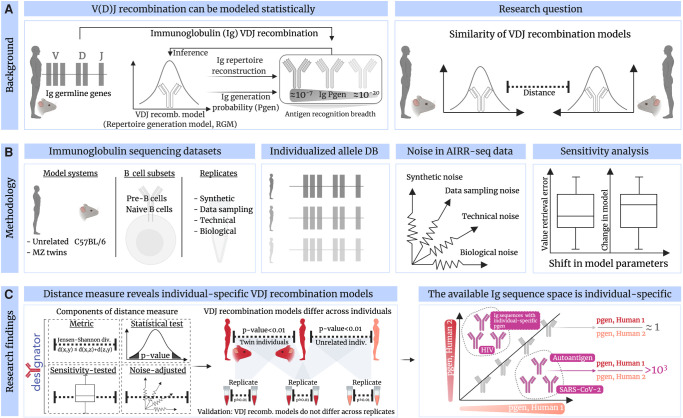
Abstract
The process of recombination between variable (V), diversity (D), and joining (J) immunoglobulin (Ig) gene segments determines an individual's naive Ig repertoire and, consequently, (auto)antigen recognition. VDJ recombination follows probabilistic rules that can be modeled statistically. So far, it remains unknown whether VDJ recombination rules differ between individuals. If these rules differed, identical (auto)antigen-specific Ig sequences would be generated with individual-specific probabilities, signifying that the available Ig sequence space is individual specific. We devised a sensitivity-tested distance measure that enables inter-individual comparison of VDJ recombination models. We discovered, accounting for several sources of noise as well as allelic variation in Ig sequencing data, that not only unrelated individuals but also human monozygotic twins and even inbred mice possess statistically distinguishable immunoglobulin recombination models. This suggests that, in addition to genetic, there is also nongenetic modulation of VDJ recombination. We demonstrate that population-wide individualized VDJ recombination can result in orders of magnitude of difference in the probability to generate (auto)antigen-specific Ig sequences. Our findings have implications for immune receptor–based individualized medicine approaches relevant to vaccination, infection, and autoimmunity.
The diversity, and thus antigen recognition breadth, of adaptive immune receptor (AIR) repertoires (AIRR) is influenced by the statistics of V, D, and J gene (and allele) segment recombination (Chi et al. 2020). Specifically, germline genes (and alleles), as well as their frequencies, have been linked to antibody neutralization breadth in infection (Avnir et al. 2016; Sangesland et al. 2020; Mikocziova et al. 2021a), the occurrence of precursor sequences of broadly neutralizing antibodies in the context of vaccine genetics (Lee et al. 2021), and autoantigen-specific binding in autoimmunity (Raposo et al. 2014; Parks et al. 2017).
With the advent of adaptive high-throughput AIRR sequencing (Weinstein et al. 2009), it has been observed that certain germline genes, and consequently recombined AIRs, occur more often than others (Weinstein et al. 2009; Rubelt et al. 2016; Greiff et al. 2017a; Elhanati et al. 2018; Dupic et al. 2021). It has been shown that the occurrence of naive AIRs could be predicted using a mathematical (explicit Bayesian or deep generative) model of VDJ recombination (Elhanati et al. 2018; Marcou et al. 2018; Olson and Matsen 2018; Davidsen et al. 2019; Remmel and Ackerman 2021)—hereafter called repertoire generation model (RGM). The Bayesian RGM parameters (RGMPs) correspond largely to those biological parameters that determine the biological mechanisms of VDJ recombination (Fig. 1A). Importantly, RGMPs enable computing the generation probability (Pgen) of a given AIR sequence. Although previous reports suggested that RGMP values differ across individuals (Marcou et al. 2018; Briney et al. 2019), the extent of this potential variation was neither quantified nor statistically verified (Fig. 1B). Inter-individual RGMP variation would imply that Pgens for identical AIR sequences differed across individuals. If this hypothesis is correct, it will implicate that each individual is biased toward exploring different AIR sequence spaces (Fig. 1C), which in turn has implications for the susceptibility to autoimmunity, cancer, and infectious diseases. For example, potentially important precursor AIRs for vaccine responses (Sangesland et al. 2019; Lee et al. 2021) or potentially damaging autospecific AIRs would occur more or less often depending on the individual's RGM.
Figure 1.
Comparison of AIR repertoire generation models. (A) The process of recombining variable (V), diversity (D), and joining (J) immunoglobulin (Ig) gene segments determines an individual's naive Ig repertoire and, consequently, (auto)antigen recognition. VDJ recombination follows probabilistic rules that can be described statistically as repertoire generation models (RGMs). So far, it remains unknown whether VDJ recombination rules differ across individuals. We set out to resolve this question by developing a distance measure that enables the quantification of RGM parameter (RGMP) similarity. (B) Accounting for several sources of noise in murine and human Ig sequencing data (by leveraging various types of replicates), as well as allelic diversity, (C) we were able to implement a noise-aware, sensitivity-tested statistical test for comparing RGM similarity. We call our method desYgnator for DEtection of SYstematic differences in GeneratioN of Adaptive immune recepTOr Repertoires (desYgnator). Using desYgnator, we found that replicate samples of the same subject are consistently more similar to each other than to samples from other unrelated individuals or even monozygotic twins (or inbred mice) indicating that not only genetic but also nongenetic factors contribute to the individualization of an RGM. We validated desYgnator by showing that RGM did not differ across synthetic and experimental replicates. We quantified the implication of individual RGMs on Ig repertoire architecture in a data set of approximately 100 human individuals by showing that the same (antigen-annotated) Ig sequence can have different generation probabilities across individuals. Thus, the available Ig sequence space is individually biased, predisposed by the individual RGM.
In this study, we aimed to measure the magnitude of the inter-individual RGMP variation and its effect on the immunoglobulin (Ig) sequence Pgens.
Results
A method for quantifying the similarity between repertoire generation models
Several studies have compared AIRRs across individuals using features such as germline gene usage (Glanville et al. 2011; Rubelt et al. 2016; Bolen et al. 2017), clonal overlap (Weinstein et al. 2009; Madi et al. 2014; Greiff et al. 2017a), clonal expansion (Stern et al. 2014; Greiff et al. 2015), and sequence similarity (Arora et al. 2018; Miho et al. 2018, 2019). However, all these features describe post VDJ recombination characteristics. So far, there is no sample size–independent measure for comparing across individuals the entirety of RGMP, such as germline gene segment choice probabilities or deletion and insertion profiles (for exact numbers of parameters, see Methods, “Using JSD to compare RGMPs”). RGMPs can be estimated from AIRR-seq data via probabilistic modeling of VDJ recombination (Marcou et al. 2018): For each nucleotide sequence, the algorithm considers all plausible recombination scenarios (see Supplemental Fig. S13) of how this sequence could be generated, and the parameters of the model (RGMPs) are optimized to maximize the likelihood over all sequences in the sample. It is important to mention that this method, as well as previous studies on VDJ recombination modeling, implicitly assumes stability of the RGMPs within a certain time window (in the Discussion section, we consider potential limitations of our analysis implied by this assumption).
Previously, Marcou and colleagues (2018) used the Kullback–Leibler divergence (KLD) (Kullback and Leibler 1951) for comparing the RGMP values inferred from a synthetic AIRR with those used to generate that synthetic AIRR. The investigators found that the KLD decreased with increasing sample size, indicating, according to the investigators, that the more sequencing reads were used for inference, the higher the inference precision. In this work, we favored the Jensen–Shannon divergence (JSD) as to compare individually inferred RGPs, which is a smoothed symmetric version of the KLD. The square root of the JSD has the advantage of satisfying the triangle inequality (see Methods, “Using JSD to compare RGMPs”), enabling the computation of a relative distance between the RGMPs of any two repertoires (e.g., from different individuals or subsamples of the same individual's repertoire). The KLD is suited for quantifying by how much a distribution P diverges from a reference, perfectly known distribution Q (Marcou et al. 2018), whereas the JSD is designed to be used in a symmetric setting, that is, with two arbitrary distributions. The JSD has previously also been used for comparing TCR RGMPs inferred from 651 individuals with a “universal” RGM, inferred from TCR sequences randomly drawn from individual samples (Sethna et al. 2020). However, this study did not compare individual RGMP sets in a pairwise fashion.
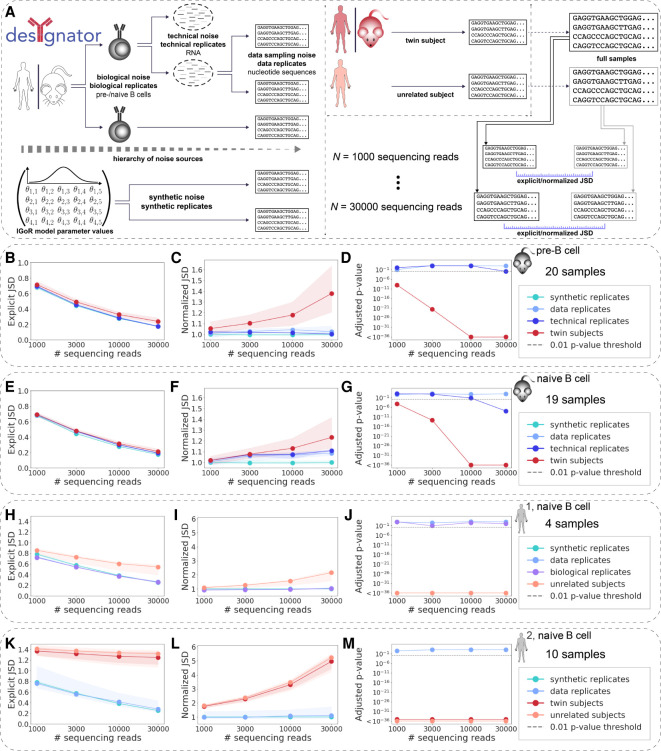
Because various sources of noise can arise in AIRR data (Puelma Touzel et al. 2020; Desponds et al. 2021; Koraichi et al. 2021), we aimed to quantify their impact on the pairwise JSD of RGMPs. We examined the selected potential sources of sample-associated noise by means of different types of replicates (Fig. 2):
Synthetic replicates (cyan curves), synthetic samples generated using the same set of IGoR parameter values; these samples differ only by the unavoidable synthetic sampling noise and thus allow for its quantification.
Data replicates (pale blue curves) subsampled without replacement from the same AIRR FASTA file; these replicates differ only by the data sampling noise.
Technical replicates (solid blue curves) for which the RNA samples were split and then sequenced independently; these samples differ owing to the technical noise and data sampling noise. The MOUSE_PRE and MOUSE_NAIVE data sets each contain a pair of technical replicates (see Methods, “Experimental immunoglobulin sequencing data”).
Biological replicates (violet curves) obtained from the same individual and differ owing to the biological noise (subsampling of the B/T cells from the actual repertoire, different expression levels), as well as owing to both technical noise and data sampling noise. The HUMAN1 data set contains a single pair of biological replicates.
Figure 2.
RGMPs are individual-specific independent of the degree of immunogenetic similarity between individuals. (A) Different sources of AIRR-seq noise may arise impacting RGMP inference. To account for these sources of noise, different kinds of replicates are necessary. Specifically, biological replicates (i.e., biological samples obtained from the same individual) allow for observing biological noise; technical replicates (an RNA sample that was split, and the parts were sequenced independently) allow for observing technical noise; and data replicates (subsamples of the same AIRR FASTA file, termed “full sample” in the figure) allow for observing data sampling noise. Samples obtained from different (either twin or unrelated) subjects incorporate all these aforementioned sources of noise along with the associated potential nongenetic or genetic individual differences between their RGMPs. Synthetic replicates (synthetic samples generated using the same RGMP sets) allow for observing synthetic noise. (B) Explicit Jensen–Shannon divergence (JSD) between RGMP inferred from samples differing by several levels of noise: synthetic replicates; data replicates; technical replicates; twin mice. We computed the explicit JSD for random subsets of [1000, 3000, 10,000, 30,000] sequencing reads taken from samples of the MOUSE_PRE data set (19 IgH pre–B cell samples from C57BL/6 mice and one technical replicate, see Methods, “Experimental immunoglobulin sequencing data”). Circles correspond to the median explicit JSD; shaded areas correspond to the whole range of the explicit JSD for the given sample size and pair type (from minimum to maximum). (C) The amount of noise that accounts for the difference between synthetic replicates is quantified using the explicit JSD. This can be considered as the lower bound of noise in our system. We then normalized the explicit JSD by this lower bound. (D) To test whether the difference between a pair of samples is significantly higher than the difference between data replicates, we adapted the Student's t-test. The adjusted P-values for data and technical replicates were above the 0.01 threshold for each sample size except 30,000 for technical replicates. The adjusted P-values for twin subjects were below the 0.01 threshold for all sample sizes, indicating that the recombination models of the twin subjects are not identical. (E–G) Same as B–D but computed for the MOUSE_NAIVE data set (19 IgH naive B cell samples from C57BL/6 mice and one technical replicate). The twin subjects are closer to each other than in the pre–B cell case. The P-values of the statistical test, as in D, indicated RGMP of cross-subject samples differed systematically. (H–J) Same as B–D but computed for the HUMAN1 data set (three IgH naive B cell samples of healthy Caucasian male donors and one biological replicate). For all samples, individually restricted germline allele databases were constructed. The considered sample pair types are synthetic replicates, data replicates, biological replicates, and unrelated subjects. P-values indicate that biological as well as technical replicates were generated with the same RGMPs and that RGMPs differed across unrelated human individuals. (K–M) Same as B–D but computed for the HUMAN2 data set (IgH naive B cell samples from five pairs of MZ twins). For all samples, individually restricted germline allele databases were constructed (Methods, “An approach to building personalized RGMs that are robust to allelic variability of IGHV genes”). The considered sample pair types are synthetic replicates, data replicates, twin subjects, and unrelated subjects. P-values indicate that RGMPs of human MZ twins differ. All P-values were adjusted using the Bonferroni correction within one data set. The significance threshold of P = 0.01 is indicated by a gray dashed line.
