Abstract
Rationale
Microbiome studies of the lower airway based on bacterial 16S rRNA gene sequencing assess microbial community structure but can only infer functional characteristics. Microbial products, such as short chain fatty acids (SCFAs), in the lower airways have significant impact on the host’s immune tone. Thus, functional approaches to the analyses of the microbiome are necessary.
Methods
Here we used upper and lower airway samples from a research bronchoscopy smoker cohort. In addition, we validated our results in an experimental mouse model.
Measurements
We extended our microbiota characterization beyond 16S rRNA gene sequencing with the use of whole genome (WGS) and RNA metatranscriptome sequencing. Short chain fatty acids (SCFA) were also measured in lower airway samples and correlated with each of the sequencing datasets. In the mouse model, 16S rRNA gene and RNA metatranscriptome sequencing were performed.
Main Results
Functional evaluations of the lower airway microbiota using inferred metagenome, WGS and metatranscriptome were dissimilar. Comparison with measured levels of SCFAs shows that the inferred metagenome from the 16S rRNA gene sequencing data was poorly correlated, while better correlations were noted when SCFAs levels were compared with WGS and metatranscriptome. Modeling lower airway aspiration with oral commensals in a mouse model showed that the metatranscriptome most efficiently captures transient active microbial metabolism, which was overestimated by 16S rRNA gene sequencing.
Conclusions
Functional characterization of the lower airway microbiota through metatranscriptome identify metabolically active organisms capable of producing metabolites with immunomodulatory capacity such as SCFAs.
Introduction
Characterization of the lower airway microbiota by 16S rRNA gene sequencing has revealed that the lower airways are frequently enriched with oral commensals in healthy subjects 1–7, most likely due to micro-aspiration. The presence of oral commensals in the lower airways has also been identified in multiple pulmonary diseases such as cystic fibrosis, bronchiectasis, chronic obstructive pulmonary disease and lung cancer 7–11. However, the viability of organisms identified in lower airway samples using targeted gene sequencing is uncertain and most investigations have been limited to just taxonomic description of the lower airway microbiota and its association with host phenotypes 1,2,7,12–14. Whole genome shotgun (WGS) and RNA metatranscriptome sequencing can directly capture gene content and active transcription, respectively. These techniques have the potential to provide a more precise functional assessment of the lower airway microbiome15–17. Due to the limited microbial biomass in the lower airways, these methods are challenging and have not yet been fully evaluated in comparison to standard microbial profiling and inferred functional content based on 16S rRNA gene sequencing.
Functionally active microbes can produce microbial products of relevance to the host and may modify host functions 13,14,18. For example, short chain fatty acids (SCFAs) cannot be produced by mammalian cells (with the exception of acetate) but are produced by facultative and obligate anaerobes in hypoxic conditions 19–24. SCFAs produced by the gut microbiota induce regulatory T cells that modify asthma, inflammatory bowel disease and cancer 19–24. These SCFAs have also been identified in the lower airways and, with 16S rRNA gene sequencing, our group has shown that its presence is associated with enrichment of the lower airway microbiota with oral commensals 13. To better characterize functional aspects of the lower airway microbiome, we explored the use of WGS and RNA metatranscriptome approaches to uncover active microbial metabolism of immunologically relevant metabolites, such as SCFAs.
Methods
Participants and samples
For this study samples were used from 21 participants, that were recruited for research bronchoscopy as part of our ongoing Chronic Obstructive Pulmonary Disease and Smoker Control cohort. All participants signed informed consent and the protocol was approved by the New York University and Bellevue Hospital Center (New York, NY) institutional review boards (IRB# S14–01546). Further details are in the Supplementary Methods.
Sample Processing
DNA was extracted from all samples using Qiagen DNA Mini Kit spin column protocol (Qiagen). RNA extraction was carried out with the miRNeasy Micro Kit (Qiagen). Bacterial burden was measured by Droplet Digital PCR. All samples had high-throughput sequencing of bacterial 16S rRNA gene amplicons, WGS and RNA metatranscriptome sequencing. Sequence data was filtered for bacteria only. Additionally, to identify active bacterial metabolism, SCFAs were measured by mass spectrometry. Further details on sample processing can be found in the Supplementary Methods.
Mouse Experiment
Three mice were inoculated with PBS while the remaining 17 mice were inoculated with a mixture of human oral commensals (MOC) consisting of Prevotella, Streptococcus and Veillonella. BAL samples were sent for 16S rRNA gene sequencing and RNA metatranscriptome sequencing. The NYU Institutional Animal Care & Use approved the animal studies (IACUC# s16–00032). Further details are in the Supplementary Methods.
Statistical Analysis
For association with discrete factors, we used non-parametric tests (Mann-Whitney or Kruskal-Wallis ANOVA). We used the vegan package in R to construct Principal Coordinate Analysis (PCoA) based on Bray-Curtis distances 25,26. To cluster microbiome communities into exclusive ‘metacommunities’ we used a Dirichlet Multinomial Mixture (DMM) Model27,28. To evaluate differences between groups within each sequence data type, we evaluated differential expression with DESeq2 29 with a false discover rate (FDR) <0.05 30. All data is publicly available in Sequence Read Archive (SRA) under accession numbers PRJNA603592, PRJNA573853 and PRJNA603675. All codes utilized for the analysis included in this manuscript are available at: https://github.com/segalmicrobiomelab/functional_microbiomics
Further details on statistical analysis are in the Supplementary Methods.
Results
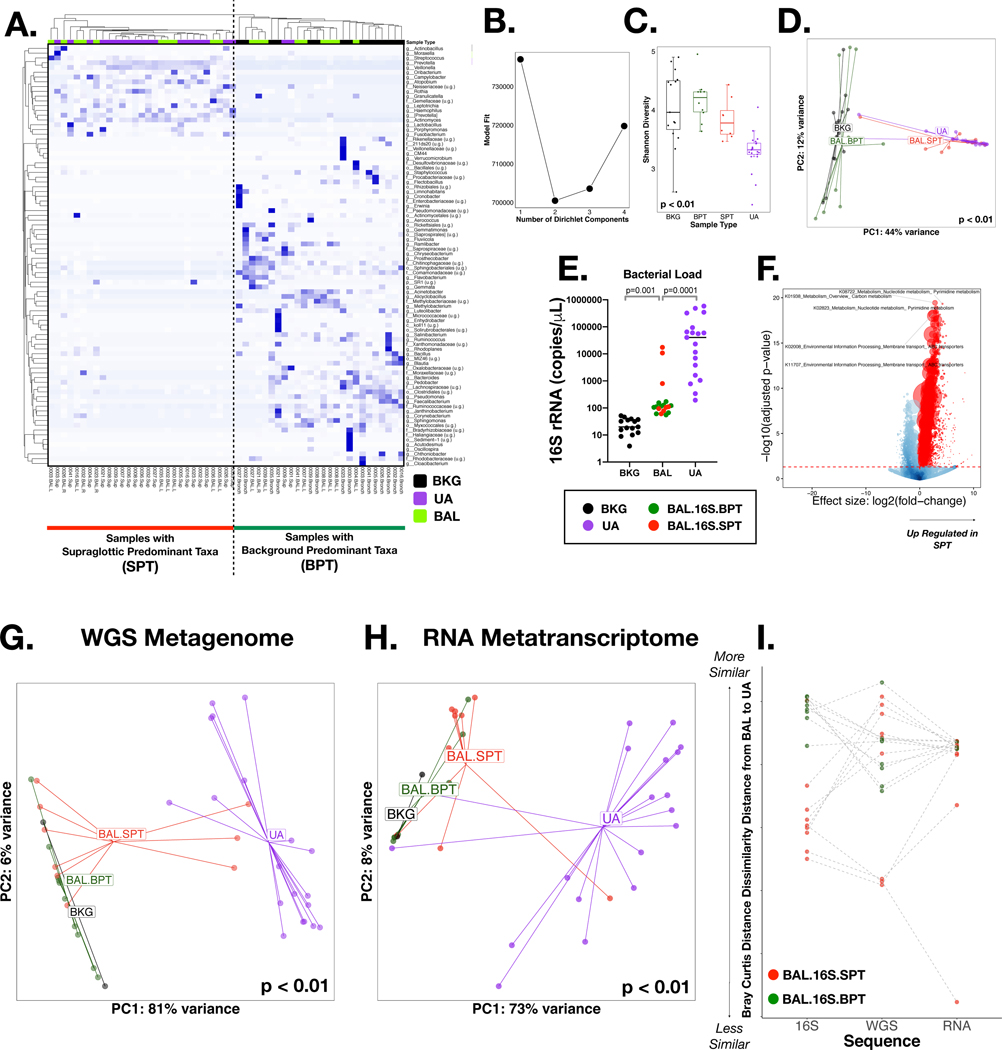
We recruited 21 smokers for this study; lower airway samples from two subjects did not yield an adequate cDNA library for metatranscriptome and were excluded from the analysis, leaving a study cohort of 19 subjects (Table 1). 16S rRNA gene sequencing characterized the microbiota present in background (BKG) controls, upper (UA) and lower [bronchoalveolar lavage (BAL)] airway samples. Hierarchical clustering of the most abundant taxa shows that the microbiota in UA and BKG samples are differentially contained within the two dominant clusters (Figure 1A). Some BAL samples were more similar to the UA, composed of taxa commonly identified as oral commensals such as Veillonella, Prevotella and Streptococcus, whereas other samples were more similar to BKG samples dominated by taxa such as Methylobacterium, Actinobacillus and Lactobacillus. We confirmed by DMM that BAL samples clustered into 2 distinct groups (Figure 1B). Samples that clustered with UA samples were enriched with Supraglottic Predominant Taxa (BAL.16S.SPT) while samples that clustered with BKG samples were enriched with Background Predominant Taxa (BAL.16S.BPT) 1,2. Significant differences between all sample types were determined by both α (Shannon Index, Figure 1C) and β diversity (Bray-Curtis distance, Figure 1D); these microbial community metrics further supported qualifying BAL.16S.SPT samples as more similar to UA samples. The median bacterial load, as determined by droplet digital (ddPCR), was ~1,000-fold higher for UA and 10-fold higher for BAL as compared to BKG samples (Figure 1E). However, three of the BAL samples clearly had higher bacterial burden, with levels similar to those found in UA samples; they were all identified as BAL.16S.SPT based on taxonomic composition (Figure 1E). To explore functional aspects using the 16S rRNA gene sequence data, we inferred metagenomic composition by PICRUSt31. Comparison of the inferred metagenome between BAL.16S.SPT and BAL.16S.BPT samples suggested that there should be several KEGGs and associated functional pathways differentially expressed between these two clusters (Figure 1F, Supplementary Data 1).
