Abstract
Kaposi's sarcoma-associated herpesvirus (KSHV) has been consistently identified in Kaposi's sarcomas, body cavity-based lymphomas, and some forms of Castleman's disease. The K9 open reading frame of KSHV encodes a viral interferon regulatory factor (vIRF) which functions as a repressor for cellular interferon-mediated signal transduction and as an oncogene to induce cell growth transformation. We demonstrate that KSHV vIRF directly interacts with cellular transcriptional coactivator p300 and displaces p300/CBP-associated factor from p300 complexes. This interaction inhibits the histone acetyltransferase activity of p300, resulting in drastic reduction of nucleosomal histone acetylation and alteration of chromatin structure. As a consequence, vIRF expression markedly alters cellular cytokine expression, which is regulated by acetylation of nucleosomal histones. These results demonstrate that KSHV vIRF interacts with and inhibits the p300 transcriptional coactivator to circumvent the host antiviral immune response and to induce a global alteration of cellular gene expression. These studies also illustrate how a cellular gene captured by a herpesvirus has evolved several functions that suit the needs of the virus.
Interferons (IFNs) are a family of cytokines that exhibit diverse biological effects that include the inhibition of cell growth and protection against viral infection (39). Newly synthesized IFN interacts with neighboring cells through cellular surface receptors, resulting in the synthesis of a group of new cellular proteins. The interaction leads to the activation of a transcriptional factor that recognizes a conserved cis-acting DNA element located within the regulatory sequences of target genes (11). This element binds to members of the IFN regulatory factor (IRF) family and the alpha IFN (IFN-α)-stimulated gene factor 3 complex (11). The p300/CBP transcriptional cofactor which contains histone acetyltransferase (HAT) activity (16, 17, 20, 30, 42, 44, 45) has been shown to interact with IRFs including IRF1 and IRF3 and STATs to form a virus-activated factor complex (4, 47). Upon viral infection, the virus-activated factor complex accumulates in the nucleus, binds to the promoters of IFN-mediated virus-inducible genes, and activates their gene expression as part of the host defense mechanism (4, 7, 44, 47). In addition, the adenovirus E1A and simian virus 40 large T-antigen oncoproteins have been shown to bind to the p300/CBP transcriptional coactivator family (13, 23). These interactions have been shown to be important for blocking IFN signaling and for inducing cell growth transformation (23, 40, 51).
Kaposi's sarcoma (KS) is a multifocal vascular tumor of mixed cellular composition that is the most common neoplasm in patients with AIDS (8). A new member of the herpesvirus group, Kaposi's sarcoma-associated herpesvirus (KSHV) or human herpesvirus 8, has been consistently identified in KS, body cavity-based lymphoma, and some forms of Castleman's disease (37). Although still limited, the presently available data provide compelling evidence that KSHV is the long-sought infectious agent of KS. The genomic sequence indicates KSHV to be a gammaherpesvirus that is closely related to herpesvirus saimiri (35), the recently isolated rhesus monkey rhadinovirus (12, 38), and retroperitoneal fibromatosis-associated herpesviruses (34). DNA sequence analysis of the entire 140.5 kbp of the KSHV genome reveals a number of cellular homologs which could possibly contribute to the pathogenesis associated with this virus (35). These include a virus-encoded interleukin-6 (IL-6) (27, 28, 29), MIP1-α/β chemokines (19, 27, 29), a bcl-2 homolog (36), v-cyclin (15, 22), vIL-8 receptor (2), and vFLIP (43) and a vN-CAM homolog.
The KSHV K9 open reading frame exhibits significant homology with cellular IRFs. We and others have demonstrated that expression of K9 dramatically represses transcriptional activation induced by IFN-α/β/γ (14, 21, 52). In addition, K9 is involved in regulation of KSHV gene expression (21). Furthermore, K9 expression leads to transformation of rodent fibroblast cells resulting in morphological change, focus formation, growth at reduced serum concentration, and tumor induction in nude mice (14, 21). Thus, the K9 gene of KSHV encodes a viral IRF (vIRF) which functions as a repressor of cellular IFN-mediated signal transduction and as an oncogene to induce cell growth transformation.
Cellular IRFs interact with and recruit the p300 transcriptional coactivator to specific promoters to induce transcriptional activation in response to virus infection. In this report, we demonstrate that KSHV vIRF directly interacts with cellular p300 transcriptional coactivator. This interaction inhibits p300 HAT activity, resulting in hypoacetylation of nucleosomal histones, alteration of chromatin structure, and perturbation of cytokine gene expression. These results suggest that KSHV employs an elaborate decoy mechanism by harboring vIRF to circumvent host antiviral defense.
MATERIALS AND METHODS
Cell culture and transfection.
HS27 and NIH 3T3 cells were grown in Dulbecco's modified Eagle medium supplemented with 10% fetal calf serum. BCBL-1 and BJAB cells were grown in RPMI 1640 medium supplemented with 10% fetal calf serum. Insect cells were maintained at 27°C in Grace's medium containing 10% fetal calf serum, yeastolate, and lactalbumin. A DEAE-dextran transfection procedure was used for transient expression in COS-1 cells. The pcDNA3.1-vIRF constructs (5 μg) were introduced into HS27 and NIH 3T3 cells by Fusin transfection (Boehringer Mannheim). After a 48-h incubation, the cells were cultured with selection medium containing 500 μg of neomycin per ml for the next 4 weeks.
Purification of flag-tagged proteins from insect cells.
Sf-9 or High-5 insect cells were infected with baculovirus containing the flag-tagged p300, vIRF, or v-cyclin for 2 to 3 days. Cells were pelleted, washed with phosphate-buffered saline (PBS), lysed with lysis buffer (0.15 M NaCl, 1% Nonidet P-40, and 50 mM HEPES buffer [pH 8.0]) containing 1 mM Na2VO3, 1 mM NaF, and protease inhibitors (leupeptin, aprotinin, phenylmethylsulfonyl fluoride, and bestatin), and briefly sonicated. Supernatants were mixed on an antiflag (M2; Sigma) antibody column at 4°C overnight. After extensive washing with Tris-buffered saline (50 mM Tris [pH 7.5], 150 mM NaCl), flag-tagged proteins were eluted by adding an excessive amount of flag peptide (100 μg/ml; Sigma) and dialyzed against PBS overnight.
Mutant constructions.
All deletion mutations in the vIRF gene were generated with PCR using oligonucleotide-directed mutagenesis. The amplified DNA fragments containing mutations in vIRF were purified and cloned into pSP72 vector. Each vIRF mutant was completely sequenced to verify the presence of the mutation and the absence of any other changes. After confirmation of the DNA sequence, DNA containing the desired vIRF mutation was recloned into the pcDNA3.1 vector.
In vitro HAT assay.
In vitro HAT assays were performed according to the manufacturer's protocol (Upstate Biotechnology Inc.). In brief, histone H4 (Sigma) was incubated with 0.25 μCi of [3H]acetyl coenzyme A (CoA) (NEN) and 30 nM purified p300 in the presence or absence of purified vIRF or v-cyclin protein in 30 μl of acetylation buffer (50 mM Tris [pH 8.0], 5% glycerol, 0.1 mM EDTA, 50 mM KCl, 1 mM dithiothreitol, and 1 mM phenylmethylsulfonyl fluoride). Reaction mixtures were incubated at 30°C for 30 min, and the reaction was stopped by addition of sodium dodecyl sulfate (SDS) sample buffer. HAT activity was quantitated by filter binding assays to measure the 3H incorporation into histone H4 or by immunoblotting assays with an antibody which detected only the acetylated form of histone H4 (Upstate Biotechnology).
Immunofluorescence tests.
