The musicatk package empowers researchers to characterize mutational signatures and tumor heterogeneity with a comprehensive set of preprocessing utilities, discovery and prediction tools, and multiple functions for downstream analysis and visualization.
Abstract
Mutational signatures are patterns of somatic alterations in the genome caused by carcinogenic exposures or aberrant cellular processes. To provide a comprehensive workflow for preprocessing, analysis, and visualization of mutational signatures, we created the Mutational Signature Comprehensive Analysis Toolkit (musicatk) package. musicatk enables users to select different schemas for counting mutation types and to easily combine count tables from different schemas. Multiple distinct methods are available to deconvolute signatures and exposures or to predict exposures in individual samples given a pre-existing set of signatures. Additional exploratory features include the ability to compare signatures to the Catalogue Of Somatic Mutations In Cancer (COSMIC) database, embed tumors in two dimensions with uniform manifold approximation and projection, cluster tumors into subgroups based on exposure frequencies, identify differentially active exposures between tumor subgroups, and plot exposure distributions across user-defined annotations such as tumor type. Overall, musicatk will enable users to gain novel insights into the patterns of mutational signatures observed in cancer cohorts.
Significance:
The musicatk package empowers researchers to characterize mutational signatures and tumor heterogeneity with a comprehensive set of preprocessing utilities, discovery and prediction tools, and multiple functions for downstream analysis and visualization.
Introduction
Somatic mutations to the genome can be caused by exposure to environmental carcinogens or aberrant cellular processes (1, 2). A “mutational signature” is a specific pattern of mutation types caused by a particular mutational process. The set of mutations observed in a single tumor genome can be the result of multiple mutational processes active during the course of tumor development. Therefore, deconvolution is needed to determine which signatures are present across a group of tumor genomes as well as the level of each signature in each individual tumor. Recently, the Pan-Cancer Analysis of Whole Genomes (PCAWG) Consortium characterized a large cohort of whole-exome and whole-genome samples with single-base-substitution (SBS), doublet-base-substitution (DBS), and small insertion-and-deletion (INDEL) mutational schemas using NMF-based methods (3). Although some software packages have been previously developed to perform mutational signature inference, they do not quantify the latest set of mutation schema from Catalogue Of Somatic Mutations In Cancer (COSMIC; ref. 3). These packages also lack functionality for comprehensive exploratory analysis or have limited functionality for predicting exposures to predefined signatures in new samples (3–9). The musicatk package provides functionality to streamline the steps of mutational inference and has several additional features to enhance exploratory analysis beyond what is available in other packages (Supplementary Fig. S1). We provide an overview of this functionality and present an exploratory analysis of tumors from The Cancer Genome Atlas (TCGA).
Materials and Methods
Importing and processing of mutations
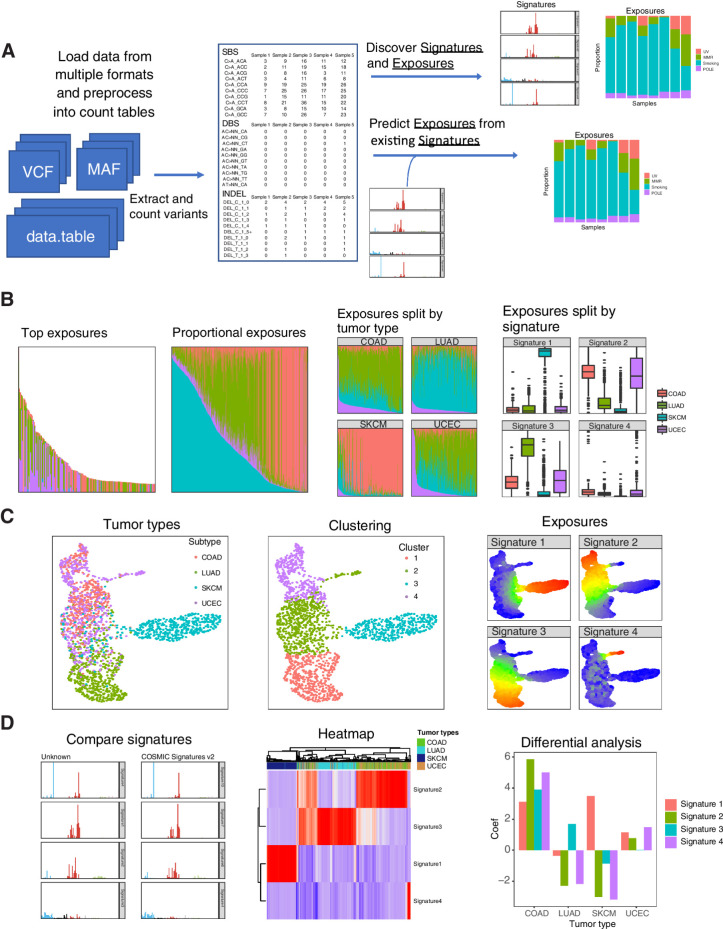
The major steps of mutational signature inference are (i) importing the variants, (ii) building and combining count tables based on different types of mutation schema, (iii) performing discovery and/or prediction of signatures and exposures, and (iv) using visualization for exploratory analysis of the results (Fig. 1A). For the first step, the musicatk package has functions to read mutations from various input formats. Mutation annotation formats (MAF) are read from files or from the R object MAF created by the maftools package. Variant call formats (VCF) can be read from files or R classes defined in the (VariantAnnotation, RRID:SCR_000074) package. In addition, variant information stored in a data.frame or data.table can also be used as input. To streamline the processing of variants, mutation profiles from multiple tumors in different formats can be automatically read and combined into the musica object (Supplementary Fig. S2).
