Abstract
More than 80% of the wheat genome consists of transposable elements (TEs), which act as major drivers of wheat genome evolution. However, their contributions to the regulatory evolution of wheat adaptations remain largely unclear. Here, we created genome-binding maps for 53 transcription factors (TFs) underlying environmental responses by leveraging DAP-seq in Triticum urartu, together with epigenomic profiles. Most TF binding sites (TFBSs) located distally from genes are embedded in TEs, whose functional relevance is supported by purifying selection and active epigenomic features. About 24% of the non-TE TFBSs share significantly high sequence similarity with TE-embedded TFBSs. These non-TE TFBSs have almost no homologous sequences in non-Triticeae species and are potentially derived from Triticeae-specific TEs. The expansion of TE-derived TFBS linked to wheat-specific gene responses, suggesting TEs are an important driving force for regulatory innovations. Altogether, TEs have been significantly and continuously shaping regulatory networks related to wheat genome evolution and adaptation.
Transposable elements (TEs) account for over 80% of the wheat genome (Luo et al. 2017; The International Wheat Genome Sequencing Consortium (IWGSC) et al. 2018; Ling et al. 2018). Although TEs are often epigenetically silenced, they are sometimes activated by internal or external changes (Slotkin and Martienssen 2007; Lisch 2009, 2013; Dubin et al. 2018). Recent genome-wide studies on wheat revealed the effects of TEs on genomic diversity (Luo et al. 2017; The International Wheat Genome Sequencing Consortium (IWGSC) et al. 2018; Ling et al. 2018; Wicker et al. 2018), chromatin architecture (Gardiner et al. 2015; Li et al. 2019; Jordan et al. 2020), and higher-order structure (Jia et al. 2021). There are also reports in wheat describing the influence of TEs in promoters on the expression of nearby genes (Kashkush et al. 2003; Ramírez-González et al. 2018). However, the extent of the contribution of TEs to the ongoing evolution of transcriptional regulation as well as the evolution of regulatory TEs during wheat adaptations is largely unknown.
Recent genome-scale studies in both animals and plants identified domesticated TEs as regulatory elements or genes (Bennetzen and Wang 2014; Chuong et al. 2017). A comprehensive survey in human revealed ∼20% of TF binding sites (TFBSs) are embedded in TEs (Sundaram et al. 2014), indicative of the considerable contribution of TEs to regulatory networks. This proportion of TE-embedded TFBSs is likely underestimated given the progressive degeneration of TE sequences. Moreover, the low tolerance of mammals to dynamic TE insertions and deletions delays the acclimation process, and ongoing TE domestication is relatively rare (Simonti et al. 2017). In contrast, the majority of the wheat genome is composed of TEs, which underwent nearly complete turnover across three subgenomes (Wicker et al. 2018); in addition, some TE families are still active in wheat populations (Moore et al. 1991; Kashkush et al. 2003). This evidence is indicative of continuously active transpositions as well as the plasticity of the wheat genome. The expansion of TE families with TFBSs or relevant precursors have produced rich raw materials for the evolution and spread of cis-regulatory elements. The recent availability of high-quality genomic, transcriptomic, and epigenomic data makes wheat a good model plant for detecting ongoing TE domestication and dynamic regulatory innovation during evolution.
The profiling of genome-wide transcription factor (TF) binding sites is important for detecting regulatory elements and clarifying TF functions. Chromatin immunoprecipitation sequencing (ChIP-seq) is an efficient method for detecting TFBSs (Park 2009). However, ChIP experiments rely on specific antibodies or genetically modified marker strains to specifically precipitate TFs. Thus, they have been conducted mainly for several model organisms. DNA affinity purification sequencing (DAP-seq) is a recently developed alternative technique for characterizing genome-wide TF binding. It involves the in vitro expression of TFs and an incubation with a DNA library to probe the specific binding of TFs (Bartlett et al. 2017). The development of DAP-seq has enabled the high-throughput profiling of TF networks. Recent research on Arabidopsis and maize revealed the binding profiles of hundreds of TFs, thereby increasing our understanding of transcriptional networks (O'Malley et al. 2016; Galli et al. 2018).
Wheat provides ∼20% of the calories consumed by humans. The major production losses in wheat are caused mostly by abiotic stresses, including drought, salinity, and heat (Dvořák et al. 1993). Triticum urartu (diploid, AA) is the progenitor of the A subgenome of tetraploid (Triticum turgidum, AABB) and hexaploid (Triticum aestivum, AABBDD) wheat (Dvořák et al. 1993; Ling et al. 2018). Elucidating the stress-responsive transcriptional network of T. urartu (Tu) is useful for studying the regulatory network changes and evolution in wheat and also facilitates the identification of cis- and trans-regulatory factors relevant for the genetic improvement of wheat. We herein systematically profiled the genome-wide TF bindings underlying abiotic stress responses and revealed the rapid regulatory innovation related to wheat adaptation mediated by linage-specific TEs.
Results
DAP-seq profiling of TFs responsive to environmental stimuli
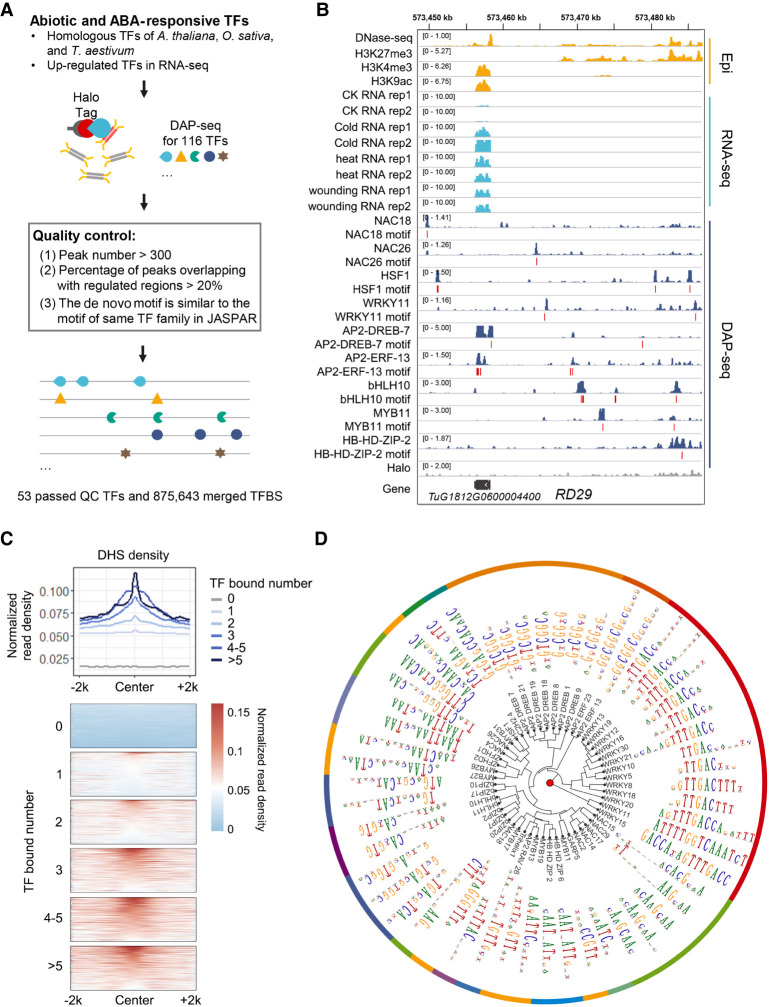
To construct the transcriptional regulatory network responsive to environmental stimuli in T. urartu, we profiled the genome-wide binding of a large spectrum of relevant TFs using DAP-seq technology (Fig. 1A; Bartlett et al. 2017). First, 107 TFs potentially responsive to environmental stimuli were collected based on publications, including TFs previously confirmed to be involved in abiotic stress responses in T. aestivum, the model dicotyledonous plant Arabidopsis thaliana, and the model monocotyledonous plant Oryza sativa (Supplemental Table S1). In addition, because abscisic acid (ABA) signaling pathways are typically triggered in response to abiotic stresses in higher plants (Cutler et al. 2010), another 77 TFs induced by both abiotic stresses and ABA revealed by an earlier RNA-seq analysis were also included (Supplemental Table S1). The binding profiles of 116 TFs from 19 families were successfully obtained; they were filtered based on the following criteria: peak number; representative motif; and distance to gene and regulatory elements as reflected by epigenetic activity. The TFs with few peaks and noncanonical representative motifs were classified as low confidence TFs. The DAP-seq data of 53 TFs (high and median quality) from 12 families were used for subsequent analyses (Supplemental Table S2). All DAP-seq data and peak files were deposited in a public database (see Data access).
Figure 1.
Genome-wide binding of wheat transcription factors underlying responses to environmental stimuli. (A) Schematic of the experimental design and filtering steps. The detailed filtering steps were listed in Supplemental Table S2. (B) Genomic tracks illustrating the targeting of RD29 by a subset of these TFs as well as locations of representative motifs. (C) DHS read density of TFBSs. TFBSs were grouped according to the number of binding TFs. DHS signal densities (bin size 50 bp) within a 4-kb window centered on merged TFBS centers. (D) Clustering of the top motif identified for each TF. Dendrogram based on motif similarity.
Figure 1B illustrates the genomic profiles of the binding of these TFs and the epigenetic architecture surrounding RD29, which is a typical marker gene used for monitoring stress-responsive pathways (Jia et al. 2012). The RD29 promoter region includes ABRE and DRE sequences, and the expression of this gene is regulated by multiple stress-responsive factors (Jia et al. 2012). The TFBSs bound by multiple TFs had higher regulatory activities as reflected by a higher level of chromatin openness (Fig. 1C). The most enriched motifs among the TFs were clustered according to sequence similarity (Fig. 1D). The motif sequences of TFs from the same family were similar and consistent with the motif associated with a given TF family in the database (Supplemental Fig. S1). These results suggested high reliability of the data. All data could be visualized through a customized genome browser (http://bioinfo.sibs.ac.cn/dap-seq_Tu_jbrowse/).
