Abstract
Lipidoid nanoparticles (LNPs) are the delivery platform in Onpattro, the first FDA-approved siRNA drug. LNPs are also the carriers in the Pfizer-BioNTech and Moderna COVID-19 mRNA vaccines. While these applications have demonstrated that LNPs effectively deliver nucleic acids to hepatic and muscle cells, it is unclear if LNPs could be used for delivery of siRNA to neural cells, which are notoriously challenging delivery targets. Therefore, the purpose of this study was to determine if LNPs could efficiently deliver siRNA to neurons. Because of their potential delivery utility in either applications for the central nervous system and the peripheral nervous system, we used both cortical neurons and sensory neurons. We prepared siRNA-LNPs using C12-200, a benchmark ionizable cationic lipidoid along with helper lipids. We demonstrated using dynamic light scattering that the inclusion of both siRNA and PEG-lipid provided a stabilizing effect to the LNP particle diameters and polydispersity indices by minimizing aggregation. We found that siRNA-LNPs were safely tolerated by primary dorsal root ganglion neurons. Flow cytometry analysis revealed that Cy5 siRNA delivered via LNPs into rat primary cortical neurons showed uptake levels similar to Lipofectamine RNAiMAX—the gold standard commercial transfection agent. However, LNPs demonstrated a superior safety profile, whereas the Lipofectamine-mediated uptake was concomitant with significant toxicity. Fluorescence microscopy demonstrated a time-dependent increase in the uptake of LNP-delivered Cy5 siRNA in a human cortical neuron cell line. Overall, our results suggest that LNPs are a viable platform that can be optimized for delivery of therapeutic siRNAs to neural cells.
Graphical Abstract
Supplementary Information
The online version contains supplementary material available at 10.1208/s12248-021-00653-2.
KEY WORDS: C12-200, lipidoid nanoparticle, neural cell, siRNA, transfection
INTRODUCTION
Delivery of small interfering RNA (siRNA) is a promising strategy to treat pathologies as it allows genetic manipulation with a greater degree of target specificity and fewer off-target effects (1, 2). Lipidoid nanoparticles (LNPs) are the carriers in Onpattro, the first LNP-based RNAi drug to gain FDA approval for the treatment of polyneuropathies caused by a rare and life-threatening disorder, hereditary transthyretin-mediated amyloidosis (3). The clinical utility of LNPs is further reiterated by the recent full and emergency FDA approvals of the Pfizer-BioNTech (Comirnaty) and Moderna COVID-19 mRNA vaccines, respectively (4, 5). Based on the clinical success of LNPs in delivering siRNAs to hepatic and non-hepatic targets (6–8), we explored if LNPs could effectively deliver siRNA to neural cells. Neural cells are targets for drug delivery in multiple CNS disorders such as Alzheimer’s disease, Parkinson’s disease, ischemic stroke, peripheral nerve injuries, and neuropathic and inflammatory pain (8–12). The key challenge associated with the delivery of siRNA to neural cells is its poor accumulation and short duration of action inside cells.
The potent delivery characteristics of LNPs may allow them to be optimized for neural cell delivery. LNPs are primarily composed of an ionizable cationic lipidoid along with helper lipids like distearoylphosphatidylcholine (DSPC), polyethylene glycol-dimyristoyl glycerol (PEG-DMG), and cholesterol (12, 13). Ionizable cationic lipidoids allow higher encapsulation efficiencies and improved intracellular release via effective endosomal escape (14). Ionizable cationic lipidoids with pKa values between 6 and 7 acquire a positive charge at acidic pH and result in electrostatic interactions with the negatively charged siRNA molecules leading to the spontaneous formation of LNPs. At physiological pH of 7–7.4, these obtain a neutral charge and thereby largely eliminate the toxicity associated with cationic lipids such as Lipofectamine (15). This key feature renders the LNPs a superior safety profile compared to cationic transfection agents. The acidic microenvironment of endosomes lend the LNPs a positive charge because of which they associate with negative anionic endosomal lipids leading to endosomal destabilization and improved siRNA release into the cytoplasm (15, 16).
Seminal studies by Langer, Anderson, and colleagues developed high-throughput lipidoid libraries consisting of thousands of ionizable cationic lipidoids and identified lead lipidoids based on their in vitro and in vivo delivery efficacies in rodents as well as non-human primates. Among those, the C12-200 lipidoid demonstrated the highest gene knockdown efficacy and uptake in vitro (6, 17, 18). Therefore, we chose C12-200 lipidoid as the ionizable cationic lipidoid component of LNPs. LNP-based siRNA delivery harnesses several key advantages of LNP chemistry such as high transfection efficiency, low toxicity and immunogenicity, protection of the payload from physiological enzymes to increase their metabolic stability, ability for ligand conjugation, and enhancing the overall pharmacokinetics of the delivered cargo (19–21). The novelty of our work lies in exploiting some of these cardinal features of LNPs to maximize siRNA uptake into neural cells, a notoriously challenging delivery target.
In this pilot study, we formulated siRNA-LNPs using C12-200 lipidoid and characterized their colloidal stability using dynamic light scattering. We characterized their cytocompatibility with a panel of breast cancer cell lines and primary dorsal root ganglion cultures, a model of sensory neurons. We studied the qualitative and quantitative uptake of siRNA delivered via LNPs into neurons using fluorescence microcopy and flow cytometry, respectively. Despite mediating similar levels of siRNA uptake, we found that LNPs are a safe carrier and show no signs of toxicity in the neuronal models compared to the commercially available cationic lipid, Lipofectamine. These findings encourage the further optimization of LNPs for knockdown of therapeutically relevant neuronal targets.
EXPERIMENTAL SECTION
Materials
1,2-Distearoyl-sn-glycero-3-phosphocholine (DSPC) (850365P) and 1,2-Dimyristoyl-rac-glycero-3-methoxypolyethylene glycol-2000 (PEG-DMG) (8801518) were obtained from Avanti Polar Lipids (Alabaster, AL). Cholesterol (8667) and Cy5-labeled siRNA were procured from Sigma-Aldrich (St. Louis, MO). 1,1′-((2-(4-(2-((2-(bis (2 hydroxy dodecyl) amino) ethyl) (2hydroxydodecyl) amino) ethyl) piperazin1yl) ethyl) azanediyl) bis(dodecan-2-ol) (C12-200), an ionizable cationic lipidoid, was a generous gift from Alnylam Pharmaceuticals (Cambridge, MA). Quant-iT RiboGreen RNA Assay Kit (R11490) was purchased from Life Technologies (Carlsbad, CA). Triton X-100 was procured from Acros Organics (Morris Plains, NJ). Heat-inactivated fetal bovine serum (FBS) and phosphate-buffered saline (PBS) were purchased from Hyclone Laboratories (Logan, UT). TrypLE express, RPMI + GlutaMAX-I (1X), and DMEM/F-12 were purchased from Life Technologies Corporation (Grand Island, NY). MCF-7, MDA-MB-231, and BT-549 cell lines were generously provided by Dr. Jane Cavanaugh (Duquesne University). Human cortical neuron cell line (HCN-2) (CRL-10742) and primary rat cortical neurons (RCN) (A10840-02) were purchased from ATCC (Manassas, VA) and Thermo Fisher Scientific (Frederick, MD) respectively. siGFP (AM4626), siGAPDH (Silencer Select GAPDH siRNA) (4390849), siTRPV1 (5′ GCGCAUCUUCUACUUCAACTT 3′, 3′ TTCGCGUAGAAGAUGAAGUUG 5′) [4390828 (Assay ID 554887)], and control, inverted siTRPV1 (5′ CAACUUCAUCUUCUACGCGTT 3′, 3′ TTGUUGAAGUAGAAGAUGCGC 5′) [4390828 (Assay ID 554886)] were procured from Thermo Fisher Scientific (Austin, TX). RNeasy Mini Kit (74104) and QIAshredder (79654) were procured from Qiagen (Qiagen, Germantown, MD). High-capacity cDNA Reverse Transcription Kit (4368813) was purchased from Thermo Fisher Scientific (Thermo Fisher, Waltham, MA). Forward (AAGGATGGAACAACGGGCTAG) and reverse primers (TCCTGGTAGTGAAGATGTGGG) (299688565) for TRPV1 were procured from Integrated DNA Technologies (Coralville, IA). All reagents were used as received unless stated otherwise.
Preparation of siRNA-Loaded LNPs (siRNA-LNPs)
LNPs were prepared using previously reported methods (7). Briefly, the ionizable cationic lipidoid C12-200, DSPC, cholesterol, and PEG-DMG were dissolved in ethanol at a molar ratio of 50/10/38.5/1.5. The concentrations and volumes used for LNP preparation are shown in Table I. A 1 mg/mL solution of siRNA was prepared in 10 mM citrate buffer, pH 4.0. The “slow mixing” and “fast mixing” protocols are defined based on the speed used during the addition of the lipid/ethanolic phase to the aqueous phase. For the “slow mixing” method, the entire volume of the lipid phase was added to the aqueous phase instantaneously (in a single shot) followed by vortexing the mixture for 30 s while maintaining the Fisher benchtop vortexer knob at position “7.” For the “fast mixing” protocol, the lipid phase was added dropwise to the aqueous phase under continuous vortexing for 30 s while maintaining the Fisher benchtop vortexer knob at position “7.” A precalculated volume of 1X PBS pH 7.4 was then added to the LNPs to adjust the final siRNA concentration to 400 nM. All the formulations were made at a final siRNA concentration of 400 nM, and cationic ionizable lipidoid/siRNA w/w ratio was maintained at 5:1, unless stated otherwise.