Samples from human monozygotic (MZ) twin subjects and inbred mice (Fig. 2, red curves) and samples from unrelated human subjects (Fig. 2, salmon curves) both incorporate all types of noise except the synthetic one. In addition, they incorporate potential nongenetic (measurable in MZ twins and inbred mice) or genetic systematic differences (measurable in unrelated human individuals). Comparing the distance between twin subjects with the distance between unrelated subjects allows for the discrimination between nongenetic and genetic factors underlying the systematic differences in AIRR generation.
Here, we developed a method for detecting such differences, DEtection of SYstematic differences in GeneratioN of Adaptive immune recepTOr Repertoires (desYgnator). To quantitatively discriminate noise from systematic differences, we first investigated how the JSD (hereafter “explicit JSD”) is impacted by each level of noise with sample sizes from 1000 to 30,000 sequencing reads (for the definition of sequencing read, see Methods, “An approach to building personalized RGMs that are robust to allelic variability of IGHV genes”; for an explanation of sample size, see Methods, “Using JSD to compare RGMPs”) on murine (Fig. 2B,E, MOUSE_PRE and MOUSE_NAIVE data sets) and human (Fig. 2H,K, HUMAN1 and HUMAN2 data sets) samples.
Consistent with previous KLD estimations (Marcou et al. 2018), we found that the explicit JSD gradually decreases with an increasing number of sequencing reads (Fig. 2B,E,H,K). Explicit JSD values for data replicates, technical replicates, and biological replicates were similar, suggesting that the noise introduced by the technological processes (and even by the biological processes, such as different expression levels) was negligible compared with the data sampling noise.
However, RGMs inferred from samples of 30,000 sequencing reads obtained from different subjects were all closer to each other (i.e., the explicit JSD between them was lower) than to models inferred from samples of 3000 sequencing reads obtained from the same subject (Fig. 2B,E,H,K). Thus, the explicit JSD between inferred RGMPs is sample size dependent (i.e., the RGMP estimates are skewed differently for different sample sizes), suggesting either that the inferred parameters are different or that the models have different levels of complexity. Therefore, it is not recommended to directly compare RGMP inferred from samples of different sizes. Furthermore, the threshold for determining the statistically significant pairwise difference of RGMP sets is also sample size dependent and thus may vary across sample sizes.
To compensate for this dependence of the thresholds on the sample size, we introduced a normalized JSD (see Methods, “Using JSD to compare RGMPs”), which is obtained by dividing the explicit JSD of two RGMPs (inferred from samples of a certain size) by the average explicit JSD between synthetic replicates (of the same sample size). For the normalization, we computed the explicit JSD between 15 independently generated pairs of synthetic replicates (we used RGMP inferred from the first sample of the HUMAN2 data set to generate human synthetic replicates and RGMP inferred from the first sample of the MOUSE_PRE data set to generate mouse synthetic replicates). To validate that JSD normalization allows compensating for potential dependencies of the explicit JSD on sample size, we repeated all explicit JSD calculations for the normalized JSD (Fig. 2C,F,I,L).
The normalized JSD, unlike the explicit one, followed a clear pattern: In the cases in which the underlying RGMP were assumed (and supposed) to be identical, that is, for all types of replicates representing the same individual (Fig. 2C,F,I,L), the normalized JSD remained on the same level with increasing sample size (cyan, pale blue, solid blue, and violet). On the contrary, it increased for the samples obtained from unrelated (Fig. 2I,L) and even from twin subjects (Fig. 2C,F,L), salmon and red, respectively. This analysis revealed (1) that the RGMP difference between individuals is distinguishable from the above-mentioned levels of noise, provided the number of sequencing reads is sufficiently high, and (2) that the normalized JSD, unlike the explicit JSD, enables the detection of this difference via a sample size–independent threshold. Of note, we expect the normalized JSD to stabilize for a high enough sample size.
To investigate the identifiability of the inter-individual RGMP difference, we derived a statistical test (see Methods, “A statistical test for comparing repertoire generation models”) that compares the JSD between two samples (technical or biological) to the expected level of noise from data replicates, and showed the associated P-values in panels (Fig. 2D,G,J,M) related to other panels (Fig. 2C,F,I,L), respectively. Altogether, using the developed statistical test (see Methods, “A statistical test for comparing repertoire generation models”), we failed to reject the null hypothesis that the explicit and normalized JSDs between RGMP sets of technical (for a sample size of [1000, 3000, 10,000] sequencing reads) or biological (for all considered sample sizes) replicates were higher than the difference between RGMP sets of data replicates for all samples. This supports the statement that both technical and biological noise were in most cases dominated by the data sampling noise. When we tested the homozygous twins and unrelated subjects in the same way against the data replicates, there was a statistically significant difference (and even for the sample size of 30,000 reads, the technical noise was negligible when compared to the inter-individual difference). The test showed that 1000 sequencing reads are sufficient to observe a significant difference (adjusted P-value < 0.01) in the case of murine pre-/naive B-cell data and human naive B cell data (Fig. 2D,G,J,M).
To measure the impact of genetic factors on the normalized JSD, we computed the pairwise normalized JSD for five pairs of MZ twins (HUMAN2 data set) (Supplemental Fig. S11). We found that the normalized JSD between the RGMPs of samples obtained from twin subjects is on average 5% lower than between samples obtained from unrelated subjects, which may indicate that nongenetic factors account for the majority of the normalized JSD difference (taking into account that some of the normalized JSD is owing to the noise). These data support the view that nongenetic factors play an important role in IgH repertoire generation, which was also noted for T cell repertoires, where MZ twins differed almost as much in their recombined repertoires as unrelated individuals (Dupic et al. 2021).
To explore the impact of different components of the RGM (e.g., V segment choice probability, V deletions, J given V choice conditional probabilities, see Methods, “Using JSD to compare RGMPs”) on the explicit and normalized JSD between the models, we reproduced the afore-described experiment using only V-choice-agnostic parameters (i.e., excluding IGHV-related parameters and only investigating J choice, J deletion, D choice, D deletion, VD insertion, DJ insertion) (Supplemental Fig. S7, columns 2 and 4) and only V-related parameters (V choice, V deletion) (Supplemental Fig. S7, columns 1 and 3). The data replicated the results obtained with the full model except that for the V-choice-agnostic models, in which the distances between samples from twin human subjects were not lower than the distances between samples from unrelated human subjects. This may indicate that genetic (genetically heritable) factors are only responsible for the difference in V-related RGMP and, consequently, that the difference in other RGMP (J segment choice, D segment choice, insertion, and deletion profiles) is entirely caused by nongenetic factors. This is consistent with the previous findings for post-recombination statistics in AIRR-seq (IgH) samples, namely, that mainly V segment usage is genetically heritable and not D or J segment usage or CDR3 similarity (Glanville et al. 2011; Rubelt et al. 2016).
The technological process and the data preprocessing workflow may introduce unavoidable bias into AIRR-seq sample generation (e.g., different choices of PCR primers may result in very different sequenced repertoires). To explore the influence of this bias on RGMP inference, we compared the average values of the normalized JSD for the HUMAN1 (where the sequences were generated with Illumina HiSeq: 2 × 125 bp; shorter reads, high sequencing depth) and HUMAN2 (Illumina MiSeq: 2 × 300 bp; longer reads, lower sequencing depth) data sets (Fig. 2I,L). Lower read length in the case of the HUMAN1 data set renders a subset of the V segment alleles indistinguishable, also introducing bias in the RGMP inference (this bias, however, cannot substantially alter the analysis results) (see Supplemental Fig. S14). The average normalized JSD differed twofold between the HUMAN1 and HUMAN2 data sets for the sample size of 30,000 sequencing reads, indicating the presence of a technological bias in RGMPs. Thus, caution and care are advised when comparing IGoR models inferred from samples generated using different experimental protocols.
To conclude, using the normalized JSD, we found that IgH RGMP differed not only across unrelated individuals but also across inbred C57BL/6 mice and human MZ twins, which may indicate that the rules of IgH VDJ recombination are governed not only by genetic factors but also by nongenetic ones.
Immunoglobulin RGM parameters are unique across human individuals
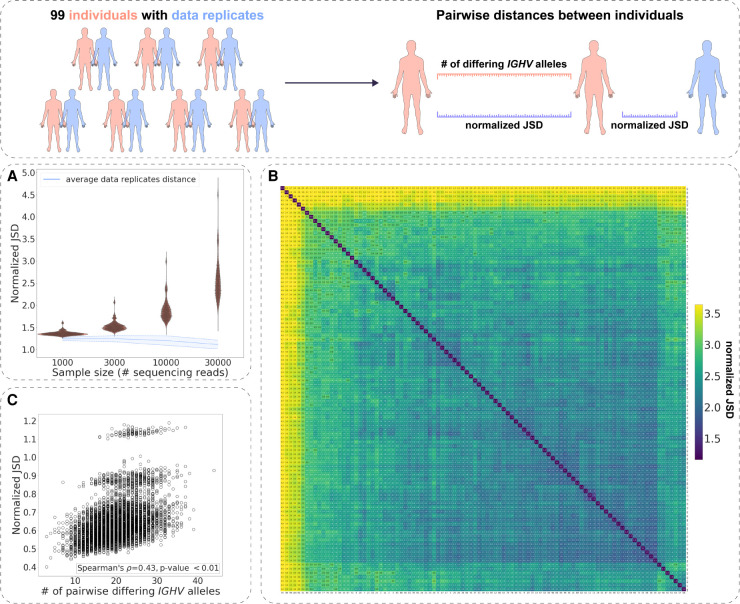
To explore the variation of VDJ recombination rules on a larger scale in human individuals (in naive B cell repertoires), we computed the normalized JSD on a cohort of 99 unrelated individuals from the HUMAN3 data set (see Methods, “Experimental immunoglobulin sequencing data”) (Fig. 3A; Gidoni et al. 2019). Analogously to the HUMAN1 and the HUMAN2 data sets (Fig. 2), we constructed individually restricted germline allele databases for all samples via merging the RGMPs corresponding to alleles of the same V/D/J gene and calculated the explicit and normalized JSD on the gene-level RGMPs (see Methods, “An approach to building personalized RGMs that are robust to allelic variability of IGHV genes”). For the sample size of 10,000 and 30,000 sequencing reads, all pairwise distances were higher than the distance between data replicates, indicating that again all individuals had different RGMPs (Fig. 2). For lower sample sizes of 1000 and 3000 sequencing reads, the distances were closer to those between data replicates but still significantly higher (Fig. 2D,G,J,M). This suggests that 1000 sequencing reads are sufficient to overcome noise when building personalized RGMs. Analogous results were obtained when investigating the IGHV-only and IGHV-agnostic explicit/normalized JSD (Supplemental Figs. S9, S10).
Figure 3.
Immunoglobulin RGM parameters are unique across human individuals. (Inset) For samples from a cohort of 99 unrelated individuals, two kinds of distance were computed: the normalized JSD between RGMPs inferred from these samples and the number of differing IGHV alleles. Additionally, for each sample, we computed the normalized JSD between its own RGMPs and RGMPs inferred from its data replicate. (A) The distribution of the pairwise normalized JSD for 99 individuals of the HUMAN3 data set was computed for subsamples of 1000, 3000, 10,000, and 30,000 sequencing reads. The blue line corresponds to the average distance between data replicates. (B) Heatmap visualization of A for the subsample size of 30,000 sequencing reads: The values on the diagonal correspond to the average distance between data replicates. (C) The number of IGHV gene alleles that differ between any two individuals as a function of the normalized JSD between their RGMP inferred from subsamples of 30,000 sequencing reads.
