Abstract
Max is a common dimerization partner for a family of transcription factors (Myc, Mad [or Mxi]), and Mnt [or Rox] proteins) that regulate cell growth, proliferation, and apoptosis. We recently characterized a novel Max-like protein, Mlx, which interacts with Mad1 and Mad4. Here we describe the cloning and functional characterization of a new family of basic helix-loop-helix–leucine zipper heterodimeric partners for Mlx termed the Mondo family. MondoA forms homodimers weakly and does not interact with Max or members of the Myc or Mad families. MondoA and Mlx associate in vivo, and surprisingly, they are localized primarily to the cytoplasm of cultured mammalian cells. Treatment of cells with the nuclear export inhibitor leptomycin B results in the nuclear accumulation of MondoA and Mlx, demonstrating that they shuttle between the cytoplasmic and nuclear compartments rather than having exclusively cytoplasmic localization. MondoA preferentially forms heterodimers with Mlx, and this heterocomplex can bind to, and activate transcription from, CACGTG E-boxes when targeted to the nucleus via a heterologous nuclear localization signal. The amino termini of the Mondo proteins are highly conserved among family members and contain separable and autonomous cytoplasmic localization and transcription activation domains. Therefore, Mlx can mediate transcriptional repression in conjunction with the Mad family and can mediate transcriptional activation via the Mondo family. We propose that Mlx, like Max, functions as the center of a transcription factor network.
The precise regulation of gene expression during cell growth and differentiation requires active repression and transactivation mechanisms. The Max network of basic helix-loop-helix–leucine zipper (BHLHZip) transcription factors clearly illustrates this point (27). Max is a dimerization partner for the Myc family of transcriptional activators (c-Myc, N-Myc, and L-Myc) and the Mad family of transcriptional repressors (Mad1, Mxi1, Mad3, Mad4, and Mnt [also called Rox]) (20, 27, 29, 38). Consistent with a role in regulating proliferation and growth, Myc-Max heterocomplexes drive cell growth and division and Myc expression is down-regulated during cellular differentiation. By contrast, Mad-Max complexes inhibit proliferation and are upregulated during cellular differentiation (1, 2, 9, 23, 37). Therefore, via the relative activities of the Myc-Max and Mad-Max heterodimers, the Max transcription factor network is thought to regulate growth and differentiation.
Considerable evidence supports the positive effects of the Myc family and the negative effects of the Mad family on cell growth and proliferation. Overexpression of Myc proteins can transform primary rat embryo fibroblasts in cooperation with a number of oncogenes, most notably activated Ras (27). Expression of Mad family genes or Mnt blocks transformation by Myc plus activated Ras (14, 16, 29, 30, 34, 52). The opposing functions of the Myc and Mad families are also observed in mice null for members of the myc or mad family and in transgenic mice overexpressing c-Myc or Mad1. Mouse embryos null for c-myc arrest development at day 9.5 postcoitum (18), and N-Myc null mice fail to fully develop the heart, lungs, and nervous system (15, 40). By contrast, mad1 null animals have defects in the differentiation capacity of granulocyte cluster-forming cells (24), and mxi1 null mice show increased proliferation in precursor cell populations of the prostatic epithelium (48). The phenotypes generated by overexpression of myc and mad family genes are reciprocal to those seen in the genetic null animals; c-myc overexpression in murine B cells results in cells that are larger than normal (31), and transgenic mice overexpressing mad1 are smaller than normal (45). Therefore, these phenotypes are entirely consistent with the proposed positive effects of the myc family and the negative effects of the mad family in regulating cell growth and differentiation. These opposing effects of the Myc and Mad families are likely also active in Drosophila melanogaster because flies homozygous for hypomorphic alleles of dmyc are smaller than normal (25) and overexpression of dmyc in wing imaginal discs yields cells that are larger than normal (32).
Myc-Max and Mad-Max heterocomplexes both recognize the CACGTG subclass of E-box elements; however, Myc-Max heterocomplexes activate transcription while Mad-Max complexes repress transcription (5, 9, 47). The opposing effects of Myc and Mad may manifest themselves by reciprocally regulating the expression of genes involved in cell growth and proliferation. Consistent with this hypothesis, Myc transcriptional targets include genes involved in metabolism (CAD, ornithine decarboxylase, lactate dehydrogenase A, and dihydrofolate reductase genes) and cell cycle progression (cyclin A, cyclin D2, cyclin E, and telomerase genes) (13, 17, 26, 44). Furthermore, Mxi1 can downregulate ornithine decarboxylase expression (55), and cyclin D2-associated kinase activity is dramatically reduced in fibroblasts that overexpress Mad1 (45).
Several experiments suggest that Myc and Mad family proteins utilize Max to activate or repress transcription. Yeast two-hybrid, DNA binding, and coprecipitation assays all demonstrate that Myc and Mad proteins form homodimers poorly but readily form heterodimers with Max. Mutations that prevent the formation of Myc-Max or Mad-Max heterodimers block the ability of Myc or Mad proteins to activate or repress transcription, respectively. Max proteins with mutations in the basic region function in a dominant-negative manner to block the transcriptional activities of both Myc and Mad proteins (for examples and review, see references 7, 11, 12, 27, 42, and 53). Therefore, Max is a cofactor utilized by the Myc and Mad proteins to bind DNA and produce changes in gene expression.
We recently characterized a new Max-like BHLHZip protein called Mlx (Max-like protein X) that also forms functional heterodimers with Mad1 (10). Database searches revealed that the Mlx BHLHZip domain is most similar to that of Max. In addition, Mlx shares many biochemical properties with Max. Like Max, Mlx forms homodimers poorly and has no intrinsic transcriptional activity. Mlx and Mad1 readily form heterodimers that bind the CACGTG subclass of E-boxes. Like Mad1-Max heterocomplexes, Mad1-Mlx heterocomplexes repress transcription by a mechanism that depends on the recruitment of the mSin3A-histone deacetylase corepressor complex. These findings, and the sequence similarity of Mlx and Max, suggest that Mlx also functions as the center of a transcription factor network. Similar to the Max network, the Mad family constitutes the negative side of the proposed Mlx network. However, Mlx does not interact with members of the Myc family (10). Therefore, we proposed that the Mlx network might have a positive side composed of new transcriptional activators.
In this study, we describe the cloning of a novel BHLHZip heterodimerization partner for Mlx that constitutes the positive side of the Mlx network. Because of the large size of the mRNA and the protein product, we have called this new protein MondoA. We find that Mlx and MondoA localize to the cytoplasm in cultured mammalian cells but shuttle through the nucleus. Despite their cytoplasmic localization, MondoA-Mlx heterocomplexes activate transcription from CACGTG-dependent reporters when targeted to the nucleus. Structure-function analysis of a Mondo family member, MondoA, identified a conserved region in the amino terminus that contains separable transcriptional activation and subcellular localization domains. Therefore, like Max, Mlx can mediate the transcriptional activities of at least two families of transcriptional regulators and as such is likely to function as the center of a transcription factor network.
MATERIALS AND METHODS
Two-hybrid screening.
The entire open reading frame of human Mlx (10) was cloned into pBTM116 to generate a LexA-Mlx fusion. LexA-Mlx was introduced into the L40 yeast strain and was used to screen a cDNA library constructed from 9.5 and 10.5 embryos (28) as previously described (8, 10).
Cloning human mondoA and invertebrate mondo homologues.
The open reading frame of human mondoA contains a GC-rich region that is not efficiently reverse transcribed. In order to obtain clones for this region, cDNA libraries were constructed with double-selected mRNA prepared from K562 cells with oligo(dT) or random hexamers as primers, and the double-stranded cDNA was size selected for molecules larger than 1 kb. The cDNA was ligated into pCDNA3 (Invitrogen), and primary clones were arrayed in 96-well plates with an average density of 100 per well. The pools were spotted in duplicate onto nylon membranes at high density and then screened using conventional 32P-labeled DNA probes. In addition, three minilibraries were constructed using gene-specific primers. A total of 6 million clones were screened to obtain three overlapping cDNAs used to reconstruct the open reading frame. The full-length MondoA cDNA was cloned into both pcDNA3 (Invitrogen) and pcDNA3.1/V5 (Invitrogen) in frame with the V5 epitope.
P1 clones for mondoA and mondoB were obtained by screening a human P1 library (Genome Systems) with random-primed 32P-labeled fragments of the respective cDNAs. The chromosomal localization of the human mondo genes was determined by in situ hybridization using the P1 clones as probes and was performed by Genome Systems. The chromosomal localization of mondoB was confirmed by the recent completion of the draft sequence of the human genome (GenBank accession no. AC073846).
