Abstract
Here, we aimed to evaluate the clinical performance of a novel automated immunoassay HISCL SARS-CoV-2 Antigen assay kit designed to detect the nucleocapsid (N) protein of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). This kit comprises automated chemiluminescence detection systems. Western blot analysis confirmed that anti-SARS-CoV antibodies detected SARS-CoV-2N proteins. The best cut-off index was determined, and clinical performance was tested using 115 nasopharyngeal swab samples obtained from 46 patients with coronavirus disease 2019 (COVID-19) and 69 individuals who tested negative for COVID-19 through reverse transcription quantitative polymerase chain reaction (RT-qPCR). The HISCL Antigen assay kit showed a sensitivity of 95.4% and 16.6% in samples with copy numbers > 100 and < 99, respectively. The kit did not cross-react with human coronaviruses causing seasonal common cold and influenza, and none of the 69 individuals without COVID-19 were diagnosed with positive results. Importantly, 81.8% of the samples with low virus load (< 50 copy numbers) were diagnosed as negative. Thus, using HISCL antigen assay kits may reduce overdiagnosis compared with RT-qPCR tests. The rapid and high-throughput HISCL SARS-CoV-2 Antigen assay kit developed here proved suitable for screening infectious COVID-19 and may help control the pandemic.
Subject terms: Virology, SARS-CoV-2
Introduction
The coronavirus disease-2019 (COVID-19) pandemic originating from Wuhan, China has caused chaos and health and economic crises across the world1–7 despite the cases of infection and death being far fewer than those of the 1918 influenza pandemic8. The World Health Organization declared COVID-19 a pandemic in March 2020.
Since severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is contagious in humans, it is important to determine the infection status of individuals accurately9. During the early phase of the pandemic, a reverse transcriptase quantitative-polymerase chain reaction (RT-qPCR)-based test was considered a gold standard. However, a meta-analysis of 51 studies reported that the overall sensitivity of PCR tests was 89.1% and the specificity was 98.9%, even though the true definition of COVID-19 remains ambiguous10. In addition, results can vary because the accuracy of PCR tests can be significantly affected by various factors, such as primer design and sample collection techniques, reagents, and laboratory equipment. Furthermore, patients can test positive for COVID-19 by PCR tests during post-infectious periods11 as the cut-off can be changed by altering the cycle threshold (Ct) values. For example, low, clinically insignificant amounts of viral RNAs can still be detected when the Ct is set to more than 35, suggesting a positive result. This indicates that the relationship between Ct values and infectious status is debatable12–14 since non-infectious COVID-19 individuals can be labeled as positive due to overdiagnosis by RT-qPCR tests15–17. This issue raises serious concerns, not only in terms of medical decisions but also with regard to the global economy and the protection of human rights18–21.
Recently, SARS-CoV-2 Antigen detection assays were developed as potential alternative tests to RT-qPCR to identify infectious patients, as stated by the U.S. Food and Drug Administration (https://www.fda.gov/consumers/consumer-updates/coronavirus-disease-2019-testing-basics). In the early phase of the COVID-19 pandemic, Ag Respi-Strip, an immunochromatographic assay, was developed (Coris BioConcept, Gembloux, Belgium) in which monoclonal antibodies against the nucleocapsid (N) protein of SARS-CoV were used. The LHUB-ULB SARS-CoV-2 working diagnostic group reported that the COVID-19 Ag Respi-Strip assay showed an overall sensitivity and specificity of 57.6% and 99.5%, respectively22. In June 2020, LUMIPULSE G1200 using a chemiluminescence enzyme immunoassay became available (Fujirebio, Tokyo, Japan), and its overall sensitivity and specificity in Japanese patients was 55.2% and 99.6%, respectively23.
We developed an automated antigen detection system, which we termed as the HISCL SARS-CoV-2 Antigen assay kit that can detect the N protein of SARS-CoV-2 using enzyme-linked immunosorbent assay (ELISA). This automated test can process 200 samples per hour, making it suitable for mass screening. In this study, we examined the feasibility and accuracy of the HISCL SARS-CoV-2 Antigen assay kit for screening COVID-19 patients in Japan.
Results
Western blot
To examine whether the previously produced anti-SARS-CoV antibodies (ab clone# 2–3 and ab clone# 2–12)24 can detect SARS-CoV-2, western blots were performed using His-tagged recombinant N proteins of SARS-CoV-2 (ACRO Biosystems, Newark, DE). Figure 1 shows that the preanti-SARS-CoV antibodies (left two strips: ab clone# 2–3 and ab clone# 2–12) and anti-His antibody (the third strip from the left) detected the SARS-CoV-2 antigens but not negative control (the far-right strip). The original stained membrane is shown in Supplemental Figure 1.
Figure 1.
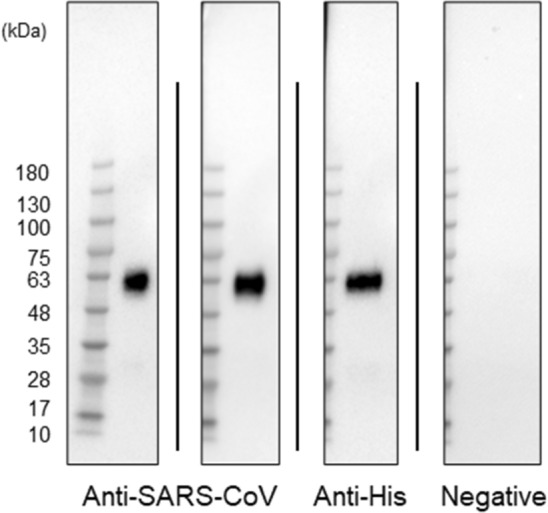
Western blot analysis. Anti-SARS-CoV antibodies (left two strips) and anti-His antibodies (third strip form the left) reacting with SARS-CoV-2 antigens. The far-right strip shows negative control (without antigens). All strips were cropped from a membrane shown in Supplemental Figure 1. SARS-CoV-2: severe acute respiratory syndrome coronavirus 2; His: histidine.
