Abstract
Brain tumors in adults may be infrequent when compared with other cancer etiologies, but they remain one of the deadliest with bleak survival rates. Current treatment modalities encompass surgical resection, chemotherapy, and radiotherapy. However, increasing resistance rates are being witnessed, and this has been attributed, in part, to cancer stem cells (CSCs). CSCs are a subpopulation of cancer cells that reside within the tumor bulk and have the capacity for self-renewal and can differentiate and proliferate into multiple cell lineages. Studying those CSCs enables an increasing understanding of carcinogenesis, and targeting CSCs may overcome existing treatment resistance. One approach to weaponize new drugs is to target these CSCs through drug repurposing which entails using drugs, which are Food and Drug Administration–approved and safe for one defined disease, for a new indication. This approach serves to save both time and money that would otherwise be spent in designing a totally new therapy. In this review, we will illustrate drug repurposing strategies that have been used in brain tumors and then further elaborate on how these approaches, specifically those that target the resident CSCs, can help take the field of drug repurposing to a new level.
Keywords: angiogenesis, brain tumors, cancer stem cells, drug repurposing, glioblastoma, meningioma, stemness, tumor heterogeneity, tumor immune infiltrate, tumor microenvironment
Introduction
Tumors of the brain are rare but deadly entities whose treatment remains challenging. 1 Glioblastoma (GBM; World Health Organization [WHO] grade IV glioma) is the most frequent and aggressive primary brain tumor in adults. Life expectancy remains bleak at around 15 to 19 months. 2 The challenges associated with treatment of brain malignancies are extensive, ranging from inability to use current cancer therapies to limited interest/diminished funding from pharmaceutical industries. 3 Due to the brain’s complexity and frailty, many tumors are unable to be completely excised, which hinders surgical treatment and worsens prognosis. 3 Furthermore, the highly selective blood–brain barrier (BBB) hinders chemotherapeutic agents from reaching the site of carcinogenesis. 4 Our limited understanding of the biology of these widely variant tumors and their microenvironment poses a grand challenge to brain cancer treatment. 4
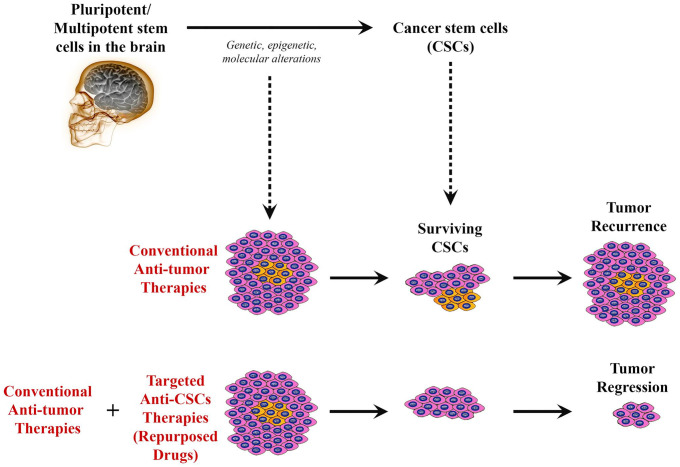
Cancer stem cells (CSCs) are a subpopulation of cancer cells that resemble normal human stem cells, having the capacity for self-renewal, differentiation, and proliferation into multiple cell lineages through symmetric and asymmetric cell division. 5 The study and investigation of CSCs (or “tumor-initiating cells”) have allowed us to further understand their role in metastasis, which accounts for 90% of cancer deaths in the world. Overall, the use of conventional therapies such as surgical resection, chemotherapy, and radiation is limited and often does not remove or target the CSCs, which are the cells responsible for tumor recurrence and metastasis, especially in the brain. Thus, targeted therapies, such as those that use cell surface markers, as well as various tumorigenic proteins found in CSCs, could provide for better patient outcomes (Fig. 1).
Figure 1.
Schematic diagram illustrating how drug repurposing to target cancer stem cells (CSCs) in brain tumors can promote tumor regression. Despite that cancer cells within a tumor might be mostly affected by conventional therapies leading to shrinkage of the tumor, surviving CSCs will remain to resist to such therapies, leading to cancer recurrence with time. However, combining traditional anticancer therapies with novel targeted therapies directed against CSCs (“repurposed drugs”) could potentially elicit tumor regression and prevent recurrence.
The field of drug repurposing (aka drug repositioning) has recently become more attractive to many researchers. It is defined as the use of preexisting therapeutics—that have been previously deemed to be safe and efficacious in a defined medical condition—for purposes different from the one that they were initially studied for. 6 The advantage of studying preexisting drugs rather than de novo drug studies can be summarized in two words: time and cost.6,7 In this review, we will consider how drug repurposing can be leveraged to provide better outcomes for patients with brain cancer.
Cancer Stem Cells in Brain Tumors
CSCs have the capacity for self-renewal, differentiation, and proliferation into multiple cell lineages through symmetric and asymmetric cell division. 5 First identified in hematological malignancies, specifically acute myeloid leukemia (AML), and then subsequently in solid malignancies, CSCs have modified our understanding of cancer initiation, progression, and metastasis. 8 The first study on CSCs was done in 1997, where Bonnet and Dick demonstrated that despite having the morphological phenotype of leukemic blasts, only a subpopulation of CD34+/CD38− leukemic blast cells, and not CD34+/CD38+ cells, were able to repopulate the entire tumor and initiate human AML in mice with non-obese diabetic/severe combined immunodeficiency disease (NOD/SCID mice). 9 Subsequently, CSCs were identified and isolated in solid tumors such as breast cancers, 10 brain tumors, 11 prostate cancers, 12 lung cancers, 13 colon cancers, 14 head and neck squamous cell carcinomas (HNSCCs), 15 and pancreatic 16 and liver cancers. 17 Increasing studies have demonstrated that CSCs are responsible for tumor heterogeneity, invasion, metastasis, and tumor recurrence via epithelial-to-mesenchymal (EMT) transformation and multidrug resistance. Therefore, identification of CSCs is crucial as it may present a key therapeutic target on which novel therapeutic agents can act.
In vivo studies have demonstrated that when a subpopulation of the original malignant cells is transplanted into immunodeficient mice, the ability to reform the parental tumor is observed, indicating the existence of CSCs.11,18–20 Based on analyses, including those that examined differential allele expression of the X-linked gene G6PD in chronic myeloid leukemia (CML) and AML and those that have shown the activation of self-renewal via the β-catenin pathway, among others, CSCs can be derived from either normal stem cell lineages or early progenitor cells that have acquired certain mutations that confer stem-like cell properties on these originally non-self-renewing cells.21,22 Of note, if the CSCs do arise from these mutated early progenitor cells, they depend on inactivation of normal inhibitory pathways of self-renewal and activation of normal stem cell pathways.23–27 The tumor microenvironment (TME) has also been demonstrated to have the capacity to generate CSCs via activation mediated through extracellular cues. 28
It is believed that CSCs are responsible for the tumorgenicity and high proliferative capacity of multiple cancers, a concept further elaborated on when comparing CSCs with other cancer cells because CSCs are able to regenerate a tumor and cause metastasis, whereas other cancer cells have limited proliferative and regenerative capacity.29,30 With this in mind, the heterogeneity observed in many cancers can be attributed to the mutagenesis that occurs within CSCs, producing different phenotypic cancer cells described. 30 In addition to CSCs being multipotent, they share several other features with normal stem cells, such as enhanced DNA damage response mechanisms, relative quiescence, decreased apoptosis, and increased immune system evasion, which amplifies the stem cell component of tumors. 29 Despite these other features, a more critical topic of concern is the drug resistance that arises from the CSCs believed to be the cause of the cancer relapse seen in several cancers. This is in part attributed to their ability to self-renew and promote greater heterogeneity as a response to the therapeutic agents, as well as the interactions between the agents and the TME.29,31–33 Relatedly, although CSCs are typically quiescent, they can also induce a cell cycle arrested state to enhance their ability to be resistant to various therapies.24,34–38
When considering the importance of targeting these cells in combating tumor relapse, it is important to first identify the unique biomarkers associated with them. However, due to the heterogeneous and plastic nature of CSCs and their inward location within tumors, molecular components of CSCs have been difficult to define. 32 There are promising cell surface markers consistently seen on CSCs that can be used as a means of targeting CSCs more readily. CSC-specific cell markers include, but are not limited to, CD24, CD26, CD29, CD34, CD44, CD133, CD166, aldehyde dehydrogenase (ALDH), and Ep-CAM (CD326).32,39,40 Marker expression on CSCs differs from one cancer to another due to their origin.41–43 This offers a point of interest to target CSCs via these stem cell markers as it may be a more precise mechanism of identifying CSCs. However, it has been noted that although normal stem cells or progenitor cells may share similar cell surface markers and enzymes, there does appear to be differences in glycosylation that would require analysis to effectively target the markers. 40 The markers’ validity requires examination as well because although the molecular component may play a role in the function of the cells, it may not be unique to the CSC and thus sufficient for identification and targeting, as seen with Lgr5 in intestinal stem cells. 41 Overall, the particular marker may assist in identifying the CSC for one type of cancer, but not another. For example, A2B5 appears to be a unique marker for CSCs in GBM, whereas CD44 is more widespread after having been observed in various cancer CSCs, including those in the breast, bladder, stomach, ovary, and GBM, among others.39,44–47
Methodologies for the Identification of CSC
CSCs can be identified by various approaches, namely, (1) cell surface markers, (2) side population study, (3) ALDH activity, and (4) molecular signatures exemplified by the expression of stemness genes and transcription factors.
Cell Surface Markers
A variety of cell surface markers/antigens have been identified in several cancers that may serve as CSC biomarkers. These include CD20, CD24, CD34, CD44, CD90, CD117, and CD133. Although a universal biomarker for CSCs does not exist, CD44 and CD133 have gained much attention for both epithelial and mesenchymal malignancies.
CD44 is a transmembrane glycoprotein that is implicated in cell adhesion, migration, differentiation, proliferation, and survival. It functions as a receptor for hyaluronan, as well as other extracellular matrix components, growth factors, and cytokines. CD44 was first described as a CSC marker in breast cancer in 2003. 10 Increasing evidence has demonstrated that CD44 serves as a CSC marker and plays an important role in regulating the functions and properties of CSCs, including self-renewal, tumorigenesis, chemoradiation resistance, and metastasis.48,49
CD133 (Prominin 1) is a transmembrane glycoprotein expressed in neural progenitor cells, epithelial cells, and non-epithelial cells. It has also been identified in stem cells in both normal and cancer cells. 50 CD133 interacts with the Wnt/β-catenin and PI3K–Akt signaling pathways, which in turn promotes angiogenesis and cancer cell growth. 50 Different CSC subtypes that are known to express CD133 include breast, 51 colon, 52 gastric, 53 pancreatic, 54 hepatocellular, 55 lungs, 56 prostate, 57 melanoma, 58 and GBM. 59 Although CD133 has been used reliably as a CSC biomarker in various cancers and correlates with worse prognosis, it is not entirely specific.
Side Population Assay
The side population (SP) assay is a flow cytometry method used to identify stem cells based on the dye efflux properties of the ATP-binding cassette (ABC) transporters. 60 Cells that have the capability to express the stemness genes (ABC multidrug transporters) also can self-renew, differentiate into multiple lineages, and therefore display stem cell properties. Similar to the CSC biomarkers mentioned above, the SP assay has been used to identify both normal cells and CSCs such as in HNSCC, 61 bladder cancer, 62 colon cancer, 63 endometrial cancer, 64 ovarian cancer, 65 hepatocellular carcinoma, 66 lung cancer, 67 osteosarcomas, 68 medulloblastoma (MB), 69 and GBM. 70
ALDH Activity
ALDHs are a group of enzymes that catalyze the oxidation of aldehyde substrates to their corresponding carboxylic acids and play an important role in cellular detoxification. 71 ALDH1A1 is one of the key ALDH isozymes that is expressed not only in normal hematopoietic progenitor cells 72 but also in various CSCs, making it a useful CSC biomarker. 73 ALDH1A1 overexpression and increased enzyme activity have been associated with chemoresistance and radioresistance and correlates with poor prognosis across different cancers such as non–small cell lung cancer, 74 stomach cancer, 75 and ductal carcinoma of the breast. 76
Molecular
Signatures Exemplified by the Expression of Stemness Genes and Transcription Factors
Increasing evidence has demonstrated that stem cell–associated molecular features are biologically important in cancer. 77 In addition, the expression of stemness genes across multiple cancers has shown association with suppressed immune response, higher intratumoral heterogeneity, and dramatically worse outcomes. 78 In a study in breast cancer, the expression of stemness genes such as OCT3, SOX2, KLF4, MYC, NOTCH1, and NANOG in tumor cells is associated with metastasis and the acquisition of non-CSC to CSC plasticity with the formation of EpCam+/CD44high/CD24low cells. 79 The expression of stemness genes, particularly OCT4, SOX2, and NANOG, is the most analyzed transcription factor in CSCs. Increased expression of these genes has been shown in gliomas, 80 lung carcinoma, 81 prostate carcinoma, 82 and bone sarcoma. 83 Other transcriptional factors to be considered include Bmi-1, Snail, and Twist. Bmi-1 was identified as a protooncogene that promoted tumorigenesis, and its overexpression confers self-renewal potential and chemoresistance and radioresistance associated with increased incidence of metastasis. 84 Snail and Twist are essential in maintaining CSCs, promoting EMT, and mediating invasiveness and metastasis in various tumors. 85 Overexpression of Twist has also been observed in breast cancer, 86 lung cancer, 87 and gastric cancer. 88
Identification of CSCs in Various Brain Tumors
Although CSCs have been identified within multiple hematological and solid tumors, malignancies of the brain require increased consideration due to the challenges faced by treatments and therapeutics used to target these areas, including limited access for surgical resection, the inability of certain treatments to cross the BBB, and their unique genetic and epigenetic characteristics allowing for resistance to those agents able to cross the BBB.3,89–91
CSCs commonly found in brain tumors like gliomas have been investigated, and studies assessing their ability to self-renew have demonstrated that when these cells are isolated from the rest of the tumor, they are able to form neurospheres—indicative of their renewal potential.11,92 This supports the notion that these cells are required for the establishment of the tumor in vivo. 11 Furthermore, the proximity of CSCs to other cells within the TME in the brain has demonstrated potentially unique environmental characteristics, leading to the success of the tumor in this location. 93 For instance, in GBM, CSCs have been noted to be in close contact with endothelial cells (ECs), suggesting that the soluble secreted materials from the ECs may maintain the self-renewal capacity and differentiation of CSCs. When CSCs and ECs have been cultured in vitro, there is enhancement of CSC function, whereas when CSCs have been transplanted into models undergoing antivascular endothelial growth factor A (VEGFA) therapy, the CSC population was reduced, emphasizing the role of this vascular niche in the brain. 93 Alongside this vascular niche, a hypoxic niche has also been observed in the brain, where hypoxia has induced the activation of hypoxia-induced factor 1α resulting in CSC expansion.94–96 Similarly, CSCs have a dynamic relationship with their respective niche, allowing for active regulation to promote niche formation and maintenance. 97
In addition, the evaluation of various tumorigenic proteins has been completed and reported in brain tumors, such as GBMs, which promote the ability of the tumor to self-renew, promote angiogenesis, resist therapeutic agents, and facilitate transcription, proliferation, differentiation, and metastasis.89,98 For instance, transforming growth factor-beta (TGF-β), PI3K/Akt, Wnt/Beta-catenin pathway, Shh signaling, and mitogen-activated protein kinase (MAPK) have been documented in brain CSCs to promote the self-renewal of the tumor.99–101 Also, the enhanced expression of forkhead box protein M1 (FOXM1) within CSCs has been shown to be metastatic inducers and one of the mechanisms used by the tumor to resist therapy. 102 The upregulation of VEGF and its respective receptor demonstrated increased angiogenic potential, alongside the use of the histone chaperone complex known as FACT to facilitate transcription of the CSC markers, enabling multiple functions including interaction with the TME. 98 Relatedly, brain CSCs have been shown to adapt to nutrient restrictions, possibly another mechanism of evading treatment, via upregulation of the high-affinity neuronal glucose transporter GLIT3. 103 Of note, pediatric brain tumors, such as MB, use signaling pathways similar to those seen in adult brain CSCs, including Shh, Notch and Wnt, which govern specification, proliferation, and survival, indicating potential widespread targeting across these tumors.104,105 Studies have also shown that particular genetic alterations have been associated with the existence of the tumor, including mutations affecting EGFR and HDM2 (gain of function), PTEN, and NF1 (loss of function), among others.106,107 Cell surface markers noted in certain brain CSCs like those seen in GBM include CD133, nestin, and A2B5, representing potential specific target sites. Noteworthy mentioning are also the stromal-derived factor-1α (SDF-1α) and its receptor C-X-C chemokine receptor type 4 (CXCR4), where SDF-1α plays a role in homing of therapy-resistant CXCR4+ GBM CSCs 108 that are shown to reside in protective niches around arterioles recapitulating hematopoietic stem cell (HSC) niches in the bone marrow. 109 Interestingly, both markers are targets of AMD3100 (plerixafor), a drug that is used to mobilize HSCs out of their niches to improve the yield of stem cell transplantation. Plerixafor is currently being investigated in a repurposing effort in GBM as an adjuvant inhibitor of vasculogenesis, 110 forcing slowly dividing CSCs out of their niches. 111 Results from a phase I/II trial using plerixafor in GBM showed that the drug was well tolerated and improves local control of tumor recurrences (ClinicalTrials.gov; NCT01977677). 112
