Abstract
The intraspecies diversity of an opportunistic yeast pathogen, Cryptococcus laurentii, was revealed by analysis of the sequences of the internal transcribed spacer regions and the 28S rRNA gene. Ten strains of C. laurentii were grouped into two major phylogenetic groups and were further divided into at least seven species. Four of the strains isolated from patients did not represent a single species but showed heterogeneity. These results suggest that C. laurentii is a genetically heterogeneous species, and this must be taken into consideration when identifying C. laurentii clinical isolates.
Cryptococcosis, which is usually caused by Cryptococcus neoformans, is considered one of the most serious fungal infections that can occur in immunocompromised patients. Non-C. neoformans species have generally been regarded as nonpathogenic saprophytes. In recent years, however, opportunistic infections associated with C. albidus, C. curvatus, C. humicolus, and C. laurentii have been reported (6, 7, 9, 14, 24) and reports of cases of infection due to C. laurentii have been increasing. Taxonomically, C. laurentii is reported to be a heterogeneous species on the basis of its nuclear DNA base composition and whole-cell protein electrophoretic fingerprints (23). The genus Cryptococcus is also polyphyletic, according to molecular phylogenies (2, 21).
In this report we reveal the intraspecies diversity of C. laurentii as determined by analyses of the internal transcribed spacer (ITS) and 28S rRNA gene (rDNA) sequences and also discuss the taxonomic position of C. laurentii clinical isolates.
MATERIALS AND METHODS
Strains used.
The 10 strains of C. laurentii listed in Table 1 were studied. They are stock strains from the Centraalbureau voor Schimmelcultures (CBS) and the Japan Collection of Microorganisms (JCM).
TABLE 1.
C. laurentii (Kufferath) Skinner strains used in this study
| Strain | Source | Co-Qab | G+C content (mol%)a |
|---|---|---|---|
| JCM 9066T (CBS 139) | Palmvine, type strain of Torula laurentii Kufferath | Q10 | 56.0, 56.4, 59.0 |
| CBS 318 | Atmosphere, type strain of Torula aurea Saito | Q10 | 55.0 |
| CBS 942 | Atmosphere, type strain of Torula flavescens Saito | Q10 | 57.2 |
| CBS 973 | Muscatel grape, type strain of Torulopsis carnescens Verona & Luchetti | ||
| CBS 2174 | Tumor | 54.7 | |
| CBS 2409 | Surface of shrimp, type strain of Rhodotorula peneaus Phaff et al. | Q10 | 51.2, 52.3 |
| CBS 2993 | Bronchus of lung patient | Q10 | 50.8 |
| CBS 6578 | Seawater | 51.8 | |
| CBS 8645 | Cerebrospinal fluid of male AIDS patient with meningitis | ||
| CBS 8648 | Lung |
From Vancanneyt et al. (23).
Co-Q, coenzyme Q (ubiquinone) homologs.
Direct DNA sequencing.
Nuclear DNA was extracted by the method of Makimura et al. (11). The D1-D2 28S rDNA and ITS sequences were directly determined by using PCR products. For D1-D2 28S rDNA, the primers NL-1 (5′-GCATATCAATAAGCGGAGGAAAAG) and NL-4 (5′-GGTCCGTGTTTCAAGACGG) were used (10). The ITS sequences were determined with the primers pITS-F (5′-GTCGTAACAAGGTTAACCTGCGG) and pITS-R (5′-TCCTCCGCTTATTGATATGC) (19). The PCR products were sequenced with an ABI PRISM Cycle Sequencing kit (Perkin-Elmer Applied Biosystems, Foster City, Calif.).
Molecular phylogenetic analysis.
The sequences were aligned with the computer program CLUSTAL W, version 1.74 (22). The evolutionary distance for the neighbor-joining method was calculated as described by Kimura (8). Sites where gaps existed in any sequences were excluded. A bootstrap analysis (1) was performed with 100 random resamplings.
Identification of strains.
Strains with 99% or more similarity in the 28S rDNA D1-D2 region and the overall ITS sequences were defined as conspecific (10, 19). The sequence data were searched by using the BLAST system (http://www.ncbi.nlm.nih.gov/BLAST/) at the National Center for Biotechnology Information Bethesda, Md.