Figure 1.
Overview of workflow for mutational signature discovery/prediction, visualization, and analysis. A, Workflow allows for loading and combining data from multiple sources, de novo discovery of signature and exposures, and prediction of exposures from existing signatures. B, The same sample exposures are plotted (subset to top samples), proportional exposures (signature exposures sum to 1), split up by tumor type, and split up by signature. C, UMAP. Tumors in a UMAP can be colored by annotations such as tumor type, clustering by methods such as K-means, or levels of exposure for each signature. D, Downstream analysis tools include automated comparison to COSMIC signatures, heatmaps, which can be used to show the relative levels of signature exposures in samples along with sample annotations, and differential analysis of exposures between groups of tumors.
We also include functions to automatically parse different types of mutation motifs from each tumor genome and create count tables that are used in downstream analysis. Tables that can be calculated by musicatk include SBS into 96 motifs, SBS with transcription strand orientation into 192 motifs, SBS with replication strand orientation into 192 motifs, DBS into 78 motifs, and INDELs into 83 motifs. Custom mutation count tables can also be defined by the user and added to the object. Importantly, multiple tables can be concatenated to create composite mutation schema tables. For example, users can combine the SBS-96 motif, DBS-78 motif, and INDEL-83 tables and perform downstream analyses, similar to process that was used to create the PCAWG composite signatures (3).
Discovery of mutational signatures
Deconvolution is the process of decomposing a matrix of mutation counts per tumor into a matrix of signatures and another matrix of exposures. The Signature matrix contains the probability of each mutation motif in each signature and the Exposure matrix contains the estimated level of each exposure in each tumor sample. musicatk supports both latent Dirichlet allocation (LDA; ref. 10) from the topicmodels package and nonnegative matrix factorization (NMF; ref. 11) from the NMF package. We observed similar accuracy and faster run times with LDA and therefore include LDA as the default discovery tool (Supplementary Fig. S3). Both algorithms can be applied to any count table. One challenging aspect of mutational signature discovery is determining the appropriate number of signatures (i.e., the value of K). To facilitate the comparison of models with different choices of K, musicatk provides a wrapper that allows users to apply deconvolution algorithms with different values of K and then compare the results with metrics such as reconstruction error (NMF and LDA), log-likelihood (LDA), or perplexity (LDA).
Prediction of mutational signatures
Prediction of exposures for existing signatures can be performed on any count table given that the mutation motif schema is the same. We include wrappers for tools such as deconstructSigs and decompTumor2Sig (4, 8). We also implement a Bayesian algorithm based on LDA where exposures are estimated using a fixed set of signatures (Supplementary File S1). To allow for prediction using previously defined signatures from catalogue of somatic mutations in cancer (COSMIC, RRID:SCR_002260; ref. 3), we include objects for COSMIC V2 signatures (SBS-96 motif schema) and COSMIC V3 signatures (SBS-96, DBS-78, and INDEL-83 motif schemas). One challenge in the prediction of existing signatures in new tumors is that not all signatures will be present in the new dataset. Including nonactive signatures may cause additional noise in the estimates for signatures that are present within the dataset. Since signatures that are present in moderate levels across many tumors or highly present in a small number of tumors are more likely to be active in the dataset (3), we implemented a two-step procedure to choose the subset of active signatures within a dataset. In the first step, exposures are estimated using all signatures and active signatures are chosen if they pass a threshold in a minimum number of samples (e.g., have an exposure of at least 0.1 in at least 30% of samples or an exposure of 0.7 in at least two samples). In the second-step, signatures are estimated using only the active signatures. Users can perform the 2-step prediction within subgroups of tumors supplying a categorical annotation such as tumor type.
