Abstract
Background
The gut microbiome is associated with the response to immunotherapy for different cancers. However, the impact of the gut microbiome on hepatobiliary cancers receiving immunotherapy remains unknown. This study aims to investigate the relationship between the gut microbiome and the clinical response to anti-programmed cell death protein 1 (PD-1) immunotherapy in patients with advanced hepatobiliary cancers.
Methods
Patients with unresectable hepatocellular carcinoma or advanced biliary tract cancers who have progressed from first-line chemotherapy (gemcitabine plus cisplatin) were enrolled. Fresh stool samples were collected before and during anti-PD-1 treatment and analyzed with metagenomic sequencing. Significantly differentially enriched taxa and prognosis associated taxa were identified. The Kyoto Encyclopedia of Genes and Genomes database and MetaCyc database were further applied to annotate the differentially enriched taxa to explore the potential mechanism of the gut microbiome influencing cancer immunotherapy.
Results
In total, 65 patients with advanced hepatobiliary cancers receiving anti-PD-1 treatment were included in this study. Seventy-four taxa were significantly enriched in the clinical benefit response (CBR) group and 40 taxa were significantly enriched in the non-clinical benefit (NCB) group. Among these taxa, patients with higher abundance of Lachnospiraceae bacterium-GAM79 and Alistipes sp Marseille-P5997, which were significantly enriched in the CBR group, achieved longer progression-free survival (PFS) and overall survival (OS) than patients with lower abundance. Higher abundance of Ruminococcus calidus and Erysipelotichaceae bacterium-GAM147 enriched in the CBR group was also observed in patients with better PFS. In contrast, worse PFS and OS were found in patients with higher abundance of Veillonellaceae, which was significantly enriched in the NCB group. Functional annotation indicated that the taxa enriched in the CBR group were associated with energy metabolism while the taxa enriched in the NCB group were associated with amino acid metabolism, which may modulate the clinical response to immunotherapy in hepatobiliary cancers. In addition, immunotherapy-related adverse events were affected by the gut microbiome diversity and relative abundance.
Conclusions
We demonstrate that the gut microbiome is associated with the clinical response to anti-PD-1 immunotherapy in patients with hepatobiliary cancers. Taxonomic signatures enriched in responders are effective biomarkers to predict the clinical response and survival benefit of immunotherapy, which might provide a new therapeutic target to modulate the response to cancer immunotherapy.
Keywords: immunotherapy, liver neoplasms, tumor biomarkers
Introduction
Hepatobiliary cancers are malignant tumors with poor prognoses. Global cancer statistics estimated that liver cancer, comprizing 75%–85% hepatocellular carcinoma (HCC) and 10%–15% intrahepatic cholangiocarcinoma, ranked sixth in incidence (905 677 new cases) and ranked third in mortality (830 180 new deaths) worldwide in 2020.1 Regarding cholangiocarcinoma, intrahepatic cholangiocarcinoma accounted for 20%, while perihilar cholangiocarcinoma and distal cholangiocarcinoma accounted for 50%–60% and 20%–30%, respectively.2 There were approximately 115 949 new cases and 84 695 new deaths from gallbladder cancer.1 For patients with unresectable HCC, atezolizumab plus bevacizumab is recommended as the first-line systemic therapy (median progression-free survival (PFS): 6.8 months; median overall survival (OS): 19.2 months).3 4 For unresectable biliary tract cancer (BTC), gemcitabine plus cisplatin is regarded as the primary treatment (median PFS: 8.0 months; median OS: 11.7 months) while lenvatinib plus pembrolizumab may be useful as subsequent-line therapy after disease progression from chemotherapy.5 Currently, more combination regimens of immune-checkpoint inhibitors (ICIs) and molecularly targeted drugs are under investigation in hepatobiliary cancers. However, the clinical response and survival benefit are limited due to a lack of effective predictive biomarkers and current biomarkers, such as tumor programmed cell death ligand 1 (PD-L1) expression, tumor lymphocyte infiltration and immune class gene signatures, warranting examination in large-scale studies.6 7 Therefore, the establishment of effective biomarkers associated with the clinical response to immunotherapy is required to identify the patient subgroup that benefits from immunotherapy and to guide treatment decisions for patients with hepatobiliary cancers.
Increasing evidence indicates that the gut microbiome mediates the host immune system and promotes carcinogenesis through several mechanisms, such as modulating inflammation by dysbiosis, inducing DNA damage by genotoxin, and influencing oncogenesis or tumor suppression by metabolic process.8 With these strategies, the gut microbiota stimulates or inhibits antitumour immunity by mediating host immune cells (including dendritic cells, macrophages, Treg cells, TH1 or TH17 cells, CD8+ T cells, B cells, natural killer T (NKT) cells, intraepithelial lymphocytes and mucosal-associated invariant T cells),9 and cytokine production (interleukin (IL)-2/6/8/10/12/17, interferon (IFN)-γ, granzyme B).10–12 The efficacy of ICI therapy appears to rely on the gut microbiome composition and favorable responses are associated with tumour-infiltrating lymphocyte enrichment and myeloid-derived suppressor cell reduction.13 Moreover, manipulation of the gut microbiome can enhance the response or influence toxicity to cancer immunotherapy in preclinical tumor models14 15 and patient cohorts.12 16 Different bacterial taxa have been reported to be enriched in responders to ICI therapy and most taxa belong to the Firmicutes, Bacteroidetes, Actinobacteria, Proteobacteria and Verrucomicrobia phyla.10 13 In patients with epithelial tumors (including melanoma, non-small cell lung cancer and renal cell carcinoma), a higher abundance of Akkermansia muciniphila correlated with increasing CCR9+CXCR3+CD4+ T lymphocytes and better clinical responses to programmed cell death protein 1 (PD-1)-based immunotherapy.17 In patients with advanced melanoma, enrichment of Ruminococcaceae, Faecalibacterium18 and several commensal bacterial species (Bifidobacterium longum, Collinsella aerofaciens and Enterococcus faecium)19 was associated with favorable responses to anti-PD-1 immunotherapy. Fecal microbiota transplantation (FMT) from donors who had achieved a complete response or partial response for a long duration to those patients refractory to immunotherapy exhibited increased CD8+ T cell activation and intratumour lymphocyte infiltration to overcome resistance to anti-PD-1 therapy in melanoma.20 21 Additionally, FMT with a consortium of 11 bacterial strains, including Bacteroides, Ruminococcaceae, Eubacterium and Alistipes, significantly enhanced the antitumour immunity by increasing tumour-infiltrating IFN-γ+CD8+ T cells and the tumor suppression effect was independent of anti-PD-1 treatment.22 This evidence strongly indicated that the gut microbiome modulated the response to cancer immunotherapy and the intervention of the gut microbiome might be a new therapeutic target.
Dysbiosis is prominently associated with the development of chronic liver disease, liver fibrosis, cirrhosis and liver cancer, via both TLR4-mediated inflammation and secondary bile acid metabolism mechanisms.23 24 The gut microbiome induced a peripheral T cell immunosuppressive phenotype in HCC, which was characterized by regulatory T cell expansion and CD8+ T cell attenuation.25 Similarly, dysbiosis promoted cholangiocarcinoma development by generating an immunosuppressive environment, which was characterized by CXCL1 expression in hepatocytes and CXCR2 +polymorphonuclear myeloid-derived suppressor cell recruitment.26 Primary-to-secondary bile acid conversion is mediated by the gut microbiome metabolites while the secondary bile acids regulated the accumulation of CXCR6+ hepatic NKT cells, which further inhibits liver cancer growth.27 Previous gut microbiome analysis of patients with HCC receiving anti-PD-1 immunotherapy observed that Akkermansia muciniphila and Ruminococcaceae spp were enriched in responders, and Proteobacteria was enriched in non-responders.28 In patients with gastrointestinal cancer (including colorectal cancer, esophageal cancer, gastric cancer) receiving anti-PD-1/PD-L1 treatment, the relative abundance of Prevotella, Ruminococcaceae and Lachnospiraceae was significantly higher in responders than non-responders.29 However, whether the gut microbiome influences the clinical response and survival benefit of ICI therapy in hepatobiliary cancers remains unclear. In this study, we aimed to conduct metagenomics analysis with stool samples to investigate the association of the gut microbiome with the clinical response to anti-PD-1 immunotherapy in patients with hepatobiliary cancers.
Methods
Patients and clinical evaluation
Patients histologically confirmed with unresectable HCC or advanced BTC who had progressed from first-line chemotherapy (gemcitabine plus cisplatin) were enrolled in the study at the Peking Union Medical College Hospital (PUMCH). All patients were administered anti-PD-1 based systemic therapy (every 3 weeks) without antibiotic application during the treatment until disease progression or intolerable toxicity. Treatment responses were assessed by radiological evaluation every 6 weeks according to the Response Evaluation Criteria in Solid Tumors (RECIST).1.1 standard. Patients with a complete response (CR), partial response (PR) or stable disease (SD) ≥6 months continuously were defined as achieving a clinical benefit response (CBR), while patients with stable disease <6 months or progression disease (PD) were defined as achieving a non-clinical benefit (NCB). Adverse events were assessed according to the Common Terminology Criteria for Adverse Events V.5.0 standard and grade ≥2 immunotherapy-related colitis (diarrhea) was regarded as a severe case.30 Clinical information, including sex, age, hepatitis history, liver function, performance status, bilirubin level, bile acid level, tumor number, tumor size and tumor stage, was collected from the medical records. The survival data and diarrhea condition were collected during follow-up.
