Abstract
During mitotic exit, thousands of nuclear pore complexes (NPCs) assemble concomitant with the nuclear envelope to build a transport‐competent nucleus. Here, we show that Nup50 plays a crucial role in NPC assembly independent of its well‐established function in nuclear transport. RNAi‐mediated downregulation in cells or immunodepletion of Nup50 protein in Xenopus egg extracts interferes with NPC assembly. We define a conserved central region of 46 residues in Nup50 that is crucial for Nup153 and MEL28/ELYS binding, and for NPC interaction. Surprisingly, neither NPC interaction nor binding of Nup50 to importin α/β, the GTPase Ran, or chromatin is crucial for its function in the assembly process. Instead, an N‐terminal fragment of Nup50 can stimulate the Ran GTPase guanine nucleotide exchange factor RCC1 and NPC assembly, indicating that Nup50 acts via the Ran system in NPC reformation at the end of mitosis. In support of this conclusion, Nup50 mutants defective in RCC1 binding and stimulation cannot replace the wild‐type protein in in vitro NPC assembly assays, whereas excess RCC1 can compensate the loss of Nup50.
Keywords: mitotic exit, nuclear pore complex assembly, Nup50, RCC1, Xenopus egg extracts
Subject Categories: Cell Cycle, Membranes & Trafficking
In addition to acting as auxiliary factor for nuclear import, vertebrate Nup50 also participates in post‐mitotic reformation of nuclear pore complexes.

Introduction
Nuclear pore complexes (NPCs) are the gatekeepers of the nucleus, controlling the exchange of proteins and nucleic acids between the cytoplasm and nucleoplasm. They are embedded in the nuclear envelope (NE). With a mass of approximately 125 MDa, they are largest protein complexes in most vertebrate cells. Despite their enormous size, they are only composed of about 30 different proteins called nucleoporins, all present in multiple copies. Because of their common function in all nucleated cells, the general structure of NPCs is evolutionary conserved (Knockenhauer & Schwartz, 2016; Lin & Hoelz, 2019). Their core structure is formed by three rings embedded in the NE, a nucleoplasmic, an inner, and a cytoplasmic ring. This symmetric core encloses a central transport channel and anchors extensions on the cytoplasmic and nuclear faces forming cytoplasmic filaments and nuclear basket, respectively.
In metazoans, NPCs form at two different stages of the cell cycle by distinct mechanisms (for review see Weberruss and Antonin (2016) and Otsuka and Ellenberg (2018)). During interphase, new NPCs assemble into the intact NE. This assembly pathway is thought to be initiated at the membranes of the NE (Vollmer et al, 2015; Otsuka et al, 2016), although the mechanistic details remain poorly defined. At the end of mitosis, NPC re‐assembly occurs concomitantly with the formation of a closed NE and is initiated by the chromatin‐binding nucleoporin MEL28/ELYS directing NPC assembly at the end of mitosis to the surface of the decondensing chromatin (Franz et al, 2007; Rotem et al, 2009). The small GTPase Ran serves as a second important spatial regulator that guides NPC assembly toward chromatin (Forbes et al, 2015). High RanGTP concentrations around chromatin, generated by the action of the chromatin‐bound Ran guanine exchange factor (GEF) RCC1, are thought to release inhibitory nuclear transport factors from key nucleoporins, which can then act in NPC assembly. The crucial functions of both MEL28/ELYS and the Ran system is probably best exemplified by experiments where tethering MEL28/ELYS and RCC1 to DNA beads is sufficient to initiate NPC assembly (Zierhut et al, 2014).
Cell‐free and cellular assays have revealed a picture of a stepwise assembly of the NPC core structure (Kutay et al, 2021). Late in the assembly process, the cytoplasmic filaments and the nuclear basket structure form. The nuclear basket is present in vertebrates in three nucleoporins, Nup153, TPR, and Nup50 (Sukegawa & Blobel, 1993; Cordes et al, 1997; Guan et al, 2000), also referred to as Npap60 (Fan et al, 1997), which have multiple roles in nuclear import and export of proteins, chromatin remodeling, control, gene expression, as well as RNA processing and export (Aksenova et al, 2020; Guglielmi et al, 2020). Both Nup50 (Guan et al, 2000; Smitherman et al, 2000; Makise et al, 2012) and TPR (Hase & Cordes, 2003) interact with Nup153. Accordingly, Nup50 NPC localization depends on Nup153 but not vice versa (Smitherman et al, 2000; Hase & Cordes, 2003; Aksenova et al, 2020), although the interdependence of Nup153 and TPR for NPC localization is controversial (Hase & Cordes, 2003; Aksenova et al, 2020). Similar to MEL28/ELYS, a fraction of the basket nucleoporins Nup50 and Nup153 is found on decondensing chromatin at the early stages of mitotic exit (Hase & Cordes, 2003; Dultz et al, 2008). Nup153 is reportedly not required for NPC assembly at the end of mitosis (Walther et al, 2001; Vollmer et al, 2015). Whether Nup50 chromatin binding, or Nup50 in general, plays a role in NPC assembly has not been studied so far.
Our current knowledge regarding Nup50 centers around its role as an auxiliary component in nuclear transport (for review, see Swaminathan and Melchior (2002) and Moore (2003)). It can interact with nuclear transport receptors (importin α, importin β, transportin, CRM1) and Ran (Guan et al, 2000; Smitherman et al, 2000; Lindsay et al, 2002; Matsuura & Stewart, 2005; Makise et al, 2012) and has been suggested to act as a platform to support the RanGTP‐mediated dissociation of complexes consisting of importin α, importin β, and import cargos in a yet ill‐defined mechanism. It thus supports the nuclear import reaction, which critically depends on the disassembly of import complexes. A similar accessory role in nuclear transport has been described for the Saccharomyces cerevisiae counterpart of Nup50, Nup2 (Solsbacher et al, 2000; Gilchrist et al, 2002; Liu & Stewart, 2005).
Here, we show that in addition to its role in nuclear transport, Nup50 has a distinct function in NPC assembly at the end of mitosis. It binds to and stimulates RCC1’s guanine nucleotide exchange activity toward Ran, which is crucial for NPC assembly. Interestingly, although this function determines a crucial role for Nup50 in NPC assembly at the end of mitosis, this does not require Nup50 to be localized to NPCs.
Results
Nup50 is required for nuclear pore complex assembly
To gain insights into Nup50 function, we generated antibodies against the Xenopus protein. The antibodies recognize two bands at a size of 50 kDa which can be efficiently depleted from egg extracts (Fig 1A and Appendix Fig S1A). In the egg extract system, nuclear assembly including the formation of a closed NE and NPCs can be faithfully recapitulated (Eisenhardt et al, 2014; Gant & Wilson, 1997). When demembranated sperm DNA is added as a substrate to the extracts, a nucleus with a functional NE and NPCs is formed (Fig 1B and Appendix Fig S1B, mock). Upon depletion of Nup50, a closed NE, indicated by the smooth membrane staining, forms which, however, lacks NPCs (Fig 1B and C). This is indicated by the absence of the NPC marker mAB414, which recognizes a subset of FG nucleoporins not including Nup50 and serves as a robust marker for intact NPCs and as proxy for NPC numbers (Davis & Blobel, 1986). Consistent with the absence of NPCs, the nuclei are not competent for nuclear import (Appendix Fig S1B) and, hence, remain of small size. Addition of recombinant Nup50, expressed and purified from bacteria, to approximately endogenous levels (Fig 1A and Appendix Fig S1C) rescues the NPC assembly phenotype (Fig 1B and C), showing the point specificity of the depletion.
Figure 1. Nup50 is crucial for NPC assembly.
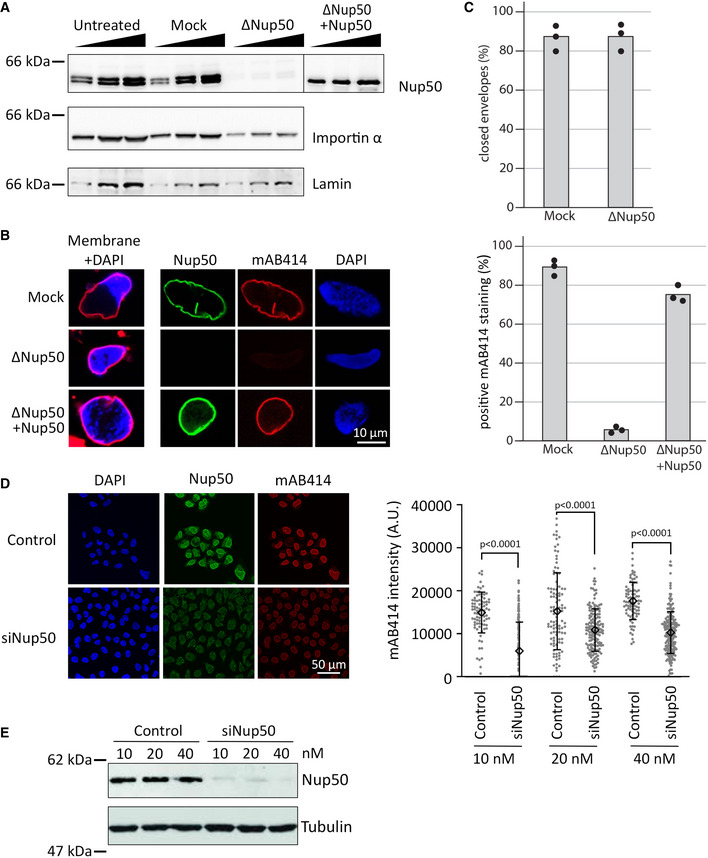
- Western blot analysis of untreated, mock‐ and Nup50‐depleted Xenopus egg extracts, with or without addition of recombinant Xenopus Nup50. 1, 2, and 4 µl of extracts were analyzed with indicated antibodies.
- Confocal microscopy images of fixed nuclei assembled for 120 min in mock‐depleted (mock) and Nup50‐depleted (ΔNup50) Xenopus egg extracts supplemented with either buffer or recombinant Nup50. In the left column, membranes were prelabeled with DiIC18 (1,1'‐dioctadecyl‐3,3,3',3'‐tetramethylindocarbocyanine perchlorate, red), and chromatin was stained with DAPI (4′,6‐diamidin‐2‐phenylindol, blue). Three right columns show the immunofluorescence staining for Nup50 (green) and NPCs (mAB414, red) on the chromatin (DAPI, blue). Scale bars: 10 µm.
- The average percentage of closed nuclear envelopes (upper panel) and mAB414‐positive nuclei (lower panel) for 100 randomly chosen chromatin substrates in each of 3 independent experiments is shown. Data points from individual experiments are indicated.
- HeLa cells were transfected with 20 nM control or Nup50 siRNA. 72‐h post‐transfection, cells were fixed with 4% PFA and stained with antibodies against Nup50 and mAB414, chromatin was labeled with DAPI. Scale bars: 50 µm. Quantitation of the mAB414 rim intensity at three different Nup50 RNAi and control oligo concentrations: 10 nM control (n = 84) or Nup50 siRNA (n = 171), 20 nM control (n = 101) or Nup50 siRNA (n = 157) and 40 nM control (n = 86) or Nup50 siRNA (n = 204). The means are indicated as diamonds; error bars show the standard deviations. P‐values have been calculated from a Student t‐test comparing the mean between the experimental conditions.
- Western blot analysis of HeLa cells treated as in (D) 72 h post transfection.
Source data are available online for this figure.
Thus, in the cell‐free system, Nup50 plays a crucial role in NPC assembly. Next, we tested whether a similar phenotype can be observed in cells. SiRNA‐mediated downregulation of Nup50 expression in human HeLa cells (Fig 1D and E) reduced mAB414 staining, consistent with a role of the protein in NPC assembly. A reduced mAB414 staining has been previously reported upon downregulation of nucleoporins with crucial functions in NPC assembly (Walther et al, 2003a; Mansfeld et al, 2006). Life cell imaging of HeLa cells stably expressing EGFP‐tagged Nup107 or Nup133 (Bolhy et al, 2011; Otsuka et al, 2016), which are both part of the Y‐complex forming the nuclear and cytoplasmic ring structures of the NPC, showed a delay in nuclear rim localization of these markers upon Nup50 downregulation (Appendix Fig S2). This indicates that loss of Nup50 affects NPC assembly at the end of mitosis.
