Abstract
Therapeutic implementation of human limb regeneration is a daring aim. Studying species that can regrow their lost appendages provides clues on how such a feat can be achieved in mammals. One of the unique features of regeneration-competent species lies in their ability to seal the amputation plane with a scar-free wound epithelium. Subsequently, this wound epithelium advances and becomes a specialized wound epidermis (WE) which is hypothesized to be the essential component of regenerative success. Recently, the WE and specialized WE terminologies have been used interchangeably. However, these tissues were historically separated, and contemporary limb regeneration studies have provided critical new information which allows us to distinguish them. Here, I will summarize tissue-level observations and recently identified cell types of WE and their specialized forms in different regeneration models.
Keywords: wound epidermis, specialized wound epidermis, AEC, single-cell transcriptomics, appendage regeneration, limb regeneration
Introduction
Limb regeneration is associated with two heterogeneous tissue types: a specialized wound epithelium that caps the amputation plane, and a blastema which forms underneath the specialized wound epidermis (WE) and contains lineage-restricted stem and progenitor cells that will give rise to the new appendage (Beck et al., 2009; Aztekin, 2021). The interaction between these two tissues leads to the outgrowth of the lost structure. Studies with newts, which can consistently perform limb regeneration throughout their lifespan, provided landmark findings governing the function of these tissues (Joven et al., 2019). Subsequent reports aimed to characterize and identify counterparts in other regeneration models.
The critical role of the WE for regenerative success stems from the observations dating back to the early 1900s (Morosow, 1938; Thornton, 1960). Mainly, newt limb amputations were found to progress with a rapid epithelial migration resulting in the closure of the amputation plane (Lash, 1955; Hay and Fischman, 1961; Repesh and Oberpriller, 1978), forming the WE. Afterward, the WE progresses into its specialized morphologically thickened epithelial form, also known as the apical epithelial cap (AEC). Repeated salamander AEC removal or blocking AEC formation halts the limb regeneration program, highlighting its essential role for limb regeneration (Morosow, 1938; Goss, 1956; Thornton, 1957, 1958; Mescher, 1976; Tassava and Garling, 1979; Tsai et al., 2020). Conversely, grafting the salamander AEC can induce ectopic limb outgrowths, with cartilage and dermal composition (Thornton, 1960; Thornton and Thornton, 1965). Owing to these features, the derivation of the AEC for grafting or identifying genes associated with the AEC holds potential for therapeutic applications. However, before delving into the properties of the WE and the AEC, it is crucial to define them clearly.
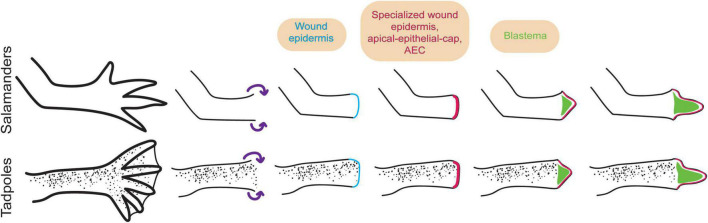
The term “AEC” was initially used to identify an epithelium covering aggregated blastema cells during limb regeneration. Nonetheless, this definition could be used for both the WE and the AEC. Moreover, investigations on different non-limb regeneration scenarios (e.g., tail regeneration) sought to identify similar tissues. However, due to unclear definitions of these tissues, the WE and the AEC terminologies were used interchangeably. To distinguish them and clarify their differences, Christensen and Tassava (2000) suggested a revision to these terminologies. Briefly, the WE is the epithelium covering the amputation plane (right after amputations), remains on the amputation plane for a short period, and has a simple morphology consisting of one to three layers of cells (Figure 1). Meanwhile, its specialized form, the AEC, appears after the WE formation. The AEC remains until proximal-distal elongation (until digit formation), and has a thickened structure of approximately 10–15 layers of cells (Figure 1). On this basis, two features were proposed to distinguish them: (1) the period coinciding with their presence during regeneration and (2) tissue morphology. Although these definitions are helpful, our current methodologies evidenced that tissue or cellular morphology could be inadequate assessments for functionally distinct populations and cell types. Moreover, the different cell types composing and identifying the WE or the AEC were not resolved. Instead, investigating their counterparts in other species or regeneration paradigms brought additional misperception to these concepts.
FIGURE 1.
The wound epidermis (WE) and the specialized wound epidermis form in a step-wise manner during amphibian limb regeneration. Limb regeneration is initiated with amputation in (top) salamanders and (bottom) tadpoles. The remaining stump epidermal cells migrate to the amputation plane (purple arrows) and form the WE (light blue). Then the WE becomes the specialized wound epidermis [apical-epithelial-cap (AEC)] (dark pink) associated with regeneration (Christensen and Tassava, 2000). Afterward, the AEC leads to blastema formation and subsequent outgrowth (Tassava and Loyd, 1977). Although the presence of the AEC in early and mid-stages of regeneration is demonstrated by tissue morphology and staining assessments (e.g., Han et al., 2001), its presence in late stages is observed only with tissue morphology assessment.
In this review, I aim to bridge historical tissue level observations of the WE and the AEC with recent single-cell transcriptomics and other cell-type focused findings, primarily in the context of vertebrate limb regeneration. Due to the interchangeable documentation of these two concepts, I will focus on the properties of the AEC and mark potential phenotypes distinguishing it from the WE. Finally, I will compare the cross-species and regeneration-model properties of the WE and the AEC.
What Does the Apical-Epithelial-Cap, Do?
The salamander AEC has been associated with multiple critical roles for successful regeneration, from influencing extracellular-matrix (ECM) organization to providing mitogenic factors. Among these undertakings, one of the early suggested AEC functions governed the question: does the blastema influence the AEC formation or vice versa? Work targeting this question proposed that the injury caused by the amputation induces morphologically identified dedifferentiated cells (Tassava and Loyd, 1977). These cells can proliferate and form a blastema in the presence of the AEC. However, without the AEC, the blastema does not form, although dedifferentiated cells can be observed. Hence, the AEC has been suggested to maintain and further instruct injury-induced dedifferentiated cells to form a blastema and patterning for regeneration. Due to the observation that injury-induced dedifferentiated cells can form without the AEC, it has been hypothesized that the lack of a specialized WE results in regeneration-incompetency in higher vertebrates (Tassava and Olsen, 1982).
In subsequent years, the AEC has been further associated with multiple essential cellular mechanisms for regenerative success. The majority of functional suggestions on the role of the AEC were concluded based on staining approaches and observed gene expression patterns. For example, the salamander AEC expresses ECM associated fn1 (Christensen and Tassava, 2000), collagen type XII (Wei et al., 1995), collagen type IV, lamb1 (Del Rio-Tsonis et al., 1992), krt5, krt17 (Moriyasu et al., 2012), and frem2 (Leigh et al., 2018); histolysis involved mmp3/10b and mmp9 (Yang et al., 1999; Vinarsky et al., 2005); mitogenic factors such as fgf1 (Boilly et al., 1991), fgf2 (Mullen et al., 1996), fgf8 (Han et al., 2001; Christensen et al., 2002), fgf10 (Christensen et al., 2002), mdk (Tsai et al., 2020), and MARCKS-like (Sugiura et al., 2016); angiogenesis-related tsp1 (Whited et al., 2011); and chemotactic factors as wnt5a (Ghosh et al., 2008). The existence of multiple AEC derived factors could either enable functional redundancy or allow for cell-type-specific interpretations. For example, some of these factors would influence muscle and some others would influence connective tissue lineages. Furthermore, several transcription factors are associated with the salamander AEC: tp63 (Kawakami et al., 2006), msx2 (Carlson et al., 1998), dlx3 (Beauchemin and Savard, 1992), id2, id3, hes1 (Shimizu-Nishikawa et al., 1999), and sp9 (Satoh et al., 2008), although further work is required to reveal their molecular role during AEC establishment or maintenance. The function of the AEC and its formation have been investigated by systemic perturbations rather than tissue or cell type-specific functional assessments. As an example, the AEC is documented to express various metalloproteinases (MMP) associated with histolysis that degrade ECM components critical for regenerative success, and MMP inhibitors can inhibit limb regeneration (Vinarsky et al., 2005). Nonetheless, it remains unclear if and how much the AEC-derived MMPs result in histolysis that could be required for regeneration. Bulk sequencing approaches to the whole AEC tissue were employed and expanded potential genes related to the AEC and identified new functionally critical gene targets for regeneration, such as mdk (Monaghan et al., 2012; Knapp et al., 2013; Tsai et al., 2019, 2020). Due to methodological limitations, bulk approaches alone could not discriminate which critical cell types within the AEC tissue are responsible for these gene expressions or if the whole AEC tissue expresses them. Overall, as the AEC has been primarily associated with secreted ligands, it is largely regarded as a signaling center orchestrating cellular mechanisms that are vital for successful regeneration.
Is the Apical Epithelial Cap Formation Re-Deployment of the Limb Development Associated Apical-Ectodermal Ridge?
