Abstract
Ubiquitin-mediated degradation plays a crucial role in many fundamental biological pathways, including the mediation of cellular responses to changes in environmental conditions. A family of ubiquitin ligase complexes, called SCF complexes, found throughout eukaryotes, is involved in a variety of biological pathways. In Saccharomyces cerevisiae, an SCF complex contains a common set of components, namely, Cdc53p, Skp1p, and Hrt1p. Substrate specificity is defined by a variable component called an F-box protein. The F- box is a ∼40-amino-acid motif that allows the F-box protein to bind Skp1p. Each SCF complex recognizes different substrates according to which F-box protein is associated with the complex. In yeasts, three SCF complexes have been demonstrated to associate with the ubiquitin-conjugating enzyme Cdc34p and have ubiquitin ligase activity. F-box proteins are not abundant and are unstable. As part of the SCFMet30p complex, the F-box protein Met30p represses methionine biosynthetic gene expression when availability of l-methionine is high. Here we demonstrate that in vivo SCFMet30p complex activity can be regulated by the abundance of Met30p. Furthermore, we provide evidence that Met30p abundance is regulated by the availability of l-methionine. We propose that the cellular responses mediated by an SCF complex are directly regulated by environmental conditions through the control of F-box protein stability.
Protein degradation is an essential regulatory mechanism used by the cell in many fundamental processes, including response to changing environmental conditions. In eukaryotes, a major proteolytic mechanism is the ubiquitin (Ub)-proteasome pathway (10, 12). Ub is a member of a family of conserved polypeptides that are covalently attached to protein substrates. Reiteration of Ub modification creates a poly-Ub chain on the substrate that is then targeted for degradation by the proteasome, a large multiprotein complex protease. The transfer of Ub to a protein substrate is a multistep process requiring at least three proteins. Free Ub is activated at the expense of ATP by a Ub-activating enzyme, or E1. Activated Ub is then transferred to a Ub-conjugating enzyme, or E2. Ub-ligases, or E3s, facilitate the transfer of Ub from E2 to protein substrate. Some E3s act as intermediary Ub carriers in the transfer of Ub from E2 to substrate (35), while other E3s act as adapters tethering the E2 to its substrate (6). A multiprotein complex called the SCF (named after the original components Skp1p, Cdc53p, and F-box protein) complex is represented in all eukaryotic taxa and has recently emerged as a major family of Ub ligases (6, 28, 42).
In budding yeast, SCF complexes are comprised of a common set of components, namely, Cdc53p (or cullin), Skp1p, and Hrt1p (or Rbx1p or Roc1p) (for reviews, see references 5, 6, 28, and 42). Each SCF complex also contains an F-box protein, which is responsible for substrate recognition (8, 37). The F box, a ∼40-amino-acid motif, is believed to link the F-box protein with the common SCF components by binding Skp1p. Although several F-box proteins exist in the cell, only one F-box protein is present within any one SCF complex (29). Thus, a family of complexes that are distinguished by the F-box protein exists. The Ub-conjugating enzyme Cdc34p is associated with several SCF complexes and is necessary for SCF-dependent ubiquitination (8, 17, 23, 29, 36, 37, 43).
Three yeast F-box proteins, Cdc4p, Grr1p, and Met30p, are present within SCF complexes, referred to as SCFCdc4p, SCFGrr1p, and SCFMet30p, respectively, and have roles in Cdc34p-mediated protein degradation events (6, 42). An emerging feature of these three SCF Ub ligases is their dual role in mediating cell cycle progression and metabolism. SCFCdc4p is required for entry into S phase by degrading the cyclin-dependent kinase-inhibitory protein Sic1p. Additionally, SCFCdc4p controls the biosynthesis of amino acids and purines (11, 13) by regulating the abundance of the transcription factor Gcn4p (25). Grr1p regulates G1 cyclin abundance (2) in addition to playing roles in heavy metal resistance, amino acid transport, cell growth, and glucose repression (4, 9, 14, 15, 20). SCFMet30p regulates methionine biosynthetic gene expression in addition to playing a positive role in cell cycle progression (16, 29, 30, 40).
A well-described role for SCFMet30p is its regulation of methionine biosynthesis (29, 32). The MET genes encode enzymes and transporters necessary for the uptake of inorganic sulfur and its assimilation into methionine and cysteine (39). A negative feedback loop regulates MET gene expression: cells supplied with l-methionine repress MET gene expression, whereas depression occurs in media lacking l-methionine. The transcription factor Met4p positively regulates methionine biosynthesis. Recently, Met4p was proposed to be a substrate for the SCFMet30p Ub ligase complex (32). How SCFMet30p activity is regulated by l-methionine is unknown. One means of regulating the activity of some SCF complexes is at the level of substrate recognition. There are well-characterized examples of substrates being phosphorylated prior to their recognition by an SCF complex (42). However, Met4p modification has not been attributed to phosphorylation, and thus, SCFMet30p complex activity may be regulated by an alternate mechanism.
Here we show that increased Met30p abundance positively regulates SCFMet30p activity. Additionally, we report that Met30p abundance is regulated by the availability of l-methionine. Therefore, SCFMet30p complex activity is, at least partially, regulated at the level of Met30p abundance. Finally, the F-box region of Met30p plays a critical role in regulating Met30p stability in a methionine-dependent manner, suggesting that this sequence may have crucial regulatory roles in addition to linking the F-box protein with the SCF complex.
MATERIALS AND METHODS
Yeast strains and manipulations.
