Abstract
Diabetes mellitus (DM) is a common feline endocrinopathy, which is similar to human type 2 diabetes (T2DM) in terms of its pathophysiology. T2DM occurs due to peripheral insulin resistance and/or β-cell dysfunction. Several studies have identified genetic and environmental factors that contribute to susceptibility to human T2DM. In cats, environmental factors such as obesity and physical inactivity have been linked with DM, although to date, the only genetic association that has been demonstrated is with a polymorphism in the feline MC4R gene. The aim of this study was to perform a genome-wide association study (GWAS) to identify polymorphisms associated with feline DM. Illumina Infinium 63k iSelect DNA arrays were used to analyse genomic DNA samples from 200 diabetic domestic shorthair cats and 399 non-diabetic control cats. Data was analysed using PLINK whole genome data analysis toolset. A linear model analysis, EMMAX, was done to test for population structure and HAPLOVIEW was used to identify haplotype blocks surrounding the significant SNPs to assist with candidate gene nomination. A total of 47,497 SNPs were available for analysis. Four SNPs were identified with genome-wide significance: chrA2.4150731 (praw = 9.94 x10-8); chrUn17.115508 (praw = 6.51 x10-8); chrUn17.394136 (praw = 2.53 x10-8); chrUn17.314128 (praw = 2.53 x10-8) as being associated with DM. The first SNP is located within chromosome A2, less than 4kb upstream of the dipeptidyl-peptidase-9 (DPP9) gene, a peptidase involved in incretin inactivation. The remaining three SNPs are located within a haplotype block towards the end of chromosome A3; within this region, genes of interest include TMEM18 and ACP1, both previously associated with T2DM. This study indicates a polygenic component to susceptibility to DM in cats and has highlighted several loci and candidate genes worthy of further investigation.
Introduction
Diabetes mellitus (DM) is a heterogeneous disease, characterised by a failure of glucose homeostasis, leading to persistent hyperglycaemia. In humans, the majority of affected individuals suffer from type 2 DM (T2DM), which is characterised by peripheral insulin resistance alongside defects in pancreatic beta cell secretion of insulin [1, 2]. Absolute or relative insulin deficiency leads to hyperglycaemia and abnormalities in carbohydrate, lipid and protein metabolism, as well as accelerated hepatic glycogenolysis and gluconeogenesis. T2DM is reaching epidemic proportions world-wide and represents a substantial proportion of the healthcare costs in many developed countries [3, 4]. Susceptibility to T2DM occurs as a consequence of a combination of environmental and lifestyle factors, mainly through obesity, inactivity, corticosteroid administration and secondary insulin resistance [5]; however, it is also known that T2DM has a strong genetic and epigenetic component [6, 7]. T2DM is a complex genetic disorder and several genetic studies, including genome-wide association studies (GWAS) have now identified more than 50 genes that are associated with the disease [8, 9]. Most of these susceptibility genes are associated with beta-cell biology, although the mechanisms through which they are linked to dysfunction and T2DM are still largely unknown.
DM is one of the most common feline endocrinopathies, with most cats suffering from a condition similar to T2DM in humans. The prevalence of DM in the overall cat population in the UK is of around 0.4%-0.58% (with an estimated cat population of 7.3 million) [10, 11], depending on the study, or 0.01% in the US with a 15 fold increase in a 30 year period [12] and 0.74% in Australia [13]. The disease is most commonly seen in domestic shorthair (DSH) and domestic longhair (DLH) cats [11, 14]. Other studies from Australia, the UK, USA and New Zealand have shown that mainly Burmese cats are overrepresented in the diabetic population [11–15]. Other breeds with increased odds ratio include Norwegian Forest, Tonkinese, Russian Blue and Oriental, whereas other pure breeds are underrepresented such as Bengal, Birman, Ragdoll, British Short Hair amongst others [11, 14]. The evidence of breed predisposition supports a genetic underlying mechanism for the development of T2DM in the cat.
Similar to the situation in humans, susceptibility to feline DM is thought to be influenced by environmental and genetic risk factors [10, 16–21]. Although little is known about the genetic risk factors in feline DM, human type 2 diabetes (T2DM) and feline DM share many of the environmental risk factors, such as obesity, physical inactivity, eating dry food and corticosteroid administration [10, 12, 22]. A polymorphism in the coding sequence of the melanocortin 4 receptor (MC4R) gene has been reported to be associated with feline DM, in a population of overweight DSH cats in the UK [21]. Recent studies in Burmese cats have also suggested the possibility that this breed has a genetically-conferred derangement of lipid metabolism, which might predispose them to developing DM, however, this study only evaluated non-diabetic cats and therefore the impact of this derangement in the diabetic Burmese population has not been determined [23].
Given the similarity in the pathophysiology, clinicopathological consequences and risk factors when comparing human T2DM and feline DM, it seems possible that feline DM could be a good model for the study of this disease in people [24] and it seems also possible that similar genetic and epigenetic susceptibility factors might be involved in development of the disease in both species.
Recently, the release of the feline genome assembly and availability of feline SNP (Single Nucleotide Polymorphism) genotyping arrays has allowed GWAS to be undertaken for breed-specific diseases such as hypokalaemia in Burmese cats [25] or breed susceptibility to feline infectious peritonitis [26], amongst others [27, 28]. For feline DM, two recent GWAS analysis in this breed identified several candidate genes [20], as well as genomic regions involved in lipid metabolism, insulin sensitivity and Beta-cell dysfunction as likely to be associated with diabetes mellitus in this breed [19]. The aim of the present study was to identify genetic factors associated with susceptibility to DM in non-obese DSH cats by using GWAS as a tool.
