Abstract
When C2C12 pluripotent mesenchymal precursor cells are treated with transforming growth factor β1 (TGF-β1), terminal differentiation into myotubes is blocked. Treatment with bone morphogenetic protein 2 (BMP-2) not only blocks myogenic differentiation of C2C12 cells but also induces osteoblast differentiation. The molecular mechanisms governing the ability of TGF-β1 and BMP-2 to both induce ligand-specific responses and inhibit myogenic differentiation are not known. We identified Runx2/PEBP2αA/Cbfa1, a global regulator of osteogenesis, as a major TGF-β1-responsive element binding protein induced by TGF-β1 and BMP-2 in C2C12 cells. Consistent with the observation that Runx2 can be induced by either TGF-β1 or BMP-2, the exogenous expression of Runx2 mediated some of the effects of TGF-β1 and BMP-2 but not osteoblast-specific gene expression. Runx2 mimicked common effects of TGF-β1 and BMP-2 by inducing expression of matrix gene products (for example, collagen and fibronectin), suppressing MyoD expression, and inhibiting myotube formation of C2C12 cells. For osteoblast differentiation, an additional effector, BMP-specific Smad protein, was required. Our results indicate that Runx2 is a major target gene shared by TGF-β and BMP signaling pathways and that the coordinated action of Runx2 and BMP-activated Smads leads to the induction of osteoblast-specific gene expression in C2C12 cells.
Transforming growth factor β (TGF-β) is a potent multifunctional regulator of cell growth and differentiation. Although nearly all cells synthesize and respond to TGF-β, bone and cartilage are particularly rich in this growth factor (6, 46). TGF-β1, the prototypic member of the TGF-β superfamily, elicits diverse cellular responses, including (i) inhibition of adipogenesis and myogenesis and (ii) stimulation of chondrogenesis and osteogenesis (31). TGF-β1 stimulates the synthesis of matrix proteins and their receptors (for example, fibronectin, fibronectin receptor, collagen, osteonectin, osteopontin, and integrins) and inhibits matrix degradation by increasing the production of protease inhibitors and decreasing the production of proteases (42). Members of the TGF-β superfamily with important effects on bone cell differentiation are bone morphogenetic proteins (BMPs) (17, 41), which were first identified as factors that induce bone formation in vivo when implanted into muscular tissues (54). Unlike TGF-β, which induces new bone formation only when injected near bone, BMPs produce bone formation even when injected into ectopic sites. TGF-β and BMPs bind to distinct receptors, TGF-β type I and II receptors for TGF-β and BMP type I and II receptors for BMPs. Following ligand binding, the receptor-associated kinase is activated and phosphorylates Smads, which move into the nucleus to stimulate the transcription of a set of target genes. Smad2 and -3 are activated by TGF-β receptors and mediate TGF-β responses, whereas Smad1, -5, and -8 are activated by BMP receptors and transduce BMP signals (15, 32, 57).
The pluripotent mesenchymal precursor cell line C2C12 provides a model system to study the early stage of osteoblast differentiation during bone formation in muscular tissues. In this model, TGF-β1 inhibits the differentiation of C2C12 cells into multinucleated myotubes without inducing osteoblast phenotypes. BMP-2 not only inhibits the terminal differentiation of C2C12 cells but also induces osteoblast phenotypes (20). Therefore, the C2C12 model is useful for analyzing both the common and specific signaling mechanisms of TGF-β and BMPs. In C2C12 cells, overexpression of Smad1 and Smad5 induced alkaline phosphatase (ALP) activity, a typical osteoblast-specific marker, and inhibited muscle-specific gene expression (11, 36, 56). These results suggested that BMP functions via either Smad1 or Smad5 and that the induction of the osteoblast phenotype and the inhibition of myogenic differentiation are regulated at the transcriptional level. However, the molecular mechanisms through which Smads block myogenic differentiation and induce osteogenic differentiation are not known.
Runx/PEBP2/Cbf (hereafter referred to as Runx) is a sequence-specific DNA binding protein that recognizes a specific DNA sequence originally identified as the binding site for polyomavirus enhancer binding protein (PEBP) (2, 38). Runx/PEBP2/Cbf was also identified as the Moloney murine leukemia virus enhancer core binding protein (53). The consensus sequence recognized by Runx was determined to be 5′-(Pu/T)ACCPuCPu-3′ or 5′-PyGPyGGT(Py/A)-3′ (3, 19, 33, 38). Each member of the Runx family is composed of two subunits, α and β. The α subunit is encoded by three distinct genes, Runx1 (PEBP2αβ/Cbfa2/AML1), Runx2 (PEBP2αA/Cbfa1/AML3), and Runx3 (PEBP2αC/Cbfa3/AML2). So far, only one gene encoding the β subunit, PEBP2β/Cbfb, has been described (4, 39, 53). The DNA binding activity of the α subunit, which binds DNA very weakly, is strongly stimulated by the β subunit. The recent identification of Runx2 as a transcription factor required for bone formation was a significant milestone in osteoblast biology. Both intramembranous and endochondral ossification were blocked owing to the maturational arrest of osteoblasts in Runx2 knockout mice (21, 40). The Runx2 gene is also involved in the human disease cleidocranial dysplasia (CCD), an autosomal dominant bone disorder. In Runx2+/− heterozygous mice, Otto et al. noticed abnormalities, the most prominent of which were hypoplasia of the clavicle and delayed development of membranous bones (40). These phenotypes are typical features of CCD. Together with these observations, deletions, insertions, or mutations that inactivated one allele of the Runx2 gene were shown to be the cause of the CCD syndrome in humans (24, 34). These results proved that Runx2 is an essential transcription factor required for bone formation. However, the underlying molecular mechanisms by which Runx2 expression is regulated and Runx2 controls osteoblast gene expression are still poorly understood.
In this study, we investigated the molecular mechanisms that block myoblast differentiation and induce osteoblast differentiation in C2C12 cells. Exogenous expression of Runx2 mimicked the common activities of TGF-β1 and BMP-2, inducing matrix gene expression, suppressing MyoD expression, and inhibiting myotube formation of C2C12 cells. However, Runx2 alone was not sufficient for osteoblast-specific gene expression. For this, the coordinate actions of Runx2 and BMP-activated Smads were required.
MATERIALS AND METHODS
Materials.
Bioactive recombinant human BMP-2 was produced and purified from the conditioned medium of CHO cells, and purity and bioactivity were checked as described previously (52). Recombinant human TGF-β1 was purchased from Sigma. Anti-Runx and anti-PEBP2β/Cbfβ polyclonal antibodies and anti-Runx2 monoclonal antibody were kind gifts from Y. Ito (Kyoto University, Kyoto, Japan). Reagents were purchased from the following vendors: restriction enzymes from Takara (Tokyo, Japan) or New England Biolabs; cell culture reagents, G418, and Lipofectamine from Gibco/BRL; human recombinant TGF-β1 from Sigma; mouse monoclonal anti-FLAG M2 antibody from IBI; enhanced chemiluminescence (ECL) Western blotting kit including goat anti-mouse horseradish peroxidase-conjugated antibody, Hybond-N+ nylon membrane, and Rediprime DNA labeling kit from Amersham; luciferase assay kit from Promega; hygromycin from Clontech; Immobilon from Millipore; poly(dI-dC) from Pharmacia; all oligonucleotides from Bioneer (Cheongiu, Korea); type I collagen gel (Cellmatrix type I-A) from Nitta Gelatin Co. (Tokyo, Japan); and collagenase from Wako (Osaka, Japan). All other chemicals of the purest grade available were obtained from commercial sources. Sense-strand sequences of the double-stranded oligonucleotides used in electrophoretic mobility shift assays (EMSAs) are as follows: TβRE (TGF-β1-responsive element), 5′-gaTCCACCACAGCCAGACCACAGGCAGACATGAgga-3′; M1, 5′-gaTCCAGGACAGCCAGACCACAGGCAGACATGAgga-3′; M2, 5′-gaTCCACCACAGCCAGAGGACAGGCAGACATGAgga-3′; and M3, 5′-gaTCCACCACAGCCAGACCACAGGCATCCATGAgga-3′. Lowercase letters indicate nucleotides not present in the immunoglobulin (Ig) Cα promoter that were added for cloning and end labeling; underlined nucleotides indicate mutations; the perfect match of the Runx binding site in TβRE is indicated in boldface.
Plasmids.
