Abstract
Embryo implantation in both humans and rodents is initiated by the attachment of a blastocyst to the uterine epithelium. For blastocyst attachment, the uterine epithelium needs to transform at both the structural and molecular levels first, and then initiate the interaction with trophectoderm. Any perturbation during this process will result in implantation failure or long-term adverse pregnancy outcomes. Endocrine steroid hormones, which function through nuclear receptors, combine with the local molecules produced by the uteri or embryo to facilitate implantation. The insulin-like growth factor (IGF) signaling has been reported to play a vital role during pregnancy. However, its physiological function during implantation remains elusive. This study revealed that mice with conditional deletion of Igf1r gene in uteri suffered from subfertility, mainly due to the disturbed uterine receptivity and abnormal embryo implantation. Mechanistically, we uncovered that in response to the nidatory estrogen on D4 of pregnancy, the epithelial IGF1R, stimulated by the stromal cell-produced IGF1, facilitated epithelial STAT3 activation to modulate the epithelial depolarity. Furthermore, embryonic derived IGF2 could activate both the epithelial ERK1/2 and STAT3 signaling through IGF1R, which was critical for the transcription of Cox2 and normal attachment reaction. In brief, our data revealed that epithelial IGF1R was sequentially activated by the uterine stromal IGF1 and embryonic IGF2 to guarantee normal epithelium differentiation during the implantation process.
Keywords: IGF1R, IGF1, IGF2, uterine epithelium, implantation, blastocyst
Introduction
In both humans and rodents, the process of embryo implantation involves the intimate interaction between the trophectoderm and uterine epithelium (Wang and Dey, 2006). Genetic studies have shown that inappropriate transformation of the epithelium barrier is one critical cause for implantation failure (Daikoku et al., 2011; Li et al., 2015; Sun et al., 2016). Uterine epithelium allows embryos to attach only in a limited time period, defined as the implantation window when the uteri are in receptivity. In mice, driven by the ovarian steroid hormones, the uteri remain prereceptive on D1‒D3 (D1 = the day that the vaginal plug was detected), turn into receptivity on D4 of pregnancy, and then become refractory to implantation by D5 (Wang and Dey, 2006). The major ovarian hormones that specify uterine receptivity are estrogen (17β-estradiol, E2) and progesterone (P4), whose functions are primarily executed by the nuclear estrogen receptor (ER) and progesterone receptor (PR), respectively (Zhang et al., 2013). On D1 of pregnancy, the preovulatory estrogen surge stimulates intense proliferation of uterine epithelial cells. On D4, the raising level of P4 together with preimplantation nidatory estrogen surge induces rapid differentiation of epithelia and proliferation of stroma, which is required for an optimal environment for blastocyst implantation (Lim and Wang, 2010). Ovariectomy conducted in D4 morning before the secretion of preimplantation estrogen results in delayed implantation, which could be terminated with E2 injection leading to blastocyst activation and the transition of uteri from the neutral state to receptive state. However, the precise mechanism by which the D4 nidatory estrogen transforms uterine epithelial cells to adapt to blastocyst attachment remains elusive.
Various locally produced signaling factors, including cytokines, growth factors, homeobox transcription factors, together with ovarian hormones, function during the establishment of uterine receptivity and embryo attachment through autocrine, paracrine, and/or juxtracrine manners (Norwitz et al., 2001; Lim et al., 2002; Paria et al., 2002). Insulin-like growth factor 1 (IGF1) is usually regarded as an estrogen-responsive gene that promotes cell proliferation in uterine epithelia in the nonpregnant mice (Murphy and Ghahary, 1990; Zhu and Pollard, 2007). A recently reported mouse model using Pgr-cre to delete the uterine Igf1 gene suggested that uterine-derived IGF1 is essential for female fertility, but not for uterine growth in response to estrogen (Hewitt et al., 2019). However, the manner of regulation and underlying mechanism for IGF1 signaling during peri-implantation are still unclear (Richards et al., 1996; Hewitt et al., 2010, 2019).
In this study, we adopted the Pgr-cre mice to explicitly delete the uterine Igf1r to explore the function of the IGF signaling during the implantation. We found that the absence of uterine Igf1r affected the pregnancy rate mainly due to the compromised uterine epithelial transformation, further impairing embryo implantation. On D4 of pregnancy with the nidatory estrogen exposure, both the endometrium and the blastocyst increase the expression of IGF ligands, which act orderly on the epithelial IGF1R to activate downstream signaling in the epithelium. First, IGF1, produced by the uterine stroma in response to nidatory estrogen, activated the epithelial STAT3 pathway, which modulates the expression of cellular junction molecules to confer the uterine epithelium depolarity. Thereafter, IGF2, produced by the late blastocyst, could be received by the epithelium, activating the ERK1/2 and STAT3 signaling to initiate the attachment reaction. In summary, we found that the epithelial IGF1R, activated sequentially by the uterine IGF1 and embryonic IGF2, was indispensable for the epithelial differentiation and normal uterine preparation for embryo implantation.
Results
IGF1R expression in the peri-implantation mouse uteri
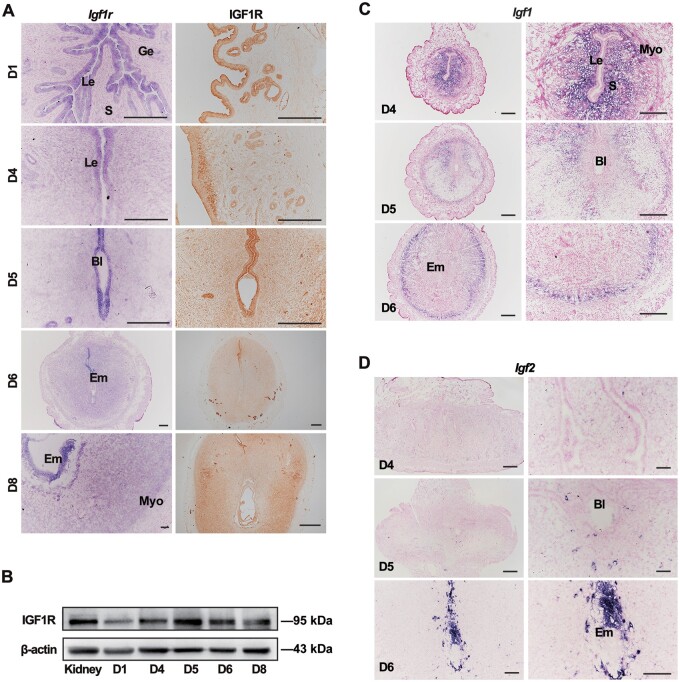
To better understand the function of the IGF signaling in early pregnancy, we first assessed the expression levels of IGF ligands and receptors in the uteri. The spatiotemporal profiles of uterine IGF1R were examined by in situ hybridization (ISH) and immunohistochemistry (IHC). On D1 of pregnancy, IGF1R was exclusively expressed in the uterine epithelium and sustained in the epithelium during the receptive phase. On D5, IGF1R expression levels were high in epithelial cells while low in stromal cells (Figure 1A). Alongside uterine decidualization progression, IGF1R remained in both the residual epithelial and stromal compartments (Figure 1B). The quantification of protein levels depicted a similar pattern, as the expression of IGF1R increased with the establishment of uterine receptivity, peaking at the implantation stage (Figure 1B). ISH results for the ligands showed that Igf1 was strongly expressed in the stroma, particularly in the subepithelial stromal cells on D4, then decreased obviously, and showed a low level in the peripheral stromal region toward the myometrium from D5 onwards (Figure 1C). In contrast, Igf2 ligand was almost undetectable in the uteri in D4 morning but exhibited a rising expression in the embryos, especially the trophoblast cell directly contacting with the uteri from D5 to D6 of pregnancy (Figure 1D).
Figure 1.
Spatiotemporal expression of IGF signaling components in mouse uteri during peri-implantation. (A) ISH (left) and IHC (right) of IGF1R in D1‒D8 uteri. Scale bar, 200 μm. (B) Western blotting analysis of IGF1R in uteri from D1 to D8. β-actin serves as a loading control. (C and D) ISH of Igf1 and Igf2 mRNA in the uteri during early pregnancy. Scale bar, 100 μm. Bl, blastocyst; Le, luminal epithelium; Ge, glandular epithelium; S, stroma; Em, embryo; Myo, myometrium.
