Abstract
The need for severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) next-generation vaccines has been highlighted by the rise of variants of concern (VoCs) and the long-term threat of emerging coronaviruses. Here, we design and characterize four categories of engineered nanoparticle immunogens that recapitulate the structural and antigenic properties of the prefusion SARS-CoV-2 spike (S), S1, and receptor-binding domain (RBD). These immunogens induce robust S binding, ACE2 inhibition, and authentic and pseudovirus neutralizing antibodies against SARS-CoV-2. A spike-ferritin nanoparticle (SpFN) vaccine elicits neutralizing titers (ID50 > 10,000) following a single immunization, whereas RBD-ferritin nanoparticle (RFN) immunogens elicit similar responses after two immunizations and also show durable and potent neutralization against circulating VoCs. Passive transfer of immunoglobulin G (IgG) purified from SpFN- or RFN-immunized mice protects K18-hACE2 transgenic mice from a lethal SARS-CoV-2 challenge. Furthermore, S-domain nanoparticle immunization elicits ACE2-blocking activity and ID50 neutralizing antibody titers >2,000 against SARS-CoV-1, highlighting the broad response elicited by these immunogens.
Keywords: SARS-CoV-2, ferritin nanoparticle, spike, receptor-binding domain, COVID-19, SARS-CoV-1, betacoronaviruses, variants of concern, B.1.1.7, B.1.351, P.1, ALFQ, neutralizing antibodies
Graphical abstract
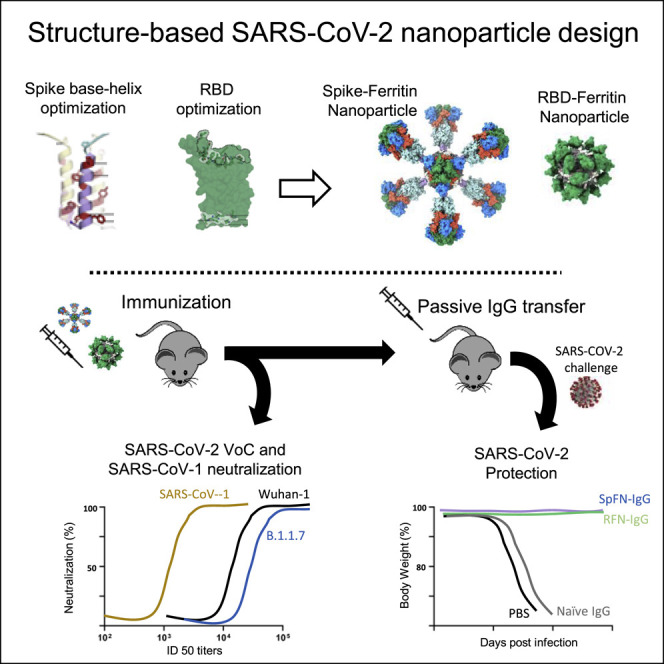
Joyce et al. generate four categories of engineered SARS-CoV-2 ferritin nanoparticle immunogens using structure-based vaccine design that recapitulate the prefusion SARS-CoV-2 spike, S1, and RBD. These immunogens induce robust and protective neutralizing antibody responses against SARS-CoV-2 and elicit potent neutralization against variants of concern and the heterologous SARS-CoV-1.
Introduction
To date, seven coronaviruses (CoVs) are known to cause disease in humans. Three of these, severe acute respiratory syndrome coronavirus 1 (SARS-CoV-1), Middle East respiratory syndrome coronavirus (MERS-CoV), and SARS-CoV-2, have emerged since 2003 (Cui et al., 2019), with high mortality rates. SARS-CoV-2 is easily transmitted by humans and created a pandemic, infecting over 100 million people, causing over 2 million deaths to date, and resulting in an urgent need for protective and durable vaccines. A rapid vaccine development effort led to the evaluation of hundreds of SARS-CoV-2 vaccine candidates and worldwide vaccine distribution and use.
The response to SARS-CoV-2 was facilitated by multiple efforts over the last decade to enable CoV pandemic preparedness, initially based on MERS-CoV vaccine design and development (Adhikari et al., 2019), phase I vaccine trials (Adney et al., 2019), and a global effort by the Coalition for Epidemic Preparedness Innovations (CEPI) to advance vaccine candidates (Gouglas et al., 2019). The elucidation of CoV spike (S) glycoprotein structures (Lee et al., 2016; Pallesen et al., 2017) allowed structure-based vaccine design of stabilized S glycoprotein immunogens from multiple CoVs (Pallesen et al., 2017), providing a blueprint for SARS-CoV-2 vaccine design (Wrapp et al., 2020b).
The CoV S protein mediates virus entry, is immunogenic (Iyer et al., 2020; Wang et al., 2021a), and encodes multiple neutralizing epitopes (Greaney et al., 2021), making it the primary target for natural and vaccine-induced CoV humoral immunity and vaccine design (Cao et al., 2020) and the target of most COVID-19 vaccines. S is a class I fusion glycoprotein consisting of an S1 attachment subunit and S2 fusion subunit that remain non-covalently associated in a metastable, heterotrimeric S on the virion surface (Walls et al., 2020b). Within the S1 subunit, there is an N-terminal domain (NTD), and a C-terminal domain (CTD) that includes the receptor-binding domain (RBD). The RBD binds to human angiotensin-converting enzyme 2 (hACE2), facilitating cell entry (Alanazi et al., 2020). Multiple antigenic sites have been identified on the S protein, including distinct sites on the RBD and the NTD supersite (Brouwer et al., 2020; Cerutti et al., 2021; Liu et al., 2020; Zost et al., 2020). Convalescent serum antibodies or monoclonal antibodies capable of potent neutralization targeting these sites can reduce disease severity or mortality in rodents, non-human primates (Barnes et al., 2020), and humans (Cao et al., 2020; Salazar et al., 2020; Shi et al., 2020).
Due to the unknown parameters of SARS-CoV-2 vaccine durability, specific age or population needs, emergence of SARS-CoV-2 variants of concern (VoCs) (Wibmer et al., 2021), and the constant threat of newly emerging human CoVs (Menachery et al., 2015), next-generation COVID-19 or pan-CoV vaccines will be needed. Iterative structure-based design for viral glycoproteins (McLellan et al., 2013; Joyce et al., 2016) that stabilize neutralizing epitopes or epitope-based vaccine design (Chen et al., 2021; Kong et al., 2019) shows that rational vaccine design can lead to the elicitation of broad immune responses. Similar broad cross-reactive responses elicited by structure-based engineered vaccines have also been advanced for influenza (Boyoglu-Barnum et al., 2020; Kanekiyo et al., 2019). In the case of CoVs, a set of cross-reactive epitopes has recently been described (Barnes et al., 2020; Joyce et al., 2020; Sauer et al., 2021; Wrapp et al., 2020a), with many of the preferred neutralizing antibodies centered on the RBD (Li et al., 2021; Pinto et al., 2020; Rappazzo et al., 2021).
Next-generation strategies to augment specific immune responses as well as enhance cross-reactivity include the use of nanoparticle vaccine technology (Cohen et al., 2021) and next-generation adjuvants. Nanoparticle technologies have been shown to improve antigen structure and stability, as well as targeted vaccine delivery and immunogenicity, with good safety profiles (Pati et al., 2018). Engineered nanoparticle vaccines can elicit broader immune responses (Darricarrère et al., 2021; Kanekiyo et al., 2013, 2019) or more efficacious immune responses (Kanekiyo et al., 2015). The repetitive array of the viral surface component allows for robust B cell activation facilitating memory B cell expansion and generation of long-lived plasma cells. More recently, in efforts to generate more effective vaccines that can prevent infection by resistant pathogens such as HIV-1 or influenza, a set of engineered nanoparticle vaccines has been developed. Utilizing naturally occurring nanoparticle molecules such as bacterial ferritin, antigens are fused to the ferritin molecule to recapitulate complex trimeric class I glycoproteins and to increase the immune response for weakly immunogenic targets. Recently designed single- and multi-component nanoparticle vaccines (Brouwer et al., 2021; Walls et al., 2020a) show promise from both an immunological (Cawlfield et al., 2019; Marcandalli et al., 2019) and a current Good Manufacturing Practice (cGMP) production perspective (Ueda et al., 2020).
Engineered nanoparticle vaccines and their capacity to generate enhanced immune responses in humans are currently being studied and include influenza (NCT03186781; NCT03814720; NCT04579250), Epstein-Barr virus (NCT04645147), malaria (NCT04296279), and a recently described SARS-CoV-2 nanoparticle vaccine (IVX-411) (Walls et al., 2020b). Use of potent adjuvants such as liposomal-saponin adjuvants can further enhance the protective immune response (Cawlfield et al., 2019; Lal et al., 2015; Om et al., 2020) even in the context of nanoparticle vaccines (Langowski et al., 2020; Kaba et al., 2018). Based on the results described herein and data from associated non-human primate experiments (Joyce et al., 2021; King et al., 2021), an S-trimer-ferritin immunogen with a liposomal adjuvant, ALFQ, is currently being assessed in a phase I clinical trial (NCT04784767).
Here we report the iterative structure-based design and pre-clinical assessment of four classes of S-domain ferritin nanoparticles including stabilized S-trimer-ferritin nanoparticles (SpFNs), RBD-ferritin nanoparticles (RFNs), S1-ferritin nanoparticles, and RBD-NTD-ferritin nanoparticles. By using a set of biophysical, structural, and antigenic assessments, combined with animal immunogenicity testing, we identify candidate immunogens that elicit potent neutralizing antibody titers against SARS-CoV-2 and robust protection against SARS-CoV-2 challenge in the K18-hACE2 mouse model. We further show that subsequent immunizations not only increase the SARS-CoV-2 neutralization titer but also expand the neutralization breadth against the heterologous SARS-CoV-1 virus and current circulating VoCs. These data provide a foundation for immunogen design strategies for pan-betacoronavirus vaccine development.
Results
Immunogen design of SARS-CoV-2 S-domain ferritin nanoparticles
Using the initial SARS-CoV-2 genome sequence from Wuhan, China (GenBank: MN9089473), we designed four classes of S-domain ferritin-fusion recombinant proteins as immunogens for expression as nanoparticles based on the major antigenic domains of the S ectodomain (Figure S1). The Helicobacter pylori ferritin molecule was genetically linked to the C-terminal region of the following S antigens: (1) S ectodomain (residues 12–1158), (2) RBD (residues 331–527), (3) RBD linked in tandem to the NTD (residues 12–303), and (4) S1 (residues 12–696) (Figures 1 and S1; Table S1). For the S-ferritin nanoparticle designs, a short linker to the ferritin molecule was used to utilize the natural three-fold axis, for display of eight spikes. For the other designs, a short region of bullfrog ferritin was utilized to allow equidistant distribution of the 24 S-domain molecules on the ferritin surface (Figure 1). Our overall approach was to compare the immunogen structure, antigenicity, and immunogenicity elicited by these different immunogens with the goal to identify the best immunogen to take forward into further development.
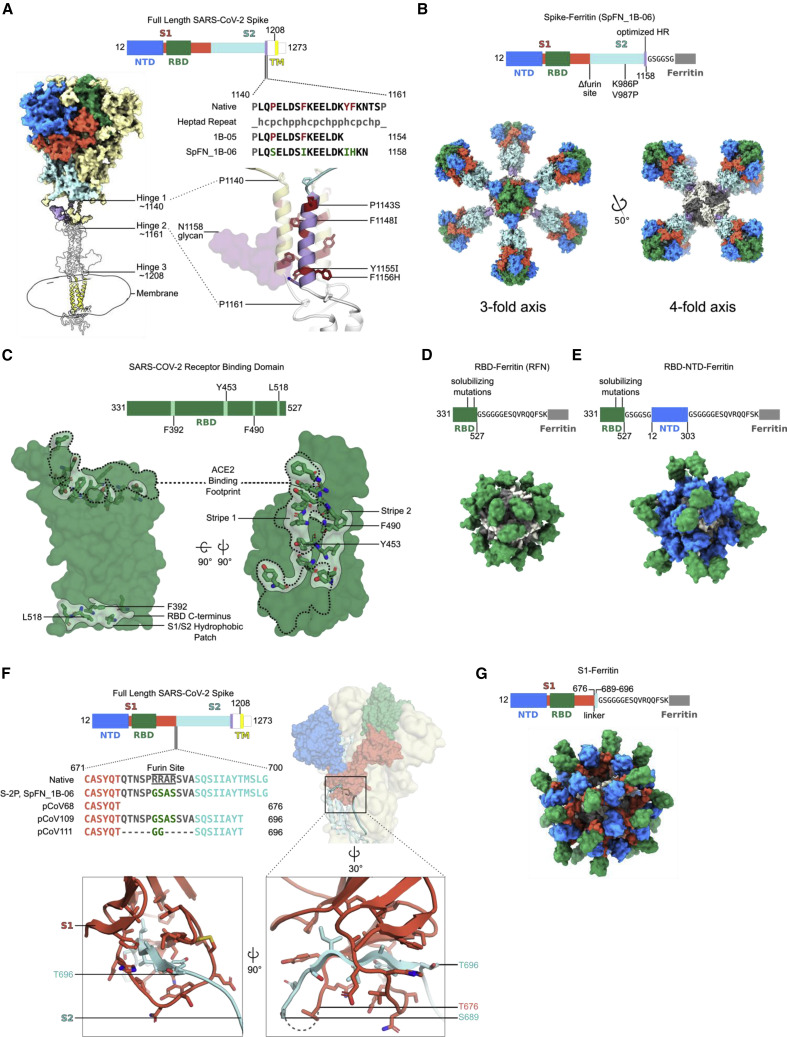
Figure 1.
Structure-based design of SARS-CoV-2 S-based ferritin nanoparticle immunogens
(A) Full-length SARS-CoV-2 S schematic and 3D structure. S hinges identified by molecular dynamics simulations and electron cryotomography are labeled on the 3D model (Turoňová et al., 2020). The structured trimeric ectodomain is colored according to the schematic with the N-terminal domain (NTD) and receptor-binding domain (RBD) of the S1 polypeptide and the C-terminal coiled coil N-terminal to hinge 1 colored blue, green, and purple, respectively. Remaining portions of the S1 and S2 polypeptides are colored in red and cyan with regions membrane proximal from hinge 2 colored in white. The transmembrane domain of all chains is depicted in yellow. To design a spike-ferritin molecule, the C-terminal heptad repeat (residues 1140–1161) between hinges 1 and 2 was aligned to an ideal heptad-repeat sequence. Residues in the native spike sequence that break this pattern are highlighted in red. These residues are also labeled and highlighted in red on the 3D structure. Two engineered designs (1B-05 and 1B-06) are shown, with the spike end residue used to link to ferritin, and heptad-repeat mutations colored green.
(B) Schematic and 3D model of a spike-ferritin nanoparticle (SpFN). Differences between the native spike sequence and the engineered nanoparticle are indicated on the schematic. A 3D model of SpFN displaying eight trimeric spikes was created using PDB: 6VXX and 3EGM with the ferritin molecule shown in alternating gray and white. The nanoparticle is depicted along the four-fold and three-fold symmetry axes of the ferritin.
(C) RBD-ferritin nanoparticle design and optimization. The RBD of SARS-CoV-2 (PDB: 6MOJ) is shown in surface representation, with the ACE2-binding site outlined in dashed lines. Three hydrophobic regions of the RBD that were mutated for nanoparticle immunogen design are shown in light green surface, with residues in stick representation. The ACE2-binding site contains two of these regions, whereas a third hydrophobic patch near the C terminus of the RBD is typically buried by S2 and part of S1 in the context of the trimer molecule.
(D) Schematic and 3D model of an RBD-ferritin nanoparticle. A modeled 24-mer nanoparticle displaying the RBD domain is depicted at the three-fold symmetry axis of ferritin and colored green. Truncation points, linkers, and alterations made to the RBD sequence are indicated on the schematic.
(E) Schematic and 3D model of an RBD-NTD-ferritin nanoparticle. A modeled nanoparticle displaying RBD and NTD epitopes is depicted and colored according to the schematic. Truncation points, linkers, and alterations made to the native spike sequence are indicated on the schematic.
(F) S1-ferritin immunogen design. The SARS-CoV-2 S1 forms a hydrophobic collar around the N-terminal β sheet of S2 (residues 689–676). S1-ferritin immunogen design required inclusion of this short stretch of S2 (colored cyan) attached by a linker. Terminal residues of the structured portions of S1 and S2 are labeled.
(G) Schematic and 3D model of an S1-ferritin nanoparticle. A modeled nanoparticle displaying RBD and NTD domains is depicted and colored according to the S1-ferritin schematic with truncation points and domain linkers indicated.
The first design category, S-trimer-ferritin nanoparticle (SpFN) designs, was based on a modified S with stabilizing prolines (K986P, V987P), removal of the furin cleavage site (RRAS to GSAS), and optimization of the coiled-coil region between hinge 1 and hinge 2 of the ectodomain stalk (Turoňová et al., 2020) to stabilize trimer formation on the ferritin scaffold (Figures 1A and S1). The designs focused on (1) modification of the c-terminal residue of the S molecule (1137, 1208, 1154, or 1158), (2) optimization of the coiled-coil region through extensions or repeats, (3) removal of the coiled-coil region, (4) removal of glycan 1158, (5) addition of heterogeneous trimerization domains (GCN4, or foldon), or (6) signal peptide sequence (Figure 1B; Table S1).
The second design category, RBD-ferritin nanoparticle (RFN) designs, used the SARS-CoV-2 RBD (residues 331–527) (Figure 1C) connected to the bullfrog-H. pylori chimeric ferritin (Kanekiyo et al., 2015) by a 6-amino acid linker (Figure 1D). The SARS-CoV-2 RBD contains a set of hydrophobic patches, including the ACE2-binding site, and a region located at about residues 517/518 that is hidden in the context of the intact S molecule. These regions were iteratively mutated to reduce hydrophobicity and increase stability of the RFN molecules, expression levels, antigenicity, and immunogenicity.