One of the main questions in regeneration biology involves revealing similarities between regeneration and development. From this perspective, the AEC has been associated with the apical ectodermal ridge (AER), which is well-established transient tissue essential for chicken and mouse limb development (Fernandez-Teran and Ros, 2008). Like the AEC, the AER also represents a heterogeneous tissue forming at the distal tips of limb buds; the AER is also covered with a periderm and marked with fgf8 at the basal layers (Nakamura and Yasuda, 1979; Aztekin et al., 2021). Moreover, multiple classical AER genes (e.g., fgf2, fgf8, sp9, and fn1) are also expressed in the AEC. The absence of the chicken AER or mutations impairing the mouse AER results in no limb development (Saunders, 1948; Fernandez-Teran and Ros, 2008), and the lack of the salamander AEC results in no limb regeneration (Goss, 1956; Thornton, 1957, 1958; Mescher, 1976; Tassava and Garling, 1979). Akin to the amphibian AEC, the amniotic AER was suggested to maintain and enable the self-renewal of underlying limb progenitors (Tassava and Mescher, 1975; Mescher, 1976; Tassava and Loyd, 1977). Although not studied in detail, the presence of the AER was suggested to influence angiogenesis (Tschumi, 1957) and muscle-cell migration for limb development (Gumpel-Pinot et al., 1984; Francis-West et al., 2003). Moreover, the AER was proposed to influence specific limb progenitor cells to express early limb progenitor markers (Watanabe et al., 1993; Mariani et al., 2008), highlighting AER-derived signals may influence progenitor cell states, although this feature has been debated (Pickering et al., 2018; McQueen and Towers, 2020).
Both the AER and the AEC were suggested to be largely mitotically inactive populations (Moriyasu et al., 2012; Storer et al., 2013; Aztekin et al., 2021), and their formation requires the activity of the well-studied signaling pathways (e.g., FGF, BMP, and WNT; Fernandez-Teran and Ros, 2008). Notably, both the AEC and the AER require signals, such as Fgf10, from their underlying mesodermal cells to form and maintain themselves (Ohuchi et al., 1997, 1999; Yokoyama et al., 2001). Ectopic application of FGF10, or B-catenin overexpression can induce both AEC and AER formation in different species (e.g., chicken and Xenopus; Yonei-Tamura et al., 1999; Kawakami et al., 2001, 2006; Yokoyama et al., 2001; McQueeney et al., 2002). Although they have many similarities, the AEC and the AER could have distinct transcriptomic signatures making the AEC a novel population with new functions forming during regeneration. Moreover, revealing the similarity between the AER and the AEC also provides new insight for our understanding of human limb regeneration. As the AER forms during human embryonic development (Kelley and Fallon, 1976), human epithelial cells may have the competency to form the AEC, and mammalian basal epidermal cells could be used to derive functional AER cells. Hence, it is critical to determine if the AEC represents a novel population or re-usage of developmental programs.
Despite all similarities, the AEC and the AER are argued to have some differences, and there are challenges to testing their equivalency. First, morphological features of the AER and the AEC were discussed as potential differences (Campbell and Crews, 2008). While the AER can be found as a stratified ridge structure in mice and chicken (Fernandez-Teran and Ros, 2008), the AEC is mainly associated with a flat epidermal tissue (Campbell and Crews, 2008; Satoh et al., 2008). Moreover, unlike the AER, the AEC is observed to be a 10- to 15-cell-thick tissue (Campbell and Crews, 2008). Hence, variations in tissue morphology do not provide a new understanding of the functional differences between these two tissues. Second, the lineage relations and cellular mechanisms enabling the formation of the AER and the AEC are still debated. Both the AER and the AEC formation have been suggested to be mediated through a boundary model where dorsal and ventral epidermal populations interact to give rise to these tissues (Fernandez-Teran and Ros, 2008; Shimokawa et al., 2012). However, numerous phenotypes cannot be explained via the boundary model for the mouse AER (discussed in Fernandez-Teran and Ros, 2008). Moreover, it remains unclear which specific cellular and molecular mechanisms and lineage-relations guide AER or AEC formation. Although several secreted factors are identified to be critical and sufficient for AER formation from specific ectodermal populations (e.g., Wnt pathway activity, Fgf10, and Fgf7; Ohuchi et al., 1997; Yonei-Tamura et al., 1999; Yokoyama et al., 2001; McQueeney et al., 2002), it remains unclear how AER cell identity is established during development. Likewise, the molecular basis for AEC induction is currently not resolved. One possibility is that the amputation plane connective tissue lineage, the WE, or both may be secreting AER inducing signals to induce an AEC.
There are different models for AER disappearance during amniotic development (Scherz et al., 2004; Verheyden and Sun, 2008; Storer et al., 2013; Pickering et al., 2018), but how the AEC disappears during regeneration remains unknown. Salamander and froglet AEC formations have been suggested to be a nerve-dependent process (Suzuki et al., 2005; Satoh et al., 2008; Stocum, 2019), while nerves are not required for the Xenopus tadpole limb regeneration (Cannata et al., 1992). Furthermore, limb explants (that are presumably devoid of nerves) can still form the AEC (Aztekin et al., 2021). Meanwhile, the nerve dependence of the AER in amniotes is not suggested. Third, critically, the AER is separated from the underlying mesoderm by a basement membrane (Saunders, 1948) whereas, the newly formed AEC and underlying cells are in direct contact (Repesh and Oberpriller, 1978; Neufeld et al., 1996), presumably enabling easier interaction between these tissues. Fourth, the AEC is mostly characterized in salamanders which are suggested not to have an AER (Tank et al., 1977; Stopper and Wagner, 2005; Purushothaman et al., 2019), and the AER is mostly characterized in amniotes, which are suggested not to have the AEC. Hence, although the AER and the AEC share many features, their detailed characterization was not done in the same animal. Nonetheless, other limb regeneration-competent species could overcome some of these issues and may enable the determination of possible similarities between the AER and the AEC.
Xenopus laevis is the only commonly used laboratory animal with a well-established AER and AEC (Stopper and Wagner, 2005; Purushothaman et al., 2019). Using single-cell transcriptomics, we revealed individual cell types defining these tissues and compared their similarities at the single-cell transcriptome level (Aztekin et al., 2021). Instead of the AER and the AEC harboring different populations, individual cells found at the basal layers of these tissues share a similar transcriptome and appear in the same computational cluster. Nevertheless, the AEC has a higher potential as a signaling center, given that the same ligands show increased expression in the AEC compared to the AER. For example, although fgf8 or fn1 are expressed in the AER, their expression is significantly higher in the AEC, presumably creating a niche with very high levels of total ligands compared to the limb development environment. Further assessments on basal fgf8 + epidermal cells during regeneration and development revealed a shared single-cell morphology, spatial organization, and common regulatory cellular mechanisms. During development and regeneration, basal epidermal cells in the AER and the AEC are seen to form from a stepwise activation of lgr5 and then fgf8 in basal epidermal cells in a spatially restricted manner, from proximal to the distal tip of a limb. However, the molecular mechanism enabling this differentiation remains unclear. Due to this very high degree of similarity, we proposed using the term “AER cells” for cells in the basal epidermal fgf8 + cells formed during limb development and regeneration. On this basis, the AEC tissue contains cells harboring the AER cell transcriptional program at its basal epidermal layer. Although a functional equivalence assay will be essential, single-cell comparison and current functional assays represent a stringent evaluation supporting the hypothesis that the AEC is re-deployment of the AER, at least in Xenopus.
Re-Assessing Morphological and Gene Expression Markers of the Apical Epithelial Cap
The salamander WE and the AEC were historically distinguished mainly based on their tissue morphology. Notably, the thickened epithelium was used to detect the AEC. Initial attempts aimed at identifying the AEC used antigen stainings (Tassava et al., 1986, 1993; Goldhamer et al., 1989; Tassava and Acton, 1989; Castilla and Tassava, 1992). These staining approaches also revealed that such antigens were enriched in secretory cells in the animal body, suggesting that the AEC might have a secretory cell phenotype (Goldhamer et al., 1989). The use of mRNA staining, particularly fn1 (Christensen and Tassava, 2000) and fgf8 (Han et al., 2001), suggested that the AEC could be a heterogeneous tissue and proposed that the basal layers may be the critical population for regeneration. AEC-associated genes are expressed at the basal layers of the AEC tissue 2–3 days after amputations but not necessarily in the middle or apical layers. These gene expressions are not found directly after amputations. Hence, cells defining the AEC are not identical to those in the WE, although there could be shared gene expressions [e.g., MARCKS-like (Sugiura et al., 2016) and tsp1 (Whited et al., 2011)] and these two tissues may have lineage relations. Moreover, as the AEC-associated genes are mainly observed at the basal layers, these results were also suggesting thickened epithelial-morphology may not be providing AEC-associated regeneration promoting functions. Overall, not all cell types and transcriptional programs are identical within the AEC itself or between the AEC and the WE.
Marker genes distinguishing the AEC from the WE have been revealed. Particularly, fgf8 has been associated with the salamander limb AEC, but not the WE (Han et al., 2001; Christensen et al., 2002). However, recent years provided inconsistent results for fgf8 expression in axolotl limb regeneration. Initial characterizations suggested fgf8 marks a population in the basal layers of the AEC (Han et al., 2001; Christensen et al., 2002). In the accessory limb model where a secondary limb is induced from an upper arm via specific experimental perturbation, fgf8 expression was not found (Nacu et al., 2016). Later work suggested that fgf8 expression is not restricted to the basal level layers (Vincent et al., 2020). Although the validity of fgf8 to detect the axolotl AEC presents certain caveats, new potential AEC markers were shown to label basal layers of the AEC [e.g., mdk (Tsai et al., 2020), wnt5a (Ghosh et al., 2008)] and are not expressed in the WE, in the initial days postamputation. While mdk and wnt5a expression can be observed in other cell populations, this gene expression pattern in basal epidermal cells labels the AEC. Overall, it remains unclear if fgf8 is expressed during axolotl limb regeneration, but there are multiple alternative markers to detect the AEC in the salamanders.
