Abstract
Astrocytes display extraordinary morphological complexity that is essential to support brain circuit development and function. Formin proteins are key regulators of the cytoskeleton; however, their role in astrocyte morphogenesis across diverse brain regions and neural circuits is unknown. Here, we show that loss of the formin protein Daam2 in astrocytes increases morphological complexity in the cortex and olfactory bulb, but elicits opposing effects on astrocytic calcium dynamics. These differential physiological effects result in increased excitatory synaptic activity in the cortex and increased inhibitory synaptic activity in the olfactory bulb, leading to altered olfactory behaviors. Proteomic profiling and immunoprecipitation experiments identify Slc4a4 as a binding partner of Daam2 in the cortex, and combined deletion of Daam2 and Slc4a4 restores the morphological alterations seen in Daam2 mutants. Our results reveal new mechanisms regulating astrocyte morphology and show that congruent changes in astrocyte morphology can differentially influence circuit function.
Keywords: astrocyte, circuit, Daam2, heterogeneity, morphology
Subject Categories: Cell Adhesion, Polarity & Cytoskeleton; Neuroscience
This study shows that loss of Daam2 leads to increased morphological complexity of mouse astrocytes in the cortex and olfactory bulb, but elicits opposing effects on astrocyte calcium dynamics, resulting in divergent synaptic activity and altered behaviors.
Introduction
Astrocytes are the most abundant glial cell in the central nervous system (CNS), playing diverse roles in metabolic support, blood–brain barrier formation, and pH regulation (Petzold & Murthy, 2011; Allen & Lyons, 2018; Theparambil et al, 2020). Although astrocytes are electrically silent, they have emerged as crucial regulators of neuronal activity and circuit function through their roles in synaptic transmission and ion homeostasis (Vesce et al, 1999; Clarke & Barres, 2013; Allen, 2014; Haim & Rowitch, 2017). Such diverse astrocytic functions are closely associated with their extraordinarily complex morphology, which is established during postnatal development in concert with synapse formation and maturation (Bushong et al, 2004; Freeman, 2010; Allen & Eroglu, 2017; Bosworth & Allen, 2017; Chen et al, 2020). Moreover, alterations in astrocytic processes alter synaptic and neuronal function, establishing an intimate link between astrocyte morphology and circuit integration (Clarke & Barres, 2013; Khakh & Sofroniew, 2015; Durkee & Araque, 2019). However, the contributions of astrocytes to brain circuits remain incompletely defined.
The morphological heterogeneity of astrocytes has been appreciated for over 100 years; however, only recently have their diverse molecular and functional properties been closely examined. Recent studies showed molecular and cellular diversity of astrocytes from distinct brain regions, which appears to be regulated by discrete transcriptional mechanisms in the adult brain (Chai et al, 2017; Huang et al, 2020). These observations indicate that local astrocytes contribute to their unique microenvironment and associated circuits, which prompted us to ask whether these region‐specific developmental mechanisms also influence associated circuit formation in the developing brain.
Cytoskeletal remodeling plays a key role in cellular morphogenesis (Wallar & Alberts, 2003). Daam2 (disheveled‐associated activator of morphogenesis 2) is a member of the formin protein family, which mediates actin assembly and cytoskeletal remodeling (Kawabata Galbraith & Kengaku, 2019). Daam2 contains a highly conserved formin‐homology domain and a GTPase binding domain, suggesting that it plays important roles in cytoskeletal regulation via downstream signaling cascades mediated by Rho GTPase binding (Lee & Deneen, 2012; Welsh et al, 2013; Ajima et al, 2015). We have previously shown that Daam2 facilitates CNS patterning by regulating myelin‐forming oligodendrocytes during development via canonical Wnt signaling (Lee & Deneen, 2012; Lee et al, 2015). Interestingly, Daam2 is also expressed in astrocyte precursors during development, raising the question of its’ role in astrocyte development and function.
Here, we defined the expression and function of Daam2 in the astrocyte lineage during brain development. Using astrocyte‐specific conditional mouse models, we discovered that loss of Daam2 enhances the morphological complexity of astrocytes in vivo. Interestingly, while we observed congruent changes in astrocyte morphology in both cortex and olfactory bulb (OB), these alterations resulted in distinct, region‐specific effects on circuit function during development. To identify mechanisms that control these region‐specific functions of Daam2, we performed an unbiased in vivo protein profiling and identified Slc4a4 (electrogenic sodium‐bicarbonates cotransporter 1, NBCe1) as a key regulator. We showed that Daam2 and Slc4a4 are co‐expressed in astrocytes and that the loss of Daam2 increased Slc4a4 expression in cortex but not in OB. Strikingly, knockout of Slc4a4 suppressed astrocyte morphological complexity and reversed the enhanced morphological complexity in the cortex caused by loss of Daam2. Together, our findings show that Daam2 is a novel regulator of astrocyte morphology and circuit formation in a region‐specific manner during development.
Results
Daam2 loss results in increased astrocyte morphological complexity during development
As a first step toward uncovering the role of Daam2 in astrocyte function, we assessed its expression throughout development. To visualize Daam2 expression in astrocytes, we first performed immunostaining in Daam2 reporter (Daam2LacZ/+ ) mice (Lee et al, 2015) that were intercrossed with an astrocyte‐specific reporter allele (Aldh1l1‐EGFP) (Doyle et al, 2008). Immunostaining revealed astrocytic expression of Daam2 in multiple brain areas, including cortex, OB, hippocampus, and striatum (Fig 1A). In cortex, approximately 80% of Aldh1l1‐EGFP expressing astrocytes co‐expressed Daam2 throughout development into adulthood (Fig 1B and C). We corroborated these expression data by performing fluorescent RNA in situ hybridization immunostaining for Daam2 alongside known astrocyte lineage markers (Fig EV1A). Importantly, no Daam2 immunostaining was observed in nestin‐positive neuronal progenitors throughout the subventricular zone and rostral migratory stream (Fig EV1B). Together, these data indicate that Daam2 is expressed in astrocyte lineage during development and throughout adulthood.
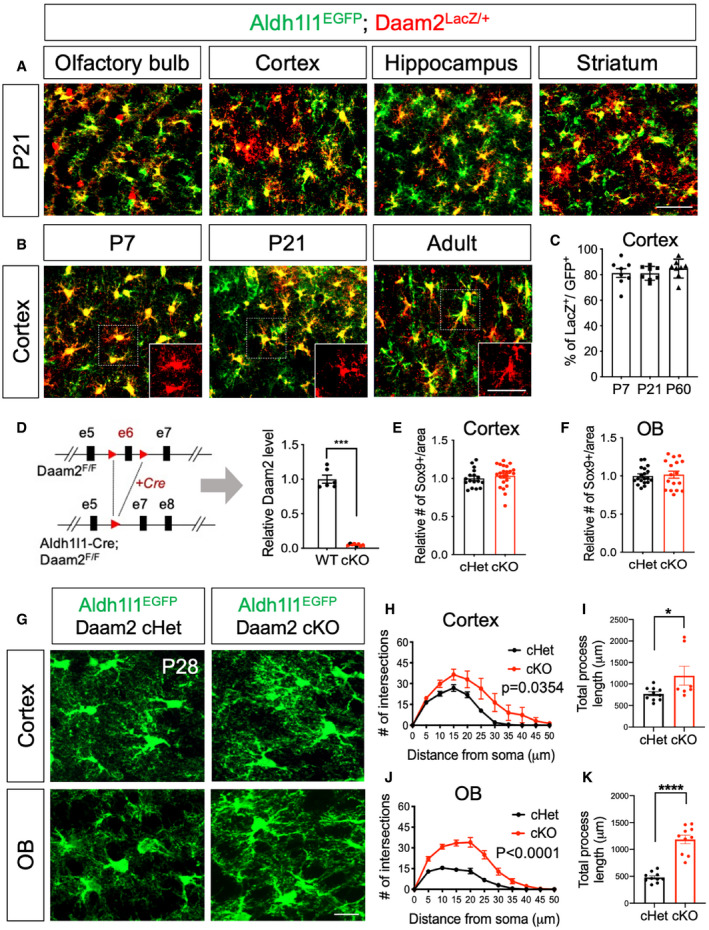
Figure 1. Loss of Daam2 increases morphological complexity of astrocytes in the brain.
-
A, BImmunostaining from Daam2LacZ/+ animals with Aldh1l1‐EGFP reporter indicates that Daam2 (LacZ+, Red) is expressed in astrocytes (Green). (A) Daam2 expression is observed in multiple brain regions at postnatal day 21. (B) Expression of Daam2 in cortex persists throughout development (P7 to adult). Scale bar: 50 μm.
-
CQuantification of Daam2‐expressing astrocytes (LacZ+) with Aldh1l1‐EGFP‐labeled astrocytes in cortex during development. Data are presented as mean ± SEM. Two brain sections, N = 4 for each age.
-
DSchematic of the generation of astrocyte‐specific deletion of Daam2 in Aldh1l1‐Cre and Daam2‐floxed mice. Quantitative RT–PCR in FAC‐sorted astrocytes confirms significant reduction of Daam2 transcript from Daam2 cKO mice. Experiments are performed as duplicates from N = 3–4. Data are presented as mean ± SEM. Student’s t‐test is used for statistical analysis. ***P < 0.001.
-
E, FQuantification of astrocyte marker, Sox9+ cells in cortex (E) and OB (F) of Daam2 cHet and Daam2 cKO mice. Data are presented as mean ± SEM. 4–6 brain sections, N = 4. Student’s t‐test is used for statistical analysis.
-
GRepresentative images of astrocytes labeled with Aldh1l1‐EGFP reporter in cortex and OB of Daam2 cHet and cKO mice at postnatal day 28 (P28). Scale bar: 50 μm.
-
H–KOverall complexity of astrocytes with Aldh1l1‐EGFP reporter was measured by Sholl analysis (two‐way ANOVA) and total process length (Student’s t‐test). Data are presented as mean ± SEM. n = 6–10 cells from N = 3–5 mice per genotype, *P < 0.05, ****P < 0.0001.
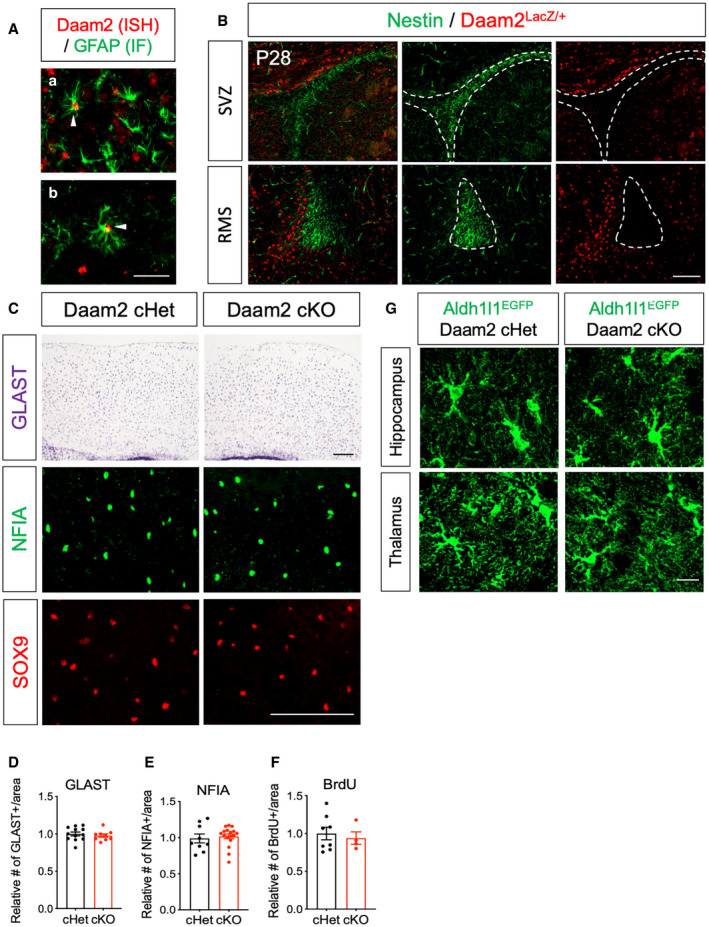
Figure EV1. Daam2 is expressed in astrocytes, and Daam2 loss regulates morphology specifically with no difference in astrocyte number.
