Abstract
Exposure to environmental toxicants can increase the risk of developing age-related neurodegenerative disorders. Exposure to the widely used organophosphate pesticide chlorpyrifos (CPF) is associated with increased risk of developing Alzheimer’s disease and Parkinson’s disease, but the cellular mechanisms underlying CPF toxicity in neurons are not completely understood. We evaluated CPF toxicity in mouse primary cortical neuronal cultures, using RNA-sequencing to identify cellular pathways modulated by CPF. CPF exposure altered the expression of genes associated with intrinsic apoptosis, significantly elevating expression of the pro-apoptotic mediator Bbc3/Puma. Bbc3 loss attenuated CPF driven neurotoxicity, induction of other intrinsic apoptosis regulatory genes including Trp53 and Pmaip1 (encoding the NOXA protein), and cleavage of apoptosis executors caspase 3 and poly (ADP-ribose) polymerase (PARP). CPF exposure was associated with enhanced expression of endoplasmic reticulum stress-related genes and proteins and the accumulation of high molecular weight protein species in primary neuronal cultures. No evidence of alterations in the ubiquitin-proteosome system were observed, however, autophagy-related proteins were upregulated in CPF-treated Bbc3−/− neuronal cultures compared with identically exposed WT cultures. Elevated autophagy-related protein expression in Bbc3−/− neuronal cultures was associated with a reduction in CPF-induced high molecular weight alpha-synuclein and tau immunoreactive protein aggregates. Studies indicate that Bbc3−/− neuronal cultures enhance the endoplasmic reticulum stress response and upregulate protein clearance mechanisms as a component of resistance to CPF-mediated toxicity.
Keywords: chlorpyrifos, neurodegeneration, apoptosis, ER stress, neurotoxicology, proteinopathy
A small proportion of neurodegenerative disease cases can be attributed to genetic causes; however, the etiology of most neurodegenerative disorders of the elderly is unknown (Chin-Chan et al., 2015; Migliore and Coppede, 2009). It is widely believed that a combination of genetic and environmental risk factors underlies the development of certain neurodegenerative disorders with epidemiological and experimental evidence clearly associating pesticide exposure with increased risk of developing Alzheimer’s disease (AD) and Parkinson’s disease (PD; Cannon and Greenamyre, 2013; Costello et al., 2009; Hatcher et al., 2008; Ritz and Yu, 2000; Tanner et al., 2011; Yan et al., 2016). Despite the growing body of evidence from epidemiological, in vitro, and in vivo studies, cellular mechanisms mediating pesticide neurotoxicity are not completely characterized. Delineating the molecular mechanisms by which environmental toxicant exposure translates to neuronal cell death is anticipated to improve our understanding of the initiation and progression of neurodegenerative disorders.
Organophosphates (OPs) are among the most commonly used classes of pesticides worldwide. Acute exposures can result from accidental poisoning, terrorist attacks, and suicidal ingestions (Blain, 2011). Of the estimated 3 million acute OP poisonings each year, approximately 1 million are due to occupational exposure (Karalliedde and Senanayake, 1989). Acute OP exposure is toxic to the nervous system, resulting in cardiorespiratory depression, and seizures that can lead to long-term cognitive abnormalities. Acute exposure to OP nerve agents, such as sarin gas, can lead to life-long neurological consequences, including electroencephalogram (EEG) alterations and convulsions (Sekijima et al., 1997), attention deficits and impaired motor coordination (Loh et al., 2010), reduced gray matter volume (Yamasue et al., 2007), and memory decline (Hood, 2001). Chronic OP exposure is observed in occupational settings, with agricultural workers being among those most at-risk (Munoz-Quezada et al., 2016). Chronic OP exposure can cause neurologic and psychiatric deficits, including anxiety, depression, psychosis, learning and memory deficits, and motor dysfunction, collectively referred to as chronic OP-induced neuropsychiatric disorders (Naughton and Terry, 2018). Epidemiologic studies link chronic OP exposure to a broad range of neurologic disorders, including developmental delay in children (Rauh et al., 2006, 2011, 2012) and neurodegenerative disorders later in life (Sanchez-Santed et al., 2016). Exposure to OPs has been shown to increase risk of developing AD (HR = 1.53; Hayden et al., 2010) and PD (OR = 1.87–5.3; Dhillon et al., 2008; Gatto et al., 2009; Manthripragada et al., 2010).
The OP pesticide chlorpyrifos (CPF) is frequently implicated in the context of neurodegenerative disease (Dhillon et al., 2008; Gatto et al., 2009; Hayden et al., 2010; Manthripragada et al., 2010). CPF enters the environment at an estimated 3.2–4.1 million kg/year in the United States alone, with corn and soybean production accounting for about 50% of its usage (Solomon et al., 2014). The most common route of CPF exposure is ingestion through well water and food residue but inhalation (eg, pesticide sprayers, rural living) and absorption (eg, dermal exposure) of CPF are also well-documented (Poet et al., 2014). CPF is an organothiophosphate metabolized by cytochromes p450 whose specificity is derived from insects’ rapid metabolism of the P = S bond in the thiophosphoryl group, forming the toxic chlorpyrifos-oxon (CPO) metabolite. The mammalian metabolic process removes the lipophilic P side groups first, oxidizing at a much slower rate overall, and favoring the formation of the less toxic metabolite, 3,5,6-trichloro-2-pyridinol (TCPy; Croom et al., 2010). Despite structural and functional selectivity to insects, human CPF exposure is of increasing concern in part because of its capability of crossing the mammalian blood-brain-barrier (Kopjar et al., 2018; Li and Ehrich, 2013; Parran et al., 2005; Williamson Leah et al., 2006). More recent studies have highlighted genetic polymorphisms that alter CPF metabolism suggesting the potential for altered susceptibility to CPF toxicity in human populations (Costa et al., 2013). CPF toxicity is typically associated with inhibition of acetylcholinesterase (AChE), the main enzyme responsible for breaking down the neurotransmitter acetylcholine, resulting in cholinergic toxicity in humans and death in insects. Interestingly, while CPF and its metabolites have been found in central nervous system (CNS) tissues of laboratory animals chronically exposed to CPF (Kopjar et al., 2018; Williamson Leah et al., 2006), AChE inhibition has not been convincingly correlated with neurologic dysfunction in humans (Rohlman et al. 2011). Several studies have indicated that AChE inhibition is neuroprotective (Ye et al., 2010; Zhang and Greenberg, 2012) and CPF toxicity is readily observed in glial cells that do not express AChE at high levels (Mense et al., 2006; Thullbery et al., 2005). Moreover, pharmacologic inhibition of AChE using agents such as donepezil and galantamine is a common strategy used to enhance cognitive function in AD (Sharma, 2019). These disconnects underscore the importance of further characterization of mechanisms of CPF-induced neuronal cell death.
To characterize mechanisms of CPF neurotoxicity, we conducted RNA-sequencing (RNA-Seq) experiments and identified Bcl-2-binding component 3/p53 upregulated modulator of apoptosis (Bbc3/Puma) as a key mediator of CPF-driven neuronal apoptosis. Using Bbc3−/− neuronal cultures, we provide evidence that suppression of Bbc3 improves survival following CPF exposure. Improved survival in Bbc3−/− cultures is associated with enhanced expression of endoplasmic reticulum (ER) stress- and autophagy-related markers, and a reduction in alpha-synuclein and microtubule-associated protein tau immunoreactive protein aggregates. Our study identifies Bbc3 signaling as a pathway of interest for neuroprotection and suggests a compensatory mechanism associated with suppression of apoptosis capable of reducing proteinopathy in pesticide-exposed neurons.
MATERIALS AND METHODS
Research Animals
All mouse experiments were conducted according to the ARRIVE guidelines and approved by the Institutional Animal Care and Use Committee at Dartmouth (Protocol No. 00002117, MCH). All mice used in these experiments were either purchased from the Jackson Laboratory (Bar Harbor, Maine) or validated on an ongoing basis using PCR-based genotyping (Supplementary Figure 5) and biochemical procedures. Genotyping primers: Bbc3 Mutant Forward: TTGACGAG TTCTTCTGAGGG, Bbc3 Mutant Reverse: CCTGGAATTACAGGCAGTTGTCAGC, Bbc3 WT Forward: AAGGGTTAAAGGTCCCTCGC, Bbc3 WT Reverse: AACTGAGGCTCCACCCATAG. Strains include C57BL/6J (Stock 000664) and Bbc3−/− (generously provided by Dr Gerard Zambetti, St. Jude Children’s Research Hospital, Nashville, Tennessee).
Cultivation of Primary Neuronal Cultures
Six or more embryos (E14.5-16.5) of mixed sexes were retrieved from wild-type (WT) and Bbc3−/− time-mated female mice, the dissected cortices were pooled and dissociated, and cortical neurons were plated on poly-D-lysine (P6407) (Sigma-Aldrich, St Louis, Missouri) and laminin-coated (L2020) (Sigma-Aldrich) tissue culture dishes. Cells were incubated in enriched neurobasal medium (21103049) (Thermo Fisher Scientific, Waltham, Massachusetts) (500 ml NB media, 200 mM L-glutamine, B-27 with vitamin A, 1 ml penicillin/streptomycin (P/S) (1%), and 340 mg glucose) to reduce glial expansion.
CPF Dose-Finding and Exposure
Analytical standard grade CPF was purchased from MilliporeSigma (No. 45395, Burlington, Massachusetts). A 100 mM stock of CPF was generated in dimethyl sulfoxide (DMSO) (No. 276855, Sigma-Aldrich). The 100 mM stock was diluted in cell culture media to achieve desired CPF concentrations, resulting in a final DMSO concentration of <0.5%. Controls were treated with equivalent DMSO concentration. Dose finding was conducted by analyzing doses ranging from 1 to 400 µM, coupled with analysis of toxicity and survival as described in the following sections. We found that the IC50 of CPF in primary neuronal cultures was 50 µM, varying doses were used as described throughout the results.
Cytotoxicity Assays
3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assays
Primary neurons were plated to a confluency of 50,000 cells/well in poly-D-lysine and laminin-coated 96-well dishes. Cells were treated with CPF 4-6 days after plating. After CPF treatment, media was removed and media containing 10% yellow tetrazolium salt (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide or MTT) (Sigma-Aldrich) was added to each well. Cells were allowed to incubate for 4 h with the MTT dye, dye-containing media was removed, and DMSO was added to each well, to solubilize formazan crystals. Colorimetric change was read on a plate reader (Bio-Rad iMark Microplate Absorbance Reader, Hercules, California) at 595 nm.
Lactate dehydrogenase (LDH) assays
Lactate dehydrogenase (LDH) was detected using a Cytotoxicity Detection Kit (LDH) (Sigma-Aldrich). Briefly, conditioned media was collected from cultures treated with CPF. Conditioned media was centrifuged at 12,000 RPM for 30 min at 4°C to remove cellular debris and frozen at −80°C until LDH analysis. Media was thawed on ice and 100 µl was added to the wells of a 96-well plate. 100 µl of LDH reaction mixture was added to the media. The assay was incubated for 30 min at room temperature protected from light. The colorimetric change was measured on a plate reader (Bio-Rad iMark Microplate Absorbance Reader) at 490 nm. Fresh media was used to calculate background LDH activity. Range was determined using vehicle-treated cell culture media as a low control and apoptosis-inducing staurosporine-treated cell culture media as a high control. Background LDH levels present in the media alone were subtracted from all samples. Cytotoxicity was measured using the following equation: (experimental value−low control)/(high control−low control) × 100.
