Abstract
Metagenomic analyses of microbial communities have revealed a large degree of interspecies and intraspecies genetic diversity through the reconstruction of metagenome assembled genomes (MAGs). Yet, metabolic modeling efforts mainly rely on reference genomes as the starting point for reconstruction and simulation of genome scale metabolic models (GEMs), neglecting the immense intra- and inter-species diversity present in microbial communities. Here, we present metaGEM (https://github.com/franciscozorrilla/metaGEM), an end-to-end pipeline enabling metabolic modeling of multi-species communities directly from metagenomes. The pipeline automates all steps from the extraction of context-specific prokaryotic GEMs from MAGs to community level flux balance analysis (FBA) simulations. To demonstrate the capabilities of metaGEM, we analyzed 483 samples spanning lab culture, human gut, plant-associated, soil, and ocean metagenomes, reconstructing over 14,000 GEMs. We show that GEMs reconstructed from metagenomes have fully represented metabolism comparable to isolated genomes. We demonstrate that metagenomic GEMs capture intraspecies metabolic diversity and identify potential differences in the progression of type 2 diabetes at the level of gut bacterial metabolic exchanges. Overall, metaGEM enables FBA-ready metabolic model reconstruction directly from metagenomes, provides a resource of metabolic models, and showcases community-level modeling of microbiomes associated with disease conditions allowing generation of mechanistic hypotheses.
INTRODUCTION
Whole metagenome shotgun sequencing and genome-resolved metagenomics allow for the exploration of personalized, context-specific microbial communities at a species or strain level resolution (1). Changes of microbiota composition are strongly linked to a range of diseases including cancer, behavioral, neurological, and metabolic disorders (2–7). However, a mechanistic understanding of the role of the human gut microbiome in disease, especially the roles of specific strains and the associated metabolic factors, remains challenging due to the vast intra- and inter-species diversity. To this end, short-read sequencing data allows for the extraction of metagenome assembled genomes (MAGs) directly from raw sequencing data, while avoiding culture-based methods or making use of the limited number of reference genomes, enabling the discovery of unknown species and the exploration of personalized microbiomes (8–10). Indeed, a number of attempts aiming to explore the human gut microbiome composition and diversity have generated hundreds of MAGs representing previously unknown or uncultured species, as well as thousands of known species MAGs (11–13). Pangenome analysis of the human gut microbiome demonstrated that the functional repertoire of gut species differ significantly, with a median core genome proportion of only 66% (14), revealing differences in metabolic potentials of individual microbiomes.
Attempts of mechanistic links between diet or inter-species interactions with microbiota composition and dynamics led to the development of gut species genome-scale metabolic models (15). GEnome-scale Metabolic models (GEMs) allow assessing species nutritional requirements (16) and their interactions in the human gut as well as in diverse environmental communities (17–20). The current paradigm of metabolic modeling typically relies on mapping identified taxa to their closest reference genomes. This limits analysis and its interpretation to the metabolic networks represented in the known reference genome space. This can cause false positives (i.e. pathways present in the reference but missing from the variant present in the community) as well as false negatives (i.e. pathways missing in the reference genomes but present in the community variant), ultimately leading to inaccurate predictions of individual species metabolism as well as that of cross-feeding interactions. Thus the current modelling attempts are likely failing to capture the specific metabolic features of a given species across different contexts, e.g. microbiota of individuals with different disease conditions. Towards overcoming this limitation, here we present metaGEM, the computational pipeline that enables reconstruction of sample specific metabolic models directly from short read metagenomics data. Instead of relying on reference genomes, metaGEM generates high quality metagenome assembled genomes, which are then used to reconstruct context-specific prokaryotic GEMs using state-of-the-art methodologies (Figure 1). Our contributions are 2-fold: (i) an end-to-end framework enabling community-level metabolic interaction simulations and (ii) a resource of >14 000 MAGs from a range of metagenomic biomes, including 3750 high quality MAGs, with corresponding FBA-ready GEMs from human gut microbiome studies (21,22) and global microbiome projects (23–25). The metaGEM pipeline is implemented in Snakemake (26), an open-source, community driven, and scalable bioinformatics workflow engine, supporting major popular high-performance-computing cluster environments as well as standalone systems (Supplementary Figure S1).
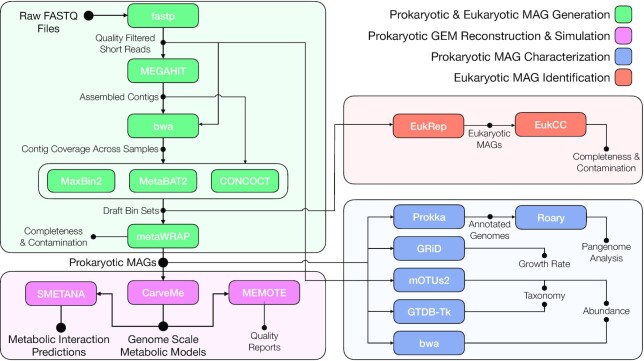
Figure 1.
Schematic of the metaGEM pipeline workflow highlighting tools, inputs, and outputs. Short reads are quality filtered and adapter trimmed using fastp (27). Quality controlled reads are assembled individually using MEGAHIT (28). Using either kallisto (31) or bwa (29), quality controlled reads are mapped to each assembly to obtain contig coverage information across samples. Coverage information and assemblies are used by CONCOCT (8), MetaBAT2 (10) and MaxBin2 (9) to generate three bin sets for each sample. The metaWRAP (32) bin_refinement module is used to dereplicate bin sets for each sample and find the highest quality version of each bin. The metaWRAP bin_reassemble module is used to extract quality controlled short reads mapping from the focal sample to the bin, which are used to generate two single genome assemblies using strict and permissive parameters. The original and reassembled versions are compared for quality, and the best version is kept. Refined and reassembled MAGs are used to generate GEMs using CarveMe (33). These models can be quality checked using MEMOTE (34). Community simulations are carried out for each sample using SMETANA (17). Other features include: taxonomic classification using mOTUs2 (36) and/or GTDB-Tk (35), a custom mapping-based abundance estimation module that does not make use of marker-gene or reference-genome based approaches, growth rate estimation for high coverage MAGs using GRiD (37), and pangenome analysis using prokka (38) and roary (39). EukRep (40) can be used to scan for eukaryotic contigs in the CONCOCT bin sets, which can then be processed by EukCC (41) to provide completeness, contamination, and taxonomic assignments for eukaryotic MAGs.
MATERIALS AND METHODS
Tools used by metaGEM
Table 1.