Cells were fixed with 4% paraformaldehyde for 15 min, permeabilized with 70% ethanol for 15 min, blocked with 10% goat serum in PBS for 30 min, and reacted with 1:100-diluted primary antibody in PBS for 30 min at room temperature. After incubation, cells were washed extensively with PBS, incubated with 1:100-diluted secondary antibody (Vector Laboratories, Burlingame, Calif.) in PBS for 30 min at room temperature, and washed three times with PBS. Protein staining was performed with 1:500-diluted Sypro (Molecular Probes) for 1 min, and DNA staining was performed with 1:5,000-diluted To-Pro 1 or 3 (Molecular Probes) for 1 min. Confocal microscopy was performed using a Leica TCS SP laser scanning microscope (Leica Microsystems, Exton, Pa.) fitted with a 100× Leica objective (Planaprochromatic; 1.4 numerical aperture) and using the Leica image software. Images were collected at 512- by 512-pixel resolution. The stained cells were optically sectioned in the z axis, and the images in the different channels (photomultiplier tubes) were collected simultaneously. The step size in the z axis varied from 0.2 to 0.5 μm to obtain 30 to 50 slices/imaged file. The images were transferred to a Macintosh G3 computer (Apple Computer, Cupertino, Calif.), and NIH Image v1.61 software was used to render the images.
Cell cycle analysis.
Cells were washed once with PBS, fixed in 70% ethanol for 15 min, and stained with staining solution (1% Triton X-100, 50 μg of propidium iodide [PI] or Hoechst 33342 per ml, 1 mg of RNase A per ml) for 30 min at room temperature. Cell cycle analysis was performed with a FACScan cell sorter (Becton Dickinson, Mountain View, Calif.).
RNase protection assays.
RNA was extracted from cells, using Trizol reagent (Gibco-BRL). The RNase protection assay was performed using 5 μg of RNA with the RiboQuant multiprobe RNase protection assay system (PharMingen, San Diego, Calif.) according to the manufacturer's specifications. In brief, RNA was individually hybridized overnight with the in vitro-transcribed 32P-labeled probe of migration inhibition factor (MIF) or glyceraldehyde-3-phosphate dehydrogenase (GAPDH) from mCK-3b cytokine template sets from PharMingen. Following hybridization, samples were treated with RNase and proteinase K, phenol-chloroform extracted, and ethanol precipitated. The protected fragments were resolved by electrophoresis on a 5% acrylamide–urea gel, and autoradiograms were developed in a Fuji Phospho Imager.
MIF promoter cloning and reporter assays.
The bp −1033 to +63 sequence of the murine MIF promoter was PCR amplified from NIH 3T3 genomic DNA using specific primers (46) and subsequently cloned into the pGL3 basic (Promega, Madison, Wis.) luciferase reporter vector (MIF-luc). An insert DNA fragment was completely sequenced to show the sequence identical to that of the previously reported MIF promoter region (46). All transfections included 2.5 μg of pGKβgal, which expresses β-galactosidase from a phosphoglucokinase promoter, and 2.5 μg of MIF-luc, which expresses luciferase from a MIF promoter together with 2.5 μg of vIRF vector, wild-type (wt) p300 vector, or p300 ΔHAT vector. At 48 h posttransfection, cells were washed once in PBS and lysed in 200 μl of reporter lysis buffer (Promega). Assays for luciferase were performed with a Luminometer using a luciferase assay (Promega). Values were normalized by β-galactosidase activity.
RESULTS
vIRF interacts with cellular p300 in vivo.
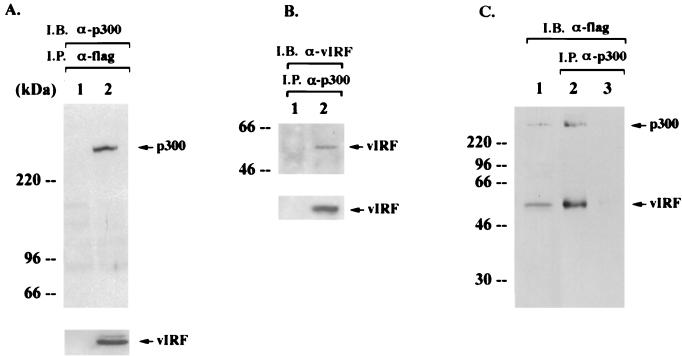
To define the molecular action of vIRF in IFN-mediated signal transduction and cell growth transformation, we investigated the potential interaction of vIRF with cellular transcriptional factors. We show that vIRF strongly interacts with p300 (Fig. 1). COS-1 cells were transfected with an expression vector containing a flag-tagged vIRF. Proteins present in antiflag immune complexes were separated by SDS-polyacrylamide gel electrophoresis (PAGE), transferred to nitrocellulose, and reacted with an anti-p300 antibody. Endogenous cellular p300 was readily detected in antiflag immune complexes from COS-1 cells with vIRF expression (Fig. 1A, lane 2), but it was not detected in antiflag immune complexes from COS-1 cells without vIRF expression (Fig. 1A, lane 1). In addition, approximately 5 to 10% of vIRF in KSHV-infected BCBL cells after tetradecanoyl phorbol acetate (TPA) stimulation was found to be interactive with p300 (Fig. 1B). To further demonstrate an interaction between vIRF and p300, purified flag-tagged p300 and vIRF proteins from insect cells were mixed in vitro. Anti-p300 immune complexes were subjected to antiflag immunoblotting analysis to detect the interaction of vIRF (Fig. 1C). These results indicated that p300 specifically interacted with vIRF in vitro. Recently, an interaction of vIRF with p300/CBP has also been demonstrated in an in vitro glutathione S-transferase (GST) pull-down assay (6) and a transient-expression assay (18). Our results further demonstrate an interaction of vIRF with p300 in various experimental settings including KSHV-infected BCBL-1 cells.
FIG. 1.
Interaction of vIRF with p300. (A) In vivo interaction of vIRF with p300. COS-1 cells were transfected with expression vector (lane 1) or the flag-tagged vIRF expression vector (lane 2). After 48 h, cell extracts were used for immunoprecipitations (I.P.) with an antiflag antibody, followed by immunoblotting assay (I.B.) with an anti-p300 antibody. vIRF expression in transfected COS-1 cells was determined by immunoblotting with an antiflag antibody (bottom panel). (B) In vivo interaction of vIRF with p300 in KSHV-infected BCBL-1 cells. Lysates of BJAB (lane 1) and TPA-induced BCBL-1 (lane 2) cells were used for immunoprecipitation with an anti-p300 antibody, followed by immunoblotting with an anti-vIRF antibody. A whole-cell lysate was used to show vIRF expression (bottom panel). (C) In vitro interaction of vIRF with p300. flag-tagged p300 and vIRF proteins were purified using an antiflag antibody column, eluted by a competing peptide, and dialyzed overnight. After 30 min of incubation of the purified flag-p300 and flag-vIRF proteins (lane 2) or the purified flag-vIRF protein alone (lane 3), an anti-p300 antibody was used to precipitate p300 complexes. Immune complexes were resolved by SDS–8% PAGE, followed by immunoblotting with an antiflag antibody. A mixture of purified flag p300 and flag vIRF protein (lane 1) was used as a control.
Identification of the region of vIRF required for p300.
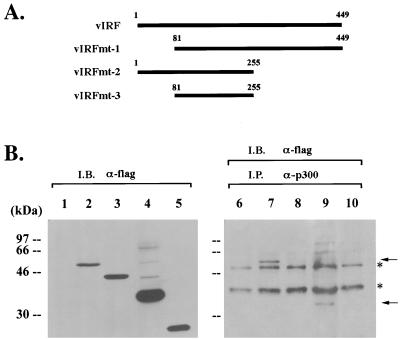
Cellular IRFs contain a conserved DNA-binding domain at the amino terminus and a divergent regulatory domain at the carboxyl terminus (10). The amino terminus of vIRF shows significant homology to the amino-terminal DNA-binding domain of IRF, while the carboxyl terminus of vIRF is divergent from the carboxyl transactivator-repressor region of IRF (27). In addition, KSHV vIRF contains 80 amino acids at the amino terminus which are not homologous to cellular IRFs. Interestingly, this region contains six repeats of a proline-rich P(X)2-3P motif. To identify the regions of vIRF required for p300 interaction in vivo, three deletion mutations were generated as follows: vIRF mt1 has a deletion of the first 80 amino acid, which contain a unique proline-rich sequence; vIRF mt2 has a deletion of amino acid residues 255 to 449, which contain the divergent carboxyl terminus; and vIRF mt3 has both deletions (Fig. 2A). To demonstrate expression of these deletion mutants, flag-tagged vIRF mutants were cloned into the pcDNA3.1 vector and expressed in COS-1 cells. After transfection, whole-cell lysates were used for immunoblotting analysis with antiflag antibody. wt vIRF and the mutants were expressed at somewhat variable but still comparable levels in COS-1 cells (Fig. 2B). The same cell lysates were used for immunoprecipitation with an anti-p300 antibody, followed by immunoblotting with an antiflag antibody to detect vIRF associated with p300 (Fig. 2B). These results demonstrated that vIRF mt2 interacted with p300, whereas vIRF mt1 and vIRF mt3 did not. In addition, repeated experiments showed that, while vIRF mt2 was expressed at a higher level in COS-1 cells than was wt vIRF, the amount of vIRF mt2 associated with p300 was lower than that of wt vIRF (Fig. 2B and data not shown). This suggests that full-length vIRF has a higher binding affinity for p300 than does vIRF mt2. Mutational analysis demonstrates that the unique amino terminus of vIRF is required for interaction with p300, and the results also provide additional evidence for the specificity of the interaction of vIRF with p300.
