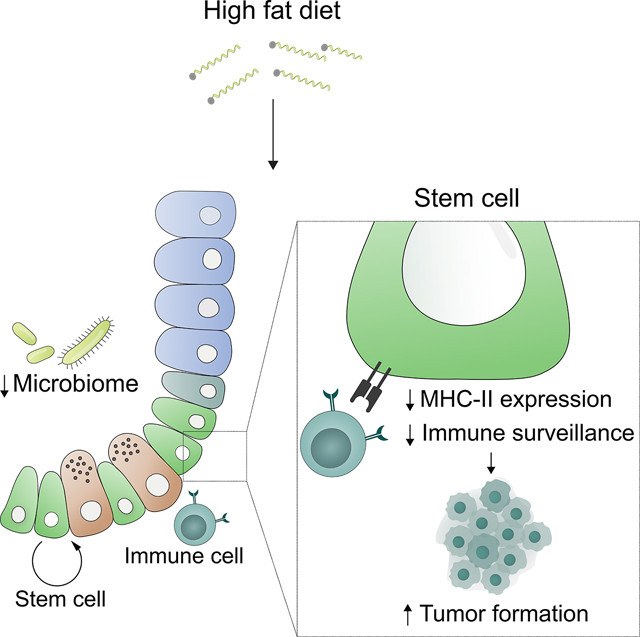
Summary
Little is known about how interactions between diet, intestinal stem cells (ISCs) and immune cells impact early stage intestinal tumorigenesis. We show that a high fat diet (HFD) reduces the expression of the major histocompatibility complex II (MHC-II) genes in intestinal epithelial cells including ISCs. This decline in epithelial MHC-II expression in a HFD correlates with reduced intestinal microbiome diversity. Microbial community transfer experiments suggest that epithelial MHC-II expression is regulated by intestinal flora. Mechanistically, pattern recognition receptor (PRR) and IFNg signaling regulate epithelial MHC-II expression. MHC-II− ISCs exhibit greater tumor-initiating capacity than their MHC-II+ counterparts upon loss of the tumor suppressor Apc coupled with a HFD, suggesting a role for epithelial MHC-II-mediated immune surveillance in suppressing tumorigenesis. ISC-specific genetic ablation of MHC-II increases tumor burden in a cell autonomous manner. Thus, HFD perturbs a microbiome–stem cell-immune cell interaction that contributes to tumor initiation in the intestine.
Keywords: Diet, Intestinal stem cells, Antigen presentation, MHC-II, Microbiome, Cancer
eTOC:
The mechanisms that link pro-obesity high fat (HFD) diets to increased colon cancer risk are not well understood. Beyaz and colleagues demonstrate that a HFD promotes intestinal tumor initiation by suppressing a microbiome – stem cell – immune cell crosstalk that is mediated by MHC-II expression on intestinal stem cells.
Graphical Abstract
Introduction
Diet is a major lifestyle factor that influences health and disease states, including cancer (Beyaz and Yilmaz, 2016; Zitvogel et al., 2017). Significant epidemiologic and preclinical studies link long-term dietary patterns such as western diets and obesity to several types of cancer, including colorectal cancer. However, how the adaptation of the intestinal epithelium to pro-obesity diets alters cancer risk remains elusive (Basen-Engquist and Chang, 2011; Calle et al., 2003; Gallagher and LeRoith, 2015). The intestinal epithelium is maintained by Lgr5+ intestinal stem cells (ISCs) that reside at the crypt base and give rise to the diverse, specialized cell types of the intestinal lining (Barker et al., 2007). These rapidly renewing ISCs coordinate intestinal adaptation in response to environmental inputs such as diet by balancing stem cell self-renewal with differentiation divisions (Beyaz et al., 2016; Wang et al., 2018; Yang et al., 2008; Yilmaz et al., 2012). ISCs are also the cells-of-origin for many early intestinal tumors and lie at the interface of dietary nutrients, commensal microbes and immune cells. Thus, understanding how diet induces changes in ISCs and their surrounding components may shed light on the early steps involved in initiation of colorrectal cancers (Barker et al., 2009; Belkaid and Hand, 2014; Clevers, 2013; Hooper et al., 2012; Thaiss et al., 2016).
Diet contributes to colorectal cancer risk through diverse cancer cell-intrinsic and -extrinsic mechanisms. While accumulating evidence demonstrates that pro-obesity or western diets enhance intestinal tumorigenesis in many ways such as through activating lipid-sensing PPAR transcription factors (Beyaz et al., 2016; Beyaz et al., 2021), bile acids (Fu et al., 2019a), altering vitamin D signaling (Li et al., 2019), inflammation (Font-Burgada et al., 2016), and the microbiome(Schulz et al., 2014), the role that immune cells play in this process is unclear. Because interactions between cancer cells and the immune system influence tumor initiation and progression, it is important to understand the crosstalk between tumor-initiating ISCs and immune cells. Recognition of antigens by T cells through antigen presentation pathways is a major mechanism for triggering anti-tumor immunity (Sade-Feldman et al., 2017; Vanneman and Dranoff, 2012). While MHC-I antigen presentation pathway-mediated activation of cytotoxic CD8+ T cells plays a major role in anti-tumor immune responses, MHC-II-mediated activation of CD4+ T cells is also pivotal for tumor immunity. CD4+ T cells can help the maintenance of CD8+ T cell responses and produce cytokines that shape the overall response against the tumor (Haabeth et al., 2016; Hirschhorn-Cymerman et al., 2012; Hung et al., 1998; Tran et al., 2014; Xie et al., 2010; Zhang et al., 2009). Although cancers develop several strategies to evade the immune system (Grasso et al., 2018; Liu et al., 2017), little is known about how diet-induced obesity impacts the immune recognition of ISCs during tumor initiation.
Results
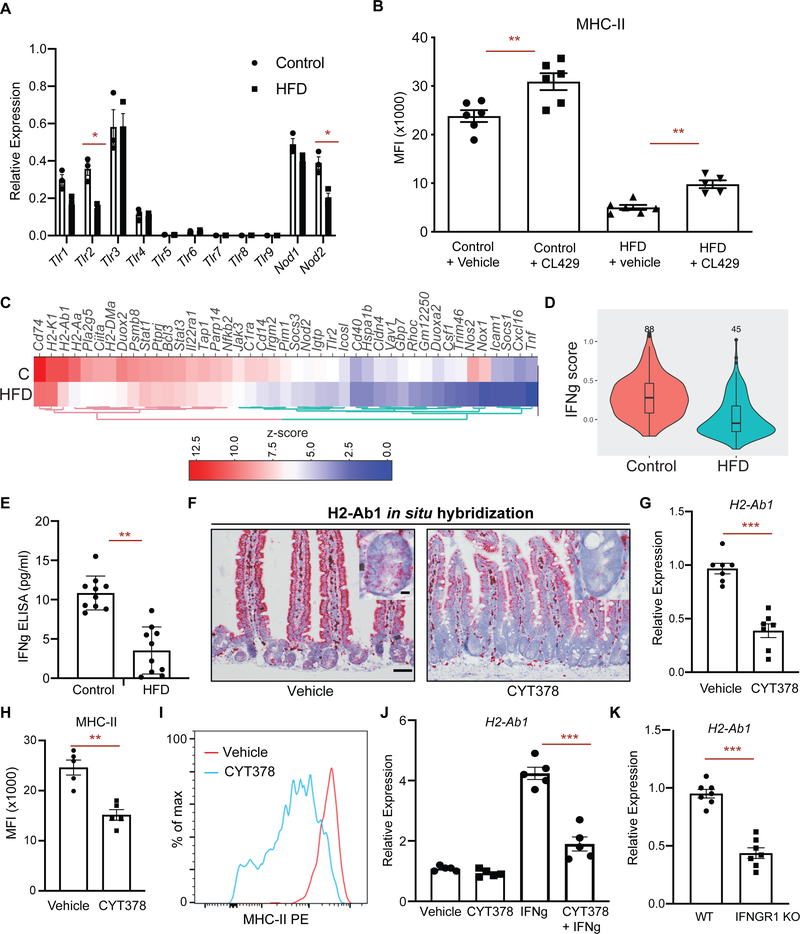
HFD dampens epithelial MHC-II expression
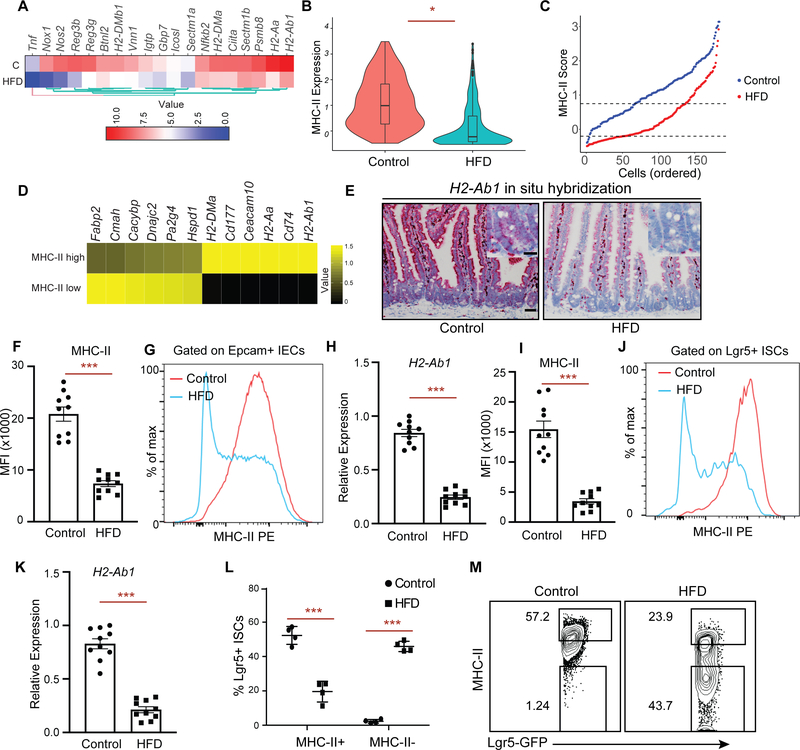
To explore how a high fat diet (HFD) perturbs immunomodulatory gene expression in ISCs, we examined our previous mRNA-sequencing (RNA-seq) dataset of Lgr5+ ISCs isolated from control and HFD-fed mice (Beyaz et al., 2016). We found that Lgr5+ ISCs derived from HFD-fed mice significantly downregulate immunomodulatory genes that are involved in the MHC-II pathway (H2-Aa, H2-Ab1, Ciita), anti-microbial response/inflammation (Reg3g, Nfkb2) and costimulation (Icosl, Sectm1a, Sectm1b) (Choi et al., 2011; Dong et al., 2001; Howie et al., 2013; Mukherjee and Hooper, 2015; Tomas et al., 2016) (Fig. 1A, S1A). Given the recently reported heterogeneity within Lgr5+ ISCs (Barriga et al., 2017; Biton et al., 2018), we next performed single-cell RNA-Seq (scRNA-Seq) of Lgr5+ ISCs derived from control or HFD mice. Consistent with the bulk RNA-seq profiles, H2-Ab1, a key component of MHC-II complex, was among the top 5 differentially expressed genes with > 3-fold higher expression in control compared to HFD ISCs (Fig. 1B, S1B, C) (see methods, MAST test, p < 1e-10)). To determine the extent of MHC-II downregulation in response to HFD in our scRNA-seq data, we ranked individual ISCs based on their expression pattern of MHC-II pathway genes (MHC-II score, Supplementary Table 1) and found that HFD Lgr5+ ISCs had consistently lower MHC-II score compared to control Lgr5+ ISCs (Fig. 1C). We then selected the upper and lower quartiles in scRNA-seq data and performed differential expression analysis between MHC-II low and MHC-II high HFD Lgr5+ ISCs. As expected, MHC-II pathway genes (H2-Ab1, Cd74, H2-Aa, H2-DMa) were among the top upregulated genes in MHC-II high cells (Fig. 1D).
Figure 1 |. High Fat Diet reduces MHC-II expression on Lgr5+ intestinal stem cells (ISCs).
A. A heat map of downregulated genes assessed by bulk RNA-seq in Lgr5-GFPhi intestinal stem cells (Lgr5+ ISCs) isolated from long-term high fat diet (HFD)-fed mice compared to control mice (n=2). Scale represents computed z-scores of log10 expression values.
B. Violin plots demonstrating MHC-II expression in single Lgr5+ ISCs isolated from control (n=171 cells, 2 independent experiments) or HFD mice by single cell RNA-seq (scRNA-seq) (n=144 cells, 2 independent experiments).
C. Control (blue) and HFD (red) Lgr5+ ISCs ranked according to their expression of MHC II pathway genes (y-axis). Dashed lines correspond to y-intercepets of −0.2 and 0.75, which are the 25th and 75th percentile of scores in HFD cells, used to define MHC-II low (score < −0.2) and MHC-II high (score > 0.75) HFD cells. In contrast, these values correspond to 1st and 35th percentile of scores in control cells.