Table I.
Representative Formulation Scheme for LNPs
| Component | w/w% in final LNP mixture | Stock concentration (mg/mL) | Volume of the stock solution required (μL) |
|---|---|---|---|
| Ethanolic phase | |||
| C12-200 | 50 | 1 | 12.5 |
| Cholesterol | 38.5 | 1 | 3.1 |
| PEG-DMG | 1.5 | 0.5 | 13.6 |
| DSPC | 10 | 0.5 | 16.9 |
| Ethanol | 78.9 | ||
| Total ethanolic phase volume | 125 | ||
| Aqueous phase | |||
| siRNA | 400 nM | 1 | 2.5 |
| 10 nM citrate buffer | 122.5 | ||
| PBS | 250 | ||
| Total aqueous phase volume | 375 | ||
| Final volume of LNPs prepared (μL) | 500 | ||
Agarose Gel Electrophoresis
Agarose gel electrophoresis was performed to qualitatively confirm the encapsulation of siRNA in LNPs. Briefly, samples were loaded in a 2% agarose gel prepared in 1× Tris-borate buffer containing 0.05% ethidium bromide. Samples pre-mixed with 1× RNA loading buffer were electrophoresed in an Owl EasyCast B2 Mini Gel Electrophoresis System (Thermo Fisher Scientific, St. Louis, MO) for 90 min. The gel was imaged under UV light using a Gel Doc system (ProteinSimple, San Jose, CA).
Determination of the Extent of siRNA Loading UsingRiboGreen RNA Assay
The extent of siRNA loading in the prepared LNPs was measured using the Quant-iT RiboGreen RNA Assay Kit by following the manufacturer’s instructions. RiboGreen is a fluorescent dye, which, in its free form, exhibits minimal fluorescence, but when bound to a nucleic acid, it fluoresces with an exceptionally high intensity that is directly proportional to the amount of nucleic acid (22). Total siRNA content is defined as the amount of encapsulated and non-encapsulated/free siRNA in the LNPs. The difference between the total and free siRNA was used to calculate the amount of siRNA encapsulated within the LNPs. A fresh sample of two µg/mL dsRNA stock solution was prepared each time with 1× Tris-EDTA (TE), and the standard curve was generated at concentrations ranging from 0 to 1000 ng/mL. An equivalent amount of free siRNA sample was prepared in 1× TE, and LNPs were lysed with 2% Triton X-100 in 1× TE to measure the “total” siRNA content. The fluorescence intensities were measured on a SYNERGY HTX multi-mode reader (Winooski, VT) at an excitation wavelength of 485 nm and an emission wavelength of 528 nm. The % encapsulation efficiency was calculated using the Eq. 1.
| 1 |
Dynamic Light Scattering
The average particle diameters, polydispersity indices, and zeta potentials of the blank and siGFP-loaded LNPs were measured by dynamic light scattering (DLS) using a Malvern Zetasizer Pro (Malvern Panalytical Inc., Westborough, PA) over a period of 7 days with intermittent storage at 2–8 °C and upon storage at 37 °C for 24 h. We also performed the colloidal stability analysis of LNPs 24-h post-preparation in 1× PBS and in samples supplemented with FBS (10%) to determine the serum stability of LNPs. The prepared LNPs were diluted to a siRNA concentration of 40 nM using 10 mM 1× PBS pH 7.4 for particle size measurements and using deionized water for zeta potential measurements. All measurements were carried out in triplicate, and the data are presented as average +/- standard deviation (SD) and are representative of at least five-six independent experiments (measurement errors <5%).
Cell Culture
MCF-7 and BT-549 cells were maintained in RPMI + GlutaMAX (1×) supplemented with 10% FBS. MDA-MB-231 cells were maintained in DMEM/F-12 1:1 medium supplemented with 2.5 mM L-glutamine + 15 mM HEPES buffer + 110 mg/L sodium pyruvate + 10% FBS. The growth medium was changed every 48 h. Primary rat dorsal root ganglion (DRG) cultures were isolated using previously reported methods (23). DRG cultures were maintained in DMEM/high glucose (4.5 g/L glucose) media supplemented with 2.5 mM L-glutamine + 15 mM HEPES buffer + 110 mg/L sodium pyruvate + 10% FBS. Human cortical neuron cell line was maintained in DMEM/high glucose (4.5 g/L glucose) medium supplemented with 10% FBS. Primary rat cortical neurons were maintained in Neurobasal Plus Medium supplemented with 200 mM GlutaMAX I and B-27 Plus supplement. All cultures were maintained in a humidified incubator at 37°C and 5% CO2.
Cytocompatibility of LNPs
MCF-7, MDA-MB-231, BT-549 cells, and primary rat DRG neurons were seeded in a clear, flat-bottom Poly-D-Lysine-coated 96-well plate (Azer Scientific, Morgantown, PA) for the cytocompatibility studies. MCF-7, MDA-MB-231, and BT-549 cells were seeded at a density of 5,000 cells/well, and DRG neurons were seeded at a density of 1,500 cells/well. Untreated cells were used as a control in all studies. Cells were transfected for 4 h with LNPs containing 50 nM of siRNA in complete growth medium in a volume of 50 μL/well. We also treated cells with increasing doses of siRNA-LNPs (10, 25, 50, 75, and 100 nM siGFP-loaded LNPs) to determine any possible effects of siRNA/lipid dose escalation on cytocompatibility. Post-transfection, the transfection mixture was replaced with 200 μL of fresh complete growth medium/well followed by a 24-h incubation at 37 °C and 5% CO2. The relative intracellular ATP levels were measured using the CellTiter-Glo Luminescence Assay (ATP assay) 24-h post-transfection using the previously described methods (24–26). The levels of ATP directly correlate with the cell numbers used here as a measure of cell viability. The ATP levels of the treatment groups were measured against those of the control, untreatedcells. Briefly, 60 μL of the complete growth medi and 60 μL of CellTiter-Glo 2.0 reagent were added to each well of the 96-well plate. The plate was incubated for 15 min in dark at room temperature in an incubator shaker (Thermo Fisher Scientific, Waltham, MA). Following incubation, 60 μL of the mixture from each well was transferred to a flat-bottom, white opaque 96-well plate (Azer Scientific, Morgantown, PA). The luminescence was measured using SYNERGY HTX multi-mode reader (BioTek Instruments, Winooski, VT). Relative ATP levels (%) were calculated for the transfection groups by normalizing them to the luminescence of the untreated cells as shown in Eq. 2.
| 2 |
Cy5 siRNA Uptake into Primary Rat Cortical Neurons Using Flow Cytometry
Primary rat cortical neurons (RCN) were plated in a clear, flat-bottom Poly-D-Lysine-coated 24-well plate (Genesee Scientific, San Diego, CA) at a density of 50,000 neurons/well in Complete Neurobasal Plus media and were allowed to acclimatize at 37 °C and 5% CO2 for 4–5 days. Cy5 siRNA-containing LNPs were prepared as described earlier and diluted with the Complete Neurobasal Plus medium to a final Cy5 siRNA concentration of 50 nM. The neurons were incubated with the transfection mixture for 24 h. The transfection mixture contained either naked Cy5 siRNA or Cy5 siRNA-LNPs with PEG-DMG (+ PEG-DMG) or Cy5 siRNA-LNPs without PEG-DMG (− PEG-DMG) or Cy5 siRNA-Lipofectamine RNAiMAX complexes diluted in complete growth medium. Post-exposure, cells were gently removed from the wells and collected in microcentrifuge tubes. The cell suspension was centrifuged at 300 g for 5 min and the pellet was resuspended in 500 µL 1 × PBS. Cells were then analyzed using an Attune NxT Acoustic Focusing Cytometer (Singapore) equipped with Attune NxT software. The fluorescence intensity of Cy5 siRNA was detected at an excitation wavelength of 638 nm and an emission wavelength of 720/30 nm. A total of 30,000 events were recorded for each sample. Histogram plots were obtained from the Attune NxT software. Cells were gated using forward vs. side scatter plots to exclude dead cells and cell debris. Untreated cells were used as negative-staining controls to set a gate for Cy5-negative and Cy5-positive populations. Data are presented as a percentage of Cy5-positive (Cy5 ( +)) events.