Faecal sample collection
Baseline fecal samples were collected at the starting point of anti-PD-1 treatment and dynamic fecal samples were continuously collected the day before each anti-PD-1 monobody infusion every 3 weeks. All fresh stool samples were stored in sterile containers and frozen at −80°C within 24 hours until DNA extraction.
Metagenomic sequencing and taxonomic profiling
Bacterial genomic DNA was extracted using a cetyltrimethylammonium bromide kit according to the manufacturer’s instructions. The DNA concentration and integrity were checked to screen qualified samples for sequencing. Individual libraries were constructed using the NEBNext Ultra DNA Library Prep Kit and DNA sequencing was performed on the Illumina NovaSeq 6000 platform using a 2×150 bp paired-end read protocol. Raw data were filtered with Trimmomatic to obtain high-quality clean reads.31 The host DNA sequence was removed by aligning to the Homo sapiens genome assembly hg3832 with Bowtie 2 to obtain metagenomic DNA sequences.33 The metagenomic DNA sequences were assigned taxonomic labels using the Kraken 2 program34 and then Bracken (Bayesian Reestimation of Abundance after Classification with KrakEN) was used to estimate the abundance of each sample at different phylogenetic levels (phylum, class, order, family, genus, and specie).35 The quantitative metagenomics analysis was finished by Wekemo Tech Group Co., Ltd. (Shenzhen, China).
Differentially abundant taxa identification
For phylogenetic diversity between the CBR group and NCB group, alpha diversity was computed using the R package vegan to evaluate the richness and evenness of each sample, and then compared with the Wilcoxon test.36 Beta diversity based on Bray-Curtis metrics was applied to compare the dissimilarities between different groups with principal coordinate analysis (PCoA).37 Common or unique taxa at each phylogenetic level between different groups were visualized using the R package VennDiagram. Based on the relative abundance, significantly differential taxa between the CBR group and NCB group were identified with the Wilcoxon test, and an alluvial diagram was used to show the association among different phylogenetic levels for the genus or species using the R package ggalluvial. Linear discriminant analysis (LDA) effect size (LEfSe) was further applied to identify significantly differentially enriched taxa between the CBR group and NCB group.38
Functional annotation of metagenomic DNA sequences
The metagenomic DNA sequences were aligned against the UniRef90 database using the DIAMOND algorithm39 and then the HUMAnN2 tiered search strategy40 was used to quantify the KEGG Orthologous (KO) groups abundance against the Kyoto Encyclopedia of Genes and Genomes (KEGG) database.41 Significantly differential KOs between the CBR group and NCB group were identified by LEfSe and potential functional species in different KOs were also identified. Metabolic pathways were further verified in the MetaCyc database.42
Statistical analyses
Fisher’s exact test was used to determine the association between the clinical characteristics of patients and clinical response groups. The Wilcoxon test was employed to compare the relative abundance at different phylogenetic levels between different groups. According to the mean value of relative abundance for different taxa, patients were stratified into a high abundance group and a low abundance group. Univariate survival analysis of PFS and OS between different groups was performed with the Kaplan-Meier method and log-rank test using the R package survminer. Redundancy analysis (RDA) with permutation test for 1000 times was applied to identify the essential clinical factors that influenced the distribution of samples or the differentially enriched taxa using the R package vegan. All tests were two-sided and p value <0.05 was considered statistically significant. All statistical analyses were conducted using R software V.4.1.0.
Results
Patient characteristics
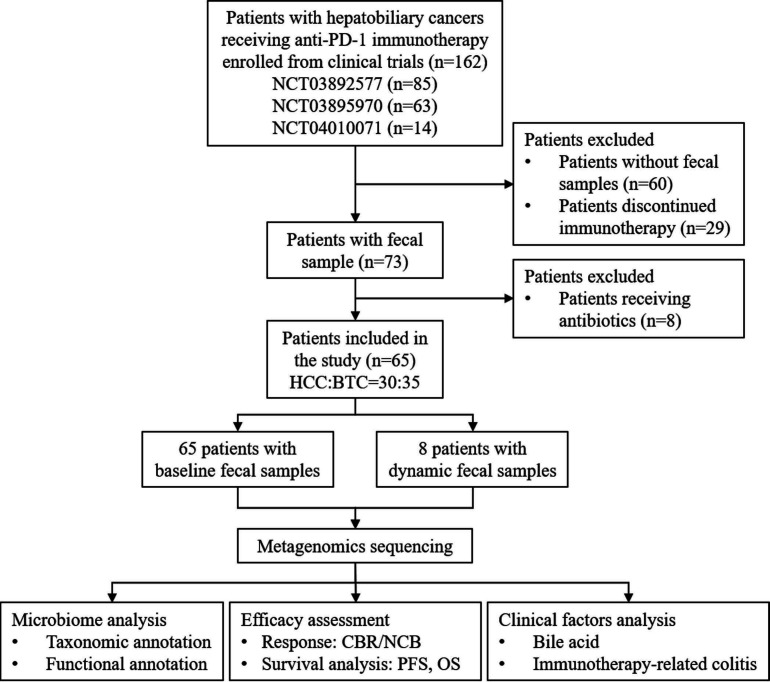
From November 2018 to December 2020, 162 patients with advanced hepatobiliary cancers were enrolled and 65 patients were ultimately included in the study (figure 1). The baseline characteristics of the patients are summarized in table 1. The study included 30 patients with HCC and 35 patients with BTC. All patients were treated with anti-PD-1 immunotherapy. According to the RECIST V.1.1 standard, 20 patients, 28 patients and 17 patients achieved PR, SD and PD, respectively. For the CBR evaluation, 32 patients were defined as having CBR and 33 patients were defined as having NCB. Approximately half of the patients had a history of hepatitis and received antiviral prophylaxis therapy during anti-PD-1 monobody medication. All the patients had preserved liver function and performance status. Bile acid elevation was found in one out of three of the patients. No significant differences were observed in sex, age, hepatitis history, bilirubin, bile acid, tumor number, tumor size and tumor stage between the CBR group and NCB group. Baseline fecal samples were collected from all the 65 patients and dynamically collected samples were obtained continuously from 8 patients (4 CBR and 4 NCB).
Figure 1.
Study workflow. BTC, biliary tract cancer; CBR, clinical benefit response; HCC, hepatocellular carcinoma; NCB, non-clinical benefit; OS, overall survival; PD-1, programmed cell death protein 1; PFS, progression-free survival.
Table 1.
Clinical manifestation of 65 patients with advanced hepatobiliary cancers receiving anti-programmed cell death protein 1 treatment
| Characteristics | Total (n=65) |
CBR (n=32) |
NCB (n=33) |
P value |
| Sex | ||||
| Male | 46 | 25 | 21 | 0.277 |
| Female | 19 | 7 | 12 | |
| Age (years) | ||||
| ≤60 | 34 | 18 | 16 | 0.622 |
| >60 | 31 | 14 | 17 | |
| Hepatitis | ||||
| Yes | 32 | 14 | 18 | 0.460 |
| No | 33 | 18 | 15 | |
| Child-Pugh score | ||||
| 5 | 45 | 22 | 23 | 0.901 |
| 6 | 9 | 4 | 5 | |
| 7 | 11 | 6 | 5 | |
| ECOG-PS | ||||
| 0 | 44 | 22 | 22 | >0.999 |
| 1 | 21 | 10 | 11 | |
| Bilirubin level | ||||
| Elevated | 8 | 4 | 4 | >0.999 |
| Normal | 57 | 28 | 29 | |
| Bile acid level | ||||
| Elevated | 21 | 10 | 11 | >0.999 |
| Normal | 44 | 22 | 22 | |
| Tumor type | ||||
| HCC | 30 | 17 | 13 | 0.267 |
| BTC | 35 | 15 | 20 | |
| Tumor number | ||||
| Single | 28 | 14 | 14 | >0.999 |
| Multiple | 37 | 18 | 19 | |
| Tumor size (cm) | ||||
| <5 | 33 | 17 | 16 | 0.806 |
| ≥5 | 32 | 15 | 17 | |
| Stage | ||||
| II | 6 | 3 | 3 | 0.993 |
| III | 26 | 13 | 13 | |
| IV | 33 | 16 | 17 |
BTC, biliary tract cancer; CBR, clinical benefit response; ECOG-PS, Eastern Cooperative Oncology Group performance status; HCC, hepatocellular carcinoma; NCB, non-clinical benefit.
