoglou C, Harris WP, Damjanov N, et al. Derazantinib (ARQ 087) in Advanced or Inoperable FGFR2 Gene Fusion-Positive Intrahepatic Cholangiocarcinoma. Br J Cancer (2019) 120(2):165–71. doi: 10.1038/s41416-018-0334-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Chen YY, Park JO, Su WC, Oh DY, Kim KP, Feng YH, et al. Preliminary Results of a Ph2a Study to Evaluate the Clinical Efficacy and Safety of Erdafitinib in Asian Patients With Biomarker-Selected Advanced Cholangiocarcinoma (CCA). Ann Oncol (2018) 29:viii209. doi: 10.1093/annonc/mdy282.008 [DOI] [Google Scholar]
- 145. Borad MJ, Bai L-Y, Chen M-H, Hubbard JM, Mody K, Rha SY, et al. Silmitasertib (CX-4945) in Combination With Gemcitabine and Cisplatin as First-Line Treatment for Patients With Locally Advanced or Metastatic Cholangiocarcinoma: A Phase Ib/II Study. J Clin Oncol (2021) 39(3_suppl):312. doi: 10.1200/JCO.2021.39.3_suppl.312 [DOI] [Google Scholar]
- 146. Demols A, Borbath I, Van Den Eynde M, Houbiers G, Peeters M, Marechal R, et al. Regorafenib After Failure of Gemcitabine and Platinum-Based Chemotherapy for Locally Advanced/Metastatic Biliary Tumors: REACHIN, a Randomized, Double-Blind, Phase II Trial. Ann Oncol (2020) 31(9):1169–77. doi: 10.1016/j.annonc.2020.05.018 [DOI] [PubMed] [Google Scholar]
- 147. Bai Y, Xu J, Sun H, Bai C, Jia R, Li Y, et al. A Single-Arm, Multicenter, Open-Label Phase 2 Trial of Surufatinib in Patients With Unresectable or Metastatic Biliary Tract Cancer. J Clin Oncol (2021) 39(15_suppl):e16123–e. doi: 10.1200/JCO.2021.39.15_suppl.e16123 [DOI] [PubMed] [Google Scholar]
- 148. Pant S, Ducreux M, Harding JJ, Javle MM, Oh D-Y, Wasan HS, et al. A Phase IIb, Open-Label, Single-Arm Study of Zanidatamab (ZW25) Monotherapy in Subjects With Advanced or Metastatic HER2-Amplified Biliary Tract Cancers. J Clin Oncol (2021) 39(3_suppl):TPS352–TPS. doi: 10.1200/JCO.2021.39.3_suppl.TPS352 [DOI] [Google Scholar]
- 149. Meric-Bernstam F, Hanna DL, El-Khoueiry AB, Kang Y-K, Oh D-Y, Chaves JM, et al. Zanidatamab (ZW25) in HER2-Positive Biliary Tract Cancers (BTCs): Results From a Phase I Study. J Clin Oncol (2021) 39(3_suppl):299. doi: 10.1200/JCO.2021.39.3_suppl.299 [DOI] [Google Scholar]
- 150. Harding JJ, Cleary JM, Quinn DI, Braña I, Moreno V, Borad MJ, et al. Targeting HER2 (ERBB2) Mutation-Positive Advanced Biliary Tract Cancers With Neratinib: Results From the Phase II SUMMIT ‘Basket’ Trial. J Clin Oncol (2021) 39(3_suppl):320. doi: 10.1200/JCO.2021.39.3_suppl.320 [DOI] [Google Scholar]
- 151. Kudo R, Kubo T, Mori Y, Harada Y, Shirota H, Hayashi H, et al. A Phase 2 Basket Trial of Combination Therapy With Trastuzumab and Pertuzumab in Patients With Solid Cancers Harboring HER2 Amplification (JUPITER Trial). J Clin Oncol (2021) 39(15_suppl):TPS3141–TPS. doi: 10.1200/JCO.2021.39.15_suppl.TPS3141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152. Lee C-K, Cheon J, Chon HJ, Kim MH, Kim JW, Lee MA, et al. A Phase II Trial of Trastuzumab Plus Modified-FOLFOX for Gemcitabine/Cisplatin Refractory HER2-Positive Biliary Tract Cancer (BTC): Multi-Institutional Study of the Korean Cancer Study Group (KCSG-HB19-14). J Clin Oncol (2021) 39(15_suppl):TPS4161–TPS. doi: 10.1200/JCO.2021.39.15_suppl.TPS4161 [DOI] [PubMed] [Google Scholar]
- 153. Inthagard J, Edwards J, Roseweir AK. Immunotherapy: Enhancing the Efficacy of This Promising Therapeutic in Multiple Cancers. Clin Sci (2019) 133(2):181–93. doi: 10.1042/cs20181003 [DOI] [PubMed] [Google Scholar]