The third design category, RBD-NTD-ferritin molecule designs, was based on addition of optimized RBD molecules in series with an NTD-ferritin construct (residues 12–303) linked to the bullfrog-H. pylori chimeric ferritin molecule. The in-series, but reversed, RBD-NTD design ensured distal displacement of the RBD molecule from the ferritin molecule (Figure 1E), promoting immune recognition of the RBD molecule with potential benefits for the production and stability of the nanoparticle.
The fourth design category, S1-ferritin designs (residues 12–676) (Figure 1F; Table S1), was initially designed based on the MERS S1 immunogen, which elicited protective immune responses (Wang et al., 2015). Subsequent designs focused on inclusion of a short region of SARS-CoV-2 S2 (residues 689–696) either using the connecting region that overlaps the furin site or by use of a short glycine-rich linker sequence (Figure 1F) to enable formation of the S1-ferritin nanoparticle (Figure 1G).
Characterization of SARS-CoV-2 S-domain ferritin nanoparticles
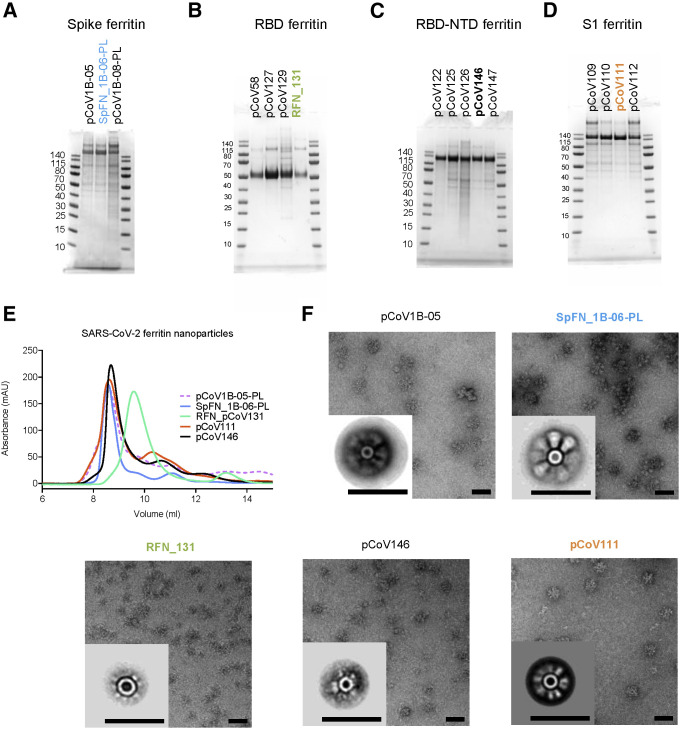
Ten SpFN constructs (Table S1) were designed and tested for expression, yield, nanoparticle formation, and antigenicity. S-ferritin nanoparticles were expressed in Expi293F cells for 3–5 days at 34°C and 37°C and purified by Galanthus nivalis lectin (GNA) affinity chromatography. A subset of these constructs showed reasonable expression levels ranging from 0.5 to 5 mg/L medium supernatant (Figure S3). SpFN constructs pCoV1B-05 and pCoV1B-06-PL (SpFN_1B-06-PL) typically yielded over 5 mg/L with expression incubation set at 34°C. Samples were assessed by SDS-PAGE, size-exclusion chromatography (SEC), dynamic light scattering (DLS), and negative-stain electron microscopy (neg-EM) to ensure intact protein was produced, and to assess nanoparticle formation and S morphology (Figures 2 and S2). In the case of SpFN and SpFN_1B-08 (Figure S2), the globular shape of the protruding S was clearly visible in both the transmission electron microscopy (TEM) images and the 2D averages. In the case of pCoV1B-05, the protruding S showed more of an “open” form in both the TEM images and the 2D averages.
Figure 2.
Biophysical characterization of SARS-CoV-2 S-based ferritin nanoparticle vaccine candidates
(A–D) SDS-PAGE of (A) spike-ferritin nanoparticles, (B) RBD-ferritin nanoparticles, (C) RBD-NTD-ferritin nanoparticles, and (D) S1-ferritin nanoparticles. Molecular weight standards are indicated in kDa.
(E) Size-exclusion chromatography on a Superdex S200 10/300 column of representative SARS-CoV-2 S-based ferritin nanoparticles.
(F) Negative-stain electron microscopy 2D class averages of purified nanoparticles. The scale bars represent 50 nm.
See also Figures S2 and S3.
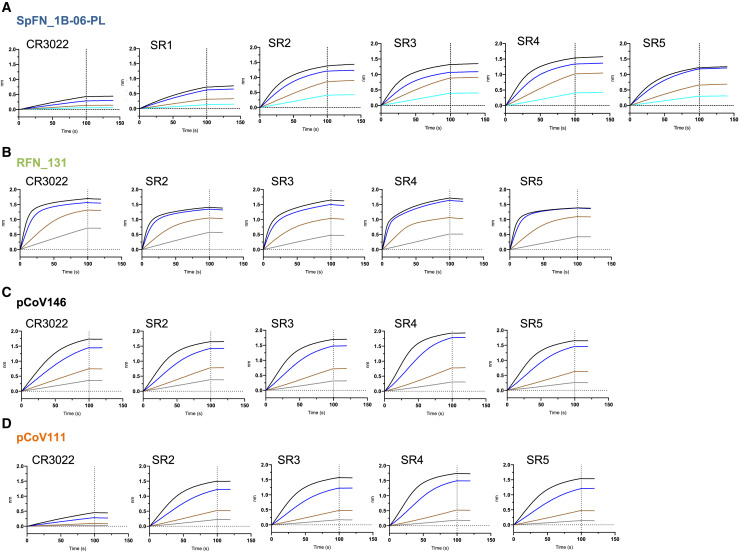
In addition, nanoparticles were assessed for antigenicity using biolayer interferometry (BLI) against a set of poorly neutralizing (CR3022, SR1) and potently neutralizing (SR2, SR3, SR4, SR5) RBD-targeting antibodies. The different SpFN designs showed variable binding to the antibodies, with SpFN_1B-06-PL having the highest binding (Figures 3 A and S1).
Figure 3.
Antigenic characterization of select SARS-CoV-2 S-based ferritin nanoparticle vaccine candidates
Binding response of SARS-CoV-2 neutralizing antibodies to each of the lead candidates from the four design categories measured by biolayer interferometry with two-fold serial dilution of each antibody starting at 30 μg/mL.
(A) Spike-ferritin nanoparticle SpFN_1B-06-PL.
(B) RBD-ferritin nanoparticle RFN_131.
(C) RBD-NTD-ferritin nanoparticle pCoV146.
(D) S1-ferritin nanoparticle pCoV111.
See also Figure S3.
Initial test expression of RBD-ferritin constructs at 37°C using either 293F or Expi293F cells showed low levels of expression. Reducing the cell expression temperature to between 30°C and 34°C following transfection rescued expression and enabled levels >20 mg/L to be purified by NiNTA purification (Figure S3). However, analysis of the constructs by SEC, DLS, and neg-EM indicated that initial RBD-ferritin constructs did not form fully intact nanoparticles (Figure S2). We hypothesized that designed variants with reduced RBD surface hydrophobicity would allow for improved nanoparticle yield. Screening through a set of variants using SDS-PAGE and SEC as primary indicators of nanoparticle formation allowed identification of constructs that readily formed nanoparticles (Figures 2, S1, and S2B). These molecules were also visualized by neg-EM, and showed clear formation of nanoparticles, with the protruding RBD visible on their surface in both TEM images and 2D class averages (Figure 2F). However, these constructs had a propensity to form aggregates and dramatically affected the ability to concentrate the samples. Addition of 5% glycerol to the NiNTA-purified material, prior to SEC or other concentration steps, mitigated the aggregation issue and increased the nanoparticle formation as judged by SEC, and was confirmed by neg-EM. The RBD-ferritin constructs showed very strong binding to the set of RBD-specific antibodies in all cases (Figure 3B). The level of binding was approximately twice that seen for the S-ferritin constructs, indicative of the exposed and accessible nature of the RBD epitopes (Figures 1D and 2F).
Due to the initial difficulty with S1-ferritin nanoparticle constructs, we developed a set of engineered S1 constructs by artificially connecting the RBD by a short linker to the NTD linked to the ferritin molecule and denoted as RBD-NTD-ferritin molecules. This multi-domain design strategy resulted in good protein expression (up to 2.5 mg/L) and clear nanoparticle formation by TEM. Using the information gained from the RBD surface optimization, we designed multiple constructs with variations in the RBD molecule to reduce surface hydrophobicity (Figure S1). Antigenic analysis of these constructs showed that pCoV146 displayed robust antibody binding (Figure 2F).
The initial S1-ferritin construct, pCoV68 (residues 12–676), yielded very low protein expression levels, even with reduced expression temperatures (Figure S3A). However, using the structure of the S-2P molecule (Wrapp et al., 2020b; Walls et al., 2020b), it was clear that a short segment of S2 formed significant interactions with the S1 domain. Addition of this short region either using the natural sequence (with the furin site removed) as in construct pCoV109, or by linking residues 689–696 with glycine-rich linkers as in construct pCoV111, allowed ∼1 mg/L of protein to be purified. Analysis of these constructs by SDS-PAGE, SEC, and neg-EM showed clear formation of the designed nanoparticles, and antigenic characterization showed binding of antibodies to the nanoparticle (Figure 3D).
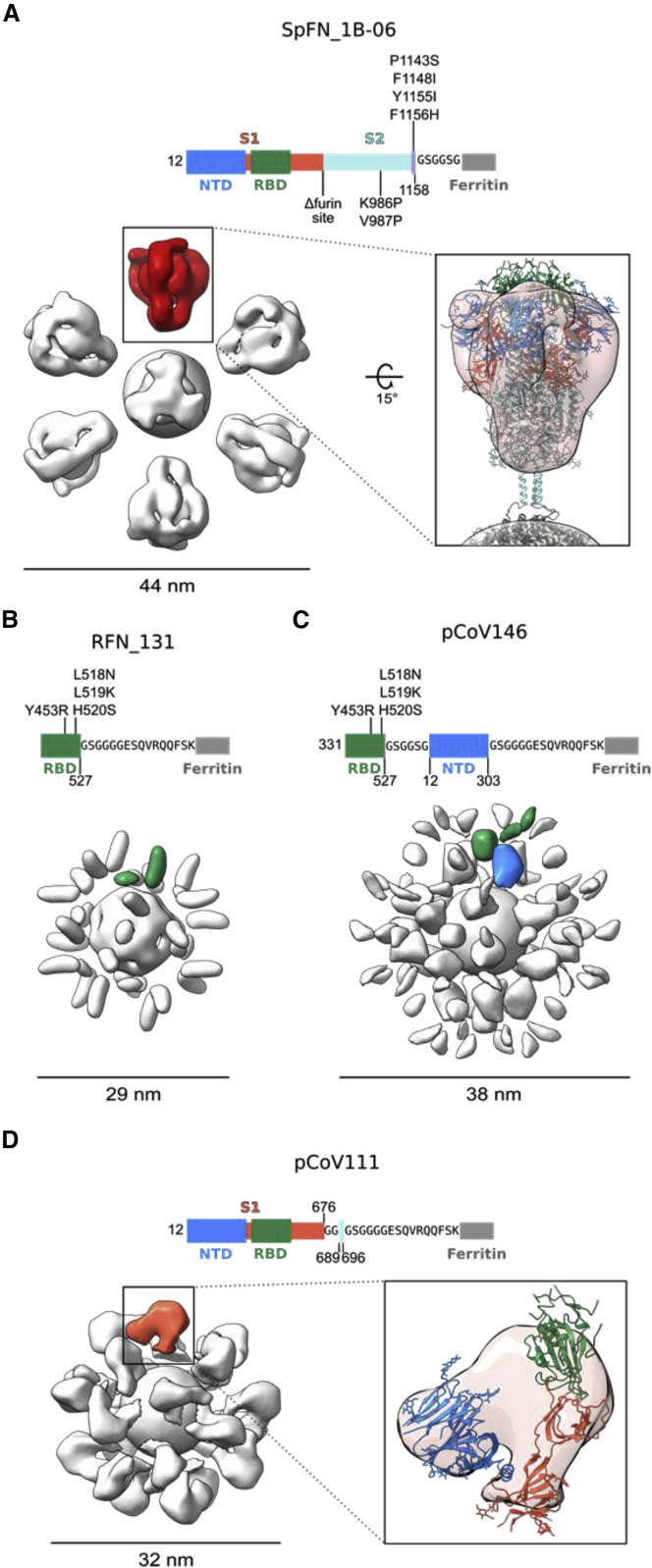
Further structural analysis of the nanoparticle immunogens from each of the four design categories was carried out by determining 3D reconstructions from negative-stain electron micrographs (Figure 4 ). For each nanoparticle, a central sphere of approximately 12 nm corresponding to ferritin was resolved. S-domain antigens were located a short distance away from the central sphere and linker regions were unresolved, likely due to their small size and flexibility. The SpFN_1B-06-PL reconstruction showed the stabilized S protruding from the ferritin molecule with a total diameter of approximately 44 nm (Figure 4A). The large size and distinct low-resolution features of the S ectodomain allowed for docking of a closed S-2P trimer model (PDB: 6VXX) into the trimer density, confirming S was in the prefusion conformation. The ferritin-distal region of the S density was slightly weaker and likely reflects the heterogeneity in RBD-up conformations or slight openings of trimer visible in raw micrographs. Additionally, although the coiled coil was unresolved, the distances between the density for S and ferritin matched the modeled coiled-coil length. Reconstruction of the RFN_131 3D EM map revealed two globular densities per asymmetric unit, suggesting that the RBD molecule was highly flexible on the surface of the ferritin sphere (Figure 4B). Similarly, the map of the RBD-NTD-ferritin nanoparticle pCoV146 showed two layers of globular densities, with a ferritin-proximal layer corresponding to the NTD and a more disordered layer for the RBD (Figure 4C). This particle was approximately 9 nm larger in diameter in both 2D and 3D than the RFN molecules. The reconstruction of the S1-ferritin fusion pCoV111 revealed a surprisingly ordered S1 density compared to the flexible RBD-NTD ferritin nanoparticle, perhaps due to geometric constraints on the surface of the ferritin particle (Figure 4D). Density similar in shape to the S1 domain in the closed S-2P trimer was resolved although it was slightly more compact, likely due to both overall flexibility of the S1 on the ferritin surface and RBD flexibility.
Figure 4.
Negative-stain electron microscopy 3D reconstructions of SARS-CoV-2 S-based ferritin nanoparticles
Modifications made to the native sequence and linkers used for each construct are shown in schematic diagrams. The size of each particle is indicated in nanometers.
(A) Negative-stain 3D reconstructions with applied octahedral symmetry are shown with an asymmetric unit of non-ferritin density colored. Spike trimer density is colored in red, and a model of a SARS-CoV-2 S-2P trimer based on PDB: 6VXX is shown docked into the negative-stain map and colored according to the schematic diagram.
(B) Two non-ferritin densities per asymmetric unit were observed for RFN_131 and are highlighted in green. These densities putatively correspond to the RBD but lack low-resolution distinguishing features due to the small, globular shape of these domains. The presence of two densities is likely due to flexibility in the linker and heterogeneity in the RBD pose.
(C) Two layers of densities were distinguishable for pCoV146, with the putative NTD density of an asymmetric unit colored blue, proximal to the ferritin, and two smaller, more flexible densities corresponding to the RBD distal to the ferritin and colored green.
(D) An asymmetric unit of non-ferritin density for pCoV111 is colored in orange and a monomer of S1 in the closed trimer state from PDB: 6VXX is shown docked into the density with domains colored as in the schematicdiagram.
Immunogenicity of SARS-CoV-2 S-domain ferritin nanoparticles in mice
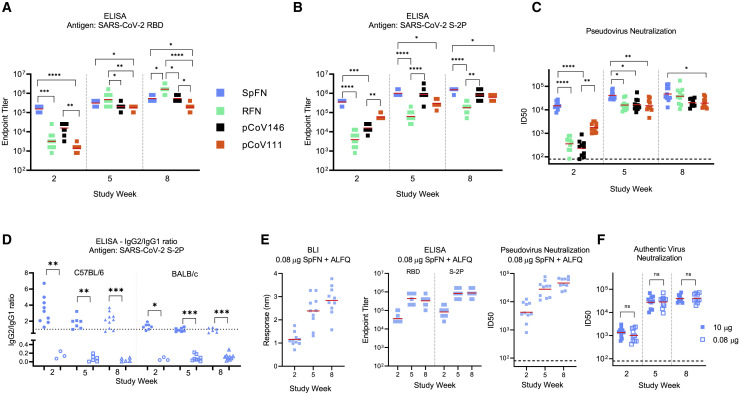
To evaluate the immunogenicity of the SARS-CoV-2 ferritin nanoparticles, we utilized two strains of mice (C57BL/6 and BALB/c) and two adjuvants (ALFQ and Alhydrogel), and immunized mice three times intramuscularly at 3-week intervals using a 10-μg dose. ALFQ consists of liposomes containing saturated phospholipids, cholesterol, and monophosphoryl lipid A as an immunostimulant, and saponin QS21 (ALFQ). In total, we assessed 14 immunogens, two S-ferritin immunogens, seven RBD-ferritin immunogens, one S1-ferritin construct, and four RBD-NTD-ferritin immunogens (Table S3). We also assessed two control proteins (S-2P and RBD), to allow comparison to the ferritin-nanoparticle platform. Assessment in immunogenicity studies was based on iterative knowledge of immunogen physical and biochemical characteristics (Figures 2, 3, and S3), in conjunction with immunogenicity results from first-generation immunogens (Figure S1). This facilitated down-selection of lead immunogen candidates. Alhydrogel and ALFQ adjuvants were selected due to their history in human vaccine trials, safety profile, and previous performance alongside nanoparticle vaccine immunogens (NCT04296279). Alhydrogel contains aluminum hydroxide gel, whereas ALFQ is a liposome-based adjuvant containing the saponin QS-21 and synthetic monophosphoryl lipid A (3D-PHAD). Following immunization, we assessed mouse sera for SARS-CoV-2 RBD and S binding, RBD-ACE2 inhibition, pseudovirus neutralizing antibody responses, and authentic SARS-CoV-2 virus neutralization at multiple time points (Figures 5 and S4). The use of a BLI assay allowed rapid characterization of immune sera and was highly informative in the early stages of the COVID-19 pandemic, as assays were being developed.
