Abstract
Schizosaccharomyces pombe pre-mRNAs are generally multi-intronic and share certain features with pre-mRNAs from Drosophila melanogaster, in which initial splice site pairing can occur via either exon or intron definition. Here, we present three lines of evidence suggesting that, despite these similarities, fission yeast splicing is most likely restricted to intron definition. First, mutating either or both splice sites flanking an internal exon in the S. pombe cdc2 gene produced almost exclusively intron retention, in contrast to the exon skipping observed in vertebrates. Second, we were unable to induce skipping of the internal microexon in fission yeast cgs2, whereas the default splicing pathway excludes extremely small exons in mammals. Because nearly quantitative removal of the downstream intron in cgs2 could be achieved by expanding the microexon, we propose that its retention is due to steric occlusion. Third, several cryptic 5′ junctions in the second intron of fission yeast cdc2 are located within the intron, in contrast to their generally exonic locations in metazoa. The effects of expanding and contracting this intron are as predicted by intron definition; in fact, even highly deviant 5′ junctions can compete effectively with the standard 5′ splice site if they are closer to the 3′ splicing signals. Taken together, our data suggest that pairing of splice sites in S. pombe most likely occurs exclusively across introns in a manner that favors excision of the smallest segment possible.
Splice site selection has been most extensively studied in higher eukaryotes (reviewed in reference 11), where abundant evidence indicates that the unit initially recognized by the splicing machinery is the exon, as proposed by Robberson et al. nearly a decade ago (53). Particularly compelling in this regard is the observation that the most common effect of a 5′ splice site mutation is skipping of the preceding exon rather than inclusion of the mutant intron (61; reviewed in reference 6). Moreover, in the subset of cases in which a 5′ junction mutation causes activation of a cryptic splice site rather than exon skipping, the new exon-intron boundary is almost invariably located within the preceding exon, again supporting the view that communication occurs across the exon rather than the intron. Finally, there are significant constraints on exon length in vertebrate pre-mRNAs, consistent with the proposal that the 3′ and 5′ splice sites on opposite sides of the exon must be recognized concurrently. Not only are the vast majority of natural internal exons in vertebrate pre-mRNAs <300 nucleotides in length (6), but expanding an exon beyond this size causes it to be skipped (53), particularly if it is surrounded by large introns (60). In contrast to the limitations on exon length, the introns in vertebrate pre-mRNAs can be extremely large (tens of kilobases [29]).
Although many questions remain to be answered, several components of the machinery responsible for exon definition have been identified. First, UV cross-linking experiments revealed that binding of the U1 snRNP to the downstream 5′ splice site stabilizes the association of U2AF65 with the polypyrimidine tract of the upstream intron (32). Likely candidates to form a bridge between these components were identified by protein-protein interaction assays, which indicated that the 70,000-Da protein of the U1 snRNP binds to members of the serine-arginine-rich (SR) family of splicing factors, which in turn bind to the small subunit of the U2AF heterodimer (5, 38, 70). The U1-70K/SR/U2AF35/U2AF65 network has also been proposed to play a role in communication across introns (70). However, because one of these components (U2AF35) is absent in Saccharomyces cerevisiae and two others (U2AF65 and SR proteins) are not highly conserved, this mode of connecting splice sites may not be ubiquitous (1, 2). A distinct network of intron-spanning interactions forms at an early stage of the splicing pathway in yeast and most likely in mammals as well (2, 7). In addition to this network, which extends from the large subunit of U2AF to the branchpoint bridging protein to a different component of the U1 snRNP, Prp40p, recent work with Drosophila melanogaster points to a third set of early intron-bridging interactions involving a divergent member of the SR protein family, SRp54 (36). The relationships among these networks of protein-protein interactions remain to be elucidated.
Extremely small exons also pose recognition problems for the vertebrate splicing machinery, leading to a default splicing pattern in which the microexon is skipped (e.g., 9, 18, 59). This phenomenon was originally proposed to result from steric interference between closely juxtaposed 3′ and 5′ splice sites (9), but it is now attributed primarily to a lack of positive interactions across the small exon (10, 13, 59). In the three examples studied most extensively, incorporation of the microexon is promoted by complex enhancer elements located in the downstream intron (10, 13, 66). In the case of c-src, it has been shown that a large assemblage of proteins, including hnRNP F (44), K-SRP (45), and hnRNP H (14), binds to the intronic enhancer and regulates microexon inclusion, possibly by promoting use of the abutting 5′ splice site.
In both budding yeast and fission yeast, as well as other unicellular eukaryotes, small introns predominate, and exon size does not appear to be constrained (15, 56, 72). These observations prompted Talerico and Berget (62) to propose that, in simple eukaryotes, the intron rather than the exon serves as the initial unit of recognition during spliceosome assembly (62; reviewed in reference 6). Consistent with this proposal, alternative exon usage has not yet been demonstrated in either yeast species. However, two well-documented instances of regulated splicing have been described in S. cerevisiae, both utilizing intron retention as an on-off switch for protein expression (19, 20). The situation is less clear in Schizosaccharomyces pombe, but a similar form of regulation at the level of splicing has been proposed for mes1 pre-mRNA during meiosis (37).
While small introns are also common in certain metazoa including Caenorhabditis elegans and D. melanogaster, these species contain large vertebratelike introns as well (22, 46). In the fruit fly, there is experimental evidence for initial splice site pairing via “intron definition,” since expansion of small introns leads either to their retention or to activation of a cryptic 3′ splice site (27, 62). On the other hand, several examples of exon skipping have been reported in Drosophila, both naturally occurring, as in the sex determination regulatory cascade (reviewed in reference 41) and experimentally induced (e.g., 47, 57), consistent with splice site pairing via exon definition. In S. cerevisiae, only a handful of pre-mRNAs harbor more than one intron (58), and the trans-acting factors implicated in exon-spanning interactions are either absent or highly divergent (1, 2, 8). In contrast, S. pombe contains all of the factors implicated in forming bridges between exons, including at least two canonical SR proteins and highly conserved homologs of both subunits of U2AF (26, 42, 49, 67). This fact, together with the ability of the Drosophila splicing machinery to utilize both the exon and intron definition modes, prompted us to ask whether communication can occur across exons in S. pombe.
To address this question, we first engineered constructs containing splice site mutations which, in mammals, would produce the outcome that is most diagnostic for this mode of initial splice site pairing, namely, exon skipping. In S. pombe, mutating the downstream 5′ splice site produced exclusively intron retention, and even in a pre-mRNA carrying severe mutations in both flanking splice sites, exon skipping was rare. To address the possibility that the lack of skipping was due to the large size of the internal exon, we turned to a different S. pombe pre-mRNA which contains a microexon. Again, the profile of products was as predicted by the intron definition model. A final indication that splice site pairing proceeds via intron definition in fission yeast is the location of several cryptic 5′ splice sites within an intron. The competition between these and the natural 5′ junction provided an opportunity to explore parameters that influence splice site pairing in S. pombe. In alleles containing deletions and insertions within the intron, as well as those with wild-type splice site spacing, we found that the pattern of cryptic splice site usage not only conformed to the predictions of the intron definition model but suggested that the fission yeast splicing machinery has a strong preference for excising the smallest intron possible.
MATERIALS AND METHODS
Plasmid construction and mutagenesis.
Construction and analysis of polypyrimidine tract variants of cdc2/pREP2, which carry the second intron of the S. pombe cdc2 gene together with flanking exon sequences under control of the nmt1 promoter, were described elsewhere (54). For the exon-skipping experiments reported here, the remainder of the third exon, as well as the third intron and the fourth exon, were incorporated into the BamHI sites of the wild-type, R-short, and R-long alleles as a PCR fragment amplified from a genomic clone (31) with Taq DNA polymerase, using the procedure suggested by the manufacturer (Gibco-BRL) (35) and the primers cdc2-Ex3-5′ and cdc2-Ex4-3′ (Table 1); these constructs are designated cdc2-Long. Site-directed mutagenesis to inactivate the 5′ splice site of intron 3 was carried out with reagents supplied commercially (Amersham Corp., Arlington Heights, Ill.), using the oligonucleotides cdc2-I3G1A, cdc2-I32nd5′SS, and cdc2I3Random (Table 1).
TABLE 1.
Oligonucleotides used for site-directed mutagenesis and PCR
| Primer | Oligonucleotidesa |
|---|---|
| cdc2-Ex3-5′ | 5′GCGGATCCGAAAAAATACATGGACCGA3′ (BamHI) |
| cdc2-Ex4-3′ | 5′CGGGATCCGATTTGTAATCTTGCAAAAGCG3′ (BamHI) |
| cdc2-I3G1A | 5′GAAAAAAAATATATCTCATGCGTATAGT3′ |
| cdc2-I32nd5′SS | 5′TTTGGGAATAAAGTSATTTAGCCAAGTAA3′ |
| cdc2I3Random | 5′GGAATAAAGTCATTTAGCCGCAGTCAGTCCTGTGTATTTCTCATGCGTATAGTTCCGC3′ |
| cgs2Int1-5′ | 5′TCCATAAGCCTACATATGCATGCAGCACTC3′ (NdeI) |
| cgs2Int1-3′ | 5′TCTTACATTGGATCCCTGTTTTCGTTAACG3′ (BamHI) |
| cgs2Int2-5′ | 5′CTAACGTTAACGACATATGGGACAAAATGTAAG3′ (NdeI) |
| cgs2Int2-3′ | 5′GGCTTTTGCGTAGGATCCATTTTCCACCG3′ (BamHI) |
| cgs2-Int2-5′pcr.mut1 | 5′GGCTCTCAATGCATAATGTTTGAATTCGATGATTTTGTCCCTGTTTTCG3′ |
| cgs2-Int2-5′pcr.mut2 | 5′GAATTCAAACATTATGCATTGAGAGCC3′ |
| cgs2-Δpcr.mut1 | 5′CTAATAAGTCAGCATGGCTCTCAAAAAAAATATTTTTCGTTTTTCATACC3′ |
| cgs2-Δpcr.mut2 | 5′TTGAGAGCCATGCTGACTTATTAG3′ |
| cgs2L/ex2Xho | 5′TGCATAATGTTTGAATTCTTACATCTCGAGCCTGTTTTCGTTAACGTTAGTCGA3′ (XhoI) |
| cgs2-22nI-5′ | 5′TCGATTGGATATTTTACATGCT3′ |
| cgs2-22nI-3′ | 5′TCGAAGCATGTAAAATATCCAA3′ |
| cgs2-49nI-5′ | 5′TCGATTGGATATTTTACATGCTGAATCAAAGTTGTATCTTGTTTTTGAG3′ |
| cgs2-49nI-3′ | 5′TCGACTCAAAAACAAGATACAACTTTGATTCAGCATGTAAA ATATCCAA3′ |
| Crypt2,4 | 5′CAATGTAAACWTACCCATGCATC3′ |
| Δ18U6X | 5′CAATGTAAACATTCCCABCTTACCGAACACAATTTGTCG3′ |
| Δ18WT | 5′AACATTCCCAACTTACCGACACAAT3′ (template Δ18U6C) |
| ∇27-5′U6X | 5′CAATGTAAACATTCCCATGCATCCTTTCTATCTGTCTCATTAAGTTGTGCTGTCAGTAAAAB CTTACCGAACACAATTTGATCG3′ |
| ∇27-5′WT | 5′GCTGTCAAGTAAAAACTTACCGAACACAAT3′ (template ∇27-5′U6A) |
| ∇27-3′U6G | 5′GGTTAGAAACAAAGTTTTTTCTATCTGTCTCATTAAGTTGTGCTTATCAATGTAAACATTC3′ (template ∇27-5′U6G) |
| nmt1-poly(A) | 5′AAACCCTAGCAGTACTGGCAAG3′ |
Restriction sites are underlined, mutant nucleotides are in boldface, and inserted nucleotides are italicized. W, 50:50 T+A; B, 33% each T, C, and G; S, 50:50 G+C.