-
ACo‐labeling of Daam2 (red) and mature astrocyte marker GFAP (green) with fluorescent in situ hybridization and immunostaining in OB (a) and cortex (b). Filled triangles indicate co‐labeled cells. Scale bar: 50 μm.
-
BImmunostaining from Daam2LacZ/+ animals indicates that Daam2 (LacZ+, Red) is not expressed in Nestin‐positive (Green) neural progenitors. SVZ: subventricular zone, RMS: rostral migratory stream. Scale bar: 100 μm.
-
C–FRepresentative images (C) and quantification (D–F) of GLAST+, NFIA+, and BrdU+ cells in Daam2 cHet and Daam2 cKO mice. Data are presented as mean ± SEM. Student’s t‐test was used for statistical comparisons. N = 4–17 images from N = 3–5 mice. Scale bar: 100 μm.
-
GRepresentative images of astrocyte with Aldh1l1‐GFP reporter in hippocampus and thalamus of Daam2 cHet and cKO mice at postnatal day 28. Scale bar: 50 μm.
To further assess the role of Daam2 in astrocyte development, we next generated astrocyte‐specific Daam2 null mice by crossing Daam2‐floxed mice (Daam2F/F) with an Aldh1l1‐Cre driver allele (Tien et al, 2012; Ding et al, 2020) (Aldh1l1‐Cre; Daam2F/F, denoted as Daam2 cKO). Daam2 cKO mice are viable, fertile, and showed no gross changes in appearance compared with controls (littermate Daam2F/F and Aldh1l1‐Cre; Daam2F/+, denoted as Daam2 cHet). We further crossed Daam2 cKO mice with the Aldhl1l1‐EGFP reporter line, allowing for confirmation of efficient Daam2 deletion via real‐time qPCR in FAC‐sorted, GFP‐positive astrocytes (Fig 1D). Using the Daam2 conditional allele alongside this reporter, we next assessed various astrocytic phenotypes via co‐labeling with cell‐specific markers. We did not observe significant changes in the overall numbers of astrocytes in Daam2 cKO mice compared with controls during development (Figs 1E and F, and EV1C–F). However, we found a profound increase in astrocytic complexity in Daam2 cKO mice with the Aldh1l1‐EGFP reporter at postnatal day 28 (P28) as noted by Sholl analysis and total process length. Notably, this increased morphological complexity was region specific. Depletion of Daam2 had an effect on morphological complexity in cortex and OB astrocytes (Fig 1G–K) whereas it had only mild effects on hippocampal and thalamic astrocytes (Fig EV1G). Given this finding, we focused our subsequent studies on enhanced morphological phenotypes in cortex and OB with more in‐depth approaches.
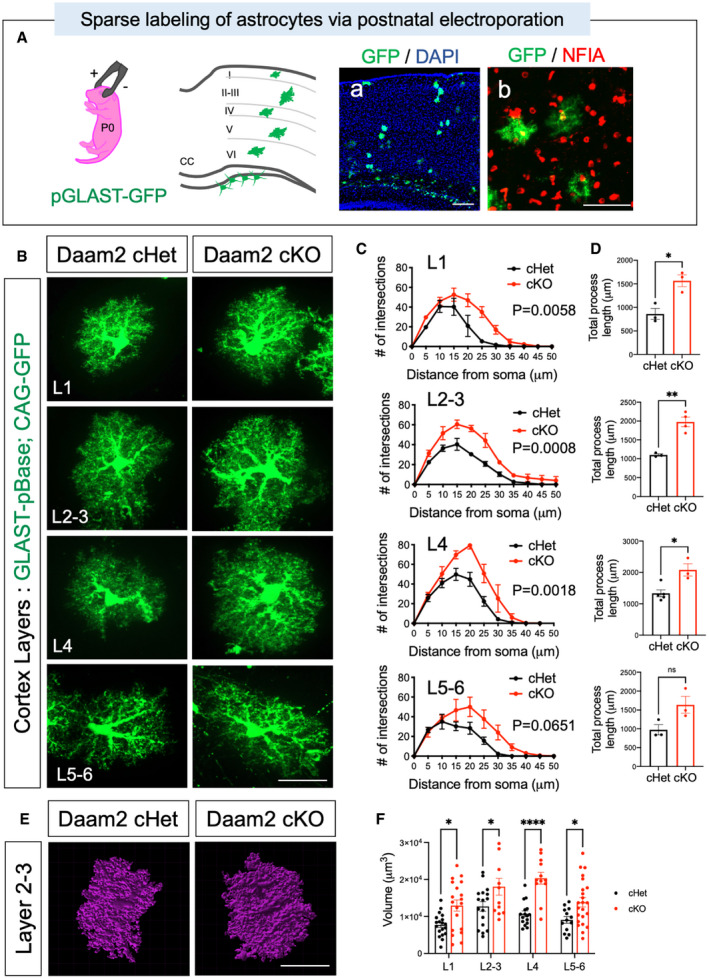
Cortical astrocytes display layer‐specific morphological and molecular differences (Lanjakornsiripan et al, 2018). To precisely define the morphological changes in astrocytes from different cortical layers, we sparsely labeled cortical astrocytes with a GFP transgene by performing postnatal electroporation (Stogsdill et al, 2017) in Daam2 cKO mice. After labeling, we performed high‐resolution confocal imaging for GFP+ astrocytes, confirmed by staining with the pan‐astrocyte marker, NFIA (Fig EV2A). Sholl analysis revealed that GFP+ astrocytes from Daam2 cKO mice had markedly increased complexity and branch lengths compared with astrocytes from Daam2 cHet controls in all layers (Fig EV2B–D and Appendix Fig S1), suggesting that increased morphological complexity of Daam2‐deficient astrocytes was not limited to any particular cortical layer. Furthermore, utilizing this high‐resolution confocal imaging approach, we found that depletion of Daam2 in cortical astrocytes significantly enhanced astrocyte volume (Fig EV2E and F).
Figure EV2. High‐resolution imaging reveals significant morphological alterations of cortical astrocytes.
-
ASchematic of postnatal electroporation for labeling single astrocytes in cortex. Representative image of GFP‐labeled astrocytes in cortical barrel with DAPI staining and immunostaining with pan‐astrocyte marker, NFIA confirmed GFP‐labeled cells as astrocytes.
-
BMorphology of single cortical astrocytes was visualized and evaluated in each cortical layer using postnatal electroporation of a GFP transgene under control of astrocyte‐specific GLAST promoter‐driven pBase. Scale bar: 20 μm.
-
C, DMorphological alterations in single astrocytes of Daam2 cHet and cKO mice were assessed and quantified as in Fig 1H–K. Data are presented as mean ± SEM. Total n = 13–14 cells from N = 5 mice per genotype were used for analysis. Two‐way ANOVA (C) and Student’s t‐test (D) are used for statistics. *P < 0.05, **P < 0.01.
-
E, FRepresentative images (E) and quantification (F) of 3D volume analysis in astrocytes of Daam2 cHet and cKO mice. Scale bar: 20 μm. (F) Data are presented as mean ± SEM. Total n = 12–23 cells from N = 5 mice per genotype were used for analysis. Two‐way ANOVA is used for statistics. *P < 0.05, ****P < 0.0001.
Prior studies have shown that newborn neurons populating the granule layer of the OB may influence astrocyte morphology (Foo & Dougherty, 2013; Winchenbach et al, 2016). Given that Aldh1l1‐Cre is also known to potentially affect neuronal progenitors, this raised the possibility that effects of Daam2 in OB astrocytes may occur secondary to unintended alterations in neurogenesis in our Daam2 cKO mice. Although our expression analysis in Daam2 reporter mice (Fig EV1B) and previously published cell type‐specific transcriptome profiling (Zhang et al, 2014; Hrvatin et al, 2018; Zeisel et al, 2018) suggests Daam2 expression is specific to glial cells, we performed additional marker analysis in Daam2 cKO mice. No changes in NeuN+ interneurons, Sox9+ astrocytes, or proliferating cell numbers (BrdU+ or Ki67+) were noted in OB of Daam2 cKO compared with Daam2 cHet controls (Fig EV3A–E), suggesting that neurogenesis was unaffected in the Daam2 cKO mouse lines. Nevertheless, to circumvent unintended effects on neurogenesis, we performed viral manipulations to delete Daam2 selectively in OB astrocytes. Toward this, we injected an adeno‐associated virus (AAV) containing Cre recombinase and RFP driven by an astrocyte‐specific GfaABC1D promoter (Srinivasan et al, 2016) into OB of adult Daam2F/F; Aldh1l1‐EGFP mice (Fig EV3F–I) and examined astrocyte morphology in OB. Similar changes in the morphological complexity of Daam2‐deficient astrocytes were observed three weeks after viral injection, suggesting that Daam2 is required to maintain proper morphology of established (post‐developmental) astrocytes in the OB. Together, these findings indicate Daam2 is expressed in astrocyte lineage cells and plays critical, region‐specific roles in development and maintenance of astrocytic morphological complexity.
Figure EV3. Temporal loss of Daam2 in adult OB exhibits increased morphological complexity without altering neurogenesis.
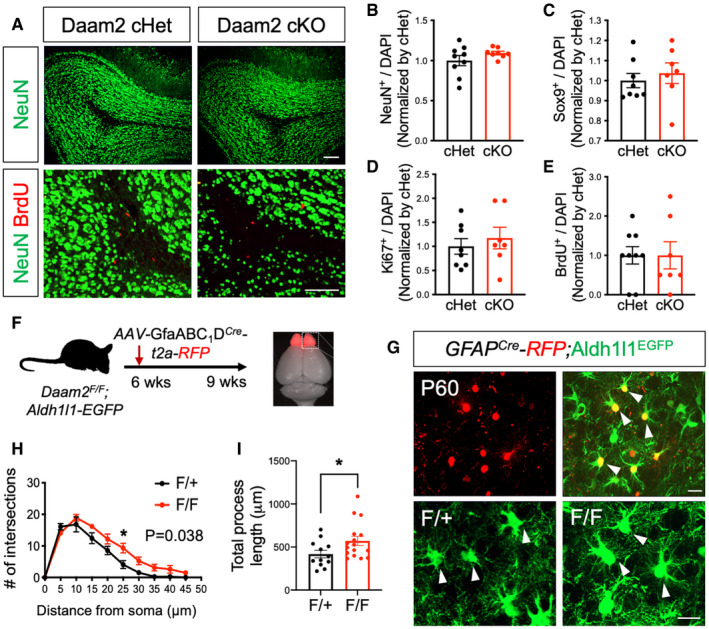
-
ARepresentative images of NeuN and BrdU staining in OB of Daam2 cHet and cKO mice. Scale bar: 100 μm.
-
B–ENo significant differences were found in numbers of NeuN+, Sox9+, BrdU+, and Ki67+ cells in OB of Daam2 cHet and Daam2 cKO mice. Data are presented as mean ± SEM. Student’s t‐test was used for statistical comparisons. n = 7–9 images from N = 3 mice per each genotype.
-
FTo visualize morphology of OB astrocytes after development, AAV‐GfaABC1D‐Cre‐t2a‐RFP was injected into 6‐week‐old Daam2F/+ and Daam2F/F mice with Aldh1l1‐EGFP reporter and analyzed three weeks later.
-
G–IVirus‐infected, RFP‐expressing, and Aldh1l1‐EGFP co‐labeled astrocytes (white triangles) were imaged and analyzed for their morphology as in Fig 1. Data are presented as mean ± SEM. n = 12, 16 cells, N = 3 mice per genotype. Two‐way ANOVA (H) and Student’s t‐test (I) were used for statistical comparisons. *P < 0.05. Scale bar: 50 μm.