TABLE 1:
Baseline characteristics of study population
| Cohort | |
|---|---|
|
| |
| n | 19 |
| Age | 53.0 [49.5–58.0] |
| Female (%) | 5 (26.3) |
| BMI | 28.4 [22.1–31.6] |
| PFT | |
| FVC Percent Predicted | 95 [86–104] |
| FEV1 Percent Predicted | 89 [79–102] |
| FEV1/FVC | 79 [69–83] |
| Smoking Status | |
| Former Smoker (%) | 17 (89) |
| Current Smoker (%) | 2(11) |
| Pack Years | 20 [11–28] |
Data expressed as Median[IQR] or counts(%)
Figure 1: 16S rRNA gene, Whole genome (WGS) and RNA sequencing:
Background (BKG), Upper Airway (UA) and Bronchoalveolar (BAL) samples were collected via bronchoscopy; 16S rRNA gene, Whole genome and RNA sequencing was performed. (A) A heatmap based on Bray-Curtis distance for the 16S rRNA gene sequencing, illustrates the top taxa for all samples. Hierarchical clustering showed two clear clusters, one with BKG samples and BAL samples similar to BKG (Background Predominant Taxa) and another with UA samples and BAL samples similar to UA (Supraglottic Predominant Taxa). (B) Dirichlet Multinomial Modelling (DMM) showed 2 clusters had the best model fit for the 16S rRNA gene sequencing. (C) α Diversity, measured by Shannon Index, showed significant (Wilcoxon) difference between all samples and lowest diversity in UA and among BAL samples that clustered to BAL.16S.SPT by DMM. (D) Beta Diversity, measured by Bray-Curtis, also indicates a significant (PERMANOVA) difference between all samples for 16S rRNA gene sequencing. (E) Bacterial load, measured by ddPCR showed highest levels in UA samples (Kruskal-Wallis). BAL Samples also had higher levels when compared to BKG samples. (F) The inferred metagenome was assessed using PICRUST highlighting several significantly enriched pathways (colored in red). (G) β Diversity for WGS, measured by Bray-Curtis, showed a significant (PERMANOVA) difference between all samples, with UA samples separate from BKG and BAL.BPT samples. Three BAL.SPT samples clustered with UA Samples. (H) β Diversity for RNA, measured by Bray-Curtis, showed a significant (PERMANOVA) difference between all sample types. Two BAL.SPT samples clustered with UA samples. (I) Z Transformed Bray-Curtis Distance between BAL samples and paired UA samples showed clear separation of BAL.16S.BPT and BAL.16S.SPT samples in 16S rRNA gene sequencing. This separation was not as clear in WGS and RNA.
Evaluation of the lower airway metagenome and metatranscriptome
To further characterize functional aspects of the airway microbiota we profiled the metagenome by WGS and the metatranscriptome by RNA sequencing. For this analysis, all BAL and UA samples were used while only 2 BKG samples had RNA sequencing libraries that passed quality control. Importantly, rarefaction analysis for the WGS and RNA data showed plateauing of the curves at a lower depth than the one accomplished in this investigation (Supplementary Figure 2). Within the WGS and RNA sequence data, UA and BKG samples were significantly different from each other, based on α and β-diversity (Figures 1G/H and Supplementary Figures 1A/B), similar to the 16S rRNA gene sequence data, while BAL samples were either similar to the UA or BKG. Using each Bray-Curtis distance matrix for 16S rRNA gene sequence, WGS, and RNA metatranscriptome data we compared the paired distances between BAL and UA samples. The 16S rRNA gene data indicated a clear separation of what we identified as BAL.16S.SPT and BAL.16S.BPT but this distinction was lost in WGS and RNA metatranscriptome data (Figure 1I). Importantly, β diversity analyses on both WGS and RNA sequencing data showed that all BAL samples identified as BAL.16S.BPT remained similar to BKG samples. However, among BAL samples identified as BAL.16S.SPT, both WGS and RNA sequencing identified a subset of samples that clearly showed greater similarity to UA samples while others showed greater similarity to BKG samples (Figures 1G-H). Interestingly, 2/3 BAL samples that had the greatest similarities with UA samples in WGS data also had the greatest similarities with UA samples in the RNA metatranscriptome data (Figure 1I).
