Analyses of ~2000 genomes of Listeria monocytogenes main clinical clone reveal its global spread and dynamics.
Abstract
The bacterial foodborne pathogen Listeria monocytogenes clonal complex 1 (Lm-CC1) is the most prevalent clonal group associated with human listeriosis and is strongly associated with cattle and dairy products. Here, we analyze 2021 isolates collected from 40 countries, covering Lm-CC1 first isolation to present days, to define its evolutionary history and population dynamics. We show that Lm-CC1 spread worldwide from North America following the Industrial Revolution through two waves of expansion, coinciding with the transatlantic livestock trade in the second half of the 19th century and the rapid growth of cattle farming and food industrialization in the 20th century. In sharp contrast to its global spread over the past century, transmission chains are now mostly local, with limited inter- and intra-country spread. This study provides an unprecedented insight into L. monocytogenes phylogeography and population dynamics and highlights the importance of genome analyses for a better control of pathogen transmission.
INTRODUCTION
Listeria monocytogenes (Lm) is a foodborne bacterial zoonotic pathogen that can cause listeriosis, a severe infection with a high case fatality rate in immunocompromised individuals (1, 2). Molecular studies have shown the clonal population structure of Lm (3, 4) and the worldwide distribution of clonal complex 1 (Lm-CC1, initially called epidemic clone ECI) (5, 6), a cosmopolitan clonal group defined by multilocus sequence typing (MLST; fig. S1), which was first isolated from an Italian soldier with meningitis during the first world war (WWI) (7, 8). Notably, Lm-CC1 is the most prevalent clinical clonal complex in several countries (9–15) and actually corresponds to 20% of all of Lm clinical isolates deposited at the National Center for Biotechnology Information (NCBI) (fig. S2). Lm-CC1 belongs to Lm major lineage I and evolved from a subgroup of serotype 4b ancestry (fig. S1) (3, 4).
While there is no proven interhuman horizontal transmission of listeriosis, it was only in 1983 that the foodborne transmission of human listeriosis was formally established (16). Since then, Lm-CC1 has been reported in different food matrices, including dairy products (17, 18), which can be heavily contaminated and constitute a major source of human listeriosis (19, 20). Previous studies have also demonstrated the hypervirulence of Lm-CC1 (9), and its higher efficiency in gut colonization and fecal shedding, compared to hypovirulent Lm clones (17, 21, 22). Moreover, increasing evidence shows that bovines, which are frequent Lm asymptomatic carriers (23–28) and contribute to Lm enrichment in soils (24), are the main source of disease (29) and constitute a reservoir for Lm-CC1 (21, 22). In addition to Lm subclinical infections that may contaminate milk (22, 25), the long-term persistence of Lm in cattle manure–amended soils (30) also poses serious risks of transmission to fresh produce.
Understanding the global evolution of Lm-CC1, which is now spread over all continents (6), as well as its emergence and dissemination across different spatial levels is critical to understand Lm population dynamics and to develop better control strategies, particularly in countries with aging and/or immunosuppressed populations who are most at risk for severe infection. However, the complex movement of livestock and food products associated with asymptomatic intestinal colonization complicates traditional epidemiological investigations aiming at deciphering Lm epidemiology by linking isolates in space and time. Here, we took a population biology approach to fill this knowledge gap and conducted the largest genomic Lm-CC1 study to date, combining genomic and evolutionary approaches to decipher its evolutionary history and pattern of emergence and spread.
RESULTS
Lm-CC1 is composed of three sublineages of uneven prevalence
We analyzed 2021 genomes, including 1230 newly sequenced isolates, originating from 40 countries in six continents and diverse sources (Fig. 1A, dataset S1, and fig. S3), including those from human patients (n = 1452, 72%), food and food processing environments (n = 477, 24%), animal and farm environments (n = 54, 3%), natural environments (n = 11, 0.5%), or unknown sources (n = 27, 1%). We covered a time span of 98 years, from the first Lm-CC1 isolation to the present time (1921–2018), and included all contemporary clinical isolates collected between 2012 and mid-2017 within the surveillance framework of seven countries over three continents (Fig. 1, A and B).
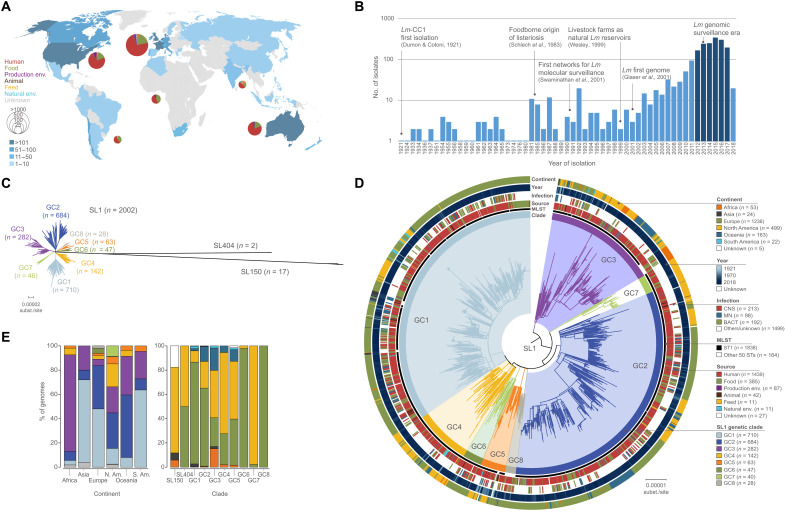
Fig. 1. Geographical and temporal distribution of the isolates used in this study (N = 2021) and phylogenetic analyses.
(A) Geographical distribution and source distribution. Sampled countries are colored in blue, with hue gradient according to the number of isolates. Pie charts are proportional to the number of isolates sampled in each continent and represent the repartition of sample source types, using the source color key indicated in (D). Eight isolates had unknown sampling location and are not shown in the map. (B) Temporal distribution of isolates collected in this study. Darker blue bars indicate the period for which exhaustive clinical sampling was obtained for seven countries spanning three continents (2012–2017; US, FR, UK, DK, NL, AU, and NZ). (C) Unrooted maximum likelihood phylogenetic tree of 2021 Lm-CC1 genomes. The tree was generated from analysis (GTR+F+G4 model, 1000 ultrafast bootstraps) of a 1.29-Mb recombination-purged core genome alignment. (D) Midpoint-rooted maximum likelihood phylogenetic tree of 2002 SL1 genomes based on a recombination-purged core genome alignment of 1.29 Mb. The four external rings indicate the world region, year, type of infection, and source type, respectively. The two inner rings indicate ST1 isolates and the eight SL1 GCs identified in this study, respectively. (E) Percentage of genomes by world region (left) and clade (right). Partitions are colored by world regions and clade, using the same color code as in (D).
Lm-CC1 genome sizes ranged from 2.77 to 3.25 million base pairs (Mbp), with an average number of 2879 ± 77 coding sequences and G+C content of 37.7 to 38.3% (fig. S4). On the basis of MLST (4), 58 sequence types (STs) could be distinguished, with ST1 representing 91% (n = 1838) of isolates. On the basis of core genome MLST (cgMLST) (31), we identified within Lm-CC1 867 cgMLST types, 92% of which were country specific (fig. S5). Rarefaction analysis based on cgMLST resampling did not reach an asymptote (fig. S5), indicating that despite the high number of sequences obtained in this study, a substantial amount of Lm-CC1 diversity remains undetected.
