Abstract
Cancer is a big group of diseases and one of the leading causes of mortality worldwide. Despite enormous studies and efforts are being carried out in understanding the cancer and developing drugs against tumorigenesis, drug resistance is the main obstacle in cancer treatments. Chemotherapeutic treatment is an important part of cancer treatment and drug resistance is getting gradually multidimensional with the advancement of studies in cancer. The underlying mechanisms of drug resistance are largely unknown. Sirtuin1 (SIRT1) is a type of the Class III histone deacetylase family that is distinctively dependent on nicotinamide adenine dinucleotide (NAD+) for catalysis reaction. SIRT1 is a molecule which upon upregulation directly influences tumor progression, metastasis, tumor cell apoptosis, autophagy, DNA repair, as well as other interlinked tumorigenesis mechanism. It is involved in drug metabolism, apoptosis, DNA damage, DNA repair, and autophagy, which are key hallmarks of drug resistance and may contribute to multidrug resistance. Thus, understanding the role of SIRT1 in drug resistance could be important. This study focuses on the SIRT1 based mechanisms that might be a potential underlying approach in the development of cancer drug resistance and could be a potential target for drug development.
Keywords: SIRT1, chemotherapeutics resistance, tumorigenesis, DNA damage repair
Introduction
With decades of potential studies of cancer development, cancer is one of the most common disease in worldwide, including in less and more developed countries, with an assessed 18.1 million new cases and 9.1 million deaths were reported in 2018 [1]. Cancer development and progression, diagnosis and cancer treatment has advanced but still cancer multidrug resistance (MDR) remains a big obstacle for effective treatment of cancer. In cancer study, chemotherapeutic based treatment is the mainstay of treatment for several malignancies at different stages. As we study cancer development and treatment, often, cancer becomes resistant to the drug that’s why investigators think about drug resistance mechanisms and then to reach this clue that drug resistance is a vast and complex mechanism. However, the chemotherapeutics resistance mechanism in cancer development has become gradually multifaceted and it is still not well understood. To overcome these limitations, we need to emphasis on how tumor cells regulate their cell signaling and metabolic pathways to inhibit drug penetration or assist the efflux of accumulated drugs in cancer cells [2]. Several studies have interlinked with the upregulation of different genes to promote the development of drug resistance that are associated with drugs penetration of cancer cells, or by altered membrane transport pathway, anti-apoptotic and enhanced DNA repair, modification of target molecules such as enzymatic deactivation that act to directly regulate gene transcription [3].
SIRT1 is a member of the histone deacetylase class III (HDAC) family that is distinctively dependent enzyme on nicotinamide adenine dinucleotide (NAD+) for catalysis reaction. The enzyme was first discovered in a yeast [4], further, it has been found in various organisms throughout evolution from bacteria to humans. Human contains different HDACs III sirtuin isoforms, SIRT1 to SIRT7, all these sirtuins have homology to the SIR2 (silent information regulator) yeast protein, which are distinguished by their various cellular enzymatic actions and sites [5]. Furthermore, sirtuins have been found in Drosophila melanogaster to increase the lifespan and control genomic instability [6]. It can regulate chromatin protein via deacetylation of histones and alterations in the methylation of histones, thereby regulating target gene expression and leading to post transcriptional modification with resultant silencing [7]. SIRT1 protein contains two export channels, one is nuclear localization signals (NLS) and another is nuclear export signals (NES), that permits the transporting of SIRT1 protein between the cytoplasm and the nucleus, depending on the type of cellular state [8]. Inclusive studies over the earlier decade have been shown SIRT1 is a big player associated in the regulation of various biological functions including cellular senescence and aging, gene expression, proliferation, differentiation, metabolism, carcinogenesis, and immune response [9-11].
SIRT1 overexpression has been identified in several tumors including lung cancer, prostate cancer, skin cancer, colon cancer, basal cell carcinoma, acute myeloid leukemia squamous cell carcinoma, actinic keratosis and Bowen’s disease [12-15]. Even though it has been a debatable issue whether it is as a tumor suppressor or an oncogene, significant evidence supports the SIRT1 overexpression in cancers acting as a tumor promoter. Most well studied of SIRT1 overexpression directly interlinked to intercellular epigenetics and coordinates with tumor progression, apoptosis, autophagy, DNA repair, homeostasis, and its function probably to regulate tumorigenesis interwoven mechanism [16]. The general mechanism of drug resistance and SIRT1 is shown in Figure 1. Moreover, SIRT1 functions to promote tumor resistance to drugs are unclear, still need intense research to improvement a better understanding of the drug resistance pathway. In this review, we highlight recent approaches of SIRT1 in multidrug resistant cancers, and how to overcome chemotherapeutics resistance and advance cancer treatment.
Figure 1.
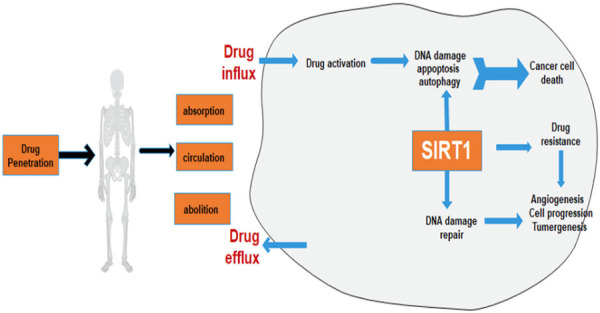
General mechanism of chemoresistance by SIRT1. Drug absorption, circulation, and abolition in cancer cells, SIRT1 regulates drug mechanism to activation of DNA damage repair machinery and downregulate DNA damage and apoptosis pathway through promoting multidrug resistance and cell proliferation.
SIRT1 mediated chemotherapeutics resistance in cancer
Cancer cells are usually misbehaved; mainly, their metabolic and cell signaling mechanism is same as in normal human cells. Tumor cells are mostly resistant, not sensitive according to tumor contexts and drugs, it means the carcinomatous state has adopted reversible changes that make the cells susceptible. British medical oncologist Adrian Harris in 1985 has been pointed out first time that cancer cells act as vulnerable than normal cells and show a oversensitive reaction to chemotherapy [17]. Moreover, neoadjuvant therapy of various solid tumors provided recent advances, when a drug enters into cells, first to shrink the tumor size or disappeared from the local site, but in some cases, especially metastases recurrent later [18,19]. Here, we discuss how SIRT1 promotes resistance to drugs and enhanced tumorigenesis, it is essential to understand the basic mechanism of SIRT1 [20]. Biological studies have been further demonstrated that SIRT1 deacetylates histone family proteins and non-histone proteins, all these proteins are regulators of numerous signaling pathways of cancer cells. All these catalytic reactions are taking place in the cytoplasm and nucleus, it monitors various cellular pathways including transcription, translation and post-translation modifications [21,22]. SIRT1 overexpression has a vital role reported by previous study in a diversity of carcinoma including breast cancer [23,24], prostate cancer [25], colorectal adenocarcinoma [26], gastroesophageal junction (GEJ) cancer [27], hepatocellular carcinoma (HCC) [28], gastric cancer [29], soft tissue sarcomas [30], pancreatic ductal adenocarcinoma [31], renal cell carcinoma [32] and non-small cell lung carcinoma [33,34].
SIRT1 to have a significant role in positively regulating intrinsically resistant to response chemotherapy and energy states which is subjected to controlling the cell death, this resistance derives from the interaction with the FOXO proteins, transcription factors and p53 can also modulate the mechanism of exogenous stress [35,36]. The association between FOXO1 and SIRT1, it is of prodigious value to understand the consequences of FOXO to cancer study overall. Here, we highlight the contributions of FOXO family in tumorigenesis is very diverse depending on the cancer state and other regulated molecules that are involved. FOXO1 act as a main regulator of multidrug resistance 1 (MDR1) protein has been reported by previous studies [37]. Most likely, ATP-binding cassette (ABC) protein is a member of membrane proteins of transporter family about 49 members of this family involved to regulate the flux proteins around the plasma membrane of several structurally and physiologically related chemotherapeutic drugs. All three proteins are multi-drug resistant proteins including P-glycoprotein and ABCB1 1 (identified as MDR1), breast cancer resistance protein and ABCG2 (BCRP) and MDR-associated protein 1. These proteins have a broad role to overlap with a substrate to promote the abolishment of several hydrophobic compounds including topoisomerase, taxanes, antimetabolites and inhibitors [38]. SIRT1 mediated chemoresistance is shown in Figures 2 and 3. In addition, SIRT1 also promotes cancer cell progression and growth expansion, which are mediated via Myc activation, p53 inactivation, and epithelial-to-mesenchymal transition. AMPKα facilitates SIRT1-dependent, hypoxia-induced chemoresistance in lung cancer and might to regulate tumor responses to other anticancer drugs reported by Shin et al. [39]. Although SIRT1 has not been widely explored in the context of chemotherapy drugs, previous reported studies have been assisted the opinion that SIRT1 plays an emerging role in tumor attainment of chemotherapeutics resistance [40,41].
Figure 2.
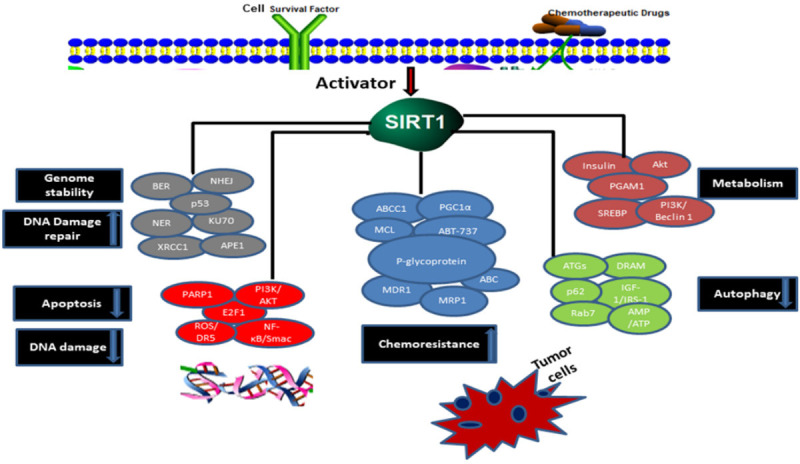
SIRT1 pathway overview in chemoresistance cancer. SIRT1 is an NAD+-dependent histone deacetylase protein that modulates several pathways of chemoresistance cancer. Overexpression of SIRT1 promotes DNA damage repair, antiapoptotic effects, confer cell proliferation and increases the function of chemoresistance in cells via protein regulators such as activators or inhibitors of SIRT1 have been described. The downstream activity targets deacetylation such as cellular metabolism, senescence, DNA repair and modulates the tumor microenvironment through multiple cell signaling pathways.