FIG. 2.
Mutational analysis of vIRF binding to p300. (A) A summary of vIRF deletion mutants. Amino-terminally flag-tagged vIRF mutants were cloned into the pcDNA3.1 vector to allow for their expression in COS-1 cells. (B) Mutational analysis of vIRF binding to p300. COS-1 cells were transfected with wt vIRF and its mutants (mt1, mt2, and mt3). Cell lysates were used for immunoprecipitation (I.P.) with an anti-p300 antibody, followed by immunoblotting (I.B.) with an antiflag antibody to detect vIRF (lanes 6 to 10). The expression level of wt vIRF and its mutants was demonstrated by immunoblotting with an antiflag antibody (lanes 1 to 5). Lanes 1 and 6, vector alone; lanes 2 and 7, wt vIRF; lanes 3 and 8, vIRF mt1; lanes 4 and 9, vIRF mt2; lanes 5 and 10, vIRF mt3. Asterisks indicate the heavy chain of immunoglobulin and background, and arrows indicate wt vIRF and vIRF mt2 protein.
Inhibition of p300 HAT activity by vIRF.
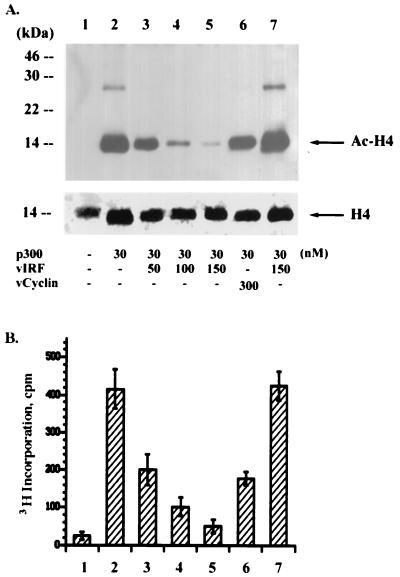
In addition to its role as a transcriptional adapter that integrates signals from many sequence-specific activators via direct interactions, p300 has HAT activity (16, 17, 42). This activity is thought to stimulate transcription by acetylating histones and disrupting nucleosomal structure when p300 is recruited to a specific promoter. To define the role of the interaction of vIRF with p300, we investigated whether this interaction altered p300 HAT activity. Purified full-length p300 was mixed with histone H4 and 3H-labeled acetyl-CoA in the presence or absence of purified vIRF protein. Purified KSHV v-cyclin protein was included as a control. HAT activity of p300 was measured by quantitating the 3H incorporation into histone H4 (Fig. 3B) and immunoblotting with an antibody that detected only the acetylated form of histone H4 (Fig. 3A). p300 HAT activity was dramatically reduced by increasing amounts of vIRF, whereas it was not significantly reduced by purified v-cyclin under the same conditions (Fig. 3). To determine if vIRF directly inhibits p300 HAT activity or instead itself has an intrinsic deacetylase activity, p300 protein was first mixed with histone H4 and 3H-labeled acetyl-CoA for 5 min, followed by incubation with vIRF protein for an additional 25 min (Fig. 3, lanes 7). Under these conditions, purified vIRF did not reduce the level of histone H4 acetylation by p300, indicating that vIRF itself does not have a deacetylase activity. These results demonstrate that, unlike interaction of cellular IRFs with p300, which leads to IFN-mediated nuclear signaling (48), the interaction of vIRF with p300 strongly inhibits p300 HAT activity.
FIG. 3.
Inhibition of p300 HAT activity by vIRF. Recombinant baculovirus containing the flag-tagged p300, vIRF, or v-cyclin was used to purify each protein from insect cells. Purified p300 protein (30 nM) was mixed with [3H]acetyl-CoA and histone H4 serving as substrates in the presence of increasing nanomolar amounts of vIRF or v-cyclin as indicated at the bottom of panel A. After 5 min, p300 HAT activity was measured by immunoblotting with an antibody specific for acetylated histone H4 (A) and quantitating radioactivity of 3H-labeled histone H4 (B). In lane 7, p300 protein was first mixed with the substrates for 5 min, followed by incubation with vIRF protein (150 nM) for an additional 25 min. The bottom panel of panel A shows the amount of histone H4 protein used in each reaction, detected by an anti-H4 antibody. The values in panel B represent the averages of three independent experiments.
vIRF competes with P/CAF for binding to p300.
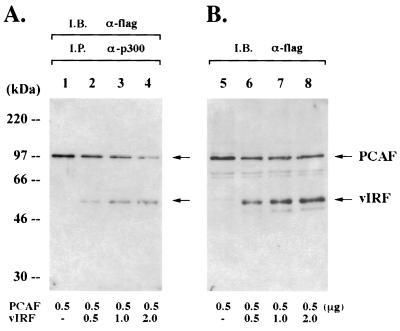
p300 not only has acetyltransferase activity but also interacts with p300/CBP-associated factor (P/CAF), another HAT. This interaction has been shown to be an important factor for cell cycle progression (1), differentiation (32), and IFN responses (24). In addition, adenovirus E1A disrupts the interaction between p300 and P/CAF, and this disruption leads to suppression of the IFN-α-activated antiviral response (4, 23, 40, 49). We further examined whether vIRF was able to displace P/CAF from p300 complexes. For this study, a constant amount of a flag-tagged P/CAF expression vector was transfected into COS-1 cells together with increasing amounts of a flag-tagged vIRF expression vector. Immune complexes of p300 were washed, separated by SDS-PAGE, and reacted with antiflag antibody to detect P/CAF and vIRF associated with p300. Consistent with previous findings (49), P/CAF strongly interacted with the endogenous p300 (Fig. 4A, lane 1). However, P/CAF binding to p300 decreased with increased vIRF binding to p300 (Fig. 4A). An antiflag immunoblotting assay of whole-cell lysates showed that P/CAF expression was not altered by the increased amount of vIRF expression (Fig. 4B). These results suggest that, similar to E1A, vIRF can compete with P/CAF for binding to p300.
FIG. 4.
Competition of vIRF with P/CAF for binding to p300. (A) The displacement of P/CAF from p300 complexes by vIRF. COS-1 cells were transfected with 0.5 μg of an expression vector containing the flag-tagged P/CAF gene together with an increasing amount of expression vector containing the flag-tagged vIRF gene as indicated at the bottom of the figure. At 48 h posttransfection, p300 complexes were precipitated with an anti-p300 antibody coupled to agarose beads, resolved by SDS-PAGE, transferred onto a nitrocellulose membrane, and reacted with an antiflag antibody. (B) Expression of P/CAF and vIRF. Whole-cell lysates of transfected COS-1 cells were used to determine the level of P/CAF and vIRF by immunoblotting with an antiflag antibody. I.B., immunoblotting; I.P., immunoprecipitation.
Alteration of chromosomal structure by vIRF.
