Abstract
Background
MicroRNAs (miRNAs) are short non-coding RNAs that play important roles in almost all biological pathways. They regulate post-transcriptional gene expression by binding to the 3’untranslated region (3’UTR) of messenger RNAs (mRNAs). MitomiRs are miRNAs of nuclear or mitochondrial origin that are localized in mitochondria and have a crucial role in regulation of mitochondrial function and metabolism. In eukaryotes, mitochondria are the major sites of oxidative metabolism of sugars, lipids, amino acids, and other bio-macromolecules. They are also the main sites of adenosine triphosphate (ATP) production.
Conclusions
In the review, we discuss the role of mitomiRs in mitochondria and introduce currently well studied mitomiRs, their target genes and functions. We also discuss their role in cancer initiation and progression through the regulation of mRNA expression in mitochondria. MitomiRs directly target key molecules such as transporters or enzymes in cell metabolism and regulate several oncogenic signaling pathways. They also play an important role in the Warburg effect, which is vital for cancer cells to maintain their proliferative potential. In addition, we discuss how they indirectly upregulate hexokinase 2 (HK2), an enzyme involved in glucose phosphorylation, and thus may affect energy metabolism in breast cancer cells. In tumor tissues such as breast cancer and head and neck tumors, the expression of one of the mitomiRs (miR-210) correlates with hypoxia gene signatures, suggesting a direct link between mitomiR expression and hypoxia in cancer. The miR-17/92 cluster has been shown to act as a key factor in metabolic reprogramming of tumors by regulating glycolytic and mitochondrial metabolism. This cluster is deregulated in B-cell lymphomas, B-cell chronic lymphocytic leukemia, acute myeloid leukemia, and T-cell lymphomas, and is particularly overexpressed in several other cancers. Based on the current knowledge, we can conclude that there is a large number of miRNAs present in mitochondria, termed mitomiR, and that they are important regulators of mitochondrial function. Therefore, mitomiRs are important players in the metabolism of cancer cells, which need to be further investigated in order to develop a potential new therapies for cancer.
Key words: microRNAs, mitomiR, mitochondria, cancer, cancer cell metabolism
Introduction
MicroRNAs (miRNAs) are short non-coding RNAs (ncRNAs) of ~18-25 nucleotides that are present in all eukaryotic cells and play important roles in almost all biological signaling pathways.1, 2, 3, 4 Since the discovery of the first miRNA (lin-4) in C. elegans5, approximately 2000 miRNAs have been annotated in the human genome.6 Data from genomic studies show that most miRNAs are highly conserved, making them very interesting targets for studying various disease states.7 They regulate post-transcriptional gene expression by binding to the 3’UTR of messenger RNAs.8, 9, 10, 11, 12, 13, 14 A single miRNA can regulate many mRNA targets, and conversely, a single mRNA target can be regulated by many miRNAs.15, 16, 17 Therefore, by regulating these fundamental target genes, miRNAs have been implicated in signaling pathways to modulate a large set of important biological processes such as cell proliferation12, metastasis18, apoptosis19, senescence12, differentiation20, autophagy21, and immune response22. Moreover, miRNAs have been found to be dysregulated in many pathological conditions, such as neurodegenerative diseases23, cardiovascular diseases24, and cancer.25, 26, 27, 28
More recently, miRNAs have been found to be specifically present in mitochondria. These mitochondrial miRNAs were named “mitomiR”.7, 29, 30, 31, 32 Most of them have a nuclear origin, but some mitomiRs originate from mRNA molecules derived from the mitochondrial genome. The association of mitomiRs with mitochondria is species- and cell type-specific.7, 33 They have been found in mitochondria in various tissues and cells and are thought to have different thermodynamic properties than miRNAs.7, 34 Mitochondria have a discrete and unique pool of mitomiRs, which has been demonstrated with various experiments.29
For the first time, in 2011, Barrey and co-workers demonstrated the presence of pre-miRNAs (precursor-miRNAs) in mitochondria and postulated that some pre-miRNA sequences could be processed into mature miRNAs that could immediately become active on mitochondrial transcripts or exported to the cytosol to disrupt genomic mRNA.35 Barrey’s group screened for 742 miRNAs using qRT-PCR and showed that 243 miRNAs had significant expression in mitochondrial RNA samples isolated from human myotubes by in situ hybridization. This study was the first to provide evidence that pre-miRNAs can be localized in mitochondria. Subsequently, a number of studies have identified “signatures” of miRNAs localized to mitochondria through various experimental approaches. Mercer et al.15 examined the human mitochondrial transcriptome and demonstrated that 3 miRNAs (miR-146a, miR-103, and miR-16) have quite high expression in the intermembrane region compared to the matrix. Latronico and Condorelli36 found 15 nuclear-encoded miRNAs in mitochondria isolated from rat liver, 20 miRNAs from mouse liver mitochondria, and 13 miRNAs from HeLa cells (isolated from human cervical cancer) by microarray. Some other groups identified novel mitomiRs from HEK293 cells (isolated from human embryonic kidneys)37, 143B cells (isolated from human bone marrow)38, mouse heart39 and HeLa cells.37, 40
MitomiRs have been shown to be important regulators of mitochondrial function.35, 38, 41 The regulation of mitochondria by mitomiRs influences the development of many diseases caused by mitochondrial dysfunction, which is responsible for the pathophysiology of numerous diseases, such as cardiovascular and neurodegenerative diseases, diabetes, obesity, and cancer.42
In the first part of this review article, we describe the biosynthesis of mitomiRs and the transport mechanisms from mitomiRs to mitochondria. The next part is dedicated to the role of these small molecules in mitochondria and the presentation of some important mitomiRs, their target genes and functions. In the last part of the review, we discuss the functions of mitomiRs in cancer cell metabolism and introduced mitomiRs in the context of cancer.
Biosynthesis of miRNA/mitomiRs
Most miRNAs/mitomiRs are produced via the canonical biosynthetic pathway, which involves transcription by RNA polymerase II (Pol II) to produce a primary transcript (pri-miRNA/mitomiR). The primary transcript is first cleaved in the nucleus by the nuclear heterodimer Drosha/DGCR8 (DiGeorge syndrome chromosomal region 8), which cleaves the pri-miRNA/mitomiR and produces a pre-miRNA/mitomiR with a hairpin structure that is much more stable than the pri-miRNA/ mitomiR due to its characteristic hairpin loop structure.43 Exportin 5 (EXP5) and GTP-binding nuclear protein (RANGTP) then form a transport machinery to export the pre-miRNA from the nucleus to the cytoplasm. After export to the cytoplasm, the pre-miRNA/mitomiR is further cleaved by the enzyme Dicer to form a double-stranded RNA (dsRNA) duplex (Figure 1). Only a single strand of the dsRNA duplex forms the mature miRNA/mitomiR and is incorporated into the RNA-induced silencing complex (RISC), which directs the binding of Argonaute (AGO) proteins in the RISC to the 3’UTR of the target mRNA to either repress protein translation or promote mRNA degradation.43, 44, 45 After incorporation into RISC, mature miRNA/mitomiRs are transported into mitochondria, back to nucleus by importin 8 (IPO-8) or extracellular environment (Figure 1).46, 47
Figure 1.
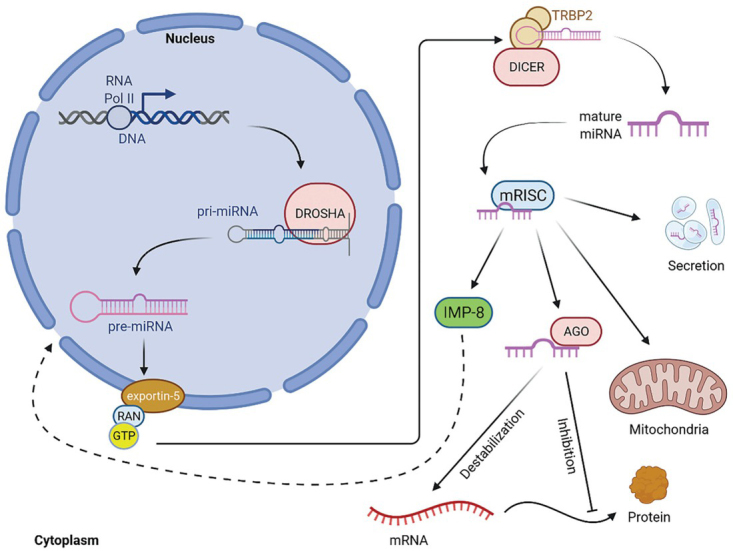
Canonical biosynthesis of miRNAs/mitomiRs (adopted from 29, 43, 45). Mature miRNA can be transported into any part of the cell; but miRNA/mitomiR regulation is possible only after incorporation into RISC. (AGO2 = argonaute 2; DGCR8 = DiGeorge critical region 8; EXP5 = exportin 5; GTP = guanosine triphosphate; IMP8 = importin 8; mRISC = RNA induced silencing complex loaded with mature miRNA; POLII = DNA polymerase II; RANGTP = binding nuclear protein RAN; RISC = RNA-induced silencing complex; TRBP2 = RISC-loading complex subunit TRBP2).