Enrichment of DAP-seq peaks in bivalent chromatin regions
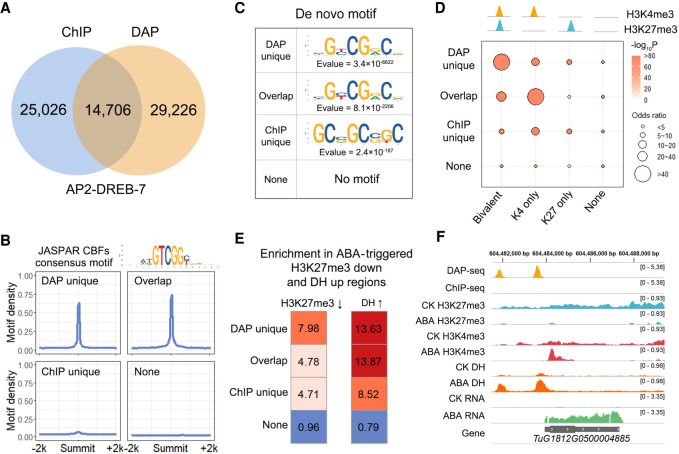
To compare the in vitro-derived DNA-binding profiles with those from in vivo experiments, we performed an AP2-DREB-7 ChIP-seq experiment involving protoplasts, the results of which were compared with the DAP-seq data. Co-occupation was observed for 37% of the ChIP-seq peaks and 34% of the DAP-seq peaks (Fig. 2A; Supplemental Fig. S2). We next compared the sequence and epigenetic features between common and unique peaks. The reported representative motif for AP2-DREB-7 binding was enriched in both DAP-seq unique and common peaks but not in the ChIP-seq unique peaks, suggestive of nondirect binding (Fig. 2B,C). A comparison with in vivo histone marks indicated that DAP-seq unique peaks were highly enriched in bivalent loci (occupied by repressive H3K27me3 and active H3K4me3 marks) (Fig. 2D). Bivalent loci are reportedly “prepared” for internal or external alterations (Ueda and Seki 2020). Under normal conditions, they are kept in a “poised” status. Following stimulation, H3K27me3 is removed, and the occupation of H3K4m3 ensures rapid activation. We hypothesized that the DAP-seq unique loci are occupied by histone marks under normal conditions but may be activated in response to external stimuli. To test this hypothesis, we performed H3K27me3 ChIP-seq and DHS-seq characterizing chromatin accessibility before and after an ABA treatment and compared H3K27me3 and DHS changes with DAP-seq and ChIP-seq binding sites. As expected, the DAP-seq unique peaks were highly enriched in H3K27me3-reduced and DHS-increased regions resulting from the ABA treatment (Fig. 2E). The genomic tracks of Figure 2F illustrated the ABA-triggered transcriptional and epigenetic changes surrounding an ABA-responsive gene, which is an AP2-DREB-7 DAP-seq unique target. In normal conditions, the gene was occupied by H3K27me3, which was removed following ABA treatment. To verify if this is also the case in other species, we obtained six Arabidopsis TF binding profiles with high-quality DAP-seq and ChIP-seq data with and without an ABA treatment. The DAP-seq unique peaks were highly enriched in the ABA-increased ChIP binding loci (Supplemental Fig. S3). These results implied that, compared with ChIP-seq, DAP-seq reflects the genome-wide direct binding potential, some of which is occupied by regulatory histone marks in vivo under normal conditions. In addition, integrating epigenetic information including chromatin openness and TF binding potential reflected by DAP-seq may help elucidate the functional significance of specific TFs.
Figure 2.
Concordance of DAP-seq and ChIP-seq peaks. (A) Venn diagrams showing the overlap between ChIP-seq peaks, DAP-seq peaks of AP2-DREB-7, and DHS. (B) Average number of AP2 motifs (bin size = 50 bp) within a 4-kb window centered on common and unique peak summits. The CBF binding motif in the JASPAR plant database is used because CBF is the orthologous gene of Tu AP2-DREB-7 in Arabidopsis. Four CBF binding motifs in the JASPAR database were merged into a consensus motif. (C) Motifs de novo identified from common and unique peaks. (D) Enrichment of histone marks in common and unique peaks. H3K4me3 and H3K27me3 overlapping (bivalent) peaks and unique peaks (K4-only and K27-only) were used for the analysis. (E) Enrichment of DAP-seq unique peaks in H3K27me3 down-regulated and DNase I hypersensitivity (DH) up-regulated regions by ABA. The MAnorm package (Shao et al. 2012) was used for the quantitative comparison of H3K27me3 ChIP-seq and DHS signals between samples. (F) Genomic tracks illustrating the coincidence between DAP-seq unique binding and ABA-induced chromatin accessibility and reduced H3K27me3. TuG1812G0500004885 is a gene with LRR domains which may relate to biotic or abiotic stress responses.
Gene-distal TFBSs are preferentially lineage-specific and embedded in TEs
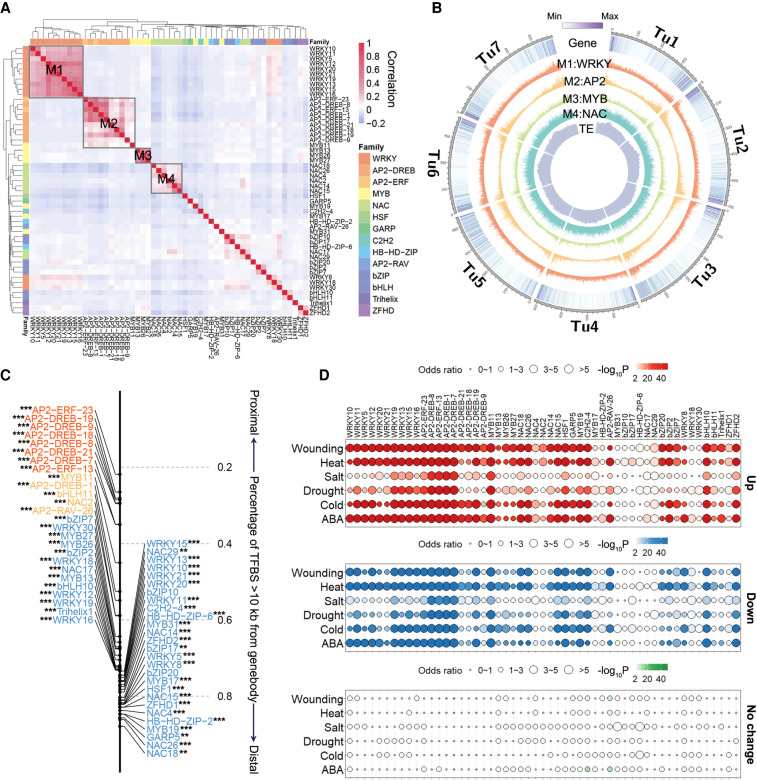
A comparison of the genome-wide TF binding patterns revealed that they are largely grouped by TF families (Fig. 3A). Different groups are preferentially localized to different chromosomal regions. The AP2 TFs mostly bind to the distal end of chromosomes, whereas NAC TFs bind across the chromosome (Fig. 3B). The distance distribution relative to the nearest genes varied substantially among TFs (Fig. 3C). Specifically, 78% of the AP2-ERF-23 peaks were localized within 10 kb of genes, whereas 76% of the NAC18 peaks were present in regions more than 10 kb from the nearest genes, possibly reflecting the remote regulation of target genes. We further integrated the expression data responsive to abiotic stresses to investigate the functional potential of these TF binding. Given the ambiguity of assigning targets to distal regulatory elements, we focused on the expression changes of the proximal targets (nearest gene within 10 kb of TF binding). The targets of most TFs were primarily stress-responsive genes (Fig. 3D). For example, the transcription factor ZFHD2 is significantly enriched near genes that are both up- and down-regulated in heat but not enriched near genes that have no significant expression change in heat (Fig. 3D).
Figure 3.
Distribution of TFBSs and stress responsiveness of TF targets. (A) Clustering of TF binding correlations based on occurrence of DAP-seq peaks shows that TFs from the same family generally have similar binding profiles. (B) Circos plot showing the genomic distribution of the four largest TFBS clusters shown in A. The visualization of genomic distribution was performed by Circos (Krzywinski et al. 2009). (C) Fraction of peaks for each TF localized to the distal regions (>10 kb from the nearest gene). For each TF peak set, the distance to the TSS of the nearest gene was compared with randomly selected regions using an unpaired Student's t-test. Almost all TFs are closer to gene body regions. (**) P < 0.01, (***) P < 0.001 (H1: expected distance > observed distance). (D) Enrichment of TF proximal targets in stress-responsive and non-stress-responsive genes. The top panel (red), middle panel (blue), and bottom panel (green) are the enrichment of TF targets in up-regulated genes, down-regulated genes, and genes with no significant expression change in response to stresses. The color range represents the enrichment P value and the circle size represents the odds ratio. Genes with FPKM > 1 were used for the analysis.
