Abstract
Malformations in the eye can be caused by either an excess or deficiency of retinoids. An early target gene of the retinoid metabolite, retinoic acid (RA), is that encoding one of its own receptors, the retinoic acid receptor β (RARβ). To better understand the mechanisms underlying this autologous regulation, we characterized the chick RARβ2 promoter. The region surrounding the transcription start site of the avian RARβ2 promoter is over 90% conserved with the corresponding region in mammals and confers strong RA-dependent transactivation in primary cultured embryonic retina cells. This response is selective for RAR but not retinoid X receptor-specific agonists, demonstrating a principal role for RAR(s) in retina cells. Retina cells exhibit a far higher sensitivity to RA than do fibroblasts or osteoblasts, a property we found likely due to expression of the orphan nuclear receptor TLX. Ectopic expression of TLX in fibroblasts resulted in increased sensitivity to RA induction, an effect that is conserved between chick and mammals. We have identified a cis element, the silencing element relieved by TLX (SET), within the RARβ2 promoter region which confers TLX- and RA-dependent transactivation. These results indicate an important role for TLX in autologous regulation of the RARβ gene in the eye.
The vitamin A derivative retinoic acid (RA) has been suggested to play important roles in vertebrate embryonic development and cell differentiation. Vitamin A deficiency and/or excessive doses of RA are known to result in a spectrum of distinct malformations during organogenesis and pattern formation (reviewed in references 8, 19, and 49). Two classes of receptors, RA receptors (RARs) and retinoid X receptors (RXRs), which belong to a large family of nuclear hormone receptors, mediate RA signaling. These receptors are capable of binding specific target DNA sequences in the regulatory regions of responsive genes, termed RA response elements (RAREs), to activate or repress transcription (5, 29).
Among vitamin A metabolites, all-trans-RA (at-RA) has been shown to bind the RARs, whereas a stereoisomer of at-RA, 9-cis-RA, acts as a high-affinity bipotential ligand for both RARs and RXRs. A RAR and RXR preferentially form a heterodimer to bind RAREs consisting of a direct repeat of canonical AGGTCA sequences separated by two or five nucleotides (28).
Three subtypes of the gene for RAR, α, β, and γ, have been identified in chick (33), mouse (58), and human (22) and are expressed in distinct spatial and temporal patterns during embryogenesis. In the chick embryo, RARβ has been shown to be prominently expressed in the developing central nervous system and RARγ expression is mainly restricted to the skin, whereas RARα expression is rather ubiquitous (32, 44, 48, 50). Several isoforms exist for each subtype. In mammals, the genes for all three subtypes can be transcribed from at least two different promoters, resulting in variation at the amino termini between isoforms. For RARβ, four distinct mRNAs result from use of alternative promoters (β1 and β2) and splicing variations (59). In chick, distinct forms of RARβ mRNAs display individual patterns of expression during embryogenesis (45, 48).
One of the parameters modulating the differential expression of RARs and RXRs is RA itself. The human and mouse RARβ2 promoters have a well-conserved RARE (the so-called βRARE) just upstream of the TATA box. As a result, the RARβ2 promoters are highly inducible by RA and are considered to be one of the earliest targets of RA action (11, 51).
In transgenic mice with the RARβ2 promoter fused to the Escherichia coli β-galactosidase (β-gal) gene, β-gal activity was present in the pigmented retina (31, 41). β-gal staining was also observed in the eye of a RARE–β-gal transgenic embryo. Staining was increased upon maternal treatment with RA, suggesting that in vivo, some of the morphogenetic effects of RA could be mediated through localized transcriptional activity controlled by the various RARs (2, 43).
In chick embryos, RARβ transcripts can be upregulated by exogenously added RA. Implantation of RA-soaked beads in limb buds causes rapid (within 4 h) accumulation of RARβ2 mRNA (39, 54). This induction was also observed by Northern analysis using mRNA isolated from facial primordia (45). Together these data strongly suggest that the mechanisms underlying the autologous regulation of RARβ gene expression are well conserved between mammals and avians.
TLX is an orphan nuclear receptor originally identified on the basis of its similarity to RXR; it is structurally and functionally (biochemically) homologous to the Drosophila terminal-gap gene tailless. Expression of TLX is restricted to the fore- and midbrain, neuroepithelium, retina, and nasal epithelium (18, 36, 57). We and others have shown that the function of TLX is necessary for the proper formation of specific eye and brain structures, but the precise molecular mechanisms of its action are still being unraveled (17, 35, 56).
Here we show that the RARβ2 promoters and TLX proteins are highly conserved during evolution and that TLX can modulate the autoregulation of the RARβ2 promoter.
MATERIALS AND METHODS
Isolation of chick RARβ promoter.
A chick 3-day embryonic cDNA library was screened with a fragment of mouse RXRβ as the probe (57). Multiple cDNA clones were found to encode the chick RARβ, including clones corresponding to mRNAs utilizing both β1 and β2 promoters. Sequence information from the RARβ2 cDNA clone containing the longest 5′ noncoding region was used to design primers for PCR. The primer sequences used for genomic PCR analyses were as follows: NMO1 (5′-GCTCTTGCAGGGCTGCTGGGAGTTT-3′), NMO2 (5′-AATCTCTCTAGAACCAGTCCCGTTCCTCAG-3′), NMO8 (5′-CCCTCAGCCATGAATAGATCCTTC-3′), and NMO9 (5′-CCTGCCTCTCTGGCTGTCTGCTTT-3′). First, for the NMO9-NMO2 primer combination, PCR amplification was carried out for 33 cycles (30 s at 94°C, 1 min at 50°C, 2 min at 72°C) with 50 ng of chick genomic DNA, 200 ng of each primer, and 2.5 U of Taq DNA polymerase (Life Technologies). A second round of amplification was carried out for 25 cycles, using 1/50 of the first PCR mixture as the template with the NMO1-NMO8 primer combination. Products from the second PCR were separated on agarose gel and purified, and a 220-bp fragment was ligated into the TA cloning vector pMOSBlue (Amersham). The DNA sequence was determined with an AutoCycle sequencing kit on an A.L.F. II DNA sequencer (Pharmacia). Sequences of at least two clones from each of three independent PCR products were determined.
Cloning of human TLX cDNA.
The National Center for Biotechnology Information expressed sequence tag database was searched for sequences related to the chick TLX, using the program BLASTN (1). Two expressed sequence tags with similarity to chick TLX, those with the GenBank accession numbers R18964 and R43976, were identified from a single human infant brain cDNA clone, am156j01. The plasmid encompassing am156j01 was used to design oligonucleotide probe NMO63 (5′-GACAACTCCGGTTAGATGC-3′). The full-length human TLX cDNA clone in the mammalian expression vector was selected using the GENETRAPPER cDNA Positive Selection System (Life Technologies) from among 4 × 1011 clones of a human fetal brain cDNA pCMV-SPORT2 library (Life Technologies).
Cell culture and transfection assay.