It has been previously reported that a Nup50 knockout mouse is not viable but that fibroblasts derived from mouse embryos can be kept in culture and show an apparent normal mAB414 staining (Smitherman et al, 2000). We speculate that the discrepancy between the human and Xenopus system versus the mouse phenotype could arise because in contrast to humans, rodents such as mouse and rats possess two Nup50‐related genes. In mouse, aside from the main locus (1700030K07Rik) on chromosome 15, named Nup50A here, a paralog (1700123L14Rik), named Nup50B here, can be found on the chromosome 6. Although referred to as a pseudogene, this locus is effectively expressed as indicated in the expression atlas (Papatheodorou et al, 2020) and the mouse genome database (Bult et al, 2019) and has been previously named Nup50rel (Smitherman et al, 2000). We propose that Nup50B might substitute the canonical Nup50 paralog at least in mouse fibroblasts. Indeed, whereas downregulation of one of the two mouse Nup50 orthologs only moderately reduced mAB414 staining in mouse NIH3T3 cells, the combined application of siRNAs against both targets reduced mAB414 staining (Fig 2A and B). Note that the canonical Nup50 paralog can be detected with antibodies in Western blotting, although we could not assay explicitly for Nup50B due to a lack of a specific antibody (Fig 2C).
Figure 2. Two Nup50 paralogs in mouse cells.
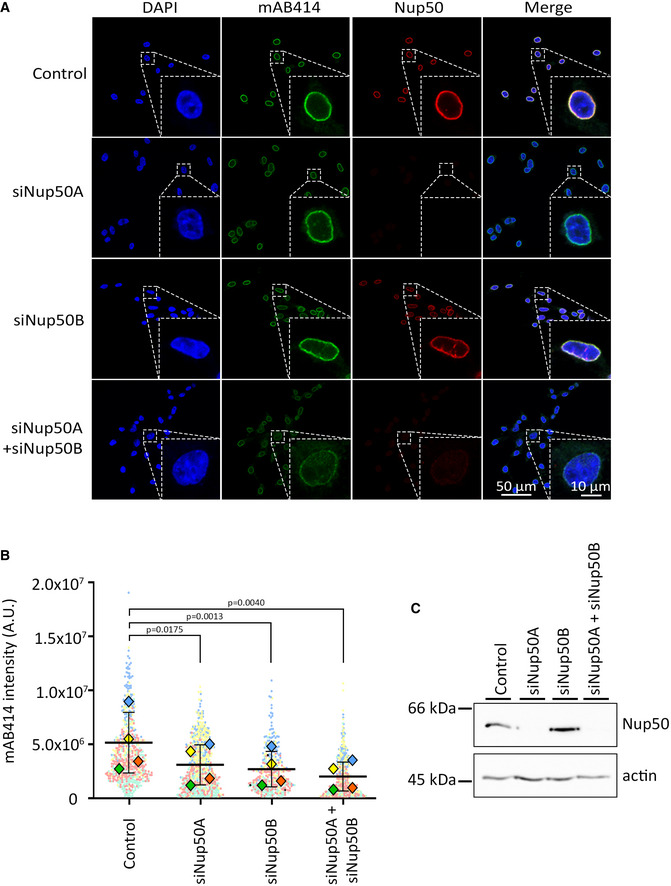
- Mouse 3T3‐NIH cells were transfected with 20 nM control, Nup50A, Nup50B, or a combination of Nup50A and Nup50B siRNA. After 72 h, cells were fixed and stained with mAB414 (green) and antibodies against mouse Nup50 (red). Chromatin is stained with DAPI (blue). The merge of the three channels is shown on the left column. Scale bar: 50 µm. An insert shows a zoom on a representative nucleus for each picture. Scale bar: 10 µm.
- Quantitation of the mAB414 rim intensity from n = 4 independent experiments done as in (A), each represented by a different color. The mean of each individual experiment and condition is represented by a diamond; the mean of all four experiments by a horizontal black line. P‐values have been calculated from a paired ratio t‐test comparing the mean between the experimental conditions. Error bars show the standard deviations of the means.
- Western blot showing the amount of Nup50A (top row) for each of the experimental condition. The actin (lower row) is shown as a loading control.
Source data are available online for this figure.
Together, these data indicate that Nup50 has a crucial function in NPC assembly, a feature which has been so far overlooked likely because of the potential redundancy of the two Nup50 paralogs in mice.
Nup50 interacts with MEL28/ELYS but both act independently in nuclear pore complex assembly at the end of mitosis
Interestingly, a similar phenotype, a closed NE without NPCs, has been previously observed upon depletion of the Y‐complex (Walther et al, 2003a) and MEL28/ELYS (Rasala et al, 2006; Franz et al, 2007), which recruits the Y‐complex to the chromatin template during NPC assembly at the end of mitosis. Nup50 and MEL28/ELYS interact in Xenopus egg extracts in co‐immunoprecipitation (data not shown), consistent with a previous report (Gillespie et al, 2007). However, although Nup50 depletion reduces the MEL28/ELYS levels by approximately 65% and MEL28/ELYS depletion reduces Nup50 by approximately 15% (Appendix Fig S3A and B), we do not think that the identical phenotypes of Nup50 and MEL28/ELYS depletion is due to a co‐depletion of the other factor: The MEL28/ELYS depletion phenotype can be reverted by expression of the protein from the corresponding mRNA (Yokoyama et al, 2014), and the Nup50 depletion is rescued by addition of recombinant Nup50 (Fig 1B and C).
Both Nup50 and MEL28/ELYS are recruited early in chromatin during NPC assembly at the end of mitosis (Dultz et al, 2008) and during in vitro nuclear assembly in the Xenopus system (Fig 3A and B (mock control), Appendix Fig S4A and B). We, therefore, tested whether chromatin recruitment of the two proteins depends on each other. Under control conditions, MEL28/ELYS is first found on the chromatin template, about 10 min after initiation of the reaction and later detected at the NE as part of NPCs. In the absence of Nup50, MEL28/ELYS is recruited to chromatin but, consistent with the lack of NPC assembly (see Fig 1), is not found at the NE at later time points and remains spread on chromatin. Similarly, MEL28/ELYS depletion does not prevent Nup50 chromatin recruitment. However, at later time points, Nup50 is not enriched at the NE due to the absence of NPCs, reported for MEL28/ELYS depletion (Rasala et al, 2006; Franz et al, 2007) and indicated by the absence of mAB414 staining.
Figure 3. Nup50 and MEL28 do not depend on each other for chromatin binding.
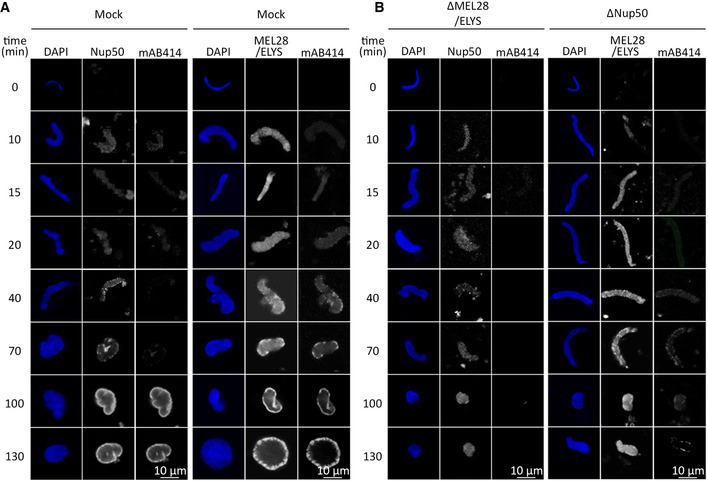
-
A, BDemembranated sperm chromatin was preincubated in Xenopus egg extract, depleted for MEL28/ELYS or Nup50 (B), or control treated (mock, A). After 10 min, membranes were added to the reaction. Reactions were stopped at the indicated time points by fixation and analyzed by confocal microscopy after immunostaining with α‐MEL28/ELYS, α‐Nup50, and mAb414. Chromatin was stained with DAPI (blue). Scale bar: 10 μm.
Nup50 binds chromatin via its N‐terminal basic motif
During mitotic exit, a relatively substantial subfraction of Nup50, particularly in comparison to other nucleoporins, is found early on the chromatin, approximately 7 min before the onset of nuclear import ((Dultz et al, 2008), see also Fig 3 and Appendix Fig S4). We, therefore, tested whether Nup50 can directly interact with chromatin. We generated recombinant EGFP‐tagged Nup50 and tested its binding to DNA‐coated magnetic beads (Fig 4). Upon preincubation with egg extracts (right panel), the DNA is rapidly chromatinized (Heald et al, 1996), whereas in the absence of egg extracts (left panel), pure DNA binding can be detected. In these assays, Nup50 can bind both chromatin and DNA similarly to a C‐terminal fragment of MEL28 (aa 2290–2408), which served as a positive control (Rasala et al, 2008).
Figure 4. Nup50 is a chromatin binding nucleoporin.
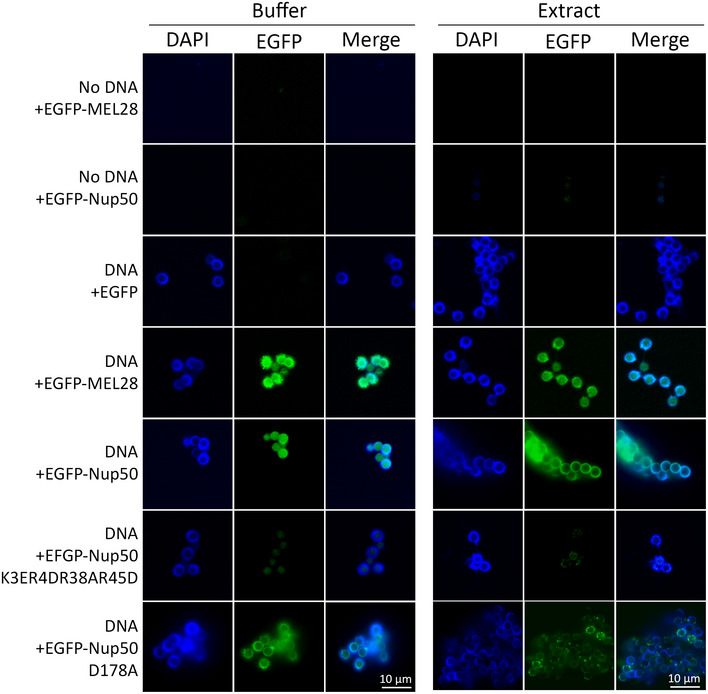
3 µM recombinant EGFP, EGFP‐tagged MEL28/ELYS (aa 2290–2408), or EGFP‐tagged Nup50 was incubated with empty or DNA‐coated magnetic beads, which were chromatinized with Xenopus egg extracts (right panel) or unchromatinized DNA beads (left panel). After 3 h, the beads were re‐isolated, washed, co‐stained with DAPI, and analyzed by confocal microscopy. Scale bar: 10 µm.
Nup50 contains an N‐terminal bipartite nuclear localization signal (Lindsay et al, 2002; Matsuura & Stewart, 2005). These motifs often serve, in addition to importin α binding, as chromatin interaction domains (Yokoyama, 2016). Indeed, mutations in this positively charged region (K3E, R4D, R38A, R45D), known to prevent importin α interaction (Matsuura & Stewart, 2005), abolishes chromatin/DNA binding of Nup50 (Fig 4).
A short 46‐residue‐long region of Nup50 is required for its nuclear pore complex localization
To gain insights into Nup50 function in NPC assembly, we aimed to identify the parts of the protein critical for its NPC localization. Nup50 is composed of an N‐terminal importin α/chromatin interaction domain and a C‐terminal Ran‐binding domain (Lindsay et al, 2002). Using EGFP‐tagged fragments of Nup50, expressed in HeLa cells, we found that neither domain is strictly required for the localization of Nup50 to NPCs (Fig 5A). Further truncations yielded a minimal 46‐amino acid region (aa 144–189) required for NPC localization. This region of Nup50 shows a higher evolutionary conservation than other parts of the proteins (Fig 5B), that is, 80.4% sequence identity between the Xenopus and human proteins versus 59.6% for the entire protein. Several residues in this domain are highly conserved, and for a number of them, if mutated in the context of the full‐length protein, NPC localization was abolished (Fig 5C). By pull‐down assays, we identified the minimal NPC‐binding region of Nup50 as a domain binding to both MEL28/ELYS and Nup153 (Fig 5D). In the context of these minimal fragments, the mutations that abolish NPC binding prevented the interaction to both MEL28/ELYS and Nup153 (Fig 5D and Appendix Fig S1D). This minimal region is included in the MAR domain of S. cerevisiae Nup2 (Chu et al, 2017) which was identified as the minimal fragment rescuing Nup2 meiotic function and its NPC localization. An alignment of S. cerevisiae MAR domain with metazoan Nup50 shows a better alignment with the 144–189 region than the rest of the protein (Appendix Fig S5). Consistently, sequence identity between X. laevis 144–189 and S. cerevisiae MAR domain is 32.6 vs 22.7% for the full‐length proteins.
