Glomerular diseases comprise 7% to 16% of chronic kidney disease (CKD) in the United States, affecting between 2.6 and 6 million Americans.1 Individuals with glomerular disease have unique risk factors for infectious complications, including prolonged exposure to immunosuppressive medications, systemic inflammation, altered immune cell function, and urinary loss of immunoglobulin and complement factors.2 As a result, individuals with glomerular disease experience rates of infection that are approximately 30 times higher than those of the general US population.3 Interventional studies typically collect and report data on adverse infectious events, which is critical to understanding patient well-being and the relative safety of therapeutic options. However, infectious events occurring in real-world clinical settings are less well studied. Accurate identification of infectious events occurring in clinical practice can be an arduous task, as recording of these events is embedded in electronic medical records (EMRs) or healthcare claims data. Diagnosis codes assigned by clinicians and coders are easily searchable but are often flawed because of time barriers, human error, and the complexities of diagnostic coding. Adjudication involving manual review of medical records is the gold standard approach but is labor and time intensive. Herein, we have developed a diagnosis code−based algorithm and then tested its internal validity to identify infection-related acute care events within a large cohort of children and adults with glomerular disease in the CureGN study.
CureGN is a prospective, multicenter, observational cohort study of patients with biopsy-proven glomerular disease, including minimal change disease, focal segmental glomerulosclerosis, membranous nephropathy, or IgA nephropathy/vasculitis. Participants are enrolled from 72 clinical sites in the United States, Canada, Italy, and Poland. Using in-person or remote visits, clinical data are collected and updated every 6 months, including details of any interval emergency room visits or hospitalizations. Detailed methods for the CureGN study have been previously published.4
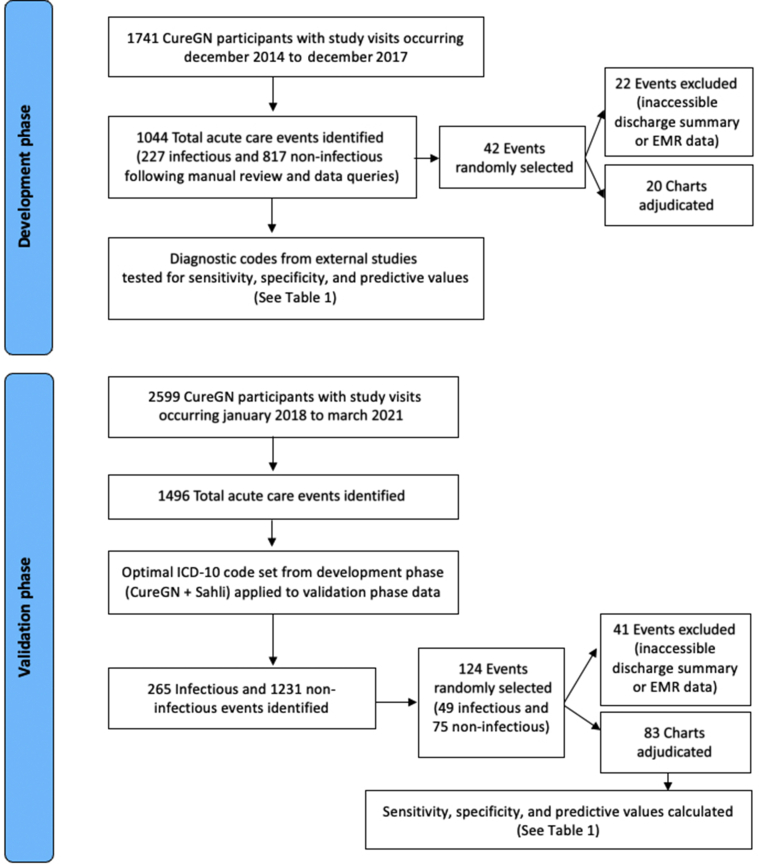
Data from CureGN visits were analyzed in 2 phases (Figure 1). In the development phase, a gold standard set of infectious and noninfectious events were identified and manually curated using information derived from CureGN study visit forms and communication with study coordinators. A random subset of events was then manually adjudicated by physician chart review. The test characteristics of multiple infection-related International Classification of Diseases, Tenth Revision (ICD-10) code lists, studied in other patient populations, were then evaluated using the manually curated events as the gold standard. In the validation phase, the test characteristics of the code list with the best test characteristics in the development phase were evaluated using more contemporary CureGN data. In the validation phase, infectious and noninfectious events adjudicated by direct chart review were considered the gold standard.
Figure 1.
Study design for developmental and validation datasets assessing the predictive ability to detect infectious versus noninfectious events. EMR, electronic medical record; ICD-10, International Classification of Diseases, Tenth Revision.