Fgf8 expressing basal epidermal cells in the AEC have been characterized during X. laevis limb regeneration (Christen and Slack, 1997; Wang and Beck, 2014; Aztekin et al., 2021). Unlike salamanders, X. laevis can regrow their lost limb in early limb bud stages but lose this ability progressively during their development (Dent, 1962). When tadpole limbs are in their secondary growth, they can only regrow a limb with a few digits. Afterward, amputations produce one out of two phenotypes toward metamorphosis: either simple wound healing or spike formation. Simple wound-healing phenotype shows epidermal covering of the amputation plane and no growth, meanwhile, the spike formation involves the growth of an unpatterned cartilaginous rod that lacks muscles and bones (Satoh et al., 2005). During regeneration-competent stages, the Xenopus AEC has been reported to contain abundant fgf8 expressing cuboidal cells in basal layers (Pearl et al., 2008; Aztekin et al., 2021). Meanwhile, when tadpoles progressively lose their limb regeneration competency, they also produce fewer fgf8 + cells in the AEC, and when they are regeneration-incompetent, there are no fgf8 + cells at the amputation plane; although these animals can still seal their amputation plane, therefore can form the WE (Christen and Slack, 1997; Aztekin et al., 2021). Unlike regeneration-incompetent tadpoles, amputated postmetamorphic froglets homogenously perform spike formation but not simple wound healing (Beck et al., 2009). There are different results for where and when the fgf8 is expressed for postmetamorphic Xenopus froglets that can consistently regrow a spike but not perform simple wound healing, for where and when the fgf8 is expressed (Endo et al., 2000; Suzuki et al., 2005; Satoh et al., 2017). Moreover, it remains unclear if the formed froglet fgf8 population is equivalent to ones in regeneration-competent limbs.
Single-Cell Studies Reveal Cell Types Defining Limb-Specific Apical Epithelial Cap
With recent advancements in cell-centric approaches, we now have an opportunity to characterize specific cell types within the WE and the AEC tissues. Although definitions of a cell type or its transient forms as a cell state have been debated (Trapnell, 2015; Arendt et al., 2016; McKinley et al., 2020), single-cell RNA sequencing (scRNA-Seq) approaches reveal transcriptome profiles for individual cells that can suggest specific functions. Moreover, with scRNA-Seq, we can now obtain more extensive gene expression profiles, expanding and clarifying the AEC-associated gene lists. Cells with highly similar transcriptomes form computationally assigned clusters, representing potential cell types or states. As in every computational method, analysis can yield artificial scenarios. Over/underclustering or analysis with low-quality or non-comprehensive datasets can suggest cell types or states that are not necessarily reflecting functionally distinct populations. Although cell types harbor stable gene regulatory networks associated with their identity and established functions, scRNA-Seq records the transcriptomes of a single time point, while cells are dynamic and change their transcriptome constantly. Moreover, scRNA-Seq can only capture transcriptional information meanwhile there could be posttranscriptional responses determining cellular functions. Therefore, it is critical to test the quality and validity of single-cell atlases, and more critically, validate the computational suggestions with experimental approaches. Secondly, contrary to traditional non-quantitative staining methods (e.g., antibody labeling, colorimetric in situ hybridization), single-cell transcriptomics and new mRNA staining approaches offer a more unbiased and quantitative methodology to discriminate lowly or highly expressed genes, which can again be an indication for specific cellular functions. These advancements allow us to identify and detect cell types in situ. On this basis, we can uncover cell types representing WE and AEC tissues and ask if they could be found in different species and regeneration paradigms.
From the initial studies in salamanders, it is well established that the salamander WE will form via the migration of remaining stump epidermal cells (Hay and Fischman, 1961). Hence, cells composing the WE can be expected to be already present before amputation in scRNA-Seq datasets as a basal epidermal cell cluster in homeostatic states of limbs. Meanwhile, based on the literature, the AEC could be reflected in these single-cell maps as a new cluster that forms during regeneration and expresses marker genes such as fn1, fgf8, wnt5a, and mdk, in addition to having a basal epidermal cell signature (e.g., tp63). Based on these criteria, the new advances in single-cell analysis provide novel insights into the cellular properties of the WE and the AEC tissues.
Several axolotl limb regeneration studies incorporated scRNA-Seq (Gerber et al., 2018; Leigh et al., 2018; Li et al., 2020; Qin et al., 2020; Rodgers et al., 2020), with some targeting the whole newly forming cells, including epidermal populations. In one of these studies, Leigh et al. detected basal epidermal clusters and validated frem2 as a new marker (Leigh et al., 2018). In this study, this frem2 expressing population was also found in the limb homeostatic state and was not suggested as an emerging population upon amputation. Qin et al. (2020) identified a separate basal epidermal population expressing known AEC markers (e.g., mdk and fn) and suggested they represent the AEC. Contrary to previous studies, this population is also seen before amputation. Hence, it remains unclear whether AEC cells are present in the axolotl before amputation or if alternatively, these studies identified a population representing a mix of basal epidermal cells due to the experimental design and/or analysis. Interestingly, these studies did not report a separate fgf8 + basal epidermal population. Further investigation on these datasets and validatory experiments will be required to pinpoint the WE and the AEC in the axolotl limb regeneration.
Single-cell investigation on Xenopus limb regeneration, focusing on the AEC, confirmed that the AEC is a heterogeneous tissue and that only the basal layers contain fgf8 + cells (Aztekin et al., 2021). Moreover, scRNA-Seq revealed that not all fgf8 + epidermal cells are equivalent. Intriguingly, the level of fgf8 expression correlates with the signaling center potential of basal epidermal cells; if fgf8 expression is high, the expression of other ligands (Fgf, Bmp, Wnt, Tgfb, and Delta) involved in major signaling pathways is also high. Moreover, Xenopus AER cells express lower levels of fgf8 in regeneration-restricted tadpoles (which can only grow back a foot with two to three digits) compared to regeneration-competent AER cells. Hence, not just the expression of fgf8, but its expression level is a new parameter to consider while determining the properties of the AEC and may indicate potentially different generated morphogen gradients generated for limb regeneration. From a technical point of view, these findings imply that caution is necessary when detecting the AEC. Methodologies that cannot reliably discriminate low and high gene expressions may not determine the signaling center potency of the formed AEC, which is indicative of how many ligands are expressed. Therefore, detection of basal epidermal fgf8 + cells relying more consistently on more user-unbiased and quantitative mRNA staining approaches [e.g., hybridization-chain-reaction (Choi et al., 2018), single amplification by exchange reaction fluorescence in situ hybridization (Kishi et al., 2019), scRNA-Seq] will be essential to study properties of the AEC. Moreover, some of the key genes detected in the salamander AEC are conserved for the Xenopus AEC (e.g., fgf8, fn1, mdk, and wnt5a). However, there are also some differences, such as, sp9 which is expressed in the salamander AEC, but its expression is not significantly detected in the Xenopus AEC (Aztekin et al., 2021). Further cellular comparisons between species can highlight conserved features of the AEC.
Divergent Molecular, Cellular, and Tissue-Level Properties of Wound Epidermis and Apical Epithelial Cap in Other Regeneration Models
The WE and the AEC have been long sought and investigated in other regeneration models. Currently, the epidermal population covering the amputation plane in different regeneration models is also named the WE or the AEC. However, due to different tissue formation kinetics in other species and regeneration paradigms, differences between two tissues are even less precisely defined than those in amphibians.