Figure 5. Minimal 46‐aa region is required for Nup50 NPC localization.
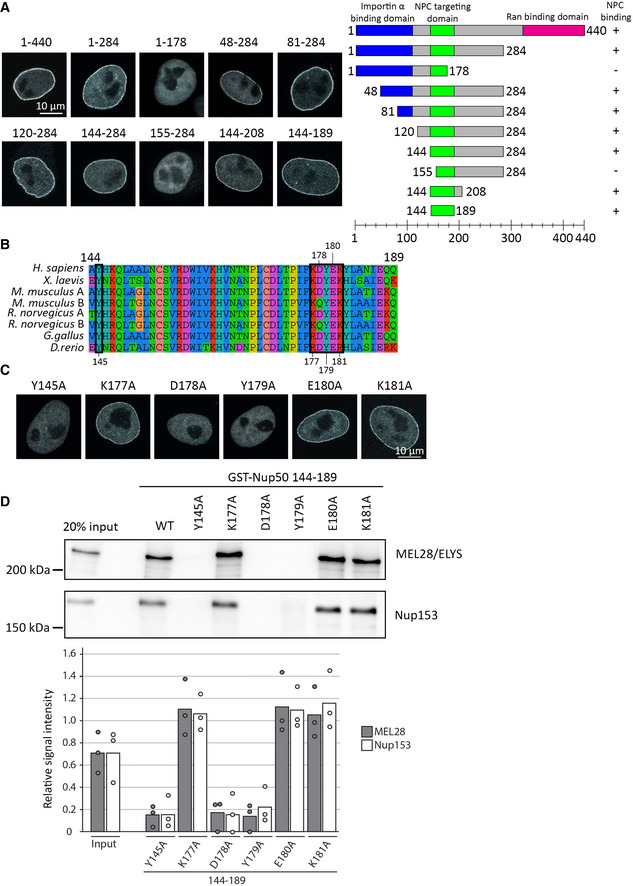
- HeLa cells were transfected with EGFP‐Nup50 and different truncations. After 24 h, cells were shortly pretreated with 0.1% Triton X‐100 in PBS, fixed with 4% PFA, and analyzed by confocal microscopy (left panel). Scale bar: 10 µm. The graph (right panel) shows the Nup50 domains (blue: importin α interaction domain, green: NPC‐targeting domain, and pink: Ran‐binding domain), and the truncation tested. Ability to bind NPCs is indicated on the right.
- Sequence alignment of Nup50 144–189, species are indicated on the left, the black boxes highlight residues tested by single‐point mutation in (C) and (D), and the residue number are shown above or below the alignment. The color scheme indicates the type of amino acids according to the alignment software default setting.
- HeLa cells were transfected with EGFP‐Nup50 comprising different point mutations in the minimal NPC‐binding region and analyzed as in (A). Scale bar: 10 µm.
- GST fusion constructs of the Xenopus Nup50 minimal NPC binding fragment (aa 144–189) comprising no or single‐point mutations were incubated with Xenopus egg extracts. Starting material as well as bound proteins were analyzed by Western blotting with indicated antibodies. The quantitation shows the average MEL28/ELYS and Nup153 bead bound signal from three independent experiments, normalized to the signal of their respective wild‐type GST fusion (see Appendix Fig S1D for a gel showing the GST baits). Data points from individual experiments are indicated.
Source data are available online for this figure.
A minimal N‐terminal fragment is required for nuclear pore complex assembly independent of NPC localization
Having identified crucial amino acids for Nup50 binding to MEL28/ELYS and Nup153, which are also required for NPC localization, we wondered whether these interactions are important for Nup50 function in NPC assembly. We used the Xenopus egg extract system to deplete Nup50 and add back the wild type or different mutant versions. Surprisingly, the mutants which abolished NPC localization rescued the NPC assembly phenotype (Fig 6A and B). Despite the rescue and consistent with our data for human cells, these mutants did not localize to NPCs (Appendix Fig S6). Also, a Nup50 version, which does not interact with importin α/chromatin (K3E, R4D, R38A, R45D) or directly with importin β (FGYG, where all five FG sequences in the Xenopus protein have were replaced by YG), was able to replace the wild‐type protein in NPC assembly, indicating that these specific protein–protein interactions are not crucial for this function.
Figure 6. A N‐terminal Nup50 fragment is required for NPC assembly.
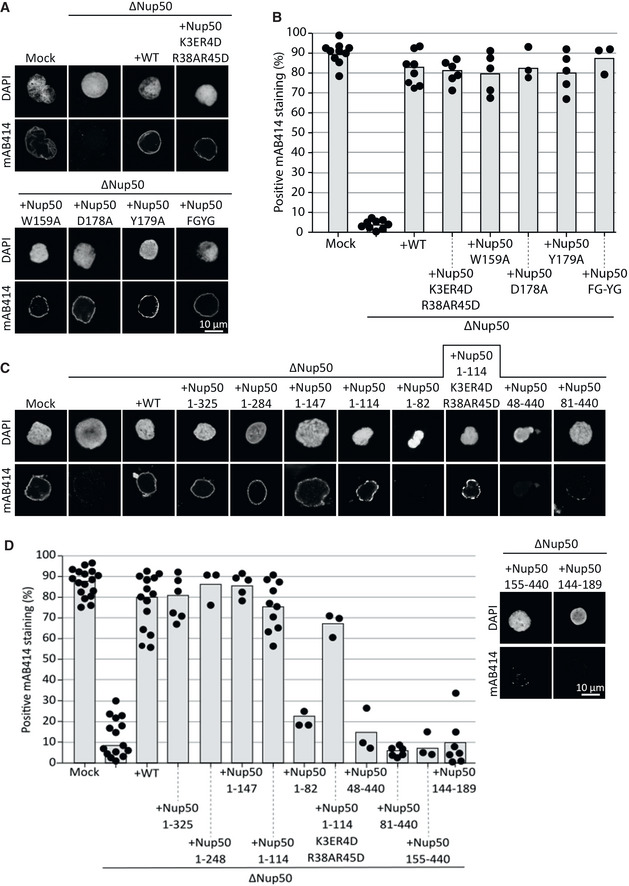
- Confocal microscopy images of nuclei assembled for 120 min in mock‐depleted, Nup50‐depleted (ΔNup50), and Nup50‐depleted Xenopus egg extracts supplemented with recombinant wild‐type Nup50 or different mutants. Nuclei were fixed in 4% PFA and 0.5% glutaraldehyde, stained for NPCs (mAB414) and the chromatin (DAPI). Scale bar: 10 µm.
- Average percentage of mAB414‐positive nuclei for 100 randomly chosen chromatin substrates in each of at least three independent experiments shown in (A). Data points from the individual experiments are indicated.
- Confocal microscopy images of nuclei assembled with N‐ and C‐terminal Nup50 truncations and an minimal NPC binding fragment (144–189). Samples were analyzed as in (A).
- Average percentage of mAB414‐positive nuclei for 100 randomly chosen chromatin substrates in each of at least three independent experiments shown in (C). Data points from the individual experiments are indicated.
To identify the regions of Nup50 crucial for NPC assembly, we employed different Nup50 fragments in the cell‐free assay. These experiments revealed that a minimal fragment comprising aa 1–114 is sufficient to substitute full‐length Nup50 in NPC assembly (Fig 6C and D). Also in the context of this minimal fragment, the mutation compromising importin α binding does not interfere with NPC formation. Thus, neither the Ran‐binding domain, located in the C‐terminus of the protein, nor MEL28/ELYS, Nup153, importin α, and importin β binding of Nup50 is required for the protein’s function in NPC assembly, consistent with the previous data.
Both Nup50 orthologs are functional in nuclear pore complex assembly
We speculated earlier that the mouse Nup50 paralogs could functionally substitute each other in the NPC assembly. To further test this hypothesis, we replaced Nup50 in egg extracts by either of the two mouse orthologs, recombinantly expressed in bacteria (Fig 7A). Interestingly, both orthologs could rescue the NPC assembly phenotype (Fig 7B and C). However, whereas Nup50A localized both to chromatin and the NE in the assembly reaction, Nup50B was only found on chromatin.
Figure 7. Both mouse Nup50 orthologs can replace the Xenopus protein.
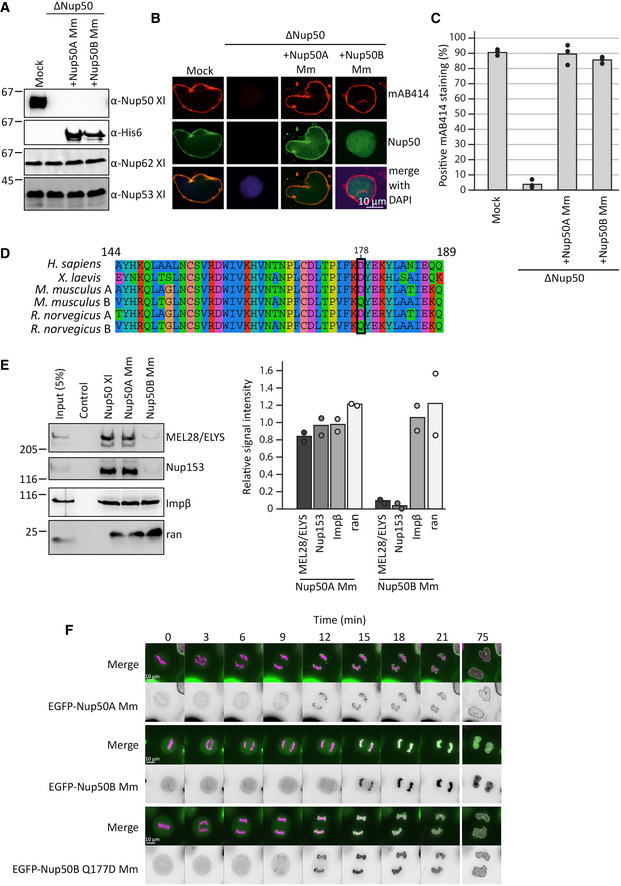
- Western blot analysis of mock‐ and Nup50‐depleted Xenopus egg extracts (ΔNup50), with or without addition of recombinant mouse Nup50 orthologs. Samples were analyzed with Xenopus Nup50 antibodies, which do not recognize the mouse proteins, which in turn were detected with an His6 antibody.
- Confocal microscopy images of fixed nuclei assembled for 120 min in mock‐depleted (mock) and Nup50‐depleted (ΔNup50) Xenopus egg extracts supplemented with recombinant mouse Nup50 orthologs. Nuclei were stained for Nup50 (Nup50 antibody for Xenopus protein, His6 for mouse proteins) and NPCs (mAB414, red) on the chromatin (DAPI). Scale bar: 10 µm.
- The average percentage of mAB414‐positive nuclei for 100 randomly chosen chromatin substrates in each of three independent experiments (performed as in B) is shown. Data points from individual experiments are indicated.
- Sequence comparison of the conserved 46‐aa fragment of the human, mouse, rat, and Xenopus laevis sequences, with the residue 178 (Xenopus numbering), crucial for NPC binding in the Xenopus protein, is highlighted (black box). The color scheme indicates the type of amino acids according to the alignment software default setting.
- GST fusion constructs of the Xenopus Nup53 RRM domain (aa 162–267, control) full‐length Xenopus Nup50, and the two mouse Nup50 orthologs were incubated with Xenopus egg extracts. Starting material as well as bound proteins were analyzed by Western blotting with indicated antibodies (see Appendix Fig S1E for a gel showing the GST‐baits). The right panel shows the average MEL28/ELYS, Nup153, and importin β (Impβ) and ran bead‐bound signal from two independent experiments normalized to their respective Xenopus signal. Data points from individual experiments are indicated.
- HeLa cells were transfected with EGFP‐tagged constructs of both mouse Nup50 orthologs and the Nup50B Q177D (ortholog to the position 178 in Xenopus) mutant. After 24 h, the cells were analyzed by live cell imaging. Panel shows cells exiting mitosis (time normalized to metaphase‐to‐anaphase transition). The merge shows histone 2B in pink and Nup50 in green, and the black‐and‐white panel shows EGFP‐Nup50 signal. Scale bar: 10 µm.