Biochemical characteristics.
The strains were tested with the ID 32C kit (bioMérieux SA, Marcy l'Etoile, France) in accordance with the manufacturer's instructions.
Nucleotide sequence accession numbers.
The nucleotide sequences discussed in this paper have been deposited in the DNA Data Bank of Japan (DDBJ), and their accession numbers are given in Table 2.
TABLE 2.
Accession numbers of sequences of 28S rDNA and ITS regions
| Species | Strain | Accession no.
|
Source | |
|---|---|---|---|---|
| ITS | 28S rDNA | |||
| Cryptococcus laurentii | CBS 318 | AB035045 | AB035041 | This study |
| Cryptococcus laurentii | CBS 942 | AB035046 | AB035042 | This study |
| Cryptococcus laurentii | CBS 973 | AB035050 | AB035054 | This study |
| Cryptococcus laurentii | CBS 2174 | AB035044 | AB035040 | This study |
| Cryptococcus laurentii | CBS 2409 | AB035047 | AB035051 | This study |
| Cryptococcus laurentii | CBS 2993 | AB035048 | AB035052 | This study |
| Cryptococcus laurentii | CBS 6578 | AB035049 | AB035053 | This study |
| Cryptococcus laurentii | CBS 8645 | AB035046 | AB035042 | This study |
| Cryptococcus laurentii | CBS 8648 | AB035044 | AB035040 | This study |
| Cryptococcus laurentii | JCM 9066 (CBS139) | AB035043 | This study | |
| Bullera unica | CBS 8290 | AF075524 | ||
| Bullera globispora | CBS 6981 | AF075509 | ||
| Bulleromyces albus | CBS 501 | AF075500 | ||
| Cystofilobasidium capitatum | CBS 6358 | AF075465 | ||
| Cryptococcus cellulolyticus | CBS 8294 | AF075525 | ||
| Cryptococcus dimennae | CBS 5770 | AF075489 | ||
| Cryptococcus flavus | CBS 331 | AF075497 | ||
| Cryptococcus heveanensis | CBS 569 | AF075467 | ||
| Cryptococcus hungaricus | CBS 4214 | AF075503 | ||
| Cryptococcus laurentii | CBS 139 | AF075469 | ||
| Cryptococcus luteolus | CBS 943 | AF075482 | ||
| Cryptococcus nodaensis | G60 | AB016234 | AB016233 | |
| Cryptococcus podzolicus | CBS 6819 | AF075481 | ||
| Cryptococcus skinneri | CBS 7890 | AF075494 | ||
| Filobasidiella neoformans var. neoformans | CBS 132 | AF075484 | ||
| Mrakia frigida | CBS 5266 | AF189849 | ||
| Trimorphomyces papilionaceus | CBS 445.92 | AF075491 | ||
RESULTS
Sequences of ITS regions and D1-D2 of 28S rDNA.
Differences in the lengths of the ITS 1 and 2 regions were observed for the strains. ITS 1 was from 110 to 139 bp long, while ITS 2 was from 140 to 179 bp long. Matrices of the overall similarities of the ITS and the D1-D2 regions of 28S rDNA are shown in Tables 3 and 4. According to the species concept based on sequence similarity, strains CBS 2174 and CBS 8648 and strains CBS 942 and CBS 8645 were identified as C. laurentii and C. nodaensis, respectively. The sequences of CBS 973 were completely identical to those of Trimorphomyces papilionaceus in both the 28S rDNA and ITS regions. The remaining strains, CBS 318, CBS 2409, CBS 2993, and CBS 6578 could not be identified as any known Cryptococcus species or other yeast species by use of the BLAST system.
TABLE 3.