- 154. Marabelle A, Le DT, Ascierto PA, Di Giacomo AM, De Jesus-Acosta A, Delord J-P, et al. Efficacy of Pembrolizumab in Patients With Noncolorectal High Microsatellite Instability/Mismatch Repair–Deficient Cancer: Results From the Phase II KEYNOTE-158 Study. J Clin Oncol (2020) 38(1):1–10. doi: 10.1200/jco.19.02105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155. Piha-Paul SA, Oh DY, Ueno M, Malka D, Chung HC, Nagrial A, et al. Efficacy and Safety of Pembrolizumab for the Treatment of Advanced Biliary Cancer: Results From the KEYNOTE -158 and KEYNOTE -028 Studies. Int J Cancer (2020) 147(8):2190–8. doi: 10.1002/ijc.33013 [DOI] [PubMed] [Google Scholar]
- 156. Kim RD, Chung V, Alese OB, El-Rayes BF, Li D, Al-Toubah TE, et al. A Phase 2 Multi-Institutional Study of Nivolumab for Patients With Advanced Refractory Biliary Tract Cancer. JAMA Oncol (2020) 6(6):888. doi: 10.1001/jamaoncol.2020.0930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157. Klein O, Kee D, Nagrial A, Markman B, Underhill C, Michael M, et al. Evaluation of Combination Nivolumab and Ipilimumab Immunotherapy in Patients With Advanced Biliary Tract Cancers. JAMA Oncol (2020) 6(9):1405. doi: 10.1001/jamaoncol.2020.2814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158. Villanueva L, Lwin Z, Chung HCC, Gomez-Roca CA, Longo F, Yanez E, et al. Lenvatinib Plus Pembrolizumab for Patients With Previously Treated Biliary Tract Cancers in the Multicohort Phase 2 LEAP-005 Study. J Clin Oncol (2021) 39(15_suppl):4080. doi: 10.1200/JCO.2021.39.15_suppl.4080 [DOI] [Google Scholar]
- 159. Valle JW, Kelley RK, Furuse J, Edeline J, Finn RS, Ren Z, et al. 78tip KEYNOTE-966 Trial in Progress: Pembrolizumab Plus Gemcitabine and Cisplatin for Advanced Biliary Tract Cancer. Ann Oncol (2020) 31:S270–S1. doi: 10.1016/j.annonc.2020.08.056 [DOI] [Google Scholar]
- 160. Sahai V, Griffith KA, Beg MS, Shaib WL, Mahalingam D, Zhen DB, et al. A Multicenter Randomized Phase II Study of Nivolumab in Combination With Gemcitabine/Cisplatin or Ipilimumab as First-Line Therapy for Patients With Advanced Unresectable Biliary Tract Cancer (BilT-01). J Clin Oncol (2020) 38(15_suppl):4582. doi: 10.1200/JCO.2020.38.15_suppl.4582 [DOI] [Google Scholar]
- 161. Ueno M, Ikeda M, Morizane C, Kobayashi S, Ohno I, Kondo S, et al. Nivolumab Alone or in Combination With Cisplatin Plus Gemcitabine in Japanese Patients With Unresectable or Recurrent Biliary Tract Cancer: A Non-Randomised, Multicentre, Open-Label, Phase 1 Study. Lancet Gastroenterol Hepatol (2019) 4(8):611–21. doi: 10.1016/s2468-1253(19)30086-x [DOI] [PubMed] [Google Scholar]
- 162. Yoo C, Oh D-Y, Choi HJ, Kudo M, Ueno M, Kondo S, et al. Phase I Study of Bintrafusp Alfa, a Bifunctional Fusion Protein Targeting TGF-β and PD-L1, in Patients With Pretreated Biliary Tract Cancer. J Immunother Cancer (2020) 8(1):e000564. doi: 10.1136/jitc-2020-000564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163. Oh D-Y, De Braud F, Bridgewater J, Furuse J, Hsu C-H, Ikeda M, et al. 79tip A Phase II/III, Randomized, Placebo-Controlled Study of Bintrafusp Alfa With Gemcitabine Plus Cisplatin as First-Line Treatment of Biliary Tract Cancer. Ann Oncol (2020) 31:S271–S2. doi: 10.1016/j.annonc.2020.08.057 [DOI] [Google Scholar]
- 164. Oh D-Y, Lee K-H, Lee D-W, Kim TY, Bang J-H, Nam A-R, et al. Phase II Study Assessing Tolerability, Efficacy, and Biomarkers for Durvalumab (D) ± Tremelimumab (T) and Gemcitabine/Cisplatin (GemCis) in Chemo-Naïve Advanced Biliary Tract Cancer (aBTC). J Clin Oncol (2020) 38(15_suppl):4520. doi: 10.1200/JCO.2020.38.15_suppl.4520 [DOI] [Google Scholar]
- 165. Ioka T, Ueno M, Oh D-Y, Fujiwara Y, Chen J-S, Doki Y, et al. Evaluation of Safety and Tolerability of Durvalumab (D) With or Without Tremelimumab (T) in Patients (Pts) With Biliary Tract Cancer (BTC). J Clin Oncol (2019) 37(4_suppl):387. doi: 10.1200/JCO.2019.37.4_suppl.387 [DOI] [Google Scholar]
