Abstract
Within the past several decades, the emergence and spread of infectious diseases with pandemic potential have endangered human lives. Coronavirus disease 2019 (COVID-19) outbreak represents an unprecedented threat for all health systems worldwide. The clinical spectrum of COVID-19 is highly heterogeneous, ranging from asymptomatic and mild upper respiratory tract illness to severe interstitial pneumonia with respiratory failure and even death. Highly age-dependent patterns of immune response potentially explain the higher rates of the severe forms of COVID-19 in elderly patients. However, genetic and epigenetic architecture can influence multiple biological processes during the lifespan, therefore as far as our knowledge shows, vulnerability to viral infection concerning telomere length and epigenetic signature is not a new idea. This review aims is to summarize the current understanding of the role of telomere length and epigenetic mechanisms on the severity of COVID-19. The current knowledge highlights the significant association between the shorter telomere length and the higher risk of developing severe COVID-19. Differential DNA methylation patterns and miRNA expression profiles imply that these hallmarks can play a pivotal role in COVID- 19 pathogenesis. Understanding the causes of inter-individual variations in COVID-19 outcomes could provide clues to the development of the personalized therapeutic intervention.
Keywords: Telomere, Epigenetic, COVID-19, SARS-CoV-2, Severity
Abbreviations: COVID-19, coronavirus disease 2019; SARS-CoV-2, severe acute respiratory coronavirus 2; COPD, chronic obstructive pulmonary disease; TMMs, telomere maintenance mechanisms; ALT, alternative lengthening of telomeres; HIV, human immunodeficiency virus; HBV, hepatitis B virus; HCV, hepatitis C virus; EBV, Epstein-Barr virus; CMV, cytomegalovirus; IFN, Interferon; COPD/IPF, obstructive pulmonary disease/ idiopathic pulmonary fibrosis; CRP, C-reactive protein level; NLR, neutrophil-to-lymphocyte ratio; MTase, DNA methyltransferase; tRNA, transfer RNA; rRNA, ribosomal RNA; miRNA, microRNA; siRNA, short-interfering RNAs; HDAC, histone deacetylases; SLE, systemic lupus erythematosus; RA, rheumatoid arthritis; MS, multiple sclerosis; SS, Sjogren’s syndrome; AITD, Autoimmune thyroid diseases; T1D, Type 1 diabetes; KSHV, Kaposi's sarcoma-associated herpesvirus; HTLV, human T-lymphotropic virus; HPV, human papillomavirus; SV40, Simian vacuolating virus 40; PBCV, paramecium bursaria chlorella virus; scATAC, single-cell assay for transposase-accessible chromatin; scRNA, single-cell RNA; AT1R, Angiotensin receptor 1; ACE2, Angiotensin-converting enzyme 2; 5′UTRs, 5′ untranslated regions; TCZ, Tocilizumab
1. Introductions
The current coronavirus disease 2019 (COVID-19) pandemic has faced the world with unprecedented challenges. COVID-19 caused by an enveloped single-stranded positive RNA virus named severe acute respiratory coronavirus 2 (SARS-CoV-2), first emerged in Wuhan City, China, in late 2019 (C. Wang et al., 2020). Gender, age and comorbidities are the main prognostic factors in patients with COVID-19. Hypertension, diabetes, cancer, heart failure, and chronic obstructive pulmonary disease (COPD) are also closely related to the severity of COVID-19. Respiratory failure and multi-organ dysfunction as the results of impaired immune response and uncontrolled inflammatory processes are the leading causes of death in COVID-19 patients (Hasan et al., 2020, Richardson et al., 2020, Sheervalilou et al., 2020; D. Wang et al., 2020). valiabl evidence point to the significant association between immunocompetence and COVID-19 vulnerability and severity. Monitoring cytokine profiles in COVID-19 patients demonstrate that the cytokine storms may be associated with impaired acquired immune responses and uncontrolled inflammatory innate responses (Hu et al., 2021, Hue et al., 2020).
Immune response patterns are highly age-dependent. Age-associated changes in immune cells can affect the efficiency of immune responses (N. ping Weng, 2006). In general, aging is described as an inevitable time-dependent accumulation of several molecular and cellular damage that contributes to the increased susceptibility to morbidity and mortality (López-Otín et al., 2013). Chronological age alone is not a sufficient reflection of the state of the physiological parameters. In the past several years, the variety of potential candidate biomarkers have been identified and developed as biological age predictors. Biological age shows a stronger association with all-cause mortality than chronological age. Genetic and epigenetic changes during lifespan can affect the biological aging processes and may also enhance vulnerability to several diseases such as infectious conditions (Jylhävä et al., 2017). According to various findings, the causal role of DNA damage accumulation in aging is not negligible and several genetic defects have been described in humans and animal models which cause accelerated biological aging. However, some specific chromosomal regions are mainly involved in age-related deterioration. In the past decade, the advent of high-throughput genomics and epigenetics technologies has led to an exponential increase in the number of potential candidate biomarkers as biological age predictors. Telomere length and epigenetic signatures are the potential well-studied predictors of biological aging that can influence genetic architecture and cellular function (Jylhävä et al., 2017, Waziry et al., 2019).
The susceptibility to viral infections with respect to telomere length and epigenetic signatures is not a new idea. Telomere length and epigenetic signatures of the host can affect the probability of viral infections (Bayarsaihan, 2011, Helby et al., 2017). Also, our current knowledge highlights the determinant effect of age on severity and mortality in patients with COVID-19 (X. Li et al., 2020). According to therapeutic strategies, Malavolta et al recently postulated that cellular senescence may be a key pathological mechanism in SARS-CoV-2 infection susceptibility (Malavolta et al., 2020).
In the present review, we have attempted to categorize the current knowledge about the effect of telomere attrition and epigenetic signatures as the main predictors of biological aging and cellular senescence, on COVID-19 severity.
2. Telomere length and epigenetic signatures as the predictors of biological age
Aging is a major risk factor for many chronic inflammatory diseases and mortality. However, there is heterogeneity in the health status in a similar age group of individuals (Lowsky et al., 2014). During the past decades, a variety of potential candidate biomarkers have been identified as biological age predictors. The potential biological age predictors have been categorized into six subtypes, including epigenetic clocks, telomere length, transcriptomic predictors, proteomic predictors, metabolomics-based predictors, and composite biomarker predictors (Jylhävä et al., 2017). The exact relationship between telomere attrition and epigenetic alterations is still elusive. However, some evidence suggests that eroded telomeres lead to differentiation instability via DNA methylation (Criqui et al., 2020).
3. Telomere attrition
Telomeres, nucleoprotein structures at the termini of mammalian chromosomes, form complexes that protect the ends of linear chromosomes from continuous shortening during DNA replication (Blackburn, 1991). Telomere maintenance mechanisms (TMMs), either the telomerase, an enzyme that synthesizes telomeric DNA, or the alternative lengthening of telomeres (ALT) are frequently activated in cancer cells but not in somatic cells. Therefore, replicative senescence as a consequence of telomere shortening is inevitable during cell proliferation. Over time, uncompensated telomere shortening subsequently leads to the loss of genomic integrity by chromosome rearrangement, cell cycle arrest, and apoptosis. Telomere attrition to a critical and threshold length can limit the proliferative capacity of cells. So, mean telomere length has emerged as a replicative clock and consequently as a biomarker for biological aging (Bekaert et al., 2005, Blackburn et al., 2015).
