Abstract
Comprehensive genetic profiling using next‐generation sequencing technologies has become an integral part of precision oncology. Variant annotation requires translating the DNA findings into protein level predictions. In this article we highlight inconsistencies in variant annotation for the MET D1228N exon 19 resistance mutations. MET D1228N and D1246N represent the same resistance mutation in MET exon 14 skipping alterations annotated on different transcripts. Additional examples of relevant variants annotated on different transcripts emphasize the importance of avoiding erroneous interpretation when realizing precision oncology.
Short abstract
Comprehensive genetic profiling using next‐generation sequencing technologies has become an integral part of precision oncology. This article highlights inconsistencies in variant annotation, focusing on the annotation for the MET D1228N exon 19 resistance mutation.
Comprehensive genetic profiling using next‐generation sequencing technologies has become an integral part of precision oncology [1]. The new wealth of genetic information underscores the need for accurate interpretation and clear communication between laboratory professionals and clinicians. Recent studies have shown marked variability in the interpretation of the pathogenicity of genomic alterations [2]. Although there are numerous reasons contributing to the complexities of interpreting genetic data, one underlying cause of propagating inconsistencies in variant annotation is the need to translate the DNA findings into protein‐level predictions. Paralleling the central dogma of molecular biology (DNA → RNA → protein), inconsistencies in protein prediction arise when mutations are named using different transcript templates resulting in different amino acid positions that represent the same DNA‐level variant.
Although there are numerous examples of inconsistencies in variant annotation, one of the most common MET resistance alterations represents a particularly interesting case study. We recently encountered inconsistencies in the annotation for the MET D1228N exon 19 resistance mutation. In our analyses, we have observed a discrepancy of the described mutation annotation in prior publications (Fig. 1A) [3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13]. Specifically, this MET point mutation confers resistance to inhibitors targeting MET exon 14 skipping mutants and can be annotated as D1228N or D1246N depending on the transcript used (Fig. 1A) [6, 7]. The D1246N annotation is based on NM_001127500.3 (MET transcript, variant 1; Fig. 1B), whereas the D1228N annotation is based on the 18 amino acid–shorter transcript, NM_000245.4 (MET transcript, variant 2; Fig. 1C).
Figure 1.
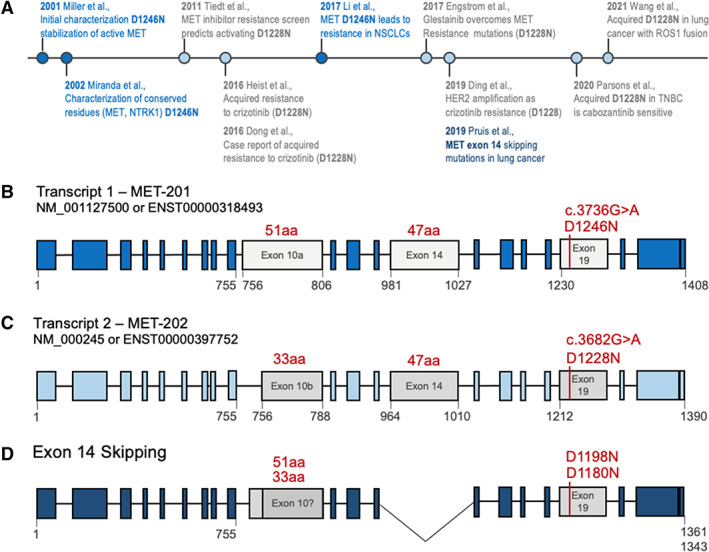
The MET D1246N and D1228N are identical variants annotated on different transcript. (A): Timeline of literature usage of MET D1246N (blue) or D1228N (gray) with key findings; references are in supplemental online Table 1. (B): Schematic of MET transcript 1, which is 6,876 nucleotides (1,408 amino acids) and uses exon 10a (gray). The exon 19 resistance mutation is denoted D1246N (c.3736G>A) (red). The formal annotation for hg19 Chr:7 Pos:116423407 Ref:G Alt:A is Ensembl: ENST00000318493: c.3736G>A; ENSP00000317272: p.Asp1246Asn, National Center for Biotechnology Information (NCBI): NM_001127500. Exon lengths in amino acids are shown in red. Relevant amino acid residues are shown in gray below exons. (C): Schematic of MET transcript 2, which is 6,822 nucleotides (1,390 amino acids) and uses the 18 amino acid–shorter exon 10b (gray). The exon 19 resistance mutation is denoted D12228N (c.3682G>A) (red). The formal annotation for hg19 Chr:7 Pos:116423407 Ref:G Alt:A is Ensembl: ENST00000397752: c.3682G>A; ENSP00000380860:p.Asp1228Asn, NCBI: NM_000245. Exon lengths in amino acids are shown in red. Relevant amino acid residues are shown in gray below exons. (D): MET exon 14 skipping transcript. In MET exon 14 skipping, the 47 amino acids encoded by exon 14 are missing, resulting in a shorter protein. This results in a shift of the C‐terminal amino acid numbering; the resistance mutations are shown in red. It is currently unknown whether all MET exon 14 skipping cases use exon 10a or 10b—therefore, the assumed position of the resistance mutation is annotated as D1198N (exon 10a) or D1180N (exon 10b). Relevant amino acid residues are shown in gray below exons. Abbreviations: aa, amino acids; MET‐201, MET transcript 1; MET‐202, MET transcript 2; NSCLC, non‐small cell lung cancer; TNBC, triple‐negative breast cancer.