D. Heatmap showing differentially expressed (DE) genes (rows) between MHC-II low and MHC-II high HFD ISCs as defined in panel C. Scale represents computed z-scores of log10 expression values.
E. Single-molecule in situ hybridization of MHC-II (H2-Ab1) in control and HFD mice (n=5).
F, G. Mean fluorescence intensity (MFI) of MHC-II in Epcam+ intestinal epithelial cells (IECs) from crypts of control and HFD mice (F, n=10 mice, mean ± s.e.m.). Representative flow cytometry histogram plots of MHC-II expression in Epcam+ IECs (G).
H. Relative expression of MHC-II (H2-Ab1) in Epcam+ IECs isolated from crypts of control and HFD mice (n=10 mice, mean ± s.e.m.).
I, J. Mean fluorescence intensity (MFI) of MHC-II in Lgr5+ ISCs from crypts of control and HFD mice (I, n=10 mice, mean ± s.e.m.). Representative flow cytometry histogram plots of MHC-II expression in Epcam+ cells (J).
K. Relative expression of MHC-II (H2-Ab1) in Lgr5+ ISCs isolated from crypts of control and HFD mice (n=10 mice, mean ± s.e.m.).
L, M. Frequency of MHC-II+ and MHC-II− Lgr5+ ISCs in control and HFD mice by flow cytometry (L, n=4, mean ± s.d). Representative flow cytometry plots of MHC-II in control and HFD ISCs (M).
*P < 0.05, **P < 0.01, ***P < 0.001 (Student’s t-tests). Scale bars, 50 μm (E) and 20 μm (E, inset).
Although MHC-II expression and function is generally considered to be restricted to professional antigen presenting cells, several studies have demonstrated that intestinal epithelial cells (IECs) express high levels of MHC-II and are able to capture, process and present antigens to CD4+ T cells, including recent work from our group on ISCs (Biton et al., 2018; Cerf-Bensussan et al., 1984; Dahan et al., 2007; Hershberg et al., 1997; Telega et al., 2000; Thelemann et al., 2014; Westendorf et al., 2009) (Fig. S1G). We validated the reduction of MHC II expression on IECs and Lgr5+ ISCs under a HFD in several ways. In situ hybridization for H2-Ab1 showed that MHC-II is expressed in the epithelium of control mice, but has diminished expression throughout the epithelium of HFD mice, including in the intestinal crypt where ISCs reside (Fig. 1E). Moreover, by flow cytometry, both Epcam+ IECs (Fig. 1F, G) and Lgr5+ ISCs (Fig. 1I, J) expressed high levels of MHC-II protein on their cell surface at steady state, and this expression was significantly downregulated in response to a HFD. Finally, we confirmed that sorted Epcam+ IECs (Fig. 1H) and Lgr5+ ISCs (Fig. 1K) from HFD-fed mice significantly downregulated MHC-II expression by qPCR. We then partitioned Lgr5+ ISCs into two subpopulations based on their MHC-II expression pattern: MHC-II+ and MHC-II− and assessed their frequencies in control or HFD-fed mice. While in control mice most Lgr5+ ISCs were MHC-II+, a HFD led to a significant reduction in the frequency of MHC-II+ Lgr5+ ISCs and to a concomitant increase in the frequency of MHC-II− Lgr5+ ISCs (Fig. 1L, M). We sorted these ISC subpopulations to confirm MHC-II expression levels. Consistent with our scRNA-seq analysis, the expression levels of H2-Ab1 mRNA were significantly reduced in response to HFD (Fig. S1D). Altogether, these results indicate that HFD leads to suppression of MHC-II expression in Epcam+ IECs including Lgr5+ ISCs.
PPAR- δ activation or obesity does not affect MHC-II expression in Lgr5+ ISCs
A HFD perturbs multiple epithelial-intrinsic and -extrinsic pathways that may influence the regulation of MHC-II expression in ISCs(Fu et al., 2019a; Schulz et al., 2014). Because our prior findings demonstrated that a PPAR-δ program mediates many of the effects of a HFD in Lgr5+ ISCs (Beyaz et al., 2016; Beyaz et al., 2021), we next assessed the activation status of a PPAR-δ program in Lgr5+ ISCs based on their MHC-II expression pattern in response to a HFD. We found no difference in the expression levels of PPAR-δ-induced signature genes Hmgcs2 and Jag1, or the stem cell marker Lgr5 in MHC-II+ and MHC-II− ISCs in either cohort (Fig. S1E). Interestingly, agonist-induced PPAR-δ activation with GW501516 also did not reduce MHC-II expression in Lgr5+ ISCs, indicating that HFD-induced MHC-II downregulation is independent of PPAR-δ activity in ISCs (Fig. S1I, J). To determine whether MHC-II expression in Lgr5+ ISCs is reduced in an independent model of obesity, we examined leptin receptor deficient (db/db) mice, an obesity model that develops on a control diet(Coleman, 1978). (Fig. S1H). Lgr5+ ISCs isolated from both lean control (db/+) and obese (db/db) mice expressed high levels of MHC-II, indicating that diet-induced alterations rather than obesity per se inhibits MHC-II expression in ISCs (Fig. S1K, L).
Epithelial MHC-II expression does not influence organoid-forming capacity of ISCs
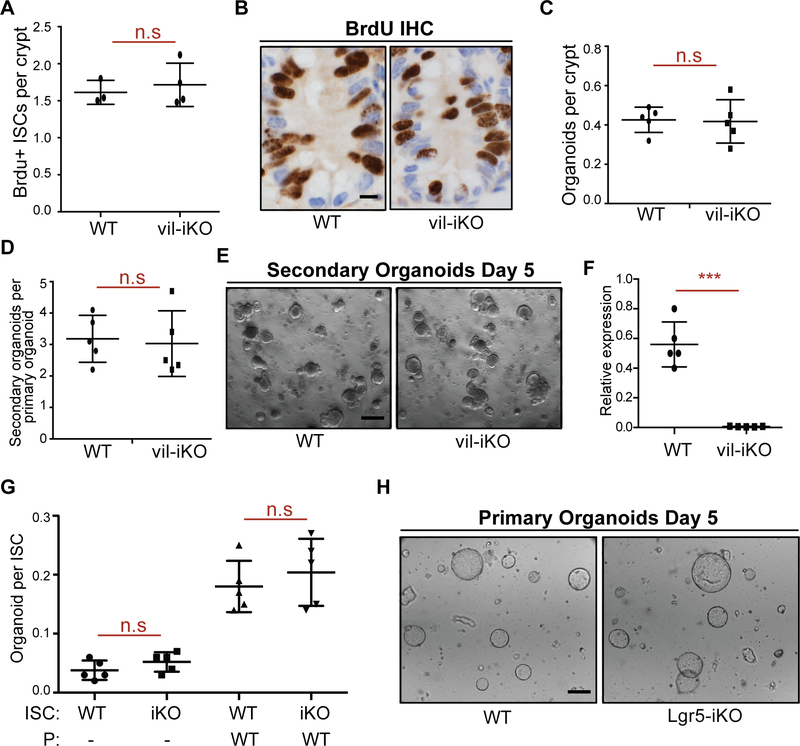
We next assessed whether HFD-mediated downregulation of MHC-II impacts stemness. First, we assayed the functional potential of MHC-II+ and MHC-II− ISCs, using an in vitro approach based on the ability of isolated Lgr5+ ISCs to form organoid bodies in 3-D culture (Beyaz et al., 2016; Sato et al., 2009b). We found that MHC-II expression levels in control and HFD Lgr5+ ISCs did not affect in vitro organoid formation (Fig. S1F). To further ascertain whether MHC-II expression regulates ISC function, we generated mice with intestinal epithelium-specific MHC-II deletion (vil-iKO; see Methods). Intestinal specific loss of MHC-II did not affect the weight and length of the small intestine or impact crypt depth and villi height (Fig. S2A–G). Furthermore, the proliferation or organoid-forming capacity of ISCs was unaltered upon loss of MHC-II in the intestinal epithelium (Fig. 2A–F). To assess how MHC-II regulates ISC function specifically, we ablated MHC-II in Lgr5+ ISCs (Lgr5-iKO; Fig. S2H, see Methods) and found that loss of MHC-II in Lgr5+ ISCs and their progenies did not affect the capacity of ISCs to initiate organoids (Fig. 2G–H). To determine how MHC-II influences Lgr5+ ISC function in vivo, we performed LacZ lineage tracing analysis and did not detect differences in the generation of LacZ+ cells between WT and Lgr5-iKO mice (Fig. S2I). While these data illustrate that the in vitro organoid-forming capacity of Lgr5+ ISCs is independent of MHC-II expression and that MHC-II loss does not alter intestinal homeostasis and ISC output (i.e LacZ+ progeny) at steady state, MHC-II likely influences ISC differentiation by engaging T helper cell cytokines in the context of inflammation, as we have previously reported (Biton et al., 2018).
Figure 2 |. Intestine-specific deletion of MHC-II does not alter the organoid forming capacity of ISCs.
A, B. Number of bromodeoxyuridine (BrdU)+ crypt base columnar cells after a 4-hour pulse in MHC-IIL/L; Villin-CreERT2 (vil-iKO) mice after tamoxifen administration (WT: n=3, vil-iKO: n=4, mean ± s.d). Representative images of BrdU immunostain in proximal small intestinal crypts (B).
C-E Organoid-initiating capacity of WT and vil-iKO crypts (C, n=5, mean ± s.d). Number of secondary organoids per dissociated crypt-derived primary organoid (D, n=5, mean ± s.d). Representative images of day-5 WT and vil-iKO primary organoids (E).
F. Relative expression of MHC-II in dissociated WT and vil-iKO primary organoids at day 5 (n=5, mean ± s.d).
G, H. Organoid-initiating capacity of ISCs from WT and MHC-IIL/L; Lgr5-EGFP-IRES-CreERT2 (Lgr5-iKO) mice with and without Paneth cells (P) from WT mice (n=5, mean ± s.d). Representative images of organoids derived from WT and Lgr5-iKO ISCs co-cultured with WT Paneth cells five days after seeding (H).
n.s.: not significant, ***P < 0.001 (Student’s t-tests). Scale bars, 100 μm (E, H) and 20 μm (B).
See also Figure S2
Epithelial MHC-II expression depends on the intestinal microbiome
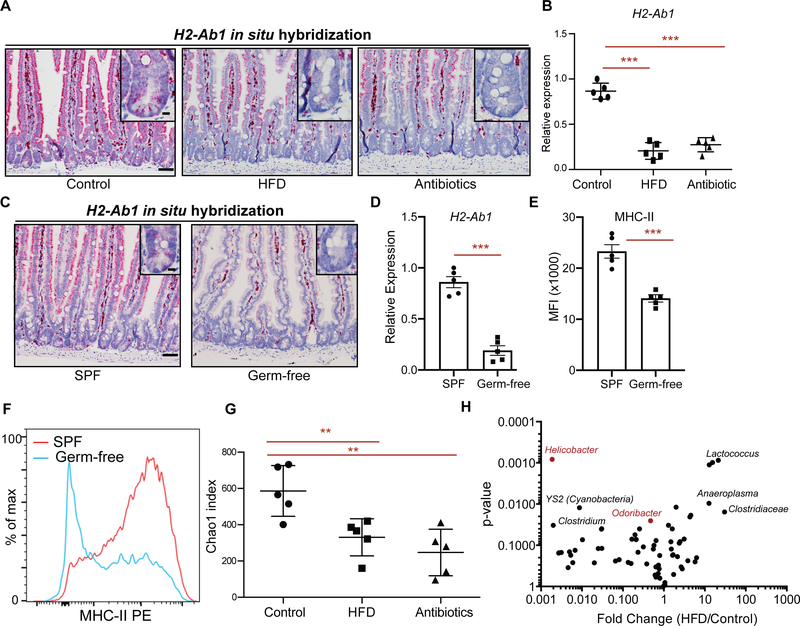
The intestinal microbiome plays a significant role in regulating intestinal immunity (Belkaid and Hand, 2014; David et al., 2014; Hooper et al., 2012; Round and Mazmanian, 2009; Thaiss et al., 2016). Since dietary perturbations are among the major external factors shaping the intestinal microbiome, we asked whether HFD-induced alterations in the microbiome influence MHC-II expression in IECs and ISCs. To determine whether the microbiome is involved in regulation of epithelial MHC-II levels, we treated mice with broad-spectrum antibiotics, which ablated bacterial diversity and massiviely altered community composition (Fig. S2J). Notably, similar to a HFD, antibiotic treatment was accompanied by decreased MHC-II expression in Lgr5+ ISCs and the intestinal epithelium (Fig. 3A, B), comparable to that observed in HFD. To further corroborate the role of the intestinal microbiome on MHC-II expression on ISCs, we generated germ-free Lgr5-GFP-IRES-CreERT2 mice. Lgr5+ ISCs from germ-free mice exhibited reduced MHC-II expression both at RNA and protein levels compared to specific pathogen-free control mice (Fig. 3C–F, S2J, K).
