Abstract
Acinetobacter baumannii has become a major challenge to clinicians worldwide due to its high epidemic potential and acquisition of antimicrobial resistance. This work aimed at investigating antimicrobial resistance determinants and their context in four extensively drug-resistant (XDR) NDM-producing A. baumannii clinical isolates collected between July and October 2020 from Kasr Al-Ainy Hospital, Cairo, Egypt. A total of 20 A. baumannii were collected and screened for acquired carbapenemases (blaNDM, blaVIM and blaIMP) using PCR. Four NDM producer A. baumannii isolates were identified and selected for whole-genome sequencing, in silico multilocus sequence typing, and resistome analysis. Antimicrobial susceptibility profiles were determined using disk diffusion and broth microdilution tests. All blaNDM-positive A. baumannii isolates were XDR. Three isolates belonged to high-risk international clones (IC), namely, IC2 corresponding to ST570Pas/1701Oxf (M20) and IC9 corresponding to ST85Pas/ST1089Oxf (M02 and M11). For the first time, we report blaNDM-1 gene on the chromosome of an A. baumannii strain that belongs to sequence type ST164Pas/ST1418Oxf. Together with AphA6, blaNDM-1 was bracketed by two copies of ISAba14 in ST85Pas isolates possibly facilitating co-transfer of amikacin and carbapenem resistance. A novel blaADC allele (blaADC-257) with an upstream ISAba1 element was identified in M19 (ST/CC164Pas and ST1418Oxf/CC234Oxf). blaADC genes harbored by M02 and M11 were uniquely interrupted by IS1008. Tn2006-associated blaOXA-23 was carried by M20. blaOXA-94 genes were preceded by ISAba1 element in M02 and M11. AbGRI3 was carried by M20 hosting the resistance genes aph(3`)-Ia, aac(6`)-Ib`, catB8, ant(3``)-Ia, sul1, armA, msr(E), and mph(E). Nonsynonymous mutations were identified in the quinolone resistance determining regions (gyrA and parC) of all isolates. Resistance to colistin in M19 was accompanied by missense mutations in lpxACD and pmrABC genes. The current study provided an insight into the genomic background of XDR phenotype in A. baumannii recovered from patients in Egypt. WGS revealed strong association between resistance genes and diverse mobile genetic elements with novel insertion sites and genetic organizations.
Keywords: healthcare-associated infections, Acinetobacter baumannii, extensive drug resistance, bla NDM , whole-genome sequencing, multilocus sequence typing
Introduction
Hospital-associated infections (HAIs) present an elevated healthcare burden in both developed and developing countries (Chng et al., 2020). Acinetobacter baumannii is implicated in a considerable fraction of difficult to treat HAIs (Ayobami et al., 2019). Antimicrobial resistance, biofilm formation, and resistance to desiccation are among the competencies contributing to the environmental persistence and the epidemic potential of this species (Antunes et al., 2014). In addition to its intrinsic resistance to multiple antimicrobial classes, effective therapeutic options are being gradually depleted by the extraordinary ability of A. baumannii to acquire and upregulate resistance genes (Di Nocera et al., 2011). The emergence of multidrug-resistant (MDR) and extensively drug-resistant (XDR) A. baumannii has been increasing worldwide as well as in Egypt (Tal-Jasper et al., 2016; Elsayed et al., 2020). This forced the WHO to declare carbapenem-resistant A. baumannii as a category 1 (critical) priority pathogen for which novel therapeutic antimicrobials are urgently required (Tacconelli et al., 2018).
The New Delhi Metallo-β-lactamase-1 (NDM-1) is a carbapenemase that has been frequently linked to the XDR phenotype owing to its association with mobile elements loaded with other resistance genes (Wailan and Paterson, 2014). A. baumannii has been long recognized as an intermediate reservoir for blaNDM-1 genes in which the harboring transposon (Tn125) was built and subsequently transmitted to other Gram-negative species (Toleman et al., 2012; Bontron et al., 2016).
Genome studies contribute significantly to better comprehend the molecular basis and evolution dynamics of antimicrobial resistance in nosocomial infectious pathogens (Hendriksen et al., 2019). Despite the large number of studies from Egypt that have discussed the epidemiology of healthcare-associated A. baumannii (Al-Hassan et al., 2019; Benmahmod et al., 2019; Wasfi et al., 2021), few studies have explored the whole-genome sequence of those circulating in Egyptian hospitals (Fam et al., 2020).
The objective of the current study was to explore the genomic features of four XDR blaNDM-positive A. baumannii clinical isolates recovered from hospitalized patients at a large tertiary hospital in Egypt by whole-genome sequencing (WGS).
Materials and Methods
Bacterial Strains and Antimicrobial Susceptibility Testing
A total of 54 nonduplicate nonfermentative Gram-negative bacterial isolates were collected from Kasr Al-Ainy University Hospital, Cairo, Egypt, between July and October 2020. Of these, 20 isolates were identified as A. baumannii using VITEK 2 (bioMérieux, Marcy l’Etoile, France). The identity of A. baumannii isolates was further confirmed using PCR amplification of the blaOXA-51-like genes (Turton et al., 2006). Bacterial isolates were recovered at the clinical pathology laboratory as part of routine clinical care of hospitalized patients. Antimicrobial resistance profiles were identified using disk diffusion test according to the recommendations of the CLSI (2018). Tigecycline susceptibility test results were interpreted according to susceptibility breakpoints recommended by the EUCAST (2021) v11.0 for Enterobacterales. For disk diffusion test, 14 antimicrobial disks (Oxoid, United Kingdom) were used including the following: ampicillin (10μg), amoxicillin/clavulanic acid (20/10μg), piperacillin/tazobactam (10/100μg), ceftriaxone (30μg), cefoxitin (30μg), cefepime (30μg), cefotaxime (30μg), levofloxacin (5μg), imipenem (10μg), meropenem (10μg), amikacin (30μg), tigecycline (15μg), and trimethoprim/sulfamethoxazole (1.25/23.75μg). The broth microdilution method was used to detect the minimum inhibitory concentration (MIC) of colistin according to CLSI guidelines. Amplification of MBL genes (blaNDM, blaVIM, and blaIMP) using polymerase chain reaction (PCR) was done for all A. baumannii isolates as previously described (Ghazawi et al., 2012). Individual A. baumannii isolates (M02, M11, M19, and M20) that harbored blaNDM were selected for WGS analysis.
Whole-Genome Sequencing, Assembly, and Annotation
DNA was extracted from all blaNDM-positive A. baumannii isolates using QIAGEN DNA purification kit (Qiagen, Valencia, CA). This was further manipulated by Nextera DNA Sample Preparation kit (Nextera, United States) for preparation of the DNA library according to the manufacturer’s recommended protocol. Sequencing was performed using the paired end 2×150bp reads sequencing technology on an Illumina MiSeq platform (Illumina Inc., San Diego, CA, United States). Reads quality was assessed using FastQC v0.11.9 (Brown et al., 2017) before trimming with Trimmomatic v0.35 to cut away remaining adaptors and low-quality reads (Bolger et al., 2014). Trimmed reads were de novo assembled using SPAdes 3.14.1 (Bankevich et al., 2012) with default parameters. The quality of genomes assembly was evaluated using QUAST v5.0.2 (Gurevich et al., 2013). Functional annotations of the draft genomes were generated during submission to the National Center for Biotechnology Information (NCBI) genome database using the NCBI Prokaryotic Genome Annotation Pipeline (PGAP; Tatusova et al., 2016). Plasmid sequences were identified using plasmidSPAdes (Antipov et al., 2016) and Unicycler (Wick et al., 2017) for raw reads assembly and Bandage (Wick et al., 2015) for visualization of circular contigs.
Multilocus Sequence Typing
Whole-genome sequencing data were used for in silico analysis of multilocus sequence types (MLSTs) of the isolates harboring blaNDM gene based on both Pasteur and Oxford schemes. Allele numbers and sequence types (STs) were assigned using PubMLST server.1 The global optimal eBURST (goeBURST) algorithm executed by PHYLOViZ V2.0 (Francisco et al., 2012) was used for constructing a complete minimum spanning tree (MST) of the sequence types of the blaNDM-positive isolates together with other STs in MLST database (accessed on March 10, 2021), and clonal complexes (CCs) were assigned accordingly.
Phylogeny Analysis
A single nucleotide polymorphism (SNP)-based phylogeny analysis of the four blaNDM-positive isolates was performed using the CSI-Phylogeny tool hosted by the CGE server (Center for Genomic Epidemiology, Lyngby, Denmark) available at http://www.genomicepidemiology.org/ (Kaas et al., 2014). The draft genomes of the isolates were compared to complete genomes of A. baumannii strains carrying blaNDM-1 gene and some A. baumannii strains that belong to high-risk international clones retrieved from the GenBank database (accessed in: October 12, 2021). In addition, draft genomes of A. baumannii strains that belong to ST1418Oxf and ST164Pas were also downloaded from PubMLST genome collection2 and included in the analysis. A. baumannii ATCC 17978 was used as a reference genome. The phylogenetic tree was visualized and edited using the interactive tree of life v3 software (Letunic and Bork, 2016) available at: https://itol.embl.de/.
