Abstract
Glycosylphosphatidylinositol (GPI)-anchored proteins (GPI-APs) play an important role in a variety of plant biological processes including growth, stress response, morphogenesis, signaling, and cell wall biosynthesis. The GPI anchor contains a lipid-linked glycan backbone that is synthesized in the endoplasmic reticulum (ER) where it is subsequently transferred to the C-terminus of proteins containing a GPI signal peptide by a GPI transamidase. Once the GPI anchor is attached to the protein, the glycan and lipid moieties are remodeled. In mammals and yeast, this remodeling is required for GPI-APs to be included in Coat Protein II-coated vesicles for their ER export and subsequent transport to the cell surface. The first reaction of lipid remodeling is the removal of the acyl chain from the inositol group by Bst1p (yeast) and Post-GPI Attachment to Proteins Inositol Deacylase 1 (PGAP1, mammals). In this work, we have used a loss-of-function approach to study the role of PGAP1/Bst1 like genes in plants. We have found that Arabidopsis (Arabidopsis thaliana) PGAP1 localizes to the ER and likely functions as the GPI inositol-deacylase that cleaves the acyl chain from the inositol ring of the GPI anchor. In addition, we show that PGAP1 function is required for efficient ER export and transport to the cell surface of GPI-APs.
The inositol deacylase AtPGAP1 mediates the first step of glycosylphosphatidylinositol (GPI) anchor-lipid remodeling and is required for efficient transport of GPI-anchored proteins
Introduction
Proteins associated with the plasma membrane are involved in a variety of essential functions in eukaryotes, including signaling, transport, and cell surface metabolism (Yeats et al., 2018). Proteins can be attached to the plasma membrane in several ways. Transmembrane proteins contain domains with hydrophobic amino acids, which are embedded within the plasma membrane lipid bilayer, while other proteins use a post-translational attachment to lipids. For instance, if a protein has to be on the intracellular face of the plasma membrane, it can be post-translationally modified by S-acylation, N-myristoylation, prenylation, or palmitoylation (Luschnig and Seifert, 2011; Hemsley, 2015). A protein can also be attached to a GPI anchor during secretion, which targets it to the outer surface of the plasma membrane.
GPI-anchored proteins (GPI-APs) have been studied from yeast and trypanosomes to mammals and plants and shown to be involved in crucial biological processes, including growth, morphogenesis, reproduction, and pathogenesis (Cheung et al., 2014). The GPI anchor is synthesized in the endoplasmic reticulum (ER) and is then attached to the C-terminus of proteins containing a GPI signal peptide by a GPI transamidase (Desnoyer et al., 2020; Kinoshita, 2020). The GPI-anchor precursor consists of a glycan core (with glycan side chains) and phosphatidylinositol (PI) with an acyl chain at the 2-position of the inositol ring. The glycan core is characterized by having a N-acetyl glucosamine and three α-linked mannoses attached to ethanolamine phosphate (EtNP). The GPI anchor is attached to the polypeptide by an amide bond between EtNP and the C-terminal of the polypeptide (Kinoshita and Fujita, 2016). The GPI-AP stays anchored to the luminal leaflet of the ER lipid bilayer by insertion of hydrocarbon chains of PI, diacyl-PI in yeast and a mixture of 1-alkyl-2-acyl PI (major form) and diacyl-PI (minor form) in mammalian cells. Once the GPI anchor is attached to the protein, the glycan and lipid moieties are remodeled and this process has been shown to be very important for ER export and transport to the cell surface of GPI-APs (Muñiz and Riezman, 2016; Kinoshita, 2020).
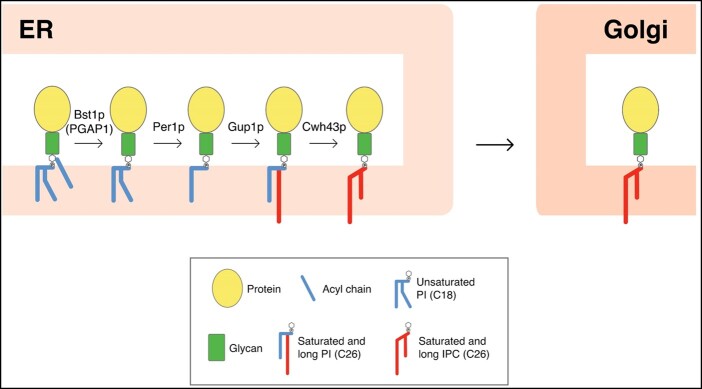
GPI-anchor remodeling has been extensively studied in yeast and mammals. In yeast, the GPI-lipid remodeling occurs entirely in the ER (Pittet and Conzelmann, 2007) and is initiated with the inositol deacylation at the 2-position of the inositol ring by the remodeling enzyme Bst1p (Post-GPI Attachment To Proteins Inositol Deacylase 1, PGAP1, in mammals; Figure 1;Tanaka et al., 2004; Fujita et al., 2006b). This makes GPI-APs sensitive to the bacterial PI-specific phospholipase C (PI-PLC; Low, 1989). Next, the short and unsaturated fatty acid (C18:1) at the sn2 position of PI is removed by the remodeling enzyme Per1p (PGAP3 in mammals; Figure 1;Fujita et al., 2006a) and is replaced with a very long-chain saturated fatty acid (C26:0) by Gup1p (Bosson et al., 2006). The C26:0 diacylglycerol generated seems to be present only in those GPI-APs destined to be transferred to the cell wall (Pittet and Conzelmann, 2007). GPI-APs that remain in the plasma membrane also contain anchors with a very long-chain saturated fatty acid (C26:0) in the form of ceramide. Cwh43p is the enzyme which carries out the addition of ceramide as the lipid moiety to the GPI anchor in yeast (Umemura et al., 2007; Yoko-o et al., 2018). The substrate for the ceramide remodeling is still not clear, but it has been described that most lipid moieties of GPI anchors are exchanged from diacylglycerol to ceramide types (Ghugtyal et al., 2007). These long-chain saturated fatty acids change the physical properties of the GPI-APs and the association with the membrane forming ordered domains at the ER lipid membrane (Silva et al., 2006) allowing these domains to be selectively concentrated at specific ER exit sites (ERES; Muñiz and Riezman, 2016; Rodriguez-Gallardo et al., 2020). The GPI-APs preassembled at ERESs are transported from the ER to the Golgi by Coat Protein II (COPII)-coated vesicles. Due to the luminal localization of GPI-APs, a cargo receptor (p24 complex) is required for incorporation of GPI-APs into nascent COPII vesicles (Castillon et al., 2011; Manzano-Lopez et al., 2015). It has been shown that GPI-anchor remodeling is required for efficient ER export of GPI-APs (Vashist et al., 2001; Kinoshita and Fujita, 2016). After leaving the ER, GPI-APs are transported along the secretory pathway, through the Golgi complex, to their final destination, the plasma membrane, or the cell wall. Proper fatty acid remodeling of the GPI anchor also allows GPI-APs to associate to membrane microdomains enriched in sphingolipids and cholesterol (lipid rafts; Fujita et al., 2006a; Maeda et al., 2007; Castillon et al., 2011; Muniz and Zurzolo, 2014).
Figure 1.
GPI-lipid remodeling in yeast. The GPI anchor is synthesized in the ER and consists of a glycan core and phosphatidylinositol (PI) with an acyl chain at the 2-position of the inositol ring (Kinoshita and Fujita, 2016). After protein attachment, the glycan and lipid moieties are remodeled and this process has been shown to be very important for the transport and final localization of the GPI-APs. The GPI-lipid undergoes a structural remodeling that has the purpose of providing saturated lipids and in yeast it occurs almost entirely at the ER where it is initiated with the inositol deacylation at the 2-position of the inositol ring by the remodeling enzyme Bst1p (PGAP1 in mammals). The rest of lipid remodeling enzymes are indicated, see text for details (modified from Muniz and Zurzolo, 2014). IPC, inositolphosphoceramide.
Around 300 GPI-APs (10% of secreted proteins) have been predicted in Arabidopsis (Arabidopsis thaliana). They play important roles in a variety of plant biological processes occurring at the interface of the plasma membrane and the cell wall, including growth regulation, transport to plasmodesmata, stress response, morphogenesis, signaling and cell wall biosynthesis, and maintenance (Borner et al., 2003; Yeats et al., 2018). To date, only one plant GPI-anchor structure has been resolved, the structure of PcAGP1, isolated from Pyrus communis (pear) cell suspension culture (Oxley and Bacic, 1999). From this structure, it can be said that the core structure of GPI anchors seems to be conserved in plant and nonplant eukaryotes. In addition, a survey of the Arabidopsis genome indicates that most of the genes involved in particular steps of GPI-anchor assembly and their remodeling have putative orthologs in Arabidopsis (Luschnig and Seifert, 2011). Disrupting GPI-anchor synthesis in Arabidopsis is lethal, as is the case in yeast and mammals (Lalanne et al., 2004; Gillmor et al., 2005; Dai et al., 2014; Bundy et al., 2016). In contrast, disruption of GPI-anchor lipid remodeling catalyzed by Bst1/PGAP1 or Per1/PGAP3 is not lethal in yeast (Elrod-Erickson and Kaiser, 1996; Fujita et al., 2006a, 2006b; Komath et al., 2018) and mammals (Ueda et al., 2007; Murakami et al., 2014; Williams et al., 2015; Kinoshita 2020). To our knowledge, no studies have reported on mutants in lipid remodeling enzymes of GPI-APs in plants. Such studies would provide important insights to understand plant GPI-anchor remodeling and its role in trafficking and function of GPI-APs, as was the case with studies in mammals and yeast (Muñiz and Riezman, 2016; Kinoshita, 2020), and may reveal plant distinct and unique characteristics. In this work, we have used a loss-of-function approach to initiate the study of the role of a putative Arabidopsis ortholog of mammalian PGAP1 and yeast Bst1p, the enzymes involved in inositol deacylation, the first step of GPI anchor remodeling.
Results
Identification of PGAP1 in Arabidopsis
Inositol deacylation of GPI-APs is mediated by mammalian PGAP1 and yeast Bst1p (Figure 1). Yeast Bst1p (1,029 amino acids) and mammalian PGAP1 (922 amino acids) are multi transmembrane ER proteins with a catalytic serine containing motif that is conserved in a number of lipases (Tanaka et al., 2004). By searching for Arabidopsis putative GPI inositol-deacylase PGAP1-like (IPR012908, pfam07819) genes using Pfam and InterPro databases (Hunter et al., 2009; Finn et al., 2010), six Arabidopsis genes were found (Supplemental Table S1). The presence of the Pfam PGAP1 motif was further checked by PfamScan and the manually curated Pfam-A database. Of the six genes, one showed no significant results with any motif, two presented other motifs from the AB_hydrolase clan (CL0028), and the rest, AT3G27325, AT4G34310.1, and AT5G17670.1 showed the presence of the PGAP1 motif, although only the AT3G27325 gene presented the complete motif. To clarify the potential redundancy of the three genes containing PGAP1 motifs, a phylogenetic maximum-likelihood tree was built using the 271 protein sequences classified as orthologs of the three genes in EnsemblPlants. The tree shows that the genes diverged early in the history of life, at least before the radiation of Viridiplantae (Supplemental Figure S1). AT3G27325 is predicted to localize to the ER (Supplemental Table S1) and its topology prediction (Dobson et al., 2015; Hofmann and Stofel, 1993; Supplemental Figure S2) indicates that the gene codifies for eight transmembrane domain protein, with a cytoplasmic N- and C-terminus, very similar to the topology of yeast Bst1p or HsPGAP1, although HsPGAP1 has only six predicted transmembrane regions. The lipase consensus sequence, located in a large hydrophilic region after the first transmembrane domain, is conserved between AT3G27325, HsPGAP1, and Bst1p. Therefore, AT3G27325 is likely the Arabidopsis ortholog of Bst1p/HsPGAP1 (Luschnig and Seifert, 2011), and thus was chosen for further investigation in this study. We chose PGAP1 (the mammalian name of the gene) to name AT3G27325 because it is the name that appears in the Arabidopsis Information Resource and because the yeast name (Bst1) had already been assigned to the Arabidopsis gene AT5G65090 (BST1, Bristled). To investigate the expression of PGAP1 in different developmental stages, we used the publicly available RNAseq expression database GENEVESTIGATOR (www.genevestigator.com;Zimmermann et al., 2004; Hruz et al., 2008). PGAP1 shows medium levels of expression in most tissues throughout plant development (Supplemental Figure S3).