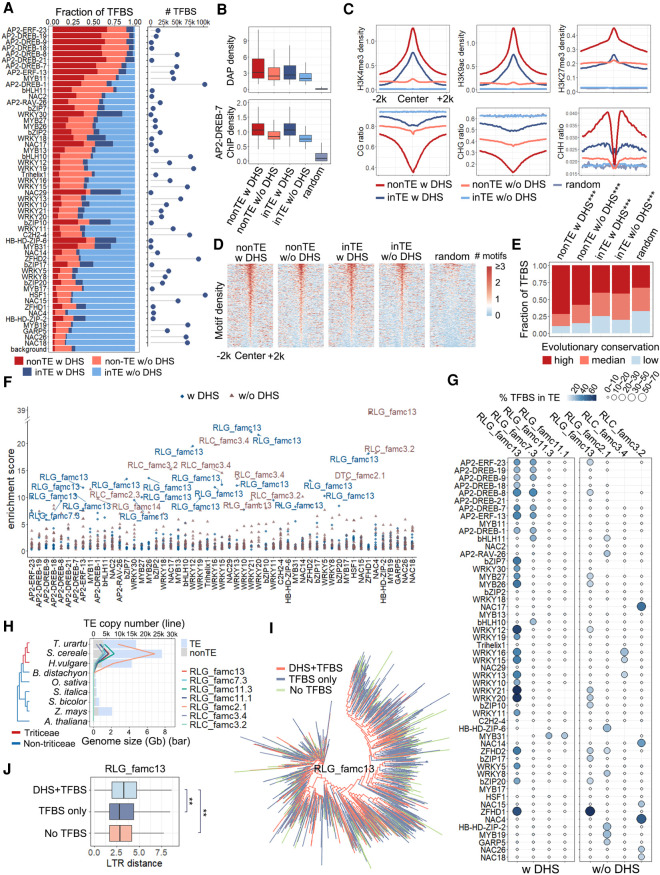
Among these TFBSs, 7%–85% were embedded in TEs (Fig. 4A). Recent studies involving animals and plants suggested that TEs have been a rich source of new TFBSs (Chuong et al. 2017; Trizzino et al. 2017; Zhao et al. 2018). The read density distributions for TFBSs in TEs and non-TE regions were very similar (Fig. 4B). Transposable elements are generally repressed by DNA methylation, with low chromatin accessibility. In this study, we observed that 40,807 collapsed TFBSs (with overlapping TFBSs merged, ∼5%) of the TE-embedded TFBSs are open and strongly associated with active chromatin signatures, including reduced DNA methylation and active histone marks (Fig. 4C). These signatures were shared between TFBSs contributed by TE sequences and by non-TE sequences, suggesting their common regulatory potential. The motif densities were highly similar between TFBSs in TEs overlapping with DHSs and those that did not overlap with DHSs (Fig. 4D). Accordingly, the differential accessibility is likely because of the relative positions and the chromatin environment, rather than the sequence context. Moreover, TE-embedded TFBS sequences were more highly conserved in wheat species than the randomly selected regions (χ2 test, P < 0.001) (Fig. 4E), reflecting the purifying selection of DNA sequences and further supporting their functional relevance. Thus, a subset of TEs have signatures of DNA regulatory elements, which contribute to gene regulatory networks by serving as TFBSs.
Figure 4.
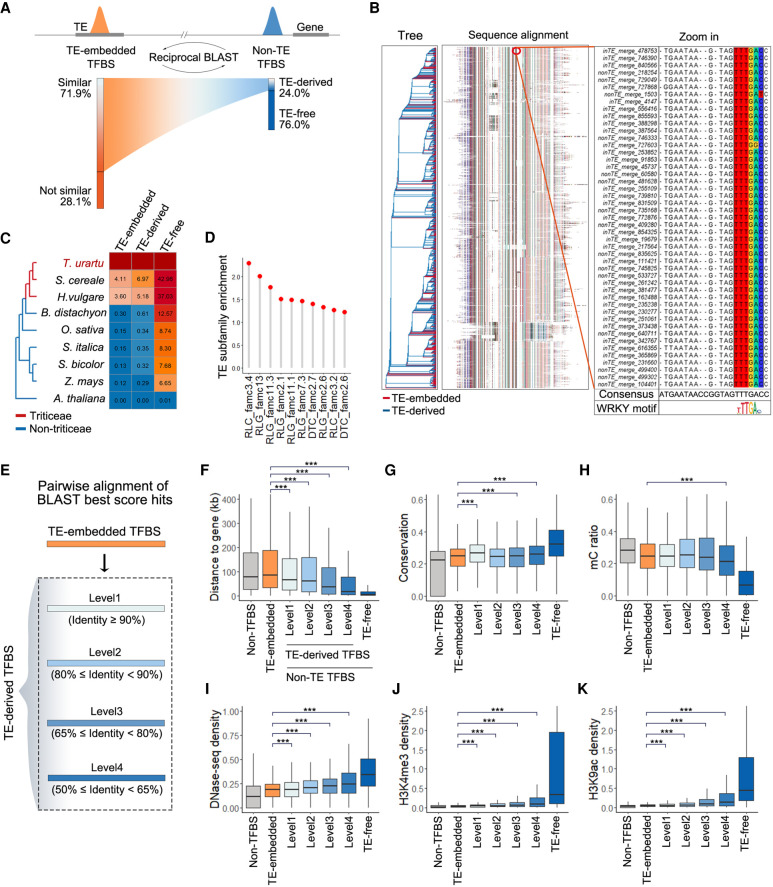
Pervasive association of TFBSs and TEs in Tu. (A) Proportion of the TFBSs that occurred in TEs with (dark blue) or without (light blue) DHSs and non-TE regions with (dark red) or without (light red) DHSs. The numbers of all TFBSs are shown on the right. (B) DAP-seq density and AP2-DREB-7 ChIP-seq density distribution of non-TE TFBSs and TE-embedded TFBSs with or without DHSs. (C) Epigenetic profiles of TE-embedded and non-TE TFBSs. All figures represent the average signal density at 50-bp resolution within a 4-kb window centered on peak summits. Top panel: Regulatory histone marks, including H3K4me3, H3K27me3, and H3K9ac. Bottom panel: DNA methylation levels in three contexts. (D,E) Distribution of motifs (D) and conservation levels (E) in non-TE TFBSs and TE-embedded TFBSs with or without DHSs. (D) The number of motif occurrences (bin size 50 bp) within a 4-kb window centered on the merged TFBS centers. The unions of the primary motifs of these TFs were used. (E) Conservation score is a measure of sequence conservation across wheat species. The 0.33 quantile (0.16) and 0.66 quantile (0.25) of the conservation score of all peaks were used to define the degree of conservation. 0 < score < 0.16 for low conservation, 0.16 ≤ score < 0.25 for median conservation, score ≥0.25 for high conservation. For each TFBS set, the number of the TFBSs in each conservation category was compared with a randomly selected set using a χ2 test. (***) P < 0.001. (F) Specific TE families enriched among TFBSs. Blue dots and brown triangles represent families contributed to TE-embedded TFBS overlapping and not overlapping with DHSs, respectively. Highly enriched TE families (enrichment score > 9) are labeled with family names. (G) Percentage of TFBSs in TE-embedded regions with or without DHSs. The color range and circle size represent the percentage of TFBSs overlapping with TEs. (H) TE copy number (line plot) of each family (represented by different colors) during evolution. The genome sizes are shown as a bar plot, light blue representing TEs and light gray representing non-TEs. (I) Dendrogram showing the sequence similarity between RLG family 13 members. (J) Age of different groups of RLG family 13 measured by sequence similarity of LTR from both ends. A Wilcoxon signed-rank test was used to compare the LTR distance of different groups. (**) P < 0.01 (H1: the LTR sequence of TEs with DHSs and TFBSs were more divergent than other TEs).
We next wondered if any TE family contributed a significant number of binding sites for specific TFs. We examined the TFBS distribution in differentially enriched TE families. A significant proportion of the binding sites were embedded in specific repeat families (Fig. 4F). For the TE-embedded TFBSs overlapping with DHSs, LTR-Gypsy family 13 is the largest contributor, accounting for 20% of the binding sites. The most enriched TFBSs contributed by this TE family include members of the WRKY and AP2 families (Fig. 4G). LTR-Gypsy family 13 is also among the top-ranked TE families contributed to TE-embedded TFBSs that did not overlap with DHSs. Among these top enriched TE families, most TFBSs localized to LTR regions (Supplemental Fig. S4). This is consistent with previous findings in mammals, whose LTRs are co-opted and acquired the host regulatory mechanisms during evolution (Jiang et al. 2004; Daron et al. 2014), which are responsible for transcriptional regulation of both TEs and host genes (Sundaram et al. 2014). Altogether, specific LTR TE families dominated the contribution of TE-embedded TFBSs.
It is likely that insertions of TEs from these families lead to TFBS expansion. Thus, we examined the emergence and expansion of these TE families. No homologous TE families were detected in non-Triticeae species, indicating the majority of binding events occurring within these TEs were amplified in Triticeae species (i.e., they are specific to Triticeae) (Fig. 4H). We observed that LTR TE families significantly contributed to TE-embeded TFBSs displayed species-specific expansion (Supplemental Fig. S5), implying they are still highly active in Triticeae species. We next asked whether the TFBSs in RLG-Gypsy family 13 have a single origin (i.e., preferentially originated from one branch of the phylogenetic tree). In contrast to our expectation, the TFBS TEs were dispersed in the TE family clusters (Fig. 4I), indicating multiple TF binding events occurred during the evolution of these TE families; alternatively, the acquired TFBSs may be lost during evolution in some subfamilies. TEs containing TFBSs overlapping with DHSs are relatively ancient (Fig. 4J), suggestive of a long period of degeneration of TEs as regulatory elements. Together, the degeneration of Triticeae-specific LTR-Gypsy families is likely subjected to a relatively long-term evolutionary selection to evolve to bona fide TFBSs.
Prevalent insertion and domestication of TE remnants to gene-proximal TFBSs contributes to ongoing regulatory expansion
Extensive TE insertions in gene-proximal regions with built-in regulatory copies may quickly rewire transcriptional patterns, leading to novel functions and increasing regulatory complexity (Sundaram et al. 2014; Chuong et al. 2017; Trizzino et al. 2017). Earlier research revealed TE bursts that predate and accompanied Triticeae divergence, leading to extremely large genomes with abundant TEs (80%–90%) (Mascher et al. 2017; The International Wheat Genome Sequencing Consortium (IWGSC) et al. 2018). To what extent have TEs promoted the ongoing evolution of wheat transcriptional regulation and how did these TE-derived TFBSs evolve?