Mice with uterine Igf1r deletion show compromised fertility due to impaired embryo attachment
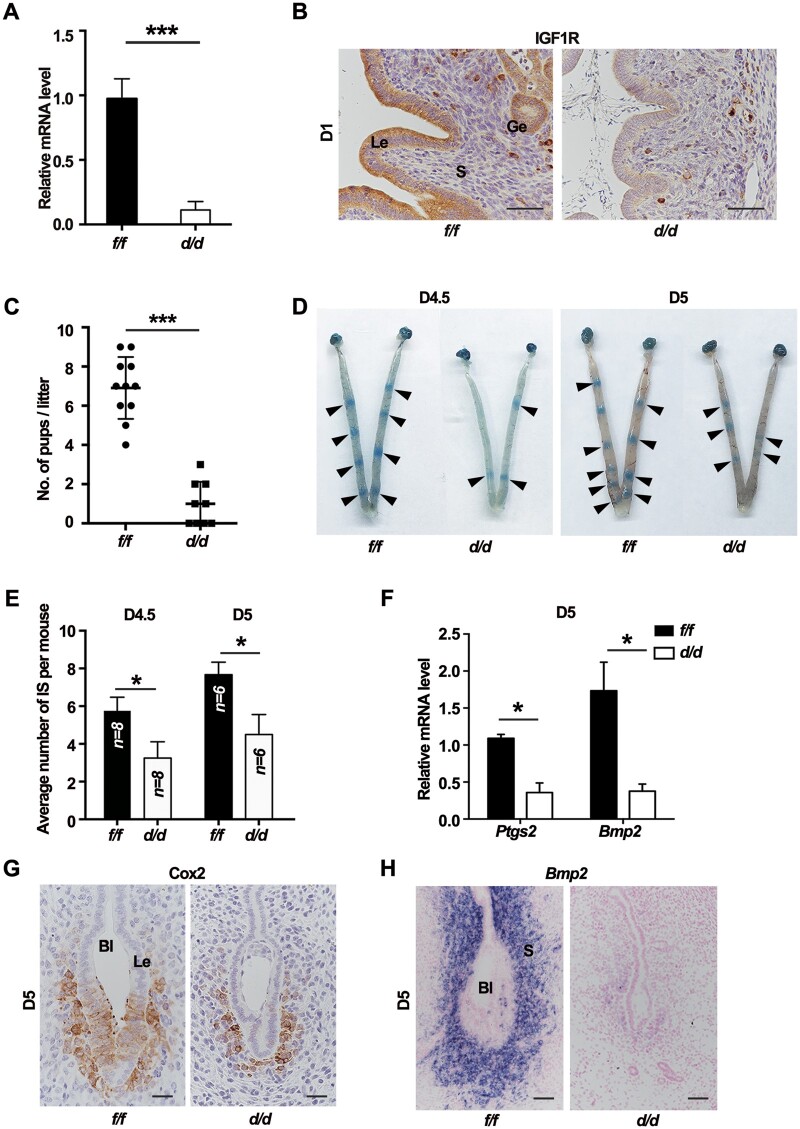
Next, females with conditional uterine inactivation of Igf1r (Igf1rd/d) were generated by crossing the Igf1r-floxed (Igf1rf/f) mice with PR-driven Cre recombinase mice. The evidence showed that Igf1r was effectively deleted in Igf1rd/d uteri at both mRNA (Figure 2A) and protein (Figure 2B) levels. Then, fertility of Igf1rf/f and Igf1rd/d females was assessed during a 6-month continuous breeding trial by mating with the wild-type (WT) males. We found that >85% of Igf1rd/d plug-positive mice produced litters less than three (Figure 2C). To ascertain the stage-specific causes accounting for this evident infertility, we first analyzed the implantation status. The implantation status from D4.5 to D5 was examined by the blue dye method, and the number of implantation sites was significantly reduced in the Igf1rd/d mice on both D4.5 and D5 (Figure 2D and E). Cox2 (Ptgs2) and Bmp2, induced by the attachment reaction, are critical for embryo implantation (Das et al., 1994; Lim et al., 1997; Paria et al., 2001; Lee et al., 2007). The induced expression of both Cox2 and Bmp2 was defective in Igf1rd/d uterine cells surrounding the blastocyst on D5 (Figure 2F‒H). Although some embryos displayed the blue band reactions, we found that some implanted embryos in knockout mice failed to develop normally when detecting the postimplantation embryo development. It seems like the growth of implanted fetus was restricted by the residual epithelium in knockout uteri, since the cytokeratin-positive epithelium surrounding the embryo failed to be eliminated and the PL-1-positive embryonic trophoblast cells could not penetrate the endometrium properly (Supplementary Figure S1). All the results revealed that the Igf1r deletion in uteri impaired the normal embryo implantation.
Figure 2.
Uterine IGF1R is indispensable for embryo implantation. (A) qPCR analysis for IGF1R mRNA level in Igf1rf/f and Igf1rd/d uteri on D1. ***P < 0.001. (B) IHC of IGF1R in uteri on D1. Scale bar, 100 μm. (C) Average litter sizes in Igf1rf/f and Igf1rd/d females. ***P < 0.001. (D) The gross morphology of implantation sites in Igf1rf/f and Igf1rd/d mice visualized by blue dye reaction. Black arrowheads indicate the implantation sites. (E) Average number of implantation sites (IS) in Igf1rf/f and Igf1rd/d mice. *P < 0.05. (F) Relative mRNA levels of marker genes in Igf1rf/f and Igf1rd/d uteri on D5. The values are normalized to Gapdh and indicated as mean ± SEM, n = 3. *P < 0.05. (G) IHC of Cox2 in D5 uteri with the blastocyst. Scale bar, 50 μm. (H) ISH of Bmp2 mRNA in D5 uteri. Scale bar, 50 μm. Le, luminal epithelium; Ge, glandular epithelium; S, stroma; Bl, blastocyst.
Since IGF1R was mainly expressed in the uterine epithelium with a relatively lower level in stroma before the implantation, we intended to explore whether the defective implantation was due to the epithelium IGF1R-mediated cell-autogenous function. Ltf-Cre mouse model was utilized to specifically delete the uterine epithelial Igf1r (Daikoku et al., 2014). The Igf1rf/f/Ltf-Cre mice demonstrated a reduced number of implantation sites (Supplementary FigureS2A and B), and the sharply decreased expression of Cox2 represented a less transformation of luminal epithelium (LE) (Supplementary Figure S2C), clarifying that the uterine epithelial IGF1R was vital for epithelial differentiation to support normal embryo implantation.
Less differentiated epithelium for implantation in the absence of uterine IGF1R
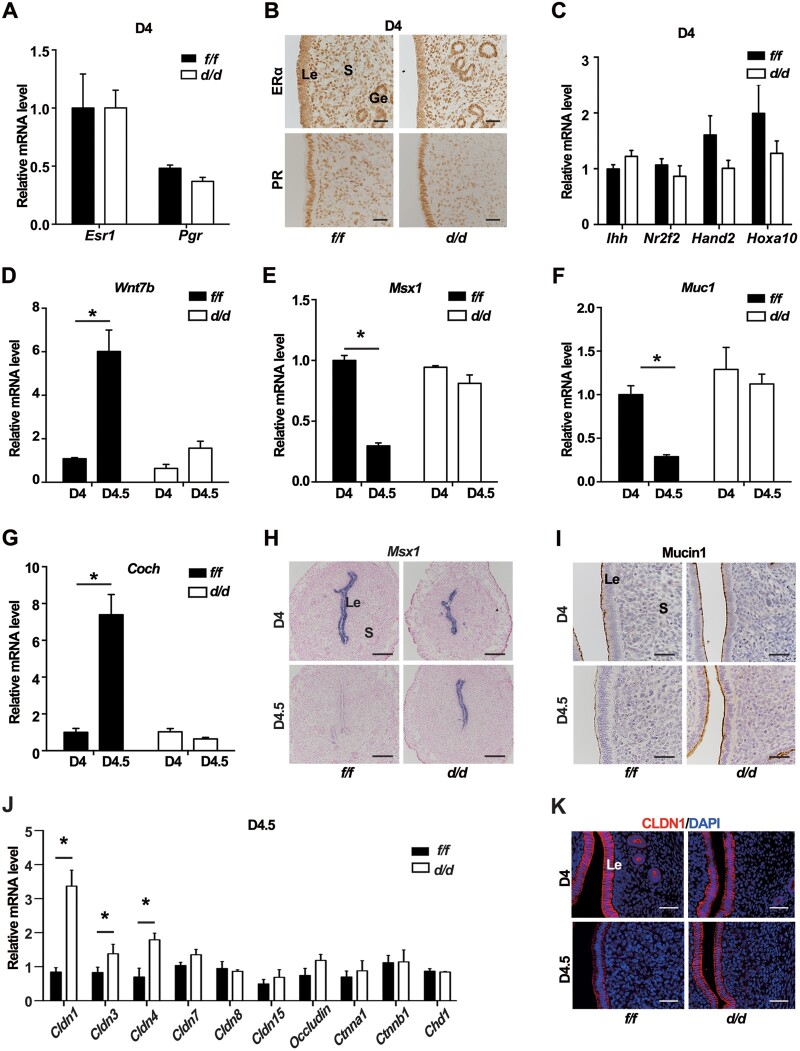
In mice, embryo implantation requires the uteri to enter a receptive state, precisely controlled by the steroid hormones E2 and P4. Since the Pgr-cre was also active in the developing corpus luteum, and IGF1R was widely expressed in the mouse ovary (Baumgarten et al., 2017), we pondered whether the levels of ovarian steroid secretion were influenced. The results showed that Igf1rd/d females exhibited comparable circulating levels of P4 and E2, accompanied by the regular expression of key steroid biosynthetic enzymes P450Scc and 3β-HSD in the corpus luteum as those in Igf1rf/f females (Supplementary FigureS3A and B). Meanwhile, considering both E2 and P4 acting through their respective nuclear receptors, we examined the mRNA and protein levels of ERα and PR in the D4 uteri but did not find significant difference (Figure 3A and B). Moreover, the expression levels of P4‒PR responsive genes, such as Ihh, Nr2f2, Hand2, and Hoxa10, were comparable between IGF1R-deficient and control uteri in D4 morning (Figure 3C).