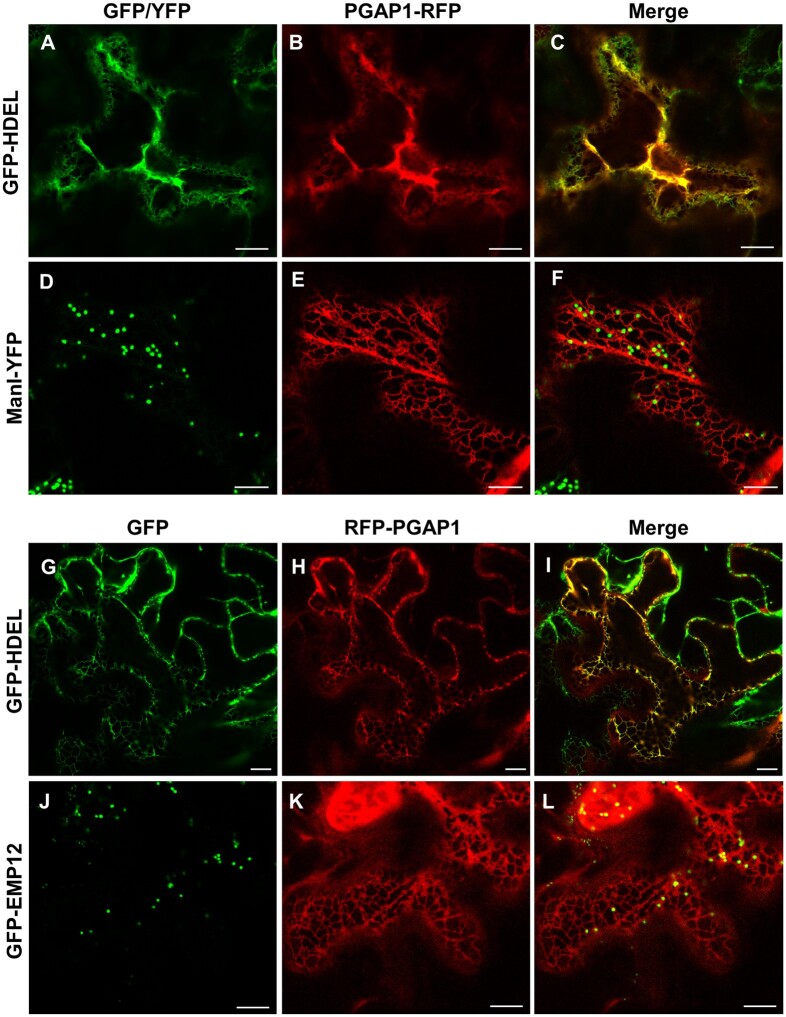
In order to determine the subcellular localization of PGAP1, PGAP1 constructs with either N- or C-terminal RFP were used for transient expression in Nicotiana benthamiana leaves. Both PGAP1-RFP and RFP-PGAP1 showed an ER-like localization pattern and extensively colocalized with the ER marker GFP-HDEL (Figure 2). No colocalization was found with markers of the Golgi apparatus (ManI-YFP or GFP-EMP12). These results clearly showed that Arabidopsis PGAP1 protein localizes to the ER, consistent with the localization of mammalian PGAP1 (Tanaka et al., 2004; Liu et al., 2018) and yeast Bst1p (Elrod-Erickson and Kaiser, 1996).
Figure 2.
Localization of PGAP1-RFP and RFP-PGAP1. Transient expression in N. benthamiana leaves of PGAP1-RFP (B and E) or RFP-PGAP1 (H and K) together with GFP-HDEL (ER marker; A and G) and the Golgi markers ManI-YFP (D) or GFP-EMP12 (J); see merged images in C, F, I, and L. Scale bars = 10 μm.
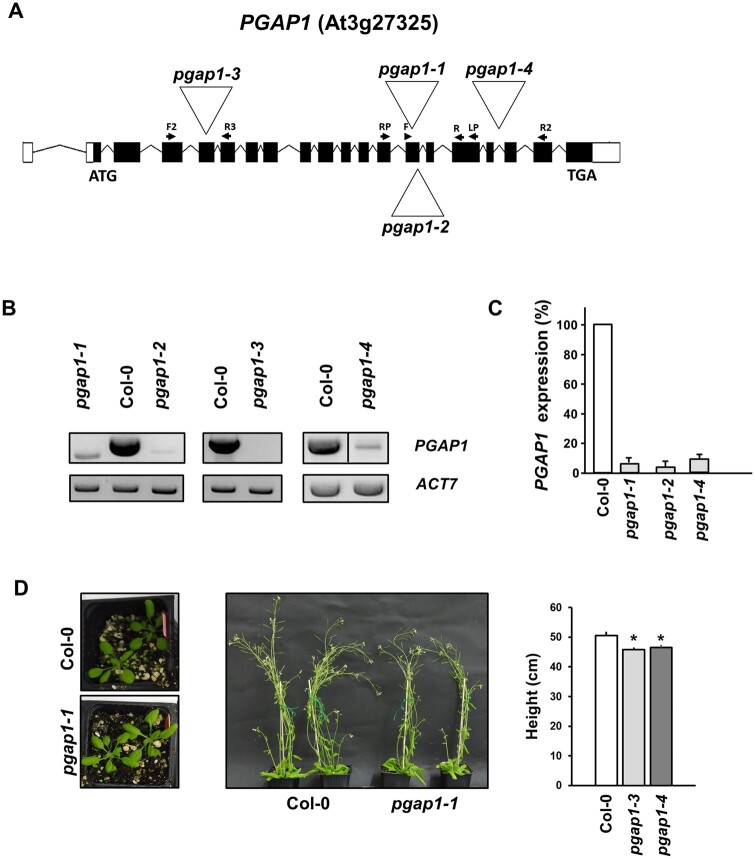
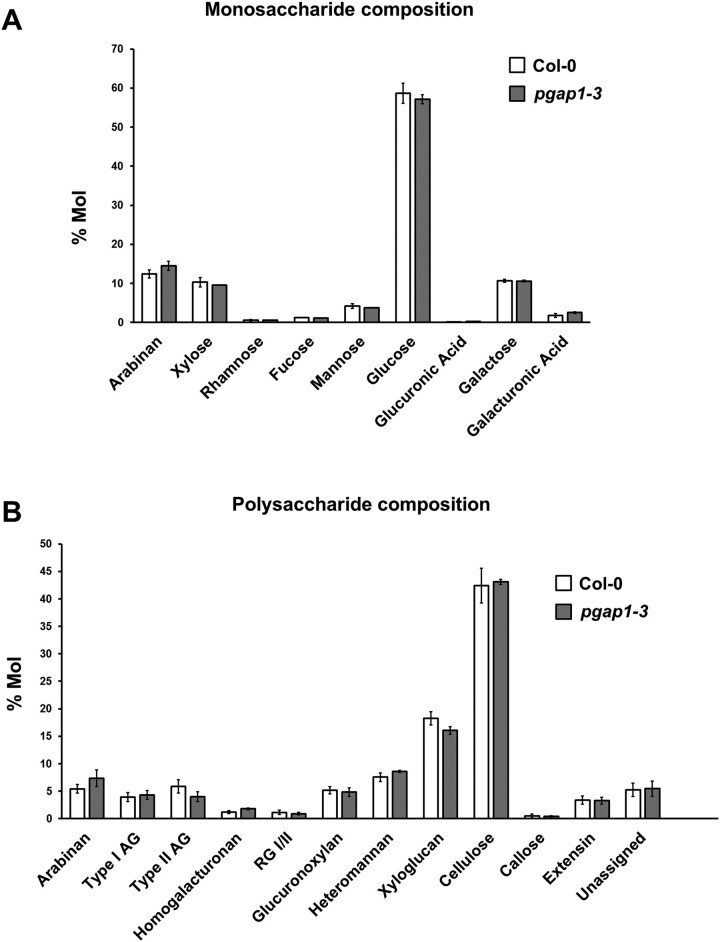
To functionally characterize Arabidopsis PGAP1, a reverse genetics approach was chosen. Several T-DNA insertion mutants of PGAP1 were found in the Arabidopsis SALK collection (http://signal.salk.edu/cgi-bin/tdnaexpress). Four mutants of PGAP1, pgap1–1 (SALK_078662), pgap1–2 (SAIL_1212_H07), pgap1–3 (SALK_027086), and pgap1–4 (SALK_004218), with T-DNA insertions in different positions within the gene, were characterized (Figure 3). Homozygous plants were selected by PCR. Reverse Transcription (RT)-semi-quantitative PCR (sqPCR) analysis indicated that pgap1–1, pgap1–2, and pgap1–4 are knockdown mutants, showing <10% of wild-type PGAP1 expression levels, while pgap1–3 is likely to be a knockout mutant (Figure 3). pgap1 mutants showed slightly reduced plant height compared to wild-type plants (Figure 3). In addition, we observed a reduction in root length in pgap1 seedlings compared to wild-type (Supplemental Figure S4). GPI-APs in plants have been shown to influence cell wall metabolism, cross-linking, and signaling (reviewed in Yeats et al., 2018). To investigate possible changes in cell walls in pgap1 mutants, cell wall compositional analysis was undertaken. Analysis of cell wall polysaccharides showed increased amounts of Arabinan and decreased content of type II AG and xyloglucan in the pgap1–3 mutant compared to wild-type (Figure 4;Supplemental Table S2). These results were confirmed by antibody labeling and immunofluorescence detection of xyloglucan (LM15) and arabinogalactan protein (AGP) (JIM8) epitopes in wild-type and pgap1 mutants. LM15 signals were stronger in cell walls of the upper and lower epidermis in wild-type plants compared to the pgap1–3 mutant (Supplemental Figure S5). JIM8 labeling of AGP epitopes showed weak fluorescence signals in upper and lower epidermal cell walls from wild-type plants, which were undetectable in epidermal cell walls from the pgap1–3 mutant (Supplemental Figure S5).
Figure 3.
Characterization of pgap1 mutants. A, Diagram of the PGAP1 gene and localization of the T-DNA insertion (triangle) in the pgap1–1 and pgap1–2, pgap1–3, and pgap1–4 mutants. Black boxes represent coding regions and white boxes represent 5′- and 3′-Untranslated region (UTR) regions and arrows indicate the positions of the primers used for the characterization of the mutants. B, RT-sqPCR analysis of PGAP1 expression in pgap1 mutants. Total RNA from pgap1–1, pgap1–2, pgap1–3, pgap1–4, and wild-type (Col-0) 4-d-old seedlings were used for PCR. PGAP1-specific primers were used (Supplemental Tables S3 and S4). Actin-7 (ACT7) was used as a control. PCR samples were collected at cycle 22 for ACT7 and at cycle 36 for PGAP1. No wild-type PGAP1 band was detected in pgap1–3. C, Quantification of the bands in the experiments shown in B from three biological samples. No band was detected in the pgap1–3 mutant and thus it is not shown in the quantification. Values were normalized against the PGAP1 fragment band intensity in wild-type that was considered to be 100%. Error bars represent standard error of the mean (SEM). D, Left, 20-d-old plants and middle panel, 42-d-old plants of wild-type and the pgap1–1 mutant. In the right, the height of 42-d-old wild-type and pgap1 mutant plants expressed as mean ± sem (n = 4). Data asterisk represents statistic differences based on Student’s t test with P < 0.05.
Figure 4.
Monosaccharide and polysaccharide compositions of wild-type (Col-0) and pgap1-3 seedlings. Methylation analysis showed no obvious differences in deduced monosaccharide (A) composition between Col-0 and pgap1–3 mutants. Polysaccharide composition (B) did not detect any statistically significant changes at P < 0.05 (Student t test, two sample assuming unequal variance). However, it showed a consistent trend of reduced Type II AG and xyloglucan and increased Arabinan content in pgap1–3 compared to Col-0. RG: rhamnogalacturonan. Data shown are the average of three biological replicates with two technical replicates each. Error bars represent sem.
