Abstract
Adipocytes are the primary cellular component within the tumor microenvironment (TME) of triple-negative breast cancer (TNBC). Increasing evidence suggests the tumor-associated adipocytes (TAA) can aggravate tumor progression, exacerbate immunosuppressive TME and compromise therapeutic efficacy. In this study, the biological effect of TAA within breast cancer TME is first investigated, and the C-C Motif Chemokine Ligand 2 (CCL2) which mainly secreted by TAA into the extracellular environment is identified as the key mediator. CCL2 recruits immune cells such as monocytes and macrophages that further differentiated into immunosuppressive myeloid-derived suppressor cells (MDSCs) and M2 macrophages, respectively. To manipulate CCL2-mediated immune response, a protein trap that binds with CCL2 with high affinity and specificity is designed. The plasmid DNA encoding the CCL2 trap (pCCL2) is specifically delivered to the TME by using targeted lipid-protamine-DNA (LPD) nanoparticles to locally express the CCL2 trap and ameliorate the immunosuppressive TME. Significantly, compared with commercially available CCL2 antibody, this strategy shows an enhanced therapeutic efficacy and appreciable tumor growth inhibition. Furthermore, the pCCL2 trap treatment successfully suppress TAA, increase T cell infiltration and decrease immunosuppressive M2 macrophage and MDSC population. Further studies show that pCCL2 trap could facilitate the PD-L1 blockade immunotherapy, demonstrating the translation potential.
Keywords: gene delivery, tumor-associated adipocyte, CCL2, lipid nanoparticle, cancer immunotherapy
Summary
In breast cancer model, we identified C-C Motif Chemokine Ligand 2 (CCL2) as the key mediator which secreted by tumor associated adipocytes and developed targeted lipid-protamine-DNA (LPD) nanoparticles to locally “trap” CCL2 and ameliorate the immunosuppressive tumor microenvironment.
Introduction
Triple-negative breast cancer (TNBC) is one of the leading killers in women with high metastasis rate and poor prognosis. Few clinical benefits have been observed in TNBC patients receiving chemotherapy and radiotherapy 1. In recent years, the development of checkpoint blockade-based immunotherapy (CTLA4, PD-1 or PD-L1 inhibitors) has greatly reshaped the landscape of cancer therapy. However, less than 20% response rate was achieved in TNBC patients owing to the immunosuppressive tumor microenvironment (TME) 1, 2. Therefore, the development of effective remedies to reverse the immunosuppressive TME is crucial for TNBC treatment.
TME is a heterogeneous and dynamic cellular milieu consisting of a variety of resident and infiltrating cells 3. Accumulated evidences have revealed the important role of TME in tumor initiation, progression, metastasis, therapeutic response and resistance 3. Breast cancer is noted to have abundant and specific resident adipocytes within TME that differ considerably from other solid tumors 4. Such distinctive cellular component difference in TME might help to better understand the mechanism that dictates the immunosuppressive TME formation of breast cancer.
Adipocyte has long been considered as a fuel tank to store energy in forms of lipid and triglycerides 5. However, recent findings unraveled the diverse aspects of adipocyte and adipocyte derived factors 4, 6. Clinical observations suggested that adipocytes residing in close proximity with cancer cells exhibit tumor-associated functions and phenotypes, including decreased sizes and overexpression of collage VI and chemokines 6. This specialized type of adipocyte is termed “tumor associated adipocyte (TAA)”. TAA may secret factors that will promote the immunosuppressive TME 6. Furthermore, TAA may trans-differentiate into fibroblasts which enhance the desmoplasia of the tumor 6, further promoting the suppressive immune microenvironment. An in-depth understanding of the biology of TAA might provide new opportunities to modify the immunosuppressive TME in TNBC and facilitate the development of more effective therapies.
C-C Motif Chemokine Ligand 2 (CCL2), which also referred to as monocyte chemoattractant protein 1 (MCP-1), is a potent inducible chemokine that recruits immune cells, in particular monocytes, to infiltrate into the inflammatory tissue region 7. The plasticity nature of monocytes enables their phenotype transition and function changes in response to TME signals Following infiltration into TME, monocytes are quickly polarized and differentiated into macrophages and myeloid-derived suppressor cells (MDSCs) accompanied with functional and phenotypical changes 8, 9. It is widely documented that tumor-associated macrophages constitutes two major subtypes, M1 and M2 macrophages 10. M1 and M2 macrophages are identified based on their surface markers and secreted cytokines/chemokines, differ considerably in terms of phenotypes and functions 10. M1 macrophages are pro-inflammatory and actively produce high levels of pro-inflammatory cytokines/chemokines to promote Th1 responses. On the contrary, M2 macrophages are anti-inflammatory and actively secret high amounts of immunosuppressive cytokines such as interleukin 10 (IL-10) and transforming growth factor beta (TGF-β). In addition, M2 macrophages repurpose arginase metabolic pathways to further promote tumor progression 10. Clinical observations suggested that increased M2 macrophage density in the tumor is closely correlated with poor prognosis whereas increased M1 macrophage proportion usually signifies a favorable clinical outcome 11. Moreover, further detailed gene expression profiles identified CCL2’s role as a potent driving factor for M2 polarization and Th2 response. Elevated CCL2 level positively correlates with enhanced M2 macrophage and MDSC population, inhibits T cell infiltration and other immunosuppressive factors that compromise the cancer immunotherapy 9. In this regard, blockade of CCL2 could be a feasible strategy to reverse the immunosuppressive TME and thereby facilitate checkpoint blockade immunotherapy.
There are two common strategies for target-protein based therapy to block CCL2-specific signaling axis: the small molecule inhibitors and the monoclonal antibodies (mAbs). Bindarit® is a small molecule inhibitor which exhibits broad inhibition of CCL2, CCL7 and CCL8, presumably by inhibiting the p65 and p65/p50 mediated MCP-1 promoter activation. Previous study revealed only moderate inhibitory ability of Bindarit® over lipopolysaccharide (LPS) induced production of CCL2 in human monocyte with IC50 over 170 μM 12. To address the clinical need of a selective inhibitor with enhanced potency and efficacy, CCL2-netralizing mAb was developed. Preclinical and clinical investigations confirmed the apparent efficacy of a systemic CCL2 mAb in breast cancers with moderate tumor inhibition effect and decrease in M2 macrophages and MDSCs in TME 13. However, discontinuation of CCL2 mAb dosing triggered a rebound effect in CCL2 expression, resulting in significant increase of M2 macrophage and MDSC infiltration and accelerated metastasis 13. Further mechanism analysis revealed that the immediate cessation of systemic CCL2 mAb resulted in increased migration and infiltration of monocytes into the primary tumor and lung metastasis 14. Therefore, a local and transient CCL2 blockade may be an effective strategy to modify the immunosuppressive TME with reduced systemic toxicity.
