Summary
The Human Genome Project opened an era of (epi)genomic research, and also provided a platform for the development of new sequencing technologies. During and after the project, several sequencing technologies continue to dominate nucleic acid sequencing markets. Currently, Illumina (short-read), PacBio (long-read), and Oxford Nanopore (long-read) are the most popular sequencing technologies. Unlike PacBio or the popular short-read sequencers before it, which, as examples of the second or so-called Next-Generation Sequencing platforms, need to synthesize when sequencing, nanopore technology directly sequences native DNA and RNA molecules. Nanopore sequencing, therefore, avoids converting mRNA into cDNA molecules, which not only allows for the sequencing of extremely long native DNA and full-length RNA molecules but also document modifications that have been made to those native DNA or RNA bases. In this review on direct DNA sequencing and direct RNA sequencing using Oxford Nanopore technology, we focus on their development and application achievements, discussing their challenges and future perspective. We also address the problems researchers may encounter applying these approaches in their research topics, and how to resolve them.
Keywords: nanopore sequencing, direct DNA sequencing, direct RNA sequencing, base modification, base-calling, long-read sequencing, tools and algorithms
Graphical abstract
Public summary
-
•
Nanopore-seq can dissect native DNA/RNA molecules from any organisms at unlimited length
-
•
A wide variety of algorithms greatly increase the accuracy of signal decoding in Nanopore-Seq
-
•
Nanopore-Seq significantly facilitates genome assembly and structural variant calling, and can simultaneously detect base modifications
-
•
These advantages ensure its great potentials in future medical and agricultural practices
Introduction
The applications of DNA and RNA sequencing have greatly promoted research in the life sciences and catapulted biological research into the genomic and post-genomic era. The success of the Human Genome Project (HGP) has promoted the development of sequencing technologies and their large-scale application. The HPG started in 1990 and its first draft genome was published in 2001,1 and finally completed and reported in 2004.2 It initiated the flourishing period of next-generation sequencing (NGS), in which diverse platforms sprang up, from short-read sequencing3 to long-read sequencing.4 In 2004, Pacific Biosciences was founded, focusing on single-molecule real-time sequencing.5 The following year, Oxford Nanopore was founded. Nanopore sequencing provides single-nucleotide detection and analytical capabilities that are achieved by electrophoretically driving molecules in solution through a nano-scale pore.6 Nevertheless, the concept behind nanopore sequencing can be traced back to 1989,4 much earlier than the NGS technologies, However, its first commercial products were not released until 2014.4 Demanding features of the sequencing methodology hindered its refinement, which slowed down its wide application. Currently, related algorithms and tools have been developed and released, and the use of nanopore sequencing is becoming increasingly frequent. Its complexity, the diversity of associated algorithms and tools, and the requirement for high-purity nucleic acids for library preparation may confuse potential users lacking sufficient background information. In the review, we focus on issues related to nanopore direct DNA sequencing and direct RNA sequencing, so that readers can understand how nanopore sequencing works and how to use the new technology to serve their interests.
A brief history of nanopore sequencing
The development of nanopore sequencing may also have been a byproduct of the HGP, which was first proposed in 1985 but was finally funded in 1990. During this period, many scientists began to imagine effective methods for sequencing human genomic DNA in its entirety. One such scientist was Dr. David Deamer, who first proposed an idea to sequence DNA with membrane electrophoresis in 1989 while vacationing in Oregon. The idea he jotted in his notebook at the time is very similar to the nanopore sequencing technology in current use. At the time, his lab was focusing on lipochemistry. Thus, the need for, and ability of, cells to pass specific molecules through membranes may have influenced his approach to sequencing, maybe the HGP budget authorized in 1988 made him think about the possibility. While Kasianowicz, who was working on DNA sequencing with protein-made pores, was invited to collaborate on the idea Deamer jotted in his notebook and published the seminal paper in PNAS in 1996, where they prosed to monitor alterations in ion current blockades to detect the sequence of bases in single molecules of DNA or RNA when they pass through a minute opening—a nanopore—in a lipid bilayer membrane.7 Afterward, Kasianowicz returned to focusing on dissecting DNA sequence through protein-comprised nanopores embedded in a lipid bilayer membrane,8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20 independent of his collaboration with Deamer on that paper.7 Meanwhile, George Church was struggling to scale up the sequencing throughput of the HPG, and thus proposed a similar idea to sequence double-stranded DNA via electronically monitoring phage packing motors embedded in a lipid bilayer. Hagan Bayley was first to artificially synthesize hemolysin nanopores in a lipid bilayer membrane to allow single-strand DNA or RNA molecules to pass through under certain voltage, the ion current signals produced from bases going through the pores could be observed,9,11,13,21, 22, 23, 24, 25, 26, 27, 28, 29 thus after he moved to Oxford University, he co-founded the Oxford Nanopore company in 2005.
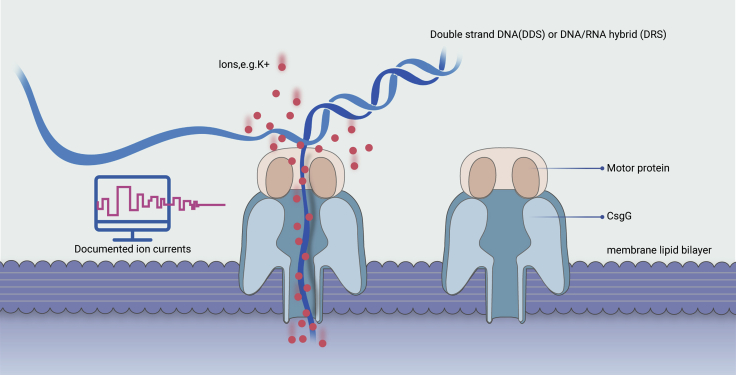
At first, the major challenge for nanopore sequencing was to control the speed of translocation from the cis- to the trans-chambers, which are separated by a lipid bilayer membrane. Finally, in 2010, phi29 DNA polymerase was found to solve this issue by effectively regulating the translocation speed of single-strand DNA.30,31 Meanwhile, the alpha-hemolysin pore was found not to be the best choice for discerning DNA nucleotides.4 Later, MspA, a Mycobacterium smegmatis porin, was found to be more suitable as the protein for nanopore, with a funnel-like geometry that narrowed to an ∼1.2-nm-diameter and a ≤0.6-nm-long tube, which is optimal for single-strand nucleic acids.32 The coupling of MspA and phi29 DNA polymerase showed significantly increased performance in the sequencing of the phi X174 genome.33 However, the porin used by Oxford Nanopore for commercial flow cells is CsgG,34 since the height (85 Å) of the channel made of CsgG35 even can set two detectors along the channel to monitor the ion current signals. In 2014, some 18 years after Deamer's initial inspiration, commercial flowcell R7.3 was finally released to the genomics community.4 Figure 1 gives an overview of how nanopore sequencing works.
Figure 1.
Schematic field application of ONT direct DNA sequencing
CsgG is the protein for current commercial nanopores in Oxford Nanopore flowcells. DNA/RNA hybrid comes from reverse-transcription, the first-strand cDNA and template RNA formed hybrid can prevent the secondary structure of native RNAs, thus facilitate direct RNA sequencing, but cDNA does not enter into the pores for sequencing. Motor protein, a kind of protein that can control the speed of nucleic acid molecules passing through nanopores, is normally used to decrease the speed.
Tools and algorithms developed for nanopore sequencing
Because nanopore sequencing detects alterations in ion current when different bases pass through protein nanometer-scale pores at a specified voltage, the device needs to be sensitive enough to discern signals reflecting different DNA nucleotides, with or without modification. In addition, DNA fragments pass through nanopores at high speed, and the device can only record signals for 5 to 6 bases as a pulse. The deduction of correct DNA bases within each pulse increases the difficulty in data analysis. Subsequently, diverse algorithms have been developed to recognize the sequenced bases; however, their accuracy was still vastly inferior to that achieved by the base-calling method of sequencing by synthesis. Although deep-learning algorism shows better performance, the relatively high error rate requires additional algorithms specific for downstream applications and analysis. Therefore, we first review the tools and algorithms developed specifically for nanopore sequencing.
Development of base-calling algorithms
Initially, Hidden Markov Model (HMM) and Viterbi decoding approaches, such as Metrichor and Nanocall, were exclusively adopted for base-calling, albeit unfortunately with low accuracy. In current years, deep neural networks have been widely implemented in base-calling tools, including convolutional neural networks, recurrent neural networks (RNN), and connectionist temporal classification (CTC) decoders, which greatly improved base-calling accuracy to more than 98% for DNA sequencing from less than 80% HMM algorithm called.36
For HMM and Viterbi decoding approaches, Timp et al. (2012)37 demonstrated the method's potential for usage in the real world, because decoding 3–base pair (bp) resolution nanopore electrical measurements into a DNA sequence using an HMM can reach around 98% accuracy. However, when the flowcell R7.3 was released, each documented ion current signal comprising 5 to 6 bases, nearly two times longer than the 3-bp length, made the algorithm dramatically decrease its accuracy. To find a better resolution for base-calling, Teng et al.38 reported an algorithm called Chiron, which was the first deep-learning model to achieve end-to-end base-calling, and not necessary to segment the ion current signals from a sequenced nucleic acid molecule to detect the corresponding bases. This means it directly translates the raw current signal to a DNA sequence without the error-prone segmentation step, which may avoid the error caused by segmentation, but may increase computing time. To further improve the quality of base-calling, Zeng et al.39 presented Causalcall, an end-to-end temporal convolution-based deep-learning model for accurate and fast nanopore base-calling. It directly identifies base sequences of varying lengths from current measurements in long time series, but still cannot decrease artificial deletion caused by low ion current signals. Zhang et al.40 also proposed a refined U-net model (thus, a UR-net model, an enhanced U-net model for 1D sequence segmentation) called URnano to improve previous end-to-end deep-learning models. Early neural-network base-callers (such as DeepNano41 and BasecRAWller42) relied on a preprocessing step that segmented the current measurements into discrete events that may artificially introduce errors when segmenting the ion current signals. To avoid such flaws, Konishi et al.43 developed an improved base-caller, Halcyon, which utilizes an encoder-decoder model incorporating neural-network techniques to improve base-calling accuracy, but may increase computing time.
Although the Nanopore company developed Guppy44 and Bonito base-callers can make the accuracy of DNA nanopore sequencing reach more than 99%, the third parties still have been attempting to develop more accurate algorithms for the nanopore sequencing community. One example comes from a trial that embedded a chip with a complementary metal-oxide-semiconductor (CMOS) base-caller alongside nanopore sensors to predict the molecule's base-pair constitution.45 Another test46 is to use an accelerator consisting of a low-power real-time field-programmable gate array (FPGA) integrated circuit for the base-calling task.46 Although these algorithms currently are not better than Nanopore company-developed base-callers, the different strategies used to develop new algorithms may benefit nanopore base-calling in the future.
For nanopore sequencing, not only can a single strand of a genomic region be sequenced, but also both strands in a genomic region can be sequenced via a hairpin adapter. Single-strand sequencing is called 1D sequencing, while double-strand sequencing was previously called 2D sequencing, because of a dispute with the Pacific Biosciences company, now the 2D sequencing is called 1D2 (1D squared). 1D2 reads reflect consensus sequence generated from the base-calling of both forward- and reverse-strand DNA connected with a hairpin adapter. Currently, 1D2 reads cannot be base-called with common open-source algorithms, e.g. Nanocall. Although Nanocall has lower base-calling accuracy (∼68%), it supports offline base-calling for Oxford Nanopore sequencing data.47 The offline base-calling may help users to save CPU/GPU when nanopore sequencing since the base-calling can be performed after nanopore sequencing.
There are further efforts to improve base-calling accuracy. In experimental design optimization, ONT has introduced a new sequencing method called 1D2 to sequentially sequence the cDNA strand after one strand has been sequenced, but without requiring ligation of a hairpin in the older ONT 2D technology, which also sequences both forward- and reverse-strand fragments but they were ligated with a hairpin adapter to sequentially sequence forward and reverse genomic fragments. In algorithm ways, Jordi Silvestre-Ryan and Ian Holmes designed a free computational approach, named Bonito base-caller.36 It allows users to use their data in a training module to generate custom models through CTC training of deep neural networks.36 With 1D2 and Bonito algorithm, the base-calling accuracy may get to 98.1%.36 Table 1 shows the tools and algorithms developed for base-calling.
Table 1.
