MON-2, which mediates Golgi-endosome trafficking, mediates mitochondrial inhibition–induced longevity by enhancing autophagy.
Abstract
The Golgi apparatus plays a central role in trafficking cargoes such as proteins and lipids. Defects in the Golgi apparatus lead to various diseases, but its role in organismal longevity is largely unknown. Using a quantitative proteomic approach, we found that a Golgi protein, MON-2, was up-regulated in long-lived Caenorhabditis elegans mutants with mitochondrial respiration defects and was required for their longevity. Similarly, we showed that DOP1/PAD-1, which acts with MON-2 to traffic macromolecules between the Golgi and endosome, contributed to the longevity of respiration mutants. Furthermore, we demonstrated that MON-2 was required for up-regulation of autophagy, a longevity-associated recycling process, by activating the Atg8 ortholog GABARAP/LGG-1 in C. elegans. Consistently, we showed that mammalian MON2 activated GABARAPL2 through physical interaction, which increased autophagic flux in mammalian cells. Thus, the evolutionarily conserved role of MON2 in trafficking between the Golgi and endosome is an integral part of autophagy-mediated longevity.
INTRODUCTION
The Golgi complex is the organelle crucial for modification and sorting of proteins destined for secretion and/or membrane localization (1, 2). First, protein cargoes are packaged into coat protein II (COPII) vesicles transported from the endoplasmic reticulum (ER). The cargoes then follow the cisternal maturation process within the Golgi complex until being associated with the trans-Golgi network and subsequently delivered to their final destinations, including the endosomal/lysosomal system or plasma membrane.
MON-2, a protein localized to the trans-Golgi network, was initially identified in a genetic screen for hypersensitivity against monensin and brefeldin A, which are inhibitors of vesicular transport in cells (3). MON-2 mediates trafficking between subcellular organelles, including the Golgi and endosome. In yeast, Mon2 forms a complex with Dopey (Dop1), an evolutionarily conserved Golgi protein (4, 5), and regulates the recycling of vesicle-associated soluble N-ethylmaleimide-sensitive-factor attachment protein receptor (v-SNARE) Snc1 from early endosomes to the Golgi (5). In Caenorhabditis elegans, MON-2 is implicated in endosome-to-Golgi retrograde transport for Wnt/β-catenin signaling, sorting of Wntless (6, 7), localization of plasma membrane proteins (8), and extracellular vesicle release with sorting nexin and PAD-1, the Dop1 ortholog (7, 9). However, the role of MON-2 in aging and longevity is currently unknown.
Autophagy is a cellular process responsible for the degradation of damaged organelles and dysfunctional components in the cytosol (10, 11). Macroautophagy (hereafter referred to as autophagy) is initiated with the activation of the Unc-51 like autophagy activating kinase 1 (ULK1)/UNC-51 complex, which is followed by vesicle nucleation at omegasomes through the Beclin 1/BEC-1– and autophagy related 14 (Atg14)-containing phosphoinositide-3 kinase complex/VPS-34 (10). Phosphatidylethanolamine-conjugated Atg8 family proteins, including gamma-aminobutyric acid receptor-associated protein (GABARAP)/LGG-1 and microtubule-associated protein 1A/1B-light chain 3 (LC3)/LGG-2 (12–15), are then associated with the elongating phagophore membrane while cargoes are recruited. The elongated phagophores become double-membrane vesicles called autophagosomes, which are subsequently fused with the lysosomes for degrading substrates (11). The transcription factor EB (TFEB)/C. elegans helix loop helix 30 (HLH-30) is responsible for the expression of genes encoding key autophagy factors, including GABARAP/LGG-1 (16). Autophagy plays crucial roles in the regulation of aging in many species (17). In C. elegans, autophagy is required for longevity conferred by various interventions, including inhibition of mitochondrial respiration, reduced insulin/insulin-like growth factor 1 (IGF-1) signaling (IIS), and dietary restriction (DR) (17).
Many factors functioning in mitochondria are important for the modulation of life span in various organisms, including C. elegans (18). However, evidence for communication between mitochondria and other organelles, including the Golgi complex, and how this communication affects organismal life span is scarce. In the present study, we identified the Golgi protein MON-2 as a key factor in mediating C. elegans longevity conferred by inhibition of mitochondrial respiration. Different from previous transcriptomic approaches toward determining the role of mitochondrial respiration in animal physiology (19–23), here, we performed an unbiased quantitative proteomic analysis. We identified MON-2 protein whose levels increased in long-lived mitochondrial respiration mutants. We showed that MON-2 enhanced autophagic activities via regulating Atg8/LGG-1, which, in turn, contributed to C. elegans longevity conferred by various interventions, including inhibition of mitochondrial respiration, reduced IIS and DR. Using cultured mammalian cells, we also demonstrated that MON2 up-regulated early autophagosome formation mediated by an Atg8/LGG-1 ortholog GABARAPL2. Thus, MON-2/MON2 appears to enhance autophagy in both C. elegans and mammals. Our study highlights the importance of organelle communication among mitochondria, the Golgi, and autophagosomes for organismal longevity.
RESULTS
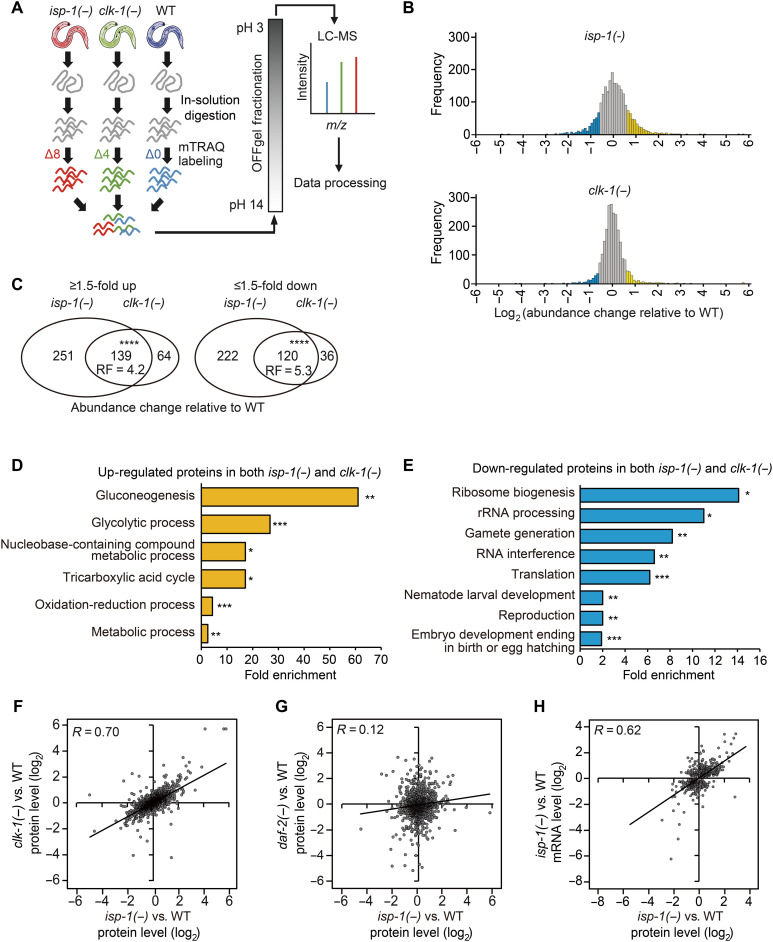
Longevity-inducing mitochondrial respiration mutations cause changes in proteomes
To identify life span–regulating proteins with levels that were altered in long-lived mitochondrial respiratory mutants of C. elegans, we used an mTRAQ (mass differential tags for relative and absolute quantitation) quantitative proteomic approach (24). We compared the proteomes of established long-lived mitochondrial respiration isp-1(qm150) and clk-1(qm30) mutants with that of wild-type (WT) animals (Fig. 1A). Using a 1% false discovery rate (FDR), we identified 2376 unique proteins, and the levels of 732 and 359 proteins in isp-1 and clk-1 mutants, respectively, were changed by ≥1.5-fold compared with those in WT (Fig. 1B and table S1A). Proteins for which levels increased (139) or decreased (120) in isp-1 and clk-1 mutants substantially overlapped (Fig. 1C and table S1A). Gene ontology (GO) analysis indicated that metabolic processes, including gluconeogenesis and nucleobase-containing compound metabolic process, were highly enriched among proteins up-regulated in isp-1 and clk-1 mutants (Fig. 1D, fig. S1A, and table S1, B and C). Proteins down-regulated in isp-1 and clk-1 mutants were enriched in GO terms, such as ribosome biogenesis, rRNA processing, and gamete generation (Fig. 1E, fig. S1B, and table S1, B and C). Overall protein levels in these two long-lived respiration mutants compared with those in WT also positively correlated (R = 0.7, P < 0.00001; Fig. 1F). In contrast, protein levels in isp-1 mutants did not significantly correlate with those in daf-2/insulin/IGF-1 receptor mutants (R = 0.12; Fig. 1G) (25). These data are consistent with the notion that isp-1 and clk-1 mutants share similarities as animals defective in mitochondrial respiration but display characteristics that differ from those of daf-2 mutants, which exhibit longevity because of reduced IIS instead of impaired respiration. By comparing our proteome lists with published microarray data (19, 20), we found that the proteins up-regulated and down-regulated in isp-1 and clk-1 mutants significantly overlapped with transcripts up-regulated and down-regulated in the respective mutants (table S1A). In addition, overall protein and transcript levels in isp-1 mutants (19) positively correlated (R = 0.62 and P < 0.0001; Fig. 1H). Together, this quantitative proteomic analysis revealed proteins whose levels were altered in longevity-promoting mitochondrial respiration mutants.
Fig. 1. Quantitative proteomic analysis identifies proteins differentially expressed in mitochondrial respiratory mutants.
(A) Schematic flow of mTRAQ proteomic analysis using isp-1(qm150) [isp-1(−)], clk-1(qm30) [clk-1(−)], and WT animal samples. (B) Histogram showing the frequency of proteins whose levels were changed in isp-1(−) and clk-1(−) mutants compared with those in WT animals (blue: ≥1.5-fold down-regulated proteins; yellow: ≥1.5-fold up-regulated proteins). (C) Up-regulated and down-regulated proteins (≥1.5-fold) significantly overlap between isp-1(−) and clk-1(−). (****P < 0.00001, chi-square test). RF, representation factor. (D and E) GO analysis [the database for annotation, visualization, and integrated discovery (DAVID)] (72) of proteins whose levels were increased (D) or decreased (E) ≥1.5-fold in both clk-1(−) and isp-1(−) mutants compared with those in WT animals (*P < 0.05, **P < 0.01, and ***P < 0.001, Benjamini P value). (F and G) Correlation between abundance changes in the protein levels in isp-1(−) and clk-1(−) (F) or isp-1(−) and daf-2(e1370) [daf-2(−)] mutants (G) relative to the levels in WT animals (R, Pearson correlation coefficient). (H) Correlation between abundance changes in the protein and mRNA levels in isp-1(−) mutants relative to those in WT animals (R). See table S1 for analysis of proteomics data.