To visualize the similarity variation of RGMP in the HUMAN3 data set, we performed hierarchical clustering (single-linkage clustering) (Müllner 2011) on pairwise computed normalized JSD (Fig. 3B). Although the range of the normalized JSD was large (min: 1.59; max: 4.71; median: 2.49), there were no clear clusters. Of note, we detected a moderate correlation of pairwise differences in RGMP values with IGH allele repertoire similarity (Spearman's correlation coefficient = 0.43, P-value < 10−36) (Fig. 3C; see also Supplemental Figs. S9, S10), suggesting that the difference in RGMP values between individuals may in part be explained by IGHV gene polymorphisms.
To conclude, by applying our analysis to a cohort of 99 human individuals, we delineated population-wide variation of RGMP values.
Generation probabilities of antigen-annotated immunoglobulin sequences vary within and among related and unrelated human individuals
A direct corollary of the variation of RGMPs across individuals is the variation of individualized generation probabilities (Pgens) for the same Ig sequence. To study and quantify Pgen variation, we analyzed generation probabilities (as computed by the RGMs corresponding to individuals from the HUMAN2 and HUMAN3 data sets) of Ig sequences with known antigen specificity.
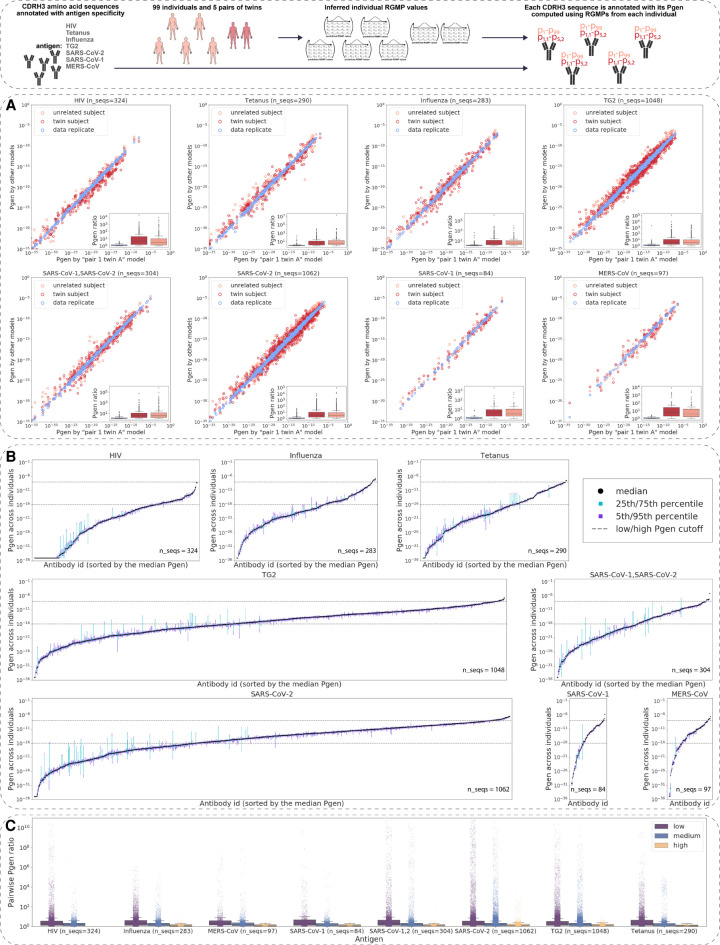
We assembled antigen-annotated IgH data from three sources (Roy et al. 2017; Swindells et al. 2017; Raybould et al. 2020), leading to 3492 unique amino acid CDRH3 sequences specific to seven antigens in total: SARS-CoV-2 (1062 sequences), transglutaminase 2 (or TG2 [autoantigen], 1048 sequences), HIV (324 sequences), tetanus (290 sequences), influenza (283 sequences), MERS-CoV (97 sequences), and SARS-CoV-1 (84 sequences). We also included 304 sequences that were specific to both SARS-CoV-1 and SARS-CoV-2. To calculate the Pgens of CDRH3 amino acid sequences, we used OLGA (Sethna et al. 2019) based on the RGMPs computed with IGoR for Figures 2 and 3 (all RGMPs were inferred from samples of 30,000 sequencing reads).
To analyze the consistency of the Pgens across the RGMPs inferred from different samples, we first used the RGMs corresponding to the individuals from the HUMAN2 data set (Fig. 4A): an RGM inferred from the sample obtained from the pair 1 twin A individual, an RGM inferred from a data replicate sample, an RGM from a twin subject (pair 1 twin B individual), and an RGM inferred from an unrelated subject (pair 2 twin A). The Pgens were highly consistent for the replicate samples: The ratio of the Pgens of the same sequence computed with the two replicate RGMs was in the overwhelming majority of cases below one order of magnitude (Fig. 4A, boxplots). However, for the RGMs inferred from different subjects, both MZ twin and unrelated, the Pgen ratio in some cases reached four or even five orders of magnitude (Fig. 4A). Of note, we did not observe that the ratios for twins were lower than those for unrelated individuals; in fact, for some of the antigens (HIV, MERS-CoV), these ratios were higher on average than for the unrelated individuals.
Figure 4.
Generation probabilities of antigen-annotated immunoglobulins (CDRH3 sequences) vary by several orders of magnitude within the human population. (Inset) For a data set of CDRH3 amino acid sequences annotated with antigen specificity, we computed Pgens using a set of RGMs corresponding to N different experimental samples. Each CDRH3 sequence is thus annotated with N Pgens. (A) Pgens of antibody CDRH3 amino acid sequences (annotated with antigen specificity) computed using RGMs corresponding to samples of different levels of immunogenetic similarity: a pair of data replicate models, a pair of models from twin individuals, and a pair of models from unrelated individuals. The x-axis always stands for the Pgen as computed with the model corresponding to the pair 1 twin A individual from the HUMAN2 data set. The y-axis corresponds to the Pgen as computed with the other model in the pair (data replicate or twin/unrelated subject). The boxplots show the distribution of the min(x,y)/max(x,y) ratios, that is, the pairwise difference of Pgens. (B) For each CDRH3 amino acid sequence, we calculated its Pgen as determined by the models corresponding to the 99 individuals from the HUMAN3 data set. The x-axis itemizes each of the CDRH3 sequences tested; the y-axis denotes the fifth, 25th, 50th, 75th, and 95th percentiles of the 99 Pgens of each CDRH3. (C) Pairwise ratios of the Pgens from B by antigen. For each antigen, we divided the CDRH3 amino acid sequences into three groups depending on the sequence's median Pgen across individuals: low (median Pgen < 10−16), medium (10−16 ≤ median Pgen < 10−8), and high (10−8 ≤ median Pgen).
To quantify the Pgen variation of antigen-annotated CDRH3s on a larger scale, we calculated the Pgen of each CDRH3 sequence (Fig. 4B) using the RGM corresponding to the 99 individuals from the HUMAN3 data set (Fig. 3). For each sequence, we calculated the fifth, the 25th, the 50th (median), the 75th, and the 95th percentiles (Fig. 4B). We found that the per-sequence Pgen variation strongly depended on the sequence itself. Variation was especially high for those CDRH3 sequences with mid to low Pgen. To investigate this further, for each CDRH3 sequence, we calculated the pairwise Pgen ratios (Fig. 4C). Then, for each antigen, we split the CDRH3 sequences into three groups according to their median Pgens: “low” group, CDRH3 sequences with median Pgen < 10−16 (i.e., sequences that are almost impossible to generate for most individuals; for instance, if a human generates approximately 3 × 1013 throughout their life, then the probability to generate a sequence with Pgen = 10−16 at least once is approximately 0.003); “medium” group, CDRH3 sequences with median Pgen between 10−16 and 10−8; and “high” group, sequences with median Pgen > 10−8 (potential public clones: if a sequence can be generated with probability 10−8 and the number of human B cells that can be represented in an AIRR-seq sample is approximately 2 × 108 [Briney et al. 2019], then the sequence will be present in more than 86% of the samples). The variation in the “high” group was lower than in the first two groups: In the “high” group, the variation in most cases stayed within one order of magnitude, whereas for the “medium” and “low” groups, it reached three orders of magnitude for more than 100 of sequences. Of note, there were no HIV-specific sequences with a median Pgen > 10−8; all tested HIV-specific CDR3 sequences belonged to either “low” or “medium” groups.
To summarize, we showed that individual RGMP variation results in individualized biases toward generating antigen-specific CDRH3 sequences that, in some cases, may lead to a difference of more than three orders of magnitude in the Pgen of an antigen-specific CDRH3 sequence.
Normalized JSD is a sensitive measure to detect subtle repertoire generation parameter differences
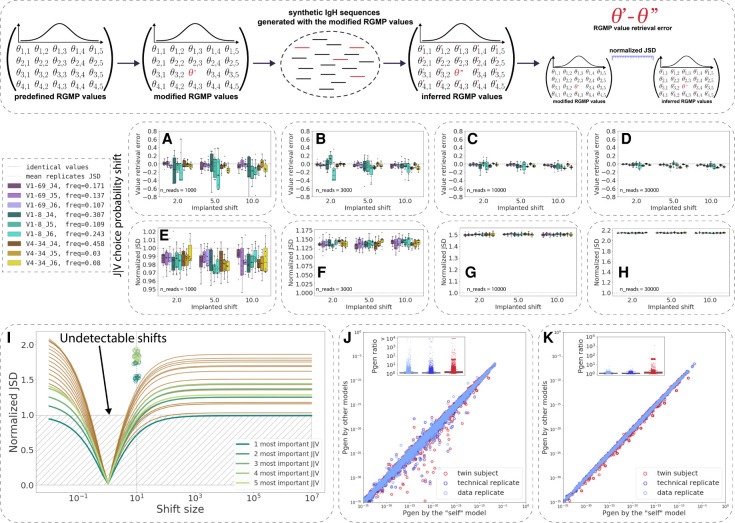
Given that the normalized JSD-based distance depends on the underlying RGMP value distribution, we sought to understand the sensitivity of RGMP parameter inference to variation in the Ig repertoire structure. To this end, we measured the sensitivity of IGoR RGMP inference following small changes in RGMP values. High/low sensitivity means that a given parameter has a great impact/negligible impact on the generated Ig repertoire. This impact can be subsequently measured with the normalized JSD between the initial and the modified RGMP sets.
Starting from an initial RGMP set, we chose a specific RGMP, changed its value, and subsequently used synthetic replicates to test if IGoR was able to correctly determine the shifted value (single-parameter focus) (Fig. 5A–D; Supplemental Fig. S12A–D) and to investigate the sensitivity of the explicit/normalized JSD to such changes (whole-repertoire focus) (Fig. 5E–H; Supplemental Fig. S12E–H). We calculated the explicit JSD between the inferred RGMP sets and the one used for generation (“ground truth RGMP”) and compared it to the explicit JSD across the inferred RGMPs (i.e., between synthetic replicates). For V and J segment (conditional) probabilities, we increased the probability of a segment and then rescaled all other ones accordingly; namely, we applied a multiplicative shift to the chosen segment probability. We chose the RGM inferred from a subsample of 30,000 sequencing reads from the pair 1 twin A sample of the HUMAN2 data set as the starting RGM.
Figure 5.