The yeast (Saccharomyces cerevisiae) strains used in this study are Y382 MATα ade2 ade3 ura3 leu2 trp1 (kindly provided by A. Bender), PY283 MATa met30-6 bar1Δ ura3Δ ade1 his2 leu2-3,112 trp1-1 (kindly provided by Steve Reed [16]), YPH1172 MATa ura3-52 trp1-Δ63 his3-Δ200 leu2-Δ1 ade2-101 skp1Δ::TRP skp1-3::LEU2 (kindly provided by Phil Hieter), C114 MATα ura3 leu2::MET25-lacZ::LEU2 (kindly provided by Yolande Surdin-Kerjan), NMmet30Δ MATα ura3-52 trp1-1 his3-Δ200 leu2-3,112 Δmet30::HIS3 (this study), NMMET30/met30Δ MATa/α ura3-52/ura3-52 leu2/leu2 his3/his3 trp1-1/trp1-1 MET30/met30::HIS3 (this study), and NMmet30ΔMET25-lacZ MATa ura3-52 his3-Δ200 trp1-1 Δmet30::HIS3 leu2::MET25-lacZ::LEU2 (this study). NMmet30Δ was created as follows. A heterozygous met30 disruption strain, NMMET30met30Δ (MET30 met30::HIS3), was transformed with pGST Met30-3. Transformants were sporulated, and asci were dissected onto yeast extract-peptone-dextrose medium. Ura+ and His+ colonies were identified, one of which we named NMmet30Δ. NMmet30ΔMET25-lacZ is a meiotic product of a cross between C114 and NMmet30Δ. Standard rich (YPD) and defined minimal SD media were prepared as described previously (31). Transformations were carried out as described previously (7). For plasmid selection, yeast cells were grown on defined minimal medium supplemented with the appropriate amino acids. For galactose induction, cells were first grown to early logarithmic phase in minimal medium containing sucrose instead of dextrose and then galactose-containing medium (2%) was added and the culture was incubated for a further 3 h. To measure protein half-lives, glutathione S-transferase (GST)–Met30p fusions were transiently induced from the GAL1 promoter for 3 h. Glucose (2%) and cycloheximide (1 mg/ml) were added to repress transcription and translation, respectively. For complementation experiments, patches derived from single colonies were grown under permissive conditions (23°C) and then replica plated and incubated further under nonpermissive conditions of 37°C.
Flow cytometry.
Y382 transformants containing pGSTMET30-3 were grown to a density of 5 × 106 cells/ml in sucrose-containing medium that lacked methionine. Galactose was added, and the culture was incubated further until cells had become arrested, which occurred within two divisions. Cells were harvested, sonicated, fixed in 70% ethanol, and stored at 4°C. To prepare for flow cytometry, cells were washed once in 10 mM Tris (pH 7.4)–15 mM NaCl and resuspended in the same buffer containing 0.1 mg of RNase A (Roche Molecular Biochemicals) per ml for 1.5 h. The cells were then harvested and resuspended in phosphate-buffered saline at 106 cells/ml, and propidium iodide was added to a final concentration of 40 μg/ml. Flow cytometry was performed using FACSvantage (Becton Dickinson, Mountain View, Calif.) with an excitation wavelength of 488 nm and monitoring of emission in the f12 channel. Data were collected in the four-parameter list mode of 20,000 cells/run.
Plasmid constructions.
Escherichia coli DH5α was used to propagate plasmids. Plasmid manipulations used standard protocols (34). The vector pEG(KG) was used for the expression of GST fusion proteins (26). Expression of the GST fusion proteins was from the GAL1 promoter. All GST-MET30 fusion constructs were created by cloning PCR-generated DNA fragments using plasmid-borne MET30 DNA as template (pP58, kindly provided by Steve Reed) as described previously (21). Full-length MET30 was generated using primers 5′-TTTTCTAGACATGAGGAGAGAGAGGCAAAGG-3′ and 5′-CCCGTCGACCTAATCATTGAGATCGAATTTG-3′. The primer annealing to the 3′ end of MET30 incorporated an XbaI restriction site, and the primer annealing to the 5′ end of MET30 incorporated a SalI restriction site. The PCR product was restricted with XbaI and SalI and ligated into pEG(KG) that had been restricted with the same enzyme to yield plasmid pGSTMet30-3. To generate MET30(Δ185–277), a PCR fragment was amplified from primers 5′-CCCTCTAGACATCTACAGAGAACGGTTCAAAG-3′ and 5′-CCCGTCGACCTAATCATTGAGATCGAATTTG-3′. This fragment contained XbaI and SalI restriction enzyme sites and was ligated into pEG(KT) digested with the same enzymes. This yielded a plasmid, pGSTMET30-6, which contained a DNA fragment encoding Met30p residues 277 to 640. A second PCR fragment was amplified using primers 5′-AAAAACCCGGGAATGAGGAGAGAGAGGCAAAGG-3′ and 5′-GGGGTCTAGAATGCTGATGAAGTCGATCTTGATC-3′ which incorporated SmaI and XbaI and was ligated into pGSTMET30-6 restricted with the same enzymes yielding plasmid pGSTMET30(Δ185–277). MT839 encoding hemagglutinin (HA)-tagged CDC53 was provided by Mike Tyers.
Protein preparation and Western immunoblotting techniques.
Yeast lysate preparations and Western immunoblotting were carried out as described previously (22). Data from Western blots detected with ECL or ECL+ were exposed to film or imaged with a Storm Phosphorimager (Amersham Pharmacia Biotech, Piscataway, N.J.) in the blue fluorescence mode. Quantification of Western blot data used Image Quant image analysis software according to the manufacturer's instructions. Antibodies raised against GST were purchased from Sigma, and antibodies raised against the HA epitope were purchased from Roche Molecular Biochemicals. Antibodies raised against Cdc34p have been previously described (22).
Assay for MET25-lacZ enzyme activity.
β-Galactosidase activity was measured as described previously (45). For repressing conditions, cultures were grown in the presence of 2 mM l-methionine, whereas for nonrepressing conditions, cultures were grown in the absence of l-methionine. Cultures were grown to 106 to 107 cells/ml in sucrose, filtered, and resuspended in medium containing galactose to induce production of GST or GST-Met30p fusion proteins. Values reported here are the averages from at least two independent assays. β-Galactosidase activities were expressed as nanomoles of o-nitrophenyl-β-d-galactopyranoside hydrolyzed per minute per milligram of protein.
RESULTS
Characterization of GST-Met30p fusion.
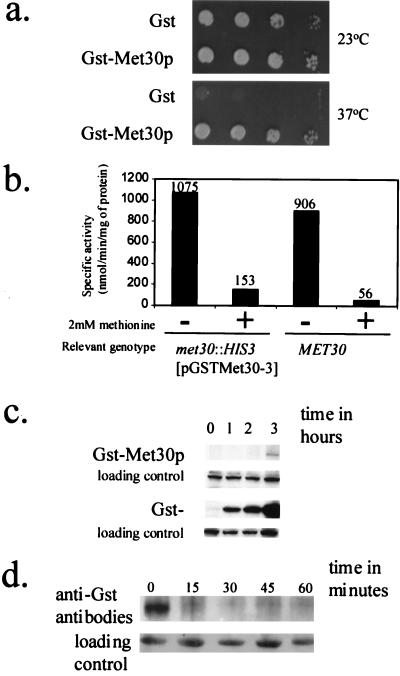
We tagged Met30p with GST, expressed from the GAL1 promoter (see Materials and Methods), which permits low-level transcription when cells are grown in the presence of dextrose and strongly induces transcription in galactose-grown cells. Therefore, we could analyze GST-Met30p activity when it was either poorly or highly produced. The plasmid pGSTMet30-3 complements the growth defect of a met30-6 temperature-sensitive mutant (Fig. 1a) and of a met30 null strain (data not shown; see Materials and Methods). Thus, the GST tag does not affect the essential function of Met30p. Since complementation of met30 temperature-sensitive and null mutations by pGSTMet30-3 was achieved on medium containing dextrose as the sole carbon source, low levels of GST-Met30p are sufficient for its function. To test whether low-level production of GST-Met30p correctly regulated MET gene expression, we examined the regulation of the MET25 promoter, whose activity is repressed by l-methionine through the SCFMet30p complex (29). In cells containing either wild-type MET30 or GST-MET30, expression of a MET25-lacZ fusion was repressed in the presence of 2 mM l-methionine and derepressed in the absence of l-methionine (Fig. 1b). Thus, the GST-Met30p fusion appears to be functional for MET30 essential function and regulation of MET gene expression.