- 166. Xie C, Duffy AG, Mabry-Hrones D, Wood B, Levy E, Krishnasamy V, et al. Tremelimumab in Combination With Microwave Ablation in Patients With Refractory Biliary Tract Cancer. Hepatology (2019) 69(5):2048–60. doi: 10.1002/hep.30482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167. Liu T, Li W, Yu Y, Guo X, Xu X, Wang Y, et al. 53p Toripalimab With Chemotherapy as First-Line Treatment for Advanced Biliary Tract Tumors: A Preliminary Analysis of Safety and Efficacy of an Open-Label Phase II Clinical Study. Ann Oncol (2020) 31:S261. doi: 10.1016/j.annonc.2020.08.031 [DOI] [Google Scholar]
- 168. Zhou J, Fan J, Shi G, Huang X, Wu D, Yang G, et al. 56p Anti-PD1 Antibody Toripalimab, Lenvatinib and Gemox Chemotherapy as First-Line Treatment of Advanced and Unresectable Intrahepatic Cholangiocarcinoma: A Phase II Clinical Trial. Ann Oncol (2020) 31:S262–S3. doi: 10.1016/j.annonc.2020.08.034 [DOI] [Google Scholar]
- 169. Zong H, Zhong Q, Zhao R, Jin S, Zhou C, Zhang X, et al. Phase II Study of Anlotinib Plus Sintlimab as Second-Line Treatment for Patients With Advanced Biliary Tract Cancers. J Clin Oncol (2021) 39(3_suppl):307. doi: 10.1200/JCO.2021.39.3_suppl.307 [DOI] [Google Scholar]
- 170. Yoshitomi M, Yutani S, Matsueda S, Ioji T, Komatsu N, Shichijo S, et al. Personalized Peptide Vaccination for Advanced Biliary Tract Cancer: IL-6, Nutritional Status and Pre-Existing Antigen-Specific Immunity as Possible Biomarkers for Patient Prognosis. Exp Ther Med (2012) 3(3):463–9. doi: 10.3892/etm.2011.424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171. Aruga A, Takeshita N, Kotera Y, Okuyama R, Matsushita N, Ohta T, et al. Phase I Clinical Trial of Multiple-Peptide Vaccination for Patients With Advanced Biliary Tract Cancer. J Trans Med (2014) 12(1):61. doi: 10.1186/1479-5876-12-61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172. Aruga A, Takeshita N, Kotera Y, Okuyama R, Matsushita N, Ohta T, et al. Long-Term Vaccination With Multiple Peptides Derived From Cancer-Testis Antigens Can Maintain a Specific T-Cell Response and Achieve Disease Stability in Advanced Biliary Tract Cancer. Clin Cancer Res (2013) 19(8):2224–31. doi: 10.1158/1078-0432.ccr-12-3592 [DOI] [PubMed] [Google Scholar]
- 173. Lepisto AJ, Moser AJ, Zeh H, Lee K, Bartlett D, McKolanis JR, et al. A Phase I/II Study of a MUC1 Peptide Pulsed Autologous Dendritic Cell Vaccine as Adjuvant Therapy in Patients With Resected Pancreatic and Biliary Tumors. Cancer Ther (2008) 6(B):955–64. [PMC free article] [PubMed] [Google Scholar]
- 174. Yamamoto K, Ueno T, Kawaoka T, Hazama S, Fukui M, Suehiro Y, et al. MUC1 Peptide Vaccination in Patients With Advanced Pancreas or Biliary Tract Cancer. Anticancer Res (2005) 25(5):3575–9. [PubMed] [Google Scholar]
- 175. Kaida M, Morita-Hoshi Y, Soeda A, Wakeda T, Yamaki Y, Kojima Y, et al. Phase 1 Trial of Wilms Tumor 1 (WT1) Peptide Vaccine and Gemcitabine Combination Therapy in Patients With Advanced Pancreatic or Biliary Tract Cancer. J Immunother (2011) 34(1):92–9. doi: 10.1097/CJI.0b013e3181fb65b9 [DOI] [PubMed] [Google Scholar]
- 176. Feng K-C, Guo Y-L, Liu Y, Dai H-R, Wang Y, Lv H-Y, et al. Cocktail Treatment With EGFR-Specific and CD133-Specific Chimeric Antigen Receptor-Modified T Cells in a Patient With Advanced Cholangiocarcinoma. J Hematol Oncol (2017) 10(1). doi: 10.1186/s13045-016-0378-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177. Tran E, Turcotte S, Gros A, Robbins PF, Lu Y-C, Dudley ME, et al. Cancer Immunotherapy Based on Mutation-Specific CD4+ T Cells in a Patient With Epithelial Cancer. Science (2014) 344(6184):641–5. doi: 10.1126/science.1251102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178. Guo Y, Feng K, Liu Y, Wu Z, Dai H, Yang Q, et al. Phase I Study of Chimeric Antigen Receptor–Modified T Cells in Patients With EGFR-Positive Advanced Biliary Tract Cancers. Clin Cancer Res (2018) 24(6):1277–86. doi: 10.1158/1078-0432.ccr-17-0432 [DOI] [PubMed] [Google Scholar]