Abstract
In immature oocytes, Balbiani bodies are conserved membraneless condensates implicated in oocyte polarization, the organization of mitochondria, and long-term organelle and RNA storage. In Xenopus laevis, Balbiani body assembly is mediated by the protein Velo1. Velo1 contains an N-terminal prion-like domain (PLD) that is essential for Balbiani body formation. PLDs have emerged as a class of intrinsically disordered regions that can undergo various different types of intracellular phase transitions and are often associated with dynamic, liquid-like condensates. Intriguingly, the Velo1 PLD forms solid-like assemblies. Here we sought to understand why Velo1 phase behavior appears to be biophysically distinct from that of other PLD-containing proteins. Through bioinformatic analysis and coarse-grained simulations, we predict that the clustering of aromatic residues and the amino acid composition of residues between aromatics can influence condensate material properties, organization, and the driving forces for assembly. To test our predictions, we redesigned the Velo1 PLD to test the impact of targeted sequence changes in vivo. We found that the Velo1 design with evenly spaced aromatic residues shows rapid internal dynamics, as probed by fluorescent recovery after photobleaching, even when recruited into Balbiani bodies. Our results suggest that Velo1 might have been selected in evolution for distinctly clustered aromatic residues to maintain the structure of Balbiani bodies in long-lived oocytes. In general, our work identifies several tunable parameters that can be used to augment the condensate material state, offering a road map for the design of synthetic condensates.
Over the past decade, biomolecular condensates have emerged as one route by which cells address the challenge of intracellular spatiotemporal organization.1−4 Defined as nonstoichiometric assemblies that locally concentrate a specific subset of biological components, condensates range in size from hundreds of molecules to micrometer-sized organelles.5,6 In many cases, condensate formation, material state, and disassembly appear well-described by the physical principles of liquid–liquid phase separation.7,8 Prominent examples of biomolecular condensates described by liquid–liquid phase separation include the P granules, stress granules, and the nucleolus.6,7,9−16 Beyond their role as naturally occurring organelles, synthetic, stimulus-responsive condensates are emerging as a new class of tools for intracellular manipulation across a wide range of length scales.17−21 As such, there is ongoing and ever-evolving interest in understanding the protein-encoded molecular grammar that determinanes condensate behavior.15,20,22−46
One feature of biomolecular condensates that has attracted substantial interest is the role of their material state. While much attention has been focused on condensates with liquid-like properties, condensates with solid-like, semiliquid, hyperviscous, or dynamically arrested properties make up a ubiquitous class of cellular assembly.47 Pioneering work by Görlich and colleagues revealed that in vitro reconstitution of phenylalanine-glycine-rich nucleoporin domains (FG-Nups) could form porous hydrogels with arrested dynamics and recapitulate nuclear transport receptor specificity.48−51 This early work provided prescient insight into how aromatic residue-dependent intermolecular interactions could drive biologically essential molecular assemblies in the context of nuclear transport.52−55 More recently, changes in condensate material state have been linked with disease14,56−59 and with altered growth and fitness in bacteria and yeast.18,59−61 Taken together, an emerging consensus suggests that the condensate material state appears to be a property that is optimized for a given molecular function.18,39,45,47,62−67
In many cases, proteins that contain intrinsically disordered regions (IDRs) are associated with biomolecular condensates. This observation likely reflects the fact that some IDRs offer a convenient platform upon which multivalent interaction sites can be encoded across a flexible scaffold.38,68−70 This hypothesis is supported by numerous studies in which specific IDRs undergo spontaneous concentration-dependent phase transitions in vitro and in cells, although we emphasize that IDRs are not required for phase transitions or biomolecular condensate formation.15,23,28,38,41,43,44,70,71
The encoding of adhesive binding sites embedded within a flexible polymeric scaffold is described well by the physics of associative polymers.72−75 In particular, the stickers-and-spacer framework has been effectively co-opted to quantitatively describe multivalent flexible polymers that can drive the formation of biomolecular condensates (Figure 1A).22,69,76,77 In the stickers-and-spacers framework, molecules can be divided into stickers and spacers. Stickers are regions or sites that contribute to the adhesive interactions that drive phase transitions. Spacers are regions between stickers and influence phase behavior primarily by tuning chain dynamics and the effective solvation volume, a parameter that reports on the total volume a polymer occupies (as determined by both steric and solvation effects).77,78 The stickers-and-spacers framework is remarkably simple yet offers a convenient first-order approximation through which the physical chemistry of a given system can be interpreted.69 An appealing feature of this framework is that it offers both a qualitative mental model and a quantitative mathematical framework through which sticker valency and strength can make predictions about phase behavior.22,23,69,76,77,79,80
Figure 1.
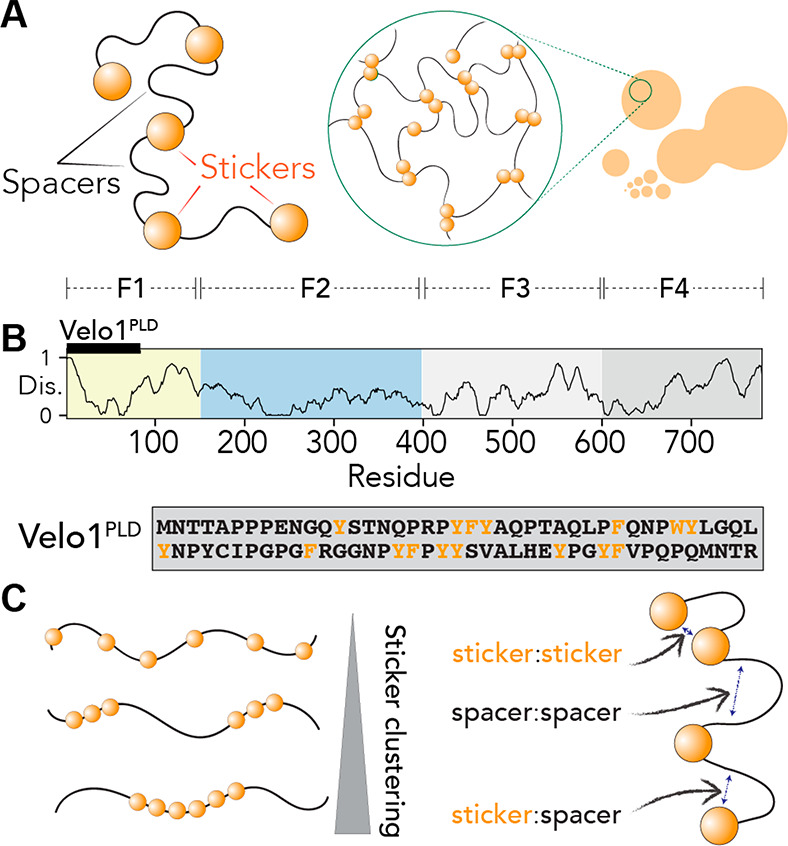
Stickers-and-spacers framework that can be used to describe Velo1PLD. (A) The stickers-and-spacers framework subdivides biomolecules into sticker regions and spacer regions, whereby stickers contribute attractive interactions that drive phase transitions through multivalent interactions. (B) The Velo1 sequence architecture contains an N-terminal prion-like domain (Velo1PLD) and four fragments (F1–F4) as originally defined by Böke et al.39 The Velo1PLD sequence is shown explicitly with aromatic residues colored orange and all other residues colored black. (C) Questions of interest in this study are how sticker clustering (left) and spacer-related interactions (right) can alter the formation and equilibrium state of condensates formed by PLDs.
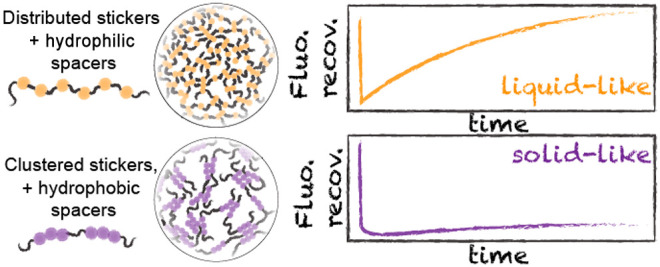
Recent work has illustrated that sticker valence, strength, and patterning are key determinants of phase separation.23,79,81,82 In this context, prion-like domains (PLDs) have emerged as a convenient domain type for understanding the molecular grammar of biological phase separation.14,22,23,28,32−34,39,43,83−89 PLDs make up a class of low-complexity IDRs characterized by an enrichment of polar residues and a depletion of charged residues.22,90,91 PLDs undergo phase transitions both in vitro and in cells, where aromatic residues are essential for their self-assembly, phase separation, gelation, and recruitment to existing biomolecular condensates.22,23,28,32,33,86,89,92 Because of their convenient sequence architecture, PLDs have been examined through the lens of the stickers-and-spacers framework, with aromatic residues demarcated as stickers and the remaining low-complexity polar context as spacers (Figure 1A). Evolutionary analysis has argued that evenly distributed hydrophobic and/or aromatic residues facilitate liquid-like condensates and prevent aggregation.23,79,93,94 In support of this hypothesis, rationally designed PLDs with clusters of aromatic or hydrophobic residues experience retarded intracellular dynamics (in the case of TDP-43) and undergo rapid aggregation in vitro (in the case of hnRNPA1).23,94
PLDs from RNA binding proteins can undergo phase separation in vitro and in cells to form dynamic, liquid-like condensates.14,22,23,32,33,43,56,85,95−98 Given the various roles of condensates with solid-like properties and our ability to rationally design PLDs that form kinetically arrested condensates, we wondered if examples of naturally occurring PLDs that formed solids in a functionally interpretable context existed. Conveniently, a counterexample to the liquid-like condensates associated with many PLDs is condensates formed by the Xenopus laevis protein Velo1.
Velo1 is responsible for scaffolding the Balbiani body, a membraneless superorganelle in X. laevis.39 The Balbiani body is characterized by a dense accumulation of mitochondria in the cytoplasm of early oocytes and is observed in many species, including humans and frogs.99,100 The Balbiani body has been proposed to play a role in protecting mitochondria and RNA in oocytes from damage,101 which is particularly important in oocytes that can remain dormant for decades before being activated and giving rise to a fertilizable egg. Velo1 undergoes amyloid-like self-assembly to form the Balbiani body, thereby providing the stable matrix for organelles to last in the Balbiani body for the duration of dormancy.39,102 Velo1 contains an N-terminal PLD (Velo1PLD) (Figure 1B) and forms superficially irreversible amyloid-positive biomolecular condensates both in vitro and in cells. Importantly, self-assembly is clearly driven by Velo1PLD.39
Velo1PLD is an unusual and unexpected outlier. Prior work by many groups has shown that PLDs taken from a wide range of other proteins robustly form liquid-like condensates in vitro and in vivo. In contrast, Velo1PLD self-assembles into a fibrous networked assembly that lacks any of the hallmarks of a liquid state. In fact, the self-assembled Velo1PLD is considered a physiological amyloid, one of the most ordered structures in protein biochemistry.102 This observation sets the stage for our work: why does Velo1PLD form solid-like assemblies in vitro and in vivo while a plethora of other PLDs drive LLPS?
To address this question, we combined proteome-wide sequence analysis with coarse-grained simulations and in-cell experiments to identify, explore, and test the determinants of liquid-like assembly in Velo1PLD (Figure 1C).
Results
Proteome-wide Bioinformatic Analysis Reveals Velo1 PLD to Be Distinct from Previously Studied Prion-like Domains
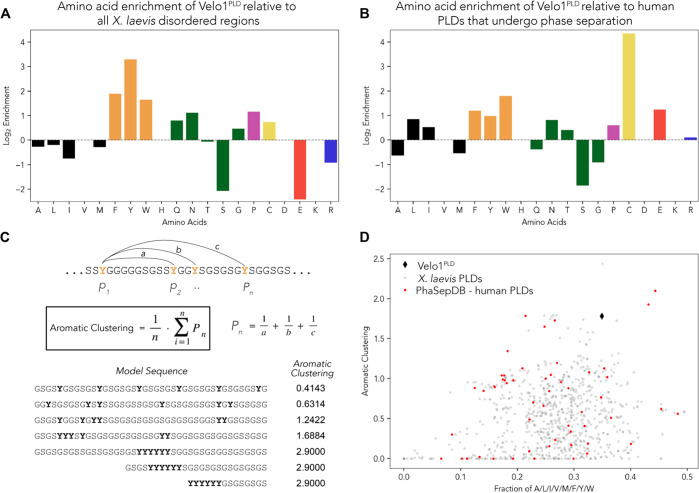
To understand why Velo1PLD is unlike other PLDs, we undertook a systematic bioinformatics analysis. To couch our analysis in the appropriate organismal context, we identified the complete set of IDRs and PLDs across the X. laevis proteome (Table S1). In comparison to all X. laevis IDRs, Velo1PLD was enriched with aromatic residues (Figure 2A), a result we interpreted to mean that like in many other PLDs, aromatic residues may act as stickers.