Sodium dodecyl-sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and Immunoblotting
Samples were prepared in ice-cold lysis buffer (150 mM NaCl, 50 mM Tris, 1% Triton X-100) containing 1× phosphatase inhibitor cocktail 2 (P5726) (Sigma-Aldrich) and 1× cOmplete Mini Protease Inhibitor Cocktail (11836153001) (Sigma-Aldrich). 4× NuPAGE LDS Sample Buffer (NP0008) diluted to 1× and 0.02% beta-mercaptoethanol was added to all samples and samples were boiled before loading into gels. The NuPAGE system was used for gel electrophoreses, with precast 4–12% Bis-Tris protein gels and MES/SDS running buffer (Thermo Fisher Scientific). Gels were transferred to Immobilon-P PVDF membranes (IPVH00010) (MilliporeSigma) for immunoblotting. Membranes were blocked in a solution of 5% skim milk powder dissolved in tris-buffered saline (TBS) and Tween 20 (TBS-T) before overnight incubation with the primary antibody. The following day, antispecies horseradish peroxidase (HRP)-conjugated secondary antibodies were incubated with the membrane before developing in Pierce ECL Western Blotting Substrate (32106) (Thermo Fisher Scientific). The following antibodies were used for apoptotic analysis: anti-Pumaα/β (sc-374223) (Santa Cruz Biotechnology, Inc., Dallas, Texas), anti-Caspase-3 (D3R6Y, No. 14220) (Cell Signaling Technology, Danvers, Massachusetts), and anti-poly (ADP-ribose) polymerase (PARP; 46D11, No. 9532) (Cell Signaling Technology). The following antibodies were used for ER stress analysis: anti-CREB-2/ Activating transcription factor 4 (ATF4; sc-200) (Santa Cruz Biotechnology, Inc.) and anti-GADD-153 (sc-7351) (Santa Cruz Biotechnology, Inc.). The following antibodies were used for autophagy analysis: anti-phospho-mammalian target of rapamycin (mTOR) (Ser2448) (D9C2, No. 5536) (Cell Signaling Technology), anti-mTOR (7C10, No. 2983) (Cell Signaling Technology), and anti-SQSTM1/p62 (No. 5114) (Cell Signaling Technology). The following antibodies were used in protein solubility analysis: anti-phospho-Alpha-Synuclein (Ser129) ([MJF-R13 (8-8)], ab168381) (Abcam, Cambridge, Massachusetts), anti-phospho-Tau (Ser202, Thr205) (AT8, No. MN1020) (Invitrogen, Thermo Fisher Scientific), anti-Tau (HT7, No. MN1000) (Invitrogen, Thermo Fisher Scientific), and anti-Ubiquitin (No. 3933) (Cell Signaling Technology). Anti-β-Actin (AC-15, A3854) (Sigma-Aldrich) was used as a loading control in all westerns.
mRNA Analysis
mRNA was isolated from cell lysates using the RNeasy Mini Kit (74104) and further processed using the QIAshredder (79654) and RNase-Free DNase Set (79254) (Qiagen, Germantown, Maryland). Total purified mRNA was quantified using the Nanodrop 2000 (NCCC Shared Instruments Resource). Equal quantities of mRNA, normalized across samples, were reversed transcribed into complementary DNA (cDNA) using the iScript cDNA Synthesis Kit (1708891) (Bio-Rad Laboratories, Hercules, California). Bbc3, Trp53, Pmaip1, and Ddit3 transcripts were analyzed and relative levels determined using the CFX96 Touch Real-Time PCR Detection System (NCCC Shared Instruments Resource) and SYBR Green reagent (Bio-Rad Laboratories). Bbc3 forward primer: GGTCTAG CCCGCGACAGT, and Bbc3 reverse primer: GCACGGGCGA CTCTAAGTG. Trp53 Bbc3 WT Forward: forward primer: CGAAGACTGGATGACTGCCA, and Trp53 reverse primer: CGTCCATGCAGTGAGGTGAT. Pmaip1 forward primer: GCCCAGATTGGGGACCTTAG, and Pmaip1 reverse primer: GTTATGTCCGGTGCACTCCA. Ddit3 forward primer: CCCTAGCTTGGCTGACAGAG, and Ddit3 reverse primer: TGCTCCTTCTCCTTCATGCG. Real-time PCR data were normalized to actin transcript.
RNA-Sequencing
Cortical neuron cultures were prepared as described previously and treated with 30 µM CPF or equivalent vehicle (DMSO) concentration for either 6 or 24 h. RNA-seq libraries were prepared using the Lexogen Quantseq FWD 3’ Workflow and 200 ng of RNA as input. Libraries were quantified, normalized and pooled onto a single NextSeq500 Mid Output to generate an average of [10M] paired-end 75 bp reads per sample. Reads were quality trimmed and aligned to the mm10 mouse reference genome and normalized using CLC Genomics Workbench v10 to produce TPM gene expression count (Liu and Di, 2020). Data were analyzed together across both time points and arbitrary p value and fold-change cutoffs were used to identify genes whose expression was persistently altered in CPF-treated cultures. Targets of interest from this discovery study were validated using real-time PCR as previously described. Access to the RNA-Seq datasets will be made available upon request. We are in the process of submitting raw data to the appropriate NIH-approved public resources.
RESULTS
Characterization of Gene Expression Changes Associated With CPF Neurotoxicity In Vitro
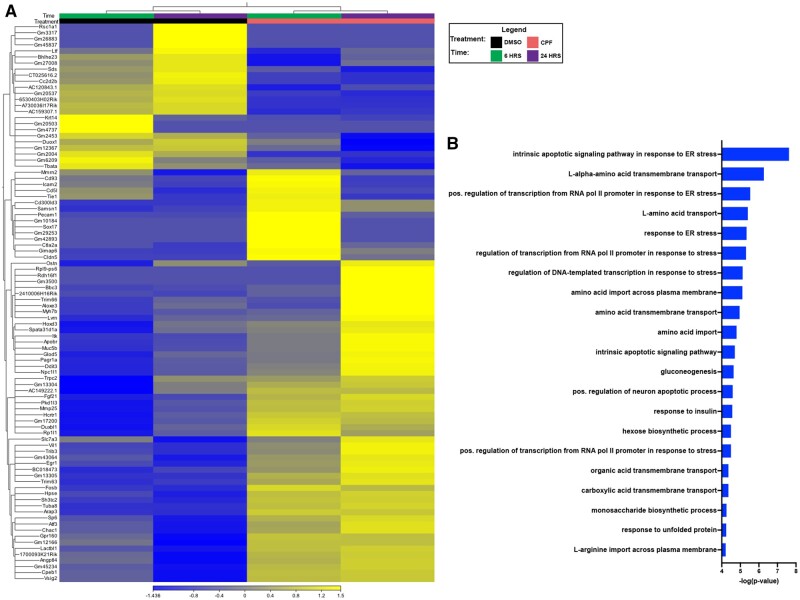
CPF neurotoxicity was characterized in vitro using primary cortical neuronal cultures established from embryonic mice. Neuronal cultures were exposed to increasing doses of CPF across a range from 0 to 120 µM for 24 h. We observed significant declines in MTT metabolism, a proxy for cellular viability (Kumar et al., 2018), at concentrations of 60 µM (**p = .0039) and 120 µM (***p = .0001) compared with vehicle-treated (0 µM) neuronal cultures (Supplementary Figure 1A). LDH assays using a time course of 48 h and the average dose for significant effects in the MTT assays (80 µM) (Supplementary Figure 1A) confirm temporal loss of viability with longer CPF incubation periods, both at 24 h (*p = .0247) and 48 h (***p = .0002; Supplementary Figure 1B). Collectively, these data show that CPF exposure induces cell death in primary cortical neuron cultures at doses >30 µM. Guided by these data, we conducted a discovery RNA-Seq screen using pooled cultures representing 6 or more embryos per group to identify changes associated with CPF exposure in viable cultures by comparing WT cultures treated with either vehicle (DMSO) or a sublethal 30 µM dose of CPF (Supplementary Figure 1A) for 6 or 24 h. Principal component analysis showed clustering of samples based on time and treatment (Supplementary Figure 2). A heatmap of the top 100 differentially expressed genes (DEGs) with minimum change of 2.5-fold and a p value cutoff of .04 shows clustering of samples based on treatment (Figure 1A). Panther Gene List Analysis was used for ontologic analysis of DEGs whose expression was modulated 1.5-fold or more with a p value of < .04 by CPF at both time points (Figure 1B and Supplementary Table 1). This analysis identified enrichment of pathways related to intrinsic apoptosis and ER stress including “intrinsic apoptotic signaling pathway in response to ER stress,” “positive regulation of transcription from RNA polymerase II promoter in response to ER stress,” “response to ER stress,” “intrinsic apoptotic signaling pathway,” “positive regulation of neuron apoptotic process,” and “response to unfolded protein.”
Figure 1.
RNA-sequencing (RNA-Seq) indicates intrinsic apoptosis-related pathways upregulated in chlorpyrifos (CPF)-exposed primary cortical neurons. Primary cortical neuron cultures were established from C57BL/6J (WT) embryos (E14.5-16.5). In order to detect the biological changes that were occurring in neurons prior to death, the cultures were treated with either vehicle (DMSO) or a sublethal 30 µM concentration of CPF (see Supplementary Figure 1) for 6 or 24 h. We conducted an unbiased transcriptomic screen using RNA-Seq and analyzed the data using CLC Genomics. A, Heat map shows top 100 differentially expressed genes (DEGs) with a minimum fold change of 2.5-fold with a p value cutoff of .04. B, Panther Gene List Analysis was used to generate ontologic data from all DEGs changed 1.5-fold or more with CPF treatment with a p value of <.04 at both timepoints.
Intrinsic Apoptotic Regulator Bbc3 Is Required for CPF Neurotoxicity
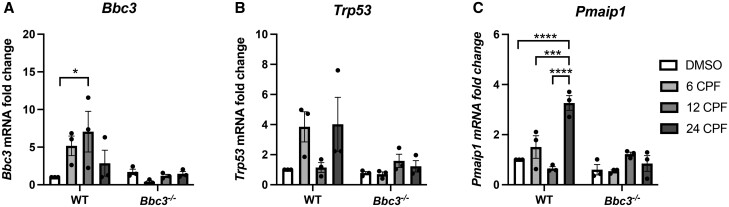
Close inspection of the pathways enriched by CPF exposure identified genes of interest that appeared in multiple ontologic sets. Among these was the pro-apoptotic mediator Bbc3 (aka Puma) (Supplementary Tables 2 and 3; fold change 2.87, p = 2.73e−3), which was observed in 4 of the top ontologic hits including “response to ER stress,” “intrinsic apoptotic signaling pathway in response to ER stress,” “intrinsic apoptotic signaling pathway,” and “positive regulation of neuron apoptotic process.” Based on its frequent appearance in cellular death-related ontologic pathways, we decided to investigate the role of Bbc3 in CPF-induced neuronal loss. The RNA-Seq screen was not powered to be a stand-alone assay so we confirmed changes detected in Bbc3 expression using real-time PCR in independently established and treated primary neuronal cultures obtained from wild type and Bbc3−/− mice (generously provided by dr Gerard Zambetti, St. Jude Children’s Research Hospital, Nashville, Tennessee; Jeffers et al., 2003). Bbc3 transcript was upregulated by CPF exposure in WT cultures treated with the IC50 of CPF (50 µM; Figure 3A) with peak expression observed at 12 h compared with vehicle-treated cells (Figure 2A). We also evaluated CPF-driven changes in related mediators of intrinsic apoptosis including transformation-related protein 53 (Trp53) (gene that encodes for p53 protein) and phorbol-12-myristate-13-acetate-induced protein 1 (Pmaip1) (gene that encodes for NOXA protein). We observed variable expression of Trp53 in WT neurons with 6 and 24 h of CPF exposure (Figure 2B) and found significant CPF-mediated upregulation of the proapoptotic mediator Pmaip1 at 24 h in WT cultures, not observed in identically treated Bbc3−/− cultures (Figure 2C). These findings verify modulation of Bbc3 by CPF in neuronal cultures and indicate an important role for intrinsic apoptosis in CPF neurotoxicity.
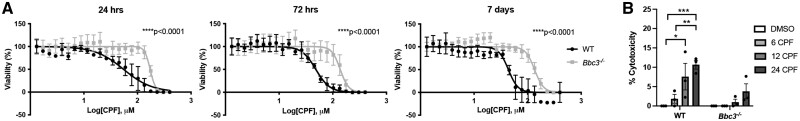
Figure 3.