Figure 3 |. Intestinal microbiome regulates epithelial MHC-II expression.
A. In situ hybridization for H2-Ab1 in control, HFD, and antibiotic-treated mice in proximal small intestine (n=5 mice).
B. Relative expression of MHC-II in Lgr5+ ISCs from control, HFD, and antibiotic-treated mice (n=5, mean ± s.d).
C. Single-molecule in situ hybridization of MHC-II (H2-Ab1) in specific pathogen-free (SPF) and germ-free mice (n=5 mice).
D. Relative expression of MHC-II (H2-Ab1) in Lgr5+ ISCs from SPF and germ-free mice (n=5, mean ± s.e.m.).
E, F. Mean fluorescence intensity (MFI) of MHC-II in Lgr5+ ISCs from SPF and germ-free mice (E, n=5, mean ± s.e.m.). Representative flow cytometry histogram plots of MHC-II expression in Lgr5+ ISCs (F).
G. Chao1 index of microbial diversity in Control, HFD, and mice treated with antibiotics for 3 months (n=5, mean ± s.d).
H. Volcano plot demonstrating significantly enriched and depleted microbial species in HFD versus control mice (n=5).
**P < 0.01, ***P < 0.001 (Student’s t-tests). Scale bars, 100 μm (A, C) and 20 μm (A, C, insets).
See also Figure S2
Helicobacter colonization correlates with epithelial MHC-II expression
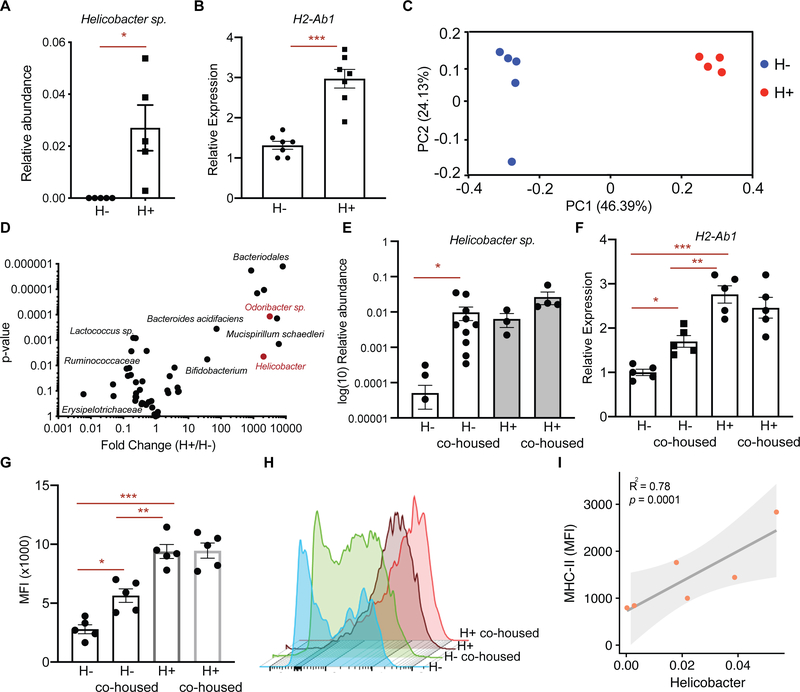
To gain insight into the spectrum of members of the bacterial microbiome capable of inducing epithelial MHC-II expression, we performed comparative 16S rDNA sequencing in fecal DNA isolated from HFD-treated mice and controls. Consistent with previous reports, HFD-induced obesity led to microbial dysbiosis with reduced bacterial diversity in the feces (Fig. 3G, S2J) (Ley et al., 2005; Schulz et al., 2014). Among the bacterial genera strongly diminished under HFD conditions was Helicobacter (Fig. 3H). To determine whether Helicobacter colonization in mice correlates with the epithelial MHC-II expression in the intestine, we surveyed our mouse facility for presence or absence of Helicobacter species in mice housed in different rooms. We determined two separate rooms with mice either naturally colonized with Helicobacter species (H+, “dirty room”) or without detectable Helicobacter species (H−, “clean room”) (Fig. 4A). Helicobacter species that naturally colonize mice in H+ room and are absent in H− room include Helicobacter spp., Helicobacter mastomyrinus and Helicobacter typhlonius (Supplementary Table 2). Interrestingly, MHC-II expression was significantly higher in the intestinal epithelium of mice that are housed in H+ room compared to the mice housed in H− room (Fig. 4B). Comparison of microbial composition between H+ mice and H− mice revealed bacteria that are more abundant in H+ mice and associate with high epithelial MHC-II expression (Fig. 4C, D). These include Helicobacter sp. (Fig. 4A, D) and Odoribacter sp. (Fig 4D, S2L), which were both reduced in response HFD (Fig. 3H). To determine whether colonization of H− mice with the microbial flora of the H+ mice enhances expression of epithelial MHC-II, we co-housed H− mice with H+ mice in the H+ room (Fig. S2M). While H− and H+ mice have distinct microbial composition at baseline as determined by principal coordinate analysis (PCoA), after co-housing the microbial profile of H− mice resembles the H+ mice (Fig. S2M). Importantly, co-housing with H+ mice led to a significant increase in both Helicobacter sp. and Odoribacter sp. abundance in the co-housed H− mice with a concomitant upregulation of epithelial MHC-II compared to mice that remained in the H− room (Fig. 4E–H, Fig. S2N). Finally, we performed fecal transplantation into gnotobiotic mice and found that Helicobacter colonization correlates with epithelial MHC-II expression levels in the intestine (Fig. 4I). These results indicate that epithelial MHC-II expression is regulated by intestinal commensal bacteria including Helicobacter sp. that is dampened in response to a HFD.
Figure 4 |. Helicobacter colonization correlates with epithelial MHC-II expression.
A. Relative abundance of Helicobacter spp. in mice housed in clean (H−) room and dirty (H+) room (n=5 mice, mean ± s.e.m.).
B. Relative expression of MHC-II (H2-Ab1) in Epcam+ cells isolated from crypts of mice housed in H− room or H+ room (n=7, mean ± s.e.m.).
C. Principal coordinate analysis (PCoA) of microbial composition in feces of mice housed in H− room or H+ room (n=5).
D. Volcano plot demonstrating significantly enriched and depleted microbial species in mice housed in H+ room versus H− room (n=5).
E. Relative abundance of Helicobacter sp. in mice housed either in H− room (n=10), H+ room (n=3), or after co-housing H− mice (n=10) with H+ mice (n=4) in H+ room (mean ± s.e.m.).
F. Relative expression of MHC-II (H2-Ab1) in Epcam+ cells isolated from crypts of mice housed either in H− room, H+ room, or after co-housing H− mice with H+ mice in H+ room (n=5, ANOVA).
G, H. Mean fluorescence intensity (MFI) of MHC-II in Epcam+ cells isolated from crypts of mice housed either in H− room, H+ room, or after co-housing H− mice with H+ mice in H+ room (G, n=5, mean ± s.e.m., ANOVA). Representative flow cytometry histogram plots of MHC-II expression in Lgr5+ ISCs (H).
I. Correlation of MHC-II expression with Helicobacter abundance in germ-free mice that were transplanted with fecal content from H− and H+ mice.
Unless otherwise indicated *P < 0.05, **P < 0.01, ***P < 0.001 (Student’s t-tests).
Pattern recognition receptor (PRR) and IFN γ signaling regulate epithelial MHC-II expression
Microbiome-induced activation of pattern recognition receptor (PRR) signaling or proinflammatory cytokines induce MHC-II expression in antigen presenting cells and control intestinal homeostasis (Abreu, 2010; Rakoff-Nahoum et al., 2004; van den Elsen, 2011). To determine the necessity of PRR signaling through the adaptor protein Myd88 in regulating MHC-II expression, we generated intestine-specific Myd88-deficient mice (Myd88 iKO) and found significant downregulation of epithelial MHC-II compared to wild-type controls (Fig. S3C, D). Next, we surveyed PRR expression patterns in ISCs in control and HFD conditions. Consistent with previous reports (Brown et al., 2014; Caruso et al., 2014; Neal et al., 2012; Price et al., 2018), ISCs expressed several Toll-like receptors (Tlr1, Tlr2, Tlr3 and Tlr4), as well as Nod-like receptors (Nod1, Nod2) (Fig. 5A). Among these PRRs, Tlr2 and Nod2 were downregulated in response to a HFD (Fig. 5A). To test whether activation of TLR2 and NOD2 pathways is sufficient to increase MHC-II expression in Lgr5+ ISCs, we treated mice with a dual TLR2/NOD2 agonist (CL429) (Pavot et al., 2014). Indeed, CL429 treatment led to significant upregulation of MHC-II in both control and HFD ISCs (Fig. 5B, S3A, B). Lastly, signaling through TLR2 and NOD2 induced MHC-II expression; however, it did not fully restore MHC-II levels of HFD-treated mice to control levels, suggesting that additional signaling pathways are needed to achieve baseline expression (Fig. 5B, S3A, B).
Figure 5 |. PRR and IFN γ signaling regulate epithelial MHC-II expression.
A. Relative expression of pattern recognition receptors (PRR) in control and HFD ISCs (n=3).
B. Mean fluorescence intensity (MFI) of MHC-II in Lgr5+ ISCs from vehicle- and TLR2/NOD2 agonist CL429− treated control and HFD mice (n=6 mice, mean ± s.e.m.).
C. A heat map of expression levels of IFN γ-induced genes between HFD and control Lgr5+ ISCs by bulk RNA-seq (n=2). Scale represents computed z-scores of log10 expression values.
D. Violin plots demonstrating the expression levels of IFN γ-induced genes in control and HFD Lgr5+ ISCs by scRNA-seq.
E. IFN γ levels in the intestines of control and HFD mice as measured by ELISA (n=10, mean ± s.e.m.).
F. In situ hybridization for H2-Ab1 in vehicle- and JAK1/2 & TBK1/IKKε inhibitor (CYT387)-treated mice in small intestine (n=3).
G. Relative expression of MHC-II (H2-Ab1) in Lgr5+ ISCs from vehicle- and CYT387-treated mice (n=7, mean ± s.e.m.).
H, I. Mean fluorescence intensity (MFI) of MHC-II in Lgr5+ ISCs from vehicle- and CYT387-treated mice (H, n=5, mean ± s.e.m.). Representative flow cytometry histogram plots of MHC-II expression in Lgr5+ ISCs (I).
J. Relative expression of MHC-II (H2-Ab1) in intestinal organoids-treated with or without CYT387 and/or IFN γ (n=5, mean ± s.e.m.).
K. Relative expression of MHC-II (H2-Ab1) in Epcam+ cells isolated from crypts of control or IFNGR1 KO (n=5, mean ± s.e.m.).
*P < 0.05, **P < 0.01, ***P < 0.001 (Student’s t-tests).
The intestinal microbiome also influences the activity of proinflammatory cytokine signaling, such as the proinflammatory cytokine interferon-gamma (IFN γ) and downstream JAK1/2 signaling pathways (van den Elsen, 2011). IFN γ is a potent inducer of MHC-II expression in antigen presenting cells and other cell types including intestinal epithelial cells (Collins et al., 1984; Thelemann et al., 2014; Wong et al., 1984). We examined the IFNγ responsiveness of MHC-II+ and MHC-II− HFD ISCs, which express similar levels of the IFNγ receptors Ifngr1 and Ifngr2 (Fig S1E). IFNγ treatment led to upregulation of MHC-II in both MHC-II+ and MHC-II− Lgr5+ ISCs (Fig. S3E). Although HFD Lgr5+ ISCs express lower levels of MHC-II, they are still responsive to IFNγ stimulation (Choi et al., 2011; Thelemann et al., 2014). To determine whether HFD leads to suppression of epithelial MHC-II expression through dampening IFN γ signaling, we assessed the expression levels of IFN γ-induced genes in control and HFD Lgr5+ ISCs. In response to HFD, we found that ISCs significantly downregulate IFN γ-induced genes, including MHC-II pathway genes (H2-Ab1, H2-Aa, Ciita, Cd74, H2-DMa) and upstream pathway genes that regulate MHC-II expression (Stat1, Stat3, Nfkb2, Jak3) (Fig. 5C, D, S3F, G). Of note, the levels of IFNγ were significantly reduced in the intestines of HFD mice compared to control mice (Fig. 5E). Notably, administration of a potent JAK1/2 & TBK1/IKKε inhibitor (CYT387) that inhibits both STAT and NFκB signaling to mice significantly decreased MHC-II expression in Lgr5+ ISCs compared to vehicle-treated controls (Fig. 5F–I, S3H, I) (Tyner et al., 2010). We next asked whether IFN γ-induced epithelial MHC-II expression is inhibited by CYT387 in in vitro organoid assays and found that CYT387 blunted the induction of MHC-II expression in response to IFN γ (Fig. 5J). We also assessed the necessity of IFN γ-signaling in regulating epithelial MHC-II expression by utilizing IFNγ receptor-1 (IFNGR1) knockout mice and found dampened epithelial MHC-II expression in IFNGR1 knockout mice compared to wild-type controls (Figure 5K). The HFD-mediated reduction in IFN γ-inducible gene expression in ISCs suggests that intestinal immune cells may be depleted in the intestinal epithelium. Indeed, we observed significant reduction of CD45+ immune infiltrates, such as CD3+, CD8+ and CD4+ T cells, involving the crypt epithelium (Figure S4A–H). Altogether, these results suggest that PRR and IFN γ signaling drive MHC-II expression in the intestinal epithelial cells, including Lgr5+ ISCs, and that HFD feeding attenuates MHC-II expression in ISCs by suppressing these pathways and are in accord with prior studies that demonstrated reduced inflammatory gene expression and immune infiltration in HFD intestinal crypts (Beyaz et al., 2016; Johnson et al., 2015; Schulz et al., 2014).