CV-1 and MC3T3-E1 cells were maintained in Dulbecco's modified Eagle's medium (DMEM) and α-MEM medium (Life Technologies), respectively, supplemented with 10% fetal bovine serum (FBS). Retina cells were isolated from day-4.5 chick embryos according to the method described previously (40). Cells were washed with phosphate-buffered saline–EDTA, treated with 0.125% trypsin, and plated on 24-well dishes (Costar). Retina cells were kept in 10% FBS–DMEM for 4 days. Then 2 h prior to transfection, medium was replaced with 10% charcoal-resin double-treated FBS–DMEM. Transfections were performed by the calcium phosphate precipitation method as previously described (53). Cells were transfected for 6 h in 24-well dishes with a total of 750 ng of DNA/well adjusted by pGEM4 plasmid together with 250 ng (or 150 ng for the thymidine kinase [tk]-driven reporter) of reporter plasmid, 350 ng of reference plasmid (pCMX-βGAL), and 50 ng of receptor plasmid. After washing out of DNA precipitates, cells were incubated with added ligand for 36 h. Cell extracts were subsequently prepared and assayed for luciferase and β-gal activities. All data points were determined in triplicate and normalized for transfection efficiency with β-gal as an internal control. at-RA (Nacalai, Kyoto, Japan) and 9-cis-RA (Wako Pure Chemical, Osaka, Japan) stock solutions were prepared in 20% dimethyl sulfoxide–80% ethanol. Synthetic retinoids LG69 and TTNPB, kindly provided by R. Heyman, were solubilized in dimethyl sulfoxide and added at the indicated concentrations.
Serum stripping.
FBS (Life Technologies) was equilibrated twice with equal volumes of n-heptane for 2 h at room temperature. After the second separation, the recovered serum was stirred twice with 50 g of resin (AG 1-X8, analytical grade, 200/400 mesh; Bio-Rad)/liter and 20 g of charcoal/liter. After centrifugation to remove particulates, the serum was passed through a 0.22-μm-pore-size filter for sterilization.
Reverse transcription-PCR analyses.
Total RNA was isolated from primary cultured chick retina cells with ISOGEN (Nippon Gene, Toyama, Japan), and cDNA template was prepared with a RNA PCR kit (Takara Shuzo, Kyoto, Japan) according to the manufacturer's instructions. Using 200 ng of each primer, PCR amplification was performed for 30 cycles (30 s at 94°C, 60 s at 68°C, 90 s at 72°C) with 2.5 U of Ex-Taq DNA polymerase (Takara Shuzo). The sequences of the 5′ primers for RARβ transcripts are as follows: NMO46 (5′-ACTGAATGGTGGTCTGAGACACGGACTAAG-3′) for β1 and NMO3 (5′-CTGAGGAACGGGACTGGTTCTAGAGAGATT-3′) for β2. NMO21 (5′-CTTGGAACAAGTTCCTCAGAACTGGTGCTC-3′) was used as the common 3′ RARβ primer. The NMO53 (5′-GATGGTCAGGTCATCACCATTGG-3′) and NMO54 (5′-CATCGTACTCCTGCTTGCTGATCC-3′) primer set was used for β-actin. The identity of each RARβ2 promoter-derived PCR product was confirmed by sequencing of at least four independent clones.
Plasmid constructions.
The expression vectors pCMX-hRXRα (55), pCMX-cTLX, pCMX-mTLX (57), pCMX-hRARα, and pCMX-βGAL and reporter plasmids tk-LUC and mβRARE-tk-LUC (53), which were used in the transfection assay, have been previously described. The coding region of human COUP-TFII/ARP-1 (24) was cloned into the EcoRI site of the pCMX vector (53) to create the pCMX-hCOUP-TFII expression vector. The cβRARE-tk-LUC, SET-tk-LUC, SET-mβRARE-tk-LUC, mβRARE-SET-tk-LUC, SETm1-mβRARE-tk-LUC, and SETm2-mβRARE-tk-LUC reporter plasmids were prepared by inserting the synthetic oligonucleotides cβRARE (agctTGGGTTCACAGAAAGTTCACTCGagct), SET (agctTGGGTCATTTGAAGGTTAGCagct), SETm1 (agctTGAACCATTTGAAGGTTAGCagct), SETm2 (agctTGGGTCATTTGAAAACTAGCagct), and mβRARE (agctTAAGGGTTCACCGAAAGTTCACTCGCATagct) into the HindIII site of a tk-LUC basal luciferase reporter or mβRARE-tk-LUC reporter. The mouse RARβ2-Δ2.2k-LUC promoter reporter plasmid was made by inserting a 2.2-kb fragment (including the transcription start site of mouse RARβ2) into the SalI/XhoI site to replace the tk promoter of the tk-LUC reporter plasmid. For the chick RARβ2-Δ119-LUC, first, the PCR-amplified fragment with NMO2 and NMO7 (5′-AAGCTCTGTGAGAATCCTGGGAG-3′), which encodes the −119 to +68 region of the chick RARβ2 promoter, was ligated into the TA cloning vector pMOSBlue (Amersham). The HindIII/BamHI fragment was excised for ligation into the HindIII/BglII site to replace the tk promoter of the tk-LUC reporter. For the chick RARβ2-Δ85-LUC and the mouse RARβ2-Δ66-LUC, first, the PCR fragment-amplified with KMO45 (5′-GTACGTCGACTGGGTCATTTGAAGGTTAGCAG-3′) and NMO2 or with NMO5 (5′-GGTGGATCCAGCAGCCCGGGAAGGGTTCACCGAA-3′) and NMO6 (5′-GTACTCGAGGCACGGGAACTCTGGTCCCCCCCTT-3′) was excised with BamHI and SalI or XhoI. The fragment was ligated into the SalI/BglII or BamHI/XhoI site, respectively, to replace the tk promoter of the tk-LUC reporter. Sequences of the PCR-amplified region were confirmed by standard methods.
Gel retardation assays.
Proteins were synthesized by an in vitro transcription-translation system using rabbit reticulocyte lysate (Promega) with plasmid pCMX-hRARα, pCMX-hRXRα, pCMX-cTLX, or pCMX-hCOUP-TFII. For binding, 4 μl of lysate was incubated first in 10 mM Tris-HCl (pH 8.0), 80 mM KCl, 1 mM dithiothreitol, 0.1% NP-40, 1 μg of poly(dI-dC), and 7.5% glycerol on ice for 20 min. Excess unlabeled competitor oligonucleotides, when included, were added during this preincubation period. Then 0.2 to 0.6 pmol of 32P-labeled oligonucleotide (3 × 105 cpm, prepared by filling in with Klenow polymerase in the presence of [α-32P]dCTP) probe was added to the reaction, followed by incubation on ice for 30 min. The protein-DNA complexes were resolved on a 5% polyacrylamide gel in 0.5× Tris-borate-EDTA buffer. Gels were subsequently dried and subjected to autoradiography. The same oligonucleotides described in plasmid constructions were used for gel retardation assays for SET, mβRARE, and SET-mβRARE. The Krüppel oligonucleotide was described previously (57). The sequence of the nonspecific competitor is agctACAAGGTTCACGAGGTTCACGTCagct.
Nucleotide sequence accession numbers.
The sequences reported in this paper have been deposited in the GenBank database (accession no. AF220160, AF220161, AF220162, and AF220163 for the chick RARβ2 promoter region and RARβ4′, RARβ4M, and RARβ4M′ cDNA and AF220532 for human TLX cDNA.
RESULTS
Induction of RARβ expression in chick retina cells.