Localization of GPI-APs in pgap1 mutants
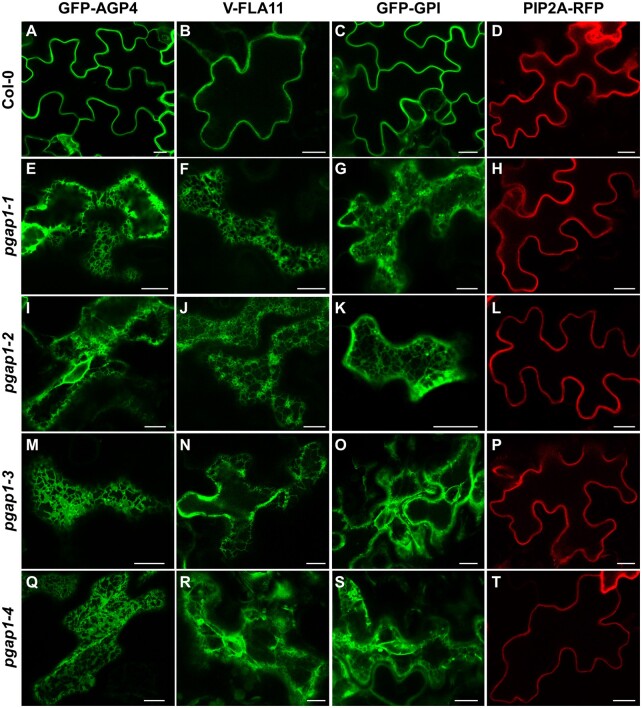
GPI-anchor remodeling has been shown to be important for efficient trafficking of yeast and mammalian GPI-APs from the ER to the plasma membrane (Tanaka et al., 2004; Fujita et al., 2006b; Castillon et al., 2009; Castillon et al., 2011). Therefore, we analyzed the localization of GPI-APs in pgap1 mutants. To this end, we used two different GPI-APs. One of them was GFP fused to arabinogalactan 4 (AGP4), GFP-AGP4. AGPs are cell surface proteoglycans that seem to be involved in diverse developmental processes such as differentiation, cell–cell recognition, embryogenesis, and programmed cell death (Ellis et al., 2010; Pereira et al., 2016; Strasser et al., 2021). GFP-AGP4 has been shown previously to localize to the plasma membrane (Martinière et al., 2012; Bernat-Silvestre et al., 2020). The second one was Venus fused to FLA11 (V-FLA11), a member of Fasciclin-Like Arabinogalactan (FLA) proteins that play important biological roles related to cell adhesion (Johnson, 2003; MacMillan et al., 2010). In addition, we also used a glycosylphosphatidylinositol-anchored GFP (GFP-GPI; Martinière et al., 2012; Bernat-Silvestre et al., 2020). As a control, we used a transmembrane plasma membrane protein, the aquaporin PIP2A-RFP (Nelson et al., 2007).
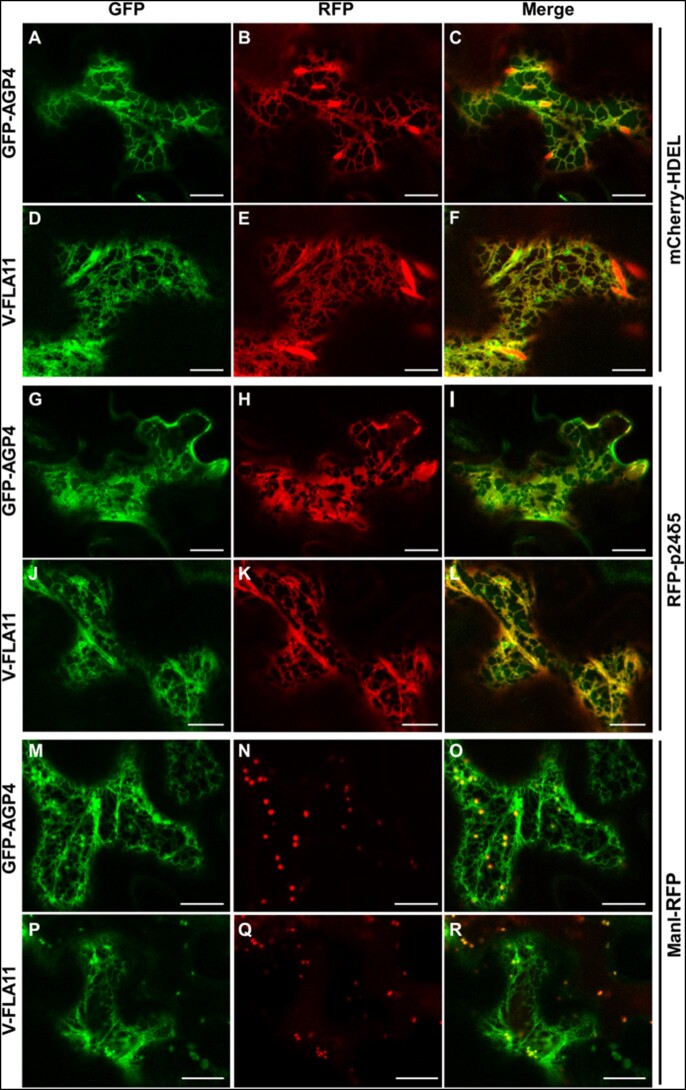
We first analyzed the localization of these proteins by transient expression in Arabidopsis seedlings. As shown in Figure 5, GFP-AGP4, V-FLA11, and GFP-GPI were exclusively localized to the plasma membrane of cotyledon cells of wild-type Arabidopsis seedlings, as was the case for the transmembrane plasma membrane protein PIP2A-RFP. This was shown previously for GFP-AGP4 and GFP-GPI (Bernat-Silvestre et al., 2020). V-FLA11 was also found to colocalize with PIP2A-RFP in wild-type seedlings (Supplemental Figure S6). In clear contrast, GFP-AGP4, V-FLA11, and GFP-GPI showed an ER-like localization pattern in pgap1 mutants, which was not the case of PIP2A-RFP, which localized to the plasma membrane in these mutants (Figure 5; Supplemental Figure S7). This suggests that PGAP1 enzyme is specifically required for transport to the plasma membrane of GPI-APs, and that loss of PGAP1 function does not affect transport from the ER to the plasma membrane of transmembrane proteins. The defect in transport of GPI-APs in pgap1 mutants was not a consequence of an alteration in the compartments of the secretory pathway, since no obvious defects were observed in the localization pattern of several organelle marker proteins, including GFP-HDEL (ER), GFP-EMP12 (Golgi apparatus), TIP1-GFP (tonoplast), SPdCt-mCherry (vacuole lumen), SCAMP1-YFP (plasma membrane), and GFP-CESA3 (TGN/plasma membrane; Supplemental Figure S8). The ER localization of GFP-AGP4 and V-FLA11 in pgap1 mutants was then confirmed by colocalization experiments. As shown in Figure 6, both GFP-AGP4 and V-FLA11 strongly colocalized with two different ER marker proteins, an ER luminal protein (mCherry-HDEL) and an ER membrane protein (RFP-p24δ5). Occasionally, GFP-AGP4 and V-FLA11 were also found in punctate structures, which colocalized with the Golgi marker ManI-RFP, suggesting that a small fraction of these GPI-APs also localize to the Golgi apparatus in pgap1 mutants. The main ER localization of GFP-AGP4 and V-FLA11 in pgap1 mutant Arabidopsis seedlings was also confirmed biochemically (see below).
Figure 5.
Localization of GFP-AGP4, V-FLA11, and GFP-GPI in wild-type and pgap1 Arabidopsis seedlings. Transient gene expression in wild-type (Col-0; A–D) or pgap1–1 (E–H), pgap1–2 (I–L), pgap1–3 (M–P), and pgap1–4 (Q–T) Arabidopsis seedlings. The three GPI-APs, GFP-AGP4, V-FLA11, and GFP-GPI mainly localized to the plasma membrane in cotyledon cells from wild-type (Col-0) seedlings, as the transmembrane plasma membrane protein PIP2A-RFP. In the four pgap1 mutants, GFP-AGP4, V-FLA11, and GFP-GPI showed a predominant ER localization pattern, in contrast to PIP2A-RFP, which mainly localized to the plasma membrane. Scale bars, 10 µm.
Figure 6.
Colocalization of GPI-APs with ER markers in pgap1–3 seedlings. Transient expression in Arabidopsis seedlings. A–F, Coexpression of GFP-AGP4 (A) and V-FLA11 (D) with the ER marker mCherry-HDEL (B and E; see merged images in C and F). G–L, Coexpression of GFP-AGP4 (G) and V-FLA11 (J) with the ER marker RFP-p24δ5 (H and K; see merged images in I and L). M–R, Coexpression of GFP-AGP4 (M) and V-FLA11 (P) with the Golgi marker ManI-RFP (N and Q; see merged images in O and R). Scale bars = 10 µm.
To test if the involvement of PGAP1 in trafficking of GPI-APs was specific for AT3G27325, we also tested the localization of GFP-AGP4, V-FLA11, and GFP-GPI in T-DNA insertion mutants of AT4G34310 (presenting a partial PGAP1 motif and expected to localize to chloroplast/mitochondria) and of other AB_hydrolases (AT2G44970 and AT3G52570; Supplemental Table S1 and Supplemental Figure S9). As shown in Supplemental Figure S10, these GPI-APs localized to the plasma membrane in these mutants, as it was the case of the transmembrane plasma membrane protein PIP2A-RFP. This suggests that these PGAP1-like genes encode proteins that are not required for transport from the ER to the plasma membrane of GPI-APs.
We next analyzed the localization of GFP-AGP4 and GFP-GPI by an alternative transient expression system, Arabidopsis protoplasts. In protoplasts from wild-type Arabidopsis plants, GFP-AGP4 and GFP-GPI mostly localized to the plasma membrane, as we have shown previously (Bernat-Silvestre et al., 2020). In contrast, GFP-AGP4 and GFP-GPI showed a predominant ER localization pattern in protoplasts from the pgap1–1 mutant (Figure 7). The ER localization of GFP-AGP4 and GFP-GPI in pgap1 mutant protoplasts was confirmed by colocalization experiments. As shown in Figure 7, both GFP-AGP4 and GFP-GPI strongly colocalized with two different ER marker proteins, RFP-calnexin and RFP-p24δ5. We could also detect the presence of both GFP-AGP4 and GFP-GPI at the plasma membrane, as shown by colocalization with Fei Mao styryl dye FM4–64, a lipid probe routinely used to label the plasma membrane (Figure 7). This suggests that a fraction of these GPI-APs can reach the plasma membrane in pgap1 mutants, as observed also biochemically (see below). To test if the lack of the PGAP1 enzyme affects the localization of other plasma membrane proteins different from GPI-APs, we used different membrane-anchoring types of minimal constructs, including a myristoylated and palmitoylated GFP (MAP-GFP) and a prenylated GFP (GFP-PAP; Martinière et al., 2012). We also used a transmembrane protein, a GFP fusion with the plasma membrane ATPase (GFP-PMA). As shown in Supplemental Figure S11, these three proteins mainly localized to the plasma membrane of pgap1–1 protoplasts, as in protoplasts from wild-type Arabidopsis plants. Therefore, PGAP1 function seems to be specifically required for ER export and transport to the plasma membrane of GPI-APs.
Figure 7.
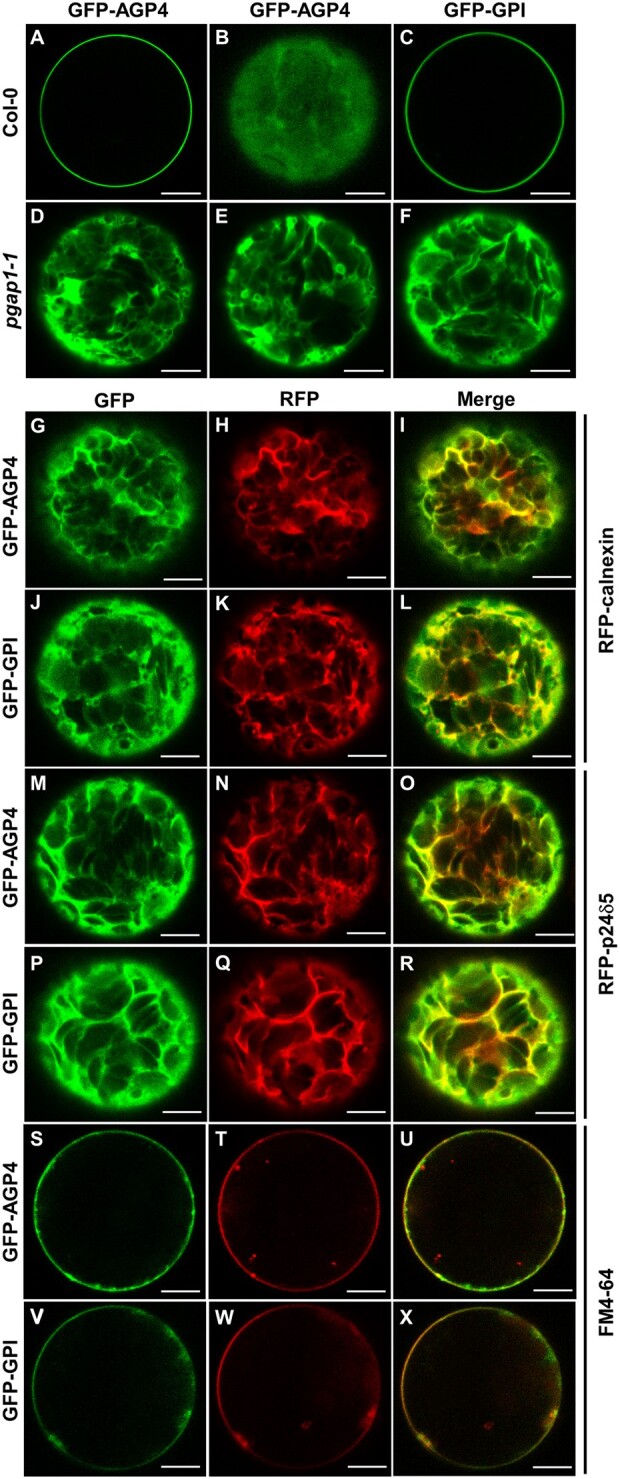
Localization of GFP-AGP4 and GFP-GPI in wild-type and pgap1–1 Arabidopsis protoplasts. Transient expression in wild-type (A–C) and pgap1–1 Arabidopsis protoplasts (D–X). A–F, GFP-AGP4 (A and B) and GFP-GPI (C) localized in the plasma membrane in wild-type protoplasts. A and B show two different confocal planes In contrast, GFP-AGP4 (D, E) and GFP-GPI (F) showed an ER-like localization pattern in pgap1–1 protoplasts (D–F). G–L, Coexpression of GFP-AGP4 (G) or GFP-GPI (J) with the ER marker RFP-calnexin (H and K; see merged images in I and L). M–R, Coexpression of GFP-AGP4 (M) or GFP-GPI (P) with the ER marker RFP- p24δ5 (N and Q; see merged images in O and R). S–X, Colocalization of GFP-AGP4 (S) or GFP-GPI (V) with the FM dye FM4–64 (T, W; see merged images in U and X). Scale bars = 10 μm.
