Abstract
Recently, circular RNAs (circRNAs) have attracted growing attention due to their pivotal roles in the complicated cellular processes of diverse human malignancies, including colorectal cancer (CRC). Phosphatase and tensin homolog (PTEN) is known as a typical tumor-suppressing gene. Nevertheless, limited investigation on the function of circRNAs generated from PTEN has been undertaken. In this research, hsa_circ_0094343 (circPTEN) was found to display low expression in CRC tissues and cells. CircPTEN is characterized with high stability due to its circular structure. Upregulation of circPTEN suppressed CRC cell proliferation, migration, and invasion but facilitated apoptosis. Data from mechanism assays revealed that circPTEN could elevate PTEN expression through sequestering microRNA-4470 (miR-4470) in CRC cells. Further, circPTEN was validated to inhibit K63-linked ubiquitination of protein kinase B (AKT) and AKT phosphorylation at Thr-308 and Ser-473 by competitively binding with tumor necrosis factor (TNF)-receptor-associated factor 6 (TRAF6). Moreover, the results of rescue assays indicated that the suppressive effect of circPTEN on CRC progression could be totally reversed by overexpression of insulin like growth factor 1 (IGF-1) or partially reversed by knockdown of PTEN. To conclude, circPTEN suppresses CRC progression via regulation of PTEN/AKT pathway.
Keywords: colorectal cancer, circPTEN, miR-4470, AKT, TRAF6
Graphical abstract
This study confirmed the circPTEN/miR-4470/PTEN regulatory mechanism in CRC. circPTEN was also validated to inhibit K63-linked AKT ubiquitination and phosphorylation at Thr-308 and Ser-473 by binding to the Ring finger domain of TRAF6 in CRC. To conclude, circPTEN suppressed CRC progression by regulating PTEN/AKT pathway.
Introduction
Colorectal cancer (CRC) originates from transformation of colon or rectum epithelial cells, which is widely diagnosed as a kind of lethal human malignancy, contributing to an increasing number of cancer-associated death cases.1, 2, 3 A wide range of germline and somatic mutations occur during CRC progression, which is associated with the dysregulation of oncogenes or anti-tumor genes.4,5 Due to the technological progress in early detection and intervention, the overall survival rate of patients with CRC has been improved to a certain degree.6 However, the prognosis of CRC patients at advanced stages remains unsatisfactory, mainly because of the recurrence and metastasis after surgery.7,8 Over the past years, distant metastasis has been recognized as one of the main factors leading to the disappointing survival rate of CRC patients.9,10 Metastasis involves the spread of cancer cells from the primary tumor to surrounding tissues and to distant organs. Activation or inactivation of diverse genes has been found to be associated with the tumor cell invasion and migration.11 Additionally, aberrantly expressed genes have been revealed to be closely correlated with CRC cellular processes.12, 13, 14 In this regard, exploration of the underlying molecular mechanisms in CRC progression is of significant value for improvement of CRC treatment.
Genomic and transcriptomic sequencing has unveiled that only a small part of human genome can be transcribed into messenger RNAs (mRNAs) with protein-coding ability, whereas the majority can be transcribed into noncoding RNAs (ncRNAs).15 Unlike protein-coding mRNAs, ncRNAs have no protein-coding ability and function in cellular processes by regulating their downstream targets.16 Circular RNAs (circRNAs) are a group of ncRNAs without 5′ and 3′ ends and featured with high stability due to their covalently closed loop structure.17,18 circRNAs are identified as more promising biomarkers for cancer treatment due to their higher stability.19 In recent years, a growing number of investigations have confirmed the critical role of circRNAs in regulating the progression of diverse human malignancies. For example, circRNA hsa_circ_0014717 is expressed at low levels in CRC and suppresses CRC tumor growth through elevating p16 expression.20 Circ-DONSON facilitates the progression of gastric cancer by recruiting NURF complex to promote SOX4 expression.21 circRNA circ-ABCB10 promotes breast cancer progression through sponging microRNA-1271 (miR-1271).22
Phosphatase and tensin homolog (PTEN) has been identified as a typical tumor suppressor in diverse cancers, including CRC.23, 24, 25 Nevertheless, circRNAs generated from PTEN hardly have been studied in human cancers. Here, we checked for the potential role of a circRNA derived from linear mRNA PTEN (circPTEN) in CRC progression. More importantly, we investigated the underlying molecular mechanism of circPTEN in CRC.
Results
circPTEN expression is significantly downregulated in CRC tissues and cells
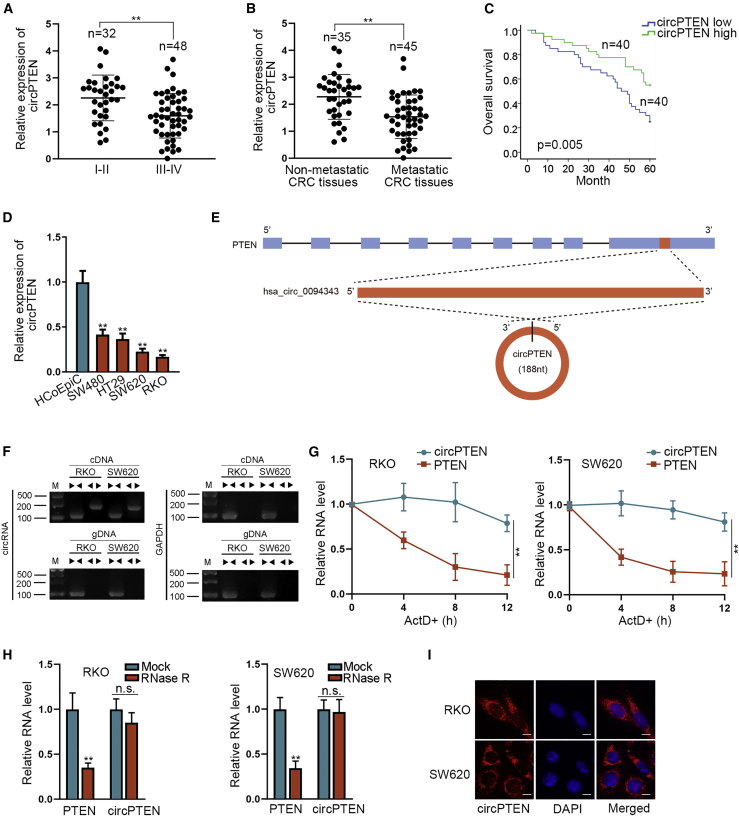
We first obtained 8 circRNAs (hsa_circ_0002232, hsa_circ_0019058, hsa_circ_0003058, hsa_circ_0094342, hsa_circ_0002934, hsa_circ_0019059, hsa_circ_0019060, and hsa_circ_0094343) whose host gene was PTEN from circBase database: http://www.circbase.org/. After detection, we observed that only hsa_circ_0094343 was notably expressed at low levels in CRC tissues in contrast to adjacent non-tumor tissues (Figure S1A). Through Pearson correlation analysis, we noticed a significantly positive relation between hsa_circ_0094343 (named as circPTEN) and PTEN (Figure S1B), and thereby, we chose it for subsequent experiments. Additionally, circPTEN expression was lower in CRC patients at advanced stages (III to IV) than those at early stages (I to II; Figure 1A). Compared with that in non-metastatic CRC tissues, circPTEN expression was markedly downregulated in metastatic CRC tissues (Figure 1B). Importantly, low expression of circPTEN was closely correlated to poor prognosis (Figure 1C). Furthermore, in comparison with that in normal human colonic epithelial cell line (HCoEpiC), circPTEN expression was low expressed in CRC cells, particularly in RKO and SW620 cells (Figure 1D). Next, the genomic location and splicing pattern of hsa_circ_0094343 (circPTEN) were illustrated (Figure 1E). As shown in Figure 1F, divergent primers could produce the circular isoform of PTEN with cDNA, but not with gDNA, whereas convergent primers could amplify the linear isoform of PTEN from both cDNA and gDNA in RKO and SW620 cells. After treatment with ActD, circPTEN presented higher stability than PTEN in RKO and SW620 cells (Figure 1G). Furthermore, linear mRNA PTEN was digested by RNase R, whereas circPTEN resisted to RNase R digestion in RKO and SW620 cells (Figure 1H). Finally, circPTEN was detected to be mainly localized in the cytoplasm of RKO and SW620 cells through fluorescence in situ hybridization (FISH) assay (Figure 1I). In brief, circPTEN, featured with circular structure, is low expressed in CRC tissues and predicts poor prognosis.