Bbc3 −/− primary cortical neurons are resistant to chlorpyrifos (CPF)-induced cell death. (A) IC50 curves for WT and Bbc3−/− primary cortical neurons treated 24 or 72 h, or 7 days with [1–400 μM] CPF. 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide assays were performed to assess viability. All data were normalized to vehicle-treated (DMSO) controls. Mean percent viability is shown for 3 biologic replicates, error bars represent standard deviation after log-transformation of CPF concentrations. p Values were determined using nonlinear regression. We saw a significantly higher IC50 value of CPF-treated Bbc3−/− neurons (172.3 μM) compared with WT neurons (54.98 μM) (****p < .0001) at 24 h, at 72 h (Bbc3−/−, 137.5 μM; WT, 48.31 μM; ****p < .0001), and at 7 days (Bbc3−/−, 134.9 μM; WT, 43.75 μM; ****p < .0001). B, A lactate dehydrogenase assay was performed on media of WT and Bbc3−/− primary cortical neurons exposed to 50 µM CPF for 6, 12, or 24 h or equivalent vehicle (DMSO) for 24 h. With increasing time exposed to the approximate WT IC50 of CPF (approximately 50 µM), there was an increase in cytotoxicity in WT cultures (2-way ANOVA; **p = .0027 (genotype), ***p = .0008 (treatment), nsp = .0936 (genotype × treatment); WT 24 DMSO to WT 12 CPF, *p = .0140; WT 24 DMSO to WT 24 CPF, ***p = .0008). An increase in cytotoxicity was seen from 6 to 24 h in WT cultures as well (WT 6 CPF to WT 12 CPF, **p = .0043). No significant difference in cytotoxicity was observed in identically exposed-Bbc3−/− primary cortical neurons. Error bars represent SEM.
Figure 2.
Intrinsic apoptosis-related transcripts are up-regulated in a Bbc3-dependent manner upon chlorpyrifos (CPF) exposure. Primary cortical neuronal cultures were established from WT and Bbc3−/− embryos (E14.5–E16.5) and treated with vehicle (DMSO) or 50 µM for 6, 12, or 24 h. RNA was collected using the RNeasy Mini Kit (Qiagen). Equal quantities of mRNA normalized across samples were reversed transcribed into cDNA using the iScript cDNA Synthesis Kit. (A) Bbc3, (B) Trp53 (encodes for the protein p53), and (C) Pmaip1 (encodes for the protein NOXA) transcripts were analyzed and relative levels determined using the CFX96 Touch Real-Time PCR Detection System. CPF treatment induced Bbc3 expression, with significant elevation observed at 12 h (2-way ANOVA, **p = .0051 [genotype], nsp = .1902 [treatment], and nsp = .0610 [genotype × treatment]; WT 24 DMSO to WT 12 CPF *p = .0190). Variable Trp53 expression with CPF treatment was observed (2-way ANOVA, *p = .0176 [genotype], nsp = .1281 [treatment], nsp = .0758 [genotype × treatment]). Treatment with CPF elevated Pmaip1 expression in a genotype-specific manner (2-way ANOVA, ***p = .0002 [genotype], ***p = .0002 [treatment], ****p < .0001 [genotype × treatment]) with the greatest induction observed at the 6 and 24 h timepoints (WT 24 DMSO to WT 24 CPF, ****p < .0001; WT 6 CPF to WT 24 CPF, ***p = .0004; WT 12 CPF to WT 24 CPF, ****p < .0001). Data are representative of 3 biologic replicates.
To further characterize the role of Bbc3 in CPF-induced neurotoxicity, we evaluated the IC50 of CPF in both WT and Bbc3−/− primary neurons. WT and Bbc3−/− neuronal cultures were treated with a range of CPF concentrations (1–400 μM) for 24 or 72 h, or 7 days. The IC50 of CPF was significantly higher in Bbc3−/− neurons than in WT cultures at 24 or 72 h, or 7 days (Figure 3A). Next, we evaluated the impact of CPF on apoptosis in primary neuronal cultures derived from WT and Bbc3−/− mice following 6, 12, and 24 h of exposure. LDH release assays confirm increasing cytotoxicity at 12 and 24 h in CPF exposed WT cultures, while this toxicity was not observed in identically exposed Bbc3−/− neuronal cultures (Figure 3B). These data indicate that Bbc3 is a key mediator of CPF neurotoxicity and that Bbc3 inactivation is neuroprotective in CPF exposed cultures.
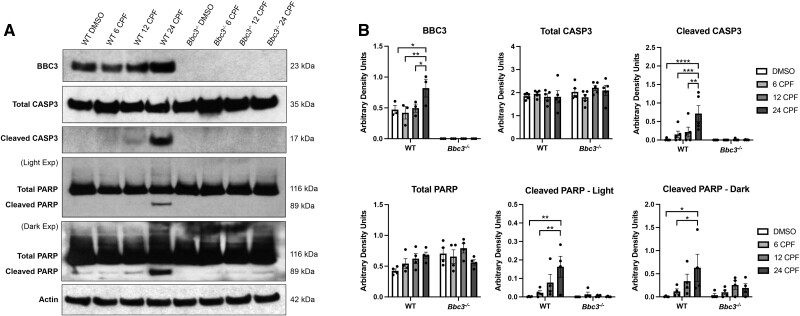
Bbc3 is a well-characterized mediator of intrinsic apoptosis (Han et al., 2001), an ontologic process we identified to be significantly modulated by CPF exposure (Figure 1B). To verify that Bbc3 functioned in intrinsic apoptosis-related processes in our CPF-exposed neuronal cultures, we performed SDS-PAGE and subsequent immunoblotting with lysates obtained from WT and Bbc3−/− neuronal cultures treated with vehicle (DMSO) or WT IC50 (50 µM) CPF for 6, 12, or 24 h (Figure 4). As expected, we found an increase in BBC3 protein expression in WT neurons exposed to CPF (Figs. 4A and 4B). We observed cleavage of canonical mediators of intrinsic apoptosis including CASP3 in WT neurons exposed to CPF that was not observed in identically treated Bbc3−/− neuronal cultures (Figs. 4A and 4B). Additionally, PARP cleavage was elevated in WT samples treated with CPF over time and only observed at long exposures in Bbc3−/− samples (Figs. 4A and 4B). Collectively, these findings indicate that in primary neuronal cultures, CPF exposure results in Bbc3-dependent canonical intrinsic apoptosis.
Figure 4.
Characterization of intrinsic apoptosis in primary cortical neurons exposed to chlorpyrifos (CPF). Primary cortical neuronal cultures were established from WT and Bbc3−/− embryos (E14.5–E16.5) and treated with vehicle (DMSO) or 50 µM CPF for 6, 12, or 24 h. A, Cortical lysate was collected and analyzed using SDS-PAGE and immunoblotting for apoptosis proteins of interest using antibodies for BBC3, Total/cleaved caspase 3(CASP3), and Total/cleaved poly (ADP-ribose) polymerase (PARP). B, BBC3 protein levels increased in WT cultures treated with CPF over time (2-way ANOVA, ****p < .0001 [genotype], *p = .0491 [treatment], *p = .0491 [genotype × treatment]; WT 24 DMSO to WT 24 CPF [*p = .0191]; WT 6 CPF to WT 24 CPF [**p = .0065]; WT 12 CPF to WT 24 CPF [*p = .0310]). Total CASP3 levels remained fairly stable across all samples (2-way ANOVA, nsp = .1234 [genotype], nsp = .8424 [treatment], nsp = .3583 [genotype × treatment]) while levels of cleaved CASP3 increased in WT samples with increasing time exposed to CPF (2-way ANOVA, ***p = .0003 [genotype], **p = .0043 [treatment], **p = .0047 [genotype × treatment]; WT 24 DMSO to WT 24 CPF [****p < .0001]; WT 6 CPF to WT 24 CPF [***p = .0010]; WT 12 CPF to WT 24 CPF [**p = .0038]). Cleavage of CASP3 was not observed in Bbc3−/− samples. Levels of total PARP remained fairly constant (2-way ANOVA, *p = .0452 [genotype], nsp = .2903 [treatment], nsp = .0844 [genotype × treatment]). PARP cleavage was elevated in WT samples treated with CPF (2-way ANOVA, **p = .0029 [genotype], *p = .0267 [treatment], *p = .0178 [genotype × treatment]; WT 24 DMSO to WT 24 CPF [**p = .0012]; WT 6 CPF to WT 24 CPF [**p = .0050]). PARP cleavage was not consistently observed at light exposures in Bbc3−/− samples. Only at dark exposures was PARP cleavage seen in Bbc3−/− cultures treated with CPF; however, it was not significant compared with Bbc3−/− 24 DMSO levels. At longer exposures, we still observe elevated PARP cleavage in WT cultures exposed to CPF (2-way ANOVA, nsp = .1737 [genotype], *p = .0228 [treatment], nsp = .2787 [genotype × treatment]; WT 24 DMSO to WT 24 CPF [*p = .0107]; WT 6 CPF to WT 24 CPF; [*p = .0438]). Two-way ANOVAs were performed on 3–5 biologic replicates. Error bars in (B) represent SEM.
Bbc3 Loss Alters Protein Homeostasis in CPF Exposed Neuronal Cultures
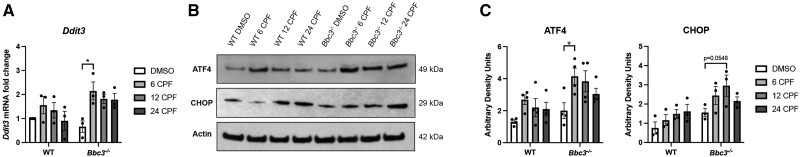
Having confirmed a role for Bbc3 in neurotoxic apoptosis resulting from CPF exposure, we shifted our attention to the related ER stress response pathway (Sano and Reed, 2013), also enriched in our ontologic analysis of DEGs in WT DMSO- and CPF-exposed cultures (Figure 1B). We analyzed DNA damage inducible transcript 3 (Ddit3) (encoding C/EBP homologous protein [CHOP] protein), a key mediator of ER stress elevated in our RNA-Seq dataset (Supplementary Tables 2 and 3; fold change 2.67, p = 2.00e−3). We saw a more significant induction of Ddit3 transcript with CPF exposure in Bbc3−/− cultures compared with WT cultures, with the highest levels found after 6 h of treatment (Figure 5A). After validating this RNA-Seq hit, we sought to evaluate ER stress-related markers at the protein level. ATF4 is a key transcription factor working upstream of Ddit3/CHOP in the ER stress response (Wortel et al., 2017) whose expression was also observed to be significantly elevated following CPF-treatment in our RNA-Seq analysis (Supplementary Tables 2 and 3; fold change 1.78, p = 8.76e−3). We treated WT and Bbc3−/− neuronal cultures with vehicle (DMSO) or 50 µM CPF for 6, 12, for 24 h and performed SDS-PAGE and immunoblotting analysis for both ATF4 and CHOP. We observed ATF4 upregulation following CPF exposure across both genotypes, which was only statistically significant in the Bbc3−/− cultures (Figs. 5B and 5C). Additionally, we observed a trending increase in CHOP protein expression with CPF treatment in Bbc3−/− cultures (Figs. 5B and 5C). These data suggest a mechanism in which Bbc3 loss, associated with enhanced survival (Figs. 3 and 4), also results in a heightened ER stress response.
Figure 5.