Tools and algorthms developed for basecalling
| Tool | Description | Algorithm | Advantages | Rate | Disadvantages | Link | Reference (PMID) |
|---|---|---|---|---|---|---|---|
| Chiron | Basecalling | deep learning | no segmentation | 2000 (bp/s) | Not suitable for large genomes | https://github.com/haotianteng/chiron | 29648610 |
| Causalcall | Basecalling | Temporal Convolutional Network | directly identifies base sequences of varying lengths | 7000 (bp/s) | base deletions | https://github.com/scutbioinformatic/causalcall | 32038706 |
| URnano | Basecalling | deep neural networks | model sequential dependencies for a one-dimensional segmentation task | 3600 (bp/s) | segmentation | https://github.com/yaozhong/URnano | 32321433 |
| DeepNano | Basecalling | Deep recurrent neural networks | open-source | 1250 (bp/s) | Not suitable for large genomes | https://github.com/jeammimi/deepnano | 28582401 |
| BasecRAWller | Basecalling | unidirectional recurrent neural networks | 1) streaming basecalling, 2) tunable ratio of insertions to deletions, and 3) potential for streaming detection of modified bases | 200 (bp/s) | non-detectable covalently modified bases | https://github.com/rrwick/Basecalling-comparison | |
| Halcyon | Basecalling | Convolutional Neural Network and recurrent neural network | no segmentation and semantic correspondence | 250 (bp/s) | decrease speed | https://github.com/relastle/halcyon | 33165508 |
| CMOS | nanopore sensors | complementary metal-oxide-semiconductor | - | - | - | - | 28269559 |
| FPGA | nanopore sensors | Field-programmable gate array | - | - | - | - | 31825872 |
| Nanocall | Basecalling | Hidden Markov Model | offline, free and private | 700 (bp/s) | not currently integrate '2D' read | https://github.com/mateidavid/nanocall | 27614348 |
| Guppy | Basecalling | taxon-specific dataset and neural network model | reduction of errors in methylation motifs and no segmentation | 120,000 (bp/s) | a custom model using a larger neural network and/or training data from the same species | https://community.nanoporetech.com | 31234903 |
| PoreOver | Basecalling | CTC-trained neural network and hidden Markov models | compatible with multiple nanopore basecallers | 450 (bp/s) | not currently integrate '2D' read | https://github.com/jordisr/poreover | 33468205 |
Tools and algorithms developed for alignment
Recent advances in nanopore sequencing technologies promise ultra-long-reads with N50 >100 kb and read lengths up to 882 kb,48 which enables production of full-length mRNA reads for direct RNA sequencing (DRS).49,50 It requires new alignment algorithms to process such long-reads. Unlike seed-and-extend algorithms in traditional alignment—e.g., BLAST and LAST use adaptive seeds, whose matches are chosen based on their rareness—these tools use fixed-length matches, thereby guaranteeing the number of matches of a given sequence length, which achieves fast and sensitive comparison of ultra-long sequences.51 Nevertheless, the runtime of LAST will increase linearly with sequence length. In addition, for those quite short DNA reads, LAST may also take a longer time than those with slightly long-reads for the same initial data. GraphMap is designed to handle high-error Nanopore reads, which progressively refine candidate alignments with a fast graph traversal to align long-reads with speed and high precision (>95%), but may need more memory resources.52 Currently, Minimap2 is a popular tool for long-read alignment. It does split-read alignment (aka chimeric read alignment), employs concave gap cost for long insertions and deletions, and introduces new heuristics to reduce spurious alignments, making the tool 30 times faster than other long-read genomic or cDNA mappers, while obtaining higher accuracy.53
To quickly find unknown DNA fragments in metagenomic sequencing data, ONT devices provide a unique solution for real-time targeted sequencing known as ReadUntil. It allows nanopore devices to selectively eject reads from pores in real time via real-time alignment while sequencing (bear in mind that Nanopore reads are typically quite long). By precluding known genomic sequences, could result in the sequencing of purely unknown genomic sequences. Kovaka et al.54 developed the UNCALLED tool, an open-source mapper that probabilistically considers k-mers that could be represented by the signal, and then prunes the candidates based on the reference encoded within a Ferragina-Manzini index. The UNCALLED alignment tool can selectively sequence and analyze targeted genomic data in metagenomic samples, also have structural variants or DNA modification information, which would be lost with conventional targeted sequencing methods.
Tools and algorithms developed for genome assembly
Genome assembly is one of the most important research topics in genome biology. Long-read sequencing can significantly improve the assembly accuracy of assembled genomes.55, 56, 57, 58 It will also play an important role in solving the assembly of (peri)centromeric genomic regions full of transposable elements (TEs) and centromeric satellites.58, 59, 60 For instance, with N50 read length around 100 Kb, ONT DDS super-long-reads can readthrough genomic repeat regions unsolvable in short-read sequencing.48,56, 57, 58 Long-reads produced from third-generation sequencing platforms, e.g., Pacific Bioscience and Oxford Nanopore Technologies, are not only long but also have relatively high error rates, which require a genome assembly strategy that differs from previous algorithms.61 However, most of at least 97 assemblers (https://bioinformaticshome.com/tools/wga/wga.html) are only suitable for NGS short-reads. By far, the relatively low base-calling accuracy of ONT DDS still needs improved algorithms and elegant strategies.62, 63, 64 Moreover, the error distribution of nanopore sequencing is more complicated than that of PacBio sequencing. The higher-error sub-sequences from ONT DDS reads are widely distributed, and sometimes even reached a 50% error rate in 1,000-bp length reads with earlier base-calling tools.65 Studies also showed that the longer the sequencing read, the more high-error subsequence regions.65, 66, 67 It limited the application of ONT DDS in complex genomes in its early developmental stages.68 Nevertheless, with optimized algorithms, the platform could still be used to re-assemble genomes relatively accurately.65 For example, Chen et al. propose an adaptive read selection method to quickly correct nanopore reads with more accuracy for assembly, the corrected ratio for low-quality reads can reach 45.85% to 99.34% of low-quality reads.65 In addition, Flye was developed by Kolmogorov et al., although the human genome assembled by Flye has a rather high error rate (1.2% for the Flye HUMAN assembly), the reduction of error rate with an order of magnitude can be reached via polishing with Illumina reads.69
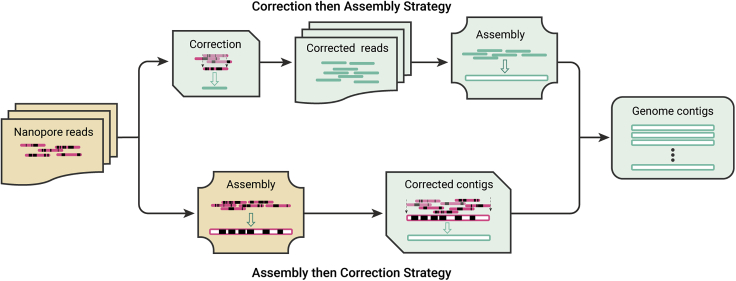
Currently, there are two strategies for long-read genome assembly of nanopore sequencing, namely “correct-then-assemble” and “assemble-then-correct” (Figure 2). For example, Falcon,70 Canu,71, and NECAT,65 employ the correct-then-assemble strategy. They first correct errors in the reads before assembling them into genomes. In contrast, other tools adopt the “assemble-then-correct” strategy, such as MiniASM,72 Flye,69 wtdbg2,73 Shasta,74 Smartdenovo,75 and Raven.76 They directly assemble uncorrected reads into genomes, and then correct the assembled genomes. The “correct-then-assemble” strategy usually takes much computing time in error correction, thus has a slower overall assembly speed than the “assemble-then-correct” strategy. However, the direct assembly of uncorrected sequencing reads may result in unsolvable errors in the assembly, which leads to assembly failures in particularly complex regions of the genome,71,77 whereas, the “correct-then-assemble” strategy can achieve accurate assembly results, and more contiguous.70,71,77 To make distinctions between repeated fragments and alleles, which are both integral to complex genome assembly,70 nanopore sequencing technology must rely on sensitive alignment and/or efficient error correction.64 The “correct-then-assemble” algorithms can distinguish regions at a distinct percentage of sequence difference, NECAT at 1%, Canu at 3%, and Falcon at about 5%. An opposite example for genome assembly is the “assemble-then-correct” algorithm MiniASM. It lacks an error correction procedure before assembly can only tell repeated fragments from each other when the sequence difference reaches 13%. This leads to the fact that MiniASM can only recognize errors when assembling large genomes, and it achieves lower assembled genome continuity than any of NECAT, CANU, or Falcon.71 Therefore, if the sequenced genome is not large enough, using the “correct-then-assemble” strategy may more easily get an accurate and continuous genome.
Figure 2.
Two genome assembly strategies for ONT
Short solid strips stand for nanopore reads, the red one means error-prone reads and the green one means corrected reads; long hollow strips represent contigs, the red one means low reliable contig and the green one means high reliable contig; the black boxes in solid strips and hollow strips indicate errors.
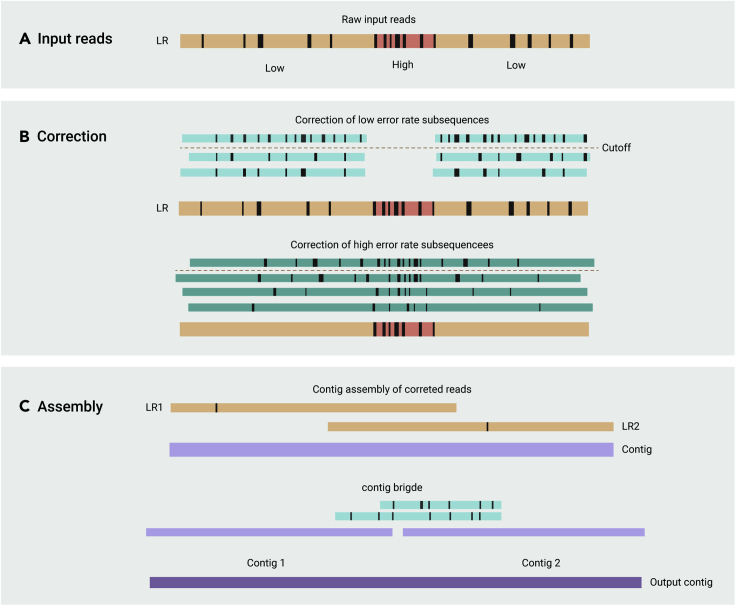
At present, most error-correcting tools of long-read-sequencing originate from those designed for the PacBio platform, have unsatisfactory performance in dealing with nanopore sequencing reads. For instance, CANU theoretically requires a CPU running 29 K hr to correct nanopore sequencing reads corresponding to 30X human genome coverage.48 Furthermore, these tools directly remove the high error regions in the datasets, significantly reducing the length and continuity of the final assembly. Therefore, assembly algorithms should be optimized for nanopore sequencing reads to improve their performance. NECAT reflects such kind of effort.65, Unlike other tools that iteratively correct reads, NECAT applies a two-step progressive method for error correction to deal with the complicated errors in nanopore sequencing reads. Briefly, it first corrects low-error-rate sub-sequences (LERS) of the reads and then corrects high-error-rate sub-sequences. In the assembly procedure, it uses a two-stage assembler (Figure 3B). First, the corrected nanopore reads are assembled into contigs (Figure 3A); next, the contigs are bridged by original raw reads to retain as many sequences for contig scaffolding, which may be lost in the error correction procedure. The progressive sequence correction and assembly strategy achieves 99% recovery of the local areas of ONT reads with high errors. Overall, NECAT not only efficiently solves the complex errors in nanopore sequencing but also ensures more reads can be used for genome assembly.
Figure 3.
NECAT two-step progressive correction and assembly
(A and B) Error correction for nanopore reads, and (C) assembly of nanopore reads. Pale yellow strips represent raw uncorrected reads; pink stripes represent the reads with corrected LERS. Green strips represent the reads with corrected RERS; the red stripes represent the error-prone reads that failed to be corrected. Purple strips represent the contigs. The block boxes in strips indicate errors and the white rectangle in strips is high-error-rate region. The black rectangle in pink strips means the high-error-rate region is shielded from correcting during first correction step. Dotted lines mean overlapping-error-rate threshold used for selecting supporting reads. The pale yellow box between two purple strips means contig bridge selected from raw reads. LR means the targeted long-read that would be corrected.
Tools and algorithms developed for small variants and structural variants
Despite the relatively low base-calling accuracy, ONT DDS reads show great performance in structural variant calling.78, 79, 80 Longshot81 utilizes an HMM to tackle the high error rate of nanopore long-reads, but it only detects SNPs. Clairvoyante was the first deep-learning–based variant-calling algorithm that supports both SNP and InDel calling using long-reads.78 Its successor, named Clair, has further improved the quality of called variants, especially SNPs.80,82 PEPPER83 phases the long-reads into two haplotype groups for calling, which effectively simplifies the long-read variant calling problem from calling both homozygous and heterozygous variants, to calling homozygous variants only, with further recombination of homozygous calls. In terms of structural variant (SV) calling, Sniffles82 works with the NGMLR aligner and uses a split-read algorithm on long-reads for accurate SV calling. SVIM84 detects five types of SVs (deletions, insertions, inversions, duplications, tandem duplications, and translocations) and is more sensitive but less accurate than Sniffles. CuteSV85 collects the signatures of various types of SVs and employs a clustering-and-refinement method to implement sensitive SV detection. NanoVar was designed to identify SVs with low-depth, long-read whole-genome sequencing data,86 while SENSV has further lowered the depth requirement to the throughput of a single MinION flow cell using algorithmic advancements.79 NanoVar and SENSV enabled cost-effective and comprehensive SV studies using ONT DDS, making it a more practical application for disease diagnosis in the future.
