Summary
Background
The outbreak of chilblain‐like lesions (CLL) during the COVID‐19 pandemic has been reported extensively, potentially related to SARS‐CoV‐2 infection, yet its underlying pathophysiology is unclear.
Objectives
To study skin and blood endothelial and immune system activation in CLL in comparison with healthy controls and seasonal chilblains (SC), defined as cold‐induced sporadic chilblains occurring during 2015 and 2019 with exclusion of chilblain lupus.
Methods
This observational study was conducted during 9–16 April 2020 at Saint‐Louis Hospital, Paris, France. All patients referred with CLL seen during this period of the COVID‐19 pandemic were included in this study. We excluded patients with a history of chilblains or chilblain lupus. Fifty patients were included.
Results
Histological patterns were similar and transcriptomic signatures overlapped in both the CLL and SC groups, with type I interferon polarization and a cytotoxic–natural killer gene signature. CLL were characterized by higher IgA tissue deposition and more significant transcriptomic activation of complement and angiogenesis factors compared with SC. We observed in CLL a systemic immune response associated with IgA antineutrophil cytoplasmic antibodies in 73% of patients, and elevated type I interferon blood signature in comparison with healthy controls. Finally, using blood biomarkers related to endothelial dysfunction and activation, and to angiogenesis or endothelial progenitor cell mobilization, we confirmed endothelial dysfunction in CLL.
Conclusions
Our findings support an activation loop in the skin in CLL associated with endothelial alteration and immune infiltration of cytotoxic and type I IFN‐polarized cells leading to clinical manifestations.
A range of cutaneous manifestations have been described in association with SARS‐CoV‐2 infection during the COVID‐19 pandemic.1 Among them, chilblain‐like lesions (CLL) have been occurring more frequently than expected. However, the link between SARS‐CoV‐2 infection and CLL is not well established.2, 3 Chilblains are cutaneous inflammatory erythematous papules mainly involving fingers and toes, which are thought to be triggered by cold. Most of them are idiopathic, and therefore called seasonal chilblains (SC), but they can be associated with connective tissue disease such as lupus erythematous, or monogenic diseases such as STING‐associated vasculopathy with onset in infancy (SAVI).4
SARS‐CoV‐2 infection strongly triggers the expression of type I interferon (IFN)‐stimulated genes, which assist in the host’s antiviral protection.5 Diseases mediated by type I IFN, such as monogenic autoinflammatory interferonopathies or lupus erythematosus, are characterized by microangiopathy leading to clinical chilblains.6
SARS‐CoV‐2 infection seems to be associated with vascular skin symptoms7 and has been shown to damage capillary endothelium and disrupt the thrombo‐protective state of endothelial cells,8 likely contributing to microvascular thrombosis.9–11 Moreover, the histopathological features of COVID‐19‐associated CLL include endothelial damage and features of microangiopathic damage.12
The aim of this study was to analyse deeply the immunological and vascular pathophysiology of CLL during the COVID‐19 outbreak compared with SC.
Patients and methods
Study design and population
All patients referred to the dermatology department of Saint‐Louis Hospital, Paris, France, with CLL during 9–16 April 2020 of the COVID‐19 pandemic were included in this noninterventional observational study. We excluded patients with a history of chilblains or chilblain lupus. Patients provided written informed consent for the collection and analysis of their data. They were assessed at day 0 and day 14. The following controls were included: (i) patients with reverse‐transcriptase polymerase chain reaction (RT‐PCR)‐proven mild COVID‐19 without chilblains, for serological, immunological and endothelial activation assessment; (ii) patients with SC before the COVID‐19 pandemic defined by typical clinical and histological presentation without antinuclear antibodies (ANA) between January 2015 and March 2019 for histological and immunological assessment; and (iii) healthy controls (HC) without COVID‐19 symptoms for serological, cytokine and endothelial activation assessment. We also included HC skin samples obtained from fresh plastic surgery waste.
Human participant declaration
All parts of the study were approved by the appropriate institutional review boards (Cochin‐Port Royal Hospital, Paris, France) and were conducted in accordance with the current ethical and legal frameworks of the Declaration of Helsinki. Informed written consent was received from participants before inclusion in this study, according to our local ethics rules.
Laboratory parameters
Blood samples were collected in ethylenediaminetetraacetic acid (EDTA), sodium heparin or 0·11 mol L−1 trisodium citrate tubes (Greiner Bio‐One, Courtaboeuf, France). The laboratory parameters recorded included whole blood count (XN3000; Sysmex, Kobe, Japan); haemostasis tests; IgA level; isotypes of IgG and IgA antineutrophil cytoplasmic antibodies (ANCA); ANA; anti‐double‐stranded DNA antibodies; cryoglobulinemia; cryofibrinogen; and anticardiolipin and anti‐β2‐glycoprotein‐I IgG antibodies (IgM and IgG). The haemostasis tests (STA R Max system; Stago, Asnières sur Seine, France) included prothrombin time (PT), activated partial thromboplastin time (aPTT), fibrinogen, D‐dimers (Liatest D‐Di Plus, Stago), antithrombin activity (Stachrom, Stago), protein C activity (Protein C Coag; Siemens, Munich, Germany) and protein S activity (Staclot PS, Stago), lupus anticoagulant (LA) using integrated diluted Russell viper venom time (dRVVT LAC Screen and LAC Confirm, Siemens) and LA‐sensitive aPTT, as described previously.13
Flow spike assay
This assay is described in Grzelak et al.14 and is detailed in Appendix S1 (see Supporting Information).
Cytokine assays
Prior to protein analysis, plasma samples were treated in a P3 laboratory for viral decontamination using a protocol validated for SARS‐CoV‐2. Briefly, samples were treated with Triton X‐100 1% (v/v) for 2 h at room temperature. IFN‐α2, IFN‐γ and IL‐17A protein plasma concentrations were quantified by a Simoa triplex assay developed with Quanterix Homebrew kits (Quanterix, Billerica, MA, USA). Interleukin (IL)‐6, tumour necrosis factor (TNF)‐α, and IL‐10 were measured with a commercial triplex assay (Quanterix). The limits of detection of these assays were 2 fg mL−1 for IFN‐α2 and 7 fg mL−1 for IFN‐γ.
Assessment of interferon‐stimulated gene expression in whole blood
Total RNA was extracted using an RNA isolation kit. Further details are provided in Appendix S1.