To better understand the phylogenetic diversity of Lm-CC1, we built maximum likelihood phylogenies and identified three sublineages (SL1, SL404, and SL150, named on the basis of their smallest ST number). These sublineages have highly uneven frequency (Fig. 1, C and D, and fig. S6), with SL1 (n = 2002, isolated worldwide) representing 99.1% of the isolates, while 0.1% are SL404 (n = 2, found in Europe and North America) and 0.8% represent SL150 (n = 17, found in North America, Africa, and Asia). Within SL1, we further identified eight distinct genetic clades (GCs), which we named GC1 to GC8 by decreasing prevalence (Fig. 1 and fig. S6). The average genetic distance was 1166 ± 134 whole-genome single-nucleotide polymorphisms (wgSNPs) (and 478 ± 20 cgMLST alleles) between Lm-CC1 sublineages and 76 ± 16 wgSNPs (and 40 ± 9 cgMLST alleles) within SL1 clades (table S1 and fig. S7). The finding that SL1 is by far the major sublineage in Lm-CC1 is consistent with its increased virulence and/or transmission [as seen at Lm species level for hypervirulent clones (32)] or may indicate that SL404 and SL150 are restricted to some yet unknown ecological niches. Within SL1, all different GCs were well represented, with strong spatial structure: GC1 is the most prevalent clade in Europe (48%, 593 of 1237), Asia (68%, 17 of 25), and South America (64%, 14 of 22); GC2 is the most prevalent clade in North America (29%, 150 of 512) and Oceania (52%, 84 of 163), while GC3 is the most prevalent clade in Africa (80%, 43 of 54) (Fig. 1E and fig. S3).
The Lm-CC1 pangenome is diverse
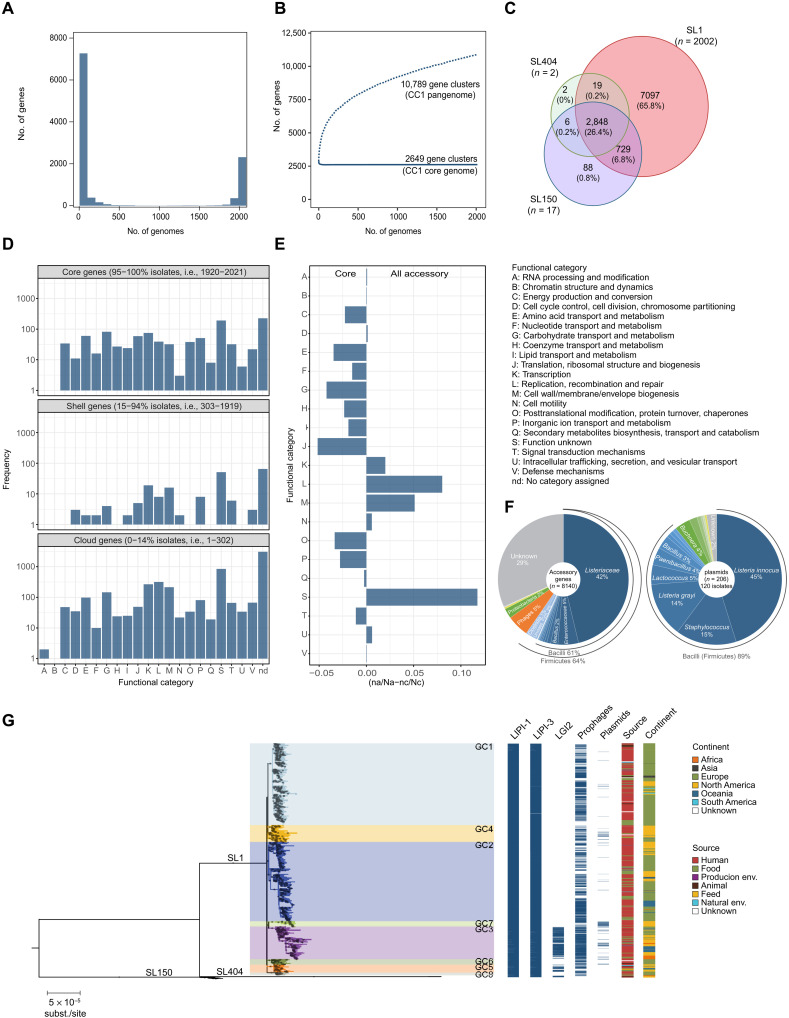
Analysis of Lm-CC1 pangenome identified 10,789 orthologous coding sequences (BlastP identity cutoff of ≥95%), 2649 of which (92% of the average isolate genome content) were present in at least 95% of isolates (core genome) (Fig. 2). The accessory genome included 8140 gene families, of which 2844 (35%) were unique to one isolate, and was enriched in transcription, replication/repair, and cell wall functions, as well as in gene families of unknown function (Fig. 2). Plasmids were present in 6% (120 of 2021) of isolates and were more prevalent in GC7 (83%; Fig. 2). The origins of most accessory genes (64%) and plasmids (89%) were attributable to the Listeriaceae family and other Firmicutes taxa (Fig. 2F). Intact prophages were present in 62% isolates (1263 of 2021) and were distributed across the breadth of Lm-CC1 phylogeny, except in SL404 (Fig. 2). In contrast to Listeria pathogenic islands LIPI-1 (33) and LIPI-3 (34), which were present in all isolates, the Listeria genomic island LGI2 (35), previously identified in Lm-CC1 isolates encoding resistance to cadmium and arsenic, was present in 14% (277 of 2021) isolates and only in GC3 (80%; 225 of 283), GC5 (60%, 38 of 63), and SL150 (82%, 14 of 17; Fig. 2). Sublineage-specific genes were detected (Fig. 2C, tables S2 and S3), and pangenome-wide association analyses identified 24 genes that are associated with a clinical origin (table S4). The impact of these traits on isolates’ differential ecology or virulence remains to be studied, yet the presence of human isolates in all sublineages and clades shows that pathogenic isolates are not restricted to a specific Lm-CC1 clade.
Fig. 2. Lm-CC1 pangenome analysis.
(A) Frequency of sampled gene families. (B) Pan- and core-gene families sampled. (C) Venn diagram showing the number of gene families present in at least one sublineage member. (D) Distribution of the functional categories of the clusters of orthologous genes across the Lm-CC1 pangenome. (E) Differential proportion of each assigned COG (cluster of orthologous group) category in core versus accessory genome, calculated as the difference between the ratio of each category (n) and the total number of hits (N) among each gene pool set, as in (naccessory/Naccessory-ncore/Ncore). (F) Taxonomic classification of accessory genes and plasmids, based on eggNOG-mapper v.2 and MOB-suite v.2, respectively. (G) Distribution of Listeria genomic islands, prophages, and plasmids across Lm-CC1 phylogeny. The midpoint-rooted maximum likelihood phylogenetic tree (GTR+F+G4 model, 1000 ultra-fast bootstraps) was inferred from the 1.29-Mb recombination-purged core genome alignment of 2021 Lm-CC1 genomes.
