Abstract
This study explores the role of the long noncoding RNA (LncRNA) CRNDE in cisplatin (CDDP) resistance of gastric cancer (GC) cells. Here, we show that LncRNA CRNDE is upregulated in carcinoma tissues and tumor‐associated macrophages (TAMs) of GC patients. In vitro experiments show that CRNDE is enriched in M2‐polarized macrophage‐derived exosomes (M2‐exo) and is transferred from M2 macrophages to GC cells via exosomes. Silencing CRNDE in M2‐exo reverses the promotional effect of M2‐exo on cell proliferation in CDDP‐treated GC cells and homograft tumor growth in CDDP‐treated nude mice. Mechanistically, CRNDE facilitates neural precursor cell expressed developmentally downregulated protein 4‐1 (NEDD4‐1)‐mediated phosphatase and tensin homolog (PTEN) ubiquitination. Silencing CRNDE in M2‐exo enhances the CDDP sensitivity of GC cells treated with M2‐exo, which is reduced by PTEN knockdown. Collectively, these data reveal a vital role for CRNDE in CDDP resistance of GC cells and suggest that the upregulation of CRNDE in GC cells may be attributed to the transfer of TAM‐derived exosomes.
Keywords: cisplatin resistance, exosome, gastric cancer, LncRNA CRNDE, tumor‐associated macrophages
Subject Categories: Cancer, Membranes & Trafficking, RNA Biology
LncRNA CRNDE is transferred from M2‐polarized macrophages to GC cells via exosomes, suppressing PTEN expression in GC cells. The latter leads to a reduced sensitivity of GC cells to cisplatin.
Introduction
Gastric cancer (GC) is one of the most common malignant carcinomas diagnosed worldwide and seriously jeopardizes human health. In China, GC is the second leading cause of cancer death after lung cancer (Chen et al, 2016; Huang et al, 2020). The first‐line therapy for GC involves surgical resection, radiotherapy, and chemotherapeutic agents (such as cisplatin [CDDP]), which typically produce good treatment outcomes in the early stages of cancer. However, resistance to CDDP occurs in advanced cancer stages is a common cause of relapse and metastasis of GC (Orditura et al, 2014). Therefore, determining the molecular mechanisms of CDDP resistance is of great importance for improving GC patient survival.
Tumor‐associated macrophages (TAMs), the major component of the tumor microenvironment (TME), play an essential role in the occurrence, development, metastasis, and chemoresistance of tumors (Qian & Pollard, 2010; Noy & Pollard, 2014). In GC patients, TAMs are polarized to the M2 phenotype and contribute to tumor cell proliferation and a poor prognosis (Yamaguchi et al, 2016; Xu et al, 2019). In addition, increasing evidence has shown that TAMs may mediate the drug resistance of cancers via secreting exosomes. For instance, TAM‐derived exosomes confer gemcitabine resistance in pancreatic adenocarcinoma by transferring microRNA (miRNA)‐365 (Binenbaum et al, 2018). TAM‐derived exosomes deliver miR‐21 to GC cells to induce CDDP resistance (Zheng et al, 2017). Except for miRNAs, several long noncoding RNAs (LncRNAs) can also be packaged into exosomes and function as messengers in cell‐to‐cell communication (Sun et al, 2018b). Takahashi et al (2014) report that exosomal Linc‐VLDLR transferred by tumor cell‐derived exosomes regulates the sorafenib resistance in hepatocellular carcinoma. However, it remains unclear whether exosomal LncRNAs transferred by TAM‐derived exosomes are involved in CDDP resistance in GC.
By detecting the expression levels of several LncRNAs that are reportedly packaged into exosomes, we found that LncRNA CRNDE is significantly upregulated in M2‐polarized macrophage‐derived exosomes (M2‐exo) compared with monocyte‐derived exosomes (monocytes‐exo). Further, utilizing human/mouse GC cell lines and a homograft mouse model, we show that LncRNA CRNDE induces CDDP resistance in GC. The upregulation of CRNDE in GC cells might be caused by the transfer of TAM‐derived exosomes.
Results
LncRNA CRNDE is enriched in TAMs of GC patients
Several LncRNAs have been reported to be packed into the exosomes secreted by cancer cells and thereby modulate the pathological process of cancer (Sun et al, 2018b). To determine whether these LncRNAs also play roles in TAM‐derived exosomes, we first assessed their expression levels in monocytes‐exo and M2‐exo. The results depicted in Fig EV1 indicated that the expression levels of LincRNA‐ROR, LncRNA MALAT1, LncRNA MEG3, LncRNA‐91H, LncRNA CRNDE, LncRNA p21, and LncRNA TUC339 changed in M2‐exo. Of these, the expression level of LncRNA CRNDE demonstrated the most significant change. Next, LncRNA CRNDE expression levels were measured in both the cancerous tissues and adjacent normal tissues obtained from 35 GC patients. The results showed that LncRNA CRNDE level was higher in GC cancerous tissues compared with adjacent normal tissues, and the high expression level of LncRNA CRNDE was positively correlated with the proportion of CD63+CD163+ M2 macrophages in GC cancerous tissues (r = 0.8434) (Fig 1A and B). The serum level of LncRNA CRNDE was higher in GC patients than in healthy controls (Fig EV2A). Compared with NTMs, TAMs showed higher expression levels of LncRNA CRNDE (Fig 1C).
Figure EV1. LncRNA CRNDE is upregulated in M2‐polarized macrophage‐derived exosomes (M2‐exo).
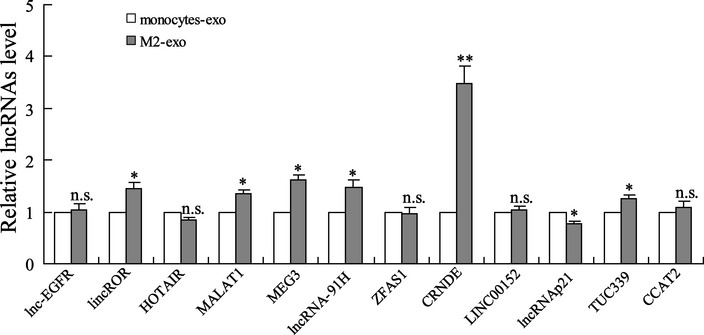
LncRNA expressions in monocytes‐derived exosomes (monocytes‐exo) and M2‐exo were measured by qRT–PCR. The results shown are from three biological replicates and are expressed as mean ± SD. Student’s t‐test. *P < 0.05, **P < 0.01 vs. monocytes‐exo.
Figure 1. LncRNA CRNDE is enriched in tumor‐associated macrophages of gastric cancer patients.
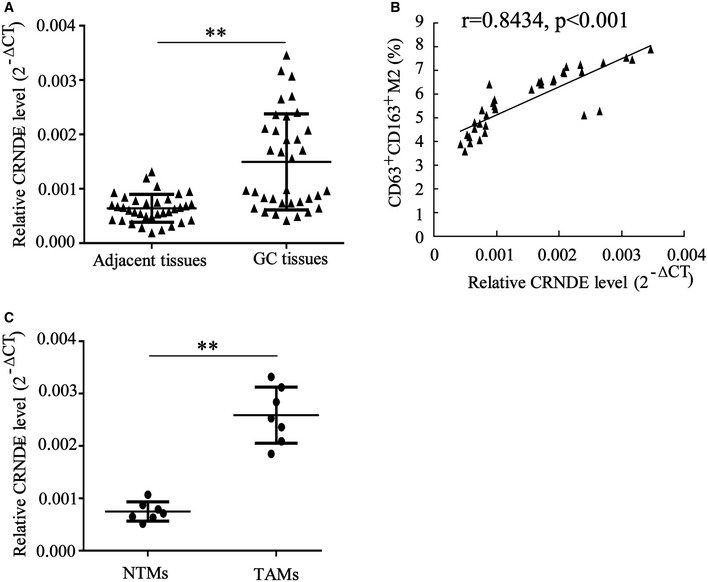
- LncRNA CRNDE expression level was measured by qRT–PCR in cancerous tissues and adjacent normal tissues obtained from 35 cases of gastric cancer (GC) patients.
- The correlation between LncRNA CRNDE expression and the proportion of CD63+CD163+ M2 macrophages in GC cancerous tissues (n = 35) was determined by the Pearson correlation coefficient.
- LncRNA CRNDE expression level was measured by qRT–PCR in adjacent normal tissue‐derived macrophages (NTMs) and tumor‐associated macrophages (TAMs) obtained from 7 cases of GC patients.
Data information: Data are expressed as mean ± SD. A and C: Student’s t‐test; B: Spearman’s correlation analysis. **P < 0.01.
Figure EV2. Overexpressed LncRNA CRNDE in M2‐exo‐treated GC cells was transferred via exosomes rather than diffusion from the media due to exosomes dissociation.