List of tools used by metaGEM
| Tool | Task | Repository |
|---|---|---|
| Snakemake v5.10.0 (26) | Workflow management | https://github.com/snakemake/snakemake |
| fastp v0.20.0 (27) | Short read QC filtering and adapter removal | https://github.com/OpenGene/fastp |
| MEGAHIT v1.2.9 (28) | Short read assembly | https://github.com/voutcn/megahit |
| bwa v0.7.17 (29) | Contig coverage | https://github.com/lh3/bwa |
| SAMtools v1.9 (30) | Contig coverage | https://github.com/samtools/samtools |
| kallisto v0.46.1 (31) | Contig coverage | https://github.com/pachterlab/kallisto |
| CONCOCT v1.1.0 (8) | Contig binning | https://github.com/BinPro/CONCOCT |
| MetaBAT2 v2.12.1 (10) | Contig binning | https://bitbucket.org/berkeleylab/metabat/src/master/ |
| MaxBin2 v2.2.5 (9) | Contig binning | https://sourceforge.net/projects/maxbin2/ |
| metaWRAP v1.2.3 (32) | Bin refinement and reassembly | https://github.com/bxlab/metaWRAP |
| CarveMe v1.2.2 (33) | GEM reconstruction | https://github.com/cdanielmachado/carveme |
| SMETANA v1.2.0 (17) | Community GEM simulation | https://github.com/cdanielmachado/smetana |
| MEMOTE v0.9.13 (34) | GEM quality report | https://github.com/opencobra/memote |
| GTDB-Tk v1.1.0 (35) | MAG taxonomy assignment | https://github.com/Ecogenomics/GTDBTk |
| mOTUs2 v2.5.1 (36) | MAG taxonomy assignment | https://github.com/motu-tool/mOTUs_v2, https://github.com/AlessioMilanese/classify-genomes |
| GRiD v1.3 (37) | MAG growth rate estimation | https://github.com/ohlab/GRiD |
| Prokka v1.14.6 (38) | MAG functional annotation | https://github.com/tseemann/prokka |
| Roary v3.13.0 (39) | Pangenome analysis | https://github.com/sanger-pathogens/Roary |
| EukRep v0.6.6 (40) | Identify eukaryotic MAGs | https://github.com/patrickwest/EukRep |
| EukCC v0.1.4.3 (41) | Eukaryotic MAG taxonomy and quality | https://github.com/Finn-Lab/EukCC |
Snakemake and HPCCs
metaGEM was implemented in Snakemake v5.10.0, and makes use of the bioinformatic tools listed in Table 1. The workflow was designed to analyze datasets independently from each other, while samples from the same datasets are processed in parallel. All Snakemake jobs were run on the Chalmers Center for Computational Science and Engineering (C3SE) and the European Molecular Biology Laboratory (EMBL) Heidelberg high performance computer clusters (HPCC).
Metagenomic samples and short read quality control
A total of 483 whole metagenome shotgun samples from five metagenomic studies (21–25) were downloaded from the NCBI SRA or EBI ENA to the HPCC. The lab culture dataset was the only single end read set, while the remaining datasets consisted of paired end read sets. The fastp tool v0.20.0 was used for short read quality filtering and adapter removal using default settings.
Short read assembly
Single sample assemblies were obtained using MEGAHIT v1.2.9. The flag ‘–presets meta-sensitive’ was used on all assemblies, which is equivalent to setting ‘–min-count 1’ ‘–k-list 21,29,39,49,59,69,79,89,99,109,119,129,141’. The parameter ‘–min-contig-len’ was set to 1000 for the TARA Oceans dataset.
Contig coverage estimation and binning
For the lab culture, gut microbiome, plant associated, and soil datasets, bwa-mem v0.7.17 was used with default settings to cross map each quality controlled set of short reads to each generated assembly within a dataset. An index was created for each assembly prior to mapping using the ‘bwa index’ command with default settings. Each mapping operation resulted in a SAM file, which was converted to BAM format using the ‘samtools view’ command with the flags ‘-Sb’, and then sorted using the ‘samtools sort’ command with default settings. The sorted BAM files were then used to create the contig coverage across samples input files for MetaBAT2 v.2.12.1 and MaxBin2 v2.2.5 with the MetaBAT2 script ‘jgi_summarize_bam_contig_depths’ (default parameters). The CONCOCT script ‘cut_up_fasta.py’ with parameters ‘-c 10000 -o 0 -m -b’ was used to generate a BEDfile for each assembly, with contigs cut into 10 kb chunks. The generated BEDfile and sorted BAM files were used by the CONCOCT script ‘concoct_coverage_table.py’ (default settings) to generate the contig coverage across samples input files for CONCOCT. CONCOCT v1.1.0 was run for each cut up assembly, using the contig coverage across samples as input and with the ‘-c 800’ parameter. The CONCOCT script ‘extract_fasta_bins.py’ script was then run with default settings to extract bins in terms of the original uncut contigs. MetaBAT2 v.2.12.1 was run using the ‘metabat2’ command for each assembly, using the contig coverage across samples as input and with default settings. MaxBin2 v2.2.5 was run using the ‘run_MaxBin.pl’ for each assembly, using the contig coverage across samples as input and with default settings.
For the TARA oceans dataset, the kallisto v0.46.1 tool was used for mapping quality controlled paired end reads to assemblies. First, each assembly was cut up into 10 kb chunks using the CONCOCT ‘cut_up_fasta.py’ script with parameters ‘-c 10000 -o 0 -m’. The cut up assembly was then used to generate a kallisto index using the ‘kallisto index’ command with default settings. Next, the ‘kallisto quant’ command was used with the ‘–plaintext’ setting to cross map each sample set of quality controlled paired end reads to each cut up assembly. Finally, the ‘kallisto2concoct.py’ script was used to summarize the mapping results across samples for each set of assembled contigs. For the TARA oceans dataset the binners were used as described above, but only CONCOCT used contig coverage across samples as input, while MetaBAT2 and MaxBin2 only used contig coverage from the assembled sample as input.
Bin refinement and reassembly
To reconcile and dereplicate the three generated binner outputs, the metaWRAP metaWRAP v1.2.3 ‘bin_refinement’ command was used with parameters ‘-x 10 -c 50’, corresponding to a maximum bin contamination threshold of 10% and a minimum bin completeness threshold of 50% (i.e. medium quality bin criteria) based on CheckM estimates. Note that CheckM makes use of a database of reference genomes for the calculation of completeness and contamination scores. The metaWRAP ‘reassemble_bins’ command was used with parameters ‘-x 10 -c 50’ to improve bin quality whenever possible by independently reassembling reads that map from the parent sample's quality controlled short reads to the refined bin. Refined and reassembled bins are hereafter referred to as metagenome assembled genomes (MAGs), although the terms may be used interchangeably.
MAG abundance quantification and taxonomic assignment
Absolute and relative abundances of MAGs were calculated using bwa v0.7.17 and SAMtools v1.9. First, all MAGs generated from the same sample were concatenated into a single fasta file, based on which an index was created using ‘bwa index’ (default parameters). Next, ‘bwa-mem’ (default parameters) was used to map the quality controlled short reads from the parent sample to the concatenation of generated MAGs. The resulting SAM file was converted to BAM format and sorted using ‘samtools view’ and ‘samtools sort’, respectively, using the same parameters as described above. Next, ‘samtools flagstat’ was used (default parameters) to extract the number of reads that mapped from the quality controlled paired end reads to the concatenation of all MAGs generated in that sample. Next, for each MAG, an index was created using ‘bwa index’ (default parameters), the quality controlled short reads were mapped to the MAG using ‘bwa-mem’ (default parameters), the resulting SAM file was converted to BAM format and sorted as described above, and the number of reads mapping to the MAG from the short reads was extracted using ‘samtools flagstat’. The abundance of a given MAG was estimated by dividing the number of quality controlled reads that map to the MAG by the number of quality controlled reads that map to the concatenation of all MAGs from that sample, divided by the megabase pair length of the MAG. For each sample, these non-normalized abundances were summed to obtain a sample specific normalizing factor. To obtain normalized relative abundances, each non-normalized abundance was divided by the sample specific normalization factor. The mOTUs2 v2.5.1 tool was also used to calculate abundances, for comparison with the above mapping based method, only in the lab culture dataset. The ‘motus profile’ command was used with default settings. GTDB-Tk v1.1.0 was used to assign taxonomic labels to the generated MAGs using the ‘gtdbtk classify_wf’ with default settings. Both mOTUs2 and GTDB-Tk make use of reference genomes and/or marker genes for taxonomic classification.