Determination of the association between Ct values and viral loads
Since RT-PCR results can be affected by different laboratory settings25, relationship between Ct values by a RT-qPCR method26 and viral copy numbers was determined in our laboratory. SARS-CoV-2 Positive Control RNA (1 × 105 copies/µL; JP-NN2-PC, Nihon Gene Research Laboratories) was sequentially diluted (50 to 5,000 copies/sample) and subjected to RT-qPCR. Figure 2 shows the standard curve of Ct values as a function of copy numbers using four RNA samples for each copy number. The plots were fitted with linear regression, yielding the slope of − 3.43 and the intercept of 50.1 (r2 = 0.999).
Figure 2.
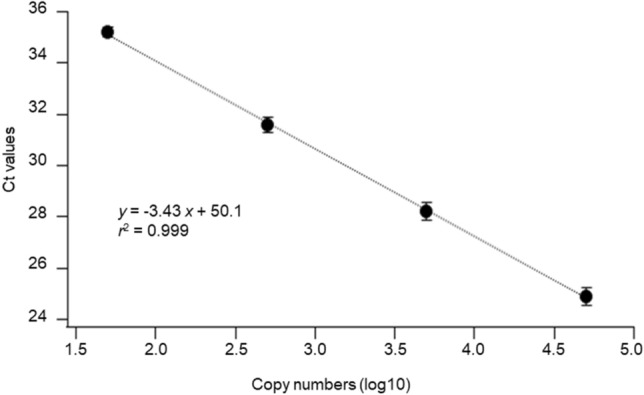
Standard curve for relations between Ct values and copy numbers. Error bars indicate SD. Ct cycle threshold, SD standard deviation.
HISCL SARS-CoV-2 Antigen assay kit
As described in the Methods section, recombinant SARS-CoV-2 proteins (ACRO Biosystems, Newark, DE) were incubated with biotinylated SARS-CoV-2 antibodies (R1: ab 2–3) and alkaline phosphatase (ALP)-bound SARS-CoV-2 antibodies (R3: ab 2–12). After washing out the unbound proteins, chemiluminescent substrates (CDP-Star, C0712, Sigma-Aldrich, St. Louis, MO, USA) were added, and fluorescence signals were measured using the photo counter of HISCL-800 (Sysmex, Kobe, Japan). The level of SARS-CoV-2 antigens was indicated as the cut-off index (COI), calculated by the difference in the luminescence intensities in the buffers with and without the SARS-CoV-2 antigens. The COI was defined as:
Cut-off index (C.O.I.) = (light intensity in a sample—light intensity with the negative control) / (light intensity at a calculated cut-off value—light intensity with the negative control).
Reproducibility
To check the reproducibility of the HISCL SARS-CoV-2 Antigen assay kit, we measured one sample containing recombinant SARS-CoV-2 antigens (109.5 ± 36.5 pg/mL, ACRO Biosystems, Newark, DE) (positive control sample) and one sample containing only buffers (negative control sample). The tests were repeated ten times for each sample and all experiments were conducted in triplicate. Table 1 shows the COI values for both samples. The negative control samples showed 100% agreement with COI of zero and the positive control samples showed the average COI of 27.7 to 28.8 with a coefficient of variation (CV) percentage of 1.3–2.5.
Table 1.
Within-run reproducibility. The COI values of samples without or with SARS-CoV-2 antigens were measured a total of 30 times (three sets of 10 consecutive measurements).
| Control panels | COI | ||
|---|---|---|---|
| Negative control | |||
| 1 | 0 | 0 | 0 |
| 2 | 0 | 0 | 0 |
| 3 | 0 | 0 | 0 |
| 4 | 0 | 0 | 0 |
| 5 | 0 | 0 | 0 |
| 6 | 0 | 0 | 0 |
| 7 | 0 | 0 | 0 |
| 8 | 0 | 0 | 0 |
| 9 | 0 | 0 | 0 |
| 10 | 0 | 0 | 0 |
| Positive control | |||
| 1 | 28.4 | 27.9 | 29 |
| 2 | 29 | 27.3 | 29 |
| 3 | 29 | 26.3 | 27.3 |
| 4 | 28.4 | 27.9 | 27.9 |
| 5 | 29 | 27.3 | 27.9 |
| 6 | 28.4 | 27.9 | 27.9 |
| 7 | 28.4 | 27.9 | 28.4 |
| 8 | 29 | 28.4 | 28.4 |
| 9 | 27.9 | 29 | 27.9 |
| 10 | 29 | 27.3 | 27.9 |
| Average | 28.7 | 27.7 | 28.2 |
| SD | 0.4 | 0.7 | 0.5 |
| CV (%) | 1.3 | 2.5 | 1.8 |
COI cutoff index, SD standard deviation, CV coefficient of variation.
To check reproducibility between measurements in five different days, we performed assays using the negative and positive control samples twice daily for 5 days. Table 2 shows that the COIs of the negative and positive control samples were consistent (CV% < 10).
Table 2.