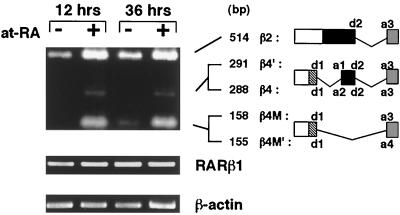
The expression of RARβs in chick embryonic retina cells was examined by reverse transcription-PCR. Three distinct bands were observed by agarose gel analysis (Fig. 1). Subcloning and subsequent DNA sequencing of the PCR products revealed the presence of five RARβ isoforms; in addition to the previously reported β2 and β4 (38, 48), three were novel (Fig. 1). These isoforms are apparently generated by alternative splicing and differ only in their amino termini (Fig. 1). The chick RARβ4M (corresponding to mouse RARβ4) isoform lacks an apparent acceptor sequence (AG) at nucleotide positions 617 and 618, suggesting the presence of an intron between nucleotides 618 and 619; an intron is present at the analogous position in mouse RARβ (37). In cultured retina cells, the expression of all RARβ2 isoforms is induced within 12 h of addition of at-RA (Fig. 1). In contrast, the RARβ1 isoform appears not to be affected by exogenous at-RA, indicating a specific role for the RARβ2 promoter in retina cells (Fig. 1).
FIG. 1.
Induction of RARβ2 isoforms by RA in chick retina cells. RNAs were reverse transcribed and amplified with chick RARβ2-, β1-, or β-actin-specific primers. Three major bands were resolved by 1.2% agarose gel in Tris-borate-EDTA buffer for RARβ2 isoforms with apparent estimated sizes of 514, 288 to 291, and 155 to 158 bp. The expression of RARβ2 isoforms is induced by 1 μM at-RA within 12 h in chick retina cells. No induction was observed 36 h after medium change in the absence of exogenous at-RA. Note that the expression of RARβ1 or β-actin transcripts is not affected by at-RA. Donor and acceptor sites for splicing are indicated in the schematic representation of chick RARβ isoforms (see Fig. 2 for details). The nomenclature of RARβ isoforms is based on correlation to mouse and human RARβ isoforms.
Determination of chick RARβ2 promoter sequence.
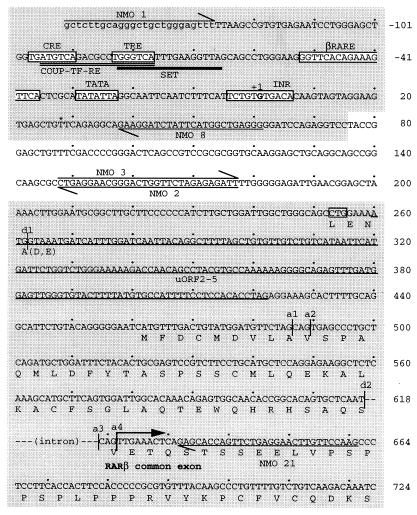
To compare the mammalian and avian RARβ2 promoters, we isolated the chick RARβ2 promoter region by genomic PCR. To accomplish this, 5′ primers were designed based upon the well-conserved human and/or mouse sequences, while 3′ primers were designed based on chick-specific sequence from within the cDNA. Over 90% sequence identity was observed among human, mouse, and chick genes in the 150-bp region surrounding the transcription start site. The high conservation in sequence also allowed prediction of the putative transcription start site for the chick gene (11). We confirmed the presence of a cyclic AMP RE, AP-1 binding site (TRE), RARE (βRARE), RNA polymerase initiator site (INR), and TATA box (12) that are conserved in mouse and human. Some short upstream open reading frames (uORFs) which have been reported to regulate the translational level of mouse RARβ2 were also found (Fig. 2) (42).
FIG. 2.
Nucleotide sequence and predicted amino acid sequence of chick RARβ. PCR primers used are indicated by arrows. The putative transcription start site is designated +1. The cAMP RE (CRE), AP-1 binding site (TRE), βRARE, TATA box, and RNA polymerase initiator site (INR) sequences are indicated by boxes. An asterisk (∗) indicates the 5′ end of the cDNA confirmed by cDNA library screening. Splicing donor and acceptor sites are indicated by “d” and “a”, respectively. The boxed CTG indicates the non-AUG initiation codon proposed for the mRARβ4 transcript (37). The large arrow indicates the common region for all RARβ transcripts. The regions of short uORFs (uORF2 to -5) reported for mouse RARβ2 (42) are underlined, and regions highly conserved between mammals and avians are indicated by shaded boxes. COUP-TF-RE (26) and SET sequences are indicated by double and bold underlines, respectively.
Induction of RARβ2 promoter by at-RA is cell type specific.
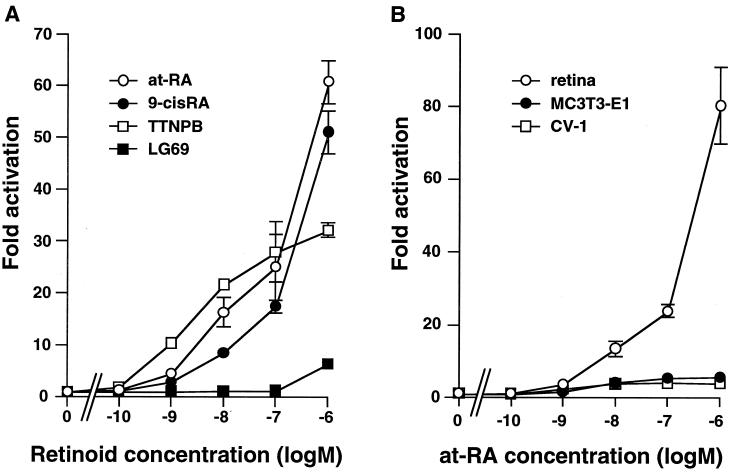
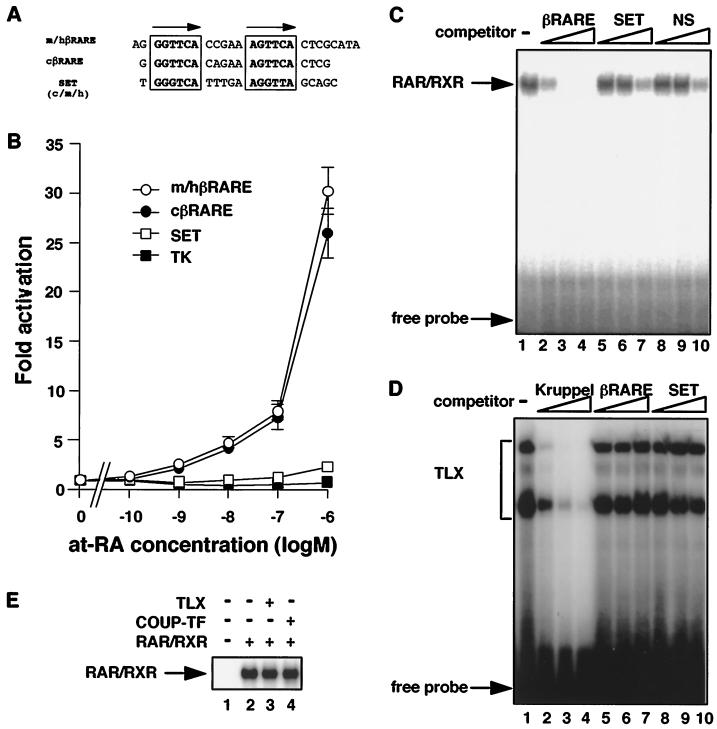
As an initial step to characterize the function of the chicken RARβ2 promoter, we generated a luciferase reporter construct and confirmed the ability of this region to direct RARβ2 transcription by transient transfection in primary cultured chick embryonic retina cells. Promoter activity could be induced by at-RA concentrations as low as 0.1 μM (Fig. 3A). Activation by 9-cis-RA or a RAR-specific agonist (TTNPB) but not by a RXR-specific agonist (LG69) was also observed. These results suggest that the induction of RARβ2 expression is mediated primarily by RAR, not RXR.