Figure 5.
SARS-CoV-2 S-domain nanoparticle vaccine candidates elicit robust binding and neutralizing antibody responses in C57BL/6 mice
Data relating to each category of immunogen are colored as follows: SpFN_1B-06-PL, blue; RFN_131, green; pCoV146, black; and pCoV111, orange. n = 10/group. ALFQ was the adjuvant used for these animal groups.
(A and B) ELISA binding of mouse sera to SARS-CoV-2 RBD or S-2P. Study week is indicated at the base of the graph. Geometric mean value is indicated by a horizontal line. Statistical comparison at each time point was carried out using a Kruskal-Wallis test followed by a Dunn’s post-test.
(C) SARS-CoV-2 pseudovirus neutralization ID50 values. Geometric mean titer values are indicated by a horizontal line. Statistical comparison at each given time point was carried out using a Kruskal-Wallis test followed by a Dunn’s post-test.
(D) ELISA analysis of antibody isotype usage following immunization with SpFN + ALFQ (solid shapes) or SpFN + Alhydrogel (open shapes). Sera collected at study week 2, 5, and 8 from immunized mice were added in quadruplicate serial dilutions to ELISA plates coated with S-2P protein. Duplicated wells were probed with anti-mouse-IgG1-HRP. Additional duplicates were probed with either anti-mouse-IgG2c-HRP or anti-mouse IgG2a-HRP for C57BL/6 and BALB/c mice, respectively. Data were interpolated to obtain the dilution factor at OD450 of 1 and plotted as ratios of IgG2/IgG1. A horizontal dotted line denotes a balanced 1:1 IgG2/IgG1 ratio. Isotype ratio values were compared between the two adjuvant groups at each timepoint for each mouse type using a Mann-Whitney unpaired two-tailed non-parametric test.
(E) Binding and pseudovirus neutralization of sera from mice immunized with 0.08 μg SpFN + ALFQ.
(F) Authentic SARS-CoV-2 virus neutralization ID50 values are shown for mice immunized with 10 μg (blue) or 0.08 μg (light blue) SpFN + ALFQ. Geometric mean titer is indicated by a horizontal line. Comparisons between dose groups at each time point were carried out using a Mann-Whitney unpaired two-tailed non-parametric test.
In (B) and (C), all groups at a given study time point were compared to each other. Only groups with significant differences are indicated by a bar; all other groups did not show statistically significant differences. ∗∗∗∗p < 0.0001, ∗∗∗p < 0.001, ∗∗p < 0.01, ∗p < 0.05. See also Figures S4–S6 and Tables S3 and S4.
All four categories of immunogens elicited robust SARS-CoV-2 immune responses. Immune responses seen in C57BL/6 mice were greater than for BALB/c mice after a single immunization, whereas binding and neutralizing antibody titers were comparable after a second or third immunization (Figure 5). In all cases, the third immunization did not substantively increase the antibody levels induced by the S-domain ferritin nanoparticles. In all cases tested, ALFQ was superior to Alhydrogel as an adjuvant for elicitation of binding and neutralizing responses (Figures 5 and S4; Table S4). In addition, Alhydrogel led to a skewed antibody isotype immune response that was TH2 in nature, as opposed to the balanced immune response seen with ALFQ-adjuvanted animals (Figure 5D). The immunoglobulin G (IgG)2a/c:IgG1 ratio observed in both C57BL/6 and BALB/c mice was consistently at 1 or greater for the SpFN+ALFQ-immunized mice, indicating a balanced or TH1 response. This antibody isotype response also matched the TH1 immune response at the cellular level (Carmen et al., 2021).
SpFN_1B-06-PL immunogen elicited rapid RBD binding and pseudovirus neutralizing antibody response with ID50 geometric mean titer (GMT) >10,000 in C57BL/6 mice and ID80 >1,000 after a single immunization (Figures 5C and S4). This rapid neutralizing immune response after one immunization was significantly higher than seen with RBD-ferritin or RBD-NTD-ferritin immunogens. Following a second immunization, both strains of SpFN-vaccinated mice showed ID50 GMT >10,000 and ID80 GMT >5,000 for both mouse types. Comparison of these neutralization responses to those reported by Powell et al. (2021) indicates that the immune responses elicited are similar, with strong immune responses seen after a single immunization, and these titers increase ∼10-fold after a boosting immunization (Table S4).
RFN immunogens elicited robust RBD- and S-binding responses and pseudovirus neutralization ID50 GMT >10,000 in both mice strains after two immunizations (Figures 5A–5C). We assessed seven RBD-ferritin immunogen designs in immunogenicity studies (Table S3), and selection for animal immunogenicity experiments was largely based on nanoparticle stability, expression level, aggregation, and antigenic profile (Figures 2, 3, S2, and S3). Based on these criteria, RFN_131 was extensively assessed and, after three immunizations, showed pseudovirus neutralization responses that were comparable to or exceeded that seen for the SpFN_1B-06-PL immunogen. Of note, the RBD-ferritin immunogens elicited substantial S-binding responses that were highly comparable to those of other immunogens that contained additional S domains (Figure 5B). When comparing RFN-elicited immune responses to the control protein RBD, the differences were striking. Using similar mouse strains and adjuvants and performing the experiments contemporaneously, it took three immunizations to elicit detectable neutralizing antibody titers using ALFQ as an adjuvant. In the Alhydrogel-adjuvanted study groups, we did not detect neutralizing titers even after the third immunization (Table S4).
In a pattern similar to that seen for the RFN immunogens, both the RBD-NTD-ferritin and the S1-ferritin immunogens elicited binding responses and detectable pseudovirus neutralization after a single immunization that was increased by the second immunization to give ID50 GMT values >10,000 and ID80 GMT titers ∼5,000 (Figures 5A–5C and S4).
Given the rapid induction of immune responses after a single immunization by SpFN_1B-06-PL, we further characterized this immunogen in a dose-ranging study (Figures 5D, 5E, and S5). In five-fold dilution steps, we reduced the SpFN_1B-06-PL immunogen from a 10-μg dose down to a 0.0032-μg dose (3,125-fold reduction) with the full ALFQ adjuvant dose. Antibody-binding responses were assessed for the full dose range by ELISA to S and RBD, and binding responses were elicited at all dose concentrations tested. We then further assessed the 0.08-μg dose (125-fold reduction from the 10-μg dose) with all our immunogenicity assays (Figures 5E and S5G). At this lower dose, the immune response was comparable to that seen for the typical 10-μg dose. In addition, we assessed both the 10- and 0.08-μg SpFN_1B-06-PL-vaccinated mouse serum for authentic SARS-CoV-2 live virus neutralization. At both doses, in both mouse strains, a single immunization elicited an ID50 GMT of ∼1,000, whereas the second immunization boosted this response more than ten-fold. The subsequent third immunization showed a modest boost effect. At each of these study time points, there was no difference between the doses in C57BL/6 mice, whereas differences in BALB/c ID80 GMT were seen at week 2 (higher for the 10-μg dose) and week 8 (higher for the 0.08-μg dose).
Further analysis of the durability of the immune responses (Figure S6) showed that the immune responses in terms of binding titers and pseudovirus neutralization titers were stably maintained for more than 10 weeks after the final immunization at ID50 GMT levels of ~10,000. This was observed for both SpFN_1B-06-PL- and RFN_131-immunized mice. Overall, these studies demonstrated robust immunogenicity of four categories of SARS-CoV-2 S-domain ferritin nanoparticles.
Vaccine-elicited broadly cross-reactive antibody responses against SARS-CoV-1 and SARS-CoV-2 VoCs
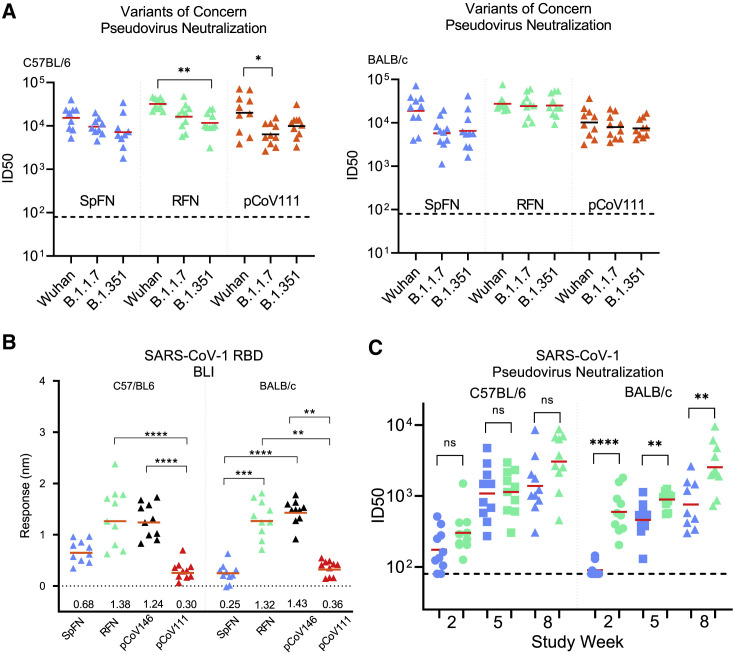
SARS-CoV-2 variants that are more transmissible and may be more lethal continue to emerge even in the midst of vaccine rollout and public health measures. Given the robust binding and pseudovirus and authentic virus neutralization titers against the original SARS-CoV-2 that were elicited by the S-domain ferritin nanoparticle immunogens, we assessed the immunized mouse sera for binding and pseudovirus neutralization to VoCs (Figure 6 ). Using study week 10 sera from mice immunized with the four categories of immunogens, SpFN_1B-06-PL, RFN_131, RBD-NTD-ferritin (pCoV146), and S1-ferritin (pCoV111), with ALFQ as the adjuvant, we assessed binding to a panel of variant RBD molecules containing K417N, E484K, N501Y, and combinations of these mutations (Figure S6). These mutations match the RBD sequence seen in the B.1.351, B.1.1.7, and P.1 SARS-CoV-2 strains. In all cases, robust binding to the RBD molecules was observed, with minimal change in overall binding when compared to the original RBD molecule. RFN_131-immunized mouse sera showed reduced binding to the E484K, N501Y double mutant but increased binding to the K417N variant. Analysis of the sera from SpFN_1B-06-PL-, RFN_131-, or pCoV111-immunized mice for pseudovirus neutralization of the VoCs B.1.1.7 (Alpha) and B.1.351 (Beta) showed minimal changes in the neutralization levels, with ID50 GMT values >2,000 for all strains (Figure 6A).
Figure 6.
SARS-CoV-2 S-domain nanoparticle vaccine candidates elicit robust antibody binding responses and neutralizing activity against SARS-CoV-2 VoCs and SARS-CoV-1
(A) Pseudovirus neutralization (ID50 values) of study week 10 mouse sera from immunized C57BL/6 and BALB/c mice to SARS-CoV-2 Wuhan-1, B.1.1.7, and B.1.351 pseudotyped viruses. Immunogens are indicated at the base of each graph. Geometric mean titer values are indicated by a horizontal line; n = 5; statistical significance for each immunogen was assessed using a Kruskal-Wallis test followed by a Dunn’s post-test.
(B) Biolayer interferometry binding of study week 10 mouse sera from immunized C57BL/6 and BALB/c mice to SARS-CoV-1 RBD. Immunogens are indicated at the base of each graph. Mean values are indicated by a horizontal line; n = 10; statistical significance was assessed using a Kruskal-Wallis test followed by a Dunn’s post-test.
(C) Pseudovirus neutralization (ID50 values) of study week 10 mouse sera from immunized C57BL/6 and BALB/c mice to SARS-CoV-1 Urbani strain pseudotyped viruses. Data related to SpFN_1B-06-PL and RFN_131 are colored blue and green, respectively. Statistical comparisons between SpFN and RFN responses at each time point were carried out using a Mann-Whitney unpaired two-tailed non-parametric test.
Immunogens are indicated at the base of each graph. Geometric mean values are indicated by a horizontal line; n = 10; ∗∗∗∗p < 0.0001, ∗∗p < 0.01, ∗p < 0.05. See also Figures S4 and S5.
Analysis of the mouse sera for binding of SARS-CoV-1 showed that RFN_131-immunized mice elicited the highest SARS-CoV-1 RBD-binding response (Figures 6B and S6). In addition to RBD binding, we also observed SARS-CoV-1 ACE2-RBD inhibitory activity with SpFN-immunized mice (Figure S5). We further assessed SpFN_1B-06-PL- or RFN_131-immunized mouse sera for neutralization against SARS-CoV-1 using the pseudovirus assay (Figure 6C). We observed robust neutralization levels with ID50 >1,000 for SpFN_1B06-PL- or RFN_131-immunized animals. In general, the RFN_131 immunogen elicited higher SARS-CoV-1 neutralizing responses compared to SpFN. Together, these data demonstrate that the SpFN_1B-06-PL and RFN_131 nanoparticles elicit broadly neutralizing and cross-reactive antibody responses against VoCs and the heterologous SARS-CoV-1. The consistency of the neutralization titers across VoCs generated by the ferritin nanoparticle immunogens is striking (Figure S6), particularly when compared to other vaccine candidates, where significantly decreased neutralization titers are seen against Beta and other variants.
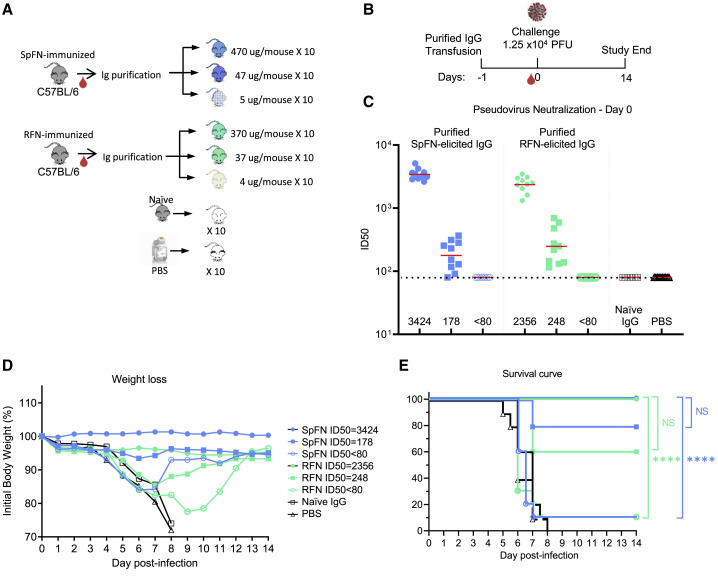
Protective immunity in K18-hACE2 transgenic mice against SARS-CoV-2 challenge
Given the neutralizing antibody responses elicited by SpFN_1B-06-PL and RFN_131, and the different components of these two immunogens, we chose to assess the protective effect of purified antibodies from these animals in a lethal SARS-CoV-2 challenge model using K18-hACE2 transgenic mice (Winkler et al., 2020). We chose to carry out passive antibody transfer to fully assess the ability of the humoral response to protect against infection, and we transferred three different amounts of purified IgG from either SpFN_1B-06-PL-ALFQ- or RFN_131-ALFQ-immunized mice to further understand whether there were significant differences between the ability of RBD-focused antibodies to provide protection compared to spike-focused antibodies. Purified IgG was transferred to the K18-hACE2 transgenic mice 24 h prior to inoculation with SARS-CoV-2 (Figures 7A and 7B). The control groups included a PBS group and a group that was passively transferred with naive mouse IgG. The dose of SARS-CoV-2 virus was titrated (1.25 × 104 PFU; plaque-forming unit) to establish significant weight loss and pathology following infection with the WA1/2020 strain (Figure S7).
Figure 7.
SpFN- and RFN-protective immunity in K18-hACE2 transgenic mice
(A) IgG was purified from SpFN- or RFN-vaccinated mouse sera and passively transferred at specific IgG amounts ranging from 4 to 470 μg/mouse in a final volume of 200 μL. Control naive mouse IgG was formulated at 2 mg/mL (n = 10/group, 5 female, 5 male).
(B) Mouse challenge study schematic. K18-hACE2 mice (n = 10/group, 5 female, 5 male) received control IgG (black), PBS (gray), and purified IgG, 1 day prior to challenge with 1.25 × 104 PFU of SARS-CoV-2.
(C) SARS-CoV-2 pseudovirus neutralization ID50 titers of mouse sera at study day 0. Pseudovirus IC50 geometric mean titer for each group is shown on the x axis. The horizontal dotted line indicates the lower limit of detection of the assay.
(D and E) Percentage of initial weight (D) and survival (E) of K18-hACE2 mice for the 8 study groups.
Groups are indicated based on original animal vaccination group and the pseudovirus ID50 neutralization values. The legend is shown in (D). Statistical comparisons were carried out using a Mantel-Cox test followed by Bonferroni correction. See also Figure S7.
