Abstract
Hydrogen peroxide (H2O2) is the most abundant reactive oxygen species (ROS) within mammalian cells. At low concentrations, H2O2 serves as a versatile cell signaling molecule that mediates vital physiological functions. Yet at higher concentrations, H2O2 can be a toxic molecule by promoting pathological oxidative stress in cells and tissues. Within normal cells, H2O2 is differentially distributed in a variety of subcellular locales. Moreover, many redox-active enzymes and their substrates are themselves differentially distributed within cells. Numerous reports have described the biological and biochemical consequences of adding exogenous H2O2 to cultured cells and tissues, but many of these observations are difficult to interpret: the effects of exogenous H2O2 do not necessarily replicate the cellular responses to endogenous H2O2. In recent years, chemogenetic approaches have been developed to dynamically regulate the abundance of H2O2 in specific subcellular locales. Chemogenetic approaches have been applied in multiple experimental systems, ranging from in vitro studies on the intracellular transport and metabolism of H2O2, all the way to in vivo studies that generate oxidative stress in specific organs in living animals. These chemogenetic approaches have exploited a yeast-derived D-amino acid oxidase (DAAO) that synthesizes H2O2 only in the presence of its D-amino acid substrate. DAAO can be targeted to various subcellular locales, and can be dynamically activated by the addition or withdrawal of its D-amino acid substrate. In addition, recent advances in the development of highly sensitive genetically encoded H2O2 biosensors are providing a better understanding of both physiological and pathological oxidative pathways. This review highlights several applications of DAAO as a chemogenetic tool across a wide range of biological systems, from analyses of subcellular H2O2 metabolism in cells to the development of new disease models caused by oxidative stress in vivo.
Keywords: D-amino acid oxidase, Hydrogen peroxide, ROS signaling, Oxidative stress
Graphical Abstract
1. The maintenance of the intracellular hydrogen peroxide balance
Reactive oxygen species (ROS) is a term that has been used to collectively describe the various O2-derived molecules that can arise within cells. Numerous reviews have described the cellular metabolism of ROS in detail [1-22]; this review will focus primarily on the stable ROS hydrogen peroxide (H2O2). It is very important to note that the various ROS are not created equal, either in terms of their chemical reactivities, cellular roles, or subcellular localizations. Some ROS–including hydroxyl (HO•), peroxyl (ROO•) or alkoxyl (RO•) radicals– which can form as by-products of cellular oxidative reactions– are so highly reactive that they are quite unlikely to subserve any physiological signaling role. Since no specific cellular defensive mechanisms exist for their inactivation, these highly reactive ROS can indiscriminately damage nearby biomolecules, leading to deleterious effects on cell function. The superoxide anion (O2•−) is less reactive than the hydroxyl, peroxyl or alkoxyl radicals. O2•− can be generated in mitochondria by the electron transport chain, and can also be synthesized in various subcellular locales by a family of membrane-bound NADPH oxidases (NOX). Other intracellular enzymes can generate O2•−, including xanthine oxidases, nitric oxide synthases, and cytochrome p450 monooxygenases. Under most cellular conditions, O2•− is rapidly converted to hydrogen peroxide (H2O2) via mitochondrial or cytosolic superoxide dismutases. Under conditions of oxidative stress, O2•− can also react with iron and form the highly reactive hydroxyl radical (HO•) via the Fenton or the Haber-Weiss reactions [1,2]. Both because of its negative charge and its high reactivity, O2•− is less likely to serve as a physiological cellular signaling molecule. The stable ROS H2O2 is recognized as the principal intracellular species responsible for redox signaling. H2O2 mediates a variety of crucial biological functions such as cell differentiation, proliferation, and migration in normal cells, yet at higher concentrations H2O2 also can cause pathological oxidative stress [13,16]. Physiological roles for the various free radical ROS have been more difficult to establish, and higher ROS concentrations have been implicated in multiple disease states including cardiovascular and neurodegenerative disorders, chronic inflammatory disease [17,18], cancer, or the aging process itself [19]. The pathophysiological progression of such diseases can be prevented or accelerated by oxidative influences from the environmental factors of the so-called “exposome”, which integrates the effects on oxidative metabolism of lifestyle, nutrition [20], and physical activity [21].
This review will focus on the intracellular metabolism of H2O2, both as an endogenous biomolecule in mammalian cells and as a product of the chemogenetic recombinant enzyme D-amino acid oxidase (DAAO). H2O2 is differentially distributed within cells, with highest H2O2 concentrations found in the peroxisomes and the endoplasmic reticulum (ER). Peroxisomes may serve as sinks for H2O2 [15,22] while the high H2O2 levels found in the ER might be required for redox-modulated protein folding and bridging disulfide bonds among cysteine residues [4]. In most cells, the overall intracellular H2O2 concentration is maintained in the low nM range by the balance of oxidative and reductive enzyme pathways [3,5]. In order to maintain this balance of H2O2, different intracellular organelles contain a distinct combination of redox-active enzymes including catalases, peroxiredoxins, thioredoxin reductases, glutathione peroxidases and reductases as well as the small redox protein thioredoxin and the antioxidative tripeptide glutathione [21]. On the one hand, this differential distribution of redox enzymes and their substrates may allow the cell to selectively counteract oxidative stress in different organelles. On the other hand, the differential subcellular distribution of enzymes involved in H2O2 metabolism facilitates dynamic physiological ROS signaling responses to discrete cell stimuli [16]. In addition, the transport of H2O2 between different subcellular organelles is itself dynamically regulated by peroxiporins, which belong to the family of aquaporin H2O transporters. Peroxiporins are differentially expressed in tissues and within cells and facilitate the transfer of H2O2 through cellular membranes and participate in physiological H2O2 homeostasis as well as in oxidative stress (see section 6) [6-8]. The wider view of redox signaling has been reviewed extensively, and will not be presented here in detail [3,9-12,14,16]. However, considering the complexity of intracellular H2O2 generation, degradation, and inter-organelle fluxes, it is challenging to dissect the various oxidative pathways within different cells and tissues. Moreover, the detection limits of many biosensor systems have confounded efforts to sensitively and specifically quantitate intracellular H2O2 levels in living cells, and the manifold roles of H2O2 in oxidative eustress and distress remain incompletely understood [15]. This review discusses the development and cellular applications of a new and promising chemogenetic approach that utilizes a H2O2-generating enzyme from yeast, D-amino acid oxidase (DAAO). Chemogenetic approaches using DAAO are successfully being used to dissect and decipher the complex pathways of ROS metabolism in health and disease.
2. DAAO: an old enzyme goes chemogenetic
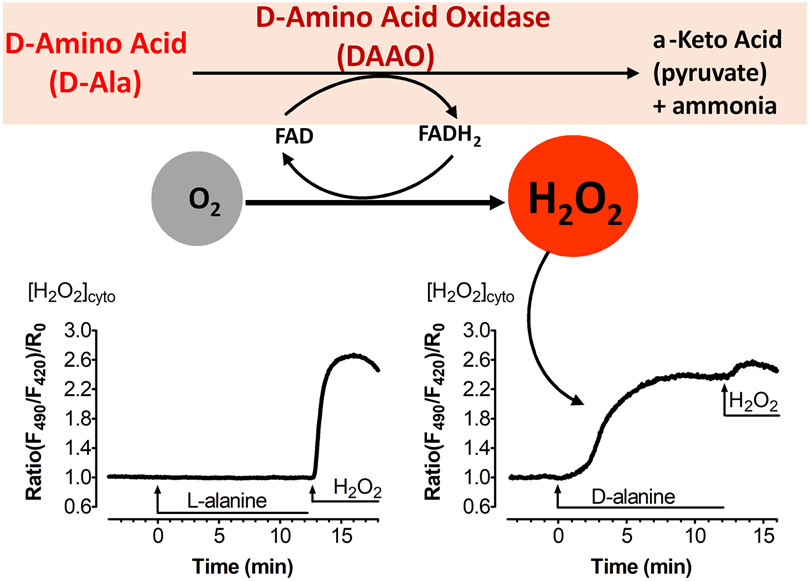
D-amino acid oxidases (DAAO) exist as distinct highly stereoselective enzyme isoforms within eukaryotic organisms. The DAAO enzyme and its cofactor flavin adenine dinucleotide (FAD) were first discovered by the metabolic pioneers Hans Adolf Krebs [23] and Otto Warburg [24] in 1935 and 1938, respectively. Over the subsequent decades, the physiological functions of DAAO were identified and described in a broad variety of organisms, from yeast to humans, whereas plants mostly lack this enzyme [25,26]. Different DAAO isoforms possess specific substrate-binding domains that catalyze the oxidation of D- but not L-amino acids with exquisite stereospecificity in the presence of the cofactor FAD. The oxidative deamination of the D-amino acid results in the generation of its corresponding α-keto acid, plus equimolar quantities of ammonia and H2O2 (Figure 1). In mammalian cells, the DAAO co-products ammonia plus α-keto acid are usually present in much higher intracellular concentrations than H2O2 [27], so there is only a nominal change in intracellular ammonia and keto acid levels, whereas H2O2 levels may increase markedly from their typically low basal levels following DAAO activation in cells [27,28].
Figure 1: Intracellular H2O2 generation by DAAO.
This figure shows stereospecific catabolism of D-amino acids to its corresponding α-keto acid, ammonia and equimolar H2O2 by DAAO. The enzyme reduces its co-factor FAD to FADH2 and converts molecular oxygen to H2O2 (upper panel). Curves are modified from ref. 98, and show H2O2 increases in endothelial cells transiently transfected with the DAAO-HyPer and treated with D-alanine (right panel), but not L-alanine (left panel); the subsequent application of extracellular H2O2 leads to full oxidation of the HyPer biosensor.