To construct cgs2-Int1/pREP1, which allows expression of the first intron and flanking exon sequences from the cgs2+ gene (17) using the nmt1 promoter and polyadenylation signal (43), we first PCR amplified the relevant sequences from a genomic clone (17) using the primers cgs2Int1-5′ and cgs2Int1-3′ (Table 1) and inserted the product between the NdeI and BamHI sites of pREP1. To generate the cgs2-Int2/pREP1 plasmid, a similar procedure was followed using the primers cgs2Int2-5′ and cgs2Int2-3′ (Table 1). To construct the plasmid cgs2-Long/pREP1, which expresses a transcript containing both the first and second introns of cgs2 together with flanking exons, the appropriate region was amplified by PCR using the primers cgs2Int1-5′ and cgs2Int2-3′. To facilitate primer extension analysis, the third and final intron in this pre-mRNA, which is located several hundred nucleotides downstream (17), was not included. To mutate the 5′ splice site of cgs2-Long, we employed recombinant PCR (30) using the primers cgs2Int1-5′, cgs2Int2-3′, cgs2-Int2-5′pcr.mut1, and cgs2-Int2-5′pcr.mut2 (Table 1). To create a hybrid intron to test splice site compatibility, recombinant PCR was performed using the primers cgs2Int1-5′, cgs2Int2-3′, cgs2-Δpcr.mut1, and cgs2-Δpcr.mut2 (Table 1).
As the first step in expanding the microexon, we introduced an XhoI site within the second exon of cgs2 by site-directed mutagenesis as described above, using the oligonucleotide cgs2L/ex2Xho (Table 1). To increase the size of exon 2 by 22 nucleotides, we used the complementary oligonucleotides cgs2-22nI-5′ and cgs2-22nI-3′; for the 49-nucleotide expansion, we used the complementary oligonucleotides cgs2-49nI-5′ and cgs2-49nI-3′. In addition to the expected products, we obtained a clone in which three copies of the 22-nucleotide fragment had been incorporated. The sequences introduced were derived from the third exon of the cdc2 gene, since they do not promote splicing in their natural context (C. M. Romfo, W. J. van Heeckeren, and J. A. Wise, unpublished data).
To analyze use of the cryptic 5′ splice site in the second intron of cdc2, we started with alleles described elsewhere (C. J. Alvarez and J. A. Wise, unpublished data), which contain mutations at position +6 of the standard 5′ splice site. Insertion and deletion alleles, as well as modifications of the cryptic 5′ splice site, were constructed by site-directed mutagenesis using the oligonucleotides Crypt2,4, Δ18U6X, Δ18WT, ∇27-5′U6X, ∇27-5′WT, and ∇27-3′U6G (Table 1).
S. pombe transformation, RNA preparation, and primer extension analysis.
The recipient S. pombe strain for assaying splicing of cdc2 and cgs2 variants was DS2 (h+ ade6-210 leu1-32 ura4-d18). Transformation and RNA preparation were as previously described (52). Primer extension reactions to assay cdc2 and cgs2 splicing were also described previously (4, 54). Quantitation was performed on a Molecular Dynamics PhosphorImager using ImageQuant software (version 3.1).
Splicing of endogenous cgs2 RNA, as well as the plasmid-borne cgs2-Int2 5′ splice site and microexon deletion mutants, was assayed by reverse transcription (RT)-PCR amplification (65) using a kit supplied by Perkin Elmer (GeneAmp RNA PCR). Reactions were carried out according to the manufacturer's instructions except that the concentration of primers was 15 μM.
To confirm the cdc2 R-Long exon-skipping product, as well as to identify the retained intron in the product derived from the cgs2-Long construct, the relevant bands were first excised and eluted from gels similar to the ones shown here. The cDNAs were PCR amplified with the outside primers that were originally used to make each construct (cdc2-Nde [54] + cdc2-Ex4-3′ and cgs2Int1-5′ + cgs2Int2-3′) and cloned into the vector pTZ19R, followed by sequencing across the splice junctions with the universal and reverse primers. The cryptic 5′ splice site activated in the Δ18U+6G mutant of cdc2 was identified by direct PCR sequence analysis as previously described (4).
Computer-assisted RNA secondary structure analysis.
Folding patterns for various RNAs mentioned in the text were analyzed using the MFold secondary structure prediction program developed by Zuker and colleagues. The program, which was run using standard parameters, is available at http://www.ibc.wustl.edu/∼zuker/.
RESULTS
Exon skipping is a rare event in pre-mRNAs derived from cdc2.
As noted in the introduction, both exon and intron definition modes of splice site pairing have been observed in Drosophila, and we wanted to test whether the S. pombe splicing machinery could also switch back and forth despite the preponderance of small introns in the genes characterized to date. The most salient prediction of the exon definition model is that an exon surrounded by weak splice sites will be ignored. Thus, to seek evidence for this mode of splice site pairing in fission yeast, we first attempted to induce exon skipping. Because strong pyrimidine tracts favor the exon mode of substrate recognition in vertebrates (62), we chose to analyze the second intron of the cdc2 gene in these experiments, since it contains the most extensive run of pyrimidines of any fission yeast intron experimentally verified to date (J. A. Wise and C. M. Romfo, unpublished observations). In earlier work, we analyzed splicing of cdc2 intron 2 alleles which contained polypyrimidine tracts of various strengths, using constructs containing a single intron (54). To provide substrates suitable for assessing exon skipping, we incorporated the third intron and fourth exon into each of our intron 2 polypyrimidine tract variants to produce the cdc2-Long constructs shown in Fig. 1A. If S. pombe recognizes any of these pre-mRNAs via exon definition, then mutating the 5′ splice site following the internal exon will result in either exon skipping or activation of an upstream cryptic 5′ splice site. On the other hand, if intron definition applies, the predicted outcome is inclusion of the downstream intron or activation of a cryptic 5′ splice site located within intron 3.
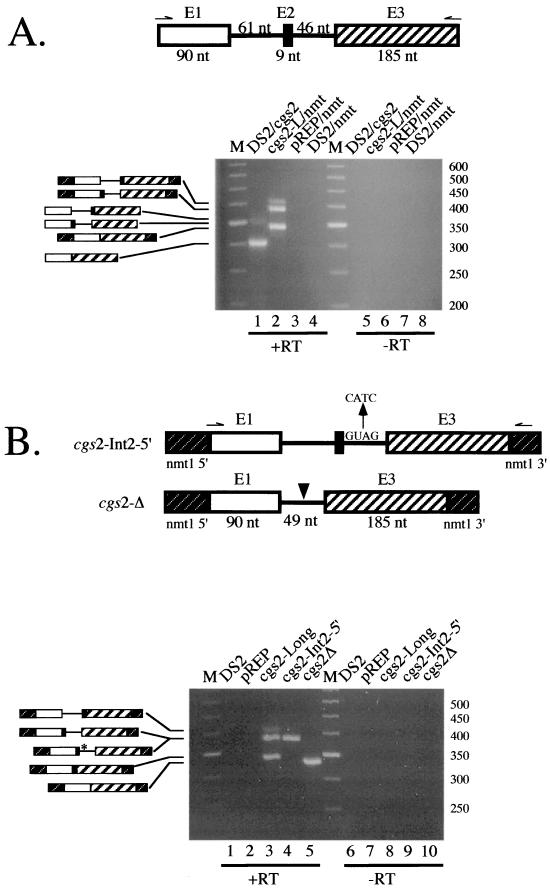
FIG. 1.
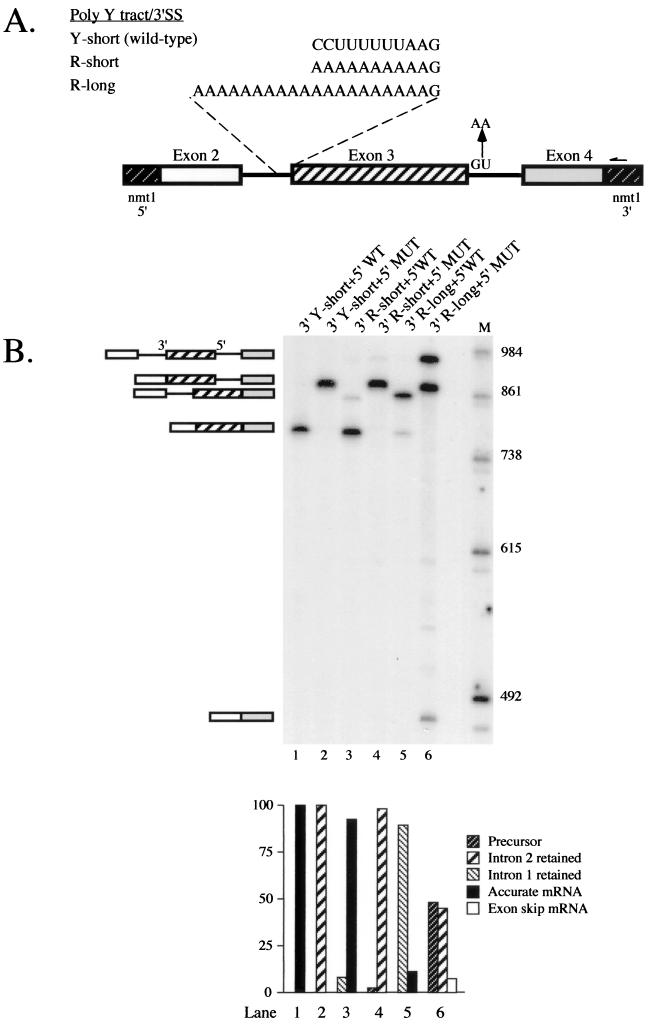
Analysis of exon skipping in cdc2 pre-mRNAs containing an internal exon flanked by mutant splicing signals. (A) Schematic representation of cdc2-Long polypyrimidine tract and 5′ splice site variants. The transcripts analyzed contained the second and third introns of the cdc2 gene and flanking exons embedded within transcription signals from the nmt1 gene (43) (see Materials and Methods for details). Sequences between the branchpoint and 3′ splice site of intron 2, as well as mutations introduced to inactivate the 5′ splice site of intron 3, are indicated. The arrow designates the position where the oligonucleotide used for primer extension [nmt1-poly(A), Table 1] hybridizes. (B) Primer extension splicing assays on cdc2-Long pre-mRNAs. Total RNA was isolated from S. pombe cells transformed with the indicated plasmid, and the relative levels of precursor, partially spliced, fully spliced, and exon-skipped RNAs were determined using primer extension analysis with an nmt1-specific oligonucleotide as described previously (4, 51). (B, top panel) Gel electrophoretic analysis. The identities and mobilities of the observed cDNA products are indicated schematically alongside the gel. The predicted sizes of the primer extension products derived from cdc2-Long are precursor, 951 nucleotides (nt); −Int1 (splicing of intron 1 only), 880 nt; −Int2 (splicing of intron 2 only), 851 nt; M (mature), 780 nt; ES (exon 2 skipping), 480 nt; intron 1 lariat, 679 nt; and intron 2 lariat, 274 nt. The positions where the lariats are expected to migrate are devoid of signal and, in the case of the intron 2 species, not shown. Lane 1, wild-type cdc2-Long; lane 2, wild-type cdc2-Long with a mutant 5′ splice site in intron 3; lane 3, R-short variant of intron 2 with wild-type intron 3; lane 4, R-short variant of intron 2 with a mutant 5′ splice site in intron 3; lanes 5 and 6, as in lanes 3 and 4 except that the constructs contain the R-long allele of intron 2; M, molecular size markers. (B, bottom panel) Quantitation of primer extension data. The levels of precursor and mature message were determined by PhosphorImager analysis and are displayed as a bar graph in which the y axis shows the percentage of each species. For each sample, pre-mRNA + mRNA totals 100%.