Recent advances in nanotechnology have opened up opportunities for the development of a local and transient in vivo mAb-like blockade with non-viral gene delivery platform 3. Nanoparticles (NP) preferentially accumulate within the tumor via the enhanced permeability and retention (EPR) effect that depends on the leaky nature of abnormal vessels in solid tumors 3. To further optimize the target efficacy, targeting ligands are often conjugated to NPs to facilitate specific uptake of NPs by cancer cells 15. Plasmid DNA encoding mAb or mAb-like protein can be encapsulated and delivered into tumor TME in a targeted manner. This approach turns the tumor into a biological factory to produce corresponding mAbs, or the likes, followed by extracellular secretion into the TME 16. In this work, we first developed a CCL2-binding single domain antibody, called “trap”, that specifically binds to mouse CCL2. We then constructed and delivered a plasmid encoding CCL2 trap (pCCL2 trap) by using a tumor targeted lipid-protamine-pDNA (LPD) NP to the TME. We hypothesized that locally expressed trap will efficiently decrease the concentration of CCL2, leading to TME remodeling. LPD NPs surface was grafted with polyethylene glycol and functionalized with the targeting ligand aminoethyl anisamide (AEAA) to reduce organ accumulation and improve tumor delivery efficacy. AEAA is a high affinity ligand (Kd = 9 nM) for the sigma-1 receptor over-expressed in both tumor cells and the tumor associated fibroblasts 17. We demonstrated in a murine orthotopic model of TNBC that pCCL2 trap delivered by LPD NPs preferentially accumulated within the TME, leading to a decrease in CCL2 level. In addition, compared with CCL2 mAb, our strategy modified the immunosuppressive TME without any rebound effect, reprogrammed TAAs into normal adipocytes, increased tumor inhibition efficacy and reduced lung metastasis. Furthermore, in combination with pPD-L1 trap-based checkpoint blockade immunotherapy, we observed a significant tumor inhibition effect in the murine TNBC model.
Results and Discussion
Tumor-associated adipocytes facilitate tumor progression and exacerbate immunosuppressive TME via secreting CCL2
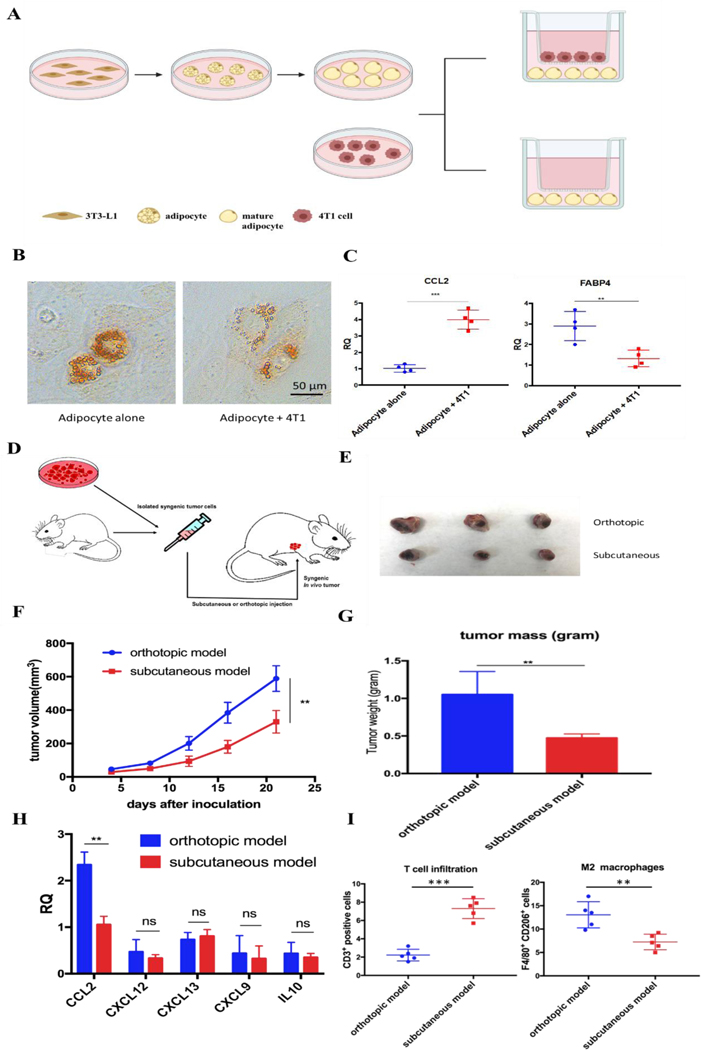
Adipocyte is one of the main resident cells that constitute the TME of TNBC 4. Previous studies suggested the interaction between adipocytes and cancer cells, while the clear role of tumor associated-adipocyte (TAA) remains undefined 6. To better understand the interaction between mature adipocytes and TNBC cells, mature adipocytes were first differentiated from 3T3-L1 preadipocytes followed by cultivation in a transwell system in the presence or absence of 4T1 murine TNBC cells (Figure 1A). Significant morphological changes were observed in adipocytes cocultured with 4T1, which exhibited a significant decrease in lipid droplet size and contents (Figure 1B). In addition, further RT-PCR analysis revealed altered gene expression in 4T1 cocultured-adipocytes. The fatty acid binding protein 4 (FABP4), a hallmark exclusively expressed in mature adipocytes with high level 6, was significantly downregulated by 2–3-fold in 4T1 cocultured-adipocytes. In contrast, cytokine CCL2, a potent chemokine for the recruitment of monocytes, was significantly upregulated (Figure 1C).
Figure 1.
Tumor-associated adipocytes facilitate tumor progression and exacerbate immunosuppressive TME via secreting CCL2. A. Cultivation of mature adipocytes in the presence or absence of 4T1 cells. Mature adipocytes were first differentiated from preadipocyte 3T3-L1 and cocultured with or without 4T1 cells in a transwell system. B. A significant decrease in lipid droplets size and contents was observed in mature adipocytes cocultured with 4T1 cells. Lipid droplets were stained by Oil-Red-O. C. RT-PCR analysis of cultivated mature adipocytes in the presence or absence of 4T1 cells. CCL2: C-C Motif Chemokine Ligand 2; FABP4: fatty acid binding protein 4. D. Establishment of subcutaneous or orthotopic 4T1 mouse model. On day 0, 1×106 4T1 cells were inoculated orthotopically on mammary fat pad or subcutaneously in six-week old female BALB/c mice. E. Representative picture of tumor burden from orthotopic and subcutaneous 4T1 mouse model on day 21. F. Tumor progression curve of orthotopic and subcutaneous 4T1 mouse models. **p<0.01. G. Quantitative analysis of tumor weight of orthotopic and subcutaneous 4T1 mouse model on day 21. **p<0.01. H. Relative mRNA expression of various chemokines and cytokines in tumors from orthotopic and subcutaneous 4T1 mouse models. CCL2: C-C Motif Chemokine Ligand 2; CXCL12: C-X-C Motif Chemokine Ligand 12; CXCL13: C-X-C Motif Chemokine Ligand 13; CXCL9: C-X-C Motif Chemokine Ligand 9; IL-10: interleukin 10. **p<0.01; ns, not significant. I. CD3+ T cell and M2 macrophage population in the tumors from orthotopic and subcutaneous 4T1 mouse models. Cells were identified by using flow cytometry. **p<0.01, ***p<0.001.