Gut microbiome composition is correlated with different clinical responses to anti-PD-1 immunotherapy
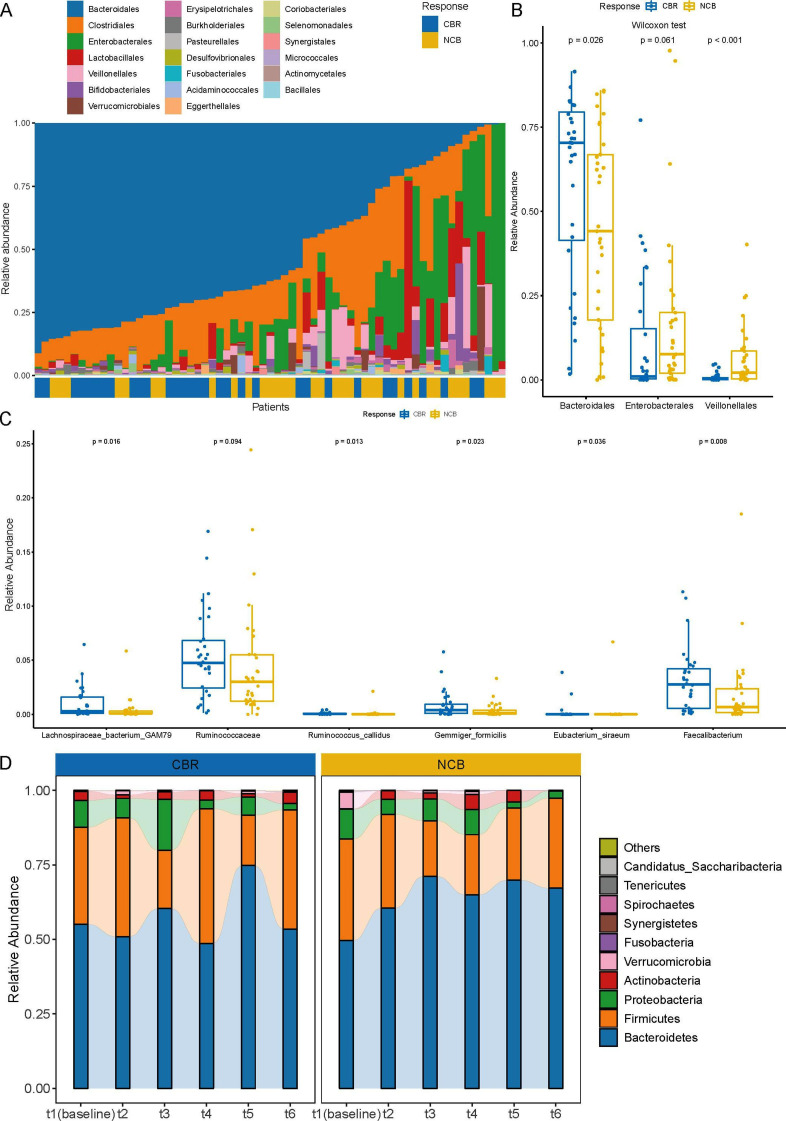
To investigate the association between the gut microbiome composition and immunotherapy response, the relative abundance at different phylogenetic levels was compared between the CBR group and the NCB group. Overall, at the phylum level, Bacteroidetes, Firmicutes and Proteobacteria dominated the gut microbial communities in both the CBR group and the NCB group, followed by Actinobacteria and Verrucomicrobia (online supplemental figure S1). A higher abundance of Bacteroidetes was observed in the CBR group (p=0.028), and a trend of a higher abundance of Proteobacteria was observed in the NCB group (p=0.067). A similar composition pattern was observed at the order level (figure 2A). The relative abundance of Bacteroidales from the Bacteroidetes phylum was significantly higher in the CBR group (p=0.028), while the relative abundance of Enterobacterales from the Proteobacteria phylum tended to be higher in the NCB group (p=0.061) (figure 2B). The Veillonellales order was predominantly found in the NCB group (figure 2A) and the relative abundance was significantly lower in the CBR group (p<0.001) (figure 2B). Furthermore, the top 20 richest species belonged mainly to the Bacteroidetes phylum (online supplemental figure S2), among which Alistipes sp Marseille-P5997, Bacteroides fluxus and Bacteroides zoogleoformans were enriched in the CBR group (p=0.024, p=0.012 and p=0.031) (online supplemental table S1). Among the statistically differently genera or species, most bacterial taxa enriched in the CBR group belonged to the Bacteroidetes and Firmicutes phyla, whereas most bacterial taxa enriched in the NCB group belonged to the Proteobacteria phylum (online supplemental figure S3 and S4). Notably, the relative abundance of Lachnospiraceae bacterium-GAM79 and several taxa from the Ruminococcaceae family (Ruminococcus callidus, Eubacterium siraeum, Gemmiger formicilis and Faecalibacterium genus) were significantly enriched in the CBR group (figure 2C). In addition, metagenomics analysis dynamic fecal samples of 8 patients revealed that the gut microbiome composition in the CBR group remained relatively stable at the phylum level along with the treatment and was similar to the gut communities of bacteria in all 65 patients, while the microbial diversity in the NCB group seemed to decrease (figure 2D). Together, baseline gut microbiome diversity is associated with favorable response to anti-PD-1 treatment in patients with hepatobiliary cancers, which supports that the gut microbiome might be an effective biomarker to predict the immunotherapy response.
Figure 2.
Gut microbiome composition of 65 patients with hepatobiliary cancers. (A) Gut microbiome composition at the order level in the 65 patients with hepatobiliary cancers (ordered by the most abundant taxa, Bacteroideles order). (B) Relative abundance comparison of Bacteroidales, Enterobacterales and Veillonellales in the CBR group and NCB group at the order level (Wilcoxon test). (C) Relative abundance comparison of Lachnospiraceae bacterium-GAM79, Ruminococcus callidus, Eubacterium siraeum, Gemmiger formicilis and Faecalibacterium genus in the CBR group and NCB group (Wilcoxon test). (D) Dynamic microbial composition in the CBR group and NCB group at the phylum level. CBR, clinical benefit response; NCB, non-clinical benefit.
jitc-2021-003334supp001.pdf (1.7MB, pdf)
jitc-2021-003334supp002.xlsx (43.7KB, xlsx)
Differentially enriched bacterial taxa are associated with the survival benefit of ICI treatment
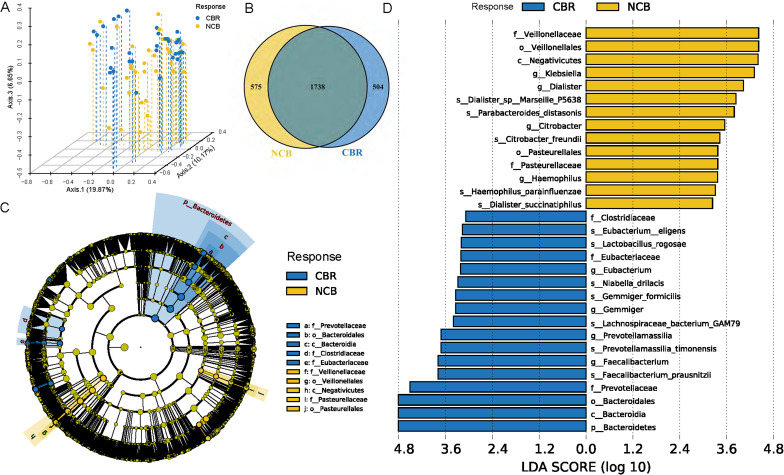
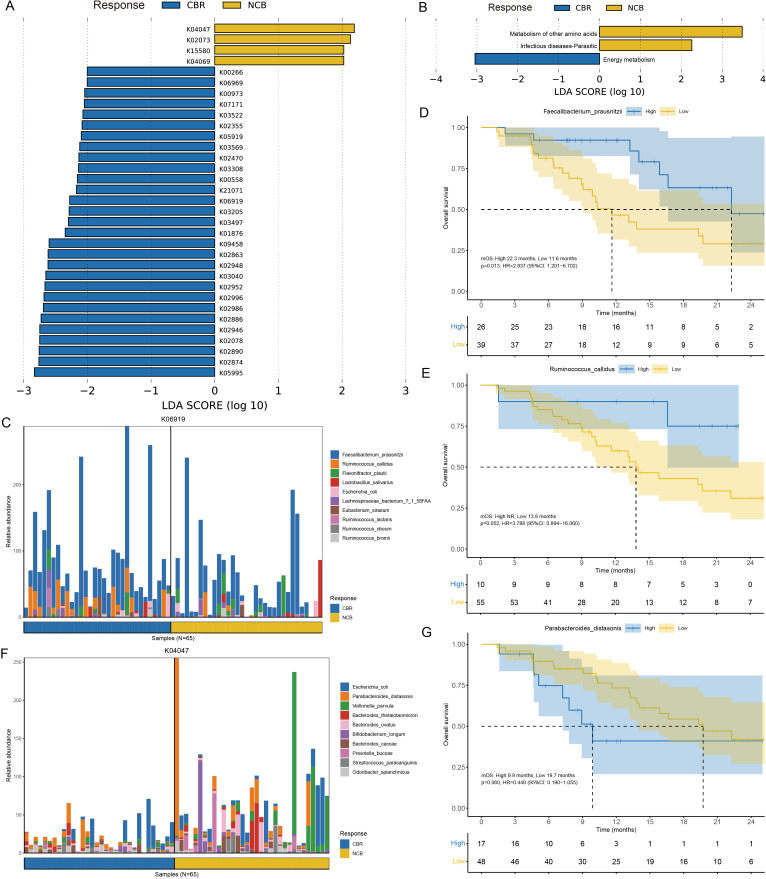
Regarding phylogenetic diversity, no significant difference was found in alpha diversity, including the Shannon index, Simpson index, Pielou index and Chao1 index, between the CBR group and NCB group (online supplemental figure S5). However, the beta diversity evaluated by Bray-Curtis distance showed that patients in the CBR group were clearly separated from those patients in the NCB group by PCoA (figure 3A). At the specie level, the CBR group and NCB group shared 1738 common species and there were 504 and 575 unique species in the CBR group and NCB group, respectively (figure 3B). We further identified 74 CBR group enriched taxa and 40 NCB group enriched taxa possibly influencing the clinical response to ICI treatment with LEfSe analysis (LDA >2, p<0.05) (online supplemental figure S6). Of note, Bacteroidetes phylum, Bacteroidia class, Bacteroidales order and Prevotellaceae family were the top enriched taxa in the CBR group while Negativicutes class, Veillonellales order and Veillonellaceae family were the top enriched taxa in the NCB group (LDA >3, p<0.05) (figure 3C–D). A similar differential enrichment pattern was observed in subgroup analysis of patients with HCC or BTC. No significant difference was found in alpha diversity or beta diversity between the patients with HCC and BTC (online supplemental figure S7A, B). The significantly different bacterial taxa between these two cancer types mainly belonged to the Bacteroidetes phylum (online supplemental table S2), and a significantly higher abundance of Bacteroidetes phylum were observed in patients with BTC than patients with HCC (LDA >3, p<0.05) (online supplemental figure S7C, D), which was also enriched in the CBR group among patients with BTC (online supplemental figure S9A, B). For patients with HCC, a higher abundance of Veillonellaceae family from the Firmicutes phylum was also observed in the NCB group (LDA >3, p<0.05) (online supplemental figure S8A, B).