Between-run reproducibility.
| COI | Run1 | Run2 | ||||
|---|---|---|---|---|---|---|
| 1 | 2 | Mean | 1 | 2 | Mean | |
| Negative control | ||||||
| Day1 | 0 | 0 | 0 | 0 | 0 | 0 |
| Day2 | 0.1 | 0.1 | 0.1 | 0 | 0.1 | 0 |
| Day3 | 0 | 0 | 0 | 0 | 0 | 0 |
| Day4 | 0 | 0 | 0 | 0 | 0 | 0 |
| Day5 | 0 | 0 | 0 | 0 | 0 | 0 |
| Positive control | ||||||
| Day1 | 26.8 | 27.3 | 27 | 26.7 | 27 | 26.8 |
| Day2 | 27.5 | 28.1 | 27.8 | 28 | 27.2 | 27.6 |
| Day3 | 28.4 | 28.5 | 28.4 | 29 | 27.7 | 28.3 |
| Day4 | 28.2 | 28 | 28.1 | 28.6 | 28.4 | 28.5 |
| Day5 | 30.7 | 30.6 | 30.6 | 30.8 | 29.8 | 30.3 |
| Average | 28.4 | Ave | 28.3 | |||
| SD | 1.3 | SD | 1.3 | |||
| CV (%) | 4.7 | CV (%) | 4.6 | |||
COI cut-off index, SD standard deviation, CV coefficient of variation.
Cross-reaction
To check for specificity, various corona and influenza viral antigens were used to measure COIs in the HISCL assay. Table 3 summarizes the COI measurements. The COI showed high values for SARS-CoV at a concentration of 0.25 ng/mL and positive values for MERS-CoV at a low concentration. In contrast, the COI values stayed in the negative range for other coronaviruses and influenza viruses, indicating that the HISCL SARS-CoV-2 Antigen assay kit does not react with seasonal common cold and flu viruses.
Table 3.
Cut-off index (COI) values in negative controls.
| Panels | Concentration (ng/mL) | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 0 | 0.25 | 0.5 | 0.75 | 1 | 25 | 50 | 75 | 100 | |
| MERS-CoV | 0 | 1.1 | 2.3 | 3.3 | 4.6 | 11.9 | 23.6 | 34.3 | 46.2 |
| HCoV-229E | 0 | – | – | – | – | 0 | 0 | 0 | 0 |
| HCoV-OC43 | 0 | – | – | – | – | 0 | 0.1 | 0.2 | 0.2 |
| HCoV-NL63 | 0 | – | – | – | – | 0 | 0 | 0 | 0.1 |
| HCoV-HKU1 | 0 | – | – | – | – | 0 | 0.1 | 0 | 0.1 |
| SARS-CoV | 0 | 26 | 53 | 75.2 | 109.8 | 2938 | 5727.1 | 8127.1 | 10,682.1 |
| Influenza H1N1 | 0 | – | – | – | – | 0.1 | 0 | 0 | 0 |
| Influenza H3N2 | 0 | – | - | – | – | 0 | 0 | 0 | 0 |
| Influenza B | 0 | – | – | – | – | 0 | 0 | 0 | 0 |
HCoV human coronavirus, MERS-CoV Middle East respiratory syndrome-related coronavirus, SARS-CoV severe acute respiratory syndrome coronavirus, SARS-CoV-2 severe acute respiratory syndrome coronavirus 2.
Determination of the cut-off value
To optimize the cut-off antigen level for accurate diagnoses, we first determined the relationship between COI and viral loads using a total of 84 nasopharyngeal swabs collected from 17 patients at Kobe City Medical Center General Hospital and 67 commercially available samples (Cantor Bioconnect, Santee, CA, USA): 30 positive samples and 54 negative samples.
Chemiluminescence counts were measured by HISCL SARS-CoV-2 Antigen assay kit using these samples and recombinant SARS-CoV-2 proteins (0, 20, and 100 pg/mL). Viral loads (copy numbers) were calculated based on the relationships between Ct values and RNA content (Supplemental Table 1). Sensitivity and specificity were calculated at different cut-off values depending upon the RT-qPCR results (i.e., copy number 0 as negative). In determination of the threshold, we put priority on obtaining best specificity according to the Antigen Testing Algorithm by the Center for Disease Control (https://www.cdc.gov/coronavirus/2019-ncov/lab/resources/Antigen_Testing_Algorithm_2020-12-14_v03_NO_DRAFT_SPW_508.pdf). When the cut-off for the antigen level was set to 3.65 pg/mL, sensitivity of 80% and specificity of 98.2% were obtained (Table 4). Thus, chemiluminescence count using 3.65 pg/mL antigen was set to a COI value of 1.0 in the HISCL SARS-CoV-2 Antigen assay kit. Figure 3 shows that the ROC analysis yielded an AUC value of 0.8988 ± 0.0464 (95% confidence interval, 0.808–0.990, p < 0.0001).
Table 4.
Sensitivity and specificity according to various cut-off values.
| Cut-off value SARS-CoV-2 Ag (pg/mL) | Sensitivity (%) | Specificity (%) |
|---|---|---|
| 1.65 | 86.7 | 81.5 |
| 2.6 | 80 | 90.7 |
| 3.65 | 80 | 98.2 |
| 4.45 | 73.3 | 98.2 |
| 6 | 63.3 | 98.2 |
SARS-CoV-2 severe acute respiratory syndrome coronavirus 2.
Figure 3.