In addition to the canonical miRNAs/mitomiRs biosynthesis pathway, there are also non-canonical, Drosha/DGCR8-independent and Dicer-independent biosynthesis pathways. Prominent classes of Drosha/DGCR8-independent miRNAs/ mitomiRs are the “mirtrons” derived from introns that, once spliced, function as pre-miRNAs and thus do not require cleavage by Drosha/DGCR8 and can be immediately exported to the cytoplasm for processing by Dicer. MiRNAs/mitomiRs can also be processed from hairpins generated directly by Pol II at specific transcription start sites. These pre-miRNAs are capped and exported via the exportin 1 (EXP1) pathway. The Dicer-independent miRNAs/mitomiRs biosynthesis pathway involves the unusually short hairpin of miR-451, which is directly cleaved by argounaute 2 (AGO2).45
MitomiRs transport to mitochondria
The discovery of mitomiRs raised the question of elucidating the underlying molecular mechanisms of their transport into mitochondria. Due to their size and charged nature, mitomiRs are unlikely to cross membranes under their own power. The molecular mechanisms of mitomiR transport into mitochondria may vary between species and are not well understood.29
Some proposals have been published on AGO2 as a potential mitomiR import protein.7, 29, 48 Due to its RNA-binding ability and dual localization in the cytosol and mitochondria, AGO2 might be involved in the trafficking of mitomiRs.7 Shepherd et al.49 showed that the exoribonuclease polyribonucleotide nucleotidyltransferase (PNPT1/ PNPase) has a major role in the import of mitomiRs. Therefore, PNPase could be part of an alternative, AGO2-independent, uptake pathway of mitochondrial miRNA. Furthermore, a possible mechanism could involve the voltage-dependent anion-selective channel protein (VDAC).34 Several studies have suggested that the instability of RISC in the cytoplasm promotes miRNA translocation to mitochondria, but the molecular components that facilitate this translocation process are not fully understood. Furthermore, the concept that mammalian mitochondria can import cytosolic ncRNAs may facilitate research in another exciting area, long ncRNAs. Clearly, these translocation mechanisms and the identification of pathway components for mitochondrial targeting require further studies.7
Roles of mitomiRs in mitochondria
Mitochondria are semi-autonomous cell organelles with their own DNA (mtDNA) encoding 22 tRNAs, 2 rRNAs, and 13 polypeptides. These polypeptides and those encoded by nuclear genes, form 4 protein complexes of the electron transport chain (ETC). Mitochondria are constantly dividing and fusing, and the balance between mitochondrial fission and fusion influences mitochondrial morphology, whose dynamics and turnover are critical for cellular homeostasis and differentiation.50 Several proteins are involved in the regulation of mitochondrial dynamics. Deregulation of mitochondrial dynamics is not only associated with deregulation of mitochondrial function, but is also closely related to several biological processes such as proliferation, cell death, apoptosis and production of reactive oxygen species (ROS), since mitochondria are the major sites of oxidative metabolism of sugars, lipids, amino acids and ATP production.1, 51, 52, 53
It’s also worth noting that the mitochondrial matrix has its own set of environmental variables. Because of its thioester bond, acetyl-coenzyme A (acetyl-CoA) is a very abundant metabolite in mitochondria and functions as a powerful acetylation reagent. Protein lysine acetylation and succinylation are caused by acetyl-CoA and mitochondrial matrix pH concentrations. Non-enzymatic acetylation occurs often in mitochondria.54 The most of mitochondrial proteins have acetyl groups, which is consistent with this hypothesis. Non-enzymatic acetylation of RNA molecules, including miRNAs, is a logical possibility for mitochondrial modification. An acetyl group covalently attached to a miRNA might change its mRNA recognition behavior. If it happens at the 2 OH group of ribose needed for the cleavage process, it could inhibit spontaneous bond cleavage and therefore increase the half-life of mRNA. Furthermore, post-transcriptional alterations can result in structural changes55 as well as changed interactions with other RNA molecules or proteins.56
As stated, mitomiRs are regulators of mitochondrial function, as shown in the following examples. In silico analysis identified miR-378, miR-24, and miR-23b in liver mitochondria (Table 1) and these mitomiRs have been shown to regulate systemic energy homeostasis, oxidative capacity, ROS, and mitochondrial lipid metabolism.35, 57, 58, 59, 60, 61, 62 Several reports have indicated that miRNAs such as miR-1291, miR-138, miR-150, miR-199a, and miR-532-5p can alter the expression of some important glycolytic enzymes (Table 1).4, 63, 64, 65, 66, 67, 68, 69, 70 miR-29a, miR-29b and miR-124 (Table 1) regulate the expression of monocarboxylate transporter 1 (SLC16A1) in pancreatic beta cells.71 miR-33a/b has been shown to regulate lipid metabolism by targeting the cholesterol transporter ATP-binding cassette transporter (ABCA1).72 miR-143 and miR-24 have also been shown to regulate mitochondrial lipid metabolism (Table 1).73, 74 On the other hand, miR-204 accelerates fatty acid oxidation by inhibiting acetyl-coenzyme A carboxylase (ACC).75 Ahmad et al. (2011) showed that miR-200 is associated with the regulation of phosphoglucose isomerase (PGI), which is an important factor in glycolysis and glucogenesis. Overexpression of miR-338 leads to downregulation of the protein level of cytochrome c oxidase IV and reduces mitochondrial oxygen consumption and ATP production.77, 78 Similarly, overexpression of miR-181c decreases mt-COX1 protein and causes remodeling of the complex IV (in vitro)48 and a dysfunctional complex IV (in vivo)79, along with increased production of ROS. It has also been reported that miR-210 modulates the function of the complex IV by targeting the nuclear-encoded mRNA, COX10.80, 81 It has also been reported that miR-15b, miR-16, miR-195 and miR-338 (Table 1) regulate ATP production by targeting several nuclear genes that play important roles in ETC.77, 82, 83 miR-101-3p regulates the expression of ATP synthase subunit beta (ATP5B) in ETC (Table 1).84 In addition, miR-210-5p reduces the expression of iron-sulfur cluster assembly enzyme (ISCU) under hypoxic conditions, which affects the proteins containing iron-sulfur clusters (Fe-S).85 It has also been reported that miR-29a-3p86 is involved in ß-oxidation of lipids (Table 1) and that miR-19b negatively regulates mitochondrial fusion by downregulating mitofusin 1 (MFN1).87
Table 1.
Summary of microRNAs and their roles in mitochondria
| miR | miR accession number | Target genes | Gene accession number | Function | Functional pathway | Location | Species | References |
|---|---|---|---|---|---|---|---|---|
| miR-378 | MI0000795 | Crat | ENSMUSG00000026853 | Downregulation | Mitochondrial oxidative metabolism | Mitochondria in liver cells | Mouse | Carrer et al., 201259 |
| miR-24 | MI0000080 | H2ax | ENSMUSG00000049932 | Downregulation | Insulin pathway signaling | Mitochondria liver cells in | Human | Jeong et al., 201761 |
| miR-23b | MI0000439 | GLS | ENSG00000115419 | Downregulation | Glutamine metabolism | LMitochondria liver cells in | Human | Gao et al., 200960 |
| SLC2A1 | ENSG00000117394 | Downregulation | ||||||
| miR-1291 | MI0006353 | CPT1C ESRRA | ENSG00000169169 ENSG00000173153 | Downregulation Downregulation | Mitochondrial | Mitochondria in | Human | Yamasaki et al., 2013; Chen et al., |
| ASS1 | ENSG00000130707 | Downregulation | metabolism | renal cells | 2020, Tu et al., 202063, 64, 65 | |||
| GLUT1 | ENSG00000117394 | Downregulation | ||||||
| miR-138 | MI0000455 | PDK1 | ENSG00000152256 | Downregulation | Glucose metabolism | Mitochondria in cardiac cells | Human | Zhu et al., 201766 |
| miR-150 | MI0000920 MI0000479 | Slc2a4 SLC2A1 | ENSRNOG00000017226 ENSG00000117394 | Downregulation Downregulation | Metabolism | Mitochondria in cardiac cells | Rat Human | Ju et al., 202067 Li et al., 201768 |
| Slc2a4 | ENSRNOG00000017226 | Upregulation | Mitochondria in | |||||
| miR-199a | MI0000941 | Hk2 | ENSRNOG00000006116 | Upregulation | Expression of | muscle cells | Rat | Esteves et al., 2018, Yan et al., 2014, Guo |
| MI0000242 | HK2 | ENSG00000159399 | Upregulation | glucose transporters | Mitochondria in liver cells | Human | et al., 20154, 69, 70 | |
| miR-532-5p | MI0006154 | SlcHk22 a4 | ENSRNOGENSRNOG00000017226 00000006116 | Upregulation Upregulation | Expression glucose transporters of | Mitochondria muscle cells in | Rat | Esteves et al., 201870 |
| Mitochondrial | Mitochondria in | |||||||
| miR-29a | MI0000576 | Slc16a1 | ENSMUSG00000032902 | Downregulation | oxidative metabolism | pancreatic betacells | Mouse | Pullen et al., 201171 |
| miR-29b | MI0000143 | Slc16a1 | ENSMUSG00000032902 | Downregulation | Mitochondrial oxidative metabolism | Mitochondria in pancreatic betacells | Mouse | Pullen et al., 201171 |
| Mitochondrial | Mitochondria in | |||||||
| miR-124 | MI0000716 | Slc16a1 | ENSMUSG00000032902 | Downregulation | oxidative | pancreatic beta- | Mouse | Pullen et al., 201171 |
| metabolism | cells | |||||||
| CROT | ENSANAG00000028065 | Downregulation | ||||||
| CPT1A | ENSANAG00000017356 | Downregulation | ||||||
| HADHB | ENSANAG00000027802 | Downregulation | ||||||
| PRKAA1 | ENSANAG00000032687 | Downregulation | ||||||
| miR-33a/b | a-MI0002684, b-MI0007603 | ABCA1 | ENSANAG00000033387 | Downregulation | Lipid metabolism | Mitochondria in liver cells | Monkey | Rayner et al., 201172 |
| SREBF1 | ENSANAG00000021477 | Upregulation | ||||||
| FASN | ENSANAG00000032055 | Upregulation | ||||||
| ACLY | ENSANAG00000036009 | Upregulation | ||||||
| ACACA | ENSANAG00000035253 | Upregulation | ||||||
| miR-143 | MI0000916 | Map2k5 | ENSRNOG00000007926 | Downregulation | Adipogenesis | Mitochondria in adipose cells | Rat | Chen et al., 201473 |
| MI0000459 | APOL6 | ENSG00000221963 | Downregulation | Adpiogenesis | Mitochondria in adipose cells | Human | Ye et al., 201374 | |
| miR-204 | MI0000284 | ACACB | ENSG00000076555 | Downregulation | Lipid metabolism | Mitochondria adipose cells in | Human | Civelek et al., 201375 |
| miR-200 | MI0000737 | ZEBZEB1 2 | ENSGENSG00000148516 00000169554 | Upregulation Upregulation | Lipid metabolism | Mitochondria breast cells in | Human | Ahmad et al., 201176 |
| miR-338 | MI0000618 | COXIV | ENSRNOG00000007827 | Downregulation | Mitochondria oxidative metabolism | Mitochondria in neural cells | Rat | Aschrafi et al., 200877 |
| miR-181c | MI0000924 | COX1 | ENSRNOG00000034234 | Downregulation | Mitochondria oxidative metabolism | Mitochondria in cardiac cells | Rat | Das et al., 201288 |
| miR-210 | MI0000268 | ISCU | ENSG00000136003 | Downregulation | Mitochondria oxidative metabolism | Mitochondria in placenta cells | Human | Colleoni et al., 2013; Qiao et al., 201381, 85 |
| miR-15b | MI0000843 | Arl2 | ENSRNOG00000021010 | Downregulation | ATP production | Mitochondria in cardiac cells | Rat | Nishi et al., 201082 |
| Bcl2 | ENSRNOG00000002791 | Downregulation | ||||||
| miR-16 | MI0000844 | Bcl2 | ENSRNOG00000002791 | Downregulation | ATP production | Mitochondria in cardiac cells | Rat | Nishi et al., 201082 |
| Arl2 | ENSRNOG00000021010 | Downregulation | ||||||
| miR-195 | MI0000939 | Arl2 | ENSRNOG00000021010 | Downregulation | ATP production | Mitochondria in cardiac cells | Rat | Nishi et al., 201082 |
| miR-29a-3p | MI0000576 | Foxa2 | ENSMUSG00000037025 | Upregulation | Lipid metabolism | Mitochondria liver cells in | Mouse | Kurtz et al., 201486 |
| miR-19b | MI0000074 | MFN1 | ENSG00000171109 | Downregulation | Apoptosis | Mitochondria in bone cells | Human | Li et al., 201487 |
| miR-101-3p | MI0000103 | ATP5B | ENSG00000110955 | Silencing | Mitochondria metabolism | Mitochondria heLa cells in | Human | Zheng et al., 201184 |
The microRNAs listed in Table 1 significantly affect mitochondrial regulation and function, which is why they are classified in the group of mitomiRs, which are crucial regulatory molecules of mitochondrial function and regulation of metabolism. In the figures (Figure 2 and Figure 3), we have shown how these mitomiRs are linked to their target genes in primates (Figure 2) and rodents (Figure 3).