Figure 1.
circPTEN expression is significantly downregulated in CRC tissues and cells
(A) qRT-PCR analysis of circPTEN expression in CRC patients at different stages. (B) circPTEN expression in non-metastatic CRC tissues and metastatic CRC tissues was detected via qRT-PCR. (C) Analysis of the overall survival of CRC patients with low or high expression of circPTEN is shown. (D) qRT-PCR analysis of circPTEN expression in CRC cell lines and HCoEpiC cells is shown. (E) The genomic location and splicing pattern of hsa_circ_0094343 (circPTEN) is shown. (F) Nucleic acid electrophoresis showed that divergent primers could produce the circular isoform of PTEN with cDNA, but not with gDNA. GAPDH was an endogenous control. (G) The resistance of circPTEN and PTEN to ActD was analyzed in RKO and SW620 cells. (H) The expression of circPTEN and PTEN mRNA was examined via qRT-PCR in RKO and SW620 cells after RNase R treatment. (I) Detection of the subcellular localization of circPTEN through FISH (bar value = 20 μm) assay is shown. ∗∗p < 0.01. n.s., no significance.
Upregulation of circPTEN restrains malignant behaviors of CRC cells
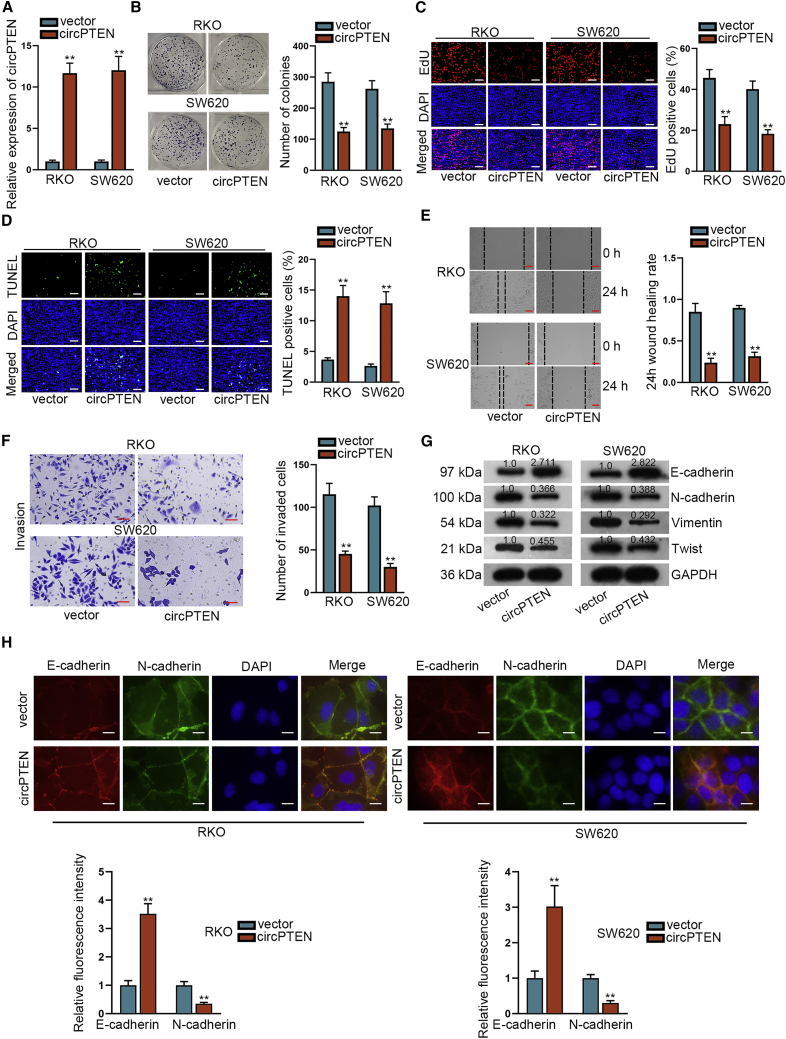
To identify the role of circPTEN in cell phenotype, gain-of-function assays were conducted. Prior to that, circPTEN overexpression efficiency in RKO and SW620 cells was determined (Figure 2A). According to results of colony formation and 5-ethynyl-2-deoxyuridine (EdU) assays, cell proliferation was repressed by circPTEN upregulation (Figures 2B and 2C). Conversely, elevated expression of circPTEN resulted in the enhancement of cell apoptosis (Figure 2D). The capabilities of RKO and SW620 cells to migrate and invade were attenuated by the upregulation of circPTEN (Figures 2E and 2F). Western blot analysis unveiled that increased level of circPTEN in RKO and SW620 cells led to upregulation of E-cadherin but downregulation of N-cadherin, vimentin, and Twist, suggesting the inhibitory effect of overexpressed circPTEN on epithelial-mesenchymal transition (EMT) process (Figure 2G). Besides, immunofluorescence (IF) demonstrated the enhanced expression of E-cadherin and reduced expression of N-cadherin after circPTEN was overexpressed in RKO and SW620 cells, further validating the suppressive effect of circPTEN upregulation on EMT process (Figure 2H). Taken together, overexpression of circPTEN restrains malignant behaviors of CRC cells.
Figure 2.
Upregulation of circPTEN restrains malignant behaviors of CRC cells
(A) qRT-PCR analysis of circPTEN overexpression efficiency in RKO and SW620 cells. (B and C) Evaluation of proliferation of transfected cells via colony formation and EdU (bar value = 150 μm) assays is shown. (D) Measurement of the apoptosis of transfected cells through TUNEL (bar value = 150 μm) assay is shown. (E and F) Analysis of the migratory and invasive capabilities of transfected RKO and SW620 cells via wound healing (bar value = 100 μm) and transwell (bar value = 70 μm) assays is shown. (G) Western blot analysis of EMT-related proteins in different groups is shown. (H) IF (bar value = 20 μm) analysis of E-cadherin and N-cadherin in transfected cells is shown. ∗∗p < 0.01.
Silencing of circPTEN promotes CRC cell growth and invasion
To further determine the functions of circPTEN in CRC cellular processes, we silenced it in SW480 and HT29 cells that possessed relative higher level of circPTEN expression (Figure S2A). Functionally, silencing of circPTEN promoted CRC cell proliferation and invasion (Figures S2B–S2D). In addition, EMT process was reduced by circPTEN silencing (Figure S2E). Therefore, we confirmed that silenced circPTEN facilitates CRC cell proliferation, invasion, and EMT process.
