Significance
Monocytes are key players in immune responses to pathogen entry. They exert a major part of their function through their differentiation into macrophages or dendritic cells, two populations with largely distinct roles in immune responses. Yet, we still have a very limited view of the factors orientating monocyte fate to become macrophages versus dendritic cells, in particular how pathogen recognition modulates this process. Here, we show that pathogen sensing skews monocyte fate decision, that is, the first steps of the differentiation process, with TLR signaling favoring macrophage development while NOD receptor signaling induces dendritic cell differentiation.
Keywords: monocyte, macrophage, dendritic cell, pathogen
Abstract
Monocytes are rapidly recruited to inflamed tissues where they differentiate into monocyte-derived macrophages (mo-mac) or dendritic cells (mo-DC). At infection sites, monocytes encounter a broad range of microbial motifs. How pathogen recognition impacts monocyte fate decision is unclear. Here, we show, using an in vitro model allowing the simultaneous differentiation of human mo-mac and mo-DC, that viruses promote mo-mac while Mycobacteria favor mo-DC differentiation. Mechanistically, we found that pathogen sensing through toll-like receptor (TLR) ligands increases mo-mac differentiation via mTORC1. By contrast, nucleotide-binding oligomerization domain (NOD) ligands favor mo-DC through the induction of TNF-α secretion and miR-155 expression. We confirmed these results in vivo, in mouse skin and by analyzing transcriptomic data from human individuals. Overall, our findings allow a better understanding of the molecular control of monocyte differentiation and of monocyte plasticity upon pathogen sensing.
Immune defense against microbial infections critically relies on the rapid mobilization of professional phagocytes, in particular macrophages and monocytes. Upon pathogen recognition, they increase their ability to engulf dying cells and produce antimicrobial molecules to control the infection. In addition, microbe sensing reprograms macrophages toward a proinflammatory state (1). However, how pathogen-derived products impact monocyte fate remains unclear.
Monocytes originate from the bone marrow and are massively recruited in tissues upon infection (2). They play an essential role in the control of infections as shown in CCR2-deficient mice, in which monocytes are unable to exit the bone marrow to circulate in the bloodstream (3, 4). In models of bacterial or viral infections, CCR2-deficient mice display higher bacterial or viral loads and lower survival (5–8). In addition to their immediate action in pathogen elimination, monocytes are involved in the immune response against infections through their capacity to differentiate into monocyte-derived macrophages (mo-mac) and monocyte-derived dendritic cells (mo-DC) (9, 10). Mo-mac can replace tissue-resident macrophages that have died upon infection (11, 12) or retain increased reactivity to subsequent inflammation (13, 14). Mo-DC can complement the action of classical DC in the induction of adaptive responses by presenting antigens to T cells directly in tissues to boost their effector functions (15–17). On the other hand, dysregulated monocyte recruitment and/or differentiation during infections can lead to increased tissue damage, high pathogen burden, and mortality (18–20). A tight control of these events therefore appears critical for mounting efficient immune responses. Yet, the factors regulating monocyte fate decision are poorly understood both at the molecular level and in a broader physiological context. In vivo differentiation of monocytes is dependent on the M-CSF receptor (21). We have shown that circulating monocytes are not precommitted to become mo-mac or mo-DC and that their fate is driven by microenvironmental cues (22). Consistent with this, sterile inflammation accelerates monocyte differentiation in the intestine in mice (23). However, how pathogen recognition impacts monocyte fate decision remains unknown.
Using an in vitro model of human monocyte differentiation, we show that pathogen recognition through toll-like receptor (TLR) promotes mo-mac differentiation via the mTORC1 pathway, which increases MAFB expression. By contrast, nucleotide-binding oligomerization domain (NOD) receptor activation favors mo-DC development through the autocrine action of TNF-α. We confirm these findings in vivo in a mouse model and in humans using Gene Set Enrichment Analysis (GSEA) analysis of skin biopsies from individuals infected with herpes simplex virus type 2 (HSV2) virus or joint biopsies from rheumatoid arthritis (RA) patients. Our results demonstrate that pathogen recognition skews monocyte fate decision, resulting in opposite outcomes depending on the nature of the pathogen.
Results
Exposure to Viruses or Mycobacteria Has Opposite Effects on Monocyte Differentiation.
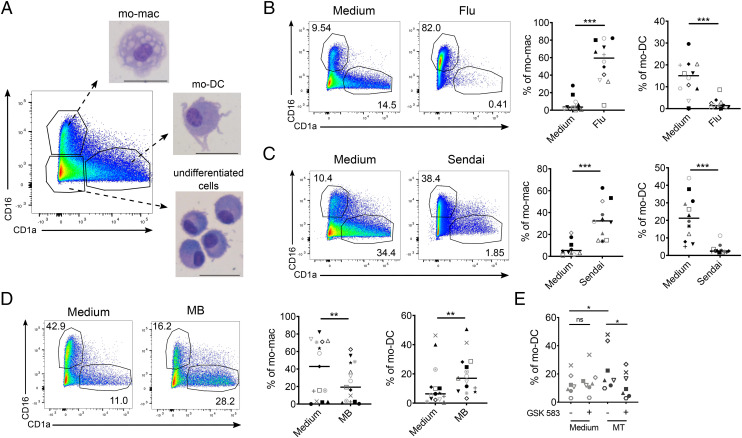
To address the impact of pathogen recognition on monocyte differentiation, we used our previously published in vitro model allowing the simultaneous differentiation of mo-mac and mo-DC (22). In this model, human monocytes cultured for 5 d with M-CSF, IL-4, and TNF-α differentiate into mo-mac (CD16+), mo-DC (CD1a+), or remain undifferentiated (double negative cells) (Fig. 1A). At the start of the culture, we exposed monocytes to inactivated or live virus, or heat-killed Mycobacteria, and assessed their differentiation after 5 d. When exposed to inactivated influenza A virus or live Sendai virus, monocytes preferentially differentiated into mo-mac, while mo-DC differentiation was almost abrogated (Fig. 1 B and C). By contrast, Mycobacterium butyricum (MB) significantly increased mo-DC differentiation (Fig. 1D). These results show that pathogen exposure impacts monocyte differentiation, with different outcomes depending on the pathogen. Mycobacteria contain motifs recognized by TLR and NOD receptors, which have been shown to antagonize each other (24, 25). To address whether NOD signaling was dominant in the observed effect on monocyte differentiation, we used GSK583, a chemical inhibitor of Receptor-interacting-serine/threonine-protein kinase 2 (RIPK2), the adapter protein required for NOD signal transduction. GSK583 inhibited the secretion by monocytes of IL-6 and IL-8 after exposure to the synthetic NOD2 ligand Murabutide, confirming the efficiency of this inhibitor (SI Appendix, Fig. S1A). Enhanced mo-DC differentiation induced by heat-killed Mycobacterium tuberculosis (MT) was abrogated in the presence of GSK583 (Fig. 1E), indicating that Mycobacteria promote mo-DC through NOD signaling.
Fig. 1.
Pathogen recognition impacts monocyte differentiation. Monocytes were cultured for 5 d with M-CSF, IL-4, and TNF-α. Monocyte-derived cells were stained for CD16 and CD1a and analyzed by flow cytometry. (A) CD16+ mo-mac, CD1a+ mo-DC, and CD16-CD1a–undifferentiated cells were sorted and stained with May–Grunwald–Giemsa solutions after cytospin. (Scale bar, 30 μM.) Representative images (n = 5). (B–D) Proportions of mo-mac and mo-DC at day 5 after monocyte exposure to inactivated influenza A virus (Flu, B, n = 12), live Sendai virus (C, n = 8), and heat-killed mycobacterium butyricum (MB) (D, n = 15). Each symbol represents one individual donor. (E) Monocytes were preincubated with GSK583, heat-killed mycobacterium tuberculosis (MT) was added, and differentiation was analyzed after 5 d of culture. Wilcoxon test. *P < 0.05; **P < 0.01; ***P < 0.001; ns, not significant.
TLR Ligands Promote mo-mac Differentiation while NOD Ligands Induce mo-DC.