Source data are available online for this figure.
A sequence comparison indicates that within the conserved region identified in the Xenopus protein as minimal NPC‐binding domain, several amino acid changes occur between the two mouse orthologs (Fig 7D), including an aspartic acid (D) to glutamine (Q) exchange in the position 177 (corresponding to the position 178 in Xenopus), which is critical for NPC binding in the Xenopus protein. Indeed, GST pull‐downs showed that the canonical Nup50A can interact with Nup153 and MEL28/ELYS, whereas mouse Nup50B shows only a very reduced interaction levels (Fig 7E and Appendix Fig S1E).
Transfection of HeLa cells with EGFP‐tagged versions of both mouse Nup50 orthologs similarly revealed that Nup50A localized to chromatin and the NE, consistent with published data (Dultz & Ellenberg, 2010), whereas Nup50B was only found on the chromatin (Fig 7F). A single point mutation, Q177D in Nup50B, was sufficient to relocalize the EGFP fusion to the NE. Together, these data demonstrate that both Nup50 orthologs can function in NPC assembly and support the notion that NPC localization of Nup50 is not required for its role in NPC re‐assembly at the end of mitosis.
A minimal N‐terminal fragment interacts with RCC1
To define the function of the N‐terminal part of Nup50, crucial for its function in NPC assembly, we performed pull‐down experiments of the corresponding human FLAG‐tagged fragment from HEK cells. By mass spectrometry, several interaction partners were identified and confirmed by Western blotting (Fig 8A and B), including importin α, RCC1, Nup93, RanBP2, and ANP32A. Although ANP32A interacted with human and Xenopus Nup50 in pull‐down assays, it only bound to the second mouse Nup50 paralog, Nup50B (Fig 8C). As both paralogs can rescue Nup50 depletion in egg extracts, it is unlikely that the ANP32A interaction is crucial for the role of Nup50 in NPC assembly. Similarly, the interaction with importin α is unlikely to be crucial for Nup50’s function in NPC assembly as a recombinant Nup50 protein defective in importin α binding can effectively replace endogenous Nup50 in the cell‐free assays (Fig 6C and D).
Figure 8. Nup50 stimulates RCC1 activity for NPC assembly.
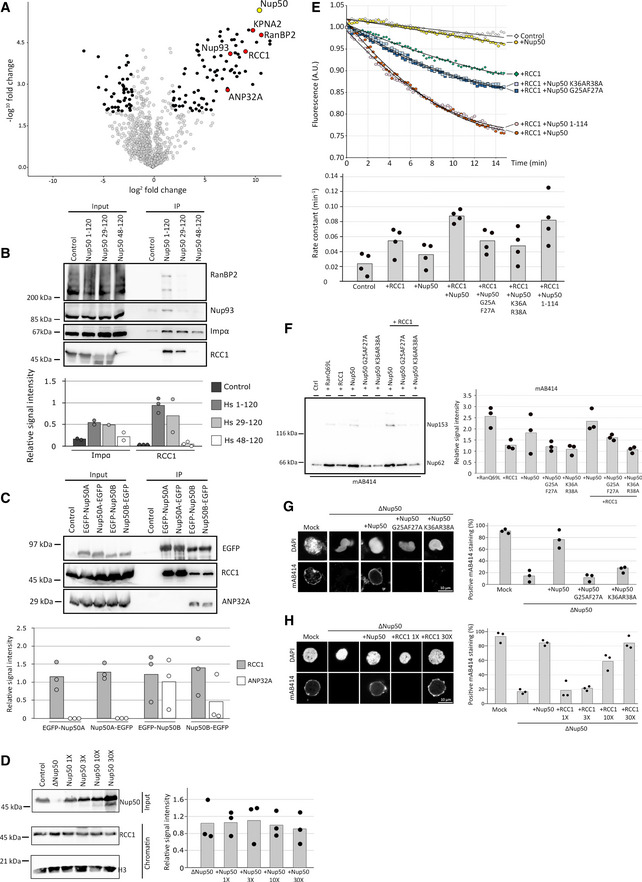
-
AHEK293T cells were transfected with empty FLAG‐tag vector as a control or FLAG‐Nup50 N‐terminal fragment (aa 1–120 of the human sequence). 24 h post‐transfection, cells were lysed, FLAG‐tagged proteins were immuno‐isolated, and analyzed by mass spectrometry with the Volcano plot showing the identified interactors (four‐fold change compared with the control and P < 0.01 in black, red if confirmed by Western blotting (B), see Dataset EV2 for full list) of Nup50 (yellow, data from three independent experiments). KPNA2 is the gene name of importin α.
-
BHEK293T cells were transfected with empty FLAG‐tag vector or FLAG‐Nup50 N‐terminal fragments (aa 1–120, 29–120 and 48–120 of the human sequence), processed as in (A) and analyzed by Western blotting, with 10% of the inputs loaded. The quantification (lower panel) shows the mean signal intensity normalized on the input of at least two independent experiments. Data points from individual experiments are indicated.
-
CHEK293T cells were transfected with both Nup50 mouse orthologs N‐ and C‐terminals tagged with EGFP. 24 h post‐transfection, cells were lysed, EGFP‐tagged proteins immuno‐isolated, and analyzed by Western blotting. The quantification of the Western blot (C, lower panel) shows the mean signal intensity normalized on EGFP of three independent experiments. Data points from individual experiments are indicated.
-
DXenopus sperm chromatin (6,000 sperm heads/µl were incubated with 120 µl of control), Nup50‐depleted or egg extracts supplemented with excess (1X = 0.07 µM final) recombinant Xenopus Nup50 and re‐purified. Total input extracts were analyzed by Western blotting against Nup50, and isolated chromatins were analyzed by Western blotting against RCC1 and histone H3 as a loading control (left panel). The quantification (right panel) shows the mean signal intensity of the RCC1 signal normalized over histone H3 from three independent experiments. Data points from individual experiments are indicated.
-
E2 µM recombinant Ran, loaded with MANT‐GDP was incubated with 2 mM GppNHp in buffer control, supplemented with 2 nM recombinant RCC1, 20 nM recombinant Xenopus Nup50 proteins, or RCC1 and Nup50 together. GDP‐to‐GppNHp exchange was monitored by the decrease in MANT fluorescence of the liberated GDP‐MANT. The lower panel shows the rate constant of each experimental condition with n = 4 independent experiment per condition, bars represent the mean, and individual data points are indicated.
-
FXenopus egg extracts were supplemented with 5 µM RanQ69L, RCC1, Xenopus Nup50 wild‐type, and RCC1‐binding mutants as well as a combination of 5 µM Nup50 and 5 µM RCC1. After 90 min, annulate lamellae were isolated by centrifugation and quantified by Western blotting with mAB414 antibody. Quantitation shows the relative Nup62 signal as a mean from three independent experiments, normalized to the buffer control. Individual data points are indicated.
-
GConfocal microscopy images of nuclei assembled for 120 min in mock‐depleted, Nup50‐depleted (ΔNup50), and Nup50‐depleted Xenopus egg extracts supplemented with recombinant Xenopus wild‐type Nup50 or RCC1 binding mutants. Nuclei were fixed in 4% PFA and 0.5% glutaraldehyde, stained for NPCs (mAB414) and the chromatin (DAPI). Scale bar: 10 µm. Quantitation shows the average percentage of mAB414‐positive nuclei for 100 randomly chosen chromatin substrates in each of three independent experiments. Individual data points are indicated.
-
HConfocal microscopy images of nuclei assembled for 120 min in mock‐depleted, Nup50‐depleted (ΔNup50), and Nup50‐depleted Xenopus egg extracts supplemented with RCC1 excess as indicated. Nuclei were fixed in 4% PFA and 0.5% glutaraldehyde and stained for NPCs (mAB414) and the chromatin (DAPI). Scale bar: 10 µm. Quantitation shows the average percentage of mAB414‐positive nuclei for 100 randomly chosen chromatin substrates in each of three independent experiments. Individual data points are indicated.
Source data are available online for this figure.
The guanine nucleotide exchange factor (GEF) of Ran, RCC1, reportedly plays a crucial role in NPC assembly (Walther et al, 2003b; Zierhut et al, 2014). RCC1 dynamically interacts with chromatin, and several factors are known to regulate this interaction. As Nup50 also interacts with chromatin, we speculated that it might affect RCC1 chromatin binding. However, RCC1 loading on re‐isolated sperm chromatin after incubation in egg extracts was neither affected by Nup50 depletion nor by the addition of excess of Nup50 (Fig 8D). Alternatively, Nup50 might affect RCC1’s GEF activity. Indeed, in GDP‐to‐GTP exchange assays, recombinant full‐length Xenopus Nup50 as well as its N‐terminal region enhanced RCC1 activity toward Ran approximately twofold (Fig 8E). Thus, Nup50 might function in NPC assembly by stimulating RCC1 activity, which in turn generates RanGTP in the vicinity of chromatin as a crucial factor for NPC formation. Ectopic addition of a high concentration of RanGTP or of the constitutively active mutant RanQ69L to egg extracts in the absence of chromatin stimulates formation of annulate lamellae (ALs) (Walther et al, 2003b), membrane stacks with tightly packed NPCs in the ER‐like membranes. Thus, AL formation in Xenopus egg extracts can serve as sensitive readout for perturbations of the Ran system. Interestingly, addition of Nup50 also increases AL formation under these conditions and co‐addition of RCC1 increases the effect (Fig 8F and Appendix Fig S7A). As AL form in the absence of chromatin, this finding is consistent with the notion that Nup50 does not act on RCC1 by recruiting it to chromatin, but rather stimulates its enzymatic activity.
To directly assess the contribution of the RCC1–Nup50 interaction to the NPC assembly, we generated Nup50 mutants defective in RCC1 binding by mutating pairs of highly conserved amino acids in the N‐terminal region. Two of them, G25A, F27A and K36A, R38A impaired RCC1 binding without preventing the importin α interaction (Appendix Fig S7B). Similarly, in the context of the N‐terminus of the Xenopus protein in egg extracts, the corresponding mutants could not bind RCC1 (Appendix Fig S7C). Accordingly, in the context of the full‐length Xenopus protein, the mutants failed to stimulate RCC1 GEF activity (Fig 8E). Moreover, addition of either mutant alone or in combination with RCC1 could not stimulate AL formation in egg extracts (Fig 8F). Furthermore, both mutants could not rescue depletion of endogenous Nup50 in the in vitro nuclear assembly assay (Fig 8G).
Thus, the Nup50‐RCC1 interaction seems crucial for the Nup50 effect on NPC reformation both in the nuclear assembly and the AL assay, presumably by stimulating RCC1 activity. If Nup50 acts by increasing RCC1’s GEF activity, we wondered whether loss of Nup50 could be compensated by an excess of RCC1. Indeed, addition of 10‐fold or 30‐fold RCC1 excess over endogenous RCC1 levels (determined as 40 nM (Zhang et al, 2014)) to Nup50‐depleted egg extracts restored NPC assembly activity (Fig 8H).
Discussion
Using cellular and cell‐free assays, we show that the nucleoporin Nup50 plays a crucial role in NPC assembly at the end of mitosis. Surprisingly, NPC localization of Nup50 is not required for its function in NPC assembly. Consistent with this conclusion, Nup50 is reportedly absent in NPCs of annulate lamellae (Hase & Cordes, 2003), ER membrane stacks in which NPCs have integrated at a distance to chromatin (Kessel, 1992). In addition, Nup50 has a short residence time at the NPC (Rabut et al, 2004) and is continuously exchanged at NPCs even in nondividing cells (D'Angelo et al, 2009), two features that are typical for peripheral nucleoporins that are not part of the NPC scaffold. Our data suggest that the N‐terminus of Nup50 binds to the RanGEF RCC1 and stimulates its GDP‐GTP exchange activity toward Ran.