The mouse Runx2-αA1 cDNA cloned into pBluescript II-KS (38) and pcDNA3.1 (Invitrogen) was used for generating the Runx2 expression construct, pcDNA3.1-Runx2-αA1. The entire coding region of mouse Runx2-αA1 cDNA was amplified by PCR with the primers 5′GGCTGGGATCCGGTATGCGTATTCCT (sense) and 5′-GGTTGAAGATCTTCAATATGGCCGCCA (antisense). Translation initiation and stop codons in the primers are underlined. The PCR product was digested with BamHI and XbaI, whose sites were provided by the primers used, and the resulting DNA fragment was ligated into pcDNA3.1/HisC treated with BamHI and XbaI. The nucleotide sequence of the entire region amplified by PCR was verified by nucleotide sequencing of both strands. The luciferase reporter plasmid, pGL3-TβRE, was constructed by inserting two copies of the TβRE (originally identified in the Ig Cα promoter [29]), separated by 72 nucleotides of DNA derived from pBluescript II-KS (Stratagene), into the BglII site of the pGL3-promoter plasmid (Promega). pGL3-M2 was constructed by the same method except that the mutant TβRE M2 was used. pcDNA3-FLAG-Smad5 expressing human Smad5 tagged with FLAG was a gift from K. Miyazono (Cancer Institute of the Japanese Foundation for Cancer Research, Tokyo, Japan).
Cell lines and culture.
The mouse pluripotent mesenchymal precursor cell line C2C12 was purchased from the American Type Culture Collection C2C12 and MC3T3-E1 cells were maintained in Dulbecco modified Eagle medium (DMEM) containing 15% fetal bovine serum (FBS), and penicillin G (100 U/ml), and streptomycin (100 μg/ml) at 37°C in a humidified atmosphere of 5% CO2 in air. The Runx2−/− calvaria cell line H1-127-30 was established as follows. Runx2+/− mice were mated with p53+/− mice, and Runx2+/− p53+/− mice were generated. Mating of these littermates produced Runx2+/− p53−/− mice. Runx2−/− p53−/− embryos were obtained at embryonic day 18.5 from the mating of these littermates. Small fragments dissected from parietal bone of Runx2−/− p53−/− embryos at embryonic day 18.5 were cultured in type I collagen gel in alpha-minimal essential medium overlaid by alpha-minimal essential medium containing 10% FBS, penicillin G (100 U/ml), and streptomycin (100 μg/ml). After 10 days of culture, the cells migrating from the explants were harvested by treatment with 0.2% collagenase. The cells were diluted on 10-cm-diameter dishes, and each colony was picked up using cloning rings. One colony, named H1-127-30, was expanded and used for further analysis.
Stable transfection.
To obtain Runx2-overexpressing C2C12 (C2C12-Rx2) cells, stable transfection of pcDNA3.1-Runx2-αA1 into C2C12 cells was done via the Lipofectamine method as specified by the manufacturer (Gibco/BRL). G418-resistant colonies were selected by adding G418 (600 μg/ml) to the medium for 2 weeks. Viable colonies were subcultured, and Runx2-overexpressing clones were screened by Western blotting. C2C12 cells stably expressing Smad5 (C2C12-Sm5 cells) were obtained by the same method except that pcDNA3-FLAG-Smad5 was used for transfection. To obtain C2C12 cells stably expressing both Smad5 and Runx2 (C2C12-Sm5-Rx2 cells), C2C12-Rx2 was subjected to a second round of stable transfection with pcDNA3-FLAG-Smad5 and pTK-Hyg vectors (Clontech). The stably transfected cells were screened for 2 weeks in selection medium containing G418 (600 μg/ml) and hygromycin B (300 μg/ml). Viable colonies were further screened by Western blotting.
Transient transfection and luciferase assays.
For the luciferase assay, cells were transfected with Lipofectamine according to the manufacturer's instructions. Briefly, 2 × 105 cells were plated in each well of a six-well plate. The next day, cells were transfected by the Lipofectamine method (Gibco/BRL). Each transfection assay was performed with various combinations of 1 μg of the luciferase construct, 1 μg of the Runx2 expression plasmid (pcDNA3.1-Runx2-αA1), and 1 μg of the constitutive active form of the TGF-β receptor I. The total amount of exogenous DNA was maintained at 5 μg/plate by adding the appropriate amount of salmon sperm DNA. All plasmid DNA was prepared using a Qiagen Midi kit. After 6 h, the medium was changed and cultured for an additional 42 h. Cells were then lysed, and luciferase activity was determined using a Dual Luciferase Reporter Assay kit as instructed by the manufacturer (Promega). Results were obtained from at least two independent experiments with triplicate samples for each experiment.
EMSA.
Nuclear protein extracts were prepared from cells stimulated with TGF-β1 or BMP-2 as described previously (45). Protein concentrations of the extracts were determined using the Bradford assay (Bio-Rad). A double-stranded DNA probe, TβRE (see above), was prepared and used for EMSA as described previously (19). All binding assays were performed at 30°C for 30 min in 20 μl of binding buffer [20 mM HEPES (pH 7.6), 4% (wt/vol) Ficoll type 400, 50 mM KCl, 2 mM EDTA, 2 μg of poly(dI-dC)] containing 1 nM 32P-end-labeled probe (20,000 to 30,000 cpm) and about 5 μg of nuclear protein extract. The competition assay contained 50 times more unlabeled double-stranded synthetic competitor DNA. For supershift experiments, monoclonal antibody (or antiserum; 0.5 μl) was added to the entire mixture. Half of each reaction mixture was loaded onto a 0.25× Tris-borate-EDTA–5% nondenaturing polyacrylamide gel, electrophoresed at 250 V for 1 h and autoradiographed for 16 h at −70°C, using two sheets of intensifying screens.
Northern blot analysis.
Total cellular RNA was prepared as described previously (44). Then 5 μg of RNA was resolved in a 1.2% formaldehyde-agarose gel and transferred to a Hybond-N+ nylon membrane using 10× SSPE buffer (0.18 M NaCl, 0.01 M NaH2PO4, 0.001 M Na2EDTA [pH 7.7]). RNA was cross-linked to the filter by UV irradiation for 1 min and stored until use. The DNA probes were either the PCR product or cloned cDNA of mouse Runx2 (38), rat ALP (37), human collagen type I (8), human fibronectin (22), rat osteocalcin (27), or mouse MyoD (9) and were all labeled with [α-32P]dCTP (3,000 Ci/mmol; NEN) using a Rediprime DNA labeling kit. The blot was prehybridized in 5× SSPE–5× Denhardt's solution–0.5% sodium dodecyl sulfate (SDS)–100 μg of salmon sperm DNA/ml at 65°C for 1 h. For hybridization, heat-denatured radioactive DNA probe (106 cpm/ml) was added, and the mixture was incubated at 65°C overnight. After hybridization, the blots were washed in 2× SSPE–0.1% SDS at room temperature for 15 min and twice in 0.1× SSPE–0.1% SDS at 65°C for 15 min for the rat and mouse probes. For the human probes, the membrane was washed in 0.5× SSPE–0.1% SDS at 42°C instead of 0.1× SSPE–0.1% SDS. The blots were exposed to Kodak XAR-5 film at −70°C with two sheets of intensifying screens.
Western blot analysis.
Proteins from cell lysates were resolved by SDS–8 to 10% polyacrylamide gel electrophoresis and transferred to Immobilon (Millipore). The blots were blocked in BP solution (50 mM Tris [pH 7.5], 150 mM NaCl, 0.1% Tween 20) containing 2% nonfat dry milk. Primary antibody (FLAG or Runx2) was added to the BP solution at a 1:2,000 dilution for 1 h at room temperature. The blots were washed three times with the BP solution and incubated with the goat anti-mouse antibody for 1 h at room temperature. After three washes with BP solution, the blots were developed by ECL and exposed on Kodak XAR-5 film.
RESULTS
The major TβRE binding protein induced by TGF-β1 and BMP-2 is Runx2.