For bioinformaticians, the genomic coordinates of the mutation is considered the ground truth and self‐explanatory—although the version (so‐called assembly) of the reference genome must also be noted for an unmistakable annotation (e.g., genome version hg19=GRCh37 from 2009 vs. hg38=GRCh38 from 2013). Translating the nucleotide alteration to the protein level enables immediate recognition of its clinical significance. For example, most oncologists readily identify BRAF V600E, EGFR L858R, and KRAS G12C as oncogenic driver alterations for which Food and Drug Administration–approved targeted therapies are available. In this MET example, the genomic alteration is hg19 Chr:7 Pos:116423407 G > A, and although it is unambiguous, the syntax is less useful than MET D1228N, the terminology used most commonly in the clinical literature. The D1228N mutation has been originally described as a relevant resistance mutation arising in lung cancers with MET exon 14 skipping mutations after treatment with MET tyrosine kinase inhibitors [14]. However, the original description of the variant years earlier used the longer transcript (D1246N). Although D1228N has become part of the clinical lexicon as a key resistance mutation, genomic standards have since evolved to annotate the longest transcript, resulting in D1246N. Thus, currently both MET D1228N and D1246N coexist, for example, when reports are received from two laboratories using different transcript isoforms for mutation annotation.
There are additional aspects further complicating this discrepancy in the setting of MET exon 14 skipping mutations. First, because of exon 14 skipping during RNA splicing, the pathogenic transcript isoform is 47 amino acids shorter. Second, it is unclear whether this transcript uses exon 10a or 10b (Fig. 1D); the exact exon makeup is currently unclear. Third, the 11‐kb genomic distance between MET exon 14 skipping mutations and the D1228N/D1246N resistance mutation in exon 19 (~11 kb) poses unique technical challenges in delineating the specific transcript isoform used. Thus, in tumors containing MET transcripts missing exon 14, neither D1246N nor D1228N truly represents the actual position at the protein level (Fig. 1D). The precise annotation on a mutant‐specific transcript, although biologically accurate, is technically problematic because of current sequencing limitations. Therefore, selection of a standard wild‐type reference transcript is necessary for consistency across laboratories.
Although we focus here on the specific MET D1246N/D1228N example, this issue is readily generalizable, as there are several other examples that share similar discrepancies (Table 1). However, there are also several precedents for successful nomenclature changes; for example, BRAF V600E (formerly reported as BRAF V599E) and H3F3A K28M/G35 (formerly reported as K27/G34) [15]. The reason for these one‐number amino acid discrepancies was that initial papers disregarded the first methionine, as it is cleaved in an early post‐translational state [16]. Variant annotation at the protein level, however, is insufficient for portraying accurate amino acid level changes, as these vary depending on the transcript used. For example, EGFR p.L858R could be annotated differently when using other isoforms. EGFR p.L858R (NM_005528) is the same variant as EGFR p.L813R (NM_001346899), EGFR p.L805R (NM_001346900), and EGFR p.L591R (NM_001346941). Other pertinent examples are highlighted in Table 1. At a minimum, the transcript ID or RefSeq ID should be included for variant annotations in clinical reports. Many clinical laboratories do report all the relevant information (nucleic acid change, transcript ID, and amino acid change); however, the synthesis of all this information is unwieldly, especially as transcript IDs are less recognizable and unlikely to be memorized.
Table 1.