3.1. Telomere dynamics and immune response
Survival in human populations is intimately related to immune system efficiency which protects the host against infections and malignancies. Recent research suggests that immunosenescence is not the only result of chronological aging, but rather it can even occur in young adults as the result of environmental and lifestyle factors (Gruver et al., 2007, Ongrádi and Kövesdi, 2010). Both the capacity to respond to antigens and maintenance of immunologic memory are altered in immunosenescence (Pawelec, 2012). So, the efficiency of immune response can be the main factor responsible for the clinical variations of diseases, especially cancer and infections.
Cellular proliferation is a fundamental feature of an effective immune response. The essential role of telomere length in cell proliferation and enormous proliferative demand in cells of the immune system leads to the vital interaction between the telomere dynamics and immune system efficiency (Effros, 2011). Therefore, telomere length can be an informative parameter for predicting the proliferative history and replicative reserve.
Considerable inter-individual variation in leukocyte telomere length in aged-matched comparison revealed that it is a highly heritable trait in humans (Hjelmborg et al., 2015). A number of cross-sectional analyses show that telomere shortening occurs in blood leukocytes of both myeloid and lymphoid lineages with age. Shortening of telomeres has been reported during naïve T cell differentiation to memory T cells, CD28 + to CD28 − CD8 T cell differentiation, and in long-term cultured T cells (Effros et al., 1994, Monteiro et al., 1996, Rufer et al., 1999; N. P. Weng et al., 1995). Comparison of leukocyte telomere length in patients with some common autoimmune syndromes and viral infections with age-matched healthy controls reveal that patients have significantly shorter telomeres (Georgin-Lavialle et al., 2010, Helby et al., 2017). Progressive telomere shortening of T cells has been observed in patients with viral infections including, HIV, HBV, HCV, EBV, and CMV. The extensive proliferation of T cells in response to viral infections is associated with telomere shortening as well as the replicative impairment and decreased immunocompetence (Bellon and Nicot, 2017, Plunkett et al., 2001, van Baarle et al., 2008, van de Berg et al., 2010, Zanet et al., 2014). Susceptibility to viral infection with respect to telomere length is not new idea. A prospective study of 75,309 individuals from the general population for up to 23 years revealed that shorter leukocyte telomere length was associated with higher risk of hospitalization due to any infection and pneumonia (Helby et al., 2017). A significant association has been reported between CD8CD28 − cell telomere length and experimentally induced upper respiratory viral infection in healthy adults (Cohen et al., 2013).
Collectively, leukocyte telomere length will affect the immunocompetence, either with normal aging or in defined pathologic conditions. So, the inevitable consequence of this complex scenario can be the inadequate immune response against newly encountered antigens. So, telomere length is positively associated with the efficiency of immune response by affecting the rapid expansion of immune cells towards fight infection or disease.
3.2. Telomere and COVID-19
Lymphopenia has been identified as a reliable predictive and prognostic marker for patients with COVID-19. Indeed, COVID-19 associated lymphopenia is marked by a low count of CD4/CD8 T cells, but not B cells. The current result proposed that lymphopenia at the initial presentation of COVID-19 is linked with poor prognosis in patients. T cells play a critical role in orchestrating responses in viral infections so, a decrease in T cells is significantly associated with increases in inflammation indicators (Diao et al., 2020, Huang and Pranata, 2020, Tan et al., 2020). Survival, proliferation, and maturation of lymphocytes in bone marrow, thymus, and secondary lymphoid organs add more complexity to the kinetics of lymphopoiesis in humans. However, lymphopoiesis is tightly coupled with telomere length. Shorter telomere length as a result of aging or inter-individual differences can induce slow proliferation of lymphocytes (Patrick & Weng, 2019; N. P. Weng et al., 1997). Drastic reduction of the T-cell count, lymphopenia, is shown to be a hallmark of severe COVID-19. Rapid expansion and recovery of the T-cell pool are essential for inducing an adequate immune response and recovery from COVID-19. Analysis of telomere length among elderly patients with COVID-19 reveal that comparatively short PBMC telomeres are associated with low lymphocyte count in older persons infected with SARS-CoV-2. T-cell proliferative capacity is telomere length-dependent, and telomeres shorten with age. So, these findings might explain the susceptibility to severe COVID-19 in older age (Benetos et al., 2021).
According to our knowledge, gender-related factors might affect COVID‑19 vulnerability, severity, and lethality. The higher capability of plasmacytoid dendritic cells to produce the antiviral response in the context of type 1 interferon (IFN-1), higher antibody response, less mitochondrial DNA instability and increased telomere length in females compared to males may explain the gender difference in response to viral infections such as SARS-CoV-2. So, it seems that the reduction of telomeric DNA reservoir in elderly male subjects may play a pivotal role in COVID-19 poor clinical prognosis (Storci et al., 2021).
Based on the above evidence, in a simplified hypothesis, shorter telomere length in human lymphocytes maybe drive susceptibility to lymphopenia and so more severe and potentially lethal SARS-CoV-2 infection (Aviv, 2020, Tsilingiris et al., 2020).
According to the recent studies, researchers hypothesize that individuals with short telomeres would have a suboptimal antiviral response upon SARS-CoV-2 infection, hence leading to more severe and progressive disease. Evaluation of different gene expression patterns in the human lung tissues of non-smokers, smokers, and obstructive pulmonary disease/ idiopathic pulmonary fibrosis (COPD/IPF) patients provides a novel direction showing a crucial and interdependent association between telomere replication/maintenance pathway and SARS-CoV-2 COVID-19 proteins. In this study, age-related gene expression alterations in cellular pathways such as telomere replication/maintenance pathway in COPD and IPF showed a significant association with the SARS-CoV-2 ACE2-TMPRSS2-Furin-DPP4 axis. These findings could open new insights into the identification of the pathogenesis process and pharmacological targets for COVID-19 (Maremanda et al., 2020).
In a prospective study, telomere length was compared by Flow-FISH in hospitalized COVID-19 patients and healthy volunteers. Compared to healthy controls, there was a significantly higher proportion of patients with short telomeres (<10th percentile) in the COVID-19 patients. Also, a significant correlation between telomere length biological factors such as C-reactive protein level (CRP), lymphocyte count, and neutrophil-to-lymphocyte ratio (NLR) was reported (Froidure et al., 2020). Analysis of telomere length in peripheral blood lymphocytes in COVID-19 patients in Spain revealed that telomeres were shorter in the groups with severe-acute severity compared to mild-moderate severity. Statistical analysis of telomere length of patients in different age groups demonstrated that the association between COVID-19 severity and short telomeres is independent of age (Sanchez-Vazquez et al., 2021).
A prospective cohort study of the survivors hospitalized with severe COVID-19 introduced the leucocyte telomere length as an independent risk factor for post-COVID lung fibrosis. A four-month follow-up of patients revealed that short blood leucocyte telomere lengths can contribute to the development of post-COVID lung fibrosis (McGroder et al., 2021). Technical and sampling details of the relevant literature papers cited in the present manuscript about the role of telomere length in COVID-19 are summarized in Table 1 .
Table 1.
Technical and sampling details of the relevant studies about the possible role of the telomere length in COVID-19 pathogenies.
| Aim of study | Sample Type | Sample Size | Method(s) | Reference |
|---|---|---|---|---|
| Analysis of association between telomere length and lymphocyte count in COVID-19 patients | Peripheral Blood | 17 COVID-19 patients, 21 non-COVID-19 patients |
TeSLA, SB |
(Benetos et al., 2021) |
| Age-Dependent Assessment of gene expression in crucial pathways of cellular maintenance |
Lung Tissue | 48 patients/controls * | qPCR | (Maremanda et al., 2020) |
| Analysis of association between telomere length and the risk of severe COVID-19 | Peripheral Blood | 70 COVID-19 patients , 491 control |
Flow-FISH | (Froidure et al., 2020) |
| Analysis of association between telomere length and COVID-19 severity | Peripheral Blood | 89 COVID-19 patients | qPCR | (Sanchez-Vazquez et al., 2021) |
| Investigation of biological risk for Pulmonary fibrosis in COVID-19 survivors | Peripheral Blood | 76 COVID-19 survivors | qPCR | (McGroder et al., 2021) |
TeSLA, Telomere Shortest Length Assay; SB, Southern Blotting; qPCR, quantitative Polymerase Chain Reaction, FISH, Fluorescent In Situ Hybridization.