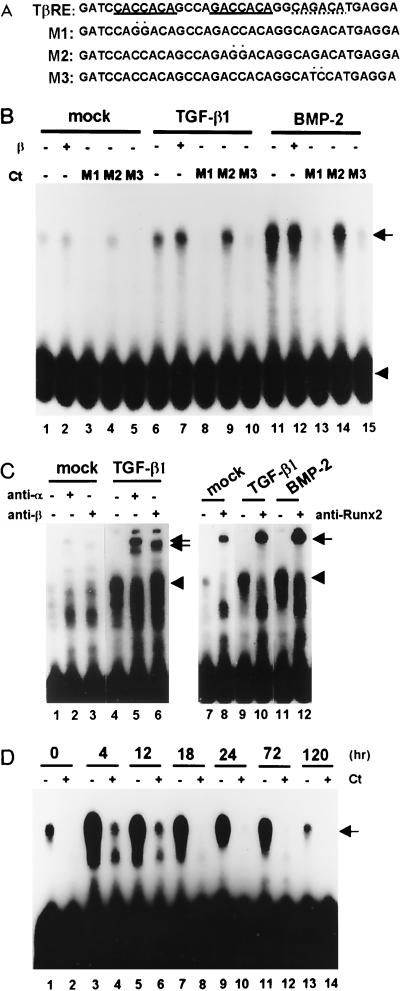
C2C12 cells differentiate into multinucleated myotubes when the serum concentration is reduced from 15% to 5%. Treatment with TGF-β1 (5 ng/ml) or BMP-2 (300 ng/ml) completely inhibits myotube formation. We studied the molecular mechanism of this inhibition of terminal differentiation. Using the TβRE present in the Ig Cα promoter (Fig. 1A) (29), the DNA binding activity of the TβRE binding protein was examined by EMSA. As shown in Fig. 1B, TβRE binding activity was detected in C2C12 cells, and the activity was significantly increased by TGF-β1 and BMP-2 (compare lanes 1, 6, and 11). The two elements present in the TβRE have been shown to be Runx and Smad binding sites (13, 47). Therefore, we tested which of these sites actually mediates the binding of the TβRE binding protein induced by TGF-β1. For this purpose, we prepared three mutant forms of the TβRE (M1, M2, and M3 [Fig. 1A]) with a base substitution in the first (M1) or second (M2) putative Runx binding site or in the putative Smad binding site (M3). Addition of M1 or M3 effectively competed out the entire TβRE-protein interaction (Fig. 1B, lanes 3, 5, 8, 10, 13, and 15), whereas M2 did not (lanes 4, 9, and 14), suggesting that the induced protein bound to the second Runx consensus sequence but not to the first (M1 site), which is imperfect (Fig. 1A). To examine whether the TGF-β1 or BMP-2-induced TβRE binding protein is actually Runx, EMSA supershift assays were performed using Runx2 or PEBP2β/Cbfβ-specific antibodies. As shown in Fig. 1C, the TβRE binding protein-DNA complex was supershifted by a polyclonal antibody that recognizes Runx1, -2, and -3 (lanes 2 and 5) and by a monoclonal antibody that recognizes only Runx2 (lanes 8, 10, and 12) as well as by PEBP2β/Cbfβ-specific antiserum (lanes 3 and 6), while no supershifted band was observed using preimmune serum (lanes 1 and 4). Since the TβRE binding protein was effectively supershifted by the Runx2-specific monoclonal antibody (lanes 8, 10, and 12), the TβRE binding protein must contain mainly Runx2. Furthermore, these results indicate that TGF-β1 or BMP-2 induces primarily the Runx2 isoform in C2C12 cells. It is worth mentioning that addition of PEBP2β/Cbfβ protein did not change the mobility or the intensity of the shifted band (Fig. 1B, lane 2, 7, and 12). However, the TβRE-bound complex was supershifted by anti-PEBP2β antibody (Fig. 1C, lanes 3 and 6). The results suggest that Runx2 bound to the TβRE was already complexed with the β subunit, as has already been suggested for all family members of the α subunit family (4, 5). A time course study showed that the induction of the TβRE binding protein (Runx2) reached a maximum as early as 4 h after TGF-β1 stimulation (about 20-fold over control at maximal level) and gradually decreased thereafter, even though the cells were continually stimulated by TGF-β1 (Fig. 1D).
FIG. 1.
Identification of the TβRE binding protein by EMSA. (A) Oligonucleotide sequences of the TβRE identified in the Ig Cα promoter (29) and three mutant DNAs, M1 (one mismatch in the putative Runx binding site [CACCACA]), M2 (a perfect match [GACCACA]), and M3 (putative Smad binding site [CAGACA]). Putative Runx2 and Smad binding sites are indicated by solid and dotted underlining, respectively, and mutated nucleotides are marked by asterisks. (B) EMSA was performed by using the TβRE probe and nuclear lysates obtained from C2C12 cells untreated or treated with TGF-β1 or BMP-2 for 24 h. The reactions were performed in the presence or absence of PEBP2β/Cbfβ protein. A 50-fold molar excess of unlabeled mutant oligonucleotide was incubated with the nuclear lysates as competitor DNA (Ct). The arrow and arrowhead indicate the TβRE binding complex and free probe, respectively. (C) EMSA was performed by using the same nuclear lysates and TβRE probes in the presence or absence of a polyclonal antibody which recognized all members of the Runx α subunit (anti-α; lane 2 and 5) or β subunit (anti-β; lane 3 and 6) or a monoclonal antibody which specifically recognized Runx2 (anti-Runx2; lanes 8, 10, and 12). The arrowheads and arrows indicate the positions of Runx2 and Runx-antibody complexes, respectively. (D) Time course induction of Runx2 was analyzed by EMSA using nuclear lysates prepared from cells treated with TGF-β1 for 0, 4, 12, 18, 24, 72, and 120 h. A nuclear extract prepared from cells cultured for 72 h in the absence of TGF-β1 was used as a control. EMSA was performed with the TβRE probe in the presence or absence of a 50-fold molar excess of unlabeled M3 as competitor. The arrow indicates the TβRE binding complex. Ct, competitor.
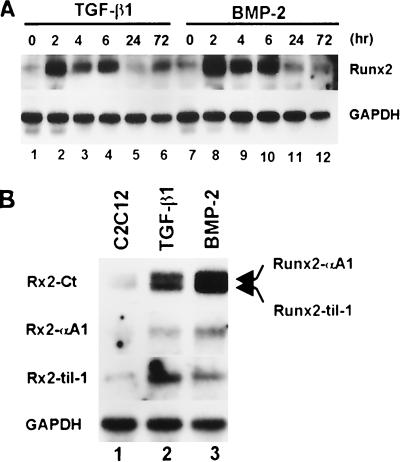
We analyzed whether the induction of Runx2 is regulated at the mRNA level. Total cellular RNA was isolated from the C2C12 cells treated with TGF-β1 or BMP-2 for the indicated time, and the level of Runx2 expression was analyzed by Northern blot hybridization. As shown in Fig. 2A, Runx2 mRNA was induced by TGF-β1 and BMP-2 stimulation. The Runx2 mRNA level reached a maximum 2 h after stimulation and decreased thereafter, even though the cells were continually stimulated with TGF-β1 or BMP-2. At least two isoforms of Runx2, which are identical except for their N-terminal regions, are known to exist. PEBP2αA1 (38), which we will call Runx2-αA1 hereafter, is a prototype of Runx2. The other (Runx2-Til-1) is known as Til-1 (48) or OSF2 (10). til-1 and OSF2 have been found to encode the same protein (49). We examined to see which form was induced by TGF-β1 and BMP-2 stimulation. Northern blot experiments using a cDNA probe covering the common region of both isoforms (Rx2-Ct) detected two bands (Fig. 2B). Rehybridization of the blot with the Runx2-αA1-specific or Runx2-til-1-specific probe revealed that the upper and lower bands corresponded to Runx2-αA1 and Runx2-Til-1, respectively. These results indicate that TGF-β1 and BMP-2 can induce both isoforms (Fig. 2B).
FIG. 2.
Induction of Runx2 mRNA by TGF-β1 and BMP2. (A) C2C12 cells were treated with TGF-β1 (5 ng/ml) or BMP-2 (300 ng/ml) for the indicated times, and total RNA was prepared. Northern blotting was performed using PEBP2αA cDNA (38), which contains the common region of Runx2-αA1 and Runx2-til-1, as a probe for Runx2 (Rx2-Ct). (B) Total RNAs were prepared from C2C12 cells treated with BMP-2 (300 ng/ml) for 6 h and analyzed by Northern blot hybridization using the Rx2-Ct probe. The same blot was stripped and rehybridized with Runx2-αA1 (38)- or Runx2-til-1 (48)-specific probes. A probe prepared from the GAPDH coding sequence was used as a loading control.
Binding of Runx2 to TβRE is required for the response to TGF-β1.
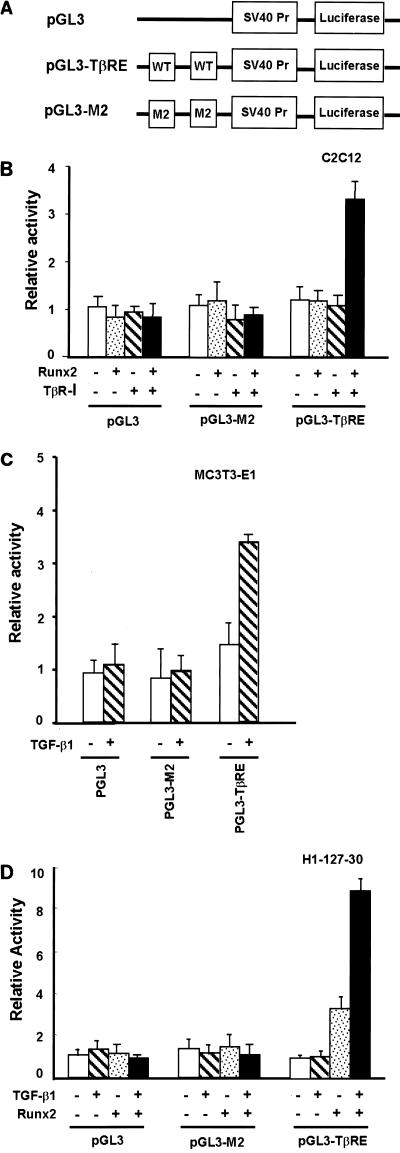
To examine the role of Runx2, we inserted two tandemly repeated TβREs or a mutant version (M2) of the Ig Cα promoter into the pGL3-promoter plasmid (Promega) (Fig. 3A) and measured cis-acting activity by the luciferase assay. In agreement with Runx2 being the major TβRE binding protein induced by TGF-β1, we observed an approximately threefold increase in the transcriptional activity of the TβRE-luciferase reporter after transfection of a constitutively active form of the TGF-β receptor I and Runx2 into C2C12 cells (Fig. 3B). A complete loss of response was observed with the M2 mutant, indicating that Runx2 is essential for transcriptional activation of target genes in response to TGF-β1. To rigorously establish the requirement for Runx2 in TβRE function, we used the cell line H1-127-30 (see Materials and Methods). When Runx2+/+ (MC3T3-E1) calvaria cells were transfected with the TβRE reporter, a two- to threefold induction was observed after TGF-β1 treatment. This TGF-β1-dependent induction was also completely abrogated by the mutation in the Runx binding site (Fig. 3C). When H1-127-30 cells were transfected with the TβRE reporter, the TβRE did not respond to TGF-β1 at all. Expression of Runx2 in H1-127-30 cells, however, strongly induced TβRE reporter activity, and treatment with TGF-β1 further induced reporter activity (Fig. 3D). It is worth noting that the expression of Runx2 by itself induced gene expression through the TβRE in the absence of TGF-β1, while TGF-β1 by itself had no effect. This result firmly establishes that Runx2 is essential for the TGF-β1 responsiveness of the TβRE.