The Caenorhabditis elegans mondo homologue is located on cosmid T20B12. The predicted open reading frame obtained from A Caenorhabditis elegans Data Base, or ACeDB, differs from the open reading frame deduced by sequencing cDNAs (kindly provided by Y. Kohara). The dmondo gene was identified by BLAST searches (using mondoA as the query) of the Berkeley Drosophila Genome Project expressed sequence tag (EST) and genomic sequence database. The open reading frame was deduced by sequencing the ESTs LD31826 (which contains the initiating ATG and stops in all three upstream reading frames) and LD27794 (which is a partial cDNA). All cDNAs were obtained from the Berkeley Drosophila Genome Project.
Expression analysis.
Northern blot analysis was performed by probing adult tissue blots (Clontech) with random-primed 32P-labeled mondoA and mondoB probes at high stringency. There was no cross-reactivity between the mondoA and mondoB probes.
Transcription assays.
Luciferase reporter assays and cell transfections were performed in triplicate, with the error given as the standard error of the mean as previously reported (10). Transfection efficiency was normalized by cotransfection of a LacZ expression plasmid. Mutant derivatives of mlx and mondoA were made using PCR with TurboPFU (Stratagene) and were verified by sequencing. Gal4 DNA binding domain (Gal4DBD) fusions were made in the vector pFA (Stratagene). The sequence of the simian virus (SV40) large T antigen is MAPKKKRKV.
Antibodies and protein interaction assays.
Antibodies against Mlx were directed against the carboxy terminus of the protein (10). Antibodies against a glutathione S-transferase (GST) fusion of the BHLHZip domain of murine MondoA were raised in rabbits. The specificity of the antiserum was tested by immunoprecipitating in vitro-translated [35S]Met-labeled MondoA with or without blocking antigen. Bleeds were also tested for their ability to immunoprecipitate MondoA from [35S]Met-labeled P19 cells. Indirect immunofluorescence was performed using standard procedures (4). Anti-FLAG (Sigma), -V5 (Invitrogen), and -Gal4 (Santa Cruz) monoclonal antibodies were used at 1:500. Rabbit polyclonal anti-MondoA and anti-Mlx antibodies were used at 1:500. Fluorescein isothiocyanate-conjugated sheep anti-mouse (Jackson ImmunoResearch Laboratories) and Alexa 594 goat anti-rabbit (Molecular Probes) antibodies were used at 1:500 to detect the primary antibodies. Cells were analyzed 24 h after transfection. To quantify staining patterns, each transfection was performed at least twice in duplicate and scored blind. The cells were counterstained with Hoechst 33342 (5 μg/ml) to discriminate nuclei, and only cells expressing both proteins were counted. A minimum of 50 cells were counted for each transfection. The cells were treated with 10 ng of leptomycin B (kindly provided by M. Yoshida)/ml for 10 h.
Immunoprecipitations and Western blotting were carried out as previously described with the following modifications (6, 36). Cytoplasmic fractions were prepared by resuspending cell pellets in buffer H (10 mM Tris-HCl [pH 7.9], 10 mM KCl, 1.5 mM MgCl2, 1 mM dithiothreitol, 0.1% NP-40, and “complete” protease inhibitor) (Boehringer Mannheim/Roche), vortexing them briefly, and incubating them for 10 min on ice. Nuclei were pelleted for 7 min at 2,000 × g and 4°C in a Sorvall tabletop centrifuge. Following a second wash in buffer H, the supernatants were pooled and centrifuged for 10 min at 10,000 × g and 4°C. The protein concentrations of the supernatants were determined by Bradford assay. Anti-Mlx immunoprecipitation was carried out on 3 mg of total cytoplasmic protein in buffer H plus 175 mM KCl. Western blots of precipitations were probed with anti-Mlx amino terminus (1:1,000) or anti-MondoA (1:500) antiserum.
Far-Western blots were performed by first transferring proteins separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) to polyvinylidene difluoride (PVDF) membranes and incubating the membranes in a blocking solution (1× phosphate-buffered saline, 1 mM dithiothreitol, 5% nonfat dry milk, 0.2% NP-40, 10% glycerol) for 1 h at room temperature. Next, the membranes were incubated in the same solution with 1% nonfat dry milk mixed with a single 50-μl TNT (Promega) in vitro translation reaction programmed to produce [35S]Met-labeled Mlx protein. The blot was incubated for 4 h at 4°C and then washed extensively (six times over an hour) with block solution without nonfat dry milk. The blot was dried and autoradiographed from overnight to 4 days with a Low Energy Biomax Transcreen (Kodak). Gel shift assays were performed as described previously (7) with a 32P-labeled CACGTG oligonucleotide and in vitro-translated Mlx or ΔLZMlx used in combination with a purified GST fusion protein containing the BHLHZip domain of MondoA.
DNA pulldown reactions were carried out in buffer H plus 175 mM KCl. Three milligrams of total cytoplasmic protein was incubated with biotinylated double-stranded oligonucleotide DNA (25 nM in a total volume of 500 μl) for 20 min at room temperature. DNA-protein complexes were precipitated by incubation with UltraLink Neutravidin beads (25 μl of a 50% slurry) (Pierce) for 30 min at 4°C with rocking. Following incubation, the beads were washed four times in 1 ml of buffer H plus 175 mM KCl and then resuspended in SDS-PAGE sample buffer. The oligonucleotide pair sequences were wild-type CM1 and Biotin (GATCCCCCCACCACGTGGTGCCTG and GATCCAGGCACCACGTGGTGGGGG), and mutant CM1 and Biotin (GATCCCCCCACCACCTGGTGCCTG and GATCCAGGCACCAGGTGGTGGGGG) (underlining marks the E-box core sequence).
Nucleotide sequence accession numbers.
GenBank accession numbers for H. sapiens MondoA, C. elegans Mondo, and D. melanogaster Mondo are AF312918, AAA19059, and AAF53988, respectively. H. sapiens MondoB is identical to the recently described putative hepatic transcription factor WBSCR14 (19). The accession number for WBSCR14 is AAF68174.
RESULTS
Cloning and characterization of a novel Mlx-binding protein.
Like Max, Mlx may function as a dimerization partner for many transcription factors. To determine if we could identify additional Mlx-interacting proteins, Mlx was immunoprecipitated from P19 cells under nondenaturing conditions and the immunoprecipitates were resolved by SDS-PAGE and transferred to PVDF membranes. The membranes were probed with [35S]methionine-labeled Mlx protein (far-Western blotting). A prominent and specific polypeptide of approximately 130 kDa was detected (Fig. 1A). Further, this protein was not detected in immunoprecipitates incubated with the cognate Mlx antigen (Fig. 1A, Block), demonstrating that it associated specifically with Mlx. Thus, Mlx appears to directly associate with at least one protein in P19 cells.
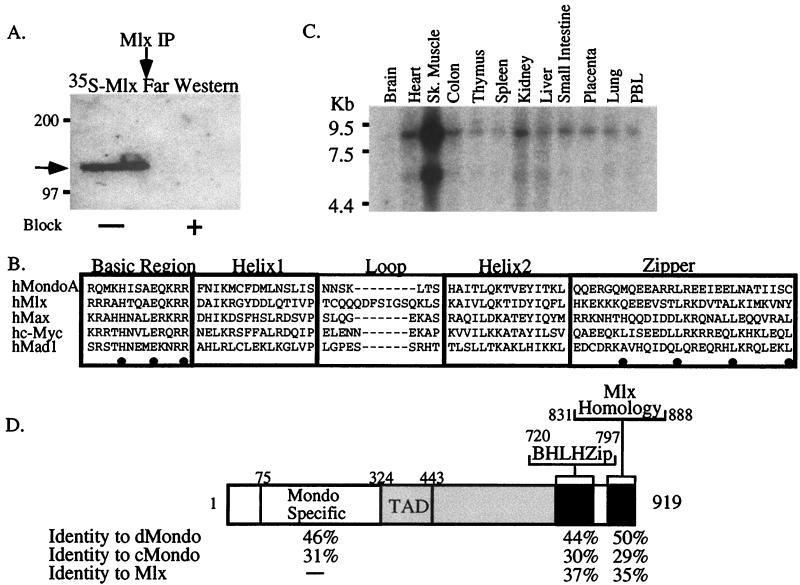
FIG. 1.