Figure 3.
Differential taxa were enriched in the CBR group and NCB group of the 65 patients with hepatobiliary cancers. (A) PCoA showed the beta diversity evaluated by Bray-Curtis distance between the CBR group and NCB group. (B) Common and unique taxa at the specie level between the CBR group and NCB group. (C) Taxonomic cladogram from LEfSe showed different taxa enriched in the CBR group and NCB group (LDA>3, p<0.05). (D) LEfSe identified significantly differentially abundant taxa in the CBR group and NCB group (LDA>3, p<0.05). CBR, clinical benefit response; LEfSe, linear discriminant analysis effect size; LDA, linear discriminant analysis; NCB, non-clinical benefit; PCoA, principal coordinate analysis.
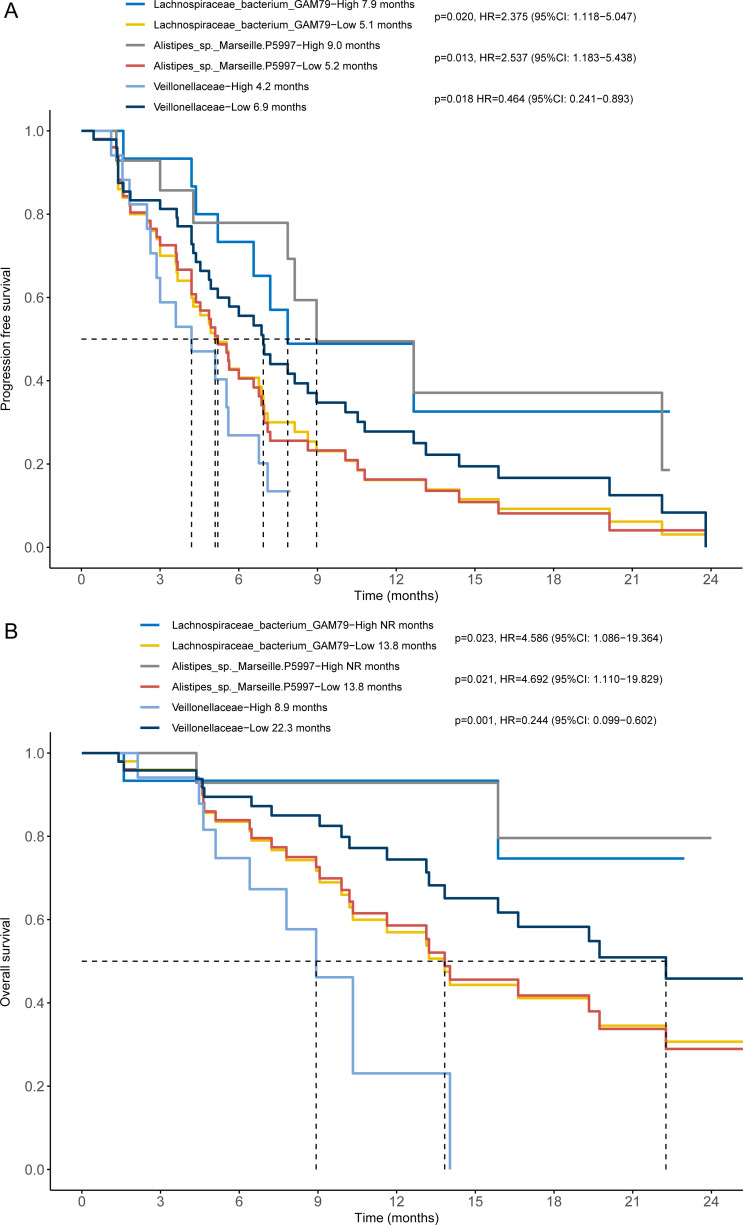
To further explore the microbial taxa associated with survival, we stratified the 65 patients with hepatobiliary cancers into a high abundance group and a low abundance group based on the mean abundance of these differentially enriched taxa in the CBR group and NCB group. Survival analysis showed that patients with a higher abundance of Lachnospiraceae bacterium-GAM79 achieved longer PFS (median PFS (mPFS): 7.9 months vs 5.1 months, p=0.020) and OS (median OS (mOS): not reached vs 13.8 months, p=0.023) than patients with a lower abundance (figure 4). Erysipelotrichaceae bacterium-GAM147 and Ruminococcus callidus enrichment were also associated with better PFS (mPFS: 10.8 months vs 4.9 months, p=0.009; mPFS: 22.1 months vs 5.5 months, p=0.006) (online supplemental figure S10A, B). Considering taxa belonging to the most abundant taxa, Bacteroidetes phylum, although only a higher abundance of Bacteroidales order showed a tendency correlated with favorable outcome (mPFS: 6.9 months vs 5.1 months, p=0.250; mOS: 32.2 months vs 13.8 months, p=0.269) (online supplemental figure S10C, D), patients harboring a higher abundance of Alistipes sp. Marseille-P5997 had significantly prolonged PFS (mPFS: 9.0 months vs 5.2 months, p=0.013) and OS (mOS: NR vs 13.8 months, p=0.021) (figure 4) and Bacteroides zoogleoformans enrichment was also positively associated with longer PFS (mPFS: 8.1 months vs 5.2 months, p=0.043) (online supplemental figure S10E, F). In contrast, the presence of the Veillonellaceae family was significantly negatively associated with PFS (mPFS: 4.2 months vs 6.9 months, p=0.018) and OS (mOS: 8.9 vs 22.3 months, p=0.001) (figure 4). Moreover, a similar association of these bacterial taxa with prognosis was observed in patients with HCC or BTC. In the 30 patients with HCC, patients enriched with Erysipelotrichaceae bacterium-GAM147 achieved better PFS (mPFS: 15.9 months vs 5.5 months, p=0.021) and OS (mOS: NR vs 15.9 months, p=0.041) although there was no significant difference of the abundance of Erysipelotrichaceae bacterium-GAM147 between the CBR group and the NCB group (online supplemental figure S8C, D). While patients with HCC enriched with Veillonellaceae family had obviously worse PFS (mPFS: 3.6 months vs 10.8 months, p=0.005) and OS (mOS: 7.8 vs NR months, p=0.030) (online supplemental figure S8E, F). In the 35 patients with BTC, patients enriched with Bacteroidales order had significantly better PFS (mPFS: 6.6 months vs 4.5 months, p=0.041) and OS (mOS: 16.6 vs 10.2 months, p=0.025) (online supplemental figure S9C, D). Furthermore, patients with BTC who tended to obtain a higher abundance of Veillonellaceae family had worse PFS (mPFS: 3.5 months vs 6.0 months, p=0.031) and OS (mOS: 7.7 vs 16.6 months, p=0.006) (online supplemental figure S9E, F). Therefore, differentially enriched gut microbial taxa are associated with the clinical response as well as the survival benefit of ICI treatment. Our results indicate that enrichment of specific taxa in gut microbiota might enhance the clinical response and prolong the survival of anti-PD-1 treatment, which can predict the survival benefit for patients with hepatobiliary cancers receiving immunotherapy.
Figure 4.