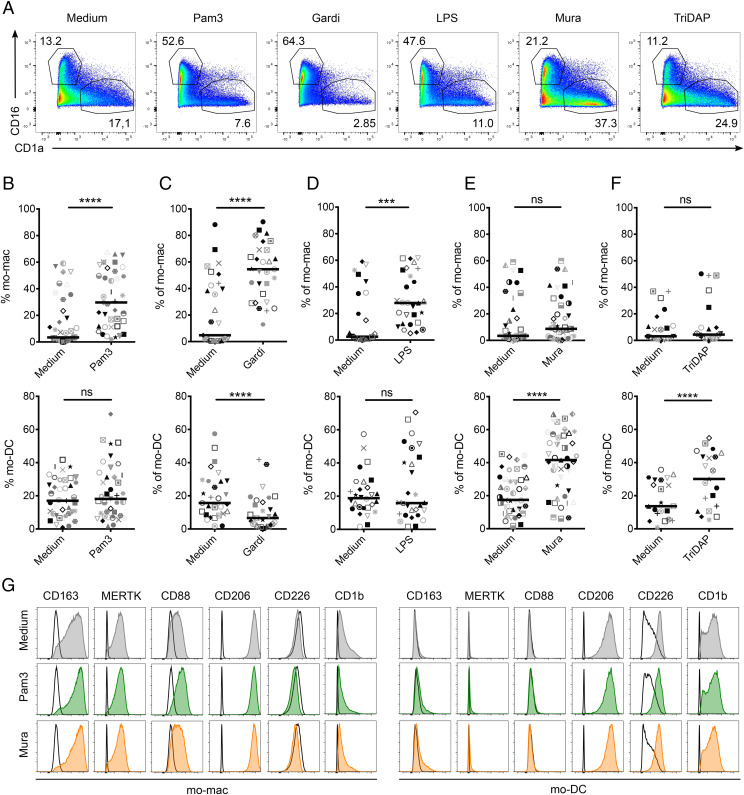
To decipher the molecular mechanisms involved, we used a reductionist approach and exposed monocytes to single ligands at the start of the culture and assessed their differentiation after 5 d (Fig. 2A). We found that monocytes exposed to Pam3 (TLR2 ligand, Fig. 2B and SI Appendix, Fig. S1B), Gardiquimod (TLR7 ligand, Fig. 2C and SI Appendix, Fig. S1B), LPS (TLR4 ligand, Fig. 2D and SI Appendix, Fig. S1B), and R848 (TLR7/8 ligand, SI Appendix, Fig. S1C) preferentially differentiated into mo-mac compared to the control condition. Gardiquimod and R848 also decreased the proportion of mo-DC, while Pam3 and LPS had no significant impact on mo-DC differentiation. By contrast, Murabutide (NOD2 ligand, Fig. 2E and SI Appendix, Fig. S1B) and TriDAP (NOD1 ligand, Fig. 2F and SI Appendix, Fig. S1B) promoted mo-DC differentiation without affecting the proportion of mo-mac. All these ligands activated monocytes to a similar extent, as shown by inflammatory cytokine secretion after 24 h (SI Appendix, Fig. S1D). In addition, TLR and NOD receptor ligands did not decrease either the percentage of live cells after 24 h (SI Appendix, Fig. S1E) or the number of live cells after 5 d (SI Appendix, Fig. S1F), suggesting that these molecules impact the differentiation of monocytes rather than their survival. Monocyte exposure to Pam3, Gardiquimod, Murabutide, and TriDAP did not modify the phenotype of mo-mac or mo-DC (Fig. 2G and SI Appendix, Fig. S1G). These results show that exposure to pathogen-derived products impacts monocyte fate decision, with TLR ligands and NOD activation having opposite effects.
Fig. 2.
TLR ligands promote mo-mac while NOD ligands induce mo-DC differentiation. Monocytes were cultured for 5 d with M-CSF, IL-4, and TNF-α with indicated lignands. Monocyte-derived cells were stained for CD16 and CD1a and analyzed by flow cytometry. (A–G) Representative results. (B–F) Proportions of mo-mac and mo-DC at day 5 after monocyte exposure to Pam3 (B, n = 39), Gardiquimod (Gardi, C, n = 29), LPS (D, n = 28), Murabutide (Mura, E, n = 41), and TriDAP (F, n = 21). Each symbol represents one individual donor. (G) Expression of the surface markers CD163, MERTK, CD88, CD206, CD226, and CD1b was analyzed on mo-mac and mo-DC at day 5 after monocyte exposure to Pam3 or Mura. Empty histograms correspond to isotype controls. Representative of four independent experiments. ***P < 0.001; ****P < 0.0001; ns, not significant.
Monocyte Exposure to TLR and NOD Receptor Ligands Modifies the Functional Properties of mo-mac and mo-DC.
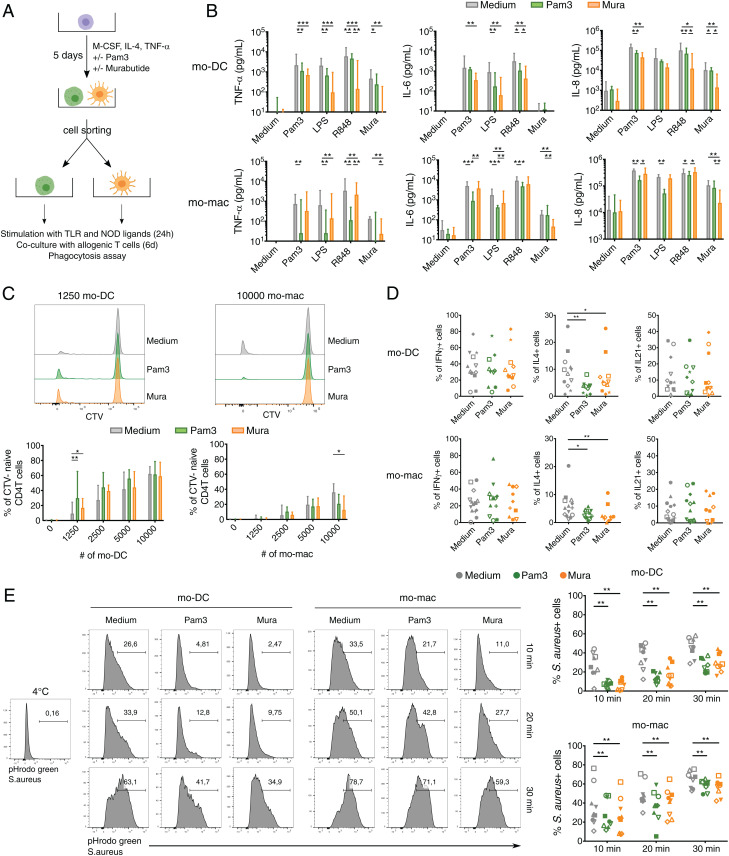
We then addressed whether pathogen recognition by monocytes affects the functional properties of resulting mo-mac and mo-DC. We cell sorted mo-mac and mo-DC differentiated from monocytes exposed or not to Pam3 or Murabutide and assessed their capacity to secrete cytokines following stimulation, to induce T cell proliferation and polarization, and to perform phagocytosis (Fig. 3A). We first analyzed cytokine secretion by mo-DC and mo-mac. We measured the secretion of TNF-α, IL-6, and IL-8 after 24 h of stimulation with Pam3, LPS, R848, or Murabutide (Fig. 3B). Mo-DC that differentiated from monocytes exposed to Pam3 or Murabutide secreted lower amounts of TNF-α, IL-6, and IL-8 following stimulation. Similar results were observed for mo-mac (Fig. 3B). To assess the ability of mo-DC and mo-mac to induce T cell proliferation and polarization, we performed a mixed leukocyte reaction with sorted mo-mac or mo-DC and allogeneic blood CD4 T cells. Mo-DC differentiated from monocytes exposed to Pam3 or Murabutide had a higher ability to stimulate naïve CD4 T cell proliferation compared to control mo-DC when using a low number of antigen-presenting cells (Fig. 3C and SI Appendix, Fig. S2A). By contrast, we did not find any significant difference for mo-DC in the induction of memory CD4 T cell proliferation (SI Appendix, Fig. S2B). Mo-mac are poorer inducers of T cell proliferation as previously observed (22). Moreover, mo-mac differentiated from monocytes exposed to Pam3 or Murabutide had a lower capacity to induce naïve CD4 T cells proliferation (Fig. 3C and SI Appendix, Fig. S2A). To analyze helper T cell polarization, we cultured isolated mo-DC and mo-mac with allogeneic blood naïve CD4 T cells for 6 d and assessed cytokine production by T cells using intracellular flow cytometry. There was no major impact of monocyte exposure to pathogen products, except for Th2 polarization where mo-DC and mo-mac differentiated from monocytes exposed to Pam3 and Murabutide induced a lower proportion of IL-4–producing T cells (Fig. 3D). A higher expression of costimulatory molecules could explain the better ability of mo-DC differentiated from monocytes exposed to Pam3 and Murabutide to induce naïve CD4 T cell proliferation. However, we did not observe higher levels of CD80, CD86, HLADR, or CD40 in comparison with control mo-DC or mo-mac (SI Appendix, Fig. S2C). Finally, we assessed the phagocytic function of mo-DC and mo-mac. We performed a phagocytic assay using pHrodo green Staphylococcus aureus bioparticles conjugate, which emits fluorescence only when exposed to an acidic pH in endocytic compartments. Mo-DC differentiated from monocytes exposed to Pam3 or Murabutide displayed lower phagocytic abilities compared to control mo-DC (Fig. 3E). We observed the same effect for mo-mac differentiated from monocytes exposed to Murabutide (Fig. 3E). Collectively, these results show that the functional properties of mo-mac and mo-DC are affected by pathogen recognition by progenitor monocytes but independently of the nature of the pathogen compounds.