Matrices of overall similarities in ITS and D1-D2 regions of 28S rDNA for C. laurentii strains belonging to phylogenetic group I
| Species | Strain | % Similaritya
|
||||||
|---|---|---|---|---|---|---|---|---|
| JCM 9066 | CBS 2174 | CBS 8648 | CBS 318 | CBS 942 | CBS 8645 | G60 | ||
| C. laurentii | JCM 9066T (CBS 139) | 98.6 | 98.6 | 80.1 | 75.3 | 75.3 | 74.0 | |
| C. laurentii | CBS 2174 | 99.8 | 100 | 80.8 | 76.0 | 76.0 | 74.7 | |
| C. laurentii | CBS 8648 | 99.8 | 100 | 81.5 | 75.7 | 75.7 | 74.7 | |
| C. laurentii | CBS 318 | 97.5 | 97.5 | 97.5 | 91.2 | 91.2 | 90.2 | |
| C. laurentii | CBS 942 | 96.3 | 96.5 | 96.5 | 98.6 | 100 | 99.0 | |
| C. laurentii | CBS 8645 | 96.3 | 96.7 | 96.7 | 98.8 | 99.1 | 99.0 | |
| C. nodaensis | G60 | 96.3 | 96.7 | 96.3 | 98.4 | 99.8 | 98.9 | |
Data above the diagonal refer to the overall similarities of the ITS sequences, and data below the diagonal refer to the overall similarities of the D1 and D2 regions of 28S rDNA. Boldface indicates strains with approximately 99% or more similarity in both 28S rDNA and ITS sequences.
TABLE 4.
Matrices of overall similarities in ITS and D1-D2 regions of 28S rDNA for C. laurentii strains belonging to phylogenetic group II
| Species | Strain | % Similaritya
|
||||
|---|---|---|---|---|---|---|
| CBS 2409 | CBS 2993 | CBS 6578 | CBS 973 | CBS 445.92 | ||
| C. laurentii | CBS 2409 | 74.8 | 74.8 | 74.8 | 74.8 | |
| C. laurentii | CBS 2993 | 98.4 | 95.0 | 80.3 | 80.3 | |
| C. laurentii | CBS 6578 | 97.2 | 98.1 | 80.5 | 80.5 | |
| C. laurentii | CBS 973 | 96.8 | 97.9 | 97.4 | 100 | |
| T. papilionaceus | CBS 445.92 | 96.8 | 97.9 | 97.4 | 100 | |
Data above the diagonal refer to the overall similarities of the ITS sequences, and data below the diagonal refer to the overall similarities of the D1 and D2 regions of 28S rDNA. Boldface indicates strains with approximately 99% or more similarity in both 28S rDNA and ITS sequences.
Molecular phylogenetic analysis on the basis of partial 28S rDNA sequences.
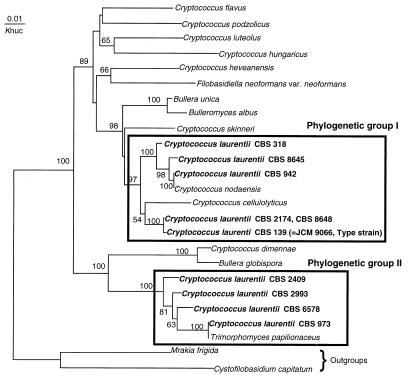
Figure 1 shows the molecular phylogenetic trees based on the D1 and D2 regions of the 28S rDNA sequences constructed by the neighbor-joining method. The 10 strains of C. laurentii fell into two major phylogenetic groups. One includes the type strain of C. laurentii, C. nodaensis, and C. cellulolyticus. The other group includes T. papilionaceus and is separated from the known Cryptococcus species. The two major groups are characterized by their nuclear DNA G+C contents (54.7 to 59.0 versus 50.8 to 52.3 mol%; Table 1).
FIG. 1.
Molecular phylogenetic tree based on the partial sequences of 28S rDNA. The tree was constructed by the neighbor-joining method. The numerals represent the confidence level from 100 replicate bootstrap samplings (frequencies less than 50% are not indicated). Knuc, Kimura's parameter (8).
Biochemical characteristics.
Table 5 shows the biochemical characteristics and biotypes of the 10 strains of C. laurentii obtained with the ID 32C kit. CBS 318 and CBS 942 were not identified as any known yeast species with this system. The biotype of CBS 973 indicated that it was C. humicolus. The remaining seven strains were identified as C. laurentii. No differences were found in the biochemical characteristics of the two major phylogenetic groups.
TABLE 5.