Animal weight was measured twice daily for 14 days after challenge, and animals that lost >25% weight during the study were euthanized. All animals that received the highest amount of antibody (470 μg SpFN_1B-06-PL-derived, or 370 μg RFN_131-derived) showed serum neutralization ID50 GMT titers of 3,424 and 2,356, respectively, at day 0 immediately prior to virus inoculation (Figure 7C). All animals in these two groups showed minimal weight loss (Figure 7D) and all survived the study (Figure 7E). In the two groups that received either 47 μg SpFN_1B-06-PL- or 37 μg RFN_131-derived antibody, neutralization ID50 GMT titers were 178 and 248, respectively. Even with these lower antibody transfer amounts and the relatively low neutralization titers, most animals were protected from weight loss and death. In the SpFN_1B-06-PL-47-μg group, only two animals showed severe weight loss, whereas in the RFN_131-37-μg group, most animals showed some weight loss during the first study week, but all recovered. In contrast, mice that received the lowest amount of purified IgG from SpFN_1B-06-PL- or RFN_131-vaccinated animals did not show any neutralizing antibody titers at the day of inoculation. The mice in these two groups showed significant weight loss, and 9/10 animals in each group were euthanized by day 9 of the study. In a similar pattern, all animals from the naive IgG and PBS groups suffered weight loss and were euthanized by study day 8. In summary, these data show that low amounts of passively transferred antibodies equivalent to 2 mg/mL from SpFN- or RFN-vaccinated animals can protect mice from a lethal challenge with SARS-CoV-2.
Discussion
Since the emergence of SARS-CoV-2 in late 2019, multiple vaccines have been developed that elicit robust and protective immune responses in small animals, non-human primates, and humans. This includes a set of mRNA-based vaccines (NIH-Moderna, Pfizer-BioNTech), viral vector vaccines (J&J, AstraZeneca), and a nanoparticle-like vaccine (Novavax) (Bangaru et al., 2020) that are starting to be distributed worldwide. In addition, next-generation SARS-CoV-2 vaccine candidates are beginning to reveal strong immunological results in pre-clinical studies (Saunders et al., 2021; Walls et al., 2020a). These protein-based nanoparticle platforms paired with powerful adjuvant systems provide multiple advantages in the ability to protect against emerging variants (Moyo-Gwete et al., 2021; Wibmer et al., 2021). Broadly protective nanoparticle vaccines that can elicit potent and durable responses may be especially important for specific high-risk professions, or in populations (Atyeo et al., 2021) including the elderly (Collier et al., 2021) or immunocompromised (Boyarsky et al., 2021), where immune responses can be attenuated. The utility of highly stable vaccines that can elicit high neutralizing antibody titers after a single immunization, or vaccines that can be easily re-purposed for specific populations or as boosting immunogens, is likely to help the long-term strategy for global COVID-19 vaccination.
Trimer-functionalized ferritin immunogens have been effective at eliciting neutralizing antibodies against class I fusion proteins, including influenza hemagglutinin (HA) (Kanekiyo et al., 2013; Kelly et al., 2020) and HIV envelope (He et al., 2016; Sliepen et al., 2015). For example, in the case of respiratory syncytial virus, a stabilized prefusion F glycoprotein vaccine based on subtype A can naturally elicit potent neutralizing antibody and T cell responses against the heterologous subtype B in animals and humans (Crank et al., 2019; Joyce et al., 2019). Here we developed a set of SARS-CoV-2 S-domain ferritin nanoparticle vaccines using structure-based vaccine design that re-creates the structural and antigenic profile of S. These immunogens elicit antibodies with potent S-binding activity, ACE2-blocking activity, and potent neutralization activity against the homologous virus. In all four immunogen design groups, we observe significant antibody responses targeting the RBD with ACE2-blocking activity, likely contributing to the neutralization responses. Additionally, dose-sparing immunization experiments show that significant antigen reduction can still elicit potent antibody responses, while simultaneously also showing robust levels of neutralizing antibodies against the heterologous SARS-CoV-1. This heterologous immune response is reminiscent of broad immune responses seen with ferritin-HA immunogens (Kanekiyo et al., 2013), and demonstrate how nanoparticle immunogens can enhance the quality of the humoral immune response. Vaccines that take advantage of a viral nanoparticle protein structure such as yellow fever 17D vaccine (Collins and Barrett, 2017) and human papillomavirus virus-like particle (VLP) vaccines (Lowy and Schiller, 2006) elicit robust and long-lived immune responses. For SARS-CoV-2 vaccine development, nanoparticles performed well in mouse (Keech et al., 2020; Walls et al., 2020a) and non-human primate studies (Brouwer et al., 2021), with a designed S-ferritin nanoparticle (Powell et al., 2021) or multi-component RBD-Spycatcher-ferritin (Wang et al., 2021b) or RBD-sortase-ferritin nanoparticles (Saunders et al., 2021) also resulting in robust immunogenicity. The need for multiple protein components within a vaccine candidate may complicate production in the short term but can be leveraged for increased breadth of immune responses if tailored appropriately.
The passive transfer of vaccine-elicited purified antibody prevented weight loss and death following SARS-CoV-2 challenge in K18-hACE2 mice at neutralizing antibody levels (ID50 < 1,000) that were routinely exceeded by ten-fold or more with SpFN_1B-06-PL or RFN_131 vaccination. To focus on understanding what levels of humoral immune response could be protective, without the additional impact of vaccine-elicited cellular responses, we selected this study design, which allowed titration of antibody amounts. We also transferred very low amounts of antibody to the K18-hACE2 mice to assess for antibody-dependent enhancement, and we saw no indication of more rapid weight loss or signs of enhanced disease. The fact that antibody amounts readily elicited after a single immunization are highly protective in this challenge model, and that low antibody levels do not enhance disease, highlights the promise of these vaccine candidates. A set of companion papers also maps out the cellular immune response and highlights the protective effect of SpFN and RFN in high-dose SARS-CoV-2 challenge studies (Joyce et al., 2021; King et al., 2021). Given the rapid induction of SARS-CoV-2 immune responses after a single immunization and the high levels of protection seen in the K18-hACE2 model, SpFN_1B-06-PL has been produced under cGMP conditions and is under assessment in a phase I clinical trial (NCT04784767).
The immune responses elicited by the ferritin nanoparticles with the adjuvant ALFQ were consistently superior to that seen with the aluminum-hydroxide-based Alhydrogel adjuvant. This result is consistent with numerous studies indicating that antigen-adjuvant assessment is critical for generating optimal immune responses (Arunachalam et al., 2021; Wørzner et al., 2021). The components of the ALFQ adjuvant including QS-21 are readily available and can be scaled up for future advanced clinical trials. The COVID-19 pandemic has set many precedents with regard to vaccine development speed and novel platforms, and should foster a new age of vaccine development utilizing advanced antigens and adjuvants to train the immune system for greater protection.
Here, we utilized structure-based design to create four categories of immunogens using the ferritin nanoparticle platform. Each of these different designs and the underlying development processes provide a greater understanding and framework for ongoing and future pan-CoV vaccine design and development. The design information outlined here can be readily transferred for emerging CoV pathogens or to pan-CoV vaccine efforts. These SARS-CoV-2 nanoparticle immunogens provided immunogenicity against VoCs and the heterologous SARS-CoV-1 with implications for vaccination efforts against putative zoonotic emergences.
Limitations of the study
One limitation of this study is the lack of high-resolution structural data to visualize the stabilizing and surface mutations on the immunogens, and how this matches the designs. Further structure-based designs focused on augmenting the SpFN molecule with additional mutations could further increase the immunogenicity. Another limitation is that this study did not assess the breadth and potency of the binding or neutralizing immune response against a broad panel of sarbecoviruses. In addition, protection studies in either rodent or non-human primate models using heterologous sarbecoviruses would provide further understanding of the ability of these immunogens to prevent CoV infection.
In addition, translation of the SpFN or RFN designs to other CoVs, or optimization of the immunogen sequence to maximize cross-sarbecovirus neutralization potency and breadth, was not assessed. The immune response elicited by these immunogens shows breadth, but the specific epitopes targeted by the antibodies have not been characterized at the molecular level, which would provide a further guide to maximize epitope targeting and enable pan-CoV vaccine design.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-SARS-CoV-2 S antibody, CR3022 | ter Meulen et al., 2006 | N/A |
| Anti-SARS-CoV-2 S antibody, SR1 | This manuscript | N/A |
| Anti-SARS-CoV-2 S antibody, SR2 | This manuscript | N/A |
| Anti-SARS-CoV-2 S antibody, SR3 | This manuscript | N/A |
| Anti-SARS-CoV-2 S antibody, SR4 | This manuscript | N/A |
| Anti-SARS-CoV-2 S antibody, SR5 | This manuscript | N/A |
| Horseradish peroxidase (HRP)-conjugated sheep anti-mouse IgG, gamma chain specific | The Binding Site | Cat# AP272 |
| HRP-conjugated AffiniPure Goat Anti-Mouse Fcγ subclass 1 | Jackson ImmunoResearch | Cat# 115-035-205 |
| HRP-conjugated AffiniPure Goat Anti-Mouse Fcγ subclass 2a | Jackson ImmunoResearch | Cat# 115-035-206 |
| HRP-conjugated AffiniPure Goat Anti-Mouse Fcγ subclass 2c | Jackson ImmunoResearch | Cat# 115-035-208 |
| anti-RBD mouse mAb, 240C | BEI Resources | NR-616 |
| Bacterial and virus strains | ||
| Stbl3 competent cells | ThermoFisher Scientific | Cat# C737303 |
| Top10 competent cells | ThermoFisher Scientific | Cat# C404010 |
| SARS-CoV-2/human/USA/WA-CDC-WA1/2020 | CDC | GenBank: MN985325.1 |
| Chemicals, peptides, and recombinant proteins | ||
| 3,5,3′5′-tetramethylbenzidine (TMB) | KPL | Cat# 5150-0021 |
| hACE2 protein | This manuscript | N/A |
| SARS-CoV-1 RBD protein | This manuscript | N/A |
| SARS-CoV-2 S-2P protein | PMID: 32075877 | N/A |
| SARS-CoV-2 S-RBD protein | PMID: 32511298 | N/A |
| SARS-CoV-2 B.1.1.7 RBD protein | This manuscript | N/A |
| SARS-CoV-2 B.1.1.351 RBD protein | This manuscript | N/A |
| ALFQ | Matyas Lab | N/A |
| Imidazole | ThermoFisher Scientific | Cat# AC122020050 |
| PBS | ThermoFisher: Scientific | Cat# 10010023 |
| Bovine serum albumin | Sigma-Aldrich | Cat# A8327 |
| Anti-Human IgG Fc Capture (AHC) Biosensors | FortéBio | Cat# 18-5063 |
| HIS1K sensors | ForteBio | Cat# 18-5120 |
| FreeStyle 293 Expression Medium | GIBCO | Cat# 12338002 |
| OPTI-MEM, Reduced Serum Medium | ThermoFisher: Scientific | Cat# 11058021 |
| Methyl-α -D mannopyranoside | Sigma Aldrich | Cat# 617-04-9 |
| Alhydrogel | Invivogen | Cat# vac-alu-250 |
| DPBS | ThermoFisher Scientific | Cat# 14-190-235 |
| Glycerol | ThermoFisher Scientific | Cat# BP229-1 |
| pCoV1B-01: S2P(1-1137)-del-4-Ferritin protein | This manuscript | N/A |
| pCoV1B-02: S2P(1-1137)-del-6-Ferritin protein | This manuscript | N/A |
| pCoV1B-03: S2P(1-1208)-del-Ferritin protein | This manuscript | N/A |
| pCoV1B-04: S2P(1-1208)-GCN4-Ferritin protein | This manuscript | N/A |
| pCoV1B-05: S2P(1-1154)-del-Ferritin protein | This manuscript | N/A |
| pCoV1B-06: S2P(1-1158)op1-del-Ferritin protein | This manuscript | N/A |
| pCoV1B-07: S2P(1-1158)op2-del-Ferritin protein | This manuscript | N/A |
| pCoV1B-08: S2P(1-1158)op1x2-del-Ferritin protein | This manuscript | N/A |
| pCoV1B-09: S2P(1-1158)op2x2-del-Ferritin protein | This manuscript | N/A |
| pCoV1B-10: S2P(1-1158)op1-fGCN4-del-Ferritin protein | This manuscript | N/A |
| pCoV1B-01-PL: PL-S2P(12-1137)-del-4-Ferritin protein | This manuscript | N/A |
| pCoV1B-02-PL: PL-S2P(12-1137)-del-6-Ferritin protein | This manuscript | N/A |
| pCoV1B-03-PL: PL-S2P(12-1208)-del-Ferritin protein | This manuscript | N/A |
| pCoV1B-04-PL: PL-S2P(12-1208)-GCN4-Ferritin protein | This manuscript | N/A |
| pCoV1B-05-PL: PL-S2P(12-1154)-del-Ferritin protein | This manuscript | N/A |
| pCoV-1B-06-PL (aka SpFN): PL-S2P(12-1158)op1-del-Ferritin protein | This manuscript | N/A |
| pCoV1B-07-PL: PL-S2P(12-1158)op2-del-Ferritin protein | This manuscript | N/A |
| pCoV1B-08-PL: PL-S2P(12-1158)op1x2-del-Ferritin protein | This manuscript | N/A |
| pCoV1B-09-PL: PL-S2P(12-1158)op2x2-del-Ferritin protein | This manuscript | N/A |
| pCoV1B-10-PL: PL-S2P(12-1158)op1-fGCN4-del-Ferritin protein | This manuscript | N/A |
| pCoV03: His8-3C-RBD(331-527)-Ferritin protein | This manuscript | N/A |
| pCoV29: His8-3C-RBD-3-Ferritin protein | This manuscript | N/A |
| pCoV30: His8-3C-RBD-3-del-Ferritin protein | This manuscript | N/A |
| pCoV31: His8-3C-RBD-6-del-Ferritin protein | This manuscript | N/A |
| pCoV1A-01: His8-3C-RBD-PPII-Ferritin protein | This manuscript | N/A |
| pCoV1A-02: His8-3C-RBD-alpha1-Ferritin protein | This manuscript | N/A |
| pCoV1A-03: His8-3C-RBD-alpha2-Ferritin protein | This manuscript | N/A |
| pCoV1A-04: His8-3C-RBD-GCN4-del-Ferritin protein | This manuscript | N/A |
| pCoV1A-05: His8-3C-RBD-1141_1158op1-del-Ferritin protein | This manuscript | N/A |
| pCoV1A-06: His8-3C-RBD-1141_1158op1x2-del-Ferritin protein | This manuscript | N/A |
| pCoV49: His8-3C-RBD-F456N/K458T-Ferritin protein | This manuscript | N/A |
| pCoV50: His8-3C-RBD-L455R/Y449K/F490R-Ferritin protein | This manuscript | N/A |
| pCoV51: His8-3C-RBD-L455R-Ferritin protein | This manuscript | N/A |
| pCoV52: His8-3C-RBD-I468R-Ferritin protein | This manuscript | N/A |
| pCoV53: His8-3C-RBD-Y453R-Ferritin protein | This manuscript | N/A |
| pCoV54: His8-3C-RBD-L452R-Ferritin protein | This manuscript | N/A |
| pCoV55: His8-3C-RBD-L492R-Ferritin protein | This manuscript | N/A |
| pCoV56: His8-3C-RBD-F490R-Ferritin protein | This manuscript | N/A |
| pCoV57: His8-3C-RBD-F490A-Ferritin protein | This manuscript | N/A |
| pCoV58: His8-3C-RBD-L517N/L518K/H519S-Ferritin protein | This manuscript | N/A |
| pCoV59: His8-3C-RBD-L518R-Ferritin protein | This manuscript | N/A |
| pCoV60: His8-3C-RBD-V367T/L335N-Ferritin protein | This manuscript | N/A |
| pCoV61: His8-3C-RBD-T385N/L387T-Ferritin protein | This manuscript | N/A |
| pCoV62: His8-3C-RBD-V382R-Ferritin protein | This manuscript | N/A |
| pCoV63: His8-3C-RBD-F377R-Ferritin protein | This manuscript | N/A |
| pCoV127: His8-3C-RBD-F490A/L517N/L518K/H519S-Ferritin protein | This manuscript | N/A |
| pCoV128: His8-3C-RBD-F490A/L518R-Ferritin protein | This manuscript | N/A |
| pCoV129: His8-3C-RBD-L455R/Y449K/F490R/L518N/L519K/H520S-Ferritin protein | This manuscript | N/A |
| pCoV130: His8-3C-RBD-L455R/Y449K/F490R/L518R-Ferritin protein | This manuscript | N/A |
| pCoV131 (aka RFN): His8-3C-RBD-Y453R/L517N/L518K/H519S-Ferritin protein | This manuscript | N/A |