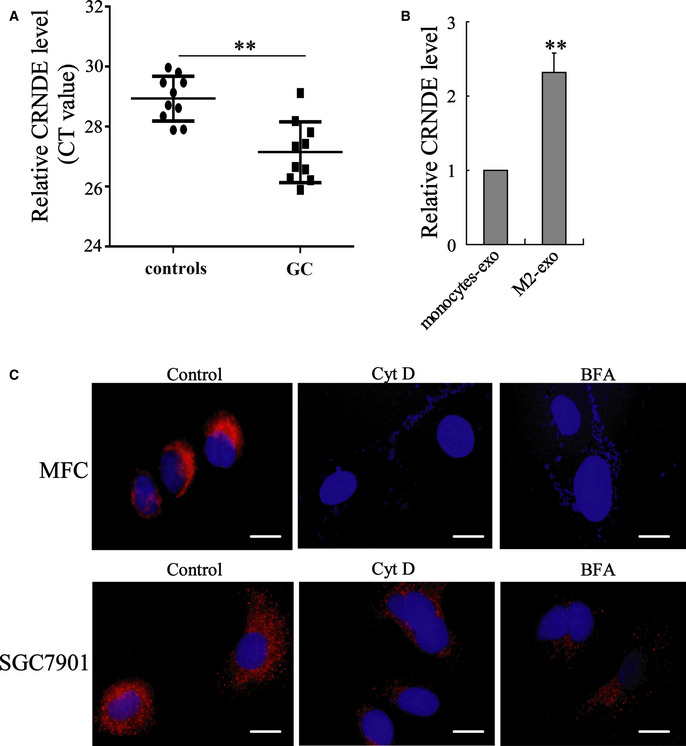
- Relative LncRNA CRNDE level in GC patients (n = 10) and healthy controls (n = 10) (the serum CRNDE levels were shown as CT values, and higher CT values indicate lower CRNDE levels). Data are expressed as mean ± SD. Student’s t‐test.
- LncRNA CRNDE expression in MGC‐803 cells incubated with monocytes‐exo or M2‐exo was measured by qRT–PCR. **P < 0.01 vs. monocytes‐exo. The results shown are from three biological replicates and are expressed as mean ± SD. Student’s t‐test.
- MFC and SGC7901 cells were incubated with Cy3‐CRNDE‐EXO (red) and cytochalasin D (Cyt D; 10 mg/ml) or brefeldin A (BFA; 20 mg/ml). Confocal microscopy was used to detect the Cy3‐labeled CRNDE in cells. The nuclei were stained with DAPI (blue). Scale bar = 20 µm.
LncRNA CRNDE is enriched in M2‐polarized macrophages and in M2‐exo
Mouse BMDMs and human peripheral blood monocytes were induced toward an M2‐polarized phenotype by the stimulation of IL‐4 plus IL‐13. As shown in Fig 2A, a significant elevation in LncRNA CRNDE expression was observed in BMDM‐polarized M2 macrophages compared with BMDMs. The results of TEM and Western blot shown in Fig 2C confirmed that the purified exosomes from the media of BMDMs or BMDM‐polarized M2 macrophages all exhibited a typical morphology and size of exosomes and expressed the specific marks of exosomes (CD63 and CD81 (Zhang & Grizzle, 2014)). In addition, more abundant LncRNA CRNDE was detected in M2‐exo compared with BMDMs‐exo (Fig 2C). Similarly, compared with monocytes and monocytes‐exo, LncRNA CRNDE expression levels were higher in monocyte‐polarized M2 macrophages and M2‐exo, respectively (Fig 2B and D).
Figure 2. LncRNA CRNDE is enriched in M2‐polarized macrophages and in M2‐polarized macrophage‐derived exosomes (M2‐exo).
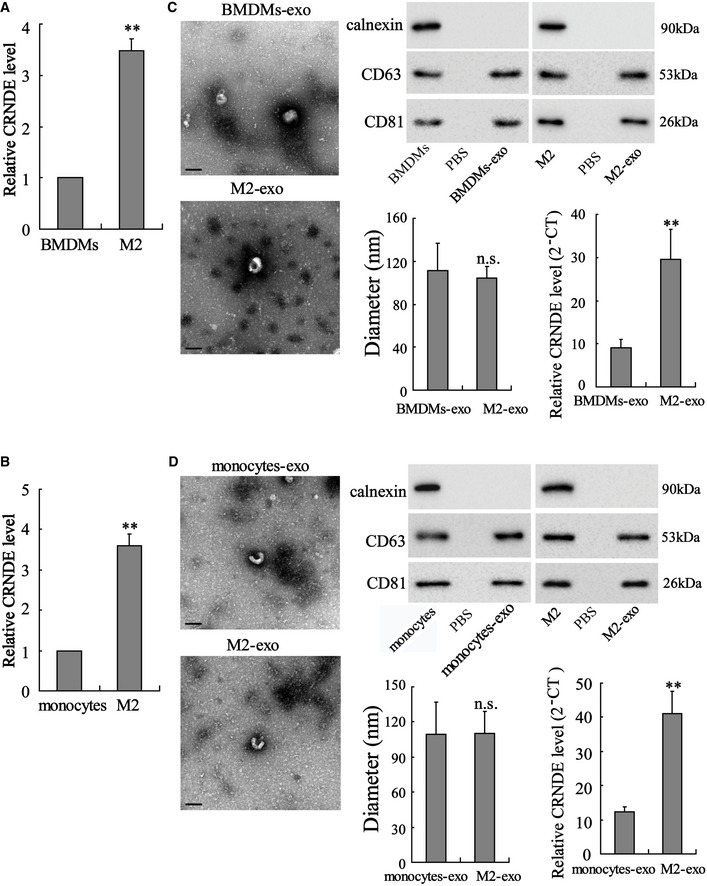
-
A, BMouse bone marrow‐derived macrophages (BMDMs) and human peripheral blood monocytes were induced toward an M2‐polarized phenotype by the stimulation of 20 ng/ml interleukin 4 (IL‐4) plus 20 ng/ml IL‐13. LncRNA CRNDE expression level in (A) BMDMs, BMDM‐polarized M2 macrophages, (B) monocytes, and monocyte‐polarized M2 macrophages was detected by qRT–PCR.
-
C, DLeft: Representative electron micrograph of exosomes isolated from the medium of BMDMs, BMDM‐polarized M2 macrophages, monocytes, and monocyte‐polarized M2 macrophages is depicted, showing the typical morphology of exosomes. Upper right: The protein levels of CD63 and CD81 were measured by Western blot. Lower left: Nanoparticle tracking analysis showed that most of the particles had a similar size of 50–150 nm in diameter with a peak at around 100 nm. Lower right: LncRNA CRNDE expression levels in different exosomes.
Data information: Scale bar =100 nm. The results shown are from three biological replicates. Data are expressed as mean ± SD. Student’s t‐test. **P < 0.01 vs. BMDMs or monocytes or BMDMs‐exo or monocytes‐exo.
Source data are available online for this figure.
Transport of exosomes might cause upregulation of LncRNA CRNDE in GC cells
The mouse GC cell line MFC was incubated with BMDMs‐exo or M2‐exo. As shown in Fig 3A, M2‐exo, rather than BMDMs‐exo, effectively upregulated LncRNA CRNDE expression in MFC cells. To further confirmed whether M2‐exo transferred LncRNA CRNDE to MFC cells, BMDM‐polarized M2 macrophages were transfected with Cy3‐labeled CRNDE, and the exosomes were isolated from the medium (Cy3‐CRNDE‐EXO). The results of Fig 3B depicted that Cy3‐labeled CRNDE could be detected in the cytoplasm of MFC cells incubated with Cy3‐CRNDE‐EXO. MFC cells incubated with Cy3‐CRNDE‐EXO showed a higher ratio of Cy3‐positive cells than MFC cells incubated with control EXO (Fig 3B) Then, we repeated these experiments with human peripheral blood monocytes, monocyte‐polarized M2 macrophages, and human GC cell lines (SGC7901 and MGC‐803). Similarly, exosomes derived from monocyte‐polarized M2 macrophages also successfully transferred LncRNA CRNDE from monocyte‐polarized M2 macrophages to human GC cell lines (Figs 3C and D, and EV2, 2B and C). In addition, to further confirm the overexpressed CRNDE in M2‐exo‐treated GC cells was transferred via exosomes rather than diffusion from the media due to disintegrating exosomes, MFC and SGC7901 cells were incubated with Cy3‐CRNDE‐EXO and Cyt D (an endocytosis inhibitor (Smani et al, 2006))/ BFA (a vesicle trafficking inhibitor; Paponov et al, 2019). As shown in Fig EV2C, both Cyt D and BFA suppressed the expression of Cy3‐labeled CRNDE in GC cells, which confirmed that exosomal LncRNA CRNDE was transferred to GC cells via exosomes. As reported, there are mainly four pathways that meditate the endocytic pathway of exosomes: clathrin‐mediated endocytosis, caveolae‐mediated endocytosis, micropinocytosis, and lipid raft‐mediated endocytosis (Tian et al, 2014). To further confirm which endocytic pathway mediated the entry of M2 macrophage‐derived exosomes to GC cells, SGC7901 cells were incubated with chlorpromazine (CPZ, the inhibitor of Clathrin‐mediated endocytosis), genistein (GEN, the inhibitor of caveolae‐mediated endocytosis), 5‐(n‐ethyl‐n‐isopropyl)‐amiloride (EIPA, the inhibitor of micropinocytosis), or methyl‐β‐cyclodextrin (MβCD, the inhibitor of lipid raft‐mediated endocytosis), following by the incubation of Cy3‐CRNDE‐EXO. The results showed that only CPZ treatment distinctly inhibited the uptake of exosomes by SGC7901 cells (Fig 3E) and the inhibitory effect of CPZ on the uptake of exosomes by cells was dose‐dependent (Fig 3F).
Figure 3. Transport of exosomes might cause upregulation of LncRNA CRNDE in GC cells.