FIG. 3.
Binding of Runx2 to the Runx binding site is essential for the TGF-β1 responsiveness of the TβRE. (A) Diagrams of luciferase reporter constructs. Two reporters for the TβRE of the Ig Cα promoter were constructed using the pGL3-promoter plasmid (Promega) containing the simian virus 40 promoter (SV40 Pr) as a backbone, i.e., one with the wild-type (WT) TβRE (pGL3-TβRE) DNA and the other with the TβRE mutated at the Runx binding site (pGL3-M2). Two copies of the element were inserted for each plasmid construct. (B) pGL3, pGL3-M2, and pGL3-TβRE reporters were transfected into C2C12 cells with the Runx2 expression plasmid (Runx2) or the constitutively active TGF-β receptor I expression plasmid (TβR-I) or with both. Cells were harvested 48 h after transfection, and luciferase activities were assayed. (C and D) The same reporters were transfected into MC3T3-E1 and H1-127-30 cells with or without the Runx2 expression plasmid (Runx2) and cultured in the presence or absence of TGF-β1 for 24 h. Luciferase activities were measured, and relative activities are shown.
Runx2 mediates the common activities of TGF-β1 and BMP-2.
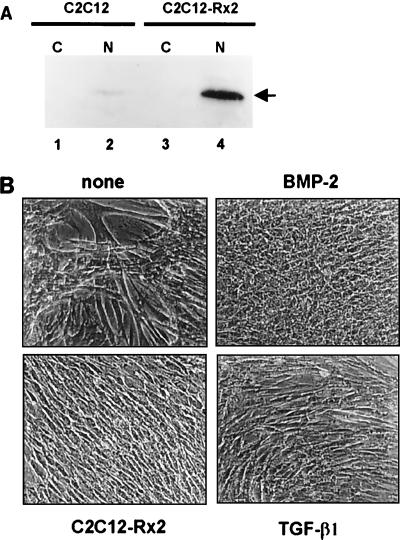
Since both TGF-β1 and BMP-2 induce Runx2, we asked whether Runx2 could mediate the common function of TGF-β1 and BMP-2. Therefore, we examined whether exogenous expression of Runx2 could block myogenic differentiation. For this purpose, C2C12-Rx2 cells were used (Fig. 4A). When control C2C12 cells, which bear only the empty vector, were cultured for 10 days in medium containing 5% serum, extensive formation of multinucleated myotubes was observed. Myotube formation was completely blocked in cells stably expressing Runx2 as well as in the TGF-β1- or BMP-2-treated cells, indicating that Runx2 by itself is sufficient to block myogenic differentiation (Fig. 4B). To confirm this result, molecular markers for TGF-β1 and BMP-2 stimulation were analyzed. Both TGF-β1 and BMP-2 are known to suppress MyoD (20) and induce collagen α1(I) and fibronectin expression (25). Up- or down-regulation of these marker genes by TGF-β1 and BMP-2 was confirmed in control C2C12 cells (Fig. 5, lanes 1 to 3); constitutively expressed Runx2 also suppressed MyoD and induced collagen α1(I) and fibronectin expression in C2C12 cells (lane 4). It is worth noting that MyoD, which is critically important for myogenic differentiation, was suppressed by exogenous expression of Runx2. Given that Runx2 blocks myogenic differentiation, it can be assumed that Runx2 expression must be suppressed for the induction of myogenic differentiation. As shown in Fig. 6, Runx2 expression was significantly suppressed during myogenic differentiation whereas MyoD expression was slightly enhanced. These results suggest that Runx2 is an essential and common target of TGF-β1 and BMP-2 signaling and that the induction of Runx2 is a key event in the inhibition of myogenic differentiation.
FIG. 4.
Morphological changes of C2C12 cells stably expressing Runx2. (A) Western blotting showing overexpression of Runx2 in C2C12-Rx2 cells. Cytoplasmic (C) and nuclear (N) protein extracts were prepared from C2C12 and C2C12-Rx2 cells, and Runx2 protein was detected by Western blotting. An arrow indicates Runx2 protein. (B) C2C12 cells containing the empty vector were cultured for 10 days in differentiation medium (5% FBS in DMEM) in the presence or absence of TGF-β1 (5 ng/ml) or BMP-2 (300 ng/ml). C2C12-Rx2 cells were cultured under the same conditions in the absence of TGF-β1 and BMP-2. After incubation, morphological changes were compared.
FIG. 5.
Pattern of gene expression following BMP-2 and TGF-β1 treatment of C2C12 and C2C12-Rx2 cells. Control C2C12 (lane 1 to 3) and C2C12-Rx2 (lane 4 to 6) cells were treated with TGF-β1 (5 ng/ml) or BMP-2 (300 ng/ml) for 3 days. Total RNA was extracted and analyzed by Northern blotting with probes homologous to osteocalcin (OC), collagen type I (Col-I), fibronectin (FN), or MyoD.
FIG. 6.
Suppression of Runx2 expression during myogenic differentiation. C2C12 cells were cultured in DMEM containing 5% FBS for 1 day (lane 1) or 7 days (lane 2). Total RNA was prepared, and the levels of myoD and Runx2 mRNA were analyzed by Northern blotting.
Conversely, the osteocalcin gene, a mineralized tissue-specific gene (28) induced by BMP-2 but not by TGF-β1, was not up-regulated by Runx2 expression alone (Fig. 5, lane 4). However, in the presence of BMP-2, overexpression of Runx2 led to significant induction of osteocalcin over the level of osteocalcin in the control of BMP-2-treated cells (compare lanes 3 and 6). Although this result suggests that Runx2 also plays an important role in osteoblast differentiation, other factors in addition to Runx2 would appear to be required.
Cooperation between Runx2- and BMP-activated Smad5 induces osteoblast-specific gene expression.
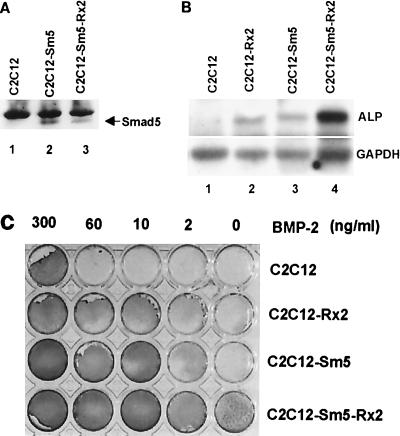
Induction of ALP activity following BMP treatment is considered to be an indicator of an early stage of osteoblast differentiation (20). TGF-β1, which can block myogenic differentiation of C2C12 cells but cannot induce osteoblast differentiation, does not induce ALP activity. Consistent with the differential effect of BMP and TGF-β, BMP-specific Smads, but not TGF-β-specific Smads, have been shown to be responsible for the induction of ALP activity in C2C12 cells (1, 36). It has been also reported that exogenous expression of Runx2 can induce ALP activity in C3H10T1/2 cells (14). We confirmed the induction of ALP activity by overexpressing Runx2 in C2C12 cells (Fig. 7B, lanes 1 and 2; Fig. 7C). However, the level of ALP activity induced by Runx2 in the absence of exogenous BMP-2 was much lower than in C2C12 cells treated with BMP-2 (300 ng/ml) alone (Fig. 7C). These observations together with the result shown in Fig. 5 (osteocalcin) indicate that Runx2 alone is not sufficient for the induction of osteoblast-specific gene expression and that an additional BMP-2-specific signal is required. Since signal-specific Smad proteins physically interact with Runx2 (13), we surmised that BMP-specific Smad proteins might be additionally required for the induction of osteoblast-specific gene expression. To prove this, we examined by Northern blot analysis the level of ALP mRNA in C2C12-Sm5 and C2C12-Sm5-Rx2 cells (Fig. 7A and B). We confirmed the low level of ALP mRNA in C2C12-Sm5 and C2C12-Rx2 cells in the absence of BMP-2 stimulation (Fig. 7B, lanes 2 and 3). Remarkably, ALP expression was strongly enhanced in C2C12-Sm5-Rx2 cells even in the absence of BMP-2 (Fig. 7B, lane 4). We further analyzed ALP expression in these cells by treating them with various concentrations of BMP-2 and measuring ALP enzyme activity. C2C12-Sm5-Rx2 cells showed quite strong ALP activity even in the absence of BMP-2 (Fig. 7C). The activity was higher than that of cells overexpressing either Smad5 or Runx2 alone. To observe ALP activity as high as that of the control cells treated with 300 ng of BMP-2/ml, C2C12-Sm5-Rx2 and C2C12-Sm5 cells required 2 and 10 ng of BMP-2/ml, respectively (Fig. 7C). Therefore, C2C12-Sm5-Rx2 cells were about 150 times more sensitive to BMP-2 than control cells and about five times more sensitive than C2C12-Sm5 cells. These results demonstrate synergistic cooperation between the functions of Smad5 and Runx2 for the induction of ALP expression.