circPTEN upregulates PTEN expression by sequestering miR-4470 in CRC cells
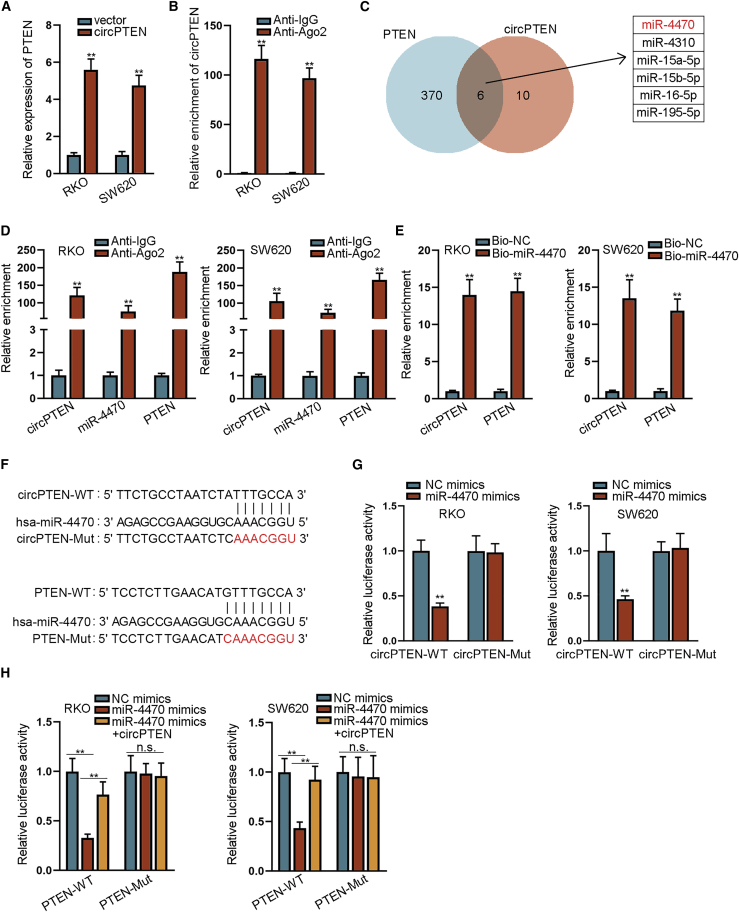
To investigate how circPTEN regulated PTEN expression in CRC, we first carried out quantitative reverse transcriptase polymerase chain reaction (qRT-PCR) to detect the expression of PTEN under circPTEN overexpression. As demonstrated in Figure 3A, circPTEN overexpression led to the elevation of PTEN expression. Then, Ago2-RNA immunoprecipitation (RIP) assay confirmed that circPTEN could bind with Ago2, indicating the existence of circPTEN in RNA-induced silencing complex (RISC) (Figure 3B). Considering that circPTEN was majorly distributed in the cytoplasm, we conjectured that circPTEN might regulate PTEN expression by sponging a certain microRNA (miRNA). After searching on miRDB database: http://mirdb.org/, 6 miRNAs were predicted to have the binding potential with circPTEN and PTEN (Figure 3C). Through qRT-PCR analysis, we noted that only miR-4470 was strikingly upregulated in CRC tissues (Figure S3A). In addition, we upregulated these miRNAs in RKO and SW620 cells and discovered only miR-4470 upregulation significantly restrained PTEN expression (Figures S3B and S3C). Besides, there was a negative correlation between miR-4470 expression and PTEN expression in CRC tissues (Figure S3D). Thus, miR-4470 was chosen for further analysis. As demonstrated in Ago2-RIP assay, circPTEN, miR-4470, and PTEN were observably enriched in anti-Ago2 group, hinting the coexistence of these three RNAs in RNA-induced silencing complex (RISC) (Figure 3D). Subsequently, RNA pull-down assay validated that miR-4470 could bind with circPTEN and PTEN in RKO and SW620 cells (Figure 3E). From miRDB, the binding sites between miR-4470 and circPTEN (or PTEN) were obtained and presented in Figure 3F. Further, data from luciferase reporter assays depicted that the miR-4470 upregulation weakened the luciferase activity of pmirGLO-circPTEN-wild type (WT) (Figure 3G). More importantly, the weakened luciferase activity of pmirGLO-PTEN-WT induced by miR-4470 upregulation could be rescued by overexpressing circPTEN in RKO and SW620 cells (Figure 3H). To sum up, circPTEN regulates PTEN expression by competitively binding with miR-4470 in CRC cells.
Figure 3.
circPTEN regulates PTEN expression by sequestering miR-4470 in CRC cells
(A) qRT-PCR analysis of PTEN expression in RKO and SW620 cells with circPTEN overexpression. (B) The enrichment of circPTEN in the immunoprecipitates conjugated to anti-Ago2 was detected via qRT-PCR in RIP assay. (C) Six miRNAs were predicted to bind with circPTEN and PTEN via miRDB. (D) The coexistence of circPTEN, miR-4470, and PTEN in RISC was testified via Ago2-RIP assay. (E) The binding capacity between miR-4470 and circPTEN (or PTEN) was validated via RNA pull-down assay. (F) The binding sites between miR-4470 and circPTEN (or PTEN) were predicted through miRDB. (G) The interaction between circPTEN and miR-4470 was demonstrated by luciferase reporter assay. (H) The competitive binding relationship among circPTEN, miR-4470, and PTEN was verified through luciferase reporter assay. ∗∗p < 0.01.
circPTEN mediates malignant behaviors of CRC cells via PTEN/AKT signaling pathway
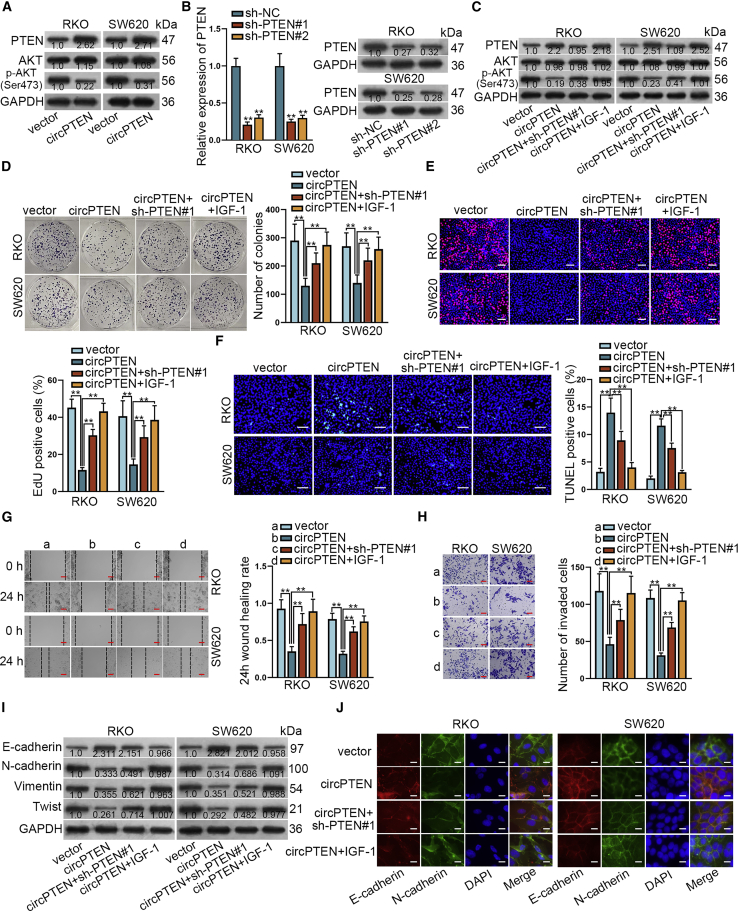
Emerging studies have revealed the significant role of PTEN/AKT pathway in CRC progression.26,27 To test whether and how circPTEN was associated with PTEN/AKT pathway, western blot analysis was performed to measure the protein levels of PTEN/AKT factors. After upregulating circPTEN, PTEN expression in RKO and SW620 cells was conspicuously augmented and p-AKT level was noticeably decreased (Figure 4A). Additionally, decline in PTEN expression but increment in p-AKT level were observed in cells with circPTEN silencing (Figure S4A). Then, PTEN was silenced in indicated CRC cells with transfection of short hairpin RNAs (shRNAs) targeting PTEN (sh-PTEN#1/2) (Figure 4B). Later on, western blot analysis indicated that PTEN silencing could completely reverse the effect of circPTEN upregulation on PTEN expression, whereas insulin growth factor 1 (IGF-1) (an activator of AKT signaling) treatment had no significant effect. However, the effect of circPTEN upregulation on p-AKT expression could only be partially recovered by PTEN silencing whereas completely restored by IGF-1 treatment (Figure 4C). Subsequently, colony formation, EdU, and terminal deoxynucleotidyl transferase (TdT)-mediated dUTP nick-end labeling (TUNEL) assays were performed and the results showed that the effect of upregulated circPTEN on cell proliferation and apoptosis could be partially rescued by PTEN depletion although entirely restored by addition of IGF-1 (Figures 4D–4F). Similarly, treatment with IGF-1 or sh-PTEN to RKO and SW620 cells could fully or partially rescue the suppressive effect of circPTEN overexpression on cell migration and invasion (Figures 4G and 4H). Likewise, we verified that the inhibitory effect on EMT process due to circPTEN upregulation could only be partly reversed by PTEN knockdown whereas completely countervailed by addition of IGF-1 (Figures 4I, 4J, and S4B). In sum, circPTEN represses malignant behaviors of CRC cells by inhibiting PTEN/AKT signaling pathway.
Figure 4.
circPTEN mediates malignant behaviors of CRC cells via AKT signaling pathway
(A) Western blot analysis of protein expression in cells with circPTEN upregulation. (B) The knockdown efficiency of sh-PTEN was evaluated via qRT-PCR and western blot. (C) Western blot analysis of protein expression after transfection with different plasmids is shown. (D and E) Evaluation of cell proliferation in different groups via colony formation and EdU (bar value = 150 μm) assays is shown. (F) TUNEL (bar value = 150 μm) assay was performed to measure cell apoptosis in different groups. (G and H) Analysis of the migration and invasion abilities via wound healing (bar value = 100 μm) and transwell (bar value = 70 μm) analyses is shown. (I and J) EMT markers in different groups were detected via western blot and IF (bar value = 20 μm). ∗∗p < 0.01.