Emergence and worldwide spread of Lm-CC1 main sublineage (SL1) occurred in the last 200 years
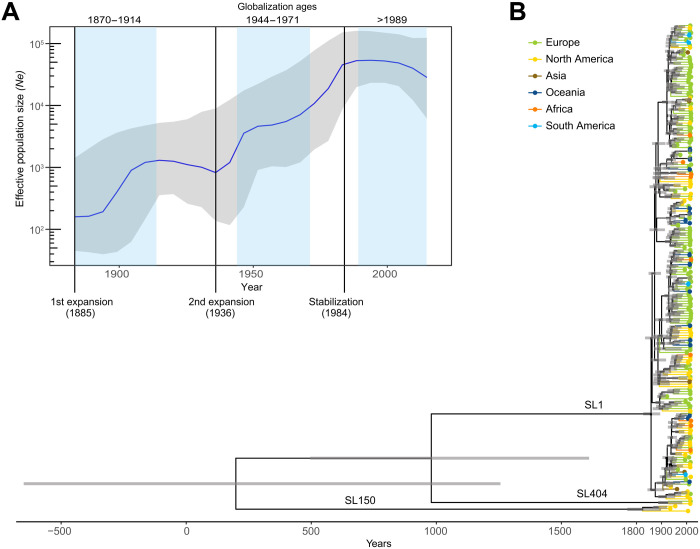
To understand Lm-CC1 evolution and spread, we performed temporal and phylogeographic analyses on the full dated dataset (1972 Lm-CC1 genomes), as well as on a subset of 200 genomes representative of Lm-CC1 genetic, temporal, and geographic diversity (see Materials and Methods; figs. S8 to S10). To control for the overrepresentation of recent isolates and geographic locations, sensitivity analyses of the evolutionary rate estimations were also performed on normalized subsets (see Materials and Methods). We estimate a core genome substitution rate of 1.95 × 10−7 substitutions per site per year [95% confidence interval (CI), 1.75 × 10−7 to 2.15 × 10−7; fig. S8], consistent with previous findings (31). We estimate that Lm-CC1 originated about 1800 years ago (date, 197 CE; 95% CI, 860 BCE to 1045 CE; Fig. 3B) and infer that its last common ancestor evolved in North America (fig. S10 and table S5), long before European colonization and the introduction of cattle in the Americas at the end of the 15th century (36). Although the low number of genomes available for Asia, Africa, and South America could bias this estimation, the estimated origin was also supported by the measures of population variability, which significantly showed higher genetic diversity within North America as compared to other world regions (P < 10−10, pairwise Wilcox test with Bonferroni correction for multiple comparisons; fig. S7 and table S1), and by the basal position of North American Lm-CC1 isolates in the phylogeny (Fig. 3B and fig. S10). The primary natural reservoir of Lm-CC1 and the events that led to its emergence remain unknown.
Fig. 3. Bayesian temporal and demographic analyses on a representative 200 isolate dataset of Lm-CC1.
(A) Bayesian skyline plot (BSP) with the estimation of Lm-CC1 effective population size (Ne). The y axis refers to the predicted number of individuals (log scale), and the x axis refers to the time scale (in years). The median population size is marked in blue, with its 95% high posterior density (HPD) in gray. Blue vertical panels delimitate the three globalization ages (1870–1914, 1944–1971, and 1989–present). (B) Bayesian time-calibrated tree. Nodes represent the estimated mean divergence times, and gray bars represent the 95% HPD CIs of node age. Scale indicates time (in years). Terminal branches and tips are colored by continents, as indicated in the key panel.
Demographic analyses performed using the Bayesian Skyline Plot method (37) (Fig. 3A) show that Lm-CC1 effective population size experienced two waves of expansion: the first in the late 1880s and the second in the 1930s, coinciding with the first and second ages of globalization, respectively. Tajima’s and Fu and Li’s D statistics (38, 39) also supported a recent Lm-CC1 population expansion and SL1 emergence (D < 0; table S1). SL1 emerged approximately 160 years ago (date, 1857; 95% CI, 1821 to 1888), thus closely following the start of the Industrial Revolution (Fig. 4). The first SL1 introductions into Europe occurred around 1868 (GC6/GC8 ancestor; 95% CI, 1827 to 1890), 1871 (GC3/GC7 ancestor; 95% CI, 1838 to 1905), and 1889 (GC2; 95% CI, 1852 to 1909), concomitant with the 1870 North Atlantic Meat trade agreement (40). Under this agreement, surplus cattle in North America were shipped to Europe, which had experienced severe livestock shortages due to widespread disease outbreaks (contagious bovine pleuropneumonia and foot and mouth disease), leading to an unprecedented man-made 1000-fold increase in cattle movement from North America to Europe (41). Within the same period, intracontinental diversification also took place, likely driven by cattle movements across North America and railway expansion in North America and Europe. The first SL1 introductions that occurred in Oceania (1903, GC2) followed the “Great Drought” of 1895–1903, which severely affected livestock (42).
Fig. 4. Phylogeography of sublineage SL1.
(A) Time-calibrated phylogeny based on the 1956 SL1 genomes (full view of Lm-CC1 in fig. S10). Pies at the nodes represent the probability of ancestral geographical locations, estimated using PastML using the MPPA method with an F81-like model. (B) Inferred spread of SL1 populations across continents. The first introductions of each phylogroup are represented by arrows from their estimated world region origin. Gray labels and arrows denote clades with uncertain ancestral origin among datasets. (C) Proportion of intercontinental transitions per 10-year bins, normalized by the total number of phylogenetic branches per bin.
In the following decades and after WWI, multiple CC1 introductions continued from North America into Europe (GC1, GC4, GC5, and GC8), Oceania (GC1 and GC4), and Asia (GC3) and from Europe to Africa (GC3) (Fig. 4, A and B), although the location of the emergence of GC2, GC4, and GC5 clades is uncertain (table S5). The rate of intercontinental bacterial movement declined after 1930s (Fig. 4C), concomitant with the protectionist trade policies that followed the “Great Depression”, which led to a sharp reduction of livestock exports from the United States during the first half of the 20th century (43). A second wave of SL1 expansion occurred after this period, likely driven by a new increase in intercontinental movements favored by the industrialization of food production and globalization of the food and cattle trade (Fig. 3A and fig. S11). Other important human pathogens that have a zoonotic reservoir such as Escherichia coli O157:H7 (44) and Campylobacter jejuni ST-61 (45) have been estimated to have most recent common ancestors (MRCAs) at similar times and to have also undergone population expansions in the context of animal trade or intensive cattle farming, respectively.
A stabilization of Lm-CC1 population is observed after 1984 (Fig. 3A), coincident with major advances in infectious disease prevention in dairy cattle (46) and with the relative decrease of the dairy cattle population in Western countries, in particular Europe (fig. S11). It also coincides with the time when human listeriosis foodborne origin was formally proven (16), which led to the implementation of surveillance programs in North America and Europe (47–49), in particular in the dairy sector following cheese- and milk-related Lm-CC1 outbreaks (50). Whether these findings can be observed in other dairy-associated Lm clonal complexes, such as CC6 (lineage I) or CC37 and CC101 (lineage II) (17, 18), will deserve future studies.
Recent SL1 transmission chains are mostly local
To further analyze more recent strain transmission dynamics, we compared the genetic diversity of SL1 isolates from 2010 to 2018 (n = 1266) across different spatial scales. To avoid oversampling isolates from outbreak investigations, we excluded all nonclinical isolates from confirmed outbreaks (n = 91 isolates from 19 outbreaks). We find that pairs of isolates present within the same 2-year period and the same country are 18.7 times (95% CI, 4.7 to 190.7) more likely to have their MRCA within the past 5 years than pairs of isolates coming from other intracontinental countries >1000 km apart (Fig. 5A). Furthermore, we observe no difference in the probability of having a recent MRCA in isolates coming from nearby intracontinental countries (<1000 km) than from further apart. Isolates coming from different continents are about 100 times less likely to have an MRCA within the past 5 years (0.2; 95% CI, 0.01 to 2.9) than isolates from the same countries (18.7; 95% CI, 4.7 to 190.7) (Fig. 5A). This strong local spatial structure persists for very long time periods, with complete mixing of isolates within a continent appearing only after 50 years (Fig. 5A). At a finer spatial scale, available for France (“départements”, subregional administrative division in France; fig. S12), a strong local spatial structure is also evident, with the proportion of genetically close pairs of clinical cases being higher between isolates coming from the same French department (4.4%; 95% CI, 1 to 10.6%) than between isolates coming from different departments (0.2%; 95% CI, 0.04 to 0.5%), with no effect of distance between them (Fig. 5B). As expected, in densely urban areas with no farming, such as the city of Paris, clinical strains are significantly less likely to share a recent MRCA than in rural areas or other departments [0.0% (95% CI, 0.0 to 4.4%) versus 3.9% (95% CI, 1.0 to 9.5%)] (Fig. 5C). This result is consistent with urban infections being driven by unrelated Lm introductions originating from across the country. Spatial dependence between French isolates persists for 20 years (fig. S13), with on average 20 (1/0.05) different sources of human infection present at any one time per department (Fig. 5B).
Fig. 5. Transmission dynamics of sublineage SL1: Evidence of local expansion after global spread.