Visualization
Visualization of mutational signatures and tumor exposures is important for exploration of mutational processes that are active in a cohort of tumors. Signature barplots can be used to show the probability of motifs in each signature and exposure barplots can be used to display the composition of exposures in each sample. Exposure barplots can be sorted by overall mutation count, by one or multiple exposures, or by sample name (Fig. 1B). They can be subsetted to show the samples with the highest total mutation counts or exposure levels. Exposures barplots can further be grouped by a sample annotation such as tumor type or by signature. The distributions of exposures can be displayed with box and/or violin plots and grouped by sample annotations. To view relationships between tumors in two dimensions, the uniform manifold approximation and projections (UMAP; ref. 12) algorithm can be used with normalized signature exposures (Fig. 1C). The UMAP can be colored by annotations (e.g., tumor type) or the levels of each exposure.
Downstream analyses
Functionality is provided for correlating sets of discovered signatures to other sets of previously defined signatures. For example, discovered signatures can be compared with COSMIC V2 and V3 signatures (Fig. 1D). Clustering of tumors into groups can be performed by applying K-means to the estimated exposure levels. Metrics such as silhouette width and total within-cluster sum of squares (wss) generated from the factoextra package can be used to identify the optimal number of clusters and cluster labels can be plotted on the UMAP (Fig. 1D). The (ComplexHeatmap, RRID:SCR_017270) package is used to plot heatmaps showing the relative levels of exposures in samples along with annotations (Fig. 1D). Differential analysis can be used to identify exposures that are significantly higher or lower between groups of tumors. Differential methods include Wilcoxon rank-sum test for two-group comparisons as well as Kruskal–Wallis and negative binomial generalized linear models (GLM) for multigroup comparisons (Fig. 1D).
Prediction of annotation labels on unknown samples
The musicatk package can predict class labels using the exposure levels of training and test cohorts. For example, tumors from TCGA can be used to predict the tumor type of samples with unknown origin. After predicting exposures using the same reference signatures in both the training and test sets (e.g., COSMIC V2), the musica objects can be combined to perform downstream analyses such as generating a UMAP for all samples. Class labels for test samples can be predicted using the median Euclidean distance to samples in each class in the training cohort. For each test sample, the class with the lowest median distance will be the predicted label.
Data availability statement: The TCGA and MSK-IMPACT data analyzed in this study were obtained from (TCGAbiolinks, RRID:SCR_017683) and (cBioPortal, RRID:SCR_014555) respectively.
The package developed for this study (musicatk, RRID:SCR_021726) is available on (Bioconductor, RRID:SCR_006442) and on Github at https://github.com/campbio/musicatk/. The code used for the generation of the analysis in this manuscript and tutorial videos are available on Github at https://github.com/campbio/Manuscripts/tree/master/musicatk/8_30_21_manuscript including a lock file that allows for exact version matching of all used packages. Additional documentation can be found at https://camplab.net/musicatk.
Results
To demonstrate how musikatk can be used to characterize and explore heterogeneity in the signatures across tumor types, we applied the LDA-based prediction method to predict COSMIC v3 SBS signatures in a Pan-Cancer dataset from TCGA. Thirty-nine of the 65 signatures were found to be active in at least one tumor type. A UMAP plot was generated to explore the patterns of signatures across tumors (Fig. 2A). Some signatures were present in nearly half of samples, some in a few tumor types, some in single tumor types, and some in subsets of multiple tumor types (Fig. 2; Supplementary Fig. S4). In addition, we generated UMAP plots for all TCGA based on DBS or IND schema from COSMIC V3 and observed similar heterogeneity, albeit to a less extent compared with the SBS signatures (Supplementary Figs. S5 and S6).