Dampening MHC-II expression in premalignant Lgr5+ ISCs increases intestinal tumorigenesis
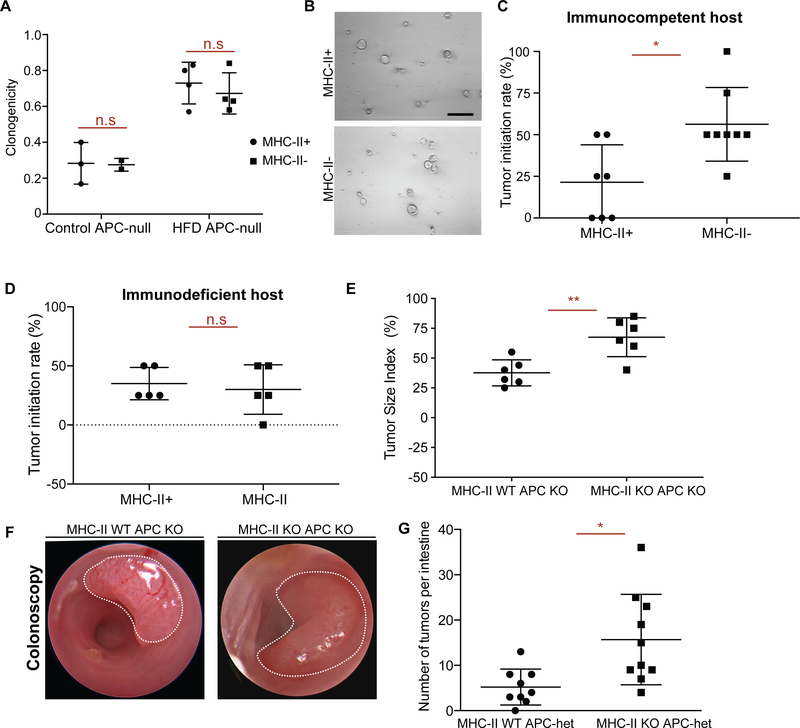
Recognition of antigens by T cells through antigen presentation pathways is a key mechanism for triggering anti-tumor immunity (Vanneman and Dranoff, 2012). Dampening the expression of genes involved in antigen presentation to evade anti-tumor immune responses is a hallmark of many cancers and correlates with poor prognosis (Lovig et al., 2002; Rimsza et al., 2004; Tarafdar et al., 2017). To test whether MHC-II expression patterns influence the tumor initiation potential of premalignant Lgr5+ ISCs, we fed Lgr5-CreERt2;ApcL/L mice control diet or HFD diet prior to tamoxifen-induced inactivation of the Apc tumor suppressor gene in Lgr5+ ISCs (APCnull). HFD led to strong downregulation of MHC-II in premalignant APCnull Lgr5+ ISCs at both the RNA and protein levels compared to controls (Fig. S5A, C). Interestingly, as we observed in the non-neoplastic intestine (Fig. 2G, H), MHC-II expression levels of APCnull Lgr5+ ISCs did not influence their ability to form adenomatous organoids in vitro (Fig. 6A, B), indicating that the cell-intrinsic oncogenic potential of premalignant APCnull ISCs in organoid cultures that lack immune cells is independent of MHC-II.
Figure 6 |. Loss of MHC-II in premalignant ISCs increases tumor initiation.
A, B. Organoid-initiating capacity of control and HFD MHC-II+ and MHC-II− Apcnull Lgr5+ ISCs at day 5 (control MHC-II+, n=3, control MHC-II-, n=3, HFD MHC-II+, n=4, HFD MHC-II-, n=4, mean ± s.d.). Representative images of HFD MHC-II+ and MHC-II− Apcnull organoids at day 5 (B).
C. Tumor initiation rate of orthotopically transplanted MHC-II+ and MHC-II− Apcnull Lgr5+ ISCs from HFD mice into immunocompetent syngeneic hosts (n=8).
D. Tumor initiation rate of orthotopically transplanted MHC-II+ and MHC-II− Apcnull Lgr5+ ISCs from HFD mice into immunodeficient (Rag2 KO) hosts (n=8).
E, F. Tumor size index in distal colon of mice that received tamoxifen through endoscopy guided tamoxifen injection to induce tumor formation upon loss of APC (E, n=6, MHC-II WT APC KO: villin-Cre-ERT2 APCL/L, MHC-IIL/+, MHC-II KO APC KO: villin-Cre-ERT2 APCL/L, MHC-IIL/L). Representative optical colonoscopy images of tumors (F).
G. Number of tumors per small intestine in Lgr5-CreERT2 MHC-IIL/+, APCL/+ (n=9, MHC-II WT APC-het) and Lgr5-CreERT2 MHC-IIL/L, APCL/+ (n=9, MHC-II KO APC KO) mice 5 months post tamoxifen injection.
Unless otherwise indicated, data are mean ± s.e.m from n independent experiments; n.s.: not significant, *P < 0.05, **P < 0.01 (Student’s t-tests).
To decipher whether HFD-mediated reduction in MHC-II expression on APCnull Lgr5+ ISCs impacts tumorigenesis in vivo, we utilized a recently developed orthotopic, syngeneic colon transplantation assay in mice (Beyaz et al., 2016; Roper et al., 2018; Roper et al., 2017a). We sorted APCnull MHC-II+ or APCnull MHC-II− Lgr5+ ISCs by flow cytometry and transplanted these populations into the colonic mucosa/submucosa of syngeneic, immune-competent mice. In contrast to the in vitro organoid assay, APCnull MHC-II− Lgr5+ ISCs exhibited 2-fold greater tumorigenicity when transplanted in vivo than their APCnull MHC-II+ counterparts (Fig. 6C, S5D–F). Remarkably, when transplanted into immunodeficient Rag2-null host colons that lack adaptive B and T immune cells, both MHC-II+ and MHC-II− APCnull premalignant cells gave rise to equal numbers of tumors, highlighting a criticial role for MHC-II recognition by adaptive immune cells in controlling intestinal tumor initiation (Fig. 6D, S5G–I). Finally, to ascertain the specific role that epithelial MHC-II plays during intestinal tumorigenesis, we generated two genetic mouse models: First, using epithelial-specific inducible deletion model of MHC-II together with tumor suppressor Apc (vilCreERT2; ApcL/L; MHC-IIL/L), we initated single tumors in the distal colon by administering 4-OHT with our endoscopy-guided injection system. We found that mice with the MHC-II-deficient allele (MHC-II KO: vilCreERT2; APCL/L, MHC-IIL/L) engendered larger tumors compared to mice with the MHC-sufficient allele (WT: vilCreERt2; APCL/L, MHC-IIL/+) (Fig 6E, F). Second, we initiated tumors using an Lgr5+ ISC-specific inducible deletion model of MHC-II together with one copy of the tumor suppressor Apc that leads to intestinal tumor formation due to loss of heterozygosity (Lgr5CreERT2; MHC-IIL/+ or L/L; APCL/+). We found that loss of MHC-II specifically in Lgr5+ ISCs and their progeny was associated with greater numbers of intestinal tumors compared to their MHC-II-proficient counterparts (Fig. 6G, S5J–L). Furthermore, we found that intestinal specific deletion of Apc resulted in the exclusion of both CD4+ and CD8+ T cells from in established tumors in both MHC-II-deficient and MHC-II-proficient setting (Fig. S5M–O, S6A–E), as has been previously reported in tumors with Wnt/β-catenin pathway activation (Kettunen et al., 2003; Luke et al., 2019). Altogether, these results illustrate that HFD-mediated reduction or genetic inactivation of MHC-II expression in premalignant ISCs enhances tumor formation in the intestine.
Discussion
Lgr5+ ISCs continuously self-renew and differentiate into cells that comprise the intestinal epithelium. (Clevers, 2013). Acquisition of oncogenic mutations in these rapidly cycling stem cells leads to tumors that are subject to clearance by T cells (Agudo et al., 2018; Barker et al., 2009; Schepers et al., 2012). Here, we find that a HFD dampens MHC-II expression in IECs and Lgr5+ ISCs, and the reduction in epithelial MHC-II expression promotes intestinal tumor initiation. While MHC-I antigen presentation pathway-mediated activation of cytotoxic CD8+ T cells is frequently studied in the context of anti-tumor immune responses, MHC-II-mediated activation of CD4+ T cells is also pivotal for tumor immunity (Hung et al., 1998; Takeuchi and Saito, 2017; Tran et al., 2014; Wang, 2001; Xie et al., 2010; Zhang et al., 2009). Indeed, recent studies have demonstrated that MHC-II-restricted CD4+ T cells are able to eradicate tumors both directly and indirectly through the licensing of dendritic cells and helping CD8+ T cell responses (Haabeth et al., 2016; Haspot et al., 2014; Hirschhorn-Cymerman et al., 2012; Kreiter et al., 2015; Lu et al., 2017; Spitzer et al., 2017; Tran et al., 2014). Consistent with prior studies demonstrating that tumors downregulate MHC-II expression to escape from immune surveillance (Park et al., 2017; Tarafdar et al., 2017), our data implicate dietary regulation of MHC-II expression in ISCs as playing a critical role in intestinal tumorigenesis. Indeed, a recent large scale genome-wide variant scan for colorectal cancer identified variants associated with cancer risk in MHC-II gene loci(Huyghe et al., 2018). Whether HFD-induced suppression of MHC-II expression in ISCs affects tumor initiation directly by eliciting anti-tumor CD4+ T cell response or indirectly through regulating CD8+ T cell response warrants further investigation.
It is becoming increasingly evident that the crosstalk between tissue stem cells and immune cells influences differentiation, homeostasis and cancer risk (Ali et al., 2017; Hoytema van Konijnenburg et al., 2017; Lindemans et al., 2015; Naik et al., 2018; Naik et al., 2017; Ordovas-Montanes et al., 2020). Together with recent studies, our findings establish a functional role for epithelial MHC-II expression in regulating epithelial – immune crostalk in the intestine and tumorigenesis (Biton et al., 2018; Fu et al., 2019b; Koyama et al., 2019; Tuganbaev et al., 2020). Importantly, we find that the intestinal microbiome is a crucial mediator of the epithelilal MHC-II expression through TLR2/Myd88 and IFNγ signaling within intestinal epithelial cells, which are perturbed in response to HFD. Among bacterial species depleted by HFD, abundance of Helicobacter spp. correlates with epithelial MHC-II expression. This observation is consistent with our previous report demonstrating that several gram-negative Helicobacter species activate TLR2 receptors in the intestine (Mandell et al., 2004). Altogether, our results highlight how diet impacts the interactions between microbes, stem cells and immune cells in the intestine and contribute to tumor initiation (Fig. S6F). Because a HFD influences cancer incidence in both mucosal and non-mucosal tissues (Beyaz et al., 2016; Chen et al., 2018; Kroenke et al., 2013; Pascual et al., 2017; Schulz et al., 2014; Yang et al., 2017; Yang et al., 2008), it will be important to explore whether MHC-II mediated immune surveillance of stem cells also takes place in other tissues and how dietary perturbations influence tissue homeostasis and tumorigenesis by altering epithelial MHC-II expression.