Biochemical characteristics of C. laurentii distinguished by the ID 32C kit
| Strain | ID 32C biotype | Reactiona
|
||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Galactose | Actidione | Saccharose | N-Acetylglucosamine | dl-Lactate | l-Arabinose | Cellobiose | Raffinose | Maltose | Trehalose | 2-Ketogluconate | α-Methyl-d-glucoside | Sorbitol | d-Xylose | Ribose | Glycerol | Rhamnose | Palatinose | Erythritol | Melibiose | Glucuronate | Melezitose | Gluconate | Levulinate | Glucose | Sorbose | Glucosamine | Mannitol | Lactose | Inositol | Esculine | ||
| JCM 9066T | 55777673 | + | − | + | + | − | + | + | + | + | + | + | + | + | + | + | + | + | + | − | + | + | + | + | + | + | + | − | + | − | + | − |
| CBS 2174 | 57777673 | + | − | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | − | + | + | + | + | + | + | + | − | + | − | + | − |
| CBS 8648 | 57777673 | + | − | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | − | + | + | + | + | + | + | + | − | + | − | + | − |
| CBS 942 | 54736663 | + | − | + | − | − | + | + | + | + | + | + | − | + | + | + | + | + | + | − | + | + | − | + | + | + | + | − | + | − | − | − |
| CBS 8645 | 54777663 | + | − | + | − | − | + | + | + | + | + | + | + | + | + | + | + | + | + | − | + | + | − | + | + | + | + | − | + | − | − | − |
| CBS 318 | 54737461 | + | − | + | − | − | + | + | + | + | + | + | − | + | + | + | + | + | + | − | − | + | − | + | + | + | − | − | + | − | − | − |
| CBS 973 | 57777777 | + | − | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | − |
| CBS 2409 | 55773673 | + | − | + | + | − | + | + | + | + | + | + | + | + | + | + | + | + | − | − | + | + | + | + | + | + | + | − | + | − | + | − |
| CBS 2993 | 55777673 | + | − | + | + | − | + | + | + | + | + | + | + | + | + | + | + | + | + | − | + | + | + | + | + | + | − | + | − | + | − | |
| CBS 6578 | 54777673 | + | − | + | − | − | + | + | + | + | + | + | + | + | + | + | + | + | + | − | + | + | + | + | + | + | + | − | + | − | − | − |
+, positive reaction; −, negative reaction.
DISCUSSION
The spectrum of opportunistic yeast pathogens appears to have become broader during the past few decades, accompanying an increase in the number of immunocompromised patients. Species considered rare pathogens or nonpathogenic yeasts have been reported as a cause of disease. For example, C. laurentii is responsible for both deep-seated infections, such as fungemia and meningitis, and superficial infections, such as keratitis (6, 7, 9, 14). Our analysis of the 28S rDNA and ITS sequences revealed the diversity of C. laurentii. Although we examined only 10 strains of C. laurentii, they consisted of seven taxonomically distinct species, and examination of a larger number of strains would probably reveal more species. The C. laurentii clinical isolates were also heterogeneous, although only four strains of this species were examined. Of the strains examined, strain CBS 8645 was the first C. laurentii strain isolated; it was isolated by Kordossis et al. (9) from an AIDS patient with meningitis in 1998. We reidentified this clinical isolate as C. nodaensis from sequence comparisons. An accurate and rapid way to identify rare yeast pathogens such as C. laurentii has not been established. The taxonomies of these species are also not generally well known. It is difficult to distinguish species of C. laurentii with the ID 32C kit, and sequence analysis of the 28S rDNA or ITS region is required for differentiation. With the reclassification of pathogenic yeast, the causative agents of mycoses can be determined only at the species level in a few cases. Trichosporon cutaneum (or Trichosporon beigelii) has long been believed to be the major causative agent of trichosporonosis. In 1992, Guého et al. (4) revised the taxonomy of the genus Trichosporon and indicated that T. cutaneum consists of more than 10 species. Using the new classification of the genus Trichosporon, some investigators (3, 5, 17, 18) subsequently reported that the major causative agent of trichosporonosis differs in each type of infection. For instance, Trichosporon asahii and Trichosporon mucoides are involved in deep-seated infections and Trichosporon asteroides and T. cutaneum are associated with superficial infections. These four species were previously classified as T. cutaneum. Sullivan et al. (20) recently described an atypical Candida albicans strain associated with oral candidiasis in human immunodeficiency virus-infected and AIDS patients as a new species, Candida dubliniensis. C. dubliniensis was recovered from the oral cavities of 27% of human immunodeficiency virus-infected individuals and 32% of AIDS patients presenting with symptoms of oral candidiasis. Moran et al. (13) reported that 20% of oral isolates (fluconazole MIC, 8 to 32 μg/ml) of C. dubliniensis recovered from AIDS patients who had previously been treated with fluconazole were fluconazole resistant. Furthermore, fluconazole-susceptible clinical isolates of this species expressed a fluconazole resistance phenotype (fluconazole MIC, 16 to 64 μg/ml) with sequential exposures to increasing concentrations of this drug. As mentioned above, the reclassification of C. albicans-C. dubliniensis and T. cutaneum is clinically significant. It is not yet known whether our finding of intraspecies diversity of C. laurentii is clinically significant, but this will become clear as more examples of this species are isolated from patients.