(A) Each point summarizes the relative risk that a pair of isolates has an MRCA within a defined time frame and between different spatial scales (within the same country, within the same continent, or within different continents), relative to the risk that a pair of isolates from countries separated by >1000 km has an MRCA in the same range (set as the reference value, “ref”). Error bars represent the 95% CIs, based on 100 bootstrap time-calibrated trees. (B) Proportion of pairs of isolates within the same country (France) sharing an MRCA of 5 or less years in function of the spatial distance within and between administrative departments (shown in the map). The green line indicates the mean proportion of genetically close strains regardless of the geographical location. (C) Relative risk for a pair of isolates to share an MRCA of 5 or less years when (left) both are coming from Paris compared to when coming from another department (P = 0.43) and (right) when coming from the same department in France, except Paris, compared to when coming from different departments (P < 0.001, see Materials and Methods for details).
DISCUSSION
Understanding pathogen evolutionary history is essential to better interpret the population dynamics and biodiversity of microbial infectious agents, and for effective disease surveillance. Here, we have shown that Lm-CC1 has spread worldwide following the Industrial Revolution through two waves of expansion coinciding with the transatlantic livestock trade in the second half of the 19th century and the rapid growth of cattle farming and food industrialization in the 20th century, respectively. The emergence of Lm-CC1 main sublineage SL1 circa 1857 (95% CI, 1821 to 1888) is concomitant with the emergence of other important widespread human pathogens that have a cattle reservoir, such as E. coli O157:H7 (1890; 95% CI, 1845 to 1925) (44) and C. jejuni ST-61 (1859; 95% CI, 1692 to 1943) (45), which have also undergone population expansions facilitated by intensive inter- and intracontinental animal trades (44) and livestock production (45). On the basis of our current dataset, North America is the most likely origin of Lm-CC1, yet this may be challenged when more genomes representing early diverged branches from undersampled regions are available.
Our results also show that, by the time Lm was first recognized as a human foodborne pathogen in 1983, Lm-CC1 had spread worldwide since long, and that genotypes are now firmly established at a local level, with decades-long localized persistence. These results are consistent with the establishment of separate, locally entrenched sources of Lm-CC1 with limited flow of bacteria either within or between countries, in line with cgMLST analyses in which 92% of clusters are country specific.
In the absence of interhuman horizontal transmission, this observation likely represents persistent infection sources, i.e., individual herds and/or production facilities, in which Lm can reside for several years (27, 51). Outbreak investigations performed at the local scale, including in farm environments, would therefore likely improve the identification of contaminating sources, which remain unknown in about 80% of clusters of human cases (52). Identifying and eradicating sources along the food chain, from the farm to the fork, could lead to significant long-term reductions in the transmission of the Lm-CC1.
Our isolates’ dataset exhibits an overrepresentation of Western Europe samples due to the relative scarcity of available genomes from Asia, Africa, and South America, and a relative lack of isolates from natural and animal reservoirs, which may miss other clades and past and current transmission chains in those regions. This limitation reflects the general global disparities in pathogen genomic sequencing. Nevertheless, this study sheds unprecedented light onto the evolutionary history, epidemiology, and population dynamics of Lm-CC1, providing critical clues on its worldwide spread. Similar approaches targeting other major globally distributed clonal complexes will allow clarifying their transmission dynamics and uncovering epidemiological specificities of Lm clones. Deciphering the dynamics and drivers of Lm sublineages across time and space will inform infection control policies to reinforce detection and genome analyses at both local and global levels to ultimately reduce the burden of listeriosis.
MATERIALS AND METHODS
Bacterial isolates and genome sequencing
A total of 2021 high-quality Lm-CC1 genomes collected by this study group (n = 1230) and from NCBI repositories (n = 791, as of 14 March 2018) were analyzed. These were part of an initial dataset of 2154 CC1 genomes, from which 133 were discarded because of low sequencing coverage (<40× after read trimming, n = 62) or low assembly quality (>200 contigs and/or N50 < 20 kb, n = 71) (31). The 2021 isolates originated from human (n = 1453; 72%) and animal hosts (n = 44; 2%), food (n = 387; 19%), food processing environments (n = 88; 4%), feed (n = 11; 0.5%), natural environments (n = 11; 0.6%), or unknown sources (n = 27; 1%) (Fig. 1 and dataset S1). Isolates were sampled in 40 countries from six continents, between 1921 and 2018 (Fig. 1 and dataset S1). Between 2012 and mid-2017, exhaustive sampling was obtained for seven countries in three continents in the context of listeriosis national surveillance programs in Australia (n = 75), Denmark (n = 42), France (n = 395), The Netherlands (n = 53), New Zealand (n = 34), the United Kingdom (n = 106), and the United States (n = 317). Sequencing reads were obtained using Illumina sequencing platforms (Illumina, San Diego, USA) and 2 × 50 bp (n = 110), 2 × 75 bp (n = 2), 2 × 100 bp (n = 233), 2 × 125 bp (n = 9), 2 × 150 bp (n = 1,145), 2 × 250 bp (n = 351), and 2 × 300 bp (n = 138) paired-end runs (dataset S1).
Sequence analysis
Whole-genome sequencing reads were available for 1988 of 2021 isolates. Reads were corrected and trimmed from adapter sequences and nonconfident bases (minimum read length of 30 bases and minimum quality Phred score of 20, i.e., 99% base call accuracy) using fqCleaner v.3.0 (https://gitlab.pasteur.fr/GIPhy/fqCleanER). Assemblies were obtained from paired-end trimmed reads of ≥75 bp (n = 1878 isolates) by using SPAdes v.3.11.0 (53) with the automatic k-mer, --only-assembler and --careful options. For paired-end trimmed reads of 50 bp (n = 111), assemblies were built using CLC Assembly Cell v.5.0.0 (Qiagen, Denmark), with estimated library insert sizes ranging from 50 to 850 bp. Contigs smaller than 500 nucleotides were discarded from both SPAdes- and CLC-generated assemblies.
Pangenome analysis
Gene prediction and annotation were carried out from the draft assemblies using Prokka v.1.12 (54). Functional and taxonomic classification was carried out with eggNOG-mapper v2 (55) using DIAMOND (Double Index Alignment of Next-generation sequencing Data). The presence of plasmids, intact prophages, and Listeria genomic regions was inferred from the assemblies using MOB-suite v.2.0.1 (56), PHASTER (https://phaster.ca/) (57), and BIGSdb-Lm (http://bigsdb.pasteur.fr/listeria/) (31, 58), respectively. Pangenome analyses were carried out using Roary v.3.12 (59) with an amino acid identity cutoff of 95% and splitting homologous groups containing paralogs into groups of true orthologs. Venn diagrams were obtained using Venny 2.1. Pangenome-wide association analyses were performed using treeWAS v.1.0 (60), to control for phylogenetic structure, using the Lm-CC1 core genome maximum likelihood phylogeny (see the “Phylogenetic analyses” section) and a significance threshold of P < 10−5.
In silico molecular typing
Polymerase chain reaction (PCR) serogrouping (5 loci) (61), MLST (7 loci) (4), and cgMLST (1748 loci) (31) profiles were extracted from draft assemblies using the BIGSdb-Lm platform (http://bigsdb.pasteur.fr/listeria/) as previously described (31). Profiles were compared using the single linkage clustering method implemented in BioNumerics v.7.6 (Applied-Maths, Belgium). cgMLST profiles were classified into cgMLST types (CTs) and sublineages (SLs) using previously defined cutoffs (7 and 150 allelic mismatches, respectively, out of 1748 loci) (31). Rarefaction curves were computed with vegan v.2.5-6 (62) R package, estimated with the rarefaction function (Joshua Jacobs, joshuajacobs.org/R/rarefaction) using 100 random samples per point.