Figure 3.
Epithelial differentiation in Igf1rd/d uteri is defective in response to nidatory estrogen. (A) Relative mRNA levels of uterine Esr1 and Pgr in Igf1rf/f and Igf1rd/d mice on D4. The values are normalized to Gapdh and indicated as mean ± SEM, n = 3. (B) IHC staining of ERα and PR in D4 Igf1rf/f and Igf1rd/d uteri. Scale bar, 100 μm. (C) Relative mRNA expression of P4 downstream genes in D4 Igf1rf/f and Igf1rd/d uteri. The values are normalized to Gapdh and indicated as mean ± SEM, n = 3. (D–G) Relative mRNA levels for estrogen target genes in Igf1rf/f and Igf1rd/d uteri in D4 morning and evening. *P < 0.5. (H) ISH of Msx1 in Igf1rf/f and Igf1rd/d uteri in D4 morning and evening. Scale bar, 200 μm. (I) IHC of Mucin1 in D4 morning and evening. Scale bar, 100 μm. (J) qPCR analysis for mRNA levels of adherent junctional and tight junctional members in D4.5 uteri. *P < 0.05. (K) Immunofluorescence analysis of uterine CLDN1 (red) expression at the indicated time. Nuclei were stained with DAPI (blue). Scale bar, 100 μm. Le, luminal epithelium; Ge, glandular epithelium; S, stroma; Bl, blastocyst.
Based on the P4 priming, the D4 nidatory estrogen induces the uterine differentiation into a receptive state, which includes epithelium depolarity and the dynamic gene expression (Daikoku et al., 2011; Zhang et al., 2013). We found that several nidatory estrogen-responsive genes, including Wnt7b, Coch, Msx1, and Mucin1 (Muc1), exhibited aberrant expression changes from D4 morning to evening in Igf1rd/d uteri, compared with the Igf1rf/f control (Figure 3D‒G). The ISH data confirmed an abnormal, persistent epithelial expression of Msx1 in the D4.5 knockout uteri (Figure 3H). The same observation was found for the Mucin1 protein (Figure 3I), which had disappeared in control D4.5 epithelium but remained an aberrantly high expression in the Igf1rd/d epithelium. The LIF signaling was reported to be a mediator of the nidatory estrogen for downregulating Msx1 (Daikoku et al., 2011). We found that both the expression level and gland localization of Lif mRNA were similar in Igf1rf/f and Igf1rd/d uteri on D4 (Supplementary Figure S4), implying that the abnormal expression of Msx1 resulting from the IGF1R deficiency was not due to the defective Lif expression.
Msx1 is crucial for embryo implantation and regulates the programming of epithelial cell polarity (Daikoku et al., 2011). Since we had observed the abnormally expressed Msx1, we wondered whether the deletion of IGF1R likewise affected epithelial differentiation. The quantification of mRNA expression depicted that while the adherens junction molecules, E-Cadherin (Chd1) and β-catenin (Ctnnb1), were comparable between the Igf1rf/f and Igf1rd/d epithelia, the tight junction molecules, such as Claudin 1 (Cldn1), Cldn3, and Cldn4, were significantly higher in Igf1rd/d epithelia after exposed to the D4 morning estrogen (Figure 3J). Consistently, the downregulation of epithelial CLDN1 protein was observed in the control uteri from D4 morning to evening, which was not occurred in the absence of Igf1r (Figure 3K), revealing that the failure of epithelial cell depolarity in the Igf1rd/d uteri.
Nidatory estrogen-induced IGF1 activates the IGF1R‒STAT3 cascade in the epithelium
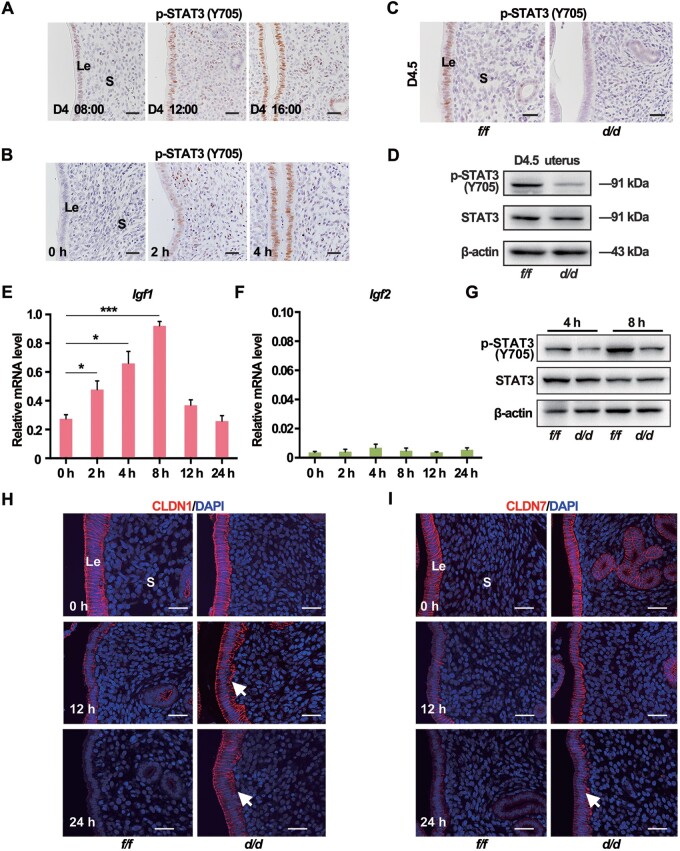
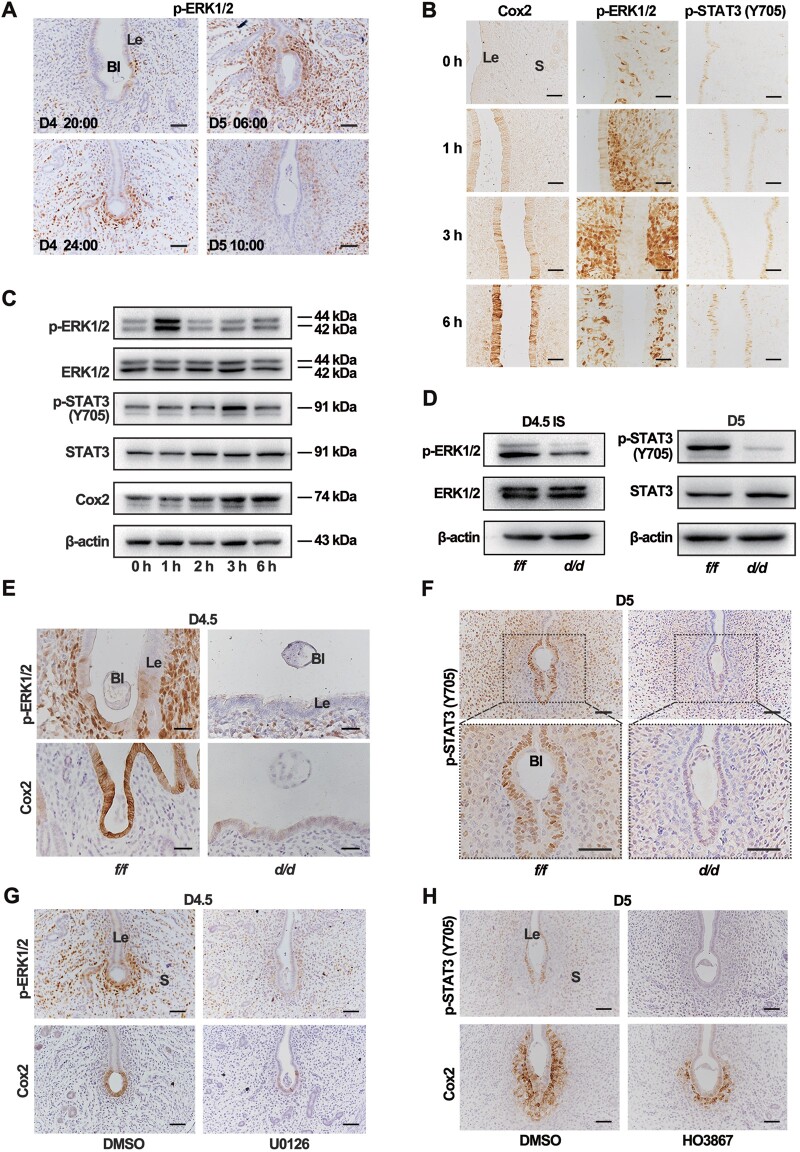
STAT3 was reported to regulate the expression of epithelial cell junction complex and direct the epithelial differentiation for implantation (Pawar et al., 2013). We examined the dynamic expression of uterine phosphorylated active STAT3 in relation with the D4 nidatory estrogen. As shown in Figure 4A, in D4 morning (08:00), the active STAT3 was low in the epithelium. Next, STAT3 was activated in the epithelium over time, consistent with the timing of D4 nidatory estrogen production. To more precisely explore the relationship between the D4 nidatory estrogen and STAT3 activation, pregnant mice were ovariectomized in D3 night and given exogenous estrogen in D4 morning. The immunoblotting and immunostaining data suggested that exogenous estrogen could induce epithelial STAT3 phosphorylation with peak activation after 4 h (Figure 4B;Supplementary Figure S5A). Consistently, on D4.5, when the D4 nidatory estrogen had challenged the uteri, active STAT3 phosphorylation positive staining was observed in the Igf1rf/f epithelium but not in the Igf1rd/d epithelium (Figure 4C), which was further confirmed by the immunoblotting data (Figure 4D). These results implied that IGF1R-mediated signaling was vital for STAT3 activation in D4.5 uteri.