Genome scale metabolic model reconstruction and quality reports
CarveMe v1.2.2 was used to automatically reconstruct genome scale metabolic models from ORF annotated protein fasta files derived from MAGs using the default CPLEX solver v12.8. The ‘carve’ command was run using the ‘–fbc2 -g’ flags to gapfill models on complete media and generate FBC2 format models. The MEMOTE v0.9.1 tool was then used to generate quality reports for each genome scale metabolic model. The ‘memote run’ command was used with the flags ‘–skip test_find_metabolites_produced_with_closed_bounds –skip test_find_metabolites_consumed_with_closed_bounds –skip test_find_metabolites_not_produced_with_open_bounds –skip test_find_metabolites_not_consumed_with_open_bounds –skip test_find_incorrect_thermodynamic_reversibility’ to avoid running time consuming tests.
Community simulations
The SMETANA v1.2.0 tool was used for simulating gut microbiome communities of reconstructed genome scale metabolic models. The ‘smetana’ command was used with the flags ‘–flavor fbc2 –detailed –mediadb media_db.tsv -m M11’ and using the default CPLEX solver v12.8. The simulation media was the same as was used for gapfilling (full media, M3) minus aromatic amino acids (M11). The media file was obtained from the authors of previous publication (16), and can be accessed from the metaGEM GitHub repository (https://github.com/franciscozorrilla/metaGEM).
Regarding the implementation specifics, SMETANA is formulated as a mixed linear integer problem (MILP) that enumerates all possible essential metabolic exchanges within a community of N species that sustain a non-zero growth of all N species subject to a mass balance constraint. SMETANA goes beyond pairwise comparison and does constraint based analysis for all members in the microbial community simultaneously. SMETANA does use any biological objective functions. Further details about implementation and specific algorithms used within the SMETANA framework are available in the original publication (17).
Additional features in the metaGEM pipeline
Although not discussed in detail, there are several additional features that were incorporated into metaGEM that may be useful to users. Growth rates for medium and high coverage MAGs can be estimated using the GRiD v1.3 tool. Prokka v1.14.6 can be used to functionally annotate MAGs and the output can be provided to Roary v3.13.0 in order to visualize the core and pangenome of a set of MAGs. Communities with suspected eukaryotic MAGs can be further probed by scanning for eukaryotic contigs in the CONCOCT bin set using EukRep v0.6.6. Identified eukaryotic bins can then be analyzed by EukCC v0.1.4.3, to obtain completeness and contamination estimates as well taxonomic lineage estimates.
RESULTS
Implementation and features of metaGEM pipeline
The metaGEM pipeline starts from quality filtering and single sample assembly of short read FASTQ data, and proceeds to the reconstruction of metagenome assembled genomes using three different binners: CONCOCT (8), MetaBAT2 (10), MaxBin2 (9). The three output draft bin sets are then refined (i.e. de-replicated) and reassembled using metaWRAP to obtain the highest quality version of each bin (32) (Supplementary Figure S2). Note that metaWRAP completeness and contamination estimates are based on a marker-gene approach used by CheckM (42). These final prokaryotic MAGs are used to automatically generate FBA-ready genome-scale metabolic reconstructions using CarveMe (33), which are quality checked using the MEMOTE (34) framework. By integrating the Species METabolic ANAlysis (SMETANA) framework (17) (https://github.com/cdanielmachado/smetana), the generated GEMs can be used for sample specific community-level metabolic interaction modeling. Other pipeline features include the automatic assignment of taxonomic classification to the reconstructed MAGs and GEMs, the calculation of relative and absolute abundance for generated MAGs, the estimation of growth rate (37) for high coverage MAGs, and pangenome analysis (39) (Figure 1). Although there is a growing number of MAG reconstruction pipelines (43–49), a simple comparison of studies that used differing methods for MAG generation revealed that metaGEM consistently recovers more high quality genomes per sample, both from gut microbiome (11–13) and ocean (50–52) metagenomes (Supplementary Figure S3). Indeed, metaGEM has already been applied in one of our recent studies to interrogate the plastic-degrading potential of the global microbiome (53). Regarding tool selection for specific tasks such as short read quality filtering, assembly, binning, etc., there are a number of published stand-alone candidate tools to choose from in the literature. Specific tools were chosen based on a combination of factors including: user-friendliness, computational efficiency, performance (i.e. quality of output), publication date, quality of documentation, development activity, technical support availability, and size of user community. The flexibility of the Snakemake framework allows easy expansion of the metaGEM toolset to add new features or alternative tools.
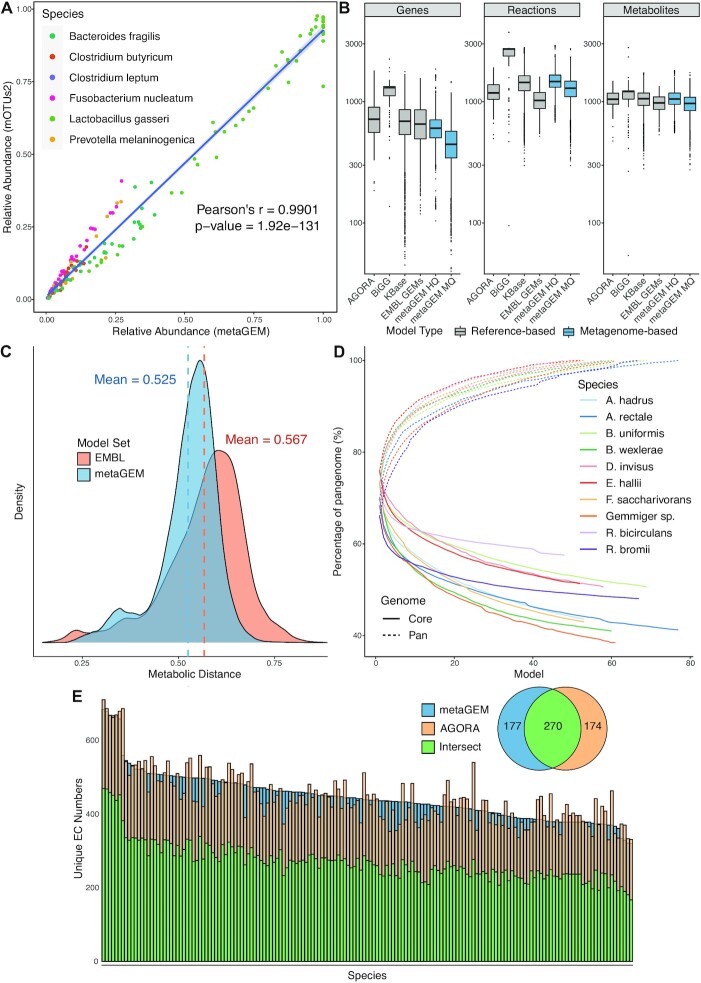
To demonstrate the versatility of metaGEM pipeline, we reconstructed genome-scale metabolic models (GEMs) from five metagenome datasets spanning different biological and technical complexity, namely: gut microbiomes (21,22), plant associated microbiomes (23), global ocean microbiomes (24), and bulk soil microbiomes (25). In total, we reconstructed 3750 high quality metagenomic assemblies (MAGs) with >90% completeness & <5% contamination and 10 349 medium quality MAGs with >50% completeness and <10% contamination (Supplementary Figure S4). We assessed the quality of MAG reconstruction by recovering genomes from a controlled multispecies lab culture experiment (21). Briefly, 7 human gut microbiome species were grown in vitro across four biological replicates with 12 time points totaling 48 metagenomic samples. A total of 154 MQ MAGs, of which 137 also meet the HQ MAG standard, were reconstructed with an average completeness of 95.4% and average contamination of 0.3%, with on average ∼3.2 MAGs per sample reflecting the growth curves shown in the original publication, which are initially dominated by very few species and ultimately just one (Supplementary Figure S5). Abundance estimates generated using a mapping-based approach (Materials and Methods) were perfectly correlated to marker gene based abundance estimates (Pearson's r = 0.99, P-value < 1e−16) well recapitulating experimental observations (Figure 2A).
Figure 2.