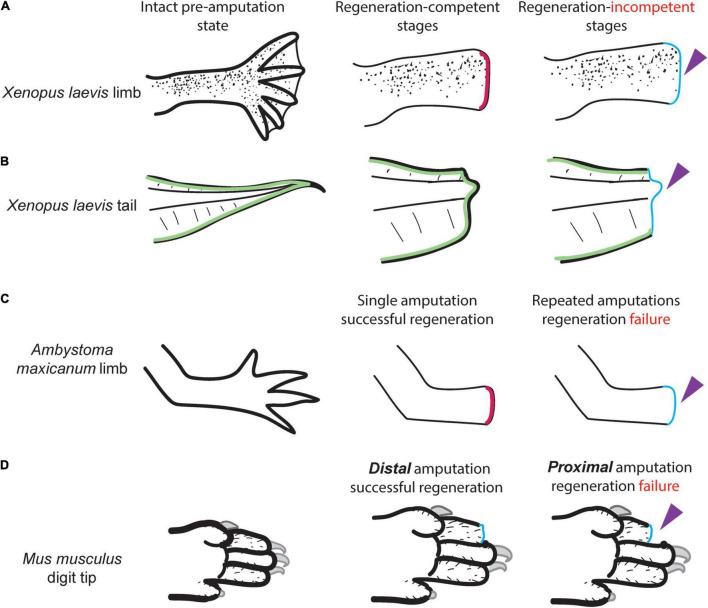
Recent research with regeneration models that show both regeneration-competency and -incompetency further stresses the need for re-evaluation to distinguish the WE and the AEC terminologies. In heavily studied appendage regeneration competent vertebrate species, the amputation plane is sealed with a simple epithelium that fits the WE definition. However, this ability can be observed even in regeneration-competent species that are exhibiting regeneration-incompetency. For example, limb amputation in regeneration-incompetent Xenopus tadpoles can result in simple wound healing and can still display the WE formation (Dent, 1962; Figure 2A). Likewise, X. laevis can regenerate its lost tail before metamorphosis except for a brief period during its development (Niewkoop & Faber Stage 46–47), referred to as the refractory period (Beck et al., 2003). During this period, amputated tails do not regenerate; however, the amputation plane is still covered with an epidermis, hence tadpoles can form the WE (Beck et al., 2003; Aztekin et al., 2019; Figure 2B). These results suggest that the WE may not have a functional role for Xenopus tail or limb regeneration, at least for regeneration purposes. Repeated amputations of the axolotl limbs result in no regeneration, yet a wound epithelium still forms (Bryant et al., 2017a; Figure 2C). In addition, in this setting, the formed WE exhibit a thickened morphology distinct from the AEC associated thickness (Bryant et al., 2017b), and it remains unclear if repeated amputations can form the AEC.
FIGURE 2.
The wound epidermis (WE) formation is not associated with regenerative success in diverse appendage regeneration models. (A, B) Xenopus laevis tadpoles lose their limb and tail regeneration abilities at specific developmental stages. Upon amputations, these animals cannot form an AEC (dark pink) at these stages but can still form the WE (light blue; Beck et al., 2009). (A) X. laevis tadpole limb regeneration ability is associated with the successful specification of AER cells to form the AEC; regeneration-incompetent tadpoles cannot specify AER cells but can still seal the amputation plane, hence they can form the WE (Aztekin et al., 2021). (B) X. laevis tail regeneration depends on the ability to relocalize their regeneration-organizing cells (ROCs; green) to amputation plane to act as the AEC (Aztekin et al., 2019). Regeneration-incompetent tadpoles cannot relocalize their ROCs, but can still seal the amputation plane, hence they can form the WE. (C) Repeated amputation of the axolotl limb results in no regeneration that can form a WE (Bryant et al., 2017b). (D) Mouse digit tip regeneration is amputation position-dependent, yet both distal regenerative and proximal regeneration deficient amputations result in the WE formation (Simkin et al., 2015).
Mice can regrow their distal digit tip throughout their life, and distal digit tip regeneration undergoes the WE formation, although this process takes much longer than amphibian limb and tail regeneration scenarios. By contrast, amputating more proximal digit tips results in no regeneration, yet an epithelium, which can be regarded as the WE, still covers the amputation plane (Dawson et al., 2021; Figure 2D). In contrast to Xenopus tail regeneration, the mouse digit tip WE seem to have a functional role in tissue histolysis by boosting the blastema formation (Simkin et al., 2015). In another appendage regeneration model, deers can regrow their lost antlers, in which the WE forms but does not necessarily influence the regrowth of the antler (Li et al., 2007, 2014). Hence, WE formation seems to be conserved across regeneration-competent species, although it does not correlate with successful appendage regeneration. Beyond these differences, tail regeneration in axolotl, zebrafish, or Xenopus, or mouse digit tip does not produce a thickened epithelium that shows morphological similarity to the amphibian limb regeneration associated AEC (Beck et al., 2003; Lehoczky et al., 2011; Pfefferli and Jaźwińska, 2015; Dolan et al., 2018). Altogether, neither the timing nor the tissue-morphology could be used to discriminate or label specific populations as the WE or the AEC across species, and their conserved role for regeneration remains ambiguous.
Gene expression profiles were tested to identify the WE and the AEC across species and regeneration paradigms, and some common gene expressions were found across certain species. For example, lef1, tp63, and msx family were expressed in the amputation plane during Xenopus tail and limb (Kawakami et al., 2006; Aztekin et al., 2019, 2021), lizard tail (Vitulo et al., 2017), and zebrafish caudal fin regeneration (Akimenko et al., 1995; Poss et al., 2000). However, these genes are not specifically seen in the WE or the AEC. Meanwhile, the canonical AEC marker fgf8 is absent in zebrafish caudal fin epidermis covering the amputation plane (Shibata et al., 2016). Interestingly, zebrafish pectoral fin amputations were suggested to form the AER due to re-expression of basal epidermal rspo2 after amputations, which is also used to detect fgf8 expressing AER (Yoshida et al., 2020). For X. laevis tail regeneration, the expression pattern of fgf8 is conflicted in different reports: while colorimetric in situ hybridizations detect fgf8 in the AEC with inconsistent patterns (Beck et al., 2006; Lin and Slack, 2008; Okumura et al., 2019), bulk-RNA-Seq and scRNA-Seq based results suggest fgf8 is not expressed in the Xenopus tail AEC (Aztekin et al., 2019; Okumura et al., 2019). Overall, currently, there is no identified specific pan-AEC marker gene found across species or regeneration paradigms.
Cross-regeneration model comparisons indicate that the AEC may not be a shared tissue type for appendage regeneration, and cell types defining the AEC may show differences. Indeed, by using bulk-RNA sequencing, Xenopus tail and limb AEC tissues are shown to exhibit gene expression differences (Okumura et al., 2019). Moving beyond tissue to the single-cell level, we identified that Xenopus tail regeneration uses an epithelial population resembling transcriptional programs similar to the AER (Aztekin et al., 2019). Due to their essential role during tail regeneration, AEC-associated gene expression, and high signaling center properties, we named this population regeneration-organizing cells (ROCs). The subsequent single-cell-based analysis demonstrated that AER cells and ROCs do not share an identical transcriptome nor single-cell morphology (Aztekin et al., 2021). Beyond transcriptomic differences, their formation on the amputation plane uses different cellular mechanisms, emphasizing that they function differently. AER cells have to be re-specified to form defining properties of the signaling center AEC for limb regeneration (Figure 2A). Meanwhile, the inability to form and maintain AER cells is observed in regeneration-incompetent tadpoles. During tail regeneration, ROCs are already present at the basal layers of midline epidermis before tail amputations, and their relocalization to the amputation plane provide the signaling center AEC for tail regeneration (Figure 2B). Meanwhile, this relocalization is not seen in regeneration-incompetent tadpoles. ROCs are found as multilayered flattened cells, while AER cells are seen as a cuboidal monolayer population (Aztekin et al., 2021). Nonetheless, they are both involved in the expression of many ligands and act as a signaling center, fitting with the acknowledged functional role of the AEC. Further cross-species single-cell transcriptomics studies and functional assessments will be required to pinpoint if there are more cell types with signaling center abilities in different species. To this aim, the detection of basal epidermal cells showing high ligand gene expressions upon amputations could be used as a valuable readout. Identifying such cell types in other species can reveal their conserved functions and potential gene regulatory networks mediating the signaling center abilities for appendage regeneration.
Conclusion
The critical functions of the AEC and its requirement for blastema formation and limb regeneration are widely accepted. Since their initial discovery, definitions of the WE and the AEC and their detection methodologies have not reached consensus across species and regeneration models. Based on current literature and initial single-cell characterizations, basal epidermal populations that migrate and seal the amputation area could be considered the WE. These cells may exhibit many similarities among all regeneration-competent species. However, the presence of the WE does not correlate with regeneration success, and their cellular identities remain elusive. Meanwhile, the presence of AEC on the amputation plane is positively correlated with regenerative success, and basal epidermal cells that form after the WE and exhibit high signaling center properties could be a more suitable description for cell types defining the AEC. Nevertheless, cell types and critical genes defining the AEC do not show conservation across species and regeneration models. For example, ROCs relocalize to form the Xenopus tail AEC, meanwhile, AER cell formation for the Xenopus limb AEC involves a differentiation event. On this basis, extra caution is required while using these terminologies and searching for their counterparts in different regeneration models. There could be diverse cell types defining the AEC tissue (as exemplified by AER cells for Xenopus limbs and ROCs for Xenopus tails). Identifying if appendage regeneration can be mediated without such populations may reveal new mechanisms for appendage regeneration.
Beyond elucidating cell fates represented in the WE or the AEC tissues, the behavioral properties of such populations and the molecular mechanisms enabling their formation on the amputation plane remain largely unknown. For example, mechanical damage has been implicated in creating Erk activation that temporarily spreads from the damaged region to the surrounding area, forming wave patterns, influencing epithelial cell migration and survival (Aoki et al., 2017; Hino et al., 2020; Gagliardi et al., 2021). Yet the implication of such Erk activity spreading for wound closure and limb regeneration remains unknown. Likewise, amputations have been indicated to promote epithelial proliferation and involve forming an actomyosin cable during zebrafish fin regeneration (Mateus et al., 2012). Moreover, in zebrafish caudal fin and tail regeneration, a transient metabolic shift to glycolysis has been shown to influence actomyosin networks in epithelial cells for successful epithelial closure, presumably forming the WE (Scott et al., 2021). How such molecular processes and metabolic changes mediate rapid wound covering during limb regeneration remains unknown. In sum, identifying cell types, characterizing their cellular behaviors and molecular regulators can promote the development of novel approaches for mammalian WE and AEC formation.