Figure 4.
Nidatory estrogen-induced IGF1 activates the STAT3 pathway to regulate the epithelial transformation. (A) IHC of uterine p-STAT3 (Y705) in different time points on D4. Scale bar, 100 μm. (B) Immunostaining of uterine p-STAT3 (Y705) in response to E2 treatment after ovariectomy on D3. Scale bar, 50 μm. (C) Immunostaining of uterine p-STAT3 (Y705) in Igf1rf/f and Igf1rd/d mice on D4.5. Scale bar, 100 μm. (D) Western blotting analysis for p-STAT3 (Y705) and total STAT3 in D4.5 uteri. (E and F) qPCR analysis for Igf1 and Igf2 mRNA expression in WT mouse uteri in response to E2 treatment after ovariectomy on D3. The values are normalized to gapdh and indicated as mean ± SEM, n = 3. (G) Western blotting analysis for expression levels of p-STAT3 (Y705) and STAT3 proteins in Igf1rf/f and Igf1rd/d mouse uteri in response to E2 treatment. (H and I) Immunostaining of CLDN1 (red) and CLDN7 (red) in Igf1rf/f and Igf1rd/d uteri. Nuclei were stained with DAPI (blue). Scale bar, 50 μm. White arrow indicates the sustained location in Igf1rd/d uteri. Le, luminal epithelium; S, stroma; Ge, glandular epithelium; Bl, blastocyst.
Since the canonical activation of IGF1R requires the IGF1 or IGF2 ligand, then we explored whether these ligands were induced in response to estrogen. As mentioned above, exogenous estrogen was given in D4 morning to the pregnant mice that were ovariectomized in D3 night. The quantitative real-time polymerase chain reaction (qPCR) analysis showed that Igf1, but not Igf2, was rapidly induced by this E2 treatment at 4 and 8 h (Figure 4E and F), similar with the response of Lif to estrogen (Supplementary Figure S5B). Consistently, this estrogen-induced IGF1 was accompanied with the STAT3 activation at 8 h after the E2 treatment in control uteri, which was significantly blocked in the absence of uterine Igf1r (Figure 4G). Furthermore, the cell junction molecules CLDN1 and CLDN7 were gradually downregulated in the control epithelium after E2 treatment, which was not occurred in the Igf1rd/d epithelium (Figure 4H and I). These data suggested that in the absence of Igf1r, the D4 nidatory estrogen-induced STAT3 activation and epithelial cell differentiation was defective.
Blastocyst-derived IGF2 is a potential ligand for uterine epithelial IGF1R during the attachment
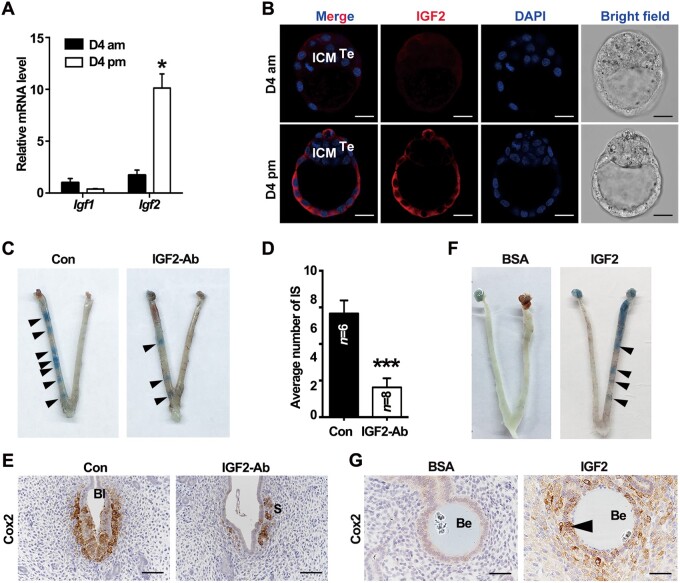
Apart from IGF1 expression in uteri, we also noticed another ligand, IGF2, mainly expressed by the trophoblast cell, most out layer of the embryo directly interacting with uteri during peri-implantation (Figure 1C and D). Initially, we quantified Igf2 expression in the blastocyst. The qPCR results illustrated that the expression of Igf2 mRNA exhibited a rapid increase in the blastocyst from D4 morning to evening (Figure 5A), with the blastocyst acquiring an implantation competence. The increased protein expression of IGF2 was further confirmed by immunofluorescence. IGF2 was slightly expressed in the trophectoderm in D4 morning and was later dramatically increased with the embryo attachment by D4 evening (Figure 5B). These implied that accompanied with the initiation of attachment, the blastocyst upregulated Igf2, which might communicate with the uterine epithelium during the attachment through the epithelial IGF1R.
Figure 5.
Embryo-derived IGF2 facilitates embryo implantation. (A) qPCR analysis for Igf1 and Igf2 mRNA expression in the blastocysts in D4 morning and evening. The values are normalized to Gapdh and indicated as mean ± SEM, n = 3. *P < 0.05. (B) Immunofluorescence analysis for IGF2 localization in the blastocysts in D4 morning and evening. Scale bar, 40 μm. (C) Blue dye reaction to detect the implantation status of blastocysts with different treatments. Black arrowheads indicate the implantation sites. (D) Average number of implantation sites (IS) in control and IGF2-specific antibody-blocked groups. ***P < 0.001. (E) Immunostaining of Cox2 in uterine sections of implantation sites from control and blocking antibody-treated groups. Scale bar, 100 μm. (F) Morphology of uteri with transferred beads coated with BSA or IGF2 recombinase protein. The implantation-like sites were visualized by blue dye reaction at 24 h after the transfer. Black arrowheads indicate the implantation-like sites induced by IGF2-coated beads. (G) Immunostaining of uterine Cox2 induced by IGF2-coated beads or BSA-treated beads. Black arrow points the Cox2 signaling in the epithelium. Scale bar, 50 μm. ICM, inner cell mass; Te, trophectoderm; Bl, blastocyst; Be, beads.
We then intended to explore the function of this potential crosstalk mediated by IGF2‒IGF1R during embryo attachment. First, blastocysts, incubated with specific anti-IGF2 antibodies to block the function of embryo-secreted IGF2 protein, were transferred into pseudopregnant receptive uteri. The number of implantation sites in the IGF2 antibody-treated group displayed an evident reduction compared to the control group (Figure 5C and D). In implantation sites with faint blue dye reaction induced by few IGF2-blocked blastocysts, Cox2 was absent in the LE with a weakly expression in the stroma around those blastocysts (Figure 5E). To further prove the vital function of the embryonic IGF2, agarose beads were coated with IGF2 recombinase protein and transferred into the pseudopregnant uteri on D4. The IGF2-coated beads could mimic the embryos to induce implantation-like reaction as visualized by the blue bands (Figure 5F). Similar with the observation in the embryo-induced attachment reaction, the Cox2 signal was noticed in the LE and sub-epithelium stroma surrounding the IGF2-coated beads on D5, while the LE cells around the bovine serum albumin (BSA)-covered beads did not exhibit obvious Cox2 expression (Figure 5G). These results indicated that embryonic IGF2 actively participated in the normal blastocyst attachment.
The embryonic derived IGF2 activates the ERK1/2 and STAT3 signaling for blastocyst attachment
Since STAT3 was active in the whole epithelia in D4 uteri after the estrogen exposure, it was speculated that the embryonic derived IGF2 probably induced additional downstream effectors to initiate the attachment reaction around the embryo just in implantation sites. We detected other well-known IGF downstream signaling, including the ERK1/2 and AKT pathways. Based on the IHC approach, we only recognized a transient ERK1/2 activation in the epithelium around the blastocyst in D4 evening (Figure 6A).