Figure 3.
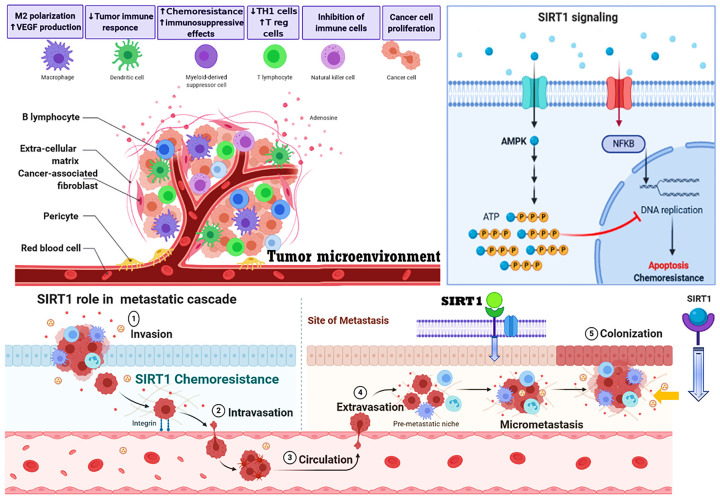
SIRT1 role in cancer development and cell progression, invasion, metastasis and MDR. SIRT1 activates many pathways in tumorigenesis such as cancer progression, cell invasion and migration, metastasis and chemoresistance. SIRT1 overexpression subjected to regulate multiple pathways of cancer including metastatic cascade and invasion, tumor suppression and cell proliferation, enhances the stemness and modulates the tumor microenvironment for multidrug resistance in cancer. Given all recent data discussed in above study about potential of SIRT1 to promote multidrug resistance mechanism and tumorigenesis has been summarized in this figure.
Mechanism of SIRT1 mediated chemotherapeutic resistance
How SIRT1 regulates metabolism?
Metabolism is well connected to longevity and cell stability, health and diseases including diabetes, cardiovascular diseases, and cancer. Increasing evidences are supporting the diverse role of SIRT1 in cellular metabolism and drug efflux, mitochondrial homeostasis, oxidative stress, PGC-1α is a known target of SIRT1-dependent deacetylation, is a transcriptional co-activator in several cell signaling pathways in metabolic activities. Previous experimental and clinical studies have exposed hypoxia to acquire resistance to chemotherapeutic drugs. As a whole, SIRT1 promotes cell progression and is challenged continually by hypoxic stress and they have their capability to ATP redeemable mode and maintain proliferation characteristics [42].
The ability of SIRT1 to modulate various transporter proteins have been directly or indirectly interlinked to drug resistance to generally induced chemotherapeutics by stimulating efflux of drug, roles of other sirtuins 2-7 in metabolic remain poorly understood [43,44]. The co-activator of SIRT1 also performs an essential role in regulating gluconeogenesis and oxidation of fatty acids metabolic pathways. SIRT1 as positively regulates Akt activation and insulin at different levels of reactions and suppress the PTPN1 transcription, which is a suppressing of the insulin signal cascade [45]. However, reported studies has provided clues that SIRT1 regulates the phosphoglycerate mutase-1 (PGAM1) which inducing suppression of glycolytic proteins expression in response to fasting condition and maintain energy balance under stress, as well as further suggesting SIRT1-AMPK pathway could be associated to overcome chemoresistance in lung cancer whether knockdown expression of SIRT1 [39,46]. Xia et al. study determined that SIRT1 was highly expressed in cervical cancer than normal cells, significantly it means SIRT1 higher in PTX-resistant cervical cancer tissues than in PTX-sensitive cervical cancer tissues. In addition, they have observed that SIRT1 revealed overexpression levels in both PTX-resistant in vitro cell lines and patients tissue [47].
More recently, lipids metabolism including cholesterols and fatty acids is a hallmark of cancer, by providing energy metabolism of structural blocks for the quickly dividing and enhancing cancer cells proliferation [48]. As a catalytic activity of SIRT1 to interrupt fatty acids regulators such as sterol regulatory element-binding protein 1 and 2 (SREBP), which is associated to the transferring and biosynthesis of lipids elements to cell elongation [49]. In metabolism NAD+ is become a stress to a cell, SIRT1 targets SREBP1 for deacetylation and promotes carcinogenesis and chemoresistance to anticancer therapy [50]. Moreover, SIRT1 is a well-known metabolic homeostasis gene that regulates metabolic syndromes, such as diabetes type 2, and is correlated to the initiation of several metabolic disorders. Consistently, in vivo and in vitro studies have been demonstrated SIRT1 overexpression by SIRT1 activators (SRT1720 and resveratrol) which can promote insulin and obesity induced liver resistance to cancer cells [51]. Furthermore, in vitro study on H460 and A549 cell lines of lung cancer indicated SIRT1 regulates apoptosis, radiation, and drug sensitization through cell signaling SIRT1/NF-κB/Smac pathway. In summary, SIRT1 has been elucidated in modulating cell progression and stress responses, and it shows to play an important role in regulating energy metabolism, tumorigenesis and resistance to chemo-radio therapy [52].
SIRT1 enhances DNA damage repair machinery and downregulate apoptosis
For a detailed understanding of SIRT1 in maintaining genomic integrity which is important for chemotherapeutics resistance to cancer and other genetic disorders. SIRT1 deacetylase is linked to various DNA damage repair genes, such as NBS1, Werner helicase, nucleotide excision repair (NER), homologous recombination (HR) non-homologous end joining (NHEJ), base excision repair (BER), mismatch repair (MMR), and inter-strand crosslink (ICL) repair. Chemotherapeutic drugs induce DNA damage either directly or indirectly to a cancer cell. Currently, we have found that SIRT1 deacetylated XRCC1 and increased expression of XRCC1 to promotes chemoresistance in H460 lung cancer cell line [53]. However, SIRT1 overexpression plays a central role into suppression of apoptosis, DNA damage, the DNA damage repair capability of tumor cells has a key role in the efficiency of anticancer agents [54-57]. Cell cycle arrest is induced by DNA damage which has switched on to various transcriptional factors to allow cell clock to repair the genetic gaps. Using genetic modified mice of adenocarcinoma and prostate (TRAMP), we also confirmed the effect of melatonin, significantly inhibited PCa tumor growth through SIRT1 inhibition [58].
In several cancers, e cell cycle regulation is disrupted by-self to gain mutational changes to oncogenes or to tumor suppressor genes. The detailed role of histone deacetylases on cell signaling mechanism of DNA damage and repair in cancer cells and genotoxin based resistance chemotherapy was reviewed by Roos et al. [57].
p53 deacetylation by SIRT1 play a significant role to regulating several checkpoints of cell cycle and inhibits apoptosis through E2F1 and promotes to suppression of DNA damage [59,60]. Previous studies have been demonstrated SIRT1 regulates all above these pathways in different tumors through deacetylating of Ku70, Nijmegen breakage syndrome protein (NBS1) and apurinic/apyrimidinic endonuclease-1 (APE1) proteins [61-63]. These pathways are highly resistant to chemotherapy in adult T-cell leukemia-lymphoma (ATL), both in vitro and in vivo. Previous findings, NHEJ and HR both are deceased due to knockout of KU70 and dephosphorylation of SIRT1, accompanied with an overexpressed level of SHP-1, which is a dephosphorylation enzyme [64]. Moreover, the DNA damage repair mechanism study in Chinese hamster ovary cells and rat Sertoli cells are treated with Microcystin-leucine arginine to induce apoptosis, suppressing acetylation of p53 and Ku70, and promoting the binding effect of Ku70 with Bax by suppression of SIRT1/p53 pathway [65].
While in vivo study of homozygous and hetrozygous SIRT1 mutation in genetic modified mice skin, increases expression of Noxa and p53 acetylation, and UVB sensitizes to the epidermis and induced apoptosis through DNA damage, whereas heterozygous SIRT1 has no significant effect on mouse skin. SIRT1 role in cell proliferation and DNA damage repair is stable with the dual functions of SIRT1 in UVB-induced tumorigenesis [66]. Further investigation has been confirmed both in vitro and in vivo that SIRT1 specific deletion in mice skin regulates invasion, cell migration, inflammation, granulation formation, re-epithelialization of epidermis, and appropriate wounds healing [67]. Downregulating DNA damage repair expression in tumor cells is a target approach to combine with DNA damaging drugs. Furthermore, carcinomas commonly have been developed due to dysfunction in at least one pathway which regulates by DNA damage machinery, which can lead to whole mechanism using a different repair pathway which is functionally deceased in normal human cells. PARP1 inhibitors could be reduced homologous recombination DNA repair pathways in BRCA1/BRCA2 mutant cells, these cells are sensitive to therapy due to their impaired [68].
It has been found that SIRT1 downregulation in T-cell leukemia-lymphoma (ATL) enhances sensitivity to chemotherapeutics drugs, apoptosis and cell cycle arrest. Further investigated SIRT1 promotes DNA damage repair by increased Ku70 and FOXO1 deacetylation [69]. We focused on SIRT1 deacetylase activity in cell genotoxic stress, such as senescence, apoptosis, and tumorigenesis, and its major function in the APE1 deacetylation. APE1 is a subunit of a multiprotein complex such as Ku70, RNA Pol II and hOGG1, which is involved as an activator on the promoter of SIRT1 to regulate functions of SIRT1 during the early metabolic redox response to stress [70]. However, the role of SIRT1 in longevity related with telomere gene and XRCC6, they have a diverse impact on the alternative pathways of telomere maintenance to aging and cell growth [71]. Recently, substantial progress has been investigated SIRT1 promotes expression of xeroderma pigmentosum complementation group A (XPA) deacetylation and enhances its binding with ataxia telangiectasia-mutated and Rad3-related (ATR) protein, increased cAMP-induced DNA repair responses mainly NER through MC1R signaling partway [72]. DNA damage repair by BER which is associated with SIRT1, SIRT1 activates its endonuclease activity of POLβ by increasing its interaction with XRCC1 through APE1 and it defends cells from base excision by H2O2 and MMS [63]. However, a recent study shown that breast cancer cells are modulating SIRT1/β-catenin signaling pathways, they are resistant to doxorubicin (DOX) effectively inhibited apoptosis and promotes cell growth and DNA damage repair [73].