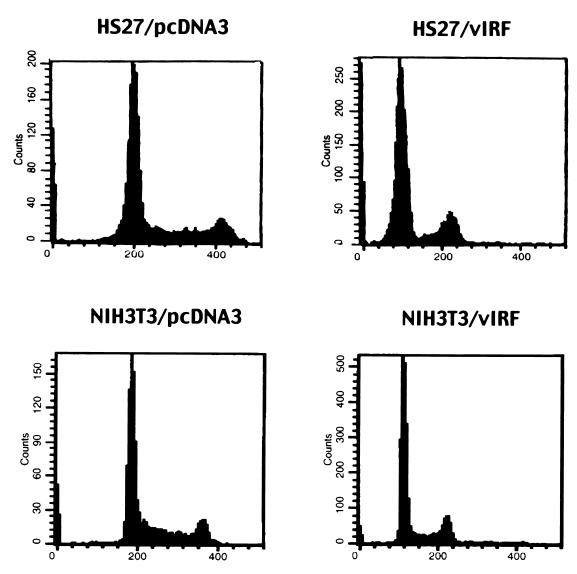
To examine the consequences of inhibition of p300 HAT activity and displacement of P/CAF from the p300 complexes by vIRF, we established human fibroblast HS27 cell lines stably expressing wt vIRF. Expression of vIRF was detected by immunoblotting assay with an anti-vIRF antibody (data not shown). Expression of vIRF resulted in morphological changes of HS27 cells. HS27 cells expressing vIRF (HS27/vIRF) were significantly larger than the control HS27 cells containing an empty expression vector (HS27/cDNA3) (data not shown). Since p300 and P/CAF have been shown to be important factors for cell cycle regulation, the effect of vIRF expression on cell cycle progression was investigated. Exponentially growing cells were stained with PI or Hoechst 33342 DNA intercalating dye and analyzed for cell cycle progression by flow cytometry. Other than a slight reduction of the cell number in S phase, no obvious alteration of cell cycle progression was detected in HS27/vIRF cells compared to control HS27/cDNA3 cells (Fig. 5). However, to our surprise, the level of chromosomal DNA staining of HS27/vIRF cells was approximately half that of the control HS27/cDNA3 cells (Fig. 5). DNA staining of HS27/cDNA3 cells at G1 phase peaked at 200 mean fluorescence intensity units (MFI), whereas that of HS27/vIRF cells at G1 phase peaked at 100 MFI; DNA staining of HS27/cDNA3 cells at G2/M phase peaked at 410 MFI, whereas that of HS27/vIRF cells at G2/M phase peaked at 210 MFI (Fig. 5). Previous experiments have shown that the expression of vIRF in mouse fibroblast NIH 3T3 cells results in the downregulation of IFN-mediated signaling and the alteration of cell growth control (21). Cell cycle analysis of these cells also showed that the expression of wt vIRF led to a marked reduction in the level of chromosomal DNA staining (Fig. 5). Cell cycle analysis with Hoechst 33342 dye produced essentially the same results as seen with PI dye (data not shown).
FIG. 5.
Cell cycle analysis of vIRF-expressing cells. Exponentially growing cells were stained for chromosomal DNA with PI and were analyzed on a FACScan flow cytometer. Results presented are representative of five individual experiments using two independently established HS27 and NIH 3T3 cell lines.
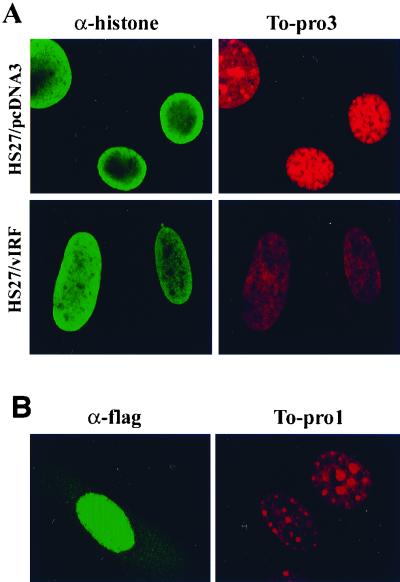
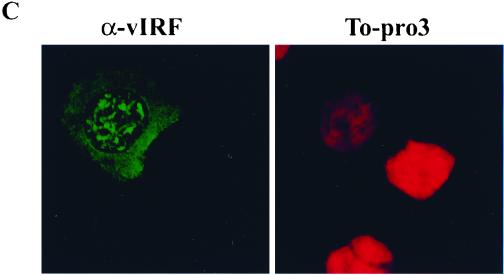
To further document the reduction of chromosomal DNA staining in vIRF-expressing cells, HS27 cells were stained with two different DNA intercalating dyes, To-Pro 1 and 3, which have a high affinity with double-stranded DNA and are detectable by confocal microscopy. To determine the localization of nucleosomal histones, cells were also costained with an antihistone antibody which reacts with all forms of histones. Consistent with flow cytometry results, confocal microscope analysis also showed that the level of DNA staining with To-Pro 1 and To-Pro 3 dyes in HS27/vIRF cells was drastically reduced compared to that in control HS27/cDNA3 cells (Fig. 6A; see also Fig. 9). Strong punctate chromosomal DNA staining was detected in HS27/cDNA3 cells, whereas weak diffuse staining was detected in HS27/vIRF cells. In addition, the localization of nucleosomal histones in HS27/vIRF cells was found to be somewhat different from that in control HS27/cDNA3 cells. The nucleosomal histones in HS27/cDNA3 cells were densely localized at the edge of the nucleus, whereas they were evenly distributed throughout the nucleus of HS27/vIRF cells (Fig. 6A). To further investigate whether this phenotype was solely attributable to vIRF expression and not to other changes introduced during the construction of these cell lines, HS27 cells were transiently transfected with an expression plasmid containing the flag-tagged vIRF gene, reacted with To-Pro 1 DNA staining dye and antiflag antibody, and examined by confocal microscopy. Immunofluorescence testing with an antiflag antibody showed the nuclear localization of vIRF in HS27 cells (left panel of Fig. 6B). The cell with vIRF expression (i.e., the left cell in Fig. 6B) showed a twofold reduction of chromosomal DNA staining in comparison to the cell without vIRF expression (i.e., the right cell in Fig. 6B). Finally, the level of chromosomal DNA staining in BCBL-1 cells with vIRF expression was also dramatically reduced compared to that in KSHV-infected BCBL-1 cells without vIRF expression (Fig. 6C). Thus, both flow cytometry and confocal microscopy showed that vIRF expression in human and mouse fibroblast cells and KSHV-infected BCBL-1 cells resulted in a significant reduction of chromosomal staining by DNA intercalating dyes.
FIG. 6.
Confocal microscopic analysis of chromosomal DNA staining. (A) Reduction of chromosomal DNA staining by the stable expression of vIRF. HS27/cDNA3 and HS27/vIRF cells were stained with To-Pro 3 dye or antihistone antibody. Stained cells were examined under the Leica confocal immunofluorescence microscope. (B) Reduction of chromosomal DNA staining by the transient expression of vIRF. HS27 cells were transfected with an expression vector containing flag-tagged vIRF. At 48 h posttransfection, HS27 cells were stained with antiflag antibody (left panel) and To-Pro 1 dye (right panel). Stained cells were examined under the Leica confocal immunofluorescence microscope. Comparison of two cells in this panel shows that the left one had vIRF expression, as shown in positive green immunofluorescence with antiflag antibody, whereas the right one did not have vIRF expression, as shown in negative immunofluorescence staining with antiflag antibody. (C) Reduction of chromosomal DNA staining by vIRF expression in BCBL-1 cells. BCBL-1 cells were stimulated with TPA for 48 h, fixed with 1% paraformaldehyde, and stained with anti-vIRF antibody (left panel) and To-Pro 3 dye (right panel). Stained BCBL-1 cells were examined under the Leica confocal immunofluorescence microscope. Among the cells in this panel, the BCBL-1 cell at left had vIRF expression, as shown in positive green immunofluorescence with anti-vIRF antibody, whereas the other cells did not have vIRF expression, as shown in negative immunofluorescence staining with anti-vIRF antibody.
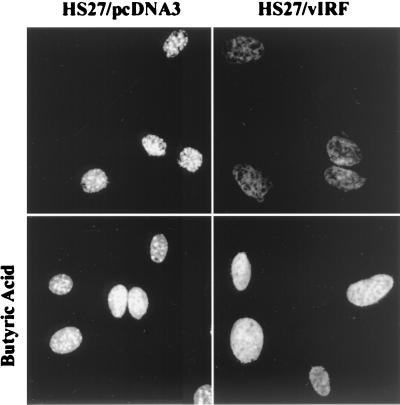
FIG. 9.