Analysis of Antimicrobial Resistance Determinants and Insertion Sequences
Acquired antimicrobial resistance genes were identified using the ResFinder 4.1 webtool on the CGE server (Center for Genomic Epidemiology, Lyngby, Denmark3; Bortolaia et al., 2020) using raw reads as an input. Assembled contigs were further analyzed using the Comprehensive Antibiotic Resistance Database server4 (Alcock et al., 2020) with coverage and identity thresholds of 80 and 95%, respectively. Genomic resistance islands were predicted using IslandViewer4 webtool5 (Bertelli et al., 2017). Gene mutations relevant to antimicrobial resistance were manually analyzed by extracting the genes of interest from genome assemblies and blasting against respective genes of the reference strain A. baumannii ATCC 19606 (Accession Number: CP045110.1). This involved analysis of gyrA and parC regions whose mutations are associated with quinolones resistance. In addition, other genes reported to affect the susceptibility of A. baumannii to colistin including those involved in lipid A biosynthesis pathway (lpxA, lpxC, and lpxD) and pmrABC operon were also analyzed in case of colistin nonsusceptibility. Insertion sequences (ISs) were identified using the online tool ISfinder6 (Siguier et al., 2006).
Characterization of the Genetic Context of Resistance Genes
Contigs containing resistance genes were extracted from the assemblies. Genetic features were obtained from PGAP annotation data. Unannotated regions were manually reannotated after blasting against the GenBank nucleotide collection. Genetic environments of resistance gene cassettes located on more than one contig were identified by mapping of raw reads to the best hits of the contigs’ blast analyses using BWA (Li and Durbin, 2009). Consensus sequences were obtained using SAMtools and bcftools v0.1.10 (Li, 2011). Finally, annotated genetic environments of resistance genes were visualized using SnapGene viewer v5.1.3.1 (from Insightful Science; available at snapgene.com) and compared to reference sequences using Easyfig v2.2.5 (Sullivan et al., 2011).
Nucleotide Sequence Accession Numbers
Raw reads obtained by WGS of the blaNDM-positive isolates were submitted to the Sequencing Read Archive7 of the NCBI. Draft genomes were submitted to the NCBI Genome database.8 Together with their BioSamples, they were submitted under the BioProject number PRJNA690827. Raw reads and draft genomes accession numbers are shown in Supplementary Table 1. The nucleotide sequence of the novel blaADC–257 gene was deposited in the NCBI GenBank database under the accession number (MZ224611.1).
Results
During the study period, a total of 20 A. baumannii isolates were recovered from 20 different hospitalized patients with age ranging between newborn (5days) and 65years old. Of these, 12 (60%) were females and 8 (40%) were males. More than half of the patients were hospitalized in intensive care units. Specimens were collected from different clinical sites (Table 1). Results are shown for the four blaNDM-positive A. baumannii isolates.
Table 1.
Clinical data of the four NDM-producing Acinetobacter baumannii.
| Isolate | Site | Age | Sex | Diagnosis | Date of isolation | Hospital ward |
|---|---|---|---|---|---|---|
| M02 | Wound | 28years | Female | Subovarian abscess removal | 2020-07-10 | ICU |
| M11 | Pleural | 20days | Female | Pneumonia | 2020-07-15 | NICU |
| M19 | Blood | 20years | Female | Fever of unknown origin | 2020-10-20 | ICU |
| M20 | Blood | 65years | Male | Splenectomy and feverish | 2020-08-2 | ICU |
ICU, intensive care unit; NICU, neonatal intensive care unit.
Acinetobacter baumannii Strains Harboring blaNDM Gene
To determine the prevalence of acquired carbapenemases in the recovered A. baumannii isolates, the presence of blaNDM, blaVIM, and blaIMP genes were assessed using PCR assay. Neither VIM- nor IMP-type carbapenemase-coding genes could be identified in the isolates. Out of 20 A. baumannii isolates, four (20%) showed amplification of 371bp PCR product corresponding to blaNDM gene.
Genome Assembly
Whole-genome sequencing of the blaNDM-positive isolates yielded total assembly lengths ranging from 3,773,846bp to 3,919,334bp with a GC content ranging from 39.19 to 39.55%. The mean number of contigs was 633. The number of coding sequences predicted by PGAP annotation of the assembled contigs ranged from 3,761 to 3,996. Post-assembly and annotation metrics of the blaNDM-positive isolates are shown in Supplementary Table 2.
MLST and Phylogenetic Analysis
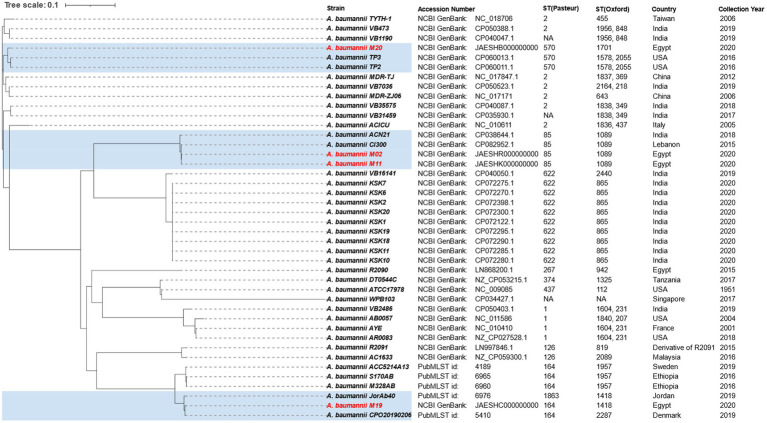
In silico MLST analysis of the blaNDM-positive isolates and goeBURST analysis of their STs together with ST data from MLST database revealed that isolate M20 (ST570Pas/1701Oxf) belongs to clonal complex (CC2Pas/546Oxf) representing international (IC) 2. Two isolates M02 and M11 had the same sequence type (ST85Pas/ST1089Oxf) that was found to belong to CC464Pas/CC1078Oxf classified within IC9. The allele profile of M19 matched ST/CC164 and ST1418/CC234, according to Pasteur and Oxford schemes, respectively. MST diagram of blaNDM-positive isolates STs together with other STs in MLST database (Pasteur scheme) is shown in Supplementary Figure 1. A SNP-based phylogenetic tree depicting the genetic relatedness of our blaNDM-positive isolates to other A. baumannii strains is shown in Figure 1.
Figure 1.
A single nucleotide polymorphism-based phylogenetic tree depicting the genetic relatedness of blaNDM-1-positive isolates sequenced in the current study to other Acinetobacter baumannii strains. A. baumannii strains sequenced in the current study together with their genetically related strains are highlighted by blue color.
Antimicrobial Susceptibility Testing and Resistance Determinants
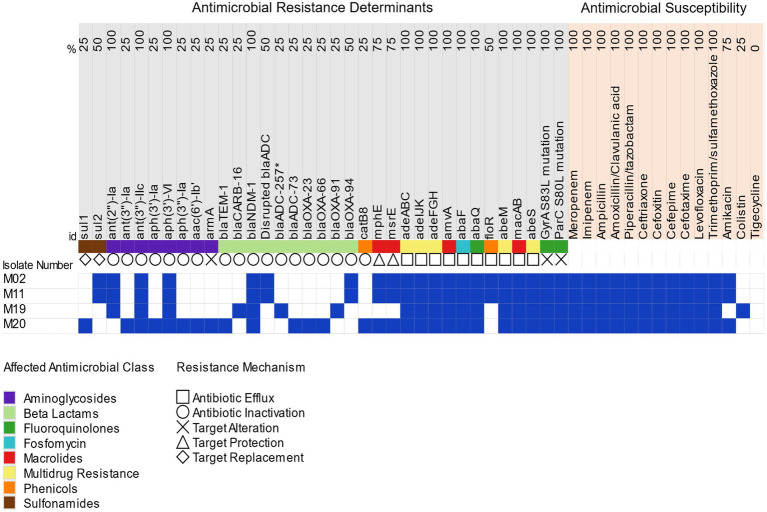
Antimicrobial susceptibility testing revealed that all isolates were extensively drug resistant (XDR) with retained susceptibility to only two antimicrobial classes (Magiorakos et al., 2012; Figure 2). All isolates were susceptible to tigecycline. MIC values of ≤0.125, 0.25, ≥128, and 0.5μg/ml were determined for colistin in M02, M11, M19, and M20, respectively. Resistance to colistin was shown by one isolate (M19) that also retained susceptibility to amikacin.
Figure 2.