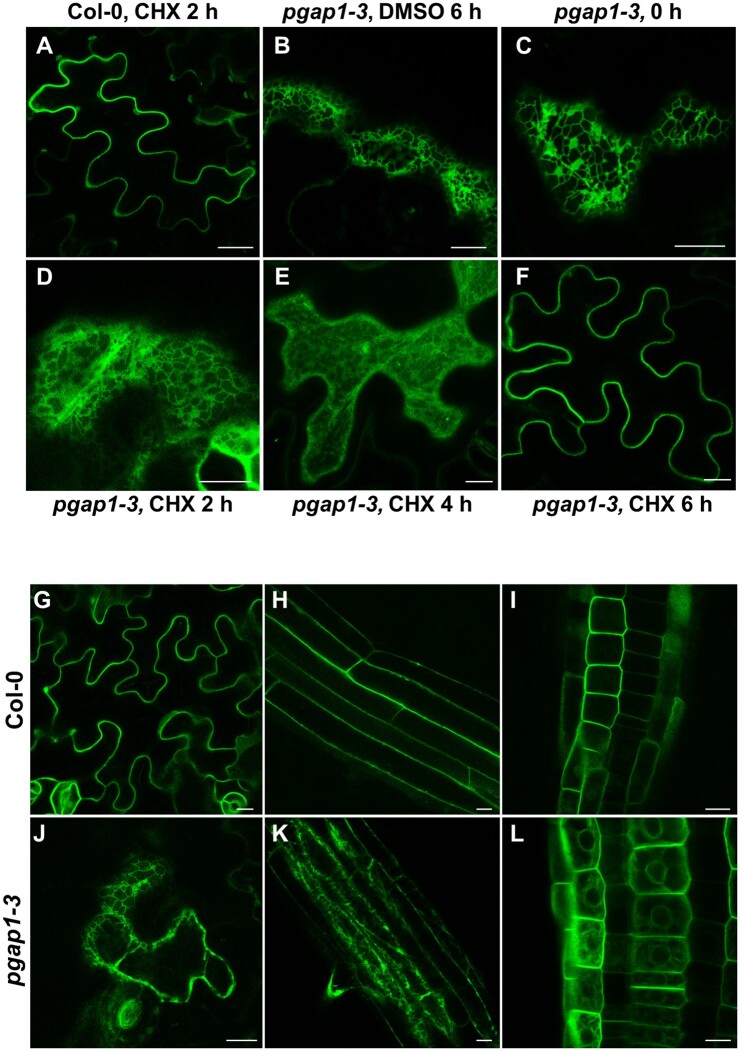
To test if GPI-APs could also reach the cell surface in pgap1 seedlings, the localization of GFP-AGP4 was also analyzed after inhibition of protein synthesis. As shown in Figure 8, A–F, treatment of pgap1–3 seedlings with 20-µM cycloheximide caused a progressive relocalization of GFP-AGP4 from the ER to the cell surface, with ER labeling being almost undetectable after 6 h. This indicates that GFP-AGP4 can reach the cell surface in the absence of PGAP1 but with a delayed kinetics, suggesting that PGAP1 is involved in efficient transport of GPI-APs from the ER to the cell surface.
Figure 8.
Localization of GFP-AGP4 in transient and stable expression systems in wild-type and pgap1–3 Arabidopsis seedlings. A–F, Transient expression of GFP-AGP4. pgap1–3 seedlings expressing GFP-AGP4 were incubated in the presence of 20 µM cycloheximide and analyzed by CLSM after 0, 2, 4, and 6 h (C–F). Controls include CHX-treated wild-type (Col-0; A) and DMSO treated pgap1–3 (B) seedlings. G–L, Stable expression of GFP-AGP4. Localization of GFP-AGP4 in cotyledon (G, J), or root cells (H, I, K, L) of wild-type (Col-0; G–I) or pgap1–3 seedlings (J–L). Scale bars, 10 µm.
Plants stably expressing GFP-AGP4 in wild-type and pgap1 mutant backgrounds were generated to confirm the localization seen in transient expression systems. As shown in Figure 8, G–L, GFP-AGP4 mainly localized at the cell surface in cotyledon and root cells from wild-type seedlings. In contrast, GFP-AGP4 showed a typical ER-localization pattern in pgap1–3 seedlings although it also localized partially to the cell surface, which is consistent with a delayed transport of GPI-APs to the cell surface in the mutant.
GPI-anchor remodeling in pgap1 mutants
We next used biochemical approaches to investigate the putative function of Arabidopsis PGAP1 in GPI anchor remodeling (in particular inositol deacylation) of GPI-APs. We have recently shown that GFP-AGP4 is a GPI-AP (Bernat-Silvestre et al., 2020). Here, we have now biochemically characterized V-FLA11. To this end, a postnuclear supernatant (PNS) from N. benthamiana leaves expressing V-FLA11 was analyzed by SDS–PAGE and immunoblotting with antibodies against GFP, to detect the Venus tag of V-FLA11 (Supplemental Figure S12). Similar to GFP-AGP4, two forms of V-FLA11 were detected, a predominant higher molecular weight smearing form (≈75 kDa), that may correspond to the mature form of the moderately glycosylated V-FLA11, and a much less abundant smaller molecular weight form (≈65 kDa) that may correspond to the ER form of V-FLA11 (Supplemental Figure S12A). To confirm localization of the two V-FLA11 forms, leaves expressing V-FLA11 were treated with brefeldin A (BFA), to accumulate newly synthesized proteins at the ER. In the absence of BFA, V-FLA11 mainly localized to the plasma membrane, as expected (Supplemental Figure S12C). However, in the presence of BFA, V-FLA11 accumulated at the ER, where it showed a high degree of colocalization with the ER membrane protein RFP-p24δ5 (Supplemental Figure S12C). As shown in Supplemental Figure S12A, BFA treatment produced a drastic reduction of the 75 kDa smearing form and a concomitant increase in the 65 kDa band. This strongly suggests that 65-kDa band corresponds to the ER form of V-FLA11, whereas the 75 kDa smear should be the glycosylated form present at the Golgi/plasma membrane. We have previously found a similar behavior for GFP-AGP4, with a 115 kDa smear form corresponding to the plasma membrane form and a 70 kDa band, which was shown to correspond to the ER form of GFP-AGP4 (Bernat-Silvestre et al., 2020).
To demonstrate that V-FLA11 is a GPI-AP, we used Triton X-114 extraction and PI-PLC treatment. PNS from N. benthamiana leaves expressing V-FLA11 was extracted with Triton X-114 (TX114) and the TX114 detergent phase (containing V-FLA11) was either treated or not with PI-PLC, as described in “Materials and Methods”. PI-PLC hydrolyzes the phosphodiester bond of the PI, thereby releasing the protein from the membrane (Low, 1989). As shown in Supplemental Figure S12B, both the plasma membrane smear and the ER form of V-FLA11 were sensitive to PI-PLC, and moved from the detergent to the aqueous phase, thus confirming the GPI anchoring of V-FLA11.
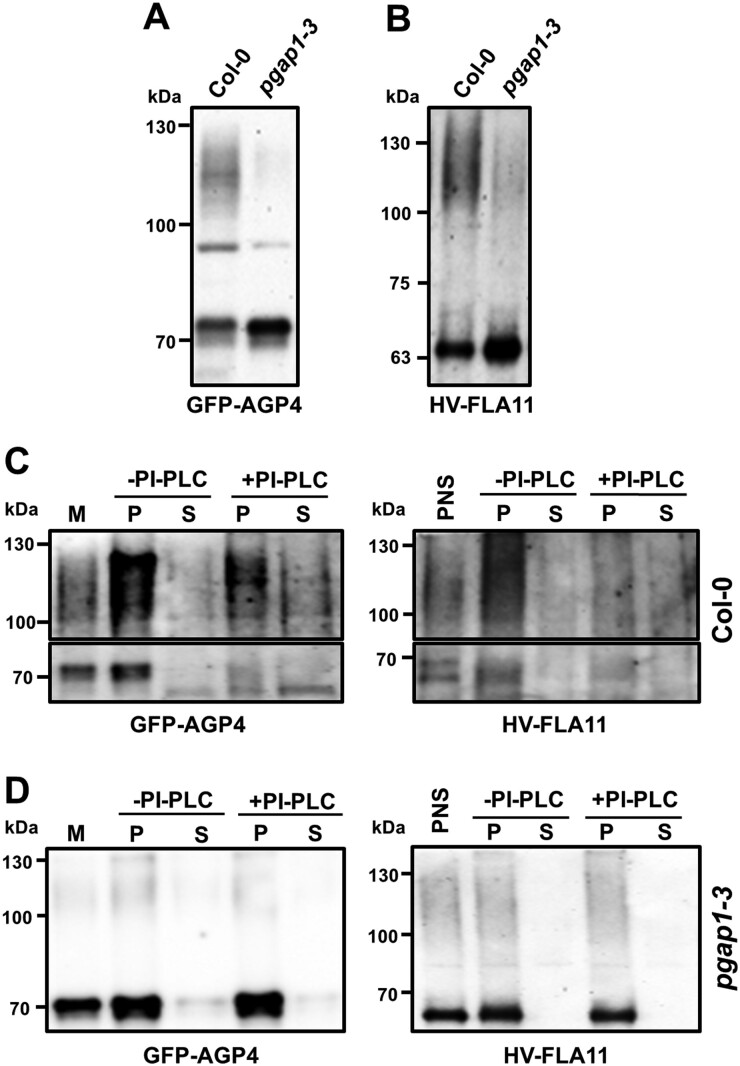
To characterize GFP-AGP4 and V-FLA11 in wild-type and pgap1 mutants, both proteins were transiently expressed in seedlings and PNSs were analyzed by SDS–PAGE and immunoblotting with GFP antibodies, as described previously. Expression of both GFP-AGP4 and V-FLA11 in wild-type seedlings resulted in detection of a higher molecular weight smear form, which represents the plasma membrane form of both proteins, and a lower molecular weight band which should correspond to their ER form (Figure 9). In the case of V-FLA11, the smeared higher molecular weight form was larger than that observed in N. benthamiana leaves, suggesting a greater degree of glycosylation. Remarkably, when both proteins were expressed in the pgap1–3 mutant we found a drastic reduction of the higher molecular weight smear forms and a concomitant increase in the lower molecular weight bands (Figure 9, A and B). This is consistent with the predominant ER localization of both GFP-AGP4 and V-FLA11 in pgap1 mutants shown previously (Figures 5 and 6).
Figure 9.