Moreover, Wei et al. found SIRT1 overexpression in MCF-7/ADR breast cancer resistant cells/which enhanced the Akt pathway to promote cell growth [74]. SIRT1 expression was positively regulated p125 and suggests that SIRT1 is a tumor promoter gene that might be involved in the development of breast cancer by downregulating of p53 and upregulating of DNA polymerase delta1 (POLD1) [75]. In short, the role of SIRT1 in the study of functional genomics and proteomic, analytical developments have concluded in a significant SIRT1 increases, in our capability to detect novel cell signaling pathways that are associated in describing the responding of tumors to specific therapeutic agent. There are sufficient evidence while deactivation of apoptosis is a hallmark of cancer and could be promote to therapy resistance, cancer cells naturally act ‘autonomous’ as to appropriately antiapoptotic proteins for their survival and progression, as long as a suitable environment for promoting these proteins as therapeutics. BCL-2 is a member of antiapoptotic family, it is most prominent in drug resistance cancer due to elevated levels of SIRT1 expression in a huge range of cancers. Clinically studies have been investigated, SIRT1 is positively linked to PRRX1in breast cancer although KLF4 is reversely correlated [76].
Mutations, genetic translocations, amplification and overexpression of the transcriptional and translational coding genes, all these proteins are involved in several diseases and related to targeted chemoresistance. In chemoresistance, death receptor (DR)-mediated apoptosis and the death ligand TRAIL has been interesting as potential endured extensive preclinical and clinical testing for cancer cells death [77-79]. In detail study of SIRT1 activation in cancer investigated that the SIRT1 role is still undefined whether SIRT1 overexpression is beneficial or harmful in cancer growth and the aging process. However, HIC1 is a tumor suppressor gene could be controlled SIRT1 transcription, they have determined in cancer cells and xenograft models, to identify, it is a ring shape like regulator among HIC1, p53 and SIRT1, in which repression of SIRT1 transcription by directly HIC1 [80]. Meanwhile, it has been indicated that SIRT1 and p53 in lung adenocarcinoma and squamous carcinoma patients, and found a stable complex among transcription regulator and SIRT1 to inhibit the expression of SIRT1, consequently persuading apoptosis in cells by oxidase [81,82]. SIRT1 might be a promising as a functional positive regulator of USP22 that may induce cell proliferation and upregulate MDR proteins to enhance resistance to chemotherapy in HCC. Notably, studies revealed SIRT1 directly interacted with USP22, both SIRT1 and USP22 significantly influenced on the AKT and MRP1 pathway. So, we found that USP22 might be promoting MDR and antiapoptotic activity in HCC by inducing AKT/MRP1/SIRT1 pathway [83]. Although, PI3K/AKT pathway and FOX O3a, EGR1 signaling cascade was enhanced and driven to apoptosis by inhibition of SIRT1 expression and AKT/p300 regulate via Bim suppression in lung cancer [84]. Further investigation has been examined mRNA of SIRT1 which positively links with growth factor, SIRT1 deacetylation leading to upregulate transcriptional and translational YAP2/TEAD4 level and cell growth progression in HCC cells [85]. Moreover, SIRT1 downregulated the apoptosis including Smac/NF-κB pathway and promotes DNA damage repair in lung cancer [52]. Lian et al. reported SIRT1 overexpression enhanced resistance to colorectal cancer due to tumor necrosis factor-related apoptosis-inducing ligand (TRAIL). It has been shown that inhibition of SIRT1 enhanced the expression of ROS and DR5 frequently through miR-128 directly targeted SIRT1 [86]. It is often stated that SIRT1 also activates anti-apoptosis protein BCL6 for the progression and survival of lymphoma [87]. However, anti-apoptotic BCL-2 family member has limited effectiveness to resistance mechanisms most notably due to another. ABT-737 weakly interacts with MCL1 and it promotes resistance which has been observed in MCL1 expressing cells [88-91]. We could restore sensitivity to ABT-737 whether downregulating of MCL1. MCL1 is a remarkably significant resistance to ABT-737, and is also resistance to decrease cytotoxicity of anticancer drug [92]. SIRT1mediated overview in chemotherapeutics resistance mechanism in cancer is shown in Figure 2.
SIRT1 is a regulator of autophagy
Autophagy is lysosomal destruction of a cellular pathway that degrades proteins and cell organelles to maintain homeostasis during metabolic stress. Significant evidence have reported autophagy has a vital role in tumorigenesis is paradoxical as its role both tumor suppressor as well chemoresistance by facilitating tumor survival and cell progression under stress condition caused by anticancer therapeutic agents [93]. Xiong et al. has been investigated the role of autophagy in chemoresistance HCC such as pirarubicin (THP), oxaliplatin and 5-fluorouracil, which is regulated by SIRT1 linked with HULC and USP22. Collectively, HULC lncRNA is overexpressed in liver carcinoma and revealed that SIRT1/HULC/USP22 is a protecting autophagy signaling pathway to overcome the sensitivity of HCC cells to chemotherapeutics [94]. SIRT1 induced resistant to doxorubicin in breast cancer due to deacetylation of p53, natural anticancer Psammaplin A (PsA) product is markedly beneficial to suppress SIRT1 and arrest cell cycle in G2/M phase. Overexpression of autophagy-related proteins has been increased significantly the expression of DNA damage-regulated autophagy modulator (DRAM) and p53, through SIRT1 in MCF-7/R breast cancer cell line [95]. SIRT1 plays an important role in the autophagy, it reduces H2O2 oxidative stress and linked ROS which is particularly dependent on metabolic reactions and mediated with class III PI3K/Beclin-1 and mTOR signaling pathways [96]. Guanglin et al. have been reported that SIRT1 is a potential marker for diagnosis because it is a mediator of drug resistance through epithelial-mesenchymal transition (EMT) pathway, cell invasion and mcell migration. Autophagy can regulate a cell-survival mechanism, the functionality of SIRT1 is defined by several features such as the tumor initiation, cell morphology and resistance to drugs. Remarkably, SIRT1 regulates the autophagy pathway in gastric cancer through the deacetylation of several genes involved in autophagy [29]. Xie et al., found klotho tumor suppressor gene epigenetically activated in gastric cancer cells, it is stimulating (IGF1/IRS1)/phospho-inositide 3-kinase (PI3K)/Akt/mTOR which is associated with autophagy, apoptosis, and cell progression pathway [97].
SIRT1 increases autophagic flux in prostate cancer and decreases p62 protein expression and also upregulate Rab7, it is a small molecule that promotes late autophagosome fusion process by the activating deacetylation of FoxO1 under stress conditions due to starvation [25,98,99]. MGC803, SGC7901 cells, and HCC are resistant to cisplatin and oxaliplatin by activating autophagy pathway to maintaining the cell stability and cell survival [100,101]. Moreover, nutrient deficiency can stimulate the metabolic pathways and phosphorylates the AMPK proteins which regulates the autophagy by phosphorylating PI3K complex and ULK1 [102,103]. Here, we emphasized briefly the role of SIRT1 mediated autophagy and observed that autophagy has emerged as a promising is a housekeeping novel approach in drug resistance, aging and age-related degeneration in cancer study.
Implications of SIRT1 inhibition in chemotherapeutics resistance
Previous findings have figured out the function of SIRT1 inhibition in chemotherapeutic resistance cancers to overcome resistance, it is a worthy approach for the development and improvement of cancer treatment. The cell cycle arrest and induction of cell senescence has been developed as a potential sponsor to the antitumor effects of chemotherapeutic agents. According to recent advances to the understanding of SIRT1 induction drug resistance molecular mechanism in cancer, it seems SIRT1 overexpression regulates metabolism, cell cycle, DNA damage repair, apoptosis, autophagy and senescence [36,104-107]. In this perspective, current studies support that SIRT1 overexpression plays a significant role in the regulation of senescence like cell growth arrest and stress responses in tumorigenesis induced by anticancer drugs. Many questions about SIRT1 remain unclear how it effects tumorigenesis and promotes multidrug resistance, in short, might be it depends on the genetic context that has arisen during oncogenesis. For better understating of SIRT1 functions the previous study found several small specific or non-specific compounds that inhibit SIRT1 expression and increases p53 acetylation in various studies including both in vitro and in vivo, but there is a need, the combination of chemotherapeutics for their potential result [108]. A significant study has been made by the previous study, SIRT1 activators or inhibitors applying to clinical practice as therapeutics, first reported SIRT1 activator resveratrol later identified other SIRT1 activators, SRT1720, SRT2183, and SRT1460. However, increased SIRT1 expression has a clear positive effect on tumorigenesis and chemoresistance, still required to be found specific activators for the better understanding of SIRT1 bio-physiology and SIRT1 associated malignancies [109-112]. There are different families of small-molecular chemical and biological inhibitors of SIRT1, one of the potentate family has been found, called tenovins that target class HDAC III. Tenovin 6 is an effective drug which inhibits SIRT1 at low concentrations and stops tumor growth in vivo and target to the p53-dependent pathway, it is more advantageous when used with inhibitors of MDM2 small molecules target P53 pathway and exhibit tumor growth suppression in several cell lines. Jin et al. also reported SIRT1-2, inhibition by tenovin-6, it promotes p53 activation and induced cell apoptosis in acute lymphoblastic leukemia (ALL) cells and clinical and pre-clinical trials [113,114]. Although, we can say SIRT1 suppression may enhance antitumor effects via independent KLB1 pathway [115]. Interestingly, tenovin-6 induced apoptosis not only wild-type TP53 either in all cell lines, rather accompanied by activation of death receptor 5 (DR5) signaling pathway and remarkably could not effect on PERK, ATF6, and CHOP. However, combination of tenovin-6 with chemotherapeutic drugs SN-38 or docetaxel to exhibit synergistic cytotoxicity effects on gastric cancer cells. Moreover, tenovin-6 and metformin synergistically downregulate SIRT1 expression in NSCLC cells, increased p53 acetylation and induced caspase-3 apoptosis activity [116].