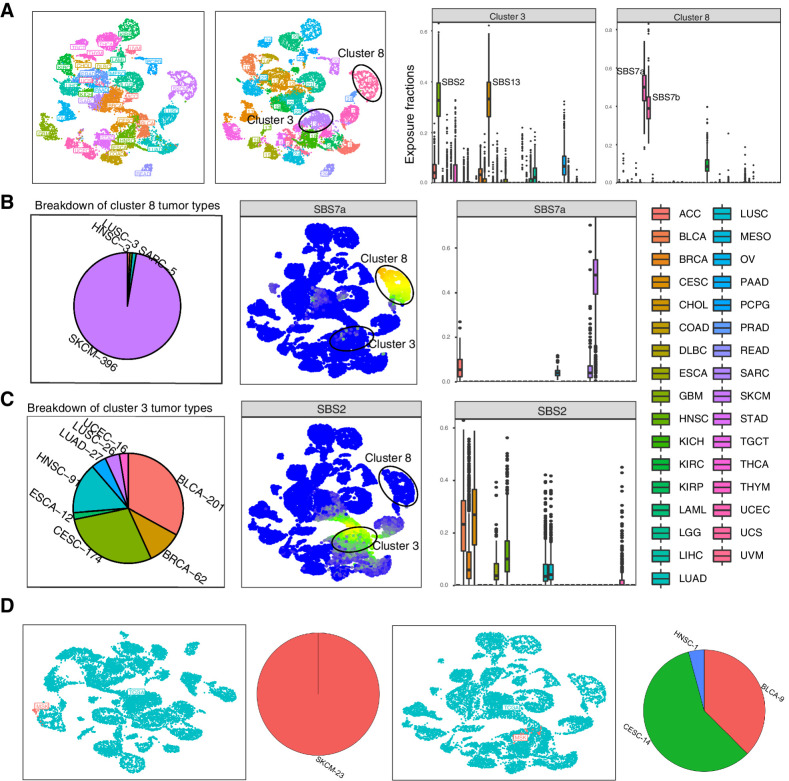
Figure 2.
Examining similarities and differences between tumor types with musicatk. Clustering of TCGA samples and inference of tumor type labels in MSK samples. A, Left to right, UMAP of TCGA samples colored by tumor types, UMAP of TCGA samples colored by cluster label, proportional exposure to COSMIC v3 signatures within cluster #3, proportional exposure to COSMIC v3 signatures within cluster #8. B, Left to right, breakdown of tumor types within cluster cluster #8, UMAP colored by exposure to SBS7a (UV), exposure of each tumor type to SBS7a. C, Left to right, breakdown of tumor types within cluster cluster #3, UMAP colored by exposure to SBS2 (APOBEC), and exposure of each tumor type to SBS2. D, Left to right, UMAP colored by cohort (TCGA, UMAP) with MSK SKCM samples, percentage of tumor types inferred for MSK SKCM samples using exposure levels from TCGA samples, UMAP colored by cohort (TCGA, UMAP) with MSK BLCA samples, percentage of tumor types inferred for MSK BLCA samples using exposure levels from TCGA samples.
Thirty clusters of tumors were identified using hierarchical K-means on the SBS exposures (Supplementary Figs. S7A and S7B). Clusters 3 and 8 were predominantly defined by a small number of signatures (Fig. 2A). Cluster 8 was dominated by exposure of the two UV signatures, SBS7a and SBS7b, almost entirely represented by skin cancer melanoma (SKCM) samples, with the exception of lung squamous cell carcinoma (LUSC) and HNSC tumors, which may represent metastatic samples from the skin to other organs (Fig. 2B; ref. 13). Cluster 3 was defined by high levels of the two APOBEC-related signatures, SBS2 and SBS13 and containing predominantly a mix of CESC (cervical), BRCA (breast), BLCA (bladder), and HNSC (head and neck; Fig. 2C).
To demonstrate the ability to map annotations from one cohort to another, we predicted exposures of SKCM and BLCA tumors profiled with the MSK-IMPACT targeted sequencing panel and then mapped these samples to the tumor types in TCGA. 100% of the MSK SKCM samples were predicted to be SKCM from TCGA (Fig. 2D). In contrast, the MSK BLCA samples mapped to several TCGA tumor types (58% CESC, 37.5% BCLA, 4% HNSC). 67% of MSK BLCA tumors assigned to CESC or HNSC had high levels of the APOBEC signatures (SBS2, SBS13) and mapped to cluster 3 in TCGA. These results show that MSK BLCA tumors exhibit similar patterns of heterogeneity as the TCGA BLCA tumors and demonstrate how the musicatk package can be used to compare sets of tumors between cohorts.