Soluble markers of endothelial activation and/or angiogenesis
Platelet‐poor plasma (PPP) was obtained from EDTA samples after centrifugation at 2500 g for 15 min at day 0 and 14. After a second centrifugation at 2500 g for 15 min, PPP was stored at −80 °C until analysis of vascular markers. Four biomarkers related to endothelial dysfunction or activation (angiopoietin‐1, angiopoietin‐2, endoglin and soluble E‐selectin) and four biomarkers related to angiogenesis or endothelial progenitor cell mobilization [vascular endothelial growth factor (VEGF)‐A, c‐Kit, basic fibroblast growth factor (b‐FGF) and VEGF receptor] were quantified in PPP with a Human Magnetic Luminex Assay from R&D Systems (Lille, France). Data were assessed with the Bio‐Plex 200 using Bio‐Plex Manager 5.0 software (Bio‐Rad, Marnes‐la‐Coquette, France) as described previously.10
Circulating endothelial cell counting after immunomagnetic separation
Peripheral venous blood samples were collected on EDTA at day 14. Circulating endothelial cells (CECs) were counted by an operator blinded to the patients’ clinical features. Immunocapture of CECs from whole blood was performed at 4 °C using magnetic beads (Dynabeads M‐450, Dynal; Invitrogen, Carlsbad, CA, USA) coated with S‐Endo 1 (Biocytex, Marseille, France), a monoclonal antibody raised against the endothelial antigen CD146. To avoid nonspecific binding of leucocytes to CD146‐coated beads, the cell suspensions were flushed vigorously through the pipette tip during the washing steps and then suspended in acridine orange (3 μg mL−1 in phosphate‐buffered saline; Sigma‐Aldrich, Saint‐Quentin Fallavier, France) before being counted under a fluorescence microscope (λexc = 490 nm). CECs were identified according to their morphological criteria, namely > 10 beads bound to > 20‐μm cells or cells with < 10 beads but with a well‐preserved and recognizable morphology (clear nucleus in a well‐delineated cytoplasm and a size consistent with that of endothelial cells). The number of cells in aggregates was determined from the number of cells with spherical rosette features. The endothelial nature of the isolated cells was confirmed by measuring lectin Ulex europaeus agglutinin‐1 expression. The number of CECs was expressed as cells per mL of blood.10, 11
Histological examinations
Skin biopsies from CLL were performed in 13 patients. Histological sections were stained with haematoxylin, eosin and safran. For each sample, histological parameters (epidermal changes, basal vacuolization, inflammatory infiltrate, and type and location of affected vessel) and immunohistochemical analysis for CD61 (integrin β3, clone 2F2; Leica Biosystems, Newcastle upon Tyne, UK), identifying intravascular platelet microthrombi, CD123 (clone 6H6; Exbio, Vestec, Czech Republic) and IgA (polyclonal; Dako, Glostrup, Denmark), were performed on a BenchMark ULTRA immunostainer (Roche‐Ventana, Basel, Switzerland). IgA staining was performed on paraffin‐embedded tissue sections as described previously.15
Gene expression
From a lesional dermoepidermal skin biopsy, 10 formalin‐fixed paraffin‐embedded (FFPE) sections were taken, each 10 μm thick. Total RNA was automatically extracted from tissue samples using the Maxwell RSC RNA FFPE Kit protocol (Promega, Charbonnières‐les‐Bains, France). We analysed 100 ng (5 μL) of total RNA from each sample using the NanoString nCounter Human Immunology V2 kit according to the manufacturer’s instructions (NanoString Technologies, Seattle, WA, USA) (Appendix S1).
Statistical analysis
GraphPad Prism v8.4.0 was used for statistical analysis (GraphPad Software, La Jolla, CA, USA). Comparisons of groups were performed using the unpaired t‐test without correction, whereas correction for variance inequality was performed using Welch’s correction when the F‐test P‐value was < 0·2. The Holm–Sidak method was used to correct for multiple testing. Correlations between quantitative variables were assessed using Spearman’s correlation coefficient and the associated P‐value. P‐values < 0·05 were considered statistically significant.
Results
Patient characteristics
Fifty patients were included; their characteristics are described in Table S1 (see Supporting Information). Twenty‐nine (58%) had suggestive extracutaneous COVID‐19 symptoms, including asthenia (n = 14), fever (n = 11), upper airway and ear–nose–throat symptoms (n = 16), cough (n = 9), dyspnoea (n = 2) and anosmia (n = 1). Twenty (40%) had been in close contact with a person with confirmed COVID‐19. The median duration between suggestive extracutaneous symptoms (when present) and CLL was 7 days [interquartile range (IQR) 2–16]. The median duration between suggestive extracutaneous symptoms and study inclusion was 24 days (IQR 16–31). The median duration between onset of CLL and study inclusion was 15 days (IQR 10–21).
Toes and fingers were involved in 86% and 24% of patients, respectively (Figure S1; see Supporting Information). Other cutaneous manifestations concurrent with CLL were extensive livedo (n = 2; 4%), arthralgia (n = 1; 2%), erythematous papules on scars (n = 2; 4%), maculopapular exanthema (n = 4; 8%) and urticaria (n = 1; 2%). CLL remission at day 15 was observed in only 16 patients (32%). Seven patients (14%) developed a new CLL flare in the meantime, and 10 patients (20%) had persistent livedo or peripheral vascular disease at day 15.
Association with SARS‐CoV‐2
Real‐time RT‐PCR testing for SARS‐CoV‐2 was performed using a nasopharyngeal swab at inclusion for all patients with CLL (n = 50), and in the skin (n = 6) and the stools (n = 3) of some patients. RT‐PCR was negative for all samples tested.
COVID‐19 serological tests were performed using three different techniques at inclusion and 14 days later: (i) IgG COVID Abbott Architect test (Abbott Laboratories, Libertyville, IL, USA); (ii) IgG and IgA enzyme‐linked immunosorbent assay (ELISA) nucleocapsid COVID EUROIMMUN tests (EUROIMMUN, Lübeck, Germany); and (iii) flow spike IgA and IgG detection.16 The serological results were compared with the data from patients with RT‐PCR‐confirmed mild COVID‐19. The serological tests were all negative in the CLL group, except for four positive and four doubtful IgA ELISA anti‐SARS‐CoV‐2 tests at the first visit (Table S2; see Supporting Information). All samples from the comparator group with confirmed COVID‐19 were positive.