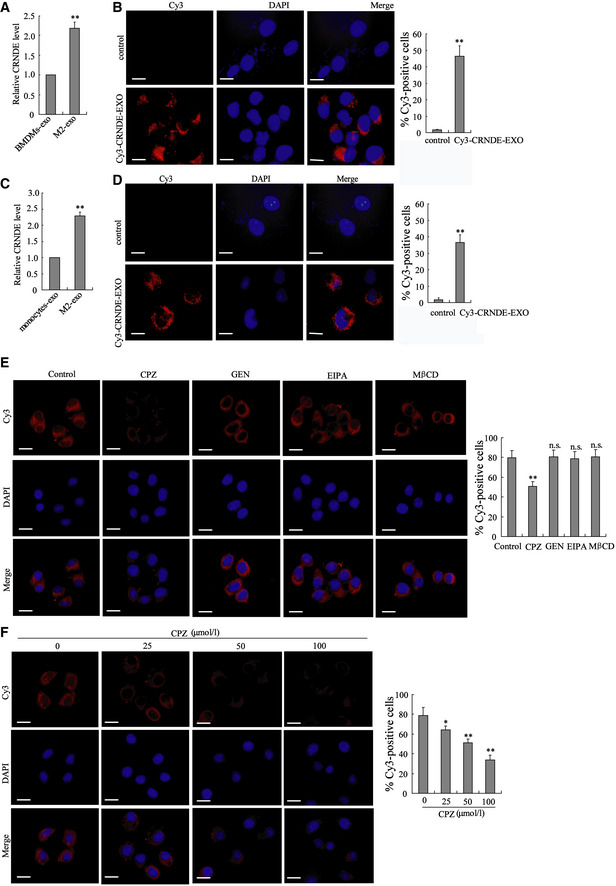
- LncRNA CRNDE expression in MFC cells incubated with BMDMs‐exo or M2‐exo was measured by qRT–PCR. **P < 0.01 vs. BMDMs‐exo.
- BMDM‐polarized M2 macrophages were transfected with Cy3‐labeled CRNDE, and exosomes were isolated from the medium (Cy3‐CRNDE‐EXO). Then, MFC cells were incubated with Cy3‐CRNDE‐EXO. Left: Confocal microscopy was used to detect the Cy3‐labeled CRNDE in MFC cells. The nuclei were stained with DAPI. Scale bar = 20 µm. Right: Flow cytometry was performed to quantitate the Cy3‐positive cells. **P < 0.01 vs. MFC cells incubated with the control EXO.
- LncRNA CRNDE expression in SGC7901 cells incubated with monocytes‐exo or M2‐exo was measured by qRT–PCR. **P < 0.01 vs. monocytes‐exo.
- Monocyte‐polarized M2 macrophages were transfected with Cy3‐labeled CRNDE, and exosomes were isolated from the medium (Cy3‐CRNDE‐EXO). Then, SGC7901 cells were incubated with Cy3‐CRNDE‐EXO. Confocal microscopy was used to detect the Cy3‐labeled CRNDE in SGC7901 cells. The nuclei were stained with DAPI. Scale bar = 20 µm. Right: Flow cytometry was performed to quantitate the Cy3‐positive cells. **P < 0.01 vs. SGC7901 cells incubated with the control EXO.
- SGC7901 cells were incubated with chlorpromazine (CPZ, 50 μM), genistein (GEN, 200 μM), 5‐(n‐ethyl‐n‐isopropyl)‐amiloride (EIPA, 100 μM), or methyl‐β‐cyclodextrin (MβCD, 10 mM), following by the incubation of Cy3‐CRNDE‐EXO. Confocal microscopy was used to detect the Cy3‐labeled CRNDE in SGC7901 cells. The nuclei were stained with DAPI. Right: Flow cytometry was performed to quantitate the Cy3‐positive cells.
- SGC7901 cells were incubated with 0, 25, 50, and 100 μM CPZ, following by the incubation of Cy3‐CRNDE‐EXO. Confocal microscopy was used to detect the Cy3‐labeled CRNDE in SGC7901 cells. The nuclei were stained with DAPI. Right: Flow cytometry was performed to quantitate the Cy3‐positive cells.
Data information: The results shown are from three biological replicates. Data are expressed as mean ± SD. A–D: Student’s t‐test. E and F: one‐way ANOVA.
LncRNA CRNDE induces CDDP resistance in GC cells
Next, we explored the influence of M2‐exo on CDDP resistance in GC. To this end, MFC cells were treated with exosomes derived from BMDM‐polarized M2 macrophages/BMDMs and CDDP. Compared with BMDMs‐exo, the M2‐exo prominently boosted the cell survival rate (Figs 4B and EV3A), accelerated colony formation (Fig 4C), and suppressed cell apoptosis (Fig 4D) in CDDP‐treated MFC cells. Considering that M2‐exo presented a higher LncRNA CRNDE level than BMDMs‐exo (Fig 4A) and that LncRNA CRNDE has been proven to be a tumor promoter in GC (Du et al, 2017), we thus wondered whether M2‐exo elicited CDDP resistance by transferring LncRNA CRNDE to GC cells. To investigate this question, BMDM‐polarized M2 macrophages were transfected with si‐control or si‐CRNDE and the exosomes (M2‐si‐control‐exo or M2‐si‐CRNDE‐exo) were isolated from the respective medium. Figure 4A proved that LncRNA CRNDE expression in M2‐si‐CRNDE‐exo was successfully repressed by si‐CRNDE transfection. Furthermore, in CDDP‐treated MFC cells, M2‐si‐CRNDE‐exo efficiently reduced the cell survival rate (Fig 4B), slowed down colony formation (Fig 4C), and boosted cell apoptosis (Fig 4D). Similarly, M2‐si‐CRNDE‐exo derived from monocyte‐polarized M2 macrophages also enhanced the sensitivity of SGC7901 and MGC‐803 cells to CDDP treatment (Figs 4E–H and EV3B and C). The aforementioned results suggest that the silencing of LncRNA CRNDE in M2‐polarized macrophages could restore the sensitivity of GC cells to CDDP.
Figure 4. LncRNA CRNDE induces cisplatin resistance in GC cells.

-
A–DExosomes were isolated from the media of BMDMs (BMDMs‐exo), BMDM‐polarized M2 macrophages (M2‐exo), BMDM‐polarized M2 macrophages transfected with si‐CRNDE (M2‐si‐CRNDE‐exo), and BMDM‐polarized M2 macrophages transfected with the negative control of si‐CRNDE (M2‐si‐control‐exo). Then, MFC cells were treated with indicated exosomes and cisplatin (CDDP; 1 μg/ml) for 48 h. (A) LncRNA CRNDE expression in indicated exosomes was detected by qRT–PCR. (B) The survival rate of MFC cells was measured by cell counting kit‐8 (CCK‐8) assay. (C) Representative images of colony formation assay performed on MFC cells. (D) Cell apoptosis was measured by flow cytometry.
-
E–HExosomes were isolated from the medium of monocytes (monocytes‐exo), monocyte‐polarized M2 macrophages (M2‐exo), monocyte‐polarized M2 macrophages transfected with si‐CRNDE (M2‐si‐CRNDE‐exo), and monocyte‐polarized M2 macrophages transfected with si‐control (M2‐si‐control‐exo). Then, SGC7901 cells were treated with indicated exosomes and CDDP (1 μg/ml). (E) LncRNA CRNDE expression, (F) cell survival rate, (G) cell proliferation, and (H) cell apoptosis were measured.
Data information: (A, D, E, and H): The results shown are from three biological replicates. (B and F): The results shown are from seven biological replicates. Data are expressed as mean ± SD. (A, B, D, E, F, and H): one‐way ANOVA. **P < 0.01.
Figure EV3. The exosomal transfer of LncRNA CRNDE induces CDDP resistance in GC cells.
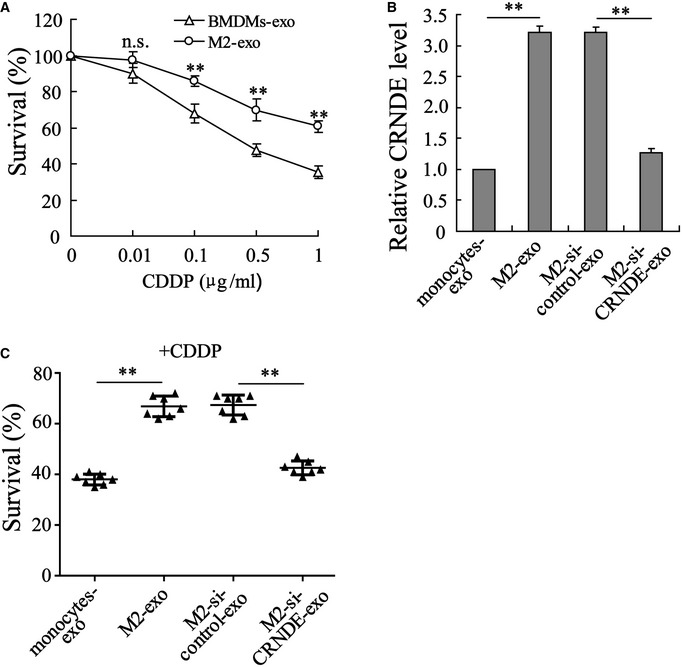
-
AMFC cells were incubated with CDDP (0, 0.01, 0.1, 0.5, and 1 µg/ml) and BMDMs‐exo/M2‐exo, and then, the cell survival rate was measured.
-
B, CExosomes were isolated from the medium of monocytes (monocytes‐exo), monocyte‐polarized M2 macrophages (M2‐exo), monocyte‐polarized M2 macrophages transfected with si‐CRNDE (M2‐si‐CRNDE‐exo), and monocyte‐polarized M2 macrophages transfected with si‐control (M2‐si‐control‐exo). Then, MGC‐803 cells were treated with indicated exosomes and CDDP (1 μg/ml) for 48 h. (B) LncRNA CRNDE expression and (C) cell survival rate were measured.