Figure 2.
Velo1PLD is enriched with aliphatic and aromatic residues and depleted of small polar residues and has highly clustered aromatic residues. (A) Log2 of fractions of different amino acids in Velo1PLD divided by equivalent fractions of the same amino acid across all X. laevis IDRs. (B) Log2 of fractions of different amino acids in Velo1PLD divided by equivalent fractions of the same amino acid across human PLDs that undergo liquid–liquid phase separation. (C) Graphical definition of the aromatic clustering parameter. (D) Assessment of aromatic clustering (y-axis) compared with the fraction of aromatic or aliphatic residues (x-axis). Among those of X. laevis PLDs, the clustering score Velo1PLD (black diamond) is higher than all but one, and among those of the PLDs from PhaseSepDB, the clustering score is higher than all but two.
We next considered the amino acid composition of a subset of previously studied human PLDs that undergo phase separation in vitro.103 On the basis of this analysis, Velo1PLD is depleted of G and S, modestly enriched with aromatic and aliphatic residues, and strongly enriched with cysteine (Figure 2B). The enrichment for hydrophobic residues explains the depressed disorder prediction for parts of Velo1 F1 (Figure 1B and Figure S5). Recent work has implicated the polar residues glycine (G) and serine (S) as chemically neutral spacer residues, in agreement with observations that (GS)n repeat sequences behave as physical instantiations of ideal (Gaussian) chains.22,79,104,105 Given the depletion of G and S (and modest enrichment with aliphatic residues), we wondered if the spacer amino acid composition might play a role in determining Velo1PLD phase behavior.
We finally considered the distribution of aromatic residues across Velo1PLD. The number and patterning of aromatic/hydrophobic residues have been shown to influence the phase behavior of prion-like domains.23,28,32,50,94 One of the conclusions from this work was the discovery that sticker patterning could influence the condensate material state.23,94 The authors suggested that there might be selection pressure for evenly distributed aromatic residues to impede (presumably) pathophysiological aggregation. Here, we wondered if natural sequences might use aromatic clustering to drive solid-like condensates. To identify sequences with well-clustered aromatic residues and motivated by Yang et al., we applied a metric that computes the average inverse distance among aromatic residues, which we term aromatic clustering (Figure 2C; see Methods).106 Of relevance to our application, this parameter is robust in the limit of small fractions of aromatic residues (<10%), a limit where other sequence patterning metrics struggle.107
We compared the fraction of aliphatic and aromatic residues versus aromatic clustering and found Velo1PLD among the PLDs with the greatest aromatic clustering scores (Figure 2D). This conclusion was true when compared against all Xenopus PLDs, but also against a set of human PLDs shown to undergo phase separation in vitro. As such, we wondered if this atypical aromatic clustering might play a part in Velo1’s assembly.
In summary, our bioinformatic analysis identified several features that may contribute to the anomalous behavior of Velo1PLD. In the context of the stickers-and-spacers model, these results can be considered in terms of altering the spacer:spacer or sticker:spacer strength (loss of G and S, gain of aliphatic hydrophobes) or altering the clustering or patterning of stickers. To examine how these potential sequence features might influence the thermodynamics, diffusivity, and assembly organization, we turned to simple coarse-grained simulations.
Sticker:Sticker and Sticker:Spacer Interactions Can Dictate Phase Behavior
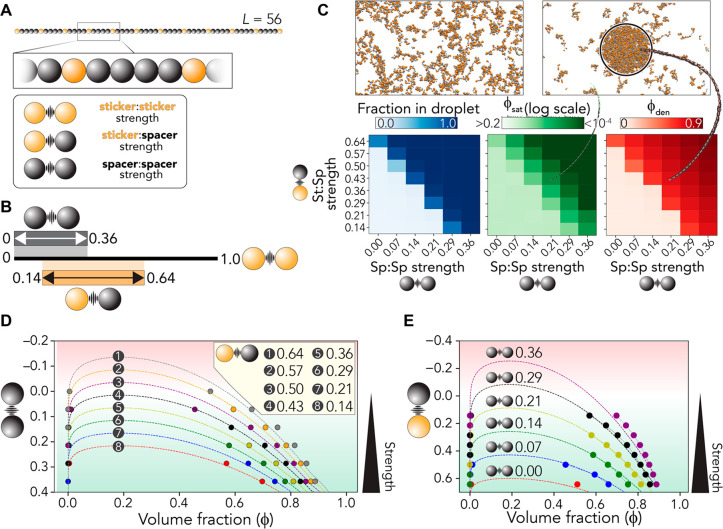
Previous work derived a simple parameter set for aromatic stickers and polar-rich spacers.23 These parameters can be qualitatively transferred across different PLDs.23 Using these same interaction parameters,23 we generated a simple 56-bead model heteropolymer with uniformly distributed stickers (Figure 3A). We used this toy system to explore how phase behavior was altered in response to changes in the sticker:sticker and sticker:spacer interaction strength (Figure 3A). To test this, we performed lattice-based Monte Carlo simulations and systematically varied the spacer:spacer and sticker:spacer interaction strengths at a fixed sticker:sticker interaction strength, concentration, and temperature (Figure 3B; see Methods).
Figure 3.
Sticker:spacer and spacer:spacer interaction strengths play a key role in determining the driving forces for phase separation. (A) Overview of the polymer models used in simulations. A 56-bead model is used (12 sticker beads, 44 spacer beads). Three parameters define the system: sticker:sticker, sticker:spacer, and spacer:spacer strength. (B) Summary of relative parameter ranges examined. The sticker:sticker strength is held fixed, and sticker:spacer and spacer:spacer interaction strengths are varied. (C) Simulations reveal that at a fixed starting volume fraction (ϕ) of 0.0168 the emergence of a two-phase regime is symmetrically dependent on the sticker:spacer and spacer:spacer interaction strength. As the interaction strength increases, ϕsat decreases, and in parallel ϕden (the concentration inside the droplet) increases. (D) Varying interaction strengths can be recast as modulating the effective Flory χ parameters or rescaling the critical temperature. As such, we can project the spacer:spacer interaction strength in the background of a fixed sticker:spacer interaction strength into a Flory–Huggins fit to analytically capture the interaction strength-dependent phase behavior. Points are simulation data, while lines are fits of data to Flory–Huggins theory. (E) Analogous analysis as in panel D but with a variable sticker:spacer strength in the background of a fixed spacer:spacer strength. Points are simulation data, while lines are fits of simulation data to Flory–Huggins theory.
Our results revealed a substantial change in the driving forces for phase separation with fractional changes in both sticker:sticker and sticker:spacer strengths (Figure 3C). We varied sticker:spacer and spacer:spacer interaction strengths by the same incremental step size (in units of sticker:sticker interaction) to assess the relative impact of each type of bead. We observed a symmetrical dependence on the driving forces for assembly in terms of the change in sticker:spacer or spacer:spacer interactions. As such, even though stickers are the “drivers” of assembly, changes that uniformly affect spacers, even by a small amount, can dramatically influence phase behavior.
To aid in the interpretation, we recast the interaction strength dependence of the phase behavior in terms of phase diagrams in the volume fraction and interaction strength plane. In doing this, we fit our data to Flory–Huggins theory to illustrate the validity of this approach and to guide the eye. Figure 3D reports how the driving force for assembly changes as a function spacer:spacer interaction strength (y-axis), while the sticker:spacer interaction strength is held fixed. Figure 3E reports the opposite of this, how phase behavior is altered when sticker:spacer interactions are varied at fixed spacer:spacer interactions.
The major takeaway from these simulations is that small changes in spacer:spacer or sticker:spacer interaction strengths can have substantial changes in the driving force for phase separation. Specifically, small changes can alter the saturation volume fraction (ϕsat) by several orders of magnitude and change the intradroplet density from 0.5 to >0.9. In the context of Velo1PLD, our results suggest that the depletion of G and S and modest enrichment with other aliphatic residues could have a substantial effect on the driving forces for phase separation, driving tighter interactions.
Intracondensate Self-Diffusion of Polymers Is Determined by the Droplet Volume Fraction
We took advantage of the ability to obtain an apparent diffusion constant (Dapp) for self-diffusion of polymers inside the droplets formed in our system (see Methods). Dapp provides a proxy for how easily polymers inside the droplet reorient themselves as a function of Monte Carlo step, offering a readout of the apparent intracondensate diffusivity (Figure 4A).
Figure 4.
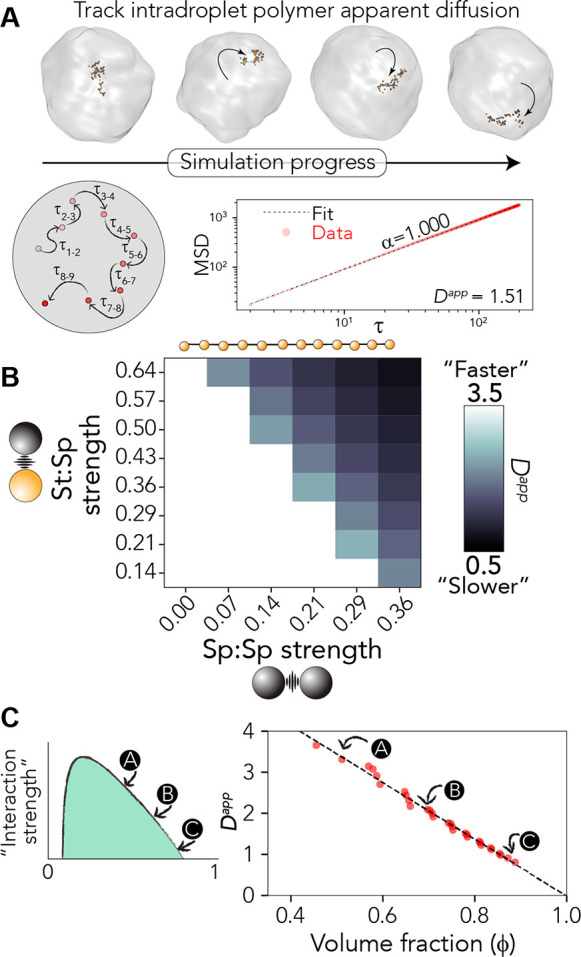
Apparent diffusion coefficient scales with droplet density. (A) Overview of the analysis approach as applied to the polymer architecture defined in Figure 3. Individual polymers are followed as they “diffuse” within a droplet and fit to extract the diffusive scaling exponent (α) and the apparent diffusion constant (Dapp). Dapp is determined only where simple Brownian diffusion is observed (i.e., α = 1), which occurs in almost all cases (see Figures S1–S3). (B) Dapp as a function of sticker:spacer and spacer:spacer strength. (C) Graphical representation of how density and Dapp relate to one another (left) and all data from panel B plotted as a single master curve of Dapp vs dense-phase volume fraction (ϕden). The linear fit to guide the eye leads to an apparent diffusion constant of 0 when the volume fraction is 1, reflecting the limit in which every lattice site is occupied such that no free sites are available for polymers to move into.
As sticker:spacer and spacer:spacer interactions become stronger, we observed a decrease in Dapp (Figure 4B). This reduction is symmetrical across the sticker:spacer and spacer:spacer interaction dimensions. A correlation of the droplet density with the apparent diffusion constant yields a linear master curve that extrapolates back to a Dapp of 0 when the volume fraction is 1.0 (Figure 4C). As such, these results suggest that for chains with an evenly distributed sticker-and-spacer architecture, changes in spacer interaction strength lead to predictable changes in material state. When cast in terms of a phase diagram, this shows the unsurprising relationship that distance from an apparent critical point tracks with decreasing condensate dynamics. In short, in this simple limiting model, the stronger the driving force for phase separation, the more solid-like a condensate is expected to appear.