The profile of pre-mRNA, mature mRNA, and partially spliced intermediates produced by each allele in vivo was assessed by primer extension analysis using an oligonucleotide complementary to the nmt1 sequences present in the expression vector (see Fig. 1A), which eliminates the signal from endogenous cdc2. To provide a baseline profile of products, we first assayed alleles containing a wild-type 5′ splice site in intron 3 in combination with each pyrimidine tract variant (see Fig. 1A). The results indicate that the ratio of partially spliced RNA (intron 2 retention product) to fully spliced message, a generally accepted measure of in vivo splicing efficiency (23, 48), is highest for the R-long variant (89:11 [Fig. 1B, lane 5]) in which the distance from the branch point to the 3′ splice site is extended and also lacks pyrimidines (Fig. 1A). At the other extreme, accumulating no detectable partially spliced RNA, is an allele in which the 3′ splice site of intron 2 is wild type (Fig. 1B, lane 1). The R-short variant, which is pyrimidine deficient but has wild-type spacing (Fig. 1A), displays only minor retention of intron 2 (partially spliced to mature ratio, 8:92 [Fig. 1B, lane 3]); the same mutations produced more dramatic splicing defects in the single-intron pre-mRNAs analyzed previously (54).
Primer extension analysis of cdc2 intron 2 polypyrimidine tract variants carrying mutations in the downstream 5′ splice site are shown in the even-numbered lanes of Fig. 1B. For the wild-type allele, mutating the 5′ splice site following exon 3 results in the exclusive accumulation of a species in which intron 2 is excised while intron 3 is retained (Fig. 1B, lane 2). Furthermore, we find no evidence of exon skipping even when the 5′ splice site mutation in intron 3 is combined with a 3′ purine tract in intron 2 in the R-short variant (Fig. 1B, lane 4). To prevent any possible recognition of a downstream 5′ splice site, we also analyzed alleles carrying more extensive mutations in intron 3, including replacement of the entire 5′ junction hexanucleotide with its complement and changes in both the natural 5′ junction and a potential cryptic site just downstream. Counter to the effects of less extreme downstream 5′ splice site mutations in mammalian cell extracts, which provided crucial evidence to support the exon definition model (40), none of the mutations we tested had any discernible effect on splicing of cdc2 intron 2 in S. pombe (data not shown). Thus, our data provide no evidence for exon-bridging interactions, at least in this fission yeast pre-mRNA, but rather they are consistent with the predictions of the intron definition model.
While there was no detectable band at the position expected for the exon-skipping product with the wild-type and R-short alleles, we did observe a band of the appropriate size upon mutating the 5′ splice site of intron 3 in the R-long allele (Fig. 1B, lane 6, bottom band). This product was confirmed by direct sequence analysis to arise from precise joining of exons 2 and 4 (data not shown; see Materials and Methods). However, two other species accumulate to levels far higher than the exon-skipping product: unspliced precursor (top band; 48% of the total) and a partially spliced product in which intron 3 is retained while intron 2 is removed (middle band; 45% of the total). Thus, we believe that the modest amount (7%) of exon 3 skipping is most accurately viewed as the result of inefficient splicing of a large intron (473 nucleotides) extending from the 5′ splice site of intron 2 to the 3′ splice site of intron 3; the size of this segment exceeds that of all but 2 of the 200 naturally occurring S. pombe introns in a database of published genes (see below). Finally, our data indicate that intron 2 is spliced from R-long transcripts to a fairly significant extent (45% of the total) when the 5′ splice site of intron 3 is incapacitated, as compared to its strong retention (88%) in combination with an intact third intron (Fig. 1B, compare lanes 5 and 6). One possible explanation for this intriguing observation is that blocking intron 3 splicing delays the transcript along its route out of the nucleus, thereby increasing the window of opportunity for intron 2 to be excised.
In cases in which mutating a 5′ splice site does not lead to exon skipping in vertebrate cells, activation of a cryptic junction that lies within the exon is generally observed, an outcome also consistent with the exon definition model (reviewed in reference 6). Therefore, the gel shown in Fig. 1B was examined for evidence of cryptic 5′ splice site activation as well as exon skipping. While we do see a few extra bands migrating in the appropriate region of the gel (between accurately spliced mRNA and the exon-skipping product), these are very faint, in contrast to the efficient use of cryptic splice sites commonly observed in mammalian cells (see, e.g., references 63 and 68). Only the uppermost of the four extra bands has the mobility expected if one of the 12 GU dinucleotides within exon 3 of cdc2 (31) were used as a 5′ splice site, and none are the correct size to arise from activation of a previously described cryptic 5′ splice site (4; C. J. Alvarez and J. A. Wise, unpublished data) (see Fig. 5A below). Because these species are most prominent in RNA prepared from a mutant in which splicing is significantly blocked before the first step (Fig. 1B, lane 6), they most likely correspond to 5′ ends generated via breakdown of full-length precursor rather than to mRNAs derived from cryptic splicing events. Taken together, these data suggest that the pairing of splice sites in cdc2 pre-mRNA is most likely restricted to the intron definition mode.
FIG. 5.
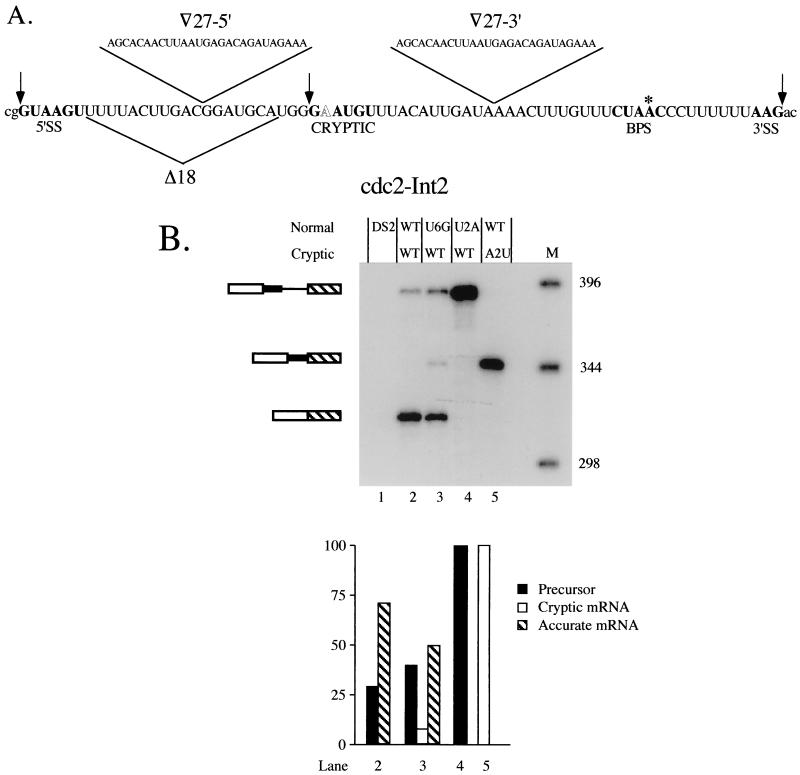
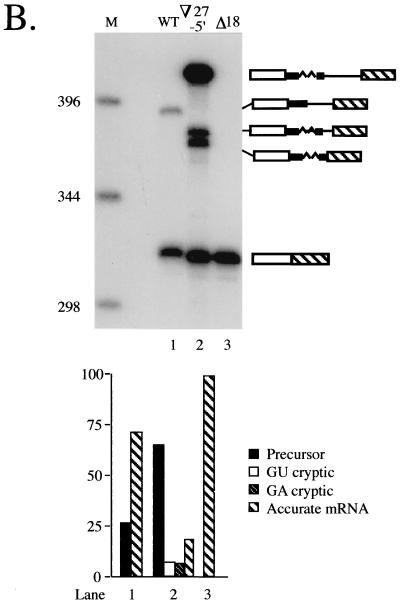
Effects of reciprocal mutations at the standard and cryptic 5′ splice sites on splicing of cdc2 intron 2. (A) Sequence of cdc2 intron 2 (31) with the splicing signals, including the nonconsensus cryptic 5′ junction discussed in the text, highlighted in bold. The 5′ and 3′ splice sites are demarcated by arrows and the branchpoint A is indicated by an asterisk. The nonconsensus nucleotide in the cryptic 5′ splice site is shown in outline. Also indicated are the locations of deletions and insertions analyzed in subsequent experiments. Shown above the sequence is a 27-nucleotide (-nt) segment derived from just downstream of the 5′ splice site in rabbit β-globin IVS 1 (3), which was inserted at the positions indicated by open triangles for the experiments shown in Fig. 7. The 18 nt bracketed by the triangle beneath the sequence were deleted for the experiment shown in Fig. 6. (B) Primer extension splicing assay on 5′ splice site mutants. The products are indicated schematically alongside the gel, with the portion of the intron retained when the cryptic 5′ splice site is used indicated by a thick line. Lane 1, control demonstrating the absence of primer extension products in RNA from the untransformed recipient strain (DS2); lane 2, primer extension products from wild-type cdc2-Int2; lane 3, primer extension products from an allele containing a U+6G substitution at the standard 5′ junction to provide a marker for the position of the cryptic band; lane 4, primer extension products from an allele containing a U+2A substitution at the standard 5′ junction; lane 5, primer extension products from an allele containing an A+2U substitution at the cryptic 5′ junction; M, molecular-size markers. The predicted sizes of the cDNA species, visualized by autoradiography, are as follows: precursor, 388 nt; mature, 317 nt; and lariat intermediate (not shown because no product was visible), 121 nt. (Lower panel) Quantitation as in Fig. 1B.
A fission yeast intron that lies downstream from a microexon is inefficiently spliced.
One possible explanation for the dearth of exon skipping in fission yeast cdc2 is the size of the internal exon in this pre-mRNA (301 nucleotides), which exceeds that of most vertebrate internal exons (6). However, because statistical analyses indicate that it is the lengths of introns, not exons, that are constrained in fission yeast (see the introduction), the size of exon 3 is more likely to pose a problem as the longest segment of the intron excised via skipping (extending from the 5′ splice site of intron 2 to the 3′ splice site of intron 3). To identify a potentially more favorable context for observing exon skipping in fission yeast, we searched a database of published gene sequences for pre-mRNAs containing a small internal exon sandwiched between introns that are also relatively small. Among several candidate genes, we selected cgs2+ (17), which contains a second intron of slightly above average size for S. pombe (see Fig. 8 below), a nine-nucleotide second exon, and an average-size third intron (72) (Fig. 2A). In addition to its potential for exon skipping, analysis of cgs2 offered an opportunity to examine similarities and differences between vertebrate and fission yeast cells in the processing of microexons.
FIG. 8.
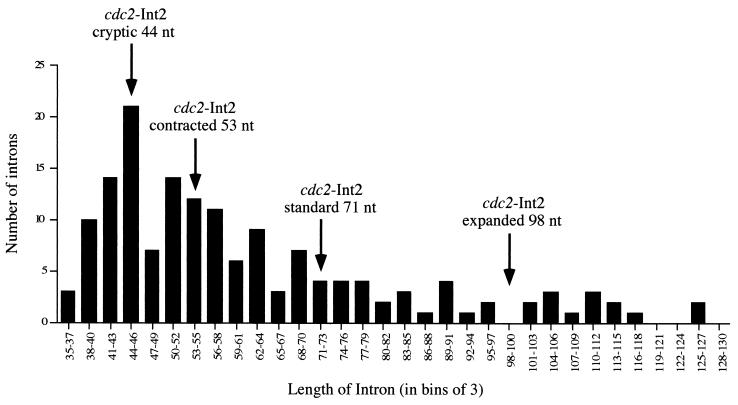
Sizes of the introns removed via use of cryptic 5′ splice sites in cdc2 intron 2 in relation to the overall distribution of intron lengths in fission yeast. The histogram shows the sizes of 156 naturally occurring fission yeast introns arranged in bins of three. The arrows denote the lengths of the segments spliced out of the alleles examined here.
FIG. 2.