Examples of relevant mutations and their preferred clinical and selected inconsistent variant annotations
| Gene symbol | Amino acid alteration | Nucleotide alteration Transcript ID(gene):coding DNA position (AA change) | Genomic COORDINATES Chromosome:position(assembly) | Clinical relevance/context |
|---|---|---|---|---|
| ABL1 | T315I | NM_005157.6(ABL1):c.944C>T (p.Thr315Ile) | Chr9:130872896C>T(GRCh38)=Chr9:133748283C>T(GRCh37) |
Imatinib resistance mutation MANE selected |
| T334I (same as T314I) | NM_007313.2(ABL1):c.1001C>T (p.Thr334Ile) | Same as above | ||
| ABL1 | M351T | NM_005157.6(ABL1):c.1052T>C (p.Met351Thr) | Chr9:130873004T>C(GRCh38)=Chr9:133748391T>C(GRCh37) |
Imatinib resistance mutation MANE selected |
| M370T (same as M351T) | NM_007313.2(ABL1):c.1109T>C (p.Met370Thr) | Same as above | ||
| ABL1 | E236K | NM_005157.6(ABL1):c.706G>A (p.Glu236Lys) | Chr9:130862919G>A(GRCh38)=Chr9:133738306G>A(GRCh37) |
Imatinib resistance mutation MANE selected |
| E255K (same as E236K) | NM_007313.2(ABL1):c.763G>A (p.Gly255Lys) | Same as above | Imatinib resistance mutation | |
| BRAF | V640E* | NM_001374258.1(BRAF):c.1919T>A (p.Val640Glu) | Chr7:140753336T>A(GRCh38)=Chr7:140453136T>A(GRCh37) |
Targetable BRAF mutation MANE selected |
| V600E | NM_001378468.1(BRAF):c.1799T>A (p.Val600Glu) | Same as above | Targetable BRAF mutation | |
| V599E (same as V600E) | NM_001378468.1(BRAF):c.1799T>A (p.Val599Glu) | Same as above | Initial numeration was disregarding the first methionine; not in use anymore | |
| EGFR | A289T | NM_005228.5(EGFR):c.865G>A (p.Ala289Thr) | Chr7:55221821G>A(GRCh37)=Chr7:55154128G>A(GRCH38) | Hotspot variant (used in COSMIC) |
| A244T (same as A289T) | NM_001346897.2(EGFR):c.730G>A (p.Ala244Thr) | Same as above | Hotspot variant (used in TCGA) | |
| EGFR | L858R | NM_005228.5(EGFR):c.2573T>G (p.Leu858Arg) | Chr7:55191822T>G(GRCh38)=Chr7:55259515 T>G(GRCh37) |
Targetable EGFR hotspot mutation MANE selected |
| L813R (same as L858R) | NM_001346897.2(EGFR):c.2438T>G (p.Leu813Arg) | Same as above | Targetable EGFR hotspot mutation | |
| L805R (same as L858R) | NM_001346900(EGFR):c.2414T>G (p.Leu805Arg) | Same as above | Targetable EGFR hotspot mutation | |
| L591R (same as L858R) | NM_001346941(EGFR):c.1772T>G (p.Leu591Arg) | Same as above | Targetable EGFR hotspot mutation | |
| EGFR | T790M | NM_005228.5(EGFR):c.2369C>T (p.Thr790Met) | Chr7:55181378C>T(GRCh38)=Chr7:55249071C>T(GRCh37) | EGFR resistance/targetable mutation |
| T745M (same as T790M) | NM_001346897.2(EGFR):c.2234C>T (p.Thr745Met) | Same as above | EGFR resistance/targetable mutation | |
| ERBB2 | V777L b | NM_004448.3(ERBB2):c.2329G>T (p.Val777Leu) b | Chr17:39724747(GRCh38)=Chr17:37881000(GRCh37) |
Oncogenic signaling No MANE selection |
| V777L b | NM_004448.3(ERBB2):c.2329G>C (p.Val777Leu) b | Same as above | Different nucleotide change converges at amino acid level | |
| V747L (same as V777L) | NM_001005862.2:c.2239G>T (p.Val747Leu) | Same as above | Oncogenic signaling | |
| V762L (same as V777L) | NM_001289936.1:c.2284G>T (p.Val762Leu) | Same as above | Oncogenic signaling | |
| FGFR2 | N549H | NM_00141.5(FGFR2):c.1645A>C (p.Asn549His) | Chr10:121498522A>C(GRCh38)=Chr10:123258036A>C(GRCh37) |
Crouzon syndrome MANE selected |
| N550H (same as N549H) | NM_001144913.1(FGFR2):c.1648A>C (p.Asn550His) | Same as above | Crouzon syndrome | |
| N437H (same as N549H) | NM_001144914.1(FGFR2):c.1309A>C (p.Asn437His) | Same as above | Crouzon syndrome | |
| H3(F3A) | K28M | NM_002107.6(H3F3A):c.83A>T (p.Lys28Met) | Chr1:226064434(GRCh38)=Chr1:226252135(GRCh37) | Diagnostic biomarker for midline glioma |
| K27 (same as K28) | NM_002107.6(H3F3A):c.83A>T (p.Lys27Met) | Same as above | Initial numeration was disregarding the first methionine; not in use anymore | |
| H3F3A | G35R | NM_002107.6(H3F3A):c.100G>C (p.Gly35Arg) | Chr1: 226064451G>C(GRCh38)=Chr1:226252152G>C(GRCh37) | Diagnostic biomarker for midline glioma |
| G34R (same as G35R) | NM_002107.6(H3F3A):c.100G>C (p.Gly34Arg) | Same as above | Initial numeration was disregarding the first methionine; not in use anymore | |