Sticker Clustering Shifts Phase Boundaries, Tunes Intradroplet Self-Diffusion, and Alters Intradroplet Organization
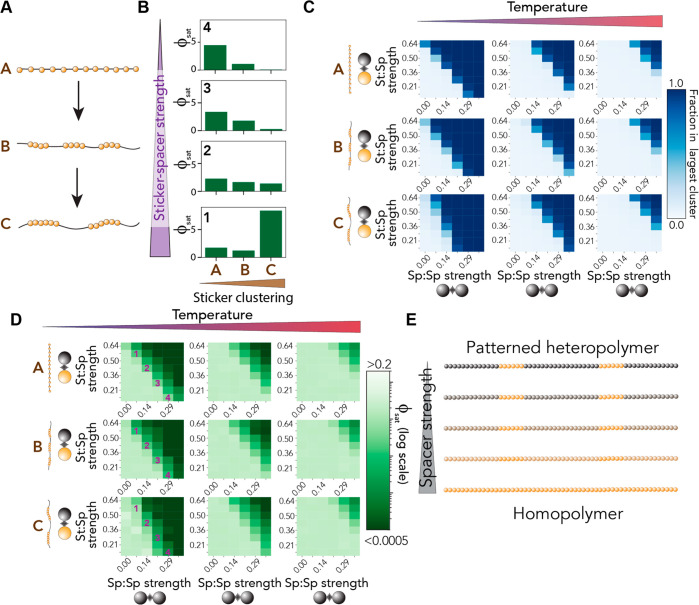
Previous experimental and theoretical work has shown that sticker patterning can determine the saturation concentration.25,36,37,81,108−111 In addition, repatterning of naturally occurring PLDs that undergo liquid–liquid phase separation revealed changes in assembly state or condensate dynamics.23,94 In agreement with this observation, recent theoretical work has shown that asymmetric sticker patterning can determine the balance between liquid–liquid phase separation and aggregation.82 To examine the interplay between sticker patterning/clustering and spacer-mediated interactions, we determined assembly behavior as a function of sticker:sticker strength, sticker:spacer strength, and sticker clustering (Figure 5A). All simulations were performed at three different temperatures to ensure that a reasonable dynamic range of behavior was observed.
Figure 5.
Impact of sticker clustering can be altered by sticker:spacer strength. (A) Three polymers of equal length with equal composition but alternative sticker clustering. (B) Saturation concentration as a function of sticker clustering (x-axis) and sticker:spacer strength (top to bottom). These specific comparisons are shown also in panel D. (C) Fraction of chains in the largest cluster shown as a function of sticker:spacer, spacer:spacer, sticker clustering (top to bottom), and system temperature (left to right). (D) Saturation concentration as a function of sticker:spacer, spacer:spacer, sticker clustering (top to bottom), and system temperature (left to right). Numbers reflect the systems examined in panel B. (E) The spacer:spacer and spacer:sticker interaction strengths can determine the impact of sticker:spacer patterning by rescaling the definition of a sticker and spacer. Spacer strength here reflects the simultaneous titration of spacer:spacer and sticker:spacer interaction strength to match that of sticker:sticker strength.
In agreement with prior work, sticker clustering reduces the ϕsat and enhances the driving force for phase separation (Figure 5B, top). The impact clustering has on ϕsat depends on the sticker:spacer interaction strength; as the sticker:spacer interaction strength increases, the impact of clustering is diminished. This behavior is also manifest in the fraction of chains found in the largest cluster (Figure 5C), where an asymmetry for the high-clustering variant is found as a function of sticker:sticker and sticker:spacer interaction strength. Finally, we also observe the same trends when ϕsat is examined across all possible combinations as opposed to just the subset shown in Figure 5B (see Figure 5D). In summary, when spacer:spacer interaction is (relatively) strong but sticker:spacer interaction is (relatively) weak, we observe the most substantial influence of clustering (Figure 5D, left column). This effect becomes weaker as the overall strength of all interactions decreases [i.e., at a higher temperature (Figure 5D, right column)].
This sticker dependence on the impact of clustering can be rationalized by recognizing that there are several extreme limits of these parameters (Figure 5D). When all three interaction strengths (sticker:sticker, sticker:spacer, and spacer:spacer) are equivalent, then sticker clustering is irrelevant; the chain is a homopolymer. As these three values become divergent, clustering effects are more strongly felt, where the sticker:spacer interaction strength acts as an effective miscibility parameter for the two residue types.
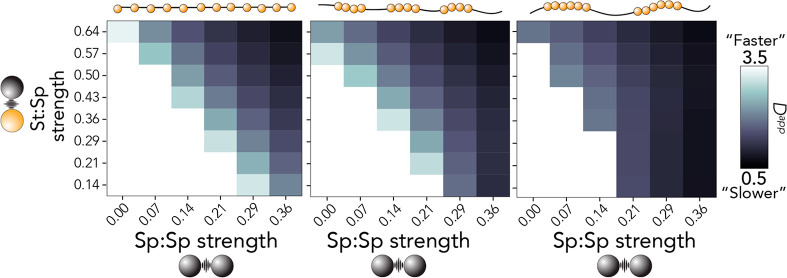
Consistent with our dependence of sticker:spacer strength on the driving force for assembly, Dapp is also influenced by sticker clustering. More clustered stickers leads to a slowing of intracondensate polymer diffusion (Figure 6), although the magnitude of this effect depends on the sticker:spacer and spacer:spacer interactions. For example, when the sticker:spacer strength is 0.14 and the spacer:spacer strength is 0.29, we observe a large value for Dapp in the chain with weak sticker clustering (Figure 6, left) but a dramatically reduced value for the chain with strong sticker clustering (Figure 6, right). If interpreted naively, these results suggest that sticker clustering can suppress liquid-like condensate dynamics for equivalently weak molecular interaction strengths.
Figure 6.
Dependence of intracondensate polymer apparent diffusion on spacer-mediated interactions that depends on sticker clustering. As the level of sticker clustering increases for polymers with a strong spacer:spacer interaction, there is little to no dependence of Dapp on sticker:spacer interactions. This suggests that in the limit of (relatively) strong spacer:spacer interactions, molecular rearrangement is dominated by spacer:spacer and sticker:sticker interactions.
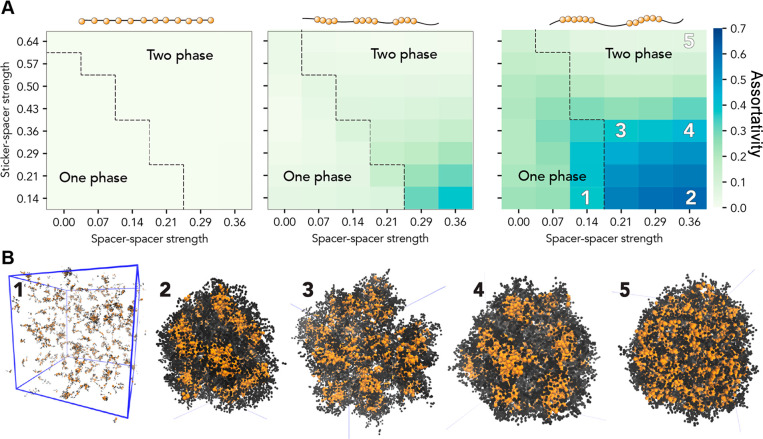
Finally, we wondered how altering sticker clustering might impact the intradroplet organization. To formally evaluate the intradroplet organization, we used the measure of assortativity to describe the spatial demixing of sticker beads and spacer beads (Figure 7). An assortativity value of 1 means stickers are in contact with only other stickers, while an assortativity value of 0 means stickers are in contact with spacers and stickers an equal amount. This analysis revealed that highly clustered sequences showed assortativity values of ≫0, revealing complex (yet labile) interdroplet organization driven by the relative strengths of stickers and spacers.
Figure 7.
Sticker clustering tunes intradroplet organization. (A) Assortativity, a measure of the spatial mixing between stickers and spacers, is measured for the entire system as a function of sticker strength, spacer strength, and sticker clustering. For each system, the phase boundary is shown as a dashed line for reference. For the well-clustered sequences, significant deviations from a value of 0 are observed. (B) To understand the origins of these large assortativity values, we generated snapshots from distinct regions in panel A (orange beads are stickers, and black beads spacers). These revealed the intradroplet organization of stickers into local clusters and subdomains. For sequences with the most well-clustered stickers, we observe an assortativity of >0 at subsaturating concentrations due to the presence of small labile clusters.
In summary, our simulations imply several general principles that we wanted to examine in the context of Velo1PLD. First, spacer-mediated interactions can tune droplet density and hence material state and molecular rearrangement. Moreover, for polymers with evenly spaced stickers, energetically equivalent changes in sticker:spacer and spacer:spacer interactions lead to approximately equivalent changes in the driving force for assembly. Second, sticker clustering can tune the driving force for assembly, intradroplet organization, and (naively interpreted) condensate dynamics, where the impact of sticker clustering is itself determined by spacer-mediated interactions or, analogously, the strength of sticker:sticker interactions.
The Rational Design of Velo1PLD Allows Us to Test the Importance of Distinct Features in the Condensate Assembly and State
While our simulations implicated several parameters in the context of determining condensate properties, our bioinformatic analysis provides a lens through which those insights can be translated into interpretable signatures in the context of protein sequence. In particular, sticker composition and clustering emerge as parameters we predict should substantially alter condensate dynamics. We took a rational design approach to test these predictions, whereby we made targeted mutations to Velo1PLD to alter these sequence features.
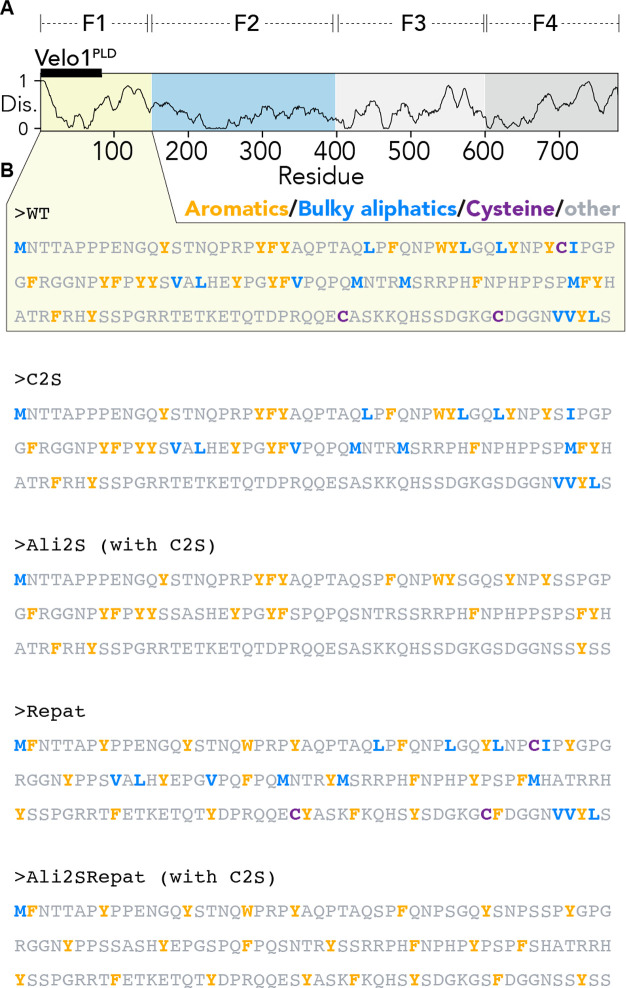
We focused on the F1 fragment of Velo1 (residues 1–150), which contains the PLD (Figures 1 and 8A). This subregion was studied extensively in previous work and sets a consistent and comparable baseline against which our rational designs can be compared.39 As such, our designs focus on this 150-residue disordered region as a model system to understand the sequence determinants of the assembly and material state.
Figure 8.
Architecture of Velo1 and amino acid sequences of F1 fragment variants. (A) Relative position of the F1 fragment. (B) Amino acid sequence of the wild type sequence and rationally designed F1 variants tested.