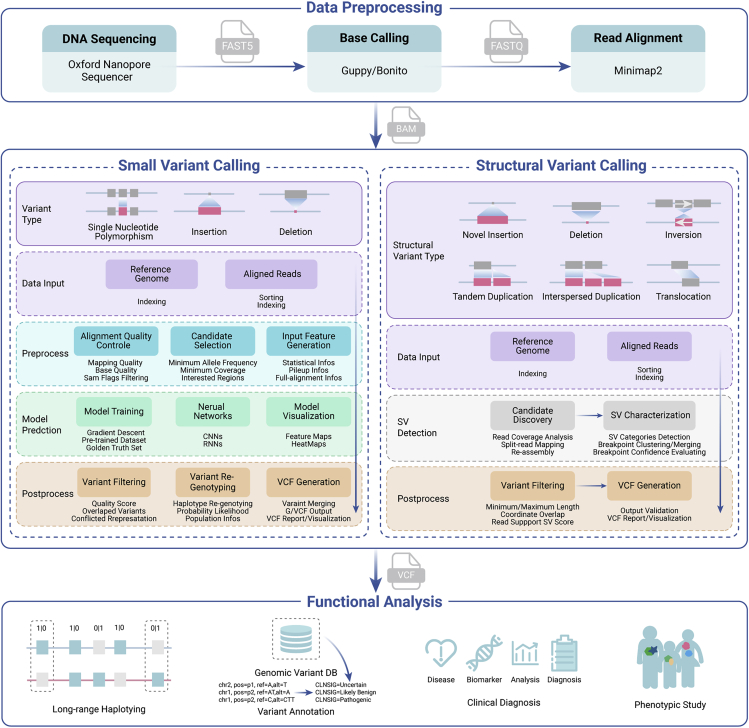
Computational solutions for SV identification should integrate more factors than those for base-calling, including split reads (SRs), depth of coverage, and local de novo assembly. Ultra-long ONT DDS reads, such as those produced by ONT DDS, can directly readthrough the entire region of SVs or have more reads covering the SV boundaries, but short-read sequencing generally does not have high confidence in the discovery of large structural variants due to limited reads from SV boundaries as evidence. Researchers have developed several tools to detect SVs in accordance with ONT DDS long-read traits (Figure 4). NanoSV, developed at the earlier stage, clusters SRs to identify SV breakpoint junctions, and has successful applications in clinical and research scenarios.87 In addition to SRs, Picky also combines the seed-and-extend process to detect SVs with ONT DDS long-reads.88 Clairvoyante represents a successful attempt to use a deep-learning algorithm to find SVs, including small SVs, using less computational time.81 Currently, an analysis pipeline named CAMPHOR can identify deletions (≥100 bp), insertions (≥100 bp), inversions, and intra-chromosomal translocations.89
Figure 4.
Schematic flow chart of small variant and SV calling with ONT DDS data
The “data preprocessing” box shows the commonly used bioinformatics tools for preprocessing the ONT sequencing data. The “small variant calling” and “Structural variant calling” boxes show the essential steps their critical parameters. The “functional analysis” box shows the applications of the detected variants. SV, structural variation; DB, database; CLISIG, clinical significance.
Tools developed for methylation identification
Various types of DNA and RNA base modifications play crucial roles in fundamental biological processes. In DNA, 5-methylcytosine (5mC) contributes to maintain genome integrity and stability,90,91 regulate pre-mRNA transcription initiation and processing, and even regulate poly(A) tail length,92 while N6-methyladenosine (m6A) modifications in mRNA could affect RNA splicing, degradation, and translation.93,94 NGS-based solutions, such as whole-genome bisulfite sequencing and methylated RNA immunoprecipitation sequencing, are widely implemented in the latest studies. Nevertheless, biases introduced in experimental procedures, particularly from the random fragmentation and DNA amplification, need to be further solved.95,96 ONT DDS and DRS sequence without converting bases and without amplification, so they could theoretically be used to directly detect DNA and RNA modifications when the trained model is sensitive enough (Figure 5).
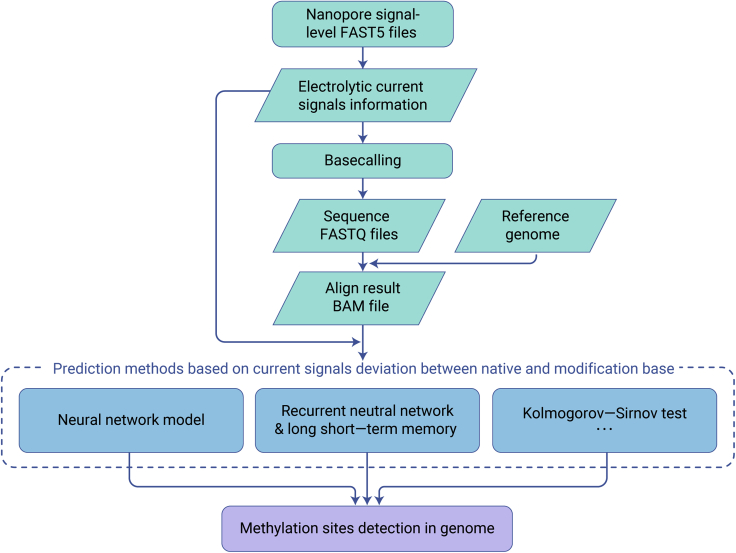
Figure 5.
The workflow of methylation identification for ONT DDS and DRS data
Raw electrolytic current signal files (FAST5) can be decoded to sequence information and electrolytic current signal information. Using the indexed electrolytic current signals and established detection models, the electrolytic current signals and sequence information can be mapped to the reference genome, then using the established detection models, we can detect methylation sites in the genome.
Several software packages are available to detect DNA and RNA modifications in nanopore sequencing data. Nanopolish97 includes an HHM trained from a 6-mer model of CpG motifs to differentiate between 5mC and unmethylated cytosine in the CpG context of ONT DDS reads of human genomic DNA.98 In the training data as initial input, mCaller utilizes four machine learning algorithms (neural network, random forest, logistic regression, and naive Bayes classifiers) with 6-mers around positions of interest to improve the accuracy of detection of m6A modifications.99 Researchers wonder if previous DNA methylation detection models may not fully utilize nanopore electric ion current signals, and Liu et al.95 trained a bidirectional RNN model with so-called long short-term memory in DeepMod. Its training data were based on bisulfite sequencing confirmed full methylated or unmethylated DNA, and it was evaluated with respect to the genomes of three different species, by comparing the DeepMod results with both naturally occurring and synthetically introduced modifications. Its average precision ranged from ∼0.9–0.99 for 5mC and N6-methyladenine (m6A, as opposed to 6mA) detection.95 In the same period, DeepSignal, another popular DNA modification detection tool, was released,100 and it is by far the best 5mC caller for human CpG modifications according to personal communication.100 Recently, the DeepSignal has a derived version, DeepSignal-plant, adjusted according to 5mC modifications in plants, which have CHG and CHH methylation in addition to CG methylation.101 However, these machine learning–based detection algorithms need prior training via extensive datasets. To address this limitation, some other tools examine the statistical difference between signals from native samples and from modification-free controls; for instance, NanoMod relies on the Kolmogorov-Smirnov test to compare raw ion current signals of sequenced samples with two sets of standard ion currents, and then judge whether the related bases are modified or not.102 NanoMod is perfect for Escherichia coli data analysis, but may not be suitable for methylation detection in eukaryotic cells.102
For RNA modification detection, there have been several widely used options. For instance, EpiNano achieves an overall accuracy of ∼90% in the identification of m6A RNA modification based on multiple trained support vector machines with m6A modified and unmodified synthetic sequencing as training data.103 ELIGOS (Epitranscriptional Landscape Inferring from Glitches of ONT Signals) detects ribonucleotide modification based on the error rates of specific bases, which are lower for unmodified RNA than for native RNA.104 Meanwhile, ELIGOS, taking various types of synthetic modified RNAs as training datasets, can be used for both rRNA and mRNA. Its accuracy reaches over 93% for the known classes of modifications in rRNA from E. coli, yeast, and human cell lines.104 Recently, nanom6A is released with an accuracy of 97%, higher than other algorithms. Its high accuracy was partially supported from the comparison between its results and those from methylated RNA immunoprecipitation sequencing and m6A-sensitive RNA-endoribonuclease–facilitated sequencing.105 The dysregulation of m6A in mRNAs may lead to tumorigenesis106; therefore, using ONT DRS to investigate the relationship between m6A modification and tumorigenesis will be clearer and easier. For instance, the inhibitor of METTL3-MTTL14–STM2457 specifically prevents m6A occurrence and thus can be used to treat acute myeloid leukemia (AML).107 Therefore, ONT DRS, can directly identify m6A modifications that contribute to AML.
Besides the tools developed for ONT DDS and DRS data analysis, ONT DRS data also contain poly(A) tail length information that can be used to evaluate mRNA stability and translation efficiency.108,109 Recently, poly(A) tail length has been confirmed to be positively correlated with DNA methylation around transcription termination sites.92 Currently, Nanopolish can be used to evaluate poly(A) tail length for each ONT DRS read.50,110 Tailfindr is another tool for evaluating poly(A) tail length.111 It uses an R package to estimate poly(A) tail length for unaligned, base-called data.
ONT DDS and its applications
The first application of ONT DDS was to sequence the model organism E. coli K-12 sub-strain MG1655 in 2014 with the R7.3 flow cell.112 Initially, however, nanopore sequencing did not get too much attention because of its very high error rate (around 30%).113 Since 2017, genome assembly using nanopore sequencing has been completed for diverse organisms, e.g., yeast,114 fish,115,116 plants,56,117, 118, 119 humans,48,120 fungi,121 Drosophila,122,123 and Caenorhabditis elegans.124 Recently, there is a rapid rise of ONT DDS for genome assembly. Over 100 publications in this field were published in 2020, and there have already been over 50 nanopore sequenced genomes reported in the first quarter of this year. The explosion of ONT DDS application is due to its recent progress, including significantly reduced cost and increased ability to provide sequence information at higher accuracy, benefiting from optimized base-calling and long-read alignment algorithms.53
One successful example for ONT DDS application is to profile centromeric DNA sequence and the related epigenetic traits in Arabidopsis.125 Centromeric DNA comprises satellite DNA with a 100- to 200-bp repeat sequence. Satellite DNA is highly variable in sequence and length among different species; therefore, it is very challenging to assemble centromeric DNA with traditional sequencing technologies. However, ONT DDS provides such a chance for de novo assembling of Arabidopsis centromeric DNA and to profile its DNA methylation. Although the Arabidopsis thaliana genome was sequenced in 2000, until recently the centromeric DNA reference was assembled with ONT DDS ultra-long-reads.125 It clearly shows that there are 66,129 centromeric satellite repeats with an approximately 180-bp sequence in the five chromosomal centromeric regions, and each chromosome possesses largely private satellite variants, higher-order CEN180 repeats are prevalent within centromeres. The investigators also found that ATHILA LTR retrotransposons interrupted centromeric genetic and epigenetic organization by invading the satellite array in centromeres. A clear picture of centromeres in Arabidopsis can be acquired mainly depending on ONT DDS.
Another example for ONT DDS application is to reprofile epigenetic patterns in a complete human genome with ONT DDS long-reads.126 In this study, the authors used T2T CHMI3 complete human genome data that were assembled with ONT DDS long-reads to further profile its epigenetic patterns, especially for those newly complete genomic regions (e.g., peri-centromeres, centromeres, acrocentric chromosome arms, sub-telomeres, segmental duplications, tandem repeats), and can distinguish epigenetically heterogeneous and homogeneous regions based on the clustered reads with methylation traits.126
An additional example of ONT DDS application is an adaptive nanopore sequencing for the golden lion tamarin (Leontopithecus rosalia) mitochondrial genome and epigenome.127 Adaptive sequencing with ONT DDS can enrich targeted reads and thus can increase the coverage of target DNA. Adaptive sequencing is particularly useful for environmental DNA, e.g., DNA from soil or feces of animals. Using ONT DDS adaptive sequencing, the authors acquired 258x coverage of the mitogenome of the closely related black lion tamarin (Leontopithecus chrysopygus) from feces of golden lion tamarin, which was successfully used to profile the mitogenome of golden lion tamarin. The differences between mitogenomes of the black lion tamarin and golden lion tamarin are 38 SNP, and in the golden lion tamarin mitogenome, some hydromethylation sites were also identified.127 For biodiversity preservation research, environmental DNA detection can be performed with ONT DDS adaptive sequencing.