Silybin is an antitumor molecule, it also inhibits SIRT1 and upregulated p53 acetylation. In addition, silybin and SIRT1 inhibitor cambinol induced in mice and used as dose and time dependent for in vitro study. Haris has been found significant results, silybin is an effective inhibitor of adenocarcinoma might be used as a therapeutic intervention in lung adenocarcinoma [117]. The progression of uveal melanoma (UM) and the diagnosis has remained pathetic, HDACs inhibitor tenovin-6 persuades apoptosis, suppression of cell migration and invasion, and elimination of cancer stem cells (CSCs) in uveal melanoma [118]. SIRT1 knockdown by another inhibitor sirtinol is induced a senescence like growth arrest, might have anticancer activity, and that impaired stimulation of Ras/MAPK pathway. Though, sirtinol tyrosine phosphorylation of the receptors for EGF and IGF-I and Akt/PKB activation was showing on effect by Sirtinol [119]. SIRT1 overexpression induced cisplatin resistance in HHUA cells accelerated tumorigenesis in nude mice, SIRT1 inhibition by EX527 significantly suppressed the tumor growth of HEC1B and HHUA endometrial carcinoma. A combination of EX527 and cisplatin might a valuable targeted therapy for curing of cisplatin-resistant malignancy [120]. EX527, Sirtinol, Nicotinamide, and Salermide are direct target of inhibitors SIRT1 and 2, having the specific inhibitory activity for SIRT1 even though, EX527 is only specific for SIRT1 rather than other sirtuin members according to computational docking analysis than other SIRTs. Furthermore, exiting approach SIRT1 inhibition by EX527 in vitro and in vivo both pancreatic tumor model, unexpectedly in vivo result is reverse to in vitro, EX527 induced tumorigenesis in SCID mice as compared with control group whether it induce apoptosis and DNA damage in in vitro [121].
Moreover, small interfering RNA analysis that targets SIRT1, observed that SIRT1 knockdown can induce cell death in MCF-7 cell line [122]. In addition, it is believed that SIRT1 accelerates cell growth, miR-29c overexpression in cisplatin-resistant cancer cells directly target SIRT1 mRNA and suppress SIRT1 expression was demonstrated by Zhang et al. to regulate cell progression, apoptosis, as well as restoring chemosensitivity of cisplatin [123], miR-34a mediated SIRT1 suppression mediates apoptotic activation and chemosensitivity [124]. Cellular mechanisms of SIRT1 in colorectal cancer, taken together clinical data with patient samples and found a mechanical approach towards regulation of p53 and FRA-1 by SIRT1 and confirmed directly linked with EMT [125].
Multidrug resistance in cancer is a complicated mechanism influenced by drug resistance proteins, tumor microenvironment, DNA damage protein and receptor alteration, and an impairment in cell to cell communication of CD9 signaling pathway [126,127]. SIRT1 overexpression in tumor cells has been associated with multidrug resistance proteins and it is known to control the traffic of extracellular vesicles in the process of chemoresistance [128-131]. Overexpression of SIRT1 might be a potential biomarker for advanced stage of osteosarcoma patients and other cancers as well [132]. Yuan et al. developed advanced novel culture model of chronic myelogenous leukemia (CML) acquired resistance based CML cell line and KCL-22 for chronic myelogenous leukemia study [133]. Hematopoietic malignancy induced by BCR-ABL oncogenic fusion gene that has many factors which is responsible for cell proliferation and DNA damage repair mechanism. While, masitinib, nilotinib and BCR-ABL tyrosine kinase inhibitor (TKI), could be a significantly extended for advance CML treatment [134,135]. Drug resistance initially facilitated by genetic alterations of BCR-ABL proteins, it remains a big challenge, particularly, for advanced cancers.
However, previous studies have been found significant role of SIRT1 in cancer development and metastasis both in vitro and in vivo. For in vivo analysis, an orthotopic xenograft model used for hepatocellular carcinoma (HCC), SIRT1 knockdown done by SIRT1 inhibitor cambinol, after treatment reduced tumor growth and an overall examined lower tumor burden, these findings were suggested that SIRT1 overexpression induced the growth of HCC [136]. Moreover, SIRT1 mechanisms in cancer resistance development, it could be more challenging lacking of well-intentioned modeling system in human cancers as well other mammalian cells [137]. SIRT1 role in chemoresistance, metastasis and tumor microenvironment is shown in Figure 3.
Future prospective
Significant advances have been made in better understanding of SIRT1 regulation and normal physiological functions in the perspective of therapeutic resistance, genomic stability and cancer progression over the past few decades. Despite all these findings, one of the major concerns of SIRT1 overexpression in oncogenic observation inactivates p53 wild type but its reverse effect in mutated p53. The relationship between SIRT1 overexpression and mutated p53 in tumorigeneses needs investigation and validation for conferring of SIRT1 as oncogenic and development of chemoresistance. However, the function of SIRT1 in cancer remains crucial and many questions need to be responded. On the other hand, Why SIRT1 overexpression could be a potential therapeutic target in various cancers but still it has suppressive effect in others. Conceivably, most demanding in the study of recent data to understand how SIRT1 is regulated stress, cellular senescence, cell growth and multidrug resistance. SIRT1 is a vital part of cell signaling pathways that play significant role in DNA damage repair, cell progression, invasion and metastasis, chemotherapeutics resistance and longevity that are significant in human cancers, it will remain a positively therapeutic study for future. Here, we discuss aforementioned evidences of SIRT1 potentials for developing new therapeutics along with SIRT1 modulators (inhibitors or activators), independently or with combination of other anticancer molecules to overcome the obstacle of treatment of advanced cancers including metastatic, chemotherapeutics as well as radio therapeutics resistance cancers.
Acknowledgements
This work was supported by NSFC, ZJNSF (LZ18H160001) and Department of Healthcare in Zhejiang.
Disclosure of conflict of interest
None.
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Harguindey S, Orive G, Luis Pedraz J, Paradiso A, Reshkin SJ. The role of pH dynamics and the Na+/H+ antiporter in the etiopathogenesis and treatment of cancer. Two faces of the same coin--one single nature. Biochim Biophys Acta. 2005;1756:1–24. doi: 10.1016/j.bbcan.2005.06.004. [DOI] [PubMed] [Google Scholar]
- 3.Housman G, Byler S, Heerboth S, Lapinska K, Longacre M, Snyder N, Sarkar S. Drug resistance in cancer: an overview. Cancers (Basel) 2014;6:1769–1792. doi: 10.3390/cancers6031769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.North BJ, Verdin E. Sirtuins: Sir2-related NAD-dependent protein deacetylases. Genome Biol. 2004;5:224. doi: 10.1186/gb-2004-5-5-224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Canto C, Houtkooper RH, Pirinen E, Youn DY, Oosterveer MH, Cen Y, Fernandez-Marcos PJ, Yamamoto H, Andreux PA, Cettour-Rose P, Gademann K, Rinsch C, Schoonjans K, Sauve AA, Auwerx J. The NAD(+) precursor nicotinamide riboside enhances oxidative metabolism and protects against high-fat diet-induced obesity. Cell Metab. 2012;15:838–847. doi: 10.1016/j.cmet.2012.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kaeberlein M, McVey M, Guarente L. The SIR2/3/4 complex and SIR2 alone promote longevity in Saccharomyces cerevisiae by two different mechanisms. Genes Dev. 1999;13:2570–2580. doi: 10.1101/gad.13.19.2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vaquero A, Scher M, Lee D, Erdjument-Bromage H, Tempst P, Reinberg D. Human SirT1 interacts with histone H1 and promotes formation of facultative heterochromatin. Mol Cell. 2004;16:93–105. doi: 10.1016/j.molcel.2004.08.031. [DOI] [PubMed] [Google Scholar]
- 8.Tanno M, Sakamoto J, Miura T, Shimamoto K, Horio Y. Nucleocytoplasmic shuttling of the NAD+-dependent histone deacetylase SIRT1. J Biol Chem. 2007;282:6823–6832. doi: 10.1074/jbc.M609554200. [DOI] [PubMed] [Google Scholar]
- 9.Saunders LR, Verdin E. Sirtuins: critical regulators at the crossroads between cancer and aging. Oncogene. 2007;26:5489–5504. doi: 10.1038/sj.onc.1210616. [DOI] [PubMed] [Google Scholar]