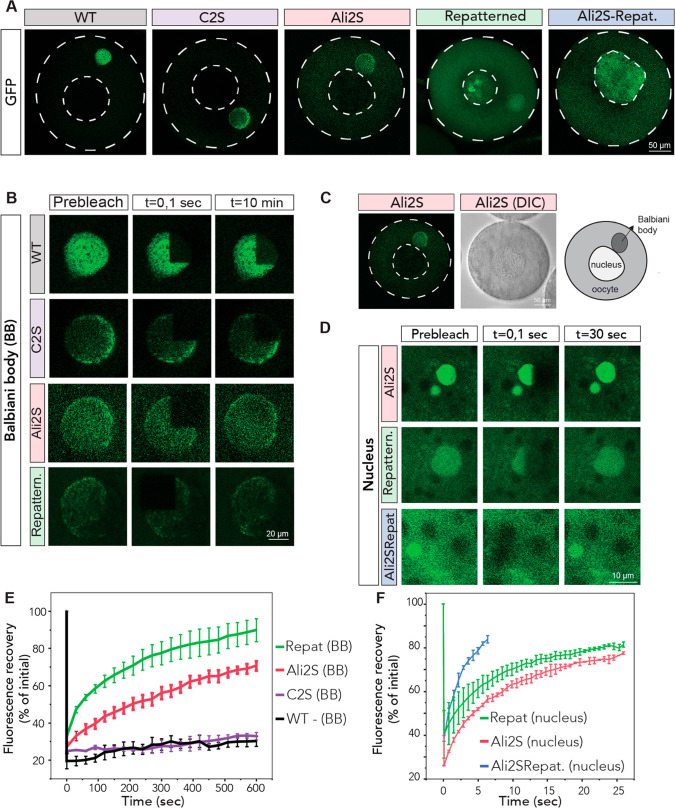
We isolated oocytes from X. laevis ovaries as described previously and injected these with mRNAs encoding our designs of Velo1 followed by GFP.39 After overnight incubation, oocytes were imaged with confocal microscopy. In agreement with previous work, wild type fragment F1 localizes to the Balbiani body and has a very slow recovery rate after photobleaching, indicating its solid-like material status39 (Figure 9A,B,D). We previously showed that disrupting Velo1PLD causes the F1 mutants to be soluble in the cytoplasm and recover much faster after photobleaching in the Balbiani body, which suggests that Velo1PLD mutants are soluble proteins.39 Thus, we used these two properties, namely, the ability of the newly translated protein to self-assemble with the endogenous Velo1 (i.e., localization to the Balbiani body) and the recovery time after photobleaching (i.e., forming solid- or liquid-like assemblies), to test the material properties of new designs of Velo1.
Figure 9.
Rationally designed Velo1 designs tune cellular localization and material state. (A) mRNAs encoding Velo1 designs and wild type Velo1 fragment F1 (Figure 8) fused to GFP were microinjected into stage I X. laevis oocytes. Oocytes were left to recover and express injected mRNAs overnight and imaged the next day. The cell membrane and nucleus are outlined in a white dashed line. (B) Internal rearrangement of fluorescent wild type or redesigned Velo1 (F1-GFP) particles after photobleaching in the Balbiani body. Note that Velo1Ali2S/Repat did not localize to the Balbiani body. (C) Overview, DIC image, and schematic of the oocyte with its Balbiani body in the Ali2S design. (D) Internal rearrangement of fluorescent redesigned Velo1 (F1-GFP) particles after photobleaching in the nucleus. Note that Velo1WT does not localize to the nucleus. (E) The fluorescent recoveries of the photobleached Velo1WT or redesigned Velo1 (F1-GFP) particles in Balbiani bodies in panel B and two other biological replicates were quantified over time. (F) The fluorescent recoveries of photobleached Velo1 designs in nuclei in panel C and two other biological replicates were quantified over time. For panels D and E, the fluorescence in the bleached region was quantified over time and normalized by an unbleached neighboring region. At least three oocytes per biological replicate were plotted. Scale bars are as indicated in the figure.
Velo1 Cysteine Residues Do Not Dictate Solid-like Properties
We first wondered if Velo1’s solid-like material state was a consequence of its high cysteine content. In principle, the reducing environment of the cell should impede disulfide bond formation. However, the chemical state within a condensate is poorly defined and could plausibly offer a microenvironment in which disulfide bond formation can occur. We generated a variant of Velo1 in which all three cysteines (C) were converted to serines (S) (Velo1C2S) (Figure 8B). Condensates formed from Velo1C2S were wild type-like in terms of morphology and dynamics (Figure 9A,B,E). As such, we conclude that intracondensate covalent cross-links formed by C do not underlie the material state of Velo1PLD in this experimental assay.
The Hydrophobicity of Velo1 Spacer Regions Can Tune the Material State
We next asked whether Velo1’s solid-like material state was a consequence of the amino acid composition of the spacer residues. If we treat aromatic residues as stickers and other residues as spacers, our bioinformatic analysis implies that Velo1PLD spacer regions are depleted of small polar amino acids (S and G) and modestly enriched with large aliphatic residues (L and I) (Figure 2). Moreover, our simulations show that tuning spacer interaction strength can alter the saturation concentration and the dynamics of intracondensate molecules (Figures 3 and 4). To test if the presence of aliphatic hydrophobic residues strengthened spacer-mediated attractive interactions, we generated a variant in which all large aliphatic side chain-containing residues (L, M, V, and I) were converted to S (Velo1Ali2S) and all C’s to S (Figure 8B).
Intriguingly, we observed that Velo1Ali2S shows two different types of condensate-associated behavior in oocytes: liquid-like recovery in condensates in the nucleus, which had spherical shapes and recovered within seconds after photobleaching, and recruitment to the Balbiani body (Figure 9A–C). We also noted that Velo1Ali2S was more soluble in the cytoplasm than the wild type, implying a reduction in the driving force for assembly. After photobleaching, Velo1Ali2S associated with the Balbiani body recovered much faster than the wild type (Figure 9E), suggesting Velo1Ali2S is more fluid than the wild type protein. Thus, we conclude that aliphatic side chains contribute to the solid-like material properties of Velo1.
Aromatic Clustering Fundamentally Changes the Dynamics and Localization of Velo1
Velo1 emerged as the PLD with the highly clustered aromatic residues in our bioinformatics analysis (Figure 2D). Our simulations also implicated sticker patterning as a critical determinant of condensate formation, material state, and intracondensate organization (Figures 5 and 6). To test if aromatic clustering mattered for intracellular solid-like condensate assembly, we generated a repatterned variant in which all aromatics were evenly spaced (i.e., minimally clustered, reducing the clustering score from 1.8 to 0.8), which we named Velo1repat.
Much like Velo1Ali2S, Velo1repat displayed two kinds of condensate-associated behavior, with assembly into liquid-like spherical nuclear condensates and recruitment to the Balbiani body where Velo1repat dynamics are slower (Figure 9). However, the Balbiani body-localized Velo1repat recovered much faster than either the wild type or Velo1Ali2S after photobleaching, displaying rapid internal dynamics (Figure 9A,B,E). This suggests that aromatic clustering (i.e., sticker patterning) plays an important role in defining the material state of Velo1, and only shuffling aromatic residues without affecting the rest of the protein makes Velo1 more dynamic and soluble.
Sticker Clustering and Spacer Composition Are Orthogonal Parameters through Which the Material State Can Be Tuned
Our simulations implied that sticker clustering and spacer interaction strength should be partially independent of one another; i.e., a design that reduced spacer interaction strength and sticker clustering would be more liquid-like than either separately. To test this, we generated a combination design that combined the features introduced in Velo1Ali2S and Velo1Repat to generate a new design, Velo1Ali2SRepat. In agreement with our expectations, Velo1Ali2SRepat fails to localize to Balbiani bodies and forms highly dynamic nuclear condensates that show the fastest recovery of all designs examined (Figure 9A,C,F). It is also the most soluble of the designs, implying the weakest driving forces for assembly.
Discussion
The stickers-and-spacers framework is a conceptually simple model through which the assembly of biomolecules can be rationally interpreted. Here, we combined bioinformatics and simple coarse-grained simulations to motivate distinct axes upon which the material state of condensates can be tuned. Using a model system that naturally forms solid-like condensates, we found that the molecular dynamics of Velo1PLD can be tuned by varying the composition of spacer residues, the patterning of sticker residues, or both. As such, we tentatively suggest our results provide an additional set of principles through which the material state and driving force for assembly of designer condensates could be encoded.17
The material state of a biomolecular condensate dictates the molecular motion and temporal progression of components enclosed within that assembly. For dynamic liquid-like condensates, there is generally a rapid exchange of molecules between the dense and dilute phases. As a result of this rapid exchange and the ability of the condensate composition to be tuned by the total concentration of components, liquid-like assemblies have been proposed to offer a means by which cells can coordinate complex stimulus-responsive function.1−3,112,113
In contrast to liquid-like condensates, in the context of protection and tolerance from abiotic stress (e.g., heat, desiccation, etc.), solid-like gels or glasses have emerged as the de facto mechanism through which molecular protection is arrived at across many kingdoms of life.114−117 Given that the Balbiani body must lie in a dormant oocyte for long periods, a speculative role for the solid-like material state of Velo1-derived assemblies is one in which the molecular motion within Balbiani bodies is sufficiently retarded that from the frame of reference of components therein time effectively slows down. In this way, upon disassembly, components can be released in a relatively unaged state. Given Velo1PLD emerges as an outlier with respect to sequence features we have shown to play a role in its material state (Figures 2 and 8), we speculate this may reflect an evolutionarily selected set of sequence features.
We applied simple stickers-and-spacers simulations to motivate the redesign of Velo1PLD. We emphasize that our stickers-and-spacers model represents a convenient tool that translates between emergent properties and molecule features but should not be taken as a literal description of the underlying physical chemistry that determines condensate assembly and properties.69,76 For example, we focused here on hydrophobic residues and aromatic clustering, but undoubtedly, additional layers of regulatory biology may be encoded by other residue types, PTMs, or solution-dependent effects.35,68,118 As such, far from representing the “end” in our understanding of how sequence-encoded emergent properties arise, a stickers-and-spacers description can be treated as the “null model”. Deviations from the simple expected behavior are hallmarks of more complex physical chemistry. This does not prevent the stickers-and-spacers model from offering a predictive framework, but it in parallel should be taken as a simplified model that is coarse-grained along the principle axes that determine the phase boundary(s) of a system.22,23,80,119
Our results suggest sticker clustering can alter condensate dynamics and that this contribution can be tuned by spacer-mediated interactions. This conclusion is in line with recent theoretical work, which similarly has suggested that the patterning of sticker residues can determine molecular rearrangement and condensate viscosity.81,119 We note that while our simulations imply an ∼4-fold change in diffusion, FRAP experiments imply an even greater dependence on sticker clustering. We speculate that this discrepancy reflects the fact that Velo1 can also form amyloids. Although our designs do not inherently impede condensate formation, they may disrupt amyloid formation, which we anticipate may be a process physically distinct from the interactions that (at least initially) drive condensate formation.
Given the prior observation that many low-complexity domains possess evenly spaced aromatic residues, it seems plausible this may reflect selection against sequences primed to undergo kinetic arrest upon self-assembly. Indeed, slow condensate dynamics can emerge either as an equilibrium phenomenon driven by a high density of molecular components (Figure 4) or through kinetic arrest of frustrated systems that become trapped, as shown in the context of prior work examining protein:RNA assemblies.40 Distinguishing between these two origins in living cells is challenging given both can be suppressed through active (energy-dependent) processes, although the two are inherently coupled, and from the perspective of biological selection, the physical basis may be irrelevant.
In summary, our results implicate both sticker clustering and spacer composition as determinants of the condensate material state and the driving forces for assembly through both simulations and live cell imaging. Although these results implicate a possible layer of sequence-encoded regulatory control of condensate material properties, our study is not without limitations. The stickers-and-spacers model is extremely simple and should be viewed as a numerical instantiation of analytical theory with finite-size effects directly captured, as opposed to a true representation of protein physical chemistry. As such, the relative contributions of stickers and/or spacers uncovered here may be different in the context of real proteins. Furthermore, additional sequence features (e.g., charged residues) are likely to also contribute and may dominate hydrophobic interactions. Finally, our live cell imaging reveals the recruitment of Velo1 and its associated designs to the Balbiani body or to nuclear condensates. In the case of nuclear assemblies, we cannot exclude the possibility that we are observing recruitment to an existing nuclear condensate. This does not fundamentally alter our conclusions but may suggest that our variants have substantially weaker driving forces for self-assembly (but stronger driving forces for heterotypic recruitment). In short, while these limitations do not undermine our general conclusions, they should be taken into consideration when future studies are considered and designed.
Methods
Reproducibility
All code, sequence information, and bioinformatics data are provided at https://github.com/holehouse-lab/supportingdata/tree/master/2021/Holehouse_velo1_2021.