Daam2 modulates GFAP expression, but not other reactive astrocyte markers
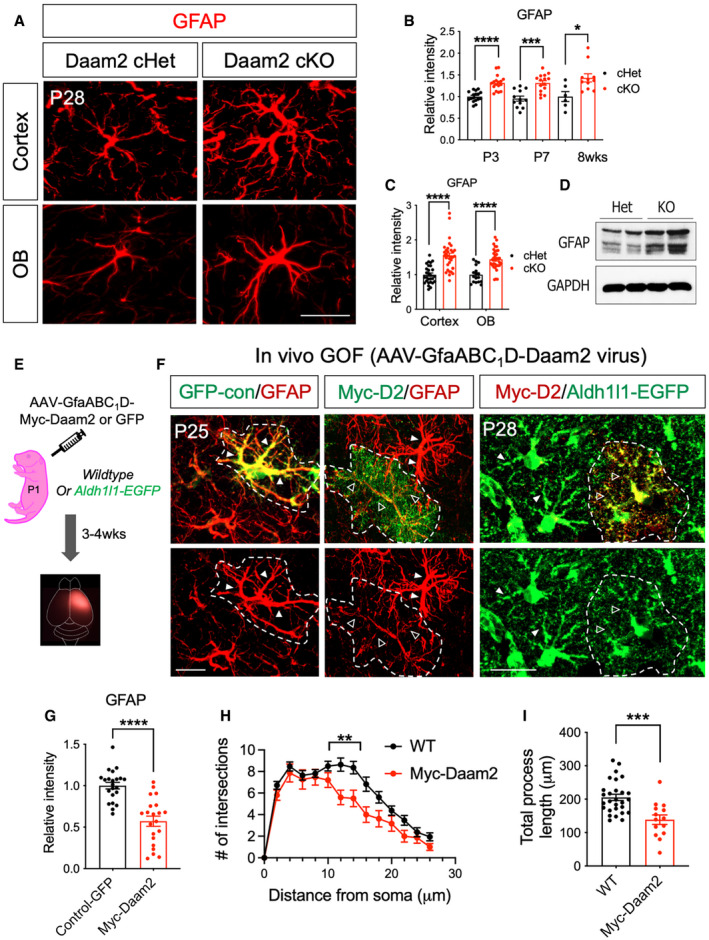
Regulation of cytoskeletal protein expression is an important determinant of astrocyte morphology and function (Molotkov et al, 2013; Schiweck et al, 2018; Zeug et al, 2018). Having established that Daam2 regulates astrocyte morphology, we next assessed changes in the expression of cytoskeletal proteins in Daam2‐deficient astrocytes. We found Daam2 loss dramatically increased levels of GFAP, a critical intermediate filament expressed in astrocytes in cortex and OB at P28 (Fig 2A and C). Enhanced expression of GFAP in cortex was maintained throughout development (Fig 2B). We also observed upregulation of GFAP and F‐actin expression in cultured primary astrocytes from Daam2 cKO mice (Figs 2D and EV4A and B), together with the in vivo observations, further suggesting that Daam2 suppresses GFAP expression. To determine whether Daam2 is sufficient to suppress GFAP expression, we next overexpressed Daam2 in astrocytes in vivo using an AAV containing Myc‐tagged Daam2 driven by GfaABC1D promoter injected in cortex (Fig 2E). We noted GFAP expression was reduced in the presence of elevated Daam2 (Fig 2F and G). Importantly, this result was coupled with a reduction in astrocyte morphological complexity (Fig 2F, H and I). While we observed increased GFAP expression in astrocytes of Daam2 cKO mice, we did not detect alterations in expression of other well‐established reactive astrocyte markers (Fig EV4C), indicating Daam2 specifically modulates astrocyte morphology via alterations in the intermediate filament GFAP.
Figure 2. Deletion of Daam2 increases intermediate filament GFAP whereas overexpression of Daam2 reduces GFAP and morphological complexity.
-
ARepresentative images of GFAP immunostaining in Daam2 cKO mice compared with controls in cortex and OB at P28. Scale bar: 20 μm.
-
BQuantification of GFAP immunostaining in Daam2 cHet and cKO mice from P3 to adult. Data are presented as mean ± SEM. n = 6–17 images from N = 4–6 mice per each genotype and age. Student's t‐test is used for statistics. *P < 0.05, ***P < 0.001, ****P < 0.0001.
-
CQuantification of GFAP immunostaining in Daam2 cHet and cKO mice in cortex and OB at P28. Data are presented as mean ± SEM. n = 17–33 images from N = 5 mice per each genotype and age. Student’s t‐test is used for statistics. ****P < 0.0001.
-
DWestern blot analysis showed a significant increase in the amount of GFAP protein in Daam2 KO primary astrocytes. N = 2.
-
ESchematic of virus injection to overexpress Daam2 in the cortex.
-
FOverexpression of Daam2 via injection of AAV‐GfaABC1D‐myc‐Daam2 in wild‐type P1 pups revealed reduced GFAP intensity compared with animals injected with control virus (AAV‐GfaABC1D‐GFP). AAV‐GfaABC1D‐myc‐Daam2 was injected into Aldh1l1‐GFP reporter mice for morphological complexity analysis, and infected cells (Myc positive) were compared with non‐infected cells. Dotted white line indicates virus‐infected cells. Filled triangles and empty triangles indicate astrocytic primary branches with the control and reduced level of GFAP or Aldh1l1‐GFP, respectively. Scale bar: 20 μm.
-
GQuantification of GFAP immunostaining in control virus‐positive cells and Daam2 overexpressing cells. Data are presented as mean ± SEM. n = 22 images from N = 3 mice per genotype. Relative fluorescent intensity was quantified, and Student’s t‐test was used, ****P < 0.0001.
-
H, ISholl analysis (H) and quantification of total length (I) of filament with overexpression of Daam2. Data are presented as mean ± SEM. n = 17–32 images from 3 to 4 mice per each. **P < 0.01, ***P < 0.001. Two‐way ANOVA and Student’s t‐test were used for (H) and (I), respectively.
Figure EV4. Daam2‐deficient astrocytes display increased GFAP in vitro and normal level of MaoB, S100b, and AldoC expression in vivo .
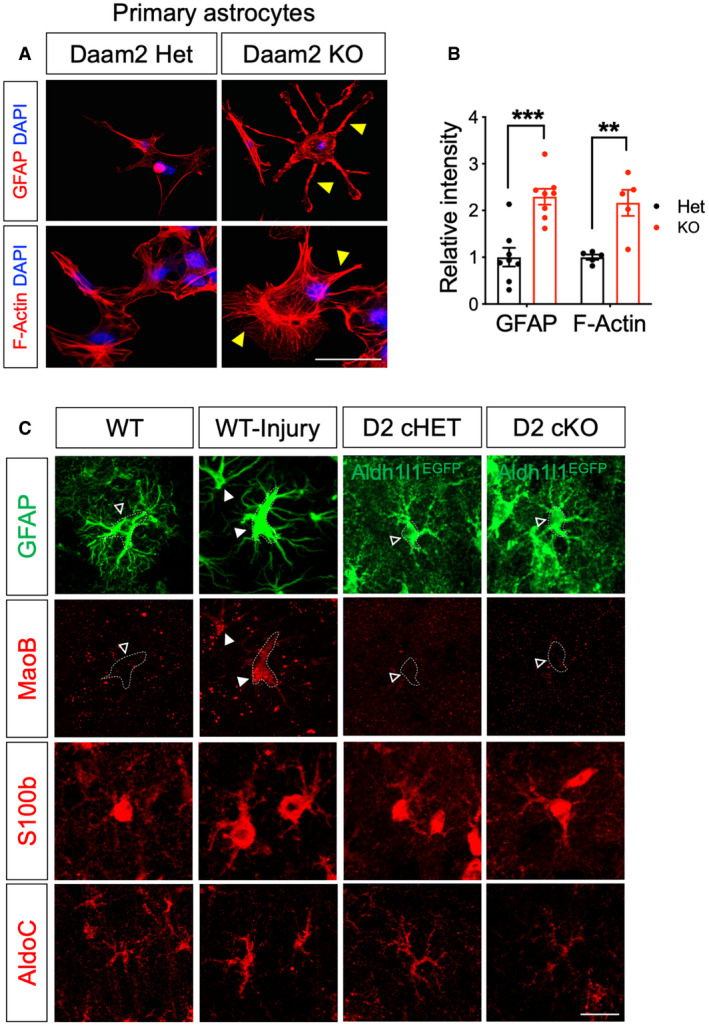
-
A, BRepresentative images and quantification of immunostaining of GFAP and F‐actin in cultured primary astrocyte of Daam2 Het and KO mice. Yellow triangles indicate increased astrocytic processes in Daam2 KO astrocytes. Scale bar: 20 μm. Experiments are repeated as triplicate, and data are presented as mean ± SEM. Student’s t‐test was used, **P < 0.01, ***P < 0.001.
-
CRepresentative images of immunostaining with reactive astrocyte markers. Photothrombotic stroke was induced in the ipsilateral cortex of wild‐type mice and used as a positive control for reactive gliosis. Dotted outline indicates astrocyte cell body labeled with GFAP or Aldh1l1‐EGFP. Filled triangles and empty triangles indicate increased expression and baseline level of expression or no changes in MaoB staining, respectively. Scale bar: 20 μm.
Loss of astrocytic Daam2 modulates spontaneous astrocytic Ca2+ activity in a region‐specific manner
The region‐specific morphological changes in astrocytes in Daam2 cKO mice prompted the question of whether loss of Daam2 affects the physiological activity of astrocytes. Astrocytes display characteristic passive conductance with a highly negative and stable membrane potential, which manifests as a characteristic linear current to voltage (I‐V) dynamic (Kressin et al, 1995). To determine whether astrocytes from Daam2 cKO mice exhibited altered passive conductance, we generated brain slices from adult mice and performed whole‐cell recordings. We found no difference in the I‐V plots compared with the littermate controls, indicating that astrocyte membrane properties and passive conductance are not impaired by loss of Daam2 in the cortex or OB (Appendix Fig S2).
Astrocytic Ca2+ levels are an important parameter of normal physiologic activity in astrocytes (Di Castro et al, 2011; Khakh & McCarthy, 2015; Semyanov et al, 2020). To address whether astrocytic Ca2+ activity was altered in the absence of Daam2, we used an astrocyte‐specific AAV virus engineered to express the genetically encoded calcium indicator GCaMP6f under the control of the GfaABC1D promoter (Haustein et al, 2014). We performed two‐photon imaging of brain slices from Daam2 cKO mice after three weeks of AAV‐GfaABC1D‐GCaMP6f expression to assess Ca2+ dynamics in cortex and OB. Using established algorithms to quantify regional Ca2+ activities (Srinivasan et al, 2015), we observed decreased Ca2+ activity as measured by fluorescence amplitudes and durations in Daam2‐deficient astrocytes within cortex, but no apparent differences in total AUC (area under the curve) of astrocyte Ca2+ signals (Fig 3A–D). Surprisingly, we observed the opposite phenomenon in OB, where Daam2 cKO astrocytes displayed increased GCaMP response amplitudes and total AUC, coupled with no differences in the duration of Ca2+ events (Fig 3E–H). These observations suggest that (i) Daam2 plays distinct roles in cortex and OB astrocytes, and (ii) the relationship between astrocyte morphology and physiology is region‐dependent and is likely a product of the unique cellular and environmental contexts in each region.
Figure 3. Deletion of Daam2 differentially alters astrocytic calcium dynamics in the cortex and olfactory bulb.
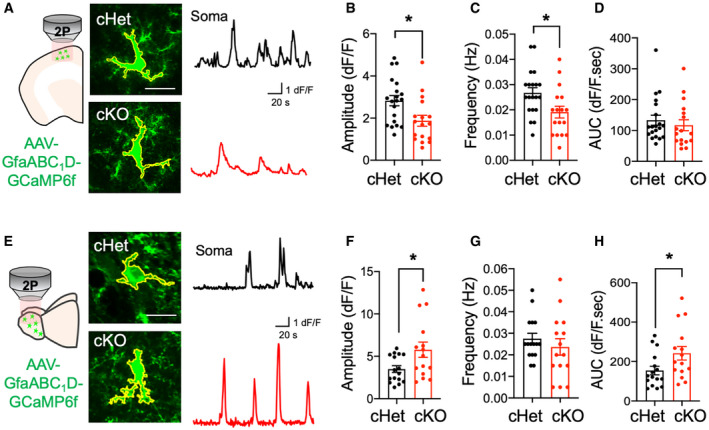
-
ASchematics of astrocytic calcium imaging in cortex from ex vivo slices and representative images of spontaneous calcium signaling (green) of Daam2 cHet and cKO mice injected with AAV‐GfaABC1D‐GCaMP6f virus. Approximate territory of astrocytes including soma and multiple microdomains is outlined in yellow. Representative traces of GCaMP6f signal in Daam2 cHet and cKO astrocytes are shown in black and red, respectively.
-
B–DQuantification of amplitude (B), frequency (C), and area under curve (AUC) (D) of ΔF/F GCaMP6f signal events in cortex. Daam2‐deficient astrocytes have lower amplitude and frequency of signal events in cortex. Data are presented as mean ± SEM. Total number of cells is n = 20, 17 from N = 3–4 mice of each genotype. Student’s t‐test was used for statistics, *P < 0.05.