Antimicrobial resistance profiles and distribution of antimicrobial resistance determinants in the blaNDM-positive isolates. Blue colors denote antimicrobial resistance and harbored antimicrobial determinants, while susceptibility to antimicrobials and lack of resistance determinants are denoted by white colors; blaADC-257*, novel blaADC allele identified in M19.
Investigating the genetic background of the XDR phenotype using WGS revealed that the isolates carried multiple acquired antimicrobial resistance determinants besides the intrinsic resistance genes (Table 2). Genes conferring resistance to β-lactams included class A β-lactamases (blaCARB-16 and blaTEM-1), one metallo-β-lactamase (blaNDM-1), class C β-lactamases (blaADC-73 and blaADC-257), and carbapenem-hydrolyzing Ambler class D β-lactamases, (blaOXA-23, blaOXA-66, blaOXA-91, and blaOXA-94). blaADC-257 is a novel allele of blaADC-52 (GenBank accession: WP_001211232.1) detected in isolate M19 with two amino acid substitutions (R2Q and G24D). Resistance to other antimicrobial agents was conferred by ant(2″)-Ia, ant(3″)-Ia, ant(3″)-IIc, aph(3′)-Ia, aph(3″)-Ia, aph(3′)-VI, aac(6′)-Ib’, and ArmA (aminoglycoside resistance), mphE and msrE (macrolide resistance), catB8 (chloramphenicol resistance), and sul1 and sul2 (sulfonamide resistance).
Table 2.
STs and antimicrobial resistance genes carried by the four blaNDM-positive isolates.
| Isolate number | MLST | Intrinsic blaOXA gene | Antimicrobial resistance genes | Efflux pumps genes | QRDRb | ||||
|---|---|---|---|---|---|---|---|---|---|
| Pasteur | Oxford | gyrA | parC | ||||||
| ST | CC | ST | CC | ||||||
| M02 | 85 | 464 | 1089 | 1078 | bla OXA-94 | aph(3′)-VI, blaNDM-1, blaADC (disrupted by IS6), mphE, msrE, sul2, ant(2″)-Ia, ant(3″)-IIc | adeABC, adeIJK, adeFGH, abeM, amvA, abeS, abaF, abaQ, floR, macAB | S83L | S80L |
| M11 | 85 | 464 | 1089 | 1078 | bla OXA-94 | aph(3′)-VI, blaNDM-1, blaADC (disrupted by IS6), mphE, msrE, sul2, ant(2″)-Ia, ant(3″)-IIc | adeABC, adeIJK, adeFGH, abeM, amvA, abeS, abaF, abaQ, floR, macAB | S83L | S80L |
| M19 | 164 | 164 | 1418 | 234 | bla OXA-91 | aph(3′)-VI, blaNDM-1, blaADC-257a, blaCARB-16, ant(2″)-Ia, ant(3″)-IIc | adeABC, adeIJK, adeFGH, abeM, amvA, abeS, abaF, abaQ, macAB | S83L | S80L |
| M20 | 570 | 2 | 1701 | 546 | bla OXA-66 | blaOXA-23, aph(3′)-VI, blaNDM-1, blaADC-73, aph(3′)-Ia, blaTEM-1, aph(3″)-I, aac(6′)-Ib’, catB8, ant(3″)-Ia, sul1, ArmA, msr(E), mph(E), ant(3″)-IIc | adeABC, adeIJK, adeFGH, abeM, amvA, abeS, abaF, abaQ, macAB | S83L | S80L |
Novel ADC allele.
QRDR, quinolone resistance determining regions.
Analysis of the nucleotide sequence of pmrABC and IpxACD genes of the colistin-resistant isolate (M19) and comparison to their wild-type alleles in A. baumannii ATCC 19606 revealed the existence of multiple mutations. These included point mutations in the histidine kinase gene pmrB (H89L) and mutations in pmrC (I42V, I212V, R323K, A354S, and V470I). Only silent mutations were identified in pmrA. Within IpxACD genes, point mutations were identified in IpxA (Y131H and Y231H), IpxC (C120R, N287D, and K130T), and lpxD (V631 and E117K). Further analysis of genomic mutations revealed that levofloxacin resistance in all isolates was promoted by amino acid substitutions in quinolone resistance determining regions (QRDRs) of both DNA gyrase (S83L) and topoisomerase (S80L) enzymes.
Multidrug efflux pumps, including members of the major facilitator superfamily (MFS) and resistance-nodulation-division (RND) family and additional multidrug efflux pumps, were identified in the isolates. Susceptibility profiles of the blaNDM-positive isolates and resistance determinants carried by each are shown in Figure 2.
Insertion Sequences
Investigating the insertion sequences using ISfinder revealed the existence of at least 24 IS elements distributed throughout the genomes. Most of them originated from A. baumannii and other Acinetobacter species. Only four IS elements were acquired from other bacterial species, such as Escherichia coli, Vibrio salmonicida, and Salmonella panama. Six types of ISs were conserved in all isolates, including ISAba1, ISAba8, ISAba10, ISAba14, ISAba33, and ISAba125. The diversity of IS content of the four genomes and their microbial origins are depicted in Figure 3.
Figure 3.
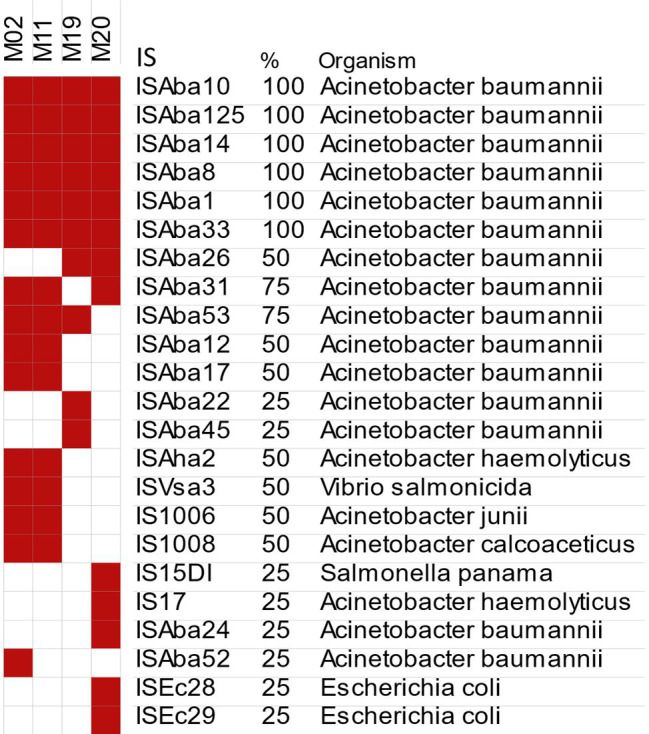
Genome-wide distribution of different IS elements in the blaNDM-positive isolates predicted by ISfinder. Red and white colors denote the presence and absence of each IS element, respectively.
Genetic Context of Resistance Genes
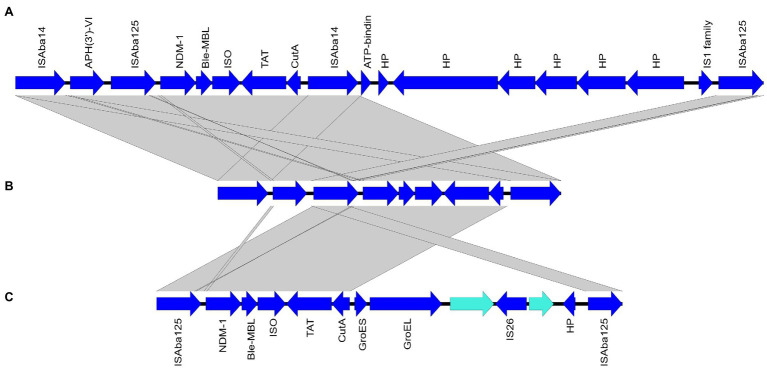
Whole-genome sequencing results revealed that blaNDM-1 genes were carried on the chromosomes of all sequenced isolates. Analysis of the immediate genetic environment of the blaNDM-1 gene revealed the existence of ISAba14 upstream to the divalent cation tolerance protein (CutA)-coding gene in the isolates M2, M11, and M20 in addition to the ISAba125 element upstream to blaNDM-1. This genetic organization is similar to that of Tn125-like transposon previously reported by Bonnin et al. (2013). BLAST analysis of the contigs harboring blaNDM-1 showed highest similarity to the chromosome of A. baumannii strain ACN21 (GenBank accession: CP038644.1; Vijayakumar et al., 2020). Using this genome as a reference for Islandviewer analysis showed an upstream amikacin resistance gene (AphA6) and another copy of ISAba14 in ST85Pas isolates (M02 and M11). This was further confirmed by mapping raw sequencing reads of such isolates against a larger segment of A. baumannii strain ACN21 chromosome. This genetic organization was shown in Figure 4 together with a comparative genetic analysis of Tn125-like transposon and Tn125 (GenBank accession: KF702386.1). Similar analysis failed to localize AphA6-ISAba14 in the upstream region of the blaNDM-1-harboring transposon in M20. Different genetic environment was noted for blaNDM-1 carried by M19 in which the upstream ISAba125 element was immediately preceded by IS1 family transposase in an organization with no similarity in the NCBI nucleotide database. Furthermore, interruption of the right hand of the transposon by ISAba14 could not be concluded.