Chilblain‐like lesions are characterized by immune cytotoxic infiltration, interferon polarization and endothelial alteration in the skin
To assess the pathophysiology of CLL we first studied all skin histological characteristics of 13 patients with CLL, compared firstly with 13 patients with SC before the COVID‐19 pandemic and secondly with HC. All CLL and SC samples showed lymphocytic infiltration around superficial and deep dermal blood vessels. Other histological patterns were frequent in both CLL and SC, namely dense perivascular lymphocytic infiltrate, lymphocytic acrosyringitis, lymphocytes around the sweat gland coil, papillary oedema, extravasation of red blood cells, presence of CD123‐positive plasmacytoid dendritic cells, presence of plump endothelial cells, and presence of CD61‐positive platelet microthrombi. CD61 staining was used to highlight platelet microthrombi. IgA staining was observed in interstitial papillary dermis only in CLL and not in SC (Table 1 and Figure 1a).
Table 1.
Histological characteristics of chilblain‐like lesions (CLL) compared with seasonal chilblain (SC)
| CLL (n = 13) | SC (n = 13) | P‐value | |
| Deep perisudoral lymphocytic infiltrate | 9 | 10 | > 0·99 |
| Dense perivascular lymphocytic infiltrate | 10 | 11 | > 0·99 |
| Lymphocytic acrosyringitis | 3 | 3 | > 0·99 |
| Papillary oedema | 10 | 9 | > 0·99 |
| Red blood cell extravasation | 8 | 6 | 0·69 |
| Plump endothelial cells | 9 | 12 | 0·32 |
| Epidermal vacuolization | 2 | 5 | 0·38 |
| Keratinocyte necrosis | 2 | 5 | 0·38 |
| Presence of microthrombi | 8 | 4 | 0·24 |
| CD61 | 7 | 7 | > 0·99 |
| CD123 (%), median (interquartile range) | 2 (2–5) | 5 (2–5) | 0·19 |
| Cytoplasmic endothelial cells IgA deposition | 13 | 8 | 0·04 |
| Vascular lumen IgA deposition | 8 | 5 | 0·43 |
| Interstitial papillary dermis IgA deposition | 8 | 0 | 0·002 |
| Epidermis IgA deposition | 8 | 3 | 0·11 |
| Positive dermis and/or perisudoral alcian blue staining | 5 | 3 | 0·67 |
The data are presented as the number, except for CD123.
Figure 1.
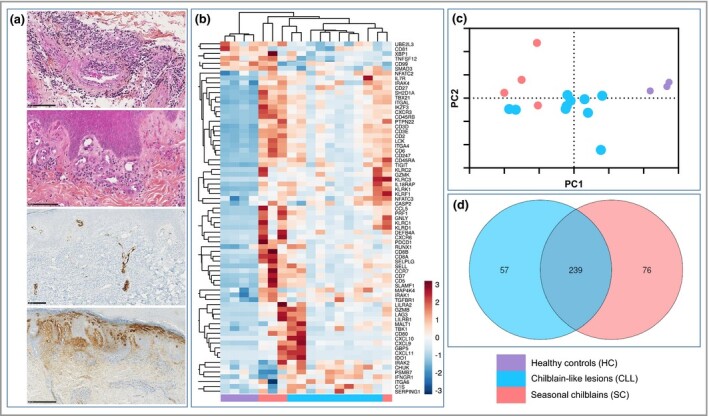
Chilblain‐like lesions (CLL) and seasonal chilblains (SC) display a common transcriptomic signature and histological pattern. (a) Histological sections of CLL skin samples representing from top to bottom: haematoxylin–eosin–safran (HES) showing dense perivascular lymphocytic infiltrate (scale bar = 100 μm); HES showing plump endothelial cells (scale bar = 100 μm); endovascular CD61 immunostaining (scale bar = 100 μm) and diffuse IgA immunostaining (scale bar = 250 μm). (b–d) Skin transcriptomic profiles of CLL (n = 10), SC (n = 4) and healthy controls (HC) (n = 4). (b) Two‐dimensional hierarchical clustering was performed on a set of 70 genes whose average expression significantly differed between CLL, SC and HC. All values were log2 transformed and mean centred across each gene. (c) Principal component analysis of gene expression data of skin samples showing the distance between CLL, SC and HC. (d) Venn diagram comparing differently expressed genes with P < 0·05 between CLL, SC and HC.
We compared the skin immune transcriptomic signature of 10 CLL, four SC and four HC skin samples (Figure 1b–d). The three groups clustered independently with an overlap between two CLL and SC samples, suggesting a common immune pathophysiology. Among 589 genes, we identified 296 differentially expressed genes (DEGs) between CLL and HC, most of them being upregulated. Common DEGs between CLL and SC included type I and II interferon pathways (upregulation of CXCL9, CXCL10, CXCL11, IDO1, TLR1, TLR2, TLR7, TLR8, MYD88, IRF5, IFNAR2, IFI16, IRAK1, IRAK4 and NFKB1); T helper 1 cell polarization (upregulation of CXCR3 and downregulation of GATA3 and RORC); cytotoxicity markers (upregulation of granzyme A, granzyme B, granzyme K, granulysin and perforin); natural killer cell signalling (upregulation of KLRC2, KLRC1, KLRD1, KLRC3, CD96, KLRC4, KLRF1, KLRB1 and KLRG1) and other activation and regulatory immune markers (LILRB1, LAG3, TIGIT, CTLA4, FOXP3, IL2RA, BTLA, HLA‐DR genes and PDCD1LG2).
Interestingly, 57 DEGs were specific to CLL, involving: (i) complement activation (upregulation in the classical pathway of C1q, C1s and C1 inhibitor, and the alternative pathway C2 and properdin, with downregulation of membrane attack complex components C5 and C6); and (ii) angiogenesis (upregulation of PDGFB and downregulation of TNFSF12) (Figure 1).
IgA antineutrophil cytoplasmic antibodies and elevated type I interferon signature characterize systemic immune response in chilblain‐like lesions
We next assessed the systemic immune response during CLL. Firstly, ANA were positive in 10 (20%) patients (titre > 160) but none had either anti‐extractable nuclear antigen or anti‐DNA antibodies. Three (6%) patients had significant levels of cryofibrinogen. Three (6%) patients had low levels of C3 (range 0·73–0·75g L−1) and 10 (20%) had low levels of C4 (range 0·12–0·15g L−1). Concerning ANCA, IgG ANCA were positive in four patients, with cytoplasmic fluorescence without specificity. Notably, IgA ANCA were positive in 34 of 46 (74%) patients with cytoplasmic fluorescence: four with proteinase 3 specificity, three with bactericidal/permeability increasing protein (BPI) specificity, one with lactoferrin specificity and three with cathepsin G specificity. IgA ANCA were positive in all five SC samples that could be analysed.