- 10.Finkel T, Deng CX, Mostoslavsky R. Recent progress in the biology and physiology of sirtuins. Nature. 2009;460:587–591. doi: 10.1038/nature08197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Houtkooper RH, Pirinen E, Auwerx J. Sirtuins as regulators of metabolism and healthspan. Nat Rev Mol Cell Biol. 2012;13:225–238. doi: 10.1038/nrm3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bradbury CA, Khanim FL, Hayden R, Bunce CM, White DA, Drayson MT, Craddock C, Turner BM. Histone deacetylases in acute myeloid leukaemia show a distinctive pattern of expression that changes selectively in response to deacetylase inhibitors. Leukemia. 2005;19:1751–1759. doi: 10.1038/sj.leu.2403910. [DOI] [PubMed] [Google Scholar]
- 13.Stunkel W, Peh BK, Tan YC, Nayagam VM, Wang X, Salto-Tellez M, Ni B, Entzeroth M, Wood J. Function of the SIRT1 protein deacetylase in cancer. Biotechnol J. 2007;2:1360–1368. doi: 10.1002/biot.200700087. [DOI] [PubMed] [Google Scholar]
- 14.Chen X, Hokka D, Maniwa Y, Ohbayashi C, Itoh T, Hayashi Y. Sirt1 is a tumor promoter in lung adenocarcinoma. Oncol Lett. 2014;8:387–393. doi: 10.3892/ol.2014.2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hida Y, Kubo Y, Murao K, Arase S. Strong expression of a longevity-related protein, SIRT1, in Bowen’s disease. Arch Dermatol Res. 2007;299:103–106. doi: 10.1007/s00403-006-0725-6. [DOI] [PubMed] [Google Scholar]
- 16.Powell MJ, Casimiro MC, Cordon-Cardo C, He X, Yeow WS, Wang C, McCue PA, McBurney MW, Pestell RG. Disruption of a Sirt1-dependent autophagy checkpoint in the prostate results in prostatic intraepithelial neoplasia lesion formation. Cancer Res. 2011;71:964–975. doi: 10.1158/0008-5472.CAN-10-3172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harris AL. DNA repair and resistance to chemotherapy. Cancer Surv. 1985;4:601–624. [PubMed] [Google Scholar]
- 18.Tsukamoto S, Akashi-Tanaka S, Shien T, Terada K, Kinoshita T. Brain metastases after achieving local pathological complete responses with neoadjuvant chemotherapy. Breast Cancer. 2007;14:420–424. doi: 10.2325/jbcs.14.420. [DOI] [PubMed] [Google Scholar]
- 19.Olive KP, Jacobetz MA, Davidson CJ, Gopinathan A, McIntyre D, Honess D, Madhu B, Goldgraben MA, Caldwell ME, Allard D, Frese KK, Denicola G, Feig C, Combs C, Winter SP, Ireland-Zecchini H, Reichelt S, Howat WJ, Chang A, Dhara M, Wang L, Ruckert F, Grutzmann R, Pilarsky C, Izeradjene K, Hingorani SR, Huang P, Davies SE, Plunkett W, Egorin M, Hruban RH, Whitebread N, McGovern K, Adams J, Iacobuzio-Donahue C, Griffiths J, Tuveson DA. Inhibition of Hedgehog signaling enhances delivery of chemotherapy in a mouse model of pancreatic cancer. Science. 2009;324:1457–1461. doi: 10.1126/science.1171362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lavu S, Boss O, Elliott PJ, Lambert PD. Sirtuins--novel therapeutic targets to treat age-associated diseases. Nat Rev Drug Discov. 2008;7:841–853. doi: 10.1038/nrd2665. [DOI] [PubMed] [Google Scholar]
- 21.Haigis MC, Guarente LP. Mammalian sirtuins--emerging roles in physiology, aging, and calorie restriction. Genes Dev. 2006;20:2913–2921. doi: 10.1101/gad.1467506. [DOI] [PubMed] [Google Scholar]
- 22.Haberland M, Montgomery RL, Olson EN. The many roles of histone deacetylases in development and physiology: implications for disease and therapy. Nat Rev Genet. 2009;10:32–42. doi: 10.1038/nrg2485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sung JY, Kim R, Kim JE, Lee J. Balance between SIRT1 and DBC1 expression is lost in breast cancer. Cancer Sci. 2010;101:1738–1744. doi: 10.1111/j.1349-7006.2010.01573.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee H, Kim KR, Noh SJ, Park HS, Kwon KS, Park BH, Jung SH, Youn HJ, Lee BK, Chung MJ, Koh DH, Moon WS, Jang KY. Expression of DBC1 and SIRT1 is associated with poor prognosis for breast carcinoma. Hum Pathol. 2011;42:204–213. doi: 10.1016/j.humpath.2010.05.023. [DOI] [PubMed] [Google Scholar]
- 25.You Z, Liu Y, Liu X. Nicotinamide N-methyltransferase enhances the progression of prostate cancer by stabilizing sirtuin 1. Oncol Lett. 2018;15:9195–9201. doi: 10.3892/ol.2018.8474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lv L, Shen Z, Zhang J, Zhang H, Dong J, Yan Y, Liu F, Jiang K, Ye Y, Wang S. Clinicopathological significance of SIRT1 expression in colorectal adenocarcinoma. Med Oncol. 2014;31:965. doi: 10.1007/s12032-014-0965-9. [DOI] [PubMed] [Google Scholar]
- 27.Zhang LH, Huang Q, Fan XS, Wu HY, Yang J, Feng AN. Clinicopathological significance of SIRT1 and p300/CBP expression in gastroesophageal junction (GEJ) cancer and the correlation with E-cadherin and MLH1. Pathol Res Pract. 2013;209:611–617. doi: 10.1016/j.prp.2013.03.012. [DOI] [PubMed] [Google Scholar]
- 28.Choi HN, Bae JS, Jamiyandorj U, Noh SJ, Park HS, Jang KY, Chung MJ, Kang MJ, Lee DG, Moon WS. Expression and role of SIRT1 in hepatocellular carcinoma. Oncol Rep. 2011;26:503–510. doi: 10.3892/or.2011.1301. [DOI] [PubMed] [Google Scholar]
- 29.Qiu G, Li X, Che X, Wei C, He S, Lu J, Jia Z, Pang K, Fan L. SIRT1 is a regulator of autophagy: implications in gastric cancer progression and treatment. FEBS Lett. 2015;589:2034–2042. doi: 10.1016/j.febslet.2015.05.042. [DOI] [PubMed] [Google Scholar]
- 30.Kim JR, Moon YJ, Kwon KS, Bae JS, Wagle S, Yu TK, Kim KM, Park HS, Lee JH, Moon WS, Lee H, Chung MJ, Jang KY. Expression of SIRT1 and DBC1 is associated with poor prognosis of soft tissue sarcomas. PLoS One. 2013;8:e74738. doi: 10.1371/journal.pone.0074738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stenzinger A, Endris V, Klauschen F, Sinn B, Lorenz K, Warth A, Goeppert B, Ehemann V, Muckenhuber A, Kamphues C, Bahra M, Neuhaus P, Weichert W. High SIRT1 expression is a negative prognosticator in pancreatic ductal adenocarcinoma. BMC Cancer. 2013;13:450. doi: 10.1186/1471-2407-13-450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Noh SJ, Kang MJ, Kim KM, Bae JS, Park HS, Moon WS, Chung MJ, Lee H, Lee DG, Jang KY. Acetylation status of P53 and the expression of DBC1, SIRT1, and androgen receptor are associated with survival in clear cell renal cell carcinoma patients. Pathology. 2013;45:574–580. doi: 10.1097/PAT.0b013e3283652c7a. [DOI] [PubMed] [Google Scholar]
- 33.Noh SJ, Baek HA, Park HS, Jang KY, Moon WS, Kang MJ, Lee DG, Kim MH, Lee JH, Chung MJ. Expression of SIRT1 and cortactin is associated with progression of non-small cell lung cancer. Pathol Res Pract. 2013;209:365–370. doi: 10.1016/j.prp.2013.03.011. [DOI] [PubMed] [Google Scholar]
- 34.Zhang T, Rong N, Chen J, Zou C, Jing H, Zhu X, Zhang W. SIRT1 expression is associated with the chemotherapy response and prognosis of patients with advanced NSCLC. PLoS One. 2013;8:e79162. doi: 10.1371/journal.pone.0079162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brunet A, Sweeney LB, Sturgill JF, Chua KF, Greer PL, Lin Y, Tran H, Ross SE, Mostoslavsky R, Cohen HY, Hu LS, Cheng HL, Jedrychowski MP, Gygi SP, Sinclair DA, Alt FW, Greenberg ME. Stress-dependent regulation of FOXO transcription factors by the SIRT1 deacetylase. Science. 2004;303:2011–2015. doi: 10.1126/science.1094637. [DOI] [PubMed] [Google Scholar]
- 36.Vaziri H, Dessain SK, Ng Eaton E, Imai SI, Frye RA, Pandita TK, Guarente L, Weinberg RA. hSIR2(SIRT1) functions as an NAD-dependent p53 deacetylase. Cell. 2001;107:149–159. doi: 10.1016/s0092-8674(01)00527-x. [DOI] [PubMed] [Google Scholar]
- 37.Chakrabarti P, English T, Karki S, Qiang L, Tao R, Kim J, Luo Z, Farmer SR, Kandror KV. SIRT1 controls lipolysis in adipocytes via FOXO1-mediated expression of ATGL. J Lipid Res. 2011;52:1693–1701. doi: 10.1194/jlr.M014647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gottesman MM, Fojo T, Bates SE. Multidrug resistance in cancer: role of ATP-dependent transporters. Nat Rev Cancer. 2002;2:48–58. doi: 10.1038/nrc706. [DOI] [PubMed] [Google Scholar]
- 39.Shin DH, Choi YJ, Park JW. SIRT1 and AMPK mediate hypoxia-induced resistance of non-small cell lung cancers to cisplatin and doxorubicin. Cancer Res. 2014;74:298–308. doi: 10.1158/0008-5472.CAN-13-2620. [DOI] [PubMed] [Google Scholar]
- 40.Yuan J, Minter-Dykhouse K, Lou Z. A c-Myc-SIRT1 feedback loop regulates cell growth and transformation. J Cell Biol. 2009;185:203–211. doi: 10.1083/jcb.200809167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Byles V, Zhu L, Lovaas JD, Chmilewski LK, Wang J, Faller DV, Dai Y. SIRT1 induces EMT by cooperating with EMT transcription factors and enhances prostate cancer cell migration and metastasis. Oncogene. 2012;31:4619–4629. doi: 10.1038/onc.2011.612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cosse JP, Michiels C. Tumour hypoxia affects the responsiveness of cancer cells to chemotherapy and promotes cancer progression. Anticancer Agents Med Chem. 2008;8:790–797. doi: 10.2174/187152008785914798. [DOI] [PubMed] [Google Scholar]