Figure 2.
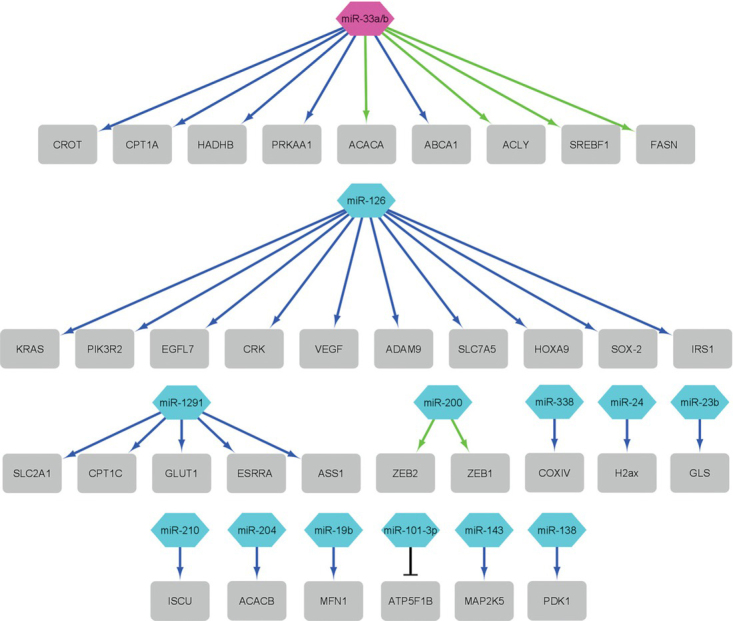
The network of the mitomiRs and their target genes (grey rectangle) in primates (data from Table 1). Blue arrows present downregulation, green arrows present upregulation and black T-line present silencing. Purple octagon shape presents monkey miRNA and cyan hexagon presents human miRNAs (figure constructed with Cytoscape Network Data Integration, Analysis, and Visualization in a Box V3.8.2).
Figure 3.
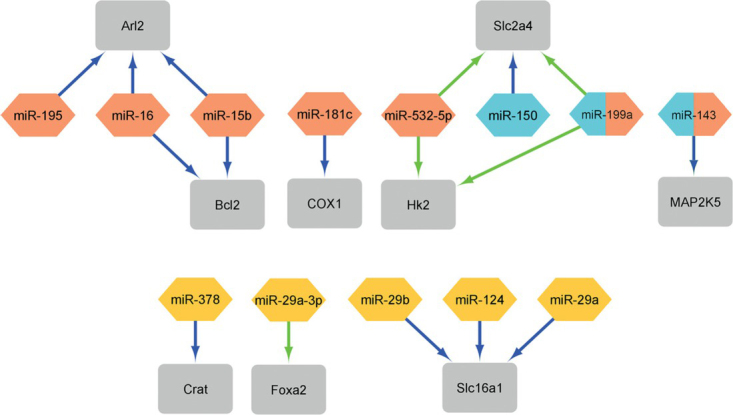
The network of the mitomiRs and their target genes (grey rectangle) in rodents (data from Table 1). Blue arrows present downregulation and green arrows present upregulation. Orange diamond shape presents rat miRNAs, yellow rectangle presents mouse miRNAs and cyan hexaon presents human miRNAs. miR-199a and miR-143 show that this two miRNAs regulate (figure constructed with Cytoscape Network Data Integration, Analysis, and Visualization in a Box V3.8.2).
In primates, there is no regulation of the same genes by different mitomiRs from Table 1 (Figure 2). Moreover, most mitomiRs target one gene and only a few mitomiRs target a larger number of genes and in most cases mitomiRs down-regulate genes.
In contrast to primates, in rodents, some genes are regulated by different mitomiRs (Figure 3). The mitomiRs miR-15b and miR-16 both regulate the Arl2 gene82, which is a nucleotide-binding gene, and the Bcl2 gene, which regulates apoptosis. In addition, the mitomiRs miR-199a69, 70 and miR-532-5p70 both regulate the Hk2 gene, which has an important function in regulating glucose metabolism, and the Slc2a4 gene, which is a glucose transmembrane transporter. It can be concluded that there is a greater overlap of mitomiRs in rodents than in primates. In most cases, mitomiRs downregulate genes.
From the figures (Figure 2 and Figure 3), we can summarize that some mitomiRs and their target genes are related in primates and rodents. MitomiR miR-199a69, 70 regulates the same gene in both primates and rodents (Figure 3), the gene Hk2, which has an important function in regulating glucose metabolism. MiR-14373, 74 regulates the same gene MAP2K5 (Figure 3), which has an important function in signal cascade involved in growth factor stimulated cell proliferation and muscle cell differentiation.
MitomiRs in cancer
Traditional cancer traits include ten biological capabilities gained during the multistage development of human tumors.89 These ten traditional cancer traits include resistance to cell death, induction of angiogenesis, maintenance of proliferative signaling, evasion of growth suppressors, activation of invasion and metastasis, facilitation of replicative immortality, altered metabolism, evasion of destruction by the immune system, tumorpromoting inflammation, and genome instability (Figure 4).89, 90
Figure 4.
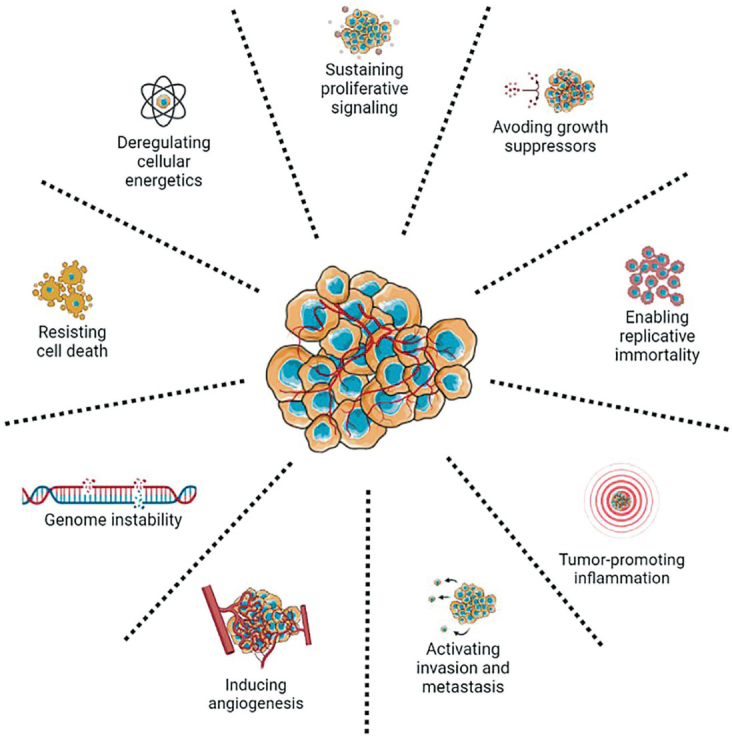
Traditional cancer traits.89
An important feature of cancer is the presence of the Warburg effect. Under aerobic conditions, normal cells generate ATP primarily in the mitochondrial oxidative phosphorylation process (OXPHOS), which utilizes the products of glycolysis and the Krebs cycle. Under anaerobic conditions, relatively little pyruvate, the end product of glycolysis, is added to the Krebs cycle and is instead converted to lactate. However, this metabolic conversion of glucose appears to be energetically detrimental. In tumor cells, ATP deficiency can be compensated to some extent by upregulation of glycolysis.91 Interestingly, it has been observed that many cancer cells prefer glycolysis over OXPHOS even in the presence of an adequate amount of oxygen. This abnormal energy metabolism is known as the Warburg effect. Reduced OXPHOS and enhanced aerobic glycolysis are the main manifestations of reprogramming of glucose metabolism in tumor cells.1, 92 Albeit the specific causes and utilitarian outcomes of this metabolic switch are as yet unclear, there is a developing agreement that the impact of Warburg effect is certifiably not an inconsequential result of carcinogenesis, yet is imperative for cancer cells to keep up with their proliferative potential and is driven by a few elements.92, 93, 94
It has been confirmed that abnormal expression of mitomiRs in mitochondria is related to the occurrence of cancer features.95 Moreover, mitomiRs play an essential role in the control of cancer cell metabolism by regulating mRNA expression. They regulate several oncogenic signaling pathways and target key transporters or enzymes in cellular metabolism. In addition, they may have a function as tumor suppressors that inhibit tumor cell proliferation or as oncogenes that induce tumorigenesis.96, 97, 98 MitomiRs can be isolated from any tissue or body fluid of any organism to study the level of expression in the organism in a diseased state, and thus can function as novel prognostic and predictive biomarkers.99
The first evidence of miRNA involvement in human cancers was provided in a study of chronic lymphocytic leukemia (CLL).100 MiR-15a and miR-16-1 localized to 13q14 were reported to be frequently deleted and/or reduced in patients with B-cell chronic lymphocytic leukemia. This finding provided the first evidence that miRNAs may be involved in the pathogenesis of human cancers, as deletion of chromosome 13q14 resulted in the loss of these two miRNAs. MiR-15a induces apoptosis by regulating mitochondrial function and affecting the activity of Bcl-2 and Mcl-1 in human (Table 2). In addition, miR-15a causes mitochondrial dysfunction, leading to the release of cytochrome c into the cytoplasm and depletion of mitochondrial membrane potential.101 MiR-15a and miR-16a have been shown to be ATP modulators correlated with cytochrome c oxidase subunit 4I2 (Cox4i2), subunit 6A2 (Cox6a2), NADH:ubiquinone oxidoreductase subunit B7 (Ndufb7), NADH:ubiquinone oxidoreductase core subunit V1 (Ndufv1) and NADH:ubiquinone oxidoreductase subunit S4 (Ndufs4) expression.102
Table 2.