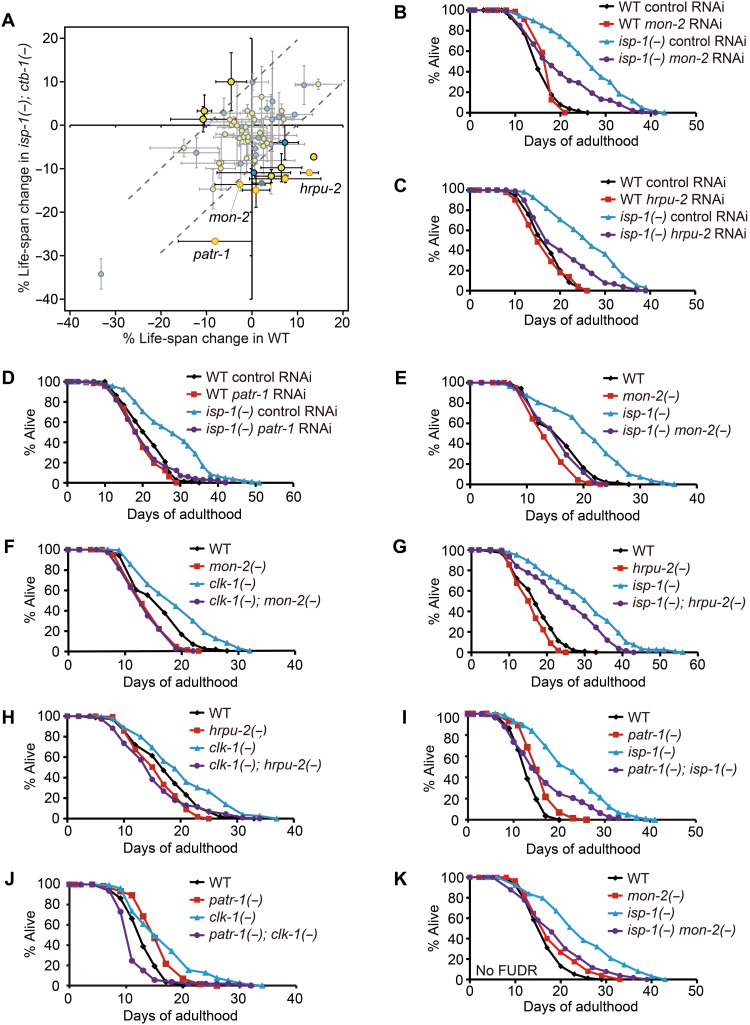
A targeted RNA interference screen identified proteins required for the longevity of respiration mutants
To identify protein factors important for the longevity of mitochondrial respiration mutants, we next performed an RNA interference (RNAi)–based life-span screen targeting 61 genes encoding proteins whose levels changed by ≥2-fold in both isp-1 and clk-1 mutants (table S2A). We used WT animals and isp-1(qm150); ctb-1(qm189) mutants for the RNAi screen (see Materials and Methods for details). We found that 14 RNAi clones had greater effects on the life span of isp-1; ctb-1 mutants than on that of WT animals (an arbitrary 10% difference in life span was applied as a cutoff value; Fig. 2A; see fig. S1 legends for further discussion). Eleven RNAi clones shortened the longevity of isp-1; ctb-1 mutants without having a significant effect on WT life span (table S2A). Eight (mon-2, gpd-2, ugt-61, F48D6.4, patr-1, alh-5, ugt-62, and hrpu-2) of these 11 RNAi clones targeted genes encoding proteins whose levels increased in isp-1 and clk-1 mutants (Fig. 2A and table S1A). In addition, knockdown of each of K01D12.15, pgp-6, and ugt-26 (the gene products of which were down-regulated in isp-1 and clk-1 mutants) reduced the longevity of isp-1; ctb-1 mutants (Fig. 2A and table S1A). Among the 11 RNAi clones, knockdown of mon-2, hrpu-2, and patr-1 also significantly decreased the long life span of isp-1 single mutants (Fig. 2, B to D, and table S2B). In addition, mon-2(xh22), hrpu-2(tm1474), and patr-1(gk155841) mutations substantially suppressed the longevity of isp-1 and clk-1 mutants (Fig. 2, E to K, and table S2C). Thus, increased levels of MON-2, heterogeneous nuclear ribonucleoprotein U 2 (HRPU-2), and protein associated with topo II related 1 (PATR-1) proteins appear to contribute to the longevity conferred by inhibition of mitochondrial respiration.
Fig. 2. A subset of proteins up-regulated by inhibition of mitochondrial respiration is required for longevity.
(A) Average percent changes in mean life span of WT and isp-1(qm150); ctb-1(qm189) [isp-1(−); ctb-1(−)] animals treated with each of 61 RNAi clones targeting genes that encode proteins whose levels were altered in both isp-1(qm150) [isp-1(−)] and clk-1(qm30) [clk-1(−)] mutants ≥2-fold. Yellow and blue circles indicate proteins with levels that were respectively increased and decreased by both isp-1(−) and clk-1(−). At least two independent life-span assays were conducted for each RNAi clone. Dotted lines, 10% life-span increase or decrease for WT or isp-1(−); ctb-1(−) animals. Circles with orange lines, significant results for both life-span trials. (B to D) RNAi targeting mon-2 (B), hrpu-2 (C), or patr-1 (D) significantly suppressed the long life span of isp-1(−) mutants. (E and F) mon-2(xh22) [mon-2(−)] mutations largely suppressed the longevity of isp-1(−) (E) and clk-1(−) (F) mutants (see the table S3 legends for the small or no effect of mon-2(−) on life span). (G to J) hrpu-2(tm1474) [hrpu-2(−)] and patr-1(gk155841) [patr-1(−)] mutations decreased the longevity of isp-1(−) (G and I) and clk-1(−) (H and J) mutants. (K) mon-2(−) suppressed the longevity of isp-1(−) animals without 5-fluoro-2′-deoxyuridine (FUDR) treatment. See also fig. S1 legends for discussion regarding the differences between our initial screen and the confirmation data. See table S2 for specific values and statistical analyses of the life-span data shown in this figure. LC-MS, liquid chromatography–mass spectrometry; m/z, mass/charge ratio.
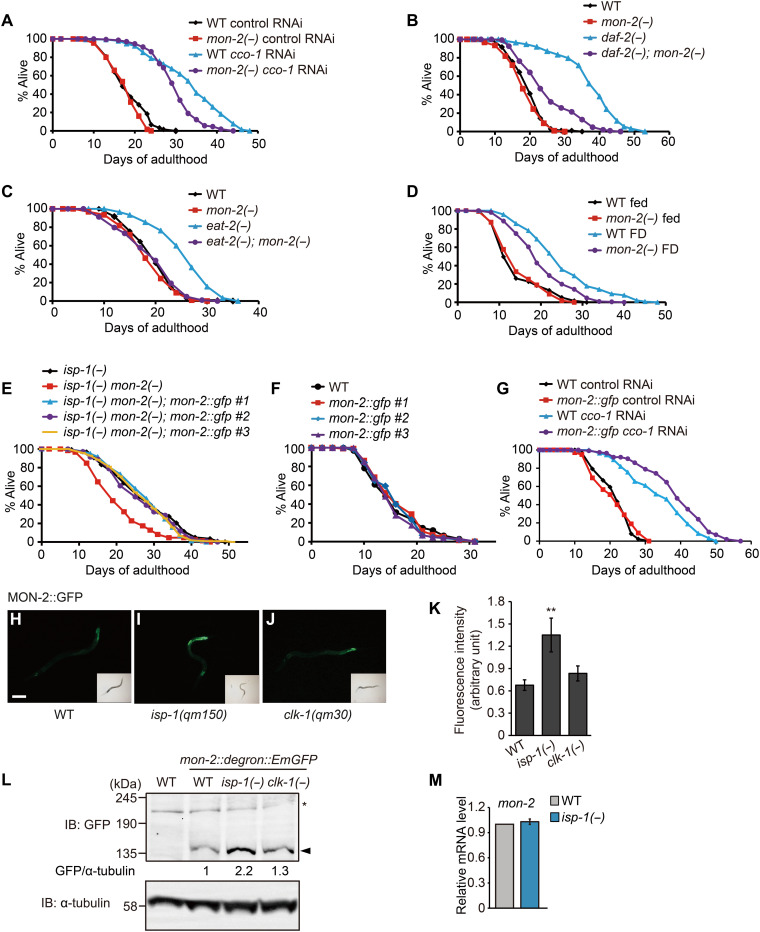
mon-2 is required for longevity conferred by various interventions
We focused our further analysis on mon-2, the genetic inhibition of which robustly suppressed the long life span resulting from isp-1 or clk-1 mutations. MON-2 has several domains, including guanine nucleotide exchange (GEF) Sec7 and dimerization and cyclophilin-binding domains (fig. S1C) and is highly conserved among eukaryotes (fig. S1, D and E) (4, 5, 26). We investigated whether MON-2 contributed to longevity conferred by interventions other than isp-1 and clk-1 respiration mutations. We found that mon-2 was at least partially required for longevity caused by RNAi targeting cco-1 (cytochrome c oxidase subunit 5B) (Fig. 3A), by mutations in the daf-2/insulin/IGF-1 receptor (Fig. 3B), and by two DR regimens, namely, mutations in eat-2 and food deprivation (starvation) (Fig. 3, C and D) (27). We next generated transgenic animals expressing mon-2::gfp under its own promoter, which fully restored longevity in isp-1 mon-2 double mutants (Fig. 3E). The transgenic expression of mon-2::gfp did not affect WT life span (Fig. 3F) but further extended the life span of cco-1 RNAi–treated animals (Fig. 3G). These data indicate that the elevation of MON-2 protein levels can increase life span in a sensitized, respiration-impaired condition. Consistent with our mass spectrometry (MS) data, green fluorescent protein (GFP)–tagged MON-2 levels were increased in isp-1 mutants, although the increase was marginal in clk-1 mutants (Fig. 3, H to L). Furthermore, isp-1 mutations did not significantly affect mon-2 mRNA levels (Fig. 3M). These data suggest that mitochondrial isp-1 mutation increases MON-2 protein levels posttranscriptionally. We also found that genetic inhibition of daf-2 or cco-1 increased the levels of MON-2::GFP (fig. S2, A and B), whereas food deprivation did not (fig. S2, C and D). Overall, mon-2 appears to be required for multiple conserved longevity paradigms.
Fig. 3. MON-2 is required for longevity conferred by various interventions.
(A to D) mon-2(−) mutations reduced the long life span caused by cco-1 RNAi (A), daf-2(e1370) [daf-2(−)] (B), eat-2(ad1116) [eat-2(−)] (C), or food deprivation (FD) (D). (E and F) Transgenic expression of three independent lines of mon-2p::mon-2::gfp rescued the short life span of isp-1(qm150) mon-2(xh22) [isp-1(−) mon-2(−)] mutants (E), while not affecting the life span of WT animals (F). (G) Transgenic expression of mon-2p::mon-2::gfp line #1 further increased the long life span of cco-1 RNAi–treated animals. (H to J) Fluorescent signals of MON-2::GFP (green fluorescent protein) in WT (H), isp-1(qm150) [isp-1(−)] (I), and clk-1(qm30) [clk-1(−)] (J) animals. L2 or L3 larvae were used for imaging. Scale bar, 100 μm. (K) Quantification of (H to J) (N > 29 animals per condition). Error bars represent the SEM (**P < 0.01, two-tailed Student’s t test). (L) Western blot analysis of MON-2 protein using mon-2::degron::EmGFP CRISPR-mediated knock-in animals in WT, isp-1(−) or clk-1(−) mutant backgrounds (n = 3). α-Tubulin was used as a loading control [note that the MON-2::degron::EmGFP band was observed at around 135 kDa, which is unexpected, as the expected size is 212 kDa; because this study is the first one for detecting an endogenously tagged C. elegans MON-2, we speculate that the 135-kDa band was derived from either a posttranslationally modified (e.g., cleaved) MON-2 or a newly identified isoform of MON-2.]. Filled arrowhead indicates MON-2::degron::EmGFP, and the asterisk indicates a nonspecific band. IB, Immunoblot. (M) isp-1(−) did not affect mon-2 mRNA levels. Error bars represent SEMs (n = 3, two-tailed Student’s t test). See tables S3 and S4 for specific values and statistical analyses of the life-span data shown in this figure.