FIG. 1.
Characterization of the GST-Met30p fusion. (a) Low-level production of GST-Met30p is sufficient for complementation of a met30-6 temperature-sensitive allele. PY283 cells containing the met30-6 temperature-sensitive mutation were transformed with either pEG(KG) or pGSTMET30-3, which allowed the production of the indicated protein. Patches were made on SD medium, and cells were incubated at the indicated temperature for 3 days. (b) NMYmet30ΔMET25-lacZ cells containing pGSTMET30-3 or a wild-type MET30 congenic strain were grown in medium in the presence (+) of a repressing concentration of l-methionine or in the absence (−) of l-methioine. The reported values represent averages of two independent assays and were expressed as nanomoles of substrate transformed per minute per milligram of protein. The individual measurements deviated from the average values shown here by 20% or less. (c) Induction time course of GST-Met30p and GST. We performed an anti-GST Western immunoblot analysis of lysate prepared from cells, grown to mid-logarithmic growth phase, containing either pEG(KG) or pGST-MET30, which produced the indicated protein that had been induced for 0, 1, 2, and 3 h by galactose. A cross-reacting band is used as a loading control. (d) Met30p is an unstable protein. We performed time course experiments to measure the stability of GST-Met30p fusion (see Materials and Methods for details). A cross-reacting band is used as a loading control.
Met30p is an unstable protein.
To confirm GST-Met30p production, Western immunoblot analysis was performed. Whereas GST was readily detected after the first time point, 60 min, GST-Met30p was detectable only after prolonged incubation of the cells in the presence of galactose (Fig. 1c), suggesting that Met30p may be an unstable protein. Indeed, a promoter shutoff experiment showed that GST-Met30p is very unstable (Fig. 1d), like the yeast F-box proteins Cdc4p, Ctf13p, and Grr1p (24, 33, 44).
Met30p abundance regulates SCFMet30p activity.
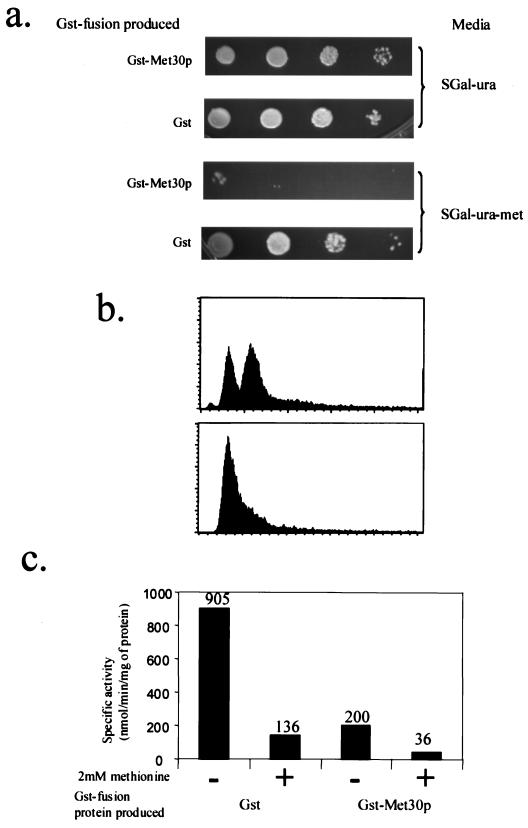
Since Met30p acts through the SCFMet30p complex to repress MET gene expression (29, 32), we tested the hypothesis that limiting Met30p abundance is a means of regulating activity of the SCFMet30p complex. We assessed the ability of methionine prototrophs to grow in the absence of l-methionine while overproducing either GST or GST-Met30p. Cells overproducing GST were able to grow in the absence of l-methionine, illustrating the fact that the host strain was indeed a methionine prototroph (Fig. 2a). The same host strain overproducing GST-Met30p, however, was unable to grow in the absence of l-methionine (Fig. 2a). Therefore, overproduction of GST-Met30p induces methionine auxotrophy in cells otherwise wild type for methionine biosynthesis.
FIG. 2.
Effect of Met30p overproduction. (a) Methionine prototrophic Y382 cells were transformed with either pEG(KG) or pGSTMET30-3, which allowed the production of the indicated protein. Patches were made on the indicated medium, and cells were incubated for 3 to 4 days. (b) Flow cytometry analysis of Y382 cells, transformed with pGSTMET30-3, grown in the absence of methionine, before production of GST-Met30p (upper panel) and after production of GST-Met30p (lower panel). (c) C114 cells, MET25-lacZ, transformed with a plasmid that allowed the production of the indicated protein, were grown in medium containing (+) a repressing concentration of l-methionine or in the absence (−) of l-methionine. The reported values represent averages of three independent assays and were expressed as nanomoles of substrate transformed per minute per milligram of protein. The individual measurements deviated from the average values shown here by 20% or less.
Methionine auxotrophs arrest growth in the G1 phase of the cell division cycle (41). The majority of the cells overproducing GST-Met30p in the absence of methionine were unbudded (data not shown), suggesting a G1 arrest. To confirm a G1 arrest, we performed flow cytometry. Y382 cells containing pGSTMET30-3 were grown in sucrose-containing medium that lacked methionine. GST-Met30p production was induced by the addition of galactose. In order to determine DNA content, samples were analyzed by flow cytometry. Induction of GST-Met30p caused cells to be uniformly arrested with 1N DNA content, consistent with our previous observations that methionine auxotrophy had been induced (Fig. 2b).