Mammalian skin wound healing has been studied to uncover ways for scar-free healing and can provide perspectives for epithelial behaviors during limb regeneration. As an example, skin wound injuries have been shown to be covered with excessive proliferation of a select number of stem cell clones, rather than using mass migratory behaviors or cell-fate switches (Aragona et al., 2017). From this perspective, during amphibian limb regeneration, epithelial behaviors seem rather distinct compared to a mouse WE formation for simple injury. Identifying such cellular features in mammals and screening them during limb regeneration in amphibians can hint at potential links for regeneration-competency. For example, the amniotic epidermis is highly stratified and exhibits a higher degree of heterogeneity with its keratin-rich layers (Blanpain and Fuchs, 2006). If and how this heterogeneity and structural complexity influence the WE formation will be important to decipher. Indeed, during metamorphosis, amphibian skin goes through specialization and extensive changes (Schreiber and Brown, 2003; Seifert et al., 2012, 2019). In a parallel, unlike premetamorphic axolotl, postmetamorphic axolotl regenerates at a slower rate with a reduced regeneration ability (Monaghan et al., 2014), and a comparison between premetamorphic and postmetamorphic axolotl showed that the wound closure is slower in postmetamorphic animals (Seifert et al., 2012). These findings bring the possibility that the dynamics of epithelial movements may be a contributor to successful regeneration and pace of regeneration. On another note, it is well established that the immune system of regeneration-incompetent mammals operates differently compared to regeneration-competent species, and this difference could be impacting epithelial cells (Julier et al., 2017; Bolaños-Castro et al., 2021). Particularly, sustained inflammation does not interfere with the wound closure and therefore the WE formation (Godwin et al., 2013; Mescher et al., 2013; Cook and Seifert, 2016). By contrast, sustained inflammation is associated with failure to form an AEC in Xenopus tail regeneration and the axolotl limb regeneration (Godwin et al., 2013; Aztekin et al., 2020). However, it remains unclear which direct and indirect immune system-mediated mechanisms impact AEC formation. Further cross-species or animal state comparisons and mammalian epidermal biology studies can guide research to elucidate WE and AEC formation.
Single-cell methods have been providing new insights on cell types mediating regeneration and divergent features of the WE and the AEC. Further cross-species systematic investigations on these tissues, cell types, and their dynamic behaviors will reveal evolutionarily conserved genetic programs of epithelial signaling centers and their association with appendage growth and regeneration.
Author Contributions
CA wrote the manuscript.
Conflict of Interest
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
I gratefully acknowledge Marian Ros and Miriam Lisci for their critical reading of the manuscript.
Funding
CA is supported by the EPFL School of Life Sciences and the Fondation Gabriella Giorgi-Cavaglieri.
References
- Akimenko M. A., Johnson S. L., Westerfield M., Ekker M. (1995). Differential induction of four msx homeobox genes during fin development and regeneration in zebrafish. Development 121 347–357. 10.1242/dev.121.2.347 [DOI] [PubMed] [Google Scholar]
- Aoki K., Kondo Y., Naoki H., Hiratsuka T., Itoh R. E., Matsuda M. (2017). Propagating wave of ERK activation orients collective cell migration. Dev. Cell 43 305–317.e5. 10.1016/j.devcel.2017.10.016 [DOI] [PubMed] [Google Scholar]
- Aragona M., Dekoninck S., Rulands S., Lenglez S., Mascré G., Simons B. D., et al. (2017). Defining stem cell dynamics and migration during wound healing in mouse skin epidermis. Nat. Commun. 8:14684. 10.1038/ncomms14684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arendt D., Musser J. M., Baker C. V. H., Bergman A., Cepko C., Erwin D. H., et al. (2016). The origin and evolution of cell types. Nat. Rev. Genet. 17 744–757. [DOI] [PubMed] [Google Scholar]
- Aztekin C. (2021). Appendage regeneration is context dependent at the cellular level. Open Biol. 11:210126. 10.1098/rsob.210126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aztekin C., Hiscock T. W., Butler R., Andino F. D. J., Robert J., Gurdon J. B., et al. (2020). The myeloid lineage is required for the emergence of a regeneration-permissive environment following Xenopus tail amputation. Development 147:dev185496. 10.1242/dev.185496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aztekin C., Hiscock T. W., Gurdon J., Jullien J., Marioni J., Simons B. D. (2021). Secreted inhibitors drive the loss of regeneration competence in Xenopus limbs. Development 148:dev199158. 10.1242/dev.199158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aztekin C., Hiscock T. W., Marioni J. C., Gurdon J. B., Simons B. D., Jullien J. (2019). Identification of a regeneration-organizing cell in the Xenopus tail. Science 364 653–658. 10.1126/science.aav9996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beauchemin M., Savard P. (1992). Two distal-less related homeobox-containing genes expressed in regeneration blastemas of the newt. Dev. Biol. 154 55–65. 10.1016/0012-1606(92)90047-k [DOI] [PubMed] [Google Scholar]
- Beck C. W., Christen B., Slack J. M. W. (2003). Molecular pathways needed for regeneration of spinal cord and muscle in a vertebrate. Dev. Cell 5 429–439. 10.1016/s1534-5807(03)00233-8 [DOI] [PubMed] [Google Scholar]
- Beck C. W., Christen B., Barker D., Slack J. M. W. (2006). Temporal requirement for bone morphogenetic proteins in regeneration of the tail and limb of Xenopus tadpoles. Mech. Dev. 123 674–688. 10.1016/j.mod.2006.07.001 [DOI] [PubMed] [Google Scholar]
- Beck C. W., Izpisúa Belmonte J. C., Christen B. (2009). Beyond early development: xenopus as an emerging model for the study of regenerative mechanisms. Dev. Dyn. 238 1226–1248. 10.1002/dvdy.21890 [DOI] [PubMed] [Google Scholar]
- Blanpain C., Fuchs E. (2006). Epidermal stem cells of the skin. Annu. Rev. Cell Dev. Biol. 22 339–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boilly B., Cavanaugh K. P., Thomas D., Hondermarck H., Bryant S. V., Bradshaw R. A. (1991). Acidic fibroblast growth factor is present in regenerating limb blastemas of axolotls and binds specifically to blastema tissues. Dev. Biol. 145 302–310. 10.1016/0012-1606(91)90128-p [DOI] [PubMed] [Google Scholar]
- Bolaños-Castro L. A., Walters H. E., Vázquez R. O. G., Yun M. H. (2021). Immunity in salamander regeneration: where are we standing and where are we headed? Dev. Dyn. 250 753–767. 10.1002/dvdy.251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant D. M., Sousounis K., Farkas J. E., Bryant S., Thao N., Guzikowski A. R., et al. (2017a). Repeated removal of developing limb buds permanently reduces appendage size in the highly-regenerative axolotl. Dev. Biol. 424 1–9. 10.1016/j.ydbio.2017.02.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant D. M., Sousounis K., Payzin-Dogru D., Bryant S., Sandoval A. G. W., Martinez Fernandez J., et al. (2017b). Identification of regenerative roadblocks via repeat deployment of limb regeneration in axolotls. NPJ Regen. Med. 2:30. 10.1038/s41536-017-0034-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell L. J., Crews C. M. (2008). Wound epidermis formation and function in urodele amphibian limb regeneration. Cell Mol. Life Sci. 65 73–79. 10.1007/s00018-007-7433-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannata S. M., Bernardini S., Di Berardino R., Filoni S. (1992). Nerve-independent DNA synthesis and mitosis in regenerating hindlimbs of larval Xenopus laevis. Roux’s Arch. Dev. Biol. 201 128–133. 10.1007/BF00188710 [DOI] [PubMed] [Google Scholar]
- Carlson M. R., Bryant S. V., Gardiner D. M. (1998). Expression of Msx-2 during development, regeneration, and wound healing in axolotl limbs. J. Exp. Zool. 282 715–723. [DOI] [PubMed] [Google Scholar]
- Castilla M., Tassava R. A. (1992). Extraction of the WE3 antigen and comparison of reactivities of mAbs WE3 and WE4 in adult newt regenerate epithelium and body tissues. Monogr. Dev. Biol. 23 116–130. [PubMed] [Google Scholar]