| MET | T992I | NM_000245.4(MET):c.2975C>T (p.Thr992Ile) | Chr7:116771936C>T(GRCh38)=Chr7:116411990C>T(GRCh37) |
SNP/activating germline variant MANE selected |
| T1010I (same as T992I) | NM_001127500.3(MET):c.3029C>T (p.Thr1010Ile) | Same as above |
Single nucleotide polymorphisms Activating germline variant |
|
| MET | D1228N | NM_000245.4(MET):c.3682G>A (Asp1228Asn) | Chr7: 116783353G>A(GRCh38)=Chr7:116423407G>A(GRCh37) |
Resistance mutation MANE selected |
| D1246N (same as D1228N) | NM_001127500.3(MET):c.3736G>A (Asp1246Asn) | Same as above | Resistance mutation |
BRAF V640E is currently MANE selected (https://www.ncbi.nlm.nih.gov/clinvar/variation/13961/, accessed July 7, 2021).
Different nucleotide changes can converge at the amino acid level.
Abbreviations: AA, amino acids; COSMIC, Catalogue of Somatic Mutations in Cancer; MANE, Matched Annotation from NCBI and EMBL‐EBI; SNP, single nucleotide polymorphism; TCGA, The Cancer Genome Atlas.
As our knowledge of clinically relevant specific mutation increases, the field is collectively tasked to converge annotations on one transcript. Large‐scale efforts are under way to unify and maintain harmonized transcript nomenclatures (curated independently by the National Center for Biotechnology Information and the European Molecular Biology Laboratories–European Bioinformatics Institute) by merging the annotations of biologically similar transcripts [17]. Importantly, in this case, the MANE Select version 0.93 transcript is NM_000245.4, which would result in D1228N (rather than D1246N).
The confusion caused by variant numbering due to alternative exons is likely only the beginning. As many of the common DNA‐level variants have been established and RNA sequencing technologies are now emerging as robust clinical diagnostics, the relevance of isoform‐specific alterations will become more apparent. For certain cancer types this has already happened. Aside from MET exon 14 skipping mutations as an emerging biomarker in lung cancer, other relevant examples include the androgen receptor splice variant ARv7 (aberrant splicing to cryptic exon 3) in castration‐resistant prostate cancer [18] and the EGFR variant III (EGFRvIII; deletion of exons 2–7) in glioblastoma [19]. These transcripts are defined by their specific exon composition, and as we identify additional oncologically relevant alternative transcripts, a harmonized transcript nomenclature becomes a foundational building block for reliable annotation and efficient integration into clinical practice.
The practical realization of precision oncology is nuanced, and accurate MET transcript annotation represents one relevant challenge. Until one reference wild‐type transcript is widely accepted and laboratories consistently follow these principles, it should be noted that D1228N and D1246N are the same variant. As we move precision oncology forward, these details will be imperative to avoid confusion in publications, avoid erroneous interpretation as a diagnostic inaccuracy, and facilitate clear communication between molecular pathologists and oncologists to improve patient care.
Disclosures
Aaron N. Hata: Pfizer, Amgen, Roche/Genentech, Novartis, Eli Lilly & Co., Blueprint Medicines, Relay Therapeutics, Nuvalent (RF), Nuvalent (C/A). The other authors indicated no financial relationships.
(C/A) Consulting/advisory relationship; (RF) Research funding; (E) Employment; (ET) Expert testimony; (H) Honoraria received; (OI) Ownership interests; (IP) Intellectual property rights/inventor/patent holder; (SAB) Scientific advisory board
Supporting information
See http://www.TheOncologist.com for supplemental material available online.
Table S1 Supporting Information.
Disclosures of potential conflicts of interest may be found at the end of this article.
No part of this article may be reproduced, stored, or transmitted in any form or for any means without the prior permission in writing from the copyright holder. For information on purchasing reprints contact commercialreprints@wiley.com. For permission information contact permissions@wiley.com.