Figure 6.
IGF2‒IGF1R signaling-induced Cox2 expression requires both ERK1/2 and STAT3 activation. (A) IHC of uterine p-ERK1/2 protein at indicated time points from D4 to D5 of pregnancy. Scale bar, 100 μm. (B) Immunostaining of Cox2, p-ERK1/2, and p-STAT3 (Y705) in the uteri after IGF2 recombinase protein treatment. Scale bar, 100 μm. (C) Western blotting analysis for different proteins with the lysates of epithelial cells of WT mice in response to E2 treatment. (D) Western blotting for D4.5 uterine lysates utilizing pERK1/2, ERK1/2, p-STAT3 (Y705), and STAT3 antibodies. (E) Immunostaining of uterine p-ERK1/2 and Cox2 in Igf1rf/f and Igf1rd/d mice on D4.5. Scale bar, 200 μm. (F) Immunostaining of p-STAT3 (Y705) in Igf1rf/f and Igf1rd/d uteri on D5. Scale bar, 200 μm. (G) Immunostaining of p-ERK1/2 and Cox2 in the uteri from both control and U0126-treated mice on D4.5. U0126, a MEK inhibitor, was intraperitoneally injected at 18:00 on D4. Scale bar, 100 μm. (H) Immunostaining of p-STAT3 (Y705) and Cox2 in the uteri on D5 from both control and U0126-treated mice. Scale bar, 100 μm. Le, luminal epithelium; Bl, blastocyst; S, stroma.
The pseudopregnant uteri, injected intraluminally with IGF2 recombinase proteins on D4, resulted in upregulation of the phosphorylated ERK1/2 and Cox2 induction in the whole uterine epithelia as revealed by the IHC analysis (Figure 6B). The IGF2 treatment also induced STAT3 phosphorylation in the epithelium, which was further confirmed by the immunoblotting assay (Figure 6C). Interestingly, when the mice were pretreated with the ERK1/2 inhibitor U0126, not only the IGF2-induced ERK1/2 activation was blocked, but also the STAT3 activation was downregulated, suggesting the potential crosstalk between the ERK1/2 and STAT3 signaling (Supplementary Figure S6). Furthermore, defective ERK1/2 and STAT3 phosphorylation was observed in the knockout uteri (Figure 6D‒F). To exclude the primary effects of aberrantly differentiated epithelium caused by the defective IGF1R‒STAT3 signaling in response to stromal IGF1 as mentioned above, pharmacological approaches were utilized to explore the function of ERK1/2 and STAT3 activation in the attachment. Intraperitoneal injection of U0126 efficiently inhibited the activation of ERK1/2, and also blocked Cox2 expression in the epithelium (Figure 6G). Similarly, STAT3 inhibitor treatment, which was conducted in the afternoon of D4 when the stromal IGF1 had activated STAT3 through IGF1R, also led to the disappearance of Cox2 expression in the epithelium (Figure 6H). All these data proved that there existed an interaction between ERK1/2 and STAT3, and both active ERK1/2 and STAT3 signals were essential for attachment reaction.
ERK1/2 and STAT3 synergistically regulate Cox2 expression
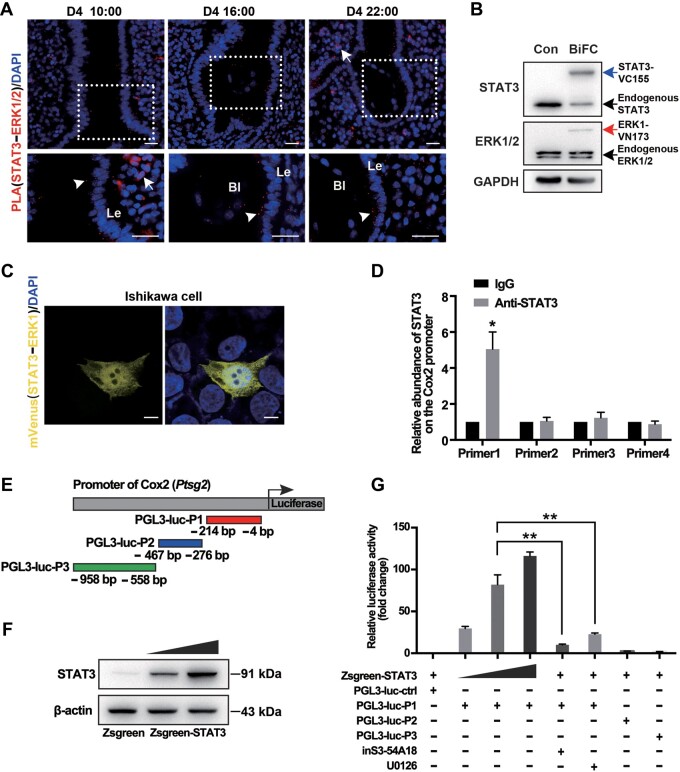
To further explore the molecular basis of the coordination between ERK1/2 and STAT3 for regulating Cox2 expression, physical interaction was first examined in vivo. The Duolink in situ proximity ligation assay (PLA) was adopted to detect ERK1/2‒STAT3 colocalization in uterine tissues. The density of the positive signals, which indicated an interaction between ERK1/2 and STAT3, gradually increased in the LE from D4 morning to D4 midnight with the blastocyst attached to the epithelium (Figure 7A), indicating that STAT3 and ERK1/2 were in close proximity at a molecular level within the LE cells around the attached embryo. Moreover, we adopted the mVenus-based bimolecular fluorescence complementation (BiFC) assay to achieve the detection and visualization of the interaction between ERK1/2 and STAT3 in endometrial Ishikawa cells. The expression levels of STAT3-VC155 and ERK1-VN173 were comparable with those of endogenous proteins (Figure 7B). The mVenus-based BiFC fluorescence was seen in cells transfected with STAT3-VC155 and ERK1-VN173 (Figure 7C), suggesting the interaction between ERK1 and STAT3.
Figure 7.
STAT3 and ERK1/2 synergistically regulate Cox2 expression. (A) Fluorescence images of Duolink in situ PLA using rabbit anti-ERK1/2 antibody and mouse anti-STAT3 antibody at the indicted time on D4. White arrowheads indicate multiple positive PLA signals (red spots). White arrow points noisy signal. The nuclei were labeled with DAPI (blue). Scale bar, 200 μm. Le, luminal epithelium; B, blastocyst. (B) Western blotting analysis for the expression of BiFC constructs. GAPDH served as the loading control. (C) BiFC assays to evaluate interactions between ERK1 and STAT3. Images were acquired 36 h after transfection. Scale bar, 5 μm. (D) ChIP‒qPCR analysis for the enrichment of STAT3 in the Cox2 promoter. IgG was used as control. *P < 0.05. (E) Schematic illustration of luciferase reporter constructs with different inserts. (F) Western blotting analysis for STAT3 in 293T cells transfected with increasing amounts of STAT3-expressing constructs. (G) Dual-luciferase reporter assay utilizing the Cox2 promoter-driven reporter combined with different plasmids. The compound inS3-54A18, an inhibitor for DNA-binding activity of STAT3, was used at 10 μM. U0126 was used at 10 μM to inhibit the activation of ERK1/2. After 36 h of culture, cell lysates were assayed for firefly and Renilla luciferase activity. The data are expressed as fold induction of luciferase activity relative to the promoter alone, and data presented are the mean of three independent experiments performed in duplicate. **P < 0.01.
Next, we tested how the interacted ERK1/2 and STAT3 regulated Cox2 expression. Previous studies had proved that STAT3 could bind directly to the Cox2 promoter (Li et al., 2018). We performed ChIP‒qPCR assay to determine the STAT3 binding on the Cox2 promotor. The IGF2-stimulated D4 uterine epithelial cells were utilized, which exhibited Cox2 expression in the whole epithelial layer (Figure 6B) and provided enough cells feasible with the ChIP assay. We determined a strong binding of STAT3 on the promoter region, specifically on this region (−214/−3 bp) of the Cox2 gene (Figure 7D), indicating that STAT3 bounded to the Cox2 promoter in vivo.
Furthermore, we tested whether STAT3 could mediate the transactivation of Cox2 via the promotor luciferase reporter assay. It was noticed that a Cox2 promoter reporter including fragment P1 (−214/−3 bp) displayed a response in a STAT3 dose-dependent manner (Figure 7E‒G), consistent with the most enrichment of this fragment in the ChIP assay. The inS3-54A18, an inhibitor of the STAT3 DNA-binding activity, or the ERK1/2 inhibitor UO126 could abolish the Cox2 promoter fragment (−214/−3 bp)-regulated luciferase activity (Figure 7G). Therefore, it seemed that STAT3 cooperated with the ERK1/2 pathway for regulating Cox2 expression in the LE by directly binding to its promoter region.
