Abstract
Hydrostatic pressure in the range of 15 to 25 MPa was found to cause arrest of the cell cycle in G1 phase in an exponentially growing culture of Saccharomyces cerevisiae, whereas a pressure of 50 MPa did not. We found that a plasmid carrying the TAT2 gene, which encodes a high-affinity tryptophan permease, enabled the cells to grow under conditions of pressure in the range of 15 to 25 MPa. Additionally, cells expressing the Tat2 protein at high levels became endowed with the ability to grow under low-temperature conditions at 10 or 15°C as well as at high pressure. Hydrostatic pressure significantly inhibited tryptophan uptake into the cells, and the Tat2 protein level was down-regulated by high pressure. The activation volume associated with tryptophan uptake was found to be a large positive value, 46.2 ± 3.85 ml/mol, indicating that there was a net volume increase in a rate-limiting step in tryptophan import. The results showing cell cycle arrest in G1 phase and down-regulation of the Tat2 protein seem to be similar to those observed upon treatment of cells with the immunosuppressive drug rapamycin. Although rapamycin treatment elicited the rapid dephosphorylation of Npr1 and induction of Gap1 expression, hydrostatic pressure did not affect the phosphorylation state of Npr1 and it decreased the level of Gap1 protein, suggesting that the pressure-sensing pathway may be independent of Npr1 function. Here we describe high-pressure sensing in yeast in comparison with the TOR-signaling pathway and discuss an important factor involved in adaptation of organisms to high-pressure environments.
Organisms respond specifically to a variety of mechanical pressure stimuli such as touch, gravity, turgor, osmotic changes, and hydrostatic pressure. In response to changes in membrane tension in Escherichia coli, MscL, a mechanosensitive ion channel of large conductance, is known to transmit the stretch force through the lipid bilayer, and it plays an essential role in mechanosensation (44). Mid1 in Saccharomyces cerevisiae, the first eukaryotic calcium-permeable stretch-activated cation channel identified, is essential for supplying calcium ions during the mating process (24, 28, 34). In bovine chromaffin cells, the large-conductance Ca2+-activated K+ (BK) channel, which is selective for potassium ions, is known to be activated by voltage and also hydrostatic pressure (31). Fluctuation of hydrostatic pressure is commonly observed in diarthroses. The articular cartilage is exposed to substantial levels of hydrostatic pressure, i.e., about 20 MPa (0.1 MPa = 1 bar = 0.9869 atm = 1.0197 kg of force/cm2; to avoid confusion, megapascals are used throughout), on standing (33). Intermittent pressure of 10 MPa has been found to increase the levels of aggrecan and type II collagen mRNA in articular chondrocytes, while both intermittent and constant hydrostatic pressures stimulate glycosaminoglycan synthesis (42). Also, in chondrocytic cells, high pressure induces Hsp70 accumulation (27).
In the ocean, hydrostatic pressure varies from 0.1 MPa at the surface to 110 MPa at the greatest depth of the deep-sea bottom. Thus, marine organisms may experience a constant hydrostatic pressure or a wide range of fluctuating pressures. The deep-sea microbial population is very complex. Piezophiles (previously referred to as barophiles) are organisms showing increased growth rates at pressures above 0.1 MPa. Numerous mesophiles whose growth rates are reduced by elevated hydrostatic pressure are present in the deep sea. These microorganisms are transported vertically from the surface of the ocean in fast-sinking particles at a rate of about 1,000 m/week (47).
The maintenance of appropriate membrane fluidity is thought to be one of the key factors for survival and growth under high-pressure conditions, as suggested by the results of studies with prokaryotes (2, 5, 14). Appropriate membrane fluidity can be achieved by increasing the ratio of unsaturated fatty acids within membrane phospholipids. Another factor involved is pressure-responsive gene expression. The transmembrane protein ToxR in Photobacterium profundum strain SS9 is known to play a role in controlling the expression of numerous genes, presumably via conformational changes under various pressure conditions (48). When the recD gene from P. profundum SS9, a homologue of a DNA recombination and repair gene, was introduced into an E. coli recD mutant, it enabled the mutant to display a normal phenotype under high-pressure conditions (elevated pressure is known to cause cell filamentation in E. coli) (10). The details of recD function at high pressure are still unclear.
In eukaryotic microorganisms, the effects of high pressure have received little attention, even in the genetically well-characterized yeast S. cerevisiae. Perhaps this is partly because the biomass of marine fungi is relatively small, and there have been few reports concerning piezophilic yeasts.
We have studied the physiological effects of hydrostatic pressure at nonlethal levels, below 100 MPa, in yeast, focusing on intracellular pH homeostasis (2, 3, 4) and a potential application in flow cytometry (1). Recently, we isolated spontaneous mutants of S. cerevisiae capable of growth under elevated pressure conditions. In the course of genetic studies using these mutants, we obtained, by chance, a striking result suggesting that the availability of tryptophan may be of primary importance for high-pressure growth in yeast.
Tryptophan is known to be transported into the cell via a high-affinity tryptophan permease encoded by TAT2 (37). TAT2 was originally identified as a gene that conferred resistance to an immunosuppressive drug, FK506 (37). Another immunosuppressant, rapamycin, is known to arrest the growth of yeast cells in early G1 phase, causing them to express several physiological properties typical of starved (G0) cells and also causing significant reduction in protein synthesis. This action has been extensively investigated as the TOR (target of rapamycin)-signaling pathway (6, 8, 9, 11, 20, 26, 35, 36, 45, 51).
In this paper, we describe an important factor involved in the adaptation of organisms to high-pressure environments and compare the requirements for the activity of this factor to the requirements for the activity of the TOR-signaling pathway. In addition, we discuss the importance of introducing the thermodynamic parameter hydrostatic pressure into the investigation of membrane protein function or protein targeting in living cells in terms of a fundamental physical parameter of a reaction, namely, volume change, as commonly used in biophysics and enzymology.
MATERIALS AND METHODS
Strains and media.
The wild-type haploid strain YPH499 (MATa his3-Δ200 leu2-Δ1 lys2-801 trp1-Δ1 ade2-101 ura3-52) (38) was used in this study. Strains containing plasmids are listed in Table 1. The compositions of yeast extract-peptone-dextrose (YPD) medium, synthetic minimal (SD) medium, and synthetic complete (SC) medium supplemented with appropriate nutrients are described in reference 19. SCv medium, which is SC medium lacking valine, was mainly used throughout this study because growth of strain YPH499 was found to be poor in SC medium containing 150 mg of valine per liter.
TABLE 1.
Plasmids and corresponding strains used in this study
| Plasmid | Description | Corresponding strain |
|---|---|---|
| YCplac33 | CEN4 URA3 | FAA3 |
| YEplac195 | URA3 | FAA4 |
| YEplac112 | TRP1 | FAA6 |
| YCplac33::TAT2 | 2.6-kb fragment containing TAT2 in YCplac33 | FAB16 |
| YEplac195::TAT2 | 2.6-kb fragment containing TAT2 in YEplac195 | FAB19 |
| YCplac33::2HA-TAT2 | TAT2 promoter–2HA-TAT2 in YCplac33 | FAB59 |
| YEplac195::2HA-TAT2 | TAT2 promoter–2HA-TAT2 in YEplac195 | FAB33 |
| pAS103 | HA-NPR1 in YEplac195 | FAE12 |
| pPL257 | HA-GAP1 in pRS316[URA3] | FAF6 |
Genetic techniques.
Yeast transformation was performed by the lithium acetate method (25). E. coli strain DH5α was used for plasmid purification and for construction of a genomic library of strain YPH499.
DNA manipulations.
Restriction endonucleases were purchased from TaKaRa and New England Biolabs, and the ligation kits and the deletion kit for kilosequencing were purchased from TaKaRa. DNA amplification was performed by the PCR method using a ThermoSequenase II dye terminator cycle sequencing premix kit (Amersham Pharmacia Biotech, Inc.). DNA sequencing was performed using a model 373A DNA sequencer (Applied Biosystems).
Plasmids.