FIG. 3.
(A) RARβ2 promoter activity is induced by RAR agonists but not by RXR-specific agonists. cRARβ2 promoter-driven luciferase reporter plasmids (cRARβ2-Δ119-LUC) were transfected into primary cultured chick retina cells. Final concentrations of retinoids are indicated. (B) Induction of cRARβ2 promoter activity by at-RA is cell type specific. cRARβ2 promoter-driven luciferase reporter plasmids were transfected as described for Fig. 3A. at-RA was added to the indicated final concentrations. Promoter activity is induced by at-RA in all cell types, but the magnitude of induction differs.
Using this promoter construct, we examined the at-RA sensitivity of the RARβ2 promoter in the monkey fibroblast cell line CV-1 and mouse osteoblast cell line MC3T3-E1. Although promoter activity can also be induced by at-RA in these cell lines, the level of induction is much lower than that observed in the retina (Fig. 3B). These results, consistent with other reports, indicate that there is cell type-specific modulation of promoter activity and/or at-RA sensitivity (10, 15, 26, 46).
Orphan receptor TLX potentiates at-RA induction of RARβ2 promoter.
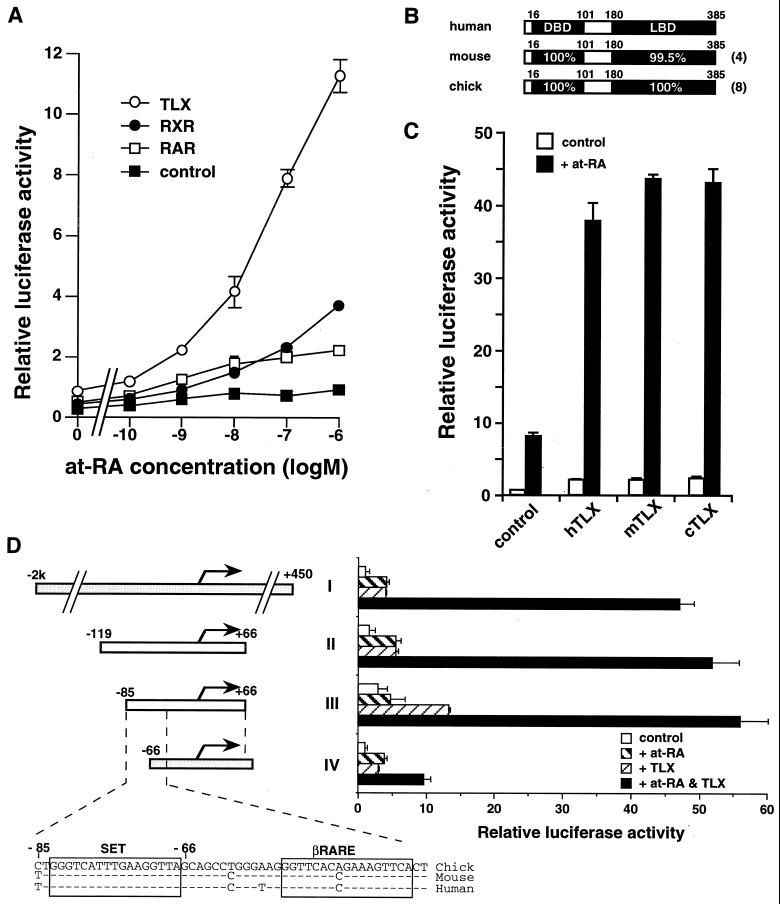
To investigate whether the above cell type specificity is a result of the amount of RARs and/or RXRs expressed in the cells, we tested the effect of overexpression of RARs or RXRs in CV-1 cells. RARα or RXRα only weakly potentiated at-RA induction of the RARβ2 promoter (Fig. 4A). RARβ, RARγ, RXRβ, RXRγ, or combinations of RARs and RXRs were also unable to significantly potentiate at-RA induction (data not shown). We examined the effect of various orphan nuclear receptors and found that TLX, which is expressed in retina, fore- and midbrain, and nasal epithelium, potentiated at-RA response in CV-1 cells (Fig. 4A). This effect was observed using chick, mouse, and human TLX constructs on both chick and mouse RARβ2 promoter reporter constructs, indicating that conservation of TLX-mediated at-RA signaling has occurred through evolution (Fig. 4B to D). These results, along with the fact that primary chick retina cells but not CV-1 or MC3T3-E1 cells (R. T. Yu, unpublished results) express high endogenous levels of TLX, suggested the involvement of TLX in activation of the RARβ2 promoter in retina cells.
FIG. 4.
TLX induces RA response to RARβ2 promoter activity in CV-1 cells. (A) cRARβ2 promoter-driven luciferase reporter plasmids were transfected as described for Fig. 3B with receptor plasmids. TLX enhances at-RA induced promoter activity better than does RAR or RXR. (B) Highly conserved structure of TLX proteins. Differences in amino acids compared to those for human TLX are indicated in parentheses. (C) The sensitizing effect of TLX is conserved during evolution. cRARβ2 promoter-driven luciferase reporter plasmids were transfected as described for Fig. 4A. Enhancement of at-RA-induced promoter activity is seen with all TLX constructs. (D) Identification of TLX-responsive element on RARβ2 promoter region. Numbering corresponds to nucleotide positions of the chick RARβ2 promoter sequence given for Fig. 2. Sequential deletion constructs were examined for at-RA-dependent transactivation with or without the presence of TLX. Chick TLX and mouse sequence (for I and IV) or chick sequence (for II and III) was used. Chick, mouse, and human RARβ2 promoter regions are aligned. Dashes indicate homology with chick sequence.
Identification of a TLX-responsive element.
To investigate the mechanism by which TLX modulates RA activation of the RARβ2 promoter, we made a series of deletion constructs. In chick retina cells, these constructs were equally induced by at-RA, thus posing difficulty for identification of the TLX-responsive element using this system. CV-1 cells, on the other hand, lack endogenous TLX and thus served as a good system for this analysis. Transient transfection of the above deletion constructs revealed that fragments as short as −85 relative to the transcription start site were sufficient and that the region between −85 and −66 was necessary to observe the TLX effect on at-RA response (Fig. 4D). Examination of this region revealed the presence of a putative RARE. Based on its function as described below, this element is referred to as the silencing element relieved by TLX (SET) and is conserved between mammalian and avian RARβ2 promoters (Fig. 4D and 5A) (10, 11, 47). To test the at-RA and TLX responsiveness of SET, we subcloned each element into a luciferase reporter driven by a tk minimal promoter. Transient transfection analysis revealed that SET does not function as a RARE in chick retina cells that express a high endogenous level of TLX (Fig. 5B). Consistent with the transfection results, gel retardation assays revealed that RAR-RXR heterodimers cannot bind to SET (Fig. 5C). TLX binds to neither SET nor βRARE (Fig. 5D) and does not appear to affect the binding activity of RAR-RXR to a composite βRARE-SET (Fig. 5E). Likewise, COUP-TFII, another orphan nuclear receptor shown to be important for the at-RA dependent activation of the RARβ2 promoter (26), did not affect the binding of RAR-RXR to βRARE-SET (Fig. 5E).
FIG. 5.