Chlorpyrifos (CPF) exposure induces endoplasmic reticulum (ER) stress in a Bbc3-dependent manner. Primary cortical neuronal cultures were established from WT and Bbc3−/− embryos (E14.5–E16.5) and treated with vehicle (DMSO) or 50 µM CPF for 6, 12, or 24 h. A, RNA was collected and equal quantities of mRNA were normalized across samples and reversed transcribed into cDNA. Ddit3 (encodes for the protein C/EBP homologous protein [CHOP]) transcripts were analyzed and relative levels determined using real-time PCR. Levels of Ddit3 transcript increased with CPF treatment in Bbc3−/− cultures (2-way ANOVA, nsp = .0704 [genotype], *p = .0200 [treatment], nsp = .2340 [genotype × treatment]; Bbc3−/− DMSO to Bbc3−/− 6 CPF *p = .0149). B, Cortical lysate was collected and analyzed using SDS-PAGE and immunoblotting for ER stress-related proteins ATF4 and CHOP was conducted. C, Levels of ATF4 protein were increased upon CPF treatment in Bbc3−/− cultures compared with treated WT cultures (2-way ANOVA, **p = .0013 [genotype], **p = .0053 [treatment], nsp = .7094 [genotype × treatment]). CHOP protein expression was elevated with CPF treatment in Bbc3−/− cultures (2-way ANOVA, ***p = .0008 [genotype], nsp = .0509 [treatment], nsp = .5421 [genotype × treatment]) and although not statistically significant, was trending (Bbc3−/− DMSO to Bbc3−/− 12 CPF nsp = .0548). All statistical analyses were performed using 2-way ANOVAs with multiple comparisons across 3 or 4 biological replicates. Error bars in A and C represent SEM.
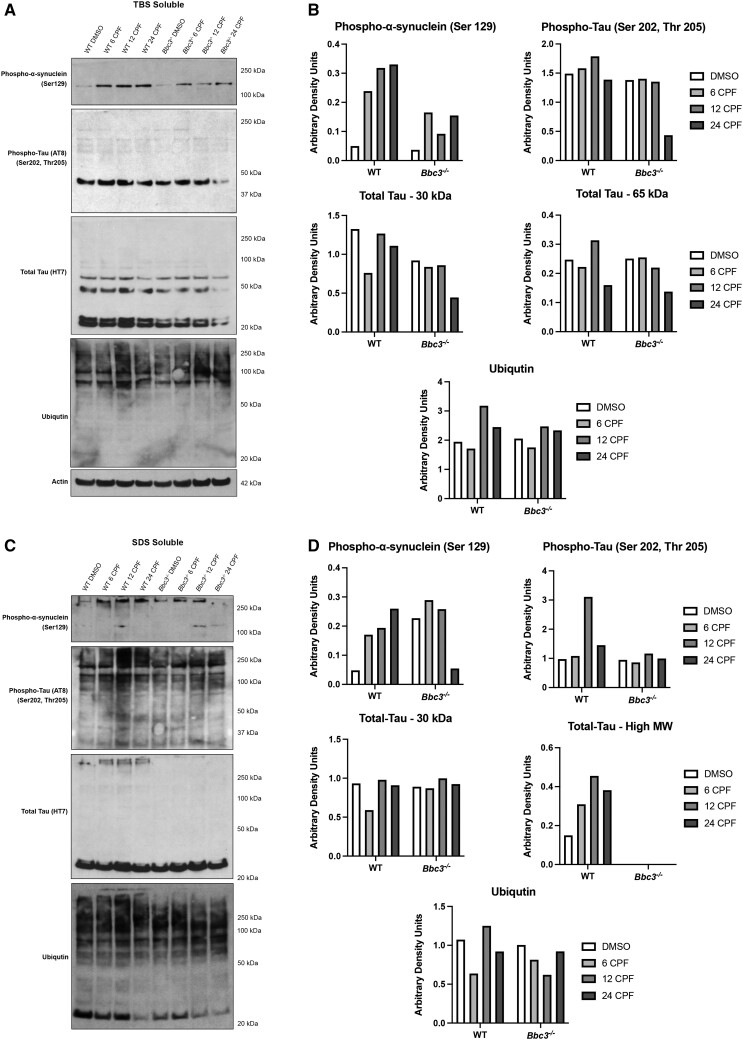
RNA-Seq findings of a CPF-induced ER stress response in WT cultures and observation of elevated ER stress response in more viable CPF-treated Bbc3−/− neuronal cultures prompted us to considered the prediction that Bbc3 loss forced cells that would otherwise undergo apoptosis to adapt and survive under ER-stress conditions. Exposure to CPF has been previously shown to cause protein aggregation, a well-characterized inducer of ER stress, in the SN56 septal basal forebrain cholinergic neuron cell line (Moyano et al., 2018). We evaluated protein solubility in WT and Bbc3−/− CPF-exposed primary neuronal culture lysates to determine (1) if CPF-induced protein aggregation and (2) if Bbc3 loss impacts this biology in association with increased ER stress. We conducted serial ultracentrifugation and immunoblotting experiments to characterize proteins known to misfold and aggregate during human neurodegenerative disease processes, including alpha-synuclein (Hijaz and Volpicelli-Daley, 2020; Spillantini et al., 1997, 1998), tau (Grundke-Iqbal et al., 1986a; Khatoon et al., 1992), and phosphorylated forms of alpha-synuclein (Anderson et al., 2006; Hasegawa et al., 2002 ) and tau (Grundke-Iqbal et al., 1986b; Kopke et al., 1993) associated with pathogenesis (Figure 6). We observed an increase in high-molecular weight phospho-alpha-synuclein (Ser129) in the TBS-soluble fraction in WT cultures treated with CPF. CPF treatment also caused an increase in phospho-alpha-synuclein in the Bbc3−/− cultures, but to a much lesser extent (Figs. 6A and 6B). A similar pattern was also observed in the SDS soluble fraction (Figs. 6C and 6D). We observed high molecular weight phospho-tau (Ser202, Thr205) and total tau in CPF-treated WT cultures which was attenuated or absent in identically treated Bbc3−/− cultures (Figs. 6C and 6D; see also Supplementary Figure 3). These changes in the abundance and solubility of disease-associated proteins suggested Bbc3−/− loss conferred enhanced protein clearance in the context of CPF-induced proteinopathy (Figure 6 and Supplementary Figure 3).
Figure 6.
Chlorpyrifos (CPF) exposure results in aberrant protein accumulation. WT and Bbc3−/− embryonic cortical neurons (E14.5–E16.5) were exposed to vehicle (DMSO) or 50 µM CPF for 6, 12, or 24 h. Lysate was harvested and protein solubility assay was performed by conducting ultracentrifugation using buffers of increasing stringency. Lysates from the tris-buffered saline (TBS) fraction (least stringent) and SDS fraction (most stringent) were analyzed using SDS-PAGE followed by immunoblotting. Membranes were probed with antibodies for proteins known to misfold and aggregate in the neurodegenerative disease process including anti-phospho-alpha-synuclein, anti-phospho-tau, anti-tau, and anti-ubiquitin. A and B, Within the TBS soluble fraction, CPF exposure increases high molecular weight (MW) phospho-alpha-synuclein (Ser129) in CPF-exposed WT cultures and, to a lesser extent, in CPF-treated Bbc3−/− cultures. C and D, In the SDS soluble fraction, high MW phospho-alpha-synuclein (Ser129), high MW phospho-tau (Ser202, Thr205), and high MW total tau is increased with CPF treatment in WT cultures, that is either slightly attenuated, or completely absent, in Bbc3−/− cultures. There does not appear to be any major differences in ubiquitination across fractions, genotypes, or treatments. (B and D) Densitometry was performed and data normalized to actin in the TBS fraction. SDS fractions vary based on the presence of insoluble protein in this original sample. Data represent 1 biologic replicate, but as noted in the Methods, each biologic replicate consists of pooled neurons from at least 6 embryos.
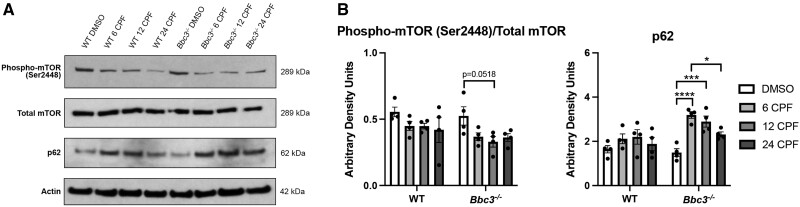
Under homeostatic conditions protein aggregation can induce ER stress, upregulating protein clearance mechanisms including proteasomal degradation (Hoseki et al., 2010) and autophagy (Rashid et al., 2015). To evaluate the ubiquitin/proteasome system in our cultures, we immunoblotted for ubiquitin to determine if CPF exposure resulted in global changes in highly ubiquitinated species in soluble (TBS) and insoluble (SDS) protein fractions. We did not identify differences in global ubiquitination across genotypes or treatments within either of the 2 fractions (Figure 6). Since global ubiquitination did not appear to be significantly altered, we evaluated indicators of autophagy. We conducted SDS-PAGE and immunoblotting of lysates obtained from WT and Bbc3−/− cultures treated with DMSO or 50 µM CPF for 6, 12, or 24 h. We identified a decline in the ratio of phospho-mTOR (Ser2448) to total mTOR, a negative regulator of autophagy (Jung et al., 2010), in both genotypes with CPF-treatment over time (Figs. 7A and 7B). Although multiple comparisons analysis following 2-way ANOVA did not withstand this more rigorous test, we found a trending decline in the ratio of phospho-mTOR (Ser2448) to total mTOR with CPF treatment in Bbc3−/− cultures that was not observed in WT cultures. Indicative of enhanced autophagic flux, we observed a robust induction of p62 in Bbc3−/− CPF-treated cultures which was significantly higher than we observed in identically exposed WT cultures (Figs. 7A and 7B). Overall, these data suggest that CPF exposure caused ER stress and protein aggregation, and that these processes were potentially counteracted by autophagy. Moreover, enhanced survival associated with Bbc3-loss augments these neuroprotective processes in association with enhanced clearance of disease-associated protein aggregates (Figs. 6 and 7).
Figure 7.
Autophagy is induced with chlorpyrifos (CPF) exposure and further increased in Bbc3−/− cultures. Primary cortical neuronal cultures were established from WT and Bbc3−/− embryos (E14.5–E16.5) and treated with vehicle (DMSO) or 50 µM CPF for 6, 12, or 24 h. A, Protein lysate was collected and analyzed using SDS-PAGE and immunoblotting for autophagy-related proteins of interest using anti-phospho-mammalian target of rapamycin (mTOR; Ser2448), anti-mTOR, and anti-p62. (B) The ratio of phospho-mTOR (Ser2448) to total mTOR modestly decreased with CPF-treatment within both genotypes (2-way ANOVA, nsp = .0531 [genotype], *p = .0165 [treatment], nsp = .8457 [genotype × treatment)]. Although none of the multiple comparisons were statistically significant, there was a trending decline in ratio of phospho-mTOR (Ser2448) to total mTOR with CPF treatment in Bbc3−/− cultures at 12 h (Bbc3−/− DMSO to Bbc3−/− 12 CPF, nsp = .0518). p62 levels were slightly increased in WT CPF-treated cultures, but significantly increased in identically treated Bbc3−/− cultures (2-way ANOVA, **p = .0036 [genotype], ***p = .0002 [treatment], nsp = .0738 [genotype × treatment]). All statistical analyses were performed using 2-way ANOVAs with multiple comparisons across 4 biological replicates. Error bars in (B) represent SEM.
DISCUSSION
This study provides experimental evidence relevant to multiple epidemiologic studies indicating that CPF exposure increases the risk of neurodegenerative disease (Costello et al., 2009; Dhillon et al., 2008; Gatto et al., 2009; Hayden et al., 2010; Manthripragada et al., 2010; Narayan et al., 2013). Data indicate that the BH3-only, pro-apoptotic mediator, Bbc3, is a key effector of CPFs cytotoxic effects (Figs. 1–4). Intrinsic apoptosis occurring in CPF-exposed cultures was associated with protein aggregation, and elevated ER stress- and autophagy-related protein expression (Figs. 5–7). The findings highlight dysregulated proteostasis as a cellular mechanism associated with CPF-induced neurotoxicity and suggest that modulation of pathways associated with intrinsic apoptosis may be useful in mitigating these effects.