Plasmids used in this study are listed in Table 1. Plasmids YCplac33[CEN4 URA3], YCplac111[CEN4 LEU2], YCplac22[CEN4 TRP1], and YEplac195[URA3] were kindly provided by H. Iida. Plasmids YCplac33::TAT2 and YEplac195::TAT2 were constructed as follows. A genomic library containing 10- to 20-kb DNA fragments from strain YPH499 was constructed using the plasmid YCplac111. Cells of strain YPH499 were transformed with the library, and then the cells were mixed with SD low-melting-temperature agar and the mixture was put into a sterilized plastic bag. After incubation at 24°C at 0.1 MPa for 24 h, the cells were incubated at a pressure of 18 or 25 MPa for 2 to 7 days to select for transformants capable of high-pressure growth. As a result, several 5- to 6-kb DNA fragments containing the TAT2 gene were obtained. The TAT2 gene was also recovered by the gap repair method from several independent clones of YPH499 and was sequenced. A 3.1-kb HgaI fragment containing TAT2 and tY (GUA)0 (tRNA for tyrosine) was applied to the deletion kit for kilo-sequencing to remove 88 bp of tY (GUA)0. The resulting 2.6-kb PstI/SphI fragment was cloned into YCplac33 and YEplac195 to obtain YCplac33::TAT2 and YEplac195::TAT2, respectively, which were used throughout the experiments. HA-TAT2, encoding an NH2-terminally hemagglutinin (HA)-tagged fully functional Tat2 protein under the control of its own promoter (YEplac195::2HA-TAT2), was constructed as described by Beck et al. (9). pAS103 (YEplac195::HA-NPR1) was kindly provided by M. N. Hall (36). p257 (pRS316::HA-GAP1) was kindly provided by G. R. Fink (29).
Culture conditions.
Cells were grown in YPD or SCv medium at 24°C with shaking (140 rpm). Cells were counted using a hemocytometer (Erma) under a phase-contrast microscope, after brief sonication for 10 s on ice to dissociate the cells. For analysis of cell growth under various pressure conditions, experiments were performed using liquid media rather than solid agar media. If the cells are grown in YPD or SD low-melting-temperature agar at high pressure, a considerable amount of CO2 generated by the cells through fermentation is released as a gas after decompression and the bubbles interfere with visualization of the colonies. Therefore, the following procedure was employed. An exponentially growing culture (3 × 106 to 5 × 106 cells/ml) was diluted to 1 × 106 cells/ml with fresh YPD or SCv medium. Aliquots of the resulting cell suspension were put into a series of sterilized polypropylene tubes, and after removal of any air bubbles present, the tubes were sealed with Parafilm. Each tube was placed in a stainless-steel hydrostatic chamber, and the appropriate hydrostatic pressure was applied using a hand pump. One to 2 min was required to reach a pressure of 50 MPa. The temperature increase in samples during pressurization was negligible, as described previously (1). The initial cell number (N0; usually in the range of 1.0 × 106 to 1.3 × 106 cells/ml) was counted just after all samples had been pressurized. Each chamber was opened at a certain time, and samples were taken, centrifuged immediately, and then kept on ice to avoid further cell proliferation after decompression. Viable cell numbers were determined on the basis of colony counts after decompression.
To examine cell growth in the presence of a high concentration of 20 individual l-amino acids (code no. LAA-21; Sigma), adenine sulfate dihydrate (code no. 013-00812; Wako), or uracil (code no. U-0750; Sigma), YPD medium supplemented with each of these test substrates at 2 g/liter was prepared. An exponentially growing culture (optical density at 600 nm [OD600] of 0.14; approximately 1.4 × 106 cells/ml) was diluted with an equal volume of YPD medium containing the test substrate (therefore, the final concentration of the test substrate was 1 g/liter), and the cells were exposed to hydrostatic pressures of 0.1 and 25 MPa at 24°C for 24 h. The OD600 was measured after appropriate dilution of the samples, except when the measured values did not exceed an OD600 of 0.5.
Flow cytometry.
Samples for flow cytometry analysis were prepared as described by Barbet et al. (6). The cells were washed once in cold distilled water and fixed in 70% ethanol for more than 5 h at 4°C with vigorous shaking. The cells were collected by centrifugation and resuspended in 50 mM Na-citrate buffer (pH 7.4). Intracellular RNA was digested with RNase A (0.25 mg/ml) at 50°C for 1 h, and thereafter the samples were treated with proteinase K (1 mg/ml) at 50°C for 1 h. Chromosomal DNA in individual cells was stained with propidium iodide (25 μg/ml). The cells were briefly sonicated prior to flow cytometry analysis, and 105 cells were analyzed using FACSCalibur (Becton Dickinson).
Tryptophan uptake assay.
Radiolabeled l-tryptophan (l-[5-3H]tryptophan, code no. TRK460, batch 109, 1.22 TBq/mmol, 37 MBq/ml, 98.2% purity; Amersham Pharmacia Biotech Inc.) was used in the assay. Cells from an exponentially growing culture in SCv medium at a cell density of 3 × 106 to 5 × 106 cells/ml were collected by centrifugation and resuspended in 15 ml of fresh SCv medium in a polypropylene tube. One milliliter of the cell suspension was placed on ice and used for determination of the cell count. Then, 14 μl of the radiolabeled tryptophan was added to 14 ml of the cell suspension and immediately mixed, and the mixture was split into seven equal aliquots (1.77 ml each) in sterilized microtubes. The concentration of unlabeled l-tryptophan in SCv medium was 98.0 μmol/liter (20 mg/liter), and that of the 3H-labeled l-tryptophan was calculated to be 30.3 nmol/liter. In our system using Scintisol EX-H (code no. 348-04013; Dojindo) as the scintillator cocktail, 1,000 cpm corresponded to 370 Bq in the case of 4.85 × 10−5 nmol of the 3H-labeled l-tryptophan. Hence, in the following procedure, 1,000 cpm per 107 cells was taken to be 153.8 pmol of l-tryptophan per 107 cells. Six tubes were carefully sealed with Parafilm. Each tube was put into a pressure chamber and then subjected to a hydrostatic pressure of 0.1, 25, or 50 MPa, for 30 or 60 min. The cells in the tube were collected by filtration on a glass filter (type GF/C, 25-mm diameter; Whatman) and washed with 12 ml of distilled water, and the radioactivity on the filter was counted using a liquid scintillation counter (Wallac model 1411). Usually, the amount of time involved from the time of addition of the labeled tryptophan to the cell suspension until the first filtration was 10 min. Therefore, the 10-min time point was taken to be the starting point in the assay of tryptophan uptake under high pressure. All experiments were performed three times, and the mean values are shown as picomoles of tryptophan incorporated per 107 cells.
Western blot analysis.
To prepare whole-cell extracts for sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis and Western blot analysis, 4 × 107 cells from cultures incubated under high-pressure conditions or cultures treated with rapamycin (code no. R-0395; Sigma) (200 ng/ml in SCv medium) were collected by centrifugation and washed once in lysis buffer (10 mM Tris-HCl, 1 mM EDTA, 2% 2-mercaptoethanol, 1 mM phenylmethylsulfonyl fluoride [pH 7.5]) (24). Sixty microliters of the lysis buffer and 0.3 g of acid-washed glass beads (code no. G-8772; Sigma) were added to the cell pellet and then vortexed at 30-s intervals for 5 min. Unbroken cells and debris were removed by centrifugation at 500 × g, and the supernatants were treated with 1% SDS at 37°C for 10 min (for HA-Tat2 and HA-Gap1) or heat-treated at 95°C for 3 min (for HA-Npr1) to denature the proteins (9, 36). Protein concentrations were determined using the Bio-Rad protein assay. For standard SDS-polyacrylamide gel electrophoresis (10% acrylamide) and Western blot analysis, a total of 8 μg of protein was loaded per lane. For detection of the HA-tagged protein, mouse anti-HA antibody (16B12; Convance) and peroxidase-labeled antimouse antibody (code no. RPN2124; Amersham Pharmacia Biotech Inc.) were used. For signal detection, an Amersham ECL Plus kit (code no. RPN2123; Amersham Pharmacia Biotech Inc.) was used.
RESULTS
Hydrostatic pressure causes arrest of the cell cycle in G1 phase.