SET does not function as a RARE. (A) Three DR-5-like elements were tested for RA responsiveness. Synthetic oligonucleotides with HindIII linkers were ligated in the tk-driven luciferase reporter plasmid. (B) Only authentic βRAREs in the RARβ2 promoter region confer the effect of at-RA induction. (C) DNA binding assays. RAR-RXR heterodimers do not bind to SET. RAR-RXR bound to labeled βRARE probe is blocked by competition from the addition of excess βRARE unlabeled probes (lanes 2 to 4) but not by SET or nonspecific (NS) probes (lanes 5 to 10). Competitor probes are indicated above, with 2-fold excess added in lanes 2, 5, and 8; 20-fold excess in lanes 3, 6, and 9; and 200-fold excess in lanes 4, 7, and 10. (D) TLX does not bind directly to SET. TLX bound to labeled Krüppel probe is blocked by competition from the addition of excess Krüppel unlabeled probes (lanes 2 to 4) but not by βRARE or SET probes (lanes 5 to 10). Competitor probes are indicated above, with 4-fold excess added in lanes 2, 5, and 8; 20-fold excess in lanes 3, 6, and 9; and 40-fold excess in lanes 4, 7, and 10. (E) TLX does not affect binding of RAR-RXR to SET-βRARE. RAR-RXR bound to labeled SET-βRARE probe (lane 2) is not affected by the addition of TLX (lane 3) or COUP-TF (lane 4). −, absence of substance; +, presence of substance.
Reconstitution of a TLX-sensitive promoter with SET and βRARE.
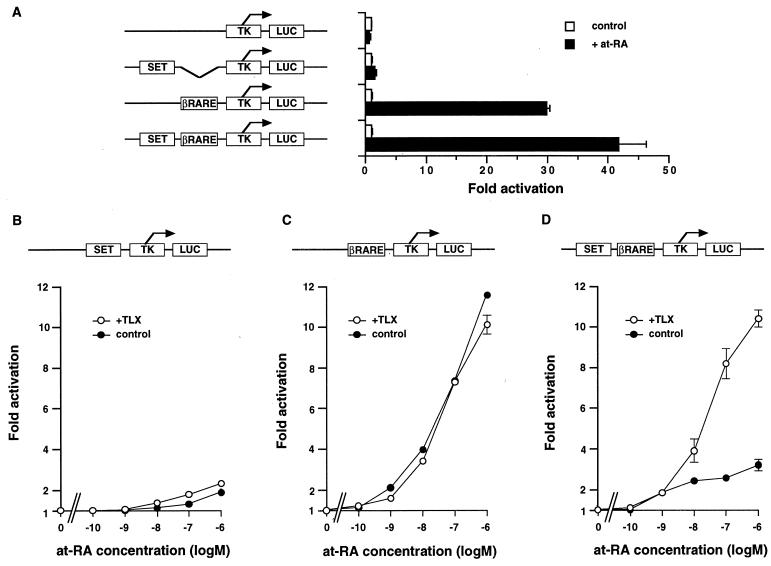
Similar to the situation observed with the deletion constructs, tk-luciferase constructs containing βRARE with or without SET showed the same level of induction by at-RA in chick retina cells (Fig. 6A). Using CV-1 cells, we could examine the effect of TLX more clearly. SET does not function as a RARE in CV-1 cells, and separately, neither SET nor βRARE confers an effect in response to TLX in CV-1 cells (Fig. 6B and C). However, when SET and βRARE are introduced together in a tk-luciferase reporter, the at-RA induction conferred by βRARE is suppressed (Fig. 6C and D) and expression of TLX relieves this silencing effect (Fig. 6C and D). This effect of SET on βRARE appears position independent; similar results were observed when the relative positions of SET and βRARE were flipped (Fig. 7B). The ability of TLX to restore the promoter activity of SET-βRARE to the same level obtained with βRARE alone but no further and the inability of TLX to directly bind either SET or βRARE suggest that SET binds a putative repressor which can be sequestered by TLX.
FIG. 6.
Reconstitution of TLX responsiveness of the RARβ2 promoter. (A) tk-luciferase reporter constructs with synthetic oligonucleotides depicted in Fig. 5A were transfected into chick retina cells. SET does not have any effect in retina cells. at-RA was added to a final concentration of 1 μM. (B to D) The same plasmids were transfected into CV-1 cells. The introduction of SET and βRARE in the same construct restores the effect of at-RA- and TLX-dependent transactivation. Final concentrations of at-RA are indicated.
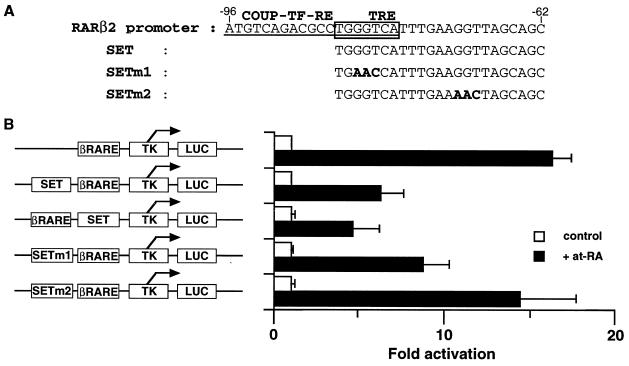
FIG. 7.
The silencing effect of SET is independent of that of TRE or COUP-TF-RE. (A) Sequences of mutated SET oligonucleotides are aligned with that of the RARβ2 promoter region. TRE, COUP-TF-RE (26), and mutated residues are boxed, underlined, and boldfaced, respectively. (B) tk-luciferase reporter constructs with synthetic oligonucleotides were transfected into CV-1 cells. The silencing effect of SET is position independent. Introducing mutations in the SET region outside TRE or COUP-TF-RE resulted in loss of the suppression effect of SET. at-RA was added to a final concentration of 1 μM.
The silencing effect of SET is independent of that of TRE or COUP-TF-RE.
To see if the silencing effect of SET is related to TRE or COUP-TF-RE (26), we introduced mutations in the region of SET overlapping with or outside those elements (Fig. 2 and 7). Mutations introduced within the overlapping region retain the silencing effect to some extent, making interpretation difficult. However, mutations introduced outside the overlapping region clearly resulted in loss of the suppression effect of SET in CV-1 cells (Fig. 7), showing that elements in the SET region independent of TRE or COUP-TF-RE are indispensable for the SET silencing effect.
DISCUSSION
Evolutionary conservation of promoter region and splicing variations of RARβ2.
Autoregulation of RARβ transcripts by at-RA has been reported in human, mouse, and chick (11, 39, 45, 51, 54). We demonstrated that in chick the RARβ2 promoter is responsible for this autoregulation and that RAR(s), not RXR(s), appears to be the primary target of RA. Comparison of the chick RARβ2 promoter region with that of human and mouse revealed a strong overall conservation of structural organization, 90% in the 150-bp region surrounding the transcription start site. As for mouse, chick embryonic retina cells also express several isoforms of RARβ mRNAs. Some of the short uORFs (uORF2 to -5) which are hypothesized to be important for regulating translation of the transcripts (42), as well as the CTG start codon proposed for mouse RARβ4, are conserved in chick (Fig. 2) (37). Together with the report that the RARβ4 is functionally tumorigenic compared to the β2 isoform (3), our results suggest a conserved indispensable role for the autologous regulation and splicing variation of the RARβ gene.
Cell type-specific regulation of RARβ2 promoter by TLX.
