Abstract
Covid-19 is a new infectious disease caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). Given the seriousness of the situation, the World Health Organization declared a global pandemic as the Covid-19 rapidly around the world. Among its applications, chest X-ray images are frequently used for an early diagnostic/screening of Covid-19 disease, given the frequent pulmonary impact in the patients, critical issue to prevent further complications caused by this highly infectious disease.
In this work, we propose 4 fully automatic approaches for the classification of chest X-ray images under the analysis of 3 different categories: Covid-19, pneumonia and healthy cases. Given the similarity between the pathological impact in the lungs between Covid-19 and pneumonia, mainly during the initial stages of both lung diseases, we performed an exhaustive study of differentiation considering different pathological scenarios. To address these classification tasks, we evaluated 6 representative state-of-the-art deep network architectures on 3 different public datasets: (I) Chest X-ray dataset of the Radiological Society of North America (RSNA); (II) Covid-19 Image Data Collection; (III) SIRM dataset of the Italian Society of Medical Radiology. To validate the designed approaches, several representative experiments were performed using 6,070 chest X-ray radiographs. In general, satisfactory results were obtained from the designed approaches, reaching a global accuracy values of 0.9706 0.0044, 0.9839 0.0102, 0.9744 0.0104 and 0.9744 0.0104, respectively, thus helping the work of clinicians in the diagnosis and consequently in the early treatment of this relevant pandemic pathology.
Keywords: Computer-aided diagnosis, Pulmonary disease detection, Covid-19, Pneumonia, X-ray imaging, Deep learning
Graphical abstract
1. Introduction
A new coronavirus SARS-CoV-2, which causes the disease commonly known as Covid-19, was first identified in Wuhan, Hubei province, China at the end of 2019 [1]. In particular, coronaviruses are a family of viruses known to contain strains capable of causing severe acute infections that typically affect the lower respiratory tract and manifests as pneumonia [2], and therefore, being potentially fatal in humans and a wide variety of animals, including birds and mammals such as camels, cats or bats [3], [4].
On 11 February 2020, the World Health Organization (WHO) declared the outbreak of Covid-19 as a pandemic, noting the more than 118,000 cases of coronavirus disease reported in more than 110 countries, resulting in 4291 deaths worldwide [5] by that extremely early moment, numbers that were deeply worsened posteriorly. In particular, this disease, among its consequences, also manifests as relevant respiratory disease, being considered a serious public health problem because it can kill healthy adults, as well as older people with underlying health problems or other recognized risk factors, such as heart disease, lung disease, hypertension and diabetes, among others [6]. Furthermore, the Covid-19 virus is transmitted quite efficiently, since an infected person is capable of transmitting the virus to 2 or 3 other people, an exponential rate of increase [7].
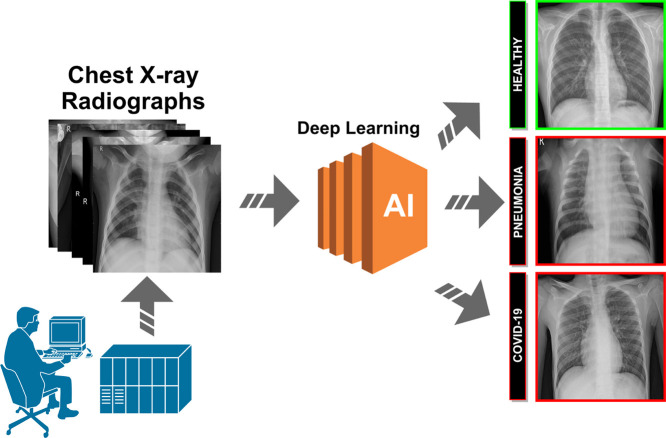
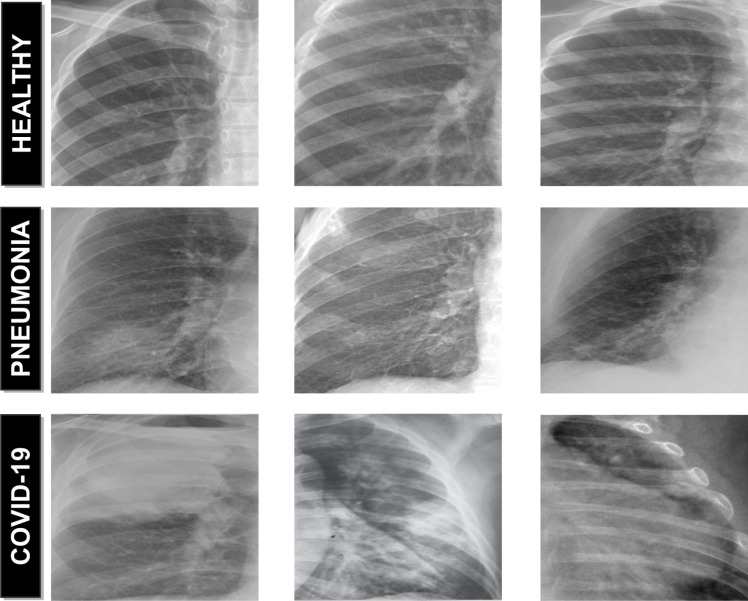
Over the years, X-ray examination of the chest plays an important clinical role in detecting or monitoring the progression of different pulmonary diseases, such as emphysema, chronic bronchitis, pulmonary fibrosis, lung cancer or pneumonia, among others [8], [9], [10]. Today, given the severity of the coronavirus pandemic, radiologists are asked to prioritize chest X-rays of patients with suspected Covid-19 infection over any other imaging studies, allowing for more appropriate use of medical resources during the initial screening process and excluding other potential respiratory diseases, a process which is extremely tedious and time-consuming. In this context, the visibility of the Covid-19 infection in the chest X-ray is complicated and requires experience of the clinical expert to analyze and understand the information and differentiate the cases from other diseases of respiratory origin with similar characteristics such as pneumonia, as illustrated in Fig. 1. For that reason, a fully automatic system for the classification of chest X-ray images between healthy, pneumonia or specific Covid-19 cases is significantly helpful as it drastically reduces the workload of the clinical staff. Complementary, it may provides a more precise identification of this highly infectious disease, reducing the subjectivity of clinicians in the early screening process, and thus also reducing healthcare costs.
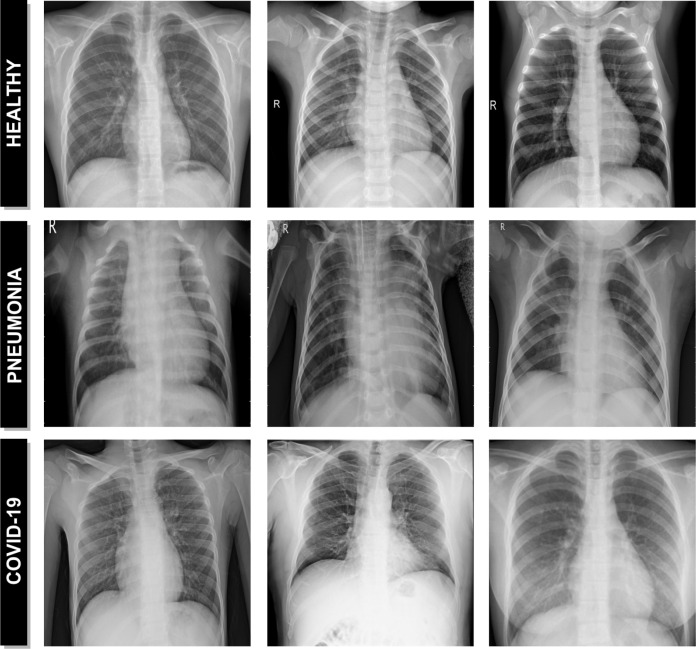
Fig. 1.
Representative examples of chest X-ray images. 1st row, chest X-ray images from healthy patients. 2nd row, chest X-ray images from patients with pneumonia. 3rd row, chest X-ray images from patients with Covid-19.