Fig. 3.
Monocyte exposure to TLR or NOD receptor ligands modifies the functional properties of mo-mac and mo-DC. Monocytes were cultured for 5 d with M-CSF, IL-4, and TNF-α in the presence or absence of Pam3 or Murabutide (Mura). At day 5, mo-mac and mo-DC were sorted and assessed for cytokine secretion, ability to induce T cell proliferation and polarization, and phagocytic properties. (A) Scheme of the experimental setup. (B) Sorted mo-DC and mo-mac were stimulated for 24 h with Pam3, LPS, R848, or Mura. TNF-α, IL-6, and IL-8 secretion was measured in supernatants. For mo-DC, n > 8 and for mo-mac, n > 9. (C and D) Sorted mo-DC and mo-mac were cocultured with blood allogenic naïve CD4 T cells for 6 d. (C) Proliferation of CD4 T cells was analyzed by measuring the dilution of a proliferation trace dye (Cell Trace Violet [CTV]). Representative histograms and percentage of CTV cells are shown. For mo-DC, n > 10 and for mo-mac, n > 9. (B and C) The median and interquartile range are represented. (D) Polarization of CD4 T cells was assessed by intracellular flow cytometry after staining for IFNy, IL-4, and IL-17. (n > 9). (E) Monocyte-derived cells were incubated with pHrodo S. aureus bioparticles conjugates for 10, 20, or 30 min at 37 °C. Phagocytosis was assessed by measuring the fluorescence emitted by phagocytosed bacteria and defined using the negative control (incubation at 4 °C). (n = 9). Wilcoxon test. *P < 0.05; **P < 0.01; ***P < 0.001. Each symbol represents an individual donor.
TLR Ligands Increase MAFB Expression through mTORC1 Activation.
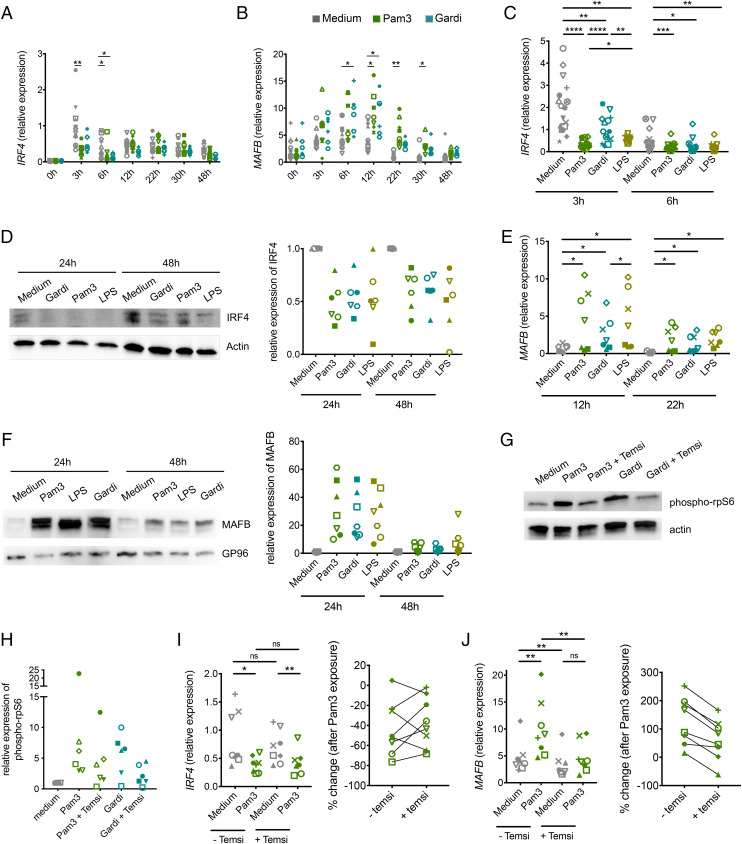
We have previously shown in this model that monocytes differentiate into mo-DC or mo-mac along two distinct pathways controlled by the transcription factors IRF4 and MAFB, respectively (22). To elucidate the mechanisms involved in monocyte fate decision upon TLR ligand recognition, we first analyzed the impact of TLR ligands on IRF4 and MAFB expression by qRT-PCR at early timepoints. Pam3 significantly decreased the expression of IRF4 at 3 and 6 h (Fig. 4A) and increased that of MAFB at 12 and 22 h (Fig. 4B). To extend these observations to other TLR ligands, we repeated this analysis using Gardiquimod and LPS at relevant timepoints. Similar to Pam3, Gardiquimod and LPS significantly decreased the early peak of IRF4 expression (Fig. 4C). To test whether IRF4 and MAFB expression were modified at the protein level, we performed Western blot analysis after 24 and 48 h. Consistent with the qRT-PCR results, TLR ligands inhibited the expression of IRF4 protein (Fig. 4D). Moreover, Gardiquimod and LPS increased the expression of MAFB at the messenger RNA (mRNA) and protein levels to the same extent as Pam3 (Fig. 4 E and F). These results indicate that exposure to TLR ligands strongly modifies the balance of IRF4 and MAFB expression by monocytes.
Fig. 4.
TLR ligands modify the balance of IRF4 and MAFB expression through mTORC1 activation. Monocytes were cultured with M-CSF, IL-4, and TNF-α in the presence or absence of indicated TLR ligands. (A–C and E) mRNA expression of IRF4 (A and C) and MAFB (B and E) were measured by qRT-PCR at indicated timepoints. (D and F) Protein expression of IRF4 (D) and MAFB (F) were measured by Western blot at indicated timepoints. Relative protein expression was assessed by densitometry as compared to actin or GP96 (n > 6). (G and H) Monocytes were cultured for 2 h in the presence or absence of Pam3, Gardiquimod (Gardi), and Temsirolimus (temsi). (G) Protein expression of phospho-rpS6 or actin was measured by Western blot. Representative of six donors. (H) Relative protein expression was assessed by densitometry as compared to actin (n = 6). (I and J) IRF4 (I) and MAFB (J) expression were measured by qRT-PCR after 3 and 12 h of culture, respectively. Percentage of change of IRF4 and MAFB expression in the Pam3 condition versus the control condition in the presence or absence of Temsirolimus were calculated. (n = 9). Each symbol represents an individual donor. Wilcoxon test. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001; ns, not significant.
TLR signaling is known to induce mammalian target of rapamycin (mTOR) activation in immune cells (26, 27), and mTORC1 inhibition has been reported to promote mo-DC differentiation (28). Therefore, we hypothesized that mTOR activation may impact monocyte differentiation upon TLR ligand exposure. To confirm that TLR ligands activate the mTOR pathway in human monocytes, we analyzed the phosphorylation of the ribosomal S6 protein, a surrogate of mTORC1 activation. Pam3 and Gardiquimod induced high amounts of ribosomal protein S6 phosphorylation (Fig. 4 G and H). This effect was inhibited by the mTORC1 inhibitor Temsirolimus, confirming that this phosphorylation was dependent on mTORC1 (Fig. 4 G and H). To address the impact of mTORC1 activation on IRF4 and MAFB expression, we performed qRT-PCR on monocytes cultured in the presence or absence of Pam3 and Temsilorimus (Fig. 4 I and J). mTORC1 inhibition had no significant impact on IRF4 expression in the presence or absence of Pam3 (Fig. 4I). By contrast, the increase of MAFB expression induced by Pam3 was abrogated by the addition of Temsirolimus (Fig. 4J). These results show that TLR ligands increase MAFB expression through mTORC1 activation.
NOD Receptor Ligands Promote mo-DC Differentiation through TNF-α Secretion.