Biochemical characterization of GFP-AGP4 and V-FLA11 in wild-type and pgap1–3 seedlings. A, B, PNS were obtained from cotyledons of wild-type (Col) and pgap1–3 mutant seedlings transiently expressing GFP-AGP4 (A) and V-FLA11 (B) and analyzed by SDS–PAGE and immunoblotting with antibodies against GFP (to detect GFP-AGP4 and V-FLA11). In wild-type seedlings, both GFP-AGP4 and V-FLA11 showed a smear with a molecular mass around 100–130 kDa, which correspond to the plasma membrane form of GFP-AGP4 (Bernat-Silvestre, 2021) and V-FLA11 (Supplemental Figure S12), and additional bands (around 70 kDa and 63 kDa) corresponding to the ER forms of GFP-GP4 (Bernat-Silvestre et al., 2020) and V-FLA11 (Supplemental Figure S12), respectively. In the pgap1–3 mutant, there was a strong decrease in the smear form (plasma membrane) of both proteins with a concomitant increase in their ER forms. C, D, Left (GFP-AGP4). Membrane fractions were obtained from PNSs of wild-type (C) or pgap1–3 mutant (D) seedlings expressing GFP-AGP4 and incubated in the absence or presence of PI-PLC. Then, membranes were pelleted by centrifugation and pellets (P) and supernatants (S) were analyzed by SDS–PAGE and immunoblotting with antibodies against GFP to detect GFP-AGP4. Right (V-FLA11). PI-PLC treatment was performed directly in the PNSs, membranes were also pelleted by centrifugation and pellets and supernatants analyzed as before. In C, the upper part of the immunoblotting shows the smear form of GFP-AGP4 (left) or V-FLA11 (right) while the lower part highlights the ER bands of GFP-AGP4 (left) or V-FLA11 (right). Notice the decrease of both the smear forms and the lower ER bands from the pellet fraction and their partial appearance in the supernatant in wild-type seedlings. The presence of both forms in the supernatant seems to be only partial, probably due to degradation upon release from the membranes. In contrast, both forms are PI-PLC resistant in the pgap1–3 mutant.
Mature GPI-APs are usually sensitive to PI-PLC because they have the 2-position of inositol free, leading to the release of the protein portions. In contrast, precursors of the GPI anchor that contain an acyl chain linked to the 2-position of inositol are resistant to PI-PLC (Kinoshita and Fujita, 2016). Therefore, PI-PLC can be used as a tool to determine whether GPI-APs in the pgap1 mutants contained or not the acyl chain in the inositol ring. Thus, GFP-AGP4 and V-FLA11 were transiently expressed in wild-type and pgap1 seedlings and PNSs were obtained. Membranes were pelleted by centrifugation, resuspended in buffer and treated or not with PI-PLC. After the treatment, membranes were pelleted again by centrifugation and pellets (membranes) and supernatants (containing released proteins) were analyzed by SDS–PAGE and immunoblotting with GFP antibodies. As shown in Figure 9C, in wild-type seedlings both the plasma membrane smear and the ER band of GFP-AGP4 and V-FLA11 were sensitive to PI-PLC, as shown previously for GFP-AGP4 (Bernat-Silvestre et al., 2020) and for V-FLA11 in N. benthamiana (Supplemental Figure S12). In the pgap1 mutant, the predominant form of both GFP-AGP4 and V-FLA11 is the lower molecular weight ER band. As shown in Figure 9D, neither GFP-AGP4 nor V-FLA11 was released from the membranes upon PI-PLC treatment. Resistance to PI-PLC treatment applied to not only the major lower molecular weight ER band, but also to the small amount of the smeared form, which corresponds to the fraction of these proteins reaching the plasma membrane. These results suggest that both GFP-AGP4 and V-FLA11 remain membrane attached in the pgap1 mutant upon PI-PLC treatment, probably due to a defect in inositol deacylation.
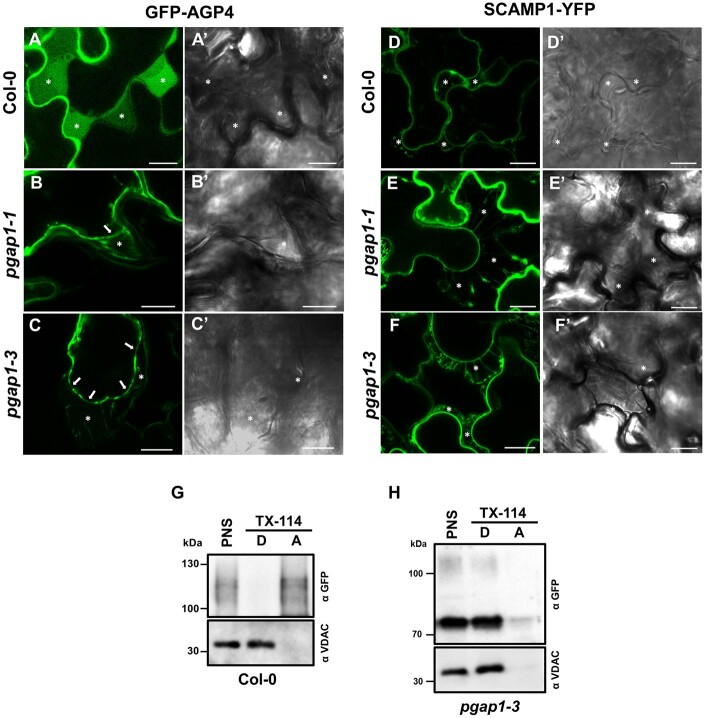
Interestingly, after plasmolysis, most of the cell surface GFP-AGP4 was detected in the apoplast in wild-type seedlings, in contrast to the transmembrane plasma membrane protein SCAMP1-YFP, which mostly appears at the plasma membrane and in Hetchian strands (attachment sites between the plasma membrane and cell wall) upon mannitol treatment (Figure 10). This is consistent with the fact that the highly glycosylated plasma membrane form of GFP-AGP4 (the 115-kDa smear) had been shown to have a high tendency to appear in the aqueous phase after Triton X-114 extraction (Bernat-Silvestre et al., 2020; Figure 10G). This localization is in agreement with the proposal that specific plasma membrane phospholipases may allow the release of some AGPs into the cell wall or the apoplast (Schultz et al., 1998; Ellis et al., 2010; Showalter and Basu, 2016; Ma et al., 2018). In pgap1 mutants, SCAMP1-YFP was still found at the plasma membrane and Hetchian strands. Interestingly, the fraction of GFP-AGP4 reaching the cell surface in pgap1 mutants was found at the plasma membrane and Hetchian strands and was not released to the apoplast (Figure 10, B and C). This was confirmed biochemically by Triton X-114 extraction. Apart from the fact that the predominant form of GFP-AGP4 in pgap1 mutants was the lower molecular weight ER band, which partitioned in the detergent phase upon Triton X-114 extraction, the plasma membrane smear of GFP-AGP4 also partitioned in the detergent phase, as the membrane marker VDAC (Figure 10H). This suggests that GFP-AGP4 remains membrane attached in pgap1 mutants and is not released to the apoplast, probably due to the presence of the acyl group in the inositol moiety of the GPI anchor in the absence of PGAP1.
Figure 10.
Localization of plasma membrane proteins following plasmolysis. A–F, Transient expression experiments in wild-type (Col-0), pgap1–1 and pgap1–3 Arabidopsis seedlings. Left panels (A–F) show CLSM images; right panels (A′–F′) show phase-contrast images. GFP-AGP4, a GPI-AP, mainly localized to the apoplast in wild-type seedlings (A), as shown after plasmolysis following 750 mM mannitol treatment (see asterisks). In the pgap1–1 (B) and pgap1–3 (C) mutants, GFP-AGP4 mainly localized to the ER, but the small fraction of GFP-AGP4 reaching the cell surface remained at the plasma membrane (see arrows) and was not released to the apoplast (asterisks). Note the Hechtian strands in the apoplast of pgap1 mutants. SCAMP1-YFP, a transmembrane plasma membrane protein, showed characteristic plasma membrane localization in a plasmolized cell in wild-type (D) and in pgap1–1 (E) and pgap1–3 (F) mutants. Note the Hechtian strands in the apoplast in all cases. G–H, PNSs from wild-type (G) and pgap1–3 (H) Arabidopsis cotyledons transiently expressing GFP-AGP4 were treated with Triton X-114, and detergent (D) and aqueous (A) phases were analyzed by SDS–PAGE and immunoblotting with GFP antibodies (to detect GFP-AGP4). In wild-type seedlings, GFP-AGP4 mainly appears in the aqueous phase, consistent to its apoplast localization (G). In contrast, GFP-AGP4 appears in the detergent phase, as the membrane marker VDAC, in the pgap1–3 mutant (H). Scale bars = 10 µm.
Discussion
Most of the genes involved in GPI anchor assembly and their remodeling have putative orthologs in Arabidopsis (Luschnig and Seifert, 2011). However, it has to be established if all these orthologs are functional and if their function is conserved in plants. Five Arabidopsis orthologs of enzymes involved in the biosynthesis and attachment of the GPI anchor have been previously studied: SETH1, SETH2, PEANUT1 (PNT1), APTG1, and AtGPI8 (Lalanne et al., 2004; Gillmor et al., 2005; Dai et al., 2014; Bundy et al., 2016; Desnoyer et al., 2020). Arabidopsis null mutants of these enzymes are either gametophytic or embryogenic lethal mutants. This indicates that GPI-APs are essential for plant growth and development. However, no previous characterization of Arabidopsis GPI anchor lipid remodeling enzymes had been reported. In this study, we have undertaken the characterization of mutants of an Arabidopsis ortholog of the enzyme involved in the first step of lipid remodeling of the GPI anchor, yeast Bst1p/mammalian PGAP1.
Arabidopsis PGAP1 (At3g27325) encodes a putative ortholog of yeast Bst1p/mammalian PGAP1, which are involved in inositol deacylation (Tanaka et al., 2004; Fujita et al., 2006b; Luschnig and Seifert, 2011). We have found that Arabidopsis PGAP1 localizes in the ER, as yeast Bst1p (Elrod-Erickson and Kaiser, 1996) and mammalian PGAP1 (Tanaka et al., 2004; Liu et al., 2018). To investigate PGAP1 function, we have characterized loss of function mutants of PGAP1. In humans, null mutations of PGAP1 are viable and result in defects in neuronal cell function (Ueda et al., 2007; Murakami et al., 2014; Williams et al., 2015). ScBst1 is not required for cell survival in yeast (Komath et al., 2018) and ScBst1 mutants grew as well as wild-type at all temperatures (Elrod-Erickson and Kaiser, 1996; Fujita et al., 2006b). In Candida albicans, deletion of Bst1 impaired host infection and caused altered cell wall polysaccharides (Liu et al., 2016). Here, we show that loss-of-function of PGAP1 caused only mild phenotypes under standard growth conditions and showed minor alterations in cell wall composition.
Importantly, trafficking of GPI-APs to the cell surface was altered in the pgap1 mutants, in both transient expression experiments and stably transformed plants. This agrees with previous results in yeast and mammals. Deletion of ScBst1 caused delay in transport of GPI-APs from the ER to the Golgi in yeast (Vashist et al., 2001; Kinoshita and Fujita, 2016) and a defect in PGAP1 caused accumulation of GPI-APs in the ER due to inefficient exit from the ER in mammalian cultured cells (Tanaka et al., 2004). However, steady-state levels of cell surface GPI-APs were only mildly affected and GPI-APs were found at the cell surface, although with unusual GPI structures (three-“footed” GPI) in mammalian cultured cells (Tanaka et al., 2004) and in cells from a patient with a PGAP1 null mutation (Murakami et al., 2014). We observed that a proportion of GPI-APs could still reach the cell surface in Arabidopsis pgap1 mutants, as in mammals and yeast, but with clearly delayed kinetics. Given the mild phenotypes of pgap1 mutants, it is possible that the small amount of GPI-APs that reach the cell surface are sufficient to perform their biological functions and prevent more severe phenotypic alterations. Consistent with this idea, a modified version of Citrin-FLA4 that lacked a GPI anchor was mostly retained in the ER, but the small amount of Citrin-FLA4 secreted to the extracellular space was sufficient to complement the fla4 mutant phenotype (Xue et al., 2017).
Over 40% of the GPI-APs predicted by bioinformatics studies contain proteins with putative AG or extensin-like glycosylation (Borner et al., 2003). Disrupted trafficking and/or glycosylation of GPI-APs could explain the altered composition of cell walls in pgap1 mutants. Changes in Type II AG levels could result from reduced/altered AGP glycosylation and this potentially impacts cell wall assembly and architecture. AG glycans are proposed to cross-link to cell wall components, as is the case for ARABINOXYLAN PECTIN ARABINOGALACTAN PROTEIN 1, shown to covalently cross-link to both pectins and arabinoxylans (Tan et al., 2013). Other GPI-APs can be involved in modulating both synthesis and remodeling of cell wall polymers, such as xyloglucan and cellulose. Members of the COBRA-like family of GPI-APs are known to regulate the deposition of cellulose into the wall (Li et al., 2013; Ben-Tov et al., 2015). A COBRA-like protein, BC1, has been shown to directly bind cellulose to affect MF crystallinity (Liu et al., 2013). Two putative GPI-anchored aspartic proteases, A36 and A39, were shown to co-localize with COBRA-LIKE 10 (Gao et al., 2017a, 2017b). In the apical cell walls of pollen tubes in double a36a39 mutants, increased levels of both highly methyl esterified homogalacturonan pectins and xyloglucans were detected. Changes in cell walls that compromise integrity, either during growth or as a result of damage from abiotic or biotic stresses, are detected by cell wall integrity sensors. GPI-APs are proposed to form part of the cell wall sensing complexes and compromised function could also contribute to changes in cell wall composition.