Abundance, quality, and diversity comparisons of reconstructions. (A) Abundance estimates generated by metaGEM using a mapping based approach compared to marker gene based approach of mOTUs2 in small lab culture communities dataset. (B) Distribution of genes, reactions and metabolites in genome scale metabolic models across AGORA (15), BiGG (54), EMBL GEMs (33), KBase (49), medium quality (MQ) metaGEM and high quality (HQ) metaGEM sets. (C) Distribution of metabolic distances between a set of 200 randomly chosen reference EMBL GEMs compared to a randomly chosen set of 800 EMBL GEMs and 800 randomly chosen metaGEMs from the gut microbiome dataset. (D) Cumulative core and pan genome curves for the top 10 most commonly reconstructed gut microbiome species based on EC numbers present in the reconstructed genome scale metabolic models. (E) Comparison of EC numbers between 165 species reconstructed from the gut microbiome dataset and also found in the AGORA collection. Inset venn diagram shows average value of EC numbers unique to the compared sets as well as their average intersect.
High-quality metabolic reconstructions directly from metagenomes
We next reconstructed a total of 14 087 GEMs from the metagenome assembled genomes (MAGs) (Materials and Methods) and compared them to highly curated reference genome-based BiGG models (54), AGORA (15), EMBL (33) and KBase models (49). In terms of the number of metabolic reactions and unique metabolites, the metagenomic GEMs show similar distributions compared to reference-based GEM reconstructions (Figure 2B). Specifically, GEMs derived from high quality MAGs had average percent differences of 1.6%, 0.3% and 10.0% in number of metabolites and 3.7%, 21.1%, 44.3% in number of reactions respectively compared to reference genome-based EMBL, AGORA and KBase models, and had average percent differences of 4.1% less metabolites and 40.1% less in reactions compared to manually curated BiGG models, which overall display a higher number of reactions and metabolites. In terms of the number of genes present in the models, GEMs derived from high quality MAGs had between 9.5% to 19.9% less genes as compared to the reference genome-based collections, with the exception of BIGG models that involved over 40% more gene annotations. Furthermore, metaGEMs reconstructed using high quality MAGs had average percent differences of 24.6%, 12.8% and 10.3% compared to medium quality MAG-based metaGEMs in terms of their number of genes, reactions, and metabolites, suggesting that the metabolic reconstruction process (33) is robust towards bin completion. Separating metagenomic models by dataset shows that the distributions of metabolites, reactions, and genes are similar respectively across datasets as well (Supplementary Figure S6).
We also evaluated whether the metabolic reaction diversity identified in metagenomes would be comparable to the expected enzymatic diversity from the reference genome reconstructions, i.e. if expected reactions and pathways would be present in metagenomes based GEMs. For this we randomly sampled 1000 metabolic models from each, reference-genome-based and metagenome-based reconstructions, and computed metabolic similarity (expressed as Jaccard index) to a random 20% held-out set of reference-genome-based reconstructions (Figure 2C). Metagenomic GEMs and reference genome based EMBL GEMs had an average 4.2% difference (Wilcoxon rank sum test P-value < 2.2e−16) in metabolic reactions compared to an independent subset of metabolic reconstructions from reference based genomes, suggesting that metabolic models derived from metagenome assembled genomes capture expected metabolic diversity and features as reconstructions from reference-based genomes (Materials and Methods). Performing PCA on the presence/absence of EC numbers across models resulted in AGORA and gut metagenomic models clustering near each other, while EMBL and ocean metaGEM models clustered closer to each other (Supplementary Figure S7).
Pan-metabolism of metagenomic GEMs uncovers metabolic diversity within species
Recent gut metagenomic microbial population studies suggest that strain-level diversity within the same species can differ by over 20% of genome content between individuals (55). With more extreme examples from environmental isolates of soil myxobacteria with only 30% conserved core part of genomes showing extreme gene diversity within the same species (56). These analyses imply that current reference based approaches (15) may not always reflect the metabolic diversity found across a species’ pangenome or pan-metabolism. To investigate whether reconstructed metagenomic GEMs are able to capture this intra-species metabolic diversity we compared intra-species EC numbers (i.e. the pan-metabolism) diversity across the top ten most prevalent species (Figure 2C). Pan-metabolism analysis confirmed that no two models were exactly the same with respect to their EC number content. Indeed, the core genome of metabolism in the analyzed species ranged between 38.5–57.6% of their respective pangenomes, in line with previously reported degree of intraspecies genetic variation in prokaryotes (14,55). We also analyzed the pangenome of 141 taxonomically undefined species, where we found the core genome to be 6.9% of the pan-species pan-genome (Supplementary Figure S8). To exemplify further, we compared unique EC numbers between metagenome based GEMs and reference genome based gut AGORA models (15) for a total of 165 matched gut microbiome species. The intersect of AGORA and metaGEM EC numbers range between 48.9% to 69% for all matched species, with metaGEM models containing more EC numbers than their corresponding AGORA model in 53.9% of cases, while AGORA models contained more EC numbers 46.1% of comparisons. Inspection of the KEGG pathway annotations of the EC numbers present exclusively in AGORA or metagenomic gut models reveals that the majority of these enzymes are associated with biosynthesis of secondary metabolites and antibiotics (Supplementary Figure S9). Finally, we note that there is little overlap between identified species based on phylogenetic marker genes and AGORA models, while we observed 10-fold greater overlap for metaGEM models (Supplementary Figure S10).
metaGEMs enables modeling of personalized human gut communities
To investigate potential microbial metabolic interactions in healthy and metabolically impaired type 2 diabetes human gut microbiomes (22), we reconstructed a total of 4127 of personalized human gut metaGEMs across 137 metagenomes that were classified according to the disease condition of participants from the original study, i.e. normal glucose tolerance (NGT, n = 42), impaired glucose tolerance (IGT, n = 42) or type 2 diabetic (T2D, n = 53). We then applied Species Metabolic Coupling Analysis (SMETANA), a constraint-based technique for modeling interspecies dependencies in microbial communities (17), to elucidate the potential microbial metabolic interactions in healthy and metabolically impared patients. Briefly, SMETANA outputs a set of scores for each community, corresponding to measures of strength of cross-feeding interactions that should take place to support the growth of community members in a given condition, i.e. a likelihood of species A growth depending on metabolite X from species B.
