Abstract
Our understanding of the composition and functions of splenic stromal cells remains incomplete. Here, based on analysis of over 20,000 single cell transcriptomes of splenic fibroblasts, we characterized the phenotypic and functional heterogeneity of these cells in healthy state and during virus infection. We describe eleven transcriptionally distinct fibroblastic cell clusters, reassuring known subsets and revealing yet unascertained heterogeneity amongst fibroblasts occupying diverse splenic niches. We further identify striking differences in innate immune signatures of distinct stromal compartments in vivo. Compared to other fibroblasts and to endothelial cells, Ly6C+ fibroblasts of the red pulp were selectively endowed with enhanced interferon-stimulated gene expression in homeostasis, upon systemic interferon stimulation and during virus infection in vivo. Collectively, we provide an updated map of fibroblastic cell diversity in the spleen that suggests a specialized innate immune function for splenic red pulp fibroblasts.
Subject terms: Spleen, Immunogenetics, Innate immunity
Joern Pezoldt et al. analyze mouse spleen fibroblasts using single cell RNA sequencing, revealing 11 distinct clusters of fibroblastic cells or subtypes. Their results collectively provide further insight into the transcriptional identities of splenic fibroblasts and innate immune signatures of distinct stromal compartments.
Introduction
The spleen is the largest secondary lymphoid organ (SLO) in the body that plays a non-redundant role in host defence. It emerged alongside with adaptive immunity at the dawn of jawed vertebrate evolution and is the only described bona fide SLO in all vertebrate classes except mammals that additionally have lymph nodes (LNs). The spleen is distinguished into two anatomically and functionally distinct compartments: the red pulp, which is filled with red blood cells and macrophages; and the white pulp, which is comprised of lymphocyte aggregates organized into a T-cell zone and B-cell follicles. The white and red pulp areas are bridged by the marginal zone, which harbours specific subsets of B-cells and macrophages1,2.
Fibroblastic cells (hereafter referred to as FC) are critical structural and functional components of all SLOs3. Splenic FC are best studied in the white pulp, where they have long been known to form three functionally specialized subsets: (i) CCL21-expressing T-zone reticular cells (TRC); (ii) CXCL13- and CR1/2-expressing follicular dendritic cells (FDC), located in the B-cell follicle as well as (iii) CXCL13-, MAdCAM1- and RANKL-expressing marginal reticular cells (MRC)4. Our current understanding of splenic FC diversity is based on the analysis of single-cell transcriptomes of 6227 ICAM1+/PDGFRβ+ FC from the spleen of naïve mice originally published by Cheng et al.5, and further explored by others6,7. In addition to corroborating the distinction of white pulp FC into TRC, FDC and MRC, analysis of the above-mentioned dataset distinguished a population of red pulp FC6; as well as provided transcriptional evidence for the existence of pericyte-like cells5 and adventitial cells7. Critically, the field still lacks a characterization of splenic FC at single-cell level with a resolution matching the detail with which we currently view the heterogeneity of FC in the LNs, where several FC subsets in addition to FRC, FDC and MRC have been resolved8,9.
Type I interferons (IFNs) are crucial antiviral cytokines produced by a broad range of, if not by all, cells upon primary contact with viruses. They signal through a heterodimeric transmembrane receptor, IFNα receptor (IFNAR), which is composed of IFNAR1 and IFNAR2 subunits. Binding of type I IFN to IFNAR triggers the Janus kinase/signal transducers and activators of transcription (JAK/STAT) pathway of signal transduction, which ultimately leads to the assembly of a trimeric STAT1/STAT2/IRF9 (ISGF3) transcription factor complex, which enhances the expression of interferon-stimulated genes (ISGs) with distinctive antiviral activities10,11. In addition to being produced upon virus infection, type I IFNs are also well recognized for their homeostatic (i.e. tonic) presence and function12. Tonic stimulation of cells by type I IFN signalling is thought to raise the cell’s antiviral potential by enhancing its responsiveness to acutely produced IFNs13 and potentially also by upregulating basal expression of antiviral genes14. Putative heterogeneity in ISG expression among splenic FC, which may imply potential differences in innate antiviral capacity of distinct FC subsets, remains unexplored.
Here, based on single-cell RNA sequencing of over 20,000 splenic FC, we provide a more detailed view of the phenotypic and functional heterogeneity among fibroblasts from this vital immune organ. Our analysis discerns eleven transcriptionally distinct FC clusters that distribute across four main fibroblastic identities, such as Bst1+ white pulp fibroblasts, Ly6c1+ red pulp fibroblasts, Mcam+ pericytes/vascular smooth muscle cells (VSMC) and Cd34+ adventitial cells. In addition to confirming the subdivision of white pulp FC into functionally specialized TRC, MRC and FDC subsets, dissection of the intra-population diversity of Bst1+ FC revealed the existence of functionally specialized Dpt+Tnfsf13b+ FC in the splenic T-cell zone. We also identify heterogeneity among FC localized in the red pulp and in the perivascular niche in the spleen. We further uncover that Ly6c1+ red pulp fibroblasts express an augmented ISG signature under homeostatic conditions, which is dependent on tonic type I IFN signalling, and upon virus infection in vivo; suggesting a potential specialized role for splenic red pulp stroma in antiviral defence. In sum, our characterization of splenic FC at single-cell level provides insight into the functional diversity of splenic stromal cells.
Results and discussion
Single-cell RNA sequencing uncovers vast heterogeneity among splenic fibroblasts
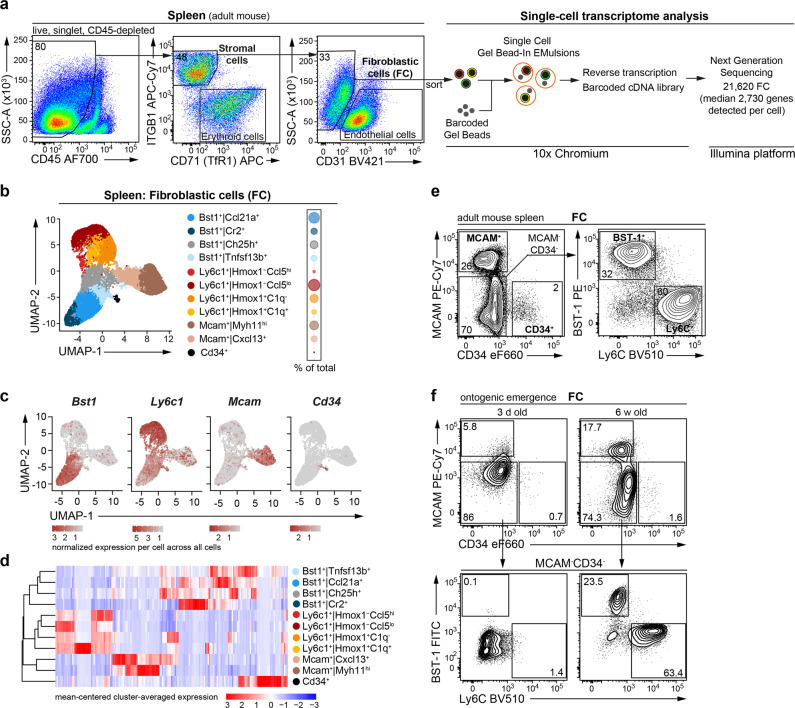
To study the phenotypic and functional heterogeneity of the splenic fibroblastic cell (FC) compartment, we performed single-cell RNA sequencing (scRNA-seq) of FC from adult mouse spleen. To obtain the full spectrum of FC, we isolated and analysed the total non-endothelial stromal cell fraction identified as CD45−ITGB1+CD31− cells in splenic digests (Fig. 1a and Supplementary Fig. 1a). Note that CD45−ITGB1− cells represent Transferrin receptor+ erythroid lineage cells (Fig. 1a). CD45−ITGB1+CD31− cells were FACS-sorted, followed by droplet-based single-cell transcriptome barcoding, reverse transcription and next generation sequencing of the resulting cDNA library (Fig. 1a). After quality control and exclusion of endothelial- (0.5 %) and mesothelial-like cells (2.1 %) as well as putative doublets (1.4 %), we retained for further analysis 21,620 FC with a median of 2730 detected genes per cell (Supplementary Fig. 1b). Following re-embedding, eleven transcriptional clusters were identified and ordered hierarchically using expression intensities of differentially expressed genes (DEGs) with the respective highest fold change per cluster (average log2(fold change) > 0.25, adjusted p-value < 0.01, fraction of expressing cells 0.1) (Fig. 1b and Supplementary Data 1). Based on overall similarity, the eleven transcriptional clusters were grouped into four fractions: (i) Bst1+ FC comprising four cell clusters, (ii) Ly6c1+ FC comprising four cell clusters, (iii) Mcam+ FC comprising two cell clusters and (iv) a single cluster of Cd34+ FC (Fig. 1c and Supplementary Fig. 1c). FC clusters belonging to the same fraction were more similar to each other than to any other cluster, indicating they may be subfractions of one main population (Fig. 1d). Consistent with the scRNA-seq data, flow cytometric analysis showed that splenic FC are comprised of four discrete populations distinguished by cell surface expression of protein markers, BST-1 (also known as CD157/BP-3), Ly6C, MCAM and CD34 (Fig. 1e). Neither BST-1+ nor Ly6C+ FC were present at birth, but emerged in the first weeks of postnatal life, suggesting that these subsets mature from neonatal precursor population/-s (Fig. 1f). We proceeded to investigate the specific features and intra-population heterogeneity of, respectively, Bst1+ FC, Mcam+ FC, Cd34+ FC and Ly6c1+ FC.