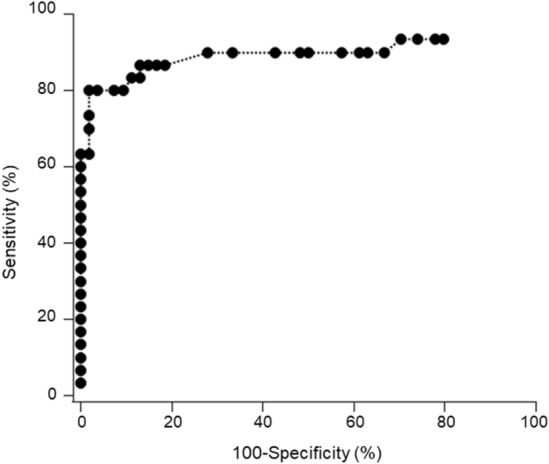
Receiver operating characteristic (ROC) curve analysis. An ROC curve was drawn using human samples with and without SARS-CoV-2.
Table 5 summarizes the concordance between the two tests in three viral load ranges using the cut-off value in validation samples.
Table 5.
Relationship between the SARS-CoV-2 viral loads and COI in the assay kit using commercially available samples.
| RT-qPCR | SARS-CoV-2 Ag test | Total | |
|---|---|---|---|
| (copies/test) | Positive | Negative | |
| 1–50 | 1 | 4 | 5 |
| 51–99 | 4 | 1 | 5 |
| > 100 | 19 | 1 | 20 |
COI cut-off-index, SARS-CoV-2 severe acute respiratory syndrome coronavirus 2, RT-qPCR reverse transcription quantitative polymerase chain reaction, Ag antigen.
Clinical performance
To examine the clinical performance, a total of 115 nasopharyngeal swab samples obtained from 115 patients at Juntendo University Hospital were subjected to the RT-qPCR and the HISCL SARS-CoV-2 Antigen assay kit. The RT-PCR test showed 46 positive and 69 negative results. The SARS-CoV-2 viral loads determined by RT-qPCR and the antigen levels detected by HISCL SARS-CoV-2 Antigen assay kit for each patient are listed in Supplemental Table 2. Figure 4 shows a dot plot of the positive COI values (> 1.0) against the viral copy numbers. The positive COI values obtained within 10 days from the onset for mild cases (n = 16) and moderate–critical cases (n = 5) had a mean [interquartile range] of 330.5 [27.6; 3849.3] and 233.9 [5.5; 1241.3], respectively (p = 0.860). Table 6 summarizes the concordance between the two tests in the two viral load ranges. The antigen tests showed 95.4% positive results in samples whose viral copy numbers were higher than 100. In samples whose copy numbers were lower than 99, the antigen test showed negative results in 20 samples and positive results in four samples.
Figure 4.
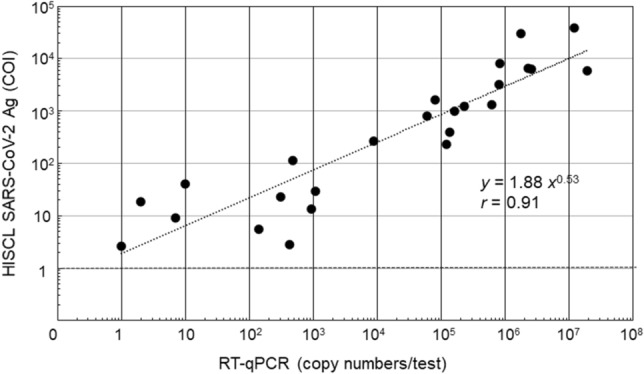
A scatter plot of HISCL SARS-CoV-2 Antigen (COI) and copy numbers measured by RT-qPCR tests. Data are plotted as logarithmic scales and the plots were fitted with a power approximation.
Table 6.
Relationship between the SARS-CoV-2 viral loads and COI by the HISCL SARS-CoV-2 Antigen assay kit.
| RT-qPCR | SARS-CoV-2 Ag test | Total | |
|---|---|---|---|
| (copies/test) | Positive | Negative | |
| 1–99 | 4 | 20 | 24 |
| > 100 | 21 | 1 | 22 |
SARS-CoV-2 severe acute respiratory syndrome coronavirus 2, RT-qPCR reverse transcription quantitative polymerase chain reaction, Ag antigen.
Discussion
In this study, we evaluated the performance of HISCL SARS-CoV-2 Antigen assay kit using chemiluminescent enzyme immunoassay in comparison with RT-qPCR test results. This test was approved by the Pharmaceuticals and Medical Device Agency of Japan in November 2020, and it is currently commercially available (Sysmex, Kobe, Japan). This automated test can process 200 samples per hour, and the run time is only 17 min. Therefore, it can be used as a high-throughput screening test.
Since the early phase of this pandemic, RT-qPCR tests have been used for diagnoses, epidemiological studies, and political decision making27–29. Severe cases with high infectivity usually show high viral loads (i.e., low Ct values) and vice versa30. However, the interpretation of positive results needs extra caution because Ct values can vary due to differences in PCR conditions31. Importantly, infectivity cannot be determined solely based on RT-qPCR results11.
To examine the correlation between RT-qPCR and our new antigen detection assay, we used SARS-CoV-2 RNAs as a standard. Our assay showed 95.4% concordance in the samples with a COI > 100, and the concordance decreased to 16.7% in samples with a COI < 99. LUMIPULSE assay (Fujirebio, Tokyo, Japan) showed 100% concordance in samples with > 100 copies23. This may be due to the small sample numbers used in both the studies, but it may also be attributed to the difference in the standard RNA (AccuPlex SARS-CoV-2 Reference Material Kit; SeraCare, Milford, MA, USA) and RT-qPCR methods (QuantiTect Probe RT-PCR Kit and Applied Biosystems 7500 Fast Real-Time PCR System versus StepOnePlus Real-Time PCR System).