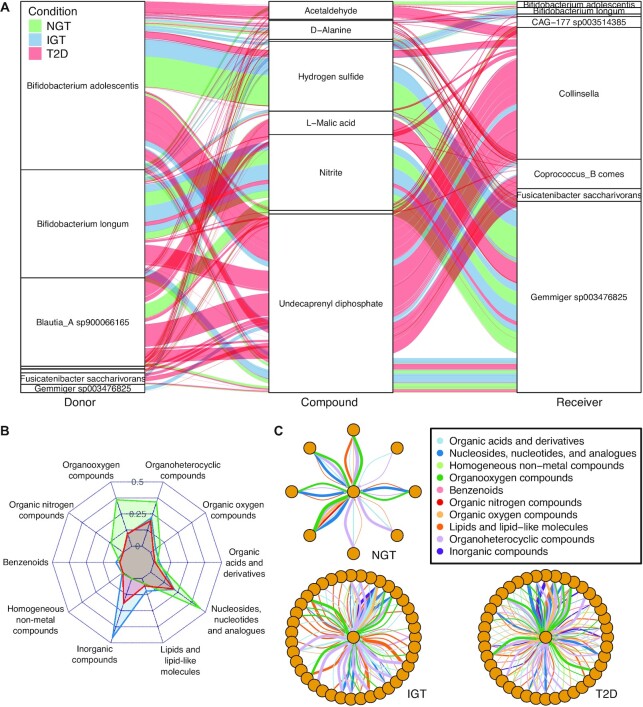
Specifically, we found 22 compound exchanges that were significantly different (Wilcoxon rank sum test, BH adjusted P-values < 0.01) between the disease groups, representing metabolites from multiple classes including organic acids and lipid-like molecules (Supplementary Figures S11 and S12). Additionally, we identified 27 donors and 27 receivers that had statistically significant distributions of SMETANA scores between disease groups (Wilcoxon rank sum test, BH adjusted P-value < 0.0001), including genera that have been associated with T2D (57) (Supplementary Figures S13 and 14) such as Bifidobacterium, Faecalibacterium, Roseburia, Ruminococcus and Blautia. Visualization of 10 compounds with the lowest P-values exchanged by the eight most frequent donors or receivers of these metabolites shows notable differences in metabolic architecture across conditions (Figure 3A). For example, in the visualized data subset, exchanges of l-malic acid undecaprenyl diphosphate were observed 2.25 and 2.75 times more frequently, and with a 115-fold and 3.9-fold higher average SMETANA score (Wilcoxon rank sum test, BH adjusted P-value = 1.8e−05 and 1.0e−03 respectively) respectively in T2D communities compared to NGT. Although exchanges of nitrite and hydrogen sulfide were observed 1.6 and 1.4 times more frequently in T2D compared to NGT communities, exchanges in the latter had a 5.5-fold and and 5.7-fold higher SMETANA score respectively compared to T2D communities (Wilcoxon rank sum test, BH adjusted P-value = 3.3e−05 and 5.8e−04 respectively). Visualization of average SMETANA scores grouped by broad metabolite class for interactions involving Faecalibacterium prausnitzii C as a receiver (Figure 3B, C) also highlight differences in metabolism, with a 3.7-fold stronger dependency on organic oxygen compounds and a 2.2-fold stronger dependency on nucleosides, nucleotides, and analogues in NGT communities compared to T2D. Also of note is the fact that Faecalibacterium prausnitzii did not demand any inorganic compounds in NGT communities, while overall it had higher dependency on inorganic compounds (nitrite) in IGT communities compared to T2D.
Figure 3.
SMETANA simulations uncover differences in metabolism across conditions. (A) Alluvial diagram showing top 10 compounds exchanged with statistical significance across conditions between eight species, representing 279 interactions (NGT n = 61, IGT n = 58, T2D n = 160) across 41 samples (NGT n = 12, IGT n = 12, T2D n = 17). Thickness of lines are proportional to magnitude of SMETANA score. (B) Radar plot of average SMETANA scores based on 543 interactions (NGT n = 50, IGT n = 161, T2D n = 249) across 18 samples (NGT n = 4, IGT n = 6, T2D n = 8) grouped by metabolite class across conditions for receiver Faecalibacterium prausnitzii C. (C) Network diagrams of interactions involving Faecalibacterium prausnitzii C (centered in each subgraph) as a receiver across conditions. Thickness of lines are proportional to magnitude of SMETANA score.
DISCUSSION
The latest genome sequencing studies suggest that bacterial genomes are highly dynamic (58), exhibiting a high exchange of genetic material between strains of the same species or between different bacterial species. With multiple examples where specific strains play key roles in disease pathogenesis (59) or niche metabolic adaptation (56), it becomes apparent that bacteria should be analyzed by the context specific functional repertoire (including metabolism) of strains and not merely by their species membership (60). The highly diverse pangenome-derived functional repertoire (61,62) of microbial communities, the interpersonal differences in gene content of human gut bacteria (55), and the strain specific plasticity of metabolic adaptation (56) cannot be captured by either amplicon sequencing nor by using reference genomes. In other words, while reference-based GEMs may ensure completeness, they only represent a single point in the pangenome landscape of a species. To address these limitations, and to further enable the interrogation of functional and metabolic diversity existing within microbial communities derived from metagenomes, here we developed metaGEM, a pipeline for reconstruction and metabolic modeling of multispecies microbial communities derived directly from metagenomic samples. In short, the pipeline generates metagenome assembled genomes from metagenomic data which are subsequently used to reconstruct and simulate genome scale metabolic models (GEMs) in their communities.
We showed the versatility and usefulness of the metaGEM pipeline by generating metagenome assembled genomes, annotating their taxonomy, calculating relative abundances, reconstructing genome scale models, and simulating metabolic interactions in microbial communities from a range of metagenomic datasets including lab cultures, human gut, plant associated, soil and ocean metagenomes. Notably, in small metagenomic communities from lab cultures nearly 90% of generated MAGs were of high quality (>90% of genome completeness and <5% contamination), and calculated MAG abundance estimates were highly correlated to marker gene-based estimates (Figure 2A). While metagenomes from more complex communities yielded more MAGs, they pose a more challenging assembly and binning scenario, resulting in a lower percentage of high quality MAGs (Supplementary Figure S4). By comparing the generated metabolic models to previously published reference based GEM collections we demonstrated that metaGEMs have a comparable number of reactions and metabolites, despite tending to have fewer genes (Figure 2B). This suggests that some of these reference based reconstructions may contain extraneous metabolic information. Furthermore, by calculating pairwise metabolic distance estimates between models, we show that metaGEMs capture a similar distribution of enzymatic diversity as compared to reference genome based reconstructions (Figure 2C). While the metagenomic models are quality checked for basic functionality using the MEMOTE test suite, passing these tests does not necessarily ensure they can correctly predict fluxes specific to the environment where they were isolated from.
We demonstrated that metaGEMs capture a large degree of intraspecies variation by analyzing the core and pangenomes of the top 10 most prevalent species from the gut microbiome dataset. Indeed, no two models from the same species were identical in terms of their metabolism, with up to 60% of metabolic diversity present within species pangenomes, demonstrating remarkable degree of intraspecies metabolic variation captured by metaGEM models (Figure 2D). Furthermore, by comparing metaGEMs with reference-based gut species metabolic AGORA models (15), we showed that reference-based models introduce metabolic reactions that may not necessarily be present in every metagenomic context, while the metaGEM models reconstruct context specific metabolism, entirely based on actual metagenomic data that would have been otherwise missing from the reference-based reconstructions (Figure 2E). Indeed, we find that the most common pathways found exclusively in either metaGEM models and AGORA correspond to the biosynthesis of antibiotics and secondary metabolites, which are known to be context specific and horizontally transferred (63).
We showed that gut metagenomes corresponding to different type 2 diabetes disease groups (NGT, IGT, T2D) (22) generate communities with different metabolic architectures. By carrying out species metabolic coupling analysis (17) for each metagenome-derived personalized community of GEMs, we identified 22 growth-related metabolic exchanges, 27 donor species, and 27 receiver species that were significantly different between at least one disease condition comparison (Supplementary Figures S11, S13, S14). Visualization of SMETANA simulation results revealed notable differences in the metabolic idiosyncrasies of species and communities as a function of their disease state condition, as reflected by scores of their metabolic dependencies (Figure 3). Seven of the twenty-two predicted metabolites (D-alanine, anhgm, murein5p5p, murein5px4p, uaagmda, udcpdp and ucdpp) are implicated in peptidoglycan and cell wall biosynthesis. While these predicted metabolite exchanges are required for microbial biomass generation, it should be noted that peptidoglycan (also known as lipopolysaccharide) exchanges in the gut microbiome have been linked to insulin resistance and increased risk of T2D (64,65). Seven predicted carbohydrate family metabolites (4-aminobutanoate, acetaldehyde, l-arabinose, carbon dioxide, fumarate, d-galactose and l-malate) of which the short chain fatty acid 4-aminobutanoate (also known as GABA), l-malate, l-arabinose and d-galactose have all been previously linked with insulin sensitivity and metabolic syndrome (4,64,66–70). Another group of predicted metabolites from the aromatic amino acid family (benzoate, chorismate and l-tryptophan) with tryptophan metabolism has been implicated in insulin resistance (71). Two of the metabolites (nitrite and ornithine) form nitrogen metabolism, in accord with the nitrite being reported to have a positive effect on insulin secretion (72). Finally, while the role of hydrogen sulfide in T2D metabolism remains to be determined, there have been several reported associations of H2S with insulin sensitivity found in the literature (73,74). The majority of the predicted differentially exchanged metabolites are thus corroborated by previous experimental findings, supporting that metaGEM is able to infer phenotype-relevant metabolic networks and metabolite exchanges within communities assembled from metagenomes.