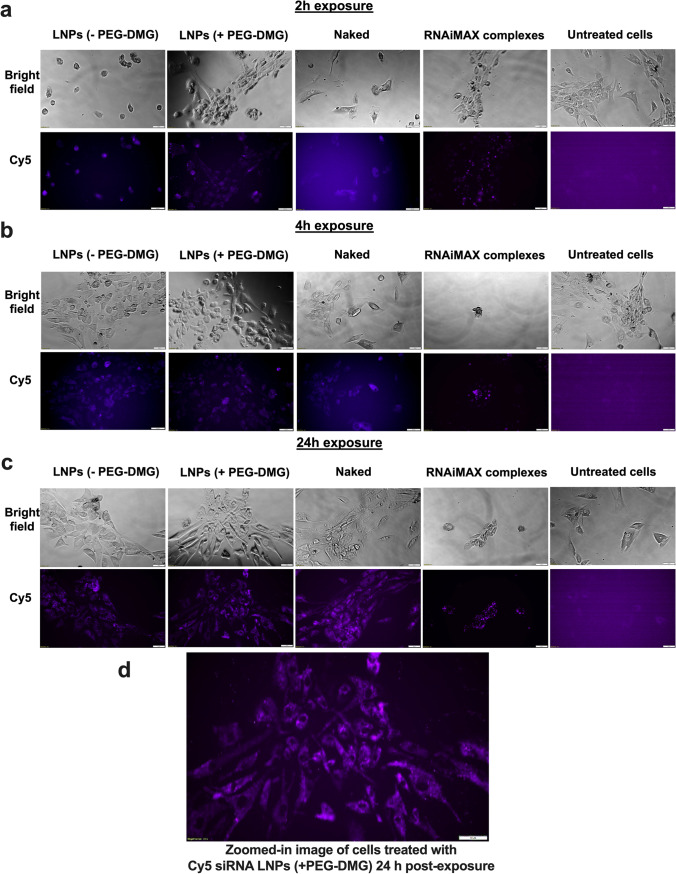
Cy5 siRNA Uptake by Fluorescence Microscopy in a Human Cortical Neuron Cell Line
We used fluorescence microscopy to determine the uptake of Cy5 siRNAinto a human neuronal cell line. Human cortical neurons (HCN-2) were seeded in a clear, flat-bottom, Poly-D-Lysine-coated 48-well plate (Genesee Scientific, San Diego, CA) at a density of 7,000 cells/well. The cells were cultured in complete growth medium in a humidified incubator at 37 °C and 5% CO2 for 1–2 weeks while replenishing the growth medium every other day. After 4–5 days of culturing, the media was replaced with 175 μL of the transfection mixture. The transfection mixture consisted of either Cy5 siRNA-LNPs or Cy5 siRNA-Lipofectamine RNAiMAX complexes containing 50 nM or 100 nM of Cy5 siRNA or naked Cy5 siRNA diluted in complete growth medium. Cells were incubated with the transfection mixture for 2, 4, and 24 h at 37 °C and 5% CO2. After the respective time points, the cells were washed once using pre-warmed PBS before adding 0.5 mL of the complete, phenol red-free growth medium/well. Untreated cells and cells treated with Cy5 siRNA-Lipofectamine RNAiMAX complexes were used as negative and positive controls, respectively. Cells were imaged using an Olympus IX 73 epifluorescent inverted microscope (Olympus, Center Valley, PA) to detect Cy5 signals at excitation and emission wavelengths of 651 nm and 670 nm, respectively.
Statistical Analysis
The data is expressed as mean ± standard deviation (SD), wherever applicable. Comparative statistical analyses were performed using either one-way, two-way ANOVA or one-sample t and Wilcoxon tests using GraphPad Prism 9 (GraphPad Software, San Diego, CA). Bonferroni’s multiple comparisons test was performed for comparative analyses using one-way ANOVA, wherever applicable. Tukey’s and Šídák’s multiple comparisons tests for statistical comparisons were performed using two-way ANOVA, wherever applicable. Alpha was set at 0.05.
RESULTS
Determination of the Extent of siRNA Loading Using the RiboGreen Assay
We used the RiboGreen assay as a quantitative measure to determine the extent of siRNA encapsulation within the LNPs (Fig. 1) and used agarose gel electrophoresis as a qualitative tool to confirm the absence of “free” or unentrapped siRNA (Supplementary Fig. 1). Triton X-100 was used to lyse the LNPs resulting in the extraction of the entrapped siRNA to measure the “total” siRNA (total siRNA = entrapped siRNA + free siRNA). Fluorescence readout from LNPs not treated with Triton X-100 is indicative of the amount of free/non-encapsulated siRNA. Two different mixing speeds used during LNP formulation, slow vs. fast, were compared for determining their effects on % siRNA encapsulation. The values increased from 74.4% at slow mixing to 83.8% at faster mixing speeds for LNPs prepared without the inclusion of PEG-DMG, whereas we did not observe a significant increase in siRNA loading for LNPs prepared with PEG-DMG at the different mixing speeds.
Fig. 1.
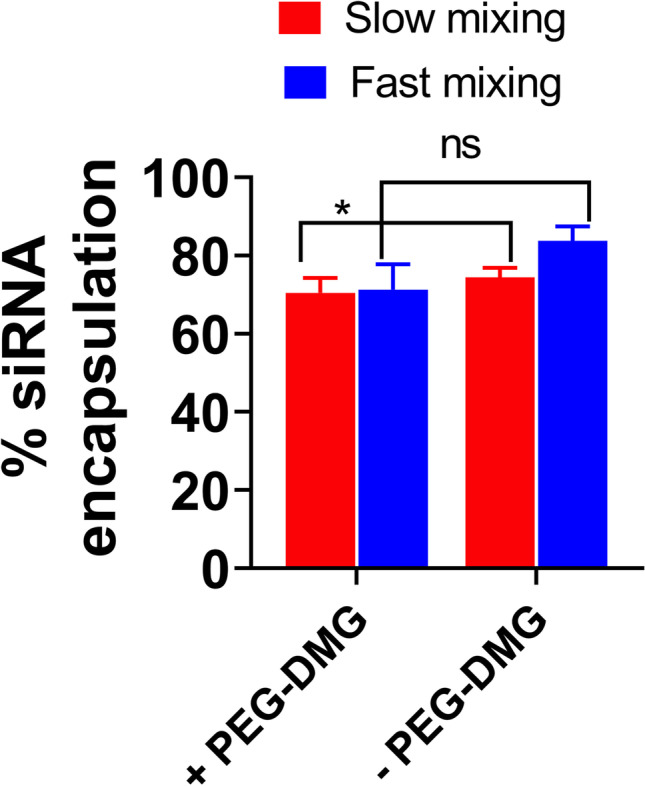
Extent of siRNA encapsulation in LNPs prepared at different mixing speeds determined by the RiboGreen assay. The extent of siRNA encapsulation in LNPs (± PEG-DMG) was calculated using the following equation: . SiRNA-LNPs were initially prepared in 10 mM citrate buffer and further diluted to a final siRNA concentration of 50 nM using 1 × PBS. Data are presented as mean ± SD of n = 3 samples. Statistical comparisons were made using two-way ANOVA. *p < 0.05 and ns, non-significant
Colloidal Stability of siRNA-Loaded LNPs
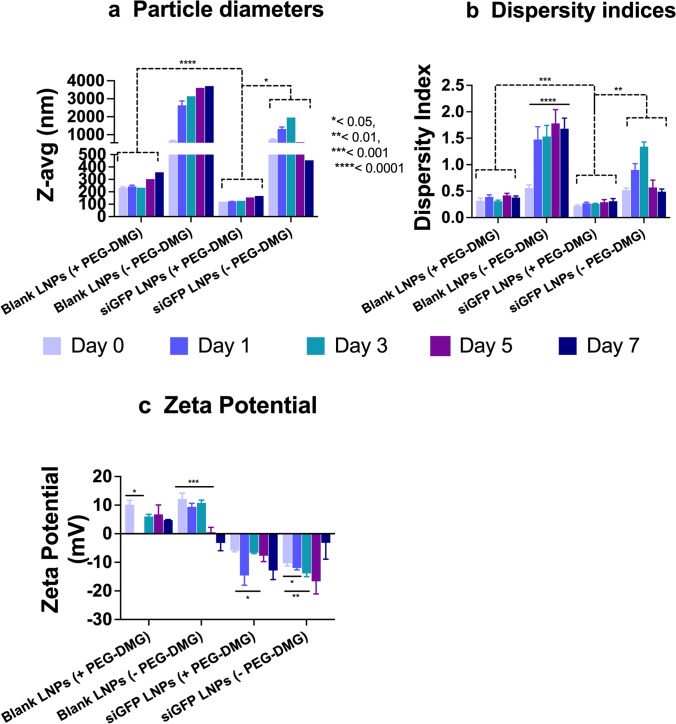
Physicochemical characteristics of nanocarriers such as particle diameter, polydispersity index (PDI), and surface charge (zeta potential) are influential determinants of their biological activity (27–29). Published studies have reported significant effects of physicochemical characteristics of nano-sized systems on the overall delivery efficacy (30–33). Particle characteristics of self-assembled systems such as LNPs can vary over time during storage potentially affecting their biological activity. We also anticipated that neural cells may show a slower rate of particle uptake compared to non-neuronal cells owing to limited endocytosis/other uptake pathways. We therefore measured the changes in particle diameters and zeta potentials of the LNPs over a period of 7 days where the samples were refrigerated interim. Surface coating of nanoparticles using PEG- DMG has been reported to improve their pharmacokinetic profile by reducing the recognition by the mononuclear phagocyte system (34–36). We studied the effect of PEG-DMG in stabilizing LNPs by comparing LNPs prepared in the presence (+) and absence (−) of PEG-DMG. We compared the particle parameters of blank LNPs and siRNA-loaded LNPs to study the effect of siRNA encapsulation on the resulting particle characteristics.
Blank LNPs (+PEG-DMG) showed average particle diameters of about 250 nm which increased to about 350 nm on day 7. In contrast, the diameters of LNPs (-PEG-DMG) were about 900-nm post-preparation and increased to 4 µm after 7 days (Figure 2a). A similar trend was observed for the PDI values of blank LNPs, i.e., an initial PDI of 0.32 increased to 0.4 after 7 days for LNPs (+PEG-DMG) and an initial PDI of 0.5 increased to 1.8 after 7 days for LNPs (−PEG-DMG) (Figure 2b). We observed a few statistically significant changes in the particle diameters, PDI values, and zeta potential of the prepared LNPs over the period of 7 days (Supplementary Table 1). The narrow polydispersity indices and smaller average particle diameters demonstrate the stabilizing effect of PEG-DMG in the LNPs. siGFP-loaded LNPs (with PEG-DMG) showed average particle diameters of about 120 nm which increased to about 200 nm on day 7 (Figure 2a). Their initial PDI values of ca. 0.2 increased to about 0.3 after 7 days (Figure 2b). From this observation, we can infer that siRNA loading in the LNPs results in an additional stabilizing effect to the respective blank LNP counterparts resulting in narrow and consistent PDI values and particle diameters.