The sensitivity of RGMP inference and their impact on Pgen values vary by RGMP. (Inset) Given a set {Θ} of RGMPs, we modified one of the parameter values, obtaining a modified RGMP set {Θ′}, generated a set of synthetic IgH sequences using {Θ′} and then inferred RGMP values from these sequences, thus obtaining RGMP set {Θ′′}. By comparing {Θ′} and {Θ′′}, we estimated the stability of the RGMP inference model in IGoR. (A–D) RGMP value retrieval error (the difference between the inferred parameter value and the ground truth one) for RGMPs inferred from synthetic samples that were generated using a modified RGMP set with increased conditional probability to observe a certain J segment given a V segment for synthetic sample sizes of 1000, 3000, 10,000, and 30,000 sequencing reads (10 synthetic samples for each sample size) based on the HUMAN2 data set. The dashed line corresponds to zero difference (i.e., no error observed). (E–H) Normalized JSD between the inferred RGMP sets and the initial modified one (boxes; each box corresponds to the same 10 synthetic samples that were used in A–D). Average normalized JSD across the inferred RGMP sets themselves equals one because it is the value used for normalization (i.e., between synthetic replicates; dotted line). (I) All J|V conditional probabilities are ranked by their importance, then the k (k in [1…20]) most important probabilities are chosen and multiplied by a coefficient from 10−2 to 106; the rest is rescaled, to sum up to one. The x-axis corresponds to this multiplicative coefficient. The y-axis corresponds to the normalized JSD between the modified model and the unmodified one. The green colors correspond to the first five most important parameters. The circles correspond to the values obtained by generating synthetic samples using the modified model and inferring the parameters back as in E–H. (J) Pgens evaluated on identical sequences using different RGM parameter values (each point corresponds to a single sequence). The x-axis corresponds to the Pgen evaluated using the model parameter values inferred from the same sequences that the Pgens were computed for (the “self” model). The y-axis corresponds to the Pgens evaluated using other RGMP values (inferred from a data replicate sample, a technical replicate sample, and a sample from a twin subject). The boxplots show the corresponding Pgen ratio distributions. (K) Analogous to J but the Pgens were computed only on a set of sequences that consisted of the most impactful combinations of V and J segments (five top pairs as computed in I).
As there are 124 human V segments in our database (for details, see Supplemental Fig. S2) and tens of thousands of parameters in total (for a detailed description of model parameters, see Methods, “Using JSD to compare RGMPs”), we restricted our focus to three representative human V gene alleles: a very rare segment (IGHV1-8*01, choice probability of 0.003 in the etalon model), a segment of moderate frequency (IGHV4-34*01, choice probability of 0.024), and a more frequent one (IGHV1-69*06, choice probability of 0.037). We defined the parameter value retrieval error as the difference between the inferred parameter value and the ground truth one used for generating the synthetic data. We calculated the parameter value retrieval error as a function of sample and shift size (Supplemental Fig. S12A–D; Fig. 5A–D): We used sample sizes of 1000 among 30,000 sequencing reads, and we shifted the parameter values multiplicatively by a factor of two, five, and 10. We chose these values to not make the modified models too distant from the models inferred from experimental samples: A difference of one order of magnitude in the usage of a given V segment can easily be observed within the population (Rubelt et al. 2016; Gidoni et al. 2019).
For the V segments (Supplemental Fig. S12, IGHV1-8*01, turquoise boxes; IGHV4-34*01, yellow boxes; IGHV1-69*06, violet boxes), the parameter value estimation was unbiased for IGHV1-8*01 and IGHV4-34*01 and was slightly biased for IGHV1-69*06: Its probability was systematically underestimated, and the bias was proportional to the multiplicative shift size (i.e., proportional to the parameter value itself). The variance of the parameter value retrieval error decreased with increasing sample size, whereas the error variance strongly decreased, as expected for a maximum likelihood estimator like the IGoR inference module. The normalized JSD distance between the inferred RGMP sets and the ground truth one (Supplemental Fig. S12E–H; Fig. 5E–H) was in most cases higher than the normalized JSD between the inferred sets themselves. This may indicate the presence of a bias in the IGoR synthetic data generation process, namely, a factor that makes synthetic replicates closer to each other than to the RGMP set used for generation.
For the J segments (Fig. 5A–D, IGHJ6, brighter boxes; IGHJ5, medium boxes; IGHJ6, darker boxes), the parameter value estimation was unbiased, and the variance of the parameter value retrieval error also showed a negative trend with increasing sample size.
To explore the boundaries of IGoR sensitivity, we iteratively modified the RGMPs by shifting 20 of the most important (i.e., of the highest value) parameters and analyzed the normalized JSD between the modified and unmodified model parameters. This time, we used the model inferred from sample 20 of the MOUSE_NAIVE data set. All V probabilities (Supplemental Fig. S12) and J|V (Fig. 5I) conditional probabilities were ranked by their importance, then the k (k in [1…20]) most important probabilities were chosen and multiplied by a coefficient from 10−2 to 106; the other probabilities were rescaled to unity. We then tested the normalized JSD values for the shift size 10 by generating five synthetic samples using the modified model. The normalized JSD values indicate that changing as few as five parameter values is sufficient to significantly change the generated repertoire.
To estimate the impact of the RGMP value variation on the Pgen of a sequence, we computed the Pgens of the same sequences using several different models (Fig. 5J): a model with parameter values inferred from the same sequences that the Pgens were computed for the “self” model (sample 20, MOUSE_NAIVE), a model inferred from a data replicate sample, a model inferred from a technical replicate sample, and a model inferred from a twin subject (sample 17, MOUSE_NAIVE). This way, the self-model Pgens are more reliable, and we can refer to them as a ground truth.
To account for how specific RGMPs impact the Pgen values, we calculated the distribution of the Pgen ratio between the self-model and the other three (Fig. 5J). The Pgens computed using the replicate models (both the data replicate and the technical one) were closer to the self-model Pgens (the ratio was close to one in most cases) than those computed using the twin subject model, for which the ratio was below three in most cases but reached almost two orders of magnitude for ∼1% of the sequences. These high Pgen differences between the twin models persisted when we limited our analysis to only those sequences that consisted of the most used V and J segments (top five V-J, i.e., to the sequences with higher Pgens) (Fig. 5K). This indicates that the difference in the Pgens is not an artifact that originates from the inference of the low-impact parameters.
Collectively, our data support the view that certain parameters impact IGoR RGMP inference and Pgen evaluation to a substantially larger extent than others and that an artificial perturbation of as few as five parameter values is sufficient to produce an observable difference in the generated repertoire. Moreover, our analysis also supports the view that the JSD is a sufficiently sensitive measure to detect this difference.
Discussion
We demonstrated that IgH VDJ recombination rules (RGM) and, consequently, IgH sequence generation probabilities differ across individuals, even between homozygous human twins and inbred mice (Fig. 1). Our approach (which we call desYgnator) relies on a hierarchy of experimental controls as well as information and statistical theories and provides new recommendations for unbiased comparison of RGM between individuals that were previously thought to be identical (Fig. 2). Our results indicate that inter-individual differences in VDJ recombination are not only influenced by genetic differences in germline gene repertoires, such as germline gene polymorphisms or structural variation (Kenter et al. 2021), but also influenced by nongenetic differences (e.g., epigenetics). Indeed, it has been previously shown that epigenetic mechanisms intervene in the regulation of VDJ recombination (Pulivarthy et al. 2016). Specifically, our work shows not only that individuals differ in their recombined expressed repertoire (Dupic et al. 2021) but also that already the individual sources (RGMs) of each expressed repertoire differ. We found that the distance between RGMs of twin subjects is on average 5% lower than that between RGMs of unrelated subjects (Supplemental Fig. S11). However, the distance between RGMs of twin subjects was not lower than between RGMs of unrelated subjects when it was measured for V-segment-agnostic RGM (Supplemental Fig. S7), suggesting that nongenetic factors account for the majority of the RGMP differences in general and for almost all of the V-segment-unrelated RGMPs. Thus, although some of the genetic factors might be masked by the limitation of the data and preprocessing pipeline (such as potentially unidentified differences in D or J gene alleles), our results suggest that V-segment-unrelated RGMPs (i.e., J and D segment choice, their deletion profiles, and nontemplated insertion profiles) are not genetically heritable (Glanville et al. 2011) for IgH. Specifically, we posit that the ability of VDJ recombination to generate a high amount of antibodies specific to a given antigen is partially genetically heritable (Venkataraman et al. 2021) when antigen binding is governed by the VH-region (e.g., influenza) (Avnir et al. 2016), whereas in cases in which most of the binding is governed by the V-agnostic portion of the Ig heavy chain (i.e., CDRH3), it may not be genetically heritable. Of note, in the largest structural antibody-antigen binding data set to date, we previously showed that the CDRH3 is the sole obligate region for antibody binding (Akbar et al. 2021a). Therefore, our analysis suggests that for a large number of antigens, large-scale antigen recognition driven by naive IgH repertoire is not genetically heritable. Future studies will need to further refine these conclusions in light of potentially unidentified D and J gene alleles as well as access to selection-free data and implementation of improved preprocessing pipelines.
We found that, for unrelated subjects, the aforementioned RGMP difference correlates with the number of differing IGHV alleles (Fig. 3C). The correlation can even be observed with V-segment-agnostic RGM (Supplemental Fig. S10C). We speculate that this may be caused by at least two reasons: First, individuals that are more different in the IGHV locus are more genetically dissimilar in general; hence, they are more likely to have polymorphisms in other parts of the IG superlocus or to differ in nongenetic factors that affect the VDJ recombination. Second, different sets of IGHV germline segments inherently affect the inference of other RGMPs: The model has to explain the data with other VDJ scenarios.
The study of AIR sequence enrichment, for instance, to identify antigen-expanded sequences, previously assumed a unique RGM shared by all individuals of a species (Marcou et al. 2018). We showed one effect of the variation in RGMPs by applying different models to a given set of CDRH3 sequences (Figs. 4, 5). We found that the extent of correlation between the Pgens was consistent with the degree of immunogenetic similarity between the models; namely, the correlation was higher for the models inferred from replicate samples than from samples obtained from different subjects. We also found that the Pgen of the same Ig sequence can differ by several orders of magnitude between individuals, and, consequently, that the effectively available Ig sequence space varies from one individual to another. Thus, future methods for identifying antigen-specific sequences may require considering individual-specific RGM.
In our work, we considered several types of replicate samples: biological replicates (DeWitt et al. 2016), technical replicates (Greiff et al. 2017a), and in silico constructed data replicates and synthetic replicates. Despite providing a reliable baseline for estimation of the RGMP variation when inferring from samples from the same subject, these types of replicates do not span all steps of AIRR-seq sample generation. For example, our analysis reveals the importance of quantifying the extent to which RGMP differs across different library preparation protocols, for example, RACE, multiplex, and influence of UMI usage (Menzel et al. 2014; Khan et al. 2016; Vázquez Bernat et al. 2019; Barennes et al. 2021; Trück et al. 2021).
Our approach is highly sensitive to subtle RGMP modifications: We showed that an artificial perturbation of as few as five parameter values can be detected using desYgnator (Fig. 5).
In addition to the biological findings and the statistical framework developed, we provide guidelines for RGM analyses. Specifically, we find that only RGMPs inferred from samples of the same size may be directly compared (Fig. 5). In contrast to current practice, we show that RGMP data sets significantly differ from one another, and we therefore suggest inferring RGMPs for each individual, especially when an increased signal-to-noise ratio is desired. Furthermore, it is commonly held that RGMPs need to be either inferred from out-of-frame sequences or very early B/T cell stages (Marcou et al. 2018). Although we agree that the ideal data for RGMP inference would be to use B/T cells unaffected by any selection (e.g., pro-B cells), we inferred RGMPs from both in- and out-of-frame sequences of either pre-B- or naive B cell repertoires. By comparing the RGMP analysis results using out-of-frame only and using all (both in- and out-of-frame) sequences for one of the data sets, we found that both approaches led to identical conclusions but that RGMPs inferred from both in- and out-of-frame sequences had lower variance (for details and reasoning, see Supplemental Fig. S3). We also provide a guideline for preprocessing human AIRR-seq data with respect to the correct set of germline alleles for each individual (Supplemental Figs. S1, S2, S8). We did not use a previously developed methodology for novel allele inference (Corcoran et al. 2016; Ralph and Matsen 2016; Zhang et al. 2016; Gadala-Maria et al. 2019) because our goal was to provide stable input for the RGMP inference step and, after that, to compare RGMP across individuals, which required a general database of validated alleles, and because there is no consensus in the field of AIRRs about inferring AIR germline genes from short-read sequencing data (Collins et al. 2021; Yang et al. 2021). We, therefore, discarded potentially novel alleles in favor of previously validated ones. This conservative approach may be shifted toward including more precise allele information once available in the future (e.g., obtained with long-read DNA sequencing) (Rodriguez et al. 2020).