Another application for ONT DDS is nanopore Cas9 targeted sequencing (nCATS), which combines the Cas9 nuclease to cut the targeted DNA from the genome, then uses an adaptor to ligate the enriched targeted DNA for ONT DDS.128 The strategy not only allows for the sequencing of genomic regions of interest for SNPs and SVs but can also acquire epigenomic information of enriched targeted genomic regions.128 In terms of using ONT DDS for small variant detection, several variant callers have been developed that could be used to provide therapeutic targets for the timely treatment of diseases related to such variation.78, 79, 80 Clinical diagnosis using both small variants (SNPs and InDels) and SVs can be a precise and effective strategy, especially for therapeutic target identification, and monitoring of hematological malignancies, etc.129
For ONT DDS, DNA quality is crucial for successful sequencing, and current commercial genomic DNA extraction kits are not enough for ONT DDS library preparation because the purity and length of ONT DDS required is higher than the standards of commercial kits. Because the ONT company has some mature protocol, readers can follow the protocol the company provides to obtain qualified genomic samples (https://community.nanoporetech.com/extraction_methods). Currently, a single MinION flow cell can produce up to 50 Gb of data if the library quality is high enough. For beginners, the main problem is how to obtain high-quality genomic DNA for ONT DDS. Currently, Oxford Nanopore provides different protocols for extracting high-quality genomic DNA for different species (https://community.nanoporetech.com/extraction_methods). To download the related protocols, interested parties need to register with the Nanopore community. To obtain high-quality long genomic DNA and avoid contamination with short genomic DNA, researchers can use BluePippin (Sage Science, Beverly, MA) to get the defined sizes of genomic DNA for sequencing,130 or use the Short-Read Eliminator Kit (Ciculomics, Baltimore, MD) (SKU SS-100-101-01). Also note that for different applications, the strategy for ONT DDS library preparation is different. For example, for assembling an unknown genome, it is better to acquire the longest reads with the Ultra-Long DNA Sequencing Kit (Oxford Nanopore Technologies, Oxford, UK) (SQK-ULK001), so that each sequencing read can cover the largest possible genomic region. However, for those species with reference genomes, if you only want to learn about SVs and/or DNA modifications, it is better to fragment genomic DNA to 8 kb with g-Tube (Covaris, Woburn, MA) (Cat# 520079), so that you can retrieve as much data as possible for downstream analysis. Readers can find more related information from the following web page: https://community.nanoporetech.com/. Published tools for alignment (genome assembly, structural variants, and methylation detection) can be found in Table 2.
Table 2.
Tools developed for analysis of Nanopore sequencing data
| Tool | Description | Algorithm | Advantages | Rate | Disadvantages | Link | Reference (PMID) |
|---|---|---|---|---|---|---|---|
| LAST | Alignment | adaptive seeds | Adaptive seeds are matches that are chosen based on their rareness, instead of using fixed-length matches | - | the running time increases linearly with sequence length and short DNA reads | https://gitlab.com/mcfrith/last | 21209072 |
| Minimap2 | Alignment | split-read alignment | DNA or long mRNA, higher accuracy, faster, and full length of reads | - | not suitable for chimeric alignments | https://github.com/lh3/minimap2 | 29750242 |
| GraphMap | Alignment | candidate alignments and fast graph traversal | long reads with speed, high sensitivity | - | large-memory | https://github.com/isovic/graphmap | 27079541 |
| UNCALLED | Alignment | Ferragina-Manzini index | mapping during sequencing and the leftmost mapping | - | not full length | https://github.com/skovaka/UNCALLED | 33257863 |
| tailfindr | poly(A) | measures poly(A) tail length | - | - | - | https://github.com/adnaniazi/tailfindr | 31266821, 33835460 |
| NaS | Assembly | illumina hybrid | entirely and with no error | - | Not suitable for large genomes | https://www.genoscope.cns.fr/externe/nas/ | 25927464 |
| LQS | Assembly | multiple-alignment corrected | corrected by a multiple-alignment and 99.5% nucleotide identity | - | Not suitable for large genomes | https://github.com/jts/nanopore-paper-analysis | 26076426 |
| Canu | Assembly | tf-idf weighted MinHash and graph construction | halves depth-of-coverage requirements, improves assembly continuity and reduces runtime on large genomes | - | accuracy depends on signal-level polishing | https://github.com/marbl/canu | 28298431 |
| Miniasm | Assembly | No correction | magnitude faster | - | error rate is as high as raw reads | https://github.com/lh3/miniasm | 27153593 |
| Nanopolish | Variant caller/Methylation detection | Hidden Markov Model | calculate an improved consensus sequence for a draft genome assembly, detect base modifications, call SNPs and indels | - | signal-level analysis | https://github.com/jts/nanopolish | 26076426 |
| Clairvoyante | Variant caller/SV caller | convolutional neural network | SV calling, small variants and genotype | - | higher sequencing depth | https://github.com/aquaskyline/Clairvoyante | 30824707 |
| Clair | Variant caller | Deep neural network | faster and complex variants with multiple alternative alleles | - | accuracy depends on pileup data and greater computational demands | https://github.com/quay/clair | |
| NanoSV | SV caller | split- and gapped-aligned reads | genotyping | - | non-detectable inversion, complex repeat regions and segmental duplications | https://github.com/mroosmalen/nanosv | 29109544 |
| Picky | SV caller | seed-and-extend process and split-read | micro-insertions and phased SV | - | high specificity | https://github.com/TheJacksonLaboratory/Picky | 29713081 |
| NanoVar | SV caller | artificial neural network | low-depth (8X) | - | the alignment profile of each read requires re-training | https://github.com/benoukraflab/nanovar | 32127024 |
| SENSV | SV caller | Deep neural network | low-depth | - | balanced translocation missed | https://github.com/HKU-BAL/SENSV | |
| CAMPHOR | SV caller | SV breakpoints | polymorphic SVs and somatic SVs | - | removed indels in short repeats, the average read length 5 kbps and non-detectable indels < 100 bp | https://github.com/afujimoto/CAMPHOR | 33910608 |
| NanoMod | Methylation detection | signal intensities | raw signal data and 5mC | - | two pair sample reads | https://github.com/WGLab/NanoMod | 30712508 |
| DeepSignal | Methylation detection | deep learning | 6mA/5mC, lower coverage, and predict methylation states | - | train DeepSignal to detect more types of base modification | https://github.com/bioinfomaticsCSU/deepsignal | 30994904 |
| mCaller | Methylation detection | neural network | 6mA and detect known or confirm suspected methyltransferase target motifs | - | only bacteria genome | https://github.com/al-mcintyre/mCaller_analysis_scripts | 30718479 |
| DeepMod | Methylation detection | recurrent neural network | 6mA/5mC,strand-sensitive and has single-base resolution | - | non-detectable other types of modifications or other different motifs, not suitable for RNA, neighboring bases inflence, elied on alignment tool to find correct reference positions of bases | https://github.com/WGLab/DeepMod | 31164644 |
| MINES | Methylation detection | random forest | m6A sites within DRACH motifs | - | lost small difference modification sites and not suitable for DNA | https://github.com/YeoLab/MINES | 31624092 |
| Nanom6A | Methylation detection | XGBoost model | m6A at single-base resolution and quantified abundance of m6A sites | - | not suitable for DNA | https://github.com/gaoyubang/nanom6A | 33413586 |
| FLAIR | Isoform detection | correct and realign | assessing 3′ poly(A) tail length, base modifications, and transcript haplotypes | - | combined short Illumina reads | https://github.com/BrooksLabUCSC/flair | 31740818 |
| TrackCluster | Isoform detection | read tracks | read classification, a transcript isoform with numerous exons, stage-specific or cell-specific expression of isoforms | - | not suitable for large genomes | https://github.com/Runsheng/trackcluster | 32024662 |
ONT DRS and its applications
Another advantage is its unrestricted read length, which breaks the limit for current commercial reverse transcription kits. ONT DRS has confirmed its ability to sequence super-long RNA molecules,49,50 after it was first demonstrated for native RNA in 2018.131 Using this technology, Zhang et al.50 provided the new splicing patterns detected by sequenced native RNA reads with experimental evidence and corrected the annotation of Arabidopsis At4g17140 splicing sites. The transcripts from At4g17140 are ultra-long RNA molecules (longer than 13,000 nt). This is beyond the ability of common commercial reverse transcriptase kits, which can only convert RNA sequences of less than 12,000 nt to complete cDNA molecules. Another great example attesting to the advantages of ONT DRS comes from a novel gene activated in met1-3 (MET1) and ddcc (DRM1DRM2CMT2CMT3) quadruple mutants. This gene covers a 17.3-kb genomic region with eight small exons interspersed within the intergenic regions of several annotated genes.92
Very interestingly, as hinted at above, ONT DRS can also be used to correct previously assembled genomes. In Arabidopsis, transcripts from At2g40980 not only contain the previously annotated Araport11 transcripts (with no additional exons found in the second intron) but also contain around 20 transcripts from a cryptic exon within the second intron. That cryptic exon in the second intron had not been annotated in the Arabidopsis genome, but the original BAC clone and follow-up experimental evidence support the existence of the cryptic exon.50
Unlike traditional NGS, ONT DRS sequences full-length transcripts without converting RNA to cDNA, and it can directly capture alternative splicing events in 15% of human hereditary diseases and cancers.132 Currently, some rare diseases, e.g., myelodysplasia syndromes, were found to be caused by aberrant splicing because of spliceosome mutation in somatic cells.133 To effectively detect aberrant splicing relating to human diseases, a pipeline-FRASER for NGS RNA-seq and ONT DRS analysis was developed.134 Besides spliceosome mutation causing aberrant splicing in human diseases, some splicing site mutations also cause alternative splicing and related human diseases.135 Actually, the SpliceDisease database was published in 2012,136 and documents most common splice-related diseases. Using ONT DRS, we can find novel isoforms in a specific disease, which may be omitted by NGS RNA-seq. However, for ONT DRS application for aberrant splicing detection or novel-isoform discovery in a disease, researchers should keep in mind that rG4 structure in some transcripts may impede the sequencing because of the huge structures, while human mRNA, at least, in vitro, easily forms the rG4 structure if no lithium ion (Li+) is present.137 To better detect the related alternative splicing events in human diseases, the purified RNAs should be treated with Li+ before making an ONT DRS library.
With the development of more base modification detection tools, epi-transcriptomes of different species have been reported.49,96,103, 104, 105,138, 139, 140, 141, 142 It has been reported that m6A modifications may have some negative connections with readthrough transcripts, because m6A writer mutant vir-1 has more readthrough transcripts,49 whereas 5mC modifications also have a high correlation with mobile mRNAs because they harbor more 5mC modifications compared with total mRNAs.50 Previously, the Kragler lab143 confirmed that in graft junction-mobile methylated mRNAs TRANSLATIONALLY CONTROLLED TUMOR PROTEIN 1 (TCTP1) and HEAT SHOCK COGNATE PROTEIN 70.1 (HSC70.1), the mRNA transport of which is diminished in mutants deficient in 5mC mRNA methylation. Furthermore, with ONT DRS, m6A modifications in adenoviral transcripts have been identified. Modification of m6A particularly affects the splicing of viral transcripts, which was confirmed with the m6A writer mutant.140 Besides m6A and 5mC modifications, other kinds of modifications of mRNA also were reported using ONT DRS reads.144,145
For modification detection models, there are two strategies. One is to use differential error calling, e.g., ELIGOS uses the error rate of specific bases between native RNA sequencing data and cDNA sequencing data of the same sample.104,106 A similar strategy was used for m6A detection in the wild-type Arabidopsis ecotype col-0, by employing an m6A writer mutant (vir-1 mutant),49 and also with the normal and m6A writer mutant, for the differential ion current associated with adenoviral transcripts.140 The other strategy adopts trained models. The EpiNano algorithm was based entirely on synthetic fully methylated and unmethylated RNAs.103 Another example using this strategy is nanom6A,105 which also used the synthetic fully methylated and unmethylated RNAs to train its detection model. Another related approach for modification detection models is to use machine or deep learning. MINES (m6A identification using nanopore sequencing) is an example, which uses a random forest classifier with miCLIP m6A sites within DRACH motifs.96 NanoCompore was also developed based on a machine learning algorithm.146
Any modification in mRNA can be detected if there are perfectly related training data. Oxford Nanopore provides the Megalodon model training software (https://nanoporetech.github.io/megalodon/model_training.html), which uses the Taiyaki algorithm (https://github.com/nanoporetech/taiyaki) to build up modified base detection models with in vitro or in vivo training data.