To date much effort has been spent to elucidate the underlying mechanism of RARβ2 promoter activation by RA (4, 7, 9, 13, 34). Proteins, such as E1A and COUP-TFs, have been shown to be involved in the cell type-specific regulation of RARβ2 promoter activity (15, 26, 46). Here we show that the orphan nuclear receptor TLX acts as a cell type-specific regulator for RARβ2 promoter activity. We identified a TLX-responsive element, SET, as a cell type-specific enhancer by deletion analysis of the RARβ2 promoter. SET acts as a cell type-specific repressor in fibroblasts, and TLX is able to relieve this silencing effect of SET.
TLX is a member of the nuclear receptor subfamily II, which consists of TLX, photoreceptor-specific nuclear receptor (PNR), COUP-TFs, RXRs, hepatocyte nuclear factor 4 (HNF-4), and TR2 (6, 14, 21). We have shown that TLX can downregulate Pax2 expression by direct binding to its promoter region (56). COUP-TFs are also known to repress the transcription of various genes through the COUP-TF binding element (52), and overexpression of COUP-TFs in CV-1 cells can repress RARβ2 promoter activity (M. Kobayashi and K. Umesono, unpublished results).
However, the expression level of COUP-TFs has been reported to be positively correlated with the at-RA responsiveness of the RARβ2 promoter in human cancer cell lines (26). COUP-TFs have also been reported to act as auxiliary cofactors for HNF-4 in the activation of the liver-specific HNF-1 promoter (23). COUP-TFs do not directly bind to the promoter but influence promoter activity through protein-protein interaction with HNF-4. TLX may similarly interact with a repressor protein that binds to SET and through this interaction relieve its repression. It is also possible that TLX and the SET binding protein may compete for a cofactor. The intact structure of TLX appears to be required, as overexpression of the TLX DNA binding domain or ligand binding domain alone does not potentiate activation (data not shown). Overexpression of the TLX DNA binding domain fused to the VP16 activation domain or engrailed repressor domain (56) likewise did not potentiate activation (data not shown). These results suggest that potentiation likely does not occur through the direct DNA binding activity of TLX. COUP-TFs as well as RAR-RXR heterodimers have been reported to enhance the activation of the estrogen receptor (ER) promoter together with ER upon the addition of estrogen (25). This interaction of COUP-TF and ER on the ER promoter resembles to some extent the relationship between TLX and RAR on the RARβ2 promoter.
Together, these observations appear to suggest that enhancement or repression by TLX and/or COUP-TFs is highly dependent on other factors specific to individual cell lines and/or each promoter context. Given that we were able to reconstruct TLX-responsive ability with SET and βRARE in the context of the tk promoter, our finding provides an important step towards revealing some of the mechanisms behind transcriptional regulation by cell-specific factors.
To further explore the transcriptional mechanisms involving TLX, the identification of its interacting protein is necessary. Recently, Lin et al. found that at-RA-dependent RARβ2 transcription requires direct binding of COUP-TFs to the element overlapping SET on the RARβ2 promoter and that COUP-TFs enhance the interaction of RAR with its coactivator, CREB-binding protein (Fig. 2) (26). However, we found that introducing mutations in the SET region outside the COUP-TF-RE resulted in loss of the suppression effect of SET in CV-1 cells (Fig. 7). Therefore, it seems that the effect of SET that we observed occurs through distinct mechanisms.
Based on analogy with other nuclear receptors, corepressor proteins, such as SMRT and Nco-R, are good candidates for TLX-interacting protein (reviewed in reference 30). Whether the effect of SET and TLX might also be applicable to different REs for other RXR heterodimers is an intriguing future question too, and such studies may reveal other aspects of TLX function.
The function of TLX and RARβ in eye development.
The importance of TLX function in vertebrate eye development has been implicated by gene knockout experiments in mice and the introduction of dominant-negative and dominant-active TLX constructs in chick and Xenopus (17, 56). These observations suggest that TLX is involved in the signaling pathways regulating various steps in retina and optic nerve formation and maintenance. Northern and in situ hybridization analyses of early chick embryos confirmed its restricted expression in fore- and midbrain and retina (57; Yu and Kobayashi, unpublished results). In this report, we showed that the signal transduction pathways controlled by RARβ can also be influenced by TLX. This finding is significant because although gene knockout and transgenic experiments in mice have suggested the importance of RARβ in eye development (16, 19, 20, 27), little is known about its upstream regulation.
Considering the high degree of structural and functional conservation of TLX and the RARβ2 promoters, it is reasonable to predict a fundamental role for TLX in the regulation of RAR in eye development. Our findings should provide a new contribution to better understanding of the complexity underlying the mechanisms of eye development.
ACKNOWLEDGMENTS
We thank H. Ohizumi, K. Nozaki, H. Otani, and H. Ono for technical assistance and members of the Yasuda Lab and Umesono Lab for valuable discussions during the study. We also thank Jochen Buck, Lonny R. Levin, Leonard P. Freedman, and David J. Mangelsdorf for critical reading of the manuscript and Richard A. Heyman and Yoshiko Ishimi for kindly providing synthetic retinoids and the MC3T3-E1 cell line, respectively.
This work was supported by Grants-in-Aid for Scientific Research from the Ministry of Education, Science, Sports, and Culture of Japan; the Research for the Future Program of the Japan Society for the Promotion of Science; and the Human Frontier Science Program.
Footnotes
This paper is dedicated to K. Umesono.
REFERENCES
- 1.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Balkan W, Colbert M, Bock C, Linney E. Transgenic indicator mice for studying activated retinoic acid receptors during development. Proc Natl Acad Sci USA. 1992;89:3347–3351. doi: 10.1073/pnas.89.8.3347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berard J, Gaboury L, Landers M, De Repentigny Y, Houle B, Kothary R, Bradley W E. Hyperplasia and tumours in lung, breast and other tissues in mice carrying a RAR β4-like transgene. EMBO J. 1994;13:5570–5580. doi: 10.1002/j.1460-2075.1994.tb06894.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bhattacharyya N, Dey A, Minucci S, Zimmer A, John S, Hager G, Ozato K. Retinoid-induced chromatin structure alterations in the retinoic acid receptor β2 promoter. Mol Cell Biol. 1997;17:6481–6490. doi: 10.1128/mcb.17.11.6481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chambon P. A decade of molecular biology of retinoic acid receptors. FASEB J. 1996;10:940–954. [PubMed] [Google Scholar]