Differentially enriched taxa were associated with the survival benefit of anti-programmed cell death protein 1 immunotherapy in patients with hepatobiliary cancers. The Kaplan-Meier method with log-rank test estimates the median progression-free survival (A) and median overall survival (B) for patients with higher or lower abundance of Lachnospiraceae bacterium-GAM79, Alistipes sp Marseille-P5997 and Veillonellaceae family.
The gut microbiome potentially influences the immunotherapy response by mediating metabolic pathways
To investigate the functional role of the gut microbiota in immunotherapy, we further annotated the metagenomic sequencing data in the KEGG database and identified 29 KOs enriched in the CBR group and 4 KOs enriched in the NCB group by LEfSe (LDA >2, p<0.05) (figure 5A). The most abundant KOs in the CBR group and NCB group were energy metabolism pathways and amino acid metabolism pathways, respectively (figure 5B). At the pathway level, the CBR group was significantly enriched with KOs that played essential functions in several KEGG pathways, including aurachin metabolism (map00999: K02078), fatty acid biosynthesis (map00061: K09458), acarbose biosynthesis (map00525: K00973) and polyketide sugar unit biosynthesis (map00523: K00973) (online supplemental figure S11). Furthermore, comparisons of differential metabolic pathways in the MetaCyc database showed that metabolic pathways dominant in the CBR group were associated with bacterial reproduction processes, such as UDP/UMP biosynthesis and queuosine biosynthesis (online supplemental figure S12, black box). Amino acid metabolism pathways such as arginine biosynthesis predominated in the NCB group (online supplemental figure S12, green box). The dominant species enriched in several CBR group associated KOs, Faecalibacterium prausnitzii and Ruminococcus callidus, which belong to the Firmicutes phylum were found to be correlated with markedly prolonged OS (mOS: 22.3 vs 11.6 months, p=0.013; mOS: NR vs 13.8 months, p=0.052) (figure 5C–E). In contrast, patients with a higher abundance of Parabacteroides distasonis enriched in the NCB group associated KOs, which belong to the Bacteroidetes phylum had poor OS compared with those patients with a lower abundance (mOS: 9.9 vs 19.7 months, p=0.060) (figure 5F–G). These findings suggested that the underlying mechanisms of the gut microbiome influencing anti-PD-1 immunotherapy efficacy and survival benefit in hepatobiliary cancers might be driven by specific bacterial species involved in different metabolic pathways.
Figure 5.
Functional annotation of the gut microbiome metagenomic sequencing data. (A) Different KOs enriched in the CBR group and NCB group identified by LEfSe (LDA>2, p<0.05). (B) Functional pathways associated with the differentially-enriched KOs identified by LEfSe (LDA>2, p<0.05). (C) The top 10 bacterial species contributed to putative DNA primase/helicase (K06919) enrichment in the CBR group. (D–E) The Kaplan-Meier method with log-rank test estimates the mOS for patients with higher or lower abundance of Faecalibacterium prausnitzii and Ruminococcus callidus enriched in the CBR group. (F) The top 10 bacterial species contributed to starvation-inducible DNA-binding protein (K04047) enrichment in the NCB group. (G) The Kaplan-Meier method with log-rank test estimates the mOS for patients with higher or lower abundance of Parabacteroides distasonis enriched in the NCB group. CBR, clinical benefit response; KEGG, Kyoto Encyclopedia of Genes and Genomes; KOs, KEGG Orthologous groups; LDA, linear discriminant analysis; LEfSe, linear discriminant analysis effect size; mOS, median overall survival; NCB, non-clinical benefit.
Immunotherapy-related adverse events are influenced by the gut microbiome
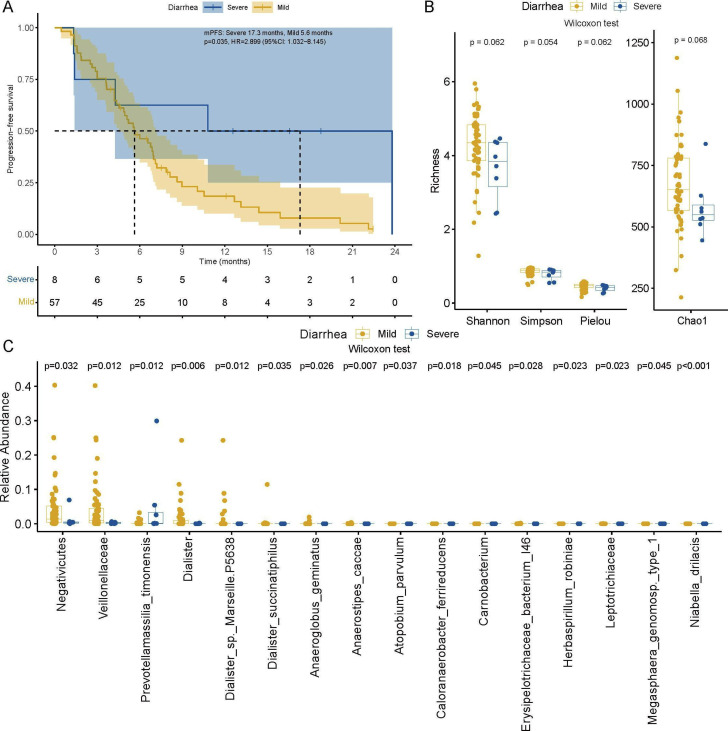
Whether immunotherapy-related adverse events (irAEs) were affected by gut microbiome composition was also explored in our study. Considering the typical adverse event, immunotherapy-related colitis, 8 patients experienced clinically severe diarrhea (grade ≥2), including 4 CBR and 4 NCB patients, and 57 patients experienced mild diarrhea (grade 1 or absent) in this study. Compared with those patients with mild diarrhea, patients who experienced clinically severe diarrhea seemed to obtain a better clinical response to anti-PD-1 treatment (Fisher’s exact test, p=0.021) and achieved longer survival (mPFS: 17.3 months vs 5.6 months, p=0.035) (figure 6A). Regarding the phylogenetic diversity of gut microbiota, patients with severe diarrhea tended to have lower richness (figure 6B), which inferred that immunotherapy-related colitis was more likely associated with decreased gut microbiome diversity and relative abundance. Specifically, a total of 16 significantly differentially enriched taxa were identified between the severe diarrhea group and mild diarrhea group, among which the Negativicutes class, Veillonellaceae family and Dialister genus all belonged to the Firmicutes phylum (figure 6C). Notably, enrichment of Prevotellamassilia timonensis from the Bacteroidetes phylum was observed in patients with more severe diarrhea, which might be a potential marker to predict severe immunotherapy-related colitis. Together, we demonstrated that the irAE profiles were influenced by the gut microbiome, and higher diversity and relative abundance of taxa from the Firmicutes phylum might be a protective factor against irAEs.
Figure 6.
Immunotherapy-related adverse events were affected by the gut microbiome. (A) The Kaplan-Meier method with log-rank test estimates the mPFS for patients with severe or mild diarrhea. (B) Gut microbiome alpha diversity comparison between the severe and mild diarrhea groups (Wilcoxon test). (C) Relative abundance comparison of significantly different taxa between patients with severe diarrhea and mild diarrhea (Wilcoxon test). mPFS, median progression-free survival.
Gut bacterial enrichment is affected by multiple clinical factors
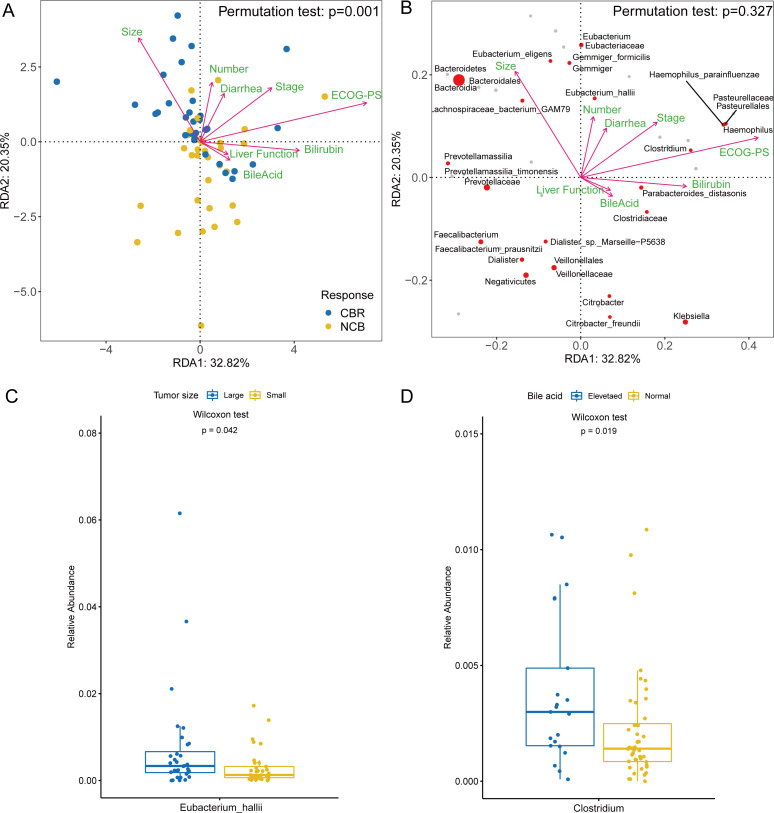
After identifying the differentially enriched taxa in the CBR group and NCB group, we addressed the relationship between the gut bacterial features and clinical factors in patients with hepatobiliary cancers. RDA showed that multiple tumour-associated factors that significantly influenced the distribution of patients were also associated with the differential enrichment of the gut microbiome between the CBR group and NCB group (figure 7A–B). The tumor size, number and stage correlated positively with the favorable response and the CBR group enriched taxa, such as Bacteroidales order, Lachnospiraceae bacterium-GAM79, Eubacterium eligens, Eubacterium hallii and Gemmiger formicilis (figure 7B). Further analysis observed that the relative abundance of Eubacterium hallii was significantly higher in patients with large tumors (≥5 cm) than those patients with small tumors (<5 cm) (p=0.042) (figure 7C). No significant difference in bacterial abundance was found among subgroups with different tumor numbers or tumor stages. In contrast, bile acid elevation, bilirubin elevation and poor liver function were associated with the NCB group enriched taxa, such as Veillonellaceae family, Dialister sp. Marseille-P5638 (belonging to Veillonellaceae family), Clostridiaceae family, Klebsiella genus and Citrobacter freundii (figure 7B). Patients with elevated bile acid levels had a higher abundance of the Clostridium genus than patients with normal bile acid levels (p=0.019) (figure 7D). In addition, patients with a higher abundance of tumor size-positively, number-positively and stage-positively associated taxa achieved better survival, while patients with a higher abundance of bile acid-positively, bilirubin-positively and liver function-positively associated taxa had worse survival (figure 4). Interestingly, the CBR group significantly enriched specie, Faecalibacterium prausnitzii, was obviously negatively associated with tumor number and stage (figure 7B). This heterogeneous impact of clinical factors on the gut microbiota enriched in the CBR group and NCB group implied that the clinical response and survival benefit of anti-PD-1 treatment relied on the whole gut microbiome diversity and differentially enriched taxa community.