We next studied circulating cytokines and the blood type I IFN signature at day 0 and day 14 in CLL compared with patients with non‐severe COVID‐19, patients with SAVI, and HC. As shown in Figure 2(a), the type I IFN gene signature in whole blood was significantly higher in CLL than in HC. Three patients reached the level observed in patients with SAVI. CLL samples also exhibited a peculiar cytokine profile with a significant elevation of IFN‐α2 and nonsignificant elevation of IFN‐γ compared with HC, and a significant decrease of IL‐10 (Figure 2b). Other cytokines (IL‐6, TNF‐α and IL‐17A) were similar in both groups (Figure 2c). When initially elevated, cytokine levels normalized at day 14.
Figure 2.

Patients with chilblain‐like lesions (CLL) display a systemic immune response with an enhanced type I interferon (IFN) signature. (a) IFN‐stimulated gene (ISG) score based on the expression of six genes (IFI44L, IFI27, RSAD2, SIGLEC1, IFIT1 and ISG15) measured with quantitative reverse‐transcriptase polymerase chain reaction in whole blood cells from patients with CLL (n = 57), healthy controls (HC) (n = 20) and patients with STING‐associated vasculopathy in infancy (SAVI) (n = 4). (b, c) Serum cytokine concentrations were measured in HC (n = 7), patients with CLL at day 0 (D0) (n = 50) and day 14 (D14) (n = 7) and patients with COVID‐19 (n = 4). The data are presented as scattered dot plots; the horizontal bar represents the median and the whiskers represent the interquartile range. P‐values were calculated using the Wilcoxon–Mann–Whitney test; P‐values < 0·05 were considered significant. IL, interleukin; TNF, tumour necrosis factor. *P < 0·05; where not specified (ns), differences are not statistically significant.
Systemic endothelial activation
Lastly, we studied the systemic modifications in haemostasis and endothelial activation during CLL. The PT ratio and aPTT were normal in all patients. Only three patients (6%) had a moderate increase in D‐dimer levels (510, 560 and 1680 ng mL−1). Antithrombin, and protein C and protein S activity levels were within the normal range in all patients, except for three (6%) with slightly decreased protein S activity (45, 48 and 49 IU dL−1). Five patients (10%) had transient positive lupus anticoagulant, which was negative 15 days later. One patient (2%) had isolated IgM anticardiolipin positivity and one (2%) had isolated IgM anti‐β2‐glycoprotein‐I positivity.
We next quantified four biomarkers related to endothelial dysfunction or activation (Figure 3a) and four biomarkers related to angiogenesis or endothelial progenitor cell mobilization (Figure 3b). We found a significant increase in all biomarkers of endothelial dysfunction in CCL compared with HC. Among angiogenic‐related biomarkers, VEGF‐A, VEGFR‐2 and c‐Kit were significantly increased, while b‐FGF was similar. Kinetics of endothelial dysfunction biomarkers (day 0 and day 14 after first sampling) suggested normalization of endothelial activation with time (Table S3; see Supporting Information). However, there was no association between the level of endothelial markers and recovery in patients with CLL.
Figure 3.
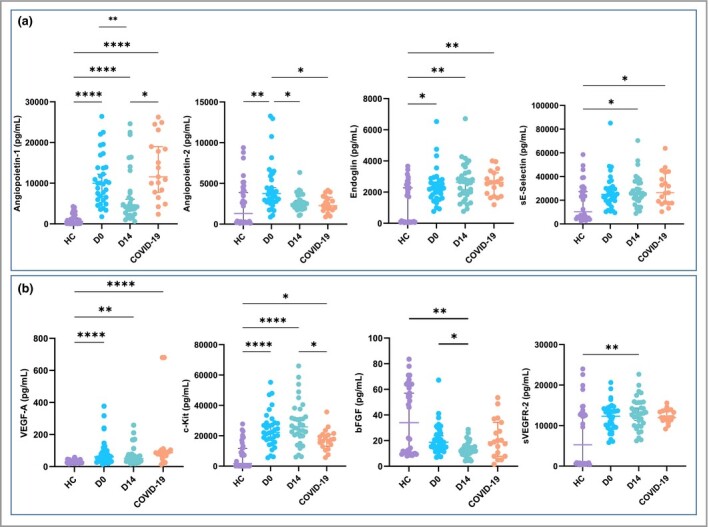
Patients with chilblain‐like lesions (CLL) display circulating markers of endothelial injury. Soluble serum markers (a) of endothelial dysfunction or activation and (b) related to angiogenesis or endothelial progenitor cell mobilization were measured in patients with CLL at day 0 (D0) (n = 34) and day 14 (D14) (n = 34), patients with COVID‐19 (n = 19) and healthy controls (HC) (n = 34). The data are presented as scattered dot plots; the horizontal bar represents the median, and the whiskers represent the interquartile range. P‐values were calculated using the Wilcoxon–Mann–Whitney test; P‐values < 0·05 were considered significant. bFGF, basic fibroblast growth factor; VEGF, vascular endothelial growth factor; VEGFR, VEGF receptor. *P < 0·05, **P < 0·01, ****P < 0·0001.
Increase of endothelial markers was associated with the delay between blood sampling and CLL onset (Table S4; see Supporting Information). In particular, angiopoietin‐1, angiopoietin‐2 and VEGF‐A were increased in patients with CLL sampled > 20 days after skin lesions had started (‘late’ CLL) compared with patients sampled at earlier timepoints (‘early’ CLL) (P = 0·05, 0·007 and 0·003, respectively). Similarly, CECs were also upregulated in ‘late’ vs. ‘early’ CLL (P = 0·03).
Discussion
Beyond classical COVID‐19 symptoms, several publications have reported CLL potentially associated with SARS‐CoV‐2 infection.2, 17 The epidemiology and clinical features of CLL have been extensively studied and published; however, little is known about the pathophysiology of CLL. This study illustrates that both the immune system and endothelial cells play a critical role in the genesis of CLL.
The relationship between SARS‐CoV‐2 infection and CLL is still controversial; however, recurrence of previous chilblain lesions during the second wave of COVID‐19 and peaks of CLLs concomitant with peaks of COVID‐19 deaths in 202018, 19 strongly suggest that this disorder is closely related to SARS‐CoV‐2 infection.20 In our study, SARS‐CoV‐2 assays were almost all negative except for four serology results at day 0.
As described partially in other studies,21–23 the present study showed skin and systemic type I IFN polarization, which could be explained by a strong antiviral response towards SARS‐CoV‐2 in CLL or other viruses in SC, involving the type I IFN pathway. Histological aspects of CLL included vascular alterations such as lymphocytic infiltrates in vessel walls and microthrombi, but we did not observe true leucocytoclastic vasculitis as seen with SAVI.6 This is potentially related to lower IFN levels in CLL.