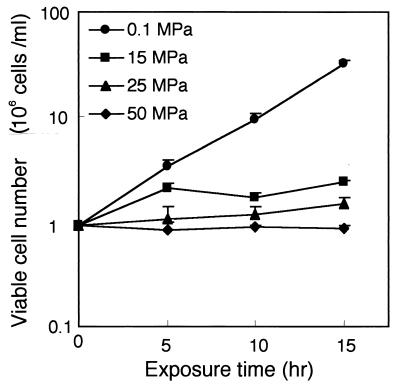
First, we examined the effect of hydrostatic pressure on exponentially growing YPH499 cells in an asynchronous culture (106 cells/ml) in YPD medium. Figure 1 shows the viable cell number derived from counting CFU. Hydrostatic pressures greater than 15 MPa considerably reduced the growth rate, and a pressure of 50 MPa completely prevented cell proliferation. Ten hours after pressurization, the cell numbers had increased 11.0-, 1.7-, 1.4-, and 0.95-fold at pressures of 0.1, 15, 25, and 50 MPa, respectively. The growth curves were almost identical to those obtained by direct counting of cells under a microscope, and cell morphology was normal under the hydrostatic pressures tested (data not shown).
FIG. 1.
Hydrostatic pressure reduces the growth rate in wild-type strain YPH499. Exponentially growing cells were subjected to a hydrostatic pressure of 0.1, 15, 25, or 50 MPa in hydrostatic chambers. The chambers were opened at 5-h intervals in the period up to 15 h after the start of incubation, and then the cells were spread on YPD agar plates and incubated for a few days at 0.1 MPa. Data presented are mean values with SD of results from three independent experiments.
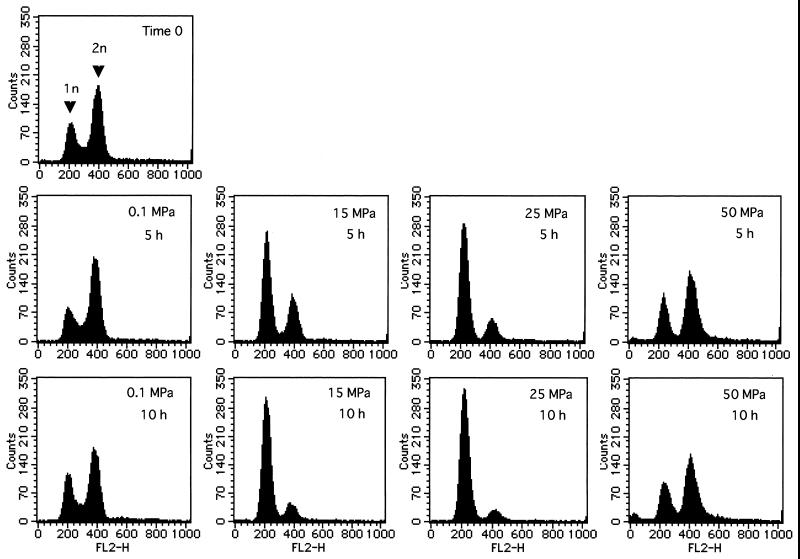
Next, we examined how elevated pressure affected progression of the cell cycle. Exponentially growing cells of strain YPH499 in YPD medium were exposed to a hydrostatic pressure of 0.1, 15, 25, or 50 MPa for 5 or 10 h. After decompression, cells were examined by flow cytometry analysis. As a result, a marked shift to 1N DNA content was observed when the cells were incubated at 15 MPa for 5 h, and moreover, after 10 h at 15 MPa, 82% of the cells contained 1N DNA (Fig. 2). Similar results were obtained for the culture incubated at 25 MPa; however, the shift to 1N DNA content was more pronounced (85% of the cells contained 1N DNA) (Fig. 2). These shifts to 1N DNA content paralleled the growth arrest shown in Fig. 1. In addition, when the cells cultured at 15 or 25 MPa were examined under a microscope, mostly unbudded cells were observed. These results indicate that hydrostatic pressure in the range of 15 to 25 MPa indeed caused G1 arrest. This fact can be applied for synchronization of the cell cycle by application of hydrostatic pressure. In contrast, a pressure of 50 MPa did not cause G1 arrest (Fig. 2). The histograms of the cell populations in cultures incubated at 50 MPa for 5 or 10 h were almost identical to those at 0.1 MPa, but there was no increase in cell number at 50 MPa (Fig. 1). Therefore, it is likely that a pressure of 50 MPa immediately stopped cell proliferation at various phases of the cell cycle. In fact, the population of budded cells after exposure to 50 MPa was essentially the same as that before the exposure.
FIG. 2.
Hydrostatic pressure causes cell cycle arrest in G1 phase in the wild-type strain YPH499. Exponentially growing cells were exposed to a hydrostatic pressure of 0.1, 15, 25, or 50 MPa for 5 or 10 h as indicated at the upper right in each graph. After decompression, samples were prepared for flow cytometry. Propidium iodide-labeled cells (approximately 105 cells) were analyzed using FACSCalibur.
A high concentration of tryptophan enables high-pressure growth.
YPH499 cells at an OD600 of 0.06 (approximately 6.0 × 105 cells/ml) were incubated in YPD medium containing a high concentration of 20 individual amino acids or other test substrates (final concentration, 1 g/liter) for 24 h at 0.1 or 25 MPa. Surprisingly only l-tryptophan enabled cell growth at 25 MPa. The increase in OD600 was 22-fold (OD600 = 1.43), whereas in the rest of the cultures, the increase in OD600 was less than 2-fold. When the concentration of each test substrate was reduced to 0.2 g/liter, again only l-tryptophan enabled cell growth at 25 MPa and the OD600 reached 0.49. d-Tryptophan, N-acetyl-l-tryptophan, and N-acetyl-d-tryptophan were not effective (data not shown). These results suggest that l-tryptophan is of primary importance for cell growth under high-pressure conditions. This observation reminded us that the immunosuppressants FK506 and rapamycin inhibit amino acid uptake, especially l-tryptophan uptake, into cells (36, 37). Consequently, these drugs lead to G1 arrest. The G1 arrest induced by these drugs can be prevented by excess l-tryptophan or expression of TAT2 on a high-copy-number plasmid (37).
TAT2 enables growth under high-pressure conditions.
The TAT2 gene encodes a high-affinity tryptophan permease which was originally discovered to be an FK506 resistance-conferring gene (37). Addition of rapamycin induces vacuolar targeting and degradation of the Tat2 protein, which results in significant reduction of tryptophan uptake activity (9, 36).
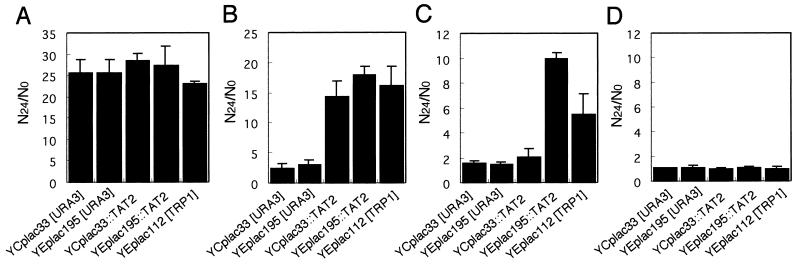
We examined whether expression of TAT2 conferred high-pressure growth in strain YPH499. The reason why we focused on TAT2 is that we have identified this gene by screening genes that can confer high-pressure growth by using a genomic DNA library that originated from strain YPH499 (Abe et al., unpublished data). This was a coincidental finding in conjunction with the result concerning the effect of excess l-tryptophan described above. Figure 3 shows the growth of TAT2-transformed cells in SCv medium under various pressure conditions. The data presented are the mean values of the ratios of the cell number after 24 h of culture (N24) to the initial cell number (N0 ranged from 1 × 106 to 1.3 × 106 cells/ml). Both the low-copy-number plasmid carrying TAT2 (YCplac33::TAT2) and the high-copy-number plasmid carrying TAT2 (YEplac195::TAT2) conferred the ability to grow at 15 MPa (Fig. 3B), whereas the vector alone (YCplac33[URA3] or YEplac195[URA3]) had no effect. At a pressure of 25 MPa, YCplac33::TAT2 was not effective but YEplac195::TAT2 still conferred the ability to grow at this elevated pressure (Fig. 3C). Therefore, it seems that the upper limit of hydrostatic pressure for growth was dependent on the copy number of the TAT2 gene, probably corresponding to the amount of Tat2 protein expressed and the extent of tryptophan uptake. YEplac112[TRP1], carrying the phosphoribosyl-anthranilate isomerase gene (TRP1), also allowed the cells to grow at 15 or 25 MPa (Fig. 3B and C). In addition, tryptophan prototrophic strains such as X2180-1A and IFO2347 (Sake yeast, no. 7) could grow at 25 MPa, but tryptophan-auxotrophic strains such as W303-1A and A364A were defective in high-pressure growth, as in the case of YPH499 (unpublished observation). Thus, the cells require a certain level of intracellular tryptophan. Our findings suggest that tryptophan uptake via Tat2 is the cellular process most sensitive to elevated hydrostatic pressure and that reduced tryptophan availability results in a decrease in protein synthesis; hence, progression of the cell cycle is impaired in the G1 phase. None of the strains examined was able to proliferate at 50 MPa; there was no change in cell numbers (Fig. 3D).