Fig. 2.
Colloidal stability of blank or siGFP-loaded LNPs measured using dynamic light scattering. SiGFP-LNPs were initially prepared in 10 mM citrate buffer and further diluted to a final siRNA concentration of 50 nM using 1 × PBS (pH 7.4). The samples were stored at 2–8 °C while not in use. Z-average particle diameters (a), dispersity indices (b), and zeta potentials (c) were measured on a Malvern Zetasizer Pro. Data are presented as mean ± SD of n = 3 measurements. Statistical comparisons were made using one-way ANOVA or one-sample t and Wilcoxon tests. *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001
The cationic lipidoid, C12-200, in the LNPs resulted in a net positive charge to the blank LNPs. Blank LNPs containing PEG-DMG showed slightly lower zeta values (6–10 mV) compared to blank LNPs without PEG-DMG (9–12 mV) (Figure 2c) that is likely explained by the insertion of PEG chains in between the C12-200 lipidoids or due to the formation of a PEG monolayer around C12-200 (37). As seen from Figure 2c, siGFP-loaded LNPs showed a negative zeta potential compared to their blank LNP counterparts. Although the changes were not statistically significant, the zeta potentials of the siGFP-loaded LNPs decreased over the 7-day period from −5.91 to −19.12 mV. Overall, the average particle diameters and polydispersity indices do not vary significantly over a period of 7 days. However, zeta potential values of siGFP-LNPs decreased over a period of 7 days. Despite the apparent numerical differences in the zeta potentials, a one-way ANOVA analysis revealed that the differences were not significant.
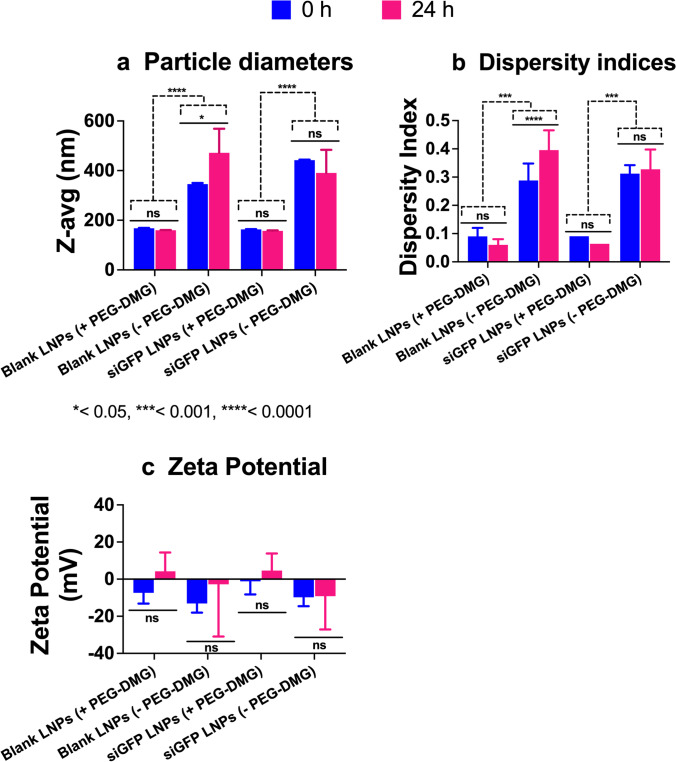
The colloidal stability of siGFP-loaded and blank-loaded LNPs was measured using dynamic light scattering at 0 (immediately after preparation) and 24-h post-preparation upon storage at 37 °C (Figure 3). We also studied the effect of PEG-DMG in stabilizing LNPs by preparing LNPs with (+) and without (−) PEG-DMG. Blank LNPs (+PEG-DMG) showed an average particle diameter of about 167.8 nm that shifted to about 159.8 nm 24-h post storage at 37 °C. In contrast, the diameters of LNPs (−PEG-DMG) were about 345.5 nm post-preparation and increased to 471.1 nm 24-h post storage at 37 °C (Figure 3a). A similar trend was observed for the PDI values of blank LNPs, i.e., an initial PDI of 0.09 decreased to 0.06 for LNPs (+PEG-DMG) and an initial PDI of 0.3 increased to 0.4 for LNPs (−PEG-DMG) 24-h post storage at 37 °C (Figure 3b). We found a similar trend for the particle diameters and dispersity indices of siGFP-loaded LNPs, both (+) and (−) PEG-DMG, 24-h post storage at 37 °C (Figure 3a and b). siGFP-loaded LNPs (+PEG- DMG) showed an average particle diameter of about 163.2 nm that shifted to about 157.2 nm 24 h-post storage at 37 °C. In contrast, the diameters of LNPs (−PEG-DMG) were about 442.2 nm post-preparation and decreased to 390.7 nm 24-h post storage at 37 °C (Figure 3a). For the dispersity indices, an initial value of 0.09 decreased to 0.06 for siGFP-loaded LNPs (+PEG-DMG), and an initial value of 0.31 shifted to 0.32 for LNPs (−PEG-DMG) 24-h post storage at 37 °C (Figure 3b). Nevertheless, the observed changes in particle diameters and dispersity indices of blank LNPs (+ PEG-DMG) and siGFP-loaded LNPs (+ and – PEG-DMG) 24-h post storage at 37 °C were statistically insignificant. However, we noted a significant increase in the particle diameters (*p < 0.05) and dispersity indices (****p < 0.0001) of blank LNPs (-PEG-DMG) likely due to the absence of PEG-DMG and the siGFP cargo—which are known to provide a stabilizing effect by inhibiting particle-particle aggregation and via complexation of the negatively charged siRNA with the positively charged C12-200, respectively. Noteworthy, we noted a similar trend of the stabilizing effects of PEG-DMG and the siGFP cargo in Figure 2.
Fig. 3.
Colloidal stability of blank- or siGFP-loaded LNPs immediately after preparation (0 h) and upon storage at 37 °C for 24 h measured using dynamic light scattering. SiGFP-LNPs were initially prepared in 10 mM citrate buffer and further diluted to a final siRNA concentration of 50 nM using 1 × PBS at a final pH of 7.4 for particle size and PDI measurements and using deionized water for zeta potential measurements. The samples were stored at 37 °C for 24 h. Z-average particle diameters (a), polydispersity indices (b), and zeta potentials (c) were measured on a Malvern Zetasizer Pro. Data are presented as mean ± SD of n = 3 measurements. Statistical significance was determined using two-way ANOVA Tukey’s and Šídák’s multiple comparisons tests. *p < 0.05, ***p < 0.001, ****p < 0.0001 and ns, non-significant
Blank LNPs containing PEG-DMG showed slightly negative zeta potentials (−5 to −7 mV) that shifted to slightly cationic values (3 to 4 mV), whereas the zeta potentials of blank LNPs without PEG-DMG were about −13 mV that increased to about −2 mV 24-h post storage at 37 °C (Figure 3c). siGFP-loaded LNPs (+ PEG-DMG) showed a zeta potential of −1.2 mV that increased to about 4.6 mV, whereas the LNPs (−PEG-DMG) counterparts showed a zeta potential of −9.8 mV that increased to about −9.1 mV 24 h- post storage at 37°C (Figure 3c). Despite the apparent numerical differences, it should be noted that the measured zeta potentials reflect electrostatically neutral samples, and a two-way ANOVA analysis revealed that the differences were not significant.
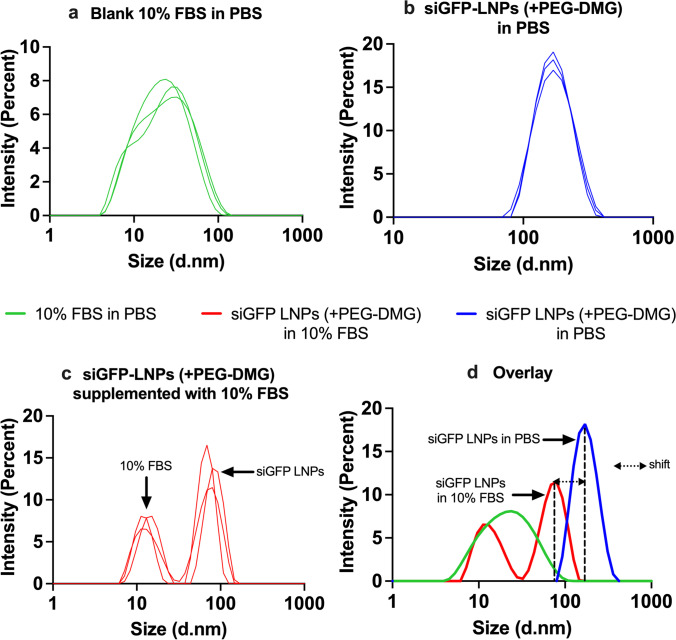
The particle diameters of siGFP-LNPs (+PEG-DMG) containing 10% FBS was measured using dynamic light scattering to determine the effect of serum proteins on the colloidal stability of LNPs. We specifically chose siGFP-LNPs (+PEG-DMG) to study the effect of serum since the presence of siRNA and PEG-DMG resulted in the maximum stability of LNPs with respect to particle diameters and dispersity indices (Figure 2a and b). We first measured the particle diameter of a control sample containing 10% FBS in PBS (Figure 4a) that showed an average particle diameter of 19.6 nm, whereas siGFP-LNPs (+PEG-DMG) in PBS showed an average particle diameter of about 170 nm (Figure 4b). We observed distinct intensity size distributionpeaks for the blank 10% FBS as well as LNP samples (Figure 4a and b). We then measured the particle diameter of siGFP-LNPs (+PEG-DMG) that was supplemented with 10% FBS. Figure 4c shows two distinct peaks denoting peaks corresponding to 10% FBS and the siGFP-LNPs (+PEG-DMG) at 20.2 nm and 80.5 nm, respectively. The overlay size distribution plot demonstrated a “shift” in the siGFP-LNPs (+PEG-DMG) peak towards the left (lower particle diameters) when the LNPs were supplemented with 10% FBS as opposed to those in PBS (80.5 nm vs 170 nm) (Figure 4d). We speculate that the observed decrease in diameter is suggestive of the additional stabilizing effects of serum (10% FBS) on the LNP diameters. This, in fact, may be beneficial/favorable for LNP uptake (Figure 7 and Figure 8) as neurons may preferentially internalize smaller compared to larger LNPs.