Much of what has been learned about DAAO catalysis derives from the work of Pollegioni and colleagues, who have characterized in detail the specificity of D-amino acids for DAAO from various species, especially those in humans and of the yeast Rhodotorula gracilis [25,29-34]. In humans, DAAO and D-amino acids are primarily present in selected regions of the brain and in some nerves, with a much more limited distribution in other tissues. However, all mammalian DAAO isoforms appear to have relatively low enzyme activity, and are predominantly localized in peroxisomes due to the presence of a peroxisomal targeting sequence; the generated H2O2 gets rapidly and effectively eliminated by catalase, which is co-localized in the mammalian peroxisome [29,33]. Thus, under physiological conditions in mammalian cells, endogenous DAAO-generated H2O2 accounts for minimal cytotoxicity even in the presence of D-amino acids. In specific regions of the mammalian brain, neuronal L-serine can be converted by the serine racemase to generate D-serine [35]. D-serine can then activate synaptic N-methyl-D-aspartate receptors (NMDAR), which may have a neuroprotective role mediating physiological effects [36-38]. In some subsets of neurons, it appears that the function of peroxisomal DAAO in neurons is to modulate the intracellular pool of D-amino acids, in particular D-serine. In selected regions of the brain, DAAO mutants lacking the enzyme’s peroxisomal targeting sequence can lead to a decrease in neuronal D-serine, which has been implicated in development of neuropathologic states such as schizophrenia [39].
In contrast to these very limited roles for DAAO in most mammalian cells, DAAO in yeast actually represents a key enzyme that plays a central role in energy metabolism. In yeast, D-amino acids are used as sources of carbon, nitrogen, and energy [40-42]. In contrast to the catalytic properties of mammalian DAAO, yeast DAAO exhibits a 400-fold higher flavin binding affinity [43,44]. Analyses of the crystal structure of mammalian and yeast DAAO reveals that for both enzymes, the apoenzyme is a monomer that forms active dimers upon binding FAD, which then binds and oxidizes the substrate D-amino acid [45-48]. FAD may be a rate limiting factor for the DAAO enzymatic reaction, especially when the recombinant enzyme is used in cultured cells. Indeed, it has been reported that the FAD concentration in many tissues can be orders of magnitude higher than the FAD levels found in cultured cells [49,50]. Thus, the supplementation of > 2.5 μM FAD has been recommended for chemogenetic in vitro approaches in cultured cells expressing recombinant yeast DAAO [51]. Moreover, the binding capacity of D-amino acids for mammalian DAAO is lower than in yeast DAAO since the yeast enzyme does not possess an inhibitory active site “lid” that is present in mammalian DAAO variants. Thus, the rate of catalytic conversion of D-alanine to H2O2 by yeast DAAO is more than 30-fold higher compared to mammalian DAAO [52,53]. The substrate selectivities of D-amino acids have been screened for several yeast DAAO isoforms, revealing a Km of 1 μM for D-alanine [54,55]; yeast DAAOs showed higher rates of H2O2 production when D-alanine was the substrate, as compared to D-serine [56]. Taken together the mechanistic and kinetic properties of yeast DAAO make it much more efficient at the generation of H2O2 compared to mammalian DAAO; indeed yeast DAAO has been used as a biotechnological tool for the synthesis of cephalosporin antibiotics [57-59]. Unless otherwise noted in this review, the term DAAO refers to the yeast enzyme [43].
The term ‘chemogenetic’ has been used to describe model systems where recombinant proteins get activated by a highly specific and unique ligand or substrate [60]. Chemogenetic approaches were used to develop “designer” receptors activated solely by a synthetic ligand (RASSLs), which now are more commonly termed designer receptors exclusively activated by designer drugs (DREADDs) [61,62]. DREADDs have been principally used to study G protein-coupled receptors in the nervous system [63,64] and enable an “on/off” receptor activation upon application of specific chemically engineered molecules or ligands. Chemogenetic approaches have also been developed using an enzyme’s unique substrate instead of a receptor’s unique ligand. DAAO represents a chemogenetic enzyme that generates H2O2 only in the presence of its substrate D-amino acids. Beside DAAO, also other enzymatic models for intracellular H2O2 production have been developed and utilized in the past such as uricase [65], glucose oxidase [66] tyrosinase [67], glycolate oxidase [68], alcohol oxidase [69], L-amino acid oxidase [70], the NADPH dependent cytochrome P450 BM3 [71] as well as light-activated probes [72,73]. But in contrast to the chemogenetic approach using DAAO and its unique D-amino acid substrate, these other enzymes can act on endogenous substrates, making it difficult to dynamically regulate intracellular H2O2 generation due to presence of endogenous substrates. Therefore, researchers developed a chemogenetic approach based on DAAO to specifically and dynamically modulate intracellular H2O2 mediated pathways.
One of several biological applications for DAAO is in the calibration and validation of H2O2 detection systems as well as for studying intra-organelle diffusion of H2O2 [74-77]. Although several chemical probes, including the redox-sensitive dye dichlorofluorescine (DCF) [78], have been used to measure H2O2 [79], these dyes lack specificity for H2O2 as they can easily be oxidized by other cellular oxidants [80] or may even generate ROS such as O2•− [81]. In contrast, the development of genetically encoded fluorescent indicators has enabled more specific and sensitive detection of H2O2. Although some fluorophores can be pH-sensitive [82], most genetically-encoded biosensors provide several advantages in live cell imaging. The commonly-used H2O2 biosensors detect H2O2 as consequence of the H2O2-dependent oxidation of vicinal cysteine thiols leading to the formation of a disulfide bond, which results in a conformational change within the sensor protein and a change in its fluorescent properties. For instance, introducing cysteines at distinct positions of a green fluorescent protein (GFP) yielded the redox sensitive biosensor, roGFP [83]. The fluorescent biosensor termed “HyPer” is composed of a circularly permutated yellow fluorescent protein N- and C-terminally flanked by the split sensory elements of the H2O2 dependent transcription factor OxyR from E. coli [84]. Ratiometric H2O2 indicators based on Foerster resonance energy transfer (FRET) have also been developed in which a H2O2 sensitive peptide is inserted between two different fluorescent proteins [85,86]. More recently, several of these genetically-encoded biosensors have been modified, leading to enhanced H2O2 sensitivity [87-89]. In particular, a novel FRET-based sensor has been developed based on peroxiredoxin-2 [90]; in addition the newest generation of Hyper sensors, HyPer7 uses the OxyR sequence from Neisseria meningitidis [88], providing enhanced H2O2 sensitivity in the low nanomolar range and also showing improved pH stability.
In contrast to most of the chemical dyes that detect intracellular biomolecules, genetically-encoded biosensors can be differentially targeted to different regions within cells, permitting the detection of H2O2 in specific subcellular organelles. DAAO can also be expressed as a fusion protein with HyPer, enabling the generation and detection of H2O2 by a single protein. The first study using HyPer fused with DAAO validated the kinetics of DAAO-mediated H2O2 generation, and demonstrated this production to be dependent on and titratable to the D-alanine concentration provided to the cell [91]. By using a cardiomyocyte-specific promoter the same construct has been successfully applied in vivo to study the effects of oxidative stress in rat hearts (see section 4) [92]. In separate studies, DAAO was fused with an ultrasensitive FRET based H2O2 probe [90] or with a genetically-encoded NADPH indicator to study the effect of oxidative stress on the mitochondrial NADPH pool [93]. The development of novel genetically-encoded H2O2 biosensors also revealed the existence of a large H2O2 gradient across the plasma membrane, showing that the ratio between extracellular and intracellular H2O2 levels was ~400 to 1 [88,94]. Indeed, plasma levels of H2O2 were also found in the micromolar range– orders of magnitude higher than those previously estimated to be present inside cells [9,95]. Since conventional experimental approaches using extracellular administration of H2O2 need to overcome cellular H2O2 gradient to study oxidative effects within cells, the high concentrations of extracellular H2O2 can result in spurious cellular responses, and should be interpreted with caution [96,97]. Instead, the applications of the DAAO system provide a more precise method to mimic endogenous H2O2 mediated pathways in oxidative eustress or distress deriving from distinct intracellular sources [98,99] as summarized below and as shown in Figure 2.
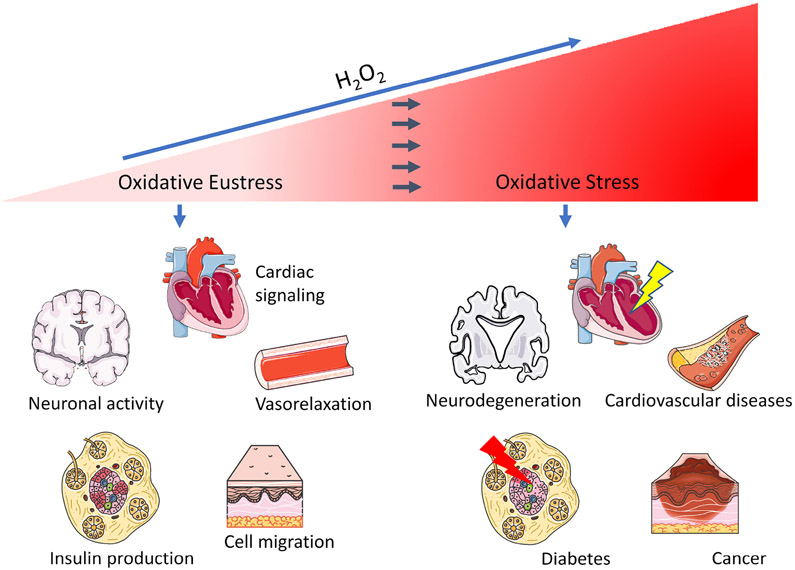
Figure 2: DAAO applications for studying H2O2-mediated pathways in various cells and tissues:
Scheme shows physiological H2O2 signaling in the heart, vasculature, nerves, β-cells and other cells and tissues (left side) or conditions of oxidative stress resulting in cardiovascular disease, neurodegeneration, diabetes and cancer induced by pathologically increased H2O2 (right side). Depicted models were modified from Servier Medical Art (https://smart.servier.com/image-set-download/), licenced under a Creative Commons Attribution 3.0 Unported License by Servier.