To test whether the induced methionine auxotrophy caused by overproduction of GST-Met30p was the result of repression of MET gene expression, we examined the regulation of the MET25 promoter. In cells overproducing GST, expression of a MET25-lacZ gene fusion was repressed in the presence of 2 mM l-methionine (Fig. 2c). In cells overproducing GST-Met30p, the MET25 promoter was repressed independently of the availability of l-methionine (Fig. 2c); however, the levels of repression were not equivalent. When cells were overproducing GST-Met30p, MET25 promoter activity repression was approximately fivefold greater in the presence of l-methionine than in the absence of l-methionine. Thus, overproducing GST-Met30p enhanced the repressing effect of medium containing 2 mM l-methionine. Taken together, these data suggest that the abundance of Met30p regulates methionine biosynthesis.
Met30p abundance is regulated by l-methionine.
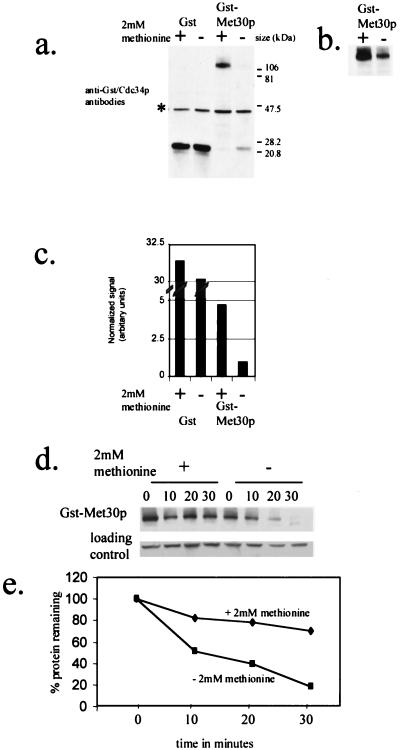
We tested whether methionine availability regulates Met30p abundance. Y382 cells were grown in medium containing 2% galactose in either the presence or absence of l-methionine. The steady-state abundance of GST-Met30p was elevated 4.5-fold after the addition of l-methionine (Fig. 3a to c).
FIG. 3.
GST-Met30p steady-state abundance and stability are dependent on methionine availability. (a) Anti-GST and anti-Cdc34p immunoblot analysis of soluble protein extracts from Y382 cells containing either pEG(KG) or pGSTMET30-3, whose expression was induced for 3 h by the addition of galactose. Cdc34p, whose abundance is unaffected by methionine (Fig. 4), is used as a loading control, and its position is indicated (*). (b) A fivefold-longer exposure time of panel a. (c) Quantitation of GST and GST-Met30p immunoblot signals of which panel a is an example. Values represent averages derived from five independent experiments. (d) Time course experiments to measure the stability of GST-Met30p fusions after promoter shutoff in medium containing or lacking methionine. Y382 cells, containing pGSTMET30-3, were grown either in the presence or in the absence of 2 mM methionine, and GST-Met30p synthesis was induced by the addition of galactose for 3 h. After the addition of dextrose and cycloheximide, samples were taken at the indicated time points. (e) Quantification of data in panel d by Storm Phosphorimager analysis. The amount of GST-Met30p protein detected at the indicated time points is represented as a percentage of GST-Met30p protein detected at time point zero.
Because both GST and GST-Met30p were expressed from the same promoter, it is unlikely that GAL1 promoter activity is affected by l-methionine availability. Therefore, in order to observe a difference in the steady-state abundance of GST-Met30p, we postulated that GST-Met30p stability is altered. To test whether the degradation rate of GST-Met30p is altered by the presence of l-methionine, we measured the half-lives of GST-Met30p in the presence and absence of 2 mM l-methionine. Although we observed that GST-Met30p was slightly unstable in the presence of 2 mM l-methionine, it had a longer half-life than in the absence of l-methionine (Fig. 3d and e). Thus, these data indicate that GST-Met30p stability is dependent upon the presence of l-methionine.
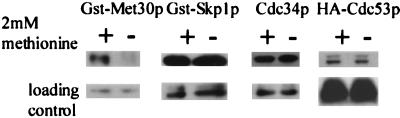
To test whether additional SCF components or F-box proteins are affected by l-methionine availability, we measured the steady-state abundances of Cdc34p, GST-Skp1p, and HA-Cdc53p by following the same regimen as described above. The steady-state abundance of Met30p, but of none of the other SCF components tested, is dependent on the availability of l-methionine (Fig. 4).
FIG. 4.
Met30p is the only component of SCFMet30p whose steady-state abundance is methionine dependent. Y382 cells transformed with the appropriate epitope-tagged plasmid were grown either in the presence (+) or in the absence (−) of 2 mM methionine. GST-Met30p and GST-Skp1p were detected using anti-GST antibodies (Sigma), Cdc34p was detected by anti-Cdc34p antibodies, and HA-Cdc53p was detected using anti-HA antibodies (Roche Molecular Biochemicals).
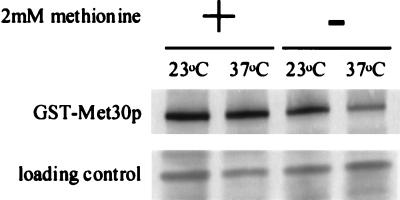
We had previously demonstrated that Cdc4p abundance was dependent on the interaction of Cdc4p with Skp1p and proposed that Skp1p shields Cdc4p from degradation events (24). To investigate whether Met30p abundance was similarly affected, we measured the steady-state level of GST-Met30p in lysate prepared from cells containing the skp1-3 temperature-sensitive mutation that were grown at either the permissive temperature (23°C) or the nonpermissive temperature (37°C) in the presence or absence of 2 mM l-methionine. Figure 5 shows that the steady-state abundance of GST-Met30p is decreased in cells lacking Skp1p activity when cells are grown in the absence of l-methionine. These data suggest that Skp1p may enhance Met30p stability. However, the steady-state abundance of GST-Met30p is unaffected in cells lacking Skp1p activity when cells are grown in the presence of l-methionine. Possibly, the decision to degrade Met30p may be regulated prior to its incorporation into an SCF complex. Alternatively, Skp1p binding to Met30p may be tighter in the presence of l-methionine.
FIG. 5.
Met30p steady-state abundance is dependent on Skp1p activity. skp1-3-containing cells, transformed with pGSTMET30-3, were grown either in the presence (+) or in the absence (−) or 2 mM methionine and induced to produce the GST fusion protein for 3 h by the addition of galactose at the indicated temperature.
Identification of an l-methionine-responsive element in the Met30p family of proteins.