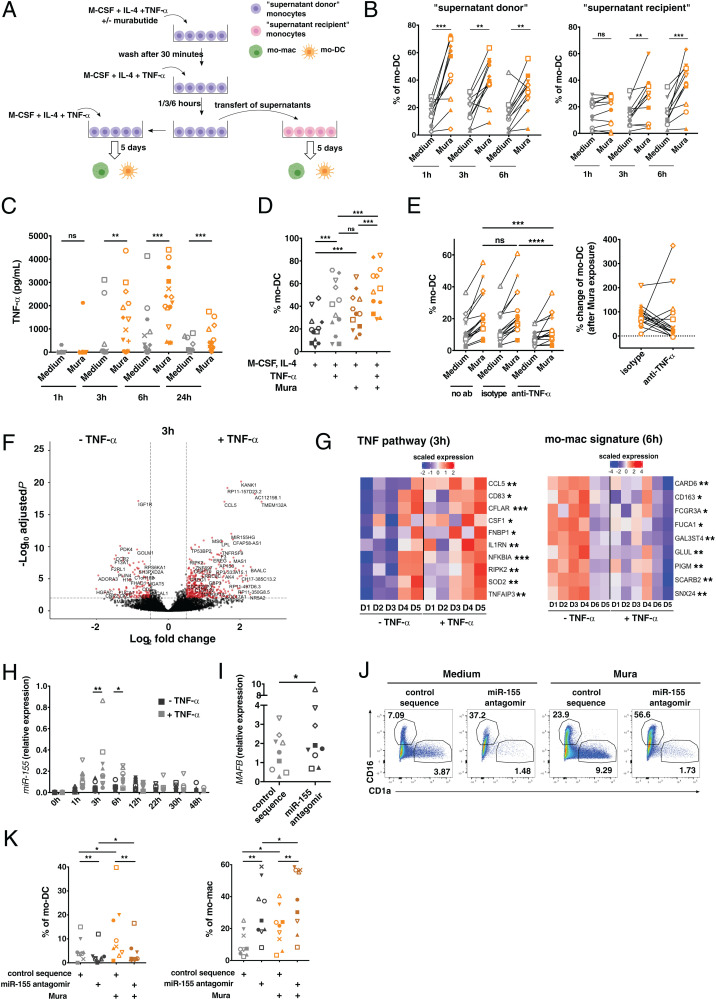
To understand how NOD receptor ligands induce mo-DC differentiation, we tested the hypothesis of a cell extrinsic mechanism relying on secreted factors. To this end, we performed supernatant transfer experiments (Fig. 5A). We stimulated or not monocytes with Murabutide for 30 min, then washed the cells and added fresh medium without Murabutide to obtain conditioned supernatant. After 1, 3, or 6 h of incubation, we transferred this supernatant from the “supernatant donor” monocytes to unstimulated “supernatant recipient” monocytes from the same blood donor and analyzed their differentiation after 5 d. As a control, we also cultured the same monocytes without manipulating the medium (“untouched cells”) and confirmed that Murabutide exposure increased mo-DC differentiation in these cells (SI Appendix, Fig. S3A). To test the potential carryover of Murabutide in the conditioned medium, we measured the overnight secretion of IL-6 and IL-8 by the “supernatant recipient” monocytes that received the conditioned medium after 1 h (SI Appendix, Fig. S3B). There was no activation of monocytes in these conditions, showing that the conditioned medium was free of Murabutide. As expected, the “supernatant donor” monocytes stimulated with Murabutide preferentially differentiated into mo-DC compared to their control counterparts (Fig. 5B). Conditioned supernatant transferred after 3 and 6 h to the “supernatant recipient” monocytes increased their differentiation into mo-DC (Fig. 5B). These results suggest that a soluble factor, secreted later than 1 h after monocyte stimulation, promotes their differentiation into mo-DC.
Fig. 5.
NOD receptor ligands promote mo-DC differentiation through TNF-α secretion and miR-155 expression. (A) Experimental setup for the supernatant transfer experiment. (B) Mo-DC percentages after 5 d in “supernatant donor” and “supernatant recipient” cultures (n = 11). (C) Monocytes were cultured with M-CSF and IL-4 in the presence or absence of Murabutide (Mura). After 1, 3, 6, and 24 h, TNF-α secretion in supernatants was measured (n > 9). (D) Monocytes were cultured for 5 d with M-CSF and IL-4 in the presence or absence of Mura or TNF-α (n = 12). (E) Monocytes were cultured for 5 d with M-CSF and IL-4 in the presence or absence of Mura, anti–TNF-α neutralizing antibodies, or isotype control antibodies (n = 17). Proportions of mo-DC (Left) and percentage of change in mo-DC proportion in the Mura condition versus the control condition with the anti–TNF-α antibody or isotype control (Right) (n = 17). (F and G) Monocytes were cultured with M-CSF and IL-4 in the presence or absence of TNF-α for 3 and 6 h and analyzed by RNA-sequencing (n = 5). (F) Differentially expressed genes in the presence of TNF-α versus control condition at 3 h. (G) Scaled expression of genes from the TNF pathway and from the mo-mac signature in each sample at indicated timepoints. Each column represents an individual donor. D, donor. Student’s t test, *P < 0.05, **P < 0.01, ***P < 0.001. (H) Monocytes were cultured with M-CSF and IL-4 in the presence or absence of TNF-α. At different timepoints, miR-155 expression was measured by qRT-PCR. (I–K) Monocytes were transfected with a miR-155 antagonist (antagomir) or a control sequence and cultured with M-CSF, IL-4, and TNF-α. (I) MAFB expression was determined by qRT-PCR after 6 h. (n = 9). (J and K) Monocytes differentiation after 5 d of culture. (J) Representative results (n = 9). (K) Proportions of mo-DC and mo-mac (n = 9). Wilcoxon test. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001. Each symbol represents one donor; ns, not significant.
IL-32 has been suggested to induce mo-DC differentiation (29). However, we were unable to measure any IL-32 secretion upon TLR ligand exposure, although other cytokines such as IL-6 were detected (SI Appendix, Fig. S3C). We have previously shown that TNF-α promotes mo-DC differentiation in our culture model (22). Moreover, monocytes secreted high amounts of TNF-α after Murabutide exposure, starting after 1 h and with a peak of secretion at 6 h (Fig. 5C), consistent with the kinetics observed in the supernatant transfer experiment. We therefore evaluated further TNF-α as a candidate factor. We hypothesized that TNF-α and Murabutide have redundant effects in our model. To test this, we cultured monocytes with or without TNF-α in the cytokine mixture (Fig. 5D). Murabutide and TNF-α induced mo-DC differentiation to the same extent, and the use of the two molecules combined resulted in an additive effect (Fig. 5D). To validate the role of TNF-α, we used neutralizing anti–TNF-α antibodies. The increase in mo-DC proportion due to Murabutide exposure was lower in the presence of anti–TNF-α antibodies compared to the isotype control (Fig. 5E). Of note, TNF-α secretion was not specific to NOD receptor stimulation, as TLR ligands also induced it (SI Appendix, Fig. S3D). This suggests that the effect of TNF-α in monocytes can be counteracted by other signaling events induced upon TLR stimulation. Collectively, these results demonstrate that NOD receptor activation promotes mo-DC differentiation through the secretion of TNF-α.
TNF-α Promotes mo-DC Differentiation by Inducing mi-R155.
To address how TNF-α promotes mo-DC differentiation, we performed transcriptomic analysis on monocytes cultured for 3 and 6 h with M-CSF and IL-4 in the presence or absence of TNF-α (Fig. 5F and SI Appendix, Fig. S3E). As an internal control, we confirmed that monocytes exposed to TNF-α up-regulated several genes of the TNF pathway at 3 h (Fig. 5G). Moreover, monocytes exposed to TNF-α down-regulated some genes of the mo-mac transcriptomic signature at 6 h, consistent with an increased differentiation into mo-DC in response to TNF-α (Fig. 5G). Among the list of differentially expressed transcripts in TNF-α–exposed monocytes versus control monocytes, we selected miR-155 as a candidate (SI Appendix, Fig. S3F), because miR-155 was previously shown in cell lines to target MAFB (30, 31). We validated by qRT-PCR that TNF-α induces miR-155 expression in monocytes with a peak at 3 and 6 h (Fig. 5H). MAFB starts being expressed after 3 h with a plateau at 6 and 12 h (Fig. 4B), making the interaction with miR-155 possible in terms of kinetics. To address directly whether miR-155 targets MAFB in human monocytes, we measured its expression by qRT-PCR 6 h after transfection of an miR-155–specific antagonist (antagomir) or a control sequence (scramble). MiR-155 antagomir increased the expression of MAFB in comparison to the control sequence, showing that miR-155 targets MAFB (Fig. 5I). Of note, Pam3 and Murabutide exposure (which induces high amounts of TNF-α) resulted in an increased expression of miR-155 (SI Appendix, Fig. S3G). To test whether miR-155 promotes mo-DC differentiation, we cultured monocytes after transfection with the miR-155 antagomir or the control sequence. miR-155 antagomir decreased mo-DC and increased mo-mac differentiation, with or without Murabutide stimulation (Fig. 5 J and K). In particular, in comparison to the control, miR-155 antagomir abrogated the increase in mo-DC differentiation induced by Murabutide. In conclusion, we found that miR-155 is necessary for monocyte differentiation into mo-DC in response to TNF-α exposure.
Pam3 and TNF-α Promote the In Vivo Differentiation of Mouse mo-mac and mo-DC, Respectively.