Machine learning is increasingly used for AIRR classification both on the sequence (Greiff et al. 2017b; Isacchini et al. 2021; Akbar et al. 2021a; Robert et al. 2021a) and repertoire level (Emerson et al. 2017; Pavlović et al. 2021; Shemesh et al. 2021; Sidhom et al. 2021), as well as for antibody generation (Friedensohn et al. 2020; Akbar et al. 2021b). Future studies will need to investigate whether differences in RGM also impact repertoire classification (Greiff et al. 2020; Rodriguez et al. 2020; Kanduri et al. 2021). Our findings depend, but do not strictly rely, on the assumption of temporal stability of RGMPs. Most of the studies on VDJ recombination models assume to some degree the stability of RGMPs—within a certain time window (Marcou et al. 2018; Davidsen et al. 2019; Sethna et al. 2019; Dupic et al. 2021; Russell et al. 2021). The question of the temporal stability of observed biological phenomena applies to the majority of biological studies (Pal and Tyler 2016; Rubelt et al. 2016; Yang et al. 2020). In our case, too, the potential temporal evolution of RGMs is of particular interest, and it warrants future investigation. Nevertheless, the identified dissimilarity of RGMPs among both MZ human twins and inbred mice supports the view that a large fraction of inter-individual Ig repertoire difference is nongenetic in nature. If epigenetic factors influence AIRR architecture, it will be useful to investigate whether, for example, aging changes the rules of VDJ recombination over time. This may be performed via analyzing longitudinal data (Mitsunaga and Snyder 2020) or pre- and post-puberty data from MZ twins (as epigenetic differences have been shown to arise after puberty) (Fraga et al. 2005). Another way to investigate the nonheritable factors that impact VDJ recombination could be by analyzing samples from different cell populations in the same individual (from pro- to naive and even memory B cells), as it will allow quantifying the influence of negative and positive selections (Nemazee 2017; Robert et al. 2021c). It will also be of interest to analyze data from individuals for whom particular VDJ recombination mechanisms are disrupted, for example, in terminal deoxynucleotidyl transferase (TdT)–deficient mice.
It is important to mention that although our study focused on B cell data sets, our approach is directly applicable to TCR repertoires, provided the availability of the appropriate high-quality data: RNA sequences of non-antigen-experienced T cells from biological and/or technical replicates and from MZ twins and unrelated subjects (Rubelt et al. 2016; Nolan et al. 2020).
In the future, it will be interesting to determine whether individual differences in RGMP lead to differences in the propensity to generate antigen-specific (e.g., auto-reactive) sequences (Shemesh et al. 2021) and, consequently, to the existence of individualized holes in the repertoire (Perelson and Oster 1979). These analyses will require large-scale naive (unselected) and disease-linked AIRR-seq data (Watson et al. 2017; Omer et al. 2021). Furthermore, an analysis linking germline polymorphisms and RGMP based on population-wide genomic data that include noncoding regions (Mikocziova et al. 2020) will be of interest in future studies. It will also be interesting to extend our analysis to account for unconventional cases of VDJ recombination, such as the absence of D segments in TRB chains (de Greef and de Boer 2021) or the occurrence of multiple D segments in IGH chains by VDDJ recombination (Safonova and Pevzner 2020).
Our work also has implications for vaccine development, because there is an increasing interest in understanding whether B cells that are to be targeted by immunogens (including broadly neutralizing antibodies) (Sangesland et al. 2019) exist within the naive B cell repertoire of most individuals in a population of interest (“public clones”) (Greiff et al. 2017b; Elhanati et al. 2018) and whether those B cells occur at a high enough precursor frequency such that they have a high likelihood to become activated in response to immunization. These considerations relate to both V gene usage and germline gene polymorphisms (Sangesland et al. 2019; Lee et al. 2021; Russell et al. 2021). Here we show that personalized VDJ recombination models contribute to the variation in naive B cell precursor frequencies. Nowadays, high-throughput AIRR sequencing technologies allow a comprehensive coverage naive Ig repertoires, thus enabling the integration of VDJ recombination models into iterative individualized immunogen design pipelines to advance vaccine discovery (Lee et al. 2021; Robert et al. 2021b).
Methods
Experimental immunoglobulin sequencing data
We analyzed five publicly available Ig experimental data sets from four sources:
MOUSE_PRE from Greiff et al. (2017a) (ArrayExpress [https://www.ebi.ac.uk/arrayexpress/] E-MTAB-5349): IgH pre–B cell samples obtained from 19 inbred SPF C57BL/6 mice, sequenced on the Illumina MiSeq platform (2 × 300 bp). For one of the mice, Greiff and colleagues prepared two technical replicates, resulting in 20 samples in total.
MOUSE_NAIVE from Greiff et al. (2017a) (ArrayExpress E-MTAB-5349): IgH naive B cell samples obtained from 20 inbred SPF C57BL/6 mice, sequenced on the Illumina MiSeq platform (2 × 300 bp). For one of the mice, Greiff and colleagues prepared two technical replicates, resulting in 21 samples in total.
HUMAN1 from DeWitt et al. (2016) (Dryad Digital Repository doi:10.5061/dryad.35ks2): IgH naive B cell samples obtained from three 25- to 40-yr-old Caucasian male donors, sequenced on the Illumina HiSeq platform (1 × 130 bp spanning CDR3). For one of the donors, DeWitt and colleagues prepared two biological replicates, resulting in four samples in total. For this data set, we performed additional preprocessing: We filtered out all the reads that spanned <70% of the V segment (for the V segment annotation to be more reliable) and then completed these reads to full length using the VJ annotation.
HUMAN2 from Rubelt et al. (2016) (Sequence Read Archive [SRA; https://www.ncbi.nlm.nih.gov/sra] SRP065626): IgH naive B cell samples obtained from five pairs of adult MZ twins (10 samples in total), sequenced on the Illumina MiSeq platform (2 × 300 bp).
HUMAN3 from Gidoni et al. (2019) (European Nucleotide Archive [ENA; https://www.ebi.ac.uk/ena/browser/home] PRJEB26509): IgH naive B cell samples obtained from 100 individuals from Norway; 48 healthy controls (out of which 28 blood bank donors and 20 healthy individuals), and 52 patients with celiac disease; sequenced on the Illumina MiSeq platform (2 × 300 bp). We discarded one of the samples (S97) owing to the poor quality of the sequencing reads.
For further experimental details on each data set, please refer to the respective publications.
An approach to building personalized RGMs that are robust to allelic variability of IGHV genes
Evidence published over the last couple of years has demonstrated an extensive polymorphic and structural diversity in the immunoglobulin germline locus (Watson et al. 2017; Yu et al. 2017; Collins et al. 2020; Lees et al. 2020; Bernat et al. 2021; Khatri et al. 2021; Martins et al. 2021; Mikocziova et al. 2021b). These findings indicate that the RGMs used to annotate each individual's AIR sequences with accurate Pgens need to be built for each individual separately with the individualized set of alleles. The human IGHV locus is highly diverse (Watson and Breden 2012; Watson et al. 2017), and there exist multiple alleles for the majority of IGHV genes in the IMGT and OGRDB databases, for example, up to 19 alleles for genes IGHV1-69 and IGHV3-30) (Lefranc 2001; Lees et al. 2020). AIRR-seq VDJ gene annotation tools (e.g., MiXCR by Bolotin et al. 2015; IgBLAST by Ye et al. 2013; the annotation module of IGoR by Marcou et al. 2018; partis by Ralph and Matsen 2016) rely solely on a given set of germline genes and alleles from a chosen germline gene database. Any sequencing read within a sample needs to be aligned to all alleles in a germline gene database. Taking a full germline database as reference might be problematic for the inference of personalized RGMP because each individual may only possess a subset of the alleles present in the germline database.
Theoretically, one human individual cannot have more than two alleles of the same gene. However, the structure of the IGHV locus allows for exceptions to the “two alleles per gene” rule (Ford et al. 2020; Mikocziova et al. 2020) owing to the existence of repeated segments. Additionally, sequencing and PCR errors may lead to misalignments (we set the upper bound for the fraction of erroneous allele assignments owing to sequencing/PCR errors to 5%) (Supplemental Fig. S6).
For each human individual of each data set in this study (HUMAN1, HUMAN2, and HUMAN3 data sets; see Methods, “Experimental immunoglobulin sequencing data”), we aimed to construct an individually restricted germline gene database that contained only those validated alleles that are present in the given individual. This restricted database is thus a subset of all available validated alleles present in IMGT and OGRDB.
In this section, we show an example of deriving such an individually restricting process via analyzing the HUMAN3 data set, as it is the most diverse one (99 unrelated individuals). The pipeline itself is described in the following.
For each IGHV (Supplemental Fig. S1A) and IGHJ (Supplemental Fig. S1B) gene and each individual, we calculated the fraction of sequencing reads (for the definition of a sequencing read, see Methods, “An approach to building personalized RGMs that are robust to allelic variability of IGHV genes”) assigned to the two most frequent alleles (top_1 and top_2), as well as the fraction of sequencing reads assigned to all remaining alleles (rest-fraction). This calculation exposed three genomic scenarios (Supplemental Fig. S1C):
Case 1: a very high fraction of reads (>90%) assigned to the most frequent allele and negligible fractions of reads assigned to the lower frequency alleles. We assumed this scenario corresponds to an individual that is homozygous for this gene. This case was found for at least four IGHV genes for 97 out of 99 individuals and for IGHJ1–IGHJ5 genes for all individuals. The IGHJ6 gene belonged to this case for 44 out of 99 individuals.
Case 2: nonnegligible fractions (>5%) of sequencing reads assigned to the first and the second most frequent alleles and a very low rest-fraction (<5%), suggesting a heterozygous individual. This case was found for at least one IGHV gene for 93 out of 99 individuals. The only IGHJ gene this case was found for was IGHJ6 (for 54 out of 99 individuals).
Case 3: a high rest-fraction (>5%), suggesting that this individual has more than two alleles of the current gene (excluding errors leading to a systematic misalignment). This case was found for at least one IGHV gene for 93 individuals. No IGHJ gene belonged to case 3 (Supplemental Fig. S1B), which may be explained by the fact that the IGHJ locus is composed of fewer genes and fewer alleles per gene (Watson and Breden 2012; Gidoni et al. 2019; Peres et al. 2019).
To illustrate case 3, we visualized the allelic complexity of the IGHV1-69 gene (Supplemental Fig. S1D–F), known to be crucial for generating potent neutralizing antibodies (Avnir et al. 2016; Brouwer et al. 2020). Specifically, in individual 20 of the HUMAN3 data set, sequencing reads were aligned to eight alleles of IGHV1-69 (gene 69 in IGHV gene family 1). IGHV1-69 has a known duplication in the IMGT database named IGHV1-69D (Giudicelli et al. 2004), and the chromosomal locations of many alleles of IGHV1-69 (D) are yet to be described. This means that the duplication and the original gene may share alleles (e.g., IGHV1-69*01) (see Supplemental Fig. S1D). It is not feasible to determine the origin of the shared allele from a recombined VDJ sequence, so it is de facto possible to observe three different alleles of IGHV1-69: two originating from the gene itself and the third one from the duplication. In the aforementioned individual 20, only three alleles out of eight had above-the-threshold fractions of the sequencing reads annotated with IGHV1-69: IGHV1-69*01, IGHV1-69*04, and IGHV1-69*06, which is exactly the case described in the previous sentence. IGHV1-69*04 and IGHV1-69*06 might have originated from the gene IGHV1-69, whereas IGHV1-69*01 might be, in fact, IGHV1-69D*01 having originated from a duplication.
IGHV1-69 has 19 alleles of this gene, some of which are very close to each other, namely, differing by a single nucleotide substitution (Supplemental Fig. S1E). There were five individuals (i.e., individuals 46, 47, 65, 83, 87) that had four alleles (Supplemental Fig. S1F) with fractions above the 5% threshold (i.e., each of these alleles accounted for >5% of the sequencing reads assigned to IGHV1-69), 37 individuals with three frequent alleles, and 34 individuals with two frequent alleles (which can signify a normal heterozygous gene or a homozygous with a mutated copy). The remaining individuals had a single IGHV1-69 allele with a frequency above the threshold. To summarize, human Ig AIRR-seq samples can have more than two alleles of the same IGHV gene, and one should treat these cases separately to avoid systematic biases in the fitted RGMs or to ensure that these systematic biases are the same for all considered AIRR-seq samples, which would allow comparing the RGMPs inferred from these samples.
Finally, we individually restricted germline gene databases as follows (see Supplemental Fig. S2): We set the maximum possible number of alleles (which we denoted by k) for each gene individually (Supplemental Table 1) based on the information on its potential copy number, duplications in the databases, and the high rest-fraction from Supplemental Figure S1A and trimmed all alleles after the second CYS, amino acid position 104 in the IMGT unique numbering scheme (Lefranc et al. 2003). Then, we determined the k most frequent alleles of the given gene in the sample in order to restrict the RGM to these alleles only.
To visualize the effect of this preprocessing pipeline on the fractions of the remaining alleles (i.e., not the two or the k most frequent ones), we recomputed the fractions from Supplemental Figure S1A after trimming the alleles and setting the per-gene k values but before restricting the set of alleles to the k most frequent ones (Supplemental Fig. S1G). This procedure led to nonzero rest-fractions <5% in the majority of cases, suggesting that we successfully eliminated (most of) the systematic alignment bias. The last step of the pipeline consisted of restricting the allele database and realigning the sequencing reads, which eliminated rest score assignments (Supplemental Fig. S5). This preprocessed data set was used for all Pgen inferences.