Enlighted by the in vitro observations, we established in vivo 4T1 tumor models in the presence or absence of adipocytes on BALB/c mice, aiming to investigate the role of TAA in 4T1 murine TNBC model. Same amounts of 4T1 cells were inoculated on mammary fat pad or subcutaneously to establish orthotopic or subcutaneous 4T1 model, respectively (Figure 1D). The only significant difference between the two models is the presence or absence of resident adipocytes in tumor environment. A significant aggressive tumor progression was observed in orthotopic 4T1 model compared with subcutaneous 4T1 model (Figure 1E-G). In addition, consistent with previous in vitro experiments, more detailed gene expression analysis by RT-PCR revealed that CCL2 mRNA was highly upregulated in orthotopic 4T1 model, whereas no significant differences in the mRNA expression levels of other tumor progression associated chemokines and cytokines between orthotopic and subcutaneous 4T1 models were found (Figure 1H). Previous studies suggested that the tumor vascularization is closely related to tumor growth and metastasis 18. As shown in Figure S1, there is no significant difference in tumor vascular density between the two models. Moreover, orthotopic model revealed an immunosuppressive TME which was characterized by abundant infiltration of M2 macrophages, presumably differentiated from the recruited monocytes via CCL2. Furthermore, activated T cells infiltration was considerably decreased, profiling an immunosuppressive TME in the orthotopic 4T1 model (Figure 1I).
We next examined the clinical database to further verify our hypothesis. Among all organs and tissues, adipose tissue is the main source of CCL2 in human. Analysis of clinical specimens also revealed the prevalence of CCL2 expression in breast cancer patients and high expression CCL2 is positively correlated with shorter survival and poor prognosis in breast cancer patients (Figure S2). Taken together, these data supported our hypothesis that TAA actively participates in TNBC progression, exacerbates immunosuppressive TME by secreting excessive CCL2. Therefore, blockade of CCL2 holds great promise to remodel the immunosuppressive TME and facilitate further immunotherapy for TNBC treatment.
Preparation and characterization of in situ CCL2 blockade depot
To develop a CCL2 trap that is ideal for local expression in TME via nanoparticle-mediated gene delivery, we screened a heavy chain variable domain (VH) library displayed on the yeast cell surface 19 and obtained a CCL2 specific trap protein. The trap was engineered with C-terminal c-Myc and Hisx6 tags to facilitate purification and signal detection. The theoretical molecular weight of the CCL2 trap is about 15.8 kDa. The recombinant trap protein was expressed in Expi 293 cells and purified by using Ni-NTA affinity chromatography. The binding affinity to murine CCL2 was measured by using immobilized trap protein on a Biacore facility. The estimated Kd was ~ 229 nM, which is relative weak. However, the binding appears to be specific. Only a weak binding with C-C Motif Chemokine Ligand 7 (CCL7) was detected, whereas its binding with C-C Motif Chemokine Ligand 8 (CCL8) and other chemokines was not detectable. The CCL2 trap gene was codon optimized for expression in mammalian cells. A plasmid (pCCL2 Trap) containing the CCL2 trap sequence was constructed in pcDNA3.1 vector under the control of CMV promoter.
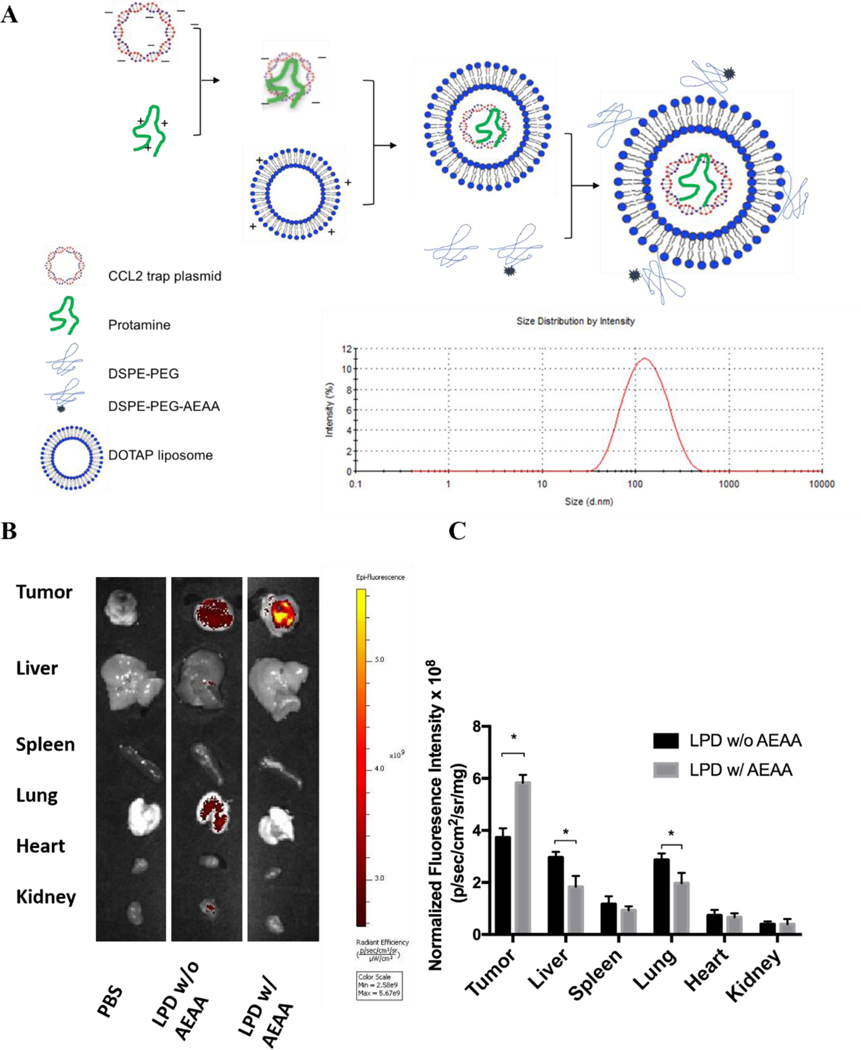
LPD NPs were constructed via a sequential self-assembly process (Figure 2A). First, blank cationic 1,2-dioleoyl-3-trimethylammonium-propane (DOTAP): cholesterol liposomes were prepared with the thin-film method 20. An anionic complex core was then formulated by mixing cationic protamine with plasmid DNA (pDNA). The anionic complex core was further coated by the blank cationic DOTAP: cholesterol liposome to formulate lipid-protamine-pDNA nanoparticle (LPD NP). To prolong the circulation time and reduce non-specific uptake by Kupffer cells, polyethylene glycol (PEG) was coated on the surface of LPD NPs. Previous studies demonstrated that sigma receptor was significantly upregulated in 4T1 murine tumor 1. A specific sigma receptor ligand, aminoethyl anisamide (AEAA), was employed to improve the local gene delivery and expression in 4T1 tumor. DSPE-PEG and target ligand DSPE-PEG-AEAA were grafted on the surface of liposomes via post-insertion method 20. Final LPD NPs exhibited a uniform size of ~100 nm and a low polydispersity index (PDI<0.2) with neutral surface charge, as suggested by dynamic light scattering (DLS) analysis (Figure 2A, Figure S3).
Figure 2.
Preparation and characterization of in situ CCL2 blockade depot. A. Preparation and characterization of pCCL2 LPD NPs. DOTAP, 1,2-dioleoyl-3-trimethylammonium propane chloride salt. AEAA, aminoethyl anisamide. B. The in vivo biodistribution of LPD NPs with or without AEAA modification in orthotopic 4T1 tumor bearing mice. Hydrophobic dye DiI was incorporated into the outer bilayer of LPD NPs as a fluorescent tracer. Twenty-four hours post intravenously injection of DiI-labeled LPD NPs, mice were sacrificed and tumors and major organs (liver, spleen, lung, heart, kidney) were harvested and examined under IVIS imaging system (n=3). C. Quantitative analysis of DiI-labeled LPD NPs biodistribution in major organs and tumors. *p<0.05.
