Abstract
Coronavirus disease 2019 (COVID-19) is an infectious disease caused by the SARS-CoV-2 betacoronavirus and has taken over 761,426 American lives as of the date of publication and will likely result in long-term, if not permanent, tissue damage for countless patients. COVID-19 presents with diverse and multisystemic pathologic processes, including a hyperinflammatory response, acute respiratory distress syndrome (ARDS), vascular injury, microangiopathy, tissue fibrosis, angiogenesis, and widespread thrombosis across multiple organs, including the lungs, heart, kidney, liver, and brain. C-X-C chemokines contribute to these pathologies by attracting inflammatory mediators, the disruption of endothelial cell integrity and function, and the initiation and propagation of the cytokine storm. Among these, CXCL10 is recognized as a critical contributor to the hyperinflammatory state and poor prognosis in COVID-19. CXCL10 is also known to regulate growth factor-induced fibrosis, and recent evidence suggests the CXCL10-CXCR3 signaling system may be vital in targeting convergent pro-inflammatory and pro-fibrotic pathways. This review will explore the mechanistic role of CXCL10 and related chemokines in fibrotic complications associated with COVID-19 and the potential of CXCL10-targeted therapeutics for early intervention and long-term treatment of COVID-19-induced fibrosis.
Keywords: Fibrosis, Chemokines, COVID-19, CXCL10
Introduction
SARS-CoV-2 infection and the subsequent COVID-19 disease manifestation involve extensive organ damage, including the lungs, heart, kidney, liver, and brain. The multi-organ complications of COVID-19 result from compromised vascular integrity, which leads to fluid retention, excessive inflammatory cell infiltration, endotheliitis, and the activation of coagulation pathways. Viral damage and the multi-organ hyperinflammatory state, often referred to as the “cytokine storm,” are thought to cause tissue damage through repetitive insult and induce a continuously remodeling fibrotic state. Fibrosis has emerged as a serious and long-lasting comorbidity of COVID-19. However, the lingering effects of the disease, often referred to as post-COVID-19 syndrome, remain elusive. An effective therapeutic has yet to sufficiently address both the underlying causes of fibrosis and the observed aberrant viral-induced tissue damage. This review will explore the intersection of the inflammatory and fibrotic state in COVID-19 and potential therapeutics to address these convergent pathologies.
A major co-morbidity of COVID-19 is acute respiratory distress syndrome (ARDS). An estimated 50% of hospitalized COVID-19 patients with COVID-19-induced pneumonia develop ARDS, which has a 52.4% mortality rate [1]. ARDS is associated with diffuse alveolar damage, fibrin-rich hyaline membranes, increased epithelial and endothelial cell permeability, fluid leakage into the pulmonary interstitium, gross disruption of gas exchange, hypoxia and respiratory failure, and subsequent pulmonary fibrosis [2]. Evidence of pulmonary fibrosis is evident in moderate to severe COVID-19 patients through lung volume loss, architectural distortion, and fibrotic bands in the late or remission stages of COVID-19 [3] and in autopsies [4]. Pulmonary fibrosis is also found in SARS patients 4 and 5 weeks after recovery and in autopsies [5–8]. A 3 year follow-up revealed that ten out of forty-six (21.74%) patients had restrictive ventilation dysfunction, but fortunately, most recovered by 15 years [9].
Endothelial cells have emerged as a central player in the pathogenesis of ARDS and multi-organ failure in patients with COVID-19. The SARS-CoV-2 virus damages the vasculature both directly and indirectly. Pathological analysis of the pulmonary vasculature of COVID-19 patients shows high levels of microthrombi and vessel growth through intussusceptive angiogenesis and, to a lesser extent, sprouting angiogenesis, which is positively correlated with hospitalization duration [10]. COVID-19 lung autopsies show diffuse alveolar damage with widespread micro- and macro-thrombi consistent with that of ARDS [11]. Inflammatory-related genes associated with intussusceptive angiogenesis have been reported to be upregulated in both ARDS and COVID-19 patients [11]. By direct infection, SARS-CoV-2 can cause endotheliitis and subsequent endothelial cell death [12]. SARS-CoV-2 can indirectly cause an increase in vasculature permeability through activation of the kallikrein–bradykinin pathway by SARS-CoV-2/ACE2 binding and subsequent ACE2 inhibition [13].
SARS-CoV-2 infection also results in pulmonary endothelial cell recruitment of neutrophils, producing reactive oxygen species (ROS) [14]. ROS increases inter-endothelial gaps by increasing endothelial cell contractility while simultaneously loosening endothelial cell junctions. Fluid retention then arises from IL-1β and TNF activation of glucuronidases which degrades glycocalyx and upregulates hyaluronic acid synthase 2 [14]. Endothelial cells simulated by IL-1β and TNF increase P-selectin, von Willebrand factor, and fibrinogen expression which bind and aggregate platelets [15]. The protein C/protein S pathway, a disintegrin and metalloproteinase with a thrombospondin type 1 motif member 13 (ADAMTS13), tissue-type plasminogen activator (t-PA), and other coagulation factors are activated and contribute to the dissolution of the fibrin-mesh. This triggers fibrinolysis through the conversion of plasminogen to plasmin [16]. The accumulation of fibrin breakdown products, such as fibrin-derived D-dimers, leads to the ischemic complications found in COVID-19 patients with approximately 76% of hospitalized adults presenting with elevated D-dimer levels [17]. Overall, widespread clotting has proven to have critical consequences as 70% of fatal cases present with disseminated intravascular coagulation (DIC) in comparison to only 1% of the surviving population [18].
Viral infection of type II epithelial cells induces alveolar macrophage activation [2], resulting in the secretion of pro-inflammatory cytokines. These pro-inflammatory cytokines recruit highly toxic neutrophils and activated platelets into the alveolar space, which contribute to ARDS, the cytokine storm, and systemic sepsis. Alveolar macrophages phagocytose alveolar debris and produce cytokines, chemokines, and growth factors involved in the repair of lung tissue and likely play a role in SARS-coronavirus-induced pulmonary fibrosis [5]; however, more work is needed to distinguish the effects of pro-inflammatory insult from that of growth factors alone.
SARS-CoV and SARS-CoV-2 Infection and Lung Fibrosis
Increased TGF-β-activity is suspected to arise, in part, from a positive-feedback loop-induced influx of TGF-β-producing immune cells and the upregulation of highly coagulative and fibrinolytic pathways apt for TGF-β activation [19]. Unfortunately, these conditions are ripe for producing rapid and massive edema and fibrosis that seeks to remodel but ultimately blocks pulmonary airways in COVD-19. TGF-β has been described as the cornerstone of tissue inflammation and increased lung collagen deposition in viral-induced pneumonia of aged individuals and is thought to be responsible for chronic lung pathology and fibrotic outcomes of viral pneumonia [20]. Pulmonary fibrosis is seen with SARS infection as early as 2 to 3 weeks [21] and persists 9 months after hospitalization in 21% of patients [22]. In relation to COVID-19, TGF-β signaling pathways were identified as an integrative pathway in the disease by miRNA analyses [23, 24]. Hence, a comprehensive treatment plan for COVID-19-induced tissue damage necessitates addressing the underlying factors of viral-induced fibrosis and biological mediators, including TGF-β.
The protective role of the ACE2 receptor in lung fibrosis is impaired by SARS-CoV-2 downregulation of ACE2. The protective role of ACE2 is evident in bleomycin models of fibrosis in which recombinant human ACE2 diminishes lung collagen amassment, while ACE2-specific small interfering RNA (siRNA) treatment increases lung collagen accrual in mouse bleomycin models [25–27]. Additionally, lung biopsies of idiopathic pulmonary fibrosis (IPF) display a 92% reduction of ACE2 mRNA and a 74% ACE2 enzymatic activity [28]. The protective role of ACE2 occurs through negative regulation of ANG-II. This is significant, as ANG-II is suspected to engage in an autocrine feedback loop to stimulate TGF-β production [29] while a decrease in ANG-II activity is accompanied by a decrease in TGF-β levels [30, 31]. This bolsters the case for SARS-CoV-2 direct growth factor-induced fibrosis, rather than the simply indirect tissue damage from the hyperinflammatory insult of COVID-19.
Moreover, Pang et al. identified elevated serum levels of TGF-β1 during the early phase of SARS infection [32]. SARS-CoV infection introduces high levels of TGF-β as observed in alveolar epithelial cells, bronchial epithelial cells, monocytes, and macrophages [33, 34] (Fig. 1). The SARS-CoV N protein can associate with the TGF-β-associated SMAD3 transcription factor to interfere with SMAD3/4 complex and can enhance the interaction between SMAD3 with downstream target genes [35]. Additionally, overexpressing the N protein in lung epithelial cells and fibroblasts enhances the TGF-β-induced expression of PAI-1 and collagen I independently of SMAD4 [35]. Interestingly, the SARS-CoV N protein shares little homology with analogous structures of other known coronaviruses [36] but shares 90.52% homology with SARS-CoV-2 [37]. More work is needed to determine if the SARS-CoV-2 N protein similarly enhances the pro-fibrotic activity of lung fibroblasts independently of TGF-β but presents a promising avenue of investigation for direct intervention in SARS-CoV-2-induced fibrosis.