Nonetheless, this difference should not be problematic in clinical practice since the viral loads in infectious patients were usually much higher than copy numbers of 100. For example, Yu et al. reported that copy number of SARS-CoV-2 in sputum samples in early-phase infection was 46.800 ± 17.272, and 1.252 ± 1.027 in the recovery phase32. Wölfel et al. reported that the pharyngeal virus shedding was high during the first week of symptoms, with a peak at 7.11 × 108 RNA copies per throat swab on day 433. Conversely, many studies also reported that there was no apparent correlation between viral loads (copy numbers) and severity of disease or mortality in patients with COVID-19 disease34,35. Indeed, the viral loads in our study were much lower than those in the previously reported cases, and mild cases showed similar COIs compared to moderate–critical cases (Supplemental Table 2). These data, including our data, indicate that infectivity and viral loads are not proportional. Thus, we propose this antigen test is considered as a useful screening test. However, diagnosis and clinical decisions should not be made upon any single test. Therapeutic strategies should be determined based on systemic physiological parameters, such as respiratory rate, blood oxygenation, heart rate, and blood pressure, along with the screening test.
There were a few limitations of this study: (1) this study was performed only in Japan with a relatively small number of patients; (2) we could not test the assay for mutated pathogens that might not be bound by our antibody; (3) cross-reactivity with unknown pathogens could not be excluded. However, diagnoses of COVID-19 may not benefit clinical practice since no proven remedies are available to date; and (4) we could not examine chronological changes of the antigen test results.
In conclusion, considering the current chaotic situation across the world due to COVID-19, which is in part caused by detecting non-infectious patients with extremely low viral load as positive cases by RT-qPCR, our antigen detection assay may be more suitable than RT-qPCR for mass screening as it would help to moderate COVID-19 overdiagnoses. Further validation of the kit with a larger number of samples and SARS-CoV-2 variants is warranted.
Methods
Patient samples
This research related to human use complied with all the relevant national regulations, institutional policies and was conducted in accordance the tenets of the Helsinki Declaration. It has been approved by the authors’ institutional review board at Juntendo University Hospital, Tokyo, Japan (IRB #20-036, Chair: Atsushi Okuzawa, MD) and Kobe City Medical Center General Hospital, Hyogo, Japan (IRB #zn200615, Chair: Yasushi Naito, MD). This study used an opt-out consent method. The Institutional Review Board of Juntendo University and the Institutional Review Board of Kobe City Medical Center General Hospital waived the need for informed consent. Therefore, written informed consent was not required.
Classification of disease severity was determined according to the Clinical Spectrum of SARS-CoV-2 Infection (https://www.covid19treatmentguidelines.nih.gov/overview/clinical-spectrum/). To determine the relationship between photo counts and viral loads, a total of 84 nasopharyngeal swabs were collected from 17 patients at Kobe City Medical Center General Hospital and 67 commercially available samples (Cantor Bioconnect, Santee, CA, USA; validation dataset). For clinical performance analysis, a total of 115 nasopharyngeal swabs were collected from 115 patients at Juntendo University Hospital (test dataset).
SARS-CoV-2 RT-qPCR
Viral RNAs were extracted using QIAamp Viral RNA Mini Kit (Qiagen, Hilden, Germany). RT-qPCR was performed following the protocol developed by the National Institute of Infectious Diseases of Japan. The primer/probe set (N2) was designed based on the N sequence of SARS-CoV-2 RNA (NC_045512.2)26. The primer/probe sequences were as following: forward primer (5′-AAATTTTGGGGACCAGGAAC-3′), reverse primer (5′-TGGCAGCTGTGTAGGTCAAC-3′), and TaqMan probe (5′-FAM-ATGTCGCGCATTGGCATGGA-BHQ-3′). The expected amplicon size was 158 bp. QuantiTect Probe RT-PCR Kit (Qiagen, Germantown, MD, USA) and Applied Biosystems 7500 Fast Real-Time PCR System (Thermo Fisher Scientific, Waltham, MA, USA) were used. The Ct values were assessed using the protocol developed by the National Institute of Infectious Diseases (version 2.9.1)26. The association between absolute viral copy numbers (viral loads) and Ct values was determined using SARS-CoV-2 Positive Control RNA (JP-NN2-PC, Nihon Gene Research Laboratories, Miyagi, Japan).
Recombinant antigen production
For antigen cross-reactivity tests, nucleocapsid antigens derived from the other coronaviruses and influenza viruses were prepared. The recombinant nucleocapsid proteins were produced based on the following sequences: SARS-CoV (YP_009825061), Middle East respiratory syndrome-related coronavirus (MERS-CoV; YP_009047211), human coronavirus HKU1 HCoV-HKU1 (YP_173242), HCoV-OC43 (YP_009555245), HCoV-NL63 (YP_003771), and HCoV-229E (NP_073556). Each recombinant protein was prepared according to previously described methods24.
Western blot
The detection of SARS-CoV-2 Antigens by SARS-CoV antibodies was confirmed via western blot analysis using the previously produced anti-N capsid SARS-CoV antibodies24. Histidine (His)-tagged SARS-CoV-2N proteins (10 ng) were loaded on NEXT Page II gel (5–20% gradient; Gellex, Tokyo, Japan), and electrophoresis was performed using iBind Western Device (Thermo Fisher Scientific). Two lots each of anti-SARS-CoV antibodies (0.5 μg/mL) and horseradish peroxidase (HRP)-conjugated anti-His antibodies (Penta-His Antibody; Cat. No. 34660, Qiagen) were used as primary antibodies. Anti-IgG antibodies (Medical & Biological Laboratories, Nagoya, Japan) were used as the secondary antibodies. Immobilon Western HRP substrate (Millipore, Burlington, MA, USA) was used for chemiluminescent detection.