Phylogenetic analyses
Core genome multiple sequence alignments were built from the 1748 cgMLST loci concatenated sequences (31). Briefly, individual allele sequences were translated into amino acids, aligned separately with MUSCLE v.3.8.31 (63), and back-translated into nucleotide sequence alignment. Concatenation of the 1748 loci alignments resulted in a multiple sequence alignment of 1.57 Mb.
In parallel, wgSNP-based alignments were built from trimmed reads and NCBI assemblies using the Snippy v.4.1.0 pipeline (https://github.com/tseemann/snippy). The closed CC1 genome F2365 (accession no. NC_002973.6), from the 1985 Canadian cheese outbreak, was used as reference in read mapping, resulting in an alignment of 2.29 Mb.
Gubbins v.2.2.0 (64) was used to detect recombination regions in both core and whole-genome alignments, using default parameters and a minimum of three base substitutions required to identify recombination. Alignment regions positive for recombination were then completely removed from the original alignments, resulting in recombination-free core- and whole-genome alignments of 1.29 and 2.28 Mb, respectively. Maximum likelihood phylogenies were obtained from the recombination-purged alignments using IQ-tree v.1.6.7.2 (65) under the determined best-fit nucleotide substitution model (GTR+F+G4, as determined by ModelFinder) and ultrafast bootstrapping of 1000 replicates. Trees were visualized and annotated with ggtree v.1.14.6 (66) and iTol v.4.2 (67).
To measure the degree of genetic variation within sublineages, GCs, and geographic locations, the pairwise allelic and SNP distance matrices were calculated from the cgMLST profiles and multiple sequence alignments, respectively. SNP distances were computed, taking into account only the ATGC polymorphic positions, extracted from the alignments using SNP-sites v.2.4.1 (68). The nucleotide diversity and the Tajima’s and Fu and Li’s D statistics per alignment were calculated using the R package PopGenome v.2.6.1 (69).
Demographic and spatiotemporal analysis
Substitution rates and demographic changes over time were estimated using a Bayesian approach in BEAST v1.10.4 (70). Two demographic models were tested: the coalescent Bayesian skyline model (nonparametric), which allows a wide range of demographic scenarios, avoiding the biases of prespecified parametric models in the estimates of demographic history (37), and the coalescent constant model (parametric), which assumes that the populations have remained constant through time. To ensure feasible computational running times, the full dated dataset (n = 1972 genomes with country and year of isolation) was down-sampled to a subset of 200 isolates randomly selected out of 421 isolates representative of genomic and geographic diversity of the full dataset (one isolate per country per cluster of 99% core genome similarity; dataset S1). To control for the overrepresentation of recent isolates and geographic locations, isolates were also divided into bins of 10 years based on the isolation dates and randomly subsampled 10 times, allowing only a maximum of 10 isolates per bin and, when possible, equal representation of continents. Sampling times positively correlated with the genetic divergence (P < 0.05, F-statistic test; fig. S8), as observed using TempEst v1.5.1 (71). To assess the significance of the temporal signatures observed, 10 randomized tip date datasets were used as controls. BEAST estimations were made using the nucleotide evolutionary model GTR+Γ4 and a default gamma prior distribution of 1, under an uncorrelated relaxed clock model, to allow each branch of the phylogenetic tree to have its own evolutionary rate (72). Runs were performed in triplicates, each consisting of Monte Carlo Markov chains of 400 million generations, with a 25% burn-in. Parameter values were sampled every 10,000 generations. The effective sample size values were confirmed to be higher than 200 for all parameters using Tracer v.1.7 (73). The tested models were compared by marginal likelihood, and stronger support was obtained for the skyline demographics model (Bayes factor = 10.179). The time of the MRCA and 95% highest posterior densities (95% HPDs) were inferred from the nodes of the maximum clade credibility tree. Estimations of the effective population size along the years were computed using Tracer v.1.7 (73).
Phylogeography analyses were then extended to the 1972 CC1 genomes for which country and year of isolation were available. Time-calibrated phylogenies were inferred from the maximum likelihood core genome trees (obtained with IQ-tree, as described above) using either BactDating v1.0.1 (74), TreeTime v0.5.2 (75), or Treedater v0.3.0 (76), assuming a relaxed clock model and the estimated substitution rate of 1.954 × 10−7 ± 2.015 × 10−8 substitutions per site per year (obtained with BEAST as described above). Cophenetic correlations between BEAST and the three alternative large-scale dating methods were evaluated, and better R2 coefficient scores were obtained for Treedater (fig. S9). For this reason, the latter dated tree was used in further downstream analyses.
Ancestral geographic reconstruction was performed with PastML (77) using the marginal posterior probabilities approximation (MPPA) method with an F81-like model, and estimated ancestral state probabilities were mapped onto the full time-calibrated phylogeny using the R package ape v5.3 (78).
SL1 global transmission dynamics
To infer the transmission dynamics at a recent time scale (Fig. 5A and fig. S13), we focused on the Lm-CC1 main sublineage, and we analyzed the genetic similarity of SL1 isolates from 2010 to 2018 (n = 1266) across different temporal and spatial scales, as described before (79). To avoid oversampling isolates from outbreak investigations, we excluded all nonclinical isolates from confirmed outbreaks (n = 91 isolates from 19 outbreaks). We computed the probability Pl that a pair of isolates that satisfy a given location criteria that were sampled within 2 years of each other had an MRCA in a specific range (0 to 5 years, 5 to 20 years, 20 to 50 years, and >50 years), relative to the probability Pref that a pair of isolates that satisfy the reference location criterion sampled within 2 years of each other had an MRCA within that particular range. The location criteria used were as follows: (i) within countries (both isolates come from the same country), (ii) between countries ≤1000 km (isolates come from distinct countries, separated by less than 1000 km, from the same continent), (iii) between countries >1000 km (isolates come from distinct countries, separated by more than 1000 km, from the same continent; used as reference), and (iv) between continents (isolates come from distinct continents). Spatial relationships between isolates were calculated using the centroid coordinates of the countries or regions of origin.
We estimated these probabilities using
Last, the relative risk (RR) was given by
To measure uncertainty, we used a combination of bootstrapping observations and sampling trees from the Treedater v0.3.0 package (76) to incorporate both sampling and tree uncertainty. Over repeated resamples, we first selected a random tree and calculate the evolutionary distance separating all pairs of sequences. Then, we resampled all the isolates with replacement and recalculate RR each time. The 95% CIs are the 2.5 and 97.5% quantiles from the resultant distribution from 1000 resampling events.
SL1 local transmission dynamics
To assess the SL1 local transmission dynamics, we used available data from France. We computed the proportion of closely related pairs of French isolates (defined as having an MRCA of <5 years) as a function of the spatial distance within and between administrative departments (Fig. 5B)
The different location criteria used are as follows: (i) within department: both isolates come from the same department; (ii) between departments: isolates come from different departments, separated by a distance from 50 to >500 km. The French departments are shown in the map in fig. S12. As shown by Salje et al. (79), the reciprocal of p(within department) represents the lower limit of the number of sources of human infection circulating within a department. To assess uncertainty, we used the bootstrapping approach as described above.
To explore possible differences between departments, we computed the relative risk that a pair of isolates share an MRCA of less than 5 years when both come from the same department compared to when coming from different departments. We looked at two different groups of departments: (i) Paris alone (Fig. 5C, left): within Paris (both isolates come from Paris) and between Paris and other departments (for each pair of isolates, one of them come from Paris, and the other one from another department); (ii) other departments, except Paris (Fig. 5C, right): with other departments (both isolates come from the same department, excluding Paris) and between all other departments (isolates come from two different departments, excluding Paris). For each group, to compute the relative risk RR, we used the same approach as explained above. We estimated
Last, the relative risk is given by
To determine uncertainty, we used the same bootstrapping approach as described above. To assess the statistical significance of each RR, we performed a one-tailed test. We set the null hypothesis (H0) as RR ≤ 1 and alternative hypothesis (H1) as RR > 1. For each group, composed of N bootstrap events, we computed
Acknowledgments
We thank all participating laboratories and the PulseNet International Network members for contributions. We are also grateful to M. Wiedmann, M. Achtman, and J. Haase for providing cultures of historical isolates; to T. Cantinelli and L. Diancourt for contributions to the initial sequencing of CC1 isolates; to K. Jolley, Y. Ghorbal, and B. Brancotte for BIGSdb-Listeria software updates and server maintenance; to E. Rocha, E. Simon-Lorière, A. Zhukova, S. Creno, E. Deveaud, G. Bayle, and E. Volz for insightful feedback on methodological issues; and to F.-X. Weill for critical reading. This work used the computational and storage services (TARS cluster) provided by the IT department at Institut Pasteur, Paris. The findings and conclusions in this report are those of the authors and do not necessarily represent the official position of the Centers for Disease Control and Prevention.