By directly modeling community level sample-specific microbiomes here we identified potential interaction differences between pathogenicity of type 2 diabetes that are based purely on metabolic capacity of the gut communities and are not dependent on species abundance estimates, thus providing additional information that would not be otherwise accessed. Moreover, the SMETANA framework is free from arbitrary assumptions of growth optimality, instead, it evaluates all scenarios of interspecies metabolic exchanges that support the growth of member species in a given community providing unbiased community level metabolism analysis (17). The framework has been validated previously (16,17,75,76) by reproducing experimentally determined interactions in well-studied microbial communities (77,78). Nevertheless, the field of metabolic modelling in ecology scale microbial communities is hypothesis-generating, aiming to propose novel insights into microbiome interactions within ecosystems. The predictions need to be supported by experimental approaches and orthogonal analyses, especially for large-scale microbiomes. Overall, our study offers an end-to-end framework to study sample-specific metabolism of complex microbial communities directly from metagenomic data without relying on reference genomes. We therefore envisage that metaGEM will become an important tool for deciphering metabolic interactions in complex microbial communities.
DATA AVAILABILITY
All FBA-ready metagenome-based metabolic reconstructions were deposited to the Zenodo repository and are available at http://doi.org/10.5281/zenodo.4407746.
The code of metaGEM can be accessed at https://github.com/franciscozorrilla/metaGEM.
Supplementary Material
ACKNOWLEDGEMENTS
Mikael Öhman and Thomas Svedberg at C3SE are acknowledged for technical assistance in making the code run on Vera C3SE resources.
Author contributions: F.Z., A.Z., K.R.P. conceptualized the project; F.Z. and A.Z. designed the computational analysis; F.Z., F.B. developed the pipeline; F.Z. performed the computational analysis; F.Z. and A.Z. interpreted the results; F.Z., A.Z. wrote the initial draft manuscript; F.Z., K.R.P. and A.Z. revised the draft and wrote the final manuscript.
Contributor Information
Francisco Zorrilla, Division of Systems and Synthetic Biology, Department of Biology and Biological Engineering, Chalmers University of Technology, Göteborg, Sweden; Structural and Computational Biology Unit, European Molecular Biology Laboratory, Heidelberg, Germany; Medical Research Council Toxicology Unit, University of Cambridge, Cambridge, UK.
Filip Buric, Division of Systems and Synthetic Biology, Department of Biology and Biological Engineering, Chalmers University of Technology, Göteborg, Sweden.
Kiran R Patil, Structural and Computational Biology Unit, European Molecular Biology Laboratory, Heidelberg, Germany; Medical Research Council Toxicology Unit, University of Cambridge, Cambridge, UK.
Aleksej Zelezniak, Division of Systems and Synthetic Biology, Department of Biology and Biological Engineering, Chalmers University of Technology, Göteborg, Sweden; Institute of Biotechnology, Life Sciences Center, Vilnius University, Vilnius, Lithuania.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
A.Z. was funded by the SciLifeLab fellows program, Formas early-career research [2019-01403] and Marius Jakulis Jason foundation; K.R.P. received support from the UK Medical Research Council [MC_UU_00025/11] and European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation programme [866028] and DD-DeCaF consortium European Union’s Horizon 2020 research and innovation programme [686070]; the computations were enabled with resources provided by EMBL and by the Swedish National Infrastructure for Computing (SNIC) at C3SE partially funded by the Swedish Research Council [2018-05973].
Conflict of interest statement. None declared.
REFERENCES
- 1. Quince C., Delmont T.O., Raguideau S., Alneberg J., Darling A.E., Collins G., Eren A.M.. DESMAN: a new tool for de novo extraction of strains from metagenomes. Genome Biol. 2017; 18:181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Durack J., Lynch S.V.. The gut microbiome: relationships with disease and opportunities for therapy. J. Exp. Med. 2019; 216:20–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Sanna S., van Zuydam N.R., Mahajan A., Kurilshikov A., Vich Vila A., Võsa U., Mujagic Z., Masclee A.A.M., Jonkers D.M.A.E., Oosting M.et al.. Causal relationships among the gut microbiome, short-chain fatty acids and metabolic diseases. Nat. Genet. 2019; 51:600–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Li Q., Chang Y., Zhang K., Chen H., Tao S., Zhang Z.. Implication of the gut microbiome composition of type 2 diabetic patients from northern China. Sci. Rep. 2020; 10:5450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Gopalakrishnan V., Helmink B.A., Spencer C.N., Reuben A., Wargo J.A.. The influence of the gut microbiome on cancer, immunity, and cancer immunotherapy. Cancer Cell. 2018; 33:570–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Vuong H.E., Hsiao E.Y.. Emerging roles for the gut microbiome in autism spectrum disorder. Biol. Psychiatry. 2017; 81:411–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cryan J.F., O’Riordan K.J., Sandhu K., Peterson V., Dinan T.G.. The gut microbiome in neurological disorders. Lancet Neurol. 2020; 19:179–194. [DOI] [PubMed] [Google Scholar]
- 8. Alneberg J., Bjarnason B.S., de Bruijn I., Schirmer M., Quick J., Ijaz U.Z., Lahti L., Loman N.J., Andersson A.F., Quince C.. Binning metagenomic contigs by coverage and composition. Nat. Methods. 2014; 11:1144–1146. [DOI] [PubMed] [Google Scholar]
- 9. Wu Y.-W., Simmons B.A., Singer S.W.. MaxBin 2.0: an automated binning algorithm to recover genomes from multiple metagenomic datasets. Bioinformatics. 2015; 32:605–607. [DOI] [PubMed] [Google Scholar]
- 10. Kang D.D., Li F., Kirton E., Thomas A., Egan R., An H., Wang Z.. MetaBAT 2: an adaptive binning algorithm for robust and efficient genome reconstruction from metagenome assemblies. PeerJ. 2019; 7:e7359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Almeida A., Mitchell A.L., Boland M., Forster S.C., Gloor G.B., Tarkowska A., Lawley T.D., Finn R.D.. A new genomic blueprint of the human gut microbiota. Nature. 2019; 568:499–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Nayfach S., Shi Z.J., Seshadri R., Pollard K.S., Kyrpides N.C.. New insights from uncultivated genomes of the global human gut microbiome. Nature. 2019; 568:505–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Pasolli E., Asnicar F., Manara S., Zolfo M., Karcher N., Armanini F., Beghini F., Manghi P., Tett A., Ghensi P.et al.. Extensive unexplored human microbiome diversity revealed by over 150,000 genomes from metagenomes spanning age, geography, and lifestyle. Cell. 2019; 176:649–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Almeida A., Nayfach S., Boland M., Strozzi F., Beracochea M., Shi Z.J., Pollard K.S., Sakharova E., Parks D.H., Hugenholtz P.et al.. A unified catalog of 204,938 reference genomes from the human gut microbiome. Nat. Biotechnol. 2020; 39:105–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Magnúsdóttir S., Heinken A., Kutt L., Ravcheev D.A., Bauer E., Noronha A., Greenhalgh K., Jäger C., Baginska J., Wilmes P.et al.. Generation of genome-scale metabolic reconstructions for 773 members of the human gut microbiota. Nat. Biotechnol. 2017; 35:81–89. [DOI] [PubMed] [Google Scholar]