Given the relevance of this topic, several approaches were recently proposed using chest X-ray images for the classification of Covid-19. As reference, Narin et al. [11] proposed an approach based on deep transfer learning using chest X-ray images for the detection of patients infected with coronavirus pneumonia. In the work of Hassanien et al. [12], the authors proposed a methodology for the automatic X-ray Covid-19 lung classification using a multi-level threshold based on Otsu algorithm and support vector machine for the prediction task. In the work of Hammoudi et al. [13], the authors proposed a deep learning strategy to automatically detect if a chest X-ray image is healthy, pneumonia (bacterial or viral), assuming that a patient infected by the Covid-19, tested during an epidemic period, has a high probability of being a true positive when the result of the classification is a virus. In the work of Feki et al. [14], the authors proposed a collaborative federated learning framework allowing multiple medical institutions screening Covid-19 from Chest X-ray images using deep learning. Khattak et al. [15] proposed a deep learning approach based on the multilayer-Spatial Convolutional Neural Network for automatic detection of Covid-19 using chest X-ray images and Computed Tomography (CT) scans. Similarly, Saygili [16] proposed an approach for computer-aided detection of Covid-19 from CT and X-ray images using machine learning methods. In the work of Saiz et al. [17], the authors proposed a system to detect Covid-19 in chest X-ray images using deep learning techniques. Sait et al. [18] proposed a deep-learning based multimodal framework for Covid-19 detection using breathing sound spectrograms and chest X-ray images. Das et al. [19] proposed an automated deep learning-based approach for detection of Covid-19 in X-ray images by using the extreme version of the Inception (Xception) model. In the work of Singh et al. [20], the authors proposed a CNN architecture to classify Covid-19 patients using X-ray images. In addition, a multi-objective adaptive differential evolution (MADE) strategy was used for the hyperparameters of the CNN architecture. Ucar et al. [21] proposed a deep SqueezeNet-based strategy with Bayes optimization for Covid-19 screening in X-ray images. Wang et al. [22] proposed a deep convolutional neural network, called COVID-Net, design tailored for the detection of Covid-19 cases from chest radiography images. Afshar et al. [23] proposed a Capsule Network-based framework, referred to as the COVID-CAPS, for diagnosis of Covid-19 from X-ray images. In the work of Chowdhury et al. [24], the authors proposed a strategy for automatic detection of Covid-19 pneumonia from chest X-rays using pre-trained models. Khan et al. [25] proposed a deep CNN model called CoroNet to automatically detect Covid-19 infection from chest X-ray images. Similarly, Sahinbas [26] proposed a deep approach for COVID-19 diagnosis, applying a deep CNN technique based on chest X-ray images of COVID-19 patients. Apostolopoulos et al. [27] proposed a study on the possible extraction of representative biomarkers of Covid-19 from X-ray images using deep learning strategies. In the work of Zulkifley et al. [28], the authors proposed a lightweight deep learning model for Covid-19 screening that is suitable for the mobile platform.
Despite the satisfactory results obtained by these works, most of them only partially address this recent and relevant problem of global interest, limiting their practical utility for usage and interpretation for support in clinical decision scenarios such as emergency triage for example.
Therefore, in order to provide a more complete methodology, we propose 4 complementary fully automatic approaches for the classification of Covid-19, pneumonia and healthy chest X-ray images. The proposed approaches allow to make predictions using complete chest X-ray images of arbitrary sizes, which is very relevant considering the great variability of X-ray devices currently available in health centers. For this purpose, we considered 6 representative deep network architectures, including DenseNet-121, DenseNet-161, ResNet-18, ResNet-34, VGG-16 and VGG-19. For each network architecture, we evaluate the separability among Covid-19, pneumonia and healthy chest X-ray images. All the proposed approaches are designed to distinguish Covid-19 not only from normal patients, but also to differentiate them from patients with lung conditions other than Covid-19. To validate our proposal, representative and comprehensive experiments were performed using chest X-ray images compiled from 3 different public image datasets.
The manuscript is organized as follows: Section 2 describes the materials and methods that were used in this research work. Section 3 presents the results and validation of the proposed approaches. Section 4 includes the discussion of the experimental results. Finally, Section 5 presents the conclusions about the proposed systems as well as possible future lines of work in this enormous topic of interest.
2. Materials and methods
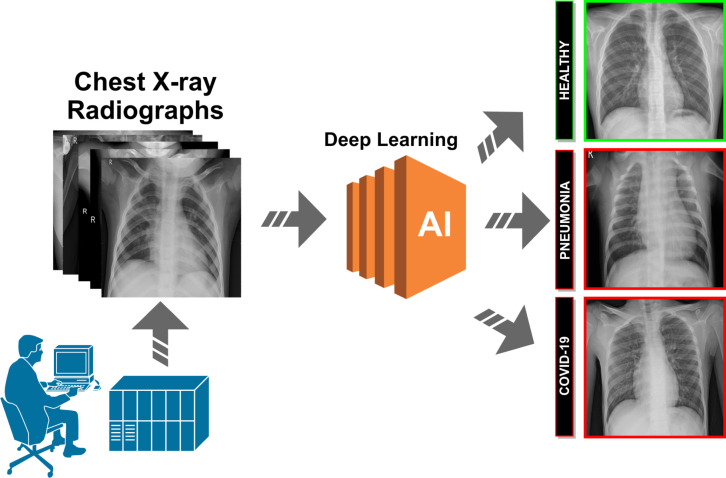
A schematic representation of the proposed paradigm of the different approaches can be seen in Fig. 2. The proposed systems receive, as input, a chest X-ray radiography. During the acquisition procedure, the patient is exposed to a small dose of ionizing radiation to produce images of the interior of the chest. The technician will usually be behind a protected wall or in an adjacent room to activate the X-ray machine. The proposed system then uses advanced artificial intelligence techniques to classify chest X-ray images into 3 different clinical categories: healthy, pneumonia or Covid-19. As a result, the system provides useful clinical information for the initial screening process and for subsequent clinical analyses.
Fig. 2.
Representative scheme of the proposed methodology.
2.1. Computational approaches for pathological X-ray image classification
Given the similarity between the pathological impact in the lungs between Covid-19 and common types of pneumonia, mainly during the initial stages of both lung diseases, we performed an exhaustive analysis of differentiation considering different pathological scenarios. In this line, we proposed 4 different and independent computational approaches for the classification of Covid-19, pneumonia and healthy chest X-ray radiographs. These computational approaches allow us to make complementary predictions using complete chest X-ray images of arbitrary sizes, which is very relevant considering the great variability of X-ray devices currently available in health centers. Each of these approaches is explained in more detail below:
2.1.1. 1st approach. Healthy vs Pneumonia, tested with Covid-19
Using as reference a consolidated public image dataset for the identification of pneumonia subjects with a considerable amount of image samples [29], we firstly used, as baseline, a trained model for the differentiation of healthy and pneumonia chest X-ray images taking advantage of this large amount of available information. Subsequently, we tested the potential of this approach to classify chest X-ray radiographs of patients diagnosed with Covid-19 and measure their similarity with both situations. In this way, we can analyze the percentage of chest X-ray images of patients with Covid-19 that may be classified as pneumonia, giver their relation in the pathological pulmonary impact.
2.1.2. 2nd approach, Healthy vs Pneumonia/Covid-19
Given the pathological similarity between pneumonia and Covid-19 subjects, subsequently we designed an screening process that analyzes the degree of separability between healthy and pathological chest X-ray radiographs, considering these both pathological scenarios. In this sense, we include the chest X-ray images of patients diagnosed with pneumonia or Covid-19 under the same class in a training of the model to predict 2 different categories: pathological and healthy cases.
2.1.3. 3rd approach, Healthy/Pneumonia vs Covid-19
Additionally, we adapted and trained a model to specifically identify Covid-19 subjects, measuring the capability of differentiation not only from healthy subjects, but also from those pathological cases with a significant similarity, as patients suffering from pneumonia. With this in mind, our system was designed to identify two different classes, including healthy and patients with pneumonia in the same category.
2.1.4. 4th Approach, Healthy vs Pneumonia vs Covid-19
Finally, we designed another approach to simultaneously determine the degree of separability between the 3 categories of chest X-ray images considered in this work. To this end, we trained a model with a set of chest X-ray radiographs of the 3 different classes: healthy subjects, patients diagnosed with pneumonia, patients diagnosed with Covid-19.