Summary of mitomiRs with roles in cancer
| miR | miR accession number | Target genes | Gene accession number | Function | Functional pathway | Type of cancer | Species | References |
|---|---|---|---|---|---|---|---|---|
| HIF-1 | ENSG00000258777 | Upregulation | ||||||
| ISCU | ENSG00000136003 | Upregulation | ||||||
| miR-210 | MI0000286 | COX10 | ENSG00000006695 | Upregulation | Hypoxia | Breast cancer, neck and head cancer, lung cancer | Human | Qin et al., 2014; Gee et al., 2010; Puissegur et al., 2011109, 110, 111 |
| SDHD | ENSG00000204370 | Upregulation | ||||||
| NDUFA4 | ENSG00000189043 | Upregulation | ||||||
| miR-200a | MI0000342 | TFAM | ENSG00000108064 | Downregulation | Mitochondrial biogenesis, cancer metabolism | Breast cancer | Human | Yao et al., 2014112 |
| miR-155 | MI0000681 | HK2 | ENSG00000159399 | Upregulation | phosphorylation Glucose | Breast cancer | Human | Jiang Fang et et al.al., , 20122012; 104 |
| miR-124 | MI0000443 | PKM | ENSG00000067225 | Upregulation | metabolism Glucose | Colorectal cancer | Human | Sun et al., 2012105 |
| miR-137 | MI0000454 | PKM | ENSG00000067225 | Upregulation | metabolism Glucose | Colorectal cancer | Human | Sun et al., 2012105 |
| miR-340 | MI0000802 | PKM | ENSG00000067225 | Upregulation | metabolism Glucose | Colorectal cancer | Human | Sun et al., 2012105 |
| miR-326 | MI0000808 | PKM2 | ENSG00000067225 | Downregulation | metabolism Glucose | Glioblastoma | Human | Kefas et al., 2010106 |
| miR-181-5p | MIMAT0000256 | RASSF6 INPP5A |
ENSG00000169435 ENSG00000068383 |
Downregulation Downregulation |
Mitogen-activated protein kinase (MAPK) signaling pathway | Gastric cancer, cervical cancer | Human | Mi et al., 2017; Zhuang et al., 2017108, 113 |
| miR-92a-1 | MI0000093 | BCL2L11 | ENSG00000153094 | Downregulation | Apoptosis | Lymphoma | Human | Mogilyansky Rigoutsos, 2013 and 94 |
| PIK3R2 | ENSG00000105647 | Downregulation | Breast cancer cells | Zhu et al., 2011114 | ||||
| PLK2 | ENSG00000260410 | Downregulation | Acute leukaemia cells | Li et al., 2008115 | ||||
| EGFL7 | ENSG00000172889 | Downregulation | Oral squamous cells | Sasahira et al., 2012116 | ||||
| CRK | ENSG00000167193 | Downregulation | Lung cancer cells | Crawford 2008117 et al., | ||||
| ADAM9 | ENSG00000168615 | Downregulation | Melanoma cells cancer | Felli et al., 2013118 | ||||
| miR-126 | MI0000471 | HOXA9 | ENSG00000078399 | Downregulation | Inflammation, angiogenesis | Acute leukaemia cells | Human | Shen et al., 2008119 |
| IRS1 | ENSG00000169047 | Downregulation | Breast cancer cells | Zhang et al., 2008120 | ||||
| SOX-2 | ENSG00000242808 | Downregulation | Gastric cells cancer | Otsubo et al., 2011121 | ||||
| SLC7A5 | ENSG00000103257 | Downregulation | Lung cancer cells | Miko et al., 2011122 | ||||
| VEGFA | ENSG00000150630 | Downregulation | Oral squamous cells | Sasahira et al., 2012116 | ||||
| MMP7 | ENSG00000137673 | Downregulation | Melanoma cells cancer | Felli et al., 2013118 | ||||
| BCL-2 | ENSG00000171791 | Downregulation | ||||||
| Gao et al., 2010101 | ||||||||
| MCL-1 | ENSG00000143384 | Downregulation | ||||||
| COX4I2 | ENSG00000131055 | Downregulation | ||||||
| miR-15a | MI0000069 | COX6A2 | ENSG00000156885 | Downregulation | Apoptosis, ATP production | B-cell chronic lymphocytic leukemia | Human | Siengdee et al., |
| NDUFB7 | ENSG00000099795 | Downregulation | 2010102 | |||||
| NDUFV1 | ENSG00000167792 | Downregulation | ||||||
| NDUFS4 | ENSG00000164258 | Downregulation | ||||||
| COX4I2 | ENSG00000131055 | Downregulation | ||||||
| COX6A2 | ENSG00000156885 | Downregulation | ||||||
| miR-16a | MI0000070 | NDUFB7 | ENSG00000099795 | Downregulation | Apoptosis, ATP production | B-lymphocytic cell chronic leukemia | Human | Siengdee et al., 2010102 |
| NDUFV1 | ENSG00000167792 | Downregulation | ||||||
| NDUFS4 | ENSG00000164258 | Downregulation |
Glycolysis is the initial step in glucose catabolism, and occurs outside of the mitochondria in the cytoplasm. In the context of miRNAs affecting cell metabolism, miR-155 (Table 2) was found to indirectly upregulate hexokinase 2 (HK2), a glucose phosphorylation enzyme that might affect energy consumption in breast cancer cells. Mir-143 appears to be one of two potential pathways regulating miR-155-dependent HK2 regulation.103, 104 Alternative splicing of pyruvate kinase isoenzyme (PKM), whose splicing proteins are regulated by miR-124, miR-137, and miR-340, is another pathway regulating glucose metabolism (Table 2). This miRNA-dependent regulation of PKM is able to influence colorectal cancer growth and counteract the Warburg effect.105 In addition, pyruvate kinase (PK) is a direct target of the tumor suppressor miR-326, making it a potential glucose metabolism regulator.94, 106, 107
In hepatocellular carcinoma, reduced mRNA levels were detected in 11 of the 13 genes encoded in the mtDNA, including the genes encoding cytochrome B (mt-CYB) and cytochrome C oxidase II (mt-CO2).108 When miR-181a-5p expression was increased, the levels of mt-CYB and mt-CO2 were reduced in hepatocellular carcinoma cells, while mitochondrial membrane potential (MMP) maintained by electron transfer chain was reduced. In vivo experiments, which were done by Zhuang et al.108, have shown to have caused glucose metabolism to reprogram and stimulated tumor growth and early lung metastasis in patients with hepatocellular carcinoma.
Several studies reported that miR-126 has an important role in different human cancers (Table 2) such as breast, lung, gastric cancers, melanoma cancer and acute leukaemia. Tomasetti et al.83 reported that miR-126 affects mitochondrial energy metabolism, resulting in malignant mesothelioma tumor suppression. This mitomiR reduce mitochondrial respiration and promote glycolysis in H28 cells, associated with IRS1 modulate ATP-citrate lyase deregulation. This leads to an increase in ATP and citrate production which is linked with reducing Akt signaling and inhibiting cytosolic sequestration of Forkhead box O1 (FoxO1), which promote the expression of genes involved in gluconeogenesis and oxidative stress defense.83
Hypoxia has previously been related to altered mitomiR expression, with hypoxia-regulated mitomiRs being found to play a key role in cell survival in oxygen-depleted settings.123 MiR-210 is one of the mitomiRs that is continuously increased in normal and transformed cells during hypoxia, suggesting that miR-210 plays a role in cells’ adaptive response to hypoxia.109 MiR-210 expression corresponds with hypoxia gene signatures in tumor tissues such as breast and head and neck cancers, demonstrating a direct connection between miR-210 expression and hypoxia in cancer.110 MiR-210 has been researched extensively and has a number of functionally significant targets in cell cycle control, cell survival, differentiation, angiogenesis, and metabolism.123 Cell metabolism switches from mitochondrial OXPHOS to glycolysis under hypoxic environments. HIF-1, a hypoxia-inducible factor that upregulates the expression of most glycolytic enzymes as well as pyruvate dehydrogenase kinase while downregulating mitochondrial respiration, plays a key role in this action. Previous research has looked into how miR-210 regulates mitochondrial metabolism under hypoxia. MiR-210 target iron-sulfur cluster assembly proteins (ISCU1/2) and inhibit the activity of iron-sulfur proteins that govern mitochondrial metabolism, such as complex I and aconitase, resulting in lower OXPHOS.123 It acts directly on cytochrome c oxidase assembly factor heme A:farnesyltransferase (COX10), succinate dehydrogenase complex subunit D (SDHD), and NADH dehydrogenase (ubiquinone) 1 alpha subcomplex 4 (NDUFA4) in regulating mitochondrial activity.123 Another study found an abnormal mitochondrial phenotype in A549 lung cells overexpressing miR-210, and mRNA expression profile analysis connecting miR-210 to mitochondrial dysfunction.112 Interestingly, HIF is rapidly destroyed upon reoxygenation of hypoxic cells due to miR-210’s high stability, whereas miR-210 stays stable to maintain the glycolytic phenotype. Under normal conditions, this slows mitochondrial metabolism and may contribute to the Warburg effect in cancer cells. This result supports miR-210’s involvement in regulating mitochondrial metabolism and promoting cancer cells’ adaptability to hypoxic environments.