Identification and cloning of an Mlx-binding protein. (A) Mlx and associated proteins were immunoprecipitated from whole-cell extract with Mlx antibodies and probed in a far-Western blot with [35S]Met-labeled Mlx. The arrow indicates an ∼130-kDa Mlx-binding protein. Block, incubation (+) of the antibody with cognate antigen as a control for the specificity of the immunoprecipitation reaction. (B) Alignment of the BHLHZip domain of MondoA with Mlx and members of the Max network. Conserved residues in the basic region predicted to dictate binding to CACGTG are marked by dots, as are the hydrophobic amino acids that make up the leucine zipper. The dashes indicate gaps in the aligned sequences. (C) Expression of mondoA in adult human tissues as determined by Northern blot analysis (Sk. muscle, skeletal muscle; PBL, peripheral blood lymphocytes). (D) Domain diagram of the MondoA protein. The percentages of amino acid identity between MondoA and D. melanogaster Mondo (dMondo), C. elegans Mondo (cMondo), and H. sapiens Mlx are shown. The serine-threonine-proline (STP) rich domain is shaded. The TAD is within the STP domain.
In order to clone Mlx-binding proteins, we carried out a yeast two-hybrid screen using full-length Mlx as bait. Approximately 107 primary transformants were obtained from a day 9.5 and 10.5 mouse embryo random-primed VP16 fusion cDNA library. Of these transformants, 40 colonies scored positive in the interaction assay. One of the colonies contained a VP16-Mad4 fusion clone, a previously known Mlx-binding partner (10). The remaining 39 clones harbored a novel BHLHZip domain. This BHLHZip domain is homologous to members of the Max family, and it is most similar to the BHLHZip domain of Mlx (Fig. 1B). The 13-amino-acid basic region contains the conserved residues required for high-affinity binding to CACGTG E-boxes (His5, Glu9, and Arg13) and is therefore predicted to bind to this DNA sequence (3, 21, 22). To determine whether this new BHLHZip domain could interact with members of the Max network, we tested it for interaction with Max, c-Myc, L-Myc, N-Myc, Mad1, Mxi, Mad3, and Mad4 in a directed two-hybrid assay. None of these Max network proteins interacted significantly with the new BHLHZip domain (data not shown), suggesting that it dimerizes specifically with Mlx but not with members of the Max network.
Northern blot analysis demonstrated that the transcript encoding this BHLHZip protein is approximately 9 kb in length, with a minor transcript of approximately 5.5 kb (Fig. 1C and data not shown). We termed this new gene mondoA. Expression of mondoA is highest in skeletal muscle, but it is expressed in most adult tissues (Fig. 1C). Subsequent database searches uncovered an EST encoding a portion of another mondo family member, mondoB. Northern blot analysis of mondoB expression revealed a broadly expressed transcript of approximately 4 kb (data not shown). Therefore, the 5.5-kb transcript detected with the mondoA probe is unlikely to be mondoB. In situ hybridization of mouse embryos with mondoA showed that the transcript is broadly expressed from day 9.5 to at least day 15 postcoitum, with slightly elevated levels in the developing central nervous system (data not shown). The chromosomal localizations of human mondoA (mondoA) and human mondoB (mondoB) were determined by in situ hybridization.
The mondoA and mondoB genes are located at 12q21.31 and 7q11.23, respectively. Williams syndrome, which has complex clinical features, including supravalvular aortic stenosis, impaired visual-spatial constructive cognition, mental retardation, and infantile hypercalcemias, is caused by a contiguous-genome deletion of greater than 1 Mb at 7q11.23 (50). This deletion results in the loss of one allele of the mondoB locus (19), suggesting that haploinsufficiency of mondoB may contribute to the disease.
We cloned and sequenced the complete open reading frame of mondoA. To facilitate the identification of functional domains, we also cloned mondo homologues from D. melanogaster and C. elegans. MondoA is approximately 23% identical and 35% similar over its entire length to its homologs from D. melanogaster and C. elegans (data not shown). A domain map of MondoA is shown in Fig. 1D. The open reading frame codes for a 919-amino-acid protein. The amino terminus is conserved among all members of the mondo family but has no sequence homology to other known proteins (Fig. 1D). We have termed this region the mondo-specific region, or MSR. A serine-threonine-proline-rich region (12% serine, 12% threonine, and 19% proline) follows the MSR in MondoA (Fig. 1D). The BHLHZip region is located in the carboxy-terminal third of the protein. The BHLHZip region is followed by a region in the carboxy terminus that has homology to the carboxy-terminal region of Mlx (10). The function of this region is not yet known, but its conservation suggests that mlx and mondo genes may have a common evolutionary ancestry. The analysis described in this report focuses on the human MondoA protein.
To determine whether MondoA associates with Mlx at normal physiological concentrations, whole-cell extracts from P19 cells were immunoprecipitated with Mlx antibodies and analyzed by far-Western blotting as shown in Fig. 1. Consistent with the previous result, a single specific band was detected (Fig. 2A, left gel). To determine whether this Mlx-binding protein was MondoA, the same membrane was reprobed with antibodies specific for the BHLHZip domain of MondoA. A band that migrated similarly to that of the Mlx-binding protein was detected by anti-MondoA antibodies (Fig. 2A, right gel), demonstrating that the Mlx-binding protein is MondoA. Together, these data demonstrate that under physiological conditions, MondoA-Mlx heterodimers exist in P19 cells.
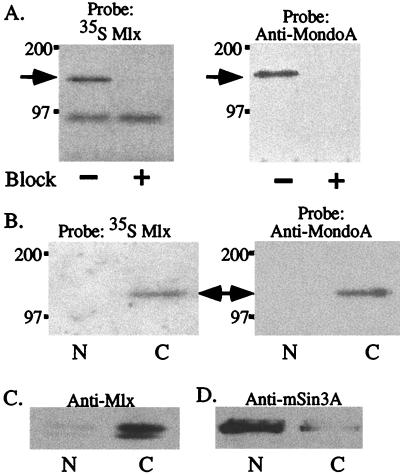
FIG. 2.
Mlx and MondoA associate in the cytoplasm. (A) Mlx immunoprecipitates were resolved on SDS gels and transferred to a PVDF membrane, and the blot was sequentially probed with 35S-labeled Mlx (left) or anti-MondoA (right) antiserum. Block, incubation of the antibody (+) with cognate antigen as a control for the specificity of the immunoprecipitation reaction. (B) Nuclear (N) and cytoplasmic (C) compartments were prepared by hypotonic lysis, immunoprecipitated with Mlx antibodies, and probed in a far-Western blot with [35S]Met-labeled Mlx (left) or anti-MondoA (right) antiserum. An ∼130-kDa band is detected in the cytoplasmic fraction (arrow). (C and D) Nuclear and cytoplasmic fractions were immunoprecipitated with Mlx (C) or mSin3 (D) antibodies, and the immunoprecipitates were examined by Western blotting with Mlx or mSin3A antibodies.
We also determined the subcellular localization of MondoA in P19 cells by far-Western blotting. Nuclear and cytoplasmic fractions were prepared and immunoprecipitated with Mlx antibody. Surprisingly, the Mlx-binding activity of MondoA was found in the cytoplasm (Fig. 2B). This result suggested that Mlx might also be localized to the cytoplasm. To test this idea, Mlx was immunoprecipitated from cytoplasmic or nuclear extracts and detected by Western blotting. The majority of Mlx protein was also in the cytoplasmic fraction, with small amounts present in the nuclear fraction (Fig. 2C). In contrast, mSin3A, a known nuclear protein, was found primarily in the nuclear fraction, with only small amounts localized in the cytoplasmic fraction (Fig. 2D). A similar analysis demonstrated that Mlx is primarily restricted to the cytoplasm of HL60, K562, and PC12 cells (data not shown).
MondoA binds CACGTG E-boxes and activates transcription.