Fig. 1. Single-cell RNA sequencing uncovers vast heterogeneity among splenic fibroblasts.
a Gating strategy used to isolate fibroblastic cells (FC) from adult mouse spleen (pre-gating shown in Supplementary Fig. 1a) and a flow-chart outlining the steps in scRNA-seq analysis. b UMAP embedding of splenic FC. c Gene expression on UMAP embedding. d Hierarchical clustering of transcriptional clusters based on the mean expression of the top 20 differentially expressed genes (DEGs) per cluster with highest fold change. a–d Data pooled from 2 independent experiments using cells sorted from pooled preparations of 3 mice per experiment. e Flow cytometric confirmation of Mcam+ FC, Cd34+ FC, Bst1+ FC and Ly6c1+ FC subsets in the adult spleen. Representative stains from 2 independent experiments with 3 mice per experiment. f Emergence of Bst1+ FC and Ly6c1+ FC subsets during spleen ontogeny. Representative stains from 2 independent experiments with 3 biological replicates per experiment using pooled cell preparations from 2 spleens/replicate. a, e, f Numbers are percentage of cells in the indicated gates.
Dissection of Bst1+ FC indicates functional specialization of FC in the splenic T-cell zone
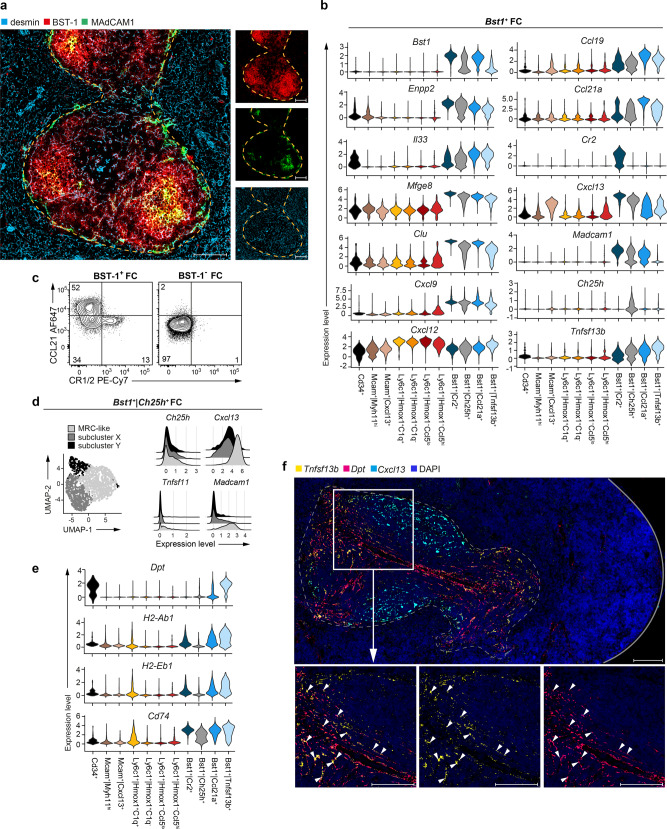
In keeping with published data on the expression pattern of BST-1 (CD157/BP-3) in the spleen15, we found that Bst1+ FC (identified as BST-1+ cells co-expressing a mesenchymal cell marker, desmin) are selectively localized in the white pulp (Fig. 2a). Consistently, scRNA-seq analysis showed that all Bst1+ FC clusters expressed white pulp stromal cell marker genes, Mfge8 and Clu5 as well as regulators of lymphocyte migration and/or differentiation, including Enpp2, a key lysophosphatidic acid (LPA)-producing enzyme linked to T-cell-high speed motility16; Il33, a critical immune modulator shaping type 1, type 2 and regulatory T-cell responses17 and Cxcl9, a chemokine ligand implicated in type 1 T-cell differentiation18 (Fig. 2b). Of the four Bst1+ clusters, two corresponded to known white pulp FC subsets, TRC and FDC, respectively represented by the clusters termed Bst1+|Ccl21a+ and Bst1+|Cr2+ (Fig. 2b). Accordingly, the Bst1+|Ccl21a+ cluster expressed highest mRNA levels of the T-zone chemokines, Ccl21 and Ccl19, while Bst1+|Cr2+ cells expressed the selective FDC-marker, Cr2 and highest mRNA levels of Cxcl13 and Madcam1 (Fig. 2b). Flow cytometric analysis confirmed that Bst1+ FC selectively contained CCL21+ TRC and CR1/2+ FDC (Fig. 2c). The third Bst1+ FC cluster, Bst1+|Ch25h+, was comprised of Ccl19loCcl21aloCxcl13+ cells that selectively expressed Ch25h (Fig. 2b). Since Ch25h expression marks MRC in the LNs8 and because the reported localization of Ch25h+ cells in the spleen (B-cell follicle perimeter)19 overlaps with the area where splenic MRC are located, we re-embedded Bst1+|Ch25h+ FC to determine if they contained MRC-like cells. Indeed, Bst1+|Ch25h+ FC encompassed cells expressing MRC-signature genes, Madcam1 and Tnfsf11 (encoding RANKL) (Fig. 2d). The remainder of Bst1+|Ch25h+ cluster, comprising Madcam1−Tnfsf11− cells, may be analogous to a previously described Ch25h+Ccl19loMadcam1− LN FC cluster located at the LN B-T-cell border8. Notably, our analysis also identified a fourth Bst1+ FC cluster, termed Bst1+|Tnfsf13b+ that did not fit TRC, FDC or MRC identity (Fig. 2b), and showed highest expression across all Bst1+ clusters for Cxcl12 and Tnfsf13b (encoding the B-cell survival factor, BAFF) (Fig. 2b). The Bst1+|Tnfsf13b+ cluster was further distinguished from other Bst1+ FC by the expression of Dpt (dermatopontin), a characteristic Bst1+|Tnfsf13b+ FC shared with Cd34+ FC, and by upregulated expression of MHC class II genes (Fig. 2e). Histological examination by in situ RNA hybridization (RNAscope) demonstrated that Bst1+|Tnfsf13b+ FC (identified as cells co-expressing mRNAs for Dpt and Tnfsf13b) reside specifically in the splenic T-cell zone (Fig. 2f). This data also suggested that Dpt-expressing cells constitute a major source of Tnfsf13b transcripts in the spleen (Fig. 2f). Notably, B-cell viability and follicular organization in the LNs appear to be maintained by BAFF produced locally by LN FC20. Whether Dpt+ FC constitute a biologically relevant source of BAFF in the spleen will require additional functional experiments. Collectively, these data resolve additional functional complexity within the FC compartment of the splenic white pulp that goes beyond subdivision into TRC, FDC and MRC.
Fig. 2. Dissection of Bst1+ FC indicates functional specialization of FC in the splenic T-cell zone.
a Confocal microscopy analysis of desmin (light blue), BST-1 (red) and MAdCAM1 (green) demonstrating that Bst1+ FC (desmin+BST-1+ cells) are localized in the white pulp. Representative stains from 2 spleens analysed in 2 independent experiments. Dashed yellow line, the white pulp/red pulp border. Scale bars are 100 μm. b Violin plots showing expression of marker genes that segregate Bst1+ FC (scaled total UMI counts per cell) across individual clusters. c Flow cytometric analysis showing that Bst1+ FC contain CCL21+ TRC and CR1/2+ FDC. Representative stains from 2 independent experiments with a single biological replicate per experiment using pooled cell preparations from 2 spleens/replicate. Numbers in the gates represent percentage of cells. d Scaled gene expression histograms for sub-clusters obtained from re-embedded Bst1+|Ch25h+ FC. e Violin plots showing expression of marker genes that segregate Bst1+|Tnfsf13b+ FC (scaled total UMI counts per cell) across individual clusters. b, e Violin plots were generated using default settings of the function VlnPlot() of the R package Seurat. f In situ RNA hybridization (RNAscope) for Dpt (pink), Tnfsf13b (yellow) and Cxcl13 (light blue). Sections were counterstained with DAPI (dark blue). Exemplary Bst1+|Tnfsf13b+ FC (Dpt+Tnfsf13b+ cells) are indicated by arrowheads. Dashed grey line, the white pulp/red pulp border. Solid grey line, the capsule. Scale bars are 100 μm. Representative stains from 2 spleens analysed in 2 independent experiments.