Discussion
As the first barrier for embryo attachment, the uterine epithelium needs to transform appropriately to accept the embryo. The transition of uterine LE from a nonreceptive to a receptive phase is accompanied by altered expression and redistribution of the catenins, claudins, and E-cadherin in uterine epithelial junctional complexes (Pawar et al., 2013). Under normal conditions, Msx1 is significantly expressed in D4 morning and remarkably downregulated with blastocyst attachment by D4 evening. During this process, E2 has been proved to inhibit Msx1 expression on D4 (Daikoku et al., 2011). Msx gene participates in the alteration of epithelial cell junction proteins during embryo implantation. Uterine double deletion of Msx1/2 led to the upregulation of CLDN1 (a tight junction protein) and small proline-rich (Sprr2) protein (a major component of cornified envelopes in keratinized epidermis) (Sun et al., 2016). In our study, we observed abnormal sustained Msx1 expression and disordered distribution of cell junction molecules in Igf1rd/d mouse uteri in D4 evening. The uterine Msx1 expression was sustained in the delayed implantation model, which is critical for the viability of dormant blastocyst (Cha et al., 2013). The observation of this study, combined with previous evidence, suggested that downregulation of epithelial Msx1 is prerequired for normal embryo implantation.
A previous paper reported that even though E2 reduced Msx1 expression in WT uteri, it failed to do so in Lif knockout mice (Daikoku et al., 2011). Thus, we tested the Lif expression by qPCR and found that it showed no difference from D4 to D5 (Supplementary Figure S3). In the uteri, LIF primarily activates the JAK‒STATs pathways (Auernhammer and Melmed, 2000; Chen et al., 2000). The LIFR antagonist can inhibit tyrosine phosphorylation of STAT3 in the LE and lead to a reduced implantation rate (Mohamet et al., 2009). Previous papers have also reported that in Lif-deficient and gp130-mutant mice, uterine epithelia are incapable of responding to embryo implantation due to dysregulation of STAT3 signaling (Stewart et al., 1992; Ernst et al., 2001). In this study, we found that in Igf1r knockout mouse model, the activation of STAT3 in the LE was defective, with the regular glandular expression of LIF. Therefore, a scenario was depicted as that the stromal cell-produced IGF1 synergized with the glandular epithelium-secreted LIF to guarantee the STAT3 activation in the D4 epithelium and subsequently directed the downstream events preparing the receptive epithelium to interact with the embryo.
STAT3 directs cellular remodeling that alters the organization of epithelial junctional complexes during implantation (Pawar et al., 2013). A recent report utilizing the mouse model with the Ltf-Cre-induced STAT3 deletion also verified the cell-autonomous function of STAT3 in the uterine epithelium (Hiraoka et al., 2020). Before the implantation occurs, the epithelium integrity primarily depends on the status of adherent and tight junctions, which form a continuous seal of junctional complexes (Singh and Aplin, 2009). There is evidence proving that STAT3 signaling is vital for suppressing the expression of CLDN1, CLDN3, and CLDN4 transcripts within the implantation window, and it also downregulates α- and β-catenin, which may contribute to the redistribution of E-cadherin away from lateral junctional complexes during implantation (Pawar et al., 2013). Here, we checked the localization of critical molecules of both tight and adherent junctions, and the results indicated that CLDN1 was altered apparently in the epithelium. The molecular relationship between the STAT3 and epithelial transformation needs further exploration.
It has been reported that E2 regulates uterine epithelial cell proliferation through stromal cell-derived IGF1, and adding specific IGF1R inhibitor PPP blocked uterine epithelial cell proliferation induced by E2 (Zhu and Pollard, 2007). However, uterine-specific knockout of IGF1 did not influence uterine growth in response to E2. Rather, it decreased the pregnancy rate of female mice (Hewitt et al., 2019), in consistence with our study that uterine IGF1R deletion caused defective epithelial depolarization and thus abnormal pregnancy. Other growth factors, such as fibroblast growth factor (FGF) members, have also been demonstrated to attend the epithelial cell proliferation (Nallasamy et al., 2012). Based on these reports, we surmise that many grow factors, such as IGF and FGF signaling, are involved in E2-induced epithelial proliferation, and the uterine-localized IGF1‒IGF1R signaling is more important for epithelial differentiation preparing for the embryo implantation. Besides IGF1R, whether the other IGF receptor IGF2R, which displays a low affinity with IGF ligands, also functions during this process needs further exploration (Martin-Kleiner and Gall Troselj, 2010).
Pregnancy is a dialogue process between the maternal uteri and fetus. IGF2, another ligand for the IGF1R, has been reported to play a vital role in fetal growth, while the altered expression of IGF2 has been determined to be related to fetal growth disorders (Constância et al., 2002). Building on a previous report that IGF2 was upregulated in the implanted embryo (Rappolee et al., 1992; Hardy and Spanos, 2002), we also found that IGF2 rapidly increased in trophectoderm before blastocyst attachment. The attachment would induce Cox2 expression in the epithelium and underlying stroma around the blastocyst. This gave us a hint that as a secretory molecule of the embryo, IGF2 might regulate uterine gene expression, such as Cox2, in a paracrine manner.
We then detected the downstream signals of IGF and uncovered that the ERK1/2 signal was transiently activated in the presence of an embryo or uterine infused with IGF2 protein. Consistently, though normal embryos were observed in the IGF1R knockout uteri, there were no ERK1/2 activation or Cox2 expression in the epithelium around the embryos. A previous study showed that HB-EGF was readily upregulated in the blastocyst, transitioning from dormancy to an activated state (Hamatani et al., 2004). Also, beads soaked in IGF1, as well as beads coated with HB-EGF, could elicit an implantation-like response when transferred into the pseudopregnant mice (Paria et al., 2001). This report implied that with a local high concentration of IGF signal, either IGF1 or IGF2 could induce an implantation-like reaction in the uterine epithelium, and the ERK1/2 might be a common downstream signal of these growth factors. In the uterine receptivity, it was found that the STAT3 activation is critical for the epithelial depolarity, and we also wonder the function of STAT3 signal during the attachment reaction. For this purpose, we utilized STAT3 inhibitor in WT mice in D4 late afternoon when uterine IGF1 and LIF had activated STAT3. It demonstrated that the activation of STAT3 was also indispensable for the epithelial Cox2 expression. Based on these results, we uncovered that ERK1/2 and STAT3 synergistically induced the epithelial Cox2 expression.
How does ERK1/2 influence STAT3 activity? A previous paper reported that ERK1/2 inhibition resulted in a decrease in the phosphorylation level of p-STAT3 (S727), but no significant change in p-STAT3 (Y705) level (Le et al., 2016). Additionally, ERK2 phosphorylated STAT3 on serine-containing peptide and decreased its tyrosine phosphorylation induced by EGF treatment (Jain et al., 1998). Here, we found that p-STAT3 (Y705) was present in the LE after the treatment with IGF2 recombinase protein (Supplementary Figure S5), indicating no mutually exclusive actions of these two different phosphorylation sites of STAT3 during implantation. However, the mechanism for the in vivo interaction and coordination between STAT3 and ERK1/2 in the uterine epithelium needs to be further explored.
In brief, in this study, we uncovered a stage-dependent function of epithelial IGF1R to guarantee the normal implantation. As depicted in Figure 8, in the morning of D4, E2 elicites uterine stromal IGF1 to activate STAT3 in the epithelium in a paracrine way, thus promoting the whole epithelial transformation for the arrival of the embryos. Afterwards, the attached embryo secretes IGF2, which acts on the local epithelia to induce the attachment reaction, such as Cox2 expression, via the synergistic ERK1/2 and STAT3 pathways.
Figure 8.
A diagram displays the sequential role of IGF1R signaling during the implantation. Nidatory estrogen peak at D4 morning induces the expression of stromal IGF1, which is involved in activating the epithelial STAT3 via a paracrine manner for depolarizing the uterine epithelium. On D4.5, the upregulated IGF2 from the late blastocyst acts on epithelial cells, synergistically activating both the ERK1/2 and STAT3 signaling to induce Cox2 expression for attachment.
Materials and methods
Animals, hormone treatments, and tissue collection
All experiments were conducted according to the approved guidelines of the Animal Welfare Committee of Research Organization (X200811) of Xiamen University. Female mice carrying floxed Igf1r (Igf1rf/f) were obtained from Jackson Lab (Dietrich et al., 2000), and crossed with PRCre/+ or LtfCre/+ mice. Implantation sites were visualized by a routine blue dye method (Paria et al., 1993). WT and knockout female mice were ovariectomized in the evening of D3 and then treated with a single dose of E2 along with P4 (2 mg in sesame oil) in D4 morning. For each treatment group, at least four mice were used at each time point. Uteri were collected and fixed in 10% formalin before IHC analysis or frozen in the liquid nitrogen.