Another important mitomiR is miR-200, which has been identified as involved in tumor progression.124, 125 One of miR-200 targets, is transcription factor mitochondria (TFAM) which is one of the most important proteins regulating mitochondrial biogenesis. TFAM has been described as a functional target of miR-200 in breast cancer cells.113 Its transcription factor activity is required for mtDNA replication and transcription. In addition to its function in replication and transcription, the presence of TFAM is necessary for mtDNA maintenance.126 It has also been implicated as a primary architectural protein of the mitochondrial genome by packaging mtDNA. In addition, TFAM expression has been reported to be involved in tumor progression, cancer cell growth, and chemoresistance.127
Regarding the role of miRNAs in cancer and metabolism, the miR-17/92 cluster is one of the best characterized oncogenic miRNAs. This cluster is also known as oncomiR-1, and there is growing evidence of its oncogenic potential.93 It has been shown that miR-17/92 suppresses apoptosis and was originally found amplified in B-cell lymphomas, where ectopically overexpressed truncated versions lacking miR-92a-1 were shown to possess oncogenic properties.110 The MiR-17/92 cluster is deregulated in B-cell lymphomas, T-cell lymphomas, B-cell chronic lymphocytic leukemia, and acute myeloid leukemia. This cluster is particularly overexpressed in several other cancers, including osteosarcoma, neuroblastoma, cervical, pancreatic, breast, lung, colorectal, ovarian, kidney, and liver cancers.93, 105 Izreig et al.128 reported that this miRNA cluster is a key factor in metabolic reprogramming of tumors. If oncomiR-1 is absent in Myc+ tumor cells, there is a global decrease in glycolytic and mitochondrial metabolism. If increased oncomiR-1 expression is present, this is sufficient for increased nutrient utilization by tumor cells. Deletion of miR-17/92 promoted changes in gene expression in Myc+ lymphoma which results in global decrease in metabolic pathways including glycolysis, the Krebs cycle, components of the electron transfer chain, amino acid metabolism, the pentose phosphate pathway, serine biosynthesis and nucleotide biosynthesis.128
Conclusions
MiRNAs have been found in the mitochondria of many cell types, as shown by an increasing number of studies and they were named mitomiRs. In general, mitomiR populations differ in various tissues and under different pathological circumstances, implying that mitomiR populations are regulated by mechanisms that remain to be discovered. Based on the available information, we can deduce that there are a significant number of miR-NAs which are present in mitochondria.7, 29, 30, 31, 32, 33
In our review, we have shown that various mitomiRs play a role in the initiation and progression of cancer via the regulation of mitochondria.
They are involved in the Warburg effect, which is necessary for cancer cells to maintain their proliferative capacity.91 MitomiRs also upregulate HK2, a glucose phosphorylation enzyme, in an indirect manner, which may impact energy consumption in breast cancer cells.103, 104 Expression of one of the mitomiRs (miR-210) corresponds with hypoxia gene signatures in tumor tissues such as breast cancer and head and neck cancers, demonstrating a clear connection between mitomiR expression and hypoxia in cancer.108, 109, 121 MiRNAs have emerged in the last decade as key regulators in cancer-related processes and are classified as either oncogenic or tumor suppressive miRNAs. The miR-17/92 cluster was first discovered to be amplified in diffuse cell lymphoma and B-cell lymphoma. This mitomiR cluster suppresses apoptosis and may act as an oncogene in B-cell lymphomas, B-cell chronic lymphocytic leukemia, acute myeloid leukemia, and T-cell lymphomas. It is also overexpressed in numerous other malignancies. This cluster is a key factor in metabolic reprogramming of tumors by regulating glycolytic and mitochondrial metabolism. Tumor-targeting treatments based on mitomiRs are emerging as a novel diagnostic and therapeutic tool.94, 106, 111, 128
Future perspectives
We have shown that mitomiRs are important players in mitochondria of cancer cell that need to be further investigated to develop a new potential therapies for cancer. Numerous studies that have been published in recent years give promising predictions that mitomirRs will receive more attention in the context of their role in cancer as possible bio-markers or targets for treatment.
Acknowledgement
This work was financially supported by the Slovenian Research Agency (ARRS grant # P3-0003).
Disclosure
No potential conflicts of interest were disclosed
References
- 1.Bienertova-Vasku J, Sana J, Slaby O. The role of microRNAs in mitochondria in cancer. Cancer Lett. 2013;336:1–7. doi: 10.1016/j.canlet.2013.05.001. [DOI] [PubMed] [Google Scholar]
- 2.Jansson MD, Lund AH. MicroRNA and cancer. Mol Oncol. 2012;6:590–610. doi: 10.1016/j.molonc.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Thomson DW, Bracken CP, Goodall GJ. Experimental strategies for microRNA target identification. Nucleic Acids Res. 2011;39:6845–53. doi: 10.1093/nar/gkr330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang X, Zuo X, Yang B, Li Z, Xue Y, zhuo Y. MicroRNA directly enhances mitochondrial translation during muscle differentiation. Cell. 2014;158:607–19. doi: 10.1016/j.cell.2014.05.047. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–54. doi: 10.1016/0092-8674(93)90529-Y. [DOI] [PubMed] [Google Scholar]
- 6.Wang JK, Wang Z, Li G. MicroRNA-125 in immunity and cancer. Cancer Lett. 2019;454:134–45. doi: 10.1016/j.canlet.2019.04.015. [DOI] [PubMed] [Google Scholar]
- 7.Macgregor-Das AM, Das S. A microRNA’s journey to the center of the mitochondria. Am J Physiol Circ Physiol. 2018;315 doi: 10.1152/ajpheart.00714.2017. H 206-H215. [DOI] [PubMed] [Google Scholar]
- 8.Fiore D, Donnrumma E, Roscigno G, Iaboni M, Russo V, Affinito A. miR-340 predicts glioblastoma survival and modulates key cancer hallmarks through down-regulation of NRAS. Oncotarget. 2016;7:19531–47. doi: 10.18632/oncotarget.6968. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pekarsky Y, Balatti V, Palamarchuk A, Rizzotto L, Veneziano D, Bigita G. Dysregulation of a family of short noncoding RNAs, tsRNAs, in human cancer. Proc Natl Acad Sci. 2016;113:5071–76. doi: 10.1073/pnas.1604266113. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Quintavalle C, Donnarumma E, Iaboni M, Roscigno G, Garofalo M, Romano G. Effect of miR-21 and miR-30b/c on TRAIL-induced apoptosis in glioma cells. Oncogene. 2013;32:4001–8. doi: 10.1038/onc.2012.410. et al. [DOI] [PubMed] [Google Scholar]
- 11.Roscigno G, Puoti I, Giordano I, Donnarumma E, Russo V, Affinito A. MiR-24 induces chemotherapy resistance and hypoxic advantage in breast cancer. Oncotarget. 2017;8:19507–21. doi: 10.18632/oncotarget.14470. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang J, He J, Zhang L. The down-regulation of microRNA-137 contributes to the up-regulation of retinoblastoma cell proliferation and invasion by regulating COX-2/PGE2 signaling. Biomed Pharmacother. 2018;106:3542. doi: 10.1016/j.biopha.2018.06.099. [DOI] [PubMed] [Google Scholar]
- 13.Gebert LFR, MacRae IJ. Regulation of microRNA function in animals. Nat Rev Mol Cell Biol. 2019;20:21–37. doi: 10.1038/s41580-018-0045-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ingenito F, Roscigno G, Affinito A, Nuzzo S, Scognamiglio I, Quintavalle C. The role of exo-miRNAs in cancer: a focus on therapeutic and diagnostic applications. Int J Mol Sci. 2019;20:4687. doi: 10.3390/ijms20194687. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mercer TR, Neph S, Dinger ME, Crawford J, Smith MA, Shearwood AMJ. The human mitochondrial transcriptome. Cell. 2011;146:645–58. doi: 10.1016/j.cell.2011.06.051. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Breving K, Esquela-Kerscher A. The complexities of microRNA regulation: mirandering around the rules. Int J Biochem Cell Biol. 2010;42:1316–29. doi: 10.1016/j.biocel.2009.09.016. [DOI] [PubMed] [Google Scholar]
- 17.Tétreault N, De Guire V. miRNAs: Their discovery, biogenesis and mechanism of action. Clin Biochem. 2013;46:842–5. doi: 10.1016/j.clinbiochem.2013.02.009. [DOI] [PubMed] [Google Scholar]
- 18.Xiong Y, Wang Y, Wang L, Huang Y, Xu Y, Xu L. et al. MicroRNA-30b targets Snail to impede epithelial-mesenchymal transition in pancreatic cancer stem cells. J Cancer. 2018;9:2147–59. doi: 10.7150/jca.25006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhu L, Xue F, Xu X, Xu J, Hu S, Liu S. MicroRNA-198 inhibition of HGF/c-MET signaling pathway overcomes resistance to radiotherapy and induces apoptosis in human non-small-cell lung cancer. J Cell Biochem. 2018;119:7873–86. doi: 10.1002/jcb.27204. et al. [DOI] [PubMed] [Google Scholar]