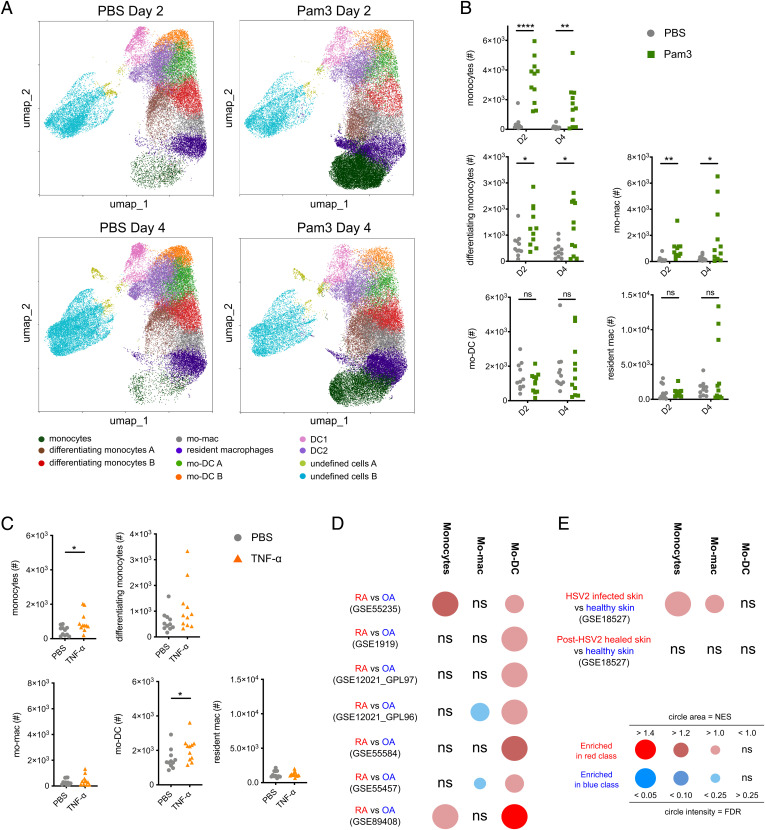
To evaluate the in vivo relevance of our findings, we analyzed the differentiation of monocytes in mouse skin. We first addressed the impact of TLR signaling by analyzing skin cells 2 and 4 d following intradermal injections of Pam3. To select a panel of phenotypic markers allowing the identification of monocytes, mo-DC, mo-mac, and their differentiation intermediates, we refined a previously published flow cytometry strategy (32). We first performed unsupervised identification of CD45+lin-EpCam-Ly6G-cells using FlowSOM (33). We found 11 cell clusters that we manually annotated based on the expression of 9 myeloid cell markers (Fig. 6A and SI Appendix, Fig. S4A). Consistent with the induction of inflammation, monocytes were increased in Pam3-treated mice at both timepoints (SI Appendix, Fig. S4B). At day 4, we observed higher numbers of mo-mac in Pam3-treated mice compared to phosphate buffered saline (PBS)-treated mice, while mo-DC were decreased at both timepoints (SI Appendix, Fig. S4B). These results suggest that Pam3 exposure preferentially increases monocyte differentiation into mo-mac. To obtain a more quantitative evaluation of monocyte differentiation, we manually gated these populations (SI Appendix, Fig. S4D). We focused on monocytes, differentiating monocytes, mo-mac, resident macrophages, mo-DC, neutrophils, Langerhans cells (LC), classical dendritic cell type 1 (DC1), and type 2 (DC2) (SI Appendix, Fig. S4D). At 2 d after Pam3 injection, monocytes and neutrophils numbers were massively increased and remained high at day 4 (Fig. 6B and SI Appendix, Fig. S4C). DC1 numbers were decreased following Pam3 injection at both times, while LC and DC2 were not changed (SI Appendix, Fig. S4C). Upon Pam3 injection, we observed higher numbers of differentiating monocytes at both timepoints in comparison with PBS-treated mice, showing that the accumulated monocytes had started to differentiate (Fig. 6B). There were more mo-mac in the skin following Pam3 injection (Fig. 6B). Neither mo-DC nor resident macrophages were affected (Fig. 6B). However, Pam3 might increase both the differentiation and migration of mo-DC, resulting in apparent constant numbers in the skin. To address this hypothesis, we analyzed DC populations in the skin-draining lymph nodes (SI Appendix, Fig. S4E). We could not detect in the lymph nodes any mo-DC population (expressing Ly6C or CD64), suggesting that mo-DC did not migrate upon Pam3-induced inflammation (SI Appendix, Fig. S4E). By contrast, both resident and migratory DC2 numbers were increased (SI Appendix, Fig. S4F). These observations are consistent with previous studies reporting poor migratory capacity for mo-DC (34–36). Collectively, these results show that Pam3 specifically increases mo-mac differentiation in vivo.
Fig. 6.
In vivo exposure to TLR ligands or TNF-α correlates with increased mo-mac or mo-DC differentiation. (A–C) Pam3, TNF-α, or PBS as control, was injected intradermally in the ears of C57B6/J mice. Single cell suspensions were prepared by mechanical and enzymatic digestion. (A) Uniform manifold approximation and projection (UMAP) representation of the 11 cell clusters identified by FlowSOM unsupervised analysis. Each dot represents an individual cell. n = 5 mice in two independent experiments. (B and C) Total cell numbers of monocytes, differentiating monocytes, mo-mac, resident macrophages (resident mac), and mo-DC are represented upon Pam3 (B, day 2 [D2] and day 4 [D4]) or TNF-α (C, day 2) injection. Each symbol corresponds to an individual mouse (three independent experiments). Student’s t test. *P < 0.05; **P < 0.01; ****P < 0.0001. (D and E) Gene expression data were extracted from indicated public datasets. GSEA was performed for monocyte, mo-DC, and mo-mac gene signatures. BubbleGUM representations of the GSEA results are shown. (D) GSEA on datasets from synovial biopsies from joints of RA or OA patients. (E) GSEA on datasets from skin biopsies from lesional or healthy skin of HSV2 infected patients. The normalized enrichment score (NES) and the false discovery rate (FDR) correspond to the strength and the significance of the result respectively. ns, not significant.
We then addressed the impact of TNF-α exposure. We employed the same strategy to analyze monocyte-derived cells 2 d following TNF-α injection. Based on the unsupervised FlowSOM analysis (SI Appendix, Fig. S5 A and B), we observed higher numbers of monocytes and mo-DC upon TNF-α injection (SI Appendix, Fig. S5C). However, we did not observe any change in the numbers of mo-mac (SI Appendix, Fig. S5C). We further confirmed these observations by manually gating the different population (SI Appendix, Fig. S5D). Monocyte and mo-DC numbers were increased while mo-mac counts were not changed, showing that monocytes preferentially differentiated into mo-DC (Fig. 6C). TNF-α exposure did not modify the other populations (Fig. 6C and SI Appendix, Fig. S5E).
In conclusion, these results confirm that TLR activation increases mo-mac differentiation while TNF-α exposure promotes mo-DC in an in vivo setting.
Enrichment for mo-mac or mo-DC in Human Skin Biopsies Correlates with In Vivo Exposure to Pathogens or TNF-α.
Finally, we assessed the validity of our findings in an in vivo setting in humans. We analyzed the enrichment of monocyte, mo-mac, and mo-DC transcriptomic signatures using GSEA (37) in relevant human samples using public microarray data sets. To determine the impact of TNF-α exposure on monocyte differentiation in vivo, we compared data sets of synovial samples from patients suffering from RA (an inflammatory disease in which TNF-α plays a major role in the pathogenesis) or osteoarthritis (OA) (a mechanical disorder that is considered independent of inflammation). Indeed, higher TNF-α levels have been reported in synovial fluid of RA patients in comparison with OA patient (38). We used our previously published gene signatures for human mo-DC (and mo-mac), defined as the genes enriched in ascites mo-DC (or mo-mac) compared to ascites mo-mac (or mo-DC) and blood CD14+ monocytes (22). The monocyte signature was enriched in RA versus OA samples in some data sets, consistent with the known recruitment of monocytes in the inflamed joints in RA (39) (Fig. 6D). The mo-DC signature was significantly enriched in RA versus OA samples in all data sets analyzed (Fig. 6D and SI Appendix, Fig. S6A). Of note, the mo-mac signature was only enriched in OA versus RA samples in some data sets. Consistent with its induction by TNF-α, miR-155 was expressed at higher levels in RA compared to OA synovial tissue (SI Appendix, Fig. S6B). Overall, these results are consistent with our finding that TNF-α promotes mo-DC differentiation.
To assess the impact of pathogen recognition, we analyzed skin biopsies from volunteers experimentally infected with HSV2 and compared infected skin with healthy skin from the same individuals, during infection or after resolution (40). We found that the monocyte and mo-mac signatures were enriched in HSV2-infected skin compared to healthy skin, while none of the three signatures was enriched in healed skin compared to healthy skin (Fig. 6E and SI Appendix, Fig. S6C). These results support our findings that viruses increase mo-mac differentiation.
Discussion
In this study, we have demonstrated that pathogen recognition skews monocyte differentiation into macrophages versus DC. We showed that viruses strongly promote mo-mac differentiation and inhibit mo-DC development. By contrast, Mycobacteria favor mo-DC development. We evidenced that TLR activation redirects monocyte differentiation toward mo-mac by inhibiting IRF4 and inducing MAFB expression. On the other hand, NOD receptor activation promotes mo-DC differentiation through autocrine TNF-α and miR-155 expression.