Bioinformatics
Prion-like domains were identified using the PLAAC with default settings.90 Disorder was predicted using metapredict.120 The X. laevis proteome (UP000186698) was obtained in May 2021.
The aromatic clustering parameter was inspired by an approach developed to cluster surface residues in the context of structural analysis.106 Similar to an inverse distance weight, functionally aromatic clustering is calculated by first summing the inverse distance between all pairs of aromatic residues for each residue. To obtain a single parameter for the sequence of interest, an average across all of the aromatic positions is taken. The result is a value that directly compares the positioning of aromatic residues relative to one another. The higher the aromatic clustering score, the more clustered the aromatics are in the sequence.
Simulations
All simulations were performed using the PIMMS simulation engine with 300 separate polymers on a 100 × 100 × 100 cubic lattice.23 Three independent simulations were run for every system, with a total of (on average) 3 × 1010 MC steps per simulation. All keyfiles, parameter files, and setup scripts for running all simulations are provided at https://github.com/holehouse-lab/supportingdata/tree/master/2021/Holehouse_velo1_2021.
The fraction of chains in the largest cluster was defined on the basis of the connected network, i.e., the set of chains in direct physical contact with one another, as described previously.23,40
The saturation concentrations were computed by taking the fraction of chains not in the largest cluster as a function of total volume, as described previously.23 In all simulations, chains partitioned into either one large cluster or monomeric/small oligomers.
The interdroplet density (Figure 3) was computed by generating radial density profiles and averaging over the central core, as described previously.23 For an example of this analysis across three independent simulations for the same system, see Figure S4.
The evolution of the intracondensate polymer position is determined by reptation-based Monte Carlo moves where each chain is perturbed an equal number of times. As such, while we do not obtain a time scale in seconds, the ability to fit the polymer mean square displacement (MSD) to the Monte Carlo step and obtain a normal diffusivity exponent (α = 1.0) reflects our ability to analyze molecular rearrangement as a proxy for bona fide molecular kinetics. In all cases, Dapp is obtained only in the limit when simple apparent diffusion is observed. Details (including every MSD vs τ fit for every simulation) and additional analyses are provided at https://github.com/holehouse-lab/supportingdata/tree/master/2021/Holehouse_velo1_2021. This approach is appropriate for identically sized symmetrical polymers with identical movesets, but we urge caution when interpreting Monte Carlo-derived molecular rearrangement through the lens of dynamics.
We used assortativity121 to quantify the propensity of polymers in our coarse-grained system to form sticker:sticker and spacer:spacer interactions versus sticker:spacer interactions. For a given frame of the simulation, we made every bead its own node in a graph using the Python package NetworkX.122 Every pair of adjacent beads in the lattice (that were not immediately next to one another on the same polymer) was considered to be an edge between nodes. We then calculated the assortativity coefficient of this graph using NetworkX’s assortativity algorithm. A perfectly assortative network (i.e., stickers interact with only stickers, and spacers with only spacers) has a coefficient value of 1. Disassortative networks have a negative coefficient, and randomly distributed networks have coefficients near zero. For each simulation, we computed and averaged the assortativity coefficients across 20 frames sampled uniformly across the last 400 frames of the simulation. The average assortativity was computed for each replica, and the average across three replicas taken. The standard error of the mean for these calculations is shown in Figure S6.
Fits of the data to Flory–Huggins theory were performed as described previously, with the three-body interaction coefficient set to zero for the sake of simplicity.23
Animal Work
X. laevis adult females were purchased from Nasco and maintained in the animal facility of the Barcelona Biomedical Research Park (PRBB, Barcelona, Spain) in water tanks with the following controlled conditions: 18–21 °C, pH 6.8–7.5, 4–20 ppm O2, conductivity of 500–1500 μs, and <0.1 ppm ammonia. All animals were sacrificed by accredited animal facility personnel before their ovaries were extracted.
Isolation, Injection, and Culturing of Oocytes
Xenopus oocytes were isolated with slight modifications to the method described in ref (39). Briefly, ovaries were digested using Collagenase IA (Sigma, C9891-1G) in MMR by gentle rocking, until most oocytes were dissociated. After several washes in MMR to remove the collagenase, stage I oocytes were separated from the rest by passing the oocyte mixture through two sets of filter meshes (Spectra/Mesh, 146424 and 146426). Later, stage I oocytes were stripped of accompanying granulosa cells by being treated with 10 mg/mL trypsin for 1 min and cultured in OCM.123
Oocytes were injected with mRNAs encoding the indicated proteins in the text by using a Femtojet Microinjector equipped with an Injectman micromanipulator (Eppendorf) such that 100 pL would be delivered in each injection. Injected oocytes were left to recover overnight and imaged the next day.
DNA and RNA Constructs
Velo1 designs were ordered from Integrated DNA Technologies in the form of gblocks and cloned into pCS2-EGFP vectors. The resulting plasmids were sequenced before any downstream application. Plasmids were linearized by NotI digestion and gel-purified before being used as templates for in vitro mRNA transcription (mMessage mMachine SP6 transcription kit, Thermo). mRNAs were cleaned via lithium acetate precipitation and suspended in RNase free water.
Confocal Live Cell Imaging
Oocytes were imaged using a 40× water immersion objective (NA 1.10, Leica, 506357) in OCM at room temperature and atmospheric air using a Leica TCS SP8 microscope with the Leica Application Suite X (LAS X) software.
Acknowledgments
E.B. thanks the Advanced Light Microscopy Unit of CRG for its support.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.biochem.1c00465.
Full sequence comparison (PDF)
Accession Codes
The UniProt accession number for Velo1 is Q7T226. This is not the canonical isoform as defined in the reference X. laevis proteome (which is B7ZQY0), although these two sequences differ by a single N569D sequence difference that falls outside of all regions relevant to this study, and as such, this is not a relevant detail; we include this explanation only for the sake of clarity and transparency.
Funding for this project was provided by the Longer Life Foundation (and RGA/Washington University Collaboration, to A.S.H.), National Science Foundation Grant DGE-2139839 (to D.G.), and an ERC Starting Grant (ERC-StG-2017 759107) and MINECO’s Proyectos de Excelencia (BFU2017-89373-P) (both to E.B.). E.B. also acknowledges support from the Spanish Ministry of Science and Innovation to the EMBL partnership, the Centro de Excelencia Severo Ochoa, and the CERCA Programme/Generalitat de Catalunya.
The authors declare the following competing financial interest(s): A.S.H. is a scientific consultant for Dewpoint Therapeutics.
Supplementary Material
References
- Lyon A. S.; Peeples W. B.; Rosen M. K. A Framework for Understanding the Functions of Biomolecular Condensates across Scales. Nat. Rev. Mol. Cell Biol. 2021, 22, 215. 10.1038/s41580-020-00303-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banani S. F.; Lee H. O.; Hyman A. A.; Rosen M. K. Biomolecular Condensates: Organizers of Cellular Biochemistry. Nat. Rev. Mol. Cell Biol. 2017, 18 (5), 285–298. 10.1038/nrm.2017.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin Y.; Brangwynne C. P. Liquid Phase Condensation in Cell Physiology and Disease. Science 2017, 357 (6357), eaaf4382. 10.1126/science.aaf4382. [DOI] [PubMed] [Google Scholar]
- Boeynaems S.; Alberti S.; Fawzi N. L.; Mittag T.; Polymenidou M.; Rousseau F.; Schymkowitz J.; Shorter J.; Wolozin B.; Van Den Bosch L.; Tompa P.; Fuxreiter M. Protein Phase Separation: A New Phase in Cell Biology. Trends Cell Biol. 2018, 28 (6), 420–435. 10.1016/j.tcb.2018.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho W.-K.; Spille J.-H.; Hecht M.; Lee C.; Li C.; Grube V.; Cisse I. I. Mediator and RNA Polymerase II Clusters Associate in Transcription-Dependent Condensates. Science 2018, 361 (6400), 412–415. 10.1126/science.aar4199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feric M.; Vaidya N.; Harmon T. S.; Mitrea D. M.; Zhu L.; Richardson T. M.; Kriwacki R. W.; Pappu R. V.; Brangwynne C. P. Coexisting Liquid Phases Underlie Nucleolar Subcompartments. Cell 2016, 165 (7), 1686–1697. 10.1016/j.cell.2016.04.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brangwynne C. P.; Eckmann C. R.; Courson D. S.; Rybarska A.; Hoege C.; Gharakhani J.; Juelicher F.; Hyman A. A. Germline P Granules Are Liquid Droplets That Localize by Controlled Dissolution/condensation. Science 2009, 324 (5935), 1729–1732. 10.1126/science.1172046. [DOI] [PubMed] [Google Scholar]
- Hyman A. A.; Weber C. A.; Jülicher F. Liquid-Liquid Phase Separation in Biology. Annu. Rev. Cell Dev. Biol. 2014, 30, 39–58. 10.1146/annurev-cellbio-100913-013325. [DOI] [PubMed] [Google Scholar]