- 20.Otto T, Candido SV, Pilarz MS, Sicinska E, Bronson RT, Bowden M. Cell cycle-targeting microRNAs promote differentiation by enforcing cell-cycle exit. Proc Natl Acad Sci. 2017;114:10660–5. doi: 10.1073/pnas.1702914114. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wen X, Han XR, Wang YJ, Wang S, Shen M, Zhang ZF. MicroRNA-421 suppresses the apoptosis and autophagy of hippocampal neurons in epilepsy mice model by inhibition of the TLR/MYD88 pathway. J Cell Physiol. 2018;233:7022–34. doi: 10.1002/jcp.26498. et al. [DOI] [PubMed] [Google Scholar]
- 22.Yang T, Ge B. miRNAs in immune responses to Mycobacterium tuberculosis infection. Cancer Lett. 2018;431:22–30. doi: 10.1016/j.canlet.2018.05.028. [DOI] [PubMed] [Google Scholar]
- 23.Rosas-Hernandez H, Chigurupati S, Raymick J, Robinson B, Cuevas E, Hanig J. Identification of altered microRNAs in serum of a mouse model of Parkinson’s disease. Neurosci Lett. 2018;687:1–9. doi: 10.1016/j.neulet.2018.07.022. et al. [DOI] [PubMed] [Google Scholar]
- 24.Islas JF, Moreno-Cuevas JE. A MicroRNA perspective on cardiovascular development and diseases: an update. Int J Mol Sci. 2018;19:2075. doi: 10.3390/ijms19072075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hosseinahli N, Aghapour M, Duijf PHG, Baradaran B. Treating cancer with microRNA replacement therapy: a literature review. J Cell Physiol. 2018;233:5574–88. doi: 10.1002/jcp.26514. [DOI] [PubMed] [Google Scholar]
- 26.Acunzo M, Romano G, Wernicke D, Croce CM. MicroRNA and cancer - a brief overview. Adv Biol Regul. 2015;57:1–9. doi: 10.1016/j.jbior.2014.09.013. [DOI] [PubMed] [Google Scholar]
- 27.Iorio MV, Croce CM. microRNA involvement in human cancer. Carcinogenesis. 2012;33:1126–33. doi: 10.1093/carcin/bgs140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tang Q, Ouyang H, He D, Yu C, Tang G. MicroRNA-based potential diagnostic, prognostic and therapeutic applications in triple-negative breast cancer. Artif Cells, Nanomedicine, Biotechnol. 2019;47:2800–9. doi: 10.1080/21691401.2019.1638791. [DOI] [PubMed] [Google Scholar]
- 29.Geiger J, Dalgaard LT. Interplay of mitochondrial metabolism and microRNAs. Cell Mol Life Sci. 2017;74:631–46. doi: 10.1007/s00018-016-2342-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Das S, Kohr M, Dunkerly-Eyring B, Lee DI, Bedja D, Kent OA. Divergent Effects of miR-181 Family members on myocardial function through protective cytosolic and detrimental mitochondrial microRNA targets. J Am Heart Assoc. 2017;6:1–17. doi: 10.1161/JAHA.116.004694. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Paramasivam A, Vijayashree Priyadharsini J. MitomiRs: new emerging microRNAs in mitochondrial dysfunction and cardiovascular disease. Hypertens Res. 2020;43:851–3. doi: 10.1038/s41440-020-0423-3. [DOI] [PubMed] [Google Scholar]
- 32.Duarte FV, Palmeira CM, Rolo AP. The role of microRNAs in mitochondria: small players acting wide. Genes. 2014;5:865–86. doi: 10.3390/genes5040865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shinde S, Bhadra U. A complex genome-microRNA interplay in human mitochondria. Biomed Res Int. 2015;2015:1–13. doi: 10.1155/2015/206382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bandiera S, Matégot R, Girard M, Demongeot J, Henrion-Caude A. MitomiRs delineating the intracellular localization of microRNAs at mitochondria. Free Radic Biol Med. 2013;64:12–19. doi: 10.1016/j.freeradbiomed.2013.06.013. [DOI] [PubMed] [Google Scholar]
- 35.Barrey E, Saint-Auret G, Bonnamy B, Damas D, Boyer O, Gidrol X. PremicroRNA and mature microRNA in human mitochondria. PLoS One. 2011;6:e20220. doi: 10.1371/journal.pone.0020220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Latronico MVG, Condorelli G. The might of microRNA in mitochondria. Circ Res. 2012;110:1540–2. doi: 10.1161/CIRCRESAHA.112.271312. [DOI] [PubMed] [Google Scholar]
- 37.Sripada L, Tomar D, Prajapati P, Singh R, Singh AK, Singh R. Systematic analysis of small RNAs associated with human mitochondria by deep sequencing: detailed analysis of mitochondrial associated miRNA. PLoS One. 2012;7:e44873. doi: 10.1371/journal.pone.0044873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mercer TR, Neph S, Dinger ME, Crawford J, Smith MA, Shearwood AMJ. The human mitochondrial transcriptome. Cell. 2011;146:645–58. doi: 10.1016/j.cell.2011.06.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jagannathan R, Thapa D, Nichols CE, Shepherd DL, Stricker JC, Croston TL. Translational regulation of the mitochondrial genome following redistribution of mitochondrial microRNA in the diabetic heart. Circ Cardiovasc Genet. 2015;8:785–802. doi: 10.1161/CIRCGENETICS.115.001067. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bandiera S, Rüberg S, Girard M, Cagnard N, Hanein S, Chretien D. Nuclear outsourcing of RNA interference components to human mitochondria. PLoS One. 2011;6:e20746. doi: 10.1371/journal.pone.0020746. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fan S, Tian T, Chen W, Lv X, Lei X, Zhang H. Mitochondrial miRNA determines chemoresistance by reprogramming metabolism and regulating mitochondrial transcription. Cancer Res. 2019;79:1069–84. doi: 10.1158/0008-5472.CAN-18-2505. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Srinivasan H, Das S. Mitochondrial miRNA (MitomiR): a new player in cardiovascular health. Can J Physiol Pharmacol. 2015;93:855–61. doi: 10.1139/cjpp-2014-0500. [DOI] [PubMed] [Google Scholar]
- 43.Di Leva G, Garofalo M, Croce CM. MicroRNAs in cancer. Annu Rev Pathol Mech Dis. 2014;9:287–314. doi: 10.1146/annurev-pathol-012513-104715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fabian MR, Sonenberg N. The mechanics of miRNA-mediated gene silencing: a look under the hood of miRISC. Nat Struct Mol Biol. 2012;19:586–93. doi: 10.1038/nsmb.2296. [DOI] [PubMed] [Google Scholar]
- 45.Treiber T, Treiber N, Meister G. Regulation of microRNA biogenesis and its crosstalk with other cellular pathways. Nat Rev Mol Cell Biol. 2019;20:5–20. doi: 10.1038/s41580-018-0059-1. [DOI] [PubMed] [Google Scholar]
- 46.Song R, Hu XQ, Zhang L. Mitochondrial miRNA in cardiovascular function and disease. Cells. 2019;8:1475. doi: 10.3390/cells8121475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Geiger J, Dalgaard LT. Interplay of mitochondrial metabolism and microRNAs. Cell Mol Life Sci. 2017;74:631–646. doi: 10.1007/s00018-016-2342-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Das S, Ferlito M, Kent OA, Fox-Talbot K, Wang R, Liu D. Nuclear miRNA regulates the mitochondrial genome in the heart. Circ Res. 2012;110:1596–603. doi: 10.1161/CIRCRESAHA.112.267732. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shepherd DL, Hathaway QC, Pinti MV, Nichols CE, Durr AJ, Sreekumar S. Exploring the mitochondrial microRNA import pathway through Polynucleotide Phosphorylase (PNPase) J Mol Cell Cardiol. 2017;110:1525. doi: 10.1016/j.yjmcc.2017.06.012. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chan DC. Fusion and fission: Interlinked processes critical for mitochondrial health. Annu Rev Genet. 2012;46:265–87. doi: 10.1146/annurevgenet-110410-132529. [DOI] [PubMed] [Google Scholar]
- 51.Kushnareva Y, Newmeyer D. Bioenergetics and cell death. Ann N Y Acad Sci. 2010;1201:50–7. doi: 10.1111/J.1749-6632.2010.05633.X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hudson G, Gomez-Duran A, Wilson IJ, Chinnery PF. Recent mitochondrial DNA mutations increase the risk of developing common late-onset human diseases. PLoS Genet. 2014;10:e1004369. doi: 10.1371/journal.pgen.1004369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dowling DK. Evolutionary perspectives on the links between mitochondrial genotype and disease phenotype. Biochim Biophys Acta - Gen Subj. 2014;1840:1393–403. doi: 10.1016/j.bbagen.2013.11.013. [DOI] [PubMed] [Google Scholar]
- 54.Wagner GR, Payne RM. Widespread and enzyme-independent Nϵacetylation and Nϵ-succinylation of proteins in the chemical conditions of the mitochondrial matrix. J Biol Chem. 2013;288:29036–45. doi: 10.1074/jbc.M113.486753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pan T. N6-methyl-adenosine modification in messenger and long non-coding RNA. Trends Biochem Sci. 2013;38:204–9. doi: 10.1016/j.tibs.2012.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Li K, Zhang J, Yu J, Liu B, Guo Y. MicroRNA-214 suppresses gluconeogenesis by targeting activating transcriptional factor 4. J Biol Chem. 2015;290:8185–95. doi: 10.1074/jbc.M114.633990. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Brunette TJ, Permeggiani F, Huand PS, Bhabha G, Ekiert DC, Tsutakawa SE. Exploring the repeat protein universe through computational protein design. Nature. 2015;528:580–4. doi: 10.1038/nature16162. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fang J, Song XW, Tian J, Chen HY, Li DF, Wang JF. Overexpression of microRNA-378 attenuates ischemia-induced apoptosis by inhibiting caspase-3 expression in cardiac myocytes. Apoptosis. 2012;17:410–23. doi: 10.1007/s10495-011-0683-0. et al. [DOI] [PubMed] [Google Scholar]