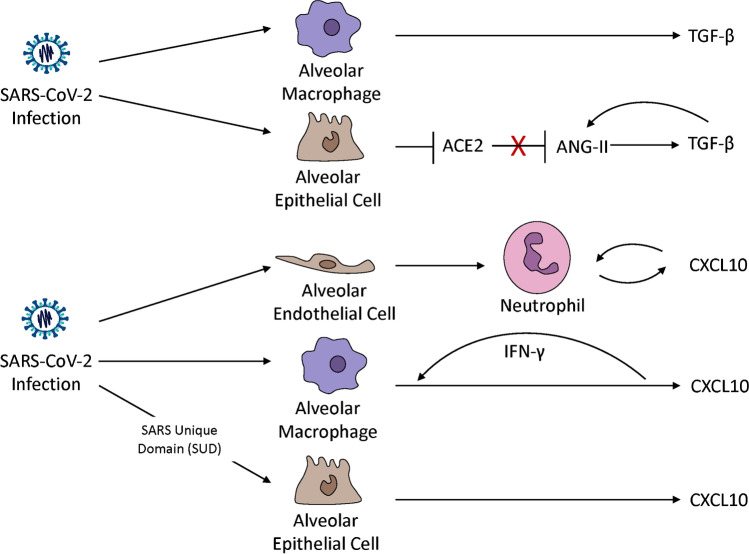
Fig. 1.
CXCL10 and TGF-β are upregulated by SARS-CoV-2 and ARDS in multiple cell types in the lung. SARS-CoV-2 infection upregulates TGF-β in alveolar macrophages and alveolar epithelial cells [33, 34, 121]. SARS-CoV-2 negates the protective role of ACE2 which occurs through negative regulation of ANG-II. ANG-II engages in an autocrine feedback loop to stimulate TGF-β production [30, 31]. Pulmonary endothelial cells recruit neutrophils which increases neutrophil oxidative burst and chemotaxis in a CXCL10-CXCR3 autocrine loop mechanism in viral and non-viral induced lung injury [87•]. SARS-CoV-2-infected macrophages significantly increase the transcription of CXCL10 [121]. CXCL10 increases IFN-γ production [88•] which, in turn, increases CXCL10 in macrophages [90]. The SARS-CoV unique domain (SUD) significantly upregulates the expression of CXCL10 in human lung epithelial cells [86•]
CXC Chemokines in Betacoronavirus Infections
C-X-C chemokines are named for the four highly conserved cysteine residues on the NH2 terminus, with the first two cysteine residues being separated by a variable, “X,” amino acid residue [38, 39]. The function of CXC chemokines in angiogenesis can be largely accredited to the presence or absence of an “ELR” (Glu-Leu-Arg) motif adjacent to the first cysteine residue [40]. ELR+ CXC chemokine ligands, such as CXCL1, CXCL2, CXCL3, CXCL5, CXCL6, CXCL7, and CXCL8, promote angiogenesis. All ELR+ chemokines signal only through CXCR2, except for CXCL6 and CXCL8, which signal through both CXCR1 and CXCR2 [41].
The ELR− chemokine family consists mainly of IFN-γ-inducible chemokines: CXCL9, CXCL10, and CXCL11. ELR− chemokines activate the CXCR3 receptor through autocrine and paracrine signaling [42]. These CXC chemokines share approximately 40% homogeny in sequence but possess different binding affinities with CXCR3 [43] and are expressed in distinct temporal and spatial patterns. Generally, CXCL9 has the lowest affinity for CXCR3, while CXCL11 has the highest [43]. CXCL10 can out-compete CXCL4, but not CCL2, CCL5, CXCL8, or MIP-1α for endothelial cell surface binding [44].
Chemokines and cytokines influence viral disease progression by modulating viral infection, orchestrating immune cell chemoattraction [45], and polarizing the growth and reduction of the microvasculature and the repair of viral-induced tissue injury. Precedence of this has been set by analysis of the previous coronavirus infections. Experts have examined the severe acute respiratory syndrome (SARS) associated coronavirus (SARS-CoV) for insight on SARS-CoV-2. Hunag et al. concluded that IFN-γ, IL-18, TGF-β, IL-6, CXCL19, CCL2, CXCL9, and IL-8, but not TNF-α, IL-2, IL-4, IL-10, IL-13, or TNFRI, are highly elevated in the acute phase of SARS infection [46]. Another group found CXCL9, CXL10, and IL-8 to have been highly correlated with adverse outcomes in SARS patients [47]. CXCL10 and CCL2 are upregulated in both severe and non-severe SARS patients compared to healthy controls. The upregulation of CXCL10 during the late phase of illness can significantly distinguish severe from non-severe SARS patients in late stages [48]. Additionally, markedly high CXCL10 levels are sustained in lung and lymphoid tissue for the duration of SARS illness [49].
Chemokine signaling pathways (hsa04062) were found to be among the most significantly involved pathways in SARS infection by KEGG analysis [50]. Protein-protein interaction (PPI) networks of SARS infected cells revealed CXCL10 to be one of the top eight genes with the most interactions of the 180 genes determined to be differentially expressed in PBMCs between SARS patients and healthy controls by microarray. CXCL10 has been identified as a promising therapeutic target for SARS [51] and was found to be among the top 10 signal transducers most closely linked to heart failure in coronavirus infections through differential gene screening analysis [52].
On the other hand, middle east respiratory syndrome (MERS) infections seem to have very different methods of transcription regulation than that of the related coronavirus, SARS-CoV-2. Microarray analysis and comparison of genes differentially regulated by MERS-CoV and SARS-CoV-2 (127 and 50, respectively), found only eight genes, including CXCL5, CXCL6, and CXCL8, to be modulated by the two coronaviruses in the same manner, either up- or downregulated [53]. Hence, alongside insight from previous coronavirus outbreaks, the roles of chemokines have proven critical in understanding COVID-19 pathogenesis but warrant further investigation.
Chemokines in the Diagnosis and Prognosis of COVID-19
Evidence suggests that chemokines may have diagnostic and prognostic significance in COVID-19. CXCL5, CXCL9, and CXCL10 are highly upregulated in the plasma of COVID-19 patients and have been suggested as potential biomarkers for disease and development of the “cytokine storm” [54]. Investigators have found a significant increase in the expression of genes encoding CXCL1, CXCL3, CXCL6, CXCL18, and CXCL17 in patients with moderate COVID-19 compared to healthy controls. Another group found CXCL8, CXCL10, and CXCL13 to be upregulated in COVID-19 patients and were the only three CXC chemokines differentially regulated against healthy controls [10]. On the other hand, severe COVID-19 patients displayed higher plasma levels of CXLC9, CXCL10, and CXCL11 than moderate COVID-19 patients [55]. Another study found bronchoalveolar cells of severely ill COVID-19 patients to express increased hypoxia-induced factor 1-alpha (HIF1α) and the downstream targets CXCL8, CXCR1, CXCR2, and CXCR4 by scRNASeq [56].
There is also data to suggest that CXC chemokine levels may be variable in the disease progression of young versus old patients. Interestingly, high CXCL9 plasma levels were found in COVID-19 infected adults, but not children [57]. However, another study found that CXCL9, CXCL10, CXCL11, and CXCR3 displayed reduced expression in older adults by shotgun RNA-sequencing profiles of nasopharyngeal swabs [58]. Angioni et al. determined CXCL8, IL-10, IL-15, IL-27, and TNF-α immune profiles to correlate with older age, longer hospitalization, and disease severity [59]. However, another study found that high IL-6, CXCL-8, and TNF-α levels were associated with adverse outcomes of COVID-19 independently of demographics and comorbidities [60] and mirrored those of critically ill ARDS and sepsis patients [61]. It is important to note that disease time course plays a role in the fluctuation of chemokine and cytokine levels along disease progression. For instance, IFN-λ3, IL-6, CXCL10, and CXCL9 levels are observed to rapidly increase before decreasing after the onset of severe pneumonia [62]. Additionally, although VEGF and CXCL10 levels appear to decrease throughout hospitalization gradually, levels remain significantly high in the severe and critical patients for the duration of hospitalization [63]. More work is needed to establish demographic differences in cytokines profiles over the course of disease to harness the full power of chemokines as distinguishable biomarkers for actionable targeting.
Chemokines in COVID-19 Disease Pathogenesis
Aydemir et al. utilized computational modeling to predict COVID-19 miRNA targets and constructed an integrative pathway network analysis of the 40 identified SARS-CoV-2 miRNAs and their genetic targets [24]. Further, KEGG analysis revealed angiogenesis and inflammation mediated by chemokine and cytokine signaling to be among the top eight differentially regulated pathways of SARS-CoV-2 infection. Additionally, investigators found CXCL1, CXCL9, CXCL10, CXCL11, and CXCL16 to be main targets of SARS-CoV-2 miRNA regulation of gene targets associated with cytotoxins and gene regulation [24]. Additionally, RNASeq analysis and KEGG enrichment of differentially expressed genes in the PBMC COVID-19 patients vs. healthy controls found “cytokine-cytokine receptor interaction” and “viral protein interaction with cytokine and cytokine receptor” related pathways to be two of the most differentially regulated [64]. Interestingly, this study found the expression of CXCL10 and CXCL8 to be significantly downregulated and upregulated in COVID-19 PMBCs, respectively.