- Choi H. M. T., Schwarzkopf M., Fornace M. E., Acharya A., Artavanis G., Stegmaier J., et al. (2018). Third-generation in situ hybridization chain reaction: multiplexed, quantitative, sensitive, versatile, robust. Development 145:dev165753. 10.1242/dev.165753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christen B., Slack J. M. W. (1997). FGF-8Is associated with anteroposterior patterning and limb regeneration inXenopus. Dev. Biol. 192 455–466. 10.1006/dbio.1997.8732 [DOI] [PubMed] [Google Scholar]
- Christensen R. N., Tassava R. A. (2000). Apical epithelial cap morphology and fibronectin gene expression in regenerating axolotl limbs. Dev. Dyn. 217 216–224. [DOI] [PubMed] [Google Scholar]
- Christensen R. N., Weinstein M., Tassava R. A. (2002). Expression of fibroblast growth factors 4, 8, and 10 in limbs, flanks, and blastemas of Ambystoma. Dev. Dyn. 223 193–203. 10.1002/dvdy.10049 [DOI] [PubMed] [Google Scholar]
- Cook A. B., Seifert A. W. (2016). Beryllium nitrate inhibits fibroblast migration to disrupt epimorphic regeneration. Development 143 3491–3505. 10.1242/dev.134882 [DOI] [PubMed] [Google Scholar]
- Dawson L. A., Schanes P. P., Marrero L., Jordan K., Brunauer R., Zimmel K. N., et al. (2021). Proximal digit tip amputation initiates simultaneous blastema and transient fibrosis formation and results in partial regeneration. Wound Repair Regen. 29 196–205. 10.1111/wrr.12856 [DOI] [PubMed] [Google Scholar]
- Del Rio-Tsonis K., Washabaugh C. H., Tsonis P. A. (1992). The mutant axolotl short toes exhibits impaired limb regeneration and abnormal basement membrane formation. Proc. Natl. Acad. Sci. USA 89 5502–5506. 10.1073/pnas.89.12.5502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dent J. N. (1962). Limb regeneration in larvae and metamorphosing individuals of the South African clawed toad. J. Morphol. 110 61–77. 10.1002/jmor.1051100105 [DOI] [PubMed] [Google Scholar]
- Dolan C. P., Dawson L. A., Muneoka K. (2018). Digit tip regeneration: merging regeneration biology with regenerative medicine. Stem Cells Transl. Med. 7 262–270. 10.1002/sctm.17-0236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endo T., Tamura K., Ide H. (2000). Analysis of gene expressions during xenopus forelimb regeneration. Dev. Biol. 220 296–306. [DOI] [PubMed] [Google Scholar]
- Fernandez-Teran M., Ros M. A. (2008). The apical ectodermal ridge: morphological aspects and signaling pathways. Int. J. Dev. Biol. 52 857–871. 10.1387/ijdb.072416mf [DOI] [PubMed] [Google Scholar]
- Francis-West P. H., Antoni L., Anakwe K. (2003). Regulation of myogenic differentiation in the developing limb bud. J. Anat. 202 69–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagliardi P. A., Dobrzyński M., Jacques M.-A., Dessauges C., Ender P., Blum Y., et al. (2021). Collective ERK/Akt activity waves orchestrate epithelial homeostasis by driving apoptosis-induced survival. Dev. Cell 56 1712–1726.e6. 10.1016/j.devcel.2021.05.007 [DOI] [PubMed] [Google Scholar]
- Gerber T., Murawala P., Knapp D., Masselink W., Schuez M., Hermann S., et al. (2018). Single-cell analysis uncovers convergence of cell identities during axolotl limb regeneration. Science 362:eaaq0681. 10.1126/science.aaq0681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh S., Roy S., Séguin C., Bryant S. V., Gardiner D. M. (2008). Analysis of the expression and function of Wnt-5a and Wnt-5b in developing and regenerating axolotl (Ambystoma mexicanum) limbs. Dev. Growth Differ. 50 289–297. 10.1111/j.1440-169X.2008.01000.x [DOI] [PubMed] [Google Scholar]
- Godwin J. W., Pinto A. R., Rosenthal N. A. (2013). Macrophages are required for adult salamander limb regeneration. Proc. Natl. Acad. Sci. U.S.A. 110 9415–9420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldhamer D. J., Tomlinson B. L., Tassava R. A. (1989). A developmentally regulated wound epithelial antigen of the newt limb regenerate is also present in a variety of secretory/transport cell types. Dev. Biol. 135 392–404. 10.1016/0012-1606(89)90188-7 [DOI] [PubMed] [Google Scholar]
- Goss R. J. (1956). Regenerative inhibition following limb amputation and immediate insertion into the body cavity. Anat. Rec. 126 15–27. 10.1002/ar.1091260103 [DOI] [PubMed] [Google Scholar]
- Gumpel-Pinot M., Ede D. A., Flint O. P. (1984). Myogenic cell movement in the developing avian limb bud in presence and absence of the apical ectodermal ridge (AER). Development 80 105–125. [PubMed] [Google Scholar]
- Han M.-J., An J.-Y., Kim W.-S. (2001). Expression patterns of Fgf-8 during development and limb regeneration of the axolotl. Dev. Dyn. 220 40–48. [DOI] [PubMed] [Google Scholar]
- Hay E. D., Fischman D. A. (1961). Origin of the blastema in regenerating limbs of the newt Triturus viridescens: an autoradiographic study using tritiated thymidine to follow cell proliferation and migration. Dev. Biol. 3 26–59. 10.1016/0012-1606(61)90009-4 [DOI] [PubMed] [Google Scholar]
- Hino N., Rossetti L., Marín-Llauradó A., Aoki K., Trepat X., Matsuda M., et al. (2020). ERK-mediated mechanochemical waves direct collective cell polarization. Dev. Cell 53 646–660.e8. 10.1016/j.devcel.2020.05.011 [DOI] [PubMed] [Google Scholar]
- Joven A., Elewa A., Simon A. (2019). Model systems for regeneration: salamanders. Development 146:dev167700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Julier Z., Park A. J., Briquez P. S., Martino M. M. (2017). Promoting tissue regeneration by modulating the immune system. Acta Biomater. 53 13–28. [DOI] [PubMed] [Google Scholar]
- Kawakami Y., Capdevila J., Büscher D., Itoh T., Esteban C. R., Belmonte J. C. I. (2001). WNT signals control FGF-dependent limb initiation and AER induction in the chick embryo. Cell 104 891–900. 10.1016/s0092-8674(01)00285-9 [DOI] [PubMed] [Google Scholar]
- Kawakami Y., Rodriguez Esteban C., Raya M., Kawakami H., Martí M., Dubova I., et al. (2006). Wnt/β-catenin signaling regulates vertebrate limb regeneration. Genes Dev. 20 3232–3237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley R. O., Fallon J. F. (1976). Ultrastructural analysis of the apical ectodermal ridge during vertebrate limb morphogenesis: i. the human forelimb with special reference to gap junctions. Dev. Biol. 51 241–256. 10.1016/0012-1606(76)90141-x [DOI] [PubMed] [Google Scholar]
- Kishi J. Y., Lapan S. W., Beliveau B. J., West E. R., Zhu A., Sasaki H. M., et al. (2019). SABER amplifies FISH: enhanced multiplexed imaging of RNA and DNA in cells and tissues. Nat. Methods 16 533–544. 10.1038/s41592-019-0404-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knapp D., Schulz H., Rascon C. A., Volkmer M., Scholz J., Nacu E., et al. (2013). Comparative transcriptional profiling of the axolotl limb identifies a tripartite regeneration-specific gene program. PLoS One 8:e61352. 10.1371/journal.pone.0061352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lash J. W. (1955). Studies on wound closure in urodeles. J. Exp. Zool. 128 13–28. 10.1002/ar.b.20082 [DOI] [PubMed] [Google Scholar]
- Lehoczky J. A., Robert B., Tabin C. J. (2011). Mouse digit tip regeneration is mediated by fate-restricted progenitor cells. Proc. Natl. Acad. Sci. U.S.A. 108 20609–20614. 10.1073/pnas.1118017108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leigh N. D., Dunlap G. S., Johnson K., Mariano R., Oshiro R., Wong A. Y., et al. (2018). Transcriptomic landscape of the blastema niche in regenerating adult axolotl limbs at single-cell resolution. Nat. Commun. 9:5153. 10.1038/s41467-018-07604-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C., Yang F., Li G., Gao X., Xing X., Wei H., et al. (2007). Antler regeneration: a dependent process of stem tissue primed via interaction with its enveloping skin. J. Exp. Zool. Part A Ecol. Genet. Physiol. 307A 95–105. 10.1002/jez.a.352 [DOI] [PubMed] [Google Scholar]
- Li C., Zhao H., Liu Z., McMahon C. (2014). Deer antler – a novel model for studying organ regeneration in mammals. Int. J. Biochem. Cell Biol. 56 111–122. 10.1016/j.biocel.2014.07.007 [DOI] [PubMed] [Google Scholar]
- Li H., Wei X., Zhou L., Zhang W., Wang C., Guo Y., et al. (2020). Dynamic cell transition and immune response landscapes of axolotl limb regeneration revealed by single-cell analysis. Protein Cell 12 57–66. 10.1007/s13238-020-00763-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin G., Slack J. M. W. (2008). Requirement for Wnt and FGF signaling in Xenopus tadpole tail regeneration. Dev. Biol. 316 323–335. [DOI] [PubMed] [Google Scholar]