3. Chemogenetic H2O2-generating approaches in vascular cells
The vascular system dynamically regulates blood flow according to the diverse metabolic needs of different tissues [100]. The endothelial layer of the vascular system is critical in regulating this homeostatic system [101]. Oxidative stress is associated with endothelial dysfunction and with pathophysiological conditions such as hypertension [102,103], vascular inflammation [104,105], diabetes [106] and adverse vascular remodeling [107-110]. Yet H2O2 also has an essential role as a signaling molecule in the vascular endothelial cell to maintain this vascular homeostasis through involvement in post-translational modifications such as phosphorylation [106,111-114], S-glutathionylation [115-118], sulfenylation (RSOH) [119,120] and other cysteine modification (cysteinylation and disulfide bond formation) [121,122]. It is critical to regulate the optimal amount of H2O2 in the endothelial cells to mediate these diverse oxidant-dependent post-translational signaling responses. Perturbed H2O2 homeostasis leads to impaired cell function, increasing the risk of disease-associated pathologies [108,123,124]. One important issue in understanding this homeostasis is determining the actual concentrations of H2O2 at physiological levels in different compartments of the endothelial cell. The recently developed Hyper7 genetically-encoded biosensor [88], is an ultrasensitive probe for the detection of H2O2, and can be combined with DAAO to modulate H2O2 levels in different organelles in vascular endothelial cells [98]. Using this chemogenetic approach along with live-cell imaging, H2O2 can both be endogenously produced and detected in an organelle-specific manner in the cytoplasm, caveolae, mitochondria, or nucleus. Saravi et al. have recently shown that compartmentalized endogenous generation of H2O2 elicits differential signaling effects (through altered Akt, AMPK and eNOS phosphorylation) compared to traditional extracellular treatment of H2O2 in endothelial cells even though levels of H2O2 achieved by both methods were quantitatively similar in the cells [98]. They also found that AMPK phosphorylation directly depends on nuclear H2O2. The inhibition of AMPK phosphorylation diminished nuclear H2O2 driven eNOS phosphorylation but had no effect on cytosol- or caveolae-generated H2O2 mediated phosphorylation response. This clearly suggests that compartmentalized H2O2 is critically essential for signaling events in endothelial cells.
Thus, spatiotemporal tracking of H2O2 levels in living cells is crucial for understanding the role of this redox molecule in vascular biology. A chemogenetic approach can be an invaluable tool for the study of vascular dysfunction caused by disturbance of H2O2 homeostasis. The opportunities that lie ahead with the newly developed HyPer7-DAAO sensors may be equally promising and should lead to a much better understanding of tissue-, cell- and subcellular-specific redox homeostasis.
4. A heart failure model based on DAAO induced oxidative stress
Oxidative stress has been implicated in the pathogenesis of heart failure and in many cardiac disease states [125]. Although these associations have been well described, it is still not clear if oxidative stress alone is a causal factor of heart failure. First insight into this question was recently provided by the application of chemogenetic approaches in vivo. Specifically, a new model of heart failure was developed by using DAAO to generate the stable ROS H2O2 in cardiac myocytes in living rats [92,126]. The DAAO construct was packaged as a fusion protein with the HyPer biosensor in the cardiotropic adeno-associated virus isotype 9 (AAV9) under control of the cardiac-specific cTnT promoter [92]. The animals were then infected with the recombinant DAAO-AAV9 virus by tail vein injection, and robust expression in the cardiac myocytes was detected within 3–4 weeks. The expression was specific to the heart, with only minimal expression in skeletal muscle [92]. Importantly, when D-alanine was added to isolated adult cardiac myocytes, there was robust generation of H2O2, detected using the HyPer biosensor. Of note, the addition of L-alanine did not result in H2O2 generation: most mammalian cells use L-amino acids and the recombinant DAAO is inactive until D-amino acids are provided [126].
To further investigate the effects of chronic oxidative stress in cardiac myocytes, animals were infected with the DAAO-AAV9 and were provided with D-alanine in their drinking water. This led to a striking dilated cardiomyopathy phenotype within 4 weeks of D-alanine treatment, characterized by significant reductions in left ventricular (LV) ejection fraction and fractional shortening along with significant increases in LV volume and heart mass [92,126]. The hemodynamic changes were accompanied by significant increases of the biomarkers atrial natriuretic peptide (ANP), brain natriuretic peptide (BNP) and cardiac troponin I (cTnI) both in the plasma and the cardiac transcriptome as has been similarly reported in humans with heart failure [92,126,127]. Despite the decline in cardiac function, there was no evidence of interstitial fibrosis by 4 weeks of D-alanine feeding. With longer provision of D-alanine, there was no further decline in cardiac function, but there was development of cardiac fibrosis at 8 weeks as seen by increased collagen deposition on histology and elevation of several transcriptional markers of fibrosis including mRNAs encoding selected collagen isoforms and transforming growth factor-β1. The robust heart failure phenotype was rapidly reversed when DAAO infected animals were treated with angiotensin-II receptor blocker valsartan alone or in combination with the neprilysin inhibitor sacubitril even in the presence of ongoing oxidative stress [126]. Interestingly, despite the recovery of cardiac function with drug treatment, the interstitial fibrosis persisted. These findings indicate that cardiac fibrosis and cardiac dysfunction should not be used interchangeably as the presence of one does not necessarily imply the presence of the other.
In conclusion, the DAAO model of chemogenetic heart failure induces a rapid, robust and tractable heart failure phenotype that is reversible and has many features in common with human heart failure. This chemogenetic approach adds new insights and establishes oxidative stress as a causal determinant of heart failure. This new model may now be utilized for the discovery and validation of new therapeutic targets for the prevention and treatment of heart failure.
5. Using DAAO in a neurobiological system
In contrast to the deleterious effects of chemogenetic oxidative stress on cardiac myocyte function [92], in other model systems the chemogenetic generation of lower H2O2 concentrations can have cytoprotective effects. ROS imbalance is also a major cause of neurodegenerative disorders such as Alzheimer’s disease, Parkinson’s disease, Huntington’s disease, and amyotrophic lateral sclerosis [128]. The molecular mechanisms, whereby alterations in redox balance lead to neurodegeneration, are incompletely understood, but several reports have implicated a neuroprotective role of the redox-modulated transcription factor nuclear factor erythroid 2-related factor 2 (Nrf2) [129-131]. However, a study using cytosolic DAAO expression in astrocytes co-cultured with neurons successfully led to a redox-regulated neuroprotection that was independent of Nrf2 [132]. Astrocytes and neurons are tightly interconnected via gap junctions to ensure crucial cell communication between the two neuronal cell types [133,134]. The exposure of DAAO-expressing astrocytes to high concentrations of D-alanine resulted in neurotoxicity, but the application of a lower D-alanine concentration enabled H2O2 transfer to neurons that led to the inhibition of protein tyrosine phosphatase within the astrocytes. Microarray analyses of the axonal transcriptome revealed an up-regulation of Interleukin-1beta (IL-1β) and Caveolin-1, but no increased gene expression of Nrf2-regulated genes. The authors could mimic the neuroprotective H2O2-induced effect with PTP inhibitors, indicating a possible neuroprotective mechanism in response to IL-1β upregulation [135]. In contrast, a subsequent publication reported that extracellular H2O2 promoted activation of Nrf2 in astrocytes under the same conditions [136]. These disparate findings again point to the divergent responses that can be elicited depending on the cellular source of H2O2 [137]. Despite the long-held view that neurons are particularly vulnerable to oxidative stress [138,139], these studies show that H2O2 can be involved in both physiological and pathological responses in neurons. Moreover, the work of Haskew-Layton et al. to date represents the only investigation in which DAAO-mediated H2O2 generation is used to elicit a physiological instead of a pathological response.
6. Visualizing inter-organelle H2O2 transfer with HyPer and DAAO in a diabetic model
Pancreatic β-cells, which produce and secrete insulin, are the largest cell population of the islets of Langerhans and are particularly vulnerable to oxidative stress [140]. This exceptional sensitivity is due to the fact that β-cells typically express low levels of antioxidant enzymes [141,142]. Catalase is almost completely absent in the peroxisomes of β-cells, and glutathione peroxidases in the mitochondria and ER are only weakly expressed [143,144]. On the other hand, β-cells generate considerable amounts of H2O2 as a by-product of disulfide formation during the synthesis of insulin [145]. Production of H2O2 is further enhanced in pathologic states. During cytokine attack by immune cells in type 1 diabetes mellitus (T1D), large amounts of H2O2 are produced in the mitochondria [146-148]. In the pathogenesis of type 2 diabetes mellitus (T2D), H2O2 is formed from the peroxisomal β-oxidation of long-chain free fatty acids [149]. Thus, increased ROS production can lead to the development of diabetes, as H2O2 cannot be sufficiently detoxified. It is therefore even more important that H2O2 can be transported to a site with a higher antioxidant capacity. Otherwise, toxic OH• may arise in the Fenton reaction, which can lead to β-cell apoptosis [150,151]. Differentially-targeted DAAO constructs (targeted to mitochondria, peroxisomes, and/or ER)– in combination with targeted HyPer constructs– allow monitoring of H2O2 transport across membranes. By expressing DAAO in the mitochondria and HyPer in the peroxisomes, it is possible to follow the path of H2O2 across two membranes from one organelle to another. This approach is used in research on peroxiporins, which are a class of aquaporins that transport not only water but also H2O2. The various peroxiporin isoforms have different H2O2 transport capacities [76,152-157]. Recombinant DAAO/HyPer approaches have been used to explore the localization and functionality of different peroxiporins by “knocking out” and/or overexpressing these peroxiporins and then using live cell imaging to quantitate H2O2 transfer rates within the cell. For example, one study documented a critical role of the peroxiporin isoform AQP8 in modulating the intracellular distribution of H2O2 in insulin-producing β-cells, which are particularly susceptible to oxidative stress [75], and indicating a potential role for AQP8 paticularly in T1D pathogenesis.