| pCoV132: His8-3C-RBD-Y453R/L518R-Ferritin protein | This manuscript | N/A |
| pCoV122: His8-3C-RBD(331-527)-GSGGSG-NTD(12-303)-Ferritin protein | This manuscript | N/A |
| pCoV123: His8-3C-RBD-F490R-NTD-Ferritin protein | This manuscript | N/A |
| pCoV124: His8-3C-RBD-F490A-NTD-Ferritin protein | This manuscript | N/A |
| pCoV125: His8-3C-RBD-L517N/L518K/H519S-NTD-Ferritin protein | This manuscript | N/A |
| pCoV126: His8-3C-RBD-L518R-NTD-Ferritin protein | This manuscript | N/A |
| pCoV146: His8-3C-RBD-Y453R-L517N/L518K/H519S-NTD-Ferrritin protein | This manuscript | N/A |
| pCoV147: His8-3C-RBD-F490A-L517N/L518K/H519S-NTD-Ferrritin protein | This manuscript | N/A |
| pCoV68: S1(12-678)-Ferritin protein | This manuscript | N/A |
| pCoV107: S1(12-655)-Ferritin protein | This manuscript | N/A |
| pCoV108: S1(12-655)-L611N/Q613T-Ferritin protein | This manuscript | N/A |
| pCoV109: S1(12-696)-Ferritin protein | This manuscript | N/A |
| pCoV110: S1(12-676)-G-S2(689-696)-Ferritin protein | This manuscript | N/A |
| pCoV111: S1(12-676)-GG-S2(689-696)-Ferritin protein | This manuscript | N/A |
| pCoV112: S1(12-676)-PG-S2(689-696)-Ferritin protein | This manuscript | N/A |
| pCoV113: S1-Y312N/Q313Y/T314T-Ferritin protein | This manuscript | N/A |
| pCoV114: S1-I651N/A653S-Ferritin protein | This manuscript | N/A |
| pCoV115: S1-S316C/V595C-Ferritin protein | This manuscript | N/A |
| pCoV116: S1-V320C/S591C-Ferritin protein | This manuscript | N/A |
| pCoV117: S1-L560Q/F562H-Ferritin protein | This manuscript | N/A |
| pCoV118: S1-F562N/Q564T-Ferritin protein | This manuscript | N/A |
| pCoV119: S1-F490R-Ferritin protein | This manuscript | N/A |
| pCoV120: S1-F490A-Ferritin protein | This manuscript | N/A |
| pCoV02: S1(16-678)-Ferritin protein | This manuscript | N/A |
| pCoV67: His8-3C-S1-Ferritin protein | This manuscript | N/A |
| Critical commercial assays | ||
| Bright-Glo Luciferase Assay System | Promega | Cat# E2610 |
| Deposited data | ||
| pCoV111, a Ferritin-based Nanoparticle Vaccine Candidate Displaying the SARS-CoV-2 Spike S1 Subunit | This manuscript | EMDB ID: EMD-25451 |
| Negative-stain EM reconstruction of SpFN_1B-06-PL, a SARS-CoV-2 spike fused to: H.pylori ferritin nanoparticle vaccine candidate | This manuscript | EMDB ID: EMD-25448 |
| RFN_131, a Ferritin-based Nanoparticle Vaccine Candidate Displaying the SARS-CoV-2: Receptor-Binding Domain | This manuscript | EMDB ID: EMD-25449 |
| pCoV146, a Ferritin-based Nanoparticle Vaccine Candidate Displaying the SARS-CoV-2: Spike Receptor-Binding and N-Terminal Domains | This manuscript | EMDB ID: EMD-25450 |
| Experimental models: Cell lines | ||
| Freestyle 293F cells | ThermoFisher Scientific | Cat# R7007 |
| Expi293F cells | ThermoFisher Scientific | Cat# A14635 |
| Vero E6 cells | PMID: 14569023 | N/A |
| Vero 81 cells | ATCC | Cat# CCL-81 |
| HEK293T/17 cells | ATCC | Cat# CRL-11268 |
| hACE2-expressing HEK293cells | Integral Molecular | N/A |
| Experimental models: Organisms/strains | ||
| Mus musculus, strain BALB/cJ | Jackson Laboratories | JAX stock #000651 |
| Mus musculus, strain C57BL/6J | Jackson Laboratories | JAX stock #000664 |
| Mus musculus, strain B6.Cg-Tg(K18-ACE2)2Prlmn/J | Jackson Laboratories | JAX stock #034860 |
| Recombinant DNA | ||
| pCMVR hACE2 | This manuscript | N/A |
| pCMVR SARS-CoV-1 RBD | This manuscript | N/A |
| pCMVR SARS-CoV-2 S-2P | PMID: 32075877 | N/A |
| pCMVR SARS-CoV-2 S-RBD | PMID: 32511298 | N/A |
| pCMVR SARS-CoV-2 B.1.1.7 RBD | This manuscript | N/A |
| pCMVR SARS-CoV-2 B.1.1.351 RBD | This manuscript | N/A |
| pCMVR pCoV1B-06: S2P(1-1158)op1-del-Ferritin | This manuscript | N/A |
| pCMVR pCoV1B-07: S2P(1-1158)op2-del-Ferritin | This manuscript | N/A |
| pCMVR pCoV1B-08: S2P(1-1158)op1x2-del-Ferritin | This manuscript | N/A |
| pCMVR pCoV1B-09: S2P(1-1158)op2x2-del-Ferritin | This manuscript | N/A |
| pCMVR pCoV1B-10: S2P(1-1158)op1-fGCN4-del-Ferritin | This manuscript | N/A |
| pCMVR pCoV1B-01-PL: PL-S2P(12-1137)-del-4-Ferritin | This manuscript | N/A |
| pCMVR pCoV1B-02-PL: PL-S2P(12-1137)-del-6-Ferritin | This manuscript | N/A |
| pCMVR pCoV1B-03-PL: PL-S2P(12-1208)-del-Ferritin | This manuscript | N/A |
| pCMVR pCoV1B-04-PL: PL-S2P(12-1208)-GCN4-Ferritin | This manuscript | N/A |
| pCMVR pCoV1B-05-PL: PL-S2P(12-1154)-del-Ferritin | This manuscript | N/A |
| pCMVR pCoV-1B-06-PL (aka SpFN): PL-S2P(12-1158)op1-del-Ferritin | This manuscript | N/A |
| pCMVR pCoV1B-07-PL: PL-S2P(12-1158)op2-del-Ferritin | This manuscript | N/A |
| pCMVR pCoV1B-08-PL: PL-S2P(12-1158)op1x2-del-Ferritin | This manuscript | N/A |
| pCMVR pCoV1B-09-PL: PL-S2P(12-1158)op2x2-del-Ferritin | This manuscript | N/A |
| pCMVR pCoV1B-10-PL: PL-S2P(12-1158)op1-fGCN4-del-Ferritin | This manuscript | N/A |
| pCMVR pCoV03: His8-3C-RBD(331-527)-Ferritin | This manuscript | N/A |
| pCMVR pCoV29: His8-3C-RBD-3-Ferritin | This manuscript | N/A |
| pCMVR pCoV30: His8-3C-RBD-3-del-Ferritin | This manuscript | N/A |
| pCMVR pCoV31: His8-3C-RBD-6-del-Ferritin | This manuscript | N/A |
| pCMVR pCoV1A-01: His8-3C-RBD-PPII-Ferritin | This manuscript | N/A |
| pCMVR pCoV1A-02: His8-3C-RBD-alpha1-Ferritin | This manuscript | N/A |
| pCMVR pCoV1A-03: His8-3C-RBD-alpha2-Ferritin | This manuscript | N/A |
| pCMVR pCoV1A-04: His8-3C-RBD-GCN4-del-Ferritin | This manuscript | N/A |
| pCMVR pCoV1A-05: His8-3C-RBD-1141_1158op1-del-Ferritin | This manuscript | N/A |
| pCMVR pCoV1A-06: His8-3C-RBD-1141_1158op1x2-del-Ferritin | This manuscript | N/A |
| pCMVR pCoV49: His8-3C-RBD-F456N/K458T-Ferritin | This manuscript | N/A |
| pCMVR pCoV50: His8-3C-RBD-L455R/Y449K/F490R-Ferritin | This manuscript | N/A |
| pCMVR pCoV51: His8-3C-RBD-L455R-Ferritin | This manuscript | N/A |
| pCMVR pCoV52: His8-3C-RBD-I468R-Ferritin | This manuscript | N/A |
| pCMVR pCoV53: His8-3C-RBD-Y453R-Ferritin | This manuscript | N/A |
| pCMVR pCoV54: His8-3C-RBD-L452R-Ferritin | This manuscript | N/A |
| pCMVR pCoV55: His8-3C-RBD-L492R-Ferritin | This manuscript | N/A |
| pCMVR pCoV56: His8-3C-RBD-F490R-Ferritin | This manuscript | N/A |
| pCMVR pCoV57: His8-3C-RBD-F490A-Ferritin | This manuscript | N/A |
| pCMVR pCoV58: His8-3C-RBD-L517N/L518K/H519S-Ferritin | This manuscript | N/A |
| pCMVR pCoV59: His8-3C-RBD-L518R-Ferritin | This manuscript | N/A |
| pCMVR pCoV60: His8-3C-RBD-V367T/L335N-Ferritin | This manuscript | N/A |
| pCMVR pCoV61: His8-3C-RBD-T385N/L387T-Ferritin | This manuscript | N/A |
| pCMVR pCoV62: His8-3C-RBD-V382R-Ferritin | This manuscript | N/A |
| pCMVR pCoV63: His8-3C-RBD-F377R-Ferritin | This manuscript | N/A |
| pCMVR pCoV127: His8-3C-RBD-F490A/L517N/L518K/H519S-Ferritin | This manuscript | N/A |
| pCMVR pCoV128: His8-3C-RBD-F490A/L518R-Ferritin | This manuscript | N/A |
| pCMVR pCoV129: His8-3C-RBD-L455R/Y449K/F490R/L518N/L519K/H520S-Ferritin | This manuscript | N/A |
| pCMVR pCoV130: His8-3C-RBD-L455R/Y449K/F490R/L518R-Ferritin | This manuscript | N/A |
| pCMVR pCoV131 (aka RFN): His8-3C-RBD-Y453R/L517N/L518K/H519S-Ferritin | This manuscript | N/A |
| pCMVR pCoV132: His8-3C-RBD-Y453R/L518R-Ferritin | This manuscript | N/A |
| pCMVR pCoV122: His8-3C-RBD(331-527)-GSGGSG-NTD(12-303)-Ferritin | This manuscript | N/A |
| pCMVR pCoV123: His8-3C-RBD-F490R-NTD-Ferritin | This manuscript | N/A |
| pCMVR pCoV124: His8-3C-RBD-F490A-NTD-Ferritin | This manuscript | N/A |
| pCMVR pCoV125: His8-3C-RBD-L517N/L518K/H519S-NTD-Ferritin | This manuscript | N/A |
| pCMVR pCoV126: His8-3C-RBD-L518R-NTD-Ferritin | This manuscript | N/A |
| pCMVR pCoV146: His8-3C-RBD-Y453R-L517N/L518K/H519S-NTD-Ferrritin | This manuscript | N/A |
| pCMVR pCoV147: His8-3C-RBD-F490A-L517N/L518K/H519S-NTD-Ferrritin | This manuscript | N/A |
| pCMVR pCoV68: S1(12-678)-Ferritin | This manuscript | N/A |
| pCMVR pCoV107: S1(12-655)-Ferritin | This manuscript | N/A |
| pCMVR pCoV108: S1(12-655)-L611N/Q613T-Ferritin | This manuscript | N/A |
| pCMVR pCoV109: S1(12-696)-Ferritin | This manuscript | N/A |
| pCMVR pCoV110: S1(12-676)-G-S2(689-696)-Ferritin | This manuscript | N/A |
| pCMVR pCoV111: S1(12-676)-GG-S2(689-696)-Ferritin | This manuscript | N/A |
| pCMVR pCoV112: S1(12-676)-PG-S2(689-696)-Ferritin | This manuscript | N/A |
| pCMVR pCoV113: S1-Y312N/Q313Y/T314T-Ferritin | This manuscript | N/A |
| pCMVR pCoV114: S1-I651N/A653S-Ferritin | This manuscript | N/A |
| pCMVR pCoV115: S1-S316C/V595C-Ferritin | This manuscript | N/A |
| pCMVR pCoV116: S1-V320C/S591C-Ferritin | This manuscript | N/A |
| pCMVR pCoV117: S1-L560Q/F562H-Ferritin | This manuscript | N/A |
| pCMVR pCoV118: S1-F562N/Q564T-Ferritin | This manuscript | N/A |
| pCMVR pCoV119: S1-F490R-Ferritin | This manuscript | N/A |
| pCMVR pCoV120: S1-F490A-Ferritin | This manuscript | N/A |
| pCMVR pCoV02: S1(16-678)-Ferritin | This manuscript | N/A |
| pCMVR pCoV67: His8-3C-S1-Ferritin | This manuscript | N/A |
| Software and algorithms | ||
| Protein Repair One-Stop Shop (PROSS) server | PMID: 27425410 | https://pross.weizmann.ac.il/step/pross-terms/ |
| Octet Data Analysis software | FortéBio | v11.1 |
| GraphPad Prism | Motulsky and Brown, 2006 | V8.0 |
| CTFFIND | Rohou and Grigorieff, 2015 | https://grigoriefflab.umassmed.edu/ctf_estimation_ctffind_ctftilt |
| SerialEM | Mastronarde, 2005 | https://bio3d.colorado.edu/SerialEM/ |
| RELION | Scheres, 2012 | https://www2.mrc-lmb.cam.ac.uk/relion |
| PyMol | Schrödinger | V2.3.2 |
| COOT | Emsley and Cowtan, 2004 | http://bernhardcl.github.io/coot/ |
| SnapGene | Insightful Science | https://www.snapgene.com/ |
| UCSF Chimera | Pettersen et al., 2004 | https://www.cgl.ucsf.edu/chimera/ |
| ChimeraX | Goddard et al., 2018 | https://www.rbvi.ucsf.edu/chimerax/ |
| cisTEM | Grant et al., 2018 | https://cistem.org/ |
| cryoSPARC | Punjani et al., 2017 | v2.15.0 |
| Other | ||
| Strep-Tactin Superflow resin | IBA Lifesciences | Cat# 2-1206-010 |
| Pierce Protein A Agarose | ThermoFisher Scientific | Cat# 20334 |
| Superdex 200 increase 10/300 GL | Cytiva | Cat# 28990944 |
| Galanthus Nivalis Lectin (GNL), Agarose bound | Vector Labs | Cat# AL-1243-5 |
| NiNTA Resin | Thermo Fisher Scientific | Cat# 88221 |
| Carbon Film 200 Mesh, Copper | Electron Microscopy Sciences | Cat# CF200-CU |
Resource availability4
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, M. Gordon Joyce (gjoyce@eidresearch.org).
Materials availability
All reagents will be made available on request after completion of a Materials Transfer Agreement.
Experimental model and subject details
Cell lines
Expi293F cells (Thermo Fisher Scientific) were maintained in Expi293 expression medium (Thermo Fisher Scientific) in a 37°C shaker supplied with 8% CO2 and 80% humidity. Vero81, and Vero E6 cells were incubated in DMEM (Thermo Fisher Scientific) at 37°C and 5% CO2.
Mouse strains
Female BALB/c mice, C57BL/6, and hACE2 K18 Tg mice mice aged 6- to 8-weeks were obtained from Jackson Laboratory (Bar Harbor, ME). All mouse experiments were conducted in compliance with the Animal Welfare Act, and other federal statutes and regulations relating to animals and experiments involving animals and adhered to the principles stated in the Guide for the Care and Use of Laboratory Animals, NRC Publication, 1996 edition. The immunization protocol was approved by the Institutional Animal Care and Use Committee of WRAIR. BALB/c and C57BL/6 mice were housed in the animal facility of WRAIR and cared for in accordance with local, state, federal, and institutional policies in a National Institutes of Health American Association for Accreditation of Laboratory Animal Care-accredited facility.
All infection experiments were done in animal biosafety level 3 (BSL-3) facilities at the Trudeau Institute and approved by the Institutional Animal Care and Use Committee of the Trudeau Institute.
Method details
Immunogen modeling and design
Following release of the SARS-CoV-2 sequence on Jan 10th 2020, initial RBD-Ferritin and S1-Ferritin immunogens were designed (Table S1). Subsequent iterative immunogen design and optimization utilized atomic models of the SARS-CoV-2 RBD molecule (Joyce et al., 2020), or the SARS-CoV-2 S-2P trimer structure PDB: 6VXX, and PDB: 3BVE for the Helicobacter pylori Ferritin, and PDB: 4LQH for the bullfrog linker sequence. Pymol (Schrödinger) was used to generate the ferritin 24- subunit particle, and a map created in UCSF Chimera (Pettersen et al., 2004) was supplied to cisTEM (Grant et al., 2018) “align_symmetry” to align the ferritin particle to an octahedral symmetry convention. This was supplied to “phenix.map_symmetry” to generate a symmetry file and PDB file, for octahedral (for monomer-fusions) and D4 (for trimer-fusions) symmetry. S-domain ferritin nanoparticle fusions were modeled using Pymol and Coot (Emsley et al., 2010) and expanded using “phenix.apply_ncs” (Liebschner et al., 2019). Visual analysis and figure generation was conducted using ChimeraX and PyMOL.
RBD-Ferritin designs were generated by assessment of the hydrophobic surface of the SARS-CoV-2 RBD surface and determining surface accessible mutations that reduced the hydrophobic surface. S1-Ferritin designs were creating using the PDB: 6VXX and including a short region of the S2 domain, which interacts with S1. Spike-Ferritin designs were created by modeling the coiled coil region between S residues 1140 and 1158 and increasing the coil-coil interaction either by mutagenesis, or by increasing the length of the interaction region. RBD-NTD-Ferritin designs utilized RBD constructs with improved properties in the context of RBD-Ferritin, which were fused to the N terminus of NTD (12 - 303)-Ferritin by a short 6 amino-acid linker.
DNA plasmid construction and preparation
SARS-CoV-2 S-domain ferritin constructs were derived from the Wuhan-Hu-1 strain genome sequence (GenBank: MN9089473), to include the following domains: RBD subunit (residues 331 - 527), NTD subunit (residues 12 - 303), RBD subunit linked to NTD subunit (residues 331 - 527 linked to residues 12 - 303 with a short GSG linker), S1 domain (residues 12 - 696) and S ectodomain (residues 12 – 1158). Constructs were modified to incorporate a N-terminal hexa-histadine tag (his) for purification of the RBD-Ferritin and RBD-NTD-Ferritin constructs.
An S-2P construct was used as a template to generate the set of Spike ferritin nanoparticles. Subsequent designs involving small deletions, additions and point mutations were generated using a modified QuikChange site-directed mutagenesis protocol (Agilent). RBD-ferritin and S1-ferritin constructs were synthesized by GenScript. For some of the RBD-ferritin constructs, gene segments (gBlocks) were synthesized by Integrated DNA Technologies to encode various linker regions between RBD and ferritin. Gene segments were stitched together with RBD- and ferritin-encoding PCR products using overlap extension PCR and were re-subcloned into the CMVR vector. The His-tagged SARS-CoV-2 RBD molecule was generated by amplifying the RBD domain from the RBD-Ferritin plasmid while encoding the 3′ purification tag and subcloned into the CMVR vector. The NTD protein subunit was generated in a similar manner, by amplifying the NTD domain from the S1-Ferritin construct. For expression of S, RBD, and NTD proteins, the S protein domains were cloned into the CMVR expression plasmid using the NotI/BamHI restriction sites. Constructs including the N-terminal region of the S protein included the native leader sequence; for constructs not including this region we utilized a prolactin leader (PL) sequence (Boyington et al., 2016).