Among the primary brain tumors, GBM is a highly aggressive and lethal cancer with frequent tumor recurrence. Accumulating evidence supports the concept that GBM harbors CSCs, which confer resistance to chemotherapy and radiotherapy, cancer recurrence, and poor prognosis. 113 Therefore, targeting CSCs emerges as a promising method to eradicate this devastating disease. To effectively identify and target CSCs, various common surface markers such as CD133/prominin, Nestin, Musashi-1, Sox2, HMG box, CD44, L1CAM (CD171), CD15 (SSEA-1), and A2B5 were studied. 114 Notably, novel therapeutic modalities specifically targeting GBM CSCs bearing CD133 and CD44 have demonstrated encouraging results both in vitro and in vivo, expanding their role as theranostic biomarkers.115,116
Repurposing of Approved Drugs in Brain Tumors
Repurposing of Approved Drugs in Cancer
The field of drug repurposing (aka drug repositioning) has been recently attractive to many researchers. It is defined as the use of preexisting therapeutics—that have been previously studied to be safe and efficacious in a defined medical condition—for purposes different from the one that they were initially indicated for. 6 Repurposed drugs comprise four categories. The first includes drugs that were discontinued due to observed toxicities but later found to be effective in certain diseases. The second group includes drugs that were studied for a particular indication but were found to have an additional disease-fighting property (off-label). The third group includes drugs that are efficacious in several diseases but only used in the treatment of one disease. The fourth category includes drugs that exhibit synergistic action with the conventional pharmacological agents used to treat a particular pathology. 117
Advantages of Drug Repurposing and Obstacles to Drug Repurposing
The approach of studying preexisting drugs carries benefits over de novo drug studies. These advantages can be summarized in two words: time and cost. A study by Kaitin et al. published in 2010 demonstrated that it takes an average of 8.3 years for an antineoplastic drug to evolve from the time of filing of the investigational drug application to the first New Drug Application/Biologic License Application submission, compared with 3 to 4 years for a drug to be studied in a drug repurposing trial.6,7 Hence, when a researcher/clinician is to follow the strategy of drug repurposing and study a particular drug, there is a wealth of published data demonstrating the drug pharmacokinetics and pharmacodynamics, reducing the time and cost needed to complete, to a certain extent, phases I and II of a clinical trial. 118 Moreover, and from an economic perspective, drug repurposing trials study drugs in a cost-effective manner, for example, bringing a repurposed drug to market with an estimated cost of $300 million compared with $2–$3 billion as an estimated cost to study a new chemical entity. 119
On the contrary, the repositioning process of a particular candidate drug might face some hurdles. The due diligence is a challenging step in the repositioning process as a repositioning candidate should have a competitive profile. 6 In addition, redirecting a preexisting drug for a new use is considered as a high-risk investment as the repurposed drug may fail to achieve a benefit–risk profile sufficient to support its use for new indications. Adding to that, the drug of interest will need to go through phases to determine the appropriate dose for the new indication and in some cases will need to repeat early phases of the study to determine the efficacy and safety, all of which can cause an increase in cost and time required for the drug to be approved.118,120
Approaches to Drug Repurposing
The challenges associated with the treatment of brain malignancies are extensive, ranging from the inability to use current cancer therapies to diminished funding from pharmaceutical entities. 3 Due to the brain’s complexity and frailty, many tumors are unable to be completely excised, which hinders surgical treatment and worsens prognosis. 3 Furthermore, the highly selective BBB hinders chemotherapy agents from reaching the site of carcinogenesis. 4 Along with our limited understanding of the biology of the widely variant tumors and their microenvironment, all these factors pose a great challenge to brain cancer treatment. 4 Interestingly, although brain malignancies are deadly, they are generally infrequent compared with other forms of cancers. As a result, the demand and interest for novel therapies from pharmaceutical industries are diminished. 3 Moreover, the discovery and development of novel drugs is an arduous process which demands high costs and extensive time for these agents to be tried and approved. 121
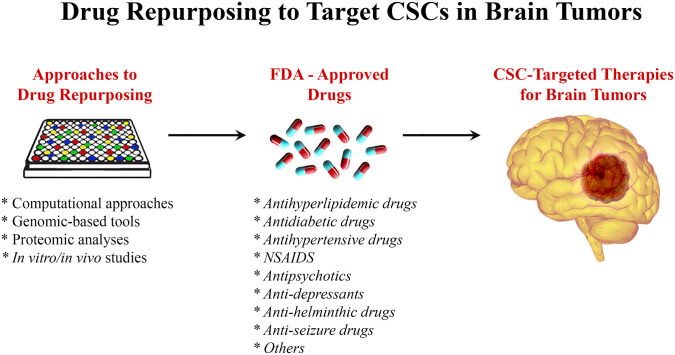
Within the field of drug repurposing, various techniques have been developed, which contribute to the discovery of compounds of interest to treat various brain malignancies (Fig. 2). The exploration of links between brain tumor advancement and medications use is facilitated by large databases containing patients’ information, such as the Clinical Practice Research Datalink. 122 In addition, these relationships can also be explored by investigating clinical trial databases and even organizations’ specific data that have already been filed. 122 However, malignancy regression cannot be attributed to drug use unless experimental analysis is first performed. Approaches in silico can be used to select drugs of interest for repurposing by using computational algorithms to recognize interactions between drugs and targets of interest. 123 The databases used vary in the drug–target interactions they explore, ranging from genomics, 124 proteomics, 125 and disease-specific and even molecular targets, among others. The idea is to identify drugs that target a certain disease mechanism, protein, or pathway that is relevant to the malignancy in question. Although this approach requires data on the disease phenotype and the targets, it has proven to be time- and labor-efficient compared with other more traditional methods. 126 A different technique to new drug recognition is activity-based repurposing. This approach uses drug targets for different diseases that have been screened previously; this is all done in the absence of concurrent structural information. 89 The library of Integrated Network–based cellular signature (LINCS) is a program that analyzes how cells respond to certain stimuli, from chemical and genetic mutations to responses to different agents such as drugs. 127 Understanding the basis of cell signaling and regulation in disease states is crucial to determining functional drugs and possible therapeutic agents. 128 Hence, activity-based drug repurposing presents a powerful tool where knowing the molecular basis of disease can serve as a way to target therapy by using drugs that are known to be effective for a certain type of molecular phenomenon and apply them to another tumor type. 129 An example of a drug group that has been repurposed in this way is the ALK inhibitors for the treatment of various brain tumors such as large cell lymphoma and neuroblastoma (NB) where the ALK kinase is overactivated. 130
Figure 2.
Drug repurposing to target CSCs in brain tumors. Using different approaches including genomics- and proteomics-based tools, computational approaches, and in vitro/in vivo studies, FDA-approved drugs could be repurposed to target CSCs in brain tumors. Abbreviation: CSC, cancer stem cell; FDA, Food and Drug Administration; NSAIDs, nonsteroidal anti-inflammatory drugs.
Targeting Cancer Stem Cells in the Treatment of Meningioma
Meningiomas are neoplasms thought to be derived from meningothelial cell arachnoid cap cells and are among the most common primary central nervous system (CNS) tumors, representing approximately 37% of all primary CNS neoplasms. 131 Current management guidelines indicate gross total resection as the primary treatment for intracranial meningiomas. 132 Clinically, the major factor in recurrence is the extent of resection, which may be complicated by adhesion or encasement of vital structures such as cranial nerves, cerebral arteries, or the brainstem.131,132 Several studies have reported the presence of CSCs in meningioma,133,134 which can be targeted using targeted therapies. Given the high cost, long development time, and technical difficulty of new drug discovery, a promising strategy to identify new therapeutic options is the repurposing of drugs, 118 which can be implicated in meningiomas.
Antiseizure Drugs
Several drugs have been proposed as candidates for repurposing in the treatment of meningiomas. Chiou et al. 135 tested the effect of valproic acid (VPA) on cultured meningioma stem cells. VPA is a drug primarily used in the treatment of seizure disorders. In the study by Chiou et al., authors demonstrated that VPA reduced the viability of meningioma CSCs and increased their sensitivity to radiation. In addition, VPA was found to reduce the expression of Oct4 and anchorage-independent growth, indicating a differentiation-promoting effect of VPA on stem cells that was more pronounced when combined with radiation. Thus, combined treatment of VPA and radiation could be proposed as a possible strategy in the treatment of meningiomas by targeting their CSC population. 135
Anthelminthic Drugs
Mebendazole (MBZ) is a wide-spectrum anthelminthic that works by inhibiting microtubule polymerization. 136 Its role as a possible agent for the treatment of CSCs is also under study. In a review of the literature by Guerini et al., 137 multiple preclinical studies showed MBZ to be an effective inhibitor of multiple mechanisms that facilitate cancer cell viability. Larsen et al. 138 described inhibition of the hedgehog (Hh) signaling pathway and Hh-dependent survival in cultured cell lines. In a study by Bai et al., 139 MBZ inhibited tumor angiogenesis in MB animal models by blocking the activity of VEGF receptor 2. Sasaki et al. also reported that MBZ caused mitotic arrest via depolymerization of tubulin and abnormal spindle cell formation. In murine models, the authors reported a significant suppression of tumor growth after treatment with MBZ. 140 In addition, Skibinski et al. 141 found a proapoptotic effect of MBZ on human meningioma cell lines and an increase in median survival in animal models treated with MBZ. They also described a synergistic effect when combining MBZ therapy with radiation.
Antidiabetic Drugs
Metformin is a biguanide antihyperglycemic agent widely used for the treatment of diabetes mellitus type 2. In vitro and in vivo studies using different CSC models, including breast,142,143 colon, 144 meningioma, 145 cholangiocarcinoma, 146 pancreatic adenocarcinoma, 142 GBM, and NB, 147 have shown inhibition of growth and proliferation of CSCs. Multiple mechanisms of action have been attributed to metformin; its major effects are largely exerted through the activation of AMP-activated protein kinase and the inhibition of mitochondrial respiratory chain (complex I).148,149 The capacity of metformin to regulate several cell signaling pathways is suggested to be the mechanism behind its anticancer properties. 142 In a study by Guo et al., 145 metformin inhibited the mammalian target of rapamycin (mTOR) signaling pathway, and significant inhibition of cell growth was seen in a concentration-dependent manner in vitro and in vivo. Metformin has also been shown to increase the efficacy of classical anticancer drugs. In meningioma and cholangiocarcinoma cell models, the anticancer effect of cisplatin was significantly enhanced when the cells were co-treated with metformin.145,146 In a study by Hirsch et al., 143 and in a similar fashion, metformin increased the therapeutic power of doxorubicin where the combination killed CSCs and non-CSCs in culture, and reduced tumor mass and prolonged remission much more effectively than either drug alone in a xenograft mouse model.
Antihypertensive Drugs
The role of the renin–angiotensin system (RAS) as a target for the treatment of CSCs is also of interest. Better known for its role in blood pressure regulation and electrolyte hemostasis, components of the RAS also mediate angiogenesis, cellular proliferation, and apoptosis. Angiotensin II (ATII) is the main effector of the RAS, via its two receptors ATII type 1 receptor (ATIIR1) and ATII type 2 receptor (ATIIR2). ATIIR1 induces angiogenesis and cellular proliferation. 150 Pro-renin, through its receptor pattern recognition receptor (PRR), is another important component of the RAS implicated in carcinogenesis through its involvement in the Wnt/β-catenin signaling pathway. 151 Cells in numerous cancer types, including brain tumors such as GBM, have been found to express both CSC markers and components of the RAS. 152 In two similar studies, Shivapathasundram et al. 150 reported the expression of PRR, ACE, ATIIR1, and ATIIR2 and Rahman et al. 153 reported the expression of cathepsin B, cathepsin D, and, to a lesser extent, cathepsin G in a putative stem cell population of low-grade meningioma. Given that the RAS is overexpressed in a wide range of tumors, numerous studies have assessed the effect of RAS inhibitors in vitro and on tumor models in vivo. These drugs, across different classes (β-blockers, angiotensin- converting enzyme inhibitors, and angiotensin receptor blockers), have been reported to reduce tumor cell growth, migration, invasion, and metastasis in numerous cancer types. 152 To the best of our knowledge, the effect of RAS inhibitors in meningioma CSCs has not been studied. The overexpression of different components of RAS suggests a potential utility for the repurposing of these drugs for the treatment of meningioma, and more research is needed to explore this possibility.
Another group of antihypertensive drugs, calcium channel blocker (CCB) is a candidate for repurposing. Cytosolic Ca2+ plays an important role in intracellular signal transduction and several processes necessary for cell proliferation and survival, making this ion a possible target in the treatment of cancer. 154 Previous studies have reported the overexpression of Ca2+ channels in various tumor types155,156 In meningioma, Jensen et al. reported a dose-dependent decrease in growth stimulation when cultured human meningioma cells exposed to serum-containing media or different growth factors were treated with verapamil, nifedipine, or diltiazem (voltage-dependent calcium channel–blocking agents).157,158 In a similar study, diltiazem and verapamil were found to decrease the growth of meningioma tumor models in mice. 158 These drugs were also found to potentiate the effects of hydroxyurea on inhibition of growth of meningioma cell models in a study by Ragel et al. 159 In CSCs, the blockade of Ca2+ channels has been reported to inhibit the proliferation, survival, and stemness features of stem-like cells in GBM and ovarian cell models. It also potentiated the effect of chemotherapeutic agents.160–162 The effect of CCB on meningioma CSCs has not been characterized to the best of our knowledge, but these observations suggest another possible treatment option worth studying.
Antihyperlipidemic Drugs
The repurposing of hydroxyl-methyl-glutaryl CoA (HMG-CoA) reductase inhibitors for the treatment of cancer has also been studied. These drugs, classically used for the treatment of hypercholesterolemia, block the conversion of HMG-CoA to mevalonate, the rate-limiting step in cholesterol synthesis. Mevalonate is necessary for normal and neoplastic cells to enter the cell cycle. 163 Several studies have found that drugs in this group inhibit cell proliferation by downregulating several intracellular signaling pathways.163,164 Wu et al. 164 reported simvastatin (SMV)-mediated downregulation of the PI3K/Akt pathway, leading to inhibition of GBM cell proliferation and induction of apoptosis via increase in caspase-3 activation in vitro and in vivo. In meningioma cells, Johnson et al. evaluated the effects of lovastatin (LVS) on meningioma cell proliferation and its influence on the activation of the MEK-1–MAPK/ERK pathway. In their study, cultured meningioma cells were exposed to three different mitogens with and without LVS, and the levels of phosphorylated MAPK kinase (MEK-1/2) and MAPK were compared. LVS significantly reduced the levels of both and inhibited cell proliferation after stimulation, suggesting inhibition of MAPK/ERK activity as one of the mechanisms that explain the effect of LVS on cell proliferation. 163 In a study by Ghering et al., cultured meningioma cell lines were treated with different combinations of statins and thiazolidinediones (TDZ). They compared the antiproliferative and cytotoxic effect of SMV, LVS, atorvastatin, pravastatin, SMV, and two TDZ, pioglitazone (PGZ) and rosiglitazone, and their combinations on various human meningioma cell lines. Among the statins, SMV was the most effective drug in the group. From the TDZ group, PGZ exhibited a significant effect when used as a single agent, and out of all the drug combinations, SMV in combination with PGZ turned out to be the most effective treatment. 165 In CSCs, SMV has been reported to inhibit proliferation and induce apoptosis in breast cancer cell models. 166 LVS has been found to have similar effects in nasopharyngeal carcinoma CSCs. 167 These findings suggest the utility of the repurposing of lipid-lowering drugs for the treatment of meningiomas.
Targeting Cancer Stem Cells in the Treatment of Glioblastoma
GBM (WHO grade IV glioma) is the most frequent and aggressive primary brain tumor in adults. Life expectancy remains bleak at around 15 to 19 months. 2 The majority of GBMs, around 90%, are primary and develop rapidly in older individuals without any proof of a precursor lesion. 168 A minority are secondary, which signifies that they arise from lower WHO grade gliomas such as II or III. 169 Incidence is on the rise but varies drastically between countries and can range between 0.59 and 5 cases per 100,000 individuals.170,171 The direct causes behind this increasing incidence have not been elucidated as of yet. However, it has been postulated that this may be due to better diagnostic modalities and an increasingly aging population as a whole. 172 Surgical resection, if possible, remains the standard of care. It may be followed by radiotherapy 173 or supplemented with chemotherapy such as temozolomide (TMZ), which is of particular benefit in patients with a methylated MGMT promoter. 174
Tumorigenic CSCs are self-renewing cells that are present in the tumor bulk of GBM. 175 They contribute to both tumor initiation and therapy resistance, including conventional chemotherapy 38 and radiotherapy. 176 Indeed, a study by Murat et al. 177 provided the first clinical evidence for the presence of a “glioma stem cell” or “self-renewal” phenotype in treatment resistance of GBM. The study analyzed the gene expression profiles of 80 GBMs and associated them with resistance to therapy, showing that the expression signature dominated by HOX genes, which comprises Prominin-1 (CD133), could serve as a predictor for poor survival in patients treated with concomitant chemoradiotherapy. 177 Another study by Pallini et al. 178 demonstrated that analyzing CSCs may predict the survival of GBM patients where in vitro CSC generation and the presence of ≥2% CD133+ cells in tumor lesions negatively correlate with overall and progression-free survival of patients. Therefore, one of the most challenging aspects of treating GBM is destroying the resident CSC population. 179 Also, it is apparent that developing novel treatment strategies that target CSCs may help advance the entire field of GBM treatment. 180 Novel treatments need to be safe, efficacious, able to penetrate the BBB, and Food and Drug Administration (FDA)-approved 181 (Table 1).
Table 1.