Mcam+ FC and Cd34+ FC represent functionally distinct components of the perivascular niche
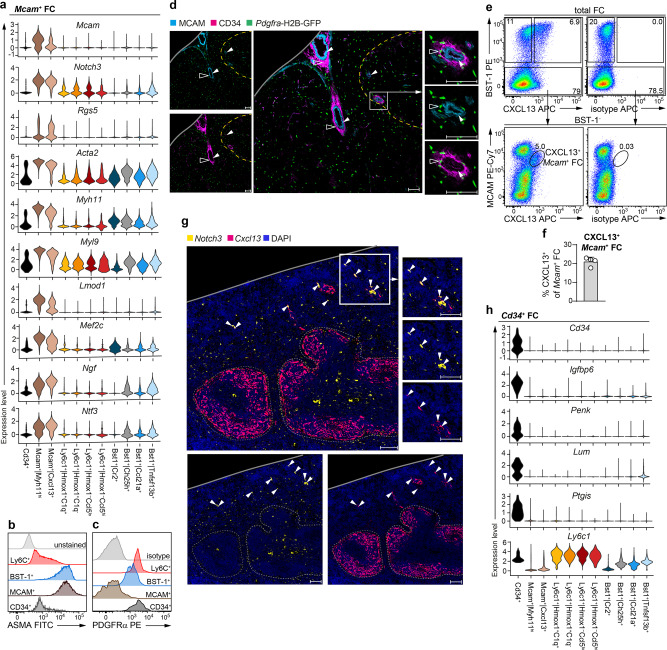
Next, we analysed Mcam+ FC and Cd34+ FC. Mcam+ FC were distinguished from other FC by selectively upregulated expression of genes associated with pericyte or vascular smooth muscle cell (VSMC)-identity, such as pericyte markers (Mcam, Rgs5, Notch3, Cspg4 (NG2) and Esam); genes associated with VSMC contractility (Acta2 (ASMA), Myh11, Myl9, Lmod1, Cald1 and Tagln (SM22)) as well as the myocyte lineage marker, Mef2c21,22 (Fig. 3a and Supplementary Data 1). Flow cytometric analysis confirmed the expression of ASMA by Mcam+ FC (Fig. 3b). Next, we assessed the localization of Mcam+ FC (note Mcam+ FC are PDGFRα−, Fig. 3c) by microscopical analysis of MCAM+PDGFRα− cells on splenic sections from Pdgfra-H2B-GFP mice, in which the nuclei of PDGFRα+ cells are labelled with GFP. Consistent with the possibility that Mcam+ FC are pericytes/VSMC, MCAM+GFP− cells were indeed associated with splenic blood vessels (Fig. 3d, filled arrowheads). Interestingly, Mcam+ FC were the main source of neurotrophic factors, such as Ngf and Ntf3 (Fig. 3a), suggesting that Mcam+ FC may support nerves which run along intrasplenic arteries and arterioles23. Our data also indicated that Mcam+ FC are heterogenous, since they encompassed two transcriptional clusters, one expressing relatively higher mRNA levels of Mcam and of VSMC markers (Mcam+|Myh11hi) and another, with lower expression of Mcam and less pronounced contractile profile that expressed higher levels of neurotrophic factors and was marked by Cxcl13 expression (Mcam+|Cxcl13+) (Fig. 3a). Flow cytometric analysis confirmed the distinction of Mcam+ FC into MCAMhiCXCL13− and MCAMloCXCL13+ subsets (Fig. 3e, f), corroborating the scRNA-seq analysis. Next, we assessed the anatomical localization of Mcam+|Myh11hi FC and Mcam+|Cxcl13+ FC. To this end, we performed in situ RNA hybridization (RNAscope) for Notch3, a pericyte/VSMC marker21 which robustly discerns both Mcam+ clusters from other FC (Fig. 3a), and for Cxcl13. In contrast to Mcam+|Myh11hi FC (identified as Notch3+Cxcl13− cells), which were found in both red and white pulp, Mcam+|Cxcl13+ FC surrounded select vessels in the red pulp (Fig. 3g, filled arrowheads). Of note, the Cxcl13 RNAscope performed in conjunction with the detection of Dpt and Tnfsf13b shown in Fig. 2f serves only to reveal the positioning of B-cell follicles and should not be used to assess the distribution of Cxcl13+ FC due to the markedly lower sensitivity of Cxcl13 detection relating to the use of a weaker fluorophore and a detection channel with a higher autofluorescence level. As far as we are aware, Mcam+Cxcl13+ cells have no apparent equivalent among previously described FC populations. Putative specialized function(-s) of the CXCL13+ mural cells in the spleen remain to be addressed by future studies. In contrast to Mcam+ FC, Cd34+ FC expressed PDGFRα (Fig. 3c) and hence could be identified as CD34+ cells with nuclear GFP signal on splenic sections from Pdgfra-H2B-GFP mice (Fig. 3d, empty arrowheads). Cd34+ FC formed an outer layer surrounding Mcam+ FC around larger splenic vessels, running inside of and close to splenic trabeculae, a localization consistent with perivascular adventitial cells22,24. Cd34+ FC expressed a gene signature composed of Igfbp6, Penk, Lum and Ptgis, similar as described for CD34+ fibroblasts in the LNs8,25 (Fig. 3h). Further of note, Cd34+ FC expressed Dpt (Fig. 2e) and Ly6c1 (Fig. 3h), suggesting they are similar to the recently described population of Cd34+Ly6c1+Dpt+ pan-tissue adventitial cells identified at transcriptional level in multiple tissues7. Collectively, the above analysis broadens our current view of the perivascular cell niche in the spleen.
Fig. 3. Mcam+ FC and Cd34+ FC are distinct components of the splenic perivascular niche.
a Violin plots showing expression of marker genes that segregate Mcam+ FC (scaled total UMI counts per cell) across individual clusters. b, c Flow cytometric analysis of b ASMA or c PDGFRα expression by the indicated FC subsets. Representative stains from 2 independent experiments with a single biological replicate per experiment using pooled cell preparations from 2 spleens/replicate. d Confocal microscopy analysis of MCAM (light blue), CD34 (pink) on splenic sections from mice expressing H2B-GFP fusion gene (i.e. nucleus-confined GFP) from the endogenous Pdgfra locus (green). Exemplary Mcam+ FC (MCAM+GFP−) are indicated by filled arrowheads. Exemplary Cd34+ FC (CD34+GFP+) are indicated by empty arrowheads. Dashed yellow line, the white pulp/red pulp border. Solid grey line, the capsule. Scale bars are 50 μm. Representative stains from 3 spleens analysed in 2 independent experiments. e, f Flow cytometric validation of Mcam+|Cxcl13+ FC. e Representative stains. Numbers are percentage of cells in the indicated gates. f Percentage of CXCL13+ cells amongst Mcam+ FC. Data are pooled from 2 independent experiments and presented as arithmetic mean ± SD of n = 4 mice (depicted as symbols). g In situ RNA hybridization (RNAscope) for Cxcl13 (pink) and Notch3 (yellow). Sections were counterstained with DAPI (dark blue). Exemplary Mcam+|Cxcl13+ FC (Notch3+Cxcl13+ cells) are indicated by arrowheads. Dashed grey line, the white pulp/red pulp border. Solid grey line, the capsule. Scale bars are 100 μm. Representative stains from 2 spleens analysed in 2 independent experiments. h Violin plots showing expression of marker genes that segregate Cd34+ FC (scaled total UMI counts per cell) across individual clusters. a, h Violin plots were generated using default settings of the function VlnPlot() of the R package Seurat.
Ly6c1+ FC are located in the splenic red pulp and the marginal zone
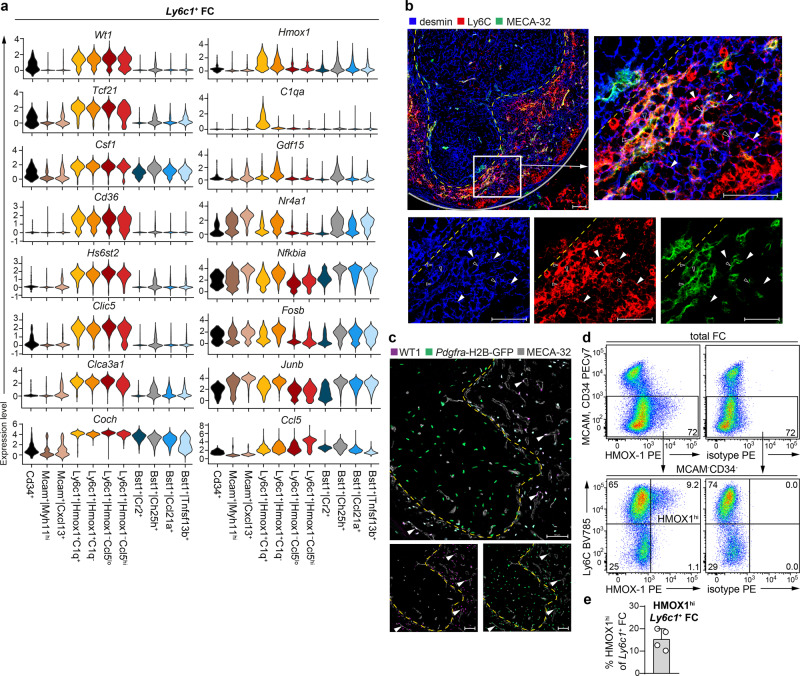
Next, we assessed the distinguishing features of Ly6c1+ FC. Ly6c1+ FC showed transcriptional similarity to previously described Csf1-expressing Wt1+ FC6 and Cxcl12-expressing Tcf21+ FC26 of the splenic red pulp. Accordingly, Ly6c1+ FC were enriched for mRNAs encoding Wt1 and Tcf21, as well as expressed highest levels of Csf1 and Cxcl12 transcripts amongst all FC (Figs. 2b and 4a). Ly6c1+ FC were further distinguished by the expression of class B scavenger receptor Cd36 and of various ECM components and regulators, such as heparan sulphate editing enzyme Hs6st2 as well as Npnt and Cadm4 (Fig. 4a and Supplementary Fig. 1c). Interestingly, Ly6c1+ FC abundantly expressed Coch (cochlin), which plays an important role in innate immune defence against bacterial infections27 (Fig. 4a). Immunohistological staining for a mesenchymal cell marker, desmin and Ly6C demonstrated that Ly6C+desmin+ FC are selectively located in the red pulp and marginal zone (Fig. 4b). Ly6C+desmin+ FC formed a network in-between MECA-32+ sinusoids (Fig. 4b, filled arrowheads) but also appeared to tightly wrap around these vessels (Fig. 4b, empty arrowheads). The existence of Ly6c1+ FC positioned in direct contact with MECA-32+ endothelium was also apparent when Ly6c1+ FC were visualized as WT1+GFP+ cells on splenic sections from Pdgfra-H2B-GFP mice (Fig. 4c, filled arrowheads). Of note, EC are PDGFRα−WT1− (Supplementary Fig. 1d–f). Whereas no apparent diversity within splenic red pulp fibroblasts could previously be discerned5,6, our analysis distinguished Ly6c1+ FC into four clusters with distinctive transcriptional profiles indicating the existence of cell subsets and/or states within the red pulp FC compartment. Ly6c1+ FC clusters showed diversity with respect to expression of Hmox1 (heme oxygenase 1) forming two Hmox1− and two Hmox1+ clusters (Fig. 4a). The two Ly6c1+|Hmox1+ clusters were distinguished from each other, respectively, by upregulated complement component 1q expression (Ly6c1+|Hmox1+C1q+) and by the expression of Gdf15 and of transcripts encoding mediators of cytokine/growth factor signalling, such as Nr4a1, Nfkbia, Fosb and Junb (Ly6c1+|Hmox1+C1q−) (Fig. 4a). The two Ly6c1+|Hmox1− clusters, termed Ly6c1+|Hmox1−Ccl5lo and Ly6c1+|Hmox1−Ccl5hi, closely resembled each other (Fig. 1d and Supplementary Fig. 1c). Flow cytometric analysis validated the existence of Ly6c1+ FC expressing high or low levels of HMOX1 protein (Fig. 4d). The fractional abundance of HMOX1hi FC was consistent with the frequency of the Ly6c1+|Hmox1+C1q+ subcluster expressing the highest level of Hmox1 mRNA (Fig. 4a, e).