2.2. Network architectures
The use of deep learning architectures has been rapidly increasing in the field of medical imaging, including computer-aided diagnosis system and medical image analysis for more accurate screening, diagnosis, prognosis and treatment of many relevant diseases [30], [31]. In this context, the present study analyzes the performance of 6 different deep network architectures such as: 2 Dense Convolutional Network Architectures (a DenseNet-121 and a DenseNet-161) [32], 2 Residual Network Architectures (a ResNet-18 and a ResNet-34) [33] as well as 2 VGG network architectures (a VGG-16 and a VGG-19, both with batch normalization) [34]. These architectures were chosen for their simplicity and adequate results for many similar classification tasks in different lung diseases [35], [36], [37]. In addition, these architectures have proven to be superior in both accuracy and predictive efficiency compared to classical machine learning techniques [38]. For each network architecture, we evaluate the separability among Covid-19, pneumonia and healthy chest X-ray images. All network architectures were initialized with the weights of a model pre-trained on ImageNet [39] dataset. On one hand, we benefit from the pre-trained model weights making the learning process much more efficient, in other words, the model converges fast given that his weights are already stabilized initially. On the other hand, it significantly reduces the amount of labeled data that is required for the model training. In addition, we have adapted the classification layer of each deep network architecture used to support the output according to the specific requirements of each proposed approach, which is to categorize the chest X-ray images into 2 or 3 different clinical classes considering healthy, pneumonia and Covid-19.
2.3. Training
Regarding the training stage of the different approaches, considering the limited amount of Covid-19 subjects, we decided that the employed chest X-ray radiographs dataset was randomly divided into 3 smaller datasets, specifically with 60% of the cases for training, 20% for validation and the remaining 20% for testing. Additionally, the classification step was performed with 5 repetitions, being calculated the mean cross-entropy loss [40] and the mean accuracy to illustrate the general performance and stability of the proposed approaches. All the architectures were trained using Stochastic Gradient Descent (SGD) with a constant learning rate of 0.01, a mini-batch size of 4 and a first-order momentum of 0.9. In particular, SGD is a simple but highly efficient approach for the discriminative learning of classifiers under convex loss functions [41].
2.4. Data augmentation
Data augmentation is a widely used strategy that enables practitioners to significantly increase the diversity of data available for training models, reducing the overfitting and making the models more robust [42], [43]. This is especially significant in our case, given the limited amount of positive Covid-19 cases that were used in the different approaches. For this purpose, we applied different configurations of affine transformations only on the training set to increase the training data and improve the performance of the neural network architectures that was used to classify chest X-rays images. In particular, we automatically generate additional training samples through a combination of scaling with horizontal flipping operations, considering the common variety of possible resolutions as well as the symmetry of the human body.
2.5. Dataset
The training and evaluation of the deep convolutional models were performed with chest X-ray radiographs that were taken from 3 different chest X-ray public datasets of reference: the Chest X-ray (Pneumonia) dataset [29], the (Covid-19) Image Data Collection dataset [44] and the (Covid-19) SIRM dataset [45].
The Chest X-ray (Pneumonia) dataset of the Radiological Society of North America (RSNA) [29] is composed by of 5863 chest X-ray radiographs. This public dataset was labeled into 2 main categories: healthy patients and patients with different types of pneumonia (viral and bacterial) presenting, therefore, a high level of heterogeneity.
Currently, public chest X-ray datasets of patients diagnosed with Covid-19 are very limited. Despite this important restriction, we have built a dataset composed of 207 radiographs, 155 were taken from (Covid-19) Image Data Collection dataset [44] and 52 were taken from the (Covid-19) SIRM dataset of the Italian Society of Medical Radiology [45].
2.6. Evaluation
In order to test its suitability, the designed paradigm of the different approaches was validated using different statistical metrics commonly used in the literature to measure the performance of computational proposals in similar medical imaging tasks. Accordingly, Precision, Recall, F1-score and Accuracy were calculated for the quantitative validation of the classification results. In particular the first three metrics are calculated for each one of the considered classes in the different experiments are they are more meaningful in that way.
Mathematically, these statistical metrics are formulated as indicated in Eqs. (1), (2), (3), (4), respectively. These performance measures use as reference the True Negatives (TN), False Negatives (FN), True Positives (TP) and False Positives (FP):
| (1) |
| (2) |
| (3) |
| (4) |
3. Experimental results
To evaluate the suitability of the different proposed approaches in the pathological classification related to Covid-19 in chest X-ray images, we conducted different complementary experiments, taking as reference the available datasets. In particular, for each experiment, we performed 5 independent repetitions, each time with a different random selection of the samples splits, specifically with 60% of the cases for training, 20% for validation and the remaining 20% for testing. Additionally, the training stage was stopped after 200 epochs given the lack of significant further improvement in both accuracy and cross-entropy loss results.
3.1. 1st experiment: Healthy vs Pneumonia, tested with Covid-19
Given the availability of a public image dataset of reference with a significant number of healthy and pneumonia chest X-ray images [29], we trained a model to obtain a consolidated approach to distinguish between healthy patients and different pathological cases of pneumonia.
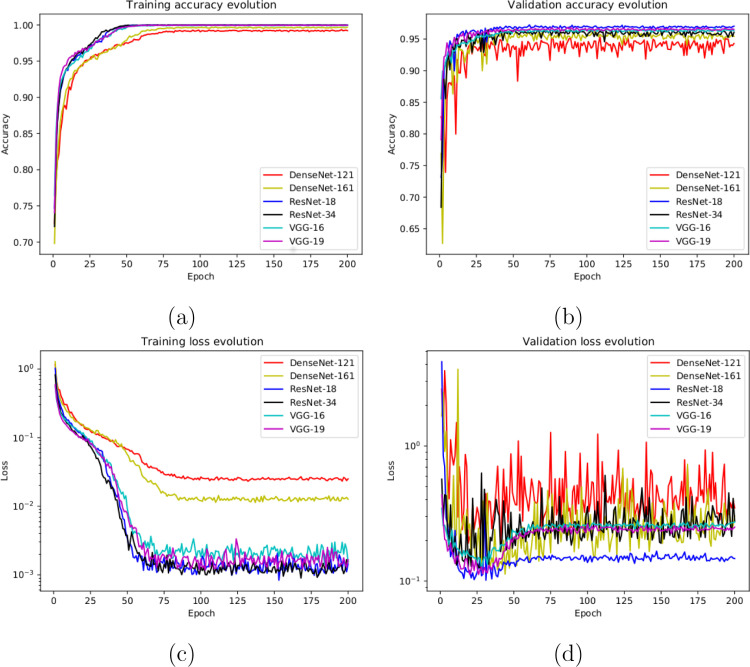
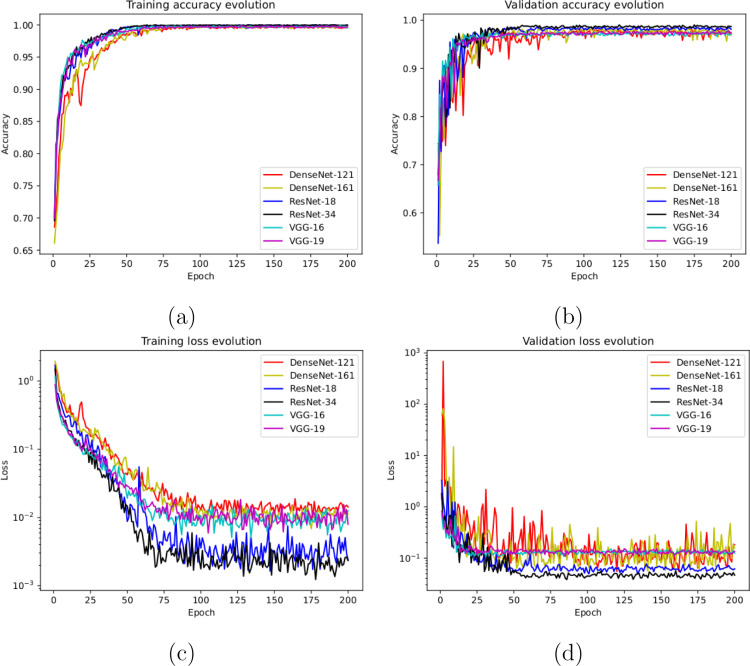
In this line, we designed an experiment with a total of 5856 X-ray images, being 1583 from healthy patients and 4273 from patients with pneumonia. Fig. 3(a) and Fig. 3(b) show the performance that was obtained using the deep learning architectures after, as previously indicated, 5 independent repetitions in the training and validation stages, respectively. Furthermore, in Fig. 3(c) and (d) we can observe that the models converge rapidly in the training and validation steps in terms of the loss cross-entropy function, demonstrating in all the cases an adequate behavior.
Fig. 3.
Results of the first experiment after 5 independent repetitions. (a) Mean standard deviation training accuracy. (b) Mean standard deviation validation accuracy. (c) Mean standard deviation training loss. (d) Mean standard deviation validation loss. A logarithmic scale has been set to correctly display the loss values for a better understanding of the results.