Examples of Some Repurposed Drugs Currently Being Clinically Investigated for the Treatment of Glioblastoma (ClinicalTrials.gov).
| Clinical Trial Number | Phase | Intervention/Treatment | Patient Population | Number of Patients Enrolled | Primary Outcome Measures | Secondary outcome measures |
|---|---|---|---|---|---|---|
| NCT02780024 | II | Metformin | GBM | 50 | To determine overall survival | To assess toxicity of the regimen |
| NCT03243851 | II | Metformin + TMZ | GBM | 108 | Comparison of progression-free survival obtained from progression-free survival curve | To determine tumor response rate, control probability, 6-month progression-free survival, and overall survival and assess quality of life |
| NCT04691960 | II | Metformin + ketogenic diet | GBM | 36 | Ability to achieve and maintain ketosis | Tolerability of metformin |
| NCT02496741 | Ib/II | Metformin + chloroquine | IDH1/2-mutated glioma | 15 | Maximum tolerated dose of metformin + chloroquine | Effect of metformin + chloroquine on tumor response and recommended dose of metformin + chloroquine |
| NCT01729260 | I | Mebendazole | High-grade glioma | 24 | To determine the maximum tolerated dose of mebendazole in combination with TMZ given after surgery and the standard radiation and TMZ treatment in patients with newly diagnosed malignant gliomas | To determine overall survival in 10 years |
| NCT01837862 | I/II | Mebendazole + vincristine, carboplatin, and TMZ | GBM | 36 | To determine the maximally tolerated dose of mebendazole in combination with vincristine, carboplatin, and temozolomide | To determine 3-year event-free survival and overall survival |
| NCT02644291 | I | Mebendazole | GBM | 21 | Adverse events attributed to mebendazole | To determine overall survival of patients |
| NCT02029573 | II | Atorvastatin + TMZ and radiotherapy | GBM | 36 | Progression free survival at 6 months | Progression free survival and overall survival at 2–3 years |
| NCT01977677 | I/II | Plerixafor | GBM | 30 | To determine the dose-limiting toxicity | To determine the progression-free survival |
Abbreviations: TMZ, temozolomide; GBM, glioblastoma.
Antidiabetic Drugs
Metformin is currently in phase II of National Institutes of Health (NIH)-approved clinical trial NCT02780024 for its use as an adjuvant component to TMZ and radiotherapy (ClinicalTrials.gov; NCT02780024). In addition, a dose-finding phase Ib/II clinical trial was done on patients with IDH1/2-mutated glioma treated with a combination of metformin and chloroquine (ClinicalTrials.gov; NCT02496741), revealing promising results and opening novel treatment avenue for those patients.182,183 In mouse xenograft neoplastic models, metformin was able to preferentially target and kill CSCs and prolong remission. 184 The mode of action is postulated to be through attenuation of the inflammatory feedback pathway, particularly on transcription factor NF-κB and interleukin 6 (IL-6). 185 Metformin and phenformin were shown to inhibit cell migration of LN229 glioma cells in vitro and in vivo. In addition, both drugs were associated with increased reactive oxygen species (ROS) production within the cell mitochondrion. 186 Phenformin is superior to metformin in terms of chemical potency and was shown to induce apoptosis of GBM CSCs and inhibit their self-renewal. Phenformin decreases stemness and mesenchymal marker expression while increasing miR-124, miR-137, and let-7 expression. 187 Silencing let-7, which normally acts as a tumor suppressor microRNA by targeting HMGA2, attenuates the effects of phenformin on inhibiting CSCs. 188 Dichloroacetate, which acts as an inhibitor of the glycolytic enzyme pyruvate dehydrogenase kinase that in turn decreases lactic acid production induced by biguanides, has been shown to potentiate the anti-CSC effect of phenformin. 187
Antihyperlipidemic Drugs
Statins are of interest in GBM therapy because they have been proposed to induce apoptosis in glioma cells. 187 SMV’s antineoplastic action is via proliferation reduction and apoptosis induction of the C6 glioma cell line. This is achieved by way of increasing phosphorylation of c-jun through activating the JNK pathway. 189 Atorvastatin, on its own and via MT1-MMP microglial decreased expression, was able to suppress both the invasive and migratory potential of glioma cells. 190 Furthermore, LVS had a synergistic effect with tumor necrosis factor (TNF)-related apoptosis-inducing ligand (TRAIL)-inducing apoptosis in 50% of cells in three human GBM malignant cells lines: M059K, M59J, and A172 in contrast to the failure of TRAIL to do so when used alone. 191 Statins could be used as an adjunct to TMZ to increase the efficacy of chemotherapy and decrease resistance. 192
Antihypertensive Drugs
The RAS system has been implicated in the hallmarks of cancer, 193 which is one explanation to as why this class of drugs may be helpful in glioma treatment. 194 ATII and ATIIR1 interaction aids in tumor angiogenesis via stimulating vascular smooth muscle cells and ECs to produce VEGF.195,196 Furthermore, the interplay of the Ang-II/AT1R system has been shown to be a contributing factor toward neoplastic immunosuppression, which may hinder the productive utilization of immunotherapy. 197
GSK3-β Inhibitors
Glycogen synthesis kinase 3-β (GSK3-β) has been implicated in many tumors including GBM, which paves the way for potential therapeutic strategies.198–200 GSK3-β is a serine/threonine kinase 201 linked to several pathways involved in tumorigenesis, such as Wnt/β-catenin 202 and PI3K/PTEN. 203 One study showed that, in vivo, infrared (IR)-induced inhibition of GSK3-β reduced the tumor load, thus extending survival. GSK3-β, at high dosages, increased GBM cell sensitivity to IR via 53BP1 phosphorylation at serine 166. Consequently, GSK3-β inhibitors such as SB216763 may become a valuable asset in treating radiotherapy-resistant GB. 204 Another study demonstrated the in vitro effects of BIO (an indirubin) and CHIR9902, both GSK3-β inhibitors, on glioma cells. Specifically, BIO decreases migration and invasion in spheroid assays, whereas CHIR9902 renders ECs more permeable, hence making the BBB more permissible to drugs. 205
In C6 glioma cells that have the isodehydrogenase 2 (IDH2) mutant, lithium chloride resulted in a decrease in proliferation and migration via inhibition of GSK3-β activity. At the same time, β-catenin accumulated less in these cells. These results illustrate a potential use of lithium chloride as a treatment modality. 206
Another promising treatment avenue is kenpaullone, a GSK3-β and cyclin-dependent kinase inhibitor. 207 Kenpaullone exhibits TMZ-enhancing activity against GBM CSCs possibly via suppressing SOX2 that attenuates stemness. This activity contrasts with the case when kenpaullone was administered alone possibly due to the increased BBB penetrance of the combined therapy. 208 An additional GSK3-β inhibitor is tideglusib which suppresses GBM tumorigeneses via downregulating KDM1A. 209
Nonsteroidal Anti-inflammatory Drugs
Nonsteroidal anti-inflammatory drugs (NSAIDs) are a drug group with anti-inflammatory, analgesic and antipyretic characteristics that act through inhibition of cyclooxygenase (COX). 210 Inflammation is a key component of tumorigenesis explaining the role of NSAIDs in anticancer therapy, specifically via decreasing tumor burden and angiogenic and metastatic potentials. 211 IL-6, an inflammatory cytokine, promotes the development of non-cancerous cells into CSCs specifically via upregulation of Oct4 gene expression by activating IL-2R/JAK/STAT3 pathway. 212 Certain prostaglandins, such as PGE2, have been shown to be implicated in cancers such as colon carcinoma. 213 This could explain the role of inflammation in persistence of the tumor burden via the resident CSCs and hence resistance to traditional GBM therapy routes. Two NSAIDs, ibuprofen and diclofenac, have been shown to potently inhibit glioma cells in vitro, albeit via distinct mechanisms; elucidating their role as potential adjuvants to GBM therapy. Ibuprofen mainly acts via a COX- and lactate-independent mechanism. On the contrary, diclofenac acts primarily via decreased STAT-3 signaling and downstream modulation of glycolysis in a more lactate-dependent manner. 214 The role of lactate is to induce TGF-β2 via its activating protein, thrombospondin 1 (THBS-1). This serves an important role in GBM pathogenesis via regulating cell migration. 215
Antipsychotic Drugs
Antipsychotics have displayed potential for utilization as anticancer therapies particularly against GBM. 216 As a general rule, antipsychotics have adequate BBB penetrance, a property that is of significant use in malignant brain gliomas. 217 Another interesting fact is that patients with schizophrenia have a lower risk of getting cancer than those without, at a rate of 1.93% to 2.97%, respectively. 218 Chlorpromazine, an FDA-approved psychotropic drug, was shown to efficaciously inhibit proliferating glioma cells which were resistant to chemotherapy. Chlorpromazine specifically induced arrest in the proliferation of cells that expressed COX4-1. 219 Thioridazine (THD), a drug belonging to the class of phenothiazine used to treat psychosis and schizophrenia, exhibits anti-GBM properties. THD, through the Wnt/β-catenin pathway, enhances p-62 mediated apoptosis in glioma cells. 220 THD also acts by upregulating AMPK, leading to apoptosis. 221 Traditionally used for its dopamine receptor antagonistic properties in schizophrenia, THD has shown a role in sensitizing GBM cells to TMZ in vivo and hence reducing tumor growth and increasing survival in mouse xenografts. The antineoplastic role is mediated by impairing autophagy, a survival mechanism for glioma cells, independent of its action on dopamine receptors. 222 Triflupherazine (TFP), another dopamine receptor antagonist in the class of phenothiazines, could be repurposed for GB treatment when combined with radiotherapy. The combination of TFP and radiotherapy targets GB CSCs and prevents the radiotherapy-induced phenotypic switch of GB into a more aggressive variant. 223
Antidepressants
The quest for finding new drugs to overcome GB resistance has also included antidepressant drugs, one of which is fluoxetine. Fluoxetine is a selective serotonin reuptake inhibitor (SSRI) that is used in the treatment of major depression. 224 In glioma cells, fluoxetine was shown to bind to α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid receptor, causing a calcium influx into the cell which consequently results in an increase in mitochondrial calcium, thus triggering apoptosis. In vivo studies showed that fluoxetine’s action was comparable with that of TMZ in suppressing glioma growth. 225
One tumorigenesis theory proposes that cancer is a “mitochondrionopathy,” meaning that mitochondria in cancer cells are both structurally and functionally diseased as opposed to those in healthy cells. 226 Those who support the “metabolic theory” propose that if one can activate the malfunctioning mitochondria in the neoplastic GB cells, cell death may progress normally and facilitate chemotherapy. 227 Tricyclic antidepressants, such as imipramine and amitriptyline, may be able to restore GB mitochondrial function and respiratory function and thus serve as potential therapeutic adjuvants. 228 In terms of mechanism of action, both drugs served to suppress p56 NF-κB, which is strongly activated in many tumors.228,229 Impramine and amitriptyline also suppressed lactate release, a potent oncogenic metabolite that induces inflammation, from GB cells.228,230 Another antidepressant drug, fluvoxamine, exhibited potential anti-GB therapeutic potential via inhibiting actin polymerization and subsequent glioma cell migration in mice models with human GB CSCs. 231
Anthelminthic Drugs
In a study by Bai et al., fenbendazole was incidentally found to decrease engraftment of brain tumors in nude mice that were being treated for a pinworm infection. This discovery led the group to explore the anticancerous effects of other benzimidazoles. They also found that MBZ was cytotoxic at IC50 concentrations ranging between 0.1 and 0.3 µM. MBZ was also shown to increase survival by 68% in mouse glioma models. 232 Other than disrupting tubulin polymerization, benzimidazoles were postulated to act via inhibiting the expression of VEGF 233 and HIF-1α. 234 MBZ-induced apoptosis may potentially occur through phosphorylation and consequent inactivation of Bcl-2. 235
“The Reverse swing-M” trial is a phase I going to phase II study conducted by Patil et al. to determine the maximal tolerable dose (MTD) of MBZ in conjunction with other treatment modalities in fighting recurrent high-grade glioma. It was shown that the MTD of MBZ is 1600 mg TDS when combined with TMZ or TMZ radiation. When using MBZ with single-agent lomustine (CCNU), the MTD was a dosage of 800 mg TDS. 236 Another NIH-approved clinical trial NCT01729260 is a phase I study whose goal is to determine the MTD of MBZ in patients who have been newly diagnosed with GB and are on TMZ (ClinicalTrials.gov; NCT01729260).
Targeting Cancer Stem Cells in the Treatment of Pediatric Brain Tumors
Brain tumors are the second most prevalent malignancies in the pediatric population after leukemia and account for the majority of the solid tumors in children. They are also counted as the leading cause of tumor-related childhood mortality. 237 In 2007, WHO initially classified brain tumors based on the histopathologic appearance. 131 With advances of biomedical research, this classification expanded in 2016 to involve molecular parameters along with histopathologic patterns to classify brain malignancies. 238
Despite the presence of a wide therapeutic armamentarium to treat brain tumors, the presence of the CSC subpopulation within the tumor bulk contributed to cancer resistance to the conventional therapy and consequent recurrence of the tumor.98,239,240 To overcome this resistance, researchers have been exploring various molecular pathways in CSCs and identifying therapeutic strategies to target this subpopulation of cells.
NB and GB cell lines demonstrated chemoresistant and radioresistant activity, respectively, of CD133+ cells with Wnt overexpression and that inhibition of this pathway decreased the viability of these cells.241,242 Moreover, targeting Wnt signaling promoted NB cell line differentiation and enhanced their sensitivity to chemotherapy. 243 GSK-3β is a serine/threonine kinase that represents a downstream switch to multiple pathways in CSCs.199,244 GSK-3β inhibition has been shown to induce apoptosis in NB cell lines via activating the Wnt signaling pathway in a p53-independent manner.199,245
MB is the most common primary brain tumor diagnosed in childhood. Studies were able to isolate CD133+ CSCs within the perivascular niche of MBs. 93 Like many other tumors as well as GB, highly aggressive MB cells have shown to upregulate CSC markers such as CD133 and Nestin. 240 Although the functional role of CD133 is not well delineated, overexpression of CD133 is associated with chemoresistance 246 and high tumor-initiating capacity. 247 Similarly, CD133 cell surface marker expression has been used to identify CSCs in NB cell lines which display more resistance to anticancer drugs. 248
Future Directions
To conclude, drug resistance is a cardinal feature of CSCs in all cancers. Currently, conventional therapies target the bulk of the tumor, mainly the non-CSC component, and patients typically experience recurrence and metastasis due to the CSC enrichment which was not tackled. 41 Some of these resistance mechanisms include dormancy, the TME, expression of multidrug resistance proteins, such as ALDH, and EMT, 249 among others, which emphasize the need to contemplate these mechanisms when considering therapies against CSCs. 250 In addition, when we are concerned with brain tumors and their respective CSCs, there are unique challenges facing treatments and therapeutics that need to be kept in mind, as discussed previously.3,89,91 The utilization of conventional therapies like chemotherapy, radiation, and surgical resection is limited and often does not remove or target the CSCs, which are the cells responsible for tumor recurrence and metastasis, especially in the brain. Therefore, innovative solutions are required to tackle the problems presently faced by CSCs in the brain: drug repurposing is a promising approach. This idea—the use of drugs in ways other than their original FDA-approved indications—could offer novel avenues to bypass the chemoresistance and recurrence seen with conventional therapy and treatment. 89 Indeed, targeted therapies, such as those that use cell surface markers, as well as various tumorigenic proteins, seen in CSCs could pave the way for better patient outcomes. Using the markers that are specific to CSCs in general, along with the markers specifically observed in brain tumor CSCs, bioinformatic and computational methods could be used to match different drug types previously indicated for different diseases to be used as treatment for CSCs.
Acknowledgments
The authors thank all members of the Arkadi M. Rywlin M.D. Department of Pathology and Laboratory Medicine, Mount Sinai Medical Center (Miami Beach, FL) and the Herbert Wertheim College of Medicine, Florida International University (Miami, FL) for their help with this work.
Footnotes
Competing Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Author Contributions: HFB was involved in conceptualization, project administration, supervision, investigation, methodology, writing—original draft preparation, writing—reviewing and editing, visualization, and validation. DD was involved in investigation, methodology, writing—original draft preparation, writing—reviewing and Editing, visualization, and validation. AAA, ME, KSO, JCAM, RD, RSu, AZ, and RA were involved in methodology, writing–original draft preparation, and validation. RSa was involved in supervision, writing—reviewing and editing, visualization, and validation. RP was involved in project administration, supervision, writing—reviewing and editing, visualization, and validation. All authors have read and approved the final manuscript.
Funding: The author(s) received no financial support for the research, authorship, and/or publication of this article.
Contributor Information
Hisham F. Bahmad, Arkadi M. Rywlin M.D. Department of Pathology and Laboratory Medicine, Mount Sinai Medical Center, Miami Beach, Florida.
Darine Daher, Faculty of Medicine, American University of Beirut, Beirut, Lebanon.
Abed A. Aljamal, Department of Internal Medicine, Mount Sinai Medical Center, Miami Beach, Florida.
Mohamad K. Elajami, Department of Internal Medicine, Mount Sinai Medical Center, Miami Beach, Florida.
Kei Shing Oh, Arkadi M. Rywlin M.D. Department of Pathology and Laboratory Medicine, Mount Sinai Medical Center, Miami Beach, Florida.
Juan Carlos Alvarez Moreno, Arkadi M. Rywlin M.D. Department of Pathology and Laboratory Medicine, Mount Sinai Medical Center, Miami Beach, Florida.
Ruben Delgado, Arkadi M. Rywlin M.D. Department of Pathology and Laboratory Medicine, Mount Sinai Medical Center, Miami Beach, Florida.
Richard Suarez, Department of Pathology, Herbert Wertheim College of Medicine, Florida International University, Miami, Florida.
Ana Zaldivar, Arkadi M. Rywlin M.D. Department of Pathology and Laboratory Medicine, Mount Sinai Medical Center, Miami Beach, Florida.
Roshanak Azimi, Arkadi M. Rywlin M.D. Department of Pathology and Laboratory Medicine, Mount Sinai Medical Center, Miami Beach, Florida.
Amilcar Castellano, Arkadi M. Rywlin M.D. Department of Pathology and Laboratory Medicine, Mount Sinai Medical Center, Miami Beach, Florida; Department of Pathology, Herbert Wertheim College of Medicine, Florida International University, Miami, Florida.