To fully evaluate the evolutionary contribution of TE-embedded TFBSs to non-TE TFBSs, particularly those with gene-proximal binding, we completed a reciprocal sequence comparison between TE-embedded TFBSs (626,865 collapsed regions) and non-TE TFBSs (248,778 collapsed) (see Methods) (Fig. 5A). Twenty-four percent of the non-TE TFBSs showed high sequence identity (≥50%) with TE-embedded TFBSs, which is significantly higher compared to random pairs (Supplemental Fig. S6). The body region of these TE-related non-TE TFBSs has high similarity with corresponding TE-embedded TFBSs, whereas the flanking region retained local similarity and no longer has typical TE structure (Fig. 5B; Supplemental Fig. S7), suggesting that these non-TE TFBSs are potentially derived from TE-embedded TFBSs. Figure 5B presents the results of a multiple sequence alignment of one cluster. The tree included both types of TFBSs, reflecting the ongoing spread of TE-embedded TFBSs to non-TE regions. This result also reflects the possibility that some TE-embedded TFBSs may originally be hijacked from non-TE TFBSs by active TEs. The TE-derived event occurred in all TFs characterized, with WRKY and AP2 family as the largest contributors (Supplemental Fig. S8). Those TE-derived TFBSs were mostly specific to Triticeae species, with almost no homologous sequences in non-Triticeae species (<0.61%) (Fig. 5C), suggesting that the transposition and degeneration are Triticeae-specific.
Figure 5.
Ongoing degeneration of remnant TEs to TFBSs in non-TE regions. (A) Left: Fraction of TE-embedded TFBSs showing high sequence similarity to non-TE TFBSs. Right: Fraction of non-TE TFBSs with high sequence similarity to TE-embedded TFBSs. (B) Multiple sequence alignment of one cluster of TE-embedded and TE-derived TFBSs based on sequence similarity (n = 2034). The alignment in the red circle is enlarged on the right. The alignment also shows a particularly high degree of sequence identity for the WRKY binding motif. (C) Fractions of homologous sequences in other species for TE-embedded TFBSs, TE-derived TFBSs in non-TE regions, and other non-TE TFBSs. (D) Enriched TE subfamilies with TFBSs showing sequence similarity to non-TE TFBSs. (E) TE-derived TFBSs were grouped according to the sequence divergence with TEs. Level 1 represents low divergence and level 4 represents high divergence. (F) Distribution of the distance between TFBSs and the proximal genes. TFBSs were classified as TE-embedded, TE-derived, and TE-free; 30,000 non-TFBS regions were randomly sampled from genomic loci without TFBSs. A Wilcoxon signed-rank test was used to compare the TE-derived TFBSs and TE-embedded TFBSs. (***) P < 0.001 (H1: TE-derived TFBSs were closer to genes than TE-embedded TFBSs). (G) Distribution of sequence conservation for different groups of TFBSs and non-TFBSs in TEs. A Wilcoxon signed-rank test was used to compare the TE-derived TFBSs and TE-embedded TFBSs. (***) P < 0.001 (H1: TE-derived TFBSs were more conservative than TE-embedded TFBSs). (H–K) Epigenetic feature distribution of TFBSs embedded in TEs or localized to non-TE regions and non-TFBSs. (H) DNA methylation. (I) DHS density. (J,K) Regulatory histone mark distribution. A Wilcoxon signed-rank test was used to compare the TE-derived TFBSs and TE-embedded TFBSs. (***) P < 0.001. (For H, H1: TE-derived TFBSs had lower methylation levels than TE-embedded TFBSs. For I–K, H1: TE-derived TFBSs had more active epigenetic signatures than TE-embedded TFBSs.)
We next traced the ancestral TEs of these TE-derived TFBSs. Figure 5D presents the most enriched TE families contributing to TE-derived TFBSs. The results are consistent with the above finding that LTR-Copia family 3.4 and LTR-Gypsy family 13 were among the most enriched TE families contributing to TE-embedded TFBSs (Fig. 4G). Accordingly, the genomic expansion of these TE families contributed to TFBS expansion in both TE and non-TE regions.
To quantitatively measure the extent of TE degeneration and TFBS regulatory activity, the TE-derived TFBSs were partitioned based on sequence similarity with corresponding TE-embedded TFBSs (Fig. 5E). In general, the TE-derived TFBSs were localized much more proximal to genes (28% within 10 kb of the nearest genes) compared with the TE-embedded TFBSs (Fig. 5F) and had more regulatory activities, as reflected by the high sequence conservation across wheat species (Fig. 5G), lower DNA methylation levels (Fig. 5H), and more active epigenetic signatures including increased chromatin accessibility (Fig. 5I–K). Moreover, extensive degeneration (i.e., decreased similarity to TE sequences) was associated with increased gene proximity and regulatory activities (Fig. 5E–K), reflecting the ongoing insertion and decay of TEs as gene-proximal regulatory elements.
Transposable elements have rewired the regulatory network
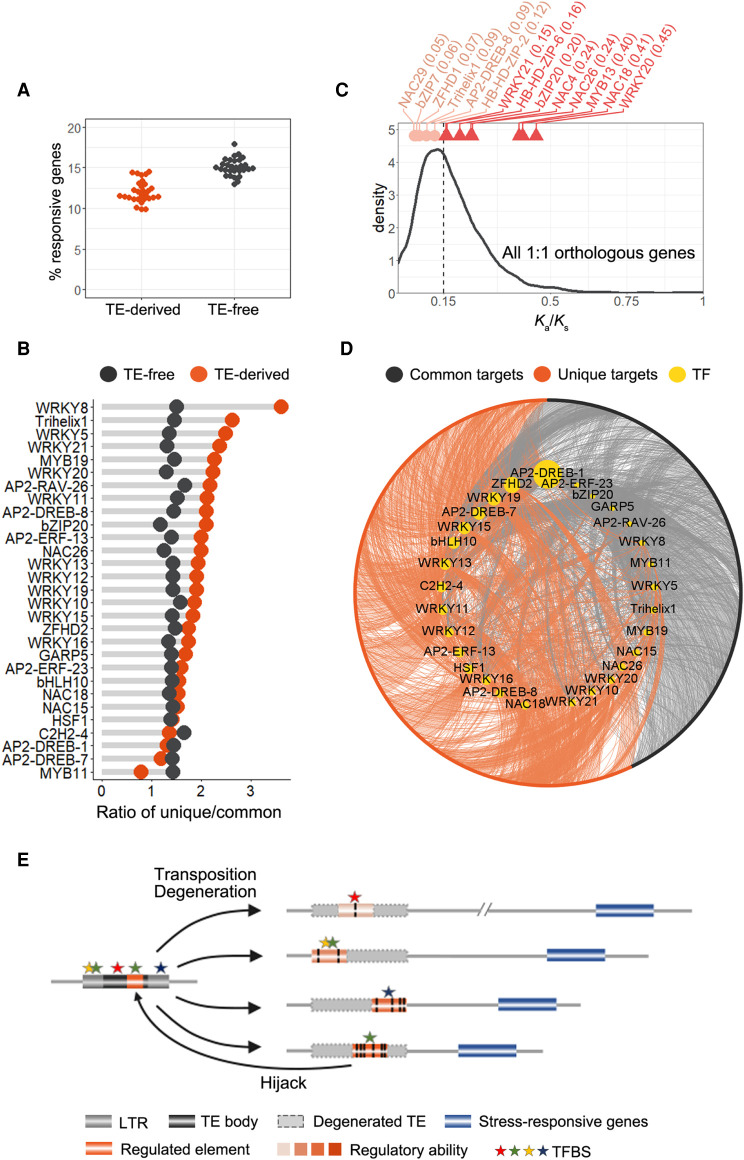
Compared with stress treatment-induced expression data sets, non-TE TFBSs, including both TE-derived and TE-free TFBSs, have comparable fractions of stress-responsive target genes (Fig. 6A). The fraction is much more similar when only TFBSs overlapping with DHSs were considered (Supplemental Fig. S9). In this regard, TE-derived TFBSs and TE-free TFBSs behave similarly. We further integrated transcriptomic data from O. sativa (Os) of similar abiotic stress treatments and identified genes commonly and uniquely induced in Os and Tu (Supplemental Table S3). We revealed that a group of TE-derived TFBS target genes in Tu have been added to the network regulating stress responses (Fig. 6B). Members of the WRKY family contributed more to the Tu-specific response genes than to the common response genes (Fig. 6B), and these TFs generally have significantly higher Ka/Ks ratios (Fig. 6C), indicating that they underwent relaxed selection in Tu. The increased binding of Tu-specific response genes by the WRKY family may be due to genetic drift, some of which would be fixed under specific stressful conditions. Despite that we do not expect that all binding events directly affect gene activity, these findings provide important evidence of the evolutionary effects of TE remnants on transcriptional regulation. In summary, our results revealed the high plasticity of the wheat stress response regulatory network as well as the importance of TEs in promoting ongoing regulatory innovation (Fig. 6D).
Figure 6.
TE-derived TFBSs have wired new genes into the regulatory network of wheat environmental responses. (A) Fraction of TE-derived TFBSs and TE-free TF targets induced by abiotic stresses. (B) Ratio of unique response genes in wheat and commonly induced genes in Tu and Os. Orange spots represent TE-derived TFBSs. Black spots represent TE-free TFBSs. The TFs with the number of targets induced by abiotic stresses greater than 20 were kept. (C) Ka/Ks ratio of TFs between Os and Tu. The values for 1:1 orthologous TFs are shown on top, and the line plot represents the background of Ka/Ks distribution for all 1:1 orthologous genes between Os and Tu. TFs with Ka/Ks ratios greater than the median of all 1:1 orthologous genes are in dark orange; other TFs displayed in light orange. (D) Network showing incorporation of new stress-responsive genes by TE-derived TFBSs. TEs in A are shown. (E) Model illustrating the rewiring of the gene regulatory network by TE-derived TFBSs. Left: Some TFBSs or TFBS precursors exist within specific TEs, transposition of which leads to expansion of corresponding TFBSs or precursors. Right: Transposed TEs were degenerated and lost typical TE structures, but some TFBSs present in TEs were evolutionarily selected for regulating nearby gene activity. The closer the TE-derived TFBS to genes, the stronger the regulatory activity. The reverse arrow at the bottom illustrates that some TE-embedded TFBSs may be hijacked from the non-TE TFBSs.