In addition to its crucial role in nuclear transport, Ran has important functions during mitosis. It promotes spindle assembly during mitotic entry and NPC re‐assembly during mitotic exit (for review, see Forbes et al (2015)). The chromatin‐bound RanGEF RCC1 is crucial for the latter function. It generates a high RanGTP concentration around chromatin, which is required to release essential spindle assembly factors such as Tpx2 or NuMA from inhibitory nuclear transport receptors (Gruss et al, 2001; Wiese et al, 2001). Although the precise RanGTP target functioning in NPC assembly is currently unknown, a similar function has been proposed for NPC formation. Our data indicate that the interaction of Nup50 with RCC1 is crucial for its function in NPC assembly at the end of mitosis. A short N‐terminal fragment interacting with RCC1 and stimulating its RanGEF activity is sufficient to replace the endogenous Nup50 in in vitro NPC assembly assays (Fig 6). Accordingly, Nup50 mutants that cannot bind and activate RCC1 are unable to substitute the wild‐type protein (Fig 8). Stimulation of RCC1 should increase RanGTP production in the proximity of chromatin and promote NPC assembly. Several additional factors are known to modulate RCC1 activity, including chromatin binding, which stimulates RCC1 (Nemergut et al, 2001), and RanBP1, which inhibits RCC1 by binding and preventing its chromatin interaction (Zhang et al, 2014). With Nup50, we now identify another regulator of RCC1 and the Ran system important for NPC re‐assembly during mitotic exit.
Consistent with the idea that Nup50 stimulates the RCC1 activity, the effect of Nup50 depletion on NPC assembly can be reverted by addition of an excess of RCC1, at least in egg extracts (Fig 8H). We attempted but failed to compensate for Nup50 depletion in the Xenopus extract system by addition of RanQ69L as appropriate titration of this system presents a significant challenge. Addition of RanQ69L is known to lead to ectopic NPC assembly in AL and block NPC assembly (Walther et al, 2003b).
Much like MEL28/ELYS, a fraction of the nucleoporin Nup50 pool is found on decondensing chromatin at the early stages of mitotic exit (Hase & Cordes, 2003; Dultz et al, 2008). This early localization is consistent with our finding that Nup50 can bind chromatin in in vitro assays (Figs 3 and 4). Nevertheless, chromatin binding of Nup50, at least in the in vitro nuclear assembly reactions, is not crucial for its function in NPC assembly. Later in the nuclear reassembly process, a larger fraction (about 80%) of Nup50 associates with NPCs in human cells (Dultz et al, 2008) and in the in vitro Xenopus assemblies (Fig 3). At later time points, a more intensive rim staining is observed for Nup50. This NPC localization depends on NPC formation because in the absence of MEL28/ELYS, which reportedly blocks NPC assembly, the rim staining is apparently lost (Fig 3B). We speculate that the first pool of Nup50 recruitment is required for RCC1 stimulation, whereas the second pool integrates into assembled NPCs.
Nup50 is localized at the nucleoplasmic side of the NPC (Guan et al, 2000). Consistent with this localization, it interacts with Nup153 (Smitherman et al, 2000; Kosako et al, 2009; Makise et al, 2012), which is similarly found in this region of the NPC. Downregulation of Nup153 abolishes the rim but not the nucleoplasmic localization of Nup50, indicating that it is no longer found at NPCs (Hase & Cordes, 2003; Kosako et al, 2009; Mackay et al, 2010; Aksenova et al, 2020). Interestingly, loss of Nup153, which leads to Nup50 displacement from the NPC, does not affect assembly of the core structure of the NPC (Hase & Cordes, 2003), which is consistent with our finding that Nup50’s NPC localization is not crucial for its role in NPC assembly. We identified a conserved 46‐amino acid region of Nup50 that is required for NPC localization and is a Nup153‐ and MEL28/ELYS‐binding domain, although we currently cannot ascertain whether Nup153 and/or MEL28/ELYS binding is direct or indirect. This minimal region is included in the Nup50 fragment (aa 1–214 of the human sequence) previously identified as Nup153 interaction site (Makise et al, 2012). It is also required for MEL28/ELYS interaction, which has been identified as Nup50 interaction partner in egg extracts (Gillespie et al, 2007). We have not been able to separate the Nup153‐ and MEL28/ELYS‐binding sites, and as all NPC‐binding mutants affect both interactions similarly, we suspect that both proteins use indeed the same sequence motifs for interaction with Nup50. In light of the published data from human cells, it is likely that Nup153 binding is the determinant for Nup50’s NPC localization. Nup50 can also interact in pull‐down experiments with Nup153 via importin α, an interaction requiring its N‐terminal importin α‐binding domain (Makise et al, 2012) (our unpublished data). However, at least in cell transfection assays, this interaction is not able to recruit Nup50 to NPCs (Fig 5).
Homozygous Nup50 knockout mice die during late embryonic development (Smitherman et al, 2000), indicating a crucial function of Nup50. The embryos are growth‐retarded and show neural tube defects. Mouse embryonic fibroblasts isolated from Nup50 null embryos exhibit no obvious defects in cell cycle control or NPC function, and nuclei possess normal mAB414 staining. These findings are surprising in light of our experiments. They may suggest that a Nup50‐related gene takes over Nup50 function (Smitherman et al, 2000). Genome and EST database searches suggest that a rodent‐specific gene, which shows 58.2 and 59.3% amino acids identity in mouse and rat, respectively, and possesses the same domain arrangement as mouse/rat canonical Nup50 (Fig 2) could compensate for the loss of Nup50. As previously noted (Smitherman et al, 2000), this gene is highly expressed in testis but additionally shows detectable expression levels in other tissues (Bult et al, 2019; Papatheodorou et al, 2020). In line with this interpretation, our RNAi experiments in HeLa cells show a marked loss of mAB414 staining upon Nup50 depletion, which is only recapitulated in mouse NIH3T3 cells if siRNAs against both potential orthologs are employed. All tested Nup50 antibodies are directed only against the canonical Nup50A ortholog and, unfortunately, did not detect the second ortholog. Using the auxin‐inducible degron system, it was recently shown that Nup50‐depleted DLD‐1 cells are viable and continued to grow, albeit somewhat more slowly than the parental cells (Aksenova et al, 2020). However, in these cells, RCC1 is expressed as fusion with a fluorescent protein and the ubiquitin ligase. This might result in expression or functional changes of RCC1 which could compensate for the loss of Nup50.
The two Nup50 paralogs described in rodents are encoded on different chromosomes and distinct from the two reported human Nup50 isoforms, Npap60L (aa 1–469) and Npap60S (aa 29–469) (Trichet et al, 1999; Patre et al, 2006) that are generated by alternative splicing. Npap60L is widely expressed from vertebrates to yeast and corresponds here to Nup2. Npap60S has been only found in humans. Npap60L and Npap60S have been suggested to differentially regulate nuclear import (Ogawa et al, 2010). Although Npap60L promotes cargo release from importin α, Npap60S would rather stabilize the importin α‐cargo complex. It should be noted that a 29–120 fragment of human Nup50 binds RCC1, arguing that both isoforms could, in theory, bind RCC1 (Fig 8B). Additionally, in Xenopus laevis, two paralogs of Nup50 are found, but this is also different from the situation observed in mouse. The two Xenopus paralogs arise from an ancestral whole‐genome duplication (Session et al, 2016), and both loci have retained their exon/intron organization (Karimi et al, 2018). Moreover, the two paralogs show a 89.5% sequence identity in Xenopus as compared with approximately 60% in rodents. In Xenopus, both paralogs can be detected by Western blotting depending on the total protein amount loaded and electrophoresis conditions (see Fig 1A and Appendix Fig S3) and are both efficiently depleted by our Nup50 antibodies. So far, we have no data indicating that the Xenopus paralogs could have different biological functions.
Although the Nup50 ortholog Nup2 is not essential in budding and fission yeast, it is vital in the fungus Aspergillus nidulans (Osmani et al, 2006; Markossian et al, 2015). During semi‐open mitosis, where NPCs only partially disassemble, Nup2 relocalizes to the mitotic chromatin. In anaphase and telophase, Nup2 attracts other nucleoporins to chromatin, thereby serving as bridge between the segregating chromatin and NPCs (Suresh et al, 2017). This prevents loss of NPCs from the NE during nuclear division and guarantees nuclear transport capabilities in G1 phase. Thus, both vertebrate Nup50 and Aspergillus nidulans Nup2 secure appropriate NPC numbers and transport competence of the nucleus during mitotic exit, yet by different mechanisms. Although for yeast Nup2, both chromatin association and interaction with NPCs matter, vertebrate Nup50 does not crucially rely on these capabilities for its function in NPC reassembly.
In summary, here, we characterize Nup50, which has been so far mainly described as a nucleoporin enhancing nuclear transport efficiency by interactions with the importin α family and Ran, as factor involved in NPC re‐assembly at the end of mitosis by enhancing RCC1 activity. This same mechanism of RCC1 activation might also contribute to the function of Nup50 in nuclear transport. Generation of RanGTP by RCC1 is critical for highly efficient and directed transport across the NPC barrier. RCC1 and the Ran system also play a crucial role early in mitosis, for example, in directing spindle assembly toward chromatin. It remains open whether Nup50 already modulates RCC1 activity during this phase of the cell cycle, akin to its newly identified role in nuclear reformation. Similarly, we cannot exclude that Nup50, in addition to its function in NPC assembly at the end of mitosis, also contributes to interphase NPC assembly, which thus remains an interesting question for further studies.
Materials and Methods
Protein expression and purification
Xenopus laevis Nup50—full length and fragments—were cloned as codon‐optimized sequences for expression in E. coli into a modified pET28a vector with an N‐terminal yeast SUMO solubility tag, which is followed by a tobacco etch virus (TEV) cleavage site, an N‐terminal GST‐tag, followed by a TEV cleavage site or an N‐terminal EGFP‐tag (see Dataset EV1 for list of all constructs used in this study). Both mouse Nup50 orthologs were cloned into an pET28a vector. Proteins were expressed in BL21de3 E. coli by autoinduction in LB medium at 18°C and purified using Ni‐NTA beads (Qiagen). When present, the SUMO‐tag was cleaved using TEV protease at 4°C overnight. SUMO‐tag and protease were removed by passing the protein mixture over Ni‐NTA beads and the protein of interest (Nup50 wild‐type and different mutants, mostly used for the addback assays) dialyzed in a sucrose buffer (250 mM sucrose, 50 mM KCl, 10 mM Hepes‐KOH, 2.5 mM MgCl2). Ran wild‐type and the Q69L mutant as well as RCC1 were purified, as described (Walther et al, 2003b). Ran, RCC1 and Nup50 proteins employed in the GEF assays (see below) were further purified by size exclusion chromatography on a Sephadex 200 increase (10/300) column (GE Healthcare).
Antibodies
A Nup50 antibody from Abcam (ab137092) was used at a 1:500 dilution for immunofluorescence in human cells and 1:1,000 dilution for Western blots, a Nup50 antibody from Abcam (ab85915) at a 1:500 dilution for immunofluorescence in mouse cells and at a 1:1,000 dilution for Western blot. An MEL28/ELYS antibody from Bio Matrix Research (BMR 00513) was used at a 1:500 dilution for immunofluorescence in human cells and 1:1,000 dilution for Western blots. The MEL28/ELYS antibody used on mouse cells was a gift from Thomas Schwartz. Mouse monoclonal antibodies mAB414 were from Covance (MMS‐120R, used 1:2,000 for immunofluorescence and 1:10,000 for detection of Nup62, Nup214 and Nup153 in Western blot), His6 antibodies from Roche (11922416001, used 1:1,000 for Western blotting), GFP antibodies from Roche (11814460001, 1:2,000), tubulin from Sigma (T61999, used at 1:10.000 for Western blots on human cells), actin from MP Biomedicals (691001, used at 1:1,000 for Western blots on mouse cells), and ANP32A from Cell Signaling (#15491, D7Z5U used 1:1,000, for Western blotting). Rabbit polyclonal Nup50 antibodies were generated in rabbits using full‐length Xenopus laevis Nup50 (see Appendix Fig S1A for specificity test). Antibodies against Xenopus Nup53 (Theerthagiri et al, 2010), Nup62 (De Magistris et al, 2018), MEL28/ELYS (Franz et al, 2007), Nup153, Importin β, and lamin B (Vollmer et al, 2015), as well as Ran, RCC1, and importin α (Walther et al, 2003b) were previously described. Antibodies were employed in a dilution of 1:1,000 for Western blotting and 1:100 for immunofluorescence. Rabbit monoclonal His6 antibodies from Cell signaling (#12698, D3I1O) were used 1:100 for immunofluorescence. Beads for immunodepletion were generated, as described (De Magistris et al, 2018).