In Table 1, we can see the precision, recall and F1-score results obtained at the test stage, providing a mean accuracy values: a 0.9529 ± 0.0185 for the DenseNet-121, a 0.9648 ± 0.0033 for the DenseNet-161, a 0.9636 ± 0.0065 for the ResNet-18, a 0.9631 ± 0.0041 for the ResNet-34, a 0.9706 ± 0.0044 for the VGG-16 and a 0.9660 ± 0.0054 for the VGG-19. In general, all the results that were obtained show the robustness of the proposed system in the classification of the different pathological cases of pneumonia and healthy patients. In particular, the VGG-16 architecture obtained slightly higher values than those obtained with the other analyzed architectures.
Table 1.
Mean precision, recall and F1-score results obtained at the test stage for the classification of chest X-ray images between Healthy vs Pneumonia cases.
| Architecture | Class | Precision | Recall | F1-score |
|---|---|---|---|---|
| DenseNet-121 | Healthy | 0.9156 ± 0.0643 | 0.9153 ± 0.0139 | 0.9142 ± 0.0298 |
| Pneumonia | 0.9684 ± 0.0050 | 0.9670 ± 0,0291 | 0.9675 ± 0,0134 | |
| DenseNet-161 | Healthy | 0.9456 ± 0.0193 | 0.9257 ± 0.0146 | 0.9353 ± 0.0057 |
| Pneumonia | 0.9720 ± 0.0059 | 0.9798 ± 0.0074 | 0.9759 ± 0.0024 | |
| ResNet-18 | Healthy | 0.9377 ± 0.0137 | 0.9271 ± 0.0208 | 0.9322 ± 0.0103 |
| Pneumonia | 0.9730 ± 0.0090 | 0.9774 ± 0.0050 | 0.9752 ± 0.0047 | |
| ResNet-34 | Healthy | 0.9401 ± 0.0208 | 0.9243 ± 0.0124 | 0.9319 ± 0.0078 |
| Pneumonia | 0.9718 ± 0.0040 | 0.9777 ± 0.0085 | 0.9747 ± 0.0030 | |
| VGG-16 | Healthy | 0.9479 ± 0.0141 | 0.9426 ± 0.0248 | 0.9449 ± 0.0070 |
| Pneumonia | 0.9789 ± 0.0101 | 0.9812 ± 0.0051 | 0.9800 ± 0.0032 | |
| VGG-19 | Healthy | 0.9396 ± 0.0162 | 0.9380 ± 0.0089 | 0.9387 ± 0.0117 |
| Pneumonia | 0.9761 ± 0.0029 | 0.9769 ± 0.0053 | 0.9765 ± 0.0036 | |
In addition to measure the capability of the adapted architecture to this pathological analysis, another relevant goal of this experimentation was to conduct a comprehensive analysis about the potential similarity of the Covid-19 subjects with pneumonia scenarios, that is, the percentage of chest X-ray images of patients with Covid-19 that are classified as pneumonia with the previously trained network.
Thus, we extended the experiments of this case using an additional blind test dataset. In particular, this dataset consisted of the used 207 X-ray images of patients diagnosed with Covid-19. For this experiment, we analyzed the behavior of the trained models that obtained the best validation values among the 5 random repetitions for each deep learning architecture. As we can see in Table 2, the proposed system achieves satisfactory results in terms of precision, recall and F1-score, considering Covid-19 cases correctly classified as pneumonia in opposition to healthy cases. In this scenario, the system provided accuracy values of 71.50%, 71.98%, 74.88%, 80.19%, 75.36% and 78.74% for the DenseNet-121, DenseNet-161, ResNet-18, ResNet-34, VGG-16 and VGG-19, respectively, demonstrating that the system is capable of correctly screening X-ray images of Covid-19 patients in the pathological pneumonia category.
Table 2.
Precision, recall and F1-score results obtained at the test stage for the classification of the chest X-ray images from Covid-19 patients between Healthy vs Pneumonia cases.
| Architecture | Class | Precision | Recall | F1-score |
|---|---|---|---|---|
| DenseNet-121 | Healthy | – | – | – |
| Pneumonia | 1.0000 | 0.7150 | 0.8338 | |
| DenseNet-161 | Healthy | – | – | – |
| Pneumonia | 1.0000 | 0.7198 | 0.8371 | |
| ResNet-18 | Healthy | – | – | – |
| Pneumonia | 1.0000 | 0.7488 | 0.8564 | |
| ResNet-34 | Healthy | – | – | – |
| Pneumonia | 1.0000 | 0.8019 | 0.8901 | |
| VGG-16 | Healthy | – | – | – |
| Pneumonia | 1.0000 | 0.7536 | 0.8595 | |
| VGG-19 | Healthy | – | – | – |
| Pneumonia | 1.0000 | 0.7874 | 0.8811 | |
3.2. 2nd experiment: Healthy vs Pneumonia/Covid-19
Under the results of the analysis of the first approach, we designed another scenario with a screening context including Covid-19 cases by separating the pathological pneumonia and Covid-19 subjects with respect to healthy images
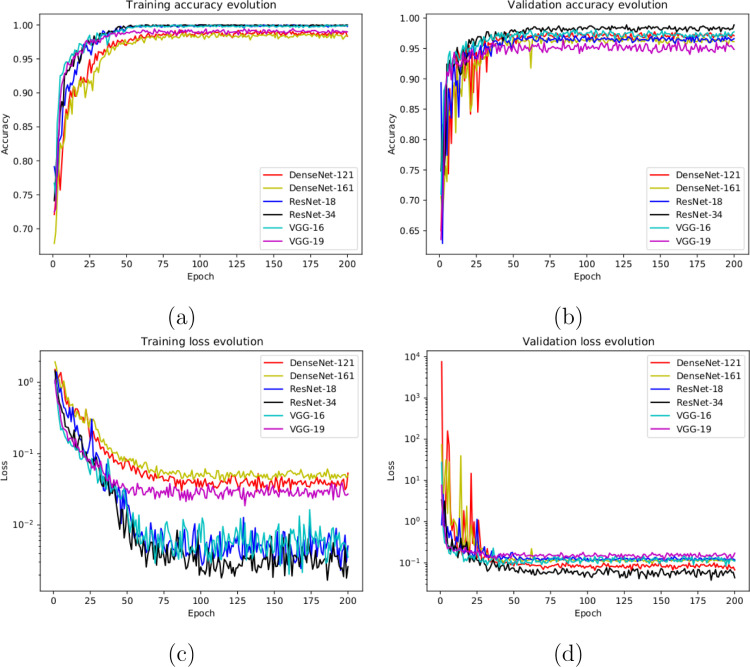
With this in mind, in this case, the designed experiment included a total of 1242 X-ray images, being 828 from healthy patients whereas 207+207 from Covid-19 and pneumonia patients, respectively. In this case, we randomly selected the 828 healthy images and the 207 pneumonia images from the total amount of the used image dataset [29]. Having the limiting factor of 207 Covid-19 images, we balanced the amount of the other cases to obtain a proportion of and between the negative and positive classes. Fig. 4(a) and (b) show the performance that was obtained from the training and validation stages using the proposed dataset through 5 independent repetitions. Moreover, as we can see in Fig. 4(c) and Fig. 4(d), the proposed approach reached stability in the cross-entropy loss function after the epoch 75 and after the epoch 50 for training and validation stages, respectively.
Fig. 4.
Results of the second experiment after 5 independent repetitions. (a) Mean standard deviation training accuracy. (b) Mean standard deviation validation accuracy. (c) Mean standard deviation training loss. (d) Mean standard deviation validation loss. A logarithmic scale has been set to correctly display the loss values for a better understanding of the results.
Table 3 shows the performance measures obtained in the test stage, in terms of precision, recall and F1-score for each class. As we can see, satisfactory results were provided, reaching a mean accuracy values of 0.9807 ± 0.0121 for the DenseNet-121, a 0.9839 ± 0.0102 for the DenseNet-161, a 0.9694 ± 0.0119 for the ResNet-18, a 0.9807 ± 0.0043 for the ResNet-34, a 0.9783 ± 0.0078 for the VGG-16 and a 0.9694 ± 0.0119 for the VGG-19. The results that were obtained can be considered statistically equivalent for some architectures with values between 96.94% and 98.39%, demonstrating that this screening approach is capable of successfully separating the pathological cases under consideration from the healthy ones.