Figure 7.
The gut microbiome distribution was affected by clinical factors. (A) RDA with permutation test showed the clinical factors associated with the distribution of patients with hepatobiliary cancers. (B) RDA with permutation test showed the clinical factors associated with the different taxa enriched in the CBR group and NCB group. (C) Relative abundance comparison of Eubacterium hallii in patients with large tumors (≥5 cm) and small tumors (≥5 cm) (Wilcoxon test). (D) Relative abundance comparison of the Clostridium genus in patients with elevated bile acid and normal bile acid (Wilcoxon test). CBR, clinical benefit response; ECOG-PS, Eastern Cooperative Oncology Group performance status; NCB, non-clinical benefit; RDA, redundancy analysis.
Discussion
Through prospective comprehensive metagenomics analysis, we demonstrated that the gut microbiota was associated with the clinical response and irAE profiles of anti-PD-1 immunotherapy in patients with hepatobiliary cancers. Differentially enriched taxa, such as a higher abundance of Lachnospiraceae bacterium-GAM79, Alistipes sp. Marseille-P5997 and a lower abundance of Veillonellaceae in the gut microbiota may be potential biomarkers predicting the favorable response and survival benefit of ICI treatment. In addition, the association between gut microbiome diversity and immunotherapy-related colitis highlights that higher diversity and relative abundance of gut microbiota are associated with a less severe irAE profile.
Several significantly differentially enriched bacterial taxa between the CBR group and NCB group were identified to be obviously associated with the favorable outcomes to anti-PD-1 treatment in our study. Enrichment of two butyrate-producing commensal bacterium, Lachnospiraceae bacterium-GAM79 and Erysipelotrichaceae bacterium-GAM147, was significantly associated with prolonged survival of immunotherapy. Lachnospiraceae and Erysipelotrichaceae are protective commensal strain that produces short-chain fatty acids such as butyrate by fermentation of dietary fiber. And short-chain fatty acids can induce protective immune responses and enhance epithelial barrier function by production of anti-inflammatory cytokines (TGF-β and IL-10) and regulation of Treg cell differentiation,43 44 which enhanced the efficacy of PD-1 blockade in colon cancer.45 Lachnospiraceae is also involved in the primary to secondary bile acid conversion process and is regarded as a candidate effective commensal bacterial strain for consortial FMT to resurrect the intestinal microbiota and restore colonization resistance.46 This evidence indicates that the impact of Lachnospiraceae and Erysipelotrichaceae on hepatobiliary cancers might be involved in short-chain fatty acid production and bile acid metabolism, which possibly modulates the immunotherapy response. In addition, several taxa from the Ruminococcaceae family were significantly enriched in the CBR group and a higher abundance of Ruminococcus callidus was positively related to the favorable response and survival improvement of anti-PD-1 treatment in hepatobiliary cancers, which supported the positive prognostic significance of the Ruminococcaceae family in immunotherapy in many other cancers. The Ruminococcaceae family involves in secondary bile acid production, and deficiency of Ruminococcaceae might result in increasing intestinal inflammation and further influence the host immune system.47 Further FMT experiments showed that the abundance of the Ruminococcaceae family significantly increased post-FMT,21 which enhanced the antitumour immunity by increasing tumor infiltrating IFN-γ+CD8+ T cells in mouse models and patients receiving immunotherapy.18 20 22 Taking together, we hypothesize that oral supplementation or FMT of Lachnospiraceae, Erysipelotrichaceae and Ruminococcaceae might enhance antitumour immunity and boost the clinical response to anti-PD-1 treatment in hepatobiliary cancers, which warrants further exploration in mice model and clinical cohorts.
It should be highlighted that different impact of the Bacteroidetes phylum and Firmicutes phylum on cancer immunotherapy was observed in our study. In our study, most bacterial taxa associated with favorable outcomes belonged to the Firmicutes phylum and most bacterial taxa with heterogeneous impact on immunotherapy belonged to the Bacteroidetes phylum. The differential analysis and survival analysis results showed that patients with HCC were more likely influenced by the taxa from the Firmicutes phylum and relevant taxa enrichment correlated with better response and survival benefit of immunotherapy. In contrast, patients with BTC tended to be enriched with bacterial taxa from the Bacteroidetes phylum and significantly higher abundance of Bacteroidetes phylum was associated with favorable response to the immunotherapy. The difference impact of the Firmicutes phylum and Bacteroidetes phylum was also observed in previous FMT experiment, which demonstrated that most significantly enriched taxa in the responders belonged to the Firmicutes phylum, whereas most decreased taxa in the responders belonged to the Bacteroidetes phylum post-FMT.21 Furthermore, there is no ideal overlap among the current bacterial taxa identified to correlate with favorable immunotherapy responses across different cancers, especially at a lower level of taxonomy.10 Two species belonging to the Bacteroides genus, Alistipes sp Marseille-P5997 and Bacteroides zoogleoformans, displayed a positive association with favorable response and survival benefit in our hepatobiliary cancer cohort. A previous study reported that Bacteroidales thetaiotaomicron or Bacteroidales fragilis was associated with better efficacy of CTLA-4 blockades in mice and patients with melanoma.15 However, a higher abundance of species in the Bacteroides genus, such as Bacteroides ovatus, Bacteroides dorei and Bacteroides massiliensis, was reported to be negatively associated with longer PFS in patients with melanoma treated with immunotherapy.48 The controversial impact of Bacteroides genus on carcinogenesis or cancer immunotherapy might be explained by different cancer types and the heterogeneous composition of Bacteroidetes phylum.
Secondary bile acid metabolism-NKT activation mediated by the gut microbiota has been proposed as another possible mechanism modulating host immunity and cancer immunotherapy in hepatobiliary cancers.23 27 The bile acid metabolites glycoursodeoxycholic acid and tauroursodeoxycholic acid were increased in intrahepatic cholangiocarcinoma and plasma tauroursodeoxycholic acid was negatively correlated with survival.49 The secondary bile acid 3β-hydroxydeoxycholic acid enhanced peripheral Treg cell differentiation by acting on dendritic cells50 and the Treg cell generation was significantly decreased by genetic abolition of bile acid metabolic pathways.51 In our results, bile acid was correlated with the Clostridiaceae family and a significantly higher abundance of the Clostridium genus was observed in patients with elevated bile acid. Enrichment of Clostridium XIVa in HCC with a high tumor burden was associated with serum bile acids.52 Clostridium species regulated primary–secondary bile acid conversion, especially the 7α-dehydroxylation reaction process, which restricted hepatic NKT cell accumulation and liver cancer inhibition effect.27 Taken together, these observations suggest that specific gut bacterial strains may be involved in bile acid metabolism and mediate the response to anti-PD-1 immunotherapy. Bile acid classification and quantitation as well as intervention of Clostridium species are required to illustrate the role of bile acid metabolism in hepatobiliary cancers receiving immunotherapy.