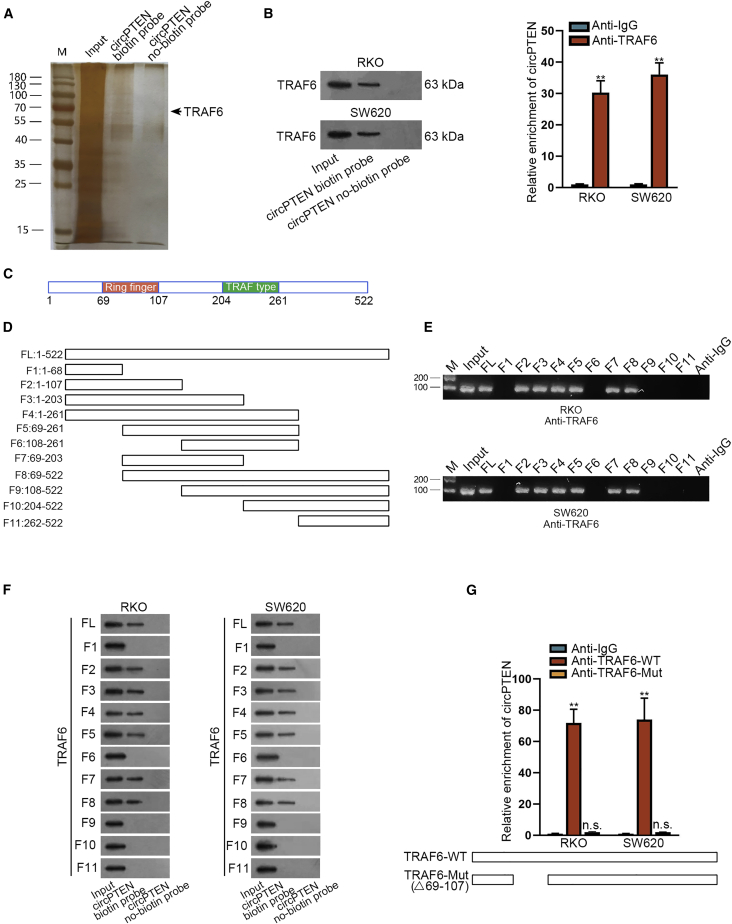
circPTEN regulates K63-linked ubiquitination of AKT through TRAF6 in CRC cells
A previous research has manifested that K63-linked ubiquitination of AKT is essential for AKT activation.28, 29, 30 Herein, we intended to explore whether circPTEN was related to K63-linked ubiquitination of AKT in CRC cells. After we overexpressed circPTEN in RKO and SW620 cells, AKT ubiquitination was inhibited and p-AKT expression was reduced, and meanwhile, MG132 caused no observable effect on AKT ubiquitination and p-AKT expression (Figure S5A). Additionally, we recognized that circPTEN could restrain the K63-linked ubiquitination of AKT, but not K48 linked (Figure S5B). Moreover, ectopic expression of circPTEN resulted in the downregulation of p-AKT at Thr-308 and Ser-473, whereas p-AKT at Tyr-474 was hardly influenced (Figure S5C). Furthermore, through RNA pull-down and mass spectrometry, tumor necrosis factor (TNF)-receptor-associated factor 6 (TRAF6) was revealed to bind with circPTEN (Figure 5A). And then, RNA pull-down and RIP assays further certified the binding relation between circPTEN and TRAF6 in RKO and SW620 cells (Figure 5B). Then, we analyzed the structure of TRAF6 (Figure 5C) and divided it into 12 segments (Figure 5D). Through RIP assay, circPTEN could be enriched by FL/F2-F5/F7/F8, hinting that circPTEN bound to the Ring finger domain of TRAF6 (Figure 5E). Besides, RNA pull-down assay further confirmed the binding capacity of circPTEN with the Ring finger domain of TRAF6 (Figure 5F). Further, after we mutated the Ring finger domain of TRAF6, TRAF6 could not bind with circPTEN (Figure 5G). All in all, circPTEN modulates K63-linked ubiquitination of AKT through binding to the Ring finger domain of TRAF6 in CRC.
Figure 5.
circPTEN regulates K63-linked ubiquitination of AKT through TRAF6 in CRC cells
(A) TRAF6 was revealed to bind with circPTEN through RNA pull-down and mass spectrometry. (B) The binding capacity between circPTEN and TRAF6 was proven by RNA pull-down and RIP assays. (C) The structure of TRAF6 is shown. (D) TRAF6 was divided into 12 segments. (E and F) The binding of circPTEN to the Ring finger domain of TRAF6 was testified via RIP and RNA pull-down assays. (G) Verification of the binding between circPTEN and the Ring finger domain of TRAF6 via RIP is shown. ∗∗p < 0.01.
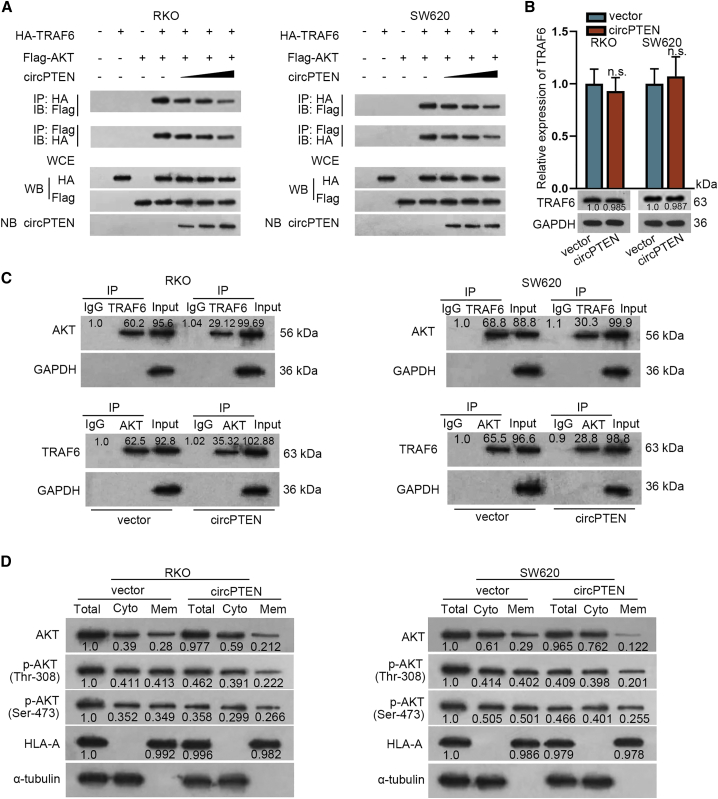
circPTEN inhibits phosphorylation of AKT in CRC cells
To proceed with this study, we first applied coimmunoprecipitation (coIP) assay and confirmed that upregulation of circPTEN repressed the binding between TRAF6 and AKT (Figure 6A). Next, qRT-PCR and western blot analyses revealed that circPTEN upregulation had no evident effect on TRAF6 expression (Figure 6B). Subsequently, coIP assay testified that overexpression of circPTEN disrupted the interaction between TRAF6 and AKT (Figure 6C). Finally, after circPTEN expression was upregulated, the expression of p-AKT (Thr-308) and p-AKT (Ser-473) in membrane was reduced, suggesting that circPTEN could suppress phosphorylation of AKT at Thr-308 and Ser-473 (Figure 6D). Briefly, circPTEN restrains phosphorylation of AKT in CRC cells.
Figure 6.
circPTEN inhibits the phosphorylation of AKT in CRC cells
(A) coIP assay confirmed that circPTEN could repress the binding between TRAF6 and AKT. WCEs were collected for immunoprecipitation. WB referred to western blot and NB represented Northern blot. (B) TRAF6 expression in transfected cells was detected via qRT-PCR and western blot analysis. (C) The effect of circPTEN on the interaction between TRAF6 and AKT was proven via coIP. (D) Western blot analysis of protein expression is shown.