CXC chemokines play a large role in the dynamic microenvironment and cellular composition of COVID-19 infections. Specifically, SARS-CoV-2-infected lungs overexpress neutrophil chemoattractants CXCL1, CXCL2, CXCL3, CXCL5, CXCL8, and CCL20 [45]. Neutrophils, which are involved in early-stage anti-viral defense, are known to cause cytotoxic lung inflammation after lysis and high neutrophil-to-lymphocyte ratios are associated with poor outcomes of patients with COVID-19 [65, 66]. COVID patients additionally displayed higher activation status of resident and non-resident macrophages. Non-resident macrophages showed an upregulation of CCL2, CCL3, CCL20, CXCL1, CXCL3, IL1β, IL8, IL18, and TNF while resident macrophages were characterized by CCL2, CCL3, and CXCL10 expression [67]. CD69high and CXCR3low mucosa-associated invariant T (MAIT) cell counts are associated with poor outcomes by unsupervised analyses [68]. Better outcomes were identified in patients with significantly higher frequency macrophages of the CD3+ CD4+ CD45RO+ CXCR3+ subsets, higher counts of CD14+ CD11C+ HLA-DR+ subset dendritic cells, and a lower neutrophil count [69]. One study found CXCL10 to be the core gene in a PPI network mapping the 29 immune-related differentially regulated genes in COVID-19 patients and could be correlated with the CD4+, CD8+, monocyte, and DC-related immune signatures [70, 71].
The cytokine storm is the leading cause of death in patients infected with COVID-19. The cytokine storm involves hyper-activated T lymphocytes and the release of pro-inflammatory cytokines that enhance vascular permeability and plasma leakage, resulting in injury to pulmonary tissue, ARDS, and multi-organ failure. These pro-inflammatory mediators include IFN-γ, IL-1RA, IL-6, IL-10, IL-19, CCL2, CCL7, CXCL2, CXCL9, CXCL10, CXCL5, ENRAGE, and poly (ADP-ribose) polymerase 1 [54]. Increased levels of IL-1β, IFN-γ, CXCL10, and CCL2 suggest that T-helper-1 (Th1) cell function is consistent with that of MERS and SARS infections.
CXCL10 in COVID-19
Particular interest has been paid to the role of IFN-γ-inducible cytokines in COVID-19 disease progression. Investigators found a marked upregulation of basal cells in nasopharyngeal and bronchial samples of COVID-19 patients, which contributed in part to immature secretory cell differentiation into ciliated cells by IFN-stimulated genes, particularly ISG15, IFIT1, and CXCL10 [67]. Interestingly, although COVID-19 monocytes displayed reduced oxidative burst, TNF and IFN-γ production remained unchanged in vitro [72]. Zhang et al. found a positive relation between CXCR3+ T follicular helper cells and neutralizing antibody titers in COVID-19-convalescent individuals [73]. CXCL9, CXCL10, CXCL11, and CXCR3 levels are also significantly increased in nasopharyngeal samples of COVID-19 patients [58].
Although IFN-γ-inducible chemokine signaling plays a critical role in pulmonary inflammation at large, CXCL10 is the most studied and has emerged as a possible contributor to the characteristically severe ARDS found with COVID-19. In regard to global transcriptome changes, one study found that several cytokine and immune-related genes such as CXCL10 were upregulated but did not find this with CXCL9 or CXCL11 [74]. On the other hand, another study found that within severe cases of COVID-19, there was strong dysregulation of CXCL9, CXCL10, and CXCL11 but the significant increase in CXCL10 was 10-fold higher than that of CXCL9 or CXCL11 [54]. CXCL10 was identified as one of four primary cytokines responsible for the cytokine storm and disease severity from SARS-CoV-2 and similar coronaviruses by a comprehensive literature review ranging from December 1, 2000, to April 4, 2020 [75], which has been corroborated by Zhang et al [70].
CXCL10 is secreted by several cell types, including endothelial cells, fibroblasts, monocytes/macrophages, and T lymphocytes, often upon INF-γ stimulation, and promotes chemoattraction for activated T lymphocytes, natural killer cells, and monocytes through CXCR3. CXCL10 has proven to be exceptionally reliable for the diagnosis and prognosis of COVID-19. Increased levels of CXCL10 are associated with a greater SARS-CoV-2 viral load in plasma, saliva, and nasopharyngeal samples [63, 76–78]. Nasopharyngeal CXCL10 levels are a sensitive predictor of COVID-19, able to identify undiagnosed cases of COVID-19, and can be used reliably as a rule-out test of the disease [79]. Single-cell analyses of nasopharyngeal and bronchial samples have revealed a major role for CXCL10 in disease severity and outcome [64, 67, 80]. Blot et al. determined CXCL10 to be associated with the number of days COVID-19 patients required mechanical ventilation [81]. CXCL10 has also been found to be highly upregulated in the plasma of ICU versus non-ICU COVID-19 patients [82] and correlated with mortality by day 28 in ICU COVID-19 patients [78]. Plasma levels of CXCL10 reliably distinguished disease severity and progression in a study of 50 COVID-19 patients, and these results were enhanced when combined with CCL7 [83].
Recent studies suggest that CXCL10 may be a distinguishing factor between COVID-19-associated pulmonary pathologies and others similar. Bronchoalveolar lavage fluid and plasma from one control, seven non-COVID-19 ARDS, and fourteen COVID-19 ARDS patients found CXCL10 highly upregulated in both plasma and epithelial lining fluid in the COVID-19 ARDS group compared with the non-COVID-19 ARDS group [81]. Similarly, CXCL10 levels were distinguishable between COVID-19-related lower respiratory tract infections (LRTIs) and non-COVID-19 LRTIs and correlated with COVID-19 disease severity [84]. ACE2 and CXCL10 are also suspected to be biomarkers of fatality in COVID-19 patients with lung cancer [85].
CXCL10 is implicated in multiple major processes associated with pulmonary inflammation (Fig. 2). The SARS-CoV unique domain (SUD) significantly upregulates the expression of CXCL10 in human lung epithelial cells [86•], modulates CXCL10-mediated pulmonary inflammation in a NLRP3 inflammasome-dependent manner [86•], and attracts and activates neutrophils in LPS, H1N1, and SARS-induced ARDS [87•, 88•, 89]. Evidence suggests that IFN-γ stimulation of CXCL10+ CCL2+ macrophages may drive severe COVID-19 [90]. CXCL10 and CCL2 are also higher in patients with the highest D-dimer levels [91], a pathology highly associated with mortality in COVID-19. Aside from the direct insult of early hyperinflammatory insults on lung tissue, this poses an equally threatening likelihood of long-term pulmonary fibrosis, as lung inflammation accumulation will increase a patient’s need for mechanical ventilation. Mechanical ventilation, although often lifesaving, greatly increases the potential for ventilator-induced lung injury (VILI). Desai et al. found a significant correlation in the duration of pressure-controlled mechanical ventilation and pulmonary fibrosis in ARDS patients at 110–267 days after extubation, with 85% displaying pulmonary fibrosis [92]. Hence, early intervention in this hyperinflammatory state is even more critical for mitigating the long-term fibrotic complications of COVID-19.
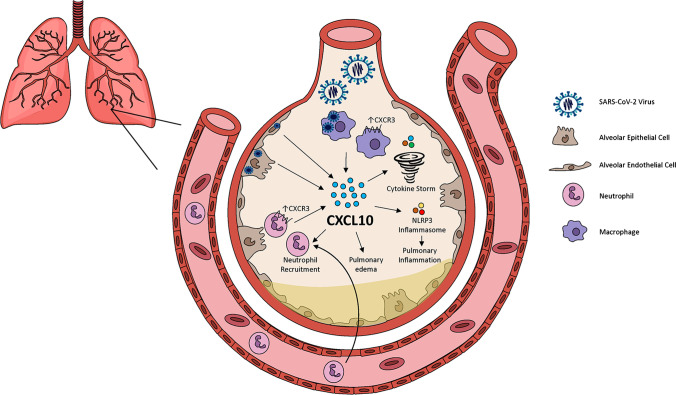
Fig. 2.
CXCL10 enhances inflammatory infiltrate, pro-inflammatory cytokine production, and pulmonary edema in SARS-CoV-2 lung infection. SARS-CoV-2 infection triggers pulmonary endothelial cell recruitment of neutrophils which produce reactive oxygen species (ROS) [14]. Viral infection of type II epithelial cells induces alveolar macrophage activation results in the secretion of proinflammatory cytokines which induce the recruitment of highly toxic neutrophils and activated platelets into the alveolar space [2]. The SARS-CoV unique domain (SUD) modulates CXCL10-mediated pulmonary inflammation in a NLRP3 inflammasome-dependent manner [86•] and attracts and activates neutrophils 87•, 88•, 89. CXCL10-CXCR3 is critical for the influx of CXCR3+ pulmonary neutrophils and increase neutrophil oxidative burst in autocrine loop mechanism [87•]. CXCL10 enhances pulmonary edema [88•], the cytokine storm [54, 70], the release of inflammatory mediators, such as IFN-γ, and the infiltration of inflammatory cells, such as neutrophils and macrophages [88•]. CXCL10 also increases neutrophil and macrophage CXCR3 expression [88•].