- 43.Zhou S, Tang X, Chen HZ. Sirtuins and insulin resistance. Front Endocrinol (Lausanne) 2018;9:748. doi: 10.3389/fendo.2018.00748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nemoto S, Fergusson MM, Finkel T. SIRT1 functionally interacts with the metabolic regulator and transcriptional coactivator PGC-1{alpha} J Biol Chem. 2005;280:16456–16460. doi: 10.1074/jbc.M501485200. [DOI] [PubMed] [Google Scholar]
- 45.Sun C, Zhang F, Ge X, Yan T, Chen X, Shi X, Zhai Q. SIRT1 improves insulin sensitivity under insulin-resistant conditions by repressing PTP1B. Cell Metab. 2007;6:307–319. doi: 10.1016/j.cmet.2007.08.014. [DOI] [PubMed] [Google Scholar]
- 46.Rodgers JT, Lerin C, Haas W, Gygi SP, Spiegelman BM, Puigserver P. Nutrient control of glucose homeostasis through a complex of PGC-1alpha and SIRT1. Nature. 2005;434:113–118. doi: 10.1038/nature03354. [DOI] [PubMed] [Google Scholar]
- 47.Xia X, Zhou X. Knockdown of SIRT1 inhibits proliferation and promotes apoptosis of paclitaxel-resistant human cervical cancer cells. Cell Mol Biol (Noisy-le-grand) 2018;64:36–41. [PubMed] [Google Scholar]
- 48.Zaidi N, Lupien L, Kuemmerle NB, Kinlaw WB, Swinnen JV, Smans K. Lipogenesis and lipolysis: the pathways exploited by the cancer cells to acquire fatty acids. Prog Lipid Res. 2013;52:585–589. doi: 10.1016/j.plipres.2013.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Armand V, Rundfeldt C, Heinemann U. Effects of retigabine (D-23129) on different patterns of epileptiform activity induced by 4-aminopyridine in rat entorhinal cortex hippocampal slices. Naunyn Schmiedebergs Arch Pharmacol. 1999;359:33–39. doi: 10.1007/pl00005320. [DOI] [PubMed] [Google Scholar]
- 50.Simmons GE Jr, Pruitt WM, Pruitt K. Diverse roles of SIRT1 in cancer biology and lipid metabolism. Int J Mol Sci. 2015;16:950–965. doi: 10.3390/ijms16010950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kim MJ, An HJ, Kim DH, Lee B, Lee HJ, Ullah S, Kim SJ, Jeong HO, Moon KM, Lee EK, Yang J, Akter J, Chun P, Moon HR, Chung HY. Novel SIRT1 activator MHY2233 improves glucose tolerance and reduces hepatic lipid accumulation in db/db mice. Bioorg Med Chem Lett. 2018;28:684–688. doi: 10.1016/j.bmcl.2018.01.021. [DOI] [PubMed] [Google Scholar]
- 52.Ji K, Sun X, Liu Y, Du L, Wang Y, He N, Wang J, Xu C, Liu Q. Regulation of apoptosis and radiation sensitization in lung cancer cells via the Sirt1/NF-kappaB/Smac pathway. Cell Physiol Biochem. 2018;48:304–316. doi: 10.1159/000491730. [DOI] [PubMed] [Google Scholar]
- 53.Yousafzai NA, Zhou Q, Xu W, Shi Q, Xu J, Feng L. SIRT1 deacetylated and stabilized XRCC1 to promote chemoresistance in lung cancer. Cell Death Dis. 2019;10:363. doi: 10.1038/s41419-019-1592-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yuan Z, Zhang X, Sengupta N, Lane WS, Seto E. SIRT1 regulates the function of the Nijmegen breakage syndrome protein. Mol Cell. 2007;27:149–162. doi: 10.1016/j.molcel.2007.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Krokan HE, Bjoras M. Base excision repair. Cold Spring Harb Perspect Biol. 2013;5:a012583. doi: 10.1101/cshperspect.a012583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Koprinarova M, Botev P, Russev G. Histone deacetylase inhibitor sodium butyrate enhances cellular radiosensitivity by inhibiting both DNA nonhomologous end joining and homologous recombination. DNA Repair (Amst) 2011;10:970–977. doi: 10.1016/j.dnarep.2011.07.003. [DOI] [PubMed] [Google Scholar]
- 57.Roos WP, Krumm A. The multifaceted influence of histone deacetylases on DNA damage signalling and DNA repair. Nucleic Acids Res. 2016;44:10017–10030. doi: 10.1093/nar/gkw922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jung-Hynes B, Schmit TL, Reagan-Shaw SR, Siddiqui IA, Mukhtar H, Ahmad N. Melatonin, a novel Sirt1 inhibitor, imparts antiproliferative effects against prostate cancer in vitro in culture and in vivo in TRAMP model. J Pineal Res. 2011;50:140–149. doi: 10.1111/j.1600-079X.2010.00823.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Enoch T, Norbury C. Cellular responses to DNA damage: cell-cycle checkpoints, apoptosis and the roles of p53 and ATM. Trends Biochem Sci. 1995;20:426–430. doi: 10.1016/s0968-0004(00)89093-3. [DOI] [PubMed] [Google Scholar]
- 60.Wang C, Chen L, Hou X, Li Z, Kabra N, Ma Y, Nemoto S, Finkel T, Gu W, Cress WD, Chen J. Interactions between E2F1 and SirT1 regulate apoptotic response to DNA damage. Nat Cell Biol. 2006;8:1025–1031. doi: 10.1038/ncb1468. [DOI] [PubMed] [Google Scholar]
- 61.Cohen HY, Lavu S, Bitterman KJ, Hekking B, Imahiyerobo TA, Miller C, Frye R, Ploegh H, Kessler BM, Sinclair DA. Acetylation of the C terminus of Ku70 by CBP and PCAF controls Bax-mediated apoptosis. Mol Cell. 2004;13:627–638. doi: 10.1016/s1097-2765(04)00094-2. [DOI] [PubMed] [Google Scholar]
- 62.Jeong J, Juhn K, Lee H, Kim SH, Min BH, Lee KM, Cho MH, Park GH, Lee KH. SIRT1 promotes DNA repair activity and deacetylation of Ku70. Exp Mol Med. 2007;39:8–13. doi: 10.1038/emm.2007.2. [DOI] [PubMed] [Google Scholar]
- 63.Yamamori T, DeRicco J, Naqvi A, Hoffman TA, Mattagajasingh I, Kasuno K, Jung SB, Kim CS, Irani K. SIRT1 deacetylates APE1 and regulates cellular base excision repair. Nucleic Acids Res. 2010;38:832–845. doi: 10.1093/nar/gkp1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yu W, Li L, Wang G, Zhang W, Xu J, Liang A. KU70 inhibition impairs both non-homologous end joining and homologous recombination DNA damage repair through SHP-1 induced dephosphorylation of SIRT1 in adult T-cell leukemia-lymphoma cells. Cell Physiol Biochem. 2018;49:2111–2123. doi: 10.1159/000493815. [DOI] [PubMed] [Google Scholar]
- 65.Liu H, Zhang S, Liu C, Wu J, Wang Y, Yuan L, Du X, Wang R, Marwa PW, Zhuang D, Cheng X, Zhang H. Resveratrol ameliorates microcystin-LR-induced testis germ cell apoptosis in rats via SIRT1 signaling pathway activation. Toxins (Basel) 2018;10:235. doi: 10.3390/toxins10060235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ming M, Soltani K, Shea CR, Li X, He YY. Dual role of SIRT1 in UVB-induced skin tumorigenesis. Oncogene. 2015;34:357–363. doi: 10.1038/onc.2013.583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Qiang L, Sample A, Liu H, Wu X, He YY. Epidermal SIRT1 regulates inflammation, cell migration, and wound healing. Sci Rep. 2017;7:14110. doi: 10.1038/s41598-017-14371-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Edwards SL, Brough R, Lord CJ, Natrajan R, Vatcheva R, Levine DA, Boyd J, Reis-Filho JS, Ashworth A. Resistance to therapy caused by intragenic deletion in BRCA2. Nature. 2008;451:1111–1115. doi: 10.1038/nature06548. [DOI] [PubMed] [Google Scholar]
- 69.Li L, Ye S, Yang M, Yu W, Fan Z, Zhang H, Hu J, Liang A, Zhang W. SIRT1 downregulation enhances chemosensitivity and survival of adult T-cell leukemia-lymphoma cells by reducing DNA double-strand repair. Oncol Rep. 2015;34:2935–2942. doi: 10.3892/or.2015.4287. [DOI] [PubMed] [Google Scholar]
- 70.Antoniali G, Lirussi L, D’Ambrosio C, Dal Piaz F, Vascotto C, Casarano E, Marasco D, Scaloni A, Fogolari F, Tell G. SIRT1 gene expression upon genotoxic damage is regulated by APE1 through nCaRE-promoter elements. Mol Biol Cell. 2014;25:532–547. doi: 10.1091/mbc.E13-05-0286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kim S, Bi X, Czarny-Ratajczak M, Dai J, Welsh DA, Myers L, Welsch MA, Cherry KE, Arnold J, Poon LW, Jazwinski SM. Telomere maintenance genes SIRT1 and XRCC6 impact age-related decline in telomere length but only SIRT1 is associated with human longevity. Biogerontology. 2012;13:119–131. doi: 10.1007/s10522-011-9360-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Jarrett SG, Carter KM, Bautista RM, He D, Wang C, D’Orazio JA. Sirtuin 1-mediated deacetylation of XPA DNA repair protein enhances its interaction with ATR protein and promotes cAMP-induced DNA repair of UV damage. J Biol Chem. 2018;293:19025–19037. doi: 10.1074/jbc.RA118.003940. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 73.Jin X, Wei Y, Liu Y, Lu X, Ding F, Wang J, Yang S. Resveratrol promotes sensitization to Doxorubicin by inhibiting epithelial-mesenchymal transition and modulating SIRT1/beta-catenin signaling pathway in breast cancer. Cancer Med. 2019;8:1246–1257. doi: 10.1002/cam4.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wei Y, Guo Y, Zhou J, Dai K, Xu Q, Jin X. Nicotinamide overcomes doxorubicin resistance of breast cancer cells through deregulating SIRT1/Akt pathway. Anticancer Agents Med Chem. 2019;19:687–696. doi: 10.2174/1871520619666190114160457. [DOI] [PubMed] [Google Scholar]
- 75.Xu Y, Qin Q, Chen R, Wei C, Mo Q. SIRT1 promotes proliferation, migration, and invasion of breast cancer cell line MCF-7 by upregulating DNA polymerase delta1 (POLD1) Biochem Biophys Res Commun. 2018;502:351–357. doi: 10.1016/j.bbrc.2018.05.164. [DOI] [PubMed] [Google Scholar]