Of note, when we applied the described workflow to the HUMAN2 data set, the individually restricted allele database of each MZ twin was slightly different owing to the presence of weakly expressed alleles. However, the allele sets of MZ twins were more similar than those of unrelated individuals.
To reduce the amount of noise caused by PCR amplification, we assembled the clone contigs using MiXCR (Bolotin et al. 2015). So as not to lose the information contained in the clone frequencies, we multiplied each clone by the number of reads used to construct it. We refer to these multiplied clone nucleotide sequences as “sequencing reads.” With this approach, RGMP inference expectedly depends on the clone size distribution, but the downstream analysis is affected only to a limited extent. By calculating clone distribution evenness profiles (Greiff et al. 2015), we showed that controlling for the noise using replicate samples allows us to separate this effect from the systematic differences in RGMP values (Supplemental Fig. S15).
Using JSD to compare RGMPs
To statistically model VDJ recombination, we used the IGoR tool (Marcou et al. 2018). IGoR allows building custom VDJ recombination pipelines (i.e., to define dependencies between recombination events); we used the following pipeline for simulation of IgH VDJ recombination, where each step defines a subset of the model parameters:
A V segment is chosen. Thus, each of the V germline segments from the database is assigned with a choice probability (forming a vector of #V parameters). For the human model, we considered 124 unique V genes. For the mouse model, we considered all 238 V gene alleles.
Depending on the V segment, a J segment is chosen. Each of the J segments is thus assigned with #V conditional choice probabilities (forming a parameter matrix of size #V × #J; we considered six unique J genes for the human model, four unique genes for the mouse model).
Depending on the V and J segments, a D segment is chosen, Each of the D segments is assigned with #V × #J conditional choice probabilities (forming a three-dimensional parameter tensor of size #V × #J × #D; 35 unique D genes for the human model, 25 unique genes for the mouse model).
Depending on the V segment, the V deletion length is determined. Thus, for each V segment, a discrete distribution is assigned—deletion profile (forming a parameter matrix of size #V × maximum deletion length = 41).
Analogously, depending on the J segment, the J deletion length is determined, and for each of the J segments, a discrete distribution is assigned (forming a parameter matrix of size #J × maximum deletion length = 41).
Depending on the D segment, the D3′ deletion length is determined, after which, depending on the D segment and the D3′ deletion length, the D5′ deletion length is determined. This way, each of the D segments is assigned with a D3′ deletion length distribution and D3′|D5′ deletion length distribution (forming a parameter matrix of size #D × maximum deletion length and a three-dimensional parameter tensor of size #D × maximum deletion length × maximum deletion length = 41).
VD insertion length and nucleotide composition are determined (length probabilities forming a vector of length maximum insertion length = 41).
VD insertion nucleotide composition is determined (via a Markov chain with 16 parameters).
DJ insertion length and nucleotide composition are determined (length probabilities forming a vector of length maximum insertion length = 41).
DJ insertion nucleotide composition is determined (via a Markov chain with 16 parameters).
This way, a whole set of IGoR model parameter values defines a multivariate discrete probability distribution.
KLD (Kullback and Leibler 1951) measures the difference between two probability distributions, P and Q. For discrete probability distributions defined on the same probability space X, the KLD is defined as . Marcou et al. (2018) calculated the KLD of the IGoR-estimated model parameter values to the ground truth ones to validate the IGoR inference module. We argue that a similar method can be used for comparing IGoR models inferred from different AIRR-seq samples. We used the JSD, which is a smoothed symmetric version of the KLD: , where .
JSD is symmetric and nonnegative, and owing to the smoothing, its square root satisfies the triangle inequality. Thus, the square root of the JSD can be used as a distance between IGoR models, as well as between AIRR-seq samples the models were inferred from.
The KLD (and, hence, the JSD) between two multivariate distributions can be decomposed into a sum of several components if components of the distributions are conditionally independent. Applied to the IGoR RGM, this means that the JSD (“explicit JSD”) between two IGoR models can be decomposed into seven additive terms.
The explicit JSD between two sets of IGoR parameter models P1 and P2 can be then written as the sum of the JSDs of the joint probabilities of conditionally independent recombination events:
We computed a lower bound for the variation of the explicit JSD introduced by the noise by recreating this variation in the most controlled setting: For each data set, for a given sample size of N sequencing reads (N in [1000, 3000, 10,000, 30,000]), we used an IGoR model M_0 (inferred from the first experimental sample of each data set), generated 10 pairs of synthetic samples of N sequencing reads each, and computed the explicit JSD of the IGoR-inferred RGMP values from these sequencing reads, within the pairs (synthetic replicates on Fig. 2A). Thus, we obtained a set of values of explicit JSD between data sets generated using the same theoretical RGMP. To obtain the normalized JSD between two RGMP sets, we divide their explicit JSD by the mean of the explicit JSDs obtained using generated synthetic samples. We used the same precomputed synthetic samples within one species: If the normalized JSD for RGMP sets inferred from samples E1 and E2 is to be calculated, we set normalizedJSD(E1, E2) = explicitJSD(E1, E2)/meani,j explicitJSD(Si, Sj), where the same synthetic replicates Si are used for any samples for a given species (human/mouse).
For the human data, we applied the allele preprocessing pipeline (Supplemental Fig. S5) to remove the systematic bias in the model inference. Subsequently, we summed the parameters corresponding to alleles of the same gene and compared different subjects on the gene, not on the allele level. We did not apply the allele preprocessing workflow for the murine data, because all subjects share by definition (inbred mice) identical germline gene sets.
In this study, we limited the maximum sample size to 30,000 sequencing reads owing to the high time and memory consumption by IGoR (inferring RGMP values from all considered samples took about 300,000 CPU hours).
A statistical test for comparing repertoire generation models
To test if the RGMP sets for two AIRR-seq samples A (containing N sequencing reads) and B (containing K sequencing reads) are indistinguishable, namely, that the explicit JSD (or the normalized JSD, because they differ by a constant) between the models inferred from subsamples of these two samples was not higher than between data replicates of A, we used the following procedure. Without loss of generality, we assume that N ≤ K.
First, we split sample A into two nonoverlapping samples A′ and A′′, both of size N/2 sequencing reads. Then, we sample N/2 sequencing reads from B to obtain samples B′. After that, for a fixed subsample size S (S in [1000, 3000, 10,000, 30,000]) sequencing reads, we performed the following:
- Subsampling (without replacement) of S sequencing reads from the samples A′ and A′′ each 30 times independently; this yields 30 nonoverlapping pairs and hence 30 (identically distributed) measurements of the JSD between data replicates of A (the explicit JSD of the models inferred from the samples within one pair).
- Subsampling of S sequencing reads from the samples A′ and B′ each 30 times independently; this yields 30 nonoverlapping pairs and hence 30 independent measurements of the explicit JSD between subsamples of A and B (the explicit JSD of the models inferred from the samples within one pair).
We used the null hypothesis that samples A and B are homogeneous, namely, that the expected values of the RGMP inferred from them are equal. Consequently, the expected values of the RGMP inferred from subsamples of A′, A′′, and B′ are equal. Under the null hypothesis, the distance between model parameters values inferred from subsamples of A′ and A′′ will not significantly differ from the distance between model parameters values inferred from subsamples of A′ and B′. Hence, the mean of the distribution of the explicit JSD between A′ and A′′ models will not differ from that of A′ and B′ models. We used the unpaired Student's t-test to check if the null hypothesis holds.
To investigate the properties of our test, we repeated the test procedure 30 times (Supplemental Fig. S4A) for pre–B cell data for data replicates and for murine twin subjects (samples 1 and 2 from the MOUSE_PRE data set) for the subsample size 3000 sequencing reads. We used the sample size 3000 owing to the high computational cost of the procedure. In the first case (i.e., when the null hypothesis was known to hold), the P-value distribution resembled a uniform distribution (Supplemental Fig. S4B); in the second case, when the null hypothesis supposedly did not hold, the P-values were highly skewed toward zero (Supplemental Fig. S4C).
Software
We used IGoR v1.4.0 (Marcou et al. 2018; https://github.com/qmarcou/IGoR) for RGMP inference, nucleotide sequence Pgen evaluation and generation of synthetic AIRR-seq data, and OLGA v1.2.3 (Sethna et al. 2019) for amino acid sequence Pgen evaluation. We used MiXCR v3.0.12 (Bolotin et al. 2015) for AIRR-seq data preprocessing and annotation.
Graphics
For visualization, we used the following Python packages: Matplotlib v3.3.2 (Hunter 2007) and Seaborn v0.11.0 (https://zenodo.org/record/4019146#.X3xdf1lRUxg).
Hardware
Computations were performed on a dedicated server as well as the high-performance computing cluster FRAM (Norwegian e-infrastructure for Research and Education sigma2.no/fram).
Data access
IGoR parameter files for all RGMs analyzed in this paper, along with the FASTA files that contain the sequences these RGMPs were inferred from, can be downloaded from the NIRD Research Data Archive (https://archive.norstore.no/) at doi.org/10.11582/2021.00089 (see also supplemental data set, https://archive.sigma2.no/pages/public/datasetDetail.jsf?id=10.11582/2021.00089 [accessed October 12, 2021]). The code used to generate the results in this paper can be found as Supplemental Code and at GitHub (https://github.com/csi-greifflab/desygnator).
Supplementary Material
Acknowledgments
We thank Dr. Corey Watson, Dr. Gur Yaari, and Dr. Julien Limenitakis for productive discussions and for their valuable feedback that has helped improve this study. We acknowledge generous support by The Leona M. and Harry B. Helmsley Charitable Trust (#2019PG-T1D011, to V.G.), UiO World-Leading Research Community (to V.G. and L.M.S.), UiO:LifeScience Convergence Environment Immunolingo (to V.G. and G.K.S.), EU Horizon 2020 iReceptorplus (#825821, to V.G. and L.M.S.), a Research Council of Norway FRIPRO project (#300740, to V.G.), and a Research Council of Norway IKTPLUSS project (#311341, to V.G. and G.K.S.).
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at https://www.genome.org/cgi/doi/10.1101/gr.275373.121.
Freely available online through the Genome Research Open Access option.
Competing interest statement
V.G. declares advisory board positions in aiNET and Enpicom. V.G. is a consultant for Roche/Genentech.