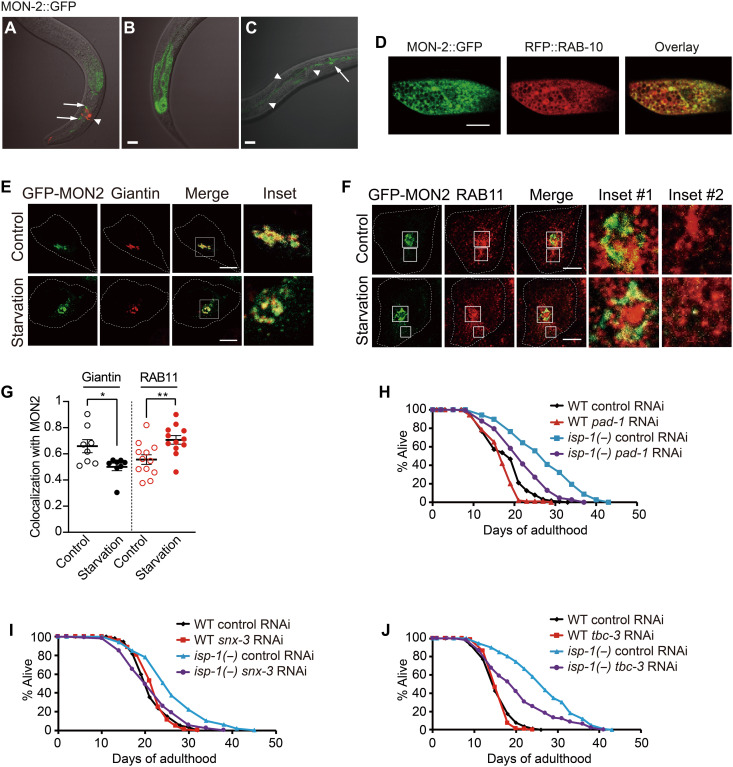
MON-2/MON2 is localized to the Golgi and recycling endosome
Next, we determined the expression patterns of MON-2. We found that mon-2::gfp was expressed in multiple cell types, including intestinal and neuronal cells and hypodermal seam cells (Fig. 4, A to C). MON-2::GFP colocalized with RFP::RAB-10, a Golgi marker (Fig. 4D) but not substantially with a marker for late endosomes and lysosomes (mCherry::RAB-7) or an early endosome marker (mCherry::RAB-5) (fig. S2, E and F). To further dissect the subcellular localization of MON-2 in endosomes and/or the Golgi, we assessed the localization of mammalian MON2 in cultured human cells. We found that mammalian MON2 colocalized with a Golgi marker (giantin) (Fig. 4, E and G), consistent with a previous report (28). MON2 also substantially colocalized with a recycling endosome marker, RAB11 (Fig. 4, F and G) (29), but not with an autophagosome marker (LC3B), a lysosomal marker (lysosome-associated membrane protein 1, LAMP1), or an early endosome marker (early endosome antigen 1, EEA1) (fig. S2, G to L); the colocalization of mammalian MON2 with the recycling endosome marker RAB11 is consistent with the colocalization of C. elegans MON-2 with RFP::RAB-10 (Fig. 4D) because C. elegans RAB-10 also substantially colocalizes with RAB-11 (30). Unexpectedly, we found that MON2 translocated to the recycling endosome from the Golgi during starvation (Fig. 4, E to G), a treatment that increased life span in a MON-2–dependent manner in C. elegans (Fig. 3D). These data raise the possibility that MON2 affects physiological processes such as aging and longevity through shuttling between the Golgi and recycling endosomes.
Fig. 4. MON-2 is required for longevity by regulating trafficking between the Golgi complex and endosomes.
(A to C) Microscopic images of transgenic animals expressing mon-2::gfp. The mon-2::gfp transgene was expressed in neurons (arrows) (A), the intestine (B), and neurons and seam cells (an arrow and arrowheads, respectively) (C). odr-1p::rfp was used as a coinjection marker (arrowhead) in (A). Scale bars, 20 μm. (D) Fluorescence microscope images of an intestinal cell in a transgenic animal expressing mon-2::gfp, rfp::rab-10 and the overlay of the two images. Scale bar, 10 μm. (E and F) Colocalization of mammalian GFP-MON2 with giantin, a Golgi marker, was decreased under starvation conditions (E), whereas that with RAB11, a recycling endosome marker, was increased (F) in HeLa cells. Scale bars, 10 μm. (G) Quantification of (E) and (F) (N = 8, 8, 12, and 12, respectively). Error bars represent SEMs (*P < 0.05 and **P < 0.01, two-tailed Student’s t test). Each dot represents a result from a single cell. (H to J) pad-1 RNAi (H), snx-3 RNAi (I), or tbc-1 RNAi (J) reduced the longevity of isp-1(qm150) [isp-1(−)] mutants. See table S5 for specific values and statistical analyses of the life-span data shown in this figure.
MON-2 appears to promote longevity by regulating trafficking between endosomes and the Golgi complex
Previous studies reported that MON-2 contributes to the transport of lipids and proteins between endosomes and the Golgi (4, 7, 28). Therefore, we sought to determine whether key components required for membrane trafficking also affected the life span of isp-1 mutants, similar to the effects of MON-2 (Fig. 4, H to J; fig. S3, A to E; and table S5). Among the factors examined, RNAi targeting pad-1 [the yeast homolog of which, Dop1, binds MON2 and regulates vesicular trafficking between endosomes and the Golgi (4, 5)] reduced the long life span of isp-1 mutants (Fig. 4H). RNAi knockdown of snx-3/sorting nexin 3, which is required for the sorting of Wntless from endosomes to the Golgi in conjunction with MON-2 (7), also decreased the long life span of isp-1 mutants (Fig. 4I). In addition, genetic inhibition of tbc-3 [an ortholog of human TBC1D22A implicated in transport between endosomes and the Golgi (6)] reduced the long life span caused by isp-1 mutation or cco-1 RNAi (Fig. 4J and fig. S3F). Collectively, these data suggest that MON-2–mediated transport between the Golgi and endosomes is crucial for longevity conferred by mitochondrial electron transport chain (ETC) inhibition.
Enhanced autophagy by MON-2 contributes to longevity
Next, we investigated whether MON-2 acted with established factors contributing to longevity conferred by mitochondrial ETC inhibition; these include Skinhead 1 (SKN-1) [an nuclear factor erythroid 2–related factor 2 (Nrf2) homolog], activating transcription factor associated with stress 1 [ATFS-1, a TF that promotes the mitochondrial unfolded protein response (UPRmt)], hypoxia-inducible factor 1 (HIF-1), and C. elegans p53-like protein 1 (CEP-1)/p53, as well as autophagy components and reactive oxygen species (31–39). We found that RNAi knockdown of mon-2 (fig. S4, A to D) did not affect the representative target gene expression levels of SKN-1, ATFS-1, HIF-1, or CEP-1 (fig. S4, E to K) in WT or isp-1 mutants.
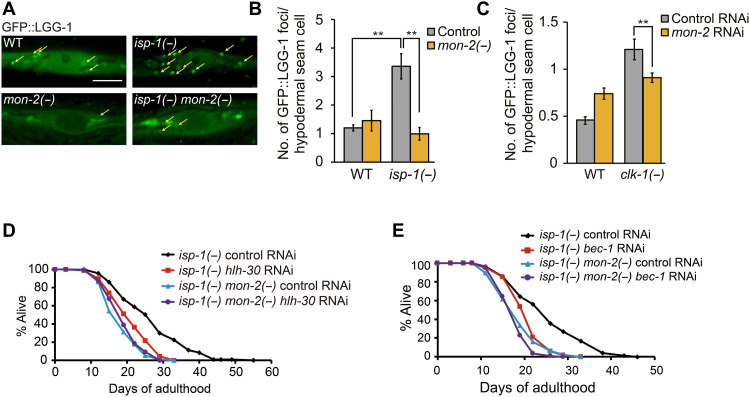
In contrast to these factors, genetic inhibition of mon-2 reduced the number of GFP::LGG-1/Atg8 puncta, known as a marker of autophagosome (11), in the hypodermal seam cells of both clk-1 and isp-1 mutants (Fig. 5, A to C). We also tested whether TFEB/HLH-30, a transcriptional regulator of multiple autophagy genes, and Atg6/Beclin 1 (BEC-1), a factor of vesicle nucleation complex required for an initial step of autophagy, acted together with MON-2 for mediating longevity. As previously reported (16), the RNAi knockdown of TFEB/hlh-30 or Atg6/bec-1 suppressed the long life span of isp-1 mutants (Fig. 5, D and E). TFEB/hlh-30 or Atg6/bec-1 RNAi had small or no effects on the short life span of isp-1 mon-2 double mutants (Fig. 5, D and E). These data are consistent with the possibility that MON-2 acts together with TFEB/HLH-30 and Atg6/BEC-1 to increase life span in respiration mutants via autophagy regulation.
Fig. 5. mon-2 is required for longevity conferred by the inhibition of mitochondrial respiration via affecting autophagy.
(A) Confocal images of GFP::LGG-1/Atg8 in hypodermal seam cells of day 1 adult animals. mon-2(xh22) [mon-2(−)] mutations suppressed the increased number of GFP::LGG-1/Atg8-positive punctae in hypodermal seam cells of day 1 adult isp-1(qm150) [isp-1(−)] mutants but not in those of WT animals. Yellow arrows indicate punctae of GFP::LGG-1/Atg8. Scale bar, 5 μm. (B) Quantification of GFP::LGG-1 punctae in hypodermal seam cells of day 1 adults (n = 3, 30 animals per condition). Error bars represent SEMs (**P < 0.01, two-tailed Student’s t test). (C) mon-2 RNAi decreased the number of GFP::LGG-1 punctae in hypodermal seam cells in clk-1(qm30) [clk-1(−)] mutants at the L2-L3 larval stage (n = 3, 30 animals per condition). Error bars represent SEMs (**P < 0.01, two-tailed Student’s t test). (D and E) hlh-30 RNAi (D) or bec-1 RNAi (E) decreased the long life span of isp-1(−) animals but did not reduce the life span of isp-1(−) mon-2(−) animals. Unexpectedly, however, hlh-30 RNAi increased the level of MON-2::GFP in WT and isp-1(−) animals (fig. S4, L and M). We speculate that decreased autophagy caused by hlh-30 RNAi may up-regulate MON-2::GFP as a compensatory response. See table S6 for specific values and statistical analyses of the life-span data shown in this figure.