We also observed a strong cutaneous upregulation of genes related to cytotoxic T cells and natural killer cells. Interestingly, granzyme B has been shown to induce dysregulated angiogenesis and vascular permeability,24 and also IFN‐γ release by skin keratinocytes.25 Altogether, these findings argue for the role of a type I IFN and cytotoxic axis, which has been previously shown to act in synergy26 and to be defective in critically ill patients with COVID‐19.27 Moreover, our results suggest a common pathophysiology between CLL and SC. Even if some patients had positive ANA or low‐level complement, none fulfilled at least four of the American College of Rheumatology revised criteria for systemic lupus erythematosus or had histologically ascertained cutaneous discoid lupus lesions.
Small‐vessel vasculitis includes ANCA‐related vasculitis, IgA‐related vasculitis and cryoprotein‐related vasculitis. These are characterized by cutaneous manifestations such as purpura, livedo or ulcers.28, 29 In this study we found a low prevalence of cryofibrinogenaemia, inconsistently with a previous study.30 However, we found a high prevalence of ANCA of the IgA isotype, seen in 73% of patients with CLL and in 100% with SC, associated with vascular and interstitial IgA deposition. The association of systemic and local IgA activation could explain the cutaneous vasculitis with a possible postviral pathophysiology, in both CLL and SC. Indeed, IgA anti‐SARS‐CoV‐2 is the first immunoglobulin detectable after SARS‐CoV‐2 infection,16 and several studies have highlighted COVID‐19‐related IgA manifestations.31–33
The pathogenicity of IgA ANCA found in our patients with CLL remains unclear, but previous reports suggest their potential implication in erythema elevatum diutinum, skin vasculitis, ulcerative colitis and granulomatosis with polyangiitis.34, 35 The vasculitis observed in our patient was mainly composed of lymphocytes, which is unusual. Indeed, IgA immune‐complex vasculitis is usually associated with neutrophil infiltration, partially explained by the fact that IgA complexes can activate neutrophils via the IgA Fc receptor FcαRI (CD89), thereby inducing neutrophil migration and activation, which ultimately cause tissue damage.36 It has been shown that ANCA can recognize vascular endothelial cell‐bound ANCA‐associated autoantigens, such as myeloperoxidase, finally leading to complement activation and vascular injury.37
Vascular and endothelial injury has been shown to be the hallmark of SARS‐CoV‐2 infection and could explain, at least in part, the pathophysiology of CLL. Our study confirms the increase of cutaneous histological and systemic markers (angiopoietin‐1, angiopoietin‐2 and VEGF‐A) of endothelial dysfunction with normalization over time. The transcriptomic analysis revealed that both the complement pathway and angiogenesis seemed specifically activated in CLL compared with SC. We previously described an increase at admission in angiopoietin‐2 and von Willebrand factor related to worsening of patients with COVID‐19,10, 38 which supports the role of microcirculatory dysfunction. Angiopoietin‐2 is a regulator of angiogenesis that can be rapidly released by activated endothelium in the presence of thrombin or inflammatory cytokines, and participates in the responsiveness of endothelium to inflammatory, hyperpermeability apoptosis. This injury could be explained (i) by activation or apoptosis of endothelial cells by the immune system, notably cytotoxic molecules, or (ii) through complement‐mediated microvascular injury.39
In conclusion, our findings support an activation loop in the skin in CLL, which associates with endothelial alteration and immune infiltration of cytotoxic and type I IFN‐polarized cells leading to clinical manifestation.
Acknowledgments
We thank all of the medical staff from the dermatology department of Saint‐Louis Hospital and particularly Drs Marie Jachiet, Antoine Petit and Anne Saussine. Our thanks also go to Marie‐Hélène Durand, Elisabeth, Julie and Alain for their valuable assistance. We are also grateful to Drs Luc Sulimovic and Michel Rybojad and all members of the SNDV (French National Union of Dermatologists‐Venereologists): Dominique Denjean, Marie‐Pierre Labarthe, Maud Bézier, Marie Risbourg, Geneviève Payan, Sabrina Alain, Frédéric Mathivon, Anny Cohen‐Letessier, Delphine Kerob, Jean‐Philippe Hellier, Christelle Comte, Fabielle Keller, Caroline Brue, Paul Lestang, Laurence Allanore, Eliane Pierkarski‐Carp, Anne Amoric, Hervé Serpier, Philippe Pruvost, Fabien Guibal, Damien Giacchero, Elisa Funck‐Brentano, Sandrine Sierra Fortuny, Isabelle Gallay, Agnès Zavarro, Caroline Bider‐Valle, Sylvie Lagrange, Etty Grynberg, Florence Weill, Dominique Penso, Marie Gomel, Jean Schneider, Anne Larabelle, Philippe Bonhomme, Marie‐Sophie Gautier, Jean Hatchuel, Imane Mourtada, Charlotte Fite, Catherine Oliveres‐Ghouti, Elisabeth Domergue, Sabrina Fourcade‐Roch, Sylvie Lecanu, Nathalie Sebban, Bruno Halioua, Anne Bellut, Fabienne Keller, Isabelle Baratte, Françoise Lejoyeux, Laurence Ollivaud, Georges Abirached, Marielle Burnouf, Beatrix Reynaud‐Mendel, Jean‐Noël Dauendorffer, Joëlle Sebaoun, Anne Larabelle, Hervé Garrat, Marie‐Martine Pomper, Anne‐Marie Heudes, Isabelle Beaulieu, Hugues Cartier, Amélie Arsouze, Dominique Lons‐Danic, Michèle Pelletier, Geneviève Payan, Valérie Gallais, Valérie Piantade, Marlène Risbourg, Georges Reuter, Serge Dahan, Murielle Creusot, Abdallah Kolli, Isabelle Egasse‐Broca, Jean‐Luc Rigon, Pascale Sabban, Hélène Flacher, Benoît Jaillard, Pierre André, Dominique Debjoux, Elodie Poirier, Bénédicte Solyga, Marc Perrussel, Sabrina Makhloufi, Bertrand Margnier, Clotilde Huzar, Laetitia Vandame, Hortense Thelu, Anne‐Claire Chollet, Frédérique Marchal, Michael Naouri, Marion Nadaud, Elodie Boissy, Abdelhamid Lameche, Charles Berdougo, Audrey Rolland, Marie‐Laure Fléchet, Gabriel Colonna, Delphine Jouannet, Isabelle Berdah, Françoise Truchot and Isabelle Lavallée. We thank all of the patients who agreed to be part of this study. The Saint‐Louis CORE (COvid REsearch) group, in alphabetic order, is G. Archer, A. Benattia, A. Bergeron, L. Bondeelle, J.D. Bouaziz, D. Bouda, D. Boutboul, I. Brindel Berthon, E. Bugnet, S. Caillat Zucman, S. Cassonnet, K. Celli Lebras, J. Chabert, S. Chevret, M. Clément, C. Davoine, N. De Castro, E. De Kerviler, C. De Margerie‐Mellon, C. Delaugerre, B. Denis, F. Depret, L. Djaghout, C. Dupin, D. Farge‐Bancel, C. Fauvaux, E. Feredj, D. Feyeux, J.P. Fontaine, V. Fremeaux‐Bacchi, L. Galicier, S. Harel, A.L. Jegu, E. Kozakiewicz, A. Lebel M Baye, J. Le Goff, P. Le Guen, E. Lengline, G. Liegeon, G. Lorillon, I. Madelaine Chambrin, G. Martin de Frémont, M. Meunier, J.M. Molina, F. Morin, E. Oksenhendler, R. Peffault de la Tour, O. Peyrony, B. Plaud, M. Salmona, J. Saussereau and J. Soret.