- 16. Tramontano M., Andrejev S., Pruteanu M., Klünemann M., Kuhn M., Galardini M., Jouhten P., Zelezniak A., Zeller G., Bork P.et al.. Nutritional preferences of human gut bacteria reveal their metabolic idiosyncrasies. Nat Microbiol. 2018; 3:514–522. [DOI] [PubMed] [Google Scholar]
- 17. Zelezniak A., Andrejev S., Ponomarova O., Mende D.R., Bork P., Patil K.R.. Metabolic dependencies drive species co-occurrence in diverse microbial communities. Proc. Natl. Acad. Sci. U.S.A. 2015; 112:6449–6454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Basile A., Campanaro S., Kovalovszki A., Zampieri G., Rossi A., Angelidaki I., Valle G., Treu L.. Revealing metabolic mechanisms of interaction in the anaerobic digestion microbiome by flux balance analysis. Metab. Eng. 2020; 62:138–149. [DOI] [PubMed] [Google Scholar]
- 19. Freilich S., Zarecki R., Eilam O., Segal E.S., Henry C.S., Kupiec M., Gophna U., Sharan R., Ruppin E.. Competitive and cooperative metabolic interactions in bacterial communities. Nat. Commun. 2011; 2:589. [DOI] [PubMed] [Google Scholar]
- 20. Stolyar S., Van Dien S., Hillesland K.L., Pinel N., Lie T.J., Leigh J.A., Stahl D.A.. Metabolic modeling of a mutualistic microbial community. Mol. Syst. Biol. 2007; 3:92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Korem T., Zeevi D., Suez J., Weinberger A., Avnit-Sagi T., Pompan-Lotan M., Matot E., Jona G., Harmelin A., Cohen N.et al.. Growth dynamics of gut microbiota in health and disease inferred from single metagenomic samples. Science. 2015; 349:1101–1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Karlsson F.H., Tremaroli V., Nookaew I., Bergström G., Behre C.J., Fagerberg B., Nielsen J., Bäckhed F.. Gut metagenome in European women with normal, impaired and diabetic glucose control. Nature. 2013; 498:99–103. [DOI] [PubMed] [Google Scholar]
- 23. Li X., Jousset A., de Boer W., Carrión V.J., Zhang T., Wang X., Kuramae E.E.. Legacy of land use history determines reprogramming of plant physiology by soil microbiome. ISME J. 2019; 13:738–751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Sunagawa S., Coelho L.P., Chaffron S., Kultima J.R., Labadie K., Salazar G., Djahanschiri B., Zeller G., Mende D.R., Alberti A.et al.. Structure and function of the global ocean microbiome. Science. 2015; 348:1261359. [DOI] [PubMed] [Google Scholar]
- 25. Bissett A., Fitzgerald A., Meintjes T., Mele P.M., Reith F., Dennis P.G., Breed M.F., Brown B., Brown M.V., Brugger J.et al.. Introducing BASE: the Biomes of Australian Soil Environments soil microbial diversity database. Gigascience. 2016; 5:21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Köster J., Rahmann S.. Snakemake—a scalable bioinformatics workflow engine. Bioinformatics. 2012; 28:2520–2522. [DOI] [PubMed] [Google Scholar]
- 27. Chen S., Zhou Y., Chen Y., Gu J.. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 2018; 34:i884–i890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Li D., Liu C.-M., Luo R., Sadakane K., Lam T.-W.. MEGAHIT: an ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics. 2015; 31:1674–1676. [DOI] [PubMed] [Google Scholar]
- 29. Li H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. 2013; arXiv doi:26 May 2013, preprint: not peer reviewedhttps://arxiv.org/abs/1303.3997.
- 30. Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R.Subgroup, 1000 Genome Project Data Processing . The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009; 25:2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bray N.L., Pimentel H., Melsted P., Pachter L.. Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol. 2016; 34:525–527. [DOI] [PubMed] [Google Scholar]
- 32. Uritskiy G.V., DiRuggiero J., Taylor J.. MetaWRAP—a flexible pipeline for genome-resolved metagenomic data analysis. Microbiome. 2018; 6:158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Machado D., Andrejev S., Tramontano M., Patil K.R.. Fast automated reconstruction of genome-scale metabolic models for microbial species and communities. Nucleic Acids Res. 2018; 46:7542–7553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lieven C., Beber M.E., Olivier B.G., Bergmann F.T., Ataman M., Babaei P., Bartell J.A., Blank L.M., Chauhan S., Correia K.et al.. MEMOTE for standardized genome-scale metabolic model testing. Nat. Biotechnol. 2020; 38:272–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Chaumeil P.-A., Mussig A.J., Hugenholtz P., Parks D.H.. GTDB-Tk: a toolkit to classify genomes with the Genome Taxonomy Database. Bioinformatics. 2019; 36:1925–1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Milanese A., Mende D.R., Paoli L., Salazar G., Ruscheweyh H.-J., Cuenca M., Hingamp P., Alves R., Costea P.I., Coelho L.P.et al.. Microbial abundance, activity and population genomic profiling with mOTUs2. Nat. Commun. 2019; 10:1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Emiola A., Oh J.. High throughput in situ metagenomic measurement of bacterial replication at ultra-low sequencing coverage. Nat. Commun. 2018; 9:4956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics. 2014; 30:2068–2069. [DOI] [PubMed] [Google Scholar]
- 39. Page A.J., Cummins C.A., Hunt M., Wong V.K., Reuter S., Holden M.T.G., Fookes M., Falush D., Keane J.A., Parkhill J.. Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics. 2015; 31:3691–3693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. West P.T., Probst A.J., Grigoriev I.V., Thomas B.C., Banfield J.F.. Genome-reconstruction for eukaryotes from complex natural microbial communities. Genome Res. 2018; 28:569–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Saary P., Mitchell A.L., Finn R.D.. Estimating the quality of eukaryotic genomes recovered from metagenomic analysiswith EukCC. Genome Biology. 2020; 21:244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Parks D.H., Imelfort M., Skennerton C.T., Hugenholtz P., Tyson G.W.. CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 2015; 25:1043–1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Murat Eren A., Esen Ö.C., Quince C., Vineis J.H., Morrison H.G., Sogin M.L., Delmont T.O.. Anvi’o: an advanced analysis and visualization platform for ‘omics data. PeerJ. 2015; 3:e1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Goecks J., Nekrutenko A., Taylor J.The Galaxy Team . Galaxy: a comprehensive approach for supporting accessible, reproducible, and transparent computational research in the life sciences. Genome Biol. 2010; 11:R86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Tamames J., Puente-Sánchez F.. SqueezeMeta, a highly portable, fully automatic metagenomic analysis pipeline. Frontiers in Microbiology. 2019; 9:3349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Clarke E.L., Taylor L.J., Zhao C., Connell A., Lee J.-J., Fett B., Bushman F.D., Bittinger K.. Sunbeam: an extensible pipeline for analyzing metagenomic sequencing experiments. Microbiome. 2019; 7:46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Murovec B., Deutsch L., Stres B.. Computational framework for high-quality production and large-scale evolutionary analysis of metagenome assembled genomes. Mol. Biol. Evol. 2019; 37:593–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Stewart R.D., Auffret M.D., Snelling T.J., Roehe R., Watson M.. MAGpy: a reproducible pipeline for the downstream analysis of metagenome-assembled genomes (MAGs). Bioinformatics. 2018; 35:2150–2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Arkin A.P., Cottingham R.W., Henry C.S., Harris N.L., Stevens R.L., Maslov S., Dehal P., Ware D., Perez F., Canon S.et al.. KBase: the United States department of energy systems biology knowledgebase. Nat. Biotechnol. 2018; 36:566–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Tully B.J., Sachdeva R., Graham E.D., Heidelberg J.F.. 290 metagenome-assembled genomes from the Mediterranean Sea: a resource for marine microbiology. PeerJ. 2017; 5:e3558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Delmont T.O., Quince C., Shaiber A., Esen Ö.C., Lee S.T.M., Rappé M.S., McLellan S.L., Lücker S., Eren A.M.. Nitrogen-fixing populations of Planctomycetes and Proteobacteria are abundant in surface ocean metagenomes. Nat. Microbiol. 2018; 3:804–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Tully B.J., Graham E.D., Heidelberg J.F.. The reconstruction of 2,631 draft metagenome-assembled genomes from the global oceans. Scientific Data. 2018; 5:170203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Zrimec J., Kokina M., Jonasson S., Zorrilla F., Zelezniak A.. Plastic-degrading potential across the global microbiome correlates with recent pollution trends. 2020; bioRxiv doi:15 December 2020, preprint: not peer reviewed 10.1101/2020.12.13.422558. [DOI] [PMC free article] [PubMed]