Fig. 4.
Intensity size distribution plots demonstrating the colloidal stability of siGFP-loaded LNPs (+ PEG-DMG) supplemented with 10% FBS. a A blank sample consisting of 10% FBS in 1 × PBS was used to understand the serum protein-mediated scattering. b SiGFP-LNPs (+ PEG-DMG) were initially prepared in 10 mM citrate buffer and further diluted to a final siRNA concentration of 50 nM using 1 × PBS. LNPs supplemented with 10% FBS were measured to determine their serum stability. d An overlay depicts the pattern of the size distribution obtained for all the LNP samples
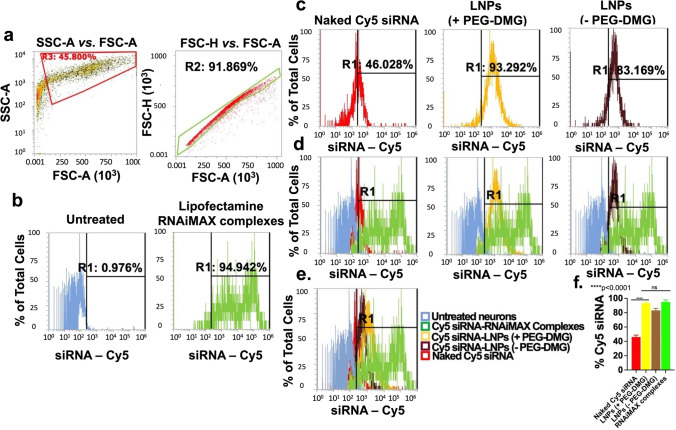
Fig. 7.
Uptake of LNPs into rat primary cortical neurons determined using flow cytometry analysis. Cells were transfected for 24 h with the indicated samples in a 24-well plate. Cy5 siRNA-Lipofectamine RNAiMAX complexes and untreated cells were used as positive and negative controls, respectively. Data are presented as percentages of positive cells for Cy5 (Cy5 ( +)). SSC-A vs. FSC-A and FSC-H vs. FSC-A plots of untreated cells demonstrating monodispersed cells (a), untreated and Cy5 siRNA/Lipofectamine RNAiMAX-treated cells (b), cells treated with the indicated samples (c), overlay histograms comparing Cy5 ( +) cells in each treatment group in comparison to the controls (d), overlay histograms (e), and % Cy5 siRNA uptake by the neurons for the respective groups (f). The histograms are representative of quadruplicate samples. Statistical comparisons were made using one-way ANOVA. ****p < 0.0001
Fig. 8.
Cellular uptake of Cy5 siRNA determined using fluorescent microscopy. HCN-2 cells (human cortical neuron cell line) were incubated for 2 h (a), 4 h (b), and 24 h (c) with the indicated samples containing 50 nM Cy5 siRNA. Panel (d) represents the zoomed-in image depicting HCN-2 cells incubated with LNPs with PEG-DMG. Scale bar = 50 µm. Cells were imaged using an Olympus IX 73 epifluorescent inverted microscope to detect Cy5 signals at excitation and emission wavelengths of 651 nm and 670 nm, respectively. Images are representative of n = 3 independent wells
The correlation function is a statistical analysis tool for measuring the non-randomness in a data set that is depicted here as a correlogram. A correlogram is a plot of the correlation coefficient G (τ) vs time (µs) (Figure 5). The Y-intercept of the correlogram is indicative of the signal to noise ratio and the presence/absence of multiple scattering. A Y-intercept of ~0.9–1.15 indicates a good signal in the absence of multiple scattering. Samples with multiple scattering show a Y-intercept of ~0.6–0.8, and samples with number fluctuations show a Y-intercept of ~1.15–1.4 (38). As observed from Table II, the Y-intercept values for all the LNP groups over a period of 7 days ranged from 0.8 to 1.1 that is indicative of a good signal-to-noise ratio in the absence of multiple scattering and number fluctuations.
Fig. 5.
Representative correlogram and intensity size distribution plots of siGFP-loaded LNPs (+ PEG-DMG). siGFP-LNPs were initially prepared in 10 mM citrate buffer and further diluted to a final siRNA concentration of 50 nM using 1 × PBS at a final pH of 7.4 prior to particle size measurements on a Malvern Zetasizer Pro
Table II.
Comparison of the Y-Intercept in the Autocorrelograms from the Colloidal Stability Data (Fig. 2) at Different Time Points
| Groups | Y-intercept | ||||
|---|---|---|---|---|---|
| Day 0 | Day 1 | Day 3 | Day 5 | Day 7 | |
| Blank LNPs (with PEG-DMG) | 0.98 | 0.98 | 0.94 | 0.89 | 0.8 |
| Blank LNPs (without PEG-DMG) | 0.99 | 1.3 | 1 | 1 | 0.9 |
| siGFP LNPs (with PEG-DMG) | 0.96 | 0.93 | 0.9 | 0.86 | 0.87 |
| siGFP LNPs (without PEG-DMG) | 1 | 1.1 | 1.1 | 0.96 | 0.88 |
Cytocompatibility of C12-200-LNPs
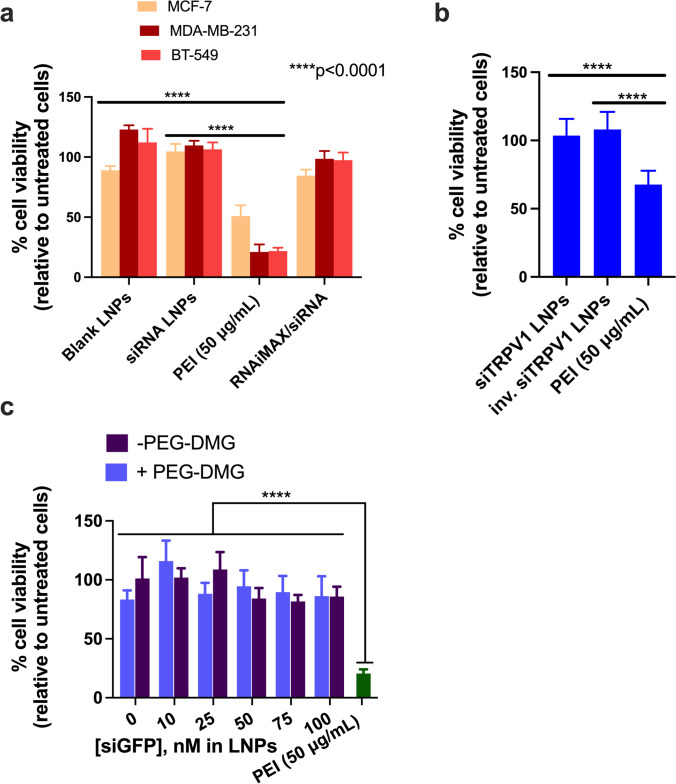
We studied the cytocompatibility of the prepared C12-200/siGAPDH LNPs with MCF-7, MDA-MB-231, and BT-549 cells as these cell models are known to overexpress TRPV1, a neuronal target for siRNA delivery (Figure 6a) (39). We also studied the compatibility of C12-200/siTRPV1 LNPs with primary rat dorsal root ganglion (DRG) neurons (Figure 6b). We specifically chose primary rat DRG neurons due to their high expression of TRPV1 (38, 39). TRPV1 is reported to be expressed in 60% of the peripheral nociceptors present in the DRG (40) and is a potential neuronal target for gene knockdown. CellTiter-Glo assay (referred henceforth as the ATP assay) was used the study cytocompatibility of LNPs. The ATP assay is a simple and straightforward technique wherein the readout in relative luminescence units (RLU) can be directly correlated with the ATP levels in the cells. RLU of the untreated cells were normalized to 100% cell viability. Untreated cells and cells treated with a synthetic polycation, polyethylenimine (PEI) served as the negative and positive controls, respectively. As we expected, MCF-7, MDA-MB-231, BT-549 cells, and primary DRG neurons treated with PEI (50 μg/mL), a synthetic polycation known to induce apoptosis, showed a 50–60% reduction in cell viability (41). However, MCF-7, MDA-MB-231, and BT-549 incubated with C12-200/siGAPDH LNPs were well-tolerated, with cell viabilities >95% 24-h post-transfection (Figure 6a). Similarly, siTRPV1-loaded C12-200 LNPs were well tolerated (cell viabilities of ca. 100%) by primary DRG neurons (Figure 6b). We tested the effect of siRNA dose escalation by incubating MDA-MB-231 cells with increasing concentrations of siRNA-LNPs (10, 25, 50, 75, and 100 nM siGFP-loaded LNPs). We noticed cell viabilities >85% 24-h post-transfection (Figure 6c) suggesting that higher siRNA and concomitantly higher lipid doses were well-tolerated by the cells. There was a significant difference (p<0.0001) in the % cell viabilities of cells treated with siRNA-LNPs compared to those treated with the positive control, PEI (20% cell viability).