Figure 4.
Graphical representation of blaNDM-1 genetic environment in isolates M02 and M11 (B) compared to the closest match sequence Tn125-like transposon of A. baumannii strain ACN21 (GenBank accession: CP038644.1) (A) and Tn125 (GenBank accession: KF702386.1) (C). ORFs orientation is indicated by arrows. Grey bands between panels indicate more than 98% sequence similarity. Genes are labelled by their protein products; NDM-1, New Delhi metal-beta-lactamase enzyme; ble-MBL, bleomycin resistance protein; ISO, phosphoribosylanthranilate isomerase; TAT, twin-arginine translocation pathway signal sequence protein; CutA, divalent cation tolerance protein; HP, hypothetical protein; GroES, co-chaperonin protein; GroEL, type I chaperonin.
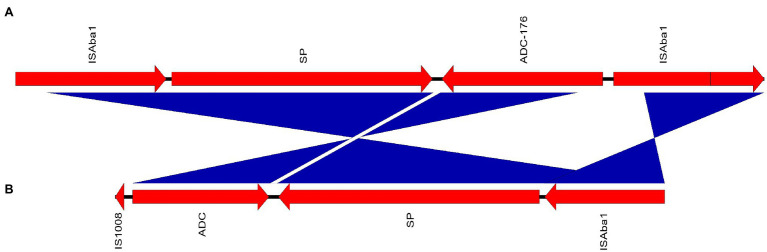
Analysis of the intrinsic blaADC genes and their association with upstream insertion elements revealed that the novel allele blaADC-257 carried by M19 was preceded by ISAba1. In the isolates M02 and M11, blaADC genes were interrupted by IS1008 family transposase leading to missing N-terminus. Hence, the Acinetobacter-derived cephalosporinase variant could not be identified. The IS1008-interrupted gene had no similarity in the NCBI nucleotide database. The context of the interrupted gene compared to the closest match sequence (A. baumannii strain ACN21 chromosome) is shown in Figure 5. Similarly, blaADC-73 with missing N-terminus was harbored by M20, while the disrupting sequence could not be identified.
Figure 5.
Gene maps showing the genetic environment of IS1008-interrupted blaADC carried by M11 (A) compared to A. baumannii ACN21 chromosome (GenBank accession: CP038644.1) (B). ORFs orientation is indicated by arrows. Blue bands between panels indicate inverted sequences with more than 98% sequence similarity. Genes are labelled by their protein products; ADC, blaADC gene disrupted by IS6; SP, signal peptide.
blaOXA-23 carried by M20 was found to be embedded within Tn2006 in which it was bracketed by ISAba1, while blaOXA-94 in M02 and M11 was preceded by ISAba1 element in a reverse orientation. On the other hand, blaOXA-91 and blaOXA-66 carried by the isolates M19 and M20 had no upstream insertion sequences.
Using A. baumannii strain MS14413 chromosome (GenBank: CP054302.1) as a reference for Islandviewer analysis, a 20,844bp genomic resistance island that showed 99.62% identity to A. baumannii genomic resistance island 3 (AbGRI3, accession number: KX011025.2) was identified in M20. The resistance island hosted the resistance genes: aph(3`)-Ia, aac(6`)-Ib`, catB8, ant(3``)-Ia, sul1, ArmA, msr(E), and mph(E) bracketed by IS26 family transposases.
In all isolates carrying ant(2``)-Ia (aadB), the gene was found on pRAY plasmid (6,076bp) derivatives. A plasmid sequence identical to pRAY*-v1 (GenBank accession: JF343536) was identified in M19, while those carried by M02 and M11 showed 100% identity to pRay* (GenBank accession: JQ904627). No other resistance plasmids were identified in our isolates.
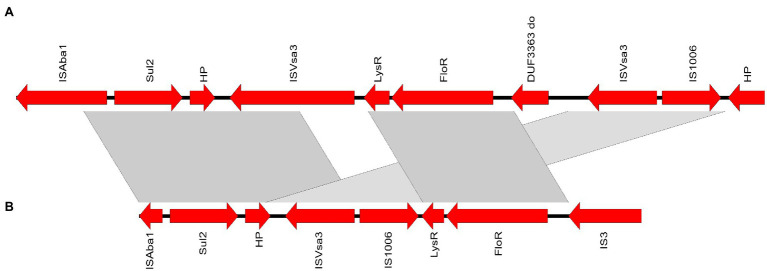
The chloramphenicol resistance gene, floR harbored by the isolates M02 and M11, was linked to a genetic structure containing sul2. Both were flanked by insertion elements with the order IS4, Sul2, hypothetical protein-coding gene, ISVsa3, IS1006, LysR, floR, and IS3. The closest match to this region was shown by Acinetobacter indicus chromosome (GenBank accession: CP071319.1). The genetic structure containing floR and sul2 genes compared to the closest match sequence is depicted in Figure 6.
Figure 6.
Depiction of the genetic structure containing the genes sul2 and floR identified in strain M11 (A) compared to Acinetobacter indicus strain GXNN62X4 chromosome, GenBank accession: CP071319 (B). ORFs orientation is indicated by arrows. Grey bands between panels indicate more than 98% sequence similarity. Genes are labelled by their protein products; LysR, LysR family transcriptional regulator.
Macrolide resistance genes msr(E) and mph(E) were flanked by an upstream ISNCY family transposase and a downstream ISAba1 element. A genetic organization that is identical to that carried by A. baumannii strain ACN21 chromosome (GenBank accession: CP038644.1).
Discussion
A threatening rise in the incidence of carbapenem-resistant A. baumannii has been increasingly reported worldwide (Levy-Blitchtein et al., 2018; Moghnieh et al., 2018; Alcantar-Curiel et al., 2019) and in Egypt as well (Al-Hassan et al., 2019; Benmahmod et al., 2019; Mabrouk et al., 2020), leaving behind a substantial number of difficult to treat infections. For a deeper insight into the molecular mechanisms underlying carbapenem resistance in this highly problematic pathogen, a collection of 20 A. baumannii clinical isolates was screened for carbapenemase-coding genes by PCR. Four NDM producers were identified in clinical specimens recovered from ICU patients with severe infections. XDR phenotype was identified in all blaNDM-positive A. baumannii with few reserved therapeutic options. These included tigecycline, colistin (for M02, M11, and M20), and amikacin (for M19) frequently associated with unfavorable pharmacokinetics and/or adverse effects particularly in critically ill patients (Spapen et al., 2011).
Draft genomes of the blaNDM-positive isolates were obtained by Illumina sequencing for subsequent MLST and resistome analysis. In silico MLST and goeBURST analysis revealed that three out of four NDM producer A. baumannii belonged to the high-risk international clones (ICs), known for outbreak potential, worldwide dissemination (Karah et al., 2012), and multidrug resistance (Diancourt et al., 2010). M02 and M11 were assigned ST85Pas/1089Oxf that belong to IC9, recently described by Müller et al. (2019). Abundance of studies reporting blaNDM-1-positive A. baumannii of ST85Pas from Middle East countries (Bonnin et al., 2013; Decousser et al., 2013; Rafei et al., 2014; Salloum et al., 2018) has drawn attention on its probable endemicity in this region. IC2 was represented only by M20 (ST570Pas/1701Oxf), whose genome was loaded by the highest share of resistance genes. The abundance of IC2 A. baumannii in Egypt was also reported by others (Al-Hassan et al., 2019; Wasfi et al., 2021). To the best of our knowledge, this is the first report of blaNDM-positive A. baumannii strain that belongs to ST164Pas/ST1418Oxf. Although MDR-resistant A. baumannii isolates that belong to ST164Pas have been increasingly reported from different parts of the world (Coelho-Souza et al., 2013; Loraine et al., 2020; Tada et al., 2020), none was reported to carry a blaNDM gene.