Limitations of Study
While our data indicate that loss of epithelial MHC-II expression enhances intestinal tumor initiation, the precise immune mechanisms that mediate this process require further investigations. Consistent with previous reports (Kettunen et al., 2003; Luke et al., 2019), our tumor initiation models that are driven by intestine-specific loss of Apc and enhanced Wnt/β-catenin pathway activity resulted in exclusion of both CD4+ and CD8+ T cells in established tumors, thus constituting a limitation for delineating the specific roles that T cell subsets play during epithelial MHC-II-mediated regulation of intestinal tumor formation. Future studies are needed to address the significance of epithelial MHC-II-mediated antigen presentation in regulating T cell responses during the very early steps of tumor formation in the intestine. In addition, the causal links between specific microbes and epithelial MHC-II expression in the context of tumorigenesis are not addressed in this study. Although we identified several microbes that correlate with epithelial MHC-II expression, it will be important to identify the precise mechanisms of how microbes such as Helicobacter and Odoribacter directly or indirectly influence epithelial MHC-II expression and control intestinal homeostasis and tumorigenesis in the intestine in future studies.
STAR METHODS
Resource Availability:
Lead contact:
Further information and requests for resources and reagents should be directed to and will be fulfilled by Semir Beyaz (beyaz@cshl.edu).
Materials availability:
All in-house generated mouse strains generated for this study are available from the Lead Contact with a completed Materials Transfer Agreement.
Data and code availability:
RNA-seq and scRNA-seq data that are used in this study are available at Gene Expression Omnibus (GEO) with the following accession numbers GSE67324 and GSE180949.
Experimental model and subject details
Animals, diet, and drug treatment
The following strains were obtained from the Jackson Laboratory: Lgr5-EGFP-IRES-CreERT2 (strain name: B6.129P2-Lgr5tm1(cre/ERT2)Cle/J, stock number 008875), Rosa26-lacZ (strain name: B6.129S4-Gt(ROSA)26Sortm1Sor/J, stock number 003474), db/db (strain name: B6.BKS(D)-Leprjb/J, stock number 000697), MhcL/L (strain name: B6.129X1-H2-Ab1tm1Koni/J, stock number 013181), IFNGR1 KO (C57BL/6N-Ifngr1tm1.2Rds/J, stock number 025545), Villin-Cre (B6.Cg-Tg(Vil1-cre)1000Gum/J Stock No: 021504), Myd88L/L (B6.129P2(SJL)-Myd88tm1Defr/J Stock No: 008888), Rag2 KO (B6(Cg)-Rag2tm1.1Cgn/J Stock No: 008449). Apcloxp exon 14 (ApcL/L) has been previously described(Colnot et al., 2004). Villin-CreERT2 was a gift from Sylvie Robine. Diet-induced obesity studies were performed by using a high fat diet consisting of 60 kcal% fat (Research Diets D12492) beginning at the age of 8–12 weeks and extending for 9 to 14 months. Control mice were age- and sex-matched and were fed matched purified control diet (Research Diets, D12450J). GW501516 (Enzo) was reconstituted in DMSO at 4.5 mg ml−1 and diluted 1:10 in a solution of 5% PEG400 (Hampton Research), 5% Tween80 (Sigma), 90% H2O (injection buffer) for a daily intraperitoneal injection of 4mg kg−1. Alleles crossed with Lgr5-CreERT2 (to generate stem cell specific knockout, Lgr5-iKO) or Villin-CreERT2 (to generate intestinal epithelium specific knockout, vil-iKO) mice were excised by administration of tamoxifen suspended in sunflower seed oil (Spectrum S1929) at a concentration of 10 mg ml−1 and 250 μl per 25g of body weight, and administered by intraperitoneal injection every other day for 4 times. BrdU (Sigma) was prepared at 10 mg ml−1 in PBS, passed through 0.22μm filter and injected at 100mg kg−1. CYT387 (SelleckChem) was reconstituted in DMSO at 10mg ml−1 and diluted 1:100 in injection buffer for a daily gavage of 25mg kg−1 for 7 days. CL429 (InvivoGen) was reconstituted in DMSO at 5mg ml−1 and diluted 1:20 in a solution of injection buffer for a daily gavage of 2mg kg−1 for 7 days. For broad-spectrum antibiotic treatment, mice received a mixture of vancomycin (0.5g/l), ampicillin (1g/l), metronidazole (1g/l) and neomycin (1g/l) in the drinking water. Sex- and age-matched animals between 8 and 12 weeks of age were used for experiments unless otherwise specified. To estimate proper number of animals, preliminary experiments were performed. Mice were allocated at random to experimental groups. The Institutional Animal Care and Use Committee (IACUC) at CSHL, Boston Children’s Hospital and Massachusetts Institute of Technology approved all animal experiments.
Method details
Immunohistochemistry (IHC)
As previously described(Yilmaz et al., 2012), tissues were fixed in 10% formalin, paraffin embedded and sectioned. Antigen retrieval was performed with Borg Decloaker RTU solution (Biocare Medical) in a pressurized Decloaking Chamber (Biocare Medical) for 3 minutes. Antibodies used: rat anti-BrdU (1:2000, Abcam 6326), mouse monoclonal β-catenin (1:100, BD Biosciences 610154), rabbit monoclonal OLFM4 (1:10,000, gift from CST, clone PP7), rat anti-CD3 (1:200, eBioscience, clone 145–2C11), rat anti-CD4, (1:200, Biolegend, clone H129.19), rat anti-CD8 (1:200, Biolegend, clone 53–6.7), anti-Ki67 (1:200, Thermo Fisher, MA5–14520). Biotin-conjugated secondary donkey anti-rabbit or anti-rat antibodies were used from Jackson ImmunoResearch. The Vectastain Elite ABC immunoperoxidase detection kit (Vector Labs) followed by Dako Liquid DAB+ Substrate (Dako) was used for visualization. All antibody incubations involving tissue or sorted cells were performed with Common Antibody Diluent (Cell Signaling).
In situ hybridization
Single-molecule in situ hybridization was performed to detect MHC-II (H2-Ab1, #414731) using Advanced Cell Diagnostics RNAscope 2.5 HD Detection Kit following manufacturer’s instructions.
Intestinal crypt isolation and flow cytometry
As previously reported(Beyaz et al., 2016) and briefly summarized here, small intestines and colons were removed, washed with cold PBS−/−, opened laterally and cut into 3–5mm fragments. Pieces were washed multiple times with cold PBS−/− until clean, washed 2–3 with PBS−/−/EDTA (10mM), and incubated on ice for 90–120 minutes while mixing at 30-minute intervals. Crypts were then mechanically separated from the connective tissue by shaking, and filtered through a 70-μm mesh into a 50-ml conical tube to remove villus material (for small intestine) and tissue fragments. Crypts were removed from this step for crypt culture experiments and embedded in Matrigel™ with crypt culture media. For ISC isolation, the crypt suspensions were dissociated to individual cells with TrypLE Express (Invitrogen) and stained for flow cytometry. ISCs were isolated as Lgr5-EGFPhiEpcam+ CD45−7-AAD− with a BD FACS Aria II SORP cell sorter into supplemented crypt culture medium for culture.
Organoid culture for crypts and isolated cells
Isolated crypts were counted and embedded in Matrigel™ (Corning 356231 growth factor reduced) at 5–10 crypts per μl and cultured in a modified form of medium as described previously(Sato et al., 2009a). Unless otherwise noted, Advanced DMEM (Gibco) was supplemented by EGF 40 ng ml−1 (R&D), Noggin 200 ng ml−1 (Peprotech), R-spondin 500 ng ml−1 (R&D or Sino Biological), N-acetyl-L-cysteine 1 μM (Sigma-Aldrich), N2 1X (Life Technologies), B27 1X (Life Technologies), Chiron 10 μM (Stemgent), Y-27632 dihydrochloride monohydrate 20 ng ml−1 (Sigma-Aldrich). 25 μL drops of Matrigel™ with crypts were plated onto a flat bottom 48-well plate (Corning 3548) and allowed to solidify for 20 minutes in a 37°C incubator. Three hundred microliters of crypt culture medium were then overlaid onto the Matrigel™, changed every three days, and maintained at 37°C in fully humidified chambers containing 5% CO2. Clonogenicity (colony-forming efficiency) was calculated by plating 50–300 crypts per well and assessing organoid formation 3–7 days or as specified after initiation of cultures. Organoids were propagated for perturbation experiments as previously described(Beyaz et al., 2016).
Isolated ISCs or progenitor cells were centrifuged for 5 minutes at 250g, re-suspended in the appropriate volume of crypt culture medium (500–1,000 cells μl−1), then seeded onto 25–30 μl Matrigel™ (Corning 356231 growth factor reduced) containing 1 μM Jagged (Ana-Spec) in a flat bottom 48-well plate (Corning 3548). Alternatively, ISCs and Paneth cells were mixed after sorting in a 1:1 ratio, centrifuged, and then seeded onto Matrigel™. The Matrigel™ and cells were allowed to solidify before adding 300 μl of crypt culture medium. The crypt media was changed every second or third day. Organoid bodies were quantified on days 3, 7 and 10 of culture, unless otherwise specified. In secondary experiments, individual primary organoids were mechanically dissociated and replated, or organoids were dissociated for 10 minutes in TrypLE Express at 32°C, resuspended with SMEM (Life Technologies), centrifuged and resuspended in cold SMEM with viability dye 7-AAD. Live cells were sorted and seeded onto Matrigel™ as previously described(Beyaz et al., 2016).
DQ-Ovalbumin Assay
Sorted Lgr5+ intestinal stem cells were plated in Matrigel and crypt media for one day. The following day, the crypt media was replaced with crypt media containing 20 μg/mL DQ-ovalbumin (Thermo Fisher, D12053) and cells were incubated at 4C and 37C for 24 hours. Cells were harvested by washing three times in PBS, removing the Matrigel using Cell Recovery Solution (Corning) and filtered through a 40 μm mesh. Mean fluorescent intensity was analyzed using CellSimple (Cell Signaling).
Bulk RNA-Seq Analysis
Processing of RNA-seq reads and measuring expression level.
Tha raw data processed using RNA-Seq pipeline in DolphinNext with following steps(Yukselen et al., 2020). Raw stranded reads (40 nt) were trimmed to remove adapter and bases with quality scores below 20, and reads shorter than 35 nt were excluded. High-quality reads were mapped to the mouse genome (mm10) with TopHat version 1.4.1 (Trapnell et al., 2009), using known splice junctions from Ensembl Release 70 and allowing at most 2 mismatches. Genes were quantified with htseq-count (with the “intersect strict” mode) using Ensembl Release 70 gene models. Gene counts were normalized across all samples using estimateSizeFactors() from the DESeq R/Bioconductor package(Anders and Huber, 2010). Differential expression analysis made with DESeq2 package using DEBrowser(Kucukural et al., 2019).
qRT-PCR
Cells were sorted into Tri Reagent (Life Technologies) and total RNA was isolated according to the manufacturer’s instructions with following modification: the aqueous phase containing total RNA was purified using RNeasy plus kit (Qiagen). RNA was converted to cDNA with cDNA synthesis kit (Bio-Rad). qRT-PCR was performed with SYBR green master mix (Bio-Rad) on Bio-Rad iCycler RT-PCR detection system. For low cell numbers (<1000), qRT-PCR was performed after sequence specific pre-amplification as described previously(Beyaz et al., 2017). Primers are listed in Table S3.
Single cell RNA-Seq
Cell sorting
FACS (Astrios) was used to sort one single cell into each well of a 96-well PCR plate containing 5μl of TCL buffer with 1% 2-mercaptoethanol. The cells were stained for 7AAD− (Life Technologies), CD45− (eBioscience), CD31− (eBioscience), Ter119− (eBioscience), EpCAM+ (eBioscience). To enrich for specific IEC populations, cells were isolated from control or HFD Lgr5-GFP mice, stained with the antibodies mentioned above and gated for GFP-high (stem cells). A population control of 200 cells was sorted into one well and a no-cell control was sorted into another well. After sorting, the plate was sealed tightly with a Microseal F and centrifuged at 800g for 1 min. The plate was immediately frozen on dry ice and kept at −80ºC until ready for the lysate cleanup.
Plate-based scRNA-seq
Libraries were prepared using a modified SMART-Seq2 protocol as previously reported (Picelli et al., 2014). Briefly, RNA lysate cleanup was performed using RNAClean XP beads (Agencourt), followed by reverse transcription with Maxima Reverse Transcriptase (Life Technologies) and whole transcription amplification (WTA) with KAPA HotStart HIFI 2× ReadyMix (Kapa Biosystems) for 21 cycles. WTA products were purified with Ampure XP beads (Beckman Coulter), quantified with Qubit dsDNA HS Assay Kit (ThermoFisher), and assessed with a high sensitivity DNA chip (Agilent). RNA-seq libraries were constructed from purified WTA products using Nextera XT DNA Library Preparation Kit (Illumina). On each plate, the population and no-cell controls were processed using the same method as the single cells. The libraries were sequenced on an Illumina NextSeq 500.