Fig. 6.
C12-200 LNPs are cytocompatible with TRPV1-expressing breast cancer cell line panel (a), rat primary DRG neurons (b), and MDA-MB-231 cells (c). Cells were incubated with LNPs containing 50 nM siGAPDH (a), 50 nM siTRPV1 (b), and the indicated concentrations of siGFP-LNPs (c) for 4 h. Untreated cells and cells treated with PEI (50 μg/mL) were used as negative and positive controls, respectively. Cell viability was determined 24 h post-transfection using a CellTiter-Glo luminescence viability assay, and the data were normalized to untreated cells. Data represents mean + SD (n = 6)
Flow Cytometry Analysis of Cy5 siRNA Uptake into Primary Rat Cortical Neurons
After confirming that LNPs had little-to-no effect on cell viability, we next wanted to determine if LNPs were taken up by neurons. Because of their commercial availability as well as their non-mitotic nature similar to that of the primary rat DRG neurons, primary rat cortical neurons were used to quantify the uptake levels of Cy5 siRNA using flow cytometry. Lipofectamine RNAiMAX, a cationic lipid, was used as a positive control. Flow cytometry is a quantitative tool for single-cell analysis to measure thefluorescence emitted from a single cell (42). The dead cells/debris were likely removed at the centrifugation step during the sample processing, and as a result, we did not observe any dead cells or debris (Figure 7a, left) that are typically found at the bottom left corner of the SSC-A vs. FSC-A plot (43). We then analyzed whether the cells were present in a monodisperse cell suspension to allow for single cell analysis. The forward scatter distributions for height vs. area (FSC-A v/s FSC-H) showed a linear profile for about 92% of the recorded events which indicated a single-cell suspension (Figure 7a, right). We analyzed untreated cells and gated out the auto fluorescence (Figure 7b, left). Thus, a shift of signal to the right of the histogram gate (R1 region) was considered to signify that the cell was Cy5-positive (+). The proportion of Cy5 (+) cells directly corresponds to the % uptake of Cy5 siRNA in the neurons as represented by the individual histogram plots for the different groups (Figure 7c).
We then analyzed the % uptake of Cy5 siRNA encapsulated in LNPs prepared with (+) and without (−) the inclusion of PEG-DMG. Around 92–94% (Figure 7c (yellow)) and 81–83% (Figure 7c (brown)) cells were Cy5 (+) when transfected with Cy5 siRNA-LNPs prepared with and without the inclusion of PEG-DMG, respectively. Figure 7d shows the relative Cy5+ cells in each of the indicated groups in reference to the untreated (blue) and positive control-treated (green) groups. The rightward shift in intensities of Cy5 (+) cells for all the groups in comparison with the controls are demonstrated in the overlay histogram plot in Figure 7e. As expected, the % uptake of Cy5 siRNA was least in cells treated with free Cy5 siRNA and greatest in cells treated with LNPs (+PEG-DEMG) and Cy5 siRNA/Lipofectamine RNAiMAX complexes (Figure 7e and f). Furthermore, as seen in Figure 7e, we saw a greater uptake of LNPs with PEG-DMG as compared to those without PEG-DMG. Particle sizes of LNPs play one of the major roles in determining uptake levels, and therefore, lower diameter particles may allow efficient cellular internalization. Furthermore, a significant difference in the uptake of Cy5 siRNA was observed when transfected with naked Cy5 siRNA and Cy5 siRNA-LNPs (± PEG-DMG) (Figure 7e and f). This reiterates the need for a safe and efficient transfection agent to maximize the uptake of siRNA in neurons, a hard-to-transfect cell type.
Around 94–96% of the cells were Cy5 (+) for the Lipofectamine RNAiMAX group indicating efficient uptake (Figure 7b right and f). Charge-based interactions of cationic lipids with the anionic cell membranes allow for higher uptake as compared to neutral or negatively-charged particles. About 44–46% of the cells were Cy5 (+) for the naked Cy5 siRNA transfection group (Figure 7c (red)). The difference between the uptake mediated by LNPs (+ PEG-DMG) and Lipofectamine RNAiMAX was statistically non-significant (Figure 7f). Despite the similar levels of uptake among the LNP(+PEG-DMG) and Lipofectamine groups, it must also be pointed out that the uptake mediated by Lipofectamine RNAiMAX was accompanied with noticeable cell stress/toxicity upon visual observation under a microscope, whereas LNPs showed a superior safety profile.
Fluorescent Microscopy Analysis of Cy5 siRNA Uptakein a Human Cortical Neuron Cell Line
We studied the effect of exposure time on the uptake of LNPs by incubating the cells for 2, 4, or 24 h. PEGylation is known to regulate the uptake kinetics of LNPs into cells (44). We compared the differences in uptake for LNPs prepared with/without PEG-DMG. Although Lipofectamine RNAiMAX is a gold standard transfection agent for RNA molecules, it is also quite toxic to cells owing to its strong cationic nature. Therefore, it was unsurprising when we saw changes in morphology indicating cell death, just 2 h after Lipofectamine RNAiMAX was added to cortical neurons, while untreated cells continued to appear spindle-shaped and healthy (Figure 8a).
For this experiment, cells were incubated either with naked Cy5 siRNA, Cy5 siRNA LNP with PEG-DMG, with Cy5 siRNA LNP without PEG-DMG, with Lipofectamine RNAiMAX Cy5 siRNA, or were left untreated. We qualitatively compared the fluorescence intensity among the cells treated with the above samples. Cells treated with naked Cy5 siRNA showed less intense fluorescent signals as compared to cells treated with Cy5 siRNA-LNPs (± PEG-DMG) for the 2- and 4-h incubation time points (Figure 8a, b). However, we saw a noticeable increase in siRNA uptake 24-h post-transfection (Figure 8c). The next pivotal observation was the difference in uptake from the LNPs (± PEG-DMG) groups. As mentioned earlier, PEG-DMG plays a key role in determining the physical stability of LNPs as it allows to maintain lower particle diameters by inhibiting particle aggregation (35).LNPs formulated with PEG-DMG showed a time-dependent increase in the uptake of Cy5 siRNA as opposed to LNPs. We observed thata few cells did not show Cy5 siRNA signals when treated with Cy5 siRNA-LNPs (+ PEG-DMG) 2- and 4-h post-transfection, whereas 24 h post-treatment, almost all the cells showed signals corresponding to Cy5 siRNA.A zoomed in image of cells treated with LNPs containing 50 nM Cy5 siRNA is presented in Figure 8d to show image resolution. This particularly depicts the uptake of Cy5 siRNA LNPs associated with diffuse cytoplasmic fluorescence.
DISCUSSION
The goal of this pilot study was to determine if LNPs based on the benchmark C12-200 lipidoid can be used for siRNA transfection into neurons, a notoriously challenging yet clinically unmet/interesting delivery target. We propose that the well-documented delivery advantages of LNPs including their high levels of siRNA encapsulation, favorable pharmacokinetics, limited immune activation, uptake into target cells, and efficient endosomal escape for siRNA into the cytoplasm (13, 42, 43) make them interesting candidates for neural delivery applications.
In this pilot study, we prepared and characterized C12-200 LNPs using DLS. The RiboGreen assay is a well-validated quantitative method to determine the % siRNA loading in LNPs (44, 45). Encapsulation of siRNA in the LNPs for the slow and fast mixing (without PEG-DMG) speeds was about 74.4% and 83.8%, respectively. LNPs with and without PEG-DMG (for fast mixing speed) showed encapsulation efficiencies of about 71.4% and 83.8%, respectively (Figure 1) which were comparable to the entrapment of siRNA reported for some of the top LNP candidates (46). Although we did not find a significant difference in the encapsulation efficiencies of LNPs prepared with and without PEG-DMG, the inclusion of PEG-DMG serves other vital roles, i.e., maintaining lower particle diameters by inhibiting particle aggregation and providing a stealth effect for longer circulation times in vivo (34, 35). We further studied the physicochemical stability of the prepared LNPs and compared the stabilizing effect provided by the inclusion of PEG-DMG and siRNA in the LNPs. We utilized siGFP as the model siRNA to compare the siRNA-loaded and blank LNP counterparts prepared both with and without PEG-DMG. We observed a non-significant increase or change in the particle diameters, polydispersity indices, and zeta potential over a 1-week storage period at 2–8 °C for all the samples (Figure 2). Nevertheless, the particle diameters differed significantly for blank vs. siRNA-loaded LNPs and LNPs with vs. LNPs without PEG-DMG (Figure 2a). A similar trend was also observed for the polydispersity indices of blank and siRNA-loaded LNPs, both with and without PEG-DMG (Figure 2b). Our data demonstrated that both siRNA loading and PEG-DMG provided a stabilizing effect to the LNPs by maintaining lower particle diameters and uniform dispersity indices over a period of 7 days. The stabilizing effects of siRNA and PEG-DMG on the resulting particle diameters and dispersity indices were noted when the LNPs were stored for 24 h at 37°C (Figure 3). LNPs showed further lower particle diameters (ca. 80.5 nm) when sizes were measured in the presence of serum (10% FBS) suggestive of the additional serum-mediated stabilization (Figure 4).