The SNP-based phylogeny analysis (Figure 1) showed that the isolates M02 and M11 were genetically related to two blaNDM-positive A. baumannii strains of the same sequence type (1089Oxf/85Pas). These included A. baumannii strain Cl300 isolated in Lebanon in 2015 and strain ACN21 isolated in India in 2018. M20 was found to be genetically related to two NDM producer A. baumannii strains isolated in United States in 2016 (TP2 and TP3). Both TP2 and TP3 had the Oxford ST1578 a double locus variant of ST1701 to which M20 belongs. On the other hand, M19 showed no genetic relatedness to any of the NDM producer A. baumannii strains for which complete genomes were available in the NCBI. Inclusion of four draft genomes that belong to ST164Pas and ST1418Oxf retrieved from PubMLST genome collection revealed that M19 was most genetically related to A. baumannii strain CPO20190206 isolated from Denmark (ST164Pas) and A. baumannii strain JorAb40 isolated from Jordan (ST1418Oxf). Both strains were isolated in 2019 and, interestingly, none was found to carry a blaNDM gene.
Resistome analysis disclosed a wide arsenal of resistance genes presented in Table 2 and correlated with the susceptibility profiles in Figure 2. Both intrinsic and acquired resistance mechanisms contributed to β-lactams resistance. The carbapenem-hydrolyzing class D β-lactamases (oxacillinases) provide both intrinsic (blaOXA51-like genes) and acquired (blaOXA-23, 40, 58, 143,235-like genes) resistance to β-lactams including carabapenems (Poirel and Nordmann, 2006; Ghaith et al., 2017). Overexpression of OXA-type β-lactamases has been linked to an upstream IS element, most frequently ISAba1, through which an additional promotor is provided (Evans and Amyes, 2014). blaOXA-94 preceded by ISAba1 element was identified in M02 and M11 while no IS elements could be identified upstream to blaOXA-91 or blaOXA-66 carried by M19 and M20, respectively.
In addition to intrinsic OXA-type β-lactamases, the IC2 isolate (M20) also carried blaOXA-23 the most widely disseminated oxacillinase acquired by carbapenem-resistant A. baumannii (Mugnier et al., 2010). Association of blaOXA-23 with IC2 A. baumannii has been reported worldwide (Hamidian and Nigro, 2019). As with other IC2 isolates, blaOXA-23 carried by M20 was found to reside in Tn2006 in which the gene is bracketed by two inversely oriented ISAba1 elements. Tn2006 is the most common structure harboring blaOXA-23 either alone or incorporated into AbGRIs (Hamidian and Nigro, 2019).
Although the association of blaOXA-91 and blaNDM-1 in A. baumannii was not previously described in Egypt, co-existence of blaOXA-51-like, blaOXA-23 and blaNDM-1 was reported by Wasfi et al. (2021).
Analysis of the genetic environment of blaNDM-1 in the sequenced isolates showed different environments in different sequence types. ISAba14 element was inserted upstream to the cutA gene in M02, M11, and M20. This was previously documented by Bonnin et al. (2013) who failed to identify a downstream second copy of ISAba125 by PCR and suggested loss of functionality of this truncated transposon (ΔTn125). WGS of blaNDM-positive isolates by a later study (Vijayakumar et al., 2020) uncovered the existence of a second copy of ISAba125 downstream to the ISAba14-interrupted transposon. Interestingly, analysis of the upstream region to the truncated transposon revealed the existence of the amikacin resistance gene AphA6 preceded by another copy of ISAba14 in ST85Pas isolates (Figure 4). The two ISAba14 elements were thus thought to form an alternative composite transposon in which two resistance genes were enclosed for transposition (blaNDM-1 and AphA6) rather than the widely known Tn125 in which blaNDM-1 was hosted as the sole antimicrobial resistance gene. Transposition of this composite transposon might, therefore, favor the co-transfer of resistance to two of the last-line antimicrobial treatment options for MDR and XDR A. baumannii. Nevertheless, experimental analysis is required to examine the transposition potential of this transposon. In M19, IS1 family transposase was identified immediately upstream to ISAba125 that precedes the blaNDM-1 gene. Insertion of IS1 element in this location was not identified in the nucleotide database of the NCBI.
Intrinsic to all A. baumannii, cephalosporin resistance is mediated by ADC (formerly known as blaAmpC). In addition to the incomplete blaADC-73 carried by M20, a novel blaADC allele (blaADC-257) with an upstream ISAba1 element was identified in M19 recovered from a blood culture of a female patient admitted to the ICU with fever of unknown origin. With no similarity in the NCBI nucleotide database, blaADC genes carried by M02 and M11 were interrupted by an IS1008 element (Figure 5). No alternative intact copies of blaADC were identified in M02, M11, or M20. Other β-lactamases identified here included class A β-lactamases, more efficiently capable of hydrolyzing penicillins and cephalosporins than carbapenems (Jeon et al., 2015). These were coded by blaTem-1 carried by M20 and blaCARB-16 in M19. However, their association with mobile elements could not be clearly determined.
In addition to the intrinsic aminoglycoside resistance gene ant(3``)-IIc (Zhang et al., 2017), the amikacin-modifying enzyme-coding gene aph(3`)-VIa (aphA6) was found in all isolates. The predominance of aph(3`)-VIa among the aminoglycoside modifying enzymes-coding genes was also reported by others (Aghazadeh et al., 2013; Sheikhalizadeh et al., 2017). Notably, the gene was also identified in the amikacin-sensitive isolate M19. Identification of aph(3`)-VIa in amikacin-susceptible isolates was also reported before (Aghazadeh et al., 2013; Sheikhalizadeh et al., 2017). In ant(2``)-Ia-positive isolates, the gene was found in pRAY plasmid variants. pRAY is a 6 Kb plasmid widely distributed in Acinetobacter species comprising the most common resistance mechanism to gentamicin and tobramycin (Hamidian et al., 2012).
Acquired 16S rRNA methyltransferases constitute the most important aminoglycoside resistance mechanism conferring resistance to most of the clinically important aminoglycosides (Galimand et al., 2003). Of them, armA has been widely reported from A. baumannii particularly those of the IC2 (Blackwell et al., 2017). Within a 20,844bp genomic resistance island closely similar to A. baumannii genomic resistance island 3 (AbGRI3; Blackwell et al., 2017), armA gene was identified in M20 (IC2). Other resistance genes hosted by the genomic island include aph(3`)-Ia, aac(6`)-Ib`, catB8, ant(3``)-Ia, sul1, msr(E), and mph(E). Another unique genetic structure in which genes coding resistance to two different antimicrobial classes was identified in M02 and M11 (Figure 6). This included the chloramphenicol efflux pump (FloR)-coding gene and sul2, conferring resistance to sulfamethoxazole/trimethoprim, enclosed by insertion elements. The closest match to this region was shown by Acinetobacter indicus chromosome (GenBank accession: CP071319.1) from which it may have been acquired with some genetic rearrangement.
In the absence of plasmid-mediated quinolones resistance genes, nonsusceptibility to levofloxacin in all NDM producers investigated here was mediated by target site mutations. These affected the QRDRs within GyrA (S83L) and ParC (S80L) enzymes. The mutation pattern identified in our isolates was commonly reported as the predominant mechanism responsible for fluoroquinolones resistance in A. baumannii (Hamed et al., 2018; Nodari et al., 2020; Roy et al., 2021).
Resistance to colistin, the last line of defense against XDR A. baumannii, was evident in only one isolate (M19) that, fortunately, retained susceptibility to amikacin and tigecycline. Colistin resistance in M19 was accompanied by multiple nonsynonymous mutations affecting pmrABC and IpxACD genes. Missense mutations identified in pmrB (H89L) and pmrC (I42V) genes carried by M19 were also reported in colistin-resistant A. baumannii studied by Nurtop et al. (2019) in Turkey. It is worth mentioning that the amino acid affected by pmrB mutation identified here is located outside the histidine kinase domain, the main determinant of colistin resistance in A. baumannii (Arroyo et al., 2011; Beceiro et al., 2011; Lesho et al., 2013). Moreover, all lpxACD mutations identified here were previously reported in both colistin-susceptible and colistin-resistant isolates (Oikonomou et al., 2015; Haeili et al., 2018; Nurtop et al., 2019). Accordingly, novel unidentified resistance mechanisms might stand behind the high-level resistance (MIC≥128μg/ml) of M19 to colistin. Further investigations including gene expression analysis are therefore required to confirm or role out the impact of such mutations on colistin susceptibility.
Diverse efflux pumps, whose overexpression has been linked to multidrug resistance, were identified in the sequenced isolates. RND efflux pumps known by their broad substrate profiles (Coyne et al., 2011), including AdeABC, AdeIJK, and AdeFGH. were identified in all isolates. RND efflux pumps contribute to intrinsic resistance of A. baumannii to several classes of antimicrobials. Other multidrug efflux pumps carried by all isolates included AbeM, a member of the multidrug and toxic compound extrusion family efflux pumps and the small multidrug resistance efflux pump AbeS (Coyne et al., 2011). Except for FloR conferring resistance to phenicols in M02 and M11 only, efflux pumps of the MFS were disseminated in all sequenced genomes. With narrow substrate profiles, AmvA, AbaF, and AbaQ are known to extrude erythromycin, fosfomycin, and quinolones, respectively (Coyne et al., 2011; Perez-Varela et al., 2018). The macrolide-specific ABC pump MacAB was also found in all isolates.