Enhanced DNA staining of vIRF-expressing cells by butyric acid treatment. HS27/cDNA3 and HS27/vIRF cells were treated with butyric acid (5 mM) overnight or mock treated. Cells were stained with To-Pro 1 dye for 1 min and examined under the Leica confocal microscope.
Hypoacetylation of nucleosomal histones by vIRF expression.
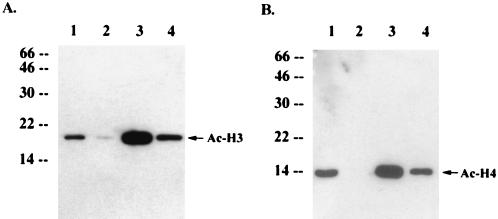
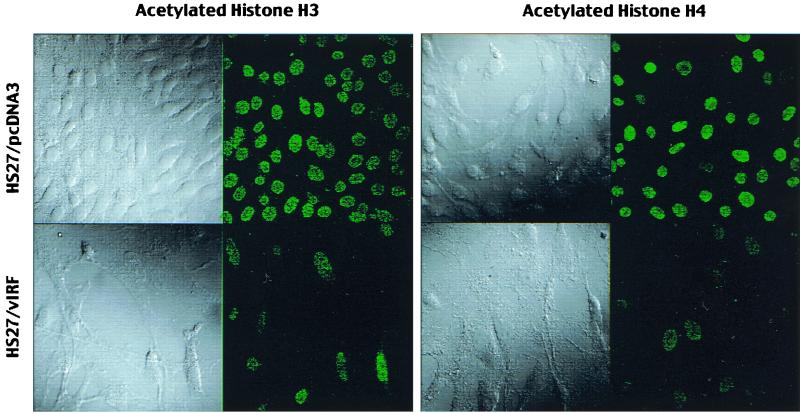
Spectrophotometry and agarose DNA gel analysis with purified chromosomal DNA showed that HS27/vIRF cells contained an amount of chromosomal DNA per cell equivalent to that of HS27/cDNA3 cells (data not shown). This suggests that the reduction of the level of chromosomal staining is due to factors other than DNA content in vIRF-expressing cells. We hypothesized that inhibition of p300 HAT by vIRF led to the reduction of cellular histone acetylation, resulting in an alteration of chromatin structure which limited accessibility of DNA staining dyes. To test this hypothesis, we first examined in vivo acetylation of the nucleosomal histones H3 and H4 using immunoblotting analysis with antibodies that specifically reacted with the acetylated forms of histones H3 and H4. These experiments revealed that the levels of in vivo acetylation of nucleosomal histones H3 and H4 were drastically reduced in HS27/vIRF cells compared to that in control HS27/cDNA3 cells (Fig. 7, lanes 1 and 2). This result was confirmed by immunofluorescence tests using the same antibodies (Fig. 8). These results demonstrate that vIRF expression leads to drastic hypoacetylation of nucleosomal histones.
FIG. 7.
Alteration of in vivo histone H3 and H4 acetylation by vIRF expression or butyric acid treatment. Identical amounts of proteins from HS27/cDNA3 cells (lanes 1 and 3) and HS27/vIRF cells (lanes 2 and 4) treated with butyric acid overnight (lanes 3 and 4) or mock treated (lanes 1 and 2) were used for immunoblotting analysis with antibodies specific for the acetylated histone H3 (A) or H4 (B). Arrows indicate acetylated histones H3 (Ac-H3) and H4 (Ac-H4). Numbers at left of each panel show sizes in kilodaltons.
FIG. 8.
Immunofluorescence test of in vivo histone H3 and H4 acetylation. HS27/cDNA3 and HS27/vIRF cells were stained with antibodies which specifically reacted with the acetylated forms of histones H3 and H4. Cells were visualized with Nomarski optics. Immunofluorescence testing was performed with a Leica confocal immunofluorescence microscope.
Treatment of cells with butyric acid and trichostatin specifically inhibits histone deacetylase activity and leads to a general hyperacetylation of nucleosomes and selective activation of cellular genes (33, 50). To recreate chromatin acetylation in vivo, HS27/cDNA3 and HS27/vIRF cells were treated with butyric acid overnight and cell lysates were reacted with antibodies specific for the acetylated forms of histones H3 and H4. Butyric acid treatment of HS27/vIRF cells not only restored in vivo acetylation of nucleosomal histones H3 and H4 (Fig. 7, lanes 4) but also resulted in a marked increase of chromosomal DNA staining (Fig. 9). Treatment with trichostatin produced essentially the same results as those shown with butyric acid (data not shown).
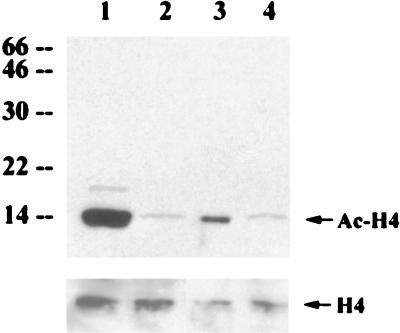
To demonstrate the role of p300 in histone acetylation, an expression vector containing wt p300 HAT or a p300 ΔHAT mutant containing a deletion of the HAT catalytic domain was transfected into HS27/vIRF cells. Overexpression of wt p300 significantly increased the level of histone H4 acetylation in HS27/vIRF cells, whereas overexpression of p300 ΔHAT mutant did not (Fig. 10, lanes 3 and 4). Thus, these results support the notion that vIRF expression leads to hypoacetylation of nucleosomal histones H3 and H4, resulting in global alteration of the chromatin structure which limits accessibility of DNA dyes. This effect could be reversed by specific deacetylase inhibitors, butyric acid and trichostatin, or overexpression of wt p300 HAT.
FIG. 10.
Recovery of histone acetylation by overexpression of wt p300 HAT. HS27/vIRF cells were transfected with wt p300 (lane 3) or p300 ΔHAT mutant (lane 4). HS27/cDNA3 (lane 1) and HS27/vIRF (lane 2) were included as controls. Identical amounts of proteins from cell lysates were used for immunoblotting analysis with an antibody specific for the acetylated histone H4. The bottom panel shows the amount of cellular histone H4 protein in each lane, detected by an anti-H4 antibody. Arrows indicate the acetylated form of histone H4 (Ac-H4) or total histone H4 (H4). Numbers at left are molecular masses in kilodaltons.
Alteration of MIF gene expression by vIRF.
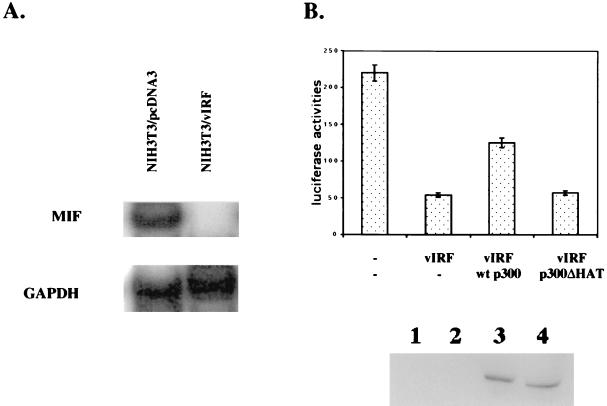
p300/CBP plays a role in expression of cellular cytokine genes including macrophage MIF (46). To investigate the effect of vIRF on expression of MIF, mRNA was extracted from NIH 3T3/cDNA3 and NIH 3T3/vIRF cells and subjected to an RNase protection assay with the in vitro-transcribed 32P-labeled MIF probe or GAPDH probe (Fig. 11A). It showed that MIF expression was drastically reduced by vIRF, whereas GAPDH expression was not altered under the same conditions (Fig. 11A). To further investigate the effect of vIRF on the promoter activity of the MIF gene, we cloned the bp −1033 to +63 sequence of the MIF promoter region into a luciferase reporter vector. Computer analysis of the 5′-flanking sequence of the MIF gene has revealed that it contains multiple potential transcriptional factor binding sequences (46). The MIF promoter-luciferase reporter was transfected into NIH 3T3 cells together with an expression vector containing wt vIRF. As seen in the RNase protection assay (Fig. 11A), wt vIRF expression suppressed the MIF promoter activity by four- to fivefold (Fig. 11B). To further examine the role of p300 HAT in the promoter activity of the MIF gene, an expression vector containing wt p300 HAT or a p300 ΔHAT mutant containing a deletion of the HAT catalytic domain was included in the reporter assay. Expression of wt p300 partially reversed the vIRF effect on the MIF promoter activity, whereas expression of p300 ΔHAT mutant did not (Fig. 11B). wt p300 and the p300 ΔHAT mutant were expressed at equivalent levels in these cells (Fig. 11B). Thus, these results suggest that hypoacetylation of nucleosomal histones H3 and H4 by vIRF likely results in suppression of MIF gene expression. Furthermore, this effect can be partially reversed by overexpression of wt p300, but not by that of the p300 ΔHAT mutant.