CXCL10 has emerged as a critical contributor to COVID-19 disease pathologies across organs [81]. CXCL10-CXCR3 is hypothesized to play a major role in the neurological symptoms of COVID-19, including the loss of taste and smell [93]. CXCL10 activity is also likely to play a role in other COVID-19-induced tissue injury and fibrosis in other organs, including the liver and heart. COVID-19 autopsies have revealed endotheliitis, vasculitis, dilated cardiomyopathy, hypertrophic cardiomyopathy, multifocal fibrosis, and myocardial interstitial fibrosis [94–96]. CXCL10 has been identified as a promising therapeutic target for SARS and was found to be among the top 10 signal transducers most closely linked to heart failure in coronavirus infections through differential gene screening analysis [97]. CXCL10 is upregulated in the infarcted myocardium [98] and inhibits basic fibroblast growth factor (bFGF)-induced fibroblast migration in the infarcted heart [99]. Although more work is needed to understand the role of CXCL10 in SARS-CoV-2-induced myocardial injury, fibroblast-CXCL0 targeting presents a promising line of investigation.
Conversely, CXCL10 exasperates fibrosis in the liver. Therefore, the observed SARS-CoV-2 downregulation of CXCL10 in the liver may contribute to the portal fibrosis seen in 60% of autopsies from severe COVID-19 patients [100]. McCord et al. found that Nrf2 activation downregulates ACE2 and TMPRSS2, the receptor and activator, of the SARS-CoV-2 spike protein while also downregulating IFN-γ-induced genes, including CXCL9, CXCL10, and CXCL11 in human liver-derived HepG2 cells [101]. Taken together, both agonism and antagonism of the CXCL10 signaling system should be taken into account to properly address the multi-faceted role of this chemokine in COVID-19-induced organ fibrosis.
CXCL10-CXCR3-Targeted Therapeutics in COVID-19
Current therapeutic targets for COVID-19 can be separated into several large categories, including RNA synthesis and replication interference proteins, SARS-CoV-2-specific structural proteins, virulence factors [102], and SARS-CoV2-human interacting proteins identified by PPI [103]. Other avenues include ACE2 receptor inhibitors [104], or modulation of immune mediators, such as TLR5 [105] or high mobility group box 1 (HMGB1) [106]. Rigorous research and human clinical trials are needed to adequately test both the efficacy and safety of new and repurposed therapeutics. These are needed for patients to overcome the infection with minimal organ damage and to mitigate long-term consequences. Data shows that 52% of those recovered from SARS had long-term lung abnormalities 2 years post-infection and 4% displayed persistent diffusing capacity impairment and reduced exercise capacity at 15 years post-infection [107]. Unfortunately, those with long-term complications from COVID-19 may be even greater and are expected to have broader effects on function in the lungs, heart, liver, GI tract, and brain [108]. The CXCL10-CXCR3 signaling axis presents as a promising therapeutic target. It is an underlying cause of abnormalities in the lungs, heart, liver, and brain and contributes to the instigation and ravaging aftermath of the cytokine storm.
Current strategies for targeting the CXCL10-CXCR3 signaling system include both transcriptional regulation and direct inhibition. PPAR agonists may be suitable for transcriptional regulation of these inflammatory cytokines. The PPARγ agonist, Rosiglitazone, lowered CXCL9, CXCL10, and CXCL11 released by orbital fibroblasts, preadipocytes in a dose-dependent manner [109], and has been suggested as a possible therapeutic for COVID-19 [110]. Additionally, the PPARα agonist, fenofibrate, decreased the expression of IL-17 and IFN-γ and reduced CXCL10 promoter activity in TNF-α-stimulated HT-29 colon epithelial cells and, hence, it has been postulated to repurpose Fenofibrate as a treatment for COVID-19 [111, 112]. Sitagliptin, a dipeptidyl peptidase-4 (DDP4, human CD26) inhibitor, is currently under investigation for the treatment of COVID-19. The S1 domain of the SARS-CoV-2 spike protein targets DDP4 [113] and is an inhibitor of CXCL10 secretion [114]. Sitagliptin was shown to improve clinical outcomes and reduce mortality in COVID-19 patients with type 2 diabetes [115, 116], although the legitimacy of these findings has been challenged by others [117]. Moreover, others have postulated that the benefits of COVID-19 patients treated with corticosteroids, which inhibit the Th1 pathway and reduce CXCL10 secretion [118], may be attributed to the accompanying decrease of CXCL10 levels, as found with SARS patients [119]. More work is needed to prove the therapeutic benefit of these repurposed drugs and evidence of a direct effect on SARS-CoV-2-induced CXCL10 upregulation would bolster enthusiasm.
Ichikawa et al. has shown that eliminating CXCL10 and/or CXCR3 activity in mice diminishes lung injury and improves survival rates in both viral and non-viral lung injury [87•]. Researchers found CXCR3+ neutrophil accumulation to be upregulated by viral lung injury-induced Toll-like receptor 4 (TLR4)-TRIF pathway stimulation [87•] and propose CXCL10-CXCR3 autocrine signaling to be responsible for the increased oxidative burst and chemotaxis of inflamed neutrophils in the lung by a feedforward autocrine loop mechanism [87•]. To complement these findings, others have shown that CXCL10 neutralization significantly reduces pulmonary edema, the release of inflammatory mediators (IFN-γ, IL-6, and ICAM-1), and inflammatory cells (neutrophils, macrophages, CD8+ T cells) infiltration into the lung in an LPS-induced ARDS rat model [88•]. CXCL10 neutralization also accompanied reduced neutrophil and macrophage CXCR3 expression [88•] and CXCR3 antagonism significantly reduced CXCL10-CXCR3-mediated T lymphocyte migration in vitro [120]. These promising findings increase support for direct targeting of the CXCL10-CXCR3 signaling axis to treat COVID-19.
Pro-Fibrotic and Pro-Inflammatory Pathway Convergence in COVID-19
CXCL10-CXCR3-specific targeting may lead to a more direct impact on the multi-organ inflammatory and fibrotic manifestations in COVID-19, due to the opportune dual-mechanistic anti-inflammatory and anti-fibrotic arms of this pathway. As previously discussed, CXCL10 has been identified as a critical contributor to the presence and severity of the SARS-CoV-2-induced cytokine storm, which itself is a contributor to the multi-organ fibrosis found in COVID-19 “long-haulers.” Interestingly, SARS-CoV-2-infected macrophages significantly increase the transcription of both CXCL10 and TGFB1 genes [121], and CXCL10 secretion by activated macrophages has been associated with increased disease severity [67]. Moreover, CXCL10-CXCR3 signaling has been found to interface with pro-fibrotic TGF-β pathways to modulate both fibrosis and inflammation [122, 123]. From these and similar findings of CXCL10/TGF-β signaling pathway convergence [124–126], CXCL10-targeted therapeutics present a potential avenue for tempering the cytokine storm and activity of pro-fibrotic mediators, such as TGF-β.
CXCL10-derived agonistic and antagonistic biomimetic peptides are currently being tested to dually target multiple mechanisms of CXCL10 in hyperinflammatory and fibrotic disease states across organ systems. Among the broad spectrum of cellular effects, our data demonstrate that these CXCL10-derived peptides can reduce α-smooth muscle actin (α-SMA) expression and mRNA and protein secretion levels of major matrix molecules including collagen I, laminin, fibronectin, and tenascin-C in dermal and cardiac fibroblasts after induction by pro-fibrotic TGF-β [127]. Furthermore, these CXCL10-derived peptides inhibit TGF-β1-mediated epithelial–mesenchymal transition (EMT) in keratinocytes and dermal fibroblast co-cultures, a known feature of fibrosis. One CXCL10-derived antagonistic peptide, named IP-10p, has revealed powerful angiostatic biomimetic capabilities by preventing VEGF-initiated endothelial cell motility on one front, and on the other, inhibiting vessel formation and promoting the regression of naïve vessels [128]. IP-10p reduced endothelial cell migration, dissociation, and tube formation to the same degree, or to greater degree, than that of the mature chemokine in vitro and in vivo [128]. These CXCL10-derived peptides present an opportunistic avenue for early intervention in SARS-CoV-2 tissue fibrosis by mitigating the hyperinflammatory state and addressing fibrotic complications of COVID-19 “long-haulers.”
In conclusion, chronic inflammation is often regarded as a primary contributor in lung fibrosis, and evidence from emerging SARS-CoV and SARS-CoV-2 studies suggests the direct upregulation of the TGF-β and CXCL10 signaling systems exasperate fibrotic pathologies in COVID-19, which warrant direct intervention [129] (Fig. 3). Further investigation is needed to determine the efficacy of CXCL10 as a primary therapy, or as adjunct treatment (antagonists) to antibody therapy in high-risk patients and patient survivors to inhibit coronavirus infection, disease progression, and/or the life-threatening fibrotic sequelae.