References
- Akbar R, Robert PA, Pavlović M, Jeliazkov JR, Snapkov I, Slabodkin A, Weber CR, Scheffer L, Miho E, Haff IH, et al. 2021a. A compact vocabulary of paratope-epitope interactions enables predictability of antibody-antigen binding. Cell Rep 34: 108856. 10.1016/j.celrep.2021.108856 [DOI] [PubMed] [Google Scholar]
- Akbar R, Robert PA, Weber CR, Widrich M, Frank R, Pavlović M, Scheffer L, Chernigovskaya M, Snapkov I, Slabodkin A, et al. 2021b. In silico proof of principle of machine learning-based antibody design at unconstrained scale. bioRxiv 10.1101/2021.07.08.451480 [DOI] [PMC free article] [PubMed]
- Arora R, Burke HM, Arnaout R. 2018. Immunological diversity with similarity. bioRxiv 10.1101/483131 [DOI]
- Avnir Y, Watson CT, Glanville J, Peterson EC, Tallarico AS, Bennett AS, Qin K, Fu Y, Huang C-Y, Beigel JH, et al. 2016. IGHV1-69 polymorphism modulates anti-influenza antibody repertoires, correlates with IGHV utilization shifts and varies by ethnicity. Sci Rep 6: 20842. 10.1038/srep20842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barennes P, Quiniou V, Shugay M, Egorov ES, Davydov AN, Chudakov DM, Uddin I, Ismail M, Oakes T, Chain B, et al. 2021. Benchmarking of T cell receptor repertoire profiling methods reveals large systematic biases. Nat Biotechnol 39: 236–245. 10.1038/s41587-020-0656-3 [DOI] [PubMed] [Google Scholar]
- Bernat NV, Corcoran M, Nowak I, Kaduk M, Dopico XC, Narang S, Maisonasse P, Dereuddre-Bosquet N, Murrell B, Hedestam GBK. 2021. Rhesus and cynomolgus macaque immunoglobulin heavy-chain genotyping yields comprehensive databases of germline VDJ alleles. Immunity 54: 355–366.e4. 10.1016/j.immuni.2020.12.018 [DOI] [PubMed] [Google Scholar]
- Bolen CR, Rubelt F, Vander Heiden JA, Davis MM. 2017. The repertoire dissimilarity index as a method to compare lymphocyte receptor repertoires. BMC Bioinformatics 18: 155. 10.1186/s12859-017-1556-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolotin DA, Poslavsky S, Mitrophanov I, Shugay M, Mamedov IZ, Putintseva EV, Chudakov DM. 2015. MiXCR: software for comprehensive adaptive immunity profiling. Nat Methods 12: 380–381. 10.1038/nmeth.3364 [DOI] [PubMed] [Google Scholar]
- Briney B, Inderbitzin A, Joyce C, Burton DR. 2019. Commonality despite exceptional diversity in the baseline human antibody repertoire. Nature 566: 393–397. 10.1038/s41586-019-0879-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brouwer PJM, Caniels TG, van der Straten K, Snitselaar JL, Aldon Y, Bangaru S, Torres JL, Okba NMA, Claireaux M, Kerster G, et al. 2020. Potent neutralizing antibodies from COVID-19 patients define multiple targets of vulnerability. Science 369: 643–650. 10.1126/science.abc5902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi X, Li Y, Qiu X. 2020. V(D)J recombination, somatic hypermutation and class switch recombination of immunoglobulins: mechanism and regulation. Immunology 160: 233–247. 10.1111/imm.13176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins AM, Yaari G, Shepherd AJ, Lees W, Watson CT. 2020. Germline immunoglobulin genes: disease susceptibility genes hidden in plain sight? Curr Opin Syst Biol 24: 100–108. 10.1016/j.coisb.2020.10.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins AM, Peres A, Corcoran MM, Watson CT, Yaari G, Lees WD, Ohlin M. 2021. Commentary on Population matched (pm) germline allelic variants of immunoglobulin (IG) loci: relevance in infectious diseases and vaccination studies in human populations. Genes Immun 10.1038/s41435-021-00152-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corcoran MM, Phad GE, Bernat NV, Stahl-Hennig C, Sumida N, Persson MAA, Martin M, Hedestam GBK. 2016. Production of individualized V gene databases reveals high levels of immunoglobulin genetic diversity. Nat Commun 7: 13642. 10.1038/ncomms13642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidsen K, Olson BJ, DeWitt WS III, Feng J, Harkins E, Bradley P, Matsen FA IV. 2019. Deep generative models for T cell receptor protein sequences. eLife 8: e46935. 10.7554/eLife.46935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Greef PC, de Boer RJ. 2021. TCRβ rearrangements without a D segment are common, abundant, and public. Proc Natl Acad Sci 118: e2104367118. 10.1073/pnas.2104367118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desponds J, Mayer A, Mora T, Walczak AM. 2021. Population dynamics of immune repertoires. In Mathematical, computational and experimental T cell immunology (ed. Molina-París C, Lythe G), pp. 203–221. Springer International Publishing, Cham, Switzerland. [Google Scholar]
- DeWitt WS III, Lindau P, Snyder TM, Sherwood AM, Vignali M, Carlson CS, Greenberg PD, Duerkopp N, Emerson RO, Robins HS. 2016. A public database of memory and naive B-cell receptor sequences. PLoS One 11: e0160853. 10.1371/journal.pone.0160853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupic T, Koraichi MB, Minervina AA, Pogorelyy MV, Mora T, Walczak AM. 2021. Immune fingerprinting through repertoire similarity. PLoS Genet 17: e1009301. 10.1371/journal.pgen.1009301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elhanati Y, Sethna Z, Callan CG, Mora T, Walczak AM. 2018. Predicting the spectrum of TCR repertoire sharing with a data-driven model of recombination. Immunol Rev 284: 167–179. 10.1111/imr.12665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emerson RO, DeWitt WS III, Vignali M, Gravley J, Hu JK, Osborne EJ, Desmarais C, Klinger M, Carlson CS, Hansen JA, et al. 2017. Immunosequencing identifies signatures of cytomegalovirus exposure history and HLA-mediated effects on the T cell repertoire. Nat Genet 49: 659–665. 10.1038/ng.3822 [DOI] [PubMed] [Google Scholar]
- Ford M, Haghshenas E, Watson CT, Sahinalp SC. 2020. Genotyping and copy number analysis of immunoglobin heavy chain variable genes using long reads. iScience 23: 100883. 10.1016/j.isci.2020.100883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraga MF, Ballestar E, Paz MF, Ropero S, Setien F, Ballestar ML, Heine-Suñer D, Cigudosa JC, Urioste M, Benitez J, et al. 2005. Epigenetic differences arise during the lifetime of monozygotic twins. Proc Natl Acad Sci 102: 10604–10609. 10.1073/pnas.0500398102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedensohn S, Neumeier D, Khan TA, Csepregi L, Parola C, de Vries ARG, Erlach L, Mason DM, Reddy ST. 2020. Convergent selection in antibody repertoires is revealed by deep learning. bioRxiv 10.1101/2020.02.25.965673 [DOI]
- Gadala-Maria D, Gidoni M, Marquez S, Vander Heiden JA, Kos JT, Watson CT, O'Connor KC, Yaari G, Kleinstein SH. 2019. Identification of subject-specific immunoglobulin alleles from expressed repertoire sequencing data. Front Immunol 10: 129. 10.3389/fimmu.2019.00129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gidoni M, Snir O, Peres A, Polak P, Lindeman I, Mikocziova I, Sarna VK, Lundin KEA, Clouser C, Vigneault F, et al. 2019. Mosaic deletion patterns of the human antibody heavy chain gene locus shown by Bayesian haplotyping. Nat Commun 10: 628. 10.1038/s41467-019-08489-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giudicelli V, Chaume D, Lefranc M-P. 2004. IMGT/GENE-DB: a comprehensive database for human and mouse immunoglobulin and T cell receptor genes. Nucleic Acids Res 33: D256–D261. 10.1093/nar/gki010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glanville J, Kuo TC, von Büdingen H-C, Guey L, Berka J, Sundar PD, Huerta G, Mehta GR, Oksenberg JR, Hauser SL, et al. 2011. Naive antibody gene-segment frequencies are heritable and unaltered by chronic lymphocyte ablation. Proc Natl Acad Sci 108: 20066–20071. 10.1073/pnas.1107498108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greiff V, Bhat P, Cook SC, Menzel U, Kang W, Reddy ST. 2015. A bioinformatic framework for immune repertoire diversity profiling enables detection of immunological status. Genome Med 7: 49. 10.1186/s13073-015-0169-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greiff V, Menzel U, Miho E, Weber C, Riedel R, Cook S, Valai A, Lopes T, Radbruch A, Winkler TH, et al. 2017a. Systems analysis reveals high genetic and antigen-driven predetermination of antibody repertoires throughout B cell development. Cell Rep 19: 1467–1478. 10.1016/j.celrep.2017.04.054 [DOI] [PubMed] [Google Scholar]
- Greiff V, Weber CR, Palme J, Bodenhofer U, Miho E, Menzel U, Reddy ST. 2017b. Learning the high-dimensional immunogenomic features that predict public and private antibody repertoires. J Immunol 199: 2985–2997. 10.4049/jimmunol.1700594 [DOI] [PubMed] [Google Scholar]
- Greiff V, Yaari G, Cowell LG. 2020. Mining adaptive immune receptor repertoires for biological and clinical information using machine learning. Curr Opin Syst Biol 24: 109–119. 10.1016/j.coisb.2020.10.010 [DOI] [Google Scholar]
- Hunter JD. 2007. Matplotlib: a 2D graphics environment. Comput Sci Eng 9: 90–95. 10.1109/MCSE.2007.55 [DOI] [Google Scholar]
- Isacchini G, Walczak AM, Mora T, Nourmohammad A. 2021. Deep generative selection models of T and B cell receptor repertoires with soNNia. Proc Natl Acad Sci 118: e2023141118. 10.1073/pnas.2023141118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanduri C, Pavlović M, Scheffer L, Motwani K, Chernigovskaya M, Greiff V, Sandve GK. 2021. Profiling the baseline performance and limits of machine learning models for adaptive immune receptor repertoire classification. bioRxiv 10.1101/2021.05.23.445346 [DOI] [PMC free article] [PubMed]
- Kenter AL, Watson CT, Spille J-H. 2021. Igh locus polymorphism may dictate topological chromatin conformation and V gene usage in the Ig repertoire. Front Immunol 12: 682589. 10.3389/fimmu.2021.682589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan TA, Friedensohn S, de Vries ARG, Straszewski J, Ruscheweyh H-J, Reddy ST. 2016. Accurate and predictive antibody repertoire profiling by molecular amplification fingerprinting. Sci Adv 2: e1501371. 10.1126/sciadv.1501371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khatri I, Berkowska MA, van den Akker EB, Teodosio C, Reinders MJT, van Dongen JJM. 2021. Population matched (pm) germline allelic variants of immunoglobulin (IG) loci: relevance in infectious diseases and vaccination studies in human populations. Genes Immun 22: 172–186. 10.1038/s41435-021-00143-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koraichi MB, Touzel MP, Mora T, Walczak AM. 2021. NoisET: Noise learning and Expansion detection of T-cell receptors with Python. arXiv:2102.03568 [q-bio.GN]. [DOI] [PubMed]
- Kullback S, Leibler RA. 1951. On information and sufficiency. Ann Math Stat 22: 79–86. 10.1214/aoms/1177729694 [DOI] [Google Scholar]
- Lee JH, Toy L, Kos JT, Safonova Y, Schief WR, Havenar-Daughton C, Watson CT, Crotty S. 2021. Vaccine genetics of IGHV1-2 VRC01-class broadly neutralizing antibody precursor naïve human B cells. NPJ Vaccines 6: 113. 10.1038/s41541-020-00265-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lees W, Busse CE, Corcoran M, Ohlin M, Scheepers C, Matsen FA IV, Yaari G, Watson CT, Collins A, Shepherd AJ. 2020. OGRDB: a reference database of inferred immune receptor genes. Nucleic Acids Res 48: D964–D970. 10.1093/nar/gkz822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefranc M-P. 2001. IMGT, the International imMunoGeneTics database. Nucleic Acids Res 29: 207–209. 10.1093/nar/29.1.207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefranc M-P, Pommié C, Ruiz M, Giudicelli V, Foulquier E, Truong L, Thouvenin-Contet V, Lefranc G. 2003. IMGT unique numbering for immunoglobulin and T cell receptor variable domains and Ig superfamily V-like domains. Dev Comp Immunol 27: 55–77. 10.1016/S0145-305X(02)00039-3 [DOI] [PubMed] [Google Scholar]
- Madi A, Shifrut E, Reich-Zeliger S, Gal H, Best K, Ndifon W, Chain B, Cohen IR, Friedman N. 2014. T-cell receptor repertoires share a restricted set of public and abundant CDR3 sequences that are associated with self-related immunity. Genome Res 24: 1603–1612. 10.1101/gr.170753.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcou Q, Mora T, Walczak AM. 2018. High-throughput immune repertoire analysis with IGoR. Nat Commun 9: 561. 10.1038/s41467-018-02832-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martins FR, de Melo Pontes LA, de Oliveira Mendes TA, Felicori LF. 2021. Discovery of 10,828 new putative human immunoglobulin heavy chain IGHV variants. bioRxiv 10.1101/2021.01.15.426262 [DOI]
- Menzel U, Greiff V, Khan TA, Haessler U, Hellmann I, Friedensohn S, Cook SC, Pogson M, Reddy ST. 2014. Comprehensive evaluation and optimization of amplicon library preparation methods for high-throughput antibody sequencing. PLoS One 9: e96727. 10.1371/journal.pone.0096727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miho E, Yermanos A, Weber CR, Berger CT, Reddy ST, Greiff V. 2018. Computational strategies for dissecting the high-dimensional complexity of adaptive immune repertoires. Front Immunol 9: 224. 10.3389/fimmu.2018.00224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miho E, Roškar R, Greiff V, Reddy ST. 2019. Large-scale network analysis reveals the sequence space architecture of antibody repertoires. Nat Commun 10: 1321. 10.1038/s41467-019-09278-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikocziova I, Gidoni M, Lindeman I, Peres A, Snir O, Yaari G, Sollid LM. 2020. Polymorphisms in human immunoglobulin heavy chain variable genes and their upstream regions. Nucleic Acids Res 48: 5499–5510. 10.1093/nar/gkaa310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikocziova I, Greiff V, Sollid LM. 2021a. Immunoglobulin germline gene variation and its impact on human disease. Genes Immun 22: 205–217. 10.1038/s41435-021-00145-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikocziova I, Peres A, Gidoni M, Greiff V, Yaari G, Sollid LM. 2021b. Germline polymorphisms and alternative splicing of human immunoglobulin light chain genes. iScience 24: 103192. 10.1016/j.isci.2021.103192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsunaga EM, Snyder MP. 2020. Deep characterization of the human antibody response to natural infection using longitudinal immune repertoire sequencing. Mol Cell Proteomics 19: 278–293. 10.1074/mcp.RA119.001633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müllner D. 2011. Modern hierarchical, agglomerative clustering algorithms. arXiv:1109.2378 [stat.ML].