7. Targeting cancer cells with DAAO
Excessive H2O2 can lead to pathological oxidative stress and cause cellular apoptosis [158,159]. One way to harness this oxidative damage for therapeutic purposes is to specifically target cancer cells for enhanced ROS production in order to induce cancer cell death. Many small molecule redox chemotherapeutics have been investigated for this purpose, but often have shown off-target effects [160]. The use of DAAO as a gene-directed enzyme prodrug represents a potentially effective approach since D-amino acids would not have a cytotoxic effect in cells that are not expressing DAAO. Moreover, some cancer cells appear to be characterized by increased ROS levels [161]. In a recent pioneering study, DAAO was stably expressed in rat 9L gliosarcoma cells, and the addition of D-alanine had a cytotoxic effect [162]. The main drawback of using this chemogenetic approach to kill cancer cells is the reliance on proper targeting of DAAO to tumors in vivo since ROS are involved in almost every stage of cancer development [163-165]. To overcome this problem, the conjugation of macromolecules to DAAO has been used to achieve more specific targeting of DAAO to cancer cells; one combination that has been used to accomplish this was polyethylene glycol conjugated (“pegylated”) DAAO (PEG-DAAO). Tumor targeting by pegylation led to the accumulation of PEG-DAAO in solid tumors. Several studies in mice showed enrichment of PEG-DAAO in tumor cells, and the addition of D-proline resulted in marked inhibition of tumor growth as a consequence of H2O2 generation– without affecting non-cancerous cells [166]. Moreover, the tumor-killing effects of PEG-DAAO could be improved by the co-application of pegylated zinc protoporphyrin (PEG-ZnPP), a potent heme oxygenase-1 (HO-1) inhibitor [167]. Compared to normal cells, most cancer cells contain a more limited set of antioxidant enzymes [166]. Although these first applications of DAAO for cancer therapy appear promising, the method does have important limitations, e.g. the low O2 levels in tumor cells led to the development of a DAAO variant that has a higher affinity for molecular oxygen. A mutated variant of a yeast DAAO was developed that showed a lower Km for D-alanine even under low oxygen condition [168]. Recently, DAAO-mediated chemotherapy has been further developed by coating magnetic nanoparticles with DAAO, enabling the use of an external magnetic field [169-172] to target the enzyme to tumors [173,174]. Sikes and colleagues have established a model in HeLa cells that is based on mitochondria-targeted DAAO as an anti-cancer cell therapy [74,175-178]. The local generation of H2O2 by targeting DAAO to mitochondria in HeLa cells showed more cytotoxicity than cytosolic DAAO [74,77,93,99]. Taken together, several approaches using DAAO have been developed to try and target redox-based chemotherapies to tumors, which may have the potential to serve as novel tools to combat cancer [179].
8. Perspectives on the applications of chemogenetics in redox biology
Chemogenetic approaches using DAAO and HyPer represent an example of “synthetic biology”, in which engineering principles are applied to create novel biological systems that are not typically found in nature. Thus, a yeast enzyme (DAAO) and a bacterial transcription factor (OxyR, which is a key component of HyPer biosensors) have been cloned as a fusion construct and used both to generate (DAAO) and detect (HyPer) H2O2 in cultured mammalian cells or in animals. While these approaches are informative as long as proper controls are included (Figure 1), there are potentially important differences between the biological effects of H2O2 generated by recombinant DAAO and the H2O2 that comes from endogenous enzymes and organelles. These differences are easier to control in the in vitro setting, in which real-time imaging of (using HyPer or other biosensors) can be used to dissect intracellular pathways of H2O2 diffusion and signaling following the addition (or removal) of D-amino acids. However, the results of these experiments can be influenced by the differential transport of D-amino acids in different organelles and in different cell types [180-183]. Moreover, the rate of reduction of oxidized HyPer might vary in different regions of the cell, and could thereby influence cellular responses. The H2O2 generated by, for example, Nox4 in the endoplasmic reticulum, cannot be viewed as being equivalent to the H2O2 molecules that are generated by ER-targeted DAAO: there will be differences both in local H2O2 flux and in the protein microenvironment that exert influence the cellular response. It can be helpful to use a HyPer control construct (as control for the HyPer-DAAO fusion protein) to avoid potential confounding by HyPer serving as a sink for the H2O2 in the cell. In general, however, genetically-encoded biosensors are expressed at a much lower level than the analytes that they detect, so HyPer itself is unlikely to independently change basal intracellular H2O2 levels. It is also important to include negative controls that include the use of L-amino acids instead of D-amino acids. All of these considerations also apply in the in vivo setting, but once again, confounding can be minimized by the use of proper controls. In vivo, the consequences of D-amino acid addition and removal may transpire over a time period of days, and differential D-amino acid uptake between different cell types can also influence the response to chemogenetic H2O2 generation. It is important to note that the DAAO-HyPer fusion protein is able to induce chronic oxidative stress in vivo, indicating that DAAO generates much more H2O2 than HyPer is able to consume [92]. D-amino acid feeding to animals expressing targeted DAAO in different cell types permits the study of oxidative stress in distinct tissues to be correlated with disease phenotype. These in vivo approaches can provide important information of the differential roles of specific cells in ROS-dependent disease states. Despite these limitations, properly-designed and carefully-controlled chemogenetic approaches using DAAO can provide novel information on the effects of differentially-generated H2O2 both in cultured cells and in vivo.
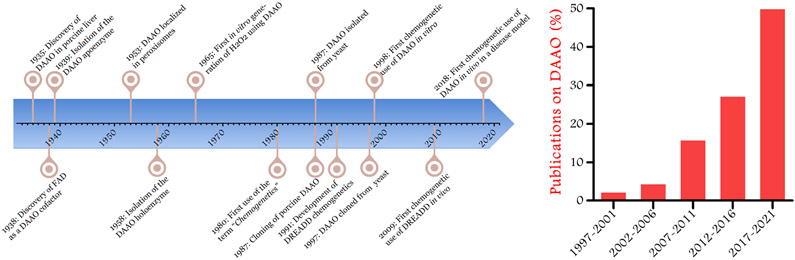
Many decades passed from the discovery of mammalian DAAO in porcine liver to the exploitation of the yeast DAAO as a chemogenetic enzyme to produce H2O2 in mammalian cells. Figure 3 shows important milestones on DAAO research from the discovery to its use as a chemogenetic tool [23,24,43,60,92,162,184-190]. At the time of its discovery, the presence of DAAO in mammalian cells seemed highly unusual since D-amino acids are found at very low levels in most mammalian tissues [33]. It has been speculated that mammalian DAAO functions in peroxisomes to catabolize the rare D-amino acids that can found in regions of the brain and in metabolically active tissues such as liver and kidney. While the yeast DAAO follows the same overall catalytic scheme as its mammalian homologues, the yeast enzyme subserves an entirely different physiological role: yeast DAAO metabolizes D-amino acids to generate energy and provide key metabolites for the cell [41]. The divergent cellular functions of mammalian and yeast DAAO are not only interesting from evolutionary point of view [25], but the distinctively robust generation of H2O2 by the yeast enzyme permits the use of yeast DAAO as a chemogenetic tool that permits the modeling of oxidative distress and oxidative eustress. These key features of yeast DAAO have led to exciting new discoveries on the physiological and pathophysiological roles of H2O2 in mammalian cells and tissues.
Figure 3: Chronological development of DAAO and chemogenetic research leading to chemogenetic DAAO applications.
The timeline shows historical milestones in research on DAAO and chemogenetics (left panel). The graph indicates the percentage of publications using DAAO as a chemogenetic tool for H2O2 generation in 5 year intervals (right panel).
9. Conclusions
H2O2 is a key signaling molecule involved in numerous physiological processes in diverse cell types. However, like other ROS, H2O2 at high concentrations is deleterious for cells. Over the last century, diseases characterized by excessive ROS accumulation– including diseases of the cardiovascular and nervous system as well as diabetes and cancer– have emerged as the leading causes of death [191]. Chemogenetic approaches using DAAO for H2O2 generation allow disease states associated with oxidative stress to be effectively modeled, enabling the more precise understanding of the pathological processes caused by redox stress. To date, relatively few studies have used chemogenetic tools to modulate ROS in vivo, but the number of studies utilizing DAAO has been increasing steadily over the last decade (Figure 3). The generation of transgenic animal models with tissue-specific expression of DAAO will enable future investigations of ROS-related pathological processes in diverse tissues and organ systems and lead to the identification of novel therapeutic targets.
Highlights.
Hydrogen peroxide (H2O2) is a key molecule that is involved in both oxidant signaling and oxidative stress.
D-amino acid oxidase (DAAO) serves as a powerful chemogenetic tool to generate intracellular H2O2 and to decipher oxidant-modulated pathways in normal cells and in disease states.
Chemogenetic applications are increasingly being used to study the roles of H2O2 in cardiovascular disease, neurodegeneration, diabetes and cancer.
Acknowledgements
We regret that we have been unable to cite all the primary literature on redox signaling and oxidative stress due to length limitations for this article; this has focused in particular on the enzyme DAAO as a chemogenetic tool for H2O2 generation from its discovery to cell applications. This work was supported by funds from the National Institutes of Health grants R21AG063073, R01HL152173, and R33HL157918, and from the Brigham Research Institute Fund to Sustain Research Excellence (to T.M.); CK is funded by the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) by grant No. 231396381/GRK1947. M.W.-W. is funded by an Erwin Schroedinger Abroad Followship from the Austrian Science Fund J4466-B and F.S. is funded by grants T32HD-098061 and 5T32HL007609-34.