Data information: The results shown are from three biological replicates and are expressed as mean ± SD. (A and B): one‐way ANOVA; (C): Student’s t‐test. (A): **P < 0.01 vs. BMDMs‐exo; (B, C): **P < 0.01
LncRNA CRNDE induces CDDP resistance in vivo
Mouse GC cell line MFC was used to establish a nude mouse model of subcutaneous GC. BMDMs‐exo, M2‐exo, M2‐si‐control‐exo, or M2‐si‐CRNDE‐exo was injected into the homograft tumors of nude mice, followed by CDDP treatment. Figure 5D indicated that LncRNA CRNDE levels were higher in M2‐exo‐injected homograft tumors whereas lower in M2‐si‐CRNDE‐exo‐injected homograft tumors. Interestingly, before the CDDP and exosome treatment, the homograft tumors grew at a similar rate in each mouse (the tumor volume of each mouse was around 400 mm3 on Day 0; Fig 5A). However, in response to the M2‐exo injection, the growth rate of the homograft tumors was clearly facilitated, and the weight of homograft tumors increased (Fig 5A–C), which suggested that M2‐exo‐treated homograft tumors exhibited lower sensitivity to CDDP treatment than BMDMs‐exo‐treated homograft tumors. Of note, the promotion effect of M2‐exo on tumor growth during CDDP treatment was eliminated after down‐regulating LncRNA CRNDE expression (Fig 5A–C). In addition, the results of the PDX model also showed that compared with M2‐si‐control‐exo, M2‐si‐CRNDE‐exo lessened the volume of xenografts tumors after CDDP treatment (Fig EV4). The nude mouse model of subcutaneous GC received the injection of si‐CRNDE (in PBS media) or its negative control (PBS) before CDDP treatment. As shown in Fig EV5A–C, si‐CRNDE injection failed to decrease tumor volume, tumor weight, and CRNDE expression in the homograft tumors of CDDP‐treated mice.
Figure 5. LncRNA CRNDE induces CDDP resistance in vivo .
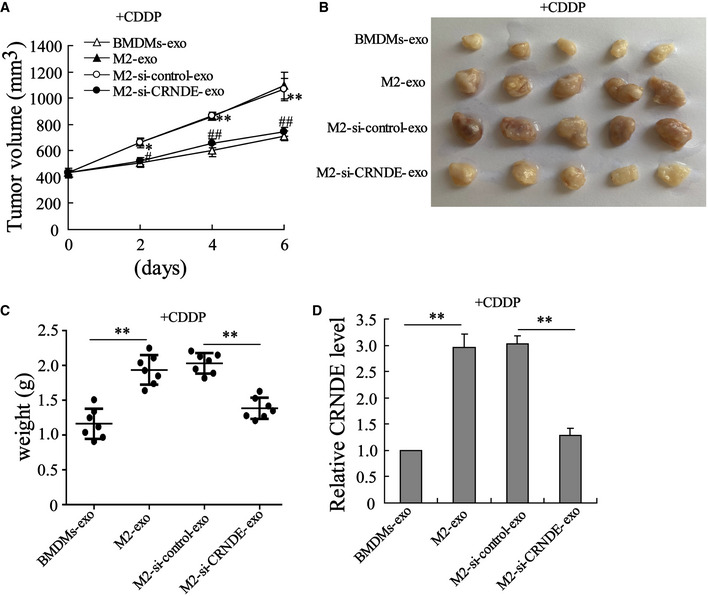
-
AGrowth curves of MFC subcutaneous homograft tumors (The homograft tumor sizes were recorded from the day of CDDP treatment). *P < 0.05, **P < 0.01 vs. BMDMs‐exo; #P < 0.05, ##P < 0.01 vs. M2‐si‐control‐exo.
-
B, CThe homograft tumors (B) photos and (C) weight of each mouse on day 6 after CDDP injection. **P < 0.01.
-
DLncRNA CRNDE expression in homograft tumors of mice. **P < 0.01.
Data information: Data are expressed as mean ± SD. (A, C and D): one‐way ANOVA.
Figure EV4. Patient‐derived xenograft model.
The tumor photos of xenografts tumors treated with CDDP + BMDMs‐exo, CDDP + M2‐exo, CDDP + M2‐si‐control‐exo, or CDDP + M2‐si‐CRNDE‐exo.
Figure EV5. si‐CRNDE fails to affect CDDP sensitivity of cancer cells in vivo and the interplay between LncRNA CRNDE, PTEN, and NEDD4‐1.
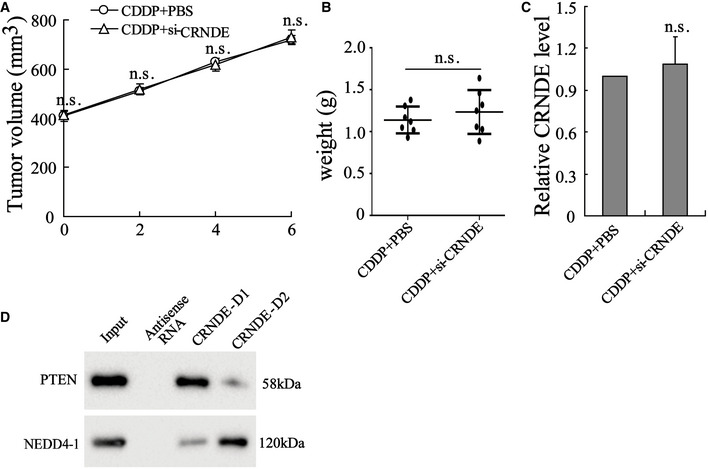
Mice were divided into CDDP + PBS and CDDP + si‐CRNDE groups. 3 × 105 MFC cells resuspended in 200 µl PBS were subcutaneously injected into nude mice; ten days after injection, si‐CRNDE (in PBS media) or its negative control (PBS) was injected into the center of the homograft tumors of mice, followed by a single intraperitoneal injection of CDDP. The (A) tumor volume, (B) tumor weight, and (C) CRNDE expression in the homograft tumors of CDDP‐treated mice. (D) Biotinylated CRNDE‐D1 (1–500 nt)/CRNDE‐D2 (501–1,059 nt) RNAs were incubated with SGC7901 cell lysates, followed by Western blot. The results shown are from three biological replicates and are expressed as mean ± SD.
Source data are available online for this figure.
LncRNA CRNDE boosts NEDD4‐1‐mediated PTEN ubiquitylation
As reported, PTEN, a well‐known tumor suppresser gene, plays a crucial role in the development of CDDP resistance in GC (Yang et al, 2013; Li et al, 2020). Using an online bioinformatic database (RPISeq), we found that LncRNA CRNDE may bind to PTEN and NEDD4‐1, an E3 ubiquitin ligase for PTEN (Wang et al, 2007) (Table 1). Therefore, we wondered whether LncRNA CRNDE regulated PTEN expression by modulating NEDD4‐1‐mediated PTEN ubiquitylation, thus influencing the CDDP resistance in GC. First, we investigated the interplay between LncRNA CRNDE, PTEN, and NEDD4‐1. As shown in Fig 6A, LncRNA CRNDE was enriched in both PTEN‐immune complex and NEDD4‐1‐immune complex. The RNA‐pull down assay showed that both PTEN and NEDD4‐1 could be pulled down by biotinylated CRNDE (Fig 6B). Subsequently, two fragments of LncRNA CRNDE (CRNDE‐D1:1–500 nt and CRNDE‐D2: 601–955 nt) were synthesized. The RNA‐pull down assay revealed that PTEN was abundant in the complex pulled by biotinylated CRNDE‐D1 whereas NEDD4‐1 was abundant in the complex pulled by biotinylated CRNDE‐D2 (Fig EV5D). What’s more, the results of Fig 6C depicted that LncRNA CRNDE overexpression enhanced the combination of PTEN and NEDD4‐1. These data indicated that LncRNA CRNDE served as a scaffold for NEDD4‐1 and PTEN in human GC cells.
Table 1.
The interaction probabilities between CRNDE and PTEN and between CRNDE and NEDD4‐1.
| CRNDE and PTEN | CRNDE and NEDD4‐1 | |
|---|---|---|
| Prediction using RF classifier | 0.75 | 0.85 |
| Prediction using SVM classifier | 0.89 | 0.95 |
Figure 6. LncRNA CRNDE negatively regulated PTEN expression.
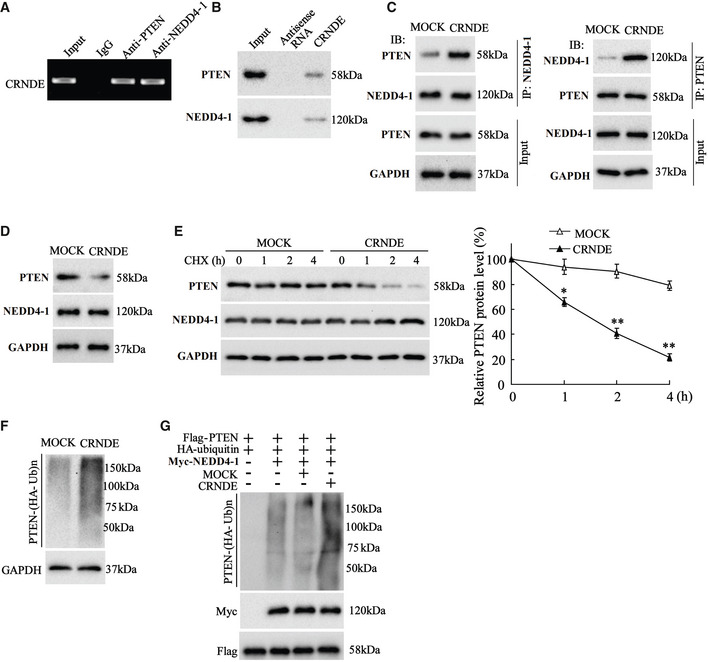
-
A, B(A) RNA Immunoprecipitation (RIP) and (B) RNA‐pull down assays were conducted to detect the association of LncRNA CRNDE and phosphatase and tensin homolog (PTEN)/neural precursor cell expressed developmentally downregulated protein 4‐1 (NEDD4‐1).