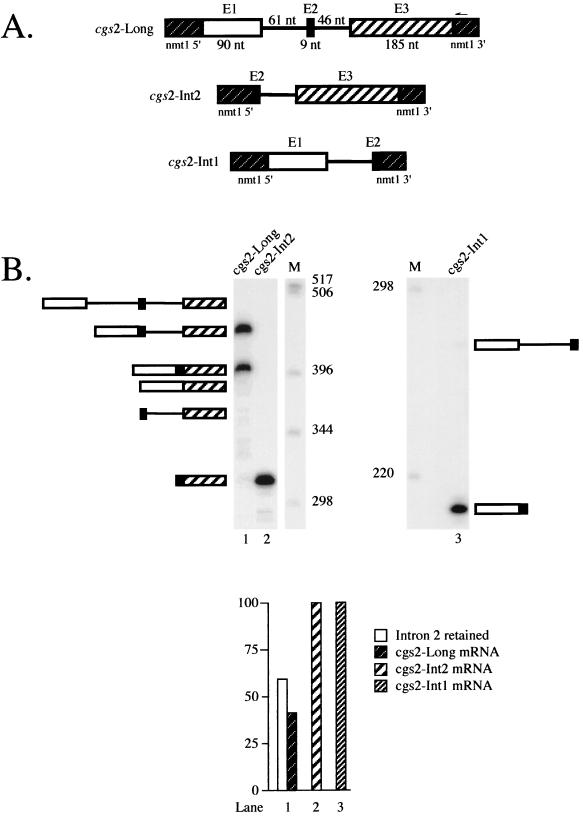
Analysis of the profile of products from a pre-mRNA containing an internal microexon. (A) Schematic representation of cgs2 transcripts containing intron 1, intron 2, or both. The construct designated cgs2-Long (top) contains both the first and second introns of the cgs2 gene and flanking exons embedded within transcription signals from the nmt1 gene (43) (see Materials and Methods for details). The constructs designated cgs2-Int2 (middle) and cgs2-Int1 (bottom) contain either the second or the first intron from the cgs2 gene and flanking exons, respectively. (B) Primer extension splicing assays on cgs2-Long, cgs2-Int2, and cgs2-Int1 pre-mRNAs. RNA extraction and analysis were performed as described in the legend to Fig. 1B. Because the products from the cgs2-Int1 pre-mRNA are much smaller than those from cgs2-Int2 and cgs2-Long, they are shown in a separate panel even though they were run on the same gel. The identities and mobilities of observed as well as some potential cDNAs derived from all three pre-mRNAs are indicated schematically alongside each gel. Lane 1, cgs2-Long; lane 2, cgs2-Int2; M, molecular size markers (1-kb ladder); lane 3, cgs2-Int1. The predicted sizes of the possible primer extension products derived from cgs2-Long are as follows: precursor, 510 nucleotides (nt); intermediate in which only intron 1 is spliced, 449 nt; intermediate in which only intron 2 is spliced, 464 nt (not indicated); mature mRNA, 403 nt; product derived from exon 2 skipping, 394 nt; intron 1 lariat, 295 nt; intron 2 lariat, 233 nt. The predicted sizes of the primer extension products derived from cgs2-Int1 are as follows: precursor, 270 nt; mature, 209 nt; lariat intermediate, 60 nt. Those from cgs2-Int2 are as follows: precursor, 359 nt; mature, 313 nt; lariat intermediate, 233 nt.
Figure 2B (lane 1) shows the result of a primer extension splicing assay on RNA isolated from fission yeast cells harboring the construct designated cgs2-Long, which contains the first two introns together with their flanking exons (Fig. 2A). Most notably, only a single band is visible in the region of the gel where mRNA is expected to migrate even after prolonged exposure of this and similar gels (data not shown). To determine whether the most rapidly migrating species includes the microexon, we cloned and sequenced it following PCR amplification (see Materials and Methods). The results (not shown) confirm that the mRNA produced by our cgs2 construct contains the nine-nucleotide exon 2 accurately spliced to exons 1 and 3. Thus, despite the common presence of a microexon in cgs2 and metazoan pre-mRNAs for which the default splicing pattern is exon skipping, an mRNA derived from such an event is not observed in S. pombe.
Although we found no evidence for microexon skipping in the cgs2 precursor, we did observe a second major cDNA, accounting for 59% of the total products, in addition to fully spliced mRNA. Based on its electrophoretic mobility, we hypothesized that this species arose via removal of only one of the two introns. To ascertain which one was retained, we amplified the cDNA using PCR followed by subcloning and sequence analysis (see Materials and Methods). The results (not shown) revealed the presence of the second intron. We did not observe a second intermediate-sized band on this gel, although a minor product that most likely corresponds to a partially spliced intermediate containing intron 1 was observed in the experiments shown in Fig. 3 and 4; in no case was a band corresponding to the full-length precursor observed. Taken together, these results indicate that intron 1 is efficiently spliced from cgs2-Long pre-mRNA, while intron 2 is not.
FIG. 3.
(A) RT-PCR assay comparing the profiles of splicing products from chromosomal and plasmid-borne cgs2 genes. (A, top panel) Schematic representation of the relevant region of cgs2 pre-mRNA expressed from the endogenous locus, with arrows indicating the cgs2Int1-5′ and cgs2Int2-3′ primers (Table 1) used for reverse transcription and PCR amplification. For RNA from strains harboring a plasmid, the same 5′ primer was employed but nmt1-poly(A) (see Fig. 2A; Table 1) was used as the 3′ primer to prevent endogenous cgs2 from contributing to the signal. (A, bottom panel) Total RNA was subjected to RT-PCR as described in Materials and Methods, and the products were displayed on a 4% Nu-Sieve agarose gel stained with ethidium bromide. To allow a semiquantitative comparison of the different species, the number of cycles was limited to 22 for all samples. The identity of each species is indicated schematically alongside the gel. Lane 1, RNA from untransformed strain DS2 cells was subjected to RT-PCR using cgs2Int2-3′ as the 3′ primer; lane 2, RNA from cells harboring the cgs2-Long plasmid was subjected to RT-PCR using nmt1-poly(A) as the 3′ primer; lane 3, as in lane 2 except that the cells harbored the empty vector; lane 4, as in lane 2 except that the RNA was from untransformed DS2 cells. The predicted sizes of the possible RT-PCR products derived from chromosomal cgs2 are as follows: precursor, 420 nucleotides (nt); intermediate in which only intron 2 is spliced, 374 nt; intermediate in which only intron 1 is spliced, 359 nt; mature mRNA, 313 nt. The predicted sizes of the possible RT-PCR products derived from cgs2-Long are as follows: precursor, 447 nt; intermediate in which only intron 2 is spliced, 401 nt; intermediate in which only intron 1 is spliced, 386 nt; mature mRNA, 340 nt. (B) Design and RT-PCR analysis of cgs2-Long mutants. (B, top panel) Schematic representation of cgs2-Long mutants in which the indicated base substitutions have been introduced at the downstream 5′ splice site or the microexon and surrounding sequences have been deleted (indicated by an arrowhead). (B, bottom panel) RT-PCR assays of wild-type and mutant cgs2 splicing were performed as in panel A, using cgs2-Int1-5′ and nmt1-poly(A) as primers. The identity of each species is indicated schematically alongside the gel, with an asterisk denoting the 5′ splice site mutations. Lane 1, untransformed DS2; lane 2, empty vector control; lane 3, wild-type cgs2-Long; lane 4, 5′-splice-site mutant; lane 5, deletion mutant. The predicted sizes of the possible RT-PCR products derived from cgs2-Int25′ are the same as for wild-type cgs2-Long. The predicted sizes of the possible RT-PCR products derived from cgs2-Δ are as follows: precursor, 380 nt; mature mRNA, 331 nt.
FIG. 4.
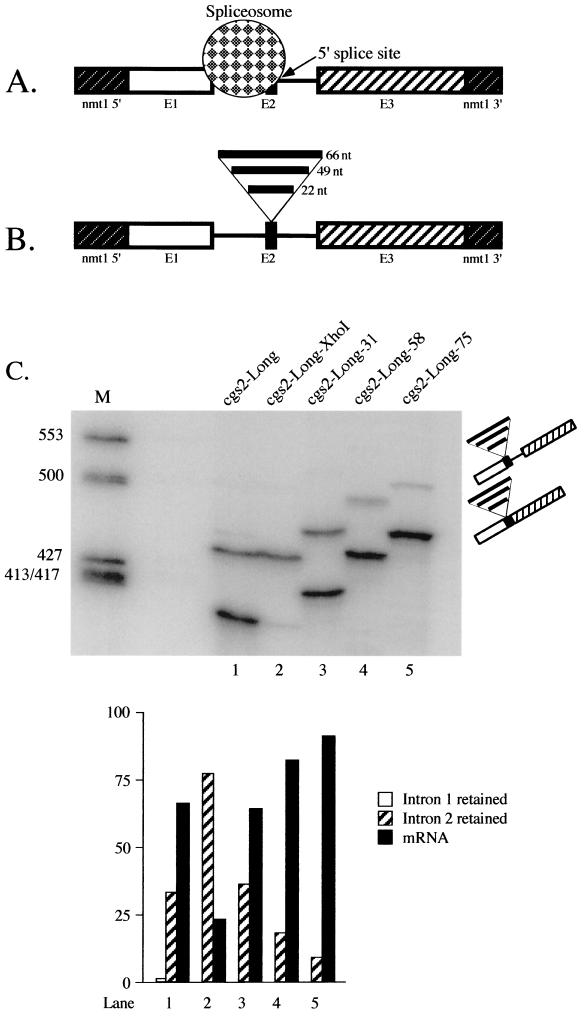
Effect of expanding the microexon in cgs2. (A) Model to account for the inefficient splicing of the second intron in cgs2-Long pre-mRNA. See text for details. (B) Schematic representations of cgs2-Long RNAs in which exon 2 (E2) is expanded. The fragments introduced to increase the size of exon 2 (see Materials and Methods for details) are indicated by thick horizontal lines above E2. (C) Primer extension splicing assays on expansion alleles of cgs2-Long. RNA extraction and analysis were performed as described in the legend to Fig. 1B. M, molecular size markers (ΦX174-HinfI); lane 1, wild-type cgs2-Long pre-mRNA; lane 2, cgs2-Long-XhoI; lane 3, cgs2-Long-31; lane 4, cgs2-Long-58; lane 5, cgs2-Long-75. In each lane, the top band corresponds to the intron 2 retention product and the bottom band to mature mRNA; the segments inserted into exon 2 are indicated as in panel B. The predicted sizes of the primer extension products derived from cgs2-Long-XhoI are the same as for wild-type cgs2-Long (see the legend to Fig. 2); for cgs2-Long-31, the predicted cDNA sizes are as follows: precursor, 532 nucleotides (nt); intermediate in which only intron 1 is spliced, 471 nt; and mature mRNA, 425 nt. For cgs2-Long-58, the sizes are as follows: precursor, 559 nt; intermediate in which only intron 1 is spliced, 498 nt; and mature mRNA, 452 nt. For cgs2-Long-75, the sizes are as follows: precursor, 576 nt; intermediate in which only intron 1 is spliced, 515 nt; and mature mRNA, 469 nt. (Lower panel) Quantitation as in Fig. 1B.
A potential explanation for the incomplete removal of intron 2 from the cgs2-Long pre-mRNA is that it contains defective splicing signals. To examine this possibility, we assayed splicing in S. pombe cells of a transcript containing only the second intron (cgs2-Int2; Fig. 2A, middle). As illustrated in Fig. 2B (lane 2), primer extension analysis indicates that virtually no unspliced RNA is detectable. Thus, the signals present in intron 2 can support efficient splicing in the absence of intron 1. This result also argues against another potential explanation for the preferential retention of intron 2 in RNA expressed from the plasmid-borne construct, namely, that its proximity to the polyadenylation signal due to truncation of the gene might interfere with splicing. As expected, the first intervening sequence of cgs2, when present as a solo intron (Fig. 2A, bottom), is also removed nearly quantitatively (Fig. 2B, lane 3). The efficient excision of each intervening sequence when expressed from a single-intron construct, in contrast to the incomplete processing of the cgs2-Long transcript, implies that it is the close proximity of the two introns that inhibits splicing in S. pombe.