The final LPD NPs were labeled with a fluorescent lipophilic dye, DiI, to track its biodistribution in 4T1 orthotopic tumor bearing mice. Twenty-four hours post intravenous injection of DiI-labeled LPD NPs, 4T1 tumor bearing mice were sacrificed, and major organs/tumors were subject to In Vivo Imaging Systems (IVIS) imaging. LPD NPs modified with target ligand AEAA predominantly accumulated within the breast tumor site, whereas non-modified LPD NPs showed a non-specific biodistribution in liver and lung (Figure 2B). Based on the normalized fluorescence intensity analysis, a 1.5-fold increase of tumor specific uptake were achieved with AEAA modification, further verifying the successful tumor-targeted delivery was achieved by AEAA modified LPD NP platform (Figure 2C).
pCCL2 trap outperformed CCL2 mAb and remodeled the immunosuppressive TME in orthotopic 4T1 mouse model
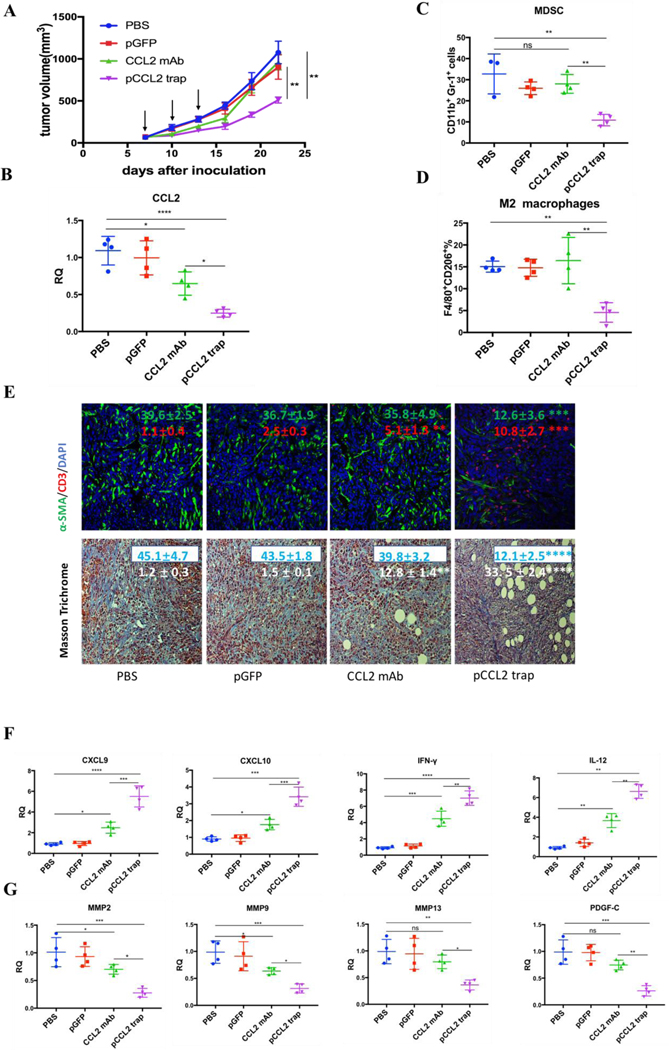
Inspired by the exciting target discovery and promising gene delivery platform, we wonder whether pCCL2 trap would generate a local and transient expression of CCL2 trap protein, remodel immunosuppressive TME and inhibit TNBC tumor progression as expected. Animal experiments were conducted to evaluate the therapeutic efficacy of pCCL2 trap in 4T1 mouse orthotopic model as follows. LPD NPs encapsulating a plasmid encoding the green fluorescence protein (pGFP) were used as negative control. In addition, CCL2 mAb control was included in the animal study aiming to investigate if pCCL2 trap could outperformed CCL mAb. Mice bearing 4T1 tumor were randomly divided into four groups and treated with different modalities shown in Figure 3A. Longitudinal tumor volume measurements were recorded every other day. In accordance with clinical studies, partial tumor inhibition effect was observed in CCL2 mAb treated group within the treatment schedule. Significantly, the CCL2 level in the tumor was the lowest in the group treated with pCCL2 trap (Figure 3B). Our gene therapy strategy to deliver the trap protein to the TME outperformed the traditional mAb approach (Figure 3). It is not surprising that this was the case because the AEAA-targeted NPs specifically delivered the trap gene to the tumor (Figure 2). In contrast, mAb distributes in the entire body. The trap protein (15.8 kDa), being much smaller than the IgG molecule (150 kDa), can readily diffuse in the TME and neutralize the target chemokine.
Figure 3.
pCCL2 trap outperformed CCL2 mAb and remodeled the immunosuppressive TME in 4T1 mouse model. A. Tumor inhibition effect of PBS, pGFP, pCCL2 trap and CCL2 mAb. 1×106 4T1 cells were orthotopically injected into mammary fat pad of six-weeks female BALB/c mice on day 0. Mice were randomly distributed into 4 groups on day 7 (n=5). Tumor volumes were recorded every other day by caliper measurement. Black arrow indicates the dosing schedule (50 μg pDNA or 200 μg mAb on day 7, 10 and 13). **p<0.01. B. The CCL2 mRNA expression in the tumor of mice received different treatments. Tumors were obtained at the end of the study (day 23, 10 days after the last treatment). *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001. C-D. The quantitative analysis of MDSC and M2 macrophage population within TME on day 23. Cells were measured by flow cytometry. MDSC, myeloid-derived suppressor cells. E. Upper panel, immunofluorescence staining of tumor samples from different treatment groups using anti-α-SMA antibody (green) and anti-CD3-antibody (red). Cell nuclei were stained as blue using DAPI. Lower panel, collagen contents quantified by Masson Trichrome staining. Five random fields were chosen for statistical analysis in each treatment groups. Images were analyzed by Image J software and quantified with GraphPad 6.0. *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001. Scale bar represents 100 μm. F. Relative mRNA expression of Th1 chemokines and cytokines in the tumor of mice received different treatments. Tumors were obtained at the end point of the study (day 23, 10 days after the last treatment). CXCL9: C-X-C Motif Chemokine Ligand 9; CXCL10: C-X-C Motif Chemokine Ligand 10; IL-12: interleukin-12; IFN-γ: interferon gamma. *p<0.05, **p<0.01, ***p<0.001. G. Relative mRNA expression of Th2 chemokines and cytokines in the tumor of mice received different treatments. Tumors were obtained at the end point of the study (day 23, 10 days after the last treatment). MMP, metallopeptidase. PDGF-C, platelet-derived growth factor C. *p<0.05, **p<0.01, ***p<0.001.