-
ESchematics of astrocytic calcium imaging and representative images of spontaneous calcium signaling (green) of Daam2 cHet and cKO mice injected with AAV‐GfaABC1D‐GCaMP6f virus in OB. Approximate territory of astrocytes including soma and multiple microdomains is outlined in yellow. Representative traces of GCaMP6f signal in Daam2 cHet and cKO astrocytes are shown in black and red, respectively.
-
F–HQuantification of amplitude (F), frequency (G), and area under curve (AUC) (H) of ΔF/F GCaMP6f signal events in OB. Amplitude and AUC of Daam2‐deficient astrocytes are significantly increased in OB. Data are presented as mean ± SEM. Total number of cells is n = 16, 15 (F–H) from N = 3–4 mice of each genotype. Student’s t‐test was used for statistics, *P < 0.05.
Congruent changes in astrocyte morphology by loss of Daam2 differentially impact neuronal activity in the cortex and olfactory bulb
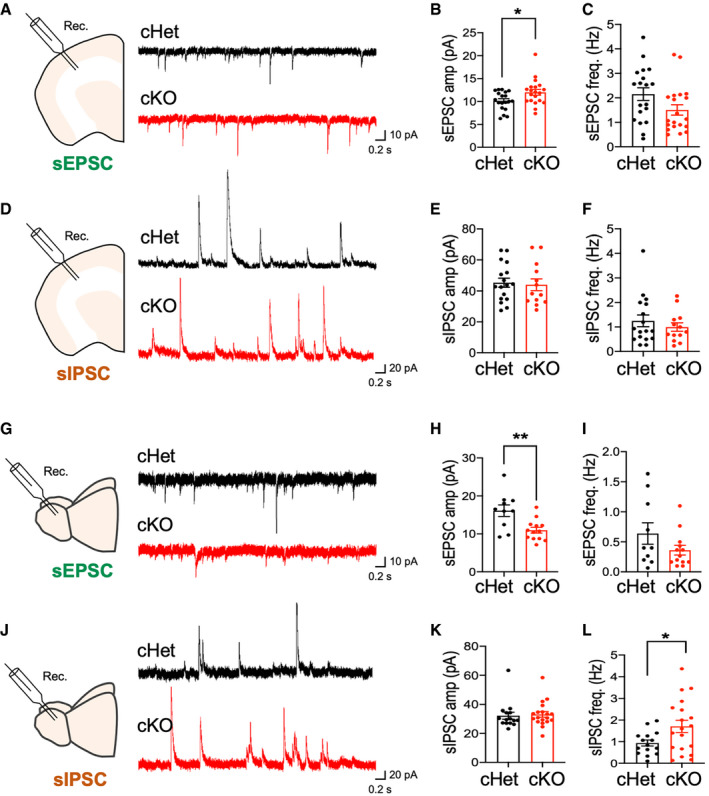
Given the intimate association between astrocytic and neuronal function, and our observation that loss of Daam2 alters morphological complexity and Ca2+ activity of astrocytes, we investigated whether synaptic transmission was similarly affected in Daam2 cKO mice using whole‐cell patch‐clamp recordings (Sakmann et al, 1989) in brain slices from adult mice. We measured electrical properties of cortical layer 2/3 pyramidal cells and OB mitral cells in Daam2 cKO mice compared with Daam2 cHet controls. Intriguingly, we found a significant increase in spontaneous excitatory postsynaptic current (sEPSCs), and no changes in spontaneous inhibitory postsynaptic currents (sIPSCs) in cortical neurons in Daam2 cKO mice (Fig 4A–F). In contrast, we observed a reduction in the amplitude of sEPSCs and increased frequencies of sIPSCs in OB neurons of Daam2 cKO mice (Fig 4G–L). These data indicate Daam2 loss has region‐specific effects on synaptic function, strengthening excitatory transmission in cortex and inhibitory synaptic transmission in OB.
Figure 4. Daam2 deletion in astrocytes results in differential synaptic activity in cortex and olfactory bulb.
-
A–F(A, D) Schematics of whole‐cell recording in ex vivo brain slices and representative traces of spontaneous EPSC and IPSC in neurons in cortex from Daam2 cHet (top, black) and cKO (bottom, red) mice. (B and C, E and F) Quantification of amplitude and frequency of sEPSC and sIPSC in each genotype. Data are presented as mean ± SEM. Total number of cells is n = 19, 20 (B and C) and 17, 13 (E and F) from N = 7 mice for each genotype. Student’s t‐test was used for comparison, *P < 0.05.
-
G–L(G, J) Schematics of whole‐cell recording in ex vivo brain slices and representative traces of spontaneous EPSC and IPSC in neurons of OB from Daam2 cHet (top, black) and cKO (bottom, red) mice. (H and I, K and L) Quantification of amplitude and frequency of sEPSC and sIPSC in each genotype. Data are presented as mean ± SEM. n = 10, 13 (H and I) and 15, 19 (K and L) from N = 6 mice. Student’s t‐test was used for comparison, *P < 0.05, **P < 0.01.
Since our data reveal decreased sEPSC amplitude and greater sIPSC frequency in OB of Daam2 cKO mice, we next examined whether the loss of astrocytic Daam2 impacts olfactory sensing after odor stimulation. To test this, we examined olfactory sensory response maps from different populations of adult OB neurons by crossing Thy1‐GCaMP3 reporter mice (Chen et al, 2012) with Daam2 KO mice. Using the Thy1‐GCaMP3 reporter, which is expressed in mitral/tufted cells, we measured odor‐evoked Ca2+ responses on the dorsal surface of OB while presenting a panel of odorants. Strikingly, we found that individual odorants activated different sets of glomeruli in Daam2 Het versus Daam2 KO mice (Appendix Fig S3). These stark differences in the odor‐specific topographical maps between the genotypes were not due to intrinsic variation between animals given that the general area of activation was consistent and reproducible in control mice (Appendix Fig S3); note that there were no obvious patterns detected in the background activity of Daam2 KO mice after various odor stimulations. Together, these findings indicate that loss of astrocytic Daam2 results in alterations in inhibitory network responses in OB, which manifests as alterations in odorant response maps.
Loss of Daam2 in astrocytes results in olfactory behavioral deficits
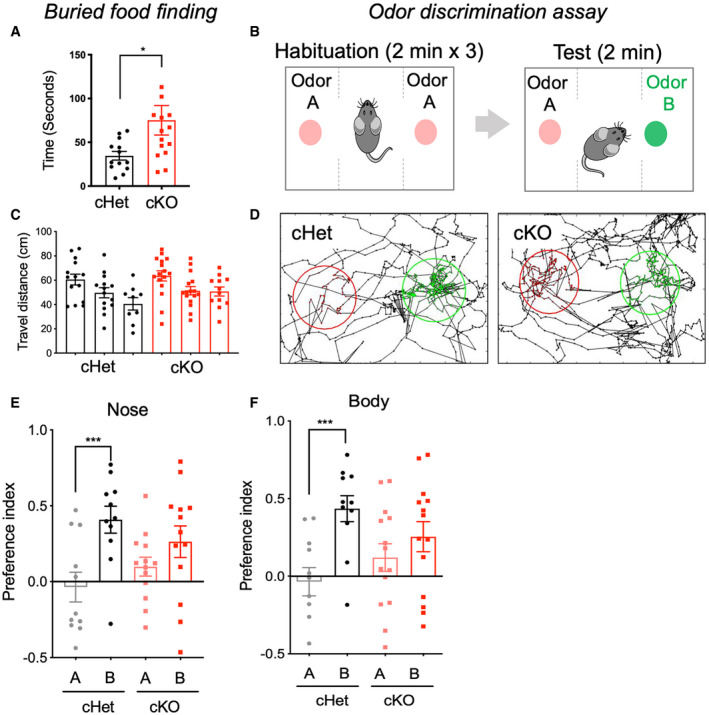
Mice rely on olfaction for navigating new environments, finding food and for executing a variety of social, sexual, and parental behaviors, and the OB serves as a critical center for recognizing and processing of olfactory information (Su et al, 2009). Given that astrocyte‐specific Daam2 loss induces both astrocytic and neuronal activity changes in OB, we performed behavioral assays to evaluate any potential effects on olfactory function. We first performed a buried food assay, which tests the ability of food‐restricted mice to detect volatile odors and uncover hidden food pellets using olfactory cues (Alberts & Galef, 1971). In this assay, the latency of Daam2 cKO mice to locate buried food was significantly longer than observed in control mice, suggesting an olfactory impairment in Daam2 cKO mice (Fig 5A). Since Daam2 KO mice displayed alterations in olfactory sensory maps with odorant stimulation (Appendix Fig S3), we further hypothesized Daam2 cKO mice may exhibit deficits in distinguishing different odorants. To test this, we next performed an olfactory discrimination (habituation/dishabituation) assay (Yang & Crawley, 2009; Takahashi et al, 2016). In the habituation phase, identical odors were presented on opposite sides of a chamber for three consecutive sessions. Following habituation, a new odor was introduced to only one side of the test chamber for dishabituation (Fig 5B). Investigation time of the novel odor was recorded and analyzed to calculate a preference index. Daam2 cKO mice showed no significant difference in the preference index between habituation and novel odors, suggesting that they failed to distinguish the novelty of the new odor. Daam2 cHet control animals, however, showed a significant increase in preference index for the novel odor (Fig 5D–F). Importantly, we observed no difference in locomotor activity or travel distance in Daam2 cKO mice compared with control animals (Fig 5C). Collectively, these results reveal that olfactory function is impaired in Daam2 cKO mice, suggesting that disruption of the astrocyte‐neuronal network by Daam2 deletion leads to defective sensory processing.
Figure 5. Astrocyte‐specific loss of Daam2 impairs olfaction behaviors.
-
AAstrocyte‐specific Daam2 cKO mice show delays in buried food finding. N = 13, 15 mice per genotype. Data are presented as mean ± SEM. Student’s t‐test was used, *P < 0.05.
-
BIllustration of odor discrimination assay. For habituation, each mouse was habituated in a chamber with identical stimulation (odor A) on both sides for 2 min, repeated three times. To test for odor discrimination, a newly introduced odor (odor B) was randomly assigned to one of the sides.
-
CTotal travel distance was measured by tracing the nose and body of each mouse in each habituation session of the odor discrimination assay. Both genotypes show no difference in total travel distance. Total N = 11, 13 mice were used for each genotype. Data are presented as mean ± SEM. Student’s t‐test was used.
-
DRepresentative traces of nose position tracking of cHet and cKO mice during the discrimination test period. Habituated (odor A) and the new odor (odor B) were placed in the center of the indicated area (odor A: red circle, odor B: green circle). Nose positions within the circled area were indicated with the corresponding color for each odor.
-
E, FPreference index was tracked and calculated using MATLAB and OptiMouse. One‐sample t‐test and Wilcoxon test were used for statistical analysis. Data are presented as mean ± SEM. ***P < 0.001. Total N = 11, 13 mice were used for each genotype.