Materials and methods
Study population
Blood samples (EDTA or clots from serum tubes) from diabetic cats (n = 200) were recruited at the Queen Mother Hospital for Animals, Royal Veterinary College or from first opinion practices, through the UK Companion Animal Diabetes Register. External samples were submitted through a standardized submission form, which included information about signalment, presence of concurrent diseases, clinical signs, body condition score (BCS) at sample submission and at/before diagnosis, diet and insulin type and dose. BCS assessment was carried out according to published 9- and 5-point BCS systems [29, 30]. Diabetic cats were selected on the basis of the presence of persistent hyperglycaemia and elevated serum fructosamine concentration with all cats requiring insulin treatment. Insulin-like growth factor-1 (IGF-1) was measured in all samples to exclude hypersomatotropism-related DM. DSH cats with an IGF-1 concentration <800 ng/ml and BCS ≤ 4/5 or 5/9 and/or BW <4kg were included in the study.
Blood samples (EDTA or clots) from non-diabetic cats (n = 399) were obtained from the Royal Veterinary College sample archive, consisting of residual samples after completion of diagnostic testing from referral and first opinion cases seen at the Queen Mother Hospital for Animals and the Beaumont Sainsbury Animal Hospital. Only DSH cats that were 10 years old or older at the time of sample collection and with no clinical or biochemical evidence of DM were included. There were no known familial relationships between any of the cats included in the study. The Royal Veterinary College’s Ethics and Welfare Committee approval was granted for use of blood samples for clinical research (URN 2011–120).
DNA extraction
Genomic DNA was extracted from blood samples using the GenElute Blood Genomic DNA Kit (Sigma-Aldrich, UK) according to the manufacturer’s instructions. The concentration of DNA was measured using a NanoDrop 1000 spectrophotometer (NanoDrop products, Wilmington, Delaware, USA). Genomic DNA samples were submitted to Geneseek Inc. (Lincoln, NE, USA) for SNP genotyping using the Illumina Infinium 63k iSelect DNA array (Illumina Inc., San Diego, USA).
Genome-wide association study
Population structure and quality control
Quality control was performed for DNA from 599 cats (200 diabetic, 399 controls) and 62897 SNPs using PLINK 1.9 [31]. Possible duplicate samples were identified using the–genome operator, which looks at the genetic relatedness of all pairs of samples. Samples with PI_HAT > 0.6 were identified as duplicates and the individual in each duplicate pair with the highest level of SNP missingness was removed. Population structure was examined using QQ plots and Multi Dimensional Scaling (MDS) plots, using PLINK 1.9 and EMMAX [32]. Quality control of the SNP was performed by removing those with minor allele frequency below 0.05 (-maf), or a variant call rate (-geno) below 0.9. Samples with a genotyping rate (-mind) of less than 0.9 were also removed.
Case-control genome-wide association analysis
The genotyping data was analysed using PLINK 1.9 whole genome data analysis toolset. A map document (S1 Table) with the chromosomal location of each SNP aligned to the latest Feline assembly (FelCat9) was used for analysis [19]. The case-control association analysis was performed on the available samples (-assoc). The association results from PLINK were plotted as -logp values (praw) and also as max(T) empirical p values (pgenome) using the–mperm 100,000 option in PLINK, to correct for multiple testing. Max(T) permuted p-values were considered significant at a pgenome value <0.05. Data for the Manhattan plots were created using PLINK. Once the analysis was finalised, those SNPs above the significance threshold were located within the latest feline genome assembly (Felis catus 9.0) to look for possible candidate genes.
Haplotype analysis
Haplotype analysis was carried out using HAPLOVIEW [33] to determine whether the significant SNPs were in linkage disequilibrium (LD) with any possible candidate genes. Using the–clump option of PLINK, with linkage disequilibrium set at r2 > 0.2, SNPs were also grouped based on LD. Significance of DM-associated haplotype blocks was measured by running 10,000 permutations in HAPLOVIEW.
Results
Study population
Genomic DNA from a total of 200 non-obese diabetic DSH cats and 399 non-diabetic DSH cats was submitted for genotyping. The mean (SD) age of the diabetic population was 11.62 (3.44) years, range 2–21 years. One hundred and twenty eight cats (64%) were male, three of them entire; seventy one cats (36%) were female (five were female entire), sex was unknown in one diabetic cat. The mean (SD) weight of the diabetics was 4.35 (1.01) kg. The mean (SD) age of the control cat population (n = 389) was 14.83 (2.06) years, range 11–24 years. One hundred and eighty two cats (46%) were female, one of them female entire and the rest were female neutered; two hundred and sixteen (54%) were male, neutering status was not known in one cat, the rest were neutered, sex was unknown in one cat. Of the control cats, two hundred and thirty nine (60%) had lean body condition score, seventy six (19%) had overweight body condition score and 84 (21%) had an unknown body condition score.
GWAS
A total of 599 cats were analysed (200 cases, 399 controls). After excluding duplicated samples (n = 8, 3 cases, 5 controls), samples without sex information (n = 2, 1 case, 1 control) and samples with insufficient genotyping rate (n = 2, 1 case, 1 control), 195 diabetic cats and 392 controls were accepted for further analysis. SNPs that did not meet the genotyping (n = 1,462 SNPs) and frequency (n = 11,279 SNPs) criteria were also excluded from further analysis, there were 47,497 SNPs for analysis.