-
CSGC7901 cells were transfected with pcDNA‐CRNDE or its negative control (Mock) before MG132 treatment (10 µM, 6 h). The co‐immunoprecipitation (Co‐IP) assay was performed to examine the combination of PTEN and NEDD4‐1.
-
DThe expressions of PTEN and NEDD4‐1 were measured using Western blot in SGC7901 cells transfected with pcDNA‐CRNDE or Mock. GAPDH was served as an internal control.
-
EThe expressions of PTEN and NEDD4‐1 were measured in SGC7901 cells transfected with pcDNA‐CRNDE or Mock at four time‐points after cycloheximide (CHX, 100 μg/ml) treatment. GAPDH was served as an internal control. Left: Representative bands. Right: Quantitative results. The results shown are from three biological replicates and are expressed as mean ± SD. Student’s t‐test. *P < 0.05, **P < 0.01 vs. Mock.
-
F, GThe ubiquitylation of PTEN was measured in (F) SGC7901 cells transfected with pcDNA‐CRNDE or Mock, and (G) 293T cells transfected with indicated vectors.
Source data are available online for this figure.
Next, we investigated the influence of LncRNA CRNDE on PTEN expression. As shown in Fig 6D and E, in response to LncRNA CRNDE overexpression, the protein level of PTEN was continuously reduced, whereas NEDD4‐1 expression did not change. The following experiments demonstrated that LncRNA CRNDE overexpression boosted NEDD4‐1‐mediated PTEN ubiquitination (Fig 6F and G). These data indicated that LncRNA CRNDE served as a scaffold to recruit NEDD4‐1 to PTEN, thus facilitating NEDD4‐1‐mediated PTEN degradation and reducing the protein level of PTEN.
PTEN mediates the modulatory effect of exosomal LncRNA CRNDE on the sensitivity of GC cells to CDDP
To further determine whether exosomal LncRNA CRNDE downregulated the sensitivity of GC cells to CDDP by suppressing PTEN expression, the following experiments were performed. Monocyte‐polarized M2 macrophages were transfected with si‐control or si‐CRNDE, and the exosomes (M2‐si‐control‐exo and M2‐si‐CRNDE‐exo) were isolated from the medium. Then, normal or si‐control‐transfected or si‐PTEN‐transfected SGC7901 cells were incubated with M2‐si‐control‐exo/M2‐si‐CRNDE‐exo and CDDP. As shown in Fig 7A, compared with the M2‐si‐control‐exo, M2‐si‐CRNDE‐exo significantly increased the PTEN protein level and decreased the protein levels of p‐PI3K, p‐AKT, p‐gp, and Bcl‐2. In addition, M2‐si‐CRNDE‐exo treatment also decreased the cell survival rate and cell proliferation and facilitated cell apoptosis in CDDP‐treated SGC7901 cells (Fig 7B–D). Of note, in response to si‐PTEN transfection, PTEN expression was reduced, the protein levels of p‐PI3K, p‐AKT, p‐gp, and Bcl‐2 were increased, the cell survival rate and cell proliferation were elevated, and cell apoptosis was reduced in M2‐si‐CRNDE‐exo+CDDP‐treated SGC7901 cells (Fig 7A–D). Similarly, the PTEN inhibitor SF1670 also abrogated the inhibitory effect of M2‐si‐CRNDE‐exo on cell viability and proliferation and removed the promotion effect of M2‐si‐CRNDE‐exo on cell apoptosis in CDDP‐treated SGC7901 cells (Fig 7E–G).
Figure 7. PTEN mediated the modulatory effect of exosomal LncRNA CRNDE on the sensitivity of GC cells to CDDP.
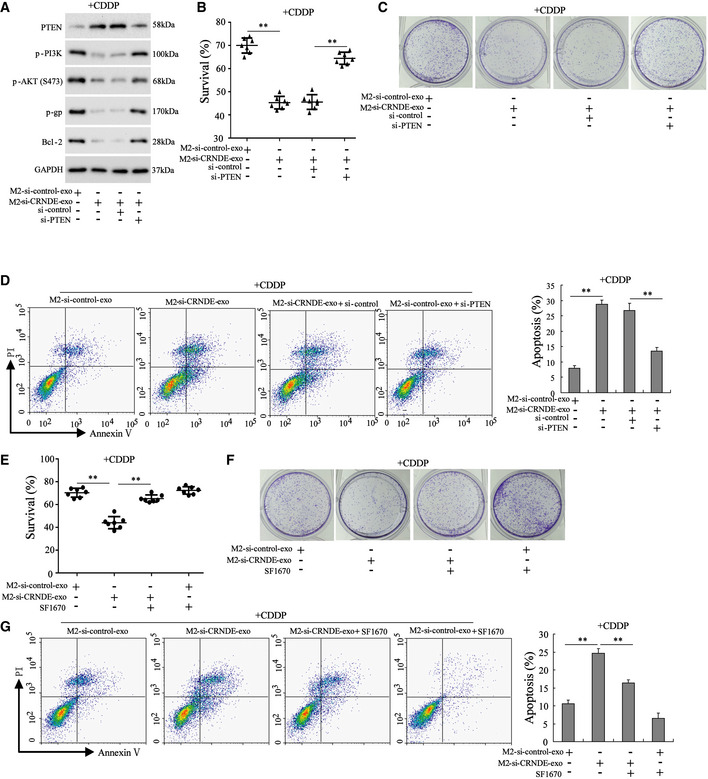
-
A–DNormal, si‐control‐transfected, or si‐PTEN‐transfected SGC7901 cells were treated with CDDP (1 µg/ml) and M2‐si‐control‐exo or M2‐si‐CRNDE‐exo. Si‐control, the negative control of si‐PTEN. (A) The protein levels of PTEN, phosphorylated phosphatidylinositol 3‐kinase (p‐PI3K), phosphorylated protein kinase B (p‐AKT), p‐glycoprotein (p‐gp), and B‐cell lymphoma 2 (Bcl‐2) were measured. GAPDH was served as an internal control. The (B) survival rate, (C) cell proliferation, (D) cell apoptosis in SGC7901 cells.
-
E–GSGC7901 cells were divided into four groups: M2‐si‐control‐exo, M2‐si‐CRNDE‐exo, M2‐si‐CRNDE‐exo +PTEN inhibitor (SF1670; 50 μmol/l), and M2‐si‐control‐exo+PTEN inhibitor. All cells were treated with CDDP. The cell survival rate, cell proliferation, and cell apoptosis were measured.
Data information: (B and E): The results shown are from seven biological replicates. (D and G): The results shown are from three biological replicates. Data are expressed as mean ± SD. (B, D, E, and G): one‐way ANOVA. **P < 0.01.
Discussion
Many studies have confirmed that the rapid development of CDDP resistance is one of the main contributors to GC treatment failure (Longley et al, 2003). Hence, a thorough understanding of the specific mechanism of CDDP resistance can greatly promote therapeutic efficacy in GC patients. In the present study, we identify a mechanism by which LncRNA CRNDE confers CDDP resistance. Our data clarify that LncRNA CRNDE reduces the sensitivity of GC cells to CDDP by suppressing PTEN expression. Moreover, our data suggest that the transport of TAM‐derived exosomes might contribute to the high expression of LncRNA CRNDE in GC cells.
TAMs are the most abundant immune cells in the TME and support tumor growth, invasion, and metastasis by boosting cancer cell proliferation and immunosuppression (Oya et al, 2020). Generally, there are two main types of macrophages: M1 and M2. M1 macrophages promote an inflammatory response against tumor cells, while M2 macrophages function as tumor‐promoting cells by enhancing tumor immunosuppression (Lin et al, 2019). Increasing studies have revealed that M2 macrophages are the prominent TAMs in several types of cancer, such as pancreatic cancer (Padoan et al, 2019), colorectal cancer (Min et al, 2020), and GC (Li et al, 2019), and they are closely associated with cancer progression. In recent years, as vital mediators of the cross‐talk between TAMs and cancer cells, exosomes have attracted the attention of many researchers. Lan et al (2019) have reported that the modulatory effect of M2 macrophages on the migration and invasion of colorectal cancer cells relied on M2‐exo. Zhu et al (2019) found that M2‐exo conferred a chemoresistant phenotype upon epithelial ovarian cancer cells. In the present study, we show that compared with BMDMs‐exo/monocytes‐exo, M2‐exo significantly reduces the sensitivity of GC cells to CDDP treatment, which facilitates cell proliferation in CDDP‐treated mouse/human GC cell lines and boosts homograft tumor growth in CDDP‐treated nude mice. Consistent with our data, Zheng et al (2017) have also suggested that M2‐exo contributes to CDDP resistance in GC cells.
Emerging evidence has revealed that the exosome‐mediated transfer of LncRNAs from TAMs to cancer cells is an important mechanism in the onset of chemotherapeutic resistance in renal cancer (Qu et al, 2016), non‑small‐cell lung cancer (Lei et al, 2018), and others. However, it remains unclear whether M2‐exo uses the same mechanism to influence CDDP resistance in GC. By examining carcinoma tissues and adjacent tissues of GC patients, we find that LncRNA CRNDE, a well‐known tumor promoter gene that is upregulated in several solid cancer types (Ishikawa et al, 2005; Chang et al, 2009; Shahab et al, 2011), is also upregulated in carcinoma tissues and TAMs of GC patients.