Footnotes
Declaration of competing interest
The authors declare no conflict of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Yin W, Robyn B, Alycia N, et al. Superoxide dismutases: Dual roles in controlling ROS damage and regulating ROS signaling. Journal of Cell Biology 2018;217:1915–1928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Collin F Chemical basis of reactive oxygen species reactivity and involvement in neurodegenerative diseases. International Journal of Molecular Sciences 2019;20:. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Sies H Hydrogen peroxide as a central redox signaling molecule in physiological oxidative stress: Oxidative eustress. Redox Biology 2017; 11:. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Rashdan NA, Pattillo CB. Hydrogen peroxide in the ER: A tale of triage. Redox Biology 2020;28:101358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Morrell CN. Reactive oxygen species: Finding the right balance. Circulation Research 2008;103:571–572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Bienert GP, Chaumont F. Aquaporin-facilitated transmembrane diffusion of hydrogen peroxide. Biochimica et Biophysica Acta - General Subjects 2014;1840:1596–1604. [DOI] [PubMed] [Google Scholar]
- [7].Azad AK, Raihan T, Ahmed J, et al. Human Aquaporins: Functional Diversity and Potential Roles in Infectious and Non-infectious Diseases. Frontiers in Genetics 2021; 12:. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Galli M, Hameed A, Żbikowski A, et al. Aquaporins in insulin resistance and diabetes: More than channels! Redox Biology 2021; 102027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Sies H Oxidative eustress: On constant alert for redox homeostasis. Redox Biology 2021;41:101867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Sies H, Jones DP. Reactive oxygen species (ROS) as pleiotropic physiological signalling agents. Nature Reviews Molecular Cell Biology 2020;21:363–383. [DOI] [PubMed] [Google Scholar]
- [11].Forman HJ, Maiorino M, Ursini F. Signaling functions of reactive oxygen species. Biochemistry 2010;49:835–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].He L, He T, Farrar S, et al. Antioxidants Maintain Cellular Redox Homeostasis by Elimination of Reactive Oxygen Species. Cellular Physiology and Biochemistry 2017;44:532–553. [DOI] [PubMed] [Google Scholar]
- [13].Sies H Oxidative stress: Eustress and distress in redox homeostasis. In: Stress: Physiology, Biochemistry, and Pathology Handbook of Stress Series, Volume 3. 2019. . [Google Scholar]
- [14].Holmström KM, Finkel T. Cellular mechanisms and physiological consequences of redox-dependent signalling. Nature Reviews Molecular Cell Biology 2014;15:411–421. [DOI] [PubMed] [Google Scholar]
- [15].Sies H, Berndt C, Jones DP. Oxidative Stress: Annual Review of Biochemistry. Annual Review of Biochemistry 2017;86:. [DOI] [PubMed] [Google Scholar]
- [16].Jones DP, Sies H. The Redox Code. Antioxidants and Redox Signaling 2015;23:734–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Forrester SJ, Kikuchi DS, Hernandes MS, et al. Reactive oxygen species in metabolic and inflammatory signaling. Circulation Research 2018;122:877–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Mittal M, Siddiqui MR, Tran K, et al. Reactive oxygen species in inflammation and tissue injury. Antioxidants and Redox Signaling 2014;20:1126–1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Madreiter-Sokolowski CT, Thomas C, Ristow M. Interrelation between ROS and Ca2+ in aging and age-related diseases. Redox Biology 2020;36:101678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Liu Z, Ren Z, Zhang J, et al. Role of ROS and nutritional antioxidants in human diseases. Frontiers in Physiology 2018;9:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Kerksick C, Willoughby D. The Antioxidant Role of Glutathione and N-Acetyl-Cysteine Supplements and Exercise-Induced Oxidative Stress. Journal of the International Society of Sports Nutrition 2005;2:38–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Fransen M, Nordgren M, Wang B, et al. Role of peroxisomes in ROS/RNS-metabolism: Implications for human disease. Biochimica et Biophysica Acta - Molecular Basis of Disease 2012; 1822:1363–1373. [DOI] [PubMed] [Google Scholar]
- [23].Krebs HA. Metabolism of amino-acids: Deamination of amino-acids. The Biochemical Journal 1935;29:. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Warburg O, Christian W. Koferment der d-Alanin-Dehydrase. Die Naturwissenschaften 1938;26:. [Google Scholar]
- [25].Pollegioni L, Piubelli L, Sacchi S, et al. Physiological functions of D-amino acid oxidases: From yeast to humans. Cellular and Molecular Life Sciences 2007;64:1373–1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Nueangaudom A, Lugsanangarm K, Pianwanit S, et al. New Aspects of the Structure of d-Amino Acid Oxidase from Porcine Kidney in Solution: Molecular Dynamics Simulation and Photoinduced Electron Transfer. In: Amino Acid - New Insights and Roles in Plant and Animal. 2017. . [Google Scholar]
- [27].Skowrońska M, Albrecht J. Oxidative and nitrosative stress in ammonia neurotoxicity. Neurochemistry International 2013;62:731–737. [DOI] [PubMed] [Google Scholar]
- [28].Adeva-Andany M, López-Ojén M, Funcasta-Calderón R, et al. Comprehensive review on lactate metabolism in human health. Mitochondrion 2014;17:76–100. [DOI] [PubMed] [Google Scholar]
- [29].Faotto L, Pollegioni L, Ceciliani F, et al. The primary structure of D-amino acid oxidase from Rhodotorula gracilis. Biotechnology Letters 1995;17:193–198. [Google Scholar]
- [30].Pollegioni L, Diederichs K, Molla G, et al. Yeast D-amino acid oxidase: Structural basis of its catalytic properties. Journal of Molecular Biology 2002;324:535–546. [DOI] [PubMed] [Google Scholar]
- [31].Caligiuri A, D’Arrigo P, Rosini E, et al. Enzymatic conversion of unnatural amino acids by yeast D-amino acid oxidase. Advanced Synthesis and Catalysis 2006;348:2183–2190. [Google Scholar]
- [32].Pollegioni L, Molla G. New biotech applications from evolved D-amino acid oxidases. Trends in Biotechnology 2011;29:276–283. [DOI] [PubMed] [Google Scholar]
- [33].Pollegioni L, Sacchi S, Murtas G. Human D-amino acid oxidase: Structure, function, and regulation. Frontiers in Molecular Biosciences 2018;5:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Kiss DJ, Ferenczy GG. A detailed mechanism of the oxidative half-reaction of d-amino acid oxidase: Another route for flavin oxidation. Organic and Biomolecular Chemistry 2019;17:7973–7984. [DOI] [PubMed] [Google Scholar]
- [35].Neame S, Safory H, Radzishevsky I, et al. The NMDA receptor activation by D-serine and glycine is controlled by an astrocytic Phgdh-dependent serine shuttle. Proceedings of the National Academy of Sciences of the United States of America 2019;116:20736–20742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Stroebel D, Casado M, Paoletti P. Triheteromeric NMDA receptors: from structure to synaptic physiology. Current Opinion in Physiology 2018;2:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].McQuate A, Barria A. Rapid exchange of synaptic and extrasynaptic NMDA receptors in hippocampal CA1 neurons. Journal of Neurophysiology 2020;123:1004–1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Papouin T, Ladépêche L, Ruel J, et al. Synaptic and extrasynaptic NMDA receptors are gated by different endogenous coagonists. Cell 2012;150:633–646. [DOI] [PubMed] [Google Scholar]
- [39].Verrall L, Burnet PWJ, Betts JF, et al. The neurobiology of D-amino acid oxidase and its involvement in schizophrenia. Molecular Psychiatry 2010;15:122–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Alonso J, Bruno LB, Melladot E, et al. D-Amino-acid oxidase gene from. 1998;1095–1101. [DOI] [PubMed] [Google Scholar]
- [41].Simonetta MP, Verga R, Fretta A, et al. Induction of D-Amino-acid Oxidase by D-Alanine in Rhodotorula gracilis Grown in Defined Medium. Microbiology 1989;135:593–600. [Google Scholar]
- [42].Molla G, Motteran L, Piubelli L, et al. Regulation of D-amino acid oxidase expression in the yeast Rhodotorula gracilis. Yeast 2003;20:1061–1069. [DOI] [PubMed] [Google Scholar]
- [43].Simonetta MP, Vanoni MA, Casalin P. Purification and properties of d-amino-acid oxidase, an inducible flavoenzyme from Rhodotorula gracilis. Biochimica et Biophysica Acta (BBA)/Protein Structure and Molecular 1987;914:136–142. [Google Scholar]
- [44].Pollegioni L, Falbo A, Pilone MS. Specificity and kinetics of Rhodotorula gracillis d-amino acid oxidase. Biochimica et Biophysica Acta (BBA)/Protein Structure and Molecular 1992;1120:11–16. [DOI] [PubMed] [Google Scholar]
- [45].Mattevi A, Vanoni MA, Todone F, et al. Crystal structure of D-amino acid oxidase: A case of active site mirror-image convergent evolution with flavocytochrome b2. Proceedings of the National Academy of Sciences of the United States of America 1996;93:7496–7501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Mizutani H, Miyahara I, Hirotsu K, et al. Three-dimensional structure of porcine kidney D-amino acid oxidase at 3.0 Å resolution. Journal of Biochemistry 1996;120:14–17. [DOI] [PubMed] [Google Scholar]
- [47].Umhau S, Pollegioni L, Molla G, et al. The x-ray structure of D-amino acid oxidase at very high resolution identifies the chemical mechanism of flavin-dependent substrate dehydrogenation. Proceedings of the National Academy of Sciences of the United States of America 2000;97:12463–12468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Subramanian K, Góra A, Spruijt R, et al. Modulating D-amino acid oxidase (DAAO) substrate specificity through facilitated solvent access. PLoS ONE 2018; 13:. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Xu Z, Reilley M, Li R, et al. Mapping absolute tissue endogenous fluorophore concentrations with chemometric wide-field fluorescence microscopy. Journal of Biomedical Optics 2017;22:. [DOI] [PubMed] [Google Scholar]
- [50].Hühner J, Ingles-Prieto Á, Neusuüß C, et al. Quantification of riboflavin, flavin mononucleotide, and flavin adenine dinucleotide in mammalian model cells by CE with LED-induced fluorescence detection. Electrophoresis 2015;36:. [DOI] [PubMed] [Google Scholar]