*Including non-smokers, smokers, and patients with COPD and IPF.
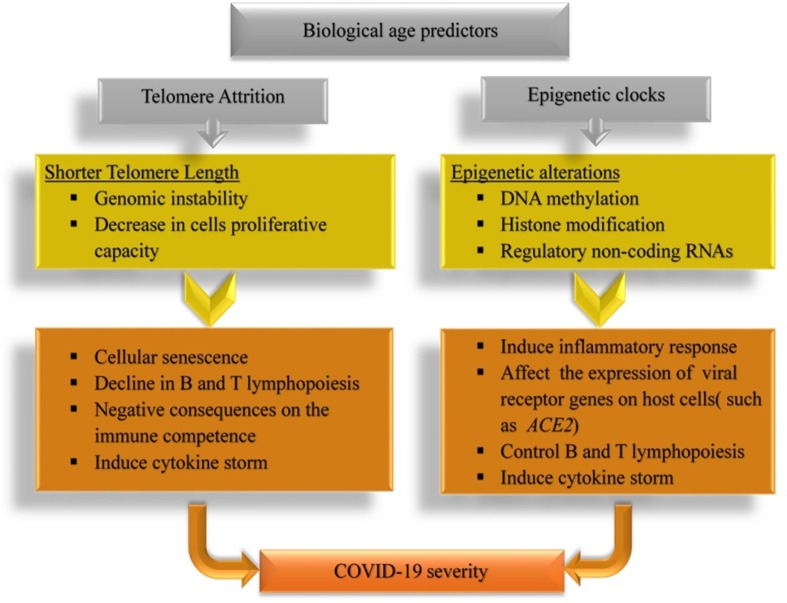
Altogether, recent findings suggest that the leucocyte telomere length, as a marker of biological age, can influence the severity of COVID-19. It seems that individuals who exhibit lymphocyte telomere shortening may be more susceptible to severe and potentially lethal SARS-CoV-2 infection. The association between the shorter telomere length and more severe clinical manifestation of COVID-19 are summarized in Fig. 1 . As short telomeres can be elongated by telomerase, new treatment strategies, such as telomerase activation-based therapies can introduce a new approach for ameliorating the deleterious complications of COVID-19.
Fig. 1.
Biological aging predictors and COVID-19. The association between shorter telomere length (right), and epigenetic clocks (left) with COVID-19 Severity.
4. Epigenetic alterations
According to a consensus definition in biology, an epigenetic trait is described as a 'stably heritable phenotype resulting from changes in a chromosome without alterations in the DNA sequence' (Berger et al., 2009). Current evidence indicates that several environmental and lifestyle factors such as nutrition, behavior, physical activity, smoking, alcohol consumption, inflammation, environmental pollutants, working habits, and occupational exposure can modify epigenetic patterns (Baccarelli & Bollati, 2009).
Epigenetic modifications are reversible and transient alterations that included DNA methylation, histone modification, and non-coding RNAs. DNA methylation is one of the main epigenetic modifications that affect the chromatin structure. CpG sites or the CpG islands are the primary targets for DNA methylation in mammalian genomes. Proximal promoters of many genes in the human genome are associated with CpG islands. CpG-island methylation by DNA methyltransferases (MTase) influences chromatin condensation by assembling methyl-binding proteins. So in general, CpG-island methylation in promoter regions is associated with gene silencing. Core histone proteins undergo major modifications such as acetylation, methylation, phosphorylation, and ubiquitination that directly or indirectly influence chromatin architecture. A large number of possible histone modifications cause an extra level of complexity for the tight control of chromatin structure. However, post-translational modification of histone proteins is a powerful epigenetic mechanism that can affect chromatin structure. Non-coding RNAs are functionally relevant RNA molecules that do not encode functional proteins. There is a wide range of non-coding RNAs such as transfer RNAs (tRNAs), ribosomal RNAs (rRNAs), microRNAs (miRNAs), and short-interfering RNAs (siRNAs). Non-coding RNAs can act as the epigenetic modifiers by interacting with DNA, other RNAs, and proteins (Dupont et al., 2009).
In general, epigenetic mechanisms act as control systems for modulating the expression regulation without affecting the actual DNA sequence of the organism. The gene expression profile in human cells mainly can be controlled by epigenetic mechanisms.
4.1. Epigenetic and immune response
A natural and effective immune system must be able to discriminate various antigens, including self from non-self and harmless non-self from dangerous non-self. Changes in tissue homeostasis as the result of invading pathogens trigger the cell's heterogeneity, and phenotypic plasticity in the innate immune system and the adaptive immune response relies on the highly diverse antigen receptor repertoire of lymphocytes. Several molecular processes such as the reprogramming of hematopoietic, random rearrangement of antigen-receptor genes, allelic exclusion, and immunologic memory guarantee the remarkable heterogeneity of immune cells and their ability to respond to pathogen challenges. This characteristic of immune cells justifies the existence of mechanisms that increase the diversification of the individual cells and maintaining the integrity of the immune system. Current evidence highlight the fundamental role of epigenetic mechanisms in regulating the immune response in both innate and adaptive immunity, as well as the regulation of the cell-fate decision and maintenance of immune cell lineages.
Epigenetic mechanisms play an important role in the control of lymphopoiesis and immunity. Early lymphopoiesis is controlled by signaling pathways for instance, Notch1, and its downstream transcription factors provide a key regulatory signal for the formation of the earliest T-cell precursors (Pui et al., 1999). Loss of function in epigenetic modifying enzymes such as MTases and histone deacetylases (HDACs) highlight the importance of epigenetic mechanisms in cell-fate decisions, cell lineage stability and functions of various immune cell subsets (Bezu et al., 2019). Also, recent genome-wide epigenetic analyses have identified immune cell-type-specific epigenetic signatures which determine the critical role of epigenetic mechanisms in the maintenance of immune homeostasis (Rahmani et al., 2019).
The large body of evidence confirms the central role of epigenetic mechanisms in the pathophysiology of autoimmune conditions. Epigenetic profiles are established during embryonic development and stably inherited to daughter cells during mitosis. So, it seems that the accumulation of epigenetic alterations could induce the inflammatory response by altering gene expression profiles (Obata et al., 2015). The pathophysiology of autoimmune diseases is closely linked to the combination of genetic susceptibility and epigenetic modifications. The advent of high-throughput epigenetics technologies is provided new insights into the pathogenesis of autoimmune diseases. Many studies over the last decade have shown that the importance of epigenetic mechanisms during the development of the most prevalent autoimmune diseases, such as systemic lupus erythematosus (SLE), rheumatoid arthritis (RA), multiple sclerosis (MS), Sjogren’s syndrome (SS), autoimmune thyroid diseases (AITD), and type 1 diabetes (T1D). Alterations in the DNA methylation and histone modification pattern, and also the abnormal expression of miRNAs can lead to aberrant gene expression, and consequently the onset of autoimmune diseases in humans (Mazzone et al., 2019). Investigation of genomic and gene-specific DNA methylation levels in patients with autoimmune diseases revealed that DNA methylation levels are altered in CD4 + T cells. Consequently, transcriptional dysregulation in several autoimmune-related genes may contribute to autoimmune disease onset and progression (Z. Wang et al., 2017).