Robert Sackstein, Department of Translational Medicine, Translational Glycobiology Institute, Herbert Wertheim College of Medicine, Florida International University, Miami, Florida.
Robert J. Poppiti, Arkadi M. Rywlin M.D. Department of Pathology and Laboratory Medicine, Mount Sinai Medical Center, Miami Beach, Florida; Department of Pathology, Herbert Wertheim College of Medicine, Florida International University, Miami, Florida.
Literature Cited
- 1. Khazaei Z, Goodarzi E, Borhaninejad V, Iranmanesh F, Mirshekarpour H, Mirzaei B, Naemi H, Bechashk SM, Darvishi I, Ershad Sarabi R, Naghibzadeh-Tahami A. The association between incidence and mortality of brain cancer and human development index (HDI): an ecological study. BMC Public Health. 2020;20(1):1696. doi: 10.1186/s12889-020-09838-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Stupp R, Hegi ME, Mason WP, van den Bent MJ, Taphoorn MJ, Janzer RC, Ludwin SK, Allgeier A, Fisher B, Belanger K, Hau P, Brandes AA, Gijtenbeek J, Marosi C, Vecht CJ, Mokhtari K, Wesseling P, Villa S, Eisenhauer E, Gorlia T, Weller M, Lacombe D, Cairncross JG, Mirimanoff RO. Effects of radiotherapy with concomitant and adjuvant temozolomide versus radiotherapy alone on survival in glioblastoma in a randomised phase III study: 5-year analysis of the EORTC-NCIC trial. Lancet Oncol. 2009;10(5):459–66. doi: 10.1016/s1470-2045(09)70025-7. [DOI] [PubMed] [Google Scholar]
- 3. Aldape K, Brindle KM, Chesler L, Chopra R, Gajjar A, Gilbert MR, Gottardo N, Gutmann DH, Hargrave D, Holland EC, Jones DTW, Joyce JA, Kearns P, Kieran MW, Mellinghoff IK, Merchant M, Pfister SM, Pollard SM, Ramaswamy V, Rich JN, Robinson GW, Rowitch DH, Sampson JH, Taylor MD, Workman P, Gilbertson RJ. Challenges to curing primary brain tumours. Nat Rev Clin Oncol. 2019;16(8):509–20. doi: 10.1038/s41571-019-0177-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bhowmik A, Khan R, Ghosh MK. Blood brain barrier: a challenge for effectual therapy of brain tumors. Biomed Res Int. 2015;2015:320941. doi: 10.1155/2015/320941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Abbaszadegan MR, Bagheri V, Razavi MS, Momtazi AA, Sahebkar A, Gholamin M. Isolation, identification, and characterization of cancer stem cells: a review. J Cell Physiol. 2017;232(8):2008–18. doi: 10.1002/jcp.25759. [DOI] [PubMed] [Google Scholar]
- 6. Ashburn TT, Thor KB. Drug repositioning: identifying and developing new uses for existing drugs. Nat Rev Drug Discov. 2004;3(8):673–83. doi: 10.1038/nrd1468. [DOI] [PubMed] [Google Scholar]
- 7. Kaitin KI, DiMasi JA. Pharmaceutical innovation in the 21st century: new drug approvals in the first decade, 2000-2009. Clin Pharmacol Ther. 2011;89(2):183–8. doi: 10.1038/clpt.2010.286. [DOI] [PubMed] [Google Scholar]
- 8. Ayob AZ, Ramasamy TS. Cancer stem cells as key drivers of tumour progression. J Biomed Sci. 2018;25(1):20. doi: 10.1186/s12929-018-0426-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bonnet D, Dick JE. Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nat Med. 1997;3(7):730–7. doi: 10.1038/nm0797-730. [DOI] [PubMed] [Google Scholar]
- 10. Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci U S A. 2003;100(7):3983–8. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Singh SK, Hawkins C, Clarke ID, Squire JA, Bayani J, Hide T, Henkelman RM, Cusimano MD, Dirks PB. Identification of human brain tumour initiating cells. Nature. 2004;432(7015):396–401. doi: 10.1038/nature03128. [DOI] [PubMed] [Google Scholar]
- 12. Maitland NJ, Frame FM, Polson ES, Lewis JL, Collins AT. Prostate cancer stem cells: do they have a basal or luminal phenotype? Horm Cancer. 2011;2(1):47–61. doi: 10.1007/s12672-010-0058-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. O’Flaherty JD, Barr M, Fennell D, Richard D, Reynolds J, O’Leary J, O’Byrne K. The cancer stem-cell hypothesis: its emerging role in lung cancer biology and its relevance for future therapy. J Thorac Oncol. 2012;7(12):1880–90. doi: 10.1097/JTO.0b013e31826bfbc6. [DOI] [PubMed] [Google Scholar]
- 14. Munro MJ, Wickremesekera SK, Peng L, Marsh RW, Itinteang T, Tan ST. Cancer stem cell subpopulations in primary colon adenocarcinoma. PLoS ONE. 2019;14(9):e0221963. doi: 10.1371/journal.pone.0221963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Furusawa J, Zhang H, Vural E, Stone A, Fukuda S, Oridate N, Fang H, Ye Y, Suen JY, Fan CY. Distinct epigenetic profiling in head and neck squamous cell carcinoma stem cells. Otolaryngol Head Neck Surg. 2011;144(6):900–9. doi: 10.1177/0194599811398786. [DOI] [PubMed] [Google Scholar]
- 16. Lee CJ, Dosch J, Simeone DM. Pancreatic cancer stem cells. J Clin Oncol. 2008;26(17):2806–12. doi: 10.1200/jco.2008.16.6702. [DOI] [PubMed] [Google Scholar]
- 17. Yang ZF, Ngai P, Ho DW, Yu WC, Ng MN, Lau CK, Li ML, Tam KH, Lam CT, Poon RT, Fan ST. Identification of local and circulating cancer stem cells in human liver cancer. Hepatology. 2008;47(3):919–28. doi: 10.1002/hep.22082. [DOI] [PubMed] [Google Scholar]
- 18. O’Brien CA, Pollett A, Gallinger S, Dick JE. A human colon cancer cell capable of initiating tumour growth in immunodeficient mice. Nature. 2007;445(7123):106–10. doi: 10.1038/nature05372. [DOI] [PubMed] [Google Scholar]
- 19. Vermeulen L, Todaro M, de Sousa Mello F, Sprick MR, Kemper K, Perez Alea M, Richel DJ, Stassi G, Medema JP. Single-cell cloning of colon cancer stem cells reveals a multi-lineage differentiation capacity. Proc Natl Acad Sci U S A. 2008;105(36):13427–32. doi: 10.1073/pnas.0805706105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Eramo A, Lotti F, Sette G, Pilozzi E, Biffoni M, Di Virgilio A, Conticello C, Ruco L, Peschle C, De Maria R. Identification and expansion of the tumorigenic lung cancer stem cell population. Cell Death Differ. 2008;15(3):504–14. doi: 10.1038/sj.cdd.4402283. [DOI] [PubMed] [Google Scholar]
- 21. Martin PJ, Najfeld V, Hansen JA, Penfold GK, Jacobson RJ, Fialkow PJ. Involvement of the B-lymphoid system in chronic myelogenous leukaemia. Nature. 1980;287(5777):49–50. doi: 10.1038/287049a0. [DOI] [PubMed] [Google Scholar]
- 22. Jamieson CHM, Ailles LE, Dylla SJ, Muijtjens M, Jones C, Zehnder JL, Gotlib J, Li K, Manz MG, Keating A, Sawyers CL, Weissman IL. Granulocyte-macrophage progenitors as candidate leukemic stem cells in blast-crisis CML. N Engl J Med. 2004;351(7):657–67. doi: 10.1056/NEJMoa040258. [DOI] [PubMed] [Google Scholar]
- 23. Clarke MF. Clinical and therapeutic implications of cancer stem cells. N Engl J Med. 2019;380(23):2237–45. doi: 10.1056/NEJMra1804280. [DOI] [PubMed] [Google Scholar]
- 24. Shlush LI, Mitchell A, Heisler L, Abelson S, Ng SWK, Trotman-Grant A, Medeiros JJF, Rao-Bhatia A, Jaciw-Zurakowsky I, Marke R, McLeod JL, Doedens M, Bader G, Voisin V, Xu C, McPherson JD, Hudson TJ, Wang JCY, Minden MD, Dick JE. Tracing the origins of relapse in acute myeloid leukaemia to stem cells. Nature. 2017;547(7661):104–8. doi: 10.1038/nature22993. [DOI] [PubMed] [Google Scholar]
- 25. Krivtsov AV, Twomey D, Feng Z, Stubbs MC, Wang Y, Faber J, Levine JE, Wang J, Hahn WC, Gilliland DG, Golub TR, Armstrong SA. Transformation from committed progenitor to leukaemia stem cell initiated by MLL-AF9. Nature. 2006;442(7104):818–22. doi: 10.1038/nature04980. [DOI] [PubMed] [Google Scholar]
- 26. Bahr C, von Paleske L, Uslu VV, Remeseiro S, Takayama N, Ng SW, Murison A, Langenfeld K, Petretich M, Scognamiglio R, Zeisberger P, Benk AS, Amit I, Zandstra PW, Lupien M, Dick JE, Trumpp A, Spitz F. A Myc enhancer cluster regulates normal and leukaemic haematopoietic stem cell hierarchies. Nature. 2018;553(7689):515–20. doi: 10.1038/nature25193. [DOI] [PubMed] [Google Scholar]
- 27. Akala OO, Park I-K, Qian D, Pihalja M, Becker MW, Clarke MF. Long-term haematopoietic reconstitution by Trp53-/-p16Ink4a-/-p19Arf-/- multipotent progenitors. Nature. 2008;453(7192):228–32. doi: 10.1038/nature06869. [DOI] [PubMed] [Google Scholar]
- 28. Dianat-Moghadam H, Heidarifard M, Jahanban-Esfahlan R, Panahi Y, Hamishehkar H, Pouremamali F, Rahbarghazi R, Nouri M. Cancer stem cells-emanated therapy resistance: implications for liposomal drug delivery systems. J Control Release. 2018;288:62–83. doi: 10.1016/j.jconrel.2018.08.043. [DOI] [PubMed] [Google Scholar]
- 29. Dean M, Fojo T, Bates S. Tumour stem cells and drug resistance. Nat Rev Cancer. 2005;5(4):275–84. doi: 10.1038/nrc1590. [DOI] [PubMed] [Google Scholar]
- 30. Pardal R, Clarke MF, Morrison SJ. Applying the principles of stem-cell biology to cancer. Nat Rev Cancer. 2003;3(12):895–902. doi: 10.1038/nrc1232. [DOI] [PubMed] [Google Scholar]
- 31. Visvader JE, Lindeman GJ. Cancer stem cells in solid tumours: accumulating evidence and unresolved questions. Nat Rev Cancer. 2008;8(10):755–68. doi: 10.1038/nrc2499. [DOI] [PubMed] [Google Scholar]
- 32. Alison MR, Lim SM, Nicholson LJ. Cancer stem cells: problems for therapy? J Pathol. 2011;223(2):148–62. doi: 10.1002/path.2793. [DOI] [PubMed] [Google Scholar]
- 33. Lenos KJ, Miedema DM, Lodestijn SC, Nijman LE, van den Bosch T, Romero Ros X, Lourenço FC, Lecca MC, van der Heijden M, van Neerven SM, van Oort A, Leveille N, Adam RS, de Sousa E, Melo F, Otten J, Veerman P, Hypolite G, Koens L, Lyons SK, Stassi G, Winton DJ, Medema JP, Morrissey E, Bijlsma MF, Vermeulen L. Stem cell functionality is microenvironmentally defined during tumour expansion and therapy response in colon cancer. Nat Cell Biol. 2018;20(10):1193–202. doi: 10.1038/s41556-018-0179-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Singh A, Settleman J. EMT, cancer stem cells and drug resistance: an emerging axis of evil in the war on cancer. Oncogene. 2010;29(34):4741–51. doi: 10.1038/onc.2010.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kiyohara MH, Dillard C, Tsui J, Kim SR, Lu J, Sachdev D, Goodglick L, Tong M, Torous VF, Aryasomayajula C, Wang W, Najafzadeh P, Gordon LK, Braun J, McDermott S, Wicha MS, Wadehra M. EMP2 is a novel therapeutic target for endometrial cancer stem cells. Oncogene. 2017;36(42):5793–807. doi: 10.1038/onc.2017.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Fang D, Kitamura H. Cancer stem cells and epithelial-mesenchymal transition in urothelial carcinoma: possible pathways and potential therapeutic approaches. Int J Urol. 2018;25(1):7–17. doi: 10.1111/iju.13404. [DOI] [PubMed] [Google Scholar]
- 37. Cojoc M, Mäbert K, Muders MH, Dubrovska A. A role for cancer stem cells in therapy resistance: cellular and molecular mechanisms. Semin Cancer Biol. 2015;31:16–27. doi: 10.1016/j.semcancer.2014.06.004. [DOI] [PubMed] [Google Scholar]
- 38. Chen J, Li Y, Yu T-S, McKay RM, Burns DK, Kernie SG, Parada LF. A restricted cell population propagates glioblastoma growth after chemotherapy. Nature. 2012;488(7412):522–6. doi: 10.1038/nature11287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Bai X, Ni J, Beretov J, Graham P, Li Y. Cancer stem cell in breast cancer therapeutic resistance. Cancer Treat Rev. 2018;69:152–63. doi: 10.1016/j.ctrv.2018.07.004. [DOI] [PubMed] [Google Scholar]
- 40. De Robertis M, Poeta ML, Signori E, Fazio VM. Current understanding and clinical utility of miRNAs regulation of colon cancer stem cells. Semin Cancer Biol. 2018;53:232–47. doi: 10.1016/j.semcancer.2018.08.008. [DOI] [PubMed] [Google Scholar]
- 41. Batlle E, Clevers H. Cancer stem cells revisited. Nat Med. 2017;23(10):1124–34. doi: 10.1038/nm.4409. [DOI] [PubMed] [Google Scholar]
- 42. Comoglio PM, Trusolino L, Boccaccio C. Known and novel roles of the MET oncogene in cancer: a coherent approach to targeted therapy. Nat Rev Cancer. 2018;18(6):341–58. doi: 10.1038/s41568-018-0002-y. [DOI] [PubMed] [Google Scholar]
- 43. Goto H, Shimono Y, Funakoshi Y, Imamura Y, Toyoda M, Kiyota N, Kono S, Takao S, Mukohara T, Minami H. Adipose-derived stem cells enhance human breast cancer growth and cancer stem cell-like properties through adipsin. Oncogene. 2019;38(6):767–79. doi: 10.1038/s41388-018-0477-8. [DOI] [PubMed] [Google Scholar]
- 44. Liu C, Kelnar K, Liu B, Chen X, Calhoun-Davis T, Li H, Patrawala L, Yan H, Jeter C, Honorio S, Wiggins JF, Bader AG, Fagin R, Brown D, Tang DG. The microRNA miR-34a inhibits prostate cancer stem cells and metastasis by directly repressing CD44. Nat Med. 2011;17(2):211–5. doi: 10.1038/nm.2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Ji C, Yang L, Yi W, Xiang D, Wang Y, Zhou Z, Qian F, Ren Y, Cui W, Zhang X, Zhang P, Wang JM, Cui Y, Bian X. Capillary morphogenesis gene 2 maintains gastric cancer stem-like cell phenotype by activating a Wnt/β-catenin pathway. Oncogene. 2018;37(29):3953–66. doi: 10.1038/s41388-018-0226-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Haspels HN, Rahman MA, Joseph JV, Gras Navarro A, Chekenya M. Glioblastoma stem-like cells are more susceptible than differentiated cells to natural killer cell lysis mediated through killer immunoglobulin-like receptors-human leukocyte antigen ligand mismatch and activation receptor-ligand interactions. Front Immunol. 2018;9:1345. doi: 10.3389/fimmu.2018.01345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Gilbertson RJ, Rich JN. Making a tumour’s bed: glioblastoma stem cells and the vascular niche. Nat Rev Cancer. 2007;7(10):733–6. doi: 10.1038/nrc2246. [DOI] [PubMed] [Google Scholar]
- 48. Yan Y, Zuo X, Wei D. Concise review: emerging role of CD44 in cancer stem cells: a promising biomarker and therapeutic target. Stem Cells Transl Med. 2015;4(9):1033–43. doi: 10.5966/sctm.2015-0048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Jacobs PP, Sackstein R. CD44 and HCELL: preventing hematogenous metastasis at step 1. FEBS Lett. 2011;585(20):3148–58. doi: 10.1016/j.febslet.2011.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Barzegar Behrooz A, Syahir A, Ahmad S. CD133: beyond a cancer stem cell biomarker. J Drug Target. 2019;27(3):257–69. doi: 10.1080/1061186x.2018.1479756. [DOI] [PubMed] [Google Scholar]
- 51. Brugnoli F, Grassilli S, Al-Qassab Y, Capitani S, Bertagnolo V. CD133 in breast cancer cells: more than a stem cell marker. J Oncol. 2019;2019:7512632. doi: 10.1155/2019/7512632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Yang ZL, Zheng Q, Yan J, Pan Y, Wang ZG. Upregulated CD133 expression in tumorigenesis of colon cancer cells. World J Gastroenterol. 2011;17(7):932–7. doi: 10.3748/wjg.v17.i7.932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Attia S, Atwan N, Arafa M, Shahin RA. Expression of CD133 as a cancer stem cell marker in invasive gastric carcinoma. Pathologica. 2019;111(1):18–23. doi: 10.32074/1591-951x-51-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Ishiwata T, Matsuda Y, Yoshimura H, Sasaki N, Ishiwata S, Ishikawa N, Takubo K, Arai T, Aida J. Pancreatic cancer stem cells: features and detection methods. Pathol Oncol Res. 2018;24(4):797–805. doi: 10.1007/s12253-018-0420-x. [DOI] [PubMed] [Google Scholar]