Table 3.
Mean precision, recall and F1-score results obtained at the test stage for the classification of chest X-ray images between Healthy vs Pneumonia/Covid-19 cases.
| Architecture | Class | Precision | Recall | F1-score |
|---|---|---|---|---|
| DenseNet-121 | Healthy | 0.9915 ± 0.0032 | 0.9794 ± 0.0207 | 0.9853 ± 0.0094 |
| Pneumonia/Covid-19 | 0.9615 ± 0.0381 | 0.9833 ± 0.0062 | 0.9719 ± 0.0173 | |
| DenseNet-161 | Healthy | 0.9869 ± 0.0113 | 0.9891 ± 0.0050 | 0.9880 ± 0.0076 |
| Pneumonia/Covid-19 | 0.9783 ± 0.0104 | 0.9734 ± 0.0235 | 0.9758 ± 0.0159 | |
| ResNet-18 | Healthy | 0.9806 ± 0.0120 | 0.9855 ± 0.0071 | 0.9830 ± 0.0082 |
| Pneumonia/Covid-19 | 0.9706 ± 0.0141 | 0.9624 ± 0.0209 | 0.9663 ± 0.0130 | |
| ResNet-34 | Healthy | 0.9888 ± 0.0092 | 0.9821 ± 0.0116 | 0.9853 ± 0.0033 |
| Pneumonia/Covid-19 | 0.9642 ± 0.0248 | 0.9798 ± 0.0161 | 0.9716 ± 0.0071 | |
| VGG-16 | Healthy | 0.9842 ± 0.0078 | 0.9831 ± 0.0045 | 0.9837 ± 0.0055 |
| Pneumonia/Covid-19 | 0.9658 ± 0.0125 | 0.9688 ± 0.0171 | 0.9672 ± 0.0131 | |
| VGG-19 | Healthy | 0.9735 ± 0.0065 | 0.9806 ± 0.0131 | 0.9770 ± 0.0088 |
| Pneumonia/Covid-19 | 0.9618 ± 0.0257 | 0.9477 ± 0.0152 | 0.9546 ± 0.0182 | |
3.3. 3rd experiment: Healthy/Pneumonia vs Covid-19
The third scenario was designed to evaluate the performance of the proposed approach to specifically distinguish between cases of patients with Covid-19 from other similar cases such as pneumonia or also from healthy patients. Thus, we measure the potential separability between those potentially similar pathological cases of Covid-19 from pneumonia.
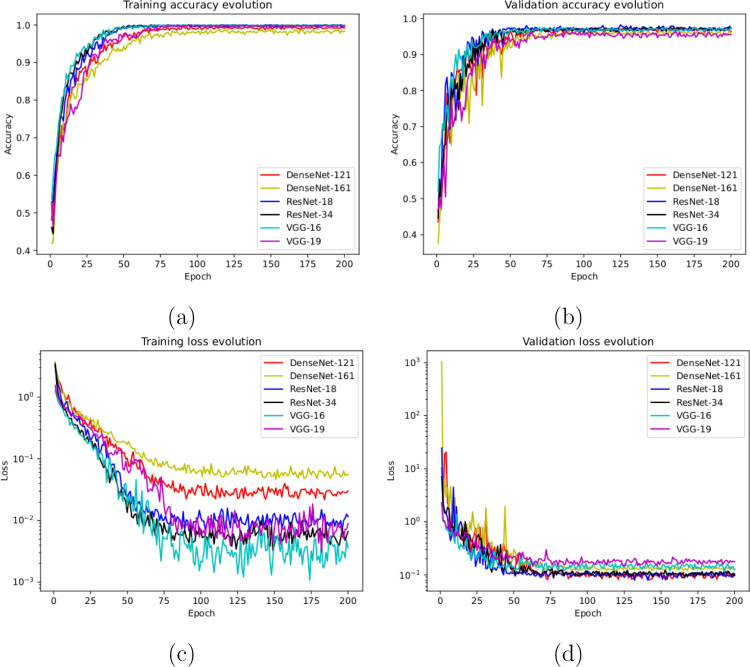
To do so, we used a total of 621 X-ray images, being 207 from Covid-19, 207 from patients with pneumonia and 207 from healthy patients. Once again, those subjects from pneumonia and healthy patients were randomly selected to obtain a proportion of and between negative and positive classes, respectively. Fig. 5(a) and Fig. 5(b) show the performance that was obtained from all the architectures after 5 independent repetitions in the training and validation stages, respectively. In particular, our models achieve high stability after 75 epochs and 50 epochs in the training and validation stages, respectively. Complementary, in Fig. 5(c) and (d) we can observe a similar behavior with the cross-entropy loss function.
Fig. 5.
Results of the third experiment after 5 independent repetitions. (a) Mean standard deviation training accuracy. (b) Mean standard deviation validation accuracy. (c) Mean standard deviation training loss. (d) Mean standard deviation validation loss. A logarithmic scale has been set to correctly display the loss values for a better understanding of the results.
In Table 4 we present the quantitative performance of the proposed system in the test dataset, in terms of precision, recall and F1-score. Our method shows an suitable performance for both categories, providing a mean accuracy value of 0.9584 ± 0.0285 for the DenseNet-121, a 0.9712 ± 0.0145 for the DenseNet-161, a 0.9744 ± 0.0104 for the ResNet-18, a 0.9744 ± 0.0153 for the ResNet-34, a 0.9728 ± 0.0121 for the VGG-16 and a 0.9504 ± 0.0249 for the VGG-19, demonstrating the robustness of the proposed system to distinguish between cases of patients with Covid-19 from other cases such as pneumonia or health. In this experiment, as we can see, no significant differences were observed between the results obtained by the DenseNet-161, ResNet-18, ResNet-34 and VGG-16 architectures. This result is specially significant considering the separation of similar pathological scenarios as pneumonia and Covid-19.
Table 4.
Mean precision, recall and F1-score results obtained at the test stage for the classification of chest X-ray images between Healthy/Pneumonia vs Covid-19 cases.
| Architecture | Class | Precision | Recall | F1-score |
|---|---|---|---|---|
| DenseNet-121 | Healthy/Pneumonia | 0.9740 ± 0.0219 | 0.9620 ± 0.0444 | 0.9680 ± 0.0259 |
| Covid-19 | 0.9380 ± 0.0665 | 0.9500 ± 0.0374 | 0.9420 ± 0.0356 | |
| DenseNet-161 | Healthy/Pneumonia | 0.9700 ± 0.0255 | 0.9880 ± 0.0110 | 0.9800 ± 0.0100 |
| Covid-19 | 0.9780 ± 0.0148 | 0.9360 ± 0.0513 | 0.9560 ± 0.0251 | |
| ResNet-18 | Healthy/Pneumonia | 0.9740 ± 0.0089 | 0.9880 ± 0.0130 | 0.9820 ± 0.0084 |
| Covid-19 | 0.9780 ± 0.0228 | 0.9500 ± 0.0071 | 0.9640 ± 0.0134 | |
| ResNet-34 | Healthy/Pneumonia | 0.9720 ± 0.0239 | 0.9900 ± 0.0000 | 0.9820 ± 0.0084 |
| Covid-19 | 0.9760 ± 0.0055 | 0.9500 ± 0.0469 | 0.9620 ± 0.0277 | |
| VGG-16 | Healthy/Pneumonia | 0.9840 ± 0.0134 | 0.9760 ± 0.0230 | 0.9780 ± 0.0084 |
| Covid-19 | 0.9520 ± 0.0455 | 0.9660 ± 0.0207 | 0.9580 ± 0.0205 | |
| VGG-19 | Healthy/Pneumonia | 0.9660 ± 0.0167 | 0.9680 ± 0.0277 | 0.9660 ± 0.0167 |
| Covid-19 | 0.9180 ± 0.0705 | 0.9140 ± 0.0351 | 0.9180 ± 0.0497 | |
3.4. 4Th experiment: Healthy vs Pneumonia vs Covid-19
In this last scenario, considering the satisfactory results of the other considered approaches, we also trained another model to directly analyze the performance of the proposed system to separate the chest X-ray radiographs into 3 different categories: healthy, pneumonia and Covid-19. To do so, we designed a complete experiment using a total of 621 X-ray radiographs, being 207 from Covid-19, 207 from patients with pneumonia and 207 from healthy patients. In this case, again, we randomly selected the healthy and pneumonia images to balance the amount of available images for the 3 considered classes. Fig. 6(a) and (b) show the performance of the proposed system using the training and validation sets after 5 independent repetitions, where we can appreciate a good behavior of the deep learning architectures. In the same line, the proposed system achieved its stability in the loss cross-entropy function after the epoch 100 for training and after the epoch 50 for validation, as we can see in Fig. 6(c) and (d).