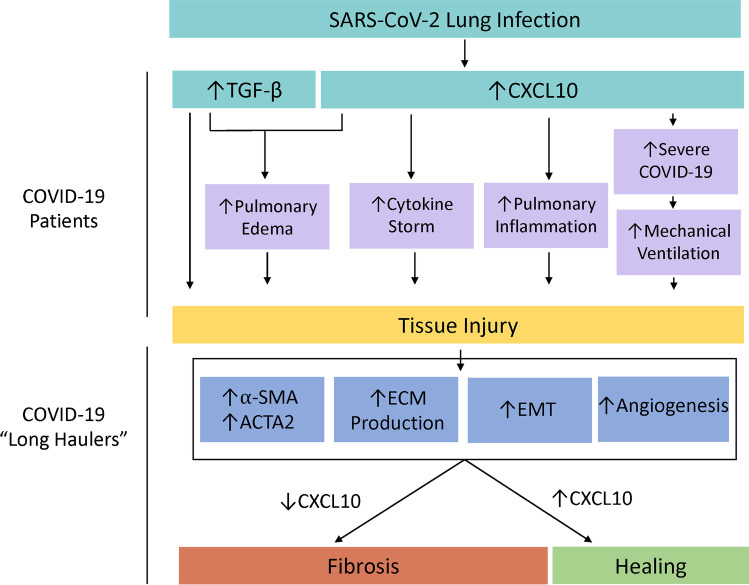
Fig. 3.
A diagram displaying the adherent pathologies and outcomes associated with SARS-CoV-2-induced TGF-β and CXCL10 upregulation in the lung which culminate in tissue injury in COVID-19 patients and subsequently manifests fibrosis in COVID-19 long-haulers which can uniquely be targeted by CXCL10-centric therapeutics. COVID-19-related tissue injury is thought to be twofold and results from both direct upregulation of the pro-fibrotic growth factor, TGF-β, but also from tissue damage initiated by a highly inflammatory state. CXCL10 and TGF-β both enhance pulmonary edema [19, 88•]. CXCL10 plays a critical role by upregulation of the cytokine storm [54, 70] and increasing pulmonary inflammation [86•]. Furthermore, CXCL10 is associated with COVID-19 disease severity which increases the likely need for mechanical ventilation, which leads to pulmonary fibrosis [92]. However, in COVID-19 long-haulers, amplifying the CXCL10-CXCR3 signaling system may in fact benefit patients by inhibiting many pro-fibrotic pathologies expected clinically [130, 131], such as through the upregulation of α-SMA, ACTA2, ECM protein, EMT, and angiogenesis [127, 128]
Acknowledgements
We kindly thank David Gansberger and Rachel Jackson (Yates Laboratory) for editorial assistance.
Author Contribution
D.R.J. contributed to this work by way of conceptualization, manuscript preparation, writing, and editing. M.A.K. assisted in manuscript conceptualization, writing, and editing. A.P. contributed to editing of this manuscript, the intellectual content described herein, and to the conceptualization of this work. J.J. and M.W. revised it critically for important intellectual content and contributed to the conceptualization. C.C.Y. contributed to conceptualization, manuscript preparation, writing, and editing, and approved the version to be published and agreed to be accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Funding
Research reported in this publication was supported by funding from the National Institute of Arthritis and Musculoskeletal and Skin Diseases of the National Institutes of Health under Award Number 5R01AR068317 (CCY) and from the University of Pittsburgh School of Nursing (CCY).
Availability of Data and Material
Not applicable
Code Availability
Not applicable
Declarations
Conflict of Interest
CCY is the Corporate Founder and CSO of FibroKine. TM, DRJ, MAK, AP, JJ, and MSW declare that they don’t have conflict of interest.
Human and Animal Rights and Informed Consent
This article does not contain any studies with human or animal subjects performed by any of the authors.
Footnotes
This article is part of the Topical Collection on Wound Healing and Tissue Repair
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
Papers of particular interest, published recently, have been highlighted as: • Of importance
- 1.Xu Z, et al. Pathological findings of COVID-19 associated with acute respiratory distress syndrome. Lancet Respir Med. 2020;8(4):420–422. doi: 10.1016/S2213-2600(20)30076-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Morris G, et al. The pathophysiology of SARS-CoV-2: a suggested model and therapeutic approach. Life Sci. 2020;258:118166. doi: 10.1016/j.lfs.2020.118166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shaw B, Daskareh M, Gholamrezanezhad A. The lingering manifestations of COVID-19 during and after convalescence: update on long-term pulmonary consequences of coronavirus disease 2019 (COVID-19) Radiol Med. 2021;126(1):40–46. doi: 10.1007/s11547-020-01295-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ding X, Xu J, Zhou J, Long Q. Chest CT findings of COVID-19 pneumonia by duration of symptoms. Eur J Radiol. 2020;127:109009. doi: 10.1016/j.ejrad.2020.109009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Venkataraman T, Frieman MB. The role of epidermal growth factor receptor (EGFR) signaling in SARS coronavirus-induced pulmonary fibrosis. Antiviral Res. 2017;143:142–150. doi: 10.1016/j.antiviral.2017.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wong KT, et al. Severe acute respiratory syndrome: thin-section computed tomography features, temporal changes, and clinicoradiologic correlation during the convalescent period. J Comput Assist Tomogr. 2004;28(6):790–5. doi: 10.1097/00004728-200411000-00010. [DOI] [PubMed] [Google Scholar]
- 7.Gu J, Korteweg C. Pathology and pathogenesis of severe acute respiratory syndrome. Am J Pathol. 2007;170(4):1136–47. doi: 10.2353/ajpath.2007.061088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tse GM, et al. Pulmonary pathological features in coronavirus associated severe acute respiratory syndrome (SARS) J Clin Pathol. 2004;57(3):260–5. doi: 10.1136/jcp.2003.013276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang P, et al. Long-term bone and lung consequences associated with hospital-acquired severe acute respiratory syndrome: a 15-year follow-up from a prospective cohort study. Bone Res. 2020;8:8. doi: 10.1038/s41413-020-0084-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ackermann M, et al. Pulmonary vascular endothelialitis, thrombosis, and angiogenesis in Covid-19. N Engl J Med. 2020;383(2):120–128. doi: 10.1056/NEJMoa2015432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hariri L, Hardin CC. Covid-19, Angiogenesis, and ARDS endotypes. N Engl J Med. 2020;383(2):182–183. doi: 10.1056/NEJMe2018629. [DOI] [PubMed] [Google Scholar]
- 12.Varga Z, et al. Endothelial cell infection and endotheliitis in COVID-19. Lancet. 2020;395(10234):1417–1418. doi: 10.1016/S0140-6736(20)30937-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hoffmann M, et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181(2):271–280. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Janardhan V, Janardhan V, Kalousek V. COVID-19 as a blood clotting disorder masquerading as a respiratory illness: a cerebrovascular perspective and therapeutic implications for stroke thrombectomy. J Neuroimaging. 2020;30(5):555–561. doi: 10.1111/jon.12770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grobler C, et al. Covid-19: The rollercoaster of fibrin(ogen), D-dimer, Von Willebrand factor, P-selectin and their interactions with endothelial cells, platelets and erythrocytes. Int J Mol Sci. 2020; 21(14). 10.3390/ijms21145168. [DOI] [PMC free article] [PubMed]
- 16.Yau JW, Teoh H, Verma S. Endothelial cell control of thrombosis. BMC Cardiovasc Disord. 2015;15:130. doi: 10.1186/s12872-015-0124-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Berger JS, et al. Prevalence and outcomes of D-dimer elevation in hospitalized patients with COVID-19. Arterioscler Thromb Vasc Biol. 2020;40(10):2539–2547. doi: 10.1161/ATVBAHA.120.314872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang F, et al. The laboratory tests and host immunity of COVID-19 patients with different severity of illness. JCI Insight. 2020;5(10). 10.1172/jci.insight.137799. [DOI] [PMC free article] [PubMed]
- 19.Chen W. A potential treatment of COVID-19 with TGF-beta blockade. Int J Biol Sci. 2020;16(11):1954–1955. doi: 10.7150/ijbs.46891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goplen NP, et al. Tissue-resident CD8(+) T cells drive age-associated chronic lung sequelae after viral pneumonia. Sci Immunol. 2020;5(53). 10.1126/sciimmunol.abc4557. [DOI] [PMC free article] [PubMed]
- 21.Hwang DM, et al. Pulmonary pathology of severe acute respiratory syndrome in Toronto. Mod Pathol. 2005;18(1):1–10. doi: 10.1038/modpathol.3800247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xie L, et al. Dynamic changes of serum SARS-coronavirus IgG, pulmonary function and radiography in patients recovering from SARS after hospital discharge. Respir Res. 2005;6:5. doi: 10.1186/1465-9921-6-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yousefi H, et al. SARS-CoV infection crosstalk with human host cell noncoding-RNA machinery: an in-silico approach. Biomed Pharmacother. 2020;130:110548. doi: 10.1016/j.biopha.2020.110548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Aydemir MN, et al. Computationally predicted SARS-COV-2 encoded microRNAs target NFKB, JAK/STAT and TGFB signaling pathways. Gene Rep. 2021;22:101012. doi: 10.1016/j.genrep.2020.101012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Imai Y, Kuba K, Penninger JM. The discovery of angiotensin-converting enzyme 2 and its role in acute lung injury in mice. Exp Physiol. 2008;93(5):543–8. doi: 10.1113/expphysiol.2007.040048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kuba K, Imai Y, Penninger JM. Angiotensin-converting enzyme 2 in lung diseases. Curr Opin Pharmacol. 2006;6(3):271–6. doi: 10.1016/j.coph.2006.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kuba K, Imai Y, Rao S, Jiang C, Penninger JM. Lessons from SARS: control of acute lung failure by the SARS receptor ACE2. J Mol Med (Berl) 2006;84(10):814–20. doi: 10.1007/s00109-006-0094-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li X, et al. Angiotensin converting enzyme-2 is protective but downregulated in human and experimental lung fibrosis. Am J Physiol Lung Cell Mol Physiol. 2008;295(1):L178–85. doi: 10.1152/ajplung.00009.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Molteni A, et al. Effect of an angiotensin II receptor blocker and two angiotensin converting enzyme inhibitors on transforming growth factor-beta (TGF-beta) and alpha-actomyosin (alpha SMA), important mediators of radiation-induced pneumopathy and lung fibrosis. Curr Pharm Des. 2007;13(13):1307–16. doi: 10.2174/138161207780618777. [DOI] [PubMed] [Google Scholar]