In the case of GFP-AGP4, we have found that in wild-type plants most of this AGP was found in the apoplast (and not at the plasma membrane), supporting the idea that AGP4 may be secreted into the extracellular matrix perhaps upon the action of plasma membrane phospholipases, as proposed for some of these proteins (Schultz et al., 1998; Ellis et al., 2010; Ma et al., 2018; Yeats et al., 2018). A similar behavior has been recently described for the AGP21, which is also found at the plasma membrane and the apoplast (Borassi et al., 2020). In contrast, other AGPs remain attached to the plasma membrane, such as LeAGP1 (Sun et al., 2004), AGP17 (Sun et al., 2005; Zhang et al., 2011) and AGP18 (Yang and Showalter, 2007; Zhang et al., 2011). In animals, substrate-specific mechanisms have been proposed to release GPI-APs from the cell surface, including the PI-PLCs and PI-PLDs (Yeats et al., 2018). In contrast, no GPI-specific phospholipases have been yet characterized in plants. The identification of both specific and general GPI-cleavage mechanisms in plants may be an interesting area of future studies to understand the function of specific AGPs. The fact that GFP-AGP4 remained attached to the plasma membrane in pgap1 mutants suggests a possible role of PGAP1 in secretion of certain GPI-APs, which could be important for signaling or cell wall localization of GPI-APs.
Finally, the ER localization of PGAP1 and the PI-PLC resistance of GPI-APs in pgap1 mutants suggest that PGAP1 indeed functions in Arabidopsis as the GPI inositol-deacylase, which cleaves the acyl chain from the inositol ring of the GPI. Therefore, the results obtained indicate that PGAP1 is involved in GPI inositol deacylation and that inositol deacylation is important for efficient ER-to-Golgi transport of GPI-APs. This provides functional characterization of an enzyme involved in remodeling of the GPI anchor in plants. The characterization of other enzymes of the remodeling pathway should provide clues on the GPI anchor remodeling pathway in plants, to elucidate whether the pathway is completed at the ER, as in yeast, or if GPI anchor remodeling is completed in the Golgi apparatus (as in mammals). Future studies should also aim to elucidate more plant GPI-anchor structures and the role of GPI-anchor remodeling in sorting of GPI-APs into specific ER export sites and COPII vesicles or in their fate at the cell surface (plasma membrane versus apoplast or cell wall localization).
Materials and methods
Plant material
Arabidopsis (A. thaliana ecotype Col-0) was used as wild-type. T-DNA insertion mutants used in this study were obtained from the Nottingham Arabidopsis Stock Center. A. thaliana plants were grown in growth chambers as previously described (Ortiz-Masia et al., 2007). The T-DNA insertion mutants were characterized by PCR (Supplemental Table S3). Wild-type N. benthamiana plants were grown from surface-sterilized seeds on soil in the greenhouse at 24°C with 16-h daylength.
RT-sqPCR
Total RNA was extracted from seedlings by using a Qiagen RNeasy plant mini kit, and 3 μg of the RNA solution was reverse-transcribed using the maxima first-strand cDNA synthesis kit for RT-sqPCR (Fermentas) according to the manufacturer’s instructions. sqPCRs were performed on 3 μl of cDNA template using Emerald Amp Max PCR Master Mix (Takara). The sequences of the primers used for PCR amplifications are included in Supplemental Table S4.
Constructs and antibodies
The coding sequences of RFP-PGAP1 and PGAP1-RFP were commercially synthesized de novo (Geneart AG) based on the sequence of RFP and that of Arabidopsis PGAP1 (At3g27325) and inserted into pCHF3 (Ortiz-Masia et al., 2007) through the sites KpnI and SalI. The coding sequence of GFP-CESA3 was commercially synthesized de novo (Geneart AG) based on the sequence of CESA3 (At5g05170) and GFP and inserted into pCHF3 through the sites XmaI and PstI.
A pGreenII0179 vector backbone (Hellens et al., 2000) was used for constructing V-FLA11 driven by pro35S: pGreen0179-35S-spFLA11-His-YFP-FLA11. The FLA11 (AT5G03170) coding sequence was amplified from Arabidopsis cDNA and assembled into a pGreen0179-35S-spFLA11-His-YFP vector using NEBuilder HiFi DNA Assembly kit (NEW ENGLAND Biolabs) according to the manufacturer’s instructions. Primers for amplification of FLA11 without a signal peptide are as follows: YMV101-FLA11-F, CAGGCGGAGGTGGGTCACCTAGGCAGGCTCCAGCTCCAGGC; YMV101-FLA11-R, CATTAAAGCAGGACTCTAGATTATAT-CCACAGAGAAGAAGAAGCAG. The constructs used for transient expression experiments (with 35S promoter) were: GFP-AGP4, GFP-GPI, MAP-GFP and GFP-PAP (Martinière et al., 2012; Bernat-Silvestre et al., 2020), GFP-PMA (Kim et al., 2001), PIP2A-RFP (Nelson et al., 2007), and RFP-calnexin (Künzl et al., 2016). Other constructs have been described previously: RFP‐p24δ5 (Langhans et al., 2008; Montesinos et al., 2012); ManI-YFP and ManI-RFP (Nebenführ et al., 1999); GFP‐HDEL (Pain et al., 2019); mCherry-HDEL (Nelson et al., 2007); OsSCAMP1-YFP (Lam et al., 2007), GFP-EMP12 (Gao et al., 2012), TIP1.1-GFP (Gattolin et al., 2011), SPΔCt-mCherry (Pereira et al., 2013). Antibodies against RFP and GFP were obtained from Rockland Immunochemicals, Inc. (USA).
Transient gene expression in Arabidopsis protoplasts, Arabidopsis seedlings, and N. benthamiana leaves
To obtain mesophyll protoplasts from Arabidopsis plants, the Tape-Arabidopsis Sandwich method was used, as described (Wu et al., 2009). Protoplasts were isolated from 4-week-old rosette leaves. For transient expression, we used the PEG transformation method (Yoo et al., 2007). Transient expression of Arabidopsis seedlings by vacuum infiltration (Bernat-Silvestre et al., 2021) and N. benthamiana leaves mediated by Agrobacterium tumefaciens (Lerich et al., 2011) were performed as described previously.
Stable transformation of Arabidopsis plants
Arabidopsis wild-type (Col-0) or pgap1–3 mutant plants were transformed with a GFP-AGP4 construct via Agrobacterium using the floral dip method according to standard procedures (Clough and Bent, 1998). T1 plants were analyzed by Confocal laser scanning microscopy (CLSM).
Preparation of protein extracts, PI-PLC treatment, SDS–PAGE, and immunoblotting
Nicotiana benthamiana leaves or cotyledons from Arabidopsis seedlings expressing XFP-Proteins were frozen in liquid N2 and then grinded in homogenization buffer (HB, 0.3 M sucrose; 1 mM EDTA; 20 mM KCl; 20 mM HEPES pH 7.5), supplemented with 1 mM DTT and a Protease Inhibitor Cocktail (Sigma), using a mortar and a pestle. The homogenate was centrifuged for 10 min at 1,200g and 4°C, and the PNS was collected.
For treatment with PI-PLC following transient expression of V-FLA11 in N. benthamiana leaves, PNS was incubated in the presence of 2% v/v TX-114 for 30 min at 4°C and then centrifuged 5 min at 16,000g to pellet insoluble material. The supernatant was collected and incubated for 10 min at 37°C to achieve phase partitioning. The mixture was centrifuged 10 min at 20,000 ×g and 25°C and the upper aqueous phase (A) and lower detergent phases (D) were collected. The detergent phase was diluted with TBS and incubated in the absence or presence of two U PI-PLC (from Bacillus cereus, 100 U mL−1, Invitrogen) for 1 h at 37°C. After this, samples were centrifuged again 10 min at 20,000g and 25°C to separate aqueous and detergent phases. Aqueous and detergent fractions were analyzed by SDS–PAGE and immunoblotting with GFP antibodies (to detect GFP-AGP4 and V-FLA11). For treatment with PI-PLC following transient expression of GFP-AGP4 or V-FLA11 in Arabidopsis seedlings, a PNS was obtained. In the case of GFP-AGP4, membranes were pelleted by centrifugation of the PNS for 1 h at 150,000g. PNS (V-FLA11) or membrane fractions (GFP-AGP4) were then incubated in the absence or presence of PI-PLC, as before, and centrifuged for 1 h at 150,000g and 4°C, in order to separate membranes (pellet) from proteins released from the membranes upon PI-PLC treatment (supernatant). Membrane fractions were extracted with a lysis buffer containing 150 mM NaCl, 0.5 mM DTT, 0.5% Triton X-100 and 50 mM Tris–HCl pH 7.5. After a 5 min centrifugation at 16,000g to remove detergent-insoluble material, membrane extracts were analyzed by SDS–PAGE and immunoblotting. Immunoblots were developed using the SuperSignal West Pico chemiluminescent substrate (Pierce,ThermoScientific) and analyzed using the ChemiDoc XRS + imagingsystem (Bio-Rad, http://www.bio-rad.com/). Immunoblots in the linear range of detection were quantified using Quantity One software (Bio-Rad Laboratories).
Confocal microscopy
Confocal fluorescent images were collected using an Olympus FV1000 confocal microscope with 603 water lens. The GFP signal was visualized with laser excitation at 488 nm and emission at 496–518 nm. The RFP signal was visualized with laser excitation at 543 nm and emission at 593–636 nm. Sequential scanning was used to avoid any interference between fluorescence channels. Post-acquisition image processing was performed using the FV10-ASW 4.2 Viewer and ImageJ (v.1.45).
Immunofluorescence labeling
Seven-day-old seedlings were fixed with 2.5% (v/v) glutaraldehyde in potassium phosphate buffer (0.025 M, pH 7), dehydrated in graded ethanol series, infiltrated in a graded LR White resin and polymerized according to Wilson and Bacic (2012). Two hundred and fifty nanometer thin sections were obtained with a Leica Ultracut R microtome (Leica Microsystems, Germany) and placed on glass microscope slides. Cell wall antibodies used were LM15 (for xyloglucan; Marcus et al., 2008) and JIM8 (for AGP; Pennell et al., 1991). Secondary antibody used was Alexa Fluor 488 goat anti-rat IgG (H + L; Life Technology; # A48262). Images were acquired with an Olympus BX53 microscope under GFP channel.
Cell wall linkage analysis
De-starched AIR samples were prepared from 7-d-old seedlings grown on MS media with 1% sucrose. AIR was carboxyl reduced and methylated for linkage analysis according to the method outlined in Pettolino et al., (2012). The resulting permethylated alditol acetates were separated and quantified by gas chromatography–mass spectrometry (GC-MS) as described in Pettolino et al., (2012). Polysaccharide composition was deduced from the linkage analyses. Two biological replicates with two technical replicates each were measured. Data are shown as average.
PGAP1 phylogeny analysis
Putative Arabidopsis PGAP1-like genes were selected based on whether they were annotated with the motif IPR012908 and/or pfam07819 in Pfam and InterPro databases. Additionally, the presence, significance, and completeness of the motifs were checked using PfamScan (http://europepmc.org/article/MED/30976793). Ortholog sequences of Arabidopsis AT3G27325, AT4G34310, and AT5G17670 genes were obtained from Ensemble Plants (https://doi.org/10.1093/nar/gkz890). A total of 271 protein sequences from 91 different plant species were retrieved. Multiple sequence alignment was performed using T-Coffee v.13.44 (Notredame et al., 2000). A maximum-likelihood tree was reconstructed using IQ-TREE (version 1.5) and branch supports were calculated with ultrafast bootstrap and SH-aLRT test (1,000 replicates; Hoang et al., 2018). Best fitting evolutionary model was determined to be JTT+R7 using ModelFinder (Kalyaanamoorthy et al., 2017).
Accession numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers: AT5G10430 (AGP4), AT5G03170 (FLA11), and AT3G27325 (PGAP1).
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1. Phylogenetic analysis of PGAP1-like genes.
Supplemental Figure S2. Alignment of N-terminal sequences and topology prediction of AtPGAP1, HsPGAP1, and Bst1p.