It is worth mentioning that the current study suffers from some limitations, most importantly is using short-read sequencing technology instead of a hybrid long- and short-read sequencing approach known to produce more accurate genome organization. Consistent with other studies (Leal et al., 2020), resistance to some antimicrobials could not be correlated to known resistance genes highlighting the need for further investigations including gene expression analysis or identification of novel resistance determinants. Finally, only four genomes were sequenced here thus correlating resistance genes with particular STs could not be fully achieved.
Conclusion
The current study is one of the few studies reporting WGS of A. baumannii clinical isolates from Egypt. The isolates showed XDR phenotype and were recovered from ICU patients. High-risk international clones were identified, predominantly IC9 (ST85Pas) widely reported from Middle East countries. Diverse mobile elements were associated with resistance genes with novel insertion sites and genetic organizations. Co-existence of amikacin and carbapenem resistance genes on an ISAba14-bracketed transposon was uniquely identified in ST85Pas/ST1089Oxf. blaNDM-1 gene was identified, for the first time, on the chromosome of an A. baumannii strain that belongs to sequence type ST164Pas/S1418Oxf. WGS of the highly problematic MDR and XDR pathogens may aid in the identification of emerging resistance genes and their dissemination dynamics. Co-existence of resistance genes within mobile genetic elements could also be identified. This may aid in optimizing treatment guidelines to avoid selection of resistance to last-line antimicrobials. WGS also permits monitoring the emergence of novel global MDR clones and facilitates comparative genomic analysis and developing cheaper molecular techniques for routine screening.
Ethical Approval
The study was performed in accordance with relevant guidelines and regulations, and no experiments were performed on humans and/or human tissue samples. The study was approved by the local Ethical Committee of clinical and chemical pathology department, Kasr Al-Aini Hospital, Cairo university. Only bacterial isolates were collected for the routine laboratory work to ensure patient care and informed consents were not required.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
MZ, AH, MA, HR, and SH contributed to the study design, performance of experiments, and data analysis. SH performed the genomes assembly and bioinformatic analysis. MZ wrote the first draft of the manuscript. All authors read and approved the final version of manuscript.
Funding
The authors thank the Deanship of Scientific Research at King Saud University for funding this work through Project No. RGP-038.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
The authors extend their appreciation to Genomic unit at The Children’s Cancer Hospital Egypt 57357 (CCHE) for performing part of the experiments.
Footnotes
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.736982/full#supplementary-material
References
- Aghazadeh M., Rezaee M. A., Nahaei M. R., Mahdian R., Pajand O., Saffari F., et al. (2013). Dissemination of aminoglycoside-modifying enzymes and 16S rRNA methylases among acinetobacter baumannii and Pseudomonas aeruginosa isolates. Microb. Drug Resist. 19, 282–288. doi: 10.1089/mdr.2012.0223, PMID: [DOI] [PubMed] [Google Scholar]
- Alcantar-Curiel M. D., Rosales-Reyes R., Jarillo-Quijada M. D., Gayosso-Vazquez C., Fernandez-Vazquez J. L., Toledano-Tableros J. E., et al. (2019). Carbapenem-resistant Acinetobacter baumannii in three tertiary care hospitals in Mexico: virulence profiles, innate immune response and clonal dissemination. Front. Microbiol. 10:2116. doi: 10.3389/fmicb.2019.02116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alcock B. P., Raphenya A. R., Lau T. T. Y., Tsang K. K., Bouchard M., Edalatmand A., et al. (2020). CARD 2020: antibiotic resistome surveillance with the comprehensive antibiotic resistance database. Nucleic Acids Res. 48, D517–D525. doi: 10.1093/nar/gkz935, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Hassan L., Zafer M. M., El-Mahallawy H. (2019). Multiple sequence types responsible for healthcare-associated Acinetobacter baumannii dissemination in a single Centre in Egypt. BMC Infect. Dis. 19:829. doi: 10.1186/s12879-019-4433-1, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antipov D., Hartwick N., Shen M., Raiko M., Lapidus A., Pevzner P. A. (2016). plasmidSPAdes: assembling plasmids from whole genome sequencing data. Bioinformatics 32, 3380–3387. doi: 10.1093/bioinformatics/btw493, PMID: [DOI] [PubMed] [Google Scholar]
- Antunes L. C., Visca P., Towner K. J. (2014). Acinetobacter baumannii: evolution of a global pathogen. Pathog Dis. 71, 292–301. doi: 10.1111/2049-632X.12125, PMID: [DOI] [PubMed] [Google Scholar]
- Arroyo L. A., Herrera C. M., Fernandez L., Hankins J. V., Trent M. S., Hancock R. E. (2011). The pmrCAB operon mediates polymyxin resistance in Acinetobacter baumannii ATCC 17978 and clinical isolates through phosphoethanolamine modification of lipid A. Antimicrob. Agents Chemother. 55, 3743–3751. doi: 10.1128/AAC.00256-11, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayobami O., Willrich N., Harder T., Okeke I. N., Eckmanns T., Markwart R. (2019). The incidence and prevalence of hospital-acquired (carbapenem-resistant) Acinetobacter baumannii in Europe, eastern Mediterranean and Africa: a systematic review and meta-analysis. Emerg. Microbes. Infect. 8, 1747–1759. doi: 10.1080/22221751.2019.1698273, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bankevich A., Nurk S., Antipov D., Gurevich A. A., Dvorkin M., Kulikov A. S., et al. (2012). SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19, 455–477. doi: 10.1089/cmb.2012.0021, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beceiro A., Llobet E., Aranda J., Bengoechea J. A., Doumith M., Hornsey M., et al. (2011). Phosphoethanolamine modification of lipid A in colistin-resistant variants of Acinetobacter baumannii mediated by the pmrAB two-component regulatory system. Antimicrob. Agents Chemother. 55, 3370–3379. doi: 10.1128/AAC.00079-11, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benmahmod A. B., Said H. S., Ibrahim R. H. (2019). Prevalence and mechanisms of Carbapenem resistance Among Acinetobacter baumannii clinical isolates in Egypt. Microb. Drug Resist. 25, 480–488. doi: 10.1089/mdr.2018.0141, PMID: [DOI] [PubMed] [Google Scholar]
- Bertelli C., Laird M. R., Williams K. P., Simon Fraser University Research Computing Group. Lau B. Y., Hoad G., et al. (2017). IslandViewer 4: expanded prediction of genomic islands for larger-scale datasets. Nucleic Acids Res. 45, W30–W35. doi: 10.1093/nar/gkx343, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackwell G. A., Holt K. E., Bentley S. D., Hsu L. Y., Hall R. M. (2017). Variants of AbGRI3 carrying the armA gene in extensively antibiotic-resistant Acinetobacter baumannii from Singapore. J. Antimicrob. Chemother. 72, 1031–1039. doi: 10.1093/jac/dkw542, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger A. M., Lohse M., Usadel B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnin R. A., Cuzon G., Poirel L., Nordmann P. (2013). Multidrug-resistant Acinetobacter baumannii clone, France. Emerg. Infect. Dis. 19, 822–823. doi: 10.3201/eid1905.121618, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bontron S., Nordmann P., Poirel L. (2016). Transposition of Tn125 encoding the NDM-1 Carbapenemase in Acinetobacter baumannii. Antimicrob. Agents Chemother. 60, 7245–7251. doi: 10.1128/AAC.01755-16, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bortolaia V., Kaas R. S., Ruppe E., Roberts M. C., Schwarz S., Cattoir V., et al. (2020). ResFinder 4.0 for predictions of phenotypes from genotypes. J. Antimicrob. Chemother. 75, 3491–3500. doi: 10.1093/jac/dkaa345, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown J., Pirrung M., Mccue L. A. (2017). FQC dashboard: integrates FastQC results into a web-based, interactive, and extensible FASTQ quality control tool. Bioinformatics 33, 3137–3139. doi: 10.1093/bioinformatics/btx373, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chng K. R., Li C., Bertrand D., Ng A. H. Q., Kwah J. S., Low H. M., et al. (2020). Cartography of opportunistic pathogens and antibiotic resistance genes in a tertiary hospital environment. Nat. Med. 26, 941–951. doi: 10.1038/s41591-020-0894-4, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- CLSI (2018). Performance Standards for Antimicrobial Susceptibility Testing: Twenty-Second Informational Supplement. CLSI Supplement M100. Wayne, PA: Clinical and Laboratory Standards Institute. [Google Scholar]
- Coelho-Souza T., Reis J. N., Martins N., Martins I. S., Menezes A. O., Reis M. G., et al. (2013). Longitudinal surveillance for meningitis by Acinetobacter in a large urban setting in Brazil. Clin. Microbiol. Infect. 19, E241–E244. doi: 10.1111/1469-0691.12145, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coyne S., Courvalin P., Perichon B. (2011). Efflux-mediated antibiotic resistance in Acinetobacter spp. Antimicrob. Agents Chemother. 55, 947–953. doi: 10.1128/AAC.01388-10, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decousser J. W., Jansen C., Nordmann P., Emirian A., Bonnin R. A., Anais L., et al. (2013). Outbreak of NDM-1-producing Acinetobacter baumannii in France, January to may 2013. Euro Surveill. 18:20547. doi: 10.2807/1560-7917.ES2013.18.31.20547, PMID: [DOI] [PubMed] [Google Scholar]
- Di Nocera P. P., Rocco F., Giannouli M., Triassi M., Zarrilli R. (2011). Genome organization of epidemic Acinetobacter baumannii strains. BMC Microbiol. 11:224. doi: 10.1186/1471-2180-11-224, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diancourt L., Passet V., Nemec A., Dijkshoorn L., Brisse S. (2010). The population structure of Acinetobacter baumannii: expanding multiresistant clones from an ancestral susceptible genetic pool. PLoS One 5:e10034. doi: 10.1371/journal.pone.0010034, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elsayed E., Elarabi M. A., Sherif D. A., Elmorshedi M., El-Mashad N. (2020). Extensive drug resistant Acinetobacter baumannii: a comparative study between non-colistin based combinations. Int. J. Clin. Pharm. 42, 80–88. doi: 10.1007/s11096-019-00940-1, PMID: [DOI] [PubMed] [Google Scholar]
- EUCAST (2021). Breakpoint tables for interpretation of MICs and zone diameters, version 11.0. Available at: http://www.eucast.org (Accessed May 1, 2021).