During infections, monocytes encounter pathogens containing various pathogen-associated molecular patterns (PAMPs) that can potentially trigger many different signaling pathways. In the face of such complex stimulation, cells make signaling decisions to either integrate multiple signals, resulting in synergy or antagonism, or activate signaling pathways independently without any cross talk (41, 42). We found that Mycobacteria, which contain both TLR and NOD ligands, increase mo-DC differentiation. This suggests that, during Mycobacteria recognition, TLR signaling is counteracted by NOD activation in monocytes. This is consistent with previous studies showing that NOD receptors negatively regulate TLR-induced NF-κB signaling in mouse bone marrow–derived macrophages and splenic myeloid cells (24, 25). Simplification of signaling integration has also been suggested in DC exposed to triple PAMPs stimulations, where the sensing outcome was similar to that of single compounds or pairs (43). Our observation that TLR activation favors mo-mac differentiation despite high secretion of TNF-α suggests that signaling priorization may also extend to the simultaneous activation of separate NF-κB–dependent pathways, such as TLR and TNF-receptor signaling. Finally, pathogens may activate other receptors such as RIG-I like, AIM2-like, and C-type lectin receptors. The impact of these signaling pathways on monocyte differentiation remains open for future investigation.
In the past few years, numerous studies have described the ability of innate immune cells to acquire a memory response following PAMP sensing, which has been termed “trained immunity.” This phenomenon has first been demonstrated in the context of monocyte to macrophage differentiation. Indeed, macrophages differentiated from β-glucan–exposed monocytes secreted higher levels of proinflammatory cytokines following stimulation with various PAMPs or whole pathogen (44, 45). By contrast, macrophages exposed to LPS secreted lower levels of proinflammatory cytokines following a second stimulation with LPS (46), which is referred to as “LPS tolerance.” For both trained immunity and tolerance, only mo-mac have been studied and whether the same phenomenon occurs in mo-DC has remained unclear. Here, we show that mo-mac and mo-DC differentiated from Pam3- and Murabutide-exposed monocytes secreted decreased levels of proinflammatory cytokines after stimulation with various PAMPs. Our results show that, similar to mo-mac, mo-DC can be imprinted for tolerance. LPS tolerance has been associated with enhanced phagocytic capacities and reduced antigen presentation abilities in macrophages derived from monocytes (47). However, we found that mo-DC differentiated from Pam3- and Murabutide-exposed monocytes were more efficient for stimulating T cell proliferation. Moreover, mo-DC and mo-mac differentiated from monocytes activated with Pam3 and Murabutide displayed decreased phagocytic capacities.
Recent studies have demonstrated a role for mTOR during myelopoiesis. The deletion of mTOR or of the mTORC1 component Raptor in mice strongly reduces the development of monocytes and neutrophils (48–50). Here, we evidenced a role for mTORC1 in the differentiation of monocytes into mo-mac. We found that mTORC1 activation induces MAFB expression, which is essential for mo-mac differentiation (22). This is in contrast with the up-regulation of MAFB detected in granulocyte-monocyte progenitors from mTOR knock-out (KO) mice (50). However, osteoclast treatment with rapamycin increased MAFB showing that mTOR inhibits MAFB expression in these cells (51). This suggests that the effect of mTOR on MAFB expression may be cell specific.
It has been demonstrated that following TLR stimulation with LPS, poly-IC or CpG, bone marrow–derived macrophages (52) and human mo-DC (53) up-regulate miR-155 expression. Here, we show that TNF-α exposure induces the expression of miR-155 in human monocytes. This suggests that increased miR-155 expression following TLR activation is mediated by TNF-α. miR-155 has pleiotropic effects in immune cells. It is involved in granulocyte/monocyte proliferation in mouse bone marrow (54), polarization of bone marrow–derived macrophages toward a proinflammatory state (55), and maturation of bone marrow–derived DC (56). Here, we demonstrate that in addition to its role in the proliferation and function of myeloid cells, miR-155 is also essential for monocyte differentiation toward mo-DC. We further show that MAFB is a target of miR-155 in human monocytes. We have previously observed that MAFB silencing in monocytes favors mo-DC differentiation (22). Thus, increased mo-DC differentiation in response to TNF-α could be due to MAFB targeting by miR-155. However, we cannot exclude that other targets of miR-155 could be involved in this process. Finally, miR-155 expression has been associated with RA severity. Synovial fluid monocytes and synovial tissue macrophages from RA patients express higher levels of miR-155 than that of OA patients (55, 57). This is consistent with the induction of miR-155 expression by TNF-α, which is highly secreted in inflamed RA joints. In monocytes from RA patients and healthy donors, miR-155 expression also induces proinflammatory cytokine secretion including TNF-α (55, 58). This suggests the existence of an amplification loop between TNF-α and miR-155 potentially exacerbating the immunopathology.
Manipulating the differentiation of monocytes into mo-DC versus mo-mac represents an appealing strategy to dampen inflammation or promote immune responses, depending on the context. This strategy is hindered by a limited knowledge of the mechanisms driving monocyte fate decision and differentiation. Here, we identified mTORC1 and mir-155 as modulators of this process. By providing a better understanding of the molecular regulation of monocyte differentiation, these results should open up possibilities for therapeutic opportunities.
Materials and Methods
Human Blood, Monocyte Isolation, and Culture.
Buffy coats from healthy donors were obtained from Etablissement du Sang Francais in accordance with the Institut National de la Santé et de la Recherche Médicale (France) (INSERM) ethical guidelines. According to French Public Health Law (art L 1121–1-1, art L 1121–1-2), written consent and Institutional Review Board approval are not required for human noninterventional studies. Peripheral blood monocuclear cells were obtained by centrifugation on a ficoll gradient (Lymphoprep, StemCell). Blood monocytes were then positively isolated using CD14+ microbeads (Miltenyi) according to the manufacturer’s recommendations. Monocytes (2.106/mL) were cultured for 5 d with Roswell Park Memorial Institute medium-Glutamax (Gibco) supplemented with antibiotics (penicillin and streptomicin) and 10% fetal calf serum (FCS) in the presence of 100 ng/mL M-CSF (Miltenyi), 5 ng/mL IL-4 (Miltenyi), and 5 ng/mL TNF-α (Miltenyi). Where indicated, 1 μg/mL Pam3 (Invivogen), 1.25 μg/mL LPS (Invivogen), 2 μg/mL Gardiquimod (Invivogen), 100 ng/mL R848 (Invivogen), 10 μg/mL Murabutide (Invivogen), 10 μg/mL TriDAP (Invivogen), 20 μg/mL formalin inactivated influenza A (Charles River), Sendai virus (Charles River), and heat-killed MB (Fisher Scientific) were added at the beginning of the culture and were extensively washed after 30 min or not washed. For the inhibition of mTORC1 signaling, monocytes were cultured with 25 nM Temsirolimus (Sigma). For the neutralization of TNF-α, monocytes were cultured in the presence of 2 μg/mL anti–TNF-α neutralizing ab (R&D Biotechne) or mouse IgG1 isotype control (R&D Biotechne). For the inhibition of miR-155, monocytes were transfected with TransIT-X2 (Mirus) and 5 pmol miR-155 antagomir (Thermo Fisher Scientific) or control sequence (Thermo Fisher Scientific) and cultured with the cytokines and Murabutide as previously described for 5 d. For the inhibition of RIPK2 signaling, monocytes were preincubated with 1 μM GSK583 (R&D Biotechne) for 30 min, and Murabutide or heat-killed MT (Invivogen) was added.
Flow Cytometry of Monocyte-Derived Cells.
After 5 d of culture, cells were detached using cold PBS containing 0.5% human serum and 2 mM EDTA. Staining details can be found in SI Appendix. Cells were acquired on a FACSVerse instrument (BD Biosciences). In the monocyte differentiation assay, we set for each individual donor the CD1a+ and CD16+ gates on the medium condition and then applied these gates to the different test conditions. To quantify the expression of activation markers, the Stain Index was calculated as follows: (mean fluorescence intensity [MFI]sample -MFIisotype) /(SDisotype)). MFI represents the median of fluorescence and SD the standard deviation.
Cytokine Secretion.
Culture details can be found in SI Appendix. Secretion of IL-8, IL-6, and TNF-α was measured by cytometric bead array (BD Biosciences).
qRT-PCR.
Details on sample preparation can be found in SI Appendix. The second derivative method was used to determine each Cp and the expression of genes of interest relative to the housekeeping genes.
Western Blot.
Details on membrane preparation can be found in SI Appendix. Primary antibodies were against IRF4 (Cell signaling), MAFB (NovusBio), gp96 (clone 9G10, Enzo Life Sciences), and actin (Millipore, clone C4). Images were acquired using a chemidoc instrument (Bio-Rad) and quantified with Fiji software.