Because the retention of intron 2 was unexpected, we wanted to determine whether it also occurs in RNA expressed from the chromosomal cgs2+ locus. To this end, the profiles of RNAs produced from endogenous and plasmid-borne genes were compared using an RT-PCR assay, which is more sensitive than primer extension. As illustrated in Fig. 3A, the same three products are observed in both transformed and untransformed cells (compare lanes 1 and 2). These products result from amplification of fully spliced mRNA, a partially spliced intermediate that retains intron 2, and a partially spliced intermediate that retains intron 1; the size differences reflect the different 3′ primers employed (see Fig. 3A legend for details). A band corresponding to unspliced RNA was not observed in RNA expressed from either the plasmid or the chromosome. One difference, however, is that the relative yield of fully spliced mRNA is higher in RNA expressed from the single-copy chromosomal locus, suggesting that a component required for splicing of cgs2 pre-mRNA has become limiting due to high-level expression from the plasmid. A notable similarity is that, in both samples, the intron 2 retention product is far more abundant than the intron 1 retention product (which is barely visible in the analysis of chromosomally expressed RNAs). Taken together, these data suggest that removal of intron 2 may require a positively acting factor in addition to the constitutive splicing machinery (see Discussion).
While it was satisfying that the chromosomal and plasmid-borne cgs2 genes gave a similar profile of products, the question remained whether skipping of the microexon might be observed under other circumstances. In an effort to induce skipping of the microexon, we used the same strategy employed above with cdc2, mutation of the downstream 5′ splice site. An RT-PCR assay on RNA from cells transformed with this construct (Fig. 3B, lane 4) revealed a single band that comigrates with the intron 2 retention product present in the adjacent sample (lane 3). Since the intron that would be excised if the microexon had been skipped in this experiment is only 116 nucleotides long, this result raised the possibility that the 5′ splice site of intron 1 and the 3′ splice site of intron 2 are somehow incompatible. To test whether this might account for our failure to induce exon skipping, we assayed splicing of a construct in which the microexon and flanking intron sequences had been deleted. The results (Fig. 3B, lane 5) indicate that the failure to skip the microexon even after mutating the downstream 5′ junction is not due to splice site incompatibility, since the hybrid intron was excised very efficiently. Taken together, these results provide strong evidence that cgs2 splicing is restricted to the intron definition mode.
Expanding the microexon allows efficient splicing of cgs2 pre-mRNA.
The findings presented in the preceding section suggest that the inefficient removal of the second intron in the cgs2-Long pre-mRNA is primarily due to the close juxtaposition of the first intron. A plausible model to explain this observation is steric occlusion of the downstream 5′ splice site by an upstream spliceosome (Fig. 4A), as originally proposed to explain the exclusion of the mouse c-src microexon in non-neuronal cells (9). To test the resulting hypothesis that increasing the distance separating the relevant 3′ and 5′ splice sites will allow efficient splicing, we first engineered an XhoI site in exon 2. Unexpectedly, the substitution of four bases at positions −3, −5, −7, and −8 relative to the 5′ splice site of intron 2 to create the restriction site virtually abolished splicing (Fig. 4C, compare lanes 1 and 2). These changes affect nucleotides which are not highly conserved through evolution (33), and thus they seem unlikely to diminish snRNA binding. Computer-assisted RNA secondary structure analysis indicates that the new sequence can form a local hairpin (W. J. van Heeckeren and J. A. Wise, unpublished data) which may interfere with splicing, as observed previously in cdc2 (4). Another possibility is that the sequence changes might affect binding of a protein factor to the microexon. For example, in rat γ2 pre-mRNA, it was shown that some of the nucleotides required for optimal inclusion of a small internal neuron-specific exon reside within the microexon itself and function in an unpaired state (71).
To expand the second exon of cgs2-Long, we ligated two pairs of complementary oligonucleotides (see Table 1 for sequences) into the XhoI site, producing three variants: cgs2-Long-31, which contains a 22-nucleotide fragment; cgs2-Long-58, which contains a 49-nucleotide fragment; and cgs2-Long-75, which contains three copies of the 22-nucleotide fragment (Fig. 4B). The profile of primer extension products from each of these substrates was compared to those produced by both the wild-type and XhoI alleles. The data shown in Fig. 4C indicate that expansion of exon 2 to 31 nucleotides improves splicing of intron 2 from the level observed with the XhoI parent to approximately wild-type efficiency (compare lanes 1 and 3; partially spliced to mature ratio, 34:66 and 36:64, respectively). Further increases in the fraction of primer extension products corresponding to mRNA are seen with the larger insertions; for the construct in which the microexon was expanded to 58 nucleotides, the ratio of partially spliced to mature RNA is 18:82, while for the 75-nucleotide exon, the ratio is 9:91 (Fig. 4C, lanes 4 and 5). Because increasing the size of exon 2 results in improved splicing even when the wild-type pre-mRNA is used as a baseline, it seems unlikely that the effects are due solely to disruption of an inhibitory structure produced in the XhoI mutant. In aggregate, the effects of expanding the microexon in cgs2 are consistent with the steric occlusion model shown in Fig. 4A.
When comparing the results of this experiment to the previous one, we noted an inversion in the ratios of partially spliced to mature mRNA for cgs2-Long wild type; in Fig. 4C, the values are 34:66 (lane 1), while in Fig. 2B, they are 59:41 (lane 1). One potential explanation for this discrepancy is that the cells used to extract the RNA assayed in Fig. 4C were grown in rich medium, whereas for the experiment shown in Fig. 2B, RNA was isolated from cells grown in minimal medium. To determine whether the change in growth conditions accounts for the diminished splicing defect, we repeated the entire set of assays shown in Fig. 4C with cells grown in minimal medium. The results confirm that nutritional state influences splicing of this pre-mRNA, since the partially spliced-mRNA ratio for the cgs2-Long wild type in this experiment mirrored that obtained with the independent transformant examined earlier, and splicing of the exon expansion alleles was also less efficient than when the cells were grown in rich medium (C. Romano, L. Lackner, and J. A. Wise, unpublished data). Notably, the ratios of partially spliced to mature mRNA in the wild-type and 31-nucleotide expansion alleles were nearly identical regardless of whether the cells were grown in rich or minimal medium, suggesting that they are spliced via the same pathway. The influence of nutritional state on splicing of cgs2-Long suggests a model for regulation (see Discussion).
Activation of an unusual cryptic 5′ splice in cdc2 intron 2 is likely to reflect its favorable location for splicing.
In addition to the incidence of exon skipping versus intron retention, the exon and intron definition models predict different locations for cryptic 5′ splice sites. In the course of analyzing the contribution of U1 snRNA to 5′ splice site selection in S. pombe (C. J. Alvarez and J. A. Wise, unpublished data), we discovered that an unusual cryptic 5′ splice site is activated by certain mutations at the standard exon 1-intron 2 boundary in fission yeast cdc2. The location of this cryptic 5′ splice site, which lies within the intron, 27 nucleotides downstream from the standard 5′ junction (Fig. 5A) provides a third indication that splice site pairing in S. pombe proceeds via intron definition. As noted in the introduction, cryptic 5′ splice sites in vertebrates generally lie upstream, within the preceding exon (6). The cryptic 5′ splice site in cdc2 intron 2 is used to a significant extent, accounting for 8% of the total primer extension products when the standard 5′ junction is mutated at position +6 (Fig. 5B, lane 3) (C. J. Alvarez and J. A. Wise, unpublished data) despite a deviation from consensus (an A at position +2) that would normally render it inactive (3, 64).
The use of a cryptic 5′ splice site containing an A at the normally invariant second position is not due to a higher tolerance for a nonconsensus nucleotide in fission yeast than in other organisms, since a U+2A mutation at the standard 5′ splice site of cdc2 intron 2 as well as in two other S. pombe introns leads to complete retention (Fig. 5B, lane 4; C. J. Alvarez and J. A. Wise, unpublished data). Intriguingly, the cryptic junction is not activated in the U+2A mutant, in contrast to the U+6G mutant, suggesting that the standard 5′ splice site must be partially active in order for the aberrant 5′ splice site to be utilized. Data described elsewhere suggest that the dependence of splicing at the cryptic junction on the natural 5′ splice site relates to the binding of U1 snRNA (C. J. Alvarez and J. A. Wise, unpublished data). To determine which 5′ junction is preferred when both contain a consensus nucleotide at the second position, we mutated the noncanonical A in the cryptic site to a U. This experiment gave a dramatic and, at first glance, unexpected result; the cryptic site is used exclusively, despite the presence of the wild-type sequence at the exon-intron boundary employed under normal circumstances (Fig. 5B, lane 5). We conclude that, when its nonconsensus second nucleotide is mutated to consensus, the cryptic site is strongly preferred over the standard 5′ splice site. Moreover, compared to the partial retention observed when the natural 5′ junction is used for splicing in a wild-type intron (precursor-mature mRNA, 26:74) (Fig. 5B, lane 2), the mutant allele containing a consensus sequence at the cryptic site accumulates no detectable precursor (lane 5).
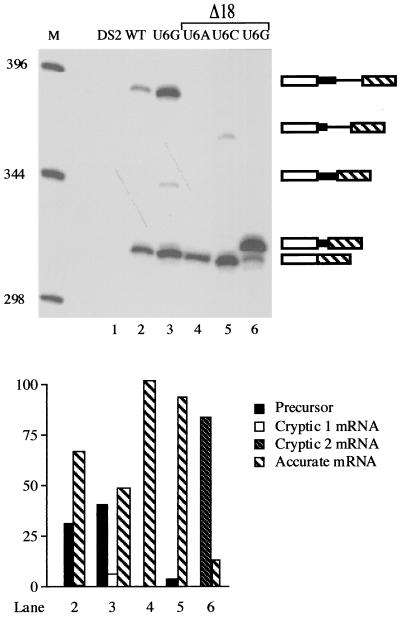
The ability of the modified cryptic junction to overwhelm the standard 5′ splice site in what essentially amounts to a cis-competition assay suggests that it is situated in a context more favorable for splicing. To determine whether this might reflect a constraint on intron size, we reduced the distance between the wild-type 5′ junction and the 3′ splicing signals by 18 nucleotides (Fig. 5A), leaving just 3 nucleotides between position +6 of the standard 5′ junction and position +1 of the cryptic site. Because our earlier work (C. J. Alvarez and J. A. Wise, unpublished data) indicated that the unusual 5′ junction is used most efficiently when the standard 5′ splice site is mutated at position +6, the effect of the deletion was initially assessed on alleles carrying each of the three possible substitutions at this nucleotide. Primer extension splicing assays revealed first that the contracted (Δ18) alleles show nearly undetectable use of the GA dinucleotide as a 5′ splice site (Fig. 6, lanes 4 to 6), in contrast to the 8% cryptic splicing product observed in an allele with normal spacing and a U to G mutation at position +6 of the standard 5′ splice site (Fig. 6, lane 3). Thus, deleting nucleotides between the normal and cryptic 5′ splice sites of cdc2 intron 2 allows the former to compete more effectively. Second, the overall efficiency of splicing is improved dramatically in the contracted alleles; only a small amount (6%) of precursor is observed for the U+6C allele, and no full-length RNA is detectable for the U+6A and U+6G alleles, in contrast to the nearly equal amounts of precursor and mature message observed for the U+6G allele with wild-type spacing (42 and 50%, respectively) (Fig. 6, lane 3). Thus, moving the standard 5′ splice site closer to the 3′ splicing signals increases splicing efficiency.
FIG. 6.
Effect of decreasing the size of cdc2 intron 2 in alleles containing position +6 changes at the standard 5′ junction. Top: M, molecular size markers; lane 1, recipient strain control; lane 2, wild-type cdc2-Int2; lane 3, U+6G allele with normal spacing between the standard and cryptic 5′ splice sites. The last three lanes show primer extension products from alleles containing an 18-nucleotide deletion between the standard and cryptic 5′ splice sites (see Fig. 5A; designated Δ18) and either a U+6A (lane 4), U+6C (lane 5), or U+6G (lane 6) substitution at the standard 5′ junction. Products are shown schematically as in Fig. 5B. (Lower panel) Quantitation as in Fig. 1B.