FIG. 11.
Inhibition of cellular MIF gene expression by vIRF. (A) Inhibition of cellular MIF gene expression by vIRF. Total RNA was extracted from NIH 3T3/cDNA3 and NIH 3T3/vIRF cells and used for RNase protection assays with the in vitro-transcribed 32P-labeled MIF or GAPDH probe. The protected fragments were resolved by electrophoresis on a 5% acrylamide–urea gel, and autoradiograms were developed in a Fuji Phospho Imager. Detailed procedures are described in Materials and Methods. (B) Alteration of the promoter activity of MIF by vIRF. MIF-luc reporter plasmid (0.25 μg) and pGKβgal reporter plasmid (0.25 μg) were transfected into NIH 3T3 cells together with 0.25 μg of vIRF, wt p300, or p300 ΔHAT expression vector as indicated. Luciferase was measured at 48 h posttransfection, and luciferase values were normalized by β-galactosidase activity. Values for luciferase activity represent the averages of three independent experiments. Lysates of transfected NIH 3T3 cells were used to determine the level of p300 and p300 ΔHAT mutant by immunoblotting with an anti-p300 antibody. Lane 1, NIH 3T3 cells transfected with vector alone; lane 2, NIH 3T3 cells transfected with vIRF; lane 3, NIH 3T3 cells transfected with vIRF and p300; lane 4, NIH 3T3 cells transfected with vIRF and p300 ΔHAT.
DISCUSSION
In this report, we demonstrate that KSHV vIRF interacts with the p300 transcriptional coactivator and that this interaction significantly inhibits p300 HAT activity. p300 is a global transcriptional adapter which contains HAT activity (17, 30). To date, four families of nuclear proteins have been described that possess HAT activity. These include GCN5 and P/CAF (5), SRC1 and ACTR (9, 41), p300 and CBP (3, 49), and TAFII250 (26). Virus infection leads to an unusually stable association of IRFs with p300, which is important for the expression of IFN-α/β genes as well as a subset of genes involved in antiviral responses (25, 47). IFN-γ induces rapid activation of the latent transcription factor, STAT, which translocates to the nucleus and also forms a complex with p300 to participate in IFN-γ-induced transcription (51). The adenovirus E1A and simian virus 40 large T-antigen oncoproteins bind to the p300/CBP transcriptional coactivator family (13, 23). Specifically, interaction of E1A with p300 has been shown to be important for blocking IFN signaling (23, 40, 51). Thus, our results suggest that disruption of the interaction between cellular factors and p300 and the inactivation of p300 HAT activity are likely common mechanisms employed by adenovirus E1A and KSHV vIRF to circumvent IFN-mediated host defense functions.
Recently, two other reports have also demonstrated an interaction of vIRF with p300/CBP (6, 18). In contrast to our study, in which the amino-terminal region of vIRF is found to be required for binding to p300 in vivo, Burysek et al. have demonstrated that the carboxyl-terminal region of vIRF is required for binding to p300 in vitro GST pull-down assays (6). Although it needs to be further characterized, this discrepancy may be derived from different assay conditions: in vivo versus in vitro and/or full-length protein versus GST fusion protein. Regardless of this discrepancy, it is now clear that KSHV vIRF targets the p300/CBP transcriptional cofactor and inhibits its HAT activity. While both adenovirus E1A and KSHV vIRF inhibit HAT activity of p300, they interact with the different domains of p300; E1A interacts with the HAT domain of p300 (7) and vIRF interacts with the C/H2 domain of p300 (18). This suggests that the interaction of vIRF with the C/H2 domain of p300 may stearically hinder HAT enzymatic activity of p300. Thus, the continuously growing list of viral proteins which apparently interact and interfere with p300/CBP cellular transcriptional cofactor suggests that p300/CBP is an important cellular factor in controlling viral infection and propagation.
Gene activation in response to extracellular signals, environmental stresses, or infection by pathogens requires highly integrated signal transduction pathways that direct the transcriptional machinery to the appropriate sets of genes. Specifically, histone acetylation plays an important role in the modulation of chromatin structure associated with signal-dependent transcriptional activation (42). Here, we provide biochemical and functional evidence demonstrating that KSHV vIRF is a potent inhibitor of the HAT activity of p300. A simple but attractive hypothesis is that vIRF suppresses the targeted nucleosomal histone acetylation achieved by recruitment of acetyltransferases to the signal-responsive promoters. Removal of acetyl moieties from nucleosomal histones by vIRF leads to an alteration of chromatin structure, thereby modulating cellular gene expression. Recent reports (6, 18) and our unpublished study show that expression of cellular IL-6 and IFNs is significantly reduced by vIRF, whereas expression of transforming growth factors β1 and β3 and myc is drastically induced by vIRF. In fact, some of these genes have been shown to be regulated by acetylation of nucleosomal histone (31, 45). Furthermore, our microarray analysis has found that vIRF expression alters the transcriptional profile of 7% of genes among 4,500 cellular genes by at least twofold (unpublished results). These results also indicate that the hypoacetylation of nucleosomal histones by vIRF leads to a global effect on cellular gene expression. Characterization of these cellular genes whose expression is altered by vIRF will further increase our understanding of a role of p300/CBP in transcriptional regulation and will identify the cellular targets of p300/CBP.
Antiviral responses include apoptosis, cell cycle arrest, and enhanced immunological surveillance, which are all focused toward preventing tumor cell growth. Cellular IRFs interact with and recruit the p300 transcriptional coactivator to specific promoters to induce transcriptional activation in response to virus infection. It is now apparent that KSHV employs an elaborate decoy mechanism by harboring vIRFs to circumvent these antiviral defenses and to facilitate uncontrolled cell proliferation. In addition to this activity, vIRF has been shown to play an important role in KSHV viral gene expression (21). We have found that vIRF induces drastic alteration of viral gene expression of KSHV and herpesvirus saimiri (unpublished data). Further studies of the detailed role of vIRFs in the regulation of cellular and viral gene expression are likely to be important clues for a better understanding of the regulation of the KSHV life cycle and pathogenesis.
ACKNOWLEDGMENTS
We thank R. Desrosiers, K. Williams, B. Means, and L. Alexander for critical reading of the manuscript. We also thank B. Roy for manuscript preparation, K. Toohey for photography support, and M. DeMaria for flow cytometry analysis.
This work was supported by U.S. Public Health Service grants CA31363, CA82057, CA86841, AI38131, and RR00168 and ACS grant RPG001102. Jae Jung is a Leukemia and Lymphoma Society Scholar, and B. Damania is a fellow of the Cancer Research Institute.