Morphological Analysis of mo-mac and mo-DC.
Cells were placed on slides using a cytospin centrifuge and were stained with May-Grünwald (Sigma) and Giemsa (Merck) solutions. Pictures were acquired using a Leica DM 4000 B microscope with a ProgRes SpeedXTcore 5 camera.
CD4 T Cell Proliferation and Polarization.
Naïve or memory CD4 T cells were isolated from peripheral blood mononuclear cells using negative isolation kits (Stemcell) according to the manufacturer’s recommendations. Following 5 d of culture, mo-mac and mo-DC were sorted on a FACSAria instrument (BD Biosciences) after staining with anti-CD16 fluorescein isothiocyanate (FITC) and anti-CD1a APC. To assess T cell proliferation, T cells were stained with CellTrace Violet (Thermo Fisher Scientific). In total, 1,250, 2,500, 5,000, or 10,000 APC were cultured with 50,000 allogenic naïve or memory T cells in Yssel Medium supplemented with 10% FCS. After 6 d, T cells were stained with fixable viability efluor 780 dye (Thermo Fisher Scientific) for 10 min at 4 °C and counted using counting beads on a FACSVerse instrument.
To assess Th cell polarization, 20,000 APC were cultured with 50,000 allogenic naïve T cells in Yssel Medium supplemented with 10% FCS. After 6 d, T cells were washed and stimulated in X-Vivo TM 15 (Lonza) without FCS in the presence of PMA (50 ng/mL), ionomycin (1 μg/mL), and BFA (4 μg/mL; Sigma-Aldrich) for 5 h at 37 °C. Cells were then washed, stained with fixable viability efluor 780 dye, fixed, and permeabilized (Intracellular Fixation and Permeabilization Buffer Set, ebioscience). The cells were then stained for intracellular cytokines in a buffer containing 2% mouse serum (Thermo fisher Scientific) and human Fcblock (BD Biosciences) for 1 h at room temperature with the following antibodies: anti–IL-4 APC (ebiosciences, 8D4-8), anti-IL21 PE (BD Biosciences, clone 3A3-N2.1), and anti-IFNg PeCy7 (ebiosciences, clone 4S.B3). Samples were acquired on a FACSVerse instrument.
Phagocytosis.
Monocytes were cultured in RPMI (Gibco) supplemented with 10% FCS and antibiotics in the presence of M-CSF, IL-4, TNF-α, and 1 μg/mL Pam3 or 10 μg/mL Murabutide. After 5 d, cells were detached and plated in 96-well plates. After 30 min, pHrodo green S. aureus (Fischer Scientific) were added to the cells. Cells were washed with cold PBS and stained with fixable zombie NIR (Biolegend) for 10 min at 4 °C. Cells were then stained with TruStain blocking solution, anti-CD16 FITC, and anti-CD1a APC and fixed with 4% PFA for 10 min at 4 °C. The samples were acquired on a FACSVerse instrument.
GSEA.
GSEA was performed using the GSEA software (version 4.0.3) with the default parameters, except for the number of permutations that we fixed at n = 1,000. Datasets were downloaded from Gene Expression Omnibus (GEO) (GSE18527, GSE55235, GSE1919, GSE12021, GSE55584, GSE55457, GSE89408). Count matrix from RNA sequencing (RNA-seq) studies were first normalized using DESeq2. Gene signatures of blood monocytes, mo-DC and mo-mac were designed from microarray data (22).
RNA-seq Analysis.
Monocytes were culture in RPMI supplemented with 10% FCS in the presence of MSCF, IL-4, and in the presence or absence of TNF-α. Monocytes cultured for 3 and 6 h were lysed in RLT buffer (Qiagen). Details on library preparation can be found in SI Appendix. Differential gene expression analysis was performed using DESeq2 (59). Details on the analysis pipeline can be found in SI Appendix. Data are accessible through GEO Series accession no. GSE166043.
Analysis of Mouse Skin and Lymph Nodes.
In both ears of C57B6/J mice, 50 μg Pam3, 150 ng TNF-α, or PBS as control were injected intradermally. Ears were harvested and analyzed as previously described (60). Briefly, the two layers of each ear were separated and incubated overnight at 4 °C in 0.2 mg/mL dispase II (Roche). Ears were then cut in small pieces and incubated 90 min at 37 °C in digestion mix containing 0.5 mg/mL DNase I (Sigma) and 1.5 mg/mL collagenase IV (Worthington Biochemical Corporation). Ear-draining lymph nodes were harvested, cut in small pieces, and incubated for 30 min at 37 °C with 2 mg/mL Collagenase D (Roche) and 0.1 mg/mL DNase I (Roche). Cell suspensions were filtered using 40-μM cell strainers. Staining details can be found in SI Appendix. Cells were acquired on a spectral flow cytometer (Aurora instrument, Cytek). Details on data analysis can be found in SI Appendix.
Software and Statistical Analysis.
Flow cytometry data were analyzed using FlowJo software version 10 (Tree Star). Statistical analyses were performed using Prism software version 7 (GraphPad). For all analyses, paired Wilcoxon tests were used.
Acknowledgments
This work was funded by INSERM, Institut Curie (CIC IGR-Curie 1428), and Agence Nationale de la Recherche (ANR-10-LABX-0043, ANR-10-IDEX-0001-02 PSL and ANR-17-CE15-0011-01). A.C. is a fellow of Fondation pour la Recherche Médicale (FDT202001010805). We wish to thank the Flow Cytometry Platform of Institut Curie, the Next Generation Sequencing Platform of Institut Curie, and the In vivo Experiments Platform of Institut Curie, B. Manoury and M. Burbage for critical reading of the manuscript, and Javiera Villar for technical assistance.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2109225118/-/DCSupplemental.
Data Availability
References
- 1.Davies L. C., Jenkins S. J., Allen J. E., Taylor P. R., Tissue-resident macrophages. Nat. Immunol. 14, 986–995 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shi C., Pamer E. G., Monocyte recruitment during infection and inflammation. Nat. Rev. Immunol. 11, 762–774 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Serbina N. V., Pamer E. G., Monocyte emigration from bone marrow during bacterial infection requires signals mediated by chemokine receptor CCR2. Nat. Immunol. 7, 311–317 (2006). [DOI] [PubMed] [Google Scholar]
- 4.Tsou C.-L., et al., Critical roles for CCR2 and MCP-3 in monocyte mobilization from bone marrow and recruitment to inflammatory sites. J. Clin. Invest. 117, 902–909 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shi C., et al., Monocyte trafficking to hepatic sites of bacterial infection is chemokine independent and directed by focal intercellular adhesion molecule-1 expression. J. Immunol. 184, 6266–6274 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lim J. K., et al., Chemokine receptor Ccr2 is critical for monocyte accumulation and survival in West Nile virus encephalitis. J. Immunol. 186, 471–478 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Peters W., et al., Chemokine receptor 2 serves an early and essential role in resistance to Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. U.S.A. 98, 7958–7963 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Haist K. C., Burrack K. S., Davenport B. J., Morrison T. E., Inflammatory monocytes mediate control of acute alphavirus infection in mice. PLoS Pathog. 13, e1006748 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jakubzick C. V., Randolph G. J., Henson P. M., Monocyte differentiation and antigen-presenting functions. Nat. Rev. Immunol. 17, 349–362 (2017). [DOI] [PubMed] [Google Scholar]
- 10.Guilliams M., Mildner A., Yona S., Developmental and functional heterogeneity of monocytes. Immunity 49, 595–613 (2018). [DOI] [PubMed] [Google Scholar]
- 11.Blériot C., et al., Liver-resident macrophage necroptosis orchestrates type 1 microbicidal inflammation and type-2-mediated tissue repair during bacterial infection. Immunity 42, 145–158 (2015). [DOI] [PubMed] [Google Scholar]
- 12.Machiels B., et al., A gammaherpesvirus provides protection against allergic asthma by inducing the replacement of resident alveolar macrophages with regulatory monocytes. Nat. Immunol. 18, 1310–1320 (2017). [DOI] [PubMed] [Google Scholar]
- 13.Aegerter H., et al., Influenza-induced monocyte-derived alveolar macrophages confer prolonged antibacterial protection. Nat. Immunol. 21, 145–157 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guilliams M., Svedberg F. R., Does tissue imprinting restrict macrophage plasticity? Nat. Immunol. 22, 118–127 (2021). [DOI] [PubMed] [Google Scholar]
- 15.Wakim L. M., Waithman J., van Rooijen N., Heath W. R., Carbone F. R., Dendritic cell-induced memory T cell activation in nonlymphoid tissues. Science 319, 198–202 (2008). [DOI] [PubMed] [Google Scholar]
- 16.Ma Y., et al., Anticancer chemotherapy-induced intratumoral recruitment and differentiation of antigen-presenting cells. Immunity 38, 729–741 (2013). [DOI] [PubMed] [Google Scholar]