MON-2/MON2 contributes to up-regulation of LGG-1/GABARAPL2–dependent autophagy in C. elegans and mammalian cells
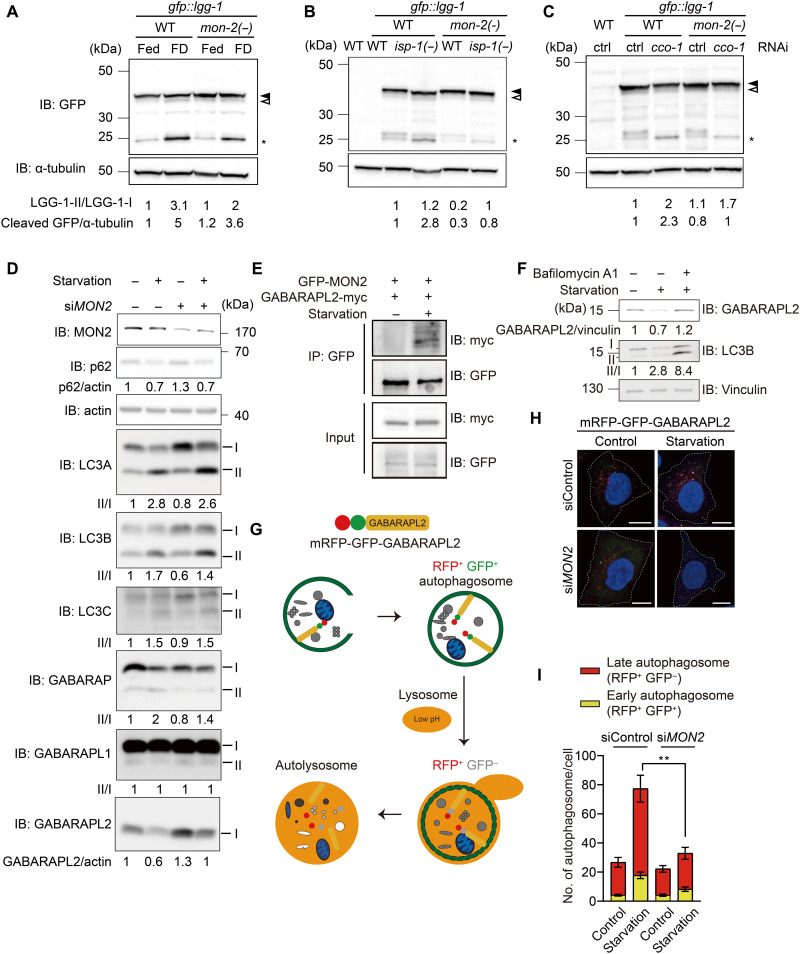
To determine the mechanisms by which MON-2 regulates autophagy, we used food deprivation (starvation), an established paradigm that activates autophagy in both C. elegans and mammalian cells (40, 41). Food deprivation increased the levels of cleaved GFP generated from GFP::LGG-1, indicating autophagy activation in a MON-2–dependent manner (Fig. 6A). In addition, mon-2 mutations suppressed the increased levels of cleaved GFP in isp-1 mutants or cco-1 RNAi–treated animals (Fig. 6, B and C). In contrast, mon-2 mutations did not consistently decrease the basal level of autophagy in WT animals (Figs. 5, A to C, and 6, A to C). To determine how MON2 regulates autophagy, we used mammalian cells to assess autophagy processes by measuring the levels of p62, an autophagy substrate, and ATG8 family members (LC3A/B/C and GABARAP/L1/L2), homologs of LGG-1 (12–14) (Fig. 6D). We found that depletion of MON2 substantially increased the level of GABARAPL2, compared with small effects on the levels of other tested autophagy factors (Fig. 6D). In addition, we showed that MON2 bound to GABARAPL2 under starvation conditions using coimmunoprecipitation assays (Fig. 6E). The level of GABARAPL2, which was reduced by starvation, was increased by inhibition of autophagosome maturation (Fig. 6F), indicating that the GABARAPL2 band is a nonlipidated (inactive) form (42, 43). We further assessed the role of MON2 in autophagy using cultured human cells expressing mRFP-GFP-GABARAPL2, in which the GFP signal is quenched at low pH in lysosomes (Fig. 6G) (44). We found that depletion of MON2 reduced the number of both RFP+GFP+ and RFP+GFP− puncta upon starvation, consistent with MON2 increasing autophagic flux (Fig. 6, H and I). In contrast, knockdown of MON2 did not affect the number of LC3B-positive puncta as determined by using an mRFP-GFP-LC3B reporter (fig. S4, N and O) (44). Thus, mammalian MON2 appears to promote autophagosome formation specifically through GABARAPL2. Together, these data suggest that MON-2/MON2 mediates up-regulation of autophagy through LGG-1/GABARAPL2 in both C. elegans and mammalian cells.
Fig. 6. MON-2/MON2 appears to increase autophagy by activating LGG-1/GABARAPL2 in C. elegans and mammalian cells.
(A to C) mon-2(xh22) [mon-2(−)] consistently decreased the high level of cleaved GFP caused by food deprivation (FD) (A), cco-1 RNAi (B), and isp-1(qm150) [isp-1(−)] mutations (C) (n = 3). Different from consistent results for cleaved GFP, LGG-1–II/I ratio was somewhat variable likely because of subtle separation of LGG-1–I and II bands. Filled arrowhead, LGG-1–I. Open arrowhead, LGG-1–II. Asterisk, cleaved GFP. α-Tubulin, loading control. WT without a transgene, negative control. IB, immunoblot. (D) Small interfering RNA (siRNA) targeting MON2 (siMON2) substantially increased the level of GABARAPL2 while having small effects on those of other tested autophagosomal markers (p62, LC3A, LC3B, LC3C, GABARAP, and GABARAP L1) under basal or starvation conditions (n = 2). One endogenous GABARAPL2 band was detected, which was decreased under starvation and increased by siMON2. In contrast, phosphatidylethanolamine-conjugated (II) and nonconjugated proteins (I) were separated for the other ATG8/LC3 family members. (E) GABARAPL2 was coimmunoprecipitated with MON2 under starvation conditions (n = 3). IP, immunoprecipitation. (F) Immunoblot for GABARAPL2, LC3B, and vinculin in HeLa cells after starvation or starvation with bafilomycin A1 treatment for 6 hours (n = 2). (G) Illustrated fluorescence changes in mRFP-GFP-GABARAPL2 based on its subcellular location in early or late autophagosomes, or autolysosomes. (H) siMON2 in HeLa cells stably transfected with mRFP-GFP-GABARAPL2 under basal or starvation conditions altered the flux of GABARAPL2. Scale bars, 10 μm. (I) The numbers of RFP+GFP+ puncta and RFP+GFP− puncta were counted for each cell and analyzed for statistical differences (N = 21, 21, 22, and 16). Error bars represent SEMs **P < 0.01, Mann-Whitney test).
DISCUSSION
MON-2 is a highly conserved regulator of intracellular trafficking in various organisms. Although the function of MON-2 in membrane trafficking has been reported, its role in life-span regulation or autophagy remains unknown. In the present study, we identified MON-2 from an unbiased proteomic analysis using two long-lived mitochondrial respiration mutants of C. elegans. MON-2 was up-regulated in mitochondrial ETC–impaired clk-1 and isp-1 mutants and was required for longevity. We further showed that MON-2 was required for long life span caused by various regimens, including reduced IIS and DR. MON-2 appeared to mediate trafficking between the Golgi and endosomes and enhanced autophagy. Furthermore, we showed that MON-2/MON2 up-regulated autophagosome formation through LGG-1/GABARAPL2 in both C. elegans and cultured mammalian cells. Our findings support the concept that proper communication among subcellular organelles, including the Golgi, endosomes, autophagosomes, and mitochondria, is essential for life-span extension.
We found that MON-2 was increased at the protein level by inhibition of mitochondrial respiration. In addition, our data showing that MON-2 was not altered at the mRNA level by isp-1 mutations raise the possibility that mon-2 may not be directly regulated by TFs that are up-regulated by mitochondrial inhibition, e.g., ATFS-1, HIF-1, C. elegans homeobox 23 (CEH-23), and defective proventriculus in Drosophila homolog 1 (DVE-1). MON-2 is also required for proper localization of membrane proteins, including Wntless/MIG-14, delivered from endosomes to the Golgi by a retromer complex and thereby secretes neuronal Wnt/EGL-20 to the intestine (6, 7, 29). The neuronal secretion of Wnt/EGL-20 activates DVE-1 and ATFS-1 TFs in the intestine to induce a cell-nonautonomous activation of UPRmt (45). Therefore, the Wnt and UPRmt axis may act with MON-2 in contributing to the longevity of isp-1 mutants. However, we showed that genetic inhibition of mon-2 did not affect ETC inhibition–induced activation of UPRmt. Thus, mon-2 does not seem to act with UPRmt, but this possibility should be further tested in future studies.
One plausible explanation for the up-regulation of MON-2 by inhibition of respiration might be the increase in MON-2 protein stability by mitochondrial chaperones. Our group previously showed that the mitochondrial chaperone heat shock protein 60 (HSP-60) stabilizes cytosolic protein mitogen-activated protein kinase kinase/SEK-1 (46). In addition, cytosolic Hsp60 in yeast inhibits proteasome activity (47), possibly leading to up-regulation of proteasome target proteins. It will be interesting to test whether MON-2 is stabilized by the chaperones induced by mitochondrial inhibition. It is also possible that the degree of decrease in MON-2 protein levels by genetic inhibition of mon-2 is higher in isp-1 mutants than in WT animals. Future studies determining whether partial knockdown of mon-2 in isp-1 mutants to a level similar to that of WT is sufficient to shorten life span in isp-1 mutants will be required to further test this possibility.
Our finding demonstrating that one protein, MON-2, was broadly required for the long life span caused by daf-2 mutations and DR, and mitochondrial inhibition is not unprecedented. For example, genetic inhibition of TFEB/hlh-30 or Myc/Mondo A/mml-1 shortens long life span caused by mutations in daf-2 and isp-1, reduced TOR signaling, and germline removal (16, 48). Because both HLH-30 and MML-1 promote longevity conferred by various regimens by up-regulating autophagy, the requirement of MON-2 for multiple longevity paradigms is actually consistent with our results demonstrating that MON-2 mediates longevity via effect on autophagy.
Because MON-2 is crucial for protein trafficking, it seems likely that various autophagy-related gene (Atg) products are sorted in a MON-2–dependent manner. In yeast, Atg9 mediates the shuttling of membranes between pre-autophagosomal structures and non–pre-autophagosomal structures (cytoplasmic vesicles) (49). Mammalian Atg9 transits between endosomes and the Golgi complex under sated conditions but colocalizes with LC3 puncta under starved conditions (50). Atg27, a component necessary for Atg9 cycling, resides in both mitochondria and the Golgi (51). Thus, it seems possible that MON-2, being a Golgi protein, assists in the proper localization of Atg9 and/or Atg27, which results in autophagosome formation. Moreover, GABARAPL2, also known as Golgi-associated adenosine triphosphatase enhancer of 16 kDa or GATE-16, localizes to the Golgi complex as well (52). On the basis of our data showing the contribution of MON2 to autophagy via GABARAPL2, we speculate that GABARAPL2 is translocated from the Golgi to recycling endosomes by binding to MON2, in particular, under starvation conditions. From recycling endosomes, GABARAPL2 may contribute to autophagosome formation in a manner similar to that of LC3 (53, 54). These findings raise the possibility that MON-2 directs intracellular trafficking of various proteins required for autophagosome formation.
Accumulating evidence suggests that subcellular organelles, including the ER, endosomes, plasma membranes, mitochondria, and the Golgi complex, contribute to autophagosome formation in many species (49, 55, 56). Autophagosomes form at ER-mitochondria contact sites in cultured mammalian cells, and disrupting a SNARE protein, STX17, blocks autophagy (57). The ER-Golgi intermediate compartment (ERGIC) also serves as a membrane source for the lipidation of Atg8/LC3, mediated by the anterograde protein COPII (58, 59). Furthermore, Atg6/Beclin1, a key autophagy component, localizes to the trans-Golgi network and mediates endosome-to-Golgi membrane trafficking (60). In addition, Sec7, a late Golgi GEF protein, is essential for autophagy in yeast (61). Therefore, we propose that MON-2 acts as a scaffold protein between the Golgi complex and endosomes to regulate membrane trafficking required for proper autophagic processes.