- 55. Piao LS, Hur W, Kim TK, Hong SW, Kim SW, Choi JE, Sung PS, Song MJ, Lee BC, Hwang D, Yoon SK. CD133+ liver cancer stem cells modulate radioresistance in human hepatocellular carcinoma. Cancer Lett. 2012;315(2):129–37. doi: 10.1016/j.canlet.2011.10.012. [DOI] [PubMed] [Google Scholar]
- 56. Wang S, Xu ZY, Wang LF, Su W. CD133+ cancer stem cells in lung cancer. Front Biosci (Landmark Ed). 2013;18:447–53. doi: 10.2741/4113. [DOI] [PubMed] [Google Scholar]
- 57. Kanwal R, Shukla S, Walker E, Gupta S. Acquisition of tumorigenic potential and therapeutic resistance in CD133+ subpopulation of prostate cancer cells exhibiting stem-cell like characteristics. Cancer Lett. 2018;430:25–33. doi: 10.1016/j.canlet.2018.05.014. [DOI] [PubMed] [Google Scholar]
- 58. Kozovska Z, Gabrisova V, Kucerova L. Malignant melanoma: diagnosis, treatment and cancer stem cells. Neoplasma. 2016;63(4):510–7. doi: 10.4149/neo_2016_403. [DOI] [PubMed] [Google Scholar]
- 59. Zhang W, Chen H, Lv S, Yang H. High CD133 expression is associated with worse prognosis in patients with glioblastoma. Mol Neurobiol. 2016;53(4):2354–60. doi: 10.1007/s12035-015-9187-1. [DOI] [PubMed] [Google Scholar]
- 60. Shimoda M, Ota M, Okada Y. Isolation of cancer stem cells by side population method. Methods Mol Biol. 2018;1692:49–59. doi: 10.1007/978-1-4939-7401-6_5. [DOI] [PubMed] [Google Scholar]
- 61. Tabor MH, Clay MR, Owen JH, Bradford CR, Carey TE, Wolf GT, Prince ME. Head and neck cancer stem cells: the side population. Laryngoscope. 2011;121(3):527–33. doi: 10.1002/lary.21032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Yin B, Zeng Y, Liu G, Wang X, Wang P, Song Y. MAGE-A3 is highly expressed in a cancer stem cell-like side population of bladder cancer cells. Int J Clin Exp Pathol. 2014;7(6):2934–41. [PMC free article] [PubMed] [Google Scholar]
- 63. Xiong B, Ma L, Hu X, Zhang C, Cheng Y. Characterization of side population cells isolated from the colon cancer cell line SW480. Int J Oncol. 2014;45(3):1175–83. doi: 10.3892/ijo.2014.2498. [DOI] [PubMed] [Google Scholar]
- 64. Kato K, Takao T, Kuboyama A, Tanaka Y, Ohgami T, Yamaguchi S, Adachi S, Yoneda T, Ueoka Y, Kato K, Hayashi S, Asanoma K, Wake N. Endometrial cancer side-population cells show prominent migration and have a potential to differentiate into the mesenchymal cell lineage. Am J Pathol. 2010;176(1):381–92. doi: 10.2353/ajpath.2010.090056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Szotek PP, Pieretti-Vanmarcke R, Masiakos PT, Dinulescu DM, Connolly D, Foster R, Dombkowski D, Preffer F, Maclaughlin DT, Donahoe PK. Ovarian cancer side population defines cells with stem cell-like characteristics and Mullerian Inhibiting Substance responsiveness. Proc Natl Acad Sci U S A. 2006;103(30):11154–9. doi: 10.1073/pnas.0603672103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Huang CS, Zhai JM, Zhu XX, Cai JP, Chen W, Li JH, Yin XY. BTG2 is down-regulated and inhibits cancer stem cell-like features of side population cells in hepatocellular carcinoma. Dig Dis Sci. 2017;62(12):3501–10. doi: 10.1007/s10620-017-4829-y. [DOI] [PubMed] [Google Scholar]
- 67. Shi Y, Fu X, Hua Y, Han Y, Lu Y, Wang J. The side population in human lung cancer cell line NCI-H460 is enriched in stem-like cancer cells. PLoS ONE. 2012;7(3):e33358. doi: 10.1371/journal.pone.0033358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Murase M, Kano M, Tsukahara T, Takahashi A, Torigoe T, Kawaguchi S, Kimura S, Wada T, Uchihashi Y, Kondo T, Yamashita T, Sato N. Side population cells have the characteristics of cancer stem-like cells/cancer-initiating cells in bone sarcomas. Br J Cancer. 2009;101(8):1425–32. doi: 10.1038/sj.bjc.6605330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Liu J, Chi N, Zhang JY, Zhu W, Bian YS, Chen HG. Isolation and characterization of cancer stem cells from medulloblastoma. Genet Mol Res. 2015;14(2):3355–61. doi: 10.4238/2015. [DOI] [PubMed] [Google Scholar]
- 70. Fukaya R, Ohta S, Yamaguchi M, Fujii H, Kawakami Y, Kawase T, Toda M. Isolation of cancer stem-like cells from a side population of a human glioblastoma cell line, SK-MG-1. Cancer Lett. 2010;291(2):150–7. doi: 10.1016/j.canlet.2009.10.010. [DOI] [PubMed] [Google Scholar]
- 71. Jackson B, Brocker C, Thompson DC, Black W, Vasiliou K, Nebert DW, Vasiliou V. Update on the aldehyde dehydrogenase gene (ALDH) superfamily. Hum Genomics. 2011;5(4):283–303. doi: 10.1186/1479-7364-5-4-283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Kastan MB, Schlaffer E, Russo JE, Colvin OM, Civin CI, Hilton J. Direct demonstration of elevated aldehyde dehydrogenase in human hematopoietic progenitor cells. Blood. 1990;75(10):1947–50. [PubMed] [Google Scholar]
- 73. Tanaka K, Tomita H, Hisamatsu K, Nakashima T, Hatano Y, Sasaki Y, Osada S, Tanaka T, Miyazaki T, Yoshida K, Hara A. ALDH1A1-overexpressing cells are differentiated cells but not cancer stem or progenitor cells in human hepatocellular carcinoma. Oncotarget. 2015;6(28):24722–32. doi: 10.18632/oncotarget.4406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Li X, Wan L, Geng J, Wu CL, Bai X. Aldehyde dehydrogenase 1A1 possesses stem-like properties and predicts lung cancer patient outcome. J Thorac Oncol. 2012;7(8):1235–45. doi: 10.1097/JTO.0b013e318257cc6d. [DOI] [PubMed] [Google Scholar]
- 75. Li XS, Xu Q, Fu XY, Luo WS. ALDH1A1 overexpression is associated with the progression and prognosis in gastric cancer. BMC Cancer. 2014;14:705. doi: 10.1186/1471-2407-14-705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Ginestier C, Hur MH, Charafe-Jauffret E, Monville F, Dutcher J, Brown M, Jacquemier J, Viens P, Kleer CG, Liu S, Schott A, Hayes D, Birnbaum D, Wicha MS, Dontu G. ALDH1 is a marker of normal and malignant human mammary stem cells and a predictor of poor clinical outcome. Cell Stem Cell. 2007;1(5):555–67. doi: 10.1016/j.stem.2007.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Kreso A, Dick JE. Evolution of the cancer stem cell model. Cell Stem Cell. 2014;14(3):275–91. doi:101016/j.stem.2014.02.006. [DOI] [PubMed] [Google Scholar]
- 78. Miranda A, Hamilton PT, Zhang AW, Pattnaik S, Becht E, Mezheyeuski A, Bruun J, Micke P, de Reynies A, Nelson BH. Cancer stemness, intratumoral heterogeneity, and immune response across cancers. Proc Natl Acad Sci U S A. 2019;116(18):9020–9. doi: 10.1073/pnas.1818210116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Litviakov N, Ibragimova M, Tsyganov M, Kazantseva P, Deryusheva I, Pevzner A, Doroshenko A, Garbukov E, Tarabanovskaya N, Slonimskaya E. Amplifications of stemness genes and the capacity of breast tumors for metastasis. Oncotarget. 2020;11(21):1988–2001. doi: 10.18632/oncotarget.27608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Lu Y, Li L, Chen L, Gao Y, Chen X, Cao Y. TRIB3 confers glioma cell stemness via interacting with β-catenin. Environ Toxicol. 2020;35(6):697–706. doi: 10.1002/tox.22905. [DOI] [PubMed] [Google Scholar]
- 81. Eramo A, Haas TL, De Maria R. Lung cancer stem cells: tools and targets to fight lung cancer. Oncogene. 2010;29(33):4625–35. doi: 10.1038/onc.2010.207. [DOI] [PubMed] [Google Scholar]
- 82. Dunning NL, Laversin SA, Miles AK, Rees RC. Immunotherapy of prostate cancer: should we be targeting stem cells and EMT? Cancer Immunol Immunother. 2011;60(8):1181–93. doi: 10.1007/s00262-011-1065-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Tirino V, Desiderio V, Paino F, De Rosa A, Papaccio F, Fazioli F, Pirozzi G, Papaccio G. Human primary bone sarcomas contain CD133+ cancer stem cells displaying high tumorigenicity in vivo. Faseb J. 2011;25(6):2022–30. doi: 10.1096/fj.10-179036. [DOI] [PubMed] [Google Scholar]
- 84. Chen YC, Chang CJ, Hsu HS, Chen YW, Tai LK, Tseng LM, Chiou GY, Chang SC, Kao SY, Chiou SH, Lo WL. Inhibition of tumorigenicity and enhancement of radiochemosensitivity in head and neck squamous cell cancer-derived ALDH1-positive cells by knockdown of Bmi-1. Oral Oncol. 2010;46(3):158–65. doi: 10.1016/j.oraloncology.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 85. Ouyang G, Wang Z, Fang X, Liu J, Yang CJ. Molecular signaling of the epithelial to mesenchymal transition in generating and maintaining cancer stem cells. Cell Mol Life Sci. 2010;67(15):2605–18. doi: 10.1007/s00018-010-0338-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Vesuna F, Bergman Y, Raman V. Genomic pathways modulated by twist in breast cancer. BMC Cancer. 2017;17(1):52. doi: 10.1186/s12885-016-3033-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Li M, Zhang X, Xu X, Wu J, Hu K, Guo X, Zhang P. Clinicopathological and prognostic significance of Twist overexpression in NSCLC. Oncotarget. 2018;9(18):14642–51. doi: 10.18632/oncotarget.24489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Gao XH, Yang XQ, Wang BC, Liu SP, Wang FB. Overexpression of twist and matrix metalloproteinase-9 with metastasis and prognosis in gastric cancer. Asian Pac J Cancer Prev. 2013;14(9):5055–60. doi: 10.7314/apjcp.2013.14.9.5055. [DOI] [PubMed] [Google Scholar]
- 89. Bahmad HF, Elajami MK, El Zarif T, Bou-Gharios J, Abou-Antoun T, Abou-Kheir W. Drug repurposing towards targeting cancer stem cells in pediatric brain tumors. Cancer Metastasis Rev. 2020;39(1):127–48. doi: 10.1007/s10555-019-09840-2. [DOI] [PubMed] [Google Scholar]
- 90. Hammoud H, Saker Z, Harati H, Fares Y, Bahmad HF, Nabha S. Drug repurposing in medulloblastoma: challenges and recommendations. Curr Treat Options Oncol. 2020;22(1):6. doi: 10.1007/s11864-020-00805-0. [DOI] [PubMed] [Google Scholar]
- 91. Mackay A, Burford A, Carvalho D, Izquierdo E, Fazal-Salom J, Taylor KR, Bjerke L, Clarke M, Vinci M, Nandhabalan M, Temelso S, Popov S, Molinari V, Raman P, Waanders AJ, Han HJ, Gupta S, Marshall L, Zacharoulis S, Vaidya S, Mandeville HC, Bridges LR, Martin AJ, Al-Sarraj S, Chandler C, Ng H-K, Li X, Mu K, Trabelsi S, Brahim DHm-B, Kisljakov AN, Konovalov DM, Moore AS, Carcaboso AM, Sunol M, de Torres C, Cruz O, Mora J, Shats LI, Stavale JN, Bidinotto LT, Reis RM, Entz-Werle N, Farrell M, Cryan J, Crimmins D, Caird J, Pears J, Monje M, Debily M-A, Castel D, Grill J, Hawkins C, Nikbakht H, Jabado N, Baker SJ, Pfister SM, Jones DTW, Fouladi M, von Bueren AO, Baudis M, Resnick A, Jones C. Integrated molecular meta-analysis of 1,000 pediatric high-grade and diffuse intrinsic pontine glioma. Cancer Cell. 2017;32(4):520–37. doi: 10.1016/j.ccell.2017.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Galli R, Binda E, Orfanelli U, Cipelletti B, Gritti A, De Vitis S, Fiocco R, Foroni C, Dimeco F, Vescovi A. Isolation and characterization of tumorigenic, stem-like neural precursors from human glioblastoma. Cancer Res. 2004;64(19):7011–21. doi: 10.1158/0008-5472.Can-04-1364. [DOI] [PubMed] [Google Scholar]
- 93. Calabrese C, Poppleton H, Kocak M, Hogg TL, Fuller C, Hamner B, Oh EY, Gaber MW, Finklestein D, Allen M, Frank A, Bayazitov IT, Zakharenko SS, Gajjar A, Davidoff A, Gilbertson RJ. A perivascular niche for brain tumor stem cells. Cancer Cell. 2007;11(1):69–82. doi: 10.1016/j.ccr.2006.11.020. [DOI] [PubMed] [Google Scholar]
- 94. Bar EE, Lin A, Mahairaki V, Matsui W, Eberhart CG. Hypoxia increases the expression of stem-cell markers and promotes clonogenicity in glioblastoma neurospheres. Am J Pathol. 2010;177(3):1491–502. doi: 10.2353/ajpath.2010.091021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Hashimoto O, Shimizu K, Semba S, Chiba S, Ku Y, Yokozaki H, Hori Y. Hypoxia induces tumor aggressiveness and the expansion of CD133-positive cells in a hypoxia-inducible factor-1α-dependent manner in pancreatic cancer cells. Pathobiology. 2011;78(4):181–92. doi: 10.1159/000325538. [DOI] [PubMed] [Google Scholar]
- 96. Soeda A, Park M, Lee D, Mintz A, Androutsellis-Theotokis A, McKay RD, Engh J, Iwama T, Kunisada T, Kassam AB, Pollack IF, Park DM. Hypoxia promotes expansion of the CD133-positive glioma stem cells through activation of HIF-1α. Oncogene. 2009;28(45):3949–59. doi: 10.1038/onc.2009.252. [DOI] [PubMed] [Google Scholar]
- 97. Codrici E, Enciu A-M, Popescu I-D, Mihai S, Tanase C. Glioma stem cells and their microenvironments: providers of challenging therapeutic targets. Stem Cells Int. 2016;2016:5728438. doi: 10.1155/2016/5728438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Abou-Antoun TJ, Hale JS, Lathia JD, Dombrowski SM. Brain cancer stem cells in adults and children: cell biology and therapeutic implications. Neurotherapeutics. 2017;14(2):372–84. doi: 10.1007/s13311-017-0524-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Lee J, Kotliarova S, Kotliarov Y, Li A, Su Q, Donin NM, Pastorino S, Purow BW, Christopher N, Zhang W, Park JK, Fine HA. Tumor stem cells derived from glioblastomas cultured in bFGF and EGF more closely mirror the phenotype and genotype of primary tumors than do serum-cultured cell lines. Cancer Cell. 2006;9(5):391–403. doi: 10.1016/j.ccr.2006.03.030. [DOI] [PubMed] [Google Scholar]
- 100. Ghosh D, Ulasov IV, Chen L, Harkins LE, Wallenborg K, Hothi P, Rostad S, Hood L, Cobbs CS. TGFβ-responsive HMOX1 expression is associated with stemness and invasion in glioblastoma multiforme. Stem Cells. 2016;34(9):2276–89. doi: 10.1002/stem.2411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Jin X, Jeon H-M, Jin X, Kim E-J, Yin J, Jeon H-Y, Sohn Y-W, Oh S-Y, Kim J-K, Kim S-H, Jung J-E, Kwak S, Tang K-F, Xu Y, Rich JN, Kim H. The ID1-CULLIN3 axis regulates intracellular SHH and WNT signaling in glioblastoma stem cells. Cell Rep. 2016;16(6):1629–41. doi: 10.1016/j.celrep.2016.06.092. [DOI] [PubMed] [Google Scholar]
- 102. Maachani UB, Shankavaram U, Kramp T, Tofilon PJ, Camphausen K, Tandle AT. FOXM1 and STAT3 interaction confers radioresistance in glioblastoma cells. Oncotarget. 2016;7(47):77365–77. doi: 10.18632/oncotarget.12670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Flavahan WA, Wu Q, Hitomi M, Rahim N, Kim Y, Sloan AE, Weil RJ, Nakano I, Sarkaria JN, Stringer BW, Day BW, Li M, Lathia JD, Rich JN, Hjelmeland AB. Brain tumor initiating cells adapt to restricted nutrition through preferential glucose uptake. Nat Neurosci. 2013;16(10):1373–82. doi: 10.1038/nn.3510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Raffel C, Jenkins RB, Frederick L, Hebrink D, Alderete B, Fults DW, James CD. Sporadic medulloblastomas contain PTCH mutations. Cancer Res. 1997;57(5):842–5. [PubMed] [Google Scholar]
- 105. Reifenberger J, Wolter M, Weber RG, Megahed M, Ruzicka T, Lichter P, Reifenberger G. Missense mutations in SMOH in sporadic basal cell carcinomas of the skin and primitive neuroectodermal tumors of the central nervous system. Cancer Res. 1998;58(9):1798–803. [PubMed] [Google Scholar]