Funding: This study was supported financially by Institut Pasteur, Inserm, Santé Publique France, the European Research Council, the Swiss National Science Foundation (Project SINERGIA, Grant No. CRSII3_147692), the Investissement d’Avenir program Laboratoire d’Excellence “Integrative Biology of Emerging Infectious Diseases” (grant ANR-10-LABX-62-IBEID), and the Advanced Molecular Detection (AMD) initiative at CDC. M.L. is a member of Institut Universitaire de France.
Author contributions: M.L. coordinated the project. M.L. and S.B. conceived and designed the study. A.M., N.L., T.W., S.C., and H.S. analyzed the data, together with S.B. and M.L. A.L., V.B., B.G., T.J.D., J.F., E.F., E.M.N., J.T., A.P., B.P.H., C.L.T., P.G.-S., S.B., M.L., and the Listeria CC1 Study Group obtained the isolates and acquired metadata data collection and genome sequences. A.M., H.S., and M.L. wrote the manuscript. All authors commented on and edited the final version of the manuscript.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
The Listeria CC1 Study Group
Tables S1 to S5
Figs. S1 to S13
Other Supplementary Material for this manuscript includes the following:
Dataset S1
REFERENCES AND NOTES
- 1.Swaminathan B., Gerner-Smidt P., The epidemiology of human listeriosis. Microbes Infect. 9, 1236–1243 (2007). [DOI] [PubMed] [Google Scholar]
- 2.Charlier C., Perrodeau É., Leclercq A., Cazenave B., Pilmis B., Henry B., Lopes A., Maury M. M., Moura A., Goffinet F., Dieye H. B., Thouvenot P., Ungeheuer M.-N. N., Tourdjman M., Goulet V., de Valk H., Lortholary O., Ravaud P., Lecuit M.; MONALISA Study Group , Clinical features and prognostic factors of listeriosis: The MONALISA national prospective cohort study. Lancet Infect. Dis. 17, 510–519 (2017). [DOI] [PubMed] [Google Scholar]
- 3.Orsi R. H., den Bakker H. C., Wiedmann M., Listeria monocytogenes lineages: Genomics, evolution, ecology, and phenotypic characteristics. Int. J. Med. Microbiol. 301, 79–96 (2011). [DOI] [PubMed] [Google Scholar]
- 4.Ragon M., Wirth T., Hollandt F., Lavenir R., Lecuit M., Le Monnier A., Brisse S., A new perspective on Listeria monocytogenes evolution. PLOS Pathog. 4, e1000146 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cantinelli T., Chenal-Francisque V., Diancourt L., Frezal L., Leclercq A., Wirth T., Lecuit M., Brisse S., “Epidemic clones” of Listeria monocytogenes are widespread and ancient clonal groups. J. Clin. Microbiol. 51, 3770–3779 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chenal-Francisque V., Lopez J., Cantinelli T., Caro V., Tran C., Leclercq A., Lecuit M., Brisse S., Worldwide distribution of major clones of Listeria monocytogenes. Emerg. Infect. Dis. 17, 1110–1112 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dumont J., Cotoni L., Bacille semblable au bacielle du Rouget du porc rencontré dans le liquide céphalo-rachidien d’un méningitique. Ann. Inst. Pasteur 35, 625–633 (1921). [Google Scholar]
- 8.Hyden P., Pietzka A., Allerberger F., Springer B., Sensen C., Ruppitsch W., Draft genome sequence of a 94-year-old Listeria monocytogenes isolate, SLCC208. Genome Announc. 4, e01572-15 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Maury M., Tsai Y.-H., Charlier C., Touchon M., Chenal-Francisque V., Leclercq A., Criscuolo A., Gaultier C., Roussel S., Brisabois A., Disson O., Rocha E. P. C., Brisse S., Lecuit M., Uncovering Listeria monocytogenes hypervirulence by harnessing its biodiversity. Nat. Genet. 48, 308–313 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kwong J. C., Mercoulia K., Tomita T., Easton M., Li H. Y., Bulach D. M., Stinear T. P., Seemann T., Howden B. P., Prospective whole-genome sequencing enhances national surveillance of Listeria monocytogenes. J. Clin. Microbiol. 54, 333–342 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bertrand S., Ceyssens P. J., Yde M., Dierick K., Boyen F., Vanderpas J., Vanhoof R., Mattheus W., Diversity of Listeria monocytogenes strains of clinical and food chain origins in Belgium between 1985 and 2014. PLOS ONE 11, e0164283 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Toledo V., Den Bakker H. C., Hormazábal J. C., González-Rocha G., Bello-Toledo H., Toro M., Moreno-Switt A. I., Genomic diversity of Listeria monocytogenes isolated from clinical and non-clinical samples in Chile. Genes (Basel) 9, 396 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hilliard A., Leong D., O’Callaghan A., Culligan E. P., Morgan C. A., DeLappe N., Hill C., Jordan K., Cormican M., Gahan C. G. M., Genomic characterization of Listeria monocytogenes isolates associated with clinical listeriosis and the food production environment in Ireland. Genes 9, 171 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Scaltriti E., Bolzoni L., Vocale C., Morganti M., Menozzi I., Re M. C., Pongolini S., Population structure of Listeria monocytogenes in Emilia-Romagna (Italy) and implications on whole genome sequencing surveillance of listeriosis. Front. Public Health 8, 519293 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rivas L., Paine S., Dupont P.-Y., Tiong A., Horn B., Moura A., Gilpin B. J., Genome typing and epidemiology of human listeriosis in New Zealand, 1999 to 2018. J. Clin. Microbiol. 59, e0084921 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schlech W. F. III, Lavigne P. M., Bortolussi R. A., Allen A. C., Haldane E. V., Wort A. J., Hightower A. W., Johnson S. E., King S. H., Nicholls E. S., Broome C. V., Epidemic listeriosis—Evidence for transmission by food. N. Engl. J. Med. 308, 203–206 (1983). [DOI] [PubMed] [Google Scholar]
- 17.Maury M. M., Bracq-Dieye H., Huang L., Vales G., Lavina M., Thouvenot P., Disson O., Leclercq A., Brisse S., Lecuit M., Hypervirulent Listeria monocytogenes clones’ adaption to mammalian gut accounts for their association with dairy products. Nat. Commun. 10, 2488 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Painset A., Björkman J. T., Kiil K., Guillier L., Mariet J. F., Felix B., Amar C., Rotariu O., Roussel S., Perez-Reche F., Brisse S., Moura A., Lecuit M., Forbes K., Strachan N., Grant K., Møller-Nielsen E., Dallman T. J., Liseq – Whole-genome sequencing of a cross-sectional survey of Listeria monocytogenes in ready-to-eat foods and human clinical cases in Europe. Microb. Genom. 5, e000257 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Costard S., Espejo L., Groenendaal H., Zagmutt F. J., Outbreak-related disease burden associated with consumption of unpasteurized cow’s milk and cheese, United States, 2009–2014. Emerg. Infect. Dis. 23, 957–964 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Filipello V., Mughini-Gras L., Gallina S., Vitale N., Mannelli A., Pontello M., Decastelli L., Allard M. W., Brown E. W., Lomonaco S., Attribution of Listeria monocytogenes human infections to food and animal sources in Northern Italy. Food Microbiol. 89, 103433 (2020). [DOI] [PubMed] [Google Scholar]