Mass‐spec proteome analysis reveals region‐specific differential protein expression in the absence of Daam2
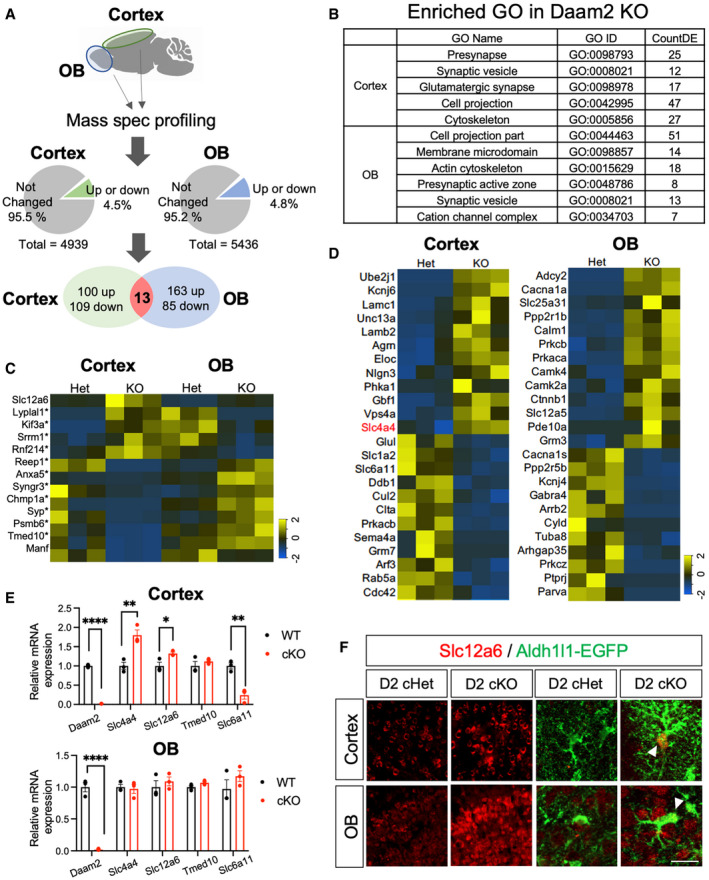
To understand how loss of Daam2 differentially impacts multiple aspects of CNS physiology in a region‐specific manner, we performed proteome profiling in both cortex and OB of Daam2 KO mice and Daam2 Het controls (Fig 6A). We generated proteome libraries for each region and each genotype with three biological replicates via nano‐HPLC‐MS/MS analysis. A total of 4939 and 5436 peptides/gene products were detected in cortex and OB, respectively. Among these, we found that 222 gene products (4.49%) in cortex and 261 gene products (4.8%) in OB showed differential expression between the Daam2 KO and control animals using cutoff criteria of at least 1.5‐fold change and P value < 0.05 (Fig 6A, C and D). Gene Ontology (GO) enrichment analysis revealed that commonly dysregulated genes in both cortex and OB are overwhelmingly linked to synaptic function and cytoskeleton modification, which correlate with the functional changes we observed in Daam2 cKO mice (Fig 6B). Among these include glutamatergic synaptic components in the cortex, and potassium channel complexes and calcium‐dependent process/calcium‐dependent kinase signaling in the OB (Fig 6B and D). Next, we focused on the cohort of genes that are commonly regulated by Daam2 in both regions and identified 13 candidates, 11 of which showed changes in the opposite direction with respect to relative abundance between cortex and OB when comparing Daam2 KO and Daam2 Het mice (Fig 6C). For example, in Daam2 cKO mice, Kif3a (kinesin family member 3A) shows a 1.9‐fold increase in expression in the cortex, whereas it was downregulated 2‐fold in OB; Slc4a4 showed a 2‐fold increase in cortical astrocytes, but no difference in OB; while Slc12a6 is up‐regulated in both regions (Fig 6C–F). Differential expression profiles of selected candidates were validated by measuring transcript and protein expression levels in both cortex and OB via real‐time qPCR and immunostaining, respectively (Fig 6E and F). Together these data indicate that loss of Daam2 differentially influences the proteome of two different brain regions, which further supporting the notion that Daam2 operates in a region‐specific manner to influence circuit function.
Figure 6. Proteome profiling reveals region‐specific regulation by Daam2.
-
ASchematic of mass‐spec (MS) proteome profiling. Cortex and OB were isolated from Daam2 Het and KO mice (N = 3 each) and digested for peptide library generation. Total numbers of detected and differentially expressed proteins in each brain region are indicated in pie charts. Venn diagram shows differentially expressed genes in each region and genes differentially expressed in both.
-
BGene ontology (GO) analysis of differentially expressed genes in each region.
-
CHeat maps of commonly regulated genes in both regions. Genes differentially regulated in opposite directions in the cortex and OB are marked with *.
-
DHeat maps of uniquely up‐ and downregulated genes in the cortex and OB.
-
E, FqRT–PCR (E) and immunostaining (F) confirm differential expression of genes found in MS profiling. qRT–PCR is repeated as triplicate and is presented as mean ± SEM. *P < 0.05, **P < 0.01, ****P < 0.0001. Student’s t‐test was used. Scale bar: 20 μm.
Slc4a4 is critical for astrocyte morphology and maturation, and it reverses phenotype by loss of Daam2 in cortex but not OB
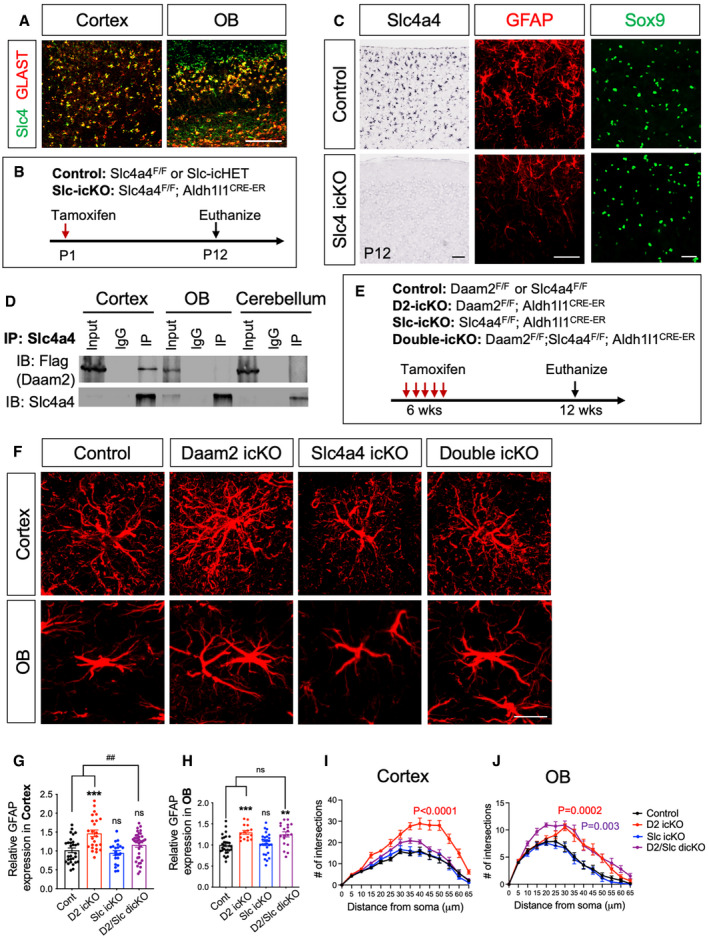
Next, we prioritized genes for further analysis with the following criteria: (i) enriched or exclusively expressed in astrocytes, and (ii) differentially regulated between cortex and OB in Daam2 cKO mice. Further, we compared a list of differentially expressed genes by Daam2 loss with the previous immunoprecipitation‐mass spec screen that identified genes have strong affinity for binding to Daam2 (Ding et al, 2020). Among genes meeting these criteria, we identified Slc4a4, an electrogenic Na+/HCO3 ‐ cotransporter which is known to play critical roles in regulating intra‐ and extra‐cellular pH, metabolic activity, and cell‐to‐cell communication (Ruminot et al, 2011; Theparambil et al, 2014, 2020; Schrödl‐Häußel et al, 2015; Khakipoor et al, 2019). Slc4a4 is exclusively expressed in astrocytes, and previous data from our laboratory indicate Slc4a4 displays a strong affinity for binding to Daam2. Interestingly, Slc4a4 relative abundance was noted to be increased only in cortex following Daam2 loss at both the transcriptional and translational levels (Fig 6D and E). In order to characterize the relationship between Daam2 and Slc4a4 in astrocyte development, we first confirmed Slc4a4 expression in the astrocyte lineage (GLAST+) in both cortex and OB (Fig 7A). While Slc4a4 function has been implicated in astrocyte physiology, its prospective role in regulating astrocyte morphology is unknown. We next examined whether Slc4a4 is critical for astrocyte morphology and function in vivo. Toward this, we generated astrocyte‐specific knockout of Slc4a4 by crossing Slc4a4‐floxed mice with the Aldh1l1‐Cre line (Aldh1l1‐Cre+/−; Slc4a4F/F, denoted Slc4a4 cKO). Slc4a4 cKO mice showed a smaller body weight and size when compared to littermate controls (wild‐type and heterozygous control) and exhibited early postnatal lethality from P3 to P14 (Appendix Fig S4A and B). We also found a significant decrease in the number of astrocyte precursors labeled with Sox9 in Slc4a4 cKO at P0 compared to littermate controls (Appendix Fig S4C). To overcome the postnatal lethality of Slc4a4 cKO, we next generated a temporally controlled astrocyte‐specific conditional null allele by crossing Slc4a4‐floxed mice with the Aldh1l1‐CreER line (Aldh1l1‐CreER+/−; Slc4a4F/F, denoted Slc4a4 icKO). For postnatal deletion of Slc4a4 in the astrocyte lineage, we injected tamoxifen into P1 pups (Fig 7B). After confirming loss of Slc4a4 by immunostaining and in situ hybridization (Appendix Fig S4D and E), we assessed cellular phenotypes with various astrocyte markers. Unlike Slc4a4 cKO mice at P0, we did not observe a reduction in the number of Sox9+ cells two weeks after postnatal deletion (Fig 7C and Appendix Fig S4F). However, we found a dramatic decrease in the expression of GFAP in Slc4a4 icKO mice compared with littermate controls at P12 (Fig 7C), suggesting that Slc4a4 is important for astrocyte development.
Figure 7. Slc4a4 regulates astrocyte maturation by interacting with Daam2.
-
ADouble fluorescence in situ hybridization images indicating co‐labeling of Slc4a4 (green) and GLAST (red). Scale bar: 100 μm.
-
BSchematics showing induction of temporally controlled astrocyte‐specific conditional null alleles.
-
CIn situ hybridization confirms deletion of Slc4a4 in the cortex. GFAP immunostaining intensity was decreased significantly in Slc4a4 icKO at P12 whereas Sox9+ cell number remained unchanged. Scale bar: 100 μm.
-
DCo‐immunoprecipitation revealing that Slc4a4 associates with Daam2 in vivo.
-
ESchematics showing induction of temporally controlled astrocyte‐specific conditional null alleles.
-
F–JRepresentative images of GFAP immunostaining (F) and quantification (G–J) in controls, Daam2 icKO, Slc4a4 icKO, and double icKO 6 weeks after tamoxifen injection. Scale bar: 20 μm. Data are presented as mean ± SEM. n = 10–24 images, N = 3–4 mice per genotype. Scale bar: 20 μm. Two‐way ANOVA is used for statistics. [*] and [#] indicates comparison with control and double icKO, respectively. **P < 0.01, ***P < 0.001, ## P < 0.01.
To dissect the functional relationship between Daam2 and Slc4a4 in cortex and OB, we first evaluated the association between Daam2 and Slc4a4 in the brain using a Daam2‐Flag‐knock‐in reporter mouse line (Appendix Fig S4G). Immunoprecipitation of endogenous Slc4a4 revealed Daam2 strongly associates with Slc4a4 in the adult cortex but not in OB or cerebellum (Fig 7D). These data indicate that Daam2 selectively interacts with Slc4a4 in a region‐specific manner.
Having established that Slc4a4 expression was elevated in the cortex of the Daam2 cKO mice and that Slc4a4 associates with Daam2, we next explored the possibility that increased expression of Slc4a4 is necessary for the increased astrocytic morphological complexity noted in cortex of the Daam2 cKO mice. To test this, we next assessed the genetic interactions between Daam2 and Slc4a4. To circumvent lethality caused by embryonic and early postnatal loss of Slc4a4, we generated temporally controlled astrocyte‐specific conditional null alleles in the astrocyte lineage by crossing our existing floxed alleles (Aldh1l1‐CreER+/−; Daam2F/F (Daam2 icKO), Aldh1l1‐CreER+/−; Slc4a4F/F (Slc4a4 icKO), Aldh1l1‐CreER+/−; Daam2F/F; Slc4a4F/F (Daam2‐Scl4a4 idcKO), Fig 7E). Notably, we observed increased numbers of GFAP+ branches in astrocytes of adult Daam2 icKO mice, similar to our developmental knockout (Daam2 cKO). However, loss of both Slc4a4 and Daam2 in idcKO mice significantly reversed the Daam2 icKO phenotype in the adult cortex, but not in OB (Fig 7F–J). These findings provide compelling evidence that the genetic interaction that occurs between Daam2 and Slc4a4 influences astrocyte morphology in a region‐specific manner. Furthermore, our findings define a novel role for the Daam2‐Slc4a4 axis in regulating astrocyte morphology and neural circuit development in a region‐specific manner.