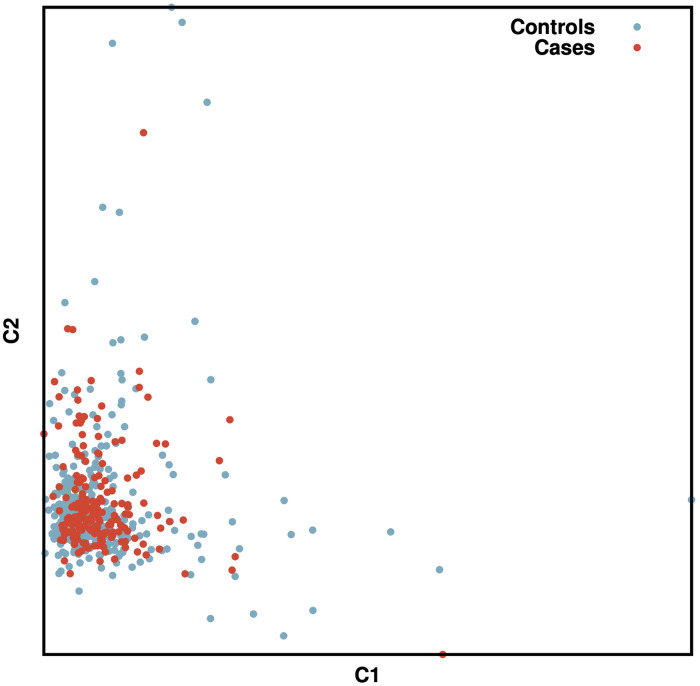
An MDS plot was performed to test for population stratification. This revealed that most cats were within one main cluster, and that both cases and controls seemed to be randomly distributed within the cluster (Fig 1).
Fig 1. MDS plot of all cats included in the GWAS.
On the X axis is the first dimension of the IBS calculation, the Y axis contains the second dimension. The blue dots represent the control cats, while the red dots represent the DM cases.
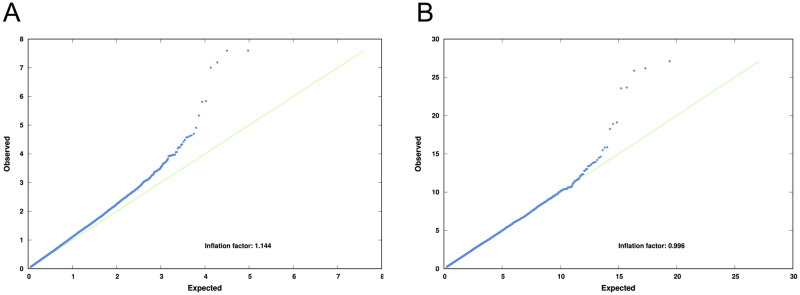
A quantile-quantile (QQ) plot of the data revealed an inflation factor of 1.146, suggesting that there may be some relatedness or structure within the population. After the data was processed using linear mixed model association (EMMAX) the inflation factor was 0.995, demonstrating that EMMAX adjustment had largely coped with any population structure (Fig 2).
Fig 2. Quantile-quantile plots of Chi-Squared values before (A) and after (B) correction by EMMAX.
The blue dots are the SNPs. On the Y axis, the observed Chi-Square value for each SNP, on the X axis is the expected value for each SNP. The green line represents when both expected and observed values coincide (inflation factor = 1).
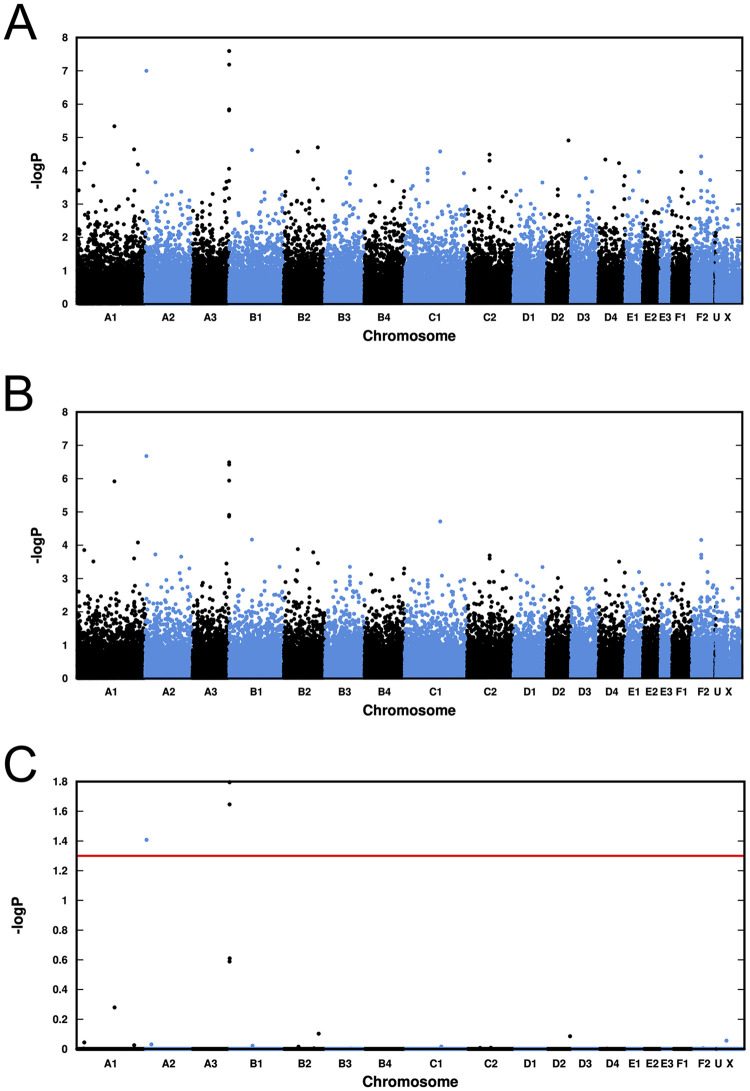
Four SNPs are significant genome-wide at the p<0.05 level after mperm correction (pgenome), one on chromosome A2 and three on chromosome A3: chrA2.4150731 (pgenome = 0.039); chrUn17.115508 (pgenome = 0.022); chrUn17.394136 (pgenome = 0.016); chrUn17.314128 (pgenome = 0.016) (Table 1).