Numerous ways guarantee and facilitate the viral hijacking of host cellular machinery for the completion of their life cycle. The heritable nature of epigenetic signatures indicates that they could play important roles in the viral hijacking of host cellular processes. Virus-induced overexpression of MTase has been reported in diverse viruses such as KSHV, EBV, HBV, HIV-1, HTLV-1, HPV, SV40, and adenoviruses. At least one virus, PBCV-1, and perhaps many bacteria, including C. pneumoniae, target the chromatin architecture of its host cell by methylation of various histone and non-histone proteins (Paschos & Allday, 2010). Generally, it seems that viruses and bacteria can manipulate epigenetic processes to complete their life cycle and influence host responses associated with immunity and inflammation.
4.2. Epigenetic and COVID-19
Epigenetic mechanisms play a crucial role in the maintenance of cellular homeostasis and genome integrity throughout viral infections. Epigenetic changes act as silent modulators of the immune system that can direct both the production of chemical mediators of inflammation and control the magnitude of the host response by regulating the chromatin structure. Several studies suggest that multiple layers of epigenetic mechanisms, including DNA methylation, histone modifications, and non-coding RNA-mediated regulatory events, are involved in the adaptation of the immune system, which determines the outcome of the host-pathogen interactions during SARS-CoV-2 infection (Chlamydas et al., 2020, Pruimboom, 2020, Sen et al., 2021). Based on our current knowledge, epigenetic signatures play a critical role in the crosstalk between SARS-CoV-2 and the host cell. Hence, elucidation of the epigenetic landscape can facilitate the identification of the diagnostic biomarkers and therapeutic targets to combat COVID-19.
An integrated analysis of the data from single-cell assay for transposase-accessible chromatin (scATAC) and single-cell RNA (scRNA) sequencing revealed that chromatin epigenetic status and transcriptomic immune profiles of T cells are functionally relevant to the pathogenesis stage of COVID-19 (S. Li et al., 2021). Distinct patterns of DNA methylation were confirmed using genome-wide DNA methylation analysis of patients with severe COVID-19. Hypermethylation of IFN related genes and hypomethylation of inflammatory genes, as well as epigenetic clock analyses, revealed that alteration in epigenetic architecture can affect the severity and mortality of COVID-19. So, elucidation of the exact epigenetic signatures of COVID-19 can be useful for the understanding of molecular mechanisms of pathogenicity and potential therapeutic targets against SARS-CoV-2 (M. J. Corley et al., 2021).
Early reports after the pandemic outbreak revealed that angiotensin receptor 1 (AT1R) blockers could reduce the aggressiveness and mortality of SARS-CoV-2 infections (Sun et al., 2020). SARS-CoV-2, similar to the other coronavirus strains, uses the host receptor angiotensin-converting enzyme 2 (ACE2) on the surface of the target cell for entry inside the host (Wan et al., 2020). Leung and colleges showed that upregulated expression of ACE2 in lung tissue of active cigarette smoking and COPD patients may explain the increased risk of severe COVID-19 in these populations (Leung et al., 2020). Previous findings demonstrated that the expression level of ACE2 is controlled by the methylation pattern of the several CpG islands in the promoter of ACE2 (Zill et al., 2012). DNA methylation profiling of the ACE2 gene showed that the methylation rate in lung epithelial cells is the lowest compared with the other tissues. There was also evidence that age and gender differences can affect the DNA methylation pattern of ACE2 gene (M. Corley & Ndhlovu, 2020). Analysis of 700 lung transcriptome samples of patients with comorbidities and suffering from severe COVID-19 revealed that ACE2 was highly expressed in these patients compared to control individuals. Correlation and network analyses also offered new insight into the epigenetic regulation of ACE2 gene in the lung (Pinto et al., 2020). The recent In silico analysis of ACE2 expression pattern suggests that epigenetic dysregulation might cause the overexpression of ACE2 in the lung tissue. It seems that overexpression of ACE2 facilitates the viral entry to host cells, which might be the cause of the excessive immune response to SARS-CoV-2 and severe form of COVID-19 (Sang et al., 2021, Sawalha et al., 2020).
Non-coding miRNAs are involved in cytokine modulation in the immune system, so miRNAs might have deeply inhibitory effects on cytokine storm and represent a novel and emerging targets for therapeutic intervention against SARS-CoV-2 (Gasparello et al., 2021, Hum et al., 2021). Baldassarre and colleges analyzed the common structural features of 5′ untranslated regions (5′UTRs) of SARS-CoV-2 genome. Their results highlighted the potential importance of non-coding RNAs for the optimization of RNA-based drugs in the development of novel therapeutic strategies (Baldassarre et al., 2020). Computational analysis of the host miRNA binding profiles of different SARS-CoV-2 genomes from different geographical locations suggested that miRNAs of host and SARS-CoV-2 can indeed play roles as the pivotal epigenetic modulator. Also, disease severity, symptoms, and mortality rate can be influenced by miRNA clusters of host and virus amongst the COVID-19 patients (Khan et al., 2020). Evaluation of the levels of the circulating inflammatory miRNAs in COVID-19 patients who did not respond to the anti-IL-6 receptor drug Tocilizumab (TCZ) showed that serum levels of miR-146a-5p were significantly lower in COVID-19 patients after the treatment compared to age- and gender-matched healthy control subjects. Therefore, identification of blood-based biomarkers, such as miRNAs, can allow for a better understanding of molecular pathways involved in the pathogenesis and treatment of SARS-CoV-2 (Sabbatinelli et al., 2021). High-throughput sequencing of miRNAs in peripheral blood of patients with COVID-19 and healthy controls revealed the dysregulated expression of miRNAs in the peripheral blood of patients. These findings suggested that differential expression of miRNAs can also open new insights into the potential targets for novel treatments (C. Li et al., 2020). Also, altered expression levels of circulating miRNAs in hospitalized patients proposed that circulating miRNA profiles are associated with the severity of COVID-19 (de Gonzalo-Calvo et al., 2021). Table 2 summarized the technical and sampling details of the relevant studies about the role of epigenetic mechanisms in COVID-19.
Table 2.
Technical and sampling details of the relevant studies about the possible role of the epigenetic mechanisms in COVID-19 pathogenies.
| Aim of study | Sample Type | Sample Size | Method(s) | Reference |
|---|---|---|---|---|
| Genome-wide DNA methylation profiling of peripheral blood of patients with severe COVID-19 | Peripheral Blood | 41 patients/controls * |
Infinium MethylationEPIC array, ddPCR |
(M. J. Corley et al., 2021) |
| Expression analysis of ACE2 gene , as the entry receptor for SARS-CoV-2, in small airway epithelia of smokers and COPD patients | Lung Tissue | 27 patients/controls ¶ |
RNA Sequencing | (Leung et al., 2020) |
| Analysis the plasma level of circulating inflamma-miRs in COVID-19 patients treated with TCZ | Serum | 30 COVID-19 patients, 29 controls |
RT-PCR ddPCR |
(Sabbatinelli et al., 2021) |
| Analysis of miRNAs expression in the peripheral blood from human patients with COVID‐19 | Peripheral Blood | 10 COVID-19 patients, 4 controls |
RNA Sequencing | (C. Li et al., 2020) |
| Analysis the circulating miRNAs profile of hospitalized COVID-19 patients | Peripheral Blood | 84 COVID-19 patients | RT-qPCR | (de Gonzalo-Calvo et al., 2021) |
ddPCR, droplet digital Polymerase Chain Reaction; TCZ, Tocilizumab; RT-PCR, Reverse Transcription Polymerase Chain Reaction PCR; RT-qPCR, Reverse Transcription- quantitative Polymerase Chain Reaction.