Fig. 4. Ly6c1+ FC are localized in the splenic red pulp and the marginal zone.
a Violin plots showing expression of marker genes that segregate Ly6c1+ FC (scaled total UMI counts per cell) across individual clusters. Violin plots were generated using default settings of the function VlnPlot() of the R package Seurat. b Confocal microscopy analysis of Ly6C (red), desmin (blue) and MECA-32 (green). Filled arrowheads point to exemplary Ly6c1+ FC (Ly6C+desmin+) localized in-between MECA-32+ sinusoids. Empty arrowheads point to exemplary Ly6c1+ FC (Ly6C+desmin+) which appear in direct contact with MECA-32+ sinusoids. Dashed yellow line, the white pulp/red pulp border. Grey line, the capsule. Scale bars are 50 μm. Representative stains from 2 spleens analysed in 2 independent experiments. c Confocal microscopy analysis of WT1 (purple) and MECA-32 (grey) on splenic sections from mice expressing H2B-GFP fusion gene (i.e. nucleus-confined GFP) from the endogenous Pdgfra locus (green). Arrowheads point to exemplary Ly6c1+ FC (WT1+GFP+) in direct contact with MECA-32+ sinusoids. Dashed yellow line, the white pulp/red pulp border. Scale bars are 50 μm. Representative stains from 3 spleens analysed in 3 independent experiments. d, e Flow cytometric validation of Ly6c1+ FC expressing high levels of HMOX-1 (HMOX-1hi). d Representative stains. HMOX1hi Ly6c1+ FC are indicated in the plot. Numbers shown are percentage of cells in the indicated gates. e Percentage of HMOX1hi cells amongst Ly6c1+ FC. Data are pooled from 2 independent experiments and presented as arithmetic mean ± SD of n = 4 mice (depicted as symbols).
Ly6c1+ FC are enriched for antiviral gene expression at steady state
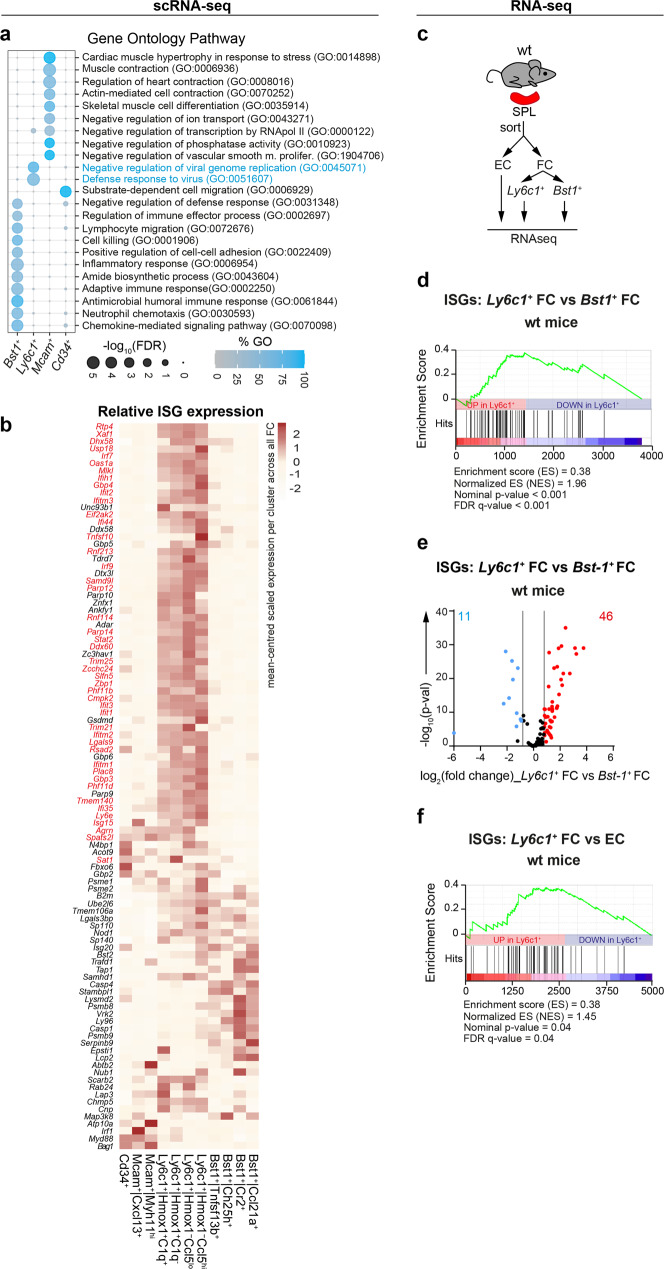
Next, to further explore functions of splenic FC, we averaged single-cell transcriptomes of the main FC fractions, and applied gene ontology analysis of biological processes based on the top 300 cell type-specific DEGs (log2(fold change) > 0.25, p-val < 0.05). Notably, Ly6c1+ FC showed specific over-representation of genes involved in the inhibition of virus replication and defence response to virus infection (Fig. 5a). To assess this antiviral propensity in more detail on the level of individual FC clusters, we next analysed the expression of 104 ISGs, common to both human and mouse lymphoid and myeloid cells28 across all eleven FC clusters. In agreement with the gene ontology analysis, the expression levels of a major proportion of ISGs were elevated in all four Ly6c1+ FC clusters compared to other FC clusters (Fig. 5b). ISGs with upregulated expression in Ly6c1+ FC encompassed a marked variety of genes with important roles in antiviral defence, including Eif2ak2 (Protein kinase R), Mlkl, Ifi44, members of the Oas, Gbp, Ifit and Ifitm families as well as mediators of viral sensing, such as Irf7, Zbp1 (DNA sensor, DAI), Dhx58, Ddx60 and Ifih1 (RNA sensor, Mda5)11 (Fig. 5b). To independently confirm as well as further quantify the observed phenomenon, we next analysed the bulk transcriptomes of Ly6c1+ FC, Bst1+ FC and splenic endothelial cells (EC) by RNA-seq (Fig. 5c). Consistent with the results obtained on the single-cell level, geneset enrichment analysis demonstrated significant enrichment for ISGs in Ly6c1+ FC compared to Bst1+ FC (Fig. 5d). Accordingly, Ly6c1+ FC significantly overexpressed 46 ISGs (log2(fold change) > 0.8, p-val < 0.05) while only 11 ISGs were overexpressed by Bst1+ FC (Fig. 5e). It deserves to be mentioned that ISGs upregulated in Ly6c1+ FC over Bst1+ FC matched between the RNA-seq and scRNA-seq data sets (Fig. 5b, ISGs upregulated in Ly6c1+ FC based on the RNA-seq analysis are indicated in red). Importantly, Ly6c1+ FC were enriched for ISG expression not only relative to other FC, but also relative to splenic EC, indicating that the observed antiviral profile of Ly6c1+ FC does not simply reflect their closeness to the blood stream but rather is regulated by organ-specific extrinsic and/or intrinsic mechanisms (Fig. 5f). In sum, these results suggest that Ly6c1+ FC may show enhanced antiviral activity under homeostatic conditions.
Fig. 5. Ly6c1+ FC are enriched for antiviral gene expression.
a GO analysis of biological processes for top 300 upregulated DEGs in scRNA-seq dataset across all cells per indicated FC subset. Shown are significantly enriched GO identifiers. GO identifiers significantly enriched in Ly6c1+ FC that relate to antiviral defence are highlighted in blue. b Heatmap showing mean-centred scaled expression of individual ISGs per cluster across all FC based on scRNA-seq analysis. Indicated in red are ISGs with upregulated expression (log2(fold change) > 0.8, adj. p-val < 0.05) in bulk Ly6c1+ FC versus Bst1+ FC based on RNA-seq analysis of these subsets isolated from the spleens of wild-type (wt) mice (as explained in c–f). c–f RNA-seq analysis of Ly6c1+ FC, Bst1+ FC and EC isolated from the spleens of wt mice. Data are from 3 biological replicates, with respective subsets sorted from pooled cell preparations of 2–4 mice/replicate. d Geneset enrichment analysis for ISGs performed on DEGs (log2(fold change) > 0.8, adj. p-val < 0.05) from the comparison between Ly6c1+ FC versus Bst1+ FC. e Volcano plot comparing fold change of ISG expression between Ly6c1+ FC versus Bst1+ FC. ISGs with upregulated expression in Ly6c1+ FC or Bst1+ FC (log2(fold change) > 0.8, adj. p-val < 0.05) are highlighted respectively in red and blue. f Geneset enrichment analysis for ISGs performed on DEGs (log2(fold change) > 0.8, adj. p-val < 0.05) from the comparison between Ly6c1+ FC versus EC.