Discussion
This work focuses on critical functions of Daam2 in astrocytes during CNS development. Using a combination of complementary loss‐ and gain‐of‐function studies, in vivo calcium imaging, electrophysiological recordings, and behavioral assays, we found that Daam2 plays key roles in morphological maturation and electrophysiologic properties of astrocytes. In addition, the ramified astrocyte morphologies observed in Daam2 cKO mice were associated with divergent trends in astrocytic calcium dynamics and altered neuronal activity in a region‐specific manner, along with alterations in sensory‐related behavior. Our mechanistic studies identified a critical interaction between Daam2 and Slc4a4 in maintaining proper astrocyte morphology. Together, our studies reveal novel mechanisms regulating astrocyte morphogenesis and delineate how these mechanisms alter glia–neuron interactions at the circuit level during brain development.
Developmental maturation of astrocytes requires a significant amount of cytoskeletal remodeling (Schiweck et al, 2018; Zeug et al, 2018; Zhou et al, 2019). Although formin family proteins are predominantly expressed in the developing CNS and display a wide range of activities to regulate the actin cytoskeleton, the exact mechanisms by which formin proteins influence astrocyte morphology and function remain unknown (Wallar & Alberts, 2003; Kawabata Galbraith & Kengaku, 2019). Our study reveals how the formin protein, Daam2, modulates astrocyte morphology, synapse formation, and physiological function of CNS circuitry during development. Furthermore, our study provides the first evidence that a formin protein plays a pivotal role in structure and function of astrocytes during brain development and does so in a region‐specific manner. Previous studies on the formin protein Daam1, a member of the Daam family, show that it may aid F‐actin nucleation, bundling, and elongation in the development of the heart and CNS (Lu et al, 2007; Salomon et al, 2008; Li et al, 2011; Jaiswal et al, 2013; Ajima et al, 2015). Interestingly, we found an increase in F‐actin filaments in cultured primary astrocytes upon the loss of Daam2, suggesting a distinctive role of Daam2 compared with Daam1. Moreover, many formin proteins interact with small GTPases, and Daam2 contains a well‐conserved GTPase binding domain, which is functionally important for its association with interacting molecules (Lee et al, 2015). Small GTPases such as RhoA, Cdc42, and Rac1 are strongly implicated in regulation of astrocyte morphology, although many of these roles have been defined using in vitro systems, and thus, their in vivo function requires further exploration. It would be intriguing to clarify a function of Daam2 in regulation of actin filaments via small GTPases during astrocyte morphogenesis in vivo.
Furthermore, it is also possible that Daam2 may exert its effect on astrocyte morphology through the Wnt signaling pathway, which is a critical signaling cascade during development. We previously characterized Daam2 as a positive regulator of canonical Wnt signaling by stimulating signalosome formation during early spinal cord development and in oligodendrocytes (Lee & Deneen, 2012; Lee et al, 2015). There are few in vivo studies showing the role of Wnt signaling in astrocytes during development. It has previously been shown that inhibition of Wnt/β‐catenin signaling induces precocious astrogliogenesis in the early developing spinal cord, whereas excessive activation of Wnt/β‐catenin leads to disruption of astrogliogenesis (Sun et al, 2019). At the behavioral level, it has also been reported that loss of β‐catenin in GLAST expressing astrocytes enhances anxiety‐like behavior, while stress increases Wnt signaling in astrocytes (Vidal et al, 2019). Although further studies are required to fully understand whether and how Daam2 modulates Wnt signaling in astrocytes, our study clearly supports critical roles for Daam2 in maintaining proper astrocyte morphology, function, and circuit integrity.
Many studies have found reductions in the size and complexity of astrocytes upon loss of essential genes (Srinivasan et al, 2015; Stogsdill et al, 2017; Holt et al, 2019; Huang et al, 2020). Unexpectedly, loss of Daam2 results in cell size enlargement and increased complexity, whereas overexpression of Daam2 reduces morphological elaboration. One important remaining question in the field is how morphological alterations in astrocytes might influence synaptic activity, neural circuitry, and subsequent behavior. To this end, previous studies have defined a functionally important role for astrocytic coverage of synapses and variable coverage between synapses in a region‐specific manner (Ventura & Harris, 1999; Oliet et al, 2001; Bernardinelli et al, 2014; Perez‐Alvarez et al, 2014). It remains unclear whether changes in morphology directly lead to downstream changes in synaptic function in our model. Increased accessibility and the coverage of astrocytic processes in Daam2 cKO mice could reshape synapses. Alternatively, Daam2 might control synaptic connectivity by modifying the release of synaptogenic factors, or by modulating the expression and localization of neuron‐astrocyte adhesion molecules (Farhy‐Tselnicker et al, 2017; Stogsdill et al, 2017). Our finding that deletion of Slc4a4 restored morphological changes associated with Daam2 loss raises questions regarding whether double loss of Daam2 and Slc4a4 might also restore physiological functions of astrocytes including calcium transients and synaptic transmission. It is also possible the function of Slc4a4 is restricted solely to astrocyte morphogenesis. Clearly, further studies will be necessary to delineate exactly how Daam2 loss alters synapse formation, maintenance, and circuit function. In addition to changing physical interactions with synapses, alterations in morphological complexity may also modify astrocytic contacts with the cerebrovasculature through endfeet processes (Mulligan & MacVicar, 2004) and/or gap‐junction coupling to other astrocytes (Theis & Giaume, 2012; Fujii et al, 2017). Considering the role of Daam2 in regulating astrocyte morphology, it will be important to decipher whether loss of Daam2 alters BBB integrity and/or gap‐junction‐coupled networks.
The distinct effects of Daam2 deletion on astrocytic calcium transients and neuronal signaling in the cortex and OB are intriguing, given that loss of Daam2 led to ramified morphology in both regions. These findings raise the question of whether intrinsic mechanisms controlling astrocytic calcium transients and synaptic functions are different in astrocytes from different regions of brain. Recent studies provided evidence for heterogeneity in astrocytic calcium signaling (Khakh & McCarthy, 2015; Clarke et al, 2021), and distinct calcium dynamics in different cortical layers have been reported with astrocytic manipulations (Takata & Hirase, 2008). Moreover, a study comparing astrocytes from two different brain regions showed distinct mechanisms for maintaining basal astrocytic calcium level (Chai et al, 2017). Our proteome profiling data show that distinct sets of proteins are affected in cortex and OB by loss of Daam2, which may help explain the unique phenomena observed in each brain region. Furthermore, we characterized differential regulation of Slc4a4 by Daam2 in a region‐dependent manner in cortex and OB. Despite both Slc4a4 and Daam2 being expressed in astrocytes from both cortex and OB, their interaction is specific to cortex and deletion of Slc4a4 restored Daam2 loss only in cortex (Fig 7). This finding raises a question how Daam2 interacts with and regulates Slc4a4. Previously, we characterized Daam2's functional expression in Wnt signalosome near the membrane. One intriguing possibility is that Daam2 regulates proper localization of Slc4a4 into the membrane for its proper function as a channel. It is also possible there are region‐specific differences in other genes associated with Daam2‐Slc4a4 axis. Given the Daam2–Slc4a4 axis exhibits regional specificity, it will be important to decipher mechanisms underlying functional specialization of astrocytes from different brain regions.
Materials and Methods
Animals
All mice were maintained and studied according to protocols approved by the Institutional Animal Care and Use Committee of Baylor college of Medicine.
All genotypes were on the C57BL/6J strain and blinded for data acquisition and analysis and experiments included both genders and age‐matched controls. Aldh1l1cre and Aldh1l1‐EGFP transgenic mice were used for astrocyte‐specific expression of Cre and labeling (Anthony & Heintz, 2007; Tien et al, 2012). For spatiotemporal expression of Cre in astrocytes, Aldh1l1‐CreER (The Jackson Laboratory, stock number 029655) was used. Daam2‐floxed animal was generated from Daam2tm1a(KOMP)Wtsi (KOMP) mouse by crossing them with the FLP mouse line. Slc4a4‐floxed mice were generated and provided by Dr. Shull (Vairamani et al, 2018). Thy1‐GCaMP3 mice were (The Jackson Laboratory, stock number 029860) used for odor‐evoked Ca2+ response. To induce deletion of Daam2 and Slc4a4, tamoxifen (Sigma, T5648) was dissolved in corn oil (Sigma, C8267) with 10% ethanol and injected intraperitoneally into 6‐week‐old mice with a dose of 100 mg/kg body weight for five consecutive days. For postnatal deletion, tamoxifen is injected one time intraperitoneally. To generate mouse lines with flag‐tagged Daam2, CRISPR‐Cas9 genome editing via homology‐directed repair was performed with followings small guide RNAs: 5’‐GGCCTTTCATGTTGAGCTAA TGG‐3’, 5’‐GCTGTCCCGGAGGTCGATCT CGG‐3’.
Immunostaining
Mice were anesthetized and intracardially perfused with phosphate‐buffered saline (PBS) and then with 4% paraformaldehyde in PBS. The brain was kept in 4% paraformaldehyde in PBS for overnight and cryoprotected in 20% sucrose for 24 h. The brain was embedded in OCT compound (Tissue TEK) and stored at −80°C until further use. Tissue sections were prepared with a cryostat (Leica) at a thickness of 15–100 μm. Brain sections were washed with PBS, treated with 0.3% Triton X‐100 in PBS (PBST) for 5 min, and then blocked with 10% goat‐serum in PBST for 1 h at room temperature (RT). Primary antibody solution was added to tissue sections at 4°C for overnight, washed for three times with PBS, and then incubated with secondary antibodies for 1 h at RT. After washing with PBS for three times, tissue was stained for DAPI and mounted with VECTASHIELD mounting medium. The following primary antibodies were used as follows: rabbit anti‐GFP (1:1,000; Chromotek; PABG1–100), rabbit anti‐GFAP (1:1,000; Agilent Dako; SIS), rabbit anti‐NFIA (gift from Dr. Benjamin Deneen), rabbit anti‐Sox9 (1:1,000; Millipore; AB5535), rat anti‐brdU (1:1,000; Abcam; ab6326), mouse anti‐NeuN (1:1,000; Milipore; MAB377), mouse and rabbit anti‐Slc4a4 (1:1,000, Sigma; WH0008671M1 and Abcam; AB187511), rabbit anti‐S100b (1:1,000; Agilent Dako; Z0311), mouse anti‐AldoC (1:1,000; EnCor; MCA‐4A9), mouse anti‐Nestin (1:1,000; BD Bioscience; 556309), rabbit anti‐MaoB (1:100; ProteinTech; 12602‐1‐AP), and mouse anti‐Slc12a6 (1:500, Abnova Corporation; H00009990A01). Secondary antibodies conjugated with Alexa Fluor 488 and 568 (Thermo Fisher Scientific) were used at 1:1,000 dilution. Images were obtained by Zeiss Imager.M2m equipped with ApoTome.2, Axiocam 506 mono, and Zeiss 880 Airyscan confocal microscope system.
Morphology analysis
Morphological analysis was performed using the Filament module in IMARIS software 9.3.0 (Oxford Instruments). Maximum orthogonal projections of Z‐stack images were imported to IMARIS, and astrocyte branches were outlined using the Filament Autopath algorithm with dendrite starting point diameter of 5.00 μm and a dendrite seed point diameter of 0.216 μm. Following automated tracing, filament structures were then manually pruned to eliminate false‐positive branches. For volume analysis, the fluorescence signal was reconstructed with the surface tool and the volume from each astrocyte was measured. Cells with all branches within the field of view were selected for analysis, and all analyses were performed using a blinding procedure. Statistical outputs from IMARIS were exported, analyzed using a custom R pipeline, and plotted using GraphPad Prism. All statistical tests were performed using GraphPad Prism.
Primary astrocyte culture
For astrocyte culture, cortical tissues of P0 brains were dissected and digested with papain/DNase in DMEM/F12 medium (Gibco) to obtain single‐cell suspension. Cells were then plated onto poly‐D‐lysine‐coated T75 culture flasks, and media were changed every 3–4 days. After 7 days, flasks were placed on an orbital shaker at 120 rpm overnight to remove microglia and oligodendrocyte precursors. For measuring GFAP and actin filaments, cells were trypsinized and reseeded on polyethylenimine‐coated cover glasses with Neurobasal medium containing B27 supplement and heparin‐binding EGF‐like growth factors for 2 days, followed by immunostaining. Alternatively, astrocytes were seeded on polyethylenimine‐coated 6‐well plates for Western blot analysis.