Johnson et al. (6) reviewed the literature on C. laurentii infection. They investigated three cases of fungemia and five cases of nonbloodstream infection. All the patients were treated with amphotericin B or fluconazole and had good prognoses. There are not many reports on the MICs of antifungal drugs for C. laurentii. Ryder et al. (15) reported that the MIC of fluconazole was 1 to 4 μg/ml (n = 7), and that that of terbinafine was 0.125 to 0.25 μg/ml (n = 7) for C. laurentii. These MICs are similar to those for C. neoformans. The MIC of amphotericin B (n = 3) was 0.037 to 0.25 μg/ml, while C. laurentii was resistant to flucytosine (n = 2), although it is not known whether the C. laurentii clinical isolates tested were true C. laurentii species or other yeast species. It appears that fungal infection due to this species does not progress and become life threatening. Infections are potentially transmitted via pulmonary routes or intravenous catheters. Recently, Mattsson et al. (12) isolated C. laurentii from feral pigeons at a high frequency (69%; 24 of 35), suggesting that these birds are a possible reservoir of infection.
In conclusion, C. laurentii is a genetically heterogeneous species, and this must be taken into consideration when identifying C. laurentii clinical isolates.
REFERENCES
- 1.Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 2.Guého E, Improvisi L, Christen R, de Hoog G S. Phylogenetic relationships of Cryptococcus neoformans and some related basidiomycetous yeasts determined from partial large subunit rRNA sequences. Antonie Leeuwenhoek. 1993;63:75–89. doi: 10.1007/BF00872392. [DOI] [PubMed] [Google Scholar]
- 3.Guého E, Improvisi L, Hoog de G S, Dupont B. Trichosporon on humans: a practical account. Mycoses. 1994;37:3–10. doi: 10.1111/j.1439-0507.1994.tb00277.x. [DOI] [PubMed] [Google Scholar]
- 4.Guého E, Smith M T, Hoog de G S, Grand G B, Christen R, Batenburg-van der Vegte W H. Contributions to a revision of the genus Trichosporon. Antonie Leeuwenhoek. 1992;61:289–316. doi: 10.1007/BF00713938. [DOI] [PubMed] [Google Scholar]
- 5.Herbrecht R, Koening H, Waller K, Liu L, Guého E. Trichosporon infections: clinical manifestations and treatment. J Mycol Med. 1993;3:129–136. [Google Scholar]
- 6.Johnson L B, Suzanne F B, Kauffman A. Fungaemia due to Cryptococcus laurentii and a review of non-neoformans cryptococcaemia. Mycoses. 1998;41:277–280. doi: 10.1111/j.1439-0507.1998.tb00338.x. [DOI] [PubMed] [Google Scholar]
- 7.Kamalam A, Yesudian P, Thambiah A S. Cutaneous infection by Cryptococcus laurentii. Br J Dermatol. 1977;97:221–223. doi: 10.1111/j.1365-2133.1977.tb15070.x. [DOI] [PubMed] [Google Scholar]
- 8.Kimura M. A simple method for estimation evolutionary rate of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1980;16:111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- 9.Kordossis T, Avlami A, Velegraki A, Stefanou I, Georgakopoulos G, Papalambrou C, Legakis N J. First report of Cryptococcus laurentii meningitis and a fatal case of Cryptococcus albidus cryptococcaemia in AIDS patients. Med Mycol. 1998;36:335–339. [PubMed] [Google Scholar]
- 10.Kurtzman C P, Robnett C J. Identification of clinically important ascomycetous yeasts based on nucleotide divergence in the 5′ end of the large-subunit (26S) ribosomal DNA gene. J Clin Microbiol. 1997;35:1216–1223. doi: 10.1128/jcm.35.5.1216-1223.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Makimura K, Murayama Y S, Yamaguchi H. Detection of a wide range of medically important fungal species by polymerase chain reaction (PCR) J Med Microbiol. 1994;40:358–364. doi: 10.1099/00222615-40-5-358. [DOI] [PubMed] [Google Scholar]