The influence of the gut microbiome on irAEs was also preliminarily explored in our study. A higher relative abundance of Prevotellamassilia timonensis from the Bacteroidetes phylum correlated with more severe immunotherapy-related colitis in hepatobiliary cancers, which might be a potential predictor of severe irAEs. Most taxa significantly enriched in the mild diarrhea group belonged to the Firmicutes phylum, such as Negativicutes class, Veillonellaceae family and Dialister genus, which inferred that taxa from the Firmicutes phylum might be a protective factor against immunotherapy-induced toxicity. Consistent with our result, a higher abundance of Bacteroides intestinalis in gut microbiota was associated with upregulation of mucosal IL-1β and severe irAEs in patients with melanoma and pre-clinical models treated with combined immune checkpoint blockade.53 In contrast, another study reported that increased bacterial species belonging to the Bacteroidetes phylum were resistant to ipilimumab-induced colitis in melanoma.54 Enrichment of taxa in the Firmicutes phylum was associated with more frequent occurrence of ipilimumab-induced colitis in metastatic melanoma55 and more severe immunotherapy-related colitis in non-small cell lung cancer.56 These different findings showed that the relationship between the gut microbiome and irAEs remains unclear. And more researches across other cancer types are required to explore the impact of the gut microbiota on irAEs and underlying mechanisms. However, a higher abundance of the gut microbiome was observed in patients who experienced mild diarrhea, suggesting that whole gut microbiota community diversity instead of specific specie enrichment might play a more important role in preventing irAEs development. These results inferred that the gut microbiome might serve as a new biomarker to predict irAEs incidence and severity. Furthermore, manipulating the gut microbial composition and diversity might protect against severe irAEs, which provided a potential new therapeutic target for modulating immunotherapy-induced toxicity.
There were some limitations in our study. First, several confounding factors, such as dietary habits and geographical differences, were confirmed to affect the gut microbiome composition, which were not controlled in our study. A recent study reported that concomitant medication of antibiotics, corticosteroids and proton pump inhibitors was associated with worse outcome of pembrolizumab monotherapy in non-small cell lung cancer,57 which indicated that concomitant medication should be focused on in further gut microbial analysis on cancer immunotherapy. Second, the sample size of our cohort was limited. Some of the high or low abundance categories had a few patients in survival analysis and large-sample study is required to validate our findings and hypothesis. The dynamic analysis of gut microbiome in our study was also small, which warranted more studies with large sample to explore whether the favorable response to immunotherapy depends on the baseline gut microbiota or some specific bacterial species harbored during treatment. Third, some different findings in patients with HCC and BTC were not exactly consistent. Whether the difference originated from cancer type or was due to the small sample size in the subgroup remains unclear, and further studies with large-scale sample is required to explore the difference between HCC and BTC. Fourth, host immune activation was not evaluated. The cross-talk between the host immune and gut microbiome may be the key mechanism modulating the antitumour immune response. Immunological assessments, such as peripheral immunophenotype and intratumour CD8 +T cell infiltration, are crucial to elucidate the relationship between the gut microbiome and the cancer immune response. Finally, FMT in mouse model or patient cohorts with CBR group-enriched taxa or feces is expected to confirm that the gut microbiome can modulate the clinical response to immunotherapy in hepatobiliary cancers.
Conclusions
In conclusion, we demonstrated that gut microbiome diversity and relative abundance might be an effective biomarker predicting the clinical benefit response and irAE profile of anti-PD-1 immunotherapy in patients with hepatobiliary cancers, which indicated that manipulation of the gut microbiome to modulate cancer immunotherapy might be a new therapeutic option for systemic treatment of hepatobiliary cancers.
Acknowledgments
We would like to appreciate all the participants gratefully, and tkank the Wekemo Tech Group Co., Ltd. Shenzhen China for metagenomics analysis.
Footnotes
JM, DW, JL and XY contributed equally.
Contributors: JM, DW, JL, XuY, JiL and HZ designed the study. JM, DW, XuY, YiS, FX, ZX, YaW, YuW, JX, BZ, JZ, JB and TZ collected stool samples and clinical data. JM, DW, XuY, JuL, FX, YL, HS and YaS performed the statistical analyses. JM, DW, JuL and XuY drafted the manuscript. JiL, XiY, LZ, XiS and HZ reviewed and revised the manuscript. HZ served as the guarantor in this study.
Funding: This work was supported by the Fundamental Research Funds for the Central Universities (3332020084 and 3332018032), the International Science and Technology Cooperation Projects (2016YFE0107100), the Capital Special Research Project for Health Development (2014-2-4012), the Beijing Natural Science Foundation (L172055 and 7192158), the National Ten-thousand Talent Program, the CAMS Innovation Fund for Medical Science (CIFMS) (2017-I2M-4-003 and 2018-I2M-3-001), the Shenzhen Science and Technology Plan (CKCY20180323174659823), and the National Science Foundation for Young Scientists of China (81802735). The funders had no role in the study design, data collection and analysis, decision to publish or preparation of the manuscript.
Competing interests: None declared.
Provenance and peer review: Not commissioned; externally peer reviewed.
Supplemental material: This content has been supplied by the author(s). It has not been vetted by BMJ Publishing Group Limited (BMJ) and may not have been peer-reviewed. Any opinions or recommendations discussed are solely those of the author(s) and are not endorsed by BMJ. BMJ disclaims all liability and responsibility arising from any reliance placed on the content. Where the content includes any translated material, BMJ does not warrant the accuracy and reliability of the translations (including but not limited to local regulations, clinical guidelines, terminology, drug names and drug dosages), and is not responsible for any error and/or omissions arising from translation and adaptation or otherwise.
Data availability statement
Data are available upon reasonable request.
Ethics statements
Patient consent for publication
Consent obtained directly from patient(s).
Ethics approval
This study was approved by the Ethics Committee of Peking Union Medical College Hospital (JS-2296). All participants provided written informed consent.
References
- 1.Sung H, Ferlay J, Siegel RL, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2021;71:209–49. 10.3322/caac.21660 [DOI] [PubMed] [Google Scholar]
- 2.Rizvi S, Khan SA, Hallemeier CL, et al. Cholangiocarcinoma - evolving concepts and therapeutic strategies. Nat Rev Clin Oncol 2018;15:95–111. 10.1038/nrclinonc.2017.157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Finn RS, Qin S, Ikeda M, et al. Atezolizumab plus bevacizumab in unresectable hepatocellular carcinoma. N Engl J Med 2020;382:1894–905. 10.1056/NEJMoa1915745 [DOI] [PubMed] [Google Scholar]
- 4.Finn RS, Qin S, Ikeda M, et al. IMbrave150: updated overall survival (OS) data from a global, randomized, open-label phase III study of atezolizumab (atezo) + bevacizumab (bev) versus sorafenib (SOR) in patients (PTS) with unresectable hepatocellular carcinoma (HCC). Journal of Clinical Oncology 2021;39:267. 10.1200/JCO.2021.39.3_suppl.267 [DOI] [Google Scholar]
- 5.NCCN Clinical Practice Guidelines in Oncology . Hepatobiliary cancers. Version 2 2021.