- 76.Shi L, Tang X, Qian M, Liu Z, Meng F, Fu L, Wang Z, Zhu WG, Huang JD, Zhou Z, Liu B. A SIRT1-centered circuitry regulates breast cancer stemness and metastasis. Oncogene. 2018;37:6299–6315. doi: 10.1038/s41388-018-0370-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kruyt FA. TRAIL and cancer therapy. Cancer Lett. 2008;263:14–25. doi: 10.1016/j.canlet.2008.02.003. [DOI] [PubMed] [Google Scholar]
- 78.Micheau O, Shirley S, Dufour F. Death receptors as targets in cancer. Br J Pharmacol. 2013;169:1723–1744. doi: 10.1111/bph.12238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Arlt A, Muerkoster SS, Schafer H. Targeting apoptosis pathways in pancreatic cancer. Cancer Lett. 2013;332:346–358. doi: 10.1016/j.canlet.2010.10.015. [DOI] [PubMed] [Google Scholar]
- 80.Deng CX. SIRT1, is it a tumor promoter or tumor suppressor? Int J Biol Sci. 2009;5:147–152. doi: 10.7150/ijbs.5.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Tseng RC, Lee CC, Hsu HS, Tzao C, Wang YC. Distinct HIC1-SIRT1-p53 loop deregulation in lung squamous carcinoma and adenocarcinoma patients. Neoplasia. 2009;11:763–770. doi: 10.1593/neo.09470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Liu X, Ehmed E, Li B, Dou J, Qiao X, Jiang W, Yang X, Qiao S, Wu Y. Breast cancer metastasis suppressor 1 modulates SIRT1-dependent p53 deacetylation through interacting with DBC1. Am J Cancer Res. 2016;6:1441–1449. [PMC free article] [PubMed] [Google Scholar]
- 83.Ling S, Li J, Shan Q, Dai H, Lu D, Wen X, Song P, Xie H, Zhou L, Liu J, Xu X, Zheng S. USP22 mediates the multidrug resistance of hepatocellular carcinoma via the SIRT1/AKT/MRP1 signaling pathway. Mol Oncol. 2017;11:682–695. doi: 10.1002/1878-0261.12067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Jeung YJ, Kim HG, Ahn J, Lee HJ, Lee SB, Won M, Jung CR, Im JY, Kim BK, Park SK, Son MJ, Chung KS. Shikonin induces apoptosis of lung cancer cells via activation of FOXO3a/EGR1/SIRT1 signaling antagonized by p300. Biochim Biophys Acta. 2016;1863:2584–2593. doi: 10.1016/j.bbamcr.2016.07.005. [DOI] [PubMed] [Google Scholar]
- 85.Mao B, Hu F, Cheng J, Wang P, Xu M, Yuan F, Meng S, Wang Y, Yuan Z, Bi W. SIRT1 regulates YAP2-mediated cell proliferation and chemoresistance in hepatocellular carcinoma. Oncogene. 2014;33:1468–1474. doi: 10.1038/onc.2013.88. [DOI] [PubMed] [Google Scholar]
- 86.Lian B, Yang D, Liu Y, Shi G, Li J, Yan X, Jin K, Liu X, Zhao J, Shang W, Zhang R. miR-128 targets the SIRT1/ROS/DR5 pathway to sensitize colorectal cancer to TRAIL-induced apoptosis. Cell Physiol Biochem. 2018;49:2151–2162. doi: 10.1159/000493818. [DOI] [PubMed] [Google Scholar]
- 87.Bereshchenko OR, Gu W, Dalla-Favera R. Acetylation inactivates the transcriptional repressor BCL6. Nat Genet. 2002;32:606–613. doi: 10.1038/ng1018. [DOI] [PubMed] [Google Scholar]
- 88.Oltersdorf T, Elmore SW, Shoemaker AR, Armstrong RC, Augeri DJ, Belli BA, Bruncko M, Deckwerth TL, Dinges J, Hajduk PJ, Joseph MK, Kitada S, Korsmeyer SJ, Kunzer AR, Letai A, Li C, Mitten MJ, Nettesheim DG, Ng S, Nimmer PM, O’Connor JM, Oleksijew A, Petros AM, Reed JC, Shen W, Tahir SK, Thompson CB, Tomaselli KJ, Wang B, Wendt MD, Zhang H, Fesik SW, Rosenberg SH. An inhibitor of Bcl-2 family proteins induces regression of solid tumours. Nature. 2005;435:677–681. doi: 10.1038/nature03579. [DOI] [PubMed] [Google Scholar]
- 89.Konopleva M, Contractor R, Tsao T, Samudio I, Ruvolo PP, Kitada S, Deng X, Zhai D, Shi YX, Sneed T, Verhaegen M, Soengas M, Ruvolo VR, McQueen T, Schober WD, Watt JC, Jiffar T, Ling X, Marini FC, Harris D, Dietrich M, Estrov Z, McCubrey J, May WS, Reed JC, Andreeff M. Mechanisms of apoptosis sensitivity and resistance to the BH3 mimetic ABT-737 in acute myeloid leukemia. Cancer Cell. 2006;10:375–388. doi: 10.1016/j.ccr.2006.10.006. [DOI] [PubMed] [Google Scholar]
- 90.Lin X, Morgan-Lappe S, Huang X, Li L, Zakula DM, Vernetti LA, Fesik SW, Shen Y. ‘Seed’ analysis of off-target siRNAs reveals an essential role of Mcl-1 in resistance to the small-molecule Bcl-2/Bcl-XL inhibitor ABT-737. Oncogene. 2007;26:3972–3979. doi: 10.1038/sj.onc.1210166. [DOI] [PubMed] [Google Scholar]
- 91.van Delft MF, Wei AH, Mason KD, Vandenberg CJ, Chen L, Czabotar PE, Willis SN, Scott CL, Day CL, Cory S, Adams JM, Roberts AW, Huang DC. The BH3 mimetic ABT-737 targets selective Bcl-2 proteins and efficiently induces apoptosis via Bak/Bax if Mcl-1 is neutralized. Cancer Cell. 2006;10:389–399. doi: 10.1016/j.ccr.2006.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Chen S, Dai Y, Harada H, Dent P, Grant S. Mcl-1 down-regulation potentiates ABT-737 lethality by cooperatively inducing Bak activation and Bax translocation. Cancer Res. 2007;67:782–791. doi: 10.1158/0008-5472.CAN-06-3964. [DOI] [PubMed] [Google Scholar]
- 93.White E. Deconvoluting the context-dependent role for autophagy in cancer. Nat Rev Cancer. 2012;12:401–410. doi: 10.1038/nrc3262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Xiong H, Ni Z, He J, Jiang S, Li X, He J, Gong W, Zheng L, Chen S, Li B, Zhang N, Lyu X, Huang G, Chen B, Zhang Y, He F. LncRNA HULC triggers autophagy via stabilizing Sirt1 and attenuates the chemosensitivity of HCC cells. Oncogene. 2017;36:3528–3540. doi: 10.1038/onc.2016.521. [DOI] [PubMed] [Google Scholar]
- 95.Kim TH, Kim HS, Kang YJ, Yoon S, Lee J, Choi WS, Jung JH, Kim HS. Psammaplin A induces Sirtuin 1-dependent autophagic cell death in doxorubicin-resistant MCF-7/adr human breast cancer cells and xenografts. Biochim Biophys Acta. 2015;1850:401–410. doi: 10.1016/j.bbagen.2014.11.007. [DOI] [PubMed] [Google Scholar]
- 96.Ou X, Lee MR, Huang X, Messina-Graham S, Broxmeyer HE. SIRT1 positively regulates autophagy and mitochondria function in embryonic stem cells under oxidative stress. Stem Cells. 2014;32:1183–1194. doi: 10.1002/stem.1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Xie B, Zhou J, Shu G, Liu DC, Zhou J, Chen J, Yuan L. Restoration of klotho gene expression induces apoptosis and autophagy in gastric cancer cells: tumor suppressive role of klotho in gastric cancer. Cancer Cell Int. 2013;13:18. doi: 10.1186/1475-2867-13-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Hariharan N, Maejima Y, Nakae J, Paik J, Depinho RA, Sadoshima J. Deacetylation of FoxO by Sirt1 plays an essential role in mediating starvation-induced autophagy in cardiac myocytes. Circ Res. 2010;107:1470–1482. doi: 10.1161/CIRCRESAHA.110.227371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Salminen A, Kaarniranta K. SIRT1: regulation of longevity via autophagy. Cell Signal. 2009;21:1356–1360. doi: 10.1016/j.cellsig.2009.02.014. [DOI] [PubMed] [Google Scholar]
- 100.Zhang HQ, He B, Fang N, Lu S, Liao YQ, Wan YY. Autophagy inhibition sensitizes cisplatin cytotoxicity in human gastric cancer cell line SGC7901. Asian Pac J Cancer Prev. 2013;14:4685–4688. doi: 10.7314/apjcp.2013.14.8.4685. [DOI] [PubMed] [Google Scholar]
- 101.Ding ZB, Hui B, Shi YH, Zhou J, Peng YF, Gu CY, Yang H, Shi GM, Ke AW, Wang XY, Song K, Dai Z, Shen YH, Fan J. Autophagy activation in hepatocellular carcinoma contributes to the tolerance of oxaliplatin via reactive oxygen species modulation. Clin Cancer Res. 2011;17:6229–6238. doi: 10.1158/1078-0432.CCR-11-0816. [DOI] [PubMed] [Google Scholar]
- 102.Amaravadi RK, Lippincott-Schwartz J, Yin XM, Weiss WA, Takebe N, Timmer W, DiPaola RS, Lotze MT, White E. Principles and current strategies for targeting autophagy for cancer treatment. Clin Cancer Res. 2011;17:654–666. doi: 10.1158/1078-0432.CCR-10-2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Moruno F, Perez-Jimenez E, Knecht E. Regulation of autophagy by glucose in Mammalian cells. Cells. 2012;1:372–395. doi: 10.3390/cells1030372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Langley E, Pearson M, Faretta M, Bauer UM, Frye RA, Minucci S, Pelicci PG, Kouzarides T. Human SIR2 deacetylates p53 and antagonizes PML/p53-induced cellular senescence. EMBO J. 2002;21:2383–2396. doi: 10.1093/emboj/21.10.2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Jang J, Huh YJ, Cho HJ, Lee B, Park J, Hwang DY, Kim DW. SIRT1 enhances the survival of human embryonic stem cells by promoting DNA repair. Stem Cell Reports. 2017;9:629–641. doi: 10.1016/j.stemcr.2017.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Maiese K. Moving to the rhythm with clock (Circadian) genes, autophagy, mTOR, and SIRT1 in degenerative disease and cancer. Curr Neurovasc Res. 2017;14:299–304. doi: 10.2174/1567202614666170718092010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Rickert E, Fernandez MO, Choi I, Gorman M, Olefsky JM, Webster NJG. Neuronal SIRT1 regulates metabolic and reproductive function and the response to caloric restriction. J Endocr Soc. 2018;3:427–445. doi: 10.1210/js.2018-00318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Solomon JM, Pasupuleti R, Xu L, McDonagh T, Curtis R, DiStefano PS, Huber LJ. Inhibition of SIRT1 catalytic activity increases p53 acetylation but does not alter cell survival following DNA damage. Mol Cell Biol. 2006;26:28–38. doi: 10.1128/MCB.26.1.28-38.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Ren Y, Du C, Shi Y, Wei J, Wu H, Cui H. The Sirt1 activator, SRT1720, attenuates renal fibrosis by inhibiting CTGF and oxidative stress. Int J Mol Med. 2017;39:1317–1324. doi: 10.3892/ijmm.2017.2931. [DOI] [PubMed] [Google Scholar]