Plasmid DNA generated by subcloning (restriction digest and ligation) was amplified in and isolated from E. coli Top10 cells. The constructs resulting from site-directed mutagenesis were either amplified in and isolated from E. coli Stbl3 or Top10 cells. Large-scale DNA isolation was performed using either endo free Maxiprep, Megaprep or Gigaprep kits (QIAGEN).
Immunogen expression and purification
All expression vectors were transiently transfected into Expi293F cells (Thermo Fisher Scientific) using ExpiFectamine 293 transfection reagent (Thermo Fisher Scientific). Cells were grown in polycarbonate baffled shaker flasks at 34°C or 37°C and 8% CO2 at 120 rpm. Cells were harvested 5-6 days post-transfection via centrifugation at 3,500 x g for 30 minutes. Culture supernatants were filtered with a 0.22-μm filter and stored at 4°C prior to purification.
His-tagged proteins were purified using Ni-NTA affinity chromatography, while untagged proteins were purified with GNA lectin affinity chromatography. Briefly, 25 mL GNA-lectin resin (VectorLabs) was used to purify untagged protein from 1L of expression supernatant. GNA resin was equilibrated with 10 column volumes (CV) of phosphate buffered saline (PBS) (pH 7.4) followed by supernatant loading twice at 4°C. Unbound protein was removed by washing with 20 CV of PBS buffer. Bound protein was eluted with 250mM methyl-α -D mannopyranoside in PBS buffer (pH 7.4). 1 mL Ni-NTA resin (Thermo Scientific) was used to purify protein from 1L of expression supernatant. Ni-NTA resin was equilibrated with 5 CV of phosphate buffered saline (PBS) (pH 7.4) followed by supernatant loading 2x at 4°C. Unbound protein was removed by washing with 200 CV of PBS, followed by 50 CV 10mM imidazole in PBS. Bound protein was eluted with 220mM imidazole in PBS. For all proteins, purification purity was assessed by SDS-PAGE. RBD-Ferritin nanoparticle constructs had a propensity to form soluble or insoluble aggregates which affected the ability to concentrate the samples. Addition of 1mM EDTA and 5% glycerol to the NiNTA purified material, prior to SEC or other concentration steps, mitigated the aggregation issue, and increased the nanoparticle formation as judged by SEC, and confirmed by neg-EM. All proteins were further purified by size-exclusion chromatography using a 16/60 Superdex-200 purification column. Purification purity for all the proteins was assessed by SDS-PAGE. Removal of the His-tags for SARS-CoV-2 S-2P and RBD for use in ELISA were carried out using HRV-3C protease. Endotoxin levels for ferritin nanoparticle immunogens were assessed (Endosafe® nexgen-PTS, Charles River Laboratories) and 5% v/v glycerol was added prior to filter-sterilization with a 0.22-μm filter, flash-freezing in liquid nitrogen, and storage at −80°C. Ferritin nanoparticle formation was assessed by dynamic light scattering (DLS) by determining the hydrodynamic diameter at 25°C using a Malvern Zetasizer Nano S (Malvern, Worcestershire, UK) equipped with a 633-nm laser.
For the antibodies, plasmids encoding heavy and light chains of antibodies (CR3022, and SR1-SR5) were co-transfected into Expi293F cells (ThermoFisher) according to the manufacturer’s instructions for expression of antibodies. After 5 days, antibodies were purified from cleared culture supernatants with Protein A agarose (ThermoFisher) using standard procedures, buffer was exchanged to PBS by dialysis, and antibody concentration was quantified using calculated extinction coefficient and A280 measurements.
Negative-stain electron microscopy
Purified proteins were deposited at 0.02-0.08 mg/ml on carbon-coated copper grids and stained with 0.75% uranyl formate. Grids were imaged using a FEI T20 operating at 200 kV with an Eagle 4K CCD using SerialEM or using a Thermo Scientific Talos L120C operating at 120 kV with Thermo Scientific Ceta using EPU. All image processing steps were done using RELION 3.0.8, RELION 3.1.1, and/or cisTEM-1.0.0-beta. Particles were picked either manually or using templates generated from manually picked 2D class averages. CTF estimation was done with CTFFIND 4.1.13 and used for 2D classification. 3D reconstructions were generated using an initial reference generated from a corresponding synthetic atomic model with a low pass filter of 80-100 Å to remove distinguishable features or from a similar construct also low pass filtered to 80-100 Å. For all 3D reconstructions, O symmetry was enforced, and no explicit mask was used. Visual analysis and figure generation was conducted using UCSF Chimera and ChimeraX.
Dynamic light scattering
Spike-domain ferritin nanoparticle hydrodynamic diameter was determined at 25°C using a Malvern Zetasizer Nano S (Malvern, Worcestershire, UK) equipped with a 633-nm laser. Samples were assessed accounting for the viscosity of their respective buffers.
Octet biolayer interferometry binding and ACE2 inhibition assays
All biosensors were hydrated in PBS prior to use. All assay steps were performed at 30°C with agitation set at 1,000crpm in the Octet RED96 instrument (FortéBio). Biosensors were equilibrated in assay buffer (PBS) for 15 s before loading of IgG antibodies (30 μg/ml diluted in PBS). SARS-COV-2 antibodies were immobilized onto AHC biosensors (FortéBio) for 100 s, followed by a brief baseline in assay buffer for 15 s. Immobilized antibodies were then dipped in various antigens for 100-200 s followed by dissociation for 20-100 s.
Mouse sera binding to the SARS-CoV-2 RBD, VoC RBDs, or SARS-CoV-1 RBD were carried out as follows. HIS1K biosensors (FortéBio) were equilibrated in assay buffer for 15 s before loading of His-tagged RBD (30 μg/ml diluted in PBS) for 120 s. After briefly dipping in assay buffer (15 s in PBS), the biosensors were dipped in mouse sera (100-fold dilution) for 180 s followed by dissociation for 60 s.
SARS-CoV-2 and SARS-CoV-1 RBD hACE2 inhibition assays were carried out as follows. SARS-CoV-2 or SARS-CoV-1 RBD (30 μg/ml diluted in PBS) was immobilized on HIS1K biosensors (FortéBio) for 180 s followed by baseline equilibration for 30 s. Serum binding was allowed to occur for 180 s followed by baseline equilibration (30 s). ACE2 protein (30 μg/ml) was the allowed to bind for 120 s. Percent inhibition (PI) of RBD binding to ACE2 by serum was determined using the equation: PI = 100 − [(ACE2 binding in the presence of mouse serum)) ∕ (ACE2 binding in the absence of mouse serum)] × 100.
Mouse immunization
All research in this study involving animals was conducted in compliance with the Animal Welfare Act, and other federal statutes and regulations relating to animals and experiments involving animals and adhered to the principles stated in the Guide for the Care and Use of Laboratory Animals, NRC Publication, 1996 edition. The research protocol was approved by the Institutional Animal Care and Use Committee of WRAIR. BALB/c and C57BL/6 mice were obtained from Jackson Laboratories (Bar Harbor, ME). Mice were housed in the animal facility of WRAIR and cared for in accordance with local, state, federal, and institutional policies in a National Institutes of Health American Association for Accreditation of Laboratory Animal Care-accredited facility.
C57BL/6 or BALB/c mice (n = 10/group) were immunized intramuscularly with 10 μg of immunogen (unless stated) adjuvanted with either ALFQ or Alhydrogel® in alternating caudal thigh muscles three times, at 3-week intervals; blood was collected 2 weeks before the first immunization, the day of the first immunization, and 2 weeks following each immunization, and at week 10 (Table S1). For immunogen SpFN_1B-06-PL, mice were immunized with reduced doses of protein adjuvanted with ALFQ with immunization schedule, site of injections, and timing of bleeds as described. Mice were randomly assigned to experimental groups and were not pre-screened or selected based on size or other gross physical characteristics. Serum was stored at 4°C or −80°C until analysis. Antibody responses were analyzed by Octet Biolayer Interferometry, enzyme-linked immunosorbent assay (ELISA), pseudovirus neutralization assay, and live-virus neutralization assay.
Immunogen-adjuvant preparation
Purified research grade nanoparticle immunogens were formulated in PBS with 5% glycerol at 1 mg/ml and subsequently diluted with dPBS (Quality Biological) to provide 10 μg or lower amount per 50 μL dose upon mixing with adjuvant. ALFQ (1.5X) liposomes, containing 600 μg/mL 3D-PHAD and 300 μg ug/mL QS-21, were gently mixed by slow speed vortex prior to use. Antigen was added to the ALFQ, vortexed at a slow speed for 1 minute, mixed on a roller for 15 minutes, and stored at 4°C for 1 h prior to immunization. Nanoparticle immunogens were formulated with ALFQ to contain 20 μg 3D-PHAD and 10 μg QS21 per 50 μL dose. Alhydrogel® stock (10 mg/ml aluminum (GMP grade; Brenntag)) was diluted to 900 μg/mL (1.5X) with DPBS and gently mixed. Appropriate volume and concentration of antigen was added to the diluted Alhydrogel® before being vortexed at low speed for 1 min, mixed on a roller for 15 minutes, and stored at 4°C for at least 1 h prior to immunization. Nanoparticle immunogens were adsorbed to Alhydrogel® aluminum hydroxide at 30 μg aluminum per 50 μL dose.
Enzyme-linked immunosorbent assay (ELISA)
96-well Immulon “U” Bottom plates were coated with 1 μg/mL of RBD or S-2P antigen in Dulbecco’s PBS, pH 7.4. Plates were incubated at 4°C overnight and blocked with blocking buffer (PBS containing 0.5% milk and 0.1% Tween 20, pH 7.4), at room temperature (RT) for 2 h. Individual serum samples were serially diluted 2-fold in blocking buffer and added to triplicate wells and the plates were incubated for 1 hour (h) at RT. Horseradish peroxidase (HRP)-conjugated sheep anti-mouse IgG, gamma chain specific (The Binding Site) was added and incubated at RT for an hour, followed by the addition of 2,2′-Azinobis [3-ethylbenzothiazoline-6-sulfonic acid]-diammonium salt (ABTS) HRP substrate (KPL) for 1 h at RT. The reaction was stopped by the addition of 1% SDS per well and the absorbance (A) was measured at 405 nm (A405) using an ELISA reader Spectramax (Molecular Devices, San Jose, CA)) within 30 min of stopping the reaction. Antibody positive (anti-RBD mouse mAb; BEI resources) and negative controls were included on each plate. The results are expressed as end point titers, defined as the reciprocal dilution that gives an absorbance value that equals twice the background value (antigen-coated wells that did not contain the test sera, but had all other components added).
Mouse isotype ELISA were performed using a similar approach as above, but with the following differences. Only S-2P was used to coat the wells, the plates were blocked with PBS containing 0.2% bovine serum albumin (BSA), pH 7.4 for 30 minutes. The mouse serum samples were serially diluted in duplicates either 3- or 4-fold in PBS containing 0.2% BSA and 0.05% Tween 20, pH7.4. The secondary antibodies were HRP-conjugated AffiniPure Goat Anti-Mouse antibodies from Jackson ImmunoResearch specific for either Fcγ subclass 1, Fcγ subclass 2a, or Fcγ subclass 2c. The secondary antibodies were incubated for 30 minutes. TMB (3,3′,5,5′-Tetramethylbenzidine) substrate (Thermo) was added and the plates were incubated at RT for 5-10 minutes to allow color development. Stop solution (Thermo) was added and the A405 was measured using a VersaMax microplate reader (Molecular Devices). A titration curve of serum concentration versus A450 was created. The titration curves were interpolated to determine the dilution factor where A450 = 1.0 for each mouse sera sample and IgG subclass, and the resulting values were used to calculate the IgG2a/IgG1 ratio (for BALB/c mice) or IgG2c/IgG1 ratio (for C57BL/6 mice). In the animal groups immunized with SpFN_1B-06-PL with the adjuvant Alhydrogel®, many of the mouse IgG usage ratio could not be calculated due to insufficient signal for either IgG2a or IgG2c in the mouse sera, particularly in the week 2 samples.
SARS-CoV-2 and SARS-CoV-1 pseudovirus neutralization assay
The S expression plasmid sequences for SARS-CoV-2 (Wuhan1, B.1.1.7, and B.1.351) and SARS-CoV-1 were codon optimized and modified to remove an 18 amino acid endoplasmic reticulum retention signal in the cytoplasmic tail in the case of SARS-CoV-2, and a 28 amino acid deletion in the cytoplasmic tail in the case of SARS-CoV. This allowed increased S incorporation into pseudovirions (PSV) and thereby improve infectivity. Virions pseudotyped with the vesicular stomatitis virus (VSV) G protein were used as a non-specific control. SARS-CoV-2 pseudovirions (PSV) were produced by co-transfection of HEK293T/17 cells with a SARS-CoV-2 S plasmid (pcDNA3.4) and an HIV-1 NL4-3 luciferase reporter plasmid (The reagent was obtained through the NIH HIV Reagent Program, Division of AIDS, NIAID, NIH: Human Immunodeficiency Virus 1 (HIV-1) NL4-3 ΔEnv Vpr Luciferase Reporter Vector (pNL4-3.Luc.R-E-), ARP-3418, contributed by Dr. Nathaniel Landau and Aaron Diamond). The SARS-CoV-2 S expression plasmid sequence was derived from the Wuhan seafood market pneumonia virus isolate Wuhan-Hu-1, complete genome (GenBank: MN908947), and the SARS-CoV-1 expression plasmid was derived from the Urbani S sequence.
Infectivity and neutralization titers were determined using ACE2-expressing HEK293 target cells (Integral Molecular) in a semi-automated assay format using robotic liquid handling (Biomek NXp Beckman Coulter). Test sera were diluted 1:40 in growth medium and serially diluted, then 25 μL/well was added to a white 96-well plate. An equal volume of diluted SARS-CoV-2 PSV was added to each well and plates were incubated for 1 hour at 37°C. Target cells were added to each well (40,000 cells/ well) and plates were incubated for an additional 48 hours. RLUs were measured with the EnVision Multimode Plate Reader (Perkin Elmer, Waltham, MA) using the Bright-Glo Luciferase Assay System (Promega Corporation, Madison, WI). Neutralization dose–response curves were fitted by nonlinear regression with a five-parameter curve fit using the LabKey Server® (Piehler et al., 2011), and the final titers are reported as the reciprocal of the dilution of serum necessary to achieve 50% neutralization (ID50, 50% inhibitory dilution) and 80% neutralization (ID80, 80% inhibitory dilution). Assay equivalency for SARS-CoV-2 was established by participation in the SARS-CoV-2 Neutralizing Assay Concordance Survey (SNACS) run by the Virology Quality Assurance Program and External Quality Assurance Program Oversite Laboratory (EQAPOL) at the Duke Human Vaccine Institute, sponsored through programs supported by the National Institute of Allergy and Infectious Diseases, Division of AIDS.
SARS-CoV-2 authentic virus neutralization assay
The neutralization assay has been described in detail previously (Case et al., 2020). Briefly, SARS-CoV-2 strain 2019-nCoV/USA_WA1/2020 was obtained from the Centers for Disease Control and Prevention. Virus was passaged once in Vero CCL81 cells (ATCC) and titrated by focus-forming assay on Vero E6 cells. Mouse sera were serially diluted and incubated with 100 focus-forming units of SARS-CoV-2 for 1 h at 37°C. Serum-virus mixtures were then added to Vero E6 cells in 96-well plates and incubated for 1 h at 37°C. Cells were overlayed with 1% (w/v) methylcellulose in MEM. After 30 h, cells were fixed with 4% PFA in PBS for 20 minutes at room temperature then washed and stained overnight at 4°C with 1 μg/ml of CR3022 (ter Meulen et al., 2006; Tian et al., 2020) in PBS supplemented with 0.1% saponin and 0.1% bovine serum albumin. Cells were subsequently stained with HRP-conjugated goat anti-human IgG for 2 h at room temperature. SARS-CoV-2-infected cell foci were visualized with TrueBlue peroxidase substrate (KPL) and quantified using ImmunoSpot® microanalyzer (Cellular Technologies, Shaker Heights, OH). Neutralization curves were generated using Prism software (GraphPad Prism 8.0).
Protection experiments in K18-hACE2 transgenic mice
All research in this study involving animals was conducted in compliance with the Animal Welfare Act, and other federal statutes and regulations relating to animals and experiments involving animals and adhered to the principles stated in the Guide for the Care and Use of Laboratory Animals, NRC Publication, 1996 edition. The research protocol was approved by the Institutional Animal Care and Use Committee of the Trudeau Institute. K18-hACE2 transgenic mice were obtained from Jackson Laboratories (Bar Harbor, ME). Mice were housed in the animal facility of the Trudeau Institute and cared for in accordance with local, state, federal, and institutional policies in a National Institutes of Health American Association for Accreditation of Laboratory Animal Care-accredited facility.
To determine an appropriate challenge viral stock amount and establish the K18-hACE2 SARS-CoV-2 mouse challenge model, five viral doses were used to inoculate the K18-hACE2 mice. Each study group was composed of 10 hACE2 K18 Tg mice (5 males and 5 females). Mice were infected on study day 0 doses ranging from 5 × 102 to 1 × 105 PFU of SARS-CoV-2 USA-WA1/2020 administered via intranasal instillation. All mice were monitored for clinical symptoms and body weight twice daily, every 12 hours, from study day 0 to study day 14. Mice were euthanized if they displayed any signs of pain or distress as indicated by the failure to move after stimulation, or inappetence, or if mice had greater than 25% weight loss compared to their study day 0 body weight. Hematoxylin and eosin staining of lung sections following infection with 1.25 × 104 PFU compared to control uninfected mouse lung sections are shown in Figure S6.