ONT DRS can sequence native RNA, and its good application is to profile NAD-capped RNA (Figure 6). Recently, it has been found that mRNAs not only have 7-methylguanosine (m7G) caps, but also have other caps, e.g., NAD-capped mRNA in E. coli,147 yeast,148 humans,130 and Arabidopsis.149, 150, 151 Previously, to purify NAD-capped RNA, the NAD caps should be labeled with biotin via click chemistry. Then streptavidin beads would be used to enrich the biotin-labeled RNA molecules. Finally, after elution from streptavidin beads, the RNAs were used in NGS.152 This protocol has been used to prepare RNA from different organisms.130,147, 148, 149, 150,153 However, the big problem with the protocol is eluting RNA from the streptavidin beads, which introduces false-positive results because of its huge background. Another strategy for accurately identifying NAD-capped RNAs is to use the click chemistry to introduce an RNA adaptor to the NAD caps, then use biotin-labeled DNA, which is complementary to the synthetic RNA adaptor to enrich the NAD-capped RNA. The enriched NAD-capped RNAs were used for ONT DRS, because the NAD-capped RNA molecules can be identified according to their adaptor sequences.151,154 This strategy can greatly decrease the false-positive rate of NAD-capped RNA molecules, allowing researchers to learn the characteristics of NAD-capped RNA, e.g., whether there are differences in splicing patterns, poly(A) tail length, etc., between NAD-capped RNA and normal RNA molecules. The original protocol for labeling NAD-capped RNA was to use copper ions to catalyze click chemistry. Unfortunately, this splices a lot of RNA, and thus a huge input of RNA was required. To conquer the problem, SPAAC (strain-promoted azide-alkyne cycloaddition) was used to replace copper ions, which allowed for a great reduction in input RNA. However, during the development of the protocol, it was found that the ADPRC enzyme can catalyze m7G-capped RNA, although with very low efficiency.149 However, because m7G-capped RNA is far more abundant than NAD-capped RNA, it is possible that previously identified NAD-capped RNA from eukaryotic cells may have been contaminated with m7G-capped RNA, especially those from humans and yeast. One way of removing such contamination is to eliminate m7G RNA when performing the ADPRC reaction.149 Another way is to identify NAD-capped RNA from organelles or prokaryotic cells.154 One problem that still needed to be resolved was to add poly(A) tails to the RNA before preparing ONT DRS libraries, as Zhang et al. did.154
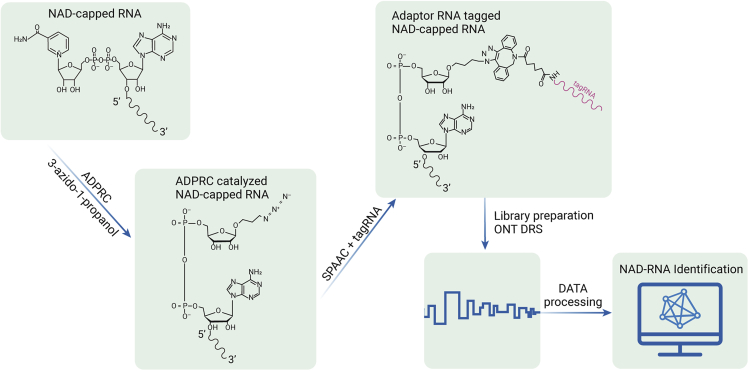
Figure 6.
Flowchart for ONT DRS of NAD-capped RNAs
The 5′and 3′ indicate the NAD-capped RNA direction. The NAD structure was illustrated and connected to the 5′ end of the NAD-capped RNA, after ADPRC catalysis, the azide moiety was linked to NAD via replacing nicotinamide, the azide contained NAD-capped RNA then can react with SPAAC, and the synthetic RNA adapter with DBCO at its 3′ end can conjugate with azide functionalized NAD-capped RNA. The tagged NAD-capped RNA then can be used for ONT DRS library preparation and further sequenced and analyzed. ADPRC, ADP-ribosyl cyclase; NAD, nicotinamide adenine dinucleotide; SPAAC, strain-promoted azide-alkyne cycloaddition; tagRNA, synthetic adaptor RNA to tag NAD-capped RNA; ONT DRS, Oxford Nanopore Technologies Direct RNA Sequencing.
Another elegant application for ONT DRS is denoting RNA secondary structure. The strategy is to label single-strand RNA bases, because the labeled bases will produce different ion currents that can be used to distinguish the positions of bases (in single- or double-stranded RNA).155,156 Stephenson et al.156 used acetylimidazole to exogenously label RNA bases, while Aw et al.155 used 2-methylnicotinic acid imidazolide azide, dimethyl sulfate, and 1-cyclohexyl-3-(2-morpholinoethyl) carbodiimide metho-p-toluenesulfonate to label RNA bases. But both papers successfully used ONT DRS to detect labeled bases and to decipher RNA structures.
For ONT DRS, to get full-length RNA reads, only RNA primary structure should remain when preparing ONT DRS libraries. Reverse transcription (RT) can destroy the secondary (and higher) structure of most RNA molecules; this is the reason that RT is necessary for preparing ONT DRS libraries, although cDNA is never used for sequencing. However, RNAs may also have G-quadruplex (G4, but rG4 when referring to RNA) secondary structures,157,158 which can cause reverse transcriptase stalling.137 It has been thought the rG4 structures are globally unfolded in eukaryotic cells,159 but RNA can quickly form rG4 structure in vivo in Na+ or K+ solution in vitro,159 while Li+ can eliminate rG4 structures.137,159 In Arabidopsis, rG4 structures are common in mRNA.158 ONT DRS library preparation with and without Li+ treatment has different average RNA molecular lengths.50
ONT DRS reads also can be used to detect poly(A) tail length.49,50,110,160,161 Poly(A) tails are homopolymers and proximal to the sequencing adapters, which can both be used to evaluate poly(A) tail length according to the dwell time of the poly(A) bases.160 Using Nanopolish to estimate poly(A) tail length, we have found that TE transcripts have longer poly(A) tails compared with other transcripts, while transcripts from housekeeping genes tend to have shorter poly(A) tails.92
Challenges for ONT DDS and DRS
Although ONT DDS and DRS were quickly applied in different research fields and greatly enhanced our understanding of genome profiling, accuracy of base-calling is still a challenge. The problem is becoming more obvious for ONT DRS data because more than 100 base modifications of RNA molecules have been discovered.162 To accurately detect all modified bases remains an enormous challenge because any modification may require a trained model for accurate detection, especially the training data used from the completely unmethylated transcriptome and fully methylated transcriptome. Although the completely unmethylated transcriptome and fully methylated transcriptome are relatively easy to get via converting the transcriptome to cDNA, and then using cDNA for in vitro transcription with modified bases or normal bases to get these training data, some RNA modifications may make nanopore sequencing impossible, e.g., base glycosylation with oligosaccharide,163 which may completely block the nanopore. For such modifications, new strategies may need to be developed.
Currently, ONT DRS is mainly used for mRNA sequencing. For other RNA sequencing, e.g., rRNA or RNAs from prokaryotic cells, the length of RNA normally is longer than 100 nt; therefore, small RNA sequencing, even for tRNA sequencing, still is impossible. To sequence small RNA molecules, especially those less than 30 nt, molecule glue or RNA adapters should be developed to ligate small RNA into long chimeric RNAs for sequencing.
Another challenge may come from ONT DRS library preparation because the RNA used for an ONT DRS library needs to have a poly(A) tail. In the case of noncoding RNA and RNA from prokaryotic cells, a poly(A) “tailing” step needs to be performed before ONT DRS library preparation.168 For ONT DRS, an additional challenge is how to get full-length RNAs, because 15 to 50 nt at the 5 ′ end of mRNAs cannot be detected because of loss of control of the speed mediated by the motor protein.51 One way to resolve the problem is to ligate an RNA adapter to the 5 ′ end of the sequenced RNAs, as was done for NAD-capped RNA with the TagSeq method.151,154,164
The relatively higher base error rate, especially in the low-complexity genomic regions, has long prevented genetic testing and microbial detection from utilizing the full power of ONT DDS. However, the miniaturization of equipment and fast turnaround time provide hope that the two applications will become more approachable. With significant efforts made to improve the performance of small variant and structural variant detection in the past 3 years, the impact is likely imminent. Nevertheless, we should also be aware of the limitations that remain. While the sensitivity and accuracy of SNPs detected from ONT DDS have surpassed those from Illumina sequencing (both sensitivity and accuracy have reached over 99.5%83), InDel detection remains an unsolved problem (sensitivity around 60% and accuracy around 90%). Most of the incorrect and missing InDels are in low-complexity genomic regions, such as tandem repeats, repetitive elements, and MHC proteins.78 One should be cautious when drawing conclusions from ONT DDS–detected InDels in these regions, and not make any medical decisions from any ONT DDS–detected InDels unless the InDels can be validated with additional experiments. The per-base cost of ONT DDS is still a few times higher than short-read sequencing, resulting in lower depth coverage per sample by some early adopters of ONT DDS with the same budget as for short-reads. Although some structural variant detection algorithms are designed to cope with the lower depth of coverage, it imposes certain limitations, such as the type of structural variants that can be reliably detected.79 As ONT DDS is still in its early development and there is no one-size-fits-all solution for variant calling, one should obtain a deeper understanding of the algorithms and their limitations before using them, especially when patients are involved.
Future perspective
ONT nanopore not only represents a new sequencing technology, but also greatly alters our strategies for answering some basic biological questions, e.g., for identification of NAD-capped RNA,151,154 and detection of the secondary structure of mRNA.155,156 Based on the merits of ONT DDS and DRS, they may also be used as biomarkers for correctly identifying species with close relatives (e.g., for identification of authentic medicinal herbs), and distinguishing different tissues from the same species (e.g., transcripts from roots and leaves, respectively, may have different m6A modification patterns in their transcripts or for some genomic regions they may have different DNA methylation patterns). In addition, because of their low cost but high output, they can be used for profiling more (epi)genomes/(epi)transcriptomes of different species. They may be especially suitable for research on the effect of the environment on the regulation of gene expression.
We have discussed the limitations of using ONT DDS for small variant and structural variant calling; however, the most significant limitation remains its high base-calling error rate. It is noteworthy that the ONT DDS error rate has improved remarkably in recent years through better chemistries and better base-calling algorithms. ONT made the new R10.3 chemistry165 available early last year for public testing. With a longer barrel and dual reader head in each nanopore, the new chemistry has reduced the base error rate from ∼8% to ∼5%. The primary base-caller “Guppy,” made by ONT, has implemented a flip-flop model along with a few deep-learning techniques with support and feedback from a large community of ONT users. With the same set of testing data, the new model reduced the base error rate from ∼9% to ∼7%.36 The most recently released ONT base-caller, “Bonito” (https://github.com/nanoporetech/bonito), has shown promising results that further reduced the base error rate to ∼5%. Focusing on genomic regions of interest to increase sequencing coverage has also shown its potential in reducing errors for using ONT DDS in the clinical context. While the community keeps developing new variant calling methods to make the most out of the ONT data, we foresee that the advancements in sequencing chemistry, base-calling, and protocol will result in a decisive moment for ONT DDS/DRS to take over more applications that are currently dominated by short-read sequencing.
Acknowledgments
This work is supported by the Key-Areas Research and Development Program of Guangdong Province (2020B020220004), the Youth Innovation Promotion Association, Chinese Academy of Sciences (2017399), the Science and Technology Program of Guangzhou (202002030097), the Hong Kong Research Grants Council Area of Excellence Scheme (AoE/M-403/16), the ECS (27204518), and TRS of the HKSAR government (T21-705/20-N).
Author contributions
M.L. and S.Z. conceived and coordinated the review. S.X., W.L., C.X., R.L., M.L., and S.Z. wrote the manuscript. S.X., Z.Z., S. Z., and D.Z. drew the figures. All authors were involved in the preparation of the final manuscript.
Declaration of interests
The authors declare no competing interests.
Published Online: August 11, 2021
Contributor Information
Chuanle Xiao, Email: xiaochuanle@126.com.
Ruibang Luo, Email: rbluo@cs.hku.hk.
Ming Luo, Email: luoming@scbg.ac.cn.
Shoudong Zhang, Email: shoudongzhang@cuhk.edu.hk.