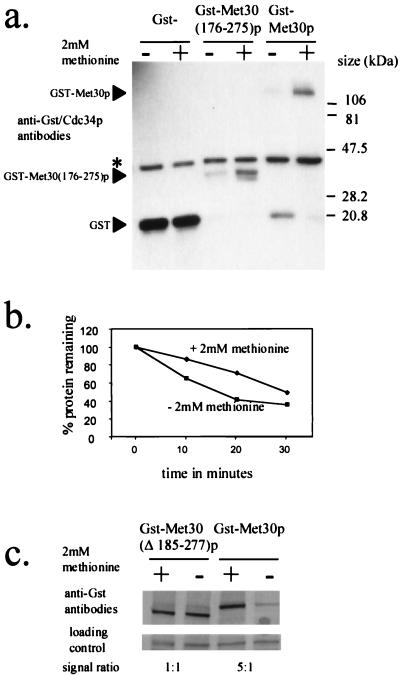
We wished to identify the Met30p element that responds to l-methionine availability. Our previous investigations of Cdc4p demonstrated that the F-box region is partly responsible for regulating Cdc4p abundance (24). During the analysis of Cdc4p degradation, we had constructed a GST-MET30 fusion containing Met30p residues 176 to 275, which include the Met30p F box. We had previously demonstrated that GST-Met30(176–275)p levels are low, which suggested that we had identified a potential Met30p degradation sequence. We, therefore, tested whether we had identified a region on Met30p that regulated the abundance of Met30p in a manner dependent on the presence of l-methionine. Cells containing plasmids encoding either GST, GST-Met30p, or GST-Met30(176–275)p were grown in the presence or absence of 2 mM l-methionine and induced by the addition of galactose to produce the GST fusion proteins. Western immunoblot analysis demonstrated that the steady-state abundance of both GST-Met30p and GST-Met30(176–275)p was dependent on the presence of l-methionine in the medium whereas the steady-state abundance of GST was unaffected (Fig. 6a). Thus, we have identified a region of Met30p that responds to the presence of l-methionine in the medium.
FIG. 6.
Identification of a methionine-responsive element in the Met30p family of proteins. (a and c) Anti-GST Western immunoblot analysis of soluble protein extracted from Y382 cells transformed with either pEG(KG), pGSTMET30-3, pGSTMET30(176–275), or pGSTMET30(Δ185–277) producing the indicated GST-Met30p fusion. Cells were grown either in the presence (+) or in the absence (−) of 2 mM methionine and induced to produce the GST fusion protein for 3 h by the addition of galactose. Either Cdc34p (∗) or a cross-reacting band was used as a loading control. (b) Quantification of the degradation rate of GST-Met30(176–275)p in the presence or absence of 2 mM methionine by Storm Phosphorimager analysis. Y382 cells transformed with pGSTMET30(176–275) were grown in the presence or absence of methionine and induced to produce GST-Met(176–275)p by the addition of galactose for 3 h. The amount of GST-Met30(176–275)p detected at the indicated time points is represented as a percentage of GST-Met30(176–275)p protein detected at time point zero.
We tested whether the degradation rate of Met30(176–275)p was altered depending upon the availability of l-methionine in the medium. Promoter shutoff experiments demonstrated that GST-Met30(176–275)p is more stable in the presence of 2 mM l-methionine (Fig. 6b). Therefore, Met30p residues 176 to 275 are sufficient to target Met30p for degradation in a manner dependent on the presence of l-methionine.
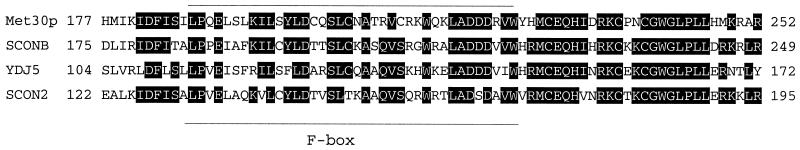
We next wanted to test whether mutations of Met30p residues 176 to 275 rendered Met30p insensitive to the availability of l-methionine in the medium. We therefore made a deletion of Met30p lacking residues 185 to 277 and measured the ability of the mutant protein to sense l-methionine availability in the medium (Fig. 6c). The steady-state abundance of the mutant protein is not affected by the availability of l-methionine in the medium (Fig. 6c). Thus, it appears that we have identified a region that is necessary for the abundance of Met30p to be modulated in a manner dependent on the presence of l-methionine. Database searches revealed that this region is highly conserved in the Met30p family of proteins (Fig. 7).
FIG. 7.
Sequence alignment of the region that regulates Met30p in a methionine-dependent manner in the Met30p family of proteins. Residues 177 to 252 in Met30p from S. cerevisiae are shown aligned with the same region of SCONB from Emericella nidulans, open reading frame YDJ5 from S. pombe, and scon2 from N. crassa. Identical residues in at least three species are highlighted. The location of the F box is indicated.
DISCUSSION
In this paper, we demonstrate that SCFMet30p activity can be modulated by changes in the abundance of Met30p. Furthermore, repressing levels of l-methionine increases Met30p stability. Finally, we have identified a region on Met30p that is degraded in a methionine-dependent manner.
Why the cell limits F-box protein abundance has been unclear. Potentially, maintaining limiting pools of F-box proteins prevents inter-F-box protein competition for common SCF components. Indeed, inter-F-box protein competition has been invoked to explain genetic interactions between mutants encoding SCF components (20, 22, 24, 29). The cell may also control F-box protein abundance as a means of regulating SCF complex formation and activity. F-box proteins appear to play an essential role in SCF complex activity, by acting as adapters bridging the Ub-conjugating machinery with the protein substrate that is to be ubiquitinated. Regulating the abundance of a particular F-box protein would be a means of regulating the activity of a particular SCF complex. In this paper, we present data that support the notion that a specific SCF activity is regulated by modulating the abundance of the respective F-box protein.
MET30 negatively regulates MET gene expression in the presence of 2 mM l-methionine. We demonstrate that Met30p abundance is limiting for SCFMet30p activity. First, ectopic expression of GST-MET30 from the GAL1 promoter results in methionine auxotrophy in a strain wild type for methionine biosynthesis (Fig. 2a). Additionally, GST-Met30p overproduction causes a cell cycle arrest when cells are grown in the absence of methionine (Fig. 2b), an additional hallmark of methionine auxotrophs (41). Finally, by measuring the activity of the MET25 promoter we demonstrate that increased Met30p abundance represses the transcriptional induction of the MET genes (Fig. 2c). Indeed, overproducing GST-Met30p in the presence of repressing concentrations of l-methionine enhances the repression of the same reporter (Fig. 2c). Together, these data demonstrate that the regulation of Met30p abundance plays a key role in regulating SCFMet30p activity. In vitro and in vivo data support the hypothesis that some SCF complexes recognize phosphorylated substrates and do not recognize nonphosphorylated substrates. However, modification of Met4p has not been attributed to phosphorylation (32). We demonstrate here that an additional means of regulating SCFMet30p complex activity is at the level of F-box protein abundance.