- [51].Alim I, Haskew-Layton RE, Aleyasin H, et al. Spatial, temporal, and quantitative manipulation of intracellular hydrogen peroxide in cultured cells. 1st ed. Elsevier Inc.; 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Porter DJT, Voet JG, Bright HJ. Mechanistic features of the D amino acid oxidase reaction studied by double stopped flow spectrophotometry. Journal of Biological Chemistry 1977;252:4464–4473. [PubMed] [Google Scholar]
- [53].Pollegioni L, Langkau B, Tischer W, et al. Kinetic mechanism of D-amino acid oxidases from Rhodotorula gracilis and Trigonopsis variabilis. Journal of Biological Chemistry 1993;268:13850–13857. [PubMed] [Google Scholar]
- [54].Gabler M, Hensel M, Fischer L. Detection and substrate selectivity of new microbial D-amino acid oxidases. Enzyme and Microbial Technology 2000;27:605–611. [DOI] [PubMed] [Google Scholar]
- [55].Tishkov VI, Khoronenkova S V. D-amino acid oxidase: Structure, catalytic mechanism, and practical application. Biokhimiya 2005;70:51–67. [PubMed] [Google Scholar]
- [56].Moussa S, Murtas G, Pollegioni L, et al. Enhancing Electrochemical Biosensor Selectivity with Engineered d -Amino Acid Oxidase Enzymes for d -Serine and d -Alanine Quantification . ACS Applied Bio Materials 2021; [DOI] [PubMed] [Google Scholar]
- [57].Pilone MS. D-amino acid oxidase: New findings. Cellular and Molecular Life Sciences 2000;57:1732–1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Pollegioni L, Caldinelli L, Molla G, et al. Catalytic properties of D-amino acid oxidase in cephalosporin C bioconversion: A comparison between proteins from different sources. Biotechnology Progress 2004;20:467–473. [DOI] [PubMed] [Google Scholar]
- [59].Obregón V, Mata I de la, Ramón F, et al. Oxidation by hydrogen peroxide of D-amino acid oxidase from Rhodotorula gracilis. Progress in Biotechnology 1998;15:89–94. [Google Scholar]
- [60].Forkmann G, Dangelmayr B. Genetic control of chalcone isomerase activity in flowers of Dianthus caryophyllus. Biochemical Genetics 1980;18:519–527. [DOI] [PubMed] [Google Scholar]
- [61].Coward P, Wada HG, Falk MS, et al. Controlling signaling with a specifically designed Gi-coupled receptor. Proceedings of the National Academy of Sciences of the United States of America 1998;95:352–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Urban DJ, Roth BL. DREADDs (designer receptors exclusively activated by designer drugs): Chemogenetic tools with therapeutic utility. Annual Review of Pharmacology and Toxicology 2015;55:399–417. [DOI] [PubMed] [Google Scholar]
- [63].Roth BL. Use of DREADDS. Neuron 2017;89:683–694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Roth BL. DREADDs for Neuroscientists. Neuron 2016;89:. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Zhao Y, Zhao L, Yang G, et al. Characterization of n uricase from Bacillus fastidious A.T.C.C. 26904 and its application to serum uric acid assay by a patented kinetic uricase method. Biotechnology and Applied Biochemistry 2006;45:. [DOI] [PubMed] [Google Scholar]
- [66].Milton RD, Giroud F, Thumser AE, et al. Hydrogen peroxide produced by glucose oxidase affects the performance of laccase cathodes in glucose/oxygen fuel cells: FAD-dependent glucose dehydrogenase as a replacement. Physical Chemistry Chemical Physics 2013;15:19371–19379. [DOI] [PubMed] [Google Scholar]
- [67].Ren Q, Henes B, Fairhead M, et al. High level production of tyrosinase in recombinant Escherichia coli. BMC Biotechnology 2013;13:. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Das S, Glenn IV JH, Subramanian M. Enantioselective oxidation of 2-hydroxy carboxylic acids by glycolate oxidase and catalase coexpressed in methylotrophic Pichia pastoris. Biotechnology Progress 2010;26:607–615. [DOI] [PubMed] [Google Scholar]
- [69].Goswami P, Chinnadayyala SSR, Chakraborty M, et al. An overview on alcohol oxidases and their potential applications. Applied Microbiology and Biotechnology 2013;97:4259–4275. [DOI] [PubMed] [Google Scholar]
- [70].Ande SR, Kommoju PR, Draxl S, et al. Mechanisms of cell death induction by L-amino acid oxidase, a major component of ophidian venom. Apoptosis 2006;11:1439–1451. [DOI] [PubMed] [Google Scholar]
- [71].Lim JB, Sikes HD. Use of a genetically encoded hydrogen peroxide sensor for whole cell screening of enzyme activity. Protein Engineering, Design and Selection 2015;28:79–83. [DOI] [PubMed] [Google Scholar]
- [72].Miller EW, Taulet N, Onak CS, et al. Light-activated regulation of cofilin dynamics using a photocaged hydrogen peroxide generator. Journal of the American Chemical Society 2010;132:. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Bulina ME, Chudakov DM, Britanova OV, et al. A genetically encoded photosensitizer. 2006; [DOI] [PubMed] [Google Scholar]
- [74].Huang BK, Langford TF, Sikes HD. Using Sensors and Generators of H2O2 to Elucidate the Toxicity Mechanism of Piperlongumine and Phenethyl Isothiocyanate. Antioxidants and Redox Signaling 2016;24:924–938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Ermakova YG, Mishina NM, Schultz C, et al. Visualization of intracellular hydrogen peroxide with the genetically encoded fluorescent probe HyPer in NIH-3T3 cells. In: Methods in Molecular Biology. 2019. . [DOI] [PubMed] [Google Scholar]
- [76].Krüger C, Waldeck-Weiermair M, Kaynert J, et al. AQP8 is a crucial H2O2 transporter in insulin-producing RINm5F cells. Redox Biology 2021;43:. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Stein KT, Moon SJ, Nguyen AN, et al. Kinetic modeling of H2O2 dynamics in the mitochondria of HeLa cells. PLoS Computational Biology 2020;16:1–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].LeBel CP, Ischiropoulos H, Bondy SC. Evaluation of the Probe 2′,7′-Dichlorofluorescin as an Indicator of Reactive Oxygen Species Formation and Oxidative Stress. Chemical Research in Toxicology 1992;5:227–231. [DOI] [PubMed] [Google Scholar]
- [79].Ashoka AH, Ali F, Tiwari R, et al. Recent Advances in Fluorescent Probes for Detection of HOCl and HNO. ACS Omega 2020;5:1730–1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Jiang X, Wang L, Carroll SL, et al. Challenges and opportunities for small-molecule fluorescent probes in redox biology applications. Antioxidants and Redox Signaling 2018;29:518–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Winterbourn CC. Toxicity of iron and hydrogen peroxide: the Fenton reaction. Toxicology Letters 1995; 82–83:. [DOI] [PubMed] [Google Scholar]
- [82].Matlashov ME, Bogdanova YA, Ermakova GV., et al. Fluorescent ratiometric pH indicator SypHer2: Applications in neuroscience and regenerative biology. Biochimica et Biophysica Acta - General Subjects 2015;1850:. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Waypa GB, Marks JD, Guzy R, et al. Nihms-172925. 2011;106:526–535. [Google Scholar]
- [84].Belousov VV, Fradkov AF, Lukyanov KA, et al. Genetically encoded fluorescent indicator for intracellular hydrogen peroxide. Nature Methods 2006; [DOI] [PubMed] [Google Scholar]
- [85].Guzy RD, Hoyos B, Robin E, et al. Mitochondrial complex III is required for hypoxia-induced ROS production and cellular oxygen sensing. Cell Metabolism 2005;1:401–408. [DOI] [PubMed] [Google Scholar]
- [86].Enyedi B, Zana M, Donkó Á, et al. Spatial and temporal analysis of NADPH oxidase-generated hydrogen peroxide signals by novel fluorescent reporter proteins. Antioxidants and Redox Signaling 2013;19:523–534. [DOI] [PubMed] [Google Scholar]
- [87].Stanford KR, Ajmo JM, Bahia PK, et al. Improving redox sensitivity of roGFP1 by incorporation of selenocysteine at position 147. BMC Research Notes 2018;11:827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Pak VV, Ezerina D, Lyublinskaya OG, et al. Ultrasensitive Genetically Encoded Indicator for Hydrogen Peroxide Identifies Roles for the Oxidant in Cell Migration and Mitochondrial Function. Cell Metabolism 2020;31:. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Yu H, Li H, Zhou Y, et al. Ultra-sensitive hydrogen peroxide sensor based on peroxiredoxin and fluorescence resonance energy transfer. Applied Sciences (Switzerland) 2020; 10:. [Google Scholar]
- [90].Langford TF, Huang BK, Lim JB, et al. Monitoring the action of redox-directed cancer therapeutics using a human peroxiredoxin-2-based probe. Nature Communications 2018;9:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Matlashov ME, Belousov VV, Enikolopov G. How Much H 2 O 2 Is Produced by Recombinant D-Amino Acid Oxidase in Mammalian Cells? . Antioxidants & Redox Signaling 2013; [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Steinhorn B, Sorrentino A, Badole S, et al. Chemogenetic generation of hydrogen peroxide in the heart induces severe cardiac dysfunction. Nature Communications 2018; [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Moon SJ, Dong W, Stephanopoulos GN, et al. Oxidative pentose phosphate pathway and glucose anaplerosis support maintenance of mitochondrial NADPH pool under mitochondrial oxidative stress. Bioengineering and Translational Medicine 2020;5:1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Lyublinskaya O, Antunes F. Measuring intracellular concentration of hydrogen peroxide with the use of genetically encoded H 2 O 2 biosensor HyPer. Redox Biology 2019;24:101200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Forman HJ, Bernardo A, Davies KJA. What is the concentration of hydrogen peroxide in blood and plasma? Archives of Biochemistry and Biophysics 2016;603:48–53. [DOI] [PubMed] [Google Scholar]
- [96].Forman HJ. Use and abuse of exogenous H2O2 in studies of signal transduction. Free Radical Biology and Medicine 2007;42:926–932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Dickinson BC, Chang CJ. Chemistry and biology of reactive oxygen species in signaling or stress responses. Nature Chemical Biology 2011;7:504–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Saeedi Saravi SS, Eroglu E, Waldeck-Weiermair M, et al. Differential endothelial signaling responses elicited by chemogenetic H2O2 synthesis. Redox Biology 2020;36:. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Stein KT, Moon SJ, Sikes HD. Mitochondrial H2O2 Generation Using a Tunable Chemogenetic Tool to Perturb Redox Homeostasis in Human Cells and Induce Cell Death. ACS Synthetic Biology 2018;7:2037–2044. [DOI] [PubMed] [Google Scholar]