For the passive immunization study, on day −1, K18-hACE2 mice were injected intravenously with purified IgG from C57BL/6 vaccinated mice, control IgG, or PBS. On study day 0, all mice were inoculated with 1.25x104 PFU of SARS-CoV-2 USA-WA1/2020 via intranasal instillation. All mice were monitored for clinical symptoms and body weight twice daily, every 12 hours, from study day 0 to study day 14. Mice were euthanized if they displayed any signs of pain or distress as indicated by the failure to move after stimulated or inappetence, or if mice have greater than 25% weight loss compared to their study day 0 body weight.
Quantification and statistical analysis
Figure legends detail all quantification and statistical analyses, inclusive of animal numbers (n), and statistical tests. Serum IgG endpoint titers and pseudovirus and authentic neutralizing antibody titers were transformed so that all values were on a log(10) scale prior to statistical analyses. For mouse sera ELISA binding level, SARS-CoV-2 pseudovirus neutralization ID50 and ID80 values, biolayer interferometry binding level comparisons were assessed with a Kruskal-Wallis test followed by a Dunn’s post-test. Authentic virus neutralization comparisons between the two dose groups at each time point were carried out using a Mann-Whitney unpaired two-tailed non-parametric test. SARS-CoV-1 pseudovirus neutralization titers elicited by SpFN and RFN immunogens were compared using a Mann-Whitney unpaired two-tailed non-parametric test. K18-hACE2 mouse challenge group comparisons were carried out using a Mantel-Cox test followed by Bonferroni correction. Serum IgG2a/c:IgG1 ratios were assessed using a Mann-Whitney unpaired two-tailed non-parametric test. P values are summarized within figures as: < 0.0001 (∗∗∗∗), < 0.001 (∗∗∗), < 0.01 (∗∗), or < 0.05 (∗). All statistical analyses were conducted using GraphPad Prism v8.0, for serum IgG endpoint titers, pseudovirus and authentic neutralization titers, biolayer interferometry binding responses, and body weight loss of mice in challenge assessments.
Acknowledgments
We thank Sandhya Vasan, Mihret Amare, Suzanne Mate, Paul Scott, and Sharon P. Daye for programmatic support and planning; Nathaniel Jackson for cell-culture maintenance; and Erin Kavusak and Jonah Heller for support with performance of the neutralization assays. Research was conducted in compliance with the Animal Welfare Act and other federal statutes and regulations relating to animals and experiments involving animals and adheres to principles stated in the Guide for the Care and Use of Laboratory Animals, NRC Publication, 1996 Edition. This work was funded by the U.S. Defense Health Agency, the U.S. Department of the Army, and a cooperative agreement between The Henry M. Jackson Foundation for the Advancement of Military Medicine, Inc. and the U.S. Department of Defense (W81XWH-18-2-0040). This study also was supported by grants from the NIH (R01 AI157155). J.B.C. is supported by a Helen Hay Whitney Foundation postdoctoral fellowship. The material has been reviewed by the Walter Reed Army Institute of Research. There is no objection to its presentation and/or publication. The views expressed are those of the authors and should not be construed to represent the positions of the U.S. Army or the Department of Defense.
Author contributions
M.G.J. and K.M. designed the study. M.G.J., P.V.T., and K.M. designed the immunogens. M.G.J., W.-H.C., M.C., R.S.S., A.H., P.V.T., R.E.C., W.C.C., C.E.P., E.B.M., E.J.M., A. Ahmed, C.S., J.B.C., Y.L., A. Anderson, J.K., T.O., L.R., A.G., C.W., J.M.C., L.M.-R., C.P.K., N.G., Z.V., D.M., Z.B., J.K., S. Shrivastava, O.J., V.D., S.M., U.T., C.B.K., S. Soman, C.K., M.Z., H.K., W.W., M.A.C., D.K.D., L.W.K., T.J.L., S.E.M., J.R.C., S.J.K., S.M., V.R.P., W.W.R., N.d.V., M.S.D., G.D.G., and M. Rao performed protein purification, biophysical assays, immunologic assays, and animal studies. Z.B., M. Rao, G.R.M., and A. Anderson designed and provided the adjuvants. S.R., P.M.M., and M.T.E. provided the SR1–SR5 antibodies. M.G.J., W.-H.C., R.S.S., A.H., P.V.T., R.E.C., C.S., A. Ahmed, L.W., Z.B., W.W., W.W.R., M. Rolland, N.d.V., M.S.D., G.D.G., M. Rao, N.L.M., and K.M. analyzed and interpreted the data. M.G.J. wrote the paper with assistance from all coauthors.
Declaration of interests
M.G.J. and K.M. are named as inventors on international patent application WO/2021/178971 A1 entitled “Vaccines against SARS-CoV-2 and other coronaviruses.” M.G.J. is named as an inventor on international patent application WO/2018/081318 and U.S. patent 10,960,070 entitled “Prefusion coronavirus spike proteins and their use.” Z.B. is named as an inventor on U.S. patent 10,434,167 entitled “Non-toxic adjuvant formulation comprising a monophosphoryl lipid A (MPLA)-containing liposome composition and a saponin.” Z.B. and G.R.M. are named as inventors on U.S. patent application 16/607,917 entitled “Compositions and methods for vaccine delivery.” M.S.D. is a consultant for Inbios, Vir Biotechnology, Fortress Biotech, and Carnival Corporation and on the scientific advisory boards of Moderna and Immunome. The Diamond laboratory has received funding support from sponsored research agreements from Moderna, Vir Biotechnology, Kaleido, and Emergent BioSolutions. S.R., P.M.M., and M.T.E. are employees of AstraZeneca and currently hold AstraZeneca stock or stock options. Z.B. is currently employed at Pfizer.
Published: December 21, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2021.110143.
Supplemental information
Data and code availability
-
•
All data supporting the findings of this study are found within the paper and its supplemental information. Negative-stain EM maps for pCOV111, SpFN_1B-06-PL, RFN_131, and pCoV146 have been deposited in the EM database under codes 25451, 25448, 25449, and 25450 respectively.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the Lead Contact author upon request.
References
- Adhikari U., Chabrelie A., Weir M., Boehnke K., McKenzie E., Ikner L., Wang M., Wang Q., Young K., Haas C.N., et al. A case study evaluating the risk of infection from Middle Eastern respiratory syndrome coronavirus (MERS-CoV) in a hospital setting through bioaerosols. Risk Anal. 2019;39:2608–2624. doi: 10.1111/risa.13389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adney D.R., Wang L., van Doremalen N., Shi W., Zhang Y., Kong W.P., Miller M.R., Bushmaker T., Scott D., de Wit E., et al. Efficacy of an adjuvanted Middle East respiratory syndrome coronavirus spike protein vaccine in dromedary camels and alpacas. Viruses. 2019;11:212. doi: 10.3390/v11030212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alanazi K.H., Abedi G.R., Midgley C.M., Alkhamis A., Alsaqer T., Almoaddi A., Algwizani A., Ghazal S.S., Assiri A.M., Jokhdar H., et al. Diabetes mellitus, hypertension, and death among 32 patients with MERS-CoV infection, Saudi Arabia. Emerg. Infect. Dis. 2020;26:166–168. doi: 10.3201/eid2601.190952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arunachalam P.S., Walls A.C., Golden N., Atyeo C., Fischinger S., Li C., Aye P., Navarro M.J., Lai L., Edara V.V., et al. Adjuvanting a subunit COVID-19 vaccine to induce protective immunity. Nature. 2021;594:253–258. doi: 10.1038/s41586-021-03530-2. [DOI] [PubMed] [Google Scholar]
- Atyeo C., Pullen K.M., Bordt E.A., Fischinger S., Burke J., Michell A., Slein M.D., Loos C., Shook L.L., Boatin A.A., et al. Compromised SARS-CoV-2-specific placental antibody transfer. Cell. 2021;184:628–642.e10. doi: 10.1016/j.cell.2020.12.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bangaru S., Ozorowski G., Turner H.L., Antanasijevic A., Huang D., Wang X., Torres J.L., Diedrich J.K., Tian J.H., Portnoff A.D., et al. Structural analysis of full-length SARS-CoV-2 spike protein from an advanced vaccine candidate. Science. 2020;370:1089–1094. doi: 10.1126/science.abe1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes C.O., West A.P., Jr., Huey-Tubman K.E., Hoffmann M.A.G., Sharaf N.G., Hoffman P.R., Koranda N., Gristick H.B., Gaebler C., Muecksch F., et al. Structures of human antibodies bound to SARS-CoV-2 spike reveal common epitopes and recurrent features of antibodies. Cell. 2020;182:828–842.e16. doi: 10.1016/j.cell.2020.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyarsky B.J., Werbel W.A., Avery R.K., Tobian A.A.R., Massie A.B., Segev D.L., Garonzik-Wang J.M. Immunogenicity of a single dose of SARS-CoV-2 messenger RNA vaccine in solid organ transplant recipients. JAMA. 2021;325:1784–1786. doi: 10.1001/jama.2021.4385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyington J.C., Joyce M.G., Sastry M., Stewart-Jones G.B., Chen M., Kong W.P., Ngwuta J.O., Thomas P.V., Tsybovsky Y., Yang Y., et al. Structure-based design of head-only fusion glycoprotein immunogens for respiratory syncytial virus. PLoS One. 2016;11:e0159709. doi: 10.1371/journal.pone.0159709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyoglu-Barnum S., Ellis D., Gillespie R.A., Hutchinson G.B., Park Y.-J., Moin S.M., Acton O., Ravichandran R., Murphy M., Pettie D., et al. Elicitation of broadly protective immunity to influenza by multivalent hemagglutinin nanoparticle vaccines. bioRxiv. 2020 doi: 10.1101/2020.05.30.125179. [DOI] [Google Scholar]
- Brouwer P.J.M., Caniels T.G., van der Straten K., Snitselaar J.L., Aldon Y., Bangaru S., Torres J.L., Okba N.M.A., Claireaux M., Kerster G., et al. Potent neutralizing antibodies from COVID-19 patients define multiple targets of vulnerability. Science. 2020;369:643–650. doi: 10.1126/science.abc5902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brouwer P.J.M., Brinkkemper M., Maisonnasse P., Dereuddre-Bosquet N., Grobben M., Claireaux M., de Gast M., Marlin R., Chesnais V., Diry S., et al. Two-component spike nanoparticle vaccine protects macaques from SARS-CoV-2 infection. Cell. 2021;184:1188–1200.e19. doi: 10.1016/j.cell.2021.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y., Su B., Guo X., Sun W., Deng Y., Bao L., Zhu Q., Zhang X., Zheng Y., Geng C., et al. Potent neutralizing antibodies against SARS-CoV-2 identified by high-throughput single-cell sequencing of convalescent patients’ B cells. Cell. 2020;182:73–84.e16. doi: 10.1016/j.cell.2020.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmen J.M., Shrivastava S., Lu Z., Anderson A., Morrison E.B., Sankhala R.S., Chen W.-H., Chang W.C., Bolton J.S., Matyas G.R., et al. A spike-ferritin nanoparticle vaccine induces robust innate immune activity and drives polyfunctional SARS-CoV-2-specific T cells. bioRxiv. 2021 doi: 10.1101/2021.04.28.441763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Case J.B., Rothlauf P.W., Chen R.E., Liu Z., Zhao H., Kim A.S., Bloyet L.M., Zeng Q., Tahan S., Droit L., et al. Neutralizing antibody and soluble ACE2 inhibition of a replication-competent VSV-SARS-CoV-2 and a clinical isolate of SARS-CoV-2. Cell Host Microbe. 2020;28:475–485.e5. doi: 10.1016/j.chom.2020.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cawlfield A., Genito C.J., Beck Z., Bergmann-Leitner E.S., Bitzer A.A., Soto K., Zou X., Hadiwidjojo S.H., Gerbasi R.V., Mullins A.B., et al. Safety, toxicity and immunogenicity of a malaria vaccine based on the circumsporozoite protein (FMP013) with the adjuvant army liposome formulation containing QS21 (ALFQ) Vaccine. 2019;37:3793–3803. doi: 10.1016/j.vaccine.2019.05.059. [DOI] [PubMed] [Google Scholar]
- Cerutti G., Guo Y., Zhou T., Gorman J., Lee M., Rapp M., Reddem E.R., Yu J., Bahna F., Bimela J., et al. Potent SARS-CoV-2 neutralizing antibodies directed against spike N-terminal domain target a single supersite. Cell Host Microbe. 2021;29:819–833.e7. doi: 10.1016/j.chom.2021.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Zhou T., Schmidt S.D., Duan H., Cheng C., Chuang G.Y., Gu Y., Louder M.K., Lin B.C., Shen C.H., et al. Vaccination induces maturation in a mouse model of diverse unmutated VRC01-class precursors to HIV-neutralizing antibodies with >50% breadth. Immunity. 2021;54:324–339.e8. doi: 10.1016/j.immuni.2020.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen A.A., Gnanapragasam P.N.P., Lee Y.E., Hoffman P.R., Ou S., Kakutani L.M., Keeffe J.R., Wu H.J., Howarth M., West A.P., et al. Mosaic nanoparticles elicit cross-reactive immune responses to zoonotic coronaviruses in mice. Science. 2021;371:735–741. doi: 10.1126/science.abf6840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collier D.A., Ferreira I.A.T.M., Kotagari P., Datir R.P., Lim E.Y., Touizer E., Meng B., Abdullahi A., Elmer A., Kingston N., et al. CITIID-NIHR BioResource COVID-19 Collaboration Age-related immune response heterogeneity to SARS-CoV-2 vaccine BNT162b2. Nature. 2021;596:417–422. doi: 10.1038/s41586-021-03739-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins N.D., Barrett A.D. Live attenuated yellow fever 17D vaccine: a legacy vaccine still controlling outbreaks in modern day. Curr. Infect. Dis. Rep. 2017;19:14. doi: 10.1007/s11908-017-0566-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crank M.C., Ruckwardt T.J., Chen M., Morabito K.M., Phung E., Costner P.J., Holman L.A., Hickman S.P., Berkowitz N.M., Gordon I.J., et al. VRC 317 Study Team A proof of concept for structure-based vaccine design targeting RSV in humans. Science. 2019;365:505–509. doi: 10.1126/science.aav9033. [DOI] [PubMed] [Google Scholar]
- Cui J., Li F., Shi Z.L. Origin and evolution of pathogenic coronaviruses. Nat. Rev. Microbiol. 2019;17:181–192. doi: 10.1038/s41579-018-0118-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darricarrère N., Qiu Y., Kanekiyo M., Creanga A., Gillespie R.A., Moin S.M., Saleh J., Sancho J., Chou T.H., Zhou Y., et al. Broad neutralization of H1 and H3 viruses by adjuvanted influenza HA stem vaccines in nonhuman primates. Sci. Transl. Med. 2021;13:eabe5449. doi: 10.1126/scitranslmed.abe5449. [DOI] [PubMed] [Google Scholar]
- Emsley P., Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Emsley P., Lohkamp B., Scott W.G., Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goddard T.D., Huang C.C., Meng E.C., Pettersen E.F., Couch G.S., Morris J.H., Ferrin T.E. UCSF ChimeraX: Meeting modern challenges in visualization and analysis. Protein Sci. 2018;27:14–25. doi: 10.1002/pro.3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouglas D., Christodoulou M., Plotkin S.A., Hatchett R. CEPI: driving progress toward epidemic preparedness and response. Epidemiol. Rev. 2019;41:28–33. doi: 10.1093/epirev/mxz012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant T., Rohou A., Grigorieff N. cisTEM, user-friendly software for single-particle image processing. eLife. 2018;7:e35383. doi: 10.7554/eLife.35383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greaney A.J., Starr T.N., Gilchuk P., Zost S.J., Binshtein E., Loes A.N., Hilton S.K., Huddleston J., Eguia R., Crawford K.H.D., et al. Complete mapping of mutations to the SARS-CoV-2 spike receptor-binding domain that escape antibody recognition. Cell Host Microbe. 2021;29:44–57.e9. doi: 10.1016/j.chom.2020.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He L., de Val N., Morris C.D., Vora N., Thinnes T.C., Kong L., Azadnia P., Sok D., Zhou B., Burton D.R., et al. Presenting native-like trimeric HIV-1 antigens with self-assembling nanoparticles. Nat. Commun. 2016;7:12041. doi: 10.1038/ncomms12041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer A.S., Jones F.K., Nodoushani A., Kelly M., Becker M., Slater D., Mills R., Teng E., Kamruzzaman M., Garcia-Beltran W.F., et al. Dynamics and significance of the antibody response to SARS-CoV-2 infection. medRxiv. 2020 doi: 10.1101/2020.07.18.20155374. [DOI] [Google Scholar]
- Joyce M.G., Zhang B., Ou L., Chen M., Chuang G.Y., Druz A., Kong W.P., Lai Y.T., Rundlet E.J., Tsybovsky Y., et al. Iterative structure-based improvement of a fusion-glycoprotein vaccine against RSV. Nat. Struct. Mol. Biol. 2016;23:811–820. doi: 10.1038/nsmb.3267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joyce M.G., Bao A., Chen M., Georgiev I.S., Ou L., Bylund T., Druz A., Kong W.P., Peng D., Rundlet E.J., et al. Crystal structure and immunogenicity of the DS-Cav1-stabilized fusion glycoprotein from respiratory syncytial virus subtype B. Pathog. Immun. 2019;4:294–323. doi: 10.20411/pai.v4i2.338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joyce M.G., Sankhala R.S., Chen W.H., Choe M., Bai H., Hajduczki A., Yan L., Sterling S.L., Peterson C.E., Green E.C., et al. A cryptic site of vulnerability on the receptor binding domain of the SARS-CoV-2 spike glycoprotein. bioRxiv. 2020 doi: 10.1101/2020.03.15.992883. [DOI] [Google Scholar]