* Including 9 severe COVID-19 patients, 9 uninfected controls, 5 hospitalized influenza, 9 untreated primary HIV infection and 9 coinfected with mild/moderate COVID-19 and HIV-1.
¶ Including 10 current smokers with COPD, 9 nonsmoker controls and 8 healthy current smokers.
Finally, our growing knowledge of COVID-19 gives a clear insight into the importance of epigenetic mechanisms in the pathogenesis of SARS-CoV-2. Epigenetic regulation mechanisms in COVID-19 severity are illustrated in Fig. 1. The epigenetic contribution in many aspects of the SARS-CoV-2 infection, including the viral life cycle and the host immune response, highlights that more studies are needed to evaluate the role of epigenetic alterations in the pathophysiology of COVID-19. A deeper understanding of the underlying mechanisms could provide critical clues about the variable clinical manifestations and improve treatment options for SARS-CoV-2.
5. Conclusion
Heterogeneous clinical manifestations of SARS-CoV-2 infection are one of the biggest challenges in COVID-19. At a glance, inter-individual variation determines the way people respond to the disease. Scientifics suggest that telomere attrition and epigenetic alterations as the main predictors of biological aging and cellular senescence (Jylhävä et al., 2017) might influence the severity of COVID-19. There is a direct correlation between telomere length and the proliferative potential of cells (Notaro et al., 1997). Significant reduction of T cells count is a hallmark of severe COVID-19 (Diao et al., 2020). Current research suggests that short telomere is associated with a higher risk of developing severe COVID-19 (Froidure et al., 2020, Sanchez-Vazquez et al., 2021). Also, recent findings demonstrated that SARS-CoV-2 likely alters the epigenetic architecture in cell-type and context-dependent fashion. Differential DNA methylation pattern and miRNA expression profiles in the peripheral blood from patients with COVID-19 and their relationships with transcriptome highlights the critical role of epigenetic mechanisms in the pathophysiology of COVID-19 (M. J. Corley et al., 2021, Gasparello et al., 2021). This review indicates that the detailed role of telomere length and epigenetic alterations in the severity of COVID-19 is still unclear. To address the gaps in current knowledge, more comprehensive studies on larger groups of the population and robust epidemiological studies population are needed to determine the role of telomere length and epigenetic mechanisms in COVID-19 severity. Understanding the exact mechanisms of COVID-19 pathophysiology can help determine the future framework against this worldwide health threat. Identification of the high-risk patient and personalized therapeutic intervention, such as telomerase activation-based therapies and epidrugs, can provide further invaluable insights into the controlling of COVID-19 outbreak.
Funding Sources
This work was supported by Tabriz University of Medical Sciences [grant code 65846].
CRediT authorship contribution statement
Ata Mahmoodpoor: Project administration, Conceptualization, Funding acquisition, Writing – review & editing. Sarvin Sanaie: Supervision, Validation, Writing – review & editing. Faranak Roudbari: Writing – review & editing. Tara Sabzevari: Writing – review & editing. Nasim Sohrabifar: Writing – review & editing. Somayeh Kazeminasab: Conceptualization, Investigation, Validation, Writing – original draft.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Edited by Andre van Wijnen
References
- Aviv A. Telomeres and COVID-19. FASEB Journal. 2020;34(6):7247–7252. doi: 10.1096/fsb2.v34610.1096/fj.202001025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baccarelli A., Bollati V. Epigenetics and environmental chemicals. Current Opinion in Pediatrics. 2009;21(2):243–251. doi: 10.1097/MOP.0b013e32832925cc. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldassarre A., Paolini A., Bruno S.P., Felli C., Tozzi A.E., Masotti A. Potential use of noncoding RNAs and innovative therapeutic strategies to target the 5’UTR of SARS-CoV-2. Epigenomics. 2020;12(15):1349–1361. doi: 10.2217/epi-2020-0162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayarsaihan D. Epigenetic mechanisms in inflammation. Journal of Dental Research. 2011;90(1):9–17. doi: 10.1177/0022034510378683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bekaert S., De Meyer T., Van Oostveldt P. Telomere attrition as ageing biomarker. Anticancer Research. 2005;25(4):3011–3022. http://www.ncbi.nlm.nih.gov/pubmed/16080560 [PubMed] [Google Scholar]
- Bellon M., Nicot C. Telomere dynamics in immune senescence and exhaustion triggered by chronic viral infection. Viruses. 2017;9(10):289. doi: 10.3390/v9100289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benetos, A., Lai, T.-P., Toupance, S., Labat, C., Verhulst, S., Gautier, S., Ungeheuer, M.-N., Perret-Guillaume, C., Levy, D., Susser, E., & Aviv, A. 2021. The Nexus Between Telomere Length and Lymphocyte Count in Seniors Hospitalized With COVID-19. The Journals of Gerontology: Series A. https://doi.org/10.1093/gerona/glab026. [DOI] [PMC free article] [PubMed]
- Berger S.L., Kouzarides T., Shiekhattar R., Shilatifard A. An operational definition of epigenetics. Genes and Development. 2009;23(7):781–783. doi: 10.1101/gad.1787609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bezu L., Wu Chuang A., Liu P., Kroemer G., Kepp O. Immunological effects of epigenetic modifiers. Cancers. 2019;11(12):1911. doi: 10.3390/cancers11121911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackburn E.H. Structure and function of telomeres. Nature. 1991;350(6319):569–573. doi: 10.1038/350569a0. [DOI] [PubMed] [Google Scholar]
- Blackburn E.H., Epel E.S., Lin J. Human telomere biology: A contributory and interactive factor in aging, disease risks, and protection. Science. 2015;350(6265):1193–1198. doi: 10.1126/science.aab3389. [DOI] [PubMed] [Google Scholar]
- Chlamydas S., Papavassiliou A.G., Piperi C. Epigenetic mechanisms regulating COVID-19 infection. Epigenetics. 2020;16(3):263–270. doi: 10.1080/15592294.2020.1796896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen S., Janicki-Deverts D., Turner R.B., Casselbrant M.L., Li-Korotky H.S., Epel E.S., Doyle W.J. Association between telomere length and experimentally induced upper respiratory viral infection in healthy adults. JAMA - Journal of the American Medical Association. 2013;309(7):699–705. doi: 10.1001/jama.2013.613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corley M.J., Pang A.P.S., Dody K., Mudd P.A., Patterson B.K., Seethamraju H., Bram Y., Peluso M.J., Torres L., Iyer N.S., Premeaux T.A., Yeung S.T., Chandar V., Borczuk A., Schwartz R.E., Henrich T.J., Deeks S.G., Sacha J.B., Ndhlovu L.C. Genome-wide DNA methylation profiling of peripheral blood reveals an epigenetic signature associated with severe COVID-19. Journal of Leukocyte Biology, JLB.5HI0720-466R. 2021;110(1):21–26. doi: 10.1002/jlb.v110.110.1002/JLB.5HI0720-466R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corley, M., & Ndhlovu, L. 2020. DNA Methylation Analysis of the COVID-19 host cell receptor, Angiotensin I Converting Enzyme 2 gene (ACE2) in the Respiratory System Reveal Age and Gender Differences. Medicine and Pharmacology, March, 19, 1–29. https://doi.org/10.20944/preprints202003.0295.v1.