The development phase included all acute care events for which participants experienced an emergency room visit or hospitalization occurring between December 2014 and December 2017. As we previously reported,3 infections during the development phase were identified using information collected by clinical site study personnel, with verification by local clinical investigators as needed. Two study investigators (DG and CH) manually reviewed all reported acute care events and classified events as infection related based on discharge ICD-10 codes, Current Procedural Terminology (CPT) codes, and data from CureGN hospitalization and study visit forms, which included information regarding antibiotic use. When these sources of data were incongruent (e.g., the visit was associated with an ICD-10 code suggesting infection, but the hospitalization form did not report an infection-related emergency department visit or hospitalization), local study site coordinators were queried to clarify the nature of the event, according to CureGN data quality protocols.5 A random sample of infectious and noninfectious events was adjudicated by physicians at 5 high-enrolling study sites using their local electronic medical record and a standardized adjudication protocol. Adjudicators were unaware of an event’s prior classification by study personnel as infectious or noninfectious. Events occurring outside of a CureGN study site were excluded from adjudication. A total of 20 events were manually adjudicated, demonstrating a high level of agreement (κ = 1.0, sensitivity = 100%, specificity = 100%) between event classification using study coordinator−derived data versus manual chart review.3
The sensitivity, specificity, positive predictive value, and negative predictive value of 4 separate ICD-10 code lists designating infection (Supplementary Table S1)6, 7, 8 were then measured using the 227 infectious and 817 noninfectious acute care events occurring in the CureGN development cohort as the gold standard. The combination of ICD-10 codes assigned by CureGN coordinators with additions from Sahli et al. was deemed to have the best overall test characteristics (preference given to higher specificity yielding more conservative effect estimates) (Table 1). To identify infectious events in the validation phase, the combined code set (CureGN + Sahil) was applied to acute care events occurring between January 2018 and March 2021 (n = 1496). A random sample of infectious (n = 49) and noninfectious (n = 75) events were again adjudicated by chart review at 4 sites, representing the gold standard. Of these, 41 events were excluded because of the absence of medical record data for validation (i.e., events occurring at outside institutions), leaving 83 events available for adjudication. The positive and negative predictive values for the selected code list were 87% (95% confidence interval [CI] = 75%−99%) and 83% (95% CI = 72%−93%), respectively (Table 1).
Table 1.
Test characteristics of ICD-10−based code lists for infection in the development and validation phases
| ICD-10 code list | Sensitivity (95% CI)a | Specificity (95% CI)a | PPV (95% CI)a | NPV (95% CI)a |
|---|---|---|---|---|
| Development phase | ||||
| CureGN3 | 87 (83–91) | 96 (94–97) | 85 (81–90) | 96 (95–98) |
| Sahli et al.8 | 71 (69–80) | 95 (94–97) | 81 (76–87) | 92 (90–94) |
| Baker et al.7 | 82 (77–87) | 87 (85–90) | 64 (58–69) | 95 (93–96) |
| USRDS6 | 74 (69–80) | 91 (89–93) | 69 (63–76) | 93 (91–94) |
| CureGN + Sahli et al. | 89 (85–93) | 93 (91–95) | 78 (73–83) | 97 (96–98) |
| CureGN + USRDS | 91 (87–95) | 88 (86–90) | 68 (63–74) | 97 (96–98) |
| CureGN + Sahli et al. + Baker et al. | 93 (90–96) | 83 (80–85) | 60 (55–65) | 98 (97–99) |
| CureGN + Sahli et al. + Baker et al. + USRDS | 94 (90–97) | 79 (76–82) | 56 (51–61) | 98 (97–99) |
| Validation phase | ||||
| CureGN + Sahli et al. | 75 (61–89) | 91 (84–99) | 87 (75–99) | 83 (72–93) |
CI, confidence interval; ICD-10, International Classification of Diseases, Tenth Revision; NPV, negative predictive value; PPV, positive predictive value; USRDS, United States Renal Data System.
The 95% confidence intervals were calculated using the Wald method.
Algorithms using EMR data to identify infections have previously shown variable accuracy depending on infection type, data source, study population, and algorithm used.9 In this era of limited funding for research, EMR-based algorithms to identify outcomes of interest using routinely collected data represent increasingly important tools for identifying adverse events in observational and pharmacoepidemiologic surveillance studies. Our data demonstrate that ICD-10 diagnosis codes can be used to efficiently identify infection-related acute care events among patients with glomerular disease. There are few studies in the literature investigating the positive predictive value of infection related ICD-10 codes. Two notable validation studies using Danish patient registries yielded similar results to those of our study (positive predictive value = 72%–98%). Lower positive predictive values were found for infections occurring in the emergency departmentS1 compared to the inpatient setting,S2 likely due to challenges in diagnostic accuracy in the emergency room setting when microbiological test results might not be immediately available. Our analysis was not powered to stratify infections by clinical setting; however, we suspect that some variability in our findings might be explained by this factor.