REFERENCES
- 1.Ait-Si-Ali S, Ramirez S, Barre F X, Dkhissi F, Magnaghi-Jaulin L, Girault J A, Robin P, Knibiehler M, Pritchard L L, Ducommun B, Trouche D, Harel-Bellan A. Histone acetyltransferase activity of CBP is controlled by cycle-dependent kinases and oncoprotein E1A. Nature. 1998;396:184–186. doi: 10.1038/24190. [DOI] [PubMed] [Google Scholar]
- 2.Arvanitakis L, Geras-Raaka E, Varma A, Gershengorn M C, Cesarman E. Human herpesvirus KSHV encodes a constitutively active G-protein-coupled receptor linked to cell proliferation. Nature. 1997;385:347–350. doi: 10.1038/385347a0. [DOI] [PubMed] [Google Scholar]
- 3.Bannister A J, Kouzarides T. The CBP co-activator is a histone acetyltransferase. Nature. 1996;384:641–643. doi: 10.1038/384641a0. [DOI] [PubMed] [Google Scholar]
- 4.Bhattacharya S, Eckner R, Grossman S, Oldread E, Arany Z, D'Andrea A, Livingston D M. Cooperation of Stat2 and p300/CBP in signalling induced by interferon-alpha. Nature. 1996;383:344–347. doi: 10.1038/383344a0. [DOI] [PubMed] [Google Scholar]
- 5.Brownell J E, Zhou J, Ranalli T, Kobayashi R, Edmondson D G, Roth S Y, Allis C D. Tetrahymena histone acetyltransferase A: a homolog to yeast Gcn5p linking histone acetylation to gene activation. Cell. 1996;84:843–851. doi: 10.1016/s0092-8674(00)81063-6. [DOI] [PubMed] [Google Scholar]
- 6.Burysek L, Yeow W S, Lubyova B, Kellum M, Schafer S L, Huang Y Q, Pitha P M. Functional analysis of human herpesvirus 8-encoded viral interferon regulatory factor 1 and its association with cellular interferon regulatory factors and p300. J Virol. 1999;73:7334–7342. doi: 10.1128/jvi.73.9.7334-7342.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chakravarti D, Ogryzko V, Kao H Y, Nash A, Chen H, Nakatani Y, Evans R M. A viral mechanism for inhibition of p300 and PCAF acetyltransferase activity. Cell. 1999;96:393–403. doi: 10.1016/s0092-8674(00)80552-8. [DOI] [PubMed] [Google Scholar]
- 8.Chang Y, Cesarman E, Pessin M S, Lee F, Culpepper J, Knowles D M, Moore P S. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi's sarcoma. Science. 1994;266:1865–1869. doi: 10.1126/science.7997879. [DOI] [PubMed] [Google Scholar]
- 9.Chen H, Lin R J, Schiltz R L, Chakravarti D, Nash A, Nagy L, Privalsky M L, Nakatani Y, Evans R M. Nuclear receptor coactivator ACTR is a novel histone acetyltransferase and forms a multimeric activation complex with P/CAF and CBP/p300. Cell. 1997;90:569–580. doi: 10.1016/s0092-8674(00)80516-4. [DOI] [PubMed] [Google Scholar]
- 10.Darnell J E, Jr, Kerr I M, Stark G R. Jak-STAT pathways and transcriptional activation in response to IFNs and other extracellular signaling proteins. Science. 1994;264:1415–1421. doi: 10.1126/science.8197455. [DOI] [PubMed] [Google Scholar]
- 11.David M. Transcription factors in interferon signaling. Pharmacol Ther. 1995;65:149–161. doi: 10.1016/0163-7258(94)00050-d. [DOI] [PubMed] [Google Scholar]
- 12.Desrosiers R C, Sasseville V G, Czajak S C, Zhang X, Mansfield K G, Kaur A, Johnson R P, Lackner A A, Jung J U. A herpesvirus of rhesus monkeys related to the human Kaposi's sarcoma-associated herpesvirus. J Virol. 1997;71:9764–9769. doi: 10.1128/jvi.71.12.9764-9769.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eckner R, Ludlow J W, Lill N L, Oldread E, Arany Z, Modjtahedi N, DeCaprio J A, Livingston D M, Morgan J A. Association of p300 and CBP with simian virus 40 large T antigen. Mol Cell Biol. 1996;16:3454–3464. doi: 10.1128/mcb.16.7.3454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gao S-J, Boshoff C, Jayachandra S, Weiss R A, Chang Y, Moore P S. KSHV ORF K9 (vIRF) is an oncogene which inhibits the interferon signaling pathway. Oncogene. 1997;15:1979–1985. doi: 10.1038/sj.onc.1201571. [DOI] [PubMed] [Google Scholar]
- 15.Godden-Kent D, Talbot S J, Boshoff C, Chang Y, Moore P, Weiss R A, Mittnacht S. The cyclin encoded by Kaposi's sarcoma-associated herpesvirus stimulates cdk6 to phosphorylate the retinoblastoma protein and histone H1. J Virol. 1997;71:4193–4198. doi: 10.1128/jvi.71.6.4193-4198.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grunstein M. Histone acetylation in chromatin structure and transcription. Nature. 1997;389:349–352. doi: 10.1038/38664. [DOI] [PubMed] [Google Scholar]
- 17.Janknecht R, Hunter T. Transcription. A growing coactivator network. Nature. 1996;383:22–23. doi: 10.1038/383022a0. [DOI] [PubMed] [Google Scholar]
- 18.Jayachandra S, Low K G, Thlick A E, Yu J, Ling P D, Chang Y, Moore P S. Three unrelated viral transforming proteins (vIRF, EBNA2, and E1A) induce the MYC oncogene through the interferon-responsive PRF element by using different transcription coadaptors. Proc Natl Acad Sci USA. 1999;96:11566–11571. doi: 10.1073/pnas.96.20.11566. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 19.Kledal T N, Rosenkilde M M, Coulin F, Simmons G, Johnsen A H, Alouani S, Power C A, Lüttichau H R, Gerstoft J, Clapham P R, Clark-Lewis I, Wells T N C, Schwartz T W. A broad-spectrum chemokine antagonist encoded by Kaposi's sarcoma-associated herpesvirus. Science. 1997;277:1656–1659. doi: 10.1126/science.277.5332.1656. [DOI] [PubMed] [Google Scholar]
- 20.Kwok R P, Lundblad J R, Chrivia J C, Richards J P, Bachinger H P, Brennan R G, Roberts S G, Green M R, Goodman R H. Nuclear protein CBP is a coactivator for the transcription factor CREB. Nature. 1994;370:223–226. doi: 10.1038/370223a0. [DOI] [PubMed] [Google Scholar]
- 21.Li M, Lee H, Guo J, Neipel F, Fleckenstein B, Ozato K, Jung J U. Kaposi's sarcoma-associated herpesvirus viral interferon regulatory factor. J Virol. 1998;72:5433–5440. doi: 10.1128/jvi.72.7.5433-5440.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li M, Lee H, Yoon D-W, Albrecht J-C, Fleckenstein B, Neipel F, Jung J U. Kaposi's sarcoma-associated herpesvirus encodes a functional cyclin. J Virol. 1997;71:1984–1991. doi: 10.1128/jvi.71.3.1984-1991.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lundblad J R, Kwok R P, Laurance M E, Harter M L, Goodman R H. Adenoviral E1A-associated protein p300 as a functional homologue of the transcriptional co-activator CBP. Nature. 1995;374:85–88. doi: 10.1038/374085a0. [DOI] [PubMed] [Google Scholar]
- 24.Masumi A, Wang I M, Lefebvre B, Yang X J, Nakatani Y, Ozato K. The histone acetylase PCAF is a phorbol-ester-inducible coactivator of the IRF family that confers enhanced interferon responsiveness. Mol Cell Biol. 1999;19:1810–1820. doi: 10.1128/mcb.19.3.1810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Merika M, Williams A J, Chen G, Collins T, Thanos D. Recruitment of CBP/p300 by the IFNβ enhanceosome is required for synergistic activation of transcription. Mol Cell. 1998;1:277–287. doi: 10.1016/s1097-2765(00)80028-3. [DOI] [PubMed] [Google Scholar]
- 26.Mizzen C A, Yang X J, Kokubo T, Brownell J E, Bannister A J, Owen-Hughes T, Workman J, Wang L, Berger S L, Kouzarides T, Nakatani Y, Allis C D. The TAF(II)250 subunit of TFIID has histone acetyltransferase activity. Cell. 1996;87:1261–1270. doi: 10.1016/s0092-8674(00)81821-8. [DOI] [PubMed] [Google Scholar]