HISCL SARS-CoV-2 Antigen assay kit
Previously produced monoclonal antibodies against the SARS-CoV N proteins24 were used to probe the SARS-CoV-2N proteins (ACRO Biosystems, Newark, DE, USA). The HISCL SARS-CoV-2 Antigen assay kit is a chemiluminescent enzyme immunoassay that uses a HISCL automatic immunoassay analyzer (Sysmex, Kobe, Japan).
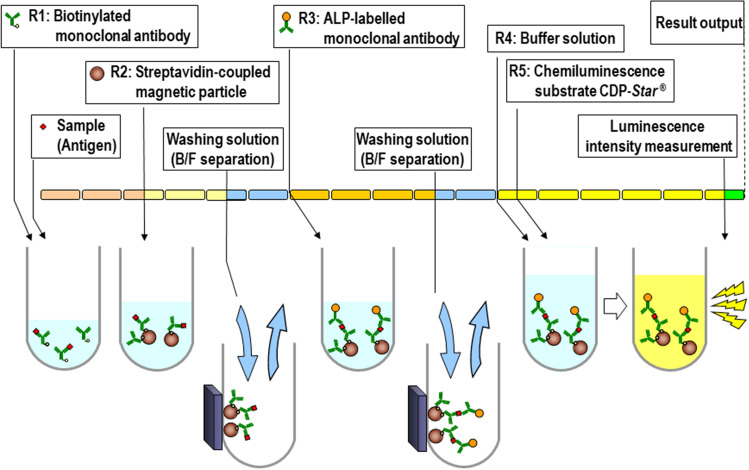
Figure 5 presents a schematic illustration of the procedure: (1) the samples were incubated with biotinylated SARS-CoV-2 antibodies (R1: antibody 2–3) at 42 °C for 3 min; (2) the mixtures were incubated with streptavidin-bonded magnetic particles at 42 °C for 2 min; (3) after protein separation and washing, alkaline phosphatase (ALP)-bound SARS-CoV-2 antibodies (R3: antibody 2–12) were added and the mixtures were incubated at 42 °C for 3 min; (4) after magnetic separation and washing again, the chemiluminescent substrates were added and the mixtures were incubated at 42 °C for 5.5 min; and (5) chemiluminescence signals (CDP-Star, C0712, Sigma-Aldrich, St. Louis, MO, USA) were measured using the photo counter of HISCL-800 (Sysmex, Kobe, Japan). The level of SARS-CoV-2 Ag was indicated as cut-off index (COI), calculated by the difference in the luminescence intensities in the buffers with and without the SARS-CoV-2 antigens.
Figure 5.
Assay protocol of the HISCL SARS-CoV-2 Antigen assay kit. The biotinylated SARS-CoV-2 Ag antibody (R1) was allowed to react with the sample at 42 °C for 3 min. Then, streptavidin-bonded magnetic particles (R2) were added and allowed to react at 42 ℃ for 2 min. After protein separation and washing, ALP-bound SARS-CoV-2 Ag antibody (R3) was added and reacted at 42 °C for 3 min. After another magnetic separation and washing, buffer solution (R4) and the chemiluminescent substrate (R5) were allowed to react at 42 °C for 5.5 min, then the luminescence intensity was measured. SARS-CoV-2: severe acute respiratory syndrome coronavirus 2; ALP: alkaline phosphatase; Ag: antigen. The illustration was drawn by A.K. using Microsoft PowerPoint 17.0 (Microsoft Corporation, Redmond, Washington: https://www.microsoft.com/en-us/microsoft-365/powerpoint).
Human sample collection
Human samples were obtained using nasopharyngeal cotton swabs following the standard method36. The swabs were immersed in 0.5 mL phosphate-buffered saline or viral transfer medium (Lampire Biological Laboratories, Pipersville, PA, USA). The suspensions were frozen at − 80 °C until antigen tests were performed. Highly viscous samples were centrifuged (2000×g for 5 min), and the supernatants were used for subsequent analyses.
Reproducibility
Within-run and between-run reproducibility were determined by running the buffers with and without SARS-CoV-2 antigens. The recombinant human antigen for SARS-CoV-2 was purchased from ACRO Biosystems (Newark, DE). To test reproducibility, buffers containing SARS-CoV-2 antigens (positive control) and those without antigens (negative control) were prepared. To test between-run reproducibility, both negative and positive controls were tested twice per day each for five consecutive days.
Cross-reaction
To determine specificity, cross-reactions were checked using measuring buffers containing various recombinant viral antigens: human SARS-CoV, MERS-CoV, HCoV-229E, HCoV-OC43, HCoV-NL63, HCoV-HKU1, influenza virus H1N1, influenza virus H3N2, and influenza virus B. Inactivated influenza viruses were purchased from Advanced Biotechnologies (Eldersburg, MD, USA). The antigens of SARS-CoV, MERS-CoV, HCoV-229E, HCoV-OC43, HCoV-NL63, and HCoV-HKU1 were prepared according to a previous established protocol24. HISCL SARS-CoV-2 antigen assays were carried out following the manufacturer’s instructions.
Statistical analyses
Statistical analyses were performed in Excel (Microsoft, Redmond, WA, USA). Receiver operating characteristic (ROC) curve analyses were conducted using GraphPad Prism 9.0.0 (GraphPad Software: https://www.graphpad.com/support/faq/prism-900-release-notes/, San Diego, USA) to evaluate the assay performance and visualize the curves. The areas under the ROC curves (AUCs), sensitivity, and specificity were calculated.