In the CCL2 mAB treatment group, the rapid discontinuation of CCL2 mAb resulted in a rebound effect and accelerated tumor progression (Figure 3A). Further detailed flow cytometry analysis suggested that no significant decrease in M2 macrophage and MDSC was observed at the endpoint of the study. In contrast, the group treated with pCCL2 trap significantly inhibited tumor progression, with obviously down-regulated M2 macrophages and MDSC population lasting for two weeks following cessation of pCCL2 trap treatment (Figure 3C-D). Additionally, immunofluorescence staining revealed a remodeled TME in mice treated with pCCL2 trap. Alpha-smooth muscle actin (α-SMA), which was highly expressed in TAA and activated fibroblasts, was reduced by 3-fold after pCCL2 trap treatment compared with pGFP control group (p<0.001) (Figure 3E). The collagen content in different treatment groups was revealed with Masson Trichrome staining, demonstrating a 4-fold decrease after pCCL2 trap treatment (p<0.0001) (Figure 3E). It is also noteworthy to mention that plentiful normalized adipocytes (cells containing large oil droplets) were observed in pCCL2 trap treated group, suggesting the restoration of TAAs into normal adipocytes (Figure 3E). Furthermore, upon CCL2 trap treatment, T cell (CD3+ cells) infiltration within TME was increased by 5-fold compared with the pGFP treated group (p<0.001), which could be attributed to the remodeled immunosuppressive TME (Figure 3E).
As an indispensable part of TME, cytokines and chemokines are important mediators and regulators for immune cell migration, infiltration and action. Highly elevated T cell recruiting cytokines including C-X-C Motif Chemokine Ligand 9 (CXCL9) and C-X-C Motif Chemokine Ligand 10 (CXCL10) are crucial for T cell infiltration. Furthermore, the activation of tumor infiltrating T cell also depends on pro-inflammatory Th1 cytokines, interleukin-12 (IL-12) and interferon gamma (IFN-γ). In accordance with previous T cell profiling, CXCL9, CXCL10, IL-12 and IFN-γ were highly upregulated in pCCL2 trap treated group (p<0.01–0.001) (Figure 3F). Conversely, common matrix metallopeptidases (MMP) that are involved in extracellular matrix (ECM) remodeling, such as MMP2, MMP9 and MMP13, were significantly downregulated compared with PBS control group, so was pro-fibrosis cytokine platelet-derived growth factor (PDGF-C) (p<0.01 – 0.001) (Figure 3G).
pCCL2 trap facilitated the checkpoint blockade therapy
RNA sequencing data derived from the Cancer Genome Atlas (TCGA) database showed up-regulated expression of programmed cell death protein ligand 1 (PD-L1) in TNBC samples in comparison with non-TNBC samples 21. PD-L1 is a checkpoint protein on tumor cells that negatively regulates the immune response by inactivating T cells 22. Monoclonal antibodies that specifically block PD-L1 is referred as a checkpoint blockade therapy. The therapy effectively blocks the inhibition signal for T cell activation and initiates effective T cell based anti-tumor response 22. However, TNBC patients do not respond well to checkpoint blockade therapy owing to the immunosuppressive TME 23. Since the data above indicated that pCCL2 trap successfully remodeled the immunosuppressive TME, we wondered if pCCL2 trap treatment would enhance the checkpoint blockade therapy efficacy in our TNBC model.
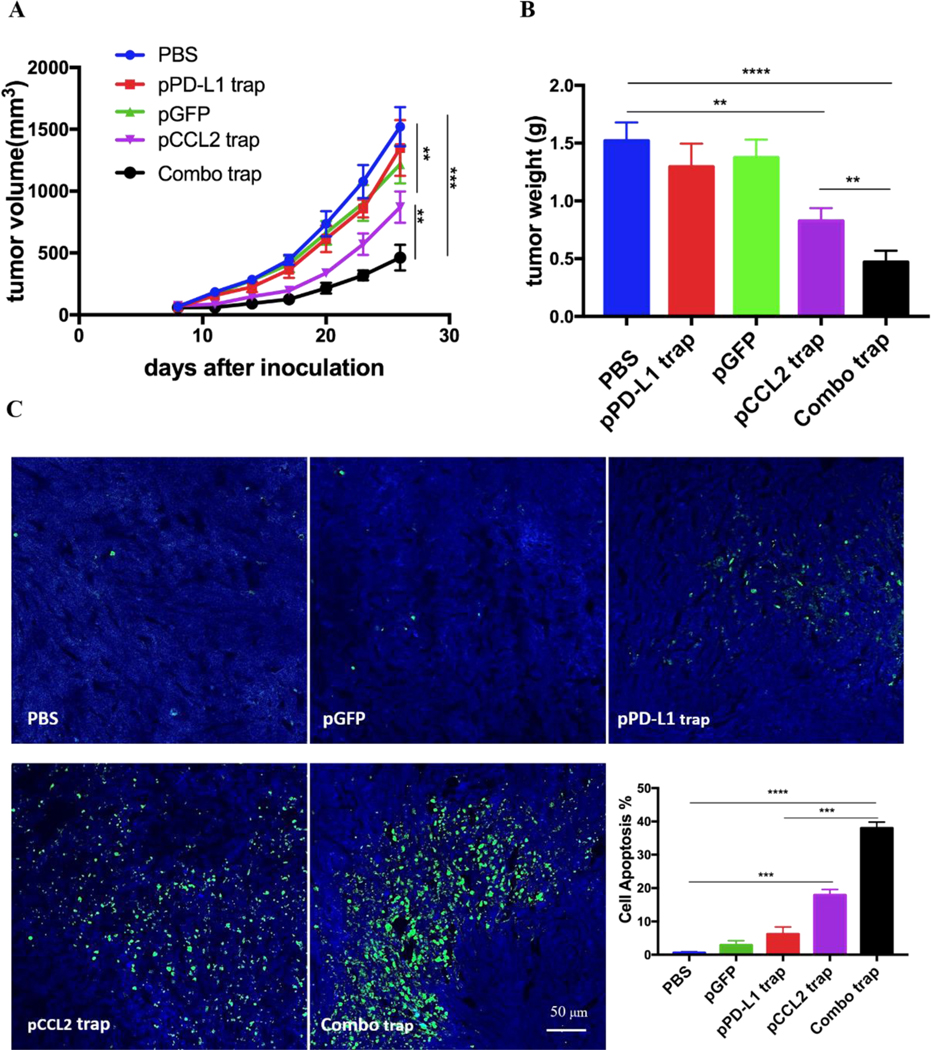
We have previously developed a trimeric protein trap that potently and specifically blocks PD-L1 with an apparent Kd ~219 pM 24. Plasmid pPD-L1 trap was formulated in LPD NPs and delivered to the orthotopic TNBC tumor in a manner similar to pCCL2 trap. Our prior studies have shown that pPD-L1 trap was efficiently expressed in a local and transient manner in TME of 4T1 model with better tolerance and lower immune-related adverse effects 25. We next conducted a combination study to investigate the possible synergistic effect of pCCL2 trap and pPD-L1 trap in the orthotopic 4T1 model. Despite the fact the pCCL2 trap or pPD-L1 trap by itself only showed a partial tumor inhibitory effect, tumor progression was significantly inhibited in the combined treatment group (Figure 4A-B). Further terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL) staining studies justified the synergistic effect of the combined treatment group (Figure 4C).