- 106. Parsons DW, Jones S, Zhang X, Lin JC-H, Leary RJ, Angenendt P, Mankoo P, Carter H, Siu I-M, Gallia GL, Olivi A, McLendon R, Rasheed BA, Keir S, Nikolskaya T, Nikolsky Y, Busam DA, Tekleab H, Diaz LA, Hartigan J, Smith DR, Strausberg RL, Marie SKN, Shinjo SMO, Yan H, Riggins GJ, Bigner DD, Karchin R, Papadopoulos N, Parmigiani G, Vogelstein B, Velculescu VE, Kinzler KW. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321(5897):1807–12. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Cancer Genome Atlas Research Network, McLendon R, Friedman A, Bigner D, Van Meir EG, Brat DJ, Mastrogianakis GM, Olson JJ, Mikkelsen T, Lehman N, Aldape K, Alfred Yung WK, Bogler O, VandenBerg S, Berger M, Prados M, Muzny D, Morgan M, Scherer S, Sabo A, Nazareth L, Lewis L, Hall O, Zhu Y, Ren Y, Alvi O, Yao J, Hawes A, Jhangiani S, Fowler G, San Lucas A, Kovar C, Cree A, Dinh H, Santibanez J, Joshi V, Gonzalez-Garay ML, Miller CA, Milosavljevic A, Donehower L, Wheeler DA, Gibbs RA, Cibulskis K, Sougnez C, Fennell T, Mahan S, Wilkinson J, Ziaugra L, Onofrio R, Bloom T, Nicol R, Ardlie K, Baldwin J, Gabriel S, Lander ES, Ding L, Fulton RS, McLellan MD, Wallis J, Larson DE, Shi X, Abbott R, Fulton L, Chen K, Koboldt DC, Wendl MC, Meyer R, Tang Y, Lin L, Osborne JR, Dunford-Shore BH, Miner TL, Delehaunty K, Markovic C, Swift G, Courtney W, Pohl C, Abbott S, Hawkins A, Leong S, Haipek C, Schmidt H, Wiechert M, Vickery T, Scott S, Dooling DJ, Chinwalla A, Weinstock GM, Mardis ER, Wilson RK, Getz G, Winckler W, Verhaak RGW, Lawrence MS, O’Kelly M, Robinson J, Alexe G, Beroukhim R, Carter S, Chiang D, Gould J, Gupta S, Korn J, Mermel C, Mesirov J, Monti S, Nguyen H, Parkin M, Reich M, Stransky N, Weir BA, Garraway L, Golub T, Meyerson M, Chin L, Protopopov A, Zhang J, Perna I, Aronson S, Sathiamoorthy N, Ren G, Yao J, Wiedemeyer WR, Kim H, Won Kong S, Xiao Y, Kohane IS, Seidman J, Park PJ, Kucherlapati R, Laird PW, Cope L, Herman JG, Weisenberger DJ, Pan F, Van Den Berg D, Van Neste L, Mi Yi J, Schuebel KE, Baylin SB, Absher DM, Li JZ, Southwick A, Brady S, Aggarwal A, Chung T, Sherlock G, Brooks JD, Myers RM, Spellman PT, Purdom E, Jakkula LR, Lapuk AV, Marr H, Dorton S, Gi Choi Y, Han J, Ray A, Wang V, Durinck S, Robinson M, Wang NJ, Vranizan K, Peng V, Van Name E, Fontenay GV, Ngai J, Conboy JG, Parvin B, Feiler HS, Speed TP, Gray JW, Brennan C, Socci ND, Olshen A, Taylor BS, Lash A, Schultz N, Reva B, Antipin Y, Stukalov A, Gross B, Cerami E, Qing Wang W, Qin L-X, Seshan VE, Villafania L, Cavatore M, Borsu L, Viale A, Gerald W, Sander C, Ladanyi M, Perou CM, Neil Hayes D, Topal MD, Hoadley KA, Qi Y, Balu S, Shi Y, Wu J, Penny R, Bittner M, Shelton T, Lenkiewicz E, Morris S, Beasley D, Sanders S, Kahn A, Sfeir R, Chen J, Nassau D, Feng L, Hickey E, Zhang J, Weinstein JN, Barker A, Gerhard DS, Vockley J, Compton C, Vaught J, Fielding P, Ferguson ML, Schaefer C, Madhavan S, Buetow KH, Collins F, Good P, Guyer M, Ozenberger B, Peterson J, Thomson E. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455(7216):1061–8. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Hira VV, Ploegmakers KJ, Grevers F, Verbovšek U, Silvestre-Roig C, Aronica E, Tigchelaar W, Turnšek TL, Molenaar RJ, Van Noorden CJ. CD133+ and nestin+ glioma stem-like cells reside around CD31+ arterioles in niches that express SDF-1α, CXCR4, osteopontin and cathepsin K. J Histochem Cytochem. 2015;63(7):481–93. doi: 10.1369/0022155415581689. [DOI] [PubMed] [Google Scholar]
- 109. Hira VVV, Wormer JR, Kakar H, Breznik B, van der Swaan B, Hulsbos R, Tigchelaar W, Tonar Z, Khurshed M, Molenaar RJ, Van Noorden CJF. Periarteriolar glioblastoma stem cell niches express bone marrow hematopoietic stem cell niche proteins. J Histochem Cytochem. 2018;66(3):155–73. doi: 10.1369/0022155417749174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Rios A, Hsu SH, Blanco A, Buryanek J, Day AL, McGuire MF, Brown RE. Durable response of glioblastoma to adjuvant therapy consisting of temozolomide and a weekly dose of AMD3100 (plerixafor), a CXCR4 inhibitor, together with lapatinib, metformin and niacinamide. Oncoscience. 2016;3(5–6):156–63. doi: 10.18632/oncoscience.311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Hira VVV, Van Noorden CJF, Molenaar RJ. CXCR4 antagonists as stem cell mobilizers and therapy sensitizers for acute myeloid leukemia and glioblastoma? Biology. 2020;9(2):31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Thomas RP, Nagpal S, Iv M, Soltys SG, Bertrand S, Pelpola JS, Ball R, Yang J, Sundaram V, Lavezo J, Born D, Vogel H, Brown JM, Recht LD. macrophage exclusion after radiation therapy (MERT): a first in human phase I/II trial using a CXCR4 inhibitor in glioblastoma. Clin Cancer Res. 2019;25(23):6948–57. doi: 10.1158/1078-0432.Ccr-19-1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Akbari-Birgani S, Paranjothy T, Zuse A, Janikowski T, Cieślar-Pobuda A, Likus W, Urasińska E, Schweizer F, Ghavami S, Klonisch T. Cancer stem cells, cancer-initiating cells and methods for their detection. Drug Discov Today. 2016;21(5):836–42. doi: 10.1016/j.drudis.2016.03.004. [DOI] [PubMed] [Google Scholar]
- 114. Sharifzad F, Ghavami S, Verdi J, Mardpour S, Mollapour Sisakht M, Azizi Z, Taghikhani A, Łos MJ, Fakharian E, Ebrahimi M, Hamidieh AA. Glioblastoma cancer stem cell biology: potential theranostic targets. Drug Resist Updat. 2019;42:35–45. doi: 10.1016/j.drup.2018.03.003. [DOI] [PubMed] [Google Scholar]
- 115. Wang C-H, Chiou S-H, Chou C-P, Chen Y-C, Huang Y-J, Peng C-A. Photothermolysis of glioblastoma stem-like cells targeted by carbon nanotubes conjugated with CD133 monoclonal antibody. Nanomedicine. 2011;7(1):69–79. doi: 10.1016/j.nano.2010.06.010. [DOI] [PubMed] [Google Scholar]
- 116. Xu Y, Stamenkovic I, Yu Q. CD44 attenuates activation of the hippo signaling pathway and is a prime therapeutic target for glioblastoma. Cancer Res. 2010;70(6):2455–64. doi: 10.1158/0008-5472.CAN-09-2505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Nowak-Sliwinska P, Scapozza L, Ruiz i, Altaba A. Drug repurposing in oncology: compounds, pathways, phenotypes and computational approaches for colorectal cancer. Biochim Biophys Acta Rev Cancer. 2019;1871(2):434–54. doi: 10.106/j.bbcan.2019.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Pushpakom S, Iorio F, Eyers PA, Escott KJ, Hopper S, Wells A, Doig A, Guilliams T, Latimer J, McNamee C, Norris A, Sanseau P, Cavalla D, Pirmohamed M. Drug repurposing: progress, challenges and recommendations. Nat Rev Drug Discov. 2019;18(1):41–58. doi: 10.1038/nrd.2018.168. [DOI] [PubMed] [Google Scholar]
- 119. Nosengo N. Can you teach old drugs new tricks? Nature. 2016;534(7607):314–6. doi: 10.1038/534314a. [DOI] [PubMed] [Google Scholar]
- 120. Breckenridge A, Jacob R. Overcoming the legal and regulatory barriers to drug repurposing. Nat Rev Drug Discov. 2019;18(1):1–2. doi: 10.1038/nrd.2018.92. [DOI] [PubMed] [Google Scholar]
- 121. McCabe B, Liberante F, Mills KI. Repurposing medicinal compounds for blood cancer treatment. Ann Hematol. 2015;94(8):1267–76. doi: 10.1007/s00277-015-2412-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Seliger C, Hau P. Drug repurposing of metabolic agents in malignant glioma. Int J Mol Sci. 2018;19(9):2768. doi: 10.3390/ijms19092768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Sohraby F, Bagheri M, Aryapour H. Performing an in silico repurposing of existing drugs by combining virtual screening and molecular dynamics simulation. In: Q. Vanhaelen, ed.: Computational methods for drug repurposing. New York: Springer; 2019. p. 23–43. [DOI] [PubMed] [Google Scholar]
- 124. Welter D, MacArthur J, Morales J, Burdett T, Hall P, Junkins H, Klemm A, Flicek P, Manolio T, Hindorff L, Parkinson H. The NHGRI GWAS catalog, a curated resource of SNP-trait associations. Nucleic Acids Res. 2013;42(D1):D1001–106. doi: 10.1093/nar/gkt1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Uhlen M, Oksvold P, Fagerberg L, Lundberg E, Jonasson K, Forsberg M, Zwahlen M, Kampf C, Wester K, Hober S, Wernerus H, Björling L, Ponten F. Towards a knowledge-based human protein atlas. Nat Biotech. 2010;28(12):1248–50. doi: 10.1038/nbt1210-1248. [DOI] [PubMed] [Google Scholar]
- 126. Shim JS, Liu JO. Recent advances in drug repositioning for the discovery of new anticancer drugs. Int J Biol Sci. 2014;10(7):654–63. doi: 10.7150/ijbs.9224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Vempati UD, Chung C, Mader C, Koleti A, Datar N, Vidović D, Wrobel D, Erickson S, Muhlich JL, Berriz G, Benes CH, Subramanian A, Pillai A, Shamu CE, Schürer SC. Metadata standard and data exchange specifications to describe, model, and integrate complex and diverse high-throughput screening data from the library of integrated network-based cellular signatures (LINCS). J Biomol Screen. 2014;19(5):803–16. doi: 10.1177/1087057114522514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Keenan AB, Jenkins SL, Jagodnik KM, Koplev S, He E, Torre D, Wang Z, Dohlman AB, Silverstein MC, Lachmann A, Kuleshov MV, Ma’ayan A, Stathias V, Terryn R, Cooper D, Forlin M, Koleti A, Vidovic D, Chung C, Schürer SC, Vasiliauskas J, Pilarczyk M, Shamsaei B, Fazel M, Ren Y, Niu W, Clark NA, White S, Mahi N, Zhang L, Kouril M, Reichard JF, Sivaganesan S, Medvedovic M, Meller J, Koch RJ, Birtwistle MR, Iyengar R, Sobie EA, Azeloglu EU, Kaye J, Osterloh J, Haston K, Kalra J, Finkbiener S, Li J, Milani P, Adam M, Escalante-Chong R, Sachs K, Lenail A, Ramamoorthy D, Fraenkel E, Daigle G, Hussain U, Coye A, Rothstein J, Sareen D, Ornelas L, Banuelos M, Mandefro B, Ho R, Svendsen CN, Lim RG, Stocksdale J, Casale MS, Thompson TG, Wu J, Thompson LM, Dardov V, Venkatraman V, Matlock A, Van Eyk JE, Jaffe JD, Papanastasiou M, Subramanian A, Golub TR, Erickson SD, Fallahi-Sichani M, Hafner M, Gray NS, Lin J-R, Mills CE, Muhlich JL, Niepel M, Shamu CE, Williams EH, Wrobel D, Sorger PK, Heiser LM, Gray JW, Korkola JE, Mills GB, LaBarge M, Feiler HS, Dane MA, Bucher E, Nederlof M, Sudar D, Gross S, Kilburn DF, Smith R, Devlin K, Margolis R, Derr L, Lee A, Pillai A. The library of integrated network-based cellular signatures NIH program: system-level cataloging of human cells response to perturbations. Cell Syst. 2018;6(1):13–24. doi: 10.1016/j.cels.2017.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Parisi D, Adasme MF, Sveshnikova A, Bolz SN, Moreau Y, Schroeder M. Drug repositioning or target repositioning: a structural perspective of drug-target-indication relationship for available repurposed drugs. Comput Struct Biotechnol J. 2020;18:1043–55. doi: 10.1016/j.csbj.2020.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Trigg RM, Turner SD. ALK in neuroblastoma: biological and therapeutic implications. Cancers. 2018;10(4):113. doi: 10.3390/cancers10040113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Louis DN, Ohgaki H, Wiestler OD, Cavenee WK, Burger PC, Jouvet A, Scheithauer BW, Kleihues P. The 2007 WHO classification of tumours of the central nervous system. Acta Neuropathol. 2007;114(2):97–109. doi: 10.1007/s00401-007-0243-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Euskirchen P, Peyre M. Management of meningioma. La Presse Médicale. 2018;47(11, Part 2):e245–52. doi: 10.1016/j.lpm.2018.05.016. [DOI] [PubMed] [Google Scholar]
- 133. Shivapathasundram G, Wickremesekera AC, Tan ST, Itinteang T. Tumour stem cells in meningioma: a review. J Clin Neurosci. 2018;47:66–71. doi: 10.1016/j.jocn.2017.10.059. [DOI] [PubMed] [Google Scholar]
- 134. Alamir H, Alomari M, Salwati AAA, Saka M, Bangash M, Baeesa S, Alghamdi F, Carracedo A, Schulten H-J, Chaudhary A, Abuzenadah A, Hussein D. In situ characterization of stem cells-like biomarkers in meningiomas. Cancer Cell Int. 2018;18(1):77. doi: 10.1186/s12935-018-0571-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Chiou H-YC, Lai W-K, Huang L-C, Huang S-M, Chueh S-H, Ma H-I, Hueng D-Y. Valproic acid promotes radiosensitization in meningioma stem-like cells. Oncotarget. 2015;6(12):9959–69. doi: 10.18632/oncotarget.3692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Abongwa M, Martin RJ, Robertson AP. A brief review on the mode of action of antinematodal drugs. Acta Vet. 2017;67(2):137–52. doi: 10.1515/acve-2017-0013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Guerini AE, Triggiani L, Maddalo M, Bonù ML, Frassine F, Baiguini A, Alghisi A, Tomasini D, Borghetti P, Pasinetti N, Bresciani R, Magrini SM, Buglione M. Mebendazole as a candidate for drug repurposing in oncology: an extensive review of current literature. Cancers. 2019;11:1284. doi: 10.3390/cancers11091284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Larsen AR, Bai RY, Chung JH, Borodovsky A, Rudin CM, Riggins GJ, Bunz F. Repurposing the antihelmintic mebendazole as a hedgehog inhibitor. Mol Cancer Ther. 2015;14(1):3–13. doi: 10.1158/1535-7163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Bai R-Y, Staedtke V, Rudin CM, Bunz F, Riggins GJ. Effective treatment of diverse medulloblastoma models with mebendazole and its impact on tumor angiogenesis. Neuro Oncol. 2015;17(4):545–54. doi: 10.1093/neuonc/nou234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Sasaki J, Ramesh R, Chada S, Gomyo Y, Roth JA, Mukhopadhyay T. The anthelmintic drug mebendazole induces mitotic arrest and apoptosis by depolymerizing tubulin in non-small cell lung cancer cells. Mol Cancer Ther. 2002;1(13):1201–9. [PubMed] [Google Scholar]
- 141. Skibinski CG, Williamson T, Riggins GJ. Mebendazole and radiation in combination increase survival through anticancer mechanisms in an intracranial rodent model of malignant meningioma. J Neuro Oncol. 2018;140(3):529–38. doi: 10.1007/s11060-018-03009-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Bao B, Azmi AS, Ali S, Zaiem F, Sarkar FH. Metformin may function as anti-cancer agent via targeting cancer stem cells: the potential biological significance of tumor-associated miRNAs in breast and pancreatic cancers. Ann Transl Med. 2014;2(6):59. doi: 10.3978/j.issn.2305-5839.2014.06.05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Hirsch HA, Iliopoulos D, Tsichlis PN, Struhl K. Metformin selectively targets cancer stem cells, and acts together with chemotherapy to block tumor growth and prolong remission. Cancer Res. 2009;69(19):7507–11. doi: 10.1158/0008-5472.CAN-09-2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Kim JH, Lee KJ, Seo Y, Kwon J-H, Yoon JP, Kang JY, Lee HJ, Park SJ, Hong SP, Cheon JH, Kim WH, Il Kim T. Effects of metformin on colorectal cancer stem cells depend on alterations in glutamine metabolism. Sci Rep. 2018;8(1):409. doi: 10.1038/s41598-017-18762-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Guo L, Cui J, Wang H, Medina R, Zhang S, Zhang X, Zhuang Z, Lin Y. Metformin enhances anti-cancer effects of cisplatin in meningioma through AMPK-mTOR signaling pathways. Mol Ther Oncolytics. 2020;20:119–31. doi: 10.1016/j.omto.2020.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Wandee J, Prawan A, Senggunprai L, Kongpetch S, Tusskorn O, Kukongviriyapan V. Metformin enhances cisplatin induced inhibition of cholangiocarcinoma cells via AMPK-mTOR pathway. Life Sci. 2018;207:172–83. doi: 10.1016/j.lfs.2018.05.046. [DOI] [PubMed] [Google Scholar]