- [100].Campinho P, Vilfan A, Vermot J. Blood Flow Forces in Shaping the Vascular System: A Focus on Endothelial Cell Behavior. Frontiers in Physiology 2020; 11:. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Eelen G, de Zeeuw P, Treps L, et al. Endothelial cell metabolism. Physiological Reviews 2018;98:. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Brandes RP. Endothelial dysfunction and hypertension. Hypertension 2014;64:. [DOI] [PubMed] [Google Scholar]
- [103].Higashi Y, Kihara Y, Noma K. Endothelial dysfunction and hypertension in aging. Hypertension Research 2012;35:. [DOI] [PubMed] [Google Scholar]
- [104].Bautista LE. Inflammation, endothelial dysfunction, and the risk of high blood pressure: Epidemiologic and biological evidence. Journal of Human Hypertension 2003;17:. [DOI] [PubMed] [Google Scholar]
- [105].Jin BY, Lin AJ, Golan DE, et al. MARCKS protein mediates hydrogen peroxide regulation of endothelial permeability. Proceedings of the National Academy of Sciences of the United States of America 2012; [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Sawada N, Jiang A, Takizawa F, et al. Endothelial PGC-1α mediates vascular dysfunction in diabetes. Cell Metabolism 2014; [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Rajendran P, Rengarajan T, Thangavel J, et al. The vascular endothelium and human diseases. International Journal of Biological Sciences 2013;9:. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Kolluru GK, Bir SC, Kevil CG. Endothelial dysfunction and diabetes: Effects on angiogenesis, vascular remodeling, and wound healing. International Journal of Vascular Medicine 2012;2012:. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Korhonen EA, Lampinen A, Giri H, et al. Tie1 controls angiopoietin function in vascular remodeling and inflammation. Journal of Clinical Investigation 2016;126:. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Sieve I, Münster-Kühnel AK, Hilfiker-Kleiner D. Regulation and function of endothelial glycocalyx layer in vascular diseases. Vascular Pharmacology 2018;100:. [DOI] [PubMed] [Google Scholar]
- [111].Eroglu E, Saeedi Saravi SS, Sorrentino A, et al. Discordance between eNOS phosphorylation and activation revealed by multispectral imaging and chemogenetic methods. Proceedings of the National Academy of Sciences of the United States of America 2019; [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Kukreja RC, Xi L. eNOS phosphorylation: A pivotal molecular switch in vasodilation and cardioprotection? Journal of Molecular and Cellular Cardiology 2007;42:. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Kalwa H, Sartoretto JL, Martinelli R, et al. Central role for hydrogen peroxide in P2Y1 ADP receptor-mediated cellular responses in vascular endothelium. Proceedings of the National Academy of Sciences of the United States of America 2014; 111:. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Kalwa H, Sartoretto JL, Sartoretto SM, et al. Angiotensin-II and Marcks: A hydrogen peroxide- and Rac1-dependent signaling pathway in vascular endothelium. Journal of Biological Chemistry 2012;287:. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [115].Pimentel D, Haeussler DJ, Matsui R, et al. Regulation of cell physiology and pathology by protein S-glutathionylation: Lessons learned from the cardiovascular system. Antioxidants and Redox Signaling 2012;16:. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [116].Lermant A, Murdoch CE. Cysteine glutathionylation acts as a redox switch in endothelial cells. Antioxidants 2019;8:. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117].Han J, Weisbrod RM, Shao D, et al. The redox mechanism for vascular barrier dysfunction associated with metabolic disorders: Glutathionylation of Rac1 in endothelial cells. Redox Biology 2016;9:. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Rashdan NA, Pattillo CB. Hydrogen peroxide in the ER: A tale of triage. Redox Biology 2020;28:101358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].Lo Conte M, Carroll KS. The redox biochemistry of protein sulfenylation and sulfinylation. Journal of Biological Chemistry 2013;288:. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [120].Sánchez-Gómez FJ, Calvo E, Bretón-Romero R, et al. NOX4-dependent Hydrogen peroxide promotes shear stress-induced SHP2 sulfenylation and eNOS activation. Free Radical Biology and Medicine 2015; [DOI] [PubMed] [Google Scholar]
- [121].Cremers CM, Jakob U. Oxidant sensing by reversible disulfide bond formation. Journal of Biological Chemistry 2013;288:. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [122].Sullivan DC, Huminiecki L, Moore JW, et al. EndoPDI, a Novel Protein-disulfide Isomerase-like Protein That Is Preferentially Expressed in Endothelial Cells Acts as a Stress Survival Factor. Journal of Biological Chemistry 2003;278:. [DOI] [PubMed] [Google Scholar]
- [123].Park Y, Capobianco S, Gao X, et al. Role of EDHF in type 2 diabetes-induced endothelial dysfunction. American Journal of Physiology - Heart and Circulatory Physiology 2008;295:. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [124].Nedeljkovic ZS, Gokce N, Loscalzo J. Mechanisms of oxidative stress and vascular dysfunction. Postgraduate Medical Journal 2003;79:. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [125].Bertero E, Maack C. Metabolic remodelling in heart failure. Nature Reviews Cardiology 2018;15:. [DOI] [PubMed] [Google Scholar]
- [126].Sorrentino A, Steinhorn B, Troncone L, et al. Reversal of heart failure in a chemogenetic model of persistent cardiac redox stress. American Journal of Physiology Heart and Circulatory Physiology 2019;317:H617–H626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [127].Kerkela R, Ulvila J, Magga J. Natriuretic peptides in the regulation of cardiovascular physiology and metabolic events. Journal of the American Heart Association 2015;4:. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [128].Singh A, Kukreti R, Saso L, et al. Oxidative stress: A key modulator in neurodegenerative diseases. Molecules 2019;24:1–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [129].Shih AY, Johnson DA, Wong G, et al. Coordinate regulation of glutathione biosynthesis and release by Nrf2-expressing glia potently protects neurons from oxidative stress. Journal of Neuroscience 2003;23:3394–3406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [130].Sigfridsson E, Marangoni M, Johnson JA, et al. Astrocyte-specific overexpression of Nrf2 protects against optic tract damage and behavioural alterations in a mouse model of cerebral hypoperfusion. Scientific Reports 2018;8:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [131].Satoh T, Okamoto SI, Cui J, et al. Activation of the Keap1/Nrf2 pathway for neuroprotection by electrophillic phase II inducers. Proceedings of the National Academy of Sciences of the United States of America 2006;103:. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [132].Haskew-Layton RE, Payappilly JB, Smirnova NA, et al. Controlled enzymatic production of astrocytic hydrogen peroxide protects neurons from oxidative stress via an Nrf2-independent pathway. Proceedings of the National Academy of Sciences of the United States of America 2010;107:17385–17390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [133].De Janeiro R, Fróes MM, Correia AHP, et al. Gap-junctional coupling between neurons and astrocytes in. Proceedings of the National Academy of Sciences of the United States of America 1999;96:7541–7546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [134].Rouach N, Glowinski J, Giaume C. Activity-dependent Neuronal Control of gap-junctional Communication in astrocytes. Journal of Cell Biology 2000;149:1513–1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [135].Zhou D, Collins CA, Wu P, et al. Protein tyrosine phosphatase SHP-1 positively regulates TLR-induced IL-12p40 production in macrophages through inhibition of phosphatidylinositol 3-kinase. Journal of Leukocyte Biology 2010;87:845–855. [DOI] [PubMed] [Google Scholar]
- [136].Bell KF, Al-Mubarak B, Fowler JH, et al. Mild oxidative stress activates Nrf2 in astrocytes, which contributes to neuroprotective ischemic preconditioning. Proceedings of the National Academy of Sciences of the United States of America 2011;108:4–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [137].Haskew-Layton RE, Ma TC, Ratan RR. Reply to Bell et al. : Nrf2-dependent and - independent mechanisms of astrocytic neuroprotection. Proceedings of the National Academy of Sciences of the United States of America 2011;108:1–2. [Google Scholar]
- [138].Saxena S, Caroni P. Selective Neuronal Vulnerability in Neurodegenerative Diseases: From Stressor Thresholds to Degeneration. Neuron 2011;71:35–48. [DOI] [PubMed] [Google Scholar]
- [139].Cobley JN, Fiorello ML, Bailey DM. 13 Reasons Why the Brain Is Susceptible To Oxidative Stress. Redox Biology 2018;15:490–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [140].Lenzen S Oxidative stress: The vulnerable β-cell. Biochemical Society Transactions 2008;36:. [DOI] [PubMed] [Google Scholar]
- [141].Lenzen S Chemistry and biology of reactive species with special reference to the antioxidative defence status in pancreatic β-cells. Biochimica et Biophysica Acta - General Subjects 2017;1861:. [DOI] [PubMed] [Google Scholar]
- [142].Lenzen S The pancreatic beta cell: an intricate relation between anatomical structure, the signalling mechanism of glucose-induced insulin secretion, the low antioxidative defence, the high vulnerability and sensitivity to diabetic stress. ChemTexts 2021;7:. [Google Scholar]
- [143].Lenzen S, Drinkgern J, Tiedge M. Low antioxidant enzyme gene expression in pancreatic islets compared with various other mouse tissues. Free Radical Biology and Medicine 1996;20:. [DOI] [PubMed] [Google Scholar]