- Wang J. T.; Smith J.; Chen B.-C.; Schmidt H.; Rasoloson D.; Paix A.; Lambrus B. G.; Calidas D.; Betzig E.; Seydoux G. Regulation of RNA Granule Dynamics by Phosphorylation of Serine-Rich, Intrinsically Disordered Proteins in C. Elegans. eLife 2014, 3, e04591. 10.7554/eLife.04591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putnam A.; Cassani M.; Smith J.; Seydoux G. A Gel Phase Promotes Condensation of Liquid P Granules in Caenorhabditis Elegans Embryos. Nat. Struct. Mol. Biol. 2019, 26 (3), 220–226. 10.1038/s41594-019-0193-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders D. W.; Kedersha N.; Lee D. S. W.; Strom A. R.; Drake V.; Riback J. A.; Bracha D.; Eeftens J. M.; Iwanicki A.; Wang A.; Wei M.-T.; Whitney G.; Lyons S. M.; Anderson P.; Jacobs W. M.; Ivanov P.; Brangwynne C. P. Competing Protein-RNA Interaction Networks Control Multiphase Intracellular Organization. Cell 2020, 181 (2), 306–324.e28. 10.1016/j.cell.2020.03.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang P.; Mathieu C.; Kolaitis R.-M.; Zhang P.; Messing J.; Yurtsever U.; Yang Z.; Wu J.; Li Y.; Pan Q.; Yu J.; Martin E. W.; Mittag T.; Kim H. J.; Taylor J. P. G3BP1 Is a Tunable Switch That Triggers Phase Separation to Assemble Stress Granules. Cell 2020, 181 (2), 325–345.e28. 10.1016/j.cell.2020.03.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillén-Boixet J.; Kopach A.; Holehouse A. S.; Wittmann S.; Jahnel M.; Schlüßler R.; Kim K.; Trussina I. R. E. A.; Wang J.; Mateju D.; Poser I.; Maharana S.; Ruer-Gruß M.; Richter D.; Zhang X.; Chang Y.-T.; Guck J.; Honigmann A.; Mahamid J.; Hyman A. A.; Pappu R. V.; Alberti S.; Franzmann T. M. RNA-Induced Conformational Switching and Clustering of G3BP Drive Stress Granule Assembly by Condensation. Cell 2020, 181 (2), 346–361.e17. 10.1016/j.cell.2020.03.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molliex A.; Temirov J.; Lee J.; Coughlin M.; Kanagaraj A. P.; Kim H. J.; Mittag T.; Taylor J. P. Phase Separation by Low Complexity Domains Promotes Stress Granule Assembly and Drives Pathological Fibrillization. Cell 2015, 163 (1), 123–133. 10.1016/j.cell.2015.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riback J. A.; Katanski C. D.; Kear-Scott J. L.; Pilipenko E. V.; Rojek A. E.; Sosnick T. R.; Drummond D. A. Stress-Triggered Phase Separation Is an Adaptive, Evolutionarily Tuned Response. Cell 2017, 168 (6), 1028–1040.e19. 10.1016/j.cell.2017.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry J.; Weber S. C.; Vaidya N.; Haataja M.; Brangwynne C. P. RNA Transcription Modulates Phase Transition-Driven Nuclear Body Assembly. Proc. Natl. Acad. Sci. U. S. A. 2015, 112 (38), E5237–E5245. 10.1073/pnas.1509317112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastings R. L.; Boeynaems S. Designer Condensates: A Toolkit for the Biomolecular Architect. J. Mol. Biol. 2021, 433 (12), 166837. 10.1016/j.jmb.2021.166837. [DOI] [PubMed] [Google Scholar]
- Lasker K.; Boeynaems S.; Lam V.; Stainton E.; Jacquemyn M.; Daelemans D.; Villa E.; Holehouse A. S.; Gitler A.; Shapiro L. A Modular Platform for Engineering Function of Natural and Synthetic Biomolecular Condensates. bioRxiv 2021, 10.1101/2021.02.03.429226. [DOI] [Google Scholar]
- Schuster B. S.; Reed E. H.; Parthasarathy R.; Jahnke C. N.; Caldwell R. M.; Bermudez J. G.; Ramage H.; Good M. C.; Hammer D. A. Controllable Protein Phase Separation and Modular Recruitment to Form Responsive Membraneless Organelles. Nat. Commun. 2018, 9 (1), 2985. 10.1038/s41467-018-05403-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dzuricky M.; Rogers B. A.; Shahid A.; Cremer P. S.; Chilkoti A. De Novo Engineering of Intracellular Condensates Using Artificial Disordered Proteins. Nat. Chem. 2020, 12 (9), 814–825. 10.1038/s41557-020-0511-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zervoudis N. A.; Obermeyer A. C. The Effects of Protein Charge Patterning on Complex Coacervation. Soft Matter 2021, 17 (27), 6637–6645. 10.1039/D1SM00543J. [DOI] [PubMed] [Google Scholar]
- Wang J.; Choi J.-M.; Holehouse A. S.; Lee H. O.; Zhang X.; Jahnel M.; Maharana S.; Lemaitre R.; Pozniakovsky A.; Drechsel D.; Poser I.; Pappu R. V.; Alberti S.; Hyman A. A. A Molecular Grammar Governing the Driving Forces for Phase Separation of Prion-like RNA Binding Proteins. Cell 2018, 174 (3), 688–699.e16. 10.1016/j.cell.2018.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin E. W.; Holehouse A. S.; Peran I.; Farag M.; Incicco J. J.; Bremer A.; Grace C. R.; Soranno A.; Pappu R. V.; Mittag T. Valence and Patterning of Aromatic Residues Determine the Phase Behavior of Prion-like Domains. Science 2020, 367 (6478), 694–699. 10.1126/science.aaw8653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brady J. P.; Farber P. J.; Sekhar A.; Lin Y.-H.; Huang R.; Bah A.; Nott T. J.; Chan H. S.; Baldwin A. J.; Forman-Kay J. D.; Kay L. E. Structural and Hydrodynamic Properties of an Intrinsically Disordered Region of a Germ Cell-Specific Protein on Phase Separation. Proc. Natl. Acad. Sci. U. S. A. 2017, 114 (39), E8194–E8203. 10.1073/pnas.1706197114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pak C. W.; Kosno M.; Holehouse A. S.; Padrick S. B.; Mittal A.; Ali R.; Yunus A. A.; Liu D. R.; Pappu R. V.; Rosen M. K. Sequence Determinants of Intracellular Phase Separation by Complex Coacervation of a Disordered Protein. Mol. Cell 2016, 63 (1), 72–85. 10.1016/j.molcel.2016.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das S.; Lin Y.-H.; Vernon R. M.; Forman-Kay J. D.; Chan H. S. Comparative Roles of Charge, π, and Hydrophobic Interactions in Sequence-Dependent Phase Separation of Intrinsically Disordered. Proc. Natl. Acad. Sci. U. S. A. 2020, 117 (46), 28795–28805. 10.1073/pnas.2008122117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernon R. M.; Chong P. A.; Tsang B.; Kim T. H.; Bah A.; Farber P.; Lin H.; Forman-Kay J. D. Pi-Pi Contacts Are an Overlooked Protein Feature Relevant to Phase Separation. eLife 2018, 7, e31486. 10.7554/eLife.31486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y.; Currie S. L.; Rosen M. K. Intrinsically Disordered Sequences Enable Modulation of Protein Phase Separation through Distributed Tyrosine Motifs. J. Biol. Chem. 2017, 292, 19110. 10.1074/jbc.M117.800466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quiroz F. G.; Chilkoti A. Sequence Heuristics to Encode Phase Behaviour in Intrinsically Disordered Protein Polymers. Nat. Mater. 2015, 14 (11), 1164–1171. 10.1038/nmat4418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng X.; Holehouse A. S.; Chilkoti A.; Mittag T.; Pappu R. V. Connecting Coil-to-Globule Transitions to Full Phase Diagrams for Intrinsically Disordered Proteins. Biophys. J. 2020, 119 (2), 402–418. 10.1016/j.bpj.2020.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P.; Banjade S.; Cheng H.-C.; Kim S.; Chen B.; Guo L.; Llaguno M.; Hollingsworth J. V.; King D. S.; Banani S. F.; Russo P. S.; Jiang Q.-X.; Nixon B. T.; Rosen M. K. Phase Transitions in the Assembly of Multivalent Signalling Proteins. Nature 2012, 483 (7389), 336–340. 10.1038/nature10879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato M.; Han T. W.; Xie S.; Shi K.; Du X.; Wu L. C.; Mirzaei H.; Goldsmith E. J.; Longgood J.; Pei J.; Grishin N. V.; Frantz D. E.; Schneider J. W.; Chen S.; Li L.; Sawaya M. R.; Eisenberg D.; Tycko R.; McKnight S. L. Cell-Free Formation of RNA Granules: Low Complexity Sequence Domains Form Dynamic Fibers within Hydrogels. Cell 2012, 149 (4), 753–767. 10.1016/j.cell.2012.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han T. W.; Kato M.; Xie S.; Wu L. C.; Mirzaei H.; Pei J.; Chen M.; Xie Y.; Allen J.; Xiao G.; McKnight S. L. Cell-Free Formation of RNA Granules: Bound RNAs Identify Features and Components of Cellular Assemblies. Cell 2012, 149 (4), 768–779. 10.1016/j.cell.2012.04.016. [DOI] [PubMed] [Google Scholar]
- Murray D. T.; Kato M.; Lin Y.; Thurber K. R.; Hung I.; McKnight S. L.; Tycko R. Structure of FUS Protein Fibrils and Its Relevance to Self-Assembly and Phase Separation of Low-Complexity Domains. Cell 2017, 171 (3), 615–627.e16. 10.1016/j.cell.2017.08.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krainer G.; Welsh T. J.; Joseph J. A.; Espinosa J. R.; Wittmann S.; de Csilléry E.; Sridhar A.; Toprakcioglu Z.; Gudiškytė G.; Czekalska M. A.; Arter W. E.; Guillén-Boixet J.; Franzmann T. M.; Qamar S.; George-Hyslop P. S.; Hyman A. A.; Collepardo-Guevara R.; Alberti S.; Knowles T. P. J. Reentrant Liquid Condensate Phase of Proteins Is Stabilized by Hydrophobic and Non-Ionic Interactions. Nat. Commun. 2021, 12 (1), 1085. 10.1038/s41467-021-21181-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y.-H.; Chan H. S. Phase Separation and Single-Chain Compactness of Charged Disordered Proteins Are Strongly Correlated. Biophys. J. 2017, 112 (10), 2043–2046. 10.1016/j.bpj.2017.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y.-H.; Forman-Kay J. D.; Chan H. S. Sequence-Specific Polyampholyte Phase Separation in Membraneless Organelles. Phys. Rev. Lett. 2016, 117 (17), 178101. 10.1103/PhysRevLett.117.178101. [DOI] [PubMed] [Google Scholar]
- Nott T. J.; Petsalaki E.; Farber P.; Jervis D.; Fussner E.; Plochowietz A.; Craggs T. D.; Bazett-Jones D. P.; Pawson T.; Forman-Kay J. D.; Baldwin A. J. Phase Transition of a Disordered Nuage Protein Generates Environmentally Responsive Membraneless Organelles. Mol. Cell 2015, 57 (5), 936–947. 10.1016/j.molcel.2015.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boke E.; Ruer M.; Wühr M.; Coughlin M.; Lemaitre R.; Gygi S. P.; Alberti S.; Drechsel D.; Hyman A. A.; Mitchison T. J. Amyloid-like Self-Assembly of a Cellular Compartment. Cell 2016, 166 (3), 637–650. 10.1016/j.cell.2016.06.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeynaems S.; Holehouse A. S.; Weinhardt V.; Kovacs D.; Van Lindt J.; Larabell C.; Van Den Bosch L.; Das R.; Tompa P. S.; Pappu R. V.; Gitler A. D. Spontaneous Driving Forces Give Rise to Protein-RNA Condensates with Coexisting Phases and Complex Material Properties. Proc. Natl. Acad. Sci. U. S. A. 2019, 116 (16), 7889–7898. 10.1073/pnas.1821038116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeynaems S.; Bogaert E.; Kovacs D.; Konijnenberg A.; Timmerman E.; Volkov A.; Guharoy M.; De Decker M.; Jaspers T.; Ryan V. H.; Janke A. M.; Baatsen P.; Vercruysse T.; Kolaitis R.-M.; Daelemans D.; Taylor J. P.; Kedersha N.; Anderson P.; Impens F.; Sobott F.; Schymkowitz J.; Rousseau F.; Fawzi N. L.; Robberecht W.; Van Damme P.; Tompa P.; Van Den Bosch L. Phase Separation of C9orf72 Dipeptide Repeats Perturbs Stress Granule Dynamics. Mol. Cell 2017, 65 (6), 1044–1055.e5. 10.1016/j.molcel.2017.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher R. S.; Elbaum-Garfinkle S. Tunable Multiphase Dynamics of Arginine and Lysine Liquid Condensates. Nat. Commun. 2020, 11 (1), 4628. 10.1038/s41467-020-18224-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke K. A.; Janke A. M.; Rhine C. L.; Fawzi N. L. Residue-by-Residue View of in Vitro FUS Granules That Bind the C-Terminal Domain of RNA Polymerase II. Mol. Cell 2015, 60 (2), 231–241. 10.1016/j.molcel.2015.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conicella A. E.; Zerze G. H.; Mittal J.; Fawzi N. L. ALS Mutations Disrupt Phase Separation Mediated by α-Helical Structure in the TDP-43 Low-Complexity C-Terminal Domain. Structure 2016, 24, 1537. 10.1016/j.str.2016.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conicella A. E.; Dignon G. L.; Zerze G. H.; Schmidt H. B.; D’Ordine A. M.; Kim Y. C.; Rohatgi R.; Ayala Y. M.; Mittal J.; Fawzi N. L. TDP-43 α-Helical Structure Tunes Liquid–liquid Phase Separation and Function. Proc. Natl. Acad. Sci. U. S. A. 2020, 117 (11), 5883–5894. 10.1073/pnas.1912055117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murthy A. C.; Dignon G. L.; Kan Y.; Zerze G. H.; Parekh S. H.; Mittal J.; Fawzi N. L. Molecular Interactions Underlying Liquid-Liquid Phase Separation of the FUS Low-Complexity Domain. Nat. Struct. Mol. Biol. 2019, 26 (7), 637–648. 10.1038/s41594-019-0250-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodruff J. B.; Hyman A. A.; Boke E. Organization and Function of Non-Dynamic Biomolecular Condensates. Trends Biochem. Sci. 2018, 43 (2), 81–94. 10.1016/j.tibs.2017.11.005. [DOI] [PubMed] [Google Scholar]