FIG. 7.
Induction of ALP by cooperation between Runx2 and Smad5. (A) Western blot showing exogenous expression of Smad5 in C2C12-Sm5 and C2C12-Sm5-Rx2 cells. The arrow indicates Smad5 protein. (B) C2C12 (control), C2C12-Rx2, C2C12-Sm5, and C2C12-Sm5-Rx2 cells were cultured in the absence of BMP-2. Total RNA was prepared, and the levels of ALP mRNA were analyzed by Northern blot hybridization. (C) The same cells were treated with the indicated concentration of BMP-2 for 3 days, and ALP enzyme activities were assayed as described by Katagiri et al. (20).
Involvement of Smad in Runx2 induction.
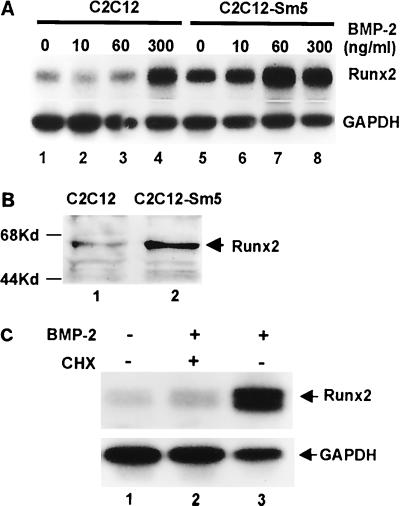
For the induction of ALP activity, C2C12-Sm5 cells appeared to be more sensitive to BMP-2 than C2C12-Rx2 cells. C2C12-Rx2 cells still required 300 ng of BMP-2/ml to obtain the same level of ALP activity as that of control C2C12 cells treated with 300 ng/ml, while C2C12-Sm5 cells required only 10 ng of BMP-2/ml (Fig. 7C). This result suggested that BMP-activated Smad5 is more rate limiting than Runx2 for the induction of ALP expression. Therefore, we examined whether Runx2 expression is under the control of Smad5. We treated C2C12-Sm5 cells with serially diluted concentrations of BMP-2 for 6 h and measured Runx2 mRNA levels by Northern blot analysis. As shown in Fig. 8A, overexpression of Smad5 by itself induced Runx2 expression even in the absence of BMP-2 (lane 5). Western blot analysis also confirmed the induced level of Runx2 protein in C2C12-Sm5 cells (Fig. 8B). Treatment of C2C12-Sm5 cells with 60 ng of BMP-2/ml induced Runx2 expression to the maximal level, while control C2C12 cells required 300 ng/ml to induce Runx2 expression (Fig. 8A). These results indicate that BMP-specific Smad proteins play an important role in the induction of Runx2 by BMP-2 stimulation.
FIG. 8.
Involvement of Smads in the induction of Runx2 expression. (A) Control C2C12 cells and C2C12-Sm5 cells were treated with the indicated concentration of BMP-2 for 6 h. Total RNA was prepared from the cells, and the Runx2 mRNA level was analyzed by Northern blotting using Rx2-Ct as a probe. (B) Protein lysates were obtained from control C2C12 (lane 1) and C2C12-Sm5 (lane 2) cells, and the level of Runx2 protein was analyzed by Western blotting. (C) C2C12 cells were cultured in the absence or presence of BMP-2 (300 ng/ml) and cycloheximide (CHX; 5 μg/ml) as indicated for 6 h. Total RNAs were prepared, and the Runx2 mRNA level was analyzed by Northern blot hybridization using the Rx2-Ct probe.
We further examined whether the receptor-activated Smads stimulate Runx2 expression directly or indirectly. C2C12 cells were treated with BMP-2 for 6 h in the presence or absence of the protein synthesis inhibitor cycloheximide (5 μg/ml), and the level of Runx2 mRNA was analyzed by Northern blotting. As shown in Fig. 8C, induction of Runx2 mRNA by BMP-2 was significantly inhibited by cycloheximide. This result indicates that Runx2 is not a direct target of receptor-activated Smads but rather an indirect target of some other factor that must be synthesized de novo.
DISCUSSION
Inactivation of the Runx2 gene in mice has been shown to result in a complete block to osteoblast differentiation (21, 40), and haploinsufficiency of Runx2 causes the CCD syndrome in humans (24, 34). In contrast to the wealth of information stressing the importance of Runx2 in osteogenesis, little is known about the molecular controls of Runx2 expression or about the mechanism through which Runx2 controls osteoblast-specific gene expression. In this study, we have shown that Runx2 acts as a common and major target of TGF-β1 and BMP-2 signaling and that there exists a functional relationship between Runx2 and Smad5 for ligand-specific transcriptional regulation.
Runx2 is the major TβRE binding protein induced by TGF-β1 and BMP-2.
To elucidate the molecular mechanism that blocks C2C12 cell differentiation after TGF-β1 and BMP-2 stimulation, we examined for TβRE binding proteins in C2C12 cells. We observed that TGF-β1 at 5 ng/ml or BMP-2 at 300 ng/ml significantly increased TβRE binding activity and that the major protein binding to the TβRE was Runx2 complexed to PEBP2β/Cbfβ. Induction of Runx2 by TGF-β1, BMP-2, BMP-7, and BMP4/7 heterodimer has been reported (10, 25, 51). However, the results have been controversial, as some showed that BMP-2 does not significantly alter Runx2 mRNA levels (51). Our analysis, which directly searched for TβRE binding protein activity, firmly establishes that Runx2 is the major and common target of TGF-β1 and BMP-2 signaling. The time course study revealed that the Runx2 mRNA level reached a maximum 2 h after stimulation and gradually decreased thereafter (Fig. 2A). This rapid and transient induction of Runx2 could be the reason why some groups failed to detect the induction of Runx2 after BMP-2 stimulation. Our analysis further establishes that Runx2 is essential for TGF-β1 and BMP-2 signaling. TGF-β1 is unable to induce transcription through TβRE in Runx2−/− calvaria cells. Interestingly, expression of Runx2 successfully activated reporter expression in the absence of TGF-β1 (Fig. 3D). This reporter assay revealed that binding of Runx2 to the Runx binding site is essential for TGF-β1 activity through the TβRE.
At least two species of Runx2 mRNA are synthesized as a result of differential promoter usage (10, 38, 48, 49). PEBP2αA1, referred to here as Runx2-αA1, is the Runx2 prototype, originally identified in ras-activated NIH 3T3 cells. The til-1 isoform gene, referred to here as Runx2-til-1, was originally identified as a frequent retrovirus integration site in virus-accelerated lymphomas of CD2-myc transgenic mice. Runx2-til-1 RNA is transcribed from a promoter located at least 20 kb upstream from the Runx2-αA1 promoter (48). OSF2 was identified as an osteoblast-specific transcription factor and as a regulator of osteoblast differentiation (10). Further analysis revealed that Runx2-til-1 and OSF2 encode identical proteins (49). Our Northern blot analysis revealed that TGF-β1 and BMP-2 induce both isoforms of Runx2 (Fig. 2B). Although it is still not clear if each isoform has a distinct function, a recent analysis failed to detect any marked differences between the two isoforms in the C3H10T1/2 fibroblast cell line (14).
Runx2 is essential for the common responses to TGF-β1 and BMP-2.
TGF-β and BMP-2 block the myogenic differentiation of C2C12 cells by suppressing the expression of master control genes, including that of MyoD (20, 35). Our data show that overexpression of Runx2 suppresses MyoD expression and stops the myogenic differentiation of C2C12 cells (Fig. 4B and 5). Osteoblasts, chondroblasts, adipoblasts, myoblasts, and fibroblasts differentiate from mesenchymal precursor cells (12). The critically important regulators of osteoblast, myocyte, and adipocyte differentiation are known to be Runx2 (10), MyoD (9), and peroxisome proliferator-activated receptor γ2 (PPARγ2) (30, 50), respectively. It is worth noting that PPARγ2 suppresses Runx2 expression during the promotion of adipocyte differentiation and inhibits osteoblast differentiation (23). Thus, exclusive expression of Runx2, MyoD, and PPARγ2 might be essential for the lineage determination of mesenchymal precursor cells. In accord with this suggestion, Runx2 expression was significantly suppressed during the myogenic differentiation of C2C12 cells (Fig. 6). These results suggest that induction of Runx2 is the key event responsible for the block of myogenic differentiation by TGF-β and BMP-2.