Supplemental Figure S3. Developmental expression of Arabidopsis PGAP1.
Supplemental Figure S4. Root length in pgap1 mutants.
Supplemental Figure S5. Localization of xyloglucan and arabinogalactan epitopes in wild-type (Col-0) and pgap1–3.
Supplemental Figure S6. Localization of V-FLA11 in wild-type (Col-0) seedlings.
Supplemental Figure S7. Localization of GFP-AGP4 and PIP2A-RFP in pgap1–3 seedlings.
Supplemental Figure S8. Localization of organelle marker proteins in wild-type (Col-0) and pgap1–3.
Supplemental Figure S9. Mutants in other AB_hydrolases different from PGAP1.
Supplemental Figure S10. Localization of plasma membrane proteins in mutants of other AB_hydrolases different from PGAP1.
Supplemental Figure S11. Localization of plasma membrane proteins without a GPI anchor in wild-type and pgap1–1 protoplasts.
Supplemental Figure S12. Biochemical characterization of V-FLA11.
Supplemental Table S1. Putative Arabidopsis GPI inositol-deacylase PGAP1-like genes.
Supplemental Table S2. Polysaccharide calculations.
Supplemental Table S3. T-DNA mutants and PCR primers used for their characterization.
Supplemental Table S4. List of primers used for RT-sqPCR.
Supplementary Material
Acknowledgments
We thank John Runions for the GFP-AGP4, GFP-GPI, MAP-GFP, and GFP-PAP constructs, Inhwan Hwang for the GFP-PMA construct, Liwen Jiang for the GFP-EMP12 and OsSCAMP1-YFP constructs, Lorenzo Frigerio for the TIP1.1-GFP construct and Claudia Pereira for the SPΔCt-mCherry construct. We thank the microscopy section and the greenhouse of SCSIE (University of Valencia). We thank Verena Kriechbaumer (Oxford Brookes University) for advice with analysis of CLSM images.
Funding
F.A. and M.J.M. were supported by the Ministerio de Economía y Competitividad (grant no BFU2016-76607-P) and Generalitat Valenciana (AICO/2020/187). C.B.S. and J.S.S. were recipient of a fellowship from Ministerio de Ciencia, Innovación y Universidades (FPU program). C.B.S. was recipient of an EMBO short-term fellowship and a short-term fellowship from Ministerio de Ciencia, Innovación y Universidades.
Conflict of interest statement. No potential conflict of interest was reported by the author(s).
F.A., K.J. and M.J.M. conceived and designed the experiments. C.B.S., J.S.S., and Y.M. performed the experiments. F.A. and M.J.M. wrote the paper with input from all other authors.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plphys/pages/general-instructions) is María Jesús Marcote (mariajesus.marcote@uv.es) and Fernando Aniento (Fernando.aniento@uv.es).
References
- Ben-Tov D, Abraham Y, Stav S, Thompson K, Loraine A, Elbaum R, de Souza A, Pauly M, Kieber JJ, Harpaz-Saad S (2015) COBRA-LIKE2, a member of the glycosylphosphatidylinositol-anchored COBRA-LIKE family, plays a role in cellulose deposition in Arabidopsis seed coat mucilage secretory cells. Plant Physiol 167: 711–724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernat-Silvestre C, De Sousa Vieira V, Sánchez-Simarro J, Aniento F, Marcote MJ (2021) Transient transformation of A. thaliana seedlings by vacuum infiltration. Methods Mol Biol 2200: 147–155 [DOI] [PubMed] [Google Scholar]
- Bernat-Silvestre C, Vieira VDS, Sanchez-Simarro J, Pastor-Cantizano N, Hawes C, Marcote MJ, Aniento F (2020) P24 Family proteins are involved in transport to the plasma membrane of GPI-anchored proteins in plants. Plant Physiol 184: 1333–1347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borassi C, Gloazzo Dorosz J, Ricardi MM, Carignani Sardoy M, Pol Fachin L, Marzol E, Mangano S, Rodríguez Garcia DR, Martínez Pacheco J, Rondón Guerrero Y, del C, et al. (2020) A cell surface arabinogalactan‐peptide influences root hair cell fate. New Phytol 227: 732–743 [DOI] [PubMed] [Google Scholar]
- Borner GHH, Lilley KS, Stevens TJ, Dupree P (2003) Identification of glycosylphosphatidylinositol-anchored proteins in Arabidopsis. A proteomic and genomic analysis. Plant Physiol 132: 568–577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosson R, Jaquenoud M, Conzelmann A (2006) GUP1 of Saccharomyces cerevisiae encodes an O-acyltransferase involved in remodeling of the GPI anchor. Mol Biol Cell 17: 2636–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bundy MGR, Kosentka PZ, Willet AH, Zhang L, Miller E, Shpak ED (2016) A mutation in the catalytic subunit of the glycosylphosphatidylinositol transamidase disrupts growth, fertility, and stomata formation. Plant Physiol 171: 974–985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castillon GA, Aguilera-Romero A, Manzano-Lopez J, Epstein S, Kajiwara K, Funato K, Watanabe R, Riezman H, Muñiz M (2011) The yeast p24 complex regulates GPI-anchored protein transport and quality control by monitoring anchor remodeling. Mol Biol Cell 22: 2924–2936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castillon GA, Watanabe R, Taylor M, Schwabe TME, Riezman H (2009) Concentration of GPI-anchored proteins upon ER exit in yeast. Traffic 10: 186–200 [DOI] [PubMed] [Google Scholar]
- Cheung AY, Li C, Zou YJ, Wu HM (2014) Glycosylphosphatidylinositol anchoring: Control through modification. Plant Physiol 166: 748–750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Dai XR, Gao X-Q, Chen GH, Tang LL, Wang H, Zhang XS (2014) ABNORMAL POLLEN TUBE GUIDANCE1, an endoplasmic reticulum-localized mannosyltransferase homolog of GLYCOSYLPHOSPHATIDYLINOSITOL10 in yeast and PHOSPHATIDYLINOSITOL GLYCAN ANCHOR BIOSYNTHESIS B in human, is required for Arabidopsis pollen tube Micropylar Gu. Plant Physiol 165: 1544–1556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desnoyer N, Howard G, Jong E, Palanivelu R (2020) AtPIG-S, a predicted glycosylphosphatidylinositol transamidase subunit, is critical for pollen tube growth in Arabidopsis. BMC Plant Biol 20:380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobson L, Reményi I, Tusnády GE (2015) CCTOP: A consensus constrained TOPology prediction web server. Nucleic Acids Res 43: W408–W412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis M, Egelund J, Schultz CJ, Bacic A (2010) Arabinogalactan-proteins: Key regulators at the cell surface? Plant Physiol 153: 403–419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elrod-Erickson MJ, Kaiser CA (1996) Genes that control the fidelity of endoplasmic reticulum to Golgi transport identified as suppressors of vesicle budding mutations. Mol Biol Cell 7: 1043–1058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn RD, Mistry J, Tate J, Coggill P, Heger A, Pollington JE, Gavin OL, Gunasekaran P, Ceric G, Forslund K, et al. (2010) The Pfam protein families database. Nucleic Acids Res 38: D211–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita M, Umemura M, Yoko-o T, Jigami Y (2006a) PER1 is required for GPI-phospholipase A2 activity and involved in lipid remodeling of GPI-anchored proteins. Mol Biol Cell 17: 5253–5264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita M, Yoko-O T, Jigami Y (2006b) Inositol deacylation by Bst1p is required for the quality control of glycosylphosphatidylinositol-anchored proteins. Mol Biol Cell 17: 834–850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao C, Yu CKY, Qu S, San MWY, Li KY, Lo SW, Jiang L (2012) The golgi-localized Arabidopsis endomembrane protein 12 contains both endoplasmic reticulum export and golgi retention signals at its C terminus. Plant Cell 24: 2086–2104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao H, Li R, Guo Y (2017a) Arabidopsis aspartic proteases A36 and A39 play roles in plant reproduction. Plant Signal Behav 12: 219–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao H, Zhang Y, Wang W, Zhao K, Liu C, Bai L, Li R, Guo Y (2017b) Two membrane-anchored aspartic proteases contribute to pollen and ovule development. Plant Physiol 173: 219–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gattolin S, Sorieul M, Frigerio L (2011) Mapping of tonoplast intrinsic proteins in maturing and germinating Arabidopsis seeds reveals dual localization of embryonic TIPs to the tonoplast and plasma membrane. Mol Plant 4: 180–189 [DOI] [PubMed] [Google Scholar]
- Ghugtyal V, Vionnet C, Roubaty C, Conzelmann A (2007) CWH43 is required for the introduction of ceramides into GPI anchors in Saccharomyces cerevisiae. Mol Microbiol 65: 1493–1502 [DOI] [PubMed] [Google Scholar]
- Gillmor CS, Lukowitz W, Brininstool G, Sedbrook JC, Hamann T, Poindexter P, Somerville C (2005) Glycosylphosphatidylinositol-anchored proteins are required for cell wall synthesis and morphogenesis in Arabidopsis. Plant Cell 17: 1128–1140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellens RP, Anne Edwards E, Leyland NR, Bean S, Mullineaux PM (2000) pGreen: A versatile and flexible binary Ti vector for Agrobacterium-mediated plant transformation. Plant Mol Biol 42: 819–832 [DOI] [PubMed] [Google Scholar]
- Hemsley PA (2015) The importance of lipid modified proteins in plants. New Phytol 205: 476–489 [DOI] [PubMed] [Google Scholar]
- Hoang DT, Chernomor O, Von Haeseler A, Minh BQ, Vinh LS (2018) UFBoot2: Improving the ultrafast bootstrap approximation. Mol Biol Evol 35: 518–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann K, Stoffel W (1993) TMbase - A database of membrane spanning proteins segments. Biol Chem Hoppe Seyler 347: 166 [Google Scholar]
- Hruz T, Laule O, Szabo G, Wessendorp F, Bleuler S, Oertle L, Widmayer P, Gruissem W, Zimmermann P (2008) Genevestigator V3: A reference expression database for the meta-analysis of transcriptomes. Adv Bioinformatics 2008: 1–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter S, Apweiler R, Attwood TK, Bairoch A, Bateman A, Binns D, Bork P, Das U, Daugherty L, Duquenne L, et al. (2009) InterPro: The integrative protein signature database. Nucleic Acids Res 37: D211–D215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson KL (2003) The fasciclin-like arabinogalactan proteins of Arabidopsis. A multigene family of putative cell adhesion molecules. Plant Physiol 133: 1911–1925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalyaanamoorthy S, Minh BQ, Wong TKF, Von Haeseler A, Jermiin LS (2017) ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat Methods 14: 587–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DH, Eu Y-J, Yoo CM, Kim Y-W, Pih KT, Jin JB, Kim SJ, Stenmark H, Hwang I (2001) Trafficking of phosphatidylinositol 3-phosphate from the trans-Golgi network to the lumen of the central vacuole in plant cells. Plant Cell 13: 287–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita T (2020) Biosynthesis and biology of mammalian GPI-anchored proteins. Open Biol 10: 190290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita T, Fujita M (2016) Biosynthesis of GPI-anchored proteins: Special emphasis on GPI lipid remodeling. J Lipid Res 57: 6–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komath SS, Singh SL, Pratyusha VA, Sah SK (2018) Generating anchors only to lose them: The unusual story of glycosylphosphatidylinositol anchor biosynthesis and remodeling in yeast and fungi. IUBMB Life 70: 355–383 [DOI] [PubMed] [Google Scholar]
- Künzl F, Früholz S, Fäßler F, Li B, Pimpl P (2016) Receptor-mediated sorting of soluble vacuolar proteins ends at the trans-Golgi network/early endosome. Nat Plants 2: 16017. [DOI] [PubMed] [Google Scholar]
- Lalanne E, Honys D, Johnson A, Borner GHH, Lilley KS, Dupree P, Grossniklaus U, Twell D (2004) SETH1 and SETH2, two components of the glycosylphosphatidylinositol anchor biosynthetic pathway, are required for pollen germination and tube growth in Arabidopsis. Plant Cell 16: 229–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam SK, Siu CL, Hillmer S, Jang S, An G, Robinson DG, Jiang L (2007) Rice SCAMP1 defines clathrin-coated, trans -golgi–located tubular-vesicular structures as an early endosome in tobacco BY-2 cells. Plant Cell 19: 296–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langhans M, Marcote MJ, Pimpl P, Virgili-López G, Robinson DG, Aniento F (2008) In vivo trafficking and localization of p24 proteins in plant cells. Traffic 9: 770–785 [DOI] [PubMed] [Google Scholar]