CPF exposure has been associated with the development of numerous neurological deficits (Rauh et al., 2006, 2011, 2012) and neurodegenerative disorders (Dhillon et al., 2008; Gatto et al., 2009; Manthripragada et al., 2010), but our understanding of the mechanisms of toxicity remains limited. Here, we used primary cortical neuronal cultures to begin to understand the impact of CPF on cellular populations in the CNS, in particular, neurons. Glial contamination is always a confound when analyzing neurotoxicity in primary neuron cultures. We used previously reported methods to prevent such contamination including the isolation of cortices at E14.5–16.5 and cultivation in B27-supplemented neurobasal media. Embryonic neuronal cultures isolated from rats grown in B27-supplemented neurobasal media had glial growth reduced to <0.5% of the pure neuronal population (Brewer et al., 1993). Moreover, subcortical neuronal cultures isolated from rat brains at E17 have been shown to contain <1.5% astrocytes (GFAP+ cells) at 10 days in vitro (d.i.v.) reaching only 10–15% by 22 d.i.v. (Patel et al., 1988). Our cultures were allowed to grow for 4-6 days prior to treatment and harvesting. Direct analysis of specific neuronal subtypes was not the primary goal of this mechanistic study; however, primary cultures such as those utilized here can provide mechanistic evidence relevant to neurodegenerative disorders. AD is characterized by brain atrophy, with the most severe cellular loss seen within cortical and hippocampal regions (Donev et al., 2009) of both cholinergic (Ferreira-Vieira et al., 2016) and catecholaminergic neurons (Pan et al., 2020). PD is classically defined by loss of dopaminergic neurons within the substantia nigra pars compacta, but evidence of neuronal cell death within other brains regions and diffuse Lewy body pathology, including in cortical regions, is commonly observed in the disease (Giguere et al., 2018). Additional studies are needed to dissect the impact of CPF intoxication on neurochemically distinct cell types in our cultures, and to ascertain the impact of unanticipated glial contamination. However, previous findings support the presence of multiple relevant cell types including cholinergic (Andre et al., 1994) and catecholaminergic (Iacovitti et al., 1987) neurons. These culture models represent a useful system to conduct future studies based on our findings of disease-related pathobiologies including proteinopathy and alterations in ER- and autophagy-related mechanisms.
Transcriptomic and ontologic analyses clearly implicated intrinsic apoptotic pathways in CPF-exposed cortical neuronal cultures (Figure 1B). These findings are consistent with other reports (Lee et al., 2014; Park et al., 2015) demonstrating CPF-mediated induction of p53-mediated apoptosis in both neuronal and nonneuronal cell lines. Our study extends these findings, identifying the pro-apoptotic mediator Bbc3 as a key node in CPF-associated apoptotic signaling. Prior to our studies, Bbc3, and other genes related to the intrinsic apoptotic pathway, were similarly detected in common carp exposed to CPF (Jiao et al., 2019) and zebrafish larvae exposed to the toxic metabolite CPO (Garcia-Reyero et al., 2016). To expand our understanding of related Bbc3 signaling in the context of CPF exposure, we evaluated the related genes Trp53 (encodes for p53 protein) and Pmaip1 (encodes for NOXA protein). Trp53 is a known transcriptional regulator of Bbc3 and Pmaip1 (Han et al., 2001; Oda et al., 2000), shown to induce their expression following exposure to various lethal stimuli (Fridman and Lowe, 2003). Trp53 expression was elevated following 6 h of CPF exposure, but expression normalized by the 12-h timepoint. Although unexpected, this finding may be related to the primary mechanism of p53 activation that relies on protein stabilization rather than transcriptional elevation (Shiloh and Ziv, 2013). Perhaps more interestingly, certain chemical carcinogens repress expression of TP53 in the context of oncogenesis. Condensed tobacco smoke particulate suppresses the TP53 apoptotic response (Narayanan et al., 2015). In addition to carcinogenesis, Bbc3 activity has been demonstrated in the context of p53-independent stimuli including dexamethasone and serum deprivation (Han et al., 2001). Our work is consistent with chemical modulation of p53 function and prompts further study of CPF’s interactions with intrinsic apoptotic pathways in the context of neurological disorders.
Although we observed CPF-induced upregulation of Trp53, Bbc3, and Pmaip1 in WT cultures (Figure 2), we did not detect robust modulation of Trp53 and Pmaip1 in Bbc3−/− CPF-treated cultures using real-time PCR. These findings are surprising since the current literature does not suggest that Bbc3 regulates Trp53 or Pmaip1. We postulate that this response might be cell-type and/or stimulus-specific and that Bbc3 may have an unreported function in maintaining normal levels of Trp53 and Pmaip1. It is also possible that germ-line deletion of Bbc3 resulted in a compensatory mechanism of intrinsic apoptotic regulation, possibly related to other pro-apoptotic BH3-only proteins including Bik, Bad, and Bim whose function in intoxicated Bbc3−/− mice remains to be characterized. Notably, there was unexpected temporal regulation of Trp53 and Pmaip1 within our CPF-exposed cultures, with the 12-h time point showing no induction of expression (Figs. 2B and 2C). This observation may represent the initial upregulation of DNA repair machinery (6-h timepoint), following which cells attempt to overcome CPF-related DNA damage and repair the genome (12-h timepoint; Williams and Schumacher, 2016). Over time, cultures are ultimately unable to overcome CPF insult and apoptotic programming is once again induced (24-h timepoint) (Figs. 2B and 2C). This temporal phenomenon was not observed for Bbc3 transcript in WT cultures (Figure 2A), suggesting that, similar to the discussion above, Bbc3 may be functioning in a p53-independent manner. Interestingly, it has been reported that following ATF4-mediated ER stress in primary cortical neuronal cultures, the ATF4-transcriptional target CHOP can directly bind the Bbc3 promoter and cause transcriptional upregulation independent of p53 (Galehdar et al., 2010). Consistent with this report, we observed elevations in the ATF4-CHOP axis following CPF treatment (Figure 5). Loss of Bbc3 conferred resistance to CPF-induced intrinsic apoptosis (Figure 4), thereby improving survival (Figs. 3 and 4). Prior reports by others describe similar findings in other cell types derived from Bbc3−/− mice. Bbc3 loss confers resistance to intrinsic apoptotic stressors including ionizing radiation in thymocytes (Jeffers et al., 2003), and cytokine deprivation, glucocorticoid exposure, staurosporine, and phorbol ester in thymocytes and spleen cells (Villunger et al., 2003). Further supporting our findings, Bbc3−/− mice and mesencephalic neuronal cultures derived from them are resistant to PD-associated toxicants including 6-OHDA (Bernstein et al., 2011) and MPP+ (Bernstein and O’Malley, 2013).
Ontologic analysis of RNA-Seq data also identified ER stress as an aspect of CPF toxicity (Figure 1 and Supplementary Table 1). Typically triggered as a protective mechanism during the regulation of protein folding (Sano and Reed, 2013), prolonged ER stress can result in cellular apoptosis (Xu et al., 2005). CPF exposure has been reported to cause ER stress in a nonneuronal cell line (Reyna et al., 2017). We confirmed upregulation of transcripts and proteins involved in the ER stress response within our primary neuronal cultures (Figure 5). Unexpectedly, evaluation of markers of ER stress indicated that this stress response was enhanced in Bbc3−/− cultures, as measured by elevations in ATF4 and Ddit3/CHOP (Figure 5). This was surprising because induction of an ER stress response is typically associated with apoptosis rather than survival (Sano and Reed, 2013). We interpret these findings to indicate that suppression of Bbc3 in the context of pesticide exposure leads to a more robust ER stress-related response resulting from a failure of intoxicated cells to undergo programmed cell death. Such a phenomenon is of interest based on the goal of protecting neuronal cell populations in individuals facing either acute or chronic OP poisoning. Findings are consistent with our observation of ER stress-related alterations in gene expression in the RNA-Seq study, however, are not sufficient to definitively confirm ER stress. Functional characterization of ER stress and evaluation of additional markers including the phosphorylated and total forms of PERK, eIF2A, and IRE1 would strengthen these data and shed light on which branch of the unfolded protein response may be modulated by CPF exposure.
Many neurodegenerative disorders are characterized by the pathogenic misfolding and aggregation of proteins, including alpha-synuclein, tau, amyloid-beta, and huntingtin (Ross and Poirier, 2004). ER stress is associated with dysregulation of the protein lifecycle, but can be overwhelmed, augmenting protein aggregation (Lindholm et al., 2006). CPF has been shown to increase levels of pathogenic proteins species, including Aβ 1-40, Aβ 1-42, and phosphorylated (pS199) tau (Moyano et al., 2018) and CPF’s toxic metabolite, CPO, is associated with the formation of high-molecular weight protein aggregates of porcine tubulin (Schopfer and Lockridge, 2018). Consistent with these studies, CPF elicited high molecular weight protein accumulation in primary neuronal cultures. We identified high-molecular weight, alpha-synuclein and tau immunoreactive aggregates in CPF-treated cultures, with neurons lacking Bbc3−/− exhibiting far less protein aggregation (Figure 6). Protein misfolding and accumulation have been shown to elicit a feed-forward loop, with misfolded proteins triggering ER stress and prolonged ER stress causing further accumulation of aberrant proteins. When under duress, cells increase the expression of molecular chaperones to aid in protein folding, slow down protein synthesis, and/or enhance protein degradation through a variety of mechanisms (Lindholm et al., 2006). Disruptions in the ubiquitin-proteasome system lead to accumulation of highly ubiquitinated, aggregated protein complexes (Berke and Paulson, 2003). Despite these associations, we saw no major differences in ubiquitination across genotypes or treatment groups (Figure 6). This prompted us to evaluate alternative mechanisms that may account for the reduction in high-molecular weight proteins we observed in Bbc3−/− cultures treated with CPF.
ER stress has been reported to preferentially upregulate autophagy in neurons (Nijholt et al., 2011). mTOR negatively regulates autophagy (Jung et al., 2010) and we observed decreases in the phospho-mTOR (Ser2448) pool following CPF-treatment in both WT and Bbc3−/− cultures (Figure 7). We also noted induction of the classic autophagy marker p62, also known as sequestosome (SQSTM1), in CPF-treated neuronal cultures (Figure 7). These responses were heightened in Bbc3−/− cultures, indicative of enhanced autophagy-related processes, which was associated with a reduction in toxic protein accumulation (Figure 6). These findings are consistent with previous reports of a neuroprotective role for autophagy in CPF-induced cell death observed in SH-SY5Y cells pretreated with either the autophagic flux inhibitor, bafilomycin A, or rapamycin, an mTOR inhibitor (Singh et al., 2018). These findings support a model in which Bbc3−/− neurons, unable to execute a programmed cell death response, undergo prolonged upregulation of autophagic mechanisms resulting in enhanced protein clearance (Figure 8). It should be noted that p62 is typically degraded during autophagy (Yoshii and Mizushima, 2017), yet we see immediate induction with CPF treatment within Bbc3−/− cultures. This phenomenon has been previously reported with CPF-treated SH-SY5Y cells (Park et al., 2013; Singh et al., 2018) and is believed to be caused by an increased formation of autophagosomes. Consistent with this model, after the significant increase in p62 levels observed at the 6-h mark, there is a step-wise decline over time, anticipated as the result of ongoing degradation at the later timepoints. This finding has exciting implications, but additional autophagy studies, such as autophagic flux assays (Yoshii and Mizushima, 2017), will be important to fully validate these data. The links between ER stress- and autophagy-related markers we identify are supported by prior reports (Rashid et al., 2015). For example, in a phenomenon coined aggrephagy, ER stress-induced activation of the ATF4-Ddit3 axis stimulates expression of autophagy receptor genes including SQSTM1 (p62). Upregulated SQSTM1 then interacts with ubiquitinated substrates bound for autophagic degradation, enhancing their clearance (Rubio et al., 2014).
Figure 8.