In addition, Runx2 induces the expression of at least two matrix proteins, type I collagen and fibronectin, whose synthesis is up-regulated by both TGF-β1 and BMP-2 (Fig. 5). Type I collagen is the most abundant extracellular matrix protein of bone tissue and is essential for bone strength (43). Fibronectin, another major component of the extracellular matrix, plays an important role during development and wound healing by promoting cell adhesion, cell migration, and cytoskeletal organization (22). The induction of these major components of the extracellular matrix and the suppression of MyoD expression by Runx2 suggests that Runx2 mediates the common activities of TGF-β1 and BMP-2. It is worth noting that the induction patterns of type I collagen and fibronectin by Runx2 were different. Type I collagen gene expression was fully induced by the exogenous expression of Runx2 without any additional effect of TGF-β1 and BMP-2 stimulation (Fig. 5). The induction of type I collagen by TGF-β1 and BMP-2, therefore, appears to rely entirely on increased Runx2 expression. On the other hand, fibronectin gene expression, induced by exogenous Runx2, was further induced by TGF-β1 but not by BMP-2 (Fig. 5). These results suggest that TGF-β1 and BMP-2 induce fibronectin expression through Runx2 but that TGF-β1 also activates fibronectin expression through additional mechanisms that work synergistically with Runx2. It has been reported that TGF-β1-mediated fibronectin induction requires the activation of Jun N-terminal kinase (JNK) (16). Thus, the JNK pathway might be a likely candidate for the TGF-β1 synergistic effect on fibronectin gene expression that operates through Runx2.
Cooperation between Runx2 and BMP-activated Smad5 induces osteoblast-specific gene expression.
Runx2 alone did not induce osteocalcin, a mineralized tissue-specific protein, but increased the level of osteocalcin induction by BMP-2 (Fig. 5). Likewise, Runx2 induced only weakly the expression of a major phenotypic marker of the osteoblast lineage, ALP, but increased the level of ALP induced by BMP-2 (Fig. 7C). This result suggests that although Runx2 is involved in the induction of osteoblast differentiation, it is not sufficient for this process. Recently, all three Runx α subunits and receptor-activated Smad proteins (Smad1, -2, -3, and -5) were shown to interact in vitro, and synergic effects of Runx3 and Smad3 on a TβRE reporter gene were reported (13). Smad1 and Smad5 were shown to be involved in the intracellular BMP signals that inhibit myogenic differentiation and induce osteogenic differentiation in C2C12 cells (36, 56). It is worth noting that overexpression of Runx2 could also inhibit myogenic differentiation, although it was not sufficient for the further induction of osteoblast differentiation (Fig. 4 and 5). These results all point to the intriguing possibility that Runx2 can mediate TGF-β1 and BMP-2 signals for the block of myogenic differentiation but not ligand-specific responses unless signal specific signal transducers are concomitantly present in C2C12 cells. Our results provide good evidence to support this hypothesis. We showed that exogenous expression of Runx2 alone mediated the common activities of TGF-β1 and BMP-2 but failed to fully induce osteoblast-specific gene expression (Fig. 5). As an additional requirement for osteoblast-specific gene expression, we identified the BMP-specific signal transducer, Smad5 (Fig. 7).
It is worth mentioning that the time course assay showed that the induction of Runx2 expression in response to BMP-2 occurred within the 2-h period following stimulation (Fig. 2A). BMP receptor IA-dependent Smad phosphorylation and nuclear translocation begin 10 to 20 min after BMP-2 stimulation and remain elevated for up to 2 h (18). These results indicate that the receptor-activated Smad and the BMP-induced Runx2 could function together in vivo, since they shared overlapping periods of expression. These observations also explain why only BMP-2 was able to provoke the differentiation of C2C12 cells to the osteoblastic lineage, even though both TGF-β1 and BMP-2 were able to induce Runx2 and inhibit the myogenic differentiation of this cell line.
Osteocalcin gene expression is initiated late during osteoblast differentiation at the onset of extracellular matrix mineralization (28). Unlike expression of ALP, an early marker of osteoblast differentiation, osteocalcin gene expression was not induced by the coexpression of Smad5 and Runx2 (data not shown). Our results suggest that the coordinate function of Smad5 and Runx2 is still insufficient for osteogenic terminal differentiation, although both are implicated in the commitment to osteogenic differentiation.
Involvement of Smads in Runx2 induction.
Overexpression of Smad5 resulted in the induction of Runx2 even in the absence of BMP-2 stimulation (Fig. 8). This result indicates that Smad5 functions as an upstream regulator of Runx2. The molecular mechanism of Smad5 activation under these conditions remains unclear. Recent observations have suggested that the subcellular localization of Smads prior to activation is important. Treatment of the cells with reagents that disrupt microtubules induced the phosphorylation of Smad2 in the absence of TGF-β and enhanced TGF-β-dependent phosphorylation of Smad2 (55). The mislocalization or promiscuous activation of Smad5 might be responsible for the induction of Runx2. However, Smad5 did not directly induce Runx2 expression; an additional step of protein de novo synthesis was required (Fig. 8C). One of the factors mediating the induction of Runx2 by Smad5 could be AP1. junB is an immediate-early gene induced by TGF-β and BMP-2 (7). JunB expression reaches a maximum 90 min after BMP-2 stimulation and then gradually decreases. Like Runx2, overexpression of JunB inhibits expression of myoblast differentiation markers in C2C12 cells (7). Analysis of the Runx2 promoter region reveals a perfect AP1 binding site situated close to the putative Smad sites (X.-Z. Zi and S.-C. Bae, unpublished observation). Thus, it will be interesting to examine whether induction of AP1 is required for the induction of Runx2 in response to TGF-β1 and BMP-2.
Since not only Smad5 but also Smad2 and -3 physically interact with Runx2 (13), interaction of Runx2 with TGF-β-specific Smads might also be important for TGF-β-specific responses. So far, the Smads that induce Runx2 after TGF-β stimulation and function together with Runx2 have not been identified. In the ROS17/2.8 osteoblast-like cell line and primary rat calvaria cells, Runx2 expression was suppressed by about 50% after overexpression of Smad2 (26), implying that distinct relationships may exist between each Smad protein and Runx2. Further study will be required for a thorough understanding of the molecular mechanisms governing ligand-specific responses of TGF-β and BMP-2 and the relationship between Runx2 and ligand-specific Smad proteins.
In this study, we identified Runx2 as the common and major target of TGF-β1 and BMP-2 in C2C12 cells. Runx2 was found to be responsible for suppression of the expression of the myogenic master gene, myoD, and for the induction of components of the extracellular matrix but insufficient for osteoblast-specific gene expression, which depended on the additional factor, Smad5, an upstream regulator of Runx2. These results provide important insights into how the downstream components of the TGF-β1 and BMP-2 signaling pathways, which we have identified as Runx2 and receptor-activated Smads (Fig. 9), mediate the block to myogenic differentiation and induce osteoblast differentiation in C2C12 cells.
FIG. 9.
A model for the common and distinct activities of TGF-β and BMP in C2C12 cells. Runx2 is an indirect target of the Smad signaling pathway. Induction of Runx2 is essential for the common activities of TGF-β and BMP. Ligand-specific gene expression, however, requires cooperation between Runx2 and receptor-activated Smads (R-Smad).
ACKNOWLEDGMENTS
We are grateful to Y. Ito (Kyoto University, Kyoto, Japan) for valuable discussions. We also thank K. Miyazono (Cancer Institute, Tokyo, Japan) for providing Smad expression plasmids.
We acknowledge financial support from the Korea Research Foundation for the program year 1998 to S.-C. Bae and H.-M. Ryoo. This work was also supported by Korea Research Foundation grant KRF-99-042-F00014 F0210.