- 30.Otsuka M, Takahashi H, Shiratori M, Chiba H, Abe S. Reduction of bleomycin induced lung fibrosis by candesartan cilexetil, an angiotensin II type 1 receptor antagonist. Thorax. 2004;59(1):31–8. doi: 10.1136/thx.2003.000893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Waseda Y, et al. Angiotensin II type 2 receptor antagonist reduces bleomycin-induced pulmonary fibrosis in mice. Respir Res. 2008;9:43. doi: 10.1186/1465-9921-9-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Beijing Group of National Research Project for S. Dynamic changes in blood cytokine levels as clinical indicators in severe acute respiratory syndrome. Chin Med J (Engl). 2003;116(9):1283-7. [PubMed]
- 33.Baas T, Taubenberger JK, Chong PY, Chui P, Katze MG. SARS-CoV virus-host interactions and comparative etiologies of acute respiratory distress syndrome as determined by transcriptional and cytokine profiling of formalin-fixed paraffin-embedded tissues. J Interferon Cytokine Res. 2006;26(5):309–17. doi: 10.1089/jir.2006.26.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.He L, et al. Expression of elevated levels of pro-inflammatory cytokines in SARS-CoV-infected ACE2+ cells in SARS patients: relation to the acute lung injury and pathogenesis of SARS. J Pathol. 2006;210(3):288–97. doi: 10.1002/path.2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhao X, Nicholls JM, Chen YG. Severe acute respiratory syndrome-associated coronavirus nucleocapsid protein interacts with Smad3 and modulates transforming growth factor-beta signaling. J Biol Chem. 2008;283(6):3272–3280. doi: 10.1074/jbc.M708033200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.He R, et al. Analysis of multimerization of the SARS coronavirus nucleocapsid protein. Biochem Biophys Res Commun. 2004;316(2):476–83. doi: 10.1016/j.bbrc.2004.02.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zeng W, et al. Biochemical characterization of SARS-CoV-2 nucleocapsid protein. Biochem Biophys Res Commun. 2020;527(3):618–623. doi: 10.1016/j.bbrc.2020.04.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Clark-Lewis I, Dewald B, Loetscher M, Moser B, Baggiolini M. Structural requirements for interleukin-8 function identified by design of analogs and CXC chemokine hybrids. J Biol Chem. 1994;269(23):16075–81. doi: 10.1016/S0021-9258(17)33975-3. [DOI] [PubMed] [Google Scholar]
- 39.Covell DG, Smythers GW, Gronenborn AM, Clore GM. Analysis of hydrophobicity in the alpha and beta chemokine families and its relevance to dimerization. Protein Sci. 1994;3(11):2064–72. doi: 10.1002/pro.5560031119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Strieter RM, et al. The functional role of the ELR motif in CXC chemokine-mediated angiogenesis. J Biol Chem. 1995;270(45):27348–57. doi: 10.1074/jbc.270.45.27348. [DOI] [PubMed] [Google Scholar]
- 41.Charo IF, Ransohoff RM. The many roles of chemokines and chemokine receptors in inflammation. N Engl J Med. 2006;354(6):610–21. doi: 10.1056/NEJMra052723. [DOI] [PubMed] [Google Scholar]
- 42.Nawaz MI, et al. Autocrine CCL2, CXCL4, CXCL9 and CXCL10 signal in retinal endothelial cells and are enhanced in diabetic retinopathy. Exp Eye Res. 2013;109:67–76. doi: 10.1016/j.exer.2013.01.008. [DOI] [PubMed] [Google Scholar]
- 43.Lasagni L, et al. An alternatively spliced variant of CXCR3 mediates the inhibition of endothelial cell growth induced by IP-10, Mig, and I-TAC, and acts as functional receptor for platelet factor 4. J Exp Med. 2003;197(11):1537–49. doi: 10.1084/jem.20021897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Luster AD, Greenberg SM, Leder P. The IP-10 chemokine binds to a specific cell surface heparan sulfate site shared with platelet factor 4 and inhibits endothelial cell proliferation. J Exp Med. 1995;182(1):219–31. doi: 10.1084/jem.182.1.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Didangelos A. COVID-19 hyperinflammation: what about neutrophils? mSphere. 2020;5(3). 10.1128/mSphere.00367-20. [DOI] [PMC free article] [PubMed]
- 46.Huang KJ, et al. An interferon-gamma-related cytokine storm in SARS patients. J Med Virol. 2005;75(2):185–94. doi: 10.1002/jmv.20255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tang NL, et al. Early enhanced expression of interferon-inducible protein-10 (CXCL-10) and other chemokines predicts adverse outcome in severe acute respiratory syndrome. Clin Chem. 2005;51(12):2333–40. doi: 10.1373/clinchem.2005.054460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cameron MJ, Bermejo-Martin JF, Danesh A, Muller MP, Kelvin DJ. Human immunopathogenesis of severe acute respiratory syndrome (SARS) Virus Res. 2008;133(1):13–9. doi: 10.1016/j.virusres.2007.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jiang Y, et al. Characterization of cytokine/chemokine profiles of severe acute respiratory syndrome. Am J Respir Crit Care Med. 2005;171(8):850–7. doi: 10.1164/rccm.200407-857OC. [DOI] [PubMed] [Google Scholar]
- 50.Hemmat N, et al. Neutrophils, crucial, or harmful immune cells involved in coronavirus infection: a bioinformatics study. Front Genet. 2020;11:641. doi: 10.3389/fgene.2020.00641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.To KF, Chan PK. Identification of human cell line model of persistent SARS coronavirus infection and studies of the response to cytokines and chemokines. Hong Kong Med J. 2009;15(Suppl 6):39–43. [PubMed] [Google Scholar]
- 52.Chen XM, et al. Exploration of omics mechanism and drug prediction of coronavirus-induced heart failure based on clinical bioinformatics. Zhonghua Xin Xue Guan Bing Za Zhi. 2020;48:E013. doi: 10.3760/cma.j.cn112148-20200308-00172. [DOI] [PubMed] [Google Scholar]
- 53.Jang Y, Seo SH. Gene expression pattern differences in primary human pulmonary epithelial cells infected with MERS-CoV or SARS-CoV-2. Arch Virol. 2020;165(10):2205–2211. doi: 10.1007/s00705-020-04730-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sims JT, et al. Characterization of the cytokine storm reflects hyperinflammatory endothelial dysfunction in COVID-19. J Allergy Clin Immunol. 2021;147(1):107–111. doi: 10.1016/j.jaci.2020.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tincati C, et al. Heightened circulating interferon-inducible chemokines, and activated pro-cytolytic Th1-cell phenotype features Covid-19 aggravation in the second week of illness. Front Immunol. 2020;11:580987. doi: 10.3389/fimmu.2020.580987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Taniguchi-Ponciano K, et al. Increased expression of hypoxia-induced factor 1alpha mRNA and its related genes in myeloid blood cells from critically ill COVID-19 patients. Ann Med. 2021;53(1):197–207. doi: 10.1080/07853890.2020.1858234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Moratto D, et al. Immune response in children with COVID-19 is characterized by lower levels of T-cell activation than infected adults. Eur J Immunol. 2020;50(9):1412–1414. doi: 10.1002/eji.202048724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lieberman NAP, et al. In vivo antiviral host transcriptional response to SARS-CoV-2 by viral load, sex, and age. PLoS Biol. 2020;18(9):e3000849. doi: 10.1371/journal.pbio.3000849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Angioni R, et al. Age-severity matched cytokine profiling reveals specific signatures in Covid-19 patients. Cell Death Dis. 2020;11(11):957. doi: 10.1038/s41419-020-03151-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Del Valle DM, et al. An inflammatory cytokine signature predicts COVID-19 severity and survival. Nat Med. 2020;26(10):1636–1643. doi: 10.1038/s41591-020-1051-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wilson JG, et al. Cytokine profile in plasma of severe COVID-19 does not differ from ARDS and sepsis. JCI Insight. 2020;5(17). 10.1172/jci.insight.140289. [DOI] [PMC free article] [PubMed]
- 62.Sugiyama M, et al. Serum CCL17 level becomes a predictive marker to distinguish between mild/moderate and severe/critical disease in patients with COVID-19. Gene. 2021;766:145145. doi: 10.1016/j.gene.2020.145145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bermejo-Martin JF, et al. Viral RNA load in plasma is associated with critical illness and a dysregulated host response in COVID-19. Crit Care. 2020;24(1):691. doi: 10.1186/s13054-020-03398-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Xiong Y, et al. Transcriptomic characteristics of bronchoalveolar lavage fluid and peripheral blood mononuclear cells in COVID-19 patients. Emerg Microbes Infect. 2020;9(1):761–770. doi: 10.1080/22221751.2020.1747363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Peng Y, et al. Clinical characteristics and prognosis of 244 cardiovascular patients suffering from coronavirus disease in Wuhan, China. J Am Heart Assoc. 2020;9(19):e016796. doi: 10.1161/JAHA.120.016796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Jimeno S, et al. Prognostic implications of neutrophil-lymphocyte ratio in COVID-19. Eur J Clin Invest. 2021;51(1):e13404. doi: 10.1111/eci.13404. [DOI] [PubMed] [Google Scholar]