Discussion
The cistrome and epicistrome maps are a valuable resource for elucidating the transcriptional networks controlling plant adaptation. We demonstrated that the majority of the distal binding sites are embedded within TEs, which are predominantly contributed by specific LTR TE families. The abundant and dynamic turnover of TEs in wheat facilitated the detection of the ongoing domestication of TEs. We revealed that ∼24% of the non-TE TFBSs shared high sequence similarity with TE-embedded TFBSs, which were linked to wheat-specific gene responses to environmental stimuli, suggesting that TEs are an important force driving regulatory innovation for wheat adaptations (model in Fig. 6E).
The findings described herein raise the possibility that TE domestication has considerably influenced the evolution of species phenotypes. We observed that specific TE families preferentially contributed to TE-derived TFBSs (Fig. 5). Given the rapid degeneration of the sequence context of TE remnants, we have likely underestimated the effects of TE-derived regulatory elements, and they may also be transient in the context of evolution. Integrating data from other relevant species would provide a more comprehensive picture. A TE burst predating the divergence of Triticeae species resulted in extremely large genomes. Transposable elements represent <30% of the genome of Brachypodium distachyon, which is a close relative of Triticeae (The International Brachypodium Initiative 2010). The divergence of Triticeae species was accompanied by the emergence and expansion of different TE families (Middleton et al. 2013). The differential decay of these ancestral TE sequences across species may result in species-specific TF binding events. Additional comparisons of the TFBS across Triticeae species will help elucidate the mechanism underlying the gain of species-specific TFBS during Triticeae evolution as well as their contribution to genome regulation, species divergence, and phenotypic variation during domestication and cultivation. This may, in turn, facilitate the targeted manipulation of gene activity.
There are some disagreements regarding TE domestication, at least from an evolutionary perspective. Some studies suggested that these elements are pervasively co-opted for the regulation of host genes. The insertion and deletion of a large amount of “junk DNA” underwent neutral genetic drift, which occasionally can be integrated into the regulatory network (Todd et al. 2019). Other studies indicated that most of these regulatory activities can be interpreted as relics of strategies used by TEs to spread within genomes and host populations (Chuong et al. 2017). In other words, TEs hijack host regulatory components to promote self-proliferation, and TE domestication was an adaptation to evolutionary conflicts between TEs and the host. We determined the TE-derived TFBSs are under purifying selection (Fig. 5G), whereas members from the WRKY family, which have a large proportion of binding sites in TEs, underwent diversifying selection (Fig. 6C). This cannot be explained by one point of view. Alternatively, the relaxed selection of these TF families is merely an adaptation to diversifying environmental stresses and has no relationship with TE binding. It was also proposed that some stress-responsive TFs, including WRKYs, are likely derived from TE components (Joly-Lopez and Bureau 2018), which may confer these TFs with TE-binding potential. Future studies should elucidate whether the selection helped promote or inhibit TE binding.
Altogether, the comprehensive stress-responsive TF binding catalog and epigenomic profiles not only provide valuable resources to elucidate transcriptional networks controlling wheat adaptation but also propose abundant indications of the widespread ongoing regulatory innovation and expansion introduced by lineage-specific TE insertion, degeneration, and domestication.
Methods
Plant materials and growth conditions
Tu seeds were surface-sterilized via a 10-min incubation in 30% H2O2 and then thoroughly washed five times with distilled water. The seeds were germinated in water for 3 d at 22°C, after which the germinated seeds with residual endosperm were transferred to soil. The seedlings (above-ground parts) were harvested after a 9-d incubation under long-day conditions. Regarding the cold and heat stress treatments, 7-d-old seedlings grown in soil were transferred to 4°C for 5 h or 40°C for 7 h, respectively. To assess the effects of drought, 7-d-old seedlings were cultivated in soil for another 2 wk without watering. For the NaCl and ABA treatments, 7-d-old seedlings grown in soil were treated with 250 mM NaCl for 7 h or 100 μm ABA for 2 wk. For wounding stress, 7-d-old seedlings were injured on leaves by scissors and samples were taken 2.5 h later. The harvested samples were either frozen in liquid nitrogen for an RNA isolation and DAP-seq assay or directly vacuum-infiltrated with a formaldehyde cross-linking solution for use in the ChIP-seq and DHS assay.
DAP-seq assay
DAP-seq was performed as previously described (Bartlett et al. 2017). Genomic DNA was extracted from wheat leaves using Plant DNAzol Reagent) and fragmented. DNA was then end-repaired using the End-It kit (Lucigen) and A-tailed using Klenow (3′–5′ exo-; NEB). Truncated Illumina Y-adapter was ligated to DNA using T4 DNA Ligase (Promega). Full-length TF was cloned into pIX-Halo vector. Halo-tagged TF was expressed in vitro using the TNT SP6 Coupled Wheat Germ Extract System (Promega). Halo-TF was immobilized by Magne HaloTag Beads (Promega) and then incubated with the DNA library. TF-specific binding DNA was eluted for 10 min at 98°C and amplified with indexed Illumina primer using Phanta Max Super-Fidelity DNA Polymerase (Vazyme). Meanwhile, to capture background DNA which captured by Halo, pIX-Halo vector without TF cloned was expressed and incubated with the DNA library as well. The PCR product was purified using VAHTS DNA Clean Beads (Vazyme) and then sequenced by Novogene with the Illumina NovaSeq 6000 system to produce 150-bp paired-end reads.
ChIP-seq and RNA sample preparation and sequencing
RNA-seq data corresponding to cold, heat, drought, salt, wounding, and ABA treatments sets were generated with biological duplicates. A ChIP-seq assay was completed as previously described (Wang et al. 2016), with antibodies specific for H3 trimethyl-Lys 27 (Millipore; 07–449), H3 trimethyl-Lys 4 (Abcam; ab8580), and H3 acetyl-Lys 9 (Millipore; 07-352). For each ChIP-seq assay, approximately 30 seedlings were pooled and ground to a powder. More than 10 ng ChIP DNA or 2 μg total RNA were used to prepare each sequencing sample. Libraries were constructed and sequenced by Berry Genomics (Beijing, China) and Novogene (Beijing, China). The libraries were sequenced with the Illumina NovaSeq 6000 system and HiSeq X Ten system to produce 150-bp paired-end reads.
Protoplast ChIP-seq assay
The ChIP assays using Tu leaf protoplasts were performed with minor modifications (Para et al. 2018). Tu plants were grown on soil under 16 h light/8 h dark conditions for 2 wk before protoplast isolation. Approximately 30 μg pMD19-T plasmids containing p35S:3flag-AP2 DNA were transfected into leaf protoplasts using the PEG-mediated transfection method. After incubating the protoplasts at room temperature for 48 h under dark conditions, the protoplasts were crosslinked with 1% formaldehyde in W5 solution for 10 min on ice and quenched with 32 μL 2 M glycine for 5 min. Protoplasts were collected by centrifuging at 600g for 2 min at 4°C, washed with 500 μL W5 solution once, and collected again. Protoplasts were lysed in 120 μL room temperature lysis buffer (50 mM Tris-HCl [pH 8.0], 10 mM EDTA, 1% [wt/vol] SDS, 1 mM PMSF, 1× protease inhibitor cocktail) by vortex. Total lysates containing chromatin were subjected to sonication by Bioruptor until the chromatin was fragmented into 300 bp–500 bp. Another 400 μL RIPA ChIP buffer (10 mM Tris-HCl [pH 7.5], 140 mM NaCl, 1 mM EDTA, 0.5 mM EGTA, 1% [vol/vol] Triton X-100, 0.1% [wt/vol] SDS, 0.1% [wt/vol] sodium deoxycholate, 1 mM PMSF, 1× protease inhibitor cocktail) was added into the lysates. The lysates were centrifuged at 12,000g for 10 min at 4°C and the supernatant was transferred to a new tube. Another 410 μL of RIPA ChIP buffer was mixed with the remaining pellet and centrifugation was performed again as above to obtain the second supernatant. The two rounds of supernatant were pooled and brought to a volume of 1 mL with RIPA ChIP buffer. One hundred microliters of chromatin was kept as 10% input. Twenty microliters of agarose beads conjugated with anti-Flag antibody (Sigma-Aldrich A2220) were added to the chromatin suspension and incubated for 2 h at 4°C. After binding with chromatin, the beads were washed subsequently with RIPA buffer twice (10 mM Tris-HCl [pH 7.5], 140 mM NaCl, 1 mM EDTA, 0.5 mM EGTA, 1% [vol/vol] Triton X-100, 0.1% [wt/vol] SDS), LiCl buffer (0.25 M LiCl, 1% [wt/vol] sodium deoxycholate, 10 mM Tris-HCl [pH 8.0], 1% NP-40, 1 mM EDTA) once, and TE (10 mM Tris-HCl [pH 8.0], 10 mM EDTA) buffer once. The protein-DNA complexes were eluted from beads by adding 150 μL of complete elution buffer (20 mM Tris-HCl [pH 7.5], 5 mM EDTA, 50 mM NaCl, 1% [wt/vol] SDS, 50 mg/mL Proteinase K) for 2 h at 68°C with agitation at 1300 rpm. The eluate was then transferred to a new tube. The beads were eluted again with 150 μL of elution buffer (20 mM Tris-HCl [pH 7.5], 5 mM EDTA, 50 mM NaCl) for 5 min, and the two rounds of eluates were combined. During the elution step, the input DNA was prepared by adding 200 μL elution buffer and 7.5 μL of Proteinase K (20 mg/mL) which was then incubated at 68°C for 2 h. ChIP DNA was extracted with phenol: chloroform (1:1), precipitated with ethanol, and resuspended in TE buffer to prepare the ChIP-seq library using the ThruPLEX DNA-seq kit. The libraries were sequenced with HiSeq-PE150 to produce 150-bp paired-end reads by Novogene.