- Nemazee D. 2017. Mechanisms of central tolerance for B cells. Nat Rev Immunol 17: 281–294. 10.1038/nri.2017.19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolan S, Vignali M, Klinger M, Dines JN, Kaplan IM, Svejnoha E, Craft T, Boland K, Pesesky M, Gittelman RM, et al. 2020. A large-scale database of T-cell receptor β (TCRβ) sequences and binding associations from natural and synthetic exposure to SARS-CoV-2. Res Sq [Preprint] rs.3.rs-51964. 10.21203/rs.3.rs-51964/v1 [DOI] [Google Scholar]
- Olson BJ, Matsen FA IV. 2018. The Bayesian optimist's guide to adaptive immune receptor repertoire analysis. Immunol Rev 284: 148–166. 10.1111/imr.12664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omer A, Peres A, Rodrigues OL, Watson CT, Lees W, Polak P, Collins AM, Yaari G. 2021. T cell receptor β (TRB) germline variability is revealed by inference from repertoire data. bioRxiv 10.1101/2021.05.17.444409 [DOI] [PMC free article] [PubMed]
- Pal S, Tyler JK. 2016. Epigenetics and aging. Sci Adv 2: e1600584. 10.1126/sciadv.1600584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parks T, Mirabel MM, Kado J, Auckland K, Nowak J, Rautanen A, Mentzer AJ, Marijon E, Jouven X, Perman ML, et al. 2017. Association between a common immunoglobulin heavy chain allele and rheumatic heart disease risk in Oceania. Nat Commun 8: 14946. 10.1038/ncomms14946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlović M, Scheffer L, Motwani K, Kanduri C, Kompova R, Vazov N, Waagan K, Bernal FLM, Costa AA, Corrie B, et al. 2021. The immuneML ecosystem for machine learning analysis of adaptive immune receptor repertoires. Nat Mach Intell 10.1038/s42256-021-00413-z [DOI] [PMC free article] [PubMed]
- Perelson AS, Oster GF. 1979. Theoretical studies of clonal selection: minimal antibody repertoire size and reliability of self-non-self discrimination. J Theor Biol 81: 645–670. 10.1016/0022-5193(79)90275-3 [DOI] [PubMed] [Google Scholar]
- Peres A, Gidoni M, Polak P, Yaari G. 2019. RAbHIT: R antibody haplotype inference tool. Bioinforma Oxf Engl 35: 4840–4842. 10.1093/bioinformatics/btz481 [DOI] [PubMed] [Google Scholar]
- Puelma Touzel M, Walczak AM, Mora T. 2020. Inferring the immune response from repertoire sequencing. PLoS Comput Biol 16: e1007873. 10.1371/journal.pcbi.1007873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulivarthy SR, Lion M, Kuzu G, Matthews AGW, Borowsky ML, Morris J, Kingston RE, Dennis JH, Tolstorukov MY, Oettinger MA. 2016. Regulated large-scale nucleosome density patterns and precise nucleosome positioning correlate with V(D)J recombination. Proc Natl Acad Sci 113: E6427–E6436. 10.1073/pnas.1605543113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ralph DK, Matsen FA IV. 2016. Likelihood-based inference of B cell clonal families. PLoS Comput Biol 12: e1005086. 10.1371/journal.pcbi.1005086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raposo B, Dobritzsch D, Ge C, Ekman D, Xu B, Lindh I, Förster M, Uysal H, Nandakumar KS, Schneider G, et al. 2014. Epitope-specific antibody response is controlled by immunoglobulin VH polymorphisms. J Exp Med 211: 405–411. 10.1084/jem.20130968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raybould MIJ, Kovaltsuk A, Marks C, Deane CM. 2020. CoV-AbDab: the coronavirus antibody database. Bioinformatics 35: 734–735. 10.1093/bioinformatics/btaa739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remmel JL, Ackerman ME. 2021. Rationalizing random walks: replicating protective antibody trajectories. Trends Immunol 42: 186–197. 10.1016/j.it.2021.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert PA, Akbar R, Frank R, Pavlović M, Widrich M, Snapkov I, Chernigovskaya M, Scheffer L, Slabodkin A, Mehta BB, et al. 2021a. One billion synthetic 3D-antibody-antigen complexes enable unconstrained machine-learning formalized investigation of antibody specificity prediction. bioRxiv 10.1101/2021.07.06.451258 [DOI]
- Robert PA, Arulraj T, Meyer-Hermann M. 2021b. Ymir: a 3D structural affinity model for multi-epitope vaccine simulations. iScience 24: 102979. 10.1016/j.isci.2021.102979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert PA, Kunze-Schumacher H, Greiff V, Krueger A. 2021c. Modeling the dynamics of T-cell development in the thymus. Entropy 23: 437. 10.3390/e23040437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez OL, Gibson WS, Parks T, Emery M, Powell J, Strahl M, Deikus G, Auckland K, Eichler EE, Marasco WA, et al. 2020. A novel framework for characterizing genomic haplotype diversity in the human immunoglobulin heavy chain locus. Front Immunol 11: 2136. 10.3389/fimmu.2020.02136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy B, Neumann RS, Snir O, Iversen R, Sandve GK, Lundin KEA, Sollid LM. 2017. High-throughput single-cell analysis of B cell receptor usage among autoantigen-specific plasma cells in celiac disease. J Immunol 199: 782–791. 10.4049/jimmunol.1700169 [DOI] [PubMed] [Google Scholar]
- Rubelt F, Bolen CR, McGuire HM, Heiden JAV, Gadala-Maria D, Levin M, Euskirchen GM, Mamedov MR, Swan GE, Dekker CL, et al. 2016. Individual heritable differences result in unique cell lymphocyte receptor repertoires of naïve and antigen-experienced cells. Nat Commun 7: 11112. 10.1038/ncomms11112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell ML, Souquette A, Levine DM, Allen EK, Kuan G, Simon N, Balmaseda A, Gordon A, Thomas P, Matsen FA, et al. 2021. Combining genotypes and T cell receptor distributions to infer genetic loci determining V(D)J recombination probabilities. bioRxiv 10.1101/2021.09.17.460747 [DOI] [PMC free article] [PubMed]
- Safonova Y, Pevzner PA. 2020. V(DD)J recombination is an important and evolutionarily conserved mechanism for generating antibodies with unusually long CDR3s. Genome Res 30: 1547–1558. 10.1101/gr.259598.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sangesland M, Ronsard L, Kazer SW, Bals J, Boyoglu-Barnum S, Yousif AS, Barnes R, Feldman J, Quirindongo-Crespo M, McTamney PM, et al. 2019. Germline-encoded affinity for cognate antigen enables vaccine amplification of a human broadly neutralizing response against influenza virus. Immunity 51: 735–749.e8. 10.1016/j.immuni.2019.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sangesland M, Yousif AS, Ronsard L, Kazer SW, Zhu AL, Gatter GJ, Hayward MR, Barnes RM, Quirindongo-Crespo M, Rohrer D, et al. 2020. A single human VH-gene allows for a broad-spectrum antibody response targeting bacterial lipopolysaccharides in the blood. Cell Rep 32: 108065. 10.1016/j.celrep.2020.108065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sethna Z, Elhanati Y, Callan CG, Walczak AM, Mora T. 2019. OLGA: fast computation of generation probabilities of B- and T-cell receptor amino acid sequences and motifs. Bioinformatics 35: 2974–2981. 10.1093/bioinformatics/btz035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sethna Z, Isacchini G, Dupic T, Mora T, Walczak AM, Elhanati Y. 2020. Population variability in the generation and selection of T-cell repertoires. PLoS Comput Biol 16: e1008394. 10.1371/journal.pcbi.1008394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shemesh O, Polak P, Lundin KEA, Sollid LM, Yaari G. 2021. Machine learning analysis of naïve B-cell receptor repertoires stratifies celiac disease patients and controls. Front Immunol 12: 627813. 10.3389/fimmu.2021.627813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidhom J-W, Larman HB, Pardoll DM, Baras AS. 2021. DeepTCR is a deep learning framework for revealing sequence concepts within T-cell repertoires. Nat Commun 12: 1605. 10.1038/s41467-021-21879-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern JNH, Yaari G, Vander Heiden JA, Church G, Donahue WF, Hintzen RQ, Huttner AJ, Laman JD, Nagra RM, Nylander A, et al. 2014. B cells populating the multiple sclerosis brain mature in the draining cervical lymph nodes. Sci Transl Med 6: 248ra107. 10.1126/scitranslmed.3008879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swindells MB, Porter CT, Couch M, Hurst J, Abhinandan KR, Nielsen JH, Macindoe G, Hetherington J, Martin ACR. 2017. Abysis: integrated antibody sequence and structure—management, analysis, and prediction. J Mol Biol 429: 356–364. 10.1016/j.jmb.2016.08.019 [DOI] [PubMed] [Google Scholar]
- Trück J, Eugster A, Barennes P, Tipton CM, Luning Prak ET, Bagnara D, Soto C, Sherkow JS, Payne AS, Lefranc M-P, et al. 2021. Biological controls for standardization and interpretation of adaptive immune receptor repertoire profiling. eLife 10: e66274. 10.7554/eLife.66274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vázquez Bernat N, Corcoran M, Hardt U, Kaduk M, Phad GE, Martin M, Karlsson Hedestam GB. 2019. High-quality library preparation for NGS-based immunoglobulin germline gene inference and repertoire expression analysis. Front Immunol 10: 660. 10.3389/fimmu.2019.00660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkataraman T, Valencia C, Mangino M, Morgenlander W, Clipman SJ, Liechti T, Valencia A, Christofidou P, Spector T, Roederer M, et al. 2021. Antiviral antibody epitope selection is a heritable trait. bioRxiv 10.1101/2021.03.25.436790 [DOI] [PMC free article] [PubMed]
- Wardemann H, Busse CE. 2019. Expression cloning of antibodies from single human B cells. In Lymphoma: methods and protocols: methods in molecular biology (ed. Küppers R), pp. 105–125. Springer, New York. [DOI] [PubMed] [Google Scholar]
- Watson CT, Breden F. 2012. The immunoglobulin heavy chain locus: genetic variation, missing data, and implications for human disease. Genes Immun 13: 363–373. 10.1038/gene.2012.12 [DOI] [PubMed] [Google Scholar]
- Watson CT, Glanville J, Marasco WA. 2017. The individual and population genetics of antibody immunity. Trends Immunol 38: 459–470. 10.1016/j.it.2017.04.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinstein JA, Jiang N, White RA, Fisher DS, Quake SR. 2009. High-throughput sequencing of the zebrafish antibody repertoire. Science 324: 807–810. 10.1126/science.1170020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Wang W, Chen Z, Lu S, Yang F, Bi Z, Bao L, Mo F, Li X, Huang Y, et al. 2020. A vaccine targeting the RBD of the S protein of SARS-CoV-2 induces protective immunity. Nature 586: 572–577. 10.1038/s41586-020-2599-8 [DOI] [PubMed] [Google Scholar]
- Yang X, Zhu Y, Chen S, Zeng H, Guan J, Wang Q, Lan C, Sun D, Yu X, Zhang Z. 2021. Novel allele detection tool benchmark and application with antibody repertoire sequencing dataset. Front Immunol 12: 739179. 10.3389/fimmu.2021.739179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye J, Ma N, Madden TL, Ostell JM. 2013. IgBLAST: an immunoglobulin variable domain sequence analysis tool. Nucleic Acids Res 41: W34–W40. 10.1093/nar/gkt382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y, Ceredig R, Seoighe C. 2017. A database of human immune receptor alleles recovered from population sequencing data. J Immunol 198: 2202–2210. 10.4049/jimmunol.1601710 [DOI] [PubMed] [Google Scholar]
- Zhang W, Wang I-M, Wang C, Lin L, Chai X, Wu J, Bett AJ, Dhanasekaran G, Casimiro DR, Liu X. 2016. IMPre: an accurate and efficient software for prediction of T- and B-cell receptor germline genes and alleles from rearranged repertoire data. Front Immunol 7: 457. 10.3389/fimmu.2016.00457 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.