- Frey S.; Görlich D. A Saturated FG-Repeat Hydrogel Can Reproduce the Permeability Properties of Nuclear Pore Complexes. Cell 2007, 130 (3), 512–523. 10.1016/j.cell.2007.06.024. [DOI] [PubMed] [Google Scholar]
- Ribbeck K.; Görlich D. The Permeability Barrier of Nuclear Pore Complexes Appears to Operate via Hydrophobic Exclusion. EMBO J. 2002, 21 (11), 2664–2671. 10.1093/emboj/21.11.2664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frey S.; Richter R. P.; Görlich D. FG-Rich Repeats of Nuclear Pore Proteins Form a Three-Dimensional Meshwork with Hydrogel-like Properties. Science 2006, 314 (5800), 815–817. 10.1126/science.1132516. [DOI] [PubMed] [Google Scholar]
- Labokha A. A.; Gradmann S.; Frey S.; Hülsmann B. B.; Urlaub H.; Baldus M.; Görlich D. Systematic Analysis of Barrier-Forming FG Hydrogels from Xenopus Nuclear Pore Complexes. EMBO J. 2012, 32 (2), 204–218. 10.1038/emboj.2012.302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt H. B.; Görlich D. Transport Selectivity of Nuclear Pores, Phase Separation, and Membraneless Organelles. Trends Biochem. Sci. 2016, 41 (1), 46–61. 10.1016/j.tibs.2015.11.001. [DOI] [PubMed] [Google Scholar]
- Schmidt H. B.; Görlich D. Nup98 FG Domains from Diverse Species Spontaneously Phase-Separate into Particles with Nuclear Pore-like Permselectivity. eLife 2015, 4, e04251. 10.7554/eLife.04251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milles S.; Mercadante D.; Aramburu I. V.; Jensen M. R.; Banterle N.; Koehler C.; Tyagi S.; Clarke J.; Shammas S. L.; Blackledge M.; Gräter F.; Lemke E. A. Plasticity of an Ultrafast Interaction between Nucleoporins and Nuclear Transport Receptors. Cell 2015, 163 (3), 734–745. 10.1016/j.cell.2015.09.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celetti G.; Paci G.; Caria J.; VanDelinder V.; Bachand G.; Lemke E. A. The Liquid State of FG-Nucleoporins Mimics Permeability Barrier Properties of Nuclear Pore Complexes. J. Cell Biol. 2020, 219 (1), e201907157. 10.1083/jcb.201907157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel A.; Lee H. O.; Jawerth L.; Maharana S.; Jahnel M.; Hein M. Y.; Stoynov S.; Mahamid J.; Saha S.; Franzmann T. M.; Pozniakovski A.; Poser I.; Maghelli N.; Royer L. A.; Weigert M.; Myers E. W.; Grill S.; Drechsel D.; Hyman A. A.; Alberti S. A Liquid-to-Solid Phase Transition of the ALS Protein FUS Accelerated by Disease Mutation. Cell 2015, 162 (5), 1066–1077. 10.1016/j.cell.2015.07.047. [DOI] [PubMed] [Google Scholar]
- Kim H. J.; Kim N. C.; Wang Y.-D.; Scarborough E. A.; Moore J.; Diaz Z.; MacLea K. S.; Freibaum B.; Li S.; Molliex A.; Kanagaraj A. P.; Carter R.; Boylan K. B.; Wojtas A. M.; Rademakers R.; Pinkus J. L.; Greenberg S. A.; Trojanowski J. Q.; Traynor B. J.; Smith B. N.; Topp S.; Gkazi A.-S.; Miller J.; Shaw C. E.; Kottlors M.; Kirschner J.; Pestronk A.; Li Y. R.; Ford A. F.; Gitler A. D.; Benatar M.; King O. D.; Kimonis V. E.; Ross E. D.; Weihl C. C.; Shorter J.; Taylor J. P. Mutations in Prion-like Domains in hnRNPA2B1 and hnRNPA1 Cause Multisystem Proteinopathy and ALS. Nature 2013, 495 (7442), 467–473. 10.1038/nature11922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niaki A. G.; Sarkar J.; Cai X.; Rhine K.; Vidaurre V.; Guy B.; Hurst M.; Lee J. C.; Koh H. R.; Guo L.; Fare C. M.; Shorter J.; Myong S. Loss of Dynamic RNA Interaction and Aberrant Phase Separation Induced by Two Distinct Types of ALS/FTD-Linked FUS Mutations. Mol. Cell 2020, 77 (1), 82–94.e4. 10.1016/j.molcel.2019.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mateju D.; Franzmann T. M.; Patel A.; Kopach A.; Boczek E. E.; Maharana S.; Lee H. O.; Carra S.; Hyman A. A.; Alberti S. An Aberrant Phase Transition of Stress Granules Triggered by Misfolded Protein and Prevented by Chaperone Function. EMBO J. 2017, 36, 1669. 10.15252/embj.201695957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber S. C.; Brangwynne C. P. Getting RNA and Protein in Phase. Cell 2012, 149 (6), 1188–1191. 10.1016/j.cell.2012.05.022. [DOI] [PubMed] [Google Scholar]
- Franzmann T. M.; Jahnel M.; Pozniakovsky A.; Mahamid J.; Holehouse A. S.; Nüske E.; Richter D.; Baumeister W.; Grill S. W.; Pappu R. V.; Hyman A. A.; Alberti S. Phase Separation of a Yeast Prion Protein Promotes Cellular Fitness. Science 2018, 359 (6371), eaao5654. 10.1126/science.aao5654. [DOI] [PubMed] [Google Scholar]
- Hubstenberger A.; Noble S. L.; Cameron C.; Evans T. C. Translation Repressors, an RNA Helicase, and Developmental Cues Control RNP Phase Transitions during Early Development. Dev. Cell 2013, 27 (2), 161–173. 10.1016/j.devcel.2013.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodruff J. B.; Wueseke O.; Viscardi V.; Mahamid J.; Ochoa S. D.; Bunkenborg J.; Widlund P. O.; Pozniakovsky A.; Zanin E.; Bahmanyar S.; Zinke A.; Hong S. H.; Decker M.; Baumeister W.; Andersen J. S.; Oegema K.; Hyman A. A. Centrosomes. Regulated Assembly of a Supramolecular Centrosome Scaffold in Vitro. Science 2015, 348 (6236), 808–812. 10.1126/science.aaa3923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sankaranarayanan M.; Emenecker R. J.; Jahnel M.; Trussina I. R. E.; Wayland M.; Alberti S.; Holehouse A. S.; Weil T. T. The Arrested State of Processing Bodies Supports mRNA Regulation in Early Development. Dev. Cell 2021, 10.1101/2021.03.16.435709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powers S. K.; Holehouse A. S.; Korasick D. A.; Schreiber K. H.; Clark N. M.; Jing H.; Emenecker R.; Han S.; Tycksen E.; Hwang I.; Sozzani R.; Jez J. M.; Pappu R. V.; Strader L. C. Nucleo-Cytoplasmic Partitioning of ARF Proteins Controls Auxin Responses in Arabidopsis Thaliana. Mol. Cell 2019, 76 (1), 177–190.e5. 10.1016/j.molcel.2019.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emenecker R. J.; Holehouse A. S.; Strader L. C. Sequence Determinants of in Cell Condensate Morphology, Dynamics, and Oligomerization as Measured by Number and Brightness Analysis. Cell Commun. Signaling 2021, 19 (1), 65. 10.1186/s12964-021-00744-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber S. C. Sequence-Encoded Material Properties Dictate the Structure and Function of Nuclear Bodies. Curr. Opin. Cell Biol. 2017, 46, 62–71. 10.1016/j.ceb.2017.03.003. [DOI] [PubMed] [Google Scholar]
- Martin E. W.; Holehouse A. S. Intrinsically Disordered Protein Regions and Phase Separation: Sequence Determinants of Assembly or Lack Thereof. Emerg Top Life Sci. 2020, 4 (3), 307–329. 10.1042/ETLS20190164. [DOI] [PubMed] [Google Scholar]
- Choi J.-M.; Holehouse A. S.; Pappu R. V. Physical Principles Underlying the Complex Biology of Intracellular Phase Transitions. Annu. Rev. Biophys. 2020, 49, 107–133. 10.1146/annurev-biophys-121219-081629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y.; Protter D. S. W.; Rosen M. K.; Parker R. Formation and Maturation of Phase-Separated Liquid Droplets by RNA-Binding Proteins. Mol. Cell 2015, 60 (2), 208–219. 10.1016/j.molcel.2015.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbaum-Garfinkle S.; Kim Y.; Szczepaniak K.; Chen C. C.-H.; Eckmann C. R.; Myong S.; Brangwynne C. P. The Disordered P Granule Protein LAF-1 Drives Phase Separation into Droplets with Tunable Viscosity and Dynamics. Proc. Natl. Acad. Sci. U. S. A. 2015, 112 (23), 7189–7194. 10.1073/pnas.1504822112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubinstein M.; Dobrynin A. V. Solutions of Associative Polymers. Trends Polym. Sci. 1997, 5 (6), 181–186. [Google Scholar]
- Semenov A. N.; Rubinstein M. Thermoreversible Gelation in Solutions of Associative Polymers. 1. Statics. Macromolecules 1998, 31 (4), 1373–1385. 10.1021/ma970616h. [DOI] [Google Scholar]
- Rubinstein M.; Semenov A. N. Thermoreversible Gelation in Solutions of Associating Polymers. 2. Linear Dynamics. Macromolecules 1998, 31 (4), 1386–1397. 10.1021/ma970617+. [DOI] [Google Scholar]
- Cates M. E.; Witten T. A. Chain Conformation and Solubility of Associating Polymers. Macromolecules 1986, 19 (3), 732–739. 10.1021/ma00157a042. [DOI] [Google Scholar]
- Choi J.-M.; Hyman A. A.; Pappu R. V. Generalized Models for Bond Percolation Transitions of Associative Polymers. Phys. Rev. E: Stat. Phys., Plasmas, Fluids, Relat. Interdiscip. Top. 2020, 102 (4), 042403. 10.1103/PhysRevE.102.042403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi J.-M.; Dar F.; Pappu R. V. LASSI: A Lattice Model for Simulating Phase Transitions of Multivalent Proteins. PLoS Comput. Biol. 2019, 15 (10), e1007028. 10.1371/journal.pcbi.1007028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmon T. S.; Holehouse A. S.; Rosen M. K.; Pappu R. V. Intrinsically Disordered Linkers Determine the Interplay between Phase Separation and Gelation in Multivalent Proteins. eLife 2017, 6, e30294. 10.7554/eLife.30294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bremer A.; Farag M.; Borcherds W. M.; Peran I.; Martin E. W.; Pappu R. V.; Mittag T. Deciphering How Naturally Occurring Sequence Features Impact the Phase Behaviors of Disordered Prion-like Domains. bioRxiv 2021, 10.1101/2021.01.01.425046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y.; Jones H. B.; Dao T. P.; Castañeda C. A. Single Amino Acid Substitutions in Stickers, but Not Spacers, Substantially Alter UBQLN2 Phase Transitions and Dense Phase Material Properties. J. Phys. Chem. B 2019, 123 (17), 3618–3629. 10.1021/acs.jpcb.9b01024. [DOI] [PubMed] [Google Scholar]
- Weiner B. G.; Meir Y.; Wingreen N. S. Sequence Dependence of Biomolecular Phase Separation. bioRxiv 2021, 10.1101/2020.09.24.312330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rana U.; Brangwynne C. P.; Panagiotopoulos A. Z. Phase Separation vs Aggregation Behavior for Model Disordered Proteins. J. Chem. Phys. 2021, 155 (12), 125101. 10.1063/5.0060046. [DOI] [PubMed] [Google Scholar]