MATERIALS AND METHODS
Strains
C. elegans were grown on Escherichia coli OP50-seeded plates following standard laboratory culture conditions as previously described (62). Some of the C. elegans strains used in this study were obtained from Caenorhabditis Genetics Center, which is funded by the National Institutes of Health Office of Research Infrastructure (P40 OD010440). The strains that were used are as follows: WT, Bristol strain N2, CF2172 isp-1(qm150) IV, CF2354 clk-1(qm30) III, MQ989 isp-1(qm150) IV; ctb-1(qm189), IJ572 mon-2(xh22) IV, IJ596 isp-1(qm150) mon-2(xh22) IV, IJ598 clk-1(qm30) III; mon-2(xh22) IV, IJ645 hrpu-2(tm1474) IV, IJ659 hrpu-2(tm1474) isp-1(qm150) IV, IJ660 clk-1(qm30) III; hrpu-2(tm1474) IV, IJ755 patr-1(gk155841) II, IJ856 patr-1(gk155841) II; isp-1(qm150) IV, IJ889 patr-1(gk155841) II; clk-1(qm30) III, IJ904 yhIs51[mon-2p::mon-2cDNA::GFP, odr-1::RFP], IJ906 yhIs53[mon-2p::mon-2cDNA::GFP, odr-1::RFP], IJ907 yhIs54[mon-2p::mon-2cDNA::GFP, odr-1::RFP], IJ997 isp-1(qm150) mon-2(xh22) IV; yhIs51[mon-2p::mon-2cDNA::GFP, odr-1::RFP], IJ999 isp-1(qm150) mon-2(xh22) IV; yhIs53[mon-2p::mon-2cDNA::GFP, odr-1::RFP], IJ1000 isp-1(qm150) mon-2(xh22) IV; yhIs54[mon-2p::mon-2cDNA::GFP, odr-1::RFP], IJ1165 clk-1(qm30) III; mon-2(xh22) IV; yhIs53[mon-2p::mon-2cDNA::GFP; odr-1p::RFP], IJ1366 yhIs51[mon-2p::mon-2cDNA::gfp; odr-1p::rfp]; pwIs430[vha-6p::mCherry::rab-7], IJ1367 yhIs51[mon-2p::mon-2cDNA::gfp; odr-1p::rfp]; pwIs493[vha-6p::mCherry::rab-5], IJ1368 yhIs51[mon-2p::mon-2cDNA::gfp; odr-1p::rfp]; pwIs414[vha-6p::rfp::rab-10; Cbr-unc-119(+)], IJ573 tbc-3(xh23) IV, DA2123 adIs2122[lgg-1p::gfp:: lgg-1 + rol-6(su1006)], IJ1279 clk-1(qm30) III; adIs2122[lgg-1p::gfp:: lgg-1; rol-6(su1006)], IJ1380 mon-2(xh22) IV; adIs2122[lgg-1p::gfp:: lgg-1; rol-6(su1006)], IJ1381 clk-1(qm30) III; mon-2(xh22) IV;adIs2122[lgg-1p::gfp:: lgg-1; rol-6(su1006)], IJ1722 isp-1(qm150) IV; adIs2122[lgg-1p::gfp:: lgg-1 + rol-6(su1006)], IJ1723 isp-1(qm150) mon-2(xh22) IV; adIs2122[lgg-1p::gfp:: lgg-1+ rol-6(su1006)], IJ173 eat-2(ad1116) II, CF1041 daf-2(e1370) III, IJ1068 eat-2(ad1116) II; mon-2(xh22) IV, IJ1069 daf-2(e1370) III; mon-2(xh22) IV, IJ32 Iszc[hsp-6p::gfp], IJ31 isp-1(qm150) IV; Iszc[hsp-6p::gfp], ZG120 iaIs7[nhr-57p::GFP; unc-119(+)], IJ2 isp-1(qm150) IV; iaIs7[nhr-57p::GFP; unc-119(+)], CL2166 dvIs19[pAF15(gst-4::GFP::NLS)], IJ945 isp-1(qm150) IV; dvIs19[pAF15(gst-4::GFP::NLS)], IJ1825 syb563[mon-2::degron::EmGFP] IV, IJ52 isp-1(qm150) syb563[mon-2::degron::EmGFP] IV, and IJ49 clk-1(qm30) III; syb563[mon-2::degron::EmGFP] IV. IJ1825 was used for analyzing GFP-tagged endogenous MON-2 proteins.
Sample preparation for quantitative protein analysis
clk-1(qm30) and isp-1(qm150) mutants and WT animals were synchronized and harvested when most of the animals reached the L4 stage; L4-stage animals were used to avoid any complications associated with reproduction. In total, 1 mg of worm samples was collected by pooling three to four independent biological repeats of each sample. The samples were harvested with M9 buffer including 0.01% polyethylene glycol (PEG) and then immediately frozen in liquid nitrogen. The samples were thawed on ice, pooled, and then washed with 50 mM tris-HCl (pH 8.0) five times to eliminate residual PEG from the buffer. Worm samples were pelleted and sonicated in 2% SDS with 10% glycerol and 50 mM tris-HCl (pH 6.9) (63). The protein solution was reduced with 5 mM dithiothreitol (DTT; from 0.5 M DTT diluted in double distilled water) for 1 hour at 56°C and then alkylated with 14 mM indole-3-acetic acid (from a freshly made 0.7 M solution) for 1 hour at room temperature in the dark. Subsequently, the protein solution was subjected to methanol chloroform precipitation to remove residual SDS. The final protein pellet was resuspended in 6 M urea with 50 mM tris-HCl and 1 mM EDTA (pH 7.5) for mTRAQ labeling (24). In total, 20 μg of protein was used for SDS–polyacrylamide gel electrophoresis (SDS-PAGE), and the gel was stained with Coomassie brilliant blue to visualize proteins. The sample quality was examined by visual inspection to confirm the lack of abnormal protein bands.
Trypsin digestion and mTRAQ
Two hundred micrograms of dissolved proteins from WT, isp-1(qm150), and clk-1(qm30) animals was diluted 10-fold with 50 mM tris-HCl (pH 7.5) and then digested with sequencing-grade trypsin (1:10 ratio of trypsin:proteins; Promega, Madison, WI, USA) at 37°C overnight. Tryptic digests were desalted with C18 solid-phase extraction (SPE) cartridges and dried in vacuo. The dried samples were reconstituted by adding 500 mM triethylammonium bicarbonate and labeled with the mTRAQ 3-plex reagent (AB Sciex, Framingham, MA, USA). Tryptic peptides obtained from WT animals were labeled with Δ0, whereas clk-1(qm30) and isp-1(qm150) mutants were labeled with Δ4 and Δ8, respectively. After a reaction time of 1 hour, the mTRAQ-labeled peptides were combined and subsequently separated using the isoelectric point (pI) of peptides. For pI-based peptide separation, a 3100 OFFGEL fractionator (Agilent Technologies, Santa Clara, CA, USA) and immobilized pH gradient strips (pH 3 to 10, linear, 24 cm; GE Healthcare) were used with a 24-well setup following the manufacturer’s protocol; twenty-four fractions were collected. The OFFGEL fractions were desalted using a C18 SPE cartridge and then dried in vacuo.
Liquid chromatography and tandem MS
OFFGEL fractions of mTRAQ-labeled samples were reconstituted in 0.4% acetic acid, and a ~1-μg aliquot was injected into a reversed-phase Magic C18 aq column (15 cm by 75 μm) on an Eksigent MDLC system at a flow rate of 300 nl/min. The column was equilibrated with 95% buffer A (0.1% formic acid in H2O) + 5% buffer B (0.1% formic acid in acetonitrile) before use. Peptides were eluted with a linear gradient of 10 to 40% buffer B over 90 min. The high-performance liquid chromatography (LC) system was coupled to an LTQ XL-Orbitrap MS (Thermo Fisher Scientific, San Jose, CA, USA). The spray voltage was set to 1.9 kV, while the heated capillary temperature was set to 250°C. Survey full-scan MS spectra [mass/charge ratio (m/z), 300 to 2000] were acquired in the Orbitrap with one microscan and a resolution of 100,000, allowing preview mode for precursor selection and charge-state determination. Tandem MS (MS/MS) spectra for the 10 most intense ions from the preview survey scan were acquired concurrently in the ion trap for full-scan acquisition with the following options: isolation width, 2 m/z; normalized collision energy, 35%; and dynamic exclusion duration, 30 s. Precursors with a single charge and unassigned charge states were discarded during data-dependent acquisition. Data were acquired using Xcalibur software v2.0.7. All fractions of samples were analyzed twice via LC-MS/MS. The first run for every sample was performed by data-dependent analysis, whereas the second run was performed by exclusion-based data acquisition (64). Precursor m/z lists were constructed by analyzing prior LC-MS/MS runs. RAPID software was used for detection, deisotoping, and integration of precursor peaks (65). Peaks were detected in at least two subsequent MS1 scans with a minimum of two isotopic peaks considered. Exclusion lists were formulated by considering the masses of all validated peptides from prior analyses, in which the validated peptides were defined as peptides identified from a Mascot search. The final exclusion lists were implemented into the MS acquisition software.
MS data analysis
The RAW files from the LTQ XL-Orbitrap MS were first converted into mzXML files using ReAdW 4.3.1 with the centroid option and then into mgf files using the Trans-Proteomic Pipeline (TPP; version 4.5 RAPTURE rev 2) with the following parameters: minimum peak count, 5; the minimum threshold for peak intensity, 0.01; and MH+ mass range, 600 to 4200. Peptide and protein identification was accomplished by comparing the mgf files with the UniProtKB C. elegans database (25,831 entries, July 2012) on a Mascot search engine with the following parameters: protease: trypsin; fixed modification: carbamidomethyl (C); variable modifications: mTRAQ Δ0 (N-term, K), mTRAQ Δ4 (N-term, K), mTRAQ Δ8 (N-term, K), and oxidation (M); taxonomy: C. elegans; missed cleavages: 2; peptide tolerance: 15 ppm; and MS/MS tolerance: ±0.5 Da. Peptide and protein assignment was performed with TPP. The Mascot result files were used as an input for the pepXML module, allowing trypsin restriction and “monoisotopic masses” options. Peptide-Prophet was then applied with the “accurate mass binning” option. Peptides with probabilities >0.9 (FDR < 0.02) were included in the subsequent Protein-Prophet analysis, and proteins with protein probabilities >0.9 (FDR < 0.01; ≥2 unique and quantified peptides) were selected. Quantification of the mTRAQ samples was performed using the in-house software Quad-Quant (66, 67). The MS data have been deposited to the PRIDE Archive (www.ebi.ac.uk/pride/archive/) with the dataset identifier PXD026411.
Targeted RNAi life-span screen
The life span of WT and isp-1(qm150); ctb-1(qm189) mutant animals treated with each of 61 RNAi clones was measured; the ctb-1(qm189) mutation, which suppresses the severe developmental delay and small brood size of isp-1(qm150) mutants without affecting their longevity (68), was used in combination with isp-1(qm150) for amenability of the screen. HT115 bacteria expressing double-stranded RNA (dsRNA) that targeted each gene were cultured in liquid Luria broth with ampicillin (50 μg/ml) at 37°C overnight. The dsRNA-expressing bacteria were seeded on nematode growth medium (NGM) containing ampicillin (50 μg/ml) at 37°C overnight and subsequently treated with 1 mM isopropyl-β-d-1-thiogalactopyranoside to induce dsRNA. Worms that were cultured on the plates and synchronized at the prefertile young adult stage were transferred to new plates containing 5 μM 5-fluoro-2′-deoxyuridine (FUDR; Sigma-Aldrich) to prevent progeny hatching. Empty vector and GFP RNAi clones were used for control RNAi. In total, 61 RNAi clones were tested for the life-span screen (tables S1 and S2). RNAi clones that targeted more than two genes redundantly because of off-target effects are described in table S1.
Phylogenetic tree and sequence comparison
Amino acid sequences of MON2/MON-2 in Homo sapiens (human, isoform 1), Mus musculus (mouse, isoform 1), Drosophila melanogaster, C. elegans (isoform a), and Saccharomyces cerevisiae (yeast) were obtained by using UniProt (www.uniprot.org/). A domain search was performed using InterPro Scan Search (www.ebi.ac.uk/interpro/search/sequence/). Amino acid sequences of the Sec7 domain from humans, mice, D. melanogaster, C. elegans, and yeast were aligned using Clustal X 2.1 (69) with the “iterate each alignment step” function. To produce a phylogenetic tree, Clustal Omega (www.ebi.ac.uk/Tools/msa/clustalo/) was used to align and draw a neighbor-joining tree without distance correction based on the sequences of MON2/MON-2 from humans, mice, D. melanogaster, C. elegans, and yeast.
Life-span assays
Starting from day 1 of adulthood, life-span assays were conducted at 20°C on NGM plates seeded with E. coli OP50 for mutants or HT115 for RNAi experiments. Briefly, synchronized prefertile young adult animals were transferred to plates containing 5 to 10 μM FUDR (Sigma-Aldrich) to prevent progeny from hatching, unless stated otherwise. All life-span assays were conducted by at least two independent researchers at least twice per condition, and a minimum of four plates were used for each condition, except where plates were discarded because of contamination. Animals that ruptured, displayed internal hatching, or crawled off the plates were censored but included in the life-span analysis as censored animals.