- [144].Grankvist K, Marklund SL, Taljedal IB. CuZn-superoxide dismutase, Mn-superoxide dismutase, catalase and glutathione peroxidase in pancreatic islets and other tissues in the mouse. Biochemical Journal 1981; 199:. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [145].Mehmeti I, Lortz S, Avezov E, et al. ER-resident antioxidative GPx7 and GPx8 enzyme isoforms protect insulin-secreting INS-1E β-cells against lipotoxicity by improving the ER antioxidative capacity. Free Radical Biology and Medicine 2017;112:. [DOI] [PubMed] [Google Scholar]
- [146].Gurgul E, Lortz S, Tiedge M, et al. Mitochondrial catalase overexpression protects insulin-producing cells against toxicity of reactive oxygen species and proinflammatory cytokines. Diabetes 2004;53:. [DOI] [PubMed] [Google Scholar]
- [147].Gurgul-Convey E, Mehmeti I, Lortz S, et al. Cytokine toxicity in insulin-producing cells is mediated by nitro-oxidative stress-induced hydroxyl radical formation in mitochondria. Journal of Molecular Medicine 2011;89:. [DOI] [PubMed] [Google Scholar]
- [148].Krüger C, Jörns A, Kaynert J, et al. The importance of aquaporin-8 for cytokine-mediated toxicity in rat insulin-producing cells. Free Radical Biology and Medicine 2021;174:. [DOI] [PubMed] [Google Scholar]
- [149].Elsner M, Gehrmann W, Lenzen S. Peroxisome-generated hydrogen peroxide as important mediator of lipotoxicity in insulin-producing cells. Diabetes 2011;60:. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [150].Jörns A, Arndt T, Zu Vilsendorf AM, et al. Islet infiltration, cytokine expression and beta cell death in the NOD mouse, BB rat, Komeda rat, LEW.1AR1-iddm rat and humans with type 1 diabetes. Diabetologia 2014;57:. [DOI] [PubMed] [Google Scholar]
- [151].Cnop M, Welsh N, Jonas JC, et al. Mechanisms of pancreatic β-cell death in type 1 and type 2 diabetes: Many differences, few similarities. Diabetes 2005;54:. [DOI] [PubMed] [Google Scholar]
- [152].Montiel V, Bella R, Michel LYM, et al. Inhibition of aquaporin-1 prevents myocardial remodeling by blocking the transmembrane transport of hydrogen peroxide. Science Translational Medicine 2020;12:. [DOI] [PubMed] [Google Scholar]
- [153].Rodrigues C, Pimpão C, Mósca AF, et al. Human aquaporin-5 facilitates hydrogen peroxide permeation affecting adaption to oxidative stress and cancer cell migration. Cancers 2019;11:. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [154].Bienert GP, Møller ALB, Kristiansen KA, et al. Specific aquaporins facilitate the diffusion of hydrogen peroxide across membranes. Journal of Biological Chemistry 2007;282:. [DOI] [PubMed] [Google Scholar]
- [155].Bestetti S, Galli M, Sorrentino I, et al. Human aquaporin-11 guarantees efficient transport of H2O2 across the endoplasmic reticulum membrane. Redox Biology 2020;28:. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [156].Bienert GP, Chaumont F. Aquaporin-facilitated transmembrane diffusion of hydrogen peroxide. Biochimica et Biophysica Acta - General Subjects 2014;1840:. [DOI] [PubMed] [Google Scholar]
- [157].Watanabe S, Moniaga CS, Nielsen S, et al. Aquaporin-9 facilitates membrane transport of hydrogen peroxide in mammalian cells. Biochemical and Biophysical Research Communications 2016;471:. [DOI] [PubMed] [Google Scholar]
- [158].Clément MV, Ponton A, Pervaiz S. Apoptosis induced by hydrogen peroxide is mediated by decreased superoxide anion concentration and reduction of intracellular milieu. FEBS Letters 1998;440:. [DOI] [PubMed] [Google Scholar]
- [159].Singh M, Sharma H, Singh N. Hydrogen peroxide induces apoptosis in HeLa cells through mitochondrial pathway. Mitochondrion 2007;7:. [DOI] [PubMed] [Google Scholar]
- [160].Wondrak GT. Redox-directed cancer therapeutics: Molecular mechanisms and opportunities. Antioxidants and Redox Signaling 2009;11:3013–3069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [161].Nogueira V, Hay N. Molecular pathways: Reactive oxygen species homeostasis in cancer cells and implications for cancer therapy. Clinical Cancer Research 2013;19:4309–4314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [162].Stegman LD, Zheng H, Neal ER, et al. Induction of cytotoxic oxidative stress by D-alanine in brain tumor cells expressing Rhodotorula gracilis D-amino acid oxidase: A cancer gene therapy strategy. Human Gene Therapy 1998;9:185–193. [DOI] [PubMed] [Google Scholar]
- [163].Dreher D, Junod AF. Role of Oxygen Free Radicals in Cancer Development. European Journal of Cancer 1996;32:30–38. [DOI] [PubMed] [Google Scholar]
- [164].Sosa V, Moliné T, Somoza R, et al. Oxidative stress and cancer: An overview. Ageing Research Reviews 2013;12:376–390. [DOI] [PubMed] [Google Scholar]
- [165].Helfinger V, Schröder K. Redox control in cancer development and progression. Molecular Aspects of Medicine 2018;63:88–98. [DOI] [PubMed] [Google Scholar]
- [166].Fang J, Sawa T, Akaike T, et al. Tumor-targeted delivery of polyethylene glycol-conjugated D-amino acid oxidase for antitumor therapy via enzymatic generation of hydrogen peroxide. Cancer Research 2002;62:3138–3143. [PubMed] [Google Scholar]
- [167].Fang J, Sawa T, Akaike T, et al. Enhancement of chemotherapeutic response of tumor cells by a heme oxygenase inhibitor, pegylated zinc protoporphyrin. International Journal of Cancer 2004;109:1–8. [DOI] [PubMed] [Google Scholar]
- [168].Rosini E, Pollegioni L, Ghisla S, et al. Optimization of d-amino acid oxidase for low substrate concentrations - Towards a cancer enzyme therapy. FEBS Journal 2009;276:4921–4932. [DOI] [PubMed] [Google Scholar]
- [169].Bava A, Gornati R, Cappellini F, et al. D-amino acid oxidase-nanoparticle system: A potential novel approach for cancer enzymatic therapy. Nanomedicine 2013;8:1797–1806. [DOI] [PubMed] [Google Scholar]
- [170].Balzaretti R, Meder F, Monopoli MP, et al. Synthesis, characterization and programmable toxicity of iron oxide nanoparticles conjugated with D-amino acid oxidase. RSC Advances 2017;7:1439–1442. [Google Scholar]
- [171].Cappellini F, Recordati C, De Maglie M, et al. New synthesis and biodistribution of the D-amino acid oxidase-magnetic nanoparticle system. Future Science OA 2015;1:. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [172].Divakaran SA, Sreekanth KM, Rao KV., et al. D-Aminoacid Oxidase-Fe<sub>2</sub>0<sub>3</sub> Nanoparticle Complex Mediated Antitumor Activity in Swiss Albino Mice. Journal of Cancer Therapy 2011;02:666–674. [Google Scholar]
- [173].Fuentes-Baile M, Bello-Gil D, Pérez-Valenciano E, et al. CLyta-DAAO, free and immobilized in magnetic nanoparticles, induces cell death in human cancer cells. Biomolecules 2020;10:. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [174].El Sayed SM, Abou El-Magd RM, Shishido Y, et al. D-amino acid oxidase gene therapy sensitizes glioma cells to the antiglycolytic effect of 3-bromopyruvate. Cancer Gene Therapy 2012;19:1–18. [DOI] [PubMed] [Google Scholar]
- [175].Raj L, Ide T, Gurkar AU, et al. Selective killing of cancer cells by a small molecule targeting the stress response to ROS. Nature 2011;475:231–234. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- [176].Silic-Benussi M, Scattolin G, Cavallari I, et al. Selective killing of human T-ALL cells: an integrated approach targeting redox homeostasis and the OMA1/OPA1 axis. Cell Death and Disease 2018;9:. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [177].Wang H, Feng Z, Wang Y, et al. Integrating Enzymatic Self-Assembly and Mitochondria Targeting for Selectively Killing Cancer Cells without Acquired Drug Resistance. Journal of the American Chemical Society 2016;138:16046–16055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [178].Vyas S, Zaganjor E, Haigis MC. Leading Edge Review Mitochondria and Cancer. Cell 2016;166:555–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [179].Sporn MB. War on cancer 25. The Lancet 1996;347:1377–1381. [DOI] [PubMed] [Google Scholar]
- [180].Bröer S Amino acid transport across mammalian intestinal and renal epithelia. Physiological Reviews 2008;88:249–286. [DOI] [PubMed] [Google Scholar]
- [181].Kaur J, Olkhova E, Malviya VN, et al. A L-lysine transporter of high stereoselectivity of the amino acid-polyamine-organocation (apc) superfamily production, functional characterization, and structure modeling. Journal of Biological Chemistry 2014;289:1377–1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [182].Kandasamy P, Gyimesi G, Kanai Y, et al. Amino acid transporters revisited: New views in health and disease. Trends in Biochemical Sciences 2018;43:752–789. [DOI] [PubMed] [Google Scholar]
- [183].Gauthier-Coles G, Vennitti J, Zhang Z, et al. Quantitative modelling of amino acid transport and homeostasis in mammalian cells. Nature Communications 2021;12:1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [184].Strader CD, Gaffney T, Sugg EE, et al. Allele-specific activation of genetically engineered receptors. Journal of Biological Chemistry 1991;266:. [PubMed] [Google Scholar]
- [185].Pollegioni L, Molla G, Campaner S, et al. Cloning, sequencing and expression in E. coli of a D-amino acid oxidase cDNA from Rhodotorula gracilis active on cephalosporin C. Journal of Biotechnology 1997;58:. [DOI] [PubMed] [Google Scholar]
- [186].Alexander GM, Rogan SC, Abbas AI, et al. Remote control of neuronal activity in transgenic mice expressing evolved G protein-coupled receptors. Neuron 2009;63:. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [187].Negelein E, Broehmel H. Protein der d-Aminosaeureoxidase. Biochem Zeitschrift 1939;239–255. [Google Scholar]
- [188].Kubo H, Yamano T, Iwatsubo M, et al. Interactions between the enzyme and the coenzyme in glutamic acid dehydrogenase and amino acid oxidase systems. Bull Soc Chim Biol 1958;40:431–447. [PubMed] [Google Scholar]
- [189].Dixon M, Kleppe K. d-amino acid oxidase I. Dissociation and recombination of the holoenzyme. BBA Section Nucleic Acids And Protein Synthesis 1965;96:357–367. [DOI] [PubMed] [Google Scholar]
- [190].Fukui K, Watanabe F, Shibata T, et al. Molecular Cloning and Sequence Analysis of cDNAs Encoding Porcine Kidney D-Amino Acid Oxidase. Biochemistry 1987;26:. [DOI] [PubMed] [Google Scholar]
- [191].Sharifi-Rad M, Anil Kumar NV, Zucca P, et al. Lifestyle, Oxidative Stress, and Antioxidants: Back and Forth in the Pathophysiology of Chronic Diseases. Frontiers in Physiology 2020;11:1–21. [DOI] [PMC free article] [PubMed] [Google Scholar]