Isolation of mouse uterine epithelial cells
Uterine horns were excised, trimmed of fat, and dissected longitudinally to expose uterine lumen. Dissected horns were then cut into 4- to 5-mm-long pieces and washed in Hanks balanced salt solution (HBSS) as previously reported (Ramathal et al., 2010). Uterine tissue pieces were placed into HBSS containing 25 g/L trypsin (Sigma, T8003) for 1 h at room temperature, followed by separation by tweezers. Trizol was used to lyse tissues for RNA extraction.
Measurement of E2 and P4 levels
Mouse blood samples were collected on D4 of pregnancy. Serum levels of E2 and P4 were measured by radioimmunoassay.
qPCR analysis
Total RNA was isolated from uterine cells by standard Trizol-based protocols and converted to cDNA as described previously (Zhang et al., 2019). The cDNA was amplified by qPCR to quantify gene expression using gene-specific primers and SYBR Green. Analysis of variance single-factor analysis was conducted on the grouped means to determine statistical significance at a significance level of P < 0.05. All primers for qPCR are listed in Supplementary Table S1. Assays were performed at least three times with each in duplicate.
Immunostaining
The 10% neutral buffer formalin-fixed tissue was processed for paraffin embedding, sectioned in 5 μm thickness, and performed immunostaining using IGF1R, Muc1, p-STAT3, STAT3, p-ERK1/2, Ki67, Cox2, 3β-HSDII, CYP11A1, and p-AKT antibodies, respectively. The signal was developed with DAB solution and counterstained with Mayer’s hematoxylin. For immunofluorescence staining, 4% formaldehyde-fixed frozen tissue section (10 μm) was incubated with anti-Claudin 1 and anti-Claudin 7, respectively. Specific secondary antibodies were used to detect the antigen and 4ʹ,6ʹ-diamidino-2-phenylindole (DAPI) was applied to identify the cell nucleus.
Embryo immunostaining
The zona pellucida was removed using acid Tyrode’s solution (Sigma) described previously (Lin et al., 2017). Embryos were fixed in 4% paraformaldehyde (PFA) in phosphate-buffered saline (PBS) with 0.1% Tween 20 and 0.01% Triton X-100 overnight at 4°C, permeabilized in 0.5% Triton X-100 in PBS for 15 min, and blocked in 10% fetal bovine serum in PBS for 1 h. The anti-IGF2 antibody was used as a primary antibody.
Beads transfer into the pseudopregnant mice
The preparation of IGF2-carrying beads and transferring beads into uteri lumen were performed as previously described (Paria et al., 2001). Affi-Gel Blue Gel (Bio-Rad; 100‒200 mesh, no. 153-7302) beads about the size of a blastocyst were washed six times with sterile PBS and then incubated with IGF2 recombinase protein (100 ng/μl) (R&D Systems, 792) or similar concentrations of BSA, in 20 μl PBS at 37°C for 1 h. After incubation, beads were washed in PBS several times and used immediately. Siliconized pipette tips and dished were used for handling IGF2. Loaded beads (7 beads/horn) were transferred into one uterine horn of D4 pseudopregnant mice. Mice were sacrificed on D5. Uteri without blue bands were flushed to recover beads.
Duolink PLA in uterine tissue
Uterine sections (10 μm) deposited on glass slides and pretreated concerning fixation, retrieval, blocking, and incubation with primary ERK1/2 and STAT3 antibodies were used for IHC at room temperature for overnight. After washing in 1× Wash Buffer at room temperature twice, the slides were incubated with secondary antibodies (PLA probe PLUS and MINUS, conjugated with oligonucleotides) in a preheated humidity chamber for 1 h at 37°C and then washed in 1× Wash Buffer A at room temperature twice. Enzymatic ligation depending on the proximity of PLA probe PLUS and MINUS was performed for 30 min at 37°C. After washing the slides in 1× Wash Buffer at room temperature twice, circle amplification was performed with polymerase for 100 min at 37°C. After the final washes with 1× Wash Buffer B and nuclear counterstaining, the processed slides were viewed using a Leica fluorescence microscope.
BiFC assay
Venus N-terminal (1‒173 amino acid residues) was in frame fused with the ERK1, and Venus C-terminal (174‒328 amino acid residues) was in frame fused with the STAT3. The correct expression of fusion proteins was confirmed by immunoblotting. Both the N-terminal and C-terminal fused plasmids were cotransfected into Ishikawa cells. Twenty-four hours after the transfection, the immunofluorescence signal for Venus protein was detected using the fluorescence microscope.
Luciferase reporter assay
Cells grown to 50%‒70% confluence were transfected with vectors using Lipofectamine™ 3000 transfection reagent (Invitrogen), with 50 ng pRL-TK vector as an internal standard, for a dual-luciferase assay (firefly and Renilla luciferases). Luciferase activity was measured at 48 h using a dual-luciferase reporter assay kit (Promega). Results were shown as relative luciferase activity. The primer sequences for constructing the vectors are listed in Supplementary Table S2. All experiments were repeated three times in triplicate.
ISH
ISH was performed as previously described (Tang et al., 2015). Frozen sections (10 μm) were mounted onto poly-L-lysine-coated slides and fixed in 4% PFA solution in PBS at 4°C. Mouse-specific cRNA probes for Igf1r, Igf1, Igf2, Bmp2, and Lif were used for hybridization. Cryo-sections hybridized with sense probes were served as negative controls. Primer sequences to amplify the templates for probe are listed in Supplementary Table S3.
Western blotting
Western blotting analysis was performed as previously described (Ran et al., 2017). Antibodies against IGF1R, p-STAT3 (Y705), STAT3, p-ERK1/2, ERK1/2, AKT, Claudin 1, Claudin 7, and β-actin were used. β-actin served as a loading control.
ChIP‒qPCR
ChIP analyses were performed as previously described (Zhang et al., 2019). The binding of STAT3 to the Cox2 promotor was assessed by ChIP assay using epithelial cells in the IGF1-infused uteri. In brief, the cells were fixed with formaldehyde for 10 min at room temperature, and the chromatin was sonicated using the Bioruptor® Pico sonication device (Diagenode) to fragment the DNA‒protein complex. This supernatant was then complexed overnight at 4°C with the STAT antibody, and then incubated with protein A/G beads to pulldown the complex. After several washes, the resulting protein/DNA complexes were subjected to cross-link reversal and DNA extraction. Specific primers were used to detect immunoprecipitated chromatin fragments, as well as input chromatin, using the qPCR assay (Supplementary Table S4).
Confocal microscopy
Fluorescent images were obtained using a confocal laser-scanning microscope (FV1000; Olympus).
Statistical analysis
Statistical analysis was performed with the SPSS11.5 program. A comparison of means was performed using the independent-sample Student’s t-test. The data are shown as mean ± SEM.
Supplementary material
Supplementary material is available at Journal of Molecular Cell Biology online.
Supplementary Material
Acknowledgements
The authors are grateful to Francesco DeMayo (National Institute of Environmental Health Sciences) and Sudhansu K. Dey (Cincinnati Children’s Hospital Medical Center) for providing us with the PRCre/+ and LtfCre/+ mice, respectively. We thank lab member Faiza Rao for proofreading the manuscript.
Funding
This work was supported by the National Key R&D Program of China (2017YFC1001402 to H.W.; 2018YFC1004401 to S.K.), the National Natural Science Foundation of China (81830045 and 82030040 to H.W.; 81971388 to S.K.), and Institution of Higher Education Projects of Building First-class Discipline Construction in Ningxia Region (NXYLXK2017B05 to G.X.).
Conflict of interest: none declared.
Author contributions: C.Z., S.K., C.W., G.X., and H.W. designed the research; C.Z., M.L., P.W., C.G., Z.N., H.B., Y.T., and H.C. performed experiments; C.Z., S.K., J.L., W.D., X.Y., and H.W. analyzed data; and C.Z., S.K., and H.W. wrote the paper.