Supplementary Information
Acknowledgements
The authors are grateful to the participants of this study. The authors would also like to thank Dr. Keisuke Tomii, Dr. Atsushi Nakagawa, and Masaaki Eto at Kobe City Medical Center General Hospital for providing patient samples, and Koji Tsuchiya, Maiko Yuri, Nobuyuki Ide, and Ayako Okabe for their technical support. Finally, we thank all our medical staff who conducted their duties diligently during the pandemic.
Author contributions
T.A. and Y.T. designed the study and wrote the paper. K.S., A.K., J.M., Y.F., N.K., T.K., M.C., T.K., and Y.H. performed experiments. H.T., Y.M., and M.H. contributed to sample collection. S.M., A.O., S.H., T.N., and T.M. contributed to data interpretation. K.T. supervised the study. All authors have accepted responsibility for the contents of this manuscript and approved its submission.
Data availability
The datasets supporting the conclusions of this article is(are) included within the article (and its additional files).
Competing interests
Sysmex Corporation provided reagents for HISCL SARS-CoV-2 Antigen assay kit measurements free of charge. Akinori Kawai, Jun Matsui, Yoshiyuki Fukushima, Norihiro Kikukawa, and Takuya Kyoutou are employees of Sysmex Corporation. The other authors (Kaori Saito, Tomohiko Ai, Masayoshi Chonan, Takeaki Kawakami, Yoshie Hosaka, Shigeki Misawa, Haruhi Takagi, Yasushi Matsushita, Makoto Hiki, Atsushi Okuzawa, Satoshi Hori, Toshio Naito, Takashi Miida, Kazuhisa Takahashi, and Yoko Tabe) do not have COI to disclose. This study was performed following strict adherence to academic standards.
Footnotes
The original online version of this Article was revised: “The best cut-off index was determined, and clinical performance was tested using 115 serum samples obtained from 46 patients with coronavirus disease 2019 (COVID-19) and 69 individuals who tested negative for COVID-19 through reverse transcription quantitative polymerase chain reaction (RT-qPCR).” now reads: “The best cut-off index was determined, and clinical performance was tested using 115 nasopharyngeal swab samples obtained from 46 patients with coronavirus disease 2019 (COVID-19) and 69 individuals who tested negative for COVID-19 through reverse transcription quantitative polymerase chain reaction (RT-qPCR).”
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Kaori Saito and Tomohiko Ai.
Change history
12/8/2021
A Correction to this paper has been published: 10.1038/s41598-021-03518-y
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-021-02636-x.
References
- 1.Zdravkovic M, Berger-Estilita J, Zdravkovic B, Berger D. Scientific quality of COVID-19 and SARS CoV-2 publications in the highest impact medical journals during the early phase of the pandemic: A case control study. PLoS ONE. 2020;15:e0241826. doi: 10.1371/journal.pone.0241826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fleming N. Coronavirus misinformation, and how scientists can help to fight it. Nature. 2020;583:155–156. doi: 10.1038/d41586-020-01834-3. [DOI] [PubMed] [Google Scholar]
- 3.Paterlini M. 'Closing borders is ridiculous': the epidemiologist behind Sweden's controversial coronavirus strategy. Nature. 2020;580:574. doi: 10.1038/d41586-020-01098-x. [DOI] [PubMed] [Google Scholar]
- 4.Greene CM, Murphy G. Individual differences in susceptibility to false memories for COVID-19 fake news. Cogn. Res. Princ. Implic. 2020;5:63. doi: 10.1186/s41235-020-00262-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kenworthy N, Koon A, Mendenhall E. On symbols and scripts: The politics of the American COVID-19 response. Glob. Public Health. 2021 doi: 10.1080/17441692.2021.1902549. [DOI] [PubMed] [Google Scholar]
- 6.Tokuda Y, et al. Authors' reply: Independent investigation may be more reliable in the event with political nature. J. Gen. Fam. Med. 2021;22:115. doi: 10.1002/jgf2.402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clarke EJR, Klas A, Dyos E. The role of ideological attitudes in responses to COVID-19 threat and government restrictions in Australia. Pers. Ind. Differ. 2021;175:110734. doi: 10.1016/j.paid.2021.110734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Taubenberger JK, Kash JC, Morens DM. The 1918 influenza pandemic: 100 years of questions answered and unanswered. Sci. Transl. Med. 2019 doi: 10.1126/scitranslmed.aau5485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bchetnia M, Girard C, Duchaine C, Laprise C. The outbreak of the novel severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2): A review of the current global status. J. Infect. Public Health. 2020;13:1601–1610. doi: 10.1016/j.jiph.2020.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mustafa Hellou M, et al. Nucleic acid amplification tests on respiratory samples for the diagnosis of coronavirus infections: a systematic review and meta-analysis. Clin. Microbiol. Infect. 2020 doi: 10.1016/j.cmi.2020.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mina MJ, Parker R, Larremore DB. Rethinking Covid-19 test sensitivity: A strategy for containment. N. Engl. J. Med. 2020;383:e120. doi: 10.1056/NEJMp2025631. [DOI] [PubMed] [Google Scholar]