The experiments that followed suggest that LncRNA CRNDE could be successfully transferred from M2 macrophages to GC cells via exosomes. Silencing LncRNA CRNDE expression in M2‐exo efficiently reverses M2‐exo‐induced CDDP resistance in GC cells both in vitro and in vivo. CRNDE knockdown does not affect the type I interferon pathway, as the unchanged mRNA levels of 2',5'‐oligoadenylate synthetase (OAS1a), interferon regulatory factor 7 (IRF7), and interferon‐alpha (IFN‐α) in GC cells treated with M2‐si‐control‐exo compared with GC cells treated with M2‐si‐CRNDE‐exo (Appendix Fig S1J) show. Collectively, our results indicate that in GC cells exosomal LncRNA CRNDE influences CDDP resistance. Though a previous study has observed high expression levels of LncRNA CRNDE in GC tissues and determined its carcinogenic effect in GC (Du et al, 2017), our study clarifies that the high expression of LncRNA CRNDE in GC tissues may depend on M2‐exo transport and also demonstrates a role for LncRNA CRNDE in CDDP resistance of GC cells.
To assess the effect of LncRNA CRNDE on CDDP resistance of GC cells in vivo, we used a homograft mouse model and a PDX model, two widely used models for cancer pathology research (Chen et al, 2019). However, these models do not proof the horizontal exosome‐mediated transfer of CRNDE in vivo. In follow‐up studies, a homograft mouse model will be established using fluorescently labeled MFC cells, and the small animal living imaging system will be employed to evaluate the tumor burden of mice treated with M2‐exo or M2‐si‐CRNDE‐exo. Moreover, the expression of PTEN will be detected in the homograft tumors of mice to further confirm the role of the CRNDE/PTEN axis on the CDDP resistance of GC cells in vivo.
Finally, the underlying mechanism by which LncRNA CRNDE may reduce the sensitivity of GC cells to CDDP was investigated. Our data show that LncRNA CRNDE serves as a scaffold to recruit NEDD4‐1 to PTEN, thus facilitating NEDD4‐1‐mediated PTEN degradation. PTEN is a dual phosphatase with both lipid phosphatase and protein activities. Relying on its lipid phosphatase activity, PTEN dephosphorylates phosphatidylinositol‐3, 4, 5‐triphosphate (PIP3), which suppresses the activation of the PI3K/Akt signaling pathway (Chen et al, 2018) and ultimately represses PI3K/Akt pathway‐mediated cancer cell proliferation, invasion, and drug resistance (Gao et al, 2019; Liu et al, 2019). In GC, the function and/or expression of PTEN is decreased, overexpressing PTEN could reverse CDDP resistance in GC by suppressing the PI3K/Akt pathway (Oki et al, 2005; Sun et al, 2018a), which suggests that PTEN is an important target for chemotherapeutic resistance in GC. Similarly, we demonstrate here that in response to M2‐si‐CRNDE‐exo treatment, PTEN expression is elevated, the PI3K/Akt pathway is inactivated, and the sensitivity to CDDP is enhanced in GC cells, whereas PTEN knockdown reverses the above events.
In summary, our study reveals that LncRNA CRNDE is upregulated in GC cells, thereby inducing CDDP resistance by decreasing PTEN expression. Moreover, our data suggest that the high levels of LncRNA CRNDE in GC cells are caused most likely by horizontal transfer of TAM‐derived exosomes.
Materials and Methods
Study participants
With the approval of the Ethics Committee of The Second Affiliated Hospital of Nanchang University, 35 pairs of cancerous tissues and adjacent normal tissues were obtained from GC patients (n = 35) who experienced curative resection at The Second Affiliated Hospital of Nanchang University. The clinicopathological characteristics of the enrolled GC patients are shown in Table 2. In addition, healthy volunteers were also enrolled in our study, and peripheral blood was obtained for the isolation of peripheral blood monocytes. All patients and healthy donors signed informed consent forms before the study began.
Table 2.
Clinicopathological characteristics of patients with GC (n = 35).
| Characteristics | Cases | Percent % |
|---|---|---|
| Age | ||
| <70 | 19 | 52.3 |
| ≥70 | 16 | 47.7 |
| Sex | ||
| Male | 25 | 71.4 |
| Female | 10 | 29.6 |
| Tumor location | ||
| Proximal | 15 | 42.9 |
| Distal | 20 | 57.1 |
| Histological type | ||
| Intestinal type | 17 | 48.6 |
| Diffuse type | 18 | 51.4 |
| Pathological T category | ||
| pT1/2 | 13 | 37.1 |
| pT3/4 | 22 | 62.9 |
| Lymph node metastasis | ||
| N0 | 12 | 34.2 |
| N1 | 23 | 65.8 |
| Peritoneal metastasis | ||
| P0 | 27 | 77.1 |
| P1 | 8 | 22.9 |
| Distant metastasis | ||
| M0 | 23 | 65.7 |
| M1 | 12 | 34.3 |
| Tumor size | ||
| <5.5 cm | 18 | 51.4 |
| ≥5.5 cm | 17 | 48.6 |
| UICC TNM classification | ||
| Stage I | 5 | 14.3 |
| Stage II | 10 | 28.6 |
| Stage III | 11 | 31.4 |
| Stage IV | 9 | 25.7 |
TNM, tumor, nodal‐involvement, metastasis; UICC, International Union against Cancer.
Isolation of adjacent normal tissue‐derived macrophages and TAMs
Normal tissue‐derived macrophages (NTMs) and TAMs were isolated from seven pairs of GC cancerous tissues and adjacent normal tissues according to a previously described method (Zheng et al, 2018). Briefly, fresh adjacent normal tissues and cancerous tissues were cut into small pieces and then immersed in an RPMI 1640 medium containing Liberase (1.67 U/ml) and DNase (0.2 mg/ml) for 30 min. The mixture was filtered using Falcon® 70‐µm Cell Strainer (Corning, USA). The red blood cells in the mixture were removed using a red blood cell lysis buffer (Solarbio, China). Subsequently, the samples were resuspended in a fluorescence activating cell sorter buffer and then underwent flow cytometry.
Macrophage generation and differentiation
Mouse bone marrow‐derived macrophages (BMDMs) were obtained as previously described (Trouplin et al, 2013) and maintained in a bone macrophage medium supplemented with macrophage colony‐stimulating factor (MC‐SF; 50 ng/ml). Human peripheral blood monocytes were obtained from healthy donors as previously described (Rahal et al, 2018) and maintained in RPMI 1640 medium containing 20% fetal bovine serum (FBS) and 50 ng/ml MC‐SF. A week later, 20 ng/ml interleukin (IL)‐4 and 20 ng/ml IL‐13 were added to the culture medium to induce cell polarization toward the M2 phenotype.
Exosome collection and characterization
Exosomes were isolated from the medium of BMDMs (BMDMs‐exo), monocytes (monocytes‐exo), BMDM‐polarized M2 macrophages (M2‐exo), monocyte‐polarized M2 macrophages (M2‐exo), BMDM‐polarized M2 macrophages transfected with si‐control or si‐CRNDE (M2‐si‐control‐exo or M2‐si‐CRNDE‐exo), and monocyte‐polarized M2 macrophages transfected with si‐control or si‐CRNDE (M2‐si‐control‐exo or M2‐si‐CRNDE‐exo) as described previously (Zhu et al, 2019). Briefly, 20 ml of cell culture medium was collected, and the exosomes were extracted utilizing the ExoQuick‐TC kit purchased from System Bioscience (USA). Then, the isolated exosomes were resuspended in phosphate‐buffered saline (PBS). Transmission electron microscopy (TEM) was adopted to visualize exosomes, and Western blot was carried out to examine the exosome‐specific markers (CD63 and CD81).
Cell culture, treatment, and transfection
Procell Life Science and Technology Co., Ltd. (China) provided the mouse GC cell line MFC and human GC cell line SGC7901 that were used in this study. Shunran Biology Co., Ltd. (China) supplied the human GC cell line MGC‐803 for this study. All cell lines were cultured in RPMI 1640 medium containing 10% FBS at 37°C. In some experiments, the MFC/SGC7901/MGC‐803 cells were incubated with the indicated exosomes (20 µg/ml, quantified by BCA protein analysis) and/or 1 μg/ml CDDP for 48 h.
To silence LncRNA CRNDE in M2‐exo, BMDM‐polarized or monocyte‐polarized M2 macrophages were transfected with si‐CRNDE using Lipofectamine 3000 (Thermo Fisher, USA). The conditioned exosomes were isolated after the transfected cells were cultured for 48 h (Du et al, 2020). To silence phosphatase and tensin homolog (PTEN) or overexpress LncRNA CRNDE in SGC7901 cells or 293T cells, si‐PTEN or pcDNA3.1‐CRNDE (CRNDE) was transfected into cells in the same manner. Si‐CRNDE, si‐PTEN, CRNDE, and their negative controls (si‐control and mock) were all produced by Guangzhou RiboBio Co., Ltd. (China).
qRT–PCR
Total RNA samples of the cells and tissues were extracted using TRIzol. The exosomal RNA samples were extracted from exosomes (400 µg) using a BasicExoRNA™ Basic RNA Extraction Kit (Biovision, USA). The qRT–PCR assay was performed using SYBR‐Green PCR Master Mix (Takara). The relative transcriptional levels of the target genes were calculated using the 2−ΔΔCT or 2−ΔCT or 2−CT method. The sequences of primers used in this study are shown in Table 3.
Table 3.