Fig. 6.
Results of the fourth experiment after 5 independent repetitions. (a) Mean standard deviation training accuracy. (b) Mean standard deviation validation accuracy. (c) Mean standard deviation training loss. (d) Mean standard deviation validation loss. A logarithmic scale has been set to correctly display the loss values for a better understanding of the results.
In Table 5, we can see the precision, recall and F1-score results obtained at the test stage, providing a mean accuracy of 0.9632 ± 0.0200 for the DenseNet-121, a 0.9536 ± 0.0143 for the DenseNet-161, a 0.9744 ± 0.0104 for the ResNet-18, a 0.9664 ± 0.0118 for the ResNet-34, a 0.9504 ± 0.0173 for the VGG-16 and a 0.9536 ± 0.0207 for the VGG-19. As we can observe, in this experiment, the best result was obtained using the ResNet-18 architecture. However, all the trained models are able to predict with adequate accuracy values, demonstrating the robustness of the proposed system in the classification of the 3 categories of chest X-ray images considered in this work.
Table 5.
Mean precision, recall and F1-score results obtained at the test stage for the classification of chest X-ray images between Healthy vs Pneumonia vs Covid-19 cases.
| Architecture | Class | Precision | Recall | F1-score |
|---|---|---|---|---|
| DenseNet-121 | Healthy | 0.9669 ± 0.0397 | 0.9637 ± 0.0175 | 0.9647 ± 0.0177 |
| Pneumonia | 0.9579 ± 0.0160 | 0.9545 ± 0.0646 | 0.9550 ± 0.0294 | |
| Covid-19 | 0.9634 ± 0.0304 | 0.9773 ± 0.0224 | 0.9700 ± 0.0196 | |
| DenseNet-161 | Healthy | 0.9535 ± 0.0236 | 0.9553 ± 0.0341 | 0.9539 ± 0.0152 |
| Pneumonia | 0.9705 ± 0.0084 | 0.9374 ± 0.0256 | 0.9534 ± 0.0098 | |
| Covid-19 | 0.9407 ± 0.0453 | 0.9701 ± 0.0213 | 0.9546 ± 0.0234 | |
| ResNet-18 | Healthy | 0.9562 ± 0.0183 | 0.9759 ± 0.0292 | 0.9656 ± 0.0149 |
| Pneumonia | 0.9807 ± 0.0212 | 0.9763 ± 0.0232 | 0.9783 ± 0.0169 | |
| Covid-19 | 0.9903 ± 0.0134 | 0.9683 ± 0.0128 | 0.9791 ± 0.0065 | |
| ResNet-34 | Healthy | 0.9387 ± 0.0465 | 0.9765 ± 0.0347 | 0.9563 ± 0.0247 |
| Pneumonia | 0.9645 ± 0.0174 | 0.9734 ± 0.0220 | 0.9686 ± 0.0100 | |
| Covid-19 | 0.9884 ± 0.0160 | 0.9520 ± 0.0176 | 0.9698 ± 0.0144 | |
| VGG-16 | Healthy | 0.9507 ± 0.0425 | 0.9412 ± 0.0156 | 0.9454 ± 0.0203 |
| Pneumonia | 0.9374 ± 0.0420 | 0.9480 ± 0.0397 | 0.9417 ± 0.0237 | |
| Covid-19 | 0.9662 ± 0.0373 | 0.9663 ± 0.0237 | 0.9659 ± 0.0248 | |
| VGG-19 | Healthy | 0.9674 ± 0.0206 | 0.9450 ± 0.0299 | 0.9558 ± 0.0179 |
| Pneumonia | 0.9480 ± 0.0371 | 0.9441 ± 0.0350 | 0.9457 ± 0.0298 | |
| Covid-19 | 0.9436 ± 0.0483 | 0.9751 ± 0.0257 | 0.9581 ± 0.0202 | |
4. Discussion
The current gold standard for detection of Covid-19 disease is the reverse transcription-polymerase chain reaction (RT-PCR), which is usually performed on a sample of nasopharyngeal and oropharyngeal secretions. However, RT-PCR is believed to be highly specific, but its sensitivity can range from 60%–70% [46] to 95%–97% [47], making false negatives a very critical problem in the diagnosis of Covid-19, especially in the early stages of this highly infectious disease. On the other hand, chest radiography is considered the first-line imaging study in suspected or confirmed Covid-19 patients, being recommended by the American College of Radiology (ACR) [48]. However, one of the main limitations of chest radiography, like RT-PCR, is the high false-negative rate [4]. In this regard, the chest radiography may be normal in mild cases or in the early stage of the Covid-19 infection, but patients with moderate or severe symptoms are unlikely to have a normal chest radiography. Findings are most extensive 10–12 days after the onset of symptoms [49].
In this work, we analyzed different and complementary fully automatic approaches for the classification of healthy, pneumonia and Covid-19 chest X-ray radiographs. All the experimental results demonstrate that the proposed system is capable of successfully distinguishing healthy patients from different pathological cases of pneumonia and Covid-19, although in some cases, the trained models present a more stable behavior during the training and validation stages. To accurately explain this behavior, it would be interesting to perform a more extensive analysis of these architectures using a larger and more complete dataset.
Regarding the obtained results, the pathological differentiation is understandable given the abnormality of the pathological scenarios with respect to normal patients and also the pathological impact in the lungs of both pneumonia and Covid-19 diseases. But also, it is significant the accurate capability of differentiation also of Covid-19 patients from other with pneumonia, which were correctly separated in the proposed third and fourth approaches, also corroborated by the experiments with the first approach. In addition, the proposed system allows to make accurate predictions using chest X-ray images of arbitrary sizes, which is very relevant considering the great variability of X-ray devices currently available in the healthcare centers.
We compared the proposed approaches with the most representative state-of-the-art methods on this topic. These methods were previously introduced and described in Section 1. In this regard, Table 6 presents the results obtained from state-of-the-art methods and our proposal. Although many methods were tested with different image datasets and under different conditions (image size, X-ray devices, pixel-level resolution, different distribution and balance of data, etc.), our method offers a competitive performance, outperforming in most cases the rest of the approaches. Another important aspect to take into account is that most of the works presented by the state of the art only consider a single scenario (2 or 3 categories), whereas in our proposal we analyze 4 different scenarios in a comprehensive way. Particularly, in our proposal, several experiments were conducted considering 6 representative state-of-the-art deep CNN architectures (DenseNet-121, DenseNet-161, ResNet-18, ResNet-34, VGG-16 and VGG-19) to analyze the degree of separability of COVID-19 samples, representing a comprehensive analysis of this relevant problem. Experimental validation shows accurate and robust results, obtaining high values (above 97% of global accuracy) for all computational approaches considered.
Table 6.
Comparison of performance between state of the art and proposed approaches.