FIG. 3.
TAT2 and TRP1 confer high-pressure growth. Using cultures of exponentially growing cells as inocula (the initial cell density [N0] was approximately 106 cells/ml), several transformants were grown at 0.1 MPa (A), 15 MPa (B), 25 MPa (C), or 50 MPa (D) for 24 h in SCv medium. All data are presented as ratios of the cell density at 24 h (N24) to the cell density at N0 and are mean values with SD of results from three independent experiments.
TAT2 enables low-temperature growth as well as high-pressure growth.
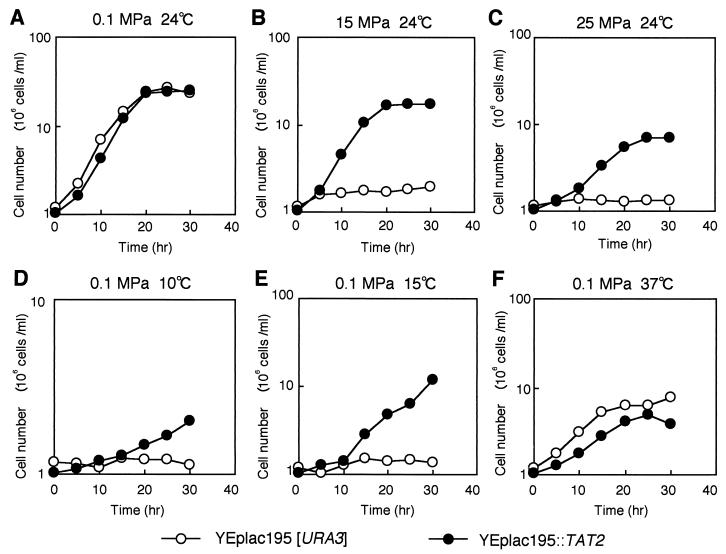
Next, we precisely analyzed the time course of growth of a transformant expressing high levels of TAT2. Strain FAA4 (transformed with YEplac195[URA3]) and strain FAB19 (transformed with YEplac195::TAT2) were used. At 0.1 MPa and 24°C, no significant difference in levels of cell growth was observed when strains FAA4 and FAB19 were compared (Fig. 4A). At 15 MPa and 24°C, strain FAB19 grew as well as at 0.1 MPa; however, strain FAA4 was unable to grow under these conditions (Fig. 4B). At 25 MPa and 24°C, strain FAB19 still grew, although the extent of cell growth was decreased (Fig. 4C), and it also grew at 35 MPa (data not shown).
FIG. 4.
Expression of TAT2 confers high-pressure growth as well as low-temperature growth. Strains FAA4 (YEplac195) and FAB19 (YEplac195::TAT2) were grown under various pressure conditions at 24°C (A to C) or under various temperature conditions at 0.1 MPa (A, D, E, and F) in SCv medium.
A striking result was that strain FAB19 became endowed with the ability to grow under low-temperature conditions such as at 10 and 15°C at 0.1 MPa (Fig. 4D and E). Under the same conditions, strain FAA4 never grew. It is well documented that an increase in hydrostatic pressure is similar to a decrease in temperature in terms of causing a decrease in membrane fluidity and that the molecular motion of phospholipids and membrane proteins becomes highly restricted (30). In contrast to the superior growth of strain FAB19 at low temperature, its rate of growth at 37°C was lower than that of strain FAA4 (Fig. 4F).
Therefore, we hypothesize that the most critical target impaired by increasing hydrostatic pressure in growing cells is the tryptophan permease required for tryptophan uptake, and this impairment might result from a decrease in membrane fluidity and/or conformational change in the Tat2 protein related to its function. As a decrease in membrane fluidity can be caused by either an increase in pressure or a decrease in temperature, this decrease in tryptophan uptake is a very important matter to be considered in relation to the survival strategies employed by microorganisms.
Hydrostatic pressure decreases and down-regulates tryptophan uptake activity.
We next analyzed tryptophan uptake under various pressure conditions. Figure 5A shows the levels of tryptophan incorporation into cells of strain FAA4 under several pressure conditions. As expected, tryptophan uptake decreased to an extent that depended on the magnitude of the pressure applied. The degree of inhibition of tryptophan uptake at 25 MPa was similar to that observed with the immunosuppressive drug FK506 (21, 36). The drug inhibited tryptophan uptake by approximately 50% in a diploid strain, JK9-3. The rate constants for tryptophan uptake at 0.1, 25, and 50 MPa were 4.20 ± 1.15, 2.61 ± 0.778, and 1.65 ± 0.444 pmol 107 cells−1 min−1 (means ± standard deviations [SD]), respectively (Table 2). All pressure effects accompany system volume changes in biochemical processes, and the response of the system is governed by the principle of Le Chatelier. The fundamental relationship describes the pressure effects in reactions as follows: (∂ lnk/∂ lnp)T = −ΔV≠/RT, where k is the rate constant, p is the pressure (atmosphere), T is the absolute temperature (kelvin), and R is the gas constant (milliliters of atmosphere per kelvin per mole). ΔV≠ is the apparent volume change of activation (activation volume, in milliliters per mole) and represents the difference in volume between the reactants and the transition state. When a reaction is accompanied by a volume increase, it is inhibited by elevated pressure. When a reaction in accompanied by a volume decrease, it is enhanced by elevated pressure. The activation volume associated with the overall process of tryptophan uptake (i.e., ΔV*) was a large positive value, 46.2 ± 3.85 ml/mol (Table 2), indicating that there was a net volume increase in a rate-limiting step in tryptophan import. In our preliminary results, uptake of other amino acids was also impaired by elevated pressure and the order of severity of inhibition was tryptophan > lysine > histidine > leucine. It is surprising that this order was the same as that reported for the inhibition of amino acid uptake by FK506 (order of severity of inhibition is tryptophan > histidine > leucine) (21, 36).
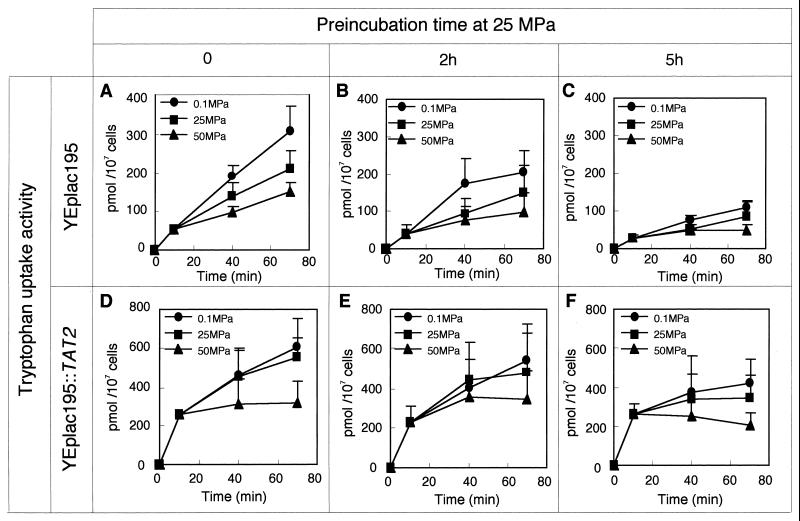
FIG. 5.