DNase I hypersensitive site sequencing library preparation and sequencing
The harvested seedlings used to prepare ChIP-seq samples were also subjected to a DNase I treatment. Specifically, to prepare DNase I hypersensitive sites sequencing (DNase-seq) libraries, approximately 20 seedlings were fixed with 1% formaldehyde in HEPES buffer (50 mM HEPES, 1 mM EDTA [pH 8.0], 0.1 M NaCl, and 1 mM PMSF). The fixed seedlings were ground to a fine powder in liquid nitrogen. Wheat nuclei were extracted with H1B buffer (20 mM Tris-HCl [pH 8.0], 50 mM EDTA, 5 mM spermidine, 0.15 mM spermine, 40% glycerol, and 0.1% mercaptoethanol). The extracted nuclei were purified with H1B buffer supplemented with 0.5% Triton X-100. The purified nuclei were washed once with RSB buffer (10 mM Tris-HCl [pH 7.4], 10 mM NaCl, and 3 mM MgCl2). The pelleted nuclei were resuspended with 2 mL RSB buffer and then divided equally into five 1.5-mL Eppendorf tubes. The aliquoted nuclei were digested with DNase I (0, 0.01, 0.03, 0.05, and 0.08 units). The resulting digested nuclei were extracted using one volume of phenol, phenol:chloroform, and chloroform, after which the DNA from each digestion was resuspended in two volumes of cold ethanol and then pelleted. A DNase-seq library was prepared from 0.03 U DNase I-digested nuclei. Approximately 2 µg DNA were separated by 1.5% agarose gel electrophoresis, and DNA fragments (50–300 bp) were cut and purified to prepare the DNase-seq library with the NEBNext Ultra II DNA Library Prep kit for Illumina (NEB). Two biological replicates of the libraries were prepared. The quality of the final DNase-seq libraries was checked, after which the libraries were sequenced with the 150-bp paired-end mode of the Illumina NovaSeq platform.
Bisulphite sequencing and data analysis
Bisulphite sequencing samples were prepared with 2.2 μg DNA extracted from the harvested seedlings that were also used to prepare the ChIP-seq and DNase-seq samples. The bisulphite sequencing libraries were constructed and the subsequent deep sequencing was completed by Genenergy Biotechnology Co. Ltd. The libraries were sequenced with the HiSeq 3000 system (Illumina) to produce 150-bp paired-end reads, which were cleaned with the Trim Galore (version 0.4.4, https://github.com/FelixKrueger/TrimGalore), Trimmomatic (version 0.36) (Bolger et al. 2014) and Sickle programs (https://github.com/najoshi/sickle). The clean reads were then aligned to the Tu reference sequence (IGDBv1.0) (Ling et al. 2018) with the default settings of the Bismark program (version 0.19.0) (Krueger and Andrews 2011). The default settings were strict, with only the best unique alignments reported, and all nonunique alignments were removed (Krueger and Andrews 2011). Thus, we applied only two additional filtering steps, namely the removal of reads with a mapping quality < 20, followed by the removal of PCR duplicates with the deduplicate_bismark implemented in the Bismark program. The extent of the cytosine methylation was determined with the bismark_methylation_extractor implemented in the Bismark program. Next, the methylation ratio of a cytosine was calculated as the number of mCs divided by the number of reads covering the position. Bases covered by fewer than three reads were considered low-confidence positions whose methylation ratios were not recorded.
Processing of DAP-seq, ChIP-seq, RNA-seq, and DHS data
Sequencing reads were cleaned with the fastp (version 0.20.0) (Chen et al. 2018) and Trim Galore (version 0.4.4), which eliminated bases with low quality scores (<25) and irregular GC contents, sequencing adapters, and short reads. The remaining cleaned reads were mapped to the Tu genome with the Burrows–Wheeler Aligner (version 0.7.17-r1188) (Li and Durbin 2010) for the DAP-seq, ChIP-seq, and DNase-seq data. The HISAT2 program (version 2.2.1) (Kim et al. 2015) was used for mapping the RNA sequencing (RNA-seq) reads to the reference sequences. For histone ChIP-seq, DNase-seq, and RNA-seq, the multimapped reads were directly removed. For DAP-seq and ChIP-seq of AP2-DREB-7, reads with mapping quality <20 were removed.
The MACS (version 2.2.6) (Zhang et al. 2008) program was used to identify the read-enriched regions (peaks) of the DAP-seq, ChIP-seq, and DHS data with the cutoff P < 1 × 10−10. For DAP-seq, the peaks detected from samples introduced with the Halo tag only were considered as nonspecific bindings, and TF peaks overlapping with peaks detected from Halo samples were removed for subsequent analysis. To quantify gene expression levels, the featureCount program of the Subread package (version 2.0.0) (Liao et al. 2013) was used to determine the RNA-seq read density for the genes. To compare expression levels across samples and genes, the RNA-seq read density of each gene was normalized based on the exon length in the gene and the sequencing depth (i.e., fragments per kilobase of exon model per million mapped reads). To quantify histone markers across genes for the figure prepared with Integrative Genomics Viewer (Robinson et al. 2011), the number of reads at each position was normalized against the total number of reads (reads per million mapped reads). The DESeq2 program (Love et al. 2014) was used for detecting differentially expressed genes based on the combined criteria: |log2 fold-change| > 1 and P < 0.05. The MAnorm package (Shao et al. 2012) was used for the quantitative comparison of ChIP-seq and DHS signals between samples with the following criteria: |M value| > 1 and P < 0.05.
Detection and enrichment analysis of transcription factor binding motifs
For de novo motif discovery, the peaks were sorted by Q-value and then by fold enrichment. The 600-bp sequence centered on the top 3000 peak summits was used to detect motifs by MEME-ChIP (Machanick and Bailey 2011) of the MEME software toolkit (version 5.1.1). All peaks were used when comparing DAP-seq and ChIP-seq.
The de novo motifs detected above or JASPAR CBFs motif were used to scan individual motif occurrences in the genome with the FIMO program (Grant et al. 2011) of the MEME software toolkit.
Motif logos were drawn by the R package motifStack (version 1.34.0) (Ou et al. 2018; R Core Team 2020) and universalmotif (version 1.4.0). Meanwhile, R package universalmotif were used to combine multiple motifs into a consensus motif.
Calculation of the sequence conservation score
We completed a pair-wise comparison of the genome sequences from T. urartu (AA sub-genome, IGDBv1.0), Aegilops tauschii (DD sub-genome, ASM34733 version 2), T. turgidum (AABB sub-genome, WEWSeq version 1.0), and T. aestivum (AABBDD sub-genome, IWGSC version 1.0) with the NUCmer tool implemented in the MUMmer package (Kurtz et al. 2004). The minimum sequence identity was set to 90 and each subgenome was treated as an individual genome. Next, ROAST (http://www.bx.psu.edu/~cathy/toast-roast.tmp/README.toast-roast.html) was used to integrate pair-wise sequence alignments into a multiple sequence alignment. The multiple sequence alignment and tree data were fitted by phyloFit, after which the conservation score was calculated with phastCons from the PHAST package (Hubisz et al. 2011).
Enrichment of specific TE family contributions to TF binding
TE annotation of Tu was performed as previously described (Wicker et al. 2018). TE subfamilies accounting for more than 1% length of all TEs in the genome were selected, and the enrichment scores (ES) between 34 TE subfamilies and 53 TFs were calculated. Enrichment of TE subfamilies for each TF was defined as
Evolutionary analysis of enriched TE subfamilies
LTRharvest (Ellinghaus et al. 2008) was used to identify the full-length LTR of Tu. BLASTN algorithm (version 2.9.0) was used to reciprocally compare the full-length LTR of each enriched LTR subfamily in Tu and LTR in other species. We used E-value <1 × 10−30, identity > 80%, and query coverage > 70% to define homologous TEs in Tu and other species. The 5′ and 3′ LTR of full-length RLG_famc13 were combined and aligned with MAFFT (version v7.149b) (Katoh and Standley 2013). FastTree (version 2.1.10) was used to build the phylogenetic tree. The tree was visualized with R package ggtree (version 2.4.1) (Yu 2020). The insertion time was based on the divergence between the 5′ and 3′ LTRs and calculated with distmat from EMBOSS (version 6.6.0.0) (Rice et al. 2000).
Definition of homologous genes, detection of syntenic region, and Ka/Ks calculation
OrthoFinder (Emms and Kelly 2015) was applied to detect orthogroups for all homologous genes and build a species tree across Triticum urartu, Secale cereale, Hordeum vulgare, Brachypodium distachyon, Oryza sativa, Setaria italica, Sorghum bicolor, Zea mays, and Arabidopsis thaliana. MCScan (Tang et al. 2008) was used to infer the synteny between Tu and other species. The alignment of 1:1 orthologous genes between Tu and Os was performed with ParaAT (version 2.0) (Zhang et al. 2012). The resulting alignment was input into codeml of the PAML package (version 4.9) (Yang 2007) to calculate Ka/Ks.
RNA-seq data of six stress treatments in Os, including cold, heat, drought, salt, ABA, and wounding, were published previously and are publicly available in the NCBI Gene Expression Omnibus (GEO; https://www.ncbi.nlm.nih.gov/geo/) under accession numbers GSE6901 (cold, salt), GSE14275 (heat), GSE80811 (drought), GSE92989 (drought), GSE37557 (ABA), and GSE77097 (wounding).
Sequence comparison of TFBSs
We collapsed the 1,546,777 DAP-seq peaks of 53 TFs into a merged TFBS set by merging the peak summits within 300 bp from each other and taking the center point. Of the merged TFBSs, 70.63% come from one TF (Supplemental Fig. S10). Then, we extended each center point 300 bp up- and downstream to form a merged TFBS set. The merged set contained 875,643 regions with length 600 bp. BLASTN algorithm was used to make reciprocal BLAST of TE-embedded TFBSs and non-TE TFBSs with the following parameters: E-value < 1 × 10−30 and identity > 80%. The sequences of TE-embedded TFBSs and TE-derived TFBSs were clustered by MCL (version 14.137) (Van Dongen 2008) with the parameter “-I = 2.5”. Sequences from one of the clusters was aligned by MAFFT and visualized with Jalview (version 2.11.1.3) (Waterhouse et al. 2009).