- 59.Carrer M, Liu N, Grueter CE, Williams AH, Frisard MI, Hulver MW. Control of mitochondrial metabolism and systemic energy homeostasis by microRNAs 378 and 378*. Proc Natl Acad Sci. 2012;109:15330–5. doi: 10.1073/pnas.1207605109. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gao P, Tchernyshyov I, Chang TC, Lee YS, Kita K, Ochi T. c-Myc suppression of miR-23a/b enhances mitochondrial glutaminase expression and glutamine metabolism. Nature. 2009;458:762–5. doi: 10.1038/nature07823. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jeong JH, Cheol Kang Y, Piao Y, Kang S, Pak YK. miR-24-mediated knockdown of H2AX damages mitochondria and the insulin signaling pathway. Exp Mol Med. 2017;49:e313. doi: 10.1038/emm.2016.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Serguienko A, Grad I, Wennerstrom A, Meza-Zepeda LA, Thiede B, Stratford EW. Metabolic reprogramming of metastatic breast cancer and melanoma by let-7a microRNA. Oncotarget. 2015;6:2451–65. doi: 10.18632/oncotarget.3235. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yamasaki T, Seki N, Yoshino H, Itesako T, Yamada Y, Tatarano S. Tumor-suppressive microRNA-1291 directly regulates glucose transporter 1 in renal cell carcinoma. Cancer Sci. 2013;104:1411–9. doi: 10.1111/cas.12240. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chen Y, Zhou Y, Han F, Zhao Y, Tu M, Wang Y. A novel miR-1291-ERRαCPT1C axis modulates tumor cell proliferation, metabolism and tumorigenesis. Theranostics. 2020;10:7193–210. doi: 10.7150/thno.44877. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tu MJ, Duan Z, Liu Z, Zhang C, Bold RJ, Gonzalez FJ. MicroRNA-1291-5p sensitizes pancreatic carcinoma cells to arginine deprivation and chemotherapy through the regulation of arginolysis and glycolysis. Mol Pharmacol. 2020;98:686–94. doi: 10.1124/molpharm.120.000130. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhu H, Xue H, Jin QH, Guo J, Chen YD. MiR-138 protects cardiac cells against hypoxia through modulation of glucose metabolism by targetting pyruvate dehydrogenase kinase 1. Biosci Rep. 2017;37:1–9. doi: 10.1042/BSR20170296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ju J, Xiao D, Shen N, Zhou T, Che H, Li X. miR-150 regulates glucose utilization through targeting GLUT4 in insulin-resistant cardiomyocytes. Acta Biochim Biophys Sin (Shanghai) 2020;52:1111–9. doi: 10.1093/abbs/gmaa094. et al. [DOI] [PubMed] [Google Scholar]
- 68.Li SJ, Liu HL, Tang SL, Li XJ, Wang XY. MicroRNA-150 regulates glycolysis by targeting von Hippel-Lindau in glioma cells. Am J Transl Res. 2017;9:1058–66. [PMC free article] [PubMed] [Google Scholar]
- 69.Guo W, Qiu Z, Wang Z, Wang Q, Tan N, Chen T. MiR-199a-5p is negatively associated with malignancies and regulates glycolysis and lactate production by targeting hexokinase 2 in liver cancer. Hepatology. 2015;62:1132–44. doi: 10.1002/hep.27929. et al. [DOI] [PubMed] [Google Scholar]
- 70.Esteves JV, Yonamine CY, Pinto-Junior DC, Gerlinger-Romero F, Enguita FJ, Machado UF. Diabetes modulates microRNAs 29b-3p, 29c-3p, 199a-5p and 532-3p expression in muscle: Possible role in GLUT4 and HK2 repression. Front Endocrinol. 2018;9:1–12. doi: 10.3389/fendo.2018.00536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Pullen TJ, da Silva Xavier G, Kelsey G, Rutter GA. miR-29a and miR-29b contribute to pancreatic β-cell-specific silencing of monocarboxylate transporter 1 (Mct1) Mol Cell Biol. 2011;31:3182–94. doi: 10.1128/MCB.01433-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Rayner KJ, Esau CC, Hussain FN, McDaniel AL, Marshall SM, van Gils JM. Inhibition of miR-33a/b in non-human primates raises plasma HDL and lowers VLDL triglycerides. Nature. 2011;478:404–7. doi: 10.1038/nature10486. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chen L, Hou J, Ye L, Chen Y, Cui J, Tian W. MicroRNA-143 regulates adipogenesis by modulating the MAP2K5-ERK5 signaling. Sci Rep. 2015;4:3819. doi: 10.1038/srep03819. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ye Q, Zhao X, Xu K, Li Q, Cheng J, Gao Y. Polymorphisms in lipid metabolism related miRNA binding sites and risk of metabolic syndrome. Gene. 2013;528:132–8. doi: 10.1016/j.gene.2013.07.036. et al. [DOI] [PubMed] [Google Scholar]
- 75.Civelek M, Hagopian R, Pan C, Che N, Yang W, Kayne PS. Genetic regulation of human adipose microRNA expression and its consequences for metabolic traits. Hum Mol Genet. 2013;22:3023–37. doi: 10.1093/hmg/ddt159. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ahmad A, Aboukameel A, Kong D, Wang Z, Sethi S, Chen W. Phosphoglucose isomerase/autocrine motility factor mediates epithelialmesenchymal transition regulated by miR-200 in breast cancer cells. Cancer Res. 2011;71:3400–9. doi: 10.1158/0008-5472.CAN-10-0965. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Aschrafi A, Schwechter AD, Mameza MG, Natera-Naranji O, Gioio AE, Kaplan BB. MicroRNA-338 regulates local cytochrome c oxidase IV mRNA levels and oxidative phosphorylation in the axons of sympathetic neurons. J Neurosci. 2008;28:12581–90. doi: 10.1523/JNEUROSCI.3338-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Li P, Jiao J, Gao G, Prabhakar BS. Control of mitochondrial activity by miR-NAs. J Cell Biochem. 2012;113:1104–10. doi: 10.1002/jcb.24004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Barman RK, Saha S, Das S. Prediction of interactions between viral and host proteins using supervised machine learning methods. PLoS One. 2014;9:e112034. doi: 10.1371/journal.pone.0112034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Chen H, Vermulst M, Wang YE, Chomyn A, Prolla TA, McCaffery JM. Mitochondrial fusion is required for mtDNA stability in skeletal muscle and tolerance of mtDNA mutations. Cell. 2010;141:280–9. doi: 10.1016/j.cell.2010.02.026. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Colleoni F, Padmanabhan N, Wung HW, Watson ED, Cetin I, Tissot van Patot M. Suppression of mitochondrial electron transport chain function in the hypoxic human placenta: a role for miRNA-210 and protein synthesis inhibition. PLoS One. 2013;8:e55194. doi: 10.1371/journal.pone.0055194. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Nishi H, Ono K, Iwanaga Y, Horie T, Nagao T, Takemura G. MicroRNA-15b modulates cellular ATP levels and degenerates mitochondria via Arl2 in neonatal rat cardiac myocytes. J Biol Chem. 2010;285:4920–30. doi: 10.1074/jbc.M109.082610. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Tomasetti M, Neuzil J, Dong L. MicroRNAs as regulators of mitochondrial function: role in cancer suppression. Biochim Biophys Acta. 2014;1840:1441–53. doi: 10.1016/j.bbagen.2013.09.002. [DOI] [PubMed] [Google Scholar]
- 84.Zheng S, Li Y, Zhang Y, Li X, Tang H. MiR-101 regulates HSV-1 replication by targeting ATP5B. Antiviral Res. 2011;89:219–226. doi: 10.1016/j.antiviral.2011.01.008. [DOI] [PubMed] [Google Scholar]
- 85.Qiao A, Khechaduri A, Mutharasan RK, Wu R, Nagpal V, Ardehali H. MicroRNA-210 Decreases heme levels by targeting ferrochelatase in cardiomyocytes. J Am Heart Assoc. 2013;2:1–12. doi: 10.1161/JAHA.113.000121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kurtz CL, Peck BCE, Fannin EE, Beysen C, Miao J, Landstreet SR. MicroRNA-29 Fine-tunes the expression of key FOXA2-activated lipid metabolism genes and is dysregulated in animal models of insulin resistance and diabetes. Diabetes. 2014;63:3141–8. doi: 10.2337/db13-1015. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Li X, Wang FS, Wu ZY, Lin JL, Lan WB, Lin JH. MicroRNA-19b targets Mfn1 to inhibit Mfn1-induced apoptosis in osteosarcoma cells. Neoplasma. 2014;61:265–73. doi: 10.4149/neo_2014_034. [DOI] [PubMed] [Google Scholar]
- 88.Das S, Ferlito M, Kent OA, Fox-Talbot K, Wang R, Liu D. Nuclear miRNA regulates the mitochondrial genome in the heart. Circ Res. 2012;110:1596–603. doi: 10.1161/CIRCRESAHA.112.267732. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Hanahan D, Weinberg RA. Hallmarks of cancer: The next generation. Cell. 2011;144:646–74. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 90.Zhong Z, Yu J, Virshup DM, Madan B. Wnts and the hallmarks of cancer. Cancer Metastasis Rev. 2020;39:625–45. doi: 10.1007/S10555-020-09887-6. [DOI] [PubMed] [Google Scholar]
- 91.Courtnay R, Ngo DC, Malik N, Ververis K, Tortorella SM, Karagiannis TC. Cancer metabolism and the Warburg effect: the role of HIF-1 and PI3K. Mol Biol Rep. 2015;42:841–51. doi: 10.1007/s11033-015-3858-x. [DOI] [PubMed] [Google Scholar]