| State-of-the-art | Methods | Computational approaches | Accuracy (%) |
|---|---|---|---|
| Das et al. [19] | Xception | Pneumonia vs Covid-19 vs Other | 97.40 |
| Singh et al. [20] | MADE-based CNN | non-Covid-19 vs Covid-19 | 94.48 |
| Ucar et al. [21] | Deep Bayes-SqueezeNet | Healthy vs Pneumonia vs Covid-19 | 98.26 |
| Wang et al. [22] | Covid-net | Healthy vs Pneumonia vs Covid-19 | 93.30 |
| Afshar et al. [23] | Covid-caps | non-Covid-19 vs Covid-19 | 95.70 |
| Chowdhury et al. [24] | MobileNetv2, SqueezeNet, ResNet-18, ResNet-101, | Normal vs Covid-19 | 99.70 |
| DenseNet-201, CheXNet, Inception-v3 and VGG-19 | Normal vs Covid-19 vs Pneumonia Viral | 97.90 | |
| Khan et al. [25] | CoroNet | Covid-19 vs Pneumonia Bacterial vs Pneumonia Viral vs Normal | 89.60 |
| Covid-19 vs Pneumonia vs Normal | 95.00 | ||
| Sahinbas et al. [26] | ResNet, DenseNet, InceptionV3, VGG-16 and VGG-19 | non-Covid-19 vs Covid-19 | 80.00 |
| Apostolopoulos et al. [27] | MobileNetv2 | non-Covid-19 vs Covid-19 | 99.18 |
| Zulkifley et al. [28] | LightCovidNet | Healthy vs Pneumonia vs Covid-19 | 96.97 |
| Our proposal | DenseNet-121, DenseNet-161, ResNet-18, | Healthy vs Pneumonia, tested with Covid-19 | 97.06 |
| ResNet-34, VGG-16 and VGG-19 | |||
| Our proposal | DenseNet-121, DenseNet-161, ResNet-18, | Healthy vs Pneumonia/Covid-19 | 98.39 |
| ResNet-34, VGG-16 and VGG-19 | |||
| Our proposal | DenseNet-121, DenseNet-161, ResNet-18, | Healthy/Pneumonia vs Covid-19 | 97.44 |
| ResNet-34, VGG-16 and VGG-19 | |||
| Our proposal | DenseNet-121, DenseNet-161, ResNet-18, | Healthy vs Pneumonia vs Covid-19 | 97.44 |
| ResNet-34, VGG-16 and VGG-19 | |||
Despite the attained satisfactory performance, the proposed system obtain an acceptably small number of misclassified cases. In particular, some misclassification is caused by the poor contrast of the X-ray images used in this work. Other times, in some cases, there is a great similarity between Covid-19 and pneumonia, mainly during the initial stages of both diseases. In Fig. 7, we can see representative examples illustrating the significant variability of the possible scenarios that are represented in this research work.
Fig. 7.
Representative examples of lung regions. 1st row, lung healthy regions. 2nd row, lung regions affected by the pneumonia disease. 3rd row, lung regions affected by the Covid-19 disease.
As no exhaustive classification method for X-ray Covid-19 images has been published to date, we cannot make any comparison with other state-of-the-art approaches. Instead, we use different public datasets for the evaluation of our proposed system, validating their accuracy in comparison with manual annotations from different clinical experts.
5. Conclusions
Coronaviruses are a large family of viruses that can cause disease in both animals and humans. In particular, the new coronavirus SARS-CoV-2, also know Covid-19, was firstly detected in December 2019 in Wuhan City, Hubei Province, China. Given its drastic spread, the WHO declared a global pandemic as the Covid-19 rapidly spreads across the world. In this context, chest X-ray images are widely used for early screening by clinical experts, allowing for more appropriate use of other medical resources during initial screening.
In this work, we proposed complementary fully automatic approaches for the classification of Covid-19, pneumonia and healthy chest X-ray radiographs. For this purpose, several experiments were performed considering 6 deep network architectures representative of the state of the art. In this way, we exploit the potential of deep learning architectures and their capabilities to describe complex, hierarchically unstructured data at higher levels of abstraction in the analysis of chest X-rays for the Covid-19 screening task. We evaluated the robustness and accuracy of the different classification approaches, obtaining satisfactory results for all the experiments that were proposed using different public image datasets of reference. Despite the complex and challenging scenario, the proposed approaches has proven to be robust and reliable, facilitating a more complete and precise analysis of the pathological lung regions and, consequently, the production of more adjusted treatments of this highly infectious disease.
As future work, we plan to expand the proposed methodology with the incorporation of other relevant lung diseases, such as chronic bronchitis, emphysema, or lung cancer. In addition, we plan to perform a comparative analysis of the proposed methodology with other techniques that use sophisticated image processing algorithms in combination with classical machine learning techniques. Finally, further analysis with larger and maybe more comprehensive chest X-ray datasets should be done in order to reinforce the conclusions of this work.
CRediT authorship contribution statement
Joaquim de Moura: Conceptualization, Methodology, Software, Formal analysis, Investigation, Data curation, Writing - original draft, Writing - review & editing, Visualization. Jorge Novo: Methodology, Validation, Investigation, Data curation, Writing - review & editing, Supervision, Project administration. Marcos Ortega: Methodology, Validation, Investigation, Data curation, Writing - review & editing, Supervision, Project administration, Funding acquisition.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This research was funded by Instituto de Salud Carlos III, Government of Spain, DTS18/00136 research project; Ministerio de Ciencia e Innovación Universidades, Government of Spain, RTI2018-095894-B-I00 research project; Ministerio de Ciencia e Innovación, Government of Spain through the research project with reference PID2019-108435RB-I00; Consellería de Cultura, Educación e Universidade, Xunta de Galicia, Spain through the postdoctoral grant contract ref. ED481B-2021-059; and Grupos de Referencia Competitiva, Spain, grant ref. ED431C 2020/24; Axencia Galega de Innovación (GAIN), Xunta de Galicia, Spain, grant ref. IN845D 2020/38; CITIC, as Research Center accredited by Galician University System, is funded by “Consellería de Cultura, Educación e Universidade from Xunta de Galicia, Spain”, supported in an 80% through ERDF Funds, ERDF Operational Programme Galicia 2014–2020, Spain, and the remaining 20% by “Secretaría Xeral de Universidades, Spain ” (Grant ED431G 2019/01). Funding for open access charge: Universidade da Coruña/CISUG.
References
- 1.She J., Jiang J., Ye L., Hu L., Bai C., Song Y. 2019 Novel coronavirus of pneumonia in wuhan, China: emerging attack and management strategies. Clin. Transl. Med. 2020;9(1):1–7. doi: 10.1186/s40169-020-00271-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stavrinides J., Guttman D.S. Mosaic evolution of the severe acute respiratory syndrome coronavirus. J. Virol. 2004;78(1):76–82. doi: 10.1128/JVI.78.1.76-82.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chan J.F., Lau S.K., To K.K., Cheng V.C., Woo P.C., Yuen K.-Y. Middle east respiratory syndrome coronavirus: another zoonotic betacoronavirus causing SARS-like disease. Clin. Microbiol. Rev. 2015;28(2):465–522. doi: 10.1128/CMR.00102-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Calisher C.H., Childs J.E., Field H.E., Holmes K.V., Schountz T. Bats: important reservoir hosts of emerging viruses. Clin. Microbiol. Rev. 2006;19(3):531–545. doi: 10.1128/CMR.00017-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Roser M., Ritchie H., Ortiz-Ospina E., Hasell J. 2020. Coronavirus disease (COVID-19) - statistics and research. Our World in Data https://ourworldindata.org/coronavirus. [Google Scholar]
- 6.Haffner S.M., Lehto S., Rönnemaa T., Pyörälä K., Laakso M. Mortality from coronary heart disease in subjects with type 2 diabetes and in nondiabetic subjects with and without prior myocardial infarction. N. Engl. J. Med. 1998;339(4):229–234. doi: 10.1056/NEJM199807233390404. [DOI] [PubMed] [Google Scholar]
- 7.Gates B. Responding to Covid-19 a once-in-a-century pandemic? N. Engl. J. Med. 2020 doi: 10.1056/NEJMp2003762. [DOI] [PubMed] [Google Scholar]
- 8.Wielpütz M.O., Heuß el C.P., Herth F.J., Kauczor H.-U. Radiological diagnosis in lung disease: factoring treatment options into the choice of diagnostic modality. Deutsches Ärzteblatt Int. 2014;111(11):181. doi: 10.3238/arztebl.2014.0181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Black W.C., Welch H.G. Advances in diagnostic imaging and overestimations of disease prevalence and the benefits of therapy. N. Engl. J. Med. 1993;328(17):1237–1243. doi: 10.1056/NEJM199304293281706. [DOI] [PubMed] [Google Scholar]