References
- 1.Lander E.S., Linton L.M., Birren B., et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 2.International Human Genome Sequencing C. Finishing the euchromatic sequence of the human genome. Nature. 2004;431:931–945. doi: 10.1038/nature03001. [DOI] [PubMed] [Google Scholar]
- 3.Barba M., Czosnek H., Hadidi A. Historical perspective, development and applications of next-generation sequencing in plant virology. Viruses. 2014;6:106–136. doi: 10.3390/v6010106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Deamer D., Akeson M., Branton D. Three decades of nanopore sequencing. Nat. Biotechnol. 2016;34:518–524. doi: 10.1038/nbt.3423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dunne K.A., Chaudhuri R.R., Rossiter A.E., et al. Sequencing a piece of history: complete genome sequence of the original Escherichia coli strain. Microb. Genom. 2017;3:mgen000106. doi: 10.1099/mgen.0.000106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Branton D., Deamer D.W., Marziali A., et al. The potential and challenges of nanopore sequencing. Nat. Biotechnol. 2008;26:1146–1153. doi: 10.1038/nbt.1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kasianowicz J.J., Brandin E., Branton D., Deamer D.W. Characterization of individual polynucleotide molecules using a membrane channel. Proc. Natl. Acad. Sci. U S A. 1996;93:13770–13773. doi: 10.1073/pnas.93.24.13770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bezrukov S.M., Kasianowicz J.J. The charge state of an ion channel controls neutral polymer entry into its pore. Eur. Biophys. J. 1997;26:471–476. doi: 10.1007/s002490050101. [DOI] [PubMed] [Google Scholar]
- 9.Braha O., Walker B., Cheley S., et al. Designed protein pores as components for biosensors. Chem. Biol. 1997;4:497–505. doi: 10.1016/s1074-5521(97)90321-5. [DOI] [PubMed] [Google Scholar]
- 10.Butler T.Z., Gundlach J.H., Troll M. Ionic current blockades from DNA and RNA molecules in the alpha-hemolysin nanopore. Biophys. J. 2007;93:3229–3240. doi: 10.1529/biophysj.107.107003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kasianowicz J.J., Burden D.L., Han L.C., et al. Genetically engineered metal ion binding sites on the outside of a Channel's transmembrane beta-barrel. Biophys. J. 1999;76:837–845. doi: 10.1016/S0006-3495(99)77247-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Henrickson S.E., Misakian M., Robertson B., et al. Driven DNA transport into an asymmetric nanometer-scale pore. Phys. Rev. Lett. 2000;85:3057–3060. doi: 10.1103/PhysRevLett.85.3057. [DOI] [PubMed] [Google Scholar]
- 13.Howorka S., Cheley S., Bayley H. Sequence-specific detection of individual DNA strands using engineered nanopores. Nat. Biotechnol. 2001;19:636–639. doi: 10.1038/90236. [DOI] [PubMed] [Google Scholar]
- 14.Kasianowicz J.J., Henrickson S.E., Weetall H.H., et al. Simultaneous multianalyte detection with a nanometer-scale pore. Anal. Chem. 2001;73:2268–2272. doi: 10.1021/ac000958c. [DOI] [PubMed] [Google Scholar]
- 15.Halverson K.M., Panchal R.G., Nguyen T.L., et al. Anthrax biosensor, protective antigen ion channel asymmetric blockade. J. Biol. Chem. 2005;280:34056–34062. doi: 10.1074/jbc.M507928200. [DOI] [PubMed] [Google Scholar]
- 16.Merzlyak P.G., Capistrano M.F., Valeva A., et al. Conductance and ion selectivity of a mesoscopic protein nanopore probed with cysteine scanning mutagenesis. Biophys. J. 2005;89:3059–3070. doi: 10.1529/biophysj.105.066472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hromada L.P., Nablo B.J., Kasianowicz J.J., et al. Single molecule measurements within individual membrane-bound ion channels using a polymer-based bilayer lipid membrane chip. Lab. Chip. 2008;8:602–608. doi: 10.1039/b716388f. [DOI] [PubMed] [Google Scholar]
- 18.Kasianowicz J.J., Robertson J.W., Chan E.R., et al. Nanoscopic porous sensors. Annu. Rev. Anal. Chem. (Palo Alto Calif.) 2008;1:737–766. doi: 10.1146/annurev.anchem.1.031207.112818. [DOI] [PubMed] [Google Scholar]
- 19.Nablo B.J., Halverson K.M., Robertson J.W., et al. Sizing the Bacillus anthracis PA63 channel with nonelectrolyte poly(ethylene glycols) Biophys. J. 2008;95:1157–1164. doi: 10.1529/biophysj.107.121715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reiner J.E., Balijepalli A., Robertson J.W., et al. The effects of diffusion on an exonuclease/nanopore-based DNA sequencing engine. J. Chem. Phys. 2012;137:214903. doi: 10.1063/1.4766363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Walker B., Krishnasastry M., Zorn L., et al. Assembly of the oligomeric membrane pore formed by Staphylococcal alpha-hemolysin examined by truncation mutagenesis. J. Biol. Chem. 1992;267:21782–21786. [PubMed] [Google Scholar]
- 22.Walker B., Krishnasastry M., Bayley H. Functional complementation of staphylococcal alpha-hemolysin fragments. Overlaps, nicks, and gaps in the glycine-rich loop. J. Biol. Chem. 1993;268:5285–5292. [PubMed] [Google Scholar]
- 23.Walker B., Bayley H. Restoration of pore-forming activity in staphylococcal alpha-hemolysin by targeted covalent modification. Protein Eng. 1995;8:491–495. doi: 10.1093/protein/8.5.491. [DOI] [PubMed] [Google Scholar]
- 24.Walker B., Bayley H. Key residues for membrane binding, oligomerization, and pore forming activity of staphylococcal alpha-hemolysin identified by cysteine scanning mutagenesis and targeted chemical modification. J. Biol. Chem. 1995;270:23065–23071. doi: 10.1074/jbc.270.39.23065. [DOI] [PubMed] [Google Scholar]
- 25.Song L., Hobaugh M.R., Shustak C., et al. Structure of staphylococcal alpha-hemolysin, a heptameric transmembrane pore. Science. 1996;274:1859–1866. doi: 10.1126/science.274.5294.1859. [DOI] [PubMed] [Google Scholar]
- 26.Cheley S., Malghani M.S., Song L., et al. Spontaneous oligomerization of a staphylococcal alpha-hemolysin conformationally constrained by removal of residues that form the transmembrane beta-barrel. Protein Eng. 1997;10:1433–1443. doi: 10.1093/protein/10.12.1433. [DOI] [PubMed] [Google Scholar]
- 27.Maglia G., Restrepo M.R., Mikhailova E., et al. Enhanced translocation of single DNA molecules through alpha-hemolysin nanopores by manipulation of internal charge. Proc. Natl. Acad. Sci. U S A. 2008;105:19720–19725. doi: 10.1073/pnas.0808296105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Japrung D., Henricus M., Li Q., et al. Urea facilitates the translocation of single-stranded DNA and RNA through the alpha-hemolysin nanopore. Biophys. J. 2010;98:1856–1863. doi: 10.1016/j.bpj.2009.12.4333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stoddart D., Heron A.J., Klingelhoefer J., et al. Nucleobase recognition in ssDNA at the central constriction of the alpha-hemolysin pore. Nano Lett. 2010;10:3633–3637. doi: 10.1021/nl101955a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lieberman K.R., Cherf G.M., Doody M.J., et al. Processive replication of single DNA molecules in a nanopore catalyzed by phi29 DNA polymerase. J. Am. Chem. Soc. 2010;132:17961–17972. doi: 10.1021/ja1087612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cherf G.M., Lieberman K.R., Rashid H., et al. Automated forward and reverse ratcheting of DNA in a nanopore at 5-A precision. Nat. Biotechnol. 2012;30:344–348. doi: 10.1038/nbt.2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Niederweis M., Ehrt S., Heinz C., et al. Cloning of the mspA gene encoding a porin from Mycobacterium smegmatis. Mol. Microbiol. 1999;33:933–945. doi: 10.1046/j.1365-2958.1999.01472.x. [DOI] [PubMed] [Google Scholar]
- 33.Laszlo A.H., Derrington I.M., Ross B.C., et al. Decoding long nanopore sequencing reads of natural DNA. Nat. Biotechnol. 2014;32:829–833. doi: 10.1038/nbt.2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Deamer D., Akeson M., Branton D. Author response to John Kasianowicz and Sergey Bezrukov. Nat. Biotechnol. 2016;34:482. doi: 10.1038/nbt.3561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Goyal P., Krasteva P.V., Van Gerven N., et al. Structural and mechanistic insights into the bacterial amyloid secretion channel CsgG. Nature. 2014;516:250–253. doi: 10.1038/nature13768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Silvestre-Ryan J., Holmes I. Pair consensus decoding improves accuracy of neural network basecallers for nanopore sequencing. Genome Biol. 2021;22:38. doi: 10.1186/s13059-020-02255-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Timp W., Comer J., Aksimentiev A. DNA base-calling from a nanopore using a Viterbi algorithm. Biophys. J. 2012;102:L37–L39. doi: 10.1016/j.bpj.2012.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Teng H., Cao M.D., Hall M.B., et al. Chiron: translating nanopore raw signal directly into nucleotide sequence using deep learning. Gigascience. 2018;7:giy037. doi: 10.1093/gigascience/giy037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zeng J., Cai H., Peng H., et al. Causalcall: nanopore basecalling using a temporal convolutional network. Front. Genet. 2019;10:1332. doi: 10.3389/fgene.2019.01332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang Y.Z., Akdemir A., Tremmel G., et al. Nanopore basecalling from a perspective of instance segmentation. BMC Bioinformatics. 2020;21:136. doi: 10.1186/s12859-020-3459-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Boza V., Brejova B., Vinar T. DeepNano: deep recurrent neural networks for base calling in MinION nanopore reads. PLoS One. 2017;12:e0178751. doi: 10.1371/journal.pone.0178751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stoiber M., Brown J. BasecRAWller: Streaming nanopore basecalling directly from raw signal. BioRxiv. 2017 doi: 10.1101/133058. [DOI] [Google Scholar]
- 43.Konishi H., Yamaguchi R., Yamaguchi K., et al. Halcyon: an accurate basecaller exploiting an encoder-decoder model with monotonic attention. Bioinformatics. 2020 doi: 10.1093/bioinformatics/btaa953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wick R.R., Judd L.M., Holt K.E. Performance of neural network basecalling tools for Oxford Nanopore sequencing. Genome Biol. 2019;20:129. doi: 10.1186/s13059-019-1727-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chengjie W., Junli Z., Magierowski S., et al. Embedded CMOS basecalling for nanopore DNA sequencing. Annu. Int. Conf. IEEE Eng. Med. Biol. Soc. 2016;2016:5745–5748. doi: 10.1109/EMBC.2016.7592032. [DOI] [PubMed] [Google Scholar]
- 46.Wu Z., Hammad K., Ghafar-Zadeh E., et al. FPGA-accelerated 3rd generation DNA sequencing. IEEE Trans. Biomed. Circuits Syst. 2020;14:65–74. doi: 10.1109/TBCAS.2019.2958049. [DOI] [PubMed] [Google Scholar]
- 47.David M., Dursi L.J., Yao D., et al. Nanocall: an open source basecaller for Oxford Nanopore sequencing data. Bioinformatics. 2017;33:49–55. doi: 10.1093/bioinformatics/btw569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jain M., Koren S., Miga K.H., et al. Nanopore sequencing and assembly of a human genome with ultra-long reads. Nat. Biotechnol. 2018;36:338–345. doi: 10.1038/nbt.4060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Parker M.T., Knop K., Sherwood A.V., et al. Nanopore direct RNA sequencing maps the complexity of Arabidopsis mRNA processing and m(6)A modification. Elife. 2020;9 doi: 10.7554/eLife.49658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang S., Li R., Zhang L., et al. New insights into Arabidopsis transcriptome complexity revealed by direct sequencing of native RNAs. Nucleic Acids Res. 2020;48:7700–7711. doi: 10.1093/nar/gkaa588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kielbasa S.M., Wan R., Sato K., et al. Adaptive seeds tame genomic sequence comparison. Genome Res. 2011;21:487–493. doi: 10.1101/gr.113985.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sovic I., Sikic M., Wilm A., et al. Fast and sensitive mapping of nanopore sequencing reads with GraphMap. Nat. Commun. 2016;7:11307. doi: 10.1038/ncomms11307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li H. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics. 2018;34:3094–3100. doi: 10.1093/bioinformatics/bty191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kovaka S., Fan Y., Ni B., et al. Targeted nanopore sequencing by real-time mapping of raw electrical signal with UNCALLED. Nat. Biotechnol. 2021;39:431–441. doi: 10.1038/s41587-020-0731-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Seo J.S., Rhie A., Kim J., et al. De novo assembly and phasing of a Korean human genome. Nature. 2016;538:243–247. doi: 10.1038/nature20098. [DOI] [PubMed] [Google Scholar]