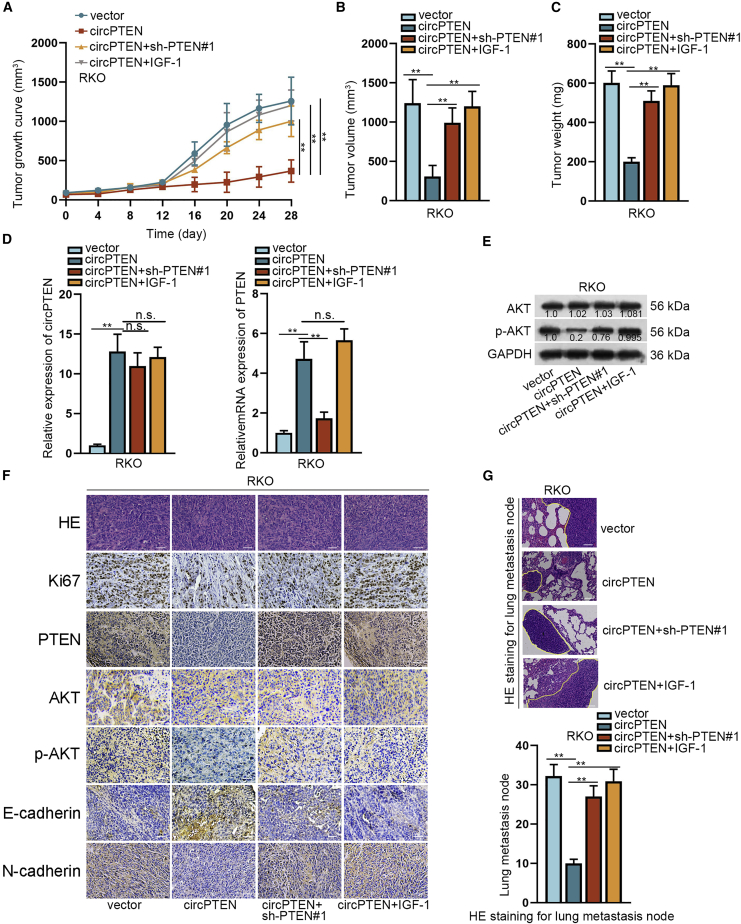
Upregulation of circPTEN inhibits CRC progression via PTEN/AKT pathway
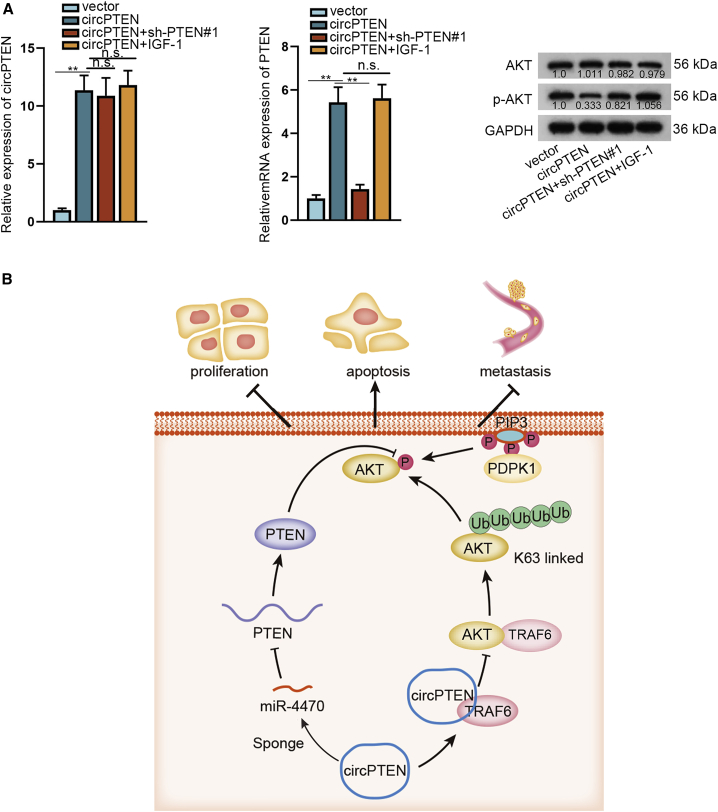
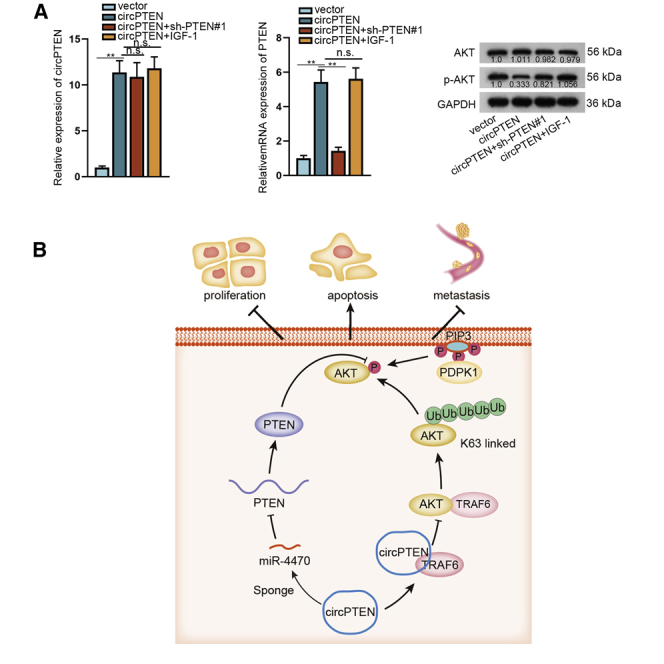
In order to further verify the regulatory effect of circPTEN on CRC, we performed in vivo assays. As shown in Figures 7A–7C, the suppressive effect of circPTEN upregulation on tumor growth could be partially rescued by PTEN knockdown whereas completely restored by adding IGF-1. qRT-PCR analysis disclosed that circPTEN expression was upregulated by transfection with circPTEN, and co-transfection with sh-PTEN#1 or treatment with IGF-1 induced no obvious changes of circPTEN expression (Figure 7D, left). However, co-transfection with sh-PTEN#1 could reverse the facilitation of PTEN expression in response to circPTEN upregulation, whereas addition of IGF-1 had no obvious impact on circPTEN-mediated PTEN expression (Figure 7D, right). Besides, we determined that the restraining effect of upregulated circPTEN on p-AKT expression could be completely restored by adding IGF-1 although partially rescued by PTEN silencing (Figure 7E). Likewise, the effect of circPTEN overexpression on the expression of Ki67, PTEN, p-AKT, E-cadherin, and N-cadherin could be entirely rescued by addition of IGF-1 whereas partly recovered by silenced PTEN (Figure 7F). Further, hematoxylin and eosin (H&E) staining for lung metastasis node further validated the complete rescue effect of IGF-1 on circPTEN upregulation-mediated lung metastasis, together with the incomplete rescue effect of downregulated PTEN (Figure 7G). As shown in Figure 8A, circPTEN expression was strikingly elevated in circPTEN overexpression group in contrast to control group, which could not be affected by co-transfection of sh-PTEN#1 or treatment with IGF-1. Nevertheless, PTEN depletion could restore the restraining effect of circPTEN upregulation on PTEN expression, although IGF-1 supplement resulted in no obvious changes. Additionally, the suppressive effect of overexpressed circPTEN on p-AKT level could be partially countervailed by PTEN deficiency whereas entirely reversed by adding IGF-1. All these aforementioned findings uncover that upregulated circPTEN suppresses CRC progression by regulating PTEN/AKT pathway (Figure 8B).
Figure 7.
Upregulation of circPTEN inhibits CRC progression via PTEN/AKT pathway
(A–C) Transfected RKO cells were subcutaneously injected into nude mice, and later, tumor growth, volume, and weight were analyzed. (D) qRT-PCR analysis of circPTEN or PTEN expression in different groups is shown. (E) Western blot analysis of AKT or p-AKT expression in different groups is shown. (F) IF (bar value = 80 μm) analysis of proteins (Ki67, PTEN, AKT, p-AKT, E-cadherin, and N-cadherin) in different groups is shown. (G) H&E (bar value = 100 μm) staining was utilized for evaluation of metastasis in different groups. ∗∗p < 0.01.
Figure 8.
The regulatory role of circPTEN in CRC cells
(A) Analyses of the expression of circPTEN, PTEN, AKT, or p-AKT in tissues removed from mice in four different groups via qRT-PCR and western blot. (B) A graphical concept of the molecular mechanism of circPTEN in CRC cells is shown. ∗∗p < 0.01.