- Criqui M., Qamra A., Chu T.W., Sharma M., Tsao J., Henry D.A., Barsyte-Lovejoy D., Arrowsmith C.H., Winegarden N., Lupien M., Harrington L. Telomere dysfunction cooperates with epigenetic alterations to impair murine embryonic stem cell fate commitment. ELife. 2020;9 doi: 10.7554/eLife.47333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Gonzalo-Calvo, D., Benítez, I. D., Pinilla, L., Carratalá, A., Moncusí-Moix, A., Gort-Paniello, C., Molinero, M., González, J., Torres, G., Bernal, M., Pico, S., Almansa, R., Jorge, N., Ortega, A., Bustamante-Munguira, E., Gómez, J. M., González-Rivera, M., Micheloud, D., Ryan, P., … Barbé, F. 2021. Circulating microRNA profiles predict the severity of COVID-19 in hospitalized patients. Translational Research. https://doi.org/10.1016/j.trsl.2021.05.004. [DOI] [PMC free article] [PubMed]
- Diao B., Wang C., Tan Y., Chen X., Liu Y., Ning L., Chen L., Li M., Liu Y., Wang G., Yuan Z., Feng Z., Zhang Y., Wu Y., Chen Y. Reduction and Functional Exhaustion of T Cells in Patients With Coronavirus Disease 2019 (COVID-19) Frontiers in Immunology. 2020;11(827) doi: 10.3389/fimmu.2020.00827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupont C., Armant D.R., Brenner C.A. Epigenetics: Definition, mechanisms and clinical perspective. Seminars in Reproductive Medicine. 2009;27(5):351–357. doi: 10.1055/s-0029-1237423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Effros R.B. Telomere/telomerase dynamics within the human immune system: Effect of chronic infection and stress. Experimental Gerontology. 2011;46(2–3):135–140. doi: 10.1016/j.exger.2010.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Effros R.B., Boucher N., Porter V., Zhu X., Spaulding C., Walford R.L., Kronenberg M., Cohen D., Schächter F. Decline in CD28+ T cells in centenarians and in long-term T cell cultures: A possible cause for both in vivo and in vitro immunosenescence. Experimental Gerontology. 1994;29(6):601–609. doi: 10.1016/0531-5565(94)90073-6. [DOI] [PubMed] [Google Scholar]
- Froidure A., Mahieu M., Hoton D., Laterre P.F., Yombi J.C., Koenig S., Ghaye B., Defour J.P., Decottignies A. Short telomeres increase the risk of severe COVID-19. Aging. 2020;12(20):19911–19922. doi: 10.18632/aging.104097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasparello J., Finotti A., Gambari R. Tackling the COVID-19 “cytokine storm” with microRNA mimics directly targeting the 3’UTR of pro-inflammatory mRNAs. Medical Hypotheses. 2021;146:110415. doi: 10.1016/j.mehy.2020.110415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgin-Lavialle S., Aouba A., Mouthon L., Londono-Vallejo J.A., Lepelletier Y., Gabet A.S., Hermine O. The telomere/telomerase system in autoimmune and systemic immune-mediated diseases. Autoimmunity Reviews. 2010;9(10):646–651. doi: 10.1016/j.autrev.2010.04.004. [DOI] [PubMed] [Google Scholar]
- Gruver A.L., Hudson L.L., Sempowski G.D. Immunosenescence of ageing. Journal of Pathology. 2007;211(2):144–156. doi: 10.1002/(ISSN)1096-989610.1002/path.v211:210.1002/path.2104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasan, S. S., Capstick, T., Ahmed, R., Kow, C. S., Mazhar, F., Merchant, H. a., & Zaidi, S. T. R. (2020). Mortality in COVID-19 patients with acute respiratory distress syndrome and corticosteroids use: a systematic review and meta-analysis. Expert Review of Respiratory Medicine, 14(11), 1149–1163. https://doi.org/10.1080/17476348.2020.1804365. [DOI] [PMC free article] [PubMed]
- Helby J., Nordestgaard B.G., Benfield T., Bojesen S.E. Shorter leukocyte telomere length is associated with higher risk of infections: A prospective study of 75,309 individuals from the general population. Haematologica. 2017;102(8):1457–1465. doi: 10.3324/haematol.2016.161943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hjelmborg J.B., Dalgård C., Möller S., Steenstrup T., Kimura M., Christensen K., Kyvik K.O., Aviv A. The heritability of leucocyte telomere length dynamics. Journal of Medical Genetics. 2015;52(5):297–302. doi: 10.1136/jmedgenet-2014-102736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Biying, Huang Shaoying, Yin Lianghong. The cytokine storm and COVID-19. Journal of Medical Virology. 2021;93(1):250–256. doi: 10.1002/jmv.v93.110.1002/jmv.26232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang I., Pranata R. Lymphopenia in severe coronavirus disease-2019 (COVID-19): Systematic review and meta-analysis. Journal of Intensive Care. 2020;8(1):36. doi: 10.1186/s40560-020-00453-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hue S., Beldi-Ferchiou A., Bendib I., Surenaud M., Fourati S., Frapard T., Rivoal S., Razazi K., Carteaux G., Delfau-Larue M.H., Mekontso-Dessap A., Audureau E., de Prost N. Uncontrolled Innate and Impaired Adaptive Immune Responses in Patients with COVID-19 Acute Respiratory Distress Syndrome. American Journal of Respiratory and Critical Care Medicine. 2020;202(11):1509–1519. doi: 10.1164/rccm.202005-1885OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hum C., Loiselle J., Ahmed N., Shaw T.A., Toudic C., Pezacki J.P. MicroRNA Mimics or Inhibitors as Antiviral Therapeutic Approaches Against COVID-19. Drugs. 2021;81(5):517–531. doi: 10.1007/s40265-021-01474-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jylhävä Juulia, Pedersen Nancy L., Hägg Sara. Biological Age Predictors. EBioMedicine. 2017;21:29–36. doi: 10.1016/j.ebiom.2017.03.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan, M. A.-A.-K., Sany, M. R. U., Islam, M. S., & Islam, A. B. M. M. K. (2020). Epigenetic Regulator miRNA Pattern Differences Among SARS-CoV, SARS-CoV-2, and SARS-CoV-2 World-Wide Isolates Delineated the Mystery Behind the Epic Pathogenicity and Distinct Clinical Characteristics of Pandemic COVID-19. Frontiers in Genetics, 11. https://doi.org/10.3389/fgene.2020.00765. [DOI] [PMC free article] [PubMed]
- Leung Janice M., Yang Chen X., Tam Anthony, Shaipanich Tawimas, Hackett Tillie-Louise, Singhera Gurpreet K., Dorscheid Delbert R., Sin Don D. ACE-2 expression in the small airway epithelia of smokers and COPD patients: Implications for COVID-19. European Respiratory Journal. 2020;55(5):2000688. doi: 10.1183/13993003.00688-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, C., Hu, X., Li, L., & Li, J. hui. 2020. Differential microRNA expression in the peripheral blood from human patients with COVID-19. Journal of Clinical Laboratory Analysis, 34(10). https://doi.org/10.1002/jcla.23590. [DOI] [PMC free article] [PubMed]