References
- Auernhammer C.J., Melmed S. (2000). Leukemia-inhibitory factor—neuroimmune modulator of endocrine function. Endocr. Rev. 21, 313–345. [DOI] [PubMed] [Google Scholar]
- Baumgarten S.C., Armouti M., Ko C., et al. (2017). IGF1R expression in ovarian granulosa cells is essential for steroidogenesis, follicle survival, and fertility in female mice. Endocrinology 158, 2309–2318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cha J., Sun X., Bartos A., et al. (2013). A new role for muscle segment homeobox genes in mammalian embryonic diapause. Open Biol. 3, 130035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J.R., Cheng J.G., Shatzer T., et al. (2000). Leukemia inhibitory factor can substitute for nidatory estrogen and is essential to inducing a receptive uterus for implantation but is not essential for subsequent embryogenesis. Endocrinology 141, 4365–4372. [DOI] [PubMed] [Google Scholar]
- Constância M., Hemberger M., Hughes J., et al. (2002). Placental-specific IGF-II is a major modulator of placental and fetal growth. Nature 417, 945–948. [DOI] [PubMed] [Google Scholar]
- Daikoku T., Cha J., Sun X., et al. (2011). Conditional deletion of Msx homeobox genes in the uterus inhibits blastocyst implantation by altering uterine receptivity. Dev. Cell 21, 1014–1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daikoku T., Ogawa Y., Terakawa J., et al. (2014). Lactoferrin-iCre: a new mouse line to study uterine epithelial gene function. Endocrinology 155, 2718–2724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das S.K., Wang X.N., Paria B.C., et al. (1994). Heparin-binding EGF-like growth factor gene is induced in the mouse uterus temporally by the blastocyst solely at the site of its apposition: a possible ligand for interaction with blastocyst EGF-receptor in implantation. Development 120, 1071–1083. [DOI] [PubMed] [Google Scholar]
- Dietrich P., Dragatsis I., Xuan S., et al. (2000). Conditional mutagenesis in mice with heat shock promoter-driven cre transgenes. Mamm. Genome 11, 196–205. [DOI] [PubMed] [Google Scholar]
- Ernst M., Inglese M., Waring P., et al. (2001). Defective gp130-mediated signal transducer and activator of transcription (STAT) signaling results in degenerative joint disease, gastrointestinal ulceration, and failure of uterine implantation. J. Exp. Med. 194, 189–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamatani T., Daikoku T., Wang H., et al. (2004). Global gene expression analysis identifies molecular pathways distinguishing blastocyst dormancy and activation. Proc. Natl Acad. Sci. USA 101, 10326–10331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardy K., Spanos S. (2002). Growth factor expression and function in the human and mouse preimplantation embryo. J. Endocrinol. 172, 221–236. [DOI] [PubMed] [Google Scholar]
- Hewitt S.C., Li Y., Li L., et al. (2010). Estrogen-mediated regulation of Igf1 transcription and uterine growth involves direct binding of estrogen receptor alpha to estrogen-responsive elements. J. Biol. Chem. 285, 2676–2685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewitt S.C., Lierz S.L., Garcia M., et al. (2019). A distal super enhancer mediates estrogen-dependent mouse uterine-specific gene transcription of Insulin-like growth factor 1 (Igf1). J. Biol. Chem. 294, 9746–9759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiraoka T., Hirota Y., Fukui Y., et al. (2020). Differential roles of uterine epithelial and stromal STAT3 coordinate uterine receptivity and embryo attachment. Sci. Rep. 10, 15523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain N., Zhang T., Fong S.L., et al. (1998). Repression of Stat3 activity by activation of mitogen-activated protein kinase (MAPK). Oncogene 17, 3157–3167. [DOI] [PubMed] [Google Scholar]
- Le J., Zhang D.Y., Zhao Y., et al. (2016). ITF promotes migration of intestinal epithelial cells through crosstalk between the ERK and JAK/STAT3 pathways. Sci. Rep. 6, 33014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K.Y., Jeong J.W., Wang J., et al. (2007). Bmp2 is critical for the murine uterine decidual response. Mol. Cell. Biol. 27, 5468–5478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li A., Chen P., Leng Y., et al. (2018). Histone deacetylase 6 regulates the immunosuppressive properties of cancer-associated fibroblasts in breast cancer through the STAT3‒COX2-dependent pathway. Oncogene 37, 5952–5966. [DOI] [PubMed] [Google Scholar]
- Li Y., Sun X., Dey S.K. (2015). Entosis allows timely elimination of the luminal epithelial barrier for embryo implantation. Cell Rep. 11, 358–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim H., Paria B.C., Das S.K., et al. (1997). Multiple female reproductive failures in cyclooxygenase 2-deficient mice. Cell 91, 197–208. [DOI] [PubMed] [Google Scholar]
- Lim H., Song H., Paria B.C., et al. (2002). Molecules in blastocyst implantation: uterine and embryonic perspectives. Vitam. Horm. 64, 43–76. [DOI] [PubMed] [Google Scholar]
- Lim H.J., Wang H. (2010). Uterine disorders and pregnancy complications: insights from mouse models. J. Clin. Invest. 120, 1004–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J., Khan M., Zapiec B., et al. (2017). PDGFRA is not essential for the derivation and maintenance of mouse extraembryonic endoderm stem cell lines. Stem Cell Rep. 9, 1062–1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Kleiner I., Gall Troselj K. (2010). Mannose-6-phosphate/insulin-like growth factor 2 receptor (M6P/IGF2R) in carcinogenesis. Cancer Lett. 289, 11–22. [DOI] [PubMed] [Google Scholar]
- Mohamet L., Heath J.K., Kimber S.J. (2009). Determining the LIF-sensitive period for implantation using a LIF-receptor antagonist. Reproduction 138, 827–836. [DOI] [PubMed] [Google Scholar]
- Murphy L.J., Ghahary A. (1990). Uterine insulin-like growth factor-1: regulation of expression and its role in estrogen-induced uterine proliferation. Endocr. Rev. 11, 443–453. [DOI] [PubMed] [Google Scholar]
- Nallasamy S., Li Q., Bagchi M.K., et al. (2012). Msx homeobox genes critically regulate embryo implantation by controlling paracrine signaling between uterine stroma and epithelium. PLoS Genet. 8, e1002500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norwitz E.R., Schust D.J., Fisher S.J. (2001). Implantation and the survival of early pregnancy. N. Engl. J. Med. 345, 1400–1408. [DOI] [PubMed] [Google Scholar]
- Paria B.C., Huet-Hudson Y.M., Dey S.K. (1993). Blastocyst's state of activity determines the ‘window’ of implantation in the receptive mouse uterus. Proc. Natl Acad. Sci. USA 90, 10159–10162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paria B.C., Ma W., Tan J., et al. (2001). Cellular and molecular responses of the uterus to embryo implantation can be elicited by locally applied growth factors. Proc. Natl Acad. Sci. USA 98, 1047–1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paria B.C., Reese J., Das S.K., et al. (2002). Deciphering the cross-talk of implantation: advances and challenges. Science 296, 2185–2188. [DOI] [PubMed] [Google Scholar]
- Pawar S., Starosvetsky E., Orvis G.D., et al. (2013). STAT3 regulates uterine epithelial remodeling and epithelial‒stromal crosstalk during implantation. Mol. Endocrinol. 27, 1996–2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramathal C., Bagchi I.C., Bagchi M.K. (2010). Lack of CCAAT enhancer binding protein β (C/EBPβ) in uterine epithelial cells impairs estrogen-induced DNA replication, induces DNA damage response pathways, and promotes apoptosis. Mol. Cell. Biol. 30, 1607–1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran H., Kong S., Zhang S., et al. (2017). Nuclear Shp2 directs normal embryo implantation via facilitating the ERalpha tyrosine phosphorylation by the Src kinase. Proc. Natl Acad. Sci. USA 114, 4816–4821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappolee D.A., Sturm K.S., Behrendtsen O., et al. (1992). Insulin-like growth factor II acts through an endogenous growth pathway regulated by imprinting in early mouse embryos. Genes Dev. 6, 939–952. [DOI] [PubMed] [Google Scholar]
- Richards R.G., DiAugustine R.P., Petrusz P., et al. (1996). Estradiol stimulates tyrosine phosphorylation of the insulin-like growth factor-1 receptor and insulin receptor substrate-1 in the uterus. Proc. Natl Acad. Sci. USA 93, 12002–12007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh H., Aplin J.D. (2009). Adhesion molecules in endometrial epithelium: tissue integrity and embryo implantation. J. Anat. 215, 3–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart C.L., Kaspar P., Brunet L.J., et al. (1992). Blastocyst implantation depends on maternal expression of leukaemia inhibitory factor. Nature 359, 76–79. [DOI] [PubMed] [Google Scholar]
- Sun X., Park C.B., Deng W., et al. (2016). Uterine inactivation of muscle segment homeobox (Msx) genes alters epithelial cell junction proteins during embryo implantation. FASEB J. 30, 1425–1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang X., Chen Y., Ran H., et al. (2015). Systemic morphine treatment derails normal uterine receptivity, leading to embryo implantation failure in mice. Biol. Reprod. 92, 118. [DOI] [PubMed] [Google Scholar]
- Wang H., Dey S.K. (2006). Roadmap to embryo implantation: clues from mouse models. Nat. Rev. Genet. 7, 185–199. [DOI] [PubMed] [Google Scholar]
- Zhang S., Kong S., Lu J., et al. (2013). Deciphering the molecular basis of uterine receptivity. Mol. Reprod. Dev. 80, 8–21. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Meng N., Bao H., et al. (2019). Circadian gene PER1 senses progesterone signal during human endometrial decidualization. J. Endocrinol. 243, 229–242. [DOI] [PubMed] [Google Scholar]
- Zhu L., Pollard J.W. (2007). Estradiol-17β regulates mouse uterine epithelial cell proliferation through insulin-like growth factor 1 signaling. Proc. Natl Acad. Sci. USA 104, 15847–15851. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.