- 17.Honda T., et al., Tuning of antigen sensitivity by T cell receptor-dependent negative feedback controls T cell effector function in inflamed tissues. Immunity 40, 235–247 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Aldridge J. R. Jr., et al., TNF/iNOS-producing dendritic cells are the necessary evil of lethal influenza virus infection. Proc. Natl. Acad. Sci. U.S.A. 106, 5306–5311 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ellis G. T., et al., TRAIL+ monocytes and monocyte-related cells cause lung damage and thereby increase susceptibility to influenza-Streptococcus pneumoniae coinfection. EMBO Rep. 16, 1203–1218 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guilliams M., et al., IL-10 dampens TNF/inducible nitric oxide synthase-producing dendritic cell-mediated pathogenicity during parasitic infection. J. Immunol. 182, 1107–1118 (2009). [DOI] [PubMed] [Google Scholar]
- 21.Greter M., et al., GM-CSF controls nonlymphoid tissue dendritic cell homeostasis but is dispensable for the differentiation of inflammatory dendritic cells. Immunity 36, 1031–1046 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Goudot C., et al., Aryl hydrocarbon receptor controls monocyte differentiation into dendritic cells versus macrophages. Immunity 47, 582–596.e6 (2017). [DOI] [PubMed] [Google Scholar]
- 23.Desalegn G., Pabst O., Inflammation triggers immediate rather than progressive changes in monocyte differentiation in the small intestine. Nat. Commun. 10, 3229 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Watanabe T., Kitani A., Murray P. J., Strober W., NOD2 is a negative regulator of Toll-like receptor 2-mediated T helper type 1 responses. Nat. Immunol. 5, 800–808 (2004). [DOI] [PubMed] [Google Scholar]
- 25.Udden S. M. N., et al., NOD2 suppresses colorectal tumorigenesis via downregulation of the TLR pathways. Cell Rep. 19, 2756–2770 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Weichhart T., et al., The TSC-mTOR signaling pathway regulates the innate inflammatory response. Immunity 29, 565–577 (2008). [DOI] [PubMed] [Google Scholar]
- 27.Schmitz F., et al., Mammalian target of rapamycin (mTOR) orchestrates the defense program of innate immune cells. Eur. J. Immunol. 38, 2981–2992 (2008). [DOI] [PubMed] [Google Scholar]
- 28.Erra Díaz F., et al., Extracellular acidosis and mTOR inhibition drive the differentiation of human monocyte-derived dendritic cells. Cell Rep. 31, 107613 (2020). [DOI] [PubMed] [Google Scholar]
- 29.Schenk M., et al., NOD2 triggers an interleukin-32-dependent human dendritic cell program in leprosy. Nat. Med. 18, 555–563 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang L., Li R., Xiang S., Xiao W., MafB, a target of microRNA-155, regulates dendritic cell maturation. Open Life Sci. 11, 46–54 (2016). [Google Scholar]
- 31.Kim H., The transcription factor MafB promotes anti-inflammatory M2 polarization and cholesterol efflux in macrophages. Sci. Rep. 7, 7591 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tamoutounour S., et al., Origins and functional specialization of macrophages and of conventional and monocyte-derived dendritic cells in mouse skin. Immunity 39, 925–938 (2013). [DOI] [PubMed] [Google Scholar]
- 33.Van Gassen S., et al., FlowSOM: Using self-organizing maps for visualization and interpretation of cytometry data. Cytometry A 87, 636–645 (2015). [DOI] [PubMed] [Google Scholar]
- 34.Bosteels C., et al., Inflammatory type 2 cDCs acquire features of cDC1s and macrophages to orchestrate immunity to respiratory virus infection. Immunity 52, 1039–1056.e9 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nakano H., et al., Blood-derived inflammatory dendritic cells in lymph nodes stimulate acute T helper type 1 immune responses. Nat. Immunol. 10, 394–402 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Langlet C., et al., CD64 expression distinguishes monocyte-derived and conventional dendritic cells and reveals their distinct role during intramuscular immunization. J. Immunol. 188, 1751–1760 (2012). [DOI] [PubMed] [Google Scholar]
- 37.Subramanian A., et al., Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U.S.A. 102, 15545–15550 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Manicourt D. H., et al., Synovial fluid levels of tumor necrosis factor alpha and oncostatin M correlate with levels of markers of the degradation of crosslinked collagen and cartilage aggrecan in rheumatoid arthritis but not in osteoarthritis. Arthritis Rheum. 43, 281–288 (2000). [DOI] [PubMed] [Google Scholar]
- 39.Kawanaka N., et al., CD14+,CD16+ blood monocytes and joint inflammation in rheumatoid arthritis. Arthritis Rheum. 46, 2578–2586 (2002). [DOI] [PubMed] [Google Scholar]
- 40.Peng T., et al., Evasion of the mucosal innate immune system by herpes simplex virus type 2. J. Virol. 83, 12559–12568 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thaiss C. A., Levy M., Itav S., Elinav E., Integration of innate immune signaling. Trends Immunol. 37, 84–101 (2016). [DOI] [PubMed] [Google Scholar]
- 42.Dorrington M. G., Fraser I. D. C., NF-κB signaling in macrophages: Dynamics, crosstalk, and signal integration. Front. Immunol. 10, 705 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pandey S., et al., Pairwise stimulations of pathogen-sensing pathways predict immune responses to multi-adjuvant combinations. Cell Syst. 11, 495–508.e10 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Quintin J., et al., Candida albicans infection affords protection against reinfection via functional reprogramming of monocytes. Cell Host Microbe 12, 223–232 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Saeed S., et al., Epigenetic programming of monocyte-to-macrophage differentiation and trained innate immunity. Science 345, 1251086 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Foster S. L., Hargreaves D. C., Medzhitov R., Gene-specific control of inflammation by TLR-induced chromatin modifications. Nature 447, 972–978 (2007). [DOI] [PubMed] [Google Scholar]
- 47.del Fresno C., et al., Potent phagocytic activity with impaired antigen presentation identifying lipopolysaccharide-tolerant human monocytes: Demonstration in isolated monocytes from cystic fibrosis patients. J. Immunol. 182, 6494–6507 (2009). [DOI] [PubMed] [Google Scholar]
- 48.Karmaus P. W. F., et al., Critical roles of mTORC1 signaling and metabolic reprogramming for M-CSF-mediated myelopoiesis. J. Exp. Med. 214, 2629–2647 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lee P. Y., et al., The metabolic regulator mTORC1 controls terminal myeloid differentiation. Sci. Immunol. 2, eaam6641 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhao Y., et al., mTOR masters monocyte development in bone marrow by decreasing the inhibition of STAT5 on IRF8. Blood 131, 1587–1599 (2018). [DOI] [PubMed] [Google Scholar]
- 51.Smink J. J., et al., Transcription factor C/EBPbeta isoform ratio regulates osteoclastogenesis through MafB. EMBO J. 28, 1769–1781 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.O’Connell R. M., Taganov K. D., Boldin M. P., Cheng G., Baltimore D., MicroRNA-155 is induced during the macrophage inflammatory response. Proc. Natl. Acad. Sci. U.S.A. 104, 1604–1609 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ceppi M., et al., MicroRNA-155 modulates the interleukin-1 signaling pathway in activated human monocyte-derived dendritic cells. Proc. Natl. Acad. Sci. U.S.A. 106, 2735–2740 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.O’Connell R. M., et al., Sustained expression of microRNA-155 in hematopoietic stem cells causes a myeloproliferative disorder. J. Exp. Med. 205, 585–594 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kurowska-Stolarska M., et al., MicroRNA-155 as a proinflammatory regulator in clinical and experimental arthritis. Proc. Natl. Acad. Sci. U.S.A. 108, 11193–11198 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dunand-Sauthier I., et al., Silencing of c-Fos expression by microRNA-155 is critical for dendritic cell maturation and function. Blood 117, 4490–4500 (2011). [DOI] [PubMed] [Google Scholar]
- 57.Stanczyk J., et al., Altered expression of MicroRNA in synovial fibroblasts and synovial tissue in rheumatoid arthritis. Arthritis Rheum. 58, 1001–1009 (2008). [DOI] [PubMed] [Google Scholar]
- 58.Elmesmari A., et al., MicroRNA-155 regulates monocyte chemokine and chemokine receptor expression in Rheumatoid Arthritis. Rheumatology (Oxford) 55, 2056–2065 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Love M. I., Huber W., Anders S., Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Malosse C., Henri S., Isolation of mouse dendritic cell subsets and macrophages from the skin. Methods Mol. Biol. 1423, 129–137 (2016). [DOI] [PubMed] [Google Scholar]
- 61.E. Segura, A. De Juan, A. Coillard, RNAseq analysis of human CD14+ monocytes exposed or not to TNFa. GEO. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE166043. Deposited 2 February 2021. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.