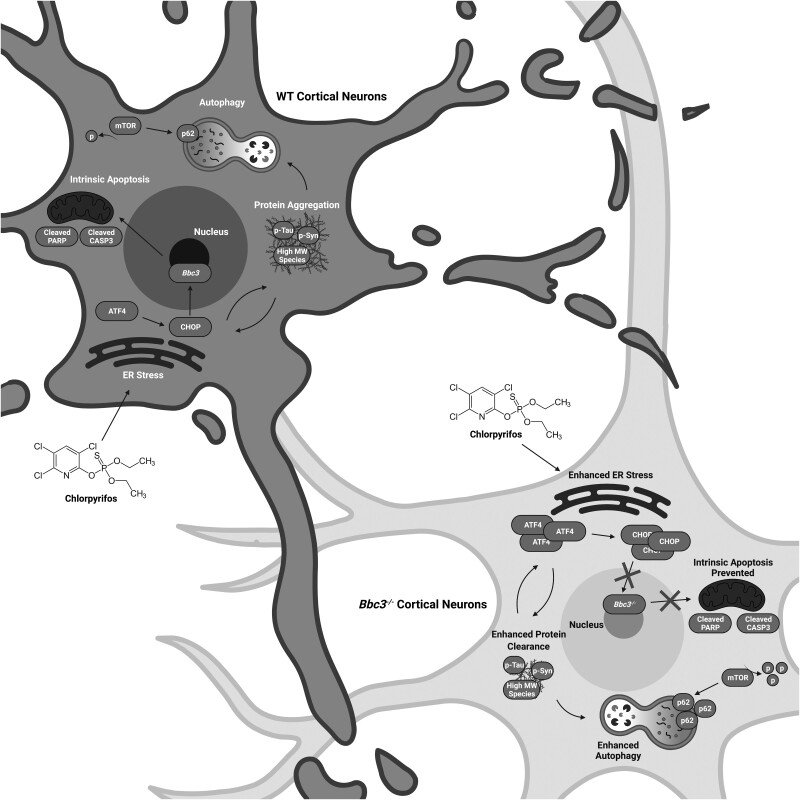
Proposed model of chlorpyrifos (CPF)-induced molecular changes impacting neuronal survival. CPF exposure results in protein aggregation and endoplasmic reticulum (ER) stress in WT cultures. Although autophagic mechanisms are slightly enhanced, presumably in order to clear these misfolded proteins, WT neurons are unable to overcome this stress and ultimately undergo Bbc3-mediated intrinsic apoptosis, as is evidenced by increased cytotoxicity and elevation of apoptotic executioners, cleaved CASP3 and cleaved PARP. Similar events occur in Bbc3−/−cortical neurons; however, the lack of Bbc3 prevents these cells from undergoing intrinsic apoptosis, protecting them from cell death as shown by reduced cleavage of apoptotic substrates. Therefore, CPF exposure leads to continued ER stress, shown by elevations in ATF4 and CHOP, and autophagy, suggested by enhanced mTOR dephosphorylation and increased p62 expression, in Bbc3−/− cultures resulting in enhanced protein clearance. Therefore, Bbc3 appears to be a key node in the cytotoxic effects of CPF exposure as Bbc3−/− neurons are protected from CPF-induced neuronal cell death as they lack of intrinsic apoptotic machinery and clear pathogenic, aggregated protein species. Created with BioRender.
Forward looking studies will confirm and investigate other genes identified in our RNA-Seq experiment, likely to be relevant to CPF’s toxic effects. For example, the most significantly upregulated gene in our study was the relatively uncharacterized gene Pagr1a (also known as PA1) (Supplementary Tables 2 and 3; fold change 44.61, p = 2.95e−6). Although little is known about its function, it has been shown to interact with PAXIP1 (also known as PTIP) at sites of DNA damage. The PTIP/PA1 complex promotes cell survival after DNA damage (Gong et al., 2009). CPF exposure has previously been associated with DNA damage both in vitro (Li et al., 2015) and in vivo (Mehta et al., 2008; Rahman et al., 2002). Interestingly, DNA damage is a well-characterized trigger of apoptosis (Norbury and Zhivotovsky, 2004). Therefore, the possibility of convergent mechanisms of CPF-induced molecular changes leading to apoptosis should be considered. In addition to this example, Supplementary Tables 2–4 list multiple novel genes presenting numerous opportunities to improve our understanding of CPF toxicity in neuronal cells.
AChE inhibition is a widely reported outcome of CPF exposure, caused by the toxic CPF metabolite, CPO (Kousba et al., 2004). Although we treated our cells with the parent compound, CPF, evidence suggests that CPF is rapidly hydrolyzed in aqueous solutions, such as water (Racke, 1993). Our own experimental evidence suggests that following addition of CPF into water, a proportion of it is immediately converted to CPO and the inert metabolite TCPy (Supplementary Figure 4). Although levels of CPF steadily drop over the course of a 7-day time period, levels of CPO and TCPy remain quite stable (Supplementary Figure 4). We believe these findings can extend to conditions within our liquid cell culture media, likely occurring at equivalent levels in WT and Bbc3−/− cultures. Therefore, we cannot rule out the possibility that the toxicity we observed is due to not only CPF, but potentially CPF’s metabolites as well. Interestingly however, we believe that the cell death we observed is not due to the potential AChE inhibition occurring, as prior reports indicate that AChE is upregulated during apoptosis, and that suppression of AChE, such as that accompanied by AD drugs like donepezil, inhibits cell death (Park et al., 2004; Zhang and Greenberg, 2012; Zhang et al., 2002). This study suggests a mechanism that may be AChE independent, and suggests the use of Bbc3−/− cultures to discover novel pathways of interest related to OP exposure and disease-related compensatory neuroprotective processes.
Others have suggested negative regulation of Bbc3 as a potential therapy for both cardiovascular and neurodegenerative disease (Tichy et al., 2018) and although loss of Bbc3 proves beneficial in our model, there is a potential for deleterious off target effects because Bbc3 is utilized for homeostatic processes across virtually all cell types. In addition, suppression of Bbc3 or related tumor suppressor genes like Trp53 have the potential to cause neoplasia (Yu et al., 2006). Although more work is needed, our unexpected findings of Bbc3-related transcriptional modulation are important because they may provide inroads towards characterizing novel cell-type specific functions and identify new, safer targets that can be evaluated for suppressing proteinopathy in the context of neuroprotection. During aging, protein clearance is compromised, such that ER stress can no longer elicit as robust an adaptive response, thought to be a function of reduced component expression and failure of chaperone systems (Naidoo, 2009). For example, levels of the molecular chaperone binding immunoglobulin protein (BiP) are found at reduced levels within the brains of aged rats compared with their younger counterparts (Paz Gavilan et al., 2006; Hussain and Ramaiah, 2007), and BiP deletion accelerates prion propagation in vivo (Park et al., 2017). Although long-term ER stress can be deleterious, resulting in cellular apoptosis (Hetz and Saxena, 2017), data from our study suggests that mild ER stress promotes neuronal survival in the face of proteinaceous insult. Supportive of this finding, deletion of Atf6α, an important mediator of one branch of the ER stress response, resulted in increased neuronal degeneration and ubiquitin accumulation within the brains of 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine (MPTP)-injected mice (Hashida et al., 2012). Further, mild ER stress induction has been shown be beneficial to reduce misfolded protein accumulation and increase longevity in C. elegans (Matai et al., 2019). We believe our data highlight a complex interplay between a widely used toxicant, ER stress, and protein clearance mechanisms, potentially including autophagy. These data inform our understanding of neuroprotective cellular biologies and suggest Bbc3-inactivated cells and animals as useful models in which to explore these processes to aid in our understanding of aging, exposure, and neurodegenerative disease.
SUPPLEMENTARY DATA
Supplementary data are available at Toxicological Sciences online.
Supplementary Material
ACKNOWLEDGMENTS
The authors acknowledge the following Shared Resources facilities: Genomics and Molecular Biology Shared Resources (GMBSR) and Pathology Shared Resource (PSR) at the Norris Cotton Cancer Center (NCCC) at Dartmouth with NCI Cancer Center Support Grant (5P30CA023108-37). They thank Dr Fred Kolling IV and the members of GMBSR within the NCCC for library preparation and sequencing. They thank the Center for Comparative Medicine and Research (CCMR) at Dartmouth for their expertise in animal husbandry and their assistance with the maintenance of our animal colonies. They sincerely thank the laboratories of Dr Scott Gerber, Dr Todd Miller, and Dr TY Chang for their reagent contributions. They appreciate those within the Office of Environmental Health and Safety (EHS) for their support of the proper handling and disposal of environmental toxicants. They are deeply appreciative of Dr Gerard Zambetti and his laboratory at St. Jude Children’s Research Hospital (Nashville, TN) for their donation of the Bbc3−/− mice.
FUNDING
The National Institute of Environmental and Health Sciences (NIEHS; R01 ES024745 to M.C.H. and F31 ES030982-01 to F.L.A.).
DECLARATION OF CONFLICTING INTERESTS
The authors declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
REFERENCES
- Anderson J. P., Walker D. E., Goldstein J. M., de Laat R., Banducci K., Caccavello R. J., Barbour R., Huang J., Kling K., Lee M., et al. , (2006). Phosphorylation of Ser-129 is the dominant pathological modification of alpha-synuclein in familial and sporadic Lewy body disease. J. Biol. Chem. 281, 29739–29752. [DOI] [PubMed] [Google Scholar]
- Andre C., Dos Santos G., Koulakoff A. (1994). Cultured neurons from mouse brain reproduce the muscarinic receptor profile of their tissue of origin. Eur. J. Neurosci. 6, 1691–1701. [DOI] [PubMed] [Google Scholar]
- Berke S. J., Paulson H. L. (2003). Protein aggregation and the ubiquitin proteasome pathway: Gaining the UPPer hand on neurodegeneration. Curr. Opin. Genet. Dev. 13, 253–261. [DOI] [PubMed] [Google Scholar]
- Bernstein A. I., O’Malley K. L. (2013). MPP+-induces PUMA- and p53-dependent, but ATF3-independent cell death. Toxicol. Lett. 219, 93–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein A. I., Garrison S. P., Zambetti G. P., O’Malley K. L. (2011). 6-OHDA generated ROS induces DNA damage and p53- and PUMA-dependent cell death. Mol. Neurodegener. 6, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blain P. G. (2011). Organophosphorus poisoning (acute). BMJ Clin. Evid. 2011, [PMC free article] [PubMed] [Google Scholar]
- Brewer G. J., Torricelli J. R., Evege E. K., Price P. J. (1993). ‘Optimized survival of hippocampal neurons in B27-supplemented Neurobasal, a new serum-free medium combination. J. Neurosci. Res. 35, 567–576. [DOI] [PubMed] [Google Scholar]
- Cannon J. R., Greenamyre J. T. (2013). Gene-environment interactions in Parkinson’s disease: Specific evidence in humans and mammalian models. Neurobiol. Dis. 57, 38–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin-Chan M., Navarro-Yepes J., Quintanilla-Vega B. (2015). Environmental pollutants as risk factors for neurodegenerative disorders: Alzheimer and Parkinson diseases. Front. Cell. Neurosci. 9, 124–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa L. G., Giordano G., Cole T. B., Marsillach J., Furlong C. E. (2013). Paraoxonase 1 (PON1) as a genetic determinant of susceptibility to organophosphate toxicity. Toxicology 307, 115–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costello S., Cockburn M., Bronstein J., Zhang X., Ritz B. (2009). Parkinson‘s disease and residential exposure to maneb and paraquat from agricultural applications in the central valley of California. Am. J. Epidemiol. 169, 919–926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croom E. L., Wallace A. D., Hodgson E. (2010). Human variation in CYP-specific chlorpyrifos metabolism. Toxicology 276, 184–191. [DOI] [PubMed] [Google Scholar]
- Dhillon A. S., Tarbutton G. L., Levin J. L., Plotkin G. M., Lowry L. K., Nalbone J. T., Shepherd S. (2008). Pesticide/environmental exposures and Parkinson’s disease in East Texas. J. Agromedicine 13, 37–48. [DOI] [PubMed] [Google Scholar]
- Donev R., Kolev M., Millet B., Thome J. (2009). Neuronal death in Alzheimer’s disease and therapeutic opportunities. J. Cell. Mol. Med. 13, 4329–4348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira-Vieira T. H., Guimaraes I. M., Silva F. R., Ribeiro F. M. (2016). Alzheimer’s disease: Targeting the Cholinergic System. Curr. Neuropharmacol. 14, 101–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fridman J. S., Lowe S. W. (2003). Control of apoptosis by p53. Oncogene 22, 9030–9040. [DOI] [PubMed] [Google Scholar]