For immunofluorescence, Alexa 488‐coupled goat anti‐mouse IgG and Alexa 546‐coupled goat anti‐rabbit IgG (Invitrogen) were used. For Western blotting, goat anti‐rabbit IgG‐HRP and goat anti‐mouse IgG‐HRP (Calbiochem) were used. For analysis of immunoprecipitations by Western blotting, protein‐A‐HRP (Calbiochem) was used to avoid cross‐reactivity with the heavy and light chains of the immunoglobulins. Signals were detected using Western Bright Quantum (Advansta) as ECL substrate on a ImageQuant LAS‐4000 system (Fuji). Intensities of nonsaturated signal were analyzed using FiJi software, an open‐source platform for biological image analysis (Schindelin et al, 2012). The bands to be measured were defined as the region of interest, and the mean intensity signals of these regions were extracted. Background intensity was subtracted in order to calculate the band signal.
Pull‐down experiments
For pull‐down assays, 0.5 µM GST bait proteins (Nup50 full‐length or fragments, Nup53 aa 162–267 served as control (Vollmer et al, 2012)) were incubated with in 400 µl of egg extracts for 1 h at 4°C, supplemented with 60 µl of 50% slurry of GSH‐Sepharose 4B (GE Healthcare), and incubated for another 2 h. The Sepharose was washed five times with PBS, and bead‐bound proteins eluted in 30 µl total volume by TEV protease cleavage (0.5 mg/ml) for 1 h at 25°C. The TEV protease cleaves the GST fusions between the GST moiety and the bait proteins. The input and elutions were analyzed by Western blotting using the indicated antibodies. For quantitation, ECL signals were determined using FiJi. Background signals were subtracted, and the signal ratio of eluate/input was calculated.
Nup50 fragments aa 1–120, 29–120, and 48–120 of human Nup50 were cloned into the pCMV‐3Tag‐1B vector. Both mouse Nup50 paralogs were cloned into pEGFP‐N3 and pEGFP‐C3 vectors (labeled in the figures as EGFP‐Nup50 or Nup50‐EGFP to indicate the N‐ or C‐terminal positions of the tag). Xenopus laevis Nup50 and point mutants were cloned into pEGFP‐C3 vector. HEK293T cells were cultivated in DMEM with 10% fetal bovine serum and 1% penicillin/streptomycin. For FLAG‐tag‐IPs, pCMV‐3Tag‐1B vectors including an empty control vector were transfected into the cells by using JetPRIME transfection reagent (PolyPlus Transfection, Illkirch, France) according to the manufacturer indication. After 24‐h transfection, the cells were lysed with ice‐cold NP‐40 lysis buffer (20 mM Hepes‐KOH, pH 7.4, 150 mM NaCl, 0.5% NP‐40, 2 mM EDTA) containing protease inhibitor. The cell lysates were incubated with Anti‐FLAG M2 Magnetic Beads (Sigma, Taufkirchen, Germany) end‐over‐end rocking for 2 h at 4°C. After washing, the beads were used for mass spectrometry‐based proteomic analysis or Western blotting. Similarly, pEGFP‐N3‐ and pEGFP‐C3‐based vectors were transfected into HEK293T cells by using JetPRIME transfection reagent. 24 h later, the cells were lysed with ice‐cold NP‐40 lysis buffer (see above), and cell lysates were incubated with GFP‐Trap Magnetic Beads (ChromoTek, Planegg‐Martinsried, Germany) end‐over‐end rocking for 2 h at 4°C. After washing, proteins were analyzed by SDS–PAGE and immunoblotting.
Mass spectrometry
FLAG‐Nup50 aa1–120 interactome analysis was performed similar to a published study (Düsterhöft et al, 2021). Co‐IP samples were prepared in biological triplicates as described before, with the exception that after the final wash with NP‐40 lysis buffer, the beads were washed three times with lysis buffer without detergent. The dried beads were then frozen at −80°C until sample preparation. The bead‐bound proteins were then treated 1 h with 5 μg/ml trypsin (in 2 M urea, 50 mM Tris‐HCl pH 7.5) at room temperature. The resulting supernatant was transferred to a new tube, and the beads were washed twice with 2 M urea, 50 mM Tris‐HCl pH 7.5, and 1 mM DTT. All individual supernatants were subsequently combined and subjected to overnight digestion at room temperature. The resulting peptide solution was then treated with iodoacetamide, acidified, desalted (using home‐made C18‐tips), and lyophilized. The peptides were then resuspended in 15 µl 3% formic acid (FA)/5% acetonitrile (ACN) and loaded onto a nanoLC system (RSLCnano, Thermo Scientific). Trapping of the peptides was performed for 10 min on a precolumn (Acclaim PepMap100, C18, 5 µm, 100 Å, 300 µm i.d. × 5 mm, Thermo Scientific), which was then followed by separation on an analytical column (Easyspray 50 cm column (ES803) at 45°C; Thermo Scientific) using a 125‐min gradient (0–10 min: 5% buffer B (buffer A: 0.1% FA; buffer B: 80% acetonitrile, 0.1% FA), 10–60 min: 5–20% buffer B, 60–98 min: 20–35% buffer B, 98–101 min: 35–99% buffer B, 101–106 min: 99% buffer B, 106–109 min: 99–5% buffer B, 109–125 min: 5% buffer B) with a spray voltage of 2 kV and the capillary temperature set at 250°C. Analysis of the samples was performed on a Q Exactive plus mass spectrometer (Thermo Scientific) in data‐dependent mode. Full MS settings: 70,000 resolution; AGC target, 1e6; maximum injection time, 50 ms; scan range, 350–1,600 m/z. dd‐MS2 settings were: 17,500 resolution; AGC target: 1e5; maximum injection time: 55 ms; top 20 precursor fragmentation; isolation window, 2.0 m/z; collision energy, 27. dd settings were: minimum AGC, 5e2; 20 s dynamic exclusion; only 2+ to 5+ peptides were allowed.
The MaxQuant software suite (using the built‐in Andromeda search engine) was used for analysis of the raw data (Tyanova et al, 2016a), which was searched against the human UniProt database version 12/2020 (only containing reviewed and canonical sequences): MaxQuant default settings (including the mass tolerance) were used. Specific settings: trypsin as the specific protease (two missed cleavages); carbamidomethylation (Cys) as fixed modification; and oxidation (Met) and N‐terminal protein acetylation as variable modifications. The false discovery rate was 0.01 on both peptide and protein levels, and the minimum peptide length was seven amino acids. Quantification was done using the label‐free quantitation algorithm from MaxQuant.
The resulting proteinGroups.txt file (Dataset EV2) from the MaxQuant search was then used for statistical analysis employing the Perseus software suite (version 1.6.14.0) (Tyanova et al, 2016b). The LFQ intensities from all biological replicates were used as the main columns. First, contaminant, reversed, and “only identified by site” entries were removed from the protein list. Prior to subsequent analysis, only entries with a minimum of 1 unique peptide and 2 total peptides (razor + unique) were left in the protein list. The three individual biological replicates were grouped as NUP and ctrl, and the data were log2‐transformed. Proteins were only included in the final data set if they were identified in all replicates in at least one group (min 3 in one group). Missing data points (“NaN”) were then imputed by replacing them with values based on normal distribution (using the default settings of Perseus). Further analysis was done by performing a two sample test (Perseus default settings). The resulting file was used for generation of the Volcano plots (Fig 8A). The settings for the volcano plot were P‐value < 0.01 and a ratio of > 4‐fold change (> 2 (–log (10) P‐value) and > 2 (log (2) ratio)).
In vitro nuclear assembly
Preparation of high‐speed interphase extracts, sperm heads, floated labelled and unlabeled membranes required for in vitro nuclear assembly, as well as immunofluorescence experiments were carried out, as described in (Eisenhardt et al, 2014), nuclear transport assays as in (De Magistris et al, 2018). For immunodepletion, Protein‐A Sepharose FF (GE Healthcare) was incubated with saturating amounts of Nup50 antisera or unspecific rabbit control sera. After washing, bound antibodies were cross‐linked to protein‐A by incubation with 10 mM dimethyl pimelimidate (Thermo Fisher Scientific) twice for 10 min, beads were blocked with 3% BSA in PBS. For immunodepletion, high‐speed egg extracts were incubated with the 0.8 volume of antibody beads for twice 20 min (Eisenhardt et al, 2014). For addback experiments, recombinant Nup50 proteins were added to Nup50‐depleted extracts to approximately endogenous levels, based on the determined protein concentration (BCA method) and confirmed by Western blotting. In all depletion‐addback experiments, full‐length Nup50 was added to depleted extracts as positive control. If this reaction did not rescue the NPC assembly phenotype to 50% or the mock depletion did not yield more than 70% of chromatin templates with full rim NPC staining the experiment was discarded.
Fluorescence images were acquired using a confocal microscope [FV1000; Olympus; equipped with a photomultiplier (model R7862; Hamamatsu)] using 405‐, 488‐ and 559‐nm laser lines and a 60× NA 1.35 immersion oil objective lens or a Zeiss LSM710 confocal microscope equipped with a Plan‐Apochromat 63×/1.4 immersion oil objective and 405, 488 and 561 nm lasers, using ZEN software. Annulate lamellae assays were performed in 45 µl of Xenopus egg extract, 5 µl of floated membrane, and an energy‐regenerating system containing (Eisenhardt et al, 2014) the protein of interest were added at 10 µM final concentration in a reaction volume of 60 µl and incubated for 4 h at 20°C. Processing of the samples were carried out, as previously described (Vollmer et al, 2015). Reactions were stopped by adding 1 ml of sucrose buffer (250 mM sucrose, 50 mM KCl, 10 mM Hepes‐KOH, 2.5 mM MgCl2), spun for 10 min at 7,500 g. The pellet was resuspended in 10 µl SDS sample buffer and analyzed by SDS–PAGE and Western blotting using mAB414 antibodies at 1:10,000 dilution. ECL signals were quantified using FiJi, measuring the mean intensity of the Nup62 signal in each condition, subtracting the background signal and normalizing to the control.
Cellular experiments
HeLa, HeLa stably expressing H2B‐mCherry, EGFP‐Nup107 (Otsuka et al, 2016) or EGFP‐Nup133, the latter expressing a triple EGFP‐tag fused to mouse Nup133 (Bolhy et al, 2011). HEK293T and NIH‐3T3 cells were grown in Dulbecco's modified Eagle's medium (DMEM, Gibco) with pyruvate supplemented with 10% of decomplemented fetal bovine serum (Gibco) and 100 U/ml of penicillin (Gibco) and 100 μg/ml of streptomycin (Gibco). Cells were incubated at 37°C in a 5% CO2 atmosphere. siRNA against Nup50 (SI00663236) was obtained from Qiagen. The siRNA against mouse Nup50 (156930) was purchased from Ambion Life Technologies. The siRNA against the mouse Nup50B was custom‐made with the following sequence: 5′ GAAGCCAGCAUCUGCCAAAtt 3′ and purchased from Ambion Life Technologies. Cell transfection was performed in a 24‐well plate with Lipofectamin RNAimax (Thermo Fisher scientific). Lipofectamine and siRNA were mixed in OptiMEM (Gibco), incubated at least 20 min at room temperature, and put in a well. 520 μl of cell suspension in growing medium was added on the top of 80 μl of siRNA solution to obtain a final concentration of 20 nM of siRNA and 20,000 cells per well. Cells were incubated 72 h at 37°C in a 5% CO2 atmosphere.
After immunofluorescence, the mAB414 signal was quantified using FiJi. Total nuclear areas were automatically selected using the threshold option, and inner nuclear areas were manually drawn and subtracted to obtain the nuclear rim area and calculated its mean signal intensity. For each experiment, the mAB414 signal was plotted and the average calculated using GraphPad Prism. The Student t‐test (Fig 1D) or paired ratio test (Fig 2B) was used for the statistical analysis.
For localizing different EGFP‐Nup50 fragments and mutants in cells, HeLa cells at a confluence of 20–30% were transfected with the respective EGFP‐C3 constructs using FuGENE6 transfection reagent (Roche) following the manufacturer’s instructions. After 36 h, the cells were washed three times with PBS and for 1 s pre‐extracted using PBS supplemented with 0.1% (v/v) Triton‐X100 (Merck) to reduce the nucleoplasmic EGFP signals. After fixation with 4% paraformaldehyde in PBS for 20 min, the chromatin was stained with RedDot™2 Far‐Red Nuclear Stain (Biotium, 1:100 diluted in PBS), and the samples were mounted using Vectashield Mounting Medium and analyzed on an FV1000; Olympus confocal microscopy (see above).