Table 1. Summary of results after association and haplotype analysis.
| Chr | Locus | Type | Position | praw | pgenome | Alleles in locus | Minor allele | Case/contr freqs minor allele | Genes within haplotype block |
|---|---|---|---|---|---|---|---|---|---|
| A3 | chrUn17.394136 | SNP | 142804428 | 2.53 x10-8 | 0.016 | C/A | C | 0.31, 0.48 | 1 |
| TMEM1 | |||||||||
| ACP-18 | |||||||||
| A3 | chrUn17.314128 | SNP | 142885267 | 2.53 x10-8 | 0.016 | G/A | G | 0.31. 0.48 | TMEM18 |
| ACP-1 | |||||||||
| SHY3YL1 | |||||||||
| A3 | chrUn17.115508 | SNP | 143084579 | 6.51x10-7 | 0.022 | A/G | A | 0.31, 0.48 | TPO |
| TMEM18AACP1 | |||||||||
| SHY3YL1 | |||||||||
| FAM110C | |||||||||
| A3 | GAGAA | Haplotype block | 142804428–143195425 | 5.13 x10-8 | 0.0002 | 0.30, 0.47 | TMEM18 | ||
| ACP1 | |||||||||
| SHY3YL1 | |||||||||
| FAM110C | |||||||||
| A2 | chrA2.4150731 | SNP | 3535683 | 9.94 x10-8 | 0.039 | A/C | A | 0.31, 0.17 | DPP9 |
The top most significant loci, with their positions in the FelCat9 assembly, as well as the frequencies in cases and controls, praw and pgenome values and genes in proximity (within 200kb of the SNP) are displayed. Also shown is the haplotype block identified in HAPLOVIEW, and the p-value after running a haplotype association test in Haploview, with and without permutations.
Fig 3 shows a comparison of the association plots of the data before and after correction for population structure by EMMAX, and also a plot of the pgenome values calculated by PLINK mperm 100,000. The top SNP in chromosome A2 has almost maintained its pre-correction value. The “peak” of SNPs in chromosome A3 is also still clearly present, though at a slightly lower value.
Fig 3. Association plots of our case-control genome-wide association analysis.
A) Standard association plot of praw values. B) Plot of EMMAX-corrected association values. (C) Plot of pgenome values after correction for multiple testing by the mperm option of PLINK. The red horizontal line shows the p = 0.05 level of significance. In all plots the chromosome marked “U” contains SNPs which are mapped to the new assembly but not to one of the standard chromosomes.
Haplotype analysis
A haplotype block enclosing the three top SNPs was identified (see Table 1 and Fig 4). This was located in chromosome A3 (praw 5.13 x10-8, pgenome 0.0002) and is present in 30% of the cases and 47% of the controls.
Fig 4. Haplotype blocks around the significant region on chromosome A3.
The block identified by PLINK–clump, and the block identified by HAPLOVIEW are shown in blue. SNP variants are plotted as -log10 praw, and are shown with colours indicating the amount of LD with the most significant SNP, as follows: red (r2 > 0.8), orange (r2 > 0.2) and blue (r2 < 0.2). The rightmost SNP is at the end of the chromosome.
The significant SNP (chrA2.4150731) on chromosome A2 was not part of any haplotype detected, but it does lie within 4kb of the start of the DPP9 gene, so is likely to act as a good proxy marker for the gene.
PLINK–clump and HAPLOVIEW identify slightly different haplotype blocks, but both are overlapping with the top SNP. The two closest genes in this area of LD are TMEM18 and ACP1.
Discussion
This study aimed to identify novel loci associated with Feline DM in a population of lean DSH cats in the United Kingdom. The Illumina 63k feline SNP array was used for this purpose. In this GWAS, a group of 195 lean DSH diabetic cats was compared with 392 DSH controls. In this type of study, where phenotypic classification of cases and controls is of high importance [34], it is possible that selection of cats could have influenced the results. Within the DSH population, cases and controls were identified on the basis of their electronic patient record or information provided on sample submission forms by first opinion veterinary practitioners. Although careful selection took place, according to the information available, the authors did not have full access to the patient’s medical records. With regards to the control cats, the main limitation is that DM is a disease of late onset and therefore some of the control cats chosen for this study might have developed diabetes later in life. In order to minimise this, an attempt was made to select cats as old as possible, and with a diagnosis as robust as possible. All control samples were selected from a population of relatively geriatric cats with a multitude of diseases and this could also have influenced the results. The diabetic population in this study was chosen to have lean body condition. Records of body weight in the control population were not consistently present, but body condition score was available for a majority of them and 60% of control cats were found to be lean, which makes it unlikely that this study could have detected markers for lean body condition as well as for diabetes.