In order to assess the divergence level between TE-embedded TFBSs and TE-derived TFBSs, the BLAST best score hit of each TE-derived TFBS was used to define 1:1 relationship between TE-embedded TFBSa and TE-derived TFBSa. Needle from EMBOSS was used to make pairwise alignment of 1:1 pair to obtain the identity score.
In order to obtain the homologous sequences of TFBSs in other species, TFBSs were broken into 100-bp bins with a step size of 50 bp to map to other species genomes with BWA-MEM. Aligned regions were required to be located in a syntenic region between Tu and other species.
B. distachyon, S. italica, and S. bicolor genomes were obtained from Phytozome (v12) (Goodstein et al. 2012), O. sativa from RAP-DB (Sakai et al. 2013), Z. mays from MaizeGDB (Portwood et al. 2019), A. thaliana from TAIR (Berardini et al. 2015), H. vulgare from the Plant Genomics and Phenomics Research Data Repository (Arend et al. 2016; Mascher 2019), and S. cereale from the Chinese National Genomics Data Center (Li et al. 2021).
Data access
The DAP-seq, ChIP-seq, RNA-seq, DNase-seq, and bisulphite sequencing data generated in this study have been submitted to the NCBI Gene Expression Omnibus (GEO; https://www.ncbi.nlm.nih.gov/geo/) under accession number GSE167229. Tracks for all sequencing data can be visualized through our local genome browser (http://bioinfo.sibs.ac.cn/dap-seq_Tu_jbrowse/).
Supplementary Material
Acknowledgments
This study was supported by the National Science Fund for Excellent Young Scholars (32022012), the Strategic Priority Research Program of the Chinese Academy of Sciences (XDA26030302 and XDB27010302), and National Natural Science Foundation of China (32070561). We thank Dr. Jizeng Jia from the Chinese Academy of Agricultural Sciences and Dr. Yang Zhao from the Shanghai Center for Plant Stress Biology for insightful comments. We thank Huang Tao for his help in maintaining the high-performance computing server.
Author contributions: Y.J.Z., Z.B.L., and Y.B.X. conceived and designed the experiments. W.L.Z., Y.P.T., Z.J.L., K.L., L.Y., Y.L.Z., Y.P., W.T., and Y.E.Z. performed the experiments. Y.Y.Z., M.Y.W., Y.L.X., J.Y.G., and Y.J.Z. analyzed the data. Y.J.Z. wrote the manuscript with input from all authors.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at https://www.genome.org/cgi/doi/10.1101/gr.275658.121.
Competing interest statement
The authors declare no competing interests.
References
- Arend D, Junker A, Scholz U, Schüler D, Wylie J, Lange M. 2016. PGP repository: a plant phenomics and genomics data publication infrastructure. Database (Oxford) 2016: baw033. 10.1093/database/baw033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartlett A, O'Malley RC, Huang SC, Galli M, Nery JR, Gallavotti A, Ecker JR. 2017. Mapping genome-wide transcription-factor binding sites using DAP-seq. Nat Protoc 12: 1659–1672. 10.1038/nprot.2017.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennetzen JL, Wang H. 2014. The contributions of transposable elements to the structure, function, and evolution of plant genomes. Annu Rev Plant Biol 65: 505–530. 10.1146/annurev-arplant-050213-035811 [DOI] [PubMed] [Google Scholar]
- Berardini TZ, Reiser L, Li D, Mezheritsky Y, Muller R, Strait E, Huala E. 2015. The Arabidopsis information resource: making and mining the “gold standard” annotated reference plant genome. Genesis 53: 474–485. 10.1002/dvg.22877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30: 2114–2120. 10.1093/bioinformatics/btu170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Zhou Y, Chen Y, Gu J. 2018. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34: i884–i890. 10.1093/bioinformatics/bty560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuong EB, Elde NC, Feschotte C. 2017. Regulatory activities of transposable elements: from conflicts to benefits. Nat Rev Genet 18: 71–86. 10.1038/nrg.2016.139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutler SR, Rodriguez PL, Finkelstein RR, Abrams SR. 2010. Abscisic acid: emergence of a core signaling network. Annu Rev Plant Biol 61: 651–679. 10.1146/annurev-arplant-042809-112122 [DOI] [PubMed] [Google Scholar]
- Daron J, Glover N, Pingault L, Theil S, Jamilloux V, Paux E, Barbe V, Mangenot S, Alberti A, Wincker P, et al. 2014. Organization and evolution of transposable elements along the bread wheat chromosome 3B. Genome Biol 15: 546. 10.1186/s13059-014-0546-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubin MJ, Mittelsten Scheid O, Becker C. 2018. Transposons: a blessing curse. Curr Opin Plant Biol 42: 23–29. 10.1016/j.pbi.2018.01.003 [DOI] [PubMed] [Google Scholar]
- Dvořák J, Terlizzi P, Zhang HB, Resta P. 1993. The evolution of polyploid wheats: identification of the A genome donor species. Genome 36: 21–31. 10.1139/g93-004 [DOI] [PubMed] [Google Scholar]
- Ellinghaus D, Kurtz S, Willhoeft U. 2008. LTRharvest, an efficient and flexible software for de novo detection of LTR retrotransposons. BMC Bioinformatics 9: 18. 10.1186/1471-2105-9-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emms DM, Kelly S. 2015. OrthoFinder: solving fundamental biases in whole genome comparisons dramatically improves orthogroup inference accuracy. Genome Biol 16: 157. 10.1186/s13059-015-0721-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galli M, Khakhar A, Lu Z, Chen Z, Sen S, Joshi T, Nemhauser JL, Schmitz RJ, Gallavotti A. 2018. The DNA binding landscape of the maize AUXIN RESPONSE FACTOR family. Nat Commun 9: 4526. 10.1038/s41467-018-06977-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardiner LJ, Quinton-Tulloch M, Olohan L, Price J, Hall N, Hall A. 2015. A genome-wide survey of DNA methylation in hexaploid wheat. Genome Biol 16: 273. 10.1186/s13059-015-0838-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodstein DM, Shu S, Howson R, Neupane R, Hayes RD, Fazo J, Mitros T, Dirks W, Hellsten U, Putnam N, et al. 2012. Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res 40: D1178–D1186. 10.1093/nar/gkr944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant CE, Bailey TL, Noble WS. 2011. FIMO: scanning for occurrences of a given motif. Bioinformatics 27: 1017–1018. 10.1093/bioinformatics/btr064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubisz MJ, Pollard KS, Siepel A. 2011. PHAST and RPHAST: phylogenetic analysis with space/time models. Brief Bioinformatics 12: 41–51. 10.1093/bib/bbq072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- The International Brachypodium Initiative. 2010. Genome sequencing and analysis of the model grass Brachypodium distachyon. Nature 463: 763–768. 10.1038/nature08747 [DOI] [PubMed] [Google Scholar]
- The International Wheat Genome Sequencing Consortium (IWGSC), Appels R, Eversole K, Feuillet C, Keller B, Rogers J, Stein N, Pozniak CJ, Choulet F, Distelfeld A, et al. 2018. Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science 361: eaar7191. 10.1126/science.aar7191 [DOI] [PubMed] [Google Scholar]
- Jia H, Zhang S, Ruan M, Wang Y, Wang C. 2012. Analysis and application of RD29 genes in abiotic stress response. Acta Physiologiae Plantarum 34: 1239–1250. 10.1007/s11738-012-0969-z [DOI] [Google Scholar]
- Jia J, Xie Y, Cheng J, Kong C, Wang M, Gao L, Zhao F, Guo J, Wang K, Li G, et al. 2021. Homology-mediated inter-chromosomal interactions in hexaploid wheat lead to specific subgenome territories following polyploidization and introgression. Genome Biol 22: 26. 10.1186/s13059-020-02225-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang N, Bao Z, Zhang X, Eddy SR, Wessler SR. 2004. Pack-MULE transposable elements mediate gene evolution in plants. Nature 431: 569–573. 10.1038/nature02953 [DOI] [PubMed] [Google Scholar]
- Joly-Lopez Z, Bureau TE. 2018. Exaptation of transposable element coding sequences. Curr Opin Genet Dev 49: 34–42. 10.1016/j.gde.2018.02.011 [DOI] [PubMed] [Google Scholar]
- Jordan KW, He F, de Soto MF, Akhunova A, Akhunov E. 2020. Differential chromatin accessibility landscape reveals structural and functional features of the allopolyploid wheat chromosomes. Genome Biol 21: 176. 10.1186/s13059-020-02093-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashkush K, Feldman M, Levy AA. 2003. Transcriptional activation of retrotransposons alters the expression of adjacent genes in wheat. Nat Genet 33: 102–106. 10.1038/ng1063 [DOI] [PubMed] [Google Scholar]