- 54. King Z.A., Lu J., Dräger A., Miller P., Federowicz S., Lerman J.A., Ebrahim A., Palsson B.O., Lewis N.E.. BiGG models: a platform for integrating, standardizing and sharing genome-scale models. Nucleic Acids Res. 2015; 44:D515–D522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Zhu A., Sunagawa S., Mende D.R., Bork P.. Inter-individual differences in the gene content of human gut bacterial species. Genome Biol. 2015; 16:82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Livingstone P.G., Morphew R.M., Whitworth D.E.. Genome sequencing and pan-genome analysis of 23 Corallococcus spp. strains reveal unexpected diversity, with particular plasticity of predatory gene sets. Front. Microbiol. 2018; 9:3187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Gurung M., Li Z., You H., Rodrigues R., Jump D.B., Morgun A., Shulzhenko N.. Role of gut microbiota in type 2 diabetes pathophysiology. EBioMedicine. 2020; 51:102590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Garud N.R., Pollard K.S.. Population genetics in the human microbiome. Trends Genet. 2020; 36:53–67. [DOI] [PubMed] [Google Scholar]
- 59. Peña-Gonzalez A., Soto-Girón M.J., Smith S., Sistrunk J., Montero L., Páez M., Ortega E., Hatt J.K., Cevallos W., Trueba G.et al.. Metagenomic signatures of gut infections caused by different Escherichia coli pathotypes. Appl. Environ. Microbiol. 2019; 85:e01820-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Frioux C., Singh D., Korcsmaros T., Hildebrand F.. From bag-of-genes to bag-of-genomes: metabolic modelling of communities in the era of metagenome-assembled genomes. Comput. Struct. Biotechnol. J. 2020; 18:1722–1734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Zou Y., Xue W., Luo G., Deng Z., Qin P., Guo R., Sun H., Xia Y., Liang S., Dai Y.et al.. 1,520 reference genomes from cultivated human gut bacteria enable functional microbiome analyses. Nat. Biotechnol. 2019; 37:179–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Kim C.Y., Lee M., Yang S., Kim K., Yong D., Kim H.R., Lee I.. Human reference gut microbiome comprising 5,414 prokaryotic species, including newly assembled genomes from under-represented Asian metagenomes. Genome Medicine. 13:134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Versluis D., D’Andrea M.M., Ramiro Garcia J., Leimena M.M., Hugenholtz F., Zhang J., Öztürk B., Nylund L., Sipkema D., van Schaik W.et al.. Mining microbial metatranscriptomes for expression of antibiotic resistance genes under natural conditions. Sci. Rep. 2015; 5:11981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Harsch I.A., Konturek P.C.. The role of gut microbiota in obesity and type 2 and type 1 diabetes mellitus: new insights into ‘old’ diseases. Med. Sci. 2018; 6:32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Zugasti O., Tavignot R., Royet J.. Gut bacteria-derived peptidoglycan induces a metabolic syndrome-like phenotype via NF-κB-dependent insulin/PI3K signaling reduction in Drosophila renal system. Sci. Rep. 2020; 10:14097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Patterson E., Ryan P.M., Wiley N., Carafa I., Sherwin E., Moloney G., Franciosi E., Mandal R., Wishart D.S., Tuohy K.et al.. Gamma-aminobutyric acid-producing lactobacilli positively affect metabolism and depressive-like behaviour in a mouse model of metabolic syndrome. Sci. Rep. 2019; 9:16323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Soto M., Herzog C., Pacheco J.A., Fujisaka S., Bullock K., Clish C.B., Kahn C.R.. Gut microbiota modulate neurobehavior through changes in brain insulin sensitivity and metabolism. Mol. Psychiatry. 2018; 23:2287–2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Aydin Ö., Nieuwdorp M., Gerdes V.. The gut microbiome as a target for the treatment of type 2 diabetes. Curr. Diab. Rep. 2018; 18:55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Liang S., Hou Z., Li X., Wang J., Cai L., Zhang R., Li J.. The fecal metabolome is associated with gestational diabetes mellitus. RSC Adv. 2019; 9:29973–29979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Zhao L., Wang Y., Zhang G., Zhang T., Lou J., Liu J.. L-Arabinose elicits gut-derived hydrogen production and ameliorates metabolic syndrome in C57BL/6J mice on High-Fat-Diet. Nutrients. 2019; 11:3054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Agus A., Clément K., Sokol H.. Gut microbiota-derived metabolites as central regulators in metabolic disorders. Gut. 2020; 70:1174–1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Ghasemi A., Jeddi S.. Anti-obesity and anti-diabetic effects of nitrate and nitrite. Nitric Oxide. 2017; 70:9–24. [DOI] [PubMed] [Google Scholar]
- 73. Tanase D.M., Gosav E.M., Neculae E., Costea C.F., Ciocoiu M., Hurjui L.L., Tarniceriu C.C., Maranduca M.A., Lacatusu C.M., Floria M.et al.. Role of gut microbiota on onset and progression of microvascular complications of type 2 diabetes (T2DM). Nutrients. 2020; 12:3719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Canfora E.E., Meex R.C.R., Venema K., Blaak E.E.. Gut microbial metabolites in obesity, NAFLD and T2DM. Nat. Rev. Endocrinol. 2019; 15:261–273. [DOI] [PubMed] [Google Scholar]
- 75. Ponomarova O., Gabrielli N., Sévin D.C., Mülleder M., Zirngibl K., Bulyha K., Andrejev S., Kafkia E., Typas A., Sauer U.et al.. Yeast creates a niche for symbiotic lactic acid bacteria through nitrogen overflow. Cell Syst. 2017; 5:345–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Blasche S., Kim Y., Mars R.A.T., Machado D., Maansson M., Kafkia E., Milanese A., Zeller G., Teusink B., Nielsen J.et al.. Metabolic cooperation and spatiotemporal niche partitioning in a kefir microbial community. Nat Microbiol. 2021; 6:196–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Hom E.F.Y., Murray A.W.. Niche engineering demonstrates a latent capacity for fungal-algal mutualism. Science. 2014; 345:94–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Miller L.D., Mosher J.J., Venkateswaran A., Yang Z.K., Palumbo A.V., Phelps T.J., Podar M., Schadt C.W., Keller M.. Establishment and metabolic analysis of a model microbial community for understanding trophic and electron accepting interactions of subsurface anaerobic environments. BMC Microbiol. 2010; 10:149. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All FBA-ready metagenome-based metabolic reconstructions were deposited to the Zenodo repository and are available at http://doi.org/10.5281/zenodo.4407746.
The code of metaGEM can be accessed at https://github.com/franciscozorrilla/metaGEM.