The SNP arrays used for these GWAS were the first manufactured for the feline species. However, the overall number of SNPs is somewhat limited, when compared to the ones used in human studies and other veterinary species such as dogs [35, 36]. This could have influenced the results [34], as it is possible that regions of interest might not have sufficient coverage within this array and therefore some potentially important gene associations could have been missed. Although the relatively low number and quality of SNPs in the array could have influenced the results, several GWAS have been published using the same SNP array for the research of feline diabetes [19, 20], albeit in a more inbred population such as the Burmese cats. Some of the limitations of the current SNP arrays have been overcome by performing haplotype analysis to further limit the presence of type 1 errors and to allow genes of interest that are in linkage disequilibrum (LD) to be determined.
Population structure was evaluated by MDS plots generated for each analysis. The initial MDS plot showed a large cluster with both cases and controls randomly distributed within it. Although the DSH is considered an outbred population, the genotyping data was also assessed using EMMAX, to investigate the effect of hidden relatedness and population stratification on the results. EMMAX, uses a linear model approach (LMM) and is currently preferred to traditional family-based association tests [32]. Quality control criteria for genotyping rate and testing for duplicates was also performed prior to the association analysis.
This GWAS has identified several SNPs that have reached genome-wide significance: 3 in the same region of Chromosome A3: A3:142,804,428, (pgenome: 0.016); A3:142,885,267 (pgenome: 0.016) and A3:143,084,579 (pgenome: 0.022). All three are included in a haplotype block that includes genes linked to human T2DM, including TMEM18 [37–39] and ACP1 [40, 41]. Although not much information is available about the exact role that these genes play in T2DM in humans, they have been reported to be associated with the disease in different populations. In the case of TMEM18, it has been linked to obesity in multiple studies, which would seem an interesting contrast with the results of this study, as SNPs near this gene were identified in our cohort of non-obese diabetic cats. However, more recent studies have reported that the association of TMEM18 with T2DM in humans is independent of BMI [38, 42].
Also in this region of chromosome A3 is the ACP1 gene, encoding a phosphatase that is thought to play a role in regulating glycolytic rate through control of insulin receptor activities [40]. ACP1 polymorphisms have been linked to insulin resistance. The increased risk of T2DM associated with ACP1 seems to be mediated through increased risk of obesity in humans [41], although one study found an association with a polymorphism in this gene with insulin resistance in males, irrespective of BMI [43]. It is possible that these two genes are not associated with feline DM and other genes within this region of chromosome A3 are involved (Table 1), however, given their established link with T2DM in humans, TMEM18 and ACP1 should be assessed more closely in future investigations.
The significant SNP in chromosome A2 (A2: 3535683; pgenome 0.039) was not part of any haplotype block detected, but it does lie within 4kb of the start of the DPP9 gene, so is likely to be inherited with mutations in this gene. DPP9 belongs to a family of serine-proteases, the dipeptidyl-peptidases, and is highly similar in sequence to dipeptidyl-peptidase 4 (DPP4). Although their specific function is not fully described, it has been shown to play a role in post-prandrial insulin secretion as well as in adipogenesis [44, 45]. DPP4 inhibitors are currently being used as part of the medical management of T2DM in humans, as they improve glycaemic control through increasing GLP-1 concentrations, thus stimulating pancreatic insulin secretion, inhibiting glucagon release and reducing appetite [46]. Many of these DPP4 inhibitors also have an inhibitory effect on DPP8 and DPP9 [47], which is again suggestive of similar biological function. Interestingly, DPP4 inhibitors have been investigated recently for the management of DM in cats, with somewhat less success than in humans [48–50]. It would therefore be interesting to further investigate the role of the DPP9 gene in feline DM and whether this could be linked to the relative lack of success obtained with these drugs, when compared to humans with T2DM. Up until now, no variants of DPP9 have been linked to T2DM, but it is possible that this study could lead to the identification of novel genetic associations for T2DM and feline DM.
The lack of a haplotype block around the DPP9 gene may be affected by the fact that the density of SNPs in the array is relatively low in this specific region: the SNP upstream of the current significant SNP is 83kb away, and the next SNP downstream is 38kb away. When comparing this with the SNP density in the region of chromosome A3, for the most significant SNPs near the ACP1 and TMEM18 genes, the upstream SNP is 18kb away and the next downstream SNP is 8kb away, showing that the SNP density is higher in the region of interest on chromosome A3 than in our region of interest in chromosome A2. The DSH breed used in this study is considered to be relatively outbred, which implies shorter haplotype blocks. Although no current studies have specifically looked at the degree of LD present in the DSH population in the UK, a recent study looked into the degree of LD in several cat breeds, including a random bred population [51]. The extent of LD in the random bred population was of approximately 18Kb. This fact, together with the sparcity of SNPs in this region could also explain why no haplotype blocks were identified in Chromosome A2.
Conclusions
Several SNPs have been identified which provide moderate to strong evidence of association to DM in the cat being located within a haplotype block spanning genes that have been associated with human diabetes (ACP1 and TMEM18). An independent significant SNP has also been identified within 4Kb of a gene that could have a pathophysiological link with DM, based on its function (such as DPP9). Further investigations could involve more detailed investigation of the regions and genes of interest highlighted in this GWAS. Additionally, further genetic analysis, preferably in an independent population and making use of higher density SNP genotyping arrays to assess for repeatability of these findings might help to confirm the associations identified in the present study and to detect further associations.