- 147. Mouhieddine TH, Nokkari A, Itani MM, Chamaa F, Bahmad H, Monzer A, El-Merahbi R, Daoud G, Eid A, Kobeissy FH, Abou-Kheir W. Metformin and Ara-a effectively suppress brain cancer by targeting cancer stem/progenitor cells. Front Neurosci. 2015;9:442. doi: 10.3389/fnins.2015.00442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. Rena G, Pearson ER, Sakamoto K. Molecular mechanism of action of metformin: old or new insights? Diabetologia. 2013;56(9):1898–906. doi: 10.1007/s00125-013-2991-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Saini N, Yang X. Metformin as an anti-cancer agent: actions and mechanisms targeting cancer stem cells. Acta Biochim Biophys Sin. 2018;50(2):133–43. doi: 10.1093/abbs/gmx106. [DOI] [PubMed] [Google Scholar]
- 150. Shivapathasundram G, Wickremesekera AC, Brasch HD, van Schaijik B, Marsh RW, Tan ST, Itinteang T. Expression of components of the renin-angiotensin system by the putative stem cell population within WHO grade I meningioma. Front Surg. 2019;6:23. doi: 10.3389/fsurg.2019.00023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Masaaki K, Yuki S, Daisuke O, Keisuke M, Akira N, Takashi T. (Pro)renin receptor is crucial for glioma development via the Wnt/β-catenin signaling pathway. J Neurosurg. 2017;127(4):819–28. doi: 10.3171/2016.9.JNS16431. [DOI] [PubMed] [Google Scholar]
- 152. Roth IM, Wickremesekera AC, Wickremesekera SK, Davis PF, Tan ST. Therapeutic targeting of cancer stem cells via modulation of the renin-angiotensin system. Front Oncol. 2019;9:745. doi: 10.3389/fonc.2019.00745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153. Gross S, Mallu P, Joshi H, Schultz B, Go C, Soboloff J. Ca(2+) as a therapeutic target in cancer. Adv Cancer Res. 2020;148:233–317. doi: 10.1016/bs.acr.2020.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154. Antal L, Martin-Caraballo M. T-type calcium channels in cancer. Cancers. 2019;11(2):134. doi: 10.3390/cancers11020134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155. Cheng Q, Chen A, Du Q, Liao Q, Shuai Z, Chen C, Yang X, Hu Y, Zhao J, Liu S, Wen GR, An J, Jing H, Tuo B, Xie R, Xu J. Novel insights into ion channels in cancer stem cells. Int J Oncol. 2018;53(4):1435–41. doi: 10.3892/ijo.2018.4500. [DOI] [PubMed] [Google Scholar]
- 156. Jensen RL, Origitano TC, Lee YS, Weber M, Wurster RD. In vitro growth inhibition of growth factor-stimulated meningioma cells by calcium channel antagonists. Neurosurgery. 1995;36(2):365–74. doi: 10.1227/00006123-199502000-00017. [DOI] [PubMed] [Google Scholar]
- 157. Jensen RL, Lee YS, Guijrati M, Origitano TC, Wurster RD, Reichman OH. Inhibition of in vitro meningioma proliferation after growth factor stimulation by calcium channel antagonists: part II—additional growth factors, growth factor receptor immunohistochemistry, and intracellular calcium measurements. Neurosurgery. 1995;37(5):937–47. doi: 10.1227/00006123-199511000-00013. [DOI] [PubMed] [Google Scholar]
- 158. Jensen RL, Wurster RD. Calcium channel antagonists inhibit growth of subcutaneous xenograft meningiomas in nude mice. Surg Neurol. 2001;55(5):275–83. doi: 10.1016/S0090-3019(01)00444-X. [DOI] [PubMed] [Google Scholar]
- 159. Ragel BT, Gillespie DL, Kushnir V, Polevaya N, Kelly D, Jensen RL. Calcium channel antagonists augment hydroxyurea- and ru486-induced inhibition of meningioma growth in vivo and in vitro. Neurosurgery. 2006;59(5):1109–21. doi: 10.1227/01.Neu.0000245597.46581.Fb. [DOI] [PubMed] [Google Scholar]
- 160. Zhang Y, Cruickshanks N, Yuan F, Wang B, Pahuski M, Wulfkuhle J, Gallagher I, Koeppel AF, Hatef S, Papanicolas C, Lee J, Bar EE, Schiff D, Turner SD, Petricoin EF, Gray LS, Abounader R. Targetable T-type calcium channels drive glioblastoma. Cancer Res. 2017;77(13):3479–90. doi: 10.1158/0008-5472.CAN-16-2347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161. Maklad A, Sharma A, Azimi I. Calcium signaling in brain cancers: roles and therapeutic targeting. Cancers. 2019;11(2):145. doi: 10.3390/cancers11020145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162. Lee H, Kim JW, Kim DK, Choi DK, Lee S, Yu JH, Kwon O-B, Lee J, Lee D-S, Kim JH, Min S-H. Calcium channels as novel therapeutic targets for ovarian cancer stem cells. Int J Mol Sci. 2020;21(7):2327. doi: 10.3390/ijms21072327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163. Johnson MD, Woodard A, Okediji EJ, Toms SA, Allen GS. Lovastatin is a potent inhibitor of meningioma cell proliferation: evidence for inhibition of a mitogen associated protein kinase. J Neurooncol. 2002;56(2):133–42. doi: 10.1023/a:1014588214966. [DOI] [PubMed] [Google Scholar]
- 164. Wu H, Jiang H, Lu D, Xiong Y, Qu C, Zhou D, Mahmood A, Chopp M. Effect of simvastatin on glioma cell proliferation, migration, and apoptosis. Neurosurgery. 2009;65(6):1087–97. doi: 10.1227/01.NEU.0000360130.52812.1D. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165. Afzali M, Vatankhah M, Ostad SN. Investigation of simvastatin-induced apoptosis and cell cycle arrest in cancer stem cells of MCF-7. J Cancer Res Ther. 2016;12(2):725–30. doi: 10.4103/0973-1482.146127. [DOI] [PubMed] [Google Scholar]
- 166. Gehring S, Tapia-Pérez JH, Kirches E, Firsching R, Keilhoff G, Schneider T, Mawrin C. Cytotoxic effects of statins and thiazolidinediones on meningioma cells. J Neurooncol. 2011;102(3):383–93. doi: 10.1007/s11060-010-0351-1. [DOI] [PubMed] [Google Scholar]
- 167. Peng Y, He G, Tang D, Xiong L, Wen Y, Miao X, Hong Z, Yao H, Chen C, Yan S, Lu L, Yang Y, Li Q, Deng X. Lovastatin inhibits cancer stem cells and sensitizes to chemo- and photodynamic therapy in nasopharyngeal carcinoma. J Cancer. 2017;8(9):1655–64. doi: 10.7150/jca.19100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168. Ohgaki H, Kleihues P. The definition of primary and secondary glioblastoma. Clinical Cancer Res. 2013;19(4):764–72. doi: 10.1158/1078-0432.CCR-12-3002. [DOI] [PubMed] [Google Scholar]
- 169. Wirsching HG, Galanis E, Weller M. Glioblastoma. Handb Clin Neurol. 2016;134:381–97. doi: 10.1016/B978-0-12-802997-8.00023-2. [DOI] [PubMed] [Google Scholar]
- 170. Grech N, Dalli T, Mizzi S, Meilak L, Calleja N, Zrinzo A. Rising incidence of glioblastoma multiforme in a well-defined population. Cureus. 2020;12(5):e8195. doi: 10.7759/cureus.8195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171. Miranda-Filho A, Piñeros M, Soerjomataram I, Deltour I, Bray F. Cancers of the brain and CNS: global patterns and trends in incidence. Neuro Oncol. 2017;19(2):270–80. doi: 10.1093/neuonc/now166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172. Davis DL, Hoel D, Percy C, Ahlbom A, Schwartz J. Is brain cancer mortality increasing in industrial countries? Ann N Y Acad Sci. 1990;609:191–204. doi: 10.1111/j.1749-6632.1990.tb32067.x. [DOI] [PubMed] [Google Scholar]
- 173. Weller M, van den Bent M, Hopkins K, Tonn JC, Stupp R, Falini A, Cohen-Jonathan-Moyal E, Frappaz D, Henriksson R, Balana C, Chinot O, Ram Z, Reifenberger G, Soffietti R, Wick W. EANO guideline for the diagnosis and treatment of anaplastic gliomas and glioblastoma. Lancet Oncol. 2014;15(9):e395–403. doi: 10.1016/S1470-2045(14)70011-7. [DOI] [PubMed] [Google Scholar]
- 174. Hegi ME, Diserens AC, Gorlia T, Hamou MF, de Tribolet N, Weller M, Kros JM, Hainfellner JA, Mason W, Mariani L, Bromberg JE, Hau P, Mirimanoff RO, Cairncross JG, Janzer RC, Stupp R. MGMT gene silencing and benefit from temozolomide in glioblastoma. N Engl J Med. 2005;352(10):997–1003. doi: 10.1056/NEJMoa043331. [DOI] [PubMed] [Google Scholar]
- 175. Galli R, Binda E, Orfanelli U, Cipelletti B, Gritti A, De Vitis S, Fiocco R, Foroni C, Dimeco F, Vescovi A. Isolation and characterization of tumorigenic, stem-like neural precursors from human glioblastoma. Cancer Res. 2004;64(19):7011–21. doi: 10.1158/0008-5472.Can-04-1364. [DOI] [PubMed] [Google Scholar]
- 176. Bao S, Wu Q, McLendon RE, Hao Y, Shi Q, Hjelmeland AB, Dewhirst MW, Bigner DD, Rich JN. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature. 2006;444(7120):756–60. doi: 10.1038/nature05236. [DOI] [PubMed] [Google Scholar]
- 177. Murat A, Migliavacca E, Gorlia T, Lambiv WL, Shay T, Hamou M-F, Tribolet Nd, Regli L, Wick W, Kouwenhoven MCM, Hainfellner JA, Heppner FL, Dietrich P-Y, Zimmer Y, Cairncross JG, Janzer R-C, Domany E, Delorenzi M, Stupp R, Hegi ME. Stem cell-related “self-renewal” signature and high epidermal growth factor receptor expression associated with resistance to concomitant chemoradiotherapy in glioblastoma. J Clin Oncol. 2008;26(18):3015–24. doi: 10.1200/jco.2007.15.7164. [DOI] [PubMed] [Google Scholar]
- 178. Pallini R, Ricci-Vitiani L, Banna GL, Signore M, Lombardi D, Todaro M, Stassi G, Martini M, Maira G, Larocca LM, De Maria R. Cancer stem cell analysis and clinical outcome in patients with glioblastoma multiforme. Clin Cancer Res. 2008;14(24):8205–12. doi: 10.1158/1078-0432.Ccr-08-0644. [DOI] [PubMed] [Google Scholar]
- 179. Clarke J, Penas C, Pastori C, Komotar RJ, Bregy A, Shah AH, Wahlestedt C, Ayad NG. Epigenetic pathways and glioblastoma treatment. Epigenetics. 2013;8(8):785–95. doi: 10.4161/epi.25440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180. Lathia JD, Mack SC, Mulkearns-Hubert EE, Valentim CL, Rich JN. Cancer stem cells in glioblastoma. Genes Dev. 2015;29(12):1203–17. doi: 10.1101/gad.261982.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181. Tan SK, Jermakowicz A, Mookhtiar AK, Nemeroff CB, Schürer SC, Ayad NG. Drug repositioning in glioblastoma: a pathway perspective. Front Pharmacol. 2018;9:218. doi: 10.3389/fphar.2018.00218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182. Molenaar RJ, Coelen RJ, Khurshed M, Roos E, Caan MW, van Linde ME, Kouwenhoven M, Bramer JA, Bovée JV, Mathôt RA, Klümpen H-J, van Laarhoven HW, van Noorden CJ, Vandertop WP, Gelderblom H, van Gulik TM, Wilmink JW. Study protocol of a phase IB/II clinical trial of metformin and chloroquine in patients with IDH1-mutated or IDH2-mutated solid tumours. BMJ Open. 2017;7(6):e014961. doi: 10.1136/bmjopen-2016-014961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183. Molenaar RJ, Maciejewski JP, Wilmink JW, van Noorden CJF. Wild-type and mutated IDH1/2 enzymes and therapy responses. Oncogene. 2018;37(15):1949–60. doi: 10.1038/s41388-017-0077-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184. Iliopoulos D, Hirsch HA, Struhl K. Metformin decreases the dose of chemotherapy for prolonging tumor remission in mouse xenografts involving multiple cancer cell types. Cancer Res. 2011;71(9):3196–201. doi: 10.1158/0008-5472.Can-10-3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185. Hirsch HA, Iliopoulos D, Struhl K. Metformin inhibits the inflammatory response associated with cellular transformation and cancer stem cell growth. Proc Natl Acad Sci U S A. 2013;110(3):972–7. doi: 10.1073/pnas.1221055110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186. Wang Y, Meng Y, Zhang S, Wu H, Yang D, Nie C, Hu Q. Phenformin and metformin inhibit growth and migration of LN229 glioma cells in vitro and in vivo. Onco Targets Ther. 2018;11:6039–48. doi: 10.2147/OTT.S168981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187. Jiang W, Finniss S, Cazacu S, Xiang C, Brodie Z, Mikkelsen T, Poisson L, Shackelford DB, Brodie C. Repurposing phenformin for the targeting of glioma stem cells and the treatment of glioblastoma. Oncotarget. 2016;7(35):56456–70. doi: 10.18632/oncotarget.10919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188. Chien CS, Wang ML, Chu PY, Chang YL, Liu WH, Yu CC, Lan YT, Huang PI, Lee YY, Chen YW, Lo WL, Chiou SH. Lin28B/Let-7 regulates expression of Oct4 and Sox2 and reprograms oral squamous cell carcinoma cells to a stem-like state. Cancer Res. 2015;75(12):2553–65. doi: 10.1158/0008-5472.Can-14-2215. [DOI] [PubMed] [Google Scholar]
- 189. Koyuturk M, Ersoz M, Altiok N. Simvastatin induces proliferation inhibition and apoptosis in C6 glioma cells via c-jun N-terminal kinase. Neurosci Lett. 2004;370(2):212–7. doi: 10.1016/j.neulet.2004.08.020. [DOI] [PubMed] [Google Scholar]
- 190. Yongjun Y, Shuyun H, Lei C, Xiangrong C, Zhilin Y, Yiquan K. Atorvastatin suppresses glioma invasion and migration by reducing microglial MT1-MMP expression. J Neuroimmunol. 2013;260(1):1–8. doi: 10.1016/j.jneuroim.2013.04.020. [DOI] [PubMed] [Google Scholar]
- 191. Chan DYL, Chen GG, Poon WS, Liu PC. Lovastatin sensitized human glioblastoma cells to TRAIL-induced apoptosis. J Neuro Oncology. 2008;86(3):273–83. doi: 10.1007/s11060-007-9475-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192. Shojaei S, Alizadeh J, Thliveris J, Koleini N, Kardami E, Hatch GM, Xu F, Hombach-Klonisch S, Klonisch T, Ghavami S. Statins: a new approach to combat temozolomide chemoresistance in glioblastoma. J Investig Med. 2018;66(8):1083–7. doi: 10.1136/jim-2018-000874. [DOI] [PubMed] [Google Scholar]
- 193. Wegman-Ostrosky T, Soto-Reyes E, Vidal-Millán S, Sánchez-Corona J. The renin-angiotensin system meets the hallmarks of cancer. J Renin Angiotensin Aldosterone Syst. 2013;16(2):227–33. doi: 10.1177/1470320313496858. [DOI] [PubMed] [Google Scholar]
- 194. Arrieta O, Guevara P, Escobar E, García-Navarrete R, Pineda B, Sotelo J. Blockage of angiotensin II type I receptor decreases the synthesis of growth factors and induces apoptosis in C6 cultured cells and C6 rat glioma. Br J Cancer. 2005;92(7):1247–52. doi: 10.1038/sj.bjc.6602483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195. Williams B, Baker AQ, Gallacher B, Lodwick D. Angiotensin II increases vascular permeability factor gene expression by human vascular smooth muscle cells. Hypertension. 1995;25(5):913–7. doi: 10.1161/01.hyp.25.5.913. [DOI] [PubMed] [Google Scholar]
- 196. Egami K, Murohara T, Shimada T, Sasaki K, Shintani S, Sugaya T, Ishii M, Akagi T, Ikeda H, Matsuishi T, Imaizumi T. Role of host angiotensin II type 1 receptor in tumor angiogenesis and growth. J Clin Invest. 2003;112(1):67–75. doi: 10.1172/jci16645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197. Pinter M, Jain RK. Targeting the renin-angiotensin system to improve cancer treatment: implications for immunotherapy. Sci Transl Med. 2017;9(410):eaan5616. doi: 10.1126/scitranslmed.aan5616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198. Kotliarova S, Pastorino S, Kovell LC, Kotliarov Y, Song H, Zhang W, Bailey R, Maric D, Zenklusen JC, Lee J, Fine HA. Glycogen synthase kinase-3 inhibition induces glioma cell death through c-MYC, nuclear factor-kappaB, and glucose regulation. Cancer Res. 2008;68(16):6643–51. doi: 10.1158/0008-5472.Can-08-0850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199. Bahmad HF, Chalhoub RM, Harati H, Bou-Gharios J, Assi S, Ballout F, Monzer A, Msheik H, Araji T, Elajami MK, Ghanem P, Chamaa F, Kadara H, Abou-Antoun T, Daoud G, Fares Y, Abou-Kheir W. Tideglusib attenuates growth of neuroblastoma cancer stem/progenitor cells in vitro and in vivo by specifically targeting GSK-3β. Pharmacol Rep. 2021;73(1):211–26. doi: 10.1007/s43440-020-00162-7. [DOI] [PubMed] [Google Scholar]