MondoA and Mlx heterodimerize and are predicted, based on primary amino acid sequence, to bind CACGTG E-box sequences. To determine whether P19 cells contained E-box-binding activity associated with MondoA-Mlx heterodimers, P19 cytoplasmic extracts were incubated with double-stranded CACGTG oligonucleotides immobilized on beads and, following extensive washing, retention of MondoA-Mlx heterodimers on the DNA beads was determined by Western blotting. Both Mlx and MondoA were retained on the wild-type oligonucleotide beads (Fig. 3A, lanes 2 and 6) but not on the mutant control oligonucleotide beads (Fig. 3A, lanes 3 and 7). In addition, a positive control immunoprecipitation of cytoplasmic extracts with Mlx antibody also coimmunoprecipitated Mlx and MondoA (Fig. 3A, lanes 1 and 5). Therefore, MondoA-Mlx heterodimers can be isolated by immunoprecipitation, and both MondoA and Mlx can be isolated by using immobilized CACGTG binding sites. These results suggest that the DNA binding activity is a heterocomplex of Mlx and MondoA; however, it is possible that Mlx and MondoA homodimers are also present in the cytoplasm and the observed DNA binding resulted from homodimer rather than heterodimer binding.
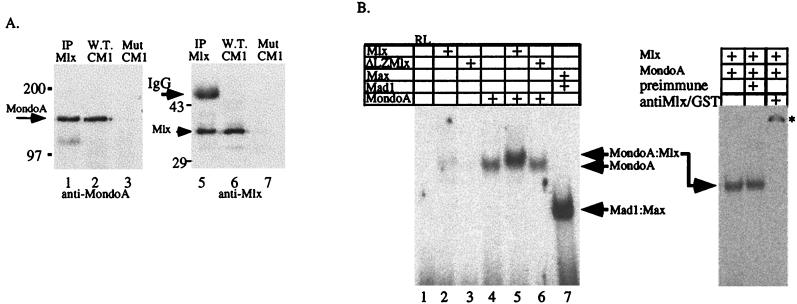
FIG. 3.
Mlx and MondoA interact in vivo and bind CACGTG sequences as heterodimers. (A) Cytoplasmic extracts of P19 cells were incubated with either CACGTG E-box (W.T. CM1) or mutant E-box CACCTG (Mut CM1) oligonucleotides immobilized on agarose beads. As a control, cell extracts were also precipitated with α-Mlx (IP Mlx). The arrow indicates MondoA (left) or Mlx (right). (B) EMSA was used to show specific dimerization and DNA binding by MondoA and Mlx. Proteins in the indicated combinations were bound to a 32P-labeled oligonucleotide containing a single CACGTG binding site. ΔLZMlx lacks the leucine zipper and cannot heterodimerize with Mondo. RL, reticulocyte lysate alone. On the right, MondoA-Mlx heterocomplexes were incubated with either preimmune or antiMlx/GST serum. The asterisk indicates the supershifted heterocomplex. +, present.
To test whether MondoA-Mlx heterodimers are favored over homodimers of either protein, an electrophoretic mobility shift assay (EMSA) was performed using a recombinant GST fusion protein containing the BHLHZip domain of MondoA and in vitro-translated Mlx. Previous results have demonstrated that Mlx homodimerizes poorly (10), and we obtained similar results (Fig. 3B, lane 2). Under the conditions used, MondoA formed homodimers weakly on the CACGTG probe (Fig. 3B, lane 4). However, Mlx and MondoA formed a specific complex that migrated with lower mobility than MondoA homodimers, indicating the formation of heterodimers (Fig. 3B, lane 5). Antibodies against Mlx and GST, which recognize the GST portion of the GST-Mondo fusion protein, completely supershift the heterodimer, confirming the presence of both proteins in the complex (Fig. 3B, right, and data not shown). A mutant Mlx protein lacking its leucine zipper (ΔLZMlx) could not heterodimerize with MondoA (Fig. 3B, lane 6). Thus, Mlx and MondoA bind DNA as heterodimers, and heterodimerization requires the leucine zipper of Mlx. Furthermore, the EMSA data suggest that MondoA-Mlx heterodimers are preferred over homodimers of either protein. Together, these experiments suggest that the cytoplasmic DNA binding activity is composed primarily of MondoA-Mlx heterodimers as opposed to homodimers of either MondoA or Mlx.
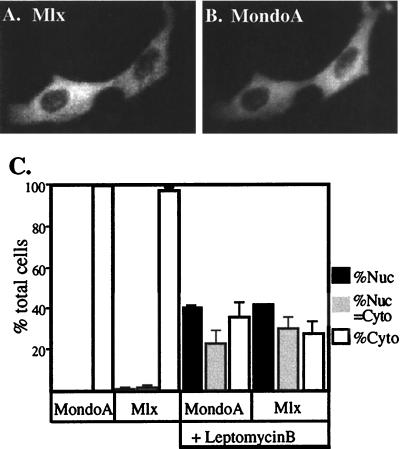
Our biochemical fractionation (Fig. 2B and data not shown) suggests that Mlx and MondoA are localized primarily to the cytoplasm. The high degree of evolutionary conservation in the BHLHZip domains of both MondoA and Mlx, however, strongly suggests that they function in the nucleus to regulate gene expression. It is possible that MondoA-Mlx heterodimers are retained in the cytoplasm and only translocate to the nucleus in response to extracellular signaling or that they enter the nucleus but are rapidly exported. If the latter is true, it would be expected that the heterodimer would not accumulate to high levels in the nucleus at steady state. To test whether MondoA-Mlx heterodimers are actively exported from the nucleus, we transfected NIH 3T3 cells, which do not express significant amounts of MondoA or Mlx (data not shown), with expression vectors encoding FLAG-tagged Mlx and MondoA and treated them with the nuclear export inhibitor leptomycin B (35, 54). MondoA and Mlx subcellular localization was monitored by indirect immunofluorescence, and we quantified the effect of leptomycin B by determining the percentage of cells that displayed exclusively cytoplasmic, nuclear, or equivalent cytoplasmic and nuclear staining. As expected, MondoA and Mlx were localized almost exclusively to the cytoplasm in untreated cells (Fig. 4). By contrast, leptomycin B treatment resulted in a dramatic relocalization of MondoA and Mlx to the nucleus (Fig. 4C). Therefore, consistent with a role for MondoA-Mlx heterodimers in transcriptional regulation, they are not exclusively cytoplasmic proteins but rather are present transiently in the nucleus.
FIG. 4.
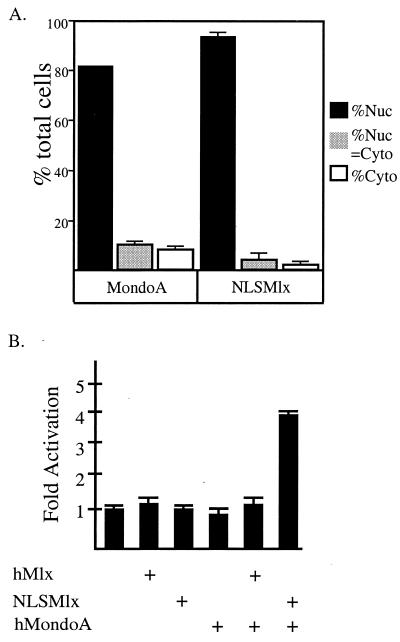
Mlx and Mondo shuttle between the cytoplasmic and nuclear compartments. Expression vectors encoding FLAG-tagged Mlx and MondoA were transfected into NIH 3T3 cells. Their subcellular localization was determined by indirect immunofluorescence using anti-FLAG and anti-MondoA antibodies. (A and B) Two representative cells coexpressing Mlx and Mondo, respectively. (C) The subcellular distribution of Mlx and MondoA was quantified in the absence or presence (+) of leptomycin B. Nuc, nuclear; Cyto, cytoplasmic. The error bars indicate standard deviations.
To test the transcriptional activity of MondoA-Mlx heterodimers, we sought to artificially target them to the nucleus. To do so, we fused the strong nuclear localization signal (NLS) of SV40 large T antigen to the amino terminus of Mlx (NLSMlx). We initially determined the subcellular localization of MondoA and NLSMlx by immunofluorescence as described above. In contrast to wild-type MondoA and Mlx (Fig. 4), NLSMlx and coexpressed MondoA localized almost exclusively to the nucleus (Fig. 5A). The nuclear colocalization of MondoA and NLSMlx provides further evidence for their in vivo interaction. To determine the transcriptional activity of MondoA-Mlx heterodimers, we transfected NIH 3T3 cells with MondoA, Mlx, or NLSMlx expression vectors along with a luciferase reporter gene containing four CACGTG binding sites. Expression of Mlx, NLSMlx, MondoA alone, or the combination of Mlx and MondoA did not appreciably affect reporter gene expression. However, coexpression of MondoA with NLSMlx resulted in a fourfold increase in reporter gene expression (Fig. 5B). Mlx does not possess intrinsic transcriptional activity (10); hence, the transcriptional activation domain (TAD) must be supplied by MondoA. Transcriptional activation depended on the presence of E-box sites in the reporter, the DNA binding activity of Mlx, and the leucine zipper of Mlx (data not shown). Therefore, even though MondoA-Mlx heterodimers are predominantly cytoplasmic, they activate transcription when redistributed to the nucleus.