Strengths of our study include its multicenter design, large size, diverse patient population, and standardized data collection procedure. We recognize several limitations. First, our approach to determining a “gold standard” differed somewhat between the development and validation cohorts (i.e., rigorous curation of events vs. manual electronic medical record adjudication, respectively). Nonetheless, we have confidence in both approaches, given high levels of agreement in a subset of adjudicated events occurring in the development phase. Second, the development and validation cohorts were derived from the same overall cohort of patients, albeit at a later stage in their disease course. In addition, the CureGN infection diagnosis code list was derived from, and subsequently tested within, the same population, potentially resulting in overestimation of its test characteristics. To improve the external validity of our final code list for future studies, we selected the combination of CureGN-derived ICD-10 codes and diagnosis codes from a validated external cohort (Sahli et al.8) for final validation testing. Finally, our study did not aim to validate specific types of infections, but rather the presence or absence of an infection. Previous studies have shown variability in the accuracy of medical billing codes by infection type.9 Future studies should validate our findings within other cohorts of patients with glomerular disease and for specific infection types of high severity or burden. Incorporating microbiological or radiographic data in future validation studies might further enhance the validity and test characteristics of ICD-10−based diagnostic algorithms. In summary, these data demonstrate that ICD-10 diagnosis codes can be used to efficiently identify infection-related acute care events among patients with glomerular disease.
Disclosure
LAG has received grant support from Advicenne, Alexion, Apellis, Reata, and Vertex; he has received consulting fees from Abbvie, Advicenne, Akebia, Alexion, Leadiant, Otsuka, Ra Pharma, and Vifor; he has served on the data safety monitoring committee for studies sponsored by Alnylam, Relypsa, Retrophin, and University of California, San Diego. MD discloses grant support from Mallinckrodt Pharmaceuticals. AM has clinical trial contracts with Amgen, Boehringer Ingelheim, Calliditas, Duke Clinical Research Institute, and Pfizer; and has had consultancy agreements with Bayer. LM reports grants from NIH-NIDDK and personal fees from Reata Pharmaceuticals, Calliditas Therapeutics, and Travere Therapeutics. KG reports honorarium received as a member of the Reata CKD Advisory Board and clinical trial contracts with Aurinia and Retrophin. All the other authors declare no competing interests.
Acknowledgments
Funding for the CureGN consortium is provided by U24DK100845 (formerly UM1DK100845), U01DK100846 (formerly UM1DK100846), U01DK100876 (formerly UM1DK100876), U01DK100866 (formerly UM1DK100866), and U01DK100867 (formerly UM1DK100867) from the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK). Patient recruitment is supported by NephCure Kidney International.
Dates of funding for first phase of CureGN was 9/16/2013-5/31/2019.
Footnotes
Table S1. International Statistical Classification of Diseases and Related Health Problems 10th Revision (ICD-10) codes used in the Development Phase.
Supplementary References
Acknowledgments
Supplementary Material
Table S1. International Statistical Classification of Diseases and Related Health Problems 10th Revision (ICD-10) codes used in the Development Phase.
Supplementary References
Acknowledgments
References
- 1.United States Renal Data System . National Institutes of Health, National Institute of Diabetes and Digestive and Kidney Diseases; Bethesda, MD: 2016. Annual Data Report: Epidemiology of Kidney Disease in the United States. [Google Scholar]
- 2.McCaffrey J., Lennon R., Webb N.J.A. The non-immunosuppressive management of childhood nephrotic syndrome. Pediatr Nephrol. 2016;31:1383–1402. doi: 10.1007/s00467-015-3241-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Glenn D.A., Henderson C.D., O’Shaughnessy M., et al. Infection-related acute care events among patients with glomerular disease. Clin J Am Soc Nephrol. 2020;15:1749–1761. doi: 10.2215/CJN.05900420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mariani L.H., Bomback A.S., Canetta P.A., et al. CureGN study rationale, design, and methods: establishing a large prospective observational study of glomerular disease. Am J Kidney Dis. 2019;73:218–229. doi: 10.1053/j.ajkd.2018.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gillespie B.W., Laurin L.-P., Zinsser D., et al. Improving data quality in observational research studies: report of the Cure Glomerulonephropathy (CureGN) network. Contemp Clin Trials Commun. 2021;22:100749. doi: 10.1016/j.conctc.2021.100749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.United States Renal Data System . 2018. Volume 2: ESRD Analytical Methods. Bethesda, MD; p. 661. [Google Scholar]
- 7.Baker M.G., Barnard L.T., Kvalsvig A., et al. Increasing incidence of serious infectious diseases and inequalities in New Zealand: a national epidemiological study. Lancet. 2012;379:1112–1119. doi: 10.1016/S0140-6736(11)61780-7. [DOI] [PubMed] [Google Scholar]
- 8.Sahli L., Lapeyre-Mestre M., Derumeaux H., Moulis G. Positive predictive values of selected hospital discharge diagnoses to identify infections responsible for hospitalization in the French national hospital database: PPV of Hospitalization for Infection Codes in France. Pharmacoepidemiol Drug Saf. 2016;25:785–789. doi: 10.1002/pds.4006. [DOI] [PubMed] [Google Scholar]
- 9.Barber C., Lacaille D., Fortin P.R. Systematic review of validation studies of the use of administrative data to identify serious infections: administrative data to identify infections. Arthritis Care Res. 2013;65:1343–1357. doi: 10.1002/acr.21959. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.