Calcium imaging
AAV‐GfaABC1D‐GCaMP6f (Addgene #52925) was injected into cortex and olfactory bulb 2˜3 weeks before the experiment using a nanoinjector with stereotaxic coordination. On the day of imaging, animals were anesthetized with isoflurane and isolated brains were submerged in ice‐cold ACSF solution (130 mM NaCl, 24 mM NaHCO3, 1.25 mM NaH2PO4, 3.5 mM KCl, 1.5 mM CaCl2, 1.5 mM MgCl2, and 10 mM D(+)‐glucose, pH 7.4). 300 μm slices were cut using a vibratome (DSK Linear Slicer, Kyoto, Japan) oxygenated in ACSF at room temperature for 1 h and then acclimated at room temperature with continuous perfusion with ASCF solution (2 ml/min). Calcium activity was collected at 1 Hz for 5 min using a two‐photon resonant microscope (LSM 7MP, Zeiss) with a Coherent Chameleon Ultra (II) Ti‐sapphire laser tuned to 900 nm. Relative change in fluorescence of soma and primary branches of each astrocyte was quantified using GECIquant with ImageJ, and Clampfit 10.7 software was used to quantify amplitude, frequency, and area under curve.
Electrophysiological recording
Brain slices were obtained identically as in calcium imaging. Slices were placed in recording chamber, and target cells were identified via upright Olympus microscope with a 60X water immersion objective with infrared differential interference contrast optics. Whole‐cell recording was performed with pCLAMP10 and MultiClamp 700B amplifier (Axon Instrument, Molecular Devices) at room temperature from layer II‐III cortical neurons and mitral cells in olfactory bulb. Recording pipettes were filled with internal solutions—action potential and passive conductance measurements: 140 mM K‐gluconate, 10 mM HEPES, 7 mM NaCl, and 2 mM MgATP (pH 7.4); EPSC and IPSC measurement: 135 mM CsMeSO4, 30 mM QX‐314, 10 mM HEPES, 8 mM NaCl, 1 mM MgATP, 0.25 mM EGTA, 0.25 mM Na2GTP, pH was adjusted to 7.2 with CsOH (278–285 mOsmol). Throughout the experiment, holding potential was −60 mV for passive conductance, −70 mV for EPSC, and 0 mV for IPSC. Electrical signals were digitized and sampled at 50‐μs intervals with Digidata 1550B and MultiClamp 700B amplifier (Molecular Devices, CA, USA) using pCLAMP 10.7 software. Data were filtered at 2 kHz. The recorded current was analyzed with Clampfit 10.7 software.
Behavior tests
Animals were habituated in a chamber with paper bedding for 15 min, three days in a row for all tests. For buried food finding assay, animals were starved for 24 h with a water‐only supply. Starved animals were placed in a chamber with dry food pellet, buried underneath, and covered with bedding to eliminate visual cue. Latency to find food and take a bite was then measured. Between animals, the behavior chamber was wiped with 70% isopropyl alcohol and paper towel. For odor discrimination assay, each animal was placed in chamber and habituated with mineral oil on filter paper in tea diffuser on both sides of the chamber for 5 min. Then, fresh acetophenone diluted in mineral oil was given in both sides for 2 min, three times in succession. For last session, anisole diluted in mineral oil was placed randomly on one side. Animals were video recorded by a Logitech C615 camera with Debut Video Capture Software for entire sessions. Time that animal spent in each side was calculated by tracking nose‐body position using MATLAB and OptiMouse software. Preference index was calculated as (Zone 1 − Zone 2)/ (Zone 1 + Zone 2).
Mass‐spec protein profiling
Mouse was deeply anesthetized, and intra‐cardiac perfusion was performed with PBS. Brains were removed, dissected, and then snap‐frozen with liquid nitrogen until further analysis. Ten sample volumes of lysis buffer (50 mM ammonium bicarbonate, 1 mM CaCl2) was added, dispersed by pipetting, and boiled at 95°C for 3 min. All denaturing procedures were repeated two times. Peptide supernatant was obtained by digesting 20 μg of proteins with trypsin (T9600, GenDEPOT) followed by extraction with 50% acetonitrile and 2% formic acid. Vacuum‐dried peptides were dissolved in pH 10 buffer (10 mM ammonium bicarbonate, pH adjusted by NH4OH) and were subjected to reverse phase column chromatography with a micro‐pipette tip C18 column. Then, it was fractionated with stepwise ACN gradient into 15 elution groups. Eluent was pooled into five pools and vacuum‐dried for nano‐HPLC‐MS/MS. Peptide was resuspended in the loading solution (5% methanol containing 0.1% formic acid) and was subjected to analysis with a nano‐LC 1000 coupled with Orbitrap FusionTM mass spectrometer (Thermo Fisher Scientific). Data were analyzed with proteome discoverer 2.1 interface (Thermo Fisher Scientific), and detected peptides were assigned into gene products by Py Grouper and Tackle analysis platform from iSpec (Saltzman et al, 2018).
In vivo co‐immunoprecipitation
Mice were prepared as described in Mass spec protein profiling method. Tissue was homogenized with lysis buffer (150 mM NaCl, 50 mM Tris–HCl, 1 mM EDTA, 1% Triton X‐100, 1% NP‐40) containing 1X protease inhibitor cocktail (GenDEPOT, P3100). Further lysis was performed by passing tissue lysate through a syringe with a 26G needle. The tissue lysate was incubated with agarose beads and Slc4a4 antibody (1:1,000, Sigma, WH0008671M1) overnight. Immunoprecipitated samples were loaded on SDS–PAGE for electrophoresis, transferred onto a nitrocellulose membrane, and incubated with anti‐Flag antibody (1:2,000, Sigma, F1804) for overnight. Membrane was washed three times for 5 min with TBST (20 mM Tris–HCl, 137 mM NaCl, 0.1% Tween 20) and incubated with HRP‐conjugated secondary antibody. After washing with TBST three times, the membrane was incubated with ECL reagent (GenDEPOT, W3653) and imaged with Bio‐Rad Gel Imager ChemiDoc E.
Statistics
GraphPad Prism v9 was used for generating graphs and statistical analysis. Data were reported as mean ± SEM. Significance was calculated using two‐tailed, unpaired Student’s t‐test. Two‐way ANOVA was used for Sholl analysis, and one‐sample t‐test and Wilcoxon test were used for discrimination behavioral test. The data were tested for variance using the F‐test, and Sidak's multiple comparison test was used for two‐way ANOVA. *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001 were considered to indicate statistical significance. Sample size for each experiment is indicated in the corresponding figure legend. Number of biological replicates, either animals (N) and/or cells/images (n), used for quantifications of each experiment were indicated in the respective figure legends.
Author contributions
JJ and HKL conceived the project and design the experiments; JJ, JW, JMC, C‐YW, QY, KU, and GL performed the experiments; CDC performed quantitative analysis. SYJ, BRA, and HKL provided reagent; DC performed technical support; JJ, JAS, and HKL wrote and edit the manuscript.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting information
Appendix
Expanded View Figures PDF
Source Data for Expanded View and Appendix
Acknowledgements
This work was supported by grants from the National Multiple Sclerosis Society (RG‐1907‐34551 to H.K.L), NIH/NINDS (R01NS110859 to H.K.L), and the Mark A. Wallace Endowment established by an anonymous donor (to H.K.L). This work was also supported by the Cytometry and Cell Sorting Core, and Mass Spectrometry Proteomics Core at Baylor College of Medicine, Microscopy Core, and the Eunice Kennedy Shriver National Institute of Child Health & Human Development of the National Institutes of Health under award number P50HD103555 for the use of the BCM IDDRC Neurobehavior and Neurovisualization Cores.
EMBO reports (2021) 22: e53200.
Data availability
No data were deposited in public databases.
References
- Ajima R, Bisson JA, Helt J‐C, Nakaya M‐A, Habas R, Tessarollo L, He X, Morrisey EE, Yamaguchi TP, Cohen ED (2015) DAAM1 and DAAM2 are co‐required for myocardial maturation and sarcomere assembly. Dev Biol 408: 126–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alberts JR, Galef BG (1971) Acute anosmia in the rat: a behavioral test of a peripherally‐induced olfactory deficit. Physiol Behav 6: 619–621 [DOI] [PubMed] [Google Scholar]
- Allen NJ (2014) Astrocyte regulation of synaptic behavior. Annu Rev Cell Dev Biol 30: 439–463 [DOI] [PubMed] [Google Scholar]
- Allen NJ, Eroglu C (2017) Cell biology of astrocyte‐synapse interactions. Neuron 96: 697–708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen NJ, Lyons DA (2018) Glia as architects of central nervous system formation and function. Science 362: 181–185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anthony TE, Heintz N (2007) The folate metabolic enzyme ALDH1L1 is restricted to the midline of the early CNS, suggesting a role in human neural tube defects. J Comp Neurol 500: 368–383 [DOI] [PubMed] [Google Scholar]
- Bernardinelli Y, Randall J, Janett E, Nikonenko I, König S, Jones E, Flores C, Murai K, Bochet C, Holtmaat A et al (2014) Activity‐dependent structural plasticity of perisynaptic astrocytic domains promotes excitatory synapse stability. Curr Biol 24: 1679–1688 [DOI] [PubMed] [Google Scholar]
- Bosworth AP, Allen NJ (2017) The diverse actions of astrocytes during synaptic development. Curr Opin Neurobiol 47: 38–43 [DOI] [PubMed] [Google Scholar]
- Bushong EA, Martone ME, Ellisman MH (2004) Maturation of astrocyte morphology and the establishment of astrocyte domains during postnatal hippocampal development. Int J Dev Neurosci 22: 73–86 [DOI] [PubMed] [Google Scholar]
- Chai H, Diaz‐Castro B, Shigetomi E, Monte E, Octeau JC, Yu X, Cohn W, Rajendran PS, Vondriska TM, Whitelegge JP et al (2017) Neural circuit‐specialized astrocytes: transcriptomic, proteomic, morphological, and functional evidence. Neuron 95: 531–549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q, Cichon J, Wang W, Qiu LI, Lee S‐J, Campbell N, DeStefino N, Goard M, Fu Z, Yasuda R et al (2012) Imaging neural activity using Thy1‐GCaMP transgenic mice. Neuron 76: 297–308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Poskanzer KE, Freeman MR, Monk KR (2020) Live‐imaging of astrocyte morphogenesis and function in zebrafish neural circuits. Nat Neurosci 23: 1297–1306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke LE, Barres BA (2013) Emerging roles of astrocytes in neural circuit development. Nat Rev Neurosci 14: 311–321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke BE, Taha DM, Tyzack GE, Patani R (2021) Regionally encoded functional heterogeneity of astrocytes in health and disease: a perspective. Glia 69: 20–27 [DOI] [PubMed] [Google Scholar]
- Di Castro MA, Chuquet J, Liaudet N, Bhaukaurally K, Santello M, Bouvier D, Tiret P, Volterra A (2011) Local Ca2+ detection and modulation of synaptic release by astrocytes. Nat Neurosci 14: 1276–1284 [DOI] [PubMed] [Google Scholar]
- Ding X, Jo J, Wang C‐Y, Cristobal CD, Zuo Z, Ye QI, Wirianto M, Lindeke‐Myers A, Choi JM, Mohila CA et al (2020) The Daam2–VHL–Nedd4 axis governs developmental and regenerative oligodendrocyte differentiation. Genes Dev 34: 1177–1189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle JP, Dougherty JD, Heiman M, Schmidt EF, Stevens TR, Ma G, Bupp S, Shrestha P, Shah RD, Doughty ML et al (2008) Application of a translational profiling approach for the comparative analysis of CNS cell types. Cell 135: 749–762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durkee CA, Araque A (2019) Diversity and specificity of astrocyte–neuron communication. Neuroscience 396: 73–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farhy‐Tselnicker I, van Casteren ACM, Lee A, Chang VT, Aricescu AR, Allen NJ (2017) Astrocyte‐secreted glypican 4 regulates release of neuronal pentraxin 1 from axons to induce functional synapse formation. Neuron 96: 428–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foo LC, Dougherty JD (2013) Aldh1L1 is expressed by postnatal neural stem cells in vivo . Glia 61: 1533–1541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman MR (2010) Specification and morphogenesis of astrocytes. Science 330: 774–778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii Y, Maekawa S, Morita M (2017) Astrocyte calcium waves propagate proximally by gap junction and distally by extracellular diffusion of ATP released from volume‐regulated anion channels. Sci Rep 7: 13115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haim LB, Rowitch DH (2017) Functional diversity of astrocytes in neural circuit regulation. Nat Rev Neurosci 18: 31–41 [DOI] [PubMed] [Google Scholar]