Hydrostatic pressure inhibits and down-regulates tryptophan uptake. Tryptophan uptake was assayed as described in Materials and Methods. Hydrostatic pressure was applied at the 10-min point in each experiment. Strain FAA4 was incubated at 25 MPa for 0 h (A), 2 h (B), or 5 h (C), and subsequently the tryptophan uptake assay was performed. Strain FAB33 was incubated at 25 MPa for 0 h (D), 2 h (D), or 5 h (F), and subsequently the tryptophan uptake assay was performed. Data presented are mean values with SD of results from three independent experiments.
TABLE 2.
Effect of hydrostatic pressure on tryptophan uptake
| Parameter | Mean ± SDa after preincubation at 25 MPa for:
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| 0 h and at a pressure (MPa) of:
|
2 h and at a pressure (MPa) of:
|
5 h and at a pressure (MPa) of:
|
|||||||
| 0.1 | 25 | 50 | 0.1 | 25 | 50 | 0.1 | 25 | 50 | |
| Rate constant (pmol of tryptophan 107 cells−1 min−1) | 4.20 ± 1.15 | 2.61 ± 0.778 | 1.65 ± 0.444 | 2.74 ± 0.614 | 1.80 ± 0.872 | 0.937 ± 0.526 | 1.33 ± 0.368 | 0.963 ± 0.534 | 0.326 ± 0.175 |
| Activation vol (ΔV*)b (ml/mol) | 46.2 ± 3.85 | 57.8 ± 19.3 | 83.0 ± 28.4 | ||||||
Data presented are mean values with SD of results from three independent experiments.
Activation volume for overall process of tryptophan uptake.
To examine how exposure to elevated pressure for a long period affected tryptophan uptake in strain FAA4, the cells were incubated in SCv medium at 25 MPa for 2 or 5 h. After decompression, the cells were immediately collected by centrifugation and resuspended in fresh SCv medium, and then the assay of radiolabeled tryptophan uptake was performed. It was found that exposure to a pressure of 25 MPa for 2 h markedly down-regulated tryptophan uptake in strain FAA4 (Fig. 5B; Table 2). The rate constants for tryptophan uptake at 0.1, 25, and 50 MPa were 2.73 ± 0.614, 1.80 ± 0.872, and 0.937 ± 0.526 pmol 107 cells−1 min−1, respectively, as determined 2 h after the exposure, and the activation volume was increased to 57.8 ± 19.3 ml/mol (Table 2). After 5 h of incubation at 25 MPa, the rate constants decreased and, moreover, the activation volume increased to 83.0 ± 28.4 ml/mol (Table 2). These results indicate that tryptophan uptake became impaired during exposure to a pressure of 25 MPa. Therefore, we conclude that (i) hydrostatic pressure decreases the rate constant for tryptophan uptake activity and (ii) hydrostatic pressure down-regulates the activity of tryptophan uptake. Presumably, if down-regulation of Tat2 activity was simply due to a decrease in the quantity of the Tat2 protein (caused by inhibition of transcription or translation), the activation volume should have remained unchanged (i.e., 46.2 ± 3.85 ml/mol), even though the rate constant was decreased. However, the activation volume increased. Therefore, we speculate that down-regulation of tryptophan uptake may involve modification of the Tat2 protein under high-pressure conditions, likely via ubiquitination (9, 17, 22, 43).
We next examined tryptophan uptake in strain FAB33 (YEplac195::2HA-TAT2) expressing a substantial amount of the Tat2 protein. To compare the level of tryptophan uptake and the amount of the Tat2 protein, Western blot analysis was performed and strain FAB33 expressing HA-tagged Tat2 was used. Strain FAB33 exhibited almost the same growth rate and almost the same level of tryptophan uptake as those of strain FAB19 (YEplac195::TAT2) under several pressure conditions tested (data not shown). As expected, the level of tryptophan uptake in strain FAB33 was greater than that in control strain FAA4, indicating that a functional Tat2 protein was expressed in strain FAB33 under high-pressure conditions (Fig. 5). Strain FAB33 showed approximately twofold-greater uptake activity than that of strain FAA4. In addition, the rate of tryptophan uptake was not significantly affected by a pressure of 25 MPa (Fig. 5D). Even though down-regulation of tryptophan uptake occurred upon exposure of the cells to a pressure of 25 MPa, the level of tryptophan uptake in strain FAB33 still remained two- to fourfold higher than that observed in strain FAA4 (Fig. 5E and F). Therefore, it seems evident that efficient tryptophan uptake was accomplished as a result of sufficient expression of Tat2 and that this enabled cell growth under high-pressure conditions. The data obtained in assays with strain FAB33 varied substantially, showing larger SD than with the data obtained in assays with strain FAA4 (Fig. 5A to C). It seems that the amount of Tat2 encoded by the gene on the plasmid YEplac195::2HA-TAT2 may vary from one experiment to another for unknown reasons.
Hydrostatic pressure decreases the amount of the Tat2 protein.
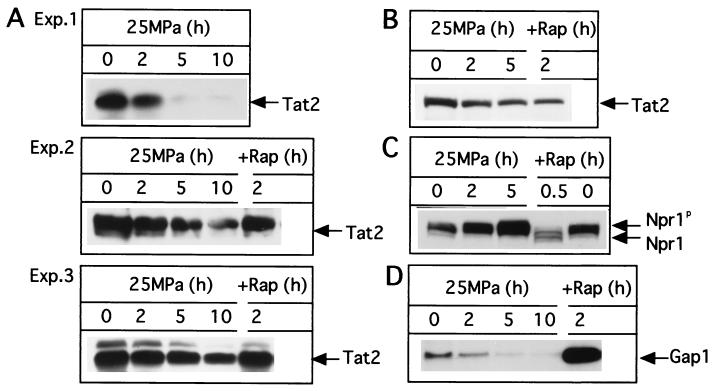
It is known that a rapamycin-FK506 binding protein complex binds Tor1 and Tor2, after which TAP42 is inactivated, and that this inactivation is followed by dephosphorylation of Npr1 (36). Dephosphorylated Npr1 is active, and it induces Tat2 degradation. We examined the levels of Tat2 expressed in cells exposed to elevated hydrostatic pressure. As seen in Fig. 6A, Tat2 levels in strain FAB33 decreased during the period of incubation at 25 MPa. The extent of the decrease in Tat2 levels caused by high pressure varied somewhat from one experiment to another (Fig. 6A, experiments 1 to 3). This resulted in variation of data, as seen in the tryptophan uptake assay shown in Fig. 5D to F. A similar result with respect to a decrease in Tat2 levels was obtained with strain FAB59 harboring the low-copy-number plasmid YCplac33::2HA-TAT2 (Fig. 6B). It is discussed below whether the pressure-induced down-regulation of Tat2 expression was due to inhibition of Tat2 protein synthesis and/or stimulation of Tat2 degradation. In previous reports, rapamycin was shown to decrease the level of Tat2 to less than 20% of the control level within 1 h in strain JK9-3d (9, 36). In our experiment using strain YPH499 in SCv medium at 24°C, rapamycin action was less pronounced (Fig. 6A, experiments 2 and 3, and 6B).
FIG. 6.
Hydrostatic pressure down-regulates intracellular levels of Tat2 protein. Exponentially growing cells of several transformants were exposed to a pressure of 25 MPa for 2, 5, or 10 h or treated with rapamycin (200 ng/ml), and Western blot analysis was performed. (A) Down-regulation of the Tat2 protein in strain FAB33 (YEplac195::2HA-TAT2); (B) down-regulation of the Tat2 protein in strain FAB59 (YCplac33::2HA-TAT2); (C) phosphorylation state of the Npr1 protein (Npr1p) in strain FAE12 (pRS103); (D) down-regulation of the Gap1 protein in strain FAF6 (pPL257). Exp., experiment; Rap, rapamycin.
Pressure-induced down-regulation of Tat2 may be independent of the function of Npr1.