Live cell imaging
HeLa cells stably expressing H2B‐mCherry generated as in Schooley et al (2015) and cultured as before were seeded in 8‐well µ‐slide chambers (Ibidi), and the next day, they were transfected with 9 ng/µl from the indicated EGFP fusions (pEGFP‐C3 constructs expressing wild‐type and mutant mouse Nup50 proteins) using jetPRIME (Polyplus, Illkirch, France). The cells were imaged 24 h post‐transfection with a spinning disk confocal microscope Ti2 Eclipse (Nikon) equipped with an LED light engine SpectraX (Lumecor, Beaverton, Oregon, USA) and GFP/mCherry filter sets, a Lambda Oil 60× NA 1.4 objective, and environmental control system UNO‐T‐H‐CO2 (Okölab, Ottaviano, NA, Italy). AR‐Elements software (Nikon) equipped with a software‐based autofocus module was used to perform confocal fluorescence imaging of the single best‐in‐focus optical section of cells in metaphase every 3 min during approximately 2 h. Image galleries from cell trajectories during mitotic exit were assembled using FiJi and mounted for figures using Inkscape (Free Software Foundation, Inc. Boston, USA).
HeLa cells stably expressing EGFP‐Nup107 (Otsuka et al, 2016) or EGFP‐Nup133 (Bolhy et al, 2011) were transfected with the indicated siRNAs. After 48 h, chromatin was stained with 200 nM of SiR‐Hoechst (Spirochrome) for 90 min and the cell subjected to live cell imaging as before. Image galleries from mitotic cell trajectories were extracted using the Nikon Elements software, assembled, and analyzed using FiJi excluding cells with a tilted axis of cell division or severely impaired chromatin segregation as well as multinucleated cells. The time of EGFP‐Nup107 or EGFP‐Nup133 appearance on the chromatin surface was determined by visual inspection of the image galleries. Additionally, the EGFP signal at the rim of their segregated chromatin masses 15 min after the last metaphase frame was measured. For this, ROIs of the chromatin border and cytoplasm were manually sampled using the FiJi “segmented line” tool with a width value of five. From these ROIs, the mean EGFP signal intensity at the chromatin border was determined after subtraction of the cytoplasmic signal. To compare cells with different EGFP‐fusion expression levels, signals from cells with similar cytoplasm mean intensities were normalized to the mean value of control cells in this range (Samwer et al, 2017). Data were tested for normality, and a two‐tailed t‐test was performed using GraphPad Prism software.
DNA beads experiments
The MCP1 plasmid (Heald et al, 1996) was digested by BamHI and NotI, and the digestion product was purified. And 30 μg of DNA was biotinylated using 0.36 U of Klenow fragment, 63 μM of biotin‐18‐dUTP, 63 μM of biotin‐14‐dATP, 90 μM of thio‐CTP, and 90 μM of thio‐GTP. The DNA was purified in a CHROMA SPIN column according to the manufacturer’s instructions. The biotinylated DNA was coupled to Dynabeads Kilobinders (Thermofisher Scientific) following the manufacturer’s instructions. DNA‐coupled magnetic beads were incubated with protein in a tabletop shaker for 3 h at 20°C at 350 rpm. After washing, beads were collected in 10 μl of egg extract or sucrose buffer (250 mM sucrose, 50 mM KCl, 10 mM Hepes‐KOH, 2.5 mM MgCl2) and incubated with DAPI (Roth) at 1 μg/ml 10 min at room temperature. Then 3 μl was sampled on a 3‐well Diagnostika slides (X1XER303B, Thermofisher Scientific) for observation on an Zeiss LSM710 confocal microscope equipped with a Plan‐Apochromat 63×/1.4 Oil objective and 405 and 488 nm lasers, using ZEN software.
RCC1 guanine nucleotide exchange assay
GEF assays were adapted from Kanie and Jackson (2018). Recombinant Ran was loaded with MANT‐GDP (Jena Bioscence, NU‐204) in a low magnesium loading buffer (20 mM Hepes‐KOH, 50 mM NaCl, 0.5 mM MgCl2, 10 mM EDTA, 2 mM DTT and 20 fold molar excess of MANT‐GDP over Ran). Exchange for GppNHp was performed. Ran MANT‐GDP to GppNHp (Jena Bioscence, NU‐401) exchange experiments were carried out with 2 µM of Ran in a volume of 50 µl in a black 96‐well glass‐bottom plate in a nucleotide exchange buffer (40 mM Hepes‐KOH, 50 mM NaCl, 10 mM MgCl2 and 2 mM DTT) in the presence of 2 mM GppNHp excess. The assays were performed with an excitation wavelength of 360 nm and emission at 440 nm in a SpectraMax iD3 spectrophotometer from Molecular Devices every 15 s. The rate constant K was calculated using GraphPad Prism software with a nonlinear regression model and the “dissociation‐one phase exponential decay” analysis.
Sequence alignment
Human (ENST00000347635.9), Xenopus laevis (BC077201.1), Xenopus tropicalis (NM_001016628.2), mouse (A: ENSMUST00000165443.4; B: ENSMUST00000090061.6), rat (A: ENSRNOT00000018155.6; B: ENSRNOG00000024560), chicken (ENSGALT00000022998.6), zebrafish (ENSDART00000004739.8), fruitfly (FBtr0088783), red flour beetle (TC007383_001), S. cerevisiae (YLR335W), S. pombe (SPCC18B5.07c.1) and S. japonicus (EEB09106) transcript sequences were retrieved from Ensembl or Genebank and aligned by ClustalO with Seaview software (Gouy et al, 2010). Sequence identity scores were calculated using the sequence identity and similarity (SIAS) webpage of the computational university of Madrid with the default settings.
Animal husbandry
Female Xenopus laevis used in this study were accommodated und used according to the German legislation regarding animal care and husbandry, approved by the animal protection commissioners of RWTH Aachen University.
Author contributions
Conceptualization: GH, UK, and WA; formal analysis: GM, PDM, CG, RS, AS, DM‐A CP, and ML; investigation: GM, PDM, CG, RS, AS, DM‐A CP, DM‐A, HL, GH, AS, HL, AKS AM, MT, and MW; resources: BL and HL; writing—original draft preparation: GH and WA; writing—review and editing: GH and WA; visualization: GH, DM‐A, and CP; supervision: DM‐A,UK, and WA; project administration: WA; funding acquisition: UK and WA All authors have read and agreed to the published version of the manuscript.
Conflict of interest
The authors declare that they have no conflict of interests.
Supporting information
Appendix
Dataset EV1
Dataset EV2
Source Data for Appendix
Source Data for Figure 1
Source Data for Figure 2
Source Data for Figure 5
Source Data for Figure 7
Source Data for Figure 8
Acknowledgements
Mass spectrometry was done at the Proteomic Facility, and confocal images were acquired at the Confocal Microscopy Facility, core facilities of the Interdisciplinary Center for Clinical Research (IZKF), Aachen, within the Faculty of Medicine at RWTH Aachen University. This work was supported by grants from the German Research Foundation (to WA AN377/7‐1), the Swiss National Science Foundation (to UK 310030_184801).
The EMBO Journal (2021) 40: e108788.
Data availability
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository (Perez‐Riverol et al, 2019) with the dataset identifier PXD028701.
References
- Aksenova V, Smith A, Lee H, Bhat P, Esnault C, Chen S, Iben J, Kaufhold R, Yau KC, Echeverria C et al (2020) Nucleoporin TPR is an integral component of the TREX‐2 mRNA export pathway. Nat Commun 11: 4577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolhy S, Bouhlel I, Dultz E, Nayak T, Zuccolo M, Gatti X, Vallee R, Ellenberg J, Doye V (2011) A Nup133‐dependent NPC‐anchored network tethers centrosomes to the nuclear envelope in prophase. J Cell Biol 192: 855–871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bult CJ, Blake JA, Smith CL, Kadin JA, Richardson JE, Anagnostopoulos A, Asabor R, Baldarelli RM, Beal JS, Bello SM et al (2019) Mouse Genome Database (MGD) 2019. Nucleic Acids Res 47: D801–D806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu DB, Gromova T, Newman TAC, Burgess SM (2017) The nucleoporin Nup2 contains a meiotic‐autonomous region that promotes the dynamic chromosome events of meiosis. Genetics 206: 1319–1337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordes VC, Reidenbach S, Rackwitz HR, Franke WW (1997) Identification of protein p270/Tpr as a constitutive component of the nuclear pore complex‐attached intranuclear filaments. J Cell Biol 136: 515–529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Angelo MA, Raices M, Panowski SH, Hetzer MW (2009) Age‐dependent deterioration of nuclear pore complexes causes a loss of nuclear integrity in postmitotic cells. Cell 136: 284–295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis LI, Blobel G (1986) Identification and characterization of a nuclear pore complex protein. Cell 45: 699–709 [DOI] [PubMed] [Google Scholar]
- De Magistris P, Tatarek‐Nossol M, Dewor M, Antonin W (2018) A self‐inhibitory interaction within Nup155 and membrane binding are required for nuclear pore complex formation. J Cell Sci 131: jcs208538 [DOI] [PubMed] [Google Scholar]
- Dultz E, Ellenberg J (2010) Live imaging of single nuclear pores reveals unique assembly kinetics and mechanism in interphase. J Cell Biol 191: 15–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dultz E, Zanin E, Wurzenberger C, Braun M, Rabut G, Sironi L, Ellenberg J (2008) Systematic kinetic analysis of mitotic dis‐ and reassembly of the nuclear pore in living cells. J Cell Biol 180: 857–865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Düsterhöft S, Kahveci‐Türköz S, Wozniak J, Seifert A, Kasparek P, Ohm H, Liu S, Kopkanova J, Lokau J, Garbers C et al (2021) The iRhom homology domain is indispensable for ADAM17‐mediated TNFα and EGF receptor ligand release. Cell Mol Life Sci 78: 5015–5040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenhardt N, Schooley A, Antonin W (2014) Xenopus in vitro assays to analyze the function of transmembrane nucleoporins and targeting of inner nuclear membrane proteins. Methods Cell Biol 122: 193–218 [DOI] [PubMed] [Google Scholar]
- Fan F, Liu CP, Korobova O, Heyting C, Offenberg HH, Trump G, Arnheim N (1997) cDNA cloning and characterization of Npap60: a novel rat nuclear pore‐associated protein with an unusual subcellular localization during male germ cell differentiation. Genomics 40: 444–453 [DOI] [PubMed] [Google Scholar]
- Forbes DJ, Travesa A, Nord MS, Bernis C (2015) Nuclear transport factors: global regulation of mitosis. Curr Opin Cell Biol 35: 78–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franz C, Walczak R, Yavuz S, Santarella R, Gentzel M, Askjaer P, Galy V, Hetzer M, Mattaj IW, Antonin W (2007) MEL‐28/ELYS is required for the recruitment of nucleoporins to chromatin and postmitotic nuclear pore complex assembly. EMBO Rep 8: 165–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gant TM, Wilson KL (1997) Nuclear assembly. Annu Rev Cell Dev Biol 13: 669–695 [DOI] [PubMed] [Google Scholar]
- Gilchrist D, Mykytka B, Rexach M (2002) Accelerating the rate of disassembly of karyopherin cargo complexes. J Biol Chem 277: 18161–18172 [DOI] [PubMed] [Google Scholar]
- Gillespie PJ, Khoudoli GA, Stewart G, Swedlow JR, Blow JJ (2007) ELYS/MEL‐28 chromatin association coordinates nuclear pore complex assembly and replication licensing. Curr Biol 17: 1657–1662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouy M, Guindon S, Gascuel O (2010) SeaView version 4: a multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol Biol Evol 27: 221–224 [DOI] [PubMed] [Google Scholar]
- Gruss OJ, Carazo‐Salas RE, Schatz CA, Guarguaglini G, Kast J, Wilm M, Le Bot N, Vernos I, Karsenti E, Mattaj IW (2001) Ran induces spindle assembly by reversing the inhibitory effect of importin alpha on TPX2 activity. Cell 104: 83–93 [DOI] [PubMed] [Google Scholar]