Supporting information
(XLSX)
Acknowledgments
The authors would like to thank Georgina Samaha for providing help with the alignment of the SNPs in the array with the most recent feline assembly.
Data Availability
All relevant data are within the manuscript and its Supporting information files. https://figshare.com/s/3bded2a6bb69bda9f1d7.
Funding Statement
YF received the SNP arrays as a grant from the Morris Animal Foundation. Grant title and code: D12FE-512: Genome-Wide Association Study to Identify Susceptibility Genes for Diabetes Mellitus in Cats. https://www.morrisanimalfoundation.org/ The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.2. Classification and Diagnosis of Diabetes. Diabetes Care. 2015. Jan 1;38(Supplement_1). doi: 10.2337/dc15-S005 [DOI] [PubMed] [Google Scholar]
- 2.American Diabetes Association ADA. 2. Classification and Diagnosis of Diabetes: Standards of Medical Care in Diabetes—2021. Diabetes Care [Internet]. 2021. Jan 9;44(Supplement 1):S15–33. Available from: http://care.diabetesjournals.org/lookup/doi/10.2337/dc21-S00233298413 [Google Scholar]
- 3.Ogurtsova K, da Rocha Fernandes JD, Huang Y, Linnenkamp U, Guariguata L, Cho NH, et al. IDF Diabetes Atlas: Global estimates for the prevalence of diabetes for 2015 and 2040. Diabetes Research and Clinical Practice. 2017. Jun;128. doi: 10.1016/j.diabres.2017.03.024 [DOI] [PubMed] [Google Scholar]
- 4.Khan MAB, Hashim MJ, King JK, Govender RD, Mustafa H, al Kaabi J. Epidemiology of Type 2 Diabetes–Global Burden of Disease and Forecasted Trends. Journal of Epidemiology and Global Health. 2019;10(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.DeFronzo RA, Ferrannini E, Groop L, Henry RR, Herman WH, Holst JJ, et al. Type 2 diabetes mellitus. Nature reviews Disease primers. 2015;1(15019). [DOI] [PubMed] [Google Scholar]
- 6.Lawlor N, Khetan S, Ucar D, Stitzel ML. Genomics of Islet (Dys)function and Type 2 Diabetes. Trends in genetics: TIG. 2017;33(4):244–55. doi: 10.1016/j.tig.2017.01.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lecoutre S, Maqdasy S, Breton C. Maternal obesity as a risk factor for developing diabetes in offspring: An epigenetic point of view. World Journal of Diabetes. 2021. Apr 15;12(4). doi: 10.4239/wjd.v12.i4.366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fuchsberger C, Flannick J, Teslovich TM, Mahajan A, Agarwala V, Gaulton KJ, et al. The genetic architecture of type 2 diabetes. Nature. 2016. Aug 11;536(7614). doi: 10.1038/nature18642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Himanshu D, Ali W, Wamique M. Type 2 diabetes mellitus: pathogenesis and genetic diagnosis. Journal of Diabetes & Metabolic Disorders. 2020. Dec 22;19(2). doi: 10.1007/s40200-020-00641-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McCann TM, Simpson KE, Shaw DJ, Butt JA, Gunn-Moore DA. Feline diabetes mellitus in the UK: the prevalence within an insured cat population and a questionnaire-based putative risk factor analysis. J Feline Med Surgery. 2007;9(4):289–99. doi: 10.1016/j.jfms.2007.02.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.O’Neill DG, Gostelow R, Orme C, Church DB, Niessen SJM, Verheyen K, et al. Epidemiology of Diabetes Mellitus among 193,435 Cats Attending Primary‐Care Veterinary Practices in England. Journal of Veterinary Internal Medicine. 2016. Jul 29;30(4). doi: 10.1111/jvim.14365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Prahl A, Guptill L, Glickman NW, Tetrick M, Glickman LT. Time trends and risk factors for diabetes mellitus in cats presented to veterinary teaching hospitals. Journal of Feline Medicine and Surgery. 2007. Oct 1;9(5). doi: 10.1016/j.jfms.2007.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lederer R, Rand JS, Jonsson NN, Hughes IP, Morton JM. Frequency of feline diabetes mellitus and breed predisposition in domestic cats in Australia. The Veterinary Journal. 2009. Feb;179(2). doi: 10.1016/j.tvjl.2007.09.019 [DOI] [PubMed] [Google Scholar]
- 14.Öhlund M, Fall T, Ström Holst B, Hansson‐Hamlin H, Bonnett B, Egenvall A. Incidence of Diabetes Mellitus in Insured Swedish Cats in Relation to Age, Breed and Sex. Journal of Veterinary Internal Medicine. 2015. Sep 14;29(5). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rand JS, Bobbermien LM, Hendrikz JK, Copland M. Over representation of Burmese cats with diabetes mellitus. Australian Veterinary Journal. 1997. Jun;75(6):402–5. doi: 10.1111/j.1751-0813.1997.tb14340.x [DOI] [PubMed] [Google Scholar]
- 16.Rand JS, Fleeman LM, Farrow HA, Appleton DJ, Lederer R. Canine and Feline Diabetes Mellitus: Nature or Nurture? The Journal of Nutrition. 2004. Aug 1;134(8). doi: 10.1093/jn/134.8.2072s [DOI] [PubMed] [Google Scholar]