FIG. 5.
Nuclear-targeted MondoA-Mlx is a transcriptional activator. (A) V5 epitope-tagged Mondo and NLSMlx were transfected into NIH 3T3 cells, and their subcellular localization was determined and quantified by immunofluorescence. MondoA and Mlx were detected using anti-V5 and anti-Mlx, respectively, as the primary antibodies. (B) Transient-transfection assays were performed to determine the transcriptional activities of NLSMlx and MondoA on CACGTG E-boxes in NIH 3T3 cells. The cells were transfected with 0.5 μg of Mlx, NLSMlx, or MondoA in the combinations shown, along with a luciferase reporter containing four CACGTG binding sites. The results are shown as fold activation, and the error is expressed as the standard error of the mean. +, present; Nuc, nuclear; Cyto, cytoplasmic.
Identification of a CLD in MondoA.
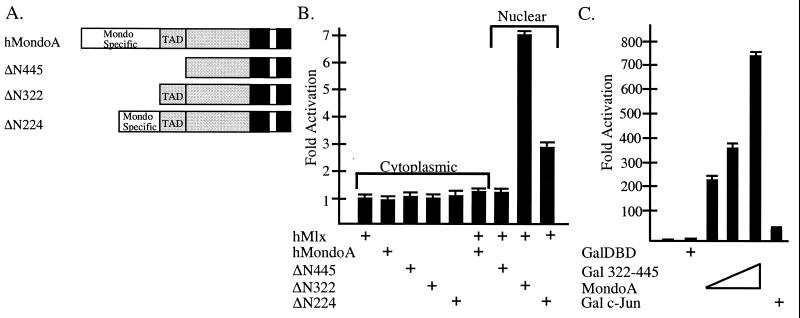
To investigate the subcellular localization of MondoA-Mlx heterodimers, we sought to identify domains in MondoA that regulate their cytoplasmic localization. Mondo proteins contain a unique and highly conserved amino terminus (MSR [Fig. 1D]); therefore, we thought it might be involved in regulating subcellular localization. A series of MondoA amino-terminal truncation mutants, ΔN224, ΔN322, and ΔN445 (diagramed in Fig. 6A), were each coexpressed with Mlx in NIH 3T3 cells, and the subcellular localization of both proteins was quantified by immunofluorescence as described above. Data from a minimum of 100 cells were obtained from two independent transfections. As before, both Mlx and MondoA localized to the cytoplasm of coexpressing cells (Table 1 and Figure 4). However, when Mlx was coexpressed with either ΔN445MondoA or ΔN322MondoA, Mlx and each MondoA protein localized to the nucleus (Table 1). Therefore, the sequences in MondoA that regulate localization of MondoA-Mlx heterocomplexes are located amino terminal to residue 322. By contrast, coexpression of Mlx and ΔN224MondoA resulted in their equivalent distribution to both the nucleus and cytoplasm (Table 1). Thus, the ΔN224MondoA mutant partially restores cytoplasmic localization. Therefore, these data suggest that amino acids 224 to 322 of MondoA function as a cytoplasmic localization domain (CLD), although additional sequences upstream of amino acid 224 are required for full cytoplasmic localization.
FIG. 6.
The amino terminus of MondoA contains an autonomous transcriptional domain. (A) Diagram illustrating the amino-terminal MondoA deletions. hMondoA, human MondoA. (B) The amino-terminal deletion series of MondoA was tested for transcriptional activation when coexpressed with Mlx. The subcellular localization of coexpressed Mlx and MondoA is indicated. (C) Amino acids 322 to 445 of MondoA were fused to the Gal4DBD and tested for activity in NIH 3T3 cells using a luciferase reporter gene containing the thymidine kinase promoter and four Gal4 binding sites. Gal4DBD (450 ng), Gal4–c-Jun (450 ng), or increasing amounts (50, 150, and 450 ng) of Gal4DBD(332-445)MondoA were transfected. The results are shown as fold activation, and the error is expressed as the standard error of the mean. +, present; −, absent.
TABLE 1.
The amino terminus of human MondoA regulates its subcellular localizationa
| Coexpressed proteins | % Nuc | % Nuc = cyto | % Cyto |
|---|---|---|---|
| hMondoA | 0 | 0 | 100 |
| Mlx | 4 ± 1.4 | 6 ± 1.4 | 90 ± 2.8 |
| ΔN445hMondoA | 100 ± .4 | 0 | 0 |
| Mlx | 100 ± 3 | 0 | 0 |
| ΔN322hMondoA | 100 | 0 | 0 |
| Mlx | 98.0 ± 17.3 | 2 ± .01 | 0 |
| ΔN224hMondoA | 21.7 ± 2.7 | 68.8 ± 13 | 9.5 ± 6.7 |
| Mlx | 53.2 ± 6.3 | 42.1 ± 1.2 | 4.8 ± 3.4 |
Percentages are given for the different staining patterns for Mlx and Mondo. Cells were transfected with the indicated combinations of expression plasmids, and the subcellular localization was scored in cells that expressed both proteins. Nuc, exclusively nuclear staining; Cyto, exclusively cytoplasmic staining; Nuc = Cyto, equivalent nuclear and cytoplasmic staining; hMondoA, human MondoA.
To determine if the amino terminus of MondoA could function as a CLD in the context of a heterologous protein, different regions of the amino terminus were fused to the Gal4DBD. Consistent with the finding that the Gal4DBD has an NLS, it localized to the nucleus in approximately 85% of cells (Table 2). By contrast, a fusion of the Gal4DBD to amino acids 82 to 445 of MondoA [Gal4DBD(82-445)MondoA] localized to the cytoplasm in 100% of the expressing cells. Therefore, amino acids 82 to 445 can completely override the activity of the Gal4 NLS, demonstrating that the CLD of MondoA is portable. Furthermore, fusion of the SV40 large-T-antigen NLS to Gal4DBD(82-445)MondoA did not result in a predominant nuclear localization of the chimera. Therefore, even multiple NLSs cannot completely overcome the activity of the CLD. A fusion between amino acids 125 to 321 of MondoA and Gal4DBD was almost completely cytoplasmic, further delineating the CLD. Amino acids 125 and 321 are in the evolutionarily conserved MSR (Fig. 1D), suggesting that cytoplasmic localization of the Mondo family is a conserved aspect of their function.
TABLE 2.
Residues 125 to 321 of human MondoA function as an autonomous CLDa
| Expressed protein | % Nuc | % Nuc = cyto | % Cyto |
|---|---|---|---|
| Gal4 | 85 ± 4.9 | 8 ± 4.9 | 7 ± 2.1 |
| Gal4DBD(82-445)hMondoA | 0 | 0 | 100 |
| Gal4NLS(82-445)hMondoA | 13 ± 3.6 | 43 ± 4.5 | 44 ± 1.4 |
| Gal4DBD(125-321)hMondoA | 0 | 3 ± 1.2 | 97 ± 4.9 |
Categories of staining patterns are the same as those shown in Table 1. hMondoA, human MondoA.
Identification of an autonomous activation domain in MondoA.
MondoA-Mlx heterodimers activated transcription when targeted to the nucleus (Fig. 5B). In order to map the transactivation domain, we tested the MondoA amino-terminal deletions, which, when coexpressed with Mlx, localized to the nucleus, for their ability to regulate a CACGTG E-box reporter gene. Expression of each of these proteins alone did not significantly affect expression of the reporter gene (Fig. 6B). Similarly, expression of either MondoA or ΔN445MondoA with Mlx did not alter the expression of the reporter gene (Fig. 6B). As before (Fig. 5), Mlx and full-length MondoA localized to the cytoplasm and did not activate transcription. By contrast, Mlx and ΔN445MondoA localized to the nucleus (Table 1), suggesting that the first 445 amino acids of MondoA contain both a transactivation domain and the CLD. Strikingly, nuclear localization of Mlx and ΔN322MondoA (Table 1) resulted in an approximately sevenfold increase in reporter gene expression (Fig. 6B), demonstrating the presence of an activation domain between amino acids 322 and 445. Coexpression of Mlx and ΔN224MondoA resulted in approximately threefold activation of the reporter gene, lower than that of ΔN322MondoA (Fig. 6B). Coexpressed Mlx and ΔN224MondoA localized to the nucleus and cytoplasm (Table 1), suggesting that the reduction in transactivation seen for ΔN224MondoA results from a decrease in nuclear localization. Therefore, the amino terminus of MondoA contains a transactivation domain between amino acids 322 and 445.