Functional overlap and differences across sequencing data types
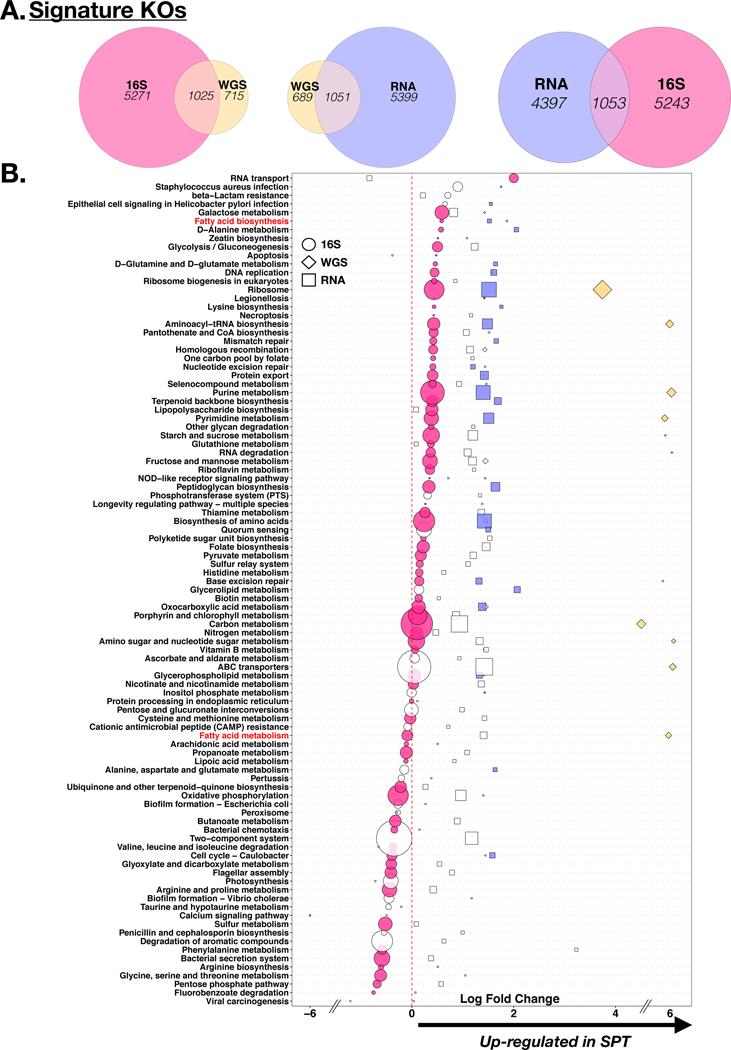
Using GSEA to compare the functional annotations across the sequencing data types, we identified significant overlap between the data obtained (>1000 overlapping KOs for each comparison, Figure 3A). In order to compare the differentially enriched pathways identified (with DESeq2) between BAL.16S.SPT and BAL.16S.BPT we overlapped the fold change of the functional pathways (summarized to Level 3 of annotation). We identified some concordance in the directionality of the fold change, most identified as enriched in BAL.16S.SPT (Figure 3B). For example, fatty acid biosynthesis, as well as purine and pyrimidine metabolism were significantly enriched in all three sequence datasets. However, the presence of statistical significance (identified in the figure by the presence of color) differed by the method used. Further, other functional pathways showed discordant directionality. For example, genes belonging to the fatty acid metabolism pathway appeared to be significantly depleted in BAL.16S.SPT samples by the inferred metagenome but significantly enriched in WGS and non-significantly enriched in RNA sequencing data (Figure 3B). Since several genes annotated to fatty acid biosynthesis and fatty acid metabolism are part of the production of SCFAs (end products of microbial metabolism associated with enrichment of the lower airway microbiota with oral anaerobes13) we measured the levels of these products directly using mass spectrometry.
Figure 3: Functional annotation of all 3 sequencing data types.
(A) Gene Set Enrichment Analysis (GSEA) comparing functional signatures identified across the different sequence data types as distinctly enriched in BAL.16S.SPT vs. BAL. 16S.BPT samples based on KEGG Orthology (KO) annotation (differential enrichment performed based on DESEQ2 analysis). (B) KOs were summarized to associated pathways and differential expression between BAL.16S.SPT and BAL.16S.BPT are displayed as circles for 16S rRNA gene sequencing, diamonds for WGS and squares for RNA. Coloring indicates statistical significance (DESeq2 p<0.05) for each sequence data type and size is relative to the amount of KOs contributing to that pathway. Two pathways highlighted in red include Fatty Acid Biosynthesis, which shows concordance of directionality between the three sequence data types and Fatty Acid Metabolism, which shows discordance.
Further differences in functional and taxonomic signatures between sequencing data types are discussed in the Supplementary Results.
SCFA levels are different in upper and lower airways
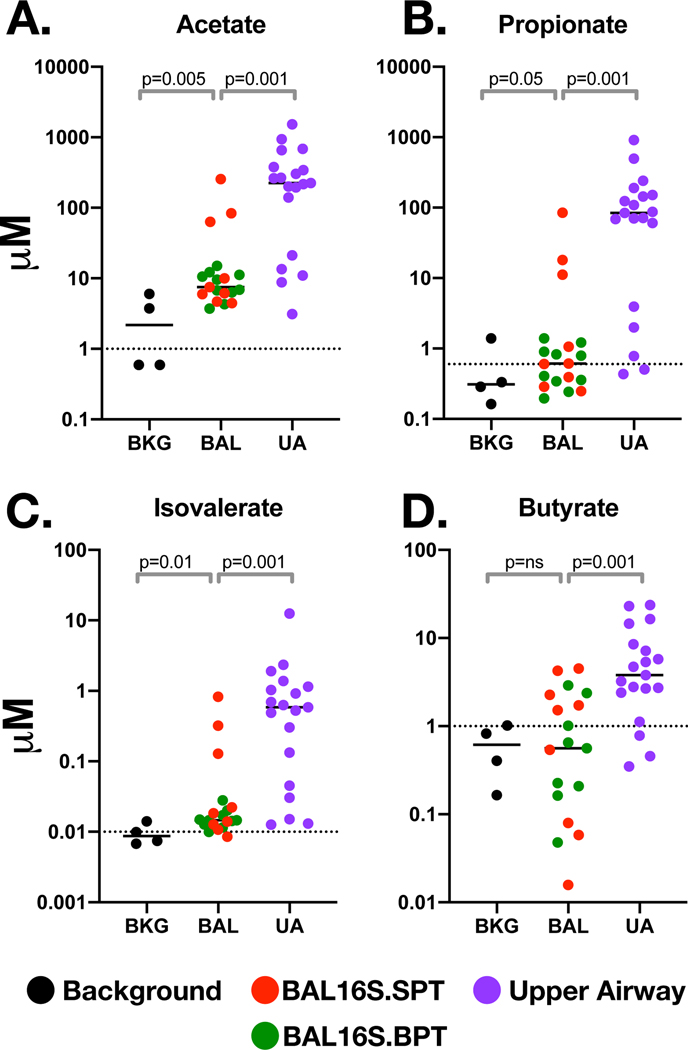
SCFA levels in ex vivo cultures is discussed in the Supplementary Results. We then evaluated SCFA levels in the 19 UA and BAL samples and 4 BKG samples. The levels of 4/7 SCFAs were significantly higher in UA samples when compared to BKG samples: acetate, propionate, isovalerate, and butyrate (Figure 4). However, the levels of 3 other SCFAs measured were similar when comparing UA with BKG samples: hexanoate, valerate and octanoate (data not shown). These data suggest that some SCFAs are produced by oral commensals or that their measurement lack a dynamic range above BKG. Among BAL samples, there were 3 SCFAs with levels statistically higher than BKG samples: acetate, propionate, and isovalerate—all of which were exponentially higher in UA samples. However, three BAL samples identified as BAL.16S.SPT had significantly higher levels of these 3 SCFAS (Figure 4). These concentrations are comparable to what we have previously published as measurable in the lower airways of a separate cohort and found to be correlated with a T-reg phenotype, blunted IL-17 and IFN gamma response.13 The remaining BAL samples had similar levels to BKG (regardless of their 16S.SPT/16S.BPT grouping). Importantly, the levels of these SCFAs in BAL samples did not correlate with levels in UA samples (p=ns for all comparisons, Supplementary Figure 7) suggesting that microaspiration of upper airway secretions containing these metabolites was not the main source of SCFAs in the lung.
Figure 4: Concentrations of Short Chain Fatty Acid (SCFA) in bronchoscopy samples:
A panel of SCFAs were measured and compared (Kruskal-Wallis) in Background (BKG), Upper Airway (UA) and Bronchoalveolar (BAL) samples by GC-MS. SCFA were derived from the linear phase of the standard curve leading to the following cutoffs values (dotted line): (A) 1μM for Acetate, (B) 0.6 μM for Propionate (C) 0.01 μM for Isovalerate and (D) 1μM for Butyrate.