In the case of the U+6G mutation at the standard 5′ splice site, the deletion junction produces a new GU dinucleotide, and the in vivo splicing assays indicate that this allele yields a predominant cDNA (85% of the total primer extension products) of slightly slower mobility than the product derived from mRNA spliced at the standard 5′ splice site (Fig. 6, lane 6). PCR sequencing of the cDNA confirms that it arises via splicing at the newly created GU (data not shown). Notably, the cryptic 5′ splice site used in this case deviates from the S. pombe consensus at positions +3, +4, and +6 downstream from the exon-intron boundary (+3GGGA+6 versus +3AAGU+6), yet is used almost exclusively. This result suggests that a 3′ proximal 5′ splice site is so strongly favored by the fission yeast splicing machinery that a nonconsensus sequence is used in preference to a consensus site just a few nucleotides upstream. However, note that this bias is still not sufficiently strong to allow use of the GA-containing cryptic 5′ junction once the intron has been shortened to bring sites with a consensus 5′ dinucleotide into the preferred range. These data imply that the fission yeast splicing machinery uses a combination of sequence and spatial cues to pair splice sites.
The effects of expanding cdc2 intron 2 are also consistent with the intron definition model.
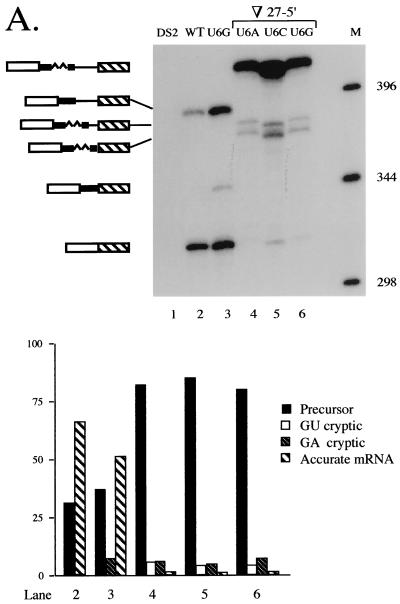
The fact that decreasing the size of the second intron in cdc2 stimulates its removal, consistent with splice site pairing via intron definition, prompted us to test the converse prediction that an increase in size will diminish splicing. To this end, we doubled the interval between the standard and cryptic 5′ splice sites from 27 to 54 nucleotides via the insertion of a segment from intron 1 of rabbit β-globin (3; see Fig. 5A, top left; designated ∇27-5′); as for the deletion constructs, the first alleles examined also contained substitutions at position +6 of the standard 5′ splice junction. Primer extension splicing assays (Fig. 7A, lanes 4 to 6) indicate that increasing the size of cdc2 intron 2 dramatically reduces splicing at the standard 5′ junction relative to an allele in which the spacing is normal; in the expanded alleles, most (from 82 to 88% in the three +6 mutants examined) of the RNA detected is linear precursor, whereas approximately equal quantities of precursor and mature mRNA are again observed for an allele with normal spacing (Fig. 7A, lane 3). In an expanded allele containing the wild-type sequence at the standard 5′ junction, splicing at the original exon-intron boundary is readily detectable, but still quite inefficient (20%; Fig. 7B, lane 2) compared to either a fully wild-type (74%; lane 1) or a contracted allele (100%; lane 3).
FIG. 7.
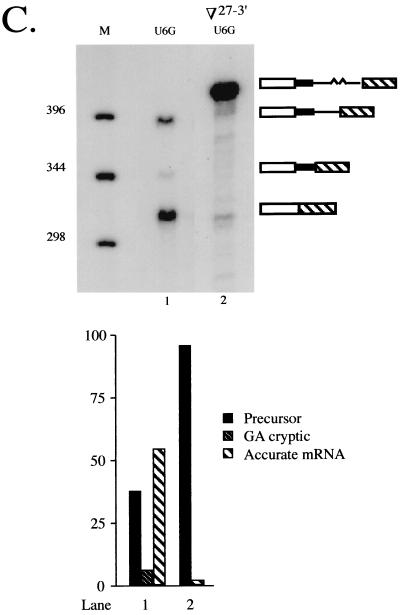
Effect of increasing the size of cdc2 intron 2. (A) Primer extension analysis of alleles containing position +6 mutations at the standard 5′ splice site. Products are shown schematically as in Fig. 4B, with the inserted nucleotides derived from rabbit β-globin IVS 1 indicated by a wavy line. Lane 1, recipient strain control; lane 2, wild-type cdc2-Int2; lane 3, U+6G allele with standard spacing. The last three lanes show products derived from alleles containing a 27-nucleotide insertion between the standard and cryptic 5′ splice sites (Fig. 5A; designated ∇27-5′) and either a U+6A (lane 4), U+6C (lane 5), or U+6G (lane 6) substitution at the standard 5′ junction. M, molecular-size markers. The more rapid mobility of the bands in lane 5 is due to salt in the sample. (B) Primer extension analysis of alleles containing the wild-type sequence at the standard 5′ junction. M, molecular-size markers; lane 1, wild-type cdc2-Int2; lane 2, primer extension products from an otherwise wild-type allele containing a 27-nucleotide insertion between the standard and cryptic 5′ splice sites; lane 3, primer extension products from an otherwise wild-type allele containing an 18-nucleotide deletion between the standard and cryptic 5′ splice sites. (C) Primer extension analysis of an allele in which the distance between the cryptic 5′ splice sites and the branch point is increased. M, molecular size markers; lane 1, U+6G allele with standard spacing; lane 2, products from an allele containing the 27-nucleotide insertion between the cryptic 5′ junctions and the branch point (Fig. 5A; designated ∇27-3′) and a U+6G substitution at the standard 5′ junction. (Lower panel) Quantitation as in Fig. 1B.
In the expanded introns, the noncanonical cryptic 5′ splice site is used to a similar extent regardless of whether position +6 of the standard 5′ splice site is mutated (5 to 8%; Fig. 7A, lanes 4 to 6; Fig. 7B, lane 2). In addition, a second cryptic junction is activated upon increasing the size of the intron, and its use is also unaffected by the sequence at the standard 5′ splice site (7 to 9%; Fig. 7A, lanes 4 to 6; Fig. 7B, lane 2). The mobility of the corresponding primer extension product as compared to a sequencing ladder run in an adjacent lane indicates that, in this case, the exon-intron boundary precedes the GU dinucleotide located at positions +5 and +6 of the original cryptic 5′ splice site (data not shown). This observation suggests that the lack of a nearby authentic 5′ splice site renders splicing more dependent on the presence of a consensus dinucleotide at the exon-intron boundary. The deviation of the second cryptic 5′ splice site from the S. pombe consensus at all four downstream nucleotides (+3UUAC+6 versus +3AAGU+6), as well as at position −1 (50, 72), may account for its limited use despite its being closer to the 3′ splicing signals.
As an additional test of the intron definition model for splicing of cdc2 intron 2, we moved the two deviant cryptic 5′ splice sites further away from the branchpoint by inserting the same sequence introduced between the standard and cryptic 5′ splice sites in the preceding experiment at the location indicated in Fig. 5A (top right; designated ∇27-3′). As expected, increasing the distance between the branchpoint and cryptic 5′ junctions abolishes the use of both aberrant splice sites (Fig. 7C, lane 2); an allele with wild-type spacing is included on this gel as a marker for the position of the cryptic splicing product (Fig. 7C, lane 1). Finally, the ratio of precursor to mRNA spliced at the standard 5′ splice site in this experiment is similar to that observed for an allele which is identical except for the location of the insertion (3%; compare Fig. 7C, lane 2, and Fig. 7A, lane 6). This is as expected, since the overall size of the intron removed is the same in both cases.
The pattern of cryptic splice site utilization in cdc2 reflects the natural distribution of intron sizes in S. pombe.
The foregoing analyses of cryptic splice site utilization are consistent with the notion that intron size is an important determinant of splicing efficiency in S. pombe. In Fig. 8, the sizes of the introns excised via use of each cryptic 5′ splice site cdc2 intron 2 are superimposed on a histogram displaying the distribution of lengths for 156 naturally occurring introns from fission yeasts. Intriguingly, the length of the intron excised via use of the unusual (GA-containing) 5′ splice site in cdc2 (44 nucleotides) coincides with the peak of this histogram, and the intron removed via use of the standard 5′ splice site in the contracted alleles (53 nucleotides) also lies in a zone that is quite densely populated. In contrast, the intron removed via splicing at the standard 5′ junction lies in a relatively barren region of the graph, and even fewer naturally occurring introns correspond in size to the very inefficiently spliced expansion alleles. Also consistent with the strong preference for small introns in S. pombe is our finding that the proximal 3′ splice site is strongly preferred in alleles of cdc2 intron 2 carrying duplications of the 3′ splicing signals (C. M. Romfo and J. A. Wise, unpublished data). As in our analysis of 5′ splice site utilization, deletions of intronic sequences improved splicing efficiency in these pre-mRNAs (data not shown).
DISCUSSION
The fission yeast splicing machinery is most likely restricted to the intron definition mode of splice site pairing.
In this report, we present three lines of evidence which, in aggregate, strongly suggest that the splicing machinery in fission yeast pairs splice sites exclusively across introns, in contrast to vertebrate cells. First, even when the splicing signals on both sides of an exon were mutated, the exon skipping product represented only a minor fraction of the total RNA (Fig. 1), while mutating the downstream 5′ splice site alone was sufficient to produce exon skipping in vertebrate pre-mRNAs (reviewed in reference 6). Second, exon skipping did not occur at a detectable level during splicing of a wild-type fission yeast pre-mRNA that contains a microexon (Fig. 2), whereas this was the default splicing pattern in pre-mRNAs containing internal exons of comparable size in mammals (e.g., 9, 18, 59). Moreover, it was not possible to induce skipping of the microexon by mutating the downstream 5′ splice site. Finally, the locations of cryptic splice sites, as well as the effects of expanding and contracting an intron (Fig. 5 through 7), are as predicted by the intron definition model, and contrast with the patterns of cryptic splice site usage observed in vertebrate pre-mRNAs (reviewed in reference 6). Our finding that increasing the size of cdc2 intron 2 compromises splicing confirms and extends the results of earlier experiments with an artificial intron in S. pombe, in which expansions also reduced splicing efficiency (24).
Despite the common occurrence of multi-intronic pre-mRNAs in S. pombe (where the majority of interrupted genes contain two or more introns) (72) versus their paucity in S. cerevisiae (where all but four interrupted genes contain only a single intron) (58), the effects of 5′ splice site mutations are similar between the two yeasts, that is, the observed outcome is generally intron retention rather than exon skipping (e.g., 23, 24, 48, 64; C. J. Alvarez and J. A. Wise, unpublished data). However, in budding yeast, it was possible to experimentally induce exon skipping by taking advantage of the fact that the 5′ and 3′ splice sites in many introns from this organism are brought into closer proximity via naturally occurring complementary sequences (25, 34 and references therein). Thus, in variants of the twice-interrupted S. cerevisiae YL8A pre-mRNA, the creation of potential pairing between sequences near the 5′ end of the first intron and the 3′ end of the second intron caused the embedded exon to be ignored (34). Similar experiments are unlikely to be illuminating in S. pombe, since computer-assisted secondary structure analysis does not support the existence of an analogous mechanism for juxtaposing 5′ and 3′ splice sites even in relatively large introns (C. J. Alvarez and J. A. Wise, unpublished data).