Microscopy and quantification of fluorescence
Images of transgenic animals were captured using AxioCam (Zeiss Corporation, Germany) attached to an HRc Zeiss Axioscope A.1 (Zeiss Corporation, Germany). Levamisole (2 mM) was used to immobilize the animals on 2% agarose pads before obtaining microphotographs. Quantification of GFP intensity was performed by using ImageJ (Rasband, W. S., ImageJ, U.S. National Institutes of Health, Bethesda, Maryland, USA, http://rsbweb.nih.gov/ij/). Mean fluorescence intensity was normalized by subtracting the intensity of background images obtained with the same exposure time. Statistical differences were determined using an unpaired Student’s t test (two-tailed). To capture the fluorescence images of animals coexpressing the mon-2::gfp transgene and each marker for the Golgi, lysosomes, or endosomes, the animals were anesthetized and transferred onto 2% agarose pads before imaging using a Nikon A1si/Ni-E upright confocal microscope. Confocal microscopy images were obtained at the Advanced Neural Imaging Center in Korea Brain Research Institute, Daegu, Korea. For the quantification of the GFP::LGG-1 puncta in the hypodermal seam cells of C. elegans, the animals were immobilized using 100 mM sodium azide. To capture confocal microscopy images for representative images of LGG-1 puncta, LSM 880 (Carl Zeiss, Germany) at KAIST Analysis Center for Research Advancement was used. The images were adjusted by linearly changing “Black,” “White,” and “Gamma” in ZEM blue lite (version 3.1).
Quantitative real-time PCR analysis
Quantitative polymerase chain reaction (PCR) using cDNA was performed using a 7300 Real-Time PCR System with SYBR Green (Applied Biosystems) and analyzed with the comparative Ct method, as described in the manufacturer’s manual. Synchronized L4 or young adult animals grown at 20°C were used for RNA extraction, cDNA synthesis, and quantitative real-time PCR (qRT-PCR) analyses. RNAiso (Takara) was used to extract total RNA, and cDNA was synthesized using ImProm-II Reverse Transcriptase (Promega). The average mRNA levels of the ama-1 gene were used for normalization. The average of at least two technical repeats was applied for each biological data point.
RNAi vector cloning
To generate F08A8.2, F57H12.6, hmt-1, cdr-2, ugt-62, alh-4, alh-5, and patr-1 RNAi clones, the coding regions or the cDNAs of these genes were PCR-amplified using the primers described below. L4440 vector was digested with Hind III and Kpn I, and amplified PCR products were inserted into the L4440 vector using In-Fusion cloning (Clontech), following the manufacturer’s instructions.
RNAi vector cloning primers
The primers used for RNAi vector cloning were as follows: (i) F-F08A8.2, GGAATTCGATATCAAGCTATGGCGAACCGATCAATTCGT; (ii) R-F08A8.2, CTATAGGGCGAATTGGGTTAAAGCTTCGCCTTTTCC; (iii) F-F57H12.6, GGAATTCGATATCAAGCTCAGCCATGAAGATTCTTTTCGC; (iv) R-F57H12.6, CTATAGGGCGAATTGGTTAGTGCTTTCCTCCACGATGT; (v) F-hmt-1, GGAATTCGATATCAAGCTCCGATGGGCTTTTCACCA; (vi) R-hmt-1, CTATAGGGCGAATTGGCTACGGAAGCTCCTCGC; (vii) F-cdr-2, GGAATTCGATATCAAGCTCATGGTGTGCTCGTGCACA; (viii) R-cdr-2, CTATAGGGCGAATTGGGGAGTTGCCTCCTCATTTG; (ix) F-ugt-62, GGAATTCGATATCAAGCTCAGAAATGAAATTGATTGCTC; (x) R-ugt-62, CTATAGGGCGAATTGGTACCTCTGTATGCATAGCAA; (xi) F-alh-4, GGAATTCGATATCAAGCTATGGCATTCACCGAACTAG; (xii) R-alh-4, CTATAGGGCGAATTGGGATCAATGAAGATTGCGAG; (xiii) F-alh-5, GGAATTCGATATCAAGCTATGTCACTGCATCGAATCG; (xiv) R-alh-5, CTATAGGGCGAATTGGTTAAGCAAAAACTCTTTGAAGGAC; (xv) F-patr-1, GGAATTCGATATCAAGCTCTGCCGGGTATGGAAGAAG; and (xvi) R-patr-1, CTATAGGGCGAATTGGGTCATCTCTCTGGTTAGGATG.
Generation of transgenic animals
The mon-2p::mon-2::gfp expression vector (25 ng/μl) was co-injected with an injection marker, odr-1p::rfp (75 ng/μl), into the gonad of day 1 adult WT animals.
qRT-PCR primers
The qRT-PCR primers used in this study were as follows: (i) ama-1 F, TGGAACTCTGGAGTCACACC; (ii) ama-1 R, CATCCTCCTTCATTGAACGG; (iii) ftn-1 F: GTCAATAAACAGATTAACGTAGAAC; (iv) ftn-1 R, CGAAGTGCGATATCATCACGATCG; (v) mon-2 F, TCTCTATAAAGTTTGCTCACTTTC; and (vi) mon-2 R, ATGATTGCACACCTTGATAAAATG.
Western blot assay using C. elegans
For measuring MON-2 protein levels using mon-2::degron::EmGFP knock-in strains, synchronized animals grown on E. coli OP50 were harvested at L4 or prefertile young adult stage, washed three times with M9 buffer, and flash-frozen. The animals were thawed, mixed with 4× Bolt lithium dodecyl sulfate (LDS) sample buffer supplemented with fresh DTT, boiled for 10 min at 90°C, vortexed for 10 min, and centrifuged for 30 min at 21,130g at 4°C, and subsequently, the supernatant was collected. Proteins were transferred to polyvinylidene fluoride (PVDF) membranes (Thermo Fisher Scientific) following electrophoresis using Bolt 8% Bis-Tris Plus gels (Thermo Fisher Scientific). Membranes were blocked for 1 hour at room temperature with tris-buffered saline [TBS; 150 mM NaCl and 50 mM tris-HCl (pH 7.6)] supplemented with 0.1% Tween 20 (TBS-T) and 5% bovine serum albumin and incubated for 1 hour at room temperature with anti–α-tubulin–horseradish peroxidase (HRP; DM1A, Ab40742, Abcam) or rabbit anti-GFP polyclonal (Ab290, Abcam) antibodies. The membranes that were treated with rabbit anti-GFP antibody were incubated with goat anti-rabbit secondary antibody (IRDye 800CW, Ab216773, Abcam) at room temperature for 1 hour. The membranes were then washed three times with TBS-T. The membranes were imaged using the ChemiDoc Imaging System (Model MP, Bio-Rad). Quantification of Western blot was performed by using ImageJ (Rasband, W. S., ImageJ, U.S. National Institutes of Health, Bethesda, Maryland, USA; http://rsbweb.nih.gov/ij/). For autophagy assays, transgenic animals expressing gfp::lgg-1 were synchronized to obtain >25,000 animals using bleach methods (62). For the preparation of animals under food-deprived conditions, L4-stage animals were first treated with 50 μM FUDR, and after 2 days, the animals were harvested and washed twice with M9 buffer. For the preparation of isp-1 mutant or cco-1 RNAi–treated animals, the animals were cultured until reaching the prefertile young adult stage, harvested, and washed twice with M9 buffer. The animals were then frozen in liquid nitrogen and stored at −80°C until use. Sample buffer [4× NuPAGE LDS sample buffer, Invitrogen; 106 mM tris-HCl, 141 mM tris base, 2% LDS, 10% glycerol, 0.51 mM EDTA, 0.22 mM SERVA Blue G250, and 0.175 mM phenol red (pH 8.5)] and NuPAGE Sample Reducing Agent (10×, Invitrogen; 500 mM DTT) were added to the animals to a final concentration, and proteins were extracted by heating samples at 70°C for 10 min, followed by centrifugation (15,800g for 30 min at 4°C). Supernatants were then transferred to new 1.5-ml tubes. Bis-tris gels (10%; GenScript) were used for separation of proteins, which were subsequently transferred to PVDF membranes. NuPAGE MOPS SDS Running Buffer (Invitrogen) with 0.2% NuPAGE Antioxidant (Invitrogen) and NuPAGE Transfer Buffer (Invitrogen) with 10 or 20% methanol (Daejung) and 0.1% NuPAGE Antioxidant (Invitrogen) were used for running gels and transferring to membranes, respectively. Five percent bovine serum albumin (Sigma-Aldrich) or skim milk (LPS solution) in TBS [24.7 mM tris base, 137 mM NaCl, and 2.7 mM potassium chloride (pH 7.3); Enzynomics] with 0.1% Tween 20 (Daejung) were used for blocking and diluting primary antibodies (anti-GFP and anti–α-tubulin, respectively) and secondary antibodies. Primary antibodies against GFP (11814460001, Roche) and α-tubulin (sc-32293, Santa Cruz Biotechnology) were incubated overnight at 4°C. Membranes were washed three times for 10 min using TBS-T before being incubated with secondary antibody [goat anti-mouse immunoglobulin G (IgG); 31430, Thermo Fisher Scientific] for 90 min at room temperature. Subsequently, the membranes were washed three times for 20 min with TBS-T. To visualize immunoblotting results, the membranes were treated with enhanced chemiluminescence (Bio-Rad). After blotting anti-GFP, the membranes were stripped using stripping buffer [Thermo Fisher Scientific or laboratory-made stripping buffer (200 μl of 10% SDS; 62.5 μl of 1 M tris-HCl, pH 6.8; and 70 μl β-mercaptoethanol with double distilled water to 10 ml) at 55°C for 15 min] and washed twice or thrice using TBS-T to detect α-tubulin. Images of Western blot assays were captured using ChemiDoc XRS+ (Bio-Rad) and Image Lab software (version 5.2.1). Quantification of band intensities was performed by using Image Lab 6.1 software (Bio-Rad).
HeLa cell culture and transfection
The HeLa cell line was obtained from American Type Culture Collection. The cells were cultured in Dulbecco’s modified Eagle’s medium (Lonza) with 10% fetal bovine serum (WELGENE) at 37°C with 5% CO2. For knockdown experiments, small interfering RNA (siRNA) was transfected with Lipofectamine 2000 (Invitrogen). The sequence of the siRNA targeting human MON2 was 5′-GGCAGUGGGUCAACCUUUA-3′. Plasmids were transfected with polyethylenimine 25000 (Polysciences) following the manufacturer’s protocol.
Plasmid construction
A plasmid encoding mRFP-GFP-LC3B was obtained from Addgene (ptf-LC3, no. 21074). The plasmid was digested with Bgl II and Eco RI and ligated with human GABARAPL2 cDNA to generate a construct expressing mRFP-GFP-GABARAPL2.
Fluorescence imaging with immunostaining using cultured mammalian cells
HeLa cells were fixed using 4% paraformaldehyde or ice-cold methanol for 10 min. The cells were then stained using antibodies against giantin (ab80864, Abcam), RAB11 (ab128913, Abcam), EEA1 (ab2900, Abcam), and LC3B (no. 2775, Cell signaling Technology) overnight at 4°C. Subsequently, cells were incubated with donkey anti-mouse IgG–Alexa Fluor 568 (A10037, Invitrogen) or donkey anti-rabbit IgG–Alexa Fluor 568 (A10042, Invitrogen). To determine autophagic flux, the colored puncta in cells expressing mRFP-GFP-GABARAPL2 and mRFP-GFP-LC3B were counted with the ImageJ macro “Red and Green Puncta Colocalization Macro.”
Immunoblot and immunoprecipitation using cultured mammalian cells
For immunoblotting, HeLa cells were lysed using radioimmunoprecipitation assay lysis buffer [150 mM NaCl, 1% Triton X-100, 25 mM tris-HCl (pH 7.5), 0.5% deoxycholic acid, 0.1% SDS, 1 mM DTT, pepstatin (2 μg/ml), phenylmethylsulfonyl fluoride (0.1 mg/ml), aprotinin (5 μg/ml), leupeptin (5 μg/ml), and 1 mM benzamidine]. After centrifugation at 15,800g for 30 min, the supernatant was subjected to the SDS-PAGE for Western blotting. The primary antibodies used were p62 (610832, BD Biosciences), MON2 (ab206685, Abcam), vinculin (SC-55465, Santa Cruz Biotechnology), β-actin (bs-0061R, Bioss), and the Autophagy Atg8 Family Antibody Sampler Kit (no. 64459, Cell Signaling Technology) for LC3A, LC3C, and GABARAP/L1/L2. The secondary antibodies used were HRP-mouse IgG (31430, Thermo Fisher Scientific) and HRP-rabbit IgG (31460, Thermo Fisher Scientific). For immunoprecipitation, HeLa cells were cotransfected with GABARAP-L2-myc and GFP-MON2 for 2 days using polyethylenimine 25000. For starvation, cells were incubated in Dulbecco’s phosphate-buffered saline with 10% fetal bovine serum for 2 hours. After cells were lysed using lysis buffer [50 mM tris-HCl (pH 7.4), 150 mM NaCl, and 1% NP-40], GFP-MON2 was immunoprecipitated using anti-GFP-antibody (sc-9996, Santa Cruz Biotechnology) on Protein G PLUS-agarose beads (sc-2002; Santa Cruz Biotechnology). The interaction of GFP-MON2 with GABARAPL2 was determined by Western blotting using anti-myc antibody (9E10).