References
- 1. Malone ER, Oliva M, Sabatini PJB et al. Molecular profiling for precision cancer therapies. Genome Med 2020;12:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Katsoulakis E, Duffy JE, Hintze B et al. Comparison of annotation services for next‐generation sequencing in a large‐scale precision oncology program. JCO Precis Oncol 2020;4:PO.19.00118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ding G, Wang J, Ding P et al. Case report: HER2 amplification as a resistance mechanism to crizotinib in NSCLC with MET exon 14 skipping. Cancer Biol Ther 2019;20:837–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Dong HJ, Li P, Wu CL et al. Response and acquired resistance to crizotinib in Chinese patients with lung adenocarcinomas harboring MET Exon 14 splicing alternations. Lung Cancer 2016;102:118–121. [DOI] [PubMed] [Google Scholar]
- 5. Engstrom LD, Aranda R, Lee M et al. Glesatinib exhibits antitumor activity in lung cancer models and patients harboring MET exon 14 mutations and overcomes mutation‐mediated resistance to type I MET inhibitors in nonclinical models. Clin Cancer Res 2017;23:6661–6672. [DOI] [PubMed] [Google Scholar]
- 6. Heist RS, Sequist LV, Borger D et al. Acquired resistance to crizotinib in NSCLC with MET exon 14 skipping. J Thorac Oncol 2016;11:1242–1245. [DOI] [PubMed] [Google Scholar]
- 7. Li A, Yang JJ, Zhang XC et al. Acquired MET Y1248H and D1246N mutations mediate resistance to MET inhibitors in non‐small cell lung cancer. Clin Cancer Res 2017;23:4929–4937. [DOI] [PubMed] [Google Scholar]
- 8. Miller M, Ginalski K, Lesyng B et al. Structural basis of oncogenic activation caused by point mutations in the kinase domain of the MET proto‐oncogene: Modeling studies. Proteins 2001;44:32–43. [DOI] [PubMed] [Google Scholar]
- 9. Miranda C, Zanotti G, Pagliardini S et al. Gain of function mutations of RTK conserved residues display differential effects on NTRK1 kinase activity. Oncogene 2002;21:8334–8339. [DOI] [PubMed] [Google Scholar]
- 10. Parsons BM, Meier DR, Richmond CS et al. Acquisition of cabozantinib‐sensitive MET D1228N mutation during progression on crizotinib in MET‐amplified triple‐negative breast cancer. Clin Breast Cancer 2020;20:e433–e438. [DOI] [PubMed] [Google Scholar]
- 11. Pruis MA, Geurts‐Giele WRR, von der TJH et al. Highly accurate DNA‐based detection and treatment results of MET exon 14 skipping mutations in lung cancer. Lung Cancer 2020;140:46–54. [DOI] [PubMed] [Google Scholar]
- 12. Tiedt R, Degenkolbe E, Furet P et al. A drug resistance screen using a selective MET inhibitor reveals a spectrum of mutations that partially overlap with activating mutations found in cancer patients. Cancer Res 2011;71:5255–5264. [DOI] [PubMed] [Google Scholar]
- 13. Wang Y, Chen Z, Han X et al. Acquired MET D1228N mutations mediate crizotinib resistance in lung adenocarcinoma with ROS1 fusion: A case report. The Oncologist 2021;26:178–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Heist RS, Shim HS, Gingipally S et al. MET exon 14 skipping in non‐small cell lung cancer. The Oncologist 2016;21:481–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Leske H, Rushing E, Budka H et al. K27/G34 versus K28/G35 in histone H3‐mutant gliomas: A note of caution. Acta Neuropathol 2018;136:175–176. [DOI] [PubMed] [Google Scholar]
- 16. Iwai K, Ishikawa K, Hayashi H. Amino‐acid sequence of slightly lysine‐rich histone. Nature 1970;226:1056–1058. [DOI] [PubMed] [Google Scholar]
- 17. Matched Annotation from NCBI and EMBL‐EBI (MANE) . 2021. Available at https://www.ncbi.nlm.nih.gov/refseq/MANE/. Accessed June 7, 2021.
- 18. Sharp A, Coleman I, Yuan W et al. Androgen receptor splice variant‐7 expression emerges with castration resistance in prostate cancer. J Clin Invest 2019;129:192–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gan HK, Cvrljevic AN, Johns TG. The epidermal growth factor receptor variant III (EGFRvIII): Where wild things are altered. FEBS J 2013;280:5350–5370. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
See http://www.TheOncologist.com for supplemental material available online.
Table S1 Supporting Information.