- 6.Chen F, Figueroa D J, Marmorstein A D, Zhang Q, Petrukhin K, Caskey C T, Austin C P. Retina-specific nuclear receptor: a potential regulator of cellular retinaldehyde-binding protein expressed in retinal pigment epithelium and Muller glial cells. Proc Natl Acad Sci USA. 1999;96:15149–15154. doi: 10.1073/pnas.96.26.15149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen J Y, Clifford J, Zusi C, Starrett J, Tortolani D, Ostrowski J, Reczek P R, Chambon P, Gronemeyer H. Two distinct actions of retinoid-receptor ligands. Nature. 1996;382:819–822. doi: 10.1038/382819a0. [DOI] [PubMed] [Google Scholar]
- 8.Conlon R A. Retinoic acid and pattern formation in vertebrates. Trends Genet. 1995;11:314–319. doi: 10.1016/s0168-9525(00)89089-7. [DOI] [PubMed] [Google Scholar]
- 9.Davis K D, Berrodin T J, Stelmach J E, Winkler J D, Lazar M A. Endogenous retinoid X receptors can function as hormone receptors in pituitary cells. Mol Cell Biol. 1994;14:7105–7110. doi: 10.1128/mcb.14.11.7105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Davis K D, Lazar M A. Induction of retinoic acid receptor-β by retinoic acid is cell specific. Endocrinology. 1993;132:1469–1474. doi: 10.1210/endo.132.4.8384988. [DOI] [PubMed] [Google Scholar]
- 11.de The H, Vivanco-Ruiz M M, Tiollais P, Stunnenberg H, Dejean A. Identification of a retinoic acid responsive element in the retinoic acid receptor β gene. Nature. 1990;343:177–180. doi: 10.1038/343177a0. [DOI] [PubMed] [Google Scholar]
- 12.Dey A, Minucci S, Ozato K. Ligand-dependent occupancy of the retinoic acid receptor β2 promoter in vivo. Mol Cell Biol. 1994;14:8191–8201. doi: 10.1128/mcb.14.12.8191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dilworth F J, Fromental-Ramain C, Remboutsika E, Benecke A, Chambon P. Ligand-dependent activation of transcription in vitro by retinoic acid receptor α/retinoid X receptor α heterodimers that mimics transactivation by retinoids in vivo. Proc Natl Acad Sci USA. 1999;96:1995–2000. doi: 10.1073/pnas.96.5.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Escriva H, Safi R, Hanni C, Langlois M C, Saumitou-Laprade P, Stehelin D, Capron A, Pierce R, Laudet V. Ligand binding was acquired during evolution of nuclear receptors. Proc Natl Acad Sci USA. 1997;94:6803–6808. doi: 10.1073/pnas.94.13.6803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Folkers G E, van der Burg B, van der Saag P T. Promoter architecture, cofactors, and orphan receptors contribute to cell-specific activation of the retinoic acid receptor β2 promoter. J Biol Chem. 1998;273:32200–32212. doi: 10.1074/jbc.273.48.32200. [DOI] [PubMed] [Google Scholar]
- 16.Ghyselinck N B, Dupe V, Dierich A, Messaddeq N, Garnier J M, Rochette-Egly C, Chambon P, Mark M. Role of the retinoic acid receptor β (RARβ) during mouse development. Int J Dev Biol. 1997;41:425–447. [PubMed] [Google Scholar]
- 17.Hollemann T, Bellefroid E, Pieler T. The Xenopus homologue of the Drosophila gene tailless has a function in early eye development. Development. 1998;125:2425–2432. doi: 10.1242/dev.125.13.2425. [DOI] [PubMed] [Google Scholar]
- 18.Jackson A, Panayiotidis P, Foroni L. The human homologue of the Drosophila tailless gene (TLX): characterization and mapping to a region of common deletion in human lymphoid leukemia on chromosome 6q21. Genomics. 1998;50:34–43. doi: 10.1006/geno.1998.5270. [DOI] [PubMed] [Google Scholar]
- 19.Kastner P, Mark M, Chambon P. Nonsteroid nuclear receptors: what are genetic studies telling us about their role in real life? Cell. 1995;83:859–869. doi: 10.1016/0092-8674(95)90202-3. [DOI] [PubMed] [Google Scholar]
- 20.Kastner P, Mark M, Ghyselinck N, Krezel W, Dupe V, Grondona J M, Chambon P. Genetic evidence that the retinoid signal is transduced by heterodimeric RXR/RAR functional units during mouse development. Development. 1997;124:313–326. doi: 10.1242/dev.124.2.313. [DOI] [PubMed] [Google Scholar]
- 21.Kobayashi M, Takezawa S, Hara K, Yu R T, Umesono Y, Agata K, Taniwaki M, Yasuda K, Umesono K. Identification of a photoreceptor cell-specific nuclear receptor. Proc Natl Acad Sci USA. 1999;96:4814–4819. doi: 10.1073/pnas.96.9.4814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Krust A, Kastner P, Petkovich M, Zelent A, Chambon P. A third human retinoic acid receptor, hRAR-γ. Proc Natl Acad Sci USA. 1989;86:5310–5314. doi: 10.1073/pnas.86.14.5310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ktistaki E, Talianidis I. Chicken ovalbumin upstream promoter transcription factors act as auxiliary cofactors for hepatocyte nuclear factor 4 and enhance hepatic gene expression. Mol Cell Biol. 1997;17:2790–2797. doi: 10.1128/mcb.17.5.2790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ladias J A, Karathanasis S K. Regulation of the apolipoprotein AI gene by ARP-1, a novel member of the steroid receptor superfamily. Science. 1991;251:561–565. doi: 10.1126/science.1899293. [DOI] [PubMed] [Google Scholar]
- 25.Lazennec G, Kern L, Valotaire Y, Salbert G. The nuclear orphan receptors COUP-TF and ARP-1 positively regulate the trout estrogen receptor gene through enhancing autoregulation. Mol Cell Biol. 1997;17:5053–5066. doi: 10.1128/mcb.17.9.5053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lin B, Chen G Q, Xiao D, Kolluri S K, Cao X, Su H, Zhang X K. Orphan receptor COUP-TF is required for induction of retinoic acid receptor β, growth inhibition, and apoptosis by retinoic acid in cancer cells. Mol Cell Biol. 2000;20:957–970. doi: 10.1128/mcb.20.3.957-970.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Luo J, Sucov H M, Bader J A, Evans R M, Giguere V. Compound mutants for retinoic acid receptor (RAR) beta and RAR alpha 1 reveal developmental functions for multiple RAR beta isoforms. Mech Dev. 1996;55:33–44. doi: 10.1016/0925-4773(95)00488-2. [DOI] [PubMed] [Google Scholar]
- 28.Mangelsdorf D J, Evans R M. The RXR heterodimers and orphan receptors. Cell. 1995;83:841–850. doi: 10.1016/0092-8674(95)90200-7. [DOI] [PubMed] [Google Scholar]
- 29.Mangelsdorf D J, Thummel C, Beato M, Herrlich P, Schütz G, Umesono K, Blumberg B, Kastner P, Mark M, Chambon P, Evans R M. The nuclear receptor superfamily: the second decade. Cell. 1995;83:835–839. doi: 10.1016/0092-8674(95)90199-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McKenna N J, Lanz R B, O'Malley B W. Nuclear receptor coregulators: cellular and molecular biology. Endocr Rev. 1999;20:321–344. doi: 10.1210/edrv.20.3.0366. [DOI] [PubMed] [Google Scholar]
- 31.Mendelsohn C, Ruberte E, LeMeur M, Morriss-Kay G, Chambon P. Developmental analysis of the retinoic acid-inducible RAR-β2 promoter in transgenic animals. Development. 1991;113:723–734. doi: 10.1242/dev.113.3.723. [DOI] [PubMed] [Google Scholar]
- 32.Michaille J J, Blanchet S, Kanzler B, Garnier J M, Dhouailly D. Characterization of cDNAs encoding the chick retinoic acid receptor γ2 and preferential distribution of retinoic acid receptor γ transcripts during chick skin development. Dev Dyn. 1994;201:334–343. doi: 10.1002/aja.1002010405. [DOI] [PubMed] [Google Scholar]