The RNA metatranscriptome correlates with measured SCFAs in the lower airways
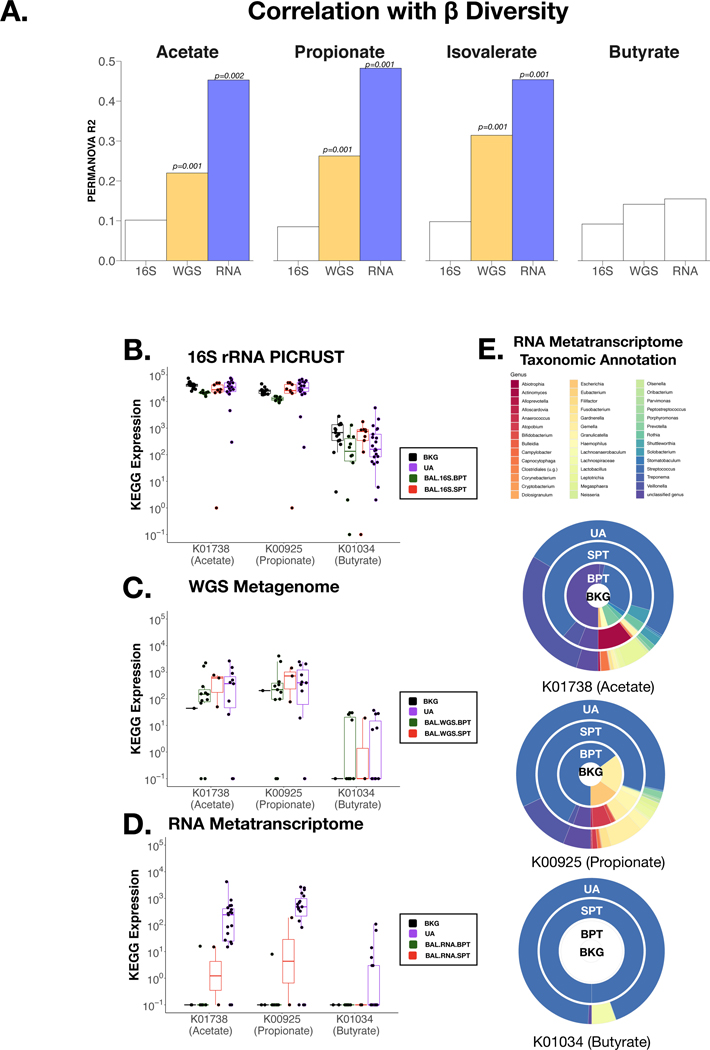
We then evaluated the associations between the 4 SCFAs found to be present at different levels across samples with the sequencing data. At a global compositional level (β-diversity), the levels of these 4 SCFAs were not statistically significantly associated with the 16S rRNA gene sequencing data but ¾ were statistically significantly associated with the WGS and RNA metatranscriptome data (Figure 5A). This is likely to be driven by the 3 BAL samples with high levels of measured SCFAs identified by mass spectrometry (Figure 4), as this correlation is not seen with BAL.BPT samples. Furthermore, rarefying the three datasets lead to no significant change in the correlations.
Figure 5: Diversity correlations with SCFA measurements:
(A) Levels of SCFAs with Acetate, Propionate, Isovalerate and Butyrate were tested (PERMANOVA) against Beta Diversity distribution of data from all three sequencing techniques in BAL samples. Relative abundance of three KOs with direct annotation to measured SCFAs were compared across sample types: K01738 (Acetate), K00925 (Propionate) and K01034 (Butyrate) with (B) 16S rRNA gene sequencing, (C) Whole Genome Sequencing and (D) RNA metatranscriptome sequencing. (E) RNA metatranscriptome taxonomic annotation for these three SCFAs-associated KOs in UA, BAL.RNA.SPT, BAL.RNA.BPT and BKG samples are represented here. Each circle represents a different sample type and colors indicate a different taxonomic annotation.
Since DMM clustering on 16S rRNA gene sequencing data did not distinguish BAL samples with high and low SCFAs levels, we performed DMM analysis of the WGS and RNA metatranscriptome data (see Supplementary Results).
As validation, we focused on 3 KOs with direct SCFA annotation: K01738 for acetate, K00925 for propionate and K01034 for butyrate. KO enrichment differences could be identified between sample types with the RNA metatranscriptome but not with the inferred metagenome (16S rRNA data) or the WGS data (Figure 5B-D). KOs in the RNA metatranscriptome were significantly elevated in UA samples and at very low levels in BKG samples. Importantly, the BAL samples identified in DMM clustering as being compositionally similar to UA had significantly higher levels of these KOs when compared with the remaining BAL samples which clustered with BKG samples (p<0.03 for all comparisons, Figure 5D). We then evaluated the taxonomic annotation available for these 3 KOs in the RNA metatranscriptome data and noted that the taxonomic source for these genes was predominantly oral commensals such as Streptococcus and Veillonella (Figure 5E). Thus, the RNA metatranscriptome has better resolution (when compared to 16S rRNA gene sequencing) for the identification of enriched genes involved in the metabolism of SCFAs present in oral anaerobes and supports the presence of viable (RNA measurable) and metabolically active (metabolites elevated above BKG) bacteria in the lower airway airways.
To ensure that these microbial patterns were related to signals present in the lower airways, we identified potential contaminants (coming from DNA/RNA present in bronchoscope or added through sample processing) within each sequencing method using the decontam package.32 (Supplementary Results)
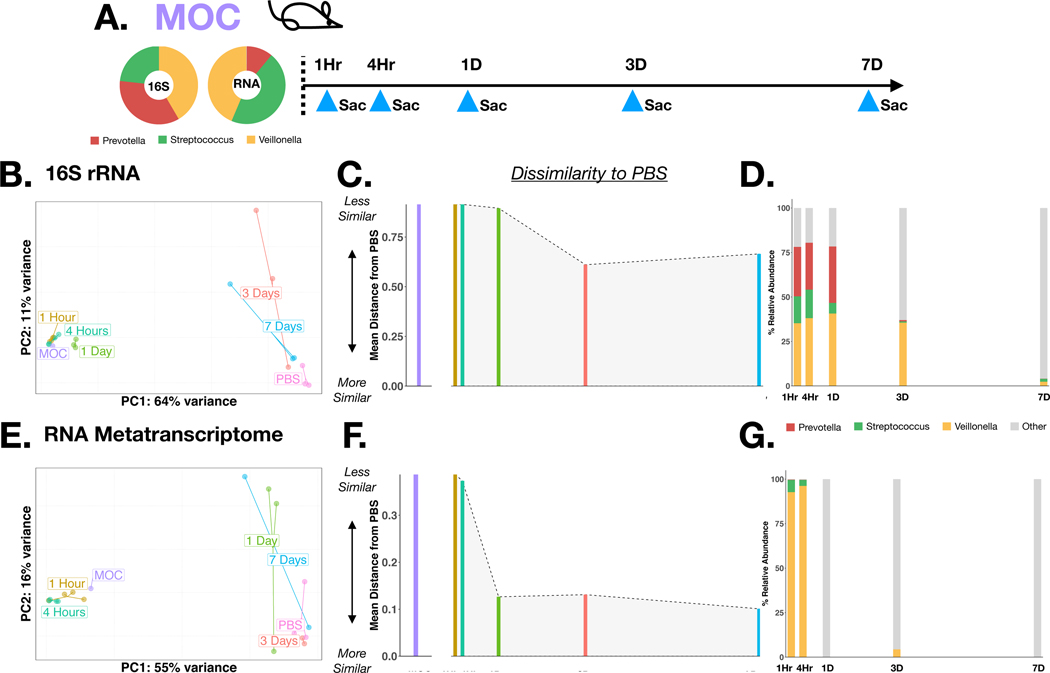
The RNA signature is lost earlier than the DNA signature in a mouse model of aspiration
It is possible that the improved resolution of RNA metatranscriptome in the identification of active microbial metabolism in the lower airways is due to differences in DNA and RNA clearance over time. To evaluate the stability and functional dynamics of aspirated oral commensals in the lower airways, we used a mouse model. For this, mice were inoculated with a mixture of human oral commensals (MOC) consisting of Prevotella melaninogenica, Streptococcus mitis and Veillonella parvula cohoused with a PBS control group. Mice were sacrificed at 1Hr, 4Hr, 1Day, 3Day and 7Day post-inoculation (Figure 6A), and BAL samples were sent for 16S rRNA gene and RNA metatranscriptome sequencing. β diversity analysis on 16S rRNA gene sequencing data shows that BAL samples remain similar to MOC for at least 1 day and become more similar to PBS by day 3 with a concordant decrease in the relative abundance of oral commensals (Figure 6B-C-D). However, in the analysis based on RNA metatranscriptome, BAL samples remain similar to MOC until the 4-hour timepoint and become more similar to PBS by day 1 with a concordant rapid loss of the RNA signal from oral commensals (Figure 6E-F-G). These data support that discrepancies between these sequencing data can be time dependent and likely reflect the loss of viable (and metabolically active) microbes.