- Haustein M, Kracun S, Lu X‐H, Shih T, Jackson‐Weaver O, Tong X, Xu JI, Yang X, O’Dell T, Marvin J et al (2014) Conditions and constraints for astrocyte calcium signaling in the hippocampal mossy fiber pathway. Neuron 82: 413–429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt LM, Hernandez RD, Pacheco NL, Torres Ceja B, Hossain M, Olsen ML (2019) Astrocyte morphogenesis is dependent on BDNF signaling via astrocytic TrkB.T1. eLife 8: e44667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hrvatin S, Hochbaum DR, Nagy MA, Cicconet M, Robertson K, Cheadle L, Zilionis R, Ratner A, Borges‐Monroy R, Klein AM et al (2018) Single‐cell analysis of experience‐dependent transcriptomic states in the mouse visual cortex. Nat Neurosci 21: 120–129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang AY‐S, Woo J, Sardar D, Lozzi B, Bosquez Huerta NA, Lin C‐CJ, Felice D, Jain A, Paulucci‐Holthauzen A, Deneen B (2020) Region‐specific transcriptional control of astrocyte function oversees local circuit activities. Neuron 106: 992–1008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaiswal R, Breitsprecher D, Collins A, Corrêa IR, Xu M‐Q, Goode BL (2013) The formin Daam1 and fascin directly collaborate to promote filopodia formation. Curr Biol 23: 1373–1379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawabata Galbraith K, Kengaku M (2019) Multiple roles of the actin and microtubule‐regulating formins in the developing brain. Neurosci Res 138: 59–69 [DOI] [PubMed] [Google Scholar]
- Khakh BS, McCarthy KD (2015) Astrocyte calcium signaling: from observations to functions and the challenges therein. Cold Spring Harb Perspect Biol 7: a020404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khakh BS, Sofroniew MV (2015) Diversity of astrocyte functions and phenotypes in neural circuits. Nat Neurosci 18: 942–952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khakipoor S, Giannaki M, Theparambil SM, Zecha J, Küster B, Heermann S, Deitmer JW, Roussa E (2019) Functional expression of electrogenic sodium bicarbonate cotransporter 1 (NBCe1) in mouse cortical astrocytes is dependent on S255–257 and regulated by mTOR. Glia 67: 2264–2278 [DOI] [PubMed] [Google Scholar]
- Kressin K, Kuprijanova E, Jabs R, Seifert G, Steinhäuser C (1995) Developmental regulation of Na+ and K+ conductances in glial cells of mouse hippocampal brain slices. Glia 15: 173–187 [DOI] [PubMed] [Google Scholar]
- Lanjakornsiripan D, Pior B‐J, Kawaguchi D, Furutachi S, Tahara T, Katsuyama Y, Suzuki Y, Fukazawa Y, Gotoh Y (2018) Layer‐specific morphological and molecular differences in neocortical astrocytes and their dependence on neuronal layers. Nat Commun 9: 1623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HK, Deneen B (2012) Daam2 is required for dorsal patterning via modulation of canonical Wnt signaling in the developing spinal cord. Dev Cell 22: 183–196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HK, Chaboub LS, Zhu W, Zollinger D, Rasband MN, Fancy SPJ, Deneen B (2015) Daam2‐PIP5K is a regulatory pathway for Wnt signaling and therapeutic target for remyelination in the CNS. Neuron 85: 1227–1243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D, Hallett MA, Zhu W, Rubart M, Liu Y, Yang Z, Chen H, Haneline LS, Chan RJ, Schwartz RJ et al (2011) Dishevelled‐associated activator of morphogenesis 1 (Daam1) is required for heart morphogenesis. Development 138: 303–315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J, Meng W, Poy F, Maiti S, Goode BL, Eck MJ (2007) Structure of the FH2 domain of Daam 1: implications for formin regulation of actin assembly. J Mol Biol 369: 1258–1269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molotkov D, Zobova S, Arcas JM, Khiroug L (2013) Calcium‐induced outgrowth of astrocytic peripheral processes requires actin binding by Profilin‐1. Cell Calcium 53: 338–348 [DOI] [PubMed] [Google Scholar]
- Mulligan SJ, MacVicar BA (2004) Calcium transients in astrocyte endfeet cause cerebrovascular constrictions. Nature 431: 195–199 [DOI] [PubMed] [Google Scholar]
- Oliet SHR, Piet R, Poulain DA (2001) Control of glutamate clearance and synaptic efficacy by glial coverage of neurons. Science 292: 923–926 [DOI] [PubMed] [Google Scholar]
- Perez‐Alvarez A, Navarrete M, Covelo A, Martin ED, Araque A (2014) Structural and functional plasticity of astrocyte processes and dendritic spine interactions. J Neurosci 34: 12738–12744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petzold GC, Murthy VN (2011) Role of astrocytes in neurovascular coupling. Neuron 71: 782–797 [DOI] [PubMed] [Google Scholar]
- Ruminot I, Gutierrez R, Pena‐Munzenmayer G, Anazco C, Sotelo‐Hitschfeld T, Lerchundi R, Niemeyer MI, Shull GE, Barros LF (2011) NBCe1 mediates the acute stimulation of astrocytic glycolysis by extracellular K+ . J Neurosci 31: 14264–14271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakmann B, Edwards F, Konnerth A, Takahashi T (1989) Patch clamp techniques used for studying synaptic transmission in slices of mammalian brain. Q J Exp Physiol 74: 1107–1118 [DOI] [PubMed] [Google Scholar]
- Salomon SN, Haber M, Murai KK, Dunn RJ (2008) Localization of the Diaphanous‐related formin Daam1 to neuronal dendrites. Neurosci Lett 447: 62–67 [DOI] [PubMed] [Google Scholar]
- Saltzman AB, Leng M, Bhatt B, Singh P, Chan DW, Dobrolecki L, Chandrasekaran H, Choi JM, Jain A, Jung SY et al (2018) gpGrouper: a peptide grouping algorithm for gene‐centric inference and quantitation of bottom‐up proteomics data. Mol Cell Proteomics 17: 2270–2283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiweck J, Eickholt BJ, Murk K (2018) Important shapeshifter: mechanisms allowing astrocytes to respond to the changing nervous system during development, injury and disease. Front Cell Neurosci 12: 261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrödl‐Häußel M, Theparambil SM, Deitmer JW, Roussa E (2015) Regulation of functional expression of the electrogenic sodium bicarbonate cotransporter 1, NBCe1 ( SLC4A4), in mouse astrocytes. Glia 63: 1226–1239 [DOI] [PubMed] [Google Scholar]
- Semyanov A, Henneberger C, Agarwal A (2020) Making sense of astrocytic calcium signals — from acquisition to interpretation. Nat Rev Neurosci 21: 551–564 [DOI] [PubMed] [Google Scholar]
- Srinivasan R, Huang BS, Venugopal S, Johnston AD, Chai H, Zeng H, Golshani P, Khakh BS (2015) Ca2+ signaling in astrocytes from Ip3r2−/− mice in brain slices and during startle responses in vivo . Nat Neurosci 18: 708–717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan R, Lu T‐Y, Chai H, Xu J, Huang BS, Golshani P, Coppola G, Khakh BS (2016) New transgenic mouse lines for selectively targeting astrocytes and studying calcium signals in astrocyte processes in situ and in vivo . Neuron 92: 1181–1195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stogsdill JA, Ramirez J, Liu D, Kim YH, Baldwin KT, Enustun E, Ejikeme T, Ji RR, Eroglu C (2017) Astrocytic neuroligins control astrocyte morphogenesis and synaptogenesis. Nature 551: 192–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su C‐Y, Menuz K, Carlson JR (2009) Olfactory perception: receptors, cells, and circuits. Cell 139: 45–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun S, Zhu X‐J, Huang H, Guo W, Tang T, Xie B, Xu X, Zhang Z, Shen Y, Dai Z‐M et al (2019) WNT signaling represses astrogliogenesis via Ngn2‐dependent direct suppression of astrocyte gene expression. Glia 67: 1333–1343 [DOI] [PubMed] [Google Scholar]
- Takahashi H, Ogawa Y, Yoshihara S‐I, Asahina R, Kinoshita M, Kitano T, Kitsuki M, Tatsumi K, Okuda M, Tatsumi K et al (2016) A subtype of olfactory bulb interneurons is required for odor detection and discrimination behaviors. J Neurosci 36: 8210–8227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takata N, Hirase H (2008) Cortical layer 1 and layer 2/3 astrocytes exhibit distinct calcium dynamics in vivo . PLoS One 3: e2525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theis M, Giaume C (2012) Connexin‐based intercellular communication and astrocyte heterogeneity. Brain Res 1487: 88–98 [DOI] [PubMed] [Google Scholar]
- Theparambil SM, Ruminot I, Schneider H‐P, Shull GE, Deitmer JW (2014) The electrogenic sodium bicarbonate cotransporter NBCe1 is a high‐affinity bicarbonate carrier in cortical astrocytes. J Neurosci 34: 1148–1157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theparambil SM, Hosford PS, Ruminot I, Kopach O, Reynolds JR, Sandoval PY, Rusakov DA, Barros LF, Gourine AV (2020) Astrocytes regulate brain extracellular pH via a neuronal activity‐dependent bicarbonate shuttle. Nat Commun 11: 5073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tien A‐C, Tsai H‐H, Molofsky AV, McMahon M, Foo LC, Kaul A, Dougherty JD, Heintz N, Gutmann DH, Barres BA et al (2012) Regulated temporal‐spatial astrocyte precursor cell proliferation involves BRAF signalling in mammalian spinal cord. J Cell Sci 125: e1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vairamani K, Prasad V, Wang Y, Huang W, Chen Y, Medvedovic M, Lorenz JN, Shull GE (2018) NBCe1 Na+ ‐HCO3 ‐ cotransporter ablation causes reduced apoptosis following cardiac ischemia‐reperfusion injury in vivo . World J Cardiol 10: 97–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ventura R, Harris KM (1999) Three‐dimensional relationships between hippocampal synapses and astrocytes. J Neurosci 19: 6897–6906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vesce S, Bezzi P, Volterra A (1999) The active role of astrocytes in synaptic transmission. Cell Mol Life Sci 56: 991–1000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidal R, Garro‐Martínez E, Díaz Á, Castro E, Florensa‐Zanuy E, Taketo MM, Pazos Á, Pilar‐Cuéllar F (2019) Targeting β‐catenin in GLAST‐expressing cells: impact on anxiety and depression‐related behavior and hippocampal proliferation. Mol Neurobiol 56: 553–566 [DOI] [PubMed] [Google Scholar]
- Wallar BJ, Alberts AS (2003) The formins: active scaffolds that remodel the cytoskeleton. Trends Cell Biol 13: 435–446 [DOI] [PubMed] [Google Scholar]
- Welsh IC, Thomsen M, Gludish DW, Alfonso‐Parra C, Bai Y, Martin JF, Kurpios NA (2013) Integration of left‐right Pitx2 transcription and Wnt signaling drives asymmetric gut morphogenesis via Daam2. Dev Cell 26: 629–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winchenbach J, Düking T, Berghoff SA, Stumpf SK, Hülsmann S, Nave K‐A, Saher G (2016) Inducible targeting of CNS astrocytes in Aldh1l1‐CreERT2 BAC transgenic mice. F1000Res 5: 2934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang M, Crawley JN (2009) Simple behavioral assessment of mouse olfaction. Curr Protoc Neurosci 48: 8.24.1–8.24.12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeisel A, Hochgerner H, Lönnerberg P, Johnsson A, Memic F, van der Zwan J, Häring M, Braun E, Borm LE, La Manno G et al (2018) Molecular architecture of the mouse nervous system. Cell 174: 999–1014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeug A, Müller FE, Anders S, Herde MK, Minge D, Ponimaskin E, Henneberger C (2018) Control of astrocyte morphology by Rho GTPases. Brain Res Bull 136: 44–53 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Chen K, Sloan SA, Bennett ML, Scholze AR, O'Keeffe S, Phatnani HP, Guarnieri P, Caneda C, Ruderisch N et al (2014) An RNA‐sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. J Neurosci 34: 11929–11947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou B, Zuo Y, Jiang R (2019) Astrocyte morphology: Diversity, plasticity, and role in neurological diseases. CNS Neurosci Ther 25: 665–673 [DOI] [PMC free article] [PubMed] [Google Scholar]