- 6.Llovet JM, Kelley RK, Villanueva A, et al. Hepatocellular carcinoma. Nat Rev Dis Primers 2021;7:6. 10.1038/s41572-020-00240-3 [DOI] [PubMed] [Google Scholar]
- 7.Valle JW, Kelley RK, Nervi B, et al. Biliary tract cancer. Lancet 2021;397:428–44. 10.1016/S0140-6736(21)00153-7 [DOI] [PubMed] [Google Scholar]
- 8.Schwabe RF, Jobin C. The microbiome and cancer. Nat Rev Cancer 2013;13:800–12. 10.1038/nrc3610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Skelly AN, Sato Y, Kearney S, et al. Mining the microbiota for microbial and metabolite-based immunotherapies. Nat Rev Immunol 2019;19:305–23. 10.1038/s41577-019-0144-5 [DOI] [PubMed] [Google Scholar]
- 10.Gopalakrishnan V, Helmink BA, Spencer CN, et al. The influence of the gut microbiome on cancer, immunity, and cancer immunotherapy. Cancer Cell 2018;33:570–80. 10.1016/j.ccell.2018.03.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Matson V, Chervin CS, Gajewski TF. Cancer and the microbiome-influence of the commensal microbiota on cancer, immune responses, and immunotherapy. Gastroenterology 2021;160:600–13. 10.1053/j.gastro.2020.11.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Routy B, Gopalakrishnan V, Daillère R, et al. The gut microbiota influences anticancer immunosurveillance and general health. Nat Rev Clin Oncol 2018;15:382–96. 10.1038/s41571-018-0006-2 [DOI] [PubMed] [Google Scholar]
- 13.Helmink BA, Khan MAW, Hermann A, et al. The microbiome, cancer, and cancer therapy. Nat Med 2019;25:377–88. 10.1038/s41591-019-0377-7 [DOI] [PubMed] [Google Scholar]
- 14.Sivan A, Corrales L, Hubert N, et al. Commensal Bifidobacterium promotes antitumor immunity and facilitates anti-PD-L1 efficacy. Science 2015;350:1084–9. 10.1126/science.aac4255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vétizou M, Pitt JM, Daillère R, et al. Anticancer immunotherapy by CTLA-4 blockade relies on the gut microbiota. Science 2015;350:1079–84. 10.1126/science.aad1329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bhatt AP, Redinbo MR, Bultman SJ. The role of the microbiome in cancer development and therapy. CA Cancer J Clin 2017;67:326–44. 10.3322/caac.21398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Routy B, Le Chatelier E, Derosa L, et al. Gut microbiome influences efficacy of PD-1-based immunotherapy against epithelial tumors. Science 2018;359:91–7. 10.1126/science.aan3706 [DOI] [PubMed] [Google Scholar]
- 18.Gopalakrishnan V, Spencer CN, Nezi L, et al. Gut microbiome modulates response to anti-PD-1 immunotherapy in melanoma patients. Science 2018;359:97–103. 10.1126/science.aan4236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Matson V, Fessler J, Bao R, et al. The commensal microbiome is associated with anti-PD-1 efficacy in metastatic melanoma patients. Science 2018;359:104–8. 10.1126/science.aao3290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Baruch EN, Youngster I, Ben-Betzalel G, et al. Fecal microbiota transplant promotes response in immunotherapy-refractory melanoma patients. Science 2021;371:602–9. 10.1126/science.abb5920 [DOI] [PubMed] [Google Scholar]
- 21.Davar D, Dzutsev AK, McCulloch JA, et al. Fecal microbiota transplant overcomes resistance to anti-PD-1 therapy in melanoma patients. Science 2021;371:595–602. 10.1126/science.abf3363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tanoue T, Morita S, Plichta DR, et al. A defined commensal consortium elicits CD8 T cells and anti-cancer immunity. Nature 2019;565:600–5. 10.1038/s41586-019-0878-z [DOI] [PubMed] [Google Scholar]
- 23.Schwabe RF, Greten TF. Gut microbiome in HCC - mechanisms, diagnosis and therapy. J Hepatol 2020;72:230–8. 10.1016/j.jhep.2019.08.016 [DOI] [PubMed] [Google Scholar]
- 24.Yu L-X, Schwabe RF. The gut microbiome and liver cancer: mechanisms and clinical translation. Nat Rev Gastroenterol Hepatol 2017;14:527–39. 10.1038/nrgastro.2017.72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Behary J, Amorim N, Jiang X-T, et al. Gut microbiota impact on the peripheral immune response in non-alcoholic fatty liver disease related hepatocellular carcinoma. Nat Commun 2021;12:187. 10.1038/s41467-020-20422-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang Q, Ma C, Duan Y, et al. Gut microbiome directs hepatocytes to recruit MDSCs and promote cholangiocarcinoma. Cancer Discov 2021;11:1248–67. 10.1158/2159-8290.CD-20-0304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ma C, Han M, Heinrich B, et al. Gut microbiome-mediated bile acid metabolism regulates liver cancer via NKT cells. Science 2018;360 10.1126/science.aan5931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zheng Y, Wang T, Tu X, et al. Gut microbiome affects the response to anti-PD-1 immunotherapy in patients with hepatocellular carcinoma. J Immunother Cancer 2019;7:193. 10.1186/s40425-019-0650-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Peng Z, Cheng S, Kou Y, et al. The gut microbiome is associated with clinical response to anti-PD-1/PD-L1 immunotherapy in gastrointestinal cancer. Cancer Immunol Res 2020;8:1251–61. 10.1158/2326-6066.CIR-19-1014 [DOI] [PubMed] [Google Scholar]
- 30.Health NIo . Common terminology criteria for adverse events (CTCAE). Version 5.0 2017.
- 31.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 2014;30:2114–20. 10.1093/bioinformatics/btu170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rosenbloom KR, Armstrong J, Barber GP, et al. The UCSC genome browser database: 2015 update. Nucleic Acids Res 2015;43:D670–81. 10.1093/nar/gku1177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods 2012;9:357–9. 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wood DE, Lu J, Langmead B. Improved metagenomic analysis with Kraken 2. Genome Biol 2019;20:257. 10.1186/s13059-019-1891-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lu J, Breitwieser FP, Thielen P, et al. Bracken: estimating species abundance in metagenomics data. PeerJ Comput Sci 2017;3:e104. 10.7717/peerj-cs.104 [DOI] [Google Scholar]
- 36.Jari Oksanen FGB, Friendly M, et al. Package vegan. community ecology package (version2) 2020:5–7.
- 37.Liu Y-X, Qin Y, Chen T, et al. A practical guide to amplicon and metagenomic analysis of microbiome data. Protein Cell 2021;12:315–30. 10.1007/s13238-020-00724-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Segata N, Izard J, Waldron L, et al. Metagenomic biomarker discovery and explanation. Genome Biol 2011;12:R60. 10.1186/gb-2011-12-6-r60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Buchfink B, Xie C, Huson DH. Fast and sensitive protein alignment using DIAMOND. Nat Methods 2015;12:59–60. 10.1038/nmeth.3176 [DOI] [PubMed] [Google Scholar]
- 40.Franzosa EA, McIver LJ, Rahnavard G, et al. Species-level functional profiling of metagenomes and metatranscriptomes. Nat Methods 2018;15:962–8. 10.1038/s41592-018-0176-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kanehisa M, Furumichi M, Tanabe M, et al. KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res 2017;45:D353–61. 10.1093/nar/gkw1092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Caspi R, Billington R, Ferrer L, et al. The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of pathway/genome databases. Nucleic Acids Res 2016;44:D471–80. 10.1093/nar/gkv1164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rooks MG, Garrett WS. Gut microbiota, metabolites and host immunity. Nat Rev Immunol 2016;16:341–52. 10.1038/nri.2016.42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Furusawa Y, Obata Y, Fukuda S, et al. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature 2013;504:446–50. 10.1038/nature12721 [DOI] [PubMed] [Google Scholar]
- 45.Roberti MP, Yonekura S, Duong CPM, et al. Chemotherapy-induced ileal crypt apoptosis and the ileal microbiome shape immunosurveillance and prognosis of proximal colon cancer. Nat Med 2020;26:919–31. 10.1038/s41591-020-0882-8 [DOI] [PubMed] [Google Scholar]
- 46.Sorbara MT, Littmann ER, Fontana E, et al. Functional and genomic variation between human-derived isolates of Lachnospiraceae reveals inter- and intra-species diversity. Cell Host Microbe 2020;28:134–46. 10.1016/j.chom.2020.05.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sinha SR, Haileselassie Y, Nguyen LP, et al. Dysbiosis-induced secondary bile acid deficiency promotes intestinal inflammation. Cell Host Microbe 2020;27:659–70. 10.1016/j.chom.2020.01.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Peters BA, Wilson M, Moran U, et al. Relating the gut metagenome and metatranscriptome to immunotherapy responses in melanoma patients. Genome Med 2019;11:61. 10.1186/s13073-019-0672-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jia X, Lu S, Zeng Z, et al. Characterization of gut microbiota, bile acid metabolism, and cytokines in intrahepatic cholangiocarcinoma. Hepatology 2020;71:893–906. 10.1002/hep.30852 [DOI] [PubMed] [Google Scholar]
- 50.Campbell C, McKenney PT, Konstantinovsky D, et al. Bacterial metabolism of bile acids promotes generation of peripheral regulatory T cells. Nature 2020;581:475–9. 10.1038/s41586-020-2193-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Song X, Sun X, Oh SF, et al. Microbial bile acid metabolites modulate gut RORγ+ regulatory T cell homeostasis. Nature 2020;577:410–5. 10.1038/s41586-019-1865-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Huang H, Ren Z, Gao X, et al. Integrated analysis of microbiome and host transcriptome reveals correlations between gut microbiota and clinical outcomes in HBV-related hepatocellular carcinoma. Genome Med 2020;12:102. 10.1186/s13073-020-00796-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Andrews MC, Duong CPM, Gopalakrishnan V, et al. Gut microbiota signatures are associated with toxicity to combined CTLA-4 and PD-1 blockade. Nat Med 2021;27:1432–41. 10.1038/s41591-021-01406-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dubin K, Callahan MK, Ren B, et al. Intestinal microbiome analyses identify melanoma patients at risk for checkpoint-blockade-induced colitis. Nat Commun 2016;7:10391. 10.1038/ncomms10391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chaput N, Lepage P, Coutzac C, et al. Baseline gut microbiota predicts clinical response and colitis in metastatic melanoma patients treated with ipilimumab. Ann Oncol 2017;28:1368–79. 10.1093/annonc/mdx108 [DOI] [PubMed] [Google Scholar]
- 56.Hakozaki T, Richard C, Elkrief A, et al. The gut microbiome associates with immune checkpoint inhibition outcomes in patients with advanced non-small cell lung cancer. Cancer Immunol Res 2020;8:1243–50. 10.1158/2326-6066.CIR-20-0196 [DOI] [PubMed] [Google Scholar]
- 57.Cortellini A, Di Maio M, Nigro O, et al. Differential influence of antibiotic therapy and other medications on oncological outcomes of patients with non-small cell lung cancer treated with first-line pembrolizumab versus cytotoxic chemotherapy. J Immunother Cancer 2021;9:e002421. 10.1136/jitc-2021-002421 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
jitc-2021-003334supp001.pdf (1.7MB, pdf)
jitc-2021-003334supp002.xlsx (43.7KB, xlsx)
Data Availability Statement
Data are available upon reasonable request.