- 33.Michaille J J, Kanzler B, Blanchet S, Garnier J M, Dhouailly D. Characterization of cDNAs encoding two chick retinoic acid receptor α isoforms and distribution of retinoic acid receptor α, β and γ transcripts during chick skin development. Int J Dev Biol. 1995;39:587–596. [PubMed] [Google Scholar]
- 34.Minucci S, Horn V, Bhattacharyya N, Russanova V, Ogryzko V V, Gabriele L, Howard B H, Ozato K. A histone deacetylase inhibitor potentiates retinoid receptor action in embryonal carcinoma cells. Proc Natl Acad Sci USA. 1997;94:11295–11300. doi: 10.1073/pnas.94.21.11295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Monaghan A P, Bock D, Gass P, Schwäger A, Wolfer D P, Lipp H-P, Schütz G. Defective limbic system in mice lacking the tailless gene. Nature. 1997;390:515–517. doi: 10.1038/37364. [DOI] [PubMed] [Google Scholar]
- 36.Monaghan A P, Grau E, Bock D, Schutz G. The mouse homolog of the orphan nuclear receptor tailless is expressed in the developing forebrain. Development. 1995;121:839–853. doi: 10.1242/dev.121.3.839. [DOI] [PubMed] [Google Scholar]
- 37.Nagpal S, Zelent A, Chambon P. RAR-β4, a retinoic acid receptor isoform, is generated from RAR-β2 by alternative splicing and usage of a CUG initiator codon. Proc Natl Acad Sci USA. 1992;89:2718–2722. doi: 10.1073/pnas.89.7.2718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nohno T, Muto K, Noji S, Saito T, Taniguchi S. Isoforms of retinoic acid receptor β expressed in the chicken embryo. Biochim Biophys Acta. 1991;1089:273–275. doi: 10.1016/0167-4781(91)90024-g. [DOI] [PubMed] [Google Scholar]
- 39.Noji S, Nohno T, Koyama E, Muto K, Ohyama K, Aoki Y, Tamura K, Ohsugi K, Ide H, Taniguchi S, Saito T. Retinoic acid induces polarizing activity but is unlikely to be a morphogen in the chick limb bud. Nature. 1991;350:83–86. doi: 10.1038/350083a0. [DOI] [PubMed] [Google Scholar]
- 40.Okada T S, Yasuda K, Araki M, Eguchi G. Possible demonstration of multipotential nature of embryonic neural retina by clonal cell culture. Dev Biol. 1979;68:600–617. doi: 10.1016/0012-1606(79)90230-6. [DOI] [PubMed] [Google Scholar]
- 41.Reynolds K, Mezey E, Zimmer A. Activity of the β-retinoic acid receptor promoter in transgenic mice. Mech Dev. 1991;36:15–29. doi: 10.1016/0925-4773(91)90068-h. [DOI] [PubMed] [Google Scholar]
- 42.Reynolds K, Zimmer A M, Zimmer A. Regulation of RARβ2 mRNA expression: evidence for an inhibitory peptide encoded in the 5′-untranslated region. J Cell Biol. 1996;134:827–835. doi: 10.1083/jcb.134.4.827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rossant J, Zirngibl R, Cado D, Shago M, Giguere V. Expression of a retinoic acid response element-hsplacZ transgene defines specific domains of transcriptional activity during mouse embryogenesis. Genes Dev. 1991;5:1333–1344. doi: 10.1101/gad.5.8.1333. [DOI] [PubMed] [Google Scholar]
- 44.Rowe A, Richman J M, Brickell P M. Development of the spatial pattern of retinoic acid receptor-β transcripts in embryonic chick facial primordia. Development. 1992;114:805–813. doi: 10.1242/dev.114.3.805. [DOI] [PubMed] [Google Scholar]
- 45.Rowe A, Richman J M, Brickell P M. Retinoic acid treatment alters the distribution of retinoic acid receptor-β transcripts in the embryonic chick face. Development. 1991;111:1007–1016. doi: 10.1242/dev.111.4.1007. [DOI] [PubMed] [Google Scholar]
- 46.Sanguedolce M V, Leblanc B P, Betz J L, Stunnenberg H G. The promoter context is a decisive factor in establishing selective responsiveness to nuclear class II receptors. EMBO J. 1997;16:2861–2873. doi: 10.1093/emboj/16.10.2861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shen S, Kruyt F A E, Hertog J D, van der Saag P T. Mouse and human retinoic acid receptor β2 promoters: sequence comparison and localization of retinoic acid responsiveness. DNA Sequence. 1991;2:111–119. doi: 10.3109/10425179109039679. [DOI] [PubMed] [Google Scholar]
- 48.Smith S M. Retinoic acid receptor isoform β2 is an early marker for alimentary tract and central nervous system positional specification in the chicken. Dev Dyn. 1994;200:14–25. doi: 10.1002/aja.1002000103. [DOI] [PubMed] [Google Scholar]
- 49.Smith S M, Dickman E D, Power S C, Lancman J. Retinoids and their receptors in vertebrate embryogenesis. J Nutr. 1998;128:467S–470S. doi: 10.1093/jn/128.2.467S. [DOI] [PubMed] [Google Scholar]
- 50.Smith S M, Eichele G. Temporal and regional differences in the expression pattern of distinct retinoic acid receptor-β transcripts in the chick embryo. Development. 1991;111:245–252. doi: 10.1242/dev.111.1.245. [DOI] [PubMed] [Google Scholar]
- 51.Sucov H M, Murakami K K, Evans R M. Characterization of an autoregulated response element in the mouse retinoic acid receptor type β gene. Proc Natl Acad Sci USA. 1990;87:5392–5396. doi: 10.1073/pnas.87.14.5392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tsai S Y, Tsai M-J. Chick ovalbumin upstream promoter-transcription factors (COUP-TFs): coming of age. Endocr Rev. 1997;18:229–240. doi: 10.1210/edrv.18.2.0294. [DOI] [PubMed] [Google Scholar]
- 53.Umesono K, Murakami K K, Thompson C C, Evans R M. Direct repeats as selective response elements for the thyroid hormone, retinoic acid, and vitamin D3 receptors. Cell. 1991;65:1255–1266. doi: 10.1016/0092-8674(91)90020-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wanek N, Gardiner D M, Muneoka K, Bryant S V. Conversion by retinoic acid of anterior cells into ZPA cells in the chick wing bud. Nature. 1991;350:81–83. doi: 10.1038/350081a0. [DOI] [PubMed] [Google Scholar]
- 55.Yao T P, Forman B M, Jiang Z, Cherbas L, Chen J D, McKeown M, Cherbas P, Evans R M. Functional ecdysone receptor is the product of EcR and Ultraspiracle genes. Nature. 1993;366:476–479. doi: 10.1038/366476a0. [DOI] [PubMed] [Google Scholar]
- 56.Yu R T, Chiang M Y, Tanabe T, Kobayashi M, Yasuda K, Evans R M, Umesono K. The orphan nuclear receptor Tlx regulates Pax2 and is essential for vision. Proc Natl Acad Sci USA. 2000;97:2621–2625. doi: 10.1073/pnas.050566897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yu R T, McKeown M, Evans R M, Umesono K. Relationship between Drosophila gap gene tailless and a vertebrate nuclear receptor Tlx. Nature. 1994;370:375–379. doi: 10.1038/370375a0. [DOI] [PubMed] [Google Scholar]
- 58.Zelent A, Krust A, Petkovich M, Kastner P, Chambon P. Cloning of murine α and β retinoic acid receptors and a novel receptor γ predominantly expressed in skin. Nature. 1989;339:714–717. doi: 10.1038/339714a0. [DOI] [PubMed] [Google Scholar]
- 59.Zelent A, Mendelsohn C, Kastner P, Krust A, Garnier J M, Ruffenach F, Leroy P, Chambon P. Differentially expressed isoforms of the mouse retinoic acid receptor β are generated by usage of two promoters and alternative splicing. EMBO J. 1991;10:71–81. doi: 10.1002/j.1460-2075.1991.tb07922.x. [DOI] [PMC free article] [PubMed] [Google Scholar]