REFERENCES
- 1.Akiyama S, Katagiri T, Namiki M, Yamaji N, Yamamoto N, Miyama K, Shibuya H, Ueno N, Wozney J M, Suda T. Constitutively active BMP type I receptors transduce BMP-2 signals without the ligand in C2C12 myoblasts. Exp Cell Res. 1997;235:362–369. doi: 10.1006/excr.1997.3680. [DOI] [PubMed] [Google Scholar]
- 2.Bae S C, Yamaguchi-Iwai Y, Ogawa E, Maruyama M, Inuzuka M, Kagoshima H, Shigesada K, Satake M, Ito Y. Isolation of PEBP2 αB cDNA representing the mouse homolog of human acute myeloid leukemia gene, AML1. Oncogene. 1993;8:809–814. [PubMed] [Google Scholar]
- 3.Bae S C, Ogawa E, Maruyama M, Oka H, Satake M, Shigesada K, Jenkins N A, Gilbert D J, Copeland N G, Ito Y. PEBP2αB/mouse AML1 consists of multiple isoforms that possess differential transactivation potentials. Mol Cell Biol. 1994;14:3242–3252. doi: 10.1128/mcb.14.5.3242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bae S C, Ito Y. Regulation mechanisms for the heterodimeric transcription factor, PEBP2/CBF. Histol Histopathol. 1999;14:1213–1221. doi: 10.14670/HH-14.1213. [DOI] [PubMed] [Google Scholar]
- 5.Banerjee C, McCabe L R, Choi J Y, Hiebert S W, Stein J L, Stein G S, Lian J B. Runt homology domain proteins in osteoblast differentiation: AML3/CBFA1 is a major component of a bone specific complex. J Cell Biochem. 1997;66:1–8. doi: 10.1002/(sici)1097-4644(19970701)66:1<1::aid-jcb1>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- 6.Bonewald L F, Mundy G R. Role of transforming growth factor-beta in bone remodeling. Clin Orthop. 1990;January:261–276. [PubMed] [Google Scholar]
- 7.Chalaux E, Lopez-Rovira T, Rosa J L, Bartrons R, Ventura F. JunB is involved in the inhibition of myogenic differentiation by bone morphogenetic protein-2. J Biol Chem. 1998;273:537–543. doi: 10.1074/jbc.273.1.537. [DOI] [PubMed] [Google Scholar]
- 8.Chu M L, Myers J C, Bernard M P, Ding J F, Ramirez F. Cloning and characterization of five overlapping cDNAs specific for the human pro alpha 1(I) collagen chain. Nucleic Acids Res. 1982;10:5925–5934. doi: 10.1093/nar/10.19.5925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Davis R L, Weintraub H, Lassar A B. Expression of a single transfected cDNA converts fibroblasts to myoblasts. Cell. 1987;51:987–1000. doi: 10.1016/0092-8674(87)90585-x. [DOI] [PubMed] [Google Scholar]
- 10.Ducy P, Zhang R, Geoffroy V, Ridall A L, Karsenty G. Osf2/Cbfa1: a transcriptional activator of osteoblast differentiation. Cell. 1997;89:747–754. doi: 10.1016/s0092-8674(00)80257-3. [DOI] [PubMed] [Google Scholar]
- 11.Fujii M, Takeda K, Imamura T, Aoki H, Sampath T K, Enomoto S, Kawabata M, Kato M, Ichijo H, Miyazono K. Roles of bone morphogenetic protein type I receptors and Smad proteins in osteoblast and chondroblast differentiation. Mol Biol Cell. 1999;10:3801–3813. doi: 10.1091/mbc.10.11.3801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grigoriadis A E, Heersche J N, Aubin J E. Continuously growing bipotential and monopotential myogenic, adipogenic, and chondrogenic subclones isolated from the multipotential RCJ 3.1 clonal cell line. Dev Biol. 1990;142:313–318. doi: 10.1016/0012-1606(90)90352-j. [DOI] [PubMed] [Google Scholar]
- 13.Hanai J, Chen L F, Kanno T, Ohtani-Fujita N, Kim W Y, Guo W H, Imamura T, Ishidou Y, Fukuchi M, Shi M J, Stavnezer J, Kawabata M, Miyazono K, Ito Y. Interaction and functional cooperation of PEBP2/CBF with Smads. Synergistic induction of the immunoglobulin germline Calpha promoter. J Biol Chem. 1999;274:31577–31582. doi: 10.1074/jbc.274.44.31577. [DOI] [PubMed] [Google Scholar]
- 14.Harada H, Tagashira S, Fujiwara M, Ogawa S, Katsumata T, Yamaguchi A, Komori T, Nakatsuka M. Cbfa1 isoforms exert functional differences in osteoblast differentiation. J Biol Chem. 1999;274:6972–6978. doi: 10.1074/jbc.274.11.6972. [DOI] [PubMed] [Google Scholar]
- 15.Heldin C H, Miyazono K, ten Dijke P. TGF-beta signalling from cell membrane to nucleus through SMAD proteins. Nature. 1997;390:465–471. doi: 10.1038/37284. [DOI] [PubMed] [Google Scholar]
- 16.Hocevar B A, Brown T L, Howe P H. TGF-beta induces fibronectin synthesis through a c-Jun N-terminal kinase-dependent, Smad4-independent pathway. EMBO J. 1999;18:1345–1356. doi: 10.1093/emboj/18.5.1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hogan B L. Bone morphogenetic proteins: multifunctional regulators of vertebrate development. Genes Dev. 1996;10:1580–1594. doi: 10.1101/gad.10.13.1580. [DOI] [PubMed] [Google Scholar]
- 18.Hoodless P A, Haerry T, Abdollah S, Stapleton M, O'Connor M B, Attisano L, Wrana J L. MADR1, a MAD-related protein that functions in BMP2 signaling pathways. Cell. 1996;85:489–500. doi: 10.1016/s0092-8674(00)81250-7. [DOI] [PubMed] [Google Scholar]
- 19.Kamachi Y, Ogawa E, Asano M, Ishida S, Murakami Y, Satake M, Ito Y, Shigesada K. Purification of a mouse nuclear factor that binds to both the A and B cores of the polyomavirus enhancer. J Virol. 1990;64:4808–4819. doi: 10.1128/jvi.64.10.4808-4819.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Katagiri T, Yamaguchi A, Komaki M, Abe E, Takahashi N, Ikeda T, Rosen V, Wozney J M, Fujisawa-Sehara A, Suda T. Bone morphogenetic protein-2 converts the differentiation pathway of C2C12 myoblasts into the osteoblast lineage. J Cell Biol. 1994;127:1755–1766. doi: 10.1083/jcb.127.6.1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Komori T, Yagi H, Nomura S, Yamaguchi A, Sasaki K, Deguchi K, Shimizu Y, Bronson R T, Gao Y H, Inada M, Sato M, Okamoto R, Kitamura Y, Yoshiki S, Kishimoto T. Targeted disruption of Cbfa1 results in a complete lack of bone formation owing to maturational arrest of osteoblasts. Cell. 1997;89:755–764. doi: 10.1016/s0092-8674(00)80258-5. [DOI] [PubMed] [Google Scholar]
- 22.Kornblihtt A R, Pesce C G, Alonso C R, Cramer P, Srebrow A, Werbajh S, Muro A F. The fibronectin gene as a model for splicing and transcription studies. FASEB J. 1996;10:248–257. doi: 10.1096/fasebj.10.2.8641558. [DOI] [PubMed] [Google Scholar]
- 23.Lecka-Czernik B, Gubrij I, Moerman E J, Kajkenova O, Lipschitz D A, Manolagas S C, Jilka R L. Inhibition of Osf2/Cbfa1 expression and terminal osteoblast differentiation by PPARgamma2. J Cell Biochem. 1999;74:357–371. [PubMed] [Google Scholar]
- 24.Lee B, Thirunavukkarasu K, Zhou L, Pastore L, Baldini A, Hecht J, Geoffroy V, Ducy P, Karsenty G. Missense mutations abolishing DNA binding of the osteoblast-specific transcription factor OSF2/CBFA1 in cleidocranial dysplasia. Nat Genet. 1997;16:307–310. doi: 10.1038/ng0797-307. [DOI] [PubMed] [Google Scholar]
- 25.Lee M H, Javed A, Kim H J, Shin H I, Gutierrez S, Choi J Y, Rosen V, Stein J L, van Wijnen A J, Stein G S, Lian J B, Ryoo H M. Transient upregulation of CBFA1 in response to bone morphogenetic protein-2 and transforming growth factor beta 1 in C2C12 myogenic cells coincides with suppression of the myogenic phenotype but is not sufficient for osteoblast differentiation. J Cell Biochem. 1999;73:114–125. [PubMed] [Google Scholar]
- 26.Li J, Tsuji K, Komori T, Miyazono K, Wrana J L, Ito Y, Nifuji A, Noda M. Smad2 overexpression enhances Smad4 gene expression and suppresses CBFA1 gene expression in osteoblastic osteosarcoma ROS17/2.8 cells and primary rat calvaria cells. J Biol Chem. 1998;273:31009–31015. doi: 10.1074/jbc.273.47.31009. [DOI] [PubMed] [Google Scholar]