To examine whether pressure-induced down-regulation of expression of the Tat2 protein was dependent on the TOR-signaling pathway, we analyzed the effect of high pressure on the phosphorylation state of functional HA-tagged Npr1. In the presence of nutrients, Npr1 is known to be kept inactive via phosphorylation by TAP42, and hence Tat2 degradation does not occur (36). Starvation or rapamycin treatment induces Tat2 degradation via active dephosphorylated Npr1. As shown in Fig. 6C, Npr1 was dephosphorylated as a result of rapamycin treatment for 2 h in cells incubated at 0.1 MPa, consistent with the previous report (36). However, Npr1 was maintained in a phosphorylated state in cells incubated at 25 MPa (Fig. 6C). We observed a slight increase in levels of phosphorylated Npr1 during the period of incubation at 25 MPa, and the biological significance of this observation is unclear.
Disruption of NPR1 (npr1) is known to result in weak resistance to rapamycin at 30°C (36). We constructed npr1-disrupted mutants of strain YPH499 and confirmed that the mutants exhibited weak rapamycin resistance (data not shown). If Npr1 is involved in pressure-induced down-regulation of Tat2, the npr1-disrupted mutants should display high-pressure growth. However, the mutants failed to grow at pressures of 15 and 25 MPa as far as they were tested in SCv medium (data not shown). These findings suggest that the pressure-induced down-regulation of the Tat2 protein may be independent of the function of Npr1.
Tat2 and the broad-range amino acid permease Gap1 are known to be regulated in an inverse manner by Tor in response to nutrients (9, 11, 20). As a result of nutrient deprivation or rapamycin treatment, Tor is inactivated and Tat2 is targeted to the vacuole for degradation whereas Gap1 is routed to the plasma membrane. A functional HA-tagged Gap1 was expressed in strain FAF6, and the protein level was measured by Western blot analysis. The level of expression of Gap1 in exponentially growing cells was very low, but it dramatically increased upon addition of rapamycin (Fig. 6D). In contrast to the increase in the Gap1 level with the addition of rapamycin, the Gap1 level decreased at a pressure of 25 MPa in a manner dependent on the period of pressurization. Therefore, it is evident that both Tat2 and Gap1 are down-regulated under elevated hydrostatic pressure conditions.
DISCUSSION
Studies with 0.1-MPa-adapted microorganisms have demonstrated that elevated hydrostatic pressure inhibits cell division; nutrient uptake; biosynthesis of DNA, RNA, and proteins; and association of multimeric proteins (reviewed in references 2 and 7). The mechanisms of adaptation to high pressure are thought to be very complex because these organisms are likely to encounter all of the inhibitory effects listed above. Perhaps the presumed complexity is the reason why no attempt has been made to isolate mutants with high growth rates or to identify the gene(s) allowing high-pressure growth in mesophiles. However, our results indicate that only one gene is sufficient to confer high-pressure growth in yeast. We also demonstrated that the availability of tryptophan was the key factor in high-pressure growth. These findings indicate a novel aspect of yeast in investigations of microbial adaptation to high-pressure environments as well as in studies of cellular responses to environmental stresses.
In recent unpublished work, we have isolated mutants capable of growth at high pressure. Genetic analysis indicated that a DNA fragment containing the RSP5 gene from one of these mutants enabled wild-type YPH499 to grow at high pressure, whereas the wild-type RSP5 DNA fragment did not. The mutant RSP5 gene was found to possess a base substitution of G to A, which resulted in an amino acid substitution of Cys to Tyr in the Rsp5 protein (F. Abe et al., unpublished data). Since the defect in Rsp5 is known to result in a marked induction of Tat2 (9), the mutant capable of high-pressure growth may express a considerable amount of the Tat2 protein. The above-mentioned observation supports the idea that Tat2 plays an important role in high-pressure growth. In our study, rapamycin regulated Tat2 and Gap1 inversely in strain YPH499. However, exposure of the cells to a pressure of 25 MPa down-regulated both Tat2 and Gap1 and high pressure did not cause dephosphorylation of Npr1. Therefore, the effect of high pressure is similar to that of rapamycin treatment in terms of causing cell cycle arrest in the G1 phase, inhibition of tryptophan uptake, and Tat2 degradation. However, the pressure-signaling pathway involved seems to be different from the TOR-signaling pathway with respect to Npr1 function. One possible explanation is that Tat2 and Gap1 are degraded, likely via ubiquitination, upon exposure of the cells to elevated hydrostatic pressure or that targeting of both permeases to the plasma membrane may be extremely sensitive to elevated pressure. It seems unlikely that high pressure prevents the traffic of all plasma membrane proteins, because the defect in growth at high pressure can be counteracted by excess tryptophan or enhanced Tat2 expression. Similar phenotypes with respect to tryptophan auxotrophy and rescue by excess tryptophan, TAT2, or TRP1 have been described in studies of sphingolipid toxicity (13, 15, 41), and there may in fact be a general endocytosis with respect to many membrane permeases under stress conditions. Apparently, the tryptophan permease is the Achilles' heel in yeast physiology and under a variety of seemingly disperse toxic conditions becomes limiting for yeast cell growth. For example, erg6 mutants with defects in ergosterol biosynthesis are tryptophan auxotrophs (16). Moreover, many cold-sensitive mutants in yeast are tryptophan auxotrophs or have mutations in tryptophan permeases or tryptophan biosynthesis (12, 39, 40). At low temperatures, tryptophan permease is not active, possibly because of changes in membrane fluidity or composition.
Tor is known to control translation initiation via TAP42 in response to nutrient availability. Elevated hydrostatic pressure is likely to inhibit Tor functions, because Tor is a large membrane protein. Inhibition of Tor proteins results in reduction of protein synthesis. Therefore, the pressure-induced down-regulation of Tat2 might be a consequence of decreased Tor function, which is caused by elevated hydrostatic pressure. In our preliminary results of Northern blot analysis, TAT2 mRNA was decreased during incubation at 25 MPa, even though ACT1 mRNA was not changed. Therefore, TAT2 also seemed to be down-regulated at the transcriptional level. At present, it is difficult to determine whether the first effect of elevated pressure is Tat2 down-regulation, inhibition of TAT2 transcription, inhibition of translation initiation, or inactivation of Tor proteins.
A pressure study yielded a fundamental physical parameter of a reaction, volume change (32). When pressure is applied, the rates of reactions change and the activation volume serves as an indicator of the kinetics, providing quantitative physical information about transition states in physicochemistry. The volume change associated with tryptophan uptake was found to be a large positive value (46.2 ± 3.85 ml/mol). This means that there is a net volume increase in a rate-limiting step in the transport of tryptophan mediated by the Tat2 protein. This step might be the association-dissociation interaction between tryptophan and the Tat2 protein or a conformational change in the Tat2 protein related to its function. Also, it is likely that the Tat2 protein functions as a multimer, because multimeric proteins can be dissociated by elevated pressure accompanied by negative volume changes. The activation volume increased during the period of incubation of the cells at 25 MPa. This finding indicates that long exposure to elevated pressure impaired tryptophan uptake into the cells, which might result from modification of the Tat2 protein, such as ubiquitination by Rsp5. To investigate the step involved in the change in volume particularly at the molecular level, further experimentation should be done using purified Tat2 protein in membrane vesicles. Thorne et al. reported that glucose transport in human erythrocytes was accompanied by a volume increase of 74.0 ± 2.0 ml/mol in its equilibrium exchange (46).
One of our striking results was that cells expressing a substantial level of Tat2 became endowed with the ability to grow at low temperatures as well as at high hydrostatic pressures. It is well documented that the lipid bilayer is one of the structures in biological systems most sensitive to elevated hydrostatic pressure (30, 32). The main phase transition is known to change by 20 to 30°C between 0.1 and 100 MPa in artificial phospholipids (23, 49). As shown in Fig. 4, the growth of strain FAB33 (YEplac195::TAT2) at 25 MPa and 24°C is roughly similar to that at 0.1 MPa and 15°C. It seems that the temperature decrease by 9°C (from 24 to 15°C) and the pressure increase by 25 MPa (from 0.1 to 25 MPa) may have an equivalent effect on growth, which is roughly correspondent to a change in the main phase transition temperature of 20 to 30°C at 100 MPa. Therefore, we can say that tryptophan uptake is sensitive to a decrease in membrane fluidity, whether it is caused by a decrease in temperature or an increase in hydrostatic pressure. Although strain FAB33 showed the ability to grow at low temperature, it exhibited relatively poor growth at 37°C and 0.1 MPa. Since tryptophan is one of the bulky amino acids, it is possible that the permease Tat2 may undergo a considerable conformational change during tryptophan transport. It seems reasonable that the sensitivity of Tat2 to changes in membrane fluidity may be a consequence of the requirement for such a putative dramatic conformational change. Microorganisms display an optimal growth temperature and an optimal growth pressure (50), but the molecular basis of these profiles is totally unknown. Our findings concerning the influence of tryptophan availability on cell growth may serve as one of the means of explaining such poorly understood characteristics of microorganisms.