- Galehdar Z., Swan P., Fuerth B., Callaghan S. M., Park D. S., Cregan S. P. (2010). Neuronal apoptosis induced by endoplasmic reticulum stress is regulated by ATF4-CHOP-mediated induction of the Bcl-2 homology 3-only member PUMA. J. Neurosci. 30, 16938–16948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Reyero N., Escalon L., Prats E., Faria M., Soares A. M. V. M., Raldúa D. (2016). Targeted gene expression in zebrafish exposed to chlorpyrifos-oxon confirms phenotype-specific mechanisms leading to adverse outcomes. Bull. Environ. Contam. Toxicol. 96, 707–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatto N. M., Cockburn M., Bronstein J., Manthripragada A. D., Ritz B. (2009). Well-water consumption and Parkinson’s disease in rural California. Environ. Health Perspect. 117, 1912–1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giguere N., Burke Nanni S., Trudeau L. E. (2018). On cell loss and selective vulnerability of neuronal populations in Parkinson’s disease. Front. Neurol. 9, 455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong Z., Cho Y. W., Kim J. E., Ge K., Chen J. (2009). Accumulation of Pax2 transactivation domain interaction protein (PTIP) at sites of DNA breaks via RNF8-dependent pathway is required for cell survival after DNA damage. J. Biol .Chem. 284, 7284–7293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grundke-Iqbal I., Iqbal K., Quinlan M., Tung Y. C., Zaidi M. S., Wisniewski H. M. (1986). Microtubule-associated protein tau. A component of Alzheimer paired helical filaments. J. Biol. Chem. 261, 6084–6089. [PubMed] [Google Scholar]
- Grundke-Iqbal I., Iqbal K., Tung Y. C., Quinlan M., Wisniewski H. M., Binder L. I. (1986). Abnormal phosphorylation of the microtubule-associated protein tau (tau) in Alzheimer cytoskeletal pathology. Proc. Natl. Acad. Sci. U.S.A. 83, 4913–4917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J., Flemington C., Houghton A. B., Gu Z., Zambetti G. P., Lutz R. J., Zhu L., Chittenden T. (2001). Expression of bbc3, a pro-apoptotic BH3-only gene, is regulated by diverse cell death and survival signals. Proc. Natl. Acad. Sci. U.S.A. 98, 11318–11323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasegawa M., Fujiwara H., Nonaka T., Wakabayashi K., Takahashi H., Lee V. M.-Y., Trojanowski J. Q., Mann D., Iwatsubo T. (2002). alpha-Synuclein is phosphorylated in synucleinopathy lesions. Nat. Cell Biol. 277, 49071–49074. [Google Scholar]
- Hashida K., Kitao Y., Sudo H., Awa Y., Maeda S., Mori K., Takahashi R., Iinuma M., Hori O. (2012). ATF6alpha promotes astroglial activation and neuronal survival in a chronic mouse model of Parkinson’s disease. PLoS One 7, e47950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatcher J. M., Pennell K. D., Miller G. W. (2008). Parkinson’s disease and pesticides: A toxicological perspective. Trends Pharmacol. Sci. 29, 322–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayden K. M., Norton M. C., Darcey D., Ostbye T., Zandi P. P., Breitner J. C., Welsh-Bohmer K. A.; Investigators Cache County Study. (2010). Occupational exposure to pesticides increases the risk of incident AD: The Cache County study. Neurology 74, 1524–1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hetz C., Saxena S. (2017). ER stress and the unfolded protein response in neurodegeneration. Nat. Rev. Neurol. 13, 477–491. [DOI] [PubMed] [Google Scholar]
- Hijaz B. A., Volpicelli-Daley L. A. (2020). Initiation and propagation of alpha-synuclein aggregation in the nervous system. Mol. Neurodegener. 15, 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hood E. (2001). The Tokyo attacks in retrospect: Sarin leads to memory loss. Environ. Health Perspect. 109, A542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoseki J., Ushioda R., Nagata K. (2010). Mechanism and components of endoplasmic reticulum-associated degradation. J. Biochem. 147, 19–25. [DOI] [PubMed] [Google Scholar]
- Hussain S. G., Ramaiah K. V. (2007). Reduced eIF2alpha phosphorylation and increased proapoptotic proteins in aging. Biochem. Biophys. Res. Commun. 355, 365–370. [DOI] [PubMed] [Google Scholar]
- Iacovitti L., Lee J., Joh T. H., Reis D. J. (1987). Expression of tyrosine hydroxylase in neurons of cultured cerebral cortex: Evidence for phenotypic plasticity in neurons of the CNS. J. Neurosci. 7, 1264–1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffers J. R., Parganas E., Lee Y., Yang C., Wang J., Brennan J., MacLean K. H., Han J., Chittenden T., Ihle J. N., et al. , (2003). Puma is an essential mediator of p53-dependent and -independent apoptotic pathways. Cancer Cell 4, 321–328. [DOI] [PubMed] [Google Scholar]
- Jiao W., Han Q., Xu Y., Jiang H., Xing H., Teng X. (2019). Impaired immune function and structural integrity in the gills of common carp (Cyprinus carpio L.) caused by chlorpyrifos exposure: Through oxidative stress and apoptosis. Fish Shellfish Immunol. 86, 239–245. [DOI] [PubMed] [Google Scholar]
- Jung C. H., Ro S. H., Cao J., Otto N. M., Kim D. H. (2010). mTOR regulation of autophagy. FEBS Lett. 584, 1287–1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karalliedde L., Senanayake N. (1989). Organophosphorus insecticide poisoning. Br. J. Anaesth. 63, 736–750. [DOI] [PubMed] [Google Scholar]
- Khatoon S., Grundke-Iqbal I., Iqbal K. (1992). Brain levels of microtubule-associated protein tau are elevated in Alzheimer’s disease: A radioimmuno-slot-blot assay for nanograms of the protein. J. Neurochem. 59, 750–753. [DOI] [PubMed] [Google Scholar]
- Kopjar N., Žunec S., Mendaš G., Micek V., Kašuba V., Mikolić A., Lovaković B. T., Milić M., Pavičić I., Čermak A. M. M., et al. , (2018). Evaluation of chlorpyrifos toxicity through a 28-day study: Cholinesterase activity, oxidative stress responses, parent compound/metabolite levels, and primary DNA damage in blood and brain tissue of adult male Wistar rats. Chem. Biol. Interact. 279, 51–63. [DOI] [PubMed] [Google Scholar]
- Kopke E., Tung Y. C., Shaikh S., Alonso A. C., Iqbal K., Grundke-Iqbal I. (1993). Microtubule-associated protein tau. Abnormal phosphorylation of a non-paired helical filament pool in Alzheimer disease. J. Biol. Chem. 268, 24374–24384. [PubMed] [Google Scholar]
- Kousba A. A., Sultatos L. G., Poet T. S., Timchalk C. (2004). Comparison of chlorpyrifos-oxon and paraoxon acetylcholinesterase inhibition dynamics: Potential role of a peripheral binding site. Toxicol. Sci. 80, 239–248. [DOI] [PubMed] [Google Scholar]
- Kumar P., Nagarajan A., Uchil P. D. (2018). Analysis of cell viability by the MTT assay. Cold Spring. Harb. Protoc. 2018, 469–471. [DOI] [PubMed] [Google Scholar]
- Lee J. E., Lim M. S., Park J. H., Park C. H., Koh H. C. (2014). Nuclear NF-kappaB contributes to chlorpyrifos-induced apoptosis through p53 signaling in human neural precursor cells. NeuroToxicology 42, 58–70. [DOI] [PubMed] [Google Scholar]
- Li W., Ehrich M. (2013). Transient alterations of the blood-brain barrier tight junction and receptor potential channel gene expression by chlorpyrifos. J. Appl. Toxicol. 33, 1187–1191. [DOI] [PubMed] [Google Scholar]
- Li D., Huang Q., Lu M., Zhang L., Yang Z., Zong M., Tao L. (2015). The organophosphate insecticide chlorpyrifos confers its genotoxic effects by inducing DNA damage and cell apoptosis. Chemosphere 135, 387–393. [DOI] [PubMed] [Google Scholar]
- Lindholm D., Wootz H., Korhonen L. (2006). ER stress and neurodegenerative diseases. Cell Death Differ. 13, 385–392. [DOI] [PubMed] [Google Scholar]
- Liu C. H., Di Y. P. (2020). Analysis of RNA sequencing data using CLC genomics workbench. Methods Mol. Biol. 2102, 61–113. [DOI] [PubMed] [Google Scholar]
- Loh Y., Swanberg M. M., Ingram M. V., Newmark J. (2010). Case report: Long-term cognitive sequelae of sarin exposure. NeuroToxicology 31, 244–246. [DOI] [PubMed] [Google Scholar]
- Manthripragada A. D., Costello S., Cockburn M. G., Bronstein J. M., Ritz B. (2010). Paraoxonase 1, agricultural organophosphate exposure, and Parkinson disease. Epidemiology 21, 87–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matai L., Sarkar G. C., Chamoli M., Malik Y., Kumar S. S., Rautela U., Jana N. R., Chakraborty K., Mukhopadhyay A. (2019). Dietary restriction improves proteostasis and increases life span through endoplasmic reticulum hormesis. Proc. Natl. Acad. Sci. U.S.A., 116, 17383–17392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta A., Verma R. S., Srivastava N. (2008). Chlorpyrifos-induced DNA damage in rat liver and brain. Environ. Mol. Mutagen. 49, 426–433. [DOI] [PubMed] [Google Scholar]
- Mense S. M., Sengupta A., Lan C., Zhou M., Bentsman G., Volsky D. J., Whyatt R. M., Perera F. P., Zhang L. (2006). The common insecticides cyfluthrin and chlorpyrifos alter the expression of a subset of genes with diverse functions in primary human astrocytes. Toxicol. Sci. 93, 125–135. [DOI] [PubMed] [Google Scholar]
- Migliore L., Coppede F. (2009). Genetics, environmental factors and the emerging role of epigenetics in neurodegenerative diseases. Mutat. Res. 667, 82–97. [DOI] [PubMed] [Google Scholar]
- Moyano P., Frejo M. T., Anadon M. J., Garcia J. M., Diaz M. J., Lobo M., Sola E., Garcia J., Del Pino J. (2018). SN56 neuronal cell death after 24h and 14days chlorpyrifos exposure through glutamate transmission dysfunction, increase of GSK-3beta enzyme, beta-amyloid and tau protein levels. Toxicology 402–403, 17–27. [DOI] [PubMed] [Google Scholar]
- Munoz-Quezada M. T., Lucero B. A., Iglesias V. P., Munoz M. P., Cornejo C. A., Achu E., Baumert B., Hanchey A., Concha C., Brito A. M., et al. , (2016). Chronic exposure to organophosphate (OP) pesticides and neuropsychological functioning in farm workers: A review. Int. J. Occup. Environ. Health 22, 68–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naidoo N. (2009). ER and aging-Protein folding and the ER stress response. Ageing Res. Rev. 8, 150–159. [DOI] [PubMed] [Google Scholar]
- Narayan S., Liew Z., Paul K., Lee P.-C., Sinsheimer J. S., Bronstein J. M., Ritz B. (2013). Household organophosphorus pesticide use and Parkinson’s disease. Int. J. Epidemiol. 42, 1476–1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayanan K. B., Ali M., Barclay B. J., Cheng Q. S., D’Abronzo L., Dornetshuber-Fleiss R., Ghosh P. M., Gonzalez Guzman M. J., Lee T. J., Leung P. S., et al. , (2015). Disruptive environmental chemicals and cellular mechanisms that confer resistance to cell death. Carcinogenesis 36(Suppl. 1), S89–S110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naughton S. X., Terry A. V. Jr. (2018). Neurotoxicity in acute and repeated organophosphate exposure. Toxicology 408, 101–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nijholt D. A., de Graaf T. R., van Haastert E. S., Oliveira A. O., Berkers C. R., Zwart R., Ovaa H., Baas F., Hoozemans J. J., Scheper W. (2011). Endoplasmic reticulum stress activates autophagy but not the proteasome in neuronal cells: Implications for Alzheimer’s disease. Cell Death Differ. 18, 1071–1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norbury C. J., Zhivotovsky B. (2004). DNA damage-induced apoptosis. Oncogene 23, 2797–2808. [DOI] [PubMed] [Google Scholar]