Figure 4.
pCCL2 trap facilitates the checkpoint blockade-based therapy. A. Tumor inhibition study of various treatments. Combo trap, the combination of pCCL2 trap and pPD-L1 trap (25 μg pCCL2 combined with 25 μg pPD-L1 on day 7, 10 and 13). *p<0.05, **p<0.01, ***p<0.001. B. Quantitative analysis of tumor weight of various treatment groups on day 26. *p<0.05, **p<0.01, ***p<0.00, ****p<0.0001. C. TUNEL assay and corresponding quantitative analysis of tumor tissue from various treatment groups. TUNEL, terminal deoxynucleotidyl transferase dUTP nick end labeling. Tumors were obtained on day 26. **p<0.01, ***p<0.001, ****p<0.0001.
Safety evaluation of different treatment groups
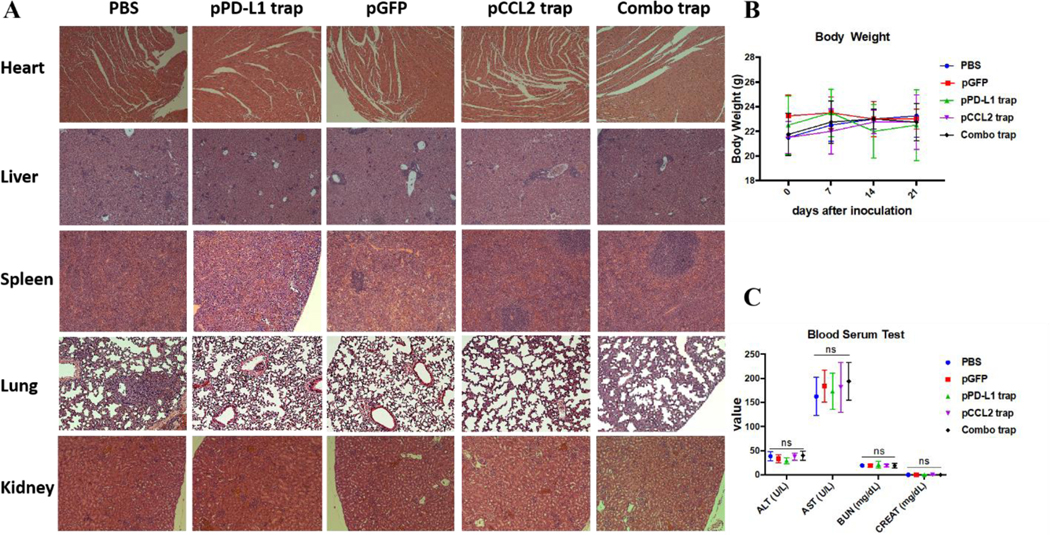
Major organ, including heart, liver, spleen, lung and kidney, and blood samples were obtained from different treatment groups at the end of the experiment described in Figure 4. In further safety and toxicity evaluations, we found that the pCCL2/pPD-L1 trap alone and the combo trap treated groups did not exhibit significant morphology damage in any of the major organs (Figure 5A). In addition, the blood serum and whole blood test suggested that there was no significant difference in levels of alanine aminotransferase (ALT), aspartate aminotransferase (AST), blood urea nitrogen (BUN) and creatinine (CREAT), suggesting the absence of liver or kidney associated toxicities (Figure 5C, Table S1). Furthermore, no significant weight loss was observed in different treatment groups (Figure 5B). Taken together, our therapeutic strategy provided a tumor-targeted, local and transient gene therapy to remodel the tumor immunosuppressive tumor microenvironment in a safe and feasible manner.
Figure 5.
Safety evaluation of different treatment groups. A. H&E staining of major organs from different treatment groups. At the endpoint of the study, major organs were collected, sectioned and stained with H&E solution and observed under microscope. B. Body weight of mice from different treatment groups. C. Blood serum test of ALT, AST, BUN and CREAT among different treatment groups. alanine aminotransferase (ALT), aspartate aminotransferase (AST), blood urea nitrogen (BUN) and creatinine (CREAT). ns, not significant.
Conclusion
We have developed a small trap protein with a relatively high affinity to bind mouse CCL2 in a specific manner. This single domain antibody was delivered to the TNBC tumor by using a gene therapy approach, resulting in a persistent decrease of CCL2 level in the tumor. Local delivery of the trap protein triggered a significant remodeling of the immune microenvironment and inhibited tumor growth. Since CCL2 is mainly produced by the TAAs in TNBC, our study demonstrated the usefulness in targeting TAAs to improve immunotherapy. In addition, the NPs used in the study is rather simple to prepare, potentially in large scale, and non-toxic 20, the approach appears to be translatable into clinic.
Materials and Methods
Materials
1,2-dioleoyl-3-trimethylammonium propane chloride salt (DOTAP), 1,2-distearoyl-sn-glycero-3-phosphoethaolamine-N-[amino(polyethylene glycol)-2000 (DSPE-PEG-2000), 3-(N-succinimidyloxyglutaryl)aminopropyl, polyethyleneglycol-carbamyl distearoylphosphatidyl-ethanolamine (DSPE-PEG-NHS) were purchased from Avanti Polar Lipids (Alabaster, AL). DSPE-PEG-aminoethyl anisamide (DSPE-PEG-AEAA) was synthesized following previous reported protocol 26.
Development of the CCL2-binding trap and expression of recombinant CCL2 trap
CCL2-binding trap was developed by screening a human single domain antibody library using yeast surface display against biotinylated mouse CCL2 protein. Yeast cells with the highest binding and expression ratio were selected. The selection was performed until a homogenous population was obtained and no further enrichment was possible. The expression vector encoding the selected CCL2 trap was generated by inserting synthesized DNAs (codon optimized for expression in mammalian cells) into the pcDNA3.1 vector. For protein expression, 25 micrograms of plasmid DNA were transfected to 25 mL of ExpiCHO-S cells (1.5× 108 cells). Ten days post transfection, the supernatant was collected and the protein was purified with Ni-NTA column. The protein was eluted with 500 mM imidazole in 1×PBS buffer. The eluted protein was dialyzed against PBS buffer and assessed with SDS-PAGE.
Affinity measurement and specificity analysis
Binding affinity between the recombinant CCL2 trap and mouse CCL2 was determined using Biacore X100 (Cytiva). Biotinylated mouse CCL2 trap was immobilized on a Biacore Biotin CAPture chip (Cytiva) at 100 nM. Mouse CCL2 analyte (from Shenandoah Biotechnology) was injected into the sample channel at different concentrations in a single-cycle kinetics mode. The data was processed by Biacore analytical software.
Synthesis of DSPE-PEG and DSPE-PEG-AEAA
DSPE-PEG-AEAA was synthesized following previously reported methods 26. Briefly, 4-Methoylbenzoyl chloride and 2-bromoethylamine hydrobromide were mixed for 6 h at room temperature. Then the DSPE-PEG-NH2 was added into the mixture and stirred at 65–70 °C overnight in oil bath. The obtained product was further precipitated and purified with diethyl ether, lyophilized and dissolved in DI water.
Preparation and Characterization of LPD NPs
LPD NPs were constructed via a sequential self-assembly process. Briefly, blank liposomes were first prepared by thin film dispersion method. DOTAP and cholesterol (1:1, molar ratio) were first dissolved in chloroform and rotary evaporator was employed to remove the organic solvent. The thin film was further hydrated with distilled water to make the final DOTAP/Chol blank liposomes with a concentration of 10 mmol/L. Unilamellar DOTAP/Chol liposomes were obtained followed by successive extrusion of the resuspended lipid mixture. 100 μL 0.5 mg/mL pDNA and 100 μL 0.2 mg/mL protamine were mixed to formulate the LPD polyplex cores. Then, 60 μL of blank DOTAP/Chol liposomes were mixed with 200 μL LPD polyplex cores. 10 μL DSPE-PEG and 10 μL DSPE-PEG-AEAA at a concentration of 10 mg/mL were added with a post-insertion methods to formulate the final LPD NPs. Glucose solution were added into the system to adjust the osmotic pressure. Size distribution and ζ potential of LPD NPs was determined by Malvern ZetaSizer Nano Series (Westborough, MA).