- Joyce M.G., King H.A.D., Elakhal Naouar I., Ahmed A., Peachman K.K., Cincotta C.M., Subra C., Chen R.E., Thomas P.V., Chen W.-H., et al. Efficacy of a broadly neutralizing SARS-CoV-2 ferritin nanoparticle vaccine in nonhuman primates. bioRxiv. 2021 doi: 10.1101/2021.03.24.436523. [DOI] [Google Scholar]
- Kaba S.A., Karch C.P., Seth L., Ferlez K.M.B., Storme C.K., Pesavento D.M., Laughlin P.Y., Bergmann-Leitner E.S., Burkhard P., Lanar D.E. Self-assembling protein nanoparticles with built-in flagellin domains increases protective efficacy of a Plasmodium falciparum based vaccine. Vaccine. 2018;36:906–914. doi: 10.1016/j.vaccine.2017.12.001. [DOI] [PubMed] [Google Scholar]
- Kanekiyo M., Wei C.J., Yassine H.M., McTamney P.M., Boyington J.C., Whittle J.R., Rao S.S., Kong W.P., Wang L., Nabel G.J. Self-assembling influenza nanoparticle vaccines elicit broadly neutralizing H1N1 antibodies. Nature. 2013;499:102–106. doi: 10.1038/nature12202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanekiyo M., Bu W., Joyce M.G., Meng G., Whittle J.R., Baxa U., Yamamoto T., Narpala S., Todd J.P., Rao S.S., et al. Rational design of an Epstein-Barr virus vaccine targeting the receptor-binding site. Cell. 2015;162:1090–1100. doi: 10.1016/j.cell.2015.07.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanekiyo M., Joyce M.G., Gillespie R.A., Gallagher J.R., Andrews S.F., Yassine H.M., Wheatley A.K., Fisher B.E., Ambrozak D.R., Creanga A., et al. Mosaic nanoparticle display of diverse influenza virus hemagglutinins elicits broad B cell responses. Nat. Immunol. 2019;20:362–372. doi: 10.1038/s41590-018-0305-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keech C., Albert G., Cho I., Robertson A., Reed P., Neal S., Plested J.S., Zhu M., Cloney-Clark S., Zhou H., et al. Phase 1-2 trial of a SARS-CoV-2 recombinant spike protein nanoparticle vaccine. N. Engl. J. Med. 2020;383:2320–2332. doi: 10.1056/NEJMoa2026920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly H.G., Tan H.X., Juno J.A., Esterbauer R., Ju Y., Jiang W., Wimmer V.C., Duckworth B.C., Groom J.R., Caruso F., et al. Self-assembling influenza nanoparticle vaccines drive extended germinal center activity and memory B cell maturation. JCI Insight. 2020;5:e136653. doi: 10.1172/jci.insight.136653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King H.A.D., Joyce M.G., Naouar I.E., Ahmed A., Cincotta C.M., Subra C., Peachman K.K., Hack H.H., Chen R.E., Thomas P.V., et al. Efficacy and breadth of adjuvanted SARS-CoV-2 receptor-binding domain nanoparticle vaccine in macaques. bioRxiv. 2021 doi: 10.1101/2021.04.09.439166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong R., Duan H., Sheng Z., Xu K., Acharya P., Chen X., Cheng C., Dingens A.S., Gorman J., Sastry M., et al. NISC Comparative Sequencing Program Antibody lineages with vaccine-induced antigen-binding hotspots develop broad HIV neutralization. Cell. 2019;178:567–584.e19. doi: 10.1016/j.cell.2019.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lal H., Cunningham A.L., Godeaux O., Chlibek R., Diez-Domingo J., Hwang S.J., Levin M.J., McElhaney J.E., Poder A., Puig-Barberà J., et al. ZOE-50 Study Group Efficacy of an adjuvanted herpes zoster subunit vaccine in older adults. N. Engl. J. Med. 2015;372:2087–2096. doi: 10.1056/NEJMoa1501184. [DOI] [PubMed] [Google Scholar]
- Langowski M.D., Khan F.A., Bitzer A.A., Genito C.J., Schrader A.J., Martin M.L., Soto K., Zou X., Hadiwidjojo S., Beck Z., et al. Optimization of a Plasmodium falciparum circumsporozoite protein repeat vaccine using the tobacco mosaic virus platform. Proc. Natl. Acad. Sci. USA. 2020;117:3114–3122. doi: 10.1073/pnas.1911792117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J., Kim D., Chang C.Y. Two new species of the genus Monstrillopsis Sars, 1921 (Copepoda: Monstrilloida: Monstrillidae) from South Korea. Zootaxa. 2016;4174:410–423. doi: 10.11646/zootaxa.4174.1.25. [DOI] [PubMed] [Google Scholar]
- Li D., Edwards R.J., Manne K., Martinez D.R., Schafer A., Alam S.M., Wiehe K., Lu X., Parks R., Sutherland L.L., et al. The functions of SARS-CoV-2 neutralizing and infection-enhancing antibodies in vitro and in mice and nonhuman primates. bioRxiv. 2021 doi: 10.1101/2020.12.31.424729. [DOI] [Google Scholar]
- Liebschner D., Afonine P.V., Baker M.L., Bunkóczi G., Chen V.B., Croll T.I., Hintze B., Hung L.W., Jain S., McCoy A.J., et al. Macromolecular structure determination using X-rays, neutrons and electrons: recent developments in Phenix. Acta Crystallogr. D Struct. Biol. 2019;75:861–877. doi: 10.1107/S2059798319011471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L., Wang P., Nair M.S., Yu J., Rapp M., Wang Q., Luo Y., Chan J.F., Sahi V., Figueroa A., et al. Potent neutralizing antibodies against multiple epitopes on SARS-CoV-2 spike. Nature. 2020;584:450–456. doi: 10.1038/s41586-020-2571-7. [DOI] [PubMed] [Google Scholar]
- Lowy D.R., Schiller J.T. Prophylactic human papillomavirus vaccines. J. Clin. Invest. 2006;116:1167–1173. doi: 10.1172/JCI28607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcandalli J., Fiala B., Ols S., Perotti M., de van der Schueren W., Snijder J., Hodge E., Benhaim M., Ravichandran R., Carter L., et al. Induction of potent neutralizing antibody responses by a designed protein nanoparticle vaccine for respiratory syncytial virus. Cell. 2019;176:1420–1431.e17. doi: 10.1016/j.cell.2019.01.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastronarde D.N. Automated electron microscope tomography using robust prediction of specimen movements. J. Struct. Biol. 2005;152:36–51. doi: 10.1016/j.jsb.2005.07.007. [DOI] [PubMed] [Google Scholar]
- McLellan J.S., Chen M., Joyce M.G., Sastry M., Stewart-Jones G.B.E., Yang Y., Zhang B., Chen L., Srivatsan S., Zheng A., et al. Structure-based design of a fusion glycoprotein vaccine for respiratory syncytial virus. Science. 2013;342:592–598. doi: 10.1126/science.1243283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menachery V.D., Yount B.L., Jr., Debbink K., Agnihothram S., Gralinski L.E., Plante J.A., Graham R.L., Scobey T., Ge X.Y., Donaldson E.F., et al. A SARS-like cluster of circulating bat coronaviruses shows potential for human emergence. Nat. Med. 2015;21:1508–1513. doi: 10.1038/nm.3985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motulsky H.J., Brown R.E. Detecting outliers when fitting data with nonlinear regression - a new method based on robust nonlinear regression and the false discovery rate. BMC Bioinformatics. 2006;7:123. doi: 10.1186/1471-2105-7-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moyo-Gwete T., Madzivhandila M., Makhado Z., Ayres F., Mhlanga D., Oosthuysen B., Lambson B.E., Kgagudi P., Tegally H., Iranzadeh A., et al. SARS-CoV-2 501Y.V2 (B.1.351) elicits cross-reactive neutralizing antibodies. bioRxiv. 2021 doi: 10.1101/2021.03.06.434193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Om K., Paquin-Proulx D., Montero M., Peachman K., Shen X., Wieczorek L., Beck Z., Weiner J.A., Kim D., Li Y., et al. Adjuvanted HIV-1 vaccine promotes antibody-dependent phagocytic responses and protects against heterologous SHIV challenge. PLoS Pathog. 2020;16:e1008764. doi: 10.1371/journal.ppat.1008764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pallesen J., Wang N., Corbett K.S., Wrapp D., Kirchdoerfer R.N., Turner H.L., Cottrell C.A., Becker M.M., Wang L., Shi W., et al. Immunogenicity and structures of a rationally designed prefusion MERS-CoV spike antigen. Proc. Natl. Acad. Sci. USA. 2017;114:E7348–E7357. doi: 10.1073/pnas.1707304114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pati R., Shevtsov M., Sonawane A. Nanoparticle vaccines against infectious diseases. Front. Immunol. 2018;9:2224. doi: 10.3389/fimmu.2018.02224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E. UCSF Chimera--a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Piehler B., Nelson E.K., Eckels J., Ramsay S., Lum K., Wood B., Greene K.M., Gao H., Seaman M.S., Montefiori D.C., Igra M. LabKey Server NAb: a tool for analyzing, visualizing and sharing results from neutralizing antibody assays. BMC Immunol. 2011;12:33. doi: 10.1186/1471-2172-12-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto D., Park Y.J., Beltramello M., Walls A.C., Tortorici M.A., Bianchi S., Jaconi S., Culap K., Zatta F., De Marco A., et al. Cross-neutralization of SARS-CoV-2 by a human monoclonal SARS-CoV antibody. Nature. 2020;583:290–295. doi: 10.1038/s41586-020-2349-y. [DOI] [PubMed] [Google Scholar]
- Powell A.E., Zhang K., Sanyal M., Tang S., Weidenbacher P.A., Li S., Pham T.D., Pak J.E., Chiu W., Kim P.S. A single immunization with spike-functionalized ferritin vaccines elicits neutralizing antibody responses against SARS-CoV-2 in mice. ACS Cent. Sci. 2021;7:183–199. doi: 10.1021/acscentsci.0c01405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Punjani A., Rubinstein J.L., Fleet D.J., Brubaker M.A. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods. 2017;14:290–296. doi: 10.1038/nmeth.4169. [DOI] [PubMed] [Google Scholar]
- Rappazzo C.G., Tse L.V., Kaku C.I., Wrapp D., Sakharkar M., Huang D., Deveau L.M., Yockachonis T.J., Herbert A.S., Battles M.B., et al. Broad and potent activity against SARS-like viruses by an engineered human monoclonal antibody. Science. 2021;371:823–829. doi: 10.1126/science.abf4830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohou A., Grigorieff N. CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J. Struct. Biol. 2015;192:216–221. doi: 10.1016/j.jsb.2015.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salazar E., Perez K.K., Ashraf M., Chen J., Castillo B., Christensen P.A., Eubank T., Bernard D.W., Eagar T.N., Long S.W., et al. Treatment of coronavirus disease 2019 (COVID-19) patients with convalescent plasma. Am. J. Pathol. 2020;190:1680–1690. doi: 10.1016/j.ajpath.2020.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer M.M., Tortorici M.A., Park Y.-J., Walls A.C., Homad L., Acton O.J., Bowen J.E., Wang C., Xiong X., de van der Schueren W., et al. Structural basis for broad coronavirus neutralization. Nat. Struct. Mol. Biol. 2021;28:478–486. doi: 10.1038/s41594-021-00596-4. [DOI] [PubMed] [Google Scholar]
- Saunders K.O., Lee E., Parks R., Martinez D.R., Li D., Chen H., Edwards R.J., Gobeil S., Barr M., Mansouri K., et al. SARS-CoV-2 vaccination induces neutralizing antibodies against pandemic and pre-emergent SARS-related coronaviruses in monkeys. bioRxiv. 2021 doi: 10.1101/2021.02.17.431492. [DOI] [Google Scholar]
- Scheres S.H. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 2012;180:519–530. doi: 10.1016/j.jsb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi R., Shan C., Duan X., Chen Z., Liu P., Song J., Song T., Bi X., Han C., Wu L., et al. A human neutralizing antibody targets the receptor-binding site of SARS-CoV-2. Nature. 2020;584:120–124. doi: 10.1038/s41586-020-2381-y. [DOI] [PubMed] [Google Scholar]
- Sliepen K., Ozorowski G., Burger J.A., van Montfort T., Stunnenberg M., LaBranche C., Montefiori D.C., Moore J.P., Ward A.B., Sanders R.W. Presenting native-like HIV-1 envelope trimers on ferritin nanoparticles improves their immunogenicity. Retrovirology. 2015;12:82. doi: 10.1186/s12977-015-0210-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ter Meulen J., van den Brink E.N., Poon L.L., Marissen W.E., Leung C.S., Cox F., Cheung C.Y., Bakker A.Q., Bogaards J.A., van Deventer E., et al. Human monoclonal antibody combination against SARS coronavirus: synergy and coverage of escape mutants. PLoS Med. 2006;3:e237. doi: 10.1371/journal.pmed.0030237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian X., Li C., Huang A., Xia S., Lu S., Shi Z., Lu L., Jiang S., Yang Z., Wu Y., Ying T. Potent binding of 2019 novel coronavirus spike protein by a SARS coronavirus-specific human monoclonal antibody. Emerg. Microbes Infect. 2020;9:382–385. doi: 10.1080/22221751.2020.1729069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turoňová B., Sikora M., Schürmann C., Hagen W.J.H., Welsch S., Blanc F.E.C., von Bülow S., Gecht M., Bagola K., Hörner C., et al. In situ structural analysis of SARS-CoV-2 spike reveals flexibility mediated by three hinges. Science. 2020;370:203–208. doi: 10.1126/science.abd5223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda G., Antanasijevic A., Fallas J.A., Sheffler W., Copps J., Ellis D., Hutchinson G.B., Moyer A., Yasmeen A., Tsybovsky Y., et al. Tailored design of protein nanoparticle scaffolds for multivalent presentation of viral glycoprotein antigens. eLife. 2020;9:e57659. doi: 10.7554/eLife.57659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walls A.C., Fiala B., Schäfer A., Wrenn S., Pham M.N., Murphy M., Tse L.V., Shehata L., O’Connor M.A., Chen C., et al. Elicitation of potent neutralizing antibody responses by designed protein nanoparticle vaccines for SARS-CoV-2. Cell. 2020;183:1367–1382.e17. doi: 10.1016/j.cell.2020.10.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walls A.C., Park Y.J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;181:281–292.e6. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Shi W., Joyce M.G., Modjarrad K., Zhang Y., Leung K., Lees C.R., Zhou T., Yassine H.M., Kanekiyo M., et al. Evaluation of candidate vaccine approaches for MERS-CoV. Nat. Commun. 2015;6:7712. doi: 10.1038/ncomms8712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Zhou T., Zhang Y., Yang E.S., Schramm C.A., Shi W., Pegu A., Oloninyi O.K., Ransier A., Darko S., et al. Antibodies with potent and broad neutralizing activity against antigenically diverse and highly transmissible SARS-CoV-2 variants. bioRxiv. 2021 doi: 10.1101/2021.02.25.432969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Huang B., Zhu Y., Tan W., Zhu M. Ferritin nanoparticle-based SARS-CoV-2 RBD vaccine induces a persistent antibody response and long-term memory in mice. Cell. Mol. Immunol. 2021;18:749–751. doi: 10.1038/s41423-021-00643-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wibmer C.K., Ayres F., Hermanus T., Madzivhandila M., Kgagudi P., Oosthuysen B., Lambson B.E., de Oliveira T., Vermeulen M., van der Berg K., et al. SARS-CoV-2 501Y.V2 escapes neutralization by South African COVID-19 donor plasma. Nat. Med. 2021;27:622–625. doi: 10.1038/s41591-021-01285-x. [DOI] [PubMed] [Google Scholar]
- Winkler E.S., Bailey A.L., Kafai N.M., Nair S., McCune B.T., Yu J., Fox J.M., Chen R.E., Earnest J.T., Keeler S.P., et al. SARS-CoV-2 infection of human ACE2-transgenic mice causes severe lung inflammation and impaired function. Nat. Immunol. 2020;21:1327–1335. doi: 10.1038/s41590-020-0778-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wørzner K., Sheward D.J., Schmidt S.T., Hanke L., Zimmermann J., McInerney G., Karlsson Hedestam G.B., Murrell B., Christensen D., Pedersen G.K. Adjuvanted SARS-CoV-2 spike protein elicits neutralizing antibodies and CD4 T cell responses after a single immunization in mice. EBioMedicine. 2021;63:103197. doi: 10.1016/j.ebiom.2020.103197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrapp D., De Vlieger D., Corbett K.S., Torres G.M., Wang N., Van Breedam W., Roose K., van Schie L., Hoffmann M., Pöhlmann S., et al. VIB-CMB COVID-19 Response Team Structural basis for potent neutralization of betacoronaviruses by single-domain camelid antibodies. Cell. 2020;181:1004–1015.e15. doi: 10.1016/j.cell.2020.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C.L., Abiona O., Graham B.S., McLellan J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zost S.J., Gilchuk P., Case J.B., Binshtein E., Chen R.E., Nkolola J.P., Schäfer A., Reidy J.X., Trivette A., Nargi R.S., et al. Potently neutralizing and protective human antibodies against SARS-CoV-2. Nature. 2020;584:443–449. doi: 10.1038/s41586-020-2548-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
All data supporting the findings of this study are found within the paper and its supplemental information. Negative-stain EM maps for pCOV111, SpFN_1B-06-PL, RFN_131, and pCoV146 have been deposited in the EM database under codes 25451, 25448, 25449, and 25450 respectively.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the Lead Contact author upon request.