- 67.Chua RL, et al. COVID-19 severity correlates with airway epithelium-immune cell interactions identified by single-cell analysis. Nat Biotechnol. 2020;38(8):970–979. doi: 10.1038/s41587-020-0602-4. [DOI] [PubMed] [Google Scholar]
- 68.Parrot T, et al. MAIT cell activation and dynamics associated with COVID-19 disease severity. Sci Immunol. 2020;5(51). 10.1126/sciimmunol.abe1670. [DOI] [PMC free article] [PubMed]
- 69.Wei LL, Wang WJ, Chen DX, Xu B. Dysregulation of the immune response affects the outcome of critical COVID-19 patients. J Med Virol. 2020;92(11):2768–2776. doi: 10.1002/jmv.26181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhang N, Zhao YD, Wang XM. CXCL10 an important chemokine associated with cytokine storm in COVID-19 infected patients. Eur Rev Med Pharmacol Sci. 2020;24(13):7497–7505. doi: 10.26355/eurrev_202007_21922. [DOI] [PubMed] [Google Scholar]
- 71.Rydyznski Moderbacher C, et al. Antigen-specific adaptive immunity to SARS-CoV-2 in acute COVID-19 and associations with age and disease severity. Cell. 2020;183(4):996–1012. doi: 10.1016/j.cell.2020.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gibellini L, et al. Altered bioenergetics and mitochondrial dysfunction of monocytes in patients with COVID-19 pneumonia. EMBO Mol Med. 2020;12(12):e13001. doi: 10.15252/emmm.202013001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhang J, et al. Spike-specific circulating T follicular helper cell and cross-neutralizing antibody responses in COVID-19-convalescent individuals. Nat Microbiol. 2021;6(1):51–58. doi: 10.1038/s41564-020-00824-5. [DOI] [PubMed] [Google Scholar]
- 74.Jain R, et al. Host transcriptomic profiling of COVID-19 patients with mild, moderate, and severe clinical outcomes. Comput Struct Biotechnol J. 2021;19:153–160. doi: 10.1016/j.csbj.2020.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Quirch M, Lee J, Rehman S. Hazards of the cytokine storm and cytokine-targeted therapy in patients with COVID-19: review. J Med Internet Res. 2020;22(8):e20193. doi: 10.2196/20193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Silva J, et al. Saliva viral load is a dynamic unifying correlate of COVID-19 severity and mortality. medRxiv. 2021. 10.1101/2021.01.04.21249236.
- 77.Kwon JS, et al. Factors of severity in patients with COVID-19: cytokine/chemokine concentrations, viral load, and antibody responses. Am J Trop Med Hyg. 2020;103(6):2412–2418. doi: 10.4269/ajtmh.20-1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hue S, et al. Uncontrolled innate and impaired adaptive immune responses in patients with COVID-19 acute respiratory distress syndrome. Am J Respir Crit Care Med. 2020;202(11):1509–1519. doi: 10.1164/rccm.202005-1885OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Cheemarla NR, et al. Host response-based screening to identify undiagnosed cases of COVID-19 and expand testing capacity. medRxiv. 2020. 10.1101/2020.06.04.20109306.
- 80.Haroun RA, Osman WH, Eessa AM. Interferon-gamma-induced protein 10 (IP-10) and serum amyloid A (SAA) are excellent biomarkers for the prediction of COVID-19 progression and severity. Life Sci. 2021;269:119019. doi: 10.1016/j.lfs.2021.119019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Blot M, et al. CXCL10 could drive longer duration of mechanical ventilation during COVID-19 ARDS. Crit Care. 2020;24(1):632. doi: 10.1186/s13054-020-03328-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Huang C, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395(10223):497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Yang Y, et al. Plasma IP-10 and MCP-3 levels are highly associated with disease severity and predict the progression of COVID-19. J Allergy Clin Immunol. 2020;146(1):119–127. doi: 10.1016/j.jaci.2020.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Laing AG, et al. A dynamic COVID-19 immune signature includes associations with poor prognosis. Nat Med. 2020;26(10):1623–1635. doi: 10.1038/s41591-020-1038-6. [DOI] [PubMed] [Google Scholar]
- 85.Mahmood TB, et al. Evaluation of the susceptibility and fatality of lung cancer patients towards the COVID-19 infection: a systemic approach through analyzing the ACE2, CXCL10 and their co-expressed genes. Curr Res Microb Sci. 2021;2:100022. doi: 10.1016/j.crmicr.2021.100022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86•.Chang YS, et al. SARS Unique Domain (SUD) of severe acute respiratory syndrome coronavirus induces NLRP3 inflammasome-dependent CXCL10-mediated pulmonary inflammation. Int J Mol Sci. 2020;21(9). 10.3390/ijms21093179. Readers would benefit from reading Chang et al. (2020) for insight into mechanistic measures by which the SARS-CoV Unique Domain (SUD) upregulates CXCL10 upregulation to cause increased pulmonary monocyte and macrophage accumulation and severe acute respiratory distress in an NLRP3-inflammasome-dependent manner in mice. [DOI] [PMC free article] [PubMed]
- 87.•.Ichikawa A, et al. CXCL10-CXCR3 enhances the development of neutrophil-mediated fulminant lung injury of viral and nonviral origin. Am J Respir Crit Care Med. 2013;187(1):65–77. doi: 10.1164/rccm.201203-0508OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.•.Lang S, et al. CXCL10/IP-10 neutralization can ameliorate lipopolysaccharide-induced acute respiratory distress syndrome in rats. PLoS One. 2017;12(1):e0169100. doi: 10.1371/journal.pone.0169100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Yen YT, et al. Modeling the early events of severe acute respiratory syndrome coronavirus infection in vitro. J Virol. 2006;80(6):2684–93. doi: 10.1128/JVI.80.6.2684-2693.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zhang F, et al. IFN- gamma and TNF- alpha drive a CXCL10 + CCL2 + macrophage phenotype expanded in severe COVID-19 and other diseases with tissue inflammation. bioRxiv. 2020. 10.1101/2020.08.05.238360. [DOI] [PMC free article] [PubMed]
- 91.Chen Y, et al. IP-10 and MCP-1 as biomarkers associated with disease severity of COVID-19. Mol Med. 2020;26(1):97. doi: 10.1186/s10020-020-00230-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Desai SR, Wells AU, Rubens MB, Evans TW, Hansell DM. Acute respiratory distress syndrome: CT abnormalities at long-term follow-up. Radiology. 1999;210(1):29–35. doi: 10.1148/radiology.210.1.r99ja2629. [DOI] [PubMed] [Google Scholar]
- 93.Oliviero A, de Castro F, Coperchini F, Chiovato L, and Rotondi M. COVID-19 pulmonary and olfactory dysfunctions: is the chemokine CXCL10 the common denominator? Neuroscientist. 2020:1073858420939033. 10.1177/1073858420939033. [DOI] [PubMed]
- 94.Fox SE, Lameira FS, Rinker EB, Vander Heide RS. Cardiac endotheliitis and multisystem inflammatory syndrome after COVID-19. Ann Intern Med. 2020;173(12):1025–1027. doi: 10.7326/l20-0882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Buja LM, et al. The emerging spectrum of cardiopulmonary pathology of the coronavirus disease 2019 (COVID-19): report of 3 autopsies from Houston, Texas, and review of autopsy findings from other United States cities. Cardiovasc Pathol. 2020;48:107233. doi: 10.1016/j.carpath.2020.107233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Babapoor-Farrokhran S, et al. Myocardial injury and COVID-19: possible mechanisms. Life Sci. 2020;253:117723. doi: 10.1016/j.lfs.2020.117723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Chen XM, et al. Exploration of omics mechanism and drug prediction of coronavirus-induced heart failure based on clinical bioinformatics. Zhonghua Xin Xue Guan Bing Za Zhi. 2020;48(7):587–592. doi: 10.3760/cma.j.cn112148-20200308-00172. [DOI] [PubMed] [Google Scholar]
- 98.Frangogiannis NG, et al. Induction and suppression of interferon-inducible protein 10 in reperfused myocardial infarcts may regulate angiogenesis. FASEB J. 2001;15(8):1428–30. doi: 10.1096/fj.00-0745fje. [DOI] [PubMed] [Google Scholar]
- 99.Bujak M, et al. Induction of the CXC chemokine interferon-gamma-inducible protein 10 regulates the reparative response following myocardial infarction. Circ Res. 2009;105(10):973–83. doi: 10.1161/CIRCRESAHA.109.199471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Sonzogni A, et al. Liver histopathology in severe COVID 19 respiratory failure is suggestive of vascular alterations. Liver Int. 2020;40(9):2110–2116. doi: 10.1111/liv.14601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.McCord JM, Hybertson BM, Cota-Gomez A, Geraci KP, and Gao B. Nrf2 activator PB125((R)) as a potential therapeutic agent against COVID-19. Antioxidants (Basel). 2020;9(6). 10.3390/antiox9060518. [DOI] [PMC free article] [PubMed]
- 102.Wu C, et al. Analysis of therapeutic targets for SARS-CoV-2 and discovery of potential drugs by computational methods. Acta Pharm Sin B. 2020;10(5):766–788. doi: 10.1016/j.apsb.2020.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Gordon DE, et al. A SARS-CoV-2-human protein-protein interaction map reveals drug targets and potential drug-repurposing. bioRxiv. 2020. 10.1101/2020.03.22.002386.