- 92.Zheng J. Energy metabolism of cancer: glycolysis versus oxidative phosphorylation (Review) Oncol Lett. 2012;4:1151–7. doi: 10.3892/ol.2012.928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Fogg VC, Lanning NJ, MacKeigan JP. Mitochondria in cancer: at the crossroads of life and death. Chin J Cancer. 2011;30:526–39. doi: 10.5732/cjc.011.10018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Mogilyansky E, Rigoutsos I. The miR-17/92 cluster: a comprehensive update on its genomics, genetics, functions and increasingly important and numerous roles in health and disease. Cell Death Differ. 2013;20:1603–14. doi: 10.1038/cdd.2013.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Chen B, Li H, Yenh X, Yang P, Liu X, Yhao X. Roles of microRNA on cancer cell metabolism. J Transl Med. 2012;10:228. doi: 10.1186/14795876-10-228. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Li L, Yuan L, Luo J, Gao J, Guo J, Xie X. MiR-34a inhibits proliferation and migration of breast cancer through down-regulation of Bcl-2 and SIRT1. Clin Exp Med. 2013;13:109–17. doi: 10.1007/s10238-012-0186-5. [DOI] [PubMed] [Google Scholar]
- 97.Buechner J, Tømte E, Haug BH, Henriksen JR, Løkke C, Flægstad T. Tumour-suppressor microRNAs let-7 and mir-101 target the proto-oncogene MYCN and inhibit cell proliferation in MYCN-amplified neuroblastoma. Br J Cancer. 2011;105:296–303. doi: 10.1007/s10238-012-0186-5. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Zhang L, Liao Y, Tang L. MicroRNA-34 family: a potential tumor suppressor and therapeutic candidate in cancer. J Exp Clin Cancer Res. 2019;38:1–13. doi: 10.1186/S13046-019-1059-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Endo K, Naito Y, Ji X, Nakanishi M, Noguchi T, Goto Y. MicroRNA 210 as a biomarker for congestive heart failure. Biol Pharm Bull. 2013;36:4854. doi: 10.1248/bpb.b12-00578. et al. [DOI] [PubMed] [Google Scholar]
- 100.Calin GA, Dumitru CD, Shimizu M, Bichi R, Zupo S, Noch E. Nonlinear partial differential equations and applications: Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci. 2002;99:15524–9. doi: 10.1073/pnas.242606799. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Gao S, Chen C, Wu J, Tan Y, Yu K, Xing CY. Synergistic apoptosis induction in leukemic cells by miR-15a/16-1 and arsenic trioxide. Biochem Biophys Res Commun. 2010;403:203–8. doi: 10.1016/J.BBRC.2010.10.137. et al. [DOI] [PubMed] [Google Scholar]
- 102.Siengdee P, Trakooljul N, Murani E, Schwerin M, Wimmers K, Ponsuksili S. MicroRNAs regulate cellular ATP levels by targeting mitochondrial energy metabolism genes during C2C12 myoblast differentiation. 2015. [DOI] [PMC free article] [PubMed]
- 103.Fang R, Xiao T, Fang Z, Sun Y, Li F, Gao Y. MicroRNA-143 (miR-143) regulates cancer glycolysis via targeting hexokinase 2 gene. J Biol Chem. 2012;287:23227–23235. doi: 10.1074/jbc.M112.373084. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Jiang S, Zhang LF, Zhang HW, Hu S, Lu MH, Liang S. A novel miR-155/ miR-143 cascade controls glycolysis by regulating hexokinase 2 in breast cancer cells. EMBO J. 2012;31:1985–98. doi: 10.1038/emboj.2012.45. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Sun Y, Zhao X, Zhou Y, Hu Y. miR-124, miR-137 and miR-340 regulate colorectal cancer growth via inhibition of the Warburg effect. Oncol Rep. 2012;28:1346–52. doi: 10.3892/or.2012.1958. [DOI] [PubMed] [Google Scholar]
- 106.Kefas B, Comeau L, Erdle N, Montgomery E, Amos S, Purow B. Pyruvate kinase M2 is a target of the tumor-suppressive microRNA-326 and regulates the survival of glioma cells. Neuro Oncol. 2010;12:1102–12. doi: 10.1093/neuonc/noq080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Olive V, Jiang I, He L. mir-17-92, a cluster of miRNAs in the midst of the cancer network. Int J Biochem Cell Biol. 2010;42:1348–54. doi: 10.1016/j.biocel.2010.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Zhuang X, Chen Y, Wu Z, Xu Q, Chen M, Shao M. Mitochondrial miR181a-5p promotes glucose metabolism reprogramming in liver cancer by regulating the electron transport chain. Carcinogenesis. 2020;41:972–83. doi: 10.1093/CARCIN/BGZ174. et al. [DOI] [PubMed] [Google Scholar]
- 109.Qin Q, Furong W, Baosheng L. Multiple functions of hypoxia-regulated miR-210 in cancer. J Exp Clin Cancer Res. 2014;33:50. doi: 10.1186/17569966-33-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Gee HE, Camps C, Buffa FM, Patiar S, Winter SC, Betts G. hsamiR-210 is a marker of tumor hypoxia and a prognostic factor in head and neck cancer. Cancer. 2010;116:2148–58. doi: 10.1002/cncr.25009. et al. [DOI] [PubMed] [Google Scholar]
- 111.Puisségur MP, Mazure NM, Bertero T, Pradelli L, Grosso S, Robbe-Sermesant K. miR-210 is overexpressed in late stages of lung cancer and mediates mitochondrial alterations associated with modulation of HIF-1 activity. Cell Death Differ. 2011;18:465–78. doi: 10.1038/cdd.2010.119. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Yao J, Zhou E, Wang Y, Xu F, Zhang D, Zhong D. microRNA-200a inhibits cell proliferation by targeting mitochondrial transcription factor A in breast cancer. DNA Cell Biol. 2014;33:291–300. doi: 10.1089/dna.2013.2132. [DOI] [PubMed] [Google Scholar]
- 113.Mi Y, Zhang D, Jiang W, Weng J, Zhou C, Huang K. miR-181a-5p promotes the progression of gastric cancer via RASSF6-mediated MAPK signalling activation. Cancer Lett. 2017;389:11–22. doi: 10.1016/j.canlet.2016.12.033. et al. [DOI] [PubMed] [Google Scholar]
- 114.Zhu N, Zang D, Xie H, Zhou Z, Chen H, Hu T. Endothelial-specific intron-derived miR-126 is down-regulated in human breast cancer and targets both VEGFA and PIK3R2. Mol Cell Biochem. 2011;351:157–64. doi: 10.1007/s11010-011-0723-7. et al. [DOI] [PubMed] [Google Scholar]
- 115.Li Z, Lu J, Sun M, Mi S, Zhang H, Luo RT. Distinct microRNA expression profiles in acute myeloid leukemia with common translocations. Proc Natl Acad Sci. 2008;105:15535–40. doi: 10.1073/pnas.0808266105. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Sasahira T, Kurihara M, Bhawal UK, Ueda N, Shimomoto T, Yamamoto K.. Downregulation of miR-126 induces angiogenesis and lymphangiogenesis by activation of VEGF-A in oral cancer. Br J Cancer. 2012;107:700–6. doi: 10.1038/bjc.2012.330. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Crawford M, Brawner E, Batte K, Yu L, Hunter MG, Otterson GA. MicroRNA-126 inhibits invasion in non-small cell lung carcinoma cell lines. Biochem Biophys Res Commun. 2008;373:607–12. doi: 10.1016/j.bbrc.2008.06.090. et al. [DOI] [PubMed] [Google Scholar]
- 118.Felli N, Felicetti F, Lustri AM, Errico MC, Bottero L, Cannistraci A. miR-126&126* restored expressions play a tumor suppressor role by directly regulating ADAM9 and MMP7 in melanoma. PLoS One. 2013;8:e56824. doi: 10.1371/journal.pone.0056824. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Shen WF, Hu YL, Uttarwar L, Passegue E, Largman C. MicroRNA-126 regulates HOXA9 by binding to the homeobox. Mol Cell Biol. 2008;28:4609–19. doi: 10.1128/MCB.01652-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Zhang J, Du YY, Lin Y, Chen Y, Yang L, Wang H. The cell growth suppressor, mir-126, targets IRS-1. Biochem Biophys Res Commun. 2008;377:136–40. doi: 10.1016/j.bbrc.2008.09.089. et al. [DOI] [PubMed] [Google Scholar]
- 121.Otsubo T, Akiyama Y, Hashimoto Y, Shimada S, Goto K, Tuasa Y. MicroRNA-126 inhibits SOX2 expression and contributes to gastric carcinogenesis. PLoS One. 2011;6:e16617. doi: 10.1371/journal.pone.0016617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Miko E, Margitai Z, Czimmerer Z, Varkonyi I, Dezso B, Lanyi A. miR-126 inhibits proliferation of small cell lung cancer cells by targeting SLC7A5. FEBS Lett. 2011;585:1191–1196. doi: 10.1016/j.febslet.2011.03.039. et al. [DOI] [PubMed] [Google Scholar]
- 123.Gee HE, Ivan C, Calin GA, Ivan M. HypoxamiRs and cancer: From biology to targeted therapy. Antioxid Redox Signal. 2014;21:1220–38. doi: 10.1089/ars.2013.5639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Roybal JD, Zang Y, Ahn YH, Yang Y, Gibbson DL, Baird BN. miR-200 inhibits lung adenocarcinoma cell invasion and metastasis by targeting Flt1/VEGFR1. Mol Cancer Res. 2011;9:25–35. doi: 10.1158/1541-7786.MCR-10-0497. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Rasheed SAK, Teo CR, Beillard EJ, Voorhoeve PM, Casey PJ. MicroRNA-182 and microRNA-200a control G-protein subunit α-13 (GNA13) expression and cell invasion synergistically in prostate cancer cells. J Biol Chem. 2013;288:7986–95. doi: 10.1074/jbc.M112.437749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Renčelj A, Škrlep M, Čandek-Potokar M, Dovč P. Tissue specific splicing of pre-mRNA porcine mitochondrial transcription factor A. Czech J Anim Sci. 2018;63:43–50. doi: 10.17221/29/2017-CJAS. [DOI] [Google Scholar]
- 127.Han B, Izumi H, Yasuniwa Y, Akiyama M, Yamaguchi T, Fujimoto N. Human mitochondrial transcription factor A functions in both nuclei and mitochondria and regulates cancer cell growth. Biochem Biophys Res Commun. 2011;408:45–51. doi: 10.1016/j.bbrc.2011.03.114. et al. [DOI] [PubMed] [Google Scholar]
- 128.Izreig S, Samborska B, Johnson RM, Sergushichev A, Ma EH, Lussier C. The miR-17-92 microRNA cluster is a global regulator of tumor metabolism. Cell Rep. 2016;16:1915–28. doi: 10.1016/j.celrep.2016.07.036. et al. [DOI] [PubMed] [Google Scholar]