Elevated pressure, 15 or 25 MPa, caused cell cycle arrest in G1 phase. This can be a new method for synchronization of the cell cycle upon application of hydrostatic pressure. Hydrostatic pressure does not introduce new components to the system; it merely changes the equilibria among preexisting components. We consider this arrest to be due to lack of tryptophan availability for protein synthesis and/or inhibition of translation initiation, especially of G1 cyclins, which are essential for G1 progression. However, a pressure of 50 MPa did not cause G1 arrest. Gross et al. have reported that a pressure of 60 MPa causes dramatic dissociation of ribosomes without association of tRNA or mRNA in E. coli (18). If the effect of pressure on yeast ribosomes is similar, a pressure of 50 MPa is likely to cause dissociation of yeast ribosomes, leading to significant inhibition of protein synthesis. As a consequence of the significant inhibition of protein synthesis at 50 MPa, the cells would not be able to place themselves in a state of suspended animation in the G1 phase of the cell cycle. Further experimentation will be needed to fully describe G1 arrest and cell cycle progression under high-pressure conditions.
The high-pressure growth mutants, including the RSP5 mutant, are being classified into several complementation groups, and cloning of the affected genes is in progress. It is likely that the products of these genes are involved in processes influencing tryptophan availability, translation initiation, Tor functions, or maintenance of the fluidity of the cell membrane, such as fatty acid desaturases.
Perhaps the lack of familiarity with pressure compared with our familiarity with temperature comes from our direct experience of hot and cold. In our studies of piezophysiology, we try to adopt the thermodynamic parameter hydrostatic pressure as a tool for understanding complex physiological events via the volume change associated with reactions or equilibria (2, 4).
ACKNOWLEDGMENTS
We thank H. Iida and M. N. Hall for generously providing plasmids and for valuable comments in the preparation of the manuscript, G. R. Fink for generously providing plasmids and information, and T. Matsuno for technical assistance in Western blot analysis.
REFERENCES
- 1.Abe F. Hydrostatic pressure enhances vital staining with carboxyfluorescein or carboxydichlorofluorescein in Saccharomyces cerevisiae: efficient detection of labeled yeast by flow cytometry. Appl Environ Microbiol. 1998;64:1139–1142. doi: 10.1128/aem.64.3.1139-1142.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abe F, Kato C, Horikoshi K. Pressure-regulated metabolism in microorganisms. Trends Microbiol. 1999;7:447–453. doi: 10.1016/s0966-842x(99)01608-x. [DOI] [PubMed] [Google Scholar]
- 3.Abe F, Horikoshi K. Vacuolar acidification in Saccharomyces cerevisiae induced by elevated hydrostatic pressure is transient and is mediated by vacuolar H+-ATPase. Extremophiles. 1997;1:89–93. doi: 10.1007/s007920050019. [DOI] [PubMed] [Google Scholar]
- 4.Abe F, Horikoshi K. Analysis of intracellular pH in the yeast Saccharomyces cerevisiae under elevated hydrostatic pressure: a study in baro-(piezo-)physiology. Extremophiles. 1998;2:223–228. doi: 10.1007/s007920050064. [DOI] [PubMed] [Google Scholar]
- 5.Allen E E, Facciotti D, Bartlett D H. Monounsaturated but not polyunsaturated fatty acids are required for growth of the deep-sea bacterium Photobacterium profundum SS9 at high pressure and low temperature. Appl Environ Microbiol. 1999;65:1710–1720. doi: 10.1128/aem.65.4.1710-1720.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barbet N C, Schneider U, Helliwell S B, Stansfield I, Tuite M F, Hall M N. TOR controls translation initiation and early G1 progression in yeast. Mol Biol Cell. 1996;7:25–42. doi: 10.1091/mbc.7.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bartlett D H. Microbiol life at high pressure. Sci Prog. 1992;76:479–496. [PubMed] [Google Scholar]
- 8.Beck T, Hall M N. The TOR signaling pathway controls nuclear localization of nutrient-regulated transcription factors. Nature. 1999;402:689–692. doi: 10.1038/45287. [DOI] [PubMed] [Google Scholar]
- 9.Beck T, Schmidt A, Hall M N. Starvation induces vacuolar targeting and degradation of the tryptophan permease in yeast. J Cell Biol. 1999;146:1227–1237. doi: 10.1083/jcb.146.6.1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bidle K, Bartlett D H. RecD function is required for high-pressure growth of a deep-sea bacterium. J Bacteriol. 1999;181:2330–2337. doi: 10.1128/jb.181.8.2330-2337.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cardenas M, Cutler N S, Lorenz M C, Di Como C J. The TOR signaling cascade regulates gene expression in response to nutrients. Genes Dev. 1999;13:3271–3279. doi: 10.1101/gad.13.24.3271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen X H, Xiao Z, Fitzgerald-Hayes M. SCM2, a tryptophan permease in Saccharomyces cerevisiae, in important for cell growth. Mol Gen Genet. 1994;244:260–268. doi: 10.1007/BF00285453. [DOI] [PubMed] [Google Scholar]
- 13.Chung N, Jenkins G, Hannun Y A, Heitman J, Obeid L M. Sphingolipids signal heat stress-induced ubiquitin-dependent proteolysis. J Biol Chem. 2000;275:17229–17232. doi: 10.1074/jbc.C000229200. [DOI] [PubMed] [Google Scholar]
- 14.DeLong E F, Yayanos A A. Adaptation of the membrane lipids of a deep-sea bacterium to changes in hydrostatic pressure. Science. 1985;228:1101–1103. doi: 10.1126/science.3992247. [DOI] [PubMed] [Google Scholar]
- 15.Friant S, Zanolari B, Riezman H. Increased protein kinase or decreased PP2A activity bypasses sphingoid base requirement in endocytosis. EMBO J. 2000;19:2834–2844. doi: 10.1093/emboj/19.12.2834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gaber R F, Copple D M, Kennedy B K, Vidal M, Bard M. The yeast gene ERG6 is required for normal membrane function but is not essential for biosynthesis of the cell-cycle-sparking sterol. Mol Cell Biol. 1989;9:3447–3456. doi: 10.1128/mcb.9.8.3447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Galan J M, Moreau V, Andre B, Volland C, Haguenauer-Tsapis R. Ubiquitination mediated by the Npi1p/Rsp5p ubiquitin-protein ligase is required for endocytosis of the yeast uracil permease. J Biol Chem. 1996;271:10946–10952. doi: 10.1074/jbc.271.18.10946. [DOI] [PubMed] [Google Scholar]
- 18.Gross M, Lehle K, Jaenicke R, Nierhaus K H. Pressure-induced dissociation of ribosomes and elongation cycle intermediates. Stabilizing conditions and identification of the most sensitive state. Eur J Biochem. 1993;218:463–468. doi: 10.1111/j.1432-1033.1993.tb18397.x. [DOI] [PubMed] [Google Scholar]
- 19.Guthrie C, Fink G R. Guide to yeast genetics and molecular biology. San Diego, Calif: Academic Press, Inc.; 1991. [Google Scholar]
- 20.Hardwick J S, Kuruvilla F G, Tong J K, Shamji A F, Schreiber S L. Rapamycin-modulated transcription defines the subset of nutrient-sensitive signaling pathways directly controlled by the Tor proteins. Proc Natl Acad Sci USA. 1999;96:14866–14870. doi: 10.1073/pnas.96.26.14866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Heitman J, Koller A, Kunz J, Henriquez R, Schmidt A, Movva N R, Hall M N. The immunosuppressant FK506 inhibits amino acid import in Saccharomyces cerevisiae. Mol Cell Biol. 1993;13:5010–5019. doi: 10.1128/mcb.13.8.5010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hicke L. Ubiquitin-dependent internalization and down-regulation of plasma membrane proteins. FASEB J. 1997;11:1215–1226. doi: 10.1096/fasebj.11.14.9409540. [DOI] [PubMed] [Google Scholar]