- 200. Bou-Gharios J, Assi S, Bahmad HF, Kharroubi H, Araji T, Chalhoub RM, Ballout F, Harati H, Fares Y, Abou-Kheir W. The potential use of tideglusib as an adjuvant radio-therapeutic treatment for glioblastoma multiforme cancer stem-like cells. Pharmacol Rep. 2021;73(1):227–39. doi: 10.1007/s43440-020-00180-5. [DOI] [PubMed] [Google Scholar]
- 201. Doble BW, Woodgett JR. GSK-3: tricks of the trade for a multi-tasking kinase. J Cell Sci. 2003;116(Pt. 7):1175–86. doi: 10.1242/jcs.00384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202. Yook JI, Li XY, Ota I, Hu C, Kim HS, Kim NH, Cha SY, Ryu JK, Choi YJ, Kim J, Fearon ER, Weiss SJ. A Wnt-Axin2-GSK3beta cascade regulates snail1 activity in breast cancer cells. Nat Cell Biol. 2006;8(12):1398–406. doi: 10.1038/ncb1508. [DOI] [PubMed] [Google Scholar]
- 203. Mulholland DJ, Dedhar S, Wu H, Nelson CC. PTEN and GSK3beta: key regulators of progression to androgen-independent prostate cancer. Oncogene. 2006;25(3):329–37. doi: 10.1038/sj.onc.1209020. [DOI] [PubMed] [Google Scholar]
- 204. Yang Y, Lei T, Du S, Tong R, Wang H, Yang J, Huang J, Sun M, Wang Y, Dong Z. Nuclear GSK3β induces DNA double-strand break repair by phosphorylating 53BP1 in glioblastoma. Int J Oncol. 2018;52(3):709–20. doi: 10.3892/ijo.2018.4237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205. Handley MV. GSK-3 inhibitors in glioblastoma therapy: mechanisms of action. Boston: Boston University; 2015. [Google Scholar]
- 206. Fu Y, Zheng Y, Chan KG, Liang A, Hu F. Lithium chloride decreases proliferation and migration of C6 glioma cells harboring isocitrate dehydrogenase 2 mutant via GSK-3β. Mol Biol Rep. 2014;41(6):3907–13. doi: 10.1007/s11033-014-3258-7. [DOI] [PubMed] [Google Scholar]
- 207. Leost M, Schultz C, Link A, Wu YZ, Biernat J, Mandelkow EM, Bibb JA, Snyder GL, Greengard P, Zaharevitz DW, Gussio R, Senderowicz AM, Sausville EA, Kunick C, Meijer L. Paullones are potent inhibitors of glycogen synthase kinase-3beta and cyclin-dependent kinase 5/p25. Eur J Biochem. 2000;267(19):5983–94. doi: 10.1046/j.1432-1327.2000.01673.x. [DOI] [PubMed] [Google Scholar]
- 208. Kitabayashi T, Dong Y, Furuta T, Sabit H, Jiapaer S, Zhang J, Zhang G, Hayashi Y, Kobayashi M, Domoto T, Minamoto T, Hirao A, Nakada M. Identification of GSK3β inhibitor kenpaullone as a temozolomide enhancer against glioblastoma. Sci Rep. 2019;9(1):10049. doi: 10.1038/s41598-019-46454-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209. Zhou A, Lin K, Zhang S, Chen Y, Zhang N, Xue J, Wang Z, Aldape KD, Xie K, Woodgett JR, Huang S. Nuclear GSK3β promotes tumorigenesis by phosphorylating KDM1A and inducing its deubiquitylation by USP22. Nat Cell Biol. 2016;18(9):954–66. doi: 10.1038/ncb3396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210. Bacchi S, Palumbo P, Sponta A, Coppolino MF. Clinical pharmacology of non-steroidal anti-inflammatory drugs: a review. Antiinflamm Antiallergy Agents Med Chem. 2012;11(1):52–64. doi: 10.2174/187152312803476255. [DOI] [PubMed] [Google Scholar]
- 211. Zappavigna S, Cossu AM, Grimaldi A, Bocchetti M, Ferraro GA, Nicoletti GF, Filosa R, Caraglia M. Anti-inflammatory drugs as anticancer agents. Int J Mol Sci. 2020;21(7):2605. doi: 10.3390/ijms21072605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212. Landskron G, De la Fuente M, Thuwajit P, Thuwajit C, Hermoso MA. Chronic inflammation and cytokines in the tumor microenvironment. J Immunol Res. 2014;2014:149185. doi: 10.1155/2014/149185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213. Bennett A, Del Tacca M. Proceedings: prostaglandins in human colonic carcinoma. Gut. 1975;16(5):409. [PubMed] [Google Scholar]
- 214. Leidgens V, Seliger C, Jachnik B, Welz T, Leukel P, Vollmann-Zwerenz A, Bogdahn U, Kreutz M, Grauer OM, Hau P. Ibuprofen and diclofenac restrict migration and proliferation of human glioma cells by distinct molecular mechanisms. PLoS ONE. 2015;10(10):e0140613. doi: 10.1371/journal.pone.0140613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215. Seliger C, Leukel P, Moeckel S, Jachnik B, Lottaz C, Kreutz M, Brawanski A, Proescholdt M, Bogdahn U, Bosserhoff A-K, Vollmann-Zwerenz A, Hau P. Lactate-modulated induction of THBS-1 activates transforming growth factor (TGF)-beta2 and migration of glioma cells in vitro. PLoS ONE. 2013;8(11):e78935. doi: 10.1371/journal.pone.0078935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216. Lee JK, Nam DH, Lee J. Repurposing antipsychotics as glioblastoma therapeutics: potentials and challenges. Oncol Lett. 2016;11(2):1281–6. doi: 10.3892/ol.2016.4074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217. Javaid JI. Clinical pharmacokinetics of antipsychotics. J Clin Pharmacol. 1994;34(4):286–95. doi: 10.1002/j.1552-4604.1994.tb01995.x. [DOI] [PubMed] [Google Scholar]
- 218. Chou FH, Tsai KY, Su CY, Lee CC. The incidence and relative risk factors for developing cancer among patients with schizophrenia: a nine-year follow-up study. Schizophr Res. 2011;129(2–3):97–103. doi: 10.1016/j.schres.2011.02.018. [DOI] [PubMed] [Google Scholar]
- 219. Oliva CR, Zhang W, Langford C, Suto MJ, Griguer CE. Repositioning chlorpromazine for treating chemoresistant glioma through the inhibition of cytochrome c oxidase bearing the COX4-1 regulatory subunit. Oncotarget. 2017;8(23):37568–83. doi: 10.18632/oncotarget.17247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220. Chu CW, Ko HJ, Chou CH, Cheng TS, Cheng HW, Liang YH, Lai YL, Lin CY, Wang C, Loh JK, Cheng JT, Chiou SJ, Su CL, Huang CF, Hong YR. Thioridazine enhances P62-mediated autophagy and apoptosis through Wnt/β-catenin signaling pathway in glioma cells. Int J Mol Sci. 2019;20(3):473. doi: 10.3390/ijms20030473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221. Cheng HW, Liang YH, Kuo YL, Chuu CP, Lin CY, Lee MH, Wu ATH, Yeh CT, Chen EIT, Whang-Peng J, Su CL, Huang CYF. Identification of thioridazine, an antipsychotic drug, as an antiglioblastoma and anticancer stem cell agent using public gene expression data. Cell Death Dis. 2015;6(5):e1753. doi: 10.1038/cddis.2015.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222. Johannessen T-C, Hasan-Olive MM, Zhu H, Denisova O, Grudic A, Latif MA, Saed H, Varughese JK, Røsland GV, Yang N, Sundstrøm T, Nordal A, Tronstad KJ, Wang J, Lund-Johansen M, Simonsen A, Janji B, Westermarck J, Bjerkvig R, Prestegarden L. Thioridazine inhibits autophagy and sensitizes glioblastoma cells to temozolomide. Int J Cancer. 2019;144(7):1735–45. doi: 10.1002/ijc.31912. [DOI] [PubMed] [Google Scholar]
- 223. Bhat K, Saki M, Vlashi E, Cheng F, Duhachek-Muggy S, Alli C, Yu G, Medina P, He L, Damoiseaux R, Pellegrini M, Zemke NR, Nghiemphu PL, Cloughesy TF, Liau LM, Kornblum HI, Pajonk F. The dopamine receptor antagonist trifluoperazine prevents phenotype conversion and improves survival in mouse models of glioblastoma. Proc Natl Acad Sci U S A. 2020;117(20):11085–96. doi: 10.1073/pnas.1920154117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224. Alboni S, van Dijk RM, Poggini S, Milior G, Perrotta M, Drenth T, Brunello N, Wolfer DP, Limatola C, Amrein I, Cirulli F, Maggi L, Branchi I. Fluoxetine effects on molecular, cellular and behavioral endophenotypes of depression are driven by the living environment. Mol Psychiatry. 2017;22(4):552–61. doi: 10.1038/mp.2015.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225. Liu K-H, Yang S-T, Lin Y-K, Lin J-W, Lee Y-H, Wang J-Y, Hu C-J, Lin E-Y, Chen S-M, Then C-K, Shen S-C. Fluoxetine, an antidepressant, suppresses glioblastoma by evoking AMPAR-mediated calcium-dependent apoptosis. Oncotarget. 2015;6(7):5088–101. doi: 10.18632/oncotarget.3243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226. Czarnecka AM, Czarnecki JS, Kukwa W, Cappello F, Scińska A, Kukwa A. Molecular oncology focus—is carcinogenesis a “mitochondriopathy”? J Biomed Sci. 2010;17(1):31. doi: 10.1186/1423-0127-17-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227. Deighton RF, Le Bihan T, Martin SF, Gerth AMJ, McCulloch M, Edgar JM, Kerr LE, Whittle IR, McCulloch J. Interactions among mitochondrial proteins altered in glioblastoma. J Neurooncol. 2014;118(2):247–56. doi: 10.1007/s11060-014-1430-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228. Bielecka-Wajdman AM, Ludyga T, Machnik G, Gołyszny M, Obuchowicz E. Tricyclic antidepressants modulate stressed mitochondria in glioblastoma multiforme cells. Cancer Control. 2018;25(1):1073274818798594. doi: 10.1177/1073274818798594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229. Troib A, Azab AN. Effects of psychotropic drugs on nuclear factor kappa B. Eur Rev Med Pharmacol Sci. 2015;19(7):1198–208. [PubMed] [Google Scholar]
- 230. Haas R, Smith J, Rocher-Ros V, Nadkarni S, Montero-Melendez T, D’Acquisto F, Bland EJ, Bombardieri M, Pitzalis C, Perretti M, Marelli-Berg FM, Mauro C. Lactate regulates metabolic and pro-inflammatory circuits in control of T cell migration and effector functions. PLoS Biol. 2015;13(7):e1002202. doi: 10.1371/journal.pbio.1002202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231. Hayashi K, Michiue H, Yamada H, Takata K, Nakayama H, Wei FY, Fujimura A, Tazawa H, Asai A, Ogo N, Miyachi H, Nishiki T, Tomizawa K, Takei K, Matsui H. Fluvoxamine, an anti-depressant, inhibits human glioblastoma invasion by disrupting actin polymerization. Sci Rep. 2016;6:23372. doi: 10.1038/srep23372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232. Bai R-Y, Staedtke V, Aprhys CM, Gallia GL, Riggins GJ. Antiparasitic mebendazole shows survival benefit in 2 preclinical models of glioblastoma multiforme. Neuro Oncology. 2011;13(9):974–82. doi: 10.1093/neuonc/nor077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233. Pourgholami MH, Yan Cai Z, Lu Y, Wang L, Morris DL. Albendazole: a potent inhibitor of vascular endothelial growth factor and malignant ascites formation in OVCAR-3 tumor-bearing nude mice. Clin Cancer Res. 2006;12(6):1928–35. doi: 10.1158/1078-0432.Ccr-05-1181. [DOI] [PubMed] [Google Scholar]
- 234. Pourgholami MH, Cai ZY, Badar S, Wangoo K, Poruchynsky MS, Morris DL. Potent inhibition of tumoral hypoxia-inducible factor 1alpha by albendazole. BMC Cancer. 2010;10:143. doi: 10.1186/1471-2407-10-143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235. Doudican N, Rodriguez A, Osman I, Orlow SJ. Mebendazole induces apoptosis via Bcl-2 inactivation in chemoresistant melanoma cells. Mol Cancer Res. 2008;6(8):1308–15. doi: 10.1158/1541-7786.MCR-07-2159. [DOI] [PubMed] [Google Scholar]
- 236. Patil VM, Bhelekar A, Menon N, Bhattacharjee A, Simha V, Abhinav R, Abhyankar A, Sridhar E, Mahajan A, Puranik AD, Purandare N, Janu A, Ahuja A, Krishnatry R, Gupta T, Jalali R. Reverse swing-M, phase 1 study of repurposing mebendazole in recurrent high-grade glioma. Cancer Med. 2020;9(13):4676–85. doi: 10.1002/cam4.3094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin. 2020;70(1):7–30. doi: 10.3322/caac.21590. [DOI] [PubMed] [Google Scholar]
- 238. Louis DN, Perry A, Reifenberger G, von Deimling A, Figarella-Branger D, Cavenee WK, Ohgaki H, Wiestler OD, Kleihues P, Ellison DW. The 2016 World Health Organization classification of tumors of the central nervous system: a summary. Acta Neuropathol. 2016; 131(6):803–20. doi: 10.1007/s00401-016-1545-1. [DOI] [PubMed] [Google Scholar]
- 239. Bahmad HF, Chamaa F, Assi S, Chalhoub RM, Abou-Antoun T, Abou-Kheir W. Cancer stem cells in neuroblastoma: expanding the therapeutic frontier. Front Mol Neurosci. 2019;12:131. doi: 10.3389/fnmol.2019.00131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240. Bahmad HF, Poppiti RJ. Medulloblastoma cancer stem cells: molecular signatures and therapeutic targets. J Clin Pathol. 2020;73(5):243–9. doi: 10.1136/jclinpath-2019-206246. [DOI] [PubMed] [Google Scholar]
- 241. Vangipuram SD, Buck SA, Lyman WD. Wnt pathway activity confers chemoresistance to cancer stem-like cells in a neuroblastoma cell line. Tumour Biol. 2012;33(6):2173–83. doi: 10.1007/s13277-012-0478-0. [DOI] [PubMed] [Google Scholar]
- 242. Kim Y, Kim KH, Lee J, Lee YA, Kim M, Lee SJ, Park K, Yang H, Jin J, Joo KM, Lee J, Nam DH. Wnt activation is implicated in glioblastoma radioresistance. Lab Invest. 2012;92(3):466–73. doi: 10.1038/labinvest.2011.161. [DOI] [PubMed] [Google Scholar]
- 243. Suebsoonthron J, Jaroonwitchawan T, Yamabhai M, Noisa P. Inhibition of WNT signaling reduces differentiation and induces sensitivity to doxorubicin in human malignant neuroblastoma SH-SY5Y cells. Anticancer Drugs. 2017;28(5):469–79. doi: 10.1097/cad.0000000000000478. [DOI] [PubMed] [Google Scholar]
- 244. Stambolic V, Woodgett JR. Mitogen inactivation of glycogen synthase kinase-3 beta in intact cells via serine 9 phosphorylation. Biochem J. 1994;303(Pt. 3):701–4. doi: 10.1042/bj3030701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245. Duffy DJ, Krstic A, Schwarzl T, Higgins DG, Kolch W. GSK3 inhibitors regulate MYCN mRNA levels and reduce neuroblastoma cell viability through multiple mechanisms, including p53 and Wnt signaling. Mol Cancer Ther. 2014;13(2):454–67. doi: 10.1158/1535-7163.Mct-13-0560-t. [DOI] [PubMed] [Google Scholar]
- 246. Glumac PM, LeBeau AM. The role of CD133 in cancer: a concise review. Clin Transl Med. 2018;7(1):18. doi: 10.1186/s40169-018-0198-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247. Beier D, Hau P, Proescholdt M, Lohmeier A, Wischhusen J, Oefner PJ, Aigner L, Brawanski A, Bogdahn U, Beier CP. CD133(+) and CD133(-) glioblastoma-derived cancer stem cells show differential growth characteristics and molecular profiles. Cancer Res. 2007;67(9):4010–5. doi: 10.1158/0008-5472.Can-06-4180. [DOI] [PubMed] [Google Scholar]
- 248. Vangipuram SD, Wang ZJ, Lyman WD. Resistance of stem-like cells from neuroblastoma cell lines to commonly used chemotherapeutic agents. Pediatr Blood Cancer. 2010;54(3):361–8. doi: 10.1002/pbc.22351. [DOI] [PubMed] [Google Scholar]
- 249. Cheaito KA, Bahmad HF, Hadadeh O, Saleh E, Dagher C, Hammoud MS, Shahait M, Mrad ZA, Nassif S, Tawil A, Bulbul M, Khauli R, Wazzan W, Nasr R, Shamseddine A, Temraz S, El-Sabban ME, El-Hajj A, Mukherji D, Abou-Kheir W. EMT markers in locally-advanced prostate cancer: predicting recurrence? Front Oncol. 2019;9:131. doi: 10.3389/fonc.2019.00131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250. Phi LTH, Sari IN, Yang Y-G, Lee S-H, Jun N, Kim KS, Lee YK, Kwon HY. Cancer Stem Cells (CSCs) in drug resistance and their therapeutic implications in cancer treatment. Stem Cells Int. 2018;2018:5416923. doi: 10.1155/2018/5416923. [DOI] [PMC free article] [PubMed] [Google Scholar]