In addition to providing evidence for splice site pairing exclusively by intron definition, the data presented in this report, specifically the correlation between the locations of cryptic splice sites and the natural distribution of intron sizes in S. pombe (Fig. 8), suggest the existence of distance constraints. One possible explanation for limiting the linear length of RNA between factors bound at the 5′ and 3′ splice sites is that the need to loop out a segment may compromise splicing efficiency in fission yeast; further studies of the U1 snRNP, U2AF, and the branchpoint bridging protein (4, 49, 55, 67) should illuminate the mechanistic basis for the trend toward small introns. Notably, the peak of the histogram displaying the distribution of intron sizes in S. pombe (44 nucleotides) (Fig. 8) is barely over half the minimum size required for splicing of an intron in a HeLa cell extract (80 nucleotides) (69). Nevertheless, the existence of size constraints on fission yeast introns is likely to be of general significance, since diminutive intervening sequences predominate not only in S. pombe and other unicellular eukaryotes, including Tetrahymena and Neurospora (reference 15 and references therein), but also in certain multicellular organisms, including the nematode C. elegans (12) and the fruit fly D. melanogaster (46). In Drosophila, it has been shown that specific sequence elements dictate the size constraints (28), and it will be interesting to determine whether the same is true in S. pombe. Despite the apparent lack of an upper limit on intron size in vertebrates, several studies show that, in extracts from mammalian cells, the splicing machinery also preferentially selects the proximal 5′ junction from a pair of duplicated splice sites (16, 21, 51). Thus, the interactions that underlie the spatial constraints described here are also likely to be important in higher eukaryotes.
Do microexons serve a regulatory role in fission yeast?
While microexon recognition has been subjected to intense experimental scrutiny in vertebrates, splicing of pre-mRNAs containing extremely small exons has not been previously investigated in a unicellular eukaryote. We could envision a priori, three possible profiles of splicing products from the cgs2-Long pre-mRNA: efficient removal of both introns, skipping of the microexon, or retention of one intron. The fact that only the third outcome was observed is consistent with the view that the fission yeast splicing machinery is restricted to the intron definition mode of splice site pairing. The apparent inability of the S. pombe splicing machinery to switch back and forth between intron and exon definition modes of splice site pairing contrasts with the situation in Drosophila, and indicates that fission yeast is unlikely to provide a simpler model system in which to study alternative splicing of the exon-skipping variety. On the other hand, the data presented here are consistent with the idea that S. pombe may be capable of modulating splicing efficiency to regulate the amount, if not the precise structure, of the mRNA produced from a particular gene.
A significant factor in our choice to focus first on cgs2 among several fission yeast genes that contain experimentally verified microexons was the fact that it encodes cyclic AMP phosphodiesterase, a critical component of the highly regulated meiotic and protein kinase cascades (17). The retention of intron 2 in both chromosomally and ectopically expressed transcripts suggests that the small size of exon 2 renders removal of the downstream intron inherently inefficient, thereby allowing for the possibility that splicing of this pre-mRNA may be subject to positive regulation. It is unlikely that this occurs by a mechanism analogous to the one used by vertebrates to stimulate microexon inclusion, since the small size of the second intron in cgs2 would most likely preclude the presence of an intronic splicing enhancer (10, 13, 45). We suggest, instead, that the downstream exon may contain a positively acting element. Consistent with this hypothesis, we have found that removal of intron 2 can be stimulated by incorporating purine-rich exonic splicing enhancers that normally function in vertebrate cells into the downstream exon (C. M. Romfo, W. J. van Heeckeren and J. A. Wise, unpublished data). Because excision of intron 2 is more efficient in cells grown in rich versus minimal medium (Fig. 2 through 4 and data not shown), the natural exon may contain an element that responds to a signaling pathway involved in sensing the nutritional state of the cell. We are currently examining splicing of cgs2 in cells grown under a variety of conditions in order to test this idea.
Could the presence of extremely small exons in S. pombe pre-mRNAs provide a general strategy for achieving on-off regulation of splicing? Consistent with this idea, internal microexons (operationally defined as ≤30 nucleotides) are quite common in this organism. Of the 48 multi-intronic pre-mRNAs included in our database of published genes, 7 contain a microexon (C. M. Romfo and J. A. Wise, unpublished observations). The fission yeast genome project has uncovered an additional 71 open reading frames with this architectural feature, several of which contain more than one microexon (M. Lyne, K. Rutherford, and V. Wood, personal communication). Furthermore, similar to our observations with cgs2, other investigators have presented evidence for retention of the intron following a microexon in fission yeast hus1 pre-mRNA, whose product is involved in checkpoint control of the cell cycle (39). Finally, we have recently found that splicing of two other fission yeast pre-mRNAs containing internal microexons is inefficient (L. Lackner, C. Romano, J. F. Sun, and J. A. Wise, unpublished observations), lending further support to the view that this architecture may be exploited for regulation of splicing in S. pombe.
Experiments are currently under way to identify both the cis-acting sequences and the trans-acting factors that influence splicing of fission yeast pre-mRNAs containing microexons. Of particular interest will be determining whether the purine-rich sequences often found downstream of retained introns (J. A. Wise, unpublished observations) represent splicing enhancers. Our finding that heterologous exonic enhancers can stimulate the removal of intron 2 from cgs2 pre-mRNA, in combination with the conservation of enhancer complex constituents in S. pombe, strongly suggests that fission yeast cells employ naturally occurring elements similar to those found in metazoa to modulate splicing efficiency.
ACKNOWLEDGMENTS
We are grateful to Mike Lyne, Kim Rutherford, and Valerie Wood of the Sanger Center S. pombe Genome Project for sharing data prior to publication. We appreciate the excellent assistance of Carissa Romano in preparing the figures. Thanks are also due to Roger VanHoy for extracting the data used to generate the histogram shown in Figure 8 from GenBank and to Maureen McLeod (Downstate Medical Center, Brooklyn, N.Y.) for providing a plasmid encoding the cgs2 gene. We gratefully acknowledge Helen Salz and Sujata Reddy for critical comments on the manuscript.
This research was supported by a grant to J.A.W. from the National Institutes of Health; C.J.A. was supported in part by a predoctoral fellowship from the Fulbright LASPAU Program (USIA).
REFERENCES
- 1.Abovich N, Liao X C, Rosbash M. The yeast MUD2 protein: an interaction with PRP11 defines a bridge between commitment complexes and U2 snRNP addition. Genes Dev. 1994;8:843–854. doi: 10.1101/gad.8.7.843. [DOI] [PubMed] [Google Scholar]
- 2.Abovich N, Rosbash M. Cross-intron bridging interactions in the yeast commitment complex are conserved in mammals. Cell. 1997;89:403–412. doi: 10.1016/s0092-8674(00)80221-4. [DOI] [PubMed] [Google Scholar]
- 3.Aebi M, Hornig H, Padgett R A, Reiser J, Weissmann C. Sequence requirements for splicing of higher eukaryotic nuclear pre-mRNA. Cell. 1986;47:555–565. doi: 10.1016/0092-8674(86)90620-3. [DOI] [PubMed] [Google Scholar]
- 4.Alvarez C J, Romfo C M, VanHoy R W, Porter G L, Wise J A. Mutational analysis of U1 function in S. pombe: pre-mRNAs differ in the extent and nature of their requirements for this snRNA in vivo. RNA. 1996;2:404–418. [PMC free article] [PubMed] [Google Scholar]
- 5.Amrein H, Hedley M L, Maniatis T. The role of specific protein-RNA and protein-protein interactions in positive and negative control of pre-mRNA splicing by transformer 2. Cell. 1994;76:735–746. doi: 10.1016/0092-8674(94)90512-6. [DOI] [PubMed] [Google Scholar]
- 6.Berget S M. Exon recognition in vertebrate splicing. J Biol Chem. 1995;270:2411–2414. doi: 10.1074/jbc.270.6.2411. [DOI] [PubMed] [Google Scholar]
- 7.Berglund J A, Chua K, Abovich N, Reed R, Rosbash M. The splicing factor BBP interacts specifically with the pre-mRNA branchpoint sequence UACUAAC. Cell. 1997;89:781–787. doi: 10.1016/s0092-8674(00)80261-5. [DOI] [PubMed] [Google Scholar]
- 8.Birney E, Kumar S, Krainer A R. Analysis of the RNA-recognition motif and RS and RGG domains: conservation in metazoan pre-mRNA splicing factors. Nucleic Acids Res. 1993;21:5803–5816. doi: 10.1093/nar/21.25.5803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Black D L. Does steric interference between splice sites block the splicing of a short c-src neuron-specific exon in non-neuronal cells? Genes Dev. 1991;5:389–402. doi: 10.1101/gad.5.3.389. [DOI] [PubMed] [Google Scholar]
- 10.Black D L. Activation of c-src neuron-specific splicing by an unusual RNA element in vivo and in vitro. Cell. 1992;69:795–807. doi: 10.1016/0092-8674(92)90291-j. [DOI] [PubMed] [Google Scholar]
- 11.Black D L. Finding splice sites within a wilderness of RNA. RNA. 1995;1:763–771. [PMC free article] [PubMed] [Google Scholar]
- 12.Blumenthal T, Thomas J. Cis and trans splicing in C. elegans. Trends Genet. 1988;4:305–308. doi: 10.1016/0168-9525(88)90107-2. [DOI] [PubMed] [Google Scholar]
- 13.Carlo T, Sterner D A, Berget S M. An intron splicing enhancer containing a G-rich repeat facilitates inclusion of a vertebrate micro-exon. RNA. 1996;2:342–343. [PMC free article] [PubMed] [Google Scholar]
- 14.Chou M-Y, Rooke N, Turck C W, Black D L. hnRNP H is a component of a splicing enhancer complex that activates a c-src alternative exon in neuronal cells. Mol Cell Biol. 1999;19:69–77. doi: 10.1128/mcb.19.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Csank C F, Taylor M, Martindale D W. Nuclear pre-mRNA introns: analysis and comparison of intron sequences from Tetrahymena thermophila and other eukaryotes. Nucleic Acids Res. 1990;18:5133–5141. doi: 10.1093/nar/18.17.5133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cunningham S A, Else A J, Potter B V L, Eperon I C. Influences of separation and adjacent sequences on the use of alternative 5′ splice sites. J Mol Biol. 1991;217:265–281. doi: 10.1016/0022-2836(91)90541-d. [DOI] [PubMed] [Google Scholar]
- 17.DeVoti J, Seydoux G, Beach D, McLeod M. Interaction between ran1+ protein kinase and cAMP dependent protein kinase as negative regulators of fission yeast meiosis. EMBO J. 1991;10:3759–3768. doi: 10.1002/j.1460-2075.1991.tb04945.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dominski Z, Kole R. Selection of splice sites in pre-mRNAs with short internal exons. Mol Cell Biol. 1991;11:6075–6083. doi: 10.1128/mcb.11.12.6075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eng F J, Warner J R. Structural basis for the regulation of splicing of a yeast messenger RNA. Cell. 1991;65:797–804. doi: 10.1016/0092-8674(91)90387-e. [DOI] [PubMed] [Google Scholar]
- 20.Engebrecht J, Voelkel-Meiman K, Roeder G S. Meiosis-specific splicing in yeast. Cell. 1991;66:1257–1268. doi: 10.1016/0092-8674(91)90047-3. [DOI] [PubMed] [Google Scholar]