- 12.Mattsson R, Haemig P D, Olsen B. Feral pigeons as carriers Cryptococcus laurentii, Cryptococcus uniguttulatus and Debaryomyces hansenii. Med Mycol. 1999;37:367–369. doi: 10.1046/j.1365-280x.1999.00241.x. [DOI] [PubMed] [Google Scholar]
- 13.Moran G P, Sullivan D J, Henman M C, McCreary C E, Harrington B J, Shanley D B, Coleman D C. Antifungal drug susceptibilities of oral Candida dubliniensis isolates from human immunodeficiency virus (HIV)-infected and non-HIV-infected subjects and generation of stable fluconazole-resistant derivatives in vitro. Antimicrob Agents Chemother. 1997;41:617–623. doi: 10.1128/aac.41.3.617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ritterband D C, Seedor J A, Shah M K, Waheed S, Schorr I. A unique case of Cryptococcus laurentii keratitis spread by a rigid gas permeable contact lens in a patient with onychomycosis. Cornea. 1998;17:115–118. doi: 10.1097/00003226-199801000-00017. [DOI] [PubMed] [Google Scholar]
- 15.Ryder N S, Wagner S, Leitner I. In vitro activities of terbinafine against cutaneous isolates of Candida albicans and other pathogenic yeasts. Antimicrob Agents Chemother. 1998;42:1057–1061. doi: 10.1128/aac.42.5.1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Saitou N, Nei M. Neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 17.Sugita T, Nishikawa A, Shinoda T, Kume H. Taxonomic position of deep-seated, mucosa-associated, and superficial isolates of Trichosporon cutaneum from trichosporonosis patients. J Clin Microbiol. 1995;33:1368–1370. doi: 10.1128/jcm.33.5.1368-1370.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sugita T, Nishikawa A, Shinoda T, Kusunoki T. Taxonomic studies on clinical isolates from superficial trichosporonosis patients by DNA relatedness. Jpn J Med Mycol. 1996;37:107–110. [Google Scholar]
- 19.Sugita T, Nishikawa A, Ikeda R, Shinoda T. Identification of medically relevant Trichosporon species based on sequences of internal transcribed spacer regions and construction of a database for Trichosporon identification. J Clin Microbiol. 1999;37:1985–1993. doi: 10.1128/jcm.37.6.1985-1993.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sullivan D J, Westerneng T J, Haynes K A, Bennett D E, Coleman D C. Candida dubliniensis sp. nov.: phenotypic and molecular characterization of a novel species associated with oral candidosis in HIV-infected individuals. Microbiology. 1995;141:1507–1521. doi: 10.1099/13500872-141-7-1507. [DOI] [PubMed] [Google Scholar]
- 21.Takashima M, Nakase T. Molecular phylogeny of the genus Cryptococcus and related species based on the sequences of 18S rDNA and internal transcribed spacer regions. Microbiol Cult Coll. 1999;15:33–45. [Google Scholar]
- 22.Thompson J, Hompson J D, Higgins D G, Gibson T J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vancanneyt M, Coopman R, Tytgat R, Hennebert G L, Kersters K. Whole-cell protein patterns, DNA base compositions and coenzyme Q types in the yeast genus Cryptococcus Kützing and related taxa. Syst Appl Microbiol. 1994;17:65–75. [Google Scholar]
- 24.Velez A, Fernandez-Roldan J C, Linares M, Casal M. Melanonychia due to Candida humicola. Br J Dermatol. 1996;134:375–376. doi: 10.1111/j.1365-2133.1996.tb07639.x. [DOI] [PubMed] [Google Scholar]