- 17.Slingerland LI, Fazilova VV, Plantinga EA, Kooistra HS, Beynen AC. Indoor confinement and physical inactivity rather than the proportion of dry food are risk factors in the development of feline type 2 diabetes mellitus. The Veterinary Journal. 2009. Feb;179(2). [DOI] [PubMed] [Google Scholar]
- 18.Sallander M, Eliasson J, Hedhammar Å. Prevalence and risk factors for the development of diabetes mellitus in Swedish cats. Acta Veterinaria Scandinavica. 2012. Dec 31;54(1). doi: 10.1186/1751-0147-54-61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Samaha G, Wade CM, Beatty J, Lyons LA, Fleeman LM, Haase B. Mapping the genetic basis of diabetes mellitus in the Australian Burmese cat (Felis catus). Scientific Reports. 2020. Dec 5;10(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Balmer L, O’Leary CA, Menotti-Raymond M, David V, O’Brien S, Penglis B, et al. Mapping of Diabetes Susceptibility Loci in a Domestic Cat Breed with an Unusually High Incidence of Diabetes Mellitus. Genes. 2020. Nov 19;11(11). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Forcada Y, Holder A, Church DB, Catchpole B. A Polymorphism in the Melanocortin 4 Receptor Gene (MC4R:c.92C>T) Is Associated with Diabetes Mellitus in Overweight Domestic Shorthaired Cats. Journal of Veterinary Internal Medicine. 2014. Mar;28(2). doi: 10.1111/jvim.12275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Öhlund M, Egenvall A, Fall T, Hansson-Hamlin H, Röcklinsberg H, Holst BS. Environmental Risk Factors for Diabetes Mellitus in Cats. Journal of Veterinary Internal Medicine. 2017. Jan;31(1). doi: 10.1111/jvim.14618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee P, Mori A, Coradini M, Mori N, Sagara F, Yamamoto I, et al. Potential predictive biomarkers of obesity in Burmese cats. The Veterinary Journal. 2013. Feb;195(2). doi: 10.1016/j.tvjl.2012.06.027 [DOI] [PubMed] [Google Scholar]
- 24.Henson MS, O’Brien TD. Feline Models of Type 2 Diabetes Mellitus. ILAR Journal [Internet]. 2006. Jan 1;47(3):234–42. Available from: https://academic.oup.com/ilarjournal/article-lookup/doi/10.1093/ilar.47.3.234 [DOI] [PubMed] [Google Scholar]
- 25.Gandolfi B, Gruffydd-Jones TJ, Malik R, Cortes A, Jones BR, Helps CR, et al. First WNK4-Hypokalemia Animal Model Identified by Genome-Wide Association in Burmese Cats. PLoS ONE. 2012. Dec 28;7(12). doi: 10.1371/journal.pone.0053173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Golovko L, Lyons LA, Liu H, Sørensen A, Wehnert S, Pedersen NC. Genetic susceptibility to feline infectious peritonitis in Birman cats. Virus Research. 2013. Jul;175(1). doi: 10.1016/j.virusres.2013.04.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Filler S, Alhaddad H, Gandolfi B, Kurushima JD, Cortes A, Veit C, et al. Selkirk Rex: Morphological and Genetic Characterization of a New Cat Breed. Journal of Heredity. 2012;103(5). doi: 10.1093/jhered/ess039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Alhaddad H, Gandolfi B, Grahn RA, Rah H-C, Peterson CB, Maggs DJ, et al. Genome-wide association and linkage analyses localize a progressive retinal atrophy locus in Persian cats. Mammalian Genome. 2014. Aug 29;25(7–8). doi: 10.1007/s00335-014-9517-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kronfeld DS, Donoghue S, Glickman LT. Body Condition of Cats. The Journal of Nutrition. 1994. Dec 1;124(suppl_12). doi: 10.1093/jn/124.suppl_12.2683S [DOI] [PubMed] [Google Scholar]
- 30.LaFlamme DP. Development and validation of a body condition score system for cats: a clinical tool. Feline Practice 1997;25(5–6):13–8. 1997;25(5–6):13–8. [Google Scholar]
- 31.Chang CC, Chow CC, Tellier LC, Vattikuti S, Purcell SM, Lee JJ. Second-generation PLINK: rising to the challenge of larger and richer datasets. GigaScience. 2015. Dec 25;4(1). doi: 10.1186/s13742-014-0042-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Eu-ahsunthornwattana J, Miller EN, Fakiola M, Jeronimo SMB, Blackwell JM, Cordell HJ. Comparison of Methods to Account for Relatedness in Genome-Wide Association Studies with Family-Based Data. PLoS Genetics. 2014. Jul 17;10(7). doi: 10.1371/journal.pgen.1004445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005. Jan 15;21(2). doi: 10.1093/bioinformatics/bth457 [DOI] [PubMed] [Google Scholar]
- 34.Seng KC, Seng CK. The success of the genome-wide association approach: a brief story of a long struggle. European Journal of Human Genetics. 2008. May 20;16(5). doi: 10.1038/ejhg.2008.12 [DOI] [PubMed] [Google Scholar]
- 35.Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature [Internet]. 2007. Jun;447(7145):661–78. Available from: http://www.nature.com/articles/nature05911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lequarré A-S, Andersson L, André C, Fredholm M, Hitte C, Leeb T, et al. LUPA: A European initiative taking advantage of the canine genome architecture for unravelling complex disorders in both human and dogs. The Veterinary Journal. 2011. Aug;189(2). doi: 10.1016/j.tvjl.2011.06.013 [DOI] [PubMed] [Google Scholar]