Author Contribution
Laure Frumholtz: Formal analysis (equal); Investigation (equal); Methodology (equal); Validation (equal); Visualization (equal); Writing‐original draft (equal). Jean David Bouaziz: Funding acquisition (equal); Project administration (equal); Supervision (equal); Validation (equal); Writing‐original draft (equal); Writing‐review & editing (equal). Maxime Battistella: Conceptualization (equal); Investigation (equal); Methodology (equal); Software (equal); Validation (equal); Writing‐original draft (equal); Writing‐review & editing (equal). Jérome Hadjadj: Data curation (equal); Methodology (equal); Visualization (equal); Writing‐review & editing (equal). Richard Chocron: Data curation (equal); Formal analysis (equal); Software (equal); Validation (equal); Writing‐review & editing (equal). Djaouida Bengoufa: Methodology (equal); Resources (equal); Visualization (equal); Writing‐review & editing (equal). Hélène Le Buanec: Methodology (equal); Supervision (equal); Writing‐review & editing (equal). Laura Barnabei: Investigation (equal); Resources (equal); Writing‐review & editing (equal). Sonia Meynier: Investigation (equal); Resources (equal); Writing‐review & editing (equal). Olivier Schwartz: Formal analysis (equal); Funding acquisition (equal); Supervision (equal); Writing‐review & editing (equal). Ludivine Grzelak: Investigation (equal); Methodology (equal); Software (equal); Writing‐review & editing (equal). Nikaia Smith: Data curation (equal); Investigation (equal); Methodology (equal); Writing‐review & editing (equal). Bruno Charbit: Investigation (equal); Methodology (equal); Resources (equal); Validation (equal); Writing‐review & editing (equal). Darragh Duffy: Formal analysis (equal); Software (equal); Supervision (equal); Validation (equal); Writing‐review & editing (equal). Nader Yatim: Conceptualization (equal); Data curation (equal); Methodology (equal); Writing‐review & editing (equal). Andreea Calugareanu: Methodology (equal); Resources (equal); Software (equal); Writing‐review & editing (equal). Aurélien Philippe: Formal analysis (equal); Investigation (equal); Methodology (equal); Writing‐review & editing (equal). Coralie Guerin: Investigation (equal); Software (equal); Writing‐review & editing (equal). Berangere Joly: Investigation (equal); Resources (equal); Supervision (equal); Writing‐review & editing (equal). Virginie Siguret: Funding acquisition (equal); Methodology (equal); Supervision (equal); Writing‐review & editing (equal). Léa Jaume: Investigation (equal); Writing‐review & editing (equal). Herve Bachelez: Funding acquisition (equal); Supervision (equal); Writing‐review & editing (equal). Martine Bagot: Funding acquisition (equal); Supervision (equal); Writing‐review & editing (equal). Frédéric Rieux‐Laucat: Funding acquisition (equal); Investigation (equal); Supervision (equal); Writing‐review & editing (equal). Sarah Maylin: Data curation (equal); Methodology (equal); Validation (equal); Writing‐review & editing (equal). Jérôme Le Goff: Funding acquisition (equal); Supervision (equal); Writing‐review & editing (equal). Constance Delaugerre: Funding acquisition (equal); Methodology (equal); Supervision (equal); Writing‐review & editing (equal). Nicolas Gendron: Conceptualization (equal); Formal analysis (equal); Funding acquisition (equal); Investigation (equal); Writing‐review & editing (equal). David Smadja: Conceptualization (equal); Funding acquisition (equal); Methodology (equal); Software (equal); Supervision (equal); Writing‐original draft (equal); Writing‐review & editing (equal). Charles Cassius: Conceptualization (equal); Data curation (equal); Formal analysis (equal); Investigation (equal); Methodology (equal); Project administration (equal); Writing‐original draft (equal); Writing‐review & editing (equal).
Supplementary Material
Appendix S1 Supplementary materials and methods.
Figure S1 Clinical presentation of chilblain‐like lesions.
Table S1 Epidemiological and clinical characteristics of the patients.
Table S2 COVID serology at day 0.
Table S3 Follow‐up of circulating markers of endothelial dysfunction.
Table S4 Circulating markers of endothelial dysfunction according to onset of chilblain‐like lesions.
Powerpoint S1 Journal Club Slide Set.
Contributor Information
L. Frumholtz, Dermatology Department AP‐HP Hôpital Saint‐Louis F‐75010 Paris France
J.‐D. Bouaziz, Dermatology Department AP‐HP Hôpital Saint‐Louis F‐75010 Paris France Université de Paris Human Immunology Pathophysiology Immunotherapy INSERM U976 Institut de Recherche Saint‐Louis F‐75010 Paris France.
M. Battistella, Université de Paris Human Immunology Pathophysiology Immunotherapy INSERM U976 Institut de Recherche Saint‐Louis F‐75010 Paris France Pathology Department AP‐HP Hôpital Saint‐Louis F‐75010 Paris France.
J. Hadjadj, Université de Paris Imagine Institute Laboratory of Immunogenetics of Pediatric Autoimmune Diseases INSERM U1163 F‐75015 Paris France Department of Internal Medicine National Reference Centre for Rare Systemic Autoimmune Diseases AP‐HP, Hôpital Cochin F‐75014 Paris France.
R. Chocron, Université de Paris PARCC INSERM F‐75006 Paris France Emergency Department AP‐HP Georges Pompidou European Hospital F‐75015 Paris France.