- 23.Ichimori H, Hata T, Matsuki H, Kaneshina S. Barotropic phase transitions and pressure-induced intergitation on bilayer membranes of phospholipids with varying acyl chain lengths Biochim. Biophys Acta. 1998;1414:165–174. doi: 10.1016/s0005-2736(98)00165-5. [DOI] [PubMed] [Google Scholar]
- 24.Iida H, Nakamura H, Ono T, Okumura M S, Anraku Y. MID1, a novel Saccharomyces cerevisiae gene encoding a plasma membrane protein, is required for Ca2+ influx and mating. Mol Cell Biol. 1994;14:8259–8271. doi: 10.1128/mcb.14.12.8259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ito H, Fukuda Y, Murata K, Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983;153:163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jiang Y, Broach J R. Tor proteins and protein phosphatase 2A reciprocally regulate Tap42 in controlling cell growth in yeast. EMBO J. 1999;18:2782–2792. doi: 10.1093/emboj/18.10.2782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kaarniranta K, Elo M, Sironen R, Lammi M J, Goldring M B, Eriksson J E, Sistonen L, Helminen H J. Hsp70 accumulation in chondrocytic cells exposed to high continuous hydrostatic pressure coincides with mRNA stabilization rather than transcriptional activation. Proc Natl Acad Sci USA. 1998;95:2319–2324. doi: 10.1073/pnas.95.5.2319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kanzaki M, Nagasawa M, Kojima I, Sato C, Naruse K, Sokabe M, Iida H. Molecular identification of a eukaryotic, stretch-activated nonselective cation channel. Science. 1999;285:882–886. doi: 10.1126/science.285.5429.882. [DOI] [PubMed] [Google Scholar]
- 29.Ljungdahl P O, Gimeno C J, Styles C A, Fink G R. SHR3: a novel component of the secretory pathway specifically required for localization of amino acid permeases in yeast. Cell. 1992;71:463–478. doi: 10.1016/0092-8674(92)90515-e. [DOI] [PubMed] [Google Scholar]
- 30.Macdonald A G. The role of membrane fluidity in complex processes under high pressure. In: Marquis R E, Zimmerman A M, Jannasch H W, editors. Current perspectives in high pressure biology. London, England: Academic Press; 1987. pp. 207–223. [Google Scholar]
- 31.Macdonald A G. Effect of high hydrostatic pressure on the BK channel in bovine chromaffin cells. Biophys J. 1997;73:1866–1873. doi: 10.1016/S0006-3495(97)78217-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Markley J L, Northrop D B, Royer C A. High-pressure effects in molecular biophysics and enzymology. Oxford, United Kingdom: Oxford University Press, Inc.; 1996. [Google Scholar]
- 33.Muir H. The chondrocyte, architect of cartilage. Biomechanics, structure, function and molecular biology of cartilage matrix macromolecules. Bioessays. 1995;17:1039–1048. doi: 10.1002/bies.950171208. [DOI] [PubMed] [Google Scholar]
- 34.Paidhungat M, Garrett S. A homolog of mammalian, voltage-gated calcium channels mediates yeast pheromone-stimulated Ca2+ uptake and exacerbates the cdc1(Ts) growth defect. Mol Cell Biol. 1997;17:6339–6347. doi: 10.1128/mcb.17.11.6339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Powers T, Walter P. Regulation of ribosome biogenesis by the rapamycin-sensitive TOR-signaling pathway in Saccharomyces cerevisiae. Mol Biol Cell. 1999;10:987–1000. doi: 10.1091/mbc.10.4.987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schmidt A, Beck T, Koller A, Kunz J, Hall M N. The TOR nutrient signalling pathway phosphorylates NPR1 and inhibits turnover of the tryptophan permease. EMBO J. 1998;17:6924–6931. doi: 10.1093/emboj/17.23.6924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schmidt A, Hall M N, Koller A. Two FK506 resistance-conferring genes in Saccharomyces cerevisiae, TAT1 and TAT2, encode amino acid permeases mediating tyrosine and tryptophan uptake. Mol Cell Biol. 1994;14:6597–6606. doi: 10.1128/mcb.14.10.6597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sikorski R S, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Singh A, Manney T R. Genetic analysis of mutations affecting growth of Saccharomyces cerevisiae at low temperature. Genetics. 1974;77:651–659. doi: 10.1093/genetics/77.4.651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Singh A, Manney T R. Suppression of two missense alleles of the TRP5 locus of Saccharomyces cerevisiae. Genetics. 1974;77:661–670. doi: 10.1093/genetics/77.4.661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Skrzypek M S, Nagiec M M, Lester R L, Dickson R C. Inhibition of amino acid transport by sphingoid long chain bases in Saccharomyces cerevisiae. J Biol Chem. 1998;273:2829–2834. doi: 10.1074/jbc.273.5.2829. [DOI] [PubMed] [Google Scholar]
- 42.Smith R L, Rusk S F, Ellison B E, Wessells P, Tsuchiya K, Carter D R, Caler W E, Sandell L J, Schurman D J. In vitro stimulation of articular mRNA and extracellular matrix synthesis by hydrostatic pressure. J Orthop Res. 1996;14:53–60. doi: 10.1002/jor.1100140110. [DOI] [PubMed] [Google Scholar]
- 43.Springael J Y, Andre B. Nitrogen-regulated ubiquitination of the Gap1 permease of Saccharomyces cerevisiae. Mol Biol Cell. 1998;9:1253–1263. doi: 10.1091/mbc.9.6.1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sukharev S I, Blount P, Martinac B, Kung C. Mechanosensitive channels of Escherichia coli: MscL gene, protein, and activities. Annu Rev Physiol. 1997;59:633–657. doi: 10.1146/annurev.physiol.59.1.633. [DOI] [PubMed] [Google Scholar]
- 45.Thomas G, Hall M N. TOR signaling and control of cell growth. Curr Opin Cell Biol. 1997;9:782–787. doi: 10.1016/s0955-0674(97)80078-6. [DOI] [PubMed] [Google Scholar]
- 46.Throne S D, Hall A C, Lowe A G. Effects of pressure on glucose transport in human erythrocytes. FEBS Lett. 1992;301:299–302. doi: 10.1016/0014-5793(92)80261-e. [DOI] [PubMed] [Google Scholar]
- 47.Turley C M, Lochte K, Lampitt R S. Transformations of biogenic particles during sedimentation in the northern Atlantic. Philos Trans R Soc Lond B. 1995;348:179–189. [Google Scholar]
- 48.Welch T J, Bartlett D H. Identification of a regulatory protein required for pressure-responsive gene expression in the deep-sea bacterium Photobacterium species strain SS9. Mol Microbiol. 1998;27:977–985. doi: 10.1046/j.1365-2958.1998.00742.x. [DOI] [PubMed] [Google Scholar]
- 49.Wong P T T, Mantsch H H. Effects of hydrostatic pressure on the molecular structure and endothermic phase transition of phosphatidylcholine bilayers: a Raman scattering study. Biochemistry. 1985;24:4091–4096. doi: 10.1021/bi00336a043. [DOI] [PubMed] [Google Scholar]
- 50.Yayanos A A. Evolutional and ecological implications of the properties of deep-sea barophilic bacteria. Proc Natl Acad Sci USA. 1986;83:9542–9546. doi: 10.1073/pnas.83.24.9542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zaragoza D, Ghavidel A, Heitman J, Schultz M C. Rapamycin induces the G0 program of transcriptional repression in yeast by interfering with the TOR signaling pathway. Mol Cell Biol. 1998;18:4463–4470. doi: 10.1128/mcb.18.8.4463. [DOI] [PMC free article] [PubMed] [Google Scholar]