To test whether amino acids 322 to 445 of MondoA could function as an autonomous activation domain, they were fused to the Gal4DBD and this chimera was tested for its ability to activate a herpes simplex virus thymidine kinase promoter that contains four Gal4 binding sites. Expression of the Gal4DBD alone resulted in little transcriptional activation, while Gal4–c-Jun, a control transcriptional activator, activated transcription approximately 50-fold. Gal4DBD(322-445)MondoA activated transcription 200- to 700-fold, depending on the amount of expression plasmid transfected (Fig. 6C). This Gal4- MondoA fusion also activated transcription to a similar extent from a minimal promoter under the control of four Gal4 binding sites (data not shown). Therefore, the amino-terminal region of MondoA contains an autonomous activation domain between amino acids 322 and 445.
DISCUSSION
Previously, we identified the novel Max-like protein Mlx. Here we describe the novel partner protein for Mlx, termed Mondo. The predicted primary amino acid sequence of the MondoA protein reveals a large BHLHZip protein with two regions of sequence similarity at the amino- and carboxy-terminal regions that are conserved among Homo sapiens, D. melanogaster, and C. elegans mondo homologues. The carboxy-terminal conserved domain is striking because it is conserved among all of the mondo and mlx family members. This suggests that mlx and mondo genes may have a common evolutionary ancestor and that this region may be crucial for aspects of Mlx and Mondo function. The amino terminus of the MondoA protein has at least two functional domains, one that regulates cytoplasmic localization of MondoA-Mlx heterocomplexes and another involved in transactivation (see below). Thus, the mlx and mondo genes form a new family of cytoplasmically localized BHLHZip transcription factors.
Several lines of evidence support the physiological relevance of the interaction between MondoA and Mlx. First, 39 of 40 positive clones in our two-hybrid screen to detect Mlx-interacting proteins included the BHLHZip domain of murine MondoA. Second, murine MondoA did not interact with other members of the Max network in a directed two-hybrid screen. Third, MondoA-Mlx heterodimers bound specifically to CACGTC sites and activated transcription from CACGTG-dependent reporters in a manner that required dimerization and DNA binding. Fourth, MondoA and Mlx localize to the cytoplasm and redistribute to the nucleus together. Finally, we have detected association between MondoA and Mlx in cytoplasmic extracts of P19 cells by coimmunoprecipitation and DNA pulldown assays. Together, these experiments provide compelling evidence that MondoA-Mlx heterocomplexes exist in vivo.
Regulated nuclear import of many transcription factors is a well-established biological control mechanism (51). MondoA-Mlx heterodimers localize predominantly to the cytoplasm; however, we propose that these proteins function in the nucleus. Our strongest experimental support for this proposal comes from targeting the heterocomplex to the nucleus with a strong NLS grafted onto the amino terminus of Mlx. The targeted heterocomplex activated transcription from CACGTG-driven reporter genes. We have not yet identified the conditions or cell types in which wild-type MondoA-Mlx heterocomplexes accumulate in the nucleus, and therefore it is possible that the transcriptional activation attributed to MondoA resulted from artificial nuclear targeting. However, as we observed both nuclear localization and transcriptional activation with wild-type Mlx and a MondoA mutant that lacks the CLD (Δ322MondoA), it is unlikely that the 9-amino-acid NLS supplies an artificial activation domain to Mlx. Furthermore, additional evidence supports a nuclear function for MondoA-Mlx heterodimers: (i) MondoA-Mlx heterodimers shuttle through the nucleus and therefore are not constitutively cytoplasmic proteins, (ii) the DBDs of both MondoA and Mlx are highly conserved across species and are capable of specific CACGTG binding, and (iii) the amino terminus of MondoA has a potent autonomous TAD. Therefore, we propose that MondoA-Mlx heterocomplexes function as transcriptional activators whose nuclear activity is under tight control via their cytoplasmic localization. A signal(s), as yet unidentified, may be required to redistribute the heterocomplex from the cytoplasm to the nucleus. Alternatively, MondoA-Mlx heterodimers may shuttle rapidly through the nucleus and not reach high levels at steady state. We are attempting to distinguish between these two mechanisms.
While we propose a nuclear function for Mondo-Mlx heterodimers, we cannot rule out the possibility that the heterocomplex has additional activities in the cytoplasm. A cytoplasmic function for MondoA might include the sequestration of Mlx, which would limit the access of Mlx to other nuclear dimerization partners, such as Mad1 and Mad4. When expressed on its own, however, Mlx localized to the cytoplasm (A. L. Eilers, unpublished data), suggesting that cytoplasmic localization of Mlx is an intrinsic property and is independent of Mondo. However, at this time we cannot rule out a dominant-negative role for MondoA in regulating the access of Mlx to as-yet-unidentified transcription factors. Mondo-Mlx heterodimers may have cytoplasmic functions, but as our evidence points strongly to a nuclear function for the heterocomplex, it seems unlikely that it has exclusively cytoplasmic functions.
Other members of the Max network BHLHZip superfamily are primarily nuclear but can also be regulated by subcellular localization, and thus this type of regulation is not unique to Mlx and Mondo. For example, though c-Myc is usually nuclear, it is sequestered in the cytoplasm of Purkinje cells via an interaction with the CDR-2 protein. This interaction is thought to block c-Myc-induced cell death in Purkinje cells (43).
To decipher the regulation of cytoplasmic localization by Mondo-Mlx heterodimers, we identified a CLD in MondoA. The CLD (amino acids 125 to 321) provides a strong cytoplasmic localization signal that is capable of overriding the activity of both Gal4 and the SV40 large-T-antigen NLS. Treatment of cells with leptomycin B resulted in the nuclear accumulation of Mlx and MondoA. Leptomycin B is an inhibitor of CRM-dependent nuclear export (54), suggesting that the CLD of MondoA functions via a known export pathway to maintain MondoA-Mlx heterocomplexes in the cytoplasm. The CLD does not show obvious sequence similarity to known export signals (39, 41) and likely constitutes a novel regulatory domain.
Wild-type MondoA, N-terminal deletions of MondoA lacking the CLD, and Mlx all localize to the cytoplasm when expressed individually (data not shown), suggesting that additional sequences in both MondoA and Mlx regulate their subcellular localization. When coexpressed, wild-type Mlx and MondoA also localize to the cytoplasm. However, when Mlx is coexpressed with any of the MondoA deletions lacking the CLD, they localize predominantly to the nucleus. Therefore, in the absence of the CLD, heterodimerization of MondoA and Mlx results in their nuclear localization. It is possible that heterodimerization blocks the cytoplasmic localization functions of monomeric MondoA and Mlx, or heterodimerization may unmask a dominant NLS(s) in either MondoA, Mlx, or both proteins. It is also possible that monomers of MondoA and Mlx are rapidly degraded in the nucleus. However, as we can detect NLSMlx in the nucleus when it is expressed alone (data not shown), we consider this possibility less likely. Therefore, our data suggest that in order for MondoA and Mlx to accumulate in the nucleus multiple negative regulatory steps must be overcome; the function of the CLD of MondoA must be inactivated, perhaps by extracellular signaling, and the two proteins must heterodimerize. Interestingly, a similar dependence on heterodimerization for nuclear accumulation has been observed for the Drosophila clock proteins PER and TIM (46).
The amino terminus of MondoA also contains a TAD between amino acids 322 and 445 which is extremely active when fused to the Gal4DBD. This activity is much greater than that seen with the wild-type MondoA protein, possibly due to the high affinity of the Gal4 homodimer for DNA. The MondoA TAD is rich in serine, threonine, and proline, a characteristic shared with TADs of the PAX family (49). In spite of the other functional similarities between Myc and Mondo, their TADs have little specific sequence similarity. However, a segment of c-Myc's activation domain, amino acids 41 to 103, is proline rich (33), suggesting that there may be functional similarities shared by Myc and Mondo transactivation.