Figure 6: Mouse experiment with 16S rRNA gene and RNA metatranscriptome sequencing:
(A) Visual schematic of the experiment, mice (n=17) were inoculated with a mixture of Prevotella, Streptococcus and Veillonella (MOC) and sacrificed at specific time intervals: 1 Hour, 4 Hours, 1 Day, 3 Days, 7 Days. BAL samples were analyzed by 16S rRNA gene sequencing (B-D) and RNA metatranscriptome sequencing (E-G): Principle coordinate analysis was performed with Bray-Curtis Distances by time point (B, E). Mean inter-group distance between sample time point and PBS was calculated (C, F). Relative abundance for taxa annotated to Prevotella, Streptococcus and Veillonella were calculated for each time point (D, G)
Discussion
Functional characterization of the lower airway microbiota has been attempted in a limited number of studies. In most of these, the inferred metagenome was used 2,33. Few have attempted metagenomic analyses 15. The purpose of this study was to evaluate different sequence data types in the evaluation of the functional microbiome of the lower airways and to use the measurement of SCFAs as an independent biological outcome, a direct measure of bacterial metabolism. Our analysis showed that, among lower airway samples with dysbiosis, characterized by the enrichment with oral commensals determined by taxonomic assignment of 16S rRNA gene sequencing, the use of WGS or RNA metatranscriptomic sequencing provide a distinct representation of functional aspects of the lower airway microbiota. Importantly, by pairing our sequence data with SCFA measurement we showed that for samples with lower airway dysbiosis based on 16S rRNA gene sequencing, there is a subset with evidence of active microbial metabolism indicative of viability of the lower airway microbiota. This active microbial metabolism in the lower airways has been shown to influence lower airway immunity34. Further support for dissimilarity between 16S rRNA gene and RNA metatranscriptomic sequencing is provided with a mouse model of aspiration of oral commensal, demonstrating time dependent differences likely related to loss of metabolically active microbes as the lower airways clear them.
With the introduction of next generation sequencing we have discovered complex microbial communities within several different mucosae 35–38. For each of these environments, the microbial-host interplay has an impact on mucosal homeostasis in health and disease 37–42. Within the lower airways, several studies have shown that complex microbial communities significantly impact the mucosal host immune tone 1,2,14,43,44. For example, we have previously shown that lower airway enrichment with oral commensals leads to an increased lower airway inflammatory tone, characterized by a Th-17-like inflammatory phenotype 2. Thus, it is increasingly important to describe these environments beyond just the presence/absence of bacteria but to look at the functional implications of these bacteria. A common technique used to evaluate bacterial function is to infer the metagenome from 16S rRNA gene sequencing data. Major concerns associated with this approach is the poor strain resolution of 16S rRNA gene sequencing and the dependence on existing reference databases of annotated microbes, which can bias the results. Direct measurement of microbial genes can be accomplished by WGS and RNA metatranscriptome sequencing. In this study, we used all three methods to evaluate taxonomic and functional signatures in the lower airways. As previously described, we identified a subset of subjects that had a lower airway microbiota enriched with oral commensals such as Prevotella, Veillonella, and Streptococcus 2. Enrichment with oral commensals in a subset of samples that were identified as BAL.SPT based on 16S rRNA gene sequencing, were also found in WGS and RNA sequencing data but, importantly, not in all of them. This enrichment with supraglottic taxa in the lower airways and its impact on host immune tone also has implications in disease states, as we have previously shown2. Thus, the information we glean from each of these data types is variable and potentially important when combined in a multi-omic approach. In addition, we have shown performing multi-omic analysis on lower airways samples is a feasible approach that provides deeper insight into the lower airway micro-environment.
As validation for such an approach we focused on SCFAs. Several papers have identified SCFAs as the products of bacterial metabolism45,46; the role these metabolites play in disease has been extensively studied in the gastrointestinal microbiota 20,21,23,24 and thought to be beneficial in inflammatory bowel disease and bowel cancer 47–50. Within the lower airways, we have described that levels of these metabolites are associated with oral commensal enrichment (as defined by 16S rRNA gene sequencing) and have significant immunomodulatory effects on T cells 13. In our prior investigation we also noted that not all subjects that had enrichment of the lower airway microbiota with oral commensals had elevated levels of SCFAs13. This suggests that functional characterization of the lower airway microbiota cannot be fully determined based solely on 16S rRNA gene sequencing data. We therefore integrated our WGS and RNA data with the measurement of SCFAs in UA, BAL and BKG samples. As expected, SCFAs were highest in UA samples consistent with the presence of oral anaerobes in these high biomass samples. Within BAL samples, a small subset of samples had detectable concentrations of acetate, propionate and isovalerate levels similar to the UA samples, identified as Supraglottic Predominant Taxa (BAL.SPT) samples based on 16S rRNA gene sequencing. Other BAL samples also identified as BAL.SPT based on 16S rRNA gene sequencing had low/below the limit of detection SCFA levels that were comparable with BAL.BPT and BKG samples. In contrast, the RNA metatranscriptome showed better sample type differentiation concordant with detected levels of SCFAs. Importantly, taxonomic evaluation of these KOs identified that the bacteria expressing these genes were oral commensals such as Streptococcus, Prevotella and Veillonella. Thus, these data suggest that in these samples, although oral commensals might have reached the lower airways and left traces of their genomic DNA, these bacteria have been cleared and are neither transcriptionally active nor capable of producing SCFAs at the time of sampling. This is further supported with a preclinical model of aspiration, where mice were exposed to a mixture of human oral commensals and sacrificed at different time points post exposure showing rapid loss of an RNA metatranscriptome signal from these microbes and longer persistence of 16S rRNA gene signal. Considering the known immunomodulatory effects of SCFAs and other microbial metabolites, both possibly beneficial and detrimental depending on the different human conditions, improved understanding of the value of different sequencing methods will be key to gain functional insights of the lower airway microbiome.
There are several limitations to this study. Firstly, in our analysis we did not remove any potential contaminants, which we found as the predominant taxa identified in many of the lower airway samples. Removing taxa identified as contaminant is frequently done in many microbiome studies hoping to remove contamination. However, there are many sources of noise that includes DNA contamination from the reagents/bronchoscope and stochastic sequencing noise 51,52. Further, in low biomass samples background removal can be quite variable within different sequencing datasets and its effect on the resulting new dataset is unclear. A recent guideline on lung microbiome research has not recommended background removal53 so our analysis was limited to just identifying possible contaminants. Importantly, none of the oral commensals associated with active microbial metabolism were identified as background contaminant. In addition, our sample size was small and further validation will require a larger cohort. We also recognize that this approach could impose a significant increase in sequencing cost compared with the traditional 16S rRNA gene sequencing. However, improved accuracy in identifying active microbial metabolism in the lower airways can potentially lead to novel mechanistic insights about microbial metabolites with significant potential effects on the host, such as SCFAs. Future investigations should focus on determining the value of this improved accuracy by evaluating the potential implications for the host immune tone, an undertake that should be designed with a larger cohort. Thus, we acknowledge that in the current investigation we did not attempt to evaluate host factors. Instead, we focused on functional evaluation of the lower airway microbiota using SCFAs as proof of concept. It is important to note that we are already facing an increase in literature suggesting that “near bedside” metagenomics is feasible (both technically and computationally) and have potential clinical implications in terms of rapid detection of pathogens when compared with culture-based approaches and ability to detect resistant genes 54,55. In this setting, our data supports that RNA sequencing could provide a better resolution of what microbial functions are active at a given time and may therefore contribute to the development of more targeted therapy. We also acknowledge that concentrations of acetate, which are not specific to microbes, can be influenced by the host and the environment, including water. For our assay, we used freshly opened HPLC-grade water which did not have detectable acetate above that in the BKG samples. Dilution of BAL can affect the levels of SCFAs but should not affect compositional data such as metagenome/metatranscriptome. Future investigation may consider estimating BAL dilution factors, noting that there is still controversy in the literature about the accuracy the best method for this56,57. Also, variability between sequence data type may be due to differences in measured targets (target amplicon vs. WGS vs. RNA) as well as technical differences. For example, it is expected that deeper sequence depth will be needed to characterize the whole genome than to characterize taxonomic composition based on 16S rRNA gene sequencing and infer the metagenome using that data. The low bacterial biomass of the lower airway environment represents a critical challenge for the evaluation of the lower airway microbiome, both taxonomically and functionally. It is likely that this is an exponentially bigger problem for the WGS metagenome and RNA metatranscriptome. Although we aimed to achieve more sequence depth for our WGS metagenome and RNA metatranscriptome data, it is not surprising that differences may be more difficult to assess and that there is a greater level of background intrusion in these samples using these methods. However, the correlation of these data with SCFA levels suggest a more accurate functional evaluation can be achieved with WGS metagenome and RNA metatranscriptome data than with 16S rRNA gene sequencing data. Finally, the analyses presented here focused on fatty acid metabolism as a surrogate for bacterial activity. The highest level of precision and differential functional expression for lower airway samples identified by using RNA metatranscriptome data suggest that this might be a preferable functional method. However, it is likely that other microbial functional pathways may be important to study in health and disease and future investigations should focus on experimental approaches to expand the observations made as proof of concept here.