- 104.Zhang H, Penninger JM, Li Y, Zhong N, Slutsky AS. Angiotensin-converting enzyme 2 (ACE2) as a SARS-CoV-2 receptor: molecular mechanisms and potential therapeutic target. Intensive Care Med. 2020;46(4):586–590. doi: 10.1007/s00134-020-05985-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Golonka RM, et al. Harnessing innate immunity to eliminate SARS-CoV-2 and ameliorate COVID-19 disease. Physiol Genomics. 2020;52(5):217–221. doi: 10.1152/physiolgenomics.00033.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Street ME. HMGB1: A possible crucial therapeutic target for COVID-19? Horm Res Paediatr. 2020;93(2):73–75. doi: 10.1159/000508291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Ngai JC, et al. The long-term impact of severe acute respiratory syndrome on pulmonary function, exercise capacity and health status. Respirology. 2010;15(3):543–50. doi: 10.1111/j.1440-1843.2010.01720.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Higgins V, Sohaei D, Diamandis EP, and Prassas I. COVID-19: from an acute to chronic disease? Potential long-term health consequences. Crit Rev Clin Lab Sci. 2020;1-23. 10.1080/10408363.2020.1860895. [DOI] [PubMed]
- 109.Antonelli A, et al. Interferon-gamma-inducible alpha-chemokine CXCL10 involvement in Graves’ ophthalmopathy: modulation by peroxisome proliferator-activated receptor-gamma agonists. J Clin Endocrinol Metab. 2006;91(2):614–20. doi: 10.1210/jc.2005-1689. [DOI] [PubMed] [Google Scholar]
- 110.Carboni E, Carta AR, Carboni E. Can pioglitazone be potentially useful therapeutically in treating patients with COVID-19? Med Hypotheses. 2020;140:109776. doi: 10.1016/j.mehy.2020.109776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Buschard K. Fenofibrate increases the amount of sulfatide which seems beneficial against Covid-19. Med Hypotheses. 2020;143:110127. doi: 10.1016/j.mehy.2020.110127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Heffernan KS, Ranadive SM, Jae SY. Exercise as medicine for COVID-19: on PPAR with emerging pharmacotherapy. Med Hypotheses. 2020;143:110197. doi: 10.1016/j.mehy.2020.110197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Vankadari N, Wilce JA. Emerging WuHan (COVID-19) coronavirus: glycan shield and structure prediction of spike glycoprotein and its interaction with human CD26. Emerg Microbes Infect. 2020;9(1):601–604. doi: 10.1080/22221751.2020.1739565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Dube MP, et al. A randomized, double-blinded, placebo-controlled trial of Sitagliptin for reducing inflammation and immune activation in treated and suppressed human immunodeficiency virus infection. Clin Infect Dis. 2019;69(7):1165–1172. doi: 10.1093/cid/ciy1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Solerte SB, et al. Sitagliptin treatment at the time of hospitalization was associated with reduced mortality in patients with type 2 diabetes and COVID-19: a multicenter, case-control, retrospective, observational study. Diabetes Care. 2020;43(12):2999–3006. doi: 10.2337/dc20-1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Mirani M, et al. Impact of comorbidities and glycemia at admission and dipeptidyl peptidase 4 inhibitors in patients with type 2 diabetes with COVID-19: a case series from an academic hospital in Lombardy, Italy. Diabetes Care. 2020;43(12):3042–3049. doi: 10.2337/dc20-1340. [DOI] [PubMed] [Google Scholar]
- 117.Nauck MA, Meier JJ. Reduced COVID-19 mortality with Sitagliptin treatment? Weighing the dissemination of potentially lifesaving findings against the assurance of high scientific standards. Diabetes Care. 2020;43(12):2906–2909. doi: 10.2337/dci20-0062. [DOI] [PubMed] [Google Scholar]
- 118.Rotondi M, et al. Elevated serum interferon-gamma-inducible chemokine-10/CXC chemokine ligand-10 in autoimmune primary adrenal insufficiency and in vitro expression in human adrenal cells primary cultures after stimulation with proinflammatory cytokines. J Clin Endocrinol Metab. 2005;90(4):2357–63. doi: 10.1210/jc.2004-1062. [DOI] [PubMed] [Google Scholar]
- 119.Cinatl J, Jr, Michaelis M, Morgenstern B, Doerr HW. High-dose hydrocortisone reduces expression of the pro-inflammatory chemokines CXCL8 and CXCL10 in SARS coronavirus-infected intestinal cells. Int J Mol Med. 2005;15(2):323–7. [PubMed] [Google Scholar]
- 120.Fang J, et al. The expression of CXCL10/CXCR3 and effect of the axis on the function of T lymphocyte involved in oral Lichen Planus. Inflammation. 2019;42(3):799–810. doi: 10.1007/s10753-018-0934-0. [DOI] [PubMed] [Google Scholar]
- 121.Boumaza A, et al. Monocytes and macrophages, targets of SARS-CoV-2: the clue for Covid-19 immunoparalysis. J Infect Dis. 2021. 10.1093/infdis/jiab044 [DOI] [PMC free article] [PubMed]
- 122.Xie H, Feng C, Fu Q, Sa YL, Xu YM. Crosstalk between TGF-beta1 and CXCR3 signaling during urethral fibrosis. Mol Cell Biochem. 2014;394(1–2):283–90. doi: 10.1007/s11010-014-2104-5. [DOI] [PubMed] [Google Scholar]
- 123.Ferreira C, et al. Type 1 Treg cells promote the generation of CD8(+) tissue-resident memory T cells. Nat Immunol. 2020;21(7):766–776. doi: 10.1038/s41590-020-0674-9. [DOI] [PubMed] [Google Scholar]
- 124.McCarron MJ, Irla M, Serge A, Soudja SM, Marie JC. Transforming growth factor-beta signaling in alphabeta thymocytes promotes negative selection. Nat Commun. 2019;10(1):5690. doi: 10.1038/s41467-019-13456-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Suga H, et al. CXCR3 deficiency prolongs Th1-type contact hypersensitivity. J Immunol. 2013;190(12):6059–70. doi: 10.4049/jimmunol.1201606. [DOI] [PubMed] [Google Scholar]
- 126.Gunderson AJ, et al. TGFbeta suppresses CD8(+) T cell expression of CXCR3 and tumor trafficking. Nat Commun. 2020;11(1):1749. doi: 10.1038/s41467-020-15404-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Yates CCJ, Z I, Willis MS, Jayne J. FIBROKINE™ peptides: a broad-spectrum of anti-fibrotic chemokine peptides to treat organ fibrosis. FASEB J. 2018;32. 10.1096/fasebj.2018.32.1_supplement.414.5.
- 128.Yates-Binder CC, et al. An IP-10 (CXCL10)-derived peptide inhibits angiogenesis. PLoS One. 2012;7(7):e40812. doi: 10.1371/journal.pone.0040812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Pei F, et al. Lung pathology and pathogenesis of severe acute respiratory syndrome: a report of six full autopsies. Zhonghua Bing Li Xue Za Zhi. 2005;34(10):656–60. [PubMed] [Google Scholar]
- 130.Leviner S. Recognizing the clinical sequelae of COVID-19 in adults: COVID-19 long-haulers. J Nurse Pract. 2021 doi: 10.1016/j.nurpra.2021.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Higgins V, Sohaei D, Diamandis EP, Prassas I. COVID-19: from an acute to chronic disease? Potential long-term health consequences. Crit Rev Clin Lab Sci. 2021;58(5):297–310. doi: 10.1080/10408363.2020.1860895. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable
Not applicable