- Oda E., Ohki R., Murasawa H., Nemoto J., Shibue T., Yamashita T., Tokino T., Taniguchi T., Tanaka N. (2000). Noxa, a BH3-only member of the Bcl-2 family and candidate mediator of p53-induced apoptosis. Science 288, 1053–1058. [DOI] [PubMed] [Google Scholar]
- Pan X., Kaminga A. C., Jia P., Wen S. W., Acheampong K., Liu A. (2020). Catecholamines in Alzheimer’s disease: A systematic review and meta-analysis. Front. Aging Neurosci. 12, 184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J. H., Ko J., Hwang J., Koh H. C. (2015). Dynamin-related protein 1 mediates mitochondria-dependent apoptosis in chlorpyrifos-treated SH-SY5Y cells. NeuroToxicology 51, 145–157. [DOI] [PubMed] [Google Scholar]
- Park J. H., Lee J. E., Shin I. C., Koh H. C. (2013). Autophagy regulates chlorpyrifos-induced apoptosis in SH-SY5Y cells. Toxicol. Appl. Pharmacol. 268, 55–67. [DOI] [PubMed] [Google Scholar]
- Park K. W., Eun Kim G., Morales R., Moda F., Moreno-Gonzalez I., Concha-Marambio L., Lee A. S., Hetz C., Soto C. (2017). The endoplasmic reticulum chaperone GRP78/BiP modulates prion propagation in vitro and in vivo. Sci. Rep. 7, 44723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S. E., Kim N. D., Yoo Y. H. (2004). Acetylcholinesterase plays a pivotal role in apoptosome formation. Cancer Res. 64, 2652–2655. [DOI] [PubMed] [Google Scholar]
- Parran D. K., Magnin G., Li W., Jortner B. S., Ehrich M. (2005). Chlorpyrifos alters functional integrity and structure of an in vitro BBB model: Co-cultures of bovine endothelial cells and neonatal rat astrocytes. NeuroToxicology 26, 77–88. [DOI] [PubMed] [Google Scholar]
- Patel A. J., Seaton P., Hunt A. (1988). A novel way of removing quiescent astrocytes in a culture of subcortical neurons grown in a chemically defined medium. Brain Res. 470, 283–288. [DOI] [PubMed] [Google Scholar]
- Paz Gavilan M., Vela J., Castano A., Ramos B., del Rio J. C., Vitorica J., Ruano D. (2006). Cellular environment facilitates protein accumulation in aged rat hippocampus. Neurobiol. Aging 27, 973–982. [DOI] [PubMed] [Google Scholar]
- Poet T. S., Timchalk C., Hotchkiss J. A., Bartels M. J. (2014). Chlorpyrifos PBPK/PD model for multiple routes of exposure. Xenobiotica 44, 868–881. [DOI] [PubMed] [Google Scholar]
- Racke K. D. (1993). Environmental fate of chlorpyrifos. Rev. Environ. Contam. Toxicol. 131, 1–150. [DOI] [PubMed] [Google Scholar]
- Rahman M. F., Mahboob M., Danadevi K., Saleha Banu B., Grover P. (2002). Assessment of genotoxic effects of chloropyriphos and acephate by the comet assay in mice leucocytes. Mutat. Res. 516, 139–147. [DOI] [PubMed] [Google Scholar]
- Rashid H. O., Yadav R. K., Kim H. R., Chae H. J. (2015). ER stress: Autophagy induction, inhibition and selection. Autophagy 11, 1956–1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauh V. A., Garfinkel R., Perera F. P., Andrews H. F., Hoepner L., Barr D. B., Whitehead R., Tang D., Whyatt R. W. (2006). Impact of prenatal chlorpyrifos exposure on neurodevelopment in the first 3 years of life among inner-city children. Pediatrics 118, e1845–e1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauh V. A., Perera F. P., Horton M. K., Whyatt R. M., Bansal R., Hao X., Liu J., Barr D. B., Slotkin T. A., Peterson B. S. (2012). Brain anomalies in children exposed prenatally to a common organophosphate pesticide. Proc. Natl. Acad. Sci. U.S.A. 109, 7871–7876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauh V., Arunajadai S., Horton M., Perera F., Hoepner L., Barr D. B., Whyatt R. (2011). Seven-year neurodevelopmental scores and prenatal exposure to chlorpyrifos, a common agricultural pesticide. Environ. Health Perspect. 119, 1196–1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyna L., Flores-Martin J., Ridano M. E., Panzetta-Dutari G. M., Genti-Raimondi S. (2017). Chlorpyrifos induces endoplasmic reticulum stress in JEG-3 cells. Toxicol. In Vitro 40, 88–93. [DOI] [PubMed] [Google Scholar]
- Ritz B., Yu F. (2000). Parkinson’s disease mortality and pesticide exposure in California 1984-1994. Int. J. Epidemiol. 29, 323–329. [DOI] [PubMed] [Google Scholar]
- Rohlman D. S., Anger W. K., Lein P. J. (2011). Correlating neurobehavioral performance with biomarkers of organophosphorous pesticide exposure. NeuroToxicology 32, 268–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross C. A., Poirier M. A. (2004). Protein aggregation and neurodegenerative disease. Nat. Med. 10(Suppl.), S10–S17. [DOI] [PubMed] [Google Scholar]
- Rubio N., Verrax J., Dewaele M., Verfaillie T., Johansen T., Piette J., Agostinis P. (2014). p38(MAPK)-regulated induction of p62 and NBR1 after photodynamic therapy promotes autophagic clearance of ubiquitin aggregates and reduces reactive oxygen species levels by supporting Nrf2-antioxidant signaling. Free Radic. Biol. Med. 67, 292–303. [DOI] [PubMed] [Google Scholar]
- Sanchez-Santed F., Colomina M. T., Herrero Hernandez E. (2016). Organophosphate pesticide exposure and neurodegeneration. Cortex 74, 417–426. [DOI] [PubMed] [Google Scholar]
- Sano R., Reed J. C. (2013). ER stress-induced cell death mechanisms. Biochim. Biophys. Acta 1833, 3460–3470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schopfer L. M., Lockridge O. (2018). Chlorpyrifos oxon promotes tubulin aggregation via isopeptide cross-linking between diethoxyphospho-Lys and Glu or Asp: Implications for neurotoxicity. J. Biol. Chem. 293, 13566–13577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekijima Y., Morita H., Yanagisawa N. (1997). Follow-up of sarin poisoning in Matsumoto. Ann. Intern. Med. 127, 1042. [DOI] [PubMed] [Google Scholar]
- Sharma K. (2019). Cholinesterase inhibitors as Alzheimer’s therapeutics (Review). Mol. Med. Rep. 20, 1479–1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiloh Y., Ziv Y. (2013). The ATM protein kinase: Regulating the cellular response to genotoxic stress, and more. Nat. Rev. Mol. Cell Biol. 14, 197–210. [PubMed] [Google Scholar]
- Singh N., Lawana V., Luo J., Phong P., Abdalla A., Palanisamy B., Rokad D., Sarkar S., Jin H., Anantharam V., et al. , (2018). Organophosphate pesticide chlorpyrifos impairs STAT1 signaling to induce dopaminergic neurotoxicity: Implications for mitochondria mediated oxidative stress signaling events. Neurobiol. Dis. 117, 82–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solomon K. R., Williams W. M., Mackay D., Purdy J., Giddings J. M., Giesy J. P. (2014). Properties and uses of chlorpyrifos in the United States. Rev. Environ. Contam. Toxicol. 231, 13–34. [DOI] [PubMed] [Google Scholar]
- Spillantini M. G., Crowther R. A., Jakes R., Hasegawa M., Goedert M. (1998). alpha-Synuclein in filamentous inclusions of Lewy bodies from Parkinson’s disease and dementia with lewy bodies. Proc. Natl. Acad. Sci. U.S.A. 95, 6469–6473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spillantini M. G., Schmidt M. L., Lee V. M., Trojanowski J. Q., Jakes R., Goedert M. (1997). Alpha-synuclein in Lewy bodies. Nature 388, 839–840. [DOI] [PubMed] [Google Scholar]
- Tanner C. M., Kamel F., Ross G. W., Hoppin J. A., Goldman S. M., Korell M., Marras C., Bhudhikanok G. S., Kasten M., Chade A. R., et al. , (2011). Rotenone, paraquat, and Parkinson’s disease. Environ. Health Perspect. 119, 866–872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thullbery M. D., Cox H. D., Schule T., Thompson C. M., George K. M. (2005). Differential localization of acetylcholinesterase in neuronal and non-neuronal cells. J. Cell Biochem. 96, 599–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tichy A., Marek J., Havelek R., Pejchal J., Seifrtova M., Zarybnicka L., Filipova A., Rezacova M., Sinkorova Z. (2018). New light on an old friend: Targeting PUMA in radioprotection and therapy of cardiovascular and neurodegenerative diseases. Curr. Drug Targets 19, 1943–1957. [DOI] [PubMed] [Google Scholar]
- Villunger A., Michalak E. M., Coultas L., Mullauer F., Bock G., Ausserlechner M. J., Adams J. M., Strasser A. (2003). p53- and drug-induced apoptotic responses mediated by BH3-only proteins puma and noxa. Science 302, 1036–1038. [DOI] [PubMed] [Google Scholar]
- Williams A. B., Schumacher B. (2016). p53 in the DNA-damage-repair process. Cold Spring Harb. Perspect. Med. 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson Leah N., Terry Alvin V., Bartlett Michael G. (2006). Determination of chlorpyrifos and its metabolites in rat brain tissue using coupled‐column liquid chromatography/electrospray ionization tandem mass spectrometry. Rapid Commun in Mass Spectrom. 20, 2689–2695. [DOI] [PubMed] [Google Scholar]
- Wortel I. M. N., T. van der Meer L., Kilberg M. S., van Leeuwen F. N. (2017). Surviving stress: Modulation of ATF4-mediated stress responses in normal and malignant cells. Trends Endocrinol. Metab. 28, 794–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C., Bailly-Maitre B., Reed J. C. (2005). Endoplasmic reticulum stress: Cell life and death decisions. J. Clin. Invest. 115, 2656–2664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamasue H., Abe O., Kasai K., Suga M., Iwanami A., Yamada H., Tochigi M., Ohtani T., Rogers M. A., Sasaki T., et al. , (2007). Human brain structural change related to acute single exposure to sarin. Ann. Neurol. 61, 37–46. [DOI] [PubMed] [Google Scholar]
- Yan D., Zhang Y., Liu L., Yan H. (2016). Pesticide exposure and risk of Alzheimer’s disease: A systematic review and meta-analysis. Sci. Rep. 6, 32222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye W., Gong X., Xie J., Wu J., Zhang X., Ouyang Q., Zhao X., Shi Y., Zhang X. (2010). AChE deficiency or inhibition decreases apoptosis and p53 expression and protects renal function after ischemia/reperfusion. Apoptosis 15, 474–487. [DOI] [PubMed] [Google Scholar]
- Yoshii S. R., Mizushima N. (2017). Monitoring and measuring autophagy. Int. J. Mol. Sci. 18, [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J., Yue W., Wu B., Zhang L. (2006). PUMA sensitizes lung cancer cells to chemotherapeutic agents and irradiation. Clin. Cancer Res. 12, 2928–2936. [DOI] [PubMed] [Google Scholar]
- Zhang X. J., Greenberg D. S. (2012). Acetylcholinesterase involvement in apoptosis. Front. Mol. Neurosci. 5, 40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X. J., Yang L., Zhao Q., Caen J. P., He H. Y., Jin Q. H., Guo L. H., Alemany M., Zhang L. Y., Shi Y. F. (2002). Induction of acetylcholinesterase expression during apoptosis in various cell types. Cell Death Differ. 9, 790–800. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.