- Li Shun, Wu Bin, Ling Yun, Guo Mingquan, Qin Boyin, Ren Xiaonan, Wang Chao, Yang Hua, Chen Lixiang, Liao Yixin, Liu Yang, Peng Xiuhua, Xu Chunhua, Wang Zhenyan, Shen Yinzhong, Chen Jun, Liu Li, Niu Bowen, Zhu Mengmin, Liu Lingling, Li Feng, Zhu Tongyu, Zhu Zhaoqin, Zhou Xiaohui, Lu Hongzhou. Epigenetic Landscapes of Single-Cell Chromatin Accessibility and Transcriptomic Immune Profiles of T Cells in COVID-19 Patients. Frontiers in Immunology. 2021;12 doi: 10.3389/fimmu.2021.625881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Xu S., Yu M., Wang K., Tao Y., Zhou Y., Shi J., Zhou M., Wu B., Yang Z., Zhang C., Yue J., Zhang Z., Renz H., Liu X., Xie J., Xie M., Zhao J. Risk factors for severity and mortality in adult COVID-19 inpatients in Wuhan. Journal of Allergy and Clinical Immunology. 2020;146(1):110–118. doi: 10.1016/j.jaci.2020.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Otín Carlos, Blasco Maria A., Partridge Linda, Serrano Manuel, Kroemer Guido. The hallmarks of aging. Cell. 2013;153(6):1194–1217. doi: 10.1016/j.cell.2013.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowsky D.J., Olshansky S.J., Bhattacharya J., Goldman D.P. Heterogeneity in Healthy Aging. The Journals of Gerontology Series A: Biological Sciences and Medical Sciences. 2014;69(6):640–649. doi: 10.1093/gerona/glt162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malavolta M., Giacconi R., Brunetti D., Provinciali M., Maggi F. Exploring the Relevance of Senotherapeutics for the Current SARS-CoV-2 Emergency and Similar Future Global Health Threats. Cells. 2020;9(4):909. doi: 10.3390/cells9040909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maremanda Krishna P., Sundar Isaac K., Li Dongmei, Rahman Irfan. Age-Dependent Assessment of Genes Involved in Cellular Senescence, Telomere, and Mitochondrial Pathways in Human Lung Tissue of Smokers, COPD, and IPF: Associations With SARS-CoV-2 COVID-19 ACE2-TMPRSS2-Furin-DPP4 Axis. Frontiers in Pharmacology. 2020;11 doi: 10.3389/fphar.2020.584637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazzone R., Zwergel C., Artico M., Taurone S., Ralli M., Greco A., Mai A. The emerging role of epigenetics in human autoimmune disorders. Clinical. Epigenetics. 2019;11(1) doi: 10.1186/s13148-019-0632-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGroder, C. F., Zhang, D., Choudhury, M. A., Salvatore, M. M., D’Souza, B. M., Hoffman, E. A., Wei, Y., Baldwin, M. R., & Garcia, C. K. 2021. Pulmonary fibrosis 4 months after COVID-19 is associated with severity of illness and blood leucocyte telomere length. Thorax, thoraxjnl-2021-217031. https://doi.org/10.1136/thoraxjnl-2021-217031. [DOI] [PMC free article] [PubMed]
- Monteiro, J., Batliwalla, F., Ostrer, H., & Gregersen, P. K. 1996. Shortened telomeres in clonally expanded CD28-CD8+ T cells imply a replicative history that is distinct from their CD28+CD8+ counterparts. Journal of Immunology (Baltimore, Md. : 1950), 156(10), 3587–3590. http://www.ncbi.nlm.nih.gov/pubmed/8621891. [PubMed]
- Notaro R., Cimmino A., Tabarini D., Rotoli B., Luzzatto L. In vivo telomere dynamics of human hematopoietic stem cells. Proceedings of the National Academy of Sciences. 1997;94(25):13782–13785. doi: 10.1073/pnas.94.25.13782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obata Y., Furusawa Y., Hase K. Epigenetic modifications of the immune system in health and disease. Immunology and Cell Biology. 2015;93(3):226–232. doi: 10.1038/icb.2014.114. [DOI] [PubMed] [Google Scholar]
- Ongrádi Joseph, Kövesdi Valéria. Factors that may impact on immunosenescence: An appraisal. Immunity and Ageing. 2010;7(1) doi: 10.1186/1742-4933-7-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paschos K., Allday M.J. Epigenetic reprogramming of host genes in viral and microbial pathogenesis. Trends in Microbiology. 2010;18(10):439–447. doi: 10.1016/j.tim.2010.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patrick, M., & Weng, N. ping. (2019). Expression and regulation of telomerase in human T cell differentiation, activation, aging and diseases. Cellular Immunology, 345, 103989. https://doi.org/10.1016/j.cellimm.2019.103989. [DOI] [PMC free article] [PubMed]
- Pawelec G. Hallmarks of human “immunosenescence” : Adaptation or dysregulation? Immunity and Ageing. 2012;9 doi: 10.1186/1742-4933-9-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto B.G.G., Oliveira A.E.R., Singh Y., Jimenez L., Gonçalves A.N.A., Ogava R.L.T., Creighton R., Peron J.P.S., Nakaya H.I. ACE2 expression is increased in the lungs of patients with comorbidities associated with severe COVID-19. Journal of Infectious Diseases. 2020;222(4):556–563. doi: 10.1093/infdis/jiaa332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plunkett F.J., Soares M.V.D., Annels N., Hislop A., Ivory K., Lowdell M., Salmon M., Rickinson A., Akbar A.N. The flow cytometric analysis of telomere length in antigen-specific CD8+ T cells during acute Epstein-Barr virus infection. Blood. 2001;97(3):700–707. doi: 10.1182/blood.V97.3.700. [DOI] [PubMed] [Google Scholar]
- Pruimboom L. Methylation Pathways and SARS-CoV-2 Lung Infiltration and Cell Membrane-Virus Fusion Are Both Subject to Epigenetics. Frontiers in Cellular and Infection Microbiology. 2020;10 doi: 10.3389/fcimb.2020.00290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pui J.C., Allman D., Xu L., DeRocco S., Karnell F.G., Bakkour S., Lee J.Y., Kadesch T., Hardy R.R., Aster J.C., Pear W.S. Notch1 expression in early lymphopoiesis influences B versus T lineage determination. Immunity. 1999;11(3):299–308. doi: 10.1016/S1074-7613(00)80105-3. [DOI] [PubMed] [Google Scholar]
- Rahmani E., Schweiger R., Rhead B., Criswell L.A., Barcellos L.F., Eskin E., Rosset S., Sankararaman S., Halperin E. Cell-type-specific resolution epigenetics without the need for cell sorting or single-cell biology. Nature. Communications. 2019;10(1) doi: 10.1038/s41467-019-11052-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson Safiya, Hirsch Jamie S., Narasimhan Mangala, Crawford James M., McGinn Thomas, Davidson Karina W., Barnaby Douglas P., Becker Lance B., Chelico John D., Cohen Stuart L., Cookingham Jennifer, Coppa Kevin, Diefenbach Michael A., Dominello Andrew J., Duer-Hefele Joan, Falzon Louise, Gitlin Jordan, Hajizadeh Negin, Harvin Tiffany G., Hirschwerk David A., Kim Eun Ji, Kozel Zachary M., Marrast Lyndonna M., Mogavero Jazmin N., Osorio Gabrielle A., Qiu Michael, Zanos Theodoros P. Presenting Characteristics, Comorbidities, and Outcomes among 5700 Patients Hospitalized with COVID-19 in the New York City Area. JAMA - Journal of the American Medical Association. 2020;323(20):2052. doi: 10.1001/jama.2020.6775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rufer N., Brümmendorf T.H., Kolvraa S., Bischoff C., Christensen K., Wadsworth L., Schulzer M., Lansdorp P.M. Telomere fluorescence measurements in granulocytes and T lymphocyte subsets point to a high turnover of hematopoietic stem cells and memory T cells in early childhood. Journal of Experimental Medicine. 1999;190(2):157–167. doi: 10.1084/jem.190.2.