In summary, the evaluation of the lower airway microbiome with 16S rRNA gene sequencing is limited in assessing bacterial function and therefore in assessing the potential impact on disease/host. The use of functional microbiome approaches that measure bacterial genes (WGS) and bacterial transcripts (RNA metatranscriptome) provide evidence of viable and active bacterial metabolism in the lower airways and will likely define subgroups of lower airway microbiota with different implications for the host.
Supplementary Material
Supplementary Figure 1: α Diversity for WGS Metagenome and RNA Metatranscriptome data. Shannon Index of (A) WGS Metagenome and (B) RNA Metatranscriptome data.
Supplementary Figure 2: Rarefaction analysis for each sequencing datasets. Each line represents a sample and the dash line represents the mean for all samples. Sequence depth (x axis) is compared with OTU or KO annotation (y axis). (A) 16S rRNA gene sequencing (B) Whole Genome Sequencing (C) RNA Metatranscriptome.
Supplementary Figure 3: Comparison of global taxonomic composition across different sequencing data types. PROCRUSTES analysis, using Bray-Curtis Distance matrices, was used to compare taxonomic annotation for the three data types. Monte-Carlo simulation test was used for statistical significance. (A) Comparison between 16S rRNA gene sequencing and WGS Metagenome (Monte-Carlo p-value=ns). (B) Comparison between 16S rRNA gene sequencing and RNA Metatranscriptome (Monte-Carlo p-value=ns). (C) Comparison between WGS Metagenome and RNA Metatranscriptome (Monte-Carlo p-value <0.01).
Supplementary Figure 4: Comparison of global functional composition across different sequencing data types. PROCRUSTES analysis, with Bray-Curtis Distance matrices, was used to compare functional annotation across the sequencing data types. Monte-Carlo simulation test was used for statistical significance. (A) Comparison between 16S rRNA gene sequencing and WGS Metagenome (Monte-Carlo p-value<0.01). (B) Comparison between 16S rRNA gene sequencing and RNA Metatranscriptome (Monte-Carlo p-value<0.01). (C) Comparison between WGS Metagenome and RNA Metatranscriptome (Monte-Carlo p-value <0.01).
Supplementary Figure 5: Functional signatures identified as differentially enriched in BAL.SPT on WGS Metagenome and RNA Metatranscriptome data. DESEQ2 analysis of WGS Metagenome (A) and RNA Metatranscriptome (B) functional annotation identified KOs differentially enriched in BAL.16S.SPT vs. BAL.BPT.16S samples (FDR<0.05). Circle size is representative of KEGG Expression.
Supplementary Figure 6: Ex-Vivo measurement of Short Chain Fatty Acids (SCFAs) levels. (A) Levels of SCFAs in supernatant of Veillonella parvula in culture media were compared with media alone. Longitudinal change in levels of Butyrate (B) or Propionate (C) after ex vivo addition to BAL cells.
Supplementary Figure 7: Comparison of levels of SCFAs in UA and BAL samples. Correlation analysis using spearman rho showed no significant association between levels of SCFAs detected in UA and BAL samples.
Supplementary Figure 8: Clustering of WGS Metagenome and RNA Metatranscriptome: Dirichlet Multinomial Modelling (DMM) of all samples was performed separately for WGS Metagenome (A) and RNA Metatranscriptome (C). In both cases, 2 clusters were identified as the best model fit. Principle Co-ordinate analysis of WGS metagenome (B), based on Bray-Curtis Distance, using DMM clustering of BAL samples showed that only three BAL Samples clustered with UA samples. Principle Co-ordinate analysis of RNA metatranscriptome (C), based on Bray-Curtis Distance, using DMM clustering of BAL samples showed that only two BAL Samples clustered with UA samples.
Supplementary Figure 9: Identifying Potential Contaminants. Using decontam with a threshold of 0.7, contaminants were identified based on their prevalence in BAL and Upper Airway (UA) samples when compared to Background (BKG) samples. The top taxa, identified as contaminants, median relative abundance, is displayed for the (A) 16S rRNA gene sequencing (B) Whole Genome Sequencing and (C) RNA Metatranscriptome.
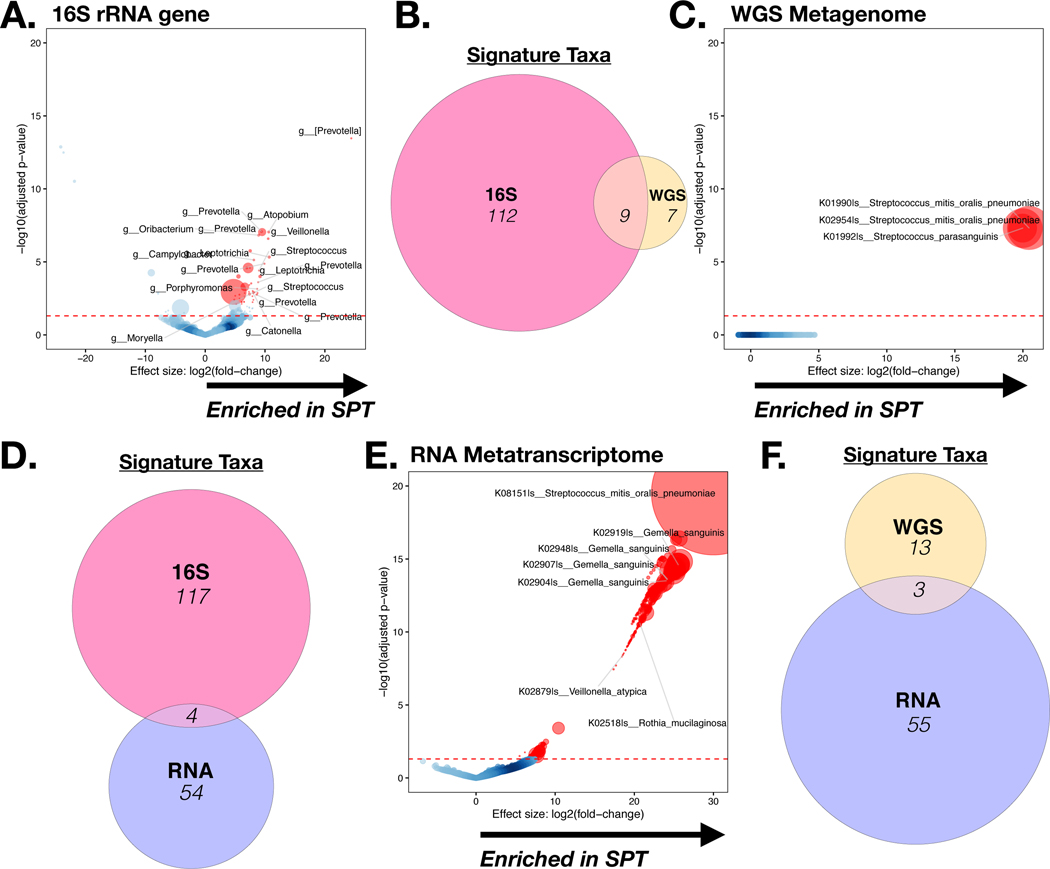
Figure 2: Taxonomic annotation of all three sequencing data types.
DESEQ2 analysis of taxonomic annotation (at the genus level) between BAL.16S.SPT versus BAL. 16S.BPT samples (FDR <0.05) was performed on 16S rRNA gene sequencing data (A), WGS data (C) and RNA metatranscriptome data (E). Circle size is representative of relative abundance. Gene Set Enrichment Analysis (GSEA) was used to compare the taxonomic signatures identified as distinctly enriched in BAL.16S.SPT vs. BAL. 16S.BPT samples across the different sequencing platforms (B, D, F).
Acknowledgments
Research support funding: R37 CA244775 (LNS, NIH/NCI); R01 HL125816 (SBK, LNS, NIH/NHLBI); R01 GM111400 (KAS, NIH/NIGMS), Stony Wold (IS), Chest Foundation Grant (IS), FAMRI (BW), PACT grant (LNS, FNIH), R01AI129958 (YJH).