- 21.Dreyer M., Aguilar-Bultet L., Rupp S., Guldimann C., Stephan R., Schock A., Otter A., Schüpbach G., Brisse S., Lecuit M., Frey J., Oevermann A., Listeria monocytogenes sequence type 1 is predominant in ruminant rhombencephalitis. Sci. Rep. 6, 36419 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Papić B., Pate M., Félix B., Kušar D., Genetic diversity of Listeria monocytogenes strains in ruminant abortion and rhombencephalitis cases in comparison with the natural environment. BMC Microbiol. 19, 299 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.L. Hafner, M. Pichon, C. Burucoa, S. H.A. Nusser, A. Moura, M. Garcia-Garcera, M. Lecuit, Listeria monocytogenes faecal carriage is common and depends on the gut microbiota. Nat. Comm. 12, 6826 (2021). [DOI] [PMC free article] [PubMed]
- 24.Nightingale K. K., Schukken Y. H., Nightingale C. R., Fortes E. D., Ho A. J., Her Z., Grohn Y. T., Mcdonough P. L., Wiedmann M., Ecology and transmission of Listeria monocytogenes infecting ruminants and in the farm environment. Appl. Environ. Microbiol. 70, 4458–4467 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Esteban J. I., Oporto B., Aduriz G., Juste R. A., Hurtado A., Faecal shedding and strain diversity of Listeria monocytogenes in healthy ruminants and swine in Northern Spain. BMC Vet. Res. 5, 2 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lyautey E., Hartmann A., Pagotto F., Tyler K., Lapen D. R., Wilkes G., Piveteau P., Rieu A., Robertson W. J., Medeiros D. T., Edge T. A., Gannon V., Topp E., Characteristics and frequency of detection of fecal Listeria monocytogenes shed by livestock, wildlife, and humans. Can. J. Microbiol. 53, 1158–1167 (2007). [DOI] [PubMed] [Google Scholar]
- 27.Borucki M. K., Gay C. C., Reynolds J., McElwain K. L., Kim S. H., Call D. R., Knowles D. P., Genetic diversity of Listeria monocytogenes strains from a high-prevalence dairy farm. Appl. Environ. Microbiol. 71, 5893–5899 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.C. Palacios-Gorba, A. Moura, J. Gomis, A. Leclercq, Á. Gómez-Martín, H. Bracq-Dieye, M. L. Mocé, N. Tessaud-Rita, E. Jiménez-Trigos, G. Vales, Á. García-Muñoz, P. Thouvenot, E. García-Roselló, M. Lecuit, J. J. Quereda. Ruminant-associated Listeria monocytogenes isolates belong preferentially to dairy-related hypervirulent clones: A longitudinal study in 19 farms. bioRxiv 2021.07.29.454412 [Preprint]. 3 August 2021. 10.1101/2021.07.29.454412. [DOI] [PubMed]
- 29.Nielsen E. M., Björkman J. T., Kiil K., Grant K., Dallman T., Painset A., Amar C., Roussel S., Guillier L., Félix B., Rotariu O., Perez-Reche F., Forbes K., Strachan N., Closing gaps for performing a risk assessment on Listeria monocytogenes in ready-to-eat (RTE) foods: Activity 3, the comparison of isolates from different compartments along the food chain, and from humans using whole genome sequencing (WGS) analysis. EFSA Support. Publ. 14, 1151E (2017). [Google Scholar]
- 30.Jiang X., Islam M., Morgan J., Doyle M. P., Fate of Listeria monocytogenes in bovine manure-amended soil. J. Food Prot. 67, 1676–1681 (2004). [DOI] [PubMed] [Google Scholar]
- 31.Moura A., Criscuolo A., Pouseele H., Maury M. M., Leclercq A., Tarr C., Björkman J. T., Dallman T., Reimer A., Enouf V., Larsonneur E., Carleton H., Bracq-Dieye H., Katz L. S., Jones L., Touchon M., Tourdjman M., Walker M., Stroika S., Cantinelli T., Chenal-Francisque V., Kucerova Z., Rocha E. P. C., Nadon C., Grant K., Nielsen E. M., Pot B., Gerner-Smidt P., Lecuit M., Brisse S., Whole genome-based population biology and epidemiological surveillance of Listeria monocytogenes. Nat. Microbiol. 2, 16185 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vázquez-Boland J. A., Wagner M., Scortti M., Why are some Listeria monocytogenes genotypes more likely to cause invasive (brain, placental) infection? MBio 11, e03126-20 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kuenne C., Billion A., Mraheil M. A., Strittmatter A., Daniel R., Goesmann A., Barbuddhe S., Hain T., Chakraborty T., Reassessment of the Listeria monocytogenes pan-genome reveals dynamic integration hotspots and mobile genetic elements as major components of the accessory genome. BMC Genomics 14, 47 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cotter P. D., Draper L. A., Lawton E. M., Daly K. M., Groeger D. S., Casey P. G., Ross R. P., Hill C., Listeriolysin S, a novel peptide haemolysin associated with a subset of lineage I Listeria monocytogenes. PLOS Pathog. 4, e1000144 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee S., Ward T. J., Jima D. D., Parsons C., Kathariou S., The arsenic resistance associated Listeria genomic island LGI2 exhibits sequence and integration site diversity and a propensity for three Listeria monocytogenes clones with enhanced virulence. Appl. Environ. Microbiol. 83, e01189-17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bowling G. A., The introduction of cattle into colonial North America. J. Dairy Sci. 25, 129–154 (1942). [Google Scholar]
- 37.Drummond A. J., Rambaut A., Shapiro B., Pybus O. G., Bayesian coalescent inference of past population dynamics from molecular sequences. Mol. Biol. Evol. 22, 1185–1192 (2005). [DOI] [PubMed] [Google Scholar]
- 38.Tajima F., Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123, 585–595 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fu Y. X., Li W. H., Statistical tests of neutrality of mutations. Genetics 133, 693–709 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.World Trade Organization, World Trade Report (World Trade Organization, 2013); www.wto.org.