- 56.Michael T.P., Jupe F., Bemm F., et al. High contiguity Arabidopsis thaliana genome assembly with a single nanopore flow cell. Nat. Commun. 2018;9:541. doi: 10.1038/s41467-018-03016-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kuderna L.F.K., Lizano E., Julia E., et al. Selective single molecule sequencing and assembly of a human Y chromosome of African origin. Nat. Commun. 2019;10:4. doi: 10.1038/s41467-018-07885-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jain M., Olsen H.E., Turner D.J., et al. Linear assembly of a human centromere on the Y chromosome. Nat. Biotechnol. 2018;36:321–323. doi: 10.1038/nbt.4109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Eichler E.E., Clark R.A., She X. An assessment of the sequence gaps: unfinished business in a finished human genome. Nat. Rev. Genet. 2004;5:345–354. doi: 10.1038/nrg1322. [DOI] [PubMed] [Google Scholar]
- 60.Fiddes I.T., Lodewijk G.A., Mooring M., et al. Human-specific NOTCH2NL genes affect notch signaling and cortical neurogenesis. Cell. 2018;173:1356–1369.e22. doi: 10.1016/j.cell.2018.03.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lin Y., Yuan J., Kolmogorov M., et al. Assembly of long error-prone reads using de Bruijn graphs. Proc. Natl. Acad. Sci. U S A. 2016;113:E8396–E8405. doi: 10.1073/pnas.1604560113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Weirather A., Cesare M., Wang Y., et al. Comprehensive comparison of Pacific Biosciences and Oxford nanopore technologies and their applications to transcriptome analysis. F1000Res. 2017 doi: 10.12688/f1000research.10571.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Eid J., Fehr A., Gray J., et al. Real-time DNA sequencing from single polymerase molecules. Science. 2009;323:133–138. doi: 10.1126/science.1162986. [DOI] [PubMed] [Google Scholar]
- 64.Schneider G.F., Dekker C. DNA sequencing with nanopores. Nat. Biotechnol. 2012;30:326–328. doi: 10.1038/nbt.2181. [DOI] [PubMed] [Google Scholar]
- 65.Chen Y., Nie F., Xie S.Q., et al. Efficient assembly of nanopore reads via highly accurate and intact error correction. Nat. Commun. 2021;12:60. doi: 10.1038/s41467-020-20236-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Magi A., Giusti B., Tattini L. Characterization of MinION nanopore data for resequencing analyses. Brief Bioinformatics. 2017;18:940–953. doi: 10.1093/bib/bbw077. [DOI] [PubMed] [Google Scholar]
- 67.Rang F.J., Kloosterman W.P., de Ridder J. From squiggle to basepair: computational approaches for improving nanopore sequencing read accuracy. Genome Biol. 2018;19:90. doi: 10.1186/s13059-018-1462-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Loman N.J., Quick J., Simpson J.T. A complete bacterial genome assembled de novo using only nanopore sequencing data. Nat. Methods. 2015;12:733–735. doi: 10.1038/nmeth.3444. [DOI] [PubMed] [Google Scholar]
- 69.Kolmogorov M., Yuan J., Lin Y., et al. Assembly of long, error-prone reads using repeat graphs. Nat. Biotechnol. 2019;37:540–546. doi: 10.1038/s41587-019-0072-8. [DOI] [PubMed] [Google Scholar]
- 70.Chin C.S., Peluso P., Sedlazeck F.J., et al. Phased diploid genome assembly with single-molecule real-time sequencing. Nat. Methods. 2016;13:1050–1054. doi: 10.1038/nmeth.4035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Koren S., Walenz B.P., Berlin K., et al. Canu: scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res. 2017;27:722–736. doi: 10.1101/gr.215087.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Li H. Minimap and miniasm: fast mapping and de novo assembly for noisy long sequences. Bioinformatics. 2016;32:2103–2110. doi: 10.1093/bioinformatics/btw152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ruan J., Li H. Fast and accurate long-read assembly with wtdbg2. Nat. Methods. 2020;17:155–158. doi: 10.1038/s41592-019-0669-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Shafin K., Pesout T., Lorig-Roach R., et al. Nanopore sequencing and the Shasta toolkit enable efficient de novo assembly of eleven human genomes. Nat. Biotechnol. 2020;38:1044–1053. doi: 10.1038/s41587-020-0503-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Liu H., Wu S., Li A., et al. SMARTdenovo: a de novo assembler using long noisy reads. Gigabyte. 2021;1:2021. doi: 10.46471/gigabyte.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Vaser R., Sikić M. Raven: a de novo genome assembler for long reads. BioRxiv. 2020 doi: 10.1101/2020.08.07.242461. [DOI] [Google Scholar]
- 77.Xiao C.L., Chen Y., Xie S.Q., et al. MECAT: fast mapping, error correction, and de novo assembly for single-molecule sequencing reads. Nat. Methods. 2017;14:1072–1074. doi: 10.1038/nmeth.4432. [DOI] [PubMed] [Google Scholar]
- 78.Luo R., Sedlazeck F.J., Lam T.W., et al. A multi-task convolutional deep neural network for variant calling in single molecule sequencing. Nat. Commun. 2019;10:998. doi: 10.1038/s41467-019-09025-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Leung H., Yu H., Zhang Y., et al. SENSV: detecting structural variations with precise breakpoints using low-depth WGS data from a single Oxford nanopore MinION flowcell. BioRxiv. 2021 doi: 10.1101/2021.04.20.440583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Luo R., Wong C., Wong Y., et al. Exploring the limit of using a deep neural network on pileup data for germline variant calling. Nat. Mach. Intell. 2020;2:8. [Google Scholar]
- 81.Edge P., Bansal V. Longshot enables accurate variant calling in diploid genomes from single-molecule long read sequencing. Nat. Commun. 2019;10:4660. doi: 10.1038/s41467-019-12493-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sedlazeck F.J., Rescheneder P., Smolka M., et al. Accurate detection of complex structural variations using single-molecule sequencing. Nat. Methods. 2018;15:461–468. doi: 10.1038/s41592-018-0001-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Shafin K., Pesout T., Chang P., et al. Haplotype-aware variant calling enables high accuracy in nanopore long-reads using deep neural networks. BioRxiv. 2021 doi: 10.1101/2021.03.04.433952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Heller D., Vingron M. SVIM: structural variant identification using mapped long reads. Bioinformatics. 2019;35:2907–2915. doi: 10.1093/bioinformatics/btz041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Jiang T., Liu Y., Jiang Y., et al. Long-read-based human genomic structural variation detection with cuteSV. Genome Biol. 2020;21:189. doi: 10.1186/s13059-020-02107-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Tham C.Y., Tirado-Magallanes R., Goh Y., et al. NanoVar: accurate characterization of patients' genomic structural variants using low-depth nanopore sequencing. Genome Biol. 2020;21:56. doi: 10.1186/s13059-020-01968-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Cretu Stancu M., van Roosmalen M.J., Renkens I., et al. Mapping and phasing of structural variation in patient genomes using nanopore sequencing. Nat. Commun. 2017;8:1326. doi: 10.1038/s41467-017-01343-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Gong L., Wong C.H., Cheng W.C., et al. Picky comprehensively detects high-resolution structural variants in nanopore long reads. Nat. Methods. 2018;15:455–460. doi: 10.1038/s41592-018-0002-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Fujimoto A., Wong J.H., Yoshii Y., et al. Whole-genome sequencing with long reads reveals complex structure and origin of structural variation in human genetic variations and somatic mutations in cancer. Genome Med. 2021;13:65. doi: 10.1186/s13073-021-00883-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Robertson K.D., Wolffe A.P. DNA methylation in health and disease. Nat. Rev. Genet. 2000;1:11–19. doi: 10.1038/35049533. [DOI] [PubMed] [Google Scholar]
- 91.Bergman Y., Cedar H. DNA methylation dynamics in health and disease. Nat. Struct. Mol. Biol. 2013;20:274–281. doi: 10.1038/nsmb.2518. [DOI] [PubMed] [Google Scholar]
- 92.Li Q., Chen S., Leung A., et al. DNA methylation affects pre-mRNA transcriptional initiation and processing in Arabidopsis. BioRxiv. 2021 doi: 10.1101/2021.04.29.441938. [DOI] [Google Scholar]
- 93.Cao G., Li H.B., Yin Z., et al. Recent advances in dynamic m6A RNA modification. Open Biol. 2016;6:160003. doi: 10.1098/rsob.160003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zhang H., Lang Z., Zhu J.K. Dynamics and function of DNA methylation in plants. Nat. Rev. Mol. Cell Biol. 2018;19:489–506. doi: 10.1038/s41580-018-0016-z. [DOI] [PubMed] [Google Scholar]
- 95.Liu Q., Fang L., Yu G., et al. Detection of DNA base modifications by deep recurrent neural network on Oxford Nanopore sequencing data. Nat. Commun. 2019;10:2449. doi: 10.1038/s41467-019-10168-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Lorenz D.A., Sathe S., Einstein J.M., et al. Direct RNA sequencing enables m(6)A detection in endogenous transcript isoforms at base-specific resolution. RNA. 2020;26:19–28. doi: 10.1261/rna.072785.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Quick J., Loman N.J., Duraffour S., et al. Real-time, portable genome sequencing for Ebola surveillance. Nature. 2016;530:228–232. doi: 10.1038/nature16996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Simpson J.T., Workman R.E., Zuzarte P.C., et al. Detecting DNA cytosine methylation using nanopore sequencing. Nat. Methods. 2017;14:407–410. doi: 10.1038/nmeth.4184. [DOI] [PubMed] [Google Scholar]
- 99.McIntyre A.B.R., Alexander N., Grigorev K., et al. Single-molecule sequencing detection of N6-methyladenine in microbial reference materials. Nat. Commun. 2019;10:579. doi: 10.1038/s41467-019-08289-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Ni P., Huang N., Zhang Z., et al. DeepSignal: detecting DNA methylation state from Nanopore sequencing reads using deep-learning. Bioinformatics. 2019;35:4586–4595. doi: 10.1093/bioinformatics/btz276. [DOI] [PubMed] [Google Scholar]
- 101.Ni P., Huang N., Fan N., et al. Genome-wide detection of cytosine methylations in plant from nanopore sequencing data using deep learning. BioRxiv. 2021 doi: 10.1101/2021.02.07.430077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Liu Q., Georgieva D.C., Egli D., et al. NanoMod: a computational tool to detect DNA modifications using Nanopore long-read sequencing data. BMC Genomics. 2019;20:78. doi: 10.1186/s12864-018-5372-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Liu H., Begik O., Lucas M.C., et al. Accurate detection of m(6)A RNA modifications in native RNA sequences. Nat. Commun. 2019;10:4079. doi: 10.1038/s41467-019-11713-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Jenjaroenpun P., Wongsurawat T., Wadley T.D., et al. Decoding the epitranscriptional landscape from native RNA sequences. Nucleic Acids Res. 2021;49:e7. doi: 10.1093/nar/gkaa620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Gao Y., Liu X., Wu B., et al. Quantitative profiling of N(6)-methyladenosine at single-base resolution in stem-differentiating xylem of Populus trichocarpa using Nanopore direct RNA sequencing. Genome Biol. 2021;22:22. doi: 10.1186/s13059-020-02241-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Gu C., Shi X., Dai C., et al. RNA m6A modification in cancers: molecular mechanisms and potential clinical applications. Innovation. 2020;1:100066. doi: 10.1016/j.xinn.2020.100066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Cully M. METTLing with RNA methylation in leukaemia. Nat. Rev. Drug Discov. 2021;20:423. doi: 10.1038/d41573-021-00071-1. [DOI] [PubMed] [Google Scholar]
- 108.Lima S.A., Chipman L.B., Nicholson A.L., et al. Short poly(A) tails are a conserved feature of highly expressed genes. Nat. Struct. Mol. Biol. 2017;24:1057–1063. doi: 10.1038/nsmb.3499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Zlotorynski E. RNA metabolism: the short tail that wags the mRNA. Nat. Rev. Mol. Cell Biol. 2018;19:2–3. doi: 10.1038/nrm.2017.120. [DOI] [PubMed] [Google Scholar]
- 110.Li R., Ren X., Ding Q., et al. Direct full-length RNA sequencing reveals unexpected transcriptome complexity during Caenorhabditis elegans development. Genome Res. 2020;30:287–298. doi: 10.1101/gr.251512.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Krause M., Niazi A.M., Labun K., et al. tailfindr: alignment-free poly(A) length measurement for Oxford Nanopore RNA and DNA sequencing. RNA. 2019;25:1229–1241. doi: 10.1261/rna.071332.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Quick J., Quinlan A.R., Loman N.J. A reference bacterial genome dataset generated on the MinION portable single-molecule nanopore sequencer. Gigascience. 2014;3:22. doi: 10.1186/2047-217X-3-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Madoui M.A., Engelen S., Cruaud C., et al. Genome assembly using Nanopore-guided long and error-free DNA reads. BMC Genomics. 2015;16:327. doi: 10.1186/s12864-015-1519-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Liem M., Jansen H.J., Dirks R.P., et al. De novo whole-genome assembly of a wild type yeast isolate using nanopore sequencing. F1000Res. 2017;6:618. doi: 10.12688/f1000research.11146.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Jansen H.J., Liem M., Jong-Raadsen S.A., et al. Rapid de novo assembly of the European eel genome from nanopore sequencing reads. Sci. Rep. 2017;7:7213. doi: 10.1038/s41598-017-07650-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Bian L., Li F., Ge J., et al. Chromosome-level genome assembly of the greenfin horse-faced filefish (Thamnaconus septentrionalis) using Oxford Nanopore PromethION sequencing and Hi-C technology. Mol. Ecol. Resour. 2020;20:1069–1079. doi: 10.1111/1755-0998.13183. [DOI] [PubMed] [Google Scholar]