Financial support for the PACT project are made possible through funding support provided to the FNIH by: AbbVie Inc., Amgen Inc., Boehringer-Ingelheim Pharma GmbH & Co. KG, Bristol-Myers Squibb, Celgene Corporation, Genentech Inc., Gilead, GlaxoSmithKline plc, Janssen Pharmaceutical Companies of Johnson & Johnson, Novartis Institutes for Biomedical Research, Pfizer Inc., and Sanofi.
Footnotes
Financial Disclosure: None
References:
- 1.Segal LN et al. Enrichment of lung microbiome with supraglottic taxa is associated with increased pulmonary inflammation. Microbiome 1, 19, doi: 10.1186/2049-2618-1-19 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Segal LN et al. Enrichment of the lung microbiome with oral taxa is associated with lung inflammation of a Th17 phenotype. Nat Microbiol 1, 16031, doi: 10.1038/nmicrobiol.2016.31 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beck JM et al. Multicenter Comparison of Lung and Oral Microbiomes of HIV-infected and HIV-uninfected Individuals. Am J Respir Crit Care Med 192, 1335–1344, doi: 10.1164/rccm.201501-0128OC (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huffnagle GB, Dickson RP & Lukacs NW The respiratory tract microbiome and lung inflammation: a two-way street. Mucosal immunology 10, 299–306, doi: 10.1038/mi.2016.108 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dickson RP, Erb-Downward JR, Martinez FJ & Huffnagle GB The Microbiome and the Respiratory Tract. Annual review of physiology 78, 481–504, doi: 10.1146/annurev-physiol-021115-105238 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lozupone C. et al. Widespread Colonization of the Lung by Tropheryma whipplei in HIV Infection. Am J Respir Crit Care Med 187, 1110–1117, doi: 10.1164/rccm.201211-2145OC (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Molyneaux PL et al. Host-Microbial Interactions in Idiopathic Pulmonary Fibrosis. Am J Respir Crit Care Med 195, 1640–1650, doi: 10.1164/rccm.201607-1408OC (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sulaiman I. et al. Evaluation of the airway microbiome in nontuberculous mycobacteria disease. Eur Respir J 52, doi: 10.1183/13993003.00810-2018 (2018). [DOI] [PubMed] [Google Scholar]
- 9.Tsay JJ et al. Airway Microbiota Is Associated with Upregulation of the PI3K Pathway in Lung Cancer. Am J Respir Crit Care Med 198, 1188–1198, doi: 10.1164/rccm.201710-2118OC (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Erb-Downward JR et al. Analysis of the lung microbiome in the “healthy” smoker and in COPD. PLoS One 6, e16384, doi: 10.1371/journal.pone.0016384 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sze MA et al. The lung tissue microbiome in chronic obstructive pulmonary disease. Am J Respir Crit Care Med 185, 1073–1080, doi: 10.1164/rccm.201111-2075OC (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Durack J. et al. Bacterial biogeography of adult airways in atopic asthma. Microbiome 6, 104, doi: 10.1186/s40168-018-0487-3 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Segal LN et al. Anaerobic Bacterial Fermentation Products Increase Tuberculosis Risk in Antiretroviral-Drug-Treated HIV Patients. Cell host & microbe 21, 530–537 e534, doi: 10.1016/j.chom.2017.03.003 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Segal LN et al. Randomised, double-blind, placebo-controlled trial with azithromycin selects for anti-inflammatory microbial metabolites in the emphysematous lung. Thorax 72, 13–22, doi: 10.1136/thoraxjnl-2016-208599 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Clarke EL et al. Microbial Lineages in Sarcoidosis. A Metagenomic Analysis Tailored for Low-Microbial Content Samples. Am J Respir Crit Care Med 197, 225–234, doi: 10.1164/rccm.201705-0891OC (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu N. et al. Metagenomic next-generation sequencing diagnosis of peripheral pulmonary infectious lesions through virtual navigation, radial EBUS, ultrathin bronchoscopy, and ROSE. J Int Med Res, 300060519866953, doi: 10.1177/0300060519866953 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Qi C. et al. Detection of respiratory pathogens in clinical samples using metagenomic shotgun sequencing. J Med Microbiol 68, 996–1002, doi: 10.1099/jmm.0.000968 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cribbs SK et al. Correlation of the lung microbiota with metabolic profiles in bronchoalveolar lavage fluid in HIV infection. Microbiome 4, 3, doi: 10.1186/s40168-016-0147-4 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Trompette A. et al. Gut microbiota metabolism of dietary fiber influences allergic airway disease and hematopoiesis. Nat Med 20, 159–166, doi: 10.1038/nm.3444 (2014). [DOI] [PubMed] [Google Scholar]
- 20.Chambers ES, Preston T, Frost G. & Morrison DJ Role of Gut Microbiota-Generated Short-Chain Fatty Acids in Metabolic and Cardiovascular Health. Curr Nutr Rep 7, 198–206, doi: 10.1007/s13668-018-0248-8 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Katsidzira L. et al. Differences in Fecal Gut Microbiota, Short-Chain Fatty Acids and Bile Acids Link Colorectal Cancer Risk to Dietary Changes Associated with Urbanization Among Zimbabweans. Nutr Cancer, 1–12, doi: 10.1080/01635581.2019.1602659 (2019). [DOI] [PubMed] [Google Scholar]
- 22.Ohigashi S. et al. Changes of the intestinal microbiota, short chain fatty acids, and fecal pH in patients with colorectal cancer. Dig Dis Sci 58, 1717–1726, doi: 10.1007/s10620-012-2526-4 (2013). [DOI] [PubMed] [Google Scholar]
- 23.Parada Venegas D. et al. Short Chain Fatty Acids (SCFAs)-Mediated Gut Epithelial and Immune Regulation and Its Relevance for Inflammatory Bowel Diseases. Front Immunol 10, 277, doi: 10.3389/fimmu.2019.00277 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sanna S. et al. Causal relationships among the gut microbiome, short-chain fatty acids and metabolic diseases. Nat Genet 51, 600–605, doi: 10.1038/s41588-019-0350-x (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dray S. a. D., A.B. The ade4 package: implementing the duality diagram for ecologists. Journal of Statistical Software 22, 1–20 (2007). [Google Scholar]
- 26.Lozupone C, Lladser ME, Knights D, Stombaugh J. & Knight R. UniFrac: an effective distance metric for microbial community comparison. The ISME journal 5, 169–172, doi: 10.1038/ismej.2010.133 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Holmes I, Harris K. & Quince C. Dirichlet multinomial mixtures: generative models for microbial metagenomics. PLoS One 7, e30126, doi: 10.1371/journal.pone.0030126 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.DirichletMultinomial: Dirichlet-Multinomial Mixture Model Machine Learning for Microbiome Data v. R package version 1.20.0 (2017). [Google Scholar]
- 29.Love MI, Huber W. & Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15, 550, doi: 10.1186/s13059-014-0550-8 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reiner A, Yekutieli D. & Benjamini Y. Identifying differentially expressed genes using false discovery rate controlling procedures. Bioinformatics 19, 368–375 (2003). [DOI] [PubMed] [Google Scholar]
- 31.Langille MG et al. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat Biotechnol 31, 814–821, doi: 10.1038/nbt.2676 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Davis NM, Proctor DM, Holmes SP, Relman DA & Callahan BJ Simple statistical identification and removal of contaminant sequences in marker-gene and metagenomics data. Microbiome 6, 226, doi: 10.1186/s40168-018-0605-2 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huang YJ et al. The airway microbiome in patients with severe asthma: Associations with disease features and severity. J Allergy Clin Immunol 136, 874–884, doi: 10.1016/j.jaci.2015.05.044 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McAleer JP & Kolls JK Contributions of the intestinal microbiome in lung immunity. Eur J Immunol 48, 39–49, doi: 10.1002/eji.201646721 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Byrd AL, Belkaid Y. & Segre JA The human skin microbiome. Nat Rev Microbiol 16, 143–155, doi: 10.1038/nrmicro.2017.157 (2018). [DOI] [PubMed] [Google Scholar]