Although LNPs are deemed to be safe and are well-tolerated by most of the cells, determining their safety and tolerability in neural cells was our primary aim. As shown in Figure 6, LNPs were deemed to be well-tolerated by primary DRG neurons as evident by >95% cell viabilities. Increasing the dose of siRNA (and concomitantly the dose of the lipids) did not alter the safety profile of LNPs (Figure 6c). Establishing the safety profile of LNPs with neural cells allowed further evaluation of their uptake in neural cell lines. We studied the uptake of Cy5 siRNA-loaded LNPs in primary rat cortical neurons and a human neuronal cell line using flow cytometry and fluorescence microscopy, respectively. Previously, Howard et al. and Thiramanas et al. studied the cellular uptake of Cy5 siRNA using flow cytometry (47, 48). As PEG-DMG is known to stabilize the LNPs with resulting lower particle diameters and thus have an impact on their cellular uptake, we compared LNPs prepared with (+) and without (−) PEG-DMG. We used the siRNA complexes of the cationic transfection agent, Lipofectamine RNAiMAX, as a positive control. The cell-associated fluorescence (Cy5 (+) neurons) for the Cy5 siRNA-LNPs (+ PEG-DMG) group was about 94%—2-fold greater than the neurons transfected with naked Cy5 siRNA group (Figure 7 c, e, and f). As expected, despite its transfection-induced toxicity, Cy5 siRNA-Lipofectamine RNAiMAX-treated cells demonstrated the highest proportion of Cy5 (+) cells (Figure 7 c, e, and f). On the other hand, cells treated with LNPs appeared healthy and showed no visible signs of toxicity, highlighting the safety of LNPs over cationic transfection agents like Lipofectamine RNAiMAX. There was a significant difference in the uptake of neurons transfected with naked Cy5 siRNA as compared to the neurons transfected with the LNPs (both with and without PEG-DMG) emphasizing the need for an effective transfection agent for neural cell uptake (Figure 7f).
We also performed fluorescence microscopy to qualitatively study the uptake of Cy5 siRNA into neural cells. A technical caveat of such qualitative assessments is rooted in the fact that the observed/apparent fluorescent intensities are not normalized to cell number. The cells appeared to be healthy and spindle-shaped in all the treatment groups except for the Lipofectamine RNAiMAX group where cells appeared stressed and rounded as early as at the 2 h timepoint (Figure 8a). As described earlier, we speculate that this is due strong cationic nature of Lipofectamine RNAiMAX. We did not observe a greater uptake for cells treated with LNPs at 100 nM siRNA concentration (Supplementary Figure 2). Nearly all the cells in the field showed Cy5 fluorescent signals 24-h post-exposure (Figure 8c) compared to the 2 and 4 h time points (Figure 8 a and b), suggesting that neural cells require a longer transfection time compared to non-neural cells. LNPs formulated with PEG-DMG showed a time-dependent increase in the uptake of Cy5 siRNA. The time dependent uptake of LNPs formulated with PEG-DMG was in agreement with the PEG-DMG dissociation kinetics as predicted by Mui et al. (49). The PEG-lipid component of the LNPs undergoes desorption from the surface of LNPs which results in efficient uptake of the cargo into the cells (50). Studies have reported desorption rates of PEG-DMG from the LNPs at a rate of 45%/h only after which LNPs were taken up by cells (49). We observed a similar trend of Cy5 siRNA uptake when neurons were transfected with LNPs (+ PEG-DMG). At the 2 and 4 h time points, most (but not all) of the cells had efficiently taken up the LNPs. However, 24-h post-treatment, almost all the cells in the field of view showed fluorescent puncta corresponding to Cy5 siRNA.The effects of RNAiMAX-mediated toxicity were evident via altered cell morphology seen in the phase contrast images.
Overall, both flow cytometry and fluorescent microscopy analysis in cortical neurons demonstrated efficient uptake of siRNA delivered using LNPs in the absence of noticeable toxicity, while the cationic Lipofectamine RNAiMAX-mediated uptake was concomitant with significant cellular toxicity. Based on these findings, we conclude that LNPs are a safe carrier for siRNA delivery to neural cells. We are currently screening a pre-existing LNP library prepared using different lipidoid chemistries to identify LNP candidates for safe and efficient neuronal gene knockdown. We anticipate that the results of these studies will set the foundation for using LNPs for neural cell transfection in a variety of CNS diseases. While our approach validates using LNPs for uptake of siRNA into neural cells, their knockdown efficacy and therapeutic index remain an open question which we are currently investigating.
CONCLUSION
SiRNA-loaded LNPs remained stable over a period of 7 days and were well-tolerated by neural cell models, allowing their further exploration for gene silencing. Although the LNPs are comparable to Lipofectamine RNAiMAX in facilitating siRNA uptake into cortical neurons, the ionizable cationic LNPs are safer and well-tolerated compared to the cationic Lipofectamine RNAiMAX. PEG-DMG served a crucial role in maintaining lower particle diameters that likely resulted in efficient uptake into primary rat cortical neurons. Our findings suggest that LNPs hold promise for further silencing of therapeutically relevant neuronal targets like TRPV1.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
We are grateful to Dr. Kathryn A. Whitehead (Carnegie Mellon University) and her group for their support. We are grateful to Drs. Jane E. Hartung and Michael S. Gold (University of Pittsburgh) for providing the DRG cultures. We are also thankful to Dr. Wilson Meng and express our special appreciation to Mr. Nevil Abraham (Duquesne University) for his help with the qPCR experiments. We express our appreciation to Mr. Duncan Dobbins (Duquesne University) for his assistance with the graphical abstract.
Funding
This work was supported via start-up funds for the Manickam laboratory from Duquesne University (DU), a 2021 Faculty Development Fund (Office of Research, DU) and a 2021 Charles Henry Leach II Fund to the PI (Duquesne University).
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Wu SY, McMillan NAJ. Lipidic systems for in vivo siRNA delivery. AAPS J. 2009;11(4):639–652. doi: 10.1208/s12248-009-9140-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang J, Lu Z, Wientjes MG, Au JLS. Delivery of siRNA therapeutics: barriers and carriers. AAPS J. 2010;12(4):492–503. doi: 10.1208/s12248-010-9210-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Akinc A, Maier M, Manoharan M, Fitzgerald K, Jayaraman M, Barros S, et al. The Onpattro story and the clinical translation of nanomedicines containing nucleic acid-based drugs. Nat Nanotechnol. 2019;1(14):1084–1087. doi: 10.1038/s41565-019-0591-y. [DOI] [PubMed] [Google Scholar]
- 4.FDA. FDA Approves First COVID-19 Vaccine. FDA News Release. 2021; Available from: https://www.fda.gov/news-events/press-announcements/fda-approves-first-covid-19-vaccine.
- 5.Cross R. Without these lipid shells, there would be no mRNA vaccines for COVID-19. Chem Eng News. 2021;16–9.