- Evans B. A., Amyes S. G. (2014). OXA beta-lactamases. Clin. Microbiol. Rev. 27, 241–263. doi: 10.1128/CMR.00117-13, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fam N. S., Gamal D., Mohamed S. H., Wasfy R. M., Soliman M. S., El-Kholy A. A., et al. (2020). Molecular characterization of Carbapenem/Colistin-resistant Acinetobacter baumannii clinical isolates from Egypt by whole-genome sequencing. Infect. Drug Resist. Volume 13, 4487–4493. doi: 10.2147/IDR.S288865, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francisco A. P., Vaz C., Monteiro P. T., Melo-Cristino J., Ramirez M., Carrico J. A. (2012). PHYLOViZ: phylogenetic inference and data visualization for sequence based typing methods. BMC Bioinformatics 13:87. doi: 10.1186/1471-2105-13-87, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galimand M., Courvalin P., Lambert T. (2003). Plasmid-mediated high-level resistance to aminoglycosides in Enterobacteriaceae due to 16S rRNA methylation. Antimicrob. Agents Chemother. 47, 2565–2571. doi: 10.1128/AAC.47.8.2565-2571.2003, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghaith D. M., Zafer M. M., Al-Agamy M. H., Alyamani E. J., Booq R. Y., Almoazzamy O. (2017). The emergence of a novel sequence type of MDR Acinetobacter baumannii from the intensive care unit of an Egyptian tertiary care hospital. Ann. Clin. Microbiol. Antimicrob. 16:34. doi: 10.1186/s12941-017-0208-y, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghazawi A., Sonnevend A., Bonnin R. A., Poirel L., Nordmann P., Hashmey R., et al. (2012). NDM-2 carbapenemase-producing Acinetobacter baumannii in the United Arab Emirates. Clin. Microbiol. Infect. 18, E34–E36. doi: 10.1111/j.1469-0691.2011.03726.x, PMID: [DOI] [PubMed] [Google Scholar]
- Gurevich A., Saveliev V., Vyahhi N., Tesler G. (2013). QUAST: quality assessment tool for genome assemblies. Bioinformatics 29, 1072–1075. doi: 10.1093/bioinformatics/btt086, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haeili M., Kafshdouz M., Feizabadi M. M. (2018). Molecular mechanisms of Colistin resistance Among Pandrug-resistant isolates of Acinetobacter baumannii with high case-fatality rate in intensive care unit patients. Microb. Drug Resist. 24, 1271–1276. doi: 10.1089/mdr.2017.0397, PMID: [DOI] [PubMed] [Google Scholar]
- Hamed S. M., Elkhatib W. F., El-Mahallawy H. A., Helmy M. M., Ashour M. S., Aboshanab K. M. A. (2018). Multiple mechanisms contributing to ciprofloxacin resistance among gram negative bacteria causing infections to cancer patients. Sci. Rep. 8:12268. doi: 10.1038/s41598-018-30756-4, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamidian M., Nigro S. J. (2019). Emergence, molecular mechanisms and global spread of carbapenem-resistant Acinetobacter baumannii. Microb. Genom. 5:306. doi: 10.1099/mgen.0.000306, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamidian M., Nigro S. J., Hall R. M. (2012). Variants of the gentamicin and tobramycin resistance plasmid pRAY are widely distributed in Acinetobacter. J. Antimicrob. Chemother. 67, 2833–2836. doi: 10.1093/jac/dks318, PMID: [DOI] [PubMed] [Google Scholar]
- Hendriksen R. S., Bortolaia V., Tate H., Tyson G. H., Aarestrup F. M., Mcdermott P. F. (2019). Using genomics to track global antimicrobial resistance. Front. Public Health 7:242. doi: 10.3389/fpubh.2019.00242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon J. H., Lee J. H., Lee J. J., Park K. S., Karim A. M., Lee C. R., et al. (2015). Structural basis for carbapenem-hydrolyzing mechanisms of carbapenemases conferring antibiotic resistance. Int. J. Mol. Sci. 16, 9654–9692. doi: 10.3390/ijms16059654, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaas R. S., Leekitcharoenphon P., Aarestrup F. M., Lund O. (2014). Solving the problem of comparing whole bacterial genomes across different sequencing platforms. PLoS One 9:e104984. doi: 10.1371/journal.pone.0104984, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karah N., Sundsfjord A., Towner K., Samuelsen O. (2012). Insights into the global molecular epidemiology of carbapenem non-susceptible clones of Acinetobacter baumannii. Drug Resist. Updat. 15, 237–247. doi: 10.1016/j.drup.2012.06.001, PMID: [DOI] [PubMed] [Google Scholar]
- Leal N. C., Campos T. L., Rezende A. M., Docena C., Mendes-Marques C. L., De Sa Cavalcanti F. L., et al. (2020). Comparative genomics of Acinetobacter baumannii clinical strains From Brazil reveals polyclonal dissemination and selective exchange of Mobile genetic elements associated With resistance genes. Front. Microbiol. 11:1176. doi: 10.3389/fmicb.2020.01176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesho E., Yoon E. J., Mcgann P., Snesrud E., Kwak Y., Milillo M., et al. (2013). Emergence of colistin-resistance in extremely drug-resistant Acinetobacter baumannii containing a novel pmrCAB operon during colistin therapy of wound infections. J. Infect. Dis. 208, 1142–1151. doi: 10.1093/infdis/jit293, PMID: [DOI] [PubMed] [Google Scholar]
- Letunic I., Bork P. (2016). Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 44, W242–W245. doi: 10.1093/nar/gkw290, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy-Blitchtein S., Roca I., Plasencia-Rebata S., Vicente-Taboada W., Velasquez-Pomar J., Munoz L., et al. (2018). Emergence and spread of carbapenem-resistant Acinetobacter baumannii international clones II and III in Lima, Peru. Emerg. Microbes Infect. 7:119. doi: 10.1038/s41426-018-0127-9, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. (2011). A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics 27, 2987–2993. doi: 10.1093/bioinformatics/btr509, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Durbin R. (2009). Fast and accurate short read alignment with burrows-wheeler transform. Bioinformatics 25, 1754–1760. doi: 10.1093/bioinformatics/btp324, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loraine J., Heinz E., Soontarach R., Blackwell G. A., Stabler R. A., Voravuthikunchai S. P., et al. (2020). Genomic and phenotypic analyses of Acinetobacter baumannii isolates From three tertiary care hospitals in Thailand. Front. Microbiol. 11:548. doi: 10.3389/fmicb.2020.00548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mabrouk S. S., Abdellatif G. R., El-Ansary M. R., Aboshanab K. M., Ragab Y. M. (2020). Carbapenemase producers Among extensive drug-resistant gram-negative pathogens recovered from febrile neutrophilic patients in Egypt. Infect. Drug Resist. 13, 3113–3124. doi: 10.2147/IDR.S269971, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magiorakos A. P., Srinivasan A., Carey R. B., Carmeli Y., Falagas M. E., Giske C. G., et al. (2012). Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 18, 268–281. doi: 10.1111/j.1469-0691.2011.03570.x, PMID: [DOI] [PubMed] [Google Scholar]
- Moghnieh R. A., Kanafani Z. A., Tabaja H. Z., Sharara S. L., Awad L. S., Kanj S. S. (2018). Epidemiology of common resistant bacterial pathogens in the countries of the Arab league. Lancet Infect. Dis. 18, e379–e394. doi: 10.1016/S1473-3099(18)30414-6, PMID: [DOI] [PubMed] [Google Scholar]
- Mugnier P. D., Poirel L., Naas T., Nordmann P. (2010). Worldwide dissemination of the blaOXA-23 carbapenemase gene of Acinetobacter baumannii. Emerg. Infect. Dis. 16, 35–40. doi: 10.3201/eid1601.090852, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller C., Stefanik D., Wille J., Hackel M., Higgins P. G., Siefert H. (2019). “Molecular epidemiology of carbapenem-resistant Acinetobacter baumannii clinical isolates and identification of the novel international clone IC9: results from a worldwide surveillance study (2012–2016)”, in Paper Presented at the ECCMID 2019: Proceeding of the 29th European Congress of Clinical Microbiology & Infectious Diseases. (Amsterdam, Netherlands: ) April 13–16, 2019. [Google Scholar]