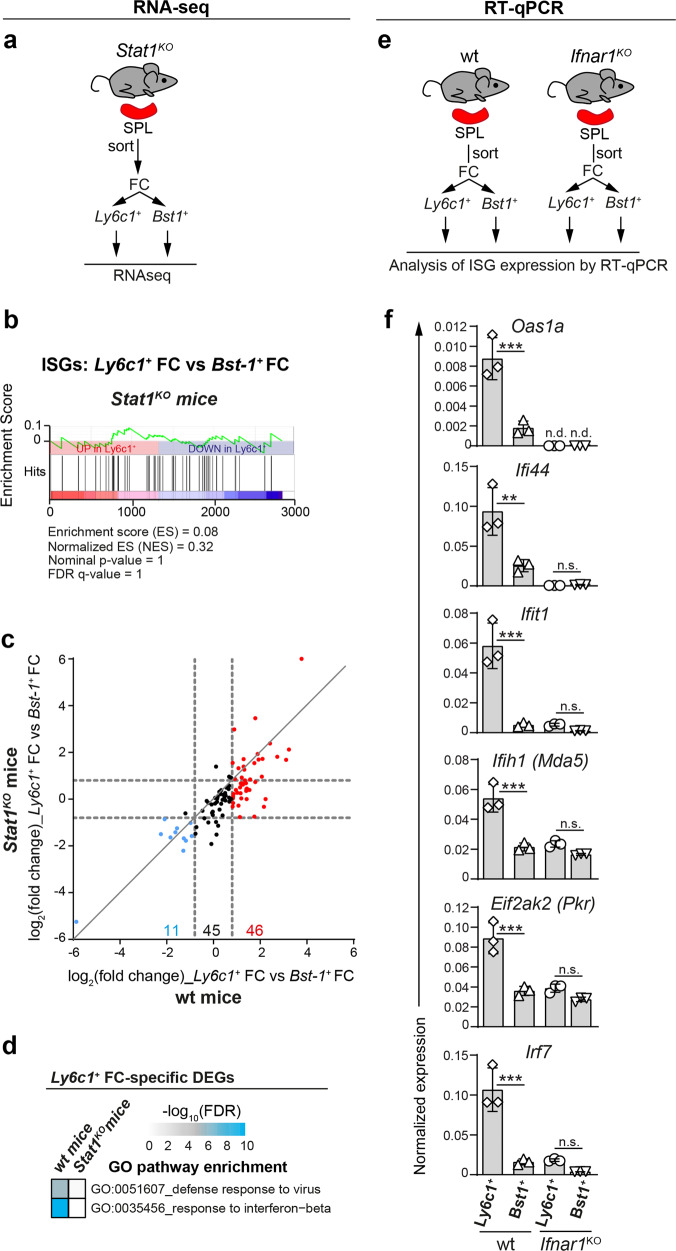
Antiviral signature of Ly6c1+ FC is dependent on Stat1
Next, we investigated whether the enhanced antiviral gene signature of Ly6c1+ FC is regulated by tonic IFN signalling. Firstly, we examined the impact of Stat1, which is required to sustain tonic IFN-dependent ISG expression29, on the relative difference in ISG expression between Ly6c1+ FC and Bst1+ FC. To this end, Ly6c1+ FC and Bst1+ FC were sorted from Stat1KO mice and analysed by RNA-seq (Fig. 6a). To ensure correct subset identification (note Ly6c1 is a type I IFN-inducible gene30 expressed by Ly6c1+ FC of Stat1KO mice at a modestly reduced level), Ly6c1+ FC were sorted as MCAM−CD34−BST-1−PDGFRα+ FC (Supplementary Fig. 2a). Notably, geneset enrichment analysis performed on bulk transcriptomes of Ly6c1+ FC and Bst1+ FC from Stat1KO mice revealed that in the absence of Stat1, Ly6c1+ FC were no longer enriched for ISG expression compared to Bst1+ FC (Fig. 6b). Accordingly, loss of Stat1 resulted in overall lower fold change difference in ISG expression between Ly6c1+ FC versus Bst1+ FC (Fig. 6c). Specifically, Stat1 was responsible for the overexpression of 26 of 46 genes that constituted the ISG signature of Ly6c1+ FC but affected none of the 11 ISGs that were overexpressed by Bst1+ FC (Supplementary Table 1). Stat1-dependent enrichment for ISG expression in Ly6c1+ FC was corroborated using an independent ISG set collated for primary fibroblasts (extracted from the Interferome database v2.031 using the following search criteria: max. 6 h post stimulation with IFNβ, fold change > 2.5; p-val < 0.05) (Supplementary Fig. 2b). Finally, in the absence of Stat1, Ly6c1+ FC were also no longer enriched for the expression of genes involved in defence response to virus as revealed by gene ontology analysis (Fig. 6d). Thus, the selectively augmented antiviral gene signature of Ly6c1+ FC is Stat1-dependent. These results were consistent with a role for tonic IFN signalling in sustaining the transcriptionally enhanced antiviral profile of Ly6c1+ FC. To corroborate this, we assessed the impact of IFNAR loss on the expression of select ISGs, which are overexpressed in Ly6c1+ FC in wt mice. To this end, Ly6c1+ FC and Bst1+ FC were sorted from wt and Ifnar1KO mice and analysed by RT-qPCR (Fig. 6e). Indeed, loss of IFNAR expression equalized expression levels of all tested ISGs between Ly6c1+ FC and Bst1+ FC (Fig. 6f). Furthermore, Ifnar1 and Stat1 similarly affected the expression of ISGs involved in IFN signalling (Supplementary Fig. 2c, d). Collectively, the presented evidence indicates that the augmented antiviral signature of Ly6c1+ FC is induced and/or sustained by tonic type I IFN signalling.
Fig. 6. Antiviral signature of Ly6c1+ FC is dependent on Stat1.
a–d RNA-seq analysis of Ly6c1+ FC and Bst1+ FC purified from the spleens of Stat1KO mice. Data are from 3 biological replicates, with respective subsets sorted from pooled cell preparations of 2 mice/replicate. b Geneset enrichment analysis for ISGs performed on DEGs (log2(fold change) > 0.8, adj. p-val < 0.05) from the comparison between Ly6c1+ FC versus Bst1+ FC in Stat1KO mice. c Scatter plot depicting fold change in the expression of individual ISGs between Ly6c1+ FC versus Bst1+ FC in (x-axis) wt and (y-axis) Stat1KO mice. ISGs with upregulated expression in Ly6c1+ FC or Bst1+ FC (log2(fold change) > 0.8, adj. p-val < 0.05) in the wt condition are highlighted respectively in red and blue. d GO analysis of biological processes for top 1188 DEGs with upregulated expression in Ly6c1+ FC versus Bst1+ FC (log2(fold change) > 0.8, adj. p-val < 0.05) in the wt condition or in Stat1KO mice. Shown are significantly enriched GO identifiers that relate to antiviral defence. e, f RT-qPCR analysis of ISG expression in Ly6c1+ FC and Bst1+ FC isolated from the spleens of wt and Ifnar1KO mice. f Data are pooled from 2 independent experiments and presented as arithmetic mean ± SD of n = 3 biological replicates (depicted as symbols) using cells sorted from pooled cell preparations of 2 mice/replicate. Statistical significance was calculated using one-way ANOVA with Sidak’s multiple comparison test. **p < 0.01; ***p < 0.001; n.s. denotes p > 0.05; n.d. denotes not detected.
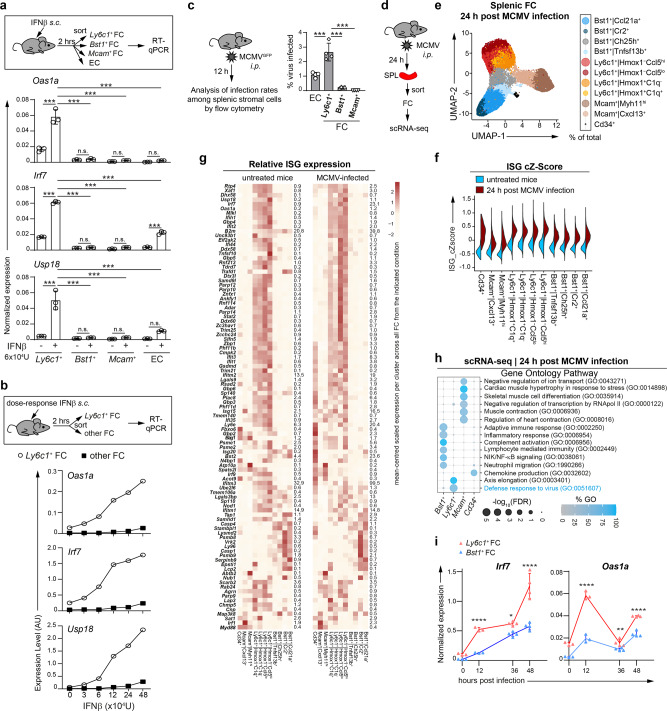
Antiviral gene expression by splenic FC following immune stimulation in vivo
Next, we addressed the question whether in the presence of acutely produced type I IFNs, Ly6c1+ FC would also express ISGs on a relatively higher level. To study the response of splenic FC to type I IFNs disseminating from a peripheral site, we analysed ISG expression in splenic FC subsets and EC following a single dose of IFN-β delivered subcutaneously (Fig. 7a). Two hours after stimulation, which is sufficient to achieve maximal ISG response in cells residing in the white pulp of the spleen28, indicated subsets were sorted and analysed by RT-qPCR. As shown in Fig. 7a, Ly6c1+ FC of IFN-treated mice attained the highest expression of all tested ISGs. Ly6c1+ FC expressed these ISGs at a higher level independently of the strength of the stimulus, as underscored by dose-response analysis (Fig. 7b). Next, we studied antiviral gene expression by splenic FC subsets in response to the infection with mouse cytomegalovirus, MCMV in vivo. Based on histological analysis, it has previously been indicated that MCMV administered intraperitoneally infects and undergoes the first round of replication in stromal cells located in the splenic marginal zone and the red pulp32, a localization corresponding to Ly6c1+ FC. Flow cytometric analysis of splenic stromal cells 12 h post intraperitoneal infection with 106 PFU of MCMVGFP, demonstrated that Ly6c1+ FC were infected at the frequency of ca 3% whereas infection rates of other analysed FC subsets, Bst1+ FC and Mcam+ FC, were substantially lower (Fig. 7c). To obtain a snapshot of antiviral gene expression across all FC subsets in MCMV-infected mice, we performed scRNA-seq analysis of splenic FC 24 h post intraperitoneal infection with 106 PFU of MCMV (Fig. 7d). In keeping with the flow cytometric analysis (Fig. 7c), MCMV-infected cells (3.1%) resided in a distinct cluster that expressed markers of cellular stress and clustered closely with Ly6c1+ FC (Supplementary Fig. 3a). Virus-infected cells were removed from subsequent analysis as we aimed to assess the bystander immune response of FC and not their cell-intrinsic response to the virus. After quality control and exclusion of mesothelial, endothelial, and virus-infected cells, we retained for further analysis 25,419 FC with 2815 median detected genes per cell (Supplementary Fig. 3b). The identity of the eleven FC clusters described in the steady-state condition was preserved among splenic FC from infected mice (Fig. 7e, Supplementary Fig. 3c). The relative abundance of individual clusters was similar between both conditions, except for a reciprocal shift between the two most similar Ly6c1+ clusters, Ly6c1+|Hmox1−Ccl5lo and Ly6c1+|Hmox1−Ccl5hi (these cells were not proliferating), suggesting they may represent alternative activation states of the same subset (Figs. 1b and 7e). Based on the comparison of cZ-scores for ISG expression calculated for individual subsets in the untreated versus infected condition, all FC clusters upregulated ISGs following MCMV infection, with the highest cZ-scores noted for Ly6c1+ FC and for a fraction of Cd34+ FC (Fig. 7f). A more detailed evaluation on the level of individual ISGs revealed that even though some Cd34+ FC reached a similar ISG cZ-score as Ly6c1+ FC, Ly6c1+ FC were the only FC subset uniquely overexpressing a sizeable array of ISGs in virus-infected mice (Fig. 7g). Notably, ISGs selectively overexpressed by Ly6c1+ FC in virus-infected mice overlapped with ISGs overexpressed by these cells in the steady state (Fig. 7g). Further importantly, Ly6c1+ FC in virus-infected mice were, like in the steady state, selectively enriched for antiviral gene expression, as underscored by gene ontology analysis (Fig. 7h). Finally, we complemented the scRNA-seq analysis performed at 24 h post infection with a time-resolved profile of ISG expression by splenic FC in virus-infected mice. To this end, we performed a kinetic analysis of the expression of Irf7 and Oas1a, which are overexpressed in Ly6c1+ FC at 24 h post MCMV infection (Fig. 7g), in Ly6c1+ FC and Bst1+ FC at 12, 36 and 48 h post intraperitoneal infection with 106 PFU of MCMV by RT-qPCR. The ISG response of splenic FC was biphasic, matching the biphasic kinetic of type I IFN production upon MCMV infection, which peaks in the first 24 h and then again at 48 h post infection33. Importantly, Ly6c1+ FC expressed the tested ISGs at a higher level compared to Bst1+ FC, with highest differences at 12 h and at 48 h (Fig. 7i). In sum, the transcriptionally augmented antiviral profile of Ly6c1+ FC across various immune conditions, suggests a specialized innate immune function of these cells. The putative importance of Ly6c1+ FC in innate antiviral defence remains to be directly addressed by future studies using conditional knockout models.