- Lerich A, Langhans M, Sturm S, Robinson DG (2011) Is the 6 kDa tobacco etch viral protein a bona fide ERES marker? J Exp Bot 62: 5013–5023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Ge FR, Xu M, Zhao XY, Huang GQ, Zhou LZ, Wang JG, Kombrink A, McCormick S, Zhang XS, et al. (2013) Arabidopsis COBRA-LIKE 10, a GPI-anchored protein, mediates directional growth of pollen tubes. Plant J 74: 486–497 [DOI] [PubMed] [Google Scholar]
- Liu L, Shang-Guan K, Zhang B, Liu X, Yan M, Zhang L, Shi Y, Zhang M, Qian Q, Li J, et al. (2013) Brittle Culm1, a COBRA-like protein, functions in cellulose assembly through binding cellulose microfibrils. PLoS Genet 9: e1003704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, Zou Z, Huang X, Shen H, He LJ, Chen SM, Li LP, Yan L, Zhang SQ, Zhang JD, et al. (2016) Bst1 is required for Candida albicans infecting host via facilitating cell wall anchorage of Glycosylphosphatidyl inositol anchored proteins. Sci Rep 6: 34854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu YS, Guo XY, Hirata T, Rong Y, Motooka D, Kitajima T, Murakami Y, Gao XD, Nakamura S, Kinoshita T, et al. (2018) N-Glycan-dependent protein folding and endoplasmic reticulum retention regulate GPI-anchor processing. J Cell Biol 217: 585–599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Low MG (1989) The glycosyl-phosphatidylinositol anchor of membrane proteins. BBA-Rev Biomembr 988: 427–454 [DOI] [PubMed] [Google Scholar]
- Luschnig C, Seifert GJ (2011) Posttranslational modifications of plasma membrane proteins and their implications for plant growth and development. Plant Cell Monogr 19: 109–128 [Google Scholar]
- Ma Y, Zeng W, Bacic A, Johnson K (2018) AGPs through time and space. Annu Plant Rev Online 1: 767–804 [Google Scholar]
- MacMillan CP, Mansfield SD, Stachurski ZH, Evans R, Southerton SG (2010) Fasciclin-like arabinogalactan proteins: Specialization for stem biomechanics and cell wall architecture in Arabidopsis and Eucalyptus. Plant J 62: 689–703 [DOI] [PubMed] [Google Scholar]
- Maeda Y, Tashima Y, Yoko-o T, Jigami Y, Fujita M, Kinoshita T, Houjou T, Taguchi R (2007) Fatty acid remodeling of GPI-anchored proteins is required for their raft association. Mol Biol Cell 18: 1497–1506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manzano-Lopez J, Perez-Linero AM, Aguilera-Romero A, Martin ME, Okano T, Silva DV, Seeberger PH, Riezman H, Funato K, Goder V, et al. (2015) COPII coat composition is actively regulated by luminal cargo maturation. Curr Biol 25: 152–162 [DOI] [PubMed] [Google Scholar]
- Marcus SE, Verhertbruggen Y, Hervé C, Ordaz-Ortiz JJ, Farkas V, Pedersen HL, Willats WG, Knox JP (2008) Pectic homogalacturonan masks abundant sets of xyloglucan epitopes in plant cell walls. BMC Plant Biol 8: 60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinière A, Lavagi I, Nageswaran G, Rolfe DJ, Maneta-Peyret L, Luu D-T, Botchway SW, Webb SED, Mongrand S, Maurel C, et al. (2012) Cell wall constrains lateral diffusion of plant plasma-membrane proteins. Proc Natl Acad Sci U S A 109: 12805–12810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montesinos JC, Sturm S, Langhans M, Hillmer S, Marcote MJ, Robinson DG, Aniento F (2012) Coupled transport of Arabidopsis p24 proteins at the ER-Golgi interface. J Exp Bot 63: 4243–4261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muñiz M, Riezman H (2016) Trafficking of glycosylphosphatidylinositol anchored proteins from the endoplasmic reticulum to the cell surface. J Lipid Res 57: 352–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muniz M, Zurzolo C (2014) Sorting of GPI-anchored proteins from yeast to mammals - common pathways at different sites? J Cell Sci 127: 2793–2801 [DOI] [PubMed] [Google Scholar]
- Murakami Y, Tawamie H, Maeda Y, Büttner C, Buchert R, Radwan F, Schaffer S, Sticht H, Aigner M, Reis A, et al. (2014) Null mutation in PGAP1 impairing Gpi-anchor maturation in patients with intellectual disability and encephalopathy. PLoS Genet 10: e1004320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nebenführ A, Gallagher LA, Dunahay TG, Frohlick JA, Mazurkiewicz AM, Meehl JB, Staehelin LA (1999) Stop-and-go movements of plant golgi stacks are mediated by the acto-myosin system. Plant Physiol 121: 1127–1141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson BK, Cai X, Nebenführ A (2007) A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. Plant J 51: 1126–1136 [DOI] [PubMed] [Google Scholar]
- Notredame C, Higgins DG, Heringa J (2000) T-coffee: A novel method for fast and accurate multiple sequence alignment. J Mol Biol 302: 205–217 [DOI] [PubMed] [Google Scholar]
- Ortiz-Masia D, Perez-Amador MA, Carbonell J, Marcote MJ (2007) Diverse stress signals activate the C1 subgroup MAP kinases of Arabidopsis. FEBS Lett 581: 1834–1840 [DOI] [PubMed] [Google Scholar]
- Oxley D, Bacic A (1999) Structure of the glycosylphosphatidylinositol anchor of an arabinogalactan protein from Pyrus communis suspension-cultured cells. Proc Natl Acad Sci U S A 96: 14246–14251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pain C, Kriechbaumer V, Kittelmann M, Hawes C, Fricker M (2019) Quantitative analysis of plant ER architecture and dynamics. Nat Commun 10: 984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennell RI, Janniche L, Kjellbom P, Scofield GN, Peart JM, Roberts K (1991) Developmental regulation of a plasma membrane arabinogalactan protein epitope in oilseed rape flowers. Plant Cell 3: 1317–1326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira AM, Lopes AL, Coimbra S (2016) JAGGER, an AGP essential for persistent synergid degeneration and polytubey block in Arabidopsis. Plant Signal Behav 11: e1209616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira C, Pereira S, Satiat-Jeunemaitre B, Pissarra J (2013) Cardosin A contains two vacuolar sorting signals using different vacuolar routes in tobacco epidermal cells. Plant J 76: 87–100 [DOI] [PubMed] [Google Scholar]
- Pettolino FA, Walsh C, Fincher GB, Bacic A (2012) Determining the polysaccharide composition of plant cell walls. Nat Protoc 7: 1590–1607 [DOI] [PubMed] [Google Scholar]
- Pittet M, Conzelmann A (2007) Biosynthesis and function of GPI proteins in the yeast Saccharomyces cerevisiae. Biochim Biophys Acta 1771: 405–420 [DOI] [PubMed] [Google Scholar]
- Rodriguez-Gallardo S, Kurokawa K, Sabido-Bozo S, Cortes-Gomez A, Ikeda A, Zoni V, Aguilera-Romero A, Perez-Linero AM, Lopez S, Waga M, et al. (2020) Ceramide chain length-dependent protein sorting into selective endoplasmic reticulum exit sites. Sci Adv 6: eaba8237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz C, Gilson P, Oxley D, Youl J, Bacic A (1998) GPI-anchors on arabinogalactan-proteins: Implications for signalling in plants. Trends Plant Sci 3: 426–431 [Google Scholar]
- Showalter AM, Basu D (2016) Glycosylation of arabinogalactan-proteins essential for development in Arabidopsis. Commun Integr Biol 9: e1177687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva L, De Almeida RFM, Fedorov A, Matos APA, Prieto M (2006) Ceramide-platform formation and -induced biophysical changes in a fluid phospholipid membrane. Mol Membr Biol 23: 137–148 [DOI] [PubMed] [Google Scholar]
- Strasser R, Seifert G, Doblin MS, Johnson KL, Ruprecht C, Pfrengle F, Bacic A, Estevez JM (2021) Cracking the “Sugar Code”: A snapshot of N- and O-glycosylation pathways and functions in plants cells. Front Plant Sci 12: 157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun W, Xu J, Yang J, Kieliszewski MJ, Showalter AM (2005) The lysine-rich arabinogalactan-protein subfamily in Arabidopsis: Gene expression, glycoprotein purification and biochemical characterization. Plant Cell Physiol 46: 975–984 [DOI] [PubMed] [Google Scholar]
- Sun W, Zhao ZD, Hare MC, Kieliszewski MJ, Showalter AM (2004) Tomato LeAGP-1 is a plasma membrane-bound, glycosylphosphatidylinositol-anchored arabinogalactan-protein. Physiol Plant 120: 319–327 [DOI] [PubMed] [Google Scholar]
- Tan L, Eberhard S, Pattathil S, Warder C, Glushka J, Yuan C, Hao Z, Zhu X, Avci U, Miller JS, et al. (2013) An Arabidopsis cell wall proteoglycan consists of pectin and arabinoxylan covalently linked to an arabinogalactan protein. Plant Cell 25: 270–287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka S, Maeda Y, Tashima Y, Kinoshita T (2004) Inositol deacylation of glycosylphosphatidylinositol-anchored proteins is mediated by mammalian PGAP1 and yeast Bst1p. J Biol Chem 279: 14256–14263 [DOI] [PubMed] [Google Scholar]
- Ueda Y, Yamaguchi R, Ikawa M, Okabe M, Morii E, Maeda Y, Kinoshita T (2007) PGAP1 knock-out mice show otocephaly and male infertility. J Biol Chem 282: 30373–30380 [DOI] [PubMed] [Google Scholar]
- Umemura M, Fujita M, Yoko-O T, Fukamizu A, Jigami Y (2007) Saccharomyces cerevisiae CWH43 is involved in the remodeling of the lipid moiety of GPI anchors to ceramides. Mol Biol Cell 18: 4304–4316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vashist S, Kim W, Belden WJ, Spear ED, Barlowe C, Ng DTW (2001) Distinct retrieval and retention mechanisms are required for the quality control of endoplasmic reticulum protein folding. J Cell Biol 155: 355–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams C, Jiang YH, Shashi V, Crimian R, Schoch K, Harper A, McHale D, Goldstein D, Petrovski S (2015) Additional evidence that PGAP1 loss of function causes autosomal recessive global developmental delay and encephalopathy. Clin Genet 88: 597–599 [DOI] [PubMed] [Google Scholar]
- Wilson SM, Bacic A (2012) Preparation of plant cells for transmission electron microscopy to optimize immunogold labeling of carbohydrate and protein epitopes. Nat Protoc 7: 1716–1727 [DOI] [PubMed] [Google Scholar]
- Wu F-H, Shen S-C, Lee L-Y, Lee S-H, Chan M-T, Lin C-S (2009) Tape-Arabidopsis Sandwich - a simpler Arabidopsis protoplast isolation method. Plant Methods 5: 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue H, Veit C, Abas L, Tryfona T, Maresch D, Ricardi MM, Estevez JM, Strasser R, Seifert GJ (2017) Arabidopsis thaliana FLA4 functions as a glycan-stabilized soluble factor via its carboxy-proximal Fasciclin 1 domain. Plant J 91: 613–630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Showalter AM (2007) Expression and localization of AtAGP18, a lysine-rich arabinogalactan- protein in Arabidopsis. Planta 226: 169–179 [DOI] [PubMed] [Google Scholar]
- Yeats TH, Bacic A, Johnson KL (2018) Plant glycosylphosphatidylinositol anchored proteins at the plasma membrane-cell wall nexus. J Integr Plant Biol 60: 649–669 [DOI] [PubMed] [Google Scholar]
- Yoko-o T, Umemura M, Komatsuzaki A, Ikeda K, Ichikawa D, Takase K, Kanzawa N, Saito K, Kinoshita T, Taguchi R, et al. (2018) Lipid moiety of glycosylphosphatidylinositol-anchored proteins contributes to the determination of their final destination in yeast. Genes Cells 23: 880–892 [DOI] [PubMed] [Google Scholar]
- Yoo SD, Cho YH, Sheen J (2007) Arabidopsis mesophyll protoplasts: A versatile cell system for transient gene expression analysis. Nat Protoc 2: 1565–1572 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Yang J, Showalter AM (2011) AtAGP18 is localized at the plasma membrane and functions in plant growth and development. Planta 233: 675–683 [DOI] [PubMed] [Google Scholar]
- Zimmermann P, Hirsch-Hoffmann M, Hennig L, Gruissem W (2004) GENEVESTIGATOR. Arabidopsis microarray database and analysis toolbox. Plant Physiol 136: 2621–2632 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.