- 12.Singanayagam A, et al. Duration of infectiousness and correlation with RT-PCR cycle threshold values in cases of COVID-19, England, January to May 2020. Euro Surveill. 2020 doi: 10.2807/1560-7917.Es.2020.25.32.2001483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rhee C, Kanjilal S, Baker M, Klompas M. Duration of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infectivity: When is it safe to discontinue isolation? Clin. Infect. Dis. 2020 doi: 10.1093/cid/ciaa1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.La Scola B, et al. Viral RNA load as determined by cell culture as a management tool for discharge of SARS-CoV-2 patients from infectious disease wards. Eur. J. Clin. Microbiol. Infect. Dis. 2020;39:1059–1061. doi: 10.1007/s10096-020-03913-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Roy S. Physicians' dilemma of false-positive RT-PCR for COVID-19: a case report. SN Compr. Clin. Med. 2021 doi: 10.1007/s42399-020-00655-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ndwandwe D, Mathebula L, Adetokunboh O, Kamadjeu R, Wiysonge CS. Cochrane corner: universal screening for SARS-CoV-2 infection. Pan Afr. Med. J. 2020;37:48. doi: 10.11604/pamj.supp.2020.37.48.26881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.van Kasteren PB, et al. Comparison of seven commercial RT-PCR diagnostic kits for COVID-19. J. Clin. Virol. 2020;128:104412. doi: 10.1016/j.jcv.2020.104412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Surkova E, Nikolayevskyy V, Drobniewski F. False-positive COVID-19 results: Hidden problems and costs. Lancet Respir. Med. 2020;8:1167–1168. doi: 10.1016/S2213-2600(20)30453-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Piltch-Loeb R, Jeong KY, Lin KW, Kraemer J, Stoto MA. Interpreting COVID-19 test results in clinical settings: It depends! J. Am. Board Fam. Med. 2021;34:S233–S243. doi: 10.3122/jabfm.2021.S1.200413. [DOI] [PubMed] [Google Scholar]
- 20.Gray N, et al. Is "no test is better than a bad test"? Impact of diagnostic uncertainty in mass testing on the spread of COVID-19. PLoS ONE. 2020;15:e0240775. doi: 10.1371/journal.pone.0240775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cohen A, Kessel B, Milgroom M. Diagnosing SARS-CoV-2 infection: the danger of over-reliance on positive test results. medRxiv. 2020 doi: 10.1101/2020.04.26.20080911. [DOI] [Google Scholar]
- 22.Mertens P, et al. Development and potential usefulness of the COVID-19 Ag Respi-Strip diagnostic assay in a pandemic context. Front. Med. (Lausanne) 2020;7:225. doi: 10.3389/fmed.2020.00225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hirotsu Y, et al. Comparison of automated SARS-CoV-2 antigen test for COVID-19 infection with quantitative RT-PCR using 313 nasopharyngeal swabs, including from seven serially followed patients. Int. J. Infect. Dis. 2020;99:397–402. doi: 10.1016/j.ijid.2020.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fujimoto K, et al. Sensitive and specific enzyme-linked immunosorbent assay using chemiluminescence for detection of severe acute respiratory syndrome viral infection. J. Clin. Microbiol. 2008;46:302–310. doi: 10.1128/JCM.01006-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Munne K, et al. Detection of SARS-CoV-2 infection by RT-PCR test: factors influencing interpretation of results. Virusdisease. 2021 doi: 10.1007/s13337-021-00692-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shirato K, et al. Development of genetic diagnostic methods for detection for novel coronavirus 2019(nCoV-2019) in Japan. Jpn. J. Infect. Dis. 2020;73:304–307. doi: 10.7883/yoken.JJID.2020.061. [DOI] [PubMed] [Google Scholar]
- 27.Islam KU, Iqbal J. An update on molecular diagnostics for COVID-19. Front. Cell Infect. Microbiol. 2020;10:560616. doi: 10.3389/fcimb.2020.560616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nuccetelli M, et al. SARS-CoV-2 infection serology: a useful tool to overcome lockdown? Cell Death Discov. 2020;6:38. doi: 10.1038/s41420-020-0275-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Leuzinger K, et al. Epidemiology and precision of SARS-CoV-2 detection following lockdown and relaxation measures. J. Med. Virol. 2020 doi: 10.1002/jmv.26731. [DOI] [PubMed] [Google Scholar]
- 30.Bustos P, et al. Quantitative Detection of SARS-CoV-2 RNA in nasopharyngeal samples from infected patients with mild disease. J. Med. Virol. 2020 doi: 10.1002/jmv.26761. [DOI] [PubMed] [Google Scholar]
- 31.Service. R One number could help reveal how infectious a COVID-19 patient is. Should test results include it? Science. 2020 doi: 10.1126/science.abf0366. [DOI] [Google Scholar]
- 32.Yu F, et al. Quantitative detection and viral load analysis of SARS-CoV-2 in infected patients. Clin. Infect. Dis. 2020;71:793–798. doi: 10.1093/cid/ciaa345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wolfel R, et al. Virological assessment of hospitalized patients with COVID-2019. Nature. 2020;581:465–469. doi: 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
- 34.KarahasanYagci A, et al. Relationship of the cycle threshold values of SARS-CoV-2 polymerase chain reaction and total severity score of computerized tomography in patients with COVID 19. Int. J. Infect. Dis. 2020;101:160–166. doi: 10.1016/j.ijid.2020.09.1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shah S, Singhal T, Davar N, Thakkar P. No correlation between Ct values and severity of disease or mortality in patients with COVID 19 disease. Indian J. Med. Microbiol. 2021;39:116–117. doi: 10.1016/j.ijmmb.2020.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Marty FM, Chen K, Verrill KA. How to obtain a nasopharyngeal swab specimen. N. Engl. J. Med. 2020;382:e76. doi: 10.1056/NEJMvcm2010260. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets supporting the conclusions of this article is(are) included within the article (and its additional files).