Fig. 7. Antiviral gene expression by splenic FC following immune stimulation in vivo.
a, b RT-qPCR analysis of ISG expression in indicated splenic subsets following a 2 h stimulation of mice with the indicated dose of IFNβ s.c. a Data from singular in vivo experiment is presented as arithmetic mean ± SD of n = 3 biological replicates (depicted as symbols) using cells sorted from pooled cell preparations of 2 mice/replicate. b Data from singular dose-response experiment with one mouse per each indicated IFNβ dose. Symbols depict ISG expression in indicated subsets sorted from a single mouse per IFNβ dose. c Percentage of GFP+ cells among indicated splenic subsets 12 h post infection of mice with 106 PFU of MCMVGFP i.p. Data from one representative experiment of 2 independent experiments is presented as arithmetic mean ± SD of n = 4 mice (depicted as symbols). d–h scRNA-seq analysis of splenic FC 24 h post infection of mice with 106 PFU of MCMV i.p. Data pooled from 2 independent experiments using cells sorted from pooled splenic preparations of 3 mice. e UMAP embedding of splenic FC. f cZ-score of ISG expression per cell across all FC clusters in the untreated or MCMV-infected condition. g Heatmaps showing mean-centred scaled expression of individual ISGs per cluster across FC in the untreated or MCMV-infected condition. Numbers indicate the average expression level of indicated ISGs across all FC in the indicated condition. h GO analysis of biological processes for top 300 upregulated DEGs across all cells per indicated FC subset in the infected condition. Shown are significantly enriched GO identifiers. GO identifiers significantly enriched in Ly6c1+ FC that relate to antiviral defence are highlighted in blue. i Kinetic assessment of ISG expression in Ly6c1+ FC and Bst1+ FC following infection of mice with 106 PFU of MCMV i.p. determined by RT-qPCR. Data from singular in vivo experiment is presented as arithmetic mean ± SD of n = 3 (0, 12 h) and n = 4 (36, 48 h) mice (depicted as symbols). a, c, i Statistical significance was calculated using one-way ANOVA with Sidak’s multiple comparison test. *p < 0.05; *p < 0.01; ***p < 0.001; ****p < 0.0001; n.s. denotes p > 0.05.
Conclusions
Here, we performed single-cell transcriptional analysis of FC from adult mouse spleen, identifying eleven transcriptionally distinct FC clusters distributed across four main cell identities, such as (i) Bst1+ white pulp fibroblasts comprising four clusters; (ii) Ly6c1+ red pulp fibroblasts comprising four clusters; (iii) Mcam+ mural cells with pericyte/VSMC identity comprising two clusters and (iv) a single cluster of Cd34+ adventitial fibroblasts. In addition to confirming the subdivision of white pulp FC into TRC, MRC and FDC, our analysis resolved a fourth, functionally specialized Dpt+Tnfsf13b+ FC subset localized in the splenic T-cell zone. Detailed characterization of the FC components of the splenic perivascular niche discerned a subset of CXCL13-expressing Mcam+ pericyte/VSMC present around select vessels in the splenic red pulp. We further uncover that Ly6c1+ red pulp FC overexpress a transcriptional signature composed of genes involved in defence response to viral infection, underpinned by overexpression of ISGs, under homeostatic conditions and upon virus infection in vivo; suggesting a potential specialized role for splenic red pulp stroma in antiviral defence. In sum, our characterization of splenic FC at single-cell level provides insight into the functional diversity of splenic stromal cells.
Methods
Mice
Unless otherwise indicated, experiments were performed with age- and sex-matched 2–5-m-old mice on C57BL/6 background. C57BL/6JrJ mice were purchased from Janvier Labs. Ifnar1KO34, Stat1KO35 and PdgfraH2B-GFP36 mice were bred and maintained under specific pathogen free (SPF) conditions according to Federation of European Laboratory Animal Science Associations (FELASA) guidelines, respectively, at central animal facility of HZI Braunschweig, Germany; University of Veterinary Medicine Vienna, Austria and San Raffaele Scientific Institute, Milan, Italy. Animal procedures were approved by the responsible state office (Lower Saxony State Office of Consumer Protection and Food Safety).
Viral infections
Infection of mice was performed by intraperitoneal administration of 106 PFU of either BAC-derived MCMV (clone pSM3fr-MCK-2fl 3.337) or BAC-derived MCMV-GFP-P2A-ie1/3, MCMVGFP. MCMV-GFP-P2A-ie1/3 was generated by en passant BAC mutagenesis38 using the BAC pSM3fr-MCK-2fl 3.337. Briefly, the gene encoding GFP was fused with the MCMV gene encoding immediate early transcripts 1 and 3 (m122/123)). The P2A peptide-encoding sequence was inserted after GFP removing its stop codon generating a bicistronic ORF. Cloning design was carried out using SnapGene software (GSL Biotech, USA). The recombinant BAC was transfected into NIH3T3 cells using FuGene HD (Promega, USA) and reconstituted viral particles were passaged five times before generating a stock from infected M2-10B4 mouse fibroblastic cells (ATCC CRL-1972).
Cell Isolation
Spleens were digested with collagenase P (0.4 mg/ml), dispase II (2 mg/ml) and DNase I (50 µg/ml) in RPMI-1640 supplemented with 1 mM sodium pyruvate, 100 U/ml penicillin, 100 U/ml streptomycin, 10 mM HEPES and 5% fetal bovine serum (FBS). Spleens were pre-incubated with the above digestion solution injected into the organ using a 26 G needle for 5 min at RT, then minced using scissors and digested for 30 min at 37 °C. Enzymatic treatment was repeated for additional 20 min followed by incubation with 5 mM EDTA at RT for 5 min. The resulting cell suspensions were passed through a 70 µm filter, followed by immunomagnetic depletion of CD45+ cells using MACS (Miltenyi Biotech). Briefly, cells from one spleen were incubated with 60 µl anti-CD45 microbeads in 600 µl of PBS containing 2 mM EDTA and 2% FBS for 20 min on ice, washed, and depleted on LS columns according to manufacturer’s instructions (Miltenyi Biotech).
Flow cytometry and cell sorting
Flow cytometry was performed with antibodies listed in Supplementary Table 2. Dead cells (identified using 7-AAD Viability Staining Solution or Zombie NIR Fixable Viability Kit; both from BioLegend) and cell aggregates (identified on FSC-A versus FSC-H scatter plots) were excluded from all analyses. For intracellular staining, surface-labelled cell suspensions were fixed using eBioscience Foxp3/Transcription Factor Staining Buffer Set or eBioscience IC Fixation Buffer (both from Thermo Fisher). HMOX-1 expressing cells were detected with anti-HMOX-1 antibody coupled to PE using the Lightning-Link conjugation kit (abcam). Data acquisition was performed on an Aria-II SORP, ARIA-Fusion or LSR-Fortessa (BD Biosciences) and analysed using FlowJo software (BD Biosciences). Sorting was performed on an Aria-II SORP or ARIA-Fusion (BD Biosciences).
Immunofluorescence staining of splenic sections
Spleens were fixed in 4% (wt/vol) paraformaldehyde (PFA) for 10 min, saturated overnight in 30% (wt/vol) sucrose at 4 °C, and embedded in Tissue-Tek optimum cutting temperature compound (Sakura) followed by freezing in −80 °C. For detection of Ly6C and BST-1, sections were fixed prior to staining with ice-cold acetone for 10 min. Sections (7 μm) were blocked with 10% (vol/vol) goat serum. Endogenous peroxidase and biotin activities were quenched respectively with 3 % (vol/vol) hydrogen peroxide solution and Endogenous Biotin-Blocking Kit (Thermo Fisher). Antibodies (listed in Supplementary Table 2) were diluted in PBS containing 0.05% (vol/vol) Tween-20 and 2% (vol/vol) goat serum. Primary biotinylated antibodies were visualized with HRP-conjugated streptavidin followed by TSA Plus Cyanine 3 or Cyanine 5 System (Akoya). BST-1 expressing cells were detected with anti-BST-1 antibody coupled to Alexa647 using the Lightning-Link conjugation kit (abcam). Images were acquired with ZEISS LSM 980 confocal microscope and analysed using ZEN software (both from Carl Zeiss MicroImaging).