D. Bengoufa, Immunobiology Department AP‐HP Hôpital Saint‐Louis F‐75010 Paris France
H. Le Buanec, Université de Paris Human Immunology Pathophysiology Immunotherapy INSERM U976 Institut de Recherche Saint‐Louis F‐75010 Paris France
L. Barnabei, Université de Paris Imagine Institute Laboratory of Immunogenetics of Pediatric Autoimmune Diseases INSERM U1163 F‐75015 Paris France
S. Meynier, Université de Paris Imagine Institute Laboratory of Immunogenetics of Pediatric Autoimmune Diseases INSERM U1163 F‐75015 Paris France
O. Schwartz, Institut Pasteur Virus and Immunity Unit F‐75015 Paris France
L. Grzelak, Institut Pasteur Virus and Immunity Unit F‐75015 Paris France
N. Smith, Institut Pasteur Translational Immunology Lab F‐75015 Paris France
B. Charbit, Institut Pasteur, Cytometry and Biomarkers UTechS CRT F‐75015 Paris France
D. Duffy, Institut Pasteur Translational Immunology Lab F‐75015 Paris France Institut Pasteur, Cytometry and Biomarkers UTechS CRT F‐75015 Paris France.
N. Yatim, Dermatology Department AP‐HP Hôpital Saint‐Louis F‐75010 Paris France Institut Pasteur Translational Immunology Lab F‐75015 Paris France.
A. Calugareanu, Dermatology Department AP‐HP Hôpital Saint‐Louis F‐75010 Paris France Université de Paris Human Immunology Pathophysiology Immunotherapy INSERM U976 Institut de Recherche Saint‐Louis F‐75010 Paris France.
A. Philippe, Institut Pasteur, Cytometry and Biomarkers UTechS CRT F‐75015 Paris France
C.L. Guerin, Université de Paris Innovative Therapies in Haemostasis INSERM F‐75006 Paris France Institut Curie Cytometry Platform F‐75006 Paris France.
B. Joly, Biological Haematology Department AP‐HP Hôpital Lariboisière F‐75010 Paris France Université de Paris EA3518 Institut de Recherche Saint‐Louis F‐75010 Paris France.
V. Siguret, Biological Haematology Department AP‐HP Hôpital Lariboisière F‐75010 Paris France Université de Paris INSERM UMR S1140 F‐75010 Paris France.
L. Jaume, Dermatology Department AP‐HP Hôpital Saint‐Louis F‐75010 Paris France
H. Bachelez, Dermatology Department AP‐HP Hôpital Saint‐Louis F‐75010 Paris France Université de Paris Imagine Institute Laboratory of Genetics of Skin Diseases INSERM U1163 F‐75015 Paris France.
M. Bagot, Dermatology Department AP‐HP Hôpital Saint‐Louis F‐75010 Paris France Université de Paris Human Immunology Pathophysiology Immunotherapy INSERM U976 Institut de Recherche Saint‐Louis F‐75010 Paris France.
F. Rieux‐Laucat, Université de Paris Imagine Institute Laboratory of Immunogenetics of Pediatric Autoimmune Diseases INSERM U1163 F‐75015 Paris France
S. Maylin, Virology Department AP‐HP Hôpital Saint‐Louis F‐75010 Paris France
J. Legoff, Virology Department AP‐HP Hôpital Saint‐Louis F‐75010 Paris France Université de Paris Team Insight INSERM U976 Institut de Recherche Saint‐Louis F‐75010 Paris France.
C. Delaugerre, Virology Department AP‐HP Hôpital Saint‐Louis F‐75010 Paris France
N. Gendron, Université de Paris Innovative Therapies in Haemostasis INSERM F‐75006 Paris France Hematology Department and Biosurgical Research Lab (Carpentier Foundation) Assistance Publique Hôpitaux de Paris‐Centre Université de Paris (APHP‐CUP) F‐75015 Paris France.
D.M. Smadja, Université de Paris Innovative Therapies in Haemostasis INSERM F‐75006 Paris France Hematology Department and Biosurgical Research Lab (Carpentier Foundation) Assistance Publique Hôpitaux de Paris‐Centre Université de Paris (APHP‐CUP) F‐75015 Paris France.
C. Cassius, Dermatology Department AP‐HP Hôpital Saint‐Louis F‐75010 Paris France; Université de Paris Human Immunology Pathophysiology Immunotherapy INSERM U976 Institut de Recherche Saint‐Louis F‐75010 Paris France.
References
- Galván Casas C, Catalá A, Carretero Hernández G et al. Classification of the cutaneous manifestations of COVID‐19: a rapid prospective nationwide consensus study in Spain with 375 cases. Br J Dermatol 2020; 183:71–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Masson A, Bouaziz J‐D, Sulimovic L et al. Chilblains are a common cutaneous finding during the COVID‐19 pandemic: a retrospective nationwide study from France. J Am Acad Dermatol 2020; 83:667–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hébert V, Duval‐Modeste AB, Joly P et al. Lack of association between chilblains outbreak and severe acute respiratory syndrome coronavirus 2: histologic and serologic findings from a new immunoassay. J Am Acad Dermatol 2020; 83:1434–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viguier M, Pinquier L, Cavelier‐Balloy B et al. Clinical and histopathologic features and immunologic variables in patients with severe chilblains. A study of the relationship to lupus erythematosus. Medicine (Baltimore) 2001; 80:180–8. [DOI] [PubMed] [Google Scholar]
- Hadjadj J, Yatim N, Barnabei L et al. Impaired type I interferon activity and inflammatory responses in severe COVID‐19 patients. Science 2020; 369:718–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Jesus AA, Marrero B et al. Activated STING in a vascular and pulmonary syndrome. N Engl J Med 2014; 371:507–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouaziz JD, Duong T, Jachiet M et al. Vascular skin symptoms in COVID‐19: a French observational study. J Eur Acad Dermatol Venereol 2020; 34:e451–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varga Z, Flammer AJ, Steiger P et al. Endothelial cell infection and endotheliitis in COVID‐19. Lancet 2020; 395:1417–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ackermann M, Verleden SE, Kuehnel M et al. Pulmonary vascular endothelialitis, thrombosis, and angiogenesis in Covid‐19. N Engl J Med 2020; 383:120–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smadja DM, Guerin CL, Chocron R et al. Angiopoietin‐2 as a marker of endothelial activation is a good predictor factor for intensive care unit admission of COVID‐19 patients. Angiogenesis 2020; 23:611–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khider L, Gendron N, Goudot G et al. Curative anticoagulation prevents endothelial lesion in COVID‐19 patients. J Thromb Haemost 2020; 18:2391–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colmenero I, Santonja C, Alonso‐Riaño M et al. SARS‐CoV‐2 endothelial infection causes COVID‐19 chilblains: histopathological, immunohistochemical and ultrastructural study of seven paediatric cases. Br J Dermatol 2020; 183:729–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siguret