- Nodari C. S., Cayo R., Streling A. P., Lei F., Wille J., Almeida M. S., et al. (2020). Genomic analysis of Carbapenem-resistant Acinetobacter baumannii isolates belonging to major endemic clones in South America. Front. Microbiol. 11:584603. doi: 10.3389/fmicb.2020.584603, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nurtop E., Bayindir Bilman F., Menekse S., Kurt Azap O., Gonen M., Ergonul O., et al. (2019). Promoters of Colistin resistance in Acinetobacter baumannii infections. Microb. Drug Resist. 25, 997–1002. doi: 10.1089/mdr.2018.0396, PMID: [DOI] [PubMed] [Google Scholar]
- Oikonomou O., Sarrou S., Papagiannitsis C. C., Georgiadou S., Mantzarlis K., Zakynthinos E., et al. (2015). Rapid dissemination of colistin and carbapenem resistant Acinetobacter baumannii in Central Greece: mechanisms of resistance, molecular identification and epidemiological data. BMC Infect. Dis. 15:559. doi: 10.1186/s12879-015-1297-x, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Varela M., Corral J., Aranda J., Barbe J. (2018). Functional characterization of AbaQ, a novel efflux pump mediating quinolone resistance in Acinetobacter baumannii. Antimicrob. Agents Chemother. 62:e00906-18. doi: 10.1128/AAC.00906-18, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poirel L., Nordmann P. (2006). Carbapenem resistance in Acinetobacter baumannii: mechanisms and epidemiology. Clin. Microbiol. Infect. 12, 826–836. doi: 10.1111/j.1469-0691.2006.01456.x, PMID: [DOI] [PubMed] [Google Scholar]
- Rafei R., Dabboussi F., Hamze M., Eveillard M., Lemarie C., Mallat H., et al. (2014). First report of blaNDM-1-producing Acinetobacter baumannii isolated in Lebanon from civilians wounded during the Syrian war. Int. J. Infect. Dis. 21, 21–23. doi: 10.1016/j.ijid.2014.01.004, PMID: [DOI] [PubMed] [Google Scholar]
- Roy S., Chatterjee S., Bhattacharjee A., Chattopadhyay P., Saha B., Dutta S., et al. (2021). Overexpression of efflux pumps, mutations in the Pumps' regulators, chromosomal mutations, and AAC(6′)-Ib-cr are associated With fluoroquinolone resistance in diverse sequence types of neonatal Septicaemic Acinetobacter baumannii: A 7-year single center study. Front. Microbiol. 12:602724. doi: 10.3389/fmicb.2021.602724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salloum T., Tannous E., Alousi S., Arabaghian H., Rafei R., Hamze M., et al. (2018). Genomic mapping of ST85 blaNDM-1 and blaOXA-94 producing Acinetobacter baumannii isolates from Syrian civil war victims. Int. J. Infect. Dis. 74, 100–108. doi: 10.1016/j.ijid.2018.07.017, PMID: [DOI] [PubMed] [Google Scholar]
- Sheikhalizadeh V., Hasani A., Ahangarzadeh Rezaee M., Rahmati-Yamchi M., Hasani A., Ghotaslou R., et al. (2017). Comprehensive study to investigate the role of various aminoglycoside resistance mechanisms in clinical isolates of Acinetobacter baumannii. J. Infect. Chemother. 23, 74–79. doi: 10.1016/j.jiac.2016.09.012, PMID: [DOI] [PubMed] [Google Scholar]
- Siguier P., Perochon J., Lestrade L., Mahillon J., Chandler M. (2006). ISfinder: the reference Centre for bacterial insertion sequences. Nucleic Acids Res. 34, D32–D36. doi: 10.1093/nar/gkj014, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spapen H., Jacobs R., Van Gorp V., Troubleyn J., Honore P. M. (2011). Renal and neurological side effects of colistin in critically ill patients. Ann. Intensive Care 1:14. doi: 10.1186/2110-5820-1-14, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan M. J., Petty N. K., Beatson S. A. (2011). Easyfig: a genome comparison visualizer. Bioinformatics 27, 1009–1010. doi: 10.1093/bioinformatics/btr039, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tacconelli E., Carrara E., Savoldi A., Harbarth S., Mendelson M., Monnet D. L., et al. (2018). Discovery, research, and development of new antibiotics: the WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect. Dis. 18, 318–327. doi: 10.1016/S1473-3099(17)30753-3, PMID: [DOI] [PubMed] [Google Scholar]
- Tada T., Uchida H., Hishinuma T., Watanabe S., Tohya M., Kuwahara-Arai K., et al. (2020). Molecular epidemiology of multidrug-resistant Acinetobacter baumannii isolates from hospitals in Myanmar. J. Glob. Antimicrob. Resist. 22, 122–125. doi: 10.1016/j.jgar.2020.02.011, PMID: [DOI] [PubMed] [Google Scholar]
- Tal-Jasper R., Katz D. E., Amrami N., Ravid D., Avivi D., Zaidenstein R., et al. (2016). Clinical and epidemiological significance of Carbapenem resistance in Acinetobacter baumannii infections. Antimicrob. Agents Chemother. 60, 3127–3131. doi: 10.1128/AAC.02656-15, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatusova T., Dicuccio M., Badretdin A., Chetvernin V., Nawrocki E. P., Zaslavsky L., et al. (2016). NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res. 44, 6614–6624. doi: 10.1093/nar/gkw569, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toleman M. A., Spencer J., Jones L., Walsh T. R. (2012). blaNDM-1 is a chimera likely constructed in Acinetobacter baumannii. Antimicrob. Agents Chemother. 56, 2773–2776. doi: 10.1128/AAC.06297-11, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turton J. F., Woodford N., Glover J., Yarde S., Kaufmann M. E., Pitt T. L. (2006). Identification of Acinetobacter baumannii by detection of the blaOXA-51-like carbapenemase gene intrinsic to this species. J. Clin. Microbiol. 44, 2974–2976. doi: 10.1128/JCM.01021-06, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijayakumar S., Wattal C., J K. O., Bhattacharya S., Vasudevan K., Anandan S., et al. (2020). Insights into the complete genomes of carbapenem-resistant Acinetobacter baumannii harbouring Bla OXA-23, Bla OXA-420 and Bla NDM-1 genes using a hybrid-assembly approach. Access Microbiol. 2:acmi000140. doi: 10.1099/acmi.0.000140, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wailan A. M., Paterson D. L. (2014). The spread and acquisition of NDM-1: a multifactorial problem. Expert Rev. Anti-Infect. Ther. 12, 91–115. doi: 10.1586/14787210.2014.856756, PMID: [DOI] [PubMed] [Google Scholar]
- Wasfi R., Rasslan F., Hassan S. S., Ashour H. M., Abd El-Rahman O. A. (2021). Co-existence of Carbapenemase-encoding genes in Acinetobacter baumannii from cancer patients. Infect. Dis. Ther. 10, 291–305. doi: 10.1007/s40121-020-00369-4, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wick R. R., Judd L. M., Gorrie C. L., Holt K. E. (2017). Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput. Biol. 13:e1005595. doi: 10.1371/journal.pcbi.1005595, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wick R. R., Schultz M. B., Zobel J., Holt K. E. (2015). Bandage: interactive visualization of de novo genome assemblies. Bioinformatics 31, 3350–3352. doi: 10.1093/bioinformatics/btv383, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G., Leclercq S. O., Tian J., Wang C., Yahara K., Ai G., et al. (2017). A new subclass of intrinsic aminoglycoside nucleotidyltransferases, ANT(3″)-II, is horizontally transferred among Acinetobacter spp. by homologous recombination. PLoS Genet. 13:e1006602. doi: 10.1371/journal.pgen.1006602, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.