Biodistribution of LPD NPs
LPD NPs were labeled with DiI by incorporation of 0.1% lipophilic dye Dil into the blank DOTAP: cholesterol liposomes. Twenty-four hours post intravenous injection of DiI-labeled LPD NPs, mice were sacrificed and the biodistribution of DiI-labeled liposomes were visualized with the IVIS Kinetics Optical System (PerkinElmer, CA). Excitation wavelength of 520 nm and emission wavelength of 560 nm were used for IVIS imaging.
Cell Lines
Murine triple negative breast cancer cells 4T1 were obtained from UNC Tissue Culture Facility. Following ATCC instructions, 4T1 cells were cultured with RPMI 1640 medium supplemented with 10% fetal bovine serum (V/V, Gibco) and 1% antibiotic-antimycotic (V/V, Gibco) in a humified atmosphere at 37 °C and 5% CO2 conditions.
Orthotopic Animal Mouse Cancer Model
Six-week old female BALB/c mice were purchased from Jackson Laboratories and maintained under germ-free conditions. Orthotopic 4T1 model was established by orthotopically injection of 1×106 4T1 cells in 50 μL PBS into the inguinal mammary fat pad of female BALB/c mice. Subcutaneous 4T1 model was established by subcutaneous injection of 1×106 4T1 cells in 100 μL PBS into the right flank of female BALB/c mice. All animal procedures were approved by the Institutional Animal Care and Use Committee in University of North Carolina at Chapel Hill. All animal procedures were conducted in compliance with federal regulations ((NIH/PHS Animal Welfare Assurance No. D16–00256; USDA Animal Research Facility Registration No. 55-R-0004; AAALAC Institutional No. #329) and ethical regulations approved by the Institutional Animal Care and Use Committee of North Carolina at Chapel Hill.
Tumor Growth Inhibition Study
The 4T1 tumor bearing mice were randomly distributed into different treatment groups when the tumor volume reaches 100mm3. Mice were treated with different modalities according to the treatment schedule (50 μg pDNA or 200 μg mAb on day 7, 10 and 13). Tumor progression was recorded by caliper, and the tumor volume was calculated as V=0.5× length ×width ×width, where the length represents the long axis and the width represents the short axis.
Flow Cytometry Assay
Fresh tissues were obtained and prepared as single cell suspension. Tumor infiltrating lymphocytes were characterized and quantified by flow cytometry assay. In brief, fresh isolated tumors were first incubated with collagenase and DNase for 1h at 37 °C to obtain single cell suspensions. Then the prepared single cell suspensions were stained with a cocktail of fluorescently labeled antibodies for surface marker expression analysis. After 0.5 h staining, cells were fixed with 4% PFA solution and analyzed with FACS (BD LSR II). The analysis was conducted with FlowJo software (TreeStar, Ashland, OR). Fluorescently labeled antibodies used for flow cytometry are listed in Table S2.
Immunofluorescence Staining
Fresh tumor tissues were first resected and rinsed in PBS upon mice sacrifice and fixed with 4% PFA solution at 4 °C for 48 h. Tissues were then dehydrated with 15% and 30% sucrose solution. Dehydrated tissues were embedded into O.C.T solution for the preparation of frozen section. Obtained cryostat sections were permeabilized and blocked with 5% goat serum for 1 h at room temperature. Then the sections were incubated with fluorescently conjugated antibodies for 12 h at 4 °C and mounted with Prolong Diamond Antifade Mountant with DAPI. IF images were taken on a confocal microscope (Zeiss LSM 710) and quantified with Image J. Five randomly chosen fields were used for statistical analysis. Antibodies used for immunofluorescence staining are listed in Table S2.
TUNEL Assay
TUNEL assay were conducted using a DeadEnd Fluorometric TUNEL System (Promega, Madison, WI) following the manufacturer’s protocol. Tumor frozen section slides were prepared and the fragmented DNA of apoptotic cells were stained with fluorescent dye. Cell nuclei that were stained to be fluorescent green were defined as TUNEL-positive nuclei. Slides were further mounted with prolonged diamond antifade mountant DAPI (ThermoFisher Scientific) and examined under Confocal microscopy (Zeiss, LSM 710). Five microscopic fields were randomly selected for quantification and statistical analysis using Image J.
Quantitative Real-Time PCR Assay
RNeasy Microarray Tissue Mini Kit (Qiagen) were used to extract the total RNA from fresh tissues. cDNA was obtained by reverse transcription using the iScript cDNA Synthesis Kit (BIO-RAD) and amplified with Taqman gene expression master mix. Mouse specific primers were used and GAPDH was used as the endogenous control. Reactions and corresponding data analysis were completed with the 7500 Real-Time PCR system. Primers used in Real-Time PCR assay are listed in Table S3.
Safety Evaluation
Mouse body weight were measured every other day following the first dose treatment. At the experiment endpoint, mice were sacrificed, whole blood, serum and major organs were collected for safety evaluation. Aspartate aminotransferase (AST), alanine aminotransferase (ALT), blood urea nitrogen (BUN) and creatinine (CRE) levels in the serum were analyzed as biomarkers for renal and hepatic function. Red blood cells (RBC), white blood cells (WBC), platelets (PLT), hemoglobin (HGB), neutrophils (NEUT), lymphocytes (LYMPH) and monocytes (MONO) in whole blood were measured to quantify myelosuppression.
To evaluate the toxicity in major organs, heart, liver, spleen, lung and kidney from different treatment groups were harvested, fixed, stained with hematoxylin and eosin (H&E) and examined under microscope.
Statistical Analysis
Results are presented as mean ± SD unless otherwise specified. GraphPad 6.0 was used for statistical analysis. For comparison between two groups, student’s t test was used. For comparison within three or more groups, one-way analysis of variance (ANOVA) was used. Kaplan-Meier estimator was used to analysis the survival studies. A p value less than 0.05 was considered to be significant. *, **, *** and **** denotes p<0.05, 0.01, 0.001, and 0.0001, respectively.
Supplementary Material
Scheme 1.