Discussion
CRC is widely reported as a prevalent type of lethal human malignancies and causes approximately 694,000 deaths annually, accounting for a large proportion in cases of cancer-related death.31 Over the past years, researchers have been dedicated to studying the pathology and molecular mechanisms underlying CRC. As a result, a number of abnormally expressed RNAs are unveiled to be implicated in the CRC progression.32, 33, 34 PTEN has been manifested to be low expressed in multiple human cancers and exert restraining function on the development of these cancers, including CRC. For example, downregulated PTEN by miR-21 promotes cervical cancer cell proliferation.35 The miR-130 family downregulates PTEN expression to drive bladder cancer cell migration and invasion.36 miR-21 motivates CRC cell growth and invasion through inhibiting PTEN expression.24 Accumulating evidence has revealed that circRNAs play significant roles in tumor progression via regulating mRNA expression.37,38 In this study, we observed significantly low expression of hsa_circ_0094343 (a circRNA derived from linear PTEN, which was later named as circPTEN) in CRC tissues and cells. Besides, circPTEN was characterized with high stability in contrast to PTEN mRNA due to its particular loop structure. Moreover, upregulation of circPTEN repressed CRC cell proliferation, migration, invasion, and EMT process whereas inducing cell apoptosis. To our knowledge, this is the first time to explore the function of circPTEN in CRC progression.
Based on previous findings, circRNAs could mediate multiple cellular processes of cancer progression by serving as a competing endogenous RNA (ceRNA). For instance, circ_0056618 acts as a ceRNA to promote gastric cancer progression by sponging miR-206 to upregulate CXCR4 expression.39 Hsa_circ_0071589 facilitates the carcinogenesis of CRC by targeting miR-600/EZH2 axis.40 In current research, we also explored whether circPTEN regulated PTEN through a ceRNA pathway. After application of bioinformatics prediction and mechanism assays, miR-4470 was uncovered to be highly expressed and negatively regulated PTEN expression in CRC cells. Additionally, miR-4470 was testified to bind with circPTEN and PTEN. Collectively, we proved that circPTEN regulated PTEN through sequestering miR-4470 in CRC.
Abundant evidence has elucidated that AKT phosphorylation was closely related to the tumorigenesis and development of multiple human cancers, such as gastric cancer,41 cervical cancer,42 and CRC.26 Interestingly, PTEN/AKT pathway has been revealed to play significant roles in human malignancies, including CRC.26,43 In this study, through a series of rescue assays, we determined that PTEN silence could completely restore the circPTEN upregulation-mediated PTEN expression. PTEN knockdown could only partially rescue the restraining effect of upregulated circPTEN on AKT phosphorylation and CRC progression, whereas addition of IGF-1 could elicit complete rescue effects. Herein, we further detected another pathway through which circPTEN influenced AKT phosphorylation.
Protein ubiquitination is considered as a crucial posttranslational modification, which can regulate a variety of biological functions in diverse human cancers.28,29. In spite of the fact that ubiquitination generally leads to the degradation of protein, ubiquitination is also significant for signaling activation as well as protein trafficking.28,29 Data from previous studies clarified that ubiquitination through lysine 48 (K48) of the ubiquitin chain usually targeted proteins for degradation although ubiquitination through K63 contributed to signaling activation and protein trafficking.28,29 Besides, TRAF6 has been reported to bind with AKT to promote AKT ubiquitination through K63 and thus induce the phosphorylation of AKT.30 In our study, circPTEN was validated to inhibit K63-linked AKT ubiquitination and phosphorylation at Thr-308 and Ser-473 by binding to the Ring finger domain of TRAF6 in CRC. Final rescue assays further proved the involvement of PTEN/AKT pathway in circPTEN-mediated CRC progression. In conclusion, circPTEN suppresses CRC progression by regulating PTEN/AKT pathway, which provides novel insights for CRC therapies.
Materials and methods
Tissue samples
Eighty matched CRC and adjacent non-tumor colorectal tissue samples were collected from CRC patients between 2013 and 2018, with the ethical approval of the Ethics Committee of the First Affiliated Hospital of Xi’an Jiaotong University. Before surgery resection, all CRC patients received histological diagnosis. Patients undergoing radiotherapy or chemotherapy before operation were excluded, as well as those with more than one type of malignant tumors. Additionally, patients with colitis were also excluded from this study. Informed consents were signed by all participants. Immediately after surgery, both tissue samples were frozen at −80°C in liquid nitrogen for use.
Cell culture and treatment
HCoEpiC and human CRC cell lines (SW480, HT29, SW620, and RKO) were procured from ATCC Company (Manassas, VA). DMEM, containing 1% penicillin/streptomycin and 10% fetal bovine serum (FBS), was purchased from Invitrogen (Carlsbad, CA) and used for cell culture under the condition of 5% CO2 and 37°C. The actinomycin D (ActD) (2 μg/mL) and IGF-1 (100 ng/mL) were purchased from Sigma-Aldrich (St. Louis, MO). RNase R (3 U mg−1) was acquired from Epicenter Technologies (Madison, WI). All were used for treating RKO and SW620 cells. Besides, 10 μM of MG132, the common proteasome inhibitor, was acquired from Selleck Chemicals (Houston, TX).
Total RNA isolation and qRT-PCR
Total RNAs were isolated from tissue samples or cells with Trizol reagent (Invitrogen) for cDNA synthesis using reverse transcription kit (Takara Bio, Shiga, Japan). qRT-PCR was performed for detection of gene expression on StepOne Real-Time PCR System (Applied Biosystems, Carlsbad, CA). Results were calculated with the 2−ΔΔCt method and standardized to small nuclear RNA (snRNA) U6 or glyceraldehyde-3-phosphate dehydrogenase (GAPDH). Primer sequences and circRNA sequences were listed in Tables S1 and S2.
Nucleic acid electrophoresis
The cDNA and gDNA PCR products of circPTEN were measured utilizing agarose gels with TE buffer (Thermo Scientific, Waltham, MA, USA), with DL600 as DNA marker. Subsequent to electrophoresis, DNA was separated and subjected to UV irradiation.
RNA FISH
RKO and SW620 cells were fixed by 4% formaldehyde, rinsed in PBS, and then dehydrated in ethanol. After hybridization with circPTEN-FISH probe (Ribobio, Guangzhou, China), cell nuclei were stained with DAPI solution and confocal microscope (Olympus, Tokyo, Japan) was employed for observation.
Plasmid transfection
The pcDNA3.1 vector for circPTEN overexpression and empty vector as the negative control (NC) were available from Genepharma (Shanghai, China). In addition, the specific shRNAs targeting PTEN or circPTEN and NC-shRNAs, as well as the miRNAs mimics and NC mimics, were all designed by Genepharma. RKO and SW620 cells were transfected for 48 h as per the instruction of Lipofectamine 2000 (Invitrogen).
Colony formation assay
CRC cells were prepared after transfection in 6-well culture plate with 500-cell density in each well. 14 days later, cells were fixed before staining with 0.1% crystal violet solution and then counted manually.
EdU staining
Transfected CRC cells in 96-well plate were first cultured with 0.5% Troxin X-100 after fixation and then with 1× Apollo 488 solution. Subsequent to nuclear counter-staining in DAPI solution, cells were examined under Olympus microscope.
TUNEL staining
After fixing, transfected RKO and SW620 cells were rinsed in cold PBS and permeabilized with 0.5% Triton X-100. Cell apoptosis was examined using In Situ Cell Apoptosis Detection Kit (Roche, Basel, Switzerland) as required. After DAPI staining, TUNEL-positive cells were assayed under Olympus microscope.
Wound healing assay
After transfection, RKO and SW620 cells were seeded in serum-free medium for 24 h and then cells were scratched by pipette tips for wound-healing assay. The wound closures were imaged at 0 and 24 h after scratching.
Transwell assay
Matrigel-coated transwell chamber (Corning, Corning, NY) was commercially acquired for cell invasion assay. After transfection, CRC cells were seeded in upper chamber at a cell density of 1 × 105, with lower chamber supplied with complete culture medium. 24 h later, invaded cells to bottom were fixed for staining in crystal violet solution, followed by observation under microscope.
Western blot
Total cellular protein extracts from CRC cells and tissues were subjected to 12% SDS-PAGE and then shifted to polyvinylidene fluoride (PVDF) membranes and cultured in 5% skim milk. The primary antibodies against GAPDH, ꞵ-actin, Twist, vimentin, N-cadherin, E-cadherin, PTEN, AKT, p-AKT, human leukocyte antigen A (HLA-A), α-tubulin, and TRAF6 were produced by Abcam (Cambridge, MA) and then diluted at 1:2,000 for use. After rinsing in PBS, the horseradish peroxidase (HRP)-tagged secondary antibodies were added for 2-h cultivation. Signals of all proteins were detected by enhanced chemiluminescence (ECL) system (Bio-Rad lab, Hercules, CA, USA).
IF
RKO and SW620 cells on coverslips were fixed for 30 min and then probed with the specific antibodies to N-cadherin or E-cadherin overnight. After incubation with fluorescein-conjugated secondary antibodies for 1 h, DAPI solution was added, and fluorescent staining was analyzed under Olympus microscope.