- 110.Milne JC, Lambert PD, Schenk S, Carney DP, Smith JJ, Gagne DJ, Jin L, Boss O, Perni RB, Vu CB, Bemis JE, Xie R, Disch JS, Ng PY, Nunes JJ, Lynch AV, Yang H, Galonek H, Israelian K, Choy W, Iffland A, Lavu S, Medvedik O, Sinclair DA, Olefsky JM, Jirousek MR, Elliott PJ, Westphal CH. Small molecule activators of SIRT1 as therapeutics for the treatment of type 2 diabetes. Nature. 2007;450:712–716. doi: 10.1038/nature06261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Huber JL, McBurney MW, Distefano PS, McDonagh T. SIRT1-independent mechanisms of the putative sirtuin enzyme activators SRT1720 and SRT2183. Future Med Chem. 2010;2:1751–1759. doi: 10.4155/fmc.10.257. [DOI] [PubMed] [Google Scholar]
- 112.Pacholec M, Bleasdale JE, Chrunyk B, Cunningham D, Flynn D, Garofalo RS, Griffith D, Griffor M, Loulakis P, Pabst B, Qiu X, Stockman B, Thanabal V, Varghese A, Ward J, Withka J, Ahn K. SRT1720, SRT2183, SRT1460, and resveratrol are not direct activators of SIRT1. J Biol Chem. 2010;285:8340–8351. doi: 10.1074/jbc.M109.088682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Brooks CL, Gu W. p53 ubiquitination: Mdm2 and beyond. Mol Cell. 2006;21:307–315. doi: 10.1016/j.molcel.2006.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Jin Y, Cao Q, Chen C, Du X, Jin B, Pan J. Tenovin-6-mediated inhibition of SIRT1/2 induces apoptosis in acute lymphoblastic leukemia (ALL) cells and eliminates ALL stem/progenitor cells. BMC Cancer. 2015;15:226. doi: 10.1186/s12885-015-1282-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Lee BB, Kim Y, Kim D, Cho EY, Han J, Kim HK, Shim YM, Kim DH. Metformin and tenovin-6 synergistically induces apoptosis through LKB1-independent SIRT1 down-regulation in non-small cell lung cancer cells. J Cell Mol Med. 2019;23:2872–2889. doi: 10.1111/jcmm.14194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Hirai S, Endo S, Saito R, Hirose M, Ueno T, Suzuki H, Yamato K, Abei M, Hyodo I. Antitumor effects of a sirtuin inhibitor, tenovin-6, against gastric cancer cells via death receptor 5 up-regulation. PLoS One. 2014;9:e102831. doi: 10.1371/journal.pone.0102831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Liang Z, Yang Y, Wang H, Yi W, Yan X, Yan J, Li Y, Feng Y, Yu S, Yang J, Jin Z, Duan W, Chen W. Inhibition of SIRT1 signaling sensitizes the antitumor activity of silybin against human lung adenocarcinoma cells in vitro and in vivo. Mol Cancer Ther. 2014;13:1860–1872. doi: 10.1158/1535-7163.MCT-13-0942. [DOI] [PubMed] [Google Scholar]
- 118.Dai W, Zhou J, Jin B, Pan J. Class III-specific HDAC inhibitor Tenovin-6 induces apoptosis, suppresses migration and eliminates cancer stem cells in uveal melanoma. Sci Rep. 2016;6:22622. doi: 10.1038/srep22622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Ota H, Tokunaga E, Chang K, Hikasa M, Iijima K, Eto M, Kozaki K, Akishita M, Ouchi Y, Kaneki M. Sirt1 inhibitor, Sirtinol, induces senescence-like growth arrest with attenuated Ras-MAPK signaling in human cancer cells. Oncogene. 2006;25:176–185. doi: 10.1038/sj.onc.1209049. [DOI] [PubMed] [Google Scholar]
- 120.Asaka R, Miyamoto T. Sirtuin 1 promotes the growth and cisplatin resistance of endometrial carcinoma cells: a novel therapeutic target. Lb Invest. 2015;95:1363–1373. doi: 10.1038/labinvest.2015.119. [DOI] [PubMed] [Google Scholar]
- 121.Oon CE, Strell C, Yeong KY, Östman A, Prakash J. SIRT1 inhibition in pancreatic cancer models: contrasting effects in vitro and in vivo. Eur J Pharmacol. 2015;757:59–67. doi: 10.1016/j.ejphar.2015.03.064. [DOI] [PubMed] [Google Scholar]
- 122.Peck B, Chen CY, Ho KK, Di Fruscia P, Myatt SS, Coombes RC, Fuchter MJ, Hsiao CD, Lam EW. SIRT inhibitors induce cell death and p53 acetylation through targeting both SIRT1 and SIRT2. Mol Cancer Ther. 2010;9:844–855. doi: 10.1158/1535-7163.MCT-09-0971. [DOI] [PubMed] [Google Scholar]
- 123.Zhang W, Luo P. MicroRNA-29c restores cisplatin sensitivity in liver cancer through direct inhibition of sirtuin 1 expression. Oncol Lett. 2018;16:1543–1550. doi: 10.3892/ol.2018.8801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Herbert KJ, Cook AL, Snow ET. SIRT1 inhibition restores apoptotic sensitivity in p53-mutated human keratinocytes. Toxicol Appl Pharmacol. 2014;277:288–297. doi: 10.1016/j.taap.2014.04.001. [DOI] [PubMed] [Google Scholar]
- 125.Cheng F, Su L, Yao C, Liu L, Shen J, Liu C, Chen X, Luo Y, Jiang L, Shan J, Chen J, Zhu W, Shao J, Qian C. SIRT1 promotes epithelial-mesenchymal transition and metastasis in colorectal cancer by regulating Fra-1 expression. Cancer Lett. 2016;375:274–283. doi: 10.1016/j.canlet.2016.03.010. [DOI] [PubMed] [Google Scholar]
- 126.Ullah M, Qiao Y, Concepcion W, Thakor AS. Stem cell-derived extracellular vesicles: role in oncogenic processes, bioengineering potential, and technical challenges. Stem Cell Res Ther. 2019;10:347. doi: 10.1186/s13287-019-1468-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Ullah M, Akbar A, Thakor AS. An emerging role of CD9 in stemness and chemoresistance. Oncotarget. 2019;10:4000–4001. doi: 10.18632/oncotarget.27021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Ullah M, Akbar A, Yannarelli G. Applications of artificial intelligence in, early detection of cancer, clinical diagnosis and personalized medicine. Artificial Intelligence in Cancer. 2020;1:39–44. [Google Scholar]
- 129.Ullah M, Ng NN, Concepcion W, Thakor AS. Emerging role of stem cell-derived extracellular microRNAs in age-associated human diseases and in different therapies of longevity. Ageing Res Rev. 2020;57:100979. doi: 10.1016/j.arr.2019.100979. [DOI] [PubMed] [Google Scholar]
- 130.Ullah M, Akbar A. Clinical relevance of RNA editing to early detection of cancer in human. Int J Stem Cell Res Ther. 2020;7:066. doi: 10.23937/2469-570x/1410066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Ullah M, Akbar A, Ng NN, Concepcion W, Thakor AS. Mesenchymal stem cells confer chemoresistance in breast cancer via a CD9 dependent mechanism. Oncotarget. 2019;10:3435–3450. doi: 10.18632/oncotarget.26952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Zhang N, Xie T, Xian M, Wang YJ, Li HY, Ying MD, Ye ZM. SIRT1 promotes metastasis of human osteosarcoma cells. Oncotarget. 2016;7:79654–79669. doi: 10.18632/oncotarget.12916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Yuan H, Wang Z, Gao C, Chen W, Huang Q, Yee JK, Bhatia R, Chen W. BCR-ABL gene expression is required for its mutations in a novel KCL-22 cell culture model for acquired resistance of chronic myelogenous leukemia. J Biol Chem. 2010;285:5085–5096. doi: 10.1074/jbc.M109.039206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Druker BJ, Guilhot F, O’Brien SG, Gathmann I, Kantarjian H, Gattermann N, Deininger MW, Silver RT, Goldman JM, Stone RM, Cervantes F, Hochhaus A, Powell BL, Gabrilove JL, Rousselot P, Reiffers J, Cornelissen JJ, Hughes T, Agis H, Fischer T, Verhoef G, Shepherd J, Saglio G, Gratwohl A, Nielsen JL, Radich JP, Simonsson B, Taylor K, Baccarani M, So C, Letvak L, Larson RA. Five-year follow-up of patients receiving imatinib for chronic myeloid leukemia. N Engl J Med. 2006;355:2408–2417. doi: 10.1056/NEJMoa062867. [DOI] [PubMed] [Google Scholar]
- 135.Kreys ED, Frei CR, Villarreal SM, Bollinger MJ, Jones X, Koeller JM. Evaluation of long-term chronic myeloid leukemia treatment practices with tyrosine kinase inhibitors in a national cohort of veterans. Pharmacotherapy. 2017;37:278–286. doi: 10.1002/phar.1893. [DOI] [PubMed] [Google Scholar]
- 136.Portmann S, Fahrner R, Lechleiter A, Keogh A, Overney S, Laemmle A, Mikami K, Montani M, Tschan MP, Candinas D, Stroka D. Antitumor effect of SIRT1 inhibition in human HCC tumor models in vitro and in vivo. Mol Cancer Ther. 2013;12:499–508. doi: 10.1158/1535-7163.MCT-12-0700. [DOI] [PubMed] [Google Scholar]
- 137.Lambert G, Estevez-Salmeron L, Oh S, Liao D, Emerson BM, Tlsty TD, Austin RH. An analogy between the evolution of drug resistance in bacterial communities and malignant tissues. Nat Rev Cancer. 2011;11:375–382. doi: 10.1038/nrc3039. [DOI] [PMC free article] [PubMed] [Google Scholar]