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabbatinelli J., Giuliani A., Matacchione G., Latini S., Laprovitera N., Pomponio G., Ferrarini A., Svegliati Baroni S., Pavani M., Moretti M., Gabrielli A., Procopio A.D., Ferracin M., Bonafè M., Olivieri F. Decreased serum levels of the inflammaging marker miR-146a are associated with non-clinical response to tocilizumab in COVID-19 patients. Mechanisms of Ageing and Development. 2021;193 doi: 10.1016/j.mad.2020.111413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Vazquez R., Guío-Carrión A., Zapatero-Gaviria A., Martínez P., Blasco M.A. Shorter telomere lengths in patients with severe COVID-19 disease. Aging. 2021;13(1):1–15. doi: 10.18632/aging.202463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sang E.R., Tian Y., Miller L.C., Sang Y. Epigenetic evolution of ACE2 and IL-6 genes: Non-canonical interferon-stimulated genes correlate to COVID-19 susceptibility in vertebrates. Genes. 2021;12(2):1–20. doi: 10.3390/genes12020154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawalha Amr H., Zhao Ming, Coit Patrick, Lu Qianjin. Epigenetic dysregulation of ACE2 and interferon-regulated genes might suggest increased COVID-19 susceptibility and severity in lupus patients. Clinical Immunology. 2020;215:108410. doi: 10.1016/j.clim.2020.108410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen R., Garbati M., Bryant K., Lu Y. Epigenetic mechanisms influencing COVID-19. Genome. 2021;64(4):372–385. doi: 10.1139/gen-2020-0135. [DOI] [PubMed] [Google Scholar]
- Sheervalilou Roghayeh, Shirvaliloo Milad, Dadashzadeh Nahid, Shirvalilou Sakine, Shahraki Omolbanin, Pilehvar‐Soltanahmadi Younes, Ghaznavi Habib, Khoei Samideh, Nazarlou Ziba. COVID-19 under spotlight: A close look at the origin, transmission, diagnosis, and treatment of the 2019-nCoV disease. Journal of Cellular Physiology. 2020;235(12):8873–8924. doi: 10.1002/jcp.v235.1210.1002/jcp.29735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storci Gianluca, Bonifazi Francesca, Garagnani Paolo, Olivieri Fabiola, Bonafè Massimiliano. The role of extracellular DNA in COVID-19: Clues from inflamm-aging. Ageing Research Reviews. 2021;66:101234. doi: 10.1016/j.arr.2020.101234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun M.L., Yang J.M., Sun Y.P., Su G.H. [Inhibitors of RAS Might Be a Good Choice for the Therapy of COVID-19 Pneumonia]. Zhonghua Jie He He Hu Xi Za Zhi = Zhonghua Jiehe He Huxi Zazhi = Chinese Journal of Tuberculosis and Respiratory Diseases. 2020;43(3):219–222. doi: 10.3760/cma.j.issn.1001-0939.2020.03.016. [DOI] [PubMed] [Google Scholar]
- Tan L., Wang Q., Zhang D., Ding J., Huang Q., Tang Y.Q., Wang Q., Miao H. Lymphopenia predicts disease severity of COVID-19: a descriptive and predictive study. Signal Transduction and Targeted Therapy. 2020;5(1):1–3. doi: 10.1038/s41392-020-0148-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsilingiris Dimitrios, Tentolouris Anastasios, Eleftheriadou Ioanna, Tentolouris Nikolaos. Telomere length, epidemiology and pathogenesis of severe COVID-19. European Journal of Clinical Investigation. 2020;50(10) doi: 10.1111/eci.v50.1010.1111/eci.13376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Baarle Debbie, Nanlohy Nening M., Otto Sigrid, Plunkett Fiona J., Fletcher Jean M., Akbar Arne N. Progressive telomere shortening of Epstein-Barr virus-specific memory T cells during HIV infection: Contributor to exhaustion? Journal of Infectious Diseases. 2008;198(9):1353–1357. doi: 10.1086/59569610.1086/592170. [DOI] [PubMed] [Google Scholar]
- van de Berg P.J.E.J., Griffiths S.J., Yong S.-L., Macaulay R., Bemelman F.J., Jackson S., Henson S.M., ten Berge I.J.M., Akbar A.N., van Lier R.A.W. Cytomegalovirus Infection Reduces Telomere Length of the Circulating T Cell Pool. The Journal of Immunology. 2010;184(7):3417–3423. doi: 10.4049/jimmunol.0903442. [DOI] [PubMed] [Google Scholar]
- Wan Yushun, Shang Jian, Graham Rachel, Baric Ralph S., Li Fang, Gallagher Tom. Receptor Recognition by the Novel Coronavirus from Wuhan: an Analysis Based on Decade-Long Structural Studies of SARS Coronavirus. Journal of Virology. 2020;94(7) doi: 10.1128/JVI.00127-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C., Horby P.W., Hayden F.G., Gao G.F. A novel coronavirus outbreak of global health concern. The Lancet. 2020;395(10223):470–473. doi: 10.1016/S0140-6736(20)30185-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D., Hu B., Hu C., Zhu F., Liu X., Zhang J., Wang B., Xiang H., Cheng Z., Xiong Y., Zhao Y., Li Y., Wang X., Peng Z. Clinical Characteristics of 138 Hospitalized Patients with 2019 Novel Coronavirus-Infected Pneumonia in Wuhan, China. JAMA - Journal of the American Medical Association. 2020;323(11):1061–1069. doi: 10.1001/jama.2020.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Chang C., Lu Q. Epigenetics of CD4 + T cells in autoimmune diseases. Current Opinion in Rheumatology. 2017;29(4):361–368. doi: 10.1097/BOR.0000000000000393. [DOI] [PubMed] [Google Scholar]
- Waziry R., Gras L., Sedaghat S., Tiemeier H., Weverling G.J., Ghanbari M., Klap J., de Wolf F., Hofman A., Ikram M.A., Goudsmit J. Quantification of biological age as a determinant of age-related diseases in the Rotterdam Study: a structural equation modeling approach. European Journal of Epidemiology. 2019;34(8):793–799. doi: 10.1007/s10654-019-00497-3. [DOI] [PubMed] [Google Scholar]
- Weng N.P., Levine B.L., June C.H., Hodes R.J. Human naive and memory T lymphocytes differ in telomeric length and replicative potential. Proceedings of the National Academy of Sciences of the United States of America. 1995;92(24):11091–11094. doi: 10.1073/pnas.92.24.11091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng Nan-ping, Palmer Larry D., Levine Bruce L, Lane H. Clifford, June Carl H., Hodes Richard J. Tales of tails: Regulation of telomere length and telomerase activity during lymphocyte development, differentiation, activation, and aging. Immunological Reviews. 1997;160(1):43–54. doi: 10.1111/j.1600-065X.1997.tb01026.x. [DOI] [PubMed] [Google Scholar]
- Weng, N. ping. (2006). Aging of the Immune System: How Much Can the Adaptive Immune System Adapt? Immunity, 24(5), 495–499. https://doi.org/10.1016/j.immuni.2006.05.001. [DOI] [PMC free article] [PubMed]
- Zanet D.A.L., Thorne A., Singer J., Maan E.J., Sattha B., Le Campion A., Soudeyns H., Pick N., Murray M., Money D.M., Côté H.C.F. Association between short leukocyte telomere length and HIV infection in a cohort study: No evidence of a relationship with antiretroviral therapy. Clinical Infectious Diseases. 2014;58(9):1322–1332. doi: 10.1093/cid/ciu051. [DOI] [PubMed] [Google Scholar]
- Zill Peter, Baghai Thomas C., Schüle Cornelius, Born Christoph, Früstück Clemens, Büttner Andreas, Eisenmenger Wolfgang, Varallo-Bedarida Gabriella, Rupprecht Rainer, Möller Hans-Jürgen, Bondy Brigitta, Mazza Marianna. Dna methylation analysis of the angiotensin converting enzyme (ACE) gene in major depression. PLoS ONE. 2012;7(7):e40479. doi: 10.1371/journal.pone.0040479. [DOI] [PMC free article] [PubMed] [Google Scholar]