In situ RNA hybridization (RNAscope)
In situ RNA hybridization was performed using the RNAscope Multiplex Fluorescent Detection Kit v2 (Advanced Cell Diagnostics) according to manufacturer’s instructions. The following target probes were used: Mm-Dpt (Cat. #561511-C3), Mm-Tnfsf13b (Cat. #414891), Mm-Notch3 (Cat. #425171), Mm-Cxcl13 (Cat. #406311-C2). In brief, spleens were fixed in 10% formalin for 24 h at RT and embedded in paraffin. 3 μm-thick sections were baked in an oven at 60 °C for 1 h, then deparaffinized and dehydrated. Following rehydration, endogenous peroxidase activity was quenched with hydrogen peroxide for 10 min at RT. Target retrieval was carried out in a steamer (Braun, Type 3216) for 15 min at >98 °C. Protease treatment was performed with Protease Plus (Advanced Cell Diagnostics) for 20 min at 40 °C. Target probes were allowed to hybridize for 2 h at 40 °C, followed by signal amplification according to the ACD protocol. After the final amplification step, signal was detected with Opal520, Opal570 or Opal650-conjugated tyramide (Perkin Elmer). Sections incubated with a negative control probe (DapB) were analysed in parallel and a mix of positive control probes (Polr2a, Ppib, Ubc) was utilized to confirm RNA integrity. Images were acquired with an Olympus VS120 slide scanner fluorescence microscope using the VS-ASW-FL software (Olympus).
RT-qPCR
Total RNA was extracted using RNeasy Plus Micro Kit (Qiagen), reverse transcribed with SuperScript IV and a 1:1 mixture of oligo-dT and random oligonucleotide hexamers (all from Thermo Fisher). Quantitative PCR was performed using Forget-Me-Not EvaGreen qPCR Master Mix (Biotium) in a LightCycler 480 Instrument II (Roche). Relative gene expression was calculated using the 2−ΔCT method with normalization to the expression of Gapdh. Primers (listed in Supplementary Table 3) were validated for the use of the 2−ΔCT method by determining the efficiency of each primer pair in the corresponding expression range.
Single-cell RNA sequencing analysis
Cells were sorted into DMEM medium containing 10% FBS and adjusted to a density of 1000 cells/μl. Chromium Controller (10x Genomics) was used for partitioning single cells into Gel Bead-In-EMulsions (GEMs) and Chromium Single Cell 3′ GEM, Library & Gel Bead Kit v3 (10x Genomics) for reverse transcription, cDNA amplification and library construction (all performed according to manufacturer’s instructions). Libraries were sequenced on a NovaSeq 6000 sequencer (Illumina) using NovaSeq 6000 S1 Reagent Kit (100 cycles, 28 bp read 1, 89 bp read 2) and attained approximately 34,100 reads per single cell across all replicates. The GEO accession number for the scRNA-seq data reported in this paper is GSE156162. Raw base call (BCL) files were demultiplexed to generate Fastq files using the cellranger mkfastq pipeline within Cell Ranger 3.0.3 (10x Genomics). Whole transcriptome Fastq files were processed using the standard cellranger pipeline within Cell Ranger 3.0.3 (10x Genomics) against the merged genomes of GRCm38 (Ensembl v90) and MCMV (BAC-derived wild-type Smith strain, Accession Identifier: NC_004065). The expected cell parameter was set to 10,000 for all libraries. Briefly, cellranger performs alignment, filtering, barcode counting, and UMI counting to obtain read count matrices (RCM). In total four RCMs were obtained, with two independent experiments performed for both steady state and virally infected condition. RCMs were further processed via R (version 3.6.2) using Seurat (version 3.1.5)39 and uwot (version 0.1.8)40. Cells with at least 1000 detected genes and <7.5% mitochondrial reads were retained in the analysis. Analysis was performed in a stepwise manner to successively identify and focus on splenic FC. In total, 50,401 cells with 16,426 detected genes were processed. Genes were included if they were expressed in at least 1 % of the cells. Initially, the four RCMs were merged using FindIntegration Anchors (list(experimental_batches), anchor. features = 1000, dims = 1:15, k. filter = 200, k. anchor = 10) and Integrate Data(). Data was scaled and regressed against the variables: percent mitochondrial UMIs, percent ribosomal protein UMIs, percent viral UMIs and the total number of UMIs per cell. PCAs were computed using default settings. Uniform Manifold Approximation and Projection (UMAP) dimensional reduction via RunUMAP() and FindNeighbors() were performed using the first 15 PCA dimensions as input features. Find Clusters() was computed at resolution 0.4. The respective RCMs are deposited under the GEO accession number GSE156162. Bst1+ FC, Ly6c1+ FC, Mcam+ FC and Cd34+ FC were identified based on the expression of the respective cell type markers and all cells residing within these clusters were re-embedded as detailed above with the following parameters: anchor. features = 1000, dims = 1:17, k. filter = 200, k.anchor = 10, resolution = 0.45 and the first 17 dimensions used as input features. Merged data was visualized using the Seurat intrinsic functions VlnPlot(), FeaturePlot(), DotPlot(), DimPlot(). Differentially expressed genes per cluster were identified using FindAllMarkers() or FindMarkers(). From two independent experiments performed for both steady state and virally infected condition, Bst1+ FC, Ly6c1+ FC, Mcam+ FC and Cd34+ FC collectively amounted to 47,039 cells with 14,306 detected genes were processed. Cells with a proportion of >1% viral UMIs were excluded. When analysing distinct subsets, all cells residing within the respective cluster were re-embedded as detailed above with the following parameters: anchor. features = 500, dims = 1:4, k. filter = 200, k. anchor = 10, resolution = 0.1 and the first 4 dimensions used as input features. Differentially expressed genes per cluster were identified using Find All Markers(). Cumulative Z-scores were calculated based on the scaled expression per gene across all clusters for the given comparison or all cells and summed for the defined gene signatures. Gene ontology analysis was performed for the indicated sets of differentially expressed genes using topGO41. Pie-chart, bubble-plot and bar graph visualizations were carried out with ggplot2.
RNA sequencing analysis
Cells were sorted into RLT-Plus lysis buffer (Qiagen) containing 10 µl/ml β-mercaptoethanol. Total RNA was extracted using RNeasy Plus Micro kit (Qiagen) according to manufacturer’s instructions. Quality and integrity of total RNA was controlled using 5200 Fragment Analyzer System (Agilent Technologies). For the experiment comparing Ly6c1+ FC, Bst1+ FC and EC from wt mice, cDNA conversion was carried out using SMART‑Seq v4 Ultra Low Input RNA Kit (Takara Clontech Laboratories) according to manufacturer’s instructions, followed by library generation using Nextera XT DNA Library Prep Kit (Illumina). Libraries were then sequenced on a HiSeq2500 sequencer (Illumina) using paired-end run (read 1–50 bp, read 2–30 bp) with an average of 5 × 107 reads per RNA sample. For the experiment comparing FC subsets from Stat1KO mice, the RNA sequencing libraries were generated using NEB Next Single Cell/Low Input RNA Library Prep Kit for Illumina (NEB) according to manufacturer’s protocol and sequenced on a NovaSeq 6000 sequencer (Illumina) using NovaSeq 6000 S1 Reagent Kit (100 cycles, paired-end run 2 × 50 bp) with an average of 5 × 107 reads per RNA sample. The GEO accession number for all RNA-seq data reported in this paper is GSE156162. Read quality of sequenced libraries was evaluated with FastQC. Sequencing reads were aligned to the reference mouse genome assembly GRCm38 using STAR42. Reads aligned to annotated genes were quantified with htseq-count43. Raw read counts were converted to RPKM (reads per kilobase of exon length per million mapped reads) values. Protein-coding genes with at least 20 reads in at least two replicates were included in the analysis. The calculated read counts were further processed with DESeq2 for quantification of differential gene expression44. Geneset enrichment analysis was performed in the pre-ranked mode using the GSEA desktop application v4.1.0 (https://www.gsea-msigdb.org/gsea). Analysis was performed on DEGs (log2(fold change) > 0.8, p-val < 0.05) from the comparison between Ly6c1+ FC and Bst1+ FC from the wild-type or Stat1KO condition with fold change serving as the ranking metric. The number of permutations was set to 1000.
Statistics and reproducibility
No statistical methods were used to predetermine sample size. One-way ANOVA with Sidak’s multiple comparison test were performed with GraphPad Prism 8 to calculate statistical significance. P-values <0.05 were considered significant. *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001. Error bars denote mean ± SD. The sample size for each experiment is reported in the Figure Legends. No data were removed from statistical analysis as outliers. Results have been confirmed in at least two independent experiments.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Supplementary information
Description of Additional Supplementary Files
Acknowledgements
This study was funded by the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation)—Projektnummer 158989968—SFB 900, the German Centre for Infection Research (DZIF) of the German Federal Ministry of Science and Education (BMBF) through TTU 07.834 (to L.C-S.), the European Union’s Horizon 2020 research and innovation program under Grant Agreement 793858 (to K.M.S.) and the Austrian Science Fund FWF SFB F6101 und F6106 (to B.S., M.M.). We thank Inge Hollatz-Rangosch and Ayse Barut for technical assistance. We acknowledge Lothar Gröbe and Maria Höxter from HZI core flow cytometry facility as well as Robert Geffers from HZI genomics platform.
Author contributions
Concept and study design, K.M.S.; Bioinformatical analysis, J.P., F.E. and M.B.; Investigation, K.M.S., C.W., U.R. and T.B.; Methodology, K.M.S., J.P. and F.E; Resources, L.C-S., J.H., B.D., M.M., B.S., A.B. and U.K.; Funding acquisition, L.C-S., K.M.S., M.M., B.S.; Writing of the manuscript, K.M.S. and J.P. with input from other authors; Supervision, K.M.S. and L.C-S.
Data availability
RNA-seq and scRNA-seq data generated during this study were deposited in the NCBI Gene Expression Omnibus (GEO) database with accession number GSE156162. All source data underlying the graphs shown in the main and supplementary figures are presented in Supplementary Data 2.