Computational analysis of scRNA-seq
We profiled Lgr5-high ISCs sorted from control (n=192 cells) and HFD (n=192 cells) using a full length scRNA-seq method (Picelli et al., 2014). Each condition included two replicate 96-well plates from 2 different mice. Expression levels of gene loci were quantified using RNA-seq by Expectation Maximization (RSEM) (Li and Dewey, 2011). Raw reads were mapped to a mouse transcriptome index (mm10 UCSC build) using Bowtie 2 (Langmead and Salzberg, 2012), as required by RSEM in its default mode. On average, 90% of the reads mapped to the genome in every sample. and 55% of the reads mapped to the transcriptome. RSEM yielded an expression matrix (genes x samples) of inferred gene counts, which was converted to TPM (transcripts per million) values and then log-transformed after the addition of 1 to avoid zeros. After filtering cells with low QC metrics (< 400,000 mapped reads, transcriptomic mapping rate < 35% and < 1500 genes detected), we selected 171 control cells and 144 HFD cells for further analysis. We identified 379 highly variable genes using Seurat’s MeanVarPlot function. H2-Ab1, a key component of MHC-II complex, was among the top 5 differentially expressed genes with > 3-fold higher expression in control compared to HFD ISCs as assessed using MAST test.(Finak et al., 2015) A Gene Ontology analysis of these genes against a background of genes matched in average expression levels showed an enrichment of terms consistent with intestinal biology such as arachidonic acid metabolic process (GO:0019369), intestinal absorption (GO:0050892), as well as immune response, such as “antigen processing and presentation of exogenous peptide antigen” (GO:0002478) and “defense response to Gram-negative (GO:0050829), and Gram-positive (GO:0050830) bacterium”. We performed principal component analysis (PCA) on the data based on the variable genes, and embedded 10 statistically significant PCs identified using a permutation test (Shekhar et al., 2016) on a 2D map using t-distributed stochastic neighbor embedding (tSNE).
Taxonomic Microbiota Analysis
Frozen fecal samples were processed for DNA isolation using the Qiagen PowerSoil kit according to the manufacturer’s instructions. 1 ng of purified fecal DNA was used for PCR amplification. Amplicons spanning the variable region 1/2 (V1/2) of the 16S rRNA gene were generated by using the following barcoded primers: Fwd 5’-xrefXXAGAGTTTGATCCTGGCTCAG-3’, Rev 5’-TGCTGCCTCCCGTAGGAGT-3’, where X represents a barcode base. The reactions were subsequently pooled and cleaned, and the PCR products were then sequenced on an Illumina MiSeq with 500 bp paired-end reads. The reads were then processed using the QIIME (Quantitative Insights Into Microbial Ecology, http://www.qiime.org) analysis pipeline. Rarefaction was used to exclude samples with insufficient count of reads per sample. Sequences sharing 97% nucleotide sequence identity in the 16S region were binned into operational taxonomic units (97% ID OTUs). For beta-diversity, unweighted UniFrac measurements were plotted according to the two principal coordinates based on >10,000 reads per sample.
Endoscopy-guided orthotopic tumor transplantation and injections
ApcL/L; Lgr5-EGFP-IRES-CreERT2 mice were injected with two doses of tamoxifen I.P. Four days later, Apc-null Lgr5-GFPhi ISCs and Lgr5-GFPlow progenitors were sorted by flow cytometry, as described above. For primary cell transplantations, 10,000 Apc-null Lgr5-GFPhi, MHChi and MHClow ISCs were resuspended into 90% crypt culture media (as described) and 10% Matrigel™, and then transplanted into the colonic lamina propria of C57BL/6 recipient mice as previously described (Beyaz et al., 2016; Roper et al., 2017b). Mice then underwent colonoscopy eight weeks later to assess tumor formation. Colonoscopy videos and images were saved for offline analysis. Following sacrifice, the distal colons were excised and fixed in 10% formalin, then examined by histology to identify adenomas. In addition, colon tumors were induced in ApcL/L;VillinCreERT2 mice and ApcL/L;H2-Ab1L/L;VillinCreERT2 mice, as previously described (Roper et al., 2018; Roper et al., 2017b). Tumors were monitored with optical colonoscopy for two weeks. Offline images were analyzed using ImageJ to calculate Tumor Size Index, as previously described(Roper et al., 2011). Finally, Lgr5CreERT2; MHC-IIL/+; APCL/+ and Lgr5CreERT2; MHC-IIL/+; APCL/+ mice were treated with tamoxifen suspended in sunflower seed oil (Spectrum S1929) at a concentration of 10 mg ml−1 and 250 μl per 25g of body weight by intraperitoneal injection every other day for 4 times and followed for 5 months to assess tumor formation by histology. All tumor histology images were reviewed by gastrointestinal pathologists who were blinded to treatment groups.
Quantification and statistical analysis
Unless otherwise specified, all experiments reported in this study were repeated at least five independent times and sample number (n) represents biological replicates. For murine organoid assays, 2–5 wells per mouse per ex vivo treatment were analyzed. All center values shown in graphs refer to the mean. No sample or animals were excluded from analysis and sample size estimates were not used. Animals were randomly assigned to groups. Experiments used roughly equivalent male and female mice to account for sex as a biological variable. Studies were not conducted blind with the exception of all histological analyses.
Supplementary Material
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Chicken anti-β-galactosidase | Abcam | ab9361 |
| Rat anti-BrdU | Abcam | ab6326 |
| Mouse anti-β catenin | BD Biosciences | 610154 |
| Rat anti-CD3ε, clone 145-2C11 | eBioscience | 100302 |
| Rat anti-CD4, clone H129.19 | Biolegend | 130302 |
| Rat anti-CD8a, clone 53-6.7 | Biolegend | 100702 |
| CD45-PE, clone 30-F11 | eBioscience | 12-0451-83 |
| CD24-Pacfic Blue, clone M1/69 | Biolegend | 101820 |
| EpCAM Apc, clone G8.8 | eBioscience | 17-5791-82 |
| PE I-A/I-E, clone M5/114.15.2 | Biolegend | 107608 |
| 7-AAD | Thermo Fisher | A1310 |
| Anti-Ki67 (SP6) | Thermo Fisher | MA5-14520 |
| CD3e (D4V8L) rabbit monoclonal antibody | CST | 99940 |
| CD8a (D4W2Z) rabbit monoclonal antibody | CST | 98941 |
| CD3 FITC, clone 145-2C11 | Biolegend | 100306 |
| CD4 PE-Cy7, clone GK1.5 | Biolegend | 100422 |
| CD8a APC-Cy7, clone 53-6.7 | ||
| Chemicals, Peptides, and Recombinant Proteins | ||
| CYT387 | Selleckchem | S2219 |
| CL429 | InvivoGen | tlrl-c429 |
| GW501516 | Enzo | ALX-420-032 |
| DQ-ovalbumin | Thermo Fisher | D12053 |
| Ifng | Peprotech | 315-05 |
| 5-Bromo-2’-deoxyuridine | Sigma Aldrich | 19-160 |
| Metronidazole | Sigma Aldrich | M3761 |
| Vancomycin | Sigma Aldrich | V2002 |
| Ampicillin | Sigma Aldrich | A9518 |
| Neomycin | Sigma Aldrich | N1142 |
| PEG400 | Sigma Aldrich | P4338 |
| Tween-80 | Sigma Aldrich | P4780 |
| Tamoxifen | Sigma Aldrich | T5648 |
| Sunflower Seed Oil | Spectrum | S1929 |
| Matrigel™ | Corning | 356231 |
| Advanced DMEM | GIBCO | 12491015 |
| EGF | Peprotech | 315-09 |
| Noggin | Peprotech | 250-38 |
| R-spondin | Peprotech | 315-32 |
| N-acetyl-l-cystine | Sigma Aldrich | A9165 |
| B27 | Life Technologies | 17504044 |
| Chir99021 dihydrochloride | LC Laboratories | C-6556 |
| Y-27632 | Sigma Aldrich | Y0503 |
| TrypLE™ Express Enzyme | Thermo Fisher | 12604013 |
| 48 well plates | Olympus | 25-108 |
| 96 well plates | Olympus | 25-109 |
| JAG-1 | Anaspec | AS-61298 |
| TRI Reagent | Sigma Aldrich | 93289 |
| Critical Commercial Assays | ||
| Elite ABC HRP Kit | Vector Laboratories | PK6100 |
| Signalstain® DAB Substrate Kit | CST | 8049S |
| Signalstain® Antibody Diluent | CST | 8112L |
| RNAscope 2.0 HD Detection Kit | ACD RNAscope® | 323110 |
| RNAscope probe: Mm-H2-Ab1 | ACD RNAscope® | Ref#414731 |
| qScript cDNA SuperMix | Quantabio | 95048 |
| PerfeCTa SYBR green fast mix | Quantabio | 95072 |
| Deposited Data | ||
| Bulk RNA sequencing data | (Beyaz et al., 2016) | GSE67324 |
| scRNA sequencing data | This study | GSE180949 |
| Experimental Models: Mouse | ||
| MHCL/L | The Jackson Laboratory | 013181 |
| Villin-CreERT2 | Dr. Sylvie Robine | N/A |
| Lgr5-EGFP-IRES-CreERT2 | The Jackson Laboratory | 008875 |
| Rosa26-LSL-lacZ | The Jackson Laboratory | 003474 |
| APCL/L | Dr. Raju Kucherplati | N/A |
| Db/Db | The Jackson Laboratory | 000697 |
| IFNGR1 KO | The Jackson Laboratory | 025545 |
| Myd88L/L | The Jackson Laboratory | 008888 |
| Villin-Cre | The Jackson Laboratory | 021504 |
| MHCL/L; APCL/L; Lgr5-EGFP-IRES-CreERT2 | First presented in this work | N/A |
| MHCL/L; APCL/+; Lgr5-EGFP-IRES-CreERT2 | First presented in this work | N/A |
| MHCL/L; APCL/L; Villin-CreERT2 | First presented in this work | N/A |
| MHCL/L; Lgr5-EGFP-IRES-CreERT2, Rosa26-LSL-lacZ | First presented in this work | N/A |
| Mouse Diets | ||
| Control Diet | Research Diets | D12450J |
| High Fat Diet | Research Diets | D12492 |
| Software and Algorithms | ||
| FlowJo v10. | FlowJo LLC | https://www.flowjo.com/ |
| GraphPad Prism 8 | GraphPad Software | https://www.graphpad.com/scientificsoftware/prism/ |
| Aperio | Leica Biosystems | https://www.leicabiosystems.com |
| ImageJ-Fiji | NIH | https://fiji.sc/ |
| Oligonucleotides | ||
| Primers for qRT-PCR, see Table S3 | IDT | N/A |
Highlights:
HFD dampens MHC-II expression in IECs including ISCs
HFD perturbs intestinal microbes that correlate with MHC-II expression in ISCs
PRR and IFN γ signaling mediate epithelial MHC-II expression
Loss of MHC-II on premalignant ISCs enhances tumor initiation
Acknowledgements
We thank CSHL Cancer Center Shared Resources (Animal, Flow Cytometry and Histology Core Facilities) supported by NCI Cancer Center Support grant 5P30CA045508. We also thank the Whitehead Institute Flow Cytometry Core and the Koch Institute Flow Cytometry and Histology Core facilities (NCI P30-CA14051). S.B. is supported by The Oliver S. and Jennie R. Donaldson Charitable Trust, Mathers Foundation, STARR Cancer Consortium (I13-0052), The Mark Foundation for Cancer Research (20-028-EDV), NIH (P30CA045508-33). Ö.H.Y is supported by NIH (R01 CA211184, CA254314, DK126545, U01CA250554, and P30CA14051-43), The MIT Stem Cell Initiative through Foundation MIT, the Pew Foundation. S.H.O is supported by the Howard Hughes Medical Institute. M.F. is supported by by The American Association of Immunologists through a Career Reentry Fellowship.
Footnotes
Decleration of interests
The authors declare no competing financial interests. S.H.O serves as an advisory board member for Cell Stem Cell. S.B. received research funding from Elstar Therpauetics and Revitope Oncology for research that is not related to this study.