- Kwon I.; Kato M.; Xiang S.; Wu L.; Theodoropoulos P.; Mirzaei H.; Han T.; Xie S.; Corden J. L.; McKnight S. L. Phosphorylation-Regulated Binding of RNA Polymerase II to Fibrous Polymers of Low-Complexity Domains. Cell 2013, 155 (5), 1049–1060. 10.1016/j.cell.2013.10.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshizawa T.; Ali R.; Jiou J.; Fung H. Y. J.; Burke K. A.; Kim S. J.; Lin Y.; Peeples W. B.; Saltzberg D.; Soniat M.; Baumhardt J. M.; Oldenbourg R.; Sali A.; Fawzi N. L.; Rosen M. K.; Chook Y. M. Nuclear Import Receptor Inhibits Phase Separation of FUS through Binding to Multiple Sites. Cell 2018, 173 (3), 693–705.e22. 10.1016/j.cell.2018.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami T.; Qamar S.; Lin J. Q.; Schierle G. S. K.; Rees E.; Miyashita A.; Costa A. R.; Dodd R. B.; Chan F. T. S.; Michel C. H.; Kronenberg-Versteeg D.; Li Y.; Yang S.-P.; Wakutani Y.; Meadows W.; Ferry R. R.; Dong L.; Tartaglia G. G.; Favrin G.; Lin W.-L.; Dickson D. W.; Zhen M.; Ron D.; Schmitt-Ulms G.; Fraser P. E.; Shneider N. A.; Holt C.; Vendruscolo M.; Kaminski C. F.; St George-Hyslop P. ALS/FTD Mutation-Induced Phase Transition of FUS Liquid Droplets and Reversible Hydrogels into Irreversible Hydrogels Impairs RNP Granule Function. Neuron 2015, 88 (4), 678–690. 10.1016/j.neuron.2015.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qamar S.; Wang G.; Randle S. J.; Ruggeri F. S.; Varela J. A.; Lin J. Q.; Phillips E. C.; Miyashita A.; Williams D.; Ströhl F.; Meadows W.; Ferry R.; Dardov V. J.; Tartaglia G. G.; Farrer L. A.; Kaminski Schierle G. S.; Kaminski C. F.; Holt C. E.; Fraser P. E.; Schmitt-Ulms G.; Klenerman D.; Knowles T.; Vendruscolo M.; St George-Hyslop P. FUS Phase Separation Is Modulated by a Molecular Chaperone and Methylation of Arginine Cation-π Interactions. Cell 2018, 173 (3), 720–734.e15. 10.1016/j.cell.2018.03.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofweber M.; Hutten S.; Bourgeois B.; Spreitzer E.; Niedner-Boblenz A.; Schifferer M.; Ruepp M.-D.; Simons M.; Niessing D.; Madl T.; Dormann D. Phase Separation of FUS Is Suppressed by Its Nuclear Import Receptor and Arginine Methylation. Cell 2018, 173 (3), 706–719.e13. 10.1016/j.cell.2018.03.004. [DOI] [PubMed] [Google Scholar]
- Xiang S.; Kato M.; Wu L. C.; Lin Y.; Ding M.; Zhang Y.; Yu Y.; McKnight S. L. The LC Domain of hnRNPA2 Adopts Similar Conformations in Hydrogel Polymers, Liquid-like Droplets, and Nuclei. Cell 2015, 163 (4), 829–839. 10.1016/j.cell.2015.10.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes M. P.; Sawaya M. R.; Boyer D. R.; Goldschmidt L.; Rodriguez J. A.; Cascio D.; Chong L.; Gonen T.; Eisenberg D. S. Atomic Structures of Low-Complexity Protein Segments Reveal Kinked β Sheets That Assemble Networks. Science 2018, 359 (6376), 698–701. 10.1126/science.aan6398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancaster A. K.; Nutter-Upham A.; Lindquist S.; King O. D. PLAAC: A Web and Command-Line Application to Identify Proteins with Prion-like Amino Acid Composition. Bioinformatics 2014, 30 (17), 2501–2502. 10.1093/bioinformatics/btu310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alberti S.; Halfmann R.; King O.; Kapila A.; Lindquist S. A Systematic Survey Identifies Prions and Illuminates Sequence Features of Prionogenic Proteins. Cell 2009, 137 (1), 146–158. 10.1016/j.cell.2009.02.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorone Y.; Boeynaems S.; Flores E.; Jin B.; Hateley S.; Bossi F.; Lazarus E.; Pennington J. G.; Michiels E.; De Decker M.; Vints K.; Baatsen P.; Bassel G. W.; Otegui M. S.; Holehouse A. S.; Exposito-Alonso M.; Sukenik S.; Gitler A. D.; Rhee S. Y. A Prion-like Protein Regulator of Seed Germination Undergoes Hydration-Dependent Phase Separation. Cell 2021, 184 (16), 4284–4298.e27. 10.1016/j.cell.2021.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dasmeh P.; Doronin R.; Wagner A. The Length Scale of Multivalent Interactions Is Evolutionarily Conserved in Fungal and Vertebrate Phase-Separating Proteins. bioRxiv 2021, 10.1101/2021.05.04.442641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt H. B.; Barreau A.; Rohatgi R. Phase Separation-Deficient TDP43 Remains Functional in Splicing. Nat. Commun. 2019, 10 (1), 4890. 10.1038/s41467-019-12740-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolognesi B.; Faure A. J.; Seuma M.; Schmiedel J. M.; Tartaglia G. G.; Lehner B. The Mutational Landscape of a Prion-like Domain. Nat. Commun. 2019, 10 (1), 4162. 10.1038/s41467-019-12101-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt H. B.; Rohatgi R. In Vivo Formation of Vacuolated Multi-Phase Compartments Lacking Membranes. Cell Rep. 2016, 16 (5), 1228–1236. 10.1016/j.celrep.2016.06.088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y.; Zolotarev N.; Yang C.-Y.; Rambold A.; Mittler G.; Grosschedl R. A Prion-like Domain in Transcription Factor EBF1 Promotes Phase Separation and Enables B Cell Programming of Progenitor Chromatin. Immunity 2020, 53 (6), 1151–1167.e6. 10.1016/j.immuni.2020.10.009. [DOI] [PubMed] [Google Scholar]
- Boulay G.; Sandoval G. J.; Riggi N.; Iyer S.; Buisson R.; Naigles B.; Awad M. E.; Rengarajan S.; Volorio A.; McBride M. J.; Broye L. C.; Zou L.; Stamenkovic I.; Kadoch C.; Rivera M. N. Cancer-Specific Retargeting of BAF Complexes by a Prion-like Domain. Cell 2017, 171 (1), 163–178.e19. 10.1016/j.cell.2017.07.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hertig A. T. The Primary Human Oocyte: Some Observations on the Fine Structure of Balbiani’s Vitelline Body and the Origin of the Annulate Lamellae. Am. J. Anat. 1968, 122 (1), 107–137. 10.1002/aja.1001220107. [DOI] [PubMed] [Google Scholar]
- Kloc M.; Bilinski S.; Etkin L. D. The Balbiani Body and Germ Cell Determinants: 150 Years Later. Curr. Top. Dev. Biol. 2004, 59, 1–36. 10.1016/S0070-2153(04)59001-4. [DOI] [PubMed] [Google Scholar]
- Jamieson-Lucy A.; Mullins M. C. The Vertebrate Balbiani Body, Germ Plasm, and Oocyte Polarity. Curr. Top. Dev. Biol. 2019, 135, 1–34. 10.1016/bs.ctdb.2019.04.003. [DOI] [PubMed] [Google Scholar]
- Boke E.; Mitchison T. J. The Balbiani Body and the Concept of Physiological Amyloids. Cell Cycle 2017, 16 (2), 153–154. 10.1080/15384101.2016.1241605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You K.; Huang Q.; Yu C.; Shen B.; Sevilla C.; Shi M.; Hermjakob H.; Chen Y.; Li T. haSepDB: A Database of Liquid-Liquid Phase Separation Related Proteins. Nucleic Acids Res. 2020, 48 (D1), D354–D359. 10.1093/nar/gkz847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sørensen C. S.; Kjaergaard M. Effective Concentrations Enforced by Intrinsically Disordered Linkers Are Governed by Polymer Physics. Proc. Natl. Acad. Sci. U. S. A. 2019, 116 (46), 23124–23131. 10.1073/pnas.1904813116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moses D.; Yu F.; Ginell G. M.; Shamoon N. M.; Koenig P. S.; Holehouse A. S.; Sukenik S. Revealing the Hidden Sensitivity of Intrinsically Disordered Proteins to Their Chemical Environment. J. Phys. Chem. Lett. 2020, 11 (23), 10131–10136. 10.1021/acs.jpclett.0c02822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z.; Deng X.; Liu Y.; Gong W.; Li C. Analyses on Clustering of the Conserved Residues at Protein-RNA Interfaces and Its Application in Binding Site Identification. BMC Bioinf. 2020, 21 (1), 1–14. 10.1186/s12859-020-3398-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holehouse A. S.; Das R. K.; Ahad J. N.; Richardson M. O. G.; Pappu R. V. CIDER: Resources to Analyze Sequence-Ensemble Relationships of Intrinsically Disordered Proteins. Biophys. J. 2017, 112 (1), 16–21. 10.1016/j.bpj.2016.11.3200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Statt A.; Casademunt H.; Brangwynne C. P.; Panagiotopoulos A. Z. Model for Disordered Proteins with Strongly Sequence-Dependent Liquid Phase Behavior. J. Chem. Phys. 2020, 152 (7), 075101. 10.1063/1.5141095. [DOI] [PubMed] [Google Scholar]
- Schuster B. S.; Dignon G. L.; Tang W. S.; Kelley F. M.; Ranganath A. K.; Jahnke C. N.; Simpkins A. G.; Regy R. M.; Hammer D. A.; Good M. C.; Mittal J. Identifying Sequence Perturbations to an Intrinsically Disordered Protein That Determine Its Phase-Separation Behavior. Proc. Natl. Acad. Sci. U. S. A. 2020, 117 (21), 11421–11431. 10.1073/pnas.2000223117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarty J.; Delaney K. T.; Danielsen S. P. O.; Fredrickson G. H.; Shea J.-E. Complete Phase Diagram for Liquid–Liquid Phase Separation of Intrinsically Disordered Proteins. J. Phys. Chem. Lett. 2019, 10 (8), 1644–1652. 10.1021/acs.jpclett.9b00099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das S.; Eisen A.; Lin Y.-H.; Chan H. S. A Lattice Model of Charge-Pattern-Dependent Polyampholyte Phase Separation. J. Phys. Chem. B 2018, 122 (21), 5418–5431. 10.1021/acs.jpcb.7b11723. [DOI] [PubMed] [Google Scholar]
- Yoo H.; Triandafillou C.; Drummond D. A. Cellular Sensing by Phase Separation: Using the Process, Not Just the Products. J. Biol. Chem. 2019, 294 (18), 7151–7159. 10.1074/jbc.TM118.001191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holehouse A. S.; Pappu R. V. Functional Implications of Intracellular Phase Transitions. Biochemistry 2018, 57 (17), 2415–2423. 10.1021/acs.biochem.7b01136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koster K. L. Glass Formation and Desiccation Tolerance in Seeds. Plant Physiol. 1991, 96 (1), 302–304. 10.1104/pp.96.1.302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu T.; Kanamori Y.; Furuki T.; Kikawada T.; Okuda T.; Takahashi T.; Mihara H.; Sakurai M. Desiccation-Induced Structuralization and Glass Formation of Group 3 Late Embryogenesis Abundant Protein Model Peptides. Biochemistry 2010, 49 (6), 1093–1104. 10.1021/bi901745f. [DOI] [PubMed] [Google Scholar]
- Boothby T. C.; Tapia H.; Brozena A. H.; Piszkiewicz S.; Smith A. E.; Giovannini I.; Rebecchi L.; Pielak G. J.; Koshland D.; Goldstein B. Tardigrades Use Intrinsically Disordered Proteins to Survive Desiccation. Mol. Cell 2017, 65 (6), 975–984.e5. 10.1016/j.molcel.2017.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munder M. C.; Midtvedt D.; Franzmann T.; Nüske E.; Otto O.; Herbig M.; Ulbricht E.; Müller P.; Taubenberger A.; Maharana S.; Malinovska L.; Richter D.; Guck J.; Zaburdaev V.; Alberti S. A pH-Driven Transition of the Cytoplasm from a Fluid- to a Solid-like State Promotes Entry into Dormancy. eLife 2016, 5, e09347. 10.7554/eLife.09347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snead W. T.; Gladfelter A. S. The Control Centers of Biomolecular Phase Separation: How Membrane Surfaces, PTMs, and Active Processes Regulate Condensation. Mol. Cell 2019, 76 (2), 295–305. 10.1016/j.molcel.2019.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranganathan S.; Shakhnovich E. I. Dynamic Metastable Long-Living Droplets Formed by Sticker-Spacer Proteins. eLife 2020, 9, e56159. 10.7554/eLife.56159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emenecker R. J.; Griffith D.; Holehouse A. S. Metapredict: A Fast, Accurate, and Easy-to-Use Predictor of Consensus Disorder and Structure. Biophys. J. 2021, 10.1016/j.bpj.2021.08.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman M. E. J. Mixing Patterns in Networks. Phys. Rev. E: Stat. Phys., Plasmas, Fluids, Relat. Interdiscip. Top. 2003, 67 (2), 026126. 10.1103/PhysRevE.67.026126. [DOI] [PubMed] [Google Scholar]
- Hagberg A.; Swart P.; Chult D. S.. Exploring Network Structure, Dynamics, and Function Using NetworkX; Los Alamos National Laboratory: Los Alamos, NM, 2008. [Google Scholar]
- Mir A.; Heasman J. How the Mother Can Help: Studying Maternal Wnt Signaling by Anti-Sense-Mediated Depletion of Maternal mRNAs and the Host Transfer Technique. Methods Mol. Biol. 2008, 469, 417–429. 10.1007/978-1-60327-469-2_26. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.