- 27.Lian J, Stewart C, Puchacz E, Mackowiak S, Shalhoub V, Collart D, Zambetti G, Stein G. Structure of the rat osteocalcin gene and regulation of vitamin D-dependent expression. Proc Natl Acad Sci USA. 1989;86:1143–1147. doi: 10.1073/pnas.86.4.1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lian J B, Stein G S, Stein J L, van Wijnen A J. Osteocalcin gene promoter: unlocking the secrets for regulation of osteoblast growth and differentiation. J Cell Biochem Suppl. 1998;30–31:62–72. doi: 10.1002/(SICI)1097-4644(1998)72:30/31+<62::AID-JCB10>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 29.Lin Y C, Stavnezer J. Regulation of transcription of the germ-line Ig alpha constant region gene by an ATF element and by novel transforming growth factor-beta 1-responsive elements. J Immunol. 1992;149:2914–2925. [PubMed] [Google Scholar]
- 30.Lowell B B. PPARγ: an essential regulator of adipogenesis and modulator of fat cell function. Cell. 1999;99:239–242. doi: 10.1016/s0092-8674(00)81654-2. [DOI] [PubMed] [Google Scholar]
- 31.Massague J. The transforming growth factor-beta family. Annu Rev Cell Biol. 1990;6:597–641. doi: 10.1146/annurev.cb.06.110190.003121. [DOI] [PubMed] [Google Scholar]
- 32.Massague J. TGF-beta signal transduction. Annu Rev Biochem. 1998;67:753–791. doi: 10.1146/annurev.biochem.67.1.753. [DOI] [PubMed] [Google Scholar]
- 33.Melnikova I N, Crute B E, Wang S, Speck N A. Sequence specificity of the core-binding factor. J Virol. 1993;67:2408–2411. doi: 10.1128/jvi.67.4.2408-2411.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mundlos S, Otto F, Mundlos C, Mulliken J B, Aylsworth A S, Albright S, Lindhout D, Cole W G, Henn W, Knoll J H, Owen M J, Mertelsmann R, Zabel B U, Olsen B R. Mutations involving the transcription factor CBFA1 cause cleidocranial dysplasia. Cell. 1997;89:773–779. doi: 10.1016/s0092-8674(00)80260-3. [DOI] [PubMed] [Google Scholar]
- 35.Namiki M, Akiyama S, Katagiri T, Suzuki A, Ueno N, Yamaji N, Rosen V, Wozney J M, Suda T. A kinase domain-truncated type I receptor blocks bone morphogenetic protein-2-induced signal transduction in C2C12 myoblasts. J Biol Chem. 1997;272:22046–22052. doi: 10.1074/jbc.272.35.22046. [DOI] [PubMed] [Google Scholar]
- 36.Nishimura R, Kato Y, Chen D, Harris S E, Mundy G R, Yoneda T. Smad5 and DPC4 are key molecules in mediating BMP-2-induced osteoblastic differentiation of the pluripotent mesenchymal precursor cell line C2C12. J Biol Chem. 1998;273:1872–1879. doi: 10.1074/jbc.273.4.1872. [DOI] [PubMed] [Google Scholar]
- 37.Noda M, Rodan G A. Type-beta transforming growth factor inhibits proliferation and expression of alkaline phosphatase in murine osteoblast-like cells. Biochem Biophys Res Commun. 1986;140:56–65. doi: 10.1016/0006-291x(86)91057-0. [DOI] [PubMed] [Google Scholar]
- 38.Ogawa E, Maruyama M, Kagoshima H, Inuzuka M, Lu J, Satake M, Shigesada K, Ito Y. PEBP2/PEA2 represents a family of transcription factors homologous to the products of the Drosophila runt gene and the human AML1 gene. Proc Natl Acad Sci USA. 1993;90:6859–6863. doi: 10.1073/pnas.90.14.6859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ogawa E, Inuzuka M, Maruyama M, Satake M, Naito-Fujimoto M, Ito Y, Shigesada K. Molecular cloning and characterization of PEBP2 beta, the heterodimeric partner of a novel Drosophila runt-related DNA binding protein PEBP2 alpha. Virology. 1993;194:314–331. doi: 10.1006/viro.1993.1262. [DOI] [PubMed] [Google Scholar]
- 40.Otto F, Thornell A P, Crompton T, Denzel A, Gilmour K C, Rosewell I R, Stamp G W, Beddington R S, Mundlos S, Olsen B R, Selby P B, Owen M J. Cbfa1, a candidate gene for cleidocranial dysplasia syndrome, is essential for osteoblast differentiation and bone development. Cell. 1997;89:765–771. doi: 10.1016/s0092-8674(00)80259-7. [DOI] [PubMed] [Google Scholar]
- 41.Reddi A H. Bone and cartilage differentiation. Curr Opin Genet Dev. 1994;4:737–744. doi: 10.1016/0959-437x(94)90141-o. [DOI] [PubMed] [Google Scholar]
- 42.Roberts A B, Flanders K C, Kondaiah P, Thompson N L, Obberghen-Schilling E, Wakefield L, Rossi P, de Crombrugghe B, Heine U, Sporn M B. Transforming growth factor beta: biochemistry and roles in embryogenesis, tissue repair and remodeling, and carcinogenesis. Recent Prog Horm Res. 1988;44:157–197. doi: 10.1016/b978-0-12-571144-9.50010-7. [DOI] [PubMed] [Google Scholar]
- 43.Rossert J, Crombrugghe B. Principles of bone biology. San Diego, Calif: Academic Press; 1996. pp. 127–142. [Google Scholar]
- 44.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1988. [Google Scholar]
- 45.Schreiber E, Matthias P, Muller M M, Schaffner W. Rapid detection of octamer binding proteins with ‘mini-extracts’, prepared from a small number of cells. Nucleic Acids Res. 1989;17:6419. doi: 10.1093/nar/17.15.6419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Seyedin S M, Thompson A Y, Bentz H, Rosen D M, McPherson J M, Conti A, Siegel N R, Galluppi G R, Piez K A. Cartilage-inducing factor-A. Apparent identity to transforming growth factor-beta. J Biol Chem. 1986;261:5693–5695. [PubMed] [Google Scholar]
- 47.Shi M J, Stavnezer J. CBF alpha3 (AML2) is induced by TGF-beta1 to bind and activate the mouse germline Ig alpha promoter. J Immunol. 1998;161:6751–6760. [PubMed] [Google Scholar]
- 48.Stewart M, Terry A, Hu M, O'Hara M, Blyth K, Baxter E, Cameron E, Onions D E, Neil J C. Proviral insertions induce the expression of bone-specific isoforms of PEBP2alphaA (CBFA1): evidence for a new myc collaborating oncogene. Proc Natl Acad Sci USA. 1997;94:8646–8651. doi: 10.1073/pnas.94.16.8646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Thirunavukkarasu K, Mahajan M, McLarren K W, Stifani S, Karsenty G. Two domains unique to osteoblast-specific transcription factor Osf2/Cbfa1 contribute to its transactivation function and its inability to heterodimerize with Cbfβ. Mol Cell Biol. 1998;18:4197–4208. doi: 10.1128/mcb.18.7.4197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tontonoz P, Hu E, Spiegelman B M. Stimulation of adipogenesis in fibroblasts by PPAR gamma 2, a lipid-activated transcription factor. Cell. 1994;79:1147–1156. doi: 10.1016/0092-8674(94)90006-x. [DOI] [PubMed] [Google Scholar]
- 51.Tsuji K, Ito Y, Noda M. Expression of the PEBP2alphaA/AML3/CBFA1 gene is regulated by BMP4/7 heterodimer and its overexpression suppresses type I collagen and osteocalcin gene expression in osteoblastic and nonosteoblastic mesenchymal cells. Bone. 1998;22:87–92. doi: 10.1016/s8756-3282(97)00267-6. [DOI] [PubMed] [Google Scholar]
- 52.Wang E A, Rosen V, Cordes P, Hewick R M, Kriz M J, Luxenberg D P, Sibley B S, Wozney J M. Purification and characterization of other distinct bone-inducing factors. Proc Natl Acad Sci USA. 1988;85:9484–9488. doi: 10.1073/pnas.85.24.9484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang S, Wang Q, Crute B E, Melnikova I N, Keller S R, Speck N A. Cloning and characterization of subunits of the T-cell receptor and murine leukemia virus enhancer core-binding factor. Mol Cell Biol. 1993;13:3324–3339. doi: 10.1128/mcb.13.6.3324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wozney J M, Rosen V, Celeste A J, Mitsock L M, Whitters M J, Kriz R W, Hewick R M, Wang E A. Novel regulators of bone formation: molecular clones and activities. Science. 1988;242:1528–1534. doi: 10.1126/science.3201241. [DOI] [PubMed] [Google Scholar]
- 55.Wrana J L. Regulation of Smad activity. Cell. 2000;100:189–192. doi: 10.1016/s0092-8674(00)81556-1. [DOI] [PubMed] [Google Scholar]
- 56.Yamamoto N, Akiyama S, Katagiri T, Namiki M, Kurokawa T, Suda T. Smad1 and smad5 act downstream of intracellular signalings of BMP-2 that inhibits myogenic differentiation and induces osteoblast differentiation in C2C12 myoblasts. Biochem Biophys Res Commun. 1997;238:574–580. doi: 10.1006/bbrc.1997.7325. [DOI] [PubMed] [Google Scholar]
- 57.Zhang Y, Derynck R. Regulation of Smad signalling by protein associations and signalling crosstalk. Trends Cell Biol. 1999;9:274–279. doi: 10.1016/s0962-8924(99)01579-2. [DOI] [PubMed] [Google Scholar]