Because SCFMet30p complex activity is required to repress MET gene expression in the presence of l-methionine (29, 32), we investigated whether Met30p abundance is regulated by the availability of l-methionine. We demonstrate that the steady-state abundance and stability of Met30p are dependent on the availability of l-methionine in the growth medium (Fig. 3). In the absence of methionine, GST-Met30p has a low abundance and is unstable, whereas in the presence of 2 mM l-methionine, Met30p is more abundant and more stable. Thus, we propose that the availability of l-methionine regulates SCFMet30p activity by regulating Met30p stability (Fig. 8). These data appear to be at odds with those of Rouillon et al. (32), who did not detect changes in the stability of Met30p according to the availability of l-methionine. Furthermore, Rouillon et al. did not report methionine auxotrophy upon MET30 overexpression. Interestingly, these authors used homocysteine rather than sulfate as a nonrepressing sulfur source. Cells mutant for MET4 can grow on homocysteine (39), suggesting that full derepression of MET genes is not required for homocysteine metabolism. Additionally, we measured Met30p half-life when cells were in steady-state sulfate or methionine levels, a situation that may be different from that when cells make the transition from a nonpreferred sulfur source to a preferred sulfur source.
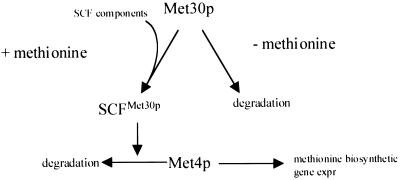
FIG. 8.
Proposed model for the regulation of SCFMet30p complex activity. Met30p abundance is regulated by methionine availability. In the presence of methionine, Met30p stability increases to permit SCFMet30p complex formation and subsequent repression of methionine biosynthetic gene expression by mediating Met4p degradation. In the absence of methionine, Met30p abundance is decreased, lowering the activity of SCFMet30p and, thus, derepressing methionine biosynthetic gene expression (expr) by allowing the accumulation of Met4p.
How does the methionine-dependent effect on Met30p relate to SCF complex formation? We reveal that the interaction of Met30p with Skp1p is important in order to maintain Met30p stability (Fig. 5). This is similar to our results with Cdc4p, which demonstrated that the Skp1p–F-box interaction was important to stabilize Cdc4p (24). Possibly, Skp1p stabilizes F-box proteins by masking adjacent degradation motifs. Our data reported here are consistent with previous reports that have similarly shown that the F-box–Skp1p interaction is important for maintaining Met30p stability (29, 32). When cells are grown in the absence of methionine, Skp1p may be removed from SCFMet30p, resulting in Met30p degradation. However, in the presence of l-methionine, growth at 37°C did not result in a loss of Met30p. There are a number of possible explanations for this effect. Possibly, the decision to degrade Met30p is made before Met30p enters the SCF complex. Alternatively, the presence of l-methionine could change the interaction between Skp1p and Met30p such that Skp1p is less susceptible to inactivation. Finally, in the presence of l-methionine, an increase in Met30p might be sufficient to increase Skp1p function, similar to what is seen when CDC4 levels are increased in skp1 mutants (1).
Paradoxically, we have also demonstrated that removal of a region containing the F box of Met30p results in a protein whose abundance is unaffected by the availability of l-methionine. How can the apparently contradictory nature of these observations be reconciled? Possibly, the region of Met30p that contains the F box carries out two functions. One function is to bind Skp1p and thus link Met30p with the Ub ligase machinery. A second function may be to target Met30p for degradation. In the absence of l-methionine, loss of Skp1p activity results in Met30p degradation. However, removal of the region containing the F box will also remove the Met30p degradation sequence and result in a protein whose stability is unaffected by the availability of l-methionine. A similar phenomenon is seen in Neurospora crassa. Loss of SCON1, a homologue of Skp1p, results in constitutive activation of the sulfate assimilation pathway. However, mutations of conserved F-box residues within SCON2, the Met30p homologue, lead to constitutive repression of the sulfate assimilation pathway (19). Our data support the notion that the F-box region is important in regulating Met30p abundance in addition to its role of coordinating SCF complex formation.
The SCFMet30p complex is not the only SCF complex that regulates metabolic processes in yeast. SCFGrr1p mediates glucose repression, and SCFCdc4p mediates general control of amino acid biosynthesis by targeting the transcriptional activator Gcn4p for degradation (15, 20). Additionally, both SCFGrr1p and SCFCdc4p are also involved in regulating cell cycle progression. Potentially, by regulating F-box protein abundance, environmental conditions would impact on both metabolism and cell cycle progression. Indeed, Met30p has a positive influence in G1 (29; C. Dixon and N. Mathias, unpublished data), and thus methionine availability could determine the length of the G1 phase of the cell cycle by regulating Met30p stability.
Finally, we have identified a region on Met30p residues which is necessary and sufficient to alter the abundance of the protein dependent upon on the availability of l-methionine. Although GST-Met30(Δ184–277)p confers methionine auxotrophy when overproduced, we were unable to demonstrate that this mutant more efficiently repressed MET gene expression than did wild-type Met30p. Possibly, repression by wild-type Met30p could not be improved upon. Another likely reason is that GST-Met30(Δ185–277)p lacks the F-box motif which is necessary for Skp1p binding and proper complex architecture. Thus, GST-Met30(Δ185–277)p activity may be slightly compromised. Indeed, GST-Met30(Δ185–277)p can complement the met30-6 temperature-sensitive strain only when overproduced and is unable to complement a MET30 null strain (data not shown). The region identified as regulating Met30p stability in a methionine-dependent manner is highly conserved in the Met30p family of proteins (Fig. 7). In N. crassa and Aspergillus nidulans, Met30p activity is most likely conferred by SCON2 and sconB, respectively (18, 27). In silico analysis revealed that these proteins, as well as an uncharacterized Schizosaccharomyces pombe protein, contain a motif highly similar to the region of Met30p that imparts l-methionine-regulated changes in Met30p stability. Thus, we have described a novel mechanism for SCF regulation by the environment; methionine-dependent stabilization of an F-box component of an SCF complex. This mechanism may be conserved for methionine biosynthesis among fungi and may have implications for SCFs controlling glucose and amino acid biosynthesis and transport.