- 36.Ma B, Forney LJ & Ravel J. Vaginal microbiome: rethinking health and disease. Annu Rev Microbiol 66, 371–389, doi: 10.1146/annurev-micro-092611-150157 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.O’Dwyer DN, Dickson RP & Moore BB The Lung Microbiome, Immunity, and the Pathogenesis of Chronic Lung Disease. J Immunol 196, 4839–4847, doi: 10.4049/jimmunol.1600279 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shreiner AB, Kao JY & Young VB The gut microbiome in health and in disease. Curr Opin Gastroenterol 31, 69–75, doi: 10.1097/MOG.0000000000000139 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Beri K. Skin microbiome & host immunity: applications in regenerative cosmetics & transdermal drug delivery. Future Sci OA 4, FSO302, doi: 10.4155/fsoa-2017-0117 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bull MJ & Plummer NT Part 1: The Human Gut Microbiome in Health and Disease. Integr Med (Encinitas) 13, 17–22 (2014). [PMC free article] [PubMed] [Google Scholar]
- 41.Lazar V. et al. Aspects of Gut Microbiota and Immune System Interactions in Infectious Diseases, Immunopathology, and Cancer. Front Immunol 9, 1830, doi: 10.3389/fimmu.2018.01830 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lozupone CA Unraveling Interactions between the Microbiome and the Host Immune System To Decipher Mechanisms of Disease. mSystems 3, doi: 10.1128/mSystems.00183-17 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Segal LN, Rom WN & Weiden MD Lung microbiome for clinicians. New discoveries about bugs in healthy and diseased lungs. Ann Am Thorac Soc 11, 108–116, doi: 10.1513/AnnalsATS.201310-339FR (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shenoy MK et al. Immune Response and Mortality Risk Relate to Distinct Lung Microbiomes in Patients with HIV and Pneumonia. Am J Respir Crit Care Med 195, 104–114, doi: 10.1164/rccm.201603-0523OC (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Roediger WE Role of anaerobic bacteria in the metabolic welfare of the colonic mucosa in man. Gut 21, 793–798, doi: 10.1136/gut.21.9.793 (1980). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Eftimiadi C. et al. Short-chain fatty acids produced by anaerobic bacteria alter the physiological responses of human neutrophils to chemotactic peptide. The Journal of infection 14, 43–53, doi: 10.1016/s0163-4453(87)90808-5 (1987). [DOI] [PubMed] [Google Scholar]
- 47.Gomes SD et al. The Role of Diet Related Short-Chain Fatty Acids in Colorectal Cancer Metabolism and Survival: Prevention and Therapeutic Implications. Curr Med Chem, doi: 10.2174/0929867325666180530102050 (2018). [DOI] [PubMed] [Google Scholar]
- 48.Hinnebusch BF, Meng S, Wu JT, Archer SY & Hodin RA The effects of short-chain fatty acids on human colon cancer cell phenotype are associated with histone hyperacetylation. J Nutr 132, 1012–1017, doi: 10.1093/jn/132.5.1012 (2002). [DOI] [PubMed] [Google Scholar]
- 49.Scheppach W, Bartram HP & Richter F. Role of short-chain fatty acids in the prevention of colorectal cancer. Eur J Cancer 31A, 1077–1080, doi: 10.1016/0959-8049(95)00165-f (1995). [DOI] [PubMed] [Google Scholar]
- 50.Silva JPB et al. Protective Mechanisms of Butyrate on Inflammatory Bowel Disease. Curr Pharm Des 24, 4154–4166, doi: 10.2174/1381612824666181001153605 (2018). [DOI] [PubMed] [Google Scholar]
- 51.Erb-Downward JR et al. Critical Relevance of Stochastic Effects on Low-Bacterial-Biomass 16S rRNA Gene Analysis. mBio 11, doi: 10.1128/mBio.00258-20 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Salter SJ et al. Reagent and laboratory contamination can critically impact sequence-based microbiome analyses. BMC Biol 12, 87, doi: 10.1186/s12915-014-0087-z (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Carney SM et al. Methods in Lung Microbiome Research. Am J Respir Cell Mol Biol, doi: 10.1165/rcmb.2019-0273TR (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Charalampous T. et al. Nanopore metagenomics enables rapid clinical diagnosis of bacterial lower respiratory infection. Nat Biotechnol 37, 783–792, doi: 10.1038/s41587-019-0156-5 (2019). [DOI] [PubMed] [Google Scholar]
- 55.Pendleton KM et al. Rapid Pathogen Identification in Bacterial Pneumonia Using Real-Time Metagenomics. Am J Respir Crit Care Med 196, 1610–1612, doi: 10.1164/rccm.201703-0537LE (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Marcy TW, Merrill WW, Rankin JA & Reynolds HY Limitations of using urea to quantify epithelial lining fluid recovered by bronchoalveolar lavage. Am Rev Respir Dis 135, 1276–1280, doi: 10.1164/arrd.1987.135.6.1276 (1987). [DOI] [PubMed] [Google Scholar]
- 57.Rennard SI et al. Estimation of volume of epithelial lining fluid recovered by lavage using urea as marker of dilution. J Appl Physiol (1985) 60, 532–538, doi: 10.1152/jappl.1986.60.2.532 (1986). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1: α Diversity for WGS Metagenome and RNA Metatranscriptome data. Shannon Index of (A) WGS Metagenome and (B) RNA Metatranscriptome data.
Supplementary Figure 2: Rarefaction analysis for each sequencing datasets. Each line represents a sample and the dash line represents the mean for all samples. Sequence depth (x axis) is compared with OTU or KO annotation (y axis). (A) 16S rRNA gene sequencing (B) Whole Genome Sequencing (C) RNA Metatranscriptome.
Supplementary Figure 3: Comparison of global taxonomic composition across different sequencing data types. PROCRUSTES analysis, using Bray-Curtis Distance matrices, was used to compare taxonomic annotation for the three data types. Monte-Carlo simulation test was used for statistical significance. (A) Comparison between 16S rRNA gene sequencing and WGS Metagenome (Monte-Carlo p-value=ns). (B) Comparison between 16S rRNA gene sequencing and RNA Metatranscriptome (Monte-Carlo p-value=ns). (C) Comparison between WGS Metagenome and RNA Metatranscriptome (Monte-Carlo p-value <0.01).
Supplementary Figure 4: Comparison of global functional composition across different sequencing data types. PROCRUSTES analysis, with Bray-Curtis Distance matrices, was used to compare functional annotation across the sequencing data types. Monte-Carlo simulation test was used for statistical significance. (A) Comparison between 16S rRNA gene sequencing and WGS Metagenome (Monte-Carlo p-value<0.01). (B) Comparison between 16S rRNA gene sequencing and RNA Metatranscriptome (Monte-Carlo p-value<0.01). (C) Comparison between WGS Metagenome and RNA Metatranscriptome (Monte-Carlo p-value <0.01).
Supplementary Figure 5: Functional signatures identified as differentially enriched in BAL.SPT on WGS Metagenome and RNA Metatranscriptome data. DESEQ2 analysis of WGS Metagenome (A) and RNA Metatranscriptome (B) functional annotation identified KOs differentially enriched in BAL.16S.SPT vs. BAL.BPT.16S samples (FDR<0.05). Circle size is representative of KEGG Expression.
Supplementary Figure 6: Ex-Vivo measurement of Short Chain Fatty Acids (SCFAs) levels. (A) Levels of SCFAs in supernatant of Veillonella parvula in culture media were compared with media alone. Longitudinal change in levels of Butyrate (B) or Propionate (C) after ex vivo addition to BAL cells.
Supplementary Figure 7: Comparison of levels of SCFAs in UA and BAL samples. Correlation analysis using spearman rho showed no significant association between levels of SCFAs detected in UA and BAL samples.
Supplementary Figure 8: Clustering of WGS Metagenome and RNA Metatranscriptome: Dirichlet Multinomial Modelling (DMM) of all samples was performed separately for WGS Metagenome (A) and RNA Metatranscriptome (C). In both cases, 2 clusters were identified as the best model fit. Principle Co-ordinate analysis of WGS metagenome (B), based on Bray-Curtis Distance, using DMM clustering of BAL samples showed that only three BAL Samples clustered with UA samples. Principle Co-ordinate analysis of RNA metatranscriptome (C), based on Bray-Curtis Distance, using DMM clustering of BAL samples showed that only two BAL Samples clustered with UA samples.
Supplementary Figure 9: Identifying Potential Contaminants. Using decontam with a threshold of 0.7, contaminants were identified based on their prevalence in BAL and Upper Airway (UA) samples when compared to Background (BKG) samples. The top taxa, identified as contaminants, median relative abundance, is displayed for the (A) 16S rRNA gene sequencing (B) Whole Genome Sequencing and (C) RNA Metatranscriptome.