Competing interests
The authors declare no competing interests.
Footnotes
Peer review information Communications Biology thanks the anonymous reviewers for their contribution to the peer review of this work. Primary Handling Editors: Simona Chera and George Inglis. Peer reviewer reports are available.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Luka Čičin-Šain, Email: luka.cicin-sain@helmholtz-hzi.de.
Katarzyna M. Sitnik, Email: katarzyna.sitnik@vetmeduni.ac.at
Supplementary information
The online version contains supplementary material available at 10.1038/s42003-021-02882-9.
References
- 1.Mebius RE, Kraal G. Structure and function of the spleen. Nat. Rev. Immunol. 2005;5:606–616. doi: 10.1038/nri1669. [DOI] [PubMed] [Google Scholar]
- 2.Neely HR, Flajnik MF. Emergence and evolution of secondary lymphoid organs. Annu. Rev. Cell Dev. Biol. 2016;32:693–711. doi: 10.1146/annurev-cellbio-111315-125306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Malhotra D, Fletcher AL, Turley SJ. Stromal and hematopoietic cells in secondary lymphoid organs: partners in immunity. Immunol. Rev. 2013;251:160–176. doi: 10.1111/imr.12023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Golub R, Tan J, Watanabe T, Brendolan A. Origin and immunological functions of spleen stromal cells. Trends Immunol. 2018;39:503–514. doi: 10.1016/j.it.2018.02.007. [DOI] [PubMed] [Google Scholar]
- 5.Cheng HW, et al. Origin and differentiation trajectories of fibroblastic reticular cells in the splenic white pulp. Nat. Commun. 2019;10:1739. doi: 10.1038/s41467-019-09728-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bellomo A, et al. Reticular fibroblasts expressing the transcription factor WT1 define a stromal niche that maintains and replenishes splenic red pulp macrophages. Immunity. 2020;53:127–142. doi: 10.1016/j.immuni.2020.06.008. [DOI] [PubMed] [Google Scholar]
- 7.Buechler MB, et al. Cross-tissue organization of the fibroblast lineage. Nature. 2021;593:575–579. doi: 10.1038/s41586-021-03549-5. [DOI] [PubMed] [Google Scholar]
- 8.Rodda LB, et al. Single-cell RNA sequencing of lymph node stromal cells reveals niche-associated heterogeneity. Immunity. 2018;48:1014–1028. doi: 10.1016/j.immuni.2018.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pikor NB, et al. Remodeling of light and dark zone follicular dendritic cells governs germinal center responses. Nat. Immunol. 2020;21:649–659. doi: 10.1038/s41590-020-0672-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ivashkiv LB, Donlin LT. Regulation of type I interferon responses. Nat. Rev. Immunol. 2014;14:36–49. doi: 10.1038/nri3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schoggins JW, Rice CM. Interferon-stimulated genes and their antiviral effector functions. Curr. Opin. Virol. 2011;1:519–525. doi: 10.1016/j.coviro.2011.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Taniguchi T, Takaoka A. A weak signal for strong responses: interferon-alpha/beta revisited. Nat. Rev. Mol. Cell Biol. 2001;2:378–386. doi: 10.1038/35073080. [DOI] [PubMed] [Google Scholar]
- 13.Gough DJ, Messina NL, Clarke CJ, Johnstone RW, Levy DE. Constitutive type I interferon modulates homeostatic balance through tonic signaling. Immunity. 2012;36:166–174. doi: 10.1016/j.immuni.2012.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bradley KC, et al. Microbiota-driven tonic interferon signals in lung stromal cells protect from influenza virus infection. Cell Rep. 2019;28:245–256. doi: 10.1016/j.celrep.2019.05.105. [DOI] [PubMed] [Google Scholar]
- 15.Katakai T, et al. Organizer-like reticular stromal cell layer common to adult secondary lymphoid organs. J. Immunol. 2008;181:6189–6200. doi: 10.4049/jimmunol.181.9.6189. [DOI] [PubMed] [Google Scholar]
- 16.Katakai T, Kondo N, Ueda Y, Kinashi T. Autotaxin produced by stromal cells promotes LFA-1-independent and Rho-dependent interstitial T cell motility in the lymph node paracortex. J. Immunol. 2014;193:617–626. doi: 10.4049/jimmunol.1400565. [DOI] [PubMed] [Google Scholar]
- 17.Liew FY, Girard JP, Turnquist HR. Interleukin-33 in health and disease. Nat. Rev. Immunol. 2016;16:676–689. doi: 10.1038/nri.2016.95. [DOI] [PubMed] [Google Scholar]
- 18.Groom JR, et al. CXCR3 chemokine receptor-ligand interactions in the lymph node optimize CD4+ T helper 1 cell differentiation. Immunity. 2012;37:1091–1103. doi: 10.1016/j.immuni.2012.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yi T, et al. Oxysterol gradient generation by lymphoid stromal cells guides activated B cell movement during humoral responses. Immunity. 2012;37:535–548. doi: 10.1016/j.immuni.2012.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cremasco V, et al. B cell homeostasis and follicle confines are governed by fibroblastic reticular cells. Nat. Immunol. 2014;15:973–981. doi: 10.1038/ni.2965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Armulik A, Genove G, Betsholtz C. Pericytes: developmental, physiological, and pathological perspectives, problems, and promises. Dev. Cell. 2011;21:193–215. doi: 10.1016/j.devcel.2011.07.001. [DOI] [PubMed] [Google Scholar]
- 22.Sitnik KM, et al. Context-dependent development of lymphoid stroma from adult CD34(+) adventitial progenitors. Cell Rep. 2016;14:2375–2388. doi: 10.1016/j.celrep.2016.02.033. [DOI] [PubMed] [Google Scholar]
- 23.Verlinden TJM, et al. Innervation of the human spleen: a complete hilum-embedding approach. Brain Behav. Immun. 2019;77:92–100. doi: 10.1016/j.bbi.2018.12.009. [DOI] [PubMed] [Google Scholar]
- 24.Corselli M, et al. The tunica adventitia of human arteries and veins as a source of mesenchymal stem cells. Stem Cells Dev. 2012;21:1299–1308. doi: 10.1089/scd.2011.0200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pezoldt J, et al. Neonatally imprinted stromal cell subsets induce tolerogenic dendritic cells in mesenteric lymph nodes. Nat. Commun. 2018;9:3903. doi: 10.1038/s41467-018-06423-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Inra CN, et al. A perisinusoidal niche for extramedullary haematopoiesis in the spleen. Nature. 2015;527:466–471. doi: 10.1038/nature15530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Py BF, et al. Cochlin produced by follicular dendritic cells promotes antibacterial innate immunity. Immunity. 2013;38:1063–1072. doi: 10.1016/j.immuni.2013.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mostafavi S, et al. Parsing the interferon transcriptional network and its disease associations. Cell. 2016;164:564–578. doi: 10.1016/j.cell.2015.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Platanitis E, et al. A molecular switch from STAT2-IRF9 to ISGF3 underlies interferon-induced gene transcription. Nat. Commun. 2019;10:2921. doi: 10.1038/s41467-019-10970-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee PY, Wang JX, Parisini E, Dascher CC, Nigrovic PA. Ly6 family proteins in neutrophil biology. J. Leukoc. Biol. 2013;94:585–594. doi: 10.1189/jlb.0113014. [DOI] [PubMed] [Google Scholar]
- 31.Rusinova I, et al. Interferome v2.0: an updated database of annotated interferon-regulated genes. Nucleic Acids Res. 2013;41:1040–1046. doi: 10.1093/nar/gks1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hsu KM, Pratt JR, Akers WJ, Achilefu SI, Yokoyama WM. Murine cytomegalovirus displays selective infection of cells within hours after systemic administration. J. Gen. Virol. 2009;90:33–43. doi: 10.1099/vir.0.006668-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schneider K, et al. Lymphotoxin-mediated crosstalk between B Cells and splenic stroma promotes the initial type I interferon response to cytomegalovirus. Cell Host Microbe. 2008;3:67–76. doi: 10.1016/j.chom.2007.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Muller U, et al. Functional role of type I and type II interferons in antiviral defense. Science. 1994;264:1918–1921. doi: 10.1126/science.8009221. [DOI] [PubMed] [Google Scholar]
- 35.Durbin JE, Hackenmiller R, Simon MC, Levy DE. Targeted disruption of the mouse Stat1 gene results in compromised innate immunity to viral disease. Cell. 1996;84:443–450. doi: 10.1016/s0092-8674(00)81289-1. [DOI] [PubMed] [Google Scholar]
- 36.Hamilton TG, Klinghoffer RA, Corrin PD, Soriano P. Evolutionary divergence of platelet-derived growth factor alpha receptor signaling mechanisms. Mol. Cell Biol. 2003;23:4013–4025. doi: 10.1128/MCB.23.11.4013-4025.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jordan S, et al. Virus progeny of murine cytomegalovirus bacterial artificial chromosome pSM3fr show reduced growth in salivary Glands due to a fixed mutation of MCK-2. J. Virol. 2011;85:10346–10353. doi: 10.1128/JVI.00545-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tischer BK, Smith GA, Osterrieder N. En passant mutagenesis: a two step markerless red recombination system. Methods Mol. Biol. 2010;634:421–430. doi: 10.1007/978-1-60761-652-8_30. [DOI] [PubMed] [Google Scholar]
- 39.Butler A, Hoffman P, Smibert P, Papalexi E, Satija R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 2018;36:411–420. doi: 10.1038/nbt.4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stuart T, et al. Comprehensive integration of single-cell data. Cell. 2019;177:1888–1902. doi: 10.1016/j.cell.2019.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Alexa, A. & Rahnenfuhrer, J. topGO: Enrichment Analysis for Gene Ontology. R package version 2.40.010.18129/B9.bioc.topGO (2020).
- 42.Dobin A, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Anders S, Pyl PT, Huber W. HTSeq–a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Files
Data Availability Statement
RNA-seq and scRNA-seq data generated during this study were deposited in the NCBI Gene Expression Omnibus (GEO) database with accession number GSE156162. All source data underlying the graphs shown in the main and supplementary figures are presented in Supplementary Data 2.