- Guan T, Kehlenbach RH, Schirmer EC, Kehlenbach A, Fan F, Clurman BE, Arnheim N, Gerace L (2000) Nup50, a nucleoplasmically oriented nucleoporin with a role in nuclear protein export. Mol Cell Biol 20: 5619–5630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guglielmi V, Sakuma S, D'Angelo MA (2020) Nuclear pore complexes in development and tissue homeostasis. Development 147: dev183442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hase ME, Cordes VC (2003) Direct interaction with nup153 mediates binding of Tpr to the periphery of the nuclear pore complex. Mol Biol Cell 14: 1923–1940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heald R, Tournebize R, Blank T, Sandaltzopoulos R, Becker P, Hyman A, Karsenti E (1996) Self‐organization of microtubules into bipolar spindles around artificial chromosomes in Xenopus egg extracts. Nature 382: 420–425 [DOI] [PubMed] [Google Scholar]
- Kanie T, Jackson PK (2018) Guanine nucleotide exchange assay using fluorescent MANT‐GDP. Bio‐Protocol 8: e2795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi K, Fortriede JD, Lotay VS, Burns KA, Wang DZ, Fisher ME, Pells TJ, James‐Zorn C, Wang Y, Ponferrada V g et al (2018) Xenbase: a genomic, epigenomic and transcriptomic model organism database. Nucleic Acids Res 46: D861–D868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessel RG (1992) Annulate lamellae: a last frontier in cellular organelles. Int Rev Cytol 133: 43–120 [DOI] [PubMed] [Google Scholar]
- Knockenhauer KE, Schwartz TU (2016) The nuclear pore complex as a flexible and dynamic gate. Cell 164: 1162–1171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosako H, Yamaguchi N, Aranami C, Ushiyama M, Kose S, Imamoto N, Taniguchi H, Nishida E, Hattori S (2009) Phosphoproteomics reveals new ERK MAP kinase targets and links ERK to nucleoporin‐mediated nuclear transport. Nat Struct Mol Biol 16: 1026–1035 [DOI] [PubMed] [Google Scholar]
- Kutay U, Jühlen R, Antonin W (2021) Mitotic disassembly and reassembly of nuclear pore complexes. Trends Cell Biol 10.1016/j.tcb.2021.06.011 [DOI] [PubMed] [Google Scholar]
- Lin DH, Hoelz A (2019) The structure of the nuclear pore complex (an update). Annu Rev Biochem 88: 725–783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsay ME, Plafker K, Smith AE, Clurman BE, Macara IG (2002) Npap60/Nup50 is a tri‐stable switch that stimulates importin‐alpha:beta‐mediated nuclear protein import. Cell 110: 349–360 [DOI] [PubMed] [Google Scholar]
- Liu SM, Stewart M (2005) Structural basis for the high‐affinity binding of nucleoporin Nup1p to the Saccharomyces cerevisiae importin‐beta homologue, Kap95p. J Mol Biol 349: 515–525 [DOI] [PubMed] [Google Scholar]
- Mackay DR, Makise M, Ullman KS (2010) Defects in nuclear pore assembly lead to activation of an Aurora B‐mediated abscission checkpoint. J Cell Biol 191: 923–931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makise M, Mackay DR, Elgort S, Shankaran SS, Adam SA, Ullman KS (2012) The Nup153‐Nup50 protein interface and its role in nuclear import. J Biol Chem 287: 38515–38522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansfeld J, Güttinger S, Hawryluk‐Gara LA, Panté N, Mall M, Galy V, Haselmann U, Mühlhäusser P, Wozniak RW, Mattaj IW et al (2006) The conserved transmembrane nucleoporin NDC1 is required for nuclear pore complex assembly in vertebrate cells. Mol Cell 22: 93–103 [DOI] [PubMed] [Google Scholar]
- Markossian S, Suresh S, Osmani AH, Osmani SA (2015) Nup2 requires a highly divergent partner, NupA, to fulfill functions at nuclear pore complexes and the mitotic chromatin region. Mol Biol Cell 26: 605–621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuura Y, Stewart M (2005) Nup50/Npap60 function in nuclear protein import complex disassembly and importin recycling. EMBO J 24: 3681–3689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore MS (2003) Npap60: a new player in nuclear protein import. Trends Cell Biol 13: 61–64 [DOI] [PubMed] [Google Scholar]
- Nemergut ME, Mizzen CA, Stukenberg T, Allis CD, Macara IG (2001) Chromatin docking and exchange activity enhancement of RCC1 by histones H2A and H2B. Science 292: 1540–1543 [DOI] [PubMed] [Google Scholar]
- Ogawa Y, Miyamoto Y, Asally M, Oka M, Yasuda Y, Yoneda Y (2010) Two isoforms of Npap60 (Nup50) differentially regulate nuclear protein import. Mol Biol Cell 21: 630–638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osmani AH, Davies J, Liu HL, Nile A, Osmani SA (2006) Systematic deletion and mitotic localization of the nuclear pore complex proteins of Aspergillus nidulans. Mol Biol Cell 17: 4946–4961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otsuka S, Bui KH, Schorb M, Hossain MJ, Politi AZ, Koch B, Eltsov M, Beck M, Ellenberg J (2016) Nuclear pore assembly proceeds by an inside‐out extrusion of the nuclear envelope. eLife 5: e19071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otsuka S, Ellenberg J (2018) Mechanisms of nuclear pore complex assembly ‐ two different ways of building one molecular machine. FEBS Lett 592: 475–488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papatheodorou I, Moreno P, Manning J, Fuentes AM, George N, Fexova S, Fonseca NA, Füllgrabe A, Green M, Huang N et al (2020) Expression Atlas update: from tissues to single cells. Nucleic Acids Res 48: D77–D83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patre M, Tabbert A, Hermann D, Walczak H, Rackwitz HR, Cordes VC, Ferrando‐May E (2006) Caspases target only two architectural components within the core structure of the nuclear pore complex. J Biol Chem 281: 1296–1304 [DOI] [PubMed] [Google Scholar]
- Perez‐Riverol Y, Csordas A, Bai J, Bernal‐Llinares M, Hewapathirana S, Kundu DJ, Inuganti A, Griss J, Mayer G, Eisenacher M et al (2019) The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res 47: D442–d450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabut G, Doye V, Ellenberg J (2004) Mapping the dynamic organization of the nuclear pore complex inside single living cells. Nat Cell Biol 6: 1114–1121 [DOI] [PubMed] [Google Scholar]
- Rasala BA, Orjalo AV, Shen Z, Briggs S, Forbes DJ (2006) ELYS is a dual nucleoporin/kinetochore protein required for nuclear pore assembly and proper cell division. Proc Natl Acad Sci U S A 103: 17801–17806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasala BA, Ramos C, Harel A, Forbes DJ (2008) Capture of AT‐rich chromatin by ELYS recruits POM121 and NDC1 to initiate nuclear pore assembly. Mol Biol Cell 19: 3982–3996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rotem A, Gruber R, Shorer H, Shaulov L, Klein E, Harel A (2009) Importin beta regulates the seeding of chromatin with initiation sites for nuclear pore assembly. Mol Biol Cell 20: 4031–4042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samwer M, Schneider MWG, Hoefler R, Schmalhorst PS, Jude JG, Zuber J, Gerlich DW (2017) DNA cross‐bridging shapes a single nucleus from a set of mitotic chromosomes. Cell 170: 956–972.e923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J, Arganda‐Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B et al (2012) Fiji: an open‐source platform for biological‐image analysis. Nat Methods 9: 676–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schooley A, Moreno‐Andres D, De Magistris P, Vollmer B, Antonin W (2015) The lysine demethylase LSD1 is required for nuclear envelope formation at the end of mitosis. J Cell Sci 128: 3466–3477 [DOI] [PubMed] [Google Scholar]
- Session AM, Uno Y, Kwon T, Chapman JA, Toyoda A, Takahashi S, Fukui A, Hikosaka A, Suzuki A, Kondo M et al (2016) Genome evolution in the allotetraploid frog Xenopus laevis. Nature 538: 336–343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smitherman M, Lee K, Swanger J, Kapur R, Clurman BE (2000) Characterization and targeted disruption of murine Nup50, a p27(Kip1)‐interacting component of the nuclear pore complex. Mol Cell Biol 20: 5631–5642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solsbacher J, Maurer P, Vogel F, Schlenstedt G (2000) Nup2p, a yeast nucleoporin, functions in bidirectional transport of importin alpha. Mol Cell Biol 20: 8468–8479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sukegawa J, Blobel G (1993) A nuclear pore complex protein that contains zinc finger motifs, binds DNA, and faces the nucleoplasm. Cell 72: 29–38 [DOI] [PubMed] [Google Scholar]
- Suresh S, Markossian S, Osmani AH, Osmani SA (2017) Mitotic nuclear pore complex segregation involves Nup2 in Aspergillus nidulans. J Cell Biol 216: 2813–2826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swaminathan S, Melchior F (2002) Nucleocytoplasmic transport: more than the usual suspects. Dev Cell 3: 304–306 [DOI] [PubMed] [Google Scholar]
- Theerthagiri G, Eisenhardt N, Schwarz H, Antonin W (2010) The nucleoporin Nup188 controls passage of membrane proteins across the nuclear pore complex. J Cell Biol 189: 1129–1142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trichet V, Shkolny D, Dunham I, Beare D, McDermid HE (1999) Mapping and complex expression pattern of the human NPAP60L nucleoporin gene. Cytogenet Cell Genet 85: 221–226 [DOI] [PubMed] [Google Scholar]
- Tyanova S, Temu T, Cox J (2016a) The MaxQuant computational platform for mass spectrometry‐based shotgun proteomics. Nat Protoc 11: 2301–2319 [DOI] [PubMed] [Google Scholar]
- Tyanova S, Temu T, Sinitcyn P, Carlson A, Hein MY, Geiger T, Mann M, Cox J (2016b) The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat Methods 13: 731–740 [DOI] [PubMed] [Google Scholar]
- Vollmer B, Lorenz M, Moreno‐Andres D, Bodenhofer M, De Magistris P, Astrinidis SA, Schooley A, Flotenmeyer M, Leptihn S, Antonin W (2015) Nup153 recruits the Nup107‐160 complex to the inner nuclear membrane for interphasic nuclear pore complex assembly. Dev Cell 33: 717–728 [DOI] [PubMed] [Google Scholar]
- Vollmer B, Schooley A, Sachdev R, Eisenhardt N, Schneider AM, Sieverding C, Madlung J, Gerken U, Macek B, Antonin W (2012) Dimerization and direct membrane interaction of Nup53 contribute to nuclear pore complex assembly. EMBO J 31: 4072–4084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walther TC, Alves A, Pickersgill H, Loiodice I, Hetzer M, Galy V, Hülsmann BB, Köcher T, Wilm M, Allen T et al (2003a) The conserved Nup107‐160 complex is critical for nuclear pore complex assembly. Cell 113: 195–206 [DOI] [PubMed] [Google Scholar]
- Walther TC, Askjaer P, Gentzel M, Habermann A, Griffiths G, Wilm M, Mattaj IW, Hetzer M (2003b) RanGTP mediates nuclear pore complex assembly. Nature 424: 689–694 [DOI] [PubMed] [Google Scholar]
- Walther TC, Fornerod M, Pickersgill H, Goldberg M, Allen TD, Mattaj IW (2001) The nucleoporin Nup153 is required for nuclear pore basket formation, nuclear pore complex anchoring and import of a subset of nuclear proteins. EMBO J 20: 5703–5714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weberruss M, Antonin W (2016) Perforating the nuclear boundary ‐ how nuclear pore complexes assemble. J Cell Sci 129: 4439–4447 [DOI] [PubMed] [Google Scholar]
- Wiese C, Wilde A, Moore MS, Adam SA, Merdes A, Zheng Y (2001) Role of importin‐beta in coupling Ran to downstream targets in microtubule assembly. Science 291: 653–656 [DOI] [PubMed] [Google Scholar]
- Yokoyama H (2016) Chromatin‐binding proteins moonlight as mitotic microtubule regulators. Trends Cell Biol 26: 161–164 [DOI] [PubMed] [Google Scholar]
- Yokoyama H, Koch B, Walczak R, Ciray‐Duygu F, Gonzalez‐Sanchez JC, Devos DP, Mattaj IW, Gruss OJ (2014) The nucleoporin MEL‐28 promotes RanGTP‐dependent gamma‐tubulin recruitment and microtubule nucleation in mitotic spindle formation. Nat Commun 5: 3270 [DOI] [PubMed] [Google Scholar]
- Zhang MS, Arnaoutov A, Dasso M (2014) RanBP1 governs spindle assembly by defining mitotic Ran‐GTP production. Dev Cell 31: 393–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zierhut C, Jenness C, Kimura H, Funabiki H (2014) Nucleosomal regulation of chromatin composition and nuclear assembly revealed by histone depletion. Nat Struct Mol Biol 21: 617–625 [DOI] [PMC free article] [PubMed] [Google Scholar]