Statistical analysis
OASIS and OASIS 2 (online application for the survival analysis; http://sbi.postech.ac.kr/oasis and http://sbi.postech.ac.kr/oasis2) were used for statistical analysis of life-span assays (70, 71). P values were calculated using the log-rank (Mantel-Cox) test. To examine colocalization, Mander’s coefficient was calculated using ImageJ with the JACop plugin. Other t tests were calculated by using two-tailed Student’s t test.
Acknowledgments
We thank Caenorhabditis Genetics Center and National BioResource Project for providing C. elegans strains and all Lee laboratory members for discussion.
Funding: This study is supported as described below. National Research Foundation of Korea (NRF) grants NRF-2019R1A3B2067745 from the Korean Government (Ministry of Science and Information and Communications Technology) (S.-J.V.L.), NRF-2017R1A5A1015366 (J.-Y.Y., S.K.P., S.-Y.P., and S.-J.V.L.), Korea Institute of Science and Technology (KIST) intramural grant (C.L.), KBRI research program through Korea Brain Research Institute funded by the Ministry of Science, ICT & Future Planning (No. 21-BR-03-02) (C.M.H.), and NIH grant AG028664 (M.H.).
Author contributions: Conceptualization: Y.J., M.A., N.K., J.Y., A.B.H., C.L., S.-Y.P., and S.-J.V.L. Formal analysis: Y.J., M.A., N.K., J.Y., and A.B.H. Funding acquisition: J.-Y.Y., S.K.P., M.H., C.L., S.-Y.P., and S.-J.V.L. Investigation: Y.J., M.A., N.K., J.Y., A.B.H., D.-E.J., Ö.A., K.S., M.S., D.L., W.H., Y.L., J.S., E.J.E.K., S.J., S.K.H., L.A., Y.R., D.J.M., and S.-J.V.L. Project administration: C.L., S.-Y.P., and S.-J.V.L. Supervision: C.K., J.-Y.Y., S.K.P., C.M.H., M.H., S.K., C.L., S.-Y.P., and S.-J.V.L. Validation: Y.J., M.A., N.K., J.Y., A.B.H., D.-E.J., Ö.A., K.S., M.S., D.L., W.H., Y.L., E.J.E.K., S.J., C.L., S.-Y.P., and S.-J.V.L. Visualization: Y.J., M.A., N.K., A.B.H., S.J., and L.A. Writing (original draft): Y.J., M.A., N.K., J.Y., A.B.H., and S.-J.V.L.; Writing (review and editing): Y.J., M.A., N.K., J.Y., D.L., C.K., M.H., C.L., S.-Y.P., and S.-J.V.L.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. The MS data are available in the PRIDE Archive (www.ebi.ac.uk/pride/archive/) with the dataset identifier PXD026411.
Supplementary Materials
This PDF file includes:
Figs. S1 to S4
Tables S1 to S6
References
REFERENCES AND NOTES
- 1.Glick B. S., Nakano A., Membrane traffic within the Golgi apparatus. Annu. Rev. Cell Dev. Biol. 25, 113–132 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nakano A., Luini A., Passage through the Golgi. Curr. Opin. Cell Biol. 22, 471–478 (2010). [DOI] [PubMed] [Google Scholar]
- 3.Murén E., Oyen M., Barmark G., Ronne H., Identification of yeast deletion strains that are hypersensitive to brefeldin A or monensin, two drugs that affect intracellular transport. Yeast 18, 163–172 (2001). [DOI] [PubMed] [Google Scholar]
- 4.Efe J. A., Plattner F., Hulo N., Kressler D., Emr S. D., Deloche O., Yeast Mon2p is a highly conserved protein that functions in the cytoplasm-to-vacuole transport pathway and is required for Golgi homeostasis. J. Cell Sci. 118, 4751–4764 (2005). [DOI] [PubMed] [Google Scholar]
- 5.Gillingham A. K., Whyte J. R. C., Panic B., Munro S., Mon2, a relative of large Arf exchange factors, recruits Dop1 to the Golgi apparatus. J. Biol. Chem. 281, 2273–2280 (2006). [DOI] [PubMed] [Google Scholar]
- 6.Kanamori T., Inoue T., Sakamoto T., Gengyo-Ando K., Tsujimoto M., Mitani S., Sawa H., Aoki J., Arai H., β-Catenin asymmetry is regulated by PLA1 and retrograde traffic in C. elegans stem cell divisions. EMBO J. 27, 1647–1657 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McGough I. J., de Groot R. E. A., Jellett A. P., Betist M. C., Varandas K. C., Danson C. M., Heesom K. J., Korswagen H. C., Cullen P. J., SNX3-retromer requires an evolutionary conserved MON2:DOPEY2:ATP9A complex to mediate Wntless sorting and Wnt secretion. Nat. Commun. 9, 3737 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhu M., Wu G., Li Y. X., Stevens J. K., Fan C. X., Spang A., Dong M. Q., Serum- and glucocorticoid-inducible kinase-1 (SGK-1) plays a role in membrane trafficking in Caenorhabditis elegans. PLOS ONE 10, e0130778 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Beer K. B., Rivas-Castillo J., Kuhn K., Fazeli G., Karmann B., Nance J. F., Stigloher C., Wehman A. M., Extracellular vesicle budding is inhibited by redundant regulators of TAT-5 flippase localization and phospholipid asymmetry. Proc. Natl. Acad. Sci. U.S.A. 115, E1127–e1136 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hurley J. H., Young L. N., Mechanisms of autophagy initiation. Annu. Rev. Biochem. 86, 225–244 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.A. Meléndez, B. Levine, Autophagy in C. elegans, in WormBook: The Online Review of C. elegans Biology (2009). [DOI] [PubMed]
- 12.Alberti A., Michelet X., Djeddi A., Legouis R., The autophagosomal protein LGG-2 acts synergistically with LGG-1 in dauer formation and longevity in C. elegans. Autophagy 6, 622–633 (2010). [DOI] [PubMed] [Google Scholar]
- 13.Manil-Ségalen M., Lefebvre C., Jenzer C., Trichet M., Boulogne C., Satiat-Jeunemaitre B., Legouis R., The C. elegans LC3 acts downstream of GABARAP to degrade autophagosomes by interacting with the HOPS subunit VPS39. Dev. Cell 28, 43–55 (2014). [DOI] [PubMed] [Google Scholar]
- 14.Jenzer C., Manil-Ségalen M., Lefebvre C., Largeau C., Glatigny A., Legouis R., Human GABARAP can restore autophagosome biogenesis in a C. elegans lgg-1 mutant. Autophagy 10, 1868–1872 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu F., Watanabe Y., Guo X.-Y., Qi X., Wang P., Zhao H.-Y., Wang Z., Fujioka Y., Zhang H., Ren J.-Q., Fang T.-C., Shen Y.-X., Feng W., Hu J.-J., Noda N. N., Zhang H., Structural basis of the differential function of the two C. elegans Atg8 homologs, LGG-1 and LGG-2, in autophagy. Mol. Cell 60, 914–929 (2015). [DOI] [PubMed] [Google Scholar]
- 16.Lapierre L. R., De Magalhaes Filho C. D., McQuary P. R., Chu C. C., Visvikis O., Chang J. T., Gelino S., Ong B., Davis A. E., Irazoqui J. E., Dillin A., Hansen M., The TFEB orthologue HLH-30 regulates autophagy and modulates longevity in Caenorhabditis elegans. Nat. Commun. 4, 2267 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hansen M., Rubinsztein D. C., Walker D. W., Autophagy as a promoter of longevity: Insights from model organisms. Nat. Rev. Mol. Cell Biol. 19, 579–593 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hwang A. B., Jeong D.-E., Lee S.-J., Mitochondria and organismal longevity. Curr. Genomics 13, 519–532 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yee C., Yang W., Hekimi S., The intrinsic apoptosis pathway mediates the pro-longevity response to mitochondrial ROS in C. elegans. Cell 157, 897–909 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cristina D., Cary M., Lunceford A., Clarke C., Kenyon C., A regulated response to impaired respiration slows behavioral rates and increases lifespan in Caenorhabditis elegans. PLOS Genet. 5, e1000450 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Senchuk M. M., Dues D. J., Schaar C. E., Johnson B. K., Madaj Z. B., Bowman M. J., Winn M. E., Van Raamsdonk J. M., Activation of DAF-16/FOXO by reactive oxygen species contributes to longevity in long-lived mitochondrial mutants in Caenorhabditis elegans. PLOS Genet. 14, e1007268 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dues D. J., Schaar C. E., Johnson B. K., Bowman M. J., Winn M. E., Senchuk M. M., Van Raamsdonk J. M., Uncoupling of oxidative stress resistance and lifespan in long-lived isp-1 mitochondrial mutants in Caenorhabditis elegans. Free Radic. Biol. Med. 108, 362–373 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Park S., Artan M., Han S. H., Park H.-H., Jung Y., Hwang A. B., Shin W. S., Kim K.-T., Lee S.-V., VRK-1 extends life span by activation of AMPK via phosphorylation. Sci. Adv. 6, eaaw7824 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kang U. B., Yeom J., Kim H., Lee C., Quantitative analysis of mTRAQ-labeled proteome using full MS scans. J. Proteome Res. 9, 3750–3758 (2010). [DOI] [PubMed] [Google Scholar]
- 25.Walther D. M., Kasturi P., Zheng M., Pinkert S., Vecchi G., Ciryam P., Morimoto R. I., Dobson C. M., Vendruscolo M., Mann M., Hartl F. U., Widespread proteome remodeling and aggregation in aging C. elegans. Cell 161, 919–932 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jochum A., Jackson D., Schwarz H., Pipkorn R., Singer-Krüger B., Yeast Ysl2p, homologous to Sec7 domain guanine nucleotide exchange factors, functions in endocytosis and maintenance of vacuole integrity and interacts with the Arf-Like small GTPase Arl1p. Mol. Cell. Biol. 22, 4914–4928 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kapahi P., Kaeberlein M., Hansen M., Dietary restriction and lifespan: Lessons from invertebrate models. Ageing Res. Rev. 39, 3–14 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mahajan D., Boh B. K., Zhou Y., Chen L., Cornvik T. C., Hong W., Lu L., Mammalian Mon2/Ysl2 regulates endosome-to-Golgi trafficking but possesses no guanine nucleotide exchange activity toward Arl1 GTPase. Sci. Rep. 3, 3362 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhao S. B., Dean N., Gao X. D., Fujita M., MON2 guides Wntless transport to the Golgi through recycling endosomes. Cell Struct. Funct. 45, 77–92 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen C. C.-H., Schweinsberg P. J., Vashist S., Mareiniss D. P., Lambie E. J., Grant B. D., RAB-10 is required for endocytic recycling in the Caenorhabditis elegans intestine. Mol. Biol. Cell 17, 1286–1297 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ventura N., Rea S. L., Schiavi A., Torgovnick A., Testi R., Johnson T. E., p53/CEP-1 increases or decreases lifespan, depending on level of mitochondrial bioenergetic stress. Aging Cell 8, 380–393 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Torgovnick A., Schiavi A., Testi R., Ventura N., A role for p53 in mitochondrial stress response control of longevity in C. elegans. Exp. Gerontol. 45, 550–557 (2010). [DOI] [PubMed] [Google Scholar]