- 117.Hoang P.N.T., Michael T.P., Gilbert S., et al. Generating a high-confidence reference genome map of the Greater Duckweed by integration of cytogenomic, optical mapping, and Oxford Nanopore technologies. Plant J. 2018;96:670–684. doi: 10.1111/tpj.14049. [DOI] [PubMed] [Google Scholar]
- 118.Mondal T.K., Rawal H.C., Gaikwad K., et al. First de novo draft genome sequence of Oryza coarctata, the only halophytic species in the genus Oryza. F1000Res. 2017;6:1750. doi: 10.12688/f1000research.12414.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Deschamps S., Zhang Y., Llaca V., et al. A chromosome-scale assembly of the sorghum genome using nanopore sequencing and optical mapping. Nat. Commun. 2018;9:4844. doi: 10.1038/s41467-018-07271-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Cai R., Dong Y., Fang M., et al. De novo genome assembly of a Han Chinese male and genome-wide detection of structural variants using Oxford Nanopore sequencing. Mol. Genet. Genomics. 2020;295:871–876. doi: 10.1007/s00438-020-01672-y. [DOI] [PubMed] [Google Scholar]
- 121.Luo R., Zimin A., Workman R., et al. First draft genome sequence of the pathogenic fungus lomentospora prolificans (formerly Scedosporium prolificans) G3 (Bethesda) 2017;7:3831–3836. doi: 10.1534/g3.117.300107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Solares E.A., Chakraborty M., Miller D.E., et al. Rapid low-cost assembly of the Drosophila melanogaster reference genome using low-coverage, long-read sequencing. G3 (Bethesda) 2018;8:3143–3154. doi: 10.1534/g3.118.200162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Miller D.E., Staber C., Zeitlinger J., et al. Highly contiguous genome assemblies of 15 Drosophila species generated using nanopore sequencing. G3 (Bethesda) 2018;8:3131–3141. doi: 10.1534/g3.118.200160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Tyson J.R., O'Neil N.J., Jain M., et al. MinION-based long-read sequencing and assembly extends the Caenorhabditis elegans reference genome. Genome Res. 2018;28:266–274. doi: 10.1101/gr.221184.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Naish M., Alonge M., Wlodzimierz P., et al. The genetic and epigenetic landscape of the Arabidopsis centromeres. BioRxiv. 2021 doi: 10.1101/2021.05.30.446350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Gershman A., Sauria M., Hook P., et al. Epigenetic patterns in a complete human genome. BioRxiv. 2021 doi: 10.1101/2021.05.26.443420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Wanner N., Larsen P., McLain A., et al. The mitochondrial genome and epigenome of the golden lion tamarin from fecal DNA using nanopore adaptive sequencing. BioRxiv. 2021 doi: 10.1101/2021.05.27.446055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Gilpatrick T., Lee I., Graham J.E., et al. Targeted nanopore sequencing with Cas9-guided adapter ligation. Nat. Biotechnol. 2020;38:433–438. doi: 10.1038/s41587-020-0407-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Au C.H., Ho D.N., Ip B.B.K., et al. Rapid detection of chromosomal translocation and precise breakpoint characterization in acute myeloid leukemia by nanopore long-read sequencing. Cancer Genet. 2019;239:22–25. doi: 10.1016/j.cancergen.2019.08.005. [DOI] [PubMed] [Google Scholar]
- 130.Jiao X., Doamekpor S.K., Bird J.G., et al. 5' end nicotinamide adenine dinucleotide cap in human cells promotes RNA decay through DXO-mediated deNADding. Cell. 2017;168:1015–1027.e10. doi: 10.1016/j.cell.2017.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Garalde D.R., Snell E.A., Jachimowicz D., et al. Highly parallel direct RNA sequencing on an array of nanopores. Nat. Methods. 2018;15:201–206. doi: 10.1038/nmeth.4577. [DOI] [PubMed] [Google Scholar]
- 132.Jiang W., Chen L. Alternative splicing: human disease and quantitative analysis from high-throughput sequencing. Comput. Struct. Biotechnol. J. 2021;19:183–195. doi: 10.1016/j.csbj.2020.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Shiozawa Y., Malcovati L., Galli A., et al. Aberrant splicing and defective mRNA production induced by somatic spliceosome mutations in myelodysplasia. Nat. Commun. 2018;9:3649. doi: 10.1038/s41467-018-06063-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Mertes C., Scheller I.F., Yepez V.A., et al. Detection of aberrant splicing events in RNA-seq data using FRASER. Nat. Commun. 2021;12:529. doi: 10.1038/s41467-020-20573-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Zhang S., Wang J., Zhang A., et al. A SNP involved in alternative splicing of ABCB1 is associated with clopidogrel resistance in coronary heart disease in Chinese population. Aging (Albany NY) 2020;12:25684–25699. doi: 10.18632/aging.104177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Wang J., Zhang J., Li K., et al. SpliceDisease database: linking RNA splicing and disease. Nucleic Acids Res. 2012;40:D1055–D1059. doi: 10.1093/nar/gkr1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Kwok C.K., Marsico G., Sahakyan A.B., et al. rG4-seq reveals widespread formation of G-quadruplex structures in the human transcriptome. Nat. Methods. 2016;13:841–844. doi: 10.1038/nmeth.3965. [DOI] [PubMed] [Google Scholar]
- 138.Wang Y., Wang H., Xi F., et al. Profiling of circular RNA N(6) -methyladenosine in moso bamboo (Phyllostachys edulis) using nanopore-based direct RNA sequencing. J. Integr. Plant Biol. 2020;62:1823–1838. doi: 10.1111/jipb.13002. [DOI] [PubMed] [Google Scholar]
- 139.Viehweger A., Krautwurst S., Lamkiewicz K., et al. Direct RNA nanopore sequencing of full-length coronavirus genomes provides novel insights into structural variants and enables modification analysis. Genome Res. 2019;29:1545–1554. doi: 10.1101/gr.247064.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Price A.M., Hayer K.E., McIntyre A.B.R., et al. Direct RNA sequencing reveals m(6)A modifications on adenovirus RNA are necessary for efficient splicing. Nat. Commun. 2020;11:6016. doi: 10.1038/s41467-020-19787-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Furlan M., Tanaka I., Leonardi T., et al. Direct RNA sequencing for the study of synthesis, processing, and degradation of modified transcripts. Front. Genet. 2020;11:394. doi: 10.3389/fgene.2020.00394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Ding H., Bailey A.D., Jain M., et al. Gaussian mixture model-based unsupervised nucleotide modification number detection using nanopore-sequencing readouts. Bioinformatics. 2020;36:4928–4934. doi: 10.1093/bioinformatics/btaa601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Yang L., Perrera V., Saplaoura E., et al. m(5)C methylation guides systemic transport of messenger RNA over graft junctions in plants. Curr. Biol. 2019;29:2465–2476. doi: 10.1016/j.cub.2019.06.042. [DOI] [PubMed] [Google Scholar]
- 144.Ramasamy S., Sahayasheela V., Yu Z., et al. Chemical probe-based nanopore sequencing to selectively assess the RNA modifications. BioRxiv. 2021 doi: 10.1101/2020.05.19.105338. [DOI] [PubMed] [Google Scholar]
- 145.Hassan D., Acevedo D., Daulatabad S.V., et al. Penguin: a tool for predicting pseudouridine sites in direct RNA nanopore sequencing data. BioRxiv. 2021 doi: 10.1101/2021.03.31.437901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Leger A., Amaral P., Pandolfini L., et al. RNA modifications detection by comparative Nanopore direct RNA sequencing. BioRxiv. 2019 doi: 10.1101/843136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Cahova H., Winz M.L., Hofer K., et al. NAD captureSeq indicates NAD as a bacterial cap for a subset of regulatory RNAs. Nature. 2015;519:374–377. doi: 10.1038/nature14020. [DOI] [PubMed] [Google Scholar]
- 148.Walters R.W., Matheny T., Mizoue L.S., et al. Identification of NAD+ capped mRNAs in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U S A. 2017;114:480–485. doi: 10.1073/pnas.1619369114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Hu H., Flynn N., Zhang H., et al. SPAAC-NAD-seq, a sensitive and accurate method to profile NAD(+)-capped transcripts. Proc. Natl. Acad. Sci. U S A. 2021;118 doi: 10.1073/pnas.2025595118. e2025595118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Wang Y., Li S., Zhao Y., et al. NAD(+)-capped RNAs are widespread in the Arabidopsis transcriptome and can probably be translated. Proc. Natl. Acad. Sci. U S A. 2019;116:12094–12102. doi: 10.1073/pnas.1903682116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Zhang H., Zhong H., Zhang S., et al. NAD tagSeq reveals that NAD(+)-capped RNAs are mostly produced from a large number of protein-coding genes in Arabidopsis. Proc. Natl. Acad. Sci. U S A. 2019;116:12072–12077. doi: 10.1073/pnas.1903683116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Winz M.L., Cahova H., Nubel G., et al. Capture and sequencing of NAD-capped RNA sequences with NAD captureSeq. Nat. Protoc. 2017;12:122–149. doi: 10.1038/nprot.2016.163. [DOI] [PubMed] [Google Scholar]
- 153.Bird J.G., Basu U., Kuster D., et al. Highly efficient 5' capping of mitochondrial RNA with NAD(+) and NADH by yeast and human mitochondrial RNA polymerase. Elife. 2018;7:e42179. doi: 10.7554/eLife.42179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Zhang H., Zhong H., Wang X., et al. Use of NAD tagSeq II to identify growth phase-dependent alterations in E. coli RNA NAD(+) capping. Proc. Natl. Acad. Sci. U S A. 2021;118 doi: 10.1073/pnas.2026183118. e2026183118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Aw J.G.A., Lim S.W., Wang J.X., et al. Determination of isoform-specific RNA structure with nanopore long reads. Nat. Biotechnol. 2021;39:336–346. doi: 10.1038/s41587-020-0712-z. [DOI] [PubMed] [Google Scholar]
- 156.Stephenson W., Razaghi R., Busan S., et al. Direct detection of RNA modifications and structure using single molecule nanopore sequencing. BioRxiv. 2020 doi: 10.1101/2020.05.31.126763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Yang S.Y., Lejault P., Chevrier S., et al. Transcriptome-wide identification of transient RNA G-quadruplexes in human cells. Nat. Commun. 2018;9:4730. doi: 10.1038/s41467-018-07224-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Mullen M.A., Olson K.J., Dallaire P., et al. RNA G-Quadruplexes in the model plant species Arabidopsis thaliana: prevalence and possible functional roles. Nucleic Acids Res. 2010;38:8149–8163. doi: 10.1093/nar/gkq804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Guo J.U., Bartel D.P. RNA G-quadruplexes are globally unfolded in eukaryotic cells and depleted in bacteria. Science. 2016;353:aaf5371. doi: 10.1126/science.aaf5371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Workman R.E., Tang A.D., Tang P.S., et al. Nanopore native RNA sequencing of a human poly(A) transcriptome. Nat. Methods. 2019;16:1297–1305. doi: 10.1038/s41592-019-0617-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Roach N.P., Sadowski N., Alessi A.F., et al. The full-length transcriptome of C. elegans using direct RNA sequencing. Genome Res. 2020;30:299–312. doi: 10.1101/gr.251314.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Shen L., Liang Z., Wong C.E., et al. Messenger RNA modifications in plants. Trends Plant Sci. 2019;24:328–341. doi: 10.1016/j.tplants.2019.01.005. [DOI] [PubMed] [Google Scholar]
- 163.Flynn R., Pedram K., Malaker S., et al. Small RNAs are modified with N-glycans and displayed on the surface of living cells. Cell. 2021 doi: 10.1016/j.cell.2021.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Shao X., Zhang H., Yang Z., et al. NAD tagSeq for transcriptome-wide identification and characterization of NAD(+)-capped RNAs. Nat. Protoc. 2020;15:2813–2836. doi: 10.1038/s41596-020-0363-z. [DOI] [PubMed] [Google Scholar]
- 165.Karst S.M., Ziels R.M., Kirkegaard R.H., et al. High-accuracy long-read amplicon sequences using unique molecular identifiers with Nanopore or PacBio sequencing. Nat. Methods. 2021;18:165–169. doi: 10.1038/s41592-020-01041-y. [DOI] [PubMed] [Google Scholar]