Inclusion and diversity
We worked to ensure sex balance in the selection of non-human subjects. One or more of the authors of this paper self-identifies as an underrepresented ethnic minority in science.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Abreu MT (2010). Toll-like receptor signalling in the intestinal epithelium: how bacterial recognition shapes intestinal function. Nat Rev Immunol 10, 131–144. [DOI] [PubMed] [Google Scholar]
- Agudo J, Park ES, Rose SA, Alibo E, Sweeney R, Dhainaut M, Kobayashi KS, Sachidanandam R, Baccarini A, Merad M, et al. (2018). Quiescent Tissue Stem Cells Evade Immune Surveillance. Immunity 48, 271–285 e275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali N, Zirak B, Rodriguez RS, Pauli ML, Truong HA, Lai K, Ahn R, Corbin K, Lowe MM, Scharschmidt TC, et al. (2017). Regulatory T Cells in Skin Facilitate Epithelial Stem Cell Differentiation. Cell 169, 1119–1129 e1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S, and Huber W (2010). Differential expression analysis for sequence count data. Genome biology 11, R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker N, Ridgway RA, van Es JH, van de Wetering M, Begthel H, van den Born M, Danenberg E, Clarke AR, Sansom OJ, and Clevers H (2009). Crypt stem cells as the cells-of-origin of intestinal cancer. Nature 457, 608–611. [DOI] [PubMed] [Google Scholar]
- Barker N, van Es JH, Kuipers J, Kujala P, van den Born M, Cozijnsen M, Haegebarth A, Korving J, Begthel H, Peters PJ, et al. (2007). Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature 449, 1003–1007. [DOI] [PubMed] [Google Scholar]
- Barriga FM, Montagni E, Mana M, Mendez-Lago M, Hernando-Momblona X, Sevillano M, Guillaumet-Adkins A, Rodriguez-Esteban G, Buczacki SJA, Gut M, et al. (2017). Mex3a Marks a Slowly Dividing Subpopulation of Lgr5+ Intestinal Stem Cells. Cell Stem Cell 20, 801–816 e807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basen-Engquist K, and Chang M (2011). Obesity and cancer risk: recent review and evidence. Curr Oncol Rep 13, 71–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belkaid Y, and Hand TW (2014). Role of the microbiota in immunity and inflammation. Cell 157, 121–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beyaz S, Kim JH, Pinello L, Xifaras ME, Hu Y, Huang J, Kerenyi MA, Das PP, Barnitz RA, Herault A, et al. (2017). The histone demethylase UTX regulates the lineage-specific epigenetic program of invariant natural killer T cells. Nat Immunol 18, 184–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beyaz S, Mana MD, Roper J, Kedrin D, Saadatpour A, Hong SJ, Bauer-Rowe KE, Xifaras ME, Akkad A, Arias E, et al. (2016). High-fat diet enhances stemness and tumorigenicity of intestinal progenitors. Nature 531, 53–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beyaz S, Mana MD, and Yilmaz OH (2021). High-fat diet activates a PPAR-delta program to enhance intestinal stem cell function. Cell Stem Cell 28, 598–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beyaz S, and Yilmaz OH (2016). Molecular Pathways: Dietary Regulation of Stemness and Tumor Initiation by the PPAR-delta Pathway. Clin Cancer Res 22, 5636–5641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biton M, Haber AL, Rogel N, Burgin G, Beyaz S, Schnell A, Ashenberg O, Su CW, Smillie C, Shekhar K, et al. (2018). T Helper Cell Cytokines Modulate Intestinal Stem Cell Renewal and Differentiation. Cell 175, 1307–1320 e1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown M, Hughes KR, Moossavi S, Robins A, and Mahida YR (2014). Toll-like receptor expression in crypt epithelial cells, putative stem cells and intestinal myofibroblasts isolated from controls and patients with inflammatory bowel disease. Clin Exp Immunol 178, 28–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calle EE, Rodriguez C, Walker-Thurmond K, and Thun MJ (2003). Overweight, obesity, and mortality from cancer in a prospectively studied cohort of U.S. adults. N Engl J Med 348, 1625–1638. [DOI] [PubMed] [Google Scholar]
- Caruso R, Warner N, Inohara N, and Nunez G (2014). NOD1 and NOD2: signaling, host defense, and inflammatory disease. Immunity 41, 898–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerf-Bensussan N, Quaroni A, Kurnick JT, and Bhan AK (1984). Intraepithelial lymphocytes modulate Ia expression by intestinal epithelial cells. J Immunol 132, 2244–2252. [PubMed] [Google Scholar]
- Chen M, Zhang J, Sampieri K, Clohessy JG, Mendez L, Gonzalez-Billalabeitia E, Liu XS, Lee YR, Fung J, Katon JM, et al. (2018). An aberrant SREBP-dependent lipogenic program promotes metastatic prostate cancer. Nat Genet 50, 206–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi NM, Majumder P, and Boss JM (2011). Regulation of major histocompatibility complex class II genes. Curr Opin Immunol 23, 81–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clevers H (2013). The intestinal crypt, a prototype stem cell compartment. Cell 154, 274–284. [DOI] [PubMed] [Google Scholar]
- Coleman DL (1978). Obese and diabetes: two mutant genes causing diabetes-obesity syndromes in mice. Diabetologia 14, 141–148. [DOI] [PubMed] [Google Scholar]
- Collins T, Korman AJ, Wake CT, Boss JM, Kappes DJ, Fiers W, Ault KA, Gimbrone MA Jr., Strominger JL, and Pober JS (1984). Immune interferon activates multiple class II major histocompatibility complex genes and the associated invariant chain gene in human endothelial cells and dermal fibroblasts. Proc Natl Acad Sci U S A 81, 4917–4921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colnot S, Niwa-Kawakita M, Hamard G, Godard C, Le Plenier S, Houbron C, Romagnolo B, Berrebi D, Giovannini M, and Perret C (2004). Colorectal cancers in a new mouse model of familial adenomatous polyposis: influence of genetic and environmental modifiers. Laboratory investigation; a journal of technical methods and pathology 84, 1619–1630. [DOI] [PubMed] [Google Scholar]
- Dahan S, Roth-Walter F, Arnaboldi P, Agarwal S, and Mayer L (2007). Epithelia: lymphocyte interactions in the gut. Immunol Rev 215, 243–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David LA, Maurice CF, Carmody RN, Gootenberg DB, Button JE, Wolfe BE, Ling AV, Devlin AS, Varma Y, Fischbach MA, et al. (2014). Diet rapidly and reproducibly alters the human gut microbiome. Nature 505, 559–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong C, Juedes AE, Temann UA, Shresta S, Allison JP, Ruddle NH, and Flavell RA (2001). ICOS co-stimulatory receptor is essential for T-cell activation and function. Nature 409, 97–101. [DOI] [PubMed] [Google Scholar]
- Finak G, McDavid A, Yajima M, Deng J, Gersuk V, Shalek AK, Slichter CK, Miller HW, McElrath MJ, Prlic M, et al. (2015). MAST: a flexible statistical framework for assessing transcriptional changes and characterizing heterogeneity in single-cell RNA sequencing data. Genome Biol 16, 278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Font-Burgada J, Sun B, and Karin M (2016). Obesity and Cancer: The Oil that Feeds the Flame. Cell Metab 23, 48–62. [DOI] [PubMed] [Google Scholar]
- Fu T, Coulter S, Yoshihara E, Oh TG, Fang S, Cayabyab F, Zhu Q, Zhang T, Leblanc M, Liu S, et al. (2019a). FXR Regulates Intestinal Cancer Stem Cell Proliferation. Cell 176, 1098–1112 e1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu YY, Egorova A, Sobieski C, Kuttiyara J, Calafiore M, Takashima S, Clevers H, and Hanash AM (2019b). T Cell Recruitment to the Intestinal Stem Cell Compartment Drives Immune-Mediated Intestinal Damage after Allogeneic Transplantation. Immunity 51, 90–103 e103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallagher EJ, and LeRoith D (2015). Obesity and Diabetes: The Increased Risk of Cancer and Cancer-Related Mortality. Physiol Rev 95, 727–748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grasso CS, Giannakis M, Wells DK, Hamada T, Mu XJ, Quist M, Nowak JA, Nishihara R, Qian ZR, Inamura K, et al. (2018). Genetic Mechanisms of Immune Evasion in Colorectal Cancer. Cancer Discov 8, 730–749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haabeth OA, Tveita A, Fauskanger M, Hennig K, Hofgaard PO, and Bogen B (2016). Idiotype-specific CD4(+) T cells eradicate disseminated myeloma. Leukemia 30, 1216–1220. [DOI] [PubMed] [Google Scholar]
- Haspot F, Li HW, Lucas CL, Fehr T, Beyaz S, and Sykes M (2014). Allospecific rejection of MHC class I-deficient bone marrow by CD8 T cells. Am J Transplant 14, 49–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershberg RM, Framson PE, Cho DH, Lee LY, Kovats S, Beitz J, Blum JS, and Nepom GT (1997). Intestinal epithelial cells use two distinct pathways for HLA class II antigen processing. J Clin Invest 100, 204–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirschhorn-Cymerman D, Budhu S, Kitano S, Liu C, Zhao F, Zhong H, Lesokhin AM, Avogadri-Connors F, Yuan J, Li Y, et al. (2012). Induction of tumoricidal function in CD4+ T cells is associated with concomitant memory and terminally differentiated phenotype. J Exp Med 209, 2113–2126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooper LV, Littman DR, and Macpherson AJ (2012). Interactions between the microbiota and the immune system. Science 336, 1268–1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howie D, Garcia Rueda H, Brown MH, and Waldmann H (2013). Secreted and transmembrane 1A is a novel co-stimulatory ligand. PLoS One 8, e73610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoytema van Konijnenburg DP, Reis BS, Pedicord VA, Farache J, Victora GD, and Mucida D (2017). Intestinal Epithelial and Intraepithelial T Cell Crosstalk Mediates a Dynamic Response to Infection. Cell 171, 783–794 e713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung K, Hayashi R, Lafond-Walker A, Lowenstein C, Pardoll D, and Levitsky H (1998). The central role of CD4(+) T cells in the antitumor immune response. J Exp Med 188, 2357–2368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huyghe JR, Bien SA, Harrison TA, Kang HM, Chen S, Schmit SL, Conti DV, Qu C, Jeon J, Edlund CK, et al. (2018). Discovery of common and rare genetic risk variants for colorectal cancer. Nature Genetics 51, 76–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson AM, Costanzo A, Gareau MG, Armando AM, Quehenberger O, Jameson JM, and Olefsky JM (2015). High fat diet causes depletion of intestinal eosinophils associated with intestinal permeability. PloS one 10, e0122195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kettunen HL, Kettunen AS, and Rautonen NE (2003). Intestinal immune responses in wild-type and Apcmin/+ mouse, a model for colon cancer. Cancer Res 63, 5136–5142. [PubMed] [Google Scholar]
- Koyama M, Mukhopadhyay P, Schuster IS, Henden AS, Hulsdunker J, Varelias A, Vetizou M, Kuns RD, Robb RJ, Zhang P, et al. (2019). MHC Class II Antigen Presentation by the Intestinal Epithelium Initiates Graft-versus-Host Disease and Is Influenced by the Microbiota. Immunity 51, 885–898 e887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreiter S, Vormehr M, van de Roemer N, Diken M, Lower M, Diekmann J, Boegel S, Schrors B, Vascotto F, Castle JC, et al. (2015). Mutant MHC class II epitopes drive therapeutic immune responses to cancer. Nature 520, 692–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroenke CH, Kwan ML, Sweeney C, Castillo A, and Caan BJ (2013). High- and low-fat dairy intake, recurrence, and mortality after breast cancer diagnosis. J Natl Cancer Inst 105, 616–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kucukural A, Yukselen O, Ozata DM, Moore MJ, and Garber M (2019). DEBrowser: interactive differential expression analysis and visualization tool for count data. BMC Genomics 20, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ley RE, Backhed F, Turnbaugh P, Lozupone CA, Knight RD, and Gordon JI (2005). Obesity alters gut microbial ecology. Proc Natl Acad Sci U S A 102, 11070–11075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Zimmerman SE, Peregrina K, Houston M, Mayoral J, Zhang J, Maqbool S, Zhang Z, Cai Y, Ye K, et al. (2019). The nutritional environment determines which and how intestinal stem cells contribute to homeostasis and tumorigenesis. Carcinogenesis 40, 937–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindemans CA, Calafiore M, Mertelsmann AM, O’Connor MH, Dudakov JA, Jenq RR, Velardi E, Young LF, Smith OM, Lawrence G, et al. (2015). Interleukin-22 promotes intestinal-stem-cell-mediated epithelial regeneration. Nature 528, 560–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Nishihara R, Qian ZR, Tabung FK, Nevo D, Zhang X, Song M, Cao Y, Mima K, Masugi Y, et al. (2017). Association Between Inflammatory Diet Pattern and Risk of Colorectal Carcinoma Subtypes Classified by Immune Responses to Tumor. Gastroenterology 153, 1517–1530 e1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovig T, Andersen SN, Thorstensen L, Diep CB, Meling GI, Lothe RA, and Rognum TO (2002). Strong HLA-DR expression in microsatellite stable carcinomas of the large bowel is associated with good prognosis. Br J Cancer 87, 756–762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu YC, Parker LL, Lu T, Zheng Z, Toomey MA, White DE, Yao X, Li YF, Robbins PF, Feldman SA, et al. (2017). Treatment of Patients With Metastatic Cancer Using a Major Histocompatibility Complex Class II-Restricted T-Cell Receptor Targeting the Cancer Germline Antigen MAGE-A3. J Clin Oncol 35, 3322–3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luke JJ, Bao R, Sweis RF, Spranger S, and Gajewski TF (2019). WNT/beta-catenin Pathway Activation Correlates with Immune Exclusion across Human Cancers. Clin Cancer Res 25, 3074–3083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandell L, Moran AP, Cocchiarella A, Houghton J, Taylor N, Fox JG, Wang TC, and Kurt-Jones EA (2004). Intact gram-negative Helicobacter pylori, Helicobacter felis, and Helicobacter hepaticus bacteria activate innate immunity via toll-like receptor 2 but not toll-like receptor 4. Infect Immun 72, 6446–6454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee S, and Hooper LV (2015). Antimicrobial defense of the intestine. Immunity 42, 28–39. [DOI] [PubMed] [Google Scholar]