- Mariani F. V., Ahn C. P., Martin G. R. (2008). Genetic evidence that FGFs have an instructive role in limb proximal–distal patterning. Nature 453 401–405. 10.1038/nature06876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mateus R., Pereira T., Sousa S., Lima J. E., Pascoal S., Saúde L., et al. (2012). In vivo cell and tissue dynamics underlying zebrafish fin fold regeneration. PLoS One 7:e51766. 10.1371/journal.pone.0051766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinley K. L., Castillo-Azofeifa D., Klein O. D. (2020). Tools and concepts for interrogating and defining cellular identity. Cell Stem Cell 26 632–656. 10.1016/j.stem.2020.03.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McQueen C., Towers M. (2020). Establishing the pattern of the vertebrate limb. Development 147:dev177956. [DOI] [PubMed] [Google Scholar]
- McQueeney K., Soufer R., Dealy C. N. (2002). β-Catenin-dependent Wnt signaling in apical ectodermal ridge induction and FGF8 expression in normal and limbless mutant chick limbs. Dev. Growth Differ. 44 315–325. 10.1046/j.1440-169x.2002.00647.x [DOI] [PubMed] [Google Scholar]
- Mescher A. L. (1976). Effects on adult newt limb regeneration of partial and complete skin flaps over the amputation surface. J. Exp. Zool. 195 117–127. 10.1002/jez.1401950111 [DOI] [PubMed] [Google Scholar]
- Mescher A. L., Neff A. W., King M. W. (2013). Changes in the inflammatory response to injury and its resolution during the loss of regenerative capacity in developing xenopus limbs. PLoS One 8:e80477. 10.1371/journal.pone.0080477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monaghan J. R., Athippozhy A., Seifert A. W., Putta S., Stromberg A. J., Maden M., et al. (2012). Gene expression patterns specific to the regenerating limb of the Mexican axolotl. Biol. Open 1 937–948. 10.1242/bio.20121594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monaghan J. R., Stier A. C., Michonneau F., Smith M. D., Pasch B., Maden M., et al. (2014). Experimentally induced metamorphosis in axolotls reduces regenerative rate and fidelity. Regeneration 1 2–14. 10.1002/reg2.8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriyasu M., Makanae A., Satoh A. (2012). Spatiotemporal regulation of keratin 5 and 17 in the axolotl limb. Dev. Dyn. 241 1616–1624. 10.1002/dvdy.23839 [DOI] [PubMed] [Google Scholar]
- Morosow J. (1938). The inhibition and restoration of the regeneration process of the extremities in the axolotl. C. R. Acad. Sci. URSS 20 207–210. [Google Scholar]
- Mullen L. M., Bryant S. V., Torok M. A., Blumberg B., Gardiner D. M. (1996). Nerve dependency of regeneration: the role of Distal-less and FGF signaling in amphibian limb regeneration. Development 122 3487–3497. 10.1242/dev.122.11.3487 [DOI] [PubMed] [Google Scholar]
- Nacu E., Gromberg E., Oliveira C. R., Drechsel D., Tanaka E. M. (2016). FGF8 and SHH substitute for anterior–posterior tissue interactions to induce limb regeneration. Nature 533 407–410. 10.1038/nature17972 [DOI] [PubMed] [Google Scholar]
- Nakamura H., Yasuda M. (1979). An electron microscopic study of periderm cell development in mouse limb buds. Anat. Embryol. 157 121–132. 10.1007/bf00305153 [DOI] [PubMed] [Google Scholar]
- Neufeld D. A., Day F. A., Settles H. E. (1996). Stabilizing role of the basement membrane and dermal fibers during newt limb regeneration. Anat. Rec. 245 122–127. [DOI] [PubMed] [Google Scholar]
- Ohuchi H., Nakagawa T., Itoh N., Noji S. (1999). FGF10 can induce Fgf8 expression concomitantly with En1 and R-fng expression in chick limb ectoderm, independent of its dorsoventral specification. Dev. Growth Differ. 41 665–673. 10.1046/j.1440-169x.1999.00466.x [DOI] [PubMed] [Google Scholar]
- Ohuchi H., Nakagawa T., Yamamoto A., Araga A., Ohata T., Ishimaru Y., et al. (1997). The mesenchymal factor, FGF10, initiates and maintains the outgrowth of the chick limb bud through interaction with FGF8, an apical ectodermal factor. Development 124 2235–2244. 10.1242/dev.124.11.2235 [DOI] [PubMed] [Google Scholar]
- Okumura A., Hayashi T., Ebisawa M., Yoshimura M., Sasagawa Y., Nikaido I., et al. (2019). Cell type-specific transcriptome analysis unveils secreted signaling molecule genes expressed in apical epithelial cap during appendage regeneration. Dev. Growth Differ. 61 447–456. 10.1111/dgd.12635 [DOI] [PubMed] [Google Scholar]
- Pearl E. J., Barker D., Day R. C., Beck C. W. (2008). Identification of genes associated with regenerative success of Xenopus laevis hindlimbs. BMC Dev. Biol. 8:66. 10.1186/1471-213X-8-66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfefferli C., Jaźwińska A. (2015). The art of fin regeneration in zebrafish. Regeneration 2 72–83. 10.1002/reg2.33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickering J., Rich C. A., Stainton H., Aceituno C., Chinnaiya K., Saiz-Lopez P., et al. (2018). An intrinsic cell cycle timer terminates limb bud outgrowth. ELife 7:e37429. 10.7554/eLife.37429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poss K. D., Shen J., Keating M. T. (2000). Induction of lef1 during zebrafish fin regeneration. Dev. Dyn. 219 282–286. [DOI] [PubMed] [Google Scholar]
- Purushothaman S., Elewa A., Seifert A. W. (2019). Fgf-signaling is compartmentalized within the mesenchyme and controls proliferation during salamander limb development. ELife 8:e48507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin T., Fan C.-M., Wang T.-Z., Sun H., Zhao Y.-Y., Yan R.-J., et al. (2020). Single-cell RNA-seq reveals novel mitochondria-related musculoskeletal cell populations during adult axolotl limb regeneration process. Cell Death Differ. 28 111–1125. 10.1038/s41418-020-00640-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Repesh L. A., Oberpriller J. C. (1978). Scanning electron microscopy of epidermal cell migration in wound healing during limb regeneration in the adult newt, Notophthalmus viridescens. Am. J. Anat. 151 539–555. [DOI] [PubMed] [Google Scholar]
- Rodgers A. K., Smith J. J., Voss S. R. (2020). Identification of immune and non-immune cells in regenerating axolotl limbs by single-cell sequencing. Exp. Cell Res. 394:112149. 10.1016/j.yexcr.2020.112149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoh A., Graham G. M. C., Bryant S. V., Gardiner D. M. (2008). Neurotrophic regulation of epidermal dedifferentiation during wound healing and limb regeneration in the axolotl (Ambystoma mexicanum). Dev. Biol. 319 321–335. 10.1016/j.ydbio.2008.04.030 [DOI] [PubMed] [Google Scholar]
- Satoh A., Ide H., Tamura K. (2005). Muscle formation in regenerating Xenopus froglet limb. Dev. Dyn. 233 337–346. 10.1002/dvdy.20349 [DOI] [PubMed] [Google Scholar]
- Satoh A., Mitogawa K., Saito N., Suzuki M., Suzuki K. T., Ochi H., et al. (2017). Reactivation of larval keratin gene (krt62.L) in blastema epithelium during Xenopus froglet limb regeneration. Dev. Biol. 432 265–272. 10.1016/j.ydbio.2017.10.015 [DOI] [PubMed] [Google Scholar]
- Saunders J. W. (1948). The proximo-distal sequence of origin of the parts of the chick wing and the role of the ectoderm. J. Exp. Zool. 108 363–403. 10.1002/jez.1401080304 [DOI] [PubMed] [Google Scholar]
- Scherz P. J., Harfe B. D., McMahon A. P., Tabin C. J. (2004). The limb bud Shh-Fgf feedback loop is terminated by expansion of former ZPA cells. Science 305 396–399. 10.1126/science.1096966 [DOI] [PubMed] [Google Scholar]
- Schreiber A. M., Brown D. D. (2003). Tadpole skin dies autonomously in response to thyroid hormone at metamorphosis. Proc. Natl. Acad. Sci. U.S.A. 100 1769–1774. 10.1073/pnas.252774999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott C. A., Carney T. J., Amaya E. (2021). Aerobic glycolysis is important for zebrafish larval wound closure and tail regeneration. BioRxiv Available Online at: 10.1101/2021.04.23.441208 (accessed September 2, 2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seifert A. W., Cook A. B., Shaw D. (2019). Inhibiting fibroblast aggregation in skin wounds unlocks developmental pathway to regeneration. Dev. Biol. 455 60–72. 10.1016/j.ydbio.2019.07.001 [DOI] [PubMed] [Google Scholar]
- Seifert A. W., Monaghan J. R., Voss S. R., Maden M. (2012). Skin regeneration in adult axolotls: a blueprint for scar-free healing in vertebrates. PLoS One 7:e32875. 10.1371/journal.pone.0032875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata E., Yokota Y., Horita N., Kudo A., Abe G., Kawakami K., et al. (2016). Fgf signalling controls diverse aspects of fin regeneration. Development 143 2920–2929. 10.1242/dev.140699 [DOI] [PubMed] [Google Scholar]