- 27.Moore P S, Boshoff C, Weiss R A, Chang Y. Molecular mimicry of human cytokine and cytokine response pathway genes by KSHV. Science. 1996;274:1739–1744. doi: 10.1126/science.274.5293.1739. [DOI] [PubMed] [Google Scholar]
- 28.Neipel F, Albrecht J-C, Ensser A, Huang Y-Q, Li J J, Friedman-Kien A E, Fleckenstein B. Human herpesvirus 8 encodes a homolog of interleukin-6. J Virol. 1997;71:839–842. doi: 10.1128/jvi.71.1.839-842.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nicholas J, Ruvolo V R, Burns W H, Sandford G, Wan X, Ciufo D, Hendrickson S B, Guo H-G, Hayward G S, Reitz M S. Kaposi's sarcoma-associated human herpesvirus-8 encodes homologues of macrophage inflammatory protein-1 and interleukin-6. Nat Med. 1997;3:287–292. doi: 10.1038/nm0397-287. [DOI] [PubMed] [Google Scholar]
- 30.Ogryzko V V, Schiltz R L, Russanova V, Howard B H, Nakatani Y. The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell. 1996;87:953–959. doi: 10.1016/s0092-8674(00)82001-2. [DOI] [PubMed] [Google Scholar]
- 31.Parekh B S, Maniatis T. Virus infection leads to localized hyperacetylation of histones H3 and H4 at the IFN-beta promoter. Mol Cell. 1999;3:125–129. doi: 10.1016/s1097-2765(00)80181-1. [DOI] [PubMed] [Google Scholar]
- 32.Puri P L, Sartorelli V, Yang X J, Hamamori Y, Ogryzko V V, Howard B H, Kedes L, Wang J Y, Graessmann A, Nakatani Y, Levrero M. Differential roles of p300 and PCAF acetyltransferases in muscle differentiation. Mol Cell. 1997;1:35–45. doi: 10.1016/s1097-2765(00)80005-2. [DOI] [PubMed] [Google Scholar]
- 33.Riggs M G, Whittaker R G, Neumann J R, Ingram V M. n-Butyrate causes histone modification in HeLa and Friend erythroleukaemia cells. Nature. 1977;268:462–464. doi: 10.1038/268462a0. [DOI] [PubMed] [Google Scholar]
- 34.Rose T M, Strand K B, Schultz E R, Schaefer G, Rankin G W, Jr, Thouless M E, Tsai C C, Bosch M L. Identification of two homologs of the Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) in retroperitoneal fibromatosis of different macaque species. J Virol. 1997;71:4138–4144. doi: 10.1128/jvi.71.5.4138-4144.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Russo J J, Bohenzxy R A, Chien M-C, Chen J, Yan M, Maddalena D, Parry J P, Peruzzi D, Edelman I S, Chang Y, Moore P S. Nucleotide sequence of the Kaposi's sarcoma-associated herpesvirus (HHV8) Proc Natl Acad Sci USA. 1996;93:14862–14867. doi: 10.1073/pnas.93.25.14862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sarid R, Sato T, Bohenzky R A, Russo J J, Chang Y. Kaposi's sarcoma-associated herpesvirus encodes a functional Bcl-2 homologue. Nat Med. 1997;3:293–298. doi: 10.1038/nm0397-293. [DOI] [PubMed] [Google Scholar]
- 37.Schulz T F, Chang Y, Moore P S. Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) In: McCance D J, editor. Human tumor viruses. Washington, D.C.: ASM Press; 1998. pp. 87–134. [Google Scholar]
- 38.Searles R P, Bergquam E P, Axthelm M K, Wong S W. Sequence and genomic analysis of a rhesus macaque rhadinovirus with similarity to Kaposi's sarcoma-associated herpesvirus/human herpesvirus 8. J Virol. 1999;73:3040–3053. doi: 10.1128/jvi.73.4.3040-3053.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sen G C, Ransohoff R M. Interferon-induced antiviral actions and their regulation. Adv Virus Res. 1992;42:57–102. doi: 10.1016/s0065-3527(08)60083-4. [DOI] [PubMed] [Google Scholar]
- 40.Somasundaram K, El-Deiry W S. Inhibition of p53-mediated transactivation and cell cycle arrest by E1A through its p300/CBP-interacting region. Oncogene. 1997;14:1047–1057. doi: 10.1038/sj.onc.1201002. [DOI] [PubMed] [Google Scholar]
- 41.Spencer T E, Jenster G, Burcin M M, Allis C D, Zhou J, Mizzen C A, McKenna N J, Onate S A, Tsai S Y, Tsai M J, O'Malley B W. Steroid receptor coactivator-1 is a histone acetyltransferase. Nature. 1997;389:194–198. doi: 10.1038/38304. [DOI] [PubMed] [Google Scholar]
- 42.Struhl K. Histone acetylation and transcriptional regulatory mechanisms. Genes Dev. 1998;12:599–606. doi: 10.1101/gad.12.5.599. [DOI] [PubMed] [Google Scholar]
- 43.Thome M, Schneider P, Hofmann K, Fickenscher H, Meinl E, Neipel F, Mattmann C, Burns K, Bodmer J-L, Schröter M, Scaffidi C, Krammer P H, Peter M E, Tschopp J. Viral FLICE-inhibitory proteins (FLIPs) prevent apoptosis induced by death receptors. Nature. 1997;386:517–521. doi: 10.1038/386517a0. [DOI] [PubMed] [Google Scholar]
- 44.Utley R T, Ikeda K, Grant P A, Cote J, Steger D J, Eberharter A, John S, Workman J L. Transcriptional activators direct histone acetyltransferase complexes to nucleosomes. Nature. 1998;394:498–502. doi: 10.1038/28886. [DOI] [PubMed] [Google Scholar]
- 45.Vanden Berghe W, De Bosscher K, Boone E, Plaisance S, Haegeman G. The nuclear factor-kappaB engages CBP/p300 and histone acetyltransferase activity for transcriptional activation of the interleukin-6 gene promoter. J Biol Chem. 1999;274:32091–32098. doi: 10.1074/jbc.274.45.32091. [DOI] [PubMed] [Google Scholar]
- 46.Waeber G, Thompson N, Chautard T, Steinmann M, Nicod P, Pralong F P, Calandra T, Gaillard R C. Transcriptional activation of the macrophage migration-inhibitory factor gene by the corticotropin-releasing factor is mediated by the cyclic adenosine 3′,5′-monophosphate responsive element-binding protein CREB in pituitary cells. Mol Endocrinol. 1998;12:698–705. doi: 10.1210/mend.12.5.0109. [DOI] [PubMed] [Google Scholar]
- 47.Wathelet M G, Lin C H, Parekh B S, Ronco L V, Howley P M, Maniatis T. Virus infection induces the assembly of coordinately activated transcription factors on the IFN-β enhancer in vivo. Mol Cell. 1998;1:507–518. doi: 10.1016/s1097-2765(00)80051-9. [DOI] [PubMed] [Google Scholar]
- 48.Weaver B K, Kumar K P, Reich N C. Interferon regulatory factor 3 and CREB-binding protein/p300 are subunits of double-stranded RNA-activated transcription factor DRAF1. Mol Cell Biol. 1998;18:1359–1368. doi: 10.1128/mcb.18.3.1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yang X J, Ogryzko V V, Nishikawa J, Howard B H, Nakatani Y. A p300/CBP-associated factor that competes with the adenoviral oncoprotein E1A. Nature. 1996;382:319–324. doi: 10.1038/382319a0. [DOI] [PubMed] [Google Scholar]
- 50.Yoshida M, Kijima M, Akita M, Beppu T. Potent and specific inhibition of mammalian histone deacetylase both in vivo and in vitro by trichostatin A. J Biol Chem. 1990;265:17174–17179. [PubMed] [Google Scholar]
- 51.Zhang J J, Vinkemeier U, Gu W, Chakravarti D, Horvath C M, Darnell J E J. Two contact regions between Stat1 and CBP/p300 in interferon gamma signaling. Proc Natl Acad Sci USA. 1996;93:15092–15096. doi: 10.1073/pnas.93.26.15092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zimring J C, Goodbourn S, Offermann M K. Human herpesvirus 8 encodes an interferon regulatory factor (IRF) homolog that represses IRF-1-mediated transcription. J Virol. 1998;72:701–707. doi: 10.1128/jvi.72.1.701-707.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]