RIP
RIP assay was conducted in RKO and SW620 cells in line with the manual of EZ-Magna RIP RNA Binding Protein Immunoprecipitation Kit (Millipore, Bedford, MA). Cell lysates were prepared for precipitation with the specific antibodies to Ago2 and TRAF6. Normal immunoglobulin G (IgG) antibody was used for control group. After adding magnetic beads, RNAs were enriched and analyzed after 1 h by qRT-PCR.
RNA pull-down
RNA pull-down assay in RKO and SW620 cells was accomplished using the Pierce Magnetic RNA-Protein Pull-Down Kit following the direction (Thermo Fisher Scientific, Waltham, MA). Protein extracts were incubated with magnetic beads and biotinylated RNA probes of miR-4470 and circPTEN. The final pull-downs were analyzed by western blot or qRT-PCR.
Luciferase reporter assay
The luciferase reporter vector pmirGLO covering the miR-4470 target sequences (WT or mutated) within circPTEN or PTEN 3′ UTR fragment were termed circPTEN-WT/Mut or PTEN-WT/Mut and co-transfected with NC mimics, miR-4470 mimics, or miR-4470 mimics+circPTEN in RKO and SW620 cells. 48 h later, Luciferase Reporter Assay System (Promega, Madison, WI) was used for detection of luciferase activity.
coIP
After lysing in IP lysis buffer, cell lysates were collected and cultured with indicated antibodies and normal control anti-IgG overnight at 4°C on a rotator. After mixing with magnetic beads, the mixture was rinsed thrice in IP lysis buffer and then eluted for western blot analysis.
Animal experiments
The 6-week-old male BALB/C nude mice were available from Beijing Vital River Laboratory Animal Technology (Beijing, China). All animal-related experiments were approved by the Animal Research Ethics Committee of the First Affiliated Hospital of Xi’an Jiaotong University. For in vivo xenograft tumor analysis, 1 × 106 transfected RKO cells were injected subcutaneously to nude mice for 28 days, with tumor growth monitored every 4 days. Tumor volume was recorded as 1/2 length × width.2 The mice were sacrificed by cervical decapitation and then tumors were dissected for weight assessment. For in vivo lung metastasis analysis, 1 × 106 transfected RKO cells were used for the tail vein injection in nude mice. 8 weeks later, mice were sacrificed and lungs were collected and fixed for further investigation. Lung metastasis node on lung surface was counted under microscope via H&E staining.
Immunohistochemistry (IHC)
Tumor samples dissected in in vivo xenograft tumor analysis were fixed in 4% paraformaldehyde and then embedded in paraffin. Thereafter, the 4-μm-thick sections were prepared for IHC assay using the specific antibodies against N-cadherin, E-cadherin, p-AKT, AKT, PTEN, and Ki67.
Statistical analyses
Experimental results of more than three independent bio-replications were analyzed by SPSS 18.0 software (SPSS, Chicago, IL) and demonstrated as the mean ± standard deviation (SD). The correlations between genes were analyzed via Pearson correlation analysis. Student’s t test was applied to analyze difference between two groups, although one-way or two-way ANOVA was applied to analyze differences among three or more groups. p < 0.05 was taken to indicate statistical significance.
Acknowledgments
We appreciate the support of the First Affiliated Hospital of Xi’an Jiaotong University.
Author contributions
C.L. accomplished the writing of this manuscript. X.L. was in charge of the statistical analysis.
Declaration of interests
The authors declare no competing interests.
References
- 1.Brenner H., Kloor M., Pox C.P. Colorectal cancer. Lancet. 2014;383:1490–1502. doi: 10.1016/S0140-6736(13)61649-9. [DOI] [PubMed] [Google Scholar]
- 2.Shike M., Winawer S.J., Greenwald P.H., Bloch A., Hill M.J., Swaroop S.V., The WHO Collaborating Centre for the Prevention of Colorectal Cancer Primary prevention of colorectal cancer. Bull. World Health Organ. 1990;68:377–385. [PMC free article] [PubMed] [Google Scholar]
- 3.Eheman C., Henley S.J., Ballard-Barbash R., Jacobs E.J., Schymura M.J., Noone A.M., Pan L., Anderson R.N., Fulton J.E., Kohler B.A., et al. Annual Report to the Nation on the status of cancer, 1975-2008, featuring cancers associated with excess weight and lack of sufficient physical activity. Cancer. 2012;118:2338–2366. doi: 10.1002/cncr.27514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.He D., Ma L., Feng R., Zhang L., Jiang Y., Zhang Y., Liu G. Analyzing large-scale samples highlights significant association between rs10411210 polymorphism and colorectal cancer. Biomed. Pharmacother. 2015;74:164–168. doi: 10.1016/j.biopha.2015.08.023. [DOI] [PubMed] [Google Scholar]
- 5.Lévy J., Cacheux W., Bara M.A., L’Hermitte A., Lepage P., Fraudeau M., Trentesaux C., Lemarchand J., Durand A., Crain A.M., et al. Intestinal inhibition of Atg7 prevents tumour initiation through a microbiome-influenced immune response and suppresses tumour growth. Nat. Cell Biol. 2015;17:1062–1073. doi: 10.1038/ncb3206. [DOI] [PubMed] [Google Scholar]
- 6.Bray C., Bell L.N., Liang H., Collins D., Yale S.H. Colorectal cancer screening. WMJ. 2017;116:27–33. [PubMed] [Google Scholar]
- 7.Sadanandam A., Lyssiotis C.A., Homicsko K., Collisson E.A., Gibb W.J., Wullschleger S., Ostos L.C., Lannon W.A., Grotzinger C., Del Rio M., et al. A colorectal cancer classification system that associates cellular phenotype and responses to therapy. Nat. Med. 2013;19:619–625. doi: 10.1038/nm.3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nishihara R., Wu K., Lochhead P., Morikawa T., Liao X., Qian Z.R., Inamura K., Kim S.A., Kuchiba A., Yamauchi M., et al. Long-term colorectal-cancer incidence and mortality after lower endoscopy. N. Engl. J. Med. 2013;369:1095–1105. doi: 10.1056/NEJMoa1301969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sasaki H., Miura K., Horii A., Kaneko N., Fujibuchi W., Kiseleva L., Gu Z., Murata Y., Karasawa H., Mizoi T., et al. Orthotopic implantation mouse model and cDNA microarray analysis indicates several genes potentially involved in lymph node metastasis of colorectal cancer. Cancer Sci. 2008;99:711–719. doi: 10.1111/j.1349-7006.2008.00725.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Siegel R.L., Miller K.D., Jemal A. Cancer statistics, 2016. CA Cancer J. Clin. 2016;66:7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 11.Chaffer C.L., Weinberg R.A. A perspective on cancer cell metastasis. Science. 2011;331:1559–1564. doi: 10.1126/science.1203543. [DOI] [PubMed] [Google Scholar]
- 12.Depeille P., Henricks L.M., van de Ven R.A., Lemmens E., Wang C.Y., Matli M., Werb Z., Haigis K.M., Donner D., Warren R., Roose J.P. RasGRP1 opposes proliferative EGFR-SOS1-Ras signals and restricts intestinal epithelial cell growth. Nat. Cell Biol. 2015;17:804–815. doi: 10.1038/ncb3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Markowitz S.D., Bertagnolli M.M. Molecular origins of cancer: molecular basis of colorectal cancer. N. Engl. J. Med. 2009;361:2449–2460. doi: 10.1056/NEJMra0804588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yu J., Han Z., Sun Z., Wang Y., Zheng M., Song C. LncRNA SLCO4A1-AS1 facilitates growth and metastasis of colorectal cancer through β-catenin-dependent Wnt pathway. J. Exp. Clin. Cancer Res. 2018;37:222. doi: 10.1186/s13046-018-0896-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Djebali S., Davis C.A., Merkel A., Dobin A., Lassmann T., Mortazavi A., Tanzer A., Lagarde J., Lin W., Schlesinger F., et al. Landscape of transcription in human cells. Nature. 2012;489:101–108. doi: 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Esteller M. Non-coding RNAs in human disease. Nat. Rev. Genet. 2011;12:861–874. doi: 10.1038/nrg3074. [DOI] [PubMed] [Google Scholar]