- 10.Candemir S., Antani S. A review on lung boundary detection in chest X-rays. Int. J. Comput. Assist. Radiol. Surg. 2019;14(4):563–576. doi: 10.1007/s11548-019-01917-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Narin A., Kaya C., Pamuk Z. Automatic detection of coronavirus disease (covid-19) using x-ray images and deep convolutional neural networks. Pattern Anal. Appl. 2021:1–14. doi: 10.1007/s10044-021-00984-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hassanien A.E., Mahdy L.N., Ezzat K.A., Elmousalami H.H., Ella H.A. 2020. Automatic X-ray COVID-19 lung image classification system based on multi-level thresholding and support vector machine. MedRxiv. [DOI] [Google Scholar]
- 13.Hammoudi K., Benhabiles H., Melkemi M., Dornaika F., Arganda-Carreras I., Collard D., Scherpereel A. 2020. Deep learning on chest X-ray images to detect and evaluate pneumonia cases at the era of COVID-19. arXiv preprint arXiv:2004.03399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Feki I., Ammar S., Kessentini Y., Muhammad K. Federated learning for COVID-19 screening from chest X-ray images. Appl. Soft Comput. 2021;106 doi: 10.1016/j.asoc.2021.107330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Khattak M.I., Al-Hasan M., Jan A., Saleem N., Verdú E., Khurshid N. Automated detection of COVID-19 using chest X-Ray images and CT scans through multilayer-spatial convolutional neural networks. Int. J. Interact. Multimedia Artif. Intell. 2021;6(6) [Google Scholar]
- 16.Saygılı A. A new approach for computer-aided detection of coronavirus (COVID-19) from CT and X-ray images using machine learning methods. Appl. Soft Comput. 2021;105 doi: 10.1016/j.asoc.2021.107323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Saiz F.A., Barandiaran I.n. Covid-19 detection in chest X-ray images using a deep learning approach. Int. J. Interact. Multimed. Artif. Intell. 2020;6(2):1–4. [Google Scholar]
- 18.Sait U., KV G.L., Shivakumar S., Kumar T., Bhaumik R., Prajapati S., Bhalla K., Chakrapani A. A deep-learning based multimodal system for Covid-19 diagnosis using breathing sounds and chest X-ray images. Appl. Soft Comput. 2021 doi: 10.1016/j.asoc.2021.107522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Das N.N., Kumar N., Kaur M., Kumar V., Singh D. Automated deep transfer learning-based approach for detection of COVID-19 infection in chest X-rays. IRBM. 2020 doi: 10.1016/j.irbm.2020.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Singh D., Kumar V., Yadav V., Kaur M. Deep neural network-based screening model for COVID-19-infected patients using chest X-ray images. Int. J. Pattern Recognit. Artif. Intell. 2021;35(03) [Google Scholar]
- 21.Ucar F., Korkmaz D. Covidiagnosis-net: Deep Bayes-SqueezeNet based diagnosis of the coronavirus disease 2019 (COVID-19) from X-ray images. Med. Hypotheses. 2020;140 doi: 10.1016/j.mehy.2020.109761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang L., Lin Z.Q., Wong A. Covid-net: A tailored deep convolutional neural network design for detection of covid-19 cases from chest x-ray images. Sci. Rep. 2020;10(1):1–12. doi: 10.1038/s41598-020-76550-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Afshar P., Heidarian S., Naderkhani F., Oikonomou A., Plataniotis K.N., Mohammadi A. Covid-caps: A capsule network-based framework for identification of covid-19 cases from x-ray images. Pattern Recognit. Lett. 2020;138:638–643. doi: 10.1016/j.patrec.2020.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chowdhury M.E., Rahman T., Khandakar A., Mazhar R., Kadir M.A., Mahbub Z.B., Islam K.R., Khan M.S., Iqbal A., Al Emadi N., et al. Can AI help in screening viral and COVID-19 pneumonia? IEEE Access. 2020;8:132665–132676. [Google Scholar]
- 25.Khan A.I., Shah J.L., Bhat M.M. CoroNet: A deep neural network for detection and diagnosis of COVID-19 from chest x-ray images. Comput. Methods Programs Biomed. 2020;196 doi: 10.1016/j.cmpb.2020.105581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sahinbas K., Catak F.O. Data Science for COVID-19. Elsevier; 2021. Transfer learning-based convolutional neural network for COVID-19 detection with X-ray images; pp. 451–466. [Google Scholar]
- 27.Apostolopoulos I., Aznaouridis S., Tzani M. 2020. Extracting possibly representative COVID-19 biomarkers from X-Ray images with deep learning approach and image data related to pulmonary diseases. arXiv preprint arXiv:2004.00338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zulkifley M.A., Abdani S.R., Zulkifley N.H. Covid-19 screening using a lightweight convolutional neural network with generative adversarial network data augmentation. Symmetry. 2020;12(9):1530. [Google Scholar]
- 29.Radiological Society of North America (RSNA) M.A. 2019. Pneumonia detection challenge. https://www.kaggle.com/c/rsna-pneumonia-detection-challenge ( Accessed 04 October 2020) [DOI] [PubMed] [Google Scholar]
- 30.de Moura J., Novo J., Ortega M. 2019 International Joint Conference on Neural Networks, IJCNN. IEEE; 2019. Deep feature analysis in a transfer learning-based approach for the automatic identification of diabetic macular edema; pp. 1–8. [Google Scholar]
- 31.Vidal P.L., de Moura J., Novo J., Ortega M. 2019 International Joint Conference on Neural Networks, IJCNN. IEEE; 2019. Cystoid fluid color map generation in optical coherence tomography images using a densely connected convolutional neural network; pp. 1–8. [Google Scholar]
- 32.Huang G., Liu S., Van der Maaten L., Weinberger K.Q. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition. 2018. Condensenet: An efficient densenet using learned group convolutions; pp. 2752–2761. [Google Scholar]
- 33.He K., Zhang X., Ren S., Sun J. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition. 2016. Deep residual learning for image recognition; pp. 770–778. [Google Scholar]
- 34.Simonyan K., Zisserman A. 2014. Very deep convolutional networks for large-scale image recognition. arXiv preprint arXiv:1409.1556. [Google Scholar]
- 35.Dey R., Lu Z., Hong Y. 2018 IEEE 15th International Symposium on Biomedical Imaging, ISBI 2018. IEEE; 2018. Diagnostic classification of lung nodules using 3D neural networks; pp. 774–778. [Google Scholar]
- 36.Liu Y., Hao P., Zhang P., Xu X., Wu J., Chen W. Dense convolutional binary-tree networks for lung nodule classification. IEEE Access. 2018;6:49080–49088. [Google Scholar]
- 37.Guo W., Xu Z., Zhang H. Interstitial lung disease classification using improved DenseNet. Multimed. Tools Appl. 2019;78(21):30615–30626. [Google Scholar]
- 38.Tseng H.-H., Wei L., Cui S., Luo Y., Ten Haken R.K., El Naqa I. Machine learning and imaging informatics in oncology. Oncology. 2018:1–19. doi: 10.1159/000493575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Deng J., Dong W., Socher R., Li L.-J., Li K., Fei-Fei L. 2009 IEEE Conference on Computer Vision and Pattern Recognition. Ieee; 2009. Imagenet: A large-scale hierarchical image database; pp. 248–255. [Google Scholar]
- 40.Rusiecki A. Vol. 55. IET; 2019. Trimmed categorical cross-entropy for deep learning with label noise; pp. 319–320. [Google Scholar]
- 41.Sutskever I., Martens J., Dahl G., Hinton G. International Conference on Machine Learning. 2013. On the importance of initialization and momentum in deep learning; pp. 1139–1147. [Google Scholar]
- 42.Van Dyk D.A., Meng X.-L. The art of data augmentation. J. Comput. Graph. Statist. 2001;10(1):1–50. [Google Scholar]
- 43.Mikołajczyk A., Grochowski M. 2018 International Interdisciplinary PhD Workshop, IIPhDW. IEEE; 2018. Data augmentation for improving deep learning in image classification problem; pp. 117–122. [Google Scholar]
- 44.Cohen J.P., Morrison P., Dao L. 2020. COVID-19 Image data collection. [Google Scholar]
- 45.Italian Society of Medical Radiology (SIRM) J.P. 2020. COVID-19 DATABASE. https://www.sirm.org/category/senza-categoria/covid-19/ ( Accessed 04 October 2020) [Google Scholar]
- 46.Kanne J.P., Little B.P., Chung J.H., Elicker B.M., Ketai L.H. 2020. Essentials for radiologists on COVID-19: an update—radiology scientific expert panel. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mossa-Basha M., Meltzer C.C., Kim D.C., Tuite M.J., Kolli K.P., Tan B.S. 2020. Radiology department preparedness for COVID-19: radiology scientific expert review panel. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Manna S., Wruble J., Maron S.Z., Toussie D., Voutsinas N., Finkelstein M., Cedillo M.A., Diamond J., Eber C., Jacobi A., et al. COVID-19: a multimodality review of radiologic techniques, clinical utility, and imaging features. Radiol. Cardiothoracic Imaging. 2020;2(3) doi: 10.1148/ryct.2020200210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chamorro E.M., Tascón A.D., Sanz L.I., Vélez S.O., Nacenta S.B. Radiologic diagnosis of patients with COVID-19. RadiologíA (English Edition) 2021;63(1):56–73. doi: 10.1016/j.rx.2020.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]