We have described a new transcription factor network whose center is Mlx. Like Max, Mlx is a common partner for both transcriptional activators, the Mondo proteins, and repressors, the Mad proteins. Given the similarities between the Max network and the Mlx network, it is likely that the Mlx network will also function in the regulation of cell proliferation and differentiation. Our preliminary data suggest that transcriptional cross talk between the Max network and the Mlx network regulates their activity. For example, in Mad1 null mice, Mlx is upregulated in the granulosa cells of the ovary, and Drosophila Myc appears to regulate Drosophila Mondo mRNA levels (data not shown). Thus, it is likely that a web of regulatory interactions between the Max and Mlx networks regulates the functions of these two transcription factor families. Future experiments will be directed at understanding the complicated interplay of these two networks.
ACKNOWLEDGMENTS
We thank Jennifer Phillips for technical assistance, Barbara Graves for suggestions on the manuscript, M. Yoshida for the gift of leptomycin B, and Jim Reamey, of the Department of Human Genetics Robotics Core Facility, for taming the robot.
A.N.B. was supported by Cancer Center Training grant 3P30CA42014, K.L.C. was supported by NRSA 5F32HL09548, and D.E.A. is supported by NIH grant GM55668-04 and is a Scholar of The Leukemia and Lymphoma Society.
REFERENCES
- 1.Amati B, Alevizopoulos K, Vlach J. Myc and the cell cycle. Front Biosci. 1998;3:D250–D268. doi: 10.2741/a239. [DOI] [PubMed] [Google Scholar]
- 2.Amati B, Land H. Myc-Max-Mad: a transcription factor network controlling cell cycle progression, differentiation and death. Curr Opin Genet Dev. 1994;4:102–108. doi: 10.1016/0959-437x(94)90098-1. [DOI] [PubMed] [Google Scholar]
- 3.Atchley W R, Fitch W M. A natural classification of the basic helix-loop-helix class of transcription factors. Proc Natl Acad Sci USA. 1997;94:5172–5176. doi: 10.1073/pnas.94.10.5172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley and Sons, Inc.; 1995. [Google Scholar]
- 5.Ayer D E. Histone deacetylases: transcriptional repression with SINers and NuRDs. Trends Cell Biol. 1999;9:193–198. doi: 10.1016/s0962-8924(99)01536-6. [DOI] [PubMed] [Google Scholar]
- 6.Ayer D E, Eisenman R N. A switch from Myc:Max to Mad:Max heterocomplexes accompanies monocyte/macrophage differentiation. Genes Dev. 1993;7:2110–2119. doi: 10.1101/gad.7.11.2110. [DOI] [PubMed] [Google Scholar]
- 7.Ayer D E, Kretzner L, Eisenman R N. Mad: a heterodimeric partner for Max that antagonizes Myc transcriptional activity. Cell. 1993;72:211–222. doi: 10.1016/0092-8674(93)90661-9. [DOI] [PubMed] [Google Scholar]
- 8.Ayer D E, Lawrence Q A, Eisenman R N. Mad-Max transcriptional repression is mediated by ternary complex formation with mammalian homologs of yeast repressor Sin3. Cell. 1995;80:767–776. doi: 10.1016/0092-8674(95)90355-0. [DOI] [PubMed] [Google Scholar]
- 9.Bernards R. Transcriptional regulation. Flipping the Myc switch. Curr Biol. 1995;5:859–861. doi: 10.1016/s0960-9822(95)00173-4. [DOI] [PubMed] [Google Scholar]
- 10.Billin A N, Eilers A L, Queva C, Ayer D E. Mlx, a novel max-like BHLHZip protein that interacts with the max network of transcription factors. J Biol Chem. 1999;274:36344–36350. doi: 10.1074/jbc.274.51.36344. [DOI] [PubMed] [Google Scholar]
- 11.Blackwood E M, Eisenman R N. Max: a helix-loop-helix zipper protein that forms a sequence-specific DNA-binding complex with Myc. Science. 1991;251:1211–1217. doi: 10.1126/science.2006410. [DOI] [PubMed] [Google Scholar]
- 12.Blackwood E M, Luscher B, Eisenman R N. Myc and Max associate in vivo. Genes Dev. 1992;6:71–80. doi: 10.1101/gad.6.1.71. [DOI] [PubMed] [Google Scholar]
- 13.Bouchard C, Thieke K, Maier A, Saffrich R, Hanley-Hyde J, Ansorge W, Reed S, Sicinski P, Bartek J, Eilers M. Direct induction of cyclin D2 by Myc contributes to cell cycle progression and sequestration of p27. EMBO J. 1999;18:5321–5333. doi: 10.1093/emboj/18.19.5321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cerni C, Bousset K, Seelos C, Burkhardt H, Henriksson M, Luscher B. Differential effects by Mad and Max on transformation by cellular and viral oncoproteins. Oncogene. 1995;11:587–596. [PubMed] [Google Scholar]
- 15.Charron J, Malynn B A, Fisher P, Stewart V, Jeannotte L, Goff S P, Robertson E J, Alt F W. Embryonic lethality in mice homozygous for a targeted disruption of the N-myc gene. Genes Dev. 1992;6:2248–2257. doi: 10.1101/gad.6.12a.2248. [DOI] [PubMed] [Google Scholar]
- 16.Chin L, Schreiber-Agus N, Pellicer I, Chen K, Lee H W, Dudast M, Cordon-Cardo C, DePinho R A. Contrasting roles for Myc and Mad proteins in cellular growth and differentiation. Proc Natl Acad Sci USA. 1995;92:8488–8492. doi: 10.1073/pnas.92.18.8488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dang C V. c-Myc target genes involved in cell growth, apoptosis, and metabolism. Mol Cell Biol. 1999;19:1–11. doi: 10.1128/mcb.19.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Davis A C, Wims M, Spotts G D, Hann S R, Bradley A. A null c-myc mutation causes lethality before 10.5 days of gestation in homozygotes and reduced fertility in heterozygous female mice. Genes Dev. 1993;7:671–682. doi: 10.1101/gad.7.4.671. [DOI] [PubMed] [Google Scholar]
- 19.de Luis O, Valero M C, Jurado L A. WBSCR14, a putative transcription factor gene deleted in Williams-Beuren syndrome: complete characterisation of the human gene and the mouse ortholog. Eur J Hum Genet. 2000;8:215–222. doi: 10.1038/sj.ejhg.5200435. [DOI] [PubMed] [Google Scholar]
- 20.Facchini L M, Penn L Z. The molecular role of Myc in growth and transformation: recent discoveries lead to new insights. FASEB J. 1998;12:633–651. [PubMed] [Google Scholar]
- 21.Ferre D A A R, Prendergast G C, Ziff E B, Burley S K. Recognition by Max of its cognate DNA through a dimeric b/HLH/Z domain. Nature. 1993;363:38–45. doi: 10.1038/363038a0. [DOI] [PubMed] [Google Scholar]
- 22.Fisher F, Goding C R. Single amino acid substitutions alter helix-loop-helix protein specificity for bases flanking the core CANNTG motif. EMBO J. 1992;11:4103–4109. doi: 10.1002/j.1460-2075.1992.tb05503.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Foley K P, Eisenman R N. Two MAD tails: what the recent knockouts of Mad1 and Mxi1 tell us about the MYC/MAX/MAD network. Biochim Biophys Acta. 1999;1423:M37–M47. doi: 10.1016/s0304-419x(99)00012-8. [DOI] [PubMed] [Google Scholar]
- 24.Foley K P, McArthur G A, Queva C, Hurlin P J, Soriano P, Eisenman R N. Targeted disruption of the MYC antagonist MAD1 inhibits cell cycle exit during granulocyte differentiation. EMBO J. 1998;17:774–785. doi: 10.1093/emboj/17.3.774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gallant P, Shiio Y, Cheng P F, Parkhurst S M, Eisenman R N. Myc and Max homologs in Drosophila. Science. 1996;274:1523–1527. doi: 10.1126/science.274.5292.1523. [DOI] [PubMed] [Google Scholar]
- 26.Grandori C, Eisenman R N. Myc target genes. Trends Biochem Sci. 1997;22:177–181. doi: 10.1016/s0968-0004(97)01025-6. [DOI] [PubMed] [Google Scholar]
- 27.Henriksson M, Lüscher B. Proteins of the Myc network: essential regulators of cell growth and differentiation. Adv Cancer Res. 1996;68:109–182. doi: 10.1016/s0065-230x(08)60353-x. [DOI] [PubMed] [Google Scholar]