- 21.Eperon I C, Ireland D C, Smith R A, Mayeda A, Krainer A R. Pathways for selection of 5′ splice sites by U1 snRNPs and SF2/ASF. EMBO J. 1993;9:3607–3617. doi: 10.1002/j.1460-2075.1993.tb06034.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fields C. Information content of Caenorhabditis elegans splice site sequences varies with intron length. Nucleic Acids Res. 1990;18:1509–1512. doi: 10.1093/nar/18.6.1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fouser L A, Friesen J D. Mutations in a yeast intron demonstrate the importance of specific conserved nucleotides for the two stages of nuclear pre-mRNA splicing. Cell. 1986;45:81–93. doi: 10.1016/0092-8674(86)90540-4. [DOI] [PubMed] [Google Scholar]
- 24.Gatermann K B, Hoffman A, Rosenberg G H, Käufer N F. Introduction of functional artificial introns into the naturally intronless ura4 gene of Schizosaccharomyces pombe. Mol Cell Biol. 1989;9:1526–1535. doi: 10.1128/mcb.9.4.1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Goguel V, Rosbash M. Splice site choice and splicing efficiency are positively influenced by pre-mRNA intramolecular base pairing in yeast. Cell. 1993;72:893–901. doi: 10.1016/0092-8674(93)90578-e. [DOI] [PubMed] [Google Scholar]
- 26.Gross T, Richert K, Mierke C, Lützelberger M, Kaüfer N F. Identification and characterization of srp1, a gene of fission yeast encoding a RNA binding domain and a RS domain typical of SR splicing factors. Nucleic Acids Res. 1998;26:505–511. doi: 10.1093/nar/26.2.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guo M, Lo P C, Mount S M. Species-specific signals for the splicing of a short Drosophila intron in vitro. Mol Cell Biol. 1993;13:1104–1118. doi: 10.1128/mcb.13.2.1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guo M, Mount S M. Localization of sequences required for size-specific splicing of a small Drosophila intron in vitro. J Mol Biol. 1995;253:426–437. doi: 10.1006/jmbi.1995.0564. [DOI] [PubMed] [Google Scholar]
- 29.Hawkins J D. A survey on intron and exon lengths. Nucleic Acids Res. 1988;16:9893–9908. doi: 10.1093/nar/16.21.9893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Higuchi R. Recombinant PCR. In: Innis M A, Gelfand D M, Sinisky J J, White T J, editors. PCR protocols: a guide to methods and applications. San Diego, Calif: Academic Press, Inc.; 1990. pp. 177–183. [Google Scholar]
- 31.Hindley J, Phear G A. Sequence of the cell division gene CDC2 from Schizosaccharomyces pombe; patterns of splicing and homology to protein kinases. Gene. 1984;31:129–134. doi: 10.1016/0378-1119(84)90203-8. [DOI] [PubMed] [Google Scholar]
- 32.Hoffman B E, Grabowski P J. U1 snRNP targets an essential splicing factor, U2AF65, to the 3′ splice site by a network of interactions spanning the exon. Genes Dev. 1992;6:2554–2568. doi: 10.1101/gad.6.12b.2554. [DOI] [PubMed] [Google Scholar]
- 33.Horowitz D S, Krainer A R. Mechanisms for selecting 5′ splice sites in mammalian pre-mRNA splicing. Trends Genet. 1994;10:100–106. doi: 10.1016/0168-9525(94)90233-x. [DOI] [PubMed] [Google Scholar]
- 34.Howe K J, Ares M., Jr Intron self-complementarity enforces exon inclusion in a yeast pre-mRNA. Proc Natl Acad Sci USA. 1997;94:12467–12472. doi: 10.1073/pnas.94.23.12467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Innis M A, Gelfand D H. Optimization of PCR. In: Innis M A, Gelfand D H, Sinisky J J, White T J, editors. PCR protocols: a guide to methods and applications. San Diego, Calif: Academic Press, Inc.; 1990. pp. 3–12. [Google Scholar]
- 36.Kennedy C F, Krämer A, Berget S M. A role for SRp54 during intron bridging of small introns with pyrimidine tracts upstream of the branchpoint. Mol Cell Biol. 1998;18:5425–5434. doi: 10.1128/mcb.18.9.5425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kishida M, Nagai T, Nakaseko Y, Shimoda C. Meiosis-dependent mRNA splicing of the fission yeast Schizosaccharomyces pombe mes1+ gene. Curr Genet. 1994;25:497–503. doi: 10.1007/BF00351668. [DOI] [PubMed] [Google Scholar]
- 38.Kohtz J D, Jamison S F, Will C L, Zuo P, Lührmann R, Garcia-Blanco M A, Manley J L. Protein-protein interactions and 5′ splice site recognition in mammalian mRNA precursors. Nature. 1994;368:119–124. doi: 10.1038/368119a0. [DOI] [PubMed] [Google Scholar]
- 39.Kostrub C F, Al-Khodary F, Ghazizadeh H, Carr A M, Enoch T. Molecular analysis of hus1+, a fission yeast gene required for S-M and DNA damage checkpoints. Mol Gen Genet. 1997;254:389–399. doi: 10.1007/pl00008606. [DOI] [PubMed] [Google Scholar]
- 40.Kuo H-C, Nasim F H, Grabowski P J. Control of alternative splicing by the differential binding of U1 small nuclear ribonucleoprotein particle. Science. 1991;251:1045–1050. doi: 10.1126/science.1825520. [DOI] [PubMed] [Google Scholar]
- 41.Lopez A J. Alternative splicing of pre-mRNA: developmental consequences and mechanisms of regulation. Annu Rev Genet. 1998;32:279–305. doi: 10.1146/annurev.genet.32.1.279. [DOI] [PubMed] [Google Scholar]
- 42.Lützelberger M, Gross T, Kaüfer N F. Srp2, an SR protein family member of fission yeast: in vivo characterization of its modular domains. Nucleic Acids Res. 1999;27:2618–2626. doi: 10.1093/nar/27.13.2618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Maundrell K. nmt1 of fission yeast. J Biol Chem. 1990;265:10857–10864. [PubMed] [Google Scholar]
- 44.Min H, Chan R C, Black D L. The generally expressed hnRNP F is involved in a neural-specific pre-mRNA splicing event. Genes Dev. 1995;9:2659–2671. doi: 10.1101/gad.9.21.2659. [DOI] [PubMed] [Google Scholar]
- 45.Min H, Turck C W, Nokolic J M, Black D L. A new regulatory protein, K-SRP, mediates exon inclusion through an intronic splicing enhancer. Genes Dev. 1997;11:1023–1036. doi: 10.1101/gad.11.8.1023. [DOI] [PubMed] [Google Scholar]
- 46.Mount S M, Burks C, Hertz G, Stormo G D, White O, Fields C. Splicing signals in Drosophila: intron size, information content, and consensus sequences. Nucleic Acids Res. 1992;10:4255–4262. doi: 10.1093/nar/20.16.4255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Peng X, Mount S M. Genetic enhancement of RNA-processing defects by a dominant mutation in B52, the Drosophila gene for an SR protein splicing factor. Mol Cell Biol. 1995;15:6273–6282. doi: 10.1128/mcb.15.11.6273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pikielny C W, Rosbash M. mRNA splicing efficiency in yeast and the contribution of nonconserved sequences. Cell. 1985;41:119–126. doi: 10.1016/0092-8674(85)90066-2. [DOI] [PubMed] [Google Scholar]
- 49.Potashkin J, Naik K, Wentz-Hunter K. U2AF homolog required for splicing in vivo. Science. 1993;262:573–575. doi: 10.1126/science.8211184. [DOI] [PubMed] [Google Scholar]
- 50.Prabhala G, Rosenberg G H, Käufer N F. Architectural features of pre-mRNA introns in the fission yeast Schizosaccharomyces pombe. Yeast. 1992;8:171–182. doi: 10.1002/yea.320080303. [DOI] [PubMed] [Google Scholar]
- 51.Reed R, Maniatis T. A role for exon sequences and splice site proximity in splice site selection. Cell. 1986;46:681–690. doi: 10.1016/0092-8674(86)90343-0. [DOI] [PubMed] [Google Scholar]
- 52.Reich C I, VanHoy R W, Porter G L, Wise J A. Mutations at the 3′ splice site can be suppressed by compensatory base changes in U1 snRNA in fission yeast. Cell. 1992;69:1159–1169. doi: 10.1016/0092-8674(92)90637-r. [DOI] [PubMed] [Google Scholar]
- 53.Robberson B L, Cote G J, Berget S M. Exon definition may facilitate splice site selection in RNAs with multiple exons. Mol Cell Biol. 1990;10:84–94. doi: 10.1128/mcb.10.1.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Romfo C M, Wise J A. Both the polypyrimidine tract and the 3′ splice site function prior to the first step of splicing in fission yeast. Nucleic Acids Res. 1997;25:4658–4665. doi: 10.1093/nar/25.22.4658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Romfo C M, Lakhe-Reddy S, Wise J A. Molecular genetic analysis of U2AF59 in Schizosaccharomyces pombe: differential sensitivity of introns to mutational inactivation. RNA. 1999;5:49–65. doi: 10.1017/s1355838299981323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rymond B C, Rosbash M. Yeast pre-mRNA splicing. In: Broach J R, Pringle J R, Jones E W, editors. The molecular and cellular biology of the yeast Saccharomyces: gene expression. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1992. pp. 143–192. [Google Scholar]
- 57.Shen J, Zu K, Cass C L, Beyer A L, Hirsch J. Exon skipping by overexpression of a Drosophila heterogeneous nuclear ribonucleoprotein in vivo. Proc Natl Acad Sci USA. 1995;92:1822–1825. doi: 10.1073/pnas.92.6.1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Spingola M, Grate L, Haussler D, Ares M., Jr Genome-wide bioinformatic and molecular analysis of introns in Saccharomyces cerevisiae. RNA. 1999;5:221–234. doi: 10.1017/s1355838299981682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sterner D A, Berget S M. In vivo recognition of a vertebrate micro-exon as an exon-intron-exon unit. Mol Cell Biol. 1993;13:2677–2687. doi: 10.1128/mcb.13.5.2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sterner D A, Carlo T, Berget S M. Architectural limits on split genes. Proc Natl Acad Sci USA. 1996;93:15081–15085. doi: 10.1073/pnas.93.26.15081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Talerico M, Berget S M. Effect of 5′ splice site mutations on splicing of the preceding intron. Mol Cell Biol. 1990;10:6299–6305. doi: 10.1128/mcb.10.12.6299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Talerico M, Berget S M. Intron definition in splicing of small Drosophila introns. Mol Cell Biol. 1994;14:3434–3445. doi: 10.1128/mcb.14.5.3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Treisman R, Orkin S H, Maniatis T. Specific transcription and RNA splicing defects in five cloned β-thalassemia genes. Nature. 1983;302:591–596. doi: 10.1038/302591a0. [DOI] [PubMed] [Google Scholar]
- 64.Vijayraghavan U, Parker R, Tamm J, Iimura Y, Rossi J, Abelson J, Guthrie C. Mutations in conserved intron sequences affect multiple steps in the yeast splicing pathway, particularly assembly of the spliceosome. EMBO J. 1986;5:1683–1695. doi: 10.1002/j.1460-2075.1986.tb04412.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang A M, Mark D F. Quantitative PCR. In: Innis M A, Gelfand D M, Sinisky J J, White T J, editors. PCR protocols: a guide to methods and applications. San Diego, Calif: Academic Press Inc.; 1990. pp. 70–75. [Google Scholar]
- 66.Wei N, Lin C Q, Modafferi E F, Gomes W A, Black D L. A unique intronic splicing enhancer controls the inclusion of the agrin Y exon. RNA. 1997;3:1275–1288. [PMC free article] [PubMed] [Google Scholar]
- 67.Wentz-Hunter K, Potashkin J. The small subunit of the splicing factor U2AF is conserved in fission yeast. Nucleic Acids Res. 1996;24:1849–1854. doi: 10.1093/nar/24.10.1849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wieringa B, Meyer F, Reiser J, Weissman C. Unusual splice sites revealed by mutagenic inactivation of an authentic splice site of the rabbit β-globin gene. Nature. 1983;301:38–43. doi: 10.1038/301038a0. [DOI] [PubMed] [Google Scholar]
- 69.Wieringa B, Hofer E, Weissman C. A minimal intron length but no specific internal sequence is required for splicing in the large rabbit β-globin intron. Cell. 1984;37:915–925. doi: 10.1016/0092-8674(84)90426-4. [DOI] [PubMed] [Google Scholar]
- 70.Wu J Y, Maniatis T. Specific interactions between proteins implicated in splice site selection and regulated alternative splicing. Cell. 1993;75:1061–1070. doi: 10.1016/0092-8674(93)90316-i. [DOI] [PubMed] [Google Scholar]
- 71.Zhang L, Ashiya M, Sherman T G, Grabowski P J. Essential nucleotides direct neuron-specific splicing of γ2 pre-mRNA. RNA. 1996;2:682–698. [PMC free article] [PubMed] [Google Scholar]
- 72.Zhang M Q, Marr T G. Fission yeast gene structure and recognition. Nucleic Acids Res. 1994;22:1750–1759. doi: 10.1093/nar/22.9.1750. [DOI] [PMC free article] [PubMed] [Google Scholar]