Primer sequences for qRT–PCR
| Gene | Sequences (5’–3’) | |
|---|---|---|
| Forward | Reverse | |
| Lnc‐EGFR | CAGCAGCCCTGCAATTAAAC | GGGTCCTCATGTAATGGTAATAGG |
| LincROR | GGCTGGAACTCTTGCTGGAG | GGCTGGAACTCTTGCTGGAG |
| HOTAIR | GGTAGTAAAAGCAACCACGAAGC | ACATAAACCTCTGTCTGTGAGTGCC |
| MALAT1 | TTGTAGACTGGAGAAGATAGG | ACTGAAGAGCATTGGAGAT |
| MEG3 | CAGCCAAGCTTCTTGAAAGG | TTCCACGGAGTAGAGCGAGT |
| LncRNA‐91H | GCTTGTCAGTAGAGTGCGCC | CATCCAGTTGACCGAGCTTG |
| ZFAS1 | GCGGGTACAGAATGGATTTTGG | CAACACCCGCATTCATCCTG |
| CRNDE | TGA AGG AAG GAA GTG GTG CA | TCC AGT GGC ATC CTA CAA GA |
| LINC00152 | TCACTTCAGAGCCAAGGCAG | AAATGCCTACCGCCAGTTCA |
| LncRNA p21 | TGTTGCATTGTTGCATCATC | TTTCTTCCAGTGGTGAGTGG |
| TUC339 | GATGAGGCCCCGAGTTTAAT | AGATGGAGGATCGGTGTGAA |
| CCAT2 | AGACAGTGCCAGCCAACC | TGCCAAACCCTTCCCTTA |
| GAPDH | GGTCTCCTCTGACTTCAACA | GTGAGGGTCTCTCTCTTCCT |
Measurement of cell survival, proliferation, and apoptosis
Cell viability was examined using a cell counting kit‐8 (CCK‐8) assay kit (Solarbio, China) according to the manufacturer’s protocol.
Cell proliferation was measured by colony formation. The cells were seeded into six‐well plates at a density of 500 cells/well and cultured under normal condition. Two weeks later, the cells were fixed and then stained with 0.5% crystal violet.
Cell apoptosis was determined with an Annexin V‐FITC/PI Apoptosis Detection Kit (Procell, China). Briefly, 3 × 105 cells were resuspended in 500 µl Annexin V binding buffer, followed by interacting with 5 µl Annexin V‐FITC and 5 µl PI for 15 min. Then, the cells were analyzed using a flow cytometer.
Western blot
Protein samples were prepared using a RIPA buffer. After quantitation, 25 µg protein samples were separated by SDS–PAGE gels and then electro‐transferred to PVDF membranes. Next, the membranes were incubated with 5% bovine serum albumin (BSA, room temperature, 0.5 h), primary antibodies (4°C, overnight), secondary antibodies (room temperature, 1 h), and the enhanced chemiluminescence reagent. The protein bands were visualized using a Bio‐Rad ChemiDoc XRS+ System. The primary antibodies used in this study were as follows: anti‐calnexin (1 µg/ml; Abcam, UK), anti‐CD63 (0.15 µg/ml, Abcam), anti‐CD81 (0.04 µg/ml; Abcam), anti‐PTEN (0.01 µg/ml, Abcam), anti‐neural precursor cell expressed developmentally downregulated protein 4‐1 (NEDD4‐1) (1:1,000; Sigma‐Aldrich, USA), anti‐phosphorylated phosphatidylinositol 3‐kinase (p‐PI3K) (0.5 µg/ml; Abcam), anti‐phosphorylated protein kinase B (p‐AKT) (1:2000; Cell Signaling), anti‐p‐glycoprotein (p‐gp) (0.4 µg/ml; Abcam), anti‐B‐cell lymphoma 2 (Bcl‐2) (0.6 µg/ml; Abcam), and anti‐GAPDH (0.1 µg/ml; Abcam).
RNA immunoprecipitation assay
Before assay, the magnetic beads were resuspended in 100 µl RNA immunoprecipitation (RIP) wash buffer and incubated with 5 µg of anti‐PTEN/anti‐NEDD4‐1/IgG antibody for 30 min. Then, the cell lysates were obtained from SGC7901 cells using RIP lysate and incubated with the bead‐antibody complex at 4°C overnight. The next day, the mixture was centrifuged, and the precipitate was collected. The RNA samples were purified from the precipitate and reversely transcribed into cDNA. The expression of LncRNA CRNDE in PTEN‐immune complex and NEDD4‐1‐immune complex was evaluated by electrophoresis on 1% agarose gel.
Bioinformatic analysis
The combination of LncRNA CRNDE and PTEN/NEDD4‐1 was evaluated by the bioinformatic database RPISeq ( http://pridb.gdcb.iastate.edu/RPISeq/ ). First, the amino acid sequence of PTEN/ NEDD4‐1 protein was input. Second, input the nucleotide sequence of CRNDE and click “submit”. Then, the interaction probability can be obtained a few minutes later.
RNA‐pull down assay
Biotinylated LncRNA CRNDE and antisense RNA were produced by RiboBio Co., Ltd. (China). The SGC7901 cell lysates were reacted with biotinylated LncRNA CRNDE or antisense RNA and streptavidin‐coupled magnetic beads (Thermo Fisher, USA) for one hour at 25°C. Then, the biotin‐coupled RNA complexes were isolated and the expression levels of PTEN and NEDD4‐1 in the complexes were determined by Western blot.
Co‐immunoprecipitation assay
Before the assay, SGC7901 cells were transfected with CRNDE or Mock. Forty‐eight hours later, SGC7901 cell lysates were incubated with anti‐PTEN/anti‐NEDD4‐1 antibody. Then, the immune complexes were obtained using protein A/G agarose beads (Thermo Fisher, USA). The expression levels of PTEN and NEDD4‐1 in the immune complexes were determined by Western blot.
IP assay
To evaluate the effect of LncRNA CNRDE on the ubiquitylation of PTEN, the 293T cells were transfected with the indicated vectors and treated with 10 μM MG132 for 6 h. The 293T cell lysates were incubated with anti‐FLAG antibody at 4°C. The next day, the protein complexes were captured with protein A/G agarose beads. The expression levels of PTEN‐(HA‐Ub)n in the immune complexes were determined by Western blot using an anti‐HA antibody.
Tumorigenicity in nude mice
In total, 28 BALB/c nude mice (4–6 weeks old; male; Charles River, China) were used in our study. First, 3 × 105 MFC cells resuspended in 200 µl PBS were subcutaneously injected into nude mice. Ten days after injection, BMDMs‐exo, M2‐exo, M2‐si‐control‐exo, or M2‐si‐CRNDE‐exo (10 µg) was twice injected into the center of the homograft tumors of mice (n = 7 in each group), followed by a single intraperitoneal injection of CDDP (10 mg/kg). The homograft tumor sizes were recorded from the first day of CDDP treatment. Six days after CDDP treatment began, all mice were sacrificed, and the homograft tumors were collected.
To establish the patient‐derived xenograft (PDX) model, fresh GC tumor tissues were cut into small pieces of ˜1 mm3 and subcutaneously transplanted into the flank region of BALB/c nude mice. When the tumor size reached 100 mm3, mice were treated with BMDMs‐exo + CDDP, M2‐exo+CDDP, M2‐si‐control‐exo+CDDP, or M2‐si‐CRNDE‐exo+CDDP as described above.
All animal procedures were approved by the Ethics Committee of The Second Affiliated Hospital of Nanchang University.
Statistical analysis
Statistical analyses were performed using GraphPad Prism 6.0. Data were expressed as mean ± SD. Statistical significance between the two experimental groups was analyzed using Student’s t‐test. Results were deemed to be statistically significant when P < 0.05.
Author contributions
LX proposed the conception, performed the experiments, analyzed the data, and wrote the paper; L‐QZ performed the experiments, analyzed the data, and wrote the paper; CL, FZ, Y‐WY, and QZ contributed significantly to analysis; S‐HL, YW, J‐LW, and D‐ZW participated in the experiments of the study; HL checked the manuscript.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting information
Review Process File
Appendix
Expanded View Figures PDF
Source Data for Expanded View
Source Data for Figure 2
Source Data for Figure 6
Acknowledgements
This work was supported by The National Natural Science Foundation of China (Nos. 81872480, 81760549, and 81560492) and Jiangxi Province Academic and Technical Leaders Training Program for Major Disciplines (Leading Talents Program).
EMBO reports (2021) 22: e52124.
Data availability
No primary datasets have been generated or deposited.