ACKNOWLEDGMENTS
We are indebted to Yolande Surdin-Kerjan, Phil Hieter, Steve Reed, and Mike Tyers for plasmids and yeast strains. We thank Kelly Tatchell and Lucy Robinson for reading the manuscript and for comments. We also thank an anonymous reviewer for significant editorial changes to the manuscript.
This work was supported by start-up funds to N.M. from Louisiana State University Health Sciences Center. Initial studies were funded by NSF award MCB 9728069 to M.G.G.
REFERENCES
- 1.Bai C, Sen P, Hofmann K, Ma L, Goebl M, Harper J W, Elledge S J. SKP1 connects cell cycle regulators to the ubiquitin proteolysis machinery through a novel motif, the F-box. Cell. 1996;86:263–274. doi: 10.1016/s0092-8674(00)80098-7. [DOI] [PubMed] [Google Scholar]
- 2.Barral Y, Jentsch S, Mann C. G1 cyclin turnover and nutrient uptake are controlled by a common pathway in yeast. Genes Dev. 1995;9:399–409. doi: 10.1101/gad.9.4.399. [DOI] [PubMed] [Google Scholar]
- 3.Burton E G, Metzenberg R L. Novel mutation causing derepression of several enzymes of sulfur metabolism in Neurospora crassa. J Bacteriol. 1972;109:140–151. doi: 10.1128/jb.109.1.140-151.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Conklin D S, Kung C, Culbertson M R. The COT2 gene is required for glucose-dependent divalent cation transport in Saccharomyces cerevisiae. Mol Cell Biol. 1993;13:2041–2049. doi: 10.1128/mcb.13.4.2041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Craig K L, Tyers M. The F-box: a new motif for ubiquitin dependent proteolysis in cell cycle regulation and signal transduction. Prog Biophys Mol Biol. 1999;72:299–328. doi: 10.1016/s0079-6107(99)00010-3. [DOI] [PubMed] [Google Scholar]
- 6.Deshaies R J. SCF and cullin/ring H2-based ubiquitin ligases. Annu Rev Cell Dev Biol. 1999;15:435–467. doi: 10.1146/annurev.cellbio.15.1.435. [DOI] [PubMed] [Google Scholar]
- 7.Elbe R. A simple and efficient procedure for transformation of yeasts. BioTechniques. 1992;13:18–20. [PubMed] [Google Scholar]
- 8.Feldman R M, Correll C C, Kaplan K B, Deshaies R J. A complex of Cdc4p, Skp1p, and Cdc53p/cullin catalyzes ubiquitination of the phosphorylated CDK inhibitor Sic1p. Cell. 1997;91:221–230. doi: 10.1016/s0092-8674(00)80404-3. [DOI] [PubMed] [Google Scholar]
- 9.Flick J S, Johnston M. GRR1 of Saccharomyces cerevisiae is required for glucose repression and encodes a protein with leucine-rich repeats. Mol Cell Biol. 1991;11:5101–5112. doi: 10.1128/mcb.11.10.5101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hershko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- 11.Hinnebusch A G, Fink G R. Positive regulation in the general control of Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1983;80:5347–5378. doi: 10.1073/pnas.80.17.5374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hochstrasser M. Ubiquitin-dependent protein degradation. Annu Rev Genet. 1996;30:405–439. doi: 10.1146/annurev.genet.30.1.405. [DOI] [PubMed] [Google Scholar]
- 13.Hope I A, Struhl K. GCN4 protein, sunthesized in vitro, binds HIS3 regulatory sequences: implications for general control of amino acid biosynthetic genes in yeast. Cell. 1985;43:177–188. doi: 10.1016/0092-8674(85)90022-4. [DOI] [PubMed] [Google Scholar]
- 14.Iraqui I, Vissers S, Bernard F, de Craene J O, Boles E, Urrestarazu A, Andre B. Amino acid signaling in Saccharomyces cerevisiae: a permease-like sensor of external amino acids and F-box protein Grr1p are required for transcriptional induction of the AGP1 gene, which encodes a broad-specificity amino acid permease. Mol Cell Biol. 1999;19:989–1001. doi: 10.1128/mcb.19.2.989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jaquenoud M, Gulli M P, Peter K, Peter M. The Cdc42p effector Gic2p is targeted for ubiquitin-dependent degradation by the SCFGrr1 complex. EMBO J. 1998;17:5360–5373. doi: 10.1093/emboj/17.18.5360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kaiser P, Sia R A, Bardes E G, Lew D J, Reed S I. Cdc34 and the F-box protein Met30 are required for degradation of the Cdk-inhibitory kinase Swe1. Genes Dev. 1998;12:2587–2597. doi: 10.1101/gad.12.16.2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kamura T, Koepp D M, Conrad M N, Skowyra D, Moreland R J, Iliopoulos O, Lane W S, Kaelin W G, Jr, Elledge S J, Conaway R C, Harper J W, Conaway J W. Rbx1, a component of the VHL tumor suppressor complex and SCF ubiquitin ligase. Science. 1999;284:657–661. doi: 10.1126/science.284.5414.657. [DOI] [PubMed] [Google Scholar]
- 18.Kumar A, Paietta J V. The sulfur controller-2 negative regulatory gene of Neurospora crassa encodes a protein with beta-transducin repeats. Proc Natl Acad Sci USA. 1995;92:3343–3347. doi: 10.1073/pnas.92.8.3343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kumar A, Paietta J V. An additional role for the F-box motif: gene regulation within the Neurospora crassa sulfur control network. Proc Natl Acad Sci USA. 1998;95:2417–2422. doi: 10.1073/pnas.95.5.2417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li F, Johnston M. Grr1 of Saccharomyces cerevisiae is connected to the ubiquitin proteolysis machinery through Skp1: coupling glucose sensing to gene expression and the cell cycle. EMBO J. 1997;16:5625–5638. doi: 10.1093/emboj/16.18.5629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu Y, Mathias N, Steussy C N, Goebl M G. Intragenic suppression among CDC34 (UBC3) mutations defines a class of ubiquitin-conjugating catalytic domains. Mol Cell Biol. 1995;15:5635–5644. doi: 10.1128/mcb.15.10.5635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mathias N, Johnson S L, Winey M, Adams A E M, Goetsch L, Pringle J R, Byers B, Goebl M G. Cdc53p acts in concert with Cdc4p and Cdc34p to control the G1-to-S-phase transition and identifies a conserved family of proteins. Mol Cell Biol. 1996;16:6634–6643. doi: 10.1128/mcb.16.12.6634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mathias N, Steussy C N, Goebl M G. An essential domain within Cdc34p is required for binding to a complex containing Cdc4p and Cdc53p in Saccharomyces cerevisiae. J Biol Chem. 1998;273:4040–4045. doi: 10.1074/jbc.273.7.4040. [DOI] [PubMed] [Google Scholar]