- 41.Zimmerman W. D., Live cattle export trade between United States and Great Britain, 1868–1885. Agric. Hist. 36, 46–52 (1962). [Google Scholar]
- 42.W. J. Burroughs, World Meteorological Organization, Climate: Into the 21st Century (Cambridge Univ. Press, 2003). [Google Scholar]
- 43.B. Groombridge, Global Biodiversity: Status of the Earth’s Living Resources (Springer Netherlands, 1992). [Google Scholar]
- 44.Franz E., Rotariu O., Lopes B., MacRae M., Bono J., Laing C., Gannon V., Söderlund R., van Hoek A., Friesema I., French N., George T., Biggs P., Jaros P., Rivas M., Chinen I., Campos J., Jernberg C., Gobius K., Mellor G., Chandry P., Perez-Reche F., Forbes K., Strachan N., Phylogeographic analysis reveals multiple international transmission events have driven the global emergence of Escherichia coli O157:H7. Clin. Infect. Dis. 69, 428–437 (2019). [DOI] [PubMed] [Google Scholar]
- 45.Mourkas E., Taylor A. J., Méric G., Bayliss S. C., Pascoe B., Mageiros L., Calland J. K., Hitchings M. D., Ridley A., Vidal A., Forbes K. J., Strachan N. J. C., Parker C. T., Parkhill J., Jolley K. A., Cody A. J., Maiden M. C. J., Kelly D. J., Sheppard S. K., Agricultural intensification and the evolution of host specialism in the enteric pathogen Campylobacter jejuni. Proc. Natl. Acad. Sci. U.S.A. 117, 11018–11028 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.LeBlanc S. J., Lissemore K. D., Kelton D. F., Duffield T. F., Leslie K. E., Major advances in disease prevention in dairy cattle. J. Dairy Sci. 89, 1267–1279 (2006). [DOI] [PubMed] [Google Scholar]
- 47.Cartwright E. J., Jackson K. A., Johnson S. D., Graves L. M., Silk B. J., Mahon B. E., Listeriosis outbreaks and associated food vehicles, United States, 1998–2008. Emerg. Infect. Dis. 19, 1–9 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.De Valk H., Vaillant V., Jacquet C., Rocourt J., Le Querrec F., Stainer F., Quelquejeu N., Pierre O., Pierre V., Desenclos J. C., Goulet V., Two consecutive nationwide outbreaks of listeriosis in France, October 1999–February 2000. Am. J. Epidemiol. 154, 944–950 (2001). [DOI] [PubMed] [Google Scholar]
- 49.Tappero J. W., Schuchat A., Deaver K. A., Mascola L., Wenger J. D.; The Listeriosis Study Group , Reduction in the incidence of human listeriosis in the United States. Effectiveness of prevention efforts? JAMA 273, 1118–1122 (1995). [DOI] [PubMed] [Google Scholar]
- 50.S. Kathariou, in Microbial Food Safety in Animal Agriculture (Blackwell Publishing, 2008), pp. 243–256. [Google Scholar]
- 51.Castro H., Jaakkonen A., Hakkinen M., Korkeala H., Lindström M., Occurrence, persistence, and contamination routes of Listeria monocytogenes genotypes on three Finnish dairy cattle farms: A longitudinal study. Appl. Environ. Microbiol. 84, e02000-17 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Moura A., Tourdjman M., Leclercq A., Hamelin E., Laurent E., Fredriksen N., Van Cauteren D., Bracq-Dieye H., Thouvenot P., Vales G., Tessaud-Rita N., Maury M., Alexandruu A., Criscuolo A., Quevillon E., Donguy M.-P., Enouf V., de Valk H., Brisse S., Lecuit M., Real-time whole-genome sequencing for surveillance of Listeria monocytogenes, France. Emerg. Infect. Dis. 23, 1462–1470 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bankevich A., Nurk S., Antipov D., Gurevich A. A., Dvorkin M., Kulikov A. S., Lesin V. M., Nikolenko S. I., Pham S., Prjibelski A. D., Pyshkin A. V., Sirotkin A. V., Vyahhi N., Tesler G., Alekseyev M. A., Pevzner P. A., SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19, 455–477 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Seemann T., Prokka: Rapid prokaryotic genome annotation. Bioinformatics 30, 2068–2069 (2014). [DOI] [PubMed] [Google Scholar]
- 55.Huerta-Cepas J., Forslund K., Coelho L. P., Szklarczyk D., Jensen L. J., Von Mering C., Bork P., Fast genome-wide functional annotation through orthology assignment by eggNOG-mapper. Mol. Biol. Evol. 34, 2115–2122 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Robertson J., Nash J. H. E., MOB-suite: Software tools for clustering, reconstruction and typing of plasmids from draft assemblies. Microb. Genom. 4, e000206 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Arndt D., Grant J. R., Marcu A., Sajed T., Pon A., Liang Y., Wishart D. S., PHASTER: A better, faster version of the PHAST phage search tool. Nucleic Acids Res. 44, W16–W21 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jolley K. A., Maiden M. C. J., BIGSdb: Scalable analysis of bacterial genome variation at the population level. BMC Bioinformatics 11, 595 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Page A. J., Cummins C. A., Hunt M., Wong V. K., Reuter S., Holden M. T. G., Fookes M., Falush D., Keane J. A., Parkhill J., Roary: Rapid large-scale prokaryote pan genome analysis. Bioinformatics 31, 3691–3693 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Collins C., Didelot X., A phylogenetic method to perform genome-wide association studies in microbes that accounts for population structure and recombination. PLOS Comput. Biol. 14, e1005958 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Doumith M., Buchrieser C., Glaser P., Jacquet C., Martin P., Differentiation of the major Listeria monocytogenes serovars by multiplex PCR. J. Clin. Microbiol. 42, 3819–3822 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dixon P., VEGAN, a package of R functions for community ecology. J. Veg. Sci. 14, 927–930 (2003). [Google Scholar]
- 63.Edgar R. C., MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Croucher N. J., Page A. J., Connor T. R., Delaney A. J., Keane J. A., Bentley S. D., Parkhill J., Harris S. R., Rapid phylogenetic analysis of large samples of recombinant bacterial whole genome sequences using Gubbins. Nucleic Acids Res. 43, e15 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Nguyen L. T., Schmidt H. A., Von Haeseler A., Minh B. Q., IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 32, 268–274 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yu G., Smith D. K., Zhu H., Guan Y., Lam T. T.-Y., ggtree: An R package for visualization and annotation of phylogenetic trees with their covariates and other associated data. Meth. Ecol. Evol. 8, 28–36 (2017). [Google Scholar]
- 67.Letunic I., Bork P., Interactive tree of life (iTOL) v3: An online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 44, W242–W245 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Page A. J., Taylor B., Delaney A. J., Soares J., Seemann T., Keane J. A., Harris S. R., SNP-sites: Rapid efficient extraction of SNPs from multi-FASTA alignments. Microb. Genom. 2, e000056 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pfeifer B., Wittelsbürger U., Ramos-Onsins S. E., Lercher M. J., PopGenome: An efficient Swiss army knife for population genomic analyses in R. Mol. Biol. Evol. 31, 1929–1936 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Suchard M. A., Lemey P., Baele G., Ayres D. L., Drummond A. J., Rambaut A., Bayesian phylogenetic and phylodynamic data integration using BEAST 1.10. Virus Evol. 4, vey016 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Rambaut A., Lam T. T., Max Carvalho L., Pybus O. G., Exploring the temporal structure of heterochronous sequences using TempEst (formerly Path-O-Gen). Virus Evol. 2, vew007 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Drummond A. J., Ho S. Y. W., Phillips M. J., Rambaut A., Relaxed phylogenetics and dating with confidence. PLOS Biol. 4, e88 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Rambaut A., Drummond A. J., Xie D., Baele G., Suchard M. A., Posterior summarization in Bayesian phylogenetics using Tracer 1.7. Syst. Biol. 67, 901–904 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Didelot X., Croucher N. J., Bentley S. D., Harris S. R., Wilson D. J., Bayesian inference of ancestral dates on bacterial phylogenetic trees. Nucleic Acids Res. 46, e134 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Himmelmann L., Metzler D., TreeTime: An extensible C++ software package for Bayesian phylogeny reconstruction with time-calibration. Bioinformatics 25, 2440–2441 (2009). [DOI] [PubMed] [Google Scholar]
- 76.Volz E. M., Frost S. D. W., Scalable relaxed clock phylogenetic dating. Virus Evol. 3, vex025 (2017). [Google Scholar]
- 77.Ishikawa S. A., Zhukova A., Iwasaki W., Gascuel O., A fast likelihood method to reconstruct and visualize ancestral scenarios. Mol. Biol. Evol. 36, 2069–2085 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Paradis E., Schliep K., ape 5.0: An environment for modern phylogenetics and evolutionary analyses in R. Bioinformatics 35, 526–528 (2019). [DOI] [PubMed] [Google Scholar]
- 79.Salje H., Lessler J., Berry I. M., Melendrez M. C., Endy T., Kalayanarooj S., A-Nuegoonpipat A., Chanama S., Sangkijporn S., Klungthong C., Thaisomboonsuk B., Nisalak A., Gibbons R. V., Iamsirithaworn S., MacAreo L. R., Yoon I.-K., Sangarsang A., Jarman R. G., Cummings D. A. T., Dengue diversity across spatial and temporal scales: Local structure and the effect of host population size. Science 355, 1302–1306 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The Listeria CC1 Study Group
Tables S1 to S5
Figs. S1 to S13
Dataset S1