References
- Binenbaum Y, Fridman E, Yaari Z, Milman N, Schroeder A, Ben David G, Shlomi T, Gil Z (2018) Transfer of miRNA in macrophage‐derived exosomes induces drug resistance in pancreatic adenocarcinoma. Can Res 78: 5287–5299 [DOI] [PubMed] [Google Scholar]
- Chang Q, Chen J, Beezhold KJ, Castranova V, Shi X, Chen F (2009) JNK1 activation predicts the prognostic outcome of the human hepatocellular carcinoma. Mol Cancer 8: 64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CY, Chen J, He L, Stiles BL (2018) PTEN: tumor suppressor and metabolic regulator. Front Endocrinol 9: 338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D, An X, Ouyang X, Cai J, Zhou D, Li QX (2019) In vivo pharmacology models for cancer target research. Methods Mol Biol 1953: 183–211 [DOI] [PubMed] [Google Scholar]
- Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, Jemal A, Yu XQ, He J (2016) Cancer statistics in China, 2015. CA Cancer J Clin 66: 115–132. [DOI] [PubMed] [Google Scholar]
- Du DX, Lian DB, Amin BH, Yan W (2017) Long non‐coding RNA CRNDE is a novel tumor promoter by modulating PI3K/AKT signal pathways in human gastric cancer. Eur Rev Med Pharmacol Sci 21: 5392–5398 [DOI] [PubMed] [Google Scholar]
- Du J, Liang Y, Li J, Zhao J‐M, Wang Z‐N, Lin X‐Y (2020) Gastric cancer cell‐derived exosomal microRNA‐23a promotes angiogenesis by targeting PTEN. Front Oncol 10: 326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X, Qin T, Mao J, Zhang J, Fan S, Lu Y, Sun Z, Zhang Q, Song B, Li L (2019) PTENP1/miR‐20a/PTEN axis contributes to breast cancer progression by regulating PTEN via PI3K/AKT pathway. J Exp Clin Cancer Res 38: 256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Z, Xie C, Huang Z, Guo Y, Zhang X (2020) Anti‐tumor effect of pcDNA3.1(‐)/rMETase‐shUCA1 hyaluronic acid‐modified G5 polyamidoamine nanoparticles on gastric cancer. Aging Pathobiol Ther 2: 86–95 [Google Scholar]
- Ishikawa M, Yoshida K, Yamashita Y, Ota J, Takada S, Kisanuki H, Koinuma K, Choi YL, Kaneda R, Iwao T et al (2005) Experimental trial for diagnosis of pancreatic ductal carcinoma based on gene expression profiles of pancreatic ductal cells. Cancer Sci 96: 387–393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan J, Sun LI, Xu F, Liu LU, Hu F, Song DA, Hou Z, Wu W, Luo X, Wang J et al (2019) M2 macrophage‐derived exosomes promote cell migration and invasion in colon cancer. Can Res 79: 146–158 [DOI] [PubMed] [Google Scholar]
- Lei Y, Guo W, Chen B, Chen L, Gong J, Li W (2018) Tumor‐released lncRNA H19 promotes gefitinib resistance via packaging into exosomes in non‐small cell lung cancer. Oncol Rep 40: 3438–3446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Ma X, Yang D, Suo Z, Dai R, Liu C (2020) PCAT‐1 contributes to cisplatin resistance in gastric cancer through epigenetically silencing PTEN via recruiting EZH2. J Cell Biochem 121: 1353–1361 [DOI] [PubMed] [Google Scholar]
- Li W, Zhang X, Wu F, Zhou Y, Bao Z, Li H, Zheng P, Zhao S (2019) Gastric cancer‐derived mesenchymal stromal cells trigger M2 macrophage polarization that promotes metastasis and EMT in gastric cancer. Cell Death Dis 10: 918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y, Xu J, Lan H (2019) Tumor‐associated macrophages in tumor metastasis: biological roles and clinical therapeutic applications. J Hematol Oncol 12: 76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Wang J, Tao Y, Li X, Qin J, Bai Z, Chi B, Yan W, Chen X (2019) Curcumol inhibits colorectal cancer proliferation by targeting miR‐21 and modulated PTEN/PI3K/Akt pathways. Life Sci 221: 354–361 [DOI] [PubMed] [Google Scholar]
- Longley DB, Harkin DP, Johnston PG (2003) 5‐fluorouracil: mechanisms of action and clinical strategies. Nat Rev Cancer 3: 330–338 [DOI] [PubMed] [Google Scholar]
- Min AKT, Mimura K, Nakajima S, Okayama H, Saito K, Sakamoto W, Fujita S, Endo H, Saito M, Saze Z et al (2021) Therapeutic potential of anti‐VEGF receptor 2 therapy targeting for M2‐tumor‐associated macrophages in colorectal cancer. Cancer Immunol Immunother 70: 289–298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noy R, Pollard JW (2014) Tumor‐associated macrophages: from mechanisms to therapy. Immunity 41: 49–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oki E, Baba H, Tokunaga E, Nakamura T, Ueda N, Futatsugi M, Mashino K, Yamamoto M, Ikebe M, Kakeji Y et al (2005) Akt phosphorylation associates with LOH of PTEN and leads to chemoresistance for gastric cancer. Int J Cancer 117: 376–380 [DOI] [PubMed] [Google Scholar]
- Orditura M, Galizia G, Sforza V, Gambardella V, Fabozzi A, Laterza MM, Andreozzi F, Ventriglia J, Savastano B, Mabilia A et al (2014) Treatment of gastric cancer. World J Gastroenterol 20: 1635–1649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oya Y, Hayakawa Y, Koike K (2020) Tumor microenvironment in gastric cancers. Cancer Sci 111: 2696–2707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padoan A, Plebani M, Basso D (2019) Inflammation and pancreatic cancer: focus on metabolism, cytokines, and immunity. Int J Mol Sci 20: 676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paponov IA, Friz T, Budnyk V, Teale W, Wüst F, Paponov M, Al‐Babili S, Palme K (2019) Natural Auxin Does not inhibit brefeldin A induced PIN1 and PIN2 internalization in root cells. Front Plant Sci 10: 574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian BZ, Pollard JW (2010) Macrophage diversity enhances tumor progression and metastasis. Cell 141: 39–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu LE, Ding J, Chen C, Wu Z‐J, Liu B, Gao YI, Chen W, Liu F, Sun W, Li X‐F et al (2016) Exosome‐transmitted lncARSR promotes sunitinib resistance in renal cancer by acting as a competing endogenous RNA. Cancer Cell 29: 653–668 [DOI] [PubMed] [Google Scholar]
- Rahal OM, Wolfe AR, Mandal PK, Larson R, Tin S, Jimenez C, Zhang D, Horton J, Reuben JM, McMurray JS et al (2018) Blocking interleukin (IL)4‐ and IL13‐mediated phosphorylation of STAT6 (Tyr641) decreases M2 polarization of macrophages and protects against macrophage‐mediated radioresistance of inflammatory breast cancer. Int J Radiat Oncol Biol Phys 100: 1034–1043 [DOI] [PubMed] [Google Scholar]
- Shahab SW, Matyunina LV, Mezencev R, Walker LD, Bowen NJ, Benigno BB, McDonald JF (2011) Evidence for the complexity of microRNA‐mediated regulation in ovarian cancer: a systems approach. PLoS One 6: e22508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smani Y, Faivre B, Fries I, Labrude P, Vigneron C (2006) Potential mechanism of dextran‐conjugated hemoglobin penetration inside arterial wall. Arch Mal Coeur Vaiss 99: 722–726 [PubMed] [Google Scholar]
- Sun MY, Sun J, Tao J, Yuan YX, Ni ZH, Tang QF (2018a) Zuo Jin Wan reverses DDP resistance in gastric cancer through ROCK/PTEN/PI3K signaling pathway. Evid Based Complement Alternat Med 2018: 4278568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Z, Yang S, Zhou Q, Wang G, Song J, Li Z, Zhang Z, Xu J, Xia K, Chang Y et al (2018b) Emerging role of exosome‐derived long non‐coding RNAs in tumor microenvironment. Mol Cancer 17: 82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Yan IK, Wood J, Haga H, Patel T (2014) Involvement of extracellular vesicle long noncoding RNA (linc‐VLDLR) in tumor cell responses to chemotherapy. Mol Cancer Res 12: 1377–1387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian T, Zhu YL, Zhou YY, Liang GF, Wang YY, Hu FH, Xiao ZD (2014) Exosome uptake through clathrin‐mediated endocytosis and macropinocytosis and mediating miR‐21 delivery. J Biol Chem 289: 22258–22267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trouplin V, Boucherit N, Gorvel L, Conti F, Mottola G, Ghigo E (2013) Bone marrow‐derived macrophage production. J Vis Exp e50966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Trotman LC, Koppie T, Alimonti A, Chen Z, Gao Z, Wang J, Erdjument‐Bromage H, Tempst P, Cordon‐Cardo C et al (2007) NEDD4‐1 is a proto‐oncogenic ubiquitin ligase for PTEN. Cell 128: 129–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Yu Y, He X, Niu N, Li X, Zhang R, Hu J, Ma J, Yu X, Sun Y et al (2019) Tumor‐associated macrophages induce invasion and poor prognosis in human gastric cancer in a cyclooxygenase‐2/MMP9‐dependent manner. Am J Transl Res 11: 6040–6054 [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi T, Fushida S, Yamamoto Y, Tsukada T, Kinoshita J, Oyama K, Miyashita T, Tajima H, Ninomiya I, Munesue S et al (2016) Tumor‐associated macrophages of the M2 phenotype contribute to progression in gastric cancer with peritoneal dissemination. Gastric Cancer 19: 1052–1065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang SM, Huang C, Li XF, Yu MZ, He Y, Li J (2013) miR‐21 confers cisplatin resistance in gastric cancer cells by regulating PTEN. Toxicology 306: 162–168 [DOI] [PubMed] [Google Scholar]
- Zhang HG, Grizzle WE (2014) Exosomes: a novel pathway of local and distant intercellular communication that facilitates the growth and metastasis of neoplastic lesions. Am J Pathol 184: 28–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng P, Chen L, Yuan X, Luo Q, Liu Y, Xie G, Ma Y, Shen L (2017) Exosomal transfer of tumor‐associated macrophage‐derived miR‐21 confers cisplatin resistance in gastric cancer cells. J Exp Clin Cancer Res 36: 53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng P, Luo Q, Wang W, Li J, Wang T, Wang P, Chen L, Zhang P, Chen H, Liu YI et al (2018) Tumor‐associated macrophages‐derived exosomes promote the migration of gastric cancer cells by transfer of functional Apolipoprotein E. Cell Death Dis 9: 434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X, Shen H, Yin X, Yang M, Wei H, Chen Q, Feng F, Liu Y, Xu W, Li Y (2019) Macrophages derived exosomes deliver miR‐223 to epithelial ovarian cancer cells to elicit a chemoresistant phenotype. J Exp Clin Cancer Res 38: 81 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Review Process File
Appendix
Expanded View Figures PDF
Source Data for Expanded View
Source Data for Figure 2
Source Data for Figure 6
Data Availability Statement
No primary datasets have been generated or deposited.