- 28.Hollenberg S M, Sternglanz R, Cheng P F, Weintraub H. Identification of a new family of tissue-specific basic helix-loop-helix proteins with a two-hybrid system. Mol Cell Biol. 1995;15:3813–3822. doi: 10.1128/mcb.15.7.3813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hurlin P J, Queva C, Eisenman R N. Mnt, a novel Max interacting protein is coexpressed with Myc in proliferating cells and mediates repression at Myc binding sites. Genes Dev. 1997;11:44–58. doi: 10.1101/gad.11.1.44. [DOI] [PubMed] [Google Scholar]
- 30.Hurlin P J, Queva C, Koskinen P J, Steingrimsson E, Ayer D E, Copeland N G, Jenkins N A, Eisenman R N. Mad3 and Mad4: novel Max-interacting transcriptional repressors that suppress c-Myc-dependent transformation and are expressed during neural and epidermal differentiation. EMBO J. 1995;14:5646–5659. doi: 10.1002/j.1460-2075.1995.tb00252.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Iritani B M, Eisenman R N. c-Myc enhances protein synthesis and cell size during B lymphocyte development. Proc Natl Acad Sci USA. 1999;96:13180–13185. doi: 10.1073/pnas.96.23.13180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Johnston L A, Prober D A, Edgar B A, Eisenman R N, Gallant P. Drosophila myc regulates cellular growth during development. Cell. 1999;98:779–790. doi: 10.1016/s0092-8674(00)81512-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kato G J, Barrett J, Villa-Garcia M, Dang C V. An amino-terminal c-myc domain required for neoplastic transformation activates transcription. Mol Cell Biol. 1990;10:5914–5920. doi: 10.1128/mcb.10.11.5914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Koskinen P J, Ayer D E, Eisenman R N. Repression of Myc-Ras cotransformation by Mad is mediated by multiple protein-protein interactions. Cell Growth Differ. 1995;6:623–629. [PubMed] [Google Scholar]
- 35.Kudo N, Khochbin S, Nishi K, Kitano K, Yanagida M, Yoshida M, Horinouchi S. Molecular cloning and cell cycle-dependent expression of mammalian CRM1, a protein involved in nuclear export of proteins. J Biol Chem. 1997;272:29742–29751. doi: 10.1074/jbc.272.47.29742. [DOI] [PubMed] [Google Scholar]
- 36.Laherty C D, Yang W M, Sun J M, Davie J R, Seto E, Eisenman R N. Histone deacetylases associated with the mSin3 corepressor mediate mad transcriptional repression. Cell. 1997;89:349–356. doi: 10.1016/s0092-8674(00)80215-9. [DOI] [PubMed] [Google Scholar]
- 37.McArthur G A, Laherty C D, Queva C, Hurlin P J, Loo L, James L, Grandori C, Gallant P, Shiio Y, Hokanson W C, Bush A C, Cheng P F, Lawrence Q A, Pulverer B, Koskinen P J, Foley K P, Ayer D E, Eisenman R N. The Mad protein family links transcriptional repression to cell differentiation. Cold Spring Harbor Symp Quant Biol. 1998;63:423–433. doi: 10.1101/sqb.1998.63.423. [DOI] [PubMed] [Google Scholar]
- 38.Meroni G, Reymond A, Alcalay M, Borsani G, Tanigami A, Tonlorenzi R, Nigro C L, Messali S, Zollo M, Ledbetter D H, Brent R, Ballabio A, Carrozzo R. Rox, a novel bHLHZip protein expressed in quiescent cells that heterodimerizes with Max, binds a non-canonical E box and acts as a transcriptional repressor. EMBO J. 1997;16:2892–2906. doi: 10.1093/emboj/16.10.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Michael W M. Nucleocytoplasmic shuttling signals: two for the price of one. Trends Cell Biol. 2000;10:46–50. doi: 10.1016/s0962-8924(99)01695-5. [DOI] [PubMed] [Google Scholar]
- 40.Moens C B, Stanton B R, Parada L F, Rossant J. Defects in heart and lung development in compound heterozygotes for two different targeted mutations at the N-myc locus. Development. 1993;119:485–499. doi: 10.1242/dev.119.2.485. [DOI] [PubMed] [Google Scholar]
- 41.Moroianu J. Nuclear import and export pathways. J Cell Biochem. 1999;(Suppl. 32–33):76–83. doi: 10.1002/(sici)1097-4644(1999)75:32+<76::aid-jcb10>3.3.co;2-h. [DOI] [PubMed] [Google Scholar]
- 42.Mukherjee B, Morgenbesser S D, DePinho R A. Myc family oncoproteins function through a common pathway to transform normal cells in culture: cross-interference by Max and trans-acting dominant mutants. Genes Dev. 1992;6:1480–1492. doi: 10.1101/gad.6.8.1480. [DOI] [PubMed] [Google Scholar]
- 43.Okano H J, Park W Y, Corradi J P, Darnell R B. The cytoplasmic Purkinje onconeural antigen cdr2 down-regulates c-Myc function: implications for neuronal and tumor cell survival. Genes Dev. 1999;13:2087–2097. doi: 10.1101/gad.13.16.2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Perez-Roger I, Kim S H, Griffiths B, Sewing A, Land H. Cyclins D1 and D2 mediate Myc-induced proliferation via sequestration of p27(Kip1) and p21(Cip1) EMBO J. 1999;18:5310–5320. doi: 10.1093/emboj/18.19.5310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Queva C, McArthur G A, Ramos L S, Eisenman R N. Dwarfism and dysregulated proliferation in mice overexpressing the MYC antagonist MAD1. Cell Growth Differ. 1999;10:785–796. [PubMed] [Google Scholar]
- 46.Saez L, Young M W. Regulation of nuclear entry of the Drosophila clock proteins period and timeless. Neuron. 1996;17:911–920. doi: 10.1016/s0896-6273(00)80222-6. [DOI] [PubMed] [Google Scholar]
- 47.Schreiber-Agus N, DePinho R A. Repression by the Mad(Mxi1)-Sin3 complex. Bioessays. 1998;20:808–818. doi: 10.1002/(SICI)1521-1878(199810)20:10<808::AID-BIES6>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 48.Schreiber-Agus N, Meng Y, Hoang T, Hou H, Jr, Chen K, Greenberg R, Cordon-Cardo C, Lee H W, DePinho R A. Role of Mxi1 in ageing organ systems and the regulation of normal and neoplastic growth. Nature. 1998;393:483–487. doi: 10.1038/31008. [DOI] [PubMed] [Google Scholar]
- 49.Tang H K, Singh S, Saunders G F. Dissection of the transactivation function of the transcription factor encoded by the eye developmental gene PAX6. J Biol Chem. 1998;273:7210–7221. doi: 10.1074/jbc.273.13.7210. [DOI] [PubMed] [Google Scholar]
- 50.Towbin J A, Casey B, Belmont J. The molecular basis of vascular disorders. Am J Hum Genet. 1999;64:678–684. doi: 10.1086/302303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Turpin P, Ossareh-Nazari B, Dargemont C. Nuclear transport and transcriptional regulation. FEBS Lett. 1999;452:82–86. doi: 10.1016/s0014-5793(99)00533-5. [DOI] [PubMed] [Google Scholar]
- 52.Vastrik I, Kaipainen A, Penttila T L, Lymboussakis A, Alitalo R, Parvinen M, Alitalo K. Expression of the mad gene during cell differentiation in vivo and its inhibition of cell growth in vitro. J Cell Biol. 1995;128:1197–1208. doi: 10.1083/jcb.128.6.1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wenzel A, Cziepluch C, Hamann U, Schurmann J, Schwab M. The N-Myc oncoprotein is associated in vivo with the phosphoprotein Max(p20/22) in human neuroblastoma cells. EMBO J. 1991;10:3703–3712. doi: 10.1002/j.1460-2075.1991.tb04938.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wolff B, Sanglier J J, Wang Y. Leptomycin B is an inhibitor of nuclear export: inhibition of nucleo-cytoplasmic translocation of the human immunodeficiency virus type 1 (HIV-1) Rev protein and Rev-dependent mRNA. Chem Biol. 1997;4:139–147. doi: 10.1016/s1074-5521(97)90257-x. [DOI] [PubMed] [Google Scholar]
- 55.Wu S, Pena A, Korcz A, Soprano D R, Soprano K J. Overexpression of Mxi1 inhibits the induction of the human ornithine decarboxylase gene by the Myc/Max protein complex. Oncogene. 1996;12:621–629. [PubMed] [Google Scholar]