- 6.Uemura Y, Naoi T, Kanai Y, Kobayashi K. The efficiency of lipid nanoparticles with an original cationic lipid as a siRNA delivery system for macrophages and dendritic cells. Pharm Dev Technol. 2018;24(24):1–22. doi: 10.1080/10837450.2018.1469149. [DOI] [PubMed] [Google Scholar]
- 7.Whitehead KA, Dorkin JR, Vegas AJ, Chang PH, Veiseh O, Matthews J, et al. Degradable lipid nanoparticles with predictable in vivo siRNA delivery activity. Nat Commun. 2014;5. [DOI] [PMC free article] [PubMed]
- 8.Lopes CDF, Gonçalves NP, Gomes CP, Saraiva MJ, Pêgo AP. BDNF gene delivery mediated by neuron-targeted nanoparticles is neuroprotective in peripheral nerve injury. Biomaterials. 2017;121:83–96. doi: 10.1016/j.biomaterials.2016.12.025. [DOI] [PubMed] [Google Scholar]
- 9.Haney MJ, Klyachko NL, Zhao Y, Gupta R, Plotnikova EG, He Z, et al. Exosomes as drug delivery vehicles for Parkinson’s disease therapy. J Control Release. 2015;207:18–30. [DOI] [PMC free article] [PubMed]
- 10.Wolfe D, Mata M, Fink DJ. Targeted drug delivery to the peripheral nervous system using gene therapy. Neurosci Lett. 2012;527(2):85–9. [DOI] [PMC free article] [PubMed]
- 11.Ferreira-Vieira HT, Guimaraes MI, Silva RF, Ribeiro MF. Alzheimer’s disease: targeting the Cholinergic System. Curr Neuropharmacol. 2016;14(1):101–15. doi: 10.2174/1570159X13666150716165726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang L, Han B, Zhang Z, Wang S, Bai Y, Zhang Y, et al. Extracellular vesicle-mediated delivery of circular RNA SCMH1 promotes functional recovery in rodent and nonhuman primate ischemic stroke models. Circulation. 2020;142(6):556–574. doi: 10.1161/CIRCULATIONAHA.120.045765. [DOI] [PubMed] [Google Scholar]
- 13.Kim J, Jozic A, Sahay G. Naturally derived membrane lipids impact nanoparticle-based messenger RNA delivery. Cell Mol Bioeng. 2020;13(5):463–474. doi: 10.1007/s12195-020-00619-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rehman ZU, Zuhorn IS, Hoekstra D. How cationic lipids transfer nucleic acids into cells and across cellular membranes: Recent advances. J Control Release. 2013;166(1):46–56. [DOI] [PubMed]
- 15.Soenen SJH, Brisson AR, De Cuyper M. Addressing the problem of cationic lipid-mediated toxicity: The magnetoliposome model. Biomaterials. 2009;30(22):3691–701. [DOI] [PubMed]
- 16.Dahlman J, Kauffman K, Langer R, Anderson D. Nanotechnology for in vivo targeted siRNA delivery. Adv Genet. 2014;21(88):37–69. doi: 10.1016/B978-0-12-800148-6.00003-1. [DOI] [PubMed] [Google Scholar]
- 17.Wan C, Allen T, Cullis P. Lipid nanoparticle delivery systems for siRNA-based therapeutics. Drug Deliv Transl Res. 2014;1:4. doi: 10.1007/s13346-013-0161-z. [DOI] [PubMed] [Google Scholar]
- 18.Love KT, Mahon KP, Levins CG, Whitehead KA, Querbes W, Dorkin JR, et al. Lipid-like materials for low-dose, in vivo gene silencing. Proc Natl Acad Sci U S A. 2010;107(5):1864–1869. doi: 10.1073/pnas.0910603106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Blakney AK, McKay PF, Yus BI, Aldon Y, Shattock RJ. Inside out: optimization of lipid nanoparticle formulations for exterior complexation and in vivo delivery of saRNA. Gene Ther. 2019;26(9):363–72. [DOI] [PMC free article] [PubMed]
- 20.Naseri N, Valizadeh H, Zakeri-Milani P. Solid lipid nanoparticles and nanostructured lipid carriers: structure preparation and application. Adv Pharm Bull. 2015;5(3):305–13. [DOI] [PMC free article] [PubMed]
- 21.Cullis PR, Hope MJ. Lipid nanoparticle systems for enabling gene therapies. Mol Ther. 2017;25(7):1467–75. [DOI] [PMC free article] [PubMed]
- 22.Jones JL, Yue TS, Cheung Ching-Ying L, Singer V. RNA Quantitation by fluorescence-based solution assay- RiboGreen reagent characterization. Anal Biochem. 1998;374(265):368–74. [DOI] [PubMed]
- 23.Hartung JE, Gold MS. GCaMP as an indirect measure of electrical activity in rat trigeminal ganglion neurons. Cell Calcium. 2020;89(April):102225. [DOI] [PMC free article] [PubMed]
- 24.Dave KM, Ali L, Manickam DS. Characterization of the SIM-A9 cell line as a model of activated microglia in the context of neuropathic pain. PLoS One. 2020;15(4):1–28. [DOI] [PMC free article] [PubMed]
- 25.Dave KM, Han L, Jackson MA, Kadlecik L, Duvall CL, S Manickam D. DNA polyplexes of a phosphorylcholine-based zwitterionic polymer for gene delivery. Pharm Res. 2020;37(9). [DOI] [PMC free article] [PubMed]
- 26.Dave KM, Zhao W, Hoover C, D’Souza A, S Manickam D. Extracellular vesicles derived from a human brain endothelial cell line increase cellular ATP levels. AAPS PharmSciTech. 2021;22(1). [DOI] [PMC free article] [PubMed]
- 27.Honary S, Zahir F. Effect of zeta potential on the properties of nano-drug delivery systems- a review (Part 1) Trop J Pharm Res. 2013;12(2):255–264. [Google Scholar]
- 28.Ing LY, Zin NM, Sarwar A, Katas H. Antifungal activity of chitosan nanoparticles and correlation with their physical properties. Int J Biomater. 2012; Article ID 632698. [DOI] [PMC free article] [PubMed]
- 29.Li X, Sloat BR, Yanasarn N, Cui Z. Relationship between the size of nanoparticles and their adjuvant activity: data from a study with an improved experimental design. Eur J Pharm Biopharm. 2011;78(1):107–116. doi: 10.1016/j.ejpb.2010.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Breznan D, Das DD, Mackinnon-Roy C, Bernatchez S, Sayari A, Hill M, et al. Physicochemical properties can be key determinants of mesoporous silica nanoparticle potency in vitro. ACS Nano. 2018;12(12):12062–12079. doi: 10.1021/acsnano.8b04910. [DOI] [PubMed] [Google Scholar]
- 31.Shin SW, Song IH, Um SH. Role of physicochemical properties in nanoparticle toxicity. Nanomaterials. 2015;5(3):1351–1365. doi: 10.3390/nano5031351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Navya PN, Daima HK. Rational engineering of physicochemical properties of nanomaterials for biomedical applications with nanotoxicological perspectives. Nano Converg. 2016;3(1):1–14. doi: 10.1186/s40580-016-0064-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Braakhuis HM, Park MVDZ, Gosens I, De Jong WH, Cassee FR. Physicochemical characteristics of nanomaterials that affect pulmonary inflammation. Part Fibre Toxicol. 2014;11(1). [DOI] [PMC free article] [PubMed]
- 34.Witzigmann D, Kulkarni JA, Leung J, Chen S, Cullis PR, Meel R Van Der. Adv Drug Deliv Rev. 2020;159:344–63. [DOI] [PMC free article] [PubMed]
- 35.Chen S, Tam YYC, Lin PJC, Leung AKK, Tam YK, Cullis PR. Development of lipid nanoparticle formulations of siRNA for hepatocyte gene silencing following subcutaneous administration. J Control Release. 2014;196:106–12. [DOI] [PubMed]
- 36.Romberg B, Hennink WE, Storm G. Sheddable coatings for long-circulating nanoparticles. Pharm Res. 2008;25(1):55–71. doi: 10.1007/s11095-007-9348-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kulkarni JA, Darjuan MM, Mercer JE, Chen S, Van Der Meel R, Thewalt JL, et al. On the Formation and Morphology of Lipid Nanoparticles Containing Ionizable Cationic Lipids and siRNA. ACS Nano. 2018;12(5):4787–4795. doi: 10.1021/acsnano.8b01516. [DOI] [PubMed] [Google Scholar]
- 38.Malvern Panalytical. Correlation intercept – what is it and what does it mean? https://www.materials-talks.com/blog/2020/03/10/correlation-intercept-what-is-it-and-what-does-it-mean/.
- 39.Pecze L, Jósvay K, Blum W, Petrovics G, Vizler C, Oláh Z, et al. Activation of endogenous TRPV1 fails to induce overstimulation-based cytotoxicity in breast and prostate cancer cells but not in pain-sensing neurons. Biochim Biophys Acta - Mol Cell Res. 2016;1863(8):2054–64. [DOI] [PubMed]
- 40.Xu Q, Zhang XM, Duan KZ, Gu XY, Han M, Liu BL, et al. Peripheral tgf-β1 signaling is a critical event in bone cancer-induced hyperalgesia in rodents. J Neurosci. 2013;33(49):19099–19111. doi: 10.1523/JNEUROSCI.4852-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Moghimi SM, Symonds P, Murray JC, Hunter AC, Debska G, Szewczyk A. A two-stage poly(ethylenimine)-mediated cytotoxicity: implications for gene transfer/therapy. Mol Ther. 2005;11(6):990–995. doi: 10.1016/j.ymthe.2005.02.010. [DOI] [PubMed] [Google Scholar]
- 42.Davey HM, Kell DB. Flow cytometry and cell sorting of heterogeneous microbial populations: the importance of single-cell analyses. Microbiol Rev. 1996;60(4):641–696. doi: 10.1128/mr.60.4.641-696.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.BIO-RAD. Flow Cytometry Data Analysis. https://www.bio-rad-antibodies.com/flow-cytometry-gating-strategies.html.
- 44.Ryals RC, Patel S, Acosta C, McKinney M, Pennesi ME, Sahay G. The effects of PEGylation on LNP based mRNA delivery to the eye. PLoS One. 2020;15(10 October):1–17. [DOI] [PMC free article] [PubMed]
- 45.Chen S, Tam YYC, Lin PJC, Sung MMH, Tam YK, Cullis PR. Influence of particle size on the in vivo potency of lipid nanoparticle formulations of siRNA. J Control Release. 2016;235:236–44. [DOI] [PubMed]
- 46.Ball R, Bajaj P, Whitehead KA. Achieving long-term stability of lipid nanoparticles: examining the effect of pH, temperature, and lyophilization. Int J Nanomedicine. 2017;12:305–315. doi: 10.2147/IJN.S123062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Buyens K, Lucas B, Raemdonck K, Braeckmans K, Vercammen J, Hendrix J, et al. A fast and sensitive method for measuring the integrity of siRNA-carrier complexes in full human serum. J Control Release. 2008;126(1):67–76. doi: 10.1016/j.jconrel.2007.10.024. [DOI] [PubMed] [Google Scholar]
- 48.Walsh C, Ou K, Belliveau N, Leaver T, Wild A, Huft J, et al. Microfluidic-based manufacture of siRNA-lipid nanoparticles for therapeutic applications. Methods Mol Biol. 2014;25(1141):109–120. doi: 10.1007/978-1-4939-0363-4_6. [DOI] [PubMed] [Google Scholar]
- 49.Mui BL, Tam YK, Jayaraman M, Ansell SM, Du X, Yi Y, Tam CPJL, Chen S, Narayanannair JK, Rajeev KG, Manoharan M, Akinc A, Maier MA, Madden TD, et al. Influence of polyethylene glycol lipid desorption rates on pharmacokinetics and pharmacodynamics of siRNA lipid nanoparticles. Mol Ther - Nucleic Acids. 2013;2(12):1–8. doi: 10.1038/mtna.2013.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kulkarni JA, Cullis PR, Van Der Meel R. Lipid nanoparticles enabling gene therapies: from concepts to clinical utility. Nucleic Acid Ther. 2018;28(3):146–157. doi: 10.1089/nat.2018.0721. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.