- Naik S, Larsen SB, Cowley CJ, and Fuchs E (2018). Two to Tango: Dialog between Immunity and Stem Cells in Health and Disease. Cell 175, 908–920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naik S, Larsen SB, Gomez NC, Alaverdyan K, Sendoel A, Yuan S, Polak L, Kulukian A, Chai S, and Fuchs E (2017). Inflammatory memory sensitizes skin epithelial stem cells to tissue damage. Nature 550, 475–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neal MD, Sodhi CP, Jia H, Dyer M, Egan CE, Yazji I, Good M, Afrazi A, Marino R, Slagle D, et al. (2012). Toll-like receptor 4 is expressed on intestinal stem cells and regulates their proliferation and apoptosis via the p53 up-regulated modulator of apoptosis. J Biol Chem 287, 37296–37308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ordovas-Montanes J, Beyaz S, Rakoff-Nahoum S, and Shalek AK (2020). Distribution and storage of inflammatory memory in barrier tissues. Nat Rev Immunol 20, 308–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park IA, Hwang SH, Song IH, Heo SH, Kim YA, Bang WS, Park HS, Lee M, Gong G, and Lee HJ (2017). Expression of the MHC class II in triple-negative breast cancer is associated with tumor-infiltrating lymphocytes and interferon signaling. PLoS One 12, e0182786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pascual G, Avgustinova A, Mejetta S, Martin M, Castellanos A, Attolini CS, Berenguer A, Prats N, Toll A, Hueto JA, et al. (2017). Targeting metastasis-initiating cells through the fatty acid receptor CD36. Nature 541, 41–45. [DOI] [PubMed] [Google Scholar]
- Pavot V, Rochereau N, Resseguier J, Gutjahr A, Genin C, Tiraby G, Perouzel E, Lioux T, Vernejoul F, Verrier B, et al. (2014). Cutting edge: New chimeric NOD2/TLR2 adjuvant drastically increases vaccine immunogenicity. J Immunol 193, 5781–5785. [DOI] [PubMed] [Google Scholar]
- Price AE, Shamardani K, Lugo KA, Deguine J, Roberts AW, Lee BL, and Barton GM (2018). A Map of Toll-like Receptor Expression in the Intestinal Epithelium Reveals Distinct Spatial, Cell Type-Specific, and Temporal Patterns. Immunity 49, 560–575 e566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakoff-Nahoum S, Paglino J, Eslami-Varzaneh F, Edberg S, and Medzhitov R (2004). Recognition of commensal microflora by toll-like receptors is required for intestinal homeostasis. Cell 118, 229–241. [DOI] [PubMed] [Google Scholar]
- Rimsza LM, Roberts RA, Miller TP, Unger JM, LeBlanc M, Braziel RM, Weisenberger DD, Chan WC, Muller-Hermelink HK, Jaffe ES, et al. (2004). Loss of MHC class II gene and protein expression in diffuse large B-cell lymphoma is related to decreased tumor immunosurveillance and poor patient survival regardless of other prognostic factors: a follow-up study from the Leukemia and Lymphoma Molecular Profiling Project. Blood 103, 4251–4258. [DOI] [PubMed] [Google Scholar]
- Roper J, Richardson MP, Wang WV, Richard LG, Chen W, Coffee EM, Sinnamon MJ, Lee L, Chen PC, Bronson RT, et al. (2011). The dual PI3K/mTOR inhibitor NVP-BEZ235 induces tumor regression in a genetically engineered mouse model of PIK3CA wild-type colorectal cancer. PLoS One 6, e25132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roper J, Tammela T, Akkad A, Almeqdadi M, Santos SB, Jacks T, and Yilmaz OH (2018). Colonoscopy-based colorectal cancer modeling in mice with CRISPR-Cas9 genome editing and organoid transplantation. Nat Protoc 13, 217–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roper J, Tammela T, Cetinbas NM, Akkad A, Roghanian A, Rickelt S, Almeqdadi M, Wu K, Oberli MA, Sanchez-Rivera F, et al. (2017a). In vivo genome editing and organoid transplantation models of colorectal cancer and metastasis. Nature biotechnology 35, 569–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roper J, Tammela T, Cetinbas NM, Akkad A, Roghanian A, Rickelt S, Almeqdadi M, Wu K, Oberli MA, Sanchez-Rivera FJ, et al. (2017b). In vivo genome editing and organoid transplantation models of colorectal cancer and metastasis. Nat Biotechnol 35, 569–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Round JL, and Mazmanian SK (2009). The gut microbiota shapes intestinal immune responses during health and disease. Nat Rev Immunol 9, 313–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sade-Feldman M, Jiao YJ, Chen JH, Rooney MS, Barzily-Rokni M, Eliane JP, Bjorgaard SL, Hammond MR, Vitzthum H, Blackmon SM, et al. (2017). Resistance to checkpoint blockade therapy through inactivation of antigen presentation. Nat Commun 8, 1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato T, Vries RG, Snippert HJ, van de Wetering M, Barker N, Stange DE, van Es JH, Abo A, Kujala P, Peters PJ, et al. (2009a). Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature. [DOI] [PubMed] [Google Scholar]
- Sato T, Vries RG, Snippert HJ, van de Wetering M, Barker N, Stange DE, van Es JH, Abo A, Kujala P, Peters PJ, et al. (2009b). Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature 459, 262–265. [DOI] [PubMed] [Google Scholar]
- Schepers AG, Snippert HJ, Stange DE, van den Born M, van Es JH, van de Wetering M, and Clevers H (2012). Lineage tracing reveals Lgr5+ stem cell activity in mouse intestinal adenomas. Science 337, 730–735. [DOI] [PubMed] [Google Scholar]
- Schulz MD, Atay C, Heringer J, Romrig FK, Schwitalla S, Aydin B, Ziegler PK, Varga J, Reindl W, Pommerenke C, et al. (2014). High-fat-diet-mediated dysbiosis promotes intestinal carcinogenesis independently of obesity. Nature 514, 508–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spitzer MH, Carmi Y, Reticker-Flynn NE, Kwek SS, Madhireddy D, Martins MM, Gherardini PF, Prestwood TR, Chabon J, Bendall SC, et al. (2017). Systemic Immunity Is Required for Effective Cancer Immunotherapy. Cell 168, 487–502 e415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi A, and Saito T (2017). CD4 CTL, a Cytotoxic Subset of CD4(+) T Cells, Their Differentiation and Function. Front Immunol 8, 194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarafdar A, Hopcroft LE, Gallipoli P, Pellicano F, Cassels J, Hair A, Korfi K, Jorgensen HG, Vetrie D, Holyoake TL, et al. (2017). CML cells actively evade host immune surveillance through cytokine-mediated downregulation of MHC-II expression. Blood 129, 199–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Telega GW, Baumgart DC, and Carding SR (2000). Uptake and presentation of antigen to T cells by primary colonic epithelial cells in normal and diseased states. Gastroenterology 119, 1548–1559. [DOI] [PubMed] [Google Scholar]
- Thaiss CA, Itav S, Rothschild D, Meijer M, Levy M, Moresi C, Dohnalova L, Braverman S, Rozin S, Malitsky S, et al. (2016). Persistent microbiome alterations modulate the rate of post-dieting weight regain. Nature. [DOI] [PubMed] [Google Scholar]
- Thelemann C, Eren RO, Coutaz M, Brasseit J, Bouzourene H, Rosa M, Duval A, Lavanchy C, Mack V, Mueller C, et al. (2014). Interferon-gamma induces expression of MHC class II on intestinal epithelial cells and protects mice from colitis. PLoS One 9, e86844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomas J, Mulet C, Saffarian A, Cavin JB, Ducroc R, Regnault B, Kun Tan C, Duszka K, Burcelin R, Wahli W, et al. (2016). High-fat diet modifies the PPAR-gamma pathway leading to disruption of microbial and physiological ecosystem in murine small intestine. Proc Natl Acad Sci U S A 113, E5934–E5943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran E, Turcotte S, Gros A, Robbins PF, Lu YC, Dudley ME, Wunderlich JR, Somerville RP, Hogan K, Hinrichs CS, et al. (2014). Cancer immunotherapy based on mutation-specific CD4+ T cells in a patient with epithelial cancer. Science 344, 641–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Pachter L, and Salzberg SL (2009). TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25, 1105–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuganbaev T, Mor U, Bashiardes S, Liwinski T, Nobs SP, Leshem A, Dori-Bachash M, Thaiss CA, Pinker EY, Ratiner K, et al. (2020). Diet Diurnally Regulates Small Intestinal Microbiome-Epithelial-Immune Homeostasis and Enteritis. Cell 182, 1441–1459 e1421. [DOI] [PubMed] [Google Scholar]
- Tyner JW, Bumm TG, Deininger J, Wood L, Aichberger KJ, Loriaux MM, Druker BJ, Burns CJ, Fantino E, and Deininger MW (2010). CYT387, a novel JAK2 inhibitor, induces hematologic responses and normalizes inflammatory cytokines in murine myeloproliferative neoplasms. Blood 115, 5232–5240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Elsen PJ (2011). Expression regulation of major histocompatibility complex class I and class II encoding genes. Front Immunol 2, 48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanneman M, and Dranoff G (2012). Combining immunotherapy and targeted therapies in cancer treatment. Nat Rev Cancer 12, 237–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Rong X, Palladino END, Wang J, Fogelman AM, Martin MG, Alrefai WA, Ford DA, and Tontonoz P (2018). Phospholipid Remodeling and Cholesterol Availability Regulate Intestinal Stemness and Tumorigenesis. Cell stem cell 22, 206–220 e204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang RF (2001). The role of MHC class II-restricted tumor antigens and CD4+ T cells in antitumor immunity. Trends Immunol 22, 269–276. [DOI] [PubMed] [Google Scholar]
- Westendorf AM, Fleissner D, Groebe L, Jung S, Gruber AD, Hansen W, and Buer J (2009). CD4+Foxp3+ regulatory T cell expansion induced by antigen-driven interaction with intestinal epithelial cells independent of local dendritic cells. Gut 58, 211–219. [DOI] [PubMed] [Google Scholar]
- Wong GH, Bartlett PF, Clark-Lewis I, Battye F, and Schrader JW (1984). Inducible expression of H-2 and Ia antigens on brain cells. Nature 310, 688–691. [DOI] [PubMed] [Google Scholar]
- Xie Y, Akpinarli A, Maris C, Hipkiss EL, Lane M, Kwon EK, Muranski P, Restifo NP, and Antony PA (2010). Naive tumor-specific CD4(+) T cells differentiated in vivo eradicate established melanoma. J Exp Med 207, 651–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang JJ, Yu D, Takata Y, Smith-Warner SA, Blot W, White E, Robien K, Park Y, Xiang YB, Sinha R, et al. (2017). Dietary Fat Intake and Lung Cancer Risk: A Pooled Analysis. J Clin Oncol 35, 3055–3064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang K, Kurihara N, Fan K, Newmark H, Rigas B, Bancroft L, Corner G, Livote E, Lesser M, Edelmann W, et al. (2008). Dietary induction of colonic tumors in a mouse model of sporadic colon cancer. Cancer Res 68, 7803–7810. [DOI] [PubMed] [Google Scholar]
- Yilmaz OH, Katajisto P, Lamming DW, Gultekin Y, Bauer-Rowe KE, Sengupta S, Birsoy K, Dursun A, Yilmaz VO, Selig M, et al. (2012). mTORC1 in the Paneth cell niche couples intestinal stem-cell function to calorie intake. Nature 486, 490–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yukselen O, Turkyilmaz O, Ozturk AR, Garber M, and Kucukural A (2020). DolphinNext: a distributed data processing platform for high throughput genomics. BMC Genomics 21, 310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Zhang H, and Zhao J (2009). The role of CD4 T cell help for CD8 CTL activation. Biochem Biophys Res Commun 384, 405–408. [DOI] [PubMed] [Google Scholar]
- Zitvogel L, Pietrocola F, and Kroemer G (2017). Nutrition, inflammation and cancer. Nat Immunol 18, 843–850. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-seq and scRNA-seq data that are used in this study are available at Gene Expression Omnibus (GEO) with the following accession numbers GSE67324 and GSE180949.