- 33.Baruah A., Chang H., Hall M., Yuan J., Gordon S., Johnson E., Shtessel L. L., Yee C., Hekimi S., Derry W. B., Lee S. S., CEP-1, the Caenorhabditis elegans p53 homolog, mediates opposing longevity outcomes in mitochondrial electron transport chain mutants. PLOS Genet. 10, e1004097 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee S.-J., Hwang A. B., Kenyon C., Inhibition of respiration extends C. elegans life span via reactive oxygen species that increase HIF-1 activity. Curr. Biol. 20, 2131–2136 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hwang A. B., Ryu E. A., Artan M., Chang H. W., Kabir M. H., Nam H. J., Lee D., Yang J. S., Kim S., Mair W. B., Lee C., Lee S. S., Lee S. J., Feedback regulation via AMPK and HIF-1 mediates ROS-dependent longevity in Caenorhabditis elegans. Proc. Natl. Acad. Sci. U.S.A. 111, E4458–E4467 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zarse K., Schmeisser S., Groth M., Priebe S., Beuster G., Kuhlow D., Guthke R., Platzer M., Kahn C. R., Ristow M., Impaired insulin/IGF1 signaling extends life span by promoting mitochondrial L-proline catabolism to induce a transient ROS signal. Cell Metab. 15, 451–465 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Curtis R., O’Connor G., DiStefano P. S., Aging networks in Caenorhabditis elegans: AMP-activated protein kinase (aak-2) links multiple aging and metabolism pathways. Aging Cell 5, 119–126 (2006). [DOI] [PubMed] [Google Scholar]
- 38.Yang W., Hekimi S., A mitochondrial superoxide signal triggers increased longevity in Caenorhabditis elegans. PLOS Biol. 8, e1000556 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Walter L., Baruah A., Chang H.-W., Pace H. M., Lee S. S., The homeobox protein CEH-23 mediates prolonged longevity in response to impaired mitochondrial electron transport chain in C. elegans. PLOS Biol. 9, e1001084 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Springhorn A., Hoppe T., Western blot analysis of the autophagosomal membrane protein LGG-1/LC3 in Caenorhabditis elegans. Methods Enzymol. 619, 319–336 (2019). [DOI] [PubMed] [Google Scholar]
- 41.Mizushima N., Yoshimori T., Levine B., Methods in mammalian autophagy research. Cell 140, 313–326 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang Z., Gu H., Li Q., Zheng J., Cao S., Weng C., Jia H., GABARAPL2 is critical for growth restriction of Toxoplasma gondii in HeLa cells treated with gamma interferon. Infect. Immun. 88, e00054-20 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pascall J. C., Rotondo S., Mukadam A. S., Oxley D., Webster J., Walker S. A., Piron J., Carter C., Ktistakis N. T., Butcher G. W., The immune system GTPase GIMAP6 interacts with the Atg8 homologue GABARAPL2 and is recruited to autophagosomes. PLOS ONE 8, e77782 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kimura S., Noda T., Yoshimori T., Dissection of the autophagosome maturation process by a novel reporter protein, tandem fluorescent-tagged LC3. Autophagy 3, 452–460 (2007). [DOI] [PubMed] [Google Scholar]
- 45.Zhang Q., Wu X., Chen P., Liu L., Xin N., Tian Y., Dillin A., The mitochondrial unfolded protein response is mediated cell-non-autonomously by retromer-dependent Wnt signaling. Cell 174, 870–883.e17 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jeong D.-E., Lee D., Hwang S.-Y., Lee Y., Lee J.-E., Seo M., Hwang W., Seo K., Hwang A. B., Artan M., Son H. G., Jo J.-H., Baek H., Oh Y. M., Ryu Y., Kim H.-J., Ha C. M., Yoo J.-Y., Lee S.-J. V., Mitochondrial chaperone HSP-60 regulates anti-bacterial immunity via p38 MAP kinase signaling. EMBO J. 36, 1046–1065 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kalderon B., Kogan G., Bubis E., Pines O., Cytosolic Hsp60 can modulate proteasome activity in yeast. J. Biol. Chem. 290, 3542–3551 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nakamura S., Karalay Ö., Jäger P. S., Horikawa M., Klein C., Nakamura K., Latza C., Templer S. E., Dieterich C., Antebi A., Mondo complexes regulate TFEB via TOR inhibition to promote longevity in response to gonadal signals. Nat. Commun. 7, 10944 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Noda T., Autophagy in the context of the cellular membrane-trafficking system: The enigma of Atg9 vesicles. Biochem. Soc. Trans. 45, 1323–1331 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Young A. R. J., Chan E. Y. W., Hu X. W., Köchl R., Crawshaw S. G., High S., Hailey D. W., Lippincott-Schwartz J., Tooze S. A., Starvation and ULK1-dependent cycling of mammalian Atg9 between the TGN and endosomes. J. Cell Sci. 119, 3888–3900 (2006). [DOI] [PubMed] [Google Scholar]
- 51.Yen W. L., Legakis J. E., Nair U., Klionsky D. J., Atg27 is required for autophagy-dependent cycling of Atg9. Mol. Biol. Cell 18, 581–593 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sagiv Y., Legesse-Miller A., Porat A., Elazar Z., GATE-16, a membrane transport modulator, interacts with NSF and the Golgi v-SNARE GOS-28. EMBO J. 19, 1494–1504 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang J., Zhang X., Liu G., Chang D., Liang X., Zhu X., Tao W., Mei L., Intracellular trafficking network of protein nanocapsules: Endocytosis, exocytosis and autophagy. Theranostics 6, 2099–2113 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Puri C., Vicinanza M., Ashkenazi A., Gratian M. J., Zhang Q., Bento C. F., Renna M., Menzies F. M., Rubinsztein D. C., The RAB11A-positive compartment is a primary platform for autophagosome assembly mediated by WIPI2 recognition of PI3P-RAB11A. Dev. Cell 45, 114–131.e8 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tooze S. A., Yoshimori T., The origin of the autophagosomal membrane. Nat. Cell Biol. 12, 831–835 (2010). [DOI] [PubMed] [Google Scholar]
- 56.Rubinsztein D. C., Shpilka T., Elazar Z., Mechanisms of autophagosome biogenesis. Curr. Biol. 22, R29–R34 (2012). [DOI] [PubMed] [Google Scholar]
- 57.Hamasaki M., Furuta N., Matsuda A., Nezu A., Yamamoto A., Fujita N., Oomori H., Noda T., Haraguchi T., Hiraoka Y., Amano A., Yoshimori T., Autophagosomes form at ER-mitochondria contact sites. Nature 495, 389–393 (2013). [DOI] [PubMed] [Google Scholar]
- 58.Ge L., Zhang M., Schekman R., Phosphatidylinositol 3-kinase and COPII generate LC3 lipidation vesicles from the ER-Golgi intermediate compartment. eLife 3, e04135 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ge L., Melville D., Zhang M., Schekman R., The ER-Golgi intermediate compartment is a key membrane source for the LC3 lipidation step of autophagosome biogenesis. eLife 2, e00947 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ruck A., Attonito J., Garces K. T., Núnez L., Palmisano N. J., Rubel Z., Bai Z., Nguyen K. C. Q., Sun L., Grant B. D., Hall D. H., Meléndez A., The Atg6/Vps30/Beclin 1 ortholog BEC-1 mediates endocytic retrograde transport in addition to autophagy in C. elegans. Autophagy 7, 386–400 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bento C. F., Puri C., Moreau K., Rubinsztein D. C., The role of membrane-trafficking small GTPases in the regulation of autophagy. J. Cell Sci. 126, 1059–1069 (2013). [DOI] [PubMed] [Google Scholar]
- 62.T. Stiernagle, Maintenance of C. elegans, in WormBook: The Online Review of C. elegans Biology (2006). [DOI] [PMC free article] [PubMed]
- 63.Fredens J., Engholm-Keller K., Giessing A., Pultz D., Larsen M. R., Højrup P., Møller-Jensen J., Færgeman N. J., Quantitative proteomics by amino acid labeling in C. elegans. Nat. Methods 8, 845–847 (2011). [DOI] [PubMed] [Google Scholar]
- 64.Yeom J., Kabir M. H., Lee C., Impact of data-dependent exclusion list based mass spectrometry on label-free proteomic quantification. Rapid Commun. Mass Spectrom. 29, 128–134 (2015). [DOI] [PubMed] [Google Scholar]
- 65.Park K., Yoon J. Y., Lee S., Paek E., Park H., Jung H. J., Lee S. W., Isotopic peak intensity ratio based algorithm for determination of isotopic clusters and monoisotopic masses of polypeptides from high-resolution mass spectrometric data. Anal. Chem. 80, 7294–7303 (2008). [DOI] [PubMed] [Google Scholar]
- 66.Yoon J. Y., Lim K. Y., Lee S., Park K., Paek E., Kang U. B., Yeom J., Lee C., Improved quantitative analysis of mass spectrometry using quadratic equations. J. Proteome Res. 9, 2775–2785 (2010). [DOI] [PubMed] [Google Scholar]
- 67.Yoon J. Y., Yeom J., Lee H., Kim K., Na S., Park K., Paek E., Lee C., High-throughput peptide quantification using mTRAQ reagent triplex. BMC Bioinformatics 12 Suppl 1, S46 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Feng J., Bussière F., Hekimi S., Mitochondrial electron transport is a key determinant of life span in Caenorhabditis elegans. Dev. Cell 1, 633–644 (2001). [DOI] [PubMed] [Google Scholar]
- 69.Larkin M. A., Blackshields G., Brown N. P., Chenna R., McGettigan P. A., McWilliam H., Valentin F., Wallace I. M., Wilm A., Lopez R., Thompson J. D., Gibson T. J., Higgins D. G., Clustal W and clustal X version 2.0. Bioinformatics 23, 2947–2948 (2007). [DOI] [PubMed] [Google Scholar]
- 70.Yang J. S., Nam H. J., Seo M., Han S. K., Choi Y., Nam H. G., Lee S. J., Kim S., OASIS: Online application for the survival analysis of lifespan assays performed in aging research. PLOS ONE 6, e23525 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Han S. K., Lee D., Lee H., Kim D., Son H. G., Yang J. S., Lee S.-J. V., Kim S., OASIS 2: Online application for survival analysis 2 with features for the analysis of maximal lifespan and healthspan in aging research. Oncotarget 7, 56147–56152 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Huang D. W., Sherman B. T., Lempicki R. A., Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57 (2009). [DOI] [PubMed] [Google Scholar]
- 73.Holdorf A. D., Higgins D. P., Hart A. C., Boag P. R., Pazour G. J., Walhout A. J. M., Walker A. K., WormCat: An online tool for annotation and visualization of Caenorhabditis elegans genome-scale data. Genetics 214, 279–294 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Munkácsy E., Khan M. H., Lane R. K., Borror M. B., Park J. H., Bokov A. F., Fisher A. L., Link C. D., Rea S. L., DLK-1, SEK-3 and PMK-3 are required for the life extension induced by mitochondrial bioenergetic disruption in C. elegans. PLOS Genet. 12, e1006133 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Suthammarak W., Morgan P. G., Sedensky M. M., Mutations in mitochondrial complex III uniquely affect complex I in Caenorhabditis elegans. J. Biol. Chem. 285, 40724–40731 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Son H. G., Seo K., Seo M., Park S., Ham S., An S. W. A., Choi E.-S., Lee Y., Baek H., Kim E., Ryu Y., Ha C. M., Hsu A.-L., Roh T.-Y., Jang S. K., Lee S.-J. V., Prefoldin 6 mediates longevity response from heat shock factor 1 to FOXO in C. elegans. Genes Dev. 32, 1562–1575 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Schaaf M. B. E., Keulers T. G., Vooijs M. A., Rouschop K. M. A., LC3/GABARAP family proteins: Autophagy-(un)related functions. FASEB J. 30, 3961–3978 (2016). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S4
Tables S1 to S6
References