- Shimizu-Nishikawa K., Tazawa I., Uchiyama K., Yoshizato K. (1999). Expression of helix-loop-helix type negative regulators of differentiation during limb regeneration in urodeles and anurans. Dev. Growth Differ. 41 731–743. 10.1046/j.1440-169x.1999.00477.x [DOI] [PubMed] [Google Scholar]
- Shimokawa T., Yasutaka S., Kominami R., Shinohara H. (2012). Wound epithelium function in axolotl limb regeneration. Okajimas Folia Anat. Jpn. 89 75–81. 10.2535/ofaj.89.75 [DOI] [PubMed] [Google Scholar]
- Simkin J., Sammarco M. C., Dawson L. A., Tucker C., Taylor L. J., Van Meter K., et al. (2015). Epidermal closure regulates histolysis during mammalian (Mus) digit regeneration. Regeneration 2 106–119. 10.1002/reg2.34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stocum D. L. (2019). Nerves and proliferation of progenitor cells in limb regeneration. Dev. Neurobiol. 79 468–478. 10.1002/dneu.22643 [DOI] [PubMed] [Google Scholar]
- Stopper G. F., Wagner G. P. (2005). Of chicken wings and frog legs: a smorgasbord of evolutionary variation in mechanisms of tetrapod limb development. Dev. Biol. 288 21–39. [DOI] [PubMed] [Google Scholar]
- Storer M., Mas A., Robert-Moreno A., Pecoraro M., Ortells M. C., Di Giacomo V., et al. (2013). Senescence is a developmental mechanism that contributes to embryonic growth and patterning. Cell 155 1119–1130. 10.1016/j.cell.2013.10.041 [DOI] [PubMed] [Google Scholar]
- Sugiura T., Wang H., Barsacchi R., Simon A., Tanaka E. M. (2016). MARCKS-like protein is an initiating molecule in axolotl appendage regeneration. Nature 531 237–240. 10.1038/nature16974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki M., Satoh A., Ide H., Tamura K. (2005). Nerve-dependent and -independent events in blastema formation during Xenopus froglet limb regeneration. Dev. Biol. 286 361–375. 10.1016/j.ydbio.2005.08.021 [DOI] [PubMed] [Google Scholar]
- Tank P. W., Carlson B. M., Connelly T. G. (1977). A scanning electron microscopic comparison of the development of embryonic and regenerating limbs in the axolotl. J. Exp. Zool. 201 417–429. 10.1002/jez.1402010308 [DOI] [PubMed] [Google Scholar]
- Tassava R. A., Acton R. D. (1989). Distribution of a wound epithelium antigen in embryonic tissues of newts and salamanders. Ohio J. Sci. 89 12–15. [Google Scholar]
- Tassava R. A., Garling D. J. (1979). Regenerative responses in larval axolotl limbs with skin grafts over the amputation surface. J. Exp. Zool. 208 97–109. 10.1002/jez.1402080111 [DOI] [PubMed] [Google Scholar]
- Tassava R. A., Loyd R. M. (1977). Injury requirement for initiation of regeneration of newt limbs which have whole skin grafts. Nature 268 49–50. 10.1038/268049a0 [DOI] [PubMed] [Google Scholar]
- Tassava R. A., Mescher A. L. (1975). The roles of injury, nerves, and the wound epidermis during the initiation of amphibian limb regeneration. Differentiation 4 23–24. 10.1111/j.1432-0436.1975.tb01439.x [DOI] [PubMed] [Google Scholar]
- Tassava R. A., Olsen C. L. (1982). Higher vertebrates do not regenerate digits and legs because the wound epidermis is not functional: a hypothesis. Differentiation 22 151–155. 10.1111/j.1432-0436.1982.tb01242.x [DOI] [PubMed] [Google Scholar]
- Tassava R. A., Castilla M., Arsanto J. P., Thouveny Y. (1993). The wound epithelium of regenerating limbs of Pleurodeles waltl and Notophthalmus viridescens: studies with mAbs WE3 and WE4, phalloidin, and DNase 1. J. Exp. Zool. 267 180–187. 10.1002/jez.1402670211 [DOI] [PubMed] [Google Scholar]
- Tassava R. A., Johnson-Wint B., Gross J. (1986). Regenerate epithelium and skin glands of the adult newt react to the same monoclonal antibody. J. Exp. Zool. 239 229–240. 10.1002/jez.1402390210 [DOI] [PubMed] [Google Scholar]
- Thornton C. S. (1957). The effect of apical cap removal on limb regeneration in Amblystoma larvae. J. Exp. Zool. 134 357–381. 10.1002/jez.1401340209 [DOI] [PubMed] [Google Scholar]
- Thornton C. S. (1958). The inhibition of limb regeneration in urodele larvae by localized irradiation with ultraviolet light. J. Exp. Zool. 137 153–179. 10.1002/jez.1401370108 [DOI] [PubMed] [Google Scholar]
- Thornton C. S. (1960). Influence of an eccentric epidermal cap on limb regeneration in Amblystoma larvae. Dev. Biol. 2 551–569. 10.1016/0012-1606(60)90054-3 [DOI] [PubMed] [Google Scholar]
- Thornton C. S., Thornton M. T. (1965). The regeneration of accessory limb parts following epidermal cap transplantation in urodeles. Experientia 21 146–148. 10.1007/BF02141984 [DOI] [PubMed] [Google Scholar]
- Trapnell C. (2015). Defining cell types and states with single-cell genomics. Genome Res. 25 1491–1498. 10.1101/gr.190595.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai S. L., Baselga-Garriga C., Melton D. A. (2019). Blastemal progenitors modulate immune signaling during early limb regeneration. Development 146:dev169128. 10.1242/dev.169128 [DOI] [PubMed] [Google Scholar]
- Tsai S. L., Baselga-Garriga C., Melton D. A. (2020). Midkine is a dual regulator of wound epidermis development and inflammation during the initiation of limb regeneration. ELife 9:e50765. 10.7554/eLife.50765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tschumi P. A. (1957). The growth of the hindlimb bud of Xenopus laevis and its dependence upon the epidermis. J. Anat. 91 (Pt 2) 149–173. [PMC free article] [PubMed] [Google Scholar]
- Verheyden J. M., Sun X. (2008). An Fgf/gremlin inhibitory feedback loop triggers termination of limb bud outgrowth. Nature 454 638–641. 10.1038/nature07085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinarsky V., Atkinson D. L., Stevenson T. J., Keating M. T., Odelberg S. J. (2005). Normal newt limb regeneration requires matrix metalloproteinase function. Dev. Biol. 279 86–98. 10.1016/j.ydbio.2004.12.003 [DOI] [PubMed] [Google Scholar]
- Vincent E., Villiard E., Sader F., Dhakal S., Kwok B. H., Roy S. (2020). BMP signaling is essential for sustaining proximo-distal progression in regenerating axolotl limbs. Development 147:dev170829. 10.1242/dev.170829 [DOI] [PubMed] [Google Scholar]
- Vitulo N., Dalla Valle L., Skobo T., Valle G., Alibardi L. (2017). Transcriptome analysis of the regenerating tail vs. the scarring limb in lizard reveals pathways leading to successful vs. unsuccessful organ regeneration in amniotes: tail and limb transcriptome in regenerating lizard. Dev. Dyn. 246 116–134. 10.1002/dvdy.24474 [DOI] [PubMed] [Google Scholar]
- Wang Y.-H., Beck C. W. (2014). Distal expression of sprouty (spry) genes during Xenopus laevis limb development and regeneration. Gene Expr. Patterns 15 61–66. 10.1016/j.gep.2014.04.004 [DOI] [PubMed] [Google Scholar]
- Watanabe A., Ohsugi K., Ide H. (1993). Formation of distal structures from stumps of chick wing buds at stages 24-25 following the grafting of quail tissue from X-irradiated distal limb buds. J. Exp. Zool. 267 447–453. 10.1002/jez.1402670410 [DOI] [PubMed] [Google Scholar]
- Wei Y., Yang E. V., Klatt K. P., Tassava R. A. (1995). Monoclonal antibody MT2 identifies the urodele alpha 1 chain of type XII collagen, a developmentally regulated extracellular matrix protein in regenerating newt limbs. Dev. Biol. 168 503–513. 10.1006/dbio.1995.1098 [DOI] [PubMed] [Google Scholar]
- Whited J. L., Lehoczky J. A., Austin C. A., Tabin C. J. (2011). Dynamic expression of two thrombospondins during axolotl limb regeneration. Dev. Dyn. 240 1249–1258. 10.1002/dvdy.22548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang E. V., Gardiner D. M., Carlson M. R., Nugas C. A., Bryant S. V. (1999). Expression of Mmp-9 and related matrix metalloproteinase genes during axolotl limb regeneration. Dev. Dyn. 216 2–9. [DOI] [PubMed] [Google Scholar]
- Yokoyama H., Ide H., Tamura K. (2001). FGF-10 stimulates limb regeneration ability in Xenopus laevis. Dev. Biol. 233 72–79. 10.1006/dbio.2001.0180 [DOI] [PubMed] [Google Scholar]
- Yonei-Tamura S., Endo T., Yajima H., Ohuchi H., Ide H., Tamura K. (1999). FGF7 and FGF10 directly induce the apical ectodermal ridge in chick embryos. Dev. Biol. 211 133–143. 10.1006/dbio.1999.9290 [DOI] [PubMed] [Google Scholar]
- Yoshida K., Kawakami K., Abe G., Tamura K. (2020). Zebrafish can regenerate endoskeleton in larval pectoral fin but the regenerative ability declines. Dev. Biol. 463 110–123. 10.1016/j.ydbio.2020.04.010 [DOI] [PubMed] [Google Scholar]