- 17.Jeck W.R., Sorrentino J.A., Wang K., Slevin M.K., Burd C.E., Liu J., Marzluff W.F., Sharpless N.E. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2013;19:141–157. doi: 10.1261/rna.035667.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen L.L., Yang L. Regulation of circRNA biogenesis. RNA Biol. 2015;12:381–388. doi: 10.1080/15476286.2015.1020271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Meng S., Zhou H., Feng Z., Xu Z., Tang Y., Li P., Wu M. CircRNA: functions and properties of a novel potential biomarker for cancer. Mol. Cancer. 2017;16:94. doi: 10.1186/s12943-017-0663-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang F., Wang J., Cao X., Xu L., Chen L. Hsa_circ_0014717 is downregulated in colorectal cancer and inhibits tumor growth by promoting p16 expression. Biomed. Pharmacother. 2018;98:775–782. doi: 10.1016/j.biopha.2018.01.015. [DOI] [PubMed] [Google Scholar]
- 21.Ding L., Zhao Y., Dang S., Wang Y., Li X., Yu X., Li Z., Wei J., Liu M., Li G. Circular RNA circ-DONSON facilitates gastric cancer growth and invasion via NURF complex dependent activation of transcription factor SOX4. Mol. Cancer. 2019;18:45. doi: 10.1186/s12943-019-1006-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liang H.F., Zhang X.Z., Liu B.G., Jia G.T., Li W.L. Circular RNA circ-ABCB10 promotes breast cancer proliferation and progression through sponging miR-1271. Am. J. Cancer Res. 2017;7:1566–1576. [PMC free article] [PubMed] [Google Scholar]
- 23.Nan H., Han L., Ma J., Yang C., Su R., He J. STX3 represses the stability of the tumor suppressor PTEN to activate the PI3K-Akt-mTOR signaling and promotes the growth of breast cancer cells. Biochim. Biophys. Acta Mol. Basis Dis. 2018;1864(5 Pt A):1684–1692. doi: 10.1016/j.bbadis.2018.01.031. [DOI] [PubMed] [Google Scholar]
- 24.Wu Y., Song Y., Xiong Y., Wang X., Xu K., Han B., Bai Y., Li L., Zhang Y., Zhou L. MicroRNA-21 (Mir-21) promotes cell growth and invasion by repressing tumor suppressor PTEN in colorectal cancer. Cell. Physiol. Biochem. 2017;43:945–958. doi: 10.1159/000481648. [DOI] [PubMed] [Google Scholar]
- 25.Feng Y., Zou W., Hu C., Li G., Zhou S., He Y., Ma F., Deng C., Sun L. Modulation of CASC2/miR-21/PTEN pathway sensitizes cervical cancer to cisplatin. Arch. Biochem. Biophys. 2017;623-624:20–30. doi: 10.1016/j.abb.2017.05.001. [DOI] [PubMed] [Google Scholar]
- 26.Zhang X., Li X., Tan F., Yu N., Pei H. STAT1 inhibits MiR-181a expression to suppress colorectal cancer cell proliferation through PTEN/Akt. J. Cell. Biochem. 2017;118:3435–3443. doi: 10.1002/jcb.26000. [DOI] [PubMed] [Google Scholar]
- 27.Lin Y., Chen Q., Liu Q.X., Zhou D., Lu X., Deng X.F., Yang H., Zheng H., Qiu Y. High expression of DJ-1 promotes growth and invasion via the PTEN-AKT pathway and predicts a poor prognosis in colorectal cancer. Cancer Med. 2018;7:809–819. doi: 10.1002/cam4.1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mukhopadhyay D., Riezman H. Proteasome-independent functions of ubiquitin in endocytosis and signaling. Science. 2007;315:201–205. doi: 10.1126/science.1127085. [DOI] [PubMed] [Google Scholar]
- 29.Pickart C.M. Mechanisms underlying ubiquitination. Annu. Rev. Biochem. 2001;70:503–533. doi: 10.1146/annurev.biochem.70.1.503. [DOI] [PubMed] [Google Scholar]
- 30.Yang W.L., Wang J., Chan C.H., Lee S.W., Campos A.D., Lamothe B., Hur L., Grabiner B.C., Lin X., Darnay B.G., Lin H.K. The E3 ligase TRAF6 regulates Akt ubiquitination and activation. Science. 2009;325:1134–1138. doi: 10.1126/science.1175065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Torre L.A., Bray F., Siegel R.L., Ferlay J., Lortet-Tieulent J., Jemal A. Global cancer statistics, 2012. CA Cancer J. Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 32.Li X.N., Wang Z.J., Ye C.X., Zhao B.C., Li Z.L., Yang Y. RNA sequencing reveals the expression profiles of circRNA and indicates that circDDX17 acts as a tumor suppressor in colorectal cancer. J. Exp. Clin. Cancer Res. 2018;37:325. doi: 10.1186/s13046-018-1006-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu T., Yu T., Hu H., He K. Knockdown of the long non-coding RNA HOTTIP inhibits colorectal cancer cell proliferation and migration and induces apoptosis by targeting SGK1. Biomed. Pharmacother. 2018;98:286–296. doi: 10.1016/j.biopha.2017.12.064. [DOI] [PubMed] [Google Scholar]
- 34.Liang Q., Tang C., Tang M., Zhang Q., Gao Y., Ge Z. TRIM47 is up-regulated in colorectal cancer, promoting ubiquitination and degradation of SMAD4. J. Exp. Clin. Cancer Res. 2019;38:159. doi: 10.1186/s13046-019-1143-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Peralta-Zaragoza O., Deas J., Meneses-Acosta A., De la O-Gómez F., Fernández-Tilapa G., Gómez-Cerón C., Benítez-Boijseauneau O., Burguete-García A., Torres-Poveda K., Bermúdez-Morales V.H., et al. Relevance of miR-21 in regulation of tumor suppressor gene PTEN in human cervical cancer cells. BMC Cancer. 2016;16:215. doi: 10.1186/s12885-016-2231-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Egawa H., Jingushi K., Hirono T., Ueda Y., Kitae K., Nakata W., Fujita K., Uemura M., Nonomura N., Tsujikawa K. The miR-130 family promotes cell migration and invasion in bladder cancer through FAK and Akt phosphorylation by regulating PTEN. Sci. Rep. 2016;6:20574. doi: 10.1038/srep20574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yang L., Wang J., Fan Y., Yu K., Jiao B., Su X. Hsa_circ_0046264 up-regulated BRCA2 to suppress lung cancer through targeting hsa-miR-1245. Respir. Res. 2018;19:115. doi: 10.1186/s12931-018-0819-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liang M., Huang G., Liu Z., Wang Q., Yu Z., Liu Z., Lin H., Li M., Zhou X., Zheng Y. Elevated levels of hsa_circ_006100 in gastric cancer promote cell growth and metastasis via miR-195/GPRC5A signalling. Cell Prolif. 2019;52:e12661. doi: 10.1111/cpr.12661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li H., Yao G., Feng B., Lu X., Fan Y. Circ_0056618 and CXCR4 act as competing endogenous in gastric cancer by regulating miR-206. J. Cell. Biochem. 2018;119:9543–9551. doi: 10.1002/jcb.27271. [DOI] [PubMed] [Google Scholar]
- 40.Yong W., Zhuoqi X., Baocheng W., Dongsheng Z., Chuan Z., Yueming S. Hsa_circ_0071589 promotes carcinogenesis via the miR-600/EZH2 axis in colorectal cancer. Biomed. Pharmacother. 2018;102:1188–1194. doi: 10.1016/j.biopha.2018.03.085. [DOI] [PubMed] [Google Scholar]
- 41.Zheng C.-H., Wang J.-B., Lin M.-Q., Zhang P.-Y., Liu L.-C., Lin J.-X., Lu J., Chen Q.-Y., Cao L.-L., Lin M., et al. CDK5RAP3 suppresses Wnt/β-catenin signaling by inhibiting AKT phosphorylation in gastric cancer. J. Exp. Clin. Cancer Res. 2018;37:59. doi: 10.1186/s13046-018-0716-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zheng J., Dai X., Chen H., Fang C., Chen J., Sun L. Down-regulation of LHPP in cervical cancer influences cell proliferation, metastasis and apoptosis by modulating AKT. Biochem. Biophys. Res. Commun. 2018;503:1108–1114. doi: 10.1016/j.bbrc.2018.06.127. [DOI] [PubMed] [Google Scholar]
- 43.Wang X., Lyu J., Ji A., Zhang Q., Liao G. Jarid2 enhances the progression of bladder cancer through regulating PTEN/AKT signaling. Life Sci. 2019;230:162–168. doi: 10.1016/j.lfs.2019.05.053. [DOI] [PubMed] [Google Scholar]