Discussion
While processing somatic variants and performing deconvolution is a major part of mutational signature analysis, the flexible and simple framework of the musicatk package can empower researchers to discover additional heterogeneity in mutational patterns with extra tools for visualization, clustering, and statistical analysis. Overall, the musicatk package provides a comprehensive set of preprocessing utilities, access to several discovery and prediction tools, and functions for downstream analysis allowing deeper characterization mutational signatures across cohorts of tumors.
Authors' Disclosures
J.D. Campbell reports personal fees from Numera Bioscience outside the submitted work. No disclosures were reported by the other authors.
Supplementary Material
Supplementary Figures and Legends in the order cited in the manuscript
Plate diagram and derivation of bayesian prediction algorithm
Acknowledgments
This work was funded by the NCI's Informatics Technology for Cancer Research (ITCR) R21 CA226188 (to J.D. Campbell and M. Yajima) and by the National Institute of General Medical Sciences of the NIH T32GM100842 (to Z. Khurshid, N. Sahelijo, and T. Tong).
Footnotes
Note: Supplementary data for this article are available at Cancer Research Online (http://cancerres.aacrjournals.org/).
Authors' Contributions
A. Chevalier: Conceptualization, data curation, software, formal analysis, validation, investigation, visualization, methodology, writing–original draft. S. Yang: Conceptualization, software, formal analysis, supervision, funding acquisition, methodology, project administration, writing–review and editing. Z. Khurshid: Conceptualization, software, formal analysis, methodology. N. Sahelijo: Conceptualization, software, formal analysis, methodology. T. Tong: Conceptualization, software, formal analysis, methodology. J.H. Huggins: Conceptualization, formal analysis. M. Yajima: Conceptualization, formal analysis, methodology. J.D. Campbell: Conceptualization, software, supervision, funding acquisition, project administration, writing–review and editing.
References
- 1. Nik-Zainal S, Davies H, Staaf J, Ramakrishna M, Glodzik D, Zou X, et al. Landscape of somatic mutations in 560 breast cancer whole-genome sequences. Nature 2016;534:47–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Alexandrov LB, Nik-Zainal S, Wedge DC, Aparicio SAJR, Behjati S, Biankin AV, et al. Signatures of mutational processes in human cancer. Nature 2013;500:415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Alexandrov LB, Kim J, Haradhvala NJ, Huang MN, Tian Ng AW, Wu Y, et al. The repertoire of mutational signatures in human cancer. Nature 2020;578:94–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Rosenthal R, McGranahan N, Herrero J, Taylor BS, Swanton C. deconstructSigs: delineating mutational processes in single tumors distinguishes DNA repair deficiencies and patterns of carcinoma evolution. Genome Biol 2016;17:31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Teresa Przytycka Research Page. Available at: https://www.ncbi.nlm.nih.gov/CBBresearch/Przytycka/index.cgi#signatureestimation. Accessed 16 November 2020.
- 6. Blokzijl F, Janssen R, van Boxtel R, Cuppen E. MutationalPatterns: Comprehensive genome-wide analysis of mutational processes. Genome Med 2018;10:33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. YAPSA: Yet Another Package for Signature Analysis version 1.16.0 from Bioconductor. Available at: https://rdrr.io/bioc/YAPSA/.Accessed: 16th November 2020.
- 8. Krüger S, Piro RM. DecompTumor2Sig: identification of mutational signatures active in individual tumors. BMC Bioinformatics 2019;20:152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gori K, Baez-Ortega A. sigfit: flexible Bayesian inference of mutational signatures. bioRxiv 2018;372896.doi: 10.1101/372896 bioRxiv bioRxiv. [Google Scholar]
- 10. Blei DM, Ng AY, Jordan MI. Latent dirichlet allocation. J Mach Learn Res 2003;3:993–1022. [Google Scholar]
- 11. Lee DD, Seung HS. Learning the parts of objects by non-negative matrix factorization. Nature 1999;401:788–91. [DOI] [PubMed] [Google Scholar]
- 12. McInnes L, Healy J, Melville J. UMAP: Uniform manifold approximation and projection for dimension reduction. arXiv 2018. [Google Scholar]
- 13. Campbell JD, Alexandrov A, Kim J, Wala J, Berger AH, Pedamallu CS, et al. Distinct patterns of somatic genome alterations in lung adenocarcinomas and squamous cell carcinomas. Nat Genet 2016;48:607–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures and Legends in the order cited in the manuscript
Plate diagram and derivation of bayesian prediction algorithm