- 37.Dusatkova L, Zamrazilova H, Sedlackova B, Vcelak J, Hlavaty P, Aldhoon Hainerova I, et al. Association of obesity susceptibility gene variants with metabolic syndrome and related traits in 1,443 Czech adolescents. Folia Biol (Praha). 2013;59(3):123–33. [PubMed] [Google Scholar]
- 38.Kalnina I, Zaharenko L, Vaivade I, Rovite V, Nikitina-Zake L, Peculis R, et al. Polymorphisms in FTO and near TMEM18 associate with type 2 diabetes and predispose to younger age at diagnosis of diabetes. Gene. 2013. Sep;527(2). doi: 10.1016/j.gene.2013.06.079 [DOI] [PubMed] [Google Scholar]
- 39.Takeuchi F, Yamamoto K, Katsuya T, Nabika T, Sugiyama T, Fujioka A, et al. Association of genetic variants for susceptibility to obesity with type 2 diabetes in Japanese individuals. Diabetologia. 2011. Jun 3;54(6). doi: 10.1007/s00125-011-2086-8 [DOI] [PubMed] [Google Scholar]
- 40.Bottini N, Gloria-Bottini F, Borgiani P, Antonacci E, Lucarelli P, Bottini E. Type 2 diabetes and the genetics of signal transduction: A study of interaction between adenosine deaminase and acid phosphatase locus 1 polymorphisms. Metabolism. 2004. Aug;53(8). doi: 10.1016/j.metabol.2004.03.006 [DOI] [PubMed] [Google Scholar]
- 41.Gloria-Bottini F, Magrini A, di Renzo L, de Lorenzo A, Bergamaschi A, Bottini E. Body mass index and acid phosphatase locus 1 in diabetic disorders. Acta Diabetologica. 2010. Dec 24;47(S1). [DOI] [PubMed] [Google Scholar]
- 42.Thomsen M, Dahl M, Tybjærg-Hansen A, Nordestgaard BG. β2-Adrenergic Receptor Thr164Ile Polymorphism, Obesity, and Diabetes: Comparison with FTO, MC4R, and TMEM18 Polymorphisms in More Than 64,000 Individuals. The Journal of Clinical Endocrinology & Metabolism. 2012. Jun 1;97(6). [DOI] [PubMed] [Google Scholar]
- 43.Shu Y-H, Hartiala J, Xiang AH, Trigo E, Lawrence JM, Allayee H, et al. Evidence for Sex-Specific Associations between Variation in Acid Phosphatase Locus 1 (ACP1) and Insulin Sensitivity in Mexican-Americans. The Journal of Clinical Endocrinology & Metabolism. 2009. Oct 1;94(10). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Han R, Wang X, Bachovchin W, Zukowska Z, Osborn JW. Inhibition of dipeptidyl peptidase 8/9 impairs preadipocyte differentiation. Scientific Reports. 2015. Dec 5;5(1). doi: 10.1038/srep12348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sell H, Bluher M, Kloting N, Schlich R, Willems M, Ruppe F, et al. Adipose Dipeptidyl Peptidase-4 and Obesity: Correlation with insulin resistance and depot-specific release from adipose tissue in vivo and in vitro. Diabetes Care. 2013. Dec 1;36(12). doi: 10.2337/dc13-0496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Deacon CF. DPP-4 inhibitor therapy: new directions in the treatment of type 2 diabetes. Frontiers in Bioscience. 2008;13(13). doi: 10.2741/2799 [DOI] [PubMed] [Google Scholar]
- 47.Huan Y, Jiang Q, Liu J, Shen Z. Establishment of a dipeptidyl peptidases (DPP) 8/9 expressing cell model for evaluating the selectivity of DPP4 inhibitors. Journal of Pharmacological and Toxicological Methods. 2015. Jan;71. doi: 10.1016/j.vascn.2014.11.002 [DOI] [PubMed] [Google Scholar]
- 48.Gilor C, Rudinsky AJ, Hall MJ. New Approaches to Feline Diabetes Mellitus. Journal of Feline Medicine and Surgery. 2016. Sep 25;18(9). doi: 10.1177/1098612X16660441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Padrutt I, Lutz TA, Reusch CE, Zini E. Effects of the glucagon-like peptide-1 (GLP-1) analogues exenatide, exenatide extended-release, and of the dipeptidylpeptidase-4 (DPP-4) inhibitor sitagliptin on glucose metabolism in healthy cats. Research in Veterinary Science. 2015. Apr;99. [DOI] [PubMed] [Google Scholar]
- 50.Riederer A, Zini E, Salesov E, Fracassi F, Padrutt I, Macha K, et al. Effect of the Glucagon‐like Peptide‐1 Analogue Exenatide Extended Release in Cats with Newly Diagnosed Diabetes Mellitus. Journal of Veterinary Internal Medicine. 2016. Jan 24;30(1). doi: 10.1111/jvim.13817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Alhaddad H, Khan R, Grahn RA, Gandolfi B, Mullikin JC, Cole SA, et al. Extent of Linkage Disequilibrium in the Domestic Cat, Felis silvestris catus, and Its Breeds. Ellegren H, editor. PLoS ONE [Internet]. 2013. Jan 7;8(1):e53537. Available from: https://dx.plos.org/10.1371/journal.pone.0053537 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(XLSX)
Data Availability Statement
All relevant data are within the manuscript and its Supporting information files. https://figshare.com/s/3bded2a6bb69bda9f1d7.