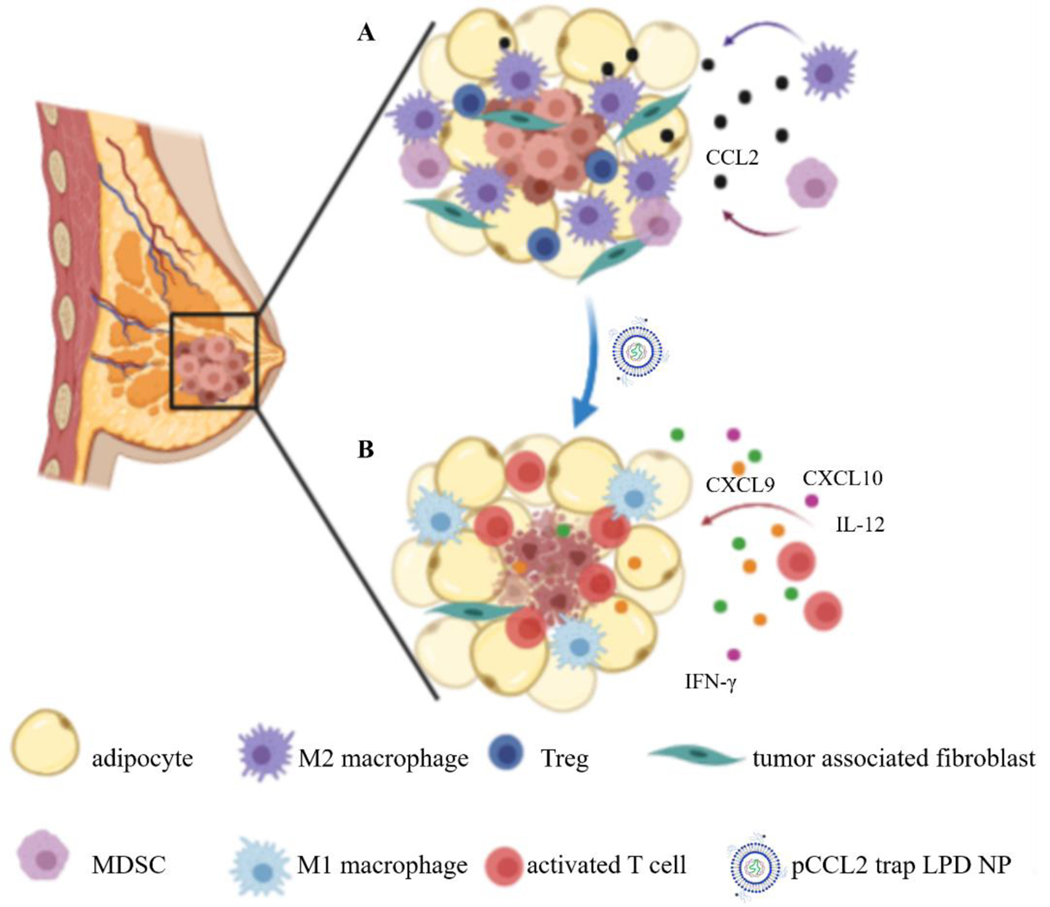
Schematic illustration of the mechanism of pCCL2 trap for the treatment of TNBC in murine model of TNBC. A. Primary mammary breast tumor immunosuppressive TME constitutes of surrounding tumor associated adipocyte (TAA) and infiltrated immunocytes. B. pCCL2 trap successfully remodels the immunosuppressive TME of TNBC in the tumor model.
Acknowledgement
The work was supported by NIH grant CA198999 (to LH) and Eshelman Institute for Innovation grant RX03812125 (to RL).
Footnotes
Conflict of Interest
LH is a consultant for PDS Biotechnology, Samyang Biopharmaceuticals, Stemirna and Beijing Inno Medicine. RH is a co-founder of OncoTrap. All other co-authors declare no conflict.
References
- 1.Liu Y, Qiu N, Shen L, Liu Q, Zhang J, Cheng YY, Lee KH and Huang L, J Control Release, 2020, 323, 431–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wahba HA and El-Hadaad HA, Cancer Biol Med, 2015, 12, 106–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liu Y, Guo J. and Huang L, Theranostics, 2020, 10, 3099–3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wu Q, Li B, Li Z, Li J, Sun S. and Sun S, J Hematol Oncol, 2019, 12, 95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cohen P. and Spiegelman BM, Mol Biol Cell, 2016, 27, 2523–2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cai Z, Liang Y, Xing C, Wang H, Hu P, Li J, Huang H, Wang W. and Jiang C, Oncol Rep, 2019, 42, 2537–2549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhu X, Fujita M, Snyder LA and Okada H, J Neurooncol, 2011, 104, 83–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pogoda K, Pyszniak M, Rybojad P. and Tabarkiewicz J, Oncol Lett, 2016, 12, 4785–4794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sierra-Filardi E, Nieto C, Dominguez-Soto A, Barroso R, Sanchez-Mateos P, Puig-Kroger A, Lopez-Bravo M, Joven J, Ardavin C, Rodriguez-Fernandez JL, Sanchez-Torres C, Mellado M. and Corbi AL, J Immunol, 2014, 192, 3858–3867. [DOI] [PubMed] [Google Scholar]
- 10.Martinez FO and Gordon S, F1000Prime Rep, 2014, 6, 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Takeya M. and Komohara Y, Pathol Int, 2016, 66, 491–505. [DOI] [PubMed] [Google Scholar]
- 12.Chen W, Foo SS, Taylor A, Lulla A, Merits A, Hueston L, Forwood MR, Walsh NC, Sims NA, Herrero LJ and Mahalingam S, J Virol, 2015, 89, 581–593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bonapace L, Coissieux MM, Wyckoff J, Mertz KD, Varga Z, Junt T. and Bentires-Alj M, Nature, 2014, 515, 130–133. [DOI] [PubMed] [Google Scholar]
- 14.Kitamura T, Qian BZ, Soong D, Cassetta L, Noy R, Sugano G, Kato Y, Li J. and Pollard JW, J Exp Med, 2015, 212, 1043–1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu Q, Das M, Liu Y. and Huang L, Adv Drug Deliv Rev, 2018, 127, 208–221. [DOI] [PubMed] [Google Scholar]
- 16.Shen L, Li J, Liu Q, Song W, Zhang X, Tiruthani K, Hu H, Das M, Goodwin TJ, Liu R. and Huang L, ACS Nano, 2018, 12, 9830–9841. [DOI] [PubMed] [Google Scholar]
- 17.Miao L, Liu Q, Lin CM, Luo C, Wang Y, Liu L, Yin W, Hu S, Kim WY and Huang L, Cancer Res, 2017, 77, 719–731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lugano R, Ramachandran M. and Dimberg A, Cell Mol Life Sci, 2020, 77, 1745–1770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cherf GM and Cochran JR, Methods Mol Biol, 2015, 1319, 155–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang S, Zhi D. and Huang L, J Drug Target, 2012, 20, 724–735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mittendorf EA, Philips AV, Meric-Bernstam F, Qiao N, Wu Y, Harrington S, Su X, Wang Y, Gonzalez-Angulo AM, Akcakanat A, Chawla A, Curran M, Hwu P, Sharma P, Litton JK, Molldrem JJ and Alatrash G, Cancer Immunol Res, 2014, 2, 361–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen DS and Mellman I, Immunity, 2013, 39, 1–10. [DOI] [PubMed] [Google Scholar]
- 23.Kwa MJ and Adams S, Cancer, 2018, 124, 2086–2103. [DOI] [PubMed] [Google Scholar]
- 24.Miao L, Li J, Liu Q, Feng R, Das M, Lin CM, Goodwin TJ, Dorosheva O, Liu R. and Huang L, ACS Nano, 2017, 11, 8690–8706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Song W, Shen L, Wang Y, Liu Q, Goodwin TJ, Li J, Dorosheva O, Liu T, Liu R. and Huang L, Nat Commun, 2018, 9, 2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Banerjee R, Tyagi P, Li S. and Huang L, Int J Cancer, 2004, 112, 693–700. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.