V, Voicu S, Neuwirth M et al. Are antiphospholipid antibodies associated with thrombotic complications in critically ill COVID‐19 patients? Thromb Res 2020; 195:74–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grzelak L, Temmam S, Planchais C et al. A comparison of four serological assays for detecting anti‐SARS‐CoV‐2 antibodies in human serum samples from different populations. Sci Transl Med 2020; 12:eabc3103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasr SH, Fidler ME, Said SM. Paraffin immunofluorescence: a valuable ancillary technique in renal pathology. Kidney Int Rep 2018; 3:1260–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterlin D, Mathian A, Miyara M et al. IgA dominates the early neutralizing antibody response to SARS‐CoV‐2. Sci Transl Med 2021; 13:eabd2223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wollina U, Karadağ AS, Rowland‐Payne C et al. Cutaneous signs in COVID‐19 patients: a review. Dermatol Ther 2020; 33:e13549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraud‐Kerleroux L, Mongereau M, Cassius C et al. Detection of a second outbreak of chilblain‐like lesions during COVID‐19 pandemic through teledermatology. J Eur Acad Dermatol Venereol 2021; 71:2020–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moghadam P, Frumholtz L, Jaume L et al. Frequency of relapse and persistent cutaneous symptoms after a first episode of chilblain‐like lesion during the COVID‐19 pandemic. J Eur Acad Dermatol Venereol 2021; 35:e566–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Signa S, Sementa AR, Coccia MC et al. Recurrence of previous chilblain lesions during the second wave of COVID‐19: can we still doubt the correlation with SARS‐CoV‐2? J Eur Acad Dermatol Venereol 2021; 35:e475–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aschoff R, Zimmermann N, Beissert S, Günther C. Type I interferon signature in chilblain‐like lesions associated with the COVID‐19 pandemic. Dermatopathology (Basel) 2020; 7:57–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubiche T, Cardot‐Leccia N, Le Duff F et al. Clinical, laboratory, and interferon‐alpha response characteristics of patients with chilblain‐like lesions during the COVID‐19 pandemic. JAMA Dermatol 2021; 157:202–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battesti G, El Khalifa J, Abdelhedi N et al. New insights in COVID‐19‐associated chilblains: a comparative study with chilblain lupus erythematosus. J Am Acad Dermatol 2020; 83:1219–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendel A, Hsu I, Granville DJ. Granzyme B releases vascular endothelial growth factor from extracellular matrix and induces vascular permeability. Lab Invest 2014; 94:716–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omoto Y, Yamanaka K, Tokime K et al. Granzyme B is a novel interleukin‐18 converting enzyme. J Dermatol Sci 2010; 59:129–35. [DOI] [PubMed] [Google Scholar]
- Kwaa AKR, Talana CAG, Blankson JN. Interferon alpha enhances NK cell function and the suppressive capacity of HIV‐specific CD8+ T cells. J Virol 2019; 93:e01541–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao C, Bora SA, Parimon T et al. Cell‐type‐specific immune dysregulation in severely ill COVID‐19 patients. Cell Rep 2021; 34:108590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frumholtz L, Laurent‐Roussel S, Aumaître O et al. Clinical and pathological significance of cutaneous manifestations in ANCA‐associated vasculitides. Autoimmun Rev 2017; 16:1138–46. [DOI] [PubMed] [Google Scholar]
- Michaud M, Moulis G, Balardy L et al. [Cryofibrinogenemia: a single‐centre study at the University Hospital of Toulouse, France]. Rev Med Interne 2015; 36:237–42 (in French). [DOI] [PubMed] [Google Scholar]
- Gómez‐Fernández C, López‐Sundh AE, González‐Vela C et al. High prevalence of cryofibrinogenemia in patients with chilblains during the COVID‐19 outbreak. Int J Dermatol 2020; 59:1475–84. [DOI] [PubMed] [Google Scholar]
- Allez M, Denis B, Bouaziz J‐D et al. COVID‐19‐related IgA vasculitis. Arthritis Rheumatol 2020; 72:1952–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandhu S, Chand S, Bhatnagar A et al. Possible association between IgA vasculitis and COVID‐19. Dermatol Ther 2021; 34:e14551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toubiana J, Poirault C, Corsia A et al. Kawasaki‐like multisystem inflammatory syndrome in children during the covid‐19 pandemic in Paris, France: prospective observational study. BMJ 2020; 369:m2094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rovel‐Guitera P, Diemert MC, Charuel JL et al. IgA antineutrophil cytoplasmic antibodies in cutaneous vasculitis. Br J Dermatol 2000; 143:99–103. [DOI] [PubMed] [Google Scholar]
- Kelley JM, Monach PA, Ji C et al. IgA and IgG antineutrophil cytoplasmic antibody engagement of Fc receptor genetic variants influences granulomatosis with polyangiitis. Proc Natl Acad Sci U S A 2011; 108:20736–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heineke MH, Ballering AV, Jamin A et al. New insights in the pathogenesis of immunoglobulin A vasculitis (Henoch–Schönlein purpura). Autoimmun Rev 2017; 16:1246–53. [DOI] [PubMed] [Google Scholar]
- Savage CO, Gaskin G, Pusey CD, Pearson JD. Anti‐neutrophil cytoplasm antibodies can recognize vascular endothelial cell‐bound anti‐neutrophil cytoplasm antibody‐associated autoantigens. Exp Nephrol 1993; 1:190–5. [PubMed] [Google Scholar]
- Philippe A, Chocron R, Gendron N et al. Circulating Von Willebrand factor and high molecular weight multimers as markers of endothelial injury predict COVID‐19 in‐hospital mortality. Angiogenesis 2021; 24:505–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magro C, Mulvey JJ, Berlin D et al. Complement associated microvascular injury and thrombosis in the pathogenesis of severe COVID‐19 infection: a report of five cases. Transl Res 2020; 220:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix S1 Supplementary materials and methods.
Figure S1 Clinical presentation of chilblain‐like lesions.
Table S1 Epidemiological and clinical characteristics of the patients.
Table S2 COVID serology at day 0.
Table S3 Follow‐up of circulating markers of endothelial dysfunction.
Table S4 Circulating markers of endothelial dysfunction according to onset of chilblain‐like lesions.
Powerpoint S1 Journal Club Slide Set.