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30: 772–780. 10.1093/molbev/mst010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Langmead B, Salzberg SL. 2015. HISAT: a fast spliced aligner with low memory requirements. Nat Methods 12: 357–360. 10.1038/nmeth.3317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krueger F, Andrews SR. 2011. Bismark: a flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics 27: 1571–1572. 10.1093/bioinformatics/btr167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krzywinski M, Schein J, Birol I, Connors J, Gascoyne R, Horsman D, Jones SJ, Marra MA. 2009. Circos: an information aesthetic for comparative genomics. Genome Res 19: 1639–1645. 10.1101/gr.092759.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurtz S, Phillippy A, Delcher AL, Smoot M, Shumway M, Antonescu C, Salzberg SL. 2004. Versatile and open software for comparing large genomes. Genome Biol 5: R12. 10.1186/gb-2004-5-2-r12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. 2010. Fast and accurate long-read alignment with Burrows–Wheeler transform. Bioinformatics 26: 589–595. 10.1093/bioinformatics/btp698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Wang M, Lin K, Xie Y, Guo J, Ye L, Zhuang Y, Teng W, Ran X, Tong Y, et al. 2019. The bread wheat epigenomic map reveals distinct chromatin architectural and evolutionary features of functional genetic elements. Genome Biol 20: 139. 10.1186/s13059-019-1746-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Wang L, Yang J, He H, Jin H, Li X, Ren T, Ren Z, Li F, Han X, et al. 2021. A high-quality genome assembly highlights rye genomic characteristics and agronomically important genes. Nat Genet 53: 574–584. 10.1038/s41588-021-00808-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y, Smyth GK, Shi W. 2013. The Subread aligner: fast, accurate and scalable read mapping by seed-and-vote. Nucleic Acids Res 41: e108. 10.1093/nar/gkt214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling HQ, Ma B, Shi X, Liu H, Dong L, Sun H, Cao Y, Gao Q, Zheng S, Li Y, et al. 2018. Genome sequence of the progenitor of wheat A subgenome Triticum urartu. Nature 557: 424–428. 10.1038/s41586-018-0108-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisch D. 2009. Epigenetic regulation of transposable elements in plants. Annu Rev Plant Biol 60: 43–66. 10.1146/annurev.arplant.59.032607.092744 [DOI] [PubMed] [Google Scholar]
- Lisch D. 2013. How important are transposons for plant evolution? Nat Rev Genet 14: 49–61. 10.1038/nrg3374 [DOI] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15: 550. 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo MC, Gu YQ, Puiu D, Wang H, Twardziok SO, Deal KR, Huo N, Zhu T, Wang L, Wang Y, et al. 2017. Genome sequence of the progenitor of the wheat D genome Aegilops tauschii. Nature 551: 498–502. 10.1038/nature24486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machanick P, Bailey TL. 2011. MEME-ChIP: motif analysis of large DNA datasets. Bioinformatics 27: 1696–1697. 10.1093/bioinformatics/btr189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mascher M. 2019. Pseudomolecules and annotation of the second version of the reference genome sequence assembly of barley cv. Morex [Morex V2]. e!DAL - Plant Genomics and Phenomics Research Data Repository (PGP), IPK Gatersleben, Seeland OT Gatersleben, Germany. https://doi.ipk-gatersleben.de/DOI/83e8e186-dc4b-47f7-a820-28ad37cb176b/d1067eba-1d08-42e2-85ec-66bfd5112cd8/2. 10.5447/ipk/2019/8 [DOI] [Google Scholar]
- Mascher M, Gundlach H, Himmelbach A, Beier S, Twardziok SO, Wicker T, Radchuk V, Dockter C, Hedley PE, Russell J, et al. 2017. A chromosome conformation capture ordered sequence of the barley genome. Nature 544: 427–433. 10.1038/nature22043 [DOI] [PubMed] [Google Scholar]
- Middleton CP, Stein N, Keller B, Kilian B, Wicker T. 2013. Comparative analysis of genome composition in Triticeae reveals strong variation in transposable element dynamics and nucleotide diversity. Plant J 73: 347–356. 10.1111/tpj.12048 [DOI] [PubMed] [Google Scholar]
- Moore G, Lucas H, Batty N, Flavell R. 1991. A family of retrotransposons and associated genomic variation in wheat. Genomics 10: 461–468. 10.1016/0888-7543(91)90333-A [DOI] [PubMed] [Google Scholar]
- O'Malley RC, Huang SC, Song L, Lewsey MG, Bartlett A, Nery JR, Galli M, Gallavotti A, Ecker JR. 2016. Cistrome and epicistrome features shape the regulatory DNA landscape. Cell 166: 1598. 10.1016/j.cell.2016.08.063 [DOI] [PubMed] [Google Scholar]
- Ou J, Wolfe SA, Brodsky MH, Zhu LJ. 2018. motifStack for the analysis of transcription factor binding site evolution. Nat Methods 15: 8–9. 10.1038/nmeth.4555 [DOI] [PubMed] [Google Scholar]
- Para A, Li Y, Coruzzi GM. 2018. μChIP-Seq for genome-wide mapping of in vivo TF-DNA interactions in Arabidopsis root protoplasts. Methods Mol Biol 1761: 249–261. 10.1007/978-1-4939-7747-5_19 [DOI] [PubMed] [Google Scholar]
- Park PJ. 2009. ChIP-seq: advantages and challenges of a maturing technology. Nat Rev Genet 10: 669–680. 10.1038/nrg2641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portwood JL, Woodhouse MR, Cannon EK, Gardiner JM, Harper LC, Schaeffer ML, Walsh JR, Sen TZ, Cho KT, Schott DA, et al. 2019. MaizeGDB 2018: the maize multi-genome genetics and genomics database. Nucleic Acids Res 47: D1146–D1154. 10.1093/nar/gky1046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramírez-González RH, Borrill P, Lang D, Harrington SA, Brinton J, Venturini L, Davey M, Jacobs J, van Ex F, Pasha A, et al. 2018. The transcriptional landscape of polyploid wheat. Science 361: eaar6089. 10.1126/science.aar6089 [DOI] [PubMed] [Google Scholar]
- R Core Team. 2020. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna. https://www.R-project.org/. [Google Scholar]
- Rice P, Longden I, Bleasby A. 2000. EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet 16: 276–277. 10.1016/S0168-9525(00)02024-2 [DOI] [PubMed] [Google Scholar]
- Robinson JT, Thorvaldsdóttir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP. 2011. Integrative genomics viewer. Nat Biotechnol 29: 24–26. 10.1038/nbt.1754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai H, Lee SS, Tanaka T, Numa H, Kim J, Kawahara Y, Wakimoto H, Yang CC, Iwamoto M, Abe T, et al. 2013. Rice Annotation Project Database (RAP-DB): an integrative and interactive database for rice genomics. Plant Cell Physiol 54: e6. 10.1093/pcp/pcs183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao Z, Zhang Y, Yuan GC, Orkin SH, Waxman DJ. 2012. MAnorm: a robust model for quantitative comparison of ChIP-Seq data sets. Genome Biol 13: R16. 10.1186/gb-2012-13-3-r16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonti CN, Pavličev M, Capra JA. 2017. Transposable element exaptation into regulatory regions is rare, influenced by evolutionary age, and subject to pleiotropic constraints. Mol Biol Evol 34: 2856–2869. 10.1093/molbev/msx219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slotkin RK, Martienssen R. 2007. Transposable elements and the epigenetic regulation of the genome. Nat Rev Genet 8: 272–285. 10.1038/nrg2072 [DOI] [PubMed] [Google Scholar]
- Sundaram V, Cheng Y, Ma Z, Li D, Xing X, Edge P, Snyder MP, Wang T. 2014. Widespread contribution of transposable elements to the innovation of gene regulatory networks. Genome Res 24: 1963–1976. 10.1101/gr.168872.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang H, Bowers JE, Wang X, Ming R, Alam M, Paterson AH. 2008. Synteny and collinearity in plant genomes. Science 320: 486–488. 10.1126/science.1153917 [DOI] [PubMed] [Google Scholar]
- Todd CD, Deniz O, Taylor D, Branco MR. 2019. Functional evaluation of transposable elements as enhancers in mouse embryonic and trophoblast stem cells. eLife 8: e44344. 10.7554/eLife.44344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trizzino M, Park Y, Holsbach-Beltrame M, Aracena K, Mika K, Caliskan M, Perry GH, Lynch VJ, Brown CD. 2017. Transposable elements are the primary source of novelty in primate gene regulation. Genome Res 27: 1623–1633. 10.1101/gr.218149.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda M, Seki M. 2020. Histone modifications form epigenetic regulatory networks to regulate abiotic stress response. Plant Physiol 182: 15–26. 10.1104/pp.19.00988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Dongen S. 2008. Graph clustering via a discrete uncoupling process. SIAM J Matrix Anal. Appl. 30: 121–141. 10.1137/040608635 [DOI] [Google Scholar]
- Wang H, Liu C, Cheng J, Liu J, Zhang L, He C, Shen WH, Jin H, Xu L, Zhang Y. 2016. Arabidopsis flower and embryo developmental genes are repressed in seedlings by different combinations of Polycomb group proteins in association with distinct sets of cis-regulatory elements. PLoS Genet 12: e1005771. 10.1371/journal.pgen.1005771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse AM, Procter JB, Martin DM, Clamp M, Barton GJ. 2009. Jalview Version 2—a multiple sequence alignment editor and analysis workbench. Bioinformatics 25: 1189–1191. 10.1093/bioinformatics/btp033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicker T, Gundlach H, Spannagl M, Uauy C, Borrill P, Ramírez-González RH, De Oliveira R, International Wheat Genome Sequencing Consortium, Mayer KFX, Paux E, et al. 2018. Impact of transposable elements on genome structure and evolution in bread wheat. Genome Biol 19: 103. 10.1186/s13059-018-1479-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. 2007. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol 24: 1586–1591. 10.1093/molbev/msm088 [DOI] [PubMed] [Google Scholar]
- Yu G. 2020. Using ggtree to visualize data on tree-like structures. Curr Protoc Bioinformatics 69: e96. 10.1002/cpbi.96 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, et al. 2008. Model-based Analysis of ChIP-Seq (MACS). Genome Biol 9: R137. 10.1186/gb-2008-9-9-r137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Xiao J, Wu J, Zhang H, Liu G, Wang X, Dai L. 2012. ParaAT: a parallel tool for constructing multiple protein-coding DNA alignments. Biochem Biophys Res Commun 419: 779–781. 10.1016/j.bbrc.2012.02.101 [DOI] [PubMed] [Google Scholar]
- Zhao H, Zhang W, Chen L, Wang L, Marand AP, Wu Y, Jiang J. 2018. Proliferation of regulatory DNA elements derived from transposable elements in the maize genome. Plant Physiol 176: 2789–2803. 10.1104/pp.17.01467 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.