- 24.Mathias N, Johnson S, Byers B, Goebl M. The abundance of cell cycle regulatory protein Cdc4p is controlled by interactions between its F box and Skp1p. Mol Cell Biol. 1999;19:1759–1767. doi: 10.1128/mcb.19.3.1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Meimoun A, Holtzman T, Weissman Z, McBride H J, Stillman D J, Fink G R, Kornitzer D. Degradation of the transcription factor gcn4 requires the kinase pho85 and the SCF(CDC4) ubiquitin-ligase complex. Mol Biol Cell. 2000;3:915–927. doi: 10.1091/mbc.11.3.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mitchell D A, Marshall T K, Deschenes R J. Vectors for the inducible overexpression of glutathione S-transferase fusion proteins in yeast. Yeast. 1993;9:715–723. doi: 10.1002/yea.320090705. [DOI] [PubMed] [Google Scholar]
- 27.Natorff R, Piotrowska M, Paszewski A. The Aspergillus nidulans sulphur regulatory gene sconB encodes a protein with WD40 repeats and an F-box. Mol Gen Genet. 1998;257:255–263. doi: 10.1007/s004380050646. [DOI] [PubMed] [Google Scholar]
- 28.Patton E E, Willems A R, Tyers M. Combinatorial control in ubiquitin-dependent proteolysis: don't Skp the F-box hypothesis. Trends Genet. 1998;14:236–243. doi: 10.1016/s0168-9525(98)01473-5. [DOI] [PubMed] [Google Scholar]
- 29.Patton E E, Willems A R, Sa D, Kuras L, Thomas D, Craig K L, Tyers M. Cdc53 is a scaffold protein for multiple Cdc34/Skp1/F-box protein complexes that regulate cell division and methionine biosynthesis in yeast. Genes Dev. 1998;12:692–705. doi: 10.1101/gad.12.5.692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Patton E E, Peyraud C, Rouillon A, Surdin-Kerjan Y, Tyers M, Thomas D. SCF(Met30)-mediated control of the transcriptional activator Met4 is required for the G(1)-S transition. EMBO J. 2000;19:1613–1624. doi: 10.1093/emboj/19.7.1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rose M D, Winston F, Heiter P. Methods in yeast genetics: a laboratory course. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1990. [Google Scholar]
- 32.Rouillon A, Barbey R, Patton E E, Tyers M, Thomas D. Feedback-regulated degradation of the transcriptional activator Met4 is triggered by the SCF(Met30p) complex. EMBO J. 2000;19:282–294. doi: 10.1093/emboj/19.2.282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Russell I D, Grancell A S, Sorger P K. The unstable F-box protein p58-Ctf13 forms the structural core of the CBF3 kinetochore complex. J Cell Biol. 1999;145:933–950. doi: 10.1083/jcb.145.5.933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 35.Scheffner M, Nuber U, Huibregtse J M. Protein ubiquitination involving an E1-E2-E3 enzyme ubiquitin thioester cascade. Nature. 1995;373:81–83. doi: 10.1038/373081a0. [DOI] [PubMed] [Google Scholar]
- 36.Seol J H, Feldman R M, Zachariae W, Shevchenko A, Correll C C, Lyapina S, Chi Y, Galova M, Claypool J, Sandmeyer S, Nasmyth K, Deshaies R J. Cdc53/cullin and the essential Hrt1 RING-H2 subunit of SCF define a ubiquitin ligase module that activates the E2 enzyme Cdc34. Genes Dev. 1999;13:1614–1626. doi: 10.1101/gad.13.12.1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Skowyra D, Craig K L, Tyers M, Elledge S J, Harper J W. F-box proteins are receptors that recruit phosphorylated substrates to the SCF ubiquitin-ligase complex. Cell. 1997;91:209–219. doi: 10.1016/s0092-8674(00)80403-1. [DOI] [PubMed] [Google Scholar]
- 38.Skowyra D, Koepp D M, Kamura T, Conrad M N, Conaway R C, Conaway J W, Elledge S J, Harper J W. Reconstitution of G1 cyclin ubiquitination with complexes containing SCFGrr1 and Rbx1. Science. 1999;284:662–665. doi: 10.1126/science.284.5414.662. [DOI] [PubMed] [Google Scholar]
- 39.Thomas D, Surdin-Kerjan Y. Metabolism of sulfur amino acids in Saccharomyces cerevisiae. Microbiol Mol Biol Rev. 1997;61:503–532. doi: 10.1128/mmbr.61.4.503-532.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Thomas D, Kuras L, Barbey R, Cherest H, Blaiseau P L, Surdin-Kerjan Y. Met30p, a yeast transcriptional inhibitor that responds to S-adenosylmethionine, is an essential protein with WD40 repeats. Mol Cell Biol. 1995;15:6526–6534. doi: 10.1128/mcb.15.12.6526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Unger M W, Hartwell L H. Control of cell division in Saccharomyces cerevisiae by methionyl-tRNA. Proc Natl Acad Sci USA. 1976;73:1664–1668. doi: 10.1073/pnas.73.5.1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Willems A R, Goh T, Taylor L, Chernushevich I, Shevchenko A, Tyers M. SCF ubiquitin protein ligases and phosphorylation-dependent proteolysis. Philos Trans R Soc Lond B Biol Sci. 1999;354:1533–1550. doi: 10.1098/rstb.1999.0497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Winston J T, Strack P, Beer-Romero P, Chu C Y, Elledge S J, Harper J W. The SCFβ-TRCP-ubiquitin ligase complex associates specifically with phosphorylated destruction motifs in IkappaBalpha and beta-catenin and stimulates IkappaBalpha ubiquitination in vitro. Genes Dev. 1999;13:270–283. doi: 10.1101/gad.13.3.270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhou P, Howley P M. Ubiquitination and degradation of the substrate recognition subunits of SCF ubiquitin-protein ligases. Mol Cell. 1998;5:571–580. doi: 10.1016/s1097-2765(00)80156-2. [DOI] [PubMed] [Google Scholar]
- 45.Zhu S, Wek R C. Ribosome-binding domain of eukaryotic initiation factor-2 kinase GCN2 facilitates translation control. J Biol Chem. 1998;273:1808–1814